ABSTRACT
Techniques based on high-throughput sequencing (HTS) of environmental DNA have provided a new way of studying fungal diversity. However, these techniques suffer from a number of methodological biases which may appear at any of the steps involved in a metabarcoding study. Air is one of the most important environments where fungi can be found, because it is the primary medium of dispersal for many species. Looking ahead to future developments, it was decided to test 20 protocols, including different passive spore traps, spore recovery procedures, DNA extraction kits, and barcode loci. HTS was performed with the Illumina MiSeq platform targeting two subloci of the fungal internal transcribed spacer. Multivariate analysis and generalized linear models showed that the type of passive spore trap, the spore recovery procedure, and the barcode all impact the description of fungal communities in terms of richness and diversity when assessed by HTS metabarcoding. In contrast, DNA extraction kits did not significantly impact these results. Although passive traps may be used to describe airborne fungal communities, a study using specific real-time PCR and a mock community showed that these kinds of traps are affected by environmental conditions that may induce losses of biological material, impacting diversity and community composition results.
IMPORTANCE The advent of high-throughput sequencing (HTS) methods, such as those offered by next-generation sequencing (NGS) techniques, has opened a new era in the study of fungal diversity in different environmental substrates. In this study, we show that an assessment of the diversity of airborne fungal communities can reliably be achieved by the use of simple and robust passive spore traps. However, a comparison of sample processing protocols showed that several methodological biases may impact the results of fungal diversity when assessed by metabarcoding. Our data suggest that identifying these biases is of paramount importance to enable a correct identification and relative quantification of community members.
KEYWORDS: aerobiology, fungal dispersion, fungal diversity, metabarcoding
INTRODUCTION
Fungi are among the most diverse organisms on Earth. They can develop in almost all terrestrial ecosystems living as saprobes, mutualists, endophytes, or pathogens. Indeed, the Fungi kingdom has been estimated to include between one and five million species, of which only approximately 5% are identified (1). In recent years, knowledge of fungal diversity has improved through the use of new molecular techniques (2). Techniques based on next-generation sequencing (NGS), such as metabarcoding by high-throughput sequencing (HTS), have enabled the study of fungal communities while bypassing conventional methods, such as microscopic observation of fungal reproductive structures, isolation techniques after sampling, or cloning and Sanger sequencing of target regions of DNA, which are time-consuming and unsuitable for high-throughput analysis. NGS-based technologies therefore have great potential for community monitoring studies, becoming a complementary approach to classical inventory procedures (3). NGS involves a number of steps from sampling via laboratory handling to bioinformatic analysis (4). Each of these steps is critical and needs to be assessed to minimize methodological biases. Much work has been done to correct platform-specific sequencing problems, very often applying bioinformatic solutions (5, 6). However, there is a risk of potential distortions and information loss prior to DNA sequencing, for example, during sample collection and transport, DNA extraction and purification, or PCR amplification (4, 7).
Despite these methodological issues, mycologists and phytopathologists have taken advantage of NGS, using environmental DNA to monitor fungal and fungus-like organisms in different substrates and matrices, such as soil (8, 9), water (10, 11), plants (12, 13), or insects (14, 15). Among these substrates, air is of major importance. Indeed, spore dispersal by air (or wind) is one of the major mechanisms used by fungal pathogens to spread and reach new susceptible hosts (16). Consequently, airborne pathogens should be monitored at the local or regional scale to predict the risk of severe epidemics, allowing, for example, the detection of emerging diseases, and to improve disease warning systems (17, 18). Airborne fungal spores have been monitored extensively, the target in most cases being a single pathogen or species (19, 20). A wide variety of spore capture methods are available for these purposes (21). In general, these methods can be divided into active and passive sampling (21, 22). Active (or volumetric) samplers employ mechanical means and energy sources to capture particles onto or into a collection matrix and sample a consistent air volume (22). This category includes, for example, commonly used samplers, like Hirst-type spore traps or rotating-arm samplers (20, 23). Passive methodologies depend on gravity to deposit propagules onto a surface (22), which can be, for instance, filter papers (19, 24), wood disks (25), or coated microscope slides (26). Although less efficient in capturing bioaerosols than active samplers, the major advantages of passive traps are their low cost and easy installation, being useful for monitoring aerial inocula on a large geographical scale, but also in environments where access can be difficult (for example, by placing traps on tree crowns or at different heights) (27, 28).
This study assessed some commonly used passive traps in order to evaluate their ability to describe airborne fungal communities by metabarcoding analysis. In order to determine how technical choices influence the description of these communities, we tested different presequencing sample protocols combining various passive spore trap surfaces, protocols to recover the fungal spores, and types of DNA extraction kits. Furthermore, we assessed the influence of two primer pairs derived from the nuclear ribosomal internal transcribed spacer (ITS), the universal DNA barcode marker for fungi (29). Finally, we also evaluated the different protocols by quantitative real-time PCR (qPCR) targeting two forest pathogens responsible for major forest diseases and compared the sensitivities of qPCR and metabarcoding assays.
RESULTS
Comparison of exposed versus unexposed trap data sets.
After bioinformatic processing, the total numbers of sequences were 1,754,349 (mean ± standard deviation [SD], 16,834 ± 6,069) and 2,330,522 (mean ± SD, 19,326 ± 8,755) for the unexposed and exposed data sets, respectively. There were 3,635 operational taxonomic units (OTUs) (0.21%) represented by fewer than five sequences in the unexposed data set, whereas for the exposed data set, there were 11,399 OTUs (0.49%). Using a conservative approach, these OTUs were not included in the subsequent analysis to minimize the inclusion of sequencing artifacts (30). The total numbers of sequences per sample ranged from 3,468 to 56,570 and from 6,353 to 38,115 for the exposed and unexposed data sets, respectively. Significant differences were observed in a comparison of the total number of sequences of both data sets (z = −2.09; P = 0.036). The α-diversity indexes computed for both data sets, rarefied to 3,468 sequences, were in all cases larger for the exposed traps and significantly different to the unexposed traps, based on observed richness (z = −10.48; P < 0.001) and the Shannon index (t = −9.55; P < 0.001).
Three hundred twelve OTUs were observed in the unexposed trap data set, whereas 585 OTUs were observed in the exposed trap data set. In terms of observed richness, the two data sets differed: 23.8% of the OTUs were common to both data sets, whereas 19.3% and 56.9% were exclusive to the exposed and the unexposed trap data sets, respectively. Indeed, in a comparison of fungal diversity in terms of observed OTUs, major differences were observed. In the unexposed trap data set, the following five species of the mock community were taxonomically assigned at the species level: Diplodia corticola (37% of the total number of sequences [all samples]), Fusarium graminearum (7.2% [all samples]), Melampsora larici-populina (0.37% [49/104 samples]), Phomopsis cotoneastri (0.1% [50/104 samples]), and Hymenoscyphus fraxineus (0.07% [41/104 samples]). Three species of the mock community were assigned at the genus level, Pestalotiopsis sp. (38.3% [all samples]), Cytospora sp. (1.4% [52/104 samples]), and Colletotrichum sp. (0.25% [89/104 samples]). For the rest of the species of the mock community, taxonomic assignation was less clear. Neonectria coccinea was not observed, although one OTU was taxonomically assigned to Nectriaceae sp. (3.5% [all samples]). Neither Cryphonectria parasitica nor Cryphonectria radicalis was found, although an OTU taxonomically assigned to Cryphonectriaceae sp. was observed (0.48% [101/104 samples]). Similarly, neither Fusarium lateritium nor Fusarium circinatum was observed in the unexposed samples, although one OTU of the data set was taxonomically assigned to Fusarium sp. (0.89% [56/104 samples]). In contrast, Diplodia pinea was not found in the unexposed data set. In all, fungal species of the mock community accounted for 89.56% of the total number of sequences in the unexposed spore traps. OTUs other than the species included in the mock community were also observed in the unexposed data set. Among the most abundant OTUs observed at the species or genus level were Leptosphaerulina chartarum (1.73% [88/104 samples]), Debaryomyces prosopidis (1.72% [81/104 samples]), Davidiella tassiana (0.68% [94/104 samples]), Filobasidium floriforme (0.52% [8/104 samples]), Malassezia restricta (0.32% [26/104 samples]), and Sarocladium strictum (0.23% [26/104 samples]).
Species included in the mock community were rare in the exposed data set; OTUs were taxonomically assigned to Pestalotiopsis sp. (0.12% [106/120 samples]), Diplodia corticola (0.09% [96/120 samples]), Fusarium sp. (0.03% in 61/120 samples), Fusarium graminearum (0.01% [62/120 samples]), and Colletotrichum sp. (0.0004% [8/120 samples]). The most abundant OTUs in the exposed trap data set, represented by more than 1% of the total number of sequences, were Davidiella tassiana (29.4% [all samples]), Epicoccum nigrum (18.9% [all samples]), Leptosphaerulina chartarum (10.54% [119/120 samples]), Sporidiobolales unclassified (7.33% [102/120 samples]), Alternaria sp. (5.8% [118/120 samples]), Xenobotryosphaeria calamagrostidis (4.3% [59/120 samples]), Cryptococcus stepposus (3.2% [59/120 samples]), Cryptococcus victoriae (2.7% [59/120 samples]), Aureobasidium sp. (2.5% [118/120 samples]), Sporobolomyces sp. (2.3% [89/120 samples]), and Debaryomyces prosopidis (2.17% [67/120 samples]). For the detailed data sets, please refer to Data Sets S1 and S2 in the supplemental material.
Comparisons of technical choices by analysis of the exposed trap data set.
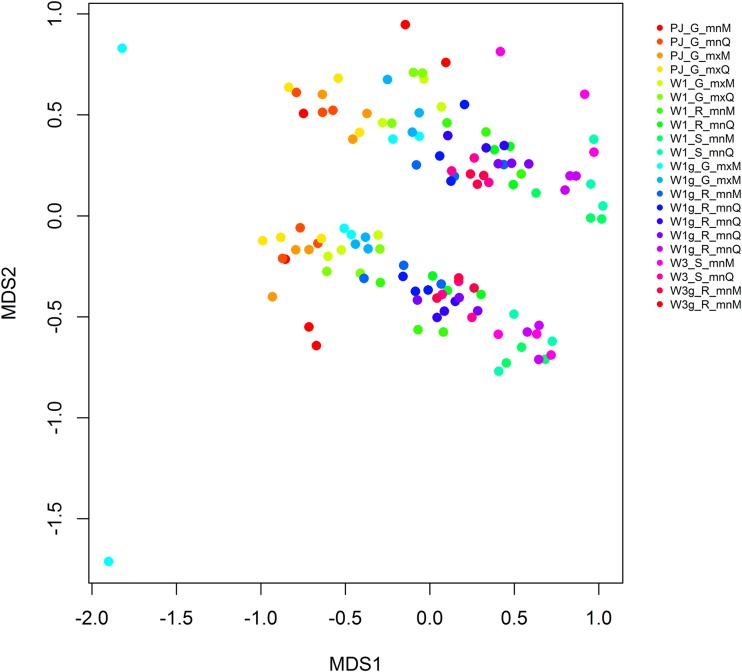
Nonmetric multidimensional scaling (NMDS) clustering of samples based on the Bray-Curtis dissimilarity using two dimensions showed acceptable values for stress (0.16) and linear fit (R2 = 0.89). NMDS clearly differentiated two groups representing genetic barcodes ITS1 and ITS2 (Fig. 1). The analysis of similarity (ANOSIM) revealed that the barcode had a significant effect on the structure of the observed communities represented by the NMDS (R = 0.32, P = 0.01). Generalized linear model (GLM) results (Fig. 2) revealed that the barcode had a significant effect on the total number of sequences (P < 0.001) and the Shannon index (P < 0.001). However, it did not have a significant effect on the observed richness (P = 0.61).
FIG 1.
NMDS representation of all the exposed trap protocols. A protocol was the combination of the type of trap, the spore recovery procedure, the DNA extraction kit, and the ITS barcode. All samples were replicated three times. The NMDS showed acceptable values for stress (0.16) and linear fit (R2 = 0.89). The codes indicate the technical procedures used in this paper. For type of trap: PJ, petri dish coated with a mix of petroleum jelly and Vaseline; W1, Whatman filter no. 1; W1g, Whatman filter no. 1 sprayed with a sticky layer; W3, Whatman filter no. 3; W3g, Whatman filter no. 3 sprayed with a sticky layer. For spore recovery protocol: G, grinding; R, rubbing; S, shaking. For DNA extraction kit: mnM, NucleoSpin plant II kit; mnQ, DNeasy plant minikit; mxM, NucleoSpin plant II maxikit; mxQ, DNeasy plant maxikit.
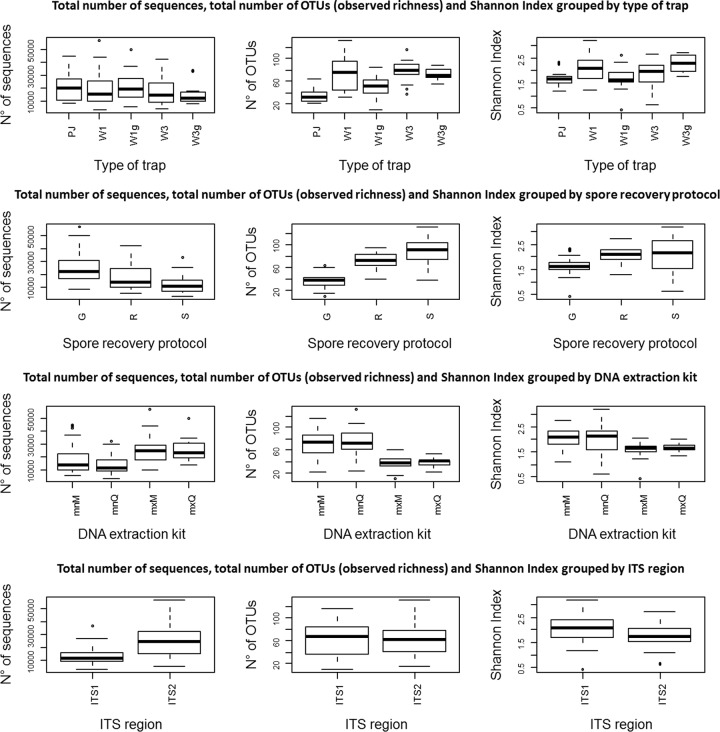
FIG 2.
Box plots representing comparisons between presequencing and sequencing technical choices: type of trap, spore recovery procedure, DNA extraction kit, and ITS subloci versus the total number of sequences, observed richness, and Shannon index. Whiskers indicate variability outside the upper and lower quantities, while outliers are represented by individual points.
GLM results (Fig. 2) showed that the type of trap had a significant effect on the total number of sequences (P = 0.03), the observed richness (P < 0.001), and the Shannon index (P < 0.001). The ANOSIM revealed a significant effect of the type of trap on the community structure represented by the NMDS (R = 0.29, P = 0.001; Fig. S1).
GLM analysis (Fig. 2) also revealed that the spore recovery protocol had a significant effect on the total number of sequences (P < 0.001), the observed richness (P < 0.001), and the Shannon index (P < 0.001). The ANOSIM revealed a significant effect of the spore recovery protocol (R = 0.43, P = 0.001; Fig. S2) on the structure represented on the NMDS.
GLM analysis did not reveal any significant effects of the DNA extraction kit (Fig. 2) on the total number of sequences (P = 0.05), on the observed richness (P = 0.95), or on the Shannon index (P = 0.27). No significant effect on the community structure was observed for the DNA extraction method after the ANOSIM (R = 0, P = 0.42; Fig. S3).
Characterization of the diversity observed in the exposed trap data set.
Some taxonomic assignments were different from one barcode to another, and only 44.3% of the OTUs were found to be common to ITS1 and ITS2 when the total data set was analyzed. When this comparison was performed for the data set containing the top 50 OTUs in terms of relative sequence abundance, we found that 84% of the OTUs were shared by ITS1 and ITS2, with 6% and 10% of OTUs exclusive to ITS1 and ITS2, respectively. Only OTUs taxonomically assigned to Davidiella tassiana and Epicoccum nigrum were present in all exposed traps independently of the barcode. These two OTUs were also the most abundant of the whole data set, with Davidiella tassiana at 29.40% and Epicoccum nigrum at 18.92%. Some of the OTUs were exclusively assigned either to ITS1 or ITS2. Examples of this were OTUs assigned to Xenobotryosphaeria calamagrostidis (found in 59 samples [4.31% of the total number of sequences]) and Pleospora sp. (found in 56 samples [0.08% of the total number of sequences]), which were exclusively detected by the ITS1 marker. In turn, OTUs assigned to Aureobasidium pullulans (found in 53 samples [0.02% of the total number of sequences]), Cryptococcus stepposus (found in 59 samples [3.2% of the total number of sequences]), Dioszegia buhagiarii (found in 42 samples [0.023% of the total number of sequences]), Filobasidium floriforme (found in 43 samples [0.04% of the total number of sequences]), Physcia dubia (found in 49 samples [0.09% of the total number of sequences]), Pleospora herbarum (found in 57 samples [0.11% of the total number of sequences]), and Spegazzinia sp. (found in 55 samples [0.04% of the total number of sequences]) were exclusively detected by the ITS2 marker.
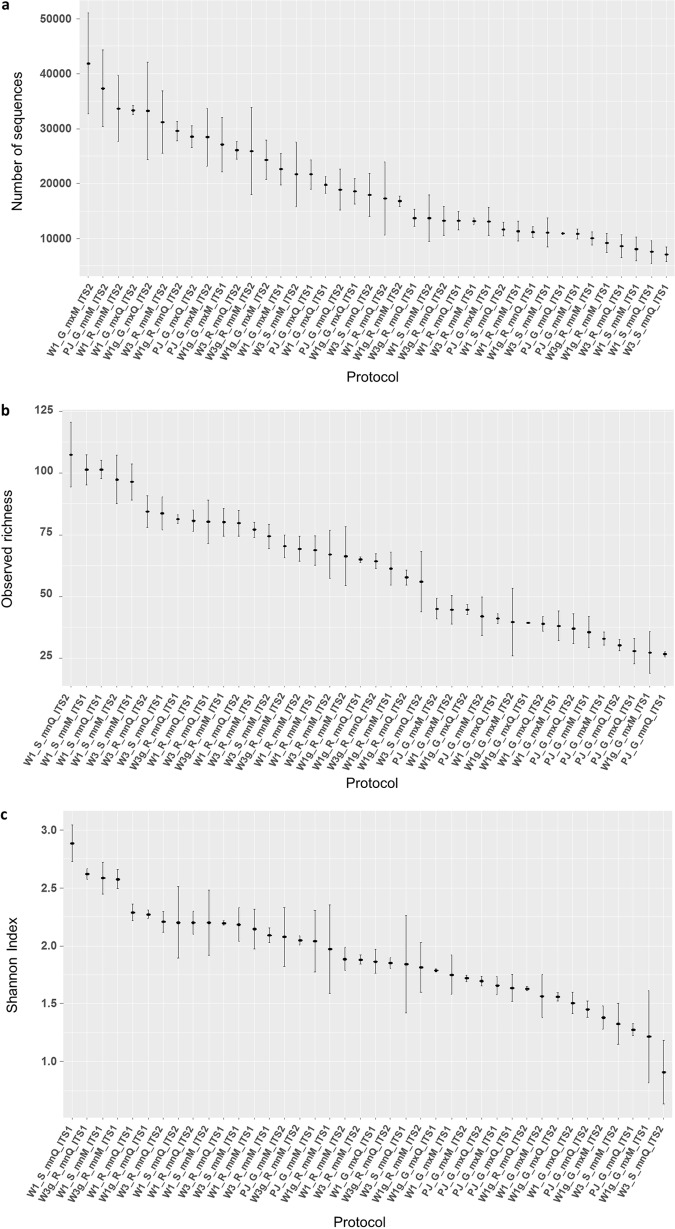
The top 50 OTUs gathered 97.9% of the total number of sequences in the data set. The heat map based on the Bray-Curtis distance generated from the relative abundance of these top 50 OTUs revealed that replicates of samples globally grouped together to form six clusters (Fig. S4). This means that sample replicates combining the same type of trap, spore recovery protocol, DNA extraction kit, and barcode were, in general, grouped in the same cluster, representing fungal communities having similar compositions. Exceptions were samples W1g_GmxM_ITS2b and W1g_GmxM_ITS1b, which were assigned to cluster I (while the other replicates were all assigned to cluster II), W1g_R_mnM_ITS1a, assigned to cluster IV (while the other replicates were assigned to cluster VI), and samples W1g_R_mnM_ITS2b and W1g_R_mnM_ITS2a, which were assigned to cluster V (while the other replicates were assigned to cluster VI). The stacked plot constructed from the relative abundance of the top OTUs of the exposed trap data set (Fig. S5) shows the differences within samples (i.e., the same sample sequenced either ITS1 or ITS2) and between samples (i.e., differences between types of sample). A comparison of both markers (Fig. 2) revealed significant differences for the number of sequences (z = 7.7; P < 0.001) and the Shannon index (z = −2.9; P < 0.001) but not for the observed richness (z = −0.095; P = 0.9). Higher mean numbers of sequences were globally observed for samples sequenced with the ITS2 barcode. Higher mean numbers of sequences were obtained for the W1_G_mxM_ITS2, PJ_G_mnM_ITS2, and W1_R_mnM_ITS2 samples (Fig. 3a). Concerning the observed richness, a higher mean number of OTUs was obtained with the ITS1 barcode (Fig. 2). Higher mean observed richness was observed in the W1_S_mnQ_ITS2, W1_S_mnM_ITS1, and W1_S_mnQ_ITS1 samples (Fig. 3b). The mean Shannon index was globally higher for samples sequenced with the ITS1 barcode, and it was higher for the W1_S_mnQ_ITS1, W3g_R_mnQ_ITS1, and W1_S_mnM_ITS1 samples (Fig. 3c). Data Set S3 presents detailed data per sample of the total number of sequences, the observed richness, and the Shannon index.
FIG 3.
Graphical representation of the mean and standard deviation by protocol in terms the total number of sequences (a), observed richness (b), and the Shannon-Wiener index (c). Means and standard deviations were computed for three replicates per protocol. The observed richness and Shannon-Wiener index values were computed after rarefaction to the smallest number of sequences per sample as a threshold.
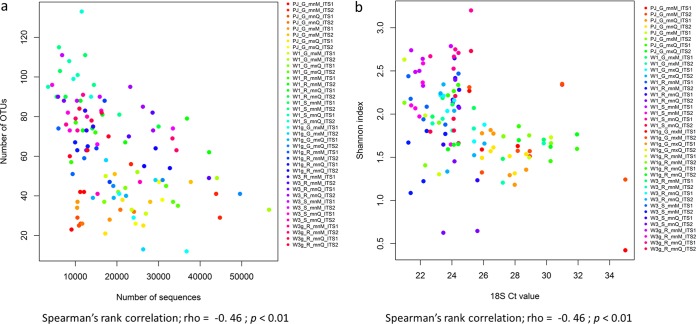
Finally, the relative abundance of each OTU, estimated by the number of sequences they represented in the data set, exhibited a negative and significant relationship with the observed richness (rho = −0.46; P < 0.001) and the Shannon index (rho = −0.46; P < 0.001) (Fig. 4a and b).
FIG 4.
Correlation between the total number of sequences and the total number of OTUs (a) and the Shannon index (b). Each color represents the three replicates of each of the samples. The codes indicate the technical procedures used in this paper. For type of trap: PJ, petri dish coated with a mix of petroleum jelly and Vaseline; W1, Whatman filter no. 1; W1g, Whatman filter no. 1 sprayed with a sticky layer; W3, Whatman filter no. 3; W3g, Whatman filter no. 3 sprayed with a sticky layer. For spore recovery protocol: G, grinding; R, rubbing; S, shaking. For DNA extraction kit: mnM, NucleoSpin plant II kit; mnQ, DNeasy plant minikit; mxM, NucleoSpin plant II maxikit; mxQ, DNeasy plant maxikit.
Real-time PCR results.
Based on mean cycle threshold values of the 18S ribosomal DNA (rDNA) real-time PCR, significant differences were observed between the unexposed and exposed traps (F = 530; P < 0.01). The mean cycle threshold (CT) values were lower for the exposed traps (mean ± SD, 25.26 ± 2.37) than for the unexposed traps (mean ± SD, 33.33 ± 1.6), showing that there was more DNA from the exposed traps than from the unexposed traps.
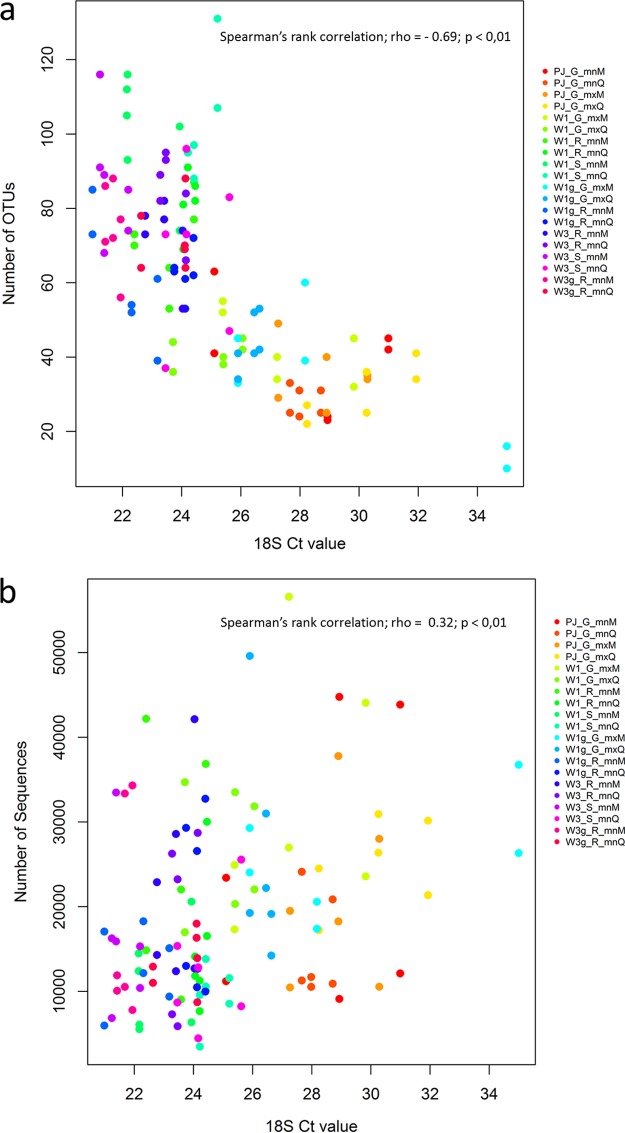
For the exposed trap data set, all the technical choices had a significant effect on the DNA quantity measured by the CT obtained with the universal 18S rDNA real-time PCR test in terms of type of trap (F = 4.45, P = 0.002), spore recovery protocol (F = 6.95, P = 0.0014), and DNA extraction kit (F = 8.20, P < 0.01). The observed richness was negatively and significantly correlated with the CT values (Spearman rank correlation, rho = −0.69; P < 0.01; Fig. 5a). The number of sequences was positively and significantly correlated with the CT values (Spearman rank correlation, rho = 0.32, P < 0.01; Fig. 5b).
FIG 5.
Plots showing the correlation between the DNA quantity measured by the cycle threshold (CT) generated by qPCR, a universal primer/barcode test targeting the 18S of plants and fungi, and the number of OTUs (a) and the total number of sequences (b). Each color represents the three replicates of each of the samples. The codes indicate the technical procedures used in this paper. For type of trap: PJ, petri dish coated with a mix of petroleum jelly and Vaseline; W1, Whatman filter no. 1; W1g, Whatman filter no. 1 sprayed with a sticky layer; W3, Whatman filter no. 3; W3g, Whatman filter no. 3 sprayed with a sticky layer. For spore recovery protocol: G, grinding; R, rubbing; S, shaking. For DNA extraction kit: mnM, NucleoSpin plant II kit; mnQ, DNeasy plant minikit; mxM, NucleoSpin plant II maxikit; mxQ, DNeasy plant maxikit.
The results for species-specific qPCR tests targeting Hymenoscyphus fraxineus and Melampsora larici-populina differed from metabarcoding in terms of successful detection. First, the rate of successful detection was higher with qPCR than with metabarcoding. H. fraxineus was detected in 63.7% of the traps by qPCR (but in 100% of the unexposed samples), whereas only 36.2% of the traps were positive for the species by metabarcoding (but in 75% of the unexposed samples). Interestingly, H. fraxineus was only detected in traps by HTS with the ITS2 barcode. Differences in the probabilities of success by group were significant (χ2 = 15.93; P < 0.001). Similar results were obtained for M. larici-populina; differences in the probabilities of success by group were significant (χ2 = 145.04; P < 0.001). The rate of successful detection was higher with qPCR (80.7% of all the traps, and 98% of the unexposed samples) than with barcodes ITS1 (4.4% of the traps) and ITS2 (30.9% of the traps).
DISCUSSION
Our study showed that technical choices impact the description of the fungal community composition in trap samples exposed at the same site when characterized by metabarcoding with Illumina MiSeq technology. It was nevertheless observed by the NMDS and heat map analysis using the Bray-Curtis dissimilarity distance that sample replicates combining the same preprocessing protocols and barcodes (type of trap plus spore recovery protocol plus DNA extraction method plus barcode) globally clustered together. This means that replicated samples combining the same protocol generally resulted in similar fungal community compositions. Further studies should, however, be performed to conclude whether physical pooling (pooling PCR products of replicates) allows a better description of fungal communities than computational pooling (each replicate is analyzed independently and results are pooled, as was the case in this study). Generally speaking, authors working with the soil matrix have suggested that it is necessary to pool between three and five replicated DNA extractions of a given sample to obtain a good representation of the microbial community (30, 31).
Our study revealed that the type of trap and spore recovery protocol had a significant impact on fungal community composition and in turn on the total number of sequences, observed richness, and Shannon index. On the other hand, the DNA extraction method did not exhibit a significant effect on the number of sequences or on any of the α-diversity indexes. Similar results were obtained by Terrat et al. (32) in a metabarcoding study assessing three DNA extraction procedures in five types of soils. The authors suggested that the soil-grinding step had an important impact on α-diversity, because some procedures are not sufficient to lyse certain fungal cells, and consequently, some taxa may be underestimated. Moreover, they suggested that α-diversity indexes may also be impacted by the physicochemical characteristics of soils (32). The present study shows that different matrices (i.e., different types of passive spore trap) and methods to recover fungal spores (i.e., different spore recovery protocols) will result in different DNA yields, which may have an impact on the description of the fungal diversity of samples exposed in the same place. Indeed, in our study, we observed significant differences in DNA quantities between methods, as measured by qPCR with the universal primer targeting the 18S region of the rRNA. Moreover, all the technical choices had a significant effect on DNA quantity when estimated by the qPCR CT value. The results of our study suggest that the qPCR CT value is negatively correlated (rho = −0.70) with the observed richness and positively correlated with the total number of sequences (rho = 0.34). In other words, this means that lower DNA yields may result in lower values of richness. Although the correlation between DNA quantity and the number of sequences was less flagrant, the relation was significant, meaning that lower DNA yields may result in a higher number of sequences. However, according to our results, samples with a higher total number of sequences do not necessarily present higher α-diversity indexes (Fig. 3 and Data Set S3). These results are not surprising considering that several studies have shown that sequence abundance does not always correlate well with the biological abundance present in a sample (12, 33, 34). Some of the factors that have been cited to explain the read abundance bias are the unequal gene copy numbers in fungal genomes and technical issues linked to PCR and sequencing (33, 34). However, despite these limitations, it has been widely suggested that read abundance may still be used as a reference for the relative proportion of the different taxa present in a sample (33–36). This relative proportion seems to be a particularly good reference when dealing with most abundant biological fungal taxa, which are often found to present a higher number of sequences in NGS data sets (12, 34).
In our work, the role of the environment in the potential loss of biological material was not assessed, yet our experimental design including unexposed and exposed traps allowed us to form some hypotheses. Although we did not estimate the proportion of biological material lost in each of the presequencing steps, we observed that in unexposed traps, the majority of species were from the mock community. However, in exposed traps, the presence of mock community fungal species was rare or inexistent. Laboratory presequencing protocols may induce material loss, but it seems that in the case of passive traps, the principal sources of biological material loss were climatic contingencies, such as wind or rain. The user of passive traps must be aware that these are the most basic devices for studying airborne microbial communities and may be more sensitive to climatic conditions than other traps. Further studies are required to assess more accurately the impact of climatic conditions on spores captured by passive traps.
Differences in fungal community composition were also observed between the ITS1 and ITS2 markers. This was clearly observed and tested statistically by the NMDS and ANOSIM, respectively. Although we did not observe significant differences in terms of observed richness, the Shannon index results for the two markers were significantly different. Our results showed that only 44% of the OTUs were shared by both barcodes when analyzing the total data set (which comprised OTUs represented by more than five sequences). This proportion increased to 84% when analyses were performed for the top 50 OTUs (97.3% of the samples). These results are in line with those of Cross et al. (12), who showed that the correlation between ITS1 and ITS2 is generally high for abundant species represented by the number of sequences, although it may vary for some specific fungal groups. Moreover, among the most abundant clusters, we observed some OTUs that were exclusively detected either by ITS1 or ITS2. Biases linked to barcodes are well documented in the literature, and generally, three types of biases are recognized: length bias, taxonomic bias, and primer mismatch bias (37–40). In particular for ITS1 and ITS2, there is not really a clear consensus on which primer pairs should be used in metabarcoding. While some authors propose a complementary strategy based on the use of both subloci (12, 31, 41), other studies suggest that both regions will provide adequate and similar answers to ecological questions (42). The choice of using one or both subloci is strongly dependent on the aims of the study, especially when focusing on a particular group (42). Bearing this in mind, it is important to emphasize that the use of the ITS will be problematic for some key fungal pathogenic groups, such as Alternaria, Fusarium, and Colletotrichum, for which species identification efficiency is low (less than 50%), as shown by Wang et al. (41). Our mock community study confirmed this for some fungal groups, such as Fusarium and Colletotrichum, but also encountered problems with Cryphonectria, Pestalotiopsis, Cytospora, Neonectria, and Diplodia. The use of genus-specific primers may be an alternative for metabarcoding short barcodes (<500 bp, like with Illumina MiSeq technology) that target some specific fungus or fungus-like group of organisms (11, 13, 43). Another alternative may entail using third-generation sequencers, such as Pacific Biosciences (PacBio RSII) or Sequel instruments, that are able to sequence the full ITS region (ITS1 and ITS2) in fungi and oomycetes, increasing taxonomical and phylogenetic information for species discrimination and taxonomic assignment (44). The application of both techniques to fungi has nevertheless shown a number of technical and bioinformatic analysis problems and biases (4, 44).
Our experimental design also revealed how the intralaboratory environment may be a source of sample contamination. In the unexposed trap set, we observed, in addition to the species included in the mock community, other fungal taxa that may be part of the indoor laboratory microbiome but may also be a product of sequencing errors not excluded after bioinformatic quality control measures. These “contaminant” OTUs accounted for approximately 10% of the total sequences of the unexposed spore traps. This may not seem like an important proportion of sequences, but the question is how to identify and handle these “contaminant” taxa in field samples. One way of identifying these OTUs is to include negative and positive controls in the different steps of the metabarcoding process (34). Using these controls, a conservative approach recommends the deletion from field samples of any OTU considered to be a “contaminant.” A less conservative approach considers subtracting the number of sequences of each OTU present in the controls from the sequence abundance of that OTU in the field samples (34). That method avoids discarding some of the most abundant OTUs. Indeed, it has been shown that some of the abundant indoor OTUs may also be present in the field (45). As a methodological paper, both data sets were kept intact in this work, and neither of these procedures were applied. However, when ecological inferences are the aim of the study, we consider that the contaminant OTUs must be taken into account in the analysis, and as a minimum, they should be reported in scientific papers. How these contaminant OTUs are to be handled should be decided beforehand depending on the study objectives.
Interesting results could be observed in a comparison of qPCR and metabarcoding results. Specific tests targeting Hymenoscyphus fraxineus and Melampsora larici-populina showed that globally for passive traps, qPCR was more sensitive than metabarcoding, either with ITS1 or ITS2. Our conclusions differed from those obtained by Castaño et al. (36), who stated that both qPCR and Illumina MiSeq can be used to detect fungal emergence. However, in their study, the authors did not work with mock communities but targeted by qPCR Lactarius vinosus, known to be present in the studied plots. Although both techniques would probably show comparable results for abundant taxa present in the environment, the detection of rare taxa may be impacted when using metabarcoding-based techniques, like Illumina MiSeq. Our results show that between 500 (in the case of H. fraxineus) and 1,000 spores (in the case of M. larici-populina) can be detected both by real-time PCR and Illumina MiSeq sequencing, as nearly all the unexposed samples gave positive results. However, it seems that these quantities of spores are close to the limit of detection in both techniques, because a large number of exposed samples were not able to detect either H. fraxineus or M. larici-populina, probably due to spore loss after outdoor exposure. However, the results of exposed traps revealed that real-time PCR performed better than Illumina MiSeq metabarcoding in detecting these two fungal pathogens. These results show that under our conditions, real-time PCR was globally more sensitive and able to detect smaller quantities of spores than Illumina MiSeq metabarcoding. Assessing the performance of each of the tested protocols was, however, not clear-cut using real-time PCR, in that none of the protocols could detect in 100% of the replicates any of the target species in the exposed traps (Data Set S4).
The comparison of results from Illumina MiSeq metabarcoding allowed us to observe some important technical patterns. In the case of the present study, this makes sense, because all the spore traps were exposed to the same conditions. We only report comparisons with the Shannon index, as it is a more complete measure of diversity than observed richness, because it integrates both abundance and evenness. We observed that in general, filter paper-based traps (Whatman no. 1 and no. 3) outperformed Vaseline (petrolatum)- and paraffin wax-based traps (Fig. 3c). Similarly, spore recovery protocols by shaking and rubbing gave higher mean diversity values than direct grinding of the spore traps. Mini DNA extraction kits gave higher mean diversity values than maxikits. On a practical level, and considering the five higher mean diversity scores estimated with the Shannon index, our study showed that either the Whatman no. 1 filter without the sticky layer coupled with a shaking protocol to recover spores, or the Whatman no. 3 filter with a sticky layer coupled with a rubbing protocol to recover spores was more suitable for metabarcoding studies than the other protocols tested. We also recommend using minikits for the DNA extraction of samples. Concerning the barcode, although we observed higher mean diversity values with ITS1 in our study, we recommend using both ITS1 and ITS2 barcodes to obtain a more complete representation of the fungal community. Further studies with more replicates and including environmental variables in the analysis could be performed to improve and technically optimize these two protocols for metabarcoding analysis of the aerial fungal microbiome. We think, however, that these simple spore traps enable, as shown here, the assessment of the diversity of airborne fungal communities but also the study of single pathogens using species-specific primers. Integrating in the analysis and reporting in scientific papers the technical biases linked to metabarcoding will allow a more precisely and accurate description of fungal communities.
MATERIALS AND METHODS
Preparation of a mock community.
Fungal cultures were obtained from the ANSES Plant Health Laboratory collection. The mock community contained 14 fungal strains, all pathogenic to forest tree species, and covering a wide range of spore sizes, shapes, and ornamentations (Table 1). Eleven of these strains were placed on malt agar (MALT) or Spezieller Nahrstoffarmer agar (SNA) medium at 22°C for between 15 and 30 days. Spores were collected by washing the surface of cultures with 10 ml of deionized water with 0.01% Tween 20 (Sigma-Aldrich, Darmstadt, Germany). Two of these fungi, Fusarium circinatum and Cryphonectria parasitica, are quarantine pests for the European Union and were therefore manipulated in level 3 biohazard containment facilities in compliance with EU Directive 2008/61/EC (46). For Melampsora larici-populina, an obligate parasite, urediniospores were produced on poplar leaf disks, as per the method of Husson et al. (47). For Hymenoscyphus fraxineus, ascospores were produced as described by Husson et al. (48). The resulting suspensions were diluted with sterile water to obtain calibrated concentrations of approximately 1,000 spores/ml for all but Hymenoscyphus fraxineus, which had approximately 500 ascospores/ml based on counts with a hemocytometer.
TABLE 1.
Fungal species included in the mock community used in this study
| Isolate | Species name | Taxonomic classification | Host | Culture medium |
|---|---|---|---|---|
| LSV M54 | Cytospora sp. | Ascomycota, Sordariomycetes, Diaporthales, Valsaceae, Cytospora | Quercus ilex | MALT |
| LSV M60 | Neonectria coccinea | Ascomycota, Sordariomycetes, Hypocreales, Nectriaceae, Neonectria | Fagus sp. | MALT |
| LSV M73 | Pestalotiopsis sp. | Ascomycota, Sordariomycetes, Xylariales, Amphisphaeriaceae, Pestalotiopsis | Rhododendron sp. | MALT |
| LSV M373 | Fusarium graminearum | Ascomycota, Sordariomycetes, Hypocreales, Nectriaceae, Fusarium | Quercus sp. | SNA |
| LSV M500 | Diplodia pinea | Ascomycota, Dothideomycetes, Botryosphaeriales, Botryosphaeriaceae, Diplodia | Pinus sp. | MALT |
| LSV M568 | Diplodia corticola | Ascomycota, Dothideomycetes, Botryosphaeriales, Botryosphaeriaceae, Diplodia | Quercus petraea | MALT |
| LSV M672 | Fusarium lateritium | Ascomycota, Sordariomycetes, Hypocreales, Nectriaceae, Fusarium | Juglans sp. | SNA |
| LSV M746 | Cryphonectria radicalis | Ascomycota, Sordariomycetes, Diaporthales, Cryphonectriaceae, Cryphonectria | Castanea sativa | MALT |
| LSV M189 | Colletotrichum fioriniae | Ascomycota, Sordariomycetes, Glomerellales, Glomereaceae, Colletotrichum | Fragariae | MALT |
| LSV M728 | Cryphonectria parasitica | Ascomycota, Sordariomycetes, Diaporthales, Cryphonectriaceae, Cryphonectria | Castanea sativa | MALT |
| LNPV 212 | Fusarium circinatum | Ascomycota, Sordariomycetes, Hypocreales, Nectriaceae, Fusarium | Pinus radiata | SNA |
| Hymenoscyphus fraxineus | Ascomycota, Leotiomycetes, Helotiales, Helotiaceae, Hymenoscyphus | Fraxinus sp. | ||
| Melampsora larici-populina | Basidiomycota, Urediniomycetes, Uredinales, Melampsoraceae, Melampsora | Populus sp. |
Types of traps.
Five types of passive spore traps were tested. The first two types were based on the work of Schweigkofler et al. (19) and Grosdidier et al. (49) with cellulose paper filters. The traps consisted of Whatman paper filters (grades no. 1 and no. 3). These two types of filter differ in their diameters and pore sizes (150 mm/11 μm and 185 mm/6 μm for the no. 1 and no. 3 paper filters, respectively). The third and fourth types of trap were variants of this type: to test a potential increase in their trapping capacity, both types of Whatman filters (no. 1 and no. 3) were sprayed with a sticky layer of Tangle-Trap sticky coating (The Tanglefoot Company, Grand Rapids, MI). The last trapping method was based on a modified protocol described by Hopkins & Dick (26) using coated microscope slides. In our study, microscope slides were replaced by 100-mm-diameter petri dishes manually coated with an approximately 1-mm-thick mix of Vaseline and paraffin wax (both from Sigma-Aldrich) using a brush. Pictures of some of the spore traps are presented in the supplemental material (Fig. S6 and S7).
Experiment design.
The five types of spore traps described previously were artificially inoculated with the calibrated spore solution (mock community) under a security hood using a 1,000-μl pipette. Each trap was inoculated at 5 or 6 points on the surface with 1 ml of calibrated spore solutions containing between 500 and 1,000 spores, as described previously (Fig. S8). A first set of traps was inoculated with the mock community and was exposed for 15 days in August 2014 in an open field in Champenoux (World Geodetic System [WGS] 48°44′ 36.96″N, 6°21′0″E) in northeastern France. The spore traps were fixed on 12-cm-high 17-cm-wide polystyrene blocks placed 1 m above ground on a metal rod planted in the ground (Fig. S9). Three replicates of each spore trap were exposed. A second set of traps was also inoculated with the calibrated spore solutions, but in this case, traps were kept at the laboratory either in plastic bags (traps using paper filters) or in a flat plastic box (petri dishes) at 4°C until processed. Two or three replicates of each spore trap were kept at the laboratory. Spore solutions containing F. circinatum and C. parasitica were only used to inoculate the unexposed set of traps. The total number of exposed and unexposed spore traps was 114. Table 2 summarizes the experimental protocol with types of traps and laboratory protocols.
TABLE 2.
Description of the presequencing technical procedures used in this studya
| Type of trap | Spore recovery | DNA extraction | No. of unexposed replicates | No. of exposed replicates | Code |
|---|---|---|---|---|---|
| Whatman no. 1 | Shaking | Macherey-Nagel minikit | 3 | 3 | W1_S_mnM |
| Whatman no. 1 | Shaking | Qiagen minikit | 3 | 3 | W1_S_mnQ |
| Whatman no. 1 | Rubbing | Macherey-Nagel minikit | 3 | 3 | W1_R_mnM |
| Whatman no. 1 | Rubbing | Qiagen minikit | 3 | 3 | W1_R_mnQ |
| Whatman no. 1 | Grinding | Macherey-Nagel maxikit | 2 | 3 | W1_G_mxM |
| Whatman no. 1 | Grinding | Qiagen maxikit | 2 | 3 | W1_G_mxQ |
| Whatman no. 3 | Shaking | Qiagen minikit | 3 | 3 | W3_S_mnQ |
| Whatman no. 3 | Shaking | Macherey-Nagel minikit | 3 | 3 | W3_S_mnM |
| Whatman no. 3 | Rubbing | Qiagen minikit | 3 | 3 | W3_R_mnQ |
| Whatman no. 3 | Rubbing | Macherey-Nagel minikit | 3 | 3 | W3_R_mnM |
| Whatman no. 1 + glue | Rubbing | Macherey-Nagel minikit | 3 | 3 | W1g_R_mnM |
| Whatman no. 1 + glue | Rubbing | Qiagen minikit | 3 | 3 | W1g_R_mnQ |
| Whatman no. 1 + glue | Grinding | Qiagen maxikit | 2 | 3 | W1g_G_mxQ |
| Whatman no. 1 + glue | Grinding | Macherey-Nagel maxikit | 2 | 3 | W1g_G_mxM |
| Whatman no. 3 + glue | Rubbing | Macherey-Nagel minikit | 3 | 3 | W3g_R_mnM |
| Whatman no. 3 + glue | Rubbing | Qiagen minikit | 3 | 3 | W3g_R_mnQ |
| Petroleum jelly + paraffin wax | Grinding | Qiagen minikit | 3 | 3 | PJ_G_mnQ |
| Petroleum jelly + paraffin wax | Grinding | Qiagen maxikit | 2 | 3 | PJ_G_mxQ |
| Petroleum jelly + paraffin wax | Grinding | Macherey-Nagel minikit | 3 | 3 | PJ_G_mnM |
| Petroleum jelly + paraffin wax | Grinding | Macherey-Nagel maxikit | 2 | 3 | PJ_G_mxM |
A sample and its replicates were composed of the combination of the type of trap, the spore recovery procedure, and the DNA extraction kit. All unexposed and exposed samples were sequenced with the Illumina MiSeq technology targeting two subloci of the fungal internal transcribed spacers, ITS1 and ITS2.
Spore recovery.
Four methods for spore recovery were assessed (Table 2). The first method, here referred to as shaking (S), consisted of placing the paper filters in 20-cm glass petri dishes containing 40 ml of hot (70°C) 4× TE buffer (40 mM Tris-HCl, 4 mM EDTA; Sigma Chemical Co., St. Louis, MO), and 1 μl of IGEPAL CA-630 (Sigma-Aldrich). The petri dishes were subsequently shaken on a VWR 3600 orbital shaker (Radnor, PA) for 2 h. The spore suspensions were collected in 50-ml Falcon tubes. The second method, here referred to as rubbing (R), was adapted from Grosdidier et al. (24). Each filter was placed in a plastic bag, and then 20 ml (for the no. 1 paper filter) or 30 ml (for the no. 3 paper filter) of the 70°C preheated 4× TE buffer and IGEPAL CA-630 mix was added to the bag, thus soaking the filter. The paper filters were rubbed manually through the bag to detach the spores from the filters' surface and collect them in the buffer. As for the shaking method, spore suspensions were collected in 50-ml Falcon tubes after opening the plastic bags at one of the bottom corners using a sterile scalpel blade. In both methods, the recovered buffer containing the fungal spores was processed according to the following protocol: the 50-ml Falcon tubes were centrifuged for 15 min at 11,000 relative centrifugal force (rcf), after which the supernatant was discarded, leaving approximately 4 ml of suspension. After vortexing, the suspension was split between two microtubes. One 2-ml tube of FastPrep lysing matrix C (MP Biomedicals, Santa Ana, CA) was filled with 1.5 ml of the spore suspension, and a 2-ml Eppendorf tube was filled with 2 ml of the same suspension. Both tubes were centrifuged at 14,000 rcf for 5 min, after which the supernatant was discarded, leaving 750 μl of spore suspension in each tube. After vortexing, all of the spore suspension from the 2-ml Eppendorf tube was transferred into the lysing matrix C tube; this was finally centrifuged for 5 min at 14,000 rcf, after which the supernatant was discarded to leave a final 400-μl spore suspension. Spore lysis was performed using a FastPrep 24 system (MP Biomedicals) with a velocity setting of 6 m · s−1 and processing for two periods of 60 s.
The third method consisted of directly grinding the traps. This method is here referred to as grinding (G). Filters with and without the sticky layer were slightly humidified with sterile water and then placed in a sterilized grinding bowl. The filters were ground for 1 min using a Microtron MB 550 mixer mill (Kinematica, Lucerne, Switzerland). It must be pointed out that this method could only be applied to the no. 1 filters for safety reasons (overheating of the mixer mill with no. 3 filters). A subsample of 1 g was taken from the ground filters, placed in a 50-ml Falcon tube, and then stored at −20°C until DNA extraction. The surface of the petri dishes coated with the Vaseline and paraffin wax mix was scraped with a clean laboratory spatula. The recovered mixture was put into either a lysing matrix C tube or a 50-ml Falcon tube. The content of the lysing matrix C tubes was ground with a FastPrep 24 system, as described above. The Falcon tubes containing the ground filters were used for DNA extraction with maxikits, as described in “DNA extraction” (below).
DNA extraction.
Four different plant DNA extraction kits were used and compared during this study: the NucleoSpin plant II kit, the NucleoSpin plant II maxikit (both from Macherey-Nagel, Düren, Germany), the DNeasy plant minikit, and the DNeasy plant maxikit (both from Qiagen, Hilden, Germany). DNA from samples processed with the shaking method were extracted only with the minikits of both marks, whereas samples treated with the grinding and rubbing spore recovery method were extracted with both minikits and maxikits of both makes (Table 2). All the extractions complied with the manufacturers' specifications.
Marker choice and high-throughput sequencing.
The impact of marker choice in metabarcoding by NGS was assessed by targeting the ITS1 and ITS2 subloci of the internal transcribed spacer (ITS) of the ribosomal DNA (rDNA). Both subloci have been widely used for metabarcoding studies of fungal communities, notwithstanding the differences that can occur when describing fungal diversity from environmental samples (39). The first objective of our work was to assess the ability of both subloci to characterize airborne fungal communities. The second objective was to compare how short reads from ITS1 and ITS2 may differ in the diversity they describe in terms of taxon assignment, species richness, and abundance. All of the samples were therefore sequenced with both ITS1 and ITS2 subloci. Partial fragments of the ITS1 and ITS2 subloci were amplified independently for each type of sample using the primer pairs ITS1F/ITS2 (CTTGGTCATTTAGAGGAAGTAA/GCTGCGTTCTTCATCGATGC) (50, 51) and ITS86/ITS4 (GTGAATCATCGAATCTTTGAA/TCCTCCGCTTATTGATATGC) (51, 52), respectively. The extracted DNA was quality controlled by PCR using both primer pairs. The goals of these PCRs were to visually check the quality of the extracted DNA and to screen out potential problems due to PCR inhibition, DNA degradation, or loss during extraction. The PCR conditions applied were those proposed by McGuire et al. (53) for primer pair ITS1F/ITS2 and those proposed by Op De Beeck et al. (31) for primer pair ITS86/ITS4. The subsequent PCR amplifications, library preparation, primary data quality control, demultiplexing, and concatenation of the forward and reverse reads were carried out by the GenoScreen sequencing service (Lille, France).
Metabarcoding data bioinformatics.
First, the ITSx 1.0.11 software (54) was used to extract the fungal ITS. The extracted ITS1 and ITS2 sequences were then analyzed using mothur version 1.38.1 (6), in accordance with a standard operating procedure geared to fungal ITS (55). Reads were screened in order to keep sequences longer than 100 bp that had neither ambiguous bases nor homopolymers longer than 8 bp. The precluster command was used to remove sequences likely to be due to PCR and/or HTS errors. Chimeric sequences were identified using the chimera.uchime command and then removed from the data set. A pairwise distance matrix between sequences was generated using the pairwise.seqs command, with a cutoff of 0.20. Sequences were clustered into operational taxonomic units (OTUs) using the average neighbor algorithm. An OTU was defined at a 97% sequence similarity level. The command get.oturep was used to generate a fasta-formatted sequence file containing a representative sequence for each OTU. Sequences were taxonomically identified using the naive Bayesian classifier implemented in the classify.seqs command, using as reference the UNITE version 6 sh dynamic database (56). Taxonomic assignment was based on an 80% confidence threshold. A consensus taxonomy of OTUs was assigned using the classify.otu command. In addition, the taxonomic assignments for all OTUs were checked with Galaxy (57) using the GenBank database and the BLAST algorithm. The following criterion was used for identification with GenBank: whenever a taxonomic assignation at the species level with the UNITE database was at the >97% confidence threshold, this assignation was kept. Under this confidence threshold, the most representative sequence of the OTU was compared against the GenBank database, excluding environmental sequences. Taxonomical assignments at the species level with the lowest E value and 100% coverage were included in the data set (replacing, if different, the original taxonomic assignment with the UNITE database). The same processing was carried out with OTU assignments using the UNITE database at taxonomic levels higher than species. If BLAST-assigned sequences at the species level fulfilled the set criteria, this taxonomic assignment was given to the OTU (replacing, if different, the original taxonomic assignment made with the UNITE database). Whenever an OTU was not assigned at the species level (i.e., genus, family, order, etc.) and the criteria were fulfilled, the BLAST assignment was kept at the lowest taxonomic level. For taxonomic-level assignments other than species, OTUs were grouped together (for example, all OTUs taxonomically assigned as Alternaria sp. were considered the same OTU).
Analysis of metabarcoding data.
Statistical tests were performed either with R package vegan (58), phyloseq (59), or betareg (60). We first built a global data set containing sequence abundance OTU results per sample (a sample was the combination of type of trap, spore recovery, DNA extraction method, and molecular marker as shown in Table 2) for unexposed and exposed traps. The total number of sequences from unexposed and exposed traps was compared by a generalized linear model (GLM). A random rarefied OTU matrix was then computed using the smallest number of sequences per sample as a threshold. Considering the hypothesis that exposed traps should present greater levels of diversity than unexposed traps, we computed the following α-diversity indexes: the observed richness and the Shannon-Wiener index. Here, the observed richness was the number of observed OTUs, and the Shannon-Wiener index (here referred to as the Shannon index) was the sum of the proportion of each species relative to the total number of species in the sample under analysis. This index takes into account both abundance and evenness (61). All the indexes for unexposed and exposed traps were compared by GLM. In this study, for all GLM statistical analyses, the total number of sequences was considered to follow negative binomial distribution, geared to counting data with overdispersion. Concerning α-diversity indexes, a negative binomial distribution was used for the observed richness, and a Gaussian distribution was used for the Shannon index.
Further analyses were performed for the exposed data set. GLMs were used to assess the impact of the presequencing methods (type of trap, spore recovery method, and DNA extraction kit) and the genetic barcode (ITS1 and ITS2) on the raw number of sequences, the observed richness, and the Shannon index. The values of the observed richness and Shannon index were obtained from the rarefied OTU matrix computed previously. We also assessed the link between the total number of sequences per sample and the α-diversity indexes by Spearman's rank correlation.
Clustering patterns between samples were visualized with a nonmetric multidimensional scaling (NMDS) plot. The Bray-Curtis dissimilarity distance was used to compute the pairwise distances between all the exposed samples. The statistical significance of each technical choice was studied using an analysis of similarity (ANOSIM). For the exposed trap data set, heat map and clustering analyses were carried out using the relative abundances of the top 50 OTUs based on the Bray-Curtis distance. To visualize the relative abundance per type of exposed trap (grouping the three replicates) at the genus level, stacked bar plots were constructed from the relative abundance of the top 50 OTUs.
Quantitative PCR.
A primer-probe combination that targets a highly conserved 18S rDNA region for a wide range of organisms, including plants and fungi (62), was used for real-time PCR. The mix and PCR conditions were those proposed by Ioos et al. (62). Real-time PCR results were compared using the cycle threshold (CT) levels, considering that these levels are inversely proportional to the amount of target nucleic acid in the sample (i.e., the lower the CT level, the greater the amount of target nucleic acid in the sample). The CT value was first used to assess the hypothesis that exposed traps should contain a greater target DNA quantity than unexposed traps, expressed by lower CT levels. Concerning the exposed traps, differences in CT values (i.e., quantity of DNA present in a sample) were analyzed by GLM, using the type of trap, the spore recovery protocol, and DNA extraction method as explanatory variables. CT values were used as a response variable following a quasi-Poisson distribution. The link between the total quantities of DNA estimated by the 18S CT value, the total number of sequences, and the observed richness per sample was assessed by Spearman's rank correlation.
Real-time PCR was also used to assess the presence of two fungal species from the mock community in both sets of samples (unexposed and exposed). Specific primer-probe combinations designed to specifically detect Hymenoscyphus fraxineus (by Ioos et al. [63]) and Melampsora larici-populina (by Guinet et al. [64]) were used according to the mix conditions and PCR parameters described by the authors. Real-time PCR was used to verify the presence or absence of these target fungi. These results were compared to metabarcoding results verifying the presence of H. fraxineus and M. larici-populina in all the samples. Presence/absence was coded as 1/0 and compared by proportion tests under the null hypothesis that the probabilities of success in the groups are the same. Real-time PCR was first compared to ITS1 and ITS2 independently and then compared to the results from both barcodes grouped together.
Accession number(s).
Raw sequence reads have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA419481.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the RESIPATH project (APR ERA-NET BIODIVERSA). The European Cooperation in Science and Technology (COST) action PINESTRENGTH (FP1406) supported this study.
We thank Vincent Hervé (Max-Planck-Institut für Terrestrische Mikrobiologie, Germany) for his help in bioinformatics. We thank the reviewers for their valuable comments and effort to improve the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02637-17.
REFERENCES
- 1.Blackwell M. 2011. The fungi: 1, 2, 3 … 5.1 million species? Am J Bot 98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 2.Desprez-Loustau M-L, Aguayo J, Dutech C, Hayden KJ, Husson C, Jakushkin B, Marçais B, Piou D, Robin C, Vacher C. 2016. An evolutionary ecology perspective to address forest pathology challenges of today and tomorrow. Ann Forest Sci 73:45–67. doi: 10.1007/s13595-015-0487-4. [DOI] [Google Scholar]
- 3.Halme P, Heilmann-Clausen J, Rämä T, Kosonen T, Kunttu P. 2012. Monitoring fungal biodiversity–towards an integrated approach. Fungal Ecol 5:750–758. doi: 10.1016/j.funeco.2012.05.005. [DOI] [Google Scholar]
- 4.Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Kõljalg U, Pennanen T, Rosendahl S, Stenlid J, Kauserud H. 2013. Fungal community analysis by high-throughput sequencing of amplified markers – a user's guide. New Phytol 199:288–299. doi: 10.1111/nph.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bálint M, Schmidt P-A, Sharma R, Thines M, Schmitt I. 2014. An Illumina metabarcoding pipeline for fungi. Ecol Evol 4:2642–2653. doi: 10.1002/ece3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bálint M, Bahram M, Eren AM, Faust K, Fuhrman JA, Lindahl B, O'Hara RB, Öpik M, Sogin ML, Unterseher M, Tedersoo L. 2016. Millions of reads, thousands of taxa: microbial community structure and associations analyzed via marker genes. FEMS Microbiol Rev 40:686–700. doi: 10.1093/femsre/fuw017. [DOI] [PubMed] [Google Scholar]
- 8.Prigigallo MI, Abdelfattah A, Cacciola SO, Faedda R, Sanzani SM, Cooke DEL, Schena L. 2015. Metabarcoding analysis of Phytophthora diversity using genus-specific primers and 454 pyrosequencing. Phytopathology 106:305–313. doi: 10.1094/PHYTO-07-15-0167-R. [DOI] [PubMed] [Google Scholar]
- 9.Coince A, Cordier T, Lengellé J, Defossez E, Vacher C, Robin C, Buée M, Marçais B. 2014. Leaf and root-associated fungal assemblages do not follow similar elevational diversity patterns. PLoS One 9:e100668. doi: 10.1371/journal.pone.0100668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurzbacher C, Warthmann N, Bourne E, Attermeyer K, Allgaier M, Powell JR, Detering H, Mbedi S, Grossart H-P, Monaghan MT. 2016. High habitat-specificity in fungal communities in oligo-mesotrophic, temperate Lake Stechlin (North-East Germany). MycoKeys 16:17–44. doi: 10.3897/mycokeys.16.9646. [DOI] [Google Scholar]
- 11.Català S, Pérez-Sierra A, Abad-Campos P. 2015. The use of genus-specific amplicon pyrosequencing to assess Phytophthora species diversity using eDNA from soil and water in northern Spain. PLoS One 10:e0119311. doi: 10.1371/journal.pone.0119311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross H, Sønstebø JH, Nagy NE, Timmermann V, Solheim H, Børja I, Kauserud H, Carlsen T, Rzepka B, Wasak K, Vivian-Smith A, Hietala AM. 2017. Fungal diversity and seasonal succession in ash leaves infected by the invasive ascomycete Hymenoscyphus fraxineus. New Phytol 213:1405–1417. doi: 10.1111/nph.14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlsson I, Edel-Hermann V, Gautheron N, Durling MB, Kolseth A-K, Steinberg C, Persson P, Friberg H. 2016. Genus-specific primers for study of Fusarium communities in field samples. Appl Environ Microbiol 82:491–501. doi: 10.1128/AEM.02748-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller KE, Hopkins K, Inward DJG, Vogler AP. 2016. Metabarcoding of fungal communities associated with bark beetles. Ecol Evol 6:1590–1600. doi: 10.1002/ece3.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malacrinò A, Schena L, Campolo O, Laudani F, Mosca S, Giunti G, Strano CP, Palmeri V. 2017. A metabarcoding survey on the fungal microbiota associated to the olive fruit fly. Microb Ecol 73:677–684. doi: 10.1007/s00248-016-0864-z. [DOI] [PubMed] [Google Scholar]
- 16.West JS, Kimber RBE. 2015. Innovations in air sampling to detect plant pathogens. Ann Appl Biol 166:4–17. doi: 10.1111/aab.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West JS, Atkins SD, Emberlin J, Fitt BDL. 2008. PCR to predict risk of airborne disease. Trends Microbiol 16:380–387. doi: 10.1016/j.tim.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Leyronas C, Halkett F, Nicot PC. 2015. Relationship between the genetic characteristics of Botrytis sp. airborne inoculum and meteorological parameters, seasons and the origin of air masses. Aerobiologia 31:367–380. doi: 10.1007/s10453-015-9370-x. [DOI] [Google Scholar]
- 19.Schweigkofler W, O'Donnell K, Garbelotto M. 2004. Detection and quantification of airborne conidia of Fusarium circinatum, the causal agent of pine pitch canker, from two California sites by using a real-time PCR approach combined with a simple spore trapping method. Appl Environ Microbiol 70:3512–3520. doi: 10.1128/AEM.70.6.3512-3520.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandelier A, Helson M, Dvorak M, Gischer F. 2014. Detection and quantification of airborne inoculum of Hymenoscyphus pseudoalbidus using real-time PCR assays. Plant Pathol 63:1296–1305. doi: 10.1111/ppa.12218. [DOI] [Google Scholar]
- 21.Jackson SL, Bayliss KL. 2011. Spore traps need improvement to fulfil plant biosecurity requirements. Plant Pathol 60:801–810. doi: 10.1111/j.1365-3059.2011.02445.x. [DOI] [Google Scholar]
- 22.Mahaffee WF, Stoll R. 2016. The ebb and flow of airborne pathogens: monitoring and use in disease management decisions. Phytopathology 106:420–431. doi: 10.1094/PHYTO-02-16-0060-RVW. [DOI] [PubMed] [Google Scholar]
- 23.Calderon C, Ward E, Freeman J, McCartney A. 2002. Detection of airborne fungal spores sampled by rotating-arm and Hirst-type spore traps using polymerase chain reaction assays. J Aerosol Sci 33:283–296. doi: 10.1016/S0021-8502(01)00179-3. [DOI] [Google Scholar]
- 24.Grosdidier M, Aguayo J, Marcais B, Ioos R. 2017. Detection of plant pathogens using real-time PCR: how reliable are late CT values? Plant Pathol 66:359–367. doi: 10.1111/ppa.12591. [DOI] [Google Scholar]
- 25.Gonthier P, Garbelotto MM, Nicolotti G. 2005. Seasonal patterns of spore deposition of Heterobasidion species in four forests of the western Alps. Phytopathology 95:759–767. doi: 10.1094/PHYTO-95-0759. [DOI] [PubMed] [Google Scholar]
- 26.Hopkins A, Dick M. 2009. An examination of assessor variation during spore counting on spore traps of Neonectria fuckeliana. N Z Plant Prot 62:250–255. [Google Scholar]
- 27.Funnell-Harris DL, Pedersen JF. 2011. Presence of Fusarium spp. in air and soil associated with sorghum fields. Plant Dis 95:648–656. doi: 10.1094/PDIS-09-10-0671. [DOI] [PubMed] [Google Scholar]
- 28.Fourie G, Wingfield MJ, Wingfield BD, Jones NB, Morris AR, Steenkamp ET. 2014. Culture-independent detection and quantification of Fusarium circinatum in a pine-producing seedling nursery. South For 76:137–143. doi: 10.2989/20702620.2014.899058. [DOI] [Google Scholar]
- 29.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium . 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, Saar I, Bahram M, Bechem E, Chuyong G, Kõljalg U. 2010. 454 pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol 188:291–301. doi: 10.1111/j.1469-8137.2010.03373.x. [DOI] [PubMed] [Google Scholar]
- 31.Op De Beeck M, Lievens B, Busschaert P, Declerck S, Vangronsveld J, Colpaert JV. 2014. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One 9:e97629. doi: 10.1371/journal.pone.0097629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terrat S, Plassart P, Bourgeois E, Ferreira S, Dequiedt S, Adele-Dit-De-Renseville N, Lemanceau P, Bispo A, Chabbi A, Maron P-A, Ranjard L. 2015. Meta-barcoded evaluation of the ISO standard 11063 DNA extraction procedure to characterize soil bacterial and fungal community diversity and composition. Microb Biotechnol 8:131–142. doi: 10.1111/1751-7915.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amend AS, Seifert KA, Bruns TD. 2010. Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol Ecol 19:5555–5565. doi: 10.1111/j.1365-294X.2010.04898.x. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen NH, Smith D, Peay K, Kennedy P. 2015. Parsing ecological signal from noise in next generation amplicon sequencing. New Phytol 205:1389–1393. doi: 10.1111/nph.12923. [DOI] [PubMed] [Google Scholar]
- 35.Taylor DL, Walters WA, Lennon NJ, Bochicchio J, Krohn A, Caporaso JG, Pennanen T. 2016. Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for Illumina amplicon sequencing. Appl Environ Microbiol 82:7217–7226. doi: 10.1128/aem.02576-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castaño C, Oliva J, de Aragón JM, Alday JG, Parladé J, Pera J, Bonet JA. 2017. Mushroom emergence detected by combining spore trapping with molecular techniques. Appl Environ Microbiol 83:e00600-. doi: 10.1128/AEM.00600-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. 2010. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blaalid R, Kumar S, Nilsson RH, Abarenkov K, Kirk P, Kauserud H. 2013. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol Ecol Resour 13:218–224. doi: 10.1111/1755-0998.12065. [DOI] [PubMed] [Google Scholar]
- 39.Monard C, Gantner S, Stenlid J. 2013. Utilizing ITS1 and ITS2 to study environmental fungal diversity using pyrosequencing. FEMS Microbiol Ecol 84:165–175. doi: 10.1111/1574-6941.12046. [DOI] [PubMed] [Google Scholar]
- 40.Mello A, Napoli C, Murat C, Morin E, Marceddu G, Bonfante P. 2011. ITS-1 versus ITS-2 pyrosequencing: a comparison of fungal populations in truffle grounds. Mycologia 103:1184–1193. doi: 10.3852/11-027. [DOI] [PubMed] [Google Scholar]
- 41.Wang X-C, Liu C, Huang L, Bengtsson-Palme J, Chen H, Zhang J-H, Cai D, Li J-Q. 2015. ITS1: a DNA barcode better than ITS2 in eukaryotes? Mol Ecol Resour 15:573–586. doi: 10.1111/1755-0998.12325. [DOI] [PubMed] [Google Scholar]
- 42.Bazzicalupo AL, Bálint M, Schmitt I. 2013. Comparison of ITS1 and ITS2 rDNA in 454 sequencing of hyperdiverse fungal communities. Fungal Ecol 6:102–109. doi: 10.1016/j.funeco.2012.09.003. [DOI] [Google Scholar]
- 43.Sapkota R, Nicolaisen M. 2015. An improved high throughput sequencing method for studying oomycete communities. J Microbiol Methods 110:33–39. doi: 10.1016/j.mimet.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Tedersoo L, Tooming-Klunderud A, Anslan S. 2017. PacBio metabarcoding of Fungi and other eukaryotes: errors, biases and perspectives. New Phytol 217:1370–1385. doi: 10.1111/nph.14776. [DOI] [PubMed] [Google Scholar]
- 45.Adams RI, Miletto M, Taylor JW, Bruns TD. 2013. Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J 7:1460–1460. doi: 10.1038/ismej.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.European Union. 2008. Commission directive 2008/61/EC of 17 June 2008 establishing the conditions under which certain harmful organisms, plants, plant products and other objects listed in Annexes I to V to Council Directive 2000/29/EC may be introduced into or moved within the Community or certain protected zones thereof, for trial or scientific purposes and for work on varietal selection. European Union, Brussels, Belgium: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32008L0061. [Google Scholar]
- 47.Husson C, Ioos R, Andrieux A, Frey P. 2013. Development and use of new sensitive molecular tools for diagnosis and detection of Melampsora rusts on cultivated poplar. For Pathol 43:1–11. [Google Scholar]
- 48.Husson C, Scala B, Caël O, Frey P, Feau N, Ioos R, Marçais B. 2011. Chalara fraxinea is an invasive pathogen in France. Eur J Plant Pathol 130:311–324. doi: 10.1007/s10658-011-9755-9. [DOI] [Google Scholar]
- 49.Grosdidier M, Aguayo J, Marçais B, Ioos R. 2016. Detection of plant pathogens using real-time PCR: how reliable are late CT values? Plant Pathol 66:359–367. doi: 10.1111/ppa.12591. [DOI] [Google Scholar]
- 50.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 51.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA. [Google Scholar]
- 52.Turenne CY, Sanche SE, Hoban DJ, Karlowsky JA, Kabani AM. 1999. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol 37:1846–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGuire KL, Payne SG, Palmer MI, Gillikin CM, Keefe D, Kim SJ, Gedallovich SM, Discenza J, Rangamannar R, Koshner JA, Massmann AL, Orazi G, Essene A, Leff JW, Fierer N. 2013. Digging the New York City skyline: soil fungal communities in green roofs and city parks. PLoS One 8:e58020. doi: 10.1371/journal.pone.0058020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, De Wit P, Sánchez-García M, Ebersberger I, de Sousa F, Amend A, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH. 2013. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919. doi: 10.1111/2041-210X.12073. [DOI] [Google Scholar]
- 55.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H. 2013. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 57.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oksanen J, Blanchet G, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2007. vegan: community ecology package. https://cran.r-project.org/web/packages/vegan/index.html.
- 59.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cribari-Neto F, Zeileis A. 2009. Beta regression in R. J Stat Softw 34:1–24. doi: 10.18637/jss.v034.i02. [DOI] [Google Scholar]
- 61.Jovel J, Patterson J, Wang W, Hotte N, O'Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, Wong GKS. 2016. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol 7:459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ioos R, Fourrier C, Iancu G, Gordon TR. 2009. Sensitive detection of Fusarium circinatum in pine seed by combining an enrichment procedure with a real-time polymerase chain reaction using dual-labeled probe chemistry. Phytopathology 99:582–590. doi: 10.1094/PHYTO-99-5-0582. [DOI] [PubMed] [Google Scholar]
- 63.Ioos R, Kowalski T, Husson C, Holdenrieder O. 2009. Rapid in planta detection of Chalara fraxinea by a real-time PCR assay using a dual-labelled probe. Eur J Plant Pathol 125:329–335. doi: 10.1007/s10658-009-9471-x. [DOI] [Google Scholar]
- 64.Guinet C, Boutigny AL, Vialle A, Hamelin RC, Frey P, Ioos R. 2016. Simultaneous monitoring and quantification of Melampsora allii-populina and Melampsora larici-populina on infected poplar leaves using a duplex real-time PCR assay. Plant Pathol 65:380–391. doi: 10.1111/ppa.12426. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.