Abstract
Ribosome biogenesis requires more than 200 trans-acting factors to achieve the correct production of the two mature ribosomal subunits. Here, we have identified Efg1 as a novel, nucleolar ribosome biogenesis factor in Saccharomyces cerevisiae that is directly linked to the surveillance of pre-40S particles. Depletion of Efg1 impairs early pre-rRNA processing, leading to a strong decrease in 18S rRNA and 40S subunit levels and an accumulation of the aberrant 23S rRNA. Using Efg1 as bait, we revealed a novel degradation pathway of the 23S rRNA. Co-immunoprecipitation experiments showed that Efg1 is a component of 90S pre-ribosomes, as it is associated with the 35S pre-rRNA and U3 snoRNA, but has stronger affinity for 23S pre-rRNA and its novel degradation intermediate 11S rRNA. 23S is cleaved at a new site, Q1, within the 18S sequence by the endonuclease Utp24, generating 11S and 17S' rRNA. Both of these cleavage products are targeted for degradation by the TRAMP/exosome complexes. Therefore, the Q1 site defines a novel endonucleolytic cleavage site of ribosomal RNA exclusively dedicated to surveillance of pre-ribosomal particles.
INTRODUCTION
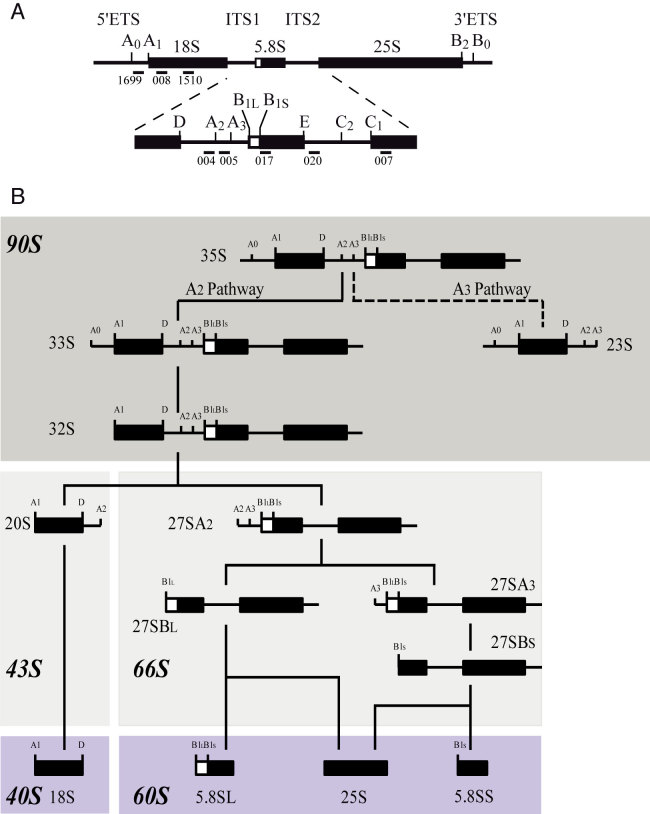
Eukaryotic ribosomes are large ribonucleoprotein (RNP) particles composed of the small (40S) and large (60S) subunits that assemble on the messenger RNA (mRNA), allowing its translation into protein. The 40S subunit is assembled around the 18S rRNA, whereas the 60S particle contains the 25S/28S, 5.8S and 5S rRNAs. Ribosome biogenesis is a highly complex process which begins in the nucleolus with the rDNA transcription by the RNA polymerase I (Pol I) leading in the yeast Saccharomyces cerevisiae to the production of the 35S pre-rRNA precursor (Figure 1A). This precursor contains the mature 18S, 5.8S and 25S rRNA sequences with external (5′-ETS and 3′-ETS) and internal (ITS1 and ITS2) transcribed spacers, which are absent from mature ribosomes. Maturation of this 35S pre-rRNA involves a well-defined series of endonucleolytic steps (Figure 1A), followed by exonucleolytic maturation (Illustrated in Figure 1B) that eliminate ETS and ITS sequences in order to release 18S, 5.8S and 25S mature rRNAs (for reviews, see (1,2)). The 5S rRNA is synthesized independently by RNA polymerase III (Pol III) and is incorporated as part of a small RNP particle (3).
Figure 1.
Simplified scheme of the pre-rRNA processing pathway in Saccharomyces cerevisiae. (A) Initial 35S pre-rRNA precursor with detailed cleavage sites as well as probes used for rRNA detection in northern blot analysis. (B) Endonucleolytic and exonucleolytic cleavages leading to the production of mature 18S, 5.8S and 25S rRNAs. Inhibition of A0, A1 and A2 cleavages (A2 pathway) leads to the non-productive A3 pathway. Resulting 23S pre-rRNA containing particles accumulate and/or are degraded. Dark and light gray boxes schematically represent 90S and 43S/66S pre-ribosomes, respectively. Mature ribosomal subunits are represented in violet.
Many trans-acting factors and some ribosomal proteins start assembling and stabilize the nascent RNA Pol I transcript leading to the formation of a large RNP complex called the small-subunit (SSU) processome or the 90S pre-ribosome (4,5); for review, see (6,7). In S. cerevisiae, more than 200 trans-acting factors, also called assembly factors, have been shown to participate directly in co- and post-transcriptional steps of ribosome biogenesis. Among these assembly factors, exo- and endonucleases are responsible for transcribed spacer processing and elimination; adenosine triphosphate (ATP)-dependent RNA helicases are involved in RNA folding or remodeling; ATPases, GTPases, kinases and other energy consuming factors are essential for the process; and the function(s) of a large class of other proteins are still unclear (7). The first particle to be assembled, the SSU processome, is a large RNP complex containing the U3 small nucleolar RNA (snoRNA). Within this pre-ribosomal complex, the three early pre-rRNA cleavages at sites A0, A1 and A2 (4,8,9), defining the ‘A2 pathway’, take place (Figure 1B). Protein-protein and protein–RNA interaction data lead to the conclusion that pre-ribosomes are sequentially assembled from multiple independently formed modules (10–12).
The SSU processome integrity is of critical importance for the early, mostly co-transcriptional, endonucleolytic cleavages, which lead to the separation of the 43S and 66S particles, precursors of the mature ribosomal subunits. In consequence, individual loss of most SSU processome factors inhibits these pre-rRNA cleavages at sites A0, A1 and A2 and leads to the accumulation of the 23S pre-rRNA (‘A3 pathway’; Figure 1B). Utp24 has been proposed as the endonuclease enzyme for the A1 and A2 sites (13). Mutation of residues in the predicted active site of Utp24 leads to reduced cell growth and defects in cleavage at the A1 and A2 sites. Consistent with a direct involvement in these cleavages, Utp24 exhibited in vitro endonuclease activity on an RNA substrate containing the A2 site. Moreover, the integrity of the Utp24 PINc domain is required for efficient cleavage at A2 site, both in yeast and human (14).
Inhibition of cleavages at sites A0, A1 and A2 leads to the accumulation of the 23S pre-rRNA (see Figure 1). The 23S pre-rRNA results from a direct cleavage of the 35S pre-rRNA at site A3, within ITS1, by RNAse MRP (Figure 1B and 8) (15,16). Depending on the yeast genetic background and growth conditions, 23S pre-rRNA is invariably present but in various amounts in wild-type (WT) cells. 23S pre-rRNA markedly accumulates in all ribosome biogenesis mutants affecting early processing events. Surprisingly, despite containing the entire 18S rRNA sequence, accumulated 23S pre-rRNA does not seem to be further processed (17,18). When it is produced, 23S pre-rRNA is efficiently targeted by a surveillance pathway (17,19,20). This quality-control mechanism involves polyadenylation of the targeted RNA by the TRAMP (TRf4/5-Air1/2-Mtr4 Polyadenylation) complex and its degradation by the nuclear exosome (reviewed in (21)). Degradation of aberrant pre-ribosomes is clearly of paramount importance to ensure the fidelity of gene expression by avoiding the production of defective ribosomes. The translation of mRNAs by such ribosomes would potentially lead to the production of mutated or truncated proteins with deleterious effects for the cell. Moreover, degradation of accumulated pre-ribosomes would also be needed for release and recycling of factors to make them available to new particles.
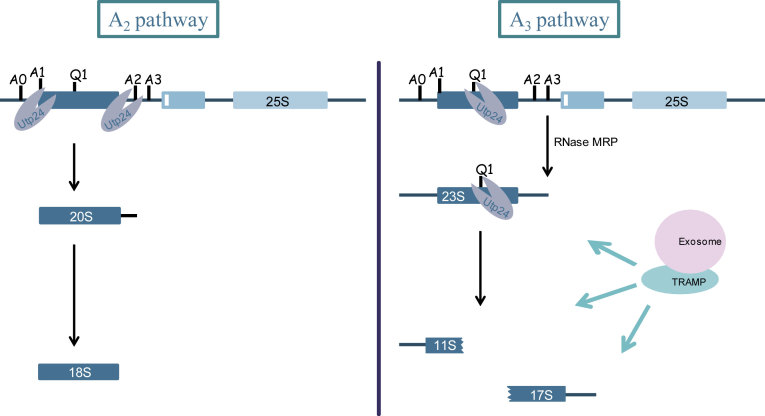
Figure 8.
Proposed model for the function of Utp24 in the ‘productive’ A2 pathway and the ‘non productive’ A3 pathway. In normal growing conditions, Utp24 cleaves at A1 and A2 sites leading to the production of the 20S pre-rRNA and subsequently the production of the mature 18S rRNA. In case of defective assembly and/or processing of early pre-ribosomal particles or if growing conditions are not optimal, A1 and A2 cleavage sites are not processed by Utp24. 35S pre-rRNA is directly cleaved at A3 site by RNase MRP. Resulting 23S pre-rRNA is targeted by Utp24 at Q1 site generating 11S and 17S'. All of them are degraded by the TRAMP/exosome pathways.
In this work, we studied EFG1, a gene encoding a non-essential and poorly characterized protein localized in the nucleolus. It was previously described that deletion of EFG1 causes slow growth at 30°C, thermosensitivity at 37°C and alters 18S/25S rRNA ratio (22,23). We present evidence that EFG1 encodes a ribosome biogenesis factor involved in 35S pre-rRNA processing at A0, A1 and A2 sites. Efg1 depletion leads to the accumulation of the 35S pre-rRNA and the aberrant 23S pre-rRNA with subsequent decrease of 18S rRNA level. We demonstrate that Efg1 is associated with the 35S pre-rRNA and U3 snoRNP, but also with 23S and a novel 11S rRNA. We propose that the 11S and the previously observed 17S' rRNAs (17,19,24) are the result of 23S pre-rRNA cleavage at site Q1 by Utp24 endonuclease. This is the first demonstration of the involvement of Utp24 in the elimination of unprocessed particles and in consequence, recycling of assembly factors.
MATERIALS AND METHODS
Strains, media, plasmids and cloning
Standard procedures were used for the propagation of yeast using YPD medium (1% yeast extract, 2% peptone and 2% glucose), YPG medium (1% yeast extract, 2% peptone and 2% galactose) or YNB medium (0.67% yeast nitrogen base, 0.5% (NH4)2SO4 and 2% glucose or galactose) supplemented with the required amino acids. Yeast strains used in this study were derivatives of S. cerevisiae S288C or BY4741, originating from the S288C background (MATa, his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). EFG1::TAP strain was purchased from Open Biosystems where insertion of the tandem affinity purification (TAP) cassette was selected using HIS3MX6 marker. The strain was grown exponentially and no growth phenotype was observed neither at 30°C nor at 37°C. EFG1Δ strain was purchased from Euroscarf haploïd system where EFG1 open reading frame was replaced by a kanamycin resistance selectable marker (kanR). The GAL1::3HA::EFG1 strain, expressing 3HA-Efg1 under the control of a GAL1 promoter was constructed as follows: a polymerase chain reaction (PCR) cassette containing the kanamycin resistance (kanR) selectable marker, GAL1 promoter and the 3HA-tag sequence was amplified by PCR from the plasmid pFA6a-KanMX6-PGAL1–3HA using primers 580 and 581 (25) (see Table 1). The PCR fragment was inserted by homologous recombination upstream of the chromosomal EFG1 open reading frame in the BY4741 strain. Transformants were selected for resistance to kanamycin and screened by immunoblotting.
Table 1. Oligonucleotides used to construct strains.
| Oligo | Sequence |
|---|---|
| 580 | GAAGCTCAAAAATATGTATTCCTTTAATTTCTGCGGTACCCTATGCCGGAGCACGAATTCGAGCTCGTTTAAAC |
| 581 | CATCTCTAGAGATGACCCAAGAGCCTTGCTTCTCTTCCTCTGTAATTTAGCCATGCACTGAGCAGCGTAATCTG |
| 700 | TCGATGAATTCGAGCTCGTT |
| 701 | GGTCGACGGATCCCCGGGTT |
| CS-447 | GGCGGTCACGCATACGTCATTGAAAAATTGCCAGATGTCTTTCGGATCCCCGGGTTAATTAA |
| CS-448 | CCAAAAACATTGCACTACATACTTAAAGAGTGAATACACAGTAGTGAATTCGAGCTCGTTTAAAC |
| CS 327 | AGCTGTCGACGTGTTTCAGTCCCAGTGGC |
| CS-332 | ACGTAGTCGACGCTGGATGGGAAGCGTACC |
| CS-599 | GAGAATTTGTATTTTCAGGGTTAGCTCAAAACCGCGGCTCTTG |
| CS-600 | CAAGAGCCGCGGTTTTGAGCTAACCCTGAAAATACAAATTCTC |
The EFG1::YFP strain containing a plasmid pUN100 NOP1::mCherry was constructed as follows: a PCR cassette containing the URA3 gene from Kluyveromyces lactis as selectable marker and YFP-tag sequence was amplified by PCR from the plasmid pFA6a-YFP-URA using primers 700 and 701. The PCR fragment was inserted by homologous recombination in the EFG1::TAP strain using the Swap-tag method (26). Transformants were selected for uracil prototrophy and screened by immunoblotting. In a second step, plasmid pUN100 NOP1::mCherry was transformed in the resulting strain (27).
The Tet::UTP24::3HA strain expressing Utp24–3HA under the control of a Tet promoter was constructed as follows: a PCR cassette containing the 3HA-tag sequence and the HIS3 gene as selectable marker was amplified by PCR from the plasmid pFA6a-3HA-His3-MX6 using CS-447 and CS-448 primers. The PCR fragment was inserted by homologous recombination in the Tet::UTP24 strain (pYDR339C::kanR-tet07-TATA, URA3::CMV-tTA, MATa, his3–1, leu2–0 met15–0; gift from Mike Tyers’s laboratory). Deletion of RRP6 was performed as previously described (28).
Plasmids expressing WT or D68N Utp24 were constructed as follows. HTP-tagged forms of Utp24 were initially amplified using CS-327 and CS-332 from UTP24::HTP strain described in (14). The PCR fragments were cloned into pRS315 plasmid cut with SalI.
Oligonucleotides used to construct UTP24::D68N mutant plasmid by site-directed mutagenesis are described in (14). HTP tag was next shortened at their C-terminus by site directed mutagenesis introducing a stop codon immediately after the TEV cleavage site using CS-599 and CS-600 oligonucleotides. The expression of the protein remains detectable with the anti-TAP antibody (CAB1001, Thermo Scientific).
Fluorescence microscopy
For fluorescence microscopy, cells were grown in YPD medium at 30°C to an OD600 of 0.6. Aliquots were collected, washed and resuspended in dextrose-containing YNB supplemented with the needed amino acids. After washing, the cells were mounted on a slide and observed using the fluorescence microscope IX-81 (Olympus) equipped with a polychrome V monochromator and a CoolSNAP HQ camera (Roper Industries). Digital pictures were processed using Photoshop software (CS3 version; Adobe).
Western analyses
Proteins from total extracts, obtained from gradient fractions after trichloroacetic acid (TCA) precipitation or from immunoprecipitated pellets, were separated on 4–12% polyacrylamide/SDS gels (Invitrogen) and transferred to hybond-ECL membranes (GE Healthcare). Nop1 was detected as described in (29). TAP-tagged proteins were detected using rabbit PAP (Sigma) diluted 10 000 fold. HA-tagged proteins were detected using anti-HA peroxydase antibodies (Roche) diluted 1000-fold.
Sucrose gradient sedimentation experiments
Sucrose gradient sedimentation experiments were performed as previously described (30).
RNA extractions and northern hybridizations
RNA extractions and northern hybridizations were performed as previously described (31). For high molecular weight RNA analysis, 2 μg of total RNA were glyoxal denatured and resolved on a 1.2% agarose gel. Low molecular weight RNA products were resolved on 8% Polyacrylamide/8.3M urea gels. The sequences of oligonucleotides used to detect the different RNA species are reported in Table 2.
Table 2. Oligonucleotides used to probe RNAs.
| Primers | Sequence | Used to detect |
|---|---|---|
| 004 | 5′-CGGTTTTAATTGTCCTA | 35S, 33/32S, 20S, 23S, 22S, 21S and 17S' rRNAs |
| 005 | 5′-ATGAAAACTCCACAGTG | 35S, 33/32S, 23S, 27SA2 and 17S' rRNAs |
| 007 | 5′-CTCCGCTTATTGATATGC | 25S rRNA |
| 008 | 5′-CATGGCTTAATCTTTGAGAC | 18S rRNA |
| 017 | 5′-GCGTTGTTCATCGATGC | 5.8S, 7S rRNAs |
| 020 | 5′-TGAGAAGGAAATGACGCT | 35S, 33/32S, 27S rRNAs |
| 041 | 5′-CTACTCGGTCAGGCTC | 5S rRNA |
| 200 | 5′-UUAUGGGACUUGUU | U3 snoRNA |
| 209 | 5′-GGAAGGTCTCTAAAGAAG | U14 snoRNA |
| 218 | 5′-CATGGGTCAAGAACGCCCCGGAGGGG | snR10 snoRNA |
| 228 | 5′-CATCCAGCTCAAGATCG | snR36 snoRNA |
| 231 | 5′-ATGTCTGCAGTATGGTTTTAC | snR30 snoRNA |
| 271 | 5′-GATTGTCCACACACTTCT | snR33 snoRNA |
| 403 | 5′-ACCGTTTGGTCTACCCAAGTGAGAAGCCAAGACA | PGK1 mRNA |
| 1000 | 5′-CAAAAGAGGCTGACGTT | snR31 snoRNA |
| 1016 | 5′-ATACCGGTCCTTGGAAT | snR189 snoRNA |
| 1510 | 5′ -GCTAATATATTCGAGCAATACG | 18S and 11S rRNA |
| 1699 | 5′-CACCTATTCCCTCTTGCTAG | 35S, 33S, 23S, 22S, 21S and 11S rRNAs |
Immunoprecipitations
Total cell extracts were prepared from strains expressing TAP-tagged proteins or untagged strains as control. Growing cells were frozen in liquid nitrogen and broken in a mortar. Immunoprecipitations and analysis of co-precipitated RNAs were performed as previously described (32). Tot/IP ratios loaded were 1/20 for agarose and acrylamide gels.
Tandem affinity purifications
Cell pellets (corresponding to about 5 × 1010 cells) frozen in liquid nitrogen were broken in a mortar and resuspended in A200 KCl buffer (20 mM Tris–Cl [pH 8.0], 5 mM MgAc, 200 mM KCl, 0.2% Triton X-100) supplemented with 1 mM dithiothreitol, 1× Complete ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor cocktail (Roche), and 0.5 U/μl RNasin (Promega).
For TAP purifications under native conditions, cell extracts were clarified by centrifugation at 14 000 rpm (21 000 × g) for 10 min at 4°C. Clarified extracts were incubated with 200 μl (bed volume) of IgG-Sepharose beads (GE Healthcare) for 4 h on a rocking table. Beads were extensively washed with A200. Columns were next equilibrated with ice-cold TEV cleavage buffer (10 mM Tris–Cl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM dithiothreitol). Beads were resuspended with 1 ml of TEV cleavage buffer and incubated for 2 h at 16°C on a rocking table in the presence of 100 units of ActTEV protease (Invitrogen). Eluted samples (about 1 ml) were mixed with 3 ml of calmodulin binding buffer (10 mM Tris–Cl [pH 8.0], 150 mM NaCl, 1 mM MgAc, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40, 10 mM ß-mercaptoethanol) and 3 μl of 1 M CaCl2 and incubated with 200 μl of calmodulin beads (Stratagene) at 4°C for 1 h on a rocking table. Beads were washed with 40 ml of calmodulin binding buffer, and proteins were eluted by the addition of 6 × 200 μl of calmodulin elution buffer (10 mM Tris–Cl [pH 8.0], 150 mM NaCl, 1 mM MgAc, 1 mM imidazole, 2 mM EGTA, 0.1% NP-40, 10 mM β-mercaptoethanol). Eluted proteins were precipitated with 20% TCA, rinsed with acetone, and resuspended with 20 μl of 1× sodium dodecyl sulphate (SDS) gel-loading buffer (40 mM Tris–Cl [pH 6.8], 2% SDS, 10% glycerol, 25 mM dithiothreitol, 0.1% bromophenol blue). Samples were loaded on 4–12% Bis-Tris gels. Gels were briefly (15 min) stained with silver staining solution (Fermentas), and pieces of gels containing the samples were excised. The proteins contained in these samples were analyzed by mass spectrometry as described (33).
Pulse chase analysis
Metabolic labeling of pre-rRNAs was performed as previously described (34) with the following modifications. Strains were pre-grown in synthetic galactose medium lacking adenine and transferred to glucose medium lacking adenine for 3 h (GAL1::3HA::EFG1). Cultures at OD600 0.8 were labeled with [2,8–3H]-adenine (NET06300 Perkin Elmer) for 2 min followed by a chase with excess cold adenine. The 1 ml samples were collected 1, 2, 5, 10, 20 and 30 min following the addition of cold adenine and cell pellets were frozen in liquid nitrogen. RNAs were then extracted and precipitated with ethanol.
RESULTS
Efg1 is a nucleolar protein associated with pre-ribosomes
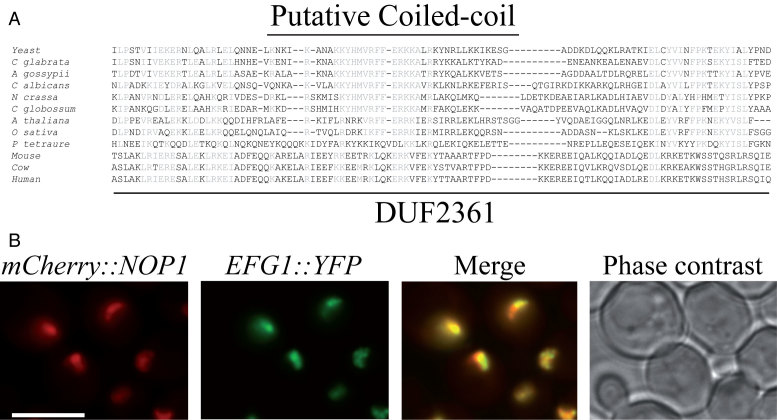
First described as two distinct open reading frames (ORF), YGR271C-A and YGR272C, EFG1 (Exit oF G1) encodes a protein of 233 amino acids and has been identified as a non essential gene in S. cerevisiae (22,23). Efg1 deletion resulted in a slow growth phenotype at all temperatures, and is fully lethal at 37°C (22); data not shown). Furthermore, S. cerevisiae strains containing a large deletion in the 5′ domain of Efg1 exhibited a 18S/25S rRNA ratio imbalance (23) suggesting a role in ribosome biogenesis. We first searched for possible Efg1 orthologs in other organisms or for conserved protein domains. Efg1 seems to be well conserved in the ascomycete phylum. Orthologs of Efg1 are also found in plants. Furthermore, it contains a domain of unknown function, DUF2361 (pfam10153), which is described as a putative coiled-coil domain, conserved through eukaryotes (Figure 2A).
Figure 2.
Efg1 is a nucleolar protein. (A) Conserved domain in Efg1. Amino acid alignment of the DUF2361 domain of Efg1 with yeast homologs, plant and mammalian orthologs. Sequence alignments were generated using Clustal W. S. cerevisiae: Saccharomyces cerevisiae; C. glabrata: Candida glabrata; A. gossypii: Ashbya gossypii; C. albicans: Candida albicans; N. crassa: Neurospora crassa; C. globossum: Candida globossum; A. thaliana: Arabidopsis thaliana; O. sativa: Oryza sativa. The putative coiled-coil region is highlighted. (B) Subcellular localization of Efg1. Yeast strain expressing Efg1-YFP and mCherry-Nop1 was grown exponentially and cell samples were used for fluorescence microscopy analysis.
Previous global analyses of yeast protein localization have shown that the GFP tagged Efg1 protein is enriched in the nucleus and/or in the nucleolus (35). To better define the protein localization, a construct expressing a C-terminal YFP fusion was integrated at the endogenous locus of EFG1, using a one step PCR strategy. The resulting strain was transformed with plasmid pUN100-mCherry-NOP1 expressing a mCherry-fused version of Nop1 (Nop1-mCherry) (27). Nop1, the yeast fibrillarin ortholog, is a core component of box C/D snoRNPs required for early, nucleolar stages of pre-rRNA processing, and is used to visualize the nucleolus. The strain was grown exponentially and no growth phenotype was observed neither at 30°C nor at 37°C. Living cells were analyzed by fluorescence microscopy. The YFP signal observed under these conditions fully co-localizes with Nop1-mCherry, in a crescent-shaped region consistent with nucleolar localization (Figure 2B). We concluded that Efg1 accumulates in the yeast nucleolus, specific locus of ribosome synthesis.
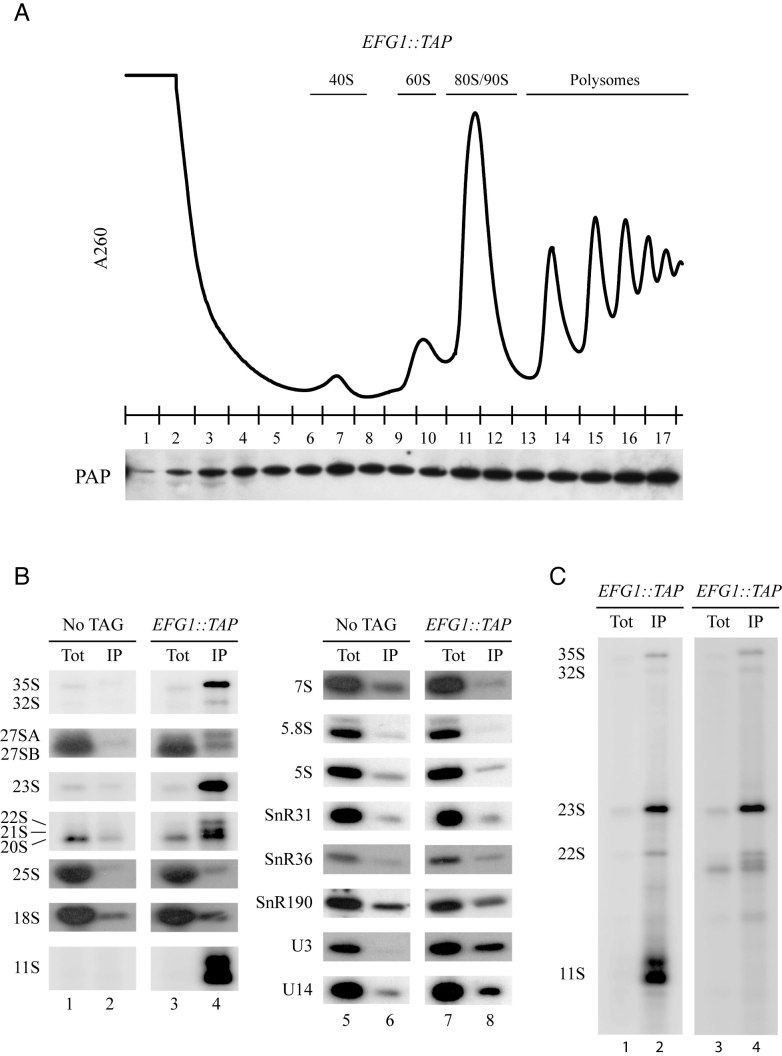
To assess the putative involvement of the Efg1 protein in ribosome synthesis, we investigated its association with ribosomal particles. To this end, sedimentation analyses on a sucrose density gradient were carried out using extracts from a strain expressing a C-terminal fusion between EFG1, at its endogenous locus, and the TAP tag. Western blot analysis of gradient fractions (Figure 3A) demonstrates that Efg1-TAP sediments all along the gradient but is enriched within high molecular mass fractions (fractions 11–13), the density of which is consistent with that of early 90S pre-ribosomal particles. Efg1-TAP also sediments within fractions containing polysomes (fractions 13–17) as it is the case for most of nucleolar proteins involved in early maturation steps in yeast (30,36,37).
Figure 3.
Efg1 is associated with pre-ribosomes. (A) Sedimentation profile of Efg1 on a sucrose gradient. A total extract prepared from EFG1::TAP cells growing exponentially was sedimented through a sucrose gradient, and 17 fractions were collected. The corresponding A260 profile is displayed with the characteristic annotated peaks. Each fraction was TCA precipitated, and Efg1-TAP was detected by western blotting using PAP antibodies. (B) The 35S and 23S pre-rRNAs are co-immunoprecipitated with Efg1. Northern blot analysis of (pre-)rRNAs co-precipitated with TAP-tagged Efg1 (lanes 3–4 and 7–8) or from control experiments using extracts of cells lacking a tagged protein (lanes 1–2 and 5–6). Immunoprecipitation was performed under native conditions using IgG-Sepharose. RNAs were extracted from the pellet after precipitation (lanes IP) or from total cell extract (lanes Tot) corresponding to 5% of the input used for the immunoprecipitation reactions. Following denaturing electrophoresis, RNAs were transferred to a nylon membrane and hybridized with anti-sense oligonucleotides corresponding to various (pre-)rRNAs and snoRNAs. (C) Full northern blot showing RNAs co-immunoprecipitated with Efg1 and detected using a probe hybridizing between A0 and A1 sites (1699, lanes 1 and 2) and between D and A2 (004, lanes 3 and 4).
To determine whether Efg1 might associate in vivo with specific pre-rRNAs and snoRNAs, immunoprecipitation experiments were performed. Total extracts from EFG1::TAP and a non-tagged WT strains were incubated with IgG-conjugated Sepharose beads. Following immunoprecipitation, total RNAs were extracted and analyzed by northern hybridization (co-immunoprecipitated proteins were analyzed independently, Supplementary Figure S1). Results show that Efg1-TAP efficiently precipitates the 35S pre-rRNA, and is strongly associated with the aberrant 21S, 22S and 23S pre-rRNAs (Figure 3B, left panel, compare lane 4 to 2). In contrast, we observed a very poor co-immunoprecipitation of the 27S and 7S pre-rRNAs as well as the mature 25S,18S, 5.8S and 5S rRNAs and a mild association with the 20S pre-rRNA. Association with Efg1 is also observed for U3 and U14 snoRNAs, but not for SnR31, SnR36 and SnR190 (Figure 3B, middle panel, compare lane 8–6). We concluded that Efg1 associates with early pre-ribosomal particles, containing U3 and U14 snoRNAs, suggesting it could play a role in early steps of ribosome synthesis. Besides, the tight association of Efg1 with the 23S pre-rRNA and the poor association with the 20S pre-rRNA might suggest that the A2 pathway, but not A3 pathway, is required for the release of the protein from pre-ribosomal particles (Figure 1B).
One striking feature of our experiment was the fact that two 1200/1300 nt long fragments were strongly co-precipitated with Efg1-TAP. We called these RNAs 11S (Figure 3B; Full lane is shown in Figure 3C). Those fragments are revealed using a 5′-ETS probe which lies 35 nt downstream from the RNA Pol I transcription start site (TSS). In contrast, no signal was detected using a probe immediately upstream from the Pol I TSS (data not shown). From these observations, we concluded that Efg1-TAP strongly co-precipitated RNA fragments starting at the Pol I TSS and ending 1200/1300 nt downstream, within the 18S rRNA sequence, ∼500/600 nt downstream of A1 site.
Efg1 is required for biogenesis of the small ribosomal subunit
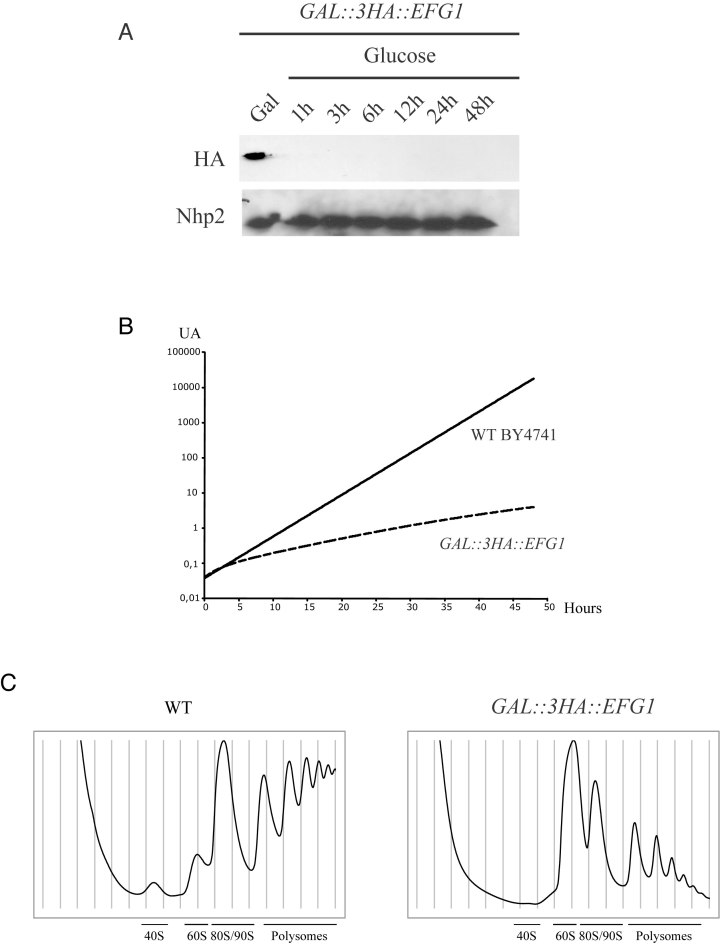
Efg1 is non-essential for cell viability (22). However, EFG1 deletion results in severely impaired cell growth. We decided to monitor ribosome biogenesis during its depletion (and thus during the onset of the phenotype) rather than in cells lacking the EFG1 gene. For that purpose, we constructed a yeast strain that conditionally expresses the Efg1 protein comprising a 3HA tag at the N-terminus (3HA-Efg1 protein), allowing its easy detection. The EFG1 open reading frame was tagged by the 3HA-encoding sequence and placed under the control of the regulated GAL1–10 promoter by homologous recombination, creating the GAL::3HA::EFG1 strain. This strain was propagated in a medium containing galactose as carbon source and was then shifted to a glucose-containing medium to allow depletion of the protein. The kinetics of Efg1 depletion were assessed by western blot using anti-HA antibodies. The abundance of 3HA-Efg1 was strongly reduced after transfer to glucose medium and became almost undetectable after <1h (see Figure 4A).
Figure 4.
Efg1 depletion affects 40S ribosomal subunit accumulation in yeast cells. (A) Western blot analysis of 3HA-Efg1 depletion. Total proteins were extracted at the times indicated and analyzed by western blot. 3HA-Efg1 and Nhp2 were detected using anti-HA and Nhp2-specific antibodies respectively. (B) Growth rate of WT and Gal::3HA::EFG1 strains following a transfer from permissive galactose medium to glucose medium for the times indicated. Cells were maintained in exponential growth throughout the time course by dilution into pre-warmed medium. (C) Ribosome profiles of Efg1-depleted cells. GAL::3HA::EFG1 and WT BY4741 strains were grown up to 0.6 (OD600) in galactose medium and shifted to glucose for 3 h. Total cell extracts were prepared, centrifuged through 4.5–45% sucrose gradients and 17 fractions were collected. The A260 absorbance profile is presented with annotated peaks.
On galactose containing medium, the growth rate of GAL1::3HA::EFG1 strain is almost identical to that of the WT strain. However, 4 h after transfer to the non-permissive glucose medium, the growth rate of GAL1::3HA::EFG1 strain was already substantially reduced compared to WT (Figure 4B). After several hours of depletion, the doubling time of GAL1::3HA::EFG1 was estimated to be 9 h. In the same conditions, the WT strain was doubling in 2.4 h. These results confirmed that even though Efg1 is not essential, the growth of strains depleted for the protein is largely impaired.
To characterize Efg1 function in ribosome biogenesis, we analyzed the sedimentation profile of ribosomal particles from a strain depleted of Efg1. Total extracts from depleted cells and WT strain were loaded on 4.5–45% sucrose gradients (Figure 4C). Compared to WT, Efg1 depletion led to a severe reduction in the relative amounts of 40S subunit, 80S ribosomes/90S pre-ribosomes and polysomes. Accordingly, 60S subunits accumulated in the cell, resulting in a large peak that partially overlaps with 80S ribosomes. This phenotype confirms a role of Efg1 in the maturation of the small ribosomal subunit.
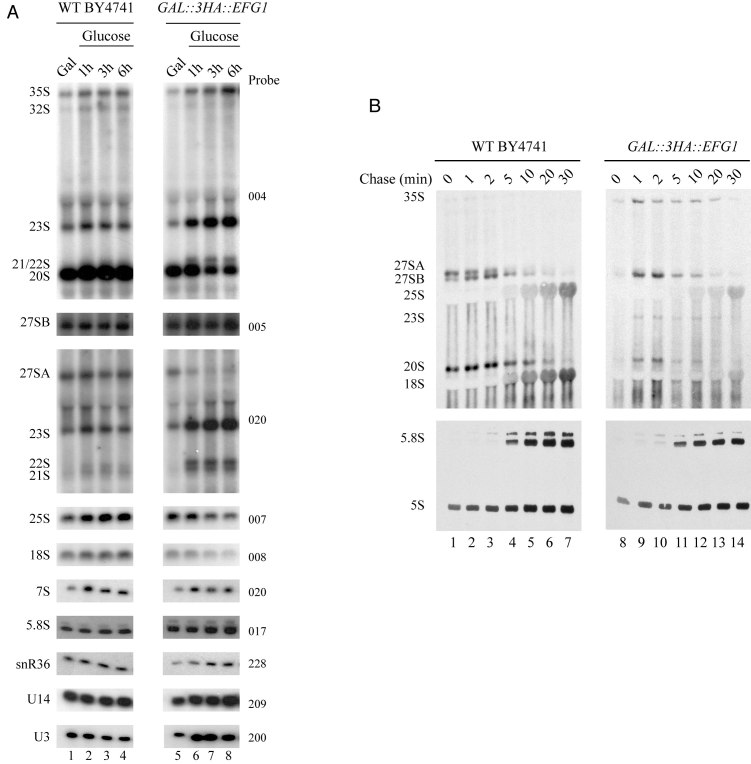
To assess which mature rRNAs require Efg1 for accumulation, we decided to analyze all ribosomal RNAs (rRNAs) from Efg1-depleted cells. Aliquots of WT and GAL1::3HA::EFG1 cells grown in galactose-containing medium or grown for 1, 3 and 6 h in glucose-containing medium were harvested. From these aliquots, total RNAs were extracted. We then compared pre-rRNA processing in WT and Efg1-depleted cells by northern analysis (Figure 5A). We observed that after Efg1 depletion, levels of the 35S pre-rRNA are highly increased. In the absence of Efg1, the 32S, 27SA2 and 20S pre-rRNAs were reduced, suggesting that A0, A1 and A2 cleavages are impaired. This is supported by the strong accumulation of the aberrant 23S pre-rRNA, produced from a direct cleavage of the nascent transcript at site A3. Note that 22S (A0–A3) and 21S (A1–A3) are also accumulated, suggesting that A0 or A1 occurs after A3. As a consequence, after 6 h of Efg1 depletion, 18S rRNA accumulation is already diminished. We also observed that depletion of Efg1 leads to a decrease of 25S rRNA level, together with the accumulation of 27SB pre-rRNA. A2 cleavage inhibition leads to a direct cleavage at A3 site. In consequence, a 27SA2 accumulation defect is observed concomitantly with an accumulation of 27SA3 pre-rRNA which is rapidly processed to produce the short form of 27SB pre-rRNA. This defect may explain a 27SBS accumulation. Altogether, these observations emphasize a role of Efg1 in early steps of ribosome biogenesis, in particular in the efficiency of A0, A1 and A2 cleavages.
Figure 5.
Efg1 depletion leads to a defect in A0, A1 and A2 cleavages. (A) WT BY4741 and GAL::3HA::EFG1 strains were shifted from a galactose to a glucose medium. Samples were collected before and at different times after the nutritional shift. Total RNAs were extracted from these cell samples, and the accumulation of the different pre-rRNAs, rRNAs and sn(o)RNAs was analyzed by northern blot using different probes. (B) Pulse-chase labeling of RNAs. WT BY4741 and GAL::3HA::EFG1 cells were grown in galactose containing medium and were next shifted to glucose for 3 h. Cells were then pulse labeled with [2,8–3H] adenine for 2 min. Samples were collected 0, 1, 2, 5, 10, 20 and 30 min after addition of an excess of cold adenine. Total RNAs were extracted from these samples, separated by gel electrophoresis and transferred to a nylon membrane.
Effects of reduced 3HA-Efg1 levels on rRNA processing were also assessed by in vivo [2,8- 3H]-adenine pulse-chase labeling (Figure 5B). As 3H-adenine can be incorporated into pre-rRNA only during ongoing transcription, any radioactive labeling indicates de novo synthesis. Following this initial pulse labeling, outcome of transcripts can be followed after chase with an excess of cold adenine. Pulse-chase labeling was performed 30 min after transfer of the GAL1::3HA::EFG1 strain to glucose medium. After depletion of Efg1, we still observe 35S pre-rRNA production. This suggests that rDNA transcription by Pol I is not affected in absence of Efg1. We also note that, compared to the WT, pre-rRNA processing is delayed during Efg1 depletion. Indeed, we see that the production of 27SA2 pre-rRNA seems greatly reduced (A2 pathway). Accordingly, we have a greater production of 27SB pre-rRNA. However, maturation of this 27SB pre-rRNA appears to be delayed as de novo synthesis of mature 25S rRNA is slightly reduced. In parallel, the absence of Efg1 strongly inhibits 18S rRNA production, which is probably the consequence of reduced 20S pre-rRNA production detected by this approach. These observations are consistent with the accumulation of newly synthesized 23S pre-rRNA, with a slow decay rate (detected from 1 to 20 min following chase). Note that 5.8S and 5S production appears mostly unaffected by Efg1 depletion. These results confirmed that cleavages at sites A0, A1 and A2 are strongly impaired in cells lacking Efg1.
23S pre-rRNA is targeted by endo- and exonucleolytic pathways for its degradation
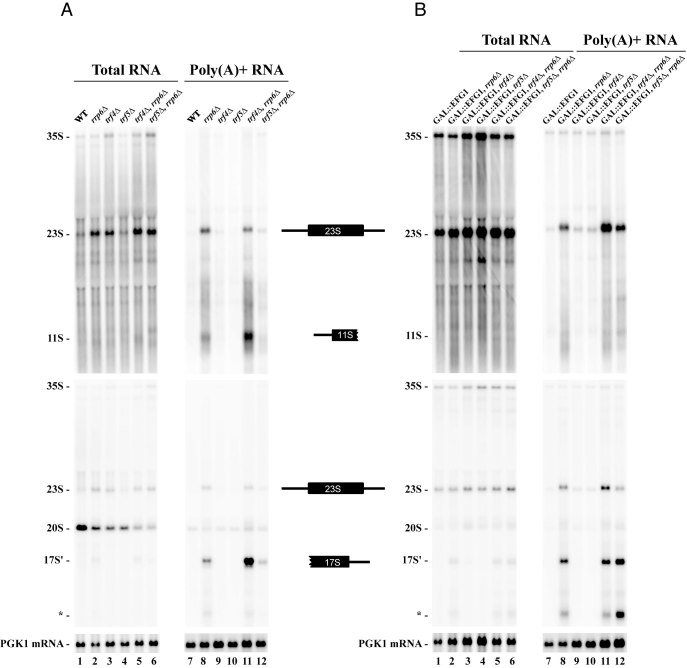
We identified two 1200/1300 nt long fragments which are strongly co-precipitated with Efg1-TAP (Figure 3). The 5′ extremity of the fragment corresponds to the Pol I TSS and the 3′ extremity lies within the 18S rRNA sequence approximately 500/600 nt downstream of A1 site, hereafter called 11S rRNA. Because Efg1 is a nucleolar protein and the 3′ extremity of the 11S rRNA ends within the sequence of the mature 18S rRNA, we hypothesized that such fragment would be a degradation product of the 35S pre-rRNA or the 23S pre-rRNA. Degradation pathways of aberrant or excised (pre-)rRNA primarily involve TRAMP and exosome complexes. The 11S rRNA 3′ extremity could be the result of incomplete exosome-dependent 3′ exonucleolytic degradation of the 23S rRNA. On the other hand, the production of this RNA could also result from an endonucleolytic cleavage within the first third of the 18S rRNA. This would lead to the production of another rRNA byproduct extending from within the 18S rRNA sequence to A3 site or Bo, when processed from 23S or 35S, respectively. To discriminate between these two possibilities, we checked the existence of this complementary fragment by northern blot in WT strains or in five strains in which exonucleolytic degradation pathways are deficient: single rrp6Δ, trf4Δ, trf5Δ and double rrp6Δ/trf4Δ, rrp6Δ/trf5Δ deletion mutants (Figure 6A). Polyadenylated forms of 23S and 11S rRNAs were observed in strains lacking Rrp6 or both Rrp6 and Trf4, suggesting that these two RNAs are targeted by the TRAMP/exosome pathway for degradation (Figure 6A, lanes 8 and 11). In the same RNA samples, using a D-A2 probe, we identified a ∼1500 nt long rRNA. Similar signal was obtained using A2–A3 probe but was not visible using a probe hybridizing downstream of A3 (data not shown) establishing that the 3′ extremity of this rRNA is A3. This particular rRNA fragment was already observed by the Tollervey and Lafontaine laboratories (17,19,24), which they called 17S'. According to the hybridization specificity, 17S' and 11S have no common sequence, and could result from the cleavage of the 23S transcript. Moreover, the size of both fragments (∼1200/1300 nt and ∼1500 nt) appeared compatible with the full size of the 23S pre-rRNA (∼2800 nt). From these results we concluded that part of the 23S pre-rRNA is endonucleolytically cleaved within the sequence of the 18S rRNA resulting in 11S and 17S' production.
Figure 6.
23S pre-rRNA is targeted by endo- and exo-nucleolytic pathways for its degradation. Pre-rRNA accumulation was monitored in (A) presence of endogenous Efg1 and compared with cells depleted for Efg1 (B). Yeast cells were grown to mid-log phase in galactose containing media. Glucose was next added within the media and cells were grown in these conditions for 3 h. Aliquots were collected and total RNAs were extracted and subjected to poly(A)+ affinity purification on oligo-dT coated beads. All RNA samples were separated on 1.2% agarose gels, transferred to a nylon membrane and hybridized with specific oligonucleotide probes. Upper panels and mid panels were hybridized with ETS1 (1699) and ITS1 (004) probes, respectively. Lower panels were hybridized with an oligonucleotide specific for PGK1 mRNA (403).
As previously shown (Figure 3B), the A2 pathway leads to the dissociation of Efg1 from pre-RNAs. In contrast, when direct A3 cleavage occurs, Efg1 stays associated with 23S and 11S (but not 17S') pre-RNAs. From this observation, we tested whether Efg1 would be required for 11S and 17S' production or alternatively, would tag these pre-rRNAs for degradation. To explore these possibilities, we decided to monitor the accumulation of the different dead-end products in absence of Efg1 (Figure 6B) and we compared with cells expressing endogenous Efg1 (Figure 6A). We introduced the GAL1::3HA::EFG1 allele in the five strains in which degradation pathways are deficient. Efg1 was depleted by adding glucose in the media for 3 h, RNAs were extracted and polyadenylated RNAs were purified. 17S' rRNA is still detected and polyadenylated in absence of Efg1. We conclude that Efg1 is not required for the endonucleolytic cleavage of 23S. However, the 17S' polyadenylation profile is different. Indeed, in the presence of Efg1, 17S' RNA is mainly targeted by the TRAMP5 complex as it was poorly polyadenylated in the absence of Trf5. In the absence of Efg1, the 17S' rRNA was polyadenylated in absence of either Trf4 or Trf5, suggesting that TRAMP4 can substitute for TRAMP5 under these conditions. We also noticed the accumulation of shorter polyadenylated rRNAs corresponding to 5′-truncated forms of the 17S' rRNA (annotated *). This modification of polyadenylated species was even more striking for 11S. Compared to cells expressing Efg1, GAL1::3HA::EFG1 cells failed to accumulate polyadenylated 11S in absence of Rrp6 and both Rrp6 and Trf4 (Figure 6B, lanes 8 and 11).
We conclude that 23S is endonucleolytically cleaved, and subsequently degraded by the TRAMP exosome pathway under the regulation of Efg1 protein.
23S pre-rRNA is cleaved at Q1 site and the PIN domain of Utp24 is required for efficient cleavage
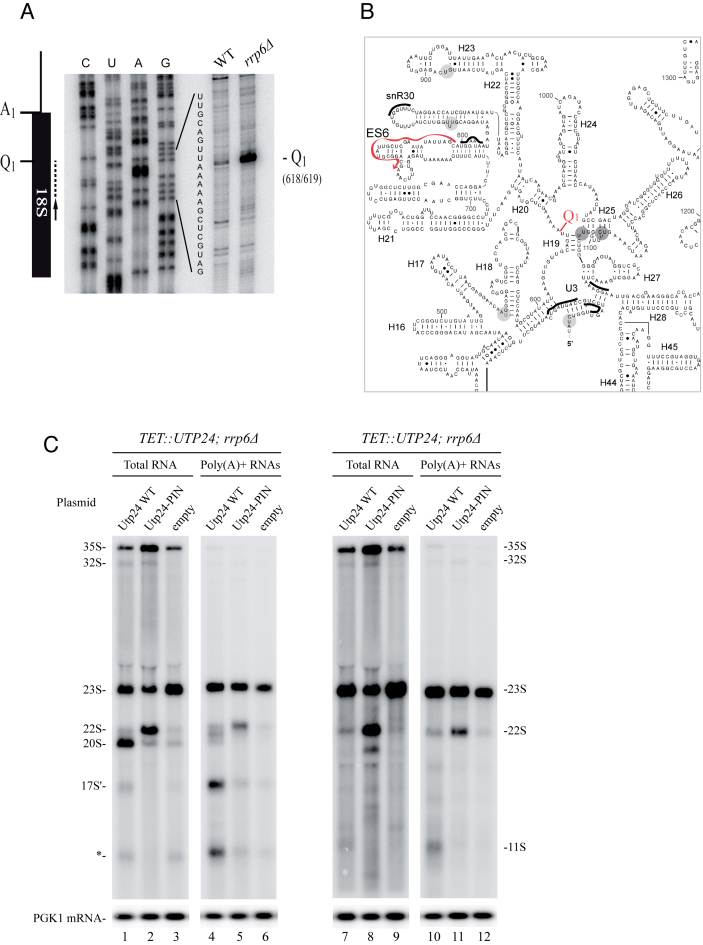
We next decided to map precisely the major 23S endonucleolytic cleavage site using the 5′ extremity of the 17S'. For this purpose, we extracted total RNAs from WT and Rrp6 deficient cells and performed serial reverse transcription assays all along the 18S rRNA sequence (data not shown). We identified one major site in the first third of the 18S rRNA, and two minor sites (data not shown). The precise 5′ extremity of the 17S' was mapped at position +618 relative to A1 site (Figure 7A). We named this cleavage site Q1 (for Quality control cleavage site 1). Q1 site is located within the central domain, between helix 19 and 20 (Figure 7B), within a very flexible A-rich sequence (10,38). This sequence is not known to be the target of any endoribonuclease activity. However, recent RNA–protein crosslinking data (CRAC analysis (39)) showed that the Utp24 endoribonuclease was predominately associated with sequences within the 18S rRNA sequence (14). The main peak of association was located around position +1103 relative to A1, in the 3′ part of the central domain. Even though the Q1 cleavage site (+618) and +1103 are separated by ∼500 nt, secondary and tertiary structure predictions position them in close vicinity, on both sides of helix 19. These observations render Utp24 a good candidate for being involved in the cleavage at Q1. To assess the putative function of Utp24 in Q1 cleavage in vivo, we decided to monitor the accumulation of 11S and 17S' rRNA in cells deficient for Utp24 enzymatic activity. For this purpose, we constructed a strain lacking the RRP6 gene, strongly accumulating 23S, in which HA-tagged Utp24 is expressed under the control of the tetracycline repressible promoter (pTetO7). This strain was next transformed with a plasmid expressing either WT, Utp24 PIN mutant allele (Utp24-D68N) (13,14) or no protein. Ten hours following addition of doxycycline, fully functional genomic Utp24 is largely depleted (Supplementary Figure S2) and the only available source of Utp24 in the cell is the version expressed from the plasmid (WT, PIN mutant or mock). In these conditions, strains expressing only WT Utp24, PIN mutant Utp24 or no protein, were grown in liquid media, harvested and total RNAs were extracted and polyadenylated RNAs were enriched. Figure 7C shows northern blot analyses using D-A2 (lanes 1–6) and 5′-ETS (lanes 7–12) probes. In cells expressing WT Utp24, 11S (lanes 7 and 10) and 17S' (lanes 1and 4) were accumulated. Cells containing empty vector showed a typical early pre-rRNA processing defect due to depletion of endogenous Utp24 with an inhibition of all early cleavage (A0, A1 and A2), leading to 35S and 23S accumulation and inhibition of 20S production (lanes 3 and 9). Note that contrary to the situation where WT Utp24 is expressed, 11S and 17S' are almost undetectable. Results are slightly different when the Utp24-D68N PINc mutant is expressed. According to previous observations (13), Utp24 D68N is enzymatically deficient but is probably still associated with an assembled SSU processome (40). In these conditions, A0 is accurately cleaved whereas A1 and A2 are blocked leading to the accumulation of the 22S pre-rRNA, an intermediate extending from A0 to A3. Cells depleted of Utp24 or expressing Utp24-D68N failed to accumulate the 11S and 17S' fragments. Note that some residual 11S and 17S' are still present in the polyadenylated fractions. This is consistent with the fact that fully functional Utp24 is not completely depleted in our system (Supplementary Figure S2: see residual amounts of Utp24 after 10 h of depletion). We concluded that the integrity of the PINc domain of Utp24 is required for the normal accumulation of the 11S and 17S' RNAs strongly suggesting that Utp24 is responsible for the cleavage at Q1.
Figure 7.
23S pre-rRNA is cleaved at Q1 site and the PIN domain of Utp24 is required for efficient cleavage. (A) RNAs extracted from WT or rrp6Δ strains were analyzed by primer extension using probe 1510 (schematized by the arrow). A PCR fragment containing 18S sequence was used to generate a sequencing ladder leading to the identification of Q1 site. (B) Predicted secondary structure of the 18S rRNA central domain in Saccharomyces cerevisiae. Utp24 crosslinking sites are marked on the sequence and shades indicate peak height with the highest peak shown in dark grey. Q1 site (18S 618/619) is indicated. Oligonucleotide used to map Q1 site is annotated (red arrow). (C) Northern analysis of pre-rRNA processing in the pTET::utp24–3HA strain transformed with a plasmid expressing either WT Utp24 or the PIN mutant (D68N) his-tagged Utp24 protein, or an empty vector. RNAs were isolated from mid log phase cells grown 8 h in presence of doxycycline to deplete endogenous Utp24 protein. Aliquots were collected and total RNAs were extracted and subjected to poly(A)+ affinity purification on oligo-dT-coated beads. Total RNAs (lanes 1–3 and 7–9) and poly(A)+ RNAs (lanes 4–6 and 10–12) were separated on an 1.2% agarose gel and detected by northern hybridization with specific oligonucleotide probes. Left panels (lanes 1–6) were hybridized with ITS1 probe (004), right panels (lanes 7–12) were hybridized with ETS1 probe (1699). Loading was assessed by hybridizing the PGK1 mRNA (lower panel) with probe 403.
DISCUSSION
Ribosome biogenesis is a very complex and error-prone pathway that is actively surveyed by quality control mechanisms. In fast growing cells, the nucleolar RNA surveillance pathway is active and the non-productive 23S pre-rRNA is rapidly targeted for degradation (41). This involves its recognition and polyadenylation by the TRAMP5 complex followed by its 3′-5′ degradation by the exosome and particularly Rrp6 (17,19,20,42). Indeed, absence of either Trf5 or Rrp6 led to a 23S pre-rRNA accumulation.
In this study, we report the characterization of EFG1, encoding a nucleolar protein involved in early cleavages of the 35S pre-rRNA leading to the production of the small ribosomal subunit. We also present the identification of a new pathway leading to the degradation of the non-productive 23S pre-rRNA. We show that 23S pre-rRNA is initially targeted by the PINc endonuclease Utp24 resulting in its cleavage at position +618 relative to A1, a new cleavage site we called Q1. Both resulting fragments (11S and 17S') are subsequently targeted by TRAMP5 and the exosome for their degradation. We also show the requirement of Efg1 for the TRAMP5 targeting of 17S' and the polyadenylation of 11S RNA by the TRAMP complex.
In S. cerevisiae, Efg1 accumulates in the nucleolus which is the specific place of early steps of ribosome biogenesis. Efg1 is a non essential protein, however upon Efg1 depletion, we observed a rapid decrease in the accumulation of the mature 18S rRNA. This decrease is the consequence of a defect in A0, A1 and A2 cleavages as Efg1 depletion resulted in the accumulation of the 35S and the 23S pre-rRNAs concomitantly with a strong decrease of the 20S pre-rRNA, the direct precursor of the mature 18S rRNA. It is noticeable that 21S and 22S pre-rRNAs are accumulated in absence of Efg1. This observation as well as the fact that residual amounts of 20S pre-rRNA persist in absence of Efg1, strongly suggest that even if the cleavages are strongly delayed, they are not fully inhibited in these conditions. Consistent with these observations, pulse chase experiments clearly revealed that small amounts of 20S pre-rRNA (leading to 18S rRNA) are still produced in absence of Efg1. We hypothesize that this residual production is sufficient to ensure cell viability. Furthermore, global genetic screens to identify Efg1 mutants alleviating or aggravating the growth phenotype did not identify other RNA processing factors (data not shown).
Co-immunoprecipitation experiments revealed that Efg1 interacts physically with numerous RNA components (including 35S pre-rRNA and U3 snoRNA) and proteins of the 90S pre-ribosomal particles (see Supplementary Figure S1). However, our results showed that Efg1 is not necessary for the assembly of the U3 processome. Indeed, gradient analyzes showed that Efg1 is not required for the association of UTP-A (Utp17p), UTP-B (Pwp2p), UTP-C (Utp22p) complexes as well as Rrp5 with the 90S pre-ribosomes suggesting that these modules assemble independently of Efg1 (Supplementary Figure S3). Conversely, components of UTP-A, UTP-B and UTP-C complexes as well as Rrp5 are not required for the association of Efg1 with the 90S pre-ribosome (Supplementary Figure S3).
Altogether, these results strongly suggest that Efg1 is not a bona fide structural component of the U3 processome as its assembly with the pre-ribosomes appears independent of the assembly of UTP modules. Therefore, Efg1 associates very early with pre-ribosomes, probably during rDNA transcription. This is in accordance with recent results showing that Efg1 associates with nascent transcripts, and more precisely with a region corresponding to the 5′ domain of 18S rRNA (10). In contrast to 35S pre-rRNA, very low levels of 20S pre-rRNA are co-immunoprecipitated with Efg1 suggesting that Efg1 is released from the particle during or immediately after A2 cleavage.
One interesting observation of the co-precipitation data was the strong recovery of the aberrant 23S pre-rRNA with Efg1. This pre-rRNA is detectable in WT cells, but at very low levels, and is strongly enriched in the Efg1-TAP immunoprecipitates. We concluded that A0, A1 and A2 cleavages are required for the release of Efg1 from pre-ribosomes. In absence of these cleavages, Efg1 stays associated with the particle. Another striking feature was the strong co-immunoprecipitation of a RNA we called 11S, extending from the 5′-TSS to the first third of the 18S rRNA sequence. The previously observed 17S' rRNA (17,19,24) starting from position +618nt relative to A1 (Q1 site) and extending to A3 site is not associated with Efg1. The 23S pre-rRNA is processed to the 17S' and 11S rRNAs through an endoribonucleolytic cleavage at Q1 site. This cleavage is achieved by the endonuclease Utp24 which binds within the central domain of the 18S rRNA (14), in close vicinity to the Q1 cleavage site, on the opposite side of helix 19.
As well as the nonproductive 23S pre-rRNA, 11S and 17S' are enriched in polyadenylated fractions and accumulated in absence of Rrp6. This observation strongly suggests that 11S and 17S' are also targeted for degradation by the TRAMP/Exosome pathway. In absence of Rrp6, the levels of these two degradation fragments are strongly diminished in poly(A+) fractions when Trf5 is absent suggesting they are mainly targeted by the TRAMP5 complex for polyadenylation. We propose that cleavage at Q1 site is likely part of a surveillance pathway that removes pre-ribosomes that have failed to undergo correct assembly and/or processing during the early steps of their biogenesis. Cleaving 23S pre-rRNA within the sequence of the mature 18S rRNA would definitely avoid the maturation of 23S containing particles. Our results suggest that 23S pre-rRNA could be either directly targeted by the TRAMP/exosome pathway to promote its 3′-5′ degradation or primarily cleaved at Q1, leading to the production of 11S and 17S' that will next undergo TRAMP/exosome elimination. This redundancy probably enhances the overall efficiency of the surveillance pathway. Indeed, taking advantage of Q1 cleavage, 5′ sequences of 23S would presumably be even more rapidly degraded, given that the degradation machinery is slowed by strong secondary structures in its 3′ sequences.
Compared to cells expressing Efg1, GAL::3HA::EFG1 cells grown in presence of glucose failed to accumulate polyadenylated 11S in absence of Rrp6 or both Rrp6 and Trf4 (Figure 5B, lanes 8, 11 and 12). These observations suggested that TRAMP was unable in these conditions to target the 11S rRNA. In contrast, the 17S' rRNA is still polyadenylated in absence of Efg1. This is correlated by the fact that Efg1 is associated with 11S and not with 17S' (this study and (10)). The presence of Efg1 within the particle appeared required to target 11S RNA to the TRAMP complex. It is possible that the absence of Efg1 leads to conformational changes at the 3′ extremity of the 11S making it poorly accessible to TRAMP. On the other hand, Efg1 could actively recruit the TRAMP complex on the 11S RNA containing particle in order to direct it to degradation. The existence of such adaptor protein has already been reported. Nop53 and Utp18 contain the same Arch Interaction Motif (AIM) consensus motif that interacts with the arch domain of Mtr4 (43). This interaction leads to the docking of TRAMP on specific substrates. Efg1 is presumably not a similar adaptor factor as such AIM motif was not found within its sequence. However, Trf5 peptides were found in Efg1 immunoprecipitates (see Supplementary Figure S2) suggesting a potential interaction.
Moreover, in Efg1 depleted cells, the global polyadenylation profile is slightly different. Indeed, in the presence of Efg1, 17S' is mainly targeted by the TRAMP5 complex as it was poorly polyadenylated in absence of Trf5. In absence of Efg1, the 17S' rRNA was polyadenylated in absence of either Trf4 or Trf5, suggesting that TRAMP4 can substitute for TRAMP5 in these conditions. Globally, TRAMP5 polyadenylation seems less efficient in absence of Efg1.
Ribosome biogenesis is an extremely complex pathway with at least 200 assembly factors, 75 snoRNPs, 79 ribosomal proteins and a multistep processing pathway leading to the production of mature ribosomes ensuring the fidelity of translation. The possibilities for errors are obviously plentiful and it seems imperative that surveillance occurs at different checkpoints during the process. A2 cleavage is presumably one of these as it splits the 90S pre-ribosome in pre-40S and pre-60S particles that will undergo independent maturation. Using Utp24 as a main checkpoint actor would make plenty of sense. Utp24 could be the key element allowing the shift between productive and non-productive pathways. Utp24 would commit 90S particles (i) to maturation by using A2 cleavage site, (ii) to degradation by cleaving at Q1 site or (iii) to accumulation of 23S containing particles by utilizing none of them (18) (see Figure 8). How Utp24 would choose between the different pathways remains unclear. The correct loading of Utp24 onto the particle may be an important feature. The environment of Q1 cleavage site appears very ‘flexible’ (38) and may differ depending on the maturation state of the pre-ribosome. Particles that would have failed to undergo correct assembly and/or processing would be in one conformation that exposes Q1 cleavage site to Utp24 instead of A1 and A2. However, we cannot exclude an active and precise modulation of Utp24 cleavage activity by specific regulation factors. During rapid exponential growth, TORC1 is active and yeast uses normal A0, A1 and A2 cleavages to produce mature ribosomes. Under unfavorable growth conditions, such as environmental stress or lack of fermentable carbon source (diauxic shift), TORC1 becomes inactive and yeast uses direct A3 cleavage instead of A0, A1 and A2 cleavages. This leads to the accumulation of 23S RNA containing particles with no further detectable production of new ribosomes (18,44). In parallel, rDNA transcription and production of ribosomal proteins or assembly factors are strongly reduced as the cells prepare their entry into quiescence (reviewed in (45)). This contributes to the general downregulation of global protein synthesis. When conditions improve, cells have to rapidly switch to exponential growth. Being very reactive in making new ribosomes would be a prerequisite to start again making proteins and resume growth. This capability should provide great evolutionary advantages to compete for resources with other organisms. Usage of the Q1 pathway would lead to disassembly of stocked 23S RNA containing particles. This would immediately release and provide most of the ribosomal proteins and assembly factors required to rapidly prime the ribosome pump and greatly increase yeast adaptability to environment.
We propose that Utp24 is a key element allowing the shift between productive and non-productive ribosome biogenesis. Fine tuning of Utp24 activity and cleavage specificity would allow the control of ribosome biogenesis at the post-transcriptional level in response to assembly defects or environmental changes.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to Yves Henry and Marie-Kerguelen Sarthou for critical reading of the manuscript. We acknowledge members of the Gadal’s lab for help, advice and discussion. This work also benefited from the assistance of the microscopy facility of the imaging platform of Toulouse TRI.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale de la Recherche (ANR) [ANR-13-BSV5–0010—ANDY]; IDEX [ATS, NudGene]; E. C. was supported by a Ph.D. Fellowship from the Ministère de l’Education Nationale Ph.D. Fellowship (E.C.); de l’Enseignement Supérieur et de la Recherche Fellowship (E.C.); ‘l’Association pour la Recherche contre le Cancer’ Fellowship (to E.C.). Royal Society [UF100666, RG110357, UF150691 to C.S.]. Funding for open access charge: ANR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Woolford J.L. Jr., Baserga S.J.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E.. An overview of pre-ribosomal RNA processing in eukaryotes. Enzymes. 2014; 41:169–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang J., Harnpicharnchai P., Jakovljevic J., Tang L., Guo Y., Oeffinger M., Rout M.P., Hiley S.L., Hughes T., Woolford J.L. Jr.. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007; 21:2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y. et al. . A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002; 417:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D. et al. . 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002; 10:105–115. [DOI] [PubMed] [Google Scholar]
- 6. Henras A.K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y.. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008; 65:2334–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Phipps K.R., Charette J.M., Baserga S.J.. The SSU processome in ribosome biogenesis—progress and prospects. WIREs RNA. 2011; 2:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gallagher J.E., Dunbar D.A., Granneman S., Mitchell B.M., Osheim Y., Beyer A.L., Baserga S.J.. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004; 18:2506–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bernstein K.A., Baserga S.J.. The small subunit processome is required for cell cycle progression at G1. Mol. Biol. Cell. 2004; 15:5038–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chaker-Margot M., Hunziker M., Barandun J., Dill B.D., Klinge S.. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat. Struct. Mol. Biol. 2015; 22:920–923. [DOI] [PubMed] [Google Scholar]
- 11. Chen W., Xie Z., Yang F., Ye K.. Stepwise assembly of the earliest precursors of large ribosomal subunits in yeast. Nucleic Acids Res. 2017; 45:6837–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perez-Fernandez J., Roman A., De Las Rivas J., Bustelo X.R., Dosil M.. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 2007; 27:5414–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bleichert F., Granneman S., Osheim Y.N., Beyer A.L., Baserga S.J.. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:9464–9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wells G.R., Weichmann F., Colvin D., Sloan K.E., Kudla G., Tollervey D., Watkins N.J., Schneider C.. The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res. 2016; 44:5399–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henry Y., Wood H., Morrissey J.P., Petfalski E., Kearsey S., Tollervey D.. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994; 13:2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmitt M.E., Clayton D.A.. Characterization of a unique protein component of yeast RNase MRP: an RNA-binding protein with a zinc-cluster domain. Genes Dev. 1994; 8:2617–2628. [DOI] [PubMed] [Google Scholar]
- 17. Wery M., Ruidant S., Schillewaert S., Lepore N., Lafontaine D.L.. The nuclear poly(A) polymerase and exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA. 2009; 15:406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kos-Braun I.C., Jung I., Kos M.. Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner. PLoS Biol. 2017; 15:e2000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Houseley J., Tollervey D.. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006; 7:205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dez C., Dlakic M., Tollervey D.. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA. 2007; 13:1516–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Houseley J., Tollervey D.. The many pathways of RNA degradation. Cell. 2009; 136:763–776. [DOI] [PubMed] [Google Scholar]
- 22. Kastenmayer J.P., Ni L., Chu A., Kitchen L.E., Au W.C., Yang H., Carter C.D., Wheeler D., Davis R.W., Boeke J.D. et al. . Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 2006; 16:365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peng W.T., Robinson M.D., Mnaimneh S., Krogan N.J., Cagney G., Morris Q., Davierwala A.P., Grigull J., Yang X., Zhang W. et al. . A panoramic view of yeast noncoding RNA processing. Cell. 2003; 113:919–933. [DOI] [PubMed] [Google Scholar]
- 24. Lepore N., Lafontaine D.L.. A functional interface at the rDNA connects rRNA synthesis, pre-rRNA processing and nucleolar surveillance in budding yeast. PLoS One. 2011; 6:e24962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Longtine M.S., McKenzie A. 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R.. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998; 14:953–961. [DOI] [PubMed] [Google Scholar]
- 26. Sung M.K., Ha C.W., Huh W.K.. A vector system for efficient and economical switching of C-terminal epitope tags in Saccharomyces cerevisiae. Yeast. 2008; 25:301–311. [DOI] [PubMed] [Google Scholar]
- 27. Berger A.B., Cabal G.G., Fabre E., Duong T., Buc H., Nehrbass U., Olivo-Marin J.C., Gadal O., Zimmer C.. High-resolution statistical mapping reveals gene territories in live yeast. Nat. Methods. 2008; 5:1031–1037. [DOI] [PubMed] [Google Scholar]
- 28. Dez C., Houseley J., Tollervey D.. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006; 25:1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dez C., Henras A., Faucon B., Lafontaine D., Caizergues-Ferrer M., Henry Y.. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 2001; 29:598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choque E., Marcellin M., Burlet-Schiltz O., Gadal O., Dez C.. The nucleolar protein Nop19p interacts preferentially with Utp25p and Dhr2p and is essential for the production of the 40S ribosomal subunit in Saccharomyces cerevisiae. RNA Biol. 2011; 8:1158–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beltrame M., Tollervey D.. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992; 11:1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dez C., Froment C., Noaillac-Depeyre J., Monsarrat B., Caizergues-Ferrer M., Henry Y.. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell. Biol. 2004; 24:6324–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lebaron S., Froment C., Fromont-Racine M., Rain J.C., Monsarrat B., Caizergues-Ferrer M., Henry Y.. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 2005; 25:9269–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E.C.. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993; 72:443–457. [DOI] [PubMed] [Google Scholar]
- 35. Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K.. Global analysis of protein localization in budding yeast. Nature. 2003; 425:686–691. [DOI] [PubMed] [Google Scholar]
- 36. Dez C., Noaillac-Depeyre J., Caizergues-Ferrer M., Henry Y.. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 2002; 22:7053–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gerus M., Bonnart C., Caizergues-Ferrer M., Henry Y., Henras A.K.. Evolutionary conserved function of RRP36 in early cleavages of the pre-ribosomal RNA and production of the 40S ribosomal subunit. Mol. Cell. Biol. 2009; 30:1130–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chaker-Margot M., Barandun J., Hunziker M., Klinge S.. Architecture of the yeast small subunit processome. Science. 2017; 355:6321–6329. [DOI] [PubMed] [Google Scholar]
- 39. Granneman S., Kudla G., Petfalski E., Tollervey D.. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tomecki R., Labno A., Drazkowska K., Cysewski D., Dziembowski A.. hUTP24 is essential for processing of the human rRNA precursor at site A1, but not at site A0. RNA Biol. 2015; 12:1010–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Allmang C., Mitchell P., Petfalski E., Tollervey D.. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000; 28:1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lacava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D.. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005; 121:713–724. [DOI] [PubMed] [Google Scholar]
- 43. Thoms M., Thomson E., Bassler J., Gnadig M., Griesel S., Hurt E.. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell. 2015; 162:1029–1038. [DOI] [PubMed] [Google Scholar]
- 44. Talkish J., Biedka S., Jakovljevic J., Zhang J., Tang L., Strahler J.R., Andrews P.C., Maddock J.R., Woolford J.L. Jr.. Disruption of ribosome assembly in yeast blocks cotranscriptional pre-rRNA processing and affects the global hierarchy of ribosome biogenesis. RNA. 2016; 22:852–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. De Virgilio C. The essence of yeast quiescence. FEMS Microbiol. Rev. 2012; 36:306–339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.