SUMMARY
We performed integrated genomic, transcriptomic and proteomic profiling of 150 pancreatic ductal adenocarcinoma (PDAC) specimens, including samples with characteristic low neoplastic cellularity. Deep whole-exome sequencing revealed recurrent somatic mutations in KRAS, TP53, CDKN2A, SMAD4, RNF43, ARID1A, TGFβR2, GNAS, RREB1 and PBRM1. KRAS wild-type tumors harbored alterations in other oncogenic drivers, including GNAS, BRAF, CTNNB1 and additional RAS pathway genes. A subset of tumors harbored multiple KRAS mutations, with some showing evidence of biallelic mutations. Protein profiling identified a favorable prognosis subset with low epithelial-mesenchymal transition and high MTOR pathway scores. Associations of non-coding RNAs with tumor-specific mRNA subtypes were also identified. Our integrated multi-platform analysis reveals a complex molecular landscape of PDAC and provides a roadmap for precision medicine.
Keywords: Pancreatic cancer, genomics, KRAS, heterogeneity, molecular subtypes, tumor cellularity, miRNA, RPPA
Graphical Abstract
This TCGA study shows that some PDAC carry multiple KRAS mutations, including biallelic mutations, that KRAS wild-type PDAC harbor alterations in other RAS pathway genes and other oncogenic drivers, and that low epithelial-mesenchymal transition and high MTOR pathway scores correlate with a favorable prognosis.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive disease that typically presents at an advanced stage and is refractory to most treatment modalities (Ryan et al., 2014; Wolfgang et al., 2013). PDAC is predicted to become the second leading cause of cancer mortality by the year 2030 (Rahib et al., 2014). Characterization of the recurrent genetic alterations in PDAC has yielded important insights into the biology of this disease, an improved understanding of familial predisposition, and a foundation for developing approaches for early detection and improved therapies. The first whole exome sequencing study of pancreatic cancer identified a large number of mutations and somatic copy number alterations (SCNAs) that alter the function of many key oncogenes and tumor suppressor genes, including KRAS, TP53, SMAD4, and CDKN2A (Jones et al., 2008). Follow-up whole exome and whole genome studies validated these findings and revealed a “long tail” of less prevalent alterations in other genes, such as those coding for regulators of axon guidance (Bailey et al., 2016; Biankin et al., 2012; Waddell et al., 2015; Witkiewicz et al., 2015). Germline alterations in DNA damage repair genes such as BRCA1, BRCA2, PALB2, or ATM give rise to genomic instability in a subset of PDACs and could make them more sensitive to platinum-based chemotherapy (Roberts et al., 2016; Sahin et al., 2016a; Waddell et al., 2015). Furthermore, recent sequencing of neoplastic cell-enriched whole genomes has demonstrated that the majority of PDACs harbor complex chromosomal rearrangement patterns, some of which are consistent with a catastrophic model of PDAC progression (Notta et al., 2016). Gene expression studies have identified subtypes of PDAC with prognostic and biological relevance (Bailey et al., 2016; Collisson et al., 2011; Moffitt et al., 2015).
PDACs are characterized by a prominent desmoplastic reaction with a dense fibrotic stroma (Iacobuzio-Donahue et al., 2002), and a typical primary pancreatic cancer often demonstrates only 5–20% neoplastic cellularity (Wood and Hruban, 2012). This low tumor cellularity has confounded the analyses of mutational and gene expression features of the actual neoplastic cells. Given this, prior genome sequencing studies have focused on tumors with neoplastic cellularity typically greater than 40% (Waddell et al., 2015), or have employed techniques that purify tumor samples, either by generating cell lines or patient-derived xenografts, or by using mechanical enrichment techniques such as macrodissection or laser capture microdissection (Jones et al., 2008; Witkiewicz et al., 2015). Consequently, samples with low neoplastic cellularity have been underrepresented in previous genome sequencing efforts, even though low cellularity cancers comprise the majority of surgically resected PDACs. Validated approaches for accurate genomic profiling in tumors with low neoplastic cellularity, such as those presented here, will be important for understanding the biology of these carcinomas and will be increasingly necessary for real-time genomic characterization of PDAC specimens to guide clinical decision making.
RESULTS
Samples, Clinical Data and Analytic Approach
Surgically resected primary infiltrating adenocarcinomas and matched germline DNA from whole blood were identified from 150 patients with mostly Stage I-III PDAC (four Stage IV patients) (Table S1). Detailed clinical and pathologic characteristics of the cohort matched those of the general population of patients with surgically resectable PDAC (He et al., 2014; Siegel et al., 2016) (Table S1). Four patients with evidence of metastatic disease (M1) at diagnosis were excluded from survival analyses. The median follow-up of the remaining 146 patients was 676 days, and 71 of these were alive at last follow-up. Among the clinical variables, only margin status (R0 vs. R1) showed a significant independent correlation with overall survival (p = 0.007, q = 0.077).
The neoplastic cellularity (or tumor purity) ranged from 0–53% (median 18%) as judged by central pathology review (Table S1). A single sample (IB-7644) was macrodissected to enrich for neoplastic cellularity. Neoplastic cellularity was evaluated independently by whole exome sequencing using the ABSOLUTE algorithm (Experimental Procedures) (Carter et al., 2012), and ranged from 9–89% (first quartile 20%; median 33%) (Table S1). Tumor purity was also evaluated using DNA methylation, which produced estimates that were strongly correlated with ABSOLUTE (R2 = 0.73, Table S1).
Landscape of Genomic Alterations
Recurrent somatic mutations
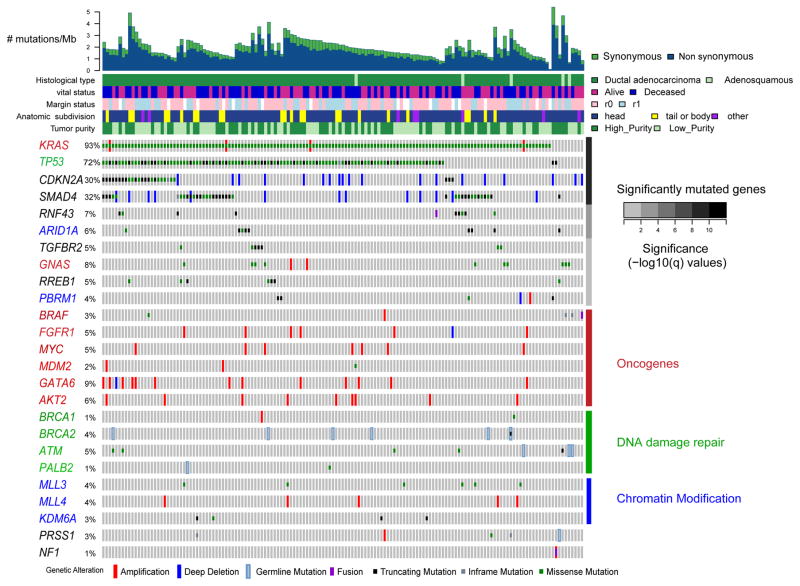
Whole exome sequencing (WES; mean coverage 405x) identified somatic DNA alterations, including single nucleotide variants (SNVs), small insertions and deletions (indels) and SCNAs. Significant recurrent mutations were identified in KRAS, TP53, CDKN2A, SMAD4, RNF43, ARID1A, TGFβR2, GNAS, RREB1 and PBRM1 (Figure 1, Figure S1A). We also observed recurrent mutations in several genes at FDRs above our threshold of q = 0.1, including mutations in other known oncogenes, DNA damage repair genes and chromatin modification genes. Except for RREB1, these genes have been previously reported as altered in PDAC (Bailey et al., 2016; Biankin et al., 2012; Jones et al., 2008; Waddell et al., 2015; Witkiewicz et al., 2015). Mutations in RREB1 included at least three predicted loss-of-function variants (Figure 1, Figure S1A). RREB1 is activated by the MAPK pathway, represses the miR-143/145 promoter, and has been reported to be downregulated in PDAC (Costello et al., 2012; Franklin et al., 2014; Kent et al., 2010; Kent et al., 2013). RREB1 is a positive regulator of the ZIP3 zinc transporter, and thus recurrent mutations in RREB1 may suggest an important role for zinc homeostasis in PDAC pathogenesis. Comparison of missense mutations in our cohort of patients to those reported in the literature using the Mutation Annotation and Genome Interpretation (MAGI) tool (Leiserson et al., 2015) highlighted mutations in CTNBB1, PIK3CA, ERBB2, POLE, SF3B1 and additional genes that have been identified in other cancer types (Table S2).
Figure 1. Landscape of genomic alterations in pancreatic ductal adenocarcinoma (PDAC).
Integrated genomic data for 149 non-hypermutated samples (columns), including: mutations (classified as truncating, in-frame or missense); high-level amplifications and homozygous deletions (“Deep Deletion”), fusions derived from analysis of mRNA data, and germline mutations for selected genes as described in the text. Overall number of mutations/Mb and clinicopathologic data for each sample are shown as tracks at the top. Significantly mutated genes (q ≤ 0.1) from exome sequencing data listed in order of q-value, followed by other recurrently altered genes organized in functional classes of oncogenes (red), DNA damage repair genes (green) and chromatin modification genes (blue). Significantly mutated genes from these classes are also colored accordingly. The percentage of PDAC samples with an alteration of any type is noted at the left. See also Figure S1, Tables S1–S3.
To increase our power to detect somatic mutations in low-purity samples, we pursued two additional sequencing strategies. First, the KRAS codon 12, 13 and 61 hotspots were sequenced using a microfluidic PCR-based approach with very deep coverage (mean ~30000x). Additionally, we designed a targeted sequencing panel that encompassed significantly mutated genes identified by MutSigCV2 analysis within the TCGA cohort, as well as a subset of additional genes across functionally relevant classes that have been identified as altered in pancreatic cancer by the International Cancer Genome Consortium (Bailey et al., 2016) (Table S2). These targeted genes were sequenced to higher coverage (~644X) compared to ~405X for WES. Through combined analysis of both the WES and targeted sequencing data, we identified many low prevalence mutations in well-annotated genes that may contribute to the pathogenesis of pancreatic cancer (Figure 1, Table S2). Several of these low prevalence mutations had potential therapeutically relevant implications (Figure S1B, see below).
Germline variants in pancreatic cancer susceptibility genes
Approximately 5–10% of PDAC occurs in patients with a family history of the disease, and several genes have been identified for which germline mutations confer susceptibility to PDAC (Roberts et al., 2016). We analyzed the matched germline exome sequencing data for alterations in known germline predisposition genes BRCA1, BRCA2, PALB2, STK11, CDKN2A, ATM, PRSS1, MLH1, MSH2, MSH6, PMS2, EPCAM and TP53. We observed predicted pathogenic germline mutations in 8% of patients in the cohort (11/149 non-hypermutated samples), including mutations in BRCA2 (n = 6), ATM (n = 3), PALB2 (n = 1) and PRSS1 (n = 1) (Figure 1). Clinical records on these 11 patients were not sufficient to fully evaluate for a family history of cancer. Evaluation of somatic mutation and copy number data on these samples with germline mutations revealed that the majority had loss or mutation of the other allele, with only the PALB2 germline mutant sample (IB-A5SP) and a single ATM mutant sample (IB-AAUT) appearing to retain the wild-type allele. The missense mutation observed in the PRSS1 cationic trypsinogen gene is a known pathogenic activating mutation (R122H) that has been associated with familial pancreatitis and a dramatically increased (> 50X) risk of pancreatic cancer (Keim et al., 2001; Whitcomb et al., 1996). Available TCGA clinical records for this case (2J-AABA) suggested that this patient had a history of chronic pancreatitis.
We observed significant enrichment for germline mutations in the predisposition genes noted above in the 10 KRAS wild-type samples (p = 0.027, Fisher’s exact test of KRAS wild-type vs. mutant).
Mutational signatures
We investigated known mutational signatures in the 150 samples and found a single primary signature of C>T transitions at CpG sites, which is associated with age of diagnosis (Alexandrov et al., 2013) (Signature A, Figure S1C). In addition, one sample with a mutation in the POLE polymerase demonstrated a hypermutator signature (Signature B). Although we detected both somatic and germline BRCA1/2 and PALB2 mutations in our cohort, we did not observe a mutational signature consistent with BRCA1/2 deficiency, perhaps because relatively few samples (n = 7) had a mutation in one of these genes. Additionally, the single somatic mutations in BRCA1 and BRCA2 were observed to have cancer cell fractions significantly less than one, suggesting that these mutations were subclonal and thus potentially less likely to exhibit a mutational signature of BRCA1/2 deficiency in WES data from bulk tumor.
Somatic copy number aberrations
Arm-level somatic copy number aberrations were identified in over a third of the tumors, using both single-nucleotide polymorphism (SNP) microarrays (whose sensitivity was constrained by low tumor purity) and WES. These included amplifications of 1q (33%) along with deletions of 6p (41%), 6q (51%), 8p (28%), 9p (48%), 17p (64%), 17q (31%), 18p (32%) and 18q (71%) (Table S3), consistent with previous studies (Bailey et al., 2016; Iacobuzio-Donahue et al., 2004; Waddell et al., 2015). GISTIC analysis of focal amplifications and deletions in the high-purity group revealed a number of recurrent events containing known oncogenic drivers (Figure S1D, Table S3)(Mermel et al., 2011). These include amplifications of GATA6 (18q11.2), ERBB2 (17q12), KRAS (12p12.1), AKT2 (19q13) and MYC (8q24.2), as well as deletions of CDKN2A (9p21.3), SMAD4 (18q21.2), ARID1A (1p36.11) and PTEN (10q23.31) (Figure 1, Figure S1D and Table S3).
Clinically relevant mutations
We assessed the clinical relevance of germline and somatic mutations, fusions, and copy number alterations in a curated list of genes (Figure S1B) using the PHIAL algorithm (Van Allen et al., 2014). Ten percent of samples harbored germline or somatic mutations in one of the DNA damage repair genes ATM, BRCA1, BRCA2 and PALB2, potentially sensitizing these tumors to platinum-based chemotherapy or poly-(ADP-ribose) polymerase (PARP) inhibition (Sahin et al., 2016b). We observed low prevalence alterations in several genes potentially amenable to other targeted therapies, including mutations in BRAF, PIK3CA, RNF43, STK11 and JAK1, as well as focal high-level amplifications in ERBB2. A single hypermutated sample harbored 19,957 mutations that included a mutation in POLE. This tumor may have a higher neo-antigen load which could have made the patient a candidate for immunotherapy approaches (Le et al., 2015). Excluding common events in KRAS or CDKN2A, 42% (63/150) of patients within this cohort had cancers with at least one genomic alteration that could potentially confer eligibility for current clinical trials, and 25% of the patients (38/150) had cancers with two or more such events, suggesting a potential basis for genotype-driven combination therapy trials.
Mutational heterogeneity of KRAS alterations in pancreatic cancer
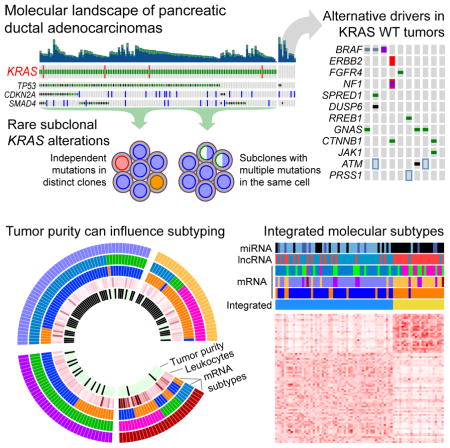
We evaluated the power to detect clonal and subclonal KRAS mutations across a range of neoplastic cellularity (Figure S2). We found that the combined depth of coverage across multiple modalities used in this project enabled high-confidence detection of KRAS mutations, including subclonal mutations that would have been missed at lower sequencing depths. We observed KRAS mutations in 93% (140/150) of the samples. Multiple oncogenic KRAS alleles were identified, including G12D (n = 62), G12V (n = 41), and G12R (n = 28), as well as numerous other hotspot codon 12 and 61 mutant alleles at a lower prevalence.
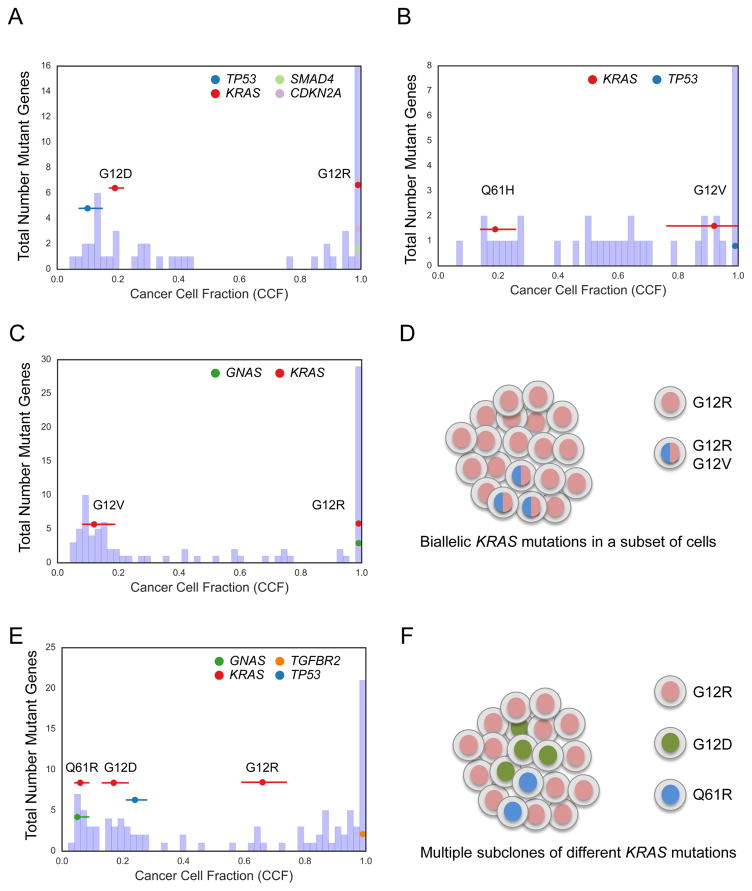
We used the ABSOLUTE algorithm for copy number and tumor purity analysis to investigate mutational heterogeneity in detail, using estimates of cancer cell fraction (CCF) for each mutation (Carter et al., 2012). Evidence of multiple distinct KRAS mutations was identified in five pancreatic cancers, including four with multiple known oncogenic hotspot mutations (Figure 2). Examination of these samples with the ContEst algorithm (Cibulskis et al., 2011) revealed very low probability of cross-individual contamination as an explanation for this observation (data not shown). We identified three examples of a clonal KRAS mutation concurrent with a subclonal KRAS mutation at a much lower CCF (Figure 2A–C), suggesting that in these samples some of the individual neoplastic cells harboured multiple KRAS mutations (Figure 2D). In each of the samples with multiple KRAS mutations, the individual mutations were observed on distinct sequencing reads, confirming that these mutations are occurring on different alleles rather than the same allele (data not shown). Notably, 3 of 4 cases with multiple hotspot KRAS mutations contained a G12R mutation as the dominant clone (p = 0.025, Fisher’s exact test of G12R vs. other hotspot codons as double mutant). Another case had multiple mutations, each of which was subclonal, and whose CCF’s complemented each other (Figure 2E), suggesting that these different KRAS mutations occurred in separate neoplastic cells in a single tumor (Figure 2F). In contrast, when we analyzed publically available TCGA data from other tumor types sequenced at conventional sequencing depths (~80–100X), we found no other evidence of multiple hotspot KRAS mutations within the same cancer (data not shown).
Figure 2. KRAS mutational heterogeneity.
(A–C) Histogram of cancer cell fraction (CCF) estimates (X-axis) for all identified mutated genes (Y-axis, blue bars) as well as point estimates and 95% confidence intervals for selected genes (colored horizontal lines) for a tumor (YB-A89D) with clonal KRASG12R mutation and clonal CDKN2A and SMAD4 mutations but also harboring a second apparent subclone with a KRASG12D and TP53 mutation (A), a tumor (XD-AAUG) with a clonal KRASG12V mutation and a subclonal KRASQ61H mutation (B), and a tumor (RB-A7B8) with a clonal KRASG12R mutation, a subclonal KRASG12V mutation, and a clonal GNAS mutation (C). (D) Schematic model of the tumor shown in (C) based on CCF evidence for biallelic KRAS mutations in a subset of cells. (E) Tumor (2J-AAB1) with CCF evidence of multiple subclonal KRAS alterations in the same tumor. (F) Schematic model of the tumor shown in (E) with evidence for multiple subclones, each harboring a different KRAS mutation. See also Figure S2.
Landscape of KRAS wild type samples
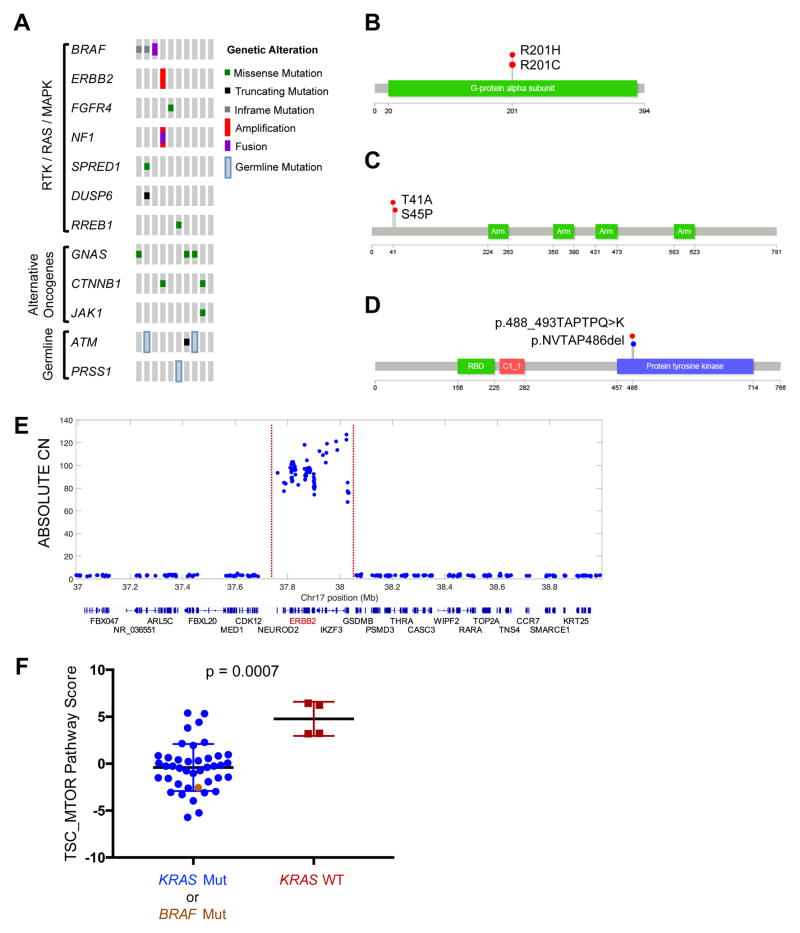
KRAS gene mutations were not identified in 10/150 samples, despite deep sequencing with three different approaches. As noted above, we observed an enrichment for germline mutations in familial risk genes within KRAS wild-type tumors. To identify other possible molecular drivers in these cancers, we conducted a thorough investigation of mutations, copy number alterations and translocation events in the RAS pathway, significantly mutated genes, and other known cancer genes (Table S4) (Figure 3A). We found a GNAS mutations in 3 of 10 KRAS wild-type samples (Figure 3A–B), as well as a known pathogenic activating mutation in JAK1 (R724H)(Flex et al., 2008). Two KRAS wild-type tumors harbored a known oncogenic missense mutation in CTNNB1 (Figure 3C).
Figure 3. Alternate drivers in KRAS wild-type samples.
(A) Co-mut plot for KRAS wild-type tumors (n = 10) displaying integrated data including mutations, copy number alterations, mRNA fusions and germline alterations as described in Figure 1. (B–D) Recurrently mutated GNAS (B), CTNNB1 (C) and BRAF (D) observed in KRAS wild-type samples. (E) Focal high-level amplification of ERBB2 in a KRAS wild-type sample. Red dotted lines indicate the boundaries of the amplicon. Chromosome position and ABSOLUTE copy number (CN) are indicated on the X- and Y-axes, respectively. Genes positioned within the genomic locus are indicated below. (F) RPPA scores for TSC/MTOR pathway in samples with KRAS mutation (blue), BRAF mutation (brown), or wild-type for both KRAS and BRAF (red). Column scatter plots show mean with standard deviation. Mann-Whitney rank-sum test, p = 0.0007. See also Table S4
In six of the ten samples, we identified somatic genetic alterations that likely activate the RAS-MAPK pathway upstream or downstream of KRAS (Figure 3A). Specifically, we discovered two in-frame deletions in BRAF that have recently been shown to activate the protein and drive MAPK signalling (Figure 3D) (Chen et al., 2016; Foster et al., 2016). A CUX1-BRAF fusion was identified in RNA-sequencing and WES data. We also observed mutations in negative regulators of the RAS-MAPK pathway, including NF1, SPRED1 and DUSP6. In a single sample, we observed a very focal high-level amplification of ERBB2 that encodes the HER2 receptor tyrosine kinase (Figure 3E). Thus, RAS pathway activation is a prominent molecular driver of pancreatic cancers, even when KRAS itself is not mutated. Several of the alternative activators of the RAS pathway are potentially targetable with existing therapies (Figure S1B).
We examined protein expression profiling with reverse phase protein arrays (RPPA) on the subset of tumors with higher neoplastic cellularity (ABSOLUTE purity ≥ 33%), including five of 10 KRAS wild-type tumors (see below, “Protein Expression”). Despite small numbers of samples examined, the KRAS wild-type tumors had significantly elevated TSC/MTOR signaling pathway activity compared to the KRAS mutant tumors (Figure 3F). Four of five KRAS wild-type tumors demonstrated elevated levels of multiple phosphorylated effector proteins in the MTOR signaling pathway including phosphorylated 4EBP1 and S6K. Notably, the TSC/MTOR pathway score was markedly elevated in the single sample (LB-A8F3-01A) for which we did not identify another putative driver event through analysis of WES data (Figure 3A, most right column). Furthermore, the only KRAS wild-type tumor that did not have an elevated TSC/MTOR pathway score harbored an activating BRAF mutation, and its pathway score tracked with those of KRAS mutant samples (Figure 3F). These data suggest that functional activation of the MTOR signaling pathway may be an alternative oncogenic driver in KRAS wild-type pancreatic cancer.
Tumor Purity Informed Analysis of Genome Characterization Platforms
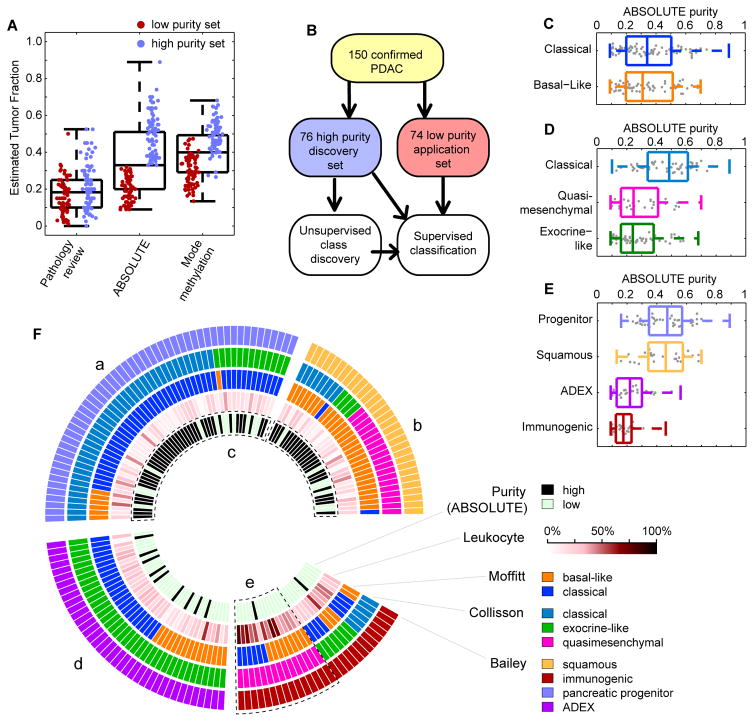
The low neoplastic cellularity of PDAC challenged analyses of messenger RNA (mRNA), long non-coding RNA (lncRNA), microRNA (miRNA), reverse phase protein array (RPPA) and DNA methylation, which were heavily confounded by tumor purity (Figure 4A and Figure S3A). Therefore, we used a two-step analysis strategy in which we split our cohort based on the median purity into a “high-purity” set of 76 samples with ABSOLUTE purity ≥33%, and a “low-purity” set of 74 samples that had ABSOLUTE purity < 33%. We clustered high-purity samples using unsupervised approaches, obtaining sets of genes/markers that were more likely to reflect the biology of the neoplastic cells, rather than that of the admixed stromal and other cells. We then used information derived from these high-purity samples, e.g. discriminatory features or trained Support Vector Machines, to classify the remaining low purity samples (Figure 4B). We found that this approach mitigated the tendency of low purity samples to cosegregate, and allowed us to achieve clustering results that were not significantly associated with purity, as discussed below.
Figure 4. Assessment and impact of purity on molecular analysis.
(A) Box plots show estimated tumor purity distributions determined by three methods for all 150 tumors. Dot plots embedded within the box plots show purity estimates for the 74 low-purity (red, purity below median) and 76 high-purity (blue, purity above median) samples used for supervised analyses. (B) Workflow of the two-stage approach for supervised clustering of 74 low purity samples using tumor-specific groups identified in the 76 high-purity samples. (C-E) Box plots of ABSOLUTE tumor purity for samples classified using the published mRNA signatures from Moffitt et al. (C), Collisson et al. (D), and Bailey et al. (E). (F) Sample overlap for mRNA subtypes from Bailey et al., Collisson et al., or Moffitt et al. (from inside to outside, respectively); DNA methylation estimated leukocyte fraction; and high/low purity based on ABSOLUTE. (a) Overlap between samples classified as ‘pancreatic progenitor’ (Bailey et al) ‘classical’ (Collisson et al.) and ‘classical’ (Moffitt et al.) mRNA subtypes. (b) Overlap between samples classified ‘squamous’ (Bailey et al.) and ‘basal-like’ (Moffitt et al.) mRNA subtypes. (c) squamous and progenitor are overrepresented in the high purity samples. (d) ADEX is a subset of exocrine-like. (e) Leukocyte fraction is elevated in Immunogenic samples, especially those also classified as quasimesenchymal. All box plots shown display full range, median, and upper and lower quartiles. See also Figures S3 and Table S5.
mRNA Subtypes
Two large studies using either PDAC (Moffitt et al., 2015) or PDAC and other types of pancreatic cancer samples (Bailey et al., 2016) recently reported gene expression subtypes of pancreatic cancer, extending the subtypes previously described by Collisson et al. (Collisson et al., 2011). We applied the clustering techniques from each of these studies to our data (Figure S3B–S3D), reproducing the 4-group classification (squamous, immunogenic, pancreatic progenitor, or aberrantly differentiated exocrine [ADEX]) of Bailey et al. (Bailey et al., 2016), the 3-group classification (classical, quasimesenchymal, or exocrine-like) of Collisson et al. (Collisson et al., 2011), and the 2-group classification (basal-like or classical) of Moffitt et al. (Moffitt et al., 2015). We found that classification of samples as basal-like or classical (Bailey et al., 2016; Moffitt et al., 2015) was independent of purity (Figure 4C). In contrast, the classifications of Collisson et al. and Bailey et al. were correlated with tumor purity in our cohort, with samples classified as exocrine-like or quasimesenchymal (Figure 4D), or samples classified as ADEX or immunogenic (Figure 4E) having lower tumor purity. We also found that, among low purity tumors, a higher estimated leukocyte fraction (Carter et al., 2012) was associated with immunogenic samples (Figure 4F). Further, the ADEX class was a subset of the exocrine-like class (Collisson et al., 2011) (Figure 4F, Figure S3E–G).
Considering only the high-purity samples in our cohort, the squamous samples of Bailey et al. showed significant overlap with the basal-like samples defined by Moffitt et al., while the Bailey et al. pancreatic progenitor and Collisson et al. classical group largely overlapped the classical samples defined by Moffitt et al. (Figure 4F, Figure S3E–G). These observations suggest that high purity tumors can be consistently classified into a basal-like/squamous group and a classical/progenitor group. The strong association of immunogenic and ADEX or exocrine-like subtypes with the low purity samples in our cohort suggests that these subtypes may reflect gene expression from non-neoplastic cells.
Analysis of genome characterization platforms
Following the schematic in Figure 4B, we identified de novo PDAC subtypes from DNA methylation, copy number, lncRNA, miRNA, and RPPA data. Using the non-coding RNA and RPPA data, all samples were classified into groups. In contrast, for DNA methylation and copy number, some samples with extremely low purity were not classified due to low signal intensity. We investigated whether classification was more feasible in higher purity tumors by measuring how well individual samples correlated to each cluster centroid (Figure S3H–J). For example, in lncRNA clusters, as purity increased, the samples classified into lncRNA group 1 became more similar to the centroid of all samples in lncRNA group 1 and less similar to the centroid of lncRNA group 2. This again demonstrates that it is easier to classify tumors into molecularly similar groups when the tumors have a high proportion of neoplastic cells.
DNA methylation
Unsupervised clustering of DNA methylation data for the 76 high-purity samples revealed two major subgroups (H1 and H2, Figure S3K). The H1 cluster (n = 41) had more extensive DNA hypermethylation than the H2 cluster (n = 35). In the low-purity sample set (n = 74), we identified three clusters (L1, L2 and L3, Figure S3K). The prevalence and level of cancer-specific DNA hypermethylation were markedly lower in the samples in the L1 cluster (n = 30), and the samples in this cluster also had significantly lower neoplastic purity than did the other clusters (p = 0.0087, median: 15% vs. 22%, 22% for clusters L2 and L3, respectively). Given this, we excluded the samples in the L1 cluster from subsequent integrative analyses. DNA hypermethylation profiles in the lower purity L2 and L3 clusters were similar to the higher purity H1 and H2 clusters respectively, even though the levels of DNA methylation were consistently weaker across CpG sites in the lower purity subgroups (Figure S3K). For the integrative multi-platform analyses described below, we merged the higher purity H1 cluster and lower purity L2 cluster to create a DNA hypermethylation subgroup 1 (n = 55), and we merged the higher purity H2 cluster and lower purity L3 cluster to form a DNA hypermethylation subgroup 2 (n = 65).
Integrated analysis of the DNA methylation and mRNA expression data revealed 98 genes that were silenced by DNA methylation, including genes that have been implicated in the development of other cancers but not previously reported to be altered in pancreatic cancer (Table S5)(Nagpal et al., 2014). Notable genes include: ZFP82, which is epigenetically silenced and suspected to function as a tumor suppressor (Xiao et al., 2014) (Yu et al., 2015) (Fan et al., 2016); PARP6 hypermethylation (Honda et al., 2016) (Qi et al., 2016); DNAJC15, which is hypermethylated in a number of tumor types (Ehrlich et al., 2002; Lindsey et al., 2006), and whose inactivation has been associated with chemotherapeutic drug resistance in breast (Fernandez-Cabezudo et al., 2016) and ovarian cancers (Rein et al., 2011). We also identified genes that were epigenetically silenced at low prevalence through manual examination of the genes known to be important in cancer, including BRCA1 and MGMT (each silenced in one case).
Copy number clustering
Clustering of SCNAs in high-purity tumors produced two major clusters, one with “high” and one with “low” levels of copy number alterations (Figure S3L, “High-Purity”). These two clusters did not significantly differ in purity (Figure S3M–N). Using a classifier generated from high-purity tumor clustering, we grouped low-purity tumors into the same clusters (Figure S3L, “Low Purity”). A smaller percentage of low-purity tumors were classified as “high” copy number variation compared to high-purity tumors (12% vs 37%, Fisher p < 0.001). In addition, 17 of the low purity tumors (22%) as well of the 1 of the high-purity tumors had few if any SCNAs, and were classified as non-aneuploid.
Non-coding RNA
miRNA
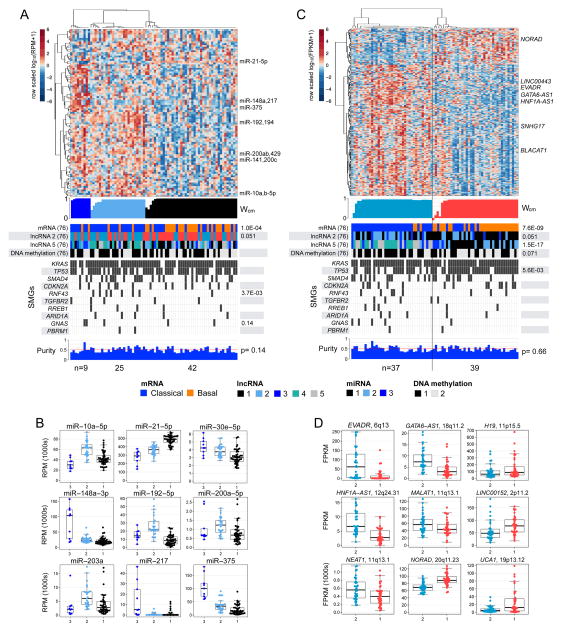
For the 76 high-purity tumour samples, we used unsupervised non-negative matrix factorization (NMF) consensus clustering (Gaujoux and Seoighe, 2010) with the most-variant 25% (n = 303) of miRNA mature strands (miRs) to obtain 3 clusters that were independent of purity (p = 0.14, Kruskal-Wallis test) (Figure 5A, Figure S4A–B). Many of the miRs that were differentially abundant across the clusters (Figure 5B, Table S6, Figure S4C) have been reported as prognostic, as differentially abundant between non-neoplastic and neoplastic tissue, or as functionally involved in signalling pathways in pancreatic cancer (Frampton et al., 2015; Halkova et al., 2015; Hernandez and Lucas, 2016; Lee et al., 2015; Lou et al., 2013; Sun et al., 2015). For example, miR-21 has been reported to be prognostic in pancreatic cancer (Frampton et al., 2015), and to be more abundant in tumors than in non-neoplastic pancreatic tissue (Halkova et al., 2015; Hernandez and Lucas, 2016). We noted that RNF43 mutations were significantly enriched (p = 3.7·10−3, Fisher exact test) in miR cluster 2 (Figure 5A). RNF43 mutations have therapeutic implications (Figure S1B) (Jiang et al., 2013; Koo et al., 2015) and frequently occur in IPMN precursor lesions (Amato et al., 2014; Wu et al., 2011a), suggesting biologic and clinical relevance for miR cluster 2.
Figure 5. Unsupervised clustering and differential abundance for miRNAs and lncRNAs, for 76 high-purity tumours.
(A) Heatmap of row-scaled, log10-transformed normalized expression for miRNA 5p and 3p mature strands (miRs) that were abundant and also differentially abundant across three consensus clusters computed using unsupervised non-negative matrix factorization clustering (NMF) (Cancer Genome Atlas Research, 2014; Gaujoux and Seoighe, 2010). Below the heatmap (top to bottom): a profile of silhouette width calculated from the consensus membership matrix (Wcm), clinical or molecular covariates with Fisher exact p values, mutation calls for significantly mutated genes, and a profile of ABSOLUTE purity (Carter et al., 2012), with a Kruskal-Wallis p value. Only p < 0.15 are shown. (B) Distributions of normalized abundance (RPM) for a subset of miRs that were scored as highly differentially abundant in a SAM multiclass analysis, or were differentially abundant (FDR < 0.05) and are known to be associated with cancers. (C, D) Results of a 2-cluster consensus clustering solution (Wilkerson and Hayes, 2010) for a subset of highly-variant lncRNAs presented similar to what shown in (A) and (B) respectively. All box plots shown display median values, and the 25th to 75th percentile, while whiskers extend up to 1.5 times the interquartile range. All data points are shown as individual dots. See also Figure S4 and Table S6.
lncRNA
We used poly(A)-selected RNA sequencing data to calculate transcript abundances for over 8000 Ensembl v82 lncRNAs, generating a comprehensive pancreatic lncRNA transcriptome. For the 76 high-purity samples, unsupervised consensus clustering (Wilkerson and Hayes, 2010), applied to expression profiles for a subset of 360 highly-variant lncRNAs, identified 2 clusters that were independent of purity (p = 0.66, Kruskal-Wallis) and concordant (p = 7.6·10−9) with the basal-like and classical mRNA subtypes (Figure 5C, Figure S4D–H). LncRNAs that were differentially expressed between the largely basal-like cluster 1 and the largely classical cluster 2 (Figure 5D, Figure S4D) included cancer-associated UCA1 (Huang et al., 2014; Li et al., 2016; Nie et al., 2016; Wang et al., 2008), HNF1A-AS1 (Muller et al., 2015; Wu et al., 2015; Yang et al., 2014), and NORAD (LINC00657)(Lee et al., 2016). We then used these differentially expressed lncRNAs to cluster all 150 of our samples, and found a stable 2-cluster solution that was concordant with the classification derived from the high-purity set alone (Figure S4H).
The most highly differentially expressed lncRNA associated with the classical mRNA subtype was EVADR, which has been reported to be specifically and abundantly expressed in adenocarcinomas, including PDAC (Gibb et al., 2015). The lncRNA DEANR1 (LINC00261) was nearly 2-fold more abundant in the classical subtype than in the basal-like subtype. This lncRNA regulates FOXA2 expression by recruiting SMAD2/3 to the FOXA2 promoter (Jiang et al., 2015). Intriguingly, DEANR1 has been implicated as having functional roles in pancreatic cancer (Muller et al., 2015) and in the formation of the pancreas (Jiang et al., 2015; Zorn and Wells, 2009). Like DEANR1, the lncRNA GATA6-AS1 was also more than 2-fold overexpressed in classical tumors; it has been shown to be transcriptionally activated when embryonic stem cells (ESCs) differentiate into endoderm (Sigova et al., 2013).
Unsupervised consensus clustering (Wilkerson and Hayes, 2010) within the 76 high-purity samples also identified a robust 5-cluster solution (Figure S4F,G,I–K), that was statistically independent of purity (p = 0.14, Kruskal-Wallis test) and overall survival (log-rank p = 0.73), and was strongly concordant with the 2-cluster solution (p = 1.5 ×10−17), and with the mRNA basal-like and classical subtypes of Moffitt et al. (p = 3.6×10−9, Fisher exact test). Distributions of abundance for certain lncRNAs varied between the largely basal-like clusters 1 and 2, and across the largely classical clusters 3–5 (Figure S4J–K), suggesting that lncRNAs, like miRNAs, may have differential effects within the classical and basal-like mRNA subtypes.
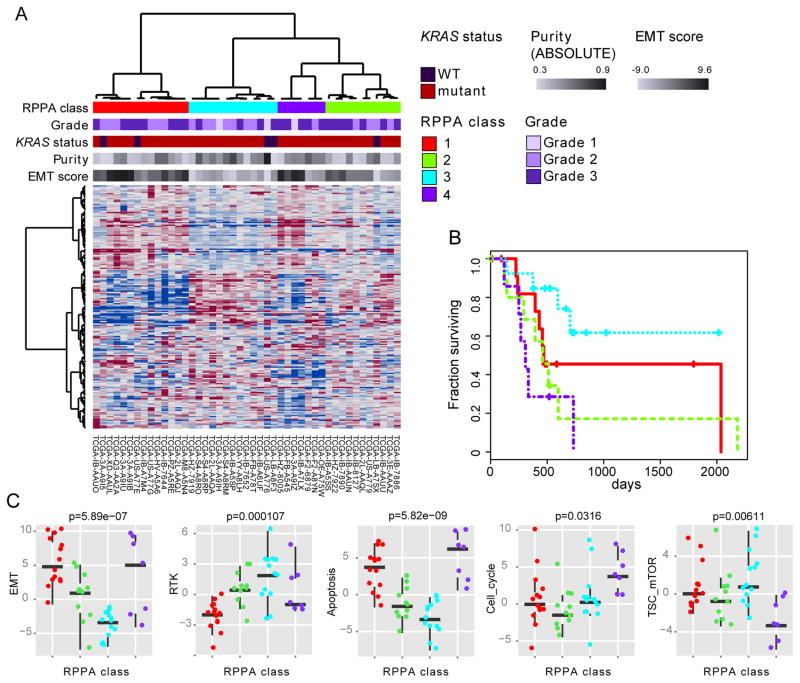
Protein expression
Unsupervised consensus clustering of protein expression measured on a 192-antibody array for 45 of the 76 high-purity samples identified four clusters (Figure 6A), which exhibited significant differences in survival (Figure 6B). We examined differences in pathway activity between clusters using 9 pathway activity scores (Akbani et al., 2014) (Table S7) identifying significantly different scores for epithelial-to-mesenchymal transition (EMT), apoptosis, TSC/mTOR, cell_cycle and RTK pathways (Figure 6C). Tumors from cluster 3, which had better survival, were characterized by low EMT and apoptosis pathway activity, but high TSC/mTOR and RTK activity. The same approach applied to the 39 low purity samples did not show significant differences in survival (p = 0.36, likelihood ratio test), suggesting, as was observed with other platforms, that low purity adversely affects the analysis.
Figure 6. RPPA profiles identify biologically distinct subsets of high purity tumors.
(A) Unsupervised consensus clustering of RPPA protein expression data for 45 of the 76 high-purity samples. (B) Cox survival analysis between clusters (p = 0.045, likelihood ratio test from Cox analysis with purity as covariate). (C) Differences in proteomic pathway activity scores across RPPA cluster/class for several pathway scores defined in (Akbani et al., 2014). Box plots indicate the median, upper and lower quartiles, with whiskers extending 1.5 times the interquartile range. Points indicate pathway scores for all 45 samples. See also Table S7.
Integrative Analysis
Cross-platform clustering
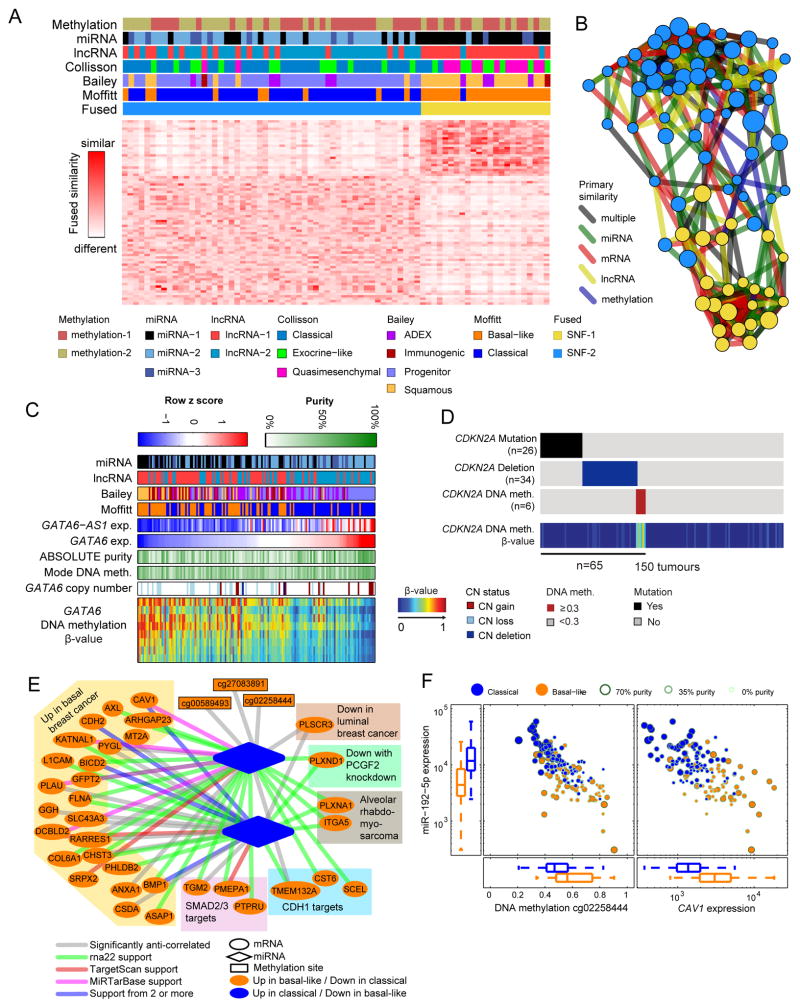
We observed a high degree of overlap between mRNA basal-like or classical subtypes and groupings produced by miRNA (p = 1.0·10−4), copy number (p = 0.014), lncRNA (p = 3.6·10−9), TP53 mutation status (83% vs. 64%, p = 0.01), and GNAS mutation status (p = 0.11) (Figure S5A). Due to the strong concordance among these data types, cluster-of-clusters analysis (Cancer Genome Atlas, 2012) favored a 2-cluster solution driven by either lncRNA or mRNA (Figure S5B).
To integrate information from multiple platforms, we performed Similarity Network Fusion (SNF), which has been shown to produce homogeneous, clinically relevant subtypes in multiple TCGA studies (Wang et al., 2014). We applied SNF to the high-purity cohort using sample-to-sample similarities derived from mRNA, miRNA, and DNA methylation. We found a two-cluster solution that was independent (p = 0.79) of tumor purity and a three-cluster (plus one outlier) solution that was associated (p = 0.025) with tumor purity. Pathology review showed that the outlier sample (US-A776) contained only a small component of invasive cancer with most of the sample being non-invasive intraductal papillary mucinous neoplasm (IPMN). The clusters defined by SNF were highly concordant with results obtained from miRNA, lncRNA, or mRNA alone (Figure 7A, B, and Figure S5C–D).
Figure 7. Integrated analysis.
(A) Integrated clustering of methylation, miRNA, lncRNA, and mRNA data using Similarity Network Fusion (SNF) on high purity samples. (B) Network fusion diagram of the two integrated clusters: each node is a sample, with node color indicating SNF cluster and node size proportional to ABSOLUTE purity. Edges are colored according to the datatype giving the strongest similarity between patients. Nodes positioned in between the top and bottom clusters generally have lower purity, reflecting the weaker signal for molecular classification. (C) DNA methylation heat map and overlapping tracks sorted by GATA6 expression. (D) CDKN2A status in all 150 cases showing mutation, deletion, or methylation in a subset of tumors. (E) Network of selected relationships between miRNA, lncRNA, mRNA, and methylation sites observed in the high purity samples, with edges indicating significant anti-correlations. Validated and predicted miRNA:mRNA associations from external sources are colored per legend. (F) Relationship of the expression of mir-192-5p with nearby DNA methylation and expression of CAV1, a predicted target of mir-192-5p. All box plots shown display full range, median, and upper and lower quartiles. See also Figures S5 and Table S8.
Activation and inactivation of genes by multiple genomic aberrations
We found that GATA6 and CDKN2A were altered by multiple mechanisms. In an integrated analysis of DNA methylation, copy number, and RNA expression, we found that GATA6 mRNA and an antisense lncRNA, GATA6-AS1, appeared to be deregulated by two distinct mechanisms (Figure 7C). Basal-like tumors exhibited higher DNA methylation near GATA6, and lower expression of both GATA6 and GATA6-AS1 mRNA; in contrast, classical tumors showed copy number gains of the GATA6 neighborhood, as well as higher expression of GATA6 and GATA6-AS1 mRNA. These results are consistent with previous reports of GATA6 amplification and elevated GATA6 mRNA expression in the classical subtype of PDAC (Collisson et al., 2011; Fu et al., 2008), as well as previous reports of GATA6 loss in basal-like tumors with poor outcome (Martinelli et al., 2016). Thus, there appears to be a subtype-associated positive or negative selective pressure on the GATA6 genomic neighborhood, confirming an important and complex role for GATA6 and possibly GATA6-AS1 in PDAC.
Cross platform examination suggested that CDKN2A is downregulated through multiple mechanisms (by DNA methylation in six samples, by deletions in 34, and by intragenic mutation in 26) (Figure 7D, Table S1). A disproportionate number of samples with CDKN2A alterations were identified in the high neoplastic cellularity group (alterations in 42/76 high-purity versus 23/74 low-purity, p = 0.003). These findings further underscore how low neoplastic cellularity may obscure genetic alterations.
RNA networks
To identify mechanisms of gene regulation in PDAC that may be contributing to the subtypes described above, we assessed correlations between DNA methylation, miRNAs, mRNAs and lncRNAs that were consistent with targeting and regulatory relationships. In the high-purity samples, we identified a network of correlations (Figure 7E) consistent with a basal-like/classical subtype model of PDAC (Figure S5E–K, Table S8). The network included many genes that were overexpressed in basal-like tumors and that we predicted were regulated by miR-192-5p and miR-194-5p; In contrast to their overexpressed mRNA targets, these miRs were underexpressed in basal-like tumors compared to classical tumors. The nomenclature ‘basal-like’ reflects similarities with basal breast and bladder cancers (Moffitt et al., 2015), and, for the genes in this correlation network, gene set analysis confirmed enrichment of genes from both ‘up in basal BRCA’ and ‘down in luminal BRCA’ sets (adjusted p = 5.2·10−55, 2.2·10−70) (Figure 7E). In high-purity tumors, the network included an anti-correlation between miR-192-5p expression and DNA hypermethylation at probe cg02258444, suggesting that miR-192-5p expression, which is high in classical tumors, may be suppressed by DNA methylation in basal-like tumors (Figure 7F). In addition, the network included anti-correlations between expression of miR-194-5p and miR-192-5p and expression of CAV1, consistent with predicted (Agarwal et al., 2015; Miranda et al., 2006) and experimentally validated miR-mRNA interactions (Chou et al., 2016a). CAV1 has been implicated in several PDAC phenotypes (Chatterjee et al., 2015) (Figure 7F). Taken together, these data suggest that regulation of a number of miRNAs by DNA methylation may contribute to the mRNA subtypes in PDAC.
DISCUSSION
We present a multiplatform molecular analysis of 150 PDAC specimens that exhibit a range of neoplastic cellularity representative of the clinico-pathologic spectrum of this disease. We demonstrated that the depth of sequencing is critical to the detection of mutations and SCNAs in low cellularity tumors, emphasizing the need for very deep sequencing of low purity samples to enable sufficient power to detect both clonal and subclonal alterations. Our analysis also highlights the importance of considering neoplastic cellularity when analyzing other molecular characterization platforms and using these to stratify samples.
We confirmed multiple previously identified driver genes in PDAC and we identified an additional driver gene, RREB1. Excluding mutations in KRAS, 42% of the patients had a cancer that harbored at least one alteration that could inform enrollment in current genotype-directed clinical trials. Germline and somatic mutations in the DNA damage repair genes BRCA2, PALB2 and ATM were observed in 8% of samples, representing a class of patients for whom platinum-based chemotherapy and/or PARP-inhibition may have therapeutic benefit. Importantly, these data highlight the potential value of clinical testing for these germline variants even in the absence of a clear cancer family history (Goggins et al., 1996; Grant et al., 2015).
Deep sequencing of KRAS enabled a high-confidence estimate that 93% of PDACs have KRAS mutation. A thorough investigation of other potential driver events in the KRAS wild-type tumors indicated that 60% of them harbour alternative RAS-MAPK pathway-activating alterations, further highlighting the importance of this pathway in this disease. We observed clinically relevant alterations with important therapeutic potential in 6 of the 10 KRAS wild-type tumors. Moreover, in a subset of these 10 KRAS wild-type tumors we observed elevated levels of phosphorylation of MTOR pathway proteins, suggesting that the MTOR pathway may be a therapeutic target in KRAS wild-type pancreatic cancers. These data support deep molecular profiling of KRAS wild-type tumors to identify drivers with potential therapeutic importance.
We also identified evidence for KRAS mutational heterogeneity that complicates our understanding of the role of KRAS in the progression of pancreatic cancer. Multiple KRAS mutations, including subclonal mutations, were identified in a small number of the specimens, including cases with apparent subclonal biallelic KRAS mutations. While the existence of multiple KRAS mutations has been previously reported in non-invasive IPMNs (Izawa et al., 2001; Tan et al., 2015; Wu et al., 2011b), we report multiple KRAS mutations occurring in invasive PDAC. The identification of multiple subclonal KRAS mutations may represent the convergent evolution of multiple clones of advanced cancer with independent KRAS mutations. Additionally, the apparent occurrence of multiple KRAS mutations within individual neoplastic cells suggests an additional selective advantage to development of a second KRAS mutation, perhaps from enhanced KRAS signalling in these cells. This observation complements other evidence that multiple RAS pathway lesions may occur in the same cancer cells to promote tumor progression, such as through amplification of the mutant allele or co-mutation of negative regulators of the pathway (Lock and Cichowski, 2015). Although the number of cancers with multiple KRAS mutations is small, the KRASG12R allele is enriched in these samples, suggesting that this allele may have distinct signalling properties that encourage selection for additional intratumoral KRAS mutations during tumor progression. Further experimental validation of this hypothesis is required. As therapeutic discovery efforts progress toward development of allele-specific small molecule inhibitors of the KRAS protein (Lito et al., 2016; Ostrem et al., 2013), the finding of multiple oncogenic KRAS mutations in the same sample may have important clinical ramifications, including the increased propensity for emergence of therapeutic resistance in these cancers.
Previous analyses of gene expression have identified mRNA subtypes of pancreatic cancer (Bailey et al., 2016; Collisson et al., 2011; Moffitt et al., 2015). Taking advantage of molecular purity estimates using the ABSOLUTE algorithm, we confirmed two tumor-specific subtypes of pancreatic ductal adenocarcinoma — basal-like/squamous and classical/pancreatic progenitor — and corroborated these across platforms. We found that GNAS mutations were enriched in classical subtype tumors, whereas TP53 mutations were more prevalent in basal-like subtype tumors. These two subtypes were also distinguished by differential regulation of gene expression via miRNA and DNA methylation. We found that the previously reported immunogenic and ADEX subtypes (Bailey et al., 2016), were associated with low neoplastic cellularity in our cohort. It is not clear from our data whether the identification of these two subtypes is driven by gene expression from the surrounding non-neoplastic tumor microenvironment or from other types of pancreatic cancer that were not included in our cohort. Further experimental characterization of these subtypes using single-cell profiling technologies is encouraged.
Examining protein expression in high purity samples revealed prognostic subtypes, including a group of tumors with improved overall prognosis and elevated RTK and MTOR signaling that may suggest therapeutic opportunity. Integrated platform analyses that also considered cellularity revealed non-coding RNA associations with tumor-specific subtypes. While biogenesis similarities for coding mRNAs and many lncRNAs (Quinn and Chang, 2016) suggest that subtypes identified from the two data types should be largely concordant, lncRNA expression can be specific for cell type and disease state (Mele et al., 2017; Nguyen and Carninci, 2016), and functionally-characterized lncRNAs can be specifically dysregulated in cancers (Huarte, 2015; Quinn and Chang, 2016). Differential expression of the EVADR, DEANR1, and GATA6-AS1 lncRNAs was associated with the classical (or pancreatic progenitor) molecular subtype of pancreatic cancer. EVADR was recently found to be associated with stomach, lung, colorectal, gastric and pancreatic adenocarcinomas (Gibb et al., 2015), while DEANR1 and GATA6-AS1 have been found to be associated with differentiation (Jiang et al., 2015). Our results suggest a potentially important relationship between non-coding RNAs and differentiation genes, including GATA6, that have previously been associated with classical/progenitor subtype tumors (Bailey et al., 2016; Collisson et al., 2011; Moffitt et al., 2015), as well as potentially new relationships between non-coding RNA and the more aggressive basal-like/squamous subtype tumors (Bailey et al., 2016; Moffitt et al., 2015).
Our integrated analysis across multiple molecular profiling platforms reveals a complex molecular landscape of PDAC and provides a roadmap for precision medicine.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Benjamin Raphael (braphael@princeton.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Tumor and normal whole blood samples were obtained from patients at contributing centers with informed consent according to their local Institutional Review Boards (IRB, see below). Biospecimens were centrally processed and DNA, RNA, and protein were distributed to TCGA analysis centers. In total, 150 evaluable primary tumors with associated clinicopathologic data were assayed on at least one molecular-profiling platform.
TCGA Project Management has collected necessary human subjects’ documentation to ensure the project complies with 45-CFR-46 (the “Common Rule”). The program has obtained documentation from every contributing clinical site to verify that IRB approval has been obtained to participate in TCGA. Such documented approval may include one or more of the following:
An IRB-approved protocol with Informed Consent specific to TCGA or a substantially similar program. In the latter case, if the protocol was not TCGA-specific, the clinical site PI provided a further finding from the IRB that the already-approved protocol is sufficient to participate in TCGA.
A TCGA-specific IRB waiver has been granted.
A TCGA-specific letter that the IRB considers one of the exemptions in 45-CFR-46 applicable. The two most common exemptions cited were that the research falls under 46.102(f)(2) or 46.101(b)(4). Both exempt requirements for informed consent, because the received data and material do not contain directly identifiable private information.
A TCGA-specific letter that the IRB does not consider the use of these data and materials to be human subjects research. This was most common for collections in which the donors were deceased.
METHOD DETAILS
Sample Processing
DNA and RNA were extracted and quality was assessed at the central BCR. RNA and DNA were extracted from tumor and adjacent non-tumor tissue specimens using a modification of the DNA/RNA AllPrep kit (Qiagen). The flow-through from the Qiagen DNA column was processed using a mirVana miRNA Isolation Kit (Ambion). This latter step generated RNA preparations that included RNA < 200 nt suitable for miRNA analysis. DNA was extracted from blood using the QiaAmp DNA Blood Midi kit (Qiagen).
RNA samples were quantified by measuring Abs260 with a UV spectrophotometer and DNA quantified by PicoGreen assay. DNA specimens were resolved by 1% agarose gel electrophoresis to confirm high molecular weight fragments. A custom Sequenom SNP panel or the AmpFISTR Identifiler (Applied Biosystems) was utilized to verify that tumor DNA and germline DNA representing a case were derived from the same patient. Five hundred nanograms of each tumor and germline DNA were sent to Qiagen (Hilden, Germany) for REPLI-g whole genome amplification using a 100 μg reaction scale. RNA was analyzed via the RNA6000 Nano assay (Agilent) for determination of an RNA Integrity Number (RIN), and only analytes with a RIN ≥ 7.0 were included in this study. Only cases yielding a minimum of 6.9 μg of tumor DNA, 5.15 μg RNA, and 4.9 μg of germline DNA were included in this study.
Sample Qualification
The BCR received tumor samples with germline controls from a total of 410 cases, of which 185 cases qualified and were sent for further genomic analysis. Of the 225 that failed to qualify, 25 cases were disqualified prior to processing, 16 failed for pathology screening, 175 cases failed due to molecular criteria, and 9 failed due to a genotype mismatch between tumor and germline.
Of the 16 that failed pathologic criteria, 12 failed for absence of tumor cells, 1 failed for necrosis, and 3 failed due to contaminating tumor in the germline control sample. The majority of the 175 cases that failed molecular screening had RNA integrity scores of < 7.0 (143 cases). The remaining 32 cases had insufficient DNA and/or RNA yields for molecular characterization.
Samples with residual tumor tissue following extraction of nucleic acids were considered for proteomics analysis. When available, a 10 to 20 mg piece of snap-frozen tumor adjacent to the piece used for molecular sequencing and characterization was submitted to MD Anderson for reverse phase protein array (RPPA analysis).
Microsatellite Instability Assay
Microsatellite instability (MSI) in qualified cases was evaluated by the Biospecimen Core Resource at Nationwide Children’s Hospital. MSI-Mono-Dinucleotide Assay was performed to test a panel of four mononucleotide repeat loci (polyadenine tracts BAT25, BAT26, BAT40, & transforming growth factor receptor type II) & three dinucleotide repeat loci (CA repeats in D2S123, D5S346, & D17S250). Two additional pentanucleotide loci (Penta D & Penta E) were included in this assay to evaluate sample identity. Multiplex fluorescent-labeled PCR & capillary electrophoresis was used to identify MSI if a variation in the number of microsatellite repeats was detected between tumor and matched non-neoplastic tissue or mononuclear blood cells. Equivocal or failed markers were re-evaluated by singleplex PCR. Tumor DNA was classified as microsatellite-stable (MSS) if zero markers were altered, low-level MSI (MSI-L) if less than 40% of markers were altered and high-level MSI (MSI-H) if greater than 40% of markers were altered. In the MSI-Mono-Dinucleotide Assay, this classification equated to MSI-L if one or two markers were altered, and MSI-H if three to seven markers were altered.
Individual markers were assigned a value of 1 through 6 based on the presence or absence of a MSI shift, allele homo/heterozygosity and loss of heterozygosity (LOH) if relevant. Markers that demonstrated MSI shift were classified as follows; 1 = homozygous alleles, 2 = heterozygous alleles with LOH and 3 = heterozygous alleles without LOH. Markers that did not demonstrate a MSI shift were classified as follows; 4 = homozygous alleles, 5 = heterozygous alleles with LOH, and 6 = heterozygous alleles without LOH. Penta D and E markers were scored in the same manner as the MSI markers; however, they did not contribute to MSI class calculation.
Analytical Approach
Samples were macrodissected to enrich for tumor purity, and characterized samples had post-dissection histologic neoplastic cellularity ranging from 0–53% (median 18%) as judged by central pathology review (Table S1). Tumor purity was independently evaluated in whole exome sequencing data on the 150 cancers that had histologically observable tumor using the ABSOLUTE algorithm (Carter et al., 2012) and ranged from 9–89%, with a first quartile of 20% and a median of 33% (Table S1). The 9 samples that were found to have < 1% neoplastic cellularity during central pathology review were held out from the tumor cohort. DNA, RNA and protein were extracted from the specimens using standard TCGA approaches. One case with high neoplastic cellularity (89% by ABSOLUTE) contained a large precursor lesion in addition to an invasive carcinoma, explaining the discordance with the histologic assessment of neoplastic cellularity, which included only an evaluation of the invasive component.
Purity estimation and two-stage clustering
Using our two-stage clustering strategy 18 samples were called non-aneuploid due to undetectable SCNA events (mean purity of 16%), and 30 samples had too little DNA methylation to be classified as either of the two subtypes (mean purity of 17%). Using the mode of DNA methylation at hypermethylated sites as an indicator of purity resulted in an estimate that correlated well with ABSOLUTE (R2 = 0.73), suggesting a low level of DNA methylation activity in stroma compared to neoplastic cells.
Whole Exome sequencing (WES)
Sample preparation and sequencing
Starting with 250 ng input DNA, samples are quantified using a PicoGreen assay and diluted to a working stock volume and concentration (2 ng/μL in 50 μL), then libraries are constructed and sequenced on Illumina HiSeq instruments with the use of 76-bp paired-end reads. Output from Illumina software is processed by the Picard data-processing pipeline to yield BAM files containing well-calibrated, aligned reads. All process steps are performed using automated liquid handling instruments, and all sample information tracking is performed by automated LIMS messaging.
Libraries are then constructed using the protocol described in Fisher et al. (Fisher et al., 2011) with several modifications. First, initial genomic DNA input into shearing has been reduced from 3 μg to 100 ng in 50 μL of solution. Second, for adapter ligation, Illumina paired end adapters have been replaced with palindromic forked adapters with unique 8 base index sequences embedded within the adapter. These index sequences enable pooling of libraries prior to sequencing. Third, custom sample preparation kits from Kapa Biosciences are now used for all enzymatic steps of the library construction process. For the majority of samples multiple libraries were generated in order to achieve sequencing depths necessary for downstream analysis.
In-solution hybrid selection was performed as previously described (Fisher et al., 2011). Following sample preparation, libraries are quantified using PicoGreen. Based on PicoGreen quantification, libraries are normalized to equal concentration and pooled by equal volume. Library pools are then quantified using a Sybr Green-based qPCR assay, with PCR primers complementary to the ends of the adapters (kit purchased from Kapa Biosciences). After qPCR quantification, library pools are normalized to 2 nM, denatured using 0.2 N NaOH, and diluted to 20 pM, the working concentration for downstream cluster amplification and sequencing. Denatured library pools are spread across the number of sequencing lanes required to achieve target coverage for all samples.
Cluster amplification and sequencing of denatured templates are performed according to the manufacturer’s protocol (Illumina) using HiSeq instruments. Read length is 76bp paired end reads, with additional cycles added to read molecular index sequences, are performed. Output from Illumina software is processed by the Picard data-processing pipeline to yield BAM files containing well-calibrated, aligned reads.
Mutation Analysis
Sequencing data-processing pipeline (“Picard pipeline”)
The “Picard” pipeline (http://picard.sourceforge.net/) generates a BAM file (http://samtools.sourceforge.net/SAM1.pdf) for each sample and was developed by the Sequencing Platform at the Broad Institute. Picard pipeline aggregates data from multiple libraries and flow cell runs into a single BAM file for a given sample. This file contains reads aligned to the human genome with quality scores recalibrated using the Table Recalibation tool from the Genome Analysis Toolkit. Reads were aligned to the Human Genome Reference Consortium build 38 (GRCh38) using BWA v0.5.9 (Li and Durbin, 2010) (http://bio-bwa.sourceforge.net/). Unaligned reads that passed the Illumina quality filter (PF reads) were also stored in the BAM file. Duplicate reads were marked such that only unique sequenced DNA fragments were used in subsequent analysis. Sequence reads corresponding to genomic regions that may harbor small insertions or deletions (indels) were jointly realigned to improve detection of indels and to decrease the number of false positive single nucleotide variations caused by misaligned reads, particularly at the 3′ end. To improve the efficiency of this step, we performed a joint local-realignment of all samples from the same individual (“co-cleaning”). All sites potentially harboring small insertions or deletions in either the tumor or the matched normal were realigned in all samples. Finally, the Picard pipeline provided summary QC metrics such as the target coverage and an estimated level of “oxo-G” artifacts (Costello et al., 2013) for each BAM that were used in subsequent processing.
Cancer genome analysis pipeline (“Firehose”)
The Firehose pipeline (http://www.broadinstitute.org/cancer/cga/Firehose) performed additional QC on the bams, mutation calling, small insertion and deletion identification, rearrangement detection, coverage calculations, annotation of detected mutations, filtering for OxoG artifacts and filtering by “panel-of-normals” and by Exome Aggregation Consortium (ExAC) dataset. The pipeline is an extensive series of tools for analyzing massively parallel sequencing data for both tumor DNA samples and their patient-matched normal DNA samples. The pipeline contains the following steps:
Quality control on BAM files: The sample cross-individual contamination levels were estimated using the ContEst program (Cibulskis et al., 2011).
Somatic point mutation calling: The MuTect algorithm (Cibulskis et al., 2013) was used to detect somatic single nucleotide variants (SNVs). SNVs were detected using a statistical analysis of the bases and qualities in the tumor and normal BAMs.
Small insertion and deletion detection: The Indelocator algorithm (https://www.broadinstitute.org/cancer/cga/indelocator) was used to detect small insertions and deletions (InDels).
SNVs and InDel annotations: SNVs and InDels detected by MuTect and Indelocator, respectively, were annotated using Oncotator (Ramos et al., 2015). Oncotator mapped somatic mutations to respective genes, transcripts, and other relevant features. These annotations correspond to the fields in the TCGA Mutation Annotation Format (MAF) files version 2.4 (https://wiki.nci.nih.gov/display/TCGA/Mutation+Annotation+Format+(MAF)+Specification).
Filtering for OxoG artifacts: 464 G>T/C>A transversions that are a consequence of heating, shearing, and oxidative damage to the DNA during genomic library preparation (Costello et al., 2013) were filtered out of the call set. These 464 transversions were found in the tumor sample BAM files of the following individuals: HZ-A77Q, IB-A7LX, IB-A7M4, S4-A8RP, XN-A8T3, YB-A89D and YY-A8LH. In addition, a tumor/normal pair whose tumor BAM file was damaged beyond recovery was removed from the final freeze list.
Filtering by “panel-of-normals”: The sites of detected SNVs and InDels were examined against a panel of 8313 normal samples (PoN). For a given SNV or InDel, a likelihood score that the allele counts are consistent with expectation of observed normals at the site is calculated. Candidate mutations with a likelihood score less than −2.5 were removed from subsequent analysis. We also removed variants outside coding regions. Additionally, any SNV or InDel that validated in either RNASeq or KRAS deep sequencing was not filtered. As a result of “panel-of-normals” filtering, 7804 SNVs and InDels were removed from the call set.
Filtering by ExAC: 60706 germline mutation calls from the ExAC database (http://exac.broadinstitute.org/) were used to screen for germline calls where coverage in normal was low, and consequently, 19 SNVs and InDels were removed from the call set.
Manual review of variants
Following Firehose processing, we performed manual review of several significantly mutated genes using the Integrative Genomics Viewer (IGV) (Thorvaldsdottir et al., 2013) for the review of sequencing evidence in the tumor and normal samples. We used a representative panel of normal WES BAMs to model a wide range of sequencing or alignment artifacts, or rare germline mutations, that might be misidentified as somatic mutations.
Multi-center calling of mutations
To strengthen confidence in mutation calls, SNV’s and InDels were called at multiple centers within the TCGA network. SNV’s were called at the Broad Institute, Baylor College of Medicine Human Genome Sequencing Center (HGSC), British Columbia Genome sequencing Center (BCGSC) and the University of California Santa Cruz (UCSC). InDels were called at the Broad Institute, HGSC and BCGSC. The final list of mutation calls for the cohort were determined as follows: 1) SNVs were accepted if called at the Broad Institute and/or 2 or more additional centers; 2) InDels were accepted if called in 2 or more centers.
Mutation Annotation Format (MAF) file
The MAF file was generated per TCGA specifications (https://wiki.nci.nih.gov/display/TCGA/Mutation+Annotation+Format+(MAF)+Specification) and has been made available with the analyses contained within this manuscript. A unique column named “usable_in_mutsig” was added to the MAF file, and this binary valued column indicates whether a given SNV or InDel was included in the downstream MutSig2CV analysis. 19956 SNVs and InDels in the hyper-mutated tumor sample, IB-7651, and 104 SNVs that were discovered in the targeted panel were not included the MutSig2CV analysis; the rest of SNVs and InDels were included (see below).
Mutation Significance Analysis
Genes with a significant excess of the number of non-synonymous mutations relative to the estimated density of background mutations were identified using the MutSig algorithm (Lawrence et al., 2014; Lawrence et al., 2013). MutSig has been previous used to identify significantly mutated genes (SMGs) in several tumor sequencing projects (Berger et al., 2012; Dulak et al., 2013; Lohr et al., 2012; Stransky et al., 2011) and the algorithm’s current version MutSig2CV (Lawrence et al., 2014) was used in this study to produce a robust list of significantly mutated genes. MutSig takes into account the background mutation rates of different mutation categories (i.e. transitions or transversions in different sequence contexts, the non-synonymous to synonymous mutation ratio for each gene, as well as the fact that different samples have different background mutation rates. It then uses convolutions of binomial distributions to calculate the p value for each gene, which represents the probability that we observe a certain configuration of mutations in a gene by chance, given the background model. Finally, it corrects for multiple hypotheses by calculating a q-value (False Discovery Rate) for each gene using the Benjamini & Hochberg procedure to produce the list of SMGs (Figure 1 and Figure S1).
KRAS Wild-type (WT) analysis
KRAS gene mutations were not identified in 10 of the 150 cancers, despite deep sequencing with three different approaches. To identify other possible molecular drivers in these cancers, we conducted a thorough investigation of mutations, copy number alterations and translocation events in a gene set (Table S4) comprised of RAS pathway, significantly mutated, and other known cancer genes (Figure 3A). RAS pathway genes were curated from the National Cancer Institute RAS pathway gene list, version 2.0. Significantly mutated genes were taken from the MutSig2CV analysis of the pancreatic cancer cohort presented in this manuscript. Additional known cancer genes were taken from the Dana-Farber Cancer Institute clinical sequencing gene set (OncoPanel v3.0). The union of these gene lists is presented in Table S4. This gene set was used to specifically interrogate for somatic mutations, germline mutations in a select set of familial risk genes as indicated in the manuscript, copy number alterations and translocation events (from RNA, as described below). RPPA data was also interrogated within KRAS wild-type samples as discussed in the text.
Mutation Clonality Assessment
To assess whether mutations are clonal (i.e. present in all cancer cells), we estimated the cancer cell fraction (CCF) of each mutation, as described (Carter et al., 2012). Mutations for which the CCF is close to 1 are considered clonal. Those mutations with lower probable CCFs are considered subclonal. To determine the CCF we first calculated the sample purity (i.e. the percentage of tumor cells in our sample) using the ABSOLUTE program to estimate sample purity and ploidy based on whole exome sequencing array data for allele specific copy number measurement and mutation allele fraction information (Carter et al., 2012).
Once we had estimated tumor purity and ploidy for the 150 samples, we then calculated the cancer cell fraction (CCF) for each mutation. The cancer cell fraction is the percentage of tumor cells harboring a given mutation. Clonal mutations have an underlying cancer cell fraction of one and subclonal mutations have an underlying cancer cell fraction of less than one. Mutations were classified as subclonal if the upper bound of the 95% confidence interval was less than 0.9 and clonal if the lower bound of the 95% confidence interval exceeded 0.9.
Copy number analysis
For copy number analysis based on exome sequencing, segmented copy data was obtained using copy number ratios. These were calculated as the ratio of tumor read depth to the average read depth observed in a panel of normal samples using the tool, RECAPSEG5. Allelic copy number analysis was done with Allelic-Capseg using B-allele frequencies from heterozygous sites ABSOLUTE (Carter et al., 2012) was used to determine purity, ploidy, and whole genome doubling status using allelic copy number data along with the allelic fraction of all somatic mutations as input. In silico admixture removal (ISAR) was used to perform purity and ploidy correction of the RECAPSEG data. We used ABSOLUTE derived copy number from WES to identify genes with loss of heterozygosity and homozygous deletions. High level amplifications were defined as those genes with three or more copies above baseline ploidy.
SCNA Significance Analysis
Significance of copy number alterations were assessed from the segmented data using GISTIC2.0 (Version 2.0.22) (Mermel et al., 2011). Briefly, GISTIC2.0 deconstructs somatic copy-number alterations into broad and focal events and applies a probabilistic framework to identify location and significance levels of somatic copy-number alterations. For the purpose of this analysis, we defined an arm-level event as any event spanning more than 50% of a chromosome arm.
SCNA Clustering
For copy number clustering, the cohort was dichotomized into one group above the median purity and one below. The high-purity tumors were clustered based on log2 copy number at regions revealed by GISTIC analysis. Clustering was done in R, with an Euclidean distance using Ward’s method. The same matrix used for the high-purity group was then applied to the low purity group. This allowed for the merger of the two by combining clusters that showed the same marker SCNAs. Of note, a group of 20 low-purity tumors had no SCNAs and were thus referred to as ‘NO’ in the clustering analysis.
Germline Variant Calling, QC, and Analysis
Germline variants were interrogated for 13 genes that are examined in patients with a significant family history of pancreatic cancer at the Dana-Farber Cancer Institute, including BRCA1, BRCA2, PALB2, STK11, CDKN2A, ATM, PRSS1, MLH1, MSH2, MSH6, PMS2, EPCAM and TP53. Briefly, germline variants were identified in these genes that occur in < 1% of the normal population, annotated for predicted functional impact and cross-referenced with the ClinVar database for prior evidence of disease linkage.
A total of 150 germline exomes from the study were called using best practices with the Genome Analysis Toolkit (GATK) HaplotypeCaller (version 3.6)(McKenna et al., 2010). The calls were then combined and jointly genotyped, and the sites were filtered through the GATK Variant Quality Score Recalibration (VQSR) workflow as recommended in GATK Best Practices (http://gatkforums.broadinstitute.org/gatk/discussion/1259/which-training-sets-arguments-should-i-use-for-running-vqsr).
Principal components analysis (PCA) was then performed on the resulting callset using a subset of 5,856 variants chosen by Purcell and others (Purcell et al., 2014) such that they were (i) on autosomal chromosomes; (ii) polymorphic across multiple ethnic populations; (iii) present in the targeted coding regions of most exome capture platforms; (iv) in approximate linkage equilibrium; and (v) in Hardy-Weinberg equilibrium. We combined the 150 PAAD germline exomes with a set of 1489 publicly available, normal population exomes with known ethnicity labels from the 1000 Genomes Project and the Exome Sequencing Project study.
Using EIGENSTRAT’s smartpca in fastmode (Price et al., 2006), we obtained 10 principal component vectors, and using the known ethnicity annotations for the normal population samples as a training set, we inferred the ethnicity of the PAAD cohort samples based on their projection onto the first five principal components (PCs). For each of the labeled ethnic groups, we calculated the center in the five-principal component space and assigned samples with unknown ethnicity based on the closest centroid (using Euclidean distance). We next examined cryptic relatedness within the PAAD cohort, running KING (Manichaikul et al., 2010) on the same set of 5,856 sites to check for duplicates and first- or second-degree relatives in the cohort. None were found.
Next, we assessed a battery of sample-level quality control (QC) metrics from the calling process, including the total number of single nucleotide variants (SNVs) and insertions/deletions (indels) called, transition-transversion ratios, and the number of singleton and novel sites. The distribution of each sample QC metric was evaluated for outliers within each ethnicity group (African American, Asian, European American, and Hispanic). None were found.
Germline variants in the 13 selected genes were extracted from the callset, and common variants (with minor allele frequency > 1% in the non-cancer ExAC normal population cohort (ftp://ftp.broadinstitute.org/pub/ExAC_release/release0.3.1/subsets/)) were removed. All genotype calls with a genotype quality score less than 20 (the phred-scaled confidence in the genotype call) were removed. We used the variant effect predictor (VEP) (http://www.ensembl.org/info/docs/tools/vep/index.html) with the LOFTEE plugin (https://github.com/konradjk/loftee) to annotate all variant sites for their expected functional impact. Missense mutations were only reported if there was prior reported evidence of functional significance in the ClinVar database.
KRAS Validation by Resequencing
Validation of KRAS mutations was performed by targeted resequencing using microfluidic PCR on the 48.48 Fluidigm Access system (Fluidigm, South San Francisco, CA) and the MiSeq sequencing system (Illumina, San Francisco, CA). Tumor samples were selected for validation based on the presence of the indicated mutations by whole exome sequencing. In addition, a subset of normals was also chosen for re-sequencing. Target-specific primers were designed to flank 2 sites of interest (chr 12: hg19 25398284–25398285 and chr 12: 25380272–25380276). Eight primer pairs were designed (five for the first target and three for the second), with target regions ranging in size from 166 to 195 bp. PCR was performed on the Fluidigm Access Array according to the manufacturer’s instructions, using the single-plex protocol. The Access Array Integrated Fluidic Circuit (IFC) enabled parallel amplification of up to 48 unique samples per chip. Every reaction combined both an amplicon-tagging PCR using tailed target-specific primers (tailed with adapter sequence), and a molecular barcoding PCR, using primers containing sequence complementary to the target-specific primer tails, a molecular barcode, and a flow cell attachment sequence that was compatible with Illumina. The Bravo Automated Liquid Handler (Agilent Technologies, Lexington, MA) was used for chip loading, PCR set-up and harvesting. Indexed libraries (pools of amplicons) were harvested for each sample from the chip into a single collection well, quantified, and quality-checked using Caliper GX (Perkin Elmer, Boston, MA). These per-sample-amplicon-pools were then normalized based on concentration, and pooled into a single tube (usually 96 samples per pool, but variable). Final amplicon library pools were quantified by qPCR using the Kapa Library Quantification Kit for NGS (Kapa Biosystems, Wilmington, MA), and sequenced on MiSeq according to manufacturer’s protocol using paired end 150-bp sequencing reads. Output from Illumina software is processed by the Picard data-processing pipeline to yield BAM files containing well-calibrated, aligned reads.
Custom Targeted Gene Panel Sequencing
Library construction was performed as described by Fisher et al. (Fisher et al., 2011) with some slight modifications. Initial genomic DNA input into shearing was reduced from 3μg to 100ng in 50μL of solution. In addition, for adapter ligation, Illumina paired end adapters were replaced with palindromic forked adapters with unique 8 base index sequences embedded within the adapter.
In-solution hybrid selection was performed using a custom design panel Illumina Rapid Capture enrichment kit with 43,164bp target territory (0.43 Mb baited). Dual-indexed libraries are pooled into groups based on library construction performance prior to hybridization. The liquid handling is automated on a Hamilton Starlet. The enriched library pools are quantified via PicoGreen after elution from streptavadin beads and then normalized to a range compatible with sequencing template denature protocols. Resulting libraries were sequenced on Illumina HiSeq2500 instruments with paired in 76bp reads. Output from Illumina software is processed by the Picard data-processing pipeline to yield BAM files containing well-calibrated, aligned reads.
RNA-sequencing (RNA-seq)
RNA library construction, sequencing, and analysis
One μg of total RNA was converted to mRNA libraries using the lllumina mRNA TruSeq kit (RS-122-2001 or RS-122-2002) following the manufacturer’s directions. Libraries were sequenced 48×7×48bp on the Illumina HiSeq 2000. FASTQ files were generated by CASAVA. RNA reads were aligned to the hg19 genome assembly using MapSplice 0.7.4 (Wang et al., 2010). Gene expression was quantified for the transcript models corresponding to the TCGA GAF2.1 (http://tcga-data.nci.nih.gov/docs/GAF/GAF.hg19.June2011.bundle/outputs/TCGA.hg19.June2011.gaf), using RSEM (Li and Dewey, 2011) and normalized within-sample to a fixed upper quartile. For further details on this processing, refer to Description file at the DCC data portal under the V2_MapSpliceRSEM workflow (https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/tumor/tgct/cgcc/unc.edu/illuminahiseq_rnaseqv2/rnaseqv2/unc.edu_PAAD.IlluminaHiSeq_RNASeqV2.mage-tab.1.0.0/DESCRIPTION.txt) or our alignment pipeline summary at CGHUB (https://cghub.ucsc.edu/docs/tcga/UNC_mRNAseq_summary.pdf).
Quantification of genes, transcripts, exons and junctions can be found at the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/).
mRNA analysis
Samples were classified into groups based on mRNA expression in three ways, based on the results in Moffitt et al. (Moffitt et al., 2015), Collisson et al. (Collisson et al., 2011), or Bailey et al. (Bailey et al., 2016). We first considered Moffitt et al.’s tumor-specific gene expression signatures, which define classical and basal-like subtypes of pancreatic ductal adenocarcinoma (PDAC). Using 50 (48 with a unique match in our data) tumor-specific transcripts from Moffitt et al., we applied consensus clustering to our mRNA cohort with Pearson correlation as the internal distance metric, seeking and reproducing two clusters of both genes and samples. We then considered the four PDAC subtypes described by Bailey et al.: squamous, pancreatic progenitor, ADEX, and immunogenic. Using the list of 613 (463 with a unique match in our data) differentially expressed transcripts from their multiclass SAM analysis, we performed consensus clustering with mRNA from our cohort, again using Pearson correlation as the internal distance metric. We verified that the four groups of samples and transcripts that we observed reflected the up/down relationships described in the t-statistics given for each gene and each class in the Bailey et al. manuscript. Using 62 (61 with a unique match in our data) transcripts identified by Collisson et al., we performed consensus clustering with mRNA from our cohort, again using Pearson correlation as the internal distance metric, seeking and verifying the presence of three clusters: classical, quasimesenchymal and exocrine-like.
RNA-seq read mapping for lncRNAs
RNA sequence reads were aligned to the human reference genome (hg38) and transcriptome (Ensembl v.82) using STAR v.2.4.2a (Dobin et al., 2013). STAR was run with the following parameters: minimum/maximum intron sizes were set to 30 and 500,000, respectively, noncanonical, unannotated junctions were removed, maximum tolerated mismatches was set to 10, and the outSAMstrandField intron motif option was enabled. The Cuffquant command included with Cufflinks v.2.2.1 (Trapnell et al., 2010) was used to quantify the read abundances per sample, with fragment bias correction and multiread correction enabled. All other options were set to default. To calculate the fragments per kilobase of exon per million fragments mapped (FPKM), the Cuffnorm command was used with default parameters. From the FPKM matrix for the 76 high-purity tumor samples, we extracted 8167 genes with Ensembl biotypes that were either “lincRNA” or “processed_transcript”.
lncRNA unsupervised and supervised clustering
For the n = 76 high-purity subset of the tumour cohort we extracted 360 lncRNAs that were robustly expressed (mean FPKM ≥1) and highly variable (≥ 95th FPKM variance percentile) from the lncRNA genes-by-samples data matrix noted above. We identified groups of samples that had similar abundance profiles by unsupervised consensus clustering with ConsensusClusterPlus (CCP) v1.24.0. Calculations were performed using Pearson correlations, partitioning around mediods (PAM), 10000 iterations, and a random 95% fraction of genes in each iteration. We selected a five-cluster solution. To generate an abundance heatmap we identified lncRNAs that had a mean FPKM of ≥ 5 and a SAM multiclass q-value of ≤ 0.01 across the unsupervised clusters (see differential abundance, below), transformed each row of the matrix by log10(FPKM + 1), then used the pheatmap R package (v1.0.2) to scale and cluster only the rows, using a Pearson correlation distance metric and Ward clustering.
We identified genes that were differentially abundant across the five unsupervised clusters using a SAM multiclass analyses (samr v2.0) (Li and Tibshirani, 2013), with an FPKM input matrix and an FDR threshold of 0.05.
We compared unsupervised clusters to clinical and molecular covariates by calculating contingency table association p values using R, with a Fisher exact or Chi-square test for categorical data (e.g. gender), and a Kruskal-Wallis test for real-valued data (e.g. purity).
For supervised clustering the full set of n = 150 tumor samples, we identified the set of lncRNA which 1) were among the 360 robustly expressed lncRNA discussed earlier, 2) had a mean abundance in the high purity subset larger than the mean abundance in the low purity subset, and 3) were differentially expressed between the 2 classes in the high purity cohort (t-test, with a B-H corrected FDR of 0.1). This resulted in 86 transcripts, which were used to perform consensus clustering on the full 150 sample data set with Pearson correlation as the internal distance metric, seeking and verifying the presence of two clusters.
mRNA analysis of fusion genes
Somatic rearrangements were detected by the STAR-Fusion Firehose tool (version STAR-Fusion5 16 based on codebase: https://github.com/STAR-Fusion Version 0.5.1) from RNA-sequencing tumor data. Three or more supporting paired-end reads were required for event detection.
miRNA sequencing
miRNA libraries and sequencing
We generated microRNA sequence (miRNA-seq) data for using methods described in (Chu et al., 2016). We aligned reads to the GRCh37/hg19 reference human genome, and annotated read count abundance to miRBase v16 stem-loops and mature strands, using only exact-match read alignments. Note that the BAM files available from the Genomic Data Commons (https://gdc.cancer.gov/) include all sequence reads. We used miRBase v20 to assign 5p and 3p mature strand (miR) names to miRBase MIMAT accession IDs.
Unsupervised and supervised clustering
For unsupervised clustering with the n = 76 high-purity tumour samples, we used unsupervised non-negative matrix factorization (NMF) consensus clustering (v0.20.5) in R 3.1.2, with default settings (Gaujoux and Seoighe, 2010). The input was a reads-per-million (RPM) data matrix for the 303 (25%) most-variant 5p or 3p mature strands. After running a rank survey with 50 iterations per solution, we chose a 3-cluster solution and performed a 500-iteration run to generate the final clustering result. To visualize typical vs. atypical cluster members, we calculated a profile of silhouette widths from the final NMF consensus membership matrix, considering samples with relatively low widths to be atypical cluster members.
To generate a heatmap for the 3-cluster solution, we first identified miRs that were differentially abundant between the unsupervised miRNA clusters, using a SAMseq multiclass analysis (samr 2.0)(Alexandrov et al., 2013) in R, with a read-count input matrix and an FDR threshold of 0.05. For the heatmap, we included miRs that had the largest SAMseq scores and median abundances greater than 25 RPM. The RPM filtering acknowledged potential sponge effects from competitive endogeneous RNAs (ceRNAs) that can make weakly abundant miRs less influential (Mullokandov et al., 2012). We transformed each row of the matrix by log10(RPM + 1), then used the pheatmap R package (v0.7.7 or v1.0.2) to scale and cluster only the rows, using a Pearson distance metric and Ward clustering.
For supervised clustering the full set of n = 150 tumor samples, we identified the set of miRNA which 1) were among the 303 robustly expressed lncRNA discussed earlier, 2) had a mean abundance in the high purity subset larger than the mean abundance in the low purity subset, and 3) were differentially expressed between the 3 classes in the high purity cohort (one class vs all t-test, with a B-H corrected FDR of 0.1). This resulted in 31 transcripts which were used to perform consensus clustering on the full 150 sample data set with Pearson correlation as the internal distance metric, seeking and verifying the presence of three clusters.
DNA methylation
Assay platform
DNA methylation data were generated using the Illumina Infinium DNA methylation platform (Bibikova et al., 2011; Bibikova et al., 2009), HumanMethylation450 (HM450). The HM450 assay analyzes the DNA methylation status of up to 482,421 CpG and 3,091 non-CpG (CpH) sites throughout the genome. It covers 99% of RefSeq genes with multiple probes per gene, as well as 96% of CpG islands from the UCSC database and their flanking regions. The assay probe sequences and information for each interrogated CpG site on Infinium DNA methylation platform are available from Illumina (www.illumina.com).
The DNA methylation score for each assayed CpG or CpH site is represented as a beta (β) value (β = (M/(M+U)) in which M and U indicate the mean methylated and unmethylated signal intensities for each assayed CpG or CpH, respectively. β-values range from zero to one, with scores of “0” indicating no DNA methylation and scores of “1” indicating complete DNA methylation. An empirically derived detection P value accompanies each data point and compares the signal intensity with an empirical distribution of signal intensities from a set of negative control probes on the array. Any data point with a corresponding p value greater than 0.05 is deemed not to be statistically significantly different from background and is thus masked as “NA” in the Level 3 data packages as described below. Further details on the Illumina Infinium DNA methylation assay technology have been described previously (Bibikova et al., 2011; Bibikova et al., 2009).
Sample and data processing
We performed bisulfite conversion on 1μg of genomic DNA from each sample using the EZ-96 DNA Methylation Kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions. We assessed the amount of bisulfite-converted DNA and completeness of bisulfite conversion using a panel of MethyLight-based quality control (QC) reactions as previously described (Campan et al., 2009). All the TCGA samples passed our QC tests and entered the Infinium DNA methylation assay pipeline. Bisulfite-converted DNAs were whole-genome-amplified (WGA) and enzymatically fragmented prior to hybridization to BeadChip arrays. BeadArrays were scanned using the Illumina iScan technology to produce IDAT files. Raw IDAT files for each sample were processed with the R/Bioconductor package methylumi. TCGA DNA methylation data packages were then generated using the EGC.tools R package which was developed internally and is publicly available on GitHub (https://github.com/uscepigenomecenter/EGC.tools).
TCGA data packages
The data levels and the files contained in each data level package are described below and are present in the NCI Genomic Data Commons (https://gdc.cancer.gov) legacy archive section (https://gdc-portal.nci.nih.gov/legacy-archive).
Level 1 data contain raw IDAT files (two per sample) as produced by the iScan system and as mapped by the Sample and Data Relationship Format (SDRF). These IDAT files were directly processed by the R/Bioconductor package methylumi. Level 2 data contain background-corrected methylated (M) and unmethylated (U) summary intensities as extracted by the R/Bioconductor package methylumi. Detection P values were computed as the minimum of the two values (one per methylation state measurement) for the empirical cumulative density function of the negative control probes in the appropriate color channel. Background correction was performed via normal-exponential deconvolution (Triche et al., 2013). Multiple-batch archives had the intensities in each of the two channels multiplicatively scaled to match a reference sample. The reference sample is defined in each array as the sample having R/G ratio of the normalization control probes closest to 1. Level 3 data contain β-value calculations with annotations for HGNC gene symbol, chromosome, and genomic coordinates (UCSC hg19, Feb 2009) for each targeted CpG/CpH site on the array. Probes having a common SNP (dbSNP build 135, Minor Allele Frequency > 1%) within 10 bp of the interrogated CpG site or having an overlap with a repetitive element (as detected by RepeatMasker and Tandem Repeat Finder based on UCSC hg19, Feb 2009) within 15 bp (from the interrogated CpG site) were masked as “NA” across all samples, and probes with a detection P value greater than 0.05 in a given sample were masked as “NA” on that array. Probes that were mapped to multiple sites in the human genome (UCSC hg19, Feb 2009) were annotated as “NA” for chromosome and 0 for CpG/CpH coordinate.
Data from the following archives were used for the analyses described in this manuscript.
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.2.11.0
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.3.11.0
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.4.11.0
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.5.11.0
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.6.11.0
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.7.11.0
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.8.11.0
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.9.11.0
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.10.11.0
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.11.11.0
jhu-usc.edu_PAAD.HumanMethylation450.Level_3.12.11.0
Leukocyte DNA methylation data
Leukocyte DNA were extracted from peripheral blood samples from two healthy 59-year-old (PBL #1) and 63-year-old (PBL #2) female subjects (HemaCare, Van Nuys, CA). DNA methylation data were then generated using the HM450 platform (Table S5).
DNA methylation analysis
We removed probes which had any “NA”-masked data points and probes that were designed for sequences on X and Y chromosomes. We split 150 tumors into two groups: those with higher purity (n = 76) and those with lower purity (n = 74) as described above. As controls for cancer-specific DNA hypermethylation we used 7 samples that were excluded from the data freeze after the expert pathology review (F2-7273-01, F2-7276-01, HZ-7920-01, HZ-7923-01, IB-AAUV-01, IB-AAUW-01, RL-AAAS-01). Those cases showed extremely low neoplastic cellularity (< 1%) and consisted essentially of stromal tissues.
Unsupervised clustering analysis of DNA methylation data
We first performed unsupervised clustering analysis using the higher purity cases. We selected CpG sites that were not methylated in the controls (mean β-value < 0.2). To minimize the influence of variable tumor purity levels on a clustering result, we dichotomized the data using a β-value of > 0.25 to define positive DNA methylation and ≤0.25 to specify lack of methylation. The dichotomization not only ameliorated the effect of tumor sample purity on the clustering, but also removed a great portion of residual batch/platform effects that are mostly reflected in small variations near the two ends of the range of β-values. Finally, we removed CpG sites that are methylated in leukocytes, which was a major source of contamination in tumor samples (mean β-value > 0.2). We then performed consensus clustering with the dichotomized data on 31,956 CpG sites that were methylated in at least 5% of the tumor samples. The optimal number of clusters was assessed based on 80% probe and tumor resampling over 1,000 iterations of hierarchical clustering for K=2,3,4,5,6 using the binary distance metric for clustering and Ward’s method for linkage as implemented in the R/Bioconductor ConsensusClusterPlus package. The heatmap was generated based on the original β-values for a subset of the most variably methylated sites. The probes and tumors were displayed based on the order of unsupervised hierarchal clustering of the dichotomous data using the binary distance metric and Ward’s linkage method. The 5,000 CpG sites that showed the most variable DNA methylation levels across the higher purity sample set were then used for unsupervised clustering of the lower purity tumor samples, after dichotomizing the data using a β-value of > 0.2 to define positive DNA methylation.
Identification of epigenetically-silenced genes
Probes that were located in a promoter region (defined as the 3 kb region spanning from 1,500 bp upstream to 1,500 bp downstream of the transcription start sites) were identified. Level 3 RNA-seq RSEM data were log2-transformed [log2 (RSEM+1)] and used to assess the expression levels associated with DNA methylation changes. DNA methylation and gene expression data were merged by Entrez Gene IDs. We removed the CpG sites that were methylated in the control samples (mean β-value > 0.2). We then dichotomized the DNA methylation data using a β-value of > 0.3 to definite positive DNA methylation, and further eliminated CpG sites methylated in fewer than 3% of the tumor samples. For each probe/gene pair, we applied the following algorithm: 1) classify the tumors as either methylated (β > 0.3) or unmethylated (β ≤0.3); 2) compute the mean expression in the methylated and unmethylated groups; 3) compute the standard deviation of the expression in the unmethylated group. We then selected probes for which the mean expression in the methylated group was lower than 1.64 standard deviations of the mean expression in the unmethylated group. We labeled each individual tumor sample as epigenetically silenced for a specific probe/gene pair selected from above if: a) it belonged to the methylated group and b) the expression of the corresponding gene was lower than the mean of the unmethylated group of samples. If there were multiple probes associated with the same gene, a sample identified as epigenetically silenced at more than half the probes for the corresponding gene was also labeled as epigenetically silenced at the gene level. Furthermore, we identified additional genes including CDKN2A and BRCA1 having evidence for epigenetic silencing at low frequencies based on manual examination of scatter plots of DNA methylation vs. expression. CDKN2A DNA methylation status was assessed based on the probe (cg13601799) located in the p16INK4 promoter CpG island. p16INK4 expression was determined by the log2(RPKM+1) level of its first exon (chr9:21974403–21975038).
Tumor purity assessments based on DNA methylation data
We identified 1,859 CpG sites that were unmethylated in controls and leukocytes (mean β-value < 0.2) but methylated (β-value > 0.25) in more than 90% of the tumors in the high purity group. We then obtained the mode DNA methylation value for these hypermethylated loci in each tumor. The mode DNA methylation values were strongly correlated with the ABSOLUTE purity estimates derived from DNA copy number data (r2 = 0.73, p < 2.2 × 10−16).
Leukocyte fraction in each tumor was estimated using the PBL DNA methylation data as described previously (Carter et al., 2012).
Reverse Phase Protein Arrays (RPPA)
RPPA experiments and data processing
Protein was extracted using RPPA lysis buffer (1% Triton X-100, 50 mmol/L Hepes (pH 7.4), 150 mmol/L NaCl, 1.5 mmol/L MgCl2, 1 mmol/L EGTA, 100 mmol/L NaF, 10 mmol/L NaPPi, 10% glycerol, 1 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L Na3VO4, and aprotinin 10 μg/mL) from human tumors and RPPA was performed as described previously (Hennessy et al., 2010; Hu et al., 2007; Liang et al., 2007; Tibes et al., 2006). Lysis buffer was used to lyse frozen tumors by Precellys homogenization. Tumor lysates were adjusted to 1 μg/μL concentration as assessed by bicinchoninic acid assay (BCA) and boiled with 1% SDS. Tumor lysates were manually serial diluted in two-fold of 5 dilutions with lysis buffer. An Aushon Biosystems 2470 arrayer (Burlington, MA) printed 1,056 samples on nitrocellulose-coated slides (Grace Bio-Labs). Slides were probed with 192 validated primary antibodies (Table S7) followed by corresponding secondary antibodies (Goat anti-Rabbit IgG, Goat anti-Mouse IgG or Rabbit anti-Goat IgG). Signal was captured using a DakoCytomation-catalyzed system and DAB colorimetric reaction. Slides were scanned in CanoScan 9000F. Spot intensities were analyzed and quantified using Microvigene software (VigeneTech Inc., Carlisle, MA), to generate spot signal intensities (Level 1 data). The software SuperCurveGUI (Hu et al., 2007) available at http://bioinformatics.mdanderson.org/Software/supercurve/, was used to estimate the EC50 values of the proteins in each dilution series (in log2 scale). Briefly, a fitted curve (“supercurve”) was plotted with the signal intensities on the Y-axis and the relative log2 concentration of each protein on the X-axis using the non-parametric, monotone increasing B-spline model (Tibes et al., 2006). During the process, the raw spot intensity data were adjusted to correct spatial bias before model fitting. A QC metric was returned for each slide to help determine the quality of the slide: if the score is less than 0.8 on a 0–1 scale, the slide was dropped. In most cases, the staining was repeated to obtain a high quality score. If more than one slide was stained for an antibody, the slide with the highest QC score was used for analysis (Level 2 data). Protein measurements were corrected for loading as described (Gonzalez-Angulo et al., 2011; Hu et al., 2007) using median centering across antibodies (level 3 data). In total, 192 antibodies and 76 samples were used. Final selection of antibodies was also driven by the availability of high quality antibodies that consistently pass a strict validation process as previously described (Hennessy et al., 2010). These antibodies are assessed for specificity, quantification and sensitivity (dynamic range) in their application for protein extracts from cultured cells or tumor tissue. Antibodies are labeled as validated and use with caution based on degree of validation by criteria previously described (Hennessy et al., 2010).
RPPA arrays were quantitated and processed (including normalization and load controlling) as described previously, using ArrayPro software (MediaCybernetics, Rockville, MD) and the R package SuperCurve (version-1.3), available at http://bioinformatics.mdanderson.org/OOMPA (Hu et al., 2007; Tibes et al., 2006). Raw data (level 1), SuperCurve nonparameteric model fitting on a single array (level 2), and loading corrected data (level 3) were deposited at the DCC.
Data Normalization
We performed median centering across all the antibodies for each sample to correct for sample loading differences. Those differences arise because protein concentrations are not uniformly distributed per unit volume. That may be due to several factors, such as differences in protein concentrations of large and small cells, differences in the amount of proteins per cell, or heterogeneity of the cells comprising the samples. By observing the expression levels across many different proteins in a sample, we can estimate differences in the total amount of protein in that sample vs. other samples. Subtracting the median protein expression level forces the median value to become zero, allowing us to compare protein expressions across samples.
Hierarchical clustering in High purity samples
For high purity samples, we used ConsensusClusterPlus to cluster the samples, as well as estimate the number of clusters. We used (1 - Pearson correlation) as the distance metric and Ward as a linkage algorithm in the unsupervised hierarchical clustering analysis. To illustrate the role of cell signaling network in pancreatic cancer, we calculated 9 pathway scores (Table S7) based on a previously described method (Akbani et al., 2014).
Integrative Quantitative Analysis (IQA)
For Integrative quantitative analysis (IQA), we analyzed tumor samples in either the high (n = 76) and low (n = 74) purity groups separately. In each of the two groups separately, the top 50% expressed mRNAs, lncRNAs and miRNAs were considered and Spearman correlation coefficients were calculated for each of the following: (a) all miRNA-mRNA and all miRNA-lncRNA pairs, (b) each miRNA with the methylation probes that are as far as 1,000 bp from the middle of the mature miRNA genomic coordinates on either strand of the genome and (c) each mRNA and lncRNA with the methylation probes that are as far as 1,000 bp from the transcription start site of the respective transcript on either strand of the genome. MiRNA’s from both miRBase and those that were previously reported (Londin et al., 2015)(Table S6) and found expressed in the PAAD cancers were considered for analysis. Only methylation probes that had a methylation value of > 0.3 in more than 3% of the samples were considered in the analysis. Calculations were done in Python 2.7 using the SciPy and NumPy packages and false discovery rate was calculated using the Benjamini-Hochberg correction procedure. The top 1,000 negative correlations (sorted by FDR) in each group are included in Table S8. For both analyses, FDR was found to be < 0.01. For each miRNA-mRNA pair further evidence of a direct interaction was sought: the rna22 (Miranda et al., 2006) and TargetScan (Agarwal et al., 2015) target prediction algorithms were used to check whether the miRNA-mRNA interaction could be predicted along with simulation data (CLIP-sim) from Argonaute HITS-CLIP from HPNE and MIA PaCa-2 model cell lines (Clark et al., 2014). Validation data from the MiRTarBase v. 6.1 (Chou et al., 2016b; Hsu et al., 2014) were also integrated in the analysis. Direct interaction evidence for miRNA-lncRNA pairs as calculated from rna22 (Loher and Rigoutsos, 2012; Miranda et al., 2006), miRcode 11 (Jeggari et al., 2012) or NPInter v3.0 (Hao et al., 2016) was also integrated. DAVID (Huang da et al., 2009) was run for the genes that were part of the network, using as background the list of genes that were initially included in the correlations, and an FDR cutoff of 10% (Table S8). Network visualization was carried out in R using the igraph package. Differential expression analyses for miRNAs, mRNAs and lncRNAs were carried using SAM (Tusher et al., 2001) with an FDR threshold of 0.0% (Table S8). These three datasets were log2-transformed before the significance analysis by SAM. Differences in the methylation status were evaluated using the non-parametric Mann-Whitney U-test and p values were corrected to FDR. To examine the cancer relevance of the differentially expressed genes between the classical and the basal mRNA-defined subtypes, their overlap with the gene sets in MsigDB v5.1 (Subramanian et al., 2005) was examined (Table S8).
Similarity Network Fusion (SNF)
Similarity network fusion (SNF)(Wang et al., 2014) was based on miRNA, mRNA, lncRNA, and DNA methylation data from 76 individuals constituting all high purity samples. RPPA data was excluded due to multiple samples with missing data. First, similarity matrices were constructed using features derived from each platform individually for the purposes of clustering: for DNA methylation, the same 5,000 CpG sites were used; for mRNA, the same 50 genes used for clustering in Moffitt et al., for lncRNA, the 86 transcripts and for miRNA, the same 31 transcripts as described above. The miRNA, mRNA and lncRNA features were log-transformed, using log(1+x), and then standardized. Euclidean distance was used on all four datasets to compute the corresponding distance matrices. Then, SNF transformed and combined the distance matrices from the different data types into a single matrix/network by performing graph diffusion across all similarities between patients. The resulting matrix captures combined similarity across all platforms. Intuitively, SNF combines all data types by keeping the strongest similarities supported by one or more types of data and the similarities supported by multiple modalities while removing similarities with weak support. We ran SNF to combine all four data types using the following parameters values: K = 10, T = 30, α = 0.5.
SNF network figures were generated using Cytoscape. From the fused similarity matrix, only the top 10% of the weights were considered for the network figure. The layout used from Cytoscape is edge-weighted spring embedded. The nodes’ sizes were scaled by the absolute purity. The edges were colored to indicate the data type most supportive of the similarity. If the weights in multiple data types are within 10% of the maximal weight we consider the edge to be supported by multiple data types.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative and statistical methods are noted above according to their respective technology and analytic approach.
DATA AND SOFTWARE AVAILABILITY
The data and analysis results can be explored through the Genomic Data Commons (https://gdc.cancer.gov), the Broad Institute GDAC FireBrowse portal (http://gdac.broadinstitute.org), the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org), and the PAAD publication page (https://tcga-data.nci.nih.gov/docs/publications/).
Supplementary Material
Table S1, related to Figure 1
Table S2, related to Figure 1
Table S3, related to Figure 1
Table S4, related to Figure 3
Table S5, related to Figure 4
Table S6, related to Figure 5
Table S7, related to Figure 6
Table S8, related to Figure 7
Table S9, related to AUTHOR CONTRIBUTIONS.
SIGNIFICANCE.
Pancreatic cancer is a devastating disease with few therapeutic options. We present a comprehensive molecular analysis of 150 pancreatic cancer specimens, including DNA alterations; DNA methylation; and mRNA, miRNA, lncRNA and protein expression profiles. We employed a rigorous approach to analyze tumors with low neoplastic cellularity, a common feature of pancreatic cancer. We uncovered evidence of KRAS mutational heterogeneity in individual pancreatic cancers, and characterized alternative driver events and pathway activation occurring in KRAS wild type tumors. We also provide a survey of clinically relevant alterations that may serve as a roadmap for genotype-directed clinical trials. The integration of diverse molecular findings supports the existence of distinct molecular subtypes of pancreatic cancer that may enhance clinical stratification of patients.
HIGHLIGHTS.
Multi-platform study of 150 pancreatic cancers accounting for neoplastic cellularity
Identify KRAS mutational heterogeneity and alternate drivers in KRAS wild-type tumors
Identify proteomic subtypes with prognostic significance and therapeutic implications
Integrated analysis of mRNA and non-coding RNA suggests consensus subtypes
Acknowledgments
We are grateful to all patients and families who contributed to this study. This work was supported by the following grants from the NIH: U54 HG003273, U54 HG003067, U54 HG003079, U24 CA143799, U24 CA143835, U24 CA143840, U24 CA143843, U24 CA143845, U24 CA143848, U24 CA143858, U24 CA143866, U24 CA143867, U24 CA143882, U24 CA143883, U24 CA144025, U24 CA211000, SPORE CA62924, P50 CA127003 and P30 CA016672.
Footnotes
See Table S9 for full listing of all analysis working group (AWG) members and other contributors to the TCGA pancreatic network.
AUTHOR CONTRIBUTIONS
The Cancer Genome Atlas Research Network contributed collectively to this study. Biospecimens were provided by the Tissue Source Sites and processed by the Biospecimen Core Resource. Data generation and analyses were performed by the Genome Sequencing Centers, Genome Characterization Centers and Genome Data Analysis Centers. All data were released through the Data Coordinating Center. The National Cancer Institute and National Human Genome Research Institute project teams coordinated project activities. The following TCGA investigators of the Pancreatic Adenocarcinoma Analysis Working Group contributed substantially to the analysis and writing of this manuscript.
Project Co-chairs: Ralph H. Hruban, Benjamin J. Raphael
Manuscript coordinator: Andrew J. Aguirre
Analysis coordinator: Richard A. Moffitt
Data coordinator: Suzi Fei
Project coordinator: Ina Felau
DNA sequence analysis: Andrew J. Aguirre, Manaswi Gupta, Chip Stewart, Mara Rosenberg, Louis Bergelson, Julian Hess, Ignaty Leshchiner, Dimitri Livitz, Grace Tiao, Carrie Cibulkis, Benjamin J. Raphael, Stacey Gabriel, Eric S. Lander, Matthew Meyerson, Gad Getz
DNA methylation analysis: Toshinori Hinoue, Hui Shen, Peter W. Laird
mRNA analysis: Richard A. Moffitt, Jen Jen Yeh, Katherine Hoadley
miRNA analysis: A. Gordon Robertson, Phillipe Loher, Eric R. Londin, Aristeidis Telonis, Isidore Rigoutsos, Richard A. Moffitt, Jen Jen Yeh
lncRNA analysis: Ewan A. Gibb, A. Gordon Robertson, Richard A. Moffitt, Jen Jen Yeh
Copy-number analysis: Andrew Cherniack, Bradley Murray, Andrew J. Aguirre, Gad Getz, Matthew Meyerson
Reverse phase protein array (RPPA): Shiyun Ling, Yiling Lu, Gordon B. Mills, Rehan Akbani
Cluster of clusters analysis: Richard A. Moffitt, Jen Jen Yeh
IQA: Phillipe Loher, Eric Londin, Isidore Rigoutsos, Aristeidis G. Telonis
SNF: Anna Goldenberg, Aziz Mezlini
Pathology review: Ralph H. Hruban, N. Volkan Adsay, David S. Klimstra
Clinical data analysis subgroup: Andrew J. Aguirre, Richard A. Moffitt, Juok Cho, Benjamin J. Raphael, Eric A. Collisson, Ralph H. Hruban, Jen Jen Yeh
Broad Institute Genome Data Analysis Center: Michael S. Noble, Hailei Zhang, David I. Heiman, Juok Cho, Nils Gehlenborg, Gordon Saksena, Douglas Voet, Pei Lin, Scott Frazer, Timothy DeFreitas, Sam Meier, Jaegil Kim, Michael S. Lawrence, Gad Getz
Manuscript writing subgroup: Andrew J. Aguirre, Richard A. Moffitt, Suzi Fei, Chip Stewart, Andrew Cherniack, Ewan Gibb, Gordon Robertson, Jen Jen Yeh, Ralph H. Hruban, Benjamin J. Raphael
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, Liu W, Yang JY, Yoshihara K, Li J, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato E, Molin MD, Mafficini A, Yu J, Malleo G, Rusev B, Fassan M, Antonello D, Sadakari Y, Castelli P, et al. Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J Pathol. 2014;233:217–227. doi: 10.1002/path.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, Gunderson KL. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- Campan M, Weisenberger DJ, Trinh B, Laird PW. MethyLight. Methods Mol Biol. 2009;507:325–337. doi: 10.1007/978-1-59745-522-0_23. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Ben-Josef E, Thomas DG, Morgan MA, Zalupski MM, Khan G, Andrew Robinson C, Griffith KA, Chen CS, Ludwig T, et al. Caveolin-1 is Associated with Tumor Progression and Confers a Multi-Modality Resistance Phenotype in Pancreatic Cancer. Sci Rep. 2015;5:10867. doi: 10.1038/srep10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Zhang Y, Van Horn RD, Yin T, Buchanan S, Yadav V, Mochalkin I, Wong SS, Yue YG, Huber L, et al. Oncogenic BRAF Deletions That Function as Homodimers and Are Sensitive to Inhibition by RAF Dimer Inhibitor LY3009120. Cancer Discov. 2016;6:300–315. doi: 10.1158/2159-8290.CD-15-0896. [DOI] [PubMed] [Google Scholar]
- Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016a;44:D239–247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CK, Chi SY, Huang CH, Chou FF, Huang CC, Liu RT, Kang HY. IRAK1, a Target of miR-146b, Reduces Cell Aggressiveness of Human Papillary Thyroid Carcinoma. J Clin Endocrinol Metab. 2016b;101:4357–4366. doi: 10.1210/jc.2016-2276. [DOI] [PubMed] [Google Scholar]
- Chu A, Robertson G, Brooks D, Mungall AJ, Birol I, Coope R, Ma Y, Jones S, Marra MA. Large-scale profiling of microRNAs for The Cancer Genome Atlas. Nucleic Acids Res. 2016;44:e3. doi: 10.1093/nar/gkv808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PM, Loher P, Quann K, Brody J, Londin ER, Rigoutsos I. Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Sci Rep. 2014;4:5947. doi: 10.1038/srep05947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello LC, Zou J, Desouki MM, Franklin RB. Evidence for changes in RREB-1, ZIP3, and Zinc in the early development of pancreatic adenocarcinoma. Journal of gastrointestinal cancer. 2012;43:570–578. doi: 10.1007/s12029-012-9378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, Fostel JL, Friedrich DC, Perrin D, Dionne D, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Jiang G, Fiala E, Dome JS, Yu MC, Long TI, Youn B, Sohn OS, Widschwendter M, Tomlinson GE, et al. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene. 2002;21:6694–6702. doi: 10.1038/sj.onc.1205890. [DOI] [PubMed] [Google Scholar]
- Fan Y, Zhan Q, Xu H, Li L, Li C, Xiao Q, Xiang S, Hui T, Xiang T, Ren G. Epigenetic identification of ZNF545 as a functional tumor suppressor in multiple myeloma via activation of p53 signaling pathway. Biochem Biophys Res Commun. 2016;474:660–666. doi: 10.1016/j.bbrc.2016.04.146. [DOI] [PubMed] [Google Scholar]
- Fernandez-Cabezudo MJ, Faour I, Jones K, Champagne DP, Jaloudi MA, Mohamed YA, Bashir G, Almarzooqi S, Albawardi A, Hashim MJ, et al. Deficiency of mitochondrial modulator MCJ promotes chemoresistance in breast cancer. JCI Insight. 2016;1 doi: 10.1172/jci.insight.86873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome biology. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flex E, Petrangeli V, Stella L, Chiaretti S, Hornakova T, Knoops L, Ariola C, Fodale V, Clappier E, Paoloni F, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205:751–758. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SA, Whalen DM, Ozen A, Wongchenko MJ, Yin J, Yen I, Schaefer G, Mayfield JD, Chmielecki J, Stephens PJ, et al. Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2. Cancer cell. 2016;29:477–493. doi: 10.1016/j.ccell.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Frampton AE, Krell J, Jamieson NB, Gall TM, Giovannetti E, Funel N, Mato Prado M, Krell D, Habib NA, Castellano L, et al. microRNAs with prognostic significance in pancreatic ductal adenocarcinoma: A meta-analysis. Eur J Cancer. 2015;51:1389–1404. doi: 10.1016/j.ejca.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Franklin RB, Zou J, Costello LC. The cytotoxic role of RREB1, ZIP3 zinc transporter, and zinc in human pancreatic adenocarcinoma. Cancer Biol Ther. 2014;15:1431–1437. doi: 10.4161/cbt.29927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B, Luo M, Lakkur S, Lucito R, Iacobuzio-Donahue CA. Frequent genomic copy number gain and overexpression of GATA-6 in pancreatic carcinoma. Cancer Biol Ther. 2008;7:1593–1601. doi: 10.4161/cbt.7.10.6565. [DOI] [PubMed] [Google Scholar]
- Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb EA, Warren RL, Wilson GW, Brown SD, Robertson GA, Morin GB, Holt RA. Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma. Genome Med. 2015;7:22. doi: 10.1186/s13073-015-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH, Kern SE. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- Gonzalez-Angulo AM, Hennessy BT, Meric-Bernstam F, Sahin A, Liu W, Ju Z, Carey MS, Myhre S, Speers C, Deng L, et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clin Proteomics. 2011;8:11. doi: 10.1186/1559-0275-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RC, Selander I, Connor AA, Selvarajah S, Borgida A, Briollais L, Petersen GM, Lerner-Ellis J, Holter S, Gallinger S. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology. 2015;148:556–564. doi: 10.1053/j.gastro.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkova T, Cuperkova R, Minarik M, Benesova L. MicroRNAs in Pancreatic Cancer: Involvement in Carcinogenesis and Potential Use for Diagnosis and Prognosis. Gastroenterol Res Pract. 2015;2015:892903. doi: 10.1155/2015/892903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Wu W, Li H, Yuan J, Luo J, Zhao Y, Chen R. NPInter v3.0: an upgraded database of noncoding RNA-associated interactions. Database (Oxford) 2016;2016 doi: 10.1093/database/baw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Ahuja N, Makary MA, Cameron JL, Eckhauser FE, Choti MA, Hruban RH, Pawlik TM, Wolfgang CL. 2564 resected periampullary adenocarcinomas at a single institution: trends over three decades. HPB: the official journal of the International Hepato Pancreato Biliary Association. 2014;16:83–90. doi: 10.1111/hpb.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, Davies MA, Liu W, Coombes K, Meric-Bernstam F, et al. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clin Proteomics. 2010;6:129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez YG, Lucas AL. MicroRNA in pancreatic ductal adenocarcinoma and its precursor lesions. World J Gastrointest Oncol. 2016;8:18–29. doi: 10.4251/wjgo.v8.i1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Minato M, Suzuki H, Fujiyoshi M, Miyagi H, Haruta M, Kaneko Y, Hatanaka KC, Hiyama E, Kamijo T, et al. Clinical prognostic value of DNA methylation in hepatoblastoma: Four novel tumor suppressor candidates. Cancer Sci. 2016;107:812–819. doi: 10.1111/cas.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23:1986–1994. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, Mo YY. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Ryu B, Hruban RH, Kern SE. Exploring the host desmoplastic response to pancreatic carcinoma: gene expression of stromal and neoplastic cells at the site of primary invasion. Am J Pathol. 2002;160:91–99. doi: 10.1016/S0002-9440(10)64353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, van der Heijden MS, Baumgartner MR, Troup WJ, Romm JM, Doheny K, Pugh E, Yeo CJ, Goggins MG, Hruban RH, Kern SE. Large-scale allelotype of pancreaticobiliary carcinoma provides quantitative estimates of genome-wide allelic loss. Cancer Res. 2004;64:871–875. doi: 10.1158/0008-5472.can-03-2756. [DOI] [PubMed] [Google Scholar]
- Izawa T, Obara T, Tanno S, Mizukami Y, Yanagawa N, Kohgo Y. Clonality and field cancerization in intraductal papillary-mucinous tumors of the pancreas. Cancer. 2001;92:1807–1817. doi: 10.1002/1097-0142(20011001)92:7<1807::aid-cncr1697>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Liu Y, Liu R, Zhang K, Zhang Y. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 2015;11:137–148. doi: 10.1016/j.celrep.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Hao HX, Growney JD, Woolfenden S, Bottiglio C, Ng N, Lu B, Hsieh MH, Bagdasarian L, Meyer R, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110:12649–12654. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Z, Liu W, Roebuck PL, Siwak DR, Zhang N, Lu Y, Davies MA, Akbani R, Weinstein JN, Mills GB, Coombes KR. Development of a robust classifier for quality control of reverse-phase protein arrays. Bioinformatics. 2015;31:912–918. doi: 10.1093/bioinformatics/btu736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim V, Bauer N, Teich N, Simon P, Lerch MM, Mossner J. Clinical characterization of patients with hereditary pancreatitis and mutations in the cationic trypsinogen gene. Am J Med. 2001;111:622–626. doi: 10.1016/s0002-9343(01)00958-5. [DOI] [PubMed] [Google Scholar]
- Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes & development. 2010;24:2754–2759. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32:2576–2585. doi: 10.1038/onc.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, van Es JH, van den Born M, Clevers H. Porcupine inhibitor suppresses paracrine Wnt-driven growth of Rnf43;Znrf3-mutant neoplasia. Proc Natl Acad Sci U S A. 2015;112:7548–7550. doi: 10.1073/pnas.1508113112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Lee JK, Choi DW, Do IG, Sohn I, Jang KT, Jung SH, Heo JS, Choi SH, Lee KT. Postoperative prognosis prediction of pancreatic cancer with seven microRNAs. Pancreas. 2015;44:764–768. doi: 10.1097/MPA.0000000000000346. [DOI] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Gramazio CC, Hu J, Wu HT, Laidlaw DH, Raphael BJ. MAGI: visualization and collaborative annotation of genomic aberrations. Nat Methods. 2015;12:483–484. doi: 10.1038/nmeth.3412. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tibshirani R. Finding consistent patterns: a nonparametric approach for identifying differential expression in RNA-Seq data. Stat Methods Med Res. 2013;22:519–536. doi: 10.1177/0962280211428386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yu K. Improved correction for population stratification in genome-wide association studies by identifying hidden population structures. Genet Epidemiol. 2008;32:215–226. doi: 10.1002/gepi.20296. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang T, Li Y, Chen D, Yu Z, Jin L, Ni L, Yang S, Mao X, Gui Y, Lai Y. Identification of long-non coding RNA UCA1 as an oncogene in renal cell carcinoma. Mol Med Rep. 2016;13:3326–3334. doi: 10.3892/mmr.2016.4894. [DOI] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Lindsey JC, Lusher ME, Strathdee G, Brown R, Gilbertson RJ, Bailey S, Ellison DW, Clifford SC. Epigenetic inactivation of MCJ (DNAJD1) in malignant paediatric brain tumours. Int J Cancer. 2006;118:346–352. doi: 10.1002/ijc.21353. [DOI] [PubMed] [Google Scholar]
- Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Cichowski K. Loss of negative regulators amplifies RAS signaling. Nat Genet. 2015;47:426–427. doi: 10.1038/ng.3299. [DOI] [PubMed] [Google Scholar]
- Loher P, Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics. 2012;28:3322–3323. doi: 10.1093/bioinformatics/bts615. [DOI] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londin E, Loher P, Telonis AG, Quann K, Clark P, Jing Y, Hatzimichael E, Kirino Y, Honda S, Lally M, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci U S A. 2015;112:E1106–1115. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou E, Subramanian S, Steer CJ. Pancreatic cancer: modulation of KRAS, MicroRNAs, and intercellular communication in the setting of tumor heterogeneity. Pancreas. 2013;42:1218–1226. doi: 10.1097/MPA.0000000000000007. [DOI] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli P, Carrillo-de Santa Pau E, Cox T, Sainz B, Jr, Dusetti N, Greenhalf W, Rinaldi L, Costello E, Ghaneh P, Malats N, et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut. 2016 doi: 10.1136/gutjnl-2015-311256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. The Ensembl Variant Effect Predictor. Genome biology. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele M, Mattioli K, Mallard W, Shechner DM, Gerhardinger C, Rinn JL. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017;27:27–37. doi: 10.1101/gr.214205.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome biology. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Raulefs S, Bruns P, Afonso-Grunz F, Plotner A, Thermann R, Jager C, Schlitter AM, Kong B, Regel I, et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94. doi: 10.1186/s12943-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal G, Sharma M, Kumar S, Chaudhary K, Gupta S, Gautam A, Raghava GP. PCMdb: pancreatic cancer methylation database. Sci Rep. 2014;4:4197. doi: 10.1038/srep04197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Q, Carninci P. Expression Specificity of Disease-Associated lncRNAs: Toward Personalized Medicine. Curr Top Microbiol Immunol. 2016;394:237–258. doi: 10.1007/82_2015_464. [DOI] [PubMed] [Google Scholar]
- Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS, Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- Notta F, Chan-Seng-Yue M, Lemire M, Li Y, Wilson GW, Connor AA, Denroche RE, Liang SB, Brown AM, Kim JC, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–382. doi: 10.1038/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kahler A, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi G, Kudo Y, Tang B, Liu T, Jin S, Liu J, Zuo X, Mi S, Shao W, Ma X, et al. PARP6 acts as a tumor suppressor via downregulating Survivin expression in colorectal cancer. Oncotarget. 2016;7:18812–18824. doi: 10.18632/oncotarget.7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, Meyerson M, Getz G. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36:E2423–2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein BJ, Gupta S, Dada R, Safi J, Michener C, Agarwal A. Potential markers for detection and monitoring of ovarian cancer. J Oncol. 2011;2011:475983. doi: 10.1155/2011/475983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts NJ, Norris AL, Petersen GM, Bondy ML, Brand R, Gallinger S, Kurtz RC, Olson SH, Rustgi AK, Schwartz AG, et al. Whole Genome Sequencing Defines the Genetic Heterogeneity of Familial Pancreatic Cancer. Cancer Discov. 2016;6:166–175. doi: 10.1158/2159-8290.CD-15-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- Sahin IH, Iacobuzio-Donahue CA, O’Reilly EM. Molecular signature of pancreatic adenocarcinoma: an insight from genotype to phenotype and challenges for targeted therapy. Expert Opin Ther Targets. 2016a;20:341–359. doi: 10.1517/14728222.2016.1094057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin IH, Lowery MA, Stadler ZK, Salo-Mullen E, Iacobuzio-Donahue CA, Kelsen DP, O’Reilly EM. Genomic instability in pancreatic adenocarcinoma: a new step towards precision medicine and novel therapeutic approaches. Expert Rev Gastroenterol Hepatol. 2016b:1–13. doi: 10.1586/17474124.2016.1153424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Mo Q, Schultz N, Seshan VE, Olshen AB, Huse J, Ladanyi M, Sander C. Integrative subtype discovery in glioblastoma using iCluster. PLoS One. 2012;7:e35236. doi: 10.1371/journal.pone.0035236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, Young RA. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Chua CY, Tian W, Zhang Z, Chiao PJ, Zhang W. MicroRNA Signaling Pathway Network in Pancreatic Ductal Adenocarcinoma. J Genet Genomics. 2015;42:563–577. doi: 10.1016/j.jgg.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Tan MC, Basturk O, Brannon AR, Bhanot U, Scott SN, Bouvier N, LaFemina J, Jarnagin WR, Berger MF, Klimstra D, Allen PJ. GNAS and KRAS Mutations Define Separate Progression Pathways in Intraductal Papillary Mucinous Neoplasm-Associated Carcinoma. Journal of the American College of Surgeons. 2015;220:845–854. e841. doi: 10.1016/j.jamcollsurg.2014.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triche TJ, Jr, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, Jane-Valbuena J, Friedrich DC, Kryukov G, Carter SL, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Mezlini AM, Demir F, Fiume M, Tu Z, Brudno M, Haibe-Kains B, Goldenberg A. Similarity network fusion for aggregating data types on a genomic scale. Nat Methods. 2014;11:333–337. doi: 10.1038/nmeth.2810. [DOI] [PubMed] [Google Scholar]
- Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS letters. 2008;582:1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, Ulrich CD, Martin SP, Gates LK, Jr, Amann ST, Toskes PP, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Hruban RH. Pathology and molecular genetics of pancreatic neoplasms. Cancer J. 2012;18:492–501. doi: 10.1097/PPO.0b013e31827459b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, Wood LD, Eshleman JR, Goggins MG, Wolfgang CL, Canto MI, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci U S A. 2011a;108:21188–21193. doi: 10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Matthaei H, Maitra A, Dal Molin M, Wood LD, Eshleman JR, Goggins M, Canto MI, Schulick RD, Edil BH, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011b;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Xiang T, Luo X, Li C, Li Q, Peng W, Li L, Li S, Wang Z, Tang L, et al. Zinc-finger protein 545 inhibits cell proliferation as a tumor suppressor through inducing apoptosis and is disrupted by promoter methylation in breast cancer. PLoS One. 2014;9:e110990. doi: 10.1371/journal.pone.0110990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Li X, Tao Q, Yu XL, Cheng ZG, Han ZY, Guo M, Liang P. Hypermethylation of ZNF545 is associated with poor prognosis in patients with early-stage hepatocellular carcinoma after thermal ablation. Gut. 2015;64:1836–1837. doi: 10.1136/gutjnl-2015-310258. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wei Q, Mao L, Liu W, Mills GB, Coombes K. Serial dilution curve: a new method for analysis of reverse phase protein array data. Bioinformatics. 2009;25:650–654. doi: 10.1093/bioinformatics/btn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, related to Figure 1
Table S2, related to Figure 1
Table S3, related to Figure 1
Table S4, related to Figure 3
Table S5, related to Figure 4
Table S6, related to Figure 5
Table S7, related to Figure 6
Table S8, related to Figure 7
Table S9, related to AUTHOR CONTRIBUTIONS.