ABSTRACT
Multidrug-resistant Pseudomonas aeruginosa presents a global medical challenge, and polymyxins are a key last-resort therapeutic option. Unfortunately, polymyxin resistance in P. aeruginosa has been increasingly reported. The present study was designed to define metabolic differences between paired polymyxin-susceptible and -resistant P. aeruginosa strains using untargeted metabolomics and lipidomics analyses. The metabolomes of wild-type P. aeruginosa strain K ([PAK] polymyxin B MIC, 1 mg/liter) and its paired pmrB mutant strains, PAKpmrB6 and PAKpmrB12 (polymyxin B MICs of 16 mg/liter and 64 mg/liter, respectively) were characterized using liquid chromatography-mass spectrometry, and metabolic differences were identified through multivariate and univariate statistics. PAKpmrB6 and PAKpmrB12, which displayed lipid A modifications with 4-amino-4-deoxy-l-arabinose, showed significant perturbations in amino acid and carbohydrate metabolism, particularly the intermediate metabolites from 4-amino-4-deoxy-l-arabinose synthesis and the methionine salvage cycle pathways. The genomics result showed a premature termination (Y275stop) in speE (encoding spermidine synthase) in PAKpmrB6, and metabolomics data revealed a decreased intracellular level of spermidine in PAKpmrB6 compared to that in PAKpmrB12. Our results indicate that spermidine may play an important role in high-level polymyxin resistance in P. aeruginosa. Interestingly, both pmrB mutants had decreased levels of phospholipids, fatty acids, and acyl-coenzyme A compared to those in the wild-type PAK. Moreover, the more resistant PAKpmrB12 mutant exhibited much lower levels of phospholipids than the PAKpmrB6 mutant, suggesting that the decreased phospholipid level was associated with polymyxin resistance. In summary, this study provides novel mechanistic information on polymyxin resistance in P. aeruginosa and highlights its impacts on bacterial metabolism.
KEYWORDS: polymyxin resistance, Pseudomonas aeruginosa, lipid A modification, l-Ara4N biosynthesis, metabolomics, glycerophospholipids, methionine salvage cycle
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic Gram-negative pathogen that can cause both acute and chronic infections, particularly in people with cystic fibrosis (1, 2). Moreover, it is notorious for developing high-level resistance to the majority of antibiotics, often leaving clinicians with few therapeutic options (2–5). Polymyxins (i.e., polymyxin B and colistin), a family of cyclic lipopeptides, are among the very few antibiotics that retain activity against multidrug-resistant P. aeruginosa (6, 7). The putative mode of action of polymyxins involves an initial electrostatic interaction between positively charged l-α,γ-diaminobutyric acid (Dab) residues of polymyxins and negatively charged phosphates within the lipid A and inner core moieties of lipopolysaccharide (LPS) in the Gram-negative outer membrane (OM) (7, 8). This is followed by hydrophobic interactions between the fatty acyl chains and positions 6 and 7 of polymyxins and the bacterial OM, which are also essential for the bacterial killing (9).
Although the current incidence of polymyxin resistance in P. aeruginosa is relatively low, suboptimal use and poor antibiotic stewardship have led to a notable increase in polymyxin-resistant clinical isolates (10–13). In particular, the recent emergence of the mcr-1 plasmid indicates that polymyxin resistance can readily spread (14). At a genetic level, polymyxin resistance in P. aeruginosa often results from mutations in the two-component regulatory systems (TCRs) PhoPQ, PmrAB, and/or ParRS (15–18). Specific mutations in these TCRs can trigger constitutive expression of the arnBCADTEF-pmrE operon, which encodes enzymes responsible for covalent attachment of 4-amino-4-deoxy-l-arabinose (l-Ara4N) to lipid A (15–17, 19, 20). ColRS and CprRS TCRs also interact with PhoPQ, presumably regulating additional genes that contribute to polymyxin resistance in P. aeruginosa (21, 22). Moreover, disruptions in galU, lptC, wapR, or ssg genes responsible for LPS biosynthesis in P. aeruginosa have been reported to affect the permeability of the OM and/or to inhibit LPS modifications, causing increased polymyxin susceptibility (19). Collectively, the current literature on polymyxin resistance is largely focused on the studies of genetic alterations and indicates elaborate mechanisms of polymyxin resistance in P. aeruginosa; unfortunately, metabolic perturbations associated with polymyxin resistance are poorly understood. The present study employed untargeted metabolomics and lipidomics to investigate the altered metabolism in P. aeruginosa due to polymyxin resistance.
RESULTS
Genomics analysis and lipid A structural characterization.
Genomes of the wild-type P. aeruginosa strain K ([PAK] polymyxin B MIC, 1 mg/liter) and its paired mutants PAKpmrB6 (polymyxin B MIC, 16 mg/liter) and PAKpmrB12 (polymyxin B MIC, 64 mg/liter) (23) were sequenced and assembled. Compared to that in PAK, pmrB mutations L243Q (locus PRK_00328) and A248V (locus PRK_00328) were observed in PAKpmrB6 and PAKpmrB12, respectively (23). Moreover, a missense mutation (P415L) in locus PRK_00106, encoding transcriptional regulator RtcR, and a premature termination (Y275stop) in speE (locus PRK_00325 in the pmrAB operon, encoding spermidine synthase) were identified in PAKpmrB6, whereas no additional mutations were observed in PAKpmrB12 compared to the wild-type PAK.
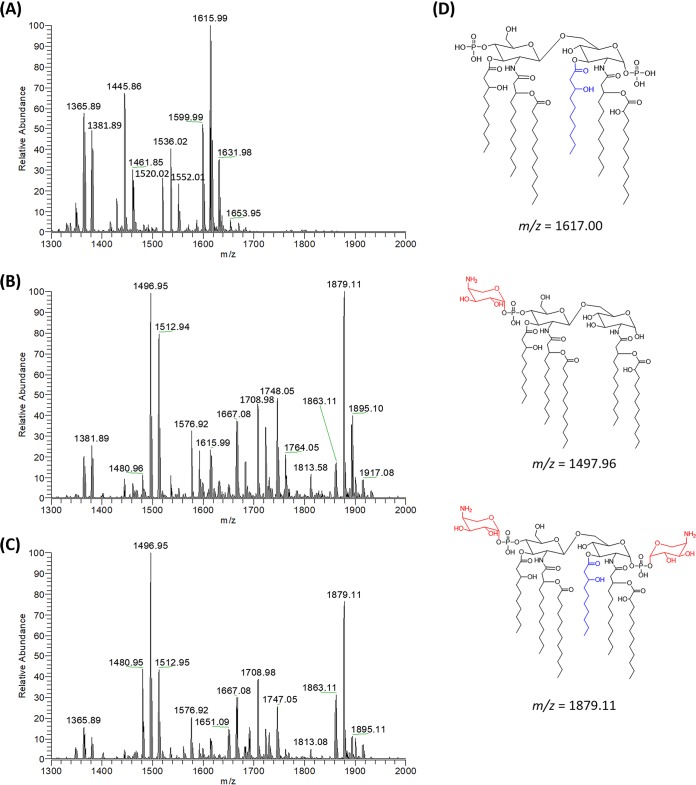
Mutations in pmrB can cause polymyxin resistance via the 4-amino-4-deoxy-l-arabinose (l-Ara4N) modification of lipid A through the activation of arn operon (17, 24). We therefore characterized lipid A profiles of each strain using electrospray ionization-mass spectrometry (ESI-MS). Penta-acylated (peaks at m/z 1445.85 and 1461.85) and hexa-acylated (peaks at m/z 1599.99, 1615.99, and 1631.98) forms of lipid A were predominant in the wild-type PAK, with a Δm/z of 170 attributable to the enzymatic removal of R-3-hydroxydecanoate at the 3 position of lipid A (Fig. 1A; see also Fig. S1 in the supplemental material). In comparison, the polymyxin-resistant PAKpmrB6 strain predominantly produced lipid A species modified with one (m/z 1576.92, 1748.05, and 1764.05) or two (m/z 1708.98, 1863.11, 1879.11, and 1895.10) l-Ara4N moieties (Δm/z = 131.05 or 262.10, respectively) (Fig. 1B; Fig. S1). The PAKpmrB12 strain displayed a very similar lipid A profile to that of PAKpmrB6 (Fig. 1C). Dephosphorylated lipid A species were detected in PAK (peaks at m/z 1365.89, 1381.89, 1520.02, 1536.02, and 1552.01) (Fig. 1A) and both mutants (peaks at m/z 1480.96, 1496.95, 1512.94, and 1667.08) (Δm/z = −79.97) (Fig. 1B and C).
FIG 1.
Mass characterization of lipid A profiles in polymyxin-susceptible and -resistant P. aeruginosa strains. Wild-type PAK (A), PAKpmrB6 (B), and PAKpmrB12 (C) mutants and the predominant structures of lipid A from the wild-type PAK and/or both pmrB mutants (D). The lipid A samples were analyzed by LC-MS/MS in negative ion mode, which obtained most lipid A peaks with the loss of one hydrogen ([M-H]−). R-3-hydroxydecanoate is shown in blue and l-Ara4N is shown in red.
Evaluation of the effects of extraction solvents on the recovery of bacterial lipidomes.
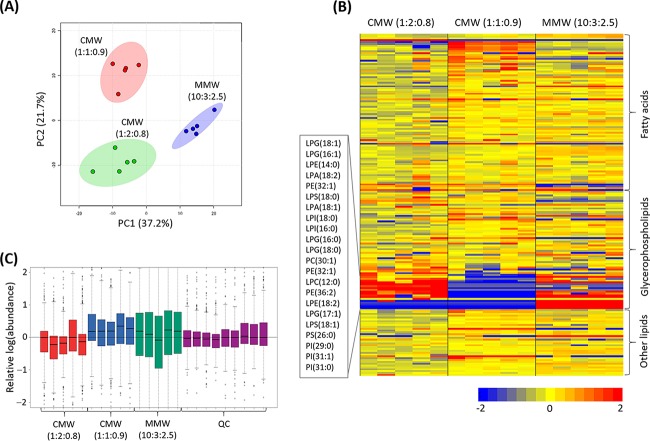
Both Bligh-Dyer (25) and methyl-tert-butyl ether (MTBE)/methanol/water (MMW) (26) extraction methods were evaluated on the basis of the recovery of lipidomes from the wild-type PAK. To avoid the loss of certain hydrophilic lipids (e.g., phospholipids), the single-phase Bligh-Dyer method was also performed in tandem. The performance of extraction solvents was assessed on the basis of the number and intensity of the peaks detected by reversed-phase liquid chromatography (RPLC) (27). Principal-component analysis (PCA) score plots were constructed to depict the impact of the solvent system used for extraction on the resultant lipidome, showing general differences among the three solvent systems (Fig. 2A) (28). As shown in the heatmap, the double-phase Bligh-Dyer method (chloroform [CHCl3]/methanol [MeOH]/water [H2O] [CMW], 1:1:0.9 [vol/vol]) yielded a greater abundance of fatty acids but without the detection of a number of lipophilic glycerophospholipids (GPLs) compared to those with single-phase CMW (CHCl3/MeOH/H2O, 1:2:0.8 [vol/vol]) and MMW (MTBE/MeOH/H2O, 10:3:2.5 [vol/vol]) methods (Fig. 2B). A minimal overall difference was apparent between the single-phase CMW and MMW methods in general; however, several specific phospholipids, putatively identified as phosphatidylserines [PS(26:0) and PS(31:0)] and phosphatidylinositols [PI(29:0) and PI(31:1)], were extracted only by the MMW method (Fig. 2B). However, the MMW extraction method displayed higher data variations in the relative abundances (log scale) from each sample than the single-phase CMW method (Fig. 2C). Therefore, the single-phase CMW method was employed in the lipidomics experiment in the present study.
FIG 2.
Method comparison in the extraction of lipidomes from P. aeruginosa strain K (PAK). (A) PCA score plot shows that metabolites are grouped due to different extraction methods: red, CHCl3/MeOH/H2O ([CMW] 1:1:0.9 [vol/vol], double-phase Bligh-Dyer); green, CMW (1:2:0.8 [vol/vol], single-phase Bligh-Dyer); and blue, MTBE/MeOH/H2O ([MMW] 10:3:2.5 [vol/vol], MTBE extraction). The colored dots on the PCA score plot represent five biological replicates. (B) The heat map illustrates the relative peak intensities of lipids with three major lipid classes extracted by three different solvents, CMW (left, 1:2:0.8 [vol/vol]), CMW (middle, 1:1:0.9 [vol/vol]), and MMW (right, 10:3:2.5 [vol/vol]). Five independent biological replicates of each extraction method are shown. Colors indicate relative abundance of lipids in each sample based on the relative peak intensity for each lipid: red, high; yellow, mean; blue, low or undetectable. (C) The bar charts show the relative abundance (log scale) of each sample between different extraction methods as well as the quality control (QC) samples. Box plots show the upper and lower quartiles (tops and bottoms of boxes), medians (lines within the boxes), and the spread of data that are not outliers (whiskers).
Perturbations of lipids and the related metabolites in the pmrB mutants.
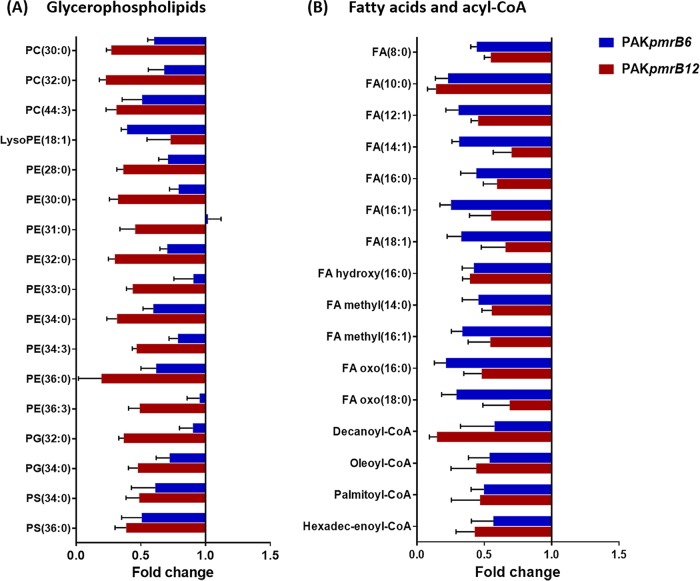
A lipidomics analysis of the wild-type PAK and the polymyxin-resistant PAKpmrB6 and PAKpmrB12 strains yielded 221 putatively identified lipids with major classes in fatty acids and glycerophospholipids (see Data Set S1). Our results revealed profound alterations (fold change of >2, P < 0.05, false discovery rate [FDR] < 0.05, one-way analysis of variance [ANOVA]) of these lipids, especially GPLs (see Data Set S2A). A high-level identification of GPLs based on molecular formula revealed significant variations in the major species, phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and phosphatidylserines (PS). In general, the relative abundances of double-chain GPLs were decreased in both pmrB mutants compared to that in the wild-type PAK (Fig. 3A). The comparison between the two mutant strains indicated a further depletion of GPLs in PAKpmrB12 relative to PAKpmrB6. In addition, the levels of fatty acids and acyl-coenzyme A (acyl-CoA) were significantly decreased in both pmrB mutants compared to that in the wild-type PAK (Fig. 3B).
FIG 3.
Lipidomic perturbations between pmrB mutants and the wild-type PAK. (A) Fold changes of GPLs in PAKpmrB6 (blue) and PAKpmrB12 (red) compared to PAK collected from the reversed-phase liquid chromatography (RPLC) method (P < 0.05, FDR < 0.05, one-way ANOVA). (B) Fold changes indicate the decreased levels of fatty acids and acyl-CoA in PAKpmrB6 (blue) and PAKpmrB12 (red) compared to PAK analyzed by the hydrophilic interaction chromatography (HILIC) method (P < 0.05, FDR < 0.05, one-way ANOVA).
Metabolomics analysis of polymyxin-susceptible and -resistant P. aeruginosa strains.
The metabolomics analysis performed for this study putatively identified 578 metabolites (see Data Set S3). The pooled quality control (QC) samples that indicate analytical reproducibility clustered tightly in the PCA score plots, reflecting minimal analytical variations (see Fig. S2A). Furthermore, the median relative standard deviation (RSD) values of the metabolites in the QC and bacterial samples were 15.7% and 25.8 to 32.3%, respectively (Fig. S2B), within the acceptable limits for metabolomics analysis (29).
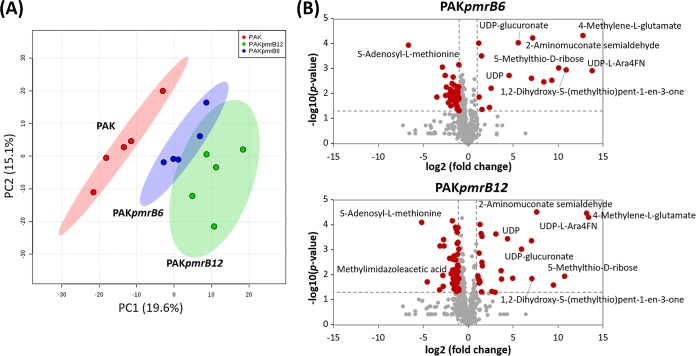
Global metabolic differences between the pmrB mutants and the wild-type PAK were analyzed by multivariate and univariate statistics (Fig. 4). The PCA score plots demonstrated that the metabolomes of the wild-type PAK and its pmrB mutants differed significantly in the first principal component, which accounts for 19.6% of the variance in the data set; however, the distance between PAKpmrB6 and PAKpmrB12 is much less dramatic (Fig. 4A). The volcano plots revealed that, compared to the wild-type PAK, 55 and 71 metabolites were dramatically perturbed in PAKpmrB6 and PAKpmrB12, respectively (>1 log2-fold, P < 0.05, FDR < 0.05, Student's t test), and more than two-thirds of the metabolites decreased significantly. It is notable that most of the significantly perturbed metabolites detected in PAKpmrB6 were also observed in PAKpmrB12. Interestingly, the results highlighted that metabolites related to amino sugars as well as methionine metabolism were significantly enriched in both PAKpmrB6 and PAKpmrB12 (Fig. 4B and Table 1). In addition, significantly depleted nucleotide levels were observed in both mutants compared to that in the wild-type strain, and this depletion was greater in PAKpmrB12 than in PAKpmrB6 (Table 1).
FIG 4.
Multivariate and univariate statistical analyses of metabolic perturbations between wild-type PAK and the pmrB mutants, PAKpmrB6 and PAKpmrB12. (A) PCA score plots of the two principle components for metabolite levels from the three strains: red, PAK; blue, PAKpmrB6; green, PAKpmrB12. (B) Volcano plots show the fold change and significance of metabolites in both pmrB mutants compared to the wild-type strain. Red plots represent metabolites having a fold change of >2 and a P value of <0.05 (Student's t test), while gray plots represent metabolites that are not significantly changed. Fold changes relative to the untreated control are based upon mean values from five biological replicates in all three strains.
TABLE 1.
Fold changes (relative intensities) in the abundance of amino acid and nucleotide metabolites in the polymyxin-resistant PAKpmrB6 and PAKpmrB12 relative to the wild-type PAK
| Pathway | Metabolitea | Formula | Mass (Da) | RTb (min) | Confidencec | Log2 fold changed |
|
|---|---|---|---|---|---|---|---|
| PAKpmrB6 | PAKpmrB12 | ||||||
| Amino acid | |||||||
| Amino sugar metabolism | N-Acetyl-d-glucosamine 6-phosphate | C8H16NO9P | 301.056 | 14.26 | 6 | −1.06 | −1.77 |
| Histidine metabolism | Hercynine | C9H15N3O2 | 197.116 | 11.98 | 6 | 1.19 | 0.79 |
| Lysine biosynthesis | l-Lysine | C6H14N2O2 | 146.105 | 18.73 | 8 | 0.53 | 1.54 |
| l-Carnitine | C7H15NO3 | 161.105 | 11.73 | 8 | 0.58 | 1.43 | |
| l-Pipecolate | C6H11NO2 | 129.079 | 18.96 | 8 | 0.59 | 1.30 | |
| Methionine metabolism | S-Methyl-5-thio-d-ribose | C6H12O4S | 180.046 | 7.41 | 6 | 10.92 | 10.71 |
| S-Methyl-5-thio-d-ribose 1-phosphate | C6H13O7PS | 260.012 | 11.57 | 8 | 6.54 | 6.11 | |
| S-Adenosyl-l-methionine | C15H22N6O5S | 398.138 | 15.84 | 8 | −6.63 | −5.81 | |
| S-Methyl-5′-thioadenosine | C11H15N5O3S | 297.090 | 7.42 | 6 | −2.56 | 0.05 | |
| 1,2-Dihydroxy-3-keto-5-methyl-thiopentene | C6H10O3S | 162.035 | 7.68 | 6 | 9.28 | 7.09 | |
| l-Methionine | C5H11NO2S | 149.051 | 11.17 | 8 | −0.56 | −1.16 | |
| S-Adenosyl-l-homocysteine | C14H20N6O5S | 384.122 | 13.13 | 6 | −0.93 | −1.34 | |
| Carbohydrate | |||||||
| Glycolysis | 2-(α-Hydroxyethyl)thiamine diphosphate | C14H22N4O8P2S | 468.064 | 13.58 | 6 | −0.53 | −1.21 |
| l-Ara4N biosynthesis | UDP-l-Ara4N | C14H23N3O15P2 | 535.060 | 16.15 | 8 | 7.18 | 6.24 |
| UDP-l-Ara4FN | C15H23N3O16P2 | 563.055 | 15.32 | 8 | 13.78 | 13.41 | |
| UDP-glucuronate | C15H22N2O18P2 | 580.035 | 18.41 | 8 | 5.56 | 5.91 | |
| Pyruvate metabolism | S-Lactoylglutathione | C13H21N3O8S | 379.105 | 15.63 | 8 | −0.57 | 3.71 |
| Nucleotide | |||||||
| Purine metabolism | GDP | C10H15N5O11P2 | 443.025 | 17.30 | 8 | −0.88 | −1.17 |
| Adenosine | C10H13N5O4 | 267.097 | 8.87 | 8 | −0.51 | −1.21 | |
| 5-Phosphoribosylamine | C5H12NO7P | 229.035 | 14.21 | 8 | −2.52 | 0.19 | |
| Adenine | C5H5N5 | 135.054 | 9.28 | 8 | −1.05 | −1.06 | |
| AMP | C10H14N5O7P | 347.063 | 13.19 | 8 | −0.46 | −1.12 | |
| Pyrimidine metabolism | CDP | C9H15N3O11P2 | 403.019 | 16.46 | 8 | −0.99 | −1.03 |
| CMP | C9H14N3O8P | 323.052 | 15.09 | 8 | −0.67 | −1.34 | |
| dTMP | C10H15N2O8P | 322.057 | 12.44 | 8 | −0.37 | −1.61 | |
| UDP | C9H14N2O12P2 | 404.002 | 15.29 | 6 | 4.56 | 4.35 | |
Putative metabolite, identified by the exact mass and retention time, with at least 2-fold differences at an FDR of <0.05 and a P value of <0.01 between pmrB mutants and the wild-type PAK. The metabolic data were collected based on five biological replicates.
RT, retention time.
Confidence, confidence level that gives an indication of the confidence of identification. Confidence level at 8 indicates that the metabolites were putatively identified in the preferred database and related peaks with the variation of standard retention time of <5%, while metabolites with confidence level at 6 were putatively identified based on preferred database and related peaks with calculated retention time within 50%.
Fold changes in bold indicate those metabolites with significant change (fold change ≥ 1 log2, P < 0.05) compared to the wild-type PAK.
Perturbations of carbohydrate metabolism associated with l-Ara4N modification of lipid A.
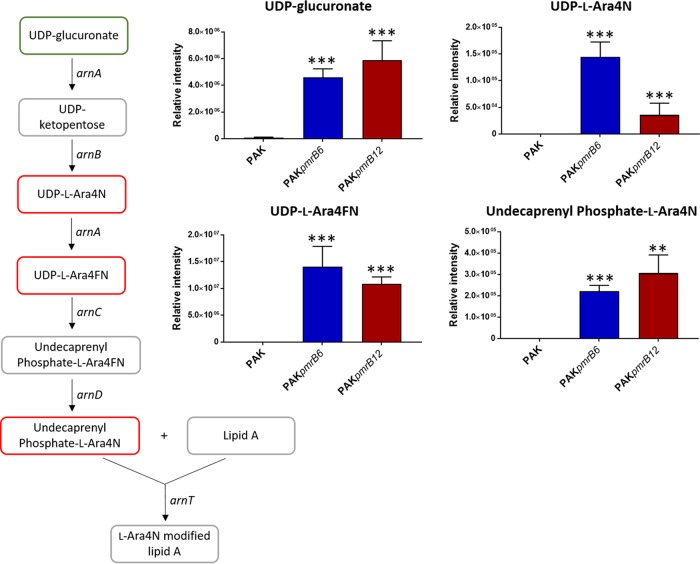
A pathway analysis of the metabolomics data revealed that the relative abundances of the intermediates in the biosynthesis of l-Ara4N were significantly increased in both pmrB mutants compared to that in the wild-type PAK (Fig. 5). In particular, the intracellular levels of the key l-Ara4N precursor UDP-glucuronate were markedly increased (>5 log2-fold) in two pmrB mutants compared to that in PAK. Notably, three key intermediates, UDP-l-Ara4N, UDP-4-deoxy-4-formamido-l-arabinose (UDP-l-Ara4FN), and the substrate for ArnT, undecaprenyl phosphate-l-Ara4N, were detected only in the two polymyxin-resistant strains. In addition, as a by-product in the generation of undecaprenyl phosphate-l-Ara4N, UDP was observed to be significantly enriched (>4 log2-fold) in both PAKpmrB6 and PAKpmrB12 compared to that in the wild-type strain (Table 1).
FIG 5.
Perturbations of the intermediates in l-Ara4N synthesis pathway. UDP-glucuronate, UDP-l-Ara4N, and UDP-l-Ara4FN were detected using the HILIC method, whereas undecaprenyl phosphate-l-Ara4N was detected using the RPLC method. The green box indicates the metabolite that was significantly increased by >2-fold, while the red boxes show metabolites that were exclusively detected in the pmrB mutants compared to that in the wild-type PAK. Metabolites in gray boxes were not detected through either the HILIC or RPLC method. **, P < 0.01; ***, P < 0.001 (Student's t test).
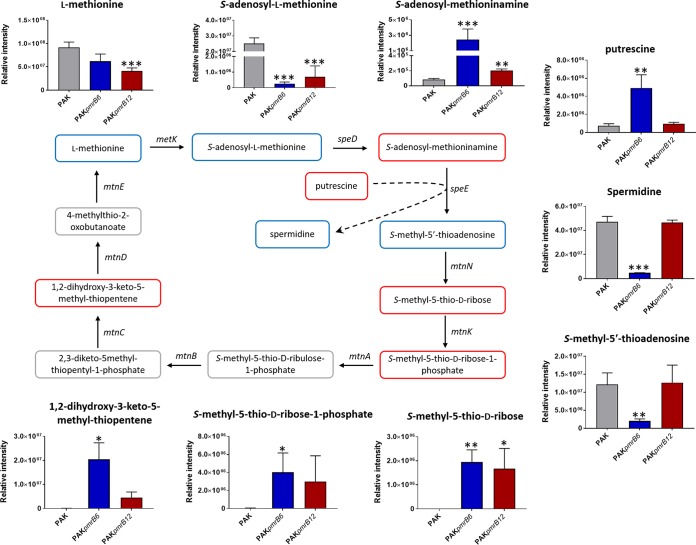
Metabolic perturbations of methionine salvage cycle and spermidine synthesis metabolism.
The intracellular levels of metabolites involved in amino sugar, histidine, lysine, methionine, and tryptophan metabolic pathways were significantly perturbed (>1 log2-fold) in the pmrB mutants compared to that in the wild-type PAK (Table 1). Remarkably, significant changes were observed in the methionine salvage cycles of both mutant strains. Specifically, the relative abundances of three metabolites, S-methyl-5-thio-d-ribose, S-methyl-5-thio-d-ribose-1-phosphate, and 1,2-dihydroxy-3-keto-5-methyl-thiopentene, were significantly higher (>7 log2-fold), while two metabolites, the universal methyl group donor S-adenosyl-l-methionine and its methyltransferase reaction product S-adenosyl-l-homocysteine, were in lower abundance (<−1 log2-fold) compared to that in the wild-type PAK (Fig. 6 and Table 1). Intriguingly, two key metabolites in the synthesis of spermidine, S-adenosylmethioninamine and putrescine, were significantly increased (4.9 log2-fold and 2.7 log2-fold, respectively). Concomitantly, the intracellular levels of spermidine and its by-product S-methyl-5-thioadenosine were dramatically decreased (2.6 log2-fold and 3.5 log2-fold, respectively) in PAKpmrB6 compared to those in both PAK and PAKpmrB12 (Fig. 6 and Table 1).
FIG 6.
Metabolic perturbations in methionine salvage cycle and spermidine synthesis pathway. S-Adenosylmethioninamine, putrescine, and spermidine were analyzed using the RPLC method, while the other metabolites were analyzed by the HILIC method. Metabolites in red boxes indicate increased abundance in PAKpmrB6 and/or PAKpmrB12, while blue boxes indicate decreased abundance compared to that in the wild-type PAK. Metabolites in gray boxes were not detected through either the HILIC or RPLC method. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test). Dashed arrow shows the conversion of putrescine to spermidine catalyzed by SpeE in the presence of the cofactor S-adenosylmethioninamine.
DISCUSSION
In view of the challenges associated with the ever-worsening spread of resistance and the development of antibiotics with novel modes of action, an understanding of the mechanisms of resistance to last-resort antibiotics such as polymyxins is paramount (30). Global metabolomics and lipidomics are of considerable utility for elucidating the mechanism of antimicrobial killing and resistance. To date, metabolomics has identified key metabolic differences between antibiotic-susceptible and -resistant strains of Acinetobacter baumannii, Mycobacterium tuberculosis, and Edwardsiella tarda (31–33). In the present study, we successfully employed metabolomics and lipidomics to elucidate key metabolic perturbations between paired polymyxin-susceptible and -resistant P. aeruginosa strains.
Mutations in the pmrB locus of P. aeruginosa result in constitutive phosphorylation of PmrA, which in turn activates the expression of the arnBCADTEF-pmrE operon and eventuates in the covalent modification of the 1 and 4′ phosphates of lipid A with l-Ara4N (20, 23). Concordantly, the lipid A profiles from both pmrB mutants PAKpmrB6 and PAKpmrB12 indicated the addition of one or two l-Ara4N moieties to the predominant hexa- and penta-acylated lipid A species (Fig. 1), which is consistent with the reported l-Ara4N-modified hexa-acylated lipid A in the pmrB mutants (23). Our metabolomics study provides an in-depth understanding of polymyxin resistance by systematically elucidating the changes in key intermediates in the l-Ara4N biosynthetic pathway (Fig. 5). Striking increases were observed in the levels of UDP-glucuronate, UDP-l-Ara4N, UDP-l-Ara4FN, and undecaprenyl phosphate-l-Ara4N in the synthesis of l-Ara4N in both pmrB mutants, which are attributed to the activated arn operon due to the mutations L243Q and A248V in pmrB. In addition, the conversion of UDP-l-Ara4FN to undecaprenyl phosphate-l-Ara4N involves the generation of UDP, which showed dramatically enriched abundances in both mutants compared to that in the wild-type strain.
The asymmetric Gram-negative OM bilayer consists of an outer leaflet predominantly composed of LPS and an inner leaflet predominantly comprising GPLs (34). The OM acts as an efficient permeability barrier against a number of antimicrobial agents. Apart from the aforementioned lipid A modifications that are associated with polymyxin resistance, we hypothesized that changes in GPLs may affect the interaction between polymyxins and the Gram-negative OM and therefore play important roles in polymyxin resistance. To investigate such perturbations, it is important to have an effective extraction procedure that reproducibly yields a broad range of total cellular lipids. Folch and Bligh-Dyer methods utilizing single- and double-phase chloroform/methanol/water (CMW) solvents are most commonly applied in lipid extraction (25, 35). The methyl-tert-butyl ether (MTBE)/methanol/water (MMW) extraction method enables a cleaner recovery of lipids (26). Our results showed that a large number of fatty acids were extracted using the double-phase CMW method, but several hydrophilic GPLs were not detected (Fig. 2B). The MMW method yielded more GPLs but was associated with higher statistical variations in the extraction than in both CMW methods, possibly due to the predrying and reconstitution steps. Overall, the single-phase CMW (1:2:0.8 [vol/vol]) method provided a high recovery and reproducibility for the extraction of total intracellular lipids and is ideal as a one-step method for untargeted lipidomics in P. aeruginosa.
Compared to that of the polymyxin-susceptible PAK, the polymyxin-resistant PAKpmrB6 and PAKpmrB12 strains produced lower levels of total cellular GPL species, including PC, PE, PG, and PS (Fig. 4A). A similar phenomenon was reported in Brucella melitensis, in which polymyxin resistance is associated with decreased PE content in the cell envelope (36). In contrast, LPS-deficient polymyxin-resistant isolate A. baumannii 19606R produces higher levels of GPLs than its paired polymyxin-susceptible wild-type strain, A. baumannii ATCC 19606 (33). Although the precise mechanism of polymyxin resistance related to the changes in GPL levels remains unclear, these observations indicate potential species-specific OM remodelling events in response to polymyxin treatment. For example, P. aeruginosa and several other Gram-negative pathogens (e.g., Salmonella enterica serovar Typhimurium) employ l-Ara4N modification of lipid A to develop resistance to polymyxins (20, 37), while Brucella is capable of either reducing divalent cations or lowering the acyl chain fluidity in its LPS to increase polymyxin resistance (38–40). In A. baumannii, a complete loss of LPS in polymyxin-resistant isolates may in turn account for the increased production of GPLs as a compensatory mechanism to fortify the OM (33, 41). Our results also showed decreased levels of fatty acids and acyl-CoA in both polymyxin-resistant mutants (Fig. 4B). It is known that fatty acids and acyl-CoA are crucial in the synthesis of phosphatidic acid (PA), which serves as a key intermediate in the synthesis of all membrane phospholipids (42). Notably, the depleted abundance of total intracellular lipids in both pmrB mutants suggests a key role in polymyxin resistance and warrants further investigation. Interestingly, PAKpmrB12 is highly resistant to polymyxins (polymyxin B MIC, 64 mg/liter) and expressed lower levels of GPLs than PAKpmrB6 (polymyxin B MIC, 16 mg/liter). Our findings support the notion that the decreased GPL level also plays a role in l-Ara4N modification-mediated polymyxin resistance.
Apart from dramatic lipidomic changes, our metabolomics results showed remarkable perturbations in the methionine salvage cycle, also called the S-methyl-5′-thioadenosine (MTA) cycle (Fig. 6) (43). This pathway is regulated in response to sulfur availability and also takes part in the regulation of polyamine synthesis (43–45). MTA and spermidine are synthesized from S-adenosylmethionine (SAM) and regulated by speE (43, 46). Our genome sequencing data revealed a nonsense mutation in speE (locus PRK_00325) in PAKpmrB6; this mutation very likely caused the dramatically depleted MTA level and enriched putrescine level and consequently led to a decreased intracellular spermidine concentration in this strain compared to those in both PAK and PAKpmrB12 (Fig. 6). Notably, the upregulation of speE homolog PA4774 mediated by PmrAB in P. aeruginosa PAO1 was suggested to contribute to reduced outer membrane permeability and increased antibacterial peptide resistance (46, 47). Therefore, the truncation of speE in PAKpmrB6 but not in PAKpmrB12 explains the difference in the levels of polymyxin resistance between the two polymyxin-resistant strains. In addition, our results showed significantly increased levels of S-adenosylmethioninamine, S-methyl-5-thio-d-ribose, S-methyl-5-thio-d-ribose-1-phosphate, and 1,2-dihydroxy-3-keto-5-methyl-thiopentene in both pmrB mutants, indicating the activation of the methionine salvage cycle. Altogether, the dramatic perturbation in the methionine salvage cycle is possibly associated with polymyxin resistance by altering the level of spermidine and therefore protecting bacteria from polymyxin killing.
In conclusion, our study is the first to use comparative metabolomics and lipidomics to elucidate significant global metabolic alterations in P. aeruginosa associated with polymyxin resistance. In addition to lipid A modifications with l-Ara4N, the pmrB mutants displayed significant metabolic perturbations in the levels of total intracellular lipids, especially GPLs, the methionine salvage cycle, and spermidine synthesis. This study provides novel insights into the multifaceted mechanisms of polymyxin resistance which may benefit the development of new-generation polymyxins.
MATERIALS AND METHODS
Chemicals and reagents.
Polymyxin B (sulfate), lipid A (L5399-5 mg), and super-DHB (50862-1g) were purchased from Sigma-Aldrich (Sydney, NSW, Australia). All other reagents and solvents were of the highest purity commercially available. The stock solution of polymyxin B was prepared in Milli-Q water (Millipore, North Ryde, NSW, Australia) and filtered through 0.22-μm syringe filters (Sartorius, Melbourne, VIC, Australia).
Bacterial strains and culture.
P. aeruginosa strain K (PAK), PAKpmrB6, and PAKpmrB12 were obtained from the Moskowitz laboratory (Massachusetts General Hospital, MA, USA) and were generated spontaneously from a late-exponential-phase culture containing polymyxin B at 20 to 50 mg/liter (23). All bacterial strains were stored at −80°C in tryptone soya broth. Prior to experiments, PAK was subcultured on Mueller-Hinton agar plates, and the mutants were subcultured on plates containing 4 mg/liter polymyxin B (Medium Preparation Unit, University of Melbourne, VIC, Australia). Overnight cultures were subsequently grown in 5 ml of cation-adjusted Mueller-Hinton broth ([CaMHB] Oxoid), from which a 1 in 100 dilution was performed in fresh medium to prepare 100 ml of mid-log-phase cultures according to an optical density at 600 nm (OD600) of 0.5. All liquid cultures were incubated at 37°C in a shaking incubator (180 rpm).
Determination of MICs.
Polymyxin B MICs of all strains were determined with the broth microdilution method (36). Briefly, experiments were performed in 96-well polypropylene microtiter plates (42) with concentrations of polymyxin B (0 to 128 mg/liter) in CaMHB. MICs were defined as the lowest concentration that inhibited visible bacterial growth after a 20-h incubation at 37°C.
DNA sequencing and genomic analysis.
A DNeasy blood and tissue kit (Qiagen) was employed to extract genomic DNA from 2 ml log-phase bacterial culture. Electrophoresis and a NanoDrop 1000 (Thermo Fisher Scientific) were used to assess the quality and quantity of genomic DNA samples. Paired-end 250-bp DNA sequencing of PAKpmrB6 and PAKpmrB12 were performed on an Illumina HiSeq 2500 platform (MicrobesNG, Birmingham, UK) with 30× coverage; paired-end 150-bp DNA sequencing of PAK was performed on a HiSeq X platform (Novogene, Hong Kong) with 35× coverage. Quality-trimmed reads of PAK were then used for de novo genome assembly with SPAdes (48), and Prokka was employed for genome annotation. The reads of PAKpmrB6 and PAKpmrB12 were aligned to the assembled PAK genome (GenBank assembly accession GCA_000408865.1) using SubRead (49). The genetic variations were determined using nesoni and annotated using snpEff (50, 51).
Isolation and structural characterization of lipid A.
Lipid A was isolated by mild acid hydrolysis as previously described (52). Briefly, cells were harvested from 200 ml of culture (OD600 of 0.5) via centrifugation at 3,220 × g for 20 min and washed twice with 5 ml phosphate-buffered saline (PBS). The cells were resuspended in 4 ml PBS, and then 5 ml chloroform and 10 ml methanol were added to the suspension, making a single-phase Bligh-Dyer mixture (chloroform/methanol/water, 1:2:0.8 [vol/vol]). The mixture was centrifuged at 3,220 × g for 15 min to remove the supernatant. LPS pellets were resuspended in 10.8 ml of hydrolysis buffer (50 mM sodium acetate [pH 4.5] with 1% sodium dodecyl sulfate [SDS]) and incubated in a boiling water bath for 45 min. To extract the lipids after hydrolysis, the SDS solution was converted into a two-phase Bligh-Dyer mixture by adding 12 ml chloroform and 12 ml methanol (25). The lower phase containing lipid A was then collected, and the samples were dried and stored at −20°C. A structural analysis of lipid A was performed using mass spectrometry in the negative mode on a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer and a Shimadzu MALDI-7090 matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) mass spectrometer. The matrix used for lipid A analysis was super-DHB (25 mg/ml in CHCl3/MeOH, 1:1 [vol/vol]).
Sample preparation for lipidomics and metabolomics experiments.
Intracellular metabolites were extracted as described previously (53). The general procedure for sample preparation was as follows. Ten milliliters of mid-log-phase culture (OD600 of 0.5) was centrifuged at 3,220 × g for 10 min at 4°C, and the supernatant was kept for footprint analysis. After washing twice with 2 ml of 0.9% NaCl, the cell pellets were resuspended in 0.5 ml of extraction solvents. Liquid nitrogen was used to permeabilize the cells and release the intracellular metabolites. The mixtures were centrifuged at 3,220 × g for 10 min at 4°C, and 300 μl of the supernatant containing the extracted metabolites was collected in 1.5-ml centrifuge tubes and stored at −80°C. The samples were further centrifuged at 14,000 × g for 10 min at 4°C, and 200 μl of particle-free supernatant was transferred into an injection vial for liquid chromatography-mass spectrometry (LC-MS) analysis.
To evaluate the effect of different extraction solvents on the recovery of lipids from whole bacterial cells, three classic extraction methods were examined. (i) For the single-phase Bligh-Dyer method (CHCl3/MeOH/H2O, 1:2:0.8 [vol/vol]), the cell pellets were resuspended in 0.5 ml solvent and 300 μl of supernatant was collected using the aforementioned method. (ii) For the double-phase Bligh-Dyer method (CHCl3/MeOH/H2O, 1:1:0.9 [vol/vol]) (25), the cell pellets were resuspended in 380 μl of the solvent from method “i” and 100 μl CHCl3 and 100 μl H2O were added after vortexing and freeze-thawing to form a double-phase Bligh-dyer mixture. The lower phase (100 μl) was finally collected after centrifugation. For LC-MS analysis, 200 μl MeOH was added to the CHCl3 extracts, which were centrifuged at 14,000 × g for 10 min at 4°C, and 200 μl of the particle-free supernatant was transferred into an LC-MS glass vial. (iii) For the method using methyl-tert-butyl ether (MTBE)/MeOH/H2O (10:3:2.5 [vol/vol]) (26), the cell pellets were resuspended in 400 μl MTBE and 120 μl MeOH, followed by vortexing and freeze-thawing to release metabolites from the bacterial cells. Milli-Q water (100 μl) was added to the suspension to separate the phases; 300 μl of the upper phase was collected after centrifugation and dried under a nitrogen stream. For further analysis, the samples were reconstituted in 300 μl CHCl3/MeOH (1:2 [vol/vol]) and centrifuged at 14,000 × g for 10 min at 4°C to obtain the particle-free supernatants.
The procedures for metabolic sample preparation were similar to those for lipidomic samples extracted by the single-phase Bligh-Dyer method. The only difference was the three-solvent system, CHCl3/MeOH/H2O (1:3:1 [vol/vol]), for extracting intracellular metabolites. For the preparation of footprint samples, an aliquot of approximately 1.5 ml of supernatant from the culture after centrifugation was rapidly filtered through a 0.22-μm filter, and the extraction solvent (250 μl) was then added. The supernatant (200 μl) was collected for LC-MS analysis after centrifugation (14,000 × g for 10 min at 4°C).
LC-MS analysis for lipids and polyamine analyses.
The lipidomics analysis was conducted on a Dionex U3000 high-performance liquid chromatography (HPLC) system in tandem with a Q Exactive Orbitrap mass spectrometer (Thermo Fisher) in both positive and negative modes with a resolution at 35,000. The mass scanning range was from 167 to 2,000 m/z. The electrospray voltage was set at 3.50 kV, and nitrogen was used as collision gas. The Ascentis Express C8 column (5 cm by 2.1 mm, 2.7 μm, 53831-U; Sigma-Aldrich) was maintained at 40°C, and the samples were controlled at 4°C. The flow rate was 0.2 ml/min for the first 24 min and then increased to 0.5 ml/min from 25 to 30 min. For lipidomics studies, the linear gradient started from 100% mobile phase A (40% isopropanol and 60% Milli-Q water with 8 mM ammonium formate and 2 mM formic acid) to a final composition of 35% mobile phase A and 65% mobile phase B (98% isopropanol and 2% Milli-Q water with 8 mM ammonium formate and 2 mM formic acid). For polyamine (mass scanning range 67 to 900 m/z) analysis, 100% mobile phase A (0.1% formic acid in Milli-Q water) was performed in the first 3 min followed by a linear gradient to 90% mobile phase B (0.1% formic acid in acetonitrile) within 5 min at a flow rate of 0.2 ml/min.
LC-MS analysis for metabolomics.
The hydrophilic interaction liquid chromatography (HILIC)-high resolution mass spectrometry (HRMS) employed in this experiment was described previously (33, 53). Samples maintained at 4°C were eluted by mobile phase A (20 mM ammonium carbonate) and B (acetonitrile) through a ZIC-pHILIC column (5 μm, polymeric, 150 mm by 4.6 mm; SeQuant, Merck). The gradient started with 80% mobile phase B at a flow rate of 0.3 ml/min, followed by a linear gradient to 50% mobile phase B over 15 min. All samples (five biological replicates each) were analyzed in a single LC-MS batch. The assessment of pooled quality control (QC) samples, internal standards, and total ion chromatograms was used for monitoring the chromatographic peaks, signal reproducibility, and the stability of analytes. Mixtures of pure metabolic standards containing over 250 metabolites were analyzed within the batch for assisting the identification of metabolites.
Data processing.
Untargeted lipidomic and metabolic analyses were performed through mzMatch (54) and IDEOM (55) (http://mzmatch.sourceforge.net/ideom.php). Raw LC-MS data files were converted to the mzXML format through a proteowizard tool, Msconvert. Automated chromatography peaks were picked by XCMS (56) and then converted to peakML files, which were combined and filtered by mzMatch on the basis of the intensity (1,000), reproducibility (RSD for all replicates, <0.8), and peak shape (codadw > 0.8). The mzMatch program was used for retrieving intensities for missing peaks and the annotation of related peaks. Unmatched peaks and noises were rejected through IDEOM. Putative metabolites were identified on the basis of the neutral exact mass within 3 ppm and confirmed by the retention time of authentic standards (<5%) and the calculated retention time (<50%). The databases used in IDEOM included KEGG, MetaCyc, and Lipidmaps, with preference for bacterial metabolites noted on PseudoCyc (57). Each metabolite was quantified by the intensity of its peak and compared between the wild-type and mutant strains. Univariate statistical analysis was performed using both one-way analysis of variance (ANOVA) and Student's t test (P < 0.05), while multivariate analysis was conducted using the metabolomics R package and MetaboAnalyst 4.0 (http://www.metaboanalyst.ca/faces/ModuleView.xhtml). Metabolic pathways were analyzed based upon the KEGG pathway (58), BioCyc (59), and Visualization and Analysis of Networks containing Experimental Data (VANTED) software (60).
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthew Johnson for technical assistance with DNA-Seq.
This research was supported by a research grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI111965 to J.L., T.V. and D.J.C.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. J.L. is an Australian National Health and Medical Research Council (NHMRC) Senior Research Fellow. T.V. and D.J.C. are Australian NHMRC Career Development Fellows.
The authors declare no conflict of interest.
J.L. conceived the project, and all authors were involved in the design of the experiments. M.-L.H. performed the experiments, and M.-L.H., Y.Z., D.J.C., and T.V. analyzed the results. All authors reviewed the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02656-17.
REFERENCES
- 1.Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. 2006. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother 50:43–48. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mesaros N, Nordmann P, Plésiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, Van Laethem Y, Jacobs F, Lebecque P, Malfroot A. 2007. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect 13:560–578. doi: 10.1111/j.1469-0691.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE, Speert DP. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Updat 3:247–255. doi: 10.1054/drup.2000.0152. [DOI] [PubMed] [Google Scholar]
- 4.Lambert P. 2002. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med 95 Suppl 4:22–26. [PMC free article] [PubMed] [Google Scholar]
- 5.Livermore DM. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis 34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 6.Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother 60:1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 7.Velkov T, Roberts KD, Nation RL, Thompson PE, Li J. 2013. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol 8:711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velkov T, Thompson PE, Nation RL, Li J. 2010. Structure-activity relationships of polymyxin antibiotics. J Med Chem 53:1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deris ZZ, Swarbrick JD, Roberts KD, Azad MA, Akter J, Horne AS, Nation RL, Rogers KL, Thompson PE, Velkov T. 2014. Probing the penetration of antimicrobial polymyxin lipopeptides into Gram-negative bacteria. Bioconjug Chem 25:750–760. doi: 10.1021/bc500094d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falagas ME, Kasiakou SK, Saravolatz LD. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 12.Gales AC, Jones R, Sader HS. 2006. Global assessment of the antimicrobial activity of polymyxin B against 54,731 clinical isolates of Gram-negative bacilli: report from the SENTRY antimicrobial surveillance programme (2001–2004). Clin Microbiol Infect 12:315–321. doi: 10.1111/j.1469-0691.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 13.Barrow K, Kwon DH. 2009. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:5150–5154. doi: 10.1128/AAC.00893-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y-Y, Wang Y, Walsh TR, Yi L- X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 15.Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Høiby N, Moskowitz SM. 2011. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP.. 2012. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 56:1019–1030. doi: 10.1128/AAC.05829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jochumsen N, Marvig RL, Damkiær S, Jensen RL, Paulander W, Molin S, Jelsbak L, Folkesson A. 2016. The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat Commun 7:13002. doi: 10.1038/ncomms13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernández L, Alvarez-Ortega C, Wiegand I, Olivares J, Kocincova D, Lam JS, Martinez JL, Hancock RE. 2013. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:110–119. doi: 10.1128/AAC.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutu AD, Sgambati N, Strasbourger P, Brannon MK, Jacobs MA, Haugen E, Kaul RK, Johansen HK, Høiby N, Moskowitz SM. 2013. Polymyxin resistance of Pseudomonas aeruginosa phoQ mutants is dependent on additional two-component regulatory systems. Antimicrob Agents Chemother 57:2204–2215. doi: 10.1128/AAC.02353-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutu AD, Rodgers NS, Park J, Moskowitz SM. 2015. Pseudomonas aeruginosa high-level resistance to polymyxins and other antimicrobial peptides requires cprA, a gene that is disrupted in the PAO1 strain. Antimicrob Agents Chemother 59:5377–5387. doi: 10.1128/AAC.00904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol 186:575–579. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 26.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. 2008. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoessel D, Nowell CJ, Jones AJ, Ferrins L, Ellis KM, Riley J, Rahmani R, Read KD, McConville MJ, Avery VM, Baell JB, Creek DJ. 2016. Metabolomics and lipidomics reveal perturbation of sphingolipid metabolism by a novel anti-trypanosomal 3-(oxazolo[4,5-b]pyridine-2-yl)anilide. Metabolomics 12:126. doi: 10.1007/s11306-016-1062-1. [DOI] [Google Scholar]
- 28.Nyamundanda G, Brennan L, Gormley IC. 2010. Probabilistic principal component analysis for metabolomic data. BMC Bioinformatics 11:571. doi: 10.1186/1471-2105-11-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirwan JA, Weber RJ, Broadhurst DI, Viant MR. 2014. Direct infusion mass spectrometry metabolomics dataset: a benchmark for data processing and quality control. Sci Data 1:140012. doi: 10.1038/sdata.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breidenstein EB, de la Fuente-Núñez C, Hancock RE. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Nandakumar M, Nathan C, Rhee KY. 2014. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun 5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 32.Peng B, Su YB, Li H, Han Y, Guo C, Tian YM, Peng XX. 2015. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab 21:249–261. doi: 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Maifiah MH, Cheah SE, Johnson MD, Han ML, Boyce JD, Thamlikitkul V, Forrest A, Kaye KS, Hertzog P, Purcell AW, Song J, Velkov T, Creek DJ, Li J. 2016. Global metabolic analyses identify key differences in metabolite levels between polymyxin-susceptible and polymyxin-resistant Acinetobacter baumannii. Sci Rep 6:22287. doi: 10.1038/srep22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikaido H, Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol Rev 49:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose HG, Oklander M. 1965. Improved procedure for the extraction of lipids from human erythrocytes. J Lipid Res 6:428–431. [PubMed] [Google Scholar]
- 36.Kerrinnes T, Young BM, Leon C, Roux CM, Tran L, Atluri VL, Winter MG, Tsolis RM. 2015. Phospholipase A1 modulates the cell envelope phospholipid content of Brucella melitensis, contributing to polymyxin resistance and pathogenicity. Antimicrob Agents Chemother 59:6717–6724. doi: 10.1128/AAC.00792-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- 38.Martinez de Tejada G, Pizarro-Cerda J, Moreno E, Moriyon I. 1995. The outer membranes of Brucella spp. are resistant to bactericidal cationic peptides. Infect Immun 63:3054–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conde-Álvarez R, Arce-Gorvel V, Iriarte M, Manček-Keber M, Barquero-Calvo E, Palacios-Chaves L, Chacón-Díaz C, Chaves-Olarte E, Martirosyan A, von Bargen K, Grilló MJ, Jerala R, Brandenburg K, Llobet E, Bengoechea JA, Moreno E, Moriyón I, Gorvel JP. 2012. The lipopolysaccharide core of Brucella abortus acts as a shield against innate immunity recognition. PLoS Pathog 8:e1002675. doi: 10.1371/journal.ppat.1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manterola L, Moriyón I, Moreno E, Sola-Landa A, Weiss DS, Koch MH, Howe J, Brandenburg K, López-Goni I. 2005. The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J Bacteriol 187:5631–5639. doi: 10.1128/JB.187.16.5631-5639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, Michael FS, Cox AD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Needham BD, Trent MS. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11:467. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M. 2013. Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem J 451:145–154. doi: 10.1042/BJ20121744. [DOI] [PubMed] [Google Scholar]
- 44.Sekowska A, Danchin A. 2002. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol 2:8. doi: 10.1186/1471-2180-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferla MP, Patrick WM. 2014. Bacterial methionine biosynthesis. Microbiology 160:1571–1584. doi: 10.1099/mic.0.077826-0. [DOI] [PubMed] [Google Scholar]
- 46.Johnson L, Mulcahy H, Kanevets U, Shi Y, Lewenza S. 2012. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J Bacteriol 194:813–826. doi: 10.1128/JB.05230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon DH, Lu CD. 2006. Polyamines induce resistance to cationic peptide, aminoglycoside, and quinolone antibiotics in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother 50:1615–1622. doi: 10.1128/AAC.50.5.1615-1622.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y, Smyth GK, Shi W. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrison P, Seemann T. 2009. From high-throughput sequencing read alignments to confident, biologically relevant conclusions with Nesoni. Victorian Bioinformatics Consortium, Monash University, Clayton, Victoria, Australia: http://www.vicbioinformatics.com/nesoni_ba2009_poster.pdf. [Google Scholar]
- 52.Nowicki EM, O'Brien JP, Brodbelt JS, Trent MS. 2014. Characterization of Pseudomonas aeruginosa LpxT reveals dual positional lipid A kinase activity and co-ordinated control of outer membrane modification. Mol Microbiol 94:728–741. doi: 10.1111/mmi.12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maifiah MH, Creek DJ, Nation RL, Forrest A, Tsuji BT, Velkov T, Li J. 2017. Untargeted metabolomics analysis reveals key pathways responsible for the synergistic killing of colistin and doripenem combination against Acinetobacter baumannii. Sci Rep 7:45527. doi: 10.1038/srep45527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheltema RA, Jankevics A, Jansen RC, Swertz MA, Breitling R. 2011. PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal Chem 83:2786–2793. doi: 10.1021/ac2000994. [DOI] [PubMed] [Google Scholar]
- 55.Creek DJ, Jankevics A, Burgess KE, Breitling R, Barrett MP. 2012. IDEOM: an Excel interface for analysis of LC-MS-based metabolomics data. Bioinformatics 28:1048–1049. doi: 10.1093/bioinformatics/bts069. [DOI] [PubMed] [Google Scholar]
- 56.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 57.Creek DJ, Jankevics A, Breitling R, Watson DG, Barrett MP, Burgess KE. 2011. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography-mass spectrometry: improved metabolite identification by retention time prediction. Anal Chem 83:8703–8710. doi: 10.1021/ac2021823. [DOI] [PubMed] [Google Scholar]
- 58.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, Walk TC, Zhang P, Karp PD. 2008. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res 36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Junker BH, Klukas C, Schreiber F. 2006. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7:109. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.