Abstract
The heterotrimeric G protein Gq regulates neuronal activity through distinct downstream effector pathways. In addition to the canonical Gq effector phospholipase Cβ, the small GTPase Rho was recently identified as a conserved effector of Gq. To identify additional molecules important for Gq signaling in neurons, we performed a forward genetic screen in the nematode Caenorhabditis elegans for suppressors of the hyperactivity and exaggerated waveform of an activated Gq mutant. We isolated two mutations affecting the MAP kinase scaffold protein KSR-1 and found that KSR-1 modulates locomotion downstream of, or in parallel to, the Gq-Rho pathway. Through epistasis experiments, we found that the core ERK MAPK cascade is required for Gq-Rho regulation of locomotion, but that the canonical ERK activator LET-60/Ras may not be required. Through neuron-specific rescue experiments, we found that the ERK pathway functions in head acetylcholine neurons to control Gq-dependent locomotion. Additionally, expression of activated LIN-45/Raf in head acetylcholine neurons is sufficient to cause an exaggerated waveform phenotype and hypersensitivity to the acetylcholinesterase inhibitor aldicarb, similar to an activated Gq mutant. Taken together, our results suggest that the ERK MAPK pathway modulates the output of Gq-Rho signaling to control locomotion behavior in C. elegans.
Keywords: Gq signaling, ERK MAPK pathway, Rho small GTPase, C. elegans, G protein
THE heterotrimeric G protein Gq is a conserved regulator of neurotransmission in metazoans. Gq is highly expressed in neurons in mammals and in the nematode Caenorhabditis elegans (Wilkie et al. 1991; Lackner et al. 1999). In its canonical signaling pathway, Gq activates phospholipase Cβ (PLCβ) to cleave the membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP2) into inositol trisphosphate (IP3) and diacylglycerol (DAG) (Rhee 2001). An increased DAG concentration at the synapse helps trigger synaptic vesicle release (Lackner et al. 1999; Miller et al. 1999).
In addition to activating PLCβ, Gq directly binds and activates the Rho guanine nucleotide exchange factor (GEF) Trio, which in turn activates the small GTPase Rho (Lutz et al. 2005, 2007; Williams et al. 2007). In mature C. elegans neurons, the Rho ortholog RHO-1 regulates synaptic activity through multiple G protein-dependent mechanisms. First, RHO-1 acts downstream of the G12-class G protein GPA-12 by binding to and inhibiting the diacylglycerol kinase DGK-1. Inhibition of DGK-1 allows DAG to accumulate at the synapse, thereby increasing synaptic vesicle release (Hiley et al. 2006; McMullan et al. 2006). Second, Gq-Rho signaling promotes neurotransmitter release by recruiting the sphingosine kinase SPHK-1 to presynaptic terminals (Chan et al. 2012). Third, Gq-Rho signaling positively regulates the NCA-1/NALCN cation channel to regulate locomotion (Topalidou et al. 2017a). Here, we identify the extracellular signal-related kinase mitogen-activated protein kinase (ERK MAPK) pathway as a positive regulator of neuronal activity acting downstream of, or in parallel to, Gq and Rho in acetylcholine neurons.
ERK MAPK signaling acts extensively in animal development, cellular proliferation, and cancer signaling (Yoon and Seger 2006; Karnoub and Weinberg 2008; Sun et al. 2015). ERKs are highly expressed in mammalian neurons (Boulton et al. 1991; Ortiz et al. 1995) and act in both the nucleus and at the synapse to regulate synaptic activity and plasticity (Sweatt 2004; Thomas and Huganir 2004; Mao and Wang 2016b). In C. elegans, the ERK pathway is required for multiple developmental events including specification of the vulva (Sternberg 2005; Sundaram 2013), and also acts in several types of neurons to control behavior. ERK signaling is activated in response to odorants in the AWC sensory neuron to regulate chemotaxis to volatile odorants and in AIY interneurons to mediate odor adaptation (Hirotsu et al. 2000; Hirotsu and Iino 2005; Chen et al. 2011; Uozumi et al. 2012). ERK is also activated in the ASER sensory neuron to regulate chemotaxis to salt (Tomioka et al. 2006; Tomida et al. 2012). ERK signaling regulates foraging behavior by acting in the IL1, OLQ, and RMD neurons (Hamakawa et al. 2015). Finally, the ERK pathway has been shown to act in interneurons to regulate the nose touch response—a mechanosensory behavior (Hyde et al. 2011).
In the canonical ERK MAPK pathway, extracellular ligand binding activates transmembrane receptor tyrosine kinases (RTKs), and adaptor proteins recruit a GEF to activate the small GTPase Ras (LET-60 in C. elegans). Upon Ras activation, LIN-45/Raf translocates to the plasma membrane, where it interacts with Ras and the scaffold protein KSR-1. KSR-1 facilitates the activation of LIN-45/Raf and the subsequent phosphorylation of the MAPK cascade consisting of LIN-45/Raf, MEK-2/MEK, and MPK-1/ERK (Sundaram 2013). In this study, we found that the ERK MAPK pathway consisting of KSR-1, LIN-45/Raf, MEK-2/MEK, and MPK-1/ERK modulates Gq-Rho signaling in acetylcholine neurons, but that, surprisingly, LET-60/Ras may not be required.
Materials and Methods
C. elegans strains
All strains were cultured using standard methods and were maintained at 20°. Table S1 contains all the strains used in this study.
Isolation and mapping of the ksr-1(ox314) and ksr-1(yak10) mutations
The ox314 and yak10 mutants were isolated from an ENU mutagenesis suppressor screen of the activated Gq mutant egl-30(tg26) (Ailion et al. 2014). We mapped the ox314 mutation by its activated Gq suppression phenotype using single nucleotide polymorphisms (SNPs) in the Hawaiian strain CB4856 as described (Davis et al. 2005).The ox314 mutation was mapped to a region of ∼709 kb in the middle of the X chromosome between SNPs on cosmids F45E1 and F53A9 (SNPs F45E1[1] and pkP6158). This region included 159 predicted protein-coding genes. A complementation test of ox314 and yak10 in the egl-30(tg26) background showed these to be alleles of the same gene. Whole genome sequencing (see below) identified these as mutations in ksr-1, and we confirmed this by performing a complementation test with the deletion allele ksr-1(ok786).
Whole genome sequencing
Strains EG4198 egl-30(tg26); ox314 and XZ1340 egl-30(tg26); yak10 were sequenced to identify candidate mutations. DNA was purified according to the Hobert laboratory protocol (http://hobertlab.org/whole-genome-sequencing/). Ion Torrent sequencing was performed at the University of Utah DNA Sequencing Core Facility. Each data set contained roughly 18,400,000 reads of a mean read length of 160 bases, resulting in ∼30× average coverage of the C. elegans genome. The sequencing data were processed on the Galaxy server at usegalaxy.org (Afgan et al. 2016). SNPs and indels were identified and annotated using the Unified Genotyper and SnpEff tools (DePristo et al. 2011; Cingolani et al. 2012). After filtering for mutations in open reading frames, we found each strain to have unique stop mutations in ksr-1, in the middle of the interval where we mapped ox314. ox314 is a G to A transition that causes a stop codon at amino acid K463, and yak10 is an A to T transversion that causes a stop codon at W254.
Locomotion assays
Track waveform and radial locomotion assays were performed on 10 cm nematode growth medium (NGM) plates seeded with 400 μl of OP50 E. coli culture and spread with sterile glass beads. Bacterial lawns were grown at 37° for 16 hr and the plates were stored at 4° until needed. For track waveform measurements, five 1st day adult animals were placed on a plate and allowed to roam for 2–5 min. We then recorded each animal’s tracks following forward locomotion. Track pictures were taken at 40× on a Nikon SMZ18 microscope with the DS-L3 camera control system. Pictures of worm tracks were processed using ImageJ. Period and 2× amplitude were measured freehand using the line tool. For each worm, we calculated the average period/amplitude ratio of five individual track bends (Figure 1C). For assays with the temperature sensitive allele sos-1(cs41), all strains were grown at 20° and shifted to the nonpermissive temperature of 25° for 24 hr before being assayed. For radial locomotion assays, 10–15 1st day adult animals were picked to the center of a plate and were then allowed to move freely for 40 min. The positions of worms were marked and the distances of the worms from the starting point were measured. For all waveform and radial locomotion assays, the experimenter was blind to the genotypes of the strains assayed.
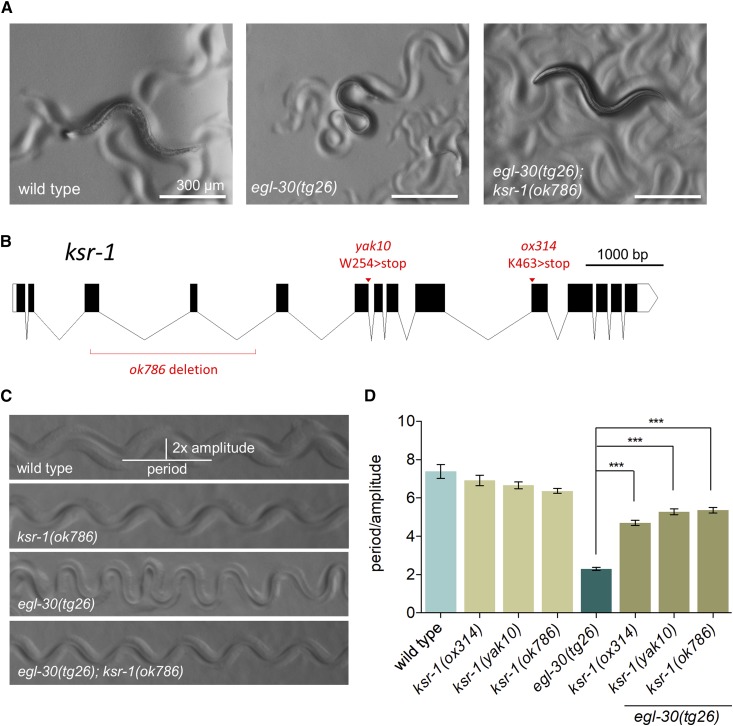
Figure 1.
ksr-1 mutations suppress activated Gq. (A) A ksr-1 mutation suppresses the coiled posture of activated Gq. The activated Gq mutant egl-30(tg26) has small size and deep body bends. ksr-1(ok786) suppresses the exaggerated body bends and small size of egl-30(tg26) worms. (B) Gene structure of ksr-1 locus. Locations of the egl-30(tg26) suppressor alleles ox314 and yak10 are indicated, as well as the position of the ok786 deletion. The gene structure was drawn using Exon-Intron Graphic Maker (www.wormweb.org/exonintron). (C) A ksr-1 mutation suppresses the exaggerated waveform of egl-30(tg26) mutants. Straightened images of tracks left in bacterial lawns show similar waveform for wild type and ksr-1(ok786) worms. egl-30(tg26) mutants have an exaggerated waveform, creating tracks with a large amplitude relative to the period. ksr-1(ok786) suppresses the exaggerated waveform of the egl-30(tg26) mutant. (D) ksr-1 mutations suppress the egl-30(tg26) exaggerated waveform. ksr-1 nonsense alleles ox314 and yak10 and the deletion allele ok786 strongly but incompletely suppress the egl-30(tg26) exaggerated waveform. N ≥ 12, *** P < 0.001, one-way ANOVA with Bonferroni’s post hoc test. All egl-30(tg26); ksr-1 double mutants were significantly different than ksr-1 single mutants (P < 0.05), indicating that suppression is incomplete.
Microscopy
Photographs of moving worms were taken at 60× on a Nikon SMZ18 microscope with the DS-L3 camera control system. The worms were age-matched as 1st day adults grown at 20°.
Constructs and transgenes
Plasmids were constructed using the three-slot multisite Gateway cloning system (Invitrogen). Plasmids and primers used are found in Tables S2 and S3. The ksr-1, lin-45, let-60, and egl-30 cDNAs were amplified by PCR from a worm cDNA library and cloned into [1–2] Gateway entry vectors. Activating Raf mutations T626E/T629D were introduced into the lin-45 cDNA vector by two sequential site-directed mutagenesis reactions (Q5 kit; NEB) with primers oBC094/095 and oBC096/097, respectively, and then confirmed by sequencing. The dominant Ras mutation S17N was introduced into the let-60 cDNA by site-directed mutagenesis with primers oBC109/110. The tg26 activating Gq mutation R243Q was introduced into the egl-30 cDNA by site-directed mutagenesis with primers oET254/255. cDNAs were cloned into expression constructs under different neuronal promoters using the multisite Gateway system. Specifics of promoter fragments used are given in Table S2. For expression in different subclasses of acetylcholine neurons, we used the Punc-17H promoter that is expressed in head acetylcholine neurons (Hammarlund et al. 2007; Topalidou et al. 2017a) and the Punc-17β promoter that is expressed in ventral cord acetylcholine motor neurons (Charlie et al. 2006; Topalidou et al. 2017a). Proper expression of ksr-1, lin-45, and let-60 was confirmed by including an operon GFP::H2B in the [2–3] slot of the expression constructs. The operon GFP template tbb-2 3′utr::gpd-2 operon::GFP::H2B:cye-1 3′utr (Frøkjær-Jensen et al. 2012) results in untagged proteins whose expression can be monitored by GFP expression.
Injections and chromosomal integrations
Worms carrying the activated lin-45 transgenes Punc-17::lin-45* and Punc-17H::lin-45* as extrachromosomal arrays were generated by injecting pBC37 or pBC44 at 20 ng/μl or 10 ng/μl, respectively, along with coinjection markers pCFJ104 (Pmyo-3::mCherry) at 5 ng/μl, pCFJ90 (Pmyo-2::mCherry) at 2.5 ng/μl, and the carrier DNA Litmus 38i to a final concentration of 100 ng/μl DNA (Mello et al. 1991). Worms carrying the dominant negative let-60 transgene Prab-3::let-60(S17N) were generated by injecting pBC50 at 20 ng/μl with the same coinjection markers. Worms carrying the activated Gq egl-30(tg26) transgene Punc-17H::egl-30(tg26) were generated by injecting pET102 at 10 ng/μl with the coinjection marker pCFJ90 (Pmyo-2::mCherry) at 1 ng/μl. MosSCI lines were generated as described (Frøkjær-Jensen et al. 2012) using an injection mix containing 10–15 ng/μl targeting vector, 50 ng/μl pCFJ601 (Peft-3::Mos1 transposase), negative selection markers pGH8 (Prab-3::mCherry) at 10 ng/μl, pCFJ104 (Pmyo-3::mCherry) at 5 ng/μl, pCFJ90 (Pmyo-2::mCherry) at 2.5 ng/μl, pMA122 (Phsp16.41::peel-1) at 10 ng/μl, and carrier DNA Litmus 38i to a final concentration of 100 ng/μl DNA.
Extrachromosomal arrays were integrated into the genome by exposure to 4000 rad of gamma irradiation. Irradiated young adult hermaphrodites were transferred to 10-cm OP50 plates (five worms/plate) and grown to starvation. The plates were chunked and grown to starvation twice more to enrich for stably expressing lines. When nearly starved, eight animals per plate were picked to individual plates. The progeny were then screened for 100% stable transmission, indicating integration into the genome. Integration was confirmed by mapping the transgene to a chromosome.
Aldicarb assays
Aldicarb assay plates (35 mm) were poured with NGM supplemented with 1 mM aldicarb. The plates were seeded with 5 μl OP50 and dried at room temperature overnight. Animals were picked onto the OP50 lawn to begin the assay (time 0) and then kept at room temperature. Every 15 min, animals were scored for paralysis by lightly touching the nose of the animal with an eyebrow hair. Animals were scored as paralyzed if the worm displayed no locomotor response to three nose touches and had no pharyngeal pumping. Animals that left the OP50 lawn were picked back onto the food.
Statistical analysis
P values were determined using GraphPad Prism 5. Normally distributed data sets were analyzed with a one-way ANOVA and Bonferroni’s post hoc test when group size was unequal, or with Tukey’s post hoc test when group size was equal. Data sets with non-normal distribution (using the Shapiro-Wilk normality test) were analyzed with a Kruskal-Wallis test and Dunn’s post hoc test. Data sets with multiple independent variables were analyzed by two-way ANOVA and Bonferroni’s post hoc test.
Data availability
Strains and plasmids are listed in Tables S1 and S2 and are available upon request. Primers are listed in Table S3. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and supplemental material. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6071009.
Results
KSR-1 regulates locomotion downstream of, or in parallel to, Gq
In C. elegans, the heterotrimeric G protein Gq regulates synaptic vesicle release (Hu et al. 2015). Gq is a key regulator of neuromuscular activity, as loss-of-function mutants in egl-30 are nearly paralyzed (Brundage et al. 1996) whereas the gain-of-function mutant egl-30(tg26) has hyperactive locomotion with an exaggerated loopy waveform (Doi and Iwasaki 2002; Bastiani et al. 2003) (Figure 1, A, C, and D). To identify pathways required for Gq signaling, we performed a forward genetic screen in C. elegans for suppressors of the activated Gq mutant egl-30(tg26). This screen has been used to identify new components of the Gq signal transduction pathway and proteins important for dense-core vesicle function (Williams et al. 2007; Ailion et al. 2014; Topalidou et al. 2016, 2017a,b). Two previously uncharacterized suppressors identified in this screen, ox314 and yak10, showed similar suppression of the loopy waveform and hyperactivity of egl-30(tg26) animals (Figure 1D). When crossed away from the egl-30(tg26) background, both mutants moved with wild-type waveform (Figure 1D), but at a slightly slower rate. We mapped the ox314 allele near the center of the X chromosome (see Materials and Methods), and a complementation test showed that ox314 and yak10 are mutations in the same gene since they failed to complement in an egl-30(tg26) background.
We used whole genome sequencing to identify ox314 and yak10 as nonsense mutations in ksr-1 (Figure 1B, see Materials and Methods). KSR-1 is a scaffold protein that facilitates the localization and interactions required for the Ras-MAPK cascade consisting of Raf, MEK, and ERK (Kornfeld et al. 1995b; Sundaram and Han 1995; Nguyen et al. 2002; Zhang et al. 2013). The deletion allele ksr-1(ok786) also suppressed the loopy waveform of the activated Gq mutant identically to ox314 and yak10. These results suggest that KSR-1 activity is required for regulation of waveform by Gq.
The ERK MAPK cascade acts to promote Gq signaling
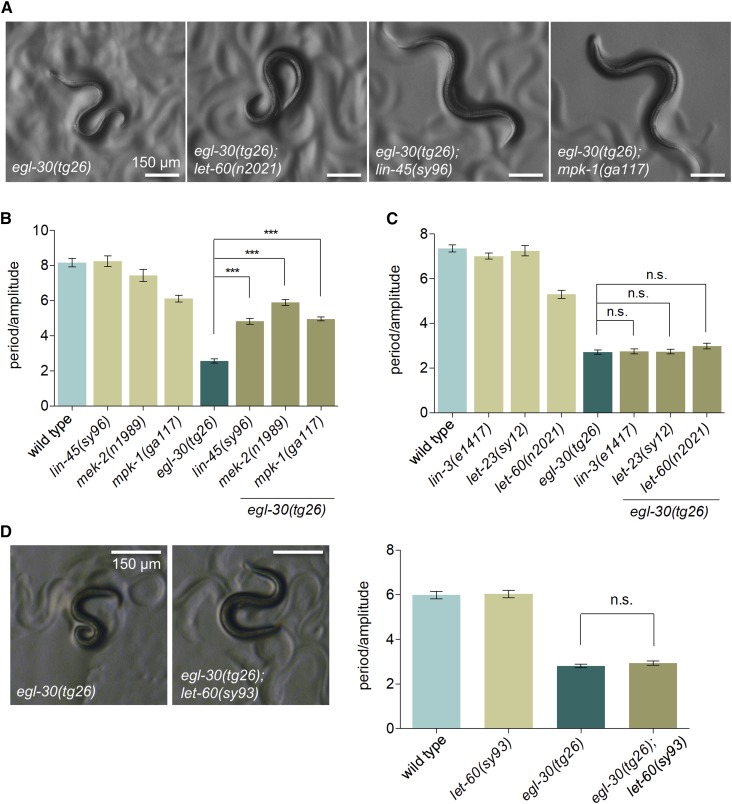
Because the loss of the MAPK scaffold ksr-1 suppresses the activated Gq mutant egl-30(tg26), we asked whether other components of the Ras-ERK pathway would also suppress. Since the core components of the Ras-ERK pathway are required for viability, we built double mutants of activated Gq with reduction-of-function mutations in genes at each step of the ERK cascade. Mutations in Raf [lin-45(sy96)], MEK [mek-2(n1989), mek-2(ku114)], and ERK [mpk-1(ga117), mpk-1(oz140)] all suppressed the loopy waveform of activated Gq animals similarly to ksr-1(ok786) (Figure 2, A and B and Figure S1, A and B). However, mutations in Ras [let-60(n2021)] and the upstream pathway activators EGF [lin-3(e1417)] and the EGF receptor [let-23(sy12)] did not suppress activated Gq (Figure 2C). Because let-60 is required for viability, most let-60 alleles including n2021 are partial loss-of-function (Beitel et al. 1990). We also analyzed the dominant negative D119N allele let-60(sy93) that disrupts Ras binding to guanine nucleotides and thus prevents Ras activation (Han and Sternberg 1991). We found that let-60(sy93) also did not suppress the loopy waveform of activated Gq (Figure 2D). Additionally, we expressed the dominant negative let-60 mutation S17N specifically in neurons and did not observe suppression of the loopy waveform of activated Gq (Figure S2D). These results support the possibility that ERK activation in this pathway occurs through a Ras-independent mechanism.
Figure 2.
Mutations in the ERK MAPK pathway suppress activated Gq. (A) Mutations in the MAPKKK lin-45/Raf and the MAPK mpk-1/ERK suppress the coiled posture of egl-30(tg26) worms. Partial loss of Ras let-60 activity does not suppress activated Gq posture. (B) Waveform quantification of ERK pathway mutants. Mutations in lin-45, mek-2, and mpk-1 suppress activated Gq egl-30(tg26) similar to ksr-1 mutations. N ≥ 12, *** P < 0.001, one-way ANOVA with Bonferroni’s post hoc test. (C) Signaling components upstream of Raf do not suppress activated Gq. Mutations in the EGF ligand (lin-3) or the EGFR (let-23) do not affect the exaggerated waveform of egl-30(tg26) animals. The Ras partial loss-of-function mutation let-60(n2021) does not suppress activated Gq waveform. N ≥ 12, *** P < 0.001, one-way ANOVA with Bonferroni’s post hoc test. (D) The let-60(sy93) dominant negative mutation in Ras does not suppress activated Gq waveform. N ≥ 13, n.s., not significant, one-way ANOVA with Bonferroni’s post hoc test.
Because partial loss-of-function mutations in the ERK MAPK pathway genes downstream of Ras showed clear suppression of activated Gq, we were surprised to find that partial loss-of-function mutations in Ras did not suppress. If LET-60/Ras is indeed not required, Gq might instead activate the ERK pathway via other Ras-subfamily proteins. To test this possibility, we made double mutants of activated Gq with putative null alleles of R-Ras/ras-1, M-Ras/ras-2, and Rap1/rap-1 and found that they also did not suppress activated Gq (Figure S2A). To further investigate whether this pathway acts independently of Ras, we tested mutations in GEFs that activate Ras. The temperature-sensitive RasGEF mutant sos-1(cs41) did not suppress activated Gq when shifted to the nonpermissive temperature (Figure S2B). Additionally, a null mutation in the neuronal RasGEF rgef-1 also did not suppress activated Gq (Figure S2C). In genetic screens for vulval induction mutants, additional factors such as the PP2A subunit sur-6 (Sieburth et al. 1999) and ion transporter sur-7 (Yoder et al. 2004) were identified as positive regulators of Ras-ERK activity. However, the sur-6(sv30) and sur-7(ku119) mutations did not suppress activated Gq locomotion (data not shown). These data suggest either that only a low level of Ras activity is needed to properly activate ERK signaling downstream of Gq, or that ERK signaling acts independently of LET-60/Ras and other known C. elegans Ras family proteins to regulate locomotion downstream of Gq.
KSR-1 and the ERK MAPK cascade modulate Rho signaling
Three classes of suppressor mutations were isolated in our forward genetic screen of activated Gq, as characterized by their molecular roles and unique suppression phenotypes (Topalidou et al. 2017a,b). We grouped together a class of suppressor mutations including ox314, yak10, and the RhoGEF Trio (unc-73 in C. elegans) by their characteristic strong suppression of the loopy waveform of activated Gq (Topalidou et al. 2017a,b), suggesting that ksr-1 might act in the same pathway as unc-73.
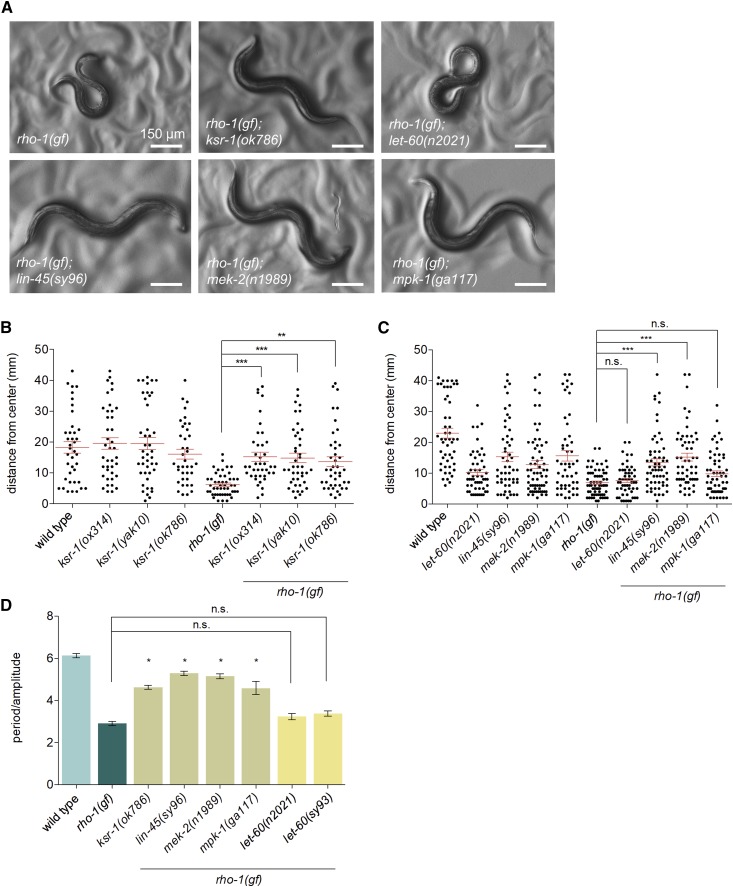
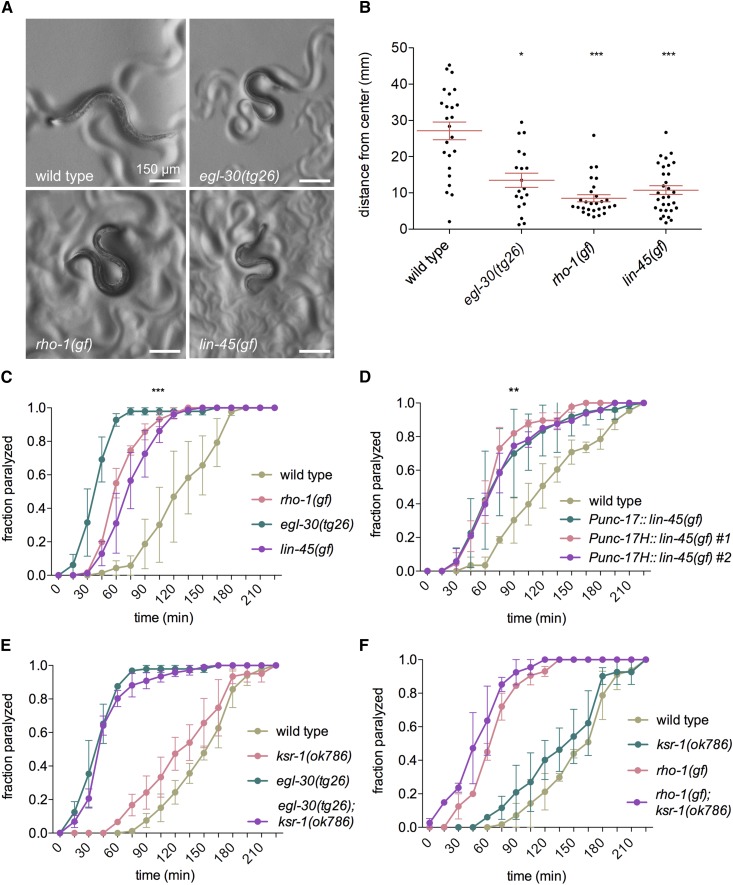
We have shown that Gq regulates locomotion via the small GTPase Rho (RHO-1 in C. elegans) (Topalidou et al. 2017a). Transgenic expression of an activated RHO-1 mutant (G14V) in acetylcholine neurons (here called “rho-1(gf)”) causes worms to have a loopy waveform and impaired locomotion (McMullan et al. 2006) (Figure 3A). To examine whether ksr-1 acts in the Gq-Rho pathway we tested whether mutations in ksr-1 suppress the phenotypes of rho-1(gf) worms. We found that the ksr-1 alleles ok786, ox314, and yak10 all suppressed the loopy waveform of rho-1(gf) worms (Figure 3, A, B, and D). Because rho-1(gf) worms have a slow locomotion rate and loopy waveform, these mutants do not efficiently travel long distances. We used radial locomotion assays (see Materials and Methods) to quantify the locomotion phenotype of rho-1(gf) worms. rho-1(gf) ksr-1 double mutants had an increased radial distance traveled compared to rho-1(gf) alone (Figure 3B). These data suggest that KSR-1 acts downstream of, or in parallel to, the Gq-Rho pathway to regulate locomotion.
Figure 3.
Mutations in ksr-1 and the ERK pathway suppress activated Rho. (A) Mutations in ERK pathway genes ksr-1, lin-45, mek-2, and mpk-1 visibly suppress the coiled posture of animals expressing an activated Rho mutant (G14V) in acetylcholine neurons (nzIs29[Punc-17::rho-1(gf)]). Reduction of LET-60/Ras activity does not suppress activated Rho. (B) The ksr-1(ok786), ksr-1(ox314), and ksr-1(yak10) mutations suppress the locomotion of activated Rho animals as shown by radial locomotion assays. N ≥ 38, ** P < 0.01, *** P < 0.001, Kruskal-Wallis test with Dunn’s post hoc test. (C) Mutations in lin-45 and mek-2 suppress the locomotion defect of activated Rho animals as shown by radial locomotion assays. The let-60(n2021) mutation does not significantly suppress activated Rho locomotion. N ≥ 50, *** P < 0.001, Kruskal-Wallis test with Dunn’s post hoc test. (D) Waveform quantification of ERK pathway mutants. Mutations in ksr-1, lin-45, mek-2, and mpk-1 show similar suppression of activated Rho, but the Ras reduction-of-function mutant let-60(n2021) or dominant negative let-60(sy93) do not suppress activated Rho. N ≥ 14, * P < 0.05, n.s., not significant, one-way ANOVA with Bonferroni’s post hoc test, compared to rho-1(gf).
Since ksr-1 mutants suppress the exaggerated waveform of both activated Gq and activated Rho animals, we expected that loss of other ERK pathway components would also suppress activated Rho. We made double mutants of activated Rho [rho-1(gf)] with reduction-of-function alleles of the Ras-ERK pathway and found that mutations in Raf, MEK, and ERK suppressed the rho-1(gf) loopy waveform phenotype (Figure 3, A, C, and D). However, the let-60(n2021) and let-60(sy93) Ras mutations did not suppress the loopy waveform of rho-1(gf) worms (Figure 3, A, C, and D). These data suggest that the ERK pathway acts downstream of, or in parallel to, the Gq-Rho pathway to regulate locomotion, possibly in a Ras-independent manner.
The ERK MAPK cascade acts in acetylcholine neurons to control locomotion
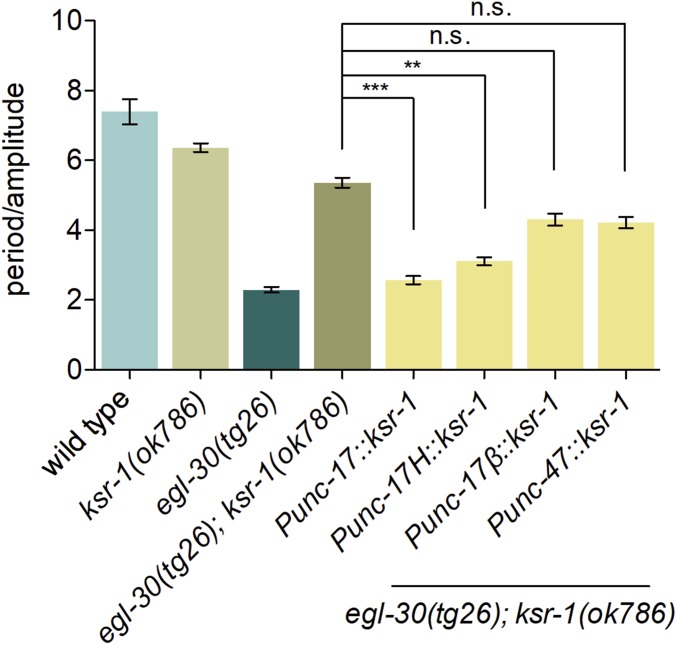
Members of the ERK pathway are expressed in neurons in C. elegans (Dent and Han 1998; Hunt-Newbury et al. 2007), and Gq and Rho act in acetylcholine neurons to promote synaptic release and regulate locomotion (Lackner et al. 1999; McMullan et al. 2006). To determine whether the ERK pathway also acts in neurons to modulate Gq signaling, we expressed the ksr-1 cDNA under promoters driving expression in specific types of neurons. Single-copy transgenic expression of ksr-1 under an acetylcholine neuron promoter (Punc-17) or under a head acetylcholine neuron promoter (Punc-17H) fully reversed the ksr-1 suppression of the loopy waveform of activated Gq worms (Figure 4). ksr-1 expression in ventral cord acetylcholine motor neurons (Punc-17β) or GABA neurons (Punc-47) did not significantly reverse the ksr-1 suppression of activated Gq (Figure 4). This suggests that ERK signaling primarily functions in the acetylcholine interneurons or motor neurons of the head to modulate Gq-dependent locomotion.
Figure 4.
KSR-1 acts in head acetylcholine neurons to modulate Gq signaling. Single-copy expression of the ksr-1 cDNA exclusively in acetylcholine neurons (Punc-17, yakSi26 transgene) or head acetylcholine neurons (Punc-17H, yakSi27 transgene) is sufficient to reverse the ksr-1 suppression of the loopy waveform of activated Gq animals. ksr-1 expression in ventral cord acetylcholine neurons (Punc-17β, yakSi28 transgene) or GABA neurons (Punc-47, yakSi29 transgene) does not reverse the ksr-1 suppression of the activated Gq exaggerated waveform. N ≥ 12, ** P < 0.01, *** P < 0.001, n.s., not significant, Kruskal-Wallis test with Dunn’s post hoc test.
We have shown that the ERK pathway is necessary for Gq-dependent effects on locomotion. To determine whether ERK signaling is sufficient to modulate locomotion, we expressed an activated form of lin-45/Raf specifically in acetylcholine neurons. Raf kinase activity is regulated via conserved phosphorylation events, and phosphomimetic mutations T626E/T629D in the kinase activation loop of lin-45 are sufficient to confer constitutive Raf activity (Chong et al. 2001). We found that expression of activated Raf in acetylcholine neurons (Punc-17) causes a loopy waveform similar to activated Gq and Rho mutants and similar limited dispersal in radial locomotion assays (Figure 5, A and B and Figure S3, A and B). Additionally, expression of activated Raf or activated Gq in head acetylcholine neurons also caused a loopy waveform (Figure S3, A–C).
Figure 5.
Increased Raf activity in acetylcholine neurons is sufficient to regulate locomotion and increase acetylcholine release. (A) Transgenic lines expressing an activated form of lin-45/Raf (T626E/T629D) in acetylcholine neurons (yakIs34[Punc-17::lin-45(gf)]) have exaggerated body bends and coiled posture similar to the activated Gq mutant egl-30(tg26) and to animals expressing activated Rho in acetylcholine neurons (nzIs29[Punc-17::rho-1(gf)]). The wild type and egl-30(tg26) photos are the same as shown in Figure 1A, and the rho-1(gf) photo is the same as the one in Figure 3A. (B) Expression of activated Rho (nzIs29[Punc-17::rho-1(gf)]) or Raf (yakIs34[Punc-17::lin-45(gf)]) in acetylcholine neurons impairs coordinated locomotion similarly to activated Gq [egl-30(tg26]). N ≥ 19, * P < 0.05, *** P < 0.001, Kruskal-Wallis test with Dunn’s post hoc test. (C) Animals expressing activated Gq, Rho, or Raf are hypersensitive to the acetylcholinesterase inhibitor aldicarb. Activated Gq [egl-30(tg26)], activated Rho expressed in acetylcholine neurons (nzIs29[Punc-17::rho-1(gf)]), and activated Raf expressed in acetylcholine neurons yakIs34[Punc-17::lin-45(gf)]) become paralyzed significantly faster than wild type animals when exposed to 1 mM aldicarb. All strains are significantly different from wild type at t = 60, 75, 90, and 105 min. N ≥ 61, *** P < 0.001, two-way ANOVA with Bonferroni’s post hoc test. (D) Animals expressing activated Raf in head acetylcholine neurons are hypersensitive to aldicarb. The yakIs34[Punc-17::lin-45(gf)] integrated array expressing activated Raf in all acetylcholine neurons and the yakEx154[Punc-17H::lin-45(gf)] (#1) and yakEx168[Punc-17H::lin-45(gf)] (#2) extrachromosomal arrays expressing activated Raf in head acetylcholine neurons show similar hypersensitivity to aldicarb. All strains are significantly different from wild type at t = 60, 75, 90, and 105 min. N ≥ 47, ** P < 0.01, two-way ANOVA with Bonferroni’s post hoc test. (E) ksr-1 is not required for the aldicarb hypersensitivity of activated Gq. Paralysis of ksr-1(ok786) animals on 1 mM aldicarb is not significantly different from wild type. The ksr-1 deletion ok786 does not suppress the aldicarb hypersensitivity of activated Gq [egl-30(tg26)]. N ≥ 53. (F) ksr-1 is not required for the aldicarb hypersensitivity of activated Rho. Paralysis of ksr-1(ok786) animals on 1 mM aldicarb is not significantly different from wild type. The ksr-1 deletion ok786 does not suppress the aldicarb hypersensitivity of the activated Rho mutant nzIs29[Punc-17::rho-1(gf)]. N ≥ 41.
Gq and Rho promote acetylcholine release at the worm neuromuscular junction (Lackner et al. 1999; Miller et al. 1999; McMullan et al. 2006). To determine if Raf activation affects acetylcholine release at the neuromuscular junction, we assayed for sensitivity to the acetylcholinesterase inhibitor aldicarb. Mutants with reduced acetylcholine secretion are resistant to aldicarb, whereas mutants with increased acetylcholine secretion are hypersensitive to aldicarb (Mahoney et al. 2006). Activated Gq and Rho mutants have increased rates of paralysis when exposed to aldicarb (Lackner et al. 1999; McMullan et al. 2006). We found that expression of activated Raf in all acetylcholine neurons or in head acetylcholine neurons also led to aldicarb hypersensitivity, similar to activated Gq and Rho mutants (Figure 5, C and D). However, we found that a ksr-1 mutation does not suppress the aldicarb hypersensitivity of activated Gq or Rho mutants, and the ksr-1 mutant on its own has similar aldicarb sensitivity to wild type (Figure 5, E and F). These results suggest that the ERK pathway is not required for synaptic transmission, but is sufficient to stimulate synaptic transmission when constitutively activated. Furthermore, these data suggest that ERK pathway activation in head acetylcholine neurons is sufficient to promote acetylcholine release by motor neurons at the neuromuscular junction.
Discussion
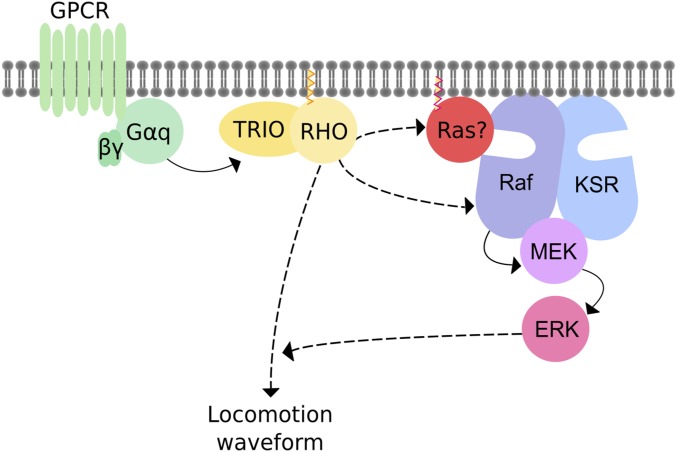
In this study, we identified KSR-1 and the ERK MAPK cascade as modulators of Gq-Rho signaling. We found that ERK signaling acts in head acetylcholine neurons (interneurons or motor neurons of the head) to modulate Gq-dependent locomotion behavior, especially the waveform of the animal. Our data support the model that Gq-Rho activation of the ERK pathway may be independent of its canonical regulator, the small GTPase Ras/LET-60 (Figure 6).
Figure 6.
Model for Gq-Rho-ERK signaling. Gq directly activates Trio and Rho (solid arrow) (Lutz et al. 2007). The core ERK cascade acts either downstream of Rho or in parallel (dashed arrows) to modulate locomotion behavior. ERK activation occurs independently of the extracellular growth factor LIN-3, its receptor LET-23, or Ras/LET-60. It is possible that Raf/LIN-45 is activated by a Ras family member other than the canonical worm Ras/LET-60. Our model suggests that Gq-Rho signaling regulates the worm waveform by acting cell autonomously in head acetylcholine neurons in an ERK-dependent pathway. By contrast, we propose that Gq-Rho signaling regulates synaptic transmission at the neuromuscular junction by acting in an ERK-independent pathway, possibly in the ventral cord motorneurons themselves.
Gq regulates multiple aspects of C. elegans locomotion behavior, including the rate of locomotion and waveform. Previous work has shown that these behaviors are at least partially genetically separable, with different pathways downstream of Gq contributing differentially to these behaviors and with Gq likely acting in several classes of neurons including both head acetylcholine neurons and ventral cord acetylcholine motor neurons (Lackner et al. 1999; Topalidou et al. 2017a). Here we focused on Gq regulation of the waveform, which is strongly dependent on Rho and ERK. The most parsimonious model supported by our findings is that Gq, Rho, and the ERK pathway function cell autonomously in the head acetylcholine neurons to regulate the waveform (Figure 6). Other components of the Gq-Rho pathway have been previously shown to act in head acetylcholine neurons to regulate the waveform (Topalidou et al. 2017a,b), and here we show that KSR-1 is required in head acetylcholine neurons to modulate Gq-dependent effects on the waveform. Additionally, expression of activated LIN-45/Raf in head acetylcholine neurons is also sufficient to regulate the waveform and cause loopy locomotion. These data suggest that ERK pathway activity in head acetylcholine neurons is both necessary and sufficient to regulate worm waveform.
Though the ERK pathway is required for Gq and Rho-dependent regulation of the waveform, it is not required for the aldicarb hypersensitivity of activated Gq and Rho mutants. This indicates that Gq and Rho regulation of acetylcholine release at the neuromuscular junction is at least partially independent of Gq and Rho regulation of the locomotion waveform. The simplest model is that Gq and Rho signaling regulates the waveform by acting in head acetylcholine neurons through an ERK-dependent pathway, but Gq and Rho signaling regulates synaptic transmission at the neuromuscular junction through an ERK-independent pathway. Overexpression of an activated Gq mutant in ventral cord acetylcholine motor neurons is sufficient to cause an aldicarb hypersensitive phenotype (Lackner et al. 1999), suggesting that Gq may act cell autonomously in the ventral cord motor neurons to promote acetylcholine release by these neurons. But, as with activated Raf, it is also possible that Gq activity in head acetylcholine neurons may contribute to acetylcholine release by downstream ventral cord motor neurons.
The ERK signaling cascade has been well-studied for its regulation of cellular proliferation and differentiation (Sun et al. 2015), but also plays important roles in mature neurons and has been associated with synaptic plasticity and memory (Impey et al. 1999). In addition to activating transcription, ERKs regulate synaptic plasticity both presynaptically and postsynaptically by phosphorylating relevant substrates. ERKs phosphorylate the presynaptic proteins synapsin I and Munc18-1, and postsynaptic proteins such as scaffolds, Kv4.2 potassium channels, and Group I metabotropic glutamate receptors (Jovanovic et al. 2000; Sweatt 2004; Thomas and Huganir 2004; Kushner et al. 2005; Boggio et al. 2007; Vara et al. 2009; Mao and Wang 2016a; Schmitz et al. 2016). Our findings suggest that the ERK pathway controls Gq-dependent locomotion in C. elegans and that activated ERK promotes synaptic transmission.
In contrast to many developmental and neuronal roles of Ras-dependent ERK signaling in C. elegans, our data suggest that Raf-MEK-ERK signaling may modulate Gq-Rho output independently of Ras/LET-60. One caveat to the conclusion that the ERK pathway acts independently of Ras to control Gq-dependent locomotion is that the alleles of let-60/Ras tested here are not null (Beitel et al. 1990; Han and Sternberg 1990, 1991; Han et al. 1990). However, other than mpk-1(ga117), the alleles we used of lin-45, mek-2, and mpk-1 are also not null (Han et al. 1993; Kornfeld et al. 1995a; Wu et al. 1995; Lackner and Kim 1998; Hsu et al. 2002), yet were able to suppress an activated Gq mutant. Furthermore, the let-60/Ras alleles used here have stronger phenotypes in vulval development than the two weak mek-2/MEK alleles we used (n1989 and ku114) (Beitel et al. 1990; Kornfeld et al. 1995a; Wu et al. 1995) and the let-60(n2021)/Ras mutant has comparable phenotypes to the lin-45(sy96)/Raf and mpk-1(ga117)/ERK alleles for chemotaxis to odorants and the regulation of foraging behavior (Hirotsu et al. 2000; Hamakawa et al. 2015). Thus, either LET-60/Ras is not required to modulate Gq signaling, or ERK signaling in the locomotor circuit requires a lower threshold of Ras activity and the let-60 mutants we used have sufficient levels of active Ras to modulate Gq signaling, but not enough for vulval induction or other behaviors.
If Ras is not required, how instead might KSR-Raf-MEK-ERK signaling be activated? Normally, active Ras recruits Raf to the plasma membrane but it has been shown that artificial recruitment of Raf to the plasma membrane in the absence of Ras is sufficient to activate Raf signaling (Stokoe et al. 1994; Marais et al. 1995). Thus, another possible way to activate the ERK pathway would be for Raf to be recruited to the membrane by proteins other than Ras. The most obvious candidates for recruiting Raf are other members of the Ras family such as Rap1 or R-Ras that have a conserved effector-binding domain (Reiner and Lundquist 2016); it has been reported that Rap1 can mediate Ras-independent activation of Raf downstream of a Gq-coupled receptor (Guo et al. 2001). However, mutations in the worm orthologs of Rap1/RAP-1 and R-Ras/RAS-1 did not suppress the activated Gq mutant, indicating that these Ras-like proteins are not required, at least individually, to activate Raf in this pathway. It is possible that more than one of these Ras family members function redundantly to activate Raf, or that Raf is activated independently of Ras family proteins. There have been other reported cases of Ras-independent activation of the Raf-MEK-ERK pathway (Robbins et al. 1992; Honda et al. 1994; Ueda et al. 1996; van Biesen et al. 1996; Drosten et al. 2014), some of which involve G protein signaling and protein kinase C, though the precise mechanisms involved are unclear.
Gq, Rho, and ERK are also required for the C. elegans behavioral and immune response to infection by the bacterium Microbacterium nematophilum (Nicholas and Hodgkin 2004; McMullan et al. 2012). Interestingly, LET-60/Ras was reported to not be required for the immune response (Nicholas and Hodgkin 2004), or only partially required (McMullan et al. 2012). Additionally, LET-60/Ras was not required for the increased sensitivity to aldicarb caused by bacterial infection (McMullan et al. 2012), at least as assayed using the let-60(n2021) allele. Thus, the same neuronal Ras-independent ERK pathway we describe here may also modulate Gq signaling in the neuronal response to bacterial infection and possibly the innate immune response as well.
Our epistasis analysis suggests that ERK signaling acts genetically downstream of, or in parallel to, Gq-Rho signaling. How might Gq-Rho signaling lead to ERK activation? ERK activation could occur via a linear pathway downstream of Gq and Rho, or ERK could signal in parallel and converge downstream of Rho to affect neuronal activity (Figure 6). There is precedence for Gq activating ERK via a linear pathway. In the pharyngeal muscle of C. elegans, ERK activity is increased by Gq-dependent signaling through protein kinase C (You et al. 2006). In the AWC olfactory neuron, ERK activity is increased downstream of Gq signaling via the RasGEF RGEF-1 (Chen et al. 2011; Uozumi et al. 2012). Protein kinase C and RGEF-1 are both activated by DAG, probably produced by the canonical Gq-PLCβ pathway. By contrast, we found that ERK signaling regulates locomotion by modulating the output of the Gq-Rho pathway that acts in parallel to Gq-PLCβ, and does not depend on RGEF-1.
How might the ERK pathway modulate neuronal activity downstream of Gq-Rho signaling? Rho regulates neuronal activity and synaptic release through several mechanisms in C. elegans neurons, any of which could be targets of ERK signaling. One possible ERK effector is the NCA/NALCN cation channel that acts genetically downstream of Gq-Rho to regulate locomotion rate and waveform (Topalidou et al. 2017a,b). Though ERK has not been directly connected to NCA/NALCN, mammalian ERK regulates neuronal excitability by directly phosphorylating voltage-gated sodium and potassium channels (Schrader et al. 2006; Stamboulian et al. 2010) and by regulating channel expression (Yang et al. 2015). Given the many possible ways that ERK may regulate excitability or synaptic transmission, C. elegans genetics is well suited to determine relevant effectors of Gq-Rho-ERK signaling.
Acknowledgments
We thank Brooke Jarvie for the isolation of ksr-1(yak10), Stephen Nurrish for the activated Rho worm strain, Jordan and Jill Hoyt for help with whole-genome sequence analysis, Laura Taylor for help with irradiation of extrachromosomal arrays, and Dana Miller for use of her Nikon SMZ18 microscope and camera. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was supported by an Ellison Medical Foundation New Scholar Award and by NIH grants R00 MH082109 and R56 NS100843 to M.A.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6071009.
Communicating editor: M. Sundaram
Literature Cited
- Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., et al. , 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44: W3–W10. 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailion M., Hannemann M., Dalton S., Pappas A., Watanabe S., et al. , 2014. Two Rab2 interactors regulate dense-core vesicle maturation. Neuron 82: 167–180. 10.1016/j.neuron.2014.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani C. A., Gharib S., Simon M. I., Sternberg P. W., 2003. Caenorhabditis elegans Gαq regulates egg-laying behavior via a PLCβ-independent and serotonin-dependent signaling pathway and likely functions both in the nervous system and in muscle. Genetics 165: 1805–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel G. J., Clark S. G., Horvitz H. R., 1990. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348: 503–509. 10.1038/348503a0 [DOI] [PubMed] [Google Scholar]
- Boggio E. M., Putignano E., Sassoè-Pognetto M., Pizzorusso T., Giustetto M., 2007. Visual stimulation activates ERK in synaptic and somatic compartments of rat cortical neurons with parallel kinetics. PLoS One 2: e604 10.1371/journal.pone.0000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton T. G., Nye S. H., Robbins D. J., Ip N. Y., Radzlejewska E., et al. , 1991. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65: 663–675. 10.1016/0092-8674(91)90098-J [DOI] [PubMed] [Google Scholar]
- Brundage L., Avery L., Katz A., Kim U. J., Mendel J. E., et al. , 1996. Mutations in a C. elegans Gqalpha gene disrupt movement, egg laying, and viability. Neuron 16: 999–1009. 10.1016/S0896-6273(00)80123-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. P., Hu Z., Sieburth D., 2012. Recruitment of sphingosine kinase to presynaptic terminals by a conserved muscarinic signaling pathway promotes neurotransmitter release. Genes Dev. 26: 1070–1085. 10.1101/gad.188003.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlie N. K., Schade M. A., Thomure A. M., Miller K. G., 2006. Presynaptic UNC-31 (CAPS) is required to activate the G alpha(s) pathway of the Caenorhabditis elegans synaptic signaling network. Genetics 172: 943–961. 10.1534/genetics.105.049577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Fu Y., Ren M., Xiao B., Rubin C. S., 2011. A RasGRP, C. elegans RGEF-1b, couples external stimuli to behavior by activating LET-60 (Ras) in sensory neurons. Neuron 70: 51–65. 10.1016/j.neuron.2011.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong H., Lee J., Guan K. L., 2001. Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 20: 3716–3727. 10.1093/emboj/20.14.3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. W., Hammarlund M., Harrach T., Hullett P., Olsen S., et al. , 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6: 118 10.1186/1471-2164-6-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent J. A., Han M., 1998. Post-embryonic expression pattern of C. elegans let-60 ras reporter constructs. Mech. Dev. 72: 179–182. 10.1016/S0925-4773(98)00026-4 [DOI] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M., Iwasaki K., 2002. Regulation of retrograde signaling at neuromuscular junctions by the novel C2 domain protein AEX-1. Neuron 33: 249–259. 10.1016/S0896-6273(01)00587-6 [DOI] [PubMed] [Google Scholar]
- Drosten M., Sum E. Y. M., Lechuga C. G., Simón-Carrasco L., Jacob H. K. C., et al. , 2014. Loss of p53 induces cell proliferation via Ras-independent activation of the Raf/Mek/Erk signaling pathway. Proc. Natl. Acad. Sci. USA 111: 15155–15160. 10.1073/pnas.1417549111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. 10.1038/nmeth.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F. F., Kumahara E., Saffen D., 2001. A CalDAG-GEFI/Rap1/B-Raf cassette couples M(1) muscarinic acetylcholine receptors to the activation of ERK1/2. J. Biol. Chem. 276: 25568–25581. 10.1074/jbc.M101277200 [DOI] [PubMed] [Google Scholar]
- Hamakawa M., Uozumi T., Ueda N., Iino Y., Hirotsu T., 2015. A role for Ras in inhibiting circular foraging behavior as revealed by a new method for time and cell-specific RNAi. BMC Biol. 13: 6 10.1186/s12915-015-0114-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M., Palfreyman M. T., Watanabe S., Olsen S., Jorgensen E. M., 2007. Open syntaxin docks synaptic vesicles. PLoS Biol. 5: e198 10.1371/journal.pbio.0050198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Sternberg P. W., 1990. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell 63: 921–931. 10.1016/0092-8674(90)90495-Z [DOI] [PubMed] [Google Scholar]
- Han M., Sternberg P. W., 1991. Analysis of dominant-negative mutations of the Caenorhabditis elegans let-60 ras gene. Genes Dev. 5: 2188–2198. 10.1101/gad.5.12a.2188 [DOI] [PubMed] [Google Scholar]
- Han M., Aroian R. V., Sternberg P. W., 1990. The let-60 locus controls the switch between vulval and nonvulval cell fates in Caenorhabditis elegans. Genetics 126: 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Golden A., Han Y., Sternberg P. W., 1993. C. elegans lin-45 raf gene participates in let-60 ras-stimulated vulval differentiation. Nature 363: 133–140. 10.1038/363133a0 [DOI] [PubMed] [Google Scholar]
- Hiley E., McMullan R., Nurrish S. J., 2006. The Galpha12-RGS RhoGEF-RhoA signalling pathway regulates neurotransmitter release in C. elegans. EMBO J. 25: 5884–5895. 10.1038/sj.emboj.7601458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu T., Iino Y., 2005. Neural circuit-dependent odor adaptation in C. elegans is regulated by the Ras-MAPK pathway. Genes Cells 10: 517–530. 10.1111/j.1365-2443.2005.00856.x [DOI] [PubMed] [Google Scholar]
- Hirotsu T., Saeki S., Yamamoto M., Iino Y., 2000. The Ras-MAPK pathway is important for olfaction in Caenorhabditis elegans. Nature 404: 289–293 [erratum: Nature 432: 653 (2004)] 10.1038/35005101 [DOI] [PubMed] [Google Scholar]
- Honda Z., Takano T., Gotoh Y., Nishida E., Ito K., et al. , 1994. Transfected platelet-activating factor receptor activates mitogen-activated protein (MAP) kinase and MAP kinase in Chinese hamster ovary cells. J. Biol. Chem. 269: 2307–2315. [PubMed] [Google Scholar]
- Hsu V., Zobel C. L., Lambie E. J., Schedl T., Kornfeld K., 2002. Caenorhabditis elegans lin-45 raf is essential for larval viability, fertility and the induction of vulval cell fates. Genetics 160: 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Vashlishan-Murray A. B., Kaplan J. M., 2015. NLP-12 engages different UNC-13 proteins to potentiate tonic and evoked release. J. Neurosci. 35: 1038–1042. 10.1523/JNEUROSCI.2825-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., et al. , 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5: e237 10.1371/journal.pbio.0050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde R., Corkins M. E., Somers G. A., Hart A. C., 2011. PKC-1 acts with the ERK MAPK signaling pathway to regulate Caenorhabditis elegans mechanosensory response. Genes Brain Behav. 10: 286–298. 10.1111/j.1601-183X.2010.00667.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S., Obrietan K., Storm D. R., 1999. Making new connections. Neuron 23: 11–14. 10.1016/S0896-6273(00)80747-3 [DOI] [PubMed] [Google Scholar]
- Jovanovic J. N., Czernik A. J., Fienberg A. A., Greengard P., Sihra T. S., 2000. Synapsins as mediators of BDNF-enhanced neurotransmitter release. Nat. Neurosci. 3: 323–329. 10.1038/73888 [DOI] [PubMed] [Google Scholar]
- Karnoub A. E., Weinberg R. A., 2008. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9: 517–531. 10.1038/nrm2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld K., Guan K. L., Horvitz H. R., 1995a The Caenorhabditis elegans gene mek-2 is required for vulval induction and encodes a protein similar to the protein kinase MEK. Genes Dev. 9: 756–768. 10.1101/gad.9.6.756 [DOI] [PubMed] [Google Scholar]
- Kornfeld K., Hom D. B., Horvitz H. R., 1995b The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell 83: 903–913. 10.1016/0092-8674(95)90206-6 [DOI] [PubMed] [Google Scholar]
- Kushner S. A., Elgersma Y., Murphy G. G., Jaarsma D., van Woerden G. M., et al. , 2005. Modulation of presynaptic plasticity and learning by the H-ras/extracellular signal-regulated kinase/synapsin I signaling pathway. J. Neurosci. 25: 9721–9734. 10.1523/JNEUROSCI.2836-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M. R., Kim S. K., 1998. Genetic analysis of the Caenorhabditis elegans MAP kinase gene mpk-1. Genetics 150: 103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M. R., Nurrish S. J., Kaplan J. M., 1999. Facilitation of synaptic transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron 24: 335–346. 10.1016/S0896-6273(00)80848-X [DOI] [PubMed] [Google Scholar]
- Lutz S., Freichel-Blomquist A., Yang Y., Rümenapp U., Jakobs K. H., et al. , 2005. The guanine nucleotide exchange factor p63RhoGEF, a specific link between Gq/11-coupled receptor signaling and RhoA. J. Biol. Chem. 280: 11134–11139. 10.1074/jbc.M411322200 [DOI] [PubMed] [Google Scholar]
- Lutz S., Shankaranarayanan A., Coco C., Ridilla M., Nance M. R., et al. , 2007. Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science 318: 1923–1927. 10.1126/science.1147554 [DOI] [PubMed] [Google Scholar]
- Mahoney T. R., Luo S., Nonet M. L., 2006. Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat. Protoc. 1: 1772–1777. 10.1038/nprot.2006.281 [DOI] [PubMed] [Google Scholar]
- Mao L.-M., Wang J. Q., 2016a Regulation of group I metabotropic glutamate receptors by MAPK/ERK in neurons. J. Nat. Sci. 2: e268. [PMC free article] [PubMed] [Google Scholar]
- Mao L.-M., Wang J. Q., 2016b Synaptically localized mitogen-activated protein kinases: local substrates and regulation. Mol. Neurobiol. 53: 6309–6315. 10.1007/s12035-015-9535-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais R., Light Y., Paterson H. F., Marshall C. J., 1995. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 14: 3136–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan R., Hiley E., Morrison P., Nurrish S. J., 2006. Rho is a presynaptic activator of neurotransmitter release at pre-existing synapses in C. elegans. Genes Dev. 20: 65–76. 10.1101/gad.359706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan R., Anderson A., Nurrish S., 2012. Behavioral and immune responses to infection require Gαq- RhoA signaling in C. elegans. PLoS Pathog. 8: e1002530 10.1371/journal.ppat.1002530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Emerson M. D., Rand J. B., 1999. Goalpha and diacylglycerol kinase negatively regulate the Gqalpha pathway in C. elegans. Neuron 24: 323–333. 10.1016/S0896-6273(00)80847-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A., Burack W. R., Stock J. L., Kortum R., Chaika O. V., et al. , 2002. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol. Cell. Biol. 22: 3035–3045. 10.1128/MCB.22.9.3035-3045.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas H. R., Hodgkin J., 2004. The ERK MAP kinase cascade mediates tail swelling and a protective response to rectal infection in C. elegans. Curr. Biol. 14: 1256–1261. 10.1016/j.cub.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Ortiz J., Harris H. W., Guitart X., Terwilliger R. Z., Haycock J. W., et al. , 1995. Extracellular signal-regulated protein kinases (ERKs) and ERK kinase (MEK) in brain: regional distribution and regulation by chronic morphine. J. Neurosci. 15: 1285–1297. 10.1523/JNEUROSCI.15-02-01285.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner D., Lundquist E. A., Small GTPases. (May 26, 2016), Wormbook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.67.2, http://www.wormbook.org.
- Rhee S. G., 2001. Regulation of phosphoinositide-specific phospholipase C. Annu. Rev. Biochem. 70: 281–312. 10.1146/annurev.biochem.70.1.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins D. J., Cheng M., Zhen E., Vanderbilt C. A., Feig L. A., et al. , 1992. Evidence for a Ras-dependent extracellular signal-regulated protein kinase (ERK) cascade. Proc. Natl. Acad. Sci. USA 89: 6924–6928. 10.1073/pnas.89.15.6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz S. K., King C., Kortleven C., Huson V., Kroon T., et al. , 2016. Presynaptic inhibition upon CB1 or mGlu2/3 receptor activation requires ERK/MAPK phosphorylation of Munc18–1. EMBO J. 35: 1236–1250. 10.15252/embj.201592244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L. A., Birnbaum S. G., Nadin B. M., Ren Y., Bui D., et al. , 2006. ERK/MAPK regulates the Kv4.2 potassium channel by direct phosphorylation of the pore-forming subunit. Am. J. Physiol. Cell Physiol. 290: C852–C861. 10.1152/ajpcell.00358.2005 [DOI] [PubMed] [Google Scholar]
- Sieburth D. S., Sundaram M., Howard R. M., Han M., 1999. A PP2A regulatory subunit positively regulates Ras-mediated signaling during Caenorhabditis elegans vulval induction. Genes Dev. 13: 2562–2569. 10.1101/gad.13.19.2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamboulian S., Choi J.-S., Ahn H.-S., Chang Y.-W., Tyrrell L., et al. , 2010. ERK1/2 mitogen-activated protein kinase phosphorylates sodium channel Na(v)1.7 and alters its gating properties. J. Neurosci. 30: 1637–1647. 10.1523/JNEUROSCI.4872-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P. W., Vulval Development (June 25, 2005), Wormbook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.6.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Stokoe D., Macdonald S. G., Cadwallader K., Symons M., Hancock J. F., 1994. Activation of Raf as a result of recruitment to the plasma membrane. Science 264: 1463–1467. 10.1126/science.7811320 [DOI] [PubMed] [Google Scholar]
- Sun Y., Liu W.-Z., Liu T., Feng X., Yang N., et al. , 2015. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. Res. 35: 600–604. 10.3109/10799893.2015.1030412 [DOI] [PubMed] [Google Scholar]
- Sundaram M., Han M., 1995. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell 83: 889–901. 10.1016/0092-8674(95)90205-8 [DOI] [PubMed] [Google Scholar]
- Sundaram M. V., Canonical RTK-Ras-ERK signaling and related alternative pathways (July 1, 2013), Wormbook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.80.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Sweatt J. D., 2004. Mitogen-activated protein kinases in synaptic plasticity and memory. Curr. Opin. Neurobiol. 14: 311–317. 10.1016/j.conb.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Thomas G. M., Huganir R. L., 2004. MAPK cascade signalling and synaptic plasticity. Nat. Rev. Neurosci. 5: 173–183. 10.1038/nrn1346 [DOI] [PubMed] [Google Scholar]
- Tomida T., Oda S., Takekawa M., Iino Y., Saito H., 2012. The temporal pattern of stimulation determines the extent and duration of MAPK activation in a Caenorhabditis elegans sensory neuron. Sci. Signal. 5: ra76 10.1126/scisignal.2002983 [DOI] [PubMed] [Google Scholar]
- Tomioka M., Adachi T., Suzuki H., Kunitomo H., Schafer W. R., et al. , 2006. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron 51: 613–625. 10.1016/j.neuron.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Topalidou I., Cattin-Ortolá J., Pappas A. L., Cooper K., Merrihew G. E., et al. , 2016. The EARP complex and its interactor EIPR-1 are required for cargo sorting to dense-core vesicles. PLoS Genet. 12: e1006074 10.1371/journal.pgen.1006074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I., Chen P.-A., Cooper K., Watanabe S., Jorgensen E. M., et al. , 2017a The NCA-1 and NCA-2 ion channels function downstream of Gq and Rho to regulate locomotion in Caenorhabditis elegans. Genetics 206: 265–282 (erratum: Genetics 206: 2225) 10.1534/genetics.116.198820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I., Cooper K., Pereira L., Ailion M., 2017b Dopamine negatively modulates the NCA ion channels in C. elegans. PLoS Genet. 13: e1007032 10.1371/journal.pgen.1007032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y., Hirai S. i., Osada S. i., Suzuki A., Mizuno K., et al. , 1996. Protein kinase C activates the MEK-ERK pathway in a manner independent of Ras and dependent on Raf. J. Biol. Chem. 271: 23512–23519. 10.1074/jbc.271.38.23512 [DOI] [PubMed] [Google Scholar]
- Uozumi T., Hirotsu T., Yoshida K., Yamada R., Suzuki A., et al. , 2012. Temporally-regulated quick activation and inactivation of Ras is important for olfactory behaviour. Sci. Rep. 2: 500 10.1038/srep00500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Biesen T., Hawes B. E., Raymond J. R., Luttrell L. M., Koch W. J., et al. , 1996. G(o)-protein alpha-subunits activate mitogen-activated protein kinase via a novel protein kinase C-dependent mechanism. J. Biol. Chem. 271: 1266–1269. 10.1074/jbc.271.3.1266 [DOI] [PubMed] [Google Scholar]
- Vara H., Onofri F., Benfenati F., Sassoè-Pognetto M., Giustetto M., 2009. ERK activation in axonal varicosities modulates presynaptic plasticity in the CA3 region of the hippocampus through synapsin I. Proc. Natl. Acad. Sci. USA 106: 9872–9877. 10.1073/pnas.0900077106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie T. M., Scherle P. A., Strathmann M. P., Slepak V. Z., Simon M. I., 1991. Characterization of G-protein alpha subunits in the Gq class: expression in murine tissues and in stromal and hematopoietic cell lines. Proc. Natl. Acad. Sci. USA 88: 10049–10053. 10.1073/pnas.88.22.10049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. L., Lutz S., Charlie N. K., Vettel C., Ailion M., et al. , 2007. Trio’s Rho-specific GEF domain is the missing Gα q effector in C. elegans. Genes Dev. 21: 2731–2746. 10.1101/gad.1592007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Han M., Guan K. L., 1995. MEK-2, a Caenorhabditis elegans MAP kinase kinase, functions in Ras-mediated vulval induction and other developmental events. Genes Dev. 9: 742–755. 10.1101/gad.9.6.742 [DOI] [PubMed] [Google Scholar]
- Yang F., Sun W., Yang Y., Wang Y., Li C.-L., et al. , 2015. SDF1–CXCR4 signaling contributes to persistent pain and hypersensitivity via regulating excitability of primary nociceptive neurons: involvement of ERK-dependent Nav1.8 up-regulation. J. Neuroinflammation 12: 219–233. 10.1186/s12974-015-0441-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J. H., Chong H., Guan K.-L., Han M., 2004. Modulation of KSR activity in Caenorhabditis elegans by Zn ions, PAR-1 kinase and PP2A phosphatase. EMBO J. 23: 111–119. 10.1038/sj.emboj.7600025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Seger R., 2006. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors 24: 21–44. 10.1080/02699050500284218 [DOI] [PubMed] [Google Scholar]
- You Y., Kim J., Cobb M., Avery L., 2006. Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 3: 237–245. 10.1016/j.cmet.2006.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Koo C. Y., Stebbing J., Giamas G., 2013. The dual function of KSR1: a pseudokinase and beyond. Biochem. Soc. Trans. 41: 1078–1082. 10.1042/BST20130042 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are listed in Tables S1 and S2 and are available upon request. Primers are listed in Table S3. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and supplemental material. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6071009.