Abstract
Flavoproteins can function as hydrophobic sites for vitamin B2 (riboflavin) or, in other structures, with cofactors for catalytic reactions such as glucose oxidation. In this contribution, we report direct observation of charge separation and recombination in two flavoproteins: riboflavin-binding protein and glucose oxidase. With femtosecond resolution, we observed the ultrafast electron transfer from tryptophan(s) to riboflavin in the riboflavin-binding protein, with two reaction times: ≈100 fs (86% component) and 700 fs (14%). The charge recombination was observed to take place in 8 ps, as probed by the decay of the charge-separated state and the recovery of the ground state. The time scale for charge separation and recombination indicates the local structural tightness for the dynamics to occur that fast and with efficiency of more than 99%. In contrast, in glucose oxidase, electron transfer between flavin-adenine-dinucleotide and tryptophan(s)/tyrosine(s) takes much longer times, 1.8 ps (75%) and 10 ps (25%); the corresponding charge recombination occurs on two time scales, 30 ps and nanoseconds, and the efficiency is still more than 97%. The contrast in time scales for the two structurally different proteins (of the same family) correlates with the distinction in function: hydrophobic recognition of the vitamin in the former requires a tightly bound structure (ultrafast dynamics), and oxidation-reduction reactions in the latter prefer the formation of a charge-separated state that lives long enough for chemistry to occur efficiently. Finally, we also studied the influence on the dynamics of protein conformations at different ionic strengths and denaturant concentrations and observed the sharp collapse of the hydrophobic cleft and, in contrast, the gradual change of glucose oxidase.
Dynamical processes in proteins play a crucial role in biological structure–function correlations (1, 2), and one such process is electron transfer (ET). In biological systems, ET reactions are ubiquitous (3, 4), especially in enzymes with redox reactions (5). Flavoproteins with flavin chromophores are examples of such enzymes and are involved in various catalytic processes (6–8). The understanding of ET reactions of flavins in proteins and their redox reactions is critical to the enzyme function.
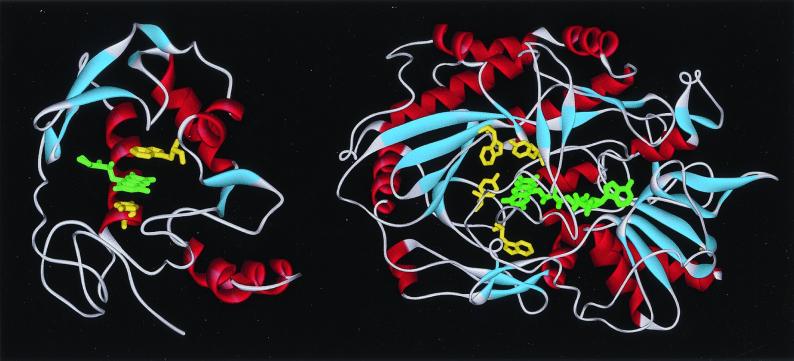
In this contribution, we report our femtosecond studies of ET dynamics of riboflavin (RF; vitamin B2) in RF-binding protein (RfBP) and flavin-adenine-dinucleotide (FAD) in glucose oxidase (GOX) (Fig. 1 and Scheme S1). High-resolution x-ray crystallographic structures of both proteins have recently been reported, and the binding structures of flavins in the proteins are well determined (9–11). RfBP is a globular monomeric protein of approximate dimensions 50 × 40 × 35 Å with a single polypeptide chain of 219 amino acids, nine disulfide bridges, six α-helices, and four series of discontinuous areas of β structure.
Figure 1.
Ribbon diagram of x-ray structures for RF-binding protein (9) (Left) and GOX (10, 11) (Right). The ligand (RF, green), cofactor (FAD, green), and aromatic amino acid residues (yellow) in the binding sites are shown; Trp-156 (Upper) and Tyr-75 (Lower) in the RfBP structure and Tyr-515, Trp-426, Tyr-68, and Trp-111 (counterclockwise from the top) in GOX. Note that a lid, two white and blue ribbons in the front, covers the cofactor FAD in the binding site.
Scheme 1.
Molecular structures of flavin chromophores.
The ligand-binding domain of RfBP is a hydrophobic cleft, ≈20 Å wide and 15 Å deep. The binding of RF occurs in the cleft with the isoalloxazine ring stacked between the parallel planes of tryptophan 156 (Trp-156) and tyrosine 75 (Tyr-75) (see Fig. 1). The isoalloxazine ring of flavins is amphipathic: the xylene portion is hydrophobic, and the pyrimidine moiety is hydrophilic. The x-ray structure reveals that the major interactions of the isoalloxazine ring with the protein are hydrophobic, and the xylene moiety is buried most deeply in the protein. There are also four tryptophan residues (Trp-54, Trp-106, Trp-120, and Trp-124) in the vicinity of the binding site. As shown later, fluorescence quenching of RF is observed when it binds to the protein, due to its interactions with the aromatic amino acid residues.
GOX is a homodimeric glycoprotein with one tight noncovalently bound FAD cofactor per monomer. The monomer with approximate dimensions of 60 × 52 × 37 Å has 583 amino acid residues, 17 α-helices and 30 β-strand structures, and consists of two separate structural domains. The cofactor FAD is situated in the first domain, which consists of two β sheets and three α-helices, and occupies a narrow channel outlined by 31 residues with a covering lid formed by residues 75–98. The main interactions of the FAD molecule with a lightly bent configuration in the channel are 23 potential hydrogen bonds, mostly involving the ribose and pyrophosphate groups. The isoalloxazine ring is rigidly fixed in a nonplanar configuration, at the pyrimidine part by three hydrogen-bonding interactions on O4, N3, and O2 (scheme 1), and at the xylene ring by hydrophobic contacts. The active site for catalysis of β-d-glucose is a large deep pocket, shaped like a funnel and formed on one side by the lid from both subunits of the dimer. In close contact with the isoalloxazine moiety, there are four aromatic residues of Tyr-515, Trp-426, Tyr-68, and Trp-111 (Fig. 1). The observed fluorescence quenching of the FAD molecule is due to its interactions with these residues.
Ultrafast spectroscopic studies have recently been reported in several flavin enzymes (12–16). The fluorescence quenching of both RfBP and GOX was observed to occur on femtosecond and picosecond time scales by measurements of the excited-state flavin (RF* or FAD*) fluorescence decay (16). All quenching dynamics are attributed to an ultrafast ET between flavins and the aromatic residues, tryptophan(s) or tyrosine(s), by a one-electron half-reduction reaction of the flavin. However, no direct evidence of the ET state was observed in these studies, and the ET reaction mechanism was deduced.
With femtosecond resolution, we observed the charge separation and recombination in RfBP and GOX by detecting the ET state and the recovery of the ground state, and studied the nature of the ET dynamics in the proteins. ET reactions of RF and FAD with aromatic residues occur on the femtosecond time scale in RfBP and on the picosecond time scale in GOX, when flavin molecules are excited in the visible region. The charge recombination in RfBP takes place in 8 ps and in GOX it takes much longer times, 30 ps and nanoseconds. These studies elucidate the nature of the interactions at the binding sites and correlate well with the spatial separation between the flavin molecules and the aromatic residues in the proteins.
We also studied the effect of different protein conformations on the ET rates and the binding ability of the proteins to the flavins by increasing the ionic strength or adding denaturants into the protein solution. We observed a decrease of ET rates with the increase of ionic strength or the increase of the protein unfolding. RfBP loses its tertiary structure and the binding ability at 3 M guanidine hydrochloride (Gdn⋅HCl), consistent with the circular dichroism and fluorescence studies, but for the large dimeric GOX, 6 M Gdn⋅HCl was required to unfold the native structure.
Methodology.
Femtosecond-resolved fluorescence up-conversion and transient absorption techniques were used in all experimental measurements reported here. The experimental setup is described in detail elsewhere (17). The pump pulse from an optical parametric amplifier (OPA) was attenuated to 0.1–0.4 μJ per pulse and fixed at 480 nm, whereas for some experiments the pump pulse was tuned to 510 nm. The probe wavelength was set at 795 nm as the gating pulse for up-conversion measurements. For transient absorption experiments, another OPA was used, and the probe pulse was tuned from 310 to 700 nm. All transients were taken at the magic angle (54.7°) of the pump polarization relative to that of the probe pulse (transient absorption), or for the fluorescence (determined by the nonlinear crystal).
Sample Preparation.
Apo RF-binding protein (i.e., without the RF) from chicken egg white and GOX from Aspergillus niger in its holo form (i.e., with the cofactor) were purchased from Sigma. All solvents, spectroscopic grade, were also purchased from Sigma. Ultrapure Gdn⋅HCl was obtained from Baker and sodium chloride (NaCl) from Mallinckrodt. All chemicals were used as supplied.
For most experiments, the lyophilized powder of apo RF-binding protein (apoRfBP) was dissolved in 30 mM phosphate buffer at pH = 7 with a concentration of 400 μM. RF was added into the solution with a concentration of 50 μM, reaching a concentration ratio of 8:1 for apoRfBP to RF. We found that more than 99% of RF molecules bind to the protein, as evidenced by the contribution to our transients of the free RF, which has a nanosecond lifetime (see below). The association constant was reported to be 107-108 M−1 (18, 19). The binding constant of RF with tryptophan alone in the buffer solution is much smaller, because we obtained no direct ET reaction up to 32 mM tryptophan; the quenching of excited-state flavins was observed at 230 mM indole in methanol solution (20).
GOX in holo form in buffer solution (pH = 4) was dialyzed before use and was prepared in 30 mM phosphate buffer at pH = 7 with a dimer concentration of ≈300 μM. For experiments involving the addition of NaCl to increase the ionic strength or Gdn⋅HCl to unfold the protein, the mixed samples were used after more than 3-h mixing to assure that the solution was at equilibrium. All samples were measured in a fused silicon cell with 1- to 3-mm path length and at ambient condition; no photobleaching was observed.
The steady-state absorption and fluorescence emission of flavins have been extensively studied (21–25). The absorption peak is at ≈450 nm, and the emission has a maximum at ≈530 nm. After the complexation of RF with apoRfBP, we found no apparent change in the absorption spectrum, and the fluorescence emission slightly shifts to the blue with a huge decrease, by several orders of magnitude, in the total intensity (at 480 nm excitation). GOX has similar spectroscopic features to those observed in RfBP.
Results and Discussion
Ultrafast Charge Separation.
All femtosecond-resolved fluorescence transients of RF in RfBP and FAD in GOX are observed to be independent of the gated fluorescence wavelengths, ranging from 510 to 670 nm. Fig. 2 shows typical temporal behaviors for two wavelengths at the blue and red ends. It is striking that all transients show ultrafast dynamics on the femtosecond to picosecond time scale with a minor component (<1–3%) in the nanosecond range. The free RF and FAD molecules in buffer water at pH = 7 have been characterized (22, 26–28). Basically, RF molecules in the first excited state have a lifetime of ≈5 ns with a quantum yield of 0.26 and the rotational anisotropy decays in ≈130 ps. With picosecond resolution, FAD shows a biexponential behavior with lifetimes of 310 ps (28%) and 2.82 ns (72%), attributed to an equilibrium between normal and closed conformations in the ground state in neutral aqueous solution; the closed form is an intramolecular complex with a stacking interaction between the isoalloxazine ring and the adenine moiety (27). With better time resolution, a ≈4 ps component was resolved by transient-absorption measurements and was shown to be due to intramolecular ET from adenine to isoalloxazine ring from studies of the folded and unfolded structures in the presence/absence of formamide (28).
Figure 2.
Femtosecond normalized fluorescence up-conversion decays of RF-binding protein (Left) and GOX (Right). Two wavelengths of detection at the blue and red ends are shown. All transients for each protein are similar, independent of the detected wavelength.
The fluorescence emission of RF and FAD depends on solvent polarity, as evidenced by the Stokes shift (22). At 480-nm excitation, our study indicates that solvation (and/or vibrational relaxation) occurs in less than 1 ps. For RF molecules, an ultrafast decay component at shorter wavelength (510 nm) and the corresponding rising components at longer wavelength (670 nm) were observed in addition to the nanosecond component. At 530 nm, we observed the dominant relaxed-state emission as the nanosecond component. Thus, the observed nanosecond component (<1%) in the protein results from the free flavin molecules in buffer water, equilibrated with bound flavins.
For the two proteins studied, the fluorescence transients (Fig. 2) at the red end do not show any apparent rise, because solvation in the proteins becomes slower, mainly due to hydrophobic interactions (29, 30). Ultrafast decay dominates the observed transients: For RfBP, the transients decay ≈100 fs (≈85%) and 700 fs (≈15%); for GOX, the decay time constants become 1.8 ps (≈72%) and 10 ps (≈26%). Studies at 410-nm excitation by Mataga's group (16) for RfBP gave 94 fs (94.7%), 1.02 ps (4%), and 10 ps (1.3%), and for GOX, 413 fs (45%), 1.85 ps (37.5%) and 6.02 ps (17.5%). Our excitation (480 nm) is at the red shoulder of the absorption, and the effect of vibrational relaxation is minimized. For all wavelengths detected, the fluorescence transients are well represented using only two decay constants.
The observed decay of the excited-state flavins is the result of the ET between the isoalloxazine ring and aromatic residues in the protein to form anionic semiquinone flavin and cationic residues. Resonance energy transfer is ruled out because of no spectral overlap between flavin emission (>480 nm) and protein absorption (<300 nm). Even in dimeric GOX, the ultrafast decay is not from the interactions between two FAD molecules, because two flavin chromophores are situated at a distance of more than 20 Å. The collision quenching by diffusion control occurs on a much longer time scale (>nanoseconds). The proton-transfer quenching mechanism is excluded, because no intermolecular proton transfer was observed in H2O.
The redox property of flavin and Trp gives a favorable driving force for the ET reaction. For example, the potential for flavin/flavin⋅− is ≈−0.8 V vs. NHE in aprotic hydrophobic environments (31), and for Trp/Trp⋅+, it is ≈+1.15 V vs. NHE (32). Knowing the 0–0 transition (S1←S0) energy of ≈2.35 eV, we obtained a net ΔG° of −0.4 eV. The observed difference in ET rates for RfBP and GOX is mainly from the spatial stacking structures between the flavins and the aromatic residues. For RfBP, RF is sandwiched between Trp-156 and Tyr-75 in a compact π-stacked structure with an equal interplanar distance of 3.7 Å. Because the oxidation potential for Tyr is higher than for Trp, the quenching dynamics observed within 100 fs dominantly results from the ET process from Trp-75 to RF. Similar ET quenching of the excited-state daunomycin (an antitumor drug) in the hydrophobic cleft of apoRfBP was observed within 1 ps (73%) and 6 ps (24%) from aromatic residues to the drug ligand (33). In GOX, the FAD molecule is surrounded by four aromatic residues, Tyr-515, Trp-426, Tyr-68, and Trp-111 (Fig. 1). None of four residues compactly stacks with FAD. Two tyrosine residues are in closer proximity, and the ET process is favorable. On the other hand, the higher oxidation potential of tyrosine results in a longer reaction time. The observed ET reaction times of 1 ps and 10 ps thus reflect the convoluted interactions between FAD and four surrounding aromatic residues.
Dynamics of Charge Recombination.
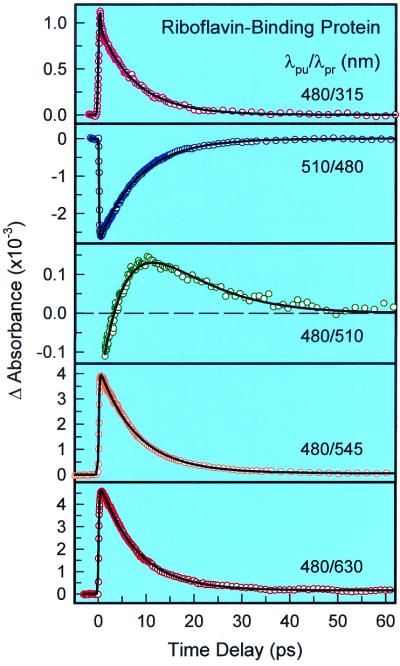
To fully elucidate the ET mechanism, we performed a series of transient absorption measurements to observe the ET state and the ground-state recovery of flavins. Fig. 3 gives five transients probed in the range of 315–630 nm for RfBP. It is very striking that all observed transients are dominated by the ET-state absorption and the ground-state recovery, not by the neutral excited-state absorption. Specifically, at 630 and 545 nm, two transients are nearly identical and can be best fitted to two rising components, 100 fs (within our resolution) and 700 fs, and a single decay component of 7.5 ps. The rise signal represents the formation of the ET state, consistent with the fluorescence up-conversion measurements. The decay component reflects the decay of the ET-state population—a charge recombination process. Clearly, at longer than 545 nm, the absorption of the ET state is dominant, consistent with the earlier observation of the anionic semiquinone flavin (34, 35) and the tryptophan cation (36) at steady state.
Figure 3.
Femtosecond transient absorption profiles of RF-binding protein probed at different wavelengths, from 315 to 630 nm. Note the positive and negative absorbance change with time.
At 510 nm, both the ET state and the ground state have comparable absorption coefficients, and the observed transient is a superposition of the ET-state dynamics and the ground-state recovery. The RF⋅−-Trp⋅+ complex decays in 7.5 ps, and the ground-state recovery of RF is about 9 ps. The slightly longer time for the recovery probably includes the ground-state vibration relaxation after charge recombination. At 480 nm (λpu = 510 nm), the ground-state absorption is dominant, and the ground-state recovery of RF was observed to occur in ≈8.5 ps. When the probe was tuned to 315 nm, we observed two decay components: one occurs in ≈100 fs, reflecting the excited-state decay of RF*-Trp, and the other decays in 7.5 ps, representing the deactivation of the RF⋅−-Trp⋅+ complex. All results observed in RfBP are well correlated with the absorption evolution of the excited-state RF*-Trp, the ET state of RF⋅−-Trp⋅+ and the recovering ground state of RF molecules.
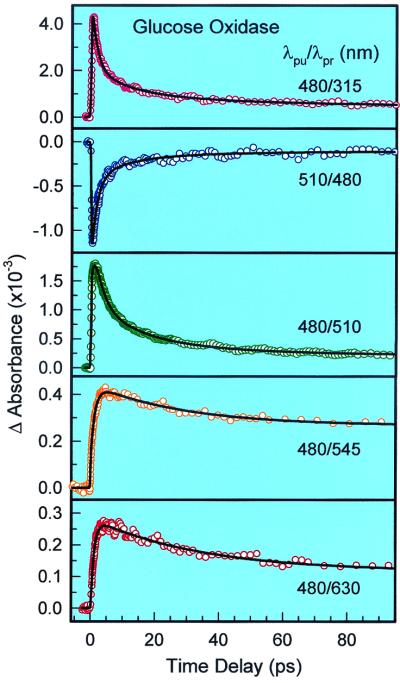
For GOX, the overall transient absorption profiles, as shown in Fig. 4, are similar to those observed in RfBP. At the longer wavelength, larger than 545 nm, the signal is dominated by the ET-state absorption. We observed a rise time of ≈1.4 ps and two decay components with time constants of ≈30 ps and nanoseconds or longer. The ET-state formation time is comparable to the excited-state FAD*-Tyr/Trp decay time; the observed new 30 ps and nanosecond components, which are present in all observed transients (Fig. 4), are the decay times of the FAD⋅−-Tyr⋅+/Trp⋅+ complexes. This disparity may reflect two sites of ET: charge recombination of the FAD⋅−-Trp⋅+ complex in ≈30 ps and the FAD⋅−-Tyr⋅+ complex in more than a nanosecond; following the initial ET, the latter can undergo a proton transfer from the oxidized tyrosine radical to the reduced FAD anion to form a neutral semiquinone FAD, and the final recombination of both electron and proton takes a much longer time. However, the presence of O2 could also be important, as discussed elsewhere (X. Qu, C. Wan, H.-C. Becker, D.Z., and A.H.Z., unpublished work) and in the accompanying paper (33).
Figure 4.
Femtosecond transient absorption profiles of GOX probed at different wavelengths, from 315 to 630 nm. Note the positive and negative absorbance change with time.
At 510 nm, the absorption is positive, and the signal represents a superposition of both excited-state FAD*-Tyr/Trp decay (≈1.7 ps) and the ET-state decay (35 ps and nanosecond decay). When the probe was tuned close to the maximum absorption of the ground state at 480 nm (λpu = 510 nm), the net signal was observed to be negative and manifests the ground-state recovery (35 ps and nanosecond formation). At 315 nm, similar to that obtained in RfBP, the absorption profile shows again both the neutral excited-state and the ET-state dynamics.
It should be noted that the contribution of the stimulated emission process to our observed transients is negligible. This is because the absorption signal from stimulated emission is negative and must rise with the lifetime of the neutral excited state (i.e., ≈100 fs for RF and ≈1.8 ps for FAD), and this was not observed in our experiments. Furthermore, when the probe is at a shorter wavelength to the pump, as in the case of 510-nm excitation and 480-nm probing, the stimulated emission process cannot occur because of energetics; note that we detected the signal on the 480-nm transmission and therefore identify the probe from the pump, as is also consistent with the pump–probe timing.
Conformation vs. ET Rate.
Because the ET rates between flavins and aromatic residues are sensitive to their relative positions in the protein, i.e., the tertiary structure of the protein, we studied the ET reaction following changes of protein conformation by adding two different salts into the protein solution. Fig. 5 shows our observed results. By increasing the ionic strength to 4 M NaCl, the transients (Fig. 5A) for both proteins show somewhat longer reaction times. But the percentage of the nanosecond components from the free flavins does not increase, indicating no flavins were repelled out of the binding site due to the conformation change. The time constants in RfBP become 170 and 800 fs at 4 M instead of ≈100 and 700 fs at 0 M; for GOX, they lengthen to 3 and 16 ps from 1.8 and 10 ps, respectively. However, the ratio of two components does not change in both proteins on adding NaCl.
Figure 5.

(A) Femtosecond fluorescence up-conversion decays of GOX and RfBP (Inset) with and without 4 M NaCl. The transients are similar except for the somewhat longer reaction times under the salt condition (see text). (B) Fluorescence decays of RfBP with a series of concentrations of the denaturant, Gdn⋅HCl, from 0 to 3 M. Note that the transient at 3 M Gdn⋅HCl is identical to that obtained for free RF molecules in buffer solution. The percentage of nanosecond components shows an abrupt increase between 2.5 and 3 M Gdn⋅HCl. (C) Fluorescence decays of GOX with a series of concentrations of Gdn⋅HCl from 0 to 6 M. The percentage of nanosecond components shows a gradual increase from 0.3 to 6 M Gdn⋅HCl, contrary to those observed in RfBP (B). All transients were detected at 530 nm (fluorescence).
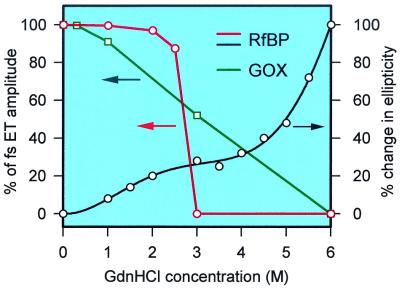
The contrast with the addition of Gdn⋅HCl is striking, and the results are shown in Fig. 5 B and C for a series of concentrations. When the concentration reaches 3 M in RfBP solution (Fig. 5B), all RF molecules behave like free molecules (nanosecond decay), and no ultrafast ET reaction was observed. It has been reported that the unfolding of the native RfBP protein occurs in two phases, from 0 to 6 M Gdn⋅HCl, as measured by circular dichroism and fluorescence spectra (37); see Fig. 6. Between 0–2 M Gdn⋅HCl, the protein loses much of its tertiary structure as well as the ability to bind RF, but retains most of its secondary structure. At the high concentration of 4–6 M Gdn⋅HCl, the protein loses its secondary structure and probably exists as an intermediate conformation between 2–4 M Gdn⋅HCl. However, our results show that with 2 M Gdn⋅HCl, only 3% free molecules of RF were observed, and the hydrophobic cleft does not completely collapse; even at 2.5 M Gdn⋅HCl, the nanosecond component accounts for only 13%. Thus, between 2.5 and 3 M Gdn⋅HCl, the observed nearly discontinuous increase of nanosecond components indicates that a partial unfolding of the protein strongly destabilizes the remaining structure. Therefore, in this critical concentration region, the collapse of the hydrophobic cleft or the loss of the tertiary structure must simultaneously occur, suggesting that the protein unfolds in a cooperative manner (Fig. 6).
Figure 6.
Amplitude percentages of electron-transfer components change with the concentration of denaturant Gdn⋅HCl. Note the dramatic difference for the two proteins (RfBP and GOX) studied here. The percentage change in ellipticity at 225 nm for RfBP, reproduced from ref. 37, is also shown. The ET measurements are sensitive to the local binding function sites, whereas the circular dichroism studies reflect the average degree of high-order structures in protein.
At 1 M Gdn⋅HCl, the transient is nearly identical to that without Gdn⋅HCl, indicating the unchanged local hydrophobic structure. Between 1 and 3 M Gdn⋅HCl, all transients are best fit by two fast decay times of 100–170 fs and 0.7–1.2 ps with a nanosecond component. The ratio of the first two components remains nearly the same, around 5–6. These results suggest that the decay times of 170 fs and 1.2 ps are the upper limits of two ET reactions for the two ET complex configurations. Beyond a certain distance between RF* and Trp/Tyr, the direct ultrafast ET reactions could not proceed. This observation is also consistent with the results observed with 4 M NaCl, as discussed above.
For GOX, the transients (Fig. 5C) gradually change with the denaturant concentrations. All transients are best fit by two fast decay times of 1.4–3 ps and 10–12 ps with a nanosecond component. The ratio of the two fast components gradually decreases from 3 at 0 M to 1 at 6 M Gdn⋅HCl. Interestingly, at 0.3 M Gdn⋅HCl, we even observed faster initial dynamics (1.4 ps) than without Gdn⋅HCl (1.8 ps), as clearly shown in Fig. 5C. These results are consistent with the large size of the protein (molecular weight ≈63,263), its dimeric state, and high-order structures. Even though the system is complex, the upper limits for ET-reaction times, 3 and 16 ps, can be determined, and the influence of its conformation change can be detected (Fig. 6).
Conclusion
The reported studies of the dynamics with femtosecond time resolution of RF-binding protein and GOX enzyme elucidate the crucial role of ET in the hydrophobic cleft, for the former, and in the cofactor, for the latter. In bulk solution, ET between flavins and tryptophan/tyrosine is diffusion-controlled, but in the protein it is determined by its native tertiary structure. The initial steps in these flavin enzymatic reactions are efficient and ultrafast in nature, a characteristic of many photoactive biological systems (X. Qu, C. Wan, H.-C. Becker, D.Z., and A.H.Z., unpublished work; refs. 38–42).
The electron-transfer and recombination rates reported here correlate well with the spatial stacking structures in the binding sites and with the function. In thermal reactions, the redox pathway involves the reported charge-separated state, in addition to others, but with slower rates; the initial photon excitation enhances the entry to these states, and, therefore, the lifetime of the ion-pair state is critical to the subsequent redox chemistry. In the case of GOX, in the dimer channel, the sugar is ultimately oxidized, and O2 is reduced to H2O2 (43).
The direct observation of charge recombination elucidates another important point, the efficient reaction cycling in the protein under repeated light irradiation. The flavoproteins studied here are stable for rather long irradiation by laser pulses in our experiments. Contrary to this observation, the flavin molecules undergo fast photodecomposition when they are irradiated in solution in presence of high concentration of quenchers, such as tryptophan (or indole) or tyrosine (or phenol) (16). Such characteristics of the stable protein photocycle, determined by the ingenious function of the native local binding structure, are most important to photoresponsive proteins working in photobiological reactions such as in photosynthesis and various photosensory actions.
As shown here, the study of the dynamics and its dependence on protein conformations, by increasing the ionic strength or the concentration of denaturants, elucidates the local ligand-binding structures and the interior properties of proteins (44) and reveals the nature of unfolding processes. Here, we observed the “sharp” and “soft” transitions as proteins unfold, and this must reflect the critical influence of ionic strength and conformations on the rates of reactions; theoretical modeling must consider these time scales of the reactions and of the conformation changes. The successful detection of the oxidized and reduced forms of flavins on the femtosecond time scale opens the door to studies of the enzymatic reactions of flavoproteins. In the accompanying paper (33), we report studies of the same protein, RF-binding protein, but now in the presence of the drug daunomycin.
Acknowledgments
We thank Prof. Hugo L. Monaco (University of Verona, Italy) for providing the RF-binding protein coordinates. Special thanks also to Drs. Chaozhi Wan and Samir Pal for their discussion and help. This work was supported by the National Science Foundation.
Abbreviations
- RF
riboflavin
- FAD
flavin-adenine-dinucleotide
- RfBP
riboflavin-binding protein
- GOX
glucose oxidase
- ET
electron transfer
- guanidine hydrochloride
Gdn⋅HCl
References
- 1.Smith T L, editor. Nat Struct Biol. 2000;7:711–743. doi: 10.1038/76741. [DOI] [PubMed] [Google Scholar]
- 2.Zewail A H. Angew Chem Int Ed. 2000;39:2587–2631. [Google Scholar]
- 3.Balzani V, editor. Electron Transfer in Chemistry. Weinheim, Germany: Wiley–VCH; 2001. [Google Scholar]
- 4.Bendall D S, editor. Protein Electron Transfer. Oxford, U.K.: Bios Scientific; 1996. [Google Scholar]
- 5.Sinnott M, editor. Comprehensive Biological Catalysis: A Mechanistic Reference. San Diego: Academic; 1998. [Google Scholar]
- 6.Müller F, editor. Chemistry and Biochemistry of Flavoenzymes. Boca Raton, FL: CRC Press; 1991. [Google Scholar]
- 7.Ghisla S, Kroneck P, Macheroux P, Sund H, editors. Flavins and Flavoproteins. Berlin: Agency for Scientific Pub.; 1999. [Google Scholar]
- 8.Massey V. Biochem Soc Transact. 2000;28(part 4):283–296. [PubMed] [Google Scholar]
- 9.Monaco H L. EMBO J. 1997;16:1475–1483. doi: 10.1093/emboj/16.7.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecht H J, Kalisz H M, Hendle J, Schmid R D, Schomburg D. J Mol Biol. 1993;229:153–172. doi: 10.1006/jmbi.1993.1015. [DOI] [PubMed] [Google Scholar]
- 11.Wohlfahrt G, Witt S, Hendle J, Schomburg D, Kalisz H M, Hecht H J. Acta Crystallogr D. 1999;55:969–977. doi: 10.1107/s0907444999003431. [DOI] [PubMed] [Google Scholar]
- 12.Kim S T, Heelis P F, Okamura T, Hirata Y, Mataga N, Sancar A. Biochemistry. 1991;30:11262–11270. doi: 10.1021/bi00111a011. [DOI] [PubMed] [Google Scholar]
- 13.van den Berg P A W, van Koek A, Walentas C D, Perham R N, Visser A J W G. Biophys J. 1998;74:2046–2058. doi: 10.1016/S0006-3495(98)77911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enescu M, Lindqvist L, Soep B. Photochem Photobiol. 1998;68:150–156. doi: 10.1562/0031-8655(1998)068<0150:esdofr>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Aubert C, Vos M H, Mathis P, Eker A P M, Brettel K. Nature (London) 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- 16.Mataga N, Chosrowjan H, Shibata Y, Tanaka F, Nishina Y, Shiga K. J Phys Chem B. 2000;104:10667–10677. [Google Scholar]
- 17.Fiebig T, Wan C, Kelley S O, Zewail A H. Proc Natl Acad Sci USA. 1999;96:1187–1192. doi: 10.1073/pnas.96.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murthy U S, Podder S K, Adiga P R. Biochim Biophys Acta. 1976;434:69–81. doi: 10.1016/0005-2795(76)90036-2. [DOI] [PubMed] [Google Scholar]
- 19.Nishikami M, Kyogoku Y. J Biochem (Tokyo) 1973;73:1233–1242. doi: 10.1093/oxfordjournals.jbchem.a130196. [DOI] [PubMed] [Google Scholar]
- 20.Karen A, Sawada M T, Tanaka F, Mataga N. Photochem Photobiol. 1987;45:49–53. [Google Scholar]
- 21.Weber G. Biochem J. 1950;47:114–121. doi: 10.1042/bj0470114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koziol J. Methods Enzymol. 1971;18(part B):253–284. [Google Scholar]
- 23.Sun M, Moore T A, Song P S. J Am Chem Soc. 1972;94:1730–1740. doi: 10.1021/ja00760a052. [DOI] [PubMed] [Google Scholar]
- 24.Ghisla S, Massey V, Lhoste J M, Mayhew S G. Biochemistry. 1974;13:589–597. doi: 10.1021/bi00700a029. [DOI] [PubMed] [Google Scholar]
- 25.Rivlin R S, editor. Riboflavin. New York: Plenum; 1975. [Google Scholar]
- 26.Visser A J W G. Fluorescent Biomolecules: Methodologies and Applications. New York: Plenum; 1989. pp. 319–342. [Google Scholar]
- 27.Barrio J R, Tolman G L, Leonard N J, Spencer R D, Weber G. Proc Natl Acad Sci USA. 1973;70:941–943. doi: 10.1073/pnas.70.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanley R J, MacFarlane IV A W. J Phys Chem A. 2000;104:6899–6906. [Google Scholar]
- 29.Nandi N, Bhattacharyya K, Bagchi B. Chem Rev. 2000;100:2013–2045. doi: 10.1021/cr980127v. [DOI] [PubMed] [Google Scholar]
- 30.Zhong D, Pal S K, Zewail A H. CHEMPHYSCHEM. 2001;2:219–227. doi: 10.1002/1439-7641(20010417)2:4<219::AID-CPHC219>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 31.Niemz A, Imbriglio J, Rotello V M. J Am Chem Soc. 1997;119:887–892. [Google Scholar]
- 32.DeFelippis M R, Murthy C P, Broitman F, Weinraub D, Faraggi M, Klapper M H. J Phys Chem. 1991;95:3416–3419. [Google Scholar]
- 33.Zhong D, Pal S K, Wan C, Zewail A H. Proc Natl Acad Sci USA. 2001;98:11873–11878. doi: 10.1073/pnas.211440298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massey V, Palmer G. Biochemistry. 1966;5:3181–3189. doi: 10.1021/bi00874a016. [DOI] [PubMed] [Google Scholar]
- 35.Land E J, Swallow A J. Biochemistry. 1969;8:2117–2125. doi: 10.1021/bi00833a050. [DOI] [PubMed] [Google Scholar]
- 36.Shida T. Electronic Absorption Spectra of Radical Ions. New York: Elsevier; 1988. [Google Scholar]
- 37.Allen S, Stevens L, Duncan D, Kelly S M, Price N C. Int J Biol Macromol. 1992;14:333–337. doi: 10.1016/s0141-8130(05)80074-1. [DOI] [PubMed] [Google Scholar]
- 38.Vos M H, Martin J-L. Biochim Biophys Acta. 1999;1411:1–20. doi: 10.1016/s0005-2728(99)00035-3. [DOI] [PubMed] [Google Scholar]
- 39.Sundström V. Prog Quant Electro. 2000;24:187–238. [Google Scholar]
- 40.Liebl U, Lipowski G, Negrerie M, Lambry J-C, Martin J-L, Vos M H. Nature (London) 1999;401:181–184. doi: 10.1038/43699. [DOI] [PubMed] [Google Scholar]
- 41.Wang Q, Schoenlein R W, Peteanu L A, Mathies R A, Shank C V. Science. 1994;266:422–424. doi: 10.1126/science.7939680. [DOI] [PubMed] [Google Scholar]
- 42.Zhu L, Sage J T, Champion P M. Science. 1994;266:629–632. doi: 10.1126/science.7939716. [DOI] [PubMed] [Google Scholar]
- 43.Gibson Q H, Swoboda B E P, Massey V. J Biol Chem. 1964;239:3927–3934. [PubMed] [Google Scholar]
- 44.Zhong D, Douhal A, Zewail A H. Proc Natl Acad Sci USA. 2000;97:14056–14061. doi: 10.1073/pnas.250491297. . (First Published December 5, 2000; 10.1073/pnas.250491297) [DOI] [PMC free article] [PubMed] [Google Scholar]