Abstract
The binding of tumor necrosis factor alpha (TNF-α) to the type-1 TNF receptor (TNFRc1) plays an important role in inflammation. Despite the clinical success of biologics (antibodies, soluble receptors) for treating TNF-based autoimmune conditions, no potent small molecule antagonists have been developed. Our screening of chemical libraries revealed that N-alkyl 5-arylidene-2-thioxo-1,3-thiazolidin-4-ones were antagonists of this protein–protein interaction. After chemical optimization, we discovered IW927, which potently disrupted the binding of TNF-α to TNFRc1 (IC50 = 50 nM) and also blocked TNF-stimulated phosphorylation of Iκ-B in Ramos cells (IC50 = 600 nM). This compound did not bind detectably to the related cytokine receptors TNFRc2 or CD40, and did not display any cytotoxicity at concentrations as high as 100 μM. Detailed evaluation of this and related molecules revealed that compounds in this class are “photochemically enhanced” inhibitors, in that they bind reversibly to the TNFRc1 with weak affinity (ca. 40–100 μM) and then covalently modify the receptor via a photochemical reaction. We obtained a crystal structure of IV703 (a close analog of IW927) bound to the TNFRc1. This structure clearly revealed that one of the aromatic rings of the inhibitor was covalently linked to the receptor through the main-chain nitrogen of Ala-62, a residue that has already been implicated in the binding of TNF-α to the TNFRc1. When combined with the fact that our inhibitors are reversible binders in light-excluded conditions, the results of the crystallography provide the basis for the rational design of nonphotoreactive inhibitors of the TNF-α–TNFRc1 interaction.
Tumor necrosis factor alpha (TNF-α) is a pleiotropic cytokine that is produced predominantly by activated macrophages and lymphocytes and plays a central role in inflammation (1). The soluble form of TNF-α exists as a homotrimeric aggregate of 17-kDa subunits, and is capable of binding to at least two different receptors: TNF Receptor-1 (TNFRc1, or p55; KD = 500 pM) and TNF Receptor-2 (TNFRc2, or p75; KD = 100 pM). Both TNF-α and its two receptors are members of two large families of proteins that contain 18 and 26 members, respectively (2). TNFRc1 is constitutively expressed on cells at low levels, whereas the amount of TNFRc2 is inducibly regulated both transcriptionally and posttranscriptionally. Because of the more rapid association and dissociation of TNF-α with TNFRc2, it has been proposed that TNFRc2 serves to increase the local concentration of TNF-α at the cell surface, thereby facilitating the binding of TNF-α to TNFRc1 (3). Binding of TNF-α to either receptor induces trimerization of that receptor. In the case of the TNFRc1, it is this TNF-mediated receptor clustering that triggers signaling pathways leading to both apoptosis (via caspase-8) and cell activation (via NF-κB) (4); the balance between these opposing signals appears to depend on the cellular context in which the stimulus occurs. From the perspective of inflammation, it is the NF-κB pathway that is relevant, because NF-κB-mediated transcription leads to the perpetuation of the inflammatory cascade through the release of proinflammatory cytokines such as IL-6 and -8. Thus, the overproduction of TNF-α by monocytes and macrophages can cause unregulated activation of the immune system, resulting in extensive tissue damage.
The physiological relevance of these cellular events has been clearly demonstrated by the striking success of biological agents (e.g., anti-TNF antibodies, soluble TNFRc2 fusion protein) that sequester TNF-α in the treatment of human autoimmune diseases (e.g., rheumatoid arthritis, Crohn's disease) (5). Although several small molecules that block the production of soluble TNF-α are in clinical development (6), the only chemical inhibitors of the interaction between TNF-α and its cognate receptors are high molecular weight peptides (ref. 7; ref. 8 and references therein) and small molecules (ref. 9 and references therein) that act by dissociating the TNF-α trimer at micromolar concentrations. The intractability of the TNF-α–TNFRc1 interaction to disruption by small organic compounds is perhaps not surprising, given that the receptor–ligand binding event capitalizes on avidity effects (10) and also utilizes a large surface area for interaction (11, 12). Herein we disclose four classes of small molecules that are potent (50–500 nM) inhibitors of the TNF-α–TNFRc1 interaction. Detailed biochemical studies revealed that these compounds are actually reversible micromolar inhibitors (40–100 μM) that covalently modify the TNFRc1 in a light-dependent fashion.
Materials and Methods
Biology.
TNF-α–TNFRc1 binding assay.
High protein-binding 96-well plates (Dynex Technologies, Chantilly, VA) were coated overnight with 20 ng of human recombinant TNFRc1 per well (R & D Systems) in 50 mM Na2CO3, pH 9.6. The following morning, wells were blocked with 1% BSA in PBS for 1 h at room temperature (RT). Europium-labeled human TNF-α (Eu-TNF-α, Wallac, Gaithersburg, MD), which had been diluted in PBS containing 0.1% BSA, was added at a final concentration of 2.4 nM. Compounds were serially diluted in DMSO and added to give a final DMSO concentration of 1%. The plates were incubated for 1 h at RT and then washed five times with PBS before adding 100 μl of enhancement solution (Wallac). Following incubation at RT for an additional 10 min, fluorescence was measured with a Victor fluorimeter (Wallac). Background values were based on TNF-α binding to wells lacking TNFRc1 and were subtracted. All compounds were tested in triplicate wells, and compound inhibition was determined relative to wells incubated with Eu-TNF-α and 1% DMSO.
Iκ-B phosphorylation assay.
Compounds were preincubated with 2 × 106 Ramos cells in 0.5 ml of serum free RPMI medium 1640 (GIBCO) for 15 min at RT before the addition of 2 ng/ml recombinant human TNF-α (R & D Systems). The samples were then incubated for 5 min at 37°C followed by centrifugation for 10 sec at 10,000 rpm (8,000 × g). The cell pellet was resuspended in 50 μl of ice-cold buffer (10 mM Tris, pH 7.2/150 mM NaCl/1% Triton X-100/1% sodium deoxycholate/0.1% SDS/1 mM PMSF/50 mM sodium fluoride/1 mM sodium orthovanadate/50 μg/ml aprotinin/50 μg/ml leupeptin). Cell debris was pelletted at 14,000 rpm (16,000 × g) for 5 min. The cell extract (20 μg) was analyzed by SDS/PAGE on a 12% Tris-glycine gel, transferred to poly(vinylidene difluoride) (PVDF) membrane, and immunoblotted with a 1:1000 dilution of the anti-phospho Iκ-B rabbit polyclonal antibody (New England Biolabs). Subsequently, membranes were incubated with a 1:2000 dilution of goat anti-rabbit IgG peroxidase conjugated antibody (New England Biolabs), followed by chemiluminescent detection (Pierce). Blots were quantitated by scanning densitometry and compound inhibition of Iκ-B phosphorylation was determined relative to TNF-stimulated controls.
TNF-α–TNFRc2 binding assay.
Binding of TNF-α to TNFRc2 was determined as described above for the TNF-α–TNFRc1 assay, except that 10 ng of recombinant human TNFRc2/Fc per well (Immunex) was used as the source of receptor.
CD40L binding assay.
Recombinant human CD40/Fc (Ancell, Bayport, MN) at a final concentration of 10 ng per well was coated overnight in 96-well plates (Dynex Technologies) as described above. The following morning, the plates were blocked for 1 h with Superblock (Pierce). Diluted compounds and europium-labeled human CD40 ligand (5 ng/ml in Superblock + 0.05% Tween-20, Wallac) were added and the plates were incubated for 2.5 h at RT. The plates were washed three times with Tris-buffered saline, pH 7.4, containing 0.05% Tween-20. Enhancement solution was added and fluorescence detected as described for the TNF-α binding assay.
MTS toxicity assay.
Ramos cells were seeded at 3 × 104 cells per well in serum free AIM V media (GIBCO/BRL) in 96-well plates with various concentrations of compound. After incubation for 24 h at 37°C, 3-(4,5-dimethylthiazol-2-yl)-5-3-carboxymethoxyphenyl)2-(4-sulfophenyl)-2H-tetrazolium salt (MTS) (Promega) was added to each well at a final concentration of 333 μg/ml. Cell viability was determined after an additional 3-h incubation at 37°C by reading the plates at 490 nM absorbance. All compounds were tested in triplicate wells and compound inhibition was determined relative to cells cultured with 1% DMSO alone.
TNF-α–TNFRc1 membrane binding assay.
Where noted, membranes isolated from Chinese hamster ovary (CHO) cells stably transfected with a C-terminal tail deletion mutant of TNFRc1 (iTNFRc1) were used as a source of TNFRc1 (13). In these experiments, plates were coated overnight with 10 ng of TNFRc1-CHO membranes per well and the assay performed as described above, using Eu-TNF-α (Wallac) at a final concentration of 1 nM.
X-ray Crystallography.
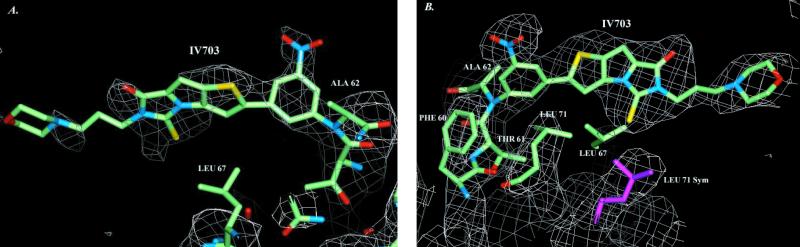
Tetragonal crystals of soluble TNFRc1 (amino acids 11–172; R & D Systems) were grown as described (14). Crystals were transferred to a stabilizing solution (100 mM Tris, pH 8.5/500 mM ammonium acetate/60% (vol/vol) 2-methyl-2,4-pentanediol/0.1% β-octyl glucoside/100 mM NaCl) and soaked with 0.5 mM IV703 (5% DMSO) exposed to light for 2 days. Crystals were then flash frozen in liquid nitrogen. Diffraction data were collected in a nitrogen cryostream (−170°C) at the DuPont Northwestern Dow–Collaborative Access Team beamline, Advanced Photon Source, Argonne National Laboratories, Argonne, IL, and were processed and scaled with hkl (15). Crystals were tetragonal and belonged to the space group P41212 with unit cell dimensions a = b = 67.8 Å and c = 190.0 Å. Data were 88% complete to 2.9 Å resolution with an Rmerge of 9.3%.¶ Molecular replacement using the program epmr (16) was carried out by using a previously determined in-house structure of TNFRc1 as an initial phasing model (J.A.M., unpublished results). Rigid body refinement followed by Powell minimization and simulated annealing was carried out by using X-PLOR (17). The 2Fo − Fc map showed electron density for IV703 near Ala-62 (Fig. 4), and clearly suggested a covalent attachment between the nitrophenyl ring of IV703 and the backbone nitrogen of Ala-62 at only one monomer of the TNFRc1 homodimer (18). Refinement (17) of a covalent model between the backbone nitrogen of Ala-62 (one monomer) and the meta carbon of the nitrophenyl ring of IV703 resulted in an Rfactor‖ of 27.5% at 2.9 Å and root-mean-square deviations (rmsd) in bond lengths and bond angles of 0.013 Å2 and 3.1°, respectively (PDB ID code 1FT4).
Figure 4.
Single crystal structure of covalent IV703–TNFRc1 complex. (A) Shown is the 2Fo − Fc electron density map (contoured to the 1.25 σ level) used to locate IV703. Fo values were from data of TNFRc1–IV703-soaked crystals, and Fc values were calculated from a TNFRc1 model without IV703. The structure shown is the TNFRc1–IV703 covalent structure to illustrate the fit between the final structural model (cf. B) and the initial electron density map. (B) A 2Fo − Fc electron density map (contoured at 1.5 σ) of the refined structure (2.9 Å resolution, Rfactor = 27.5%; PDB ID code 1FT4) rotated 180° relative to A. Labeled receptor residues make van der Waals contacts with IV703.
Results
Inhibitor Discovery and Optimization.
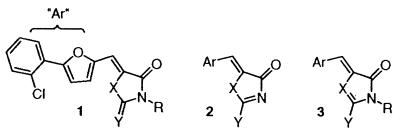
We began our search for small molecule inhibitors of the TNF-α–TNFRc1 interaction with a limited screen of our proprietary library of chemical compounds. This provided us with moderately potent leads (IC50's = 2–50 μM for blocking TNF-α binding), all of which contained the rhodanine heterocycle (e.g., Table 1, 1; ref. 19). Our efforts to understand the structure–activity relationship (SAR) profile of this series began by focusing on the heterocyclic group itself. All analogs were prepared by using known or standard chemistry. As shown in Table 1, there was some benefit derived from modifying the size of the rhodanine N-substituent (entries 1–4), such that longer substituents were favored. Replacement of the sulfur of the rhodanine thiocarbonyl with either oxygen or nitrogen abrogated the binding activity of the compounds (compare entry 3 with entries 5–7). Replacement of the “internal” sulfur of the heterocycle was more successful and revealed that both 2-thioxo-1,3-oxazolidin-4-ones (entries 8 and 9) and 2-thioxo-1,3-bisazolidin-4-ones (entry 10) could inhibit the binding of TNF-α to its receptor, albeit with weaker potency than the parent 2-thioxo-1,3-thiazolidin-4-ones. Notably, the R—N—C⩵S grouping (cf. 1) could be changed to its tautomeric N⩵C—S—R form (cf. 2), although there was more of a size restriction on the “R” group in the latter series (compare entries 2–4 with entries 11–13); the direction of tautomerization was also important, because compounds of formula 3 were inactive. Overall, the activity of compounds of formula 2 was a critical result, because it indicated that the thiocarbonyl was not strictly required for biological activity.
Table 1.
Effects of modifying the core heterocycle
 | ||||||
|---|---|---|---|---|---|---|
| Entry | Class code* | C⩵C† | X | Y | R | IC50, μM‡ |
| 1 | 1a | Z | S | S | H | 2.9 |
| 2 | 1b | Z | S | S | Me | 0.35 |
| 3 | 1c | Z | S | S | Allyl | 0.1 |
| 4 | 1d | Z | S | S | Bn | 0.1 |
| 5 | 1e | Z | S | O | Et | >100 |
| 6 | 1f | Z | S | NAc | Allyl | ≫50 |
| 7 | 1g | Z | S | NH | Allyl | >30 |
| 8 | 1h | Z | O | S | Et | 0.4 |
| 9 | 1i | E | O | S | Et | 0.4 |
| 10 | 1j | Z | NH | S | Me | 2.3 |
| 11 | 2a | Z | S | SMe | — | 3.7 |
| 12 | 2b | Z | S | Sallyl | — | 2.1 |
| 13 | 2c | Z | S | SBn | — | ≈10 |
| 14 | 3a | Z | N | SMe | Me | ≫10 |
| 15 | 3b | Z | N | S-(CH2)2− | ≫10 | |
Class refers to the structures shown above the table.
Refers to the olefin geometry (Z is illustrated).
Inhibition of TNF-α binding to a soluble form of monomeric TNFRc1.
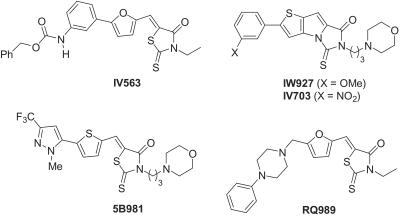
Table 2 summarizes other critical aspects of the SAR profile of this series of TNF inhibitors.** The syntheses of the key compounds are described in the supporting information on the PNAS web site, www.pnas.org. The electronics of the π-system conjugated to the heterocyclic carbonyl (C⩵O) were important for biological activity, in that the arylalkylidene had to be electron-rich. Thus, furan (IV563, RQ989), vinylfuran, thiophene (5B981), and indole were all tolerated as central aryl groups, but electron-deficient heterocycles or phenyl rings were not (see Fig. 7, which is published as supporting information on the PNAS web site). The biological activity did not depend on the relative orientation of the arylalkylidene and the heterocycle, because both E and Z isomers were active (cf. Table 1, entries 8 and 9). Consistent with this observation, methylation of the alkylidene to give the tetrasubstituted olefin was acceptable (see Fig. 7). It also proved possible to constrain the rotation of the central aromatic ring, as in IW927 and IV703 (Fig. 1). This series of compounds—representing conformationally restricted analogs of IW169 (Table 1, entry 10)—proved to be quite potent (Table 2). Although there did appear to be some correlation between structural changes in the aromatic ring distal to the rhodanine core (cf. the ortho-chlorophenyl group of 1) with biological activity, the SAR profile could not be rationalized. As will be readily appreciated from the structural diversity evident in the compounds of Fig. 1, a variety of groups were tolerated as appendages to the central electron-rich heterocycle: electron-rich aromatic (IW927), electron-poor aromatic (IV563, IV703), heterocyclic (5B981), and nonaromatic groups (RQ989).
Table 2.
Biological data for compounds shown in Fig. 1
| Code | IC50 Values (μM)
|
||||
|---|---|---|---|---|---|
| TNFRc1 Bnd* | TNF-IκB† | TNFRc2 Bnd‡ | CD40 Bnd§ | MTS Tox¶ | |
| IV563 | 0.08 | 12 | ≈100 | 3.0 | >100 |
| IW927 | 0.05 | 0.6 | >100 | >50 | >100 |
| IV703 | 0.27 | 1.4 | 28 | 9.0 | >100 |
| 5B981 | 0.5 | 3.2 | 24 | 2.3 | >100 |
| RQ989 | 0.7 | 8.5 | ≈100 | 10 | >100 |
Inhibition of TNF-α binding to a soluble form of monomeric TNFRc1.
Inhibition of TNF-induced phosphorylation of Iκ-B in Ramos cells.
Inhibition of TNF-α binding to a soluble form of monomeric TNFRc2.
Inhibition of CD40 ligand binding to a soluble form of dimeric CD40/Fc.
Inhibition of mitochondrial viability in Ramos cells (24-h exposure).
Figure 1.
Chemical structures of some optimized TNF-α inhibitors.
The most potent compound synthesized in these efforts was IW927, which exhibited an IC50 value of 50 nM for inhibiting the binding of TNF-α to the TNFRc1, and an IC50 value of 600 nM for inhibiting the TNF-α-induced phosphorylation of Iκ-B in Ramos cells (Table 2). The related compound IV703 was also effective in a 24-h functional assay that measured inhibition of TNF-stimulated IL-6 and -8 production (see Fig. 8, which is published as supporting information on the PNAS web site). In addition, IW927 exhibited >2000-fold selectivity for binding to the TNFRc1 relative to the related TNFRc2 or CD40, and was not cytotoxic at concentrations up to 100 μM (Table 2). The remaining compounds showed varying levels of selectivity for binding to the TNFRc1 over the TNFRc2 (50- to 1000-fold) and CD40 (5- to 40-fold); none of the compounds exhibited any significant cytotoxicity.
Binding Properties of the Inhibitors.
Even though these compounds exhibited low cytotoxicity and receptor-subtype selectivity in binding, the unusual nature of their SAR profiles (vide supra) was a concern, and prompted a more complete characterization of the binding properties of these inhibitors. We were unable to demonstrate reversible inhibition with these compounds by using classical pharmacological techniques. In addition, binding of these compounds was diminished when the assays were conducted either in a 37°C incubator or a 4°C refrigerator. Because molecules like 1 are readily excited/isomerized photochemically (20), we examined the interaction of these compounds with the TNFRc1 in the absence of light. Indeed, when the binding assays were performed in the dark, the potencies of all of the compounds tested were attenuated 50-fold to >1000-fold (IC50's > 30 μM). We also examined a sampling of the more potent compounds (Fig. 1) in both the cell-based functional assay and the CD40 ligand binding assay in light-excluded conditions; none of these compounds displayed any significant activity. Although most of the compounds had their binding affinities severely attenuated (IC50's > 100 μM) when the binding assay was performed in the dark, three of the more aqueous-soluble compounds still displayed measurable binding affinity in the absence of light: RQ989 (47 μM) and 5B981 (34 μM), and IV703 (50 μM).
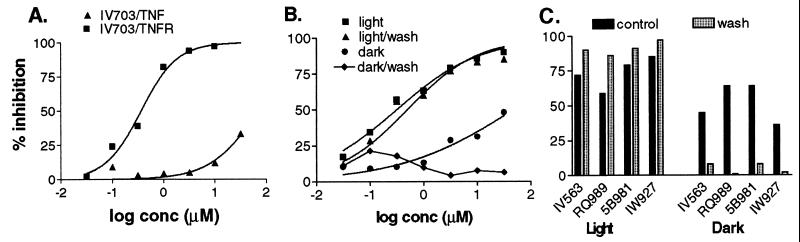
To understand better the light-dependence of the system, we performed two critical control experiments using IV703 as a representative compound (Fig. 2 A and B). In the first, we tried to determine whether the compounds were covalently modifying the TNFRc1 or TNF-α itself. When IV703 was preincubated with TNFRc1 in the light before adding TNF-α in the dark, dose-dependent inhibition of TNF-α binding was observed. However, when IV703 was preincubated with TNF-α in the light and this mixture was added to the TNFRc1 in the dark, the inhibitory properties of IV703 were attenuated (Fig. 2A). We conclude from this that IV703 covalently modifies the TNFRc1, but not TNF-α. In the second experiment, we examined whether the compounds were irreversible modifiers under both light and dark conditions. Thus, IV703 was preincubated with TNFRc1 for 5 min, the system was washed repeatedly with PBS to remove unbound IV703, and then TNF-α was added as usual (Fig. 2B). Relative to the unwashed controls (0.3 μM, light; 50 μM, dark), a change was only observed in the light-excluded conditions (0.3 μM light; no measurable inhibition in the dark). Thus, it appears that IV703 binds reversibly in the dark and irreversibly in the light. Notably, equivalent results to those for IV703 (Fig. 2B) were obtained with IW927, IV563, 5B981, and RQ989 (Fig. 2C). Taken together, these data suggest that the compounds described herein (Fig. 1) are “photochemically enhanced” inhibitors, in that they bind reversibly to the TNFRc1 with weak affinity (ca. 40–100 μM) and then covalently modify the TNFRc1 through a photochemical reaction.
Figure 2.
Binding of IV703 to TNFRc1 under light and dark conditions. (A) IV703 was incubated with TNFRc1-coated plates (20 ng per well) at RT in the light for 30 min before adding Eu-TNF-α (2.4 nM) (squares). In the same experiment, IV703 was incubated with Eu-TNF-α (2.4 nM) at RT in the light for 30 min before adding the mixture to TNFRc1-coated plates (20 ng per well) (triangles). The standard binding assay was then performed in the dark. (B) TNFRc1-coated plates were preincubated with various concentrations of IV703 for 5 min at RT in the light or in the dark. The plates were then either washed extensively with PBS (triangles, light; diamonds, dark) or left untreated (squares, light; circles, dark). Eu-TNF-α was added and the standard binding assay performed in the dark. (C) TNFRc1-coated plates were preincubated with IV563, RQ989, 5B981, or IW927 for 5 min at RT at a concentration of 1 μM in the light or 100 μM in the dark. The plates were then either washed extensively with PBS (hatched bars) or left untreated (control, dark bars). Eu-TNF-α was added and the standard binding assay performed in the light or in the dark. In all cases, the data represent the average of triplicate wells.
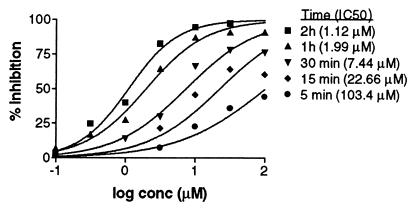
To understand better the nature of the irreversible binding, we examined the time-dependence of the phenomenon. Under the conditions of the normal TNF-α–TNFRc1 binding assay, irreversible binding was complete within 5 min (data not shown). However, in a membrane-based binding assay that used a signaling-deficient TNFRc1 mutant (iTNFRc1; ref. 13), it proved possible to document the time-dependence of the irreversible binding (Fig. 3). Thus, membrane preparations containing iTNFRc1 were exposed to varying doses of IV703, washed, and treated with Eu-TNF-α to measure irreversible binding. As expected, the extent of irreversible binding increased in a time-dependent manner (Fig. 3).
Figure 3.
The irreversible binding of IV703 with TNFRc1 is time-dependent. iTNFRc1-CHO membrane coated plates (10 ng per well) were preincubated with IV703 for various lengths of time at RT in the light. The plates were then washed extensively with PBS. Eu-TNF-α (1 nM) was added and the standard binding assay performed in the light. Data represent the average of triplicate wells.
X-ray Crystallography.
In parallel with the biological experiments described above, we also performed crystal-soaking experiments with the TNFRc1 and a limited number of compounds. The greater solubility of IV703 in aqueous media relative to the other inhibitors—a property that is presumably attributable to the pendant morpholino moiety—allowed us to obtain the best results with this compound. After soaking crystals of soluble TNFRc1 with a solution of IV703, the crystals were flash frozen and a single crystal structure was obtained. The initial electron density maps from this structure clearly revealed attachment of the nitrophenyl ring of IV703 to the backbone nitrogen of Ala-62 of the TNFRc1 (Fig. 4A); this experiment was repeated two additional times, each of which yielded the same results. Refinement of the structure provided a final model with an Rfactor of 27.5% at a resolution of 2.9 Å (Fig. 4B). This result thus confirms that it is in fact possible for compounds such as IV703 to bind irreversibly to the TNFRc1 under normal lighting conditions.
Discussion
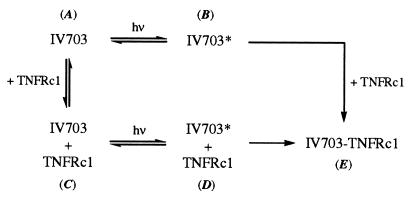
The discovery of small molecules (MW < 0.65 kDa) that potently disrupt the binding of TNF-α to the TNFRc1 has historically proven difficult. Through screening of our proprietary library of compounds, we discovered that compounds containing the N-alkyl 5-arylalkylidene-2-thioxo-1,3-thiazolidin-4-one structural motif (e.g., 1) could, in fact, inhibit this protein–protein interaction. Accordingly, we embarked on a medicinal chemistry program that led to the synthesis of a number of TNF inhibitors, including the potent compounds IV563 and IW927 (Fig. 1, Table 2). We subsequently demonstrated that the potent inhibition displayed by these compounds was related to the fact that the binding assays were run under normal lighting conditions. More detailed experiments revealed that these compounds have weak intrinsic affinity for the TNFRc1 (ca. 40–100 μM), and that this affinity is sufficient to provide for site-specific covalent modification of the TNFRc1 in the presence of light, thereby leading to apparent high-affinity binding (50–1000 nM). Because this photochemically enhanced binding mechanism proceeds through a reversible binding event, it is possible to achieve selectivity for binding to one of several receptors. This mechanism is readily distinguished from “photochemically induced” binding, which leads to the production of a highly reactive photoexcited species; one would assume that this latter mechanism should result in less discriminate covalent binding (Fig. 5).‡‡
Figure 5.
A comparison of the general mechanistic schemes for a photochemically induced inhibitor (A → B → E) and a photochemically enhanced inhibitor (A → C → D → E). In the former case, the activated inhibitor (IV703*) scavenges the protein from solution; this would lead to nonselective reactivity. In contrast, in the case of the photochemically enhanced inhibitor, the mechanistic requirement for initial protein binding imposes an element of selectivity on its reactivity profile.
We were able to obtain a crystal structure of one of the inhibitors (IV703) bound to the TNFRc1. A covalent linkage between IV703 and the main chain nitrogen of Ala-62 of the TNFRc1 was observed in this structure (Fig. 4). The crystal structure of TNF-β with the TNFRc1 indicates that this receptor residue (Ala-62) participates in a hydrogen-bonding interaction with the phenolic hydroxyl of Tyr-108 of TNF-β, and homology modeling has indicated that a similar interaction should be observed between Ala-62 of TNFRc1 and Tyr-87 of TNF-α (11, 12). The importance of both Tyr-108 (TNF-β) and Tyr-87 (TNF-α) have been confirmed by site-directed mutagenesis studies (21, 22). Because IV703 inhibited the binding of both TNF-α and -β to the TNFRc1 equally well (data not shown), we overlapped the published structure of TNFRc1–TNF-β and our own structure of TNFRc1–IV703. As shown in Fig. 6, this overlay demonstrates that the covalent binding of IV703 to Ala-62 most likely disrupts the binding of TNF-α and -β by blocking some of the interactions between the exposed Tyr-containing β-turn of the native ligands and the region near Ala-62 of the TNFRc1. The overlay also makes it clear that the covalent binding of IV703 to the TNFRc1 does not appear to induce large structural changes in the receptor.
Figure 6.
Superimposition of the crystal structures of IV703 bound to TNFRc1 (purple, this study) and that of TNF-β bound to TNFRc1 (green; ref. 11). Note that Tyr-108 (TNF-β) normally interacts with Ala-62 (TNFRc1) (11); this receptor residue is bound to IV703 in our structure.
In addition to the covalent contact with Ala-62, it also appears that IV703 makes several van der Waals contacts (ca. 3.1–4.1 Å) with the side chains of residues from Phe-60, Thr-61, Leu-67, and Leu-71 of the TNFRc1 (Fig. 4B). A more distant contact (5 Å) from a symmetry-related monomer of the TNFRc1 was also observed. Although it is not clear whether these four hydrophobic contacts are involved in the reversible binding event that precedes the formation of the observed covalent adduct, we note that there is some variation in these residues between TNFRc1, TNFRc2, and CD40: Phe-60 is Tyr in TNFRc2, Thr-61 is Leu in CD40, Ala-62 is Gln in TNFRc2 and Asp in CD40, Leu-67 is Val in TNFRc2 and Glu in CD40, and Leu-71 is His in CD40. The variation in this receptor region may at least partially explain why IV703 exhibits selective binding (Table 2). This hypothesis is supported by the observation that changes in Ser-86 of TNF-α—the residue adjacent to the critical Tyr-87 (vide supra) on the exposed 84–88 β-turn of TNF-α—alter the TNFRc1/2 binding selectivity of TNF-α (23).
Several possibilities exist for the photochemical mechanism that leads to the conversion of the noncovalent inhibitor–receptor complex to covalent inhibitor–receptor complex (Fig. 5, D → E). Based on the wide array of groups that are tolerated as “side chain” appendages to the 5-arylalkylidene-2-thioxo-1,3-thiazolidin-4-one core (Table 1, Fig. 1), it seems likely that the 5-arylalkylidene-2-thioxo-1,3-thiazolidin-4-one core itself is involved in the photochemical event. The role of the C⩵S substituent is unclear, but given the activity of analogs like 2a and 2b (Table 1, entries 11 and 12), it seems unlikely that the C⩵S is itself participating in the photochemistry; the role of the sulfur is more likely electronic. Indeed, it is known that the arylidene portion of 5-arylidene-2-thiohydantoins is readily reduced in one-electron transfer processes (24). Thus, one plausible mechanism could entail a nearby aromatic amino acid side chain (perhaps Phe-60; cf. Fig. 4B) of the TNFRc1 donating an electron to the excited state of the inhibitor to form a radical anion. Anionic abstraction of the main chain hydrogen of Ala-62 would lead to a C-radical/N-anion pair, which would then collapse, protonate, and aromatize†† to form the observed adduct. This mechanism is consistent with the regiochemical outcome of the reaction between IV703 and TNFRc1, which is presumably strongly biased by the presence of the conjugated nitrophenyl group. It may be that similar mechanisms are applicable to the other compounds illustrated in Fig. 1, although the regiochemical outcomes of these reactions will clearly be compound-specific.
The results described herein are significant for a number of reasons. First, several groups have reported compounds similar in structure to 1 as leads for unrelated biological targets (25–31). Our data suggest that some or all of these results may be attributable to the type of mechanism outlined herein, and indicate that caution should be taken when compounds containing an N-alkyl 5-arylalkylidene-2-thioxo-1,3-thiazolidin-4-one structural motif are obtained as chemical lead structures. Second, the mechanism that we have described herein (photochemically enhanced inhibition) is unique, as best we can discern. Third, given that the compounds are reversible (albeit weak) inhibitors of the TNF-α–TNFRc1 interaction in light-excluded conditions, they represent interesting lead compounds. Finally, our results suggest that it is indeed possible to inhibit the interaction of TNF-α with the TNFRc1 by blocking the region near Ala-62 of the TNFRc1, and the covalent IV703–TNFRc1 complex may thus prove helpful in the design of other classes of TNF inhibitors. Toward this end, we note that the use of a ligand–receptor covalent tethering strategy as a method for lead discovery has been advocated recently (32).
Supplementary Material
Acknowledgments
We thank Karl Hardman for data collection on the IV703–TNFRc1 crystals and acknowledge Chong-Hwan Chang, Doug Batt, Scott Priestley, and Robert Cherney for discussions.
Abbreviations
- TNF
tumor necrosis factor
- TNFRc1
type-1 TNF receptor
- TNFRc2
type-2 TNF receptor
- SAR
structure–activity relationship
- RT
room temperature
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates for the IV703–TNFRc1 crystal structure have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1FT4).
Rmerge = 100 × Σh Σ j |I h,j − 〈I h〉| /Σ h Σ j |I h,j|, where I h,j is the jth observation of reflection h and 〈Ih〉 is the mean intensity of reflection h over all measurements of I h.
Rfactor = 100 × Σ ||Fo| − |Fc|| /Σ |Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
A complete description of the SAR profiles for all of our biologically active series of TNF-α inhibitors lies outside the scope of this manuscript and will be disclosed separately.
This hypothesis is also consistent with the lack of reactivity of these compounds toward secondary amides (see Fig. 9, which is published as supporting information on the PNAS web site).
Because the inhibitors still function in the absence of oxygen (but the presence of light), the importance of the final aromatization step to the inhibitory capacity of IV703 is suspect (see Fig. 10, which is published as supporting information on the PNAS web site).
References
- 1.Kollias G, Douni E, Kassiotis G, Kontoyiannis D. Immunol Rev. 1999;169:175–194. doi: 10.1111/j.1600-065x.1999.tb01315.x. [DOI] [PubMed] [Google Scholar]
- 2.Locksley R M, Kileen N, Lenardo M J. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 3.Tartaglia L A, Pennica D, Goeddel D V. J Biol Chem. 1993;268:18542–18548. [PubMed] [Google Scholar]
- 4.Vandervoorde V, Haegeman G, Fiers W. J Cell Biol. 1997;137:1627–1638. doi: 10.1083/jcb.137.7.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreland L W. J Rheumatol. 1999;26, Suppl. 57:7–15. [PubMed] [Google Scholar]
- 6.Newton R C, Decicco C P. J Med Chem. 1999;42:2295–2314. doi: 10.1021/jm980541n. [DOI] [PubMed] [Google Scholar]
- 7.Kruszynski M, Shealy D J, Leone A O, Heavner G A. Cytokine. 1999;11:37–44. doi: 10.1006/cyto.1998.0389. [DOI] [PubMed] [Google Scholar]
- 8.Takasaki W, Kajino Y, Kajino K, Murali R, Greene M I. Nat Biotechnol. 1997;15:1266–1270. doi: 10.1038/nbt1197-1266. [DOI] [PubMed] [Google Scholar]
- 9.Mancini F, Toro C M, Mabilia M, Giannangeli M, Pinza M, Milanese C. Biochem Pharmacol. 1999;58:851–859. doi: 10.1016/s0006-2952(99)00150-1. [DOI] [PubMed] [Google Scholar]
- 10.Chan F K-M, Chun H J, Zheng L, Siegel R M, Bui K L, Lenardo M J. Science. 2000;288:2351–2354. doi: 10.1126/science.288.5475.2351. [DOI] [PubMed] [Google Scholar]
- 11.Banner D W, D'Arcy A, Janes W, Gentz R, Schoenfeld H-J, Broger C, Loetscher H, Lesslauer W. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 12.Fu Z-Q, Harrison R W, Reed C, Wu J, Xue Y-N, Chen M-J, Weber I T. Protein Eng. 1995;8:1233–1241. doi: 10.1093/protein/8.12.1233. [DOI] [PubMed] [Google Scholar]
- 13.Brakebusch C, Nophar Y, Kemper O, Engelmann H, Wallach D. EMBO J. 1992;11:943–950. doi: 10.1002/j.1460-2075.1992.tb05133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodseth L E, Brandhuber B, Devine T Q, Eck M J, Hale K, Naismith J H, Sprang S R. J Mol Biol. 1994;239:332–335. doi: 10.1006/jmbi.1994.1371. [DOI] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.Kissinger C R, Gehlhaar D K, Fogel D B. Acta Crystallogr D. 1998;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 17.Brünger A T, Kuriyan J, Karplus M. Science. 1987;234:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 18.Naismith J H, Devine T Q, Brandhuber B J, Sprang S R. J Biol Chem. 1995;270:13303–13307. doi: 10.1074/jbc.270.22.13303. [DOI] [PubMed] [Google Scholar]
- 19.Wang, J., Ramnarayan, K., Rideout, D., Mong, S., Zhu, H., Niemeyer, C. & Brady, T. P. (2000) PCT Int. Appl. WO 00/32598.
- 20.Ishida T, In Y, Inoue M, Ueno Y, Tanaka C. Tetrahedron Lett. 1989;30:959–962. [Google Scholar]
- 21.Goh C R, Loh C-S, Porter A G. Protein Eng. 1991;4:785–791. doi: 10.1093/protein/4.7.785. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X-M, Weber I, Chen M-J. J Biol Chem. 1992;267:24069–24075. [PubMed] [Google Scholar]
- 23.Loetscher H, Stueber D, Banner D, Mackay F, Lesslauer W. J Biol Chem. 1993;268:26350–26357. [PubMed] [Google Scholar]
- 24.Abou-Elenien G M, Ismail N A, Eldin A A M. Monatshefte für Chemie. 1992;123:1117–1124. [Google Scholar]
- 25.Sudo K, Matsumoto Y, Matsushima M, Fujiwara M, Konno K, Shimotohno K, Shigeta S, Yokota T. Biochem Biophys Res Commun. 1997;238:643–647. doi: 10.1006/bbrc.1997.7358. [DOI] [PubMed] [Google Scholar]
- 26.Sato, S., Shirakawa, S., Tatsui, A., Hasegawa, T., Yamada, H., Kazayama, S., Hayashi, K., Takahashi, A., Kojo, K. & Narita, S. (2000) Jpn. Kokai Tokkyo Koho, JP 2000095770.
- 27.Bailey, T. R. & Young, D. C. (2000) PCT Int. Appl. WO 00/10573.
- 28.Grant E B, Guiadeen D, Baum E Z, Foleno B D, Jin H, Montenegro D A, Nelson E A, Bush K, Hlasta D J. Bioorg Med Chem Lett. 2000;10:2179–2182. doi: 10.1016/s0960-894x(00)00444-3. [DOI] [PubMed] [Google Scholar]
- 29.Degterev A, Lugovskoy A, Cardone M, Mulley B, Wagner G, Mitchison T, Yuan J. Nat Cell Biol. 2001;3:173–182. doi: 10.1038/35055085. [DOI] [PubMed] [Google Scholar]
- 30.Scott, I. L., Biediger, R. J. & Market, R. V. (1998) PCT Int. Appl. WO 98/53790.
- 31.Kobayashi, K., Nishiyama, T. & Nakaide, S. (1999) Jpn. Kokai Tokkyo Koho, JP 11302280.
- 32.Erlanson D A, Braisted A C, Raphael D R, Randal M, Stroud R M, Gordon E M, Wells J A. Proc Natl Acad Sci USA. 2000;97:9367–9372. doi: 10.1073/pnas.97.17.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.