Abstract
Small RNAs are a major class of RNAs along with transfer RNAs, ribosomal RNAs, and messenger RNAs. They vary in size from less than 100 nucleotides to several thousand nucleotides and have been identified and characterized both in prokaryotes and eukaryotes. Small RNAs participate in a variety of cellular functions including regulating RNA synthesis, RNA processing, guiding modifications in RNA, and in transport of proteins. Small RNAs are generated by a series of posttranscriptional processing steps following transcription. While RNA 5′ end structure, 5′ cap formation, and RNA processing mechanisms have been fairly well characterized, the 3′ end processing is poorly understood. Recent data point to an emerging theme in small RNAs metabolism in which the 3′ end processing is mediated by the exosome, a large multienzyme complex. In addition to removal of nucleotides by the exosome, there is simultaneous rebuilding of the 3′ end of some small RNA by adenylation and/or uridylation. This review presents a picture of both degradative and rebuilding reactions operative on the 3′ end of some small RNA molecules in prokaryotes and eukaryotes.
Keywords: Small RNAs, 3′ End formation, 3′ Adenylation, 3′ Uridylation
THE three major types of cellular RNAs directly involved in protein synthesis are: messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). In addition to these major RNAs, many small RNAs exist that are not directly involved in protein synthesis but play important roles in diverse metabolic pathways, such as transcription, translation, mRNA and rRNA processing, protein secretion, and protein stability. In human cells, there are over 100 small RNAs that have been characterized thus far. Many small RNAs are known to participate in important cellular functions. In eukaryotes, U1, U2, U4, U5, and U6 small nuclear RNAs as a part of ribonucleoproteins (snRNPs) are required co-factors for splicing of nuclear pre-mRNAs (116,124,125,135). RNase P RNA, U7 RNA, and many small nucleolar RNAs (snoRNAs) are necessary for the site-specific cleavage of pre-tRNA (123), histone pre-mRNA (113), and site-specific methylation or pseudouridine formation of pre-ribosomal RNAs (18,67), respectively. Signal recognition particles (SRPs) that contain SRP (7SL) RNA recognize the signal peptide of the secretory and membrane proteins and participate in the translocation of these proteins (140). MRP RNA (a component of the mitochondrial RNA processing complex) and U8 snoRNA are found mostly in the nucleolus and are required for the formation of ribosomal 5.8S RNA from pre-rRNA (26,83,97). In addition, there are many other small RNAs that are believed to play important roles in both eukaryotic and prokaryotic cells (5,18,67,107,108). Therefore, small RNAs represent a diverse and functionally important class of RNAs.
The synthesis, posttranscriptional processing, and modifications in major cellular RNAs (i.e., transfer RNAs, ribosomal RNAs, and messenger RNAs) have been extensively studied and reviewed (34,49,63,84,86,111,112,116,124,125,139). This review focuses on the 3′ end formation in small RNAs from human cells and draws a comparison of this 3′ end processing with that of small RNAs from yeast and E. coli. Evolutionarily distant organisms ranging from bacteria and yeast to humans appear to be using common underlying mechanisms for the formation and maintenance of RNA 3′ ends. The most common processes are exonuclease digestion of precursor RNAs with longer 3′ ends to generate the mature 3′ ends, and addition of one or more nucleotides to rebuild and/or elongate the 3′ ends.
THE 3′ END FORMATION IN EUKARYOTIC SMALL RNAs
Most eukaryotic RNAs undergo posttranscriptional processing of their 3′ ends. These modifications include polyadenylation of eukaryotic mRNAs (36) and –CCA addition/turnover on tRNAs (34). In most eukaryotic small RNAs, the 3′ end processing is usually limited to the removal of 1–15 nucleotides from their 3′ ends. Small RNAs in eukaryotic cells are synthesized by various RNA polymerases (Table 1). Some RNAs such as U3 snoRNA and telomerase RNA are synthesized by different RNA polymerases in different species. While eukaryotic small RNAs may be synthesized by different RNA polymerases, conserved 3′ end processing steps are common to most of the small RNAs. The 3′→5′ exonucleases as part of the exosome complex appear to be the major components for this 3′ end processing (3,136).
TABLE 1.
SYNTHESIS OF SMALL RNAs BY DIFFERENT RNA POLYMERASES
| RNA Polymerase | Examples | Number of Nucleotides Removed by 3′ End Processing |
|---|---|---|
| Pol I | Yease ribosomal 5.8S RNA | 8–10 nucleotides |
| Pol II | Human U1 snRNA | 8–11 nucleotides |
| Mouse U14 snoRNA | 8–9 nucleotides | |
| Pol III | Human SRP RNA | 3 nucleotides |
| Mitochondrial | Trypanosomal guide RNAs | not known |
THE 3′ END FORMATION IN 5.8S RIBOSOMAL RNA TRANSCRIBED BY RNA POLYMERASE I (Pol I)
The 5.8S ribosomal RNA is transcribed as part of a large polycistronic transcript by RNA polymerase I. The eukaryotic pre-rRNA transcript begins with the 5′ external transcribed spacer (5′ ETS), after which is the 18S rRNA, an internal transcribed spacer (ITS1), the 5.8S rRNA, an ITS2, the 28S rRNA (25S in yeast), and finally a 3′ ETS. This precursor RNA transcript is processed first by extensive nucleotide modifications and then by nucleolytic processing into the mature 18S, 5.8S, and 28S rRNAs (122). Nucleolytic processing, to generate the mature 5′ and 3′ ends, is carried out primarily by three types of enzymes: specific endonucleases, 5′→3′ exonucleases, and 3′→5′ exonucleases.
The 5.8S rRNA processing is initiated by cleavage at a specific site in the ITS1 by RNase MRP, an endoribonuclease. In yeast further 5′→3′ processing to generate the mature 5′ end of the short form of the 5.8S RNA is through the action of two homologous exonucleases, Rat1p and Xrn1p (52). In both yeast and vertebrates, the mature 3′ end of 5.8S rRNA is formed by endonucleolytic cleavage(s) in ITS2 followed by action of 3′→5′ exonucleases. However, the processing in the two organisms differs in the number of 3′ endonucleolytic cleavages involved. In mammals, two precursor intermediates to 5.8S rRNA have been identified, the 8S and 12S pre-5.8S rRNAs, indicating that there are two endonucleolytic cleavages. In contrast, only one internal cleavage has been detected in yeast, corresponding to the accumulation of a pre-5.8S rRNA processing intermediate, termed 7S RNA. U8 snoRNA, present in both mammals and Xenopus, is required for accurate processing of the 5′ and 3′ ends of 5.8S RNA (97). The yeast homolog of the U8 snoRNA has not yet been identified.
The exonucleolytic trimming of the 3′ end of 5.8S rRNA is mediated by a complex package of 3′→5′ exoribonucleases and RNA helicases, termed the exosome (3,28,89,153). Human homologs of the exosome components have been identified and are found to be part of the PM-Scl particle, the autoantigen in the polymyositis-scleroderma overlap syndrome (4,12). The 3′ processing of small RNAs, transcribed by other RNA polymerases, is also by the exosome complex.
THE 3′ END FORMATION IN snRNAs TRANSCRIBED BY RNA POLYMERASE II (Pol II)
In human cells, the most abundant Pol II small RNA transcripts are the spliceosomal snRNAs. Two forms of spliceosomes are found in higher eukaryotes. The major spliceosomes contain the U1, U2, U4, U5, and U6 snRNAs; the minor spliceosomes contain the U11, U12, U4atac, U5, and U6atac snRNAs. All the vertebrate spliceosomal snRNAs, except U6 and U6atac RNAs, are Pol II transcripts.
Biogenesis of these small RNAs involves many steps at both the 5′ and the 3′ ends, and different steps of this maturation occur in the cytoplasm or in the nucleus. The formation of the 3′ end of vertebrate snRNAs requires transcription initiation from an snRNA promoter (54,92). The snRNA precursors are made with the m7G cap (same as in mRNAs) at their 5′ end and 9–15 extra nucleotides at their 3′ end (31). The 3′ end formation of mature snRNAs also requires a cis-acting sequence, known as the 3′ box, located 9–19 nucleotides downstream of the coding region of these snRNA genes (2,25,53,92,150). Mutation of this sequence causes the accumulation of transcripts that are not processed accurately and therefore they are longer than the mature snRNAs. The U1, U2, U4, and U5 snRNAs are transported as precursors into the cytoplasm where binding of several Sm proteins precedes the hypermethylation of the m7G cap structure to the 2,2,7-trimethylguanosine cap (85) and the trimming of their 3′ trailer sequences (Fig. 1). Complete processing to the mature 3′ end appears to occur only after the snRNA is transported back into the nucleus. It is postulated that, in the nucleus, there is an activity called the 3′ terminal processing inhibitor (TPI), which inhibits snRNA 3′ end processing when the 5′ cap of the RNA is monomethylated (i.e., newly synthesized snRNA with m7G cap). However, when the snRNA is imported into the nucleus after cap hypermethylation, the TPI cannot bind to the trimethylated cap, and the 3′ processing is thus completed (149). Recent data show that 3′ exoribonucleases, as part of the exosome, are responsible for the accurate 3′ end formation of the U-series of small RNAs synthesized by RNA polymerase II (136,138).
Figure 1.
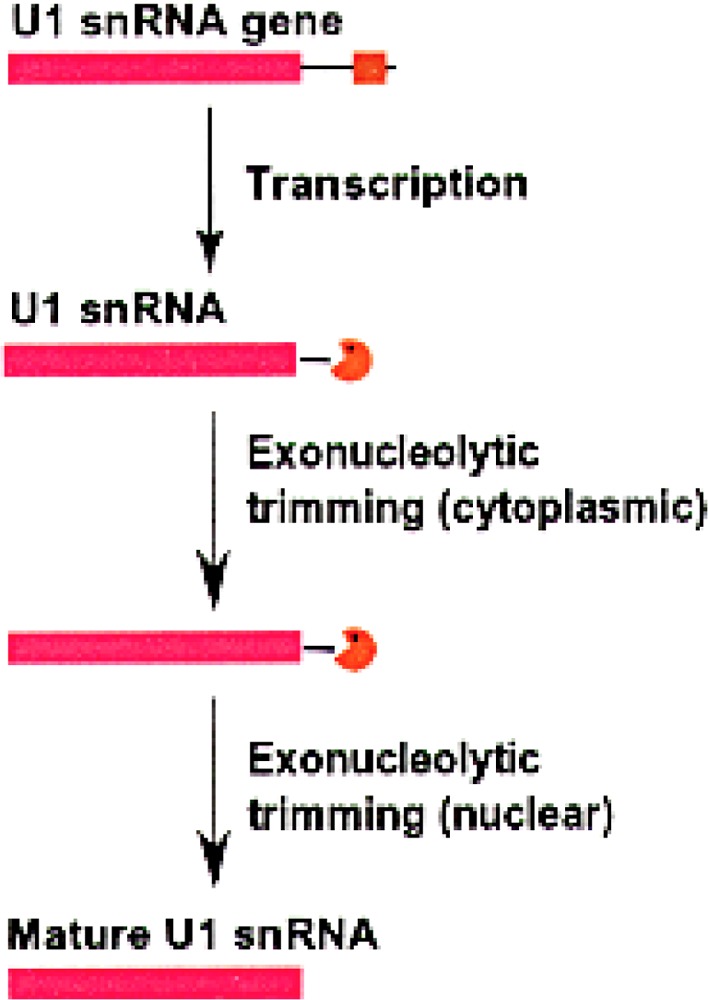
The 3′ end formation in human U1 snRNA. The sequence corresponding to the mature human U1 RNA (nucleotides 1–164) is shown in pink. The 3′ box (shown in brown) is 14 nucleotides long and is 11 nucleotides downstream of the mature 3′ end. The pacman represents the exosome complex.
The sequence requirements for correct 3′ end formation have been studied in U1 and U2 snRNAs. The mature 3′ end of U1 snRNA is formed in at least two steps (Fig. 1). The 3′ box, a 13-nucleotide-long sequence, located downstream from the U1 coding region is the only sequence required to direct the first step in the formation of the 3′ end of U1 snRNA (53). Pre-U1 snRNAs with an m7G cap are first transported to the cytoplasm, where they undergo cap hypermethylation and binding to the Sm proteins. The 3′ extra nucleotides are trimmed in the cytoplasm, leaving only one or two extra nucleotides after the mature 3′ end. The final processing then occurs in the nucleus where the one and/or two extra nucleotides are removed to generate the mature 3′ end.
Most U2 snRNA precursors detected in HeLa cells have 3′ extensions of 10–16 nucleotides (145) but, recently, transcription of the U2 snRNA has been shown to continue at least 250 nucleotides downstream of the 3′ box (30). Huang et al. (56) showed that base pairing between nucleotides in the pre-U2 RNA 3′ extension and a sequence between the Sm domain and the stem loop III of U2 snRNA are responsible for the correct 3′ end processing of pre-U2 snRNA. Accurate 3′ end processing is important for transport of the snRNA from the cytoplasm back into the nucleus. Human U2 snRNA with a mutant 3′ end, that cannot be processed, was found to be defective in import (57). Similar results have been obtained in the Xenopus oocyte system, where U1 snRNAs with longer 3′ ends were not transported into the nucleus, showing that the 3′ end structure is critical for RNA transport across the nuclear pore (93).
THE 3′ END FORMATION IN SMALL NUCLEOLAR (sno) RNAs
Another important group of small RNAs transcribed by Pol II are the small nucleolar RNAs. In eukaryotes, there are two distinct classes of snoRNAs, namely the fibrillarin-associated box C/D snoRNAs and the Gar1p-associated box H/ACA snoRNAs. Many box C/D snoRNAs direct the site-specific 2′-O-ribose methylation and many box H/ACA snoRNAs guide the pseudouridylation of the ribosomal RNAs. Human U85 possesses the box elements of both classes of snoRNAs and associates with both fibrillarin and Gar1p; it is the first example of a snoRNA that functions in both RNA pseudouridylation and 2′-O-methylation (60).
Some yeast snoRNAs and the more abundant vertebrate snoRNAs, such as U3, U8, and U13, are expressed from their own promoters (75,86). However, majority of the vertebrate and many yeast snoRNAs are expressed as part of an intron of a pre-mRNA (17,66,86,130). Host genes for most of these intronic snoRNAs code for nucleolar or ribosomal proteins. This raises the possibility that intronic location of these snoRNA genes is to facilitate coordinated regulation of the ribosomal proteins and the ribosomal RNA modifying snoRNAs. Some of the host genes belong to the 5′-terminal oligopyrimidine gene family and do not encode a functional mRNA. An example is the vertebrate UHG gene in which the snoRNAs U22 to U31 are all encoded within introns of a gene whose final spliced product has no open reading frame (133). It is likely that this gene only exists for the production of these 10 snoRNAs.
Intron-encoded snoRNAs can be matured via a major splicing-dependent pathway and a secondary splicing-independent pathway (Fig. 2). In the splicing-dependent pathway, the snoRNA-containing intron is spliced out as a lariat. Then the RNA lariat-debranching enzyme debranches the lariat, facilitating the exonucleolytic digestion of the flanking sequences to produce the accurate 5′ and 3′ ends of the snoRNA (17,66,96). In the splicing-independent pathway, endonucleolytic cleavages are made within the host intron followed by exonucleolytic trimming to produce the mature 5′ and 3′ ends (Fig. 2). The gene for the L1 ribosomal protein of Xenopus and its human homolog contain two snoRNAs, U16 and U18, which are processed in this endonuclease-dependent pathway (15). In snoRNAs transcribed from independent promoters, initiation sites for exonucleolytic trimming are often produced by the endonucleases like Rnt1p, an RNase III homolog in yeast (20). Two 5′→3′ exonucleases, Xrn1p and Rat1p, in yeast are required for the 5′ processing of several snoRNAs. These snoRNAs can be either synthesized from polycistronic pre-snoRNA transcripts or excised from the introns of pre-mRNAs following intron lariat debranching (96,102).
Figure 2.
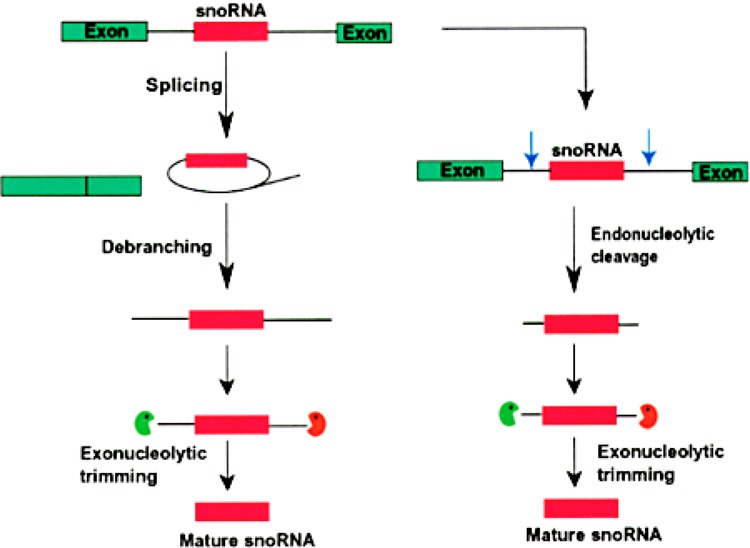
The 3′ end formation of intronic snoRNAs in eukaryotes. Intron-encoded snoRNAs (pink box) are transcribed as a part of a pre-mRNA. Green boxes indicate exons. The snoRNA is released from the rest of the intron by two possible ways. In the first pathway, the intron lariat, formed by splicing, is linearized by a debranching activity. 5′ end processing is carried out by 5′→3′ exonucleases (green pacman), while the 3′→5′ exonucleases organized in the exosome complex (orange pacman) trim the 3′ end trailer sequence to form the mature 3′ end. In the alternate pathway, endonucleolytic cleavages (blue arrows) upstream and downstream of the snoRNA release the snoRNA. The mature 5′ and 3′ ends are generated by exonucleases, similar to the first processing pathway.
Correct 3′ end processing of the box C/D snoRNAs depends on conserved structural elements located in their coding regions. Formation of the 5′, 3′-terminal stem structure and binding of protein factors to the adjacent box C and D sequences are thought to block further cleavage by exonucleases in the formation of mature snoRNA (15,17,132,143,147). A similar structural motif with the terminal stem structures and the H/ACA boxes is required for the 3′ end formation in the H/ACA box snoRNAs (11). Maturation of some yeast snoRNAs by trimming of short 3′ trailer sequences specifically requires the 3′→5′ exonuclease Rrp6p that is a nuclear component of the yeast exosome (3,4).
SnoRNAs in plants have two unique features that are in contrast to vertebrate snoRNAs. First, plants have a unique organization of snoRNA genes where multiple snoRNA genes are tightly clustered around a number of different loci (13,70). Both of the two major classes of snoRNAs (box C/D and box H/ACA) are transcribed as polycistronic pre-snoRNA transcripts from an upstream promoter (Fig. 3) and are processed by a splicing-independent mechanism that requires endonucleolytic cleavage in the spacer regions (71,117). Yeast snoR190 and U14 (151) are also transcribed as polycistronic transcripts. From yeast polycistronic pre-snoRNA transcripts, the Rnt1p endoribonuclease releases individual pre-snoRNA fragments and exonucleolytic trimming forms the correct 5′ and 3′ termini of the snoRNA (21,105). Second, U3 snoRNA is transcribed by RNA polymerase III in plants and by RNA polymerase II in animals (65). However, the precursor U3 snoRNAs in both plant and animal cells are processed by the exosome to form the mature 3′ end.
Figure 3.
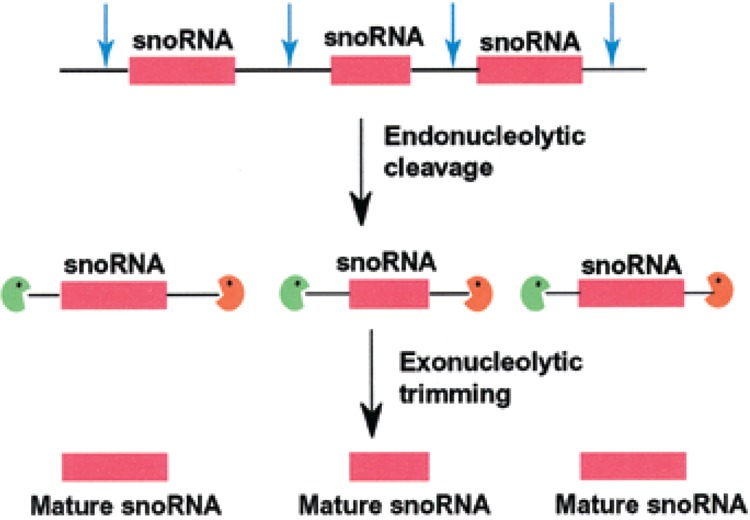
The 3′ end formation of polycistronic snoRNAs in eukaryotes. Polycistronic snoRNAs are transcribed by a common upstream promoter. Individual snoRNAs (pink box) are released by the action of endonucleases upstream and downstream of the snoRNA sequence. 5′→3′ exonucleases and 3′→5′ exonucleases trim the 5′ leader and 3′ trailer sequences to generate the mature 5′ and 3′ ends of the snoRNA.
THE 3′ END FORMATION IN snRNAs TRANSCRIBED BY RNA POLYMERASE III (Pol III)
Most eukaryotic small RNAs transcribed by RNA polymerase III, such as 5S, U6, SRP, MRP, RNaseP, 7SK, and plant U3 RNAs, terminate with 4–uridylic acid residues as their 3′ end sequence. Some pol III transcripts such as Ro (Y) RNAs retain their 3′ ends and are found in the cytoplasm with –UUUUOH or –UUUUUOH. Most other Pol III transcripts are processed at their 3′ ends and contain sequences slightly different from their original transcripts (120). In some instances, the 3′ end modifications are minor, and in other cases they are significant. Accurate in vitro systems are now available where the 3′ end processing reactions on the SRP/Alu RNA and U6 snRNA are faithfully replicated. In addition to uridylation and deuridylation of the 3′ U tail, a single adenylic acid residue is added in many small RNAs in vitro and in vivo (23,100,120).
POSTTRANSCRIPTIONAL URIDYLATION OF U6 snRNA
Among RNAs that are posttranscriptionally uridylated, the 3′ uridylation of U6 snRNA has been most extensively studied. We and other investigators have shown that when HeLa cell extracts are incubated in the presence of [α-32P]UTP, many small RNAs including U6 and 5S RNAs get labeled on their 3′ ends (55,82,106,120,129). The specific activity of [32P]phosphate in the 3′-terminal nucleotides of frog 5S rRNA (33) and human U6 snRNA (82) were found to be several-fold higher compared with the internal phosphates, indicating that the 3′ end sequences undergo posttranscriptional turnover. Benecke’s lab has partially purified a terminal uridylic acid transferase with specificity to the U6 snRNA from HeLa cells (131). In addition to the uridylation on U6 snRNA and 5S rRNA, these RNAs are processed by exonucleases in vivo as well as in vitro. The deuridylation process has been demonstrated in vitro (48) and it appears that it is during this deuridylation process that the cyclic phosphate structure is formed on the 3′ end of U6 snRNA (Fig. 4).
Figure 4.
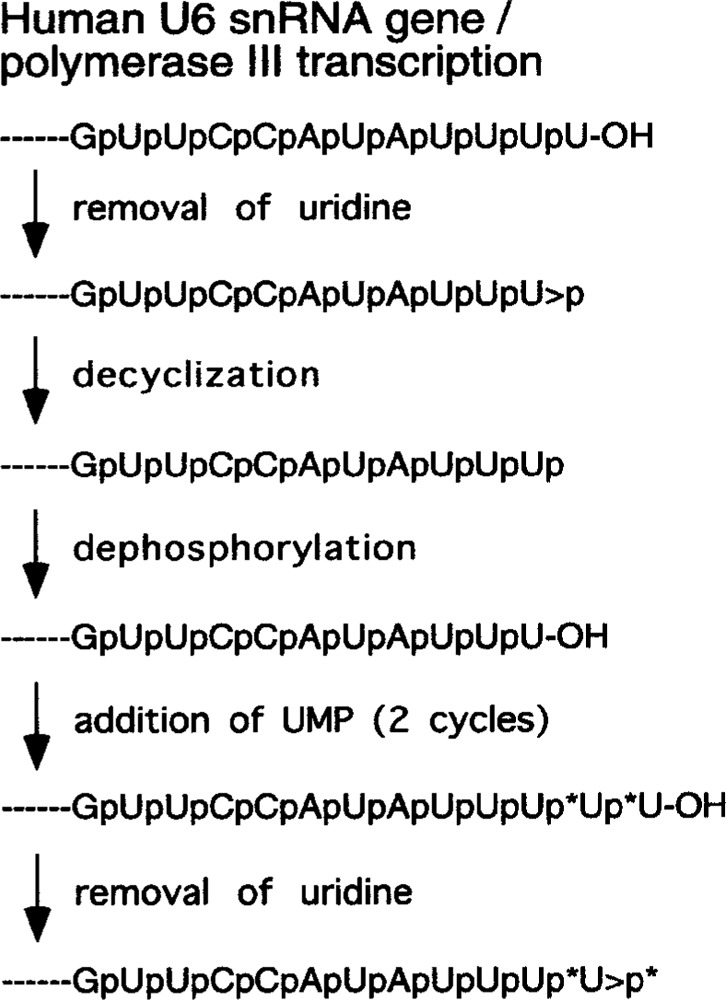
A model for the trimming, 2′,3′-cyclic phosphate (>p) formation, and addition of nucleotides to the 3′ end of U6 snRNA. The removal of nucleoside involves cleavage of ester bond between 3′-phosphate and 5′-hydroxyl group. The asterisks indicate 32P-labeled phosphate residues derived from [α-32P]UTP.
CYCLIC PHOSPHATE ON THE 3′ END OF RNAs
Lund and Dahlberg (82) showed that about 90% of the U6 snRNA in human cells contains 2′, 3′-uridine cyclic phosphate (U>p) at its 3′ end. Brunel and his coworkers concluded that this U>p formation is coupled to its involvement in splicing of pre-mRNAs (40,129). But in splicing-deficient extracts where essential spliceosomal snRNAs were specifically degraded, the cyclic phosphate formation still occurred on U6 snRNA. These data indicate that U>p formation in U6 snRNA may not coupled to pre-mRNA splicing (48). Moreover, the formation of >p is not unique to U6 snRNA. In addition to U6 snRNA, many other RNAs are known to contain 2′, 3′-cyclic phosphate structures. The autolytic products of several viral RNAs contain cyclic phosphates at their 3′ ends (14,41,58,81). The cleavage products of pre-tRNA (38,46,98) and pre-rRNA (50) contain 2′, 3′-cyclic phosphate structures. Shumyatsky et al. (119) isolated and fractionated Ehrlich ascites carcinoma (mouse) cell RNAs on sucrose density gradients and analyzed poly(A)+ RNA isolated from light, intermediate, and heavy fractions. Cyclic phosphates (pC>p and pU>p) were found in the poly(A)+ fraction obtained from the intermediate fraction. The synthesis of these cyclic phosphate-containing RNAs was inhibited by low concentrations of α-amanitin, indicating that cyclic phosphate may be present at the 3′ end of some mRNAs. It appears that cyclic phosphate-containing RNAs are intermediate products in the 3′ end metabolism of many small RNAs.
Because nascent U6 snRNA transcript contains a 3′ hydroxyl group, >p in U6 snRNA is formed post-transcriptionally. In the 3′ end formation model proposed by Gu et al. (48), the U>p appears to be an intermediate after the removal of a nucleoside from RNAs containing a 3′-OH by an exonuclease (Fig. 4). One possible candidate enzyme for the cyclic phosphate formation is the human RNA 3′ cyclase enzyme that has been extensively characterized by Filipowicz’s group. The cyclase enzyme is a nuclear enzyme that is consistent with the involvement of the cyclase in the formation of >p on the 3′ end of U6 snRNA and possibly other small RNAs. Filipowicz’s lab has purified, characterized, and cloned a RNA 3′ cyclase enzyme from many species including E. coli, yeast, plants, and human cells (9,39,42,44). In fact, the substrate that was used to identify and purify the cyclase enzyme was RNA with 3′ phosphate (37). In addition to the cyclase enzyme, there is also another enzyme that hydrolyzes 2′, 3′-cyclic phosphates on nucleotides or RNAs (43). At present, the function(s) of this highly conserved cyclase enzyme or the phosphodiesterase are not known.
ADENYLATION OF SMALL RNAs
We have characterized the 3′-terminal nucleotide of several small RNA species and found that, in every case examined, a fraction of the RNA contained a posttranscriptionally added adenylic acid residue that is not present in the corresponding gene [(120); Table 2]. In the case of human SRP and 7SK RNAs, this posttranscriptional adenylation was found in 70% of the RNA molecules (24,120,134). These data indicate that in many human small RNA molecules, one or more 3′ end nucleotides are removed and a single adenylic acid residue is added. Adenylation where 1–2 adenylic acid residues are posttranscriptionally added is known to occur in many RNAs, including some stable small RNAs of E. coli (78).
TABLE 2.
POSTTRANSCRIPTIONAL ADENYLATION/URIDYLATION OF HUMAN SMALL RNAs IN VIVO
| RNA | Gene Sequence | Primary Transcript | Posttranscriptional Adenylation/Uridylation | |||
|---|---|---|---|---|---|---|
| Trimming | % | Adenylation | % | |||
| SRP RNA | –GTCTCTTTT/GCCCCCCCTCC | –GUCUCUUUUOH | –GUCUCUUOH | 30 | –GUCUCUUA OH | 70 |
| –GUCUCUOH | –GUCUCUA OH | |||||
| 7SK RNA | –GCCTTTCTTTT/GACCCATT | –GCCUUUCUUUUOH | –GCCUUUCUUUOH | –GCCUUUCUUUA OH | ||
| –GCCUUUCUUOH | 40 | –GCCUUUCUUA OH | 60 | |||
| –GCCUUUCUOH | –GCCUUUCUA OH | |||||
| U2 snRNA | –GGTGCACCCCCTCCGGGGA/ | –GGUGCACCCCCUCCGGGGAOH | –GGUGCACCOH | 30 | –GGUGCACCA OH | 70 |
| 5S rRNA | –GGCTTTT/TCTTTGG | –GGCUUUUOH | –GGCUUUOH | –GGCUUUA OH | ||
| –GGCUUOH | 90 | –GGCUUA OH | 10 | |||
| –GGCUOH | –GGCUA OH | |||||
| Deuridylation↑↓Uridylation | ||||||
| –GGCUU OH | ||||||
| –GGCUUU OH | ||||||
| –GGCUUUU OH | ||||||
| U6 snRNA | –GTTCCATATTTT/TACATCAGG | –GUUCCAUAUUUUOH | –GUUCAUAUUUOH | –GUUCCAUAUUUA OH | ||
| –GUUCCAUAUUOH | >90 | –GUUCCAUAUUA OH | >5 | |||
| –GUUCCAUAUOH | –GUUCCAUAUA OH | |||||
| Deuridylation↑↓Uridylation | ||||||
| –GUUCCAUAUUUUU OH | ||||||
| –GUUCCAUAUUUUUUU OH | ||||||
The adenylic and uridylic acid residues shown in bold italic are added posttranscriptionally and are not encoded in the corresponding genes. The percentages of different 3′ end nucleotides were obtained by quantitating the labeled 3′ ends (120).
SRP RNA is the RNA component of the signal recognition particle, which plays an important role in translocation of membrane proteins and secretory proteins (126,140,141). SRP RNA is synthesized in the nucleus by RNA polymerase III and translocates to the nucleolus, enroute to the cytoplasm. SRP RNA in the nucleolus is already processed and adenylated, indicating that 3′ end processing and adenylation are early events in the biogenesis of the signal recognition particle (23). The SRP consists of two distinct functional domains. The first one is the Alu domain consisting of the 5′ and the 3′ end portions of the SRP RNA associated with the SRP 9/14 protein heterodimer (127). This domain has a tRNA-like structure and plays an important role in arresting elongation of the nascent polypeptide in the ribosome (146). The minimal domain necessary for binding with the SRP 9/14 protein heterodimer is an 86-nucleotide-long motif including the 5′ end, the 3′ end, and stems III and IV in the Alu portion of the SRP RNA (144). The second functional domain consists of the SRP RNA-specific S fragment and four SRP proteins. This domain is responsible for targeting the ribosome–nascent peptide chain complex to the surface of rough endoplasmic reticulum by interacting with the SRP receptor (141). Recently, Jacobson and Pederson (59) showed that SRP RNA, when injected into the nucleoplasm, first migrates to the nucleolus and then to the cytoplasm. The minimal domain necessary for migration from the nucleoplasm to the nucleolus is an 86-nucleotide-long domain consisting of the 5′ end, 3′ end, stem III, and stem IV in the Alu portion of SRP RNA. The minimal domain necessary for 3′ end processing and adenylation is again the same 87-nucleotide-long Alu motif (23). These data show that this tRNA-like domain of SRP RNA has multiple functions in the biogenesis of SRP RNA, including binding of the SRP 9/14 kDa protein heterodimer, 3′ end processing, 3′ adenylation, and transport to the nucleolus prior to exiting into the cytoplasm.
The 3′ adenylated Alu RNA as well as the adenylated SRP RNA were bound to the SRP 9/14 kDa heterodimer and can be coimmunoprecipitated by specific antibodies against SRP 9/14 proteins (23). The mutant Alu RNA that fails to bind SRP 9/14 heterodimer cannot be adenylated or processed at the 3′ end (121). These data are consistent with a model (Fig. 5) where the nascent SRP and/or Alu RNAs first bind to SRP 9/14 protein heterodimer, followed by the removal of extra nucleotides on the 3′ end and then the addition of one adenylic acid residue in the nucleus, before transport into the cytoplasm (23). The machinery capable of accurately adenylating SRP RNA is present both in the nucleus and in the cytoplasm (100). This is similar to the mRNA adenylation and the tRNA aminoacylation machinery, which are present and functional in the nucleus as well as in the cytoplasm. However, the model presented in Figure 5 does not exclude the possibility that SRP RNA without the posttranscriptionally added adenylic acid is also transported from nucleus to the cytoplasm. In fact, some cytoplasmic RNAs like human and yeast ribosomal 5S RNA and yeast SRP RNA contain a very low percentage of posttranscriptional adenylation where adenylated RNAs account for 5% or less (95,100). Therefore, it is likely that presence of a posttranscriptionally added adenylic acid is not a requirement for transport across the nuclear membrane. This 3′ end adenylation of small RNAs is carried out by an adenylating machinery different from the mRNA polyadenylation machinery (121). The enzyme responsible for the single adenylic acid addition has been characterized and is highly homologous to the poly(A) polymerase that polyadenylates mRNA (101).
Figure 5.
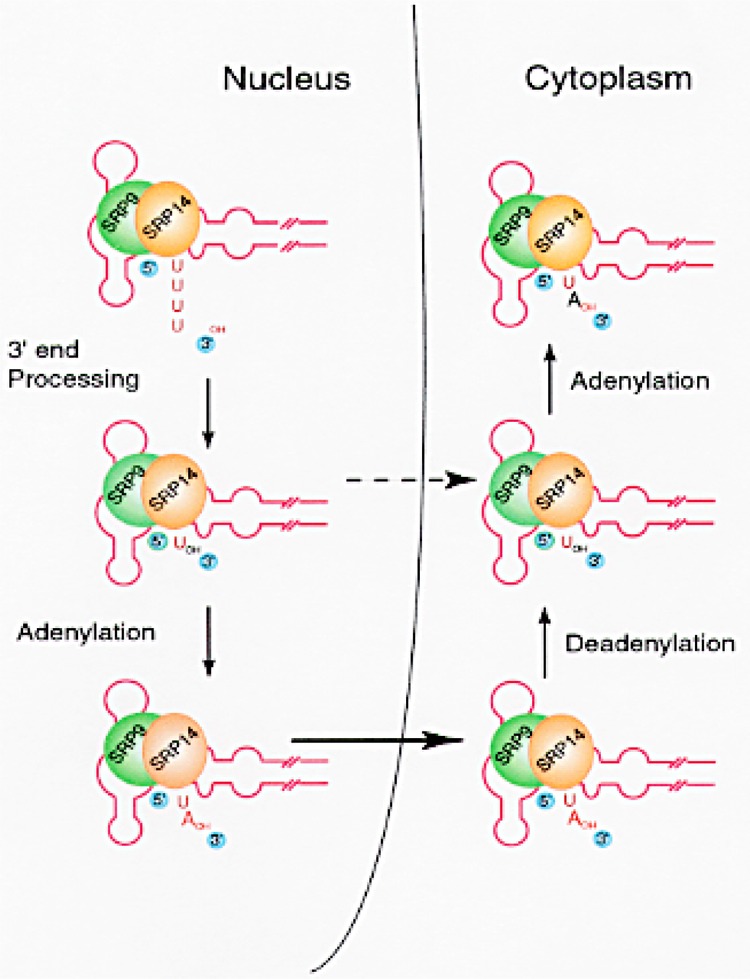
A model for the adenylation and turnover of posttranscriptionally added adenylic acid in SRP RNA. The secondary structure of SRP and Alu RNAs has been studied by several investigators (141). SRP 9/14 protein heterodimer was shown to bind in this region of SRP RNA (127,144). The relative size and sites of binding of SRP 9/14 protein heterodimer are shown only for the purpose of illustration. Broken arrow represents a possible alternate pathway for the transport of SRP RNA without the 3′ end adenylation.
URIDYLATION AND ADENYLATION ON THE 3′ ENDS OF SMALL RNAs AS MECHANISMS TO MAINTAIN THE INTEGRITY OF 3′ ENDS
U6 small nuclear RNA and ribosomal 5S RNA are examples of small RNAs that are posttranscriptionally uridylated on their 3′ ends. However, a small fraction of U6 snRNA and 5S rRNA molecules from human cells as well as Xenopus oocytes contain a single posttranscriptionally added adenylic acid residue on their 3′ ends. While the U6 snRNA with uridylic acid residue on its 3′ end was readily uridylated, U6 snRNA molecules with posttranscriptionally added adenylic acid residue on their 3′ ends were not uridylated in vivo and in vitro, or when injected into Xenopus oocytes (24). Similar results were obtained with 5S rRNA and 7SK RNA in vitro where 3′ AOH-containing RNAs were not further uridylated (24). These data demonstrate that the presence of a single post-transcriptionally added adenylic acid residue on the 3′ end of U6 snRNA, 5S rRNA or 7SK RNA prevents 3′ uridylation. A model (Fig. 6) has been proposed where adenylation and uridylation are two competing processes that add nucleotides on the 3′ end of some small RNAs. One of the functions of the 3′ adenylation could be to negatively regulate the 3′ uridylation of small RNAs. We, as well as other investigators, showed that the posttranscriptionally added 3′ adenylic acid residues and 3′ uridylic acid residues constantly turn over. In other words, the 3′ ends of these RNAs are being constantly trimmed and rebuilt. However, the 3′ adenylic acid residue turnover is slow whereas uridylation has a faster turnover (23,120). Thus, even a low percentage of 3′ adenylation may result in significant reduction of the uridylation of RNAs. It is possible that 3′ adenylation with a low turnover rate and uridylation with a high turnover rate are two intimately related processes designed to rebuild and maintain the 3′ ends of RNAs intact.
Figure 6.
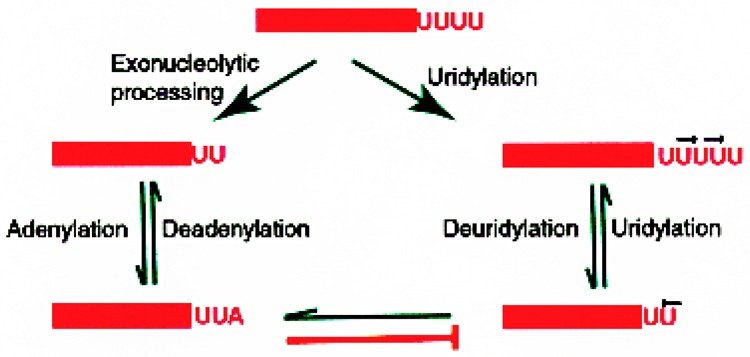
A model depicting the 3′ end deletions/additions occurring on the 3′ end of human small RNAs. All available evidence is in support of this model. The RNAs bind with appropriate proteins to form the ribonucleoprotein particles. The 3′ ends of RNAs are trimmed where one or more nucleotides are removed. These RNAs can be rebuilt by uridylation; thus, this reaction is reversible. The RNAs are also adenylated and deadenylated; this reaction also is reversible. The RNAs containing adenylic acid residues cannot be uridylated. This reaction is not reversible. RNAs containing adenylic acid on the 3′ end have to be first deadenylated before further uridylation can take place.
URIDYLATION OF GUIDE RNAs IN KINETOPLASTID MITOCHONDRIA
Guide RNAs that are involved in mRNA editing are another group of RNAs that undergo 3′ end uridylation. RNA editing is the posttranscriptional process during which the nucleotide sequence of mRNAs is altered by base modifications, substitutions, insertions, or deletions of nucleotides to produce a new coding sequence (7,8). This mRNA editing is facilitated by trans-acting guide RNAs of 55–70 nucleotides in length that act as templates for editing. The enzymatic activities required for RNA editing are in two RNP complexes (99,104) called complex I and complex II. Complex I is made up of the gRNAs, an editing site-specific endonuclease, an RNA ligase, a 3′ uridyl exonuclease, and a terminal uridyl transferase (TUTase). Complex II is essentially complex I with the mRNA to be edited (29,61,110,115).
When isolated mitochondria were incubated with [α-32P]UTP, both edited mRNA and gRNAs were labeled as a result of TUTase activity but not due to mitochondrial transcription (51,99). This is reminiscent of [α-32P]UTP labeling of U6 snRNA in higher eukaryotes and in HeLa cell extracts as a result of posttranscriptional uridylation at the 3′ end of U6 RNA (106,109). The U residues inserted during editing are derived from the cellular UTP pool and are added to the 3′ terminus of a 5′ pre-mRNA cleavage product (61). U residues are also added posttranscriptionally to the 3′ end of gRNAs to give a poly(U) tail of about 5–24 nucleotides (10).
A model in which the gRNA maturation occurs in complex II of the mRNA editing reaction has been proposed by McManus et al. (87). The gRNAs that are associated with complex I are subject to both 3′ end uridylation by TUTase and trimming of the U tail by the 3′ uridyl exonuclease. These gRNAs therefore contain a stretch of 3′ U residues. When complex II is assembled, these gRNAs pair with their cognate pre-mRNA at specific sites for editing. Subsequently, the 3′ U tail of the gRNA is then stabilized by base pairing with the purine-rich regions flanking the editing site of the pre-mRNA. Using 3′ cross-linking studies, Leung and Koslowsky (73) have shown that the U tail interacts with purine-rich mRNA sequences upstream to the editing site, thus strengthening the gRNA/cognate pre-mRNA interactions. The 3′ U tail may also act together with the 5′ anchor domain of the gRNAs to reduce secondary structure in the mRNA in the immediate editing domain, thus increasing accessibility of the editing complex to the immediate editing site. The U tail also acts as a tether and stabilizes the 5′ pre-mRNA fragment after cleavage by the endonuclease. But as editing progresses, the number of U residues that interact with the upstream sequences decreases and instead the U tail of the gRNA can interact with its own guiding region to maintain important secondary structures (74). The poly(U) tail of the gRNAs would thus be inaccessible for trimming by the 3′ terminal exonuclease.
THE 3′ END FORMATION IN TELOMERE RNA AND B2 RNA
Some of the small RNAs are processed at their 3′ ends by the mRNA adenylation pathway. The two main examples are the telomere RNA and the rodent B2 RNA.
Telomere RNA
The integral RNA subunit of telomerase contains a template region that determines the sequence added to the chromosome ends. In yeast and mammals, telomere RNA is a Pol II transcript, while in ciliates, it is a Pol III transcript. Human telomerase RNA has a H/ACA domain that is essential in vivo for its accumulation, 3′ end processing, as well as for telomerase activity (88). This RNA is also known to be associated with GAR protein that is common to H/ACA box-containing RNAs. At steady state, 5–10% of the telomerase RNA in Saccharomyces cerevisiae and Kluyveromyces lactis contains a poly(A) tail of about 80 nucleotides. The telomere RNA has the mRNA polyadenylation signal at its 3′ end and its poly(A) tail is added by the same machinery that polyadenylates mRNAs (22).
B2 RNA
B2 repeats are a group of short interspersed elements (SINEs) specific for the rodent genome. The repeats are about 180 bp long and are present at nearly 105 copies per genome. The B2 RNA is transcribed from distinct B2 genes and terminate with 3′ UUUU-OH on its 3′ end. The B2 element has a Pol III promoter and B2 genes have a consensus Pol III promoter and a 3′ polyadenylation signal (69). This polyadenylation signal is the same as the mRNA polyadenylation sequence (AAUAAA) and the B2 RNA is polyadenylated by the mRNA poly(A) polymerase. Two populations of B2 RNA exist: a polyadenylated form with poly(A) tail ranging in size from 200 to 400 nt and a nonpolyadenylated form. Both the polyadenylated and the nonpolyadenylated B2 RNAs contain the methylphosphate cap structure at their 5′ ends (119).
THE 3′ END FORMATION IN YEAST SMALL RNAs
Yeast spliceosomal RNAs, transcribed by Pol II, are processed at their 3′ ends in very similar but distinct pathways. The 3′ end formation in U1 yeast snRNA is absolutely dependent on the 3′ terminal Sm site (114). Any mutation at this site leads to accumulation of two species: a minor polyadenylated and a major nonadenylated 64–78 nt 3′ extended form (Fig. 7). At 80–81 nucleotides and 114–115 nucleotides downstream of the mature 3′ end are a pair of cleavage sites for the endonuclease Rnt1p. Following cleavage by Rnt1p, these precursor RNAs are further processed by the exosome to produce the mature 3′ end. But inactivation of the Rnt1p cleavage pathway still yields U1 snRNAs with mature 3′ ends. Therefore, yeast U1 RNA has Rnt1p-dependent and Rnt1p-independent processing pathways, both of which require the functional Sm site. Yeast U5 snRNA exists as two forms: the longer U5L and the shorter U5S. U5 precursor RNA is cleaved by Rnt1p at sites located 26–27 (pre-U5S) and 90–91 (pre-U5L) nucleotides downstream of the mature 3′ end (19). These cleaved precursors are then trimmed by the exosome to the mature 3′ end (3). Exonucleases from the RNase D family have also been implicated in this pathway (137).
Figure 7.
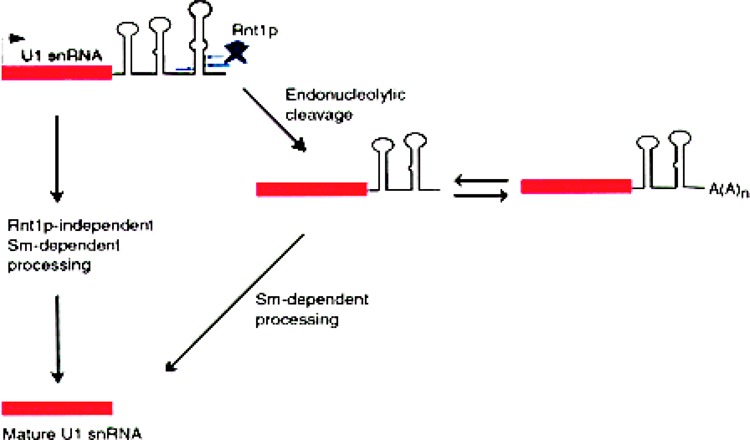
The 3′ end formation in yeast U1 snRNA. U1 snRNA in S. cerevisiae is made as a precursor with ∼120 extra nucleotides on the 3′ end. There are two alternate pathways for generating the mature 3′ end (114). One of the pathways is dependent on Rnt1p, an endonuclease with homology to RNase III, while the second pathway is independent of Rnt1p. Both of these maturation pathways are dependent on the 3′-terminal Sm site and associated proteins. In the Rnt1p-dependent pathway, there are two prominent intermediates: a nonpolyadenylated RNA, extending 64–78 nucleotides beyond the mature 3′ end, and the related polyadenylated RNA.
Unlike yeast U1 and U5 snRNAs that have alternate 3′ processing pathways to bypass the Rnt1p processing, yeast U2 processing is Rnt1p dependent. Preventing normal processing of U2 snRNA by Rnt1 results in the accumulation of extended and polyadenylated U2 RNA, which are still functional in splicing (1). The role of Rnt1 appears to be limited to the production of site-specific cleavages to generate processing intermediates, which are later trimmed to the mature ends, possibly by the exosome. Polyadenylated forms of U4 snRNA, 5S rRNA, and snoRNAs also accumulate in yeast, which are deficient in essential exosome components (72,103,138). This dynamic balance, between the exosome to trim the 3′ end and the polyadenylation machinery to elongate the 3′ end, is also seen in the prokaryotes. Recently, it has also been shown that the 3′ end of SRP RNA in yeast is also processed by the exosome; cells lacking Dis3p (Rrp44p) accumulate aberrantly processed RNA that is not transported out of the nucleus (47). This demonstrates that integrity of the 3′ end could be very essential to the function of some RNAs.
THE 3′ END FORMATION IN PROKARYOTIC SMALL RNAs
In addition to ribosomal 5S RNA and tRNAs, stable small RNAs in E. coli include many small regulatory RNAs (142). Most of these RNAs are synthesized as longer precursors and then processed at both the 5′ and 3′ ends to yield the mature RNA. Similar to eukaryotic 3′ end processing, endonucleolytic cleavage followed by exonucleolytic trimming of RNAs are the two common processes involved in the formation of the 3′ end of E. coli small RNAs. Different combinations of these two processes are used to generate the mature 3′ end. Formation of the 3′ end in two representative RNAs are discussed below (Fig. 8).
Figure 8.
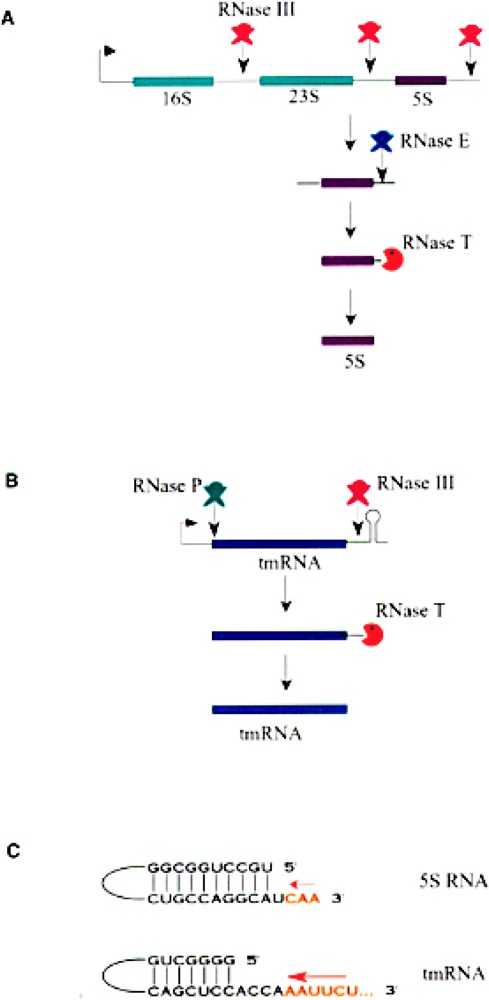
The 3′ end formation in two bacterial RNAs: ribosomal 5S RNA and tmRNA. (A) 5S rRNA (purple box) is synthesized as part of a polycistronic primary transcript that contains the other two ribosomal RNAs, 16S and the 18S rRNAs (green boxes). The endonuclease RNase III cleaves the primary transcript to separate the individual rRNAs. Another endonuclease RNase E cleaves the 5S RNA precursor at about three nucleotides away from the mature 3′ end (128). The exoribonuclease RNase T trims the final three nucleotides to generate the mature 3′ end (77). (B) Pre-tmRNA is folded into a pre-tRNA-like structure in vivo such that it can be cleaved by RNase P to generate the 5′ end of the mature tmRNA. The 3′ trailer sequence is acted upon by endoribonuclease RNase III. The final exonucleolytic trimming by RNase T yields the mature 3′ end –CCA. It has also been shown that the final 3′ end could be obtained by endonucleolytic cleavage by RNase E. (C) Sequence and structure near the 3′ end of 5S RNA and tmRNA (77). Nucleotides present in the mature RNA are shown in black letters while the nucleotides that are removed by the final exonucleolytic trimming are shown in orange letters. Arrows indicate the direction of trimming by RNase T.
Ribosomal 5S RNA is synthesized as part of a 30S precursor that contains the other ribosomal RNAs. RNase III releases the individual rRNAs from the primary transcript by a series of endonucleolytic cleavages. RNase E cleaves the E. coli 5S RNA precursor in a single-stranded region of four nucleotides, after which three nucleotides are removed to yield mature 5S RNA by RNase T (Fig. 8A) (77,128). RNase M5 cleaves the B. subtilis equivalent of 5S RNA precursor in a double-stranded region to yield mature 5S rRNA in one step (27). RNase P RNA (M1 RNA) is also processed in a similar manner (77). M1 RNA is generated as a primary transcript from a proximal promoter or as part of a long precursor RNA like the 5S RNA from a distal promoter.
The tmRNA, also known as the 10Sa RNA or the SsrA RNA, functions uniquely both as tRNA and mRNA when ribosomes pause at the 3′ end of a truncated mRNA lacking an in-frame stop codon. This process, referred to as trans-translation, leads to the addition of a short peptide tag (11 amino acids) to the carboxy-terminus of the incomplete nascent poly-peptide (62,68). The tagged polypeptide labels the truncated protein as a target for carboxy-terminal-specific proteases. The tmRNA is synthesized as a 457-nucleotide-long primary transcript from its own promoter and has a rho-dependent termination signal. Seven nucleotides from the 5′ end are removed through an endonucleolytic cleavage by RNase P to generate the mature 5′ end (Fig. 8B). The 3′ end is acted upon by an endonuclease, RNase III following which the final mature 3′ end is generated by 3′→5′ exonuclease, RNase T (77). RNase E has also been shown to generate the mature 3′ end by a single endonucleolytic cleavage (79). Regulatory RNAs like OxyS RNA, DsrA RNA, and MicF appear to require no 3′ exoribonucleolytic trimming and are functional as primary transcripts.
The 3′ end processing appears to be dictated by RNA secondary structure and the property of exoribonuclease(s). Most of the small RNAs have the 5′ and 3′ ends base paired with each other to form a stable, double-stranded stem generally followed by several unpaired 3′ nucleotides (76,77) (Fig. 8C). The 3′ extra nucleotides in the RNA precursor extend the 3′ unpaired region further, and these unpaired nucleotides are trimmed by various exoribonucleases in the final maturation step. For most exoribonucleases, this barrier serves to stop the trimming reaction when the single-stranded tail is four nucleotides long. However, RNase T appears to differ from the other RNases in that it can approach closer to the double-stranded stem. This property is the hallmark of RNase T, which is the only exoribonuclease that can trim the 3′ terminus of 5S RNA to yield the mature 3′ end with only one unpaired nucleotide. RNase T is also the only enzyme that participates in the 3′ end processing of the –CCA sequence of tRNA. This is also consistent with the fact that RNase T is generally the most active RNase for removing the extra residues closest to the mature 3′ termini of the other stable RNAs. RNase T does not act on long 3′ trailer sequences or on single-stranded substrates (77).
Polyadenylated precursor species of many small RNAs have been detected in exoribonuclease-deficient cells (78,148). Most of the mature small RNAs are not polyadenylated even in these cases, possibly because their 3′ end is protected either by secondary structures or buried inside a ribonucleoprotein complex. There appears to be a dynamic balance between trimming to generate the mature 3′ end and polyadenylation to mediate decay. The oligo(A) tails synthesized by poly(A) polymerase might facilitate RNA decay through the degradosome; a multiprotein complex containing an endoribonuclease (RNase E), an exoribonuclease (polynucleotide phosphorylase), and a DEAD box helicase (RhlB) has a central role in mRNA degradation (16,94).
EXOSOME: ENZYME COMPLEX RESPONSIBLE FOR THE 3′ END FORMATION OF SMALL RNAs
Many exonucleases have been extensively characterized from bacteria and yeast, and the RNA processing and/or RNA degradation/turnover is affected in yeast mutants defective in these exonucleases. Tollervey’s lab made the exciting discovery that many of these exonucleases are part of a large functional complex designated “exosome” (90). The E. coli counterpart of the exosome is referred to as the degradosome. The exosome is a multiprotein complex consisting of several 3′→5′ exoribonucleases, helicases, and associated factors (Table 3). It has been shown to be responsible for accurate 3′ end processing and degradation of many different cellular RNAs including mRNAs and small RNAs. The presence of multiple exoribonucleases in an exosome complex is analogous to the presence of multiple proteases in the proteasome (6,32,45). It is suggested that there might be a fundamental advantage to the compartmentalization of degradative enzymes as large complexes (136). Both exosomes and proteasomes require ATP for their functions. Similar to the proteasome, the exosome is present in both nucleus and cytoplasm. This conclusion is based on immunolocalization of core exosome subunits as well as biochemical fractionation (3,64,90,151). However, one of the known exoribunucleases Rrp6p in yeast is confined to the nucleus. In human cells, Rrp6p corresponds to the PM-Scl 100 protein that is also restricted to the nucleus (Table 3). Therefore, it appears that exosome does not have a uniform and homogeneous structure. The nuclear and cytoplasmic forms have many common components and few different subunits (91,136). It is also possible that different components can be recruited to the core exosome complex in order to assist RNA-specific 3′ end processing.
TABLE 3.
COMPONENTS OF THE YEAST EXOSOME
| Exosome Subunit (Yeast) | Proposed In Vitro Activity | Phenotype | E. coli Homolog | Mammalian Homolog |
|---|---|---|---|---|
| Core subunits | ||||
| Rrp4p | 3′ exohydrolase | essential | S1 RNA BD | hRrp4p |
| Rrp40p | essential | S1 RNA BD | hRrp40p | |
| Rrp41p/ski6p | 3′ exophosphorolase | essential | RNase PH | hRrp41p |
| Rrp42p | essential | RNase PH | hRrp42p | |
| Rrp43p | essential | RNase PH | ||
| Rrp44p/Dis3p | 3′ exohydrolase | essential | RNase II | hDis3p |
| Rrp45p | 3′ exophosphorolase | essential | RNase PH | PM-Scl75 |
| Rrp46p | essential | RNase PH | hRrp46p | |
| Mtr3p | essential | RNase PH | ||
| Cs14p | essential | S1 RNA BD | hCs14p | |
| Nuclear subunit | ||||
| Rrp6p | 3′ exohydrolase | ts lethal | RNase D | PM-Scl100 |
| Associated factors | ||||
| Mtr4p | RNA helicase | |||
| Ski2p | RNA helicase | |||
| Ski3p | TPR domains | |||
| Ski8p | WD domains | |||
The various exonucleases in the yeast exosome are listed with their proposed activity in vitro and with the phenotype of the corresponding yeast mutant (4,136). The E. coli and mammalian homologos are also listed wherever known.
All the characterized exosome exonucleases act by two well-defined mechanisms (35,80,118,136). The first category is made up of the 3′ exohydrolases, which use water to hydrolyze the 3′ nucleotide releasing it as 5′ pN 3′OH, and leaving the RNA substrate as RNA 3′OH:
The second class of exonucleases termed as phosphorolases use phosphate as the attacking nucleophilic group instead of water. The orthophosphate moiety (*Pi) is transferred to the 3′ nucleotide that is being removed. The digestion products are nucleotide 5′ diphosphates and RNA with 3′ hydroxyl group:
Both types of exonucleases have been extensively characterized from E. coli and yeast. While detailed characterization of human exonucleases has not been reported, it is likely that similar mechanisms of action will be found for human exoribonucleases as in bacteria and yeast. Exonucleases, other than those present as part of the exosome, have also been implicated in 3′ end processing of small RNAs (137). These exonucleases work in redundant pathways or sequentially with the exosome in the same 3′ end processing pathway.
CONCLUSIONS AND PERSPECTIVES
The 3′ end formation is an important metabolic step in small RNA biogenesis. Generation of the mature 3′ end appears to be very critical because multiple redundant pathways exist in a cell to ensure the process. Although the correct 3′ end formation has been shown to be important in localization and function of many RNAs, like in nuclear import for the snRNAs and in base pairing with cognate mRNA for the gRNAs, the primary role is believed to be in the maintenance of the integrity of the RNA. Stem–loop structures and long homopolymeric tails are hindrances to the 3′ exonucleolytic complexes. The 3′ ends of RNAs could be protected by their inclusion in ribonucleoproteins. Synthetic reactions like uridylation and adenylation could serve as repair processes to regenerate the 3′ end. Because small RNAs are essential components for many pathways inside the cell, future challenges lie in understanding not only the 3′ end formation mechanisms but also the 3′ end maintenance mechanisms in these RNAs.
ACKNOWLEDGMENT
Studies in our lab were supported by a grant from NIH (GM-38320).
REFERENCES
- 1. Abou-Elela S.; Ares M. Depletion of yeast RNase III blocks correct U2 3′-end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17:3738–3746; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ack R. A.; Weiner A. M. The highly conserved U small nuclear RNA 3′-end formation signal is quite tolerant to mutation. Mol. Cell. Biol. 7:2070–2079; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allmang C.; Kufel J.; Chanfreau G.; Mitchell P.; Petfalski E.; Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399–5410; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allmang C.; Petfalski E.; Podteejnikov A.; Mann M.; Tollervey D.; Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 13:2148–2158; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balakin A. G.; Smith L.; Fournier M. J. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86:823–834; 1996. [DOI] [PubMed] [Google Scholar]
- 6. Baumeister W.; Waiz J.; Zuhl F.; Seemuller E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell 92:367–380; 1998. [DOI] [PubMed] [Google Scholar]
- 7. Benne R.; Van den Burg J.; Brakenhoff J. P.; Sloof P.; Van Boom J. H.; Tromp M. C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46:819–826; 1986. [DOI] [PubMed] [Google Scholar]
- 8. Benne R. RNA editing in trypanosomes. The us(e) of guide RNAs. Mol. Biol. Rep. 1:217–227; 1992. [DOI] [PubMed] [Google Scholar]
- 9. Billy E.; Hess D.; Hofsteenge J.; Filipowicz W. Characterization and adenylation site in the RNA 3′-terminal phosphate cyclase from E. coli. J. Biol. Chem. 274:39455–39460; 1999. [DOI] [PubMed] [Google Scholar]
- 10. Blum B.; Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell 62: 391–397; 1990. [DOI] [PubMed] [Google Scholar]
- 11. Bortolin M. L.; Ganot P.; Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ri-bosomal RNAs. EMBO J. 18:457–469; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brouwer R.; Pruijn G. J.; van Venrooij W. J. The human exosome: An autoantigenic complex of exori-bonucleases in myositis and scleroderma. Arthritis Res. 3:102–106; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brown J. W.; Shaw P. J. Small nucleolar RNAs and pre-rRNA processing in plants. Plant Cell 10:649–657; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buzayan J. M.; Hampel A.; Bruening G. Nucleotide sequence and newly formed phosphodiester bond of spontaneously ligated satellite tobacco ringspot virus RNA. Nucleic Acids Res. 14:9729–9743; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Caffarelli E.; Fatica A.; Prislei S.; De Gregorio E.; Fragapane P.; Bozzoni I. Processing of the intron-encoded U16 and Ul 8 snoRNAs: The conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 15: 1121–1131; 1996. [PMC free article] [PubMed] [Google Scholar]
- 16. Carpousis A. J.; Vanzo N. F.; Raynal L. C. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 15:24–28; 1999. [DOI] [PubMed] [Google Scholar]
- 17. Cavaille J.; Bachellerie J. P. Processing of fibril-larin-associated snoRNAs from pre-mRNA introns: An exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie 78: 443–456; 1996. [DOI] [PubMed] [Google Scholar]
- 18. Cavaille J.; Nicoloso M.; Bachellerie J. P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383:732–735; 1996. [DOI] [PubMed] [Google Scholar]
- 19. Chanfreau G.; Elela S. A.; Ares M. Jr.; Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 11:2741–2751; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chanfreau G.; Legrain P.; Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284:975–988; 1998. [DOI] [PubMed] [Google Scholar]
- 21. Chanfreau G.; Rotondo G.; Legrain P.; Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17: 3726–3737; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chapon C.; Cech T. R.; Zaug A. J. Polyadenylation of telomerase RNA in budding yeast. RNA 3:1337–1351; 1997. [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y.; Sinha K.; Perumal K.; Gu J.; Reddy R. Accurate 3′-end processing and adenylation of human signal recognition particle RNA and Alu RNA in vitro. J. Biol. Chem. 273:35023–35031; 1999. [DOI] [PubMed] [Google Scholar]
- 24. Chen Y.; Sinha K.; Perumal K.; Reddy R. Presence of a single adenylic acid residue on the 3′ end of human U6 snRNA prevents 3′ uridylation in vitro and in frog oocytes. RNA 6:1277–1288; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ciliberto G.; Dathan N.; Frank R.; Philipson L.; Mattaj I. W. Formation of the 3′ end on U snRNAs requires at least three sequence elements. EMBO J. 5:2931–2937; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clayton D. A. A nuclear function for RNase MRP. Proc. Natl. Acad. Sci. USA 91:4615–4617; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Condon C.; Brechemier-Baey D.; Beltchev B.; Grunberg-Manago M.; Putzer H. Identification of the gene encoding the 5S ribosomal RNA maturase in Bacillus subtilis: Mature 5S rRNA is dispensable for ribosome function. RNA 7:242–253; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de la Cruz J.; Kressler D.; Tollervey D.; Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in S. cerevisiae . EMBO J. 17:1128–1140; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cruz-Reyes J.; Sollner-Webb B. Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc. Natl. Acad. Sci. USA 93: 8901–8906; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cuello P.; Boyd D. C.; Dye M. J.; Proudfoot N. J.; Murphy S. Transcription of the human U2 snRNA genes continues beyond the 3′ box in vivo. EMBO J. 18:2867–2877; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dahlberg J. E.; Lund E. The genes and transcription of major small nuclear RNAS. In: Birnstiel M., ed. Structure and function of major and minor snRNPs. Berlin: Springer Verlag; 1988:38–70. [Google Scholar]
- 32. DeMartino G. N.; Slaughter C. A. The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 274:22123–22126; 1999. [DOI] [PubMed] [Google Scholar]
- 33. Denis H.; Wegnez M. Biochemical research on oogenesis. Synthesis and maturation of 5S RNA in the small oocytes of Xenopus laevis . Biochimie 55:1137–1151; 1973. [DOI] [PubMed] [Google Scholar]
- 34. Deutscher M. P. Ribonucleases, tRNA nucleotidyl-transferase, and the 3′ processing of tRNA. Prog. Nucleic Acids Res. Mol. Biol. 39:209–240; 1990. [DOI] [PubMed] [Google Scholar]
- 35. Deutscher M. P. Promiscuous exoribonucleases of E. coli . J. Bacteriol. 175:4577–4583; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Edmonds M. Polyadenylate polymerases. Methods Enzymol. 181:161–170; 1990. [DOI] [PubMed] [Google Scholar]
- 37. Filipowicz W.; Konarska M.; Gross H. J.; Shatkin A. RNA 3′-terminal phosphate cyclase activity and RNA ligation in HeLa cell extract. Nucleic Acids Res. 11:1405–1418; 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Filipowicz W.; Shatkin A. J. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell 32:547–557; 1983. [DOI] [PubMed] [Google Scholar]
- 39. Filipowicz W.; Billy E.; Drabikowski K.; Genschik P. Cyclases of the 3′-terminal phosphate in RNA: A new family of RNA processing enzymes conserved in eucarya, bacteria and archaea. Acta Biochim. Pol. 45: 895–906; 1998. [PubMed] [Google Scholar]
- 40. Forne T.; Labourier E.; Antoine E.; Rossi F.; Gallouzi I.; Cathala G.; Tazi J.; Brunei C. Structural features of U6 snRNA and dynamic interactions with other spliceosomal components leading to pre-mRNA splicing. Biochimie 78:436–442; 1996. [DOI] [PubMed] [Google Scholar]
- 41. Forster A. C.; Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell 49:211–220; 1987. [DOI] [PubMed] [Google Scholar]
- 42. Genschik P.; Billy E.; Swianiewicz M.; Filipowicz W. The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in Eucarya, Bacteria and Archaea. EMBO J. 16:2955–2967; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Genschik P.; Hall J.; Filipowicz W. Cloning and characterization of Arabidopsis cyclic phosphodias-terase which hydrolyzes ADP-ribose 1′, 2-cyclic phosphate and nucleoside 2′, 3′-cyclic phosphates. J. Biol. Chem. 272:13211–13219; 1997. [DOI] [PubMed] [Google Scholar]
- 44. Genschik P.; Drabikowski K.; Filipowicz W. Characterization of the E. coli RNA 3′-terminal phosphate cyclase and its sigma54-regulated operon. J. Biol. Chem. 273:25516–25526; 1998. [DOI] [PubMed] [Google Scholar]
- 45. Gottesman S.; Maurizi M. R.; Wickner S. Regulatory subunits of energy-dependent proteases. Cell 91: 435–438; 1997. [DOI] [PubMed] [Google Scholar]
- 46. Greer C. L.; Peebles C. L.; Gegenheimer P.; Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell 32:537–546; 1983. [DOI] [PubMed] [Google Scholar]
- 47. Grosshans H.; Deinert K.; Hurt E.; Simos G. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP RNA, and Xpo1p-mediated export. J. Cell Biol. 153:745–762; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gu J.; Shumyatsky G.; Makan N.; Reddy R. Formation of 2′, 3′-cyclic phosphates at the 3′ end of human U6 snRNA in vitro. J. Biol. Chem. 272: 21989–21993; 1997. [DOI] [PubMed] [Google Scholar]
- 49. Guthrie C.; Patterson B. Spliceosomal snRNAs. Annu. Rev. Genet. 22:387–419; 1988. [DOI] [PubMed] [Google Scholar]
- 50. Hannon G. J.; Maroney P. A.; Branch A.; Benenfield B. J.; Robertson H. D.; Nilsen T. W. Accurate processing of human pre-rRNA in vitro. Mol. Cell. Biol. 9:4422–4431; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harris M. E.; Moore D. R.; Hajduk S. L. Addition of uridines to the edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem 265:11368–11376; 1990. [PubMed] [Google Scholar]
- 52. Henry Y.; Wood H.; Morrissey J. P.; Petfalski E.; Kearsey S.; Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13:2452–2463; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hernandez N. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 4:1827–1837; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hernandez N.; Weiner A. M. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell 47:249–258; 1986. [DOI] [PubMed] [Google Scholar]
- 55. Hirai H.; Lee D. I.; Natori S.; Sekimizu K. Uridylation of U6 RNA in a nuclear extract in Ehrlich ascites tumor cells. J. Biochem. (Tokyo) 104:991–994; 1988. [DOI] [PubMed] [Google Scholar]
- 56. Huang O.; Jacobson M. R.; Pederson T. 3′ processing of human pre-U2 small nuclear RNA: A base-pairing interaction between the 3′ extension of the precursor and an internal region. Mol. Cell. Biol. 17: 7178–7185; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang O.; Pederson T. A human U2 RNA mutant stalled in 3′ end processing is impaired in nuclear import. Nucleic Acids Res. 27:1025–1031; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hutchins C. J.; Rathjen P. D.; Forster A. C.; Symons R. H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14:3627–3640; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jacobson M.; Pederson T. Localization of SRP RNA in the nucleolus of mammalian cells. Proc. Natl. Acad. Sci. USA 95:7981–7986; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jady B. E.; Kiss T. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 20:541–551; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kable M. L.; Seiwert S. D.; Heidmann S.; Stuart K. RNA editing: A mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273:1189–1195; 1996. [DOI] [PubMed] [Google Scholar]
- 62. Keiler K. C.; Waller P. R.; Sauer R. T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993; 1996. [DOI] [PubMed] [Google Scholar]
- 63. Keller W. No end yet to messenger RNA 3′ processing! Cell 81:829–832; 1995. [DOI] [PubMed] [Google Scholar]
- 64. Kinoshita N.; Goebl M.; Yanagida M. The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol. Cell. Biol. 11: 5839–5847; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kiss T.; Marshalisay C.; Filipowicz W. Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell 65:517–526; 1991. [DOI] [PubMed] [Google Scholar]
- 66. Kiss T.; Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 9:1411–1424; 1995. [DOI] [PubMed] [Google Scholar]
- 67. Kiss-Laszlo Z.; Henry Y.; Bachellerie J. P.; Caizergues-Ferrer M.; Kiss T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 85:1077–1088; 1996. [DOI] [PubMed] [Google Scholar]
- 68. Komine Y.; Kitabatake M.; Yokogawa T.; Nishikawa K.; Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli . Proc. Natl. Acad. Sci. USA 91: 9223–9227; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kramerov D. A.; Tillib S. V.; Ryskov A. P.; Georgiev G. P. Nucleotide sequence of small polyadenyated B2 RNA. Nucleic Acids Res. 13:6423–6437; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Leader D. J.; Clark G. P.; Wafters J.; Seven A. F.; Shaw P. J.; Brown J. W. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 16:5742–5751; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Leader D. J.; Clark G. P.; Watters J.; Seven A. F.; Shaw P. J.; Brown J. W. Splicing-independent processing of plant box C/D and box H/ACA small nucleolar RNAs. Plant Mol. Biol. 39:1091–1100; 1999. [DOI] [PubMed] [Google Scholar]
- 72. Lee Y.; Nazar R. N. Ribosomal 5S RRNA maturation in S. cerevisiae . J. Biol. Chem. 272:15206–15212; 1997. [DOI] [PubMed] [Google Scholar]
- 73. Leung S. S.; Koslowsky D. J. Mapping contacts between gRNA and mRNA in trypanosome RNA editing. Nucleic Acids Res. 27:778–787; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Leung S. S.; Koslowsky D. J. RNA editing in Trypanosoma brucei: Characterization of gRNA U-tail interactions with partially edited mRNA substrates. Nucleic Acids Res. 29:703–709; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Leverefte R. D.; Andrews M. T.; Maxwell E. S. Mouse U14 snRNA is a processed intron of the cognate hsp7O heat shock pre-messenger RNA. Cell 71: 1215–1221; 1992. [DOI] [PubMed] [Google Scholar]
- 76. Li Z.; Deutscher M. P. Maturation pathways for E. coli tRNA precursors: A random multienzyme process in vivo. Cell 86:503–512; 1996. [DOI] [PubMed] [Google Scholar]
- 77. Li Z.; Pandit S.; Deutscher M. P. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli . Proc. Natl. Acad. Sci. USA 95:2856–2861; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li Z.; Pandit S.; Deutscher M. P. Polyadenylation of stable RNA precursors in vivo . Proc. Natl. Acad. Sci. USA 95:12158–12162; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lin-Chao S.; Wei C. L.; Lin Y. T. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl. Acad. Sci. USA 96:12406–12411; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Littauer U. Z.; Soreq H. Polynucleotide phosphorylase. In: Boyer P., ed. The enzymes, vol. 15 New York: Academic Press; 1982:518–553. [Google Scholar]
- 81. Long D. M.; Uhlenbeck O. C. Self-cleaving catalytic RNA. FASEB J. 7:25–30; 1993. [DOI] [PubMed] [Google Scholar]
- 82. Lund E.; Dahlberg J. E. Cyclic 2′, 3′-phosphates and nontemplated nucleotides at the 3′-end of spliceosomal U6 snRNAs. Science 255:327–330; 1992. [DOI] [PubMed] [Google Scholar]
- 83. Lygerou Z.; Allmang C.; Tollervey D.; Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 272:268–270; 1996. [DOI] [PubMed] [Google Scholar]
- 84. Manley J. L. A complex protein assembly catalyzes polyadenylation of mRNA precursors. Curr. Opin. Genet. Dev. 5:222–228; 1995. [DOI] [PubMed] [Google Scholar]
- 85. Mattaj I. W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46:905–911; 1986. [DOI] [PubMed] [Google Scholar]
- 86. Maxwell E. S.; Fournier M. J. The small nucleolar RNAs. Annu. Rev. Biochem. 64:897–934; 1995. [DOI] [PubMed] [Google Scholar]
- 87. McManus M. T.; Adler B. K.; Pollard V. W.; Hajduk S. L. Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol. Cell. Biol. 20:883–891; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mitchell J. R.; Cheng J.; Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19:567–576; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mitchell P.; Petfalski E.; Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 15:502–513; 1996. [DOI] [PubMed] [Google Scholar]
- 90. Mitchell P.; Petfalski E.; Shevchenko A.; Mann M.; Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91:457–466; 1997. [DOI] [PubMed] [Google Scholar]
- 91. Mitchell P.; Tollervey D. Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 7:843–846; 2000. [DOI] [PubMed] [Google Scholar]
- 92. Neuman de Vegvar H. E.; Lund E.; Dahlberg J. E. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell 47:259–266; 1986. [DOI] [PubMed] [Google Scholar]
- 93. Neuman de Vegvar H. E.; Dahlberg J. E. Nucleo-cytoplasmic transport and processing of small nuclear RNA precursors. Mol. Cell. Biol. 10:3365–3375; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nicholson A. W. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 23:371–390; 1999. [DOI] [PubMed] [Google Scholar]
- 95. O’Brien C. A.; Wolin S. L. A possible role for the 60-kDa Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes Dev. 8:2891–2903; 1994. [DOI] [PubMed] [Google Scholar]
- 96. Ooi S. L.; Samarsky D. A.; Fournier M. J.; Boeke J.eD. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: Intron length effects and activity of a precursor snoRNA. RNA 4:1096–1110; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Peculis B. A. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S RRNA maturation. Mol. Cell. Biol. 17:3702–3713; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Peebles C. L.; Gegenheimer P.; Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell 32:525–536; 1983. [DOI] [PubMed] [Google Scholar]
- 99. Peris M.; Frech G. C.; Simpson A. M.; Bringaud F.; Byrne E.; Bakker A.; Simpson L. Characterization of two classes of RNP complexes possibly involved in RNA editing from Lelishmania tarentolae mitochondria. EMBO J. 13:1664–1672; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Perumal K.; Gu J.; Reddy R. Evolutionary conservation of post-transcriptional 3′ end adenylation of small RNAs: S. cerevisiae SRP RNA and U2 snRNA are post-transcriptionally adenylated. Mol. Cell. Biochem. 208:99–109; 2000. [DOI] [PubMed] [Google Scholar]
- 101. Perumal K.; Sinha K.; Henning D.; Reddy R. Purification, characterization, and cloning of the cDNA of human signal recognition particle RNA 3′-adenylating enzyme. J. Biol. Chem. 276:21791–21796; 2001. [DOI] [PubMed] [Google Scholar]
- 102. Petfalski E.; Dandekar T.; Henry Y.; Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181–1189; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Piper P. W.; Bellatin J. A.; Lockheart A. Altered maturation of sequences at the 3′ terminus of 5S gene transcripts in a Saccharomyces cerevisiae mutant that lacks a RNA processing endonuclease. EMBO J. 3: 353–359; 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Pollard V. W.; Harris M. E.; Hajduk S. L. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11:4429–4438; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Qu L. H.; Henras A.; Lu Y. J.; Zhou H.; Zhou W. X.; Zhu Y. O.; Zhao J.; Henry Y.; Caizergues-Ferrer M.; Bachellerie J. P. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 19:1144–1158; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Reddy R.; Henning D.; Das G.; Harris M.; Wright D. The capped U6 small nuclear RNA is transcribed by RNA polymerase 111. J. Biol. Chem. 262:75–81; 1987. [PubMed] [Google Scholar]
- 107. Reddy R.; Busch H. Small nuclear RNAs: RNA sequences, structure, and modification. In: Birnstiel M., ed. Structure and function of major and minor snRNPs. Berlin: Springer Verlag; 1988:1–37. [Google Scholar]
- 108. Reddy R.; Singh R. Synthesis of small nuclear RNAs. Prog. Mol. Subcell. Biol. 12:1–36; 1991. [Google Scholar]
- 109. Rinke J.; Steitz J. A. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 13:2617–2629; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rusche L. N.; Cruz-Reyes J.; Piller K. J.; Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16:4069–4081; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sachs A.; Wahle E. Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem. 268:22955–22958; 1993. [PubMed] [Google Scholar]
- 112. Sarkar N. Polyadenylation of MRNA in prokaryotes. Annu. Rev. Biochem. 66:173–197; 1997. [DOI] [PubMed] [Google Scholar]
- 113. Schaufele F.; Gilmartin G. M.; Bannwarth W.; Birnstiel M. L. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature 323: 777–781; 1986. [DOI] [PubMed] [Google Scholar]
- 114. Seipelt R. L.; Zheng B.; Asuru A.; Rymond B. C. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 27:587–595; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Seiwert S. D.; Heidmann S.; Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84:831–841; 1996. [DOI] [PubMed] [Google Scholar]
- 116. Sharp P. A. Split genes and RNA splicing. Cell 77: 805–815; 1994. [DOI] [PubMed] [Google Scholar]
- 117. Shaw P. J.; Beven A. F.; Leader D. J.; Brown J. W. Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J. Cell Sci. 111:2121–2128; 1998. [DOI] [PubMed] [Google Scholar]
- 118. Shen V.; Schiessinger D. RNases 1, 11 and IV of E. coli . In: Boyer P., ed. The enzymes, vol. 15 New York: Academic Press; 1982:501–515. [Google Scholar]
- 119. Shumyatsky G. P.; Tillib S. V.; Kramerov D. A. B2 RNA and 7SK RNA, RNA polymersae III transcripts, have a cap-like structure at their 5′ end. Nucleic Acids Res. 18:6357–6351; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sinha K. M.; Gu J.; Chen Y.; Reddy R. Adenylation of small RNAs in human cells. J. Biol. Chem. 273:6853–6859; 1998. [DOI] [PubMed] [Google Scholar]
- 121. Sinha K.; Perumal K.; Chen Y.; Reddy R. Post-transcriptional adenylation of SRP RNA is carried out by an enzyme different from mRNA poly(A) poly-merase. J. Biol. Chem. 274:30826–30831; 1999. [DOI] [PubMed] [Google Scholar]
- 122. Sollner-Webb B.; Tycowski K. T.; Steitz J. A. Ribosomal RNA processing in eukaryotes. In: Zimmerman R.; Dahlberg A., ed. Ribosomal RNA: Structure, evolution, processing and function in protein synthesis. Boca Raton, FL: CRC Press; 1996:469–490. [Google Scholar]
- 123. Stark B. C.; Kole R.; Bowman E. J.; Altman S. Ribonuclease P: An enzyme with an essential RNA component. Proc. Natl. Acad. Sci. USA 75:3717–3721; 1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Steitz J. A.; Black D.; Gerke V.; Parker K. A.; Kramer A.; Frendeway D.; Keller W. Functions of abundant U snRNAs. In: Birnstiel M., ed. Structure and function of major and minor snRNPs. Berlin: Springer Verlag; 1988:1–37. [Google Scholar]
- 125. Steitz J. A. Splicing takes a holiday. Science 257: 888–889; 1992. [DOI] [PubMed] [Google Scholar]
- 126. Stroud R. M.; Walter P. Signal sequence recognition and protein targeting. Curr. Opin. Struct. Biol. 9:754–759; 1999. [DOI] [PubMed] [Google Scholar]
- 127. Strub K.; Moss J.; Walter P. Binding sites of the 9-and 14-kDa heterodimeric protein subunit of SRP are contained exclusively in the Alu domain and contain a sequence motif conserved in evolution. Mol. Cell. Biol. 11:3949–3959; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Szeberenyi J.; Roy M. K.; Vaidya H. C.; Apirion D. 7S RNA, containing 5S ribosomal RNA and the termination stem, is a specific substrate for the two RNA processing enzymes RNase III and RNase E. Biochemistry 23:2952–2957; 1984. [DOI] [PubMed] [Google Scholar]
- 129. Tazi J.; Forne T.; Jeanteur P.; Cathala G.; Brunel C. Mammalian U6 snRNA undergoes 3′-end modifications within the spliceosome. Mol. Cell. Biol. 13: 1641–1650; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Tollervey D.; Kiss T. Function and synthesis of small nucleolar RNAS. Curr. Opin. Cell. Biol. 9(3): 337–342; 1997. [DOI] [PubMed] [Google Scholar]
- 131. Trippe R.; Sandrock B.; Senecke B. J. A highly specific terminal uridyl transferase modifies the 3′-end of U6 snRNA. Nucleic Acids Res. 26:3119–3126; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Tycowski K. T.; Shu M. D.; Steitz J. A. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 7:1176–1190; 1993. [DOI] [PubMed] [Google Scholar]
- 133. Tycowski K. T.; Shu M. D.; Steitz J. A. A mammalian gene with introns instead of exons generating stable RNA products. Nature 379:464–466; 1996. [DOI] [PubMed] [Google Scholar]
- 134. Ullu E.; Weiner A. M. Human genes and pseudo-genes for the 7SL RNA component of signal recognition patcle. EMBO J. 3:3303–1330; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Umen J. G.; Guthrie C. The second catalytic step of pre-mRNA splicing. RNA 1:869–885; 1995. [PMC free article] [PubMed] [Google Scholar]
- 136. Van Hoof A.; Parker R. The exosome: A proteasome for RNA? Cell 99:347–350; 1999. [DOI] [PubMed] [Google Scholar]
- 137. Van Hoof A.; Lennertz P.; Parker R. Three conserved members of the RNaseD family have unique and overlapping functions in the processing of 5S, 5.8S, U4, US, RNase MRP and RnaseP RNAs in yeast. EMBO J. 19:1357–1365; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Van Hoof A.; Lennartz P.; Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 snRNA and snoRNAs. Mol. Cell. Biol. 20:441–452; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wahle E.; Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu. Rev. Biochem. 61:419–440; 1992. [DOI] [PubMed] [Google Scholar]
- 140. Walter P.; Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 299:691–698; 1982. [DOI] [PubMed] [Google Scholar]
- 141. Walter P.; Johnson A. E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10:87–119; 1994. [DOI] [PubMed] [Google Scholar]
- 142. Wassarman K. M.; Zhang A.; Storz G. Small RNAs in Escherichia coli . Trends Microbiol. 7:37–45; 1999. [DOI] [PubMed] [Google Scholar]
- 143. Watkins N. J.; Leverette R. D.; Xia L.; Andrews M. T.; Maxwell E. S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA 2:118–133; 1996. [PMC free article] [PubMed] [Google Scholar]
- 144. Weichenrieder O.; Kapp U.; Cusack S.; Strub K. Identification of minimal Alu RNA folding domain that specifically binds SRP9/14. RNA 3:1262–1274; 1997. [PMC free article] [PubMed] [Google Scholar]
- 145. Wieben E. D.; Nenninger J. M.; Pederson T. Ribonucleoprotein organization of eukaryotic RNA. U2 small nuclear RNA precursors and their accurate 3′ processing in vitro as ribonucleoprotein particles. J. Mol. Biol. 183:69–78; 1985. [DOI] [PubMed] [Google Scholar]
- 146. Wolin S. L.; Walter P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol. 109:2617–2622; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Xia L.; Watkins N. J.; Maxwell E. S. Identification of specific nucleotide sequences and structural elements required for intronic U 14 snoRNA processing. RNA 3:17–26; 1997. [PMC free article] [PubMed] [Google Scholar]
- 148. Xu F.; Cohen S. N. RNA degradation in E. coli regulated by 3′ adenylation and 5′ phosphorylation. Nature 374:180–183; 1995. [DOI] [PubMed] [Google Scholar]
- 149. Yang H.; Moss M. L.; Lund E.; Dahlberg J. E. Nuclear processing of the 3′-terminal nucleotides of pre-U1 RNA in Xenopus laevis oocytes. Mol. Cell. Biol. 12:1553–1560; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Yuo C. Y.; Ares M. Jr.; Weiner A. M. Sequences required for 3′ end formation of human U2 small nuclear RNA. Cell 42:193–202; 1985. [DOI] [PubMed] [Google Scholar]
- 151. Zagorski J.; Tollervey D.; Fournier M. J. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensable and essential small nuclear RNAs. Mol. Cell. Biol. 8:3282–3290; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Zanchin N. I.; Goldfarb D. S. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol. Cell. Biol. 19:1518–1525; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Zanchin N. I.; Goldfarb D. S. The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res. 27: 1283–1288; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


