Abstract
Proper floral patterning, including the number and position of floral organs in most plant species, is tightly controlled by the precise regulation of the persistence and size of floral meristems (FMs). In Arabidopsis, two known feedback pathways, one composed of WUSCHEL (WUS) and CLAVATA3 (CLV3) and the other composed of AGAMOUS (AG) and WUS, spatially and temporally control floral stem cells, respectively. However, mounting evidence suggests that other factors, including phytohormones, are also involved in floral meristem regulation. Here, we show that the boundary gene SUPERMAN (SUP) bridges floral organogenesis and floral meristem determinacy in another pathway that involves auxin signaling. SUP interacts with components of polycomb repressive complex 2 (PRC2) and fine‐tunes local auxin signaling by negatively regulating the expression of the auxin biosynthesis genes YUCCA1/4 (YUC1/4). In sup mutants, derepressed local YUC1/4 activity elevates auxin levels at the boundary between whorls 3 and 4, which leads to an increase in the number and the prolonged maintenance of floral stem cells, and consequently an increase in the number of reproductive organs. Our work presents a new floral meristem regulatory mechanism, in which SUP, a boundary gene, coordinates floral organogenesis and floral meristem size through fine‐tuning auxin biosynthesis.
Keywords: auxin, floral meristem, floral organogenesis, H3K27me3, polycomb repressive complexes, SUPERMAN
Subject Categories: Plant Biology
Introduction
In many angiosperms, floral patterning is tightly controlled by the precise coordination of stem cell proliferation in the floral meristem (FM), commitment of stem cell descendants to specific floral organs, and establishment of meristem‐to‐organ and organ‐to‐organ boundaries. By such mechanisms, the number and position of floral organs for a given species are well defined. Wild‐type (WT) Arabidopsis flowers consist of four types of organs arranged in a series of concentric whorls: four sepals in the outermost whorl 1, followed by four petals in whorl 2, six stamens in whorl 3, and two fused carpels in the innermost whorl 4. While three classes of homeotic genes, classes A, B, and C, function alone or in combination to determine the cell identities of floral organs (Bowman et al, 1991; Coen & Meyerowitz, 1991), the precise developmental regulations of the FM that determine the family‐ and/or species‐specific numbers of floral organs and whorls remain unknown.
In Arabidopsis, a negative feedback loop between the WUSCHEL (WUS)‐expressing organizing center and CLAVATA3 (CLV3)‐expressing stem cells maintains the appropriate size of both FMs and shoot apical meristems (SAMs; Brand et al, 2000). FM activity is associated with the number of floral organs. Mutation in WUS causes plants to lose the ability to maintain stem cells and prematurely stops organ formation (Laux et al, 1996). In contrast, stem cells accumulate in clv3 mutants due to unrestricted WUS expression, leading to the formation of more organs (Clark et al, 1995; Fletcher et al, 1999). Unlike the indeterminate SAM, the FM is determinate and ceases to maintain stem cells after the initiation of carpels. Another negative feedback between WUS and the class C gene AGAMOUS (AG) plays a central role in this termination process (Lenhard et al, 2001; Lohmann et al, 2001; Sun et al, 2009, 2014). AG is induced at floral stage 3 by WUS and the FM regulator LEAFY (LFY) in whorls 3 and 4 of floral primordia where stamens and carpels will develop in later stages (Lohmann et al, 2001). AG in turn represses WUS, both directly by affecting the recruitment of polycomb group (PcG) proteins to the WUS locus and indirectly through the C2H2 zinc finger protein KNUCKLES (KNU), to terminate stem cell maintenance at floral stage 6, approximately 2 days after AG induction (Sun et al, 2009; Liu et al, 2011). In ag and knu loss‐of‐function mutants, WUS expression remains active beyond stage 6, which is sufficient to induce FM indeterminacy, leading to the production of extra whorls of reproductive organs (Lenhard et al, 2001; Sun et al, 2009). AG also activates the YABBY family transcription factor CRABS CLAW to regulate carpel organogenesis and FM determinacy through the establishment of auxin maxima in the fourth whorl (Yamaguchi et al, 2017).
The number and position of floral organs are also controlled by boundary genes, which function through various mechanisms, including the crosstalk with the phytohormone auxin (Zadnikova & Simon, 2014). The NAC family transcription factors CUP‐SHAPED COTYLEDON1‐3 (CUC1‐3), which participate in the formation of boundaries between organs and between organs and meristems, are negatively regulated by auxin‐dependent signaling pathways (Takada et al, 2001; Daimon et al, 2003). In the Arabidopsis SAM, new floral primordia are initiated in the peripheral zone, at the region where auxin concentration is highest. As the primordium forms, auxin is depleted from the boundary separating the emerging primordium from the meristem and flows toward the incipient position of the next primordium (Heisler et al, 2005). Thus, CUC genes are restricted in the boundary regions of low auxin activity. Auxin also controls the size of the root meristem non‐cell autonomously; this auxin signaling is antagonistic to cytokinin signaling, and cytokinin negatively controls the root meristem size (Dello Ioio et al, 2007). In contrast to root meristems, cytokinin signaling and WUS activity in the SAM could reinforce each other in a positive feedback (Leibfried et al, 2005; Gordon et al, 2009; Zhao et al, 2010). Although auxin and cytokinin show opposite functions in the regulation of shoot and root meristems, the function of auxin in FMs is not well understood (Werner et al, 2003; Schaller et al, 2015).
The SUPERMAN (SUP) gene encodes a transcription factor with a C2H2‐type zinc finger motif and is proposed to function as a boundary gene to separate the stamen‐producing whorl 3 from the carpel‐producing whorl 4 (Sakai et al, 1995). Loss of function of SUP leads to an increased number of stamens, suggesting that SUP is involved in both floral patterning and FM determinacy (Bowman et al, 1992; Gaiser et al, 1995). AG is a positive regulator of SUP transcription, and SUP mRNA level is greatly reduced in ag mutants (Bowman et al, 1992). Notably, the transient and weak expression of SUP in ag mutants is sufficient for some level of function, since ag sup double mutants show strong synergistic effects on FM size, causing enlarged and fasciated FMs (Bowman et al, 1992). Although sup mutants were identified and well characterized decades ago, how SUP functions to bridge floral organogenesis and FM determinacy is still unclear. A recent study showed that SUP cell autonomously prevents the ectopic expression of class B/stamen identity genes in whorl 4, and non‐cell autonomously promotes stem cell termination in developing flowers (Prunet et al, 2017). The ectopic expression of SUP in different plant species leads to dwarf plants with organs of reduced size, which could be associated with both auxin and cytokinin signaling defects (Hiratsu et al, 2002; Nibau et al, 2011). However, it is difficult to distinguish the causal factors of the sup phenotypes from the consequence of altered morphology.
Here, we elucidate how SUP functions to control floral organogenesis and FM size non‐cell autonomously. SUP interacts with PcG proteins to exert its function as an active repressor and negatively regulates auxin biosynthesis in the stamen‐to‐carpel boundary region. In the sup mutant, the derepression of YUCCA (YUC) genes YUC1 and YUC4 leads to increased auxin accumulation and the formation of extra primordia of reproductive organs. Consistently, treatment with an anti‐auxin (p‐chlorophenoxyisobutyric acid, PCIB) can rescue the stamen number and carpel defects of sup mutants. Increased local auxin biosynthesis in the SUP‐expressing region leads to sup‐like floral phenotypes. Our work presents a new mechanism on how the boundary gene SUP coordinates floral organogenesis and FM size through fine‐tuning of auxin biosynthesis.
Results
SUP regulates floral stem cells non‐cell autonomously
We first tested whether the formation of supernumerary stamens in sup mutants is associated with WUS function in FMs. A loss of WUS activity leads to the premature termination of FMs so that both wus‐1 single‐mutant and wus‐1 sup‐5 double‐mutant flowers typically form only a single stamen and no carpels (Laux et al, 1996; Fig 1A and B). Thus, wus‐1 is fully epistatic to sup, suggesting that the sup phenotype of supernumerary stamens is dependent on WUS function. In contrast, flowers of ag‐1 sup‐5 double mutants show enhanced meristem indeterminacy (Bowman et al, 1991; Uemura et al, 2017). Taken together, these results suggest that SUP may regulate WUS in FMs and that this regulation might be at least in part independent from the known AG‐WUS feedback pathways.
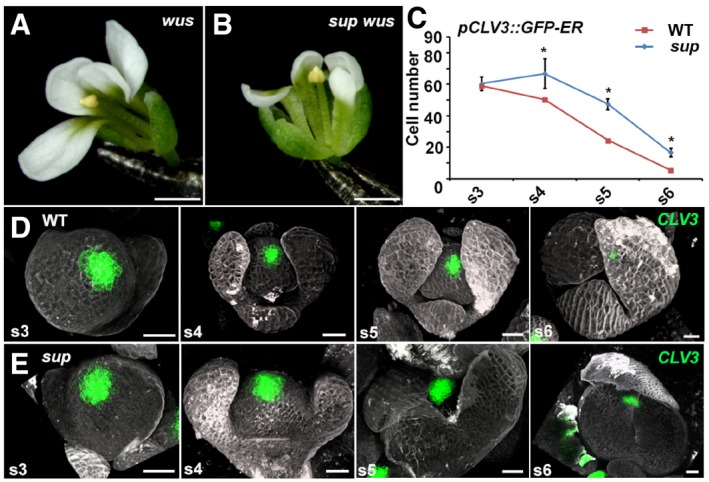
Figure 1. SUP spatially controls the FM size in a non‐cell‐autonomous manner.
-
A, Bwus‐1 (A) and sup‐5 wus‐1 (B) mutant flowers with one stamen and without carpels. Scale bars, 1 mm.
-
CThe comparison of the number of cells with the stem cell marker pCLV3::GFP‐ER signals in WT and sup‐5. The numbers of cells with the signals were counted based on the z‐stack images. From stage 4 (s4) onwards, the sup‐5 floral buds showed increased numbers of CLV3‐expressing stem cells compared with those of WT. Error bars indicate s.d. of 12–15 samples; two‐tailed Student's t‐test, *P < 0.05.
-
D, EThe pCLV3::GFP‐ER (green) in WT (D) and sup‐5 (E) floral buds at different floral stages. Scale bars, 20 μm.
To address whether SUP regulates floral stem cell activities, we monitored the expression of the stem cell marker CLV3 in sup‐5 mutant flowers (Fig 1C–E). Using a pCLV3::GFP‐ER reporter (Reddy & Meyerowitz, 2005), we determined that there is no obvious difference of fluorescence intensity between WT and sup; however, the CLV3 expression region appeared slightly broader in sup flowers from stage 4 onward (Fig 1D and E). To further test this, we counted the number of cells expressing pCLV3::GFP‐ER in sup and WT flowers at different stages and found that while the number of cells expressing pCLV3::GFP‐ER was comparable between WT and sup at stage 3, from stage 4 onward, it was significantly higher in sup floral buds (Fig 1C–E). This result suggests that there are an increased number of floral stem cells in sup mutants. To confirm this observation, we employed a floral induction system (denoted: ap1 cal p35S::AP1‐GR), which is based on the activation of a fusion protein between the APETALA1 (AP1) transcription factor and the steroid‐binding domain of the rat glucocorticoid receptor (GR) in the inflorescence‐like meristems of ap1 cauliflower (cal) double mutants by dexamethasone (DEX) treatment and allows the collection of a large number of synchronized floral buds for analysis (Wellmer et al, 2006). Using real‐time quantitative reverse transcription PCR (qRT–PCR), we detected increased transcription levels for both CLV3 and WUS in stage 6 flowers of ap1 cal p35S::AP1‐GR sup‐5 plants relative to those of ap1 cal p35S::AP1‐GR plants (Fig EV1A). We also detected pCLV3::GFP‐ER expression at later floral stages in sup‐5 than in the wild type (Fig EV1B), confirming previous reports that floral stem cell termination is delayed in sup (Prunet et al, 2017). Altogether, our data show that SUP influences floral stem cells both spatially and temporally.
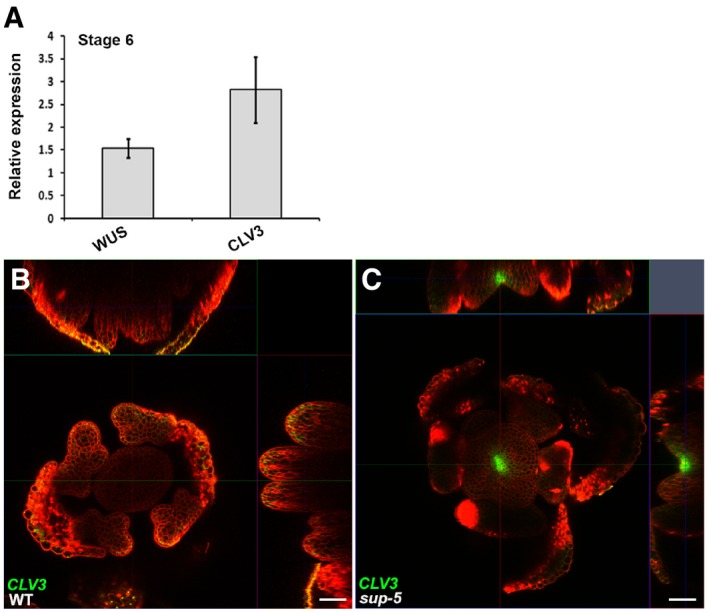
Figure EV1. WUS and CLV3 are increased at stage 6 floral buds in sup‐5 .
-
AExpression of WUS and CLV3 (in floral buds of approximately stage 6) 5 days after treatment of ap1 cal p35S::AP1‐GR sup‐5 inflorescences with 1 μM DEX. The relative values to equally treated ap1 cal p35S::AP1‐GR plants are shown. Error bars indicate standard errors from qRT–PCR experiments of four biological repeats. P = 0.028 and 0.031 for WUS and CLV3 based on a Student's t‐test, respectively.
-
B, CThe pCLV3::GFP‐ER (green) can be detected in a sup‐5 floral bud (C) but not in WT (B) at a floral stage later than 6. Scale bars, 20 μm.
We also analyzed the expression of the meristem marker SHOOT MERISTEMLESS (STM) by using a translational reporter pSTM::STM‐VENUS and a transcriptional reporter pSTM::CFP‐N7 (Fig EV2; Heisler et al, 2005; Landrein et al, 2015). Up to stage 4, STM expression appears identical in sup‐5 and WT flowers: STM is initially expressed throughout stage 1–2 flower buds, before fading from developing sepals at stage 3 (Fig EV2A and B). STM expression domain appears larger in sup than in the wild type at late stage 5 (Fig EV2A and B), which is associated with an enlarged FM in sup. By stage 6, STM expression ceases in whorls 2 and 3 in both wild‐type and sup‐5 flowers (Fig EV2A and B). STM then becomes restricted to emerging carpel primordia in the fourth whorl of wild‐type flowers, whereas in sup‐5 flowers, its expression domain in the center becomes enlarged. Later on, STM only remains expressed at the carpel margins/placenta region in the WT (Fig EV2C and D). Conversely, in sup‐5, STM is expressed in a larger domain, which encompasses the FM that keeps proliferating; STM is also transiently expressed in the emerging extra stamen primordia that form in the center of sup‐5 flowers (Fig EV2C and D). The expression domain of SUP forms a ring at the boundary between whorls 3 and 4 (Appendix Fig S1A and B) that is mostly non‐overlapping with that of CLV3 or WUS throughout flower development, indicating that SUP affects floral stem cells non‐cell autonomously (Prunet et al, 2017).
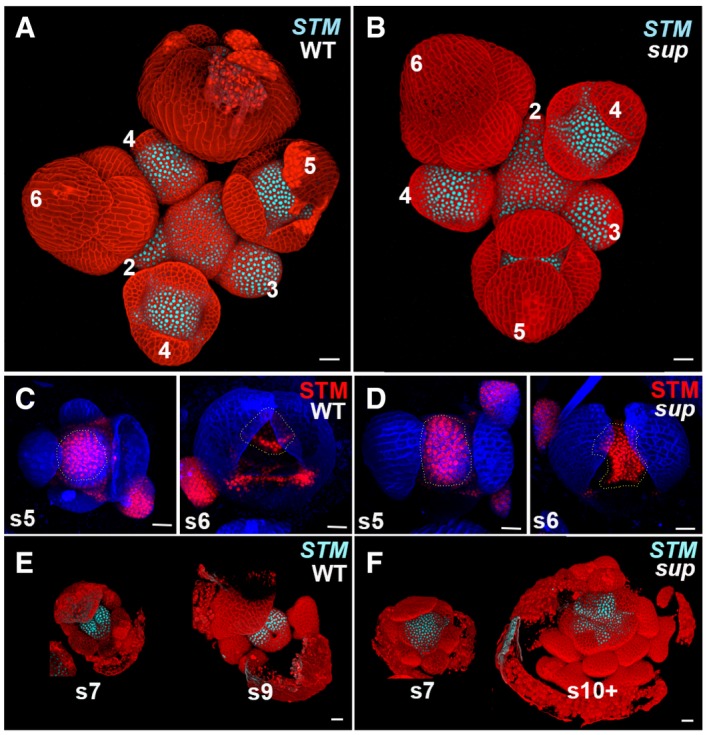
Figure EV2. STM expression is expanded in sup floral buds.
-
A, BThe expression of pSTM::CFP‐N7 (light blue) in WT and sup‐5 inflorescence. In both WT and sup‐5, the STM‐VENUS fusion protein was highly expressed in the center of floral buds younger than stage 5 as well as in the boundary regions of sepals.
-
C, DA stem cell marker pSTM::STM‐VENUS (red) in WT (C) and sup‐5 (D) floral buds at the stages 5 and 6. From late stage 5 (s5), STM was reduced in the regions with developing stamens. The STM expression domain in FM centers is relatively larger in sup‐5 (1,680 ± 167 μm2, n = 15) than that in WT (1,150 ± 160 μm2, n = 16). P < 0.05 based on a Student's t‐test. Dashed lines mark the FM regions and whorl 3/4 boundary regions with STM‐VENUS.
-
E, FThe expression of pSTM::CFP‐N7 (light blue) only remains expressed at the carpel margins/placenta region in the WT at stage 7 (s7) and greatly diminished at stage 9 (s9) (E). Conversely, in sup‐5, STM is expressed in a larger domain, which encompasses the FM that keeps proliferating as well as in emerging extra stamen primordia up to stage later than 10 (s10+) (F).
Auxin signaling is disrupted in sup mutants
To investigate how SUP regulates organ boundaries, FM size, and differentiation, we compared the expression of a CUC2 reporter in wild‐type and sup flowers. CUC genes encode closely related members of the NAC family of transcription factors, which participate in shoot meristem and boundary formation (Takada et al, 2001; Daimon et al, 2003). In situ hybridization analysis showed CUC2 mRNA accumulation in the center of FMs in sup‐1 (Breuil‐Broyer et al, 2004). As ectopic expression of CUC2 is associated with an increased number of petals (Huang et al, 2012), we monitored CUC2 expression using pCUC2::CUC2‐3xVENUS‐N7 (Heisler et al, 2005) in the sup mutant (Fig EV3A–D). CUC2 is widely expressed in stage 3 floral buds in both WT and sup (Fig EV3A and B). From stage 4 onward, clear differences in CUC2 expression were observed between WT and sup flowers. In the WT, high CUC2 expression was observed in cells at the boundary regions between the sepal primordia, and in the inner part of the whorl 3/4 boundary regions, while the central region of the FM showed no CUC2 expression (Fig EV3A and C). In sup, CUC2 expression was also observed in the FM region (Fig EV3B and D), in a domain where SUP is not normally expressed, suggesting that CUC2 is not a direct target of SUP. Since it has been shown that CUC2 is induced by low levels of auxin but repressed by high levels of auxin (Heisler et al, 2005), we hypothesized that auxin signaling or accumulation could be disturbed in sup mutant flowers. To test this hypothesis, we monitored the activity of the auxin response reporters pDR5rev::2xGFP‐N7 and pDR5rev::GFP‐ER in sup and WT flowers (Xu et al, 2006a; Liao et al, 2015). In WT stage 4 flower buds, DR5 expression occurs only at the sites of petal primordia initiation and at the tips of sepals (Fig 2A and Appendix Fig S2A). In contrast, in sup mutant stage 4 flower buds, DR5 is also expressed at the whorl 3/4 boundary, indicating an increase in auxin response in that region (Fig 2B and Appendix Fig S2B). The DII‐VENUS auxin sensor, expressed under the control of the ubiquitous RPS5A promoter, is degraded in presence of auxin (Liao et al, 2015). In WT flower buds at stage 4, DII‐VENUS is detected at the boundary between whorls 3 and 4 but not in the center of the flower, indicating that auxin is depleted at the boundary, but not in the FM region (Fig 2C). In contrast, in sup flower buds at stage 4, DII‐VENUS is observed in the center of the FM but not at the boundary between whorls 3 and 4, showing that auxin is depleted in the FM region rather than at the boundary (Fig 2D). These data imply that the loss of SUP function leads to auxin accumulation, rather than depletion, at the boundary between whorl 3 and 4, and to a reduction in auxin in the center of the FM. The increase in auxin at the whorl 3/4 boundary in sup could be due either to an increase in auxin biosynthesis or to a perturbation of auxin transport. To test whether the sup phenotype is due to the cell‐autonomous effect of an increase in auxin levels or due to perturbed auxin transport, we treated sup mutant inflorescences with p‐chlorophenoxyisobutyric acid (PCIB), which inhibits auxin action (Oono et al, 2003). PCIB treatment strongly rescued both the stamen number and carpel defects in sup‐5 (Fig 2E–G). Next, we tested the stage‐specific rescue effect of PCIB by measuring time to anthesis. Generally, stage 4–5 floral buds were rescued better than stage 6 floral buds in terms of carpel morphology and stamen numbers (Fig 2G). We also tested the effect of PCIB treatment on CUC2 and DR5 expressions in sup (Fig EV4). CUC2 expression was not reversed to a WT‐like pattern following treatment with PCIB. Instead, CUC2 was ectopically expressed through most of the flower bud (Fig EV4A and B), which may be due to CUC2 activation by low auxin levels. In contrast, DR5 expression at the whorl 3/4 boundary is almost completely absent in sup flowers 5 h after PCIB treatment (Fig EV4C and D), indicating that PCIB restores a wild‐type pattern of auxin response in sup flowers, which is consistent with the fact that PCIB treatments rescue the sup phenotype (Fig 2E–G).
Figure EV3. pCUC2::3xVENUS‐N7 expression is expanded in the epidermal cells of sup mutants.
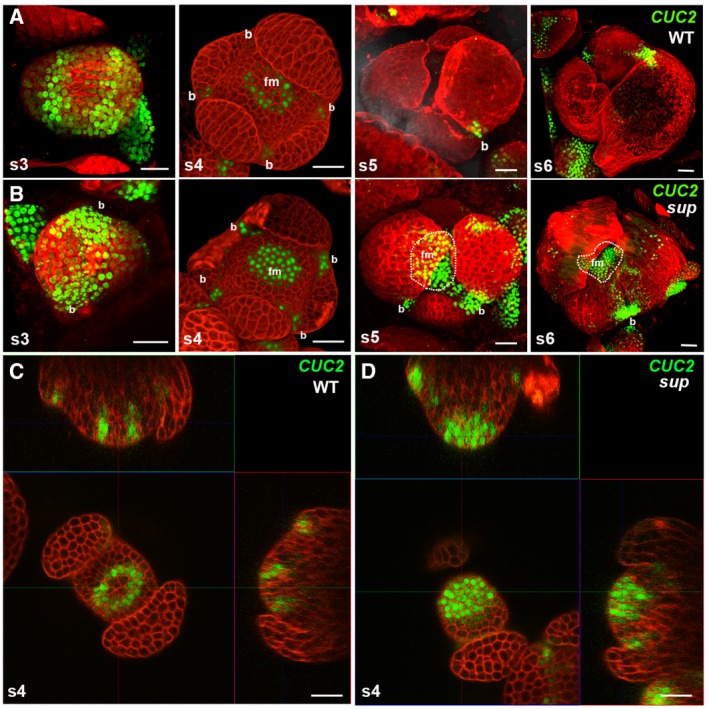
- pCUC2::3xVENUS‐N7 in WT floral buds at stages 3‐6. CUC2 was highly expressed in the floral buds of stage 3, and its expression started to be constrained to the boundary regions between the sepals after stage 3. At stage 5 and stage 6 floral buds, CUC2 was only detected in the small boundary regions at the bottom of the sepals.
- pCUC2::3xVENUS‐N7 in sup‐5 floral buds at stages 3–6. At stage 3, the CUC2 expression pattern was similar to that in WT. From stage 4, the CUC2 expression pattern in sup‐5 started to show differences from that of WT. CUC2 expression was detected in FM region of at stage 4. At stages 5 and 6, the CUC2 expression was still relatively high but was more concentrated at the bottom of the developing sepals as well as in the broad FM regions. Dashed lines mark the regions with CUC2‐3xVENUS‐N7 in the flower center. “b” indicates the boundary region between sepals; “fm” indicates the floral meristem region in the center.
- A cross section of a stage 4 WT floral bud showing a ring of CUC2‐3xVENUS‐N7 signal at the whorl 3/4 boundary.
- A cross section of a stage 4 sup‐5 floral bud showing the additional signal of CUC2‐3xVENUS‐N7 in the FM center.
Figure 2. sup mutant phenotypes are associated with perturbed auxin distribution.
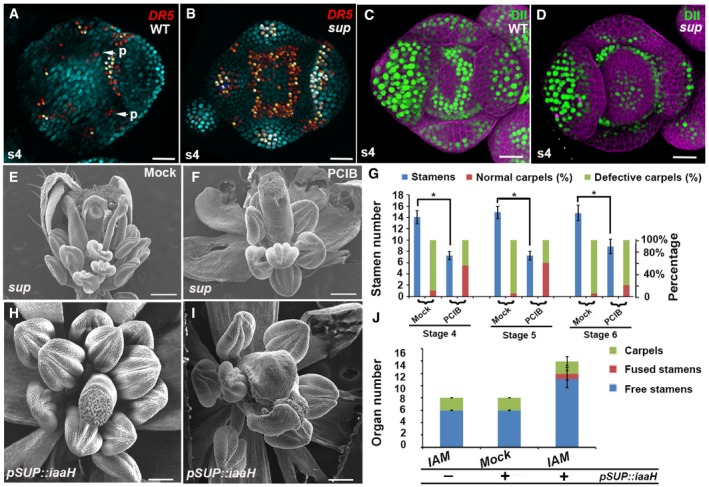
-
A, BActivity of the auxin marker pDR5rev::2xGFP‐N7 in stage 4 (s4) floral buds of WT (A) and sup‐1 (B). In WT flowers, the fluorescence signal was detected mostly at the tips of the sepals and at the sites (p, marked with the arrows) where the petal primordia would emerge at later stages (A). In sup‐1 flowers, strong reporter activity was additionally detected at the whorl 3/4 boundary.
-
C, DActivity of the auxin reporter DII‐VENUS (green) in stage 4 floral buds of WT (C) and sup‐1 (D). In WT flowers, fluorescence signals were detected at the whorl 3/4 boundary but were absent at the center of the FM and the tips of the sepals (C). In contrast, DII‐VENUS signals were absent at the whorl 3/4 boundary but were detected in the FM region (D).
-
E, Fsup‐5 flowers after treatment with the anti‐auxin PCIP (F) and mock solutions (E). While the mock treatment did not affect sup‐5 flowers (E), treatment with PCIB strongly rescued both carpels and stamen numbers.
-
GThe statistical analysis indicated that PCIB treatment strongly rescued stage 4–5 floral buds, which took approximately 9–10 days to anthesis; the floral buds of stage 6 were best rescued in terms of carpel morphology. Error bars indicate s.d. of 20 flowers from around 10 individual plants; two‐tailed Student's t‐test, *P < 0.05.
-
H–JpSUP::iaaH transgenic flowers with the IAM treatment mimicked the various sup‐like phenotypes, including increased stamen numbers and defective carpels, as shown in the SEM images of the flowers (H, I). Wild‐type plants (lacking pSUP:iaaH) were treated with IAM as a negative control. The number of free stamen, fusion stamen, and carpels was determined for a total of 20 flowers from 20 individual plants and is summarized in (J). Error bars indicate s.d.
Figure EV4. PCIB treatments induced CUC2 expression but repressed DR5 activity in sup‐5 .
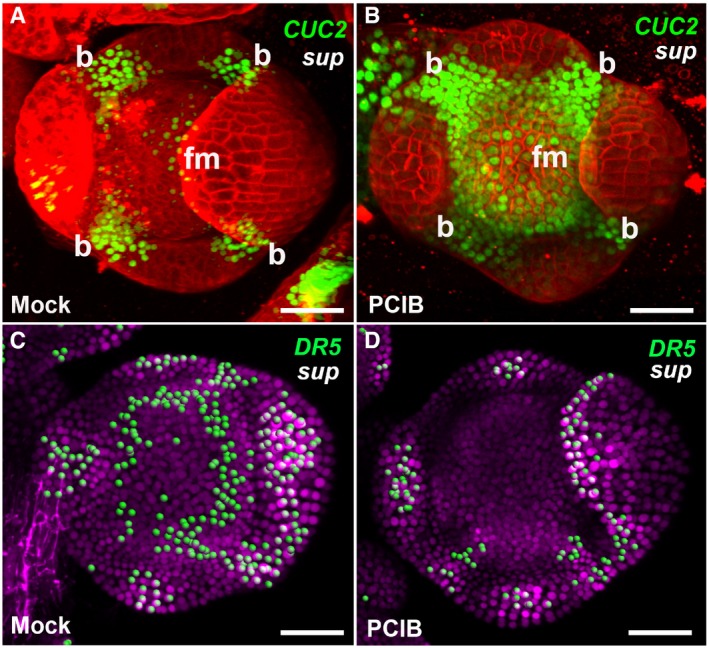
-
A, BThe expression of pCUC2::CUC2‐3xVENUS‐N7 was induced in stage 4 floral buds of sup‐5 plants 4–6 h after treatment with PCIB (B) but after a mock treatment (A).
-
C, DThe expression of pDR5rev::3xVENUS‐N7 was reduced in stage 4 floral buds of sup‐5 plants 4–6 h after treatment with PCIB (D) but not after a mock treatment (C).
To test whether an increase in auxin levels in the SUP expression domain is sufficient to cause the development of supernumerary stamens and carpel defects, we generated the transgenic line pSUP::iaaH with the bacterial auxin biosynthetic gene iaaH under the control of the SUP regulatory regions. iaaH can convert the auxin precursor, indoleacetamide (IAM) to the active form of auxin, indole‐3‐acetic acid (IAA) in Arabidopsis (Oka et al, 1999). With a 1 mM IAM treatment, flowers of the transgenic plants showed weak to strong sup‐like phenotypes. In contrast, WT flowers treated with IAM and mock‐treated pSUP::iaaH flowers were unaffected (Fig 2H–J). These results confirmed that a local increase in auxin biosynthesis at the boundary region between the 3rd and 4th whorl is sufficient to cause sup‐like phenotypes.
Auxin gradients and maxima rely on both local biosynthesis and polar transport. We therefore tested the effect of the polar auxin transport inhibitor N‐1‐naphthylphthalamic acid (NPA) on the sup mutant phenotype. The application of NPA at stages 4–6 partially rescued both carpel morphology and stamen number defects in sup (Appendix Fig S3), suggesting that polar auxin transport is also important for the expression of the sup phenotype.
Derepression of local auxin biosynthesis is responsible for sup mutant phenotypes
To identify the downstream targets of SUP that contribute to auxin biosynthesis and/or accumulation, we generated p35S::SUP‐GR and pSUP::SUP‐GR transgenes that allow the DEX‐dependent activation of SUP when expressed under either the SUP promoter or ubiquitously with the enhancer element of the cauliflower mosaic virus 35S promoter inserted in the SUP promoter. After DEX treatment, we observed a complete rescue of the sup‐5 mutant phenotype in the pSUP::SUP‐GR plants, indicating that SUP‐GR mimics the endogenous function of SUP (Appendix Fig S4). p35S::SUP‐GR plants showed reduced sizes of floral organs and increased carpel numbers (Appendix Fig S5). Once introduced into the clv3 mutant (which forms larger meristems than the WT), ectopic SUP expression caused the differentiation of FMs into leaf‐like structures in all the flowers observed (Appendix Fig S5). These results suggest that the function of SUP could be associated with organ differentiation.
To identify downstream targets of SUP at a genome‐wide scale, we performed microarray analyses with RNA isolated from p35S::SUP‐GR inflorescences 4 h after DEX and mock treatments. In these experiments, we identified 642 down‐regulated and 421 up‐regulated genes whose expression changed more than twofold after DEX treatment (Gene Expression Omnibus accession number GSE92729). While there was no significant enrichment of Gene Ontology (GO) terms among up‐regulated genes, for the down‐regulated genes, eight GO terms were over‐represented, which were classified into three superclusters based on their relatedness using REVIGO (Supek et al, 2011), including “hormone metabolism” and “response to endogenous stimulus” (Fig 3A). Our reporter assays as well as a previous study (Nibau et al, 2011) suggest that SUP may function in the auxin signaling pathway. Thus, the supercluster of “hormone metabolism”, which contains three hormone‐related GO biological processes: hormone metabolism, P FDR = 0.00039; regulation of hormone level, P FDR = 0.0011; auxin biosynthesis, P FDR = 0.0042 (Fig 3A), was further inspected. There are total 17 genes in the category of “hormone metabolism”. Among these, 12 of the genes also belong to the category of “regulation of hormone level”, and interestingly, eight of which are listed in the term of “auxin synthesis” as well (Appendix Table S1). These eight genes include three known auxin biosynthesis genes: the YUCCA (YUC) flavin monooxygenases YUC1/4 and the TRP‐a‐transferase TRYPTOPHAN AMINOTRANSFERASE RELATED 2 (TAR2) (Appendix Table S1). In addition, two auxin efflux transporters, PIN‐FORMED1/3 (PIN1/3), were also identified among the down‐regulated genes (Appendix Table S1). We confirmed these microarray results by analyzed RNA levels in p35S::SUP‐GR at 2 and 4 h after DEX treatment using qRT–PCR (Fig 3B). To further test whether these four genes are targets of SUP, we also compared their expression levels in stage 4 floral buds of the WT and sup mutants using the ap1 cal p35S::AP1‐GR floral induction system (Wellmer et al, 2006). As SUP is a strong active repressor (Hiratsu et al, 2002), we expected to see increased transcription levels of its direct targets in sup compared with those in WT. The transcription of YUC1/4 was up‐regulated in sup at approximately stage 4 (3 days after DEX treatment; Fig 3C). In contrast, PIN3/4 were down‐regulated in sup (Fig 3C), which may be due to an indirect feedback regulation. These expression comparisons suggest that auxin biosynthesis genes may be immediate targets of SUP and that their derepression is primarily responsible for the sup mutant phenotype.
Figure 3. Increased auxin biosynthesis in the SUP‐expressing region is essential for the sup mutant phenotype.
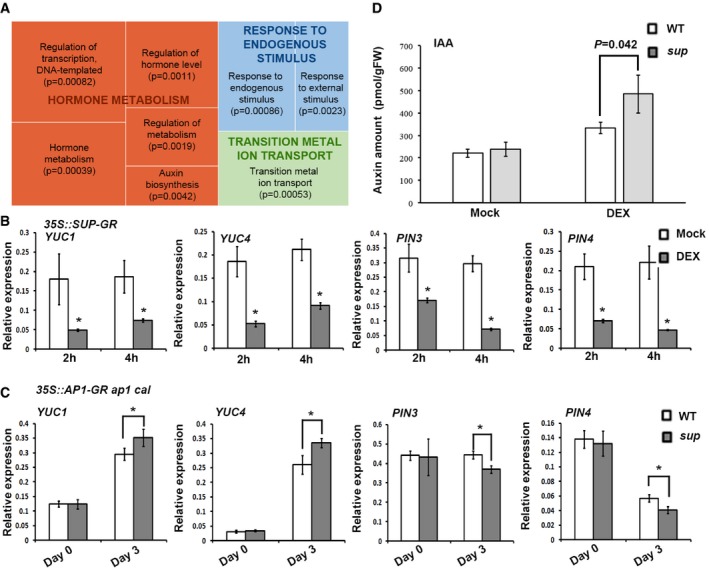
- REVIGO analysis of pathways significantly enriched among the down‐regulated genes. Each rectangle is a single cluster representative for the non‐redundant GO term, which are joined into “superclusters” of related terms, visualized with different colors. Size of the rectangles reflects the P‐value.
- YUC1/4 and PIN3/4 are reduced in p35S::SUP‐GR inflorescences 2 and 4 h after treatment with 10 μM DEX. Error bars indicate s.d. of three biological replicates; two‐tailed Student's t‐test, *P < 0.05.
- Expression levels of YUC1/4 and PIN3/4 3 days after 1 μM DEX treatment in ap1 cal p35S::AP1‐GR sup‐5 and ap1 cal p35S::AP1‐GR. Expression of YUC1/4 was increased in the ap1 cal p35S::AP1‐GR sup‐5 background, while that of PIN3/4 was reduced. Error bars indicate s.d. of three biological replicates; two‐tailed Student's t‐test, *P < 0.05.
- IAA levels in ap1 cal p35S::AP1‐GR sup‐5 and ap1 cal p35S::AP1‐GR 3 days after treatment with 1 μM DEX or a mock solution. The P‐value was calculated using one‐way ANOVA and standard errors from three biological replicates.
To test whether the increased expression of YUC flavin monooxygenases in sup leads to an over‐accumulation of auxin, we again employed the floral induction system to measure the major form of auxin IAA in WT and sup mutant flowers (Fig 3D). While in mock‐treated inflorescences before the initiation of flower formation, we did not detect any significant difference, IAA levels in stage 4 sup flowers were significantly (P = 0.042) higher than those in the WT (Fig 3D), in agreement with the observed derepression of YUC1/4 genes.
SUP directly binds to YUC1/4 genomic regions and mediates the deposition of the repressive mark H3K27me3
To test whether YUC1/4 are direct SUP target genes, we analyzed the binding profiles of SUP‐GFP at the YUC1 and YUC4 loci. As the spatial and temporal SUP‐GFP protein expression domain is quite limited in pSUP::SUP‐GFP (Appendix Fig S1B), we performed a ChIP binding assay with pSUP::SUP‐GFP in ap1 cal p35S::AP1‐GR to obtain a large amount of synchronized stage 4 floral buds. We detected enrichment of SUP‐GFP at the 5′‐proximal promoters and coding regions of both YUC1 and YUC4 compared with those of the control (Fig 4A, B, D, and E). It has been reported that the coding region of INNER NO OUTER (INO) is required for SUP regulation during ovule development (Meister et al, 2002), suggesting that it may not be unusual for SUP to bind the coding region of downstream targets.
Figure 4. SUP binds to YUC1/4 chromatin to achieve high levels of H3K27me3.
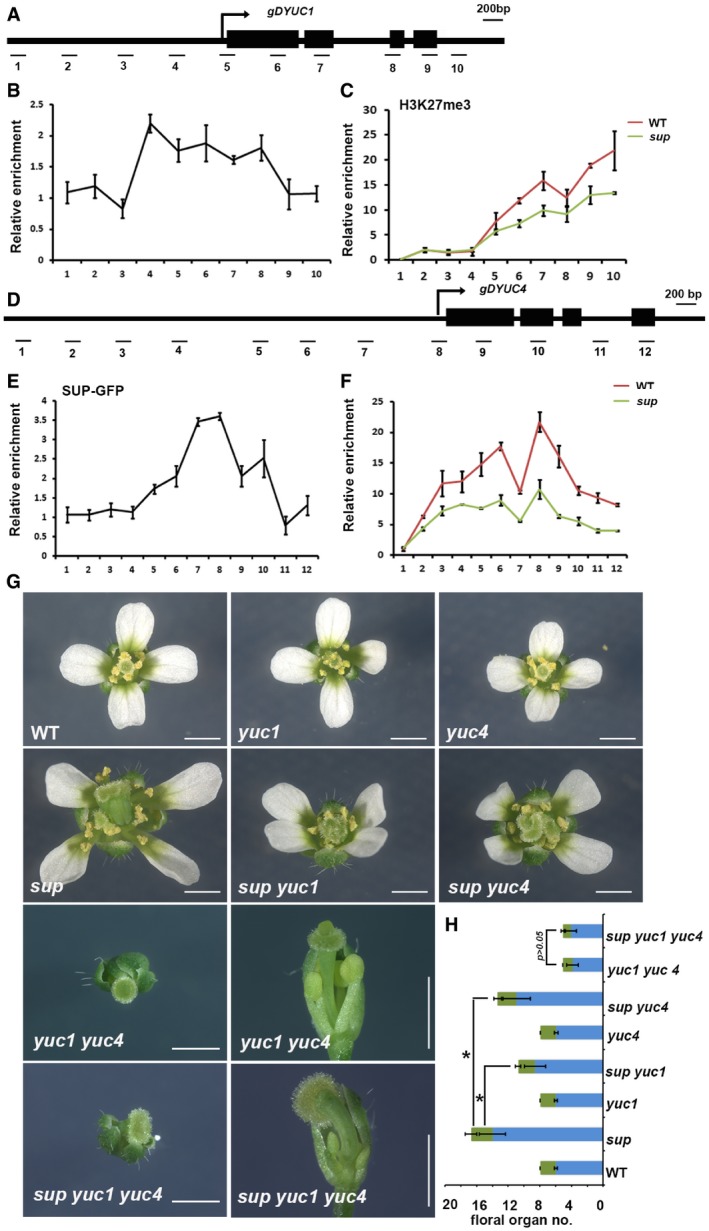
- Schematic drawing of the YUC1 genome structure showing regions amplified by primer sets used for ChIP analyses.
- ChIP binding assay of the SUP protein at the YUC1 genome. Relative enrichment of SUP‐GFP at the YUC1 locus, including the promoter and coding regions, was analyzed using the stage 4 floral buds from ap1 cal p35S::AP1‐GR sup‐5 pSUP::SUP‐GFP plants.
- Relative enrichment of H3K27me3 at the YUC1 locus is decreased in the stage 4 floral buds of ap1 cal p35S::AP1‐GR sup‐5 plants relative to ap1 cal p35S::AP1‐GR.
- Schematic drawing of the YUC4 gene structure showing regions amplified by primers used for ChIP analyses.
- ChIP binding assay of the SUP protein at the YUC4 gene. Relative enrichment of SUP‐GFP at the YUC4 locus, including the promoter and coding regions, analyzed using the stage 4 floral buds of ap1 cal p35S::AP1‐GR sup‐5 pSUP::SUP‐GFP.
- Relative enrichment of H3K27me3 is decreased at the YUC4 locus in stage 4 floral buds of ap1 cal p35S::AP1‐GR sup‐5 compared with that of the WT control. Error bars represent standard errors with three biological repeats.
- yuc1 and yuc4 mutant alleles partially rescue the stamen and carpel number defects of sup‐5, while yuc1 yuc4 is epistatic to sup‐5. The yuc4 mutation results in reduced floral organ size while yuc4 appears WT‐like. Both yuc1 and yuc4 single mutant have no defects in floral organ numbers. The yuc1 yuc4 double mutant shows strong floral defects, namely a decreased numbers of stamen and carpel together with a reduction in floral organ size. The sup‐5 yuc1 yuc4 triple mutant shows a yuc1 yuc4‐like floral morphology. A typical flower is shown for WT, yuc1, yuc4, sup‐5, sup‐5 yuc1, sup‐5 yuc4, yuc1 yuc4, and sup‐5 yuc1 yuc4, respectively. For yuc1 yuc4 and sup‐5 yuc1 yuc4, the top view and side view are shown for the same flower. Scale bars: 1 mm.
- The statistical analysis showed a reduction in total stamens and carpels in sup‐5 yuc1 (8.60 ± 1.33 for stamen, 2.13 ± 0.34 for carpel, n = 30), sup‐5 yuc4 (10.93 ± 1.74 for stamen, 2.4 ± 0.50 for carpel, n = 30) once compared with sup‐5 (14.00 ± 1.68 for stamen, 2.7 ± 0.75 for carpel, n = 30). *P < 0.05 based on Student's t‐test. There is no significant difference between yuc1 yuc4 (3.8 ± 0.77 for stamen, 1.25 ± 0.44 for carpel, n = 20) and sup‐5 yuc‐1 yuc4 (4.05 ± 0.70 for stamen, 1.05 ± 0.23 for carpel, n = 19) in floral organ number and floral organ size, P > 0.05 with a Student's t‐test.
Many genes involved in auxin synthesis and transport, including YUC1/4, are regulated by the polycomb group (PcG) complex, which can introduce an H3K27me3 repressive epigenetic mark to silence genes (Lafos et al, 2011). To address whether the repressive mark H3K27me3 is associated with transcriptional repression of SUP targets, we performed ChIP assays to look for differences in the H3K27me3 repressive mark and the H3K4me3 active mark in ap1 cal p35S::AP1‐GR with or without the sup mutation at stage 4. The ChIP assay showed that H3K27me3 is reduced at both YUC1 and YUC4 loci in sup mutant floral buds (Fig 4A, C, D, and F), while H3K4me3 is increased (Appendix Fig S6), which is consistent with the transcriptional up‐regulation of YUC1/4.
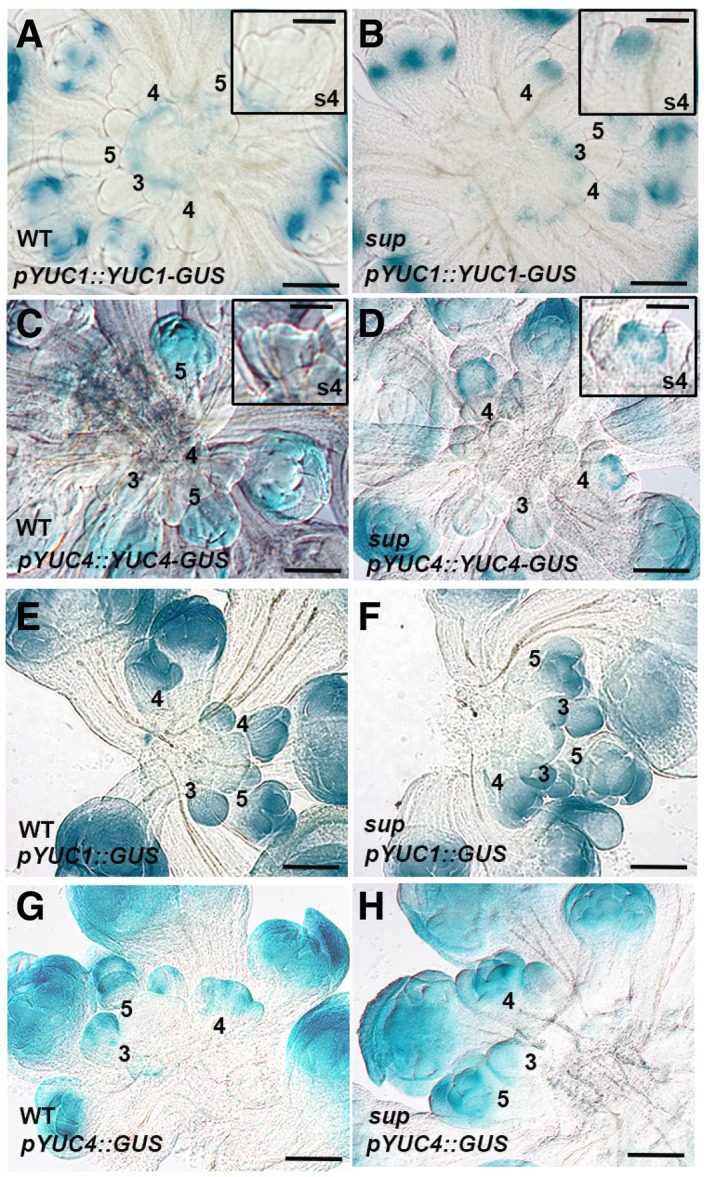
To investigate whether the SUP binding regions (including coding regions and introns) of YUC1/4 are responsible for their proper expression patterns in the inflorescence, we generated GUS reporters for YUC1 and 4, with and without the coding regions (Appendix Fig S7A–D). YUC1‐GUS shows higher staining than that of YUC4‐GUS, which is consistent with previous reports (Cheng et al, 2006). GUS staining is stronger and expands both spatially and temporally in the pYUC1::GUS and pYUC4::GUS lines compared to the full‐length reporters pYUC1::YUC1‐GUS and pYUC4::YUC4‐GUS (Appendix Fig S7). qRT–PCR assays of GUS transcripts confirmed that the lower staining in pYUC1::YUC1‐GUS and pYUC4::YUC4‐GUS is mainly due to lower transcription of the transgenes (Appendix Fig S7F). These data confirmed that the coding regions of YUC1/4, which are bound by SUP and contain high levels of H3K27me3, are important for their negative regulation (Fig 4A–F and Appendix Fig S7). However, the ectopic GUS expression observed in our transcriptional reporters is detected in sepals and is not exclusive to the whorl 3/4 boundary, suggesting that there are other regulators repressing YUC1/4 via regulatory elements in the coding region of these genes (Appendix Fig S7). We could not see any obvious difference in GUS staining between the WT and sup with the pYUC1::GUS and pYUC4::GUS reporters (Fig EV5E–G). However, ectopic GUS staining was observed in a region that resembles SUP expression domain in sup flowers at stage 4 with both full‐length pYUC1::YUC1‐GUS and pYUC4::YUC4‐GUS reporters, which confirms that SUP represses YUC1/4 at the boundary between whorl 3 and 4 in a cell‐autonomous fashion (Fig EV5A–D).
Figure EV5. The up‐regulation of YUC1 and YUC4 GUS reporter lines in young sup floral buds.
-
A, BGUS staining of YUC1 full‐length reporter pYUC1::YUC1‐GUS in WT (A) and sup‐5 (B) inflorescences. The expression of pYUC1::YUC1‐GUS is weak at floral buds earlier than stage 5. At stage 5, GUS starts to be highly expressed at the sepals, especially at the bottom of sepals at the later stages. In sup‐5, pYUC1::YUC1‐GUS is expressed highly at the SUP expression domain as well as the FM region at the stage 4. Floral buds at stage 4 with GUS staining are shown as insets.
-
C, DGUS staining of YUC4 full‐length reporter pYUC4::YUC4‐GUS in WT (C) and sup‐5 (D) inflorescences. In WT, the expression of pYUC4::YUC4‐GUS is weak at the floral buds at earlier than stage 5. From stage 5, GUS is almost expressed in the whole floral buds (C). In sup‐5, pYUC4::YUC4‐GUS is highly expressed at the SUP expression domain at the floral buds of stage 4. Floral buds at stage 4 with GUS staining are shown as insets.
-
E, FGUS staining of YUC1 promoter reporter pYUC1::GUS was detected in almost all WT (E) and sup‐5 (F) young floral buds.
-
G, HGUS staining of YUC4 promoter reporter pYUC4::GUS was detected in almost all WT (G) and sup‐5 (H) young floral buds.
Since YUC1 and YUC4 are direct targets of SUP, we tested whether the activity of YUC1 and YUC4 is essential for the expression of the sup mutant phenotype. Flowers of the yuc1 single mutant do not show any obvious morphological defect (Fig 4G; Cheng et al, 2006), while yuc4 single‐mutant flowers show a reduction in the size of all floral organs (Fig 4G), consistent with previous reports that YUC4 is broadly expressed. However, in contrast to previous reports (Cheng et al, 2006), our yuc4 allele (SALK_047083) did not exhibit any obvious decrease in floral organ numbers (Fig 4G and H). Flowers of the yuc1 yuc4 double mutant have a much stronger phenotype, with a strong reduction in both stamen and carpel numbers, along with abnormally shaped carpels (Fig 4G and H; Cheng et al, 2006). We next generated yuc1 sup and yuc4 sup double mutants as well as yuc1 yuc4 sup triple mutant plants. As expected, both yuc1 and yuc4 can partially rescue the increase in stamen and carpel number in sup, and yuc1 rescues sup to a greater extent than yuc4 (Fig 4G and H). The yuc1 yuc4 double mutant is epistatic to sup (Fig 4G and H), confirming that SUP controls stamen and carpel number through the repression of YUC1/4. We next checked DR5 expression in sup yuc1. Consistent with the partial rescue of the sup phenotype by yuc1, we observed a strong reduction in DR5 activity at the whorl 3/4 boundary in sup yuc1 flowers (Appendix Fig S8A–C).
Since our pharmacological analyses using auxin signaling and auxin transport inhibitors showed that sup mutant phenotypes depend on both auxin biosynthesis and auxin transport, we also checked the expression pattern of the auxin efflux transporter PIN3 using the pPIN3::PIN3‐GFP reporter. PIN3 transcript is slightly lower in a sup background compared to the wild type (Fig 3C), and there is no obvious difference of expression pattern between sup and the wild type (Appendix Fig S9). Based on these results, we conclude that SUP primarily functions through the control of auxin biosynthesis.
SUP forms a repressor complex with the PcG components CLF and TFL2
SUP is an active repressor with a conserved EAR motif at its C‐terminus (Hiratsu et al, 2002; Yun et al, 2002). However, how SUP executes its repressor function is still unknown. Given that the repressive mark H3K27me3 is increased at the YUC1/4 loci after the loss of function of SUP (Fig 4A–F), we explored the link between SUP and the repressive H3K27me3 modifications by performing a yeast two‐hybrid assay to examine the interaction of SUP with factors associated with repressive histone modifications, including CURLY LEAF (CLF), FERTILIZATION‐INDEPENDENT ENDOSPERM (FIE), RING1A/B, TERMINAL FLOWER 2 (TFL2), BMI1A/B, and EMBRYONIC FLOWER 2 (EMF2). We found that SUP interacts with CLF and TFL2 but not with the other proteins we tested (Appendix Fig S10A).
To define the region of SUP required for the interaction with CLF and TFL2, we tested a series of SUP truncations as baits with CLF and TFL2 AD fusion constructs. Full‐length SUP was required for its interaction with CLF, while the interaction with TFL2 is mapped to a short region of the SUP protein (amino acids 89–205; Fig 5A and B). The EAR motif is essential for proper SUP function as an active repressor, and the ectopic expression of the truncated SUP protein without the EAR motif leads to sup‐like floral phenotypes (Hiratsu et al, 2002). Thus, we also mutated the EAR motif in the full‐length SUP protein and tested whether two versions of mutated SUP proteins (SUP‐EARm1 and SUP‐EARm2) interact with CLF and TFL2. The interaction of SUP with CLF but not with TFL2 requires the intact EAR motif (Fig 5A and B). These results suggest that the active repressor function of SUP could be more dependent on its interaction with CLF. To further verify the interactions of SUP with CLF and TFL2, we carried out a bimolecular fluorescence complementation (BiFC) assay in tobacco. Fluorescence was observed in the nuclei of tobacco epidermal cells only when SUP and CLF or when SUP and TFL2 constructs were co‐infiltrated (Appendix Fig S10B). We further performed co‐immunoprecipitation analysis to test the interaction between SUP and CLF in vivo (Fig 5C). To this end, we generated a functional pCLF::HA‐CLF transgene and introgressed it into the ap1 cal p35S::AP1‐GR background with or without the pSUP::SUP‐GFP transgene (Doyle & Amasino, 2009). We found that anti‐GFP (recognizing SUP‐GFP) could pull‐down HA‐CLF in stage 4 floral buds in ap1 cal p35S::AP1‐GR pSUP::SUP‐GFP pCLF::HA‐CLF, but not in ap1 cal p35S::AP1‐GR pCLF::HA‐CLF (Fig 5C). Taken together, these results suggest that SUP can recruit the PcG complex to at least some of its target genes.
Figure 5. SUP interacts with PcG proteins CLF and TFL2.
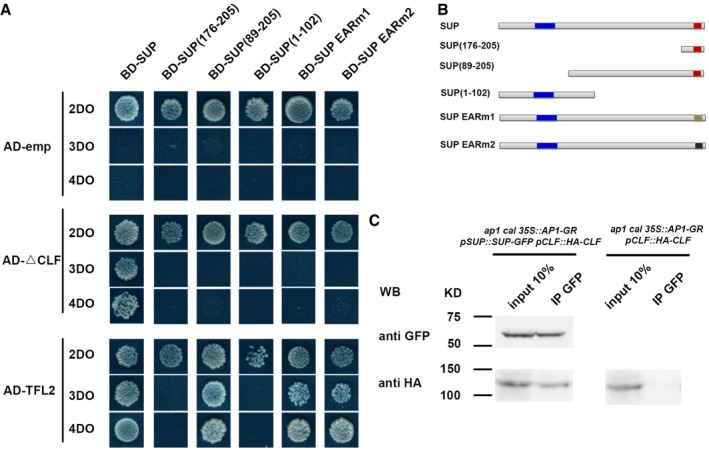
-
A, BThe yeast two‐hybrid assay using a series of truncated SUP proteins with CLF and TFL2. Full‐length, three truncated SUP proteins and SUP proteins with the two versions of the abolished EAR motif (EARm1 and EARm2) were fused to a GAL4 DNA‐binding domain (BD). Schematic structures of the full‐length, truncated, or mutated SUP protein are shown in (B). The blue region indicates the zinc finger domain; the red regions indicate an intact EAR motif; and beige and black regions indicate mutated EAR motif, respectively. The truncated CLF without its C‐terminal SET domain (CLF) and the full‐length TFL2 was fused to the GAL4 activation domain (AD). Yeast colonies harboring these fusion constructs and/or empty vectors as indicated were grown on selective media of 2DO, 3DO, and 4DO. For CLF, yeast growth was only detected when the combination of the full‐length SUP and the truncated CLF was co‐transformed. None of the truncated SUP proteins or SUP proteins with the mutated EAF motif interacted with the truncated CLF. For TFL2, a short domain of SUP (amino acids 89–205) was sufficient for the interaction, and the mutation of EAR motifs did not affect the interaction with TFL2.
-
CInteraction between SUP and CLF as determined by co‐IP. Using stage 4 floral buds of ap1 cal p35S::AP1‐GR pSUP::SUP‐GFP pCLF::HA‐CLF or of ap1 cal p35S:: AP1‐GR pCLF::HA‐CLF plants, total proteins (input) were subjected to immunoprecipitation with anti‐GFP‐conjugated beads. Immunoblotting analysis with another anti‐GFP and anti‐HA antibody were performed to detect SUP‐GFP (to test pull‐down efficiency) and HA‐CLF (protein interactions). Only in the samples containing both SUP‐GFP and HA‐CLF, HA‐CLF was pull‐down together with anti‐GFP antibody. Molecular mass of the protein ladder is indicated in kilodaltons (KD).
Source data are available online for this figure.
Discussion
WUS activity is essential for floral stem cell maintenance, and its termination at stage 6 is important to define the fixed number of whorls of floral organs (Lohmann et al, 2001). Delayed WUS termination in ag mutants leads to additional whorls of organs in the center of the flower (Bowman et al, 1991; Lenhard et al, 2001; Sun et al, 2009). The supernumerary whorls of stamens in sup mutant flowers suggest a possible delay of FM termination (Fig EV1; Gaiser et al, 1995). ag sup double‐mutant flowers show fasciated FMs, and extra petals continue to form in the center (Bowman et al, 1992; Breuil‐Broyer et al, 2016), suggesting that SUP also has a spatial function in the regulation of FMs. In agreement with this assumption, increased numbers of stem cells (marked by the pCLV3::GFP‐ER) were found in sup‐5 at floral stages 4–6 (Fig 1C–E), as compared to the wild type. Increased FM size in sup at late stage 5 was also confirmed with the meristem marker pSTM:: CFP‐N7 (Fig EV2C and D). Prolonged CLV3 and STM expressions in the center of the FM in sup (Figs EV1 and EV2) further show that the FM activity persists longer in sup than in wild type. Altogether, our data show that SUP affects floral stem cells both spatially and temporally.
There are two alternative but not mutually exclusive hypotheses to explain the increased number of stem cells in sup. Firstly, increased cell division rates could be responsible. A sup mutation did affect cell division rates at the whorl 3/4 boundary, as BrdU incorporation assays showed reduced non‐dividing domains at the boundaries between reproductive organs in both sup‐1 and sup‐5 flowers (Breuil‐Broyer et al, 2004, 2016). This increase in cell proliferation at the boundary regions in whorls 3 and 4 may explain the partial fusions between stamens and between stamens and carpels that are seen in sup (Breuil‐Broyer et al, 2004, 2016), but seem insufficient to explain the increase in stamen and carpel number. Indeed, we also observed that ectopic expression of SUP promotes the differentiation of the FM in the clv3 background (Appendix Fig S9). Together with the expanded and prolonged expression of CLV3 and STM in the FM of sup flowers (Figs 1, EV1 and EV2), this indicates that SUP promotes the differentiation of floral stem cells. Auxin affects cell division, and together with cytokinin, it also contributes to cell differentiation (Schaller et al, 2015). Our work shows that SUP cell autonomously represses auxin biosynthesis, leading to a non‐cell‐autonomous effect in the center of the FM, with a depletion in auxin, as well as an increase in the number of floral stem cells and prolonged floral stem cell maintenance. We found that SUP directly represses the YUC1/4 genes, reducing auxin biosynthesis at the whorl 3/4 boundary (Fig 4). We used different auxin reporters to compare auxin gradients in wild‐type and sup flowers (expression patterns are summarized in Appendix Fig S11). Auxin is normally depleted at the boundary between whorls 3 and 4 (Fig 2C and D), where SUP is expressed, but strong auxin signaling is observed instead in this region when SUP is mutated (Fig 2A and B, and Appendix Fig S1A and B). Treatment of the anti‐auxin PCIB and the analysis of IAM‐treated pSUP::iaaH transgenic plants confirmed that increased auxin levels at the whorl 3/4 boundary are responsible for the floral indeterminacy in sup (Fig 2E–J). SUP was also shown to repress the expression of class B/stamen identity genes AP3 and PI cell autonomously, but it remains unclear whether AP3 and PI are direct targets of SUP (Prunet et al, 2017). One possibility is that the local increase in auxin levels at the boundary between whorls 3 and 4 in sup flowers causes the ectopic expression of AP3 and PI. Conversely, auxin levels appear lower in the center of the FM in sup flowers than in the wild type (Fig 2C and D), which is consistent with ectopic CUC2 expression observed in the sup FMs (Fig EV3). The PIN1 transporter generates an auxin flow toward regions with higher auxin levels (Schaller et al, 2015), suggesting that increased auxin levels at the whorl 3/4 boundary in sup may cause auxin transport from the center of the FM to this neighboring boundary, resulting in auxin depletion at the center. Thus, changes in auxin dynamics based on polar transport may explain how SUP non‐cell autonomously affects floral stem cells. Indeed, polar auxin transport also contributes to the formation of extra stamens and carpels in sup, as NPA treatments can partially rescue the sup phenotype (Appendix Fig S3). This suggests that up‐regulation of local auxin biosynthesis at the whorl 3/4 boundary without polar auxin transport is not sufficient to cause the sup floral defects and that the extra auxin produced due to derepression of YUC1/4 may trigger dynamic changes in auxin gradients in the FM through polar auxin transport. Interestingly, PIN3 and PIN4 were identified as potential downstream targets of SUP, and they appear down‐regulated in a sup mutant background (Fig 3). However, we did not observe any obvious difference in PIN3 localization between WT and sup flowers (Appendix Fig S9). Other polar auxin transporters might contribute to the changes in auxin distribution in the center of sup mutant flowers. Both the cell‐autonomous effect of SUP on auxin biosynthesis and class B gene expression at the boundary between whorls 3 and 4, and its non‐cell‐autonomous effect on auxin levels and stem cells in the center of the FM contribute to the control of stamen number. In sup mutant flowers, local derepression of auxin biosynthesis and class B gene expression allows for the formation of a few extra stamens at the boundary between whorls 3 and 4. This is not sufficient, however, to account for the large increase in stamen number in sup flowers. The sup phenotype is iterative: As these extra stamens emerge, they form a new boundary with the FM, and the lack of SUP function in this region causes the formation of more stamens. The increase in number and prolonged maintenance of floral stem cells replenishes the center of the FM and allows for several extra whorls of stamen to form.
The direct binding of SUP to the YUC1 and YUC4 loci, together with the ectopic expression of YUC1 and YUC4 at the boundary between whorls 3 and 4 in sup mutant flowers, suggests that both YUC1 and YUC4 are directly repressed by SUP in WT (Figs 4A–F and EV5A–D, and Appendix Fig S11). We also showed that both yuc1 and yuc4 mutant can partially rescue the abnormal stamen and carpel number in sup, and the yuc1 yuc4 double mutant is epistatic to sup‐5 in flowers, confirming that YUC1 and YUC4 are major targets of SUP, and that their ectopic expression is responsible for the floral phenotype of sup (Fig 4G and H). Interestingly, we also found cytokinin‐related genes among the potential targets of SUP (Appendix Table S1), suggesting that SUP function may involve a crosstalk between auxin and cytokinins.
Our yeast two‐hybrid and BIFC assays revealed that both the PRC2 component CLF, which catalyzes H3K27 methylation (Goodrich et al, 1997), and the PRC1 component TFL2, which interacts with the core catalytic components of the PRC1 complex, AtRING1 and AtBMI1(Xu & Shen, 2008), associate with SUP (Fig 5 and Appendix Fig S10). We further validated the SUP‐CLF interaction in vivo by co‐IP assays (Fig 5C). Notably, tfl2 mutants show similar developmental defects as clf plants (Goodrich et al, 1997; Gaudin et al, 2001). Moreover, TFL2 was recently found to be a part of the PRC2 complex (Derkacheva et al, 2013; Wang et al, 2016). It will therefore be interesting to examine whether TFL2 also participates in a SUP‐CLF‐containing complex. Yeast two‐hybrid assays with SUP variants showed that the EAR motif, which is essential for the SUP repressor activity, is indispensable for SUP's interaction with CLF (Fig 5A and B), indicating that the interaction between CLF and SUP is necessary for PRC2‐mediated gene repression (Hiratsu et al, 2002). Compared with SUP, the ubiquitously expressed CLF has a broader biological function in plant development and FM activity. CLF is involved in AG‐mediated FM termination, and loss of function of clf has a weak FM indeterminacy (Liu et al, 2011). In addition, CLF has multiple roles in flower development, including repression of AG and STM, and auxin signaling (Schubert et al, 2006; Gu et al, 2014). The derepression of YUC1/4 in sup and the reduction in H3K27me3 level at the YUC1/4 genomic regions suggest that SUP could function as the recruiter of CLF/TFL2 to YUC1/4 genomic regions (Figs 4A, C, D, and F, and EV5). It is worth noting that SUP can bind both the promoter and coding region of the YUC1/4 and the coding region of YUC1/4 is important for its negative transcription regulation (Fig 4A, B, D, and E, and Appendix Fig S7). Recent genome‐wide analysis with mutants of PRC1/2 components, including clf and tfl2, revealed that many transcription factors are associated with PRC2 target specificity in flower development, and both CLF and TFL2 are involved in spread of H3K27me3 marks (Wang et al, 2016). SUP contains a C2H2 zinc finger domain that is expected to bind DNA (Dathan et al, 2002). It is worth examining in the future whether SUP participates in the spreading of H3K27me3 marks at YUC1/4 loci via this domain, and whether multiple DNA‐binding motifs are required for SUP binding.
Materials and Methods
Plant materials and growth conditions
All Arabidopsis thaliana plants used were in the background of the Landsberg erecta (Ler) ecotype, except clf‐28, yuc1 (SALK_106293), yuc4 (SALK_047083), and pDR5rev::GFP, which were from the Col‐0 background and crossed into Ler at least three times. The sup‐1, sup‐5, clf‐28, wus‐1, yuc1, yuc4, pCUC2::3xVENUS‐N7, pCLV3::GFP‐ER, pSTM::CFP‐N7, pSUP‐SUP‐3xVenusN7, pDR5rev::GFP, pDR5rev::3xVENUS‐N7, pRPS5A::DII‐VENUS, and pDR5rev::2xGFP‐N7 line were described previously (Bowman et al, 1989, 1992; Sakai et al, 1995; Laux et al, 1996; Goodrich et al, 1997; Heisler et al, 2005; Cheng et al, 2006; Xu et al, 2006a; Gordon et al, 2007; Doyle & Amasino, 2009; Landrein et al, 2015; Liao et al, 2015; Prunet et al, 2017). Plants were grown at 22°C under 24 h of continuous light. Genotyping primer sequences are shown in Appendix Table S2.
Chemical treatment and statistical analyses
For PCIB, NPA, and IAM treatments, sup‐5 or pSUP::IAAH and WT plants with inflorescence shoot of approximately 2 cm in length were dipped into concentrations of 100 μM, 100 μM, 100 μM, and 1 mM, respectively. Two open flowers from 10 individual plants (total of 20 flowers) were examined for the number of stamens and carpels on continuous days. Control mock treatments were performed using equal amounts of solvent and Silwet L‐77. SEM was performed with flowers approximately 1 day before anthesis as previously described with minor changes (Xu et al, 2006b). Statistical significance was computed using Student's t‐test.
Plasmid construction and plant transformation
For cloning, pGreen‐35S::SUP‐GR and pGreen‐pSUP::SUP‐GFP were prepared in a pGreen vector (http://www.addgene.org). pENTR‐pSUP::iaaH were prepared in a pENTR/D‐TOPO vector (Invitrogen). The full length of genomic DNA of SUP of ~ 7.3 kb (−5,370 to +1,910) was cloned into a pENTR vector, and mutagenesis PCR was performed to introduce a SfoI site after the start codon. The IAAH fragment with the stop codon was then cloned into the SfoI site. GUS constructs were prepared using the Gateway system. Genomic DNA fragments were cloned into a pENTR/D‐TOPO vector (Invitrogen) and recombined into pBGFW to fuse with the GUS coding region. pYUC1::YUC1‐GUS contained a YUC1 genomic region of ~ 4.8 kb (−2,904 to +1,904; A of the start codon was set as +1), while pYUC1::GUS only contained the promoter region (−2,904 to +10). pYUC4::YUC4‐GUS was prepared with a YUC4 genomic region of ~ 5.6 kb (−3,735 to +1,930; A of the start codon was set as +1), and pYUC4::GUS only contained the promoter region (−2,904 to +16). For pCLF::HA‐CLF, the CLF genomic region of ~ 7.8 kb (−2,128 to +5,615) was cloned into a pCR8/GW/TOPO vector (Invitrogen). The SfoI restriction site was introduced just after ATG of CLF coding region by the mutagenesis PCR, and 3xHA was subcloned with the SfoI site. pHGW (Invitrogen) was used as the destination vector for pCLF::HA‐CLF. Primer sequences are listed in Appendix Table S2. Transgenic plants were generated by floral dipping with Agrobacterium tumefaciens and the corresponding constructs. The additional pSOUP helper plasmid was co‐transfected for the pGreen‐based constructs during the transformation.
Microarray analysis
For inducible expression analysis with p35S::SUP‐GR, a microarray analysis was performed with three biological replicates as described previously (Xu et al, 2013; Gan et al, 2014). The transgenic plants were grown at 22°C under 24‐h light conditions. When the plants reached a height of around 5 cm, inflorescences containing flowers of up to stage 12 were harvested 4 h after the DEX or control mock treatment. Total RNAs were extracted using an RNeasy plant mini kit (Qiagen), and double‐stranded cDNAs were synthesized with the Superscript Double‐Stranded cDNA Synthesis Kit (Invitrogen). The microarray was performed according to NimbleGen's protocol (Roche). Gene expression was analyzed using Arraystar (DNAStar). Genes showing a 2.0‐fold change in expression within a 90% confidence interval were considered to be differentially expressed and are presented in Appendix Table S3. Gene Ontology biological process enrichment was analyzed using agriGO software version 1.2 (http://bioinfo.cau.edu.cn/agriGO/). Enriched GO terms were further refined by REVIGO (http://revigo.irb.hr/) to reduce redundancy, with a cutoff of P < 0.01. The microarray data are available at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE92729.
RNA extraction and expression analysis
To verify the microarray data, an inducible expression analysis with p35S::SUP‐GR was performed with the transgenic inflorescences without open flowers harvested 2 and 4 h after the DEX or control mock treatments. To compare the expression profile between sup and WT, the inflorescences of ap1‐1 cal‐1 p35S::AP1‐GR sup‐5 and ap1‐1 cal‐1 p35S::AP1‐GR were harvested 3 days after 1 μM DEX treatment. Approximately 2 μg of total RNA was used for reverse transcription using the Superscript III RT–PCR system (Invitrogen). Real‐time quantitative reverse transcription PCR was performed using an ABI PRISM 7900HT sequence detection system (Applied Biosystems) with KAPA SYBR FAST ABI Prism qPCR Master Mix (KAPA Biosystems). The ubiquitously expressed Tip41‐like (AT4G34270; Czechowski et al, 2005) was used as an internal reference gene. Primer sequences are shown in Appendix Table S2.
GUS staining
For the GUS expression analysis, more than 20 independent T1 plants were obtained to examine the expression pattern (Sun et al, 2009). Inflorescences were incubated with GUS staining solution at 37°C overnight after the fixation in cold 90% acetone for 20 min, and the inflorescences were rinsed with GUS staining solution without X‐Gluc. The resulting stained tissues were fixed with the fixative solution overnight before clearing by a series of ethanol solutions. The samples were then mounted on a microscope slide (Fisher) with one or two drops of Hoyer's clearing solution and observed under an Axio Scope A1 microscope (Zeiss).
Chromatin immunoprecipitation (ChIP) and co‐immunoprecipitation (co‐IP) assays
ChIP experiments were performed as previously described with minor modifications (Xu et al, 2013). Briefly, to investigate the SUP‐GFP binding profiles at stage 4, total chromatin was extracted from ap1‐1 cal‐1 p35S::AP1‐GR sup‐5 pSUP::SUP‐GFP inflorescences 3 days after 1 μM DEX treatment and immunoprecipitated using anti‐GFP (Life Technologies, #A11122); normal rabbit IgG (Santa Cruz Biotechnology, #sc‐2027) was used as the control. To investigate the epigenetic profile in sup mutants, total chromatin was extracted from ap1‐1 cal‐1 p35S::AP1‐GR sup‐5 and ap1‐1 cal1‐1 p35S::AP1‐GR inflorescences 3 days after 1 μM DEX treatment and immunoprecipitated using anti‐H3K4me3 (Millipore, #07473) and anti‐H3K27me3 (Millipore, #07449) anti‐sera. DNA fragments were recovered with phenol–chloroform extraction and ethanol precipitation. Quantitative PCR with locus‐specific primers (Appendix Table S2) was performed to measure the amounts of YUC1 and YUC4 fragments relative to those of the constitutively expressed ACTIN2 (AT3G18780) on an ABI PRISM 7900HT sequence detection system (Applied Biosystems) using KAPA SYBR FAST ABI Prism qPCR Master Mix (KAPA Biosystems).
Co‐immunoprecipitation was carried out as described previously with minor modification (Xu et al, 2014). The pCLF::HA‐CLF was first transformed into the clf‐28 loss‐of‐function mutant. The transgenic plant of a single insertion, which can fully rescue clf‐28 mutant phenotype, was picked up and back‐crossed into Ler three times. After that, pCLF::HA‐CLF in Ler background was crossed into ap1‐1 cal‐1 p35S::AP1‐GR with or without pSUP::SUP‐GFP, respectively. Total proteins were extracted from inflorescences of ap1‐1 cal‐1 p35S::AP1‐GR pSUP::SUP‐GFP pCLF::HA‐CLF or ap1‐1 cal‐1 p35S::AP1‐GR pCLF::HA‐CLF at 3 days after 1 μM DEX treatment, followed by immunoprecipitation with using anti‐GFP (Life Technologies, #A11122). Immunoblotting was conducted to detect the presence of SUP‐GFP (Santa Cruz Biotechnology, #SC‐8334; 1:2,500 dilution) and CLF‐HA (Santa Cruz Biotechnology, #SC‐7392; 1:2,500 dilution) in the precipitate.
Yeast two‐hybrid and bimolecular fluorescence complementation (BiFC) assay
For the yeast two‐hybrid assay, the full‐length coding sequences for SUP, TFL2, RING1A/B, BMI1A/B, FIE, EMF2, and the truncated CLF (without its C‐terminal domain; according to Chanvivattana et al, 2004) were cloned in a Matchmaker GAL4 Two‐Hybrid System 3 (BD Clontech) according to the manufacturer's instructions. Site mutagenesis and PCR were performed to mutate the functional amino acids of the EAR motif or to create a series of truncated SUP proteins. For the BiFC assay, the full‐length coding sequences for SUP, TFL2, and the truncated CLF without its C‐terminus were fused in frame with either the coding sequence for an N‐terminal EYFP fragment or the C‐terminal EYFP fragment of the primary pSAT1 vector (Lee et al, 2008). To detect the interaction in tobacco, leaves of 2‐ to 4‐week‐old tobacco plants were infiltrated with Agrobacterium containing the respective plasmid pairs (Sparkes et al, 2006). Epidermal cell layers were examined 3–4 days after infiltration and imaged with a Zeiss LSM 5 EXCITER upright laser scanning confocal microscope (Zeiss; Xu et al, 2013).
Measurements and image analysis
The images of the inflorescence with the pSUP::SUP‐3xVENUS were taken with a Zeiss LSM 510 upright confocal microscope with a 40× oil objective, and the projections of confocal data were exported using Zeiss LSM software. All other confocal images were taken using a Leica SP8 confocal microscope with a 63× water‐dipping objective or Zeiss LSM 710 and 780 with a 40× water‐dipping objective as described previously (Prunet, 2017; Prunet et al, 2016), and cell measurements and image analyses were performed using the Imaris software (Bitplane).
Quantification of auxin
The inflorescences from ap1‐1 cal‐1 p35S::AP1‐GR sup‐5 and ap1‐1 cal‐1 p35S::AP1‐GR were harvested at 3 days after mock or 1 μM DEX treatment. Auxins were extracted and semi‐purified as described previously (Kojima et al, 2009). IAA was quantified with ultra‐high performance liquid chromatography (UHPLC)–electrospray interface (ESI) and quadrupole–orbitrap mass spectrometer (UHPLC/Q‐Exactive™; Thermo Scientific) with an ODS column (AQUITY UPLC HSS T3, 1.8 μm, 2.1 × 100 mm; Waters; Shinozaki et al, 2015). Data collected from three biological replicates were analyzed by one‐way ANOVA test.
Author contributions
YX and TI conceived the study. TI supervised and coordinated the study. YX, NP, EMM, and TI designed the experiments. YX and YW performed the Y2H, BiFC, and Co‐IP assays. YX, NP, and TI prepared all the constructs and the transgenic lines. YX and NP took the confocal images. YX and E‐SG carried out the microarray analysis and ChIP experiments. YX and NY carried out the genetic analysis. NY, YT, MK, TK, and HS quantified the auxin amount. YX and DS did transcriptional analysis. YX performed the SEM analysis. YX and JH performed the chemical treatments. YX, TI, NP, and EMM wrote the manuscript. TPJ and FW edited the manuscript. All authors discussed the results and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 5C
Acknowledgements
The authors would like to thank Jing Han Hong for reading the draft of this manuscript, Shen Lisha for the BiFC and Y2H constructs for some PcG components, and Jian Xu for the pDR5rev::GFP seeds and the unpublished transgenic plants that contain the IAAH gene. This work was supported by grants from the NAIST Foundation, the Mitsubishi Foundation, Grant‐in‐Aid for Scientific Research on Innovative Areas (Nos. 17H05843), Grant‐in‐Aid for Scientific Research A (No. 15H02405), Temasek Life Sciences Laboratory (TLL), and the National Research Foundation Singapore under the Competitive Research Programme (CRP Award NRFCRP001‐108) to T.I. Funding in the Meyerowitz Laboratory was provided by the Howard Hughes Medical Institute, the US National Institutes of Health through grant R01 GM104244, and the Gordon and Betty Moore Foundation through Grant GBMF3406. Funding in the Jack laboratory was provided by the US National Science Foundation through grant IOS‐0926347. Funding in the Wellmer laboratory was provided by the Science Foundation Ireland through grant #10/IN.1/B2971.
The EMBO Journal (2018) 37: e97499
References
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis . Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis . Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM (1992) SUPERMAN, a regulator of floral homeotic genes in Arabidopsis . Development 114: 599–615 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Breuil‐Broyer S, Morel P, de Almeida‐Engler J, Coustham V, Negrutiu I, Trehin C (2004) High‐resolution boundary analysis during Arabidopsis thaliana flower development. Plant J 38: 182–192 [DOI] [PubMed] [Google Scholar]
- Breuil‐Broyer S, Trehin C, Morel P, Boltz V, Sun B, Chambrier P, Ito T, Negrutiu I (2016) Analysis of the Arabidopsis superman allelic series and the interactions with other genes demonstrate developmental robustness and joint specification of male‐female boundary, flower meristem termination and carpel compartmentalization. Ann Bot 117: 905–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stoc C, Moon YH, Sung ZR, Goodrich J (2004) Interaction of Polycomb‐group proteins controlling flowering in Arabidopsis . Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis . Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM (1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121: 2057–2067 [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daimon Y, Takabe K, Tasaka M (2003) The CUP‐SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol 44: 113–121 [DOI] [PubMed] [Google Scholar]
- Dathan N, Zaccaro L, Esposito S, Isernia C, Omichinski JG, Riccio A, Pedone C, Di Blasio B, Fattorusso R, Pedone PV (2002) The Arabidopsis SUPERMAN protein is able to specifically bind DNA through its single Cys2‐His2 zinc finger motif. Nucleic Acids Res 30: 4945–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana‐Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root‐meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Derkacheva M, Steinbach Y, Wildhaber T, Mozgova I, Mahrez W, Nanni P, Bischof S, Gruissem W, Hennig L (2013) Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J 32: 2073–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Amasino RM (2009) A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization‐independent, rapid flowering in Arabidopsis . Plant Physiol 151: 1688–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Gaiser JC, Robinson‐Beers K, Gasser CS (1995) The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell 7: 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan ES, Xu Y, Wong JY, Goh JG, Sun B, Wee WY, Huang J, Ito T (2014) Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis . Nat Commun 5: 5098 [DOI] [PubMed] [Google Scholar]
- Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, Lefebvre D, Grandjean O (2001) Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis . Development 128: 4847–4858 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb‐group gene regulates homeotic gene expression in Arabidopsis . Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM (2007) Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548 [DOI] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Xu T, He Y (2014) A histone H3 lysine‐27 methyltransferase complex represses lateral root formation in Arabidopsis thaliana . Mol Plant 7: 977–988 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme‐Takagi M (2002) The SUPERMAN protein is an active repressor whose carboxy‐terminal repression domain is required for the development of normal flowers. FEBS Lett 514: 351–354 [DOI] [PubMed] [Google Scholar]
- Huang T, Lopez‐Giraldez F, Townsend JP, Irish VF (2012) RBE controls microRNA164 expression to effect floral organogenesis. Development 139: 2161–2169 [DOI] [PubMed] [Google Scholar]
- Kojima M, Kamada‐Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi‐Tanaka M, Matsuoka M, Suzuki K, Sakakibara H (2009) Highly sensitive and high‐throughput analysis of plant hormones using MS‐probe modification and liquid chromatography‐tandem mass spectrometry: an application for hormone profiling in Oryza sativa . Plant Cell Physiol 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B, Kiss A, Sassi M, Chauvet A, Das P, Cortizo M, Laufs P, Takeda S, Aida M, Traas J, Vernoux T, Boudaoud A, Hamant O (2015) Mechanical stress contributes to the expression of the STM homeobox gene in Arabidopsis shoot meristems. Elife 4: e07811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jurgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis . Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lee LY, Fang MJ, Kuang LY, Gelvin SB (2008) Vectors for multi‐color bimolecular fluorescence complementation to investigate protein‐protein interactions in living plant cells. Plant Methods 4: 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin‐inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814 [DOI] [PubMed] [Google Scholar]
- Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D (2015) Reporters for sensitive and quantitative measurement of auxin response. Nat Methods 12: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim YJ, Muller R, Yumul RE, Liu C, Pan Y, Cao X, Goodrich J, Chen X (2011) AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23: 3654–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis . Cell 105: 793–803 [DOI] [PubMed] [Google Scholar]
- Meister RJ, Kotow LM, Gasser CS (2002) SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 129: 4281–4289 [DOI] [PubMed] [Google Scholar]
- Nibau C, Di Stilio VS, Wu HM, Cheung AY (2011) Arabidopsis and Tobacco superman regulate hormone signalling and mediate cell proliferation and differentiation. J Exp Bot 62: 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka M, Miyamoto K, Okada K, Ueda J (1999) Auxin polar transport and flower formation in Arabidopsis thaliana transformed with indoleacetamide hydrolase (iaaH) gene. Plant Cell Physiol 40: 231–237 [DOI] [PubMed] [Google Scholar]
- Oono Y, Ooura C, Rahman A, Aspuria ET, Hayashi K, Tanaka A, Uchimiya H (2003) p‐Chlorophenoxyisobutyric acid impairs auxin response in Arabidopsis root. Plant Physiol 133: 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Jack TP, Meyerowitz EM (2016) Live confocal imaging of Arabidopsis flower buds. Dev Biol 419: 114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N (2017) Live confocal imaging of developing Arabidopsis flowers. J Vis Exp 122: e55156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Yang W, Das P, Meyerowitz EM, Jack TP (2017) SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc Natl Acad Sci USA 114: 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GV, Meyerowitz EM (2005) Stem‐cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 310: 663–667 [DOI] [PubMed] [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM (1995) Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378: 199–203 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ (2015) The yin‐yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27: 44–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J (2006) Silencing by plant Polycomb‐group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki Y, Hao S, Kojima M, Sakakibara H, Ozeki‐Iida Y, Zheng Y, Fei Z, Zhong S, Giovannoni JJ, Rose JK, Okabe Y, Heta Y, Ezura H, Ariizumi T (2015) Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J 83: 237–251 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Sun B, Xu Y, Ng KH, Ito T (2009) A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev 23: 1791–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Looi LS, Gu S, He Z, Gan ES, Huang J, Xu Y, Wee WY, Ito T (2014) Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343: 1248559 [DOI] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M (2001) The CUP‐SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128: 1127–1135 [DOI] [PubMed] [Google Scholar]
- Uemura A, Yamaguchi N, Xu Y, Wee W, Ichihashi Y, Suzuki T, Shibata A, Shirasu K, Ito T (2017) Regulation of floral meristem activity through the interaction of AGAMOUS, SUPERMAN, and CLAVATA3 in Arabidopsis . Plant Reprod 31: 89–105 [DOI] [PubMed] [Google Scholar]
- Wang H, Liu C, Cheng J, Liu J, Zhang L, He C, Shen WH, Jin H, Xu L, Zhang Y (2016) Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of polycomb group proteins in association with distinct sets of cis‐regulatory elements. PLoS Genet 12: e1005771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Alves‐Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM (2006) Genome‐wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet 2: e117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T (2003) Cytokinin‐deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B (2006a) A molecular framework for plant regeneration. Science 311: 385–388 [DOI] [PubMed] [Google Scholar]
- Xu Y, Teo LL, Zhou J, Kumar PP, Yu H (2006b) Floral organ identity genes in the orchid Dendrobium crumenatum . Plant J 46: 54–68 [DOI] [PubMed] [Google Scholar]
- Xu L, Shen WH (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis . Curr Biol 18: 1966–1971 [DOI] [PubMed] [Google Scholar]
- Xu Y, Wang Y, Stroud H, Gu X, Sun B, Gan ES, Ng KH, Jacobsen SE, He Y, Ito T (2013) A matrix protein silences transposons and repeats through interaction with retinoblastoma‐associated proteins. Curr Biol 23: 345–350 [DOI] [PubMed] [Google Scholar]
- Xu Y, Gan ES, Zhou J, Wee WY, Zhang X, Ito T (2014) Arabidopsis MRG domain proteins bridge two histone modifications to elevate expression of flowering genes. Nucleic Acids Res 42: 10960–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Huang J, Xu Y, Tanoi K, Ito T (2017) Fine‐tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat Commun 8: 1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun JY, Weigel D, Lee I (2002) Ectopic expression of SUPERMAN suppresses development of petals and stamens. Plant Cell Physiol 43: 52–57 [DOI] [PubMed] [Google Scholar]
- Zadnikova P, Simon R (2014) How boundaries control plant development. Curr Opin Plant Biol 17: 116–125 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU (2010) Hormonal control of the shoot stem‐cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 5C


