Abstract
Background
Barley (Hordeum vulgare) is an important crop cultivated across the world. Drought is a major abiotic factor compromising barley yield worldwide, therefore in modern spring barley cultivars superior seed and malting quality characteristics should be combined with reasonable level of drought tolerance. Previously we have identified a number of barley lines demonstrating the superior yield performance under drought conditions. The aim of this work was to perform a QTL analysis of malting quality traits in a doubled haploid (DH) mapping population of two elite barley lines that differ in their reaction pattern to drought stress.
Results
A population of DH lines was developed by crossing two drought-tolerant elite breeding lines, Victoriana and Sofiara, exploiting distinct mechanism of drought tolerance, sustaining assimilation vs remobilization. The mapping population was assayed under field conditions at four distinct locations that differed in precipitation rate. DH lines were genotyped with the Illumina 9 K iSelect assay, and linkage map including 1782 polymorphic markers and covering a total map length of 1140 cM was constructed. The result of quantitative trait loci (QTL) analysis showed that majority of the traits were affected by several main effect QTL and/or QTL x environment (QE) interactions. In total, 57, 41, and 5 QTL were associated with yield-related traits, malting quality traits and seed quality traits, respectively. 11 and 29 of mapped QTL explained more than 10 and 5% of phenotypic variation, respectively. In several chromosomal regions co-localization between QTL for various traits were observed. The largest clusters were detected on chromosomes 3H and 4H.
Conclusions
Our QTL mapping results revealed several novel consistent genomic regions controlling malting quality which could be exploited in marker assisted selection. In this context, the complex QTL region on chromosome 3H seems of particular interest, as it harbors several large effect QTL.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1323-4) contains supplementary material, which is available to authorized users.
Keywords: QTL mapping, Malting quality, Drought-tolerance, Spring barley, Elite breeding lines, iSelect array
Background
Barley (Hordeum vulgare) is an important crop cultivated throughout the world with Russia, France, Germany, Australia and Canada being the major producers. It is used as human food, livestock feed and for malt products. About 30% of the global barley production (133.5 million tons) is processed as malt by the brewing or the distilling industry [1, 2]. Therefore, one of the primary breeding goals is to improve the malting quality.
Malting quality is the result of complex interactions of numerous trait components, each of which is under control of multiple genes [3, 4]. Therefore, phenotypic selection of malting quality is labor-intensive and costly [5]. Marker-assisted selection is a promising alternative to improve malting quality [6] but necessitates detailed knowledge of its genetic architecture. Several linkage and association mapping studies have been conducted and QTL controlling components of barley malting quality, such as alpha- and beta-amylase activity, malt extract, malt soluble nitrogen, diastatic power, Kolbach index, wort β-glucans, wort viscosity, friability and apparent final attenuation have been identified [7–12]. Malting quality QTL were mapped to all seven barley chromosomes, however, the distribution was uneven and in some cases the formation of QTL clusters simultaneously affecting several malting traits have been observed [13–15]. Moreover, detected QTL were frequently dependent on genetic background and environmental conditions [16–18].
A few QTL explaining a large proportion of the genotypic variance have been used in marker-assisted selection (MAS). Igartua et al. [19] applied MAS for two QTL regions on chromosome 5H affecting several malting quality traits in the Harrington/TR306 population. Selection for the Harrington allele at target regions produced DH lines with superior malting quality characteristics such as low grain protein and β-glucan content, high diastatic power and malt extract. Ayoub et al. [20] introgressed a QTL from chromosome 5H affecting alpha-amylase activity originating from the cultivar Morex into the feed barley cultivar Labelle and observed an increase in alpha-amylase activity. Laido et al. [21] reported successful marker-assisted introgression of major QTL regions for grain protein content, malt extract, friability and viscosity on chromosome 1H in a population consisting of DH lines derived from winter and spring barley cross (Nure/Tremois). MAS for malting quality QTL may greatly benefit the introduction of novel diversity into elite germplasm. This is of particular importance if complex traits such as tolerance to biotic and abiotic stresses are to be introgressed from unadapted germplasm sources.
Previously, we have identified a number of barley lines showing senescence or stay-green phenotype and demonstrating the superior yield performance under drought conditions. The stay-green and senescing lines differed in their assimilation performance under drought stress. Moreover, it was revealed that senescing lines synthesized greater levels of ABA than the stay-green lines under short-term stress, and it continued to maintain a high level of ABA and ABA catabolites under long-term stress. This suggests that a greater flux in ABA metabolism in the senescing lines negatively affected water use efficiency and assimilation [22]. The objective of the current study was to perform a QTL analysis of malting quality traits in a biparental mapping population of two elite barley lines that differ in their reaction pattern to drought stress in order to identify genetic regions associated with twenty two malting-, grain-quality and yield-related traits.
Results
Phenotypic analysis
The BLUEs of the DH population were widely distributed for all examined traits approximating in most cases a normal distribution (Additional file 1: Figure S1; Table 1). For all traits we observed transgressive variation with several DH lines possessing phenotypic values lower or higher than their parental lines. The genotypic variance components) were for all traits except seed nitrogen content (SNC) significantly (P < 0.05) larger than zero (Table 2). For 14 out of the 22 measured traits, significant (P < 0.05) variance components were detected for genotype-by-environment interaction effects ). The ratio of was minimum for grain yield (YLD) amounting to 0.27. The broad-sense heritability estimates (h2) ranged from 0.34 for SNC to 0.98 for seed length (SL).
Table 1.
Summary statistics of the evaluated quantitative traits for the parents and the Sofiara/Victoriana DH population across environments
| MMC | FGE | VIS | PRO | SNI | KOL | FLA | FRI | BGL | TML | REL | RTL | YLD | TGW | SA | SB | SL | SSC | SNC | SCC | FTa | PHa | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Victoriana | 5.96 | 80.60 | 1.50 | 11.07 | 742.72 | 42.53 | 80.89 | 75.73 | 371.27 | 9.84 | 4.97 | 4.88 | 54.14 | 51.89 | 27.81 | 4.01 | 9.18 | 26.78 | 1.71 | 40.66 | 23.67 | 68.00 |
| Sofiara | 5.82 | 83.36 | 1.46 | 11.22 | 857.44 | 47.75 | 81.53 | 88.15 | 152.66 | 11.18 | 5.71 | 5.47 | 49.32 | 53.47 | 28.56 | 4.19 | 9.09 | 31.13 | 1.72 | 40.83 | 25.42 | 82.84 |
| Maximum | 6.10 | 83.50 | 1.58 | 11.77 | 855.70 | 50.36 | 82.46 | 92.40 | 683.20 | 11.93 | 6.30 | 5.97 | 60.20 | 57.66 | 30.70 | 4.32 | 9.65 | 31.87 | 1.73 | 41.09 | 25.5 | 74.00 |
| Minimum | 5.40 | 80.84 | 1.43 | 9.80 | 683.20 | 39.45 | 79.54 | 64.64 | 88.76 | 8.78 | 4.75 | 3.93 | 48.11 | 48.13 | 25.90 | 3.94 | 8.43 | 25.43 | 1.48 | 40.46 | 20.28 | 66.00 |
| Mean | 5.67 | 82.43 | 1.49 | 10.64 | 763.50 | 45.05 | 80.96 | 81.42 | 281.90 | 10.33 | 5.37 | 4.97 | 54.03 | 52.36 | 27.67 | 4.09 | 8.96 | 28.31 | 1.60 | 40.85 | 22.83 | 70.24 |
| SD | 0.17 | 0.58 | 0.03 | 0.43 | 38.62 | 2.42 | 0.72 | 6.65 | 144.50 | 0.52 | 0.29 | 0.38 | 2.07 | 2.07 | 0.85 | 0.07 | 0.21 | 1.30 | 0.05 | 0.12 | 1.064 | 1.78 |
| CV | 2.94 | 0.71 | 2.23 | 4.00 | 5.06 | 5.37 | 0.89 | 8.17 | 51.28 | 5.07 | 5.39 | 7.64 | 3.83 | 3.95 | 3.07 | 1.74 | 2.37 | 4.60 | 3.16 | 0.30 | 4.662 | 2.53 |
| Skewness | 0.72 | −0.45 | 0.63 | 0.41 | 0.21 | −0.15 | − 0.07 | − 0.34 | 0.95 | − 0.19 | 0.27 | 0.07 | − 0.09 | 0.36 | 0.62 | 0.50 | 0.46 | 0.22 | 0.29 | − 0.58 | 0.0804 | − 0.13 |
| Kurtosis | −0.34 | − 0.45 | − 0.14 | − 0.16 | − 0.44 | − 0.49 | − 0.81 | − 0.91 | − 0.03 | 0.34 | 0.55 | 0.21 | 0.23 | − 0.49 | 0.65 | 0.33 | 0.74 | − 0.03 | − 0.30 | 0.28 | − 0.08 | − 0.34 |
MMC moisture content, FGE fine grind extract, VIS viscosity, PRO protein content, SNI soluble nitrogen, KOL kolbach index, FLA limit of attenuation, FRI friability, BGL β-glucan, TML total malting losses, PH plant height, FT flowering time, REL respiration losses, RTL rootlet losses, TGW thousand-grain weight, SA seed area, SB seed breadth, SL seed length, YLD grain yield, SSC seed starch content, SNC seed nitrogen content, SCC seed carbon content, SD standart deviation, CV coefficient of variation
atraits were measured only in some environments
Table 2.
Estimates of variance components for measured traits across environments
| Trait | Environments | σ2G | σ2E | σ2GxE | σ2e | h2 |
|---|---|---|---|---|---|---|
| MMC | 3 | 0.016*** | 0.009*** | 0.026*** | 0.014 | 0.569 |
| FGE | 3 | 0.236*** | 0.100*** | 0.118*** | 0.131 | 0.761 |
| VIS | 3 | 0.001*** | 0.000*** | 0.001*** | 0.001 | 0.575 |
| PRO | 3 | 0.084*** | 0.474*** | 0.231*** | 0.122 | 0.433 |
| SNI | 3 | 979.487*** | 55.790* | 941.560*** | 643.514 | 0.669 |
| KOL | 3 | 3.695*** | 5.793*** | 3.167*** | 2.720 | 0.675 |
| FLA | 3 | 0.163* | 0.221*** | 0.508*** | 0.442 | 0.363 |
| FRI | 3 | 33.685*** | 3.004*** | 19.203*** | 13.321 | 0.772 |
| BGL | 3 | 15,979.8*** | 272.305 | 9595.9*** | 6179.561 | 0.768 |
| TML | 3 | 0.089* | 3.887*** | 0.094 | 0.233 | 0.489 |
| REL | 3 | 0.038*** | 1.171*** | 0.032** | 0.057 | 0.593 |
| RTL | 3 | 0.046* | 0.786*** | 0.043 | 0.124 | 0.492 |
| YLD | 6 | 1.735*** | 200.854*** | 6.506*** | 6.908 | 0.536 |
| TGW | 6 | 3.687*** | 7.974*** | 2.119*** | 1.655 | 0.891 |
| SA | 4 | 0.654*** | 0.778*** | 0.002 | 0.126 | 0.974 |
| SB | 4 | 0.004*** | 0.009*** | 0.001*** | 0.002 | 0.867 |
| SL | 4 | 0.042*** | 0.058*** | 0.000 | 0.005 | 0.985 |
| SSC | 3 | 0.856*** | 8.602*** | 1.253*** | 1.588 | 0.551 |
| SNC | 3 | 0.000 | 0.016*** | 0.000 | 0.003 | 0.336 |
| SCC | 3 | 0.005* | 0.019*** | 0.006* | 0.019 | 0.449 |
| PH | 2 | 1.311** | 102.928*** | NA | 3.541 | 0.420 |
| ME | 2 | 1.993*** | 0.358*** | NA | 1.019 | 0.800 |
σ2G - genotypic variance, σ2GxE - genotype by environment interaction variance, σ2e - error variance, h2 - heritability, ***p < 0.001, **p < 0.01, *p < 0.05. Abbreviations of the traits are the same as in Table 1
Correlation estimates
Pearson correlation coefficients were estimated between all pairs of traits (Table 3). Grain yield was negatively correlated (P < 0.05) with soluble nitrogen. Correlation coefficients between malting quality traits ranged from − 0.92 to 0.86. Seed quality traits such as SNC and seed starch content (SSC) were moderately negatively correlated with fine-grind malt extract (FGE) (r = − 0.49) and rootlet losses (RTL) (r = − 0.28), respectively.
Table 3.
Pearson correlation coefficients between measured traits
| MMC | ||||||||||||||||||||||
| FGE | −0.10 | |||||||||||||||||||||
| VIS | 0.27 | −0.45 | ||||||||||||||||||||
| PRO | 0.15 | −0.52 | 0.29 | |||||||||||||||||||
| SNI | − 0.03 | 0.21 | − 0.39 | 0.31 | ||||||||||||||||||
| KOL | −0.14 | 0.57 | − 0.58 | − 0.43 | 0.72 | |||||||||||||||||
| FLA | −0.26 | 0.40 | − 0.72 | − 0.35 | 0.23 | 0.48 | ||||||||||||||||
| FRI | −0.18 | 0.57 | −0.79 | −0.28 | 0.54 | 0.72 | 0.65 | |||||||||||||||
| BGL | 0.17 | −0.49 | 0.86 | 0.24 | −0.55 | − 0.69 | − 0.67 | − 0.92 | ||||||||||||||
| TML | −0.07 | 0.10 | −0.23 | 0.07 | 0.28 | 0.22 | 0.24 | 0.11 | −0.19 | |||||||||||||
| REL | −0.17 | 0.14 | −0.17 | 0.00 | 0.29 | 0.28 | 0.19 | 0.22 | −0.28 | 0.69 | ||||||||||||
| RTL | 0.03 | 0.02 | −0.18 | 0.11 | 0.15 | 0.07 | 0.18 | −0.02 | −0.05 | 0.83 | 0.17 | |||||||||||
| YLD | 0.15 | 0.19 | 0.02 | −0.16 | −0.25 | −0.12 | −0.07 | − 0.08 | 0.04 | 0.11 | 0.02 | 0.16 | ||||||||||
| TGW | 0.20 | 0.09 | 0.11 | 0.02 | 0.09 | 0.06 | −0.28 | −0.06 | 0.06 | −0.18 | −0.06 | − 0.20 | 0.33 | |||||||||
| SA | 0.15 | 0.01 | 0.13 | 0.02 | 0.15 | 0.12 | −0.34 | −0.05 | 0.07 | − 0.35 | −0.21 | − 0.31 | 0.17 | 0.84 | ||||||||
| SB | 0.06 | 0.23 | −0.01 | 0.19 | 0.23 | 0.07 | −0.13 | 0.15 | −0.11 | − 0.13 | − 0.06 | −0.12 | 0.22 | 0.64 | 0.60 | |||||||
| SL | 0.17 | −0.08 | 0.12 | −0.10 | 0.11 | 0.17 | −0.29 | −0.09 | 0.08 | −0.35 | − 0.23 | − 0.30 | 0.10 | 0.65 | 0.88 | 0.18 | ||||||
| SSC | 0.07 | 0.24 | −0.05 | −0.12 | 0.13 | 0.18 | 0.01 | 0.13 | −0.11 | −0.22 | − 0.03 | −0.28 | 0.09 | 0.62 | 0.62 | 0.45 | 0.50 | |||||
| SNC | −0.07 | −0.49 | 0.03 | 0.54 | 0.24 | −0.16 | −0.13 | − 0.14 | 0.09 | 0.00 | −0.11 | 0.09 | −0.36 | − 0.06 | 0.06 | 0.11 | −0.03 | − 0.07 | ||||
| SCC | −0.40 | − 0.13 | −0.14 | 0.20 | 0.03 | −0.12 | 0.14 | 0.06 | −0.09 | 0.06 | 0.06 | 0.03 | 0.03 | 0.10 | 0.00 | 0.24 | −0.15 | 0.04 | 0.34 | |||
| FT | 0.11 | −0.07 | −0.01 | 0.02 | −0.26 | −0.27 | 0.07 | −0.06 | 0.07 | 0.02 | −0.07 | 0.09 | 0.15 | −0.04 | −0.15 | 0.16 | −0.26 | − 0.03 | 0.02 | 0.02 | ||
| PH | 0.03 | 0.05 | 0.08 | 0.23 | 0.07 | −0.13 | −0.07 | 0.04 | 0.04 | 0.09 | 0.10 | 0.04 | −0.06 | 0.11 | 0.12 | 0.30 | −0.06 | 0.30 | 0.09 | 0.15 | 0.20 | |
| MMC | FGE | VIS | PRO | SNI | KOL | FLA | FRI | BGL | TML | REL | RTL | YLD | TGW | SA | SB | SL | SSC | SNC | SCC | FT | PH |
Significant correlations (P < 0.05) are labeled in bold text. Abbreviations of the traits are the same as in Table 1
QTL analysis
The 1782 mapped SNP markers detected a total of 380 independent loci. Hence, for QTL mapping 380 polymorphic high-quality SNP markers were selected, i.e. one marker per locus. The number of polymorphic markers ranged from 106 for chromosome 1H to 374 for chromosome 2H (Additional file 2: Table S1). Despite the high marker density, we observed some regions such as on the chromosome 1H exhibiting a low number of polymorphic markers. It is likely that these regions are identical by descent in both parents.
Genome-wide mapping resulted in 103 QTL with additive main effects and/or additive x environment interaction effects (QE) (Tables 4, 6, Fig. 1). No QTL were detected for seed carbon content. Twenty-nine QTL explained more the 5% and 11 QTL even more than 10% of the phenotypic variation. Variation explained by main effects (QTL) ranged from 0.41 to 68.77%, which was substantially higher than that of QTL x Environment interaction effects.
Table 4.
Main-effect QTL for measured traits detected in the Sofiara/Victoriana DH population in six environments
| Trait | QTL name | Chr. | Position (cM) | Peak Marker | LOD | Effect | R2 (%) |
|---|---|---|---|---|---|---|---|
| Malt moisture content | |||||||
| QMMC-3 | 3H | 74.64 | BOPA1_2616–2560 | 4.27 | −0.04 | 2.66 | |
| QMMC-4-1 | 4H | 47.94 | BOPA1_10346–92 | 6.38 | 0.04 | 1.91 | |
| QMMC-4-2 | 4H | 111.57 | BOPA1_3652–872 | 6.52 | −0.06 | 3.67 | |
| Fine grind extract | |||||||
| QFGE-3 | 3H | 11.55 | SCRI_RS_153718 | 5.84 | 0.22 | 6.96 | |
| QFGE-7 | 7H | 1.53 | BOPA1_7970–387 | 4.52 | 0.21 | 6.76 | |
| Viscosity | |||||||
| QVIS-3 | 3H | 63.66 | BK_08 | 13.91 | 0.00 | 13.82 | |
| QVIS-4 | 4H | 0.00 | BOPA2_12_30540 | 6.83 | −0.01 | 8.48 | |
| QVIS-5 | 5H | 169.36 | BOPA1_1394–1222 | 5.92 | 0.01 | 5.50 | |
| Malt protein content | |||||||
| QPRO-4 | 4H | 116.80 | SCRI_RS_179695 | 11.75 | −0.21 | 5.06 | |
| QPRO-6 | 6H | 17.05 | BOPA1_397–288 | 7.76 | −0.18 | 4.50 | |
| Soluble protein | |||||||
| QSNI-2 | 2H | 118.20 | BOPA1_11591–265 | 4.06 | −5.85 | 1.10 | |
| QSNI-3-1 | 3H | 60.96 | BK_08 | 25.79 | −68.76 | 27.31 | |
| Kolbach index | |||||||
| QKOL-1 | 1H | 21.79 | SCRI_RS_205669 | 5.80 | 0.41 | 0.85 | |
| QKOL-3 | 3H | 57.11 | SCRI_RS_115045 | 9.93 | 0.17 | 13.74 | |
| QKOL-5-1 | 5H | 100.05 | BOPA2_12_31361 | 4.70 | −2.04 | 1.34 | |
| QKOL-5-2 | 5H | 103.41 | BOPA1_370–443 | 4.26 | 1.68 | 1.48 | |
| QKOL-7 | 7H | 0.00 | BOPA1_7970–387 | 8.00 | 0.94 | 4.05 | |
| Limit of attenuation | |||||||
| QFLA-3 | 3H | 62.76 | BK_08 | 5.26 | 0.32 | 6.85 | |
| Friability | |||||||
| QFRI-3 | 3H | 59.08 | SCRI_RS_115045 | 36.80 | 4.40 | 33.81 | |
| QFRI-4-1 | 4H | 0.00 | BOPA2_12_30540 | 5.42 | 1.64 | 3.93 | |
| QFRI-4-2 | 4H | 121.34 | BOPA1_ABC08009–1–2-304 | 7.04 | 1.80 | 4.68 | |
| QFRI-7 | 7H | 6.80 | BOPA2_12_31173 | 8.94 | 2.29 | 6.64 | |
| β-glucan | |||||||
| QBGL-3 | 3H | 60.06 | BK_08 | 38.62 | −81.46 | 37.42 | |
| QBGL-4-1 | 4H | 0.00 | BOPA2_12_30540 | 6.59 | −32.38 | 4.19 | |
| QBGL-4-2 | 4H | 115.31 | BOPA1_4160–1365 | 6.83 | −22.12 | 2.62 | |
| QBGL-5 | 5H | 169.36 | BOPA1_1394–1222 | 6.01 | 38.15 | 4.29 | |
| Total malting losses | |||||||
| QTML-2 | 2H | 175.69 | BOPA1_8586–1221 | 5.83 | 0.25 | 1.89 | |
| Respiration losses | |||||||
| QREL-2 | 2H | 162.13 | SCRI_RS_193100 | 8.13 | 0.16 | 2.10 | |
| Rootlet losses | |||||||
| QRTL-2 | 2H | 175.69 | BOPA1_8586–1221 | 5.84 | 0.13 | 1.88 | |
| QRTL-4 | 4H | 64.90 | SCRI_RS_89959 | 4.09 | −0.07 | 0.57 | |
| Grain yield | |||||||
| QYLD-3-1 | 3H | 85.25 | SCRI_RS_156056 | 3.87 | −0.88 | 0.24 | |
| QYLD-6 | 6H | 30.19 | BOPA1_2294–573 | 5.85 | 0.80 | 0.32 | |
| TGW | |||||||
| QTGW-2 | 2H | 124.98 | SCRI_RS_209622 | 7.81 | − 0.66 | 3.39 | |
| QTGW-4-1 | 4H | 36.84 | BOPA2_12_30328 | 5.68 | 0.44 | 3.41 | |
| QTGW-4-2 | 4H | 60.73 | SCRI_RS_134953 | 4.30 | 0.65 | 0.99 | |
| QTGW-4-3 | 4H | 87.41 | 12_31523 | 7.42 | −0.84 | 0.90 | |
| QTGW-5-1 | 5H | 135.55 | SCRI_RS_119308 | 4.67 | 0.54 | 5.16 | |
| QTGW-5-2 | 5H | 169.36 | BOPA1_1394–1222 | 4.04 | 0.48 | 1.17 | |
| QTGW-6-1 | 6H | 9.93 | BOPA2_12_30665 | 8.03 | 0.43 | 3.77 | |
| QTGW-6-2 | 6H | 65.50 | BOPA1_3349–759 | 3.95 | 0.35 | 0.94 | |
| QTGW-7 | 7H | 24.27 | BOPA1_3187–1073 | 10.49 | −0.43 | 4.89 | |
| Seed area | |||||||
| QSA-1-1 | 1H | 35.65 | SCRI_RS_117492 | 4.74 | −0.80 | 0.17 | |
| QSA-2 | 2H | 124.23 | SCRI_RS_209622 | 26.94 | −0.53 | 15.87 | |
| QSA-4-1 | 4H | 62.51 | SCRI_RS_140349 | 7.04 | 0.37 | 3.86 | |
| QSA-4-2 | 4H | 84.71 | BOPA2_12_31523 | 4.56 | −0.23 | 0.77 | |
| QSA-4-3 | 4H | 103.70 | BOPA2_12_30158 | 6.86 | −0.13 | 1.27 | |
| QSA-4-4 | 4H | 112.31 | SCRI_RS_148330 | 7.18 | −0.06 | 3.31 | |
| QSA-5-1 | 5H | 73.55 | SCRI_RS_205235 | 4.90 | 0.15 | 1.56 | |
| QSA-5-2 | 5H | 173.16 | SCRI_RS_102414 | 21.59 | 0.23 | 4.30 | |
| QSA-6-1 | 6H | 14.38 | BOPA1_397–288 | 4.07 | 0.10 | 1.16 | |
| QSA-6-2 | 6H | 54.57 | BOPA2_12_30698 | 4.07 | 0.09 | 0.31 | |
| QSA-7-1 | 7H | 25.02 | SCRI_RS_223021 | 7.80 | −0.45 | 2.37 | |
| QSA-7-2 | 7H | 28.05 | BOPA1_8365–454 | 5.45 | 0.28 | 0.59 | |
| QSA-7-3 | 7H | 152.86 | BOPA1_3900–611 | 4.77 | −0.03 | 0.25 | |
| QSA-7-4 | 7H | 157.25 | SCRI_RS_13570 | 8.69 | 0.16 | 0.38 | |
| Seed breadth | |||||||
| QSB-2 | 2H | 8.13 | SCRI_RS_188511 | 8.33 | −0.02 | 3.75 | |
| QSB-3 | 3H | 1.49 | SCRI_RS_155475 | 5.03 | −0.02 | 2.31 | |
| QSB-4 | 4H | 83.95 | BOPA2_12_31246 | 5.07 | −0.01 | 0.86 | |
| QSB-5-1 | 5H | 102.48 | BOPA1_370–443 | 9.93 | 0.03 | 5.31 | |
| QSB-5-2 | 5H | 138.29 | SCRI_RS_178985 | 13.44 | 0.02 | 8.58 | |
| QSB-6-1 | 6H | 17.05 | BOPA1_397–288 | 8.56 | 0.07 | 1.97 | |
| QSB-6-2 | 6H | 18.55 | SCRI_RS_136658 | 6.77 | −0.05 | 0.83 | |
| QSB-7 | 7H | 114.27 | SCRI_RS_194841 | 4.71 | 0.02 | 1.49 | |
| Seed length | |||||||
| QSL-1 | 1H | 48.06 | SCRI_RS_128285 | 18.98 | 0.06 | 1.82 | |
| QSL-2-1 | 2H | 124.23 | SCRI_RS_209622 | 41.34 | −0.10 | 18.13 | |
| QSL-2-2 | 2H | 148.64 | SCRI_RS_12444 | 4.34 | −0.04 | 0.56 | |
| QSL-2-3 | 2H | 175.69 | BOPA1_8586–1221 | 17.56 | −0.06 | 4.57 | |
| QSL-4-1 | 4H | 44.21 | BOPA2_12_30597 | 16.19 | 0.06 | 6.09 | |
| QSL-4-2 | 4H | 59.83 | SCRI_RS_134953 | 6.89 | 0.03 | 0.56 | |
| QSL-4-3 | 4H | 110.82 | 12_30987 | 19.91 | −0.08 | 3.30 | |
| QSL-5 | 5H | 173.16 | SCRI_RS_102414 | 19.46 | 0.05 | 2.14 | |
| QSL-6 | 6H | 12.60 | BOPA2_12_30665 | 10.02 | 0.04 | 1.15 | |
| QSL-7-1 | 7H | 0.00 | BOPA1_7970–387 | 8.08 | 0.05 | 1.10 | |
| QSL-7-2 | 7H | 24.27 | BOPA1_3187–1073 | 16.42 | −0.08 | 3.00 | |
| QSL-7-3 | 7H | 177.23 | BOPA1_6470–1005 | 5.17 | −0.02 | 0.29 | |
| Seed starch content | |||||||
| QSSC-2 | 2H | 126.48 | BOPA2_12_10735 | 4.10 | −0.50 | 2.39 | |
| QSSC-3 | 3H | 66.37 | SCRI_RS_168665 | 3.96 | 0.41 | 1.69 | |
| Seed nitrogen content | |||||||
| QSNC-4-1 | 4H | 87.41 | BOPA2_12_31523 | 5.33 | −0.03 | 2.77 | |
| QSNC-7 | 7H | 112.75 | SCRI_RS_104566 | 4.39 | 0.02 | 2.17 | |
| Flowering time | |||||||
| QFT-1 | 1H | 180.94 | SCRI_RS_196025 | 4.60 | −0.37 | 12.99 | |
| QFT-3 | 3H | 83.76 | BOPA1_4150–398 | 6.47 | −0.48 | 12.90 | |
| QFT-4 | 4H | 113.06 | SCRI_RS_217794 | 5.29 | 0.35 | 8.45 | |
| QFT-6 | 6H | 35.43 | BOPA1_4146–1154 | 4.27 | 0.21 | 3.63 | |
| Plant height | |||||||
| QPH-5 | 5H | 22.40 | BOPA1_6184–200 | 4.82 | 0.44 | 0.37 | |
| QPH-7 | 7H | 114.27 | SCRI_RS_194841 | 4.18 | 0.59 | 0.50 | |
Table 6.
Environments in which the Sofiara/Victoriana DH population was evaluated
| Code | Number of plot replications | Environments | Plot size |
|---|---|---|---|
| E1 | 3 | WW, Gatersleben, Germany, April–August, 2012 | 0.4 m2 |
| E2 | 4 | DS, Gatersleben, Germany, April–August, 2012 | 0.4 m2 |
| E3 | 2 | RF, Wohlde, Germany, April–August, 2011 | 5.0 m2 |
| E4 | 2 | RF, Wohlde, Germany, April–August, 2012 | 5.0 m2 |
| E5 | 2 | RF, Walewice, Poland, April–August, 2011 | 5.0 m2 |
| E6 | 2 | RF, Walewice, Poland, April–August, 2012 | 5.5 m2 |
WW, DS, and RF well watered, drought stress and rainfed conditions, respectively
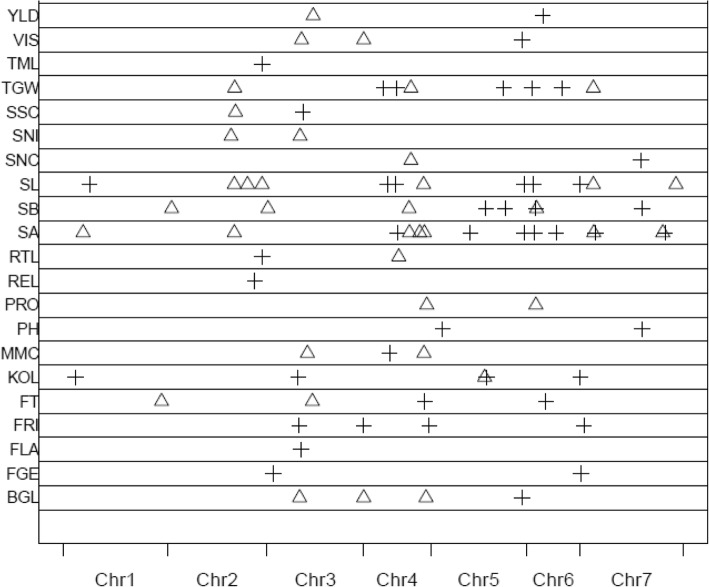
Fig. 1.
Location of main-effect QTL on chromosomes. Triangle and cross represent favorable QTL alleles contributed by Victoriana and Sofiara, respectively
QTL and environment interactions for yield-related traits
The QTL analysis identified a total of 50 QTL for five yield-related traits. Seed area (SA) yielded the highest number of QTL (14) with phenotypic variation in the range of 0.17 to 15.87%. QTL of highest LOD score for SA were mapped on chromosome 2H and 5H explaining 15.87 and 4% of phenotypic variation, respectively. One significant environmental interaction effect was found for QSA-1-2. Twelve main effect QTL were observed for SL. The phenotypic variation explained by a single QTL ranged from 0.29 to 18.13%. A major QTL with a LOD value of 41.3 was found on chromosome 2H near marker SCRI_RS_209622. None of the QTL showed significant QE interaction. Thousand grain weight (TGW) and seed breadth (SB) were also among the traits with the highest number of QTL, 9 and 8 respectively. Loci affecting TGW were found to be located on chromosomes 2H, 4H, 5H, 6H and 7H and phenotypic variance explained by individual QTL ranged from 0.90 to 5.16%. Two QTL, QTGW-3-1 and QTGW-3-2 displayed significant QE interactions. QTL for SB were located on each chromosome except 1H and explained from 0.83 to 8.58% of the phenotypic variation. QSB-3 had only significant QE interaction effect. Four significant QTL affecting YLD were detected on three chromosomes (3H, 4H and 6H) and collectively explained 1.27% of phenotypic variation (Additional file 3: Table S2). QYLD-3-2 and QYLD-4 showed QE interaction only whereas QYLD-3-1 and QYLD-6 showed main effects only. The alleles for higher yield of QYLD-3-1 and QYLD-6 were contributed by Victoriana and Sofiara, respectively.
QTL for malting quality-related traits
Overall, 41 significant QTL were identified for twelve malting quality traits, ranging from 1 to 11 QTL per trait. There were five QTL that explained more than 10% of phenotypic variance for such traits as wort viscosity (VIS), soluble nitrogen (SNI), Kolbach Index (KOL), friability (FRI) and wort β-glucan content (BGL). Among all traits, the highest numbers of main effect QTL were detected for KOL (5), which were mapped to four chromosomes. The highest number of QE interaction effects were detected for SNI (9), located to three chromosomes.
Fourteen QTL with phenotypic variation in the range of 0.62 to 27.31% were detected for SNI. They were mapped to four chromosomes (2H, 3H, 4H and 5H). There were two main-effect QTL and nine QE interaction effects which did not show significant main effect. The proportion of collective phenotypic variation explained by main-effects and QE interaction effects were 33.44 and 13.06% (Additional file 3: Table S2). This suggests that the environmental interaction effects were important for the expression of SNI. The phenotypic effects of QE demonstrated large variation across the environments both in the magnitude and direction. The largest main effect QTL (QSNI-3-1) was mapped on chromosome 3H at 60.96 cM near the marker BK_08. The superior allele was contributed by Sofiara and explained 27.31% of the phenotypic variation with a LOD of 25.79.
For KOL six QTL were identified, explaining 0.85 to 13.74% of the phenotypic variance. At five loci the Sofiara alleles had increasing effects on the trait and only at one (QKOL-5-1) caused the reduction. Only single QTL located on chromosome 3H (QKOL-3) with significant main effect showed QE interactions.
A total of 6 QTL located on four chromosomal regions 2H, 3H, 4H and 5H were associated with BGL. Collectively, these four main effect QTL and QE interactions explained 50.80 and 11.99% of the phenotypic variation, respectively. Alleles for the QBGL-3, QBGL-4-1 and QBGL-4-2 for increased BGL content were contributed by Victoriana and at QBGL-5 the Sofiara allele increased the trait value. The major QTL, explaining up to 37.42% of phenotypic variation in BGL was located on 3H chromosome at 60.06 cM.
Four loci controlling FRI were identified on three chromosomes. The phenotypic variance explained by each QTL ranged from 3.93 to 33.81%. In all cases the alleles responsible for increasing the trait value were contributed by Sofiara. Collectively, the four main effect QTL explained 50.78% of the phenotypic variation. The largest QTL (QFRI-3) with LOD value 36.80 was identified on chromosome 3H at position 59.08 cM and explained 33.81% of the phenotypic variance. QFRI-3 also showed QE effect that explained 12.41% of the phenotypic variation.
Two main effect QTL were identified for malt protein content (PRO). The QTL were located on chromosomes 4H and 6H and explained 5.1 and 4.5% of the phenotypic variation, respectively. The Victoriana allele at each locus increased the protein content.
Four QTL for VIS were identified on three chromosomes. The phenotypic variance explained by each QTL ranged from 4.57 to 13.82%. Collectively, three main effect QTL and one QE interaction effect explained 29.25 and 1.67% of the phenotypic variation, respectively. In most cases the Victoriana alleles were responsible for increased trait value. The largest QTL (QVIS-3) with LOD value 13.91 was located on chromosome 3H at position 63.66 cM and explained 13.82% of the phenotypic variance.
For FGE, a total of three QTL were detected that mapped to chromosomes 1H, 3H and 7H. QFGE-3 and QFGE-7, which were main effect QTL collectively explained 21.37% of the phenotypic variation and positives alleles at these loci were contributed by Sofiara. One QE interaction effect (QFGE-1) was detected on chromosome 1H and explained 0.99% of phenotypic variance.
Three main effect QTL were detected for MMC. One was located on chromosome 3H at position 74.64 cM and explained 2.66% of the phenotypic variation. The other two were mapped on chromosome 4H at position 47.94 and 111.57 cM and explained 1.9 and 3.67% of the phenotypic variance, respectively. At the locus QMMC-3 and QMMC-4-2 positive effects were associated with Victoriana alleles and at QMMC-4-1 the Sofiara allele increased the trait value.
Three main effect QTL affecting malting losses were identified. Two QTL for RTL were located on chromosome 2H and 4H and one QTL for respiration losses (REL) on chromosome 2H. The percentage of phenotypic variation explained by individual QTL varied between 0.57 and 2.1%. The positive alleles for the QREL-2 and QRTL-2 were contributed by Sofiara and for QRTL-4 by Victoriana.
Only one QTL was detected for limit of attenuation (FLA) with LOD value 5.26. It was located on chromosome 3H near the marker BK_08 and explained 6.85% of phenotypic variance. The Sofiara allele contributed to the increased trait value.
QTL for seed-quality traits
A total of five significant QTL located on four chromosomes (2H, 3H, 4H and 7H) were detected for three seed-quality traits (Tables 4, 6, Fig. 1). The percentage of phenotypic variation explained by these QTL varied from 1.69 to 2.77%. For SSC, two main effect QTL (QSSC-2 and QSSC-3) were detected. The first QTL was located at the position 126.5 cM on 2H, explaining of the 2.38% of phenotypic variation. The second one was found on 3H at the position 66.4 cM and explained the 1.69% of the phenotypic variance. No significant QE interaction was detected for SSC. Two main effect QTL influencing SNC were observed on the chromosome 4H and 7H at the position 87.4 and 112.7 cM, respectively. These QTL accounted for 2.17–2.77% of the phenotypic variation in the trait. QTL on chromosome 4H (QSNC-4-2) showed no significant main effect while QE interaction effect was detected, explaining 2.01% of the phenotypic variance.
QTL for plant height and flowering time
Three QTL for plant height (PH) were found. QPH-5 explained 0.37% of variance. QPH-7 was located on 7H at position 114.27 cM near the marker SCRI_RS_194841 explaining 0.5% of phenotypic variation. QE interaction effect (QPH-2) was identified on chromosome 2H and explained 1.09% of phenotypic variation.
Four main effect QTL for flowering time (FT) with an LOD > 3 that collectively accounted for 46.66% of phenotypic variance were mapped. Victoriana alleles increased the time to flowering at the QFT-1 and QFT-3 and reduced it at the QFT-4 and QFT-6.
Co-localizations of QTL
For several traits hot spots of co-locating QTL were observed on all chromosomes except for 1H. It is well known that such clustering is a consequence of either close linkage or pleiotropy. The highest level of clustering occurred in two regions: in the centromeric region of chromosome 3H and at the bottom of chromosome 4H (Additional file 4: Figure S2). The cluster on chromosome 3H mainly comprises malting quality QTL for BGL, FRI, KOL, VIS and SNI (R2 > 10%). The cluster on 4H contained QTL for BGL, FRI and PRO together with several QTL for seed morphological traits. QTL for BGL were found to be most often co-localized with loci affecting FRI and VIS. These traits were highly correlated to each other (Table 3). A higher content of one component (BGL/VIS) corresponded to a lower content of the other (FRI). This was corroborated by the observed allelic effects, with Victoriana alleles increasing BGL content and decreasing FRI. In two chromosomal regions (3H, 4H) QTL for BGL co-localized with loci affecting FRI and VIS. In two other regions on chromosome 4H and 5H BGL QTL coincided with QTL for FRI or VIS, respectively. QBGL-3 co-localized not only with QTL for VIS and FRI but also with loci affecting FLA, SSC and SNI. The region involved in BGL variation in the distal region of chromosome 4H also accounted for variation in the protein content. Here, the Victoriana allele caused an increase in BGL and PRO content (Table. 5). Protein and β-glucan metabolism are independent biological processes therefore the co-localisation of QTL in this case is likely due to linkage between different genes than due to pleiotropy. Clusters of QTL affecting yield-related traits such as TGW, SL, SB and SA were observed on chromosomes 2H, 4H, 5H, 6H and 7H. The QTL cluster on chromosome 2H was also associated with SSC and SNI, while the cluster on the chromosome 5H involved QTL influencing BGL and VIS. We also observed that two regions on chromosomes 3H and 6H harbor coinciding QTL for YLD and FT, reflecting the interaction between these traits. The positive alleles for both traits at the cluster on chromosome 3H and 4H were derived from Victoriana and Sofiara, respectively.
Table 5.
QTL x Environment interaction effect for measured traits detected in the Sofiara/Victoriana DH population in six environments
| Trait | QTL name | Chr. | Position (cM) | Nearest Marker | LOD | R2 (%)a | E1 | E2 | E3 | E4 | E5 | E6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fine grind extract | ||||||||||||
| QFGE-1 | 1H | 180.94 | SCRI_RS_196025 | 5.08 | 4.39 | −0.019 | 0.242 | NA | −0.148 | NA | NA | |
| Viscosity | ||||||||||||
| QVIS-3 | 3H | 60.06 | BK_08 | 5.17 | 4.57 | −0.008 | −0.006 | NA | −0.027 | NA | NA | |
| Soluble protein | ||||||||||||
| QSNI-3-2 | 3H | 60.06 | BK_08 | 12.85 | 9.09 | 82.810 | 69.669 | NA | 102.227 | NA | NA | |
| QSNI-3-3 | 3H | 72.37 | SCRI_RS_231801 | 7.52 | 0.73 | 17.337 | 52.080 | NA | −11.807 | NA | NA | |
| QSNI-3-4 | 3H | 73.89 | BOPA1_2616–2560 | 7.54 | 1.86 | −15.369 | −32.424 | NA | 20.908 | NA | NA | |
| QSNI-3-5 | 3H | 83.02 | SCRI_RS_225522 | 5.93 | 0.62 | 26.691 | 19.770 | NA | 2.040 | NA | NA | |
| QSNI-3-6 | 3H | 85.25 | SCRI_RS_156056 | 6.93 | 0.65 | −19.966 | −22.262 | NA | 7.317 | NA | NA | |
| QSNI-4 | 4H | 54.76 | BOPA1_ABC08788–1–1-329 | 4.76 | 1.90 | −7.178 | −7.735 | NA | 7.375 | NA | NA | |
| QSNI-5-1 | 5H | 93.27 | SCRI_RS_159611 | 5.41 | 3.60 | 43.891 | 17.155 | NA | 20.709 | NA | NA | |
| QSNI-5-2 | 5H | 97.05 | SCRI_RS_128664 | 6.04 | 2.03 | −44.480 | 9.496 | NA | 12.656 | NA | NA | |
| QSNI-5-3 | 5H | 98.54 | BOPA1_ABC14990–1–1-126 | 5.75 | 1.73 | −1.879 | −30.265 | NA | −22.775 | NA | NA | |
| Kolbach index | ||||||||||||
| QKOL-3 | 3H | 60.06 | BK_08 | 9.28 | 5.63 | 1.158 | 0.485 | NA | 2.630 | NA | NA | |
| Friability | ||||||||||||
| QFRI-3 | 3H | 59.08 | SCRI_RS_115045 | 19.48 | 11.36 | 2.622 | 9.418 | NA | 4.398 | NA | NA | |
| β-glucan | ||||||||||||
| QBGL-2 | 2H | 147.15 | SCRI_RS_156220 | 4.93 | 3.27 | −3.787 | 3.058 | NA | −57.402 | NA | NA | |
| QBGL-3 | 3H | 60.06 | BK_08 | 22.93 | 12.70 | −81.459 | −46.583 | NA | −190.728 | NA | NA | |
| Grain yield | ||||||||||||
| QYLD-3-2 | 3H | 71.61 | SCRI_RS_231801 | 6.39 | 0.37 | 1.521 | 0.302 | 0.607 | 0.126 | −1.331 | −0.322 | |
| QYLD-4 | 4H | 109.33 | BOPA1_1241–1649 | 7.73 | 0.47 | 0.696 | 0.210 | −1.231 | −0.932 | 1.241 | 0.691 | |
| TGW | ||||||||||||
| QTGW-3-1 | 3H | 60.06 | BK_08 | 8.17 | 1.64 | 0.559 | 0.228 | −0.147 | 0.359 | −0.562 | 0.157 | |
| QTGW-3-2 | 3H | 79.25 | BOPA1_10317–448 | 6.43 | 0.22 | 0.011 | 0.340 | −0.227 | 0.016 | −0.443 | −0.014 | |
| Seed area | ||||||||||||
| QSA-1-2 | 1H | 37.44 | SCRI_RS_128285 | 5.64 | 1.59 | 0.741 | 0.749 | 1.053 | 0.874 | NA | NA | |
| Seed breadth | ||||||||||||
| QSB-3 | 3H | 72.37 | SCRI_RS_231801 | 5.70 | 1.68 | −0.012 | −0.002 | −0.008 | 0.030 | NA | NA | |
| Seed nitrogen content | ||||||||||||
| QSNC-4-2 | 4H | 81.71 | SCRI_RS_238618 | 4.89 | 2.01 | −0.011 | −0.006 | NA | 0.030 | NA | NA | |
| Plant height | ||||||||||||
| QPH-2 | 2H | 124.23 | SCRI_RS_209622 | 9.00 | 1.09 | NA | NA | −0.380 | 1.060 | NA | NA | |
E1- well watered Gatersleben 2012, Germany; E2- drought stress Gatersleben 2012, Germany; E3- rainfed Wohlde 2011, Germany; E4- rainfed Wohlde 2012, Germany; E5- rainfed Walewice 2011, Poland; E6- rainfed Walewice 2012, Poland
aR2 collective percentage of phenotypic variation explained by QE interactions; NA - not analysed
Discussion
In the present study comprehensive phenotyping, genotyping and QTL analysis of a population derived from two elite barley lines, that differ for drought tolerance behavior, has led to the identification of numerous QTL for yield and yield components as well as for seed and malting quality. On all chromosomes except 1H the hot spots carrying QTL for three or more traits where identified.
Main effect QTL
Our analysis resulted in identification of 85 main effect QTL for 21 traits. Victoriana alleles increased the trait value in 36 instances, while in 49 cases they were responsible for decreasing of trait expression. For about half of the traits parental alleles of both increasing and decreasing effects were observed. This underscores on the one hand the complex inheritance and on the other hand the transgressive potential of the cross. However, for two traits (PRO and SNI) the increased trait performance was associated with Victoriana alleles; while only Sofiara alleles were responsible for increasing of the trait value for FGE, FRI, FLA, total malting losses (TML), REL and PH. We will focus on QTL which are supposed important to understand the genetic architecture of a given trait and could be useful for further improvement of malting quality.
TGW is important yield component, and many investigations to determine genomic regions controlling this trait have been performed. QTL for TGW were mapped across all seven chromosomes in various bi-parental populations and diverse panels for association mapping studies [23–26]. In our study, a total of nine QTL were detected for TGW. As expected, high positive correlations with r values in the range of 0.64 to 0.84 were observed between TGW and SL, SB and SA confirming that TGW is mainly a function of seed volume. Indeed, co-localization of each QTL for TGW and at least one of the parameters for seed volume were observed. With regard to potential improving of TGW, the two QTL localized on long arm of 5H and short arm of 7H were of special interest, because they explained about 5% of phenotypic variation. The alleles for increasing TGW at QTGW-5-1 and QTGW-7 were contributed by Sofiara and Victoriana, respectively. The QTGW-5-1 and QTGW-7 were found to correspond to QTL detected previously by Rode et al. [25] and Schnaithmann and Pillen [26], respectively; whereas QTGW-5-2, corresponding to the marker BOPA1_1394–1222 at the peak position 169.4 cM is a newly identified locus. QTGW-6-1 and QTGW-6-2, found in our study, were both derived from Sofiara. They coincided with previously reported QTL for TGW [26], suggesting that these genetic factor(s) influencing TGW could operate in a different genetic background. QTGW-6-2 could be considered of special interest, as it was previously reported that loci at this position were able to increase TGW under nitrogen deficiency [26].
A total of 34 main effect QTL were determined for seed morphological traits. A half (50%) of QTL was contributed by Sofiara alleles while a second half by Victoriana. QSL-1 was coincident with QSA-1-1 and found to correspond to the locus controlling seed length detected by Kalladan et al. [27]. In the same genomic region the gene encoding drought-responsive MYB transcription factor was mapped [28]. The largest QTL, explaining more than 5% of phenotypic variation, were mapped on chromosomes 2H, 4H and 5H. QSL-2-1, mapped to the distal part of chromosome 2H at the marker SCRI_RS_209622 co-localized with QSA-2 and QTGW-2. For all these QTL the alleles increasing the trait value were contributed by Victoriana. That could suggest the presence of the loci with pleiotropic effect in this genomic region. Of interest to note that no QTL for SL were found in this region on chromosome 2H using the set of barley introgression lines S42ILs, produced from the cross Scarlett/ISR42–8. QTL affecting SA was identified on chromosome 2H, however, it was associated with different genomic region [26]. Kalladan et al. [27] recently reported mapping QTL for SL and TGW on chromosome 2H in the DH population Brenda/HS584. Due to absence of common markers, it is difficult to conclude whether they match to the ones identified in our study. However, a previously identified QTL for SL mapped before [27] was assigned to bin 8 according to the BinMap2005 (http://wheat.pw.usda.gov/cgi-bin/cmap), while QSL-2-1 detected in our study belongs to the bin 11. Therefore it could be considered as a newly identified locus. In addition, we have identified several QTL affecting such morphological seed traits as SL, SB and SA on chromosome 4H. QTL for SL mapped on chromosome 4H were not identified in previous studies, either in the S42IL or Brenda/HS584 populations. Therefore they could be considered as newly identified loci. QTL for SB and SA on chromosome 4H have been identified in the QTL study of wild barley introgression lines S42ILs. Most likely two loci (QSB-4 and QSA-4-1) identified in our study are identical to the ones detected by Schnaithmann and Pillen [26]. However, the additional loci controlling SA (QSA-4-2, QSA-4-3 and QSA-4-4) that all were contributed by Victoriana and have been mapped to the distal part of chromosome 4H in our investigation, represent novel QTL. On chromosome 5H, five loci influencing variation of all three examined seeds morphological traits were mapped. Positive alleles were exclusively contributed by Sofiara. Two of them, QSA5–2 and QSL-5, were mapped to identical position. Recently a locus controlling variation in SL was mapped on chromosome 5H [27], however, its position differs from the one detected in our study.
Our analysis revealed 30 main effect malting quality QTL, explaining 0.6–37.4% of the phenotypic variation. Seventeen favorable QTL alleles were contributed by Sofiara and 13 by Victoriana. 56.7% of those QTL were mapped to chromosomes 3H (8 loci) and 4H (9 loci). Two clusters comprising the largest effect QTL associated with 10 different malting quality traits have been identified on these chromosomes. This finding indicates the importance of these chromosomal regions for developing superior malting phenotype.
Malt extract and fermentability
In previous mapping studies, QTL for malt extract have been mapped on all seven chromosomes [15, 21, 29, 30]. FGE in the present population was controlled by two main effect QTL positioned on the short arm of 3H and 7H with the favorable alleles of Sofiara, accounting for 7% of the phenotypic variation. Malt extract QTL on the chromosome 3H was reported by Mohammadi et al. [11]. However, that locus was assigned to the long arm of chromosome 3H (bin 16) and could not be matched with QFGE-3 detected in our study. Position of QFGE-7 did not coincide with the localization of the loci controlling malt extract on chromosome7H reported earlier either [1, 13]. However, QFGE-7 was situated in proximity (about 15 cM) to QTL detected by Wang et al. [15] in the TX9425/Naso Nijo population. Comparison based on marker bins (BinMap2005) revealed that QFGE-7 was assigned to the bin 1 while the one detected by Wang et al. [15] to the bin 2 on chromosome 7H. This finding suggests that the top of chromosome 7 contains genetic factors affecting malt extract which operate in different genetic backgrounds.
Regarding the malt wort fermentability, a single QTL for FLA has been localized on the short arm of chromosome 3H at the marker BK_08 (bin 6). Loci influencing fermentability were reported on chromosomes 4H and 6H [31]. In addition, Swanston et al. [12] mapped QTL for fermentability on 3H at the sdw1 locus (bin11) which was suggested to be determined by ß-amylase activity. QFLA-3 detected in our study was contributed by Sofiara and coincided with fermentability locus detected in the Triumph/Morex population by Elía et al. [10]. Our correlation analyses (Table 3) suggested that a number of traits like PRO, FGE, VIS, FRI and BGL influenced FLA, with VIS having the strongest effect. Moreover, QTL for FRI, BGL, SNI, and VIS co-localized to the same genomic region, providing evidence that extent of endosperm modification and wort viscosity are major factors influencing fermentability.
Viskosity, friability and ß-glucan content
Such traits as VIS, FRI and BGL showed a high correlation to each other. Co-localizations QTL for at least two of these traits were observed on 3H, 4H and 5H. The favorable alleles for FRI QTL were exclusively contributed by Sofiara that was opposed to the loci controlling VIS and BGL which in their majority have been provided by Victoriana. Observed co-localization of loci affecting FRI and VIS could be explained by strong dependence of both traits on the process of degradation of ß-glucans that represent a major component of endosperm cell walls in barley [32]. The BGL QTL with the largest effect has been mapped near the centromeric region of chromosome 3H at the marker BK_08. QBGL-3 matched with QTL for malt ß-glucan reported by Han et al. [33] and was located in close proximity to the region affecting ß-glucan in barley grains [34]. QBGL-4-1 was found to be located in the same region as a QTL for malt ß-glucan reported earlier [33] while the location of QBGL-4-2 and QBGL-5 did not match with positions of the loci controlling BGL detected either in linkage or association mapping [34, 35]. None of mapped BGL QTL coincide with the position of known genes Glb1 and Glb2 on chromosome 1H and 7H, encoding (1,3;1,4)-β-glucanases. This could be explained by lack of genetic differences between the parents at these particular loci. Two loci QVIS-3, QVIS-5, controlling more than 5% of variability in viscosity, were found to match the QTL reported before [15, 21, 31]. In addition we have identified QTL for VIS at the short arm of the chromosome 4H linked to the marker BOPA2_12_30540. Previously, a locus controlling viscosity positioned on chromosome 4H was reported in the VB9524/ND11231_12 population [9], however, its precise location could not be clarified due to lack of common markers in our studies.
A total of four QTL were detected for FRI which were all derived from Sofiara, the parent possessing the favorable alleles (higher trait value). Two of them, QFRI-4-1 and QFRI-7, could be matched to the ones reported before [31, 36, 37]. However, the FRI in our population was mainly influenced by the locus on chromosome 3H which explained 33.8% of genetic variance. QFRI-3 belongs to the bin 6 and was found to be located in close proximity to FRI locus mapped on 3H (bin7) in the population Nure/Tremois [21]. Another interesting novel QTL for increased FRI maps at a distal position of chromosome 4 near marker BOPA1_ABC08009–1–2-304 and accounted for 5% of the phenotypic variation. QFRI-4-2 coincided with QBGL-4-2 and QPRO-4, suggesting that terminal region of chromosome 4H contains closely linked or pleiotropic genes affecting a range of important malting quality traits.
Protein content, soluble nitrogen, Kohlbach index
Nine QTL were found to be associated with PRO, SNI and KOL, traits that are used for characterization of nitrogen fractions of malt. A major QTL for SNI (QSNI-3-1) was mapped on chromosome 3H near the marker BK_08 in the same chromosomal region where locus controlling variation of SNI in Harrington/Mikamo Golden population was reported by Zhou et al. [38]. A novel QTL for SNI was detected on 2H near the marker BOPA1_11591–265. This QTL explained 1% of the phenotypic variation and was found to be co-located with loci affecting TGW, SA and SL. Genomic regions affecting malt protein content are known on all seven chromosomes [15, 38, 39]. However, two QTL QPRO-4 and QPRO-6 detected in the present study could not be matched with any previous findings. A total of five QTL were detected for KOL, and at four loci the increased KOL was contributed by Sofiara. Locations of QKOL-5-1, QKOL-5-2 and QKOL-7 coincide with position for the loci reported previously [15, 16]. The major QTL, QKOL-3, was mapped on 3H with closest marker SCRI_RS_115045 (bin 6). Locus controlling variation in KOL on chromosome 3H was reported in population 88Ab536/Morex [29]. This QTL was linked with the marker 1_1328 associated with bin 14 and therefore could not be matched with the one identified in current study. In addition, a minor QTL for KOL was mapped on 1H near the marker SCRI_RS_205669 (bin 5). In a similar region, a QTL affecting KOL was reported in wild barley introgression lines S42ILs [14]. In particular, line S24IL-102 bearing an introgression of about 85 cM in size showed a strong reduction of KOL up to 11.5%. However, it has to be noted that the large size of introgressed region covering nine bins obviates a precise comparison of QTL.
Seed starch and seed nitrogen content
QSNC-4-1 and QSNC-7 were found to match the QTL for grain protein content or N% reported before in the Vlamingh/Buloke and Brenda/HS584 populations, respectively [27, 30]. Locus controlling seed starch content on chromosome 3H (QSSC-3) was co-localized with QTL for a range of important malting quality traits like KOL, FRI, SNI and VIS. It coincides with QTL controlling SSC in the same region reported before by Kalladan et al. [27]. That suggests that the short arm of chromosome 3H contains genetic factors influencing a number of malting quality characters which stay active in different genetic backgrounds and are of primary importance for breeders. In addition, we have mapped QTL for SSC on 2H chromosome contributed by Victoriana. It was linked to the marker BOPA2_12_10735 (bin 11) and could not be matched to any QTL detected in any of above mentioned studies. The same region harbors a locus for SNI at which the Victoriana allele causes an increase in SNI. High SNI content would be considered a disadvantage, because it finally results in decreasing of malt extract. Moreover, QSSC-2 co-localized with QTL affecting yield-related parameters such as SA, SL and TGW. For all three QTL alleles increasing trait value were contributed exclusively by Victoriana.
QE interaction effects
From the practical point of view it is important to know how the identified loci performed in different environmental conditions. Our analysis revealed that 5 out of 86 main effect QTL showed QE interactions, in particularly QBGL-3, QVIS-3, QFRI-3, QKOL-3 and QSB-3. Phenotypic variation explained by QE interactions amounted to 12.7, 4.6, 11.4, 5.6 and 1.7%, respectively (Table 5). Loci for malting quality traits showed differences only in magnitude, while for QSB-3 differences were observed in both the magnitudes and directions of effects. QBGL-3 showed the largest variation among environments, with QE values of − 81.5, − 46.6 and − 190.7 in E1, E2 and E4, respectively. This suggests that expression of QBGL-3 has similar effect under all tested environments. The QE interaction effects of these QTL were substantially smaller than the corresponding main effects (Table 4). This suggests that the genetic effects of these loci were largely controlled by their main effect and thus they could be considered as primary targets in future breeding programs. In addition, QE effects were observed for 18 QTL with no significant main effect. The contribution of phenotype variations explained by QE interactions for these QTL ranged from 0.2 to 9.1%. The highest number of QE interactions (8 loci) was detected for the SNI, suggesting that this trait was highly sensitive to environment. The QSNI-3-2 with the largest effect, accounting for 9.1% of the phenotypic variance, demonstrated changes in magnitude of effect; while QSNI-5-2 had differential expression between E1 and E2, demonstrating negative and positive QE effects, respectively.
Candidate genes within the 3H QTL cluster
A number of coincidental QTL affecting two seed quality traits and five malt characters were identified on the short arm of chromosome 3H. These loci explained a relatively large proportion of phenotypic variation, in particular QSNI-3-1, QFRI-3 and QBGL-3 accounted for 27, 34 and 37% of phenotypic variance, respectively. Defined QTL cluster occupied relatively small chromosomal region and comprised at once five loci controlling variation of important malting quality characteristics. That makes it valuable for using in future breeding programs through marker assisted selection. In order to reveal possible candidate genes underlying identified QTL, the marker BK_08 associated with majority of detected loci was located on the POPSEQ genetic map of Morex × Barke [40]. The morex_contig_1580005, bearing the sequence of BK_08, was mapped at position 45.4 cM. The list of all annotated genes in the region between 40.4–50.4 cM was downloaded from publicly available BARLEX database [41]. Even a rough annotation-based assay of the genes located in this region (Additional file 5: Table S3) revealed a number of well-known malting quality related genes. These are the genes involved in starch, protein, lipid and cell wall metabolism. For instance, two genes MLOC_68194.1 and MLOC_36529.1 encoding aspartyl and serine proteases were found to be present in selected region. These two proteinase types are known to be involved in storage protein solubilisation, as well as in the regulation of the starch degrading enzyme, ß-amylase [42, 43]. In addition, four genes (AK375791, MLOC_32229.1, AK355146, AK375970) coding for heat shock proteins (HSP) were present in the region of interest. HSP might play a crucial role in protection of enzymes from degradation during the malting process especially at the stage of kilning. Association of HSP genes with such malting quality traits as malt extract, ß-amylase activity, friability, free amino acids and Kolbach Index have been previously reported [44, 45]. Interestingly, the target region also contained the gene MLOC_6671.1 encoding serine carboxypeptidase (Ser-CP). Ser-CPs play an important role in hydrolysis of storage proteins in germinating barley seeds. At least 6 members in barley were previously identified, all of which were found to be active during germination [46]. Transcript profiling of the gene Cxp1 (MLOC_55542.1), mapped to long arm of chromosomes 3H, in the Steptoe/Morex population revealed that expression QTL for respective gene co-localized with QTL affecting diastatic power [47]. Moreover, the region contains 18 genes encoding different types of transcription factors (TFs). They also could be considered as probable candidate genes, as these TFs might control expression of structural or regulatory genes playing key role in malting process. The fact that the members of the MYB gene family were found to be present in the list of potential candidates is particularly interesting. It has been previously reported that a MYB protein (GAMYB) expressed in cereal aleurone cells triggered transcriptional activation of α-amylase gene through binding to the TAACAAA box in its promoter [48]. The presence of elements of gibberellin responsive complex (GARC) have been reported within promoter regions of some other hydrolytic enzymes such as (1–3, 1–4)-β-glucanase and cathepsin B-like protease [49, 50]. Moreover, Gubler et al. [51] in a series of transient expression experiments showed that GAMYB protein indeed was able to promote expression of reporter gene fused with promoters of genes encoding EII(1–3, 1–4)-β-glucanase and cathepsin B-like protease.
Conclusions
In this study we have investigated agronomic, malting and seed quality characteristics of two high yielding elite breeding lines that differ in response to drought stress. Our QTL mapping revealed several novel loci controlling malting quality which could be exploited in breeding programs. In this context, the complex QTL region on chromosome 3H seems of particular interest, as it harbors several large effect QTL, which need to be validated in different genetic backgrounds. Moreover, identification and validation of the candidate genes will help to select for intralocus recombinants, since recombination of favorable alleles is required for further optimization of the genetic make-up of this locus.
Methods
Plant materials and field experiments
Our study is based on a population of 100 unique DH lines developed from the cross between the elite spring barley lines Sofiara and Victoriana (KWS, Germany). The parental lines were selected because they largely differ with respect to YLD, TGW, SNI and, BGL, and their response to drought stress. The DH lines and their parents were evaluated in replicated field trials at three locations in 2 years (Table 6; Additional file 6: Figure S3). The experimental design followed a lattice (Wohlde, Walewice) or a randomized complete block design (Gatersleben). The plot sizes ranged from 0.4 m2 to 5.5 m2. In Gatersleben, terminal drought stress was applied 1 week after anthesis using a rain-out shelter in addition to a control variant with absence of drought stress. Soil moisture was monitored in the drought stress trials twice a week in 10, 20, 30 and 40 cm depths by using 82 evenly distributed soil moisture sensors (SM300, PR2/4; Delta T devices Ltd., England).
Yield-related traits
Plant height was measured before harvest from the base to the top of the plant and recorded in centimeters. SL, SB, SA and TGW were assessed using a sample of 400–500 grains and a digital seed analyser (Marvin; GTA Sensorik GmbH, Germany).
Starch, carbon and nitrogen content
Soluble carbohydrates were extracted three times with 80% ethanol at 60 °C from 0.4 g of homogenized grains tissue and ethanol-insoluble pellet was used for the quantification of starch content. Pellet was solubilized in 1 N KOH for 1 h at 95 °C. After neutralization with 5 N HCl, starch hydrolysis was achieved by incubation with amyloglucosidase (6u/ml) and a-amylase (10 u/ml) at 37 °C for 90 min. Glucose was quantified using enzymatic assay kit (R-Biopharm AG, Darmstadt, Germany). Carbon and nitrogen content were determined in ground and oven dried samples of grains using an elemental analyzer (vario EL III; Elementar analysensysteme GmbH Hanau, Germany).
Malting quality analysis
Grain samples (80 g per genotype) were malted in the malting facilities of the Research Institute for Raw Materials at the VLB Berlin e.V.. Micromalting conditions were the following: 5 h wet steep, 19 h air-rest, 2 h wet steep, 22 h air-rest, spray steeping until reaching a final moisture content of 45%, then germination for 96 h at 14.5 °C and 95 to 98% relative air humidity. Germinated samples were kilned with the following regime: 16 h at 50 °C, 1 h at 60 °C, 1 h at 70 °C and 5 h at 80 °C.
To assess malt quality following twelve parameters such as malt moisture content (MMC), FGE, VIS, PRO, SNI, KOL, FLA, FRI, BGL, TML, REL and RTL were determined according to the methods of MEBAK (Mitteleuropäische Brautechnische Analysenkommission). A brief outline of some malting traits is given in Potokina et al. [44].
Genotyping and construction of the genetic map
DNA samples from leaf tissue of the DH lines were extracted using the DNeasy Plant DNA miniprep kit (Qiagen, Germany), and scored with barley 9 K SNP iSelect Illumina array. Genetic map was constructed using the software package JoinMap 4.0 [52]. Linkage groups were established using minimum LOD values of 4. Monte Carlo maximum likelihood (ML) mapping algorithm was applied to determine the orders of markers within each linkage group. Recombination frequencies were converted to centimorgans (cM) using Haldane’s mapping function. The order of markers was compared with a published map [40].
Phenotypic data analysis and QTL mapping
We performed a two-stage analysis of phenotypic data [53]. The BLUEs of all genotypes in each environment were combined and a linear mixed model across environments was fitted with random genotype, environment, and residual effects. All linear mixed models were implemented using ASReml-R [54]. For QTL mapping we applied a composite mapping (CM) approach [55, 56]. For more details see Additional file 7: Data S1.
Additional files
Figure S1. Frequency distribution of the BLUEs of the 100 DH for the measured traits. (PDF 257 kb)
Table S1. Genetic positions of markers. (XLSX 48 kb)
Table S2. The collective percent of phenotypic variation explained by main-effect QTL and QE interactions for the trait. (XLSX 12 kb)
Figure S2. Locations of main-effect QTL on 3H (a) and 4H (b) chromosomes. (PDF 183 kb)
Table S3. The list of candidate genes associated with largest QTL cluster on chromosome 3H. (XLSX 34 kb)
Figure S3. Climate conditions in rainfed field located in Wohlde (a-b), Walewice (c-d) and Gatersleben (e). (PDF 409 kb)
Data S1. Experimental procedures related to phenotypic data analysis, and QTL mapping [57–61]. (DOCX 14 kb)
Acknowledgments
We are grateful to Mandy Püffeld and Jana Lorenz for excellent technical assistance. We thank two anonymous reviewers for their valuable comments on this manuscript.
Funding
This research was funded by the Bundesministerium für Bildung and Forschung (BMBF). The funding bodies had no role in the experiment design, data analysis, preparation of the manuscript and decision to publish.
Availability of data and materials
All data generated during the current study are included in this published article.
Abbreviations
- BGL
β-glucan
- FGE
Fine grind extract
- FLA
Limit of attenuation
- FRI
Friability
- FT
Flowering time
- KOL
Kolbach index
- LOD
Logarithm of odds
- MAS
Marker-assisted selection
- MMC
Moisture content
- PH
Plant height
- PRO
Protein content
- QTL
Quantitative trait locus
- REL
Respiration losses
- RTL
Rootlet losses
- SA
Seed area
- SB
Seed breadth
- SCC
Seed carbon content
- SL
Seed length
- SNC
Seed nitrogen content
- SNI
Soluble nitrogen
- SNP
Single nucleotide polymorphism
- SSC
Seed starch content
- TGW
Thousand-grain weight
- TML
Total malting losses
- VIS
Viscosity
- YLD
Grain yield
Authors’ contributions
AK performed phenotypic measurements, genotypic data analysis, statistical analysis and drafted the manuscript; YJ performed statistical analysis and gave a significant support during the phenotypic and QTL analyses; CS, KS performed phenotypic measurements and revised manuscript; SK participated in design and carried out the collection of data; JCR critically revised study and contributed to the interpretation of the results; VK and AG designed, coordinated the study and revised manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The plant materials come from germplasm resources of the KWS LOCHOW GmbH. All experimental trials were conducted according to the national legislation.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1323-4) contains supplementary material, which is available to authorized users.
Contributor Information
Andriy Kochevenko, Email: andkochevenko@gmail.com.
Yong Jiang, Email: jiang@ipk-gatersleben.de.
Christiane Seiler, Email: seiler@ipk-gatersleben.de.
Korana Surdonja, Email: surdonja@ipk-gatersleben.de.
Sonja Kollers, Email: sonja.kollers@kws.com.
Jochen Christoph Reif, Email: reif@ipk-gatersleben.de.
Viktor Korzun, Email: viktor.korzun@kws.com.
Andreas Graner, Phone: +49-394-825-219, Email: graner@ipk-gatersleben.de.
References
- 1.Newton AC, Flavell AJ, George TS, Leat P, Mullholland B, Ramsay L, Revoredo-Giha C, Russell J, Steffenson B, Swanston JS, Thomas WTB, Waugh R, White PJ Bingham IJ. Crops that feed the world 4 barley: a resilient crop? Strengths and weaknesses in the context of food security. Food Secur. 2011;3:141–178. doi: 10.1007/s12571-011-0126-3. [DOI] [Google Scholar]
- 2.Newman RK, Newman CW. Barley for food and health science, technology, and products. Hoboken: Willey; 2008. [Google Scholar]
- 3.Fox GP, Panozzo JF, Li CD, Lance RCM, Inkerman PA, Henry RJ. Molecular basis of barley quality. Aust J Agric Res. 2003;54:1081–1101. doi: 10.1071/AR02237. [DOI] [Google Scholar]
- 4.Molina-Cano JL, Francesch M, Pérez-Vendrell AM, Ramo T, Voltas J, Brufau J. Genetic and environmental variation in malting and feed barley quality. J Cereal Sci. 1997;25:37–47. doi: 10.1006/jcrs.1996.0067. [DOI] [Google Scholar]
- 5.Hayes PM, Jones BL. Malting quality from a QTL perspective. In 8th Int. Barley Genet. Symp., Adelaide Convention Centre, Adelaide, South Australia 22–27 Oct. 2000;8:99–105.
- 6.Han F, Romagosa I, Ullrich SE, Jones BL, Hayes PM, Wesenberg DM. Molecular marker-assisted selection for malting quality traits in barley. Mol Breed. 1997;3:427–437. doi: 10.1023/A:1009608312385. [DOI] [Google Scholar]
- 7.Cai SG, Yu G, Chen XH, Huang YC, Jiang XG, Zhang GP, Jin XL. Grain protein content variation and its association analysis in barley. BMC Plant Biol. 2013;13:35–45. doi: 10.1186/1471-2229-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutiérrez L, Cuesta-Marcos A, Castro AJ, von Zitzewitz J, Schmitt M, Hayes PM. Association mapping of malting quality quantitative trait loci in winter barley: positive signals from small germplasm arrays. Plant Genom. 2011;4:256–272. doi: 10.3835/plantgenome2011.07.0020. [DOI] [Google Scholar]
- 9.Emebiri LC, Moody DB, Panozzo JF, Read BJ. Mapping of QTL for malting quality attributes in barley based on a cross of parents with low grain protein concentration. Field Crops Res. 2004;87:195–205. doi: 10.1016/j.fcr.2003.11.002. [DOI] [Google Scholar]
- 10.Elía M, Swanston JS, Moralejo M, Casas A, Pérez-Vendrell AM, Ciudad FJ, Thomas WTB, Smith PL, Ullrich SE, Molina-Cano JL. A model of the genetic differences in malting quality between European and north American barley cultivars based on a QTL study of the cross triumph × Morex. Plant Breed. 2010;129:280–290. doi: 10.1111/j.1439-0523.2009.01694.x. [DOI] [Google Scholar]
- 11.Mohammadi M, Blake TK, Budde AD, Chao S, Hayes PM, Horsley RD, Obert DE, Ullrich SE, Smith KP. A genome-wide association study of malting quality across eight US barley breeding programs. Theor Appl Genet. 2015;128:705–721. doi: 10.1007/s00122-015-2465-5. [DOI] [PubMed] [Google Scholar]
- 12.Swanston JS, Thomas WTB, Powell W, Young GR, Lawrence PE, Ramsey L, Waugh R. Using molecular markers to determine barleys most suitable for malt distilling. Mol Breed. 1999;5:103–109. doi: 10.1023/A:1009606705925. [DOI] [Google Scholar]
- 13.Hayes PM, Lui BH, Knapp SJ, Chen F, Jones B, Blake T, Franckowiak J, Rasmusson D, Sorrells M, Ullrich SE, Wesenberg D, Kleinhofs A. Quantitative trait locus effects and environmental interaction in a sample of north American barley germplasm. Theor Appl Genet. 1993;87:392–401. doi: 10.1007/BF01184929. [DOI] [PubMed] [Google Scholar]
- 14.Schmalenbach I, Pillen K. Detection and verification of malting quality QTLs using wild barley introgression lines. Theor Appl Genet. 2009;118:1411–1427. doi: 10.1007/s00122-009-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Yang J, Zhang Q, Zhu J, Jia Q, Hua W, Shang Y, Li C, Zhou M. Mapping a major QTL for malt extract of barley from a cross between TX9425 x Naso Nijo. Theor Appl Genet. 2015;128:943–952. doi: 10.1007/s00122-015-2481-5. [DOI] [PubMed] [Google Scholar]
- 16.Oziel A, Hayes PM, Chen FQ, Jones B. Application of quantitative trait locus mapping to the development of winter-habit malting barley. Plant Breed. 1996;115:43–51. doi: 10.1111/j.1439-0523.1996.tb00869.x. [DOI] [Google Scholar]
- 17.Mather DE, Tinker NA, Laberge DE, Edney M, Jones BL, Rossnagel BG, Legge WG, Briggs KK, Irvine RB, Falk DE. Regions of the genome that affect grain and malt quality in a north American two-row barley cross. Crop Sci. 1997;37:544–554. doi: 10.2135/cropsci1997.0011183X003700020039x. [DOI] [Google Scholar]
- 18.Islamovic E, Obert DE, Budde AD, Schmitt M, Brunickii R, Kilian A, Chao S, Lazo GR, Marshall JM, Jellen EN, Maughan PJ, Hu G, Klos KE, Brown RH, Jackson EW. Quantitative trait loci of barley malting quality trait components in the stellar/01Ab8219 mapping population. Mol Breed. 2014;34:59–73. doi: 10.1007/s11032-014-0017-3. [DOI] [Google Scholar]
- 19.Igartua E, Edney M, Rossnagel BG, Spaner D, Legge WG, Scoles GJ, Eckstein PE, Penner GA, Tinker NA, Briggs KG, Falk DE, Mather DE. Marker-based selection of QTL affecting grain and malt quality in two-row barley. Crop Sci. 2000;40:1426–1433. doi: 10.2135/cropsci2000.4051426x. [DOI] [Google Scholar]
- 20.Ayoub M, Armstrong E, Bridger G, Fortin MG, Mather DE. Marker-based selection in barley for a QTL region affecting alpha amylase activity of malt. Crop Sci. 2003;43:556–561. doi: 10.2135/cropsci2003.0556. [DOI] [Google Scholar]
- 21.Laidò G, Barabaschi D, Tondelli A, Gianinetti A, Stanca AM, Li Destri Nicosia O, NDi F, Francia E, Pecchioni N. QTL alleles from a winter feed type can improve malting quality in barley. Plant Breed. 2009;128:598–605. doi: 10.1111/j.1439-0523.2009.01636.x. [DOI] [Google Scholar]
- 22.Seiler C, Harshavardhan VT, Reddy PS, Hensel G, Kumlehn J, Eschen-Lippold L, Rajesh K, Korzun V, Wobus U, Lee J, Selvaraj G, Sreenivasulu N. Abscisic acid flux alterations result in differential ABA signalling responses and impact assimilation efficiency in barley under terminal drought stress. Plant Physiol. 2014;164:1677–1696. doi: 10.1104/pp.113.229062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JZ, Huang XQ, Heinrichs F, Ganal MW, Röder MS. Analysis of QTLs for yield components, agronomic traits, and disease resistance in an advanced backcross population of spring barley. Genome. 2006;49:454–466. doi: 10.1139/g05-128. [DOI] [PubMed] [Google Scholar]
- 24.Schmalenbach I, Leon J, Pillen K. Identification and verification of QTLs for agronomic traits using wild barley introgression lines. Theor Appl Genet. 2009;118:483–497. doi: 10.1007/s00122-008-0915-z. [DOI] [PubMed] [Google Scholar]
- 25.Rode J, Ahlemeyer J, Friedt W, Ordon F. Identification of marker trait associations in the German winter barley breeding gene pool. (Hordeum vulgare L) Mol Breed. 2012;30:831–843. doi: 10.1007/s11032-011-9667-6. [DOI] [Google Scholar]
- 26.Schnaithmann F, Pillen K. Detection of exotic QTLs controlling nitrogen stress tolerance among wild barley introgression lines. Euphytica. 2013;189:67–88. doi: 10.1007/s10681-012-0711-3. [DOI] [Google Scholar]
- 27.Kalladan R, Worch S, Rolletschek H, Harshavardhan VT, Kuntze L, Seiler C, et al. Identification of quantitative trait loci contributing to yield and seed quality parameters under terminal drought in barley advanced backcross lines. Mol Breed. 2013;32(1):71–90. doi: 10.1007/s11032-013-9853-9. [DOI] [Google Scholar]
- 28.Worch S, Rajesh K, Harshavardhan VT, Pietsch C, Korzun V, Kuntze L, Börner A, Wobus U, Roder MS, Sreenivasulu N. Haplotyping, linkage mapping and expression analysis of barley genes regulated by terminal drought stress influencing seed quality. BMC Plant Biol. 2011;11(1) [DOI] [PMC free article] [PubMed]
- 29.Muñoz-Amatriaín M, Cistué L, Xiong Y, Bilgic H, Budde AD, Schmitt MR, Smith KP, Hayes PM, Muehlbauer GJ. Structural and functional characterization of a winter malting barley. Theor Appl Genet. 2010;120:971–984. doi: 10.1007/s00122-009-1225-9. [DOI] [PubMed] [Google Scholar]
- 30.Walker CK, Ford R, Munoz-Amatrianın M, Panozzo JF. The detection of QTLs in barley associated with endosperm hardiness, grain density, grain size and malting quality using rapid phenotyping tools. Theor Appl Genet. 2013;126:2533–2551. doi: 10.1007/s00122-013-2153-2. [DOI] [PubMed] [Google Scholar]
- 31.von Korff M, Wang H, Leon J, Pillen K. AB-QTL analysis in spring barley: III identification of exotic alleles for the improvement of malting quality in spring barley (H vulgare ssp spontaneum) Mol Breed. 2008;21:81–93. doi: 10.1007/s11032-007-9110-1. [DOI] [Google Scholar]
- 32.Fincher GB. Molecular and cellular biology associated with endosperm mobilization in germinating cereal-grains. Annu Rev Plant Physiol. 1989;40:305–346. doi: 10.1146/annurev.pp.40.060189.001513. [DOI] [Google Scholar]
- 33.Han F, Ullrich SE, Chirat S, Menteur S, Jestin L, Sarrafi A, Hayes PM, Jones BL, Blake TK, Wesenberg DM, Kleinhofs A, Kilian A. Mapping of β-glucan content and β-glucanase activity loci in barley grain and malt. Theor Appl Genet. 1995;91:921–927. doi: 10.1007/BF00223901. [DOI] [PubMed] [Google Scholar]
- 34.Mohammadi M, Endelman JB, Nair S, Chao SM, Jones SS, Muehlbauer GJ, Ullrich SE, Baik BK, Wise ML, Smith KP. Association mapping of grain hardness, polyphenol oxidase, total phenolics, amylose content, and beta-glucan in US barley breeding germplasm. Mol Breed. 2014;34:1229–1243. doi: 10.1007/s11032-014-0112-5. [DOI] [Google Scholar]
- 35.Islamovic E, Obert D, Oliver R, Harrison S, Ibrahim A, Marshall JM, Stuthman DD. Genetic dissection of grain beta-glucan and amylose content in barley (Hordeum vulgare L) Mol Breed. 2013;31:15–25. doi: 10.1007/s11032-012-9764-1. [DOI] [Google Scholar]
- 36.Edney MJ, Mather DE. Quantitative trait loci affecting germination traits and malt friability in a two-rowed by six-rowed barley cross. J Cereal Sci. 2004;39:283–290. doi: 10.1016/j.jcs.2003.10.008. [DOI] [Google Scholar]
- 37.Li JZ, Huang XQ, Heinrichs F, Ganal MW, Röder MS. Analysis of QTLs for yield, yield components, and malting quality in a BC3-DH population of spring barley. Theor Appl Genet. 2005;110:356–363. doi: 10.1007/s00122-004-1847-x. [DOI] [PubMed] [Google Scholar]
- 38.Zhou T, Takashi I, Ryouichi K, Naohiko H, Makoto K, Takehiro H, Kazuhiro S. Malting quality quantitative trait loci on a high-density map of Mikamo golden x Harrington cross in barley (Hordeum vulgare L) Mol Breed. 2012;30:103–112. doi: 10.1007/s11032-011-9602-x. [DOI] [Google Scholar]
- 39.Matthies IE, Malosetti M, Röder MS, van Eeuwijk F. Genome-wide association mapping for kernel and malting quality traits using historical european barley records. PLoS One. 2014;9(11):e110046. doi: 10.1371/journal.pone.0110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Muñoz-Amatriaín M, Close TJ, Wise RP, Schulman AH, Himmelbach A, Mayer KFX, Scholz U, Poland JA, Stein N, Waugh R. Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ) Plant J. 2013;76:718–727. doi: 10.1111/tpj.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colmsee C, Beier S, Himmelbach A, Schmutzer T, Stein N, Scholz U, Mascher M. BARLEX - the barley draft genome explorer. Mol Plant. 2015;8:964–966. doi: 10.1016/j.molp.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Jones BL, Budde AD. Various malt endoproteinase classes affect wort soluble protein levels. J Cereal Sci. 2005;41:95–106. doi: 10.1016/j.jcs.2004.09.007. [DOI] [Google Scholar]
- 43.Schmitt MR, Marinac L. Beta-amylase degradation by serine endoproteinases from green barley malt. J Cereal Sci. 2008;47:480–488. doi: 10.1016/j.jcs.2007.06.002. [DOI] [Google Scholar]
- 44.Potokina E, Caspers M, Prasad M, Kota R, Zhang H, Sreenivasulu N, Wang M, Graner A. Functional association between malting quality trait components and cDNA array based expression patterns in barley (Hordeum vulgare L) Mol Breed. 2004;14:153–170. doi: 10.1023/B:MOLB.0000038004.21342.3c. [DOI] [Google Scholar]
- 45.Lapitan NLV, Hess A, Cooper B, Botha AM, Badillo D, Iyer H, Menert J, Close TJ, Wright L, Hanning G, Tahir M, Lawrence C. Differentially expressed genes during malting and correlation with malting quality phenotypes in barley (Hordeum vulgare L) Theor Appl Genet. 2009;118:937–952. doi: 10.1007/s00122-008-0951-8. [DOI] [PubMed] [Google Scholar]
- 46.Dal Degan F, Rocher A, Cameron-Mills V, von Wettstein D. The expression of serine carboxypeptidases during maturation and germination of the barley grain. Proc Natl Acad Sci U S A. 1994;91:8209–8213. doi: 10.1073/pnas.91.17.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potokina E, Prasad M, Malysheva L, Röder MS, Graner A. Expression genetics and haplotype analysis reveal cis regulation of serine carboxypeptidase I (Cxp1), a candidate gene for malting quality in barley (Hordeum vulgare L) Funct Integr Genomics. 2006;6:25–35. doi: 10.1007/s10142-005-0008-x. [DOI] [PubMed] [Google Scholar]
- 48.Gubler F, Kalla R, Roberts JK, Jacobsen JV. Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for MYB transactivation of a high-pI α-amylase gene promoter. Plant Cell. 1995;7:1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saleski N, Fincher GB. Developmental regulation of (1–3, 1–4) -b-glucanase gene expression in barley. Plant Physiol. 1992;99:1226–1231. doi: 10.1104/pp.99.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cejudo FJ, Ghose TK, Stabel P, Baulcombe DC. Analysis of the gibberellin-responsive promoter of a cathepsin B-like gene from wheat. Plant Mol Biol. 1992;20:849–856. doi: 10.1007/BF00027156. [DOI] [PubMed] [Google Scholar]
- 51.Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 1999;17:1–9. doi: 10.1046/j.1365-313X.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- 52.Van Ooijen JW. JoinMap® 4, software for the calculation of genetic linkage maps in experimental populations. Wageningen: Kyazma BV; 2006. [Google Scholar]
- 53.Möhring J, Piepho HP. Comparison of weighting in two-stage analysis of plant breeding trials. Crop Sci. 2009;49:1977–1988. doi: 10.2135/cropsci2009.02.0083. [DOI] [Google Scholar]
- 54.Gilmour AR, Gogel B, Cullis BR, Thompson R. ASReml user guide release 3.0. Hemel Hempstead: VSN International Ltd; 2009. [Google Scholar]
- 55.Jansen RC. Controlling the type I and type II errors in mapping quantitative trait loci. Genetics. 1994;138:871–881. doi: 10.1093/genetics/138.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jansen RC, Van Ooijen JW, Stam P, Liste C, Dean C. Genotype-by-environment interaction in genetic mapping of multiple quantitative trait loci. Theor Appl Genet. 1995;91:33–37. doi: 10.1007/BF00220855. [DOI] [PubMed] [Google Scholar]
- 58.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. doi: 10.1214/aos/1176344136. [DOI] [Google Scholar]
- 59.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.SAS Institute. SAS/STAT: user’s guide, version 93 SAS Institute Inc Cary. NC. 2009;
- 61.R Core Team (2014) R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria. URL http://wwwR-projectorg.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Frequency distribution of the BLUEs of the 100 DH for the measured traits. (PDF 257 kb)
Table S1. Genetic positions of markers. (XLSX 48 kb)
Table S2. The collective percent of phenotypic variation explained by main-effect QTL and QE interactions for the trait. (XLSX 12 kb)
Figure S2. Locations of main-effect QTL on 3H (a) and 4H (b) chromosomes. (PDF 183 kb)
Table S3. The list of candidate genes associated with largest QTL cluster on chromosome 3H. (XLSX 34 kb)
Figure S3. Climate conditions in rainfed field located in Wohlde (a-b), Walewice (c-d) and Gatersleben (e). (PDF 409 kb)
Data S1. Experimental procedures related to phenotypic data analysis, and QTL mapping [57–61]. (DOCX 14 kb)
Data Availability Statement
All data generated during the current study are included in this published article.