Abstract
Regulation of transcription is fundamental to the control of cellular gene expression and function. Although recent studies have revealed a role for the oncoprotein MYC in amplifying global transcription, little is known as to how the global transcription is suppressed. Here we report that SUMO and MYC mediate opposite effects upon global transcription by controlling the level of CDK9 sumoylation. On one hand, SUMO suppresses global transcription via sumoylation of CDK9, the catalytic subunit of P-TEFb kinase essential for productive transcriptional elongation. On the other hand, MYC amplifies global transcription by antagonizing CDK9 sumoylation. Sumoylation of CDK9 blocks its interaction with Cyclin T1 and thus the formation of active P-TEFb complex. Transcription profiling analyses reveal that SUMO represses global transcription, particularly of moderately to highly expressed genes and by generating a sumoylation-resistant CDK9 mutant, we confirm that sumoylation of CDK9 inhibits global transcription. Together, our data reveal that SUMO and MYC oppositely control global gene expression by regulating the dynamic sumoylation and desumoylation of CDK9.
Introduction
Transcription initiation by RNA Polymerase (Pol) II is generally recognized as a key regulatory step in transcription at most eukaryotic genes.1–4 However, recent studies indicate that transcriptional elongation is also a key regulatory step for productive transcription.5–8 The transcription of many protein-coding genes is paused soon after initiation of transcription due to the concerted action of chromatin structure and factors that negatively regulate transcription elongation such as DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF).5,9 Positive transcription elongation factor b (P-TEFb), a complex comprising cyclin-dependent kinase (CDK) 9 and a Cyclin (Cyc) T or K subunit, is required for releasing Pol II promoter-proximal pausing by phosphorylating negative transcription elongation factors10–13 as well as the second serine residue (Ser2) of the heptapeptide (YSPTSPS) repeats within the C-terminal domain (CTD) of the largest subunit of Pol II.14 Ser2 phosphorylation (Ser2P) of the CTD serves to recruit transcription-associated proteins and is the hallmark for the transition from transcriptional initiation to productive elongation.7,15 Consistent with its key role in the control of transcriptional elongation, P-TEFb has been shown to be negatively regulated by the 7SK snRNP complex and positively regulated by bromo-domain containing protein 4 (BRD4)16–18 and to interact with other proteins to form the super elongation complex.19
In the literature, it is generally assumed that cells respond to various external or internal stimuli by regulating the expression of specific genes or sets of genes without affecting the global levels of transcription. However, there are also many examples in which global levels of gene expression are drastically affected. For instance, T cell activation is associated with a growth phase of around 24 h followed by massive clonal expansion and differentiation.20 During the growth phase, T cells increase in size and show elevated global gene expression. Similarly, cardiac hypertrophy is also associated with the up-regulation of global gene expression.21 Furthermore, MYC (also known as c-Myc), a proto-oncogenic transcription factor that has a central role in cell growth control, has been shown to amplify global transcription, a phenomenon termed transcription amplification,22,23 and does so by regulating transcriptional pause release.24 However, how MYC antagonizes the pausing of Pol II is not well understood.
Post-translational modification by the small ubiquitin-related modifier SUMO entails a cascade of enzymatic reactions similar to ubiquitination and regulates diverse cellular processes, including the cell cycle, nuclear integrity, genomic stability, and transcription.25–27 SUMO is first activated by an E1 activating enzyme; subsequently transferred to the unique E2 enzyme, UBC9; and then conjugated to substrates with or without help of E3 enzymes such as the PIAS family proteins. Vertebrate SUMO-1 shares only ~50% sequence identity with SUMO-2 and SUMO-3, which are often referred as SUMO2/3 because they have a 97% sequence identity with each other. As a dynamic modification, SUMO is removed from substrates by the SENP family isopepetidases.28 Interestingly, sumoylation of transcription factors and cofactors has a striking correlation with transcriptional repression.29 Extensive studies have linked SUMO-mediated transcriptional repression to the recruitment of multiple corepressors including the HDAC1/2-containing corepressor complexes.30,31 However, recent findings that SUMO is enriched at actively transcribed genes has led to the suggestion that SUMO is likely to regulate Pol II-associated factors.32–34
In this study we present evidence that SUMO functions as a brake controlling the global level of transcription (elongation) through modification of CDK9. Cellular CDK9 is abundantly and dynamically sumoylated. CDK9 sumoylation blocks the formation of a functional P-TEFb complex and consequently transcriptional elongation. Furthermore, we demonstrate that MYC enhances global transcription by antagonizing CDK9 sumoylation. Elevated global transcription during T cell activation is associated with reduced CDK9 sumoylation. Together, our study uncovers a novel role and mechanism by which SUMO controls global transcription and provides new insight into transcription amplification by MYC.
Results
Ectopically expressed SUMO and PIAS1 inhibit Ser2 phosphorylation of Pol II CTD
Sumoylation of the androgen receptor (AR) was shown previously to repress AR transcriptional activity.35,36 In accord with this finding, we observed that the SUMO E3 PIAS1 alone or plus SUMO-1 repressed transcriptional activation by AR from a chromatinized MMTV reporter in Xenopus oocytes (Supplementary information, Figure S1a).37,38 In line with the previous report,35 replacement of K386 and K520, two major sumoylation sites, with arginine residues virtually abolished AR sumoylation (Supplementary information, Figure S1b). However, expression of PIAS1 and SUMO-1 in Xenopus oocytes repressed transcription driven by either the wild-type or mutant AR to a similar extent (Supplementary information, Figure S1c), indicating that PIAS1/SUMO-1 represses AR transactivation largely independently of AR sumoylation. A subsequent chromatin immunopreciptation (ChIP) assay on the MMTV promoter revealed that PIAS1/SUMO-1 had no effect on AR’s chromatin binding nor histone acetylation, but resulted in a marked reduction of Ser2-phosphorylated Pol II without any significant effect on the levels of total Pol II (Supplementary information, Figure S1d). This suggests that PIAS1/SUMO-1 may repress transcription by regulating the phosphorylation of Ser2 residues in the Pol II CTD.
The observation in Xenopus oocytes prompted us to examine if PIAS1/SUMO-1 also regulated Pol II Ser2P in mammalian cells. We expressed PIAS1 and a mutant PIAS1 lacking the SUMO E3 ligase activity (PIAS1m) with SUMO-1 in HeLa cells and analyzed the effect on Pol II phosphorylation and histone acetylation by Western blotting (WB). As a positive control, HeLa cells were treated with flavopiridol, a CDK inhibitor that is known to selectively prevent Pol II Ser2P.39 We found that expression of PIAS1, but not PIAS1m, alone inhibited phosphorylation of Ser2, but not Ser5, of endogenous Pol II (Fig. 1a). Moreover, co-expression of SUMO-1 further augmented the inhibitory effect on Ser2P. Furthermore, consistent with our ChIP result from Xenopus oocytes, expression of PIAS1/SUMO-1 had no effect on the global levels of the acetylation of histones H3 and H4 (Fig. 1a). The suppression of Ser2P was dependent on the SUMO E3 ligase activity of PIAS1, as expression of PIAS1m/SUMO-1 led to an increased rather than reduced level of Ser2P, likely due to a dominant negative effect of PIAS1m in inhibiting endogenous PIAS proteins. The same results were observed when the experiments were carried out in 293 T cells (Supplementary information, Figure S2a). Thus, PIAS1 is able to markedly inhibit the phosphorylation of Pol II Ser2 in a SUMO-dependent manner.
Fig. 1.
SUMO inhibits phosphorylation of Pol II Ser2 and selectively sumoylates CDK9. a The levels of Ser2P, Ser5P, Pol II, acH3 and acH4 were determined by WB analysis upon ectopic expression of PIAS1 and PIAS1m with SUMO-1 in HeLa cells. b IF analysis showing the effect of ectopically expressed PIAS1 and PIAS1m with or without SUMO-1 on Ser2P of Pol II CTD. c WB analysis for known proteins involved in the phosphorylation of Ser2 of the Pol II CTD. Sumoylation was detected only in CDK9 upon ectopic expression of PIAS1/SUMO-1
We next used immunofluorescent (IF) staining to verify the effect of PIAS1/SUMO-1 on Ser2P of Pol II. As shown in Fig. 1b, reduced levels of Ser2P were observed in HeLa cells transfected with PIAS1 or PIAS1/SUMO-1 but not with PIAS1m and PIAS1m/SUMO-1. In accord with the WB data (Fig. 1a), we did not observe any reduction in levels of acetylated H3 and H4 (Supplementary information, Figure S2b) and Pol II Ser5 phosphorylation (Ser5P, Supplementary information, Figure S2c) in HeLa cells transfected with PIAS1 or PIAS1/SUMO-1. Similar results were also observed when HCT116 and 293 T cells were used (data not shown). Furthermore, just as with SUMO-1, both SUMO-2 and SUMO-3 also enhanced the inhibition of the phosphorylation of Ser2 by PIAS1 (Supplementary information, Figure S3a) and had no effect on the phosphorylation of Ser5 (Supplementary information, Figure S3b). In addition to PIAS1, PIASxβ and PIASy, but not PIAS3 and PIASxα, could also suppress Pol II Ser2P (Supplementary information, Figure S3c). Together these results show that multiple, but not all, members of the PIAS family of SUMO E3 ligases can inhibit the phosphorylation of Pol II at Ser2 in a SUMO-dependent manner.
Inhibition of phosphorylation of Ser2 of Pol II correlates with CDK9 sumoylation
Although CDK9 is the key kinase responsible for phosphorylating Ser2 of the Pol II CTD, CDK7,40,41 CDK12/1342 and BRD443 have also been reported to directly or indirectly phosphorylate Ser2. To unravel how SUMO inhibits Ser2P, we analyzed if any of the above proteins could be sumoylated under the conditions in which inhibition of Ser2P was observed (Fig. 1a). We found that ectopic expression of PIAS1/SUMO-1 resulted in sumoylation of endogenous CDK9. However, no sumoylation was observed for endogenous CDK7, CDK12 and BRD4 (Fig. 1c). As CDK9 and CDK7 are known to form the active P-TEFb complex with Cyc T1 and the CAK complex with MAT1 and Cyc H (part of TFIIH complex that phosphorylates Ser5 of Pol II CTD and T loop of other CDKs), we also examined whether these subunits could be sumoylated. However, we could detect no sumoylation of endogenous Cyc T1, Cyc H and MAT1 (Supplementary information, Figure S2d) under conditions where phosphorylation of Ser2 was effectively inhibited by ectopically expressed PIAS1/SUMO-1 (Supplementary information, Figure S2a). In addition, expression of PIAS1/SUMO-1 did not affect sumoylation of endogenous HDAC1 (Supplementary information, Figure S2d), in accord with the lack of effect upon global histone H3 and H4 acetylation. Furthermore, no sumoylation on either histone H3 or H4 was observed (Supplementary information, Figure S2e). Together these data suggest that PIAS1/SUMO-1 inhibits the phosphorylation of Ser2 of Pol II CTD most likely through CDK9 sumoylation.
CDK9 is readily sumoylated in cells and in vitro
We next tested if ectopically expressed CDK9 could be sumoylated. We found that co-expression of PIAS1, but not the E3-deficient PIAS1m, with SUMO-1 in 293 T cells resulted in drastically elevated sumoylation of ectopically expressed CDK9 (Fig. 2a). Co-expression of PIAS1 and SUMO-1 also elevated the overall level of CDK9, indicating that sumoylation stabilized CDK9. Similarly, PIASxβ and PIASy could also promote CDK9 sumoylation (data not shown). Protein-protein interaction analyses by co-immunoprecipitation (co-IP) revealed that CDK9 interacted with PIAS1 (Fig. 2b) and with SUMO E2 conjugating enzyme UBC9 (Fig. 2c). Furthermore, recombinant CDK9 expressed and purified from bacteria could be sumoylated in vitro even in the absence of E3 ligase (Fig. 2d).
Fig. 2.
Cellular CDK9 is abundantly sumoylated and this correlates with suppression of Ser2 phosphorylaiton. a WB analysis showing that ectopically expressed CDK9 was extensively sumoylated when co-expressed with PIAS1/SUMO-1 in HEK293T cells. b Interaction between CDK9 and PIAS1 ectopically expressed in HEK293T cells determined by co-immunoprecipitation assay. c Interaction between CDK9 and UBC9 ectopically expressed in HEK293T cells revealed by co-immunoprecipitation assay. d In vitro sumoylation assay showing that recombinant CDK9 proteins were sumoylated in the presence of SUMO E1, E2 and 6x His-SUMO proteins. e WB analysis of 293T cell extracts showing the presence of 42 kDa and multiple higher molecular weight CDK9 bands that were all substantially reduced upon knockdown of CDK9 by two different siRNAs. f, g Knockdown of UBC9 in 293T (f) and HeLa and HCT116 (g) by two different UBC9 shRNAs resulted in marked reduction of the higher molecular-mass sumoylated CDK9 bands and the increased levels of Ser2P . h WB analysis showing increased higher molecular-mass sumoylated CDK9 bands in Senp1−/− and Senp2−/− MEF cells in comparison to the wild-type MEFs. i WB analysis of stable Myc-CDK9 HeLa cell extracts showing the presence of higher molecular-weight forms of Myc-CDK9. Knockdown of UBC9 by two different shRNAs resulted in substantial reduction of the higher molecular-weight forms of Myc-CDK9. j WB analysis of HeLa cell extracts stably expressing Myc-CDK9K/R or Myc-CDK9 and transfected with PIAS1 or PIAS1/SUMO-1 as indicated. No sumoylation was detected for the Myc-CDK9K/R mutant
Endogenous CDK9 is extensively and dynamically sumoylated
Endogenous mammalian CDK9 is generally identified as a band migrating with an apparent molecular mass of 42 kDa in WB analysis. Although a CDK9 isoform of 55 kDa was reported previously,44 we detected multiple bands with apparent molecular masses of greater than 42 kDa CDK9 by WB in whole cell extracts prepared from various cell lines using various anti-CDK9 antibodies (Fig. 2e-h and Supplementary information, Figure S4). Although we could not exclude the possibility that some of the bands could be nonspecific, it seemed likely that most, if not all, of the higher-molecular bands represented sumoylated forms of CDK9 for the following reasons. First, knockdown of CDK9 in 293 T cells using two different siRNAs individually or in combination resulted in substantial reduction not only of CDK9 migrating with an apparent molecular mass of 42 kDa, but also of most of the higher-molecular mass bands detected by the anti-CDK9 antibody (Fig. 2e). Second, knockdown of UBC9, the sole mammalian SUMO E2 conjugating enzyme, in 293 T cells led to a marked reduction of the higher- molecular mass CDK9 bands but not the 42 kDa CDK9 band (Fig. 2f). Similarly, marked reduction of the higher-molecular mass CDK9 bands was observed when UBC9 was knocked down in HeLa and HCT116 cells (Fig. 2g). Third, in immortalized mouse embryonic fibroblasts (MEF) derived from Senp1 and Senp2 knockout mice (Senp1−/− and Senp2−/−),45,46 a greater number of more intensely stained higher-molecular mass CDK9 bands were detected using the anti-CDK9 antibody (Fig. 2h). It is noteworthy that the putative higher-molecular mass sumoylated CDK9 proteins were poorly immunoprecipitated by most anti-CDK9 antibodies that we have tested with the exception of an anti-CDK9 (C-20) antibody from Santa Cruz (Supplementary information, Figure S4a), suggesting that sumoylation is likely to block the access of antibodies to CDK9. In accord with the observation that CDK9 sumoylation is negatively controlled by SENP1 and SENP2 desumoylases in MEFs, we demonstrated that expression of SENP1 or SENP2 effectively abrogated the sumoylated forms of CDK9 generated by co-expressed PIAS1/SUMO-1 (Supplementary information, Figure S5). Fourth, we established a HeLa cell line stably expressing Myc-tagged CDK9 and subsequently detected, in addition to the expected Myc-CDK9 band, a series of higher molecular mass bands by WB analysis using an anti-Myc antibody (Fig. 2i, lane 2). Furthermore, knockdown of UBC9 by shRNAs substantially reduced the higher-molecular mass Myc-CDK9 bands and increased the levels of unmodified Myc-CDK9 proteins (Fig. 2i, compare lanes 3-5 with 2), indicating that most of the higher-molecular mass Myc-CDK9 bands correspond to CDK9 sumoylated to various degrees. In support of this view, we also detected higher molecular mass CDK9 bands in immunoprecipitates obtained using either anti-SUMO-1 or anti-SUMO-2/3 antibodies under denaturing condition (Supplementary information, Figure S5b-c). Fifth, we detected sumoylation on immunoprecipitated endogenous CDK9 by mass spectrometry (Supplementary information, Figure S6a-b). Sixth, sequence analysis revealed the presence of three potential consensus sumoylation sites (K18, K74 and K264, Supplementary information, Figure S6c) in CDK9. However, subsequent mutation of these and multiple additional K residues to arginines (R) failed to eliminate CDK9 sumoylation by PIAS1/SUMO-1 (Supplementary information, Figure S6d), indicating that CDK9 can be sumoylated on multiple redundant sites. To completely eliminate CDK9 sumoylation, we generated a human CDK9 mutant termed the CDK9K/R in which all 29 K residues were changed to R. Stable 293 T cell lines expressing either 3x Myc-CDK9 or 3x Myc-CDK9K/R mutant were generated and subsequent WB analysis revealed the presence of the higher-molecular mass CDK9 bands for 3x Myc-CDK9 but not for 3x Myc-CDK9K/R mutant (compare lane 2 with lane 5). Furthermore, while ectopic expression of PIAS1/SUMO-1 resulted in drastically increased sumoylation of 3x Myc-CDK9, it had no effect, as expected, on the 3x Myc-CDK9K/R mutant (Fig. 2j, compare lane 4 with lane 7). In further support, sumoylation of CDK9 was detected at multiple sites in a previous proteomic study.47 Taken together, these results provide compelling evidence that endogenous CDK9 is extensively sumoylated, most likely at multiple sites. As reduced levels of sumoylated CDK9 were observed upon knockdown of UBC9 and elevated levels of sumoylated CDK9 observed in Senp1−/− and Senp2−/− MEFs, we conclude that the endogenous CDK9 is dynamically controlled by sumoylation and desumoylation.
CDK9 sumoylation inversely correlates with Pol II Ser2 phosphorylation
We found that CDK9 sumoylation inversely correlates with the cellular level of Ser2P of Pol II CTD. Knockdown of UBC9 in 293T cells, HeLa and HCT116 cells not only led to reduced CDK9 sumoylation but also significantly increased (up to 7-fold) the levels of Pol II Ser2P (Fig. 2f, g). In contrast, no change in Ser5P was observed. In addition, reduced levels of Ser2P were observed in both Senp1−/− and Senp2−/− MEFs in comparison to control MEFs (Fig. 2h). Furthermore, immunofluorecence staining revealed increased levels of Ser2P in HeLa (Supplementary information, Figure S7a) and HCT116 cells (Supplementary information, Figure S7b) upon knockdown of UBC9. Most importantly, whereas expression of PIAS1 and PIAS1/SUMO-1 resulted in progressive suppression of Ser2P in stably transformed 3x Myc-CDK9 293T cells, this effect was not observed in the SUMO-resistant 3x Myc-CDK9K/R 293T3 cells (Fig. 2j, compare lanes 3 and 4 with lanes 6 and 7). Taken together, these results indicate that SUMO inhibits Pol II Ser2P in a CDK9- sumoylation dependent manner.
CDK9 sumoylation inhibits formation of P-TEFb complex
Having established that elevated CDK9 sumoylation correlates with a reduced level of Pol II Ser2P, we investigated how CDK9 sumoylation affects CDK9 kinase activity. Previous studies indicated that formation of the P-TEFb complex with a Cyc T or K subunit is a prerequisite for CDK9 to phosphorylate Ser2 of the Pol II CTD.48 By using purified Flag-CDK9 with or without a Strep-tagged Cyc T1 subunit, we confirmed that CDK9 could phosphorylate recombinant GST-CTD in vitro only in the presence of Cyc T1 (Supplementary information, Figure S8a-c). We therefore tested if PIAS1/SUMO-1 is able to inhibit Pol II Ser2P by inhibiting the formation of functional P-TEFb complex. Ectopic expression of PIAS1/SUMO-1 did not reduce but rather increased the level of CDK9 protein, suggesting that CDK9 sumoylation might stabilize CDK9 protein by inhibiting CDK9 ubiquitination (Fig. 2). Immunofluorescence analysis revealed that ectopically expressed PIAS1/SUMO-1 did not affect the subcellular localization of CDK9 (data not shown). However, immunoprecipitation of endogenous CDK9 using an anti-CDK9 antibody (Santa Cruz C-20) revealed that ectopic expression of PIAS1/SUMO-1 substantially inhibited the interaction of endogenous CDK9 with Cyc T1 (Fig. 3a, b). The marked reduction of the interaction between CDK9 and Cyc T1 upon expression of PIAS1/SUMO-1 was confirmed by a reciprocal IP using anti-Cyc T1 antibody (Fig. 3c). An in vitro pulldown assay also showed that sumoylation inhibited the binding of Cyc T1 to CDK9 (Supplementary information, Figure S8d). Interestingly, we found that CDK9 can interact efficiently with itself (Supplementary information, Figure S8e). However, CDK9 sumoylation did not affect the self-interaction of CDK9, but markedly inhibited the interaction between CDK9 and Cyc T1 (Supplementary information, Figure S8e). Taken together, we conclude that CDK9 sumoylation inhibits the formation of a functionally active P-TEFb complex.
Fig. 3.
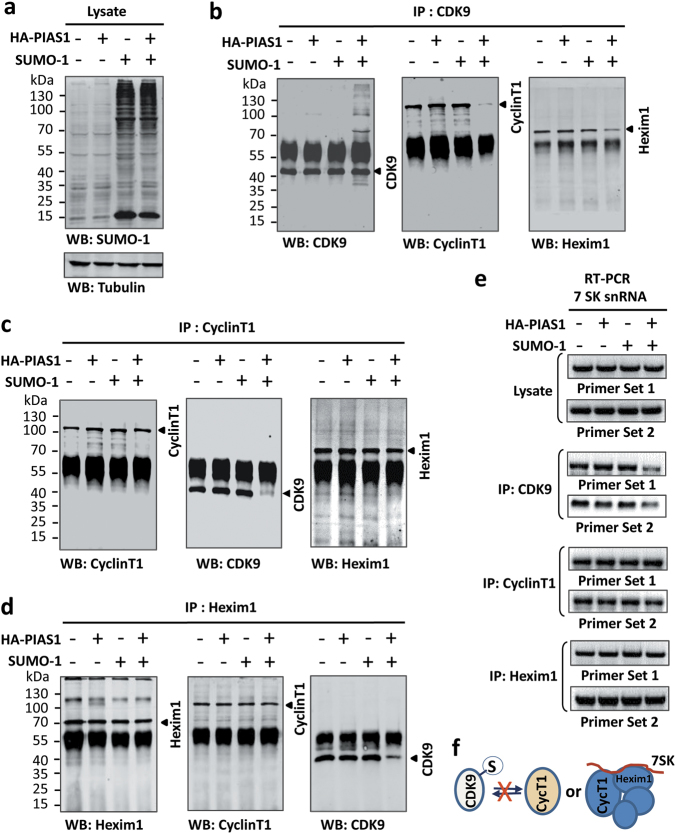
CDK9 sumoylation inhibits formation of the P-TEFb complex. a HEK293T cells were transfected with PIAS1 and/or SUMO-1 as indicated and the global levels of sumoylation were analyzed by WB blotting using an anti-SUMO-1 antibody as indicated. b-e The effect of PIAS1 and/or SUMO-1 expression on the formation of the P-TEFb complex was analyzed by IP using anti-CDK9 (b), anti-Cyc T1 (c) and anti-Hexim1 (d), followed by WB using antibodies as indicated. In addition, the effect on association of 7SK snRNA was assayed by RT-PCR (e). f Diagram illustrating that CDK9 sumoylation blocks the formation of P-TEFb complex
Previous studies have demonstrated a role of the 7SK snRNP complex in suppressing P-TEFb kinase activity.17,18 We thus also examined whether PIAS1/SUMO-1 could inhibit P-TEFb formation by influencing its interaction with the 7SK snRNP complex. We observed that ectopic expression of PIAS1/SUMO-1 reduced the interaction between CDK9 and HEXIM1 (Fig. 3b). However, PIAS1/SUMO-1 did not affect the interaction between Cyc T1 and HEXIM1 (Fig. 3c, d). Accordingly, IP followed by RT-PCR analysis of the 7SK non-coding RNA revealed that ectopic expression of PIAS1/SUMO-1 reduced the association between 7SK RNA and CDK9 but not between 7SK RNA and Cyc T1 and HEXIM1 (Fig. 3e). Taken together, we conclude that CDK9 sumoylation blocks the formation of P-TEFb complex, but not the Cyc T1/7SK/HEXIM1 complex (Fig. 3f).
SUMO inhibits global levels of transcription
Our finding that SUMO controls global levels of Pol II Ser2P promoted us to test if SUMO controls global levels of gene expression. To this end, we first examined if ectopically expressed PIAS1 and PIAS1/SUMO-1 affected the incorporation of BrUTP into newly synthesized RNAs. We found that expression of PIAS1 reduced the incorporation of BrUTP and this reduction was further enhanced by co-expression of PIAS1 and SUMO-1 (Fig. 4a). However, no repression of BrUTP incorporation was observed for PIAS1m and PIAS1m/SUMO-1. Thus, PIAS1 is able to substantially repress the global level of transcription in a SUMO E3 ligase-dependent manner. Furthermore, we found that the repression of global transcription correlated with the ability of PIAS proteins to inhibit Ser2P of Pol II because PIASxβ and PIASy, but not PIAS3 and PIASxα, were able to repress transcription globally in the same assay (Supplementary information, Figure S9a).
Fig. 4.
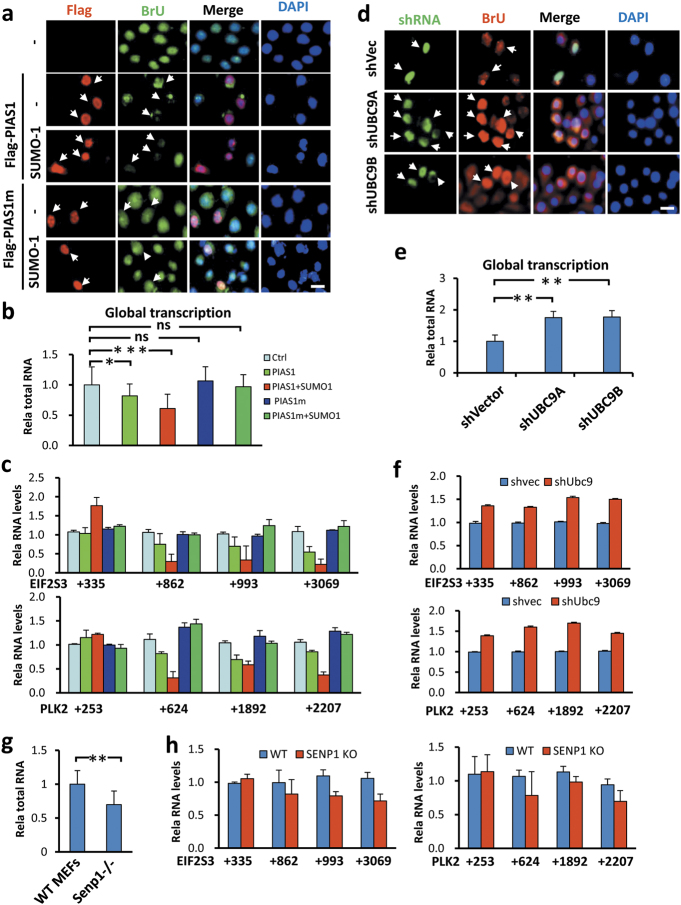
SUMO inhibits global transcription by inhibiting transcriptional elongation. a BrU incorporation assay showing that ectopic expression of PIAS1 inhibited global transcription in a SUMO-dependent manner. b Quantitative measurement of total RNAs from equivalent numbers of cells showing that PIAS1 inhibited global transcription in a SUMO-dependent manner. c Quantitative RT-PCR analysis of two representative genes EIF2S3 and PLK2 showing the inhibition of transcriptional elongation by ectopically expressed PIAS1 and more severely by PIAS1/SUMO-1. Note that ectopic expression of PIAS1/SUMO-1 significantly reduced the levels of long transcripts with the accumulation of promoter-proximal transcripts. d Knockdown of UBC9 in HeLa cells by two different shRNAs led to increased incorporation of BrU and thus increased global transcription. e Quantitative measurement of total RNAs from equivalent numbers of cells showing that knockdown of UBC9 resulted in increased global transcription. f Quantitative RT-PCR analysis of two representative genes EIF2S3 and PLK2 showing knockdown of UBC9 resulted in elevated productive transcriptional elongation. g Quantitative measurement of total RNAs from equivalent numbers of wild-type and Senp1−/− MEFs showing a reduced global transcription in Senp1−/− MEFs. h Quantitative RT-PCR analysis of two representative genes EIF2S3 and PLK2 showing reduced productive transcriptional elongation in Senp1−/− MEFs
To test further that SUMO represses global transcription, we transfected HeLa cells as above described but with an increasing amount of DNA to ensure almost 100% transfection efficiency. We then counted the cell number and prepared total RNAs from each group using the same amount of cells. This analysis revealed that expression of PIAS1 resulted in a reduced level of total RNAs and co-expression of PIAS1/SUMO-1 led to an even stronger reduction of total RNAs (Fig. 4b). In accord with the results of the BrUTP incorporation assay, expression of PIAS1m with or without co-expression of SUMO-1 had no significant influence on global levels of total RNAs.
As P-TEFb plays a key role in promoting transcriptional elongation by releasing paused Pol II,7,49 we next examined if expression of PIAS1 or PIAS1/SUMO-1 affected transcriptional elongation. For analysis we selected two representative genes that were previously shown to be regulated in pausing release by P-TEFb.50 We found that expression of PIAS1 and, more obviously, expression of PIAS1/SUMO-1 resulted in accumulation of short transcripts and progressively reduced levels of longer transcripts for EIF2S3 and PLK2 (Fig. 4c). Similar results were observed when additional genes were analyzed (data not shown). Again this effect was not observed for PIAS1m, implying it is a sumoylation-dependent event.
To test if SUMO controls global level of transcription physiologically, we knocked down UBC9 in HeLa cells by using two different shRNAs. Knockdown of UBC9 resulted in increased incorporation of BrUTP (Fig. 4d). Similarly, quantitative measurement of total RNAs prepared from the same number of cells also confirmed that UBC9 knockdown resulted in elevated levels of total RNAs (Fig. 4e). Furthermore, analysis of EIF2S3 and PLK2 transcripts revealed increased levels of the full-length mRNAs (Fig. 4f), indicating an enhancement of productive transcription upon UBC9 knockdown. The observation that UBC9 knockdown results in an elevated level of global transcription was also observed in A549 cells by both BrUTP incorporation (Supplementary information, Figure S9b) and by measurement of total RNAs (Supplementary information, Figure S9c). Thus, in the multiple cell lines we have tested, SUMO has a role in repressing global transcription.
As increased CDK9 sumoylation and decreased Pol II Ser2P were observed in Senp1−/− and Senp2−/− MEFs, we also measured if loss of Senp1 would result in a reduced level of global transcription and impaired transcriptional elongation in MEFs. By quantitatively measuring the amounts of total RNAs from the same number of wild-type and Senp1−/− MEFs, we found a reduced level of total RNAs in Senp1−/− MEFs (Fig. 4g). Furthermore, analysis of the EIF2S3 and PLK2 genes revealed moderately impaired transcriptional elongation for both genes in Senp1−/− MEFs (Fig. 4h). BrUTP incorporation assay also revealed reduced transcriptional activity in both Senp1−/− and Senp2−/− MEF cells (Supplementary information, Figure S9d). Together, these results support a model that sumoylation has an intrinsic role in repressing global cellular transcription, possibly by inhibiting transcriptional elongation by modifying CDK9.
SUMO represses global transcription by targeting actively transcribed genes
Gene expression profiling by microarray or RNA-seq, in general, aims to identify differentially expressed genes and involves normalization of the signal from different samples, with an assumption that the total level of gene expression/cell is the same. The recent finding that MYC amplifies gene expression has called for a different method for normalization using spike-in RNA standards for the analysis of global gene expression by RNA-seq.22,51 To further define whether SUMO represses global gene expression, we prepared RNA samples from the same number of control HEK293T cells, HEK293T cells transfected with PIAS1 or PIAS1/SUMO-1 and carried out RNA-seq analysis with spike-in yeast RNA standards. After normalization to the spike-in control, we found that expression of PIAS1 and PIAS1/SUMO-1 reduced the global levels of gene expression by approximately 16% and 46%, respectively (Fig. 5a). In line with the observed suppression of global gene expression, the majority of genes (15,453 and 17,662, respectively) were found to be down-regulated upon expression of PIAS1 or PIAS1/SUMO-1 (Fig. 5b). Furthermore, when gene expression was divided into five classes according to increasing levels of expression, SUMO was found to preferentially repress active to highly active expressed genes (Fig. 5c). The repression of active to highly active expressed genes by PIAS1 and PIAS1/SUMO-1 was further confirmed by qRT-PCR analysis of randomly selected genes from each class (Fig. 5d).
Fig. 5.
SUMO globally represses actively transcribed genes. a Normalized read counts in RNA-seq data of control, PIAS1-, and PIAS1/SUMO-1-expressing HEK293T cells. RNAs were prepared from equivalent numbers of each group of cells and the levels of transcription were normalized against the spike-in yeast RNAs. b The majority of genes were repressed upon ectopic expression of PIAS1 or PIAS1/SUMO-1. “Down”, “Equal” and “Up” represent genes whose reads counts in treated samples are less than, equal to and more than that in the control sample. c SUMO preferentially repressed active to highly active transcribed genes. Genes were divided into five classes according to their levels of expression and the effect of ectopically expressed PIAS1 and PIAS1/SUMO-1 was analyzed. d Quantitative RT-PCR analysis verified the inhibitory effect of PIAS1 and PIAS1/SUMO-1 on two representative genes from each class. e ChIP-seq analysis showing the average ChIP signal profile of Pol II at important genome features. f ChIP-seq analysis showing the average ChIP signal profile of Ser2P at important genome features. g ChIP-seq analysis showing the average ChIP signal profile of Flag-PIAS1 at important genome features. h Normalized RNA-seq read counts of control and HEK293T cells upon knockdown of UBC9. The RNAs were prepared from equivalent numbers of each group of cells and the levels of transcription were normalized against the spike-in yeast RNAs. i The majority of genes were activated upon knockdown of UBC9. j Knockdown of UBC9 preferentially increased transcription from active to highly active transcribed genes. k Quantitative RT-PCR analysis showing increased transcription of representative genes upon knockdown of UBC9. l Genes repressed by ectopically expressed PIAS1/SUMO-1 substantially overlap with genes activated upon UBC9 knockdown
To assess if PIAS1/SUMO-1 represses global transcription by affecting transcription elongation, chromatin immunoprecipitation sequencing (ChIP-seq) analyses were performed for Pol II, Ser2P-Pol II and Flag-PIAS1 in parallel for control and PIAS1/SUMO-1-expressing HEK293T cells. Metagene analysis showed that expression of PIAS1/SUMO-1 resulted in accumulation of Pol II at the transcriptional start site (TSS) and reduced Pol II in the gene body (Fig. 5e). Consistent with a global reduction of Ser2P detected by Western blot analysis of cells expressing PIAS1/SUMO-1, ChIP-seq analysis revealed a substantial reduction of Ser2P at all regions of genes (Fig. 5f). These effects on Pol II and Ser2P were also observed for EIF2S3 and PLK2 genes (Supplementary information, Figure S10). It is noteworthy that ChIP-seq analysis with anti-Flag antibody only detected a very weak enrichment of Flag-PIAS1 at the TSS region (Fig. 5g). Thus, sumoylation of CDK9 may not occur at the TSS site, although we could not exclude the possibility the lack of enrichment of Flag-PIAS1 is the result of poor cross-linking of Flag-PIAS1 with DNA.
As a complementary experiment, we also carried out RNA-seq analysis for HEK293T cells following knockdown of UBC9. In contrast to ectopic expression of PIAS1/SUMO-1, UBC9 knockdown led to an increased level of global gene expression (Fig. 5h), elevated expression of the majority of genes (Fig. 5i), and preferential elevation of gene expression of active to highly active expressed genes (Fig. 5j). We verified the RNA-seq results by qRT-PCR analyses of representative genes, which showed an increased level of expression of the same set of genes that were repressed by PIAS1/SUMO-1 (Fig. 5k). Furthermore, the class 5 of most genes down-regulated by PIAS1/SUMO-1 significantly overlapped with the class 5 of most genes up-regulated by UBC9 knockdown (Fig. 5l). Gene Ontology (GO) biological process term analysis revealed that overlapping genes are highly enriched for basic cellular functions including gene expression, cell cycle, protein targeting, mRNA processing and translation (Supplementary information, Figure S11a) and pathways in ribosome, splicesome, protein processing and cell cycle (Supplementary information, Figure S11b). Together, our gene expression profiling analyses via complementary gain- and loss-of-function approaches validate a role for SUMO in repression of global gene expression and demonstrate that SUMO preferentially represses transcriptional elongation of actively transcribed genes. This accords with previous findings that SUMO is enriched at constitutively transcribed genes in yeast and active promoters in mammalian cells.32,34
SUMO suppresses global transcription via CDK9 sumoylation
Thus far, we have shown that CDK9 sumoylation correlates with repression of global gene expression. To rigorously test if SUMO represses global gene expression via sumoylation of CDK9 and inhibition of the P-TEFb complex, we knocked out the endogenous CDK9 gene by CRISPR/CAS9 (see Experimental Procedures) from HEK293T cell lines stably expressing 3x Myc-CDK9 and the 3x Myc-CDK9K/R mutant, respectively. CDK9 expression in the resulting CDK9KO/Myc-CDK9 and CDK9KO/Myc-CDK9K/R cells cell lines was verified by WB analysis (Fig. 6a) and the genotype confirmed by DNA sequencing (data not shown). It should be noted that CDK9KO/Myc-CDK9 and CDK9KO/Myc-CDK9K/R HEK293T cells were indistinguishable from the parental HEK293T cells in their growth properties and morphology (data not shown), whereas the CDK9 knockout (CDK9KO) in parental HEK293T cells resulted in cell growth arrest and apoptosis. It is also noteable that the CDK9 knockout in Myc-CDK9K/R cells not only led to the loss of the endogenous 42 kDa CDK9 band but also of all higher-molecular mass CDK9 bands (Fig. 6a, Input, compare lane 2 with 1). These results provide compelling evidence that the high-molecular mass CDK9 bands observed in various cells are most likely sumoylated forms of CDK9 because CDK9 knockout resulted in their elimination. Subsequent IP-WB analysis also demonstrated that Myc-CDK9 but not Myc-CDK9K/R were sumoylated (Fig. 6a, left panel).
Fig. 6.
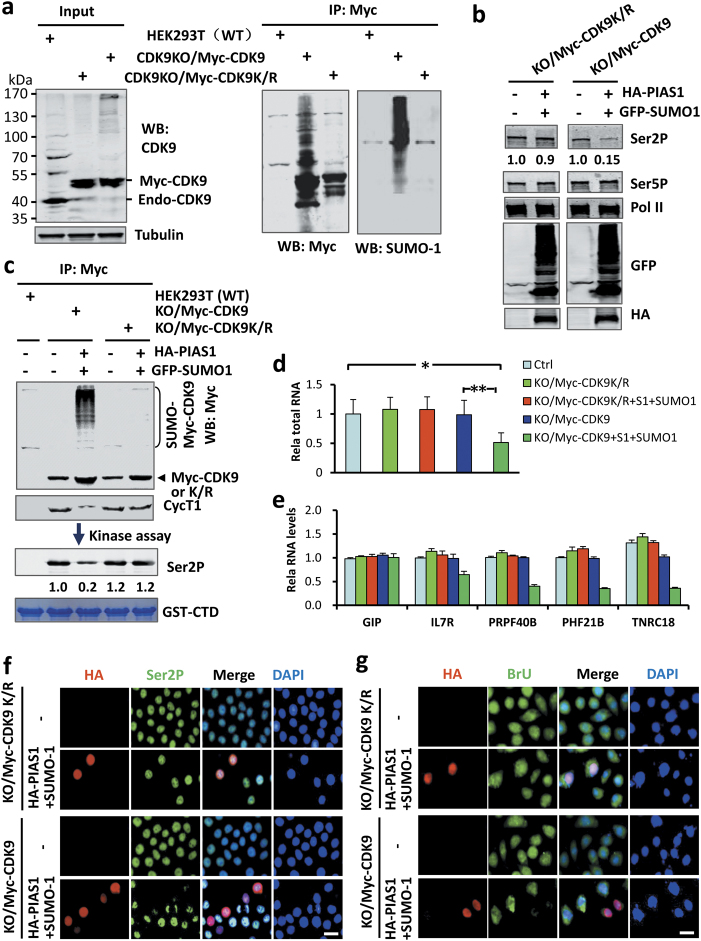
SUMO represses global transcription via CDK9 sumoylation. a Generation of HEK293T cell lines with knockout of endogenous CDK9 and expression of Myc-CDK9 or SUMO-deficient Myc-CDK9K/R mutant. The lack of endogenous CDK9 and expression of Myc-CDK9 or Myc-CDK9K/R mutant were verified by WB analysis using anti-CDK9 antibody. The cellular extracts were immunoprecipitated with anti-Myc antibody, followed by WB analysis using either Myc or SUMO-1 antibody as indicated. b WB analysis showing that ectopic expression of PIAS1/SUMO-1 inhibited Ser2P in CDK9KO/Myc-CDK9 cells but not in CDK9KO/Myc-CDK9K/R cells. c The cellular extracts in (b) were immunoprecipitated with anti-Myc antibody and the half of the resulting proteins were analyzed by WB using anti-Myc or anti-Cyc T1. The rest of half samples were used for in vitro kinase assay using GST-CTD as substrate and the levels of Ser2P were detected by WB analysis. d The measurement of total RNAs from equivalent numbers of cells showing that ectopic expression of PIAS1/SUMO-1 inhibited global transcription in CDK9KO/Myc-CDK9 cells but not in CDK9KO/Myc-CDK9K/R cells. e RT-PCR analysis of representative SUMO-regulated genes showing that ectopic expression of PIAS1/SUMO-1 inhibited transcription in CDK9KO/Myc-CDK9 cells but not in CDK9KO/Myc-CDK9K/R cells. f IF staining showing that ectopic expression of PIAS1/SUMO-1 reduced the levels of Ser2P in the CDK9KO/Myc-CDK9 but not in the CDK9KO/Myc-CDK9K/R cells. g BrU incorporation assay showing that ectopic expression of PIAS1/SUMO-1 substantially reduced the levels of BrU incorporation in CDK9KO/Myc-CDK9 but not in CDK9KO/Myc-CDK9K/R cells
Having obtained CDK9 sumoylation-resistant (CDK9KO/Myc-CDK9K/R) and sumoylation-competent (CDK9KO/Myc-CDK9) cell lines, we tested if expression of PIAS1/SUMO-1 was able to inhibit Pol II Ser2P and global gene expression in these cell lines. We found that expression of PIAS1/SUMO-1 had no effect on Pol II Ser2P in CDK9KO/Myc-CDK9K/R cells, but substantially reduced the level of Pol II Ser2P in CDK9KO/Myc-CDK9 cells (Fig. 6b). Subsequent IP-WB analysis showed that expression of PIAS1/SUMO-1 did not lead to sumoylation of Myc-CDK9K/R, as expected, whereas it resulted in substantial sumoylation of Myc-CDK9 (Fig. 6c). Furthermore, IP-WB analysis revealed that expression of PIAS1/SUMO-1 inhibited the formation of the P-TEFb complex in CDK9KO/Myc-CDK9 cells but not in CDK9KO/Myc-CDK9K/R cells (Fig. 6c). Importantly, when immunoprecipitated Myc-CDK9 and Myc-CDK9K/R were subjected to in vitro kinase activity assays using recombinant GST-CTD as substrate, we found that expression of PIAS1/SUMO-1 strongly suppressed the kinase activity derived from CDK9KO/Myc-CDK9 cells but had no effect on the kinase activity from CDK9KO/Myc-CDK9K/R (Fig. 6c, lower panel). Thus, the ability of PIAS1/SUMO-1 to inhibit P-TEFb kinase activity and phosphorylation of cellular Ser2 is entirely dependent on whether CDK9 can be sumoylated.
We next analyzed whether PIAS1/SUMO-1 expression influences global gene expression in CDK9KO/Myc-CDK9 and CDK9KO/Myc-CDK9K/R cells. Whereas expression of PIAS1/SUMO-1 repressed the global level of transcription in CDK9KO/Myc-CDK9 cells, it had no effect on global transcription in CDK9KO/Myc-CDK9K/R cells (Fig. 6d). The lack of repression for PIAS1/SUMO-1 on gene expression in CDK9KO/Myc-CDK9K/R cells was further demonstrated by quantitative analysis of representative SUMO-regulated genes (Fig. 6e). Accordingly, immunofluorescence revealed that ectopic expression of PIAS1/SUMO-1 reduced the levels of Ser2P (Fig. 6f) and BrUTP incorporation (Fig. 6g) in the CDK9KO/Myc-CDK9 but not in the CDK9KO/Myc-CDK9K/R cells. Altogether, these data demonstrate that PIAS1/SUMO-1 is unable to suppress Ser2P and global gene expression in the absence of CDK9 sumoylation.
MYC antagonizes CDK9 sumoylation
A recent seminal finding on MYC is its ability to amplify global transcription and regulate release of Pol II pausing.22,24 Having established that SUMO represses global transcription through CDK9 sumoylation, we tested whether MYC could amplify transcription by antagonizing CDK9 sumoylation. By quantitatively determining the levels of total RNAs from control and Myc-tagged MYC-expressing HeLa cells, we confirmed that ectopic MYC expression resulted in an elevated level of global gene expression (Fig. 7a). Importantly, we found that ectopic expression of Myc-MYC resulted in a substantially reduced CDK9 sumoylation and an elevated level of Pol II Ser2P (Fig. 7b). Using an immunoflurescence assay for BrUTP incorporation, we validated that expression of Myc-MYC enhanced the global levels of transcription (Fig. 7c). We also confirmed by IF staining that ectopic expression of Myc-MYC in HeLa cells strongly elevated the levels of Pol II Ser2P (Fig. 7d).
Fig. 7.
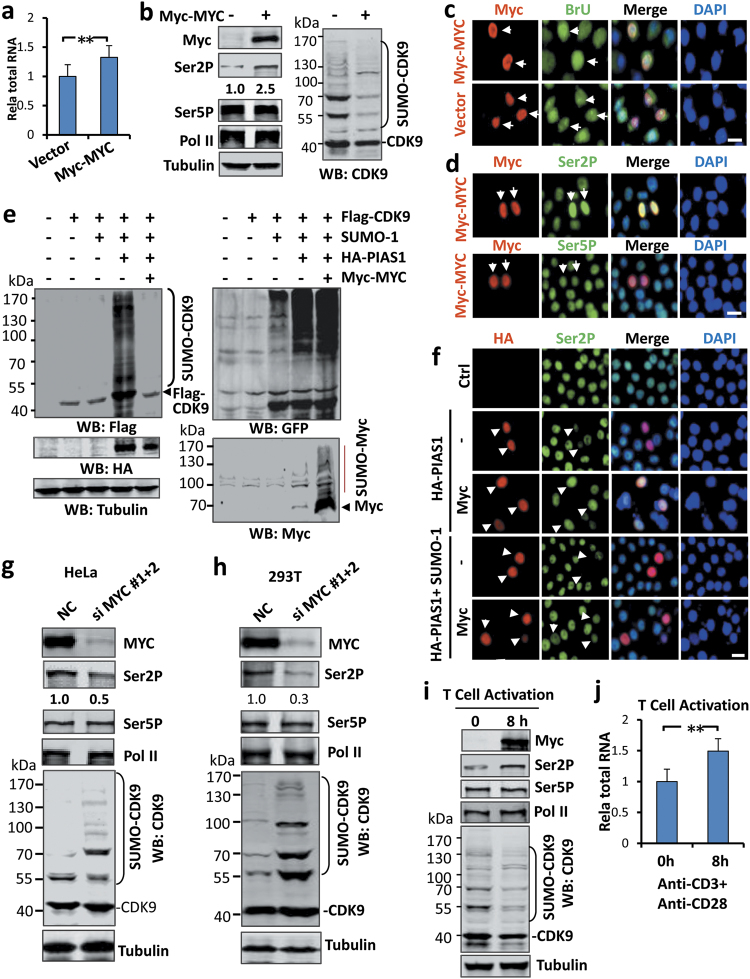
MYC enhances global transcription and phosphorylation of Ser2 of the CTD and antagonizes CDK9 sumoylation. a Ectopic expression of MYC in HEK293T cells resulted in increased global transcription. The total levels of RNAs were determined from equivalent numbers of control and Myc-MYC expressed HEK293T cells. b WB analyses showing that ectopic expression of Myc-MYC resulted in an increased level of Ser2P and reduced levels of CDK9 sumoylation. c BrU incorporation assay showing that ectopic expression of Myc-MYC resulted in elevated levels of BrU incorporation in HeLa cells. d IF staining analysis showing that ectopic expression of Myc-MYC resulted in elevated levels of Ser2P in HEK293T cells. e Expression of Myc-MYC effectively blocked CDK9 sumoylation. HeLa cells were transfected with Flag-CDK9, GFP-SUMO-1, HA-PIAS1 and Myc-MYC as indicated and sumoylation on Flag-CDK9, Myc-MYC and global sumoylation were detected by WB using anti-Flag, anti-Myc or anti-GFP as indicated. f IF staining showing that expression of Myc-MYC blocked the inhibition of PIAS1 and PIAS1/SUMO-1 on Ser2P. g WB analysis showing that knockdown of MYC in HeLa cells resulted in reduced level of Pol II Ser2P and increased CDK9 sumoylation. NC, cells treated with scrambled siRNA. h WB analysis showing that knockdown of MYC in 293T cells resulted in reduced levels of Pol II Ser2P and increased CDK9 sumoylation. NC, cells treated with scrambled siRNA. i WB analysis showing that T-cell activation induced by co-stimulation using anti-CD3 and anti-CD28 in vitro was accompanied by induction of MYC, increased Ser2P and reduced CDK9 sumoylation. j T-cell activation induced by co-stimulation using anti-CD3 and anti-CD28 in vitro was associated with increased global transcription. Total levels of RNAs were determined from the same amount of naive and T-cells treated with anti-CD3 and anti-CD28 for 8 h
Next, we directly tested if MYC was able to antagonize PIAS1-mediated sumoylation of CDK9. Expression of Myc-MYC almost completely abolished Flag-CDK9 sumyolation instigated by PIAS1/SUMO-1 without affecting the expression of HA-PIAS1 and GFP-SUMO-1 (Fig. 7e). The inhibition on CDK9 sumoylation was specific, because expression of Myc-MYC did not affect overall cellular sumoylation (Fig. 7e, right panel). We noticed that, although CDK9 sumoylation was almost completely abolished when it was co-expressed with Myc-MYC, Myc-MYC itself became extensively sumoylated (Fig. 7e, right lower panel). Consistent with the ability of MYC to antagonize CDK9 sumoylation, we found that MYC was able to reverse the inhibition of ectopically expressed PIAS1/SUMO-1 on Pol II Ser2P (Fig. 7f).
To test if endogenous MYC regulates CDK9 sumoylation, we knocked down MYC by siRNA treatment in both HeLa and 293 T cells. Subsequent WB analysis revealed that MYC knockdown in HeLa cells resulted in a reduced level of Pol II Ser2P and a substantial increase in the level of CDK9 sumoylation (Fig. 7g). Similar results were observed in 293 T cells (Fig. 7h). Thus, endogenous MYC proteins apparently antagonize CDK9 sumoylation and promote the phosphorylation of Pol II at Ser2.
To test if the antagonism of CDK9 sumoylation by MYC is physiologically relevant, we turned to the process of T-cell activation, which is highly dependent on MYC induction.20,52 Consistent with previous reports, we observed that mouse T-cell activation induced by anti-CD3 and anti-CD28 for 8 h resulted in a robust induction of Myc protein (Fig. 7i). The induction of Myc was accompanied by a marked reduction of CDK9 sumoylation and an increased level of Pol II Ser2P without affecting Ser5P and the total level of Pol II (Fig. 7i). Measurement of total RNAs from an equivalent number of naïve and activated T-cells also confirmed an increased level of total RNAs in activated T-cells (Fig. 7j). Thus, MYC induction and the accompanying increase in global gene expression during T-cell activation is associated with reduced CDK9 sumoylation and elevated Pol II Ser2P.
MYC amplifies global transcription primarily by antagonizing CDK9 sumoylation
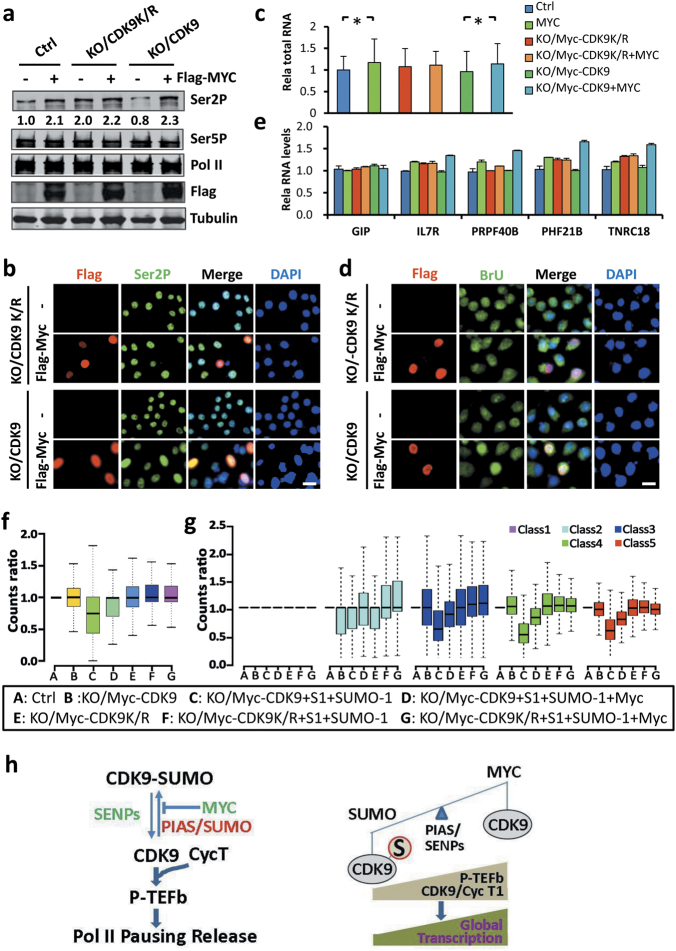
The availability of CDK9KO/Myc-CDK9 and CDK9KO/Myc-CDK9K/R cell lines also provided the opportunity to test whether amplification of transcription by MYC is dependent on its ability to antagonize CDK9 sumoylation. We first tested if ectopic expression of Flag-MYC was able to affect Ser2P in these cell lines. We found that ectopic MYC expression led to increased levels of Pol II Ser2P in parental HEK293T and CDK9KO/Myc-CDK9 cells, but not in the CDK9KO/Myc-CDK9K/R cells (Fig. 8a). However, careful examination revealed that the CDK9KO/Myc-CDK9K/R cells had an elevated level of Pol II Ser2P that was comparable to the levels of Ser2P in parental and CDK9KO/Myc-CDK9 cells with ectopic MYC expression (Fig. 8a), in line with a lack of inhibition of CDK9K/R kinase activity by the endogenous SUMO system. Similarly, analysis of Ser2P by immunofluorescence also revealed that Flag-MYC elevated the levels of Ser2P in CDK9KO/Myc-CDK9 but not in CDK9KO/Myc-CDK9K/R cells (Fig. 8b). Furthermore, measurement of total RNAs (Fig. 8c) or BrUTP incorporation by immunofluorescence (Fig. 8d) showed that global gene expression was elevated by ectopic expression of MYC in parental and/or CDK9KO/Myc-CDK9 but not in CDK9KO/Myc-CDK9K/R cells. The inability of MYC to activate global gene expression in the CDK9KO/Myc-CDK9K/R cells was further confirmed by quantitative RT-PCR analysis of a group of SUMO-regulated genes (Fig. 8e).
Fig. 8.
MYC amplifies global transcription by antagonizing CDK9 sumoylation. a WB analysis showing that ectopic expression of MYC resulted in an increased level of Ser2P in control and CDK9KO/Myc-CDK9 cells but not in CDK9KO/Myc-CDK9K/R cells. b IF staining showing that ectopic expression of MYC increased the levels of Ser2P in CDK9KO/Myc-CDK9 but not in CDK9KO/Myc-CDK9K/R cells. c The measurement of total RNAs from the same amount of cells showing that ectopic expression of MYC increased the global transcription in control and CDK9KO/Myc-CDK9 cells but not in CDK9KO/Myc-CDK9K/R cells. d BrU incorporation assay showing that ectopic expression of MYC substantially increased the levels of BrU incorporation in CDK9KO/Myc-CDK9 but not in CDK9KO/Myc-CDK9K/R cells. e RT-PCR analysis of representative SUMO-regulated genes showing that ectopic expression of MYC activated transcription in CDK9KO/Myc-CDK9 cells but not in CDK9KO/Myc-CDK9K/R cells. f The ability for PIAS1/SUMO-1 to repress and MYC to antagonize repression by PIAS1/SUMO-1 was examined in CDK9KO/Myc-CDK9 and CDK9KO/Myc-CDK9K/R cells by RNA-seq analysis. The RNAs were prepared from equivalent numbers of each group of cells and the levels of transcription were normalized against the spike-in yeast RNAs. g MYC antagonizes the repression on active to highly active transcribed genes by PIAS1/SUMO-1 in CDK9KO/Myc-CDK9 cells but not in CDK9KO/Myc-CDK9K/R cells. h Model showing that SUMO and MYC antagonistically control global gene expression through regulating CDK9 sumoylation, P-TEFb formation, and transcriptional elongation
To examine further if MYC regulates global transcription by antagonizing CDK9 sumoylation, we compared the ability of MYC to antagonize repression by PIAS1/SUMO-1 in CDK9KO/Myc-CDK9 and CDK9KO/Myc-CDK9K/R cells by RNA-seq analysis using spiked-in controls. We found that, as expected, expression of PIAS1/SUMO-1 repressed the global level of transcription in CDK9KO/Myc-CDK9 cells and co-expression of MYC substantially relieved the repression by PIAS1/SUMO-1 (Fig. 8f). However, neither expression of PIAS1/SUMO-1 led to a global repression, nor co-expression of MYC led to global activation of transcription in CDK9KO/Myc-CDK9K/R cells (Fig. 8f). The lack of an effect of PIAS1/SUMO-1 and PIAS1/SUMO-1/MYC upon global transcription in CDK9KO/Myc-CDK9K/R cells was further confirmed by analyzing the most highly expressed class 3 to class 5 genes (Fig. 8g). Detailed analysis of RNA-seq data revealed that among the PIAS1/SUMO-1-repressed genes, the majority were dependent on CDK9 sumoylation (97. 07%) (Supplementary information, Figure S12a). Of significance, the large majority of MYC-activated genes (94. 02%) belong to genes repressed in a CDK9 sumoylation-dependent manner (Supplementary information, Figure S12a). These RNA-seq data were further validated by qRT-PCR analyses of selective genes from each group (Supplementary information, Figure S12b). Taken together, we conclude that MYC activates global transcription largely by antagonizing transcriptional repression instigated by CDK9 sumoylation.
Discussion
In this study we discovered that SUMO represses the phosphorylation of Ser2 of the Pol II CTD and has an unexpected role in controlling global gene expression and that it does so by controlling the formation of active P-TEFb complex through sumoylation of CDK9. Furthermore, we provide compelling evidence that MYC amplifies gene expression primarily through its ability to antagonize CDK9’s sumoylation.
Numerous transcription factors and cofactors have been identified as sumoylated proteins and their modification by sumoylation has generally correlated with transcriptional repression.53,54 Multiple mechanisms have been proposed to explain SUMO-mediated transcriptional repression, including alteration of subcellular localization of transcription factors, inhibition of DNA binding, sumoylation of Pol II, and targeted histone deacetylation by recruitment of HDAC-containing corepressor complexes and other corepressors.30,54,55 However, it was not known prior to our study that SUMO had a role in the control of global Pol II CTD phosphorylation (Ser2P) and consequently global transcription through sumoylation of CDK9. We show that ectopic expression of PIAS1/SUMO-1 markedly inhibits, whereas knockdown of UBC9 strongly enhances phosphorylation of Ser2 but not Ser5 of the Pol II CTD (Fig. 1 and Fig. 2). This effect of SUMO involves no change in global histone acetylation or sumoylation (Fig. 1a and Supplementary information, Figure S2a, b, e). Rather, it correlates with sumoylation of CDK9 (Fig. 1e and Fig. 2). Mechanistically, sumoylation of CDK9 inhibits the formation of the P-TEFb complex (Fig. 3), which is primarily responsible for phosphorylation of Ser2 of the Pol II CTD and is critically important for productive transcriptional elongation. Indeed, we substantiated that transcriptional repression by PIAS1/SUMO-1 is associated with reduced transcriptional elongation (Fig. 4c, f, h and Fig. 5e, f). The finding that PIAS1/SUMO-1 failed to inhibit phosphorylation of Ser2 of the Pol II CTD and global transcription in the CDK9 sumoylation-resistant cell line CDK9KO/Myc-CDK9K/R provides compelling evidence that SUMO inhibits Ser2P and represses global transcription primarily through sumoylation of CDK9 (Fig. 6d, g).
A novel finding in this study is that CDK9 is extensively sumoylated in cells and its sumoylation is dynamically controlled by PIAS proteins and SENPs (Fig. 2). Although a growing number of proteins have been reported to be sumoylated,56,57 in most cases sumoylation of endogenous proteins is either undetectable or observed only for a small fraction of substrates, most likely due to highly active desumoylation by SENPs.28,45 In this regard, CDK9 is an exception since it is abundantly sumoylated in all cell lines we have tested (Fig. 2e-h). WB analysis using multiple CDK9-specific antibodies detected not only the expected 42 kDa CDK9 protein but also a series of higher molecular mass bands (Fig. 1c, Fig. 2e-h, Fig. 6a and Supplementary information, Figure S4). We believe these higher-molecular mass bands are CDK9 proteins sumoylated to various degrees, since their electrophoretic mobility changes in response to knockdown of CDK9 (Fig. 2e) and alteration of SUMO system (Fig. 2f-h). They were no longer detected, along with the 42 kDa CDK9 band, in the CDK9 KO and CDK9 sumoylation-resistant cells (CDK9KO/Myc-CDK9K/R) (Fig. 6a). The presence of multiple higher-molecular mass CDK9 bands in various cell lines implies that CDK9 sumoylation is a ubiquitous event and occurs at multiple sites. In support of this idea, the higher-molecular-mass CDK9 bands were detected in immunoprecipitations by anti-SUMO-1 and anti-SUMO-2/3 antibodies (Supplementary information, Figure S5b-c), and SUMO-1, SUMO-2 and SUMO-3 were detected by mass spectrometry in several of the immunoaffinity-purified CDK9 protein bands from gels (Supplementary information, Figure S6a-b). Moreover, our extensive mutagenesis effort consistently failed to abrogate CDK9 sumoylation (Supplementary information, Figure S6d). In fact, we were able to eliminate CDK9 sumoylation and its inhibition on P-TEFb complex formation only by generating a CDK9 K/R mutant with all 29 K residues replaced by R (Fig. 2i and Fig. 6a). This implies that sumoylation can occur at multiple sites on CDK9 and one or a small number of sumoylation events is sufficient to block the interaction of CDK9 with Cyc T1. The extensive sumoylation we observed on ectopically expressed Flag-CDK9 was also not due to sumoylation of the Flag tag, because sumoylation was not observed for Flag-CDK9K/R mutant (Supplementary information, Figure S13). The mechanisms underlying the abundant sumoylation of CDK9 remain to be elucidated, but it could be partly explained by the readily detectable interaction between CDK9 and both UBC9 and PIAS proteins (Fig. 2b, c). The abundant CDK9 sumoylation we observed in various cell lines also supports CDK9 sumoylation as a general mechanism for the control of global transcription. In support of this notion, we found that T-cell activation is associated with a substantial reduction of CDK9’s sumoylation and an increase of global transcription (Fig. 7).
We observed that CDK9 sumoylation strongly inhibits the CDK9-Cyclin T1 interaction and therefore the formation of P-TEFb complex (Fig. 3). Interestingly, the inhibition of interaction with Cyclin T1 was observed even with the presence of a considerable level of unmodified CDK9 (Fig. 3b). One likely explanation is that CDK9 desumoylation occurred during sample preparation, due to incomplete inhibition of SENP activity. In addition, we found that CDK9 interacts with itself (Supplementary information, Figure S8d). Interestingly, sumoylation of CDK9 inhibits its interaction with Cyc T1 but not with itself, suggesting that sumoylated CDK9 may sequester unsumoylated CDK9 from the process of P-TEFb complex formation (Supplementary information, Figure S8e). Together these mechanisms may explain why sumoylation of CDK9 can efficiently control the formation of the P-TEFb complex and thus transcriptional elongation. However, we cannot exclude the possibility that the PIAS family proteins may also inhibit the formation of the P-TEFb complex through their interaction with CDK9.
Previous studies have demonstrated a role of the 7SK snRNP complex in suppressing P-TEFb kinase activity.17,18 We found that sumoylated CDK9 is not associated with 7SK RNA (Fig. 3e) and HEXIM1 (Fig. 3b, d), a key protein of the 7SK snRNP. This observation is consistent with previous reports that 7SK RNA interacts with the Cyc T subunit of P-TEFb.58 Thus, CDK9 sumoylation acts to block the formation of the P-TEFb complex, whereas 7SK snRNP may further inhibit P-TEFb kinase activity by association with P-TEFb. In addition, the effect of the CDK9-K/R mutant on phosphorylation of Ser2 is unlikely due to loss of CDK9’s acetylation. SIRT7 was previously shown to deacetylate CDK9 and promote the release of P-TEFb from the inactive 7SK snRNA complex.59 While we confirmed CDK9’s deacetylation by SIRT7, SIRT7 overexpression had no effect on global Ser2P levels (Supplementary information, Figure S14), thus ruling out the possibility that the loss of CDK9 acetylation is responsible for elevated Ser2P in the CDK9KO/Myc-CDK9K/R cell line.
In this study we provide multiple lines of evidence that SUMO represses transcription globally. Three independent assays, namely assessment of BrU incorporated into newly synthesized RNAs by immunofluorescence, quantitative measurement of the total amount of RNAs from equivalent numbers of cells, and RNA-seq analysis with spike-in standards, all support the conclusion that SUMO has a repressive role on global levels of gene expression. Of these three methods, immunofluorescence assessment of BrUTP incorporation better reflects the levels of global gene expression since it represents the amount of newly synthesized RNAs within a 30 min time interval. Measurements of total RNAs and read counts from RNA-seq are likely to underestimate the effect of SUMO on global gene expression because in both cases not all RNAs analyzed are newly synthesized. Nevertheless, all three methods demonstrate a role for SUMO in the repression of global transcription. Furthermore, our RNA-seq analysis revealed that SUMO represses global transcription by suppressing the expression of actively transcribed genes. This observation is in good agreement with recent genome-wide analyses of chromatin-associated SUMO proteins and the findings that SUMO selectively represses actively transcribed genes in the stress response.32–34 It is also noteworthy, as elegantly elaborated by Richard Young and his colleagues, that normalization against spike-in RNA standards is required for RNA-seq profiling to give a quantitative analysis of effects on global gene expression.51 If RNA-seq data were not normalized against spike-in RNA standards but against total reads, the effect of ectopic expression of PIAS1/SUMO-1 or knockdown of UBC9 on global gene expression would not be detected (Supplementary information, Figure S15). Furthermore, one would mistakenly conclude that both ectopic expression of PIAS1/SUMO-1 and knockdown of UBC9 lead to up- and down-regulation of a similar number of genes (Supplementary information, Figure S12). Although we focused on global repression of Pol II transcription in this study, our preliminary data suggest that SUMO also has a role in the repression of Pol I-mediated rDNA transcription (Peng and Wong, unpublished data).
Another novel finding of our study is that MYC enhances global gene expression primarily through antagonizing CDK9’s sumoylation. As a proto-oncogene, MYC is highly associated with cell proliferation and aberrant MYC activation has been linked to many human cancers.60 In addition to its gene-specific role in transcription,61 MYC has been shown to amplify global gene expression and does so possibly through regulating the release of Pol II pausing by interaction with P-TEFb.22,24,61 We have confirmed a role for MYC in regulation of global gene expression. Importantly, we demonstrate that MYC drastically inhibits CDK9 sumoylation (Fig. 7b, e, g, h). We observed that T-cell activation, a model system for MYC-stimulated cell proliferation and activation of global gene expression, is associated with a substantial reduction of CDK9 sumoylation (Fig. 7i). Importantly, we found that MYC-stimulated global gene activation is not observed in the CDK9KO/Myc-CDK9K/R cell line (Fig. 8), indicating that MYC amplifies transcription primarily by antagonizing sumoylation of CDK9. The mechanism(s) by which MYC efficiently antagonizes the sumoylation of CDK9 need to be determined in future.
In sum, we demonstrate in this study that SUMO has a novel role in the control of global cellular transcription, via the sumoylation of CDK9 and the control of transcriptional elongation. We also demonstrate that MYC amplifies gene expression by antagonizing the sumoylation of CDK9. As both SUMO modification and P-TEFb function are evolutionally conserved processes in eukaryotic cells, we propose that the regulation of global gene expression via the sumoylation of CDK9 is also a conserved mechanism in eukaryotic cells. Although the vast majority of transcriptional studies focus on regulation of specific genes or specific transcriptional programs, our study, together with the previous studies on MYC,22,24,61 argues that regulation of global gene expression is likely a common and fundamental mechanism for the regulation of gene expression in eukaryotic cells. Furthermore, given the broad role of MYC in oncogenesis and our new finding that MYC amplifies gene expression by antagonizing the sumoylation of CDK9, modulating CDK9’s sumoylation is likely to be a feasible therapeutic approach for so-called “undruggable” MYC related cancers.
Methods
Plasmids and antibodies
The plasmids for pCDNA3.0-HA-PIAS1m(C346SC351SC356S) was derived from pCDNA3.0-HA-PIAS1 by site-directed mutagenesis. Various PIAS expression constructs were generated by sub-cloning the corresponding coding sequences into the pCDNA3.0-HA vector. The Myc-CDK9 K/R mutant (in which all lysines were mutated to arginines) was synthesized by Biosun and then inserted into the pCDNA3.1-3× Myc vector and the construct verified by DNA sequencing. For generating stable HeLa cell lines expressing Myc-CDK9 or Myc-CDK9K/R mutant, the corresponding CDK9-encoding sequence was subcloned into the pPYCAGIP vector. The antibodies used in this study were listed as follows: anti-CDK9(C-20) (Santa Cruz,#sc-484), anti-CDK9 (CY5832, Abway), anti-CDK9 (Abclonal # A1564), anti-Cyclin T1 (H-245) (SantaCruz,#sc-10750), anti-Pol II (C-21) (SantaCruz,#sc-900), anti-Pol II 8WG16 (Covance), anti-Pol II CTD Ser2P (abcam#ab5095), anti-Pol II CTD Ser2P (H5, Covance), anti-Pol II CTD Ser5P (abcam#ab5131), anti-Cyclin H (SantaCruz,#sc-855), anti-CDK7 (SantaCruz,#sc-7433), anti-MAT1 (SantaCruz,#sc-6234), anti-SUMO1 (SantaCruz,#sc-5308), anti-BRD4 (abcam#ab128874), anti-CDK12 (CST#11973), anti-HEXIM1 (homemade Polyclonal antibody), anti-Histone H3 (Huaan#M130918), anti-H4 (Proteintech#16047-1-AP), anti-BrUTP (Sigma#B-2531), anti-AcH4 (Millipore#05-1355), anti-AcH3 (homemade), anti-HA (Abmart#M2003), anti-GFP (Abmart#M2004L), anti-Flag (Sigma#F7425), anti-Myc (Abmart#M2002), anti-hSpt5 (SantaCruz,#sc-390961), and anti-NELF-E (SantaCruz,#sc-32912)
Western blotting analysis, co-immunoprecipitation, and immunofluorescence staining
The Western blot, co-immunoprecipitation and immunofluorescence staining analyses were performed essentially according to the standard protocols,62,63,9 using the indicated antibodies. For examining SUMO-conjugated proteins directly by Western blot, cells were lysed in denaturing buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 4% SDS, 1 mM EDTA, 8% glycerol, 50 mM NaF, 1 mM DTT, 1 mM PMSF and protein inhibitors) supplemented with 20 mM NEM and heated at 90 °C for 10 min. For immunoprecipitation, lysates were further diluted to 0.1% SDS and immunoprecipitated using anti-CDK9 antibody or other indicated antibodies at 4 °C overnight, followed by Western blot analysis. Relative levels of Ser2P of Pol II CTD were quantified using Image-J software, and the value of Ser2P in control samples was set as 1.0.
In vitro sumoylation and kinase activity assay
The in vitro sumoylation assay was carried out as previously described.60 In brief, 3 μg GST-CDK9 was incubated in 20 μL reactions containing 50 mM Tris-HCl pH7.5, 5 mM MgCl2, 2 mM ATP, 1 mM DTT, 200 ng SAE1/2, 400 ng Ubc9, 2.5 μg SUMO-1. The reactions were incubated at 37 °C for 2.5 h, stopped by addition of the 2× SDS loading buffer and analyzed by Western blotting with anti-CDK9 antibody. The effect of sumoylation on CDK9 kinase activity of Myc-CDK9 and Myc-CDK9K/R mutant was performed as follows: The Myc-CDK9 and Myc-CDK9K/R stable cell lines deleted for endogenous CDK9 were transfected with HA-PIAS1 and GFP-SUMO-1. Two days after transfection, Myc-CDK9 and Myc-CDK9K/R proteins were immunoaffinity-purified using anti-Myc agarose beads. The resulting beads were washed three times with ice-cold kinase activity reaction buffer (50 mM Tris-HCl pH7.5, 1 mM EGTA, 10 mM KCl, 10 mM MgCl2, 1 mM DTT) and incubated at 30 °C for 1 h in 20 μL reaction buffer containing 5 μg of GST-Pol II CTD and 2 mM ATP. The levels of Ser2P were examined by Western blotting using anti-Ser2P antibody.
Knockdown with siRNAs or shRNAs and quantitative RT-PCR
Knockdown of target RNAs by siRNAs or shRNAs was essentially as described. All siRNAs were synthesized by Genepharma. The vector for shRNAs was pLKO.1. The sequences for siRNAs and shRNAs are listed in Supplementary information, Table S1. For qRT-PCR analysis, total RNAs were extracted from the inducated cell lines and reverse transcribed using kits purchased from Transgene. The primer sequences used in the qRT-PCR are listed in Supplementary information, Table S1.
BrUTP incorporation assay
The BrUTP incorporation assay was carried out essentially as described.64 Briefly, HeLa cells were transfected with HA-PIAS1 or HA-PIAS1m mutant alone or together with SUMO-1. Two days after initial transfection, BrUTP was transfected into cells with Lipofectamine 2000 and cells were incubated at 37 °C for 30 min. Cells were then fixed with 4% paraformaldehyde and stained with anti-BrUTP antibody.
Generation of CDK9 knockout cell lines with expression of Myc-CDK9 or Myc-CDK9K/R mutant
The HEK293T cells were first transfected with Myc-CDK9 or Myc-CDK9K/R mutant to establish cell lines stably expressing Myc-CDK9 or Myc-CDK9K/R mutant. Lines with comparable expression of endogenous CDK9 and Myc-CDK9 or Myc-CDK9K/R mutant were selected and the endogenous CDK9 gene deleted using the CRISPR-Cas9 technology as described.65,2 The clones having knockouts of the endogenous CDK9 and showing comparable expression of Myc-CDK9 and the Myc-CDK9K/R mutant were selected based on Western blot analysis. The guide RNA sequences for CDK9 are listed in Supplementary information, Table S1.
Senp1−/− and Senp2−/− MEFs
The Senp1−/− and Senp2−/− MEFs and their routine culture were previously described.45,46[,4
Preparation of naïve T cells and activation by anti-CD3 and anti-CD28
Preparation of Naïve T Cells and Activation by Anti-CD3 and Anti-CD28 was as described.52
RNA-seq and ChIP-seq assay
For RNA-seq analysis of global gene expression, equivalent numbers of cells from each group were used for preparation of RNAs. The same amount of spike-in yeast RNAs was added to each RNA sample. The construction of libraries and deep sequencing analysis were carried out by BerryGenomics. Raw reads counts of all genes annotated by the UCSC database (hg19) in all samples were gained from Tophat2 (version 2.1.0)66 and htseq-count (version 0.6.0)67 softwares. Read counts for each sample were normalized against the reads of spike-in yeast RNAs using R package DESeq2 (version 1.8.2).68 GO and pathway enrichment analysis were performed by a comprehensive gene set enrichment analysis web server Enrichr. ChIP-seq peaks were identified by MACS14 (version 1.4.2)69 and ChIP enrichment profile on important gene features was calculated through package CEAS (version 1.0.2).70
Statistical analysis
All statistical tests were performed using the unpaired Student’s test and p < 0.05 was considered as statistically significant. In all the results, “*” denotes p < 0.05, “**” denotes p < 0.01, and “ns” denotes no significant difference.
Electronic supplementary material
Acknowledgements
We thank Dr.Jinqiu Zhou for providing yeast RNA and Dr. Xin-Hua Feng for PIAS constructs. We thank members of Wong’s laboratory for valuable discussion. This study is supported by grants from the National Natural Science Foundation of China (81272287 and 30971640 to J.L. and 81530078 to J.W.), the Ministry of Science and Technology of China (2015CB910402 and 2017YFA0504201 to J.W.), the National Institutes of Health (CA103867 to C.M.C.), Cancer Prevention Research Institute of Texas (RP110471 and RP140367 to C.M.C.), and the Welch Foundation (I-1805 to C.M.C.). F.Y. was also supported by “Outstanding doctoral dissertation cultivation plan of action” from ECNU (PY2015037).
Author contributions
F.Y., G.S., J.L. and J.W. conceived the idea for this project. F.Y., G.S., S.C., Z.W., Y.P., Z.W., D.W. designed and conducted the experiments and interpreted the data. J.C. and T.S. performed bioinformatic analysis of RNA-seq and ChIP-seq data. Y.Z performed T cell activation experiment. N.X. and J.C provided Senp1−/− and Senp2−/− MEFs and performed related experiments. L.L. carried out mass spectromic analysis of CDK9 sumoylation. S.-Y.W. and C.-M.C. performed in vitro kinase assay. J.W. and J.L. wrote the manuscript with inputs from all the other authors.
Competing interests
The authors declare no competing interests.
Contributor Information
Jiwen Li, Email: jwli@bio.ecnu.edu.cn.
Jiemin Wong, Email: jmweng@bio.ecnu.edu.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0023-9.
References
- 1.Sainsbury S, Bernecky C, Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:129–143. doi: 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- 2.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Bushnell DA, Kornberg RD. RNA polymerase II transcription: structure and mechanism. Biochim. Biophys. Acta. 2013;1829:2–8. doi: 10.1016/j.bbagrm.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hochheimer A, Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- 5.Guo J, Price DH. RNA polymerase II transcription elongation control. Chem. Rev. 2013;113:8583–8603. doi: 10.1021/cr400105n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi Y, Shibata H, Handa H. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta. 2013;1829:98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 11.Wada T, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi Y, et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/S0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 13.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J. Biol. Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 14.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 15.Gu B, Eick D, Bensaude O. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 2013;41:1591–1603. doi: 10.1093/nar/gks1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 19.Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 2012;13:543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- 20.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu. Rev. Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sano M, et al. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 22.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hay RT. Decoding the SUMO signal. Biochem. Soc. Trans. 2013;41:463–473. doi: 10.1042/BST20130015. [DOI] [PubMed] [Google Scholar]
- 26.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang XJ, Chiang CM. Sumoylation in gene regulation, human disease, and therapeutic action. F1000Prime Rep. 2013;5:45. doi: 10.12703/P5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nayak A, Muller S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014;15:422. doi: 10.1186/s13059-014-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Ouyang J, Gill G. SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics. 2009;4:440–444. doi: 10.4161/epi.4.7.9807. [DOI] [PubMed] [Google Scholar]
- 31.Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell. 2004;13:611–617. doi: 10.1016/S1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 32.Rosonina E, Duncan SM, Manley JL. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 2010;24:1242–1252. doi: 10.1101/gad.1917910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neyret-Kahn H, et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013;23:1563–1579. doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niskanen EA, et al. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 2015;16:153. doi: 10.1186/s13059-015-0717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc. Natl Acad. Sci. USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishida T, Yasuda H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 2002;277:41311–41317. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- 37.Wong J, Shi YB, Wolffe AP. A role for nucleosome assembly in both silencing and activation of the Xenopus TR beta A gene by the thyroid hormone receptor. Genes Dev. 1995;9:2696–2711. doi: 10.1101/gad.9.21.2696. [DOI] [PubMed] [Google Scholar]
- 38.Huang ZQ, Li J, Sachs LM, Cole PA, Wong J. A role for cofactor-cofactor and cofactor-histone interactions in targetingp300, SWI/SNF and mediator for transcription. EMBO J. 2003;22:2146–2155. doi: 10.1093/emboj/cdg219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao SH, et al. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 2000;275:28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- 40.Rossi DJ, et al. Inability to enter S phase and defective RNA polymerase II CTD phosphorylation in mice lacking Mat1. EMBO J. 2001;20:2844–2856. doi: 10.1093/emboj/20.11.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larochelle S, et al. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat. Struct. Mol. Biol. 2012;19:1108–1115. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartkowiak B, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devaiah BN, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc. Natl Acad. Sci. USA. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shore SM, Byers SA, Dent P, Price DH. Characterization of Cdk9(55) and differential regulation of two Cdk9 isoforms. Gene. 2005;350:51–58. doi: 10.1016/j.gene.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang X, et al. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016;17:581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- 48.Peng J, Marshall NF, Price DH. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 1998;273:13855–13860. doi: 10.1074/jbc.273.22.13855. [DOI] [PubMed] [Google Scholar]
- 49.Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laitem C, et al. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nat. Struct. Mol. Biol. 2015;22:396–403. doi: 10.1038/nsmb.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loven J, et al. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 54.Cubenas-Potts C, Matunis MJ. SUMO: a multifaceted modifier of chromatin structure and function. Dev. Cell. 2013;24:1–12. doi: 10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chymkowitch P, Nguea PA, Enserink JM. SUMO-regulated transcription: challenging the dogma. BioEssays. 2015;37:1095–1105. doi: 10.1002/bies.201500065. [DOI] [PubMed] [Google Scholar]
- 56.Makhnevych T, et al. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell. 2009;33:124–135. doi: 10.1016/j.molcel.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 57.Tammsalu T, et al. Proteome-wide identification of SUMO2 modification sites. Sci. Signal. 2014;7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 59.Blank MF, et al. SIRT7-dependent deacetylation of CDK9 activates RNA polymerase II transcription. Nucleic Acids Res. 2017;45:2675–2686. doi: 10.1093/nar/gkx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sabo A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi G, et al. PHD finger protein 2 (PHF2) represses ribosomal RNA gene transcription by antagonizing PHF finger protein 8 (PHF8) and recruiting methyltransferase SUV39H1. J. Biol. Chem. 2014;289:29691–29700. doi: 10.1074/jbc.M114.571653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang L, et al. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell. 2014;55:537–551. doi: 10.1016/j.molcel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 64.Haukenes G, Szilvay AM, Brokstad KA, Kanestrom A, Kalland KH. Labeling of RNA transcripts of eukaryotic cells in culture with BrUTP using a liposome transfection reagent (DOTAP) BioTechniques. 1997;22:308–312. doi: 10.2144/97222st03. [DOI] [PubMed] [Google Scholar]
- 65.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.