Abstract
AICAR (Acadesine) is a pharmacological precursor of purine nucleotide biosynthesis with anti-tumoral properties. Although recognized as an AMP mimetic activator of the protein kinase AMPK, the AICAR monophosphate derivative ZMP was also shown to mediate AMPK-independent effects. In order to unveil these AMPK-independent functions, we performed a transcriptomic analysis in AMPKα1/α2 double knockout murine embryonic cells. Kinetic analysis of the cellular response to AICAR revealed the up-regulation of the large tumor suppressor kinases (Lats) 1 and 2 transcripts, followed by the repression of numerous genes downstream of the transcriptional regulators Yap1 and Taz. This transcriptional signature, together with the observation of increased levels in phosphorylation of Lats1 and Yap1 proteins, suggested that the Hippo signaling pathway was activated by AICAR. This effect was observed in both fibroblasts and epithelial cells. Knockdown of Lats1/2 prevented the cytoplasmic delocalization of Yap1/Taz proteins in response to AICAR and conferred a higher resistance to the drug. These results indicate that activation of the most downstream steps of the Hippo cascade participates to the antiproliferative effects of AICAR.
Abbreviations: AICAR, 5-Aminoimidazole-4-carboxamide-1-β-D-ribofuranoside; AMPK, adenosine monophosphate-activated protein kinase; CTGF, connective tissue growth factor (CCN2); CYR61, cysteine-rich angiogenic protein 61; HPRT, hypoxanthine-guanine phosphoribosyl transferase; LATS, large tumor suppressor kinase; MEFs, murine embryonic fibroblasts; YAP1, Yes-associated protein 1; TAZ, transcriptional coactivator with PDZ-binding motif; ZMP, AICAR monophosphate
Introduction
AICAR (5-Aminoimidazole-4-carboxamide-1-β-D-ribofuranoside) is a pharmacologically active compound entering cells via purine transporters and being converted to its active-monophosphorylated form ZMP by adenosine kinase [1], [2]. ZMP mimics AMP as a low-energy charge signal by activating the heterotrimeric AMP-activated protein kinase (AMPK), which promotes integrated regulatory functions at the cellular and tissular levels [3].
In mammal cells, an AICAR-induced cytotoxic response is observed, leading to both apoptotic and non-apoptotic death mechanisms [4], [5], [6], [7], [8]. Interestingly, AICAR is more cytotoxic for aneuploid cells compared to their euploid counterparts [6], [9], which is of a great therapeutic interest since aneuploidy is observed in human in 90% of solid tumors and 85% of hematopoietic malignancies [10]. Moreover, AICAR showed antitumoral properties in vivo in different animal models [6], [11], [12] and is well tolerated in mice and human, even at elevated and repeated administration doses [13], [14]. AICAR has been subjected to Phase I/II clinical trials for treating chronic lymphocytic leukemia [14].
Although AMPK activation has been initially recognized as a prevalent effect of AICAR, recent data indicated that many effects mediated by this compound are in fact AMPK-independent. These include inhibition of proliferation and limited tumor growth in vivo [2], [5], [15], [16], [17]. These intriguing properties of AICAR therefore motivate the search for alternative targets of its metabolic derivative ZMP that could be exploited therapeutically.
In order to get further insight on the AMPK-independent action of AICAR, we carried out a kinetic analysis of the effects of AICAR on the transcriptome changes in murine embryonic fibroblasts (MEFs) invalidated for AMPK (α1 and α2 subunits) [18]. We identified Lats1 and Lats2 as early AICAR-induced genes and showed that the encoded proteins (herein referred as to Lats1/2) are potent mediators of the cell response to this drug. Lats1/2 proteins are downstream transducers of the core Hippo pathway known to play a pivotal role in cells and tissues [19], [20]. Lats1/2 display pleiotropic functions, which include the inhibition of the two related co-transcriptional factors Yap1 (Yes-associated protein 1) and Taz (Transcriptional coactivator with PDZ-binding motif). Knockdown of Lats1/2 established that some of the cellular effects triggered by AICAR result of the activation of this central pathway controlling cell cycle and density. These findings provide a rationale to analyze the effects of AICAR and motivate the research of derivatized compounds of AICAR with therapeutic perspectives.
Materials and Methods
Reagents
Culture media was from Invitrogen. Specific antibodies were as follows: Taz (#560235) (BD Pharmingen); actin (#A2668) (Sigma); AMPKα (#2532), PARP (#9542), LATS2 (#13646), LATS1 (#3477), phospho-(Thr1079)-LATS1 (#8654), LC3A/B (#4108), cleaved-(Asp175)-caspase-3 (#9604 and 9664), YAP1/TAZ (#8418), YAP1 (#14074 and #4912), phospho-(Ser127)-YAP1 (phospho-(Ser112)-Yap1 in mice) (#4911) were from Cell Signaling; LATS2 (#NB200–199, NovusBio); DAPI and secondary antibodies labeled with AlexaFluor488 or with AlexaFluor555 were from ThermoFisher Scientific. Primers (Table S1) and siRNA (see below) were from Sigma. Control siRNA was from Cell Signaling (#6568). AICAR (#A611700, Toronto Research Chemicals, Canada) was solubilized in water and filtered at 0.22 μm. Its concentration was determined spectrophotometrically (A269; pH = 7; εM = 12,700 M−1/cm−1).
Cell Culture
Human RPE-1 (ATCC; CRL-4000) cells were grown in DMEM, 4.5 g/L glucose supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Wild-type and knock-out mouse embryonic fibroblasts (MEFs) for both AMPKα1 and AMPKα2 subunits (MEF-dKO) were obtained from the U1016-INSERM, Paris (Laderoute et al. [18]) and the absence of AMPKα in MEF-dKO was confirmed by Western blot (see text). The WST-1 cell viability assay was performed in 96-well plates as previously described [2]. Cell proliferation assays were performed in 24-well plates and cells were counted using a Multisizer 4 Coulter Counter (Beckman). Each time point was done in triplicate, and the averages ± STD were calculated. Experiments were repeated three times.
Preparation of cRNA, Microarray Hybridization and Gene Expression Profiling
Microarray analysis was performed at the Affymetrix transcriptome Platform, CHRU-Inserm U1040, Montpellier France (http://www.chu-montpellier.fr/fr/irmb/). MEF-dKO were grown in complete DMEM medium in the presence or absence of 1 mM AICAR and harvested at the indicated times (Final density ~15,000 cells/cm2). Total RNA was extracted using the RNeasy kit (Qiagen, Santa Clarina, CA) and the quality of each sample was assessed on a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA). Affymetrix Gene Chip 3`IVT Express Kit Bundle was used to generate amplified RNA. cRNAs were hybridized to Mouse MG-430 PM Array Strip Kit (44,000 probesets). All steps were performed according to the standard protocol of the manufacturer. Fluorescence intensities were quantified and analyzed using the Expression Console v 1.2 software. The converted digital intensity values were then converted into cell intensity files using the Affymetrix GeneAtlas Software. The fold change in gene expression corresponds to the ratio between each incubation times with AICAR and control without AICAR. Significance analysis of microarrays (SAM) analysis was applied in the different samples with 400 permutations, a fold change of 2 and a false discovery rate (FDR) < 3.55% (see Table S2A). All steps were conducted according to the MIAME (Minimum Information About a Microarray Experiment) checklist [21]. Annotation of genes was performed using NetAffx (http://www.affymetrix.com). Affymetrix IDs were submitted to the DAVID analysis tool (http://david.abcc.ncifcrf.gov/home.jsp), the Netaffx Analysis Center (https://www.affymetrix.com/analysis/compare/index.affx) and the KEGG pathway database (www.genome.jp/kegg/pathway.html) to depict biological processes associated with the functions of regulated genes. Data files were deposited in the Gene Expression Omnibus (GEO) database (accession number, GSE106460; www.ncbi.nlm.nih.gov/geo).
Small Interfering RNA Knockdown
Cells were plated at a density of 5.104 cells per well in six-well plates. Small interfering RNA (siRNA) against human and mouse LATS1 (sihmLATS1): CACGGCAAGAUAGCAUGGAUU; human and mouse LATS2 (sihmLATS2-y): GAAGAUUGUAUUUAUGGUAAA; mouse AMPKα1/2_human AMPKα2 (simAMPKα1/2 sihAMPKα2): GAGAAGCAGAAGCACGACG; human AMPKα1 (sihAMPKα1): AUGAUGAAAUUACAGAAGCCA were from Sigma. Non-targeting siRNA (#D-001810-01-20) was from Dharmacon (Lafayette, CO). Transfection was performed for 6–15 h using lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocol, with total amounts of siRNA at a final concentration of 10–60 nM, as indicated in the figure legends.
Immunoblot
Cells in culture dishes were collected using a rubber policeman and snap frozen. Proteins were extracted at 4°C with RIPA buffer containing phosphatase inhibitors (PhosStop EasyPack, ROCHE) and protease inhibitors (Complete ULTRA Tablets, mini EasyPack, ROCHE). Protein content was determined by using a BCA protein assay kit (Pierce). Proteins were resolved on 7 or 10% SDS-PAGE, transferred to a PVDF blotting membrane (Hybond P 0.45 μm PVDF membrane; Amersham) and probed using primary antibodies. Secondary antibodies coupled to HRP were used for revelation. Densitometric analyses were carried out using ImageJ (http://rsbweb.nih.gov/ij/download.html) and results were normalized to the actin loading control.
Real-Time PCR
Cells in culture dishes were solubilized in Trizol (Life Technologies) and frozen at −80°C until use. RNA extraction, quantification and gene expression analyses by RT-qPCR were performed as previously reported [22]. Gene expression was normalized to actin and HPRT1. Results were the mean of triplicate determinations ± SD.
Immunofluorescence
MEFs and RPE-1 cells were seeded at the density of 7,000 cells/cm2 on 18-mm glass coverslips N° 1.5H (Marienfeld, Germany). After one day of incubation, cells were washed and incubated with AICAR for the indicated times. They were then washed with PBS, fixed for 20 min with 4% paraformaldehyde in PHEM, permeabilized with 0.1% Triton X-100 in PBS and blocked in 0.1% BSA in PBS for 15 min. Cells were then incubated first for 60 min at 37°C in the blocking buffer containing anti-Yap1 or anti-Taz antibodies, followed by incubation with AlexaFluor 488-labeled anti-rabbit antibodies (Yap1) or AlexaFluor 555-labeled anti-mouse antibodies (Taz). Nuclei were counter-stained with DAPI. Microphotographs were obtained using an Olympus Ix81 inverted microscope (Olympus, Rungis, France) with a 40× oil immersion objective, NA 1.3. Image analyses were carried out using ImageJ free software. Statistical analyses were based on the quantification of the fluorescent signal on at least 10 different fields (>50 cells per condition).
Results
AMPK-Independent Transcriptional Response to AICAR in MEF-dKO
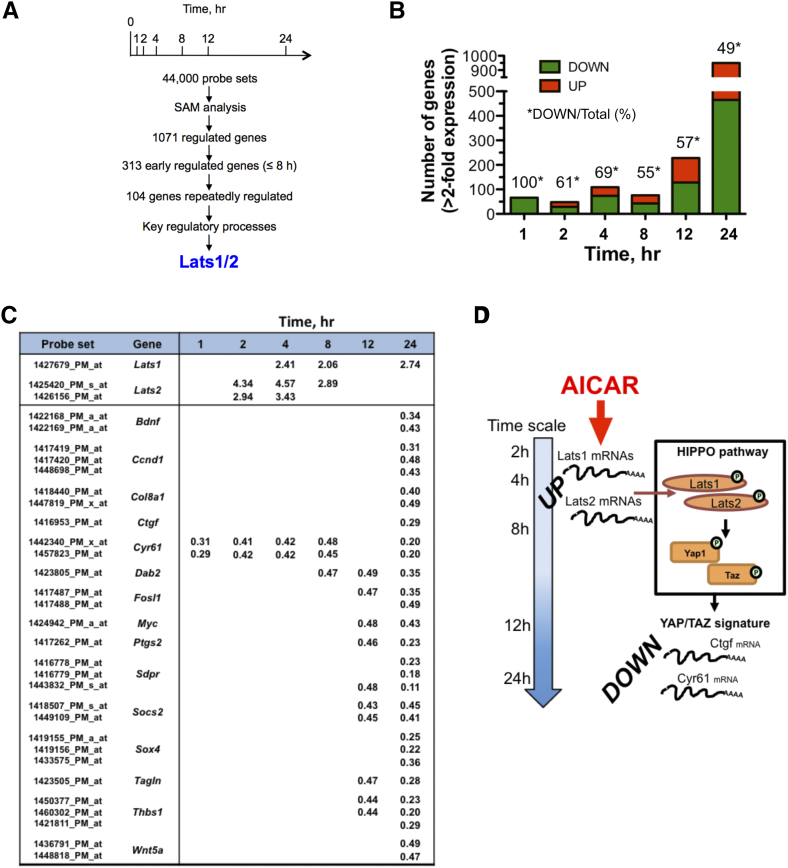
In order to get a dynamic view of the transcriptomic response to AICAR, a 24-h kinetic analysis was carried out in MEF-dKO AMPK in the presence of 1 mM AICAR, a condition resulting in robust ZMP accumulation in both MEF-dKO and MEF-wt (Figure S1). We observed that the absence of AMPK did not significantly affect the propagation of MEFs in standard culture conditions (Figure S2A). However, the MEF-dKO were slightly more sensitive than MEF-wt to the highest dose of the drug, as monitored by using a cytotoxic assay (Figure S2, B and C) and analyses of the apoptotic process using standard procedures (Figure S2, D and E). By assessing the transcriptome across seven time points in MEF-dKO (Figure 1A), significance analysis of microarray (SAM) revealed that 1325 probesets (1071 genes; 19.1% redundancy), corresponding to ~2.9% of the total number of the probesets available on the chips, were AICAR-responsive (minimum 2-fold change; FDR (%) < 3.55, values given in Table S2A). The number of genes modulated under AICAR treatment increased with time (Figure 1B). A majority of genes were down regulated at early times (100% at t = 1 h), while a similar number of genes were up- and down- regulated from 8 to 24 h of incubation. The variation of gene expression pattern indicated that 627 AICAR-responsive probesets (47.3%) were consistently up-regulated overtime whereas 697 probesets (52.6%) were strictly down-regulated (Table S2A).
Figure 1.
Gene expression analysis highlights the up-regulation of Lats1/2 under AICAR treatment. Gene profiling analyses were performed in murine embryonic fibroblasts that have been invalidated for the two catalytic subunits of the AMPK (MEF-dKO). The kinetic of AICAR-mediated gene expression was obtained from cells stimulated or not with 1 mM AICAR. Functional annotation clustering was performed by using Netaffx Analysis Center (Affymetrix database) as well as the DAVID Bioinformatics Resources v6.8. (A) General schematic of the procedure that led to a focus on the up-regulation of Lats1/2 transcripts. (B) Histogram representation of the number of genes Up- and Down-regulated as a function of time. (C) Early up-regulation of Lats1/2 mRNA in response to AICAR treatment, and late repression of reporters of Yap1/Taz nuclear functions. Values indicate the fold-increases associated with AICAR treatment. (D) Schematic depicting the model of a two-wave effect of AICAR on the gene expression pattern in MEF-dKO: Lats1 and Lats2 transcripts increased (UP) after 2 h of treatment with AICAR, and the Yap1/Taz-type signature (DOWN) was essentially observed at times 12 h and 24 h.
An early and sustained up-regulation of transcripts encoding the large tumor suppressor proteins Lats1 and Lats2 was detected along the time-course experiment (Figure 1C). Lats1/2 are protein kinases and members of the Hippo pathway core, which is critically involved in mammals in several processes including growth inhibition, survival, epithelial to mesenchymal transition and the response to stressors. A major Hippo signaling outcome is the inhibition by Lats1/2 of the two highly related oncoproteins Yap1 (Yes-associated protein 1) and Taz (transcriptional co-activator with PDZ-binding motif) [19], [20]. We therefore questioned the possible impact of AICAR on the transcriptional activities of Yap1 and Taz. Comparison of transcriptome signatures in human cells overexpressing YAP1 or TAZ [23] with transcripts responding to AICAR revealed an overlap of ~200 genes. Notably, most of these genes were detected lately (12 and 24 h) upon AICAR treatment, well after the onset of Lats1/2 up-regulation (Figure 1C). Several of these genes, including Ccnd1, Ctgf, Cyr61, Irs1 and Thbs1 (Figure C and Table S2A) are well-recognized Hippo targets [24], [25], [26]. Further combination of literature and data mining established an expandable common gene signature of AICAR and Yap1/Taz, currently representing ~19% of the total number of genes regulated by AICAR (Table S2B). The extent of this overlap is likely underestimated here, since comparisons were carried out between mice (AICAR signature) and human (YAP1/TAZ signatures), the gene symbol attribution being somewhat different in the two organisms. The GO-term enrichment profile of AICAR-responsive genes predominantly points to the regulation of gene expression, nucleotide binding and mRNA processing (Table S3). Of note, AICAR treatment could also have concurrent effects on cell proliferation by the coordinate inhibition of several genes functionally linked to the epidermal growth factor (EGF) and to the insulin/insulin-like growth factor (IGF) signaling pathways (Table S4). Repression of several of these genes is expected to contribute to the inhibition of Yap1/Taz functions [27], [28]. Most of these events occurred later in time and are likely subordinated to primary events triggered by AICAR. Overall, analysis of the gene-expression profile pointed to the Hippo signaling pathway as a major target of AICAR in MEF-dKO (Figure 1D).
The Hippo Pathway is Activated Under AICAR Treatment
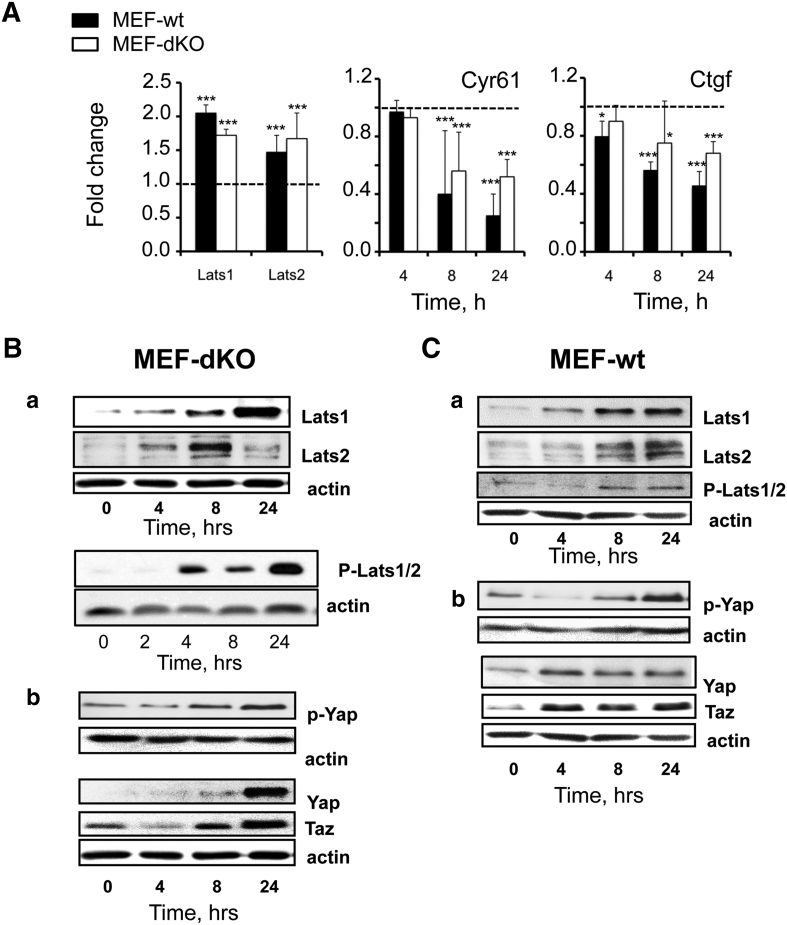
RT-PCR analysis validated the fact that Lats1 and Lats2 mRNAs were increased in MEF-dKO in response to AICAR (Figure 2A). These effects were associated to the progressive decrease in the level of both Cyr61 and Ctgf transcripts, two surrogate markers of Hippo pathway activation and Yap1/Taz inhibition. Importantly, these effects were found whether AMPK was present or not, i.e. in both MEF-dKO and MEF-wt (Figure 2A).
Figure 2.
AMPK-independent activation of the Hippo pathway under AICAR treatment. RT-qPCR and Western blot analyses of MEF-wt and MEF-dKO. Exponentially growing cells were stimulated or not with AICAR for different times and cell extracts were subjected to RT-qPCR or to Western blot analyses. Specific antibodies of the following proteins and phosphorylations were used: Lats1, Lats2, phospho-(Thr1079)-Lats1/2, Yap1, phospho-(Ser127)-Yap. A) RT-qPCR analyses: early induction (t = 4 h) of Lats1/2 transcripts and late transcripts repression of the reporter genes Cyr61 and Ctgf in MEF-wt and MEF-dKO. Dotted lines represent control values in the absence of AICAR. (*P < .05; **P < .01; ***P < .001). (B-C) Western blot: Lats1/2 were phosphorylated in MEF-dKO (B) and in MEF-wt (C) in response to AICAR. Lats1 and Lats2 proteins were induced following treatment with AICAR and phosphorylation/inhibition of Yap1/Taz activities was detected in both cell types.
Lats1/2 protein levels were also increased after 4 h of incubation in the two isogenic cell lines as measured by Western blot (Figure 2, Ba and Ca). Activation of the Hippo pathway under AICAR treatment was further confirmed by the increased phosphorylation of the proteins Lats1/2 using an antibody directed against phospho-(Thr1079)-LATS1 that also partially cross-reacts with p-LATS2 (Figure 2, Ba and Ca). Phosphorylation of Lats1/2 was accompanied in both cell lines by the increase of Yap1 phosphorylation (p-S112-Yap1) (Figure 2, Bb and Cb), a marker of Yap1 inhibition by Lats1/2 [19], [20]. The total amounts of Yap1 and Taz proteins were also increased under AICAR treatment, in agreement with results obtained by others in HepG2 cells [29]. Thus, Lats1/2 were up-regulated at both the mRNA and protein levels upon AICAR treatment, and this effect was associated with their activating phosphorylation. These events did not require the presence of AMPK, as they were observed in both MEF-dKO and MEF-wt cells.
AICAR Treatment Promoted Nuclear Delocalization of YAP1/TAZ
One reported consequence of the activation of the Hippo pathway is the nuclear exclusion of YAP1/TAZ [19], [20]. Hence, we performed immunofluorescence analyses on MEF-wt and MEF-dKO subjected or not to AICAR treatment. As shown in Figure S3, A-B, both Yap1 and Taz immunoreactivities were predominantly nuclear in control conditions. Besides, a modest but reproducible delocalization of the signal (< 50%) was measured in these cells at different incubation times with AICAR. We considered that this limited nuclear exclusion of Yap1/Taz in MEFs could be the consequence of the cellular immortalization by SV40 large T antigen [18], the Hippo pathway being perverted by viral oncoproteins [30], [31]. We therefore designed a control experiment based on the well-described subcellular delocalization of YAP1 in cell–cell contact condition at high cell density [26]. Strikingly, endogenous signals associated to a high cell density did not lead either to a substantial delocalization of Yap1 in immortalized MEFs, whereas a robust translocation was observed in human retinal epithelial (RPE-1) cells (Figure S3C). In keeping with this, we observed that primary cultured MEF readily responded to AICAR treatment by the unambiguous decrease of both Yap1 and Taz immunoreactivities in the nuclear compartment (Figure S3D). Thus, subcellular delocalization of Yap1/Taz under AICAR treatment was readily observed in primary cultured MEFs and was attenuated in SV40-immortalized MEFs. Noticeably, inhibition of Yap1 in immortalized MEFs was dissociated from its complete cytoplasmic translocation.
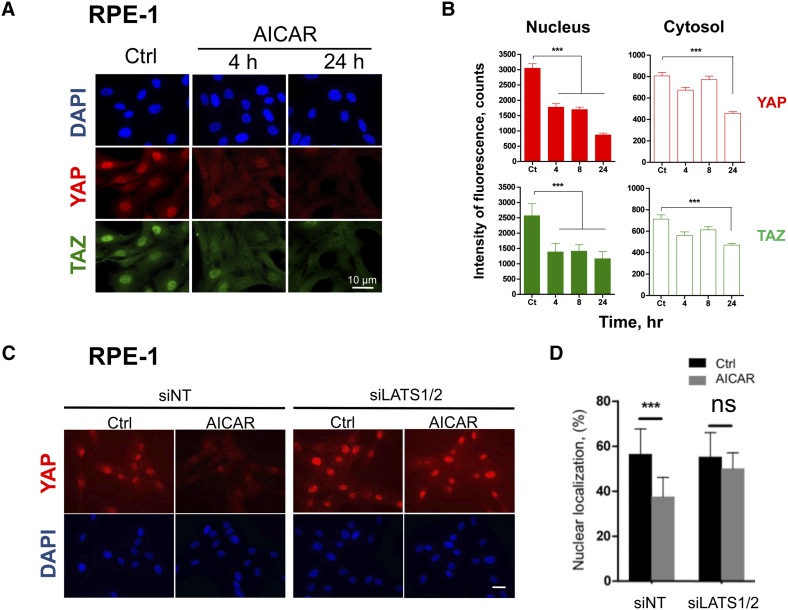
The effect of AICAR on the Hippo pathway was indeed confirmed in RPE-1 cells. AICAR induced a time-dependent redistribution of YAP1 and TAZ from the nucleus to the cytoplasm (Figure 3, A and B) due to a rapid (t < 4 h) decrease of the corresponding signals in the nucleus. This effect was followed by their late (t > 8 h) extinction in the cytosol (Figure 3B). A higher expression of LATS1 protein was consistently observed after 4 h of treatment (Figure S4A). Overall, these results indicated that AICAR regulates YAP1/TAZ subcellular localization in polarized epithelial cells as well as in fibroblasts.
Figure 3.
Subcellular redistribution of YAP1/TAZ proteins under AICAR treatment depends on LATS1/2. Human RPE-1 cells were seeded at low density and stimulated one day later with 3 mM AICAR. Subcellular localization of YAP1 and TAZ was then monitored by immunofluorescence. (A) Representative microphotographs of control RPE-1 cells (Ctrl) and of the same cells stimulated with AICAR for 4 h or 24 h. DAPI (blue) was used to stain cell nuclei. Scale bars: 10 μm. (B) Quantification of the cytosolic and nuclear fractions of YAP1 and TAZ proteins in RPE-1 cells by using the ImageJ software. At least ten images taken randomly (> 50 cells) were analyzed for each condition and the average intensity of fluorescence was reported as a function of time. (C) and (D) siRNAs-mediated knockdown of LATS1/2 impeded the subcellular redistribution of YAP1 in RPE-1 cells. siRNA-transfected RPE-1 cells were incubated for 8 h with or without 3 mM AICAR. siNT, nontarget siRNA-transfected cells. siLATS1/2, LATS1/2 siRNA-transfected cells. Scale bars: 10 μm. (C) Microphotographs of RPE-1 cells. (D) Quantification of YAP1 in nuclei of RPE-1 cells. Results are mean values ± SD (ns, P > .05; ***P < .001).
Finally, we analyzed the implication of LATS1/2 in the AICAR-mediated translocation of YAP1 by siRNA LATS1/2 knockdown approaches, which efficiently reduced the expression of the corresponding proteins in RPE-1 cells (Figure S4B). In cells treated with the non-targeting siRNA, AICAR treatment induced the cytoplasmic delocalization and decrease of the YAP1 protein (Figure 3, C and D). In comparison, delocalization of YAP1 was abolished when RPE-1 cells were treated with siRNA targeting LATS1/2 mRNA. We concluded that the decrease of the nuclear fraction of YAP1 under AICAR treatment was dependent on LATS1/2.
Knockdown of LATS1/2 Rescued Proliferation of Human and Mice Cells Under AICAR Treatment
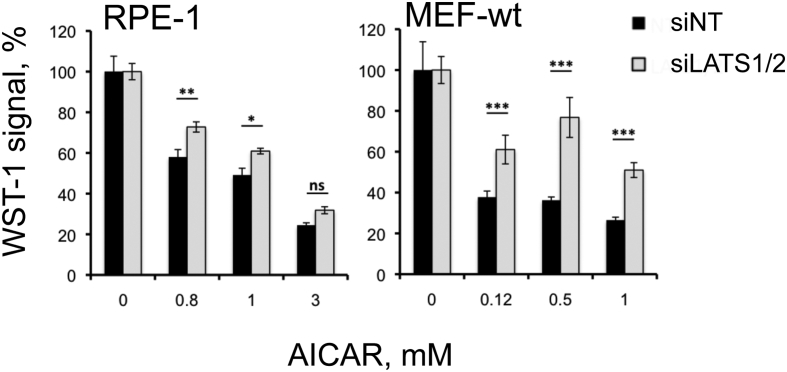
LATS1/2 being essential for the YAP1/TAZ-mediated effects of AICAR, we then used the knockdown approach to evaluate the impact of LATS1/2 activation on AICAR-mediated cytotoxicity. Cellular viability was measured in MEF-wt and RPE-1 cells treated or not with AICAR. siRNA knockdown against LATS1/2 induced a strong decrease of the level of the corresponding proteins in these cells (Figure S4, B and C) and significantly restored their growth in the presence of AICAR (Figure 4). The higher resistance of MEF and RPE-1 cells to AICAR treatment is consistent with the expected effect of LATS1/2, these protein kinases acting as potent repressors of proliferation through phosphorylation/inhibition of YAP1 and TAZ oncogenes.
Figure 4.
Viability of MEFs and RPE-1 cells is partly restored by inhibition of LATS1/2. Cells were either transfected with siRNA Lats1/2, or with a nontarget siRNA (siNT). The influence of siRNA-mediated knockdown on cell growth was then monitored by WST-1 after incubation for two (MEF-wt) or three days (RPE-1) in the presence of AICAR. Results are representative of three independent experiments and are mean ± SD of triplicates. Relative cell proliferation was set at 100%, corresponding to the signal obtained with cells transfected with siNT. (ns, P > .05; *P < .05; **P < .01; ***P < .001).
Discussion
AICAR is a purine precursor endowed with therapeutic properties [32]. Its intracellular monophosphorylated form (ZMP) mimics the binding of AMP to the low energy sensor regulatory enzyme AMPK, and is therefore widely used as an energy-stress mimicking agonist [3], [33]. Recent studies also pointed to AICAR as a potent modulator of AMPK-independent functions in mammals. Such effects converge towards cell growth inhibition and cytotoxicity [2], [5], [15], [16]. Interestingly, a higher sensitivity of aneuploid cells versus euploid cells to AICAR treatment was evidenced in different cell models, offering appealing perspectives in cancer therapy [6], [9].
By using an unbiased approach, we aimed at identifying cellular functions that could be modulated by AICAR independently of AMPK. We carried out a gene expression microarrays analysis using the AMPKα1/α2 double knockout MEF model. Time-course analysis indicated that these cells responded to AICAR treatment by up-regulating the tumor suppressor genes Lats1 and Lats2, both of which encode the most downstream protein kinases of the Hippo cascade. This effect, hereby defined as to the early cell response (~2–4 h) to AICAR, was accompanied with the increase in the corresponding proteins Lats1/2. Phosphorylation of Lats1 was also observed on its threonine residue 1079, a molecular marker of its catalytic activation. In a late (≥ 12 h) cell response to AICAR, down-regulation of a collection of gene was displayed as a signature of the repression of Yap1/Taz transcriptional coactivators. These finding were in good agreement with the fact that AICAR promoted Yap1/Taz inhibitory phosphorylation as well as their subcellular delocalization. One prominent response of mammalian cells to AICAR is therefore the downstream activation of the Hippo signaling cascade, leading to the blockade of Yap1/Taz effector functions. Remarkably, knockdown of Lats1/2 partially rescued the growth of AICAR-treated cells, which established that Lats1/2 activity is indeed involved in the antiproliferative effect of this drug.
Lats1/2 play a central role in tissue growth and size in metazoans [20], [26]. In addition, Lats1/2 (acting as tumor suppressors) and Yap1/Taz (as proto-oncogenes) are involved at critical stages of tumor progression [34], [35], [36], [37], [38]. Thus, it may come as no surprise that AICAR interferes with mechanisms linked to the cell cycle, inhibition of proliferation, decrease of cell migration/invasion, inhibition of the epithelial–mesenchymal transition (EMT) and induction of apoptosis (see for example [2], [4], [5], [39], [40]). In xenograft models, AICAR consistently slows tumor progression, inhibits tumor cell invasion and also expresses anti-angiogenic properties [12], [41].
The cellular response to AICAR was observed in MEF-dKO cells and thus did not require the presence of AMPK. Notably, AICAR activation in the absence of AMPK did not exactly replicate in MEF-wt, suggesting that the AMPK pathway could also modulate LATS and YAP1 activities. Indeed, AMPK was previously shown to inhibit YAP1 by direct phosphorylation, and was also able to activate LATS [29], [42]. In addition, AICAR-activated AMPK signaling repressed the transcription of YAP1-regulated genes CTGF and CYR61 in LATS1/2 double knockdown cells [29], [42]. Two parallel pathways, involving either AMPK or LATS kinases, may therefore converge to YAP1 inhibition under AICAR treatment. The rapid post-translational activation (in minutes) of AMPK upon AICAR binding has however to be distinguished from the transcriptional activation of LATS1/2 (≥ 2 h) reported here. Interestingly, YAP1 activity is inhibited by both AMPKα1/2 kinases and LATS1/2 kinases through phosphorylation at distinct sites (YAP-Ser61/Ser94 and YAP-Ser127, respectively) [29], [42], which suggests complementary downstream functions of AICAR through these two types of regulatory kinases. Inhibition of YAP1-dependent transcription using paired signals (i.e. AMPK- and LATS-dependent) may conform physiologically with the energetic and biosynthetic status of the cells as well as with the mechanical constraints imposed by their positioning in tissues [20], [43].
These findings broaden our knowledge of AICAR's antitumor properties. They provide a rationale for the development of derivatized pharmacological compounds by exploiting the antitumor properties of the Hippo pathway through activation of LATS1/2. Caution must be taken however when considering such approaches. Besides the most characteristic YAP1/TAZ targets, other LATS1/2 kinases substrates were identified, including AMOT, AF6, PARD3A and NUMA1 proteins whose functions are involved in the organization of the actin network or in the mitotic spindle orientation [38]. These, as well as other YAP1/TAZ-independent effects of LATS [44], arguably enlarge the contribution of AICAR as a multi-target drug for different tumor types. Besides its opposing effect on tumor progression through AMPK activation [29], [45], an awaited effect of AICAR, or of derivatized analogs, in therapy would be its ability to reactivate LATS1/2 in tissues presenting altered upstream signals in a variety of diseases [34], [35], [36], [37], [38].
In conclusion, a transcriptomic approach established that the Hippo pathway was stimulated under AICAR treatment even in the absence of AMPK. AICAR activated the tumor suppressors LATS1/2 and, consequently, inhibited the co-transcriptional activators YAP1/TAZ and downstream-regulated genes in an AMPK-independent manner. Functionally, the inhibition of cell proliferation by AICAR was due, in part, to the presence of LATS1/2.
The following are the supplementary data related to this article.
Supplementary methods.
Primers used in this study.
Table S2. (A) Expression data from Ampkα1/α2 double knockout murine embryonic fibroblasts stimulated or not with AICAR. Ampkα1/α2 double knockout murine embryonic cells were incubated with 1 mM AICAR for 0, 1, 2, 4, 8, 12 and 24 h. Hybridization was performed on Affymetrix (Mouse MG-430 PM Array Strip Kit). Analyses were performed in triplicates (except t = 8 h in duplicate). Results were expressed as fold-increases of the transcripts significantly modulated under 1 mM AICAR treatment. Initial cut-offs for differential expression were set to a fold change threshold of 2 or above and a maximum adjusted P value of .05. (B) Comparison of transcriptome signatures in human cells overexpressing YAP1 and TAZ (Zhang et al., JBC, 2009, 284, 13355-13362; see ref. [23] in the text) with transcripts responding to AICAR in MEF-dKO (this work).
Table S3. Modulation under AICAR treatment of the gene expression profile in MEF-dKO. Analysis of differentially regulated genes at a high stringency under AICAR treatment. The analysis was carried out using genes that were modulated by AICAR at all times of incubation. Cut-off of P < .05, 2.0-fold change. P-values were corrected according according to the Benjamini method.
Table S4. Fold-changes in the amounts of mRNAs of the EGF and insulin/IGF signaling pathways in response to AICAR treatment in MEF-dKO. Values indicate the fold-increase associated with AICAR treatment. Brackets on the left group together the genes related to the EGF or insulin/IGF pathways.
Figure S1. Metabolism of AICAR in MEF-wt and MEF-dko. (A) Schematic overview of the canonical de novo purine biosynthesis pathway and conversion of AICAR to ZMP. (B) Time-dependent variations of intracellular ZMP, ATP and GTP in MEF-wt (closed bars) and MEF-dKO (open bars). MEFs at the density of ~50,000 cells/cm2 in 6-well plates were subjected to 1 mM AICAR treatment for different periods of time. Analysis was carried out as reported in Supplementary Methods. Results are means ± SD of triplicate experiments. Intracellular concentration of the metabolites was determined from median cell volumes (in μm3). Median volumes (μm3) ± SD at 24 h of incubation: MEF-wt (–Zr) (3675 ± 2030), MEF-wt (+Zr) (3787 ± 1510), MEF-dKO (–Zr) (3146 ± 1840) and MEF-dKO (+Zr) (3609 ± 1660).
Figure S2. AICAR inhibited growth of murine embryonic fibroblasts. (A) Long-term propagation of MEFs in culture, showing that MEF-wt and MEF-dKO have the same division time. (B) Comparative effect of AICAR on the growth of MEF-wt (closed bars) and MEF-dKO (open bars). Results of a WST-1 cell proliferation assay at days 2 and 3 of AICAR treatment. Results are mean ± SD of two independent determinations, each made in hexaplicates. (C) AMPKα subunits were not detected in MEF-dKO. Cell extracts were prepared from MEF-wt and MEF-dKO that were stimulated or not with AICAR for 4 h. Western blot was performed using antibodies against AMPKα1/α2 and actin. (D) Flow cytometry measurements of AICAR-induced apoptosis in MEFs (see also Supplementary Methods). Cells were grown in the presence or absence of 1 mM AICAR. After one or two days of incubation, they were collected and stained with Annexin V and analyzed by flow cytometry. Left panel: cytometric profiles; right panel: percentage of apoptotic cells. The data are representative of three separate experiments. (E) AICAR promotes apoptosis in MEFs. Cell extracts were prepared from MEF-wt and MEF-dKO treated or not with 1 mM AICAR for 24 h. Proteins were resolved by SDS/PAGE and probed with either anti-PARP or anti-cleaved caspase 3 (casp-3) antibodies. Staurosporine (25 and 50 nM; 24 h) and hydrogen peroxide (0.01%; 24 h) were used as positive inducers of apoptosis. Pixel intensities of cleaved PARP and (Asp175)-caspase-3 (Casp-3) proteins were normalized to the internal control actin and the resulting values are indicated.
Figure S3. Subcellular localization of YAP1 and TAZ proteins in MEFs and in human RPE-1 cells. A-B) Immortalized MEF-wt and MEF-dKO were grown at low densities and were then stimulated with 1 mM AICAR. Yap1 and Taz subcellular localization was monitored by immunofluorescence at the indicated times. (A) Microphotographs from immunofluorescence analysis carried out in MEF-wt and MEF-dKO. DAPI-labeled nuclei are in blue. (B) Quantification of the cytosolic and nuclear fractions of Yap1 and Taz proteins using ImageJ. The average intensity of fluorescence was reported as a function of time (see also Fig. 3). The quantitative analysis of Yap1 and Taz subcellular localization showed a faint but significant decrease of the signals in the nucleus of both MEF-wt and MEF-dKO under AICAR treatment. Results are mean values ± SD (*P < .05; **P < .01; ***P < .001). Scale bar: 10 μm. (C) Effect of the cell density on the nuclear to cytoplasm delocalization of YAP in immortalized MEF-wt (SV40) and in RPE-1 cells. (D) Subcellular localization of Yap1 in primary cultured MEFs. Immunofluorescence analysis was carried out in normal MEFs. Representative microphotographs of the MEFs treated or not for 8 h with 1 mM AICAR (left panel). Quantification of the cytosolic and nuclear fractions of Yap1 and Taz proteins using ImageJ (right panel). DAPI-labeled nuclei are in blue. Results are mean values ± SD (N.S.; P > .05; **P < .01; ***P < .001). Scale bars: 5 μm.
Figure S4. Western blot analyses of LATS1/2 in MEFs and RPE-1 cells. MEF-wt and RPE-1 cells were grown at low densities and Western blot analyses were carried out on total cell lysates using antibodies directed against LATS1 and LATS2. A) RPE-1 cells were stimulated with 3 mM AICAR for different times. B-C) Knockdown experiments using non targeting siRNA (NT) or siRNAs that target LATS1 and LATS2 (see materials and methods). RPE-1 cells (B) and MEF-wt cells (C) were incubated with siRNAs as indicated in Materials and Methods and then subjected to Western blot analysis for detection of LATS proteins.
Disclosure of potential conflicts of interest
The authors declare no conflict of interests.
Acknowledgements
This work was supported by Ministère de l'Éducation Nationale de l'Enseignement Supérieur et de la Recherche (C. P.), Centre National de la Recherche Scientifique, and by grants from the Ligue Nationale Contre le Cancer, Comité des Landes (L.A.R.G.E project, M.M.), Comité de la Gironde (M. M.); Association pour la Recherche sur le Cancer (ARC SFI 201221205915, B. D-F.).
Contributor Information
Bertrand Daignan-Fornier, Email: b.daignan-fornier@ibgc.cnrs.fr.
Michel Moenner, Email: michel.moenner@u-bordeaux.fr.
References
- 1.Sabina RL, Patterson D, Holmes EW. 5-Amino-4-imidazolecarboxamide riboside (Z-riboside) metabolism in eukaryotic cells. J Biol Chem. 1985;260:6107–6114. [PubMed] [Google Scholar]
- 2.Ceschin J, Saint-Marc C, Laporte J, Labriet A, Philippe C, Moenner M, Daignan-Fornier B, Pinson B. Identification of yeast and human 5-aminoimidazole-4-carboxamide-1-beta-d-ribofuranoside (AICAr) transporters. J Biol Chem. 2014;289:16844–16854. doi: 10.1074/jbc.M114.551192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardie DG. AMPK--sensing energy while talking to other signaling pathways. Cell Metab. 2014;20:939–952. doi: 10.1016/j.cmet.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Girones DM, Moncunill-Massaguer C, Iglesias-Serret D, Cosialls AM, Perez-Perarnau A, Palmeri CM, Rubio-Patino C, Villunger A, Pons G, Gil J. AICAR induces Bax/Bak-dependent apoptosis through upregulation of the BH3-only proteins Bim and Noxa in mouse embryonic fibroblasts. Apoptosis. 2013;18:1008–1016. doi: 10.1007/s10495-013-0850-6. [DOI] [PubMed] [Google Scholar]
- 5.Santidrian AF, Gonzalez-Girones DM, Iglesias-Serret D, Coll-Mulet L, Cosialls AM, de Frias M, Campas C, Gonzalez-Barca E, Alonso E, Labi V. AICAR induces apoptosis independently of AMPK and p53 through up-regulation of the BH3-only proteins BIM and NOXA in chronic lymphocytic leukemia cells. Blood. 2010;116:3023–3032. doi: 10.1182/blood-2010-05-283960. [DOI] [PubMed] [Google Scholar]
- 6.Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardeleben C, Sharma S, Reeve JR, Bassilian S, Frost P, Hoang B, Shi Y, Lichtenstein A. Metabolomics identifies pyrimidine starvation as the mechanism of 5-aminoimidazole-4-carboxamide-1-beta-riboside-induced apoptosis in multiple myeloma cells. Mol Cancer Ther. 2013;12:1310–1321. doi: 10.1158/1535-7163.MCT-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robert G, Ben Sahra I, Puissant A, Colosetti P, Belhacene N, Gounon P, Hofman P, Bost F, Cassuto JP, Auberger P. Acadesine kills chronic myelogenous leukemia (CML) cells through PKC-dependent induction of autophagic cell death. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ly P, Kim SB, Kaisani AA, Marian G, Wright WE, Shay JW. Aneuploid human colonic epithelial cells are sensitive to AICAR-induced growth inhibition through EGFR degradation. Oncogene. 2013;32:3139–3146. doi: 10.1038/onc.2012.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Guo D, Hildebrandt IJ, Prins RM, Soto H, Mazzotta MM, Dang J, Czernin J, Shyy JY, Watson AD, Phelps M. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc Natl Acad Sci U S A. 2009;106:12932–12937. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theodoropoulou S, Brodowska K, Kayama M, Morizane Y, Miller JW, Gragoudas ES, Vavvas DG. Aminoimidazole Carboxamide Ribonucleotide (AICAR) Inhibits the Growth of Retinoblastoma In Vivo by Decreasing Angiogenesis and Inducing Apoptosis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0052852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Den Neste E, Cazin B, Janssens A, Gonzalez-Barca E, Terol MJ, Levy V, Perez de Oteyza J, Zachee P, Saunders A, de Frias M. Acadesine for patients with relapsed/refractory chronic lymphocytic leukemia (CLL): a multicenter phase I/II study. Cancer Chemother Pharmacol. 2013;71:581–591. doi: 10.1007/s00280-012-2033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent EE, Coelho PP, Blagih J, Griss T, Viollet B, Jones RG. Differential effects of AMPK agonists on cell growth and metabolism. Oncogene. 2015;34:3627–3639. doi: 10.1038/onc.2014.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Chhipa RR, Pooya S, Wortman M, Yachyshin S, Chow LM, Kumar A, Zhou X, Sun Y, Quinn B. Discrete mechanisms of mTOR and cell cycle regulation by AMPK agonists independent of AMPK. Proc Natl Acad Sci U S A. 2014;111:E435–E444. doi: 10.1073/pnas.1311121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guigas B, Taleux N, Foretz M, Detaille D, Andreelli F, Viollet B, Hue L. AMP-activated protein kinase-independent inhibition of hepatic mitochondrial oxidative phosphorylation by AICA riboside. Biochem J. 2007;404:499–507. doi: 10.1042/BJ20070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laderoute KR, Amin K, Calaoagan JM, Knapp M, Le T, Orduna J, Foretz M, Viollet B. 5'-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Mol Cell Biol. 2006;26:5336–5347. doi: 10.1128/MCB.00166-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen CG, Moroishi T, Guan KL. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 2015;25:499–513. doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 22.Auf G, Jabouille A, Guerit S, Pineau R, Delugin M, Bouchecareilh M, Magnin N, Favereaux A, Maitre M, Gaiser T. Inositol-requiring enzyme 1alpha is a key regulator of angiogenesis and invasion in malignant glioma. Proc Natl Acad Sci U S A. 2010;107:15553–15558. doi: 10.1073/pnas.0914072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cebola I, Rodriguez-Segui SA, Cho CH-H, Bessa J, Rovira M, Luengo M, Chhatriwala M, Berry A, Ponsa-Cobas J, Maestro MA. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat Cell Biol. 2015;17:615–626. doi: 10.1038/ncb3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sudol M. Neuregulin 1–activated ERBB4 as a “dedicated” receptor for the Hippo-YAP pathway. Sci Signal. 2014;7:pe29. doi: 10.1126/scisignal.aaa2710. [DOI] [PubMed] [Google Scholar]
- 28.Strassburger K, Tiebe M, Pinna F, Breuhahn K, Teleman AA. Insulin/IGF signaling drives cell proliferation in part via Yorkie/YAP. Dev Biol. 2012;367:187–196. doi: 10.1016/j.ydbio.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Mo JS, Meng Z, Kim YC, Park HW, Hansen CG, Kim S, Lim DS, Guan KL. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat Cell Biol. 2015;17:500–510. doi: 10.1038/ncb3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen HT, Hong X, Tan S, Chen Q, Chan L, Fivaz M, Cohen SM, Voorhoeve PM. Viral small T oncoproteins transform cells by alleviating hippo-pathway-mediated inhibition of the YAP proto-oncogene. Cell Rep. 2014;8:707–713. doi: 10.1016/j.celrep.2014.06.062. [DOI] [PubMed] [Google Scholar]
- 31.Shanzer M, Ricardo-Lax I, Keshet R, Reuven N, Shaul Y. The polyomavirus middle T-antigen oncogene activates the Hippo pathway tumor suppressor Lats in a Src-dependent manner. Oncogene. 2015;34:4190–4198. doi: 10.1038/onc.2014.347. [DOI] [PubMed] [Google Scholar]
- 32.Daignan-Fornier B, Pinson B. 5-Aminoimidazole-4-carboxamide-1-beta-D-ribofuranosyl 5'-Monophosphate (AICAR), a Highly Conserved Purine Intermediate with Multiple Effects. Metabolites. 2012;2:292–302. doi: 10.3390/metabo2020292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Day P, Sharff A, Parra L, Cleasby A, Williams M, Horer S, Nar H, Redemann N, Tickle I, Yon J. Structure of a CBS-domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP and ZMP. Acta Crystallogr D Biol Crystallogr. 2007;63:587–596. doi: 10.1107/S0907444907009110. [DOI] [PubMed] [Google Scholar]
- 34.Visser S, Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 2010;9:3892–3903. doi: 10.4161/cc.9.19.13386. [DOI] [PubMed] [Google Scholar]
- 35.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 37.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfleger CM. The Hippo Pathway: A Master Regulatory Network Important in Development and Dysregulated in Disease. Curr Top Dev Biol. 2017;123:181–228. doi: 10.1016/bs.ctdb.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Zheng B, Cantley LC. Regulation of epithelial tight junction assembly and disassembly by AMP-activated protein kinase. PNAS. 2007;104:819–822. doi: 10.1073/pnas.0610157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou CC, Lee KH, Lai IL, Wang D, Mo X, Kulp SK, Shapiro CL, Chen CS. AMPK reverses the mesenchymal phenotype of cancer cells by targeting the Akt-MDM2-Foxo3a signaling axis. Cancer Res. 2014;74:4783–4795. doi: 10.1158/0008-5472.CAN-14-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HS, Kim MJ, Kim EJ, Yang Y, Lee MS, Lim JS. Berberine-induced AMPK activation inhibits the metastatic potential of melanoma cells via reduction of ERK activity and COX-2 protein expression. Biochem Pharmacol. 2012;83:385–394. doi: 10.1016/j.bcp.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Xiao ZD, Li X, Aziz KE, Gan B, Johnson RL, Chen J. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat Cell Biol. 2015;17:490–499. doi: 10.1038/ncb3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santinon G, Pocaterra A, Dupont S. Control of YAP/TAZ Activity by Metabolic and Nutrient-Sensing Pathways. Trends Cell Biol. 2016;26:289–299. doi: 10.1016/j.tcb.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Britschgi A, Duss S, Kim S, Couto JP, Brinkhaus H, Koren S, De Silva D, Mertz KD, Kaup D, Varga Z. The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERalpha. Nature. 2017;541:541–545. doi: 10.1038/nature20829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, Saud SM, Young MR, Chen G, Hua B, Targeting AMPK. for cancer prevention and treatment. Oncotarget. 2015;6:7365–7378. doi: 10.18632/oncotarget.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods.
Primers used in this study.
Table S2. (A) Expression data from Ampkα1/α2 double knockout murine embryonic fibroblasts stimulated or not with AICAR. Ampkα1/α2 double knockout murine embryonic cells were incubated with 1 mM AICAR for 0, 1, 2, 4, 8, 12 and 24 h. Hybridization was performed on Affymetrix (Mouse MG-430 PM Array Strip Kit). Analyses were performed in triplicates (except t = 8 h in duplicate). Results were expressed as fold-increases of the transcripts significantly modulated under 1 mM AICAR treatment. Initial cut-offs for differential expression were set to a fold change threshold of 2 or above and a maximum adjusted P value of .05. (B) Comparison of transcriptome signatures in human cells overexpressing YAP1 and TAZ (Zhang et al., JBC, 2009, 284, 13355-13362; see ref. [23] in the text) with transcripts responding to AICAR in MEF-dKO (this work).
Table S3. Modulation under AICAR treatment of the gene expression profile in MEF-dKO. Analysis of differentially regulated genes at a high stringency under AICAR treatment. The analysis was carried out using genes that were modulated by AICAR at all times of incubation. Cut-off of P < .05, 2.0-fold change. P-values were corrected according according to the Benjamini method.
Table S4. Fold-changes in the amounts of mRNAs of the EGF and insulin/IGF signaling pathways in response to AICAR treatment in MEF-dKO. Values indicate the fold-increase associated with AICAR treatment. Brackets on the left group together the genes related to the EGF or insulin/IGF pathways.
Figure S1. Metabolism of AICAR in MEF-wt and MEF-dko. (A) Schematic overview of the canonical de novo purine biosynthesis pathway and conversion of AICAR to ZMP. (B) Time-dependent variations of intracellular ZMP, ATP and GTP in MEF-wt (closed bars) and MEF-dKO (open bars). MEFs at the density of ~50,000 cells/cm2 in 6-well plates were subjected to 1 mM AICAR treatment for different periods of time. Analysis was carried out as reported in Supplementary Methods. Results are means ± SD of triplicate experiments. Intracellular concentration of the metabolites was determined from median cell volumes (in μm3). Median volumes (μm3) ± SD at 24 h of incubation: MEF-wt (–Zr) (3675 ± 2030), MEF-wt (+Zr) (3787 ± 1510), MEF-dKO (–Zr) (3146 ± 1840) and MEF-dKO (+Zr) (3609 ± 1660).
Figure S2. AICAR inhibited growth of murine embryonic fibroblasts. (A) Long-term propagation of MEFs in culture, showing that MEF-wt and MEF-dKO have the same division time. (B) Comparative effect of AICAR on the growth of MEF-wt (closed bars) and MEF-dKO (open bars). Results of a WST-1 cell proliferation assay at days 2 and 3 of AICAR treatment. Results are mean ± SD of two independent determinations, each made in hexaplicates. (C) AMPKα subunits were not detected in MEF-dKO. Cell extracts were prepared from MEF-wt and MEF-dKO that were stimulated or not with AICAR for 4 h. Western blot was performed using antibodies against AMPKα1/α2 and actin. (D) Flow cytometry measurements of AICAR-induced apoptosis in MEFs (see also Supplementary Methods). Cells were grown in the presence or absence of 1 mM AICAR. After one or two days of incubation, they were collected and stained with Annexin V and analyzed by flow cytometry. Left panel: cytometric profiles; right panel: percentage of apoptotic cells. The data are representative of three separate experiments. (E) AICAR promotes apoptosis in MEFs. Cell extracts were prepared from MEF-wt and MEF-dKO treated or not with 1 mM AICAR for 24 h. Proteins were resolved by SDS/PAGE and probed with either anti-PARP or anti-cleaved caspase 3 (casp-3) antibodies. Staurosporine (25 and 50 nM; 24 h) and hydrogen peroxide (0.01%; 24 h) were used as positive inducers of apoptosis. Pixel intensities of cleaved PARP and (Asp175)-caspase-3 (Casp-3) proteins were normalized to the internal control actin and the resulting values are indicated.
Figure S3. Subcellular localization of YAP1 and TAZ proteins in MEFs and in human RPE-1 cells. A-B) Immortalized MEF-wt and MEF-dKO were grown at low densities and were then stimulated with 1 mM AICAR. Yap1 and Taz subcellular localization was monitored by immunofluorescence at the indicated times. (A) Microphotographs from immunofluorescence analysis carried out in MEF-wt and MEF-dKO. DAPI-labeled nuclei are in blue. (B) Quantification of the cytosolic and nuclear fractions of Yap1 and Taz proteins using ImageJ. The average intensity of fluorescence was reported as a function of time (see also Fig. 3). The quantitative analysis of Yap1 and Taz subcellular localization showed a faint but significant decrease of the signals in the nucleus of both MEF-wt and MEF-dKO under AICAR treatment. Results are mean values ± SD (*P < .05; **P < .01; ***P < .001). Scale bar: 10 μm. (C) Effect of the cell density on the nuclear to cytoplasm delocalization of YAP in immortalized MEF-wt (SV40) and in RPE-1 cells. (D) Subcellular localization of Yap1 in primary cultured MEFs. Immunofluorescence analysis was carried out in normal MEFs. Representative microphotographs of the MEFs treated or not for 8 h with 1 mM AICAR (left panel). Quantification of the cytosolic and nuclear fractions of Yap1 and Taz proteins using ImageJ (right panel). DAPI-labeled nuclei are in blue. Results are mean values ± SD (N.S.; P > .05; **P < .01; ***P < .001). Scale bars: 5 μm.
Figure S4. Western blot analyses of LATS1/2 in MEFs and RPE-1 cells. MEF-wt and RPE-1 cells were grown at low densities and Western blot analyses were carried out on total cell lysates using antibodies directed against LATS1 and LATS2. A) RPE-1 cells were stimulated with 3 mM AICAR for different times. B-C) Knockdown experiments using non targeting siRNA (NT) or siRNAs that target LATS1 and LATS2 (see materials and methods). RPE-1 cells (B) and MEF-wt cells (C) were incubated with siRNAs as indicated in Materials and Methods and then subjected to Western blot analysis for detection of LATS proteins.