Significance
Organisms must constantly make regulatory decisions in response to a change in cellular state or environment. However, while the catalog of genomes expands rapidly, we remain ignorant about how the genes in these genomes are regulated. Here, we show how a massively parallel reporter assay, Sort-Seq, and information-theoretic modeling can be used to identify regulatory sequences. We then use chromatography and mass spectrometry to identify the regulatory proteins that bind these sequences. The approach results in quantitative base pair-resolution models of promoter mechanism and was shown in both well-characterized and unannotated promoters in Escherichia coli. Given the generality of the approach, it opens up the possibility of quantitatively dissecting the mechanisms of promoter function in a wide range of bacteria.
Keywords: gene regulation, massively parallel reporter assay, quantitative models, DNA affinity chromatography, mass spectrometry
Abstract
Gene regulation is one of the most ubiquitous processes in biology. However, while the catalog of bacterial genomes continues to expand rapidly, we remain ignorant about how almost all of the genes in these genomes are regulated. At present, characterizing the molecular mechanisms by which individual regulatory sequences operate requires focused efforts using low-throughput methods. Here, we take a first step toward multipromoter dissection and show how a combination of massively parallel reporter assays, mass spectrometry, and information-theoretic modeling can be used to dissect multiple bacterial promoters in a systematic way. We show this approach on both well-studied and previously uncharacterized promoters in the enteric bacterium Escherichia coli. In all cases, we recover nucleotide-resolution models of promoter mechanism. For some promoters, including previously unannotated ones, the approach allowed us to further extract quantitative biophysical models describing input–output relationships. Given the generality of the approach presented here, it opens up the possibility of quantitatively dissecting the mechanisms of promoter function in E. coli and a wide range of other bacteria.
The sequencing revolution has left in its wake an enormous challenge: the rapidly expanding catalog of sequenced genomes is far outpacing a sequence-level understanding of how the genes in these genomes are regulated. This ignorance extends from viruses to bacteria to archaea to eukaryotes. Even in Escherichia coli, the model organism in which transcriptional regulation is best understood, we still have no indication if or how more than one-half of the genes are regulated (SI Appendix, Fig. S1) [RegulonDB (1) or EcoCyc (2)]. In other model bacteria, such as Bacillus subtilis, Caulobacter crescentus, Vibrio harveyii, or Pseudomonas aeruginosa, far fewer genes have established regulatory mechanisms (3–5).
New approaches are needed for studying regulatory architecture in these bacteria and others. Chromatin immunoprecipitation (ChIP) and other high-throughput techniques are increasingly being used to study gene regulation in E. coli (6–11), but these methods are incapable of revealing either the nucleotide-resolution location of all functional transcription factor binding sites or the way in which interactions between DNA-bound transcription factors and RNA polymerase (RNAP) modulate transcription. Although an arsenal of now classic genetic and biochemical methods has been developed for dissecting promoter function at individual bacterial promoters [reviewed in the work by Minchin and Busby (12)], these methods are not readily parallelized and often require purification of promoter-specific regulatory proteins.
In recent years, a variety of massively parallel reporter assays have been developed for dissecting the functional architecture of transcriptional regulatory sequences in bacteria, yeast, and metazoans. These technologies have been used to infer biophysical models of well-studied loci, to characterize synthetic promoters constructed from known binding sites, and to search for new transcriptional regulatory sequences (13–19). CRISPR assays have also shown promise for identifying longer-range enhancer–promoter interactions in mammalian cells (20). However, no approach for using massively parallel reporter technologies to decipher the functional mechanisms of previously uncharacterized regulatory sequences has yet been established.
Here, we take a first step toward quantitative, multipromoter dissection and describe a systematic approach for identifying the functional architecture of previously uncharacterized bacterial promoters at nucleotide resolution using a combination of genetic, functional, and biochemical measurements. A massively parallel reporter assay [Sort-Seq (13)] is performed on a promoter in multiple growth conditions to identify functional transcription factor binding sites. DNA affinity chromatography and mass spectrometry (21, 22) are then used to identify the regulatory proteins that recognize these sites. In this way, one is able to identify both the functional transcription factor binding sites and cognate transcription factors in previously unstudied promoters. Subsequent massively parallel assays are then performed in gene deletion strains to provide additional validation of the identified regulators. The reporter data thus generated are also used to infer sequence-dependent quantitative models of transcriptional regulation. In what follows, we first illustrate the overarching logic of our approach through application to four previously annotated promoters: lacZYA, relBE, marRAB, and yebG. We then apply this strategy to the previously uncharacterized promoters of purT, xylE, and dgoRKADT, showing the ability to go from regulatory ignorance to explicit quantitative models of a promoter’s input–output behavior.
Results
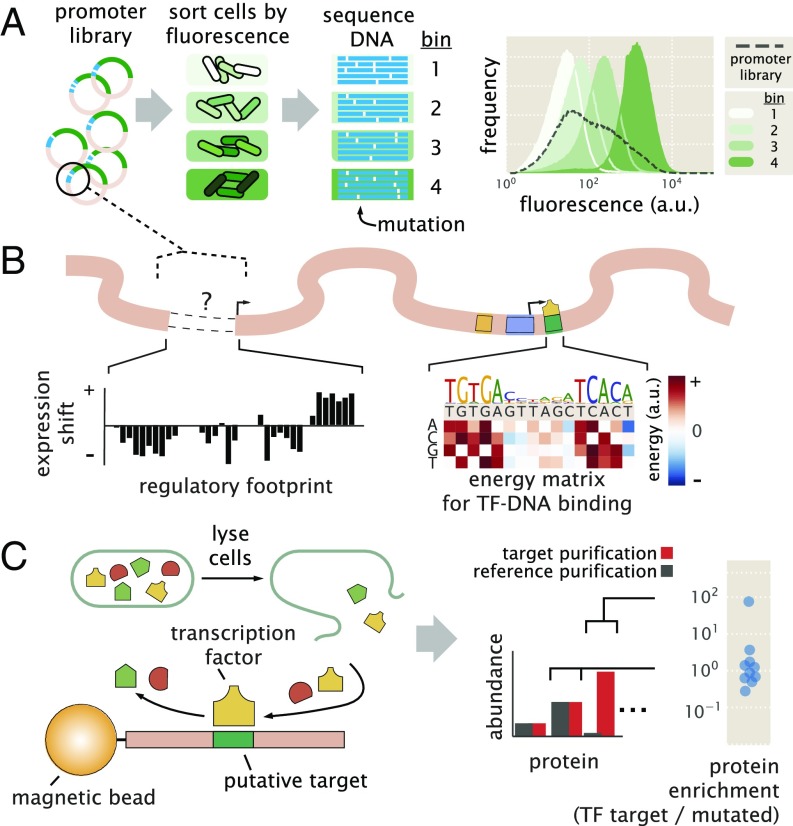
To dissect how a promoter is regulated, we begin by performing Sort-Seq (13). As shown in Fig. 1A, Sort-Seq works by first generating a library of cells, each of which contains a mutated promoter that drives expression of green fluorescent protein (GFP) from a low copy plasmid [5–10 copies per cell (23)] and provides a readout of transcriptional state. We use fluorescence-activated cell sorting (FACS) to sort cells into multiple bins gated by their fluorescence level and then sequence the mutated plasmids from each bin. We found it sufficient to sort the libraries into four bins and generated datasets of about 0.5–2 million sequences across the sorted bins (SI Appendix, Fig. S3 A–D). To identify putative binding sites, we calculate “expression shift” plots that show the average change in fluorescence when each position of the regulatory DNA is mutated (Fig. 1B, Left). Mutations to the DNA will, in general, disrupt binding of transcription factors (24), and therefore, regions with a positive shift are suggestive of binding by a repressor, while a negative shift suggests binding by an activator or RNAP.
Fig. 1.
Overview of the approach to characterize transcriptional regulatory DNA using Sort-Seq and mass spectrometry. (A) Schematic of Sort-Seq. A promoter plasmid library is placed upstream of GFP and is transformed into cells. The cells are sorted into four bins by FACS, and after regrowth, plasmids are purified and sequenced. The entire intergenic region associated with a promoter is included on the plasmid, and a separate downstream ribosomal binding site sequence is used for translation of the GFP gene. The fluorescence histograms show the fluorescence from a library of the rel promoter and the resulting sorted bins. (B) Regulatory binding sites are identified by calculating the average expression shift due to mutation at each position. In the schematic, positive expression shifts are suggestive of binding by repressors, while negative shifts would suggest binding by an activator or RNAP. Quantitative models can be inferred to describe and further interrogate the associated DNA–protein interactions. An example energy matrix that describes the binding energy between an as yet unknown transcription factor (TF) and the DNA is shown. By convention, the wild-type nucleotides have zero energy, with blue squares identifying mutations that enhance binding (negative energy) and red squares identifying mutations that reduce binding (positive energy). The wild-type sequence is written above the matrix. (C) DNA affinity chromatography and mass spectrometry are used to identify the putative transcription factor for an identified repressor site. DNA oligonucleotides containing the target binding site are tethered to magnetic beads and used to purify the target transcription factor from cell lysate. Protein abundance is determined by mass spectrometry, and a protein enrichment is calculated as the ratio in abundance relative to a second reference experiment where the target sequence is mutated away.
The identified binding sites are further interrogated by performing information-based modeling with the Sort-Seq data. Here, we generate energy matrix models (13, 25) that describe the sequence-dependent energy of interaction of a transcription factor at each putative binding site. For each matrix, we use a convention that the wild-type sequence is set to have an energy of zero (an example energy matrix is in Fig. 1B, Right). Mutations that enhance binding are identified in blue in Fig. 1B, while mutations that weaken binding are identified in red in Fig. 1B. We also use these energy matrices to generate sequence logos (26), which provide a useful visualization of the sequence specificity (Fig. 1B, above energy matrix).
To identify the putative transcription factors, we next perform DNA affinity chromatography experiments using DNA oligonucleotides containing the binding sites identified by Sort-Seq. Here, we apply a stable isotopic labeling of cell culture [SILAC (27–30)] approach, which enables us to perform a second reference affinity chromatography that is simultaneously analyzed by mass spectrometry. We perform chromatography using magnetic beads with tethered oligonucleotides containing the putative binding site (Fig. 1C). Our reference purification is performed identically, except that the binding site has been mutated away. The abundance of each protein is determined by mass spectrometry and used to calculate protein enrichment ratios, with the target transcription factor expected to exhibit a ratio greater than one. The reference purification ensures that nonspecifically bound proteins will have a protein enrichment near one. This mass spectrometry data and the energy matrix models provide insight into the identity of each regulatory factor and potential regulatory mechanisms. In certain instances, these insights then allow us to probe the Sort-Seq data further through additional information-based modeling using thermodynamic models of gene regulation. As further validation of binding by an identified regulator, we also perform Sort-Seq experiments in gene deletion strains, which should no longer show the associated positive or negative shift in expression at their binding site.
Sort-Seq Recovers the Regulatory Features of Well-Characterized Promoters.
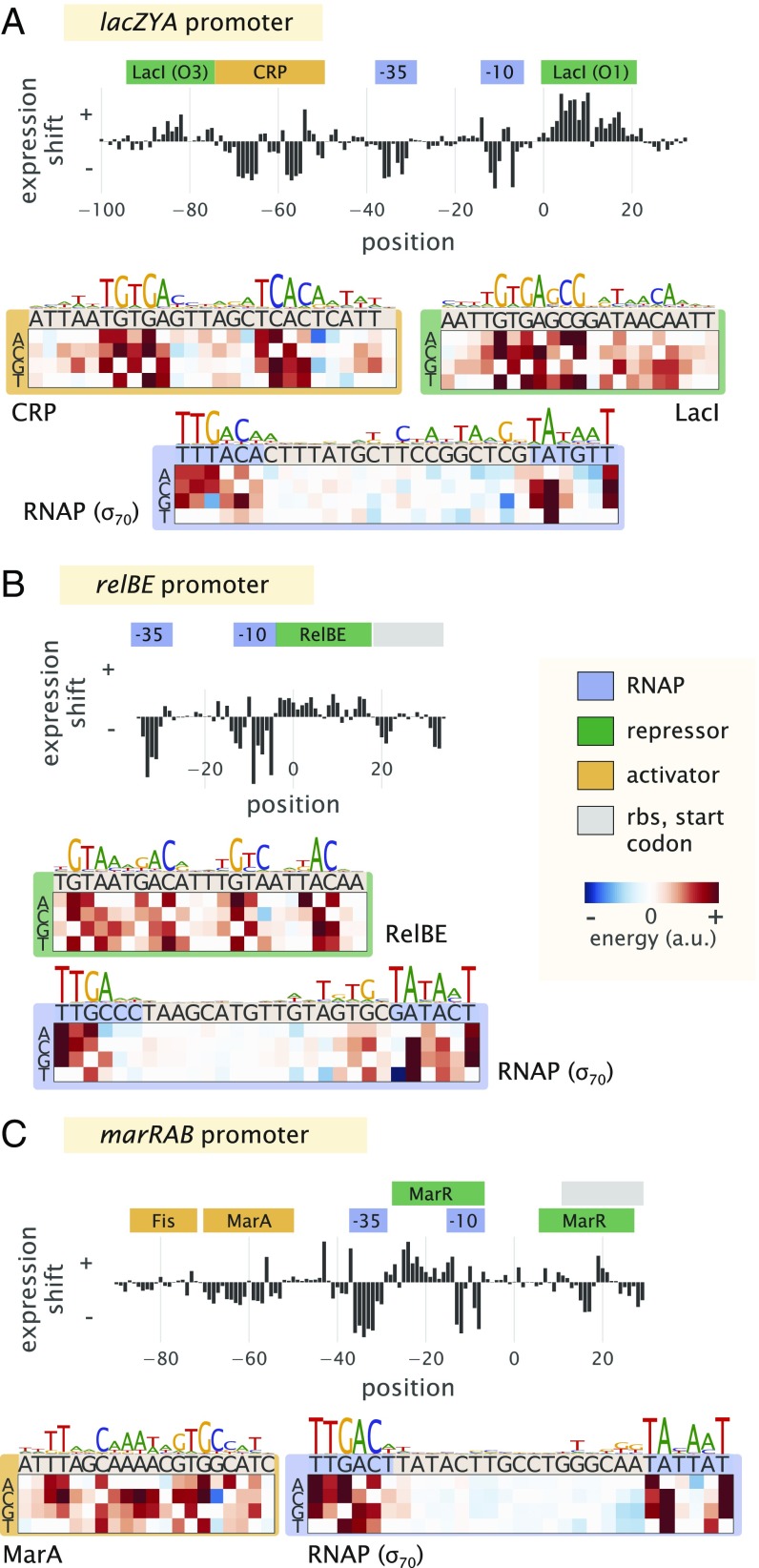
To first show Sort-Seq as a tool to discover regulatory binding sites de novo, we began by looking at the promoters of lacZYA (lac), relBE (rel), and marRAB (mar). These promoters have been studied extensively (31–33) and provide a useful testbed of distinct regulatory motifs. To proceed, we constructed libraries for each promoter by mutating their known regulatory binding sites (SI Appendix, Fig. S3 E and F shows additional characterization). We begin by considering the lac promoter, which contains three Lac repressor (LacI) binding sites, two of which we consider here, and a cAMP receptor protein (CRP) binding site. It exhibits the classic catabolic switch-like behavior that results in diauxie when E. coli is grown in the presence of glucose and lactose sugars (31). Here, we performed Sort-Seq with cells grown in M9 minimal media with 0.5% glucose. The expression shifts at each nucleotide position are shown in Fig. 2A, with annotated binding sites noted above the plot. The expression shifts reflect the expected regulatory role of each binding site, showing positive shifts for LacI and negative shifts for CRP and RNAP. The difference in magnitude at the two LacI binding sites likely reflects the different binding energies between these two binding site sequences, with LacI O3 having an in vivo dissociation constant that is almost three orders of magnitude weaker than that of the LacI O1 binding site (31, 34).
Fig. 2.
Characterization of the regulatory landscape of the lac, rel, and mar promoters. (A) Sort-Seq of the lac promoter. Cells were grown in M9 minimal media with 0.5% glucose at 37 °C. Expression shifts are shown, with annotated binding sites for CRP (activator), RNAP (−10 and −35 subsites), and LacI (repressor) noted. Energy matrices and sequence logos are shown for each binding site. (B) Sort-Seq of the rel promoter. Cells were also grown in M9 minimal media with 0.5% glucose at 37 °C. The expression shifts identify the binding sites of RNAP and RelBE (repressor), and energy matrices and sequence logos are shown for these. (C) Sort-Seq of the mar promoter. Here, cells were grown in LB at 30 °C. The expression shifts identify the known binding sites of Fis and MarA (activators), RNAP, and MarR (repressor). Energy matrices and sequence logos are shown for MarA and RNAP. Annotated binding sites are based on those in RegulonDB.
Next, we consider the rel promoter that transcribes the toxin–antitoxin pair RelE and RelB. It is 1 of about 36 toxin–antitoxin systems found on the chromosome, with important roles in cell physiology, including cellular persistence (35). When the toxin, RelE, is in excess of its cognate binding partner, the antitoxin RelB, the toxin causes cellular paralysis through cleavage of mRNA (36). Interestingly, the antitoxin protein also contains a DNA binding domain and is a repressor of its own promoter (37). We similarly performed Sort-Seq with cells grown in M9 minimal media. The expression shifts are shown in Fig. 2B and were consistent with binding by RNAP and RelBE. In particular, a positive shift was observed at the binding site for RelBE, and the RNAP binding site mainly showed a negative shift in expression.
The third promoter, mar, is associated with multiple antibiotic resistance, since its operon codes for the transcription factor MarA, which activates a variety of genes, including the major multidrug resistance efflux pump, ArcAB-TolC, and increases antibiotic tolerance (33). The mar promoter is itself activated by MarA, SoxS, and Rob (via the so-called marbox binding site) and further enhanced by Fis, which binds upstream of this marbox (38). Under standard laboratory growth, it is under repression by MarR (33). We found that the promoter’s fluorescence was quite dim in M9 minimal media and instead, grew libraries in LB at 30 °C (39). Again, the different features in the expression shift plot (Fig. 2C) seemed to be consistent with the noted binding sites. One exception was that the downstream MarR binding site was not especially apparent. Both positive and negative expression shifts were observed along its binding site, which may be due to overlap with other features present, including the native ribosomal binding site. There have also been reported binding sites for CRP, Cra, CpxR/CpxA, and AcrR (1). However, the studies associated with these annotations required overexpression of the associated transcription factor, were computationally predicted, or were shown in in vitro assays and not necessarily expected under the growth condition considered here.
While each promoter qualitatively showed the expected regulatory behavior in each expression shift plot, it was important to show that we could also recover the quantitative features of binding by each transcription factor. Here, we inferred energy matrices and associated sequence logos for the binding sites of RNAP, LacI, CRP, RelBE, MarA, and Fis. These are shown in Fig. 2 and SI Appendix, Fig. S4, and indeed, the matrices agreed well with those generated from known genomic binding sites for each transcription factor (Pearson correlation coefficient = 0.5–0.9) (SI Appendix).
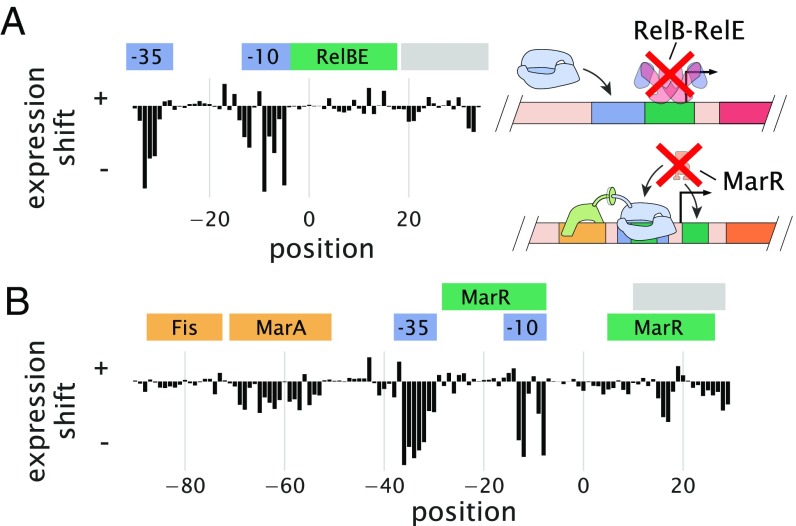
For the repressors RelBE and MarR, there were no data available that characterized their sequence specificity with which to compare. Here, instead, we validated our data by performing Sort-Seq in strains where the relBE or marR genes were deleted. In each case, this resulted in a loss of the expression shift associated with binding by these repressors (Fig. 3) and an inability of the energy matrices to explain the data in the deletion strain (SI Appendix, Fig. S7), suggesting that the observed features in the wild-type strain data are due to binding by these transcription factors.
Fig. 3.
Expression shifts reflect binding by regulatory proteins. (A) Expression shifts for the rel promoter but in a relBE genetic background. Cells were grown in conditions identical to Fig. 2B but no longer show a substantial positive expression shift across the annotated RelBE binding site. (B) Expression shifts for the mar promoter but in a marR genetic background. The positive expression shift observed where MarR is expected to bind is no longer observed. Binding site annotations are identified in blue for RNAP sites, green for repressor sites, yellow for activator sites, and gray for ribosomal binding site and start codons. These annotations refer to the binding sites noted on RegulonDB that were observed in the Sort-Seq data.
Identification of Transcription Factors with DNA Affinity Chromatography and Quantitative Mass Spectrometry.
Next, it was important to show that DNA affinity chromatography could be used to identify transcription factors in E. coli. In particular, a challenge arises in identifying transcription factors in most organisms due to their very low abundance. In E. coli, the cumulative distribution in protein copy number shows that more than one-half have a copy number less than 100 per cell, with 90% having a copy number less than 1,000 per cell. This is several orders of magnitude below that of many other cellular proteins (40).
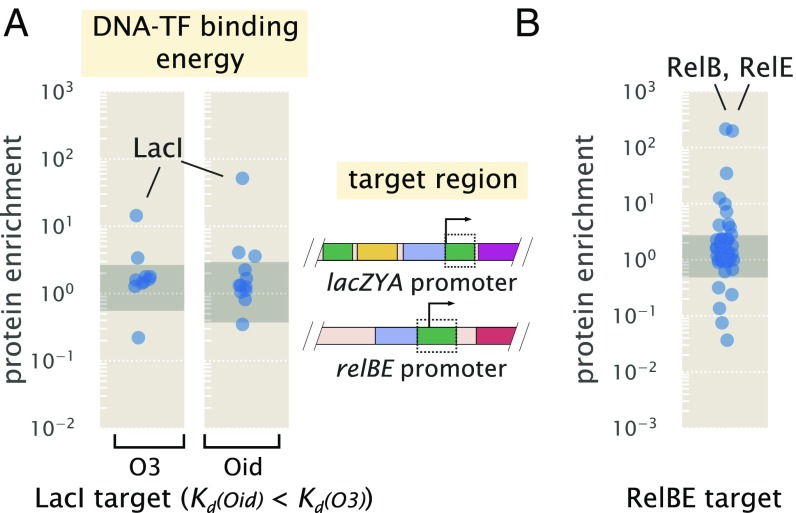
We began by applying the approach to known binding sites for LacI and RelBE. For LacI, which is present in E. coli in about 10 copies per cell, we used the strongest binding site sequence, Oid (in vivo nM), and the weakest natural binding site sequence, O3 (in vivo nM) (31, 34, 41). In Fig. 4A, we plot the protein enrichments from each transcription factor identified by mass spectrometry. LacI was found with both DNA targets, with fold enrichment greater than 10 in each case, and it was significantly higher than most of the proteins detected (indicated by the shaded region in Fig. 4A, which represents the 95% probability density region of all proteins detected, including non-DNA binding proteins). Purification of LacI with about 10 copies per cell using the weak O3 binding site sequence is near the limit of what would be necessary for most E. coli promoters.
Fig. 4.
DNA affinity purification and identification of LacI and RelBE by mass spectrometry using known target binding sites. (A) Protein enrichment using the weak O3 binding site and strong synthetic Oid binding sites of LacI. LacI was the most significantly enriched protein in each purification. The target DNA region was based on the boxed area of the lac promoter schematic but with the native O1 binding site sequence replaced with either O3 or Oid. Data points represent average protein enrichment for each detected transcription factor measured from a single purification experiment. (B) For purification using the RelBE binding site target, both RelB and its cognate binding partner RelE were significantly enriched. Data points show the average protein enrichment from two purification experiments. The target binding site is shown by the boxed region of the rel promoter schematic. Data points in each purification show the protein enrichment for detected transcription factors. The gray shaded regions show where 95% of all detected protein ratios were found.
To ensure that this success was not specific to LacI, we also applied chromatography to the RelBE binding site. RelBE provides an interesting case, since the strength of binding by RelB to DNA is dependent on whether RelE is bound in complex to RelB [with at least a 100-fold weaker dissociation constant reported in the absence of RelE (42, 43)]. As shown in Fig. 4B, we found over 100-fold enrichment of both proteins by mass spectrometry. To provide some additional intuition into these results, we also considered the predictions from a statistical mechanical model of DNA binding affinity (SI Appendix). As a consequence of performing a second reference purification, we find that fold enrichment should mostly reflect the difference in binding energy between the DNA sequences used in the two purifications and be much less dependent on whether the protein was in low or high abundance within the cell. This seemed to be the case when considering other E. coli strains with LacI copy numbers between about 10 and 1,000 copies per cell (SI Appendix, Fig. S5C). Additional characterization of the measurement sensitivity and dynamic range of this approach is noted in SI Appendix.
Sort-Seq Discovers Regulatory Architectures in Unannotated Regulatory Regions.
Given that more than one-half of the promoters in E. coli have no annotated transcription factor binding sites in RegulonDB, we narrowed our focus by using several high-throughput studies to identify candidate genes to apply our approach (44, 45). The work by Schmidt et al. (45) in particular measured the protein copy number of about one-half the E. coli genes across 22 distinct growth conditions. Using these data, we identified genes that had substantial differential gene expression patterns across growth conditions, thus hinting at the presence of regulation and even how that regulation is elicited by environmental conditions (additional details are in SI Appendix, Fig. S2). On the basis of this survey, we chose to investigate the promoters of purT, xylE, and dgoRKADT. To apply Sort-Seq in a more exploratory manner, we considered three 60-bp mutagenized windows spanning the intergenic region of each gene. While it is certainly possible that regulatory features will be outside of this window, a search of known regulatory binding sites suggests that this should be sufficient to capture just over 70% of regulatory features in E. coli and provide a useful starting point (SI Appendix, Fig. S6).
The purT promoter contains a simple repression architecture and is repressed by PurR.
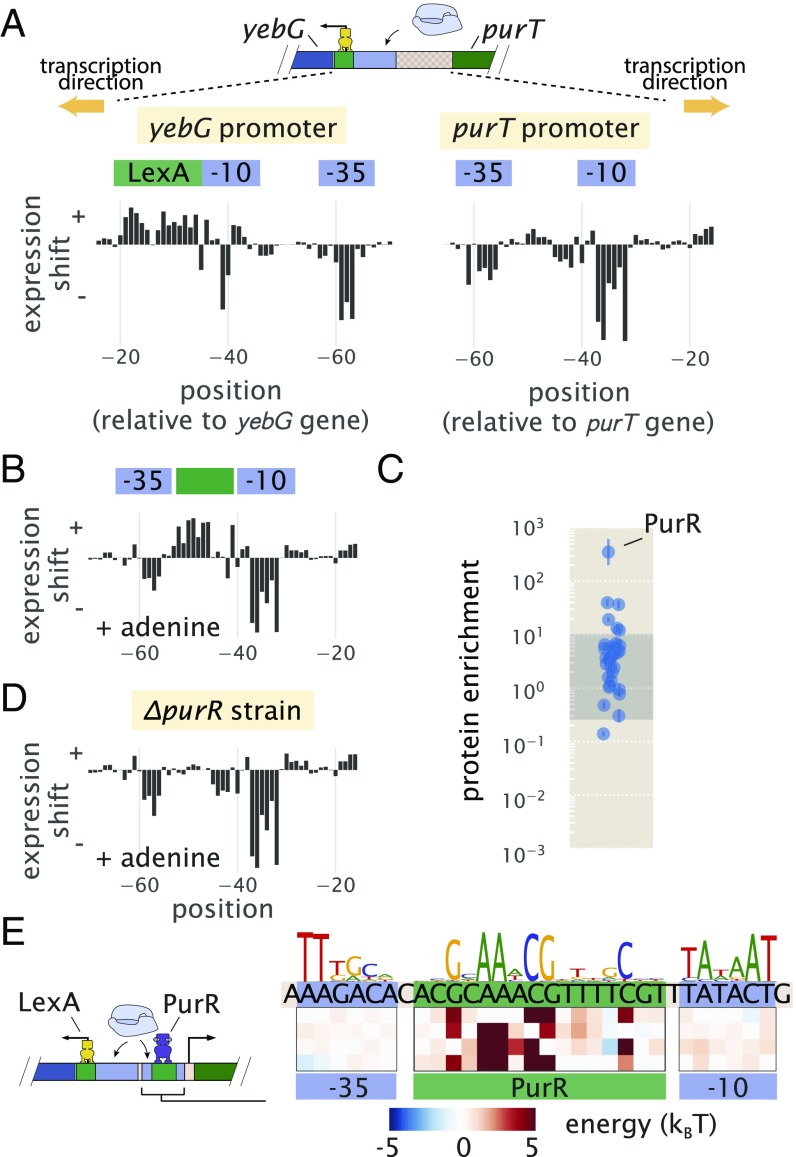
The first of our candidate promoters is associated with expression of purT, one of two genes found in E. coli that catalyze the third step in de novo purine biosynthesis (46, 47). Due to a relatively short intergenic region about 120 bp in length that is shared with a neighboring gene yebG, we also performed Sort-Seq on the yebG promoter [oriented in the opposite direction (48)] (schematic in Fig. 5A). To begin our exploration of the purT and yebG promoters, we performed Sort-Seq with cells grown in M9 minimal media with 0.5% glucose. The associated expression shift plots are shown in Fig. 5A. While we performed Sort-Seq on a larger region than shown for each promoter, we have only plotted the regions where regulation was apparent.
Fig. 5.
Sort-Seq distinguishes directional regulatory features and uncovers the regulatory architecture of the purT promoter. (A) A schematic is shown for the approximately 120-bp region between the yebG and purT genes, which code in opposite directions. Expression shifts are shown for 60-bp regions where regulation was observed for each promoter, with positions noted relative to the start codon of each native coding gene. Cells were grown in M9 minimal media with 0.5% glucose. The −10 and −35 RNAP binding sites of the purT promoter were determined through inference of an energy matrix and are identified in blue. (B) Expression shifts for the purT promoter but in M9 minimal media with 0.5% glucose supplemented with adenine (100 g/ml). A putative repressor site is annotated in green. (C) DNA affinity chromatography was performed using the identified repressor site, and protein enrichment values for transcription factors are plotted. Cell lysate was produced from cells grown in M9 minimal media with 0.5% glucose. Binding was performed in the presence of hypoxanthine (10 g/ml). Error bars represent the SEM calculated using log protein enrichment values from three replicates, and the gray shaded region represents the 95% probability density region of all protein detected. (D) Identical to B but performed with cells containing a purR genetic background. (E) Summary of regulatory binding sites and transcription factors that bind within the intergenic region between the genes of yebG and purT. Energy weight matrices and sequence logos are shown for the PurR repressor and RNAP binding sites. Data were fit to a thermodynamic of simple repression, yielding energies in units of .
For the yebG promoter, the features were largely consistent with prior work, containing binding sites for LexA and RNAP. However, we did find that the RNAP binding site is shifted 9 bp downstream from what was identified previously (48). The previous annotation was based on a computational search and not confirmed experimentally. We were also able to confirm that the yebG promoter was induced in response to DNA damage by repeating Sort-Seq in the presence of mitomycin C [a potent DNA cross-linker known to elicit the DNA damage response and proteolysis of LexA (49)] (SI Appendix, Fig. S8 A, B, and D).
Given the role of purT in the synthesis of purines and the tight control over purine concentrations within the cell (46), we performed Sort-Seq of the purT promoter in the presence or absence of the purine adenine in the growth media. In growth without adenine (Fig. 5A, Right), we observed two negative regions in the expression shift plot. Through inference of an energy matrix, these two features were identified as the −10 and −35 regions of an RNAP binding site. While these two features were still present on addition of adenine, as shown in Fig. 5B, this growth condition also revealed a putative repressor site between the −35 and −10 RNAP binding sites, indicated by a positive shift in expression (green annotation in Fig. 5B).
Following our strategy to find not only the regulatory sequences but also, their associated transcription factors, we next applied DNA affinity chromatography using this putative binding site sequence. In our initial attempt, however, we were unable to identify any substantially enriched transcription factor (SI Appendix, Fig. S8C). With repression observed only when cells were grown in the presence of adenine, we reasoned that the transcription factor may require a related ligand to bind the DNA, possibly through an allosteric mechanism. Importantly, we were able to infer an energy matrix to the putative repressor site with sequence specificity that matched that of the well-characterized repressor, PurR ( = 0.82) (SI Appendix, Fig. S4). We also noted ChIP-chip data of PurR that suggest that it might bind within this intergenic region (47). We, therefore, repeated the purification in the presence of hypoxanthine, which is a purine derivative that also binds PurR (50). As shown in Fig. 5C, we now observed a substantial enrichment of PurR with this putative binding site sequence. As further validation, we performed Sort-Seq once more in the adenine-rich growth condition but in a strain. In the absence of PurR, the putative repressor binding site disappeared (Fig. 5D), which is consistent with PurR binding at this location.
In Fig. 5E, we summarize the regulatory features between the coding genes of purT and yebG, including the features identified by Sort-Seq. With the appearance of a simple repression architecture (51) for the purT promoter, we extended our analysis by developing a thermodynamic model to describe repression by PurR. This enabled us to infer the binding energies of RNAP and PurR in absolute energies (52), and we show the resulting model in Fig. 5E (additional details are in SI Appendix).
The xylE operon is induced in the presence of xylose mediated through binding of XylR and CRP.
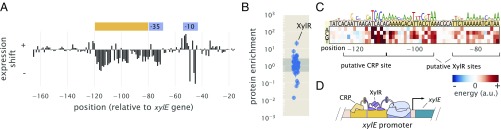
The next unannotated promoter that we considered was associated with expression of xylE, a xylose/proton symporter involved in uptake of xylose. From our analysis of the data from Schmidt et al. (45), we found that xylE was sensitive to xylose and proceeded by performing Sort-Seq in cells grown in this carbon source. Interestingly, the promoter exhibited essentially no expression in other media [the work by Schmidt et al. (45)] (SI Appendix, Fig. S8E). We were able to locate the RNAP binding site between −80 and −40 bp relative to the xylE gene (annotated in blue in Fig. 6A). In addition, the entire region upstream of the RNAP seemed to be involved in activating gene expression (annotated in orange in Fig. 6A), suggesting the possibility of multiple transcription factor binding sites.
Fig. 6.
Sort-Seq identifies a set of activator binding sites that drive expression of RNAP at the xylE promoter. (A) Expression shifts are shown for the xylE promoter, with Sort-Seq performed on cells grown in M9 minimal media with 0.5% xylose. The −10 and −35 regions of an RNAP binding site (blue) and a putative activator region (orange) are annotated. (B) DNA affinity chromatography was performed using the putative activator region, and protein enrichment values for transcription factors are plotted. Cell lysate was generated from cells grown in M9 minimal media with 0.5% xylose, and binding was performed in the presence of xylose supplemented at the same concentration as during growth. Error bars represent the SEM calculated using log protein enrichment values from three replicates. The gray shaded region represents the 95% probability density region of all proteins detected. (C) An energy matrix was inferred for the region upstream of the RNAP binding site. The associated sequence logo is shown above the matrix. Two binding sites for XylR were identified (SI Appendix, Figs. S4 and S8F) along with a CRP binding site. (D) Summary of regulatory features identified at xylE promoter, with the identification of an RNAP binding site and tandem binding sites for XylR and CRP.
We applied DNA affinity chromatography using a DNA target containing this entire upstream region. Due to the stringent requirement for xylose to be present for any measurable expression, xylose was supplemented in the lysate during binding with the target DNA. In Fig. 6B, we plot the enrichment ratios from this purification and find XylR to be most significantly enriched. From an energy matrix inferred for the entire region upstream of the RNAP site, we were able to identify two correlated 15-bp regions (dark yellow shaded regions in Fig. 6C) (Pearson correlation = 0.74 between energy matrices from each binding site). Mutations of the XylR protein have been found to diminish transport of xylose (53), which in light of our result, may be due in part to a loss of activation and expression of this xylose/proton symporter. This is in addition to the loss of activation expected by XylR of the high-affinity xylose uptake system XylFHG (53). These binding sites were also similar to those found on two other promoters known to be regulated by XylR (xylA and xylF promoters), which also exhibit tandem XylR binding sites and strong binding energy predictions with our energy matrix (SI Appendix, Fig. S8F).
Within the upstream activator region in Fig. 6A, there still appeared to be a binding site unaccounted for upstream of the tandem XylR binding sites. From the energy matrix, we were further able to identify a binding site for CRP, which is noted in Fig. 6C. While we did not observe a significant enrichment of CRP in our protein purification, the most energetically favorable sequence predicted by our model, TGCGACCNAGATCACA, closely matches the CRP consensus sequence of TGTGANNNNNNTCACA. In contrast to the lac promoter, binding by CRP here seems to depend more on the right half of the binding site sequence. CRP is known to activate promoters by multiple mechanisms (54), and CRP binding sites have been found adjacent to the activators XylR and AraC (53, 55), in line with our result. While additional work will be needed to characterize the specific regulatory mechanism here, it seems that activation of RNAP is mediated by both CRP and XylR, and we summarize this result in Fig. 6D (considered further in SI Appendix).
The promoter is autorepressed by DgoR with transcription mediated by class II activation by CRP.
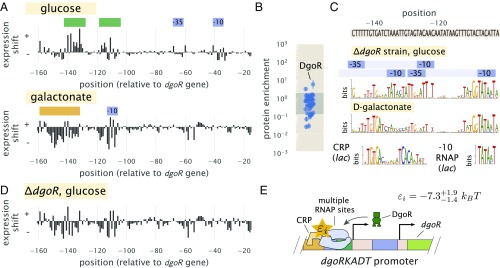
As a final illustration of the approach developed here, we considered the unannotated promoter of dgoRKADT. The operon codes for d-galactonate–catabolizing enzymes; d-galactonate is a sugar acid that has been found as a product of galactose metabolism (56). We began by measuring expression from a nonmutagenized dgoRKADT promoter reporter in response to glucose, galactose, and d-galactonate. Cells grown in galactose exhibited higher expression than in glucose as found by Schmidt et al. (45), and they exhibited even higher expression when cells were grown in d-galactonate (SI Appendix, Fig. S9A). This likely reflects the physiological role provided by the genes of this promoter, which seems necessary for metabolism of d-galactonate. We, therefore, proceeded by performing Sort-Seq with cells grown in either glucose or d-galactonate, since these appeared to represent distinct regulatory states, with expression low in glucose and high in d-galactonate. Expression shift plots from each growth conditions are shown in Fig. 7A.
Fig. 7.
The dgoRKADT promoter is induced in the presence of d-galactonate due to loss of repression by DgoR and activation by CRP. (A) Expression shifts due to mutating the dgoRKADT promoter are shown for cells grown in M9 minimal media with either 0.5% glucose (Upper) or 0.23% d-galactonate (Lower). Regions identified as RNAP binding sites (−10 and −35) are shown in blue, and putative activator and repressor binding sites are shown in orange and green, respectively. (B) DNA affinity purification was performed targeting the region between −145 bp and −110 bp of the dgoRKADT promoter. The transcription factor DgoR was found most enriched among the transcription factors plotted. Error bars represent the SEM calculated using log protein enrichment values from three replicates, and the gray shaded region represents the 95% probability density region of all proteins detected. (C) Sequence logos were inferred for the most upstream 60-bp region associated with the upstream RNAP binding site annotated in A. Multiple RNAP binding sites were identified using Sort-Seq data performed in a strain grown in M9 minimal media with 0.5% glucose (further detailed in SI Appendix, Fig. S9). Below this, a sequence logo was also inferred using data from Sort-Seq performed on wild-type cells grown in d-galactonate, identifying a CRP binding site [class II activation (54)]. (D) Expression shifts are shown for the dgoRKADT promoter when performed in a genetic background grown in 0.5% glucose. This resembles growth in d-galactonate, suggesting d-galactonate may act as an inducer for DgoR. (E) Summary of regulatory features identified at the dgoRKADT promoter, with the identification of multiple RNAP binding sites and binding sites for DgoR and CRP. The interaction energy between CRP and RNAP, , was inferred to be , where the superscripts and subscripts represent the upper and lower bounds associated with 95 percent of the inferred parameter value distribution, respectively.
We begin by considering the results from growth in glucose (Fig. 7A, Upper). Here we identified an RNAP binding site between −30 and −70 bp relative to the native start codon for dgoR (SI Appendix, Fig. S9B). Another distinct feature was a positive expression shift in the region between −140 and −110 bp, suggesting the presence of a repressor binding site. Applying DNA affinity chromatography using this target region, we observed an enrichment of DgoR (Fig. 7B), suggesting that the promoter is indeed under repression and regulated by the first coding gene of its transcript. As further validation of binding by DgoR, the positive shift in expression was no longer observed when Sort-Seq was repeated in a strain (Fig. 7D and SI Appendix, Fig. S9C). We also were able to identify additional RNAP binding sites that were not apparent due to binding by DgoR. While only one RNAP 10 motif is clearly visible in the sequence logo shown in Fig. 7C (top sequence logo; TATAAT consensus sequence), we used simulations to show that the entire sequence logo shown can be explained by the convolution of three overlapping RNAP binding sites (SI Appendix, Fig. S9F).
Next, we consider the d-galactonate growth condition (Fig. 7A, Lower). Like in the expression shift plot for the strain grown in glucose, we no longer observe the positive expression shift between −140 and −110 bp. While there are still several positions between −120 and −100 bp that are still positive, this can be attributed to a nonoptimal −10 binding site sequence for RNAP (wild type, TACATT) (Fig. 7C). The loss of the repressive feature, therefore, suggests that DgoR may be induced by d-galactonate or a related metabolite. However, in comparison with the expression shifts in the strain grown in glucose, there were some notable differences in the region between −160 and −140 bp. Here, we find evidence for another CRP binding site. The sequence logo identifies the sequence TGTGA (Fig. 7C, Lower), which matches the left side of the CRP consensus sequence. In contrast to the lac and xylE promoters, however, the right half of the binding site directly overlaps with where we would expect to find a −35 RNAP binding site. This type of interaction by CRP has been previously observed and is defined as class II CRP-dependent activation (54), although this sequence specificity has not been previously described.
To isolate and better identify this putative CRP binding site, we repeated Sort-Seq in E. coli strain JK10 grown in 500 M cAMP. Strain JK10 lacks adenlyate cyclase (cyaA) and phosphodiesterase (cpdA), which are needed for cAMP synthesis and degradation, respectively, and it is thus unable to control intracellular cAMP levels necessary for activation by CRP [derivative of TK310 (41)]. Growth in the presence of 500 M cAMP provided strong induction from the dgoRKADT promoter and resulted in a sequence logo at the putative CRP binding site that even more clearly resembled binding by CRP (SI Appendix, Fig. S9E). This is likely because expression is now dominated by the CRP-activated RNAP binding site. Importantly, these data allowed us to further infer the interaction energy between CRP and RNAP, which we estimate to be −7.3 (further detailed in SI Appendix). We summarize the identified regulatory features in Fig. 7E.
Discussion
We have established a systematic procedure for dissecting the functional mechanisms of previously uncharacterized regulatory sequences in bacteria. A massively parallel reporter assay, Sort-Seq (13), is used to first elucidate the locations of functional transcription factor binding sites. DNA oligonucleotides containing these binding sites are then used to enrich the cognate transcription factors and identify them by mass spectrometry analysis. Information-based modeling and inference of energy matrices that describe the DNA binding specificity of regulatory factors provide further quantitative insight into transcription factor identity and the growth condition-dependent regulatory architectures.
To validate this approach, we examined four previously annotated promoters of lac, rel, mar, and yebG, and our results were consistent with established knowledge (13, 31, 33, 34, 39, 43). Importantly, we find that DNA affinity chromatography experiments on these promoters were highly sensitive. In particular, LacI was unambiguously identified with the weak O3 binding site, although LacI is present in only about 10 copies per cell (34). Emboldened by this success, we then studied promoters having little or no prior regulatory annotation: purT, xylE, and dgoR. Here, our analysis led to a collection of regulatory hypotheses. For the purT promoter, we identified a simple repression architecture (51), with repression by PurR. The xylE promoter was found to undergo activation only when cells are grown in xylose, likely due to allosteric interaction between the activator XylR and xylose and activation by CRP (53, 55). Finally, in the case of dgoR, the base pair resolution allowed us to tease apart overlapping regulatory binding sites, identify multiple RNAP binding sites along the length of the promoter, and infer further quantitative detail about the interaction between the identified binding sites for CRP and RNAP. We view these results as a critical first step in the quantitative dissection of transcriptional regulation, which will ultimately be needed for a predictive understanding of how such regulation works.
While our results show the successful identification of regulatory binding sites and regulatory mechanism at previously unannotated promoters, there also remain important challenges. The uncharacterized genes were selected based on genome-wide studies (44, 45), and indeed, the hints of regulation in these data were a necessary part of our strategy to systematically dissect each promoter. Datasets that quantitate protein abundance across a number of growth conditions, like those available in E. coli (45) and yeast (57), or alternatively, transcript abundance using RNA sequencing (RNA-Seq) will provide an important starting point for the dissection of regulatory mechanism in other bacteria.
An important aspect of the presented approach is that it can be applied to any promoter sequence, and there are a number of ways that throughput can be increased further. Microarray-synthesized promoter libraries and measurement of expression from barcoded transcripts using RNA-Seq instead of flow cytometry can be used to allow multiple loci to be studied simultaneously (14, 18). Landing pad technologies for chromosomal integration (58–60) should enable massively parallel reporter assays to be performed in chromosomes instead of on plasmids. Techniques that combine these assays with transcription start site readout (61) may provide additional resolution, further allowing the molecular regulators of overlapping RNAP binding sites to be deconvolved or the contributions from separate RNAP binding sites, like those observed on the dgoR promoter, to be better distinguished. As the number of regulatory regions under study increases, it will also be important to develop additional analysis tools that provide automated identification of regulatory binding sites.
To identify transcription factors across many target binding sites, DNA affinity chromatography samples can be further mutliplexed using isobaric labeling strategies (62, 63). Continued performance improvements in mass spectrometer sensitivity and sample processing (64–66) will also make this assay less onerous to apply across many targets and different binding conditions. This will be especially important for situations where the data suggest that a small molecule effector might be acting to modulate binding of the transcription factor to its target sequence, requiring multiple binding conditions to be tested. Performing reporter assays in transcription factor deletion strains will continue to play an important role in promoter dissection as we have shown for a variety of the promoters, and it will provide a secondary means with which to identify and validate binding sites. Genome-wide gene deletion libraries are now available for a wide variety of bacteria (67–72), and it is now possible to perform genetic perturbations using CRISPR interference (73, 74) that should open up the possibility of applying such perturbation strategies more easily in less studied organisms.
Although our work was directed toward regulatory regions of E. coli, there are no intrinsic limitations that restrict the analysis to this organism. Rather, most bacteria contain small intergenic regions several hundred base pairs in length that make this approach especially suitable. The sequence specificity of most characterized prokaryotic transcription factors (75, 76) and the sigma factors that allow RNAP to recognize each promoter (54, 77) suggest that this approach will permit regulatory dissection in any bacterium that supports efficient transformation by plasmids. Additionally, although we have focused on bacteria, our general strategy should be feasible for dissecting regulation in a number of eukaryotic systems—including human cell culture—using massively parallel reporter assays (14–16) and DNA-mediated protein pull-down methods (21, 22) that have already been established.
Materials and Methods
SI Appendix has extended experimental details.
Bacterial Strains.
All E. coli strains used in this work were derived from K-12 MG1655, with deletion strains generated by the lambda red recombinase method (78). In the case of deletions for lysA (::kan), purR (::kan), and xylE (::kan), strains were obtained from the Coli Genetic Stock Center (Yale University) and transferred into a fresh MG1655 strain using P1 transduction. The others were generated in house and include the following deletion strains: , ::kan, marR::kan, and ::kan. Details on strain construction are provided in SI Appendix.
Sort-Seq.
Mutagenized single-stranded oligonucleotide pools were purchased from Integrated DNA Technologies. Library oligonucleotides were PCR amplified, inserted into the PCR-amplified plasmid backbone (i.e., vector) of pJK14 (SC101 origin) (13) by Gibson assembly, and electroporated into cells after drop dialysis in water. Cell libraries were then grown to saturation in LB and diluted 1:10,000 into the appropriate growth media for the promoter under consideration, and grown to an optical density at 600 nm of 0.2–0.4. A Beckman Coulter MoFlo XDP cell sorter was used to sort cells by fluorescence, with 500,000 cells collected into each of the four bins. Sorted cells were then regrown overnight in 10 mL of LB media under kanamycin selection. The plasmids in each bin were miniprepped (Qiagen) after overnight growth, and PCR was used to amplify the mutated region from each plasmid for Illumina sequencing. SI Appendix has additional details on library construction and Sort-Seq as well as on calculating expression shift plots and energy matrices.
DNA Affinity Chromatography and Liquid Chromatography-MS/MS.
SILAC labeling (27, 28, 30) was implemented by growing cells (MG1655 ) in either the stable isotopic form of lysine (13C6H14 15N2O2) or natural form. SI Appendix has details on lysate preparation.
DNA affinity chromatography was performed by incubating cell lysate (150 mg/mL protein) with magnetic beads (Dynabeads MyOne T1; ThermoFisher) containing tethered DNA (streptavidin-biotin linkage). ssDNA was purchased from Integrated DNA Technologies with the biotin modification on the 5′ end of the oligonucleotide sense strand. Cell lysates were incubated on a rotating wheel with the DNA tethered beads overnight at 4 °C. Elution was achieved by cleaving the DNA with the restriction enzyme PstI, and samples were then prepared for mass spectrometry by ingel digestion with endoproteinase Lys-C. Liquid chromatography tandem mass spectrometry experiments were carried out as previously described (79), and they are further detailed in SI Appendix. Thermo RAW files were processed using MaxQuant (v. 1.5.3.30) (80).
Code Availability and Data Analysis.
All code used for processing data and plotting as well as the final processed data, plasmid sequences, and primer sequences can be found on our GitHub repository (https://www.github.com/RPGroup-PBoC/sortseq_belliveau; DOI: 10.5281/zenodo.1184169). Thermo RAW files for mass spectrometry are available at the jPOSTrepo repository (81) (accession no. PXD007892). Sort-Seq sequencing files are available at the Sequence Read Archive (accession no. SRP121362).
Supplementary Material
Acknowledgments
We thank David Tirrell, Bradley Silverman, and Seth Lieblich for access to their Beckman Coulter MoFlo XDP cell sorter. Jost Vielmetter and Nina Budaeva provided access to their Cell Disruptor. We also thank Hernan Garcia, Manuel Razo-Mejia, Griffin Chure, Suzannah Beeler, Heun Jin Lee, Justin Bois, and Soichi Hirokawa for useful discussion. This work was supported by La Fondation Pierre-Gilles de Gennes; the Rosen Center at Caltech; NIH Grants DP1 OD000217 (Director’s Pioneer Award), R01 GM085286, 1R35 GM118043-01 (Maximizing Investigators’ Research Award), and 1S10RR029594-01A1; the Gordon and Betty Moore Foundation through Grant GBMF227; and the Beckman Institute. N.M.B. was supported by an HHMI International Student Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw sequencing files have been deposited on the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. SRP121362) and Sort-Seq regulatory sequencing data from Escherichia coli has been deposited in NCBI BioSample, https://www.ncbi.nlm.nih.gov/biosample (accession no. SAMN07830099). Thermo RAW mass spectrometry files have been deposited in the jPOST Repository, https://repository.jpostdb.org (accession no. PXD007892). Files containing processed data and python code association with data analysis and plotting have been deposited on GitHub and Zenodo (available at https://www.github.com/RPGroup-PBoC/sortseq_belliveau and https://doi.org/10.5281/zenodo.1184169, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722055115/-/DCSupplemental.
References
- 1.Gama-Castro S, et al. RegulonDB version 9.0: High-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016;44:D133–D143. doi: 10.1093/nar/gkv1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keseler IM, et al. EcoCyc: Fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Münch R, et al. PRODORIC: Prokaryotic database of gene regulation. Nucleic Acids Res. 2003;31:266–269. doi: 10.1093/nar/gkg037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cipriano MJ, et al. RegTransBase–A database of regulatory sequences and interactions based on literature: A resource for investigating transcriptional regulation in prokaryotes. BMC Genomics. 2013;14:213–221. doi: 10.1186/1471-2164-14-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiliç S, White ER, Sagitova DM, Cornish JP, Erill I. CollecTF: A database of experimentally validated transcription factor-binding sites in bacteria. Nucleic Acids Res. 2013;42:D156–D160. doi: 10.1093/nar/gkt1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJW. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc Natl Acad Sci USA. 2005;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vora T, Hottes AK, Tavazoie S. Protein occupancy landscape of a bacterial genome. Mol Cell. 2009;35:247–253. doi: 10.1016/j.molcel.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonocora RP, Wade JT. ChIP-Seq for Genome-Scale Analysis of Bacterial DNA-Binding Proteins. Humana; New York: 2015. pp. 327–340. [DOI] [PubMed] [Google Scholar]
- 9.Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh SS, et al. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 2014;28:214–219. doi: 10.1101/gad.234336.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wade JT. ChIP-seq for genomic-scale analysis of bacterial DNA-binding proteins. In: Artsimovitch I, Santangelo TJ, editors. Prokaryotic Systems Biology. Vol 883. Humana Press; New York: 2015. pp. 119–134. [Google Scholar]
- 12.Minchin SD, Busby SJW. Analysis of mechanisms of activation and repression at bacterial promoters. Methods. 2009;47:6–12. doi: 10.1016/j.ymeth.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Kinney JB, Murugan A, Callan CG, Cox EC. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc Natl Acad Sci USA. 2010;107:9158–9163. doi: 10.1073/pnas.1004290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kheradpour P, et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharon E, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosuri S, et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci USA. 2013;110:14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maricque BB, Dougherty JD, Cohen BA. A genome-integrated massively parallel reporter assay reveals DNA sequence determinants of cis-regulatory activity in neural cells. Nucleic Acids Res. 2017;45:e16–e16. doi: 10.1093/nar/gkw942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulco CP, et al. Systematic mapping of functional enhancer–Promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittler G, Butter F, Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2009;19:284–293. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirzaei H, et al. Systematic measurement of transcription factor-DNA interactions by targeted mass spectrometry identifies candidate gene regulatory proteins. Proc Natl Acad Sci USA. 2013;110:3645–3650. doi: 10.1073/pnas.1216918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mustonen V, Kinney J, Callan CG, Lassig M. Energy-dependent fitness: A quantitative model for the evolution of yeast transcription factor binding sites. Proc Natl Acad Sci USA. 2008;105:12376–12381. doi: 10.1073/pnas.0805909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ireland WT, Kinney JB. 2016. MPAthic: Quantitative modeling of sequence-function relationships for massively parallel assays. bioRxiv:054676.
- 26.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 28.Kerner MJ, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 29.Calloni G, et al. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Soufi B, Macek B. Stable isotope labeling by amino acids applied to bacterial cell culture. In: Warscheid B, editor. Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) Humana; New York: 2014. pp. 9–22. [DOI] [PubMed] [Google Scholar]
- 31.Oehler S, Eismann ER, Krämer H, Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerdes K, Christensen SK, Løbner-Oleson A. Prokaryotic toxin–Antitoxin stress response loci. Nat Rev Microbiol. 2005;2:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 33.Alekshun MN, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: The mar regulon. J Mol Biol. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia HG, Phillips R. Quantitative dissection of the simple repression input-output function. Proc Natl Acad Sci USA. 2011;108:12173–12178. doi: 10.1073/pnas.1015616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 36.Overgaard M, Borch J, Gerdes K. Bacterial toxin RelE: A highly efficient ribonuclease with exquisite substrate specificity using atypical catalytic residues. Biochem. 2013;52:8633–8642. doi: 10.1021/bi401325c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overgaard M, Borch J, Gerdes K. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon–helix–helix motif in RelB. J Mol Biol. 2009;394:183–196. doi: 10.1016/j.jmb.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin RG, Rosner JL. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J Bacteriol. 1997;179:7410–7419. doi: 10.1128/jb.179.23.7410-7419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seoane AS, Levy SB. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhlman T, Zhang Z, Saier MH, Hwa T. Combinatorial transcriptional control of the lactose operon of Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6043–6048. doi: 10.1073/pnas.0606717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li GY, Zhang Y, Inouye M, Ikura M. Structural mechanism of transcriptional autorepression of the Escherichia coli RelB/RelE antitoxin/toxin module. J Mol Biol. 2008;380:107–119. doi: 10.1016/j.jmb.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 43.Overgaard M, Borch J, Jørgensen MG, Gerdes K. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol Microbiol. 2008;69:841–857. doi: 10.1111/j.1365-2958.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- 44.Marbach D, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt A, et al. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol. 2016;34:104–111. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rolfes RJ. Regulation of purine nucleotide biosynthesis: In yeast and beyond. Biochem Soc Trans. 2006;34:786–790. doi: 10.1042/BST0340786. [DOI] [PubMed] [Google Scholar]
- 47.Cho BK, et al. The PurR regulon in Escherichia coli K-12 MG1655. Nucleic Acids Res. 2011;39:6456–6464. doi: 10.1093/nar/gkr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lomba MR, Vasconcelos AT, Pacheco ABF, Almeida DF. Identification of yebG as a DNA damage-inducible Escherichia coli gene. FEMS Microbiol Ecol. 1997;156:119–122. doi: 10.1111/j.1574-6968.1997.tb12715.x. [DOI] [PubMed] [Google Scholar]
- 49.Wade JT, Reppas NB, Church GM, Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi KY, Zalkin H. Structural characterization and corepressor binding of the Escherichia coli purine repressor. J Bacteriol. 1992;174:6207–6214. doi: 10.1128/jb.174.19.6207-6214.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bintu L, et al. Transcriptional regulation by the numbers: Models. Curr Opin Genet Dev. 2005;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atwal GS, Kinney JB. Learning quantitative sequence-function relationships from massively parallel experiments. J Stat Phys. 2016;162:1203–1243. [Google Scholar]
- 53.Song S, Park C. Organization and regulation of the D-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J Bacteriol. 1997;179:7025–7032. doi: 10.1128/jb.179.22.7025-7032.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Browning DF, Busby SJW. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol. 2016;14:638–650. doi: 10.1038/nrmicro.2016.103. [DOI] [PubMed] [Google Scholar]
- 55.Laikova ON, Mironov AA, Gelfand MS. Computational analysis of the transcriptional regulation of pentose utilization systems in the gamma subdivision of Proteobacteria. FEMS Microbiol Ecol. 2001;205:315–322. doi: 10.1111/j.1574-6968.2001.tb10966.x. [DOI] [PubMed] [Google Scholar]
- 56.Cooper RA. The utilisation of D-galactonate and D-2-oxo-3-deoxygalactonate by Escherichia coli K-12. Biochemical and genetical studies. Arch Microbiol. 1978;1:199–206. doi: 10.1007/BF00415730. [DOI] [PubMed] [Google Scholar]
- 57.Ho B, Baryshnikova A, Brown GW. Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Sys. 2018;6:1–14. doi: 10.1016/j.cels.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Kuhlman TE, Cox EC. Site-specific chromosomal integration of large synthetic constructs. Nucleic Acids Res. 2010;38:e92. doi: 10.1093/nar/gkp1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H, Susanto TT, Wan Y, Chen SL. Comprehensive mutagenesis of the fimS promoter regulatory switch reveals novel regulation of type 1 pili in uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2016;113:4182–4187. doi: 10.1073/pnas.1522958113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urtecho G, Tripp AD, Insigne K, Kim H, Kosuri S. Systematic dissection of sequence elements controlling 70 promoters using a genomically-encoded multiplexed reporter assay in E. coli. Biochemistry. February 1, 2018 doi: 10.1021/acs.biochem.7b01069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vvedenskaya IO, et al. Massively systematic transcript end readout, “MASTER”: Transcription start site selection, transcriptional slippage, and transcript yields. Mol Cell. 2015;60:953–965. doi: 10.1016/j.molcel.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson A, et al. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by mass spectrometry/mass spectrometry. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 63.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Erickson BK, et al. A strategy to combine sample multiplexing with targeted proteomics assays for high-throughput protein signature characterization. Mol Cell. 2017;65:361–370. doi: 10.1016/j.molcel.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 66.Hubner NC, Nguyen LN, Hornig NC, Stunnenberg HG. A quantitative proteomics tool to identify DNA–protein interactions in primary cells or blood. J Proteome Res. 2014;14:1315–1329. doi: 10.1021/pr5009515. [DOI] [PubMed] [Google Scholar]
- 67.Baba T, et al. Construction of Escherichia coli k-12 in-frame, single-gene knockout mutants: The keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koo BM, et al. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Sys. 2017;4:291–305.e7. doi: 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Berardinis V, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174–154. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Porwollik S, et al. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One. 2014;9:e99820. doi: 10.1371/journal.pone.0099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu P, et al. Genome-wide essential gene identification in Streptococcus Sanguinis. Sci Rep. 2011;1:125. doi: 10.1038/srep00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liberati NT, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larson MH, et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gordon GC, et al. CRISPR interference as a titratable, trans-acting regulatory tool for Metab Eng in the cyanobacterium Synechococcus sp. strain PCC 7002. Metab Eng. 2016;38:170–179. doi: 10.1016/j.ymben.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lässig M. From biophysics to evolutionary genetics: Statistical aspects of gene regulation. BMC Bioinformatics. 2007;8(Suppl 6):S7. doi: 10.1186/1471-2105-8-S6-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stewart AJ, Plotkin JB. Why transcription factor binding sites are ten nucleotides long. Genetics. 2012;192:973–985. doi: 10.1534/genetics.112.143370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feklístov A, Sharon BD, Darst SA, Gross CA. Bacterial sigma factors: A historical, structural, and genomic perspective. Annu Rev Microbiol. 2014;68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 78.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kalli A, Hess S. Effect of mass spectrometric parameters on peptide and protein identification rates for shotgun proteomic experiments on an LTQ-orbitrap mass analyzer. Proteomics. 2011;12:21–31. doi: 10.1002/pmic.201100464. [DOI] [PubMed] [Google Scholar]
- 80.Cox J, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 81.Okuda S, et al. jPOSTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 2017;45:D1107–D1111. doi: 10.1093/nar/gkw1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.