Abstract
Background
The genetic basis of animal domestication remains poorly understood, and systems with substantial phenotypic differences between wild and domestic populations are useful for elucidating the genetic basis of adaptation to new environments as well as the genetic basis of rapid phenotypic change. Here, we sequenced the whole genome of 78 individual ducks, from two wild and seven domesticated populations, with an average sequencing depth of 6.42X per individual.
Results
Our population and demographic analyses indicate a complex history of domestication, with early selection for separate meat and egg lineages. Genomic comparison of wild to domesticated populations suggests that genes that affect brain and neuronal development have undergone strong positive selection during domestication. Our FST analysis also indicates that the duck white plumage is the result of selection at the melanogenesis-associated transcription factor locus.
Conclusions
Our results advance the understanding of animal domestication and selection for complex phenotypic traits.
Keywords: duck, domestication, intensive selection, neuronal development, energy metabolism, plumage colouration
Background
Animal domestication was one of the major contributory factors to the agricultural revolution during the Neolithic period, which resulted in a shift in human lifestyle from hunting to farming [1]. Compared with their wild progenitors, domesticated animals showed notable changes in behavior, morphology, physiology, and reproduction [2]. Detecting domestication-mediated selective signatures is important for understanding the genetic basis of both adaptation to new environments and rapid phenotype change [3, 4]. In recent years, to characterize signatures of domestication, whole-genome resequencing studies have been performed on a wide range of agricultural animals, including pig [5], sheep [6], rabbit [7], and chicken [8, 9].
Mallards (Anas platyrhynchos) are the world's most widely distributed and agriculturally important waterfowl species and are of particular economic importance in Asia [10]. Southeast Asia, particularly southern China, is the major center of duck domestication, with records indicating duck farming in the region dating at least 2,000 years [11, 12], particularly in wet environments [13] associated with rice crops [14]. In the absence of archaeological evidence, the exact timing of domestication and the time of meat and egg type ducks split remains unknown, with the first written records of domestic ducks in central China shortly after 500 BC [15].
It is clear that the domesticated duck originated from mallards [16], and domestic ducks can be classified as those produced primarily for meat (similar to chicken broilers) or eggs (similar to chicken layer lines). Together with the timing of duck domestication, the relative separation of duck meat and egg lines is also unknown. It is unclear whether ducks were domesticated once and subsequently selected for divergent meat and egg production traits or whether meat and egg populations were derived independently in two domestication events from wild mallards.
Moreover, domesticated mallards show many important behavioral [17] and morphological [18–20] differences from their wild ancestors, particularly related to plumage and neuroanatomy. However, the genetic basis of these phenotypic differences is still poorly understood.
Data Description
In order to determine the timing of duck domestication in China, as well as identify the genomic regions under selection during domestication, we performed whole-genome resequencing from 78 individuals belonging to seven duck breeds (three for meat breeds, three for egg breeds, and one dual-purpose breed) and two geographically distinct wild populations. Using the large number of single nucleotide polymorphisms (SNPs) as well as small insertions and deletions (INDELs), we tested for population structure between domesticated and wild populations and we assessed the genome for signatures of selection associated with domestication. We tested alternative demographic scenarios with the pairwise sequential Markovian coalescent method combined with the diffusion approximation method.
Analyses
Genetic variation
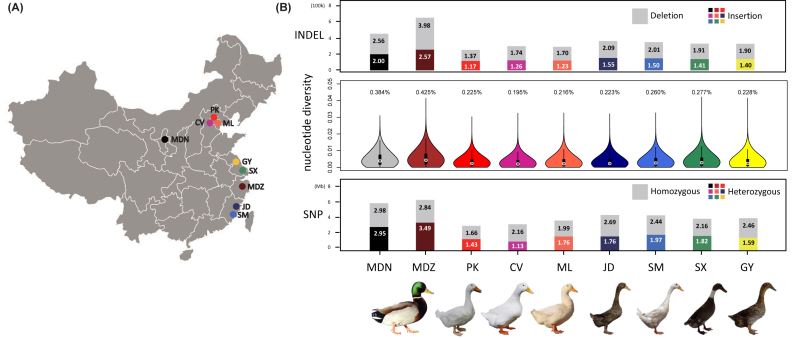
We individually sequenced 22 wild and 56 domestic ducks from two wild populations and seven domestic breeds (three meat breeds, three egg breeds, and one dual-purpose breed) from across China (Fig.1A) to an average of 6.42X coverage per individual (613.37 Gb of high-quality paired-end sequence data) after filtering and quality control, resulting in 535 billion mappable reads across 78 ducks (Supplemental Table S1).
Figure 1:
Experimental design and variants statistics. A)null Sampling sites in this study. A total of 78 ducks from two wild populations (mallard Ningxia [MDN] n = 8; mallard Zhejiang [MDZ] n = 14), three meat breeds (Pekin [PK] n = 8; Cherry Valley [CV] n = 8; maple leaf [ML] n = 8), three egg breeds (Jin Ding [JD] n = 8; Shan Ma [SM] n = 8; Shao Xing [SX] n = 8), and one dual-purpose breed (Gao You [GY] n = 8) were selected. B) Genomic variation of nine populations. Mean number of SNPs and heterozygous and homozygous SNP ratio in the nine populations are shown at the bottom. Nucleotide diversity ratios of the nine populations are shown at the middle. The nucleotide diversity ratios in wild mallards are dramatically higher than ratios in domesticated ducks. Number of insertions and deletions in the nine populations are shown at the top. The number of deletions was higher than the number of insertions in all nine populations.
Across samples, we identified 39.2 million variants, consisting of 36.1 million SNPs (average per sample = 4.5 m SNPs; range = 2.34 – 9.52 M SNPs) and 3.1 million INDELs (average per sample = 0.4 million INDELs; range = 0.21 – 0.89 million INDELs) (Fig.1B, Supplemental Figs. S1 and S2, Supplemental Table S2). Single base-pair INDELs were the most common, accounting for 38.63% of all detected INDELs (Supplemental Table S3). Our dataset covers 96.2% of the duck dbSNP database deposited in the Genome Variation Map (GVM) [21]. In general, domesticated populations showed lower number of SNPs (t test, P = 3.13 × 10−12) and nucleotide diversity (t test, P = 2.20 × 10−16) compared to wild mallards (Fig.1B). Moreover, homozygosity in domesticated ducks was significantly higher than ratios in wild mallards (t test, P = 1.35 × 10−10) consistent with the larger panmictic wild population or with the higher artificial selection and inbreeding within domesticated stocks.
Population structure and domestication
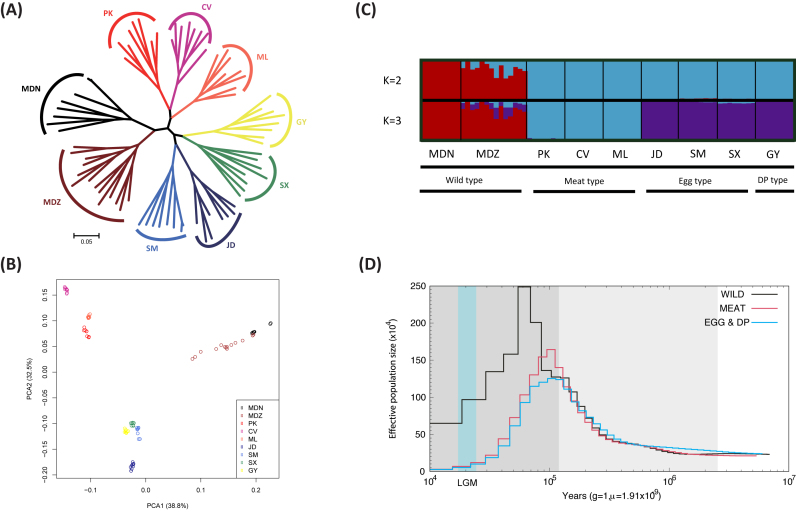
Phylogenetic relationships, based on a neighbor-joining of pairwise genetic distances of whole-genome SNPs (Fig.2A) and principal component analysis (PCA; Fig.2B), revealed strong clustering into three distinct genetic groups. In general, we observed separate clusters corresponding to wild ducks (MDN and MDZ), ducks domesticated for meat production (PK, CV, and ML), and ducks domesticated for egg production (JD, SM, and SX). The dual-purpose domesticate (GY) clustered with ducks domesticated for egg production (Fig.2B and C).
Figure 2:
Population genetic structure and demographic history of nine duck populations. A) Neighbor-joining phylogenetic tree of nine duck populations. The scale bar is proportional to genetic differentiation (p distance). B) PCA plot of duck populations. Eigenvector 1 and 2 explained 38.8% and 32.5% of the observed variance, respectively. C) Population genetic structure of 78 ducks. The length of each colored segment represents the proportion of the individual genome inferred from ancestral populations (K = 2–3). The population names and production type are at the bottom. DP type means dual-purpose type. D) Demographic history of duck populations. Examples of PSMC estimate changes in the effective population size over time, representing variation in inferred Ne dynamics. The lines represent inferred population sizes and the gray shaded areas indicate the Pleistocene period, with Last Glacial Period (LGP) shown in darker gray, and Last Glacial Maximum (LGM) shown in light blue areas.
We further performed population structure analysis using FRAPPE [22], which estimates individual ancestry and admixture proportions assuming K ancestral populations (Fig. 2C). With K = 2, a clear division was found between wild-type ducks (MDN and MDZ) and domesticated ducks (PK, CV, ML, JD, SM, SX, and GY). With K = 3, a clear division was found between meat type ducks (PK, CV, and ML) and egg type ducks mixed with dual-purpose type ducks (JD, SM, SX, and GY).
Next, we explored the demographic history of our samples to differentiate whether domestication of meat- and egg-producing ducks was the result of one or multiple events. First, we estimated changes in effective population size (Ne) in our three genetic clusters in a pairwise sequentially Markovian coalescent (PSMC) framework [23]. The meat type ducks (PK, CV, and ML) showed concordant demographic trajectories with egg and mixture dual-purpose type populations (JD, SM, SX, and GY), with one apparent expansion around the Penultimate Glaciation Period (0.30-0.13 million years ago) [4, 24] and Last Glacial Period (110–12 thousand years ago) [25, 26], followed by a subsequent contraction (Fig. 2D). Next, we tested multiple demographic scenarios related to domestication using a diffusion approximation method for the allele frequency spectrum (∂a∂i) (Supplemental Figs. S3 and S4). Among the four isolation models tested (models 1 – 4), the model of a single domestication with subsequent divergence of the domesticated breeds (model 2) was both consistent with our population structure results (Fig.2) and had the lowest Akaike information criteria (AIC) value, indicating a better overall fit to the data (log-likelihood = –33 388.43; AIC = 66 788) (Supplemental Fig. S3).
Demographic parameters estimated from the single domestication model (model 2) indicated that domestication occurred 2,228 years ago, with 95% confidence interval (CI) ± 441 years ago, followed by a rapid subsequent divergence of the meat breed from the egg/dual-purpose breeds roughly 100 years after the initial domestication event (Table1). Our results suggest that following an initial bottleneck associated with domestication, with an estimated Ne of 320 (95% CI ± 3) individuals for the ancestral domesticated population, the population has expanded to the current Ne of 5,597 (95% CI ± 1,195) and 12,988 (95% CI ± 2,877) in the meat type and egg/dual purpose breeds, respectively. Ne estimates for domesticated breeds are lower than the Ne of 88,842 (95% CI ±18,065) in wild mallards, consistent with the large panmictic wild population.
Table 1:
Maximum likelihood population demographic parameters
| Parameter | ML estimate | 95% CI |
|---|---|---|
| Ne of ancestral population after size change | 663,439 | 644,726–682,152 |
| Ne of the wild population | 88,842 | 70-778–106-907 |
| Ne of the ancestral domesticated population | 320 | 316–323 |
| Ne of the meat breed | 5,597 | 4,402–6,792 |
| Ne of the egg/dual-purpose | 12,988 | 10,111–15,865 |
| Time of size change in the ancestral population | 249,944 | 227,912–267,518 |
| Time of domestication | 2,228 | 1,787–2,669 |
| Time of breed divergence | 2,126 | 1,686–2,567 |
| Migration wild ← meat | 1.12 | 1.00–1.24 |
| Migration wild ← egg/dp | 3.92 | 3.11–4.73 |
Best fit parameter estimates for the model of a single domestication event followed by divergence of the domesticated breeds, including changes in population size. The 95% confidence intervals were obtained from 100 bootstrap datasets. Time estimates are given in years and migration are in units of number of migrants per generation.
Gene flow estimates were relatively high, with 1 and 4 migrants per generation from the meat and egg/dual-purpose breeds, respectively, into the wild population. Our results suggest duck domestication was a recent single domestication event followed by rapid subsequent selection for separate meat and egg/dual-purpose breeds.
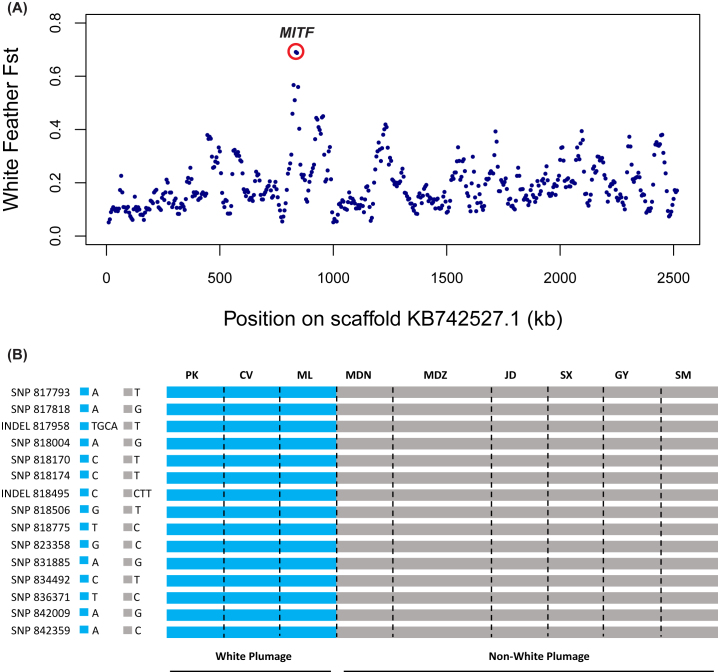
Selection for plumage color
Derived traits in domesticated animals tend to evolve in a predictable order, with color variation appearing in the earliest stages of domestication, followed by coat or plumage and structural (skeletal and soft tissue) variation, and finally behavioral differences [27, 28]. One of the simplest and most visible derived traits of ducks is white plumage color. In order to detect the signature of selection associated with white feathers, we searched the duck genome for regions with high FST between the populations of white-feather (PK, CV, and ML) and non-white-feather (MDN, MDZ, JD, SX, and GY) birds based on sliding 10-kbwindows. We identified a region of high differentiation between white-plumage and non-white-plumage ducks overlapping the melanogenesis associated transcription factor (MITF; FST = 0.69) (Fig. 3A). In the intronic region of MITF, we identified 13 homozygous SNPs and 2 homozygous INDELs present in all white-plumage breeds (n = 24) and absent in all non-white-plumage breeds (n = 54) (Fig.3B). These mutations were completely associated with the white-plumage phenotype, suggesting a causative mutation at the MITF locus. Moreover, to validate the reliability of variants detected in MITF gene, we amplified the first three SNPs (SNP817793, SNP817818, and SNP818004) and all INDELs by diagnostic polymerase chain reaction (PCR) combined with Sanger sequencing in the 78 white- and non-white-plumage ducks. The results show that the three SNPs and INDEL817958 completely match our NGS analysis (Supplemental Fig. S5). For INDEL818495, we were unable to design a suitable PCR primer to amplify this region.
Figure 3:
MITF shows different genetic signatures between white-plumage and non-white-plumage ducks. A) FST plot around the MITF locus. The FST value of MITF is highest for scaffold KB742527.1, circled in red. Each plot represent a 10-kb window. B) The 13 homozygous SNPs and 2 homozygous INDELs were identified in white-plumage ducks and absent in non-white-plumage ducks. SNPs and INDELs were named according to their position on the scaffold.
Selection for other domestication traits
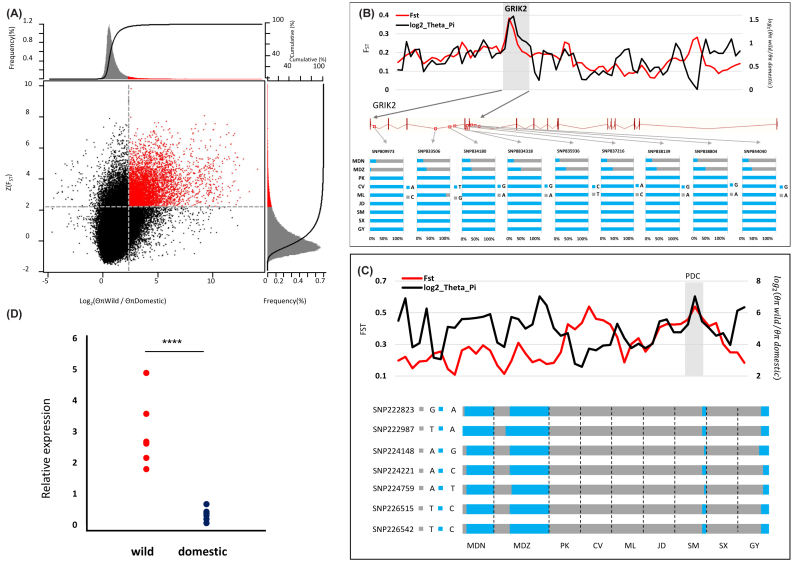
In order to detect the signature of selection for other traits associated with duck domestication, we scanned the duck genome for regions with a high coefficient of nucleotide differentiation (FST) among the populations of wild (MDN and MDZ) and domesticated (PK, CV, ML, JD, SM, SX, and GY) ducks based on 10-kbsliding windows, as well as global FST between each population (Supplemental Table S4). Owing to the complex and partly unresolved demographic history of these populations, it is difficult to define a strict threshold that distinguishes true sweeps from regions of homozygosity caused by drift. We therefore also calculated the pairwise diversity ratio (θπ(wild/domesticated)). We identified 292 genes in the top 5% of both FST and θπ scores, putatively under positive selection during domestication (Fig. 4A, Supplemental Table S5).
Figure 4:
Genomic regions with strong selective sweep signals in wild population ducks and domesticated population ducks. A) Distribution of θπ ratios θπ (wild/domesticated) and Z(FST) values, which are calculated by 10-kbwindows with 5-kbsteps. Only scaffolds >10 kbwere used for our calculation, as FST results calculated on a small scaffold are unlikely to be accurate. Red data points located to the top-right regions correspond to the 5% right tails of empirical log2 (θπ wild/θπ domestic) ratio distribution, and the top 5% empirical Z(FST) distribution are genomic regions under selection during duck domestication. The two horizontal and vertical gray lines represent the top 5% value of Z(FST) (2.216) and log2 (θπ wild/θπ domestic) (2.375), respectively. B) The log2 (θπ) ratios and FST values around the GRIK2 locus and allele frequencies of nine SNPs within the GRIK2 gene across nine duck populations. The black and red lines represent log2 (θπ wild/θπ domestic) ratios and FST values, respectively. The gray bar shows the region under strong selection in GRIK2 gene. The nine red rectangular frames correspond to the locus on gene of nine SNPs. The SNPs were named according to their position on the scaffold. C) The PDC gene showed a different genetic signature in domesticated and wild ducks. The log2 (θπ) ratios and FST values around the PDC locus. The PDC gene region is shown in gray. Allele frequencies of seven SNPs within the PDC gene across nine duck populations. The SNPs are named according to their scaffold position. D) The PDC gene expression level differs between domesticated and wild ducks. PDC mRNA expression levels in brain of wild (MDN, n = 3; MDZ, n = 4) and domesticated (PK, n = 1; CV, n = 1; ML, n = 1; JD, n = 1; SM, n = 1; SX, n = 1; GY, n = 1) ducks. ****P value from t test (P < 0.0001).
All 292 genes located in the top 5% FST regions were used for the gene ontology GO (The framework for the model of biology, which provides the most comprehensive rescoure currently available for computable knowledge regarding the functions of genes and gene products) analysis, resulting in a total of 57 GO enrichment terms (Supplementary Table S6). Because domesticated ducks are known to differ from wild ducks in body size, body fat percentage, behavior, egg productivity, growth speed, and flight capability, we focused our analysis on GO annotations of neural-related processes, lipid metabolism and energy metabolism, reproduction, and skeletal muscle contraction for our 292 putative positive selection genes. In this reduced data set, the neuro-synapse-axon and lipid-energy metabolism pathways were overrepresented (Supplemental Table S7) in our list of genes under selection.
From the highlighted GO terms, 25 neuro-synapse-axon genes were identified as being under positive selection, with six (ADGRB3, EFNA5, GRIN3A, GRIK2, SYNGAP1, and HOMER1) in the top 1% of FST and θπ (Supplemental Table S8). In particular, GRIK2 (glutamate receptor, ionotropic kainate 2) and GRIN3A (glutamate receptor, subunit 3A) both showed high FST and θπ value compared to neighboring regions, suggesting functional importance (Fig. 3B, Supplemental Tables S5, S8).
Beyond the neuronal-synapse-axon genes, 115 genes were identified in the four lipid- and energy-related pathways with high FST and θπ values, particularly related to fatty acid metabolism. Among these genes, 37 were found with both parameters yielding top 1% ranked values (Supplemental Table S8) such as phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3) and patatin-like phospholipase domain containing 8 (PNPLA8).
To infer whether selection extends beyond allelic variation and also affects gene expression, we compared individual gene expression in the brain, liver, and breast muscle between seven wild mallards and seven domesticated ducks in natural states with RNA-sequencing (RNA-seq) (Supplemental Table S9). We detected three genes (PDC, MLPH, and NID2) in the brain, two genes (MAPK12 and BST1) in the liver, and no genes in breast muscle with significantly different expression between wild and domesticated ducks. Of the five differentially expressed genes, PDC was the only gene that also showed evidence of a selective sweep at the genomic level (Supplemental Table S5, Fig.3C and D). The results suggest that the PDC gene is of substantial functional importance in phenotypic differentiation among wild and domestic ducks.
Discussion
Domesticated animals have contributed greatly to human society and human population growth by providing a stable source of animal protein, fat, and accessory products such as leather and feathers (including down). To illuminate the genetic trajectories of duck domestication, we performed whole-genome sequencing of 78 ducks including seven domesticate breeds and two wild populations. This is the first study to characterize the genetic architecture, phylogenetic relationships, and domestication history of domesticated ducks and wild mallards.
Using this powerful dataset and a suite of cutting-edge population genomic and functional genetic analyses, we observed higher mean variant numbers and nucleotide diversity for the wild mallard populations compared to the domestics, consistent with both a greater panmictic mallard population as well as recent sweeps associated with domestication.
Population structure and domestication
We observed a large expansion of the duck population at the interglacial period, which could be the result of beneficial climatic changes including rising temperatures and sea levels. In contrast, the glacial maximum coincided with a reduction in population size, consistent with harsher conditions and limited access to arctic breeding grounds [4, 29–31]. The demographic pattern we observe in wild ducks is similar to that observed in wild boars [5], wild yaks [32], and wild horses [33]. However, it is worth noting that although PSMC is a powerful method to infer changes in Ne over time, it is also sensitive to deviations from a neutral model. The effects of genetic drift and/or selection could lead to time-dependent estimates of mutation rate and could bias our estimates of population expansion [26].
We observed three genetic clusters, with wild mallard, meat breeds, and egg/dual purpose breeds each representing unique groups. These results suggest either a single domestication event followed by subsequent breed-specific selection or two separate domestication events. In order to distinguish alternative models of domestication, we modeled population demographics and found strong support for a single domestication event roughly 2,200 years ago, with the rapid subsequent selection for separate meat and egg/dual-purpose breeds roughly 100 generations later. Difficulty in differentiating between very recent divergence and high migration rates in the frequency spectrum prevented convergence between independent runs when trying to fit other migration parameters to our model. We note that the evolutionary history of wild mallards and domesticated duck breeds is likely to be more complex than the simple demographic scenarios modeled here, and further studies may be needed to fully capture the evolutionary dynamics of duck domestication. Given the recent origin of wild ducks, as well as the high levels of diversity we observe in the wild and domestic duck genomes, it is not possible to differentiate recent admixture from incomplete lineage sorting with our current data. This issue has important conservation implications and represents an interesting area for future study. Nevertheless, the time estimates obtained with our model are compatible with previous written records from 500 BC [15].
Selection for white plumage
Plumage color is an important domestication trait, and we compared breeds with white plumage to those with colored plumage. We identified high levels of divergence in the intronic region of the MITF gene, an important developmental locus with a complex regulation implicated in pigmentation and melanocyte development in several vertebrate species [34–36], including Japanese quail [37], dog [38], and duck [39, 40].
Selection for other domestication traits
In order to identify those genomic regions that have been the target of selection during domestication, we used estimates of diversity between wild and domestic samples, retaining those 292 genes in the top 5% of both FST and θπ values for further analysis. These genes were overrepresented for both neural developmental and lipid metabolism, suggesting that these functionalities were under strong selection during domestication. Two loci, GRIK2 and GRIN3A, showed particularly strong signs of selective sweeps presumably associated with domestication. GRIK2 encodes a subunit of a glutamate receptor that has a role in synaptic plasticity and is important for learning and memory. GRIN3A encodes a subunit of the N-methyl-D-aspartate receptors, which are expressed abundantly in the human cerebral cortex [41] and are involved in the development of synaptic elements.
We also identified five genes with significantly different expression in the brain and liver of domesticated ducks compared to their wild ancestor. One of these, PDC also showed evidence of selective sweeps at the genomic level. PDC encodes phosducin, a photoreceptor-specific protein that is highly expressed in the retina and the pineal gland [42], as well as the brain [43].
Our results suggest that PDC, GRIK2, and GRIN3A may have played a crucial role in duck domestication by altering functional regulation of the developing brain and nervous system. This finding is consistent with theories that behavioral traits are the most critical in the initial steps of animal domestication, allowing animals to tolerate humans and captivity [44, 45]. Indeed, compared to wild mallards, domestic ducks are more docile, less vigilant, and show important differences in brain morphology [17, 18]. Interestingly, differences between wild and domesticated animals in brain and nervous system functions due to directional selection were also observed in domestication studies of rabbits [7], dogs [46], and chickens [8]. In particular, GRIK2 was also found to play a crucial role during rabbit domestication [7].
In addition to brain- and nervous system-related genes, we also identified several genes that play an important function in lipid and energy metabolism. For example, PIK3C3 plays an important role in ATP binding but also regulates brain development and axons of cortical neurons [47–51]. PNPLA8 is involved in facilitating lipid storage in adipocyte tissue energy mobilization and maintains mitochondrial integrity [52, 53], as well as plays a role in lipid metabolism associated with neurodegenerative diseases [54–56]. PRKAR2B is associated with body weight regulation, hyperphagia, and other energy metabolism [57, 58].
Taken together, our results show that duck domestication was a relatively recent and complex process, and the genetic basis of domestication traits show many striking overlaps with other vertebrate domestication events. The whole-genome resequencing data and SNP and INDEL variant datasets are valuable resources for researchers studying evolution, domestication, and trait discovery and for breeders of Anas platyrhynchos. Furthermore, the data represent a foundation for development of new, ultrahigh-density variant screening arrays for duck population level trait analysis and genomic selection.
Methods
Sample selection
A total of 78 ducks were chosen for sequencing, seven populations of domesticated ducks and two populations of mallards from different geographic regions. The domesticated ducks include three meat type populations, i.e., Pekin duck (PK; n = 8), Cherry Valley duck (CV; n = 8), and maple leaf duck (ML; n = 8); three egg type populations, i.e., Jin Ding duck (JD; n = 8), Shao Xing duck (SX; n = 8), and Shan Ma duck (SM; n = 8); one egg and meat dual-purpose type (DP type) population, i.e., Gao You duck (GY; n = 8); and two wild populations come from two provinces in China separated by nearly 2,000 km, i.e., mallard from Ningxia Province (MDN; n = 8) and mallardform Zhejiang Province (MDZ; n = 14). The classification of production types follow the description of Animal Genetic Resources in China Poultry [59]. PK, CV, and ML ducks originated from Beijing; JD and SM ducks originated from Fujian Province; and SX and GY ducks originated from Jiangsu Province. Whole blood samples were collected from brachial veins of ducks by standard venipuncture.
In addition, 14 male ducks (MDNM, n = 3; MDZM, n = 4; PKM, n = 1; CVM, n = 1; MLM, n = 1; JDM, n = 1; SMM, n = 1; SXM, n = 1; GYM, n = 1) were chosen for RNA-seq.
Sequencing and mapping statistic of individual ducks in genome and transcriptome analyses are detailed in the Supplementary files (Supplemental Tables S1, S7).
Sequencing and library preparation
Genomic DNA was extracted using the standard phenol/chloroform extraction method. For each sample, two paired-end libraries (500 bp) were constructed according to the manufacturer's protocols (Illumina) and sequenced on the Illumina Hiseq 2500 sequencing platform. We sequenced each sample at 5X depth in order to reduce the false-negative rate of variants due to our strict filter criteria. We randomly selected one individual for 10X coverage, except for the MDN population, where we sequenced seven individuals at 5X coverage and random one at 20X coverage and the MDZ population, where we sequenced all individuals at 10X coverage. We generated 628.37 Gb of paired-end reads of 100 bp (or 150 bp; MDZ) length (Supplemental Table S1).
The mRNA from brain, liver, and breast muscle of 14 ducks were extracted using the standard trizol extraction methods. For each sample, two paired-end libraries (500 bp) were constructed according to the manufacturer's instruction (Illumina). All samples were sequenced using Illumina Hiseq 4000 sequencing platform with the coverage of 6X. We generated 278.62 Gb of paired-end reads of 150 bp length (Supplemental Table S9).
Read alignment and variant calling
To avoid low-quality reads, mainly the result of base-calling duplicates and adapter contamination, we filtered out sequences according to the default parameters of NGS QC Toolkit (v2.3.3) [60]. Those paired reads that passed Illumina's quality control filter were aligned using BWA-MEM (v0.7.12) to version 1.0 of the Anas platyrhynchos genome (BGI_duck_1.0) [10]. Duplicate reads were removed from individual sample alignments using Picard tools MarkDuplicates, and reads were merged using MergeSamFiles [61].
The Genome Analysis Toolkit v3.5 (GATK, RRID:SCR_001876), RealignerTargetCreator, and IndelRealigner protocol were used for global realignment of reads around INDELs before variant calling [62, 63]. SNPs and small INDELs (1–50 bp) were called using the GATK UnifiedGenotyper set for diploids with the parameter of a minimum quality score of 20 for both mapped reads and bases to call variants, similar to previous studies [64–68]. We filtered variants both per population and per individual using GATK according to the stringent filtering criteria. For SNPs of population filter: a) QUAL >30.0; b) QD >5.0; c) FS <60.0; d) MQ >40.0; e) MQRankSum >-12.5; and f) ReadPosRankSum >-8.0.Additionally, if there were more than 3 SNPs clustered in a 10-bpwindow, all three SNPs were considered as false positives and removed [69].
We used the following population criteria to identify INDELs: QUAL >30.0, QD >5.0, FS <200.0, ReadPosRankSum >-20.0. Of individual filters, we also removed all INDELs and SNPs where the depth of derived variants was less than half the depth of the sequence. All SNPs and INDELs were assigned to specific genomic regions and genes using SnpEff v4.0 (SnpEff, RRID:SCR_005191) [70] based on the Ensembl duck annotations. After filtering, 36,107,949 SNPs and 3,082,731 INDELs were identified (Supplemental Table S2).
SNP validation
In order to evaluate the reliability of our data, we compared our SNPs to the duck dbSNP database deposited in the GVM at the Big Data Center at the Beijing Institute of Genomics, Chinese Academy of Science [71]. A total of 7,908,722 SNPs were validated in the duck dbSNP database, which covered 96.2% of the database (Supplemental Table S2). For the 28,199,227 SNPs not confirmed by dbSNPs, 390 randomly selected nucleotide sites were further validated using diagnostic PCR combined with Sanger sequence method described in previous researchM [8, 72, 73]. The result showed 100% accuracy, indicating the high reliability of the called SNP variation identified in this study.
Population structure
We removed all SNPs with a minor allele frequency < = 0.1 and kept only SNPs that occurred in more than 90% of individuals. Vcf files were converted to hapmap format with custom perl scripts and to PLINK format file by GLU v1.0b3 [74] and PLINK v1.90 (PLINK, RRID:SCR_001757) [75, 76], when appropriate. We used GCTA (v1.25) [77] for PCA, first by generating the genetic relationship matrix from which the first 20 eigenvectors were extracted.
To estimate individual admixture assuming different numbers of clusters, the population structure was investigated using FRAPPE v1.1 [22] base on all high-quality SNPs information, with a maximum likelihood method. We increased the coancestry clusters spanning from 2 to 4 (Supplemental Fig. S6), because there are four duck types (wild, meat, egg, and dual-purpose) across the nine duck populations, with 10,000 iterations per run.
A distance matrix was generated by calculating the pairwise allele sharing distance for each pair of all high-quality SNPs. Multiple alignment of the sequences was performed with MUSCLE v3.8 (MUSCLE, RRID:SCR_011812) [78]. A neighbor-joining maximum likelihood phylogenetic tree was constructed with the DNAML program in the PHYLIP package v3.69 (PHYLIP, RRID:SCR_006244) [79] and MEGA7 [80, 81]. All implementation was performed according to the recommended manipulations of SNPhylo [82].
Demographic history reconstruction
The demographic history of both wild and domesticated ducks was inferred using a hidden Markov model approach as implemented in pairwise sequentially Markovian coalescence based on SNP distributions [23]. In order to determine which PSMC (v0.6.5) settings were most appropriate for each population, we reset the number of free atomic time intervals (-p option), upper limit of time to most recent common ancestor (-t option), and initial value of r = θ/ρ (-r option) according to previous research [26] and online suggestions by Li and Durbin [83]. Based on estimates from the chicken genome, an average mutation rate (μ) of 1.191 × 10-9 per base per generation and a generation time (g) of 1 year were used for analysis [84].
Three-population demographic inference was performed using a diffusion-based approach as implemented in the program ∂a∂i (v1.7) [85]. To minimize potential effects of selection that could interfere with demographic inference, these analyses were performed using the subset of noncoding regions across the whole genome and spanning 750,939,264 bp in length. Noncoding SNPs were then thinned to 1% to alleviate potential linkage between the markers. The final dataset consisted of 95,181 SNPs with an average distance of 7,112 bp (± 18,810 bp) between neighboring SNPs. To account for missing data, the folded allele frequency spectrum for the three populations (wild, meat, and egg/dual-purpose breeds) was projected down in ∂a∂i to the projection that maximized the number of segregating SNPs, resulting in 92,966 SNPs.
We tested four scenarios to reconstruct the demographic history of the domesticated breeds of mallards: simultaneous domestication of the meat and egg and dual-purpose breeds (model 1); a single domestication event followed by divergence of the meat and egg and dual-purpose breeds (model 2); two independent domestication events, with the meat type breed being domesticated first (model 3); and two independent domestication events, with the egg and dual-purpose breeds being domesticated first (model 4). Using the “backbone” of the best model, we then used a step-wise strategy to add parameters related with variation in population sizes and population growth, keeping a new parameter only if the AIC and log likelihood improved considerably over the previous model with fewer parameters. In cases where additional parameters resulted in negligibly improved AIC and likelihood, we retained the simpler, less parameterized model. Gene flow was modeled as continuous migration events after population divergence. Each model was run at least 10 times from independent starting values to ensure convergence to the same parameter estimates. We rejected models where we failed to obtain convergence across the replicate runs. Scaled parameters for the best-supported model were transformed into real values using the same average mutation rate (μ) and (g) as described above for the PSMC analysis. Parameter uncertainty was obtained using the Godambe information matrix [86] from 100 nonparametric bootstraps.
Selective-sweep analysis
In order to define candidate regions that have undergone directional selection during duck domestication, we calculated the coefficient of nucleotide differentiation (FST) between mallards and domesticated ducks described by Weir and Cockerham [87]. We calculated the average FST in 10-kbwindows with a 5-kbshift for all seven domesticated duck populations combined and two mallard populations combined. Only scaffolds longer than 10 kb, 2,368 of 78,488 scaffolds, were chosen for the analysis. We transformed observed FST values to Z transformation (Z(FST)) with μ = 0.1154 and σ = 0.0678 according to previously described methods [88].
To estimate levels of nucleotide diversity (π) across all sampled populations, we used the VCFtools software (v0.1.13) [89] to calculate θπ(wild/domesticated) [90], computing the average difference per locus over each pair of accessions. As the measurement of FST, averaged π ratio (θπ(wild/domesticated)) was calculated for each scaffold in 10–kbsliding windows.
Functional classification of GO categories was performed in Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.8) [91]. Statistical significance was accessed by using a modified Fisher exact test and Benjamini correction for multiple testing.
RNA-seq and data processing
To infer whether novel allelic variants located in the top 5% FST regions of genome comparison between wild mallards and domesticated ducks could also affect gene expression, we compared gene expression in brain, liver, and breast muscle between wild mallards and domesticated ducks. To make our result more universal, seven male mallards and seven male domesticated ducks were choose for RNA-seq. All samples were individually sequenced using the Illumina Highseq 4000 sequencing platform.
For each sample, adapters and primers of paired-end reads were removed using the NGSQC Tool kit (v2.3.3) [60]. For each paired-end read pair, if one of two reads had an average base quality less than 20 (PHRED quality score), then both reads were removed. If one end of a paired-end read had a percentage of high-quality base less than 70%, the two paired reads were also removed. After that, high-quality reads were mapped to the reference genome using STAR (v.2.5.3a) [92]. The featureCounts function of the Rsubread (v.1.5.2) [93, 94] was used to output the counts of reads aligning to each gene. We detected the differential expression genes with edgeR (v3.6) [95–98] using a padj <0.05 threshold.
Availability of supporting data
The 78 ducks used in whole-genome resequencing analysis and the 14 ducks used in RNA–seq analysis are accessible at the National Center for Biotechnology Information (NCBI) under BioProject accession numbers PRJNA419832 and PRJNA419583, respectively. The unassembled sequencing reads of 78 ducks and RNA-seq reads of 14 ducks have been deposited in NCBI Sequence Read Archive under accession numbers SRP125660 and SRP125529, respectively. All VCF files of SNPs and INDELs and other supporting data, such as scripts, alignments for phylogenetic trees, and sweep regions, are available via the GigaScience database GigaDB [99].
Additional file
Supplemental Figure S1: Distribution of variants in functional regions. SNPs distribution were showed on the left, and INDELs were showed on right. Most variants were synonymous mutations both in SNPs and in INDELs at genome wide across all populations.
Supplemental Figure S2: INDELs statistics of 9 population ducks. The largest INDEL detected in this study was 50 bp, and the majority of INDELs were less than 10 bp. Single base-pair INDEL was the predominant form and accounted for 38.63% of all detected INDELs. Both count and percentage were mean value of 9 population ducks.
Supplemental Figure S3: Comparison of four demographic models for the domestication of meat and egg/dual purpose breeds of mallards using ∂a∂i. The top panel shows the distribution of the log-likelihood for each one of the tested models and the middle panel the distribution of the Akaike information criterion (AIC) with outliers excluded. Model 1: simultaneous domestication of the meat and egg and dual purpose breeds; Model 2: a single domestication event followed by divergence of the meat and egg and dual purpose breeds; Model 3: two independent domestication events, with the meat type breed being domesticated first; Model 4: two independent domestication events, with the egg and dual purpose breed being domesticated first.
Supplemental Figure S4: Demographic history of meat and egg/dual purpose breed domestication using the best fit model inferred by ∂a∂i. (A) Model of single domestication event with changes in population sizes and migration. Time units are in years before present and migration are in units of number of migrants per generation. (B) Site frequency spectrum for the three populations of domesticated and wild mallards. The frequency spectrum is shown for the data (first row) and for the best fit model (second row). The last two rows show the normalized difference (i.e., residuals) between model and data for each bin in the spectrum.
Supplemental Figure S5: White plumage related variants of MITF validation by Sanger sequence in 78 ducks. Three SNPs and one INDEL of MITF was amplified by diagnostic PCR and sequenced by Sanger method, resulted completely matched with the analysis result of NGS. White plumage ducks contains PK, CV, and ML; non-white plumage ducks contains MDN, MDZ, JD, SM, SX, and GY.
Supplemental Figure S6: Population genetic structure of 78 ducks. The length of each colored segment represents the proportion of the individual genome inferred from ancestral populations (K = 2–4). The population names and production type are at the bottom. DP type means dual-purpose type. With K = 2, a clear division was found between wild type ducks (MDN and MDZ) and domesticated ducks (PK, CV, ML, JD, SM, SX, and GY). With K = 3, a clear division was found between meat type ducks (PK, CV, and ML) and egg type ducks mixed with dual-purpose type ducks (JD, SM, SX, and GY). With K=4, a clear division was found between egg type ducks (JD, SM, and SX) and dual-purpose type ducks (GY).
Supplemental Table S1: Summary of genome sequencing and mapping statistic.
Supplemental Table S2: Summary of SNPs and INDELs.
Supplemental Table S3: INDELs statistics of 9 population ducks.
Supplemental Table S4: global Fst between each population.
Supplemental Table S5: gene name in top 5% sweep regions.
Supplemental Table S6: Total GO terms of genes located in top 5% FST and θπ regions.
Supplemental Table S7: Summary of results from enrichment analysis of neuronal and lipid related in regions of top 5% FST and θπ.
Supplemental Table S8: gene name in top 1% sweep regions.
Supplemental Table S9: summary of transcriptome sequencing and mapping statistic.
Abbreviations
AIC: Akaike information criteria; CI: confidence interval; GO: gene ontology; GVM: Genome Variation Map; INDEL: insertion and deletion; MITF: melanogenesis-associated transcription factor; NCBI: National Center for Biotechnology Information; PCA: principle component analysis; PCR: polymerase chain reaction; PSMC: pairwise sequentially Markovian coalescent; RNA-seq: RNA sequencing; SNP: single-nucleotide polymorphism.
Ethics statement
The entire procedure was carried out in strict accordance with the protocol approved by the Animal Welfare Committee of China Agricultural University (permit XK622).
Funding
This work was supported by the Earmarked fund for the Beijing Innovation Team of the Modern Agro-industry Technology Research System (BAIC04–2017), European Research Council (grant agreement 680951), and Wolfson Merit Award. We gratefully acknowledge our colleagues in the Poultry Team at the National Engineering Laboratory for Animal Breeding of China Agricultural University for their assistance with sample collection and helpful comments on the manuscript.
Author contributions
Conceived and designed the experiments: L.Q. Wrote the paper: Z.Z. Revised the paper: L.Q., J.E.M., M.vanT. Analyzed the data: Z.Z., P.A., Q.W., Y.J. Performed the experiments: Z.Z., Y.J. Contributed reagents/materials: Z.J., Y.C., K.Z., S.H., Z.Zhou, H.L., F.Y., Y.H., Z.N., and N.Y.
Supplementary Material
07 Dec 2017 Reviewed
06 Feb 2018 Reviewed
11 Dec 2017 Reviewed
12 Feb 2018 Reviewed
20 Dec 2017 Reviewed
Acknowledgement
We are gratefully acknowledge our colleagues in the Poultry Team at the National Engineering Laboratory for Animal Breeding of China Agricultural University, for their assistance on sample collection and helpful comments on the manuscript.
References
- 1. Li J, Zhang Y. Advances in research of the origin and domestication of domestic animals. Biodiversity Sci. 2009;17(4):319–29. [Google Scholar]
- 2. Darwin C, Mayr E. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. John Murray: london: Harvard University Press; 1859. [Google Scholar]
- 3. Chen C, Liu Z, Pan Q, et al. Genomic analyses reveal demographic history and temperate adaptation of the newly discovered honey bee subspecies Apis mellifera sinisxinyuann. ssp. Mol Biol Evol. 2016;33(5):1337–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang J, Li WR, Lv FH, et al. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol Biol Evol. 2016;33(10):2576–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li M, Tian S, Jin L, et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 2013;45(12):1431–8. [DOI] [PubMed] [Google Scholar]
- 6. Jiang Y, Xie M, Chen W, et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science. 2014;344(6188):1168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carneiro M, Rubin CJ, Di Palma F, et al. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science. 2014;345(6200):1074–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang MS, Zhang RW, Su LY et al. Positive selection rather than relaxation of functional constraint drives the evolution of vision during chicken domestication. Cell Res. 2016;26(5):556–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rubin CJ, Zody MC, Eriksson J, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464(7288):587–91. [DOI] [PubMed] [Google Scholar]
- 10. Huang Y, Li Y, Burt DW, et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nat Genet. 2013;45(7):776–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeuner FE. A history of domesticated animals, Haper & Row: New York: 1963. [Google Scholar]
- 12. Thomson SAL, Ornithologists' Union B, Thomson AL. A New Dictionary of Birds. London: Nelson, 1964. [Google Scholar]
- 13. Crawford RD, Mason IL. Evolution of domesticated animals, 345–349., London and New York: Longman; 1984. [Google Scholar]
- 14. Bray F, Needham J. Science and Civilization in China. vol. 6, part 1 Cambridge, UK: Agriculture: Cambridge University Press, 1984. [Google Scholar]
- 15. Kiple KF. The Cambridge World History of Food. Cambridge: Cambridge University Press, 2000. [Google Scholar]
- 16. Chang H. Conspectus of Genetic Resources of Livestock. Beijing, China: Chinese Agriculture Press, 1995. [Google Scholar]
- 17. Miller DB. Social displays of mallard ducks (Anasplatyrhynchos): effects of domestication. J Comp Physiol Psychol. 1977;91(2):221–32. [Google Scholar]
- 18. Ebinger P. Domestication and plasticity of brain organization in mallards (Anasplatyrhynchos). Brain Behav Evol. 1995;45(5):286–300. [DOI] [PubMed] [Google Scholar]
- 19. Frahm H, Rehkämper G, Werner C. Brain alterations in crested versus non-crested breeds of domestic ducks (Anasplatyrhynchosf.d.). Poult Sci. 2001;80(9):1249–57. [DOI] [PubMed] [Google Scholar]
- 20. Duggan BM, Hocking PM, Schwarz T et al. Differences in hindlimb morphology of ducks and chickens: effects of domestication and selection. Genet Sel Evol. 2015;47(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genome Variation Map website. http://bigd.big.ac.cn/gvm/ Data accessed, 1st March 2018.
- 22. Tang H, Peng J, Wang P et al. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28(4):289–301. [DOI] [PubMed] [Google Scholar]
- 23. Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475(7357):493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ehlers J, Gibbard PL. The extent and chronology of Cenozoic global glaciation. Quat Int. 2007;164:6–20. [Google Scholar]
- 25. Williams MAJ, Dunkerley D, De Deckker P et al. Quaternary Environments. London: Science Press; 1997. [Google Scholar]
- 26. Nadachowska-Brzyska K, Li C, Smeds L, et al. Temporal dynamics of avian populations during pleistocene revealed by whole-genome sequences. Curr Biol. 2015;25(10):1375–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shapiro MD, Kronenberg Z, Li C, et al. Genomic diversity and evolution of the head crest in the rock pigeon. Science. 2013;339(6123):1063–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Price TD. Domesticated birds as a model for the genetics of speciation by sexual selection. Genetica. 2002;116(2/3):311–27. [PubMed] [Google Scholar]
- 29. Lorenzen ED, Nogués-Bravo D, Orlando L, et al. Species-specific responses of late quaternary megafauna to climate and humans. Nature. 2011;479(7373):359–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hewitt G. The genetic legacy of the quaternary ice ages. Nature. 2000;405(6789):907–13. [DOI] [PubMed] [Google Scholar]
- 31. Hewitt G. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society B: Biological Sciences. 2004;359(1442):183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qiu Q, Wang L, Wang K et al. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat Commun. 2015;6(1):10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Orlando L, Ginolhac A, Zhang G, et al. Recalibrating Equus evolution using the genome sequence of an early middle pleistocene horse. Nature. 2013;499(7456):74–78. [DOI] [PubMed] [Google Scholar]
- 34. Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the Microphthalmia transcription factor network. Annu Rev Genet. 2004;38(1):365–411. [DOI] [PubMed] [Google Scholar]
- 35. Hallsson JH, Haflidadottir BS, Schepsky A, et al. Evolutionary sequence comparison of the Mitf gene reveals novel conserved domains. Pigment Cell Res. 2007;20(3):185–200. [DOI] [PubMed] [Google Scholar]
- 36. Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12(9):406–14. [DOI] [PubMed] [Google Scholar]
- 37. Minvielle F, Bed'hom B, Coville JL, et al. The “silver” Japanese quail and the MITF gene: causal mutation, associated traits and homology with the “blue” chicken plumage. BMC Genet. 2010;11(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karlsson EK, Baranowska I, Wade CM et al. Efficient mapping of Mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39(11):1321–8. [DOI] [PubMed] [Google Scholar]
- 39. Li S, Wang C, Yu W, et al. Identification of genes related to white and black plumage formation by RNA-seq from white and black feather bulbs in ducks. PLoS One. 2012;7(5):e36592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sultana H, Seo D, Choi NR, et al. Identification of polymorphisms in MITF and DCT genes and their associations with plumage colors in Asian duck breeds. Asian-Australasian J Animal Sci. 2017; doi: 10.5713/ajas.17.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eriksson M, Nilsson A, Samuelsson H et al. On the role of NR3A in human NMDA receptors. Physiology & Behavior. 2007;92(1-2):54–59. [DOI] [PubMed] [Google Scholar]
- 42. Bauer PH, Muller S, Puzicha M, et al. Phosducin is a protein kinase A-regulated G-protein regulator. Nature. 1992;358(6381):73–76. [DOI] [PubMed] [Google Scholar]
- 43. Sunayashiki-Kusuzaki K, Kikuchi T, Wawrousek EF et al. Arrestin and phosducin are expressed in a small number of brain cells. Mol Brain Res. 1997;52(1):112–20. [DOI] [PubMed] [Google Scholar]
- 44. Mignon-Grasteau S, Boissy A, Bouix J, et al. Genetics of adaptation and domestication in livestock. Livestock Prod Sci. 2005;93(1):3–14. [Google Scholar]
- 45. Dugatkin LA, Trut L. How to Tame a Fox (and Build a Dog): Visionary Scientists and a Siberian Tale of Jump-Started Evolution. University of Chicago Press, 2017. [Google Scholar]
- 46. Axelsson E, Ratnakumar A, Arendt ML, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495(7441):360–4. [DOI] [PubMed] [Google Scholar]
- 47. Volinia S, Dhand R, Vanhaesebroeck B, et al. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14(14):3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Inaguma Y, Ito H, Iwamoto I, et al. Morphological characterization of class III phosphoinositide 3-kinase during mouse brain development. Med Mol Morphol. 2016;49(1):28–33. [DOI] [PubMed] [Google Scholar]
- 49. Stopkova P, Saito T, Papolos DF, et al. Identification of PIK3C3 promoter variant associated with bipolar disorder and schizophrenia. Biol Psychiatry. 2004;55(10):981–8. [DOI] [PubMed] [Google Scholar]
- 50. Tang R, Zhao X, Fang C, et al. Investigation of variants in the promoter region of PIK3C3 in schizophrenia. Neurosci Lett. 2008;437(1):42–44. [DOI] [PubMed] [Google Scholar]
- 51. Zhou X, Wang L, Hasegawa H, et al. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci. 2010;107(20):9424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilson PA, Gardner SD, Lambie NM, et al. Characterization of the human patatin-like phospholipase family. J Lipid Res. 2006;47(9):1940–9. [DOI] [PubMed] [Google Scholar]
- 53. Kienesberger PC, Oberer M, Lass A, et al. Mammalian patatin domain containing proteins: a family with diverse lipolytic activities involved in multiple biological functions. J Lipid Res. 2009;50(Supplement):S63–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tesson C, Nawara M, Salih MA et al. Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am J Hum Genet. 2012;91(6):1051–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schuurs-Hoeijmakers JH, Oh EC, Vissers LE et al. Recurrent de novo mutations in PACS1 cause defective cranial-neural-crest migration and define a recognizable intellectual-disability syndrome. Am J Hum Genet. 2012;91(6):1122–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martin E, Schüle R, Smets K, et al. Loss of function of glucocerebrosidase GBA2 is responsible for motor neuron defects in hereditary spastic paraplegia. Am J Hum Genet. 2013;92(2):238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gagliano SA, Tiwari AK, Freeman N et al. Protein kinase cAMP-dependent regulatory type II beta (PRKAR2B) gene variants in antipsychotic-induced weight gain. Hum Psychopharmacol Clin Exp. 2014;29(4):330–5. [DOI] [PubMed] [Google Scholar]
- 58. Czyzyk TA, Sikorski MA, Yang L et al. Disruption of the RII subunit of PKA reverses the obesity syndrome of agouti lethal yellow mice. Proc Natl Acad Sci. 2008;105(1):276–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Resources CNCoAG. Animal Genetic Resources in China poultry. Beijing: China Agriculture Press; 2010. [Google Scholar]
- 60. Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7(2):e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. http://broadinstitute.github.io/picard/.
- 62. McKenna A, Hanna M, Banks E et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yan Y, Yi G, Sun C et al. Genome-wide characterization of insertion and deletion variation in chicken using next generation sequencing. PLoS One. 2014;9(8):e104652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Qu Y, Tian S, Han N, et al. Genetic responses to seasonal variation in altitudinal stress: whole-genome resequencing of great tit in eastern Himalayas. Sci Rep. 2015;5(1):14256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Meyer RS, Choi JY, Sanches M, et al. Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat Genet. 2016;48(9):1083–8. [DOI] [PubMed] [Google Scholar]
- 67. Russell J, Mascher M, Dawson IK et al. Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat Genet. 2016;48(9):1024–30. [DOI] [PubMed] [Google Scholar]
- 68. Mascher M, Schuenemann VJ, Davidovich U, et al. Genomic analysis of 6000-year-old cultivated grain illuminates the domestication history of barley. Nat Genet. 2016;48(9):1089–93. [DOI] [PubMed] [Google Scholar]
- 69. Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cingolani P, Platts A, Wang LL et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. http://bigd.big.ac.cn/gvm/.
- 72. Zhang Z, Nie C, Jia Y, et al. Parallel evolution of polydactyly traits in Chinese and European chickens. PLoS One. 2016;11(2):e0149010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Van Tassell CP, Smith TP, Matukumalli LK, et al. SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat Methods. 2008;5(3):247–52. [DOI] [PubMed] [Google Scholar]
- 74. https://code.google.com/archive/p/glu-genetics/.
- 75. Purcell S, Neale B, Todd-Brown K et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chang CC, Chow CC, Tellier LC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaSci. 2015;4(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yang J, Lee SH, Goddard ME, et al. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Plotree D, Plotgram D. PHYLIP-phylogeny inference package (version 3.2). Cladistics. 1989;5(163):6. [Google Scholar]
- 80. Tamura K, Dudley J, Nei M, et al. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–9. [DOI] [PubMed] [Google Scholar]
- 81. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lee TH, Guo H, Wang X et al. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics. 2014;15(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. https://github.com/lh3/psmc.
- 84. Nam K, Mugal C, Nabholz B et al. Molecular evolution of genes in avian genomes. Genome Biol. 2010;11(6):R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gutenkunst RN, Hernandez RD, Williamson SH, et al. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5(10):e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Coffman AJ, Hsieh PH, Gravel S, et al. Computationally efficient composite likelihood statistics for demographic inference. Mol Biol Evol. 2016;33(2):591–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Weir BS, Cockerham CC. Estimating F-Statistics for the analysis of population-structure. Evolution. 1984;38(6):1358–70. [DOI] [PubMed] [Google Scholar]
- 88. Kreyszig E. Advanced Engineering Mathematics. John Wiley & Sons: FL: CRC Press; 2007. [Google Scholar]
- 89. Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105(2):437–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 92. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 95. Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23(21):2881–7. [DOI] [PubMed] [Google Scholar]
- 96. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lun AT, Chen Y, Smyth GK. It's DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Statistical Genomics: Methods and Protocols. 2016:391–416. [DOI] [PubMed] [Google Scholar]
- 99. Zhang Z, Jia Y, Almeida P et al. Supporting data for “whole-genome resequencing reveals signatures of selection and timing of duck domestication.”. GigaScience database. 2018. http://dx.doi.org/10.5524/100417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
07 Dec 2017 Reviewed
06 Feb 2018 Reviewed
11 Dec 2017 Reviewed
12 Feb 2018 Reviewed
20 Dec 2017 Reviewed