Abstract
Background
New generations of sequencing platforms coupled to numerous bioinformatics tools have led to rapid technological progress in metagenomics and metatranscriptomics to investigate complex microorganism communities. Nevertheless, a combination of different bioinformatic tools remains necessary to draw conclusions out of microbiota studies. Modular and user-friendly tools would greatly improve such studies.
Findings
We therefore developed ASaiM, an Open-Source Galaxy-based framework dedicated to microbiota data analyses. ASaiM provides an extensive collection of tools to assemble, extract, explore, and visualize microbiota information from raw metataxonomic, metagenomic, or metatranscriptomic sequences. To guide the analyses, several customizable workflows are included and are supported by tutorials and Galaxy interactive tours, which guide users through the analyses step by step. ASaiM is implemented as a Galaxy Docker flavour. It is scalable to thousands of datasets but also can be used on a normal PC. The associated source code is available under Apache 2 license at https://github.com/ASaiM/framework and documentation can be found online (http://asaim.readthedocs.io).
Conclusions
Based on the Galaxy framework, ASaiM offers a sophisticated environment with a variety of tools, workflows, documentation, and training to scientists working on complex microorganism communities. It makes analysis and exploration analyses of microbiota data easy, quick, transparent, reproducible, and shareable.
Keywords: metagenomics, metataxonomics, user-friendly, Galaxy, Docker, microbiota, training
Findings
Background
The study of microbiota and microbial communities has been facilitated by the evolution of sequencing techniques and the development of metataxonomics, metagenomics, and metatranscriptomics. These techniques are giving insight into taxonomic profiles and genomic components of microbial communities. However, meta'omic data exploitation is not trivial due to the large amount of data, their complexity, the incompleteness of reference databases, and the difficulty to find, configure, use, and combine the dedicated bioinformatics tools, etc. Hence, to extract useful information, a sequenced microbiota sample has to be processed by sophisticated workflows with numerous successive bioinformatics steps [1]. Each step may require execution of several tools or software. For example, to extract taxonomic information with the widely used QIIME [2] or Mothur [3], at least 10 different tools with at least four parameters each are needed. Designed for amplicon data, both QIIME and Mothur cannot be directly applied to shotgun metagenomics data. In addition, the tools can be complex to use; they are command-line tools and may require extensive computational resources (memory, disk space). In this context, selecting the best tools, configuring them to use the correct parameters and appropriate computational resources, and combining them together in an analysis chain is a complex and error-prone process. These issues and the involved complexity are prohibiting scientists from participating in the analysis of their own data. Furthermore, bioinformatics tools are often manually executed and/or patched together with custom scripts. These practices raise doubts about a science gold standard: reproducibility [3, 4]. Web services and automated pipelines such as MG-RAST [5] and EBI metagenomics [6] offer solutions to the accessibility issue. However, these web services work as a black box and are lacking in transparency, flexibility, and even reproducibility as the version and parameters of the tools are not always available. Alternative approaches to improve accessibility, modularity, and reproducibility can be found in open-source workflow systems such as Galaxy [6-8]. Galaxy is a lightweight environment providing a web-based, intuitive, and accessible user interface to command-line tools, while automatically managing computation and transparently managing data provenance and workflow scheduling [6-8]. More than 5,500 tools can be used inside any Galaxy environment. For example, the main Galaxy server [9] integrates many genomic tools, and the few integrated metagenomics tools such as Kraken [10] or VSearch [11] have been showcased in the published windshield splatter analysis [12]. The tools can also be selected and combined to build Galaxy flavors focusing on specific type of analysis, for example, the Galaxy RNA workbench [13] or the specialized Galaxy server of the Huttenhower lab [14]. However, none of these solutions is dedicated to microbiota data analysis in general and with the community-standard tools.
In this context, we developed ASaiM (Auvergne Sequence analysis of intestinal Microbiota, RRID:SCR_015878), an Open-Source opinionated Galaxy-based framework. It integrates more than 100 tools and several workflows dedicated to microbiota analyses with an extensive documentation [15] and training support.
Goals of ASaiM
ASaiM is developed as a modular, accessible, redistributable, sharable, and user-friendly framework for scientists working with microbiota data. This framework is unique in combining curated tools and workflows and providing easy access and support for scientists.
ASaiM is based on four pillars: (1) easy and stable dissemination via Galaxy, Docker, and Conda, (2) a comprehensive set of microbiota-related tools, (3) a set of predefined and tested workflows, and (4) extensive documentation and training to help scientists in their analyses.
A framework built on the shoulders of giants
The ASaiM framework is built on existing tools and infrastructures and combines all their forces to create an easily accessible and reproducible analysis platform.
ASaiM is implemented as a portable virtualized container based on the Galaxy framework [8]. Galaxy provides researchers with means to reproduce their own workflows analyses, rerun entire pipelines, or publish and share them with others. Based on Galaxy, ASaiM is scalable from single CPU installations to large multi-node high performance computing environments and manages efficiently job submission as well as memory consumption of the tools. Deployments can be achieved by using a pre-built ASaiM Docker image, which is based on the Galaxy Docker project [16]. This ASaiM Docker flavour is customized with a variety of selected tools, workflows, interactive tours, and data that have been added as additional layers on top of the generic Galaxy Docker instance. The containerization keeps the deployment task to a minimum. The selected Galaxy tools are automatically installed from the Galaxy ToolShed [17] using the Galaxy API BioBlend [18], and the installation of the tools and their dependencies are automatically resolved using packages available through Bioconda [19]. To populate ASaiM with the selected microbiota tools, we migrated the 12 tools/suites of tools and their dependencies to Bioconda (e.g., HUMAnN2), integrated 16 suites (>100 tools) into Galaxy (e.g., HUMAn2 or QIIME with its approximately 40 tools), and updated the already available ones (Table 1).
Table 1:
Available tools in ASaiM
| Section | Subsection | Tools |
|---|---|---|
| File and meta tools | Data retrieval | EBISearch [20], ENASearch [21], SRA Tools |
| Text manipulation | Tools from Galaxy ToolShed | |
| Sequence file manipulation | Tools from Galaxy ToolShed | |
| BAM/SAM file manipulation | SAM tools [22-24] | |
| BIOM file manipulation | BIOM-Format tools [25] | |
| Genomics tools | Quality control | FastQC [26], PRINSEQ [27], Trim Galore! [28, Trimmomatic [29], MultiQC [30] |
| Clustering | CD-Hit [31], Format CD-HIT outputs | |
| Sorting and prediction | SortMeRNA [32], FragGeneScan [33] | |
| Mapping | BWA [34], Bowtie [35] | |
| Similarity search | NCBI Blast+ [36, 37], Diamond [38] | |
| Alignment | HMMER3 [39] | |
| Microbiota dedicated tools | Metagenomics data manipulation | VSEARCH [11], Nonpareil [40] |
| Assembly | MEGAHIT [41], metaSPAdes [42], metaQUAST [43], VALET [44] | |
| Metataxonomic sequence analysis | Mothur [3], QIIME [2] | |
| Taxonomy assignation on WGS sequences | MetaPhlAn2 [45], Format MetaPhlan2, Kraken [10] | |
| Metabolism assignation | HUMAnN2 [46], Group HUMAnN2 to GO slim terms [47], Compare HUMAnN2 outputs, PICRUST [48], InterProScan | |
| Combination of functional and taxonomic results | Combine MetaPhlAn2 and HUMAnN2 outputs | |
| Visualization | Export2graphlan [49], GraPhlAn [50], KRONA [51] |
This table presents the tools, organized in sections and subsections to help users. A more detailed table of the available tools and some documentation can be found in the online documentation (http://asaim.readthedocs.io/en/latest/tools/).
Tools for microbiota data analyses
The tools integrated in ASaiM can be seen in Table 1. They are expertly selected for their relevance with regard to microbiota studies, such as Mothur (mothur, RRID:SCR_011947) [3], QIIME (QIIME, RRID:SCR_008249) [2], MetaPhlAn2 (MetaPhlAn, RRID:SCR_004915) [45], HUMAnN2 [46], or tools used in existing pipelines such as EBI Metagenomics’ one. We also added general tools used in sequence analysis such as quality control, mapping, or similarity search tools.
An effort in development was made to integrate these tools into Conda and the Galaxy environment (>100 tools integrated) with the help and support of the Galaxy community. We also developed two new tools to search and get data from EBI Metagenomics and ENA databases (EBISearch [20] and ENASearch [21]) and a tool to group HUMAnN2 outputs into Gene Ontology Slim Terms [47]. Tools inside ASaiM are documented [15] and organized to make them findable.
Diverse source of data
An easy way to upload user-data into ASaiM is provided by a web interface or more sophisticatedly via FTP or SFTP. On the top, we added specialised tools that can interact with external databases like NCBI, ENA, or EBI Metagenomics to query them and download data into the ASaiM environment.
Visualization of the data
An analysis often ends with summarizing figures that conclude and represent the findings. ASaiM includes standard interactive plotting tools to draw bar charts and scatter plots for all kinds of tabular data. Phinch visualization [52] is also included to interactively visualize and explore any BIOM file and generate different types of ready-to-publish figures. We also integrated two other tools to explore and represent the community structure: KRONA [51] and GraPhlAn [53]. Moreover, as in any Galaxy instance, other visualizations are included such as Phyloviz [54] for phylogenetic trees or the genome browser Trackster [55] for visualizing SAM/BAM, BED, GFF/GTF, WIG, bigWig, bigBed, bedGraph, and VCF datasets.
Workflows
Each tool can be used separately in an explorative manner, the Galaxy tool form helping users in setting meaningful parameters. Tools can be also orchestrated inside workflows using the powerful Galaxy workflow manager. To assist in microbiota analyses, several workflows, including a few well-known pipelines, are offered and documented (tools and their default parameters) in ASaiM. These workflows can be used as is; customized either on the fly to tune the parameters or globally to change the tools, their order, and their default parameters; or even used as subworkflows. Moreover, users can also design novel meaningful workflows via the Galaxy workflow interface using the >100 available tools.
Analysis of raw metagenomic or metatranscriptomic shotgun data
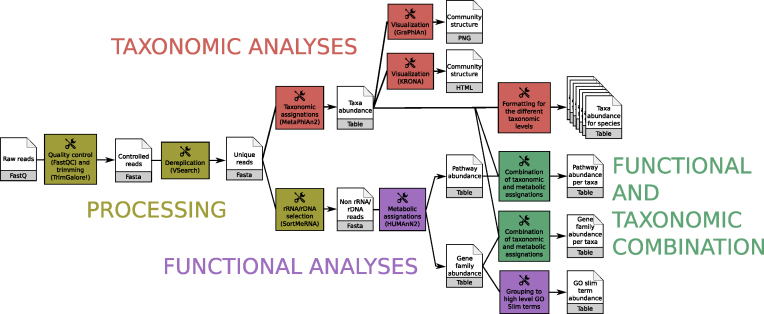
The workflow quickly produces, from raw metagenomic or metatranscriptomic shotgun data, accurate and precise taxonomic assignations, wide extended functional results, and taxonomically related metabolism information (Fig. 1). This workflow consists of (i) processing with quality control/trimming (FastQC and Trim Galore!) and dereplication (VSearch [11]); (ii) taxonomic analyses with assignation (MetaPhlAn2 [45]) and visualization (KRONA, GraPhlAn); (iii) functional analyses with metabolic assignation and pathway reconstruction (HUMAnN2 [46]); (iv) functional and taxonomic combination with developed tools combining HUMAnN2 and MetaPhlAn2 outputs.
Figure 1:
Main ASaiM workflow to analyze raw sequences. This workflow takes as input a dataset of raw shotgun sequences (in FastQ format) from microbiota, preprocess it (yellow boxes), extracts taxonomic (red boxes) and functional (purple boxes) assignations, and combines them (green boxes). Image available under CC-BY license (https://doi.org/10.6084/m9.figshare.5371396.v3).
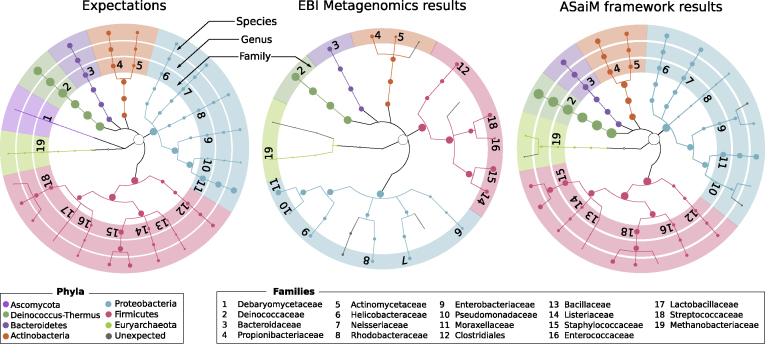
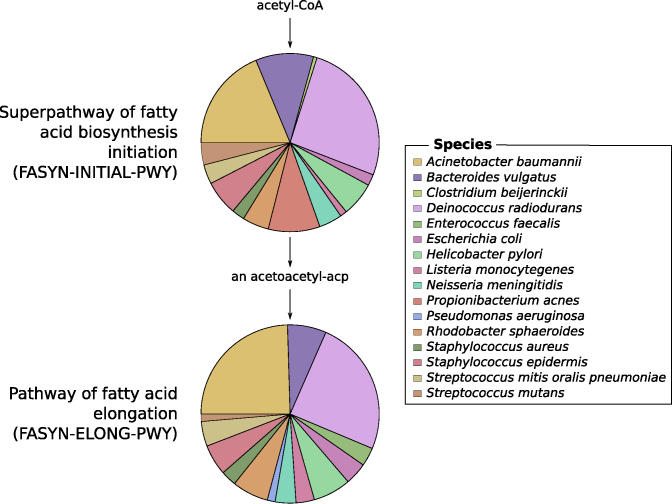
This workflow has been tested on two mock metagenomic datasets with controlled communities (Supplementary material). We have compared the extracted taxonomic and functional information to such information extracted with the EBI metagenomics’ pipeline and to the expectations from the mock datasets to illustrate the potential of the ASaiM workflow. With ASaiM, we generate accurate and precise data for taxonomic analyses (Fig. 2), and we can access information at the level of the species. More functional information (e.g., gene families, gene ontologies, pathways) are also extracted with ASaiM compared to the ones available on EBI metagenomics. With this workflow, we can go one step further and investigate which taxons are involved in a specific pathway or a gene family (e.g., involved species and their relative involvement in different step of fatty acid biosynthesis pathways, Fig. 3).
Figure 2:
Comparisons of the community structure for SRR072233. This figure compares the community structure between the expectations (mapping of the sequences on the expected genomes), data found on EBI Metagenomics database (extracted with the EBI Metagenomics pipeline), and the results of the main ASaiM workflow (Fig. 1).
Figure 3:
Example of an investigation of the relation between community structure and functions. The involved species and their relative involvement in fatty acid biosynthesis pathways have been extracted with ASaiM workflow (Fig. 1) for SRR072233.
For the tests, ASaiM was deployed on a computer with Debian GNU/Linux System, 8 cores Intel(R) Xeon(R) at 2.40 GHz and 32 Go of RAM. The workflow processed the 1,225,169 and 1,386,198 454 GS FLX Titanium reads of each datasets, with a stable memory usage, in 4h44 and 5h22 respectively (Supplementary material). The execution time is logarithmically linked to the input data size. With this workflow, it is then easy and quick to process raw microbiota data and extract diverse useful information.
Assembly of metagenomics data
Microbiota data usually come with quite short reads. To reconstruct genomes or to get longer sequences for further analysis, microbiota sequences have to be assembled with dedicated metagenome assemblers. To help in this task, two workflows have been developed in ASaiM, each one using one of the well-performing assemblers [56-62]: MEGAHIT [41] and MetaSPAdes [42]. Both workflows consists of: (1) processing with quality control/trimming (FastQC and Trim Galore!); (2) assembly with either MEGAHIT or MetaSPAdes; (3) estimation of the assembly quality statistics with MetaQUAST [43]; (4) identification of potential assembly error signature with VALET; and (5) determination of percentage of unmapped reads with Bowtie2 (Bowtie, RRID:SCR_005476) [36] combined with MultiQC [30] to aggregate the results.
Analysis of metataxonomic data
To analyze amplicon or internal transcribed spacer data, the Mothur and QIIME tool suites are available in ASaiM. We integrated the workflows described in tutorials of Mothur and QIIME as an example of metataxonomic data analyses as well as support for the training material.
Running as in EBI Metagenomics
As the tools used in the EBI Metagenomics pipeline (version 3) are also available in ASaiM, we integrate them in a workflow with the same steps as the EBI Metagenomics pipeline. Analyses made in the EBI Metagenomics website can be then reproduced locally without having to wait for availability of EBI Metagenomics or to upload any data on EBI Metagenomics. However, the parameters must be defined by the user, as we cannot find them on EBI Metagenomics documentation. In ASaiM, the entire provenance and every parameter are tracked to guarantee the reproducibility.
Documentation and training
A tool or software is easier to use if it is well documented. Hence, extensive documentation helps the users to be familiar with the tool and also prevents mis-usage. For ASaiM, we developed an extensive online documentation [15] , mainly to explain how to use it, how to deploy it, which tools are integrated with small documentation about these tools, which workflows are available, and how to use them.
In addition to this online documentation, training materials have been developed. Some Galaxy interactive tours are included inside the Galaxy instance to guide users through entire microbiota analyses in an interactive (step-by-step) way. We also developed several step-by-step tutorials to explain the concepts of microbiota analyses, the different tools and parameters, and ASaiM workflows with toy datasets. Hosted within the Galaxy Training Material [63], the tutorials are available online at [64] and also directly accessible from ASaiM and its documentation for self-training. These tutorials and ASaiM have been used during several workshops on metagenomics data analysis and some undergraduate courses to explain and use the EBI Metagenomics workflow in a reproducible way. ASaiM is also used as support for a citizen science and education project (BeerDeCoded [65]).
Installation and running ASaiM
Running the containerized ASaiM simply requires the user to install Docker and to start the ASaiM image with:
$ docker run -d -p 8080:80 quay.io/bebatut/asaim-framework:latest
As Galaxy, ASaiM is production ready and can be configured to use external accessible computer clusters or cloud environments. It is also possible and easy to install all or only a subset of tools of the ASaiM framework on existing Galaxy instances, as we did on the European Galaxy instance [66]. More details about the installation and the use of ASaiM are available on the online documentation [15].
Conclusion
ASaiM provides a powerful framework to easily and quickly analyze microbiota data in a reproducible, accessible, and transparent way. Built on a Galaxy instance wrapped in a Docker image, ASaiM can be easily deployed with its extensive set of tools and their dependencies, saving users from the hassle of installing all software. These tools are complemented with a set of predefined and tested workflows to address the main questions of microbiota research (assembly, community structure, and function). All these tools and workflows are extensively documented online [15] and supported by interactive tours and tutorials.
With this complete infrastructure, ASaiM offers a sophisticated environment for microbiota analyses to any scientist while promoting transparency, sharing, and reproducibility.
Methods
For the tests, ASaiM was deployed on a computer with Debian GNU/Linux System, 8 cores Intel(R) Xeon(R) at 2.40 GHz and 32 Go of RAM. The workflow has been run on two mock community samples of the Human Microbiome Project containing a genomic mixture of 22 known microbial strains. The details of comparison analyses are described in the Supplementary Material.
Availability of supporting data
Archival copies of the code and mock data are available in the GigaScience GigaDB repository [67].
Availability of supporting source code and requirements
Project name: ASaiM
Project home page: https://github.com/ASaiM/framework
Operating system(s): Platform independent
Other requirements: Docker
License: Apache 2
All tools described herein are available in the Galaxy Toolshed (https://toolshed.g2.bx.psu.edu). The Dockerfile to automatically deploy ASaiM is provided in the GitHub repository (https://github.com/ASaiM/framework) and a pre-built Docker image is available at https://quay.io/repository/bebatut/asaim-framework.
Supplementary Material
9/13/2017 Reviewed
10/4/2017 Reviewed
1/23/2018 Reviewed
10/9/2017 Reviewed
1/29/2018 Reviewed
ACKNOWLEDGEMENTS
The authors would like to thank EA 4678 CIDAM, UR 454 INRA, M2iSH, LIMOS, AuBi, Mésocentre, and de.NBI for their involvement in this project, as well as Réjane Beugnot, Thomas Eymard, David Parsons, and Björn Grüning for their help.
Additional files
sup_mat_1.pdf
Abbreviations
API: application programming interface; AsaiM: Auvergne Sequence analysis of intestinal Microbiota; CPU: central processing unit; Galaxy Training Network.
Competing interests
The author(s) declare that they have no competing interests.
Funding
The Auvergne Regional Council and the European Regional Development Fund supported this work.
Authors’ contributions
B.B., K.G., C.D., S.H., J.F.B., E.P., and P.P. contributed equally to the conceptualization, methodology, and writing process; J.F.B. and P.P. contributed equally to the funding acquisition; B.B., K.G., and S.H. contributed equally to the software development; and B.B., K.G., C.D., and J.F.B. contributed equally to the validation.
References
- 1. Ladoukakis E, Kolisis FN, Chatziioannou AA. Integrative workflows for metagenomic analysis. Front Cell Dev Biol. 2014;2:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caporaso JG, Kuczynski J, Stombaugh J, et al. , QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010, 7, 5, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nekrutenko A, Taylor J. Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat Rev Genet. 2012;13:667–72. [DOI] [PubMed] [Google Scholar]
- 5. Meyer F, Paarmann D, D'Souza M, et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hunter S, Corbett M, Denise H, et al. EBI metagenomics–a new resource for the analysis and archiving of metagenomic data. Nucleic Acids Res. 2014;42:D600–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goecks J, Nekrutenko A, Taylor J et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Afgan E, Baker D, van den Beek M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Main Galaxy instance, http://usegalaxy.org [Google Scholar]
- 10. Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rognes T, Flouri T, Nichols B et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kosakovsky Pond S, Wadhawan S, Chiaromonte F, et al. Windshield splatter analysis with the Galaxy metagenomic pipeline. Genome Res. 2009;19:2144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grüning BA, Fallmann J, Yusuf D, et al. The RNA workbench: best practices for RNA and high-throughput sequencing bioinformatics in Galaxy. Nucleic Acids Res. 2017;45:W560–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galaxy instance of the Huttenhower Lab, http://huttenhower.sph.harvard.edu/galaxy [Google Scholar]
- 15. ASaiM Documentation, http://asaim.readthedocs.io [Google Scholar]
- 16. Docker images tracking the stable Galaxy releases, http://bgruening.github.io/docker-galaxy-stable [Google Scholar]
- 17. Blankenberg D, Von Kuster G, Bouvier E, et al. Dissemination of scientific software with Galaxy ToolShed. Genome Biol. 2014;15:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sloggett C, Goonasekera N, Afgan E. BioBlend: automating pipeline analyses within Galaxy and CloudMan. Bioinformatics. 2013;29:1685–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grüning B, , Dale R, Sjödin A, et al. Bioconda: A sustainable and comprehensive software distribution for the life sciences. bioRxiv. 2017. http://dx.doi.org/10.1101/207092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. EBISearch,http://github.com/bebatut/ebisearch [Google Scholar]
- 21. Batut B, Grüning B. ENASearch: A Python library for interacting with ENA's API. The Journal of Open Source Software. 2017;2:418. [Google Scholar]
- 22. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H. Improving SNP discovery by base alignment quality. Bioinformatics. 2011;27:1157–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McDonald D, Clemente JC, Kuczynski J, et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience. 2012;1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. FastQC, https://www.bioinformatics.babraham.ac.uk/projects/fastqc. [Google Scholar]
- 27. Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trim Galore!, https://www.bioinformatics.babraham.ac.uk/projects/trim_galore.
- 29. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ewels P, Magnusson M, Lundin S, et al. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fu L, Niu B, Zhu Z, et al. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–7. [DOI] [PubMed] [Google Scholar]
- 33. Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38:e191–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cock PJA, Chilton JM, Grüning B, et al. NCBI BLAST+ integrated into Galaxy. Gigascience. 2015;4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. [DOI] [PubMed] [Google Scholar]
- 39. Mistry J, Finn RD, Eddy SR, et al. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41(12):e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodriguez-R LM, Konstantinidis KT. Nonpareil: a redundancy-based approach to assess the level of coverage in metagenomic datasets. Bioinformatics. 2014;30:629–35. [DOI] [PubMed] [Google Scholar]
- 41. Li D, Luo R, Liu C-M, et al. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. [DOI] [PubMed] [Google Scholar]
- 42. Nurk S, Meleshko D, Korobeynikov A, et al. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mikheenko A, Saveliev V, Gurevich A. MetaQUAST: evaluation of metagenome assemblies. Bioinformatics. 2016;32:1088–90. [DOI] [PubMed] [Google Scholar]
- 44. VALET, http://github.com/jgluck/valet.
- 45. Truong DT, Franzosa EA, Tickle TL, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–3. [DOI] [PubMed] [Google Scholar]
- 46. Abubucker S, Segata N, Goll J, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Group HUMAnN2 to GO slim terms, https://github.com/asaim/group_humann2_uniref_abundances_to_GO.
- 48. Langille MGI, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. export2graphlan, http://bitbucket.org/CibioCM/export2graphlan.
- 50. Asnicar F, Weingart G, Tickle TL, et al. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ. 2015;3:e1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bik HM, Phinch: an interactive, exploratory data visualization framework for -Omics datasets. bioRxiv. 2014; http://dx.doi.org/10.1101/009944. [Google Scholar]
- 53. GraPhlAn, http://huttenhower.sph.harvard.edu/graphlan.
- 54. Nascimento M, Sousa A, Ramirez M, et al. , PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33(1):128–129. [DOI] [PubMed] [Google Scholar]
- 55. Goecks J, Coraor N, Galaxy Team, NGS analyses by visualization with Trackster. Nat Biotechnol. 2012;30(11):1036–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Awad S, Irber L, Titus Brown C, . Evaluating metagenome assembly on a simple defined community with many strain variants, bioRxiv. 2017. http://dx.doi.org/10.1101/155358.
- 57. Greenwald WW, Klitgord N, Seguritan V, et al. Utilization of defined microbial communities enables effective evaluation of meta-genomic assemblies. BMC Genomics. 2017;18:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Olson ND, Treangen TJ, Hill CM, et al. Metagenomic assembly through the lens of validation: recent advances in assessing and improving the quality of genomes assembled from metagenomes. Brief Bioinform. 2017, bbx098; http://dx.doi.org/10.1093/bib/bbx098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Quince C, Walker AW, Simpson JT, et al. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833–44. [DOI] [PubMed] [Google Scholar]
- 60. Sczyrba A, Hofmann P, Belmann P, et al. Critical Assessment of Metagenome Interpretation-a benchmark of metagenomics software. Nat Methods. 2017;14:1063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van der Walt AJ, Van Goethem MW, Ramond J-B, et al. Assembling Metagenomes, One Community At A Time, BMC Genomics. 2017, 18:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vollmers J, Wiegand S, Kaster A-K. Comparing and evaluating metagenome assembly tools from a microbiologist's perspective - not only size matters!. PLoS One. 2017;12:e0169662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Batut B, Hiltemann S, Bagnacani A, et al. , Community-driven data analysis training for biology, bioRxiv, 2017, http://dx.doi.org/10.1101/225680. [DOI] [PMC free article] [PubMed]
- 64. Galaxy Training Material for metagenomics, http://training.galaxyproject.org/topics/metagenomics
- 65. Sobel J, Henry L, Rotman N, et al. BeerDeCoded: the open beer metagenome project. F1000Res. 2017;6:1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Metagenomics flavor of the European Galaxy instance, https://metagenomics.usegalaxy.eu
- 67. Batut B, Gravouil K, Defois C, et al. Supporting data for “ASaiM: a Galaxy-based framework to analyze microbiota data”. GigaScience Database. 2018; http://dx.doi.org/10.5524/100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
9/13/2017 Reviewed
10/4/2017 Reviewed
1/23/2018 Reviewed
10/9/2017 Reviewed
1/29/2018 Reviewed