Abstract
Purpose of Review
The objective of this review is to critically assess the contributing role of the gut microbiota in human obesity and type 2 diabetes (T2D).
Recent Findings
Experiments in animal and human studies have produced growing evidence for the causality of the gut microbiome in developing obesity and T2D. The introduction of high-throughput sequencing technologies has provided novel insight into the interpersonal differences in microbiome composition and function.
Summary
The intestinal microbiota is known to be associated with metabolic syndrome and related comorbidities. Associated diseases including obesity, T2D, and fatty liver disease (NAFLD/NASH) all seem to be linked to altered microbial composition; however, causality has not been proven yet. Elucidating the potential causal and personalized role of the human gut microbiota in obesity and T2D is highly prioritized.
Keywords: Diabetes, Gut microbiota, Metabolism, Obesity, Personalized medicine
Introduction
The current obesity pandemic is increasing worldwide, thus contributing to the rising incidence and prevalence of type 2 diabetes (T2D) mellitus. Particularly alarming is the equal trend of increase in obesity in children [1, 2]. T2D, notorious for its macrovascular and microvascular complications, presents a heavy challenge for health care systems [3]. Therefore, researchers globally are attempting to identify new preventive and therapeutic options. Obesity puts people at risk for developing metabolic syndrome, defined by increased waist circumference, hypertension, dyslipidemia, and insulin resistance [3, 4]. It has been clarified that obesity contributes to T2D by decreasing insulin sensitivity in adipose tissue, liver, and skeletal muscle, and subsequently impaired beta-cell function. So, the question arises: if insulin resistance is key in developing T2D, what are the contributing factors in its development? For example, DeFronzo describes a total of eight possible pathophysiological derangements contributing to the development of T2D [5], highlighting the multifactorial aspect of T2D and also suggesting the need of a more personalized approach in recognizing these derangements.
Increasing evidence indicates a fundamental role of excess central adiposity in developing the metabolic syndrome [6]. However, the underlying mechanism is not well understood [7]. For example, it is known that surgical removal of subcutaneous [8] or intra-abdominal [9] adipose tissue does not improve insulin sensitivity in obese human subjects despite weight loss induction. Underscoring the role of systemic factors maintaining insulin resistance, the search continues and the focus has shifted to the gut microbiota and its systemic properties for the past few years. The gut microbiota has attracted more and more attention as an underlying mechanistic driver in obesity and insulin resistance. Based on recent studies, we see an altered gut microbiota in obesity [10] and view the gut microbiota as an environmental factor contributing to adiposity and insulin resistance [11, 12].
The gut microbiota is a collective term for the microbial community in the gut [13], whereas the gut microbiome is defined as the full collection of genes in the gut microbiota [14]. This interest in gut microbiota is not illogical. Even in 460–377 B.C., the father of modern medicine Hippocrates stated his famous quote, “All disease begins in the gut”. The gut microbiome contains an immense diversity of microorganisms, varying from bacteria as well as viruses, fungi, phages, protozoa, and archaea [15–17] all colonizing our adult bodies. Archaea, bacteria and Eukarya encompass the three-domain taxonomy of life [13]. The introduction of molecular techniques using 16S rRNA gene sequencing created phylogenetic information on microbial taxa. In other words, it can be used to distinguish microbial groups in phylotypes yet lacks specificity to describe bacterial species for which more advanced techniques like shotgun metagenomic sequencing are needed [13]. Recent estimates show that our microbiota equal our total number of our somatic and germ cells [18]. The proposed view of our microbiota as a microbial (endocrine) organ living symbiotically inside our gut has led to a new perspective postulating multiple lineages capable of communicating with each other and shaping host immunometabolism in several ways [12]. This intimate co-evolution has led to an interlocked symbiotic relationship, with diverse capacities including the degradation of otherwise indigestible components of our diet, harvesting of energy and nutrients, shaping of the host immune system, maintaining the integrity of the gut mucosal barrier, and xenobiotic metabolism [12, 19–23]. Thus, gut microbiota complement our biology in ways that are mutually beneficial.
Diversity of the gut microbiota is the result of strong host selection and co-evolution, influenced by many factors. It includes thousands of species with a collective genome, the metagenome, which is close to 5 million genes. Our insight into the composition of the gut microbiota is based on culture-based studies and became more precisely characterized with high-throughput sequencing technology [12]. Several studies have shown the presence of microRNA (small non-coding RNA) extracellularly, which could be indicative for diseases such as intestinal malignancies [24, 25]. MicroRNA is a normal compound in feces in mice and humans, with intestinal epithelial cells and Hopx-expressing cells (intestinal stem cells, later derived in Goblet and Paneth cells) as the major source [26]. The concentration of microRNA is inversely correlated with microbial density, suggesting that it is absorbed by microbes and in this way is influencing the bacterial genes [27]. Sequencing technology makes it possible to analyze the intestinal metagenome based on genetic material from stool samples [28]. The development of these culture-independent genomics methods revived interest of microbiota in the etiology of diseases, allowing better understanding of the complexity of the gut microbiota [22, 29]. Much attention has been paid to identify the diverse composition of microbes in the healthy human population, creating the concept for a “personal microbiota” [30]. Despite the interpersonal variation, the gut microbiota maintain a stable relative abundance at operational taxonomic unit levels [31]. Five phyla dominate the microbial community: Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia [16]. From animal studies, we know that rapid luminal flow, high acidity, and secretion of bile acids and other compounds cause a changing microbial density along the gastrointestinal (GI) tract. The gradient starts with low density in the upper GI tract and it expands towards the rectum [32]. The proximal part of the GI tract is enriched with Lactobacillaceae (belonging to Firmicutes) and proteobacteria (Enterobacteriaceae), whereas the more distal large intestine shows higher concentrations of Firmicutes (Lachnospiraceae, Ruminococcaceae) and anaerobes such as Bacteroidetes (Bacteroidaceae, Prevotellaceae, and Rikenellaceae). The representation of Verrucomicrobia, Akkermansia muciniphila, is also mainly located distally [33–36].
Metagenomic-wide association studies combine metagenomic data with clinical features. Such studies presented significant differences on a metagenomic level between metabolically healthy versus metabolically unhealthy subjects [37, 38]. The Human Microbiome Project (HMP) [30] from the USA as well as LIFELINES [39••], and the Flemish Gut flora cohorts in Europe [40] are the largest cohorts to date (> 1000 subjects), containing high-throughput metagenomic data serving as a catalogue of the human microbiota. Although small, there seems to be a relationship between human metabolism and gut microbiota composition in otherwise healthy subjects.
Symbionts and commensals are not the only microorganisms the host encounters. The host immune system is continuously challenged by distinguishing beneficial microbes from pathogens. The interactive role of phages [41] and fungi [42] in gut microbiota composition is also gaining more attention. Although still poorly understood, the gut microbiota and the host immune system have co-evolved so profoundly that they can influence our immunological well-being [17]. This is seen in germ-free mice, where the absence of a microbiota leads to defects in the development and function of the immune system [43]. This described dynamic maturation of the gut microbiome and host immune system is determining host-microbe interactions and influencing the susceptibility to infection, inflammatory diseases, and autoimmunity [21].
There are different factors influencing the development of the microbiome in the early years of life, starting with the mode of birth [44, 45], breastfeeding or formula-feeding infants, and possibly the introduction of solid food [46]. The intestinal microbiome stabilizes about 3 years after birth, when it resembles the adult microbiome and stays relatively stable over time [47, 48]. In adulthood, the microbiome can be altered by changes in diet [49] as well as by the use of several types of medication such as antibiotics [50], metformin [51, 52], and even proton pump inhibitors [53]; this is also illustrated in Fig. 1. Nonetheless, by deep sequencing the gut microbiomes in a large Caucasian cohort, only 18.7% of the interpersonal variation in microbial composition could be explained by host characteristics (physiologic and biomedical measures), previous diseases, medication use, smoking, and dietary factors [54].
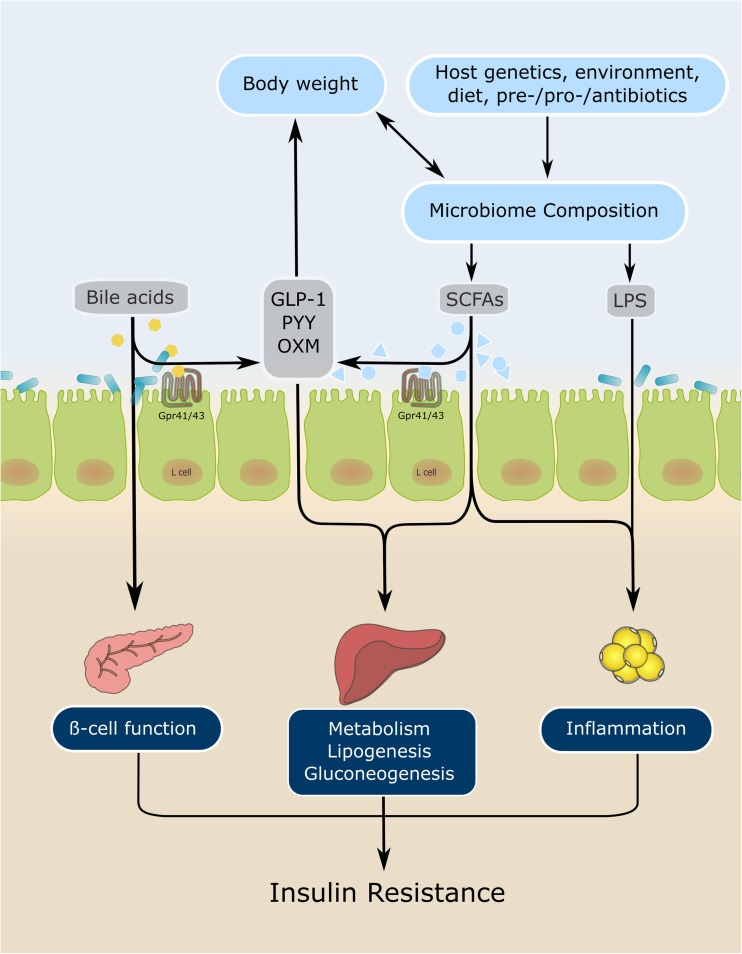
Fig. 1.
Effect of gut microbiota in liver disease, insulin resistance, and type 2 diabetes. GLP-1, glucagon like peptide 1; Gpr41, G-coupled receptor 41; Gpr43, G-coupled receptor 43; LPS, lipopolysaccharides; OXM, oxyntomodulin; PYY, protein YY; SCFA, short-chain fatty acids
The Gut Microbiota in Cardiometabolism
Diet is a modulator of the composition and the function of the gut microbiota [20]. Human intervention studies have revealed some aspects in which diet can alter the gut microbiota. First, the microbiota alters rapidly when exposed to great and fast changes in diet. Short-term dietary changes such as switching between plant- and meat-based diets, or adding more than 30 grams of fiber per day to the diet, or following a diet with a different fat/fiber content can change the human gut microbiota in function and composition significantly in 48 hours [49, 55, 56]. Fiber-enriched diets have been shown to improve insulin resistance in lean and in obese subjects with diabetes [57, 58]. However, only long-term dietary habits are most important in actually shaping the composition of the gut microbiota. Short-term dietary interventions failed to change the major features and classification of the microbiota [55]. Another aspect to consider is the high interpersonal variance in the effect of changed diets on the gut flora, thus mirroring the individualized nature of our gut microbiota [20]. Moreover, the landmark paper of Zeevi et al. elucidated the possibility of predicting individual glycemic responses to diet based on the composition and function of the individual microbiota. They measured continuous (postprandial) glycemic responses to meals during 1 week in a human cohort of 900 subjects and found high interpersonal variability to identical meals. Using machine learning algorithms, the authors were able to predict post-prandial responses to meals. Also, they effectively used and validated these algorithms to personalize dietary recommendations [59•].
In line with their finding that diet is an important driver of gut microbiota composition, Yatsunenko et al. found that gut microbiomes differ over human populations and that Malawian and Amerindian infants have more shared features than when compared to those of American infants [60].
It is thought that (aromatic) metabolites derived from gut microbiota processing of dietary compounds are important factors influencing host physiology; conversely, the responses to nutrients are influenced by the gut microbiota. Choline and trimethylamine-N-oxide (TMAO) are two of those impacting micronutrients. For example, Spencer et al. showed that a choline-deficient diet in humans modulates the gut bacteria with altered levels of Gammaproteobacteria and Erysipelotrichi and this is directly associated with fatty liver development [61]. Wang et al. experimented in atherosclerosis-prone mice, supplementing a diet with either choline or TMAO, which resulted in increased levels of TMAO in plasma; the latter enhanced atherosclerotic plaques. The previous link was negated in atherosclerosis-prone mice with antibiotic-suppressed microbiota [62], underscoring the relationship between the gut microbiota, the consumed diet, and host metabolism.
Several studies, mostly animal models, demonstrated the microbiota as a major factor in the development of obesity [10, 11, 63, 64]. This is illustrated in a study on genetically obese ob/ob mice, which had a 50% reduction of Bacteroidetes and an increase of Firmicutes in comparison to their lean siblings [10]. These differences affect the metabolic potential of the gut microbiota, and these ob/ob mice are known to have an increased capacity in harvesting energy from ingested food. Furthermore, this highly sufficient harvesting of energy is a trait which appeared to be transmissible to germ-free mice through microbiota transplantation resulting in increase of obesity [63].
A similar composition of the microbiota is seen in obese human subjects, and the microbial ecology in humans changes with weight loss [65]. The transmissible trait of the gut microbiota has also been reported in humans. A fascinating study by Ridaura et al. showed that transferred human microbiota from discordant obese twins into mice changed their phenotype, underscoring potential causality. Transplanting the gut microbiota of the human obese twin significantly increased adipose mass in the recipient mice, and this effect was reproducible. They measured increased butyrate and propionate levels in mice colonized with lean human gut microbiota, compared to obese human gut microbiota. [66]. This demonstrates that not only obesity in mice but also obesity in humans could be transmitted through fecal transplantation. We have also demonstrated that performing a fecal microbiota transplantation (FMT) from lean human donors to obese human recipients with metabolic syndrome improved insulin sensitivity [67]. Influencing the gut microbiota by treatment with probiotics containing Lactobacillus gasseri demonstrated positive effects on body weight in overweight and obese subjects treated in a double-blind randomized controlled intervention trial [68]. This also suggests that interventions in microbial composition may have impact on body weight.
Currently, the most effective treatment for obesity in the long term is bariatric surgery [69]. Intriguingly, when mainly focused on the Roux-en-Y gastric bypass (RYGB), the effects of bariatric surgery exceed weight loss alone, with favorable changes in beta cell function in patients with T2D [70]. The metabolic benefits are reported in a few days post-bariatric surgery, which suggest a weight-independent effect [70]. This effect is possibly explained by stimulating the entero-insular axis, with altered levels in GLP-1, PYY, OXM, and ghrelin. GLP-1, PYY, and OXM are all produced by enteroendocrine L cells in the distal small intestine [70]. These intestine-derived hormones have anorexic properties such as decreased hunger, decreased food intake, and delayed gastric emptying. Levels of ghrelin, a gastric hormone known for its orexigenic properties, are reported to be suppressed after bariatric surgery [71]. Moreover, these endogenous peptides are involved in the regulation of insulin sensitivity and insulin secretion.
Metabolic Action of Gut Microbiota-Derived Short-Chain Fatty Acids
Diet is a major source of substrates for the production of small molecules by the gut microbiota. After absorption, these molecules may have a direct effect on hepatocytes via uptake and transportation in the portal vein. After escaping first-pass metabolism in the liver, these small molecules circulate systemically, and in this way, may contribute to diverse effects on host physiology [22]. Dependent on the substrate (amino acids, lipids, carbohydrates, iron) presented to the intestinal lumen, gut microbes can generate specific (aromatic) metabolites. Most of the otherwise not digestible carbohydrates are fermented by the microbial community to produce short-chain fatty acids (SCFA), of which acetate, propionate, lactate, and butyrate are the most important [72]. The subsequent host response is dependent on the relative proportion of SCFA and subsequent liver clearance, where they induce de novo hepatic lipogenesis and other processes, while only a small subset enters the systemic circulation [11]. It should be highlighted that not all SCFA have similar metabolic effects [73]. The causal role of intestinal bacterial strains in the production of SCFA has been demonstrated clearly using oral antibiotic treatment that induced profound effects on the production of most of these metabolites by the gut microbiota [74]. The importance of iron as a substrate was demonstrated in rats: iron deficiency was correlated with lower levels of cecal SCFA, such as propionate and butyrate, when compared to iron sufficient rats. An increased abundance of Lactobacilli and Enterobacteriaceae was observed in addition to a significantly decreased abundance of the butyrate-producing Roseburia spp./E. rectale group in iron-deficient rats. Repletion of iron significantly increased cecal butyrate concentrations [75]. SCFA account for approximately 5–10% of the energy source of the body [76]. In this regard, butyrate is known as a key energy substrate for both colonocytes and enterocytes and has been described to play a role in energy expenditure by stimulating mitochondrial respiration and also increasing fatty acid oxidation [77, 78]. On the other hand, acetate functions as a substrate for cholesterol synthesis [79]. Indeed, composition of the gut microbiota and proportions of SCFA are correlated with obesity: obese mouse models show increased propionate levels compared to lean mice with an altered phyla ratio of Bacteroidetes-to-Firmicutes as well as an increased proportion of Bacteroidetes [79]. Conversely, oral administration of butyrate has also been reported to improve insulin sensitivity [78]. It is thought that SCFA are capable of functioning as signaling molecules by binding to G-protein-coupled receptors Gpr41 (FFAR3) and Gpr43 (FFAR2), and thus fine-tune metabolism. [80, 81] Moreover, Gpr43-deficient mice become obese when fed a normal diet, whereas mice with overexpression of Gpr43 remain lean even when consuming a high-fat diet. It was demonstrated that SCFA-mediated activation of Gpr43 led to suppression of insulin signaling in adipose tissue of these mice [82].
Finally, Gpr43 activation by SCFA in (small) intestinal enteroendocrine L cells induces GLP-1 release that can also affect insulin sensitivity via incretin response [83]. In conclusion, intestinally derived SCFA are an important source for energy, but are also capable of acting as signaling molecules in adipose tissue, thereby exerting an influence on energy homeostasis.
Gut Microbiome in Insulin Resistance and Type 2 Diabetes
As described above, the intestinal microbiota should not be viewed as an isolated community. Instead, it functions as a large metacommunity, in which microorganisms reciprocally affect each other. In this way, they can disperse and colonize new gut habitats [84]. The importance of diversity in metabolic control was shown in landmark papers by Le Chatelier et al. and Cotillard et al. who showed that low diversity in the gut microbiome associates with obesity and a higher prevalence of insulin resistance, non-alcoholic fatty liver disease (NAFLD), and low-grade inflammation [85, 86]. Furthermore, low bacterial diversity was characterized by pro-inflammatory properties, suggested by the reduction in butyrate-producing bacteria and the increase in mucin-degrading bacteria. These characteristics potentially impair the gut integrity causing low-grade inflammation through endotoxemia [76, 85, 86].
As described above, low-grade inflammation of visceral adipose tissue may provide a link between obesity and insulin resistance [87]. Adipose macrophage accumulation in crown-like structures correlates with altered gene expression involved in inflammatory control [88]. These findings indicate a profound role of the innate immune system in insulin resistance. Low-grade inflammation related to increased (translation of) Gram-negative bacterial strains has also been observed in patients with T2D. Mice and humans with diabetes have elevated plasma levels of lipopolysaccharides (LPS), a bacterial endotoxin derived from the membrane of Gram-negative bacteria. This phenomenon, also known as metabolic endotoxemia, is induced via either bacterial translocation over the intestinal wall or partly via bacterial capsule fragments that enter the bloodstream. Indeed, increased levels of circulating LPS have been shown to impair glucose metabolism in mice [89, 90]. To underscore the relevance to humans, Qin et al. used the concept of metagenomic linkage groups (MLG) and observed an abundance of Escherichia coli in the T2D MLG, a bacterial strain that is thought to be one of the major sources of LPS in humans [38]. LPS and other gut-derived molecules, such as peptidoglycans [91] and flagellins [92], activate pattern recognition receptors at the surface of immune cells, such as Toll-like receptors, triggering the production of pro-inflammatory cytokines [93].
Microbiome sequencing also revealed that butyrate-producing Clostridialis (Roseburia and Faecalibacterium prausnitzii), known for their anti-inflammatory properties [67], are less abundant in the feces of subjects with T2D compared to controls [37, 38]. In contrast, animal data that showed that Akkermansia muciniphila was associated with improved metabolic control and less obesity in mouse models of T2D [94]. Qin et al. reported an abundance of Akkermansia muciniphila in the T2D MLG in humans [38]. These findings potentially underscore the differences between human and mouse models of metabolism in relation to microbiota composition, but may also be explained by sampling errors because stools that have been exposed to the epithelial mucus layer are more likely to comprise Akkermansia muciniphila. Moreover, ethnic differences between human populations may also affect microbiota composition. Karlsson et al. compared data of T2D-associated metagenomes between Chinese and Swedish subjects with T2D, which indicated that different intestinal bacterial species were involved in similar metabolic functions [37]. The authors were also able to distinguish subjects with T2D from healthy subjects, with a predictive power exceeding that of body mass index (BMI) [37]. Nevertheless, these data should be interpreted with caution. Finucane et al. performed a meta-analysis, pooling results from four studies, including publicly available data from HMP and MetaHIT. This meta-analysis confirmed the differences in phylum-level taxonomic composition between lean and obese subjects. However, it could not reproduce the association between BMI and taxonomic composition [95]. Composition of the gut microbiota does not always translate into function. Identifying the microbiota-derived metabolites through metabolomics gives new insight into the functional capacity of the gut microbiota. Pedersen et al. performed metabolomics and microbiome profiling in subjects with T2D and without diabetes. They reported increased plasma levels of branched-chain amino acids (BCAA) as characteristic of individuals with insulin resistance, which in turn correlates with specific bacterial species (P. copri and B. vulgatus) in their gut microbiota [96]. Systems biology approaches can be used to connect microbial composition to functional capacity. The latter has been extensively reviewed by Meijnikman et al. [97].
In conclusion, in the past decade, animal and human studies have identified relevant differences in intestinal microbiota composition in subjects with obesity and T2D [10, 89, 98]. However, the causality and magnitude of effect on metabolic function remains to be proven [99].
Gut Microbiota in Lipid Metabolism
In recent decades, it has become clear that many metabolic, inflammatory, and innate immune mechanisms are also coordinated by (dietary-derived) lipids [4]. The nutritional importance of dietary lipids is unequivocal, but they also operate as (proinflammatory) ligands binding to nuclear receptors [100]. These include peroxisome-proliferator-activated receptor (PPAR) and liver X receptor (LXR) families, which are pivotal in metabolic and inflammatory pathways. Numerous fatty acids are capable of activating all three members of the PPAR family, thereby improving insulin action and suppressing production of pro-inflammatory cytokines such as TNF-α [101, 102].
G-coupled protein receptors (Gpr) are also activated by both diet-derived metabolites and lipids. For example, the activation of Gpr43 by the dietary-derived metabolite acetate directly reduces lipolysis in adipocytes leading to decreased plasma-free fatty acid levels. This suggests a potential therapeutic role for Gpr43 in the regulation of lipid metabolism [103].
Lipid accumulation in conjunction with low-grade inflammation is a pathophysiological hallmark of atherosclerosis [3]. There is emerging evidence that the pathophysiology of atherosclerosis is related to interpersonal gut microbiome differences [104]. Atherosclerosis seems to be related to TMAO, which is a new marker associated with increased risk of atherosclerosis and coronary artery disease [62]. Human studies using short-term antibiotic intervention have shown that changes in the composition of gut microbiota drives differential production of dietary choline and L-carnitine that can be converted by the gut microbiota to trimethylamine (TMA), which is subsequently oxidized by the hepatic flavin monooxygenases to form TMAO [105]. A major source of dietary choline is food rich in lipid phosphatidylcholine, also known as lecithin, which is present in milk, eggs, liver, red meat, poultry, and fish [62]. However, causality has not yet been established, and our recent fecal transplantation study failed to show altered TMAO production in humans with increased CVD risk that underwent donor FMT [106].
Other key intestinal regulators of lipid and cholesterol metabolism are bile acids, which are involved in facilitating intestinal absorption and transport of diet-derived nutrients, vitamins, and lipids. Whereas bile production takes place in the liver (and is facilitated by products derived from lipid catabolism), 95% of all bile acids will be reabsorbed in the terminal ileum and subsequently re-absorbed by the liver, constituting the so-called enterohepatic circulation. The intestinal microbiota is responsible for converting primary bile salt to secondary bile salts via bile acid de-hydroxylation [107]. Although short courses of oral antibiotics affect intestinal microbiota composition and bile acid metabolism in humans, we found differential effects on glucose metabolism [108, 109]. A more drastic intervention in the intestinal physiology is the RYGB which also has a major influence on enterohepatic bile acid circulation. For example, increased concentrations of primary plasma bile acids have been reported after RYGB, while this effect has not been seen after adjustable gastric banding [110, 111]. In murine models, bile acids promote the release of (enteroendocrine L-cell produced) GLP-1 through the activation of Takeda G-protein coupled receptor-5 (TGR5), thus affecting insulin secretion and whole-body insulin sensitivity [112]. Another important receptor for these bile acids is the farnesoid X receptor (FXR), which is mainly expressed in the liver and intestine, but also in pancreatic beta cells [113]. However, some studies could not find elevated plasma bile acid levels in the first days after bariatric surgery, when most of the metabolic benefits are seen [114, 115]. Thus, bile acids might contribute to the long-term metabolic benefits of bariatric surgery by improving intestinal hormone secretion, but do not explain the observed metabolic effects immediately after surgery.
Gut Microbiome in Appetite
Obesity is defined as an imbalance between energy intake (usually food intake) and energy expenditure [116]. The brain is a key regulator in detecting alterations in energy balance and induces behavioral and metabolic responses to correct these alterations. The hypothalamus plays an important role in regulation of both food intake as well as energy homeostasis, receiving hormonal and (vagal) neuronal information from the periphery [117, 118]. The gut-brain axis is increasingly highlighted as an important pathway connecting gut microbiota and its metabolites with central regulation of metabolism. For example, in a recent study, mice were exposed to a high-fat diet with SCFA butyrate, either intragastric or administered via intravenous injection. Interestingly, intragastric administration reduced food intake by 21% in 24 hours, while intravenous injection did not lead to any behavioral changes in food intake, suggesting that the effect of butyrate on feeding behavior is indirect through a mechanism involving gut-brain neural circuits [119].
SCFA has also been implicated in intestinal gluconeogenesis (IGN), which is thought to influence food intake and glucose metabolism in mice [120]. Recent animal work showed that SCFA such as butyrate and proprionate might exert part of their beneficial metabolic effects via induction of IGN and IGN genes. De Vadder et al. suggest direct activation of IGN genes in enterocytes by butyrate, while proprionate stimulates Gpr-41 in the periportal afferent neural system and indirectly activates IGN via a gut-brain communication axis [121]. However, this effect has not been reproduced in humans.
As seen in animal models, IGN induces release of glucose into the portal vein where glucose sensors are located and these sensors transmit signals to the brain by the peripheral nervous system, which curbs hunger and improves insulin resistance [122].
Animal studies describe IGN as an important factor in metabolic control both after protein-enriched diet [123–125], and after RYGB surgery [126]. Troy et al. presented a hepatoportal sensor pathway providing feedback to IGN regulation as a driving mechanism for the beneficial insulin-sensitizing effects after gastric bypass in mouse models [126]. On the other hand, a recent study could not detect significant differences in glucose levels between the portal venous blood and central venous blood during RYGB surgery and 6 days after surgery in human subjects with T2D [127].
Changing the gut microbiome composition with prebiotics has also been shown to affect portal vein levels of other hormones including GLP-1, which in turn affected food intake, followed by a decrease in body weight and fat mass [128]. This supports the hypothesis of the communicating property of the gut-brain axis. Experiments with prebiotic-treated mice demonstrated an increase in GLP-1 in portal venous blood, whereas the orexigenic hormone ghrelin was decreased in plasma [128]. Supporting the latter, GLP-1 receptor knock-out mice are completely insensitive to the beneficial effects of prebiotic treatment [129, 130]. This effect has also been demonstrated in humans treated with prebiotics, wherein peripheral plasma changes in GLP-1 and PYY levels were associated with increased satiety and decreased hunger scores [131]. As discussed above, Gpr41 and Gpr43 are also expressed on enteroendocrine L cells, which makes is plausible to consider a possible link between Gpr41 and Gpr43 activation on L cells by SCFA, thereby inducing secretion of GLP-1 and PYY [83, 130, 132]. Taken together, the gut microbiota is perfectly capable of interacting with other organs, such as the brain, potentially functioning as an individual endocrine organ and using metabolites and hormones as key messengers.
The Gut Microbiome: Conclusions and Future Perspective
The studies discussed in this review suggest that individuals who are obese are likely to have an imbalance in gut microbiota composition and a ‘gut signature’ varying with metabolic control [76]. This possibility is an exciting avenue for further research and possible novel treatment targets. However, because most studies have been undertaken in animals, direct translation of the findings to human is limited. Many studies have been carried out in animal models, which differ significantly in metabolism, gut microbiota composition, and immune system [133].
Moreover, conclusions on the magnitude and causal role of the intestinal microbiota in human metabolism are blurred by other confounding factors, such as (ethnic) differences in study populations, differences in sequencing and analytical techniques, and variability of diets used for the studies. Therefore, guidelines for standardized techniques are essential.
Shifting towards large prospective cohort studies will provide us with more evidence and information on clinical relevance. Similarly, mapping patients with pre-diabetes and diabetes, and observing their diet, type of obesity, metabolic parameters, medication, and inflammatory state is essential for clarifying whether microbiome composition and the presence of metabolites is a result of disease phenotype or if it is causally interlocked with the underlying pathophysiology. Identifying the most important microbiota-related metabolic pathways will enable us to develop new therapeutic agents by influencing these microbiota-related pathways.
Treatment with prebiotics or with newly identified beneficial bacterial strains are potential interventions that will be used in the near future, and it will be important to evaluate their efficacy. Likewise, interventional studies with metabolites of microbiota will be performed (including SCFA butyrate supplementation) to evaluate if this compound has similar effects on food intake, energy expenditure, and improved metabolic features in humans [119].
Newly identified bacterial strains with probiotic potential will probably be found using FMT. For example, our recent study illustrated how donor FMT can (transiently) improve insulin sensitivity in male recipients with metabolic syndrome, bringing new insight in pathophysiology [134••]. Nevertheless, FMT procedures need standardization to provide safety and similar quality between centers. It should also be considered that the therapeutic effect from FMT is not only from bacteria; other microorganisms or other features from the donor could enhance the observed effect during FMT [135].
In conclusion, the modifiable effects of the human gut microbiota on the development of metabolic syndrome make its manipulation a promising therapeutic approach. Analyzing and mapping individual microbial composition on a metagenomic level provides insight into specific targets for treatment and contributes to personalized therapeutic interventions.
Funding
This paper was supported by Le Ducq consortium grant 17CVD01 and a Novo Nordisk Foundation GUT-MMM grant. M. Nieuwdorp is supported by a ZONMW-VIDI grant 2013 (016.146.327) and CVON Young Talent grant 2012.
Compliance with Ethical Standards
Conflict of Interest
Ömrüm Aydin and Victor Gerdes declare that they have no conflict of interest.
Max Nieuwdorp is on the Scientific Advisory Board of Caelus Pharmaceuticals, The Netherlands. None of these conflicts of interest is directly related to the research currently described.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Genetics
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major Importance
- 1.Rocchini AP. Childhood Obesity and Diabetes Epidemic. The New England Journal of Medicine. 2002;346:854–855. doi: 10.1056/NEJM200203143461112. [DOI] [PubMed] [Google Scholar]
- 2.Swinburn BA, Sacks G, Hall KD, McPherson K, Finegood DT, Moodie ML, et al. The global obesity pandemic: shaped by global drivers and local environments. The Lancet. 2011;378(9793):804–814. doi: 10.1016/S0140-6736(11)60813-1. [DOI] [PubMed] [Google Scholar]
- 3.Semenkovich CF. Insulin resistance and atherosclerosis. J Clin Invest. 2006;116(7):1813–1822. doi: 10.1172/JCI29024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 5.Defronzo RA. Banting Lecture. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes. 2009;58(4):773–795. doi: 10.2337/db09-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. The Lancet. 2005;365(9468):1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- 7.Eckel RH, Grundy SM, Zimmet PZ. The Metabolic Syndrome. The Lancet. 2010;375:181–183. doi: 10.1016/S0140-6736(09)61794-3. [DOI] [PubMed] [Google Scholar]
- 8.Klein S, Fontana L, Young VL, Coggan AR, Kilo C, Patterson BW, Mohammed BS. Absence of an Effect of Liposuction on Insulin Action and Risk Factors for Coronary Heart Disease. The New England Journal of Medicine. 2004;350:2549–2557. doi: 10.1056/NEJMoa033179. [DOI] [PubMed] [Google Scholar]
- 9.Fabbrini E, Tamboli RA, Magkos F, Marks-Shulman PA, Eckhauser AW, Richards WO, et al. Surgical removal of omental fat does not improve insulin sensitivity and cardiovascular risk factors in obese adults. Gastroenterology. 2010;139(2):448–455. doi: 10.1053/j.gastro.2010.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101(44):15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bäckhed F, Sonnenburg JL, Peterson DA, Gordon JI. Host-Bacterial Mutualism in the Human Intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 13.Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol. 2006;21(9):517–523. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Reviews. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38(5):996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330(6012):1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell. 2016;164(3):337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587(Pt 17):4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16(5):559–564. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21(29):8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, Sinar DR, et al. Diagnostic MicroRNA Markers for Screening Sporadic Human Colon Cancer and Active Ulcerative Colitis in Stool and Tissue. Cancer Genomics & Proteomics. 2009;6:281–296. [PubMed] [Google Scholar]
- 25.Link A, Becker V, Goel A, Wex T, Malfertheiner P. Feasibility of fecal microRNAs as novel biomarkers for pancreatic cancer. PLoS One. 2012;7(8):e42933. doi: 10.1371/journal.pone.0042933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Weiner HL. Control of the gut microbiome by fecal microRNA. Cell Host & Microbe. 2016;3(4):176–177. doi: 10.15698/mic2016.04.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, et al. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe. 2016;19(1):32–43. doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vos WMD, Nieuwdorp M. Genomics: A Gut Prediction. Nature. 2013;498:48–49. doi: 10.1038/nature12251. [DOI] [PubMed] [Google Scholar]
- 29.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Human Microbiome Project C A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandal RS, Saha S, Das S. Metagenomic surveys of gut microbiota. Genomics Proteomics Bioinformatics. 2015;13(3):148–158. doi: 10.1016/j.gpb.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu S, Chen D, Zhang JN, Lv X, Wang K, Duan LP, et al. Bacterial community mapping of the mouse gastrointestinal tract. PLoS One. 2013;8(10):e74957. doi: 10.1371/journal.pone.0074957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14(1):20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World Journal of Gastroenterology. 2005;11(8):1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheithauer TP, Dallinga-Thie GM, de Vos WM, Nieuwdorp M, van Raalte DH. Causality of small and large intestinal microbiota in weight regulation and insulin resistance. Mol Metab. 2016;5(9):759–770. doi: 10.1016/j.molmet.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 38.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 39.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 40.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 41.Lerner A, Matthias T, Aminov R. Potential Effects of Horizontal Gene Exchange in the Human Gut. Front Immunol. 2017;8:1630. doi: 10.3389/fimmu.2017.01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Botschuijver S, Roeselers G, Levin E, Jonkers DM, Welting O, Heinsbroek SEM, et al. Intestinal Fungal Dysbiosis Is Associated With Visceral Hypersensitivity in Patients With Irritable Bowel Syndrome and Rats. Gastroenterology. 2017;153(4):1026–1039. doi: 10.1053/j.gastro.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nature reviews Immunology. 2004;4(6):478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 44.Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86(Suppl 1):13–15. doi: 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63(4):559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 46.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493(7430):45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falony G. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 51.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23(7):850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 53.Freedberg DE, Toussaint NC, Chen SP, Ratner AJ, Whittier S, Wang TC, et al. Proton Pump Inhibitors Alter Specific Taxa in the Human Gastrointestinal Microbiome: A Crossover Trial. Gastroenterology. 2015;149(4):883–885. doi: 10.1053/j.gastro.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu GD, Compher C, Chen EZ, Smith SA, Shah RD, Bittinger K, et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut. 2016;65(1):63–72. doi: 10.1136/gutjnl-2014-308209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robertson MD, Currie JM, Morgan LM, Jewell DP, Frayn KN. Prior short-term consumption of resistant starch enhances postprandial insulin sensitivity in healthy subjects. Diabetologia. 2003;46(5):659–665. doi: 10.1007/s00125-003-1081-0. [DOI] [PubMed] [Google Scholar]
- 58.Robertson MD, Wright JW, Loizon E, Debard C, Vidal H, Shojaee-Moradie F, et al. Insulin-sensitizing effects on muscle and adipose tissue after dietary fiber intake in men and women with metabolic syndrome. J Clin Endocrinol Metab. 2012;97(9):3326–3332. doi: 10.1210/jc.2012-1513. [DOI] [PubMed] [Google Scholar]
- 59.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell. 2015;163(5):1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140(3):976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 64.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Human gut microbes associated with obesity Ley Nature 2006.pdf. [DOI] [PubMed]
- 66.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–916. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 68.Kadooka Y, Sato M, Imaizumi K, Ogawa A, Ikuyama K, Akai Y, et al. Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. Eur J Clin Nutr. 2010;64(6):636–643. doi: 10.1038/ejcn.2010.19. [DOI] [PubMed] [Google Scholar]
- 69.Adams TD, Davidson LE, Litwin SE, Kim J, Kolotkin RL, Nanjee MN, et al. Weight and Metabolic Outcomes 12 Years after Gastric Bypass. N Engl J Med. 2017;377(12):1143–1155. doi: 10.1056/NEJMoa1700459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malin SK, Kashyap SR. Effects of various gastrointestinal procedures on beta-cell function in obesity and type 2 diabetes. Surg Obes Relat Dis. 2016;12(6):1213–1219. doi: 10.1016/j.soard.2016.02.035. [DOI] [PubMed] [Google Scholar]
- 71.Malin SK, Samat A, Wolski K, Abood B, Pothier CE, Bhatt DL, et al. Improved acylated ghrelin suppression at 2 years in obese patients with type 2 diabetes: effects of bariatric surgery vs standard medical therapy. Int J Obes (Lond). 2014;38(3):364–370. doi: 10.1038/ijo.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roy CC, Kien CL, Bouthillier L, Levy E. Short-Chain Fatty Acids: Ready for Prime Time? Nutrition in Clinical Practice. 2006;21(4):351–366. doi: 10.1177/0115426506021004351. [DOI] [PubMed] [Google Scholar]
- 73.Cani PD, Knauf C. How gut microbes talk to organs: The role of endocrine and nervous routes. Mol Metab. 2016;5(9):743–752. doi: 10.1016/j.molmet.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488(7413):621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dostal A, Chassard C, Hilty FM, Zimmermann MB, Jaeggi T, Rossi S, et al. Iron depletion and repletion with ferrous sulfate or electrolytic iron modifies the composition and metabolic activity of the gut microbiota in rats. J Nutr. 2012;142(2):271–277. doi: 10.3945/jn.111.148643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut. 2014;63(9):1513–1521. doi: 10.1136/gutjnl-2014-306928. [DOI] [PubMed] [Google Scholar]
- 77.Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13(5):517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58(7):1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring). 2010;18(1):190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 80.Brown AJ, Jupe S, Briscoe CP. A Family of Fatty Acid Binding Receptors. DNA and Cell Biology. 2005;24(1):54–61. doi: 10.1089/dna.2005.24.54. [DOI] [PubMed] [Google Scholar]
- 81.Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278(28):25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 82.Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61(2):364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Griffin NW, Ahern PP, Cheng J, Heath AC, Ilkayeva O, Newgard CB, et al. Prior Dietary Practices and Connections to a Human Gut Microbial Metacommunity Alter Responses to Diet Interventions. Cell Host Microbe. 2017;21(1):84–96. doi: 10.1016/j.chom.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 86.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500(7464):585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 87.Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci U S A. 2006;103(17):6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Apovian CM, Bigornia S, Mott M, Meyers MR, Ulloor J, Gagua M, et al. Adipose macrophage infiltration is associated with insulin resistance and vascular endothelial dysfunction in obese subjects. Arterioscler Thromb Vasc Biol. 2008;28(9):1654–1659. doi: 10.1161/ATVBAHA.108.170316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 90.Creely SJ, McTernan PG, Kusminski CM, Fisher FM, Da Silva NF, Khanolkar M, et al. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab. 2007;292(3):E740–E747. doi: 10.1152/ajpendo.00302.2006. [DOI] [PubMed] [Google Scholar]
- 91.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16(2):228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328(5975):228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cani PD, Delzenne NM. Gut microflora as a target for energy and metabolic homeostasis. Current Opinion in Clinical Nutrition and Metabolic Care. 2007;10:729–734. doi: 10.1097/MCO.0b013e3282efdebb. [DOI] [PubMed] [Google Scholar]
- 94.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23(1):107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 95.Finucane MM, Sharpton TJ, Laurent TJ, Pollard KS. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS One. 2014;9(1):e84689. doi: 10.1371/journal.pone.0084689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 97.Meijnikman AS, Gerdes VE, Nieuwdorp M, Herrema H. Evaluating causality of gut microbiota in obesity and diabetes in humans. Endocr Rev. 2017. [DOI] [PubMed]
- 98.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 99.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear Receptors and Lipid Physiology: Opening the X-Files. Science. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 101.Glass CK, Ogawa S. Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol. 2006;6(1):44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- 102.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115(5):1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ge H, Li X, Weiszmann J, Wang P, Baribault H, Chen JL, et al. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology. 2008;149(9):4519–4526. doi: 10.1210/en.2008-0059. [DOI] [PubMed] [Google Scholar]
- 104.Shapiro H, Suez J, Elinav E. Personalized microbiome-based approaches to metabolic syndrome management and prevention. J Diabetes. 2017;9(3):226–236. doi: 10.1111/1753-0407.12501. [DOI] [PubMed] [Google Scholar]
- 105.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kootte RS, Nieuwdorp M. [In press]. JAHA. 2018(In Press).
- 107.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50(10):1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol. 2014;60(4):824–831. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 109.Reijnders D, Goossens GH, Hermes GD, Neis EP, van der Beek CM, Most J, et al. Effects of Gut Microbiota Manipulation by Antibiotics on Host Metabolism in Obese Humans: A Randomized Double-Blind Placebo-Controlled Trial. Cell Metab. 2016;24(1):63–74. doi: 10.1016/j.cmet.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 110.Patti ME, Houten SM, Bianco AC, Bernier R, Larsen PR, Holst JJ, et al. Serum bile acids are higher in humans with prior gastric bypass: potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring). 2009;17(9):1671–1677. doi: 10.1038/oby.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pournaras DJ, Glicksman C, Vincent RP, Kuganolipava S, Alaghband-Zadeh J, Mahon D, et al. The role of bile after Roux-en-Y gastric bypass in promoting weight loss and improving glycaemic control. Endocrinology. 2012;153(8):3613–3619. doi: 10.1210/en.2011-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Düfer M, Hörth K, Wagner R, Schittenhelm B, Prowald S, Wagner TFJ, et al. Bile Acids Acutely Stimulate Insulin Secretion of Mouse b-Cells via Farnesoid X Receptor Activation and KATP Channel Inhibition. Diabetes. 2012;61:1479–1489. doi: 10.2337/db11-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jorgensen NB, Dirksen C, Bojsen-Moller KN, Kristiansen VB, Wulff BS, Rainteau D, et al. Improvements in glucose metabolism early after gastric bypass surgery are not explained by increases in total bile acids and fibroblast growth factor 19 concentrations. J Clin Endocrinol Metab. 2015;100(3):E396–E406. doi: 10.1210/jc.2014-1658. [DOI] [PubMed] [Google Scholar]
- 115.Steinert RE, Peterli R, Keller S, Meyer-Gerspach AC, Drewe J, Peters T, et al. Bile acids and gut peptide secretion after bariatric surgery: a 1-year prospective randomized pilot trial. Obesity (Silver Spring). 2013;21(12):E660–E668. doi: 10.1002/oby.20522. [DOI] [PubMed] [Google Scholar]
- 116.Schoeller DA. Insights into energy balance from doubly labeled water. Int J Obes (Lond). 2008;32(Suppl 7):S72–S75. doi: 10.1038/ijo.2008.241. [DOI] [PubMed] [Google Scholar]
- 117.Ahima RS, Antwi DA. Brain regulation of appetite and satiety. Endocrinol Metab Clin North Am. 2008;37(4):811–823. doi: 10.1016/j.ecl.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Apovian CM, Garvey WT, Ryan DH. Challenging obesity: Patient, provider, and expert perspectives on the roles of available and emerging nonsurgical therapies. Obesity (Silver Spring). 2015;23(Suppl 2):S1–S26. doi: 10.1002/oby.21140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li Z, Yi C-X, Katiraei S, Kooijman S, Zhou E, Chung CK, et al. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut. 2017;0(1):1. doi: 10.1136/gutjnl-2017-314050. [DOI] [PubMed] [Google Scholar]
- 120.Mithieux G, Andreelli F, Magnan C. Intestinal gluconeogenesis: key signal of central control of energy and glucose homeostasis. Curr Opin Clin Nutr Metab Care. 2009;12(4):419–423. doi: 10.1097/MCO.0b013e32832c4d6a. [DOI] [PubMed] [Google Scholar]
- 121.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1–2):84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 122.Delaere F, Duchampt A, Mounien L, Seyer P, Duraffourd C, Zitoun C, et al. The role of sodium-coupled glucose co-transporter 3 in the satiety effect of portal glucose sensing. Mol Metab. 2012;2(1):47–53. doi: 10.1016/j.molmet.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Duraffourd C, De Vadder F, Goncalves D, Delaere F, Penhoat A, Brusset B, et al. Mu-opioid receptors and dietary protein stimulate a gut-brain neural circuitry limiting food intake. Cell. 2012;150(2):377–388. doi: 10.1016/j.cell.2012.05.039. [DOI] [PubMed] [Google Scholar]
- 124.Mithieux G, Misery P, Magnan C, Pillot B, Gautier-Stein A, Bernard C, et al. Portal sensing of intestinal gluconeogenesis is a mechanistic link in the diminution of food intake induced by diet protein. Cell Metab. 2005;2(5):321–329. doi: 10.1016/j.cmet.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 125.Pillot B, Soty M, Gautier-Stein A, Zitoun C, Mithieux G. Protein feeding promotes redistribution of endogenous glucose production to the kidney and potentiates its suppression by insulin. Endocrinology. 2009;150(2):616–624. doi: 10.1210/en.2008-0601. [DOI] [PubMed] [Google Scholar]
- 126.Troy S, Soty M, Ribeiro L, Laval L, Migrenne S, Fioramonti X, et al. Intestinal gluconeogenesis is a key factor for early metabolic changes after gastric bypass but not after gastric lap-band in mice. Cell Metab. 2008;8(3):201–211. doi: 10.1016/j.cmet.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 127.Hayes MT, Foo J, Besic V, Tychinskaya Y, Stubbs RS. Is Intestinal Gluconeogenesis a Key Factor in the Early Changes in Glucose Homeostasis Following Gastric Bypass? Obesity Surgery. 2011;21(6):759–762. doi: 10.1007/s11695-011-0380-7. [DOI] [PubMed] [Google Scholar]
- 128.Cani PD, Dewever C, Delzenne NM. Inulin-type fructans modulate gastrointestinal peptides involved in appetite regulation (glucagon-like peptide-1 and ghrelin) in rats. British Journal of Nutrition. 2007;92(03):521. doi: 10.1079/bjn20041225. [DOI] [PubMed] [Google Scholar]
- 129.Cani PD, Knauf C, Iglesias MA, Drucker DJ, Delzenne NM, Burcelin R. Improvement of Glucose Tolerance and Hepatic Insulin Sensitivity by Oligofructose Requires a Functional Glucagon-Like Peptide 1 Receptor. Diabetes. 2006;55(5):1484–1490. doi: 10.2337/db05-1360. [DOI] [PubMed] [Google Scholar]
- 130.Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105(43):16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cani PD, Lecourt E, Dewulf EM, Sohet FM, Pachikian BD, Naslain D, et al. Gut microbiota fermentation of prebiotics increases satietogenic and incretin gut peptide production with consequences for appetite sensation and glucose response after a meal. Am J Clin Nutr. 2009;90(5):1236–1243. doi: 10.3945/ajcn.2009.28095. [DOI] [PubMed] [Google Scholar]
- 132.Nohr MK, Pedersen MH, Gille A, Egerod KL, Engelstoft MS, Husted AS, et al. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013;154(10):3552–3564. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 133.Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8(1):1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.•• Kootte RS, Levin E, Salojarvi J, Smits LP, Hartstra AV, Udayappan SD, et al. Improvement of Insulin Sensitivity after Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell Metab. 2017;26(4):611–9. e6. In this study leads to new insight in the possible therapeutic options of the gut microbiota in humans. [DOI] [PubMed]
- 135.Bouter KE, van Raalte DH, Groen AK, Nieuwdorp M. Role of the Gut Microbiome in the Pathogenesis of Obesity and Obesity-Related Metabolic Dysfunction. Gastroenterology. 2017;152(7):1671–1678. doi: 10.1053/j.gastro.2016.12.048. [DOI] [PubMed] [Google Scholar]