Abstract
Ubiquitination plays key roles in eukaryotic growth, stress adaptation, and metabolic regulation. In our previous work, ubiquitin was found to be secreted in the hypovirus-infected strain of Cryphonectria parasitica, a phytopathogenic filamentous fungus responsible for the chestnut blight. Here we report the functional and molecular characterization of a polyubiquitin gene, cpubi4, in C. parasitica. The expression of cpubi4 was upregulated by the infection of a hypovirus. Deletion of cpubi4 resulted in abnormal morphology, reduced sporulation, attenuation of virulence, and significant reduction in ubiquitination. A total of 378 sites in 236 proteins were identified to be significantly decreased in ubiquitination in the absence of cpubi4. Quantitative proteome analysis revealed that 285 in 4,776 identified proteins changed in abundance (1.5-fold, P < 0.05) in the cpubi4 null mutant, as compared with the wild-type strain.
Keywords: polyubiquitin, virulence, ubiquitination, hypovirus, chestnut blight fungus
Introduction
Ubiquitin is a highly conserved 76-amino acid protein, produced through de novo synthesis by ubiquitin coding genes and recycled from the ubiquitinated proteins by deubiquitinating enzymes (Kimura and Tanaka, 2010). Ubiquitin mediates post-translational modifications of proteins and plays critical roles in a variety of cellular processes, including transcriptional regulation, signal transduction, cell cycle, and cellular differentiation (Finley et al., 1987; Hershko and Ciechanover, 1998). There are four ubiquitin-encoding genes in mammal, UBA52 and RPS27A encode a single copy ubiquitin fused to ribosomal proteins L40 and S27a, respectively, and the other two genes encode polyubiquitin precursors with different number of ubiquitin repeats (Kimura and Tanaka, 2010). In yeast, there are three ubiquitin-related genes (UBI1, UBI2, and UBI3) each encoding a single ubiquitin that is fused to unrelated amino acid sequences (Ozkaynak et al., 1987) and the fourth gene UBI4 encodes a polyubiquitin precursor containing five ubiquitin repeats in a head-to-tail manner (Ozkaynak et al., 1984). In Candida albicans, inactivation of polyubiquitin gene UBI4 affected fungal growth, stress resistance ability, and virulence (Leach et al., 2011). In rice blast pathogenic fungi Magnaporthe oryzae, deletion of polyubiquitin-encoding gene MGG_01282, resulted in a reduction of growth and sporulation, abnormal conidia, reduced germination and appressorium formation, and lost the ability to cause disease (Oh et al., 2012).
Cryphonectria parasitica, a filamentous fungus, is the causative agent of chestnut blight, well known for its destructive effect on the chestnut forestry in North America. Hypoviruses are a group of plus-stranded RNA virus originally found in the chestnut blight fungus, known for its ability to attenuate the virulence of its host (termed hypovirulence) (Nuss, 2005). In addition to hypovirulence, hypovirus-infected C. parasitica strains exhibit a phenotype different from the virus-free wild-type strain, i.e., reduction in pigmentation, inhibition of sporulation, and female infertility. By virtue of transmissible hypovirulence, hypovirus-carrying strains have been used as biocontrol agents for chestnut blight disease (Anagnostakis, 1982). Due to the availability of genetic manipulation system for both the virus and the host fungus, hypovirus-C. parasitica interaction has evolved into a model to dissect the mechanism of virulence regulation in pathogenic fungi (Nuss, 2005; Eusebio-Cope et al., 2015). In our previous secretome analysis, ubiquitin was found to be up-regulated in the hypovirus-infected strain EP155/CHV1-EP713, indicating that ubiquitin expression may be manipulated by the hypovirus (Wang et al., 2016). To better understand the roles of ubiquitin in C. parasitica, we disrupted a polyubiquitin-encoding gene, cpubi4, and analyzed the phenotypic traits of the mutants. The cpubi4 mutants were impaired in development with diminished orange pigment, slower growth, reduced sporulation, and attenuated virulence. Ubiquitomics analysis revealed that cpubi4 was responsible for ubiquitination of a wide range of proteins, some of which are of critical importance in biological and metabolic processes.
Materials and methods
Fungal strains and growth conditions
C. parasitica EP155 (ATCC38755) is an orange pigment-producing, virulent, and highly sporulated wild-type strain. EP155/CHV1-EP713 is isogenic to EP155 but harboring hypovirus CHV1-EP713 by transfection with a synthetic infectious viral transcript (Chen et al., 1994). EP155/CHV1-EP713 is white, hypovirulent, and produces hardly any conidial spore on both PDA (Difco, Detroit, USA) or chestnut stems. DK80 was derived from EP155 but with its nonhomologous end joining DNA repair component encoding gene ku80 deleted. DK80 is highly efficient in gene homologous replacement, indistinguishable from the parental wild-type strain EP155 in colony morphology, conidiation, virulence, and ability to support hypovirus replication (Lan et al., 2008; Choi et al., 2012). All strains, including the cpubi4 deletion strains, were maintained on PDA at 25°C with a 12-h light at 1,500 lx and 12-h dark cycle (Hillman et al., 1990).
Gene manipulation
Preparation of protoplast and transformation of C. parasitica were carried out essentially as described previously (Churchill et al., 1990). The antibiotics hygromycin B (40 μg/ml) or G418 (25 μg/ml) was added to the medium for selection of transformants.
Deletion of the cpubi4 gene in C. parasitica
Disruption of C. parasitica gene was performed by homologous recombination. Primers and templates used to generate relevant fragments were listed in Supplementary Table 1. The 780-bp left flanking fragment, the 2146-bp hph cassette and the 761-bp right flanking fragment were fused to form a 3.7-kb cassette by double-joint PCR with 1:3:1 molar ratio. The PCR reaction cycles consisted of 94°C for 2 min, followed by 15 cycles of 94°C for 30 s, 58°C for 2 min, and 72°C for 4 min, with a final extension of 5 min at 72°C. A nested PCR was performed following the double-joint PCR using the primer pair cpubi4-LF/cpubi4-RR. After verification by agarose gel electrophoresis and gel extraction, 500 ng of hph cassette was used to transform DK80 protoplasts as described previously (Lan et al., 2008). Putative cpubi4 disruptants were identified by PCR, purified to nuclear homogeneity by single-spore isolation and further verified by Southern blot analysis. The confirmed transformants were designated as Δcpubi4 strains. Gene cloning, PCR, and Southern analysis were performed as described (Sambrook and Russell, 2001).
Complementation of cpubi4 mutant
To construct the complementary strains for Δcpubi4, a 4-kb genomic fragment containing the complete cpubi4 transcript region (983 bp), promoter region (1.6 kb), and terminator region (1.2 kb) from EP155 DNA was amplified and cloned into SacII and HpaI sites of vector pCPXG418 to generate plasmid pCPXG418/cpubi4. The cpubi4 complementary strains were obtained by transforming Δcpubi4 protoplast with plasmid pCPXG418/cpubi4. Selected transformants were subjected to single-spore purification with antibiotic G418 as the selection marker. PCR was performed to confirm the complementation of cpubi4 gene.
DNA and RNA analysis
The fungal strains were grown in EP complete medium (Puhalla and Anagnostakis, 1971) for 3 days at 25°C. DNA or RNA was prepared from the cultures as described (Shi et al., 2014). Total cDNA was generated with oligo (dT) and random primer, using a RevertAid RT Reverse Transcription Kit (Thermo Scientific). PCRs were performed in a Light Cycler 480 (Roche Applied Science). Relative accumulation level of a gene transcript was examined by quantitative real-time RT-PCR using the 18S rRNA values to normalize for variations in template concentrations (Livak and Schmittgen, 2001).
Protein analysis
Protein preparation and quantification
Fungal mycelia were ground into powder in liquid nitrogen and re-suspended in 1 ml of pre-chilled acetone (−20°C) containing 10% TCA and 0.07% β-mercaptoethanol. After incubation at −20°C for 2 h, the mixture was centrifuged at 13,523 g for 30 min at 4°C. The pellet was washed with ice-cooled acetone, air-dried, and then re-suspended in lysis buffer (7.5 M urea, 2.5 M thiourea, 12.5% glycerol, 50 mM Tris, and 2% protease inhibitor; Wang et al., 2014). After treated with ultrasonic at 200 W, the protein sample was centrifuged at 13,523 g for 30 min at 4°C to remove the debris. Three replicates for each strain were performed. The protein concentration was determined using Bradford Protein Assay (Bio-Rad, Hercules, CA, USA). The quality of the protein preparations was checked with SDS-PAGE using a Mini-PROTEAN system (BioRad, Hercules, USA) by loading 30 μg of protein per lane. After separation by electrophoresis, the gels were stained with Coomassie solution.
Western blotting
Simple Western system (Simon™, ProteinSimple, San Jose, CA) was used to analyze protein ubiquitination (Chen et al., 2013). The concentration of protein lysates was adjusted at 20 μg/μl with RIPA buffer (Beyotime Institute of Biotechnology, Shanghai, China) and 4 μl of the adjusted lysate (containing 80 μg of protein) was mixed with a master mix to a final concentration of 1 × sample buffer, 1 × fluorescent molecular weight marker, and 40 mM DTT. The ubiquitination of total proteins was identified using ubiquitin-specific antibody (catalog #sc-8017, Santa Cruz, USA) as primary antibody at 1:50 dilution. Compass software (ProteinSimple) was used to analyze the captured digital image.
Protein ubiquitination analysis
Ubiquitinated peptide affinity enrichment
For trypsin digestion, the protein solution was reduced with 10 mM DTT for 2.5 h at 37°C and alkylated with 50 mM IAA for 30 min at room temperature in darkness. An amount of 5 mg of protein sample was digested with trypsin, and label-free immune-affinity enrichment was performed using di-Gly-Lys antibody (Cell Signaling Technology), which is specifically recognizes the di-Gly-Lys remnant of ubiquitinated peptides (Udeshi et al., 2013). The protein samples were diluted by adding an appropriate volume of water to a urea concentration of <2 M. Trypsin was added at a 1:50 (trypsin: protein) mass ratio for digestion overnight. Digested peptides were desalted using a C18 SPE column (Waters), and freeze drying. Digested peptides were dissolved with precooled IAP buffer (50 mM MOPS/NaOH pH 7.2, 10 mM Na2HPO4, 50 mM NaCl), and then incubated with prewashed Anti-K-ε-GG antibody beads (PTM Biolabs) at 4°C overnight with gentle shaking. The beads were washed 3 times with 1 ml precooled IAP buffer and twice times with 1 ml precooled ddH2O. The bound peptides were eluted from the di-Gly-Lys antibody beads with 0.1% trifluoroacetic acid at room temperature twice for 10 min. The two elute fractions were combined, and desalted with C18 STAGE Tips (Thermo Fisher), then vacuum-dried, and thawed with 0.1% formic acid (Rappsilber et al., 2007). Three biological replicates were performed.
TMT labeling
The labeling was done following manufacturer's protocol. One hundred micro grams of digested peptides were labeled with TMT kit (Thermo Fisher Scientific, USA). A 6-plex TMT-126,−127,−128,−129,−130, and−131 were used to label protein samples of DK80 and Δcpubi4, with three independent biological replicates respectively. Labeled peptides were mixed at equal ratios and subjected to high-pH reverse-phase HPLC using an Agilent 300 Extend C18 column (5 μm particles, 4.6 mm i.d., 250 mm length) for fractionation. Fractions were collected with a gradient of 2–60% acetonitrile, and dried by vacuum centrifugation, then thawed with 0.1% formic acid for further LC-MS/MS analysis.
LC-MS/MS analysis
The peptides were analyzed using LC-MS/MS according to previously described protocols (Li et al., 2015). Briefly, peptides passed through a reverse-phase analytical column were loaded into the reverse-phase precolumn (EASY column SC001; Thermo Scientific, USA) that was connected to a reverse-phase column (EASY column SC200; Thermo Scientific, USA), separated with a linear gradient of solvent B (0.1% formic acid in 80% acetonitrile) at a constant flow rate of 300 nL/min on an EASY-nLC 1000 UPLC system. The separated peptides were analyzed with the Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, USA). Intact peptides were detected on the Orbitrap at a resolution of 70,000. Peptides of the top 20 most abundant precursor ions from the survey scan (300–1,800 m/z) were selected for HCD fragmentation. The ion fragments were detected on the Orbitrap at a resolution of 17,500. Determination of the target value was based on predictive automatic gain control.
Database search
For characterization of ubiquitinated peptides, the MS data were processed using MaxQuant software (version 1.3.0.5) and searched against the C. parasitica v2 database. The enzymatic cleavage rule was trypsin allowing a maximum of four missed cleavage sites. First, the search range was set at 5 ppm for precursor ions, and the main search range was set at 6 ppm and 0.02 D for fragment ions. Carbamidomethylation on cysteines was defined as a fixed modification. Oxidation on methionine, acetylation on protein N-terminal and ubiquitination on Lysine were defined as variable modifications for database searching. The cutoff for the global false discovery rate for peptides was set at 0.01. The intensity of the monoisotopic parent ion (precursor) was used to quantify each peptide (peptide intensity from MaxQuant software). Quantification was based on spectral characteristics (retention time, m/z ratio, and peak intensity) determined by comparing the direct mass spectrometric signal intensities for given peptides.
For characterization of proteomic peptides, the RAW data were processed using Mascot software (version 2.2) and Proteome Discoverer (version 1.4) and searched against the C. parasitica v2 database. The enzymatic cleavage rule was trypsin allowing a maximum of two missed cleavage sites. The mass tolerance was set to 20 ppm for the peptides and 0.1 Da for the fragment. Carbamidomethyl on Cys, TMT-6 plex on N-terminal, and TMT-6 plex on Lys were specified as fixed modifications, and oxidation on Met and TMT-6 plex on Tyr was specified as a variable modification. A decoy database search for determining false discovery rate (FDR) was set for a maximum of 1%. The protein ratios were calculated as the median of unique peptides of the protein.
Biological assays
Temperature stress assays
Temperature stress assays were done by keeping the 3-days-old fungal cultures on PDA at 4°C for 60 days or at 37°C for 7 days. After the treatment, the strains were re-inoculated onto PDA at 25°C to examine the survival rate. Three replicates were executed.
Virulence assays
Virulence assays were performed on stems of Chinese chestnut with three replicates per fungal strain as previously described (Yao et al., 2013). Fungal strains were cultured on PDA plates for 5 days to allow the hyphal colony to grow to about 3–5 cm in diameter. For inoculation, the bark in the stem was removed with a 5-mm cork borer (about 3 mm deep), and a 5-mm plug with fungal mycelium from the PDA plate was inserted into the shallow hole. Inoculated stems were kept at room temperature in a plastic bag to maintain moisture. Canker statistical analysis was performed with the Proc GLM procedure SAS (version 8.0), and the type I error rate was set at 0.05.
Bioinformatics
The protein sequence of yeast UBI4 (NP_013061) was used to BLAST C. parasitica genome1 for searching the ubiquitin coding gene. The protein conserved domain analyses were conducted using the NCBI database2. Statics were based on three independent experiments, and Student's test was used to compare the difference of means between two strains (Wang et al., 2016).
The Kyoto Encyclopedia of Genes and Genomes (KEGG) online tool KAAS was used to annotate protein pathways (Kanehisa and Goto, 2000). The annotated proteins were mapped on the KEGG pathway using the KEGG Mapper. KOBAS software was used to test the statistical enrichment of the proteins in KEGG pathways (Wu et al., 2006).
Results
Identification of ubiquitin-coding genes in C. parasitica
Ubiquitin is highly conserved in amino acids sequence (Ozkaynak et al., 1987). A secreted protein spot was identified as ubiquitin with increased accumulation in CHV1-EP713 infected strain in our previous work (Wang et al., 2016). To identify ubiquitin gene candidates, we used the 76 aa protein sequence of yeast ubiquitin protein (NP_013061) to blast against EP155 genome. Three ubiquitin motif-containing genes were identified in C. parasitica, designated as cpubi1 (scaffold_3:3613558-3614685), cpubi3 (scaffold_6:1195287-1196660), and cpubi4 (scaffold_8:305148-306130). Analysis of conserved domain with CD-search Tool at NCBI2 revealed that CPUBI1 contains a UBQ superfamily motif in position 1–76 and a ribosomal L40 family motif in position 78–100; CPUBI3 contains a UBQ superfamily motif in position 1–76 and a ribosomal S27 family motif in position 103–147; CPUBI4 contains three UBQ superfamily in positions 1–76, 77–152, and 153–228, respectively (Figure 1A). The cpubi1 and cpubi3 encode hybrid proteins in which ubiquitin is fused to other amino acid sequences. The third ubiquitin gene, cpubi4 encodes a polyubiquitin with three consecutive head-to-tail ubiquitin monomers. To investigate whether the transcription of ubiquitin genes are affected by the infection of CHV1-EP713, the wild-type strains EP155 and the hypovirus infected strain EP155/CHV1-EP713 were examined for the transcript accumulation of the three ubiquitin genes by quantitative real-time RT-PCR. In 7-days-old cultures on PDA, expression of cpubi1 and cpubi3 were not significantly altered upon viral infection. By contrast, cpubi4 expression was up-regulated by about 80% in hypovirus infected strain (Figure 1B).
Figure 1.

Schematic representation of ubiquitin coding genes in C. parasitica and quantification of transcript accumulation levels of ubiquitin coding genes in response to infection by hypovirus CHV1-EP713. (A) The UBQ domains were identified by searching NCBI's Conserved Domains database and are indicated by green boxes; Ribosomal L40e domain in CPUBI1 is indicated by the red box; Ribosomal S27 domain in CPUBI3 is indicated by the blue box. Solid lines represent full-length proteins. The ranges of amino acids included in the match are indicated next to and below the domain name. (B) Strains were cultured on PDA for 7 days, and total RNA was extracted. The expression of cpubi1, cpubi3, and cpubi4 transcripts was measured by quantitative real-time RT-PCR (2−ΔΔCT method). The relative transcript level was calibrated using 18S rRNA as reference, and the transcript levels of EP155 were set to a value of one (Lin et al., 2007). Three replicates were performed.
Knockout of cpubi4 affects C. parasitica phenotypes
To investigate the function of cpubi4, we designed a gene replacement cassette with hygromycin B resistance as a selection marker to disrupt cpubi4 via homologous recombination (Figure 2A). Three randomly selected single-spore derived transformants, Δcpubi4-1, Δcpubi4-2, and Δcpubi4-3, were further confirmed by Southern blot analysis (Figure 2B). The growth of Δcpubi4 mutants was slower, and the aerial hyphae were significantly less than the wild-type strain and the parental strain DK80. The conidiation level was also drastically reduced. The altered phenotype of the mutants could be fully restored by re-introducing a wild-type copy of cpubi4 (Figure 2C). More importantly, Δcpubi4 lost the ability to incite a canker on the chestnut stem, similar to the hypovirulent strain EP155/CHV1-EP713. Again, the virulence could be restored to the wild-type level after a wild-type copy of cpubi4 was reintroduced into the Δcpubi4 mutant (Figure 3).
Figure 2.
Southern blot analysis and phenotypes of C. parasitica cpubi4 gene disruption mutants. (A) The genomic DNA containing the cpubi4 coding region composed of one exon is indicated by the white box. (A) cpubi4 gene replacement construct was generated by insertion of the 3.6 kb hygromycin (hph) resistant cassette using homologous recombination strategy (see section Materials and Methods). Probes fragment on the right arm and hph were used in the Southern blot analysis to distinguish the wild-type strain and cpubi4 null mutants. (B) Southern analysis of cpubi4 disruptant strains. DNA samples were digested with HindIII/NcoI (left), or independently with SmaI (right), and blotted using the probe A, and probe B, respectively. Fragments sizes are indicated in the figure margins. (C) Colony morphologies of DK80, Δcpubi4, and the cpubi4 complementary strain Δcpubi4-com on PDA plates.
Figure 3.
Virulence assay on chestnut stems. (A) Cankers caused by the tested strains. The wild-type strain EP155, starting strain DK80, hypovirus-infected EP155/CHV1-EP713, cpubi4 disrupts strain Δcpubi4, and cpubi4-complemented strain Δcpubi4-com were inoculated onto Chinese chestnut (Castanea mollissima Blume) stems. The inoculated stems were kept at 24°C. The resulting cankers were photographed and measured at day 21 post-inoculation. (B) Statistical analysis of virulence of the cpubi4 null mutants. The assays were with three duplicates for each strain.
cpubi4 is essential for temperature stress adaption but does not affect hypovirus dsRNA accumulation in C. parasitica
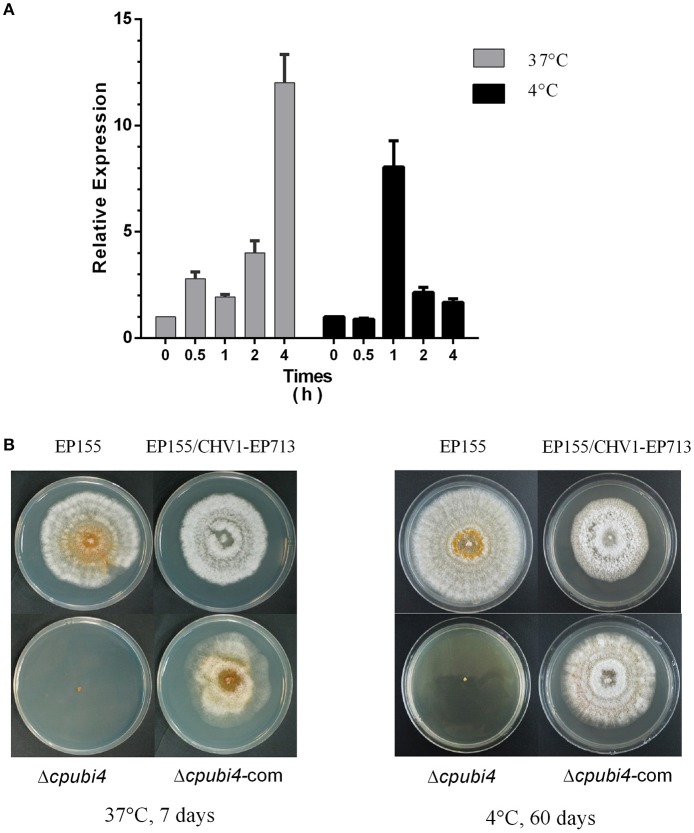
In S. cerevisiae, UBI4 expression was increased in response to elevated temperatures, and inactivation of UBI4 resulted in sensitivity to high-temperature stress (Finley et al., 1987). We reasoned that the cpubi4 gene might contribute to the temperature stress adaption in C. parasitica. To test this hypothesis, we used real-time RT-PCR to analyze the transcript accumulation of cpubi4 gene in the wild-type strain EP155 after heat and cold treatments. EP155 was first cultured on PDA plates at 25°C for 3 days, then kept at 37 and 4°C, respectively. After treatments, the strains were harvested, and total RNA was isolated for real-time RT-PCR. It was found that the expression of cpubi4 was upregulated for more than 2-fold after 37°C treatment for 0.5 h, and increased to more than 10-fold after for 4 h. Similarly, the expression of cpubi4 was also increased by 5-fold after cold treatment at 4°C for 1 h (Figure 4A). To further confirm the roles of cpubi4 in temperature adaption, we tested the viability of wild-type strain EP155, virus-harboring EP155/CHV1-EP713, Δcpubi4, and Δcpubi4-com under temperature stress conditions. The Δcpubi4 failed to grow after treatment at 37°C for 1 week and at 4°C for 2 months, respectively. In contrast, EP155, EP155/CHV1-EP713, and Δcpubi4-com were viable under the same conditions (Figure 4B). Collectively, our data indicate that polyubiquitin gene cpubi4 is required for temperature adaptation in C. parasitica.
Figure 4.
Response of C. parasitica strains to temperature stress. (A) Quantification of cpubi4 transcript. EP155 was first cultured on PDA plates at 25°C for 3 days, and then transferred to 37°C or 4°C incubators for certain periods of time. RNA quantification was done as above (see legend to Figure 1). cpubi4 transcript level at time zero was set at a value of 1.0. (B) Viability and morphology of the strains. After being kept at 37°C for 1 week or 4°C for 2 months, the strains were inoculated onto PDA plate at 25°C. Photographs were taken at day 7 post-inoculation.
To verify if the cpubi4 gene would influence the accumulation of the hypovirus, we introduced CHV1-EP713 into Δcpubi4 via anastomosis by paring hypovirus-infected strain EP155/CHV1-EP713 with Δcpubi4. Infection by the hypovirus resulted in the loss of orange pigment in Δcpubi4, but the mutant seemed to support viral replication almost equally well as the wild-type strain (Supplementary Figure 1).
cpubi4 contributes significantly to the global ubiquitination in C. parasitica
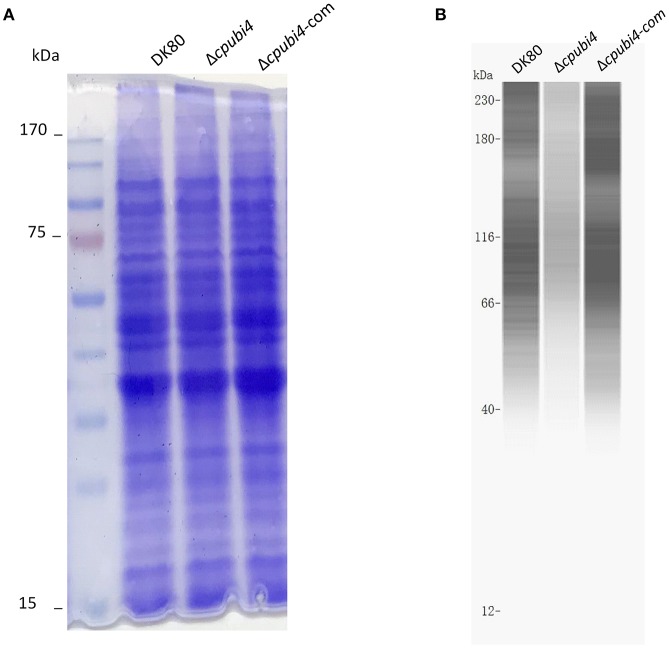
With the same amount of mycelia (0.8 g), Δcpubi4 yielded 33.9% more total protein than its parental strain Dk80 (6.87 vs. 5.13 mg, paired Student's test p = 0.009). To evaluate the global effects of polyubiquitin on proteins ubiquitination, total proteins from the wild-type and cpubi4-deleted strains were analyzed by Western blot with ubiquitin antibody. With the same amount of protein loaded, the ubiquitination proteins in the Δcpubi4 were strikingly less than its parental DK80 (Figure 5). We also compared the global ubiquitination of the wild-type strain EP155 and the hypovirus-infected strain EP155/CHV1-EP713 and found that hypovirus infection led to slightly enhanced ubiquitination (Supplementary Figure 2).
Figure 5.
Evaluation of the ubiquitome of wild-type DK80 and Δcpubi4 strains. (A) SDS-PAGE. Loading amount was 30 μg of total protein extracted from 3-days old culture in EP. The gel was stained with Coomassie solution. (B) Immuno blot analysis on a Simple Western system with ubiquitin-specific antibody. An amount of 80 μg of total protein was loaded for each lane. Signals were stronger with the high molecular weight proteins, likely due to multi ubiquitination sites in the protein. Δcpubi4-com represents the wild-type cpubi4-complemented Δcpubi4 strain.
To quantify the ubiquitinated proteins in C. parasitica, a label-free quantitative strategy involving antibody-based affinity enrichment and high-resolution LC-MS/MS were employed for both DK80 and Δcpubi4 in parallel. In three independent experiments, a total of 2,684 di-Gly-Lys ubiquitination sites that mapped to 1195 unique proteins were identified in DK80 and Δcpubi4 (Supplementary Table 2). Between 1 and 24 putative ubiquitination sites, with the majority of one site (57.9%) and two sites (18.8%), were found in proteins from DK80 (Figure 6A). The most ubiquitinated proteins were hypothetical proteins (id: 330586, 331295) with 24 and 22 lysine ubiquitination sites, and the 70 kDa heat shock protein (HSP70) with 17 lysine ubiquitination sites. Majority of the ubiquitinated proteins were enriched in pathways of key cellular processes, including metabolism of protein, fatty acid, and sugar, as well as second metabolites (Figure 6B).
Figure 6.
Characteristics of ubiquitylome of C. parasitica. (A) Distribution of ubiquitination sites identified per protein. (B) KEGG pathway enrichment of proteins from DK80. Significance was ranked according to the p-value. (C) Heat-map of the hierarchical clusters of the 429 affected ubiquitinated peptides (fold change > 2, p-value < 0.05) in the mutant stains Δcpubi4. Peptides showing similar expression patterns have been clustered. For details about peptides ubiquitination, see the Supplementary Table 2. (D) KEGG pathway enrichment of proteins with up-regulated (red) and down-regulated (blue) ubiquitination sites. Significance was ranked according to the p-value.
To identify protein substrates of the polyubiquitin, we compared the abundance of di-Gly-Lys-containing peptides between Δcpubi4 and DK80 (Supplementary Table 3). A total of 428 proteins with ubiquitination level change >2-fold (p < 0.05) were identified, among which 50 peptides in 45 proteins were increased, and 378 peptides in 236 proteins decreased in ubiquitination (Figure 6C). Among these proteins, some are essential metabolic enzymes, including enolase, peptidyl-prolyl cis-trans isomerase (CYP1); mitogen-activated protein kinases, ribosome, G-protein, ubiquitin-related protein; and stress responding such as heat shock proteins (Table 1). KEGG enrichment showed that proteins with increased ubiquitination function mainly in protein synthesis and processing and endocytosis, while proteins with decreased ubiquitination function in biosynthesis of amino acid, steroid, antibiotics, and secondary metabolites; and in carbon metabolism, glycolysis/gluconeogenesis, and pentose phosphate pathway (Figure 6D).
Table 1.
Examples of proteins with significant changes in ubiquitination.
| Proteins ID | Proteins description | Up-regulated ubiquitination sitesa | Down-regulated ubiquitination sitesb | Positions within proteinsc |
|---|---|---|---|---|
| HEAT SHOCK PROTEIN | ||||
| 227872 | Heat shock protein sti1 homolog | – | 1 | 152 |
| 331015 | Heat shock protein SSB1 | – | 2 | 326,967 |
| 253316 | Heat shock protein hsp88 | 1 | – | 755 |
| 245307 | Heat shock protein 90 homolog | – | 4 | 86,235,384,567 |
| 260193 | Heat shock 70 kDa protein 12A | – | 1 | 732 |
| 346270 | Heat shock 70 kDa protein | 3 | 5 | 323,326,450,499,511,523,556,560 |
| MAPK PATHWAY | ||||
| 262898 | 1,3-beta-glucan synthase component FKS1 | – | 1 | 951 |
| 231325 | Mitogen-activated protein kinase | – | 1 | 279 |
| 277770 | Peptidyl-prolyl cis-trans isomerase, mitochondrial | – | 2 | 129,211 |
| UBIQUITIN-MEDIATED PROTEOLYSIS | ||||
| 108221 | E3 ubiquitin-protein ligase RSP5 | – | 3 | 87,88,780 |
| 258058 | Ubiquitin-conjugating enzyme E2 N | – | 2 | 81,91 |
| 328806 | Ubiquitin-activating enzyme E1 1 | – | 2 | 568,569 |
| 245142 | Ubiquitin-conjugating enzyme E2-34 kDa | – | 1 | 15 |
| 334719 | NEDD8-conjugating enzyme UBC12 | – | 1 | 98 |
| 331066 | Ubiquitin-conjugating enzyme E2-20 kDa | 1 | – | 16 |
| 328046 | Ubiquitin-conjugating enzyme E2 14 | – | 1 | 94 |
| UBIQUITIN | ||||
| 103787 | Ubiquitin-40S ribosomal protein S27a | – | 4 | 6,29,48,63 |
| 331210 | Ubiquitin-like protein pmt3/smt3 | – | 1 | 120 |
| 263247 | Ubiquitin-like protein 1 | 1 | – | 45 |
| PROTEASOME | ||||
| 334850 | Probable proteasome subunit beta type-4 | – | 1 | 221 |
| 333003 | Probable proteasome subunit alpha type-7 | 1 | – | 64 |
| 263113 | 26S proteasome regulatory subunit RPN10 | – | 1 | 133 |
| 107783 | 26S proteasome non-ATPase regulatory subunit 1 | – | 1 | 1059 |
| 220665 | 26S protease regulatory subunit 8 homolog | – | 1 | 20 |
| 220595 | Probable 26S protease subunit rpt4 | – | 2 | 26,198 |
| G-PROTEIN | ||||
| 325103 | Guanine nucleotide-binding protein subunit gamma | – | 1 | 84 |
| 103579 | Guanine nucleotide-binding protein subunit beta-like protein | – | 1 | 280 |
| RIBOSOME | ||||
| 223537 | 60S ribosomal protein L9-B | – | 1 | 84 |
| 105502 | 60S ribosomal protein L4-A | – | 1 | 174 |
| 333723 | 60S ribosomal protein L36 | – | 1 | 50 |
| 103417 | 60S ribosomal protein L30 | – | 1 | 86 |
| 271891 | 60S ribosomal protein L17 | 1 | 1 | 52,96 |
| 281725 | 60S ribosomal protein L13 | – | 1 | 53 |
| 283348 | 60S ribosomal protein L12 | – | 1 | 48 |
| 280826 | 60S ribosomal protein L11 | – | 1 | 35 |
| 222795 | 40S ribosomal protein S3 | – | 4 | 78,168,200,205 |
| 103717 | 40S ribosomal protein S20 | – | 5 | 8,15,33,45,50 |
| 103050 | 40S ribosomal protein S2 | – | 1 | 39 |
| 242837 | 40S ribosomal protein S18 | 1 | – | 53 |
| 273821 | 40S ribosomal protein S15 | 1 | – | 85 |
| 329021 | 40S ribosomal protein S17 | – | 2 | 544,634 |
| 224177 | 40S ribosomal protein S13 | – | 3 | 27,39,43 |
| 271207 | 40S ribosomal protein S12 | 1 | 1 | 56,94 |
| 101666 | 40S ribosomal protein S10-B | – | 2 | 93,137 |
| 103787 | Ubiquitin-40S ribosomal protein S27a | – | 4 | 6,29,48,63 |
| GLYCOLYSIS/GLUCONEOGENESIS | ||||
| 104720 | Fructose-bisphosphate aldolase | – | 1 | 115 |
| 252290 | Pyruvate decarboxylase | – | 2 | 40,523 |
| 258754 | Pyruvate kinase | – | 8 | 43,99,148,246,268,426,483,516 |
| 106275 | Alcohol dehydrogenase 1 | – | 1 | 31 |
| 328301 | Phosphoglycerate kinase | – | 7 | 15,17,83,91,150,156,295 |
| 249839 | ATP-dependent 6-phosphofructokinase | – | 3 | 352,638,690 |
| 101684 | Glyceraldehyde-3-phosphate dehydrogenase | – | 4 | 185,193,214,332 |
| 221435 | Alcohol dehydrogenase | – | 1 | 211 |
| 233295 | Triosephosphate isomerase | – | 3 | 144,175,216 |
| 103155 | Enolase | – | 7 | 60,64,79,96,120,194,347 |
A portion of proteins with significant changes in ubiquitination due to lack of cpubi4.
Number of up-regulated ubiquitination sites from the protein.
Number of down-regulated ubiquitination sites from the protein.
The position of ubiquitination site in the protein.
Deletion of cpubi4 alters protein expression patterns
To see if deletion of cpubi4 would change the abundance of specific proteins, we quantified the proteomes of Δcpubi4 and DK80 using the same amount of total protein with three biological replicates. Among 4,767 proteins quantified, the vast majority of proteins remained basically unchanged (< 1.5-fold; Supplementary Table 4). Volcano plotting of proteomes of both strains identified a subset of proteins with fold-change ≥1.5 and p-value < 0.05 (Figure 7). As seen in Table 2, the up-regulated proteins contain many ubiquitin-conjugating enzymes and ubiquitin-protein ligases. Other up-regulated proteins include glutathione transporter, methyltransferases, serine/threonine- and mitogen-activated protein kinases, mitochondrial activity proteins (cytochrome P450/NADPH-P450), autophagy-related protein, and apoptosis-inducing factor 2. Among the down-regulated proteins, are many ribosomal proteins, belonging to 30S, 40S, and 60S. Besides, mitochondrion-related proteins, e.g., mitochondrial ribosomal protein, mitochondrial import inner membrane translocase subunits, and NADH-ubiquinone oxidoreductase subunit were also down-regulated (Table 2, Supplementary Table 4 for detail information). Accumulation of ubiquitin-conjugating enzymes was a sign of speeding up in ubiquitin-recycling process and reduction of essential mitochondrial proteins would jeopardize the normal function of a mitochondrion.
Figure 7.

Volcano plot of the proteomes of DK80 and Δcpubi4. Black dots represent the total proteins identified from both DK80 and Δcpubi4, red dots represent the differentially expressed proteins in Δcpubi4 with fold-change greater than 1.5 and p < 0.05. A total of 142 proteins exhibited reduced ubiquitination upon deletion of cpubi4, indicating that they are the substrates of CPUBI4.
Table 2.
Examples of proteins up-regulated and down-regulated by deletion of cpubi4a.
| Protein IDb | Proteins description | Average ratioc | t test p-valued |
|---|---|---|---|
| UP-REGULATED PROTEINS | |||
| Ubiquitin-Conjugating Enzyme | |||
| 255535 | Ubiquitin-conjugating enzyme E2Q-like protein CG4502 | 1.737895085 | 0.0042126 |
| 331066 | Ubiquitin-conjugating enzyme E2-20 kDa | 1.7595107 | 0.00755426 |
| 232237 | Ubiquitin-conjugating enzyme E2 J1 | 1.611945028 | 0.000112666 |
| 243167 | Ubiquitin-conjugating enzyme E2 8 | 1.736645576 | 0.00335787 |
| 104034 | Ubiquitin-conjugating enzyme E2 6 | 2.305704848 | 0.00129594 |
| 252753 | Ubiquitin-conjugating enzyme E2 2 | 2.168058757 | 0.000809489 |
| 328046 | Ubiquitin-conjugating enzyme E2 14 | 1.648350773 | 0.0171935 |
| Ubiquitin-Protein Ligase | |||
| 215338 | E3 ubiquitin-protein ligase ubr1 | 1.56805089 | 0.00254144 |
| 274970 | E3 ubiquitin-protein ligase itt1 | 1.567666682 | 0.00697192 |
| 341211 | E3 ubiquitin-protein ligase dbl4 | 1.75861228 | 0.00343743 |
| 266035 | Probable HECT-type ubiquitin ligase-interacting protein creD | 1.609215843 | 0.00026883 |
| 274707 | STIP1 homology and U box-containing protein 1 | 1.57155688 | 0.00569711 |
| 335876 | PHD and RING finger domain-containing protein C126.07c | 2.100016891 | 0.00467357 |
| 255228 | F-box protein YLR352W | 1.68577 | 0.000456609 |
| Transport Activity | |||
| 223105 | Vesicle-associated membrane protein 7B | 1.667315817 | 0.000254997 |
| 252500 | Transmembrane protein 184 homolog C30D11.06c | 1.50120028 | 0.00144419 |
| 220118 | UDP-galactose transporter homolog 1 | 1.702008993 | 0.000144171 |
| 325360 | Store-operated calcium entry-associated regulatory factor | 1.651085956 | 0.0013294 |
| 109037 | Probable ran guanine nucleotide release factor | 1.670196023 | 7.16E-06 |
| Transferase Activity | |||
| 345836 | Transaldolase | 1.732496465 | 0.00370331 |
| 245522 | Sterol 24-C-methyltransferase | 1.758109157 | 0.00752994 |
| 330996 | Probable S-adenosylmethionine-dependent methyltransferase CRG1 | 2.044572993 | 0.000850226 |
| 109461 | Lovastatin diketide synthase LovF | 1.501137033 | 0.00269576 |
| 351527 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase | 1.501428385 | 0.00539262 |
| 219562 | Histone-lysine N-methyltransferase, H3 lysine-79 specific | 1.730819125 | 0.0011541 |
| Kinase | |||
| 261928 | Mitogen-activated protein kinase 20 | 1.915753416 | 0.000893821 |
| 66486 | Serine/threonine-protein kinase ATG1 | 1.561655632 | 0.00359346 |
| Others | |||
| 229417 | Bifunctional cytochrome P450/NADPH–P450 reductase | 1.746372739 | 0.00361005 |
| 219948 | Apoptosis-inducing factor 2 | 1.839557188 | 0.00251934 |
| 277081 | Autophagy-related protein 8 | 1.684164422 | 0.00647872 |
| DOWN-REGULATED PROTEINS | |||
| Ribosomal-Related Protein | |||
| 332673 | 60S ribosomal protein L35 | 0.662046014 | 0.0036187 |
| 333723 | 60S ribosomal protein L36 | 0.65764289 | 0.0191438 |
| 102781 | 60S ribosomal protein L14 | 0.638642957 | 0.0006562 |
| 103140 | 60S acidic ribosomal protein P0 | 0.632333414 | 0.0134515 |
| 244514 | 40S ribosomal protein S29 | 0.618094491 | 0.0006993 |
| 224760 | 60S acidic ribosomal protein P1 | 0.6063268 | 0.0003723 |
| 100780 | 60S ribosomal protein L7 | 0.585235713 | 0.0011423 |
| 234544 | 40S ribosomal protein S19 | 0.577554815 | 0.0067663 |
| 275342 | Ubiquitin-60S ribosomal protein L40 | 0.502721352 | 0.003331 |
| 358141 | 30S ribosomal protein 2, chloroplastic | 0.552951761 | 0.0121389 |
| 103787 | Ubiquitin-40S ribosomal protein S27a | 0.571426883 | 0.0005912 |
| Mitochondrial Protein | |||
| 235111 | Mitochondrial import inner membrane translocase subunit TIM9 | 0.655945111 | 0.0020106 |
| 225024 | Mitochondrial 37S ribosomal protein S27 | 0.643542957 | 0.0014632 |
| 328740 | Altered inheritance of mitochondria protein 41, mitochondrial | 0.592556243 | 0.0001034 |
| 327534 | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | 0.591234659 | 0.0002282 |
| 357354 | Respiratory supercomplex factor 1, mitochondrial | 0.588303102 | 0.0101486 |
| 220172 | Probable proline dehydrogenase, mitochondrial | 0.626096845 | 0.0024221 |
| 221383 | Probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial | 0.58799909 | 0.0044118 |
| 224403 | ATP synthase subunit f, mitochondrial | 0.586069963 | 0.0040819 |
| 107757 | ATP synthase subunit 4, mitochondrial | 0.53900636 | 4.32E-05 |
| 245314 | Mitochondrial import inner membrane translocase subunit TIM13 | 0.532395384 | 0.0013224 |
| 329791 | Cytochrome c oxidase polypeptide 5, mitochondrial | 0.644503601 | 0.0043218 |
| 334855 | Cytochrome c oxidase subunit 6, mitochondrial | 0.643009812 | 0.0001996 |
| 238062 | 2-methylcitrate dehydratase, mitochondrial | 0.644348666 | 0.0023069 |
| 220455 | 2-oxoisovalerate dehydrogenase subunit beta, mitochondrial | 0.644098628 | 0.0105008 |
| NADH Related Protein | |||
| 261324 | NADH-cytochrome b5 reductase 2 | 0.650784426 | 0.0026476 |
| 224496 | NADH dehydrogenase | 0.638523013 | 0.000362 |
| 322879 | NADH-ubiquinone oxidoreductase 10.5 kDa subunit | 0.628958467 | 0.0001824 |
See Supplementary Table 4 for the complete list.
Accession number from Cryp80honectria parasitica database v2.0.
Parental stain DK80 was set as reference. The average ratio (Δcpubi4/DK80) were from three independent experimental data.
Scored using Student's t-test, n = 3, It could be used to see if they are significantly different from each other by student's test.
Cross-checking of the differentially accumulated proteins with the ubiquitome identifies 79 proteins that show weak correlation between protein abundance and number of ubiquitination site per protein: among the up-regulated proteins in Δcpubi4, 85.7% of them contain 1 or 2 ubiquitination sites, no protein contains more than 5 ubiquitination sites; while among the down-regulated proteins, 65.9% contain 1 or 2 ubiquitination sites, but 25% contain 5 or more ubiquitination sites (Table 3). However, since proteins identified from the ubiquitome are far fewer than those from the proteomes (about 1/4), the exact correlation between protein degradation profile and the ubiquitination sites in a protein remains to be verified in the future.
Table 3.
Number of ubiquitination site in differentially accumulated proteinsa.
| Protein ID | Description | Number of ubiquitination sitec |
|---|---|---|
| UP-REGULATED PROTEINSb | ||
| 246699 | – | 4 |
| 340305 | – | 3 |
| 331066 | Ubiquitin-conjugating enzyme E2-20 kDa | 3 |
| 245522 | Sterol 24-C-methyltransferase | 3 |
| 277081 | Autophagy-related protein 8 | 3 |
| 280355 | 30 kDa heat shock protein | 2 |
| 349821 | Uncharacterized ATP-dependent helicase C23E6.02 | 2 |
| 341211 | E3 ubiquitin-protein ligase dbl4 | 2 |
| 329877 | Protein SPT23 | 2 |
| 282187 | ATPase family AAA domain-containing protein 3B | 2 |
| 328046 | Ubiquitin-conjugating enzyme E2 14 | 2 |
| 108749 | L-ornithine N(5)-monooxygenase | 2 |
| 266035 | Probable HECT-type ubiquitin ligase-interacting protein creD | 2 |
| 269206 | Transcription elongation factor 1 | 2 |
| 325438 | Carboxypeptidase Y homolog A | 2 |
| 326539 | – | 1 |
| 344892 | – | 1 |
| 329908 | – | 1 |
| 103600 | – | 1 |
| 104034 | Ubiquitin-conjugating enzyme E2 6 | 1 |
| 252753 | Ubiquitin-conjugating enzyme E2 2 | 1 |
| 75354 | SWIRM domain-containing protein YOR338W | 1 |
| 261928 | Mitogen-activated protein kinase 20 | 1 |
| 271551 | Aldehyde dehydrogenase | 1 |
| 267009 | Cell division control protein 4 | 1 |
| 255535 | Ubiquitin-conjugating enzyme E2Q-like protein CG4502 | 1 |
| 243167 | Ubiquitin-conjugating enzyme E2 8 | 1 |
| 107003 | – | 1 |
| 219053 | Protein ssh4 | 1 |
| 223105 | Vesicle-associated membrane protein 7B | 1 |
| 333459 | – | 1 |
| 332318 | – | 1 |
| 248650 | NAD-dependent protein deacetylase SRT1 | 1 |
| 252500 | Transmembrane protein 184 homolog C30D11.06c | 1 |
| 109461 | Lovastatin diketide synthase LovF | 1 |
| DOWN-REGULATED PROTEINS | ||
| 330586 | –d | 33 |
| 67838 | – | 16 |
| 337355 | 14-3-3 protein homolog | 10 |
| 226763 | Probable 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase | 9 |
| 103787 | Ubiquitin-40S ribosomal protein S27a | 8 |
| 104921 | – | 7 |
| 101014 | – | 7 |
| 267236 | Indoleamine 2,3-dioxygenase 1 | 7 |
| 324752 | – | 6 |
| 253168 | Glycogen phosphorylase | 5 |
| 275342 | Ubiquitin-60S ribosomal protein L40 | 5 |
| 332673 | 60S ribosomal protein L35 | 4 |
| 332324 | – | 4 |
| 105888 | – | 3 |
| 102597 | Uncharacterized protein C9E9.04 | 3 |
| 103866 | – | 2 |
| 103140 | 60S acidic ribosomal protein P0 | 2 |
| 105467 | Farnesyl pyrophosphate synthase | 2 |
| 263597 | Putative HC-toxin efflux carrier TOXA | 2 |
| 332325 | – | 2 |
| 103157 | Putative daunorubicin C-13 ketoreductase DnrU | 2 |
| 328973 | – | 2 |
| 245581 | Lipid phosphate phosphatase 1 | 2 |
| 332047 | Uncharacterized protein MT3735 | 2 |
| 333723 | 60S ribosomal protein L36 | 1 |
| 325606 | – | 1 |
| 102781 | 60S ribosomal protein L14 | 1 |
| 245734 | Levodione reductase | 1 |
| 263730 | – | 1 |
| 335178 | – | 1 |
| 102161 | – | 1 |
| 244514 | 40S ribosomal protein S29 | 1 |
| 100780 | 60S ribosomal protein L7 | 1 |
| 234544 | 40S ribosomal protein S19 | 1 |
| 250085 | Elongation factor 1-gamma-A | 1 |
| 333867 | Homoserine O-acetyltransferase | 1 |
| 251895 | Lysine acetyltransferase | 1 |
| 265885 | Versicolorin B synthase | 1 |
| 253380 | – | 1 |
| 105308 | Uncharacterized protein C32A11.02c | 1 |
| 326707 | Tetracycline resistance protein from transposon Tn4351/Tn4400 | 1 |
| 255109 | 2,3-dimethylmalate lyase | 1 |
| 255822 | Uncharacterized oxidoreductase C162.03 | 1 |
| 343596 | Uncharacterized protein MT1298 | 1 |
Discussion
Hypovirus infection specifically up-regulates cpubi4 expression
There are three ubiquitin-encoding genes in C. parasitica, but only cpubi4 responds to the infection of a hypovirus at transcription level (Figure 1). This observation provides a line of evidence that hypovirus perturbs the host protein ubiquitination through regulation of transcription of a specific ubiquitin-encoding gene. A similar observation was reported in S. cerevisiae in which UBI4 was the only one among four ubiquitin-encoding genes to respond to stresses (Fraser et al., 1991), and in pea plant in which a heat-inducible polyubiquitin gene was induced by the infection of pea seed-borne mosaic virus (Aranda et al., 1996).
It has been reported that viruses hijack the host ubiquitin system to enhance viral replication, through modification of host ubiquitination to degrade the cellular proteins, or recruiting ubiquitin to modify viral proteins (Randow and Lehner, 2009). To test if CPUBI4 is required for the hypovirus replication, we introduced CHV1-EP713 into a cpubi4-null mutant. However, deletion of cpubi4 did not seem to have a significant impact on the accumulation of the hypoviral dsRNA (Supplementary Figure 1), suggesting that the increase of cpubi4 expression is not for the replication of hypovirus, but a result of responding to the hypovirus infection. Since there are two other ubiquitin genes in C. parasitica genome, the involvement of ubiquitin in the replication of hypovirus remains to be clarified. On the other side, increased accumulation of cpubi4 transcript did not seem to lead to the significant increase in ubiquitination of the host proteins (Supplementary Figure 2), indicating that hypovirus infection does not alter ubiquitination pattern at a global basis. Since the interplay between viruses and ubiquitin system are complex (Calistri et al., 2014), how hypovirus bypasses or exploits the ubiquitin system still needs further investigation.
CPUBI4 is a major contributor to C. parasitica ubiquitination
It was reported that deletion of polyubiquitin gene Ubc in mouse embryonic fibroblasts resulted in a reduction of total ubiquitin by 40% (Ryu et al., 2007). Western blotting with ubiquitin and polyubiquitin-specific antibody revealed that the total ubiquitinated proteins were dramatically reduced in Δcpubi4 (Figure 5), and comparative ubiquitome analysis further showed that cpubi4 is the major player for ubiquitination in C. parasitica (Supplementary Table 2). The fact that total amount of protein in mycelia is higher in Δcpubi4 mutant than in the wild-type strain also suggests that lack of sufficient amount of ubiquitin may result in an elevated level of cellular protein. We assume that reduction in ubiquitination level in Δcpubi4 might slow down the protein degradation process, resulting in accumulation of proteins in the cells.
Mechanism of CPUBI4 regulation of development, stress adaptation, and virulence in C. parasitica
As a protein fate controller, ubiquitin modulates gene function by control of gene product quantity. Ubiquitin homeostasis, or the maintenance of free ubiquitin above the certain threshold level, is important for cellular function and survival under normal or stress conditions (Park and Ryu, 2014). In fungi, the polyubiquitin-encoding gene is not indispensable but is required for development and adaption to environmental changes. For example, loss of UBI4 in S. cerevisiae tenders the fungus to be more sensitive to starvation, amino acid analogs, and high temperatures (Finley et al., 1987; Tanaka et al., 1988; Fraser et al., 1991). Deletion of UBI4 in C. albicans resulted in altered morphology, defects in cell cycle, and loss of virulence (Leach et al., 2011). In M. oryzae, the polyubiquitin deletion mutant exhibited defects in morphology and virulence (Oh et al., 2012). Likewise, cpubi4 gene was shown to be required for mycelial growth, sporulation, virulence, and temperature stress adaption (Figures 2–4).
Previous study had shown that metabolic enzymes were targets of ubiquitination in C. albicans (Leach et al., 2011) and in the current study, a similar conclusion was reached (Supplementary Table 3). It has been demonstrated that CYP1, MAPK2, MAPK1 are components of or related to the heterotrimeric G-protein signaling pathway and required for growth, pigmentation, conidiation, and virulence in C. parasitica (Park et al., 2004; Choi et al., 2005; Chen et al., 2011). Although slower growth in Δcpubi4 may lead to reduction in virulence and sporulation, alteration in ubiquitination level of key proteins may also contribute to virulence attenuation and lower level of sporulation in Δcpubi4, as it has been noticed that sporulation and virulence are regulated by different mechanisms in C. parasitica (Yao et al., 2013).
Heat shock proteins (Hsps) that function as molecular chaperones are crucial in adaptation to harsh conditions such as temperature stress for an organism (Richter et al., 2010). Several Hsps including Hsp70, Hsp90, STI1, and SSB1 were found to be at a significantly lower ubiquitination level in Δcpubi4 (Supplementary Table 3). It is speculated that change in ubiquitination of these heat shock proteins could account for the temperature stress sensitivity in Δcpubi4 (Figure 4). In this regard, polyubiquitin deletion has been shown to affect growth, morphogenesis, and virulence in C. albicans and Aspergillus fumigatus (Lorenz and Fink, 2001; Hwang et al., 2003; Bhabhra and Askew, 2005).
Another dimension that CPUBI4 may have an impact on is to regulate cellular protein level. As found in comparative proteome analysis, a small fraction of proteins (288 in 4767) changed in abundance due to lack of CPUBI4 (Supplementary Table 4). These include some important proteins that function in protein synthesis and degradation, enzymatic activity, mitochondrial activity, and energy generation (Table 3). Imbalanced accumulation of proteins in the cells may disturb the orchestra of cellular proteins.
In conclusion, we demonstrated that cpubi4 is the major ubiquitin gene that contributes significantly to the global ubiquitination in the chestnut blight fungus and this gene plays crucial roles in the growth, conidial development, temperature adaptation, and virulence in C. parasitica. Knowledge gained through this study provides new insights into the mechanism of cpubi4 in regulation of multiple traits of C. parasitica.
Author contributions
QC, JW, RL, and BC conceived and designed the experiments. QC and YL performed the experiments. QC analyzed the data and wrote the manuscript draft. BC supervised the project and wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Pu Li (Pfomic bioinformation corp., Nanning, China) for assistance in data analysis.
Funding. This research was supported by a grant (No. 31370173) from the National Natural Science Foundation of China to BC.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01286/full#supplementary-material
References
- Anagnostakis S. L. (1982). Biological control of chestnut blight. Science 215, 466–471. 10.1126/science.215.4532.466 [DOI] [PubMed] [Google Scholar]
- Aranda M. A., Escaler M., Wang D., Maule A. J. (1996). Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl. Acad. Sci. U.S.A. 93, 15289–15293. 10.1073/pnas.93.26.15289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabhra R., Askew D. S. (2005). Thermotolerance and virulence of Aspergillus fumigatus: role of the fungal nucleolus. Med. Mycol. 43(Suppl. 1), S87–S93. 10.1080/13693780400029486 [DOI] [PubMed] [Google Scholar]
- Calistri A., Munegato D., Carli I., Parolin C., Palù G. (2014). The ubiquitin-conjugating system: multiple roles in viral replication and infection. Cells 3, 386–417. 10.3390/cells3020386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Choi G. H., Nuss D. L. (1994). Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264, 1762–1764. 10.1126/science.8209256 [DOI] [PubMed] [Google Scholar]
- Chen J. Q., Heldman M. R., Herrmann M. A., Kedei N., Woo W., Blumberg P. M., et al. (2013). Absolute quantitation of endogenous proteins with precision and accuracy using a capillary Western system. Anal. Biochem. 442, 97–103. 10.1016/j.ab.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. M., Jiang M., Shang J., Lan X., Yang F., Huang J., et al. (2011). CYP1, a hypovirus-regulated cyclophilin, is required for virulence in the chestnut blight fungus. Mol. Plant Pathol. 12, 239–246. 10.1111/j.1364-3703.2010.00665.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E. S., Chung H. J., Kim M. J., Park S. M., Cha B. J., Yang M. S., et al. (2005). Characterization of the ERK homologue CpMK2 from the chestnut blight fungus Cryphonectria parasitica. Microbiology 151(Pt 5), 1349–1358. 10.1099/mic.0.27796-0 [DOI] [PubMed] [Google Scholar]
- Choi G. H., Dawe A. L., Churbanov A., Smith M. L., Milgroom M. G., Nuss D. L. (2012). Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 190, 113–127. 10.1534/genetics.111.133983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill A. C. L., Ciuffetti L. M., Hansen D. R., Van Etten H. D., Van Alfen N. K. (1990). Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17, 25–31. 10.1007/BF00313245 [DOI] [Google Scholar]
- Eusebio-Cope A., Sun L., Tanaka T., Chiba S., Kasahara S., Suzuki N. (2015). The chestnut blight fungus for studies on virus/host and virus/virus interactions: from a natural to a model host. Virology 477, 164–175. 10.1016/j.virol.2014.09.024 [DOI] [PubMed] [Google Scholar]
- Finley D., Ozkaynak E., Varshavsky A. (1987). The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48, 1035–1046. 10.1016/0092-8674(87)90711-2 [DOI] [PubMed] [Google Scholar]
- Fraser J., Luu H. A., Neculcea J., Thomas D. Y., Storms R. K. (1991). Ubiquitin gene expression: response to environmental changes. Curr. Genet. 20, 17–23. 10.1007/BF00312760 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hillman B. I., Shapira R., Nuss D. L. (1990). Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80, 950–956. 10.1094/Phyto-80-950 [DOI] [Google Scholar]
- Hwang C. S., Oh J. H., Huh W. K., Yim H. S., Kang S. O. (2003). Ssn6, an important factor of morphological conversion and virulence in Candida albicans. Mol. Microbiol. 47, 1029–1043. 10.1046/j.1365-2958.2003.03353.x [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y., Tanaka K. (2010). Regulatory mechanisms involved in the control of ubiquitin homeostasis. J. Biochem. 147, 793–798. 10.1093/jb/mvq044 [DOI] [PubMed] [Google Scholar]
- Lan X., Yao Z., Zhou Y., Shang J., Lin H., Nuss D. L., et al. (2008). Deletion of the cpku80 gene in the chestnut blight fungus, Cryphonectria parasitica, enhances gene disruption efficiency. Curr. Genet. 53, 59–66. 10.1007/s00294-007-0162-x [DOI] [PubMed] [Google Scholar]
- Leach M. D., Stead D. A., Argo E., MacCallum D. M., Brown A. J. (2011). Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol. Microbiol. 79, 1574–1593. 10.1111/j.1365-2958.2011.07542.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. M., Chao D. Y., Wu Y., Huang X., Chen K., Cui L. G., et al. (2015). Natural alleles of a proteasome alpha2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 47, 827–833. 10.1038/ng.3305 [DOI] [PubMed] [Google Scholar]
- Lin H., Lan X., Liao H., Parsley T. B., Nuss D. L., Chen B. (2007). Genome sequence, full-length infectious cDNA clone, and mapping of viral double-stranded RNA accumulation determinant of hypovirus CHV1-EP721. J. Virol. 81, 1813–1820. 10.1128/JVI.01625-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Fink G. R. (2001). The glyoxylate cycle is required for fungal virulence. Nature 412, 83–86. 10.1038/35083594 [DOI] [PubMed] [Google Scholar]
- Nuss D. L. (2005). Hypovirulence: mycoviruses at the fungal–plant interface. Nat. Rev. Microbiol. 3, 632–642. 10.1038/nrmicro1206 [DOI] [PubMed] [Google Scholar]
- Oh Y., Franck W. L., Han S. O., Shows A., Gokce E., Muddiman D. C., et al. (2012). Polyubiquitin is required for growth, development and pathogenicity in the rice blast fungus Magnaporthe oryzae. PLoS ONE 7:e42868. 10.1371/journal.pone.0042868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkaynak E., Finley D., Varshavsky A. (1984). The yeast ubiquitin gene: head-to-tail repeats encoding a polyubiquitin precursor protein. Nature 312, 663–666. 10.1038/312663a0 [DOI] [PubMed] [Google Scholar]
- Ozkaynak E., Finley D., Solomon M. J., Varshavsky A. (1987). The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 6, 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. W., Ryu K. Y. (2014). Cellular ubiquitin pool dynamics and homeostasis. BMB Rep. 47, 475–482. 10.5483/BMBRep.2014.47.9.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. M., Choi E. S., Kim M. J., Cha B. J., Yang M. S., Kim D. H. (2004). Characterization of HOG1 homologue, CpMK1, from Cryphonectria parasitica and evidence for hypovirus-mediated perturbation of its phosphorylation in response to hypertonic stress. Mol. Microbiol. 51, 1267–1277. 10.1111/j.1365-2958.2004.03919.x [DOI] [PubMed] [Google Scholar]
- Puhalla J. E., Anagnostakis S. L. (1971). Genetics and nutritional requirements of Endothia parasitica. Phytopathology 61, 169–173. 10.1094/Phyto-61-169 [DOI] [Google Scholar]
- Randow F., Lehner P. J. (2009). Viral avoidance and exploitation of the ubiquitin system. Nat. Cell Biol. 11, 527–534. 10.1038/ncb0509-527 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y. (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Richter K., Haslbeck M., Buchner J. (2010). The heat shock response: life on the verge of death. Mol. Cell 40, 253–266. 10.1016/j.molcel.2010.10.006 [DOI] [PubMed] [Google Scholar]
- Ryu K. Y., Maehr R., Gilchrist C. A., Long M. A., Bouley D. M., Mueller B., et al. (2007). The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J. 26, 2693–2706. 10.1038/sj.emboj.7601722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: a Laboratory Manual. New York, NY: Cold Spring Harbor press. [Google Scholar]
- Shi L., Li R., Liao S., Bai L., Lu Q., Chen B. (2014). Prb1, a subtilisin-like protease, is required for virulence and phenotypical traits in the chestnut blight fungus. FEMS Microbiol. Lett. 359, 26–33. 10.1111/1574-6968.12547 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-e A. (1988). Dual regulation of the expression of the polyubiquitin gene by cyclic AMP and heat shock in yeast. EMBO J. 7, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeshi N. D., Mertins P., Svinkina T., Carr S. A. (2013). Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 8, 1950–1960. 10.1038/nprot.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lu L., Yang Y., Chen Q., Chen B. (2014). Proteomic analysis of Cryphonectria parasitica infected by a virulence-attenuating hypovirus. Wei Sheng Wu Xue Bao 54, 803–812. 10.13343/j.cnki.wsxb.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Wang J., Shi L., He X., Lu L., Li X., Chen B. (2016). Comparative secretome analysis reveals perturbation of host secretion pathways by a hypovirus. Sci. Rep. 6:34308. 10.1038/srep34308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Mao X., Cai T., Luo J., Wei L. (2006). KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 34 W720–W724. 10.1093/nar/gkl167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Zou C., Zhou H., Wang J., Lu L., Li Y., et al. (2013). Δ-Pyrroline-5-Carboxylate/glutamate biogenesis is required for fungal virulence and sporulation. PLoS ONE 8:e73483 10.1371/journal.pone.0073483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.