ABSTRACT
The high-resolution structure of glucan binding protein C (GbpC) at 1.14 Å, a sucrose-dependent virulence factor of the dental caries pathogen Streptococcus mutans, has been determined. GbpC shares not only structural similarities with the V regions of AgI/II and SspB but also functional adherence to salivary agglutinin (SAG) and its scavenger receptor cysteine-rich domains (SRCRs). This is not only a newly identified function for GbpC but also an additional fail-safe binding mechanism for S. mutans. Despite the structural similarities with S. mutans antigen I/II (AgI/II) and SspB of Streptococcus gordonii, GbpC remains unique among these surface proteins in its propensity to adhere to dextran/glucans. The complex crystal structure of GbpC with dextrose (β-d-glucose; Protein Data Bank ligand BGC) highlights exclusive structural features that facilitate this interaction with dextran. Targeted deletion mutant studies on GbpC's divergent loop region in the vicinity of a highly conserved calcium binding site confirm its role in biofilm formation. Finally, we present a model for adherence to dextran. The structure of GbpC highlights how artfully microbes have engineered the lectin-like folds to broaden their functional adherence repertoire.
KEYWORDS: microbial adherence, salivary agglutinin, glucan binding protein, Streptococcus mutans, sucrose-dependent adhesion, sucrose-independent adhesion, lectin-like fold, Streptococcus gordonii, antigen I/II, fibrillar, polyproline type II helix
INTRODUCTION
The oral cavity harbors diverse microbes. Commensal oral streptococci such as Streptococcus mitis and Streptococcus gordonii constitute the early colonizers, and they maintain homeostasis of the microbial community (1). When disease-causing microbes become predominant, the homeostasis of this symbiotic ecosystem is disturbed and turns dysbiotic. Streptococcus mutans dominates the dental caries dysbiotic ecosystem and has been implicated in the pathogenesis of tooth decay. Our group has been studying the surface proteins of S. mutans (2, 3) in an effort to decipher the adherence mechanisms adapted by oral streptococci in the harsh but nutrient-rich oral cavity. Our ultimate goal is to identify factors that enhance bacterial colonization and community development that would offer potential therapeutic interventions to modulate onset of disease.
Previous studies on various adherence factors of S. mutans led to the classification of the sucrose-dependent and sucrose-independent adhesins (4, 5). The glucosyltransferases (Gtfs) and the glucan binding proteins (Gbps) were classified as sucrose-dependent virulence factors, whereas antigen I/II (AgI/II), which does not require sucrose for binding, was classified as a sucrose-independent adhesin (6). The Gtfs are enzymes that utilize ingested starch and sugars to produce various types of glucan polymers. Among these, GtfB produces insoluble α-1,3-linked glucans, and GtfD produces soluble α-1,6-linked glucans whereas GtfC generates insoluble as well as soluble glucans (7). These glucan polymers then form a basal scaffold onto which other microbes adhere and colonize, thus initiating the formation of biofilms. The by-products of this catalytic reaction create a localized acidic environment, which demineralizes the tooth surface, resulting in the onset of cariogenesis.
The other group of proteins in this sucrose-dependent classification are the glucan binding proteins (Gbps) of S. mutans, which were named for their ability to interact with the glucan matrix (dextran). Both GbpA and GbpB were discovered through dextran matrix adherence (8, 9), and GbpC was identified from a deficient mutant strain (GbpCΔ) that did not aggregate in the presence of dextran, a phenomenon now known as dextran-dependent aggregation (DDAG) (10). GbpD was the last glucan binding protein to be discovered from annotations of the S. mutans genome (11). Interestingly, these Gbps share limited homology in their amino acid sequences.
Among these glucan binding proteins only GbpC is a surface-anchored protein (12) that contains the signature LPXTG motif, which is processed by sortases (13) to anchor such surface proteins onto the peptidoglycan layer. Recombinant GbpC (rGbpC) bound in significantly larger amounts to the rGtfD-synthesized α-1,6 glucans than rGtfB that produces insoluble α-1,3 glucans and rGtfC that synthesizes both soluble and insoluble glucans (14). The GbpC-defective mutant was able to survive longer in the blood of rats than the parental strain, indicating that the protein is one of the targets for the immune system (15). In in vitro experiments, GbpC-deficient S. mutans exhibited reduction in biofilm formation, and the deletion of GbpC in combination with GbpA and GbpD significantly reduced cariogenicity in a rat model of tooth decay (16). It is hypothesized that GbpC plays an important role in early biofilm formation (17), while GbpA and GbpD contribute to later biofilm formation (16). Therefore, sucrose-dependent adhesins are considered important virulence factors and potential targets for therapeutic interventions.
GbpC shares sequence homology with the V region of AgI/II, another well-characterized S. mutans surface-anchored protein (12). Our studies on the crystal structure of antigen I/II elucidated two distinct regions which adhere to salivary agglutinin (SAG/Gp340), namely, the variable (V) domain and the carboxy-terminal regions. These two binding domains are far apart from each other (50 nm) and are connected through a novel fibrillar stalk formed by the association of a right-handed alpha-helix and a left-handed polyproline type II (PPII) helix (2, 3). More interestingly, this fibrillar stalk is formed through two discontinuous regions, namely, the alanine-rich (A) and the proline-rich (P) regions in the primary structure of AgI/II. In addition to the noted similarity of GbpC to AgI/II's V domain, our own sequence analysis discovered short homologous segments that resemble the A and P regions. In spite of numerous reports on GbpC, no study to date has assessed whether GbpC would share similar structural or functional attributes with AgI/II.
In this work we undertook the structural, biophysical, and genetic characterization of GbpC. Here, we report the first crystal structure of GbpC at 1.14-Å resolution. GbpC shares structural similarity with the V region and the fibrillar stalk of AgI/II. Affinity studies on GbpC demonstrate that it adheres to SAG/Gp340 and its derived scavenger receptor cysteine-rich (SRCR) domains with nanomolar affinity, whereas it binds to soluble dextran with micromolar affinity. These studies reveal a new activity of GbpC. Comparative structural analysis of S. mutans AgI/II and S. gordonii SspB show that the putative binding regions are vastly divergent. Site-directed mutagenesis confirmed this binding motif to be important for sucrose-dependent and sucrose-independent phenotypes. The cocrystal structure with dextrose (β-d-glucose) highlights the role of Ca2+, and comparative analysis of the same region in AgI/II and SspB further illustrates the specific adherence of GbpC to glucans. Taking these results together, we have determined that GbpC plays an integral role in sucrose-dependent and sucrose-independent adhesion, exhibiting the ability to interact with diverse ligands functionally overlapping with AgI/II.
RESULTS
Structure of GbpC.
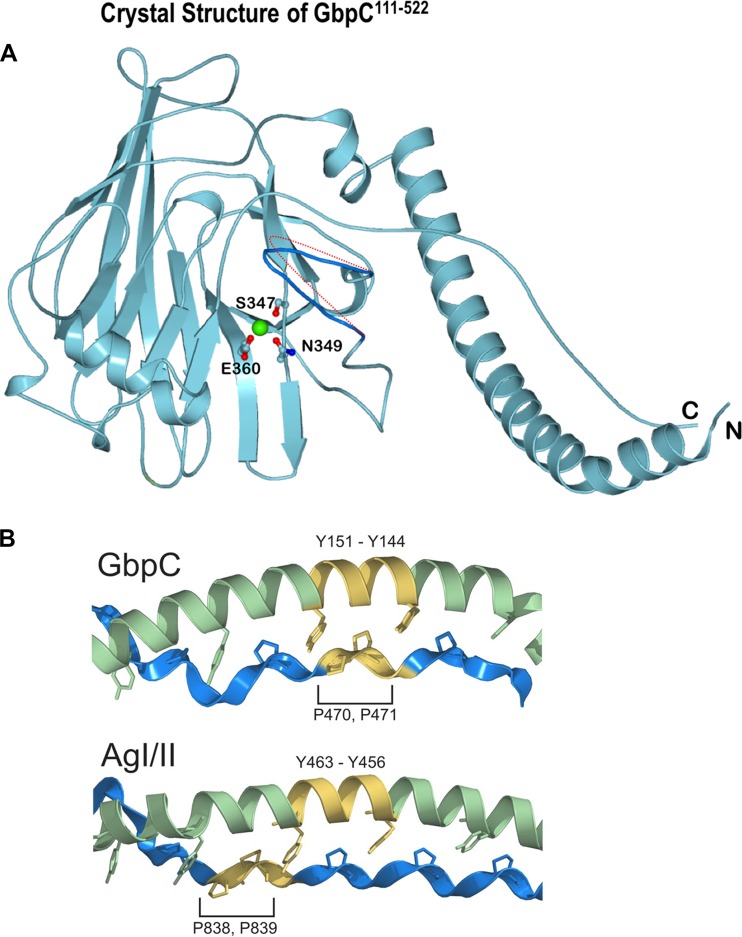
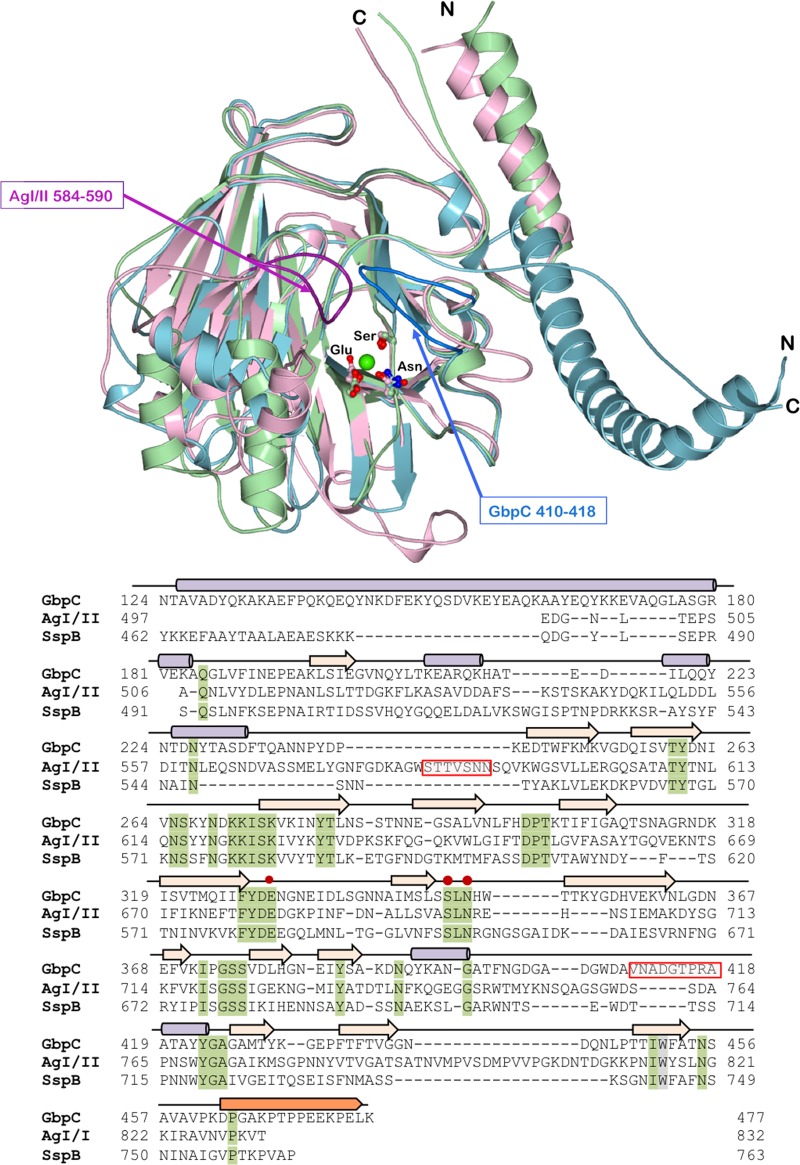
GbpC of S. mutans contains 583 amino acid residues (Fig. 1) and shares homology with AgI/II of S. mutans (PDB accession number 1JMM; 26.3% identity and 40.9% homology) and SspB of S. gordonii (PDB accession number 2WD6; 27.2% identity and 40.6% homology) (18, 19). This homology covers portions of the V and A/P regions of AgI/II. Constructs generated based on this homology were crystallized, and the crystal structure was resolved at 1.14-Å resolution (Fig. 2 and Table 1). Only residues 124 to 477 of the GbpC construct consisting of residues 111 to 522 (GbpC111–522) could be resolved in the native crystal structure. This was not surprising since we had observed proteolytic cleavage occurring in this construct over time (see Fig. S1 in the supplemental material). The structure of GbpC resembles the lectin-like beta-supersandwich fold present in the V regions of both AgI/II and SspB. The fibrillar stalk that projects the V region of AgI/II and SspB is also present in GbpC, where an N-terminal alpha-helix is complemented by a C-terminal proline-rich region that adopts the left-handed PPII helical secondary structure (3, 19). A distinctive feature of GbpC's fibrillar stalk is its orientation, whereby it is projected 180° in the opposite direction from that of AgI/II and SspB (Fig. 3). Consequently, GbpC emanating from the cell surface would project its ligand binding region in a different spatial conformation than SspB and AgI/II (Fig. S2). On closer examination of the fibrillar stalk in GbpC, an additional difference was observed such that there exists a register shift in the knobs-in-holes interaction between the proline side chains and the alpha-helical tyrosines (Fig. 2). The V region itself superposes with an root mean square (RMS) deviation of 1.80 and 1.56 Å with AgI/II and SspB (PDB accession numbers 1JMM and 2WD6), respectively, but despite these similarities there are several important variations observed between these crystal structures. Most notably, the loop regions in AgI/II (residues 584 to 590) and GbpC (residues 410 to 418) are absent in the SspB structure (Fig. 3), resulting in a flatter surface. Each of these loop regions points into a large surface area, which contains a conserved calcium binding site in all three proteins (3, 18, 19).
FIG 1.

Primary structure features of GbpC and the extent of the recombinant fragments used in this study. The equivalent of AgI/II's V region in GbpC is highlighted.
FIG 2.
(A) Crystal structure of GbpC111–522 with the lectin-like fold; the calcium binding site is highlighted. Three residues, Ser347, Glu360, and Asn349, form bipyramidal geometrical coordination for the calcium binding site. The loop region of residues 410 to 418 is highlighted in dark blue, and the potential movement of this loop region based on the B factors (see Table S2 in the supplemental material) is denoted by the dotted red line. (B) A closer examination of the A-P interaction on GbpC and AgI/II shows that there is a register shift in the interactions of the AxYxAx(L/V) motif on the A region and the PxxP motif on the P region that we had described earlier (3).
TABLE 1.
Crystallographic data and refinement statistics
| Parametera,b | GbpC-KIc | GbpC-native | GbpC-BGCd |
|---|---|---|---|
| Data collection statistics | |||
| Space group | C2 | C2 | C2 |
| Unit cell | |||
| a, b, c (Å) | 213.088, 48.917, 40.930 | 213.098, 48.596, 40.755 | 210.02, 47.94, 40.09 |
| α, β, γ (°) | 90.0, 96.2, 90.0 | 90.0, 96.3, 90.0 | 90.0, 96.3, 90.0 |
| Resolution (Å) | 1.9 | 1.15 | 1.75 |
| Rmerge | 0.055 (0.141) | 0.060 (0.607) | 0.153 (0.855) |
| Rpim | 0.023 (0.062) | 0.036 (0.375) | 0.094 (0.529) |
| CC1/2 | 0.993 (0.998) | 0.911 (0.688) | 0.992 (0.625) |
| I/σI | 12.0 | 7.5 | 8.6 (2.7) |
| Completeness (%) | 95.8 (89.5) | 97.2 (95.9) | 98.5 (96.5) |
| Redundancy | 6.6 (5.7) | 3.6 (3.5) | 3.6 (3.6) |
| Refinement statistics | |||
| Resolution (Å) | 1.9 | 1.15 | 1.75 |
| No. of reflections | 32330 | 142968 | 39632 |
| Rwork/Rfree | 12.9/14.9 | 20.7/23.5 | |
| Atoms | |||
| Protein (no.) | 2,834 | 2,752 | |
| Ion (no.) | 1 | 1 | |
| Ligand(s) | BGC1, BGC2 | ||
| Water (no.) | 528 | 181 | |
| B factors (Å2) | |||
| Wilson B | 9.3 | 16.0 | |
| Protein | 18.20 | 20.7 | |
| Ion | 9.76 | 17.3 | |
| Ligand(s) | 27.3, 37.0 | ||
| Water | 18.29 | 24.7 | |
| RMS deviation | |||
| Bond length (Å) | 0.005 | 0.014 | |
| Bond angle (°) | 1.061 | 1.64 | |
| Ramachandran statistics (no. of residues [%]) | |||
| Favored | 99 (349) | 97.7 (343) | |
| Allowed | 1 (5) | 2.3 (8) | |
| Outliers | 0 (0) | 0 (0) |
CC1/2, Pearson correlation coefficient between two random half data sets.
Rwork/Rfree, R factors calculated by the following: R = (Σ||Fobs| − | Fcalc||)/(Σ|Fobs|).
GbpC-KI, potassium iodide quick-soaked GbpC crystal data.
GbpC-BGC, crystal data of GbpC complexed with β-d-glucose ligands (BGC1 and BGC2).
FIG 3.
The figure shows the structural superposition of the V regions of AgI/II (light pink), SspB (light green), and GbpC (light blue) and their structure-based sequence alignment with alpha-helices (purple cylinders), strands (beige arrows), and the PPII helix (saffron). The residues of the conserved calcium-binding site are highlighted (red dots on the sequence). The loop regions that hover over the calcium-binding site which can be imagined to emanate from two sides of a palm are highlighted in dark pink for AgI/II (residues 584 to 590) and dark blue for GbpC (residues 410 to 418; shaded red boxes in sequence). SspB does not display either of these two loop regions.
GbpC binds to dextran as measured by ITC.
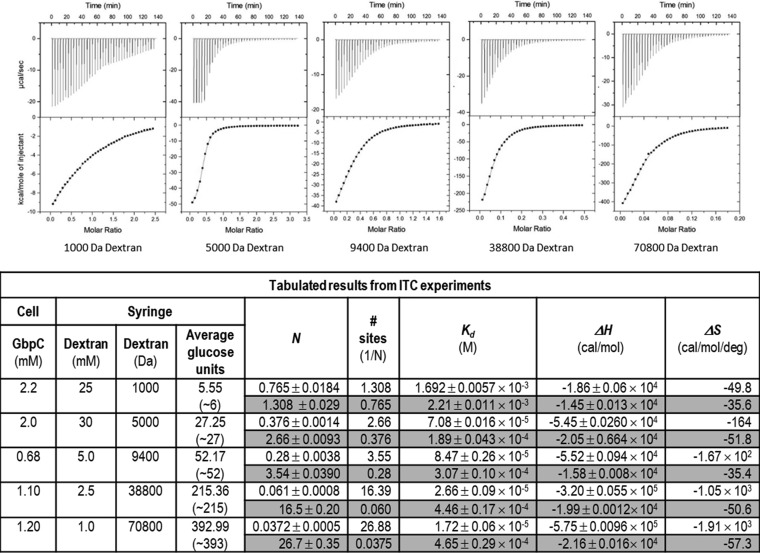
GbpC was originally identified to be a glucan binding protein, and therefore we evaluated its affinity coefficients to dextran. Various sizes of dextran (1,000, 5,000, 9,400, 38,800, and 70,800 Da) were titrated, and exothermic reactions predominated the observed micromolar interaction between GbpC and dextran (Fig. 4). The isothermal titration calorimetry (ITC) thermograms fitted with a simple one-site model resulted in stoichiometries which correspond to 1.30, 2.66, 3.55, 16.39, and 26.88 for the 1,000-, 5,000-, 9,400-, 38,800-, and the 70,800-Da dextrans, respectively. Considering dextran to be a lattice of glucose units, on average GbpC's adherence to this lattice would span 11 to 14 glucose units (Fig. 4). While the observed cumulative enthalpies (ΔH) in these experiments are −18.6, −54.5, −55.2, −320, and −575 kcal/mol for the 1,000-, 5,000-, 9,400-, 38,800-, and 70,800-Da dextrans, respectively, the reverse-mode analysis (shaded in the table of Fig. 4) which describes each individual binding event displays consistent enthalpies (−14.5, −20.5, −15.8, −19.9, and −21.6 kcal/mol, respectively). This reaction is predominantly enthalpically (ΔH) driven, involving noncovalent interactions, and the near constant entropy (ΔS) of the reverse-mode analysis (shaded in table of Fig. 4) suggests that there are only minor conformational changes associated with either GbpC or dextran (20). The linearity of the Scatchard plots (Fig. S3) indicates noncooperative binding to the dextran lattice, and this interaction appears to occur independently at each site as the affinity of subsequent binding of GbpC molecules is not affected in reverse analysis (21–23). Furthermore, the noncooperative adherence and saturation of the dextran lattice binding sites is confirmed by the decreasing slope values in the Hill plots (Fig. 3), collectively indicating the isolated binding events of GbpC to the dextran lattice, which agrees well with the observations of cumulative enthalpies and entropies (Fig. 4). In reverse titration experiments, we observed similar enthalpies, albeit we could not achieve thermograms of sufficient quality due to both machine and protein/ligand concentration limitations (Fig. S4). ITC experiments conducted with the equivalent AgI/II and SspB did not display any measurable binding to dextran (Fig. S4).
FIG 4.
Interaction of GbpC with 1,000-, 5,000-, 9,400-, 38,800-, and 70,800-Da dextran was determined using an Auto ITC 200. GbpC adheres to dextran with micromolar affinity. The stoichiometry reported here is the ratio of GbpC to dextran binding. The shaded portions of the table are values for model fits in the reverse mode in Origin, which displays the various parameters calculated for the reversed binding. In preliminary reversed ITC experiments with dextran in the cell and GbpC in the syringe, we observed very similar values (see Fig. S4 in the supplemental material). In similar experiments with AgI/II and SspB, dextran did not show any specific interaction (Fig. S4). Kd, dissociation constant; N, stoichiometry.
Binding to SRCR and salivary agglutinin.
Based on the structural similarities to AgI/II and SspB, we hypothesized that GbpC would also bind to SAG and SRCR domains. We tested this hypothesis using surface plasmon resonance (SPR) experiments, where GbpC exhibited high nanomolar affinity for SAG, recombinant iSRCR1 and iSRCR123 (insect cell-expressed recombinant SRCR domain 1 and SRCR domains 1, 2, and 3, respectively) (24) (Table 2). Additionally, in on-chip deglycosylation experiments GbpC's adherence characteristics were not affected. This trait was previously observed with the V regions of AgI/II and SspB (24), indicating a similar lack of interaction with glycosylations present on iSRCR1 and iSRCR123. As previously observed with both AgI/II and SspB, in the absence of Ca2+ there was a complete abrogation of GbpC binding to SRCRs (Fig. S5) (2, 3, 24).
TABLE 2.
Surface plasmon resonance studies on the adherence of GbpC with SRCR domains
| Liganda | ka (1/Ms)b | kd (1/s)c | Rmax (RU)d | KA (1/M)e | KD (M)f | Chi2 |
|---|---|---|---|---|---|---|
| iSRCR1 | 2.22 × 105 | 7.53 × 10−3 | 179 | 2.95 × 107 | 3.39 × 10−8 | 4.85 |
| iSRCR1 (deglycosylated) | 2.48 × 104 | 3.39 × 10−3 | 34.7 | 7.3 × 106 | 1.37 × 10−7 | 9.42 |
| iSRCR123 | 6.64 × 104 | 2.0 × 10−3 | 316 | 3.32 × 107 | 3.02 × 10−8 | 9.23 |
| iSRCR123 (deglycosylated) | 6.49 × 104 | 2.89 × 10−3 | 41.1 | 2.25 × 107 | 4.45 × 10−8 | 10.9 |
| SAG | 6.77 × 104 | 5.08 × 10−3 | 75.1 | 1.33 × 107 | 7.5 × 10−8 | 0.828 |
In all cases, the analyte was GbpC111–522.
ka, association rate constant.
kd, dissociation rate constant.
Rmax, maximum analyte binding capacity. RU, response units.
KA, equilibrium binding constant.
KD, equilibrium dissociation constant.
GbpC adherence to SRCR/SAG is inhibited by the presence of dextran.
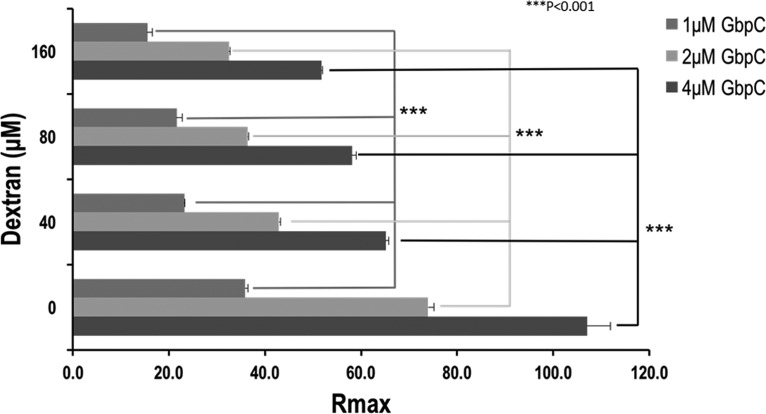
We investigated the effect of dextran on the interaction between GbpC and SRCR/SAG. The 5,000-Da dextran inhibited the binding of SRCR/SAG in a dose-dependent manner although it was not able to completely abrogate the adherence of GbpC (Fig. 5). Analysis of the binding curves in the presence and absence of dextran indicated no change in the affinity coefficients (Table S1). These results show a possible overlap of the binding sites for both dextran and SRCR/SAG. Similar experiments with AgI/II (Fig. S6) and SspB (data not shown) did not display any such phenomenon, confirming their lack of interaction with dextran.
FIG 5.
The effect of dextran on the adherence of GbpC to immobilized SRCR1 (iSRCR1) was analyzed using surface plasmon resonance. These results indicate that there is a significant dose-dependent reduction in the amount of GbpC that binds to iSRCR1. While the affinities remain unchanged (see Table S1 in the supplemental material), the presence of dextran in close proximity to the SRCR binding surface could produce steric hindrance, thus precluding the adherence of GbpC. Rmax, maximum analyte binding capacity. ***, P < 0.001.
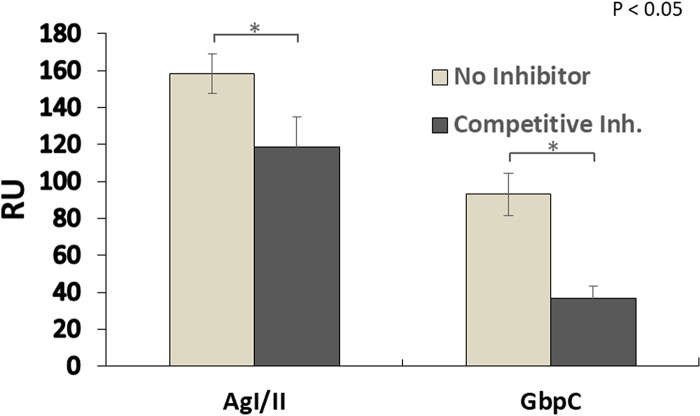
AgI/II inhibited the binding of GbpC in competition experiments.
To further characterize the binding of GbpC, we performed competitive adherence assays (Fig. 6). AgI/II significantly inhibited the binding of GbpC by 60%, whereas GbpC did not significantly alter the binding of AgI/II (25%). There are two different sites on AgI/II and SspB (2, 3, 24), one present in the V region and the other in the C-terminal region, and we have previously shown that each of these regions adheres to independent sites on SRCR domains (24). By comparison, GbpC has only one site on its homologous V region, and the results here confirm that both GbpC and AgI/II could compete for adherence to one of these sites. These results indicate that S. mutans employs diverse adhesins to interact with Gp340/SRCR and that AgI/II may be one of the primary functional receptors for Gp340.
FIG 6.
Competition assays between GbpC and AgI/II against immobilized SAG. SAG was immobilized to a CM5 chip, and either buffer (no inhibitor) or a competitive inhibitor (AgI/II or GbpC) was passed to saturate SAG receptor sites. Afterwards, the competing protein (labeled at the bottom) was passed through, and the maximal binding was measured. GbpC inhibited the binding of AgI/II (left) by 25.1% ± 10%, whereas AgI/II inhibited GbpC binding (right) by 60.3 ± 6.7%. All experiments were carried out in duplicate; error bars indicate means ± standard deviations. In control experiments, where either AgI/II or GbpC was immobilized, neither one displayed any measurable adherence to the other (data not shown). RU, response units. *, P < 0.05.
Complex crystal structure.
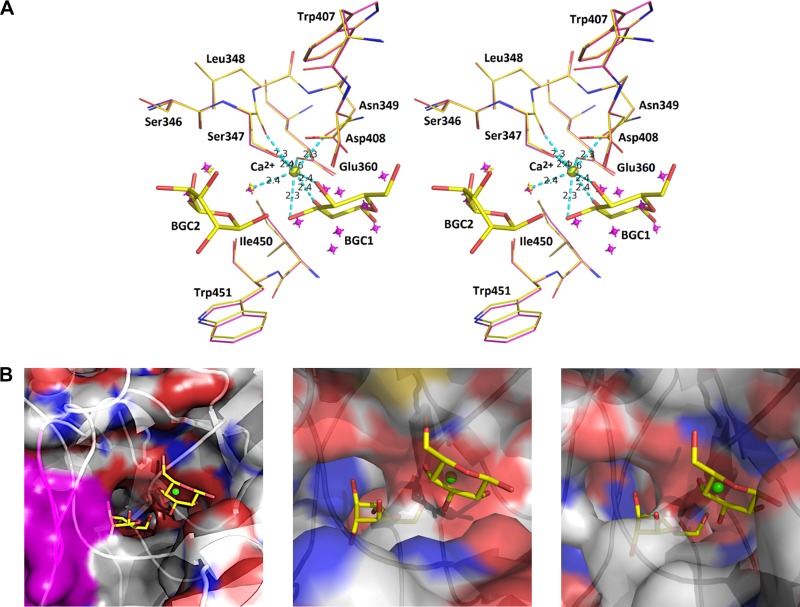
The crystal structure in complex with dextrose (β-d-glucose; PDB ligand BGC) highlights the interaction of two glucose molecules (BGC1 and BGC2). In the native structure, a metal binding site contains a Ca2+ ion coordinated through oxygen atoms from Ser347 (main chain and side chain), Asn349 (side chain), and Glu360 (side chain), where a minimum of two water molecules are within the allowed hydrogen bonding distance (2.5 Å) (Fig. 7; see also Fig. S7). The metal coordination sphere allows a maximum of eight interacting oxygens at a distance of 2.3 to 2.4 Å. The first dextrose (BGC1) directly interacts with the Ca2+ ion in the metal binding site by effectively displacing the previously observed water molecules in the native GbpC structure (Fig. 7). In close proximity to BGC1, BGC2 interacts with the glycine of the loop region (residues 410 to 418) (Fig. 7). The distance between these two BGCs is 3.4 Å and provides significant clues for the adherence of a dextran chain to perch within the confines of this binding site. The interaction of BGC1 and BGC2 is described in detail in Fig. S8 in the supplemental material. This binding region has an entirely different spatial arrangement of secondary structures in both AgI/II and SspB, and the residues within their pockets would sterically hinder the BGC's interaction (Fig. 7). Our attempts to cocrystallize the V regions of AgI/II and SspB with glucose did not produce viable electron difference densities within this pocket (data not shown), further supporting the idea that these regions of AgI/II and SspB do not interact with dextran. Based on the positions of BGC1 and BGC2, a short dextran molecule was modeled into this open pocket that accommodated six glucose units, and the ligplot diagrams (25) describing potential interactions are shown in Fig. 8.
FIG 7.
(A) Stereo diagram of the interaction of BGC1 and BGC2 in the vicinity of the Ca2+ (yellow) binding site. The superposed native structure and its displaced water molecules are shown in magenta. (B) Surface plots highlighting the binding pocket, where for GbpC the loop region of residues 410 to 418 is highlighted in magenta. For AgI/II and SspB these binding pockets are very different and would pose steric hindrances for the glucose molecules, thus explaining the propensity for GbpC to specifically adhere to dextran.
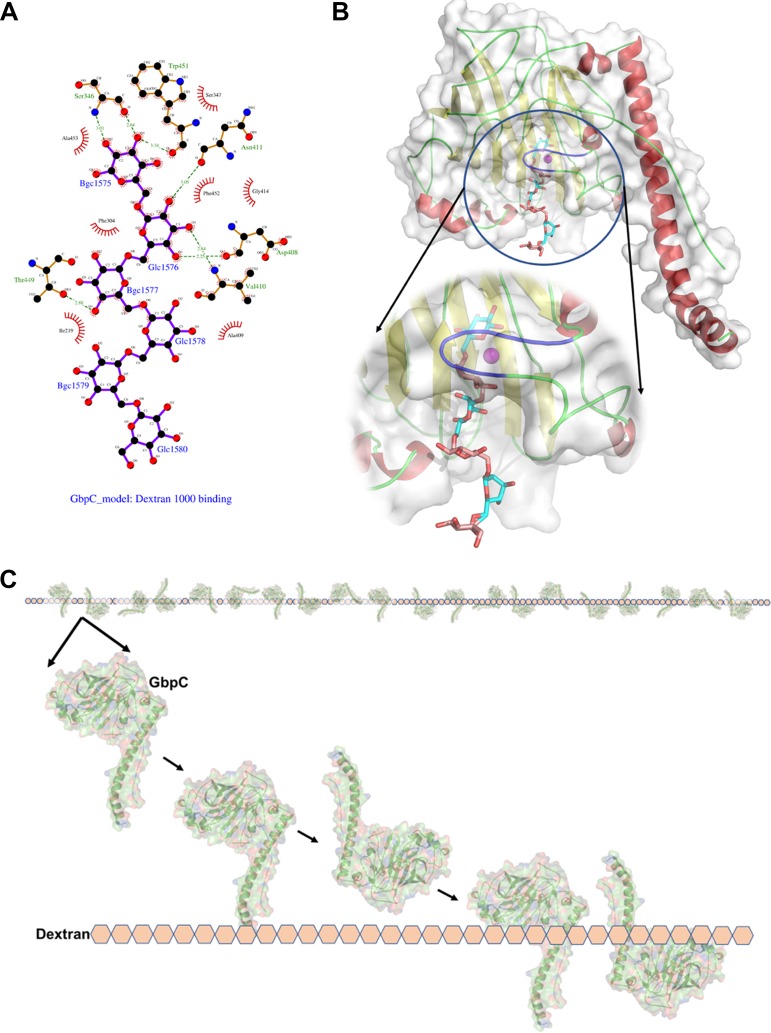
FIG 8.
Based on positions of BGC1 and BGC2 in the complex crystal structure, we have modeled dextran (6 glucose units) into the binding site. (A) Interactions of this modeled dextran with GbpC. (B) The loop region of residues 410 to 418, highlighted in purple, hovers over the Ca2+ site, which hosts the dextran (glucan) binding site. This loop region is highly flexible in the structure and could open and close to accommodate the glucose molecules thus locking into place. (C) The model for the interaction of GbpC with dextran (glucose lattice) based on ITC and crystal structure.
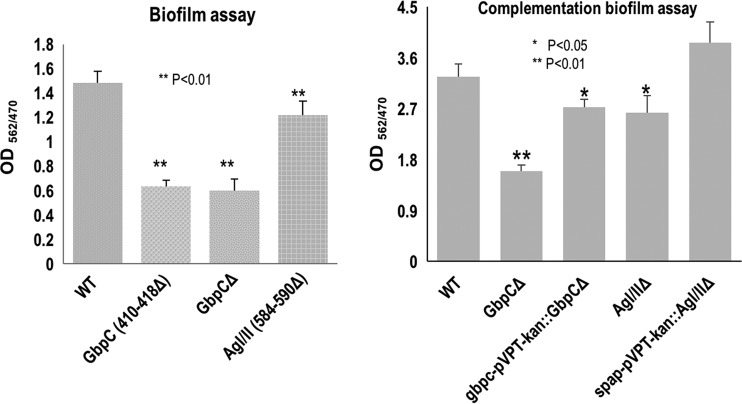
The GbpC loop region is crucial for biofilm formation.
Mutants of the uniquely displayed loop regions of both GbpC and AgI/II, which hovers over the calcium binding site and was observed interacting with that of BGC on the complex crystal structure, were generated (GbpC with a deletion of residues 410 to 418 [GbpC410–418Δ] and AgI/II584–590Δ). Neither mutant had any apparent growth defect; however, when assessed for biofilm formation in 0.5% sucrose, GbpC410–418Δ exhibited a significant reduction in its ability to form biofilms. The reduction is comparable to the level of the GbpC deletion mutant, indicating the importance of the GbpC loop region in biofilm formation (Fig. 9). In contrast, the biofilm formation of AgI/II584–590Δ was only slightly affected by the absence of the loop region. In gene complementation experiments, both GbpC and AgI/II restored the wild-type (wt) biofilm levels. These data are consistent with the ascription of greater functional importance to GbpC in the sucrose-dependent biofilm formation over AgI/II.
FIG 9.
Effects of the loop region of GbpC and AgI/II on biofilm formation of S. mutans. Wild-type S. mutans and the loop region deletion mutants (GbpC410–418Δ and AgI/II584–590Δ) (left) and their complemented strains (right) were evaluated for biofilm formation using crystal violet staining. The results show that the loop region of residues 410 to 418 of GbpC plays an important role in biofilm formation. Biofilm formation defects caused by GbpC and AgI/II deficiency can be recovered to wild-type levels by complementation.
Both GbpC and AgI/II are important for dextran-dependent aggregation.
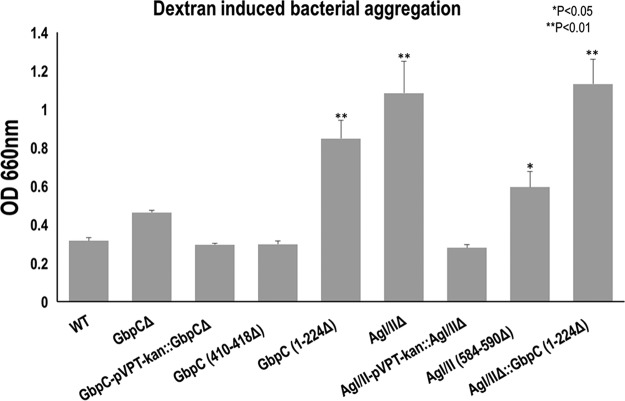
GbpC has been previously reported to mediate dextran-dependent aggregation (26, 27); therefore, we examined the effects of AgI/II and GbpC deficiency on this phenotypic behavior. Wild-type bacteria aggregated well (Fig. 10). Surprisingly, the GbpC null mutant modestly affected bacterial aggregation, while the AgI/II null mutant was significantly inhibited in its bacterial aggregation. Complementation with their respective alleles restored the wild-type levels of bacterial aggregation, demonstrating the involvement of GbpC and AgI/II in bacterial aggregation. Consistently, the loop region deletion mutant of AgI/II584–590Δ exhibited decreased bacterial aggregation, but the similar GbpC410–418Δ mutation did not affect bacterial aggregation. Furthermore, deletion of the N-terminal end of GbpC (GbpC1–224Δ) significantly reduced bacterial aggregation (Fig. 10). When both AgI/II and GbpC were deleted, S. mutans lost its aggregation propensity. These data suggest that both surface proteins are important for dextran-induced bacterial aggregation.
FIG 10.
Effects of GbpC and AgI/II deficiency on dextran dependent bacterial aggregation (DDAG) of S. mutans. Wild type (WT) S. mutans and various gbpC and AgI/II gene (pac) mutant variants were evaluated for the dextran-dependent bacterial aggregation by monitoring bacteria remaining in suspension at OD600 nm. The higher the OD reading, the lower was the bacterial aggregation. AgI/II appears to have a more profound effect on aggregation than GbpC. Particularly, the result of deleting the loop region of residues 584 to 590 of AgI/II (AgI/II584–590Δ) indicated that it is involved in DDAG, whereas the loop region of residues 410 to 418 of GbpC is not. The deletion of the N-terminal region (residues 1 to 224) of GbpC resulted in a significant reduction in bacterial aggregation, while the double mutant AgI/II and GbpC1–224Δ had the largest effect on bacterial aggregation.
DISCUSSION
GbpC homologs have been identified in all mutans streptococci (28) and are known to be an important component for both biofilm formation (17) and S. mutans-induced cariogenicity (29, 30). In this study, the high-resolution structure of GbpC at 1.14 Å (Fig. 2) revealed similarities with the V regions of AgI/II and SspB (Fig. 3). Nonetheless, on closer analysis major structural variations are observed around the conserved Ca2+ ion binding site that could be described as the palm of the hand in this lectin-like fold (Fig. 3). Particularly, in the structure of AgI/II there is a loop region (residues 584 to 590) that emanates from one side of this palm, whereas in GbpC the loop region (residues 410 to 418) hovers over the Ca2+ ion binding site from the opposite side of the palm.
Although GbpC was named based on its ability to bind glucans and has been studied over the last 2 decades, the affinity coefficients for the interaction between GbpC and dextran have never been quantified. ITC experiments with various sizes of dextran show that the number of sites to which GbpC bound increased proportionally with larger sizes of dextran. For example, 1,000-Da dextran, which has approximately six glucose residues, produced the closest to a 1:1 stoichiometry (n = 0.765 ± 0.0184), whereas for 5,000-Da dextran (28 units of glucose) the stoichiometry was 0.376 ± 0.0014, indicating binding to approximately 2.66 sites. We observed the same trend for dextrans of 9,400 (52 units of glucose), 38,800 (215 units of glucose), and 70,800 Da (393 units of glucose), which revealed that GbpC bound to approximately every 11 to 14 residues of glucose (Fig. 4). The cumulative interaction of GbpC with dextran is reflected by the total enthalpy released, which is also proportional to the size of dextran used in these experiments. The adherence of GbpC to a one-dimensional lattice of glucose units does not display cooperativity as either a positive or negative curvature, as is observed on Scatchard plots (see Fig. S3 in the supplemental material). The Hill plots (Fig. S3) show a gradual reduction in the slopes, thus indicating the saturation of sites on the linear dextran polymer (21–23). Our results here summarily indicate that GbpC adheres to dextran with micromolar affinity. Although we were expecting higher-affinity coefficients with dextran, we cannot rule out the fact that soluble dextran is only a representative of the 1,6-linked soluble glucan, and our experiments may not reflect the biological environment accurately. Previously, it was shown that larger amounts of soluble 1,6-linked glucans adhered to GbpC; however, their specificity and/or affinities were not evaluated, and this particular study also relayed their difficulties in dealing with insoluble glucans (14). Considering the presence of both insoluble and soluble glucans produced by the Gtfs on the tooth surface, GbpC could adhere with better affinity and specificity to the 1,3-linked insoluble glucans. Despite the observed micromolar affinity, multiple GbpC adhesins on the bacterial cell surface could very well bind to glucan polymers, thus resulting in a much larger avidity. The proportional increase in the number of sites with the size of the dextran molecule (Fig. 4) supports this avidity model. In a similar set of experiments, we also tested AgI/II and SspB, and neither one displayed any detectable affinity for dextran (Fig. S4), thus indicating that GbpC uniquely adheres to glucans.
Consistent with its structural similarities to AgI/II and SspB, GbpC displayed adherence to immobilized SAG/iSRCR with nanomolar affinity, thus ascribing a new activity and multidimensionality to its functional repertoire. Previously, we had shown that calcium-induced conformational changes in SRCR domains present a surface to which AgI/II-like proteins specifically adhere with high affinity (24). GbpC binding only in the presence of calcium recalls the characteristics previously observed with AgI/II and SspB (Fig. S5). Such adherence emanating from the lectin-like structural fold present on GbpC, AgI/II, and SspB confirms the existence of a common adherence mechanism to SAG/SRCR, albeit their orientations from the cell surface would project them in completely opposite spatial conformations (Fig. S2).
After rebuilding and refinement of the model, we observed higher B factors for the loop formed by residues 410 to 418 in GbpC (Table S2), indicating the presence of inherent flexibility, which we presumed could latch and lock the dextran into the binding site. In fact, the cocrystal structure shows that BGC (dextrose) interacts with this loop region and is in close proximity to the Ca2+ ion binding site (Fig. 7). In mutagenesis studies, it was further evident that the loop region of residues 410 to 418 plays a significant functional role in biofilm formation (Fig. 9). The deletion of this loop region in GbpC410–418Δ produced a biofilm-deficient phenotype strikingly similar to that of the GbpC null mutant, whereas AgI/II584–590Δ had a minimal effect on biofilm formation (Fig. 9). In dextran-induced aggregation assays, it was even more intriguing to note that AgI/II584–590Δ had reduced bacterial aggregation, while GbpC410–418Δ did not. While initially the dextran-dependent aggregation phenomenon was attributed to GbpC under certain stress and growth conditions (12, 27, 31, 32), later studies on this property had shown that DDAG is related not only to GbpC but also to AgI/II as well as WapA (26). The results from our aggregation experiments define a role for AgI/II and particularly the divergent loop region of residues 584 to 590. However, further work is needed to explain this intriguing phenomenon whereby the V region of AgI/II does not adhere to dextran but is seemingly involved in DDAG. We speculate that other components could promote this DDAG property in concert with AgI/II, similar to the previously reported head-to-tail interactions (33).
GbpC still remains unique among the structurally similar proteins for dextran affinity and biofilm formation but functionally overlaps with binding to the salivary receptor SAG/SRCR. Despite overall structural similarities, the subtle structural differences between GbpC and AgI/II or SspB are consistent with the notion that GbpC harbors an independent binding region that supports the interaction of GbpC to dextran. The fact that in the presence of dextran the binding of GbpC to SRCR is inhibited indicates that the binding sites for dextran and SRCR may be in close proximity. The absence of complete abrogation of adherence to SRCR and the observed inhibition could be the result of steric hindrances that would impede the adherence to these independent sites. The dextran (6 units of glucose) modeled within the pocket agrees well with our ITC results, according to which GbpC is expected to bind between 11 and 14 glucose units of dextran, with the necessary spacing to allow the next GbpC molecule to adhere (Fig. 8C). This study also shows how similar adhesion structures are adapted for the evolution of bacterial heterogeneity that may act as additional fail-safe mechanisms for host colonization, which we had earlier reported for the structure of AgI/II, in which two different regions were shown to bind to SAG/SRCR (2, 3). GbpC is known to be highly expressed under environmental stress conditions (27), during which we surmise that it may be necessary for bacteria to have multiple unique binding modules to interact with host receptors such as SAG/SRCRs. Further studies may shed light on how viridans/mutans streptococci regulate the expression of these surface binding proteins at different stages of colonization to adapt to the changing host environment.
Previous classification of sucrose-dependent and sucrose-independent adhesion was determined through studies that evaluated the impact of sucrose on S. mutans adhesion and colonization. These studies confirm the structural hypothesis that GbpC, being similar to the AgI/II-family, would share functionality in its adherence to SAG/SRCR. This newly discovered function and structural feature of GbpC suggest that GbpC should be reclassified more generally as a bacterial adhesin within the AgI/II family.
While the crystal structures of S. mutans adhesins have been resolved and while significant advances have been made toward understanding their biology, the exact binding site(s) for either SAG or SRCR remains elusive. The existence of a common motif/domain that adapts to accommodate different molecules is plausible in the case of GbpC and AgI/II. This would be consistent with the idea that bacterial surface proteins evolve to mutate local residues on structural folds to bind to various receptors. In the case of GbpC, it is fascinating that such minor modifications of loop regions hovering over the Ca2+ binding site in the lectin-like fold would allow for specific adherence to dextran. Such phenomena have been previously observed in Staphylococcus aureus, where most surface proteins exhibit the DE variant (DEv)-IgG fold and have adapted to bind to various host receptors such as collagen, fibrinogen, and fibronectin (34–36). The structural studies of Als3 of Candida albicans provide another example of such modifications to the DEv-IgG fold, in which C. albicans has adapted to bind to fibrinogen, laminin, casein, bovine serum albumin (BSA), cadherin, and ferritin (37). These microbes are fascinating molecular artists/engineers, which employ simple modifications to retrofit existing tools and adhere to diverse ligands. In the case of GbpC, AgI/II, and SspB, these modifications are now beginning to be understood. Further studies are warranted to determine the binding site/motif and the evolution of this lectin-like fold to bind to different molecules.
MATERIALS AND METHODS
Molecular cloning.
Two GbpC constructs were generated based on their sequence similarity with the A, V, and P regions of AgI/II of S. mutans (12, 38). PCR amplification of two fragments, GbpC171–464 and GbpC111–522 (Fig. 1), were carried out using DeepVent DNA polymerase (New England BioLabs) using forward (TATAACCATGGAAGTTGCACAAGGTTTGGCATCTGGG) and reverse (TTATACTCGAGCGGATCTTTAGGCACAGCTACAGC) primers for GbpC171–464 and, similarly, forward (TATAACCATGGCTGCAGATGCAGATAACAAAGCACAGGC) and reverse (TTATACTCGAGAGGTGATTGAGGATCCTCTGG) primers for GbpC111–522. The restriction sites XhoI and NcoI are underlined in the primers. After amplification, the PCR fragments were purified using a PCR purification kit (Qiagen, Inc.); subsequently, both the vector pET-23d and the purification products were digested, purified, and ligated with T4 ligase. The ligated products were transformed into XL1-Blue cells (Agilent, Inc.), and colonies were selected by their ampicillin resistance. DNA sequencing of the extracted plasmids confirmed the incorporation of the appropriate GbpC fragments as designed. The confirmed plasmids were then transformed into Escherichia coli BL21(DE3) cells (Invitrogen, Inc.) for protein expression.
Protein expression and purification.
BL21(DE3) cells harboring GbpC constructs within pET-23d vectors were initially grown in 20 ml of LB in 100-ml shaker flasks overnight at 37°C and then transferred into 1 liter of LB in 2,400-ml shaker flasks the next day. These LB cultures were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm (OD600) of 0.6, and induction continued for 4 h at 30°C. After 4 h, cells were pelleted by centrifugation (5,000 relative centrifugal force [rcf] for 20 min) using a Beckman Avanti JL50 centrifuge. These pellets were resuspended in the nickel affinity column binding buffer (50 mM Tris, pH 8.0, 500 mM sodium chloride, augmented with a Complete EDTA-free protease inhibitor cocktail) and sonicated (Fisher Brand Sonicator) for a total of 5 min while a maximum temperature of 10°C was maintained (using a temperature probe). The supernatant of the cell lysate was collected after centrifugation at 35,000 rpm for 1 h using a Ti70 rotor and subsequently filtered with a 0.22-μm-pore-size filter and applied onto a 20-ml HisPrep nickel column (GE Healthcare, Inc.). The bound GbpC fragments were eluted with a gradient of 0 to 300 mM imidazole after the unbound fractions were thoroughly washed with 50 mM imidazole in the binding buffer. The purest protein fractions were identified on SDS-PAGE gels, pooled, and dialyzed overnight into 20 mM Tris, pH 8.0, and 50 mM sodium chloride, and subsequently loaded onto a MonoQ column (GE Healthcare, Inc.). The purest fractions identified from SDS-PAGE gels were then concentrated under 55 lb/in2 nitrogen gas pressure using an Amicon stirred-cell concentrator. Protein quantitation was measured as described elsewhere (39). The final concentrations of GbpC171–464 and GbpC111–522 were 107.5 and 43.3 mg/ml, respectively, indicating the high solubility of the GbpC fragments.
Crystallization, data collection, structure solution, and refinement.
GbpC171–464 and GbpC111–522 were subjected to crystallization trials in 96-well-format Corning 3550 plates using an Art Robbins Gryphon crystallization robot and commercial screens. GbpC171–464 crystals were obtained after optimization of crystal screen II condition 43; similarly, index screen condition 40 was optimized for GbpC111–522. Therefore, we focused on obtaining the structure of the larger fragment, GbpC111–522. Large crystals of GbpC111–522 were routinely obtained from a droplet containing 2 μl of protein (43.3 mg/ml) and 2 μl of the well solution containing 100 mM bis-Tris, pH 6.5, and 25% (wt/vol) polyethylene glycol (PEG) 3350. Crystals were cryo-frozen in 15% glycerol, and data on both native and potassium iodide quick-soaked crystals were collected at the NE-CAT beamline of the Argonne Photon Source (APS) and subsequently processed with HKL2000 (40). The data collection parameters are summarized in Table 1. The native crystals diffracted to a usable diffraction limit of 1.14 Å. Since our attempts to seek a molecular replacement solution with the V region of AgI/II could not be refined further (every solution converged around an R factor of 35%), despite producing clear peaks, we attempted de novo SIRAS (single isomorphous replacement with anomalous scattering) phasing through the use of the iodide derivative by collecting anomalous diffraction data. SHELX identified 17 iodine sites (41) with a figure of merit (FOM) of 0.78 and a map correlation coefficient of 0.89 at 2.0-Å resolution. Automated structure building with the resulting map was carried out in Phenix (42), which resulted in rebuilding of 95% of the residues within GbpC. Manual model building was done with COOT (43), in particular for the loop region of residues 410 to 418, which was observed to be more flexible. Final refinement parameters are reported in Table 1.
Since cocrystallization with 1,000-Da dextran did not produce any crystals, we attempted soaking GbpC crystals in the presence of dextrose and maltose. The observed additional electron difference density in the metal binding site indicated the presence of ligand molecules but did not allow reliable modeling and refinement of these molecules (data not shown). Therefore, we subjected solutions of 26 mg/ml (500 μM) GbpC and glucose (final concentration, 5 mM) at a molar ratio (ligand/protein) of 10:1 in 20 mM Tris, pH 8.0, 50 mM NaCl, and 1 mM CaCl2 to two commercial screens (Crystal Screen HT and Index HT) using an Art Robbins Gryphon crystallization robot. Manual optimization of index condition 40 resulted in crystals that were grown in 0.1 M citric acid at pH 3.5 and 25% PEG 3350 as a precipitant. Initial data sets were collected on our home source (Rigaku MicroMax-007HF source equipped with a Dectris Pilatus 200K detector), but final data sets were obtained on SERCAT beamline 22ID at APS using a Rayonix MX300HS detector on crystals that were cryo-protected by adding 20% glycerol. Data were reduced using XDS (44, 45), and the structure was resolved through molecular replacement, with subsequent refinement, as described for the native structure. The data collection and final refinement parameters are reported in Table 1, and both coordinates and structure factors have been submitted to the Protein Data Bank (PDB). Both REFMAC (46) and Phenix (42) model-map correlation coefficients (CC) validated the presence of glucose molecules with a CC of 0.80 (0.88 for the entire protein). Furthermore, Polder omit maps in Phenix confirmed the placement of these glucose molecules.
Isothermal titration calorimetry.
The adherence of GbpC with dextran (Sigma-Aldrich, Inc.) was evaluated using the Auto ITC 200 (University of Alabama at Birmingham [UAB] Comprehensive Cancer Center's core facility) as described previously (3, 24). For this experiment we chose five different sizes of dextran with average molecular masses of 1,000, 5,000, 9,400, 38,800, and 70,800 Da and assessed the affinity coefficients. Briefly, the calorimetric cell contained 200 μl of GbpC (0.68 to 2.2 mM) in a buffer containing 20 mM HEPES, 150 mM NaCl, and 2.5 mM CaCl2, and the syringe was loaded with 120 μl of dextran (1 to 30 mM). Following a first trial injection of 1 μl, heat exchange was recorded for 40 injections of 1 μl of dextran at intervals of 300 s. Control injections with buffer, buffer plus calcium chloride, and buffer plus dextran did not produce any measurable heat (data not shown). The results were analyzed using Origin, version 7.0, with the MicroCal ITC analysis module, and these results are shown in Fig. 4.
Surface plasmon resonance.
The ligand salivary agglutinin (SAG) or the insect cell-expressed recombinant scavenger receptor cysteine rich domain 1 (iSRCR1) or iSRCR123, was purified as described previously (2, 24) and immobilized on a carboxymethyl dextran chip (CM5) using an amine coupling procedure, as described previously (2, 3, 24). Briefly, the chip surface was activated using N-hydroxysuccinimide (NHS) and N-ethyl-N′-(3-diethylaminopropyl) carbodiimide (EDC), and the ligand (at 50 μg/ml) in sodium acetate buffer (10 mM, pH 4.0) was injected at a flow rate of 5 μl/min until the desired resonance units were obtained. The remaining activated group and the control were blocked by 1.0 M ethanolamine. After initial trial injections confirmed adherence, the analyte GbpC (0.25 μM to 4 μM) in binding buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 2.5 mM CaCl2) was injected at 20 μl/min, and the association and dissociation phases were monitored. The surface was prepared for the next injection using a nondenaturing regeneration buffer (1 M NaCl and 20 mM EDTA, pH 7.4). SPR experiments carried out with analytes, ligands, and binding buffer devoid of calcium (2 mM EDTA) were used to determine the effect of calcium on GbpC binding with immobilized SAG or iSRCRs. The 1:1 Langmuir model was used to deduce the kinetics parameters of the obtained data from experiments using BIAevaluation software. Following the on-chip deglycosylation procedure as described earlier (24), the role of glycosylation on the adherence of GbpC was measured. All the results are summarized in Table 2, and the sensorgrams are shown in Fig. S9a and b in the supplemental material.
Competition assay.
The adherence competition between AgI/II and GbpC with SAG was determined using a competition assay as described earlier (2, 24). Briefly, the first analyte (GbpC) was flowed over an SAG-immobilized CM5 sensor chip for 1 min to saturate available binding sites. The maximal response curve of GbpC binding was recorded as RU1 (RU is response units), and then the RU1 served as the baseline for the second injected analyte (AgI/II). The response curve of AgI/II was measured as RU2, and then the adherence competition between the two was calculated (RU2 − RU1). Similarly, these analytes were injected in reverse order, and the results were plotted as bar graphs (Fig. 6). The percentage of inhibition was calculated by comparing the binding of the fragment by itself and in the presence of the competing fragment. In control experiments, where either GbpC or AgI/II was immobilized on the CM5 sensor chip and the other protein was run over as the analyte, they did not display any measurable adherence (data not shown).
GbpC, dextran, and SRCR/SAG.
Experiments were designed to determine the adherence of GbpC to SRCR/SAG in the presence of dextran. Various concentrations of 5,000-Da dextran (0, 40, 80, and 160 μM) were mixed with different concentrations of GbpC (1, 2, and 4 μM), and their adherence to immobilized SRCR1 was determined using SPR. The results were plotted as a clustered bar graph, which showed a dose-dependent inhibition by dextran (Fig. 5). When tested with higher concentrations of dextran, the effect plateaued (data not shown) and did not result in complete abrogation of binding of SRCR1.
Dextran-dependent aggregation.
Dextran-induced S. mutans aggregation was shown to be important for sucrose-dependent bacterial adherence and virulence (16). GbpC is defined as a glucan binding protein partly due to its ability to induce bacterial aggregation in the presence of dextran (glucan). AgI/II is sucrose independent but has also been implicated in dextran-induced bacterial aggregation (26). Thus, we examined the contribution of GbpC and AgI/II to this phenotype using a variety of GbpC and AgI/II mutant variants. Briefly, 100 μg/ml of 500,000-Da dextran was added to overnight cultures of S. mutans in 3-ml test tubes, mixed, and then incubated for 30 min to measure bacterial aggregation at an OD600, and the results were plotted as bar graphs (Fig. 10). Statistical analysis was done with Excel's one-way analysis of variance (ANOVA), followed by Tukey's range test to determine the significance of the difference in dextran aggregation between mutants and the wild type.
Construction of mutants and complementation of mutants.
Since the loop region 410VNADGTPRA418 on GbpC (Fig. 3) was distinctly different from the same region in AgI/II and SspB, to determine if the loop region identified in GbpC is important for any biological activity, we constructed an in-frame deletion mutant that deleted amino acid residues 410 to 418 using a well-established markerless site-directed mutagenesis scheme developed for S. mutans (47), as we described earlier (48, 49). Similarly, the loop region of residues 584 to 590 was also deleted in the V region of AgI/II. Briefly, the construction of both the counterselection cassette and mutant alleles was completed using overlapping PCR methods. In the first step of constructing the counterselection cassette, a 0.8-kb fragment upstream of the deletion gene and a 1.0-kb fragment downstream of the deletion gene were PCR amplified with appropriate primer pairs. The counterselection cassette IFDC2 contains a constitutive ldh promoter, a counterselection marker, a pheS mutant open reading frame (ORF) that renders the bacterium sensitive to p-chloro-phenylalanine (p-Cl-Phe), and an antibiotic selection erythromycin cassette (2.2 kb). IFDC2 is PCR amplified with the primer pair ldhF and ermR. Three amplicons containing overlapping regions allowed subsequent overlapping PCR using linker primers. The resulting 4-kb amplicon was transformed into S. mutans UA159, and transformants were selected on brain heart infusion (BHI) plates containing erythromycin. The IFDC2 cassette-disrupted allele in the S. mutans genome was PCR amplified and confirmed by DNA sequencing. For the second step of constructing in-frame deletion mutant alleles, the 0.7-kb upstream fragment and the 1-kb downstream fragment surrounding the deletion gene were generated by PCR using direct-linker primers. One amplicon has a region that overlaps the other amplicon; subsequent overlapping PCR yielded a 1.7-kb amplicon with a deletion of the targeting gene and linked the up- and downstream fragments together. The resulting amplicon was transformed into the counterselective mutant obtained from the first step, and markerless mutants were selected on BHI plates containing p-Cl-Phe that counterselects the mutant that contains the IFDC2 cassette, which conferred the sensitivity to the presence of p-Cl-Phe. The markerless in-frame deletion mutants were confirmed by DNA sequencing of the mutant alleles amplified from selected p-Cl-Phe-resistant mutants by PCR. The full-length genes coding for GbpC and AgI/II were cloned into pVPT E. coli-streptococcal shuttle vector (50) and transformed into either gbpC or AgI/II gene (pac) mutant variants to rescue the activity related to GbpC or AgI/II.
Biofilm assays.
Wild-type S. mutans, various GbpC and AgI/II mutants, including GbpC410–418Δ and AgI/II584–590Δ, and relevant complemented strains were grown in tryptone-soy-yeast extract (TSYE) broth with 1% sucrose for 16 h, after which biofilm biomass was quantified by staining with crystal violet dye, and the optical density was measured at 562 nm, as described previously (51, 52). Results were plotted as bar graphs (Fig. 9).
Accession number(s).
Coordinates and structure factors of native and glucose-bound GbpC were submitted to the Protein Data Bank (https://www.rcsb.org/) under accession numbers 5UQZ and 6CAM, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Jeffrey Banas, University of Iowa, for providing us with a plasmid that encoded the full-length GbpC. J.L.M., M.R.L., N.S., S.P., and C.D. are grateful to all the core facilities at the University of Alabama at Birmingham, including the Comprehensive Cancer Center's Structural Biology shared facility, Heflin Center for Genomic Sciences and Proteomics and Mass Spectrometry.
This work is based upon research conducted at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline and at the Northeastern Collaborative Access Team (NE-CAT) 24-ID-C and 24-ID-E beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). This research used resources of the Advanced Photon Source. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract number W-31-109-Eng-38. J.L.M. is a T-90 fellow and is supported by the NIH/NIDCR T-90DE022736-06; J.L.M. (previously) and C.D. were partially supported through NIH/NIAID R21AI105038. R.W. and H.W. were partially supported by NIH/NIDCR grants R01DE022350 and R01DE017954.
We declare that we have no conflicts of interest.
J.L.M. conducted all ITC and SPR experiments and resolved the cocrystal structure with N.S. and cowrote the manuscript; M.L.R. cloned, expressed, purified, and crystallized the GbpC fragments and cowrote the manuscript. S.P. initially contributed to ITC and BIAcore experiments, R.W. carried out mutational analysis and biofilm studies, K.R.R. collected X-ray data and obtained initial maps of the structure, H.W. conceived all biologically relevant data and cowrote manuscript, and C.D. conceived ideas, grew crystals, collected data, refined the structures, and prepared the manuscript for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00146-18.
REFERENCES
- 1.Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol 54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 2.Larson MR, Rajashankar KR, Crowley PJ, Kelly C, Mitchell TJ, Brady LJ, Deivanayagam C. 2011. Crystal structure of the C-terminal region of Streptococcus mutans antigen I/II and characterization of salivary agglutinin adherence domains. J Biol Chem 286:21657–21666. doi: 10.1074/jbc.M111.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larson MR, Rajashankar KR, Patel MH, Robinette RA, Crowley PJ, Michalek S, Brady LJ, Deivanayagam C. 2010. Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices. Proc Natl Acad Sci U S A 107:5983–5988. doi: 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banas JA. 2004. Virulence properties of Streptococcus mutans. Front Biosci 9:1267–1277. doi: 10.2741/1305. [DOI] [PubMed] [Google Scholar]
- 5.Haas W, Banas JA. 1997. The glucan binding domain of the Streptococcus mutans glucan binding protein. Adv Exp Med Biol 418:707–708. doi: 10.1007/978-1-4899-1825-3_165. [DOI] [PubMed] [Google Scholar]
- 6.Staat RH, Langley SD, Doyle RJ. 1980. Streptococcus mutans adherence: presumptive evidence for protein-mediated attachment followed by glucan-dependent cellular accumulation. Infect Immun 27:675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banas JA, Vickerman MM. 2003. Glucan-binding proteins of the oral streptococci. Crit Rev Oral Biol Med 14:89–99. doi: 10.1177/154411130301400203. [DOI] [PubMed] [Google Scholar]
- 8.Banas JA, Russell RR, Ferretti JJ. 1990. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutans Ingbritt. Infect Immun 58:667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell RR. 1979. Glucan-binding proteins of Streptococcus mutans serotype C. J Gen Microbiol 112:197–201. doi: 10.1099/00221287-112-1-197. [DOI] [PubMed] [Google Scholar]
- 10.Smith DJ, Akita H, King WF, Taubman MA. 1994. Purification and antigenicity of a novel glucan-binding protein of Streptococcus mutans. Infect Immun 62:2545–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah DS, Russell RR. 2004. A novel glucan-binding protein with lipase activity from the oral pathogen Streptococcus mutans. Microbiology 150:1947–1956. doi: 10.1099/mic.0.26955-0. [DOI] [PubMed] [Google Scholar]
- 12.Sato Y, Yamamoto Y, Kizaki H. 1997. Cloning and sequence analysis of the gbpC gene encoding a novel glucan-binding protein of Streptococcus mutans. Infect Immun 65:668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Igarashi T, Asaga E, Goto N. 2003. The sortase of Streptococcus mutans mediates cell wall anchoring of a surface protein antigen. Oral Microbiol Immunol 18:266–269. doi: 10.1034/j.1399-302X.2003.00076.x. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto M, Fujita K, Ooshima T. 2006. Binding of glucan-binding protein C to GTFD-synthesized soluble glucan in sucrose-dependent adhesion of Streptococcus mutans. Oral Microbiol Immunol 21:42–46. doi: 10.1111/j.1399-302X.2005.00252.x. [DOI] [PubMed] [Google Scholar]
- 15.Nomura R, Nakano K, Ooshima T. 2004. Contribution of glucan-binding protein C of Streptococcus mutans to bacteremia occurrence. Arch Oral Biol 49:783–788. doi: 10.1016/j.archoralbio.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Lynch DJ, Michalek SM, Zhu M, Drake D, Qian F, Banas JA. 2013. Cariogenicity of Streptococcus mutans glucan-binding protein deletion mutants. Oral Health Dent Manag 12:191–199. [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch DJ, Fountain TL, Mazurkiewicz JE, Banas JA. 2007. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol Lett 268:158–165. doi: 10.1111/j.1574-6968.2006.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsgren N, Lamont RJ, Persson K. 2009. Crystal structure of the variable domain of the Streptococcus gordonii surface protein SspB. Protein Sci 18:1896–1905. doi: 10.1002/pro.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Troffer-Charlier N, Ogier J, Moras D, Cavarelli J. 2002. Crystal structure of the V-region of Streptococcus mutans antigen I/II at 2.4 Å resolution suggests a sugar preformed binding site. J Mol Biol 318:179–188. doi: 10.1016/S0022-2836(02)00025-6. [DOI] [PubMed] [Google Scholar]
- 20.Perozzo R, Folkers G, Scapozza L. 2004. Thermodynamics of protein-ligand interactions: history, presence, and future aspects. J Recept Signal Transduct Res 24:1–52. doi: 10.1081/RRS-120037896. [DOI] [PubMed] [Google Scholar]
- 21.Brown A. 2009. Analysis of cooperativity by isothermal titration calorimetry. Int J Mol Sci 10:3457–3477. doi: 10.3390/ijms10083457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dam TK, Talaga ML, Fan N, Brewer CF. 2016. Measuring multivalent binding interactions by isothermal titration calorimetry. Methods Enzymol 567:71–95. doi: 10.1016/bs.mie.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Xin H, Qu Y, Wu H, Yu P, Zhang Y. 2016. Univariate and multi-variate comparisons of protein and carbohydrate molecular structural conformations and their associations with nutritive factors in typical by-products. J Sci Food Agric 96:4736–4748. doi: 10.1002/jsfa.7791. [DOI] [PubMed] [Google Scholar]
- 24.Purushotham S, Deivanayagam C. 2014. The calcium-induced conformation and glycosylation of scavenger-rich cysteine repeat (SRCR) domains of glycoprotein 340 influence the high affinity interaction with antigen I/II homologs. J Biol Chem 289:21877–21887. doi: 10.1074/jbc.M114.565507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace AC, Laskowski RA, Thornton JM. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 26.Zhu M, Ajdic D, Liu Y, Lynch D, Merritt J, Banas JA. 2009. Role of the Streptococcus mutans irvA gene in GbpC-independent, dextran-dependent aggregation and biofilm formation. Appl Environ Microbiol 75:7037–7043. doi: 10.1128/AEM.01015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biswas I, Drake L, Biswas S. 2007. Regulation of gbpC expression in Streptococcus mutans. J Bacteriol 189:6521–6531. doi: 10.1128/JB.00825-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto-Shibayama K, Sato Y, Yamamoto Y, Ohta K, Kizaki H. 2006. Identification of a glucan-binding protein C gene homologue in Streptococcus macacae. Oral Microbiol Immunol 21:32–41. doi: 10.1111/j.1399-302X.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- 29.Matsumura M, Izumi T, Matsumoto M, Tsuji M, Fujiwara T, Ooshima T. 2003. The role of glucan-binding proteins in the cariogenicity of Streptococcus mutans. Microbiol Immunol 47:213–215. doi: 10.1111/j.1348-0421.2003.tb03389.x. [DOI] [PubMed] [Google Scholar]
- 30.Nakano K, Matsumura M, Kawaguchi M, Fujiwara T, Sobue S, Nakagawa I, Hamada S, Ooshima T. 2002. Attenuation of glucan-binding protein C reduces the cariogenicity of Streptococcus mutans: analysis of strains isolated from human blood. J Dent Res 81:376–379. doi: 10.1177/0810376. [DOI] [PubMed] [Google Scholar]
- 31.Sato Y, Yamamoto Y, Kizaki H. 2000. Xylitol-induced elevated expression of the gbpC gene in a population of Streptococcus mutans cells. Eur J Oral Sci 108:538–545. doi: 10.1034/j.1600-0722.2000.00928.x. [DOI] [PubMed] [Google Scholar]
- 32.Niu G, Okinaga T, Zhu L, Banas J, Qi F, Merritt J. 2008. Characterization of irvR, a novel regulator of the irvA-dependent pathway required for genetic competence and dextran-dependent aggregation in Streptococcus mutans. J Bacteriol 190:7268–7274. doi: 10.1128/JB.00967-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heim KP, Sullan RM, Crowley PJ, El-Kirat-Chatel S, Beaussart A, Tang W, Besingi R, Dufrene YF, Brady LJ. 2015. Identification of a supramolecular functional architecture of Streptococcus mutans adhesin P1 on the bacterial cell surface. J Biol Chem 290:9002–9019. doi: 10.1074/jbc.M114.626663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deivanayagam CC, Wann ER, Chen W, Carson M, Rajashankar KR, Hook M, Narayana SV. 2002. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J 21:6660–6672. doi: 10.1093/emboj/cdf619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganesh VK, Barbu EM, Deivanayagam CC, Le B, Anderson AS, Matsuka YV, Lin SL, Foster TJ, Narayana SV, Hook M. 2011. Structural and biochemical characterization of Staphylococcus aureus clumping factor B/ligand interactions. J Biol Chem 286:25963–25972. doi: 10.1074/jbc.M110.217414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rich RL, Deivanayagam CC, Owens RT, Carson M, Hook A, Moore D, Symersky J, Yang VW, Narayana SV, Hook M. 1999. Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, α1β1 integrin and Staphylococcus aureus Cna MSCRAMM. J Biol Chem 274:24906–24913. doi: 10.1074/jbc.274.35.24906. [DOI] [PubMed] [Google Scholar]
- 37.Hoyer LL, Cota E. 2016. Candida albicans agglutinin-like sequence (Als) family vignettes: a review of Als protein structure and function. Front Microbiol 7:280. doi: 10.3389/fmicb.2016.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato Y, Senpuku H, Okamoto K, Hanada N, Kizaki H. 2002. Streptococcus mutans binding to solid phase dextran mediated by the glucan-binding protein C. Oral Microbiol Immunol 17:252–256. doi: 10.1034/j.1399-302X.2002.170408.x. [DOI] [PubMed] [Google Scholar]
- 39.Gill SC, von Hippel PH. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 40.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 41.Sheldrick GM. 2015. Crystal structure refinement with SHELXL. Acta Crystallogr C Struct Chem 71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Kabsch W. 2010. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr 66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Xie Z, Okinaga T, Qi F, Zhang Z, Merritt J. 2011. Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl Environ Microbiol 77:8025–8033. doi: 10.1128/AEM.06362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng X, Michalek S, Wu H. 2016. Effects of diadenylate cyclase deficiency on synthesis of extracellular polysaccharide matrix of Streptococcus mutans revisit. Environ Microbiol 18:3612–3619. doi: 10.1111/1462-2920.13440. [DOI] [PubMed] [Google Scholar]
- 49.Peng X, Zhang Y, Bai G, Zhou X, Wu H. 2016. Cyclic di-AMP mediates biofilm formation. Mol Microbiol 99:945–959. doi: 10.1111/mmi.13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H, Zhu F, Yang T, Ding L, Zhou M, Li J, Haslam SM, Dell A, Erlandsen H, Wu H. 2014. The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold. Nat Commun 5:4339. doi: 10.1038/ncomms5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Q, Nguyen T, McMichael M, Velu SE, Zou J, Zhou X, Wu H. 2015. New small-molecule inhibitors of dihydrofolate reductase inhibit Streptococcus mutans. Int J Antimicrob Agents 46:174–182. doi: 10.1016/j.ijantimicag.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu F, Zhang H, Yang T, Haslam SM, Dell A, Wu H. 2016. Engineering and dissecting the glycosylation pathway of a streptococcal serine-rich repeat adhesin. J Biol Chem 291:27354–27363. doi: 10.1074/jbc.M116.752998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.