The Chlamydomonas reinhardtii LTD null mutant generated by CRISPR–Cas9, displayed aberrant PSI–LHCI holocomplexes, suggesting that the LTD protein may selectively function in PSI–LHCI assembly in green microalgae
Keywords: Chlamydomonas reinhardtii, chloroplast signal recognition particle pathway, CRISPR–Cas9, LHCP translocation defect (LTD), light-harvesting chlorophyll- and carotenoid-binding proteins, photosystem I
Abstract
Nuclear-encoded light-harvesting chlorophyll- and carotenoid-binding proteins (LHCPs) are imported into the chloroplast and transported across the stroma to thylakoid membrane assembly sites by the chloroplast signal recognition particle (CpSRP) pathway. The LHCP translocation defect (LTD) protein is essential for the delivery of imported LHCPs to the CpSRP pathway in Arabidopsis. However, the function of the LTD protein in Chlamydomonas reinhardtii has not been investigated. Here, we generated a C. reinhardtii ltd (Crltd) knockout mutant by using CRISPR–Cas9, a new target-specific knockout technology. The Crltd1 mutant showed a low chlorophyll content per cell with an unusual increase in appressed thylakoid membranes and enlarged cytosolic vacuoles. Profiling of thylakoid membrane proteins in the Crltd1 mutant showed a more severe reduction in the levels of photosystem I (PSI) core proteins and absence of functional LHCI compared with those of photosystem II, resulting in a much smaller PSI pool size and diminished chlorophyll antenna size. The lack of CrLTD did not prevent photoautotrophic growth of the cells. These results are substantially different from those for Arabidopsis ltd null mutant, indicating LTD function in LHCP delivery and PSI assembly may not be as stringent in C. reinhardtii as it is in higher plants.
Introduction
Photosynthesis relies on the linear coordinate function of two photosystems (PSI and PSII), which are light-absorbing complexes in the thylakoid membrane of photosynthetic organisms (Taiz and Zeiger, 2010). Light-harvesting chlorophyll- and carotenoid-binding proteins (LHCPs) contain the photosynthetic pigments and function to absorb the energy of sunlight and transfer it to the reaction centers of the photosystems. LHCPs are also responsible for dissipating excess energy that can be harmful to the photosynthetic apparatus (Wobbe et al., 2016). LHCPs include LHCA and LHCB, which form the light-harvesting complex I (LHCI) associated with PSI and the light-harvesting complex II (LHCII) associated with PSII, respectively. Since the LHC gene family is encoded in the nuclear genome (Jansson, 1999; Stauber et al., 2003), LHCPs are synthesized in the cytosol and are then imported into the chloroplast. Once an LHCP is inside the chloroplast, it is transported across the stroma to its final destination, the thylakoid membrane assembly sites, by the so-called chloroplast signal recognition particle (CpSRP) pathway (Henry, 2010; Richter et al., 2010).
In the higher plant Arabidopsis, CpSRP pathway components such as CpSRP54 and CpSRP43 associate with imported LHCPs and form a transit complex to maintain solubility of the hydrophobic LHCPs (Li et al., 1995; Schuenemann et al., 1998; Tzvetkova-Chevolleau et al., 2007). CpSRP54 binding to the LHCPs increases the affinity of CpSRP43 for LHCP binding (Liang et al., 2016). When the transit CpSRP54–LHCP–CpSRP43 complex reaches the thylakoid membrane surface, CpSRP43 interacts with the C-terminal tail of the ALB3 translocase, causing the CpSRP43 to release the LHCP enabling insertion of the LHCP into the developing thylakoid membrane (Dünschede et al., 2011; Horn et al., 2015; Liang et al., 2016). Recruitment of ALB3 requires the CpSRP receptor CpFTSY (Moore et al., 2003; Asakura et al., 2008). CpFTSY is activated by anionic phospholipids and forms a transient complex with CpSRP54, thereby facilitating the binding of the ALB3 to the CpSRP54–LHCP–CpFTSY complex (Chandrasekar and Shan, 2017).
In addition to the above LHCP transport proteins, the LHCP translocation defect (LTD) protein has been reported to function in the stroma of chloroplasts (Cui et al., 2011; Ouyang et al., 2011). Upon the LHCP release from the Tic-Toc chloroplast envelope translocon complex, LTD first binds to the third transmembrane domain of the LHCP and delivers it to CpSRP43. An Arabidopsis ltd null mutant showed a drastic lowering of total chlorophyll (Chl) and LHCP and no growth under photoautotrophic conditions. These severe phenotypes suggested a critical role of the LTD protein in LHCP trafficking (Cui et al., 2011; Ouyang et al., 2011).
The genes of the CpSRP pathway proteins are present not only in higher plants but also in various eukaryotic photosynthetic microorganisms, ranging from chlorophytes to heterokonts. In the green microalga Chlamydomonas reinhardtii, the homologs of most CpSRP components have been identified and null mutants of each CpSRP protein have been characterized (Bellafiore et al., 2002; Ossenbühl et al., 2004; Göhre et al., 2006; Kirst et al., 2012a, b; Jeong et al., 2017). However, to the best of our knowledge, no LTD deletion mutants have been reported in green algae. Here, we generated an LTD null mutant in C. reinhardtii (Crltd) by using the CRISPR–Cas9 method, a new target-specific knockout technology that has recently been successfully applied to C. reinhardtii (Baek et al., 2016). In the Crltd1 mutant, total Chl content was decreased to 33% of the wild-type level, but the Chl a/b ratio was not changed. The level of the PSI-LHCI complex in the mutant was severely reduced, to a far greater extent than that of the PSII-LHCII complex. Concomitantly, the ultrastructure of the thylakoid membranes was drastically altered. These results suggest that the CrLTD is specifically involved in the transport and assembly of the PSI-LHCI complexes, and that the associated Chlamydomonas CpSRP pathway may act differently in microalgae than in higher plants.
Materials and methods
Cell growth conditions
Chlamydomonas reinhardtii wild-type strain CC-4349 cw15 mt− (Baek et al., 2016; Jeong et al., 2017) and CRISPR–Cas9-induced knockout mutants were cultivated mixotrophically in Tris–acetate–phosphate (TAP) medium, or photoautotrophically in Tris–bicarbonate–phosphate (TBP), or high-salt medium (Harris, 1989) with continuous air bubbling under continuous illumination (50 or 100 μmol photons m−2 s−1) at 25 °C. For the growth analysis of cells, cultures were photoautotrophically grown in 400 ml Tris–phosphate medium with light intensities ranging from 100 to 350 μmol photons m−2 s−1. A 500 ml laboratory glass bottle (Duran) was employed with continuous 3% CO2 bubbling to avoid carbon limitation. The cell growth rate (μ) was measured during the exponential growth phase from the increase in cell number as a function of time, following the method described in Levasseur et al. (1993).
CRISPR–Cas9 driven mutagenesis
All procedures were performed according to Baek et al. (2016) and Yu et al. (2017). Recombinant Cas9 protein (200 μg) and in vitro transcribed single guide RNA (sgRNA; 140 μg) were mixed and preincubated for 10 min at room temperature. Then, Chlamydomonas cells (5 × 105 cells) were transformed with the ribonucleoprotein (RNP) complex in a Gene Pulser Xcell Electroporation System (Bio-Rad). After transformation, cells were diluted and plated on TAP medium containing 1.5% agar to obtain single colonies for further investigation. For genotype characterization, genomic DNA was extracted as described in Jeong et al. (2017) and DNA segments were analysed as given by Baek et al. (2016) using Cas-Analyzer (Park et al., 2017) and Cas-OFFinder (Bae et al., 2014).
Cell counting and chlorophyll determination
Cells in liquid media were counted with a Neubauer Bright Line hemocytometer and an Olympus CH30 microscope. The Chl content was determined spectrophotometrically in 100% (v/v) methanol extracts according to Holden (1976).
Transmission electron microscopy
Cells were fixed with cold 5% glutaraldehyde buffered with 0.2 M sodium cacodylate at pH 6.8 for 1 h at 4 °C, and pre-embedded in 1% agar dissolved in distilled water. After solidification, they were post-fixed with sodium cacodylate buffer containing 1% OsO4 and 0.8% potassium ferricyanide for 1 h at 4 °C, and dehydrated at 4 °C using graded ethanol (from 50% to 100%). Specimens were then brought to room temperature and transferred through propylene oxide and Spurr’s embedding resin (Spurr, 1969) in propylene oxide. The specimens were moved to new pure resin and polymerized at 70 °C. Polymerized blocks were thin-sectioned using a PT-X ultramicrotome (RMC Boeckeler). Sections were collected on 0.25% (w/v) formvar-coated slot copper grids, stained with 3% (w/v) uranyl acetate and Reynold’s lead citrate (Reynolds, 1963), and examined and photographed using a JEM-1010 transmission electron microscope operated at 80 kV (JEOL). Images were recorded on Kodak EM Film 4489 and scanned to tagged image file format using an Epson Perfection V700 Photo scanner.
Measurements of photosynthetic activity
Oxygen evolution was measured with a Clark-type oxygen electrode following the method described in Jeong et al. (2017) with the modification of illumination with incandescent light ranging from 25 to 1200 μmol photons m−2 s−1.
Spectrophotometric and kinetic analysis
Thylakoid membranes were isolated as described in Kirst et al. (2012a). Spectrophotometric measurements of the amplitude of the light-minus-dark absorbance difference signal at 700 nm (P700) for PSI and 320 nm (QA) for PSII were used to estimate the concentration of the photosystems in thylakoid membranes (Melis and Brown, 1980; Melis, 1989). Extinction coefficients at 700 nm (P700) were used as given by Hiyama and Ke (1972) and at 320 nm (QA) as given by van Gorkom (1974). The kinetics of P700 photo-oxidation and QA photoreduction of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU)-poisoned thylakoids were measured under weak but continuous green actinic excitation. First-order photoconversion kinetics were used to estimate the functional light-harvesting Chl antenna size of PSI and PSII (Melis and Thielen, 1980; Thielen and van Gorkom 1981; Melis, 1989, 1990, 1991). More specifically, the functional light-harvesting chlorophyll antenna size of Chlamydomonas PSI and PSII was measured from the first-order rate constants of P700 photo-oxidation and QA photoreduction, conducted upon weak continuous green actinic illumination of isolated thylakoid membranes (Polle et al., 2000). For the functional PSII Chl antenna size, thylakoids were suspended in the presence of 10 μM DCMU, thereby blocking electron transport from QA to QB and the plastoquinone pool. For the functional PSI Chl antenna size, thylakoid membranes were suspended in the presence of DCMU, 200 μM potassium ferricyanide, and 100 μM methylviologen. The presence of ferricyanide ensured oxidation of the electron carriers between the two photosystems (e.g. the Rieske Fe–S center, cytochrome f, and plastocyanin), whereas methyl viologen acted as an efficient electron acceptor from the reducing side of PSI.
SDS-PAGE and western blot analysis
SDS-PAGE analysis was carried out according to Laemmli (1970). Proteins were loaded on the basis of equal cell number. After protein separation, gels were stained with Coomassie Blue or blotted onto a polyvinylidene difluoride membrane (ATTO) in a semi-dry transfer system. Membranes were probed with antibodies against thylakoid membrane proteins. Immunodetection was performed using antibodies against LHCAs (Petroutsos et al., 2011), LHCBs (Agrisera), PS core proteins (Agrisera), PetA (Agrisera), Atpβ (Agrisera), CrCpFTSY (Kirst et al., 2012a), CrCpSRP43 (Kirst et al., 2012b) and CrCpSRP54 (Jeong et al., 2017). Polyclonal antibodies specific for CrLTD were generated in rabbit against two LTD peptides, CNFFKFGKNGFDSEAAGIVGS and GIVGSQGRDEYTYDDVEQYF (Abfrontier). Signals were visualized by using the WestSaveUp ECL Reagent (Abfrontier) and exposing the membranes to X-ray film. The National Institutes of Health ImageJ 1.48 software (https://imagej.nih.gov/ij/) was used for quantification of protein bands. To measure protein concentration, the DC Protein Assay kit (Bio-Rad) was used.
Native Deriphat-PAGE and two-dimensional electrophoresis
Thylakoid membranes were solubilized at a Chl concentration of 0.5 mg ml−1 with n-dodecyl-α-D-maltoside (final concentration 1%), incubated on ice for 10 min and centrifuged at 20000 g for 10 min to remove unsolubilized material. Thylakoid membrane proteins (25 µg Chl per lane) were separated by gradient Deriphat-PAGE; the running gel had an acrylamide concentration gradient from 3.5 to 10.5% (w/v) (29:1 acrylamide–bisacrylamide) containing 12 mM Tris–HCl pH 8.5, 48 mM glycine, and a glycerol gradient from 10 to 14% (w/v). The stacking gel had 3.5% (w/v) acrylamide, 12 mM Tris–HCl pH 8.5, 48 mM glycine, and 10% (w/v) glycerol. The electrophoresis anode buffer was 12 mM Tris–HCl pH 8.3, 96 mM glycine. The cathode buffer had the same components as the anode buffer except for the addition of 0.1% (w/v) Deripat-160 (Cognis). The gel was electrophoresed at 50 V constant voltage overnight. For two-dimensional electrophoresis analysis, proteins were extracted from one-dimensional native Deriphat-PAGE strips by soaking them in SDS-PAGE stacking buffer containing 5 M urea twice for 25 min each and resolved by denaturing in a 12% SDS-polyacrylamide gel containing 2 M urea (second dimension). Acrylamide gels were stained with Coomassie Blue.
Results
Targeted ltd gene knockout using CRISPR–Cas9 technology
The Chlamydomonas reinhardtii ltd gene (Cre12.g551950) contains one exon, which is 504 bp in length, and encodes a protein of 167 amino acids including a 38-amino-acid- long chloroplast transit peptide predicted by Predalgo software (Tardif et al., 2012). The CrLTD protein shares 54% identity and 70% similarity with its Arabidopsis homolog. It also contains an ankyrin domain (amino acids 105–137), already identified in Arabidopsis LTD (Cui et al., 2011; Ouyang et al., 2011). Chlamydomonas reinhardtii LTD knockout strains (Crltd) were generated by CRISPR–Cas9 methodology, comprising small insertions and deletions (indels) in this gene.
Four kinds of single guide RNAs (sgRNA) were designed using the Cas-Designer (Park et al., 2015) to recognize and cleave the target gene (see Supplementary Table S1 at JXB online). Then, preassembled sgRNA and the Cas9 protein forming a CRISPR–Cas9 ribonucleoprotein (RNP) complex were transfected into the C. reinhardtii by electroporation. Because the Arabidopsis ltd knockout mutant has yellow leaves and lower Chl content than the wild-type (Ouyang et al., 2011), Chlamydomonas ltd deletion mutants were initially selected on the basis of pale green coloration of transformant colonies (Supplementary Fig. S1). In a second screening step, the ltd gene knockout was confirmed by Sanger sequencing. Out of 388 colonies, four ltd knockout mutants were isolated, which was calculated to represent a 1.12% transformation efficiency.
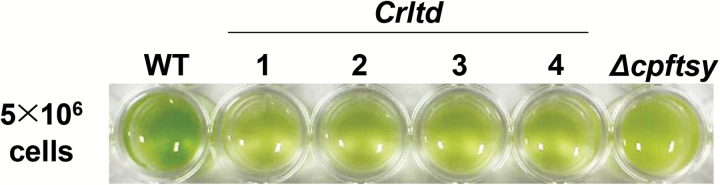
All ltd knockout mutants were isolated by using sgRNA 1 (see Supplementary Table S2). At the same cell density in liquid culture, the coloration of these mutants was lighter green compared with the wild-type and was similar to that of the Δcpftsy mutant (Baek et al., 2016), which was used as a positive control in this study (Fig. 1). A variety of indel mutations were detected in the ltd locus of the four ltd knockout mutants (Fig. 2A; Supplementary Fig. S2), specifically so in the start codon of the ltd gene, which was targeted by the sgRNA 1. To test the expression of the LTD protein in the ltd stains, western blot analysis was performed with polyclonal antibodies raised against the LTD protein, the wild-type of which is a putative 18 kD protein (Fig. 2B). No protein–antibody cross-reaction signal could be detected in the 17–28 kDa region in any of the ltd stains. Therefore, all ltd strains showed the same phenotype in terms of lower pigmentation and lack of the LTD protein (Fig. 1). Absence of off-target mutations was examined by targeted deep sequencing (Supplementary Table S3). No indels were found at potential off-target sites that differed from on-target site by up to four nucleotides. A Chlamydomonas ltd mutant, designated as Crltd1, strain was selected for further investigation in this work.
Fig. 1.
Coloration of C. reinhardtii wild-type (WT), Crltd strains 1–4, and the Δcpftsy strain. Cells were grown in TBP liquid media. At the same cell density (5 × 106 cells ml−1), Crltd and Δcpftsy mutants showed a lighter green coloration, whereas the wild-type was dark green. The Δcpftsy strain, one of the light-harvesting antenna mutants generated by CRISPR–Cas9-mediated mutagenesis in our previous study (Baek et al., 2016), was used as a control for comparison purposes.
Fig. 2.
CRISPR–Cas9-mediated ltd gene disruption in C. reinhardtii. (A) Alignment of the DNA sequences of the wild-type and the Crltd mutants at the ltd locus. All mutants induced by CRISPR–Cas9 had small insertions and deletions (indels) in the ltd gene, which disrupted the start codon. The ATG start codon of the wild-type is indicated by the red box. (B) Coomassie-stained SDS-PAGE and western blot analysis with CrLTD-specific antibodies revealed that CRISPR–Cas9-induced mutations resulted in a deletion of the LTD protein in Crltd strains. Protein loading: 10 µg per lane. (This figure is available in color at JXB online.)
Cell growth analysis of the wild-type and Crltd1 mutant
The rate of growth of the wild-type and Crltd1 mutants was measured in photoautotrophic media under different light intensities (Table 1; Supplementary Fig. S3). At 100 μmol photons m−2 s−1 irradiance, Crltd1 showed a growth rate of 1.37 ± 0.26 d−1, which was lower than that of wild-type (1.93 ± 0.03 d−1, Table 1). This retarded growth rate of the Crltd1 strain is attributed to slower rates of light absorption, as compared with that of the wild-type (see below), consistent with the possibility that the Crltd1 mutant possesses a truncated light harvesting antenna size. When the cells were exposed to a higher light intensity, i.e. 350 μmol photons m−2 s−1, the wild-type and Crltd1 strains exhibited similar growth rates of 2.22 ± 0.07 and 2.03 ± 0.02 d−1, respectively, meaning that both strains harvested enough light energy to grow with similar rates. Furthermore, after 60 h of growth, the cell densities of the Crltd1 mutant were 1.19-fold higher than that of the wild-type (Table 1).
Table 1.
Growth characteristics of the wild-type and Crltd1 mutant photoautotrophically grown at different light conditions
| 100 μmol photons m−2 s−1 | 350 μmol photons m−2 s−1 | |||
|---|---|---|---|---|
| Parameter measured | Wild-type | Crltd1 | Wild-type | Crltd1 |
| Growth rate (μ d−1) | 1.93 ± 0.03 | 1.37 ± 0.26 | 2.22 ± 0.07 | 2.03 ± 0.02 |
| Cell density after 60 h of growth (107 cells ml−1) | 2.4 ± 0.37 | 1.39 ± 0.07 | 3.56 ± 0.22 | 4.25 ± 0.16 |
Values shown are means±SD (n=3).
Chl content and composition in wild-type and the Crltd1 mutant
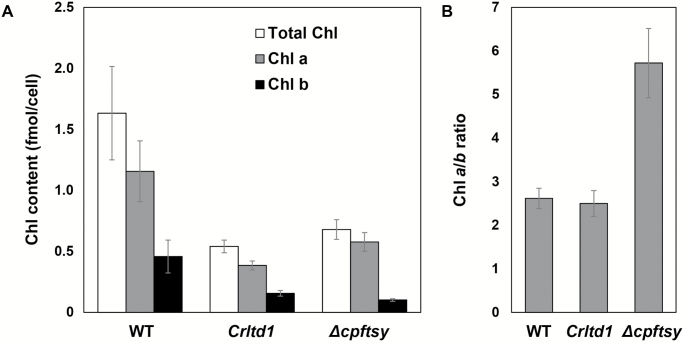
Chl content and composition of the wild-type, the Δcpftsy strain, and the Crltd1 strain were measured in cultures grown photoautotrophically (Fig. 3). Results were essentially the same when cells were grown photoheterotrophically with supplemental organic carbon (results not shown). In the Crltd1 strain, total Chl content per cell (0.54 fmol cell−1) was about 33% of that in the wild-type (1.63 fmol cell−1). However, the average Chl a/b ratio of the Crltd1 strain (2.5 ± 0.3) was similar to that of the wild-type (2.62 ± 0.24). These pigmentation characteristics are unusual and unique for the Crltd1 strain because the Δcpftsy mutant, as well as other Chl-deficient mutants examined so far (Kirst and Melis, 2014; Kirst et al., 2017), exhibited lower Chl contents and a substantially elevated Chl a/b ratio in comparison with the wild-type (Fig. 3).
Fig. 3.
Total chlorophyll (Chl) content (A), and Chl a/b ratios (B) of the wild-type (WT) and the Crltd1 and Δcpftsy strains (n≥3; values shown are means±SD). Note the lower Chl contents in Crltd1 compared with the wild-type. The Chl a/b ratio of Crltd1 was similar to that of the wild-type.
For example, the Δcpftsy mutant contained 0.58 fmol Chl a per cell (50% of the wild-type level) and 0.1 fmol Chl b per cell (22% of the wild-type level) (Fig. 3A) (see also Kirst et al., 2012a;Baek et al., 2016). The differential lowering of Chl a and b levels resulted in a much higher Chl a/b ratio in the Δcpftsy strain (Fig. 3B). In the Crltd1 strain, the content of both Chl a and b was lowered proportionally, resulting in no discernable change in the Chl a/b ratio (Fig. 3B).
Cell ultrastructure in wild-type and the Crltd1 mutant
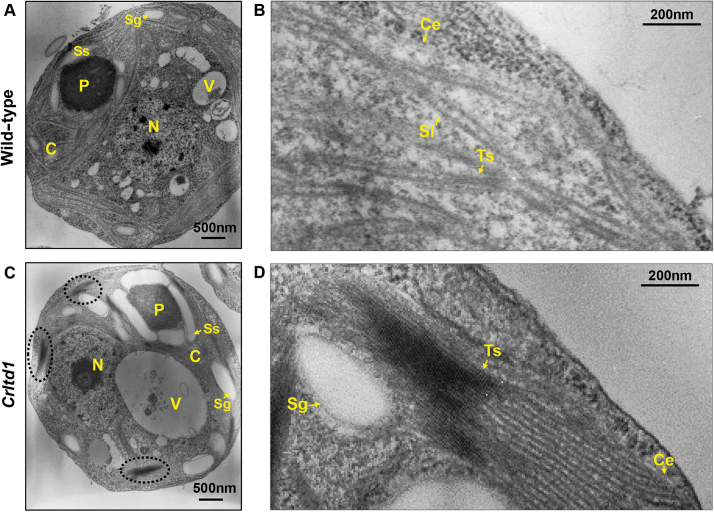
To investigate the cellular and subcellular properties of the Chlamydomonas ltd mutant, we performed transmission electron microscopy analysis. In both the wild-type and Crltd1 grown photoheterotrophically, the chloroplast was observed to surround the nucleus and vacuole (Fig. 4A, C). Interestingly, in the Crltd1 mutant, the vacuole was exceptionally large. The size of the vacuoles in seven to ten randomly selected cell images was 6 ± 2.53% of the total cell area in the wild-type and 10.45 ± 4.38% in the mutant cells. These values were significantly different from each-other (Student’s t-test, P<0.05). The wild-type thylakoid membranes formed stacks of two or more thylakoids and stroma lamellae composed of single thylakoids (Fig. 4B). On the other hand, extensive and dense grana-like thylakoid membrane layers were observed in the mutant with substantially smaller number of stroma-exposed lamellae (Fig. 4D).
Fig. 4.
Transmission electron microscopy images of the cross-sections from wild-type (A, B) and Crltd1 (C, D). N, nucleus; V, vacuole; C, chloroplast; Ce, chloroplast envelope; P, pyrenoid; Ss, starch sheath; Sg, starch granule; Ts, thylakoid stacks; Sl, stroma lamellae. Crltd1 showed over-stacked thylakoid membranes (dotted circles in C) and an exceptionally large cytosolic vacuole. (This figure is available in color at JXB online.)
Photosynthetic activity of wild-type and Crltd1 mutant
Changes in photosynthetic pigment accumulation and structure of thylakoids may have affected the photosynthesis of the mutant. To investigate the function of the photosynthetic apparatus, we measured the light-saturation curves of photosynthesis by the oxygen evolution of cells grown photoautotrophically. In the dark, the rate of respiration of the Crltd1 mutant was similar to that of the wild-type (Table 2). In the 20–100 μmol photons m−2 s−1 range of incident intensity, the mutant and the wild-type displayed similar rates of oxygen evolution per Chl per second (Fig. 5). However, on a per cell basis, the photosynthetic activity of the mutant was about 50% of that in the wild-type under the above light-limiting conditions. Therefore, under light-limiting conditions, absence of the ltd gene had no noticeable effect on the per Chl rate or quantum yield of photosynthesis but it negatively affected the photosynthetic capacity of the cells.
Table 2.
Photosynthesis and respiration characteristics of wild-type and the Crltd1 mutant grown photoautotrophically
| Parameter | Wild-type | Crltd |
|---|---|---|
| Respiration (mmol oxygen (mol Chl)−1 s−1) | 6.89 ± 2.14 | 7.48 ± 3.57 |
| Respiration (fmol oxygen (106 cells)−1 s−1) | 3.66 ± 0.46 | 2.19 ± 1.10 |
| P max (mmol oxygen (mol Chl)−1 s−1) | 47.8 ± 1.87 | 108.08 ± 11.97 |
| P max (fmol oxygen (106 cell)−1 s−1) | 33.22 ± 2.42 | 31.59 ± 2.34 |
| Half-saturation intensity (mmol photons m−2 s−1) | 289.01 ± 13.62 | 380.49 ± 20.54 |
Values shown are means±SD (n≥3).
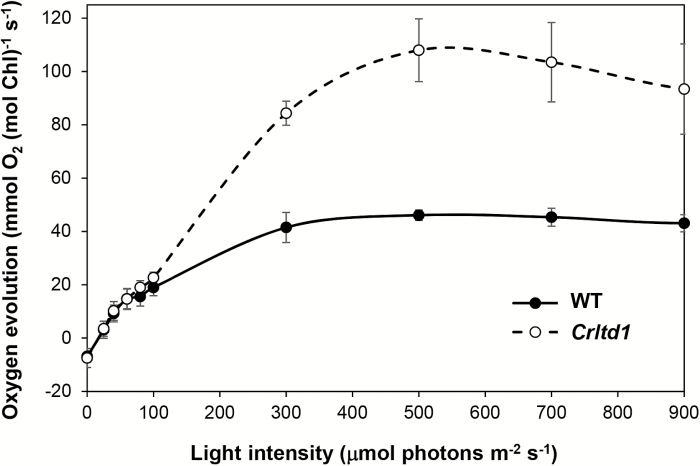
Fig. 5.
Light-saturation curves of photosynthesis in the wild-type (WT) and Crltd1 (n≥3; values shown are means±SD). Measured on a per Chl basis, the rate of respiration of the Crltd1 mutant was similar to that of the wild-type, but the light-saturated rate of oxygen evolution (Pmax) was about twice that of the wild-type. A 1 ml aliquot of cell suspension containing 2 μM Chl was loaded onto the oxygen electrode chamber.
The photosynthetic activity of the wild-type saturated at ~300 μmol photons m−2 s−1, whereas that of the mutant saturated at 500 μmol photons m−2 s−1 (Fig. 5). At saturating light intensities, the maximum photosynthetic capacity (Pmax) was 47.8 ± 1.87 mmol oxygen (mol Chl)−1 s−1 for the wild-type and 108.08 ± 11.97 mmol oxygen (mol Chl)−1 s−1 for the mutant. It was reported that mutant strains in which photosynthesis saturates at a higher light intensity could possess a smaller light-harvesting Chl antenna than the wild-type (Polle et al., 2000, 2003; Kirst et al., 2012a, b; Jeong et al., 2017). We therefore expected that the Crltd1 mutant also has a truncated Chl antenna and further characterized its photosystems and their Chl antenna size.
Photosynthetic apparatus characterization of wild-type and Crltd1 mutant
To measure the concentration of the photosystems and size of the light-harvesting antenna, we isolated thylakoid membranes from cultures grown photoautotrophically. The concentration of the photosystems was estimated spectrophotometrically by the ΔA320nm light-minus-dark absorbance change for the electron acceptor QA of PSII and the ΔA700nm light-minus-dark absorbance change for the PSI reaction center P700 (Melis and Brown, 1980). The kinetics of QA photoreduction and P700 photo-oxidation were also measured under weak green actinic light to determine the Chl antenna size of each photosystem (Melis, 1989).
The ratio of QA to total Chl was 2.13 ± 0.63 for the wild-type and 4.52 ± 0.72 (mmol:mol) in the Crltd1 mutant, that is 212% greater than that of the wild-type. The ratio of P700 to total Chl was 1.65 ± 0.08 for the wild-type and 1.97 ± 0.34 (mmol:mol) in the mutant, that is 119% greater than that of the wild-type (Table 3). These phenotypes indicated that there is less Chl relative to the photosystems in this mutant, which may explain the higher light intensity needed to saturate the photosynthesis of the mutant.
Table 3.
Photochemical apparatus characteristics of the wild-type and the Crltd1 mutant grown photoautotrophically (TBP) or photoheterotrophically (TAP)
| TBP | TAP | |||
|---|---|---|---|---|
| Parameter | Wild-type | Crltd1 | Wild-type | Crltd1 |
| QA/total Chl (mmol:mol) | 2.13 ± 0.63 | 4.52 ± 0.72 | 1.73 ± 0.25 | 3.43 ± 0.85 |
| P700/total Chl (mmol:mol) | 1.65 ± 0.08 | 1.97 ± 0.34 | 2.09 ± 0.31 | 2.29 ± 0.44 |
| QA/cell (mol (1018 cells)−1) | 3.78 ± 1.23 | 2.52 ± 0.4 | 2.66 ± 0.39 | 1.86 ± 0.46 |
| P700/cell (mol (1018 cells)−1) | 3 ± 0.14 | 1.1 ± 0.19 | 3.21 ± 0.48 | 1.24 ± 0.24 |
| PSII/PSI ratio | 1.21 | 2.30 | 0.83 | 1.47 |
| Functional PSII Chl antenna size | 345.11 ± 16.22 | 191.96 ± 40.71 | 395.02 ± 22.83 | 249.02 ± 28.16 |
| Functional PSI Chl antenna size | 191.65 ± 13.67 | 77.43 ± 16.22 | 160.05 ± 11.57 | 83.46 ± 8.68 |
Values shown are means±SD (n≥4). The size of the Chl antennae of photosystems I and II and reaction center concentrations were measured spectrophotometrically.
Interesting quantitative tendencies in the abundance of PSI and PSII were found on a per cell basis. The content of QA per cell was lowered from 3.78 ± 1.23 (wild-type) to 2.52 ± 0.4 mol per 1018 cells in the Crltd1 mutant, i.e. down to 67%. By contrast, the content of P700 per cell was lowered from 3 ± 0.14 (wild-type) to 1.1 ± 0.19 mol per 1018 cells in the mutant, i.e. down to 37%, revealing a disproportionate lowering in PSI content (Table 3). These mutation-induced changes altered the PSII/PSI ratio of the Crltd1 mutant (2.3:1) relative to that of the wild-type (1.21:1) (Table 3). This alteration in the PSII/PSI ratio was noted in cells grown either autotrophically or heterotrophically on externally provided carbon source (Table 3). The results strengthen the notion that the ratio of the two photosystems depends in this case on the ltd antenna mutation but it does not depend on the carbon source during cell growth (Polle et al., 2000). The functional light-harvesting Chl antenna size of PSII and PSI in the mutant were 55% and 40% of those in the wild-type, respectively. It was noted that, under all growth conditions, the Chl antenna size of PSI in the Crltd1 mutant was essentially that of the PSI core complex (Glick and Melis, 1988), suggesting the total absence of the peripheral LHCA antenna from PSI in this mutant (Table 3).
Western blot analysis of thylakoid membrane proteins in the Crltd1 mutant
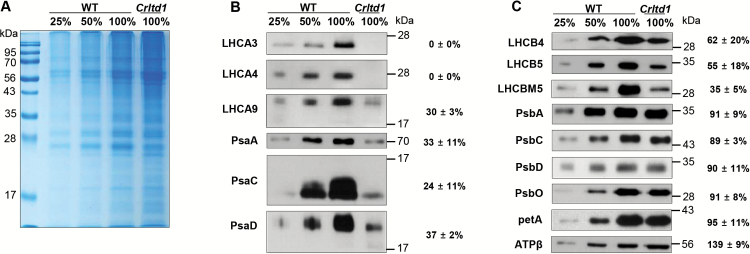
A lower photosystem content and diminished light-harvesting antenna size, especially those of PSI, in the Crltd1 mutant would inevitably have resulted in altered amounts of thylakoid membrane proteins. We examined the amount of specific proteins by western blotting using cells grown photoautotrophically. Proteins were loaded on the basis of equal cell number and, for the wild-type, samples were loaded at three different concentrations (25, 50, and 100%=8 × 105 cells per lane) to ensure linearity of the staining signal (Fig. 6A). The amount of each protein in the mutant relative to the wild-type was measured in at least three experiments, representative results of which are shown in Fig. 6B, C.
Fig. 6.
SDS-PAGE and western blot analysis of total thylakoid membrane proteins from the wild-type (WT) and Crltd1 mutant. (A) A Coomassie-stained SDS-PAGE gel of total C. reinhardtii protein extracts. (B) Western blot analysis with specific polyclonal antibodies raised against PSI subunits of C. reinhardtii. (C) Western blot analysis with specific polyclonal antibodies raised against PSII, cytochrome f (PetA) and the β-subunit of ATP synthase (ATPβ) of C. reinhardtii. Relative amounts of proteins in the Crltd1 strain compared with the wild-type are shown next to the protein bands (n≥3; values shown are means±SD). Loading of the gels: 8 × 105 cells per lane (23.4 ± 2.6 µg of protein for WT and 23.3 ± 4 µg of protein for Crltd1). (This figure is available in color at JXB online.)
In the Crltd1 mutant, no cross-reaction was observed with the LHCA3- and LHCA4-specific antibodies, and the abundance of LHCA9 was only 30 ± 3% or less of that in the wild-type (Fig. 6B). The amounts of LHCB4, LHCB5, and LHCBM5 in the mutant were 62 ± 20%, 55 ± 18%, and 35 ± 5%, respectively, of those measured in the wild-type (Fig. 6C). Overall, the content of LHCB proteins in the mutant was 50% of that in the wild-type, and the content of LHCA proteins was reduced even more severely to a very low level.
A dissimilar effect of the mutation was also noted on the photosystem content. The amounts of photosystem reaction center proteins in the mutant were lowered disproportionately for PSI than for PSII. In the mutant, levels of PSI core proteins PsaA, PsaC, and PsaD, which are associated with the LHCA, were 30% of those in the wild-type (Fig. 6B). Levels of the PSII core proteins PsbA, PsbC, PsbD, and PsbO were about 90% of those in the wild-type (Fig. 6C).
Qualitatively, the spectrophotometric QA per cell and P700 per cell measurements (Table 3) are in agreement with the western blot PsbA, PsbC, PsbD, and PsbO (for PSII) and PsaA, PsaC, and PsaD (for PSI) per cell data for wild-type and ltd mutant (Fig. 6). The small quantitative discrepancy between spectrophotometric and western blot results for PSII is probably due to the density of the bands in PsbA, PsbC, PsbD, and PsbO, which tends to minimize the difference between wild-type and ltd mutant.
To examine whether the cytochrome b6f complex was also affected in the mutant, western blot analyses with polyclonal antibodies against the cytochrome f protein (PetA) were conducted. Results showed no difference between wild-type and the ltd mutant in terms of Cyt f content (Fig. 6C). In contrast, the β-subunit of ATP synthase (ATPβ) was increased in the mutant to 139% of the level in the wild-type (Fig. 6C). This increment may be a consequence of the altered bioenergetic landscape in this strain. The levels of the CpSRP pathway proteins were similar in the wild-type and the Crltd mutant (see Supplementary Fig. S4).
Analysis of photosystem complexes in wild-type and Crltd1 mutant by Deriphat-PAGE and two-dimensional PAGE
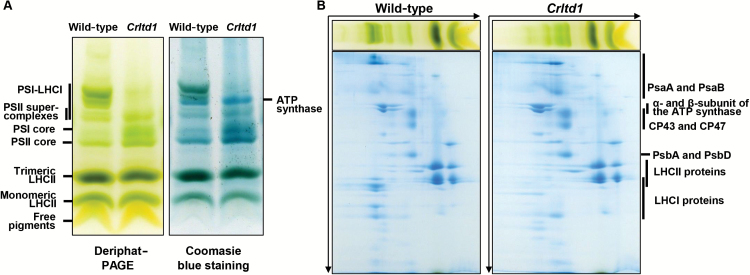
To gain further insight into the organization of photosynthetic complexes, we performed native Deriphat-PAGE analysis of the wild-type and Crltd1 mutant. Thylakoid membranes of photoautotrophically grown cells were solubilized with 1% α-dodecyl maltoside and loaded on the basis of equal Chl. In the wild-type, we observed several green bands representing free pigments, monomeric LHCII, trimeric LHCII, PSII core, and PSI core (Fig. 7A, left panel), which resulted from sequential release of the LHC proteins from the photosystems (Järvi et al., 2011). In addition, PSII supercomplexes and PSI–LHCI complexes of various sizes were observed in the slow electrophoretic mobility part of the gel. The PSI–LHCI complexes appeared to be larger and showed a broader range of electrophoretic mobility than the PSII supercomplexes.
Fig. 7.
Analysis of thylakoid membrane protein complexes in the wild-type and Crltd1 mutant by Deriphat-PAGE (A) and two-dimensional PAGE (B). Loading of the gels: 25 µg Chl per lane.
In the Crltd1 mutant, the PSI–LHCI complex was nearly absent compared with the wild-type, showing loss of PSI holocomplexes, consistent with the lower levels of PSI (Table 3) and LHCA (Fig. 6A) implying that loss of LHCI had a considerable effect on the formation of the PSI–LHCI complex. Instead, the abundance of the PSII supercomplexes, PSII core and ATP synthase complexes, which could be observed in a Coomassie-stained gel, was much higher than that in the wild-type (Fig. 7A, right panel).
Each native Deriphat-PAGE band was further characterized by western blotting with antibodies against PsaA, PsbA, LHCB, and LHCBM5 (see Supplementary Fig. S5) and two-dimensional PAGE analysis (Fig. 7B). Identification of the bands by western blotting after Deriphat-PAGE was consistent with the results of two-dimensional PAGE. For example, the presence of PSI–LHCI complexes in the mutant as faint bands on Deriphat gels agreed with a lower abundance of LHCI in two-dimensional PAGE. Keeping the α- and β-subunits of ATP synthase as migration references of the 2D-PAGE, it was clear that PsaA and PsaB in the wild-type were associated with a complex of a lower mobility than the ATP synthase in the first dimension. The opposite was instead observed with the Crltd1 PsaA and PsaB, because of their association with a complex smaller than the ATP synthase. This Crltd1 complex probably corresponds to a PSI without its peripheral antenna, because it contained the PSI core subunits but lacked the LHCI.
Discussion
In the green lineage of eukaryotic photosynthetic organisms, the light-harvesting antennae of photosystems I and II consist of chloroplast-encoded core complexes and nuclear-encoded LHCPs peripheral to the core. LHCPs are imported from the cytosol, translocated across the chloroplast envelope, and then directed to the CpSRP pathway by the LTD protein. The latter has been identified as an ankyrin domain-containing protein in Arabidopsis (Cui et al., 2011; Ouyang et al., 2011). The CpSRP pathway facilitates the post-translational transport of these proteins to photosystem assembly sites in the thylakoid membrane (Henry, 2010). The function of the CpSRP pathway in green microalgae is analogous to that in higher plants, with several distinct and important differences (Kirst and Melis, 2014). It appears from this work that the function of the LTD protein in green microalgae is slightly different from its Arabidopsis homolog.
Generation of null LTD from green algae
To elucidate the function of CrLTD and the effect of CrLTD deficiency on LHCP assembly in vivo, we generated ltd null mutants in C. reinhardtii by using CRISPR–Cas9 technology, which was recently successfully applied to C. reinhardtii (Baek et al., 2016). This method has advantages over traditional mutagenesis approaches: generation of knockout or knockdown C. reinhardtii mutants by DNA insertional mutagenesis is time-consuming and labor-intensive (Polle et al., 2003; Tetali et al., 2007; Jinkerson and Jonikas, 2015; Baek et al., 2016). RNA silencing approaches cannot completely suppress the expression of the target gene (Schroda, 2006). Generation of Crltd null mutants by transfecting a preassembled CRISPR–Cas9 complex is advantageous because this complex induces indel-type mutations at the target site, unlike random mutagenesis by vector insertion (Kim et al., 2014). The transformation efficiency of the CRISPR–Cas9 method was calculated as 1.12% (see Supplementary Fig. S1). Furthermore, our sequencing analysis of the Crltd null mutant confirmed that the RNP complex cleaved only the Crltd locus but not any other part of the genome (Supplementary Table S3). Thus, the Crltd mutant generated by the CRISPR–Cas9 method allowed a clear observation of the specific effect(s) of the absence of the CrLTD protein because there were no other genetic lesions.
Phenotype of the C. reinhardtii LTD mutant
The ankyrin domain is known to mediate protein–protein interactions (Mosavi et al., 2004). In the CpSRP pathway, such interactions occur between the ankyrin domain of the CpSRP43 protein and the L18 motif of the LHCPs, or between the ankyrin domain of the LTD protein and the T14 motif of the LHCPs in Arabidopsis (DeLille et al., 2000; Tu et al., 2000; Stengel et al., 2008; Cui et al., 2011; Ouyang et al., 2011). Therefore, it is suggested that the ankyrin domain of the LTD protein in Chlamydomonas, which has a high similarity to its Arabidopsis homolog, also participates in protein–protein interaction with the LHCPs in the chloroplast of this green microalga. Indeed, this work presented evidence that loss of LTD function quantitatively affected the LHCP assembly in the chloroplast of Chlamydomonas. This is shown in the results of Table 3, where the size of the functional PSI Chl antenna of the Crltd1 mutant is strictly limited to that of the PSI reaction center core (Glick and Melis 1988) and the antenna totally lacks LHCI (see also Fig. 6B), in spite of the assembly of significant amounts of PSI.
The Crltd1 mutant investigated in this work exhibited pale green coloration and had a lower total Chl content per cell than the wild-type (Figs 1 and 3). This phenotype is typical for null mutants lacking CpSRP pathway proteins both in Arabidopsis and C. reinhardtii and is caused by a lower level of LHCP assembly in the thylakoid membrane of photosynthesis (Amin et al., 1999; Hutin et al., 2002; Tzvetkova-Chevolleau et al., 2007; Kirst et al., 2012a, b; Jeong et al., 2017). However, the Crltd1 mutant displayed a profile of thylakoid membrane proteins (Fig. 6), which was different from the previously reported Arabidopsis ltd null mutant (Ouyang et al., 2011). In the latter, levels of both LHCI and LHCII were considerably lower, and levels of the core PSI and PSII complexes were half of those in the wild-type control (Ouyang et al., 2011). In the Crltd1 mutant, LHCI was dramatically lowered to very low levels, whereas LHCII accumulated at 50% of the wild-type. In the Crltd1 mutant, levels of the PSI core complex were 30% of that in the wild-type, but those of PSII were similar to wild-type. Consequently, the Crltd1 mutant showed very low levels of PSI–LHCI holocomplex accumulation due to the very low LHCI content (Fig. 7). This phenotype resembles that of the Arabidopsis triple knockout mutant ΔLhca, in which four LHCA subunits were deleted (Bressan et al., 2016), supporting the notion that Crltd1 has a serious defect in LHCI and probably PSI core assembly.
The differential accumulation of PSI and PSII proteins in the Crltd1 mutant is a unique and dissimilar feature in comparison to Chlamydomonas deletion mutants lacking CpSRP pathway proteins. A null mutant lacking CpFTSY, Δcpftsy, was also generated by the CRISPR–Cas9 method (Baek et al., 2016), and LHCP content of both PSI and PSII was found to be lower than that in the wild-type (see Supplementary Fig. S4). Similarly, the tla2 strain of C. reinhardtii (Kirst et al., 2012a), in which the CpFTSY gene was disrupted by DNA insertional mutagenesis, possessed lower levels of both the LHCI and LHCII proteins (Kirst et al., 2012a). The null mutants lacking CpSRP43 (tla3) and CpSRP54 (tla4) also showed uniformly lower levels of LHCI and LHCII (Kirst et al., 2012b; Jeong et al., 2017). However, ac29-3, a knockout mutant of the ALB3.1 translocase, exhibited a disproportionate lowering of LHCI compared with LHCII (Bellafiore et al., 2002) and, in this respect, its phenotype was similar to the Crltd1 mutant.
The unique profile of thylakoid membrane proteins in the Crltd1 mutant is closely linked to its Chl composition. In all other CpSRP mutants examined to date, such as tla2 (Δcpftsy), tla3 (ΔcpSRP43), and tla4 (ΔcpSRP54) (Kirst et al., 2012a, b; Jeong et al., 2017), the Chl a/b ratio was substantially elevated relative to that of the wild-type. Other Chl-deficient mutants affected in terms of their LHCP content or Chl biosynthesis also displayed a higher than wild-type Chl a/b ratio phenotype (Mussgnug et al., 2007; Beckmann et al., 2009; Bonente et al., 2011; Perrine et al., 2012). This was always attributed to the predominant loss of Chl b and the LHCB in these mutants. However, uniquely, the Chl a/b ratio of the Crltd1 mutant was similar to that of the wild-type (Fig. 3). This is attributed to the near absence of LHCA and substantial lowering of the PSI core content in the thylakoid membrane of the Crltd1 mutant. In general, the PSII core complex contains 37 Chl a molecules (Glick and Melis, 1988), and its peripheral light-harvesting antenna complex contains up to 200 Chl molecules, nearly equally divided between Chl a and Chl b. The PSI core complex contains 95 Chl a molecules (Glick and Melis, 1988) and its peripheral light-harvesting antenna complex contains an additional 100 Chl molecules, with the latter having a Chl a/b ratio of ~8:1 (Melis, 1990; Polle et al., 2000). Selective loss of the LHCA and a substantially lower level of PSI core in the Crltd1 mutant resulted in a proportionate lowering of Chl a and Chl b in this mutant and a pigment composition dominated by that of PSII, translating into a Chl a/b ratio similar to that of the wild-type.
A pleotropic effect upon deletion of LTD in C. reinhardtii
A severe reduction in the level of the PSI holocomplex (PSI core and LHCI proteins) in the Crltd1 mutant changed the overall structure of the chloroplast. Transmission electron microscopy analysis revealed substantial differences in thylakoid membrane ultrastructure between the wild-type and Crltd1 mutant (Fig. 4). The number of appressed thylakoid membranes was increased in the mutant chloroplast. Considering the diminished grana stacking in the Arabidopsis ltd mutant, it is interesting that the thylakoid membrane structure of the Crltd1 mutant changed in the opposite direction (Cui et al., 2011; Ouyang et al., 2011; Mitra et al., 2012). The structure of thylakoid membranes and distribution of the photosystems are highly related. In general, PSII and PSI are predominantly located in the grana lamellae and stroma lamellae, respectively (Andersson and Anderson, 1980; Vallon et al., 1986). Therefore, the predominance of the PSII complex in the Crltd1 mutant corresponds with the preponderance of appressed (stacked) thylakoids, as they are more likely to contain PSII than PSI (Andersson and Anderson, 1980; Vallon et al., 1986). In addition, exceptionally large vacuoles were observed in the cytosol of the Crltd1 mutant, which may be a consequence of inhibition of LHCP import in the chloroplast and the need to sequester them in the vacuole prior to degradation (Fig. 4). In this respect, a knockdown strain of C. reinhardtii lacking the ALB3.2 translocase also showed enlarged vacuoles compared with the wild-type (Göhre et al., 2006). In the latter, a lower level of ALB3.2 led to misfolding of thylakoid membrane proteins, triggering an extensive recycling of chloroplast proteins and necessitating their sequestration into vacuoles for degradation (Göhre et al., 2006). In this process, LHCPs and other proteins, including the large subunit of Rubisco and the α-subunit of the ATP-synthase (both synthesized on plastid ribosomes), were also found in cytoplasmic vacuoles (Park et al., 1999; Göhre et al., 2006). It is possible that LHCP whose import has been inhibited or imported but unassembled stromal LHCPs are transported to the vacuoles by a general degradation pathway responsible for scavenging stromal proteins. Taken together, these observations suggest that the imbalance of chloroplast proteins caused by the severe down-regulation of PSI concentration and thylakoid membrane holocomplex assembly in the Crltd1 mutant may induce this cytoplasmic vacuole enhancement. Thus, deletion of the Crltd protein appeared to have pleiotropic effects, emanating from the loss of a protein required for LHCP transport, the absence of which destabilized the entire photosynthetic complex assembly process and caused enhanced cytoplasmic vacuolation. However, the mechanism by which a lack of a single gene product elicits such far-reaching reorganization of cytosolic compartmentalization and thylakoid membrane restructuring in C. reinhardtii is not yet clear.
The requirement for LTD may not be stringent in green microalgae
The Crltd1 mutant exhibited photoautotrophic growth (Table 1; Supplementary Fig. S3), with a light-saturated rate of photosynthesis about the same as that of the wild-type (per cell basis, Table 2). This is at variance with the Arabidopsis ltd mutant, which was unable to grow photoautotrophically (Ouyang et al., 2011).
Thus, despite the apparent similarity between the Arabidopsis LTD and Chlamydomonas LTD, phenotypes of the respective mutants are distinct and different from each other. The phenotype of the knockout mutant of the LTD gene in C. reinhardtii seemed to be leaky, suggesting flexible or redundant function of the CrLTD protein, as absence of this protein did not fully alleviate transport and assembly of PSI–LHCI complexes and photoautotrophic growth. Since light-harvesting antenna complexes not only harvest light energy for photosynthesis but also protect the photosynthetic apparatus from various environmental stresses (Erickson et al., 2015; Wobbe et al., 2016), especially in green algae, maintenance of the proper LHCP assembly may be required for survival in the wild. Arabidopsis and C. reinhardtii are under a substantially different developmental program. Arabidopsis chloroplasts stop developing when cell size reaches a maximum volume and chloroplast number increase stops (Jarvis and López-Jez, 2013). In C. reinhardtii possessing a single cup-shaped chloroplast, such developmental restrictions do not apply, as the cells are in a state of continuous growth and development. This fundamental developmental difference between the two species could account for the differences in LTD function between C. reinhardtii and Arabidopsis reported here.
Intriguingly, all mutations of individual CpSRP pathway components in C. reinhardtii caused mild to severe impairment of the pathway, affecting LHCP accumulation, but did not impair photoautotrophic growth (Bellafiore et al., 2002; Ossenbühl et al., 2004; Göhre et al., 2006; Kirst et al., 2012a, b; Jeong et al., 2017). These phenotypes showed that most single-gene null mutations in the Chlamydomonas CpSRP pathway are leaky, unlike those in higher plants, in which the null mutations of CpFTSY and ALB3 proved to be lethal at the seedling stage (Asakura et al., 2004; Asakura et al., 2008). We cannot exclude the existence of an alternative pathway for LHCPs transport and assembly in C. reinhardtii, which may partially compensate for the loss of function of the CpSRPs. The presence of two C. reinhardtii ALB homologs (ALB3.1 and ALB3.2), unlike the single ALB in Arabidopsis, may support the possibility of an alternative pathway for LHCP assembly in C. reinhardtii (Bellafiore et al., 2002; Göhre et al., 2006). In this respect, generation of more than one mutant of the Chlamydomonas CpSRP pathway components may help to further dissect the mechanism(s) of LHCP transport and assembly in the thylakoid membrane of the green microalgae.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Visual examination of C. reinhardtii colonies in the course of screening for Crltd gene knockout mutant.
Fig. S2. Sanger sequencing chromatograms for CRISPR–Cas9-induced Crltd mutant strains.
Fig. S3. Growth curves of wild-type and the Crltd1 mutant at different light intensities.
Fig. S4. Coomassie-stained SDS-PAGE of total proteins and western blot analysis of LHCPs and CpSRP components in the wild-type (WT) and Crltd strains.
Fig. S5. Analysis of thylakoid membrane protein complexes in the wild-type and Crltd1 mutant.
Table S1. Target sequences of four sgRNAs used to recognize the Crltd gene.
Table S2. The mutation frequency (A) and pattern (B) of wild-type and RNP-transfected cells.
Table S3. Analysis of off-target effects in the wild-type and Crltd1.
Acknowledgements
This work was supported by the Korea CCS R&D Center (NRF-2014M1A8A1049273) funded by the Korean Government (Ministry of Science and ICT). This work was also supported by the Next Generation BioGreen 21 Program (PJ01119201) funded by the Plant Molecular Breeding Center.
Glossary
Abbreviations:
- Chl
chlorophyll
- CpSRP
chloroplast signal recognition particle
- Crltd
C. reinhardtii ltd
- indel
insertions and deletion
- LHCI
light-harvesting complex I
- LHCII
light-harvesting complex II
- LHCP
light-harvesting chlorophyll- and carotenoid-binding protein
- LTD
LHCP translocation defect
- RNP
ribonucleoprotein
- sgRNA
single guide RNA
- TAP
Tris–acetate–phosphate
- TBP
Tris–bicarbonate–phosphate.
References
- Amin P, Sy DA, Pilgrim ML, Parry DH, Nussaume L, Hoffman NE. 1999. Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiology 121, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson B, Anderson JM. 1980. Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochimica et Biophysica Acta 593, 427–440. [DOI] [PubMed] [Google Scholar]
- Asakura Y, Hirohashi T, Kikuchi S, Belcher S, Osborne E, Yano S, Terashima I, Barkan A, Nakai M. 2004. Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. The Plant Cell 16, 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Kikuchi S, Nakai M. 2008. Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. The Plant Journal 56, 1007–1017. [DOI] [PubMed] [Google Scholar]
- Bae S, Park J, Kim JS. 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K, Kim DH, Jeong J, Sim SJ, Melis A, Kim JS, Jin E, Bae S. 2016. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Scientific Reports 6, 30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J, Lehr F, Finazzi G, Hankamer B, Posten C, Wobbe L, Kruse O. 2009. Improvement of light to biomass conversion by de-regulation of light-harvesting protein translation in Chlamydomonas reinhardtii. Journal of Biotechnology 142, 70–77. [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Ferris P, Naver H, Göhre V, Rochaix JD. 2002. Loss of Albino3 leads to the specific depletion of the light-harvesting system. The Plant Cell 14, 2303–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan M, Dall’Osto L, Bargigia I, Alcocer MJ, Viola D, Cerullo G, D’Andrea C, Bassi R, Ballottari M. 2016. LHCII can substitute for LHCI as an antenna for photosystem I but with reduced light-harvesting capacity. Nature Plants 2, 16131. [DOI] [PubMed] [Google Scholar]
- Bonente G, Formighieri C, Mantelli M, Catalanotti C, Giuliano G, Morosinotto T, Bassi R. 2011. Mutagenesis and phenotypic selection as a strategy toward domestication of Chlamydomonas reinhardtii strains for improved performance in photobioreactors. Photosynthesis Research 108, 107–120. [DOI] [PubMed] [Google Scholar]
- Chandrasekar S, Shan SO. 2017. Anionic phospholipids and the Albino3 translocase activate SRP-receptor interaction during LHCP targeting. The Journal of Biological Chemistry 292, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YL, Jia QS, Yin QQ, Lin GN, Kong MM, Yang ZN. 2011. The GDC1 gene encodes a novel ankyrin domain-containing protein that is essential for grana formation in Arabidopsis. Plant Physiology 155, 130–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLille J, Peterson EC, Johnson T, Moore M, Kight A, Henry R. 2000. A novel precursor recognition element facilitates posttranslational binding to the signal recognition particle in chloroplasts. Proceedings of the National Academy of Sciences, USA 97, 1926–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünschede B, Bals T, Funke S, Schünemann D. 2011. Interaction studies between the chloroplast signal recognition particle subunit cpSRP43 and the full-length translocase Alb3 reveal a membrane-embedded binding region in Alb3 protein. The Journal of Biological Chemistry 286, 35187–35195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson E, Wakao S, Niyogi KK. 2015. Light stress and photoprotection in Chlamydomonas reinhardtii. The Plant Journal 82, 449–465. [DOI] [PubMed] [Google Scholar]
- Glick RE, Melis A. 1988. Minimum photosynthetic unit size in System I and System II of barley chloroplasts. Biochimica et Biophysica Acta 934, 151–155. [Google Scholar]
- Göhre V, Ossenbühl F, Crèvecoeur M, Eichacker LA, Rochaix JD. 2006. One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. The Plant Cell 18, 1454–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. 1989. Chlamydomonas handbook. New York: Academic Press Inc. [Google Scholar]
- Henry RL. 2010. SRP: adapting to life in the chloroplast. Nature Structural & Molecular Biology 17, 676–677. [DOI] [PubMed] [Google Scholar]
- Hiyama T, Ke B. 1972. Difference spectra and extinction coefficients of P 700. Biochimica et Biophysica Acta 267, 160–171. [DOI] [PubMed] [Google Scholar]
- Holden M. 1976. Chlorophylls. In: Goodwin TW, ed. Chemistry and biochemistry of plant pigments, Vol 2. London: Academic Press, 1–37. [Google Scholar]
- Horn A, Hennig J, Ahmed YL, Stier G, Wild K, Sattler M, Sinning I. 2015. Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction. Nature Communications 6, 8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin C, Havaux M, Carde JP, Kloppstech K, Meiherhoff K, Hoffman N, Nussaume L. 2002. Double mutation cpSRP43–/cpSRP54– is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light-harvesting chlorophyll proteins. The Plant Journal 29, 531–543. [DOI] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Paakkarinen V, Aro EM. 2011. Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. The Biochemical Journal 439, 207–214. [DOI] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E. 2013. Biogenesis and homeostasis of chloroplasts and other plastids. Nature Reviews. Molecular Cell Biology 14, 787–802. [DOI] [PubMed] [Google Scholar]
- Jansson S. 1999. A guide to the Lhc genes and their relatives in Arabidopsis. Trends in Plant Science 4, 236–240. [DOI] [PubMed] [Google Scholar]
- Jeong J, Baek K, Kirst H, Melis A, Jin E. 2017. Loss of CpSRP54 function leads to a truncated light-harvesting antenna size in Chlamydomonas reinhardtii. Biochimica et Biophysica Acta 1858, 45–55. [DOI] [PubMed] [Google Scholar]
- Jinkerson RE, Jonikas MC. 2015. Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. The Plant Journal 82, 393–412. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research 24, 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst H, Gabilly ST, Niyogi KK, Lemaux PG, Melis A. 2017. Photosynthetic antenna engineering to improve crop yields. Planta 245, 1009–1020. [DOI] [PubMed] [Google Scholar]
- Kirst H, García-Cerdán JG, Zurbriggen A, Melis A. 2012a. Assembly of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii requires expression of the TLA2-CpFTSY gene. Plant Physiology 158, 930–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst H, Garcia-Cerdan JG, Zurbriggen A, Ruehle T, Melis A. 2012b. Truncated photosystem chlorophyll antenna size in the green microalga Chlamydomonas reinhardtii upon deletion of the TLA3-CpSRP43 gene. Plant Physiology 160, 2251–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst H, Melis A. 2014. The chloroplast signal recognition particle (CpSRP) pathway as a tool to minimize chlorophyll antenna size and maximize photosynthetic productivity. Biotechnology Advances 32, 66–72. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Levasseur M, Thompson PA, Harrison PJ. 1993. Physiological acclimation of marine phytoplankton to different nitrogen sources. Journal of Phycology 29, 587–595. [Google Scholar]
- Li X, Henry R, Yuan J, Cline K, Hoffman NE. 1995. A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proceedings of the National Academy of Sciences, USA 92, 3789–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang FC, Kroon G, McAvoy CZ, Chi C, Wright PE, Shan SO. 2016. Conformational dynamics of a membrane protein chaperone enables spatially regulated substrate capture and release. Proceedings of the National Academy of Sciences, USA 113, E1615–E1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A. 1989. Spectroscopic methods in photosynthesis: photosystem stoichiometry and chlorophyll antenna size. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 323, 397–409. [Google Scholar]
- Melis A. 1990. Regulation of photosystem stoichiometry in oxygenic photosynthesis. Botanical Magazine 2, 9–28. [Google Scholar]
- Melis A. 1991. Dynamics of photosynthetic membrane composition and function. Biochimica et Biophysica Acta 1058, 87–106. [Google Scholar]
- Melis A, Brown JS. 1980. Stoichiometry of system I and system II reaction centers and of plastoquinone in different photosynthetic membranes. Proceedings of the National Academy of Sciences, USA 77, 4712–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A, Thielen AP. 1980. The relative absorption cross-sections of photosystem I and photosystem II in chloroplasts from three types of Nicotiana tabacum. Biochimica et Biophysica Acta 589, 275–286. [DOI] [PubMed] [Google Scholar]
- Mitra M, Kirst H, Dewez D, Melis A. 2012. Modulation of the light-harvesting chlorophyll antenna size in Chlamydomonas reinhardtii by TLA1 gene over-expression and RNA interference. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 367, 3430–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Goforth RL, Mori H, Henry R. 2003. Functional interaction of chloroplast SRP/FtsY with the ALB3 translocase in thylakoids: substrate not required. The Journal of Cell Biology 162, 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Science 13, 1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussgnug JH, Thomas-Hall S, Rupprecht J, Foo A, Klassen V, McDowall A, Schenk PM, Kruse O, Hankamer B. 2007. Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion. Plant Biotechnology Journal 5, 802–814. [DOI] [PubMed] [Google Scholar]
- Ossenbühl F, Göhre V, Meurer J, Krieger-Liszkay A, Rochaix JD, Eichacker LA. 2004. Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. The Plant Cell 16, 1790–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M, Li X, Ma J, Chi W, Xiao J, Zou M, Chen F, Lu C, Zhang L. 2011. LTD is a protein required for sorting light-harvesting chlorophyll-binding proteins to the chloroplast SRP pathway. Nature Communications 2, 277. [DOI] [PubMed] [Google Scholar]
- Park H, Eggink LL, Roberson RW, Hoober JK. 1999. Transfer of proteins from the chloroplast to vacuoles in Chlamydomonas reinhardtii (Chlorophyta): a pathway for degradation. Journal of Phycology 35, 528–538. [Google Scholar]
- Park J, Bae S, Kim JS. 2015. Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31, 4014–4016. [DOI] [PubMed] [Google Scholar]
- Park J, Lim K, Kim JS, Bae S. 2017. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33, 286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine Z, Negi S, Sayre RT. 2012. Optimization of photosynthetic light energy utilization by microalgae. Algal Research 1, 134–142. [Google Scholar]
- Petroutsos D, Busch A, Janssen I, Trompelt K, Bergner SV, Weinl S, Holtkamp M, Karst U, Kudla J, Hippler M. 2011. The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. The Plant Cell 23, 2950–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polle JE, Benemann JR, Tanaka A, Melis A. 2000. Photosynthetic apparatus organization and function in the wild type and a chlorophyll b-less mutant of Chlamydomonas reinhardtii. Dependence on carbon source. Planta 211, 335–344. [DOI] [PubMed] [Google Scholar]
- Polle JE, Kanakagiri SD, Melis A. 2003. tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta 217, 49–59. [DOI] [PubMed] [Google Scholar]
- Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. The Journal of Cell Biology 17, 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter CV, Bals T, Schünemann D. 2010. Component interactions, regulation and mechanisms of chloroplast signal recognition particle-dependent protein transport. European Journal of Cell Biology 89, 965–973. [DOI] [PubMed] [Google Scholar]
- Schroda M. 2006. RNA silencing in Chlamydomonas: mechanisms and tools. Current Genetics 49, 69–84. [DOI] [PubMed] [Google Scholar]
- Schuenemann D, Gupta S, Persello-Cartieaux F, Klimyuk VI, Jones JDG, Nussaume L, Hoffman NE. 1998. A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proceedings of the National Academy of Sciences, USA 95, 10312–10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AR. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. Journal of Ultrastructure Research 26, 31–43. [DOI] [PubMed] [Google Scholar]
- Stauber EJ, Fink A, Markert C, Kruse O, Johanningmeier U, Hippler M. 2003. Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryotic Cell 2, 978–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel KF, Holdermann I, Cain P, Robinson C, Wild K, Sinning I. 2008. Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science 321, 253–256. [DOI] [PubMed] [Google Scholar]
- Taiz L, Zeiger E. 2010. Plant physiology. Sunderland, MA: Sinauer Associates Inc, Ch. 7, 163–197. [Google Scholar]
- Tardif M, Atteia A, Specht M et al. . 2012. PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Molecular Biology and Evolution 29, 3625–3639. [DOI] [PubMed] [Google Scholar]
- Tetali SD, Mitra M, Melis A. 2007. Development of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii is regulated by the novel Tla1 gene. Planta 225, 813–829. [DOI] [PubMed] [Google Scholar]
- Thielen AP, van Gorkom HJ. 1981. Quantum efficiency and antenna size of photosystems IIα, IIβ and I in tobacco chloroplasts. Biochimica et Biophysica Acta 635, 111–120. [DOI] [PubMed] [Google Scholar]
- Tu CJ, Peterson EC, Henry R, Hoffman NE. 2000. The L18 domain of light-harvesting chlorophyll proteins binds to chloroplast signal recognition particle 43. The Journal of Biological Chemistry 275, 13187–13190. [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T, Hutin C, Noël LD et al. . 2007. Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. The Plant Cell 19, 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O, Wollman F-A, Olive J. 1986. Lateral distribution of the main protein complexes of the photosynthetic apparatus in Chlamydomonas reinhardtii and in spinach: an immunocytochemical study using intact thylakoid membranes and a PS II enriched membrane preparation. Photobiochemistry and Photobiophysics 12, 203–220. [Google Scholar]
- van Gorkom HJ. 1974. Identification of the reduced primary electron acceptor of photosystem II as a bound semiquinone anion. Biochimica et Biophysica Acta 347, 439–442. [DOI] [PubMed] [Google Scholar]
- Wobbe L, Bassi R, Kruse O. 2016. Multi-level light capture control in plants and green algae. Trends in Plant Science 21, 55–68. [DOI] [PubMed] [Google Scholar]
- Yu J, Baek K, Jin E, Bae S. 2017. DNA-free genome editing of Chlamydomonas reinhardtii using CRISPR and subsequent mutant analysis. Bio-Protocol 7, e2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.