Summary
Many naturalistic behaviors are built from modular components that are expressed sequentially. Although striatal circuits have been implicated in action selection and implementation, the neural mechanisms that compose behavior in unrestrained animals are not well understood. Here we record bulk and cellular neural activity in the direct and indirect pathways of dorsolateral striatum (DLS) as mice spontaneously express action sequences. These experiments reveal that DLS neurons systematically encode information about the identity and ordering of sub-second 3D behavioral motifs; this encoding is facilitated by fast-timescale decorrelations between the direct and indirect pathways. Furthermore, lesioning the DLS prevents appropriate sequence assembly during exploratory or odor-evoked behaviors. By characterizing naturalistic behavior at neural timescales, these experiments identify a code for elemental 3D pose dynamics built from complementary pathway dynamics, support a role for DLS in constructing meaningful behavioral sequences, and suggest models for how actions are sculpted over time.
Keywords: Striatum, Direct Pathway, Indirect Pathway, Ethology, Coding, Behavior, Mouse, Photometry, Microendoscopy, Machine Learning, Basal Ganglia
Introduction
The brain evolved to support the generation of naturalistic behaviors, in which animals interact with the environment by freely choosing and executing actions that reflect internal state, external cues, innate biases and past experiences (Tinbergen, 1951). Such behaviors are at once discrete and continuous: they are built out of stereotyped motifs of movement, which are concatenated to generate coherent action (Lashley, 1951). In order to create meaningful behaviors, the brain must therefore select and string together behavioral components into sequences. This implies a mechanism in which the brain uniquely encodes each motif, and then takes advantage of these representations to flexibly organize and implement behavior over time.
Multiple lines of evidence suggest that the basal ganglia — and specifically spiny projection neurons (SPNs) within its input nucleus, the striatum — play a key role in specifying both the contents and structure of behavior (Aldridge et al., 1993; Graybiel, 1998). Within the dorsal striatum, for example, neural correlates have been identified for many movement parameters, including body velocity, the speed of head movements, turn angle, and reaching kinematics (Barbera et al., 2016; Cui et al., 2013; Isomura et al., 2013; Jaeger et al., 1995; Kim et al., 2014; Klaus et al., 2017; Panigrahi et al., 2015; Rueda-Orozco and Robbe, 2015; Tecuapetla et al., 2014). These observations raise the possibility that the striatum comprehensively encodes the moment-to-moment kinematics of ongoing behavior.
In addition, SPN activity can represent more abstract aspects of behavior, like the start and stop of an action sequence, or the identity of a specific behavioral component within a sequence (Jin and Costa, 2010; Jog et al., 1999). Neural correlates have been identified in dorsolateral striatum (DLS) for components of naturalistic behaviors, such as grooming, “warm up” locomotion and juvenile play (Aldridge et al., 1993; Aldridge et al., 2004). Focal lesions of the DLS alter the sequencing of these hand-annotated patterns of action, demonstrating a causal role for the striatum in controlling the sequential structure of at least some behaviors (Berridge and Fentress, 1987). These findings suggest an additional role for the striatum in action selection, the process of deciding what to do — and not to do — next.
However, it is not clear how SPNs simultaneously encode granular information about movement parameters and higher-order information about behavioral components and sequences. The striatum influences action through two main constituent neural circuits, the direct and indirect pathways. The opposing influence of these two pathways on downstream areas like thalamus and cortex has suggested a broad model in which the direct pathway selects actions for expression, while the indirect pathway inhibits unwanted behaviors. Consistent with this model, optogenetic and circuit-level evidence supports a role for the direct pathway in initiating locomotion and the indirect pathway in arresting locomotion (Kravitz et al., 2010). Yet when pathway activity is recorded in separate mice and then temporally aligned to a turn or a lever press, the direct and indirect pathways often exhibit near-coincident activation; these observations suggest that both pathways somehow collaborate to generate specific behaviors (Cui et al., 2013).
We wished to characterize how direct and indirect pathway activity in the DLS relates to both the discrete and continuous features of spontaneous 3D behaviors. Furthermore, we wished to ask whether DLS activity during such behaviors is obligate for action implementation and/or for behavioral sequencing. To address these questions, we took advantage of a recently-developed machine vision and unsupervised machine learning technique called Motion Sequencing (“MoSeq”), which identifies a set of sub-second behavioral motifs or “syllables” that make up 3D mouse behavior within any given experiment, and captures the statistical “grammar” that governs how these syllables are sequenced over time (Berridge et al., 1987; Wiltschko et al., 2015). This approach has revealed that open field behavior can be remarkably complex, involving tens of discrete behavioral syllables that are placed probabilistically into sequences to generate an overall pattern of exploratory locomotion. Importantly, MoSeq independently identifies which syllable is expressed at any moment in time (enabling analysis of action selection), as well as the 3D pose dynamics that make up each syllable (enabling analysis of behavioral kinematics).
Here we combine MoSeq with bulk and cellular imaging technologies to assess direct and indirect pathway activity during spontaneous behavior. In these experiments pathway activity is characterized both separately and simultaneously, allowing assessment of the relative contribution of each pathway to behavioral encoding. This approach reveals that the direct and indirect pathways are frequently decorrelated at sub-second timescales, that SPN activity encodes both ongoing 3D pose dynamics and sequence probabilities, and that the DLS is required for assembling behavioral sequences. Thus, striatum does not contain purely abstract representations for syllables, nor does it simply represent behavioral kinematics. Instead, these data suggest a model in which the striatum takes advantage of kinematic information to uniquely identify each behavioral syllable, and then uses this code to organize naturalistic behavioral sequences. In principle, such a code could also support the generation of new behaviors under conditions of learning and reward.
Results
To characterize the influence of the DLS on the sub-second structure of spontaneous behavior, we developed a novel multicolor photometry system to monitor the fluorescence of green (GCaMP6s) and red (jRCaMP1b) genetically-encoded calcium indicators expressed in SPNs belonging to the direct (dSPNs) and indirect (iSPNs) pathways (Figure 1A, Figures S1A – 1D, see Methods) (Chen et al., 2013; Dana et al., 2016). Direct pathway expression was specified by delivering jRCaMP1b using a Cre-On adeno-associated virus (AAV) to Drd1a-Cre mice (Gerfen et al., 2013). Simultaneous indirect pathway expression was achieved by infecting these same animals with a Cre-Off AAV expressing GCaMP6s. Given that 95 percent of cells in striatum are SPNs, we operationally assigned signals generated by Cre-Off infection to the indirect pathway (Rymar et al., 2004; Saunders et al., 2012).
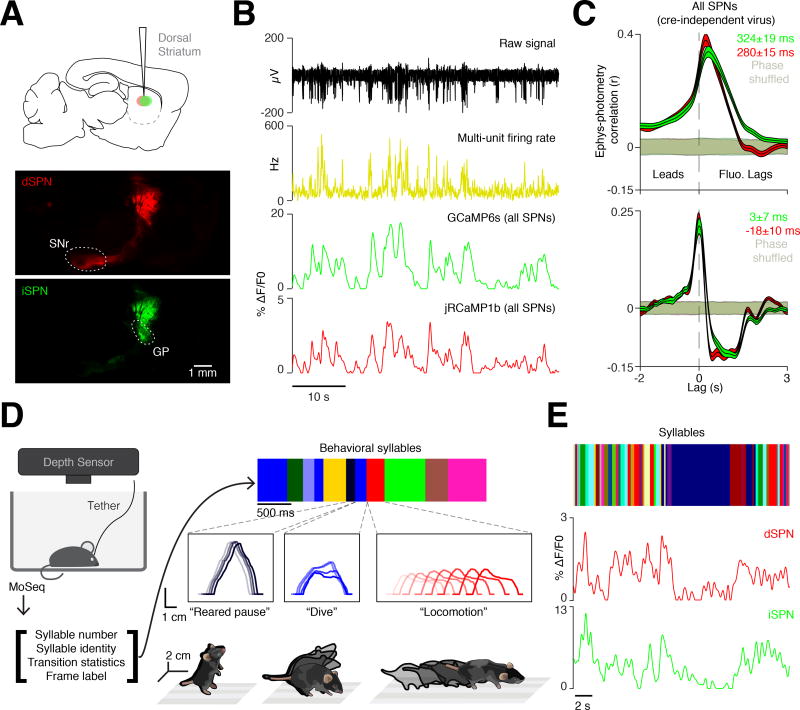
Figure 1. Motion Sequencing during neural recordings.
A. AAVs expressing Cre-On jRCaMP1b and Cre-Off GCaMP6s were injected into the DLS to assess direct (dSPN, red) and indirect (iSPN, green) pathway activity via multicolor photometry (see Methods). Top, sagittal schematic of injection site. Middle and bottom, histological verification of jRCaMP1b (dSPN, red) and GCaMP6s (iSPN, green) expression (sagittal sections, SNr = substantia nigra pars reticulata, GP = globus pallidus). Scale bar, 1 mm.
B. An example of simultaneous electrophysiological (top, black), firing rate (middle, yellow) and photometry recording (bottom, green, GCaMP6s, red, jRCaMP1b).
C. Top, correlation between electrophysiologically-acquired firing rates and cre-independent photometry signals (red, jRCaMP1b; green, GCaMP6s), compared to time-shuffled electrophysiology (shading, 95% confidence interval). Bottom, same as top panel using the derivative of the photometry signals.
D. Left, Experimental schematic. Three-dimensional (3D) imaging data is fed to the MoSeq algorithm, which outputs identified behavioral syllables and their transition statistics (color bars, lower and right, top). Middle, three examples of syllables, occurring successively over time, depicted as “spinograms,” in which the spine of the mouse is depicted as if looking at the mouse from the side, with time indicated as increasing color darkness. Bottom, isometric-view illustrations of the 3D imaging data associated with the “reared pause,” “dive,” and “locomotion” behavioral syllables.
E. Example of MoSeq-defined syllables (top, individual syllables labeled with unique colors) aligned to dSPN (middle, red) and iSPN-related (bottom, green) photometry signals.
The photometry system exhibited negligible cross-talk between channels, and low noise attributable to motion artifacts (Figures S1E and S1F). To account for the temporal lag between photometric signals and underlying electrophysiological changes, we performed simultaneous two-channel photometry and multiunit electrophysiology in awake behaving mice. While photometry signals lagged multiunit activity by circa 300 milliseconds, the derivative of the photometry signal lagged by only circa 10 milliseconds, indicating that the fluorescence derivative could be used to identify the onset of changes in the underlying neural activity (Figures 1B, 1C and S1G).
We integrated the photometry system into MoSeq, enabling us to analyze behavior and record from direct and indirect SPNs in parallel (all recordings in right DLS, Figures 1D and S2A). Probabilistic machine learning methods were used to infer the pose of the mouse when it was occluded by the photometry tether (Figure S2A, see Methods) (Roweis, 1998; Tipping and Bishop, 1999). These corrected data were then processed by MoSeq to characterize the identity and usage of individual behavioral syllables, and the grammar that connects syllables together over time (Figure 1E). As has been shown previously, each identified syllable (of the 41 discovered by MoSeq that were expressed more than 1 percent of the time) was morphologically distinct, with a median syllable duration of 347 ± 0.1 milliseconds (Bootstrap SEM, Figure S2B). Each syllable was associated with a 3D behavioral motif that could be described by a human observer, and included multiple variants of pauses, locomotion, turns, rears and sniffs (Movie S1; see isometric view examples in Figure 1D).
Differential encoding of 2D and 3D velocity by the direct and indirect pathways
Neural activity in the striatum has been shown to correlate with several performance-related behavioral variables, including average running speed, turning speed, and action velocity. We therefore asked whether our integrated system could capture the expected correlation between direct pathway activity and two-dimensional (2D) velocity. Indeed, when averaged over timescales of seconds to minutes, mouse velocity was well-correlated with direct pathway activity. The dual-channel photometry system revealed that indirect pathway activity also correlated with velocity over long timescales, although less strongly than the direct pathway (Figure 2A).
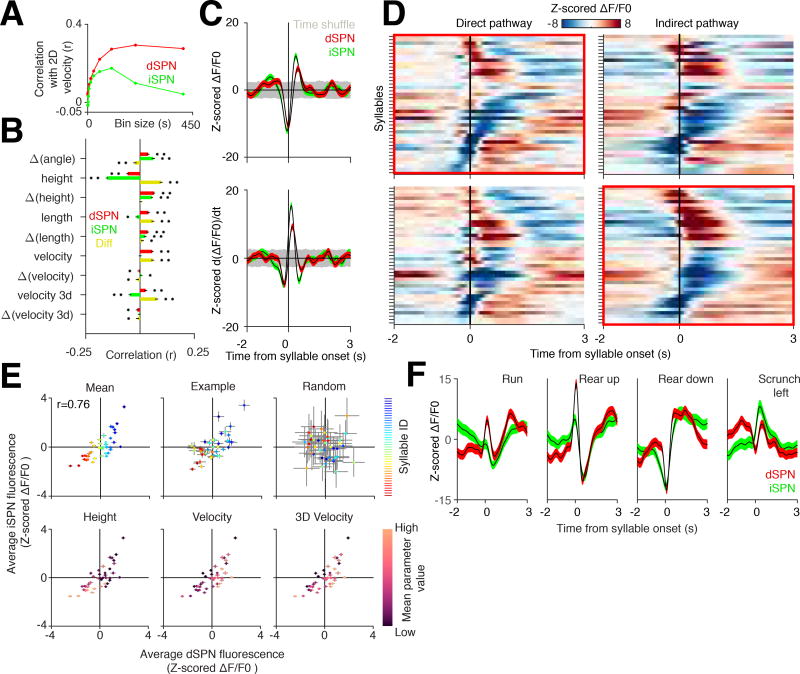
Figure 2. Direct and indirect pathway activity correlates with fast behavioral transitions.
A. Correlation between velocity, direct pathway activity (red), and indirect pathway activity (green) at indicated time bins.
B. Correlations between scalar variables and their derivatives to dSPN fluorescence signals (Pearson r, n = 189315 comparisons), iSPN fluorescence signals (n = 173577 comparisons), and to the difference between the two signals binned at the timescale of individual syllables (n = 146636 comparisons). * = p < 0.001, ** = p < 1×10e−10.
C. Top, grand-averaged, z-scored dSPN (red) and iSPN (green) activity aligned to all syllable transitions. Bottom, derivative of top panel. Shading, time-shuffled 95% confidence interval. Note that all syllable-triggered averages are z-scored relative to the time-shuffled control (see Methods).
D. Average z-scored dSPN and iSPN fluorescence levels for individual syllables. Top, syllable-triggered averages for each syllable shown for the direct and indirect pathways, sorted by the averages in the direct pathway (first positive then negative peaks). Bottom, same as Top except sorted by indirect pathway syllable-triggered averages.
E. Top left, average peak z-scored direct and indirect pathway fluorescence signal (ΔF/F0) associated with each syllable (individual syllables identified via an arbitrary color code, with coding preserved across all top panels to illustrate relative dSPN and iSPN activity across individuals; Pearson r = 0.76, p < 1×10e−7, n = 41 comparisons). Top middle, example peak z-scored dSPN and iSPN fluorescence signals (ΔF/F0) from a single mouse. Top right, result of randomly shuffling syllable onsets. Bottom, z-scored dSPN and iSPN fluorescence signal for each syllable is plotted similarly to the top left but each syllable is now instead shaded by their average height (left), 2D velocity (middle), and 3D velocity (right) during the execution of the indicated syllable.
F. Average dSPN (red) and iSPN (green) activity for four example syllables, each of whose pose dynamics were described by a human observer.
Surprisingly, however, both of these correlations weakened when behavior was binned at shorter timescales (Figure 2A). We therefore asked whether binning striatal activity at the sub-second timescale associated with each behavioral syllable would reveal correlations with speed, turn angle (which has been previously associated with near-simultaneous activity in both pathways), length or height (Tecuapetla et al., 2014). Weak but statistically-significant relationships were observed between syllable-timescale neural activity and each of these behavioral parameters. Activity in the direct pathway, but not the indirect pathway, correlated with the mouse’s syllable-binned 2D velocity (Figure 2B). While 3D velocity (which accounts for the 3D position of the mouse centroid) was also correlated with direct pathway activity, it was anti-correlated with indirect pathway activity, suggesting a role for the indirect pathway in modulating fast 3D movements. The difference in activity between the pathways was a better predictor of 3D velocity than either pathway alone, consistent with the direct and indirect pathways encoding non-redundant information relevant to 3D behavioral variables.
Average direct and indirect pathway activity is correlated across syllables, but fine-timescale decorrelations distinguish syllables
In addition to 3D velocity, strong correlations were observed between syllable-binned neural activity and the height of the mouse, consistent with the possibility that the DLS may encode 3D pose dynamics (Figure 2B). As behavioral syllables and grammar effectively encapsulate these pose dynamics, we asked whether striatal activity fluctuates systematically as mice express syllable sequences. Consistent with this possibility, direct and indirect pathway fluorescence was on average decreased immediately before and elevated immediately after a transition into a new behavioral syllable (Figure 2C). The signal derivative in both dSPNs and iSPNs increased before syllable onset, suggesting that average activity in both pathways begins to change 50–75 milliseconds before a new syllable begins. This temporal correlation, which is not present in shuffled data, demonstrates a structured relationship between striatal activity and fast behavioral transitions (representing switching amongst 3D behavioral motifs) during spontaneous behavior. Control experiments demonstrated that the strong short-timescale neural-behavioral relationships observed in DLS were not the consequence of contaminating striatal interneuron fluorescence, and were not apparent in multicolor recordings from the ventral striatum (Figures S3A–S3C).
Plotting the average fluorescence observed during each behavioral syllable revealed that each was associated with specific levels of fluorescence in the direct and indirect pathways (Figures 2D and 2E). Characteristic direct and indirect photometry waveforms also corresponded to each syllable (Figures 2F and S4A–S4C). While the direct and indirect pathways generally exhibited correlated levels of average syllable-specific fluorescence, many syllables were observed in which elevations in direct pathway fluorescence occurred in the context of decreased indirect pathway fluorescence and vice-versa (Pearson’s r = 0.76, Figure 2E). Syllables with particularly high or low velocity tended to exhibit the most decorrelated average pathway activity; interestingly, several syllables with low activity in both pathways exhibited high 3D velocities.
Inspection of syllable-specific direct and indirect waveforms revealed that, although similar, pathway dynamics often exhibited fast-timescale decorrelations (Figures 2F, S4D and S4E). These periods of decorrelation could be related anecdotally to the specific patterns of motion associated with each syllable. For example, a running syllable included an epoch in which direct pathway activity was elevated while the indirect pathway was inhibited. Similarly, the waveforms associated with “scrunching” behavior were characterized by a sharp and phasic elevation of activity in the indirect pathway, and a less pronounced phasic elevation of activity in the direct pathway (Figures 2F and S4D). Taken together, these results suggest that direct and indirect pathway activity dynamics relate to the ongoing 3D pose dynamics expressed by mice during spontaneous behaviors; although direct and indirect pathway activity was largely correlated when averaged over the timescale of individual syllables, pervasive within-syllable decorrelations were also observed.
Pathway decorrelations facilitate behavioral encoding and decoding
The observed relationships between neural activity and specific behavioral syllables were conserved across individual mice and behavioral states, consistent with different types of 3D actions being represented through an invariant neural code (Figures S5A – S5E). To address whether this invariance reflects a systematic mapping between neural activity and behavior, we developed a distance metric based upon the 3D pose trajectories that define each syllable (Figures 3A, 3B and S4A – C; see Methods). Organizing behavior hierarchically using this distance metric revealed that morphologically related behavioral syllables were often associated with similar photometry waveforms, despite the distinctiveness of each syllable (Figures 3B, S4C, and S2B). For example, two different rearing syllables were represented by simultaneous impulse-like upward-deflecting waveforms in both the direct and indirect pathways, with the dSPNs exhibiting a higher amplitude response; however, neural activity (e.g., temporal dynamics and amplitude) associated with each of the two rears varied in a syllable-specific manner (Figure 3B). Likewise, two different “scrunching” syllables were represented by similar but distinguishable phasic waveforms in both the direct and indirect pathways.
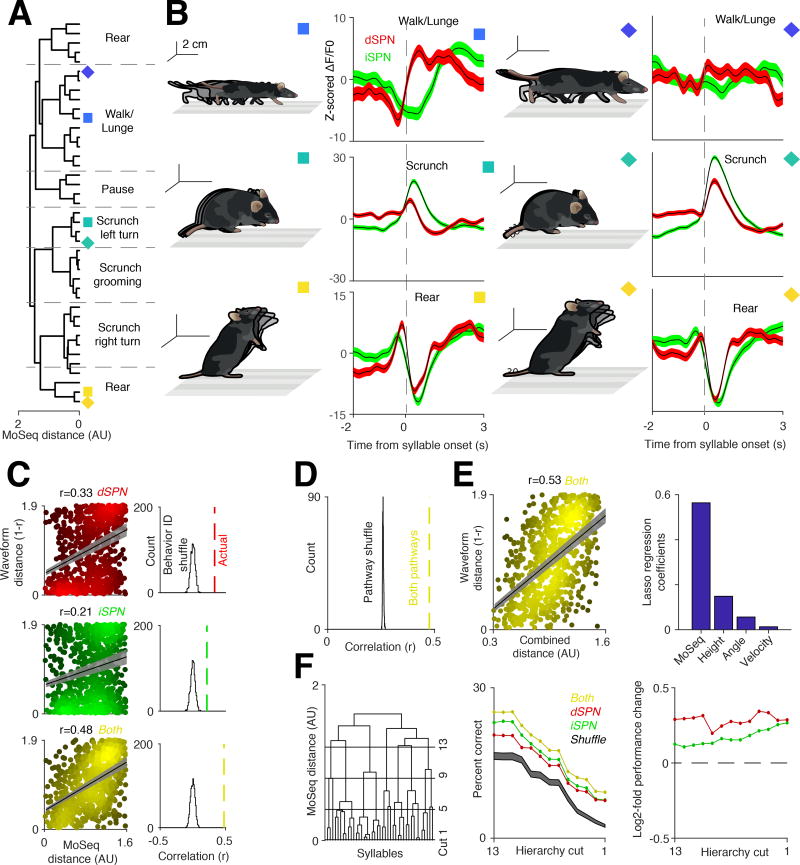
Figure 3. The direct and indirect pathways contain complementary codes for behavioral syllables.
A. Syllables hierarchically ordered by model-based distance (see Methods). Dashed lines indicate human observer-specified boundaries between syllable classes.
B. Six examples of average z-scored photometry signals, aligned to syllable onset (red, dSPNs; green, iSPNs). Colored boxes designate the syllable in panel A represented by the associated waveform.
C. Left, correlations between behavioral distances and syllable-associated dSPN (red), iSPN (green), and combined (yellow) photometry signals (Pearson r, direct pathway p < 1×10e−10; indirect pathway p < 1×10e−9; both pathways p < 1×10e−10; n = 820 comparisons). Right, histograms describing residual correlations after shuffling relationships between syllable identities and photometry signals (1000 random shuffles, see Methods).
D. Same as C bottom right, except here pathway identities were shuffled and the analysis was repeated 1,000 times (pathway shuffle, see Methods).
E. Left, correlation between photometry signals and behavioral distance (here, defined using MoSeq, height, angle, and velocity, with each parameter weighted using LASSO regression) (Pearson r = 0.53, p < 1×10e−10, n = 820 comparisons, see Methods). Right, recovered LASSO regression weights that maximize the neural-behavioral correlations in left.
F. Left, same dendrogram shown in A, with 4 example hierarchical cuts used for the decoding shown on the left displayed (see Methods). Middle, classifier decoding hit rate (y-axis) of syllable identity based upon either dSPN (red) or iSPN (green) waveforms alone, or their combination (yellow); classifier performance was evaluated at progressively deeper levels of the hierarchical clustering (left), with the outermost branch (the full set of 41 syllables) represented as cut 1. Shuffle represents the random assignment of syllable identity to the classifier. Note that the number of classes to decode decreases at higher hierarchical cuts, leading to increased chance performance. Right, ratio between the performance using both pathways and either dSPNs (red) or iSPNs (green) alone.
To quantify the degree to which neural activity systematically encodes for behaviors based upon similarity, we asked whether the behavioral distances between any two syllables relates to the distances between their corresponding neural representations. This analysis revealed that direct and indirect pathway activity each reflect behavioral relationships between syllables (Pearson’s correlation between behavior and dSPNs r = 0.33, iSPNs r = 0.21). However, stronger neural-behavioral relationships were apparent when waveforms from both pathways were considered (r = 0.48; Figure 3C), consistent with fine-timescale decorrelations in dSPN and iSPN activity conveying additional information about behavior. This improvement was only observed when both pathways were considered as independent channels, as repeating this analysis with pathway identities scrambled failed to enhance neural-behavioral correlations (Figure 3D). Performing a predictive correlation analysis using held-out data confirmed that maximal correlation was observed with information combined from the two pathways (Figure S5F). Because weak correlations were also observed between direct and indirect pathway activity and velocity, turn angle and height (Figure 2B), we used LASSO regression to consider the influence of these variables (Tibshirani, 1996). Including these parameters plus syllable-based pose trajectories increased the Pearson’s correlation between fluorescence signal in both pathways and behavior (r = 0.53), although most of this dependence still relied upon information about 3D pose dynamics rather than 2D variables like velocity (Figure 3E).
These data demonstrate a systematic and largely invariant relationship between direct and indirect pathway activity and the particular behavioral syllable being expressed at any moment in time, raising the possibility that syllable-associated waveforms can identify the syllable being expressed. We therefore asked if neural activity in DLS could be used to decode behavioral syllable type or identity, and if so whether decoding improved when considering information from both pathways compared to each pathway alone. Although the resolution of photometry is limited by strong filtering and averaging, a random forest classifier could use syllable-specific waveforms to decode 3D pose dynamics on a trial-by-trial basis across mice (Figure 3F). The classifier was most accurate when decoding a root level of a hierarchical tree describing behavior (in which multiple, related behavioral syllables were lumped), and gradually became less accurate as more behavioral details were considered (Figure 3F). Despite this decline in accuracy, the classifier successfully decoded the identity of each behavioral syllable at a rate significantly above chance (19.36 ± .12% vs. 12.58 ± .013% at chance when decoding 10 “types” of syllables, 9.17 ± .03% vs. 2.45 ± .007% at chance when decoding all 41 syllables, bootstrap SEM). This performance degraded when each pathway was considered separately, with the combined pathways consistently outperforming each pathway alone (Figure 3F). Taken together, these data demonstrate that the direct and indirect pathway waveforms associated with each syllable differentially and collectively represent the 3D pose dynamics that define each behavioral syllable.
Sequence-dependent neural representations for syllables
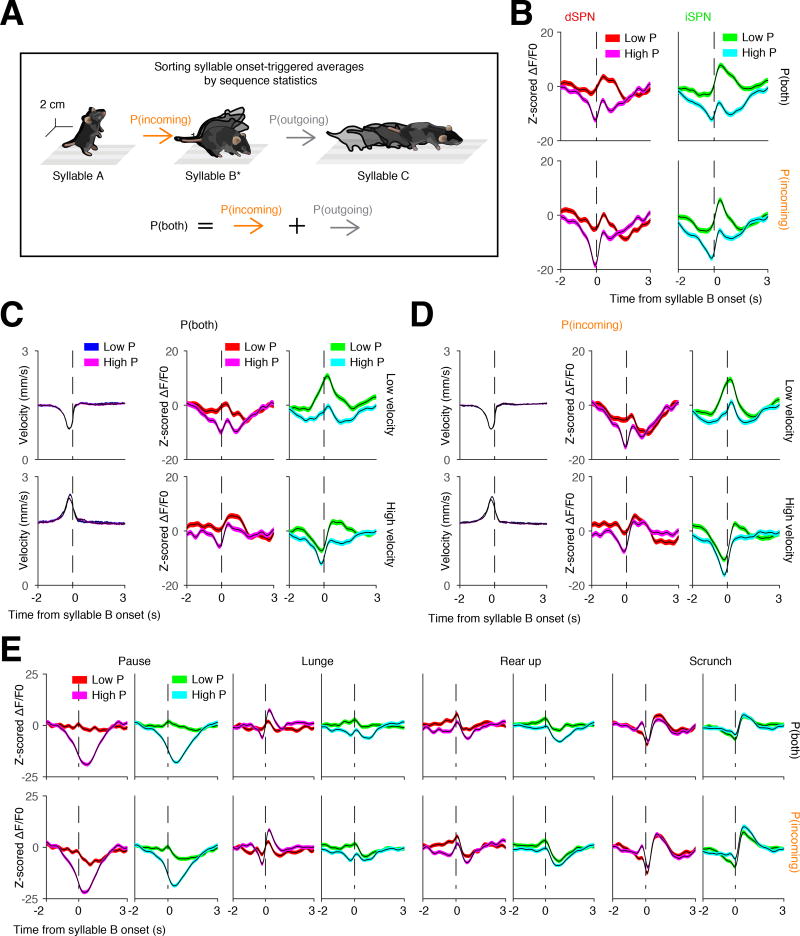
Previous experiments have suggested that the striatum can represent components of sequenced behaviors like grooming (Aldridge et al., 1993). We therefore asked whether syllable-associated DLS activity depends upon the sequential context in which a given syllable is expressed during naturalistic exploratory behavior (Figure 4A). On average, high probability transitions into a given syllable were associated with less activation of the direct and indirect pathways than was observed after low probability transitions (Figure 4B). These sequence-dependent differences were abolished by temporally shuffling the data (p < .001, shuffle test, see Methods) and did not depend upon the 3D velocities of the prior syllable, demonstrating that differences in the neural representations of high and low probability sequences were not due to quantitative differences in movement (Figures 4C and 4D). Furthermore, inspection revealed that the context-dependent modulation of average syllable-associated photometry waveforms reflected differential modulation of individual syllables (Figure 4E). Taken together, these results demonstrate that the neural code associated with a given behavioral syllable is altered by the sequential context in which that syllable is expressed. Syllable-associated waveforms therefore can be at least partially uncoupled from behavioral kinematics, consistent with a role for the DLS in action selection and sequencing.
Figure 4. Sequence-dependent syllable representations in DLS.
A. Schematic of an example syllable sequence. Throughout all panels in this figure, waveforms are aligned to syllable B (starred) and then sorted based on the likelihood of either the incoming transition, or both the incoming and outgoing transitions.
B. Grand average photometry waveforms (red colors, dSPNs; green colors, iSPNs; n = 8 mice) for all syllables, separated by high or low probability of expression (here defined as above or below the 50th percentile probability, respectively). Top, waveforms sorted based upon the summed probability of incoming and outgoing syllables. Bottom, waveforms sorted based upon the probability of the incoming syllable.
C and D. Same as B but controlling for the 3D velocity of the incoming syllable (n = 8 mice). Velocities were grouped by whether they were above or below the 50th percentile of average velocity prior to syllable onset. Syllables were sorted by either high or low incoming velocity, and the associated neural waveforms were then sorted by either high or low probability of combined (C) or incoming (D) syllable expression.
E. Individual syllables analyzed using the same scheme shown in A.
Direct and indirect pathway neural ensembles represent behavioral syllables and grammar
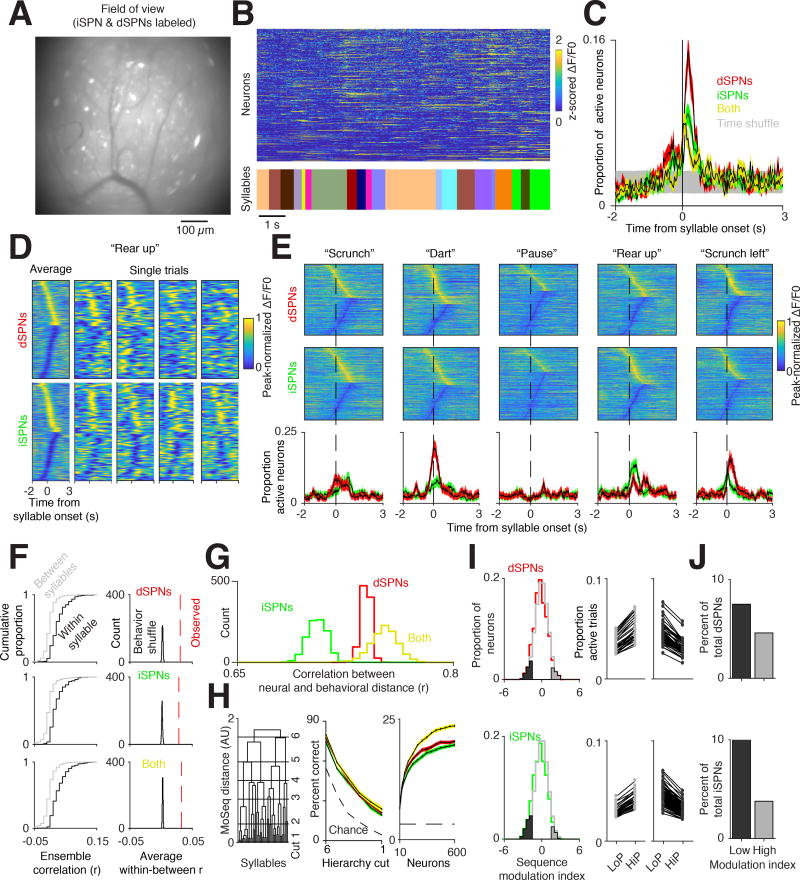
The fluctuations in activity captured by photometry represent the collective dynamics of DLS neurons during behavior; these signals may reflect the continuous evolution of DLS neural activity or switching between different neural ensembles. To explore the relationship between cellular activity in the striatum and the expression of behavioral syllables, we implanted a gradient-index (GRIN) lens over DLS neurons expressing virally-delivered GCaMP6f, and used head-mounted miniscopes to ask how the activity of individual striatal neurons relates to behaviors expressed during open field exploration (Figure 5A) (Ghosh et al., 2011). The direct and indirect pathways were labeled by infecting Drd1a-Cre and A2a-Cre mice with Cre-dependent AAVs (n = 653 dSPNs, n = 794 iSPNs) (Gerfen et al., 2013), and pathway-independent labeling was achieved by infecting mice with a Cre-independent AAV (n = 694 neurons).
Figure 5. Direct and indirect pathway neural ensembles encode syllable identity.
A. Example miniscope field of view (FOV) from a mouse with both dSPNs and iSPNs labeled. Scale bar = 100 µm.
B. Normalized fluorescence of individual neurons extracted using CNMF-e (top, see Methods) aligned to behavioral syllables (bottom, each syllable uniquely color coded).
C. Proportion of active direct (red), indirect (green), and both pathway (yellow) neurons aligned to syllable onset (see Methods). Time-shuffled data is shown in gray (95% confidence interval). Shading indicates bootstrap SEM.
D. Left columns, peak-normalized fluorescence averaged across all instances of the same syllable, in this case a “rear up.” All neurons from all mice are included in this representation. Cells with positive peaks were sorted from earliest to latest, then cells with negative-going peaks were sorted from latest to earliest. Single trials (all other columns) are shown for all neurons, merged across mice (see Methods). In each panel here (and in E) cellular traces are normalized individually to emphasize responses.
E. Top, peak-normalized fluorescence traces for five different syllables aligned to syllable onset, with human annotations above. Cells are sorted as in D. Bottom, proportion of dSPNs (red) and iSPNs (green) active aligned to syllable onset (defined as exceeding two SDs above the mean ΔF/F0). Shading indicates 95% bootstrap confidence interval.
F. Left, cumulative distribution function of ensemble correlations, computed using different examples of the same syllable (black) or examples of different syllables (gray) for the direct pathway (top), indirect pathway (middle), or both (bottom). Right, histogram of the average within syllable correlation after shuffling syllable identities.
G. Histogram of correlations between neural activity distance and Moseq-defined behavioral distance for dSPNs (red), iSPNs (green), or both cell types (yellow).
H. Left, Decoding accuracy along different cuts of the behavioral hierarchy for dSPNs (red), iSPNs (green), and both cell types (yellow) (as in Figure 3F, syllables are clustered into larger groups moving from cut 1 to cut 6). Right, decoding accuracy for individual syllables as a function of the number of neurons provided to the decoder. Shading indicates 99% bootstrap confidence interval. Chance performance increased at higher hierarchical cuts given fewer classes to decode.
I, Left, Distribution of single neuron modulation with respect to syllable sequence frequency. Lower indices indicate higher activity for low probability transitions, while higher indices indicate higher activity for high probability transitions (see Methods). Cells significantly modulated relative to a shuffle control (gray line) are highlighted in dark and light gray. Middle, Each pair of dots indicates the proportion of trials a given neuron was active across all syllables for high (HiP) and low (LoP) transition probabilities. Shown are neurons that increase activity for high transition probability examples (left, light gray) and low transition probability examples (middle, dark gray) from Left.
J. Percent of cells that have a significant modulation index (p<.05 relative to shuffle controls) reveals a greater number of inhibited than activated neurons during high probability sequences.
Inspection of cellular fluorescence traces suggested that DLS neurons preferentially exhibited calcium transients near syllable boundaries (Figure 5B). Indeed, quantifying the number of neurons active in each or both pathways at syllable transitions revealed fluctuations in syllable-associated activity that were similar to those observed by photometry (Figure 5C). Small groups of neurons were modulated during each instance of any given behavioral syllable, although averaging across all instances of a given syllable revealed the participation of a greater proportion of DLS neurons (median fraction of neurons active during each syllable = 8.88 percent, 95 percent bootstrap confidence interval 4.76 - 14 percent, median fraction active during each syllable instance = 6.51 percent, 95 percent bootstrap confidence interval 6.38 – 6.6 percent, see Methods, Figure 5D). When sorted, neurons were active throughout the expression of a given syllable, although this pattern was far less apparent during single syllable instances, consistent with sparse firing of DLS ensembles. Furthermore, dSPN and iSPN activity revealed similar syllable-specific correlations and decorrelations in amplitude and timing as observed previously via photometry (Figures 5E and S6A).
These observations suggest that the identity of the behavioral syllable being expressed at any given moment is represented in DLS by an ensemble code. Consistent with this possibility, ensemble activity patterns aligned to individual examples of the same syllable were more similar to each other than to the activity patterns occurring during different syllables (Figure 5F). The neural code for behavioral syllables was systematic, as the expression of similar behaviors correlated with activation of similar direct and indirect pathway neural ensembles; these correlations were maximized when information was combined from the two pathways (Figure 5G). Ensemble activity could also be used to decode syllable identity, with the best performance observed when neurons from the direct and indirect pathways were simultaneously fed to the classifier (Figure 5H). Classifiers based upon ensembles outperformed those based upon photometry signals, suggesting that the variability of individual neurons substantially contributes to decoding accuracy (using 300 dSPNs and 300 iSPNs, 43.1 ± .18 percent vs. 11.7 ± .05 percent at chance when decoding 10 “types” of syllables, 24.2 ± .18 percent vs. 3.2 ± .03 percent at chance when decoding all syllables, bootstrap SEM).
The membership of syllable-associated DLS ensembles was modulated by the sequence in which particular syllables were expressed (Figure 5I). These sequencing effects were not uniform across the population; rather, bidirectional modulation of neurons was observed when comparing high and low probability sequences. The net result of this modulation was the inhibition of overall neural ensemble activity associated with high probability sequences, a finding consistent with the results obtained with photometry (Figure 5J). Moreover, syllable sequences could be classified as high or low probability at above chance rates when using data from all recorded neurons (rates varied from 70 percent accuracy distinguishing the top and bottom 50th percentile to 80 percent accuracy distinguishing the top and bottom 30th percentiles, see Methods). Taken together, these data demonstrate that the DLS ensembles comprised of both direct and indirect pathway neurons systematically encode information about behavioral syllables and grammar in a manner accessible to downstream circuits.
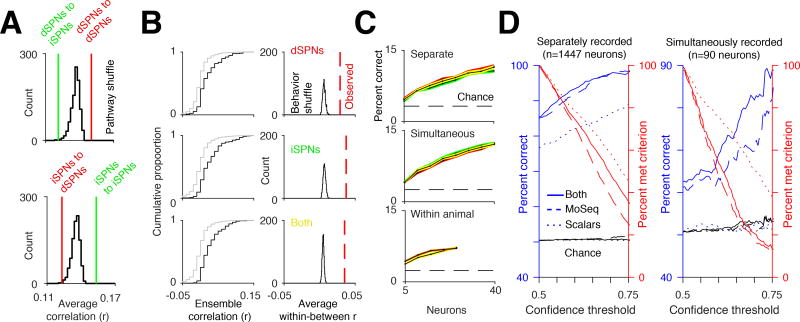
These cellular analyses were performed by assessing the activity of dSPNs and iSPNs in separate mice. To ask whether cellular activity in the two pathways was decorrelated within the same mouse, SPN activity was characterized in behaving mice without regard to pathway identity, and then multiphoton microscopy was used post hoc to assign individual neurons to the direct or indirect pathway (based upon pathway-specific expression of a dTomato marker, see Methods). By aligning miniscope and multiphoton images, we could assess functional activity and definitively assign pathway identity for a subset of neurons per mouse (4–19 neurons per pathway per mouse, n = 4 mice, n=40 total dSPNs and n=50 total iSPNs, Figure S6B).
Simultaneous analysis of syllable-associated cellular activity in both pathways revealed temporal decorrelations in the activity of individual dSPNs when compared to iSPNs, and in iSPNs when compared to dSPNs, particularly during active 3D movements (Figure 6A). Specific neural ensembles were associated with individual behavioral syllables and could be used to decode syllable identity; however, classifier performance was not substantially different to that observed when dSPNs and iSPNs were recorded separately (with the caveat that few neurons were available for this comparison, Figures 6B and 6C). These dual pathway cellular recordings validate the main conclusions drawn from experiments in which pathway-specific activity was assessed in separate mice, and suggest that the behavioral segmentation afforded by MoSeq is sufficiently precise for effective alignment of pathway-specific neural data between mice.
Figure 6. Simultaneous imaging of dSPNs and iSPNs reveals pathway decorrelations.
A. Average cross-correlations compared to the average cross-correlation of cell-type-identity-shuffled traces when the animal’s 3D velocity exceeded the 50th percentile. For all comparisons between the observed correlations and identity-shuffled traces, p<.01.
B. Left, similar to Figure 5F, cumulative distribution function of ensemble correlations for separate examples of the same syllable (black, within syllable) and examples of different syllables (gray, between syllable). Right, histogram of average within syllable correlation after shuffling syllable identities. Red line shows the observed average correlation before shuffling.
C. Top, decoding accuracy of all syllables in psuedopopulations of separately recorded iSPNs (green) and dSPNs (red) as a function of neuron number. Decoding performance using both iSPNs and dSPNs shown in yellow. Middle, decoding accuracy using psudopopulations of simultaneously recorded iSPNs and dSPNs (see Methods). Bottom, example decoding accuracy using simultaneously recorded iSPNs and dSPNs from the same mouse (orange), and from a pseudopopulation of iSPNs and dSPNs where both the iSPNs and the dSPNs used were from a single animal (yellow).
D. Left, performance of a classifier predicting pathway identity of separately recorded dSPNs and iSPNs (see Methods) using MoSeq (blue dashed line), scalars (blue dotted line), or both combined (blue solid line), as a function of the confidence criterion for the classifier (percent of neurons meeting criterion shown in red, see Methods). Right, same as left, except predicting the class of simultaneously recorded dSPNs and iSPNs. Note that the noise in percent correct is due to finite size effects (i.e. there are fewer neurons to classify in the test set).
Given that dSPN and iSPN activity is decorrelated and encodes complementary information about behavioral syllables, we asked whether we could predict if a given neuron belonged to the direct or the indirect pathway based upon its syllable-associated pattern of activity. Using merged data from experiments in which pathway-specific cellular activity was recorded separately, a trained random forest classifier could correctly assign pathway identity to a given neuron between 85 and 100 percent of the time (Figure 6D). Above chance performance was also observed when the pathways were recorded simultaneously, though the performance was degraded due to the relatively small size of the training and testing datasets. These syllable-based classifiers significantly outperformed classifiers in which individual neurons were characterized on the basis of their correlation with scalar behavioral metrics like velocity or height. Thus, behavioral syllables are more effective at highlighting differences between the direct and indirect pathways than scalar metrics for characterizing spontaneous behavior. Furthermore, syllable-specific differences in patterned activity are sufficient to identify the pathway to which a given DLS neuron belongs.
The DLS is required for moment-to-moment action selection
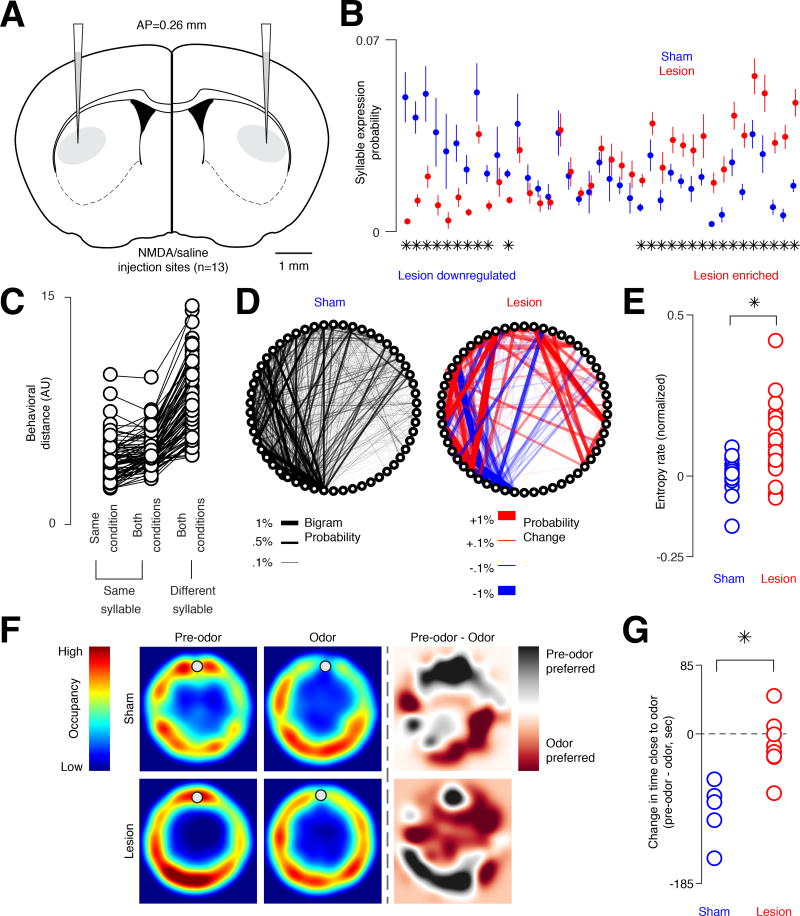
Both photometry and cellular recordings indicate that the DLS encodes information relevant to both action implementation (e.g., the fast dynamics associated with each syllable, the decorrelations between the direct and indirect pathways, and the systematic relationships between neural activity and behavior) and action selection (e.g., the systematic fluctuations of activity at syllable boundaries, and the context-specific representation of syllables). To characterize the functional influence of the DLS on the sub-second structure of spontaneous behavior, we focally lesioned the DLS, allowed the mice to recover for several days, and then assessed the pattern of exploration generated by mice in the circular open field (Figures 7A and S7A, see Methods). Control and lesioned mice exhibited similar velocity during locomotion, and inspection of 3D video revealed no obvious changes in the overall pattern of behavior (Figure S7B, Movie S2). Furthermore, MoSeq-based characterization of the underlying sub-second structure of behavior revealed that the set of behavioral syllables deployed during exploration was similar in control and lesioned mice, as were the specific 3D pose dynamics associated with each syllable (Figures 7B and 7C).
Figure 7. Excitotoxic lesions of the DLS alter the expression and sequencing (but not number or content) of behavioral syllables.
A. Coronal schematic of NMDA and saline injection sites. Scale bar = 1 mm.
B. Syllables expressed in saline and DLS lesioned mice, ordered by differential usage, sorted with the most “lesion-downregulated” syllables on the left and “lesion-enriched” syllables on the right (asterisks = p < 0.05, two-sample t-test corrected for false discovery rate.
C. Average distance between the 3D pose dynamics that define each syllable (behavioral distance) for each instance of the same syllable within a condition (i.e., sham or lesion), for each syllable instance in both conditions, and for all instances of a given syllable and for all other syllables in both conditions (see Methods). Each circle indicates a different syllable.
D. Statemap depiction of syllables (as nodes) and transition probabilities (as edges, here weighted by bigram probability, the probability of one syllable occurring after the other) from control (sham, left, n = 3 mice) injections. Heatmap depiction of the difference (red = upregulated transition, blue = downregulated transition) between this control statemap and the DLS lesion statemap shown on right (n = 5 lesioned mice).
E. The first-order transition entropy rate between syllables in sham and lesioned cases, demonstrating that DLS lesioned mice exhibit less predictable behavioral sequences (p < 0.01 ranksum, z = −2.79; n = 24 lesion trials; n = 15 sham trials). Each circle represents a trial.
F. Top, average occupancy heatmap for sham mice during a session without odor (left), during an odor presentation session (middle), and the difference between these two sessions (right). Bottom, same as top but for lesioned mice. Gray circles mark the approximate location of the odor source.
G. The difference in the amount of time spent within 10 cm of the odor source (dot in F), between pre-odor and odor trials for sham and lesioned mice (p < 0.01 ranksum; ranksum value 16; n = 5 sham trials; n = 8 lesion trials).
However, the frequency with which each syllable was used was significantly altered by DLS lesions, with a subset of syllables preferentially expressed in each condition (Figure 7B). In addition, the sequencing of these syllables was changed, with the transitions between syllables becoming significantly more random (Figures 7D, 7E and S7C). Furthermore, an intact DLS was required for mice to avoid the fox odor TMT, which has been previously demonstrated to elicit innate aversion entirely through stimulus-driven changes in syllable transition frequencies (Figures S5C, 7F and 7G, Movie S3) (Wiltschko et al., 2015). Thus, lesioning the DLS profoundly alters the grammatical microstructure of action — while preserving its underlying components — during spontaneous and motivated behaviors. Although these experiments cannot rule out a role for the DLS in implementing behavioral kinematics during normal physiology (due to the focality of the lesions, and the possibility of functional redundancy), these findings suggest a general role for the DLS in assembling context-appropriate sequences out of sub-second behavioral syllables.
Discussion
In many organisms, naturalistic behaviors are built from modular components that are organized hierarchically and expressed probabilistically (Tinbergen, 1951). Mice express sub-second 3D behavioral syllables that are placed into coherent sequences to create patterns of action (Wiltschko et al., 2015). However, the neural mechanisms that govern the expression and sequencing of these behavioral motifs are not understood. Here, by monitoring DLS activity in both the direct and indirect pathways, we show that behavioral syllables are associated with characteristic and pathway-specific neural dynamics. These dynamics represent key 2D and 3D movement parameters, and effectively encoding these parameters — which collectively encapsulate much of the behavioral diversity expressed by mice — requires both dSPN and iSPN activity. In addition, the DLS both encodes and specifies behavioral grammar, demonstrating that the striatum plays a key role in choosing which sub-second behavioral syllable to express at any given moment.
DLS encodes behavioral syllables and grammar
Recordings from SPNs in awake behaving animals, along with anatomical evidence, demonstrates that the striatum is both somatotopically mapped and preserves timing and kinematic details about motor outputs (Alexander and DeLong, 1985; Barbera et al., 2016; Isomura et al., 2013; Kim et al., 2014; Panigrahi et al., 2015; Rueda-Orozco and Robbe, 2015; Tecuapetla et al., 2014). Information about movement parameters arrives at DLS from the motor cortex and from sensory cortex, and thus DLS activity could reflect motor commands and/or detected movements. We observe pervasive and invariant relationships between sub-second DLS neural activity and behavioral syllables, reminiscent of relationships previously suggested between striatal activity and a subset of high-velocity 3D actions (Klaus et al., 2017). One possible function for these representations in the striatum is to influence pose dynamics and thereby to facilitate the implementation of behavior. Our focal excitotoxic lesion experiments suggest that the DLS is not required for the execution of spontaneously-expressed behavioral syllables, but do not rule out a physiological role for the DLS in action implementation under circumstances in which cortical-basal ganglia circuits are intact.
In addition to potentially influencing motor outputs, the striatum has been proposed to mediate action selection. This notion is supported by the broad convergence of cortical inputs onto SPNs, the integrative nature of SPN physiology, and the widespread mutual inhibition that is characteristic of SPN networks, which has led to “winner-take-all” models in which striatal ensembles representing chosen actions effectively inhibit competing actions (Carter et al., 2007; Mao and Massaquoi, 2007; Zheng and Wilson, 2002). Most work on the function of the striatum in action selection has been in the context of decision making, in which animals are rewarded for choosing from amongst a set of simple behavioral alternatives. However, the concept of action selection can include more probabilistic and naturalistic forms of behavior, such as sequence generation (Berridge and Fentress, 1987; Jin and Costa, 2010). Our demonstration that both bulk and cellular DLS activity are modulated by sequence probability, and that the DLS is required to place behavioral syllables into appropriate sequences during both exploration and odor avoidance, argues that the striatum actively selects and organizes the contents of behavior at the sub-second timescale.
The requirement for the DLS in behavioral sequencing raises an important question about neural representations for action: why does the DLS represent syllables by encoding their contents rather than more abstractly? In other neural systems charged with constructing behavioral sequences, such as the song circuit in the zebra finch and the descending motor circuit in the fruit fly, behavioral components are represented in the patterned firing of command neurons that recruit downstream circuits responsible for action implementation (Cande et al., 2017; Hahnloser et al., 2002). We speculate that the DLS uses this alternative representational strategy for two reasons. First, mice can learn to express new behaviors and, presumably therefore, new behavioral syllables. Using representations of 3D movement as a code for syllables obviates the need to generate a new abstract representation for each new syllable that is generated during a lifetime. Second, the striatum is a likely locus at which particular motor actions are reinforced. It therefore follows that the striatum should contain sufficient information to enable credit assignment of the appropriate action in a rewarding context (Fee and Goldberg, 2011). Our experiments reveal that during exploratory locomotion — a typical precursor to reinforcement in the wild — the striatum encodes all of the information required to reinforce both specific syllables and syllable sequences. This mode of behavioral representation may therefore afford the DLS the ability to flexibly modulate both syllables and grammar in response to learning cues.
Fine-timescale pathway decorrelations support neural codes for action
When averaged over timescales of single syllables, we observe strong correlations in direct and indirect pathway activity across most behavioral syllables. However, when viewed at the sub-syllable timescale, many syllables exhibit periods in which decorrelations between dSPN and iSPN activity are apparent. From both an encoding and decoding perspective these decorrelations appear to play an important role in conveying information about the identity and form of individual behavioral syllables. Indeed, syllable-specific patterns of activity are sufficient to predict whether a given SPN belongs to the direct or indirect pathway. Thus, despite the strong correlations in their dynamics, these pathways fundamentally represent complementary (rather than redundant) codes for behavior. The observed correlations between dSPN and iSPN activity at longer timescales, together with decorrelations at the sub-syllable timescale, potentially reconcile the longstanding conflict between models in which the direct and indirect pathway compete, with those suggesting that they collaborate (Cui et al., 2013; Kravitz et al., 2010; Oldenburg and Sabatini, 2015; Tecuapetla et al., 2014). Further characterization of the relationship between decorrelated pathway activity and sub-second behavior will likely require the use of behavioral segmentation methods like MoSeq, which enables temporally-precise alignment of neural data to a large number of diverse behavioral components.
Revealing neural correlates for 3D behavior in time and space
While syllable identity is encoded in the patterned activity of DLS ensembles, we were unable to perfectly decode behavior from neural activity either via photometry or cellular imaging. This may reflect our inability to record from sufficient numbers of neurons, an imperfect behavioral segmentation (which is almost certain to be the case, given the linear nature of our behavioral modeling and the inherent non-linearities in behavior itself), or a biological limit on the amount of information SPNs convey about ongoing syllables. Furthermore, although we failed to observe strong coupling to behavioral syllables in ventral striatum, it is unlikely that the DLS is unique in encoding information relevant to the underlying structure of behavior. Future experiments will almost certainly reveal important (and potentially causal) relationships between syllables and neural activity in the many cortical and subcortical structures involved in motor control, including different subregions of dorsal striatum not queried here. While the circa 300 ms timescale at which behavioral syllables are organized appears to be at least somewhat privileged in DLS, behavior is structured hierarchically across many timescales. As a consequence, there likely also exist representations for the longer-timescale structure of behavior both within DLS and elsewhere. Further development of MoSeq — which offers a powerful but limited view of behavior — will help to clarify how multiple regions of the brain cooperate to organize spontaneous behavior at multiple spatiotemporal scales, and ultimately to understand how behavioral variability and reward interact to generate precise learned behavioral sequences.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Sandeep Datta (srdatta@hms.harvard.edu).
Experimental model and subject details
Mice
All experiments were approved by and in accordance with Harvard Medical School IACUC protocol number IS00000138. Photometry and imaging were performed in 8–26 week old C57/BL6J (Jackson Laboratories) male mice harboring either the Drd1a-Cre allele (B6.FVB(Cg)-Tg(Drd1-cre)EY262Gsat/Mmucd; MMRRC #030989-UCD) or the A2a-Cre allele (B6.FVB(Cg)-Tg(Adora2a-cre)KG139Gsat/Mmucd; MMRRC #036158-UCD) (Gerfen et al., 2013). Mice were group-housed prior to virus injection and fiber implantation, and then individually-housed for the remainder of the study, on a reverse 12-hour light-dark schedule.
No randomization of animals was implemented, and experimenters were not blinded to animal genotype or condition. Sample sizes for all experiments were determined based on previously published work, and statistical significance was determined post hoc.
Method details
Stereotaxic surgery
Virus injection and optical fiber cannula implant
8 week old male Drd1a-Cre mice were anesthetized with 1–3% isoflurane in oxygen, at a flow rate of 1 L/min. Each animal was injected with 250 nL or 500 nL of a 3:1 or 2:1 mixture of AAV9.CBA.DIO.jRCaMP1b (Cre-On) and AAV9.CBA.DO(fas).GCaMP6s (Cre-Off) (UNC Vector Core), respectively (n = 10 mice) using a Nanoject II Injector (Drummond Scientific, USA) (Saunders et al., 2012). This pattern of infection is expected to simultaneously label direct pathway SPNs with the red calcium reporter jRCaMP1b, and the indirect pathway SPNs with the green calcium reporter GCaMP6s; these fluorophores were chosen due to similarities in the kinetics of their calcium responses (Dana et al., 2016). A 200 µm core 0.37 NA multimode optical fiber (Doric) was implanted in the right dorsolateral striatum, 200 µm above the injection site. Coordinates used for injections and fiber implants: AP 0.260 mm; ML 2.550 mm; DV −2.400 mm from bregma (Paxinos and Franklin, 2004). The optical setup was coupled to the fiber via a zirconia sleeve (Doric).
Due to the low number of local interneurons present in the striatum (<3%), and no known interneuron-specific tropisms for the CBA promoter or the AAV9 serotype in striatum, the contribution of fluorescence signal from interneurons infected from the Cre-Off virus is expected to be minimal (Rymar et al., 2004). We therefore heuristically throughout refer to neurons infected with the red fluorophore as “direct pathway” neurons, and neurons infected with the green fluorophore as “indirect pathway” neurons. To validate this heuristic we directly assessed the degree of interneuron signal contamination. 8 week old A2a-Cre mice were injected with 350 nL of a 3:1 mixture of AAV9.CBA.DIO.jRCaMP1b (Cre-On) and AAV9.CBA.DO(fas).GCaMP6s (Cre-Off) virus, respectively (n = 5 mice), effectively the genetic inverse of fluorophore expression with respect to the Drd1a-Cre photometry mice. The same optical fiber implant procedure and implant coordinates described in the previous paragraph were used. Photometry and behavioral acquisition occurred 3–5 weeks post-surgery, for 5–6 sessions of 20 minutes each.
For photometry in the nucleus accumbens core (NAcc), 8–26 week old Drd1a-Cre mice were injected with a 3:1 mixture of AAV9.CBA.DIO.jRCaMP1b (Cre-On) and AAV9.CBA.DO(fas).GCaMP6s (Cre-Off) virus, respectively (n = 5 mice). A 200 µm core multimode fiber was implanted 200 µm above the injection site (Doric 0.37 NA, 5.5 mm length or Thorlabs 0.39 NA, 5 mm length). Coordinates used for injections and fiber implants: AP 1.3 mm; ML 1.0 mm; DV −3.25 mm. Photometry and behavioral acquisition were performed 3–5 weeks post-surgery, for 4 sessions of 20 minutes each.
Virus injection and GRIN lens implants
To assess pathway-specific single-cell contributions to behavior, 5–12 week old mice (Drd1a-Cre to label the direct pathway; A2a-Cre to label the indirect pathway; both male and female) were unilaterally injected with 500–600 nl of AAV1.Syn.Flex.GCaMP6f.WPRE.SV40 virus (Penn Vector Core) into the right dorsal striatum (n = 4 Drd1a-Cre mice; n = 6 A2a-Cre mice). Coordinates used for injections: AP 0.5mm; ML 2.25 mm; DV −2.4 mm or AP 0.6mm; ML 2.2 mm; DV −2.5 mm from bregma. A gradient index (GRIN) lens (1 mm diameter, 4 mm length; Inscopix; PN 130-000143) was implanted into the dorsal striatum 200 µm above the injection site immediately following virus injection. Behavioral data were obtained between 2–5 weeks post-surgery by coupling the GRIN lens to a miniaturized head-mounted single photon microscope with an integrated 475 nm LED (Inscopix). In total, from these mice, we were able to record calcium activity from n = 653 dSPNs and n = 794 iSPNs,
Pathway-independent single-cell contributions to behavior were also measured. 8–21 week old Drd1a-Cre mice were unilaterally injected with 600 nl of a 1:1 mixture of AAV1.EF1a.DIO.GCaMP6s.P2A.nls.dTomato (Cre-On; Addgene #51082) and AAV9.CBA.DO(Fas).GCaMP6s (Cre-Off; UNC Vector Core), with the coordinates described above (n = 5 mice). This approach selectively labeled direct pathway SPNs with a nuclear-localized dTomato, so that the direct pathway-projecting neurons could be identified under a two-photon (2P) microscope and cross-registered with single photon miniscope data, as described below. While endoscopic data was included from all mice for the pathway independent analysis shown in Figure 5F and Figure 5G (n = 986 neurons), n = 4 of these mice had suitable dTomato expression for cell type identification. To generate additional pathway independent data, 5–11 week old Dyn-Cre mice were unilaterally injected with 500 nl of AAV1.Syn.GCaMP6f (Penn Vector Core), using the same coordinates described above (n = 3 mice). Calcium imaging and behavioral data were obtained 4–6 weeks post-surgery. We were able to identify calcium activity from n = 694 neurons from these mice.
Dorsolateral striatum lesions
C57/BL6 mice were injected with 150 nL NMDA (2mg/100µL; n = 8 mice) or PBS (saline; n = 5 mice) into the DLS. Coordinates used for injections: AP 0.260 mm; ML 2.550 mm; DV −2.500 mm from bregma. Mice were allowed to recover for 1 week. Mice were then placed in a circular open field and imaged using depth cameras during 6 sessions of 45–60 minutes each; data were then analyzed using MoSeq.
Photometry experimental methods and analysis
Photometry and electrophysiological array assembly and implantation
An optical fiber was glued in place above an electrode array (Neuronexus model number A4×8-5mm-200-703-CM32, n = 15 channels were included in this study). The end of the fiber was located in close proximity but dorsal to the conducting pads. Photo-curable cement was used to hold the optical fiber in place. A craniotomy was performed, and the electrode array and fiber combination was implanted so that the fiber was placed 200 µm above the virus injection site (same coordinates described above). The electrode ground was placed in the cerebellum while the reference was placed in visual cortex. The lag between spiking and fluorescence traces was not directly used to adjust the timing of any data in this paper.
Photometry and behavioral data acquisition
Depth-based mouse videos were acquired using a Kinect 2 for Windows (Microsoft) uing a custom user interface programmed in C#. Animals were placed in a circular open field (US Plastics, Ohio) for 20 minutes per session, with at least one day of rest between sessions. The open field was painted black with spray paint (Acryli-Quik Ultra Flat Black; 132496) to avoid image artifacts created by reflective surfaces. Frames were recorded at a rate of 30 Hz.
Photometry data were collected simultaneously with behavioral data. A lock-in amplifier was programmatically designed using a TDT RX8 digital signal processor (code available online at https://github.com/dattalab). A 475 nm LED was sinusoidally modulated at 300 Hz while a 550 nm LED was modulated at 550 Hz (Mightex). Excitation light was passed through a two-color fluorescence mini cube (Doric FMC2_GFP-RFP_FC) into a rotary joint and, in turn, through a fiber optic patch cord connected to the mouse via a zirconia sleeve. Emission was collected through the same patch cord, and then passed through the mini cube to two fiber-coupled silicon photomultipliers (SenSL MiniSM 30035). Raw photometry data were collected through the TDT interface at 6103.52 Hz. Data were de-modulated, low-pass filtered, and down sampled to 30Hz. To align photometry and behavioral data, timestamps obtained from the depth sensor were aligned to the closest timestamp obtained from the TDT acquisition. Sessions with apparent equipment failure or motion artifacts present in the reference channel were excluded from further analysis.
Photometry and electrophysiological array data acquisition
Photometry data were collected through the TDT interface described above, while the electrophysiological data were collected at 30 kHz through the supplied Intan user interface software. Timestamps for the photometry data were aligned to the Intan data through interpolation between both sets of timestamps. Electrophysiological data were common average referenced offline prior to downstream analysis.
Photometry and electrophysiological lag computation
Electrophysiological data were bandpass filtered between 300 and 3000 Hz (4th order Elliptic bandpass filter, 0.2 dB ripple, 40 dB attenuation) and the number of threshold crossings above two standard deviations from the mean were smoothed to produce a measure of multi-unit firing rate. These filtered signals were used to compute the cross-correlation between the multi-unit firing rate and photometry signals.
Photometry data pre-processing
Photometry data from the green and red silicon photomultipliers were collected at 6103.52 Hz with 24-bit sigma-delta analog to digital converters. Then data were demodulated offline by multiplying the digitized raw signals with the appropriate in-phase and quadrature reference sinusoids (300 Hz and 550 Hz for the two excitation LEDs) and then low-pass filtering with a 4th order Butterworth filter (2 Hz cutoff). After demodulation, photometry data were then anti-alias filtered and resampled at the same rate as video acquisition, 30 Hz. The baseline fluorescence component F0 was estimated using a 15 second sliding estimate of the 10th percentile. ΔF was then estimated by subtracting this baseline. For alignment to behavioral changepoints (i.e. switches between syllables, Figure 2C), photometry data were high-pass filtered with a 2nd order Elliptic filter (40 dB attenuation, 0.2 dB ripple, 0.25 Hz cutoff) to remove low-frequency content. We note that this result was robust across a variety of filtering settings, and was also apparent without any filtering at all. Only sessions where the 97.5th percentile of ΔF/F0 exceeded 1% were included. For the calculations shown in Figure 2E, to account for the lag between photometry and electrophysiology signals, syllable-associated activity was considered from 50 ms before syllable onset to 50 ms before the onset of the next syllable. Finally, for all syllable-triggered averages with the exception of those shown in Figures S4C and S5D, to normalize for the different number of trials included in any given plot, the averages were z-scored relative to their respective time-shuffled controls. That is, for each average the mean of the time shuffled averages was subtracted and then divided by the standard deviation of the time shuffled averages.
Photometry waveform distance
For comparisons of waveform distance in Figures 3C and 3E, analysis was conducted in the same manner as the decoding analysis (see below). In brief, waveforms aligned to syllable onset were clipped from syllable onset to syllable offset, then linearly time warped to a common time-base–20 samples. As with the decoding analysis, to compute waveform distance, both z-scored ΔF/F0 and the derivative of this trace were used.
Pathway shuffle
The pathway shuffle shown in Figure 3D was performed as follows. First, the pathway identity for all single trial syllable triggered waveforms was permuted. Then, the same number of trials that went into the averages for Figure 3C was used for each pathway. Finally, the correlation was computed between photometry waveform and behavior distance for each pair of syllables as in Figures 3C and 3E. The procedure was repeated 1,000 times.
Decoding and held-out regression analysis
Decoding of photometry data was performed using a random forest classifier (2000 trees, maximum number of splits 1000, minimum leaf size 1) initialized with uniform prior probabilities for each syllable ID. As with the correlation between photometry waveforms and behavioral distance, for each trial we used the z-scored ΔF/F0 and the derivative of this trace as features. Then, since syllables could have a variety of durations, waveforms were clipped from syllable onset to syllable offset and were linearly time warped to a common time-base (20 samples, same as the correlation analysis). Qualitatively similar results were found when simply clipping the waveforms from syllable onset to 300 ms after syllable onset with no time warping, though the best performance was observed using time warping along with combining both the z-scored ΔF/F0 and derivative traces; the hit rates reported represent the performance of this particular variant. Decoding accuracy was then assessed using the average hit rate measured using a five-fold cross validation (i.e. the number of correct classes predicted out of the total number of predictions). Note that similar performance was observed using alternative classification algorithms, including Naïve Bayes. Syllable labels were clustered using agglomerative clustering with complete linkage. Clusters were formed using a distance cutoff in steps of 0.1 from the minimum to the maximum linkage distance (e.g. in Figure 3A from 0 to 2). Held-out regression of behavioral distance (Figure S5F) was set up in an identical manner, except we used random forest regression. Behavioral distances were defined as the combined distance of a given syllable from syllable 1 (here the reference syllable does not matter, combined distance was constructed using the LASSO regression coefficients in Figure 3E, see below). All parameters for the regression were the same with the exception of minimum leaf size, which was set to 5.
Sequence Modulation
The statistical significance of sequence modulation was tested using the same time window that was used for imaging data–syllable onset to 350 ms after onset (see below). Here, we compared the mean absolute difference in average waveforms for high and low probability sequences (50th percentile cutoff) to the same difference computed for controls where the sequence probabilities were shuffled (n=1,000 shuffles).
Histological examination and cellular counting
Mice used for the two-color photometry were perfused first with PBS and then 4% formaldehyde, and 50 µm coronal sections were cut on a vibratome. Slices were placed under a confocal microscope with a 40X/1.3 NA oil objective, and tiled z-stacked images were acquired. Cells that were labeled with only red, only green, and both fluorophores were manually counted, and expression frequencies were recorded.
Mice used for the lesion experiment were also perfused first with PBS and then with 4% paraformaldehyde and 50 µm coronal sections were cut on a vibratome. Slices were stained with antibodies for glial fibrillary acidic protein (GFAP; Abcam; ab4674, 1/1000 dilution) and NeuN (Abcam; ab104225, 1/1000 dilution) to visualize the extent of the lesion. Slices were imaged using an Olympus VS120 Virtual Slide Microscope.
Imaging experimental methods and analysis
Miniscope data processing
Data were acquired at a rate of 30 Hz, and timestamps were aligned to the nearest depth sensor timestamp for behavioral analysis. Inscopix nVista software was used to concatenate experimental recordings from a single session. The data were motion corrected and spatially down sampled by a factor of 4 and cells were extracted using the constrained non-negative matrix factorization for microendoscopic data (CNMF-e) algorithm (Pnevmatikakis et al., 2016; Zhou et al., 2018). Extracted data underwent a manual exclusion criteria to remove non-neural objects. Only one session per animal was included for downstream analysis, so tracking of neurons across sessions was not necessary. Raw CNMF-e traces were used for all analysis except for Figure 6B, where we used the deconvolved traces. For Figure 5C, a neuron was considered active if its average ΔF/F0 exceeded two SDs above the mean. To better visualize tiling patterns in single trials for Figure 5D, the data in this panel were first smoothed with a .5 Hz low-pass filter, and the trial-average images were smoothed with a 2D Gaussian filter (3 pixel standard deviation).
Two-photon and single photon image cross-registration
In a subset of mice, GCaMP6s was expressed in all SPNs while dTomato was co-expressed in dSPNs. A custom-built 2-photon (2P) microscope was used post hoc to register dSPNs within the field-of-view (FOV) of the miniscope recordings. Mice were awake and head-fixed during 2P data acquisition. A z-stack was taken across 150–220 µm of tissue. Each frame was smoothed with a median filter, and averaged to generate a template image. Then, at least 4 reference points were selected in both the 2P template image and miniscope data to compute an affine transform to map from the miniscope FOV to the 2P FOV (shown in Figure S6B). After this, dSPNs and iSPNs were hand-picked according to a conservative red/green signal overlap threshold. A cell was considered a dSPN if there was red/green overlap that exceeded this threshold, while a cell was considered an iSPN if it only emitted green fluorescence. GCaMP6s was excited with 940–1000 nm. dTomato was excited at the highest SNR wavelength found during acquisition, at 940–1040 nm. For the cross-correlations between dSPNs and iSPNs shown in Figure 6A, we included only timepoints where the animal’s 3D velocity exceeded the 50th percentile (although significant decorrelations were observed across nearly all behaviors except when the mouse was still or nearly still). Pausing syllables increase the inter- and intra-pathway correlations due to the overall lack of activity observed when imaging both pathways at the single cell level during stillness; note that pathway decorrelations were still observed during the expression of slow syllables in data obtained using photometry, which averages activity across hundreds to thousands of neurons.
Decoding of calcium imaging data
For decoding of miniscope calcium imaging data, the ΔF/F0 trace for each neuron was averaged from each syllable onset to syllable offset (syllable binning). Then, neurons across mice were merged by including responses to syllables with a minimum number of trials for all neurons (in this case 15). The merge was used to form a pseudo-population of neurons by including the 15 trials of each neuron’s response to each syllable that met this criterion, so the number of trials per syllable was balanced. This resulted in a feature matrix with 15 × n(syllables) observations and n(neurons) dimensions. A random forest using the same parameters as with the photometry data (2000 trees, 1000 maximum splits, minimum leaf size 1) was then trained using 5-fold cross-validation, and as with the photometry decoding, performance is reported as percent correct–percent syllable labels predicted correctly out of all predictions.
To compare the performance as a function of number of neurons in the pseudopopulation, we randomly sampled neurons 100 times without replacement with a varying number of samples from 10 to 600. For comparisons between dSPNs, iSPNs, and both neuron types, we used the same number of neurons, and equal numbers of iSPNs and dSPNs were sampled to create a balanced dual-pathway population. For example, to compare performance for n=300 neurons, we compared n=300 dSPNs with n=300 iSPNs or n=150 dSPNs and iSPNs (for a total of 300). Data were pooled from n=4 Drd1a-Cre mice and n=6 A2a-Cre mice, with the number of neurons per mouse ranging from 27–336.
Additionally, to test the consistency of decoding performance across mice, we trained decoders on data from individual mice to classify syllable identity. Here, we only used mice with more than 50 neurons in their field of view, and randomly sampled 50 neurons without replacement 10 times and assessed performance using 5-fold cross-validation. Again, a random forest classifier was used with the same parameters that were used for decoding from pooled data. For the A2a-Cre animals (n=4/6 with more than 50 neurons), when classifying all 42 syllables, the decoder achieved 8.35 percent accuracy on average (8.06–8.65 percent, 95 percent bootstrap confidence interval; chance performance was 2.5%). For the Drd1a-Cre animals (n=3/4), average accuracy was 10.21 percent (9.81–10.67 percent, 95 percent bootstrap confidence interval). Thus, performance was not substantially different from decoders trained on data pooled across animals using a similar number of neurons (Figures 5H and 6D).
To decode sequence probability from the miniscope calcium imaging data, we simply merged neurons from the entire dataset into a pseudo-population and replaced syllable labels with a 0 or 1, indicating whether each syllable was in a low or high probability sequence, respectively. As with decoding of syllable identity, the ΔF/F0 trace for each neuron was averaged from each syllable onset to syllable offset. Decoding efficiencies were computed for multiple subsets of the data, including above and below the 50th percentile for both incoming and outgoing transition probabilities, and for the top and bottom 40th percentiles and 30th percentiles; performance increased from 70 percent accuracy to 80 percent accuracy as the separation between what was considered low and high probability progressively increased. Decoding was performed using a support vector machine with a 3rd order polynomial kernel, and accuracy was assessed using 5-fold cross-validation.
Decoding cell type using response properties
For decoding the molecular identity of neurons recorded from Drd1a-Cre and A2a-Cre mice, we used the following features: (1) the Pearson correlation coefficients between each neuron’s time-course and all scalars included in Figure 2B, and (2) the trial-averaged response to each of 42 syllables, from syllable onset to syllable offset. A random forest classifier (2000 trees, minimum leaf size 1) was then trained to predict whether a neuron came from a Drd1a-Cre or A2a-Cre mouse based on either a feature set with just (1), just (2) or (1) and (2) combined. To include predictions of varying confidence levels, we used the probability that a neuron was assigned to a particular class from the random forest classifier. For instance, at a confidence threshold of .6, we only included predictions assigned a probability of .6 or higher when calculating percent correct. Performance was assessed using 5-fold cross validation. The same preprocessing pipeline and classifier were used for decoding the molecularly identity of simultaneously recorded dSPNs and iSPNs, with two differences: (1) given the small size of the dataset of simultaneously recorded neurons (n=90) relative to the number of features (n=42 for the syllable-based features), each feature vector was compressed using PCA, retaining the minimum number of components required to account for 90% of the variance, (2) for the same reasons we used 100 trees instead of 2000.
Identifying significant relationships between syllables and single neuron activity
For each neuron and syllable pair, we estimated the area under the receiver operating characteristic curve (AUC) and evaluated its significance through shuffling the syllable labels and repeating the calculation 1000 times. A neuron-syllable pair was considered significant for p<.05 according to the shuffle test. Then, to determine the reliability of each neuron-syllable pair that passed this test, we computed the number of single trials for which the neuron’s peak ΔF/F0 exceeded 1 STD above the mean.
Sequence modulation index
To quantify the degree to which sequence context impacted the activity of a given neuron, we derived a simple modulation index. First we averaged each neuron’s activity to every instance of a given syllable from syllable onset to 350 ms after syllable onset, which was chosen since it approximates the modal duration of syllables in our model. Then, as with the photometry data, we computed the likelihood of incoming and outgoing transitions for each syllable, and split into low and high likelihood groups using a 50th percentile cutoff for both; that is, if the transition probabilities coming in and leaving a syllable instance exceeds the 50th percentile cutoff, it is considered a high likelihood sequence, if the same probabilities are both below their respective cutoffs, the instance is considered a low likelihood sequence. These group labels are shuffled, and their difference is computed 1000 times to create a null distribution. The mean and standard deviation of this distribution is used to z-score the observed difference, shown in Figure 5I. Cells that are greater than or less than the null distribution for more than 95% of the randomizations are considered significantly modulated.
Motion Sequencing (MoSeq)
Motion sequencing overview
Motion Sequencing (MoSeq) is a catch-all term for a combined machine vision and machine learning system that automatically identifies behavioral motifs and the order in which they occur (Wiltschko et al., 2015). In brief, the technique works as follows: mice are placed in an apparatus (in this case, a circular open field), and their 3D pose dynamics are measured at 30 Hz through the use of a depth camera (here, the Kinect 2, Microsoft). MoSeq seeks to quantify how these pose dynamics change as the mouse freely explores, and so pre-processsing code automatically identifies the mouse in the arena, centers the mouse in an 80 × 80 pixel square, and then aligns the mouse’s nose to the right and tail to the left. All processing is then done on this aligned and cropped set of images, in which the mouse is continually aligned along the virtual axis of its spine.
In the case of simultaneous neural recordings, either through electrophysiology, fiber photometry or through calcium imaging, an additional complexity occurs: the mouse wears some sort of “hat” that partially obscures its head, and there is a fiber coming out of the “hat” that swings as the animal freely behaves, thus variably obscuring the animals from the depth camera placed above. To address this challenge, the updated version of MoSeq described here identifies the hats and cables, subtracts them from the image of the mouse, and then reconstructs those missing pixels through probabilistic PCA.
After filling in missing pixels, the MoSeq algorithm then takes the 80 × 80 pixel aligned movies of each mouse, and performs PCA on this high-dimensional datastream to lower the dimensionality of the data that is being analyzed. Note that this PCA procedure is only performed to make the subsequent computations easier; MoSeq returns similar results when fed raw pixels, although at significant computational cost.
These PCA-reduced 3D imaging data are then fit to a generative model for mouse behavior through the use of computational inference techniques. The model and fitting procedure use regularities in the data to automatically discover, in any behavioral dataset, the optimal number of behavioral syllables, the identity of each of these behavioral syllables (in terms of the specific pose trajectories that define each syllable) and the frequency with which each syllable transitions from one to the other. MoSeq also labels the dataset that was used for training: for each frame of 3D data, MoSeq identifies the most likely behavioral syllable to be expressed. Thus MoSeq takes as its input 3D imaging data of mice, and returns a set of behavioral syllables that characterizes the expressed behavior of those mice, and the statistics that govern the order in which those syllables were expressed in the experiment. While MoSeq has been described previously, the main advance here is to build extensions to this technique that afford better imaging (as the Kinect 2 has better spatial resolution than the originally-used Kinect for Windows), and that enable simultaneous neural recordings. See Figure S2A for a schematic overview of the current MoSeq pipeline.
Motion sequencing
Depth data was modeled as in (Wiltschko et al., 2015) with modifications to accommodate imaging with the Kinect 2. First, raw depth frames were collected from a Microsoft Kinect V2, mounted above the arena, using custom acquisition software written in C#. Frames were collected at 30 Hz, and each frame was composed of 512 × 424 pixels, where each pixel contained a 16-bit integer specifying the distance of that pixel from the sensor in mm. After each session, frames were gzip compressed and moved to another computer for offline analysis.
If no cable was present, the mouse’s center and orientation were found using an ellipse fit. Otherwise, a simple tracking model was used to maintain the mouse’s position in the presence of noise in the depth image. The model assumes that the mouse’s body in the depth image is well fit by a 3D Gaussian with full covariance (x, y, and height from the ground). For each frame, the mean and covariance of the model were initialized using the fit from the previous frame and then refined using expectation maximization, which was run until the improvement in likelihood fell below a threshold value. The log-likelihood weighted pixels were used to then estimate the mouse’s center and orientation with an ellipse fit (similar to the case with no cable). Then, an 80 × 80 pixel box was drawn around the mouse, and the mouse was rotated to face the righthand side. Next, if the tracking model was used, missing pixels were identified by their likelihood according to the Gaussian model. Low-likelihood pixels were treated as missing data and principal components (PCs) are computed using probabilistic PCA (Roweis, 1998; Tipping and Bishop, 1999). If the experiment had no occluding artifacts in the depth video (all experiments that did not require tethering the animal), standard PCA was used. Finally, frames were projected onto the first 10 PCs, forming a 10 dimensional time series that described the mouse’s 3D pose trajectory.
These data were used to train an autoregressive hidden Markov model (AR-HMM) with 3 lags to cluster mouse behavioral dynamics into discrete “syllables,” with state number automatically identified through the use of a hierarchical Dirichlet process. Each state was comprised of a vector autoregressive process that captures the evolution of the 10 PCs over time. The model was fit using Gibbs sampling as described in (Wiltschko et al., 2015) using freely available software (https://github.com/mattjj/pybasicbayes). Model output was insensitive to all but two hyperparameters, which were set using unsupervised techniques for determining the length scales for discrete behaviors as in (Wiltschko et al., 2015). Due to differences in mouse shape and size, along with differences in equipment attached to the mouse, a separate model was fit to each experimental condition: NMDA lesions, Drd1a-Cre DLS photometry, A2a-Cre DLS photometry, NAcc photometry, and miniscope imaging.
Syllable Cross-Likelihood Computation
In order to demonstrate that the states discovered by the AR-HMM (autoregressive hidden Markov mdoel) described meaningful structure in the data, we estimated how well, on average, each state’s AR parameters can describe data segments assigned to other states. This assesses how likely the model is to confuse two given syllables based on the similarity of their 3D pose trajectories on a trial for trial basis. Cross-likelihoods near or above 1 indicate one state can explain another. Cross-likelihoods well below 1 indicate one state does not explain another well. For details see (Wiltschko et al., 2015).
Behavioral usage and transition matrix analysis
Syllable usage was calculated by summing the number of occurrences of each syllable and dividing by total syllable usage across a recording session, converting syllable usage into a percentage. The number of syllables used for each analysis was based on the syllable usage across all sessions within a condition. Syllable usage was cut off at 1% usage (Wiltschko et al., 2015) for all conditions, resulting in 41 syllables in both the lesion and Drd1a-Cre DLS photometry experiments, 42 syllables in the Inscopix experiments, 27 syllables in the NAcc experiments, and 32 syllables in the A2a-Cre experiments. Transition matrices were calculated by counting the total number of occurrences syllable A transitions into syllable B (for all syllables). Behavioral distances used in Figure 7C were computed by estimating the difference in principal component trajectories over the time course of each syllable. More specifically, the Euclidean distance between the first 10 principal components for the first 600 ms after syllable onset was used as a behavioral distance metric. The median behavioral distance value was used for comparisons within conditions and between conditions for the same syllable, and between syllables across all conditions. Note that this behavioral distance metric is a separate distance metric from MoSeq-based behavioral distances (see below), and used specifically in Figure 7C.
MoSeq-based behavioral distances
In order to ask whether similarities between waveforms or neural ensembles corresponds to similarities in behavior, we established a metric to assess how similar or different each behavioral syllable is from every other behavioral syllable. Importantly, each behavioral syllable is clearly distinct from each other (see Figure S2B); although in two-dimensional representations these differences can be hard to appreciate (in no small part because behavior occurs in 3D), from a mathematical perspective and in 3D movies each syllable appears distinct, which suggests that one can derive such a metric for behavioral similarity or difference. To do this, we took advantage of the fact that MoSeq identifies a mean 3D pose trajectory (and the variance from that mean) that is associated with each behavioral syllable. This pose trajectory is encapsulated by the autoregressive process that describes each syllable.
We therefore assessed the similarity of behavioral syllables using the distance between simulated trajectories derived from the autoregressive (AR) coefficients fit to the data. The dynamics of the AR coefficients for each of the 10 principal components (PCs) were computed over 10 time steps (corresponding to 300 ms), and represent that syllable’s prototypical 3D pose dynamics. The trajectories were initialized from the average starting position of the 10 PCs for each syllable. The correlation distance (1-r, where r is Pearson’s r) was then computed between each AR trajectory for all syllables.
LASSO regression
LASSO regression (Figure 3E) was performed using MoSeq distance between all syllables (Figure 3A), along with the correlation distances for height, angle, and 2D velocity as predictors, and the waveform distance (Figure 3C) as observations (Tibshirani, 1996). The final coefficients we selected minimized the 10-fold cross-validated mean square error on this dataset.
Striatal lesions experimental methods and analysis
Information theoretic analysis
Entropy rate was calculated using the standard formula over bigram probabilities (Wiltschko et al., 2015). Jensen-Shannon divergence, a symmetric distance measure between probability distributions (either usage distributions and first order transition distributions), was calculated as described in (Lin, 1991).
Trimethylthiazoline (TMT) exposures
Mice were subjected to one open field session consisting of a 3.5 cm petri dish with a piece of filter paper inside as a control for novel object interactions. In the control and odor experiments, the dish was sealed and holes were drilled in the top to promote odor exchange, but to prevent direct interaction with the odorant. 25% TMT diluted in dipropylene glycol was presented on a piece of filter paper within the petri dish during the next open field session (odor exposure session). In the photometry experiments, to show persistence in syllable encoding between valence states, fluorescence signals were compared between odor and control sessions. In the lesion experiments, to show differences in odor-related behavior in lesioned mice vs sham mice, animal positions were compared between odor and control sessions. To determine the amount of time spent close to the odor source in the lesion odor exposure experiments, we counted the number of frames the mouse was within 10 cm of the petri dish (Euclidean distance).
Quantification and statistical analysis
Binning
For all correlation and decoding analysis (Figures 3C–F, 5F–H, 6B–D) in order to control for Markovian dependencies in the behavior itself that could impact our analysis, we only considered neural activity from syllable onset to syllable offset. Specifically, photometry data was clipped from syllable onset to syllable offset and linearly time warped to a common timebase and calcium traces in the single cell data were averaged from onset to offset (for details see relevant sections above). Note that we also tiled bin sizes to compute neural behavioral relationships at different offsets, widened and narrowed the bin size itself, and considered warped and unwarped data across those bin sizes. In all cases, the specific choice of bin size did not substantially affect any results in the paper.
Statistical comparisons
Statistical comparisons were non-parametric, exceptions are marked throughout the manuscript. Except where noted, error bars indicate bootstrap standard error (1000 bootstraps) and multiple comparison corrections were applied using the Holm-Bonferonni step-down procedure where appropriate.
Data and software availability
All code related to Motion Sequencing will be made available upon request (see www.dattalab.org for download instructions). Data related to this study will also be made upon reasonable request to the Lead Contact (srdatta@hms.harvard.edu).
Declaration of interest
Sandeep Robert Datta is a founder of Syllable Life Sciences, Inc., and is an author on U.S. Patent 9,826,922, which related to the Motion Sequencing technology.
Supplementary Material
“Crowd” movies depicting six distinct behavioral modules automatically identified by the AR-HMM, each labeled with a descriptor; color scale bar indicates the height of any given pixel above the floor of the arena (mm). “Crowd” movies are generated by overlaying individual movies from individual mice; by collating and overlaying 40 of these individual movies into a single movie, the stereotyped nature of each behavioral syllable is made obvious. In these movies, the ongoing expression of the syllable is indicated by a dot being placed over each mouse; imaging frames before the dot onset and after the dot offset are associated with different behavioral syllables that are distinct in all six movies, as is the position of each mouse within the arena. Nevertheless, during expression of the module (when the dot is over the mouse), the mice in all six examples emit a stereotyped motif of three-dimensional motion that can be identified in these movies, and labeled by a human observer. Note that this “crowd” movie is timelocked such that all the modules are expressed at approximately the same time during the movie; this timelocking was achieved by aligning the middle frame of the module across all examples. Despite this timelocking, the duration of each instance of the behavioral module is slightly different; this is because MoSeq defines a unique duration distribution for every behavioral module, rather than a single duration. Also note the occasional appearance of a fiber over or adjacent to the mouse; this fiber represents the photometry cable, which occasionally swings over the mouse or into the camera’s field of view.
A. Left, peak-normalized trial-averages from all dSPNs shown aligned to the onsets of 3 different syllables, with annotations shown above. For each row, the trial average for a given syllable is sorted using the peaks from the trial average highlighted with the red box (e.g. in row 1 all trial averages are sorted using the order from the leftmost trial average). Peaks are sorted by positive-going peaks first from earliest to latest, and then by latest to earliest for negative-going peaks. Right, same as left but for iSPNs.
B. Workflow for registration of miniscope data to 2P data (see Methods). Top left, maximum projection of the green channel for a z-stack taken through the dorsolateral striatum. Here, both dSPNs and iSPNs express GCaMP6s. Top middle, maximum projection of the red channel, revealing dTomato expression (dSPNs only). Top right, merge of the green and red maximum projections. Bottom left, same FOV as top left, except visualized using the miniscope. Shown is the correlation image computed over a behavior session–each pixel is weighted by its correlation with its 4 nearest neighbors, then averaged across all frames in the session. Bottom middle, arrows point to four reference points used in the miniscope and 2P data to compute an affine transform for mapping the miniscope FOV to the 2P FOV. Bottom right, highlighted in red and green are an identified dSPN and iSPN, respectively. Scale bars, 100 µm. Note that while four registration points are shown here, this is the minimum number of points used for registrations; for nearly all FOVs more points were used for alignment.
A. Coronal tissue section of a mouse lesioned with NMDA, and stained with GFAP (red) and NeuN (green). White dashed perimeter marks the extent of the lesion. A depletion of cell bodies within the lesioned area is clearly visible. Scale bar, 1 mm. The extra-striatal red fluorescence is attributable to the background signal, as it was also observed in saline-injected mice.
B. Mean velocity of saline-treated (blue; n = 5 mice) and lesioned (red; n = 8 mice) mice (velocity, p = 0.37 ranksum, z = 0.8805; n = 24 lesion sessions; n = 15 saline sessions).
C. Lesioned mice display significantly more similar behavior to themselves than to saline-treated mice as revealed by Jensen-Shannon divergence, measured for syllable transition frequency and number of transitions, where lesioned mouse syllable statistics were either compared to other lesioned mice (red) or to saline mice (blue) (for both comparisons, p < 1×10e−4 signed rank, z = 4.2857; n = 24 sessions, circles indicate a single session).
Depth imaging of saline-injected (right) and DLS lesioned (left) mice. Scale bar indicates the height of any given pixel above the floor of the arena (mm).
Depth imaging of saline-injected (right) and DLS lesioned (left) mice during odor exposure. Scale bar indicates height of any given pixel above the floor of the arena (mm). White squares indicate the location of the odor dish in each condition.
A. Schematic of photometry set-up. A custom programmed digital signal processor was used to modulate the frequency of the two excitation wavelengths, and fluorescence emission was filtered and decomposed into the resulting photometry signals.
B. Coronal schematic of virus injection and optical fiber placement sites. Scale bar, 1 mm.
C. Coronal confocal image of striatal tissue after infection with Cre-On jRCaMP1b and Cre-Off GCaMP6s; scale bar, 100 µm.
D. Mean proportion of each cell category (both GCaMP/jRCaMP expressing, yellow; GCaMP expressing, green; jRCaMP expressing, red, n = 5 tissue slices from n = 1 animal).
E. Left, correlation between the red and green emission channels with either: (1) the blue LED on only (GCaMP6s excitation), (2) the lime LED on only (jRCaMP1b excitation), or (3) both LEDs on while recording from both green and red cre-independent calcium indicators. The absence of correlation when only one excitation source is activated indicates the absence of crosstalk in the system. Right, average correlation between fluorescence and firing rate, depicted for each indicator and the reference channel. The reference was estimated by calculating the red emission due to blue light excitation (i.e. filtering for the blue LED frequency on the red emission channel), which we assume to be due to autofluorescence of brain tissue and not from fluorescence of either GCaMP6s or jRCaMP1b (Croce and Bottiroli, 2014). Here, any residual bleed-through from GCaMP6s emission on the red channel from blue light excitation was removed using standard blind source separation techniques (FastICA (Hyvarinen, 1999)). This reference was not subtracted from our signal for any other analyses, but is included here to demonstrate the minimal contribution of movement artifacts to the syllable onset-triggered signal.
F. Syllable onset triggered average of the reference signal (top) and its derivative (bottom). Light gray indicates time-shuffled 95% confidence interval of the reference signal. Triggered averages of the GCaMP and jRCaMP signals are shown in main text Figure 2C.
G. Syllable onset-triggered fluorescence waveform derivative (top) appears similar to a trace deconvolved from a kernel generated using the correlation between firing rate and fluorescence activity (bottom). Deconvolution was performed using inverse filtering, i.e. division between the signal and the cross-correlation kernel (Figure 1C) in the frequency domain.
A. Illustration of the MoSeq pipeline in the presence or absence of occluding artifacts (e.g. cables; see Methods for an extended discussion of the MoSeq pipeline). First, raw depth frames are collected from a Microsoft Kinect V2 mounted above the arena. If no cable is present, the mouse’s center and orientation is estimated using an ellipse fit. Otherwise, a simple tracking model is used to establish the mouse’s position and heading in the presence of noise in the depth image. The model assumes that the mouse’s body in the depth image is well fit by a 3D Gaussian with full covariance (x, y, and height from the floor). The Gaussian model is initialized using the fit parameters from the previous frame, and then refined using expectation maximization, which is run until only a minimal improvement in likelihood is found. Then, the mouse is cropped to an 80 × 80 pixel box and rotated such that its head faces rightward and tail leftward (along the virtual axis of its spine). Next, if the tracking model was used, missing pixels are identified by their likelihood according to the Gaussian model. Then, these pixels are treated as missing data, and principal components (PCs) are computed using probabilistic PCA (Roweis, 1998; Tipping and Bishop, 1999). If no artifacts are present in the depth video (e.g. in the lesion dataset), standard PCA is used. Finally, frames are projected onto the first 10 PCs to create a 10 dimensional time series, and then an AR-HMM is fit these data using the MoSeq modeling code (Wiltschko et al., 2015). This modeling approach takes the PCA representation of the mouse’s 3D pose dynamics as input, and automatically identifies repeatedly-used motifs of action, which are characterized mathematically as autoregressive processes in PCA space. Each of these brief autoregressive processes is referred to as a behavioral syllable. Because each frame of the behavioral imaging data can be assigned to its most likely syllable, this approach also yields the observed transition probabilities from one behavioral syllable to the next. Thus, this analytical framework allows simultaneous recording of neural activity and 3D behavior, which is then transformed into a set of behavioral syllables and their sequence of expression.
B. Cross-likelihood analysis depicting the likelihood that a given data instance assigned to a particular syllable is effectively modeled by another syllable, demonstrating that each syllable is well separated from all other syllables (units are nats, where enats is the likelihood ratio, see Methods).
A. Top, model-assigned labels given during behavior. Bottom, photometry signals from the direct (green) and indirect (red) pathways in A2a-Cre animals using the same viral infection strategy as Figure 1A, but with the fluorophores assigned to each pathway now reversed.
B. Top left, dSPN (green) and iSPN (red) activity in A2a-Cre animals aligned to syllable onset. Photometry signals shuffled by time shown in gray (95% confidence interval).
Bottom left, the derivative of the top signals. Right, reference signal and derivative change in fluorescence aligned to syllable onset.
C. Response magnitude from the D1R-expressing SPNs (red) and D2R-expressing SPNs (green) in the nucleus accumbens core (NAcc; left) compared to the DLS (right) at syllable onset. Gray shading, 95% percent confidence interval from time-shuffled signals.
A. Top, mice are imaged at 30 frames per second in three-dimensions using a depth camera (see heatmap) and then MoSeq is used to identify individual syllables expressed during a given experiment. Syllables can be represented as “spinograms” (middle), in which the spine of the mouse is depicted as if looking at the mouse from the side, with time spanning the syllable indicated as increasing color darkness, or as trajectories in principal component (PC) space (bottom, see Methods). Although only two PCs are included in the depiction here, 10 PCs are used to identify behavioral syllables by MoSeq. Note also that here and throughout, the spinograms are shown to aid in human interpretation of the MoSeq-identified syllables, and are not used analytically in any way; note also that these 2D representations of individual syllables (like the isometric views of the syllables shown in e.g., Figure 1) by definition cannot completely capture the 3D behavioral dynamics of each syllable, and therefore may appear similar despite significant differences between syllables (as mathematically shown in S2B).
B. Left, Hierarchical organization of the 3D pose trajectories (computed using principal components, as in the lower panel of A) across mice, along with their associated 2D spinograms (see Methods). Scale bar, 10 mm in both x and y axes. Right, human observer-assigned labels for each syllable; note each syllable is visually distinctive, but because of the limitations of human language often the same descriptors are used (e.g., scrunch, rear up).
C. Same organization as in B with respect to the photometry waveforms, aligned to syllable onset (dashed line). Scale bar, 500 ms x-axis, 0.1 standard deviations (z-scored ΔF/F0) y-axis.
D. Average dSPN and iSPN waveforms associated with each syllable ‘class’ (each given a human observer-assigned descriptor), with waveforms aligned to syllable onset (dashed line). Note that these waveforms are largely correlated, but that differences in the two pathways are apparent (and can often be rationalized based upon the human observer-assigned descriptor).
E. Three examples of dSPN and iSPN activity aligned to MoSeq-defined syllables. Yellow bars indicate periods of high decorrelation between dSPNs and iSPNs.
A. Peak z-scored direct and indirect pathway fluorescence signal associated with each syllable (individual syllables are arbitrarily color coded to allow comparisons between graphs); given the lag between photometry and electrophysiology signals, here syllable-associated activity was considered from 50 ms before syllable onset to 50 ms before the onset of the next syllable. Data are plotted for three individual mice, the mean across these mice, and after randomly shuffling syllable onsets.
B. Average pairwise correlation between waveforms from the same syllable (each syllable-specific set of correlations plotted as a dot) compared to waveforms of other syllables (p < 1×10e−7, z = −5.5332, signed rank, n = 41).
C. To change their behavioral state, mice were exposed to the fox odor trimethylthiazoline (TMT), which induces fear-like behaviors, avoidance, freezing, and alters hormonal status (Fendt et al., 2005; Wiltschko et al., 2015). Density plot depicting the average position of the mouse in the circular open field with and without TMT odor exposure at one location (indicated by red dot; n = 11 mice for odor exposure, and n = 9 mice for pre odor). Lighter color indicates higher occupancy.
D. Average photometry signals from the top 20 most used syllables during exposure to TMT (right) compared to the same syllables with no odor exposure (left).
E. Average pairwise correlation between odor/no-odor waveforms (with data merged between the control and TMT-exposed conditions) from the same syllable compared with the correlation to other syllables (each syllable-specific set of correlations plotted as a dot, p < 1×10e−3, z = −3.5466, signed rank, n = 20 syllables from n = 7 mice).
F. A random forest regression was trained to predict the combined behavioral distance using the LASSO regression coefficients shown in Figure 3E. The best performance, quantified using the mean squared prediction error, was found when combining both iSPN and dSPN waveforms (i.e. this combination results in the minimal error between the predicted and actual behavioral distance). Note that as before, all analyses comparing the direct and indirect pathways separately and together are normalized for the total amount of data.
Table 1.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chicken Anti-GFAP | abcam | Cat# ab4674 |
| Alexa Fluor 488, Donkey Anti-Chicken IgY | Jackson Laboratory | Cat# 703-545-155 |
| Rabbit Anti-NeuN | abcam | Cat# ab104225 |
| Alex Fluor 555, Donkey Anti-Rabbit IgG | Thermo-Fisher | Cat# A-31572 |
| Bacterial and Virus Strains | ||
| AAV9-CBA-do(Fas)-GCaMP6s | UNC Vector Core | N/A |
| AAV9-CBA-DIO-jRCaMP1b | UNC Vector Core | N/A |
| AAV1.Syn.Flex.GCaMP6f.WPRE.SV40 | Penn Vector Core | Cat# AV-1-PV2819 |
| AAV1.EF1α.DIO.GCaMP6s.P2A.NLS.dTomato | Addgene | Cat# 51082 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| N-Methyl-D-aspartic acid | Sigma-Aldrich | Cat# M3262-250MG |
| Experimental Models: Organisms/Strains | ||
| Mouse: B6.FVB(Cg)-Tg(Drd1-cre)EY262Gsat/Mmucd | MMRRC-UCD | MMRRC Stock# 030989-UCD |
| Mouse: B6.FVB(Cg)-Tg(Adora2a-cre) KG139Gsat/Mmucd | MMRRC-UCD | MMRRC Stock# 036158-UCD |
| Mouse: C57BL/6J | Jackson Laboratory | Jax stock #000664 |
| Mouse: B6;129S-Pdyntm1.1(cre)Mjkr/LowlJ | Jackson Laboratory | Jax stock #027958 |
| Recombinant DNA | ||
| pAAV-CBA-DIO-jRCaMP1b | This paper | Addgene ID 110134 |
| pAAV-CBA-do(Fas)-GCaMP6s | This paper | Addgene ID 110135 |
| Software and Algorithms | ||
| MoSeq | (Wiltschko et al., 2015) | N/A |
| MATLAB | The MathWorks | https://www.mathworks.com/products/matlab.html |
| Constrained Non-negative Matrix Factorization for microendoscope data (CNMF-E) | (Zhou et al., 2018) | N/A |
| Mosaic version 1.2.0 | Inscopix | https://www.inscopix.com |
| Intan Recording System Software | Intan Technologies | http://intantech.com/downloads.html |
| Other | ||
| Kinect for Xbox One (Kinect 2) | Microsoft | https://www.xbox.com/en-US/xboxone/accessories/kinect |
| Xbox One S Kinect Adapter | Microsoft | https://www.xbox.com/en-US/xboxone/accessories/kinect/kinect-adapter |
| Shallow tank | US Plastic | Cat# 14317 |
Acknowledgments
We thank members of the Datta and Sabatini labs, Vanessa Ruta, Taralyn Tan, and John Assad for helpful comments on the manuscript, Alex Wiltschko and Matthew Johnson for discussions about MoSeq, Sigrid Knemeyer for technical illustrations, and Paneed Jalilli, Kristen Drummey and David Brann for immunohistochemistry assistance. We thank Ofer Mazor and Pavel Gorelik from the Research Instrumentation Core Facility for engineering support. Core facility support is provided by NIH grant P30 HD18655. SRD is supported by the National Institutes of Health (RO11DC011558, RO11DC016222, and U01NS094191), the Vallee Foundation, and the Simons Collaboration on the Global Brain. BLS is supported by the US National Institutes of Health (U01NS094190) and the Simons Collaboration on the Global Brain. We thank the GENIE Program and the Janelia Research Campus, specifically V. Jayaraman, R. Kerr, D. Kim, L. Looger, and K. Svoboda, for making GCaMP6 and jRCaMP1b available.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
JM, WG, BLS and SRD conceived of and designed experiments. JM and WG carried out the photometry and lesion experiments. CB, SN, KR, JM and WG performed microendoscopy experiments. JM, WG, and SL analyzed data. MH designed the Cre-On/Cre-Off constructs. EP and NB conducted histology for the photometry and lesion experiments. WG, REP and EP conducted odor behavioral experiments. JM, WG, BLS and SRD wrote the manuscript.
References
- Aldridge JW, Berridge KC, Herman M, Zimmer L. Neuronal coding of serial order: syntax of grooming in the neostriatum. Psychological Science. 1993;4:391–395. [Google Scholar]
- Aldridge JW, Berridge KC, Rosen AR. Basal ganglia neural mechanisms of natural movement sequences. Canadian journal of physiology and pharmacology. 2004;82:732–739. doi: 10.1139/y04-061. [DOI] [PubMed] [Google Scholar]
- Alexander GE, DeLong MR. Microstimulation of the primate neostriatum II. Somatotopic organization of striatal microexcitable zones and their relation to neuronal response properties. Journal of Neurophysiology. 1985;53:1417–1430. doi: 10.1152/jn.1985.53.6.1417. [DOI] [PubMed] [Google Scholar]
- Barbera G, Liang B, Zhang L, Gerfen CR, Culurciello E, Chen R, Li Y, Lin D-T. Spatially compact neural clusters in the dorsal striatum encode locomotion relevant information. Neuron. 2016;92:202–213. doi: 10.1016/j.neuron.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge KC, Fentress JC. Disruption of natural grooming chains after striatopallidal lesions. Psychobiology. 1987;15:336–342. [Google Scholar]
- Berridge KC, Fentress JC, Parr H. Natural syntax rules control action sequence of rats. Behavioural brain research. 1987;23:59–68. doi: 10.1016/0166-4328(87)90242-7. [DOI] [PubMed] [Google Scholar]
- Cande J, Berman GJ, Namiki S, Qiu J, Korff W, Card G, Shaevitz JW, Stern DL. Optogenetic dissection of descending behavioral control in Drosophila. bioRxiv. 2017 doi: 10.7554/eLife.34275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AG, Soler-Llavina GJ, Sabatini BL. Timing and location of synaptic inputs determine modes of subthreshold integration in striatal medium spiny neurons. Journal of Neuroscience. 2007;27:8967–8977. doi: 10.1523/JNEUROSCI.2798-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V. Ultra-sensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce A, Bottiroli G. Autofluorescence spectroscopy and imaging: a tool for biomedical research and diagnosis. European journal of histochemistry: EJH. 2014;58 doi: 10.4081/ejh.2014.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Jun SB, Jin X, Pham MD, Vogel SS, Lovinger DM, Costa RM. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature. 2013;494:238–242. doi: 10.1038/nature11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D, et al. Sensitive red protein calcium indicators for imaging neural activity. eLife. 2016;5:e12727. doi: 10.7554/eLife.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee MS, Goldberg JH. A hypothesis for basal ganglia-dependent reinforcement learning in the songbird. Neuroscience. 2011;198:152–170. doi: 10.1016/j.neuroscience.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt M, Endres T, Lowry CA, Apfelbach R, McGregor IS. TMT-induced autonomic and behavioral changes and the neural basis of its processing. Neurosci Biobehav Rev. 2005;29:1145–1156. doi: 10.1016/j.neubiorev.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, El Gamal A, Schnitzer MJ. Miniaturized integration of a fluorescence microscope. Nature methods. 2011;8:871–878. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel AM. The basal ganglia and chunking of action repertoires. Neurobiology of learning and memory. 1998;70:119–136. doi: 10.1006/nlme.1998.3843. [DOI] [PubMed] [Google Scholar]
- Hahnloser RH, Kozhevnikov AA, Fee MS. An ultra-sparse code underliesthe generation of neural sequences in a songbird. Nature. 2002;419:65–70. doi: 10.1038/nature00974. [DOI] [PubMed] [Google Scholar]
- Hyvarinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE transactions on Neural Networks. 1999;10:626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- Isomura Y, Takekawa T, Harukuni R, Handa T, Aizawa H, Takada M, Fukai T. Reward-modulated motor information in identified striatum neurons. Journal of Neuroscience. 2013;33:10209–10220. doi: 10.1523/JNEUROSCI.0381-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger D, Gilman S, Aldridge JW. Neuronal activity in the striatum and pallidum of primates related to the execution of externally cued reaching movements. Brain research. 1995;694:111–127. doi: 10.1016/0006-8993(95)00780-t. [DOI] [PubMed] [Google Scholar]
- Jin X, Costa RM. Start/stop signals emerge in nigrostriatal circuits during sequence learning. Nature. 2010;466:457. doi: 10.1038/nature09263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jog MS, Kubota Y, Connolly CI, Hillegaart V, Graybiel AM. Building neural representations of habits. Science. 1999;286:1745–1749. doi: 10.1126/science.286.5445.1745. [DOI] [PubMed] [Google Scholar]
- Kim N, Barter JW, Sukharnikova T, Yin HH. Striatal firing rate reflects head movement velocity. European Journal of Neuroscience. 2014;40:3481–3490. doi: 10.1111/ejn.12722. [DOI] [PubMed] [Google Scholar]
- Klaus A, Martins GJ, Paixao VB, Zhou P, Paninski L, Costa RM. The spatiotemporal organization of the striatum encodes action space. Neuron. 2017;95:1171–1180. e1177. doi: 10.1016/j.neuron.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz AV, Freeze BS, Parker PR, Kay K, Thwin MT, Deisseroth K, Kreitzer AC. Regulation of parkinsonian motor behaviors by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashley KS. The problem of serial order in behavior. 1951 doi: 10.1016/j.humov.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Lin J. Divergence measures based on the Shannon entropy. IEEE Transactions on Information theory. 1991;37:145–151. [Google Scholar]
- Mao Z-H, Massaquoi SG. Dynamics of winner-take-all competition in recurrent neural networks with lateral inhibition. IEEE transactions on neural networks. 2007;18:55–69. doi: 10.1109/TNN.2006.883724. [DOI] [PubMed] [Google Scholar]
- Oldenburg IA, Sabatini BL. Antagonistic but not symmetric regulation of primary motor cortex by basal ganglia direct and indirect pathways. Neuron. 2015;86:1174–1181. doi: 10.1016/j.neuron.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi B, Martin KA, Li Y, Graves AR, Vollmer A, Olson L, Mensh BD, Karpova AY, Dudman JT. Dopamine is required for the neural representation and control of movement vigor. Cell. 2015;162:1418–1430. doi: 10.1016/j.cell.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KB. The mouse brain in stereotaxic coordinates. Gulf Professional Publishing; 2004. [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron. 2016;89:285–299. doi: 10.1016/j.neuron.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roweis ST. EM algorithms for PCA and SPCA; Paper presented at: Advances in neural information processing systems.1998. [Google Scholar]
- Rueda-Orozco PE, Robbe D. The striatum multiplexes contextual and kinematic information to constrain motor habits execution. Nature neuroscience. 2015;18:453–460. doi: 10.1038/nn.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymar VV, Sasseville R, Luk KC, Sadikot AF. Neurogenesis and stereological morphometry of calretinin-immunoreactive GABAergic interneurons of the neostriatum. Journal of comparative neurology. 2004;469:325–339. doi: 10.1002/cne.11008. [DOI] [PubMed] [Google Scholar]
- Saunders A, Johnson CA, Sabatini BL. Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Frontiers in neural circuits. 2012;6 doi: 10.3389/fncir.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecuapetla F, Matias S, Dugue GP, Mainen ZF, Costa RM. Balanced activity in basal ganglia projection pathways is critical for contraversive movements. Nature communications. 2014;5 doi: 10.1038/ncomms5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B (Methodological) 1996:267–288. [Google Scholar]
- Tinbergen N. The study of instinct 1951 [Google Scholar]
- Tipping ME, Bishop CM. Probabilistic principal component analysis. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 1999;61:611–622. [Google Scholar]
- Wiltschko AB, Johnson MJ, Iurilli G, Peterson RE, Katon JM, Pashkovski SL, Abraira VE, Adams RP, Datta SR. Mapping sub-second structure in mouse behavior. Neuron. 2015;88:1121–1135. doi: 10.1016/j.neuron.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Wilson C. Corticostriatal combinatorics: the implications of corticostriatal axonal arborizations. Journal of neurophysiology. 2002;87:1007–1017. doi: 10.1152/jn.00519.2001. [DOI] [PubMed] [Google Scholar]
- Zhou P, Resendez SL, Rodriguez-Romaguera J, Jimenez JC, Neufeld SQ, Giovannucci A, Friedrich J, Pnevmatikakis EA, Stuber GD, Hen R. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife. 2018;7:e28728. doi: 10.7554/eLife.28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
“Crowd” movies depicting six distinct behavioral modules automatically identified by the AR-HMM, each labeled with a descriptor; color scale bar indicates the height of any given pixel above the floor of the arena (mm). “Crowd” movies are generated by overlaying individual movies from individual mice; by collating and overlaying 40 of these individual movies into a single movie, the stereotyped nature of each behavioral syllable is made obvious. In these movies, the ongoing expression of the syllable is indicated by a dot being placed over each mouse; imaging frames before the dot onset and after the dot offset are associated with different behavioral syllables that are distinct in all six movies, as is the position of each mouse within the arena. Nevertheless, during expression of the module (when the dot is over the mouse), the mice in all six examples emit a stereotyped motif of three-dimensional motion that can be identified in these movies, and labeled by a human observer. Note that this “crowd” movie is timelocked such that all the modules are expressed at approximately the same time during the movie; this timelocking was achieved by aligning the middle frame of the module across all examples. Despite this timelocking, the duration of each instance of the behavioral module is slightly different; this is because MoSeq defines a unique duration distribution for every behavioral module, rather than a single duration. Also note the occasional appearance of a fiber over or adjacent to the mouse; this fiber represents the photometry cable, which occasionally swings over the mouse or into the camera’s field of view.
A. Left, peak-normalized trial-averages from all dSPNs shown aligned to the onsets of 3 different syllables, with annotations shown above. For each row, the trial average for a given syllable is sorted using the peaks from the trial average highlighted with the red box (e.g. in row 1 all trial averages are sorted using the order from the leftmost trial average). Peaks are sorted by positive-going peaks first from earliest to latest, and then by latest to earliest for negative-going peaks. Right, same as left but for iSPNs.
B. Workflow for registration of miniscope data to 2P data (see Methods). Top left, maximum projection of the green channel for a z-stack taken through the dorsolateral striatum. Here, both dSPNs and iSPNs express GCaMP6s. Top middle, maximum projection of the red channel, revealing dTomato expression (dSPNs only). Top right, merge of the green and red maximum projections. Bottom left, same FOV as top left, except visualized using the miniscope. Shown is the correlation image computed over a behavior session–each pixel is weighted by its correlation with its 4 nearest neighbors, then averaged across all frames in the session. Bottom middle, arrows point to four reference points used in the miniscope and 2P data to compute an affine transform for mapping the miniscope FOV to the 2P FOV. Bottom right, highlighted in red and green are an identified dSPN and iSPN, respectively. Scale bars, 100 µm. Note that while four registration points are shown here, this is the minimum number of points used for registrations; for nearly all FOVs more points were used for alignment.
A. Coronal tissue section of a mouse lesioned with NMDA, and stained with GFAP (red) and NeuN (green). White dashed perimeter marks the extent of the lesion. A depletion of cell bodies within the lesioned area is clearly visible. Scale bar, 1 mm. The extra-striatal red fluorescence is attributable to the background signal, as it was also observed in saline-injected mice.
B. Mean velocity of saline-treated (blue; n = 5 mice) and lesioned (red; n = 8 mice) mice (velocity, p = 0.37 ranksum, z = 0.8805; n = 24 lesion sessions; n = 15 saline sessions).
C. Lesioned mice display significantly more similar behavior to themselves than to saline-treated mice as revealed by Jensen-Shannon divergence, measured for syllable transition frequency and number of transitions, where lesioned mouse syllable statistics were either compared to other lesioned mice (red) or to saline mice (blue) (for both comparisons, p < 1×10e−4 signed rank, z = 4.2857; n = 24 sessions, circles indicate a single session).
Depth imaging of saline-injected (right) and DLS lesioned (left) mice. Scale bar indicates the height of any given pixel above the floor of the arena (mm).
Depth imaging of saline-injected (right) and DLS lesioned (left) mice during odor exposure. Scale bar indicates height of any given pixel above the floor of the arena (mm). White squares indicate the location of the odor dish in each condition.
A. Schematic of photometry set-up. A custom programmed digital signal processor was used to modulate the frequency of the two excitation wavelengths, and fluorescence emission was filtered and decomposed into the resulting photometry signals.
B. Coronal schematic of virus injection and optical fiber placement sites. Scale bar, 1 mm.
C. Coronal confocal image of striatal tissue after infection with Cre-On jRCaMP1b and Cre-Off GCaMP6s; scale bar, 100 µm.
D. Mean proportion of each cell category (both GCaMP/jRCaMP expressing, yellow; GCaMP expressing, green; jRCaMP expressing, red, n = 5 tissue slices from n = 1 animal).
E. Left, correlation between the red and green emission channels with either: (1) the blue LED on only (GCaMP6s excitation), (2) the lime LED on only (jRCaMP1b excitation), or (3) both LEDs on while recording from both green and red cre-independent calcium indicators. The absence of correlation when only one excitation source is activated indicates the absence of crosstalk in the system. Right, average correlation between fluorescence and firing rate, depicted for each indicator and the reference channel. The reference was estimated by calculating the red emission due to blue light excitation (i.e. filtering for the blue LED frequency on the red emission channel), which we assume to be due to autofluorescence of brain tissue and not from fluorescence of either GCaMP6s or jRCaMP1b (Croce and Bottiroli, 2014). Here, any residual bleed-through from GCaMP6s emission on the red channel from blue light excitation was removed using standard blind source separation techniques (FastICA (Hyvarinen, 1999)). This reference was not subtracted from our signal for any other analyses, but is included here to demonstrate the minimal contribution of movement artifacts to the syllable onset-triggered signal.
F. Syllable onset triggered average of the reference signal (top) and its derivative (bottom). Light gray indicates time-shuffled 95% confidence interval of the reference signal. Triggered averages of the GCaMP and jRCaMP signals are shown in main text Figure 2C.
G. Syllable onset-triggered fluorescence waveform derivative (top) appears similar to a trace deconvolved from a kernel generated using the correlation between firing rate and fluorescence activity (bottom). Deconvolution was performed using inverse filtering, i.e. division between the signal and the cross-correlation kernel (Figure 1C) in the frequency domain.
A. Illustration of the MoSeq pipeline in the presence or absence of occluding artifacts (e.g. cables; see Methods for an extended discussion of the MoSeq pipeline). First, raw depth frames are collected from a Microsoft Kinect V2 mounted above the arena. If no cable is present, the mouse’s center and orientation is estimated using an ellipse fit. Otherwise, a simple tracking model is used to establish the mouse’s position and heading in the presence of noise in the depth image. The model assumes that the mouse’s body in the depth image is well fit by a 3D Gaussian with full covariance (x, y, and height from the floor). The Gaussian model is initialized using the fit parameters from the previous frame, and then refined using expectation maximization, which is run until only a minimal improvement in likelihood is found. Then, the mouse is cropped to an 80 × 80 pixel box and rotated such that its head faces rightward and tail leftward (along the virtual axis of its spine). Next, if the tracking model was used, missing pixels are identified by their likelihood according to the Gaussian model. Then, these pixels are treated as missing data, and principal components (PCs) are computed using probabilistic PCA (Roweis, 1998; Tipping and Bishop, 1999). If no artifacts are present in the depth video (e.g. in the lesion dataset), standard PCA is used. Finally, frames are projected onto the first 10 PCs to create a 10 dimensional time series, and then an AR-HMM is fit these data using the MoSeq modeling code (Wiltschko et al., 2015). This modeling approach takes the PCA representation of the mouse’s 3D pose dynamics as input, and automatically identifies repeatedly-used motifs of action, which are characterized mathematically as autoregressive processes in PCA space. Each of these brief autoregressive processes is referred to as a behavioral syllable. Because each frame of the behavioral imaging data can be assigned to its most likely syllable, this approach also yields the observed transition probabilities from one behavioral syllable to the next. Thus, this analytical framework allows simultaneous recording of neural activity and 3D behavior, which is then transformed into a set of behavioral syllables and their sequence of expression.
B. Cross-likelihood analysis depicting the likelihood that a given data instance assigned to a particular syllable is effectively modeled by another syllable, demonstrating that each syllable is well separated from all other syllables (units are nats, where enats is the likelihood ratio, see Methods).
A. Top, model-assigned labels given during behavior. Bottom, photometry signals from the direct (green) and indirect (red) pathways in A2a-Cre animals using the same viral infection strategy as Figure 1A, but with the fluorophores assigned to each pathway now reversed.
B. Top left, dSPN (green) and iSPN (red) activity in A2a-Cre animals aligned to syllable onset. Photometry signals shuffled by time shown in gray (95% confidence interval).
Bottom left, the derivative of the top signals. Right, reference signal and derivative change in fluorescence aligned to syllable onset.
C. Response magnitude from the D1R-expressing SPNs (red) and D2R-expressing SPNs (green) in the nucleus accumbens core (NAcc; left) compared to the DLS (right) at syllable onset. Gray shading, 95% percent confidence interval from time-shuffled signals.
A. Top, mice are imaged at 30 frames per second in three-dimensions using a depth camera (see heatmap) and then MoSeq is used to identify individual syllables expressed during a given experiment. Syllables can be represented as “spinograms” (middle), in which the spine of the mouse is depicted as if looking at the mouse from the side, with time spanning the syllable indicated as increasing color darkness, or as trajectories in principal component (PC) space (bottom, see Methods). Although only two PCs are included in the depiction here, 10 PCs are used to identify behavioral syllables by MoSeq. Note also that here and throughout, the spinograms are shown to aid in human interpretation of the MoSeq-identified syllables, and are not used analytically in any way; note also that these 2D representations of individual syllables (like the isometric views of the syllables shown in e.g., Figure 1) by definition cannot completely capture the 3D behavioral dynamics of each syllable, and therefore may appear similar despite significant differences between syllables (as mathematically shown in S2B).
B. Left, Hierarchical organization of the 3D pose trajectories (computed using principal components, as in the lower panel of A) across mice, along with their associated 2D spinograms (see Methods). Scale bar, 10 mm in both x and y axes. Right, human observer-assigned labels for each syllable; note each syllable is visually distinctive, but because of the limitations of human language often the same descriptors are used (e.g., scrunch, rear up).
C. Same organization as in B with respect to the photometry waveforms, aligned to syllable onset (dashed line). Scale bar, 500 ms x-axis, 0.1 standard deviations (z-scored ΔF/F0) y-axis.
D. Average dSPN and iSPN waveforms associated with each syllable ‘class’ (each given a human observer-assigned descriptor), with waveforms aligned to syllable onset (dashed line). Note that these waveforms are largely correlated, but that differences in the two pathways are apparent (and can often be rationalized based upon the human observer-assigned descriptor).
E. Three examples of dSPN and iSPN activity aligned to MoSeq-defined syllables. Yellow bars indicate periods of high decorrelation between dSPNs and iSPNs.
A. Peak z-scored direct and indirect pathway fluorescence signal associated with each syllable (individual syllables are arbitrarily color coded to allow comparisons between graphs); given the lag between photometry and electrophysiology signals, here syllable-associated activity was considered from 50 ms before syllable onset to 50 ms before the onset of the next syllable. Data are plotted for three individual mice, the mean across these mice, and after randomly shuffling syllable onsets.
B. Average pairwise correlation between waveforms from the same syllable (each syllable-specific set of correlations plotted as a dot) compared to waveforms of other syllables (p < 1×10e−7, z = −5.5332, signed rank, n = 41).
C. To change their behavioral state, mice were exposed to the fox odor trimethylthiazoline (TMT), which induces fear-like behaviors, avoidance, freezing, and alters hormonal status (Fendt et al., 2005; Wiltschko et al., 2015). Density plot depicting the average position of the mouse in the circular open field with and without TMT odor exposure at one location (indicated by red dot; n = 11 mice for odor exposure, and n = 9 mice for pre odor). Lighter color indicates higher occupancy.
D. Average photometry signals from the top 20 most used syllables during exposure to TMT (right) compared to the same syllables with no odor exposure (left).
E. Average pairwise correlation between odor/no-odor waveforms (with data merged between the control and TMT-exposed conditions) from the same syllable compared with the correlation to other syllables (each syllable-specific set of correlations plotted as a dot, p < 1×10e−3, z = −3.5466, signed rank, n = 20 syllables from n = 7 mice).
F. A random forest regression was trained to predict the combined behavioral distance using the LASSO regression coefficients shown in Figure 3E. The best performance, quantified using the mean squared prediction error, was found when combining both iSPN and dSPN waveforms (i.e. this combination results in the minimal error between the predicted and actual behavioral distance). Note that as before, all analyses comparing the direct and indirect pathways separately and together are normalized for the total amount of data.