Abstract
Cholesterol is required for maintenance of plasma membrane fluidity and integrity and for many cellular functions. Cellular cholesterol can be obtained from lipoproteins in a selective pathway of HDL-cholesteryl ester (CE) uptake without parallel apolipoprotein uptake. Scavenger receptor B type 1 (SR-B1) is a cell surface HDL receptor that mediates HDL-CE uptake. It is most abundantly expressed in liver, where it provides cholesterol for bile acid synthesis, and in steroidogenic tissues, where it delivers cholesterol needed for storage or steroidogenesis in rodents. SR-B1 transcription is regulated by trophic hormones in the adrenal gland, ovary, and testis; in the liver and elsewhere, SR-B1 is subject to posttranscriptional and posttranslational regulation. SR-B1 operates in several metabolic processes and contributes to pathogenesis of atherosclerosis, inflammation, hepatitis C virus infection, and other conditions. Here, we summarize characteristics of the selective uptake pathway and involvement of microvillar channels as facilitators of selective HDL-CE uptake. We also present the potential mechanisms of SR-B1-mediated selective cholesterol transport; the transcriptional, posttranscriptional, and posttranslational regulation of SR-B1; and the impact of gene variants on expression and function of human SR-B1. A better understanding of this unique pathway and SR-B1’s role may yield improved therapies for a wide variety of conditions.
Keywords: adrenal, liver, bile, steroids
Cholesterol is required for the maintenance of plasma membrane fluidity and integrity, as well as many cellular functions. Tissues such as liver, adrenal gland, and gonads have a special requirement for cholesterol, which is used as a substrate for product biosynthesis (1, 2). In many species, this cholesterol is obtained from plasma lipoproteins by a unique pathway in which circulating lipoproteins bind to the surface of the biosynthetic cells and contribute their cholesteryl esters (CEs) to the cells by a process known as the “selective” cholesterol uptake pathway (3). This pathway differs from the classic endocytic LDL receptor pathway (4) in that CEs, predominantly contained within HDL, are taken into the cell without the uptake and lysosomal degradation of the HDL particle itself (3, 5–10). As described, the selective pathway is a high-capacity physiologically regulated bulk-cholesterol delivery system operating in steroidogenic tissues of a variety of animals to selectively internalize cholesterol to produce steroid hormones (11–14), and in the liver to mediate the transfer of cholesterol into bile and provide cholesterol for bile acid production (15–18). The selective pathway also operates in isolated hepatocytes (7, 19, 20), fibroblasts (19, 20), adipocytes (21, 22), macrophages (23, 24), and adrenal (7, 11, 19, 25), ovarian (12, 13) and testicular Leydig (14) cells, though its function in fibroblasts is less clear. Scavenger receptor B type 1 (SR-B1) is the physiologically relevant cell surface HDL receptor responsible for selective HDL-CE uptake (26).
MOLECULAR AND BIOCHEMICAL CHARACTERISTICS OF SR-B1
The SR-B1 gene (Scarb1) was almost simultaneously cloned as a cluster determinant 36 (CD36)-related class B scavenger receptor by expression cloning using cDNA libraries from a Chinese hamster ovary (CHO) cell line, Var-261, with a mutated LDL receptor gene (27), and as CD36 and lysosomal integral membrane protein (LIMP) II analogous-I (CLA-1) from human erythroleukemia (HEL) cells based on its sequence similarity to CD36 and LIMP-2 (28). CLA-1 is a human homolog of SR-B1. Subsequently, the isolation and characterization of SR-B1 from mouse (26), rat (29), rabbit (30), pig (31), cow (32), and Northern tree shrew (33) have been described. Further studies by Monty Krieger’s group revealed that SR-B1 binds HDL and mediates the selective delivery of HDL-CE (26). This led to identification of SR-B1 as the first bona fide HDL receptor (26).
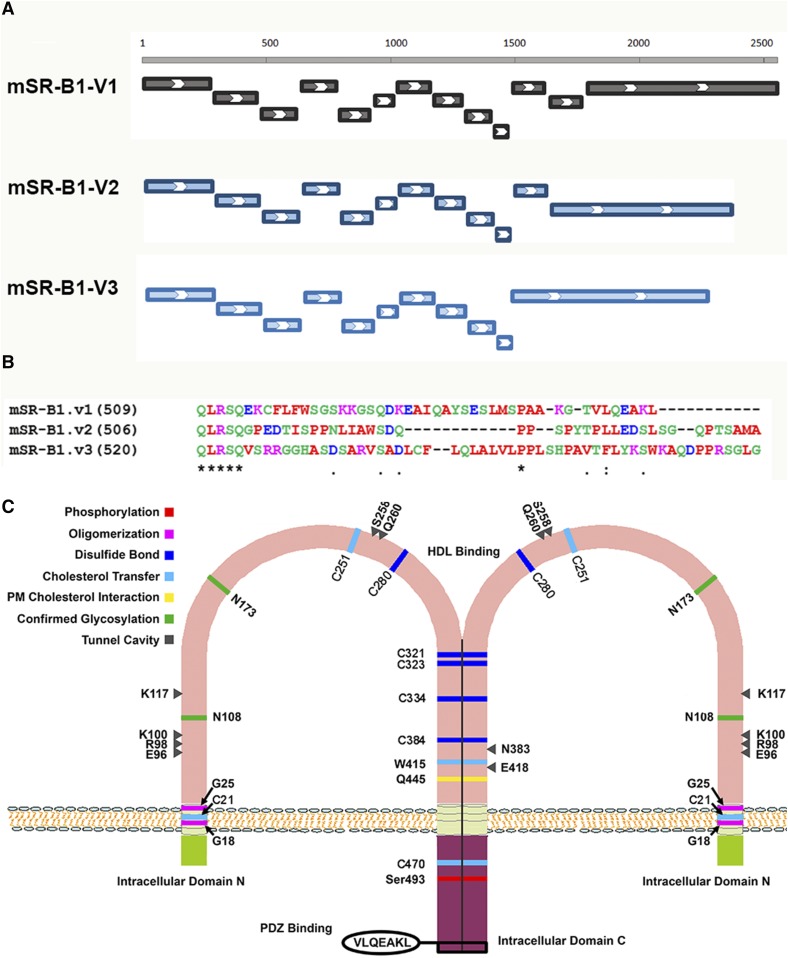
SR-B1 is a member of the class B family of scavenger receptors, which is also known as the CD36 superfamily of scavenger receptors, which are plasma membrane receptors that recognize and take up macromolecules that have a negative charge. Although classified as a group, the scavenger receptor family members are structurally heterogeneous; specifically, the scavenger receptor B family members have a very different molecular structure from other scavenger receptors by having two transmembrane domains with both the N and C termini of the proteins residing intracellularly (34). Proteins in this group are cell surface-associated glycoproteins, which include SR-B1 and its splicing variant, SR-B2 (SR-BII), its human homolog, CLA-1/human SR-B1, and spliced variant, CLA-2/human SR-B2, CD36, and LIMP-2 (35–41). Full-length SR-B1 encodes a 509 amino acid protein, which, like other family members, contains a short N-terminal cytoplasmic domain of 9 amino acids (N-terminal domain), a transmembrane spanning domain of 22 amino acids, a large extracellular domain of 408 amino acids containing a cysteine-rich region and multiple sites for N-linked glycosylation, a second transmembrane domain of 23 amino acids, and a cytoplasmic C-terminal domain of 44 amino acids (37, 42). SR-B1 is posttranslationally glycosylated, and mutational analysis showed the importance of Asn-108 and Asn-173 for plasma membrane localization of SR-B1 and for the ability to transfer lipid from HDL to cells (43). The mouse SR-B1 gene is located on chromosome 5 with 13 exons and 12 introns. Due to alternative splicing, the second isoform of SR-B1, SRB2 (SR-BII), has only 12 exons and a different C terminal with 41 amino acids (36). Using a specific antibody for SR-B2 isoform, SR-B2 protein was detected in mouse liver, testis, and adrenal glands at about 5–12% of immunodetectable SR-B1 (36, 44). Recent genomic analysis revealed a third SR-B1 variant with another different alternative splicing that results in 11 exons and 520 amino acids; however, no functional significance of this variant has been reported yet. The mouse SR-B1 gene exon organization and the C termini of the three variants, as well as a cartoon of the structural features of SR-B1, are shown in Fig. 1. The predicted molecular mass of SR-B1 is ∼57 kDa, but, because the protein is heavily glycosylated (43), it runs ∼83–84 kDa on Western blot (37, 42). SR-B1 is also myristoylated and palmitoylated, but fatty acid acylation of the protein does not affect its selective HDL-CE transport function (45).
Fig. 1.
Gene exon organization (A) and C-termini sequences (B) of mouse SR-B1 variants and structural features of SR-B1 (C). A: Exon organization. Mouse SR-B1 variant 1 has 13 exons and 509 amino acids. Variant 2 arises from alternative splicing at the C-terminal end resulting in 12 exons and 506 amino acids. Genomic analysis shows that variant 3 has 11 exons and 520 amino acids. B: C-terminal sequences of mouse SR-B1 variants. C: Structural features of SR-B1. SR-B1 has an N-terminal and a C-terminal intracellular domain and an extended extracellular ectodomain. Several structural features are important for the function of SR-B1. An N-terminal transmembrane glycine motif (G15_G18_G25) was shown to be required for oligomerization and lipid transport. The C terminal of SR-B1 also contains sequences important for oligomerization. Six conserved cysteine residues (C251, C280, C321, C323, C334, and C384) found in the ectodomain of SR-B1 were also demonstrated to be involved in dimer/oligomer formation. Analysis of human SR-B1 revealed 11 putative N-linked glycosylation sites. Mutational analysis showed the importance of Asn-108 and Asn-173 for plasma membrane localization and for the ability to transfer lipid from HDL to cells. Structural analysis of SR-B1 homolog, LIMP-2, showed that there are eight amino acids that work coordinately to form a tunnel cavity that spans the entire length of the ectodomain allowing facilitated lipid transfer. E96, R98, K100, K117, W258, Q260, N383, and E418 are the amino acids that comprise the tunnel cavity to facilitate lipid transfer of SR-B1. The C-terminal intracellular domain contains an interacting domain (VLQEAKL) for binding with PDZ domain-containing proteins, through which the function of SR-B1 is regulated in a tissue-specific manner.
SR-B1 LIGANDS
Scavenger receptors are cell surface membrane proteins that can bind chemically modified lipoproteins, including acetylated LDL (AcLDL), oxidized LDL (OxLDL), and often many other types of ligands (34, 46). SR-B1 was first identified in scavenger receptor expression cloning experiments using AcLDL as a ligand. Further studies have demonstrated that SR-B1 or its human homolog, CLA-1, bind an array of ligands (Table 1), including both native (VLDL, LDL, and HDL) (38, 47) and modified lipoproteins (OxLDL and AcLDL) (27, 47). SR-B1 can also bind maleylated BSA (38); advanced glycation end-product modified proteins, such as advanced glycation end product BSA (48); hypochlorite-modified LDL (49, 50); discoidal reconstituted phospholipid/unesterified cholesterol particles containing apoA-I, apoA-II, apoC-III, or apoE (51, 52); reconstituted HDL particles containing apoA-I or apoA-II (53); and lipid-free apoE (53). However, SR-B1 exhibits much higher affinity for native HDL (26, 37). The facts that HDL consists of particles of varying sizes and different lipid and protein compositions and densities (54), and that apoA-I is the principal component of HDL, are in accordance with observations that the physical characteristics of HDL as well as the conformation/organization of apoA-I in HDL particles are critical for optimal binding of HDL to SR-B1 (38). Indeed, it has been demonstrated that spherical α-HDL particles, which are larger in size, cholesterol rich, and lower density, bind more strongly to SR-B1 as compared with high-density lipid-poor preβ-HDL (55, 56). Interestingly, reconstituted discoidal apoA-I complexes bind SR-B1 with even much greater affinity than native α-HDL (55). Furthermore, amphipathic helices of apoA-I have been demonstrated to be critical for its binding to SR-B1 (57). The relative levels of apoA-I and apoA-II in spherical α-HDL particles also dictate the association with SR-B1 (38, 53, 58). SR-B1 can also bind anionic phospholipids (59–61), serum amyloid A (60), silica (62), oxidized phospholipids (63, 64), and bacterial cell-wall components, such as endotoxin [lipopolysaccharides (LPSs)] of gram-negative bacteria and lipoteichoic acid of gram-positive bacteria (65, 66). Additionally, SR-B1/CLA-1 can bind and internalize other bacterial and mammalian proteins containing amphipathic helices (60, 66, 67). Finally, SR-B1 has been shown to bind several types of apoptotic cells (65, 68–72). From the perspective of displaying a broad spectrum of ligand recognition, the most closely related proteins are from the LDL receptor family (73, 74). Interestingly, the structure of the SR-B1 family is distinct from that of the LDL receptor family proteins, again by having two transmembrane domains with both the N and C termini of the protein residing intracellularly, while LDL receptor family members have a single transmembrane domain.
TABLE 1.
SR-B1 ligands
| Ligands |
| Native VLDL, LDL, and HDL |
| OxLDL and AcLDL |
| Maleylated BSA |
| Liposomes containing anionic phospholipids |
| Hypochlorite-modified LDL |
| Lipoprotein (a) |
| Discoidal POPC-apoE particles |
| Discoidal reconstituted phospholipid/unesterified cholesterol particles containing apoA-I, apoA-II, or apoC-III |
| Advanced glycation end product BSA |
| Lipid free apoE |
| Oxidized phosphatidylcholine CD36 and other oxidized phospholipids |
| Myeloperoxidase-H2O2-NO2-modified LDL |
| Serum amyloid A |
| Silica |
| Endotoxin (LPSs of gram-negative bacteria and lipoteichoic acid of gram-positive bacteria) |
| Amphipathic helices containing bacterial and mammalian proteins |
| Apoptotic cells |
CELLULAR AND TISSUE DISTRIBUTION OF SR-B1
SR-B1 is expressed in a wide variety of tissues and cell types, including adipocytes, macrophages, endothelial cells, intestine, keratinocytes, epithelial cells, smooth muscle cells, monocytes, placenta, gallbladder, and ocular tissues (37–39, 75–77). However, in rodents, SR-B1 is expressed most abundantly in the liver and steroidogenic cells of the adrenal gland, testis, and ovary (26), tissues that efficiently utilize the selective pathway in deriving CEs for use in bile acid synthesis and biliary cholesterol secretion (78–81) or steroid hormone production (12, 14, 82–86). In humans, CLA-1/human SR-B1 shows a similar tissue distribution (47, 87). Interestingly, variable expression levels of SR-B2 mRNA are also detected in mouse liver, adrenal gland, and testis, with SR-B2 transcripts ranging from 13% to 40% of total SR-B1 in mouse liver (36, 88), from 6% to 30% in adrenal (36, 88), and 80% in whole testis (36). Moreover, of the total hepatic SR-B1/SR-B2 mRNA, only 12% of SR-B1/SR-B2 protein is detected in mouse liver by Western blotting, possibly because of less efficient translation of the SR-B2 protein (36).
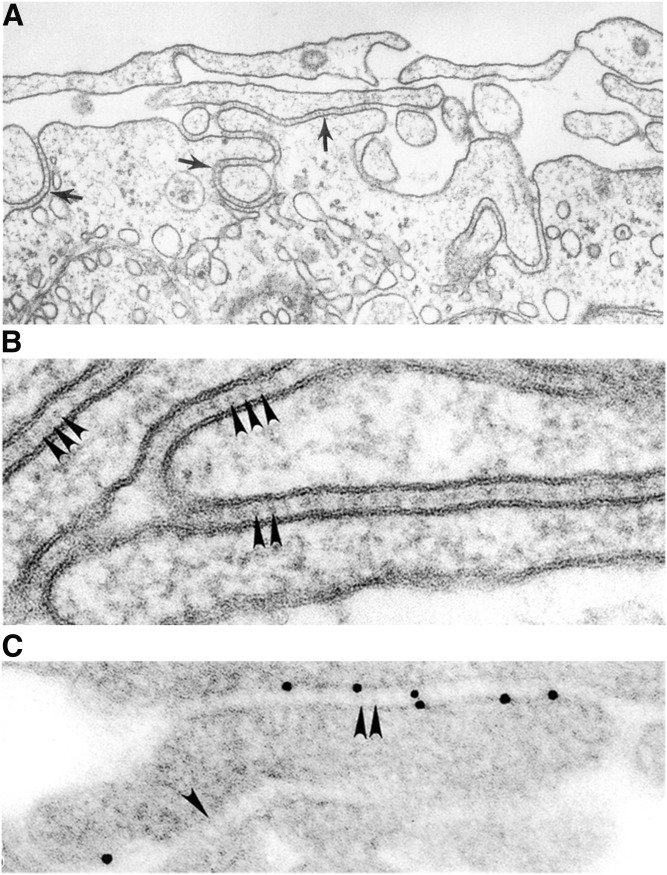
It is of interest that steroidogenic tissues, which express high levels of SR-B1 in vivo, are equipped with an intricate microvillar system for the trapping of lipoproteins (6, 8, 14, 83, 84, 89). This general region of steroidogenic cells is referred to as the microvillar compartment, and the specialized spaces created between adjacent microvilli are called microvillar channels. It is the microvillar channels where the various lipoproteins are trapped prior to the selective uptake of lipoprotein-CEs into cells. Use of electron microscopic immunocytochemistry techniques revealed heavy labeling of SR-B1 specifically in these regions (corresponding to microvilli and microvillar channels), and current evidence suggests that these microvillar compartments express high levels of SR-B1 to facilitate the selective uptake of HDL-CE in steroidogenic tissues. Figure 2 displays photomicrographs demonstrating the microvillar system. In this context, we previously demonstrated that expression of SR-B1 in heterologous (Sf9 insect cells) (90) or homologous (Y1-BS1 mouse adrenocortical cells) (91) cell systems promotes microvillar channel formation and selective HDL-CE uptake. Furthermore, examination of SR-B1 knockout mice provided further evidence that SR-B1 is required for formation of microvilli and microvillar channels. Unlike their wild-type counterparts, the adrenal zona fasciculata cells from SR-B1-null mice exhibit disorganized microvilli, devoid of microvillar channels with an absence of HDL particles (92).
Fig. 2.
Microvillar channels in murine adrenocortical cells. A: Ultrastructure appearance of microvilli (denoted by arrows). B: High magnification of microvillar channels containing HDL (denoted by arrowheads). C: Localization of immunogold-labeled HDL in microvillar channels. This research was originally published in the Journal of Lipid Research (84). © The American Society for Biochemistry and Molecular Biology.
SR-B1 expression has also been reported in several nonmammalian vertebrates, as well as in several insect species. High expression of SR-B1 is reported in the liver, ovary, heart, and blood vessels in the turtle and in the liver of chickens, frogs, goldfish, sharks, and skates (93). SR-B1 expression is reported in the liver of Atlantic salmon (Salmo salar L.) (94) and in several organs of prawns (Macrobrachium nipponense) (95). Twelve to fourteen candidate SNMP/CD36 homologs from each of the genomes of Drosophila melanogaster, Drosophila pseudoobscura, Anopheles gambiae, and Aedes aegypti (Diptera), eight candidate homologs from Apis mellifera (Hymenoptera), and 15 from Tribolium castaneum (Coleoptera) have been identified (96, 97). The 14 D. melanogaster SNMP/CD36 homologs include CG10345, CG1887, CG2736, CG31741, CG3829, CG40006, CG7227, CG7422, crq, emp, ninaD, pes, santa-maria, and Snmp (98). In the silkworm, two genes, Cameo1 and Cameo2, encode proteins homologous to SR-B1 (97, 99). There are 13 other genes homologous to Cameo1 and Cameo2. These genes are SCRB5, SCRB6, SCRB7, SCRB8, SCRB9, SCRB10, SCRB11, SCRB12, SCRB13, SCRB14, SCRB15, SNMP1, and SNMP2 (93).
REGULATION OF SR-B1 EXPRESSION
Promoter analysis
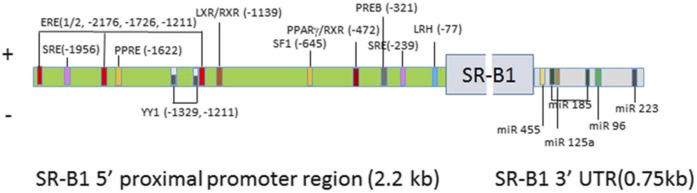
In humans, the SCARB1 gene is located on chromosome 12, and analysis of its 5′-flanking promoter region shows that it is very guanine-cytosine rich with consensus binding sites for many nuclear transcription factors, such as SREBP-1, which regulates SCARB1 expression in response to altered intracellular sterol levels (87, 100). In steroidogenic cells, trophic hormones stimulate SCARB1 expression through changes of cellular cAMP levels (101), and steroidogenic factor-1 (SF-1) is one of the major transcription factors involved in this pathway through binding to its consensus sequence in the SCARB1 promoter (101). Analysis of both the human and rat promoter 5′-flanking regions revealed the presence of many consensus sequences for binding transcription factors responsible for positive and negative regulation of the SCARB1 promoter, although the exact location of the regulatory elements varies in the human and rat promoters. Figure 3 illustrates the presence of the binding sites for positive and negative regulatory factors in the 5′ proximal 2.2 kb region of the rat promoter. There are two binding sites for SREBP-1 that mediate positive regulation of Scarb1 expression in steroidogenic tissue and parenchymal liver cells when cellular cholesterol is depleted (100). There is one SF-1 binding site for regulation mediated through alterations in cellular cAMP levels (101). There are three estrogen response element binding sites (ERE half-sites) through which estrogen receptor regulates Scarb1 expression in an E2-dependent manner in coordination with mixed-lineage leukemia histone methylases (102) in human liver and placenta cells. In the adrenal, prolactin regulatory element-binding protein (PREB) can bind to the PREB-binding core element at −321 of the human Scarb1 promoter and can induce SR-B1 protein expression under conditions that increase cellular cAMP levels (103). In both human and rodent preadipocytes and liver cells, LXRα/RXR and LXRβ/RXR were shown to bind the Scarb1 promoter and induce Scarb1 transcription (104). In the rat Scarb1 promoter, there is a PPRE motif located at −1622, and PPARα and RXRα were shown to bind to the SR-B1 PPRE motif and result in increased expression of Scarb1 in hepatic and macrophage cell lines (105). Estrogen receptors α and β bind to the three estrogen-responsive elements in the rat SR-B1 promoter and liver receptor homolog 1 binds to an LRH binding site in the promoter, both upregulating Scarb1 (106). There are two binding sites for Yin Yang 1 transcription factor in the rat Scarb1 promoter at −1329 and −1211, through which YY1 binds and represses Scarb1 promoter activity under basal conditions (107). Using two-hybrid studies, Shea-Eaton and colleagues also confirmed that YY1 can bind to SREBP-1a, therefore inhibiting the interaction of SREBP-1a binding to the SRE and negatively regulating the expression of Scarb1 (107). Other negative regulators for the Scarb1 promoter include the nuclear receptor, NR0B1 (DAX-1), a protein that plays an important role in adrenal development. DAX-1 was shown to repress the expression of the Scarb1 promoter through interaction with SREBP-1a and SF-1 (108). In addition, pregnane X receptor, which is a regulator of detoxification processes, was shown to repress the expression of human SCARB1 in response to the pregnane X receptor agonists, rifampicin and lithocholic acid (109).
Fig. 3.
Regulation of SR-B1 expression. The 5′ proximal 2.2 kb region of the rat Scarb1 promoter contains binding sites for various regulatory factors for positive and negative regulation of Scarb1 expression. The 3′ UTR of the rat Scarb1 gene contains binding sites for miR-185, miR-96, and miR-223 for negative regulation of SR-B1 expression in liver and macrophages, as well as binding sites for miR-125a and miR-455 for negative regulation of SR-B1expression in steroidogenic tissues.
The 3′ untranslated region
Recently, small noncoding RNAs, including microRNAs, were demonstrated to regulate expression of many genes. These microRNAs are about 22 nucleotides and can bind to specific motifs, mostly at the 3′ untranslated region (3′ UTR) of a gene, silencing gene expression via mRNA degradation or preventing mRNA from being translated. Analysis of the sequence of the SR-B1 3′ UTR revealed potential binding sites for several miRNAs. Further analysis demonstrated negative regulation of SR-B1 in liver and macrophages by miR-185, miR-96, and miR-223 (110). The 3′ UTR of SR-B1 also possesses binding sites for miR-125a and miR-455. Studies show that both miR-125a and miR-455 are present in steroidogenic cells and their expression was suppressed by ACTH and cAMP treatment. Mutational analysis of their binding sites in the 3′ UTR of SR-B1 revealed that both miRNA-125a and miRNA-455 bind to those specific sites in the 3′ UTR of SR-B1 mRNA and negatively regulate SR-B1 expression and inhibit steroidogenesis (111).
STRUCTURAL FEATURES OF SR-B1
Various approaches, including site-directed mutagenesis, “domain swap,” and chemical cross-linking, have been utilized and have identified several structural features to be important for the function of SR-B1 (Fig. 1C). The C-terminal intracellular domain contains an interacting domain (VLQEAKL) for binding with PDZ domain-containing protein, through which the function of SR-B1 is regulated in a tissue-specific manner (for detail see below). SR-B1 was shown to form oligomers, which display improved function (84, 112–116), with several portions involved in oligomerization. An N-terminal transmembrane glycine motif (G15_G18_G25) was shown to be required for oligomerization and lipid transport (114), whereas studies using fluorescence resonance energy transfer techniques to visualize SR-B1 homo-oligomerization revealed that the C termini of SR-B1 appeared to be involved in its homo-dimerization (113). Meanwhile, using a sulfhydryl-reactive reagent, the six conserved cysteine residues (C251, C280, C321, C323, C334, and C384) found in the ectodomain of SR-B1 were also demonstrated to be involved in dimer/oligomer formation (117). Experiments using chimeric SR-B1 and CD36 revealed that the N terminal of the extracellular domain is important for unesterified cholesterol efflux to HDL (118, 119).
Several recent studies have also examined the impact of cysteine residues on cellular trafficking and selective HDL-CE transport function (117, 120–123). These studies were achieved primarily by mutating cysteine residues in mouse, rat, or human SR-B1 individually or in certain combinations to either Ser or Gly, and subsequent expression of individual constructs in cultured cells followed by determination of receptor activity, subcellular distribution of receptor protein, or selective lipid transport function. Two reports demonstrated that four cysteine residues, C280, C321, C323, and C334, may be involved in disulfide bond formation and critically involved in the expression and function of SR-B1 (117, 122). In addition, Yu et al. (123), using a mass spectrometry technique, have reported the identification of two disulfide bonds in SR-B1 that connect cysteine residues within the conserved C321-P322-C323 (CPC) motif and connect C280 to C334 and a reduced cysteine side chain that contribute to the functional expression of SR-B1. Using a C323G mutant, a blocking antibody against C323 region, and a C323G mutant transgenic mouse model, Guo et al. (121) demonstrated that C323 plays a critical role in HDL binding to cell surface SR-B1 and SR-B1-mediated selective HDL-CE uptake. An exoplasmic cysteine, C384, of SR-B1 has also been implicated in SR-B1-mediated selective HDL-CE transport activity (120).
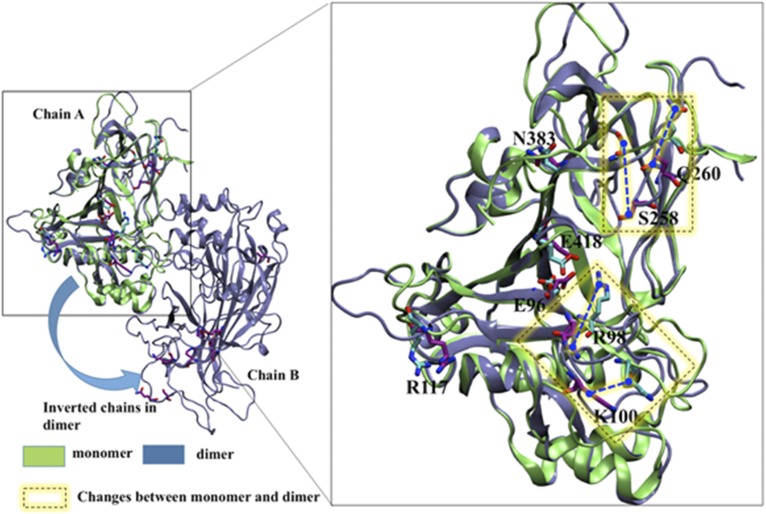
The crystal structure of LIMP-2 was obtained recently and showed that the main ectodomain of the protein contains an antiparallel β-barrel core with many short α-helical segments (124). Two disulphide bridges stabilize the fold. The disulphide bridge pattern for LIMP-2 (C274-C329 and C312-C318) is similar to that predicted for SR-B1 (C321-C323, C274-C329) and for CD36 (C313-C322, C272-C333), and is consistent with experimental data (123, 125, 126). Most of the proteins in this family have lipid transport activity, and the crystal structure of LIMP-2 showed that eight amino acids work coordinately to form a tunnel cavity that spans the entire length of the ectodomain, allowing facilitated lipid transfer. These eight acidic and basic amino acids form a network of hydrogen and ionic bonds and contribute to the lining of the cavity, which is predominately hydrophobic to accommodate lipid moieties. Figure 4 displays the structure of SR-B1, both as a monomer and as a dimer, modeled on the crystal structure of LIMP-2, with the eight amino acids comprising the tunnel cavity highlighted. As illustrated, there are conformational changes in the tunnel cavity that are predicted to occur during the transition between monomer and dimer. Further experimental studies will be needed to explore the functional significance of the structural transition between monomer to dimer.
Fig. 4.
Structure of human SR-B1, both as a monomer and as a dimer. The monomer and dimer forms of human SR-B1 were built separately through homology modeling using MODELLER (246). From 200 generated models, the top ranked models were chosen for energy minimization through Amber molecular dynamics simulation (247). The energy-minimized structures were used for the structural analysis. The residues comprising the tunnel cavity are labeled, and the regions where conformational differences are observed between monomers and dimers are highlighted in yellow squares.
Recently, a fragment of SR-B1 (residues 405-475), which includes the C-terminal transmembrane domain, was purified and reconstituted in detergent micelles (123). Analysis of this fragment using NMR has generated high-resolution structural information of the C-terminal transmembrane domain of SR-B1 (127), which revealed that there are two short α-helices (residues 409-419 and 427-436) and a long α-helix spanning the entire transmembrane domain (residues 438-469). There is a potential GXXXG glycine dimerization motif starting at G420 together with a putative leucine zipper motif spanning from amino acid 413 to amino acid 455. Mutational analysis showed that mutants L413A and L448A resulted in decreased receptor efflux of unesterified cholesterol to HDL. G420 was previously shown to be important for SR-B1 to function for CE delivery (128).When a smaller SR-B1 fragment containing residues 405-445, which lacks the hydrophobic transmembrane domain, was purified and examined by NMR, this fragment was shown to require a hydrophobic environment to fold properly, suggesting the existence of a potential membrane-interacting juxtamembrane domain.
As mentioned above, oligomerization of SR-B1 improves its function. Recently a crystal structure of LIMP-2 dimer bound with cholesterol and phosphatidylcholine was obtained. Further analysis revealed that LIMP-2 switches from a monomeric form to a dimeric form when binding to different ligands. When a monomer, it binds glucocerebrosidase; whereas as a dimer, it preferentially binds anionic phosphatidylserine (126). This shifting of ligand specificity through oligomerization could be a mechanism of regulation shared with other proteins in the family, such as SR-B1 and CD36, but awaits experimental evidence.
LIPID UPTAKE FUNCTION OF SR-B1
A major function of SR-B1 is to facilitate the transport of CEs from HDL or other lipoproteins to cells by a nonendocytic mechanism referred to as selective uptake (37–39, 75, 76, 129–131). Numerous studies have demonstrated that SR-B1-mediated selective uptake of CE does not show a specificity toward a single class of lipoproteins. It can utilize a number of lipoproteins, including human and rat HDL, reconstituted synthetic HDL particles containing apoA-I, apoC, or native or modified apoE, IDL, and LDL as donors of CE for SR-B1-mediated selective CE uptake in vitro and/or in vivo (38, 76). However, because these donor lipoproteins differ in characteristics, such as apolipoprotein conformation and apolipoprotein and lipid composition, as well as the fact that some of these lipoproteins may serve as ligands for other important pathways (e.g., human LDL and apoE-rich rodent HDL are potent ligands for the LDL receptor endocytic pathway), these lipoproteins do not deliver CEs to cells with similar efficacies. Based on the evidence obtained using CD36/SR-B1 chimeras (132, 133), SR-B1 mutant (N173Q) (43), apoA-I mutants (134), apoA-I−/− HDL particles (135, 136), and a chemical inhibitor of selective CE transport (137), it appears that initial binding of HDL to cell surface-associated SR-B1 and subsequent selective CE transport to cells are independent processes (39). The selective uptake process itself can be broadly divided into three steps: 1) binding of cholesterol-rich donor lipoprotein particles to the loop domain of SR-B1; 2) SR-B1-mediated transfer of CEs from the lipoprotein particles to the plasma membrane; and 3) the release of the cholesterol-poor lipoprotein particles back into the circulation (37, 39, 138). Among these steps, the mechanism by which SR-B1 facilitates the transfer of CEs to the plasma membrane is not fully understood. Rodrigueza et al. (139) provided evidence in favor of a model in which SR-B1 forms a nonaqueous channel between lipoprotein particles and the cell membrane through which CEs move down in a concentration gradient manner. The validity of this model is supported by the recent report of the high-resolution crystal structure of the extracellular domain of the lysosomal LIMP-2, and by modeling the structure of SR-B1 and its other family member, CD36 (124, 140). Importantly, the crystal structure shows the existence of a large cavity or tunnel that traverses the entire length of the molecule. The dimensions (with 5 × 5 Å opening and 22 × 11 × 8 Å cavity) of this hydrophobic tunnel are sufficient to accommodate CE and free cholesterol (FC) molecules, and SR-B1 mutagenesis studies provided direct support for the involvement of this hydrophobic tunnel as a facilitator of cholesterol ester transfer from bound donor HDL particles to the cell surface plasma membrane (124, 140, 141). This is further supported by the observation that an inhibitor of selective HDL-CE uptake binds covalently to Cys384, which is located in the lumen of the tunnel. It is predicted that binding of the inhibitor to this site would block cholesterol (ester) transport (120, 141). Once selectively delivered to the cell, the intact CEs are either transported to lipid droplets for storage (12) or hydrolyzed extra-lysosomally by neutral cholesteryl esterase (39, 80) (142–145) to provide cholesterol substrate for product formation or reesterification. Previously, we provided evidence that hormone-sensitive lipase serves as the neutral cholesterol esterase in adrenal cells and participates in CE turnover and steroidogenesis (131, 146).
Besides CEs, SR-B1 also mediates the selective uptake of other lipid components of receptor bound HDL particles, including FC, triglycerides (TGs), phospholipids, and α-tocopherol (38, 39, 147). Nonpolar FC, CE, and TG are transported five to ten times more efficiently than more polar phospholipid molecules. The relative selective uptake rate constants for CE, FC, TG, and phospholipid (phosphatidylcholine) have been calculated to be 1.0, 1.6, 0.7, and 0.2, respectively (147). SR-B1 also promotes the bidirectional flux of FC between cells and lipoproteins (39, 141, 147–149). In addition, in the liver, SR-B1 functions in reverse cholesterol transport (130, 141, 150), including selective uptake of CEs from HDL into the liver (one of the later steps in reverse cholesterol transport) and biliary cholesterol secretion (the last step in reverse cholesterol transport), thus promoting conversion of hepatic HDL cholesterol (HDL-C) to bile acids (16, 79, 81). SR-B1 also alters the composition of lipid domains of plasma membranes, which then leads to changes in FC flux, changes in membrane cholesterol content, or altered physical/chemical properties of membranes (151, 152).
OTHER MAJOR FUNCTIONS OF SR-B1
In addition to mediating the selective transport of HDL-CEs and other lipids, SR-B1 also performs many other essential functions. Besides facilitating selective cholesterol influx into cells, SR-B1 enhances the efflux of cellular cholesterol to HDL (149, 153–155). This property of SR-B1 in mediating the bidirectional flux of FC between cells and HDL is different from two other proteins involved in cellular cholesterol efflux, ABCA1 and ABCG1; ABCA1 mediates unidirectional efflux of cholesterol and phospholipids to apolipoprotein acceptors, such apoA-I and apoE (156), whereas ABCG1 facilitates unidirectional efflux of cholesterol to nascent HDL particles (157). Both gain-of-function and loss-of-function strategies in mice have established that SR-B1 determines the level of plasma-HDL-C by promoting selective transport of HDL-CE to the liver for transfer to bile and that SR-B1 is a key component of reverse cholesterol transport (158–161). Consistent with its importance in reverse cholesterol transport, several mouse models have shown that SR-B1 overexpression in the liver decreases atherosclerosis (162–165), whereas partial or total loss of SR-B1 increases atherosclerosis (166–170). Thus, SR-B1 can exert atheroprotective actions by impacting HDL metabolism and reverse cholesterol transport, mediating removal of cholesterol from macrophages via cholesterol efflux, and minimizing inflammation and oxidation (138, 150, 171). In addition, macrophage and endothelial cell SR-B1 could interfere with the development of atherosclerosis by altering cholesterol trafficking to suppress atherosclerotic lesion formation (171). Likewise, platelet SR-B1 has been implicated as a negative regulator in the development of thrombosis (138). SR-B1 signaling also helps to minimize inflammation and apoptosis, and promotes efferocytosis of apoptotic cells in atherosclerotic lesions, thus minimizing vulnerable plaque formation (171). In contrast, endothelial cell SR-B1 plays a more complex role, perhaps through involvement in LDL transcytosis, and may contribute to the development of early atherosclerotic lesions (172). SR-B1 has also been shown to regulate fertility (161, 166, 173), modulate erythrocyte development (174) and platelet function (138, 175), contribute to the pathogenesis of inflammation (175), regulate immunity (175), protect against nitric-oxide-induced cytotoxicity, mediate inducible glucocorticoid production and LPS clearance, protect against sepsis, and play a role in lymphocyte homeostasis and autoimmunity (175). SR-B1 has been implicated in some pathological processes as well. For instance, SR-B1, in concert with several viral and cellular factors, mediates hepatitis C virus entry into hepatocytes (33, 176), enhances dengue virus infection (177), mediates bacterial adhesion and bacterial invasion in cells (65), potentiates LPS-induced inflammation and acute liver and kidney injury in mice (178), and contributes to the pathogenesis of cancer and tumor progression.
SR-B1 SIGNALING
In view of the large variety of ligands reported to bind SR-B1 and the large number of functions attributed to SR-B1 in addition to lipid uptake and efflux, the observation that SR-B1 mediated the ability of HDL to stimulate endothelial nitric oxide synthase (eNOS) in cultured endothelial cells suggested that SR-B1 might possess signaling properties (179). Although neither cytoplasmic tail of SR-B1 possesses intrinsic signaling properties, experiments identified the interaction of c-Src with SR-B1 following HDL binding (180) with downstream activation of PI3 kinase, Akt, and MAPK, resulting in phosphorylation and activation of eNOS (181). The adaptor protein, PDZK1, which interacts with SR-B1 (see below), was reported to be required for SR-B1 signaling (180); however, a mutation in the C-terminal transmembrane domain that disrupts the ability of SR-B1 to bind plasma membrane cholesterol eliminated SR-B1 signaling to eNOS without altering HDL binding, cholesterol uptake or efflux, or interaction with PDZK1 (182), suggesting that the processes are separate. Recent work suggests that the localization of SR-B1 in lipid rafts and its ability to sense plasma membrane cholesterol link it to signal transduction (183). Whether all signaling via SR-B1 is due to its function as a lipid sensor is currently unclear. In favor of functioning as a lipid sensor, cell signaling that is induced by cellular cholesterol depletion with methyl-β-cyclodextrin is dependent on SR-B1 and, specifically, its ability to bind plasma membrane cholesterol (182). Moreover, serum amyloid A binding to SR-B1 induces activation of ERK1/2 and p38 MAPKs leading to increased cytokine production (184), while serum amyloid A also promotes SR-B1-mediated cholesterol efflux (185). In addition, SR-B1 can mediate the uptake or efflux of sphingosine-1-phosphate, along with interaction of sphingosine-1-phosphate receptors with SR-B1, leading to activation of signaling via sphingosine-1-phosphate receptors (186, 187). Likewise, SR-B1-mediated efferocytosis occurs through the binding of phosphatidylserine with induction of Src phosphorylation and activation of PI3 kinase and Rac1 leading to clearance of apoptotic cells (188). Other downstream signaling pathways have also been suggested to be involved in SR-B1 signaling because apoA-I binding to SR-B1 has been reported to stimulate AMP-activated protein kinase (189). On the other hand, CpG binds to SR-B1 and promotes pro-inflammatory cytokine production by triggering calcium entry via phospholipase C-mediated activation of TRPC3 channels in a Toll-like receptor-independent process (190) without evidence for cellular lipid changes, though it is possible that alterations in membrane lipids occur.
REGULATION OF HEPATIC SR-B1
SR-B1 mRNA and protein are primarily expressed within the liver in hepatocytes (138, 191). Hepatic SR-B1 is subject to both transcriptional and posttranscriptional regulation. Several different transcription factors, including nuclear receptors (PPARγ, HNF4α, PPARα, LXRα, LRH-1, and FXR) (191, 192), estrogen receptor (192, 193), and SREBPs, control SR-B1 transcription (see above). A variety of dietary components, hormones, metabolites, and pharmacological agents also modulate gene transcription of hepatic SR-B1 (192).
Though under transcriptional control, hepatic SR-B1 is primarily regulated posttranscriptionally by a key scaffolding protein termed PDZK1 or NHERF3. PDZK1/NHERF3 belongs to the NHERF family, which consists of three additional proteins, namely, NHERF1, NHERF2, and NHERF4 (194). All four NHERFs contain multiple “PDZ” domains [postsynaptic density (PSD-95/SAP90), the Drosophila septate junction protein disc-large (Dlg), and the tight junction protein, zona occludense-1 (ZO-1)] (193). NHERF1 (SLC9A3R1 gene, also called EBP50) and NHERF2 (SLC9A3R2 gene, also called E3KARP) consist of two tandem PDZ domains and a C-terminal MERM (merlin-ezrin-radixin-moesin) domain, which binds to the actin-associated proteins, ezrin, radixin, moesin, and merlin (194), whereas NHERF3 (PDZK1 gene, also called PDZK1, CLAMP) and NHERF4 (PDZD3 gene, also called IKEPP) contain four tandem PDZ domains (193). PDZ domains are protein-protein interaction modules that generally consist of ∼90 amino acid residues and usually bind to a specific peptide sequence located at the extreme C terminus of a target; although in some cases, the target peptide sequence has an internal location. NHERF proteins bind a variety of membrane transporters and receptors, and modulate membrane localization, interaction with other proteins, formation of signaling complexes, and maintenance of microvilli on the apical surface of endothelial cells (193, 194).
Ikemoto et al. (195) and Silver (196) initially demonstrated that the extreme C terminus of SR-B1 interacts with the first N-terminal PDZ domain of PDZK1/NHERF3/CLAMP. Gene deletion studies demonstrated that the level of hepatic SR-B1 was reduced by >95% in PDZK1-null mice and plasma HDL-C was significantly elevated (197) to a value similar to that observed in SR-B1 knockout mice (158). Further studies demonstrated that PDZK1 regulates hepatic SR-B1 stability and steady state levels, but does not directly affect the ability of SR-B1 to transport HDL-CE (197–201). Furthermore, it was demonstrated that overexpression of the first PDZ domain of PDZK1 increases hepatic SR-B1 abundance and affects localization in both wild-type and PDZK1 knockout mice, but was not completely effective in restoring normal SR-B1 function in PDZK1-null mice (199). Likewise, overexpression of the first three PDZ domains of PDZK1 in PDZK1-deficient mice partially restored cell surface localization of hepatic SR-B1 and selective HDL-CE uptake, but failed to fully restore hepatic SR-B1 abundance and cell surface localization, as compared with the control (200, 201). Only full-length native PDZK1 or a mutant missing the C-terminal PDZ domain-binding motif of PDZK1 in PDZK1-null mice fully restored normal SR-B1 abundance and selective HDL-CE transport function (199). Interestingly, steroidogenic tissues express very low levels of PDZK1 (197, 202), and its deficiency has no apparent effect on SR-B1 levels or its selective HDL-CE transport function in steroidogenic tissues (see below) (197). A small PDZK1-associated protein, SPAP/DD96/MAP17, is also involved in the regulation of PDZK1 (203), and hepatic DD96 overexpression can alter plasma HDL-C levels (204). One recent report suggests that microRNAs 185, 96, and 223 negatively regulate the expression of hepatic SR-B1 at a posttranscriptional level (110).
REGULATION OF STEROIDOGENIC SR-B1
The selective uptake pathway was shown to be the major supplier of exogenous cholesterol for storage or steroidogenesis in steroidogenic cells of the adrenal gland (3, 7, 9, 11, 142, 205–207), ovary (3, 6–8, 205, 208–210), and, under certain conditions, testicular Leydig cells (211–214) in rodents, prior to the discovery of SR-B1. SR-B1 was then cloned and identified as the receptor responsible for selective cholesterol uptake (26–28). Given this, it is not surprising that, in addition to liver (115), SR-B1 is abundantly expressed in the steroidogenic cells of the adrenal gland (84, 115), ovary (83, 115), and hormone-stimulated testis (14, 115). SR-B1 in steroidogenic tissues is regulated transcriptionally, posttranscriptionally, and posttranslationally. Trophic hormone-stimulated cAMP and activation of PKA are the key pathways responsible for regulating SR-B1 gene transcription in the adrenal gland (84, 115, 202, 215–218), ovary (12, 32, 83, 219–221), and testis (14, 86, 216, 217).
In the rodent adrenal gland, SR-B1 is localized on the cell surface of fasciculata-reticularis cells (84, 115, 215). Adrenal SR-B1 mRNA and protein expression is increased by treatment of mice or rats with ACTH and suppressed by treatment of animals with the synthetic corticosteroid, dexamethasone (which inhibits the hypothalamic pituitary axis and decreases ACTH secretion) (84, 98, 202, 215). Likewise, cAMP treatment of mouse adrenocortical cells (Y1-BS1) (217) and primary human adrenocortical cells (217) upregulates SR-B1. Besides ACTH, adrenal glomerulosa cell lines respond to angiotensin II stimulation with increased expression of SR-B1 mRNA and protein levels (222). Adrenal SR-B1 mRNA and protein levels are upregulated in apoA-I knockout mice, but not in mice deficient in apoA-II, LDL receptor, or apoE, or in CE transfer protein transgenic mice (223). Likewise, adrenal SR-B1 mRNA and protein levels are also increased and the content of stored CEs decreased in female hepatic lipase knockout mice (223). Depletion of adrenal cholesterol ester stores is associated with increased expression of SR-B1 in an ACTH-independent manner in vivo and in cultured Y1-BS1 adrenocortical cells (218). As additional evidence for non-ACTH regulation of adrenal SR-B1 expression, SR-B1 mRNA levels are increased in adrenals from corticosterone-insufficient Crh (−/−) mice, whereas corticosterone replacement by oral administration inhibited SR-B1 gene expression in these mice (224). Adrenals from mice deficient in lecithin:cholesterol acyltransferase exhibit elevated levels of SR-B1 mRNA and protein (225). A previous study (84) from our laboratory reported that treatment of rats with ACTH or 17α-ethinyl estradiol (17α-E2) leads to increased expression of SR-B1 in the adrenal gland (especially in the microvillar compartment of adrenocortical cells), whereas dexamethasone treatment results in decreased levels of SR-B1. These changes in SR-B1 levels are also accompanied by striking ultrastructural changes in the adrenocortical cell microvillar compartment, i.e., microvillar area and microvillar channel formation and complexity are dramatically increased in response to ACTH or 17α-E2 treatment, whereas these structures are decreased significantly following dexamethasone treatment.
Granulosa cells, isolated from mouse (226), rat (12), pig (221), and cow (32) and cultured under basal conditions, exhibit very little or no expression of SR-B1 mRNA and protein, but expression is increased many fold following luteinization of the cells with gonadotropins or cAMP. In the rat ovary, insulin and trophic hormone LH/hCG stimulate the expression of SR-B1 in theca-interstitial cells and increase intracellular cholesterol, which is subsequently mobilized for androgen production (220, 227). Similarly, treatment of immature rats with PMSG/eCG or PMSG/eCG + hCG also results in the induction of SR-B1 mRNA by several fold, especially in theca interna cells (29, 219), whereas prostaglandin F2α treatment downregulates SR-B1 mRNA expression (219). Estrogen treatment of normal cycling rats also induces SR-B1 protein expression in luteal cells of the ovary (115). Using a novel hormone-desensitized rat ovarian model, it was demonstrated that HDL binding, SR-B1, and selective HDL-CE uptake are tightly linked (83). Moreover, immunostaining at the light microscope level indicated strong expression of SR-B1 specifically on the surface of luteal cells in the luteinized and desensitized ovary. Immunoelectron microscopy further demonstrated that SR-B1 protein was associated with microvilli and microvillar channels of the luteal surface (83), further confirming that these structures represent a cell surface compartment that is specialized for the selective uptake of lipoprotein-derived cholesterol into steroidogenic cells for hormone production or storage. Normal rat Leydig cells under basal conditions (14, 115) show only negligible expression of SR-B1. However, both mRNA and protein levels of SR-B1 in rodent Leydig cells are dramatically induced following chronic treatment of animals with hCG (14, 115). Interestingly, in contrast to the adrenal gland and ovary, hormone-induced SR-B1 is not localized in surface microvilli, but is present in an elaborate and complex channel system within the cytoplasm of the Leydig cells (14). Leydig tumor cell lines, such as MA-10 and MLTC-1, express significant levels of basal SR-B1 and hormonal/cAMP treatment further upregulates its expression (86, 111, 228). R2C Leydig tumor cells, which constitutively secrete large amounts of steroids, also show extremely high basal levels of SR-B1 protein and mRNA levels, and cAMP treatment modestly increases their levels further (86).
It is becoming clear that the physical form of SR-B1 also plays a key role in the expression and function of SR-B1. It is well-known that hormonal changes in tissues (which alter the expression of SR-B1, alter selective CE uptake in the same tissues, and correspondingly produce architectural changes in the cell surface of affected cells) also show changes in dimerization2 of SR-B1 in cell or tissue samples. In one of the earliest demonstrations of protein-protein interaction involving SR-B1, we showed (by Western blotting) that SR-B1 exists as homodimers in 17α-E2-primed and microvilli-enriched rat adrenal plasma membranes (84). In subsequent studies, it was demonstrated that SR-B1 exists in dimeric and higher order oligomeric forms in all cells and tissues that are active in “selective” uptake of HDL-CEs [e.g., hormone-activated steroidogenic tissues (such as mouse and rat adrenals, testis, and ovary), steroidogenic-derived cells (such as rat ovarian luteal cells, mouse adrenal Y1-BS1 cells, rat Leydig R2C tumor cells, and MLTC-1 Leydig tumor cells), liver from SR-B1 transgenic mice, SR-B1-transfected nonsteroidogenic and steroidogenic cells (such as HEK-293, Y1-BS1, CHO, and COS cells), or primitive Sf9 insect cells programmed to express SR-B1] (116). Early functional evidence for SR-B1 dimerization came from the observation that, in normal rat adrenal tissue, SR-B1 exits primarily in a monomeric form with some dimer formation. ACTH stimulation increased the dimerization of SR-B1 in the adrenal along with increased selective HDL-CE uptake, and dexamethasone-induced suppression of ACTH led dramatically to the loss of SR-B1, SR-B1 dimers, and selective HDL-CE uptake (84). These results are coupled with striking architectural changes of the microvillar compartment at the adrenocortical cell surface, and suggest that SR-B1 dimers may, in a very basic way, be associated with SR-B1 sites of action and function.
As noted above, PDZK1/NHERF3 regulates hepatic SR-B1 stability and steady-state levels, but does not directly affect the ability of expressed SR-B1 to mediate HDL-CE transport. However, steroidogenic tissues express very low levels of PDZK1, and its deficiency has no apparent effect on SR-B1 levels or HDL-CE transport function in steroidogenic tissues. In contrast, NHERF1 and NHERF2 mRNA and protein are expressed at varying levels in model steroidogenic cell lines and the adrenal gland, with only low expression of NHERF4 (202). Using several different mechanistic approaches, our laboratory recently established that NHERF1 and NHERF2 specifically interact with SR-B1 in liver and steroidogenic tissues/cells and reduce SR-B1 levels via novel translational/posttranslational mechanisms and, consequently, negatively impact SR-B1-mediated selective HDL-CE uptake and HDL-supported steroidogenesis (202). Recently, a mouse line carrying a 507Ala/STOP mutation of the SR-B1 gene that produces a truncated receptor (SR-B1ΔCT) was generated (88). Results show a significant decrease in the level of SR-B1 in steroidogenic cells (adrenal cortical cells, ovarian cells, and Leydig cells), providing additional evidence for the function of non-NHERF3 adaptors for SR-B1 in steroidogenic tissues.
Salt-inducible kinase (SIK1) is a serine/threonine kinase that is a member of the stress- and energy-sensing AMPK family of kinases (229). Its expression is rapidly induced in Y1 adrenal cells in response to ACTH and was presumed to act as a repressor of adrenal steroidogenesis. Recently, it was demonstrated that SIK1 in fact stimulates adrenal steroidogenesis by modulating the selective HDL-CE transport activity of SR-B1. Evidence showed that overexpression of SIK1 increased cAMP-stimulated and SR-B1-mediated selective CE uptake in steroidogenic cell lines without impacting SR-B1 protein levels. Conversely, knockdown of SIK1 resulted in attenuation of cAMP-stimulated selective HDL-CE uptake. It was further observed that SIK1 forms a complex with SR-B1 by interacting with its cytoplasmic C-terminal domain and catalyzes the phosphorylation of this domain at Ser493. Finally, loss-of-function and gain-of-function strategies demonstrated that SIK1 increased SR-B1-mediated selective delivery of HDL-derived cholesterol. This was the first report demonstrating that SIK1 functions as a positive modulator of selective cholesterol transport function of SR-B1.
SCARB1 GENE POLYMORPHISM
Studies in animal models have established a role for SR-B1 in lipid homeostasis, specifically involving HDL-C and apoB-containing lipoprotein metabolism. However, the plasma lipoprotein profiles of rodents and humans are different in that the bulk of plasma cholesterol in adult humans is in LDL and VLDL particles, whereas mice utilize HDL as the major cholesterol-carrying lipoprotein in plasma (230). The involvement of SR-B1 in regulating lipoprotein metabolism in humans has begun to be clarified by examining the association of gene polymorphisms in SCARB1 with lipoprotein levels. In an early study in Spanish Caucasians, single-strand conformation polymorphism analysis together with direct sequencing following PCR amplification of interested regions identified five variants, one each at introns 3 and 5 and three each at exons 1, 8, and 11. Further analysis showed that the variants at exon 1 were significantly associated with increased HDL-C and lower LDL cholesterol values, as well as TGs and BMI, in men (231), consistent with the involvement of SR-B1 in the metabolism of both lipoprotein classes in humans. Subsequent population-based genetic studies of SR-B1 gene polymorphisms, including the Framingham Study (232) and the Rancho Bernardo Study (233), not only confirmed the association of common SCARB1 variations with plasma lipid profiles (234, 235), but also expanded the populations to include women (234), multi-ethnics (236–239), diabetics (232), and subjects with heterozygous familial hypercholesterolemia (240), as well as populations with subclinical atherosclerosis (241). Recently, a patient with a missense mutation for SCARB1 S129L was shown to have high levels of both HDL-C and lipoprotein (a) (242). Subsequent analysis of two cohorts identified rare or uncommon missense or splice site mutations in SCARB1. Further analysis of four SCARB1 variants in functional studies in vitro (c.386C>T, c.631-14T>G, c.4G>A, and c.631-53mC>T and c.726+55mCG>CA) demonstrated decreased receptor function, providing a functional mechanism for the association of SCARB1 gene variants with the high HDL-C and high lipoprotein (a) phenotype (242).
In humans, four more mutations in SCARB1 have now been reported. A family with a single loss-of-function mutation in P297 to S was identified in 2011 (243), and the mutant carrier had increased HDL-C and reduced cholesterol efflux from macrophages. The carriers also displayed decreased adrenal steroidogenesis and changes in platelet function; however, they did not have evidence for significant atherosclerosis. Almost at the same time, a separate study identified two novel mutations in the SCARB1 gene, S112F and T175A, and showed that these mutations are associated with high HDL-C (244). Most recently, through targeted sequencing of the coding region of lipid-modifying genes in a population with extremely high HDL-C levels, a homozygote subject harboring a P376L missense mutation in the SCARB1 gene was identified (245). Carriers of this P376L mutation have increased risk of coronary heart disease, even though they have very high plasma HDL-C levels.
CONCLUSIONS
In conclusion, SR-B1 is a multiligand membrane receptor that functions as a physiologically relevant HDL receptor to selectively deliver HDL-C to cells, but it has a number of other functions. SR-B1 is primarily expressed in liver and steroidogenic cells, where the SR-B1/selective pathway provides the bulk of the cholesterol needed for steroidogenesis in rodents. The expression of SR-B1 is regulated by hormones, and its function is regulated by many posttranslational interactions and modifications. Structurally, SR-B1 forms a hydrophobic tunnel within the plasma membrane, which appears to facilitate the selective cellular influx and efflux of lipid molecules, as well as other molecules, including vitamins, viruses, and apoptotic cells. SR-B1 polymorphism analysis and identification of mutations in human SR-B1 have confirmed the involvement of SR-B1 in regulating lipoprotein metabolism in humans.
Acknowledgments
The authors thank Mr. Alex Bittner for creating illustrations.
Footnotes
Abbreviations:
- AcLDL
- acetylated LDL
- CD36
- cluster determinant 36
- CE
- cholesteryl ester
- CLA-1
- cluster determinant 36 and lysosomal integral membrane protein II analogous-I
- eNOS
- endothelial nitric oxide synthase
- 17α-E2
- 17α-ethinyl estradiol
- FC
- free cholesterol
- HDL-C
- HDL cholesterol
- LIMP
- lysosomal integral membrane protein
- LPS
- lipopolysaccharide
- OxLDL
- oxidized LDL
- PREB
- prolactin regulatory element-binding protein
- SF-1
- steroidogenic factor-1
- SIK1
- salt-inducible kinase
- SR-B1
- scavenger receptor B type 1
- TG
- triglyceride
- 3′ UTR
- 3′ untranslated region
This work supported by National Institutes of Health, National Heart, Lung, and Blood Institute Grant R56HL122773 and VA Biomedical Laboratory Research and Development Merit Review 2I01BX001923 (S.A.), Senior Research Career Scientist (SRCS) Award IK6BX004200, and Merit Review 10BX000398 (F.B.K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Russell D. W. 2009. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 50 (Suppl.): S120–S125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller W. L., and Bose H. S.. 2011. Early steps in steroidogenesis: intracellular cholesterol trafficking. J. Lipid Res. 52: 2111–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glass C., Pittman R. C., Weinstein D. B., and Steinberg D.. 1983. Dissociation of tissue uptake of cholesterol ester from that of apoprotein A-I of rat plasma high density lipoprotein: selective delivery of cholesterol ester to liver, adrenal, and gonad. Proc. Natl. Acad. Sci. USA. 80: 5435–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown M. S., and Goldstein J. L.. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science. 232: 34–47. [DOI] [PubMed] [Google Scholar]
- 5.Quarfordt S., Hanks J., Jones R. S., and Shelburne F.. 1980. The uptake of high density lipoprotein cholesteryl ester in the perfused rat liver. J. Biol. Chem. 255: 2934–2937. [PubMed] [Google Scholar]
- 6.Reaven E., Chen Y. D., Spicher M., and Azhar S.. 1984. Morphological evidence that high density lipoproteins are not internalized by steroid-producing cells during in situ organ perfusion. J. Clin. Invest. 74: 1384–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glass C., Pittman R. C., Civen M., and Steinberg D.. 1985. Uptake of high-density lipoprotein-associated apoprotein A-I and cholesterol esters by 16 tissues of the rat in vivo and by adrenal cells and hepatocytes in vitro. J. Biol. Chem. 260: 744–750. [PubMed] [Google Scholar]
- 8.Reaven E., Chen Y. D., Spicher M., Hwang S. F., Mondon C. E., and Azhar S.. 1986. Uptake of low density lipoproteins by rat tissues. Special emphasis on the luteinized ovary. J. Clin. Invest. 77: 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azhar S., Stewart D., and Reaven E.. 1989. Utilization of cholesterol-rich lipoproteins by perfused rat adrenals. J. Lipid Res. 30: 1799–1810. [PubMed] [Google Scholar]
- 10.Stein Y., Dabach Y., Hollander G., Halperin G., and Stein O.. 1983. Metabolism of HDL-cholesteryl ester in the rat, studied with a nonhydrolyzable analog, cholesteryl linoleyl ether. Biochim. Biophys. Acta. 752: 98–105. [DOI] [PubMed] [Google Scholar]
- 11.Gwynne J. T., and Mahaffee D. D.. 1989. Rat adrenal uptake and metabolism of high density lipoprotein cholesteryl ester. J. Biol. Chem. 264: 8141–8150. [PubMed] [Google Scholar]
- 12.Azhar S., Nomoto A., Leers-Sucheta S., and Reaven E.. 1998. Simultaneous induction of an HDL receptor protein (SR-BI) and the selective uptake of HDL-cholesteryl esters in a physiologically relevant steroidogenic cell model. J. Lipid Res. 39: 1616–1628. [PubMed] [Google Scholar]
- 13.Azhar S., Tsai L., Medicherla S., Chandrasekher Y., Giudice L., and Reaven E.. 1998. Human granulosa cells use high density lipoprotein cholesterol for steroidogenesis. J. Clin. Endocrinol. Metab. 83: 983–991. [DOI] [PubMed] [Google Scholar]
- 14.Reaven E., Zhan L., Nomoto A., Leers-Sucheta S., and Azhar S.. 2000. Expression and microvillar localization of scavenger receptor class B, type I (SR-BI) and selective cholesteryl ester uptake in Leydig cells from rat testis. J. Lipid Res. 41: 343–356. [PubMed] [Google Scholar]
- 15.Scobey M. W., Johnson F. L., and Rudel L. L.. 1989. Delivery of high-density lipoprotein free and esterified cholesterol to bile by the perfused monkey liver. Am. J. Physiol. 257: G644–G652. [DOI] [PubMed] [Google Scholar]
- 16.Pieters M. N., Schouten D., Bakkeren H. F., Esbach B., Brouwer A., Knook D. L., and van Berkel T. J.. 1991. Selective uptake of cholesteryl esters from apolipoprotein-E-free high-density lipoproteins by rat parenchymal cells in vivo is efficiently coupled to bile acid synthesis. Biochem. J. 280: 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fluiter K., Vietsch H., Biessen E. A., Kostner G. M., van Berkel T. J., and Sattler W.. 1996. Increased selective uptake in vivo and in vitro of oxidized cholesteryl esters from high-density lipoprotein by rat liver parenchymal cells. Biochem. J. 319: 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guertin F., Brunet S., Lairon D., and Levy E.. 1997. Oxidative tyrosylation of high density lipoprotein impairs biliary sterol secretion in rats. Atherosclerosis. 131: 35–41. [DOI] [PubMed] [Google Scholar]
- 19.Pittman R. C., Glass C. K., Atkinson D., and Small D. M.. 1987. Synthetic high density lipoprotein particles. Application to studies of the apoprotein specificity for selective uptake of cholesterol esters. J. Biol. Chem. 262: 2435–2442. [PubMed] [Google Scholar]
- 20.Rinninger F., Kaiser T., Windler E., Greten H., Fruchart J. C., and Castro G.. 1998. Selective uptake of cholesteryl esters from high-density lipoprotein-derived LpA-I and LpA-I:A-II particles by hepatic cells in culture. Biochim. Biophys. Acta. 1393: 277–291. [DOI] [PubMed] [Google Scholar]
- 21.Despres J. P., Fong B. S., Jimenez J., Julien P., and Angel A.. 1988. Selective uptake of HDL cholesterol ester by human fat cells. Am. J. Physiol. 254: E667–E675. [DOI] [PubMed] [Google Scholar]
- 22.Schorsch F., Malle E., and Sattler W.. 1997. Selective uptake of high density lipoprotein-associated cholesterylesters by differentiated Ob1771 adipocytes is modulated by endogenous and exogenous lipoprotein lipase. FEBS Lett. 414: 507–513. [DOI] [PubMed] [Google Scholar]
- 23.Stein O., Israeli A., Leitersdorf E., Halperin G., and Stein Y.. 1987. Preferential uptake of cholesteryl ester-HDL by cultured macrophages. Atherosclerosis. 65: 151–158. [DOI] [PubMed] [Google Scholar]
- 24.Rinninger F., Deichen J. T., Jackle S., Windler E., and Greten H.. 1994. Selective uptake of high-density lipoprotein-associated cholesteryl esters and high-density lipoprotein particle uptake by human monocyte-macrophages. Atherosclerosis. 105: 145–157. [DOI] [PubMed] [Google Scholar]
- 25.Leitersdorf E., Israeli A., Stein O., Eisenberg S., and Stein Y.. 1986. The role of apolipoproteins of HDL in the selective uptake of cholesteryl linoleyl ether by cultured rat and bovine adrenal cells. Biochim. Biophys. Acta. 878: 320–329. [DOI] [PubMed] [Google Scholar]
- 26.Acton S., Rigotti A., Landschulz K. T., Xu S., Hobbs H. H., and Krieger M.. 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 271: 518–520. [DOI] [PubMed] [Google Scholar]
- 27.Acton S. L., Scherer P. E., Lodish H. F., and Krieger M.. 1994. Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J. Biol. Chem. 269: 21003–21009. [PubMed] [Google Scholar]
- 28.Calvo D., and Vega M. A.. 1993. Identification, primary structure, and distribution of CLA-1, a novel member of the CD36/LIMPII gene family. J. Biol. Chem. 268: 18929–18935. [PubMed] [Google Scholar]
- 29.Mizutani T., Sonoda Y., Minegishi T., Wakabayashi K., and Miyamoto K.. 1997. Cloning, characterization, and cellular distribution of rat scavenger receptor class B type I (SRBI) in the ovary. Biochem. Biophys. Res. Commun. 234: 499–505. [DOI] [PubMed] [Google Scholar]
- 30.Ritsch A., Tancevski I., Schgoer W., Pfeifhofer C., Gander R., Eller P., Foeger B., Stanzl U., and Patsch J. R.. 2004. Molecular characterization of rabbit scavenger receptor class B types I and II: portal to central vein gradient of expression in the liver. J. Lipid Res. 45: 214–222. [DOI] [PubMed] [Google Scholar]
- 31.Kim J. G., Vallet J. L., Nonneman D., and Christenson R. K.. 2004. Molecular cloning and endometrial expression of porcine high density lipoprotein receptor SR-BI during the estrous cycle and early pregnancy. Mol. Cell. Endocrinol. 222: 105–112. [DOI] [PubMed] [Google Scholar]
- 32.Rajapaksha W. R., McBride M., Robertson L., and O’Shaughnessy P. J.. 1997. Sequence of the bovine HDL-receptor (SR-BI) cDNA and changes in receptor mRNA expression during granulosa cell luteinization in vivo and in vitro. Mol. Cell. Endocrinol. 134: 59–67. [DOI] [PubMed] [Google Scholar]
- 33.Tong Y., Zhu Y., Xia X., Liu Y., Feng Y., Hua X., Chen Z., Ding H., Gao L., Wang Y., et al. . 2011. Tupaia CD81, SR-BI, claudin-1, and occludin support hepatitis C virus infection. J. Virol. 85: 2793–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.PrabhuDas M. R., Baldwin C. L., Bollyky P. L., Bowdish D. M. E., Drickamer K., Febbraio M., Herz J., Kobzik L., Krieger M., Loike J., et al. . 2017. A consensus definitive classification of scavenger receptors and their roles in health and disease. J. Immunol. 198: 3775–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calvo D., Dopazo J., and Vega M. A.. 1995. The CD36, CLA-1 (CD36L1), and LIMPII (CD36L2) gene family: cellular distribution, chromosomal location, and genetic evolution. Genomics. 25: 100–106. [DOI] [PubMed] [Google Scholar]
- 36.Webb N. R., de Villiers W. J., Connell P. M., de Beer F. C., and van der Westhuyzen D. R.. 1997. Alternative forms of the scavenger receptor BI (SR-BI). J. Lipid Res. 38: 1490–1495. [PubMed] [Google Scholar]
- 37.Krieger M. 1999. Charting the fate of the “good cholesterol”: identification and characterization of the high-density lipoprotein receptor SR-BI. Annu. Rev. Biochem. 68: 523–558. [DOI] [PubMed] [Google Scholar]
- 38.Rigotti A., Miettinen H. E., and Krieger M.. 2003. The role of the high-density lipoprotein receptor SR-BI in the lipid metabolism of endocrine and other tissues. Endocr. Rev. 24: 357–387. [DOI] [PubMed] [Google Scholar]
- 39.Connelly M. A., and Williams D. L.. 2004. Scavenger receptor BI: a scavenger receptor with a mission to transport high density lipoprotein lipids. Curr. Opin. Lipidol. 15: 287–295. [DOI] [PubMed] [Google Scholar]
- 40.Silverstein R. L., and Febbraio M.. 2009. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2: re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez A., Valeiras M., Sidransky E., and Tayebi N.. 2014. Lysosomal integral membrane protein-2: a new player in lysosome-related pathology. Mol. Genet. Metab. 111: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams D. L., Connelly M. A., Temel R. E., Swarnakar S., Phillips M. C., de la Llera-Moya M., and Rothblat G. H.. 1999. Scavenger receptor BI and cholesterol trafficking. Curr. Opin. Lipidol. 10: 329–339. [DOI] [PubMed] [Google Scholar]
- 43.Viñals M., Xu S., Vasile E., and Krieger M.. 2003. Identification of the N-linked glycosylation sites on the high density lipoprotein (HDL) receptor SR-BI and assessment of their effects on HDL binding and selective lipid uptake. J. Biol. Chem. 278: 5325–5332. [DOI] [PubMed] [Google Scholar]
- 44.Webb N. R., Connell P. M., Graf G. A., Smart E. J., de Villiers W. J., de Beer F. C., and van der Westhuyzen D. R.. 1998. SR-BII, an isoform of the scavenger receptor BI containing an alternate cytoplasmic tail, mediates lipid transfer between high density lipoprotein and cells. J. Biol. Chem. 273: 15241–15248. [DOI] [PubMed] [Google Scholar]
- 45.Babitt J., Trigatti B., Rigotti A., Smart E. J., Anderson R. G., Xu S., and Krieger M.. 1997. Murine SR-BI, a high density lipoprotein receptor that mediates selective lipid uptake, is N-glycosylated and fatty acylated and colocalizes with plasma membrane caveolae. J. Biol. Chem. 272: 13242–13249. [DOI] [PubMed] [Google Scholar]
- 46.Zani I. A., Stephen S. L., Mughal N. A., Russell D., Homer-Vanniasinkam S., Wheatcroft S. B., and Ponnambalam S.. 2015. Scavenger receptor structure and function in health and disease. Cells. 4: 178–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calvo D., Gomez-Coronado D., Lasuncion M. A., and Vega M. A.. 1997. CLA-1 is an 85-kD plasma membrane glycoprotein that acts as a high-affinity receptor for both native (HDL, LDL, and VLDL) and modified (OxLDL and AcLDL) lipoproteins. Arterioscler. Thromb. Vasc. Biol. 17: 2341–2349. [DOI] [PubMed] [Google Scholar]
- 48.Ohgami N., Nagai R., Miyazaki A., Ikemoto M., Arai H., Horiuchi S., and Nakayama H.. 2001. Scavenger receptor class B type I-mediated reverse cholesterol transport is inhibited by advanced glycation end products. J. Biol. Chem. 276: 13348–13355. [DOI] [PubMed] [Google Scholar]
- 49.Marsche G., Hammer A., Oskolkova O., Kozarsky K. F., Sattler W., and Malle E.. 2002. Hypochlorite-modified high density lipoprotein, a high affinity ligand to scavenger receptor class B, type I, impairs high density lipoprotein-dependent selective lipid uptake and reverse cholesterol transport. J. Biol. Chem. 277: 32172–32179. [DOI] [PubMed] [Google Scholar]
- 50.Marsche G., Zimmermann R., Horiuchi S., Tandon N. N., Sattler W., and Malle E.. 2003. Class B scavenger receptors CD36 and SR-BI are receptors for hypochlorite-modified low density lipoprotein. J. Biol. Chem. 278: 47562–47570. [DOI] [PubMed] [Google Scholar]
- 51.Xu S., Laccotripe M., Huang X., Rigotti A., Zannis V. I., and Krieger M.. 1997. Apolipoproteins of HDL can directly mediate binding to the scavenger receptor SR-BI, an HDL receptor that mediates selective lipid uptake. J. Lipid Res. 38: 1289–1298. [PubMed] [Google Scholar]
- 52.Li X., Kan H. Y., Lavrentiadou S., Krieger M., and Zannis V.. 2002. Reconstituted discoidal ApoE-phospholipid particles are ligands for the scavenger receptor BI. The amino-terminal 1-165 domain of ApoE suffices for receptor binding. J. Biol. Chem. 277: 21149–21157. [DOI] [PubMed] [Google Scholar]
- 53.de Beer M. C., Durbin D. M., Cai L., Mirocha N., Jonas A., Webb N. R., de Beer F. C., and van Der Westhuyzen D. R.. 2001. Apolipoprotein A-II modulates the binding and selective lipid uptake of reconstituted high density lipoprotein by scavenger receptor BI. J. Biol. Chem. 276: 15832–15839. [DOI] [PubMed] [Google Scholar]
- 54.Phillips M. C. 2013. New insights into the determination of HDL structure by apolipoproteins: Thematic review series: high density lipoprotein structure, function, and metabolism. J. Lipid Res. 54: 2034–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liadaki K. N., Liu T., Xu S., Ishida B. Y., Duchateaux P. N., Krieger J. P., Kane J., Krieger M., and Zannis V. I.. 2000. Binding of high density lipoprotein (HDL) and discoidal reconstituted HDL to the HDL receptor scavenger receptor class B type I. Effect of lipid association and APOA-I mutations on receptor binding. J. Biol. Chem. 275: 21262–21271. [DOI] [PubMed] [Google Scholar]
- 56.de Beer M. C., Durbin D. M., Cai L., Jonas A., de Beer F. C., and van der Westhuyzen D. R.. 2001. Apolipoprotein A-I conformation markedly influences HDL interaction with scavenger receptor BI. J. Lipid Res. 42: 309–313. [PubMed] [Google Scholar]
- 57.Williams D. L., de La Llera-Moya M., Thuahnai S. T., Lund-Katz S., Connelly M. A., Azhar S., Anantharamaiah G. M., and Phillips M. C.. 2000. Binding and cross-linking studies show that scavenger receptor BI interacts with multiple sites in apolipoprotein A-I and identify the class A amphipathic alpha-helix as a recognition motif. J. Biol. Chem. 275: 18897–18904. [DOI] [PubMed] [Google Scholar]
- 58.Pilon A., Briand O., Lestavel S., Copin C., Majd Z., Fruchart J. C., Castro G., and Clavey V.. 2000. Apolipoprotein AII enrichment of HDL enhances their affinity for class B type I scavenger receptor but inhibits specific cholesteryl ester uptake. Arterioscler. Thromb. Vasc. Biol. 20: 1074–1081. [DOI] [PubMed] [Google Scholar]
- 59.Rigotti A., Acton S. L., and Krieger M.. 1995. The class B scavenger receptors SR-BI and CD36 are receptors for anionic phospholipids. J. Biol. Chem. 270: 16221–16224. [DOI] [PubMed] [Google Scholar]
- 60.Cai L., de Beer M. C., de Beer F. C., and van der Westhuyzen D. R.. 2005. Serum amyloid A is a ligand for scavenger receptor class B type I and inhibits high density lipoprotein binding and selective lipid uptake. J. Biol. Chem. 280: 2954–2961. [DOI] [PubMed] [Google Scholar]
- 61.Kawasaki Y., Nakagawa A., Nagaosa K., Shiratsuchi A., and Nakanishi Y.. 2002. Phosphatidylserine binding of class B scavenger receptor type I, a phagocytosis receptor of testicular Sertoli cells. J. Biol. Chem. 277: 27559–27566. [DOI] [PubMed] [Google Scholar]
- 62.Tsugita M., Morimoto N., Tashiro M., Kinoshita K., and Nakayama M.. 2017. SR-B1 is a silica receptor that mediates canonical inflammasome activation. Cell Reports. 18: 1298–1311. [DOI] [PubMed] [Google Scholar]
- 63.Ashraf M. Z., Kar N. S., Chen X., Choi J., Salomon R. G., Febbraio M., and Podrez E. A.. 2008. Specific oxidized phospholipids inhibit scavenger receptor bi-mediated selective uptake of cholesteryl esters. J. Biol. Chem. 283: 10408–10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao D., Sayre L. M., and Podrez E. A.. 2015. Analysis of relationship between oxidized phospholipid structure and interaction with the class B scavenger receptors. Methods Mol. Biol. 1208: 29–48. [DOI] [PubMed] [Google Scholar]
- 65.Vishnyakova T. G., Bocharov A. V., Baranova I. N., Chen Z., Remaley A. T., Csako G., Eggerman T. L., and Patterson A. P.. 2003. Binding and internalization of lipopolysaccharide by Cla-1, a human orthologue of rodent scavenger receptor B1. J. Biol. Chem. 278: 22771–22780. [DOI] [PubMed] [Google Scholar]
- 66.Bocharov A. V., Baranova I. N., Vishnyakova T. G., Remaley A. T., Csako G., Thomas F., Patterson A. P., and Eggerman T. L.. 2004. Targeting of scavenger receptor class B type I by synthetic amphipathic alpha-helical-containing peptides blocks lipopolysaccharide (LPS) uptake and LPS-induced pro-inflammatory cytokine responses in THP-1 monocyte cells. J. Biol. Chem. 279: 36072–36082. [DOI] [PubMed] [Google Scholar]
- 67.Vishnyakova T. G., Kurlander R., Bocharov A. V., Baranova I. N., Chen Z., Abu-Asab M. S., Tsokos M., Malide D., Basso F., Remaley A., et al. . 2006. CLA-1 and its splicing variant CLA-2 mediate bacterial adhesion and cytosolic bacterial invasion in mammalian cells. Proc. Natl. Acad. Sci. USA. 103: 16888–16893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukasawa M., Adachi H., Hirota K., Tsujimoto M., Arai H., and Inoue K.. 1996. SRB1, a class B scavenger receptor, recognizes both negatively charged liposomes and apoptotic cells. Exp. Cell Res. 222: 246–250. [DOI] [PubMed] [Google Scholar]
- 69.Murao K., Terpstra V., Green S. R., Kondratenko N., Steinberg D., and Quehenberger O.. 1997. Characterization of CLA-1, a human homologue of rodent scavenger receptor BI, as a receptor for high density lipoprotein and apoptotic thymocytes. J. Biol. Chem. 272: 17551–17557. [DOI] [PubMed] [Google Scholar]
- 70.Shiratsuchi A., Kawasaki Y., Ikemoto M., Arai H., and Nakanishi Y.. 1999. Role of class B scavenger receptor type I in phagocytosis of apoptotic rat spermatogenic cells by Sertoli cells. J. Biol. Chem. 274: 5901–5908. [DOI] [PubMed] [Google Scholar]
- 71.Svensson P. A., Johnson M. S., Ling C., Carlsson L. M., Billig H., and Carlsson B.. 1999. Scavenger receptor class B type I in the rat ovary: possible role in high density lipoprotein cholesterol uptake and in the recognition of apoptotic granulosa cells. Endocrinology. 140: 2494–2500. [DOI] [PubMed] [Google Scholar]
- 72.Imachi H., Murao K., Hiramine C., Sayo Y., Sato M., Hosokawa H., Ishida T., Kodama T., Quehenberger O., Steinberg D., et al. . 2000. Human scavenger receptor B1 is involved in recognition of apoptotic thymocytes by thymic nurse cells. Lab. Invest. 80: 263–270. [DOI] [PubMed] [Google Scholar]
- 73.Herz J., and Strickland D. K.. 2001. LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 108: 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neels J. G., van Den Berg B. M., Lookene A., Olivecrona G., Pannekoek H., and van Zonneveld A. J.. 1999. The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J. Biol. Chem. 274: 31305–31311. [DOI] [PubMed] [Google Scholar]
- 75.Azhar S., and Reaven E.. 2002. Scavenger receptor class BI and selective cholesteryl ester uptake: partners in the regulation of steroidogenesis. Mol. Cell. Endocrinol. 195: 1–26. [DOI] [PubMed] [Google Scholar]
- 76.Azhar S., Leers-Sucheta S., and Reaven E.. 2003. Cholesterol uptake in adrenal and gonadal tissues: the SR-BI and ‘selective’ pathway connection. Front. Biosci. 8: s998–s1029. [DOI] [PubMed] [Google Scholar]
- 77.Provost A. C., Pequignot M. O., Sainton K. M., Gadin S., Salle S., Marchant D., Hales D. B., and Abitbol M.. 2003. Expression of SR-BI receptor and StAR protein in rat ocular tissues. C. R. Biol. 326: 841–851. [DOI] [PubMed] [Google Scholar]
- 78.Ji Y., Wang N., Ramakrishnan R., Sehayek E., Huszar D., Breslow J. L., and Tall A. R.. 1999. Hepatic scavenger receptor BI promotes rapid clearance of high density lipoprotein free cholesterol and its transport into bile. J. Biol. Chem. 274: 33398–33402. [DOI] [PubMed] [Google Scholar]
- 79.Mardones P., Quinones V., Amigo L., Moreno M., Miquel J. F., Schwarz M., Miettinen H. E., Trigatti B., Krieger M., VanPatten S., et al. . 2001. Hepatic cholesterol and bile acid metabolism and intestinal cholesterol absorption in scavenger receptor class B type I-deficient mice. J. Lipid Res. 42: 170–180. [PubMed] [Google Scholar]
- 80.Yuan Q., Bie J., Wang J., Ghosh S. S., and Ghosh S.. 2013. Cooperation between hepatic cholesteryl ester hydrolase and scavenger receptor BI for hydrolysis of HDL-CE. J. Lipid Res. 54: 3078–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wiersma H., Gatti A., Nijstad N., Oude Elferink R. P., Kuipers F., and Tietge U. J.. 2009. Scavenger receptor class B type I mediates biliary cholesterol secretion independent of ATP-binding cassette transporter g5/g8 in mice. Hepatology. 50: 1263–1272. [DOI] [PubMed] [Google Scholar]
- 82.Temel R. E., Trigatti B., DeMattos R. B., Azhar S., Krieger M., and Williams D. L.. 1997. Scavenger receptor class B, type I (SR-BI) is the major route for the delivery of high density lipoprotein cholesterol to the steroidogenic pathway in cultured mouse adrenocortical cells. Proc. Natl. Acad. Sci. USA. 94: 13600–13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reaven E., Nomoto A., Leers-Sucheta S., Temel R., Williams D. L., and Azhar S.. 1998. Expression and microvillar localization of scavenger receptor, class B, type I (a high density lipoprotein receptor) in luteinized and hormone-desensitized rat ovarian models. Endocrinology. 139: 2847–2856. [DOI] [PubMed] [Google Scholar]
- 84.Azhar S., Nomoto A., and Reaven E.. 2002. Hormonal regulation of adrenal microvillar channel formation. J. Lipid Res. 43: 861–871. [PubMed] [Google Scholar]
- 85.Connelly M. A., and Williams D. L.. 2003. SR-BI and cholesterol uptake into steroidogenic cells. Trends Endocrinol. Metab. 14: 467–472. [DOI] [PubMed] [Google Scholar]
- 86.Rao R. M., Jo Y., Leers-Sucheta S., Bose H. S., Miller W. L., Azhar S., and Stocco D. M.. 2003. Differential regulation of steroid hormone biosynthesis in R2C and MA-10 Leydig tumor cells: role of SR-B1-mediated selective cholesteryl ester transport. Biol. Reprod. 68: 114–121. [DOI] [PubMed] [Google Scholar]
- 87.Cao G., Garcia C. K., Wyne K. L., Schultz R. A., Parker K. L., and Hobbs H. H.. 1997. Structure and localization of the human gene encoding SR-BI/CLA-1. Evidence for transcriptional control by steroidogenic factor 1. J. Biol. Chem. 272: 33068–33076. [DOI] [PubMed] [Google Scholar]
- 88.Pal R., Ke Q., Pihan G. A., Yesilaltay A., Penman M. L., Wang L., Chitraju C., Kang P. M., Krieger M., and Kocher O.. 2016. Carboxy-terminal deletion of the HDL receptor reduces receptor levels in liver and steroidogenic tissues, induces hypercholesterolemia, and causes fatal heart disease. Am. J. Physiol. Heart Circ. Physiol. 311: H1392–H1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reaven E., Boyles J., Spicher M., and Azhar S.. 1988. Evidence for surface entrapment of cholesterol-rich lipoproteins in luteinized ovary. Arteriosclerosis. 8: 298–309. [DOI] [PubMed] [Google Scholar]
- 90.Reaven E., Leers-Sucheta S., Nomoto A., and Azhar S.. 2001. Expression of scavenger receptor class B type 1 (SR-BI) promotes microvillar channel formation and selective cholesteryl ester transport in a heterologous reconstituted system. Proc. Natl. Acad. Sci. USA. 98: 1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reaven E., Nomoto A., Cortez Y., and Azhar S.. 2006. Consequences of over-expression of rat Scavenger Receptor, SR-BI, in an adrenal cell model. Nutr. Metab. (Lond.). 3: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Williams D. L., Wong J. S., and Hamilton R. L.. 2002. SR-BI is required for microvillar channel formation and the localization of HDL particles to the surface of adrenocortical cells in vivo. J. Lipid Res. 43: 544–549. [PubMed] [Google Scholar]
- 93.Duggan A. E., Marie R. S. Jr., and Callard I. P.. 2002. Expression of SR-BI (scavenger receptor class B type I) in turtle (Chrysemys picta) tissues and other nonmammalian vertebrates. J. Exp. Zool. 292: 430–434. [DOI] [PubMed] [Google Scholar]
- 94.Kleveland E. J., Syvertsen B. L., Ruyter B., Vegusdal A., Jorgensen S. M., and Gjoen T.. 2006. Characterization of scavenger receptor class B, type I in Atlantic salmon (Salmo salar L.). Lipids. 41: 1017–1027. [DOI] [PubMed] [Google Scholar]
- 95.Ding Z., Luo N., Kong Y., Li J., Zhang Y., Cao F., and Ye J.. 2016. Scavenger receptor class B, type I, a CD36 related protein in Macrobrachium nipponense: characterization, RNA interference, and expression analysis with different dietary lipid sources. Int. J. Genomics. 2016: 6325927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nichols Z., and Vogt R. G.. 2008. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 38: 398–415. [DOI] [PubMed] [Google Scholar]
- 97.Vogt R. G., Miller N. E., Litvack R., Fandino R. A., Sparks J., Staples J., Friedman R., and Dickens J. C.. 2009. The insect SNMP gene family. Insect Biochem. Mol. Biol. 39: 448–456. [DOI] [PubMed] [Google Scholar]
- 98.Herboso L., Talamillo A., Perez C., and Barrio R.. 2011. Expression of the scavenger receptor class B type I (SR-BI) family in Drosophila melanogaster. Int. J. Dev. Biol. 55: 603–611. [DOI] [PubMed] [Google Scholar]
- 99.Sakudoh T., Iizuka T., Narukawa J., Sezutsu H., Kobayashi I., Kuwazaki S., Banno Y., Kitamura A., Sugiyama H., Takada N., et al. . 2010. A CD36-related transmembrane protein is coordinated with an intracellular lipid-binding protein in selective carotenoid transport for cocoon coloration. J. Biol. Chem. 285: 7739–7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lopez D., and McLean M. P.. 1999. Sterol regulatory element-binding protein-1a binds to cis elements in the promoter of the rat high density lipoprotein receptor SR-BI gene. Endocrinology. 140: 5669–5681. [DOI] [PubMed] [Google Scholar]
- 101.Lopez D., Sandhoff T. W., and McLean M. P.. 1999. Steroidogenic factor-1 mediates cyclic 3′,5′-adenosine monophosphate regulation of the high density lipoprotein receptor. Endocrinology. 140: 3034–3044. [DOI] [PubMed] [Google Scholar]
- 102.Ansari K. I., Kasiri S., Hussain I., Bobzean S. A., Perrotti L. I., and Mandal S. S.. 2013. MLL histone methylases regulate expression of HDLR-SR-B1 in presence of estrogen and control plasma cholesterol in vivo. Mol. Endocrinol. 27: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Murao K., Imachi H., Yu X., Cao W. M., Muraoka T., Dobashi H., Hosomi N., Haba R., Iwama H., and Ishida T.. 2008. The transcriptional factor prolactin regulatory element-binding protein mediates the gene transcription of adrenal scavenger receptor class B type I via 3′,5′-cyclic adenosine 5′-monophosphate. Endocrinology. 149: 6103–6112. [DOI] [PubMed] [Google Scholar]
- 104.Malerød L., Juvet L. K., Hanssen-Bauer A., Eskild W., and Berg T.. 2002. Oxysterol-activated LXRalpha/RXR induces hSR-BI-promoter activity in hepatoma cells and preadipocytes. Biochem. Biophys. Res. Commun. 299: 916–923. [DOI] [PubMed] [Google Scholar]
- 105.Lopez D., and McLean M. P.. 2006. Activation of the rat scavenger receptor class B type I gene by PPARalpha. Mol. Cell. Endocrinol. 251: 67–77. [DOI] [PubMed] [Google Scholar]
- 106.Lopez D., Sanchez M. D., Shea-Eaton W., and McLean M. P.. 2002. Estrogen activates the high-density lipoprotein receptor gene via binding to estrogen response elements and interaction with sterol regulatory element binding protein-1A. Endocrinology. 143: 2155–2168. [DOI] [PubMed] [Google Scholar]
- 107.Shea-Eaton W., Lopez D., and McLean M. P.. 2001. Yin yang 1 protein negatively regulates high-density lipoprotein receptor gene transcription by disrupting binding of sterol regulatory element binding protein to the sterol regulatory element. Endocrinology. 142: 49–58. [DOI] [PubMed] [Google Scholar]
- 108.Lopez D., Shea-Eaton W., Sanchez M. D., and McLean M. P.. 2001. DAX-1 represses the high-density lipoprotein receptor through interaction with positive regulators sterol regulatory element-binding protein-1a and steroidogenic factor-1. Endocrinology. 142: 5097–5106. [DOI] [PubMed] [Google Scholar]
- 109.Sporstøl M., Tapia G., Malerød L., Mousavi S. A., and Berg T.. 2005. Pregnane X receptor-agonists down-regulate hepatic ATP-binding cassette transporter A1 and scavenger receptor class B type I. Biochem. Biophys. Res. Commun. 331: 1533–1541. [DOI] [PubMed] [Google Scholar]
- 110.Wang L., Jia X. J., Jiang H. J., Du Y., Yang F., Si S. Y., and Hong B.. 2013. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol. Cell. Biol. 33: 1956–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hu Z., Shen W. J., Kraemer F. B., and Azhar S.. 2012. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol. Cell. Biol. 32: 5035–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sahoo D., Darlington Y. F., Pop D., Williams D. L., and Connelly M. A.. 2007. Scavenger receptor class B Type I (SR-BI) assembles into detergent-sensitive dimers and tetramers. Biochim. Biophys. Acta. 1771: 807–817. [DOI] [PubMed] [Google Scholar]
- 113.Sahoo D., Peng Y., Smith J. R., Darlington Y. F., and Connelly M. A.. 2007. Scavenger receptor class B, type I (SR-BI) homo-dimerizes via its C-terminal region: fluorescence resonance energy transfer analysis. Biochim. Biophys. Acta. 1771: 818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gaidukov L., Nager A. R., Xu S., Penman M., and Krieger M.. 2011. Glycine dimerization motif in the N-terminal transmembrane domain of the high density lipoprotein receptor SR-BI required for normal receptor oligomerization and lipid transport. J. Biol. Chem. 286: 18452–18464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Landschulz K. T., Pathak R. K., Rigotti A., Krieger M., and Hobbs H. H.. 1996. Regulation of scavenger receptor, class B, type I, a high density lipoprotein receptor, in liver and steroidogenic tissues of the rat. J. Clin. Invest. 98: 984–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reaven E., Cortez Y., Leers-Sucheta S., Nomoto A., and Azhar S.. 2004. Dimerization of the scavenger receptor class B type I: formation, function, and localization in diverse cells and tissues. J. Lipid Res. 45: 513–528. [DOI] [PubMed] [Google Scholar]
- 117.Hu J., Zhang Z., Shen W-J., Nomoto A., and Azhar S.. 2011. Differential roles of cysteine residues in the cellular trafficking, dimerization, and function of the high-density lipoprotein receptor, SR-BI. Biochemistry. 50: 10860–10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kartz G. A., Holme R. L., Nicholson K., and Sahoo D.. 2014. SR-BI/CD36 chimeric receptors define extracellular subdomains of SR-BI critical for cholesterol transport. Biochemistry. 53: 6173–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Papale G. A., Nicholson K., Hanson P. J., Pavlovic M., Drover V. A., and Sahoo D.. 2010. Extracellular hydrophobic regions in scavenger receptor BI play a key role in mediating HDL-cholesterol transport. Arch. Biochem. Biophys. 496: 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu M., Romer K. A., Nieland T. J., Xu S., Saenz-Vash V., Penman M., Yesilaltay A., Carr S. A., and Krieger M.. 2011. Exoplasmic cysteine Cys384 of the HDL receptor SR-BI is critical for its sensitivity to a small-molecule inhibitor and normal lipid transport activity. Proc. Natl. Acad. Sci. USA. 108: 12243–12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guo L., Chen M., Song Z., Daugherty A., and Li X. A.. 2011. C323 of SR-BI is required for SR-BI-mediated HDL binding and cholesteryl ester uptake. J. Lipid Res. 52: 2272–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Papale G. A., Hanson P. J., and Sahoo D.. 2011. Extracellular disulfide bonds support scavenger receptor class B type I-mediated cholesterol transport. Biochemistry. 50: 6245–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu M., Lau T. Y., Carr S. A., and Krieger M.. 2012. Contributions of a disulfide bond and a reduced cysteine side chain to the intrinsic activity of the high-density lipoprotein receptor SR-BI. Biochemistry. 51: 10044–10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Neculai D., Schwake M., Ravichandran M., Zunke F., Collins R. F., Peters J., Neculai M., Plumb J., Loppnau P., Pizarro J. C., et al. . 2013. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature. 504: 172–176. [DOI] [PubMed] [Google Scholar]
- 125.Rasmussen J. T., Berglund L., Rasmussen M. S., and Petersen T. E.. 1998. Assignment of disulfide bridges in bovine CD36. Eur. J. Biochem. 257: 488–494. [DOI] [PubMed] [Google Scholar]
- 126.Conrad K. S., Cheng T. W., Ysselstein D., Heybrock S., Hoth L. R., Chrunyk B. A., Am Ende C. W., Krainc D., Schwake M., Saftig P., et al. . 2017. Lysosomal integral membrane protein-2 as a phospholipid receptor revealed by biophysical and cellular studies. Nat. Commun. 8: 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chadwick A. C., Jensen D. R., Peterson F. C., Volkman B. F., and Sahoo D.. 2015. Expression, purification and reconstitution of the C-terminal transmembrane domain of scavenger receptor BI into detergent micelles for NMR analysis. Protein Expr. Purif. 107: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Parathath S., Sahoo D., Darlington Y. F., Peng Y., Collins H. L., Rothblat G. H., Williams D. L., and Connelly M. A.. 2004. Glycine 420 near the C-terminal transmembrane domain of SR-BI is critical for proper delivery and metabolism of high density lipoprotein cholesteryl ester. J. Biol. Chem. 279: 24976–24985. [DOI] [PubMed] [Google Scholar]
- 129.Shen W-J., Hu J., Hu Z., Kraemer F. B., and Azhar S.. 2014. Scavenger receptor class B type I (SR-BI): a versatile receptor with multiple functions and actions. Metabolism. 63: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shen W. J., Azhar S., and Kraemer F. B.. 2017. SR-B1: a unique multifunctional receptor for cholesterol influx and efflux. Annu. Rev. Physiol. 80: 95–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shen W. J., Azhar S., and Kraemer F. B.. 2016. ACTH regulation of adrenal SR-B1. Front. Endocrinol. (Lausanne). 7: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gu X., Trigatti B., Xu S., Acton S., Babitt J., and Krieger M.. 1998. The efficient cellular uptake of high density lipoprotein lipids via scavenger receptor class B type I requires not only receptor-mediated surface binding but also receptor-specific lipid transfer mediated by its extracellular domain. J. Biol. Chem. 273: 26338–26348. [DOI] [PubMed] [Google Scholar]
- 133.Connelly M. A., Klein S. M., Azhar S., Abumrad N. A., and Williams D. L.. 1999. Comparison of class B scavenger receptors, CD36 and scavenger receptor BI (SR-BI), shows that both receptors mediate high density lipoprotein-cholesteryl ester selective uptake but SR-BI exhibits a unique enhancement of cholesteryl ester uptake. J. Biol. Chem. 274: 41–47. [DOI] [PubMed] [Google Scholar]
- 134.Liu T., Krieger M., Kan H. Y., and Zannis V. I.. 2002. The effects of mutations in helices 4 and 6 of ApoA-I on scavenger receptor class B type I (SR-BI)-mediated cholesterol efflux suggest that formation of a productive complex between reconstituted high density lipoprotein and SR-BI is required for efficient lipid transport. J. Biol. Chem. 277: 21576–21584. [DOI] [PubMed] [Google Scholar]
- 135.Temel R. E., Walzem R. L., Banka C. L., and Williams D. L.. 2002. Apolipoprotein A-I is necessary for the in vivo formation of high density lipoprotein competent for scavenger receptor BI-mediated cholesteryl ester-selective uptake. J. Biol. Chem. 277: 26565–26572. [DOI] [PubMed] [Google Scholar]
- 136.Temel R. E., Parks J. S., and Williams D. L.. 2003. Enhancement of scavenger receptor class B type I-mediated selective cholesteryl ester uptake from apoA-I(−/−) high density lipoprotein (HDL) by apolipoprotein A-I requires HDL reorganization by lecithin cholesterol acyltransferase. J. Biol. Chem. 278: 4792–4799. [DOI] [PubMed] [Google Scholar]
- 137.Nieland T. J., Penman M., Dori L., Krieger M., and Kirchhausen T.. 2002. Discovery of chemical inhibitors of the selective transfer of lipids mediated by the HDL receptor SR-BI. Proc. Natl. Acad. Sci. USA. 99: 15422–15427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hoekstra M. 2017. SR-BI as target in atherosclerosis and cardiovascular disease - a comprehensive appraisal of the cellular functions of SR-BI in physiology and disease. Atherosclerosis. 258: 153–161. [DOI] [PubMed] [Google Scholar]
- 139.Rodrigueza W. V., Thuahnai S. T., Temel R. E., Lund-Katz S., Phillips M. C., and Williams D. L.. 1999. Mechanism of scavenger receptor class B type I-mediated selective uptake of cholesteryl esters from high density lipoprotein to adrenal cells. J. Biol. Chem. 274: 20344–20350. [DOI] [PubMed] [Google Scholar]
- 140.Gomez-Diaz C., Bargeton B., Abuin L., Bukar N., Reina J. H., Bartoi T., Graf M., Ong H., Ulbrich M. H., Masson J. F., et al. . 2016. A CD36 ectodomain mediates insect pheromone detection via a putative tunnelling mechanism. Nat. Commun. 7: 11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Phillips M. C. 2014. Molecular mechanisms of cellular cholesterol efflux. J. Biol. Chem. 289: 24020–24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sparrow C. P., and Pittman R. C.. 1990. Cholesterol esters selectively taken up from high-density lipoproteins are hydrolyzed extralysosomally. Biochim. Biophys. Acta. 1043: 203–210. [DOI] [PubMed] [Google Scholar]
- 143.DeLamatre J. G., Carter R. M., and Hornick C. A.. 1991. Evidence for extralysosomal hydrolysis of high-density lipoprotein cholesteryl esters in rat hepatoma cells (Fu5AH): a model for delivery of high-density lipoprotein cholesterol. J. Cell. Physiol. 146: 18–24. [DOI] [PubMed] [Google Scholar]
- 144.Shimada A., Tamai T., Oida K., Takahashi S., Suzuki J., Nakai T., and Miyabo S.. 1994. Increase in neutral cholesteryl ester hydrolase activity produced by extralysosomal hydrolysis of high-density lipoprotein cholesteryl esters in rat hepatoma cells (H-35). Biochim. Biophys. Acta. 1215: 126–132. [DOI] [PubMed] [Google Scholar]
- 145.Connelly M. A., Kellner-Weibel G., Rothblat G. H., and Williams D. L.. 2003. SR-BI-directed HDL-cholesteryl ester hydrolysis. J. Lipid Res. 44: 331–341. [DOI] [PubMed] [Google Scholar]
- 146.Kraemer F. B., Shen W. J., Harada K., Patel S., Osuga J., Ishibashi S., and Azhar S.. 2004. Hormone-sensitive lipase is required for high-density lipoprotein cholesteryl ester-supported adrenal steroidogenesis. Mol. Endocrinol. 18: 549–557. [DOI] [PubMed] [Google Scholar]
- 147.Thuahnai S. T., Lund-Katz S., Williams D. L., and Phillips M. C.. 2001. Scavenger receptor class B, type I-mediated uptake of various lipids into cells. Influence of the nature of the donor particle interaction with the receptor. J. Biol. Chem. 276: 43801–43808. [DOI] [PubMed] [Google Scholar]
- 148.Yancey P. G., Bortnick A. E., Kellner-Weibel G., de la Llera-Moya M., Phillips M. C., and Rothblat G. H.. 2003. Importance of different pathways of cellular cholesterol efflux. Arterioscler. Thromb. Vasc. Biol. 23: 712–719. [DOI] [PubMed] [Google Scholar]
- 149.Ji Y., Jian B., Wang N., Sun Y., Moya M. L., Phillips M. C., Rothblat G. H., Swaney J. B., and Tall A. R.. 1997. Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J. Biol. Chem. 272: 20982–20985. [DOI] [PubMed] [Google Scholar]
- 150.Kent A. P., and Stylianou I. M.. 2011. Scavenger receptor class B member 1 protein: hepatic regulation and its effects on lipids, reverse cholesterol transport, and atherosclerosis. Hepat. Med. 3: 29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kellner-Weibel G., de La Llera-Moya M., Connelly M. A., Stoudt G., Christian A. E., Haynes M. P., Williams D. L., and Rothblat G. H.. 2000. Expression of scavenger receptor BI in COS-7 cells alters cholesterol content and distribution. Biochemistry. 39: 221–229. [DOI] [PubMed] [Google Scholar]
- 152.Silver D. L., and Tall A. R.. 2001. The cellular biology of scavenger receptor class B type I. Curr. Opin. Lipidol. 12: 497–504. [DOI] [PubMed] [Google Scholar]
- 153.Jian B., de la Llera-Moya M., Ji Y., Wang N., Phillips M. C., Swaney J. B., Tall A. R., and Rothblat G. H.. 1998. Scavenger receptor class B type I as a mediator of cellular cholesterol efflux to lipoproteins and phospholipid acceptors. J. Biol. Chem. 273: 5599–5606. [DOI] [PubMed] [Google Scholar]
- 154.de la Llera-Moya M., Rothblat G. H., Connelly M. A., Kellner-Weibel G., Sakr S. W., Phillips M. C., and Williams D. L.. 1999. Scavenger receptor BI (SR-BI) mediates free cholesterol flux independently of HDL tethering to the cell surface. J. Lipid Res. 40: 575–580. [PubMed] [Google Scholar]
- 155.Gu X., Kozarsky K., and Krieger M.. 2000. Scavenger receptor class B, type I-mediated [3H]cholesterol efflux to high and low density lipoproteins is dependent on lipoprotein binding to the receptor. J. Biol. Chem. 275: 29993–30001. [DOI] [PubMed] [Google Scholar]
- 156.Oram J. F., Lawn R. M., Garvin M. R., and Wade D. P.. 2000. ABCA1 is the cAMP-inducible apolipoprotein receptor that mediates cholesterol secretion from macrophages. J. Biol. Chem. 275: 34508–34511. [DOI] [PubMed] [Google Scholar]
- 157.Sankaranarayanan S., Oram J. F., Asztalos B. F., Vaughan A. M., Lund-Katz S., Adorni M. P., Phillips M. C., and Rothblat G. H.. 2009. Effects of acceptor composition and mechanism of ABCG1-mediated cellular free cholesterol efflux. J. Lipid Res. 50: 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rigotti A., Trigatti B. L., Penman M., Rayburn H., Herz J., and Krieger M.. 1997. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc. Natl. Acad. Sci. USA. 94: 12610–12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Varban M. L., Rinninger F., Wang N., Fairchild-Huntress V., Dunmore J. H., Fang Q., Gosselin M. L., Dixon K. L., Deeds J. D., Acton S. L., et al. . 1998. Targeted mutation reveals a central role for SR-BI in hepatic selective uptake of high density lipoprotein cholesterol. Proc. Natl. Acad. Sci. USA. 95: 4619–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ueda Y., Royer L., Gong E., Zhang J., Cooper P. N., Francone O., and Rubin E. M.. 1999. Lower plasma levels and accelerated clearance of high density lipoprotein (HDL) and non-HDL cholesterol in scavenger receptor class B type I transgenic mice. J. Biol. Chem. 274: 7165–7171. [DOI] [PubMed] [Google Scholar]
- 161.Yesilaltay A., Morales M. G., Amigo L., Zanlungo S., Rigotti A., Karackattu S. L., Donahee M. H., Kozarsky K. F., and Krieger M.. 2006. Effects of hepatic expression of the high-density lipoprotein receptor SR-BI on lipoprotein metabolism and female fertility. Endocrinology. 147: 1577–1588. [DOI] [PubMed] [Google Scholar]
- 162.Arai T., Wang N., Bezouevski M., Welch C., and Tall A. R.. 1999. Decreased atherosclerosis in heterozygous low density lipoprotein receptor-deficient mice expressing the scavenger receptor BI transgene. J. Biol. Chem. 274: 2366–2371. [DOI] [PubMed] [Google Scholar]
- 163.Kozarsky K. F., Donahee M. H., Glick J. M., Krieger M., and Rader D. J.. 2000. Gene transfer and hepatic overexpression of the HDL receptor SR-BI reduces atherosclerosis in the cholesterol-fed LDL receptor-deficient mouse. Arterioscler. Thromb. Vasc. Biol. 20: 721–727. [DOI] [PubMed] [Google Scholar]
- 164.Ueda Y., Gong E., Royer L., Cooper P. N., Francone O. L., and Rubin E. M.. 2000. Relationship between expression levels and atherogenesis in scavenger receptor class B, type I transgenics. J. Biol. Chem. 275: 20368–20373. [DOI] [PubMed] [Google Scholar]
- 165.Zhang Y., Da Silva J. R., Reilly M., Billheimer J. T., Rothblat G. H., and Rader D. J.. 2005. Hepatic expression of scavenger receptor class B type I (SR-BI) is a positive regulator of macrophage reverse cholesterol transport in vivo. J. Clin. Invest. 115: 2870–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Trigatti B., Rayburn H., Vinals M., Braun A., Miettinen H., Penman M., Hertz M., Schrenzel M., Amigo L., Rigotti A., et al. . 1999. Influence of the high density lipoprotein receptor SR-BI on reproductive and cardiovascular pathophysiology. Proc. Natl. Acad. Sci. USA. 96: 9322–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Huszar D., Varban M. L., Rinninger F., Feeley R., Arai T., Fairchild-Huntress V., Donovan M. J., and Tall A. R.. 2000. Increased LDL cholesterol and atherosclerosis in LDL receptor-deficient mice with attenuated expression of scavenger receptor B1. Arterioscler. Thromb. Vasc. Biol. 20: 1068–1073. [DOI] [PubMed] [Google Scholar]
- 168.Covey S. D., Krieger M., Wang W., Penman M., and Trigatti B. L.. 2003. Scavenger receptor class B type I-mediated protection against atherosclerosis in LDL receptor-negative mice involves its expression in bone marrow-derived cells. Arterioscler. Thromb. Vasc. Biol. 23: 1589–1594. [DOI] [PubMed] [Google Scholar]
- 169.Van Eck M., Twisk J., Hoekstra M., Van Rij B. T., Van der Lans C. A., Bos I. S., Kruijt J. K., Kuipers F., and Van Berkel T. J.. 2003. Differential effects of scavenger receptor BI deficiency on lipid metabolism in cells of the arterial wall and in the liver. J. Biol. Chem. 278: 23699–23705. [DOI] [PubMed] [Google Scholar]
- 170.Huby T., Doucet C., Dachet C., Ouzilleau B., Ueda Y., Afzal V., Rubin E., Chapman M. J., and Lesnik P.. 2006. Knockdown expression and hepatic deficiency reveal an atheroprotective role for SR-BI in liver and peripheral tissues. J. Clin. Invest. 116: 2767–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Linton M. F., Tao H., Linton E. F., and Yancey P. G.. 2017. SR-BI: a multifunctional receptor in cholesterol homeostasis and atherosclerosis. Trends Endocrinol. Metab. 28: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Armstrong S. M., Sugiyama M. G., Fung K. Y., Gao Y., Wang C., Levy A. S., Azizi P., Roufaiel M., Zhu S. N., Neculai D., et al. . 2015. A novel assay uncovers an unexpected role for SR-BI in LDL transcytosis. Cardiovasc. Res. 108: 268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yates M., Kolmakova A., Zhao Y., and Rodriguez A.. 2011. Clinical impact of scavenger receptor class B type I gene polymorphisms on human female fertility. Hum. Reprod. 26: 1910–1916. [DOI] [PubMed] [Google Scholar]
- 174.Holm T. M., Braun A., Trigatti B. L., Brugnara C., Sakamoto M., Krieger M., and Andrews N. C.. 2002. Failure of red blood cell maturation in mice with defects in the high-density lipoprotein receptor SR-BI. Blood. 99: 1817–1824. [DOI] [PubMed] [Google Scholar]
- 175.Zheng Z., Ai J., and Li X. A.. 2014. Scavenger receptor class B type I and immune dysfunctions. Curr. Opin. Endocrinol. Diabetes Obes. 21: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Burlone M. E., and Budkowska A.. 2009. Hepatitis C virus cell entry: role of lipoproteins and cellular receptors. J. Gen. Virol. 90: 1055–1070. [DOI] [PubMed] [Google Scholar]
- 177.Li Y., Kakinami C., Li Q., Yang B., and Li H.. 2013. Human apolipoprotein A-I is associated with dengue virus and enhances virus infection through SR-BI. PLoS One. 8: e70390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Baranova I. N., Souza A. C., Bocharov A. V., Vishnyakova T. G., Hu X., Vaisman B. L., Amar M. J., Chen Z., Kost Y., Remaley A. T., et al. . 2016. Human SR-BI and SR-BII potentiate lipopolysaccharide-induced inflammation and acute liver and kidney injury in mice. J. Immunol. 196: 3135–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yuhanna I. S., Zhu Y., Cox B. E., Hahner L. D., Osborne-Lawrence S., Lu P., Marcel Y. L., Anderson R. G., Mendelsohn M. E., Hobbs H. H., et al. . 2001. High-density lipoprotein binding to scavenger receptor-BI activates endothelial nitric oxide synthase. Nat. Med. 7: 853–857. [DOI] [PubMed] [Google Scholar]
- 180.Zhu W., Saddar S., Seetharam D., Chambliss K. L., Longoria C., Silver D. L., Yuhanna I. S., Shaul P. W., and Mineo C.. 2008. The scavenger receptor class B type I adaptor protein PDZK1 maintains endothelial monolayer integrity. Circ. Res. 102: 480–487. [DOI] [PubMed] [Google Scholar]
- 181.Mineo C., Yuhanna I. S., Quon M. J., and Shaul P. W.. 2003. High density lipoprotein-induced endothelial nitric-oxide synthase activation is mediated by Akt and MAP kinases. J. Biol. Chem. 278: 9142–9149. [DOI] [PubMed] [Google Scholar]
- 182.Saddar S., Carriere V., Lee W. R., Tanigaki K., Yuhanna I. S., Parathath S., Morel E., Warrier M., Sawyer J. K., Gerard R. D., et al. . 2013. Scavenger receptor class B type I is a plasma membrane cholesterol sensor. Circ. Res. 112: 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Morel E., Ghezzal S., Lucchi G., Truntzer C., Pais de Barros J. P., Simon-Plas F., Demignot S., Mineo C., Shaul P. W., Leturque A., et al. . 2018. Cholesterol trafficking and raft-like membrane domain composition mediate scavenger receptor class B type 1-dependent lipid sensing in intestinal epithelial cells. Biochim. Biophys. Acta. 1863: 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Baranova I. N., Vishnyakova T. G., Bocharov A. V., Kurlander R., Chen Z., Kimelman M. L., Remaley A. T., Csako G., Thomas F., Eggerman T. L., et al. . 2005. Serum amyloid A binding to CLA-1 (CD36 and LIMPII analogous-1) mediates serum amyloid A protein-induced activation of ERK1/2 and p38 mitogen-activated protein kinases. J. Biol. Chem. 280: 8031–8040. [DOI] [PubMed] [Google Scholar]
- 185.van der Westhuyzen D. R., Cai L., de Beer M. C., and de Beer F. C.. 2005. Serum amyloid A promotes cholesterol efflux mediated by scavenger receptor B-I. J. Biol. Chem. 280: 35890–35895. [DOI] [PubMed] [Google Scholar]
- 186.Lee M. H., Appleton K. M., El-Shewy H. M., Sorci-Thomas M. G., Thomas M. J., Lopes-Virella M. F., Luttrell L. M., Hammad S. M., and Klein R. L.. 2017. S1P in HDL promotes interaction between SR-BI and S1PR1 and activates S1PR1-mediated biological functions: calcium flux and S1PR1 internalization. J. Lipid Res. 58: 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liu X., Ren K., Suo R., Xiong S. L., Zhang Q. H., Mo Z. C., Tang Z. L., Jiang Y., Peng X. S., and Yi G. H.. 2016. ApoA-I induces S1P release from endothelial cells through ABCA1 and SR-BI in a positive feedback manner. J. Physiol. Biochem. 72: 657–667. [DOI] [PubMed] [Google Scholar]
- 188.Tao H., Yancey P. G., Babaev V. R., Blakemore J. L., Zhang Y., Ding L., Fazio S., and Linton M. F.. 2015. Macrophage SR-BI mediates efferocytosis via Src/PI3K/Rac1 signaling and reduces atherosclerotic lesion necrosis. J. Lipid Res. 56: 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Song P., Kwon Y., Yea K., Moon H. Y., Yoon J. H., Ghim J., Hyun H., Kim D., Koh A., Berggren P. O., et al. . 2015. Apolipoprotein a1 increases mitochondrial biogenesis through AMP-activated protein kinase. Cell. Signal. 27: 1873–1881. [DOI] [PubMed] [Google Scholar]
- 190.Zhu P., Liu X., Treml L. S., Cancro M. P., and Freedman B. D.. 2009. Mechanism and regulatory function of CpG signaling via scavenger receptor B1 in primary B cells. J. Biol. Chem. 284: 22878–22887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Trigatti B. L., Krieger M., and Rigotti A.. 2003. Influence of the HDL receptor SR-BI on lipoprotein metabolism and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 23: 1732–1738. [DOI] [PubMed] [Google Scholar]
- 192.Leiva A., Verdejo H., Benitez M. L., Martinez A., Busso D., and Rigotti A.. 2011. Mechanisms regulating hepatic SR-BI expression and their impact on HDL metabolism. Atherosclerosis. 217: 299–307. [DOI] [PubMed] [Google Scholar]
- 193.Donowitz M., Cha B., Zachos N. C., Brett C. L., Sharma A., Tse C. M., and Li X.. 2005. NHERF family and NHE3 regulation. J. Physiol. 567: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bretscher A., Edwards K., and Fehon R. G.. 2002. ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 3: 586–599. [DOI] [PubMed] [Google Scholar]
- 195.Ikemoto M., Arai H., Feng D., Tanaka K., Aoki J., Dohmae N., Takio K., Adachi H., Tsujimoto M., and Inoue K.. 2000. Identification of a PDZ-domain-containing protein that interacts with the scavenger receptor class B type I. Proc. Natl. Acad. Sci. USA. 97: 6538–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Silver D. L. 2002. A carboxyl-terminal PDZ-interacting domain of scavenger receptor B, type I is essential for cell surface expression in liver. J. Biol. Chem. 277: 34042–34047. [DOI] [PubMed] [Google Scholar]
- 197.Kocher O., Yesilaltay A., Cirovic C., Pal R., Rigotti A., and Krieger M.. 2003. Targeted disruption of the PDZK1 gene in mice causes tissue-specific depletion of the high density lipoprotein receptor scavenger receptor class B type I and altered lipoprotein metabolism. J. Biol. Chem. 278: 52820–52825. [DOI] [PubMed] [Google Scholar]
- 198.Kocher O., Birrane G., Tsukamoto K., Fenske S., Yesilaltay A., Pal R., Daniels K., Ladias J. A., and Krieger M.. 2010. In vitro and in vivo analysis of the binding of the C terminus of the HDL receptor scavenger receptor class B, type I (SR-BI), to the PDZ1 domain of its adaptor protein PDZK1. J. Biol. Chem. 285: 34999–35010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Fenske S. A., Yesilaltay A., Pal R., Daniels K., Barker C., Quinones V., Rigotti A., Krieger M., and Kocher O.. 2009. Normal hepatic cell surface localization of the high density lipoprotein receptor, scavenger receptor class B, type I, depends on all four PDZ domains of PDZK1. J. Biol. Chem. 284: 5797–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kocher O., Birrane G., Yesilaltay A., Shechter S., Pal R., Daniels K., and Krieger M.. 2011. Identification of the PDZ3 domain of the adaptor protein PDZK1 as a second, physiologically functional binding site for the C terminus of the high density lipoprotein receptor scavenger receptor class B type I. J. Biol. Chem. 286: 25171–25186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tsukamoto K., Wales T. E., Daniels K., Pal R., Sheng R., Cho W., Stafford W., Engen J. R., Krieger M., and Kocher O.. 2013. Noncanonical role of the PDZ4 domain of the adaptor protein PDZK1 in the regulation of the hepatic high density lipoprotein receptor scavenger receptor class B, type I (SR-BI). J. Biol. Chem. 288: 19845–19860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hu Z., Hu J., Zhang Z., Shen W-J., Yun C. C., Berlot C. H., Kraemer F. B., and Azhar S.. 2013. Regulation of expression and function of scavenger receptor class B, type I (SR-BI) by Na+/H+ exchanger regulatory factors (NHERFs). J. Biol. Chem. 288: 11416–11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kocher O., Cheresh P., Brown L. F., and Lee S. W.. 1995. Identification of a novel gene, selectively up-regulated in human carcinomas, using the differential display technique. Clin. Cancer Res. 1: 1209–1215. [PubMed] [Google Scholar]
- 204.Silver D. L., Wang N., and Vogel S.. 2003. Identification of small PDZK1-associated protein, DD96/MAP17, as a regulator of PDZK1 and plasma high density lipoprotein levels. J. Biol. Chem. 278: 28528–28532. [DOI] [PubMed] [Google Scholar]
- 205.Green S. R., and Pittman R. C.. 1991. Selective uptake of cholesteryl esters from low density lipoproteins in vitro and in vivo. J. Lipid Res. 32: 667–678. [PubMed] [Google Scholar]
- 206.Leitersdorf E., Stein O., Eisenberg S., and Stein Y.. 1984. Uptake of rat plasma HDL subfractions labeled with [3H]cholesteryl linoleyl ether or with 125I by cultured rat hepatocytes and adrenal cells. Biochim. Biophys. Acta. 796: 72–82. [DOI] [PubMed] [Google Scholar]
- 207.Pittman R. C., Knecht T. P., Rosenbaum M. S., and Taylor C. A. Jr. 1987. A nonendocytotic mechanism for the selective uptake of high density lipoprotein-associated cholesterol esters. J. Biol. Chem. 262: 2443–2450. [PubMed] [Google Scholar]
- 208.Reaven E., Tsai L., and Azhar S.. 1995. Cholesterol uptake by the ‘selective’ pathway of ovarian granulosa cells: early intracellular events. J. Lipid Res. 36: 1602–1617. [PubMed] [Google Scholar]
- 209.Reaven E., Tsai L., and Azhar S.. 1996. Intracellular events in the “selective” transport of lipoprotein-derived cholesteryl esters. J. Biol. Chem. 271: 16208–16217. [DOI] [PubMed] [Google Scholar]
- 210.Azhar S., Tsai L., and Reaven E.. 1990. Uptake and utilization of lipoprotein cholesteryl esters by rat granulosa cells. Biochim. Biophys. Acta. 1047: 148–160. [DOI] [PubMed] [Google Scholar]
- 211.Quinn P. G., Dombrausky L. J., Chen Y. D., and Payne A. H.. 1981. Serum lipoproteins increase testosterone production in hCG-desensitized Leydig cells. Endocrinology. 109: 1790–1792. [DOI] [PubMed] [Google Scholar]
- 212.Klinefelter G. R., and Ewing L. L.. 1988. Optimizing testosterone production by purified adult rat Leydig cells in vitro. In Vitro Cell. Dev. Biol. 24: 545–549. [DOI] [PubMed] [Google Scholar]
- 213.Hedger M. P., and Risbridger G. P.. 1992. Effect of serum and serum lipoproteins on testosterone production by adult rat Leydig cells in vitro. J. Steroid Biochem. Mol. Biol. 43: 581–589. [DOI] [PubMed] [Google Scholar]
- 214.Risbridger G. P., and Hedger M. P.. 1992. Adult rat Leydig cell cultures: minimum requirements for maintenance of luteinizing hormone responsiveness and testosterone production. Mol. Cell. Endocrinol. 83: 125–132. [DOI] [PubMed] [Google Scholar]
- 215.Rigotti A., Edelman E. R., Seifert P., Iqbal S. N., DeMattos R. B., Temel R. E., Krieger M., and Williams D. L.. 1996. Regulation by adrenocorticotropic hormone of the in vivo expression of scavenger receptor class B type I (SR-BI), a high density lipoprotein receptor, in steroidogenic cells of the murine adrenal gland. J. Biol. Chem. 271: 33545–33549. [DOI] [PubMed] [Google Scholar]
- 216.Liu J., Voutilainen R., Heikkila P., and Kahri A. I.. 1997. Ribonucleic acid expression of the CLA-1 gene, a human homolog to mouse high density lipoprotein receptor SR-BI, in human adrenal tumors and cultured adrenal cells. J. Clin. Endocrinol. Metab. 82: 2522–2527. [DOI] [PubMed] [Google Scholar]
- 217.Cao G., Zhao L., Stangl H., Hasegawa T., Richardson J. A., Parker K. L., and Hobbs H. H.. 1999. Developmental and hormonal regulation of murine scavenger receptor, class B, type 1. Mol. Endocrinol. 13: 1460–1473. [DOI] [PubMed] [Google Scholar]
- 218.Sun Y., Wang N., and Tall A. R.. 1999. Regulation of adrenal scavenger receptor-BI expression by ACTH and cellular cholesterol pools. J. Lipid Res. 40: 1799–1805. [PubMed] [Google Scholar]
- 219.McLean M. P., and Sandhoff T. W.. 1998. Expression and hormonal regulation of the high-density lipoprotein (HDL) receptor scavenger receptor class B type I messenger ribonucleic acid in the rat ovary. Endocrine. 9: 243–252. [DOI] [PubMed] [Google Scholar]
- 220.Li X., Peegel H., and Menon K. M.. 2001. Regulation of high density lipoprotein receptor messenger ribonucleic acid expression and cholesterol transport in theca-interstitial cells by insulin and human chorionic gonadotropin. Endocrinology. 142: 174–181. [DOI] [PubMed] [Google Scholar]
- 221.Miranda-Jiménez L., and Murphy B. D.. 2007. Lipoprotein receptor expression during luteinization of the ovarian follicle. Am. J. Physiol. Endocrinol. Metab. 293: E1053–E1061. [DOI] [PubMed] [Google Scholar]
- 222.Cherradi N., Bideau M., Arnaudeau S., Demaurex N., James R. W., Azhar S., and Capponi A. M.. 2001. Angiotensin II promotes selective uptake of high density lipoprotein cholesterol esters in bovine adrenal glomerulosa and human adrenocortical carcinoma cells through induction of scavenger receptor class B type I. Endocrinology. 142: 4540–4549. [DOI] [PubMed] [Google Scholar]
- 223.Wang N., Weng W., Breslow J. L., and Tall A. R.. 1996. Scavenger receptor BI (SR-BI) is up-regulated in adrenal gland in apolipoprotein A-I and hepatic lipase knock-out mice as a response to depletion of cholesterol stores. In vivo evidence that SR-BI is a functional high density lipoprotein receptor under feedback control. J. Biol. Chem. 271: 21001–21004. [DOI] [PubMed] [Google Scholar]
- 224.Mavridou S., Venihaki M., Rassouli O., Tsatsanis C., and Kardassis D.. 2010. Feedback inhibition of human scavenger receptor class B type I gene expression by glucocorticoid in adrenal and ovarian cells. Endocrinology. 151: 3214–3224. [DOI] [PubMed] [Google Scholar]
- 225.Ng D. S., Francone O. L., Forte T. M., Zhang J., Haghpassand M., and Rubin E. M.. 1997. Disruption of the murine lecithin:cholesterol acyltransferase gene causes impairment of adrenal lipid delivery and up-regulation of scavenger receptor class B type I. J. Biol. Chem. 272: 15777–15781. [DOI] [PubMed] [Google Scholar]
- 226.Reaven E., Lua Y., Nomoto A., Temel R., Williams D. L., van der Westhuyzen D. R., and Azhar S.. 1999. The selective pathway and a high-density lipoprotein receptor (SR-BI) in ovarian granulosa cells of the mouse. Biochim. Biophys. Acta. 1436: 565–576. [DOI] [PubMed] [Google Scholar]
- 227.Wu Q., Sucheta S., Azhar S., and Menon K. M.. 2003. Lipoprotein enhancement of ovarian theca-interstitial cell steroidogenesis: relative contribution of scavenger receptor class B (type I) and adenosine 5′-triphosphate- binding cassette (type A1) transporter in high-density lipoprotein-cholesterol transport and androgen synthesis. Endocrinology. 144: 2437–2445. [DOI] [PubMed] [Google Scholar]
- 228.Hu Z., Shen W-J., Cortez Y., Tang X., Liu L-F., Kraemer F. B., and Azhar S.. 2013. Hormonal regulation of microRNA expression in steroid producing cells of the ovary, testis and adrenal gland. PLoS One. 8: e78040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hu Z., Hu J., Shen W. J., Kraemer F. B., and Azhar S.. 2015. A novel role of salt-inducible kinase 1 (SIK1) in the post-translational regulation of scavenger receptor class B type 1 activity. Biochemistry. 54: 6917–6930. [DOI] [PubMed] [Google Scholar]
- 230.Tsutsumi K., Hagi A., and Inoue Y.. 2001. The relationship between plasma high density lipoprotein cholesterol levels and cholesteryl ester transfer protein activity in six species of healthy experimental animals. Biol. Pharm. Bull. 24: 579–581. [DOI] [PubMed] [Google Scholar]
- 231.Acton S., Osgood D., Donoghue M., Corella D., Pocovi M., Cenarro A., Mozas P., Keilty J., Squazzo S., Woolf E. A., et al. . 1999. Association of polymorphisms at the SR-BI gene locus with plasma lipid levels and body mass index in a white population. Arterioscler. Thromb. Vasc. Biol. 19: 1734–1743. [DOI] [PubMed] [Google Scholar]
- 232.Osgood D., Corella D., Demissie S., Cupples L. A., Wilson P. W., Meigs J. B., Schaefer E. J., Coltell O., and Ordovas J. M.. 2003. Genetic variation at the scavenger receptor class B type I gene locus determines plasma lipoprotein concentrations and particle size and interacts with type 2 diabetes: the framingham study. J. Clin. Endocrinol. Metab. 88: 2869–2879. [DOI] [PubMed] [Google Scholar]
- 233.Richard E., von Muhlen D., Barrett-Connor E., Alcaraz J., Davis R., and McCarthy J. J.. 2005. Modification of the effects of estrogen therapy on HDL cholesterol levels by polymorphisms of the HDL-C receptor, SR-BI: the Rancho Bernardo Study. Atherosclerosis. 180: 255–262. [DOI] [PubMed] [Google Scholar]
- 234.Roberts C. G., Shen H., Mitchell B. D., Damcott C. M., Shuldiner A. R., and Rodriguez A.. 2007. Variants in scavenger receptor class B type I gene are associated with HDL cholesterol levels in younger women. Hum. Hered. 64: 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Cerda A., Genvigir F., Arazi S., Hirata M., Dorea E., Bernik M., Bertolami M., Faludi A., and Hirata R.. 2010. Influence of SCARB1 polymorphisms on serum lipids of hypercholesterolemic individuals treated with atorvastatin. Clin. Chim. Acta. 411: 631–637. [DOI] [PubMed] [Google Scholar]
- 236.Hong S. H., Kim Y. R., Yoon Y. M., Min W. K., Chun S. I., and Kim J. Q.. 2002. Association between HaeIII polymorphism of scavenger receptor class B type I gene and plasma HDL-cholesterol concentration. Ann. Clin. Biochem. 39: 478–481. [DOI] [PubMed] [Google Scholar]
- 237.Morabia A., Ross B. M., Costanza M. C., Cayanis E., Flaherty M. S., Alvin G. B., Das K., James R., Yang A. S., Evagrafov O., et al. . 2004. Population-based study of SR-BI genetic variation and lipid profile. Atherosclerosis. 175: 159–168. [DOI] [PubMed] [Google Scholar]
- 238.Niemsiri V., Wang X., Pirim D., Radwan Z. H., Bunker C. H., Barmada M. M., Kamboh M. I., and Demirci F. Y.. 2015. Genetic contribution of SCARB1 variants to lipid traits in African Blacks: a candidate gene association study. BMC Med. Genet. 16: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Niemsiri V., Wang X., Pirim D., Radwan Z. H., Hokanson J. E., Hamman R. F., Barmada M. M., Demirci F. Y., and Kamboh M. I.. 2014. Impact of genetic variants in human scavenger receptor class B type I (SCARB1) on plasma lipid traits. Circ Cardiovasc Genet. 7: 838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Tai E. S., Adiconis X., Ordovas J. M., Carmena-Ramon R., Real J., Corella D., Ascaso J., and Carmena R.. 2003. Polymorphisms at the SRBI locus are associated with lipoprotein levels in subjects with heterozygous familial hypercholesterolemia. Clin. Genet. 63: 53–58. [DOI] [PubMed] [Google Scholar]
- 241.Manichaikul A., Naj A., Herrington D., Post W., Rich S., and Rodriguez A.. 2012. Association of SCARB1 variants with subclinical atherosclerosis and incident cardiovascular disease: the multi-ethnic study of atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 32: 1991–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yang X., Sethi A., Yanek L. R., Knapper C., Nordestgaard B. G., Tybjærg-Hansen A., Becker D. M., Mathias R. A., Remaley A. T., and Becker L. C.. 2016. SCARB1 gene variants are associated with the phenotype of combined high high-density lipoprotein cholesterol and high lipoprotein (a). Circ Cardiovasc Genet. 9: 408–418. [DOI] [PubMed] [Google Scholar]
- 243.Vergeer M., Korporaal S. J., Franssen R., Meurs I., Out R., Hovingh G. K., Hoekstra M., Sierts J. A., Dallinga-Thie G. M., Motazacker M. M., et al. . 2011. Genetic variant of the scavenger receptor BI in humans. N. Engl. J. Med. 364: 136–145. [DOI] [PubMed] [Google Scholar]
- 244.Brunham L. R., Tietjen I., Bochem A. E., Singaraja R. R., Franchini P. L., Radomski C., Mattice M., Legendre A., Hovingh G. K., Kastelein J. J., et al. . 2011. Novel mutations in scavenger receptor BI associated with high HDL cholesterol in humans. Clin. Genet. 79: 575–581. [DOI] [PubMed] [Google Scholar]
- 245.Zanoni P., Khetarpal S. A., Larach D. B., Hancock-Cerutti W. F., Millar J. S., Cuchel M., DerOhannessian S., Kontush A., Surendran P., Saleheen D., et al. . 2016. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science. 351: 1166–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Webb B., and Sali A.. 2017. Protein structure modeling with MODELLER. Methods Mol. Biol. 1654: 39–54. [DOI] [PubMed] [Google Scholar]
- 247.Case D. A., Cheatham T. E. 3rd, Darden T., Gohlke H., Luo R., Merz K. M. Jr., Onufriev A., Simmerling C., Wang B., and Woods R. J.. 2005. The Amber biomolecular simulation programs. J. Comput. Chem. 26: 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]