Abstract
At the core of the changes characteristic of alcoholism are alterations in gene expression in the brain of the addicted individual. These changes are believed to underlie some of the neuroadaptations that promote compulsive drinking. Unfortunately, the mechanisms by which alcohol consumption produces changes in gene expression remain poorly understood. MicroRNAs (miRNAs) have emerged as important regulators of gene expression because they can coordinately modulate the translation efficiency of large sets of specific mRNAs. Here we investigate the early miRNA responses elicited by an acute sedating dose of alcohol in the Drosophila model organism. In our analysis, we combine the power of next generation sequencing with Drosophila genetics to identify alcohol-sensitive miRNAs and to functionally test them for a role in modulating alcohol sensitivity. We identified 14 known Drosophila miRNAs, and 13 putative novel miRNAs that respond to an acute sedative exposure to alcohol. Using the GeneSwitch Gal4/UAS system, a subset of these ethanol-responsive miRNAs was functionally tested to determine their individual contribution in modulating ethanol sensitivity. We identified two microRNAs that when overexpressed significantly increased ethanol sensitivity: miR-6 and miR-310. MicroRNA target prediction analysis revealed that the different alcohol-responsive miRNAs target overlapping sets of mRNAs. Alcoholism is the product of accumulated cellular changes produced by chronic ethanol consumption. Although, all of the changes described herein are extremely rapid responses evoked by a single ethanol exposure, understanding the gene expression changes that occur in the first few minutes after ethanol exposure will help us to categorize ethanol responses into those that are near instantaneous and those that are emergent responses produced only by repeated ethanol exposure.
Keywords: Alcoholism, Neuroadaptation, miRNA-seq, Genomics, miR-6, miR-310
Introduction
MicroRNAs (miRNAs) are endogenous RNAs of approximately 22 nucleotides that have been implicated in the post-transcriptional regulation of complex biological networks. These small noncoding RNAs are thought to play a crucial role in shaping the function of tissues and organs in both health and disease. In general, miRNAs suppress gene expression by repressing the translation of specific mRNAs and/or by message destabilization (Bethune et al., 2012; Gurtan & Sharp, 2013).
Canonical miRNAs are produced from long primary RNA transcripts (pri-miRNA) that by cleavage in the nucleus by the enzyme Drosha to form ∼60-70 nt precursor miRNA hairpin loops (pre-miRNA). Individual pri-miRNAs have been observed to encode as many as 6 different pre-miRNAs. Each pre-miRNA hairpin loop is then transported to the cytoplasm where they are processed into small ∼22 nt double stranded RNA duplexes by the enzyme Dicer (Bartel, 2004). Although, both strands of the duplex are necessarily produced in equal amounts by transcription, their accumulation is known to be asymmetric. The most abundant strand from each duplex will become the biologically active strand that is incorporated into the RNA-Induced Silencing Complex (RISC) and serves as the guide strand during miRNA-directed silencing. The other strand, which is considered the inactive strand and is called the miRNA* (miRNA star) or passenger strand, is typically degraded (Meijer et al., 2014). The miRNA/RISC complex then binds to specific sequences predominantly found in the 3′UTR of protein-coding mRNAs triggering inhibition of protein synthesis and mRNA degradation, depending on the degree of complementarity between the miRNA and the target mRNA (Fang & Rajewsky, 2011). Nonetheless, a strong complementarity within positions 2 to 8 of the 5′ end of the miRNA (known as the “seed”) is considered critical for recognition of target mRNA and its regulation (Bartel, 2009; Doench & Sharp, 2004; Lai, 2004). Both the mechanism of generating miRNAs and the miRNAs themselves are highly conserved from flies to mammals, therefore, analysis of miRNA function in the Drosophila model system should translate well to mammals (Ibanez-Ventoso et al. 2008).
Recent evidence suggests that some miRNAs are alcohol-responsive and play important regulatory roles in neuroadaptations that mediate the development of alcohol addiction. The best example so far of miRNA regulation that alters an alcohol response is the miR-9 regulation of slo gene expression. The slo gene encodes BK-type Ca2+-activated K+ channels whose activity is sensitive to ethanol. In mammalian hypothalamo-neurohypophysial system neurons, acute alcohol exposure up-regulates miR-9 to mediate selective destabilization of mRNA splice variants encoding ethanol-sensitive BK channels. This favors the production of more ethanol-resistant channels, thereby contributing to the development of functional alcohol tolerance (Pietrzykowski et al., 2008). Importantly, the slo gene is also involved in the development of functional ethanol tolerance in Drosophila. In this model system, ethanol affects slo transcription in a way that contributes to behavioral functional tolerance (Cowmeadow et al., 2005; Cowmeadow et al., 2006). However, miR-9 is not the only alcohol-responsive miRNA nor is slo the only protein-coding gene involved in alcohol responses. There is a growing body of evidence indicating that alcohol regulates the expression of other miRNAs and other protein-coding genes (Bala & Szabo, 2012; Li et al., 2013; Nunez et al., 2013; Tapocik et al., 2013).
Here, we use the genetically malleable Drosophila model system and next generation sequencing to identify miRNAs whose abundance changes within the first 30 minutes after a sedative alcohol exposure. Our central hypothesis is that changes in the expression of specific miRNAs induced by alcohol exposure contribute to the neuroadaptations that lead to alcohol dependence. Changes in miRNA expression that contribute to alcoholism-related phenotypes in flies will be a useful guide for identifying the relevant miRNA mechanisms that contribute to alcoholism-related phenotypes in mammals. The advantage of doing this work in Drosophila is that large numbers of candidate miRNAs can be rapidly and functionally tested for their role in producing ethanol tolerance and dependence by controlled miRNA overexpression or inhibition.
Methods
Stocks and Fly Husbandry
All flies used in this study were maintained on a standard cornmeal/molasses/agar medium under 12:12 light:dark conditions. Experiments were conducted using 3-5 day-old female flies. Only females were used because all previous directly comparable studies used only females. Canton S was the wild-type stock used for the RNA-seq analysis. All transgenic stocks were obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537). These are (stock numbers in parenthesis):
W[1118]; P{UAS-LUC-mir-1003.T}attP2 (41220)
w[1118]; P{UAS-LUC-mir-124.T}attP2 (41126)
w*; P{UAS-mir-308.D}10 (41809)
w[1118]; P{UAS-LUC-mir-6-1.mir-6-2.mir-6-3.T}attP2 (41136)
w[1118]; P{UAS-LUC-mir-304.T}attP2 (41170)
w[1118]; P{UAS-LUC-mir-310.T}attP2/TM3, Sb1 (41155)
w[1118]; P{UAS-LUC-mir-310.mir-311.mir-312.mir-313.T}attP2/TM3, Sb1 (41135)
y[1] w[*]; P{w[+mC]=elav-witch.O}GSG301 (43642)
The GeneSwitch system was used to specifically overexpress each UAS-miRNA in the nervous system (Osterwalder et al., 2001; Roman et al., 2001). Inducible transheterozygous animals were produced by crossing each of the UAS-miRNA stocks to the neuronal specific “elav-Switch” driver stock. To activate transgene expression RU486 (Cayman Chemical, Ann Arbor, MI, USA) was supplied to the transgenic flies in their food as described in Troutwine et al (2016). RU486-laced food was generated by adding 25 mM RU486 in 80% ethanol to molten fly food to a final concentration of 200 μM RU486 (after dilution in food, final ethanol concentration is below 0.64%). Control food was also melted and mixed with an equivalent amount of the carrier solution (80% ethanol). The RU486 food was distributed to fly vials and allowed to cool and dry for at least one hour before flies were allowed to feed. Flies were kept on RU486 or carrier-containing food for three consecutive days before being subjected to our ethanol resistance assay.
Ethanol treatment, RNA Isolation and Next-Generation Sequencing
Prior to Next-Generation Sequencing miRNA analysis, wild-type Canton S flies were treated with alcohol vapor using the inebriator as described previously (Cowmeadow et al., 2005; Krishnan et al., 2012). Briefly, approximately 180 flies were divided in 6 vials of ∼30 flies each and exposed to an air stream saturated with ethanol vapor until all flies were sedated (typically 12-15 minutes). After sedation, the ethanol-saturated air stream was replaced with a stream of humidified air for 30 minutes and then the animals were sacrificed for RNA extraction. An untreated group, which was exposed to an ethanol-free stream of humidified air, was prepared in parallel. This group served as control in the miRNA-seq analysis. Preparation of RNA and quality control was performed as described in Troutwine et al. (2016). Small RNA libraries were generated from each RNA sample using Illumina Small RNA Sample Preparation kit according to the manufacturer's instructions and sequenced with the Illumina HiSeq 2000 at the Beijing Genomics Institute (Shenzhen, China). Sequence depth was set to 20M reads using single-end chemistry and 50-bp cycles. Raw sequences have been deposited in the NCBI: Gene Expression Omnibus (GEO) public functional genomics data repository. Data can be found on the GEO website (http://www.ncbi.nlm.nih.gov/geo/) using accession number GSE89137. All essential sample annotation and experimental design information including sample data relationships have been included in the repository according to the Minimum Information About a Microarray Experiment (MIAME) guidelines (Brazma et al., 2001).
miRNA Sequence Analysis and Differential Expression
rRNAs, tRNAs, snRNAs and low-quality reads (containing adapters or high content of unknown bases) were filtered out by aligning to Rfam and Genbank databases (Griffiths-Jones et al., 2003). miRNA-seq reads were aligned and mapped to the Drosophila reference genome (BDGP Release 5) using SOAPaligner/SOAP2 (Li et al., 2009), allowing no more than 5 bp mismatches. Known miRNAs were identified by alignment to miRBase21.0 (Kozomara & Griffiths-Jones, 2014). Expression levels for miRNAs were calculated using the CLC Genomics Workbench (CLC bio, Boston, MA). Differential expression analysis was conducted using Cluster 3.0, and Java TreeView software (Eisen et al., 1998; Saldanha, 2004) and expressed as log2 Ratios (EtOH/Ctrl). We used FDR ≤ 0.0001 and an absolute value of log2 Ratio ≥ 1 as the threshold to judge the significance of expression difference.
The identification of putative novel miRNA were based on the analysis recommendations of the Beijing Genomics Institute (Shenzhen, China). This institute sequenced the miRNAs and provided an initial analysis. In brief the following criteria were used to identify novel miRNAs. After removing the known miRNAs, RNA sequences with a length of 19 to 26 nucleotides were examined further. Those that matched the sense strand of a mRNA, rRNA, tRNA, snoRNA, or a snRNA gene were filtered out. The remaining RNAs were mapped to the genome and the mirReap algorithm was used to show that a stem-loop structure of the appropriate loop size, without any internal bulges, could be formed by a transcript produced by the genomic locus. The putative hairpin structure of a possible precursor is relatively stable with an energy of hybridization less than or equal to -18 kcal/mol. Each predicted precursor has a 3 n depth for the Drosha/Dicer cutting site. All putative miRNAs also must have an arbitrary cutoff count of no fewer than 5 reads per million. None of the putative novel miRNAs have been functionally tested for miRNA activity. The structure of the putative precursor miRNA is shown in Supplemental Figure 1.
miRNA Target Analysis
Prediction of miRNA targets was performed using TargetScanFly (Ruby et al., 2007b). Gene ontology analysis was conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID) web-accessible tool, version 6.7 (Huang da et al., 2009). For gene ontology annotation search and clustering, significant gene categories in “Biological Process” for each cluster were identified using default High Classification Stringency parameters (Kappa Similarity Term Overlap: 3; Similarity Threshold: 0.85; Initial Group Membership: 3; Final Group Membership: 3; Multiple Linkage Threshold: 0.5) and official gene symbols as input.
Ethanol Resistance Assay
To test for ethanol resistance, all transgenic stocks tested were subjected to the inebriator as described above (Ethanol treatment, RNA Isolation and Next-Generation Sequencing). This time however, six sets of 10 flies from the RU486-treated group and six sets of 10 flies from the control group were placed in plastic vials (one set per vial) and exposed to an ethanol-saturated air stream in parallel, until all flies were sedated (Sedation was determined visually as when flies lose positional control and remain on their backs with only occasional twitching. This typically occurs within 12-15 minutes). After sedation, the ethanol-saturated air stream was replaced with a stream of humidified air and recovery from sedation was monitored over a 90-minute period in 3-minute intervals. Flies that regain postural control were considered recovered. The number of recovered flies was plotted against time to generate a recovery curve. For each vial, the 50% recovery time (RT50) was calculated using Prism 6 (GraphPad Software, Inc.) by nonlinear regression curve fit of the recovery curves to the Richard's five-parameter dose response equation. Statistically significant differences in RT50 between induced and uninduced lines were determined using Student t-tests. Error bars represent standard error of the mean. The magnitude of the difference in ethanol sensitivity between induced and uninduced transgenic lines was calculated using the following formula:
All experiments were performed between 11:00 and 16:00 (zeitgeber time 3-8) using age-matched, ethanol-naïve female flies. We did not observe death of flies after a single sedation (examples – Krishnan et al. 2012, 2016) nor did we observe death in the 14 behavioral resistance assays in Figure 2 (7 with and 7 without RU486).
Figure 2.
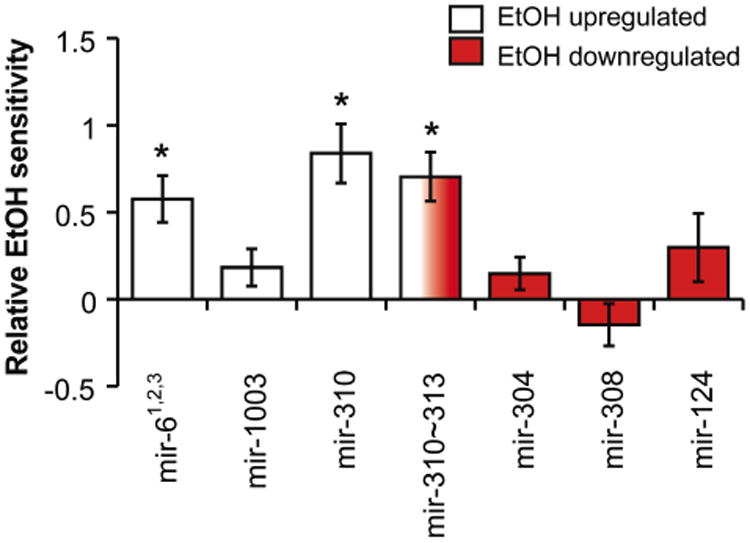
Feeding of RU486 to GeneSwitch-elav/UAS-miRNA transgenic stocks alters the ethanol sensitivity of three out of seven tested miRNA overexpression lines (relative to stock not fed RU486). Shown is the fold difference in ethanol sensitivity between the RU486-fed and not fed stocks calculated using the formula in the Methods section (error bars are SEM, * indicates a P value of < 0.05). miRNAs found to be induced by ethanol are shown by open bars while miRNAs normally suppressed by ethanol are by closed bars. The open/closed bar is a transgene that encodes four miRNAs: miR-310 was induced by ethanol and miR-313 was suppressed by ethanol. mir-311 and mir-312 were not found to be ethanol-responsive. The mir-310∼313 transgene induces all four of these miRNAs. The mir-61,2,3 transgene induces all 3 mir-6 paralogs.
Results
Identification of Ethanol Responsive miRNAs
To examine early ethanol-induced changes in miRNA abundance in the nervous system of Drosophila, we conducted a genome-wide survey of microRNAs from ethanol-treated flies. For this, we extracted total RNA from the heads of two groups of age-matched flies. One group was exposed to a single sedating dose of ethanol vapor, while the second group was left untreated (the untreated group was used for comparison purposes). RNA was extracted 30 minutes after onset of sedation (∼45 minutes from the initiation of the ethanol vapor exposure). At this point, all flies in the treated group were still sedated. miRNAs were isolated from both samples by size selection and sequenced using whole genome RNA sequencing. Differential expression between the two treatment groups was determined to identify ethanol-responsive miRNAs. Analysis of protein coding mRNAs was also performed in parallel, and has been previously reported in Troutwine et al. (2016).
We observed significant changes (P < 0.05, FDR < 0.005, log2 ratio ≥ 1) in multiple miRNAs and mRNAs. Out of 466 miRNAs annotated in the Drosophila genome (miRBase21.0, BDGP5.0), acute alcohol exposure induced abundance changes in only fourteen miRNAs. Of these, 7 miRNAs were upregulated, and 7 were downregulated by ethanol (Figure 1). In addition, we identified 13 putative novel miRNA species that were also alcohol responsive (5 upregulated, 8 downregulated). All previously annotated miRNAs with significant ethanol-induced changes in expression are listed in Table 1. In Troutwine et al. (2016), we also identified 137 protein-coding transcripts that respond to the ethanol treatment. Of these, 89 increased whereas 48 decreased in abundance after the ethanol treatment. Between miRNAs and mRNAs, a total of 166 transcripts were affected.
Figure 1.
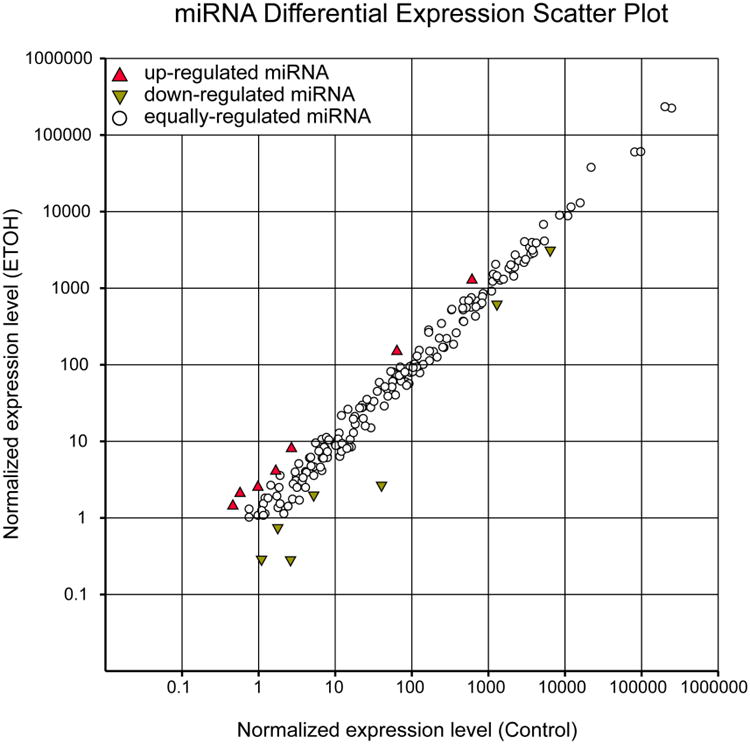
Scatter plot showing a comparison of the relative abundance of the 426 known Drosophila miRNAs. The comparison is between miRNAs isolated from heads 30 minutes post ethanol sedation and miRNAs isolated from non-ethanol treated matched controls. The normalized count is reads per million [(actual count/total reads)*106]. Open circles represent miRNAs whose abundance did not differ between the two treatments. Upward-pointing triangles represent miRNAs whose abundance increases while downward-pointing triangles represent miRNAs whose abundance decreases at least two fold. Values for the relative abundances of these miRNAs are found in Supplementary Data 1.
Table 1. Known miRNAs with significant changes after ethanol exposure.
| miRNA | Δ EtOH | Fold-change | P-value | Sequence | Guide miRNA | Behavioral EtOH test |
|---|---|---|---|---|---|---|
| miR-6-3p | Up | 1.87 | 7.65E-05 | UAUCACAGUGGCUGUUCUUUUU | Yes † | Sensitive |
| miR-310-3p | Up | 1.62 | 3.38E-03 | UAUUGCACACUUCCCGGCCUUU | Yes | Sensitive |
| miR-304-5p | Up | 1.56 | 5.66E-12 | UAAUCUCAAUUUGUAAAUGUGAG | Yes | No change |
| miR-1009-3p | Up | 1.35 | 6.18E-04 | UCUCAAAAAUUGUUACAUUUCAG | Yes | |
| miR-1008-3p | Up | 1.27 | 3.12E-05 | UCACAGCUUUUUGUGUUUACA | Yes | |
| miR-1003-3p | Up | 1.24 | 3.11E-139 | UCUCACAUUUACAUAUUCACAG | Yes | No change |
| miR-193-3p | Up | 1.08 | 0.00E+00 | UACUGGCCUACUAAGUCCCAAC | Yes | |
| miR-124-3p | Down | -1.03 | 0.00E+00 | UAAGGCACGCGGUGAAUGCCAAG | Yes | No change |
| miR-277-5p | Down | -1.07 | 0.00E+00 | CGUGUCAGGAGUGCAUUUGCA | No | |
| miR-313-5p | Down | -1.28 | 5.67E-03 | UGCUGCGGAUGGGGGCAGUACU | No | No change |
| miR-11-5p | Down | -1.40 | 2.80E-07 | CAAGAACUUUCUCUGUGACCCG | No | |
| miR-13b-1-5p | Down | -1.95 | 3.62E-03 | UCGUUAAAAUGUUUGUGAACUUAUG | No | |
| miR-4976-5p | Down | -3.19 | 1.70E-09 | UCCAGGCUGUCGUCCAUGGAGU | Yes | |
| miR-308-3p | Down | -3.94 | 8.93E-153 | AAUCACAGGAUUAUACUGUGAG | Yes † | No change |
Ethanol-induced changes are shown relative to control. Fold change is expressed as log2 values (EtOH/Ctrl). The underlined region in each sequence designates the seed region.
denotes miRNAs that share the same seed sequence. Information of whether a miRNA is known to serve as a guide miRNA is included. The effects of miRNA overexpression on alcohol sensitivity are also reported for all miRNAs tested in the present study. The relative abundance of each miRNA, measured as normalized reads per million before and after ethanol treatment, can be found in the Supplementary Data 1.
Of the fourteen annotated ethanol-responsive miRNAs, four belong to a family that share the same seed and therefore recognize the same mRNA targets. These are miR-6-3p, miR-11-5p, miR13b-1-5p, and miR-308-3p, which belong to the miR-2a-1/6/11/13/308 family. Members of this family share a common seven nucleotide seed sequence: 5′-AUCACAG-3′ (Aravin et al., 2003). However, the mature miRNAs produced from the 5′ arm of the miR-11 and miR-13b-1 identified in our study are very likely the passenger strand and are destined for degradation as they do not carry the seed sequence (Aravin et al., 2003; Ruby et al., 2007b; Stark et al., 2007). We observed that forty-five minutes after the onset of a sedating dose of alcohol vapor that miR-6-3p was increased in abundance but that the abundance of miR-308-3p was reduced. While this data may seem contradictory at first, it is possible that these two miRNAs reflect an ethanol response in different parts of the brain, and thus do not necessarily negate each other's abundance change. The miR-6-3p miRNA is a Drosophila-specific miRNA and is transcribed as a precursor containing the miRNAs mir-6-1, mir-6-2, and mir-6-3. The mature products generated from the 3′ arm of each mir-6 precursor hairpin have identical sequences, and have been experimentally validated to act as guide miRNAs. The mature miR-6 miRNAs have been shown to target apoptotic genes such as hid, grim and rpr (Lagos-Quintana et al., 2001; Ruby et al., 2007b; Sempere et al., 2003; Stark et al., 2007). Similarly, the 3′ mature product of mir-308 is a validated guide miRNA with the same targets.
A second well-conserved miRNA family represented in our ethanol-responsive data set is the miR-92/310/311/312/313 family. We observed that both miR-310-3p and miR-313-5p are affected by acute alcohol exposure. miR-310-3p is upregulated by ethanol whereas miR-313-5p is downregulated. In this case, only miR-310-3p has been validated as a guide miRNA, while miR-313-5p according to miRBase analysis is most likely to serve as a passenger strand with an unknown role (Kozomara & Griffiths-Jones, 2014).
With the exception of miR-277-5p, all the other ethanol-responsive miRNAs have been validated as guide miRNAs (Aravin et al., 2003; Berezikov et al., 2011; Lai et al., 2003; Ruby et al., 2007b; Stark et al., 2003; Stark et al., 2007). The miR-124 miRNA is well conserved and has been identified in flies, worms, mice and humans. The mature miR-124-3 originates from the 3′ arm of the precursor hairpin and is perhaps the most abundant miRNA expressed in neuronal cells. It is believed to participate in neuronal differentiation (Sun et al., 2012; Wang et al., 2014; Weng & Cohen, 2012). miR-1003-3p, miR-1009-3p and miR-1008-3p miRNAs, which are generated from intronic microRNA precursors that bypass Drosha processing, are not conserved in mammals (Ruby et al., 2007a).
In our genome-wide survey for alcohol-responsive miRNAs we also detected new microRNAs that were not reported before in the fruit flies. A temporary name was assigned to each of the putative novel miRNAs. The sequence, fold change, and chromosomal locations of these miRNAs are reported in Table 2.
Table 2. Putative novel miRNAs with significant changes after ethanol exposure.
| miRNA | Δ EtOH | Fold-change | P-value | Sequence | Chromosomal location |
|---|---|---|---|---|---|
| novel_miR_1 | Up | 10.92 | 2.93E-102 | GCGUCGGUGGUGUAAUGGUUAG | 3R 2955586:2955607 |
| novel_miR_2 | Up | 8.09 | 5.14E-15 | GUGGGCGGUGGAGAGGUGGAG | 3L 935306:935326 |
| novel_miR_3 | Up | 8.09 | 5.14E-15 | GUGGGUGGGGUGGACGGGCA | X 9176644:9176663 |
| novel_miR_4 | Up | 7.51 | 2.98E-10 | UCGGUCCACCCGGGGGCGCCA | 2L 1856619:1856599 / 2R 9632238:9632218 |
| novel_miR_5 | Up | 7.09 | 7.19E-08 | UUGGGGGGCGCGGGUGGUGG | X 9057913:9057893 |
| novel_miR_6 | Down | -1.02 | 6.81E-03 | AGAGCGACGAAGACUGAGCUG | 3L 14420650:14420630 |
| novel_miR_7 | Down | -6.78 | 1.64E-06 | CCGGGUGUGUGUGGAUGGGUA | 3R 21410603:21410623 |
| novel_miR_8 | Down | -7.11 | 4.93E-08 | UUGGGGGGCGCGGGUGGUGGC | X 9057913:9057893 |
| novel_miR_9 | Down | -7.17 | 2.45E-08 | UGUGGGCAGGCGAAUGAUGUGG | 2L 4458246:4458225 |
| novel_miR_10 | Down | -7.34 | 2.99E-09 | GUUGAACGGUCGUUGAACAAU | X 13635866:13635886 |
| novel_miR_11 | Down | -7.43 | 7.36E-10 | GCGGGUCGGAGUGGGUGGUGG | 2L 16283135:16283115 |
| novel_miR_12 | Down | -9.05 | 9.98E-29 | UUGGGCGGGGACGGGCGGCGG | 3R 3300451:3300431 |
| novel_miR_13 | Down | -10.82 | 5.54E-96 | GUGGGAGCGGUGGGCGGAGCG | 3L 4576278:4576298 |
Ethanol-induced changes are shown relative to control. Fold change is expressed as log2 values (EtOH/Ctrl). The relative abundance of each miRNA, measured as normalized reads per million before and after ethanol treatment, can be found in the Supplementary Data 1. All of these match criteria for a miRNA (see Methods) but none have been functionally tested to confirm that functionally act as a miRNA.
Functional Testing of miRNAs on alcohol sensitivity
To test the effects of ethanol-responsive miRNAs on behavioral ethanol sensitivity, we overexpressed six miRNAs for which overexpression lines were available (mir-6, mir-124, mir-304, mir-308, mir-310 and mir-1003). These overexpression lines carry UAS-controlled transgenes encoding the specific precursor miRNAs (Bejarano et al., 2012). To specifically drive expression of alcohol-responsive miRNAs in the Drosophila nervous system, we used the conditional tissue-specific transgene expression system called GeneSwitch (Osterwalder et al., 2001; Roman et al., 2001). This system makes use of a fusion of Gal4 and progesterone receptor domains to generate a transcription factor that activates the miRNA-expressing UAS transgene only in the presence of RU486 (a.k.a. mifepristone). Additionally, because we use the neuronal-specific elav-Switch driver of the Gal4-fusion transgene, expression of miRNAs is specific to the nervous system. Of the six tested miRNAs, miR-6 was available as a transgenic cluster of mir-6-1, mir-6-2 and mir-6-3. Thus, when transgene expression was activated, all 3 mir-6 paralogs were expressed together. These three paralogs contain identical mature miRNA sequences. On the other hand, miR-1003, miR-310, miR-304, miR-308, and miR-124, were all tested independently using transgenic lines individually expressing each precursor sequence. Additionally, miR-310 was also tested a second time as a cluster of four different miRNA precursor sequences (mir-310, mir-311, mir-312 and mir-313), all of which share the same seed sequence. While miR-313 is also represented in this cluster, only the passenger strand from this pre-miRNA was found to be alcohol-responsive.
All overexpression lines were induced by feeding RU486 to the transgenic flies for 3 consecutive days, before testing for their effect on alcohol sensitivity using a behavioral assay. In this assay, flies were exposed to a sedating dose of alcohol vapor until knockdown, after which the ethanol was switched to fresh air. At this point, recovery from ethanol was monitored to calculate the 50% recovery time (RT50). The RT50 of miRNA-induced flies was compared to that of their uninduced counterparts. The uninduced flies were fed food without RU486 but were otherwise treated identically. Also, because these flies share the same genetic background, a direct comparison was possible. We observed that the overexpression of the miR-6 cluster as well as the mir-310 precursor — either individually or as a cluster — significantly increased ethanol sensitivity. These two miRNAs were both induced by ethanol. However, miR-1003 — also induced by ethanol — or the ethanol-down regulated miR-304, miR-308, and miR-124, had no effect on ethanol sensitivity (Figure 2).
Gene Ontology Analysis of Predicted Targets
Predicted targets for miR-6 and miR-310 were obtained using the TargetScanFly (release 6.2) web-based algorithm (Ruby et al., 2007b). To categorize the predicted targets of each of the identified miRNA groups according to their Gene Ontology annotations, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) web-accessible tool, version 6.7 (Huang da et al., 2009). This model categorizes a set of genes based on their functional annotation and assigns each category a score that corresponds to the significance of enrichment. For miR-6, TargetScanFly predicts a total of 377 targets, and DAVID functional annotation results in a total of 34 gene ontology clusters. The first term of the top ten clusters of miR-6 targets is displayed in Table 3. A significant enrichment in genes involved in organ morphogenesis, including neuronal morphogenesis was found. This is consistent with previous reports for this miRNA. For miR-310, TargetScanFly predicts a total of 349 targets and DAVID functional annotation resulted in a total of 35 gene ontology clusters. The first term of the top ten clusters of miR-310 targets is displayed in Table 4. Again, a significant enrichment in genes involved in organ morphogenesis, including neuronal morphogenesis was found. A list of all target genes for miR-6 and miR-310, and a list of the complete GO terms clusters for miR-6 and miR-310, can be found in the supplementary information (Supplementary Data 2 and Supplementary Data 3, respectively).
Table 3. Gene ontology annotations for miR-6 targets.
| Annotation cluster | Enrichment score | GO Term: Biological Function | Gene Count | % of total | P-Value | FE | BS | FDR |
|---|---|---|---|---|---|---|---|---|
| 1 | 3.84 | instar larval or pupal morphogenesis | 23 | 6.42 | 0.00 | 2.62 | 0.00 | 0.10 |
| 2 | 3.63 | neuron projection morphogenesis | 20 | 5.59 | 0.00 | 2.76 | 0.00 | 0.17 |
| 3 | 3.46 | post-embryonic organ morphogenesis | 19 | 5.31 | 0.00 | 2.64 | 0.01 | 0.47 |
| 4 | 2.93 | imaginal disc-derived appendage morphogenesis | 16 | 4.47 | 0.00 | 2.64 | 0.02 | 1.66 |
| 5 | 2.39 | imaginal disc-derived wing morphogenesis | 14 | 3.91 | 0.00 | 2.54 | 0.05 | 5.43 |
| 6 | 1.66 | negative regulation of transcription | 12 | 3.35 | 0.02 | 2.29 | 0.16 | 22.50 |
| 7 | 1.54 | negative regulation of macromolecule biosynthetic process | 13 | 3.63 | 0.02 | 2.09 | 0.21 | 30.00 |
| 8 | 1.50 | leg segmentation | 4 | 1.12 | 0.01 | 9.29 | 0.10 | 12.63 |
| 9 | 1.37 | response to other organism | 9 | 2.51 | 0.02 | 2.73 | 0.17 | 24.13 |
| 10 | 1.35 | cell migration | 10 | 2.79 | 0.03 | 2.26 | 0.26 | 41.86 |
Significant biological function categories were clustered based on enrichment scores using DAVID. The top term in each of the top-10 cluster is listed. The ‘Gene Count’ column displays number of genes in each cluster associated with a particular GO term, as well as the % of the total number of genes in the cluster is shown. P-value, Fold Enrichment (FE), Benjamini Score (BS) and False Discovery Rate (FDR) are also shown.
Table 4. Gene ontology annotations for miR-310 targets.
| Annotation cluster | Enrichment score | GO Term: Biological Function | Gene Count | % of total | P-Value | FE | BS | FDR |
|---|---|---|---|---|---|---|---|---|
| 1 | 6.06 | morphogenesis of an epithelium | 21 | 6.33 | 0.00 | 3.86 | 0.00 | 0.00 |
| 2 | 4.69 | maginal disc-derived appendage morphogenesis | 19 | 5.72 | 0.00 | 3.29 | 0.00 | 0.03 |
| 3 | 3.43 | branching involved in open tracheal system development | 7 | 2.11 | 0.00 | 7.43 | 0.01 | 0.50 |
| 4 | 2.90 | imaginal disc-derived wing hair organization | 6 | 1.81 | 0.00 | 7.52 | 0.02 | 1.72 |
| 5 | 2.41 | neuron projection morphogenesis | 16 | 4.82 | 0.00 | 2.32 | 0.05 | 5.99 |
| 6 | 2.32 | positive regulation of cellular biosynthetic process | 11 | 3.31 | 0.00 | 2.98 | 0.04 | 6.02 |
| 7 | 2.15 | muscle cell development | 6 | 1.81 | 0.00 | 5.52 | 0.05 | 6.82 |
| 8 | 2.11 | positive regulation of transcription | 9 | 2.71 | 0.01 | 3.21 | 0.07 | 10.54 |
| 9 | 1.73 | establishment of planar polarity | 6 | 1.81 | 0.02 | 4.07 | 0.13 | 22.45 |
| 10 | 1.46 | epithelial cell migration | 4 | 1.20 | 0.03 | 5.71 | 0.22 | 41.09 |
Significant biological function categories were clustered based on enrichment scores using DAVID. The top term in each of the top-10 cluster is listed. The ‘Gene Count’ column displays number of genes in each cluster associated with a particular GO term. Percentage of the total number of genes in the cluster, P-value, Fold Enrichment (FE), Benjamini Score (BS) and False Discovery Rate (FDR) are also shown.
It is important to note that the sets of putative targets for miR-6 and miR-310 show a significant overlap. We found a group of 30 genes in common between the two datasets, 26 of which are neurally-expressed genes. A list of these genes can be found in Supplementary Data 2. When subjected to Gene Ontology Clustering analysis, this group of genes showed significant enrichment in transcription regulator activity, with 8 genes showing a direct association with the GO term (Enrichment Score: 2.63, 30%, P-Value 3.69E-05). These genes are: salm, caup, en, nub, Eip93F, aop, nab, kay. From the overlapping set of putative miRNA targets, none of them overlap with changes in mRNA abundance noted in Troutwine et al. (2016). However, this does not mean very much because 1) miRNAs can alter translatability of a mRNA before mRNA destabilization occurs, and 2) any destabilization wrought by a miRNA will take time to occur and both of these samples (Troutwine's mRNA and Ghezzi's miRNA) were taken at the same time point.
Discussion
It is generally held that changes in gene expression in the brain contribute to alcoholism. Alcohol-induced changes in miRNA expression are believed to be responsible for many changes in gene expression that underlie alcohol-response adaptations. While some of these changes have been documented in mammals, it is currently expensive and time consuming to determine the functional consequence of each change in miRNA expression in a mammalian model system. Because all mammalian miRNA families are represented in Drosophila, experiments in flies can be highly informative and applicable to the biology of higher organisms, including humans (Ibanez-Ventoso et al., 2008). Here we investigate the consequences of alcohol-induced changes in miRNA expression in the genetically tractable Drosophila melanogaster model system. We have shown that 30 minutes after ethanol sedation there are significant changes in miRNA abundance in the fly head. The sedation protocol we use has been shown to also produce significant long-term changes in alcohol related behaviors and physiological alterations. This single ethanol exposure results in functional behavioral tolerance that is detectable 4 hrs after sedation and persists for 7–10 days (Cowmeadow et al., 2005; Krishnan et al., 2016). Similarly, symptoms of ethanol withdrawal to the same dose of ethanol are detectable 24 hours after exposure and also persist for 7–10 days (Ghezzi et al., 2014). At this early 30-minute post-sedation time point we have observed changes in 14 previously identified miRNAs as well as changes in 13 novel miRNAs (27 total). While this is perhaps only a snapshot of all changes induced by ethanol sedation, we hypothesize that the immediate-early changes in miRNA abundance detected here are an important part of gene expression changes that modulate future alcohol responses. At this time, we cannot formally distinguish which of these changes are the product of ethanol exposure, ethanol sedation, or just sedation.
Among the ethanol responsive miRNAs, we identified four mature miRNA*s that have not been previously considered to serve as guide miRNAs. These are presumed to be inactive complementary passenger strands. This is intriguing because miRNA* strands are usually degraded. The fact that we observed significant accumulation changes in these miRNA*s suggest that they have regulatory potential. Evidence for functional miRNA* strands exists. In Drosophila for instance, certain miRNA* strands have been found to associate with Argonaute2 (Ago2) but not with Argonaute1 (Ago1) (Ghildiyal et al., 2010). Ago1 is the central protein component of the microRNA (miRNA) pathway, while Ago2 is central to the RNA interference pathway (siRNAs). The evidence suggests that in some instances that miRNA* strands can be recycled into the RNA interference pathway thereby increasing the complexity of regulatory networks (Okamura et al., 2008).
A key aspect of this work is that in Drosophila, alcohol-responsive miRNAs can be functionally tested to evaluate their contribution to ethanol sensitivity. Furthermore, because in this study each miRNA was expressed using a conditional tissue-specific transgene expression system, we limited the analysis to the adult nervous system. This is important because by activating the transgenes at a specific developmental stage, we could avoid occluding developmental effects. Using this system, we identified two conserved Drosophila miRNAs with a significant role in modulating ethanol sensitivity: miR-6 and miR-310. Both miRNAs were upregulated by ethanol. None of the ethanol-downregulated miRNAs had a significant effect on alcohol sensitivity. Our evidence, so far, indicates that the downregulated miRNAs do not play a role in modulating alcohol sensitivity, however, it is possible that overexpression is not effective for functionally validating the role of downregulated miRNAs. Future studies should explore this issue further by transgenically inhibiting microRNA activity. Efforts in creating collections of transgenic miRNA sponge lines should soon provide the tools needed for this purpose (Kluiver et al., 2012; Loya et al., 2009). miR-6 and miR-310 are well-studied miRNAs in Drosophila. Both have been associated with neural morphogenesis during development. Mutations in miR-6 have been shown to cause defects in the central nervous system and embryonic lethality (Ge et al., 2012). These effects have been directly linked to the regulation of the proapoptotic genes, reaper (rpr), head involution defective (hid), grim and sickle (skl). Upregulation of these genes results in elevated apoptosis within the central nervous system in the mutants. On the other hand, miR-310 has been associated with the proliferation and differentiation of early germ and somatic progenitor cells through the regulation of the Wingless pathway members arm and pangolin (Pancratov et al., 2013). mir-310 was also shown to participate in the regulation of synaptic transmission at the Drosophila larval neuromuscular junction (Tsurudome et al., 2010). Perhaps most significant with regards to the effects of alcohol, loss of the miR-310-313 cluster has been shown to lead to a significant enhancement of neurotransmitter release, as well as a significant increase in the active zone protein Bruchpilot (encoded by brp). Interestingly, brp was shown to be upregulated by alcohol and to participate in alcohol tolerance in flies (Ghezzi et al., 2013). More recently, miR-310 was also shown to orchestrate Hedgehog signaling strength in response to diet, implicating miRNA-directed silencing as an important regulator of Drosophila metabolic state (Çiçek et al., 2016). These connections between miR-310 and synaptic activity and its response to changes in diet supports our interpretation that miR-310 plays an important role in the response to alcohol.
Among the gene targets of miR-6 and miR-310, we have identified a series of interesting candidates. These include genes with known links to alcohol related behaviors. Potential miR-310 targets include: Arfip, a dynactin binding protein associated with positive regulation of synaptic growth at neuromuscular junction recently shown to increase ethanol sensitivity when mutated (Chang et al., 2013; Peru et al., 2012); CASK, a calcium/calmodulin-dependent protein kinase associated with the regulation of neurotransmitter secretion and reduced ethanol tolerance when mutated (Lloyd et al., 2000; Maiya et al., 2012); Toll, a transmembrane receptor associated with activation of the innate immune response —activation of Toll has been recently linked to increased ethanol resistance (Tauszig et al., 2000; Troutwine et al., 2016); kn, a transcription factor also associated with the innate immune response and the development of ethanol tolerance (Ghezzi et al., 2013); and tao, a protein serine/threonine kinase involved in promoting axon guidance and the ethanol-induced hyperactivity response (King et al., 2011; King & Heberlein, 2011). Meanwhile, miR-6 can potentially regulate: AcCoAS, an acetyl-coenzyme A synthetase associated with many biochemical pathways, including those for ethanol metabolism, and is thought to contribute to ethanol-induced hyperactivity (Kong et al., 2010); rho, a serine-type endopeptidase associated with axogenesis and also linked to increased ethanol sensitivity when mutant (Berger et al., 2008; Grueber et al., 2007); and Teh2, putative sodium channel and BK channel auxiliary subunit associated with the development of ethanol tolerance (Derst et al., 2006; Ghezzi et al., 2013).
Additionally, we also detected a series of immune response genes (imd, PGRP-LC, Tollo, upd3, crq), ion channels alpha and beta subunit genes (Ca-alpha1T, SK, Slob, Teh3, Ih, SKIP, Best2), synaptic genes (Syt12, Prosap, Dscam3), as well as transcription factor and co-factor genes (CrebA, kay, Sirt2, Sin3A). While these genes have not been directly linked the alcohol responses, they fall within important gene categories associated with the behavioral responses to alcohol. Among the immune genes, PGRP-LC and imd are particularly interesting candidates, as both belong to the innate immune IMD pathway that results in activation of the Drosophila NFkB homolog Rel. In a recent study we showed that overexpression of the active form of Rel increases resistance to ethanol (Troutwine et al., 2016). In the ion channel category, the gene Slob is another salient example. Slob encodes a well-known modulatory subunit of the BK channel gene slo (Shahidullah et al., 2009). In both flies and mammals the BK channel has been strongly linked to alcohol tolerance (for review, see: Ghezzi & Atkinson, 2011), and modulation of its activity by Slob can have a strong impact on alcohol sensitivity. Finally the transcriptional activator encoded by kay (kayak) is the Drosophila homolog of AP1/c-FOS in mammals (Perkins et al., 1990). In mammals, alcohol has been shown to selectively attenuate stress-induced c-fos expression (Ryabinin et al., 1995), and has also been implicated in the persistent changes underlying cocaine addiction (Larson et al., 2010; Xu, 2008). It is thus not surprising that collectively, modulation of these genes by miRNAs can severely affect the neural responses to alcohol.
A complicating factor for work of this kind is that the behaviors of interest might be the product of more than one miRNA. Furthermore, multiple miRNAs can regulate partially overlapping sets of mRNA and might all be necessary to see an effect. Furthermore, because miRNA functionality does not require strict sequence complementarity to the mRNA target for its regulation, our prediction of targets may have misidentified or not identified some targets. Although, an individualistic approach to miRNA analysis may not completely recapitulate the natural response, nonetheless this approach has already identified alcohol-responsive miRNAs that share a common biological output — the regulation of neural morphogenesis. Furthermore, we have connected these miRNAs to new interesting gene candidates with potential roles in ethanol neuroadaptation.
Supplementary Material
Acknowledgments
This study was funded by NIH grant R21AA023372-01A1 to NSA.
References
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Bala S, Szabo G. MicroRNA Signature in Alcoholic Liver Disease. Int J Hepatol. 2012;2012:498232. doi: 10.1155/2012/498232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F, Bortolamiol-Becet D, Dai Q, Sun K, Saj A, Chou YT, Raleigh DR, Kim K, Ni JQ, Duan H, Yang JS, Fulga TA, Van Vactor D, Perrimon N, Lai EC. A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development. 2012;139:2821–2831. doi: 10.1242/dev.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, Zhao Y, Zamore PD, Hannon GJ, Marra MA, Weng Z, Perrimon N, Lai EC. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Kong EC, Dubnau J, Tully T, Moore MS, Heberlein U. Ethanol sensitivity and tolerance in long-term memory mutants of Drosophila melanogaster. Alcohol Clin Exp Res. 2008;32:895–908. doi: 10.1111/j.1530-0277.2008.00659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Chang L, Kreko T, Davison H, Cusmano T, Wu Y, Rothenfluh A, Eaton BA. Normal dynactin complex function during synapse growth in Drosophila requires membrane binding by Arfaptin. Mol Biol Cell. 2013;24:1749–64. S1. doi: 10.1091/mbc.E12-09-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çiçek IÖ, Karaca S, Brankatschk M, Eaton S, Urlaub H, Shcherbata HR. Hedgehog Signaling Strength Is Orchestrated by the mir-310 Cluster of MicroRNAs in Response to Diet. Genetics. 2016;202:1167–1183. doi: 10.1534/genetics.115.185371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowmeadow RB, Krishnan HR, Atkinson NS. The slowpoke gene is necessary for rapid ethanol tolerance in Drosophila. Alcohol Clin Exp Res. 2005;29:1777–1786. doi: 10.1097/01.alc.0000183232.56788.62. [DOI] [PubMed] [Google Scholar]
- Cowmeadow RB, Krishnan HR, Ghezzi A, Al'Hasan YM, Wang YZ, Atkinson NS. Ethanol tolerance caused by slowpoke induction in Drosophila. Alcohol Clin Exp Res. 2006;30:745–753. doi: 10.1111/j.1530-0277.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- Derst C, Walther C, Veh RW, Wicher D, Heinemann SH. Four novel sequences in Drosophila melanogaster homologous to the auxiliary Para sodium channel subunit TipE. Biochem Biophys Res Commun. 2006;339:939–948. doi: 10.1016/j.bbrc.2005.11.096. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Rajewsky N. The impact of miRNA target sites in coding sequences and in 3′UTRs. PLoS One. 2011;6:e18067. doi: 10.1371/journal.pone.0018067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Chen YW, Weng R, Lim SF, Buescher M, Zhang R, Cohen SM. Overlapping functions of microRNAs in control of apoptosis during Drosophila embryogenesis. Cell Death Differ. 2012;19:839–846. doi: 10.1038/cdd.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A, Atkinson NS. Homeostatic control of neural activity: a Drosophila model for drug tolerance and dependence. Int Rev Neurobiol. 2011;99:23–50. doi: 10.1016/B978-0-12-387003-2.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A, Krishnan HR, Atkinson NS. Susceptibility to ethanol withdrawal seizures is produced by BK channel gene expression. Addict Biol. 2014;19:332–337. doi: 10.1111/j.1369-1600.2012.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A, Krishnan HR, Lew L, Prado FJ, Ong DS, Atkinson NS. Alcohol-induced histone acetylation reveals a gene network involved in alcohol tolerance. PLoS Genet. 2013;9:e1003986. doi: 10.1371/journal.pgen.1003986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Yang CH, Younger S, Borden K, Jan LY, Jan YN. Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development. 2007;134:55–64. doi: 10.1242/dev.02666. [DOI] [PubMed] [Google Scholar]
- Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425:3582–3600. doi: 10.1016/j.jmb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ibanez-Ventoso C, Vora M, Driscoll M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS One. 2008;3:e2818. doi: 10.1371/journal.pone.0002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I, Heberlein U. Tao kinases as coordinators of actin and microtubule dynamics in developing neurons. Commun Integr Biol. 2011;4:554–556. doi: 10.4161/cib.4.5.16051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I, Tsai LT, Pflanz R, Voigt A, Lee S, Jackle H, Lu B, Heberlein U. Drosophila tao controls mushroom body development and ethanol-stimulated behavior through par-1. J Neurosci. 2011;31:1139–1148. doi: 10.1523/JNEUROSCI.4416-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver J, Gibcus JH, Hettinga C, Adema A, Richter MK, Halsema N, Slezak-Prochazka I, Ding Y, Kroesen BJ, van den Berg A. Rapid generation of microRNA sponges for microRNA inhibition. PLoS One. 2012;7:e29275. doi: 10.1371/journal.pone.0029275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong EC, Allouche L, Chapot PA, Vranizan K, Moore MS, Heberlein U, Wolf FW. Ethanol-regulated genes that contribute to ethanol sensitivity and rapid tolerance in Drosophila. Alcohol Clin Exp Res. 2010;34:302–316. doi: 10.1111/j.1530-0277.2009.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HR, Al-Hasan YM, Pohl JB, Ghezzi A, Atkinson NS. A role for dynamin in triggering ethanol tolerance. Alcohol Clin Exp Res. 2012;36:24–34. doi: 10.1111/j.1530-0277.2011.01587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HR, Li X, Ghezzi A, Atkinson NS. A DNA element in the slo gene modulates ethanol tolerance. Alcohol. 2016;51:37–42. doi: 10.1016/j.alcohol.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai EC. Predicting and validating microRNA targets. Genome Biol. 2004;5:115. doi: 10.1186/gb-2004-5-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EB, Akkentli F, Edwards S, Graham DL, Simmons DL, Alibhai IN, Nestler EJ, Self DW. Striatal regulation of ΔFosB, FosB, and cFos during cocaine self-administration and withdrawal. J Neurochem. 2010;115:112–122. doi: 10.1111/j.1471-4159.2010.06907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu X, Qin S, Guan Y, Liu Y, Cheng Y, Chen X, Li W, Wang S, Xiong M, Kuzhikandathil EV, Ye JH, Zhang C. MicroRNA expression profile and functional analysis reveal that miR-382 is a critical novel gene of alcohol addiction. EMBO Mol Med. 2013;5:1402–1414. doi: 10.1002/emmm.201201900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Lloyd TE, Verstreken P, Ostrin EJ, Phillippi A, Lichtarge O, Bellen HJ. A genome-wide search for synaptic vesicle cycle proteins in Drosophila. Neuron. 2000;26:45–50. doi: 10.1016/s0896-6273(00)81136-8. [DOI] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiya R, Lee S, Berger KH, Kong EC, Slawson JB, Griffith LC, Takamiya K, Huganir RL, Margolis B, Heberlein U. DlgS97/SAP97, a neuronal isoform of discs large, regulates ethanol tolerance. PLoS One. 2012;7:e48967. doi: 10.1371/journal.pone.0048967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Smith EM, Bushell M. Regulation of miRNA strand selection: follow the leader. Biochem Soc Trans. 2014;42:1135–1140. doi: 10.1042/BST20140142. [DOI] [PubMed] [Google Scholar]
- Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA, Mayfield RD. Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC Genomics. 2013;14:725. doi: 10.1186/1471-2164-14-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancratov R, Peng F, Smibert P, Yang S, Olson ER, Guha-Gilford C, Kapoor AJ, Liang FX, Lai EC, Flaherty MS, DasgGupta R. The miR-310/13 cluster antagonizes β-catenin function in the regulation of germ and somatic cell differentiation in the Drosophila testis. Development. 2013;140:2904–2916. doi: 10.1242/dev.092817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins KK, Admon A, Patel N, Tjian R. The Drosophila Fos-related AP-1 protein is a developmentally regulated transcription factor. Genes Dev. 1990;4:822–834. doi: 10.1101/gad.4.5.822. [DOI] [PubMed] [Google Scholar]
- Peru YCDPRL, Acevedo SF, Rodan AR, Chang LY, Eaton BA, Rothenfluh A. Adult neuronal Arf6 controls ethanol-induced behavior with Arfaptin downstream of Rac1 and RhoGAP18B. J Neurosci. 2012;32:17706–17713. doi: 10.1523/JNEUROSCI.1944-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59:274–287. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007a;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007b;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabinin AE, Melia KR, Cole M, Bloom FE, Wilson MC. Alcohol selectively attenuates stress-induced c-fos expression in rat hippocampus. J Neurosci. 1995;15:721–730. doi: 10.1523/JNEUROSCI.15-01-00721.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Shahidullah M, Reddy S, Fei H, Levitan IB. In vivo role of a potassium channel-binding protein in regulating neuronal excitability and behavior. J Neurosci. 2009;29:13328–13337. doi: 10.1523/JNEUROSCI.3024-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Westholm JO, Tsurudome K, Hagen JW, Lu Y, Kohwi M, Betel D, Gao FB, Haghighi AP, Doe CQ, Lai EC. Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS Genet. 2012;8:e1002515. doi: 10.1371/journal.pgen.1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapocik JD, Solomon M, Flanigan M, Meinhardt M, Barbier E, Schank JR, Schwandt M, Sommer WH, Heilig M. Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. Pharmacogenomics J. 2013;13:286–296. doi: 10.1038/tpj.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci U S A. 2000;97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troutwine BR, Ghezzi A, Pietrzykowski AZ, Atkinson NS. Alcohol resistance in Drosophila is modulated by the Toll innate immune pathway. Genes Brain Behav. 2016;15:382–394. doi: 10.1111/gbb.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurudome K, Tsang K, Liao EH, Ball R, Penney J, Yang JS, Elazzouzi F, He T, Chishti A, Lnenicka G, Lai EC, Haghighi AP. The Drosophila miR-310 cluster negatively regulates synaptic strength at the neuromuscular junction. Neuron. 2010;68:879–893. doi: 10.1016/j.neuron.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Feng T, Wan Q, Kong Y, Yuan L. miR-124 controls Drosophila behavior and is required for neural development. Int J Dev Neurosci. 2014;38:105–112. doi: 10.1016/j.ijdevneu.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Weng R, Cohen SM. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development. 2012;139:1427–1434. doi: 10.1242/dev.075143. [DOI] [PubMed] [Google Scholar]
- Xu M. c-Fos is an intracellular regulator of cocaine-induced long-term changes. Ann N Y Acad Sci. 2008;1139:1–9. doi: 10.1196/annals.1432.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


