Abstract
The human fungal pathogen Candida albicans switches from a budding yeast form to a polarized hyphal form in response to various external signals. This morphogenetic switching has been implicated in the development of pathogenicity. We have cloned the CaCDC35 gene encoding C. albicans adenylyl cyclase by functional complementation of the conditional growth defect of Saccharomyces cerevisiae cells with mutations in Ras1p and Ras2p. It has previously been shown that these Ras homologues regulate adenylyl cyclase in yeast. The C. albicans adenylyl cyclase is highly homologous to other fungal adenylyl cyclases but has less sequence similarity with the mammalian enzymes. C. albicans cells deleted for both alleles of CaCDC35 had no detectable cAMP levels, suggesting that this gene encodes the only adenylyl cyclase in C. albicans. The homozygous mutant cells were viable but grew more slowly than wild-type cells and were unable to switch from the yeast to the hyphal form under all environmental conditions that we analyzed in vitro. Moreover, this morphogenetic switch was completely blocked in mutant cells undergoing phagocytosis by macrophages. However, morphogenetic switching was restored by exogenous cAMP. On the basis of epistasis experiments, we propose that CaCdc35p acts downstream of the Ras homologue CaRas1p. These epistasis experiments also suggest that the putative transcription factor Efg1p and components of the hyphal-inducing MAP kinase pathway depend on the function of CaCdc35p in their ability to induce morphogenetic switching. Homozygous cacdc35Δ cells were unable to establish vaginal infection in a mucosal membrane mouse model and were avirulent in a mouse model for systemic infections. These findings suggest that fungal adenylyl cyclases and other regulators of the cAMP signaling pathway may be useful targets for antifungal drugs.
INTRODUCTION
Candida albicans is a major fungal pathogen of humans, and infections with this fungus are a particular problem in immune compromised patients. C. albicans grows in several morphological forms. Under conditions of moderate temperature and low pH and in the absence of inducers such as serum or N-acetylglucosamine, the cells grow as budding yeasts (Castilla et al., 1998). Increases in temperature to 37°C, increases in pH, and the addition of inducers can stimulate the formation of filamentous forms. These filamentous forms include pseudohyphae (chains of elongated cells) as well as true hyphae, where growth involves parallel-sided walls with the cells separated by septa (Mitchell, 1998). The finding that mutant strains defective in hyphal growth are avirulent (Leberer et al., 1997; Lo et al., 1997) has implicated the yeast–hyphal transition in C. albicans pathogenicity.
Adenosine 3′:5′-cyclic monophosphate (cAMP) plays a role in the differentiation of many fungi, and dimorphic behavior has often been linked to intracellular levels of this cyclic nucleotide. In Schizosaccharomyces pombe, deletion of the adenylyl cyclase gene does not affect viability but derepresses conjugation and sporulation under conditions that normally inhibit these differentiation stages in wild-type cells (Kawamukai et al., 1991). In contrast, adenylyl cyclase mutants of Magnaporthe grisea have decreased vegetative growth rate, are sterile, and are defective in forming appresoria on an inductive surface and thus are unable to infect susceptible rice leaves (Choi and Dean, 1997). In Cryptococcus neoformans, the cAMP signaling pathway functions to sense nutritional signals that regulate mating and the induction of virulence factors such as melanin and capsules (Alspaugh et al., 1997). In Ustilago maydis, mutations in the gene encoding adenylyl cyclase cause constitutive growth (Gold et al., 1994), whereas strains with defects in the gene encoding the regulatory subunit of protein kinase A (PKA) are incapable of forming tumors in plants (Gold et al., 1997; Durrenberger et al., 1998). In addition, in Neurospora crassa the regulatory subunit of PKA has been demonstrated to play a role in polarized growth and the localization of septa (Bruno et al., 1996), whereas adenylyl cyclase has been shown to control carbon source utilization (Terenzi et al., 1979). Recently, regulation of adenylyl cyclase protein levels in N. crassa has been shown to be controlled by a G protein α subunit (Kays et al., 2000).
In the yeast Saccharomyces cerevisiae, cAMP signaling has been found to be essential for growth, and together with a mitogen activated protein (MAP) kinase signaling pathway, has been found to play a role in pseudohyphal differentiation (Kronstad et al., 1998; Thevelein and de Winde, 1999; Borges-Walmsley and Walmsley, 2000). Deletion of the CDC35/CYR1 gene encoding adenylyl cyclase causes yeast cells to arrest in G1 of the cell cycle (Matsumoto et al., 1982; Kataoka et al., 1985). The activity of adenylyl cyclase is regulated by the yeast Ras homologues Ras1p and Ras2p (DeFeo-Jones et al., 1985; Toda et al., 1985; Field et al., 1988). Simultaneous depletion of these homologues is lethal (Kataoka et al., 1984, 1985; Tatchell et al., 1985). Moreover, in addition to the Ras proteins, the G protein α subunit homologue Gpa2p appears also to modulate the activity of adenylyl cyclase (Kubler et al., 1997; Lorenz and Heitman, 1997; Colombo et al., 1998). Mutant cells defective in either RAS2 or GPA2 have normal vegetative growth, but double mutant cells exhibit very slow growth even on rich medium. This defect can be suppressed by exogenous cAMP or by mutation in the PDE2 gene encoding cAMP phosphodiesterase (Kubler et al., 1997; Xue et al., 1998). Ras2p and Gpa2p are both required for filamentous growth in S. cerevisiae, and evidence suggests that Ras2p might be the activator of both the cAMP and the MAP kinase pathways that control filamentous growth (Kronstad et al., 1998; Thevelein and de Winde, 1999; Borges-Walmsley and Walmsley, 2000).
In C. albicans, a role for cAMP in the yeast to hyphal switch has been controversial. Some research provides evidence for a transient rise in cAMP levels during germ tube formation (Niimi et al., 1980; Chattaway et al., 1981; Sabie and Gadd, 1992), but other studies support a transient decrease due to increased phosphodiesterase activity (Egidy et al., 1990) or provide no evidence at all for fluctuations in cAMP levels during the yeast-to-hyphal switch (Sullivan et al., 1983). Genetic evidence for a potential role of cAMP in dimorphic switching has come from the cloning of the CaTPK2 gene encoding a C. albicans homologue of the catalytic subunit of PKA (Sonneborn et al., 2000). Deletion of both CaTPK2 alleles interferes with morphogenesis under some environmental conditions and partially reduces virulence in a mouse model for systemic candidiasis. Biochemical characterization of a protein with PKA activity but a molecular weight size that is different from the protein encoded by CaTPK2 suggests that a second C. albicans homologue of the PKA catalytic subunit might exist (Zelada et al., 1998). Consistent with this view is the observation that a second gene encoding a homologue of the catalytic subunit of PKA is present in the C. albicans genome (http://sequence-www.stanford.edu/group/candida/index.html).
Additional support for a role of the cAMP pathway in dimorphic switching of C. albicans has involved the phenotypic characterization of a gene encoding a homologue of Ras. This homologue has been shown to be required for the yeast-to-hyphal switch (Feng et al., 1999) and to contribute to virulence in an animal model through regulation of the MAP kinase and cAMP signaling pathways (Leberer et al., 2001). In this study, we describe the isolation and functional characterization of the CaCDC35 gene encoding a homologue of adenylyl cyclase. We provide genetic evidence that signal transduction through adenylyl cyclase contributes to vegetative growth of C. albicans cells and is essential for dimorphic differentiation in response to environmental cues and for virulence in mouse models for vaginal and systemic fungal infections.
MATERIALS AND METHODS
Isolation of CaCDC35
The CaCDC35 gene was isolated in a screen searching for C. albicans genes capable of suppressing the temperature-sensitive growth defect of the ras1Δ ras2ts S. cerevisiae strain TTM3–4B (Powers et al., 1989) as described (Leberer et al., 2001). In addition to plasmids carrying CaRAS1 (Leberer et al., 2001), we isolated from a genomic C. albicans library constructed in the S. cerevisiae vector YEp352 (Boone et al., 1991) plasmids pDH222 and pDH223 carrying inserts of 2.3 and 5.2 kb, respectively. Plasmid pDH222 carried an open reading frame of 1311 bp encoding the carboxyl terminal catalytic domain of adenylyl cyclase from amino acids 1254–1690. Plasmid pDH223 contained an open reading frame of 4917 bp encoding a carboxyl terminally truncated version of adenylyl cyclase from amino acids 1–1639. To create plasmid pCR0 containing the complete coding region of CaCDC35, a 5.1-kb SphI-EcoRI fragment of plasmid pDH223 was ligated to a 1.17-kb EcoRI fragment from pDH222 in pTZ18R (Mead et al., 1986). To identify more 5′ upstream noncoding sequences of CaCDC35, we screened the C. albicans genomic library by colony hybridization under high-stringency (Feinberg and Vogelstein, 1983) and isolated plasmid pCR21 containing 2743 bp of sequence upstream of the coding region and 4743 bp of sequence within the coding region of CaCDC35.
Yeast Manipulations
The C. albicans and S. cerevisiae strains used in this study are listed in Table 1. Media and culture conditions for the growth of C. albicans and S. cerevisiae cells were as described (Rose et al., 1990). All media were supplemented with uridine (25 μg/ml) for the growth of C. albicans Ura- strains. Transformation of S. cerevisiae and C. albicans cells were performed by the lithium acetate method and spheroplast methods, respectively (Rose et al., 1990). Plasmid DNA was isolated from S. cerevisiae cells as described (Rose et al., 1990).
Table 1.
C. albicans and S. cerevisiae strains used in this study
| Strains | Genotype | Source |
|---|---|---|
| C. albicans | ||
| SC5314 | CDC35/CDC35 URA3/URA3 | Fonzi and Irwin, 1993 |
| CAI4 | ura3∷λ imm434/ura3∷1 imm434 | Fonzi and Irwin, 1993 |
| CR20 | ura3∷λ imm434/ura3∷1 imm434 CDC35/cdc35Δ∷hisG-URA3-hisG | This study |
| CR20.1 | ura3∷λ imm434/ura3∷1 imm434 CDC35/cdc35Δ∷hisG | This study |
| CR216 | ura3∷λ imm434/ura3∷1 imm434 cdc35Δ∷hisG-URA3-hisG/cdc35Δ∷hisG | This study |
| CR276 | ura3∷λ imm434/ura3∷1 imm434 cdc35Δ∷hisG/cdc35Δ∷hisG | This study |
| CR293 | ura3∷λ imm434/ura3∷1 imm434 cdc35Δ∷hisG/cdc35Δ∷hisG [pVEC-CaCDC35] | This study |
| CR323 | ura3∷λ imm434/ura3∷1 imm434 cdc35Δ∷hisG/cdc35Δ∷hisG [pVEC] | This study |
| CR340 | ura3∷λ imm434/ura3∷1 imm434 cdc35Δ∷hisG/cdc35Δ∷hisG TRP1∷CDC35 URA3 | This study |
| DH108+pCR2 | ura3∷λ imm434/ura3∷1 imm434 ras1Δ∷hisG/ras1Δ∷hisG+ [yPB1-ADHp-CaCDC35] | Leberer et al., 2001 |
| S. cerevisiae | ||
| TTM3-4B | MATa ras1Δ∷HIS3 ras2ts ura3 his3 leu2 trp1 ade8 | Powers et al., 1989 |
| Plasmids | Description | Source |
| pVEC | CaARS CaURA3 | Magee and Magee, 1997 |
| pYPB1-ADHpL | CaADH1 promoter URA3 Ca.ARS, S.c. 2 μm plasmid | Leberer et al., 1996 |
| p5921 | hisG-CaURA3-hisG | Fonzi et al., 1993 |
| pCAO1 | CaPCK1 promoter in pUC21 | Leuker et al., 1997 |
| pCR0 | CaCDC35 Sph1/EcoRI in pTZ18R | This study |
| pCR1 | 6.2 kb BamHI-KpnI CDC35 in pVEC | This study |
| pCR2 | 5.9 kb MluI CaCDC35 in pYPB1-ADHpL | This study |
| pCR3 | 5.8 kb KpnI CaCDC35 in pVEC-CaPCK1p | This study |
| pCR21 | CaCDC35 (7.5 kb) in YEp352 | This study |
| pCR4 | CaARS CaURA3 CaPCK1p | This study |
| pCRF3 | CaCDC35BamHI-BglIIhisG-CaURA3-hisGBamHI-BamHICaCDC35 | This study |
| PCRQ38 | CaCDC35BamHI-KpnI CaTRP1KpnI in pVEC | This study |
| pDH222 | CaCDC35 (2.3 kb) in YEp352 | This study |
| pDH223 | CaCDC35 (5.2 kb) in YEp352 | This study |
| pDH233 | pVEC-CaRAS1 CaURA3 | Leberer et al., 2001 |
| pDH240 | pYPB1-ADHpL-CaRAS1 | This study |
| HLB 134 | 4.2 kb HindIII EFG1 fragment in pRC2312 | Lo et al., 1997 |
| pADH-HST7 | pYPB1-ADHpT-CaHST7 | Leberer et al., 1996 |
| pLJ19 | pYPB1-ADHpL-CaCPH1 | Csank et al., 1998 |
| pLJ57 | pYPB1-ADHpL-CaRAS1G13V | This study |
| pSU1 | pVEC-CaRAS1G13V | This study |
| pJA39 | pVEC-CaTRP1 | This study |
Plasmids
All plasmids used in this study are listed in Table 1. PCRs were performed with the use of the Expand High Fidelity or Long Template PCR system (Boehringer Mannheim, Laval, Quebec, Canada). Fragments generated by PCR were always sequenced to ensure no unanticipated mutations were introduced during the amplification procedure. To construct pCR1, we amplified a 6.2-kb fragment containing the complete open reading frame of CaCDC35 and 242 bp of upstream regulatory region with the use of the oligodeoxynucleotides 5′-CGGGATCCGATCAACACATTTTTAAT-3′ and 5′-CGGGTACCTCATGTATCCTTTAGGAA-3′ (the newly created BamHI and KpnI sites, respectively, are underlined) and pCR0 as a template. The amplified fragment was then cloned into pVEC (Magee and Magee, 1997). To construct pCR2, we amplified a 5.9-kb fragment containing the complete open reading frame of CaCDC35 with the use of the oligonucleotides 5′-CGACGCGTATGAGTTT-TTTAAGGAGA-3′ and 5′-CGACGCGTTAATCCAACCAATTGA-TT-3′ (the newly created MluI sites are underlined; the start codon is in italics) and pCR0 as template, and cloned the fragment into pYPB1-ADHpL (Leberer et al., 1996). To place CaCDC35 under the control of the CaPCK1 promoter, we first constructed plasmid pCR4 by cloning a BamHI-BglII fragment with the CaPCK1 promoter from plasmid pCAO1 (Leuker et al., 1997) into pVEC. Next, we PCR-amplified the complete open reading frame of CaCDC35 with the use of the oligodeoxynucleotides 5′-CGGGTACCATGAGTTT-TTTAAGGAGA-3′ and 5′-CGGGTACCCAAATAGTATATAAAT-CG-3′ (the newly created KpnI sites are underlined; the start codon is in italics) and cloned the amplified 5.9-kb fragment into the KpnI site of pCR4.
The integration plasmid pCRQ38 contains CaCDC35, CaTRP1 for integration into the TRP1 locus, and CaURA3 as selectable marker. It was generated by cloning, into the BamHI site of pCR1, a 1.4-kb PCR fragment of the CaTRP1 gene amplified by PCR with the use of the divergent oligodeoxynucleotide primers OJA19 (5′-CG-AGATCTTAAGCCGTGCTGGCGTGAAT-3′) and OJA17 (5′-CG-AGATCTCATGAGACACTGGTCTCGCGTCTG-3′) (the newly introduced BglII sites are underlined) and with the use of plasmid pJA39 as template.
Plasmid pDH240 was constructed by cloning a SmaI fragment containing the complete open reading frame of CaRAS1 (Leberer et al., 2001) into the EcoRV site of pYPBl-ADHpL. Plasmid pSU1 was derived from pDH233, which is wild-type RAS cloned as a PstI-HindIII fragment in pVEC, by site-directed mutagenesis with the use of the QuickChange Site-Directed Mutagenesis kit from Stratagene (La Jolla, CA). The oligodeoxynucleotide primer pairs were 5′-GTTGTTGTTGGAGGAGTTGGTGTTGGTAAATCCGC-3′ and 5′-GCGGATTTACCAACACCAACTCCTCCAACAACAAC-3′ for replacing the glycine residue at position 13 into a valine residue. The mutant RAS1 allele was then cut with SmaI and transferred to EcoRV cut YPB1-ADHpL to generate pLJ57. Plasmid pJA39 consists of the C. albicans TRP1 inserted into pVEC.
Deletion of CaCDC35
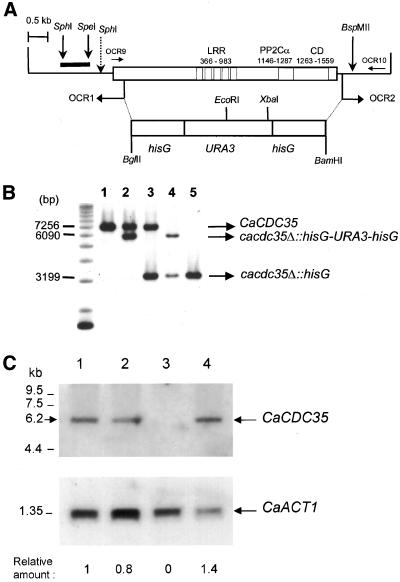
To construct a CaCDC35 deletion cassette, a DNA fragment that contained CaCDC35 flanking sequences from nucleotide positions −242–218 and 5649–6275, respectively, joined with BamHI sites was created by PCR with the use of the divergent oligodeoxynucleotide primers OCR1 (5′-CGGGATCCTTCAAATGGTGGGTAGCTGAG3′) and OCR2 (5′CGGGATCCCACCTTCAGCTGAAGCAACAC-3′; newly introduced BamHI sites are underlined) and plasmid pCR0 as template. The amplified DNA was cleaved with BamHI and ligated with a BamHI-BglII hisG-CaURA3-hisG cassette from plasmid p5921 to yield plasmid pCRF3. This plasmid was linearized with SphI and BspMII and transformed into the Ura− C. albicans strain CAI4 (Fonzi and Irwin, 1993). Transformants in which the coding region of one of the chromosomal CaCDC35 alleles was replaced by homologous recombination with the hisG-CaURA3-hisG cassette were selected on Ura− medium. Integration of the cassette into the CaCDC35 locus was confirmed by Southern blot analysis with the use of a SphI to SpeI fragment from nucleotide positions −1366 to −630 as a probe (Figure 1, A and B). Spontaneous Ura− derivatives were then selected on medium containing 5-fluoro-orotic acid as described (Fonzi and Irwin, 1993). These clones were screened by Southern blot hybridization to identify those that had lost the CaURA3 gene by intrachromosomal recombination mediated by the hisG repeats. The remaining functional allele of CaCDC35 was then deleted by repeating the same procedure. With the use of this two-step approach, we independently isolated the homozygous cacdc35Δ/cacdc35Δ strains CR153 and CR216 that showed the identical structural and phenotypic behavior.
Figure 1.
Deletion of CaCDC35 in C. albicans. (A) Restriction endonuclease map of CaCdc35p. The white rectangle indicates the coding region of the gene. The conserved central leucine rich repeat (LRR) motif, the protein phosphatase 2Cα (PP2Cα) domain, and the catalytic domain (CD) are indicated. The PCR with the divergent oligonucleotides OCR1 and OCR2 was used to delete the coding sequence of CaCDC35. A hisG-URA3-hisG cassette was then inserted, and a two-step procedure was used to delete both alleles of CaCDC35 by homologous recombination (see MATERIAL AND METHODS). (B) Southern blot analysis with the use of a 0.8-kb SphI-SpeI probe of the CaCDC35 coding region. The genomic DNA samples, digested with SphI and BspMII, were prepared from strains CAI4 (CaCDC35/CaCDC35; lane 1), CR20 (CaCDC35/cacdc35Δ::hisG-URA3-hisG, lane 2), CR20.1 (CaCDC35/ cacdc35Δ::hisG, lane 3), CR216 (cacdc35Δ::hisG-URA3-hisG/ cacdc35Δ:: hisG, lane 4), and CR276 (cacdc35Δ::hisG/cacdc35Δ::hisG, lane 5). The wild-type band was 7.25 kb, whereas the KO band with the URA3 blaster was 6.1 kb. After looping out the URA3 fragment, the band was reduced to 3.2 kb. (C) Northern blot analysis of poly(A)+ RNA isolated from strains SC5314 (CaCDC35/CaCDC35; lane 1), CR20 (CaCDC35/cacdc35Δ; lane2), CR216 (cacdc35Δ/cacdc35Δ; lane 3), and CR323 (cacdc35Δ/cacdc35Δ [CDC35]; lane 4). The blot was probed with fragments specific for CaCDC35 or the actin gene (CaACT1) and quantified by phosphorimaging. The ratios at the bottom of each lane represent the amount of the CaCDC35 transcript relative to the CaACT1 transcript in the same lane. The relative overexpression of the reintegrant construct may be due to the consequence of having only a limited region of the promoter on the plasmid as well as to the influence of the site of integration on general expression levels.
To reintegrate the CaCDC35 gene into the CaTRP1 locus, strain CR276 (cacdc35Δ::hisG/cacdc35Δ::hisG) was transformed with plasmid pCRQ38 linearized with BsiWI, and recombinants were selected on -ura medium. Correct integration of CaCDC35 at the CaTRP1 locus was then confirmed by Southern blot analysis.
Northern Blot Analysis
Northern blots of total RNA and poly(A)+ RNA from C. albicans were performed as described (Leberer et al., 1992). The probe for CaCDC35 was a 3.7-kb NheI-SacII fragment from nucleotide positions 432-4159, and the CaACT1 probe was an EcoRI-HindIII fragment of the CaACT1 gene (Losberger and Ernst, 1989).
Phenotypic Characterization of cacdc35Δ C. albicans Mutant Cells
Proliferation of C. albicans cells was determined in YPD medium at 30°C. An overnight culture was diluted to OD600 = 0.1 into fresh medium and grown at 30°C. The density at 600 nm (OD600) of each culture was determined every hour over a period of 8 h. For the analysis of colony morphology, microphotographs of single colonies were taken directly from Petri plates by phase contrast microscopy. To analyze filaments, microphotographs were taken with a Leitz Aristoplan microscope with the use of Nomarski optics at 1000× magnification (Leitz, Wetzlar, Germany).
Measurements of cAMP levels were performed as described (Lorenz et al., 2000). Briefly, C. albicans cells were grown in YPD medium at 30°C to stationary phase for 48 h, washed twice with water and once with MES buffer (10 mM, pH 6, containing 0.5 mM EDTA, pH 7.4), and then resuspended into MES buffer at OD600 of 2. After addition of 100 mM glucose, 500-μl aliquots were transferred at different time points to test tubes each containing 600 μl of acid-washed glass beads and 500 μl of 10% trichloroacetic acid. The tubes were mixed and immediately frozen in liquid nitrogen. After 30 min, the cells were thawed, disrupted by a bead-beater at 4°C, and centrifuged for 10 min at 20,000 × g. The samples were neutralized by washing the supernatant five times with water-saturated ether, lyophilized, and then resuspended in 500 μl of assay buffer (0.05 M acetate buffer, pH 5.8, 0.02% [wt/vol] bovine serum albumin). cAMP levels were determined by with the use of the EIA system–cAMP immunoassay (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's instructions.
Macrophage Cytopathology Assays
The mouse macrophage cell line RAW264.7 clone D3 (kindly provided by A. Descoteaux, IAF, Laval, Canada) was cultured in an eight-chambered Permanox slide (Lab-Tek, Naperville, IL) at a cell density of 105 cells/well in Dulbecco's modified Eagle's medium (DMEM; Life Technologies-BRL, Rockville, MD) supplemented with 10% heat-inactivated fetal bovine serum (D-10) at 37°C in a 5% CO2 atmosphere for 24 h. Before the addition of C. albicans cells, the chambers were washed once with D-10 medium.
C. albicans cells were grown in YPD medium at 30°C to stationary phase and then washed twice with phosphate-buffered saline, pH 7.4 (PBS). These cells were then added to macrophages at a ratio of 2.5:1. After incubation at 37°C in 5% CO2 atmosphere for 1–4 h, slides were washed three times with D-10 medium, incubated at 4°C for 1 h with 100 μl of rabbit anti–C. albicans antibodies (Accurate Chemical & Scientific Corp., Westbury, NY) diluted 1:200 in D-10 medium. Slides were washed four times with cold D-10 medium, fixed with the use of HEMA3 fixative (Biochemical Sciences Inc.) according to the manufacturer's instructions, washed three times with D-10 medium, and then incubated with FITC-conjugated F(ab)′2 donkey anti-rabbit antibodies (Jackson ImmunoResearch Laboratories Inc., West Grove, PA) diluted 1:100 in D-10. After 45 min of incubation at room temperature, wells were washed four times with PBS, and the slides were mounted in Prolong antifade mounting medium (Molecular Probes, Eugene, OR). Epifluorescence was monitored with the use of a Leitz Laborlux S microscope equipped with a CoolSnap CCD camera (Photometrics, Tucson, AZ) at a magnification of 1000×.
To analyze the survival rate of C. albicans cells exposed to macrophages, we used an end-point dilution survival assay. One milliliter of a saturated C. albicans culture grown in YPD was washed twice in d-PBS, sonicated, and resuspended at 107 cells/ml in cold D-10. Fifty microliters of the suspension was added to 150 μl of D-10 in 96-well plates containing medium only or macrophages. After serial fourfold dilutions, plates were incubated on ice for 30 min and subsequently for 48 h at 37°C in 5% CO2 atmosphere. Colonies were visualized with the use of a Nikon (Garden City, NY) TMS inverted microscope at 20× or 100× magnification.
Virulence Studies
Eight- to 10-week-old female BALB/c mice were obtained from Charles Rivers Breeding Laboratories (Sulzfeld, Germany). Mucosal infection of the vaginal canal was initiated by inoculating mice (5–8 for each group) intravaginally with 5 × 104 stationary phase cells of the wild-type strain CR340 or 4 × 105 stationary phase cells of the homozygous cacdc35Δ mutant strain CR323 taken up in 20 μl of PBS (Fidel et al., 1993). To induce pseudoestrus during the infection, mice were injected subcutaneously with 0.02 mg/mouse estradiol valerate (Sigma Chemical, St. Louis, MO) in 0.1 ml sesame oil 72 h before inoculation with C. albicans cells (Sobel et al., 1985; Ryley and McGregor, 1986; Fidel et al., 1993). Estradiol treatments were continued at weekly intervals thereafter. After killing, the vaginas of the animals were lavaged with 100 μl of PBS, and the vaginal fungal burden was quantified by determination of colony-forming units (CFU) as previously described (Fidel et al., 1993).
For systemic infections, groups of 10 mice were inoculated with 5 × 105 wild-type cells of strain CR340 or 4 × 106 mutant cells of strain CR323 by intravenous injection and monitored for survival as described (Csank et al., 1997, 1998; Timpel et al., 1998). The eightfold excess of mutant cells was used to compensate for the slower growth rate of these cells. Survival curves were calculated according to the Kaplan-Meier method with the use of the PRISM program (GraphPad Software, San Diego, CA) and analyzed by with the use of the log-rank test. A p value < 0.05 was considered as significant.
Accession Number
The GenBank/EMBL Data Library accession number for CaCDC35 is AF295379.
RESULTS
Isolation and Characterization of CaCDC35
The CaCDC35 gene was identified in a screen searching for C. albicans genes capable of complementing the conditional growth defect of S. cerevisiae ras1Δ ras2ts cells. In addition to clones carrying the CaRAS1 gene (Leberer et al., 2001), we isolated two overlapping clones with the potential to encode either the carboxyl terminal catalytic domain of adenylyl cyclase from amino acid positions 1253–1690 or a carboxyl terminally truncated version of adenylyl cyclase from amino acid positions 1–1639, respectively. The alignment of both sequences generated an open reading frame that contains no introns and is predicted to encode a protein of 1690 amino acids with a calculated molecular weight of 186.9 kDa. Our sequence is identical to that recently determined independently (Mallet et al., 2000).
CaCdc35p has overall sequence identities of 32, 26, and 21% with the homologues of S. cerevisiae (Kataoka et al., 1985), M. grisea (Choi and Dean, 1997), and U. maydis (Gold et al., 1994), respectively. Like the other currently known fungal homologues, CaCdc35p has a domain structure typical of peripheral membrane adenylyl cyclases lacking a transmembrane domain (Tang and Gilman, 1992). Typical for this type of adenylyl cyclases, CaCdc35p has from amino acids 366–983 a central domain composed of amphipathic leucine-rich repeats of 23 amino acids (Figure 1A). This region has 42% identity and 58% similarity to Cdc35p from S. cerevisiae. In S. cerevisiae, this region has been shown to be required for the interaction of adenylyl cyclase with the Ras homologues Ras1p and Ras2p (Suzuki et al., 1990).
The carboxyl terminal half of CaCdc35p contains the ATP-binding catalytic domain from amino acid positions 1263–1559 (Figure 1A). This domain has 26% identity and 48% similarity with the homologous domain of Cdc35p from S. cerevisiae, but shows less homology to the catalytic domains of adenylyl cyclases from mammalian cells (Tesmer et al., 1997). Like the homologue from S. cerevisiae (Tamura et al., 1989), CaCdc35p contains a protein phosphatase 2Cα-like domain in the region between the central and catalytic domains from amino acids 1146–1287 with 31% homology to human protein phosphatase 2Cα (Figure 1A; Mann et al., 1992). In comparison to the homologue from S. cerevisiae, CaCdc35p lacks a stretch of sequence of 381 amino acids at the amino terminus. The function of this 42-kDa amino terminal region is not known, although a 100-amino acid region just N-terminal to the central domain has been shown to be required for optimal regulation of S. cerevisiae adenylyl cyclase by Ras (Colicelli et al., 1990).
Chromosomal Deletion of CaCDC35
Both CaCDC35 alleles were deleted in strain CAI4 by homologous recombination in a multistep procedure (Figure 1A). The deletions were confirmed by Southern blot analysis (Figure 1B) and by PCR (our unpublished results). Northern blots showed that the level of the CaCDC35 transcript, which had a size of 6.2 kb, was reduced to ∼80% in cells containing a deletion of one allele of CaCDC35 relative to the wild-type strain, and was absent in cells deleted for both alleles (Figure 1C). This transcript was present at about a 40% increased level relative to the wild-type strain when the CaCDC35 gene was reintegrated into the CaTRP1 locus of homozygous mutant cells (Figure 1C, lane 4).
We found that cells deleted for both alleles were viable but grew ∼2.5-fold slower than wild-type cells (Table 2). This attenuated growth was observed in media containing glucose as well as in media containing galactose, glycerol, or ethanol as carbon sources (our unpublished results). The growth defect could be complemented by either reintegration of the wild-type gene into the CaTRP1 locus of homozygous mutant cells or addition of exogenous cAMP (Table 2), demonstrating that the defect in growth of mutant cells was caused by disruption of the function of adenylyl cyclase.
Table 2.
Growth at 30°C in YPD
| Strain | Doubling time (h) |
|---|---|
| SC5314 (CaCDC35/CaCDC35) | 1.46 ± 0.01 |
| CR216 (cacdc35Δ/cacdc35Δ) | 4.19 ± 0.01 |
| CR340 (cacdc35/cacdc35Δ TRP1∷CDC35) | 1.54 ± 0.02 |
| CR216 (+10 mM dibutyryl cAMP) | 1.69 ± 0.03 |
Data represent mean values ± SD of three independent experiments.
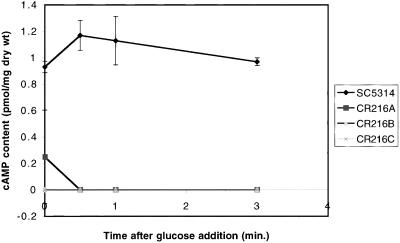
As shown in Figure 2, cAMP levels were undetectable in homozygous mutant cells demonstrating that deletion of both copies of CaCDC35 resulted in a complete loss of cAMP production. In agreement with previous work (Niimi, 1984), we found that the glucose-induced cAMP burst normally observed in S. cerevisiae cells (Mbonyi et al., 1988) is not well developed in C. albicans cells (Figure 2).
Figure 2.
Intracellular cAMP concentrations in strains SC5314 (wild-type) and three isolated colonies A, B, and C of CR216 (cacdc35Δ::hisG-URA3-hisG/cacdc35Δ). After addition of 100 mM glucose to the medium, cells were harvested at the indicated time points, and cAMP levels were determined as described in MATERIAL AND METHODS. The data represent mean values of two independent measurements.
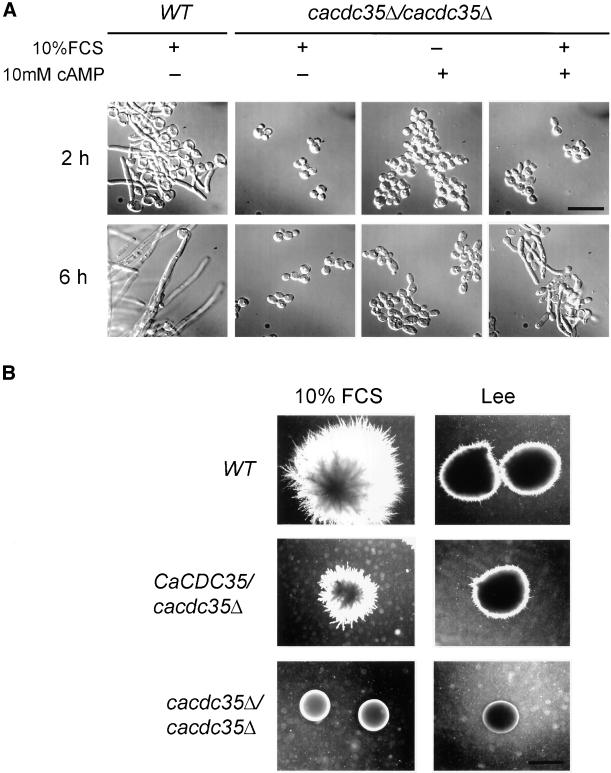
The ability of C. albicans cells to switch from a yeast-like to a hyphal mode of growth was not affected by deletion of only one allele of CaCDC35 (our unpublished results). However, deletion of both alleles completely blocked the yeast-to-hyphal transition in all media and under all conditions that we investigated. When morphological switching was induced in liquid media by either serum or Lee's medium, the homozygous mutant cells were completely defective in the formation of germ tubes or filaments (Figure 3A). On solid agar plates containing serum or solid Lee's medium (as well as SLAD or Spider media), the normal formation of hyphae was completely suppressed by deletion of both alleles of CaCDC35 (Figure 3B). These defects were reversed by either addition of exogenous cAMP (Figure 3A) or by reintegration of the wild-type CaCDC35 gene into the CaTRP1 locus of the homozygous mutant cells (our unpublished results).
Figure 3.
Defects in hyphal formation caused by deletion of both CaCDC35 alleles. (A) The CaCDC35 wild-type strain SC5314 (WT) and strain CR216 deleted for both alleles of CaCDC35 (cacdc35Δ/cacdc35Δ) were grown for 2 or 6 h at 37°C in liquid Lee's medium with or without (+/−) 10% fetal calf serum (FCS) and with or without (+/−) 10 mM dibutyryl-cAMP. Photomicrographs were taken by Nomarski optics at 1000× magnification. Scale bar, 10 μm. (B) The CaCDC35 wild-type strain SC5314 (WT), the heterozygous strain CR20 (CaCDC35/cacdc35Δ), and the homozygous strain CR216 (cacdc35Δ/cacdc35Δ) were grown for 5 d at 37°C on either solid agar medium containing 10% fetal calf serum (FCS) or on solid Lee's medium (Lee et al., 1975). The same defects in hyphal growth were observed on solid low ammonia dextrose nitrogen starvation medium (SLAD; Gimeno et al., 1992) and Spider medium (Liu et al., 1994; our unpublished results). Scale bar, 2 mm. Photomicrographs were taken with the use of phase contrast at 20× magnification.
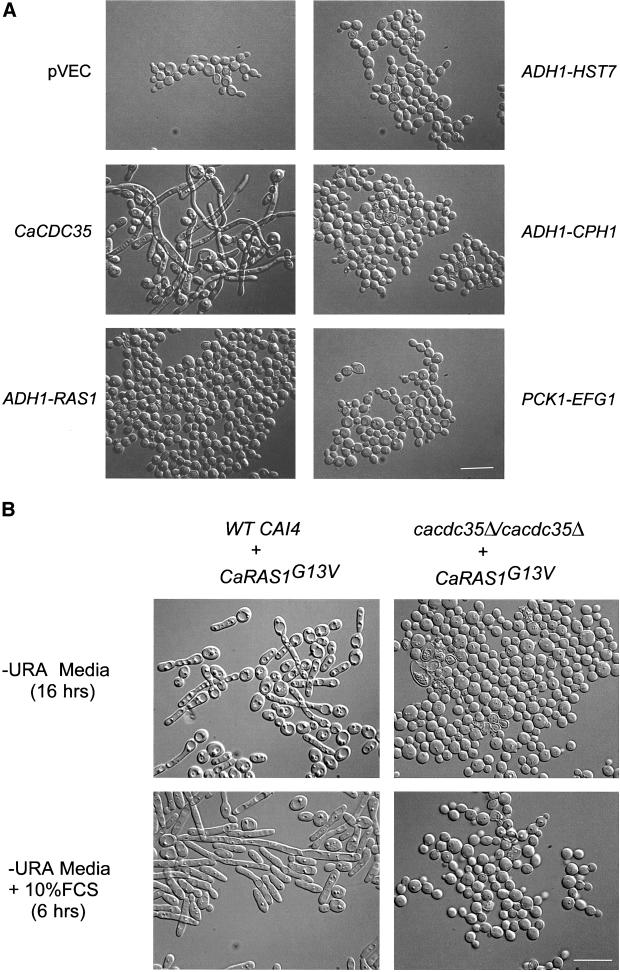
The inability of homozygous mutant cells to switch into the hyphal mode of growth was also corrected by introduction of the CaCDC35 gene on an autonomously replicating plasmid (Figure 4A). However, overexpression of either the CaRAS1, HST7, or CPH1 genes driven by the strong ADH1 promoter or moderate overexpression of the EFG1 gene driven by the CaPCK1 promoter failed to suppress the hyphal switch defect (Figure 4A).
Figure 4.
Epistasis analysis. (A) C. albicans strain CR216 deleted for both alleles of CaCDC35 was transformed with the empty control plasmid pVEC and plasmids pVEC-CaCDC35, pYPB1-ADHpL-CaRAS1, pYPBL-ADHpT-HST7, pYPBl-ADHpL-CPH1, and pRC2312–EFG1 carrying the indicated C. albicans genes. Transformants were grown for 6 h at 37°C in selective medium containing 10% fetal calf serum (FCS). To induce expression of EFG1, 2% casamino acids were added to the medium. (B) Strains CAI4 (WT) and CR216 deleted for both alleles of CaCDC35 were transformed with pYPBl-ADHpL-CaRAS1G13V carrying the hyperactive mutant version of CaRAS1 and grown for 16 h in selective medium at 30°C (two top panels). The overnight cultures were transferred to fresh -URA medium containing 10% FCS and grown for 6 h at 37°C (two bottom panels). Photomicrographs were taken by Nomarski optics at a 1000× magnification (scale bar, 10 μm) and are representative of many cells.
In agreement with previous observations (Feng et al., 1999; Leberer et al., 2001), we found that expression of the hyperactive G13V mutant version of CaRas1p in wild-type cells both induced the formation of hyphae under conditions that favor the yeast mode of growth and enhanced the formation of hyphae under inducing conditions (Figure 4B). These effects of the hyperactive CaRas1p mutant allele were completely blocked by deletion of both CaCDC35 alleles (Figure 4B).
Role of CaCdc35p in Phagocytosis of C. albicans Cells by Macrophages
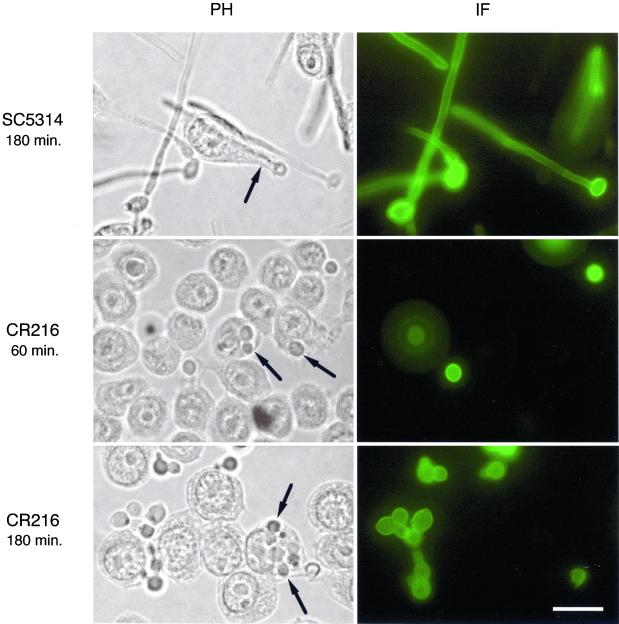
We monitored the uptake of C. albicans cells by macrophages through an immunofluorescence procedure that distinguished between C. albicans cells that were either free in the medium or attached to the external surface of macrophages from those cells that were undergoing phagocytosis. Cells that were not yet undergoing phagocytosis were accessible to antibodies and hence could be immunostained, whereas cells already engulfed by the macrophages were not stained by this procedure. Candida cells, either in yeast or hyphal form, could be phagocytosed (Figure 5 and our unpublished results). C. albicans wild-type cells developed long hyphal tubes either inside or outside of macrophages. This growth allowed them to escape the macrophage engulfment. In contrast, the homozygous cacdc35Δ/cacdc35Δ mutant cells engulfed by macrophages were completely blocked in the formation of hyphae, and this inability to form hyphae prevents them from escaping phagocytosis. Their slower growth rate may also contribute to their susceptibility. Quantification of this host–pathogen interaction with the use of an end-point dilution survival assay (see MATERIALS AND METHODS) indicated that C. albicans adenylyl cyclase defective cells were more readily killed through interaction with macrophages than were the wild-type cells (Table 3).
Figure 5.
Phagocytosis of C. albicans cells by macrophages. Macrophages incubated with wild-type C. albicans cells (SC5314) or C. albicans cells deleted for both alleles of CaCDC35 (CR216) for the indicated time periods. Micrographs were taken at a magnification of 1000× by phase contrast (left panels) or fluorescence after selective immunostaining of C. albicans cells with C. albicans–specific antibodies (right panels). The black arrows point to C. albicans cells phagocytosed by the macrophage cells. Scale bar, 10 μm.
Table 3.
Survival of Candida albicans cells cultured with macrophages (cell line RAW264.7)
| Strains | No. of colonies in presence (+) or absence (−) of macrophages
|
% Survivala | |||||
|---|---|---|---|---|---|---|---|
| Exp. 1
|
Exp. 2
|
Exp. 3
|
|||||
| − | + | − | + | − | + | ||
| SC5314 (wild type) | 272 | 191 | 153 | 109 | 436 | 281 | 68.6 ± 3.6 |
| CR216 (cacdc35Δ/cacdc35Δ) | 141 | 4 | 177 | 9 | 489 | 27 | 4.5 ± 1.5 |
Survival of C. albicans cells was determined as described in MATERIALS AND METHODS and is expressed as the number of colonies in the presence of macrophages divided by the number of colonies in the absence of macrophages.
Data represent mean value ± SD of three independent experiments.
Requirement of Adenylyl Cyclase for Virulence
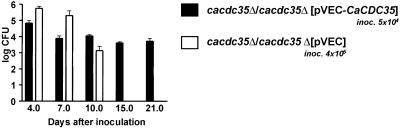
To investigate whether CaCdc35p is required for virulence in a mucosal model of murine candidiasis, we inoculated mice intravaginally with C. albicans cells and monitored for fungal survival on the vaginal mucosa. In contrast to homozygous mutant cells transformed with a plasmid carrying the wild-type CaCDC35 gene, mutant cells carrying an empty control plasmid were completely cleared from the vaginal mucosa by day 15, although the initial number of cells applied to the mucosa was eightfold higher for mutant cells than for wild-type cells to compensate for their slower growth rate (Figure 6).
Figure 6.
Vaginal infection. Mice were intravaginally inoculated with 5 × 104 cells of strain CR340 (cacdc35Δ/cacdc35Δ [pVEC-CaCDC35]; dark bars) and 4 × 105 cells of strain CR323 (cacdc35Δ/cacdc35Δ [pVEC]; open bars). C. albicans cells were then recovered from the vaginal canal at the indicated time points and colony forming units (CFU) were determined. The data represent log mean values ± SD of 10 independent experiments.
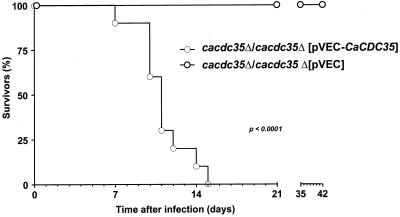
To investigate whether CaCdc35p is required for virulence in systemic candidiasis, mice were inoculated intravenously with C. albicans cells and monitored for survival. As illustrated in Figure 7, inoculation with homozygous mutant cells transformed with a plasmid carrying the CaCDC35 gene resulted in complete mortality after 16 d. However, all mice that were infected with mutant cells transformed with an empty control plasmid survived for at least 42 d, even when the number of C. albicans mutant cells injected into the animals was eightfold higher than the inoculation number of wild-type cells to compensate for their slower growth rate (Figure 7). All of the animals infected with mutant cells showed absolutely no clinical signs of disease.
Figure 7.
Survival curves of mice (10 for each group) intravenously infected with 5 × 105 cells of C. albicans strains CR340 (cacdc35Δ/cacdc35Δ [pVEC-CaCDC35]; light open circle) and 4 × 106 cells of strain CR323 (cacdc35Δ/cacdc35Δ [pVEC]; bold open circle).
DISCUSSION
We have cloned and sequenced the C. albicans CaCDC35 gene, which encodes a homologue of fungal adenylyl cyclases. In contrast to mammalian adenylyl cyclases that are characterized by two blocks of transmembrane segments, fungal adenylyl cyclases lack transmembrane domains and appear to be peripherally bound to the cell membrane (Tang and Gilman, 1992). CaCdc35p has a domain structure typical of the currently known fungal adenylyl cyclases (Figure 1A) and has some similarity to the mammalian isoforms around the catalytic core. Other similarity found with mammalian proteins is located in a region of unknown function that shows 31% homology with protein phosphatase 2Cα. In contrast to the mammalian enzymes, the fungal isoforms appear to be regulated by Ras through the interaction of this small GTP binding protein with a leucine-rich repeat domain in the central region of adenylyl cyclase (Suzuki et al., 1990). This Ras binding domain is highly conserved in CaCdc35 suggesting that the C. albicans protein has the potential to interact with the recently identified C. albicans Ras homologue CaRas1p (Feng et al., 1999).
Deletion of both CaCDC35 alleles completely abolished detectable levels of intracellular cAMP (Figure 2), suggesting that CaCdc35p is the only enzyme capable of producing cAMP in C. albicans cells. The mutant cells were viable, indicating that cAMP is not essential for vegetative growth in C. albicans cells. However, mutant cells exhibited significantly reduced growth rates (Table 2). This mutant phenotype could be complemented by addition of exogenous cAMP (Table 2), suggesting that cAMP-dependent mechanisms contribute to not yet identified steps during vegetative growth of C. albicans. In this context, C. albicans resembles M. grisea, U. maydis, S. pombe, and N. crassa, where adenylyl cyclase genes are not essential (Terenzi et al., 1976; Kawamukai et al., 1991; Gold et al., 1994; Choi and Dean, 1997) but differs from S. cerevisiae, where Cdc35p is essential for progression of cells through G1 of the cell cycle (Matsumoto et al., 1982; Kataoka et al., 1985). It is tempting to speculate that the reason for this functional diversity may be an important attribute in C. albicans for resisting stresses such as nutritional limitation. Although in S. cerevisiae the cAMP signaling pathway is primarily involved in mediating nutritional signals to the cell cycle machinery, in C. albicans the main function of this pathway could be to mediate stress signals to the morphogenetic machinery that controls the yeast-to-hyphal transition as part of a survival strategy. The observation that the highly similar cyclase proteins of C. albicans and S. cerevisiae provide an essential function in one organism and not the other suggests that it is the function of the downstream targets that determine whether cAMP formation is required for viability.
This interpretation is consistent with our finding that C. albicans cells deleted for both CaCDC35 alleles are completely deficient in the ability to switch from the yeast mode of growth into the hyphal mode under all environmental conditions that we investigated (Figure 3). This morphogenetic defect could be cured by addition of exogenous cAMP (Figure 3A), demonstrating that the catalytic function of CaCdc35p is responsible for the induction of hyphal formation in response to environmental cues. Consistent with the view that Ras is a regulator of fungal adenylyl cyclases (DeFeo-Jones et al., 1985; Toda et al., 1985; Broek et al., 1987; Field et al., 1988), the filament-inducing activity of the hyperactive G13V mutant version of CaRas1p was completely blocked in CaCdc35p-deleted mutant cells (Figure 4B). This epistatic relationship places the function of CaRas1p upstream of adenylyl cyclase.
We have previously proposed that coordinated activation of both the filament-inducing MAP kinase cascade and the cAMP signaling pathway initiates morphogenetic processes in a Ras-dependent manner (Leberer et al., 2001). The requirement for dual regulation of morphogenetic switching is corroborated by our studies reported here. In contrast to the morphogenetic switching defect of homozygous caras1Δ mutant cells (Feng et al., 1999), the switching defect of homozygous cacdc35Δ mutant cells could not be complemented by overexpression of Hst7p or Cph1p (a protein kinase and a transcription factor, respectively, in the filament inducing MAP kinase cascade; Liu et al., 1994; Kohler and Fink, 1996; Leberer et al., 1996) or Efg1p (a putative transcription factor believed to respond to the cAMP signaling pathway; Stoldt et al., 1997; Whiteway, 2000; Figure 4A). A plausible interpretation for this observation is that stimulation of the filament-inducing MAP kinase cascade is only capable of inducing hyphal formation during simultaneous elevation of cAMP levels and that Efg1p is a direct target of the cAMP pathway requiring cAMP-dependent activation. By analogy with the dual regulation of the S. cerevisiae adenylyl cyclase by Ras and Gpa2p (Thevelein and de Winde, 1999), it is tempting to speculate that in homozygous caras1Δ mutant cells this requirement is fulfilled through stimulation of CaCdc35p by a G protein α subunit homologue similar to Gpa2p in S. cerevisiae. This explanation is supported by the findings that overexpression of either components of the filament-inducing MAP kinase cascade or of Efg1p can complement the yeast-to-hyphal switching defect of homozygous caras1Δ mutant cells (Leberer et al., 2001). In addition, overexpression of Efg1p complements the switching defect of mutant cells deleted for a homologue of protein kinase A (Sonneborn et al., 2000).
Our finding that adenylyl cyclase mutants defective in the formation of hyphae are more vulnerable to phagocytosis by macrophages (Table 3) supports the hypothesis that the yeast-to-hyphal transition is part of a survival strategy of C. albicans to escape the cellular immune system (Lo et al., 1997). This view is supported by our in vivo studies in mouse models for mucosal and systemic infections. In contrast to wild-type cells, mutant cells were rapidly eradicated from the mucosa after vaginal infection (Figure 6). Mice intravenously infected with mutant cells survived without any clinical signs of disease, whereas mice infected with wild-type cells were efficiently killed (Figure 7). It is unclear whether the defects in virulence are caused by reduced growth of the mutant cells or by defects in morphogenetic switching. However, because we have used eight times more mutant cells than wild-type cells for infection of the animals, it is more likely that the defects in virulence were caused by defects in morphological transitions than by the reduced growth.
In summary, we propose that CaCdc35p represents a key regulatory element in the cAMP signaling pathway and is part of a sensory system involved in detecting changes in the environment and sending signals to a morphogenetic machinery that controls the mode of growth. These interconnected signaling and morphogenetic systems are part of a strategy of C. albicans to resist environmental stresses and thereby contribute to the virulence of this human pathogen. Because CaCdc35p and other fungal adenylyl cyclases differ significantly in their type of regulation from their mammalian counterparts, the fungal adenylyl cyclases and their regulators could represent very attractive targets for the identification of specific inhibitors with antifungal activity.
ACKNOWLEDGMENTS
The authors thank Tamara Michaeli for providing the S. cerevisiae strain TTM3–4B; C. Boone for providing the C. albicans genomic library; W.A. Fonzi, B. Magee, J.F. Ernst, and A. Brown for providing plasmids and C. albicans strains; and A. Descoteaux for providing the mouse macrophage cell lines. The authors are grateful to M. Pelletier for help with data processing in the cAMP assays and to Ursula Oberholzer for her comments on the manuscript. C.R.C.R. was supported by a postdoctoral fellowship from Fundação de Amparo à pesquisa do Estado de São Paulo (FAPESP), Brazil. This is National Research Council of Canada publication number 44794.
REFERENCES
- Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C, Sdicu A, Laroche M, Bussey H. Isolation from Candida albicans of a functional homolog of the Saccharomyces cerevisiae KRE1 gene, which is involved in cell wall beta-glucan synthesis. J Bacteriol. 1991;173:6859–6864. doi: 10.1128/jb.173.21.6859-6864.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges-Walmsley MI, Walmsley AR. cAMP signaling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 2000;8:133–141. doi: 10.1016/s0966-842x(00)01698-x. [DOI] [PubMed] [Google Scholar]
- Broek D, Toda T, Michaeli T, Levin L, Birchmeier C, Zoller M, Powers S, Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987;48:789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Bruno KS, Aramayo R, Minke PF, Metzenberg RL, Plamann M. Loss of growth polarity and mislocalization of septa in a Neurospora mutant altered in the regulatory subunit of cAMP-dependent protein kinase. EMBO J. 1996;15:5772–5782. [PMC free article] [PubMed] [Google Scholar]
- Castilla R, Passeron S, Cantore ML. N-acetyl-d-glucosamine induces germination in Candida albicans through a mechanism sensitive to inhibitors of cAMP-dependent protein kinase. Cell Signal. 1998;10:713–719. doi: 10.1016/s0898-6568(98)00015-1. [DOI] [PubMed] [Google Scholar]
- Chattaway FW, Wheeler PR, O'Reilly J. Involvement of adenosine 3′:5′-cyclic monophosphate in the germination of blastospores of Candida albicans. J Gen Microbiol. 1981;123:233–40. doi: 10.1099/00221287-123-2-233. [DOI] [PubMed] [Google Scholar]
- Choi W, Dean RA. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell. 1997;9:1973–1983. doi: 10.1105/tpc.9.11.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli J, Field J, Ballester R, Chester N, Young D, Wigler M. Mutational mapping of RAS-responsive domains of the Saccharomyces cerevisiae adenylyl cyclase. Mol Cell Biol. 1990;10:2539–2543. doi: 10.1128/mcb.10.6.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S, Ma P, Cauwenberg L, Winderickx J, Crauwels M, Teunissen A, Nauwelaers D, de Winde JH, Gorwa MF, Colavizza D, Thevelein JM. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signaling in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:3326–3341. doi: 10.1093/emboj/17.12.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C, Makris C, Meloche S, Schröppel K, Röllinghoff M, Dignard D, Thomas DY, Whiteway M. Derepressed hyphal growth and reduced virulence in a VH1 family-related protein phosphatase mutant of the human pathogen Candida albicans. Mol Biol Cell. 1997;8:2539–2551. doi: 10.1091/mbc.8.12.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C, Schroppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas DY, Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFeo-Jones D, Tatchell K, Robinson LC, Sigal IS, Vass WC, Lowy DR, Scolnick EM. Mammalian and yeast ras gene products: biological function in their heterologous systems. Science. 1985;228:179–184. doi: 10.1126/science.3883495. [DOI] [PubMed] [Google Scholar]
- Durrenberger F, Wong K, Kronstad JW. Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc Natl Acad Sci USA. 1998;95:5684–5689. doi: 10.1073/pnas.95.10.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egidy G, Paveto C, Passeron S, Galvagno MA. cAMP levels and in situ measurement of cAMP related enzymes during yeast-to-hyphae transition in Candida albicans. Cell Biol Int Rep. 1990;14:59–68. doi: 10.1016/0309-1651(90)90071-6. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Feng Q, Summers E, Guo B, Fink G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol. 1999;181:6339–6346. doi: 10.1128/jb.181.20.6339-6346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidel PL, Jr, Lynch ME, Sobel JD. Candida-specific cell-mediated immunity is demonstrable in mice with experimental vaginal candidiasis. Infect Immun. 1993;61:1990–1995. doi: 10.1128/iai.61.5.1990-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson IA, Lerner RA, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Gold S, Duncan G, Barrett K, Kronstad J. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 1994;8:2805–16. doi: 10.1101/gad.8.23.2805. [DOI] [PubMed] [Google Scholar]
- Gold SE, Brogdon SM, Mayorga ME, Kronstad JW. The Ustilago maydis regulatory subunit of a cAMP-dependent protein kinase is required for gall formation in maize. Plant Cell. 1997;9:1585–1594. doi: 10.1105/tpc.9.9.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Broek D, Wigler M. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell. 1985;43:493–505. doi: 10.1016/0092-8674(85)90179-5. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Kawamukai M, Ferguson K, Wigler M, Young D. Genetic and biochemical analysis of the adenylyl cyclase of Schizosaccharomyces pombe. Cell Regul. 1991;2:155–64. doi: 10.1091/mbc.2.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays AM, Rowley PS, Baasiri RA, Borkovich KA. Regulation of conidiation and adenylyl cyclase levels by the Galpha protein GNA-3 in Neurospora crassa. Mol Cell Biol. 2000;20:7693–7705. doi: 10.1128/mcb.20.20.7693-7705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler JR, Fink GR. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J, De Maria AD, Funnell D, Laidlaw RD, Lee N, de Sa MM, Ramesh M. Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch Microbiol. 1998;170:395–404. doi: 10.1007/s002030050659. [DOI] [PubMed] [Google Scholar]
- Kubler E, Mosch HU, Rupp S, Lisanti MP. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- Leberer E, Dignard D, Harcus D, Thomas DY, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signaling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Harcus D, Broadbent ID, Clark KL, Dignard D, Ziegelbauer K, Schmidt A, Gow NA, Brown AJ, Thomas DY. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas DY. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr Biol. 1997;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- Leberer, E., Harcus, D., Dignard, D., Johnson, L., Ushinsky, S., Thomas, D.Y., and Schröppel, L. (2001). Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42 (in press). [DOI] [PubMed]
- Lee KL, Buckley HR, Campbell CC. An Amino Acid Liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Leuker CE, Sonneborn A, Delbruck S, Ernst JF. Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene. 1997;192:235–240. doi: 10.1016/s0378-1119(97)00069-3. [DOI] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog [published erratum appears in Science 1995 Jan 6;267(5194):17] Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Pan X, Harashima T, Cardenas ME, Xue Y, Hirsch JP, Heitman J. The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losberger C, Ernst JF. Sequence of the Candida albicans gene encoding actin. Nucleic Acids Res. 1989;17:9488. doi: 10.1093/nar/17.22.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee BB, Magee PT. WO-2, a stable aneuploid derivative of Candida albicans strain WO-1, can switch from white to opaque and form hyphae. Microbiology. 1997;143:289–295. doi: 10.1099/00221287-143-2-289. [DOI] [PubMed] [Google Scholar]
- Mallet L, Renault G, Jacquet M. Functional cloning of the adenylate cyclase gene of Candida albicans in Saccharomyces cerevisiae within a genomic fragment containing five other genes, including homologues of CHS6 and SAP185. Yeast. 2000;16:959–966. doi: 10.1002/1097-0061(200007)16:10<959::AID-YEA592>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Mann DJ, Campbell DG, McGowan CH, Cohen PT. Mammalian protein serine/threonine phosphatase 2C: cDNA cloning and comparative analysis of amino acid sequences. Biochim Biophys Acta. 1992;1130:100–104. doi: 10.1016/0167-4781(92)90471-b. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Uno I, Oshima Y, Ishikawa T. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1982;79:2355–2359. doi: 10.1073/pnas.79.7.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonyi K, Beullens M, Detremerie K, Geerts L, Thevelein JM. Requirement of one functional RAS gene and inability of an oncogenic ras variant to mediate the glucose-induced cyclic AMP signal in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:3051–3057. doi: 10.1128/mcb.8.8.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead DA, Szczesna-Skorupa E, Kemper B. Single-stranded DNA ‘blue’ T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986;1:67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Mitchell AP. Dimorphism and virulence in Candida albicans. Curr Opin Microbiol. 1998;1:687–92. doi: 10.1016/s1369-5274(98)80116-1. [DOI] [PubMed] [Google Scholar]
- Niimi M. [Glucose effect in Candida albicans] Fukuoka Igaku Zasshi. 1984;75:356–365. [PubMed] [Google Scholar]
- Niimi M, Niimi K, Tokunaga J, Nakayama H. Changes in cyclic nucleotide levels and dimorphic transition in Candida albicans. J Bacteriol. 1980;142:1010–1014. doi: 10.1128/jb.142.3.1010-1014.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S, O'Neill K, Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Ryley JF, McGregor S. Quantification of vaginal Candida albicans infections in rodents. J Med Vet Mycol. 1986;24:455–460. [PubMed] [Google Scholar]
- Sabie FT, Gadd GM. Effect of nucleosides and nucleotides and the relationship between cellular adenosine 3′:5′-cyclic monophosphate (cyclic AMP) and germ tube formation in Candida albicans. Mycopathologia. 1992;119:147–156. doi: 10.1007/BF00448812. [DOI] [PubMed] [Google Scholar]
- Sobel JD, Muller G, McCormick JF. Experimental chronic vaginal candidosis in rats. Sabouraudia. 1985;23:199–206. doi: 10.1080/00362178585380301. [DOI] [PubMed] [Google Scholar]
- Sonneborn A, Bockmuhl DP, Gerads M, Kurpanek K, Sanglard D, Ernst JF. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol Microbiol. 2000;35:386–396. doi: 10.1046/j.1365-2958.2000.01705.x. [DOI] [PubMed] [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PA, Yin CY, Molloy C, Templeton MD, Shepherd MG. An analysis of the metabolism and cell wall composition of Candida albicans during germ-tube formation. Can J Microbiol. 1983;29:1514–1525. doi: 10.1139/m83-233. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Choe HR, Nishida Y, Yamawaki-Kataoka Y, Ohnishi S, Tamaoki T, Kataoka T. Leucine-rich repeats and carboxyl terminus are required for interaction of yeast adenylate cyclase with RAS proteins. Proc Natl Acad Sci USA. 1990;87:8711–8875. doi: 10.1073/pnas.87.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Lynch KR, Larner J, Fox J, Yasui A, Kikuchi K, Suzuki Y, Tsuiki S. Molecular cloning of rat type 2C (IA) protein phosphatase mRNA. Proc Natl Acad Sci USA. 1989;86:1796–800. doi: 10.1073/pnas.86.6.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WJ, Gilman AG. Adenylyl cyclases. Cell. 1992;70:869–872. doi: 10.1016/0092-8674(92)90236-6. [DOI] [PubMed] [Google Scholar]
- Tatchell K, Robinson LC, Breitenbach M. RAS2 of Saccharomyces cerevisiae is required for gluconeogenic growth and proper response to nutrient limitation. Proc Natl Acad Sci USA. 1985;82:3785–3789. doi: 10.1073/pnas.82.11.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi HF, Flawia MM, Tellez-Inon MT, Torres HN. Control of Neurospora crassa morphology by cyclic adenosine 3′, 5′- monophosphate and dibutyryl cyclic adenosine 3′, 5′-monophosphate. J Bacteriol. 1976;126:91–99. doi: 10.1128/jb.126.1.91-99.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi HF, Jorge JA, Roselino JE, Migliorini RH. Adenylyl cyclase deficient cr-1 (Crisp) mutant of Neurospora crassa: cyclic AMP-dependent nutritional deficiencies. Arch Microbiol. 1979;123:251–258. doi: 10.1007/BF00406658. [DOI] [PubMed] [Google Scholar]
- Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsalpha.GTPgammaS. Science. 1997;278:1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Timpel C, Strahl-Bolsinger S, Ziegelbauer K, Ernst JF. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J Biol Chem. 1998;273:20837–20846. doi: 10.1074/jbc.273.33.20837. [DOI] [PubMed] [Google Scholar]
- Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Whiteway M. Transcriptional control of cell type and morphogenesis in Candida albicans. Curr Opin Microbiol. 2000;3:582–588. doi: 10.1016/s1369-5274(00)00144-2. [DOI] [PubMed] [Google Scholar]
- Xue Y, Batlle M, Hirsch JP. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelada A, Passeron S, Lopes Gomes S, Cantore ML. Isolation and characterization of cAMP-dependent protein kinase from Candida albicans. Purification of the regulatory and catalytic subunits. Eur J Biochem. 1998;252:245–252. doi: 10.1046/j.1432-1327.1998.2520245.x. [DOI] [PubMed] [Google Scholar]