ABSTRACT
The genus Lactobacillus is a widespread taxon, members of which are highly relevant to functional and fermented foods, while they are also commonly present in host-associated gut and vaginal microbiota. Substantial efforts have been undertaken to disclose the genetic repertoire of all members of the genus Lactobacillus, and yet their species-level profiling in complex matrices is still undeveloped due to the poor phylotype resolution of profiling approaches based on the 16S rRNA gene. To overcome this limitation, an internal transcribed spacer (ITS)-based profiling method was developed to accurately profile lactobacilli at the species level. This approach encompasses a genus-specific primer pair combined with a database of ITS sequences retrieved from all available Lactobacillus genomes and a script for the QIIME software suite that performs all required steps to reconstruct a species-level profile. This methodology was applied to several environments, i.e., human gut and vagina and the ceca of free-range chickens, as well as whey and fresh cheese. Interestingly, the data collected confirmed a relevant role of lactobacilli present in functional and fermented foods in defining the population harbored by the human gut, while, unsurprisingly perhaps, the ceca of free-range chickens were observed to be dominated by lactobacilli characterized in birds living in natural environments. Moreover, vaginal swabs confirmed the existence of previously hypothesized community state types, while analysis of whey and fresh cheese revealed a dominant presence of single Lactobacillus species used as starters for cheese production. Furthermore, application of this ITS profiling method to a mock Lactobacillus community allowed a minimal resolution level of <0.006 ng/μl.
IMPORTANCE The genus Lactobacillus is a large and ubiquitous taxon of high scientific and commercial relevance. Despite the fact that the genetic repertoire of Lactobacillus species has been extensively characterized, the ecology of this genus has been explored by metataxonomic techniques that are accurate down to the genus or phylogenetic group level only. Thus, the distribution of lactobacilli in environmental or processed food samples is relatively unexplored. The profiling protocol described here relies on the use of the internal transcribed spacer to perform an accurate classification in a target population of lactobacilli with a <0.006-ng/μl sensitivity. This approach was used to analyze five sample types collected from both human and animal host-associated microbiota, as well as from the cheese production chain. The availability of a tool for species-level profiling of lactobacilli may be highly useful for both academic research and a wide range of industrial applications.
KEYWORDS: Lactobacillus, microbiota, ITS, next-generation sequencing, Illumina
INTRODUCTION
The genus Lactobacillus is a widespread and diverse taxon encompassing more than 170 species and 17 subspecies, which are classified as Gram-positive, non-spore-forming, and catalase-negative facultative anaerobes (1, 2). Moreover, based on their metabolic capability to produce lactic acid as the main metabolic end product of carbohydrate fermentation, lactobacilli are classified as members of the lactic acid bacteria. Notably, 16S rRNA gene-based phylogenetic analyses revealed the existence of 22 distinct phylogenetic groups of Lactobacillus species (24 when including pediococci) (2–4).
Regarding their ecological distribution, lactobacilli are found in a wide range of environments, including plants, water, soil, and silage, and at different body sites of humans and other animals as members of host-associated microbiomes, such as those colonizing the oral cavity, the vagina, and the gastrointestinal tract (4, 5). Moreover, 37 species of this genus have been granted the qualified presumption of safety (QPS) status by the European Food Safety Authority (6). Thus, they are extensively used in the food industry, in particular in fermented foods, due to their high performance in lactic acid fermentation coupled with a high tolerance for low pH, preservative and organoleptic properties, and the production of exopolysaccharides that contribute to the texture of foods (2, 7). In this context, members of the genus Lactobacillus have in recent years gained significant scientific and commercial interest as health-promoting microorganisms, as evidenced by the fact that 22 species encompass strains patented as probiotics in Europe (8).
The high commercial and scientific relevance of lactobacilli coupled with the recent introduction of next-generation sequencing technologies has recently led to genome decoding of all (then) known Lactobacillus species (3, 7). The retrieved genomic data have been exploited for comparative genomic analyses and has allowed the identification of many shared or distinct genetic features of this genus. Furthermore, this genomic information has permitted the reconstruction of their metabolic potential, has shed light on host-microbe interactions (such as adhesion to the mucus layer and modulation of the immune system of the host), and has revealed particular microbe-microbe interactions with other commensals or (opportunistic) pathogens (1, 7, 8).
Despite the large body of data concerning the physiology and genetics of lactobacilli, knowledge of the ecology and distribution in environmental or host-associated niches of individual species relies mainly on culture-dependent studies. This is partly due to the resolution limit of currently used metagenomic approaches. Although microbial profiling based on a partial 16S rRNA gene can discriminate between phylogenetic groups of lactobacilli due to the high phylogenetic diversity of this genus, it cannot provide an accurate species-level resolution. Moreover, the majority of the current exiting studies of lactobacillus populations based on 16S rRNA gene profiling do not even perform phylogenetic group-level analyses. To offer a more refined taxonomic view of lactobacilli in a given environment or sample, we developed a profiling approach based on amplification of the internal transcribed spacer (ITS) sequence. Notably, due to their high variability, ITS sequences have previously been exploited in a wide range of studies encompassing the identification of unique species-specific restriction patterns of lactobacilli, as well as the identification and characterization of Leuconostoc strains and for the genotyping of Streptococcus pneumoniae strains (9–11). The developed methodology in the current study is able to determine the composition of lactobacillus-containing communities down to the species level. The method was validated through the analysis of a sample artificially constituted by DNA of 14 lactobacillus taxa at known concentrations. Furthermore, we applied this methodology for the precise investigation of bacterial communities harbored by human-, animal-, and food-associated matrices that were previously explored down to the genus level only.
RESULTS AND DISCUSSION
Analysis of ITS variability within the Lactobacillus genus.
The genomes of 1,523 strains assigned to the genus Lactobacillus and corresponding to 176 species were retrieved from the NCBI genome database and then processed using the MEGAnnotator software (12) to predict rRNA genes in order to ensure the same high-quality standard for all sequences of ribosomal loci included in this study (see Table S1 in the supplemental material). Notably, the genomic sequences of 892 Lactobacillus strains, representing the 58.6% of the total strain pool analyzed, did not harbor complete rRNA loci, i.e., encompassing complete 5S, 16S, and 23S rRNA genes. In contrast, at least one complete ribosomal rRNA gene locus was identified for 631 of the 1,523 analyzed strains, corresponding to 70 species, and a custom script was then used to extract a total of 1,788 ITS sequences. Assembly of draft genomes generally generates the collapse of reads that correspond to rRNA genes into a single rRNA locus. However, availability of multiple draft sequences of a given Lactobacillus taxon, complemented with analysis of 217 complete genomes of Lactobacillus species, allowed us to retrieve an average of 25.5 ITS sequences per species. Interestingly, 137 of the 1,788 retrieved ITS sequences include stretches of >3 undefined (N) nucleotides, thus highlighting a high rate of assembly-related issues and/or low-quality regions in genomes deposited in the NCBI genome database. Comparative analysis of the 1,651 complete ITS sequences without multiple contiguous nucleotide ambiguities revealed that 92.5% of the ITS sequences range between 200 and 500 bp.
As previously observed for bifidobacteria (13, 14), alignment of ITS sequences from Lactobacillus genomes shows a high level of diversity, probably due to a high mutation frequency, and corresponding to a high evolutionary rate, as reflected by multiple substitutions at a given nucleotide position and indicative of mutational saturation of such ITS sequences. While this particularly high mutation frequency prevents phylogeny inference (15), it is suitable for metagenomic amplicon-based profiling below the genus level, as previously validated for members of the genus Bifidobacterium (13).
Design of a PCR primer pair for ITS profiling of the Lactobacillus genus.
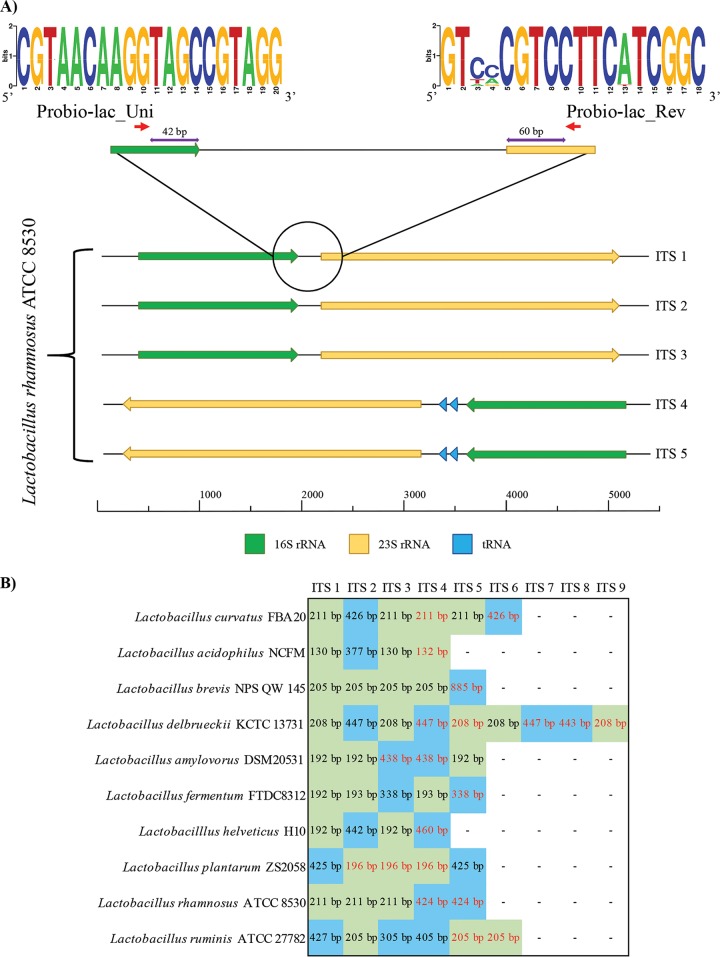
Many profiling approaches have been developed to accurately reconstruct the taxonomic composition of complex bacterial communities. These include methods based on low-coverage sequencing of full-length 16S rRNA genes and the use of technologies providing long reads, i.e., Sanger and PacBio. Nevertheless, despite the fact that full-length sequencing of the 16S rRNA gene allows high accuracy in taxonomic assignment, the low sequencing coverage permits the detection only of dominant taxa and prevents profiling of bacteria present at a low relative abundance in a given population (10). Furthermore, the use of alternative marker genes has also been proposed, though their use remains limited due to difficulties in the definition of universal primers, as well as in the lack of a complete reference database. The advent of next-generation sequencing, characterized by high coverage and short reads, facilitated the amplification and sequencing of partial 16S rRNA genes, i.e., 16S rRNA gene profiling. This metagenomic method has in recent years been used as the gold standard for taxonomic characterization of environmental and host-associated microbiomes. While this methodology covers all bacterial biodiversity, it is generally only accurate for the reconstruction of taxonomic profiles down to genus level (16) or down to phylogenetic groups in the case of genera with a high level of phylogenetic diversity, e.g., the genus Lactobacillus (3, 4) since it relies on sequencing of a small region of the whole 16S rRNA gene through next-generation sequencing. To overcome this limitation and to obtain species-level resolution, the use of the ITS sequence as an alternative molecular marker has been proposed (13). In order to develop a universal primer pair suitable for profiling of all members of the Lactobacillus genus, we aligned the 16S and 23S rRNA genes flanking the 1,651 complete ITS sequences without stretches of undefined nucleotides that were retrieved from lactobacillus genomes deposited at the NCBI database. Manual inspection of the alignments allowed the identification of “universal” primers located at the 5′ end of the 16S rRNA gene and at the 3′ end of the 23S rRNA gene, i.e., Probio-lac_Uni (CGTAACAAGGTAGCCGTAGG) and Probio-lac_Rev (GTYVCGTCCTTCWTCGSC), respectively (Fig. 1). Sequence conservation among the aligned 16S and 23S rRNA genes is reported in Fig. 1 through a WebLogo representation. These primers generate an amplicon of an average length of 380 bp covering the complete ITS region and suitable for 2×250-bp paired-end Illumina sequencing, followed by single-end bioinformatic analysis of both paired reads (see below). Analysis of single-end reads provided reliable assignment to species level even in cases where a tRNA gene was located within the ITS region (see below). Notably, the final sequence of the primers was defined after multiple iterative alignments to the Silva SSU and LSU databases (17) using the Silva TestProbe v3.0 tool (https://www.arb-silva.de/search/testprobe/). The latter approach led to the introduction of specific IUPAC bases in order to maximize alignment of the primers to all currently available 16S and 23S rRNA gene sequences of lactobacilli corresponding to all known species of this genus, while minimizing alignment to non-Lactobacillus rRNA genes. The usefulness of the Probio-lac_Uni/Probio-lac_Rev primer pair was validated in vitro through successful amplicon generation in the case of 31 lactobacillus species belonging to the 23 phylogenetic groups identified previously in the genus Lactobacillus (3, 4) (see Fig. S1 in the supplemental material). In contrast, no amplification was observed when the Probio-lac_Uni/Probio-lac_Rev primer pair was used to amplify DNA extracted from nine non-Lactobacillus taxa (see Fig. S1 in the supplemental material). Interestingly, for all tested lactobacilli, we observed two PCR fragments, each with a molecular size ranging from 300 to 350 bp and from 500 to 550 bp, corresponding to the ITS region with and without a tRNA gene (see below for details), respectively (Fig. 1; see Fig. S1 in the supplemental material). Such ITS patterns confirmed those displayed in previous studies targeting the amplification of the ITS region of lactobacilli (18). Notably, for few taxa we observed a faint amplification fragment of 500 to 550 bp, which might suggest a lower copy number of ITS regions encompassing tRNA genes in the same genome.
FIG 1.
Genetic map of the ITS region of Lactobacillus with and without tRNA genes. (A) Genetic organization of the five complete ITS regions predicted in the complete genome of Lactobacillus rhamnosus ATCC 8530, used here as a test case. Primer sequence conservation is shown through a WebLogo representation where the overall height of the stacks indicates the sequence conservation at that position, while the height of symbols within the stacks indicates the relative frequency of nucleic acids at that position. (B) Details of ITS regions identified in the complete genomes of species included in the mock sample for which a complete genome was available. ITS sequences without tRNA genes are highlighted in green, while ITS regions harboring tRNA genes are indicated in blue. Black and red text indicates forward and reverse strand orientations, respectively.
The Probio-lac_Uni/Probio-lac_Rev primer pair was used for in silico PCR amplification of the 631 genomes of the genus Lactobacillus encoding at least one rRNA gene locus. This approach facilitated the development of a database encompassing 1,651 complete ITS sequences without multiple ambiguous nucleotides and flanked by partial 16S and 23S rRNA sequences, together constituting the Lactobacillus ITS Amplicon database (LITSA database).
Cross-alignment of all retrieved LITSA sequences using MatGAT software (19) was performed in order to evaluate the level of identity between predicted amplicons (see Table S2 in the supplemental material) and to evaluate possible limits imposed by actual lactobacillus taxonomy to the proposed ITS profiling methodology. Notably, this analysis highlighted cases in which the comparison of multiple LITSA sequences from the same strain showed low identity. In-depth investigation revealed that 46 of the 62 lactobacillus species included in the LITSA database contain at least one ITS sequence that harbors two tRNA genes (for alanine and isoleucine) (Fig. 1). Notably, despite the fact that this prediction is limited due to the small number of complete genomes available, the presence of tRNA genes in one or multiple rRNA loci appears to be a common feature of genomes from members of the Lactobacillus genus.
Furthermore, cross-alignment analysis also revealed that the majority of the 62 Lactobacillus species, for which a complete LITSA sequence was available, can be discriminated (see Table S2 in the supplemental material), with the exception of putatively misclassified strains and/or species (see below). In this context, despite the fact that lactobacilli are known to possess a very high level of phylogenetic diversity (3, 4), strains corresponding to 18 species showed an average LITSA sequence identity of >97% with at least one other Lactobacillus species, thus showing a very close phylogenetic relationship between such taxa (Table 1). Among the lactobacilli, Lactobacillus casei and Lactobacillus paracasei strains possess an average LITSA sequence identity of 99%, while the amplicon sequences of Lactobacillus pentosus, Lactobacillus plantarum, and Lactobacillus paraplantarum strains show up to 100% identity (see Table S2 in the supplemental material). An in-depth analysis of each strain revealed that 23 of the 25 strains classified as L. casei share an average LITSA sequence identity of ≤96.1% with the type strain L. casei ATCC 393, while the average identity with the type strain L. paracasei ATCC 394 is ≥99.7% (see Table S2 in the supplemental material). In contrast, the putative lactobacillus species Lactobacillus sp. strain FMNP02 shares 99.7% identity with L. casei ATCC 393 (see Table S2 in the supplemental material), thus representing a possible misclassification of the latter strain.
TABLE 1.
Lactobacillus species with a LITSA sequence identity of ≥97% with another Lactobacillus speciesa
| Species | LITSA % identity with the closest species | Closest species |
|---|---|---|
| Lactobacillus acidophilus | 98 | Lactobacillus amylovorus |
| 97 | Lactobacillus crispatus | |
| Lactobacillus amylovorus | 98 | Lactobacillus acidophilus |
| 97 | Lactobacillus crispatus | |
| Lactobacillus buchneri | 99 | Lactobacillus parabuchneri |
| Lactobacillus casei | 99 | Lactobacillus paracasei |
| 100 | Lactobacillus rhamnosus | |
| Lactobacillus crispatus | 97 | Lactobacillus acidophilus |
| 97 | Lactobacillus amylovorus | |
| Lactobacillus curvatus | 98 | Lactobacillus sakei |
| Lactobacillus gallinarum | 99 | Lactobacillus helveticus |
| Lactobacillus gasseri | 99 | Lactobacillus johnsonii |
| Lactobacillus helveticus | 99 | Lactobacillus gallinarum |
| Lactobacillus johnsonii | 99 | Lactobacillus gasseri |
| Lactobacillus parabuchneri | 99 | Lactobacillus buchneri |
| Lactobacillus paracasei | 99 | Lactobacillus casei |
| 100 | Lactobacillus rhamnosus | |
| Lactobacillus paraplantarum | 100 | Lactobacillus pentosus |
| 100 | Lactobacillus plantarum | |
| Lactobacillus pentosus | 100 | Lactobacillus paraplantarum |
| 99 | Lactobacillus plantarum | |
| Lactobacillus plantarum | 100 | Lactobacillus paraplantarum |
| 99 | Lactobacillus pentosus | |
| Lactobacillus rhamnosus | 100 | Lactobacillus paracasei |
| 100 | Lactobacillus casei | |
| Lactobacillus sakei | 98 | Lactobacillus curvatus |
The percentage reported corresponds to the highest identity observed among all LITSA sequences identified in strains of the two species compared.
In our attempts to obtain insights into the phylogeny of L. pentosus, L. plantarum, and L. paraplantarum, we observed an average LITSA sequence identity of 98.9% between L. pentosus and L. plantarum strains (see Table S2 in the supplemental material). Moreover, the two strains of L. paraplantarum, for which we were able to predict an rRNA gene locus, show an average LITSA identity of 99.5% with L. plantarum strains (see Table S2 in the supplemental material), indicating that such taxa may belong to the same species and therefore cannot be discriminated using metataxonomic techniques. Nevertheless, evaluation of the average nucleotide identity is needed to confirm this hypothesis. Furthermore, we could not retrieve an in silico Probio-lac_Uni/Probio-lac_Rev-corresponding amplicon for the type strains of L. pentosus and L. paraplantarum due to absence of a complete ITS region in the deposited genomes, and we were therefore unable to evaluate their amplicon identity with the LITSA sequences of L. plantarum strains.
Notably, these observations suggest that major issues in the classification of the genus Lactobacillus still exist, resulting in the unfeasibility of distinguishing a number of species through ITS profiling. Thus, as has been proposed previously, it is desirable that a reevaluation of the taxonomy of lactobacilli is undertaken based on a phylogenomic approach (20, 21), as was also corroborated by several recent studies (3, 4, 7).
Development of a bioinformatic tool for ITS profiling of the Lactobacillus genus.
The length of the amplicon produced by the Probio-lac_Uni/Probio-lac_Rev primer pair may exceed 600 bp, particularly when tRNA-containing sequences are present in the ITS sequence. Thus, sequencing produced nonoverlapping paired-end reads even with the maximum length obtainable using next-generation Illumina sequencing, i.e., 2×250-bp paired-end reads, using MiSeq reagent kit (v3, 600 cycles) chemistry. Nevertheless, each forward and reverse read covers 42 and 60 nucleotides corresponding to the 16S rRNA gene 3′ end and the 23S rRNA gene 5′ end, respectively, which are followed by 190 to 208 bp of hypervariable ITS sequence suitable for profiling at species level (Fig. 1). Thus, we developed a package for QIIME software suite v1.9.1 (22) that encompasses the LITSA database and a bash script for analysis of both forward and reverse reads of the Lactobacillus ITS profiling data (http://probiogenomics.unipr.it/sw/Lactobacilli_ITS_profiling_tool.zip). Notably, the LITSA database will be updated regularly to include additional ITS sequences as new lactobacillus genome sequences become available, thus increasing the number of Lactobacillus species that can be profiled. The script performs quality filtering, de novo operational taxonomic unit (OTU) clustering at 100% identity, and taxonomic classification of OTU reference sequences through an RDP classifier with a confidence level of 0.80. Notably, these cutoff values permit the discrimination of closely related taxa. Due to the average size of the amplicon, the paired-end reads are not joined prior to classification. Instead, the script analyzes both the forward and the reverse reads together and provides an average profile.
Notably, the different number of rRNA loci predicted in the genomes of Lactobacillus species may generate biases in the retrieved profiles. Thus, we evaluated the average number of ITS regions present in the 217 available complete Lactobacillus genomes. This analysis provided data for the normalization of 45 of the 62 species of lactobacilli for which a LITSA sequence could be retrieved. Moreover, the average number of rRNA gene loci of the remaining 17 species with only draft genomes was set at 5.6, i.e., the average obtained for all the species with at least a complete genome. Notably, the Lactobacillus ITS profiling analysis script includes a normalization step based on the number of rRNA gene loci predicted for all the 62 Lactobacillus species for which a LITSA sequence could be retrieved. The output produced by the script is summarized in the “output” folder, which contains the predicted taxonomic profile based on the LITSA database (both non-normalized and normalized for the number of rRNA loci) and the OTU table in tabular text format that reports the reference sequence and associated taxonomy. All Lactobacillus ITS profiles reported in the manuscript correspond to the average between forward and reverse read profiles after normalization for the number of predicted rRNA gene loci.
Assessing detection sensitivity and accuracy using the Lactobacillus ITS profiling protocol.
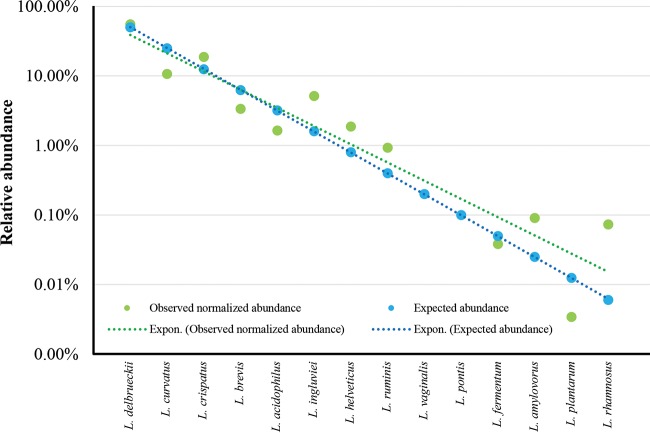
In order to provide an evaluation of the sensitivity and accuracy of the Probio-lac_Uni/Probio-lac_Rev primer pair, 14 Lactobacillus type strains were used to artificially compose a mock community (see Table S3 in the supplemental material). The DNA extracted from each taxon grown in pure culture was added to the mix at known amounts, ranging from 0.006 to 50 ng of DNA, corresponding to 0.006 to 50% of the total DNA pool (Fig. 2). Sequencing of the mock sample was performed using an Illumina MiSeq with 2×250-bp chemistry, producing 45,146 quality-filtered paired-end reads. Interestingly, Lactobacillus ITS profiling of this data set successfully profiled all Lactobacillus species included in this sample, except Lactobacillus vaginalis and Lactobacillus pontis, for which we could not retrieve a LITSA sequence from analysis of the available genome sequences (Fig. 2). Thus, even though the Probio-lac_Uni/Probio-lac_Rev primer pair produces an amplicon for these species, the latter cannot be taxonomically classified due to the absence of L. vaginalis and L. pontis in the present version of the LITSA database. This is a temporary limitation, and the LITSA database will be updated regularly to include LITSA sequences of newly sequenced genomes in order to cover all the Lactobacillus species that currently cannot be profiled. Moreover, comparison of the retrieved profile with the expected composition revealed a strong correlation for each taxon with few discrepancies (Fig. 2). The causes of such differences between expected and observed relative abundances may be imputed to the lack of sufficient information in the LITSA database, at this time, regarding the average number of rRNA loci per genome used for normalization of the ITS profiling data.
FIG 2.
Evaluation of the sensitivity and accuracy of the Lactobacillus ITS profiling protocol. The graph shows the expected and observed relative abundances of 14 Lactobacillus taxa constituting an artificial sample. An exponential trendline is reported for the expected and observed data.
Furthermore, since PCR amplicon size has been identified as a source of bias in ITS-based profiling studies of fungi (23), we evaluated the presence of possible biases introduced by amplification of lactobacillus ITS sequences of different lengths due to the presence or absence of tRNA genes (see above). The 14 Lactobacillus species that constitute the mock community (see Table S3 in the supplemental material) were subjected to manual characterization of the corresponding rRNA loci. Notably, the 10 species for which a complete genome was available confirmed what had been observed for the in vitro PCR, i.e., presence of longer ITS sequences that encompass two tRNA genes (Fig. 1; see also Fig. S1 in the supplemental material). Interestingly, the different intensities observed in the PCR fragments, i.e., 300 to 350 bp and 500 to 550 bp (see Fig. S1 in the supplemental material), did not influence the expected relative abundance of the mock community (Fig. 2). Notably, detection of Lactobacillus rhamnosus, whose concentration in the mock community is 0.006 ng/μl, indicates that the limit of detection of the lactobacillus ITS profiling is <0.006 ng/μl, corresponding to 1.85 × 103 cells/μl.
Validation of the Lactobacillus ITS profiling protocol through analysis of samples from multiple environments.
Lactobacillus is a highly diverse microbial genus, members of which are found in a wide range of environments (5). To perform a comprehensive testing of the performances of the Lactobacillus ITS profiling protocol, we analyzed a total of 25 samples encompassing five human fecal samples, five human vaginal swab samples, five free-range chicken cecal samples, five whey samples, and five parmesan cheese samples (see Table S4 in the supplemental material). Sequencing was performed with an Illumina MiSeq instrument using 2×250-bp chemistry, producing an average of 15,529 forward and 15,293 reverse quality-filtered reads per sample (see Table S4 in the supplemental material).
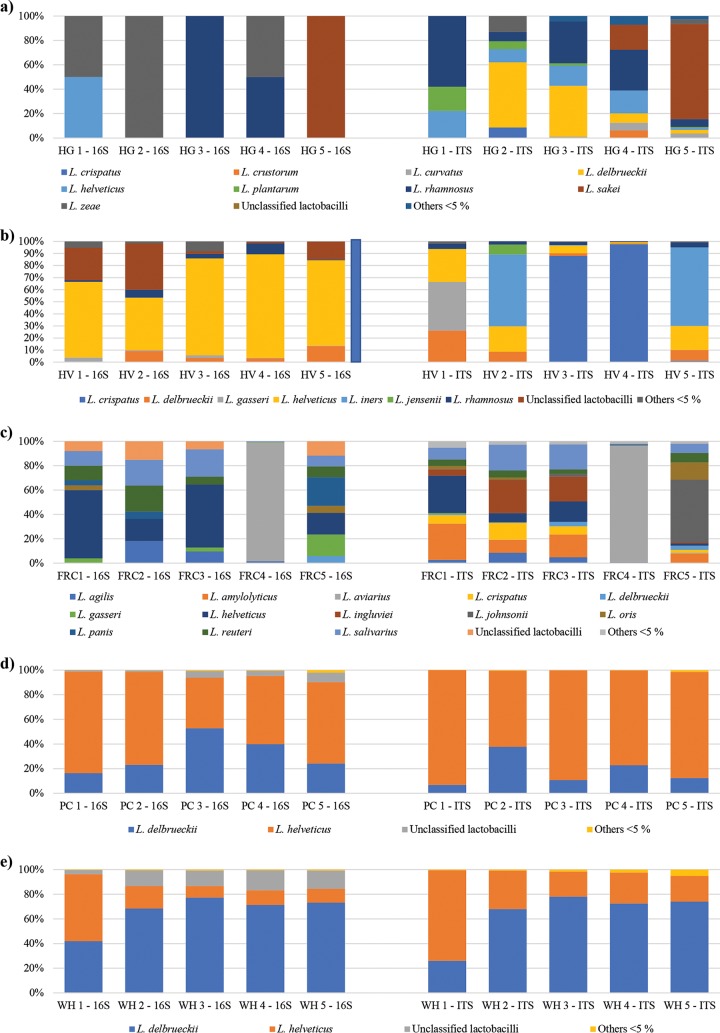
Interestingly, analysis of the human fecal samples revealed the presence of human gut colonizers, such as Lactobacillus rhamnosus, along with a range of lactobacilli used in functional or fermented foods that are typically part of the human diet, such as L. plantarum, Lactobacillus helveticus, Lactobacillus delbrueckii, and Lactobacillus sakei (Fig. 3).
FIG 3.
ITS and 16S rRNA gene profiling of Lactobacillus species in five ecological niches. The profiles of the Lactobacillus population obtained for five human fecal samples (HG) (a), five human vaginal swab samples (HV) (b), five free-range chicken fecal samples (FRC) (c), five parmesan cheese samples (PC) (d), and five whey samples (WH) (e) are depicted in the corresponding bar plots. Only species with a relative abundance of >5% in at least a sample are reported. Species below 5% are collapsed in “Others < 5%”.
Moreover, the obtained profiles of the five human vaginal swab samples confirmed the proposed existence of community state types (CSTs) of the vaginal microbiota dominated by specific Lactobacillus taxa (24, 25). In fact, HV1 is dominated by Lactobacillus gasseri, while Lactobacillus iners and Lactobacillus crispatus are the most abundant Lactobacillus taxa in HV2/HV5 and HV3/HV4, respectively (Fig. 3). Furthermore, in all five reconstructed human vaginal profiles, L. helveticus is the second most abundant Lactobacillus species, as observed in the aforementioned CSTs (24, 25) (Fig. 3). Thus, based on the classification proposed by DiGiulio et al. (24), HV1 can be classified as a CST 2, while HV2/HV5 falls within the CST 3, and HV3/HV4 can be attributed to CST 1.
To demonstrate the relevance of an efficient methodology for precise cataloguing of the Lactobacillus species for which a complete LITSA sequence is available in different environments, we analyzed five cecal samples from free-range chickens. The retrieved profiles revealed a high relative abundance (ranging from a total of 53.1 to 96.8%) of Lactobacillus species previously characterized in poultry or other birds, such as Lactobacillus salivarius, Lactobacillus reuteri, Lactobacillus ingluviei, Lactobacillus amylovorus, Lactobacillus agilis, Lactobacillus aviarius, and Lactobacillus johnsonii (26–33) (Fig. 3). Notably, samples FRC1, FRC2, and FRC3 showed a similar profile with a high abundance of L. salivarius, L. ingluviei, and L. amylovorus, reflecting the fact that they were kept in the same hen house (Fig. 3). Accordingly, samples FRC4 and FRC5, collected in two additional hen houses, showed different profiles characterized by high abundance of L. aviarius and L. johnsonii, respectively (Fig. 3).
For milk and milk-related products, profiling of five whey and five fresh parmesan cheese (at 1 day of ripening) samples revealed, as expected, similar profiles dominated by L. helveticus and L. delbrueckii (Fig. 3), which represent two Lactobacillus species typically used as starter cultures for the production of cheese (34). These data indicate that the Lactobacillus ITS profiling approach also represents a valuable tool for monitoring the population of lactobacilli across the cheese production chain.
Results obtained from ITS profiling were also compared to profiles reconstructed through analysis of OTUs generated at 99% identity from 16S rRNA profiling data (Fig. 3; see also Table S5 in the supplemental material). Notably, only OTUs classified as lactobacilli have been included in the representation; thus, the relative abundance of unclassified lactobacilli reported in the bar plot does not include additional OTUs that could not be attributed to this genus. Moreover, lactobacillus species whose relative abundance is below 5% in each sample were collapsed under “Others < 5%” in the bar plot representation. Interestingly, the ITS profiling approach provided a more accurate species-level reconstruction of the lactobacillus populations when used to analyze human fecal and vaginal samples, as well as free-range chicken fecal samples. Moreover, it confirmed and partially improved the simple lactobacillus community of whey and fresh parmesan cheese samples observed through 16S rRNA gene profiling. In fact, differences in the profiles obtained through 16S rRNA gene and ITS profiling can be observed in all cases (Fig. 3; see also Table S5 in the supplemental material). Such differences are caused by the limited number of Lactobacillus species that could be discriminated based on partial16S rRNA gene sequence respect to the ITS sequence (Fig. 3; see also Table S5 in the supplemental material). Altogether, these results confirm the performance of the Lactobacillus ITS profiling protocol observed from analysis of the artificial sample and validate its use, complementary to 16S rRNA gene profiling, for analysis of a wide range of complex environmental and host-associated matrices.
Conclusions.
We developed a newly designed method for characterization of the Lactobacillus population in complex environments based on the use of the ITS, which represents a hypervariable region located between the 16S and 23S rRNA genes that allows high-accuracy species-level profiling. The accuracy and sensitivity of this method allowed profiling of complex communities of lactobacilli with a successful identification of taxa with an abundance of 1.85 × 103 cells/μl, which is even lower than what was previously identified for a similar approach developed for the profiling of bifidobacterial communities (13). Notably, despite the fact that the current LITSA database allows the precise profiling of just 62 species, the ITS profiling approach represents a new metagenomic tool for species-level profiling of complex lactobacillus communities that complements phylogenetic group assignments that can be obtained from 16S rRNA gene profiling data. Moreover, the database will be regularly updated to represent additional Lactobacillus species as additional genomes encompassing complete LITSA sequences become available. When the ITS lactobacillus profiling method was applied to different biological samples encompassing stools from humans, as well as birds, vaginal swabs, and cheese, it allowed the reconstruction of the cataloguing of lactobacillus communities residing in these environments. Altogether, these results highlight that ITS-mediated profiling of populations of lactobacilli could be useful not only for academic purposes but also for industrial applications such as tracing the microbial composition of probiotic products based on lactobacilli, as well as of starter cultures in food manufacturing.
MATERIALS AND METHODS
Sample collection.
In the framework of a more extensive bacterial cataloguing project, this study included stool, vaginal swab, fresh parmesan cheese (1 day of ripening), whey, and cecal (from free-range chickens) samples.
Five fresh stool samples obtained from human healthy volunteers and five cecal samples retrieved from free-range chickens were immediately frozen upon collection at −80°C until processing for DNA extraction. The DNA extraction was performed using the QIAamp DNA stool minikit according to the manufacturer's instructions (Qiagen, Manchester, UK). In addition, five vaginal swab samples were collected in sterile tubes containing 1 ml of DNA-RNA shield from ZYMO Research until bacterial DNA extraction using ZymoBIOMICS DNA miniprep kit (ZYMO Research). Furthermore, 10-ml samples of whey and 2 to 4 g of fresh parmesan cheese were collected in sterile tubes, and the DNA was extracted using a DNeasy mastitis minikit (Qiagen, Ltd., Strasse, Germany) according to the manufacturer's instructions. Notably, whey samples and cheese samples at 1 day of ripening were collected from the same parmesan cheese producer in Parma, Italy.
Ethical statement.
This study was carried out in accordance with the recommendations of the ethical committee of the University of Parma and was approved by the Comitato di Etica Università degli Studi di Parma, Parma, Italy. All animal procedures were performed according to national guidelines (decreto legislativo 26/2014).
Bacterial growth conditions and DNA extraction.
Type strains of several Lactobacillus taxa (see Table S3 in the supplemental material) were growth in de Man-Rogosa-Sharpe (MRS) medium (Scharlau Chemie) supplemented with 0.05% (wt/vol) l-cysteine hydrochloride, followed by incubation in an anaerobic atmosphere (2.99% H2, 17.01% CO2, and 80% N2) in a Concept 400 chamber (Ruskin) at 37°C for 24 h. In addition, nine non-Lactobacillus microorganisms were used in this study. These included Bifidobacterium bifidum LMG11041, which was cultivated in MRS broth as Lactobacillus strains, and Collinsella intestinalis DSM 13280, Escherichia coli LMG 2092, and Klebsiella pneumoniae CECT 143, which were grown in MRS broth (Difco, Detroit, MI) supplemented with 0.05% (wt/vol) l-cysteine (MRSC; Sigma, St. Louis, MO). Prevotella copri DSM 18205 and Blautia coccoides DSM 935 were cultivated in a combination of reinforced clostridial broth (Merck, Darmstadt, Germany) and brain-heart infusion (Difco), supplemented with 5% (vol/vol) heat-inactivated fetal bovine serum (LabClinics, Barcelona, Spain), respectively. For Bacteroides thetaiotaomicron DSMZ 2079, the latter medium was supplemented with 0.005% hemin (Sigma) and 0.005% vitamin K1 (Sigma). Faecalibacterium prausnitzii DSM 17677 was grown in Wilkins-Chalgren anaerobe broth (Merck) according to the recommendations included in the DSMZ medium 339. Finally, an active culture of Methanobrevibacter smithii DSM 861 grown in Methanobacterium medium (DSMZ 119) was directly supplied by DSMZ.
Bacterial DNA was extracted using GenElute bacterial genomic DNA kits (Sigma-Aldrich) according to the manufacturer's instructions. The taxonomic identity of the microorganisms was validated by sequencing the V3 variable region of the 16S rRNA gene using the primer pair Probio_Uni and/Probio_Rev (14).
Lactobacillus mock community.
The cultures of 14 different Lactobacillus strains were grown separately on MRS medium supplemented with 0.05% (wt/vol) l-cysteine hydrochloride, followed by incubation in an anaerobic atmosphere (2.99% H2, 17.01% CO2, and 80% N2) in a Concept 400 chamber (Ruskin) at 37°C until they reached late log phase. The bacteria were enumerated by counting colonies on solid medium, and the optical density at 600 nm was determined. The final bacterial cell concentration was approximately 107 CFU/ml. Chromosomal DNA of each strains was extracted as previously described and subsequently mixed.
Specifically, the mock community consists of a pool of known concentration of 14 different Lactobacillus strains to obtain the final quantity of DNA indicated in Table S3 in the supplemental material. Furthermore, the mix was prepared by combining equal volumes (20 μl) of DNA.
The DNA from the mix was diluted to produce a final DNA concentration of 2 ng/μl, and 4 μl of these dilutions was used in each PCR. For the PCR, the primer pair Probio-lac_Uni/Probio-lac_Rev was used, and the generated amplicons were sequenced using Illumina MiSeq as described below.
Lactobacillus ITS-specific primer design and gene amplification.
The bioinformatics platforms MEGAnnotator (10) and METAnnotatorX (unpublished data) were used to perform 16S and 23S rRNA gene predictions in all 1,523 sequenced lactobacillus genomes deposited at the NCBI genome database. Primers Probio-lac_Uni (CGTAACAAGGTAGCCGTAGG) and Probio-lac_Rev (GTYVCGTCCTTCWTCGSC) were manually designed based on the alignment of all 16S and 23S rRNA sequences to generate an amplicon encompassing the 3′ end of the 16S rRNA gene, the ITS region, and the 5′ end of the 23S rRNA gene. A specificity test was performed using the Silva TestProbe v3.0 tool (https://www.arb-silva.de/search/testprobe/) that allows alignment of primer sequences to the Silva SSU and LSU databases (15). A custom bioinformatics script was then used to create a database of all the Probio-lac_Uni/Probio-lac_Rev-generated lactobacillus ITS amplicon sequences (LITSA database), encompassing the ITS region and partial 16S and 23S rRNA genes.
The PCR conditions used for Lactobacillus ITS profiling using the Probio-lac_Uni/Probio-lac_Rev primer pair were 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 58°C, and 40 s at 72°C, followed in turn by 10 min at 72°C. Amplification was carried out using a Verity thermocycler (Applied Biosystems). The integrity of the PCR amplicons was analyzed by gel electrophoresis. An additional specificity test was performed by PCR using the DNA extracted from all known Lactobacillus species, as well as Bifidobacterium bifidum ATCC 11041, Collinsella intestinalis DSM 13280, Escherichia coli LMG 2092, Klebsiella pneumoniae CECT 143, Prevotella copri DSM 18205, Blautia coccoides DSM 935, Bacteroides thetaiotaomicron DSMZ 2079, Faecalibacterium prausnitzii DSM 17677, and Methanobrevibacter smithii DSM 861. A WebLogo representation of the primer sequence conservation among the retrieved 16S and 23S rRNA genes flanking complete ITS sequences was obtained through the WebLogo website (http://weblogo.berkeley.edu/) (35).
Illumina MiSeq sequencing of ITS gene-based amplicons.
An Illumina adapter overhang nucleotide sequence was added to the PCR amplicons obtained following amplification of the ITS region, as previously described (13). The library of ITS amplicons was prepared according to the 16S metagenomic sequencing library preparation protocol (part 15044223, rev. B; Illumina). Sequencing was performed using an Illumina MiSeq sequencer with MiSeq reagent kit (v3) chemicals.
ITS-based microbiota analysis.
Fastq files obtained from metagenomic sequencing of each sample were analyzed using a custom script for QIIME software suite (22) and the LITSA database (http://probiogenomics.unipr.it/sw/Lactobacilli_ITS_profiling_tool.zip).
Input data were processed in the following steps: (i) filtering of the reads based on a length of >100 nucleotides (primers included) to avoid primer dimers, an overall quality of >25, and the presence of forward and reverse primers in the forward and reverse reads, respectively; (ii) the creation of OTUs constituted by identical sequences using the prefix_suffix method; and (iii) the removal of OTUs represented by <10 sequences. Taxonomy assignment was performed by using RDP method (RDP classifier with a confidence level of 0.80) and the LITSA database constituted by ITS sequences retrieved from the 1,523 Lactobacillus genomes available at the NCBI genome database. This script is easily modifiable to obtain a profiling based on a different sequence, though it will depend on the availability of a corresponding database.
Evaluation of the sensitivity of the Probio-lac_Uni/Probio-lac_Rev primer pair.
The artificial sample used for the evaluation of the detection sensitivity and accuracy of the Probio-lac_Uni/Probio-lac_Rev primer set was generated using known DNA amounts, ranging from 50 to 0.006 ng, of 14 different Lactobacillus taxa (see Table S3 in the supplemental material).
Microbiota identification by 16S rRNA gene amplification, sequencing, and data analysis.
Partial 16S rRNA gene sequences were amplified from extracted DNA using the primer pair Probio_Uni/Probio_Rev, which target the V3 region of the 16S rRNA gene sequence (16). 16S rRNA gene amplification and amplicon checks were carried out as previously described (16). 16S rRNA gene sequencing was performed using a MiSeq (Illumina) at the DNA sequencing facility of GenProbio Srl according to a previously reported protocol (16). After sequencing, the .fastq files were processed using a custom script based on the QIIME software suite (22). Paired-end read pairs were assembled to reconstruct complete Probio_Uni/Probio_Rev amplicons. Quality control retained sequences with a length between 140 and 400 bp and mean sequence quality score of >20, while sequences with homopolymers >7 bp and mismatched primers were omitted. 16S rRNA gene OTUs were defined at ≥99% sequence homology using uclust (36), and OTUs with fewer than 10 sequences were filtered. All reads were classified to the lowest possible taxonomic rank using QIIME (22) and a reference data set from the SILVA database (17).
Accession number(s).
The raw ITS and 16S rRNA gene sequences reported in this article have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA434072.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the EU Joint Programming Initiative “A Healthy Diet for a Healthy Life” (JPI HDHL, http://www.healthydietforhealthylife.eu/) to D.V.S. (in conjunction with Science Foundation Ireland [SFI], grant 15/JP-HDHL/3280) and to M.V. (in conjunction with MIUR, Italy). We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. This research benefits from the HPC (High Performance Computing) facility of the University of Parma, Parma, Italy. D.V.S. is a member of The APC Microbiome Ireland supported by SFI, through the Irish Government's National Development Plan (grant SFI/12/RC/2273). Furthermore, D.V.S. is a visiting professor of the University of Parma, supported by Fondazione Cariparma, under the TeachInParma Project. The authors declare that they have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00706-18.
REFERENCES
- 1.Goldstein EJ, Tyrrell KL, Citron DM. 2015. Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clin Infect Dis 60(Suppl 2):S98–S107. doi: 10.1093/cid/civ072. [DOI] [PubMed] [Google Scholar]
- 2.Salvetti E, Torriani S, Felis GE. 2012. The genus Lactobacillus: a taxonomic update. Probiotics Antimicrob Proteins 4:217–226. doi: 10.1007/s12602-012-9117-8. [DOI] [PubMed] [Google Scholar]
- 3.Zheng J, Ruan L, Sun M, Ganzle M. 2015. A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl Environ Microbiol 81:7233–7243. doi: 10.1128/AEM.02116-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Perez-Munoz ME, Leulier F, Ganzle M, Walter J. 2017. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev 41:S27–S48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 5.Walter J. 2008. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol 74:4985–4996. doi: 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Girones R, Koutsoumanis K, Herman L, Lindqvist R, Nørrung B, Robertson L, Ru G, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Klein G, Peixe L, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak J, Correia S, Fernández Escámez PS. 2017. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 5: suitability of taxonomic units notified to EFSA until September 2016. EFSA J 15:e04663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Z, Harris HM, McCann A, Guo C, Argimon S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea MC, O'Sullivan O, Ritari J, Douillard FP, Paul Ross R, Yang R, Briner AE, Felis GE, de Vos WM, Barrangou R, Klaenhammer TR, Caufield PW, Cui Y, Zhang H, O'Toole PW. 2015. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6:8322. doi: 10.1038/ncomms9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salvetti E, O'Toole PW. 2017. The genomic basis of lactobacilli as health-promoting organisms. Microbiol Spectr doi: 10.1128/microbiolspec.BAD-0011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandes SH, Alvin LB, Silva BC, Zanirati DF, Jung LR, Nicoli JR, Neumann E, Nunes AC. 2014. Lactobacillus species identification by amplified ribosomal 16S-23S rRNA restriction fragment length polymorphism analysis. Benef Microbes 5:471–481. doi: 10.3920/BM2013.0092. [DOI] [PubMed] [Google Scholar]
- 10.Barrangou R, Yoon SS, Breidt F Jr, Fleming HP, Klaenhammer TR. 2002. Identification and characterization of Leuconostoc fallax strains isolated from an industrial sauerkraut fermentation. Appl Environ Microbiol 68:2877–2884. doi: 10.1128/AEM.68.6.2877-2884.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore JE, Hirayama J, Hayashi K, Mason C, Coulter W, Matsuda M, Goldsmith CE. 2018. Examination of 16S-23S rRNA intergenic spacer region (ISR) heterogeneity in a population of clinical Streptococcus pneumoniae: a new laboratory epidemiological genotyping tool to aid outbreak analysis. Br J Biomed Sci 75:95–97. doi: 10.1080/09674845.2017.1382025. [DOI] [PubMed] [Google Scholar]
- 12.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. 2016. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363:fnw049. doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 13.Milani C, Lugli GA, Turroni F, Mancabelli L, Duranti S, Viappiani A, Mangifesta M, Segata N, van Sinderen D, Ventura M. 2014. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol Ecol 90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 14.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, Tett A, Segata N, van Sinderen D, Ventura M. 2015. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philippe H, Adoutte A. 1996. What can phylogenetic patterns tell us about the evolutionary process generating biodiversity? Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 16.Milani C, Hevia A, Foroni E, Duranti S, Turroni F, Lugli GA, Sanchez B, Martin R, Gueimonde M, van Sinderen D, Margolles A, Ventura M. 2013. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One 8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chebenova-Turcovska V, Zenisova K, Kuchta T, Pangallo D, Brezna B. 2011. Culture-independent detection of microorganisms in traditional Slovakian bryndza cheese. Int J Food Microbiol 150:73–78. doi: 10.1016/j.ijfoodmicro.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Campanella JJ, Bitincka L, Smalley J. 2003. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4:29. doi: 10.1186/1471-2105-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lugli GA, Mangifesta M, Duranti S, Anzalone R, Milani C, Mancabelli L, Alessandri G, Turroni F, Ossiprandi MC, van Sinderen D, Ventura M. 2018. Phylogenetic classification of six novel species belonging to the genus Bifidobacterium comprising Bifidobacterium anseris sp. nov., Bifidobacterium criceti sp. nov., Bifidobacterium imperatoris sp. nov., Bifidobacterium italicum sp. nov., Bifidobacterium margollesii sp. nov., and Bifidobacterium parmae sp. nov. Syst Appl Microbiol 41:173–183. doi: 10.1016/j.syapm.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. 2010. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, Sun CL, Goltsman DS, Wong RJ, Shaw G, Stevenson DK, Holmes SP, Relman DA. 2015. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A 112:11060–11065. doi: 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108(Suppl 1):S4680–S4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svetoch EA, Eruslanov BV, Levchuk VP, Perelygin VV, Mitsevich EV, Mitsevich IP, Stepanshin J, Dyatlov I, Seal BS, Stern NJ. 2011. Isolation of Lactobacillus salivarius 1077 (NRRL B-50053) and characterization of its bacteriocin, including the antimicrobial activity spectrum. Appl Environ Microbiol 77:2749–2754. doi: 10.1128/AEM.02481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobierecka PA, Wyszynska AK, Aleksandrzak-Piekarczyk T, Kuczkowski M, Tuzimek A, Piotrowska W, Gorecki A, Adamska I, Wieliczko A, Bardowski J, Jagusztyn-Krynicka EK. 2017. In vitro characteristics of Lactobacillus spp. strains isolated from the chicken digestive tract and their role in the inhibition of Campylobacter colonization. Microbiologyopen doi: 10.1002/mbo3.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merhej V, Armougom F, Robert C, Raoult D. 2012. Genome sequence of Lactobacillus ingluviei, a bacterium associated with weight gain in animals. J Bacteriol 194:5697. doi: 10.1128/JB.01205-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dec M, Puchalski A, Urban-Chmiel R, Wernicki A. 2014. Screening of Lactobacillus strains of domestic goose origin against bacterial poultry pathogens for use as probiotics. Poult Sci 93:2464–2472. doi: 10.3382/ps.2014-04025. [DOI] [PubMed] [Google Scholar]
- 30.Baele M, Devriese LA, Haesebrouck F. 2001. Lactobacillus agilis is an important component of the pigeon crop flora. J Appl Microbiol 91:488–491. doi: 10.1046/j.1365-2672.2001.01407.x. [DOI] [PubMed] [Google Scholar]
- 31.Fujisawa T, Shirasaka S, Watabe J, Mitsuoka T. 1983. Lactobacillus aviarius sp. nov.: a new species isolated from the intestine of chickens. Syst Appl Microbiol 5:414–420. doi: 10.1016/S0723-2020(84)80042-9. [DOI] [Google Scholar]
- 32.La Ragione RM, Narbad A, Gasson MJ, Woodward MJ. 2004. In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett Appl Microbiol 38:197–205. doi: 10.1111/j.1472-765X.2004.01474.x. [DOI] [PubMed] [Google Scholar]
- 33.Ventura M, Zink R. 2002. Specific identification and molecular typing analysis of Lactobacillus johnsonii by using PCR-based methods and pulsed-field gel electrophoresis. FEMS Microbiol Lett 217:141–154. doi: 10.1111/j.1574-6968.2002.tb11468.x. [DOI] [PubMed] [Google Scholar]
- 34.Gala E, Landi S, Solieri L, Nocetti M, Pulvirenti A, Giudici P. 2008. Diversity of lactic acid bacteria population in ripened Parmigiano Reggiano cheese. Int J Food Microbiol 125:347–351. doi: 10.1016/j.ijfoodmicro.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.