Abstract
Tuberculosis ranks as one of the world’s deadliest infectious diseases causing more than a million casualties annually. IL10 inhibits the function of Th1 type cells, and IL10 deficiency has been associated with an improved resistance against Mycobacterium tuberculosis infection in a mouse model. Here, we utilized M. marinum infection in the zebrafish (Danio rerio) as a model for studying Il10 in the host response against mycobacteria. Unchallenged, nonsense il10e46/e46 mutant zebrafish were fertile and phenotypically normal. Following a chronic mycobacterial infection, il10e46/e46 mutants showed enhanced survival compared to the controls. This was associated with an increased expression of the Th cell marker cd4-1 and a shift towards a Th1 type immune response, which was demonstrated by the upregulated expression of tbx21 and ifng1, as well as the down-regulation of gata3. In addition, at 8 weeks post infection il10e46/e46 mutant zebrafish had reduced expression levels of proinflammatory cytokines tnfb and il1b, presumably indicating slower progress of the infection. Altogether, our data show that Il10 can weaken the immune defense against M. marinum infection in zebrafish by restricting ifng1 response. Importantly, our findings support the relevance of M. marinum infection in zebrafish as a model for tuberculosis.
Introduction
Annually more than 10 million new tuberculosis cases are estimated to emerge, leading to over a million casualties1. The immune defense against the pathogen which causes tuberculosis, Mycobacterium tuberculosis, requires the elaborate collaboration of both the innate and adaptive immunity, as is demonstrated by the increased disease susceptibility in recipients of TNF-antagonist as well as in HIV-positive individuals1–3. Accordingly, deficient, but also excessive, macrophage mediated TNF production as well as the lack of T helper (Th) 1 type cell responses compromise the host’s ability to resist a mycobacterial infection and accelerate the disease pathogenesis4–7.
IL10 is an anti-inflammatory cytokine that was originally identified as a protein secreted by Th2 cells able to inhibit cytokine production in Th1 cells8,9. Later, it was discovered that several other cell types, including both immune and nonimmune cells, produce IL1010. Genome-wide association studies in humans have linked IL10 polymorphisms to susceptibility and resistance towards tuberculosis, although the results vary depending on the polymorphism studied and the study subjects11,12. Furthermore, in vivo mouse studies have shown that IL10 impairs the immune defense against M. tuberculosis by impeding host immunity at an early13,14, but also during later stages of an infection15. In these studies, the lack of functional IL10 signaling resulted in enhanced protection against mycobacteria and was attributed to an increased Th1 response13–16. As a consequence, for example, macrophages were more capable of presenting M. tuberculosis antigens and recruiting inflammatory cells13,14. Enhanced protection was demonstrated by a lower bacterial burden in the lungs and spleen13–15 and improved survival of the mice15. While IL10 deficiency or receptor blockade has been associated with an enhanced protection against mycobacteria in mice, it has also been reported that the lack of IL10 can eventually lead to harmful lung inflammation and to the progression of a mycobacterial disease in the mouse model16.
The zebrafish is a small teleost, which is constantly gaining popularity as a model organism. The cellular components of the zebrafish immune system, such as mononuclear phagocytes17, dendritic cells18, T cells and B cells19–23 and eosinophils24 have been described and they resemble those of humans. To date, specific transcription factors expressed by different zebrafish Th as well as Treg cells have also been identified and characterized25,26. Previously, lymphocyte marker gene expression has been used to characterize immune response in adult zebrafish27 and for example foxp3a has been validated as a Treg marker28. As for the humoral components of the zebrafish immune system, the mammalian homologs of the complement system29, as well as the immunoglobulin isotypes IgD, IgM and the bony fish specific immunoglobulin Z/T30 have been found. Overall, the zebrafish is a suitable model for immunological research (reviewed in31).
Mycobacterium marinum is a natural pathogen of the zebrafish and a close relative of Mycobacterium tuberculosis32. Comparably to M. tuberculosis, M. marinum infects macrophages33,34 and eventually causes a systemic disease in zebrafish, which shares pathological and histological features with human tuberculosis35–37. A M. marinum infection model in zebrafish larvae has been widely used to study the innate immune response in a mycobacterial infection, and it enables the real-time visualization and rapid screening of potential tuberculosis drugs38–40. Furthermore, the M. marinum infection model in adult zebrafish allows studying the adaptive response35–37.
Zebrafish il10 has a mammalian-like gene organization and conserved IL10 signature motif 41,42. Furthermore, Grayfer and Belosevic43 have found IL10 receptor 1 in zebrafish and in goldfish (Carassius Auratus L.). Among their analyses, an alignment of these protein sequences with those of other vertebrates, the expression measurements in different tissues and immune cell populations at mRNA level and in vitro binding studies of recombinant goldfish IL10 receptor 1 and IL10 proteins, indicated conservation of the IL10 system throughout evolution. In order to study the role of Il10 in the immune defense against mycobacteria, we have here characterized an il10e46/e46 mutant zebrafish strain in relation to a M. marinum infection. Also, we aim to gain more information about M. marinum infection in zebrafish as a model for human tuberculosis.
Results
A nonsense il10e46 mutation creates an early stop codon in the zebrafish il10 gene
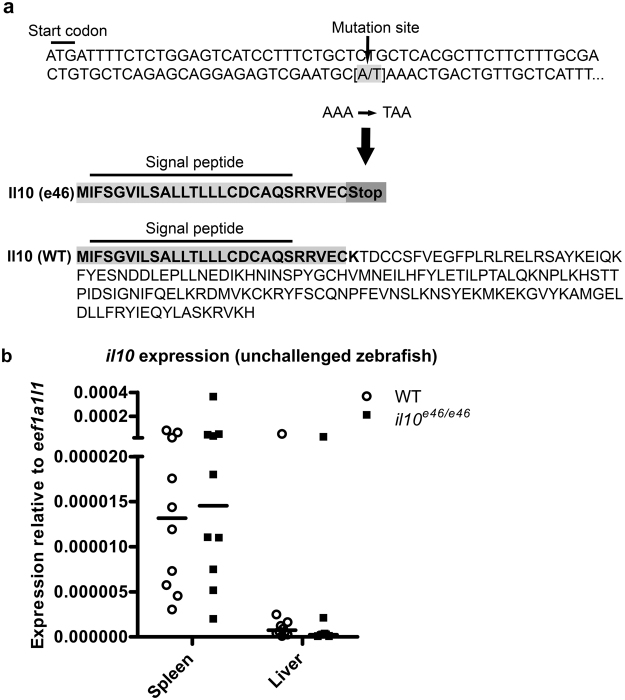
In order to study Il10 in the host response against mycobacteria the zebrafish line e46, carrying a nonsense il10 mutation, was obtained from the Wellcome Trust Sanger Institute44. In the il10e46 mutant zebrafish a specific adenosine (A) to thymidine (T) point mutation results in a stop codon (TAA) after the first 27 amino acids in the translated region of exon 1 (Fig. 1a). As nonsense mediated decay degrades mRNA molecules producing non-functional proteins45, we first studied if the il10e46 mutation affects the levels of the il10 mRNA, and quantified the expression of il10 in different organs of the abdominal cavity by quantitative PCR (qPCR) (Fig. 1b). However, in any of the studied tissues il10 mRNA expression did not differ between il10e46/e46 mutants and wild type (WT) zebrafish, suggesting that the effects of the mutation are only evident at the translational level. In fact, signal peptide prediction using SignalP 4.1 Server46 revealed that only five amino acids remain in the truncated protein, which consequently prevents the normal function of Il10 in the fish carrying the homozygous il10e46 mutation (Fig. 1a).
Figure 1.
il10e46 point mutation in the exon 1 of the il10 results in a premature stop codon but does not alter il10 expression. (a) A schematic presentation of the il10e46 mutation; a adenosine (A) to thymidine (T) point mutation results in a stop codon (TAA) after the first 27 amino acids in exon 1. (b) Relative il10 expression was measured with qPCR in unchallenged il10e46/e46 mutant and WT zebrafish, spleen and liver from the same individuals (n = 10 in both groups) and is presented as a scatter dot blot and median. Note the divided y axis. Gene expressions were normalized to the expression of eef1a1l1. A two-tailed Mann-Whitney test was used for the statistical comparison of differences between il10e46/e46 zebrafish and WT controls.
Unchallenged il10e46/e46 zebrafish are phenotypically normal and have similar blood cell populations and cytokine expression profiles compared to WT fish
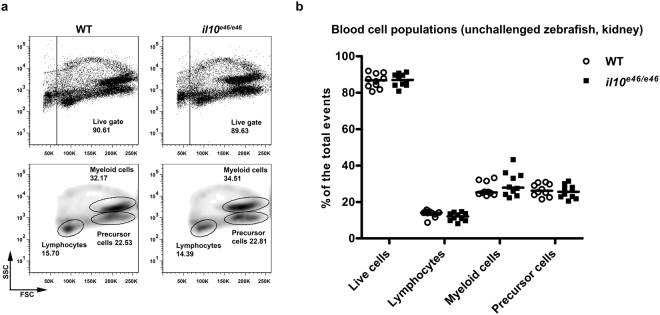
IL10 knock-out (KO) mice have growth defects and suffer from chronic intestinal inflammation leading to 30% mortality before 3 months of age47,48. Like IL10 KO mouse strains, il10e46/e46 zebrafish are fertile and can be maintained by spawning homozygous mutant siblings47. However, in contrast to mice, il10e46/e46 mutant fish are phenotypically normal and do not have increased mortality compared to WT zebrafish. Our flow-cytometric analysis of the blood cell populations in kidney blood cell isolates revealed no differences in live cell, lymphocyte, myeloid cell or blood cell precursor cell counts in unchallenged il10e46/e46 zebrafish compared to the WT control fish (Fig. 2a,b).
Figure 2.
Unchallenged il10e46/e46 zebrafish have kidney blood cell populations similar to those of WT control fish. (a,b) The relative proportions of live cells, lymphocytes, myeloid cells and precursor cells were determined with flow cytometry in il10e46/e46 mutant zebrafish and in WT fish (n = 10 in both groups) based on granularity (SSC) and cell size (FSC). Representative flow cytometry plots are shown in panel (a). Gated populations are outlined, and the cell counts inside the gates are given as the percentages of the total viable cell population. The median of the relative proportions of different blood cell populations is presented as a scatter plot in panel (b). A two-tailed Mann-Whitney test was used for the statistical comparison of differences between il10e46/e46 zebrafish and WT controls.
Colitis in IL10 KO mice is attributed to the increased production of inflammatory mediators such as TNF and IL1b as well as to a hyper-activated Th1 response47,49,50. In order to study signs of inflammation and T cell homeostasis in unchallenged il10e46/e46 zebrafish, we extracted RNA from different adult zebrafish tissues and measured the expression levels of selected proinflammatory cytokines (il1b, tnfa and tnfb), Th cell cytokines (ifng1 and il4) as well as T cell markers cd4-1 and cd8a) and a B cell marker IgM (Supplementary Fig. S1). In contrast to the IL10 KO mice, qPCR analysis from the liver and spleen revealed no differences in the relative expression levels of the studied inflammatory markers tnfa, tnfb and il1b between il10e46/e46 and WT fish. The expression levels of the Th1 cytokine ifng1 and the Th2 cytokine il4 were also comparable between the fish groups. In addition, no differences in the mRNA expression of the T cell marker genes cd8a and cd4-1, between il10e46/e46 and WT fish, were observed. The expression of IgM in the liver of il10e46/e46 fish was decreased compared to controls (P = 0.029). Collectively, these data indicate that the il10e46/e46 mutant zebrafish have no apparent immune abnormalities under unchallenged conditions.
In order to study in more detail the possible inflammation in the intestine of the Il10 deficient zebrafish, we collected the intestine from 1-year-old WT and il10e46/e46 fish and measured the expression levels of several inflammatory cytokines and immune cell markers (Supplementary Fig. S2a). No difference in the expression of the inflammatory cytokines (il1b, tnfa and tnfb), T lymphocyte markers (cd4-1 and cd8a), B lymphocyte marker IgM, Th2 and Treg markers, (gata3 and foxp3a, respectively), or in Th1 and Th2 hallmark cytokines (ifng1 and il4, or in il10) was observed between WT and il10e46/e46 fish. The expression of Th1 cell marker tbx21 was higher in WT fish compared to il10e46/e46 mutants (P = 0.023) but this is unlikely a sign towards colitis in il10e46/e46 mutants. Furthermore, the visual appearance of WT and il10e46/e46 intestine was similar with identifiable intestinal bulb, mid-intestine and posterior-intestine (Supplementary Fig. S2b).
Survival of adult il10e46/e46 mutants is improved in a low-dose M. marinum infection
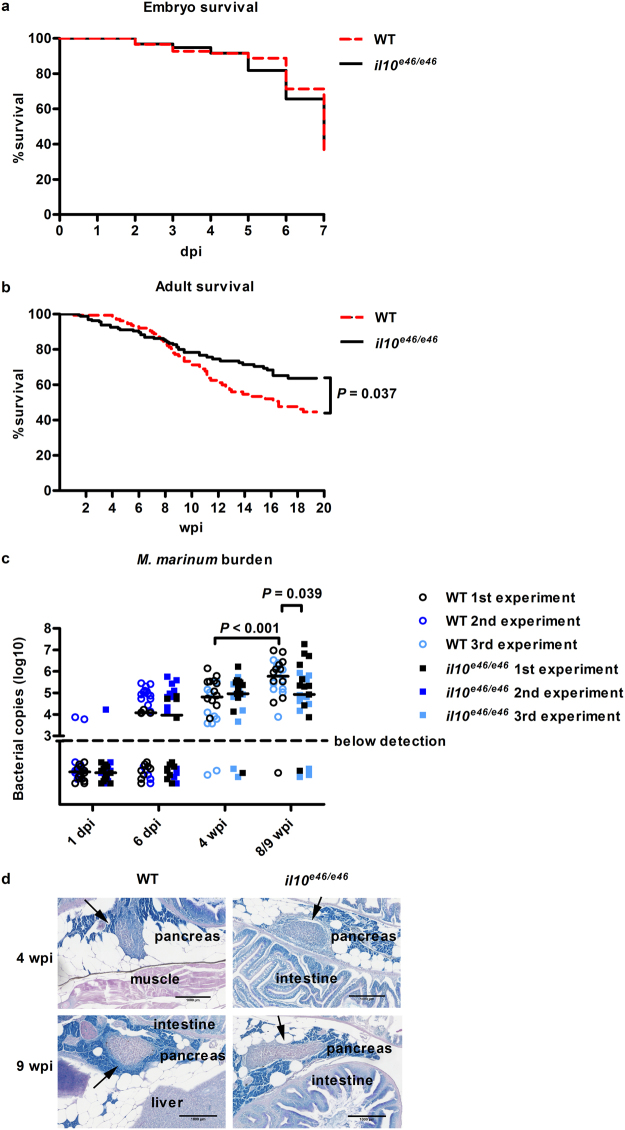
In mice, the absence of functional IL10 signaling leads to the improved control of a M. tuberculosis infection, and consequently, to better survival13,15. Other studies have, however, associated IL10 deficiency to increased inflammation leading to reduced survival16. To study the role of Il10 in the immune defense against a M. marinum infection in zebrafish, we first infected il10e46/e46 and WT embryos into the yolk at 0–6 hours post fertilization and followed the survival of the larvae for seven days (Fig. 3a). After three days, mortality was observed in both of the groups resulting in ca. 70% mortality at the end of the follow-up with no significant difference between il10e46/e46 and WT larvae.
Figure 3.
Adult il10e46/e46 zebrafish have enhanced survival compared to WT controls in a low-dose M. marinum infection. (a) il10e46/e46 (n = 158) and WT (n = 181) zebrafish larvae were microinjected before 6 hours post fertilization with M. marinum (3–29 CFU) and their survival was monitored for 7 days. The experiment was done six times and the data presented here is collected from one representative experiment. (b) The survival of adult il10e46/e46 (n = 172) and WT (n = 149) zebrafish was monitored for 16–20 weeks after a low-dose (2–156 CFU) mycobacterial infection. The data were collected from four experiments. (c) The M. marinum burden in abdominal organ blocks (including kidney in the first and the second experiment, without kidney in the third experiment) of adult il10e46/e46 mutant zebrafish (n = 22–27) and WT controls (n = 22–30 fish) was quantified with qPCR at 4 and 8/9 weeks post a low-dose infection (1–18 CFU). The bacterial load is presented as a scatter dot plot and as the median of total bacterial copies (log10). The data were collected from three experiments. (d) M. marinum granulomas were detected with Ziehl-Neelsen staining (n = 4 in both groups at both time points) at 4 and 9 weeks post a low-dose infection (2–9 CFU). Representative individuals from each group are shown. Granulomas are indicated with arrows. For panels (a) and (b) a log-rank (Mantel-Cox) and for panel (c) A two-tailed Mann-Whitney test was used for the statistical comparison of differences.
Next, we injected a low-dose of M. marinum into the abdominal cavity of adult il10e46/e46 and WT zebrafish and followed the survival of the fish for 20 weeks (Fig. 3b). During the first 8 weeks, we observed ca. 20% mortality in both of the fish groups, which was similar to our previously reported results from a low-dose mycobacterial infection35,51. However, from week eight onwards the mortality of the WT fish increased compared to the il10e46/e46 mutants with an endpoint survival of 44% and 63%, respectively (P = 0.037).
In order to differentiate between resistance and tolerance against a M. marinum infection, we collected abdominal organ block samples from infected fish during the course of a mycobacterial infection. The samples were taken at 1 day post infection (dpi), 6 dpi, 4 weeks post infection (wpi) and 8/9 wpi, and bacterial amounts were analyzed. In our qPCR based bacterial quantification, detectable amounts of M. marinum in the infected zebrafish were primarily observed for the first time at the 6 dpi time point (Fig. 3c). At 6 dpi, but also at 4 wpi, no differences in M. marinum counts between il10e46/e46 mutants and WT zebrafish were observed (at 6 dpi; bacterial copy number median 65,000 vs 74,000, at 4 wpi; median 126,000 vs 90,000, Fig. 3c). However, at 8/9wpi there was a smaller bacterial amount in the il10e46/e46 fish compared to the WT fish (median 86,000 vs 645,000, P = 0.039). In addition, bacterial counts in il10e46/e46 fish remained stable between 4 and 8/9 weeks, whereas there was a clear increase in bacteria in WT fish (median 65,000 vs 645,000, P < 0.001) (Fig. 3c). A Ziehl-Neelsen staining of formalin fixed and paraffin embedded zebrafish confirmed the presence of mycobacteria in both infected il10e46/e46 and WT fish at 4 and 9 wpi (Fig. 3d). The histopathological analysis did not reveal any differences in granuloma morphology or in their tissue distribution (Supplementary Fig. S3). In conclusion il10e46/e46 mutant zebrafish showed improved survival but no apparent changes in histopathology. In addition, there was a decrease in bacterial burden of il10e46/e46 mutants at 8/9 wpi fish indicating enhanced resistance rather than a higher tolerance against low dose M. marinum infection.
il10e46/e46 mutant zebrafish have an enhanced ifng1 response in a low-dose mycobacterial infection
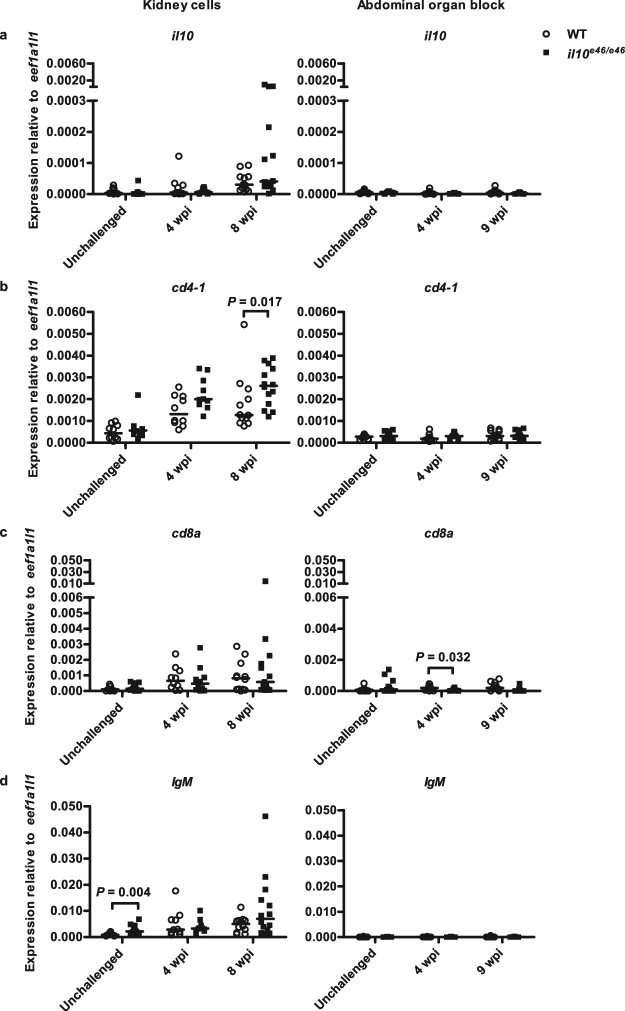
To study the mechanisms underlying enhanced survival in il10e46/e46 mutant zebrafish against a low-dose M. marinum infection we quantified the expression of selected cytokines and immune cell markers and conducted a flow cytometric analysis in the zebrafish kidney blood cells at different time points post infection (Figs 4–8). The expression levels of the studied genes in the kidney and abdominal organ block samples of unchallenged WT and il10e46/e46 fish were similar. However, the relative IgM expression in the il10e46/e46 mutant kidney was increased 2.5-fold compared to WT zebrafish (P = 0.004) but otherwise the mutants did not show any differences compared to WT.
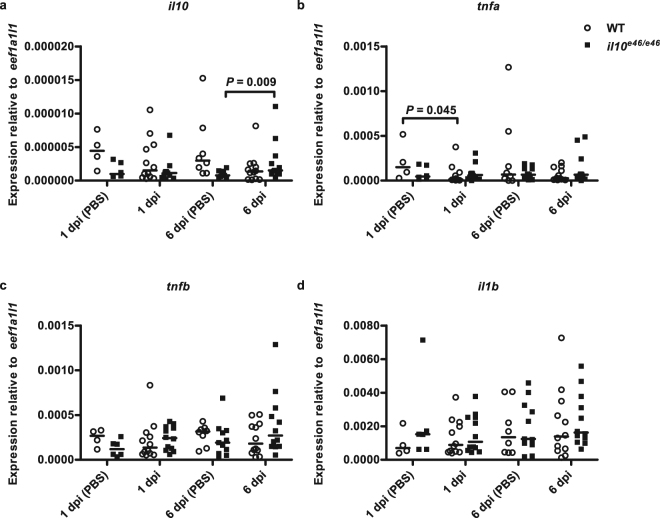
Figure 4.
Nonfunctional il10 does not increase the expression of proinflammatory cytokines at the early stages of a low-dose (2–9 CFU) mycobacterial infection in zebrafish. (a–d) The relative expression levels of il10 and proinflammatory cytokine genes (il1b, tnfa and tnfb) were measured in the abdominal organ blocks (including kidney) of il10e46/e46 mutant fish (n = 6–12) and WT controls (n = 5–12) at 1 dpi and 6 dpi and are presented as a scatter dot plot and median. Note the different scales of the y axes. Gene expressions were normalized to the expression of eef1a1l1. The data were collected from a single experiment. A two-tailed Mann-Whitney test was used for the statistical comparison of differences.
First, we quantified the expression of il10, tnfa, tnfb, and il1b in the abdominal organ blocks of zebrafish at early time points during an infection (Fig. 4). Until 6 dpi, M. marinum infection did not alter the expression of tnfb or il1b either in il10e46/e46 mutants or in WT fish compared to corresponding PBS injected controls. However, similarly to previous reports about the upregulation of Il10 upon immunogenic stimulation52,53, il10 was significantly upregulated in the il10e46/e46 mutants at 6 dpi in comparison to PBS controls (P = 0.009). WT fish, in turn, had a slight reduction in tnfa expression at 1 dpi compared to PBS injected fish (P = 0.045). As in unchallenged zebrafish, we did not see any differences in the expression levels of the cytokine genes between the il10e46/e46 mutants and WT fish at 1 dpi or 6 dpi. At these early time points, expression levels of the T cell markers cd4-1 (CD4 + cells) or cd8a (CD8 + cells) or the B cell marker IgM did not differ between il10e46/e46 and WT fish either (Supplementary Fig. S4). These data indicate that a low-dose mycobacterial infection does not cause acute systemic inflammation in zebrafish and that the il10e46/e46 zebrafish have a transcriptional innate cytokine response similar to the WT controls in the low-dose infection.
In order to study the role of Il10 later in a M. marinum infection, we conducted a flow cytometric analysis in the zebrafish kidney blood cells at 4 and 8 wpi and analyzed the relative amounts of lymphocyte, precursor cell and myeloid cell populations in il10e46/e46 mutants and WT controls (Fig. 5). There were no differences in the relative lymphocyte or precursor cell counts between the groups. However, the relative proportion of myeloid cells was significantly lower in WT fish compared to the il10e46/e46 mutants (median 21.6% vs. 26.2%, P = 0.014). Of note, the total live cell numbers were also lower in the WT controls compared to il10e46/e46 mutant fish at 8 wpi (P < 0.001). Furthermore, in the WT control group the median of the relative myeloid cell count was 14.5% lower compared to the median of unchallenged fish at 8 wpi (P = 0.001).
Figure 5.
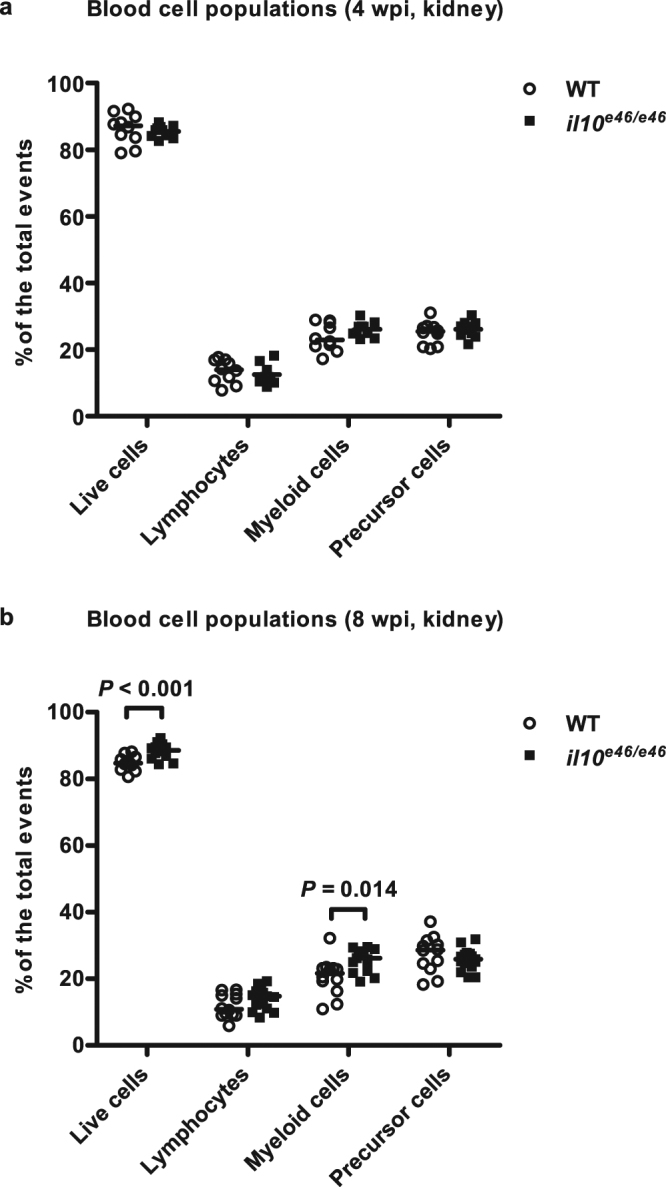
il10e46/e46 mutation associates with a higher proportion of myeloid cells at 8 weeks post a low-dose (1–18 CFU) M. marinum infection. (a,b) The relative proportions of live cells, lymphocytes, precursor cells and myeloid cells were determined with flow cytometry from the kidneys of il10e46/e46 mutants (n = 10–15) and WT control fish (n = 10–12) at 4 and 8 wpi based on granularity (SSC) and cell size (FSC). The data were collected from a single experiment and are presented as a scatter dot plot and median. A two-tailed Mann-Whitney test was used for the statistical comparison of differences between il10e46/e46 zebrafish and WT controls.
To obtain more quantitative data on T and B lymphocytes in il10e46/e46 mutant and WT fish at 4 and 8 wpi, we next extracted RNA from the unsorted kidney cell samples used for the flow cytometry and determined relative expression levels of il10, the T cell markers cd4-1 and cd8a as well as the B cell marker IgM (Fig. 6). No differences in the relative il10 expression levels between the il10e46/e46 mutant and the WT groups were observed. Interestingly, a qPCR analysis showed approximately 34.3% higher relative expression of cd4-1 in il10e46/e46 mutants compared to WT control fish at 4wpi (P = 0.052, NS) and a significantly higher expression at 8 wpi (P = 0.017). No differences in the kidney blood cell cd8a nor the B lymphocyte marker IgM expression were detected between il10e46/e46 and WT zebrafish at 4 and 8 wpi. Notably, however, cd8a expression was significantly lower in il10e46/e46 abdominal organ blocks compared to WT controls at 4 wpi (P = 0.032) (Fig. 6). Abdominal organ blocks did not have statistically significant differences in their cd4-1 or IgM expression levels between il10e46/e46 mutants and WT fish, although a trend towards the upregulation of cd4-1 was seen at 4 wpi (P = 0.053, NS, Fig. 6). Altogether, the upregulated cd4-1 expression in the il10e46/e46 mutant kidneys is a possible consequence of an enhanced Th cell response in the infected il10e46/e46 zebrafish.
Figure 6.
il10e46/e46 mutant zebrafish have an elevated Th cell marker, cd4-1, expression level at 8 weeks post a M. marinum infection. (a–d) The relative expressions of il10 and lymphocyte markers (cd4-1, cd8a and IgM) in kidneys of unchallenged il10e46/e46 mutants (n = 9–10) and WT control fish (n = 9–10) and in the unsorted kidney cell populations of il10e46/e46 mutants (n = 10–14) and WT control fish (n = 10–12) at 4 and 8 weeks post a low-dose (1–18 CFU) infection (on the left) as well as in the abdominal organ blocks (including kidney) of unchallenged il10e46/e46 mutants (n = 12) and WT control fish (n = 11–12) and at 4 and 9 weeks post a low-dose (2–9 CFU) infection (n = 9–12 and n = 11–12, respectively) (on the right) were measured with qPCR. Data are presented as a scatter dot plot and median. Note the different scales of the y axes and the divided y axis in panels a and c. Gene expressions were normalized to the expression of eef1a1l1. Each dataset was collected from a single experiment. A two-tailed Mann-Whitney was used for the statistical comparison of differences.
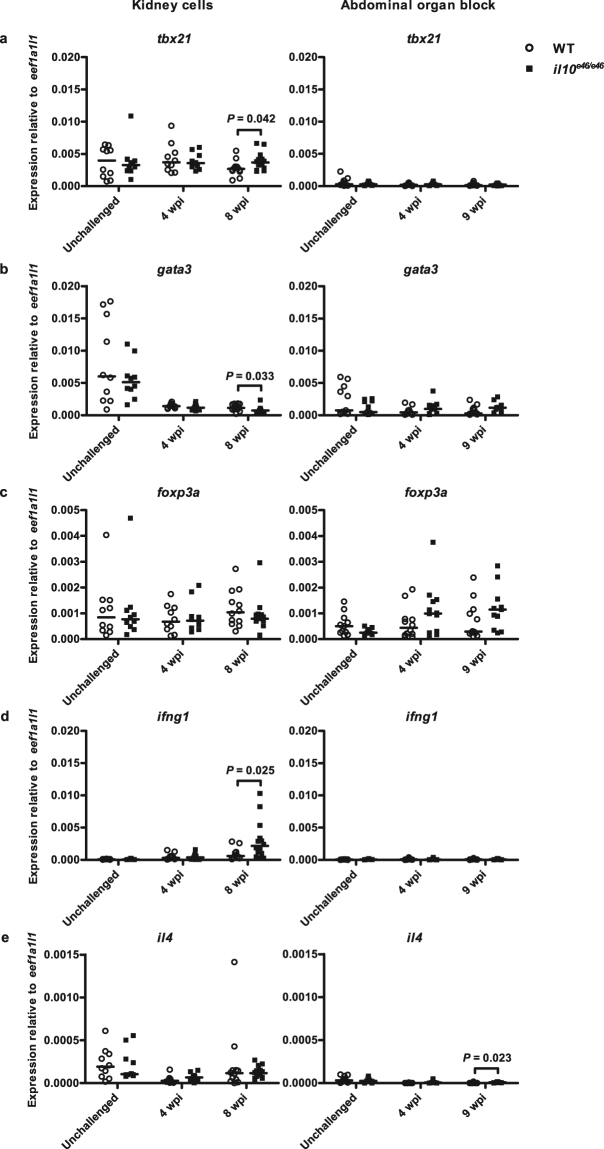
In addition to the effects of Il10 in regulating the production of proinflammatory cytokines in mice, human IL10 is known to suppress the activation of Th cells by inhibiting the production of IFNG and IL454. Hence, we quantified the relative expression of the Th1, Th2 and Treg cell transcription factors, tbx21, gata3 and foxp3a, respectively, as well as the canonical Th1 and Th2 cell cytokine genes ifng1 and il4 in the kidney blood cells (Fig. 7). Indicative of an enhanced Th1 type immune response, our qPCR analysis showed that il10e46/e46 mutant fish had higher relative expression levels of tbx21 at 8wpi (P = 0.042), whereas the expression of gata3 was lower compared to WT controls at the same time point (P = 0.033). Additionally, expression of the Th1 type cytokine gene ifng1 was upregulated in il10e46/e46 fish compared to WT zebrafish 8 wpi (P = 0.025). The expression of the canonical Th2 cytokine gene il4 was instead similar in both il10e46/e46 and WT fish the unsorted kidney cells at 4 and 8 wpi. Nor were any differences seen in the expression levels of foxp3a between the groups at either of the time points, suggesting a similar transcriptional Treg cell response in both mutants and WT fish during an infection. Mutation in il10 can also cause differential tissue and cell type specific regulation of Th type gene expression since upregulation of il4 was observed in the abdominal organ blocks of the il10e46/e46 mutants compared to WT zebrafish at 9 wpi (P = 0.023, Fig. 7).
Figure 7.
il10e46/e46 mutant zebrafish have an enhanced Th1 cell mediated immune response in a mycobacterial infection. (a–e) The relative expressions of Cd4 + lymphocyte transcription factors (tbx21, gata3, foxp3a) and Th cell cytokines (ifng1 and il4) in kidneys of unchallenged il10e46/e46 mutants (n = 10) and WT control fish (n = 10) and in the unsorted kidney cell populations of il10e46/e46 mutants (n = 10–14) and WT control fish (n = 10–12) at 4 and 8 weeks post a low-dose (1–18 CFU) infection (on the left) as well as in the abdominal organ blocks (including kidney) of unchallenged il10e46/e46 mutants (n = 12) and WT control fish (n = 11–12) and at 4 and 9 weeks post a low-dose (2–9 CFU) infection (n = 12 and n = 12, respectively) (on the right) were measured with qPCR. Data are presented as a scatter dot plot and median. Gene expressions were normalized to the expression of eef1a1l1. Each dataset was collected from a single experiment. A two-tailed Mann-Whitney was used for the statistical comparison of differences.
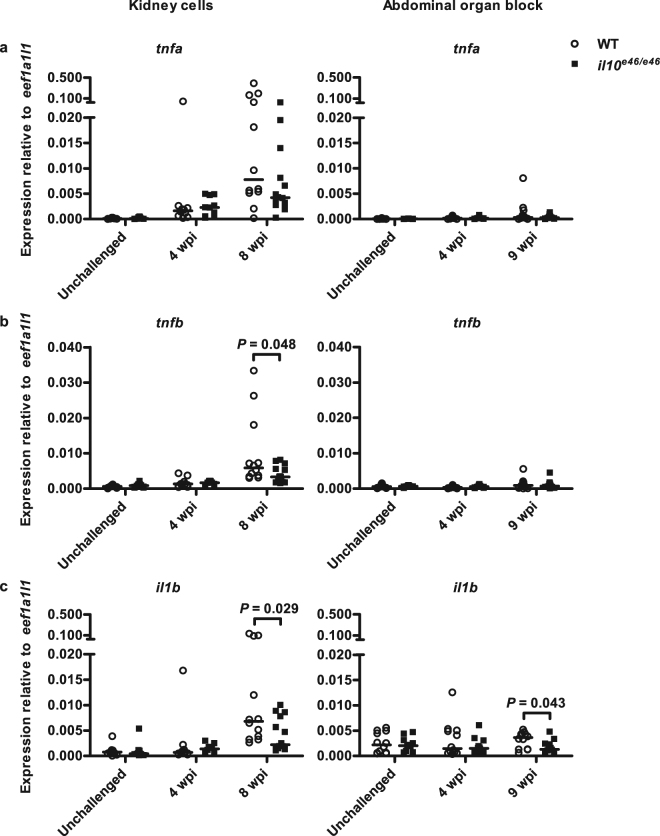
A mycobacterial infection elicits the host’s immune cells to produce humoral effectors such as complement components, reactive oxygen and nitrogen intermediates as well as proinflammatory cytokines to fight the infection55. In general, the magnitude of this response can be used to assess the severity of the prevalent bacterial disease. To compare the expression of mediators of inflammation in il10e46/e46 mutants and WT fish in a chronic M. marinum infection, we quantified the expression of tnfa, tnfb and il1b in the kidney blood cells at 4 and 8 wpi (Fig. 8). qPCR results showed that il10e46/e46 mutants had significantly lower expression levels of tnfb (P = 0.048) and il1b (P = 0.029) compared to WT fish at 8 wpi. No differences were observed in the expression of tnfa between il10e46/e46 mutants and WT fish. Similar results were seen in the abdominal organ blocks as the relative expression of il1b was downregulated in il10e46/e46 mutant fish compared to WT controls (P = 0.043, Fig. 8). Together, the elevated tbx21 and ifng1 expressions as well as the reduced expression of gata3 in the il10e46/e46 mutants indicate that a nonsense mutation in il10 leads to a Th1 cell type immune response in zebrafish. In addition, this Th1 cell response associates with lower tnfb and il1b expression presumably indicating the slower progress of a mycobacterial infection as the expression of these cytokines have been shown to associate with the bacterial burden during the reactivation of M. marinum infection56.
Figure 8.
il10e46/e46 mutant zebrafish have reduced inflammation in a chronic mycobacterial infection. (a–c) The relative expression of selected proinflammatory cytokines (il1b, tnfa, tnfb) in kidneys of unchallenged il10e46/e46 mutants (n = 10) and WT control fish (n = 10) and in the unsorted kidney cell populations of il10e46/e46 mutants (n = 10–14) and WT control fish (n = 10–12) at 4 and 8 weeks post a low-dose (1–18 CFU) infection (on the left) as well as in the abdominal organ blocks (including kidney) of unchallenged il10e46/e46 mutants (n = 12) and WT control fish (n = 12) and at 4 and 9 weeks post a low-dose (2–9 CFU) infection (n = 12 and n = 12, respectively) (on the right) were measured with qPCR. Data are presented as a scatter dot plot and median. Note the different scales of the y axes and the divided y axis in panel a. Gene expressions were normalized to the expression of eef1a1l1. Each dataset was collected from a single experiment. A two-tailed Mann-Whitney was used for the statistical comparison of differences.
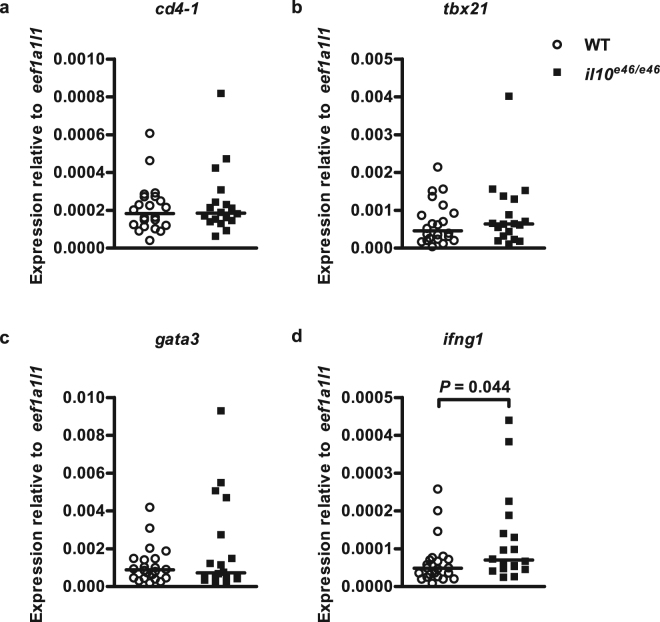
The il10e46/e46 mutants are produced by ENU mutagenesis and thus likely contain also other mutations in their background in addition to the e46 mutation. To address this, we outcrossed il10e46/e46 mutants to wild type AB zebrafish and incrossed their heterozygous progeny. Thereafter, we infected ungenotyped offspring of the heterozygous il10e46/+ mutants with a low-dose of M. marinum and collected zebrafish kidneys for gene expression analysis (cd4-1, tbx21, gata3 and ifng1) 8 wpi. Also in this setting, ifng1 expression was enhanced in il10e46/e46 compared to controls (Fig. 9). We did not see significant differences in the expression levels of cd4-1, tbx21 or gata3. However, the elevated ifng1 expression further supports the notion that a nonsense mutation in il10 leads to a Th1 cell type immune response in zebrafish.
Figure 9.
il10e46/e46 mutant progeny of il10e46/+ zebrafish have an elevated ifng1 expression level in a M. marinum infection. (a–d) il10e46/e46 mutants were outcrossed to wild type AB zebrafish and their heterozygous progeny was incrossed. The ungenotyped offspring of il10e46/+ fish was infected with a low-dose (8–17 CFU) of M. marinum. The relative expression of lymphocyte marker (cd4-1), Cd4 + lymphocyte transcription factors (tbx21, gata3) and Th1 cytokine (ifng1) was measured in the kidneys of il10e46/e46 (n = 18) and WT (n = 22) zebrafish at 8 wpi. Note the different scales of the y axes. Gene expressions were normalized to the expression of eef1a1l1. The data were collected from a single experiment. A two-tailed Mann-Whitney was used for the statistical comparison of differences.
To further evaluate if there are significant co-segregating mutations, we extracted the DNA from the abdominal organ blocks of the offspring of the heterozygous il10e46/+ mutants and performed whole genome sequencing. We compared the sequence of the coding regions of il10e46/e46 mutants and WT fish to the GRCz11 reference genome and to the sequence of WT AB zebrafish. The putative mutations found in the analysis are shown in the Supplementary Table S1. 57 mutations with a mutant allele fraction equal to or greater than 25% caused either a stop codon or a frameshift (Supplementary Table S2). One of these mutant alleles in gene si:dkey-19a16.2 had a 100% allele fraction in both il10e46/e46 mutants and WT zebrafish indicating that it is homozygous in both. The allele fraction of 17 mutations shown in the Supplementary Table S2 differed significantly (P < 0.05) between il10e46/e46 and WT zebrafish (Supplementary Table S3). As expected, e46 allele had 100% fraction in il10e46/e46 sample and 0% fraction in WT sample. Noteworthy, there were no other mutations that would cause either a nonsense or a frameshift mutation in the chromosome 11 (Supplementary Table S2). This indicates that none of these types of mutations are likely to be co-segregating with the e46 mutation. The role of the other detected mutations in the immunity or other phenotypes remains to be evaluated.
Discussion
The exact mechanisms of the immune defense against M. tuberculosis are in many ways still unknown, although disease progression has been studied in human samples, as well as with various model organisms in vivo (reviewed in57). Although spontaneous latency occurs in M. tuberculosis infected maqaque58 and rabbit59,60, these models raise serious ethical concerns. On the other hand, a M. tuberculosis infection in mouse is a well-established model for tuberculosis, but accomplishing a latent disease requires either a prior Bacillus Calmette-Guérin vaccination to boost the host’s immune response or a period of antibiotic treatment post infection in order to prevent an acute infection61. To study mycobacterial pathogenesis in the context of a natural host-pathogen interaction, a M. marinum infection in several fish species such as medaka62, goldfish43,63,64 and zebrafish35–37 is used to model tuberculosis (reviewed in65). Compared to other fish models, the zebrafish M. marinum infection has its benefits due to fast disease progression as well as the small size of the host43,62–64. In addition, a mycobacterial infection in adult zebrafish spontaneously reaches a latent state that can be reactivated with an immuno-suppressive treatment35.
IL10 KO mice develop an age-related enterocolitis, demonstrated by symptoms such as weight loss, diarrhea and abnormal gut morphology50. More specifically, the lack of IL10 leads to the excess production of proinflammatory cytokines, such as IL1b and TNF, as well as increased Th1 activation leading to the over-production of IFNG47,50. Interestingly, unchallenged il10e46/e46 zebrafish did not show any detectable signs of auto-immunity before the age of 10 months; the maximum age of the zebrafish used in the study. This is in line with the observed similar expression levels of the proinflammatory cytokine genes tnfa, tnfb and il1b, as well as the hallmark Th1 cytokine gene ifng1 in all of the studied tissues in il10e46/e46 mutants compared to WT control fish. Furthermore, no difference was observed in the relative expression levels of the T cell markers cd8a and cd4-1 or in our flow cytometric analysis of lymphocyte counts. In addition, Treg cells have been shown to be required in maintaining peripheral tolerance to prevent autoimmunity66–68 and recently, Sugimoto et al. have studied zebrafish colitis by using foxp3a mutant zebrafish28. While they found the inflammatory phenotype in these mutants, it did not lead to death as quickly as the corresponding genotype in mice. Consequently, they suggested that this may be due to the milder immune cell effector responses in aquatic vertebrates, including fish, which in turn may contribute to the ability of scarless regeneration of fish. Moreover, also in fish the microbes of the environment affect commensal bacteria, which in turn can explain the differences in enterocolitis between mice and fish69. It has also been suggested that fish maintenance in constant water circulation systems (similar to ones we have used) can prevent development of intestinal inflammation to some extent70. Noteworthy, in our studies, foxp3a expression was not altered in il10e46/e46 mutants. Since IL10 has been shown to maintain the FOXP3 expression in Treg cells in mice and humans, it points to a compensatory mechanism in fish71. Furthermore, Brugman et al. have speculated that there could be other immune mediators in addition to il1b and il10 which contribute to the development of enterocolitis in zebrafish70. Of note, we did observe increased IgM expression in the kidney as well as decreased IgM expression in the liver of il10e46/e46 zebrafish. Increased expression is in line with the previously reported higher serum immunoglobulin levels in IL10 deficient mice48,50 whereas decreased expression of IgM in the liver supports studies demonstrating carp IL10 in enhancing IgM + B cell proliferation72. Together these results indicate that, in contrast to IL10 KO mice, unchallenged il10e46/e46 mutant zebrafish have neither an overt inflammatory phenotype nor an inherently over-activated T cell response.
IL10 has been widely studied for its potent role in tuberculosis. Interestingly, mouse models of tuberculosis have not only reported that the lack of IL10 is beneficial to the host in a mycobacterial infection, but also that an IL10 deficiency can have detrimental effects on immunological control. For example, CBA/J background mice, which are susceptible to tuberculosis and whose IL10 function has been blocked, as well as Il10−/− mice, survived longer than control mice14,15, whereas Il10−/− mice with the C57BL/6 J background were all dead after ca. 27 weeks, while all the WT controls survived16. Among other things, these contradictory results have been attributed to differences in mouse and bacterial strains and to variation between mouse phenotypes (e.g. gut flora)73. Here, we first used zebrafish embryos in the context of a mycobacterial infection to specifically study the innate immune response prior to the development of the adaptive immunity. In our embryonic M. marinum infection, the mortality of il10e46/e46 mutant zebrafish larvae did not differ significantly from the WT controls. This suggests that in our model a nonsense mutation in the il10 gene does not improve, or compromise, resistance against mycobacteria during the early innate immune response. Accordingly, mouse models of tuberculosis have indicated that IL10 deficient mice show no mortality until ca. 14 wpi16 indicating a significant adaptive immune component in infection control. Using different injection route might lead to different outcome since the bacteria is able to proliferate in the yolk sac before it spreads into the different tissues74. However, Carvalho et al. have showed that the infection of M. marinum into the yolk sac leads to the formation of initial stages of granulomas, similar to ones that form when the caudal vein infection route is used38. The yolk sac infection method was also validated by showing that the decrease in the myeloid cell count lead to increased, and use of antibiotics to decreased M. marinum burden in the larvae38. Thus, since yolk sac infection is time-effective, it was chosen for the current study.
Next, we infected adult zebrafish with a low-dose of M. marinum and followed the fish for an average of 18.5 weeks. Curiously, the mortality of adult il10e46/e46 zebrafish was on average significantly lower compared to their WT controls. The impaired survival of WT zebrafish compared to il10e46/e46 mutants may be due to their inability to reach latency or a higher tendency for spontaneous reactivation. In fact, the function of IL10 has been linked to the reactivation of a chronic pulmonary M. tuberculosis infection in transgenic mice, which produce increased amounts of IL1075. In the study by Turner et al., the over-production of IL10 associated with a significantly increased bacterial burden as well as with decreased expression of Tnf and Il12p40 as signs of reactivation. Mechanistically, the higher susceptibility to mycobacterial reactivation in WT fish could be a consequence of an unfavorable inflammatory TNF/IL10 balance in the tuberculous granuloma as was previously reported in silico76.
In our low-dose infection model, we have previously demonstrated that a proinflammatory phenotype leading to an enhanced immediate immune response can associate with a decreased bacterial burden later in an infection at 9 wpi51. In addition, an appropriate TNF level has been shown to be important for tuberculosis immunity, as both the deficient and excessive production of TNF are linked to accelerated pathogenesis7. Here, we saw that, as in unchallenged zebrafish, the expression of the proinflammatory cytokine genes tnfa, il1b and tnfb in il10e46/e46 mutant fish was at the same level compared to WT controls at 1 and 6 dpi. Furthermore, at these early time points after a low-dose M. marinum infection, the expression of il1b and tnfb was not upregulated compared to the fish injected with PBS. This is consistent with our previous studies35 and with mouse studies in which TNF levels are not elevated in the serum or lungs of IL10 deficient mice before 14 and 15 dpi, respectively13. Expectedly, we did not see any changes or significant induction of the lymphocyte markers cd4, cd8 or IgM at 1 or 6 dpi. This is in line with the onset of the adaptive immune response only after two weeks post the mycobacterial infection55,77.
The immune response against tuberculosis requires an interplay between several of the host’s immune cells, the most important being macrophages, dendritic cells and CD4 + T lymphocytes78. Our flow cytometric analyses at 4 and 8 wpi exhibited similar lymphocyte amounts in il10e46/e46 mutant zebrafish compared to WT controls. Interestingly, however, il10e46/e46 mutant fish had significantly higher relative amounts of kidney myeloid cells compared to WT zebrafish at 8 wpi and further studies are warranted to understand the downstream effects of the lack of Il10 leading to increased myeloid cell counts after a mycobacterial infection. Although we could not detect any differences in total lymphocyte amounts between il10e46/e46 and WT zebrafish, our qPCR analysis on the unsorted kidney cell samples showed a trend of elevated cd4-1 expression in il10e46/e46 mutant fish compared to WT controls at 4 wpi and a significantly higher expression at 8 wpi. This may also be due to some other cell types than lymphocytes, such as dendritic cells and macrophages. However, this result is similar to IL10 deficient mice, in which elevated CD4 + T cell amounts are observed at different time points after an infection, and may imply an enhanced Th response in il10e46/e46 mutants14,15.
Th1 cells are important in attacking intracellular pathogens such as M. tuberculosis4–6. In tuberculosis, IFNG produced by Th1 cells activates macrophages, which results in the stimulation of phagocytosis, phagosome maturation, the production of reactive nitrogen intermediates and antigen presentation79. Furthermore, a M. tuberculosis infection in Il10 deficient mice has also been shown to lead to the enhanced production of IFNG by T cells13–15. il10e46/e46 mutant zebrafish had enhanced expression levels of the Th1 marker, tbx21, and decreased expression levels of the Th2 marker, gata3, at 8wpi. In addition, the Th1 cytokine ifng1 was upregulated at 8 wpi in il10e46/e46 mutants further suggesting a shift towards a Th1 cell mediated response. The upregulation of ifng1 in il10e46/e46 compared to WT control fish was also seen in the siblings from a heterozygous il10e46/+ incross. This further confirms the enhanced production of ifng1, likely from Th1 cells, in the absence of Il1014,15. However, since the whole genome sequencing revealed also other mutant alleles causing a stop codon or frameshift in addition to the e46 allele, we cannot exclude the possibility that these mutations contribute to our results. Noteworthy, with the allele fragment cut-off of 25% there were no stop codon or frameshift causing mutations in the chromosome 11 where il10 is located. In other words, our analysis suggests no significant co-segregating damaging mutations.
It has been previously shown that Th2 type response (gata3/tbx21 ratio) after four weeks of a M. marinum infection is associated with a low bacterial burden in the wild type AB fish27. In this study, adult wild type AB zebrafish were infected with a low-dose of M. marinum and were divided into three subgroups based on the bacterial burden at different time points. In the subgroup of the lowest bacterial burden, a Th2 type marker gene expression bias was observed. However, also Th1 response is induced (as indicated by elevated ifng1 expression also in this current study) in response to M. marinum infection35. Thus, for the optimal immune response both Th1 and Th2 types of responses are apparently required. Our current data indicate that lack of Il10 results in enhanced ifng1 expression and in better survival.
Although the role of IL10 in the host defense against M. tuberculosis is unambiguous, in vivo studies have concluded that in the absence of functional IL10 signaling, the Th1 response is hyper-activated resulting in improved resistance against a mycobacterial infection13,15,73. Consistent with this, our results show that a nonsense mutation in the zebrafish il10 leads to improved survival after a M. marinum infection, and associates with an enhanced Th1 response against mycobacteria. The higher Th1/Th2 ratio in the chronically infected il10e46/e46 mutant zebrafish is reflected by elevated ifng1 and tbx21 expression levels as well as decreased gata3 mRNA levels at 8 wpi. We did not detect differences in the expression of the proinflammatory cytokine genes tnfa, il1b and tnfb between unchallenged il10e46/e46 mutants and WT fish at the early stages of a M. marinum infection at 1 and 6 dpi. Taken together, these results suggest that the lack of Il10 does not enhance the early proinflammatory response after a M. marinum inoculate, but instead they highlight the importance of the Th1 response in resistance against a mycobacterial infection. Furthermore, our study validates the use of the zebrafish model of a M. marinum infection in tuberculosis studies.
Methods
Zebrafish lines and maintenance
Four to ten months old zebrafish were used in the adult experiments. The il10 mutation carrying zebrafish line e46 was obtained from Wellcome Trust Sanger Institute (Hinxton UK)44. In addition, wild type AB zebrafish from the Tampere Zebrafish Core Facility were used in whole genome sequencing experiment. Unchallenged fish were maintained in a standard flowthrough system (Aquatic Habitats, Florida, USA) with an automated light/dark cycle of 14 h and 10 h and fed with SDS 400 food (Special Diets Services, Essex, UK) twice and with in-house cultured Artemia nauplia once a day. During the time when gene expression measurements in the intestine, infection experiment with outcrossed e46 line and the whole genome sequencing were done, the fish were fed with GEMMA Micro 500 food (Skretting, Stavanger, Norway) once a day. The genotypes of the fish were confirmed by Sanger sequencing performed by our faculty’s sequencing facility. Homozygous fish and wild type (WT) siblings were spawn as separate groups and maintained separately for the experiments. M. marinum infected fish were maintained in a standard flowthrough system (Aqua Schwarz GMbH, Göttingen, Germany) in the aforementioned light/dark cycle and fed with SDS 400 food twice a day. Infected fish were monitored daily and humane endpoint criteria defined in animal experiment permits were applied. The Animal Experiment Board of Finland has approved the zebrafish housing, care, and all of the experiments (permits ESAVI/10079/04.10.06/2015, ESAVI/10823/04.10.07/2016 and ESAVI/2464/04.10.07/2017). The same ethical regulations as for all other vertebrate model animals are applied for zebrafish and all methods of this article were performed in accordance with relevant guidelines and regulations.
qPCR
Both RNA and DNA were extracted from kidney, spleen, liver, abdominal organ blocks (+/−kidney, which is the main hematopoietic tissue in zebrafish) and kidney blood cells with TRIreagent (Molecular Research Center, Ohio, USA) following the manufacturer’s coextraction protocol. The quality of the RNA was validated from several abdominal organ block (including kidney) samples of with 1.5% agarose (Bioline, London, United Kingdom) gel electrophoresis. The RNA samples were treated with the RapidOut DNA Removal Kit (Thermo Fischer Scientific, Waltham, USA) to remove genomic DNA. For the RNA samples, reverse transcription was done with the SensiFASTTM cDNA synthesis kit (BioLine, London, UK) and the relative gene expression levels of target genes were determined from cDNA with quantitative PCR (qPCR) using the PowerUp™ SYBR® master mix (Thermo Fischer Scientific). The qPCR primer sequences and the ZFIN identification codes for the analyzed genes are given in Supplementary Table S4. The expression levels of the target genes were calculated relative to the expression of eef1a1l180 using 2-ΔCt method. Total DNA was used to quantify the M. marinum colony forming units (CFU) with qPCR using SensiFAST™ SYBR® No-ROX (Bioline)35. The detection limit of this method was considered 1,000 CFU. qPCR was performed with a CFX96 qPCR machine (Bio-Rad, California, USA) and the data was analyzed with the Bio-Rad CFX Manager software v1.6 (Bio-Rad). Genomic DNA contamination was controlled by including no reverse transcriptase controls to the cDNA synthesis and to the following qPCR reaction from randomly selected RNA samples. The specificity of the qPCR for each amplified region was validated with a melt curve analysis and with a 1.5% agarose gel electrophoresis from a series of selected samples.
Imaging of zebrafish intestine
The intestine of il10e46/e46 mutants and WT zebrafish was imaged with Nikon SMZ745T microscope and DS-Vi1 camera (Nikon, Minato, Tokyo, Japan). The scale bar was measured with NIS-Elements D 4.2 software (Nikon).
Flow cytometry
Flow cytometry was performed as described previously19,51. In short, adult zebrafish were euthanized with 0.04% 3-amino benzoic acid ethyl ester and their kidneys were isolated and suspended in PBS with 0.5% fetal bovine serum (Gibco/Invitrogen, California, USA). Relative amounts of lymphocytes, blood cell precursors and myeloid cells in unchallenged and infected zebrafish were determined with a FACSCanto II (Becton, Dickinson, New Jersey, USA) and the data were analyzed with the FlowJo program (v7.5; Tree Star, Inc, Oregon, USA). After flow-cytometry, the remaining kidney cell suspensions from the M. marinum infected zebrafish were pelleted with 1,000 g for 1 min at + 4 °C, washed with 500 µl of PBS, centrifuged at 10,000 g for 1 min at + 4 °C, resuspended in 700 µl of TRIreagent (Molecular Research Center, Ohio, USA) and stored at −20 °C until RNA extraction. The remaining abdominal organ block (excluding kidney) from the infected fish were also collected and stored at −80 °C until DNA extractions. Throughout the article these remaining organs (without kidney) from flow cytometry are referred the abdominal organ block. In other experiments, abdominal organ block refers to all the organs isolated from the abdominal cavity (including kidney).
Experimental M. marinum infections
M. marinum (ATCC 927) was cultured and inoculated as described previously35. However, here M. marinum was suspended in phosphate buffered saline (PBS) rather than in potassium chloride prior to infections. In the zebrafish embryos, PBS with 2% polyvinylpyrrolidone-40 and 0.3 mg/ml phenol red (Sigma-Aldrich, Missouri, USA) was used as a mycobacterial carrier solution. A volume of 1 nl was injected 0–6 hours post fertilization into the yolk sac with aluminosilicate capillary needles (Sutter instrument Co., California, USA) using a micromanipulator (Narishige International, London UK) and a PV830 Pneumatic PicoPump (World Precision Instruments, Sarasota, Florida, USA) and visualized with a Stemi 2000 microscope (Carl Zeiss MicroImaging GmbH, Göttingen, Germany). Survival was followed daily by inspecting the larvae under a microscope. For the adult zebrafish infections, fish were anesthetized with 0.02% 3-amino benzoic acid ethyl ester, and 5 µl of M. marinum with 0.3 mg/ml phenol red (Sigma-Aldrich, Missouri, USA) was injected into the abdominal cavity with a 30 gauge Omnican 100 insulin needle (Braun, Melsungen, Germany). The M. marinum amounts (CFU) used in both the embryonic and adult infections were verified by plating bacterial inoculates on 7H10 agar (Becton Dickinson, New Jersey, USA) plates.
Ziehl-Neelsen staining
The presence of M. marinum in infected adult zebrafish was verified with Ziehl-Neelsen staining from paraffin embedded tissue sections as described previously35,81. Visualization of stained sections was performed using an Objective Imaging Surveyor virtual slide scanner (Objective Imaging, Cambridge, United Kingdom) and the scanned sections were digitized with a 20 × Plan Apochromatic microscope objective at a resolution of 0.4 μm/pixel. The image data were converted to JPEG2000 format as described previously82. The granulomas were counted and classified according to Myllymäki et al.56.
Whole genome sequencing
The DNA samples extracted from the offspring of the heterozygous il10e46/+ mutants and wild type AB fish was used for the whole genome sequencing. In each sample DNA of ten fish was pooled together. The RNA removal, Kapa Hyper Plus -library preparation and the whole genome sequencing were conducted at the Institute for Molecular Medicine Finland FIMM Technology Centre, Helsinki, Finland. The DNA libraries were sequenced with the 150 bp paired-end sequencing on the Illumina NovaSeq 6000 platform with a sequencing depth of >110 gigabases/sample.
Whole genome sequence alignment and quality control
Paired-end reads were processed prior to alignment to remove sequencing adapters and low-quality bases at the read tails. Adapters were trimmed using cutadapt-1.1183 in paired mode. Low-quality bases at read ends with a smoothed base quality <25 were trimmed using an in-house algorithm. After quality control, the paired-end reads were aligned against the GRCz11 zebrafish reference genome using Bowtie-2.3.084. Optical and PCR duplicates were removed using samblaster-0.1.2485.
Mutation analysis from the whole genome sequencing data
Mutations were called from the il10e46/e46 mutant and WT zebrafish using an in-house pipeline. This was done by identifying variants with an alternate allele fraction of at least 20% and at least 5 supporting reads. The allele fraction was also required to be 20 times higher than the alternate allele fraction of the mutation in the AB zebrafish. Mutations and their protein-level effects were annotated using ANNOVAR86. Frameshift and stop-gain mutations were curated using Integrative Genomics Viewer (IGV)87.
Statistical analyses
Web-based ClinCalc program (http://clincalc.com/Stats/SampleSize.aspx) was used for all of the sample size calculations. Based on our previous adult zebrafish survival experiments with a low-dose M. marinum infection, we estimated the end-point mortality difference between WT and il10e46/e46 zebrafish to be 40%. With a 80% statistical power, a minimum group size of 22 was determined for the survival experiments. In order to estimate the bacterial quantification sample size for the current research, our previously published M. marinum quantification with 0.5 unit difference (log10 scale, standard deviation 0.5) between study groups51 was used as a guideline. Using the desired 80% power this difference accounted for a group size of 16 fish for the bacterial quantification.
Statistical analyses of the results, except the whole genome sequencing data, were performed with the Prism program, version 5.02 (GraphPad Software, Inc, California, USA). The statistical significance from the survival experiments was determined with a log-rank (Mantel-Cox) test and from the flow cytometry and qPCR experiments with a nonparametric Mann-Whitney analysis. When analyzing the whole genome sequencing data, P values for each mutation were calculated between the il10e46/e46 mutants and WT fish using Fisher’s exact test. P values of <0.05 were considered significant.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request. The whole genome sequencing data is available at European Nucleotide Archive (https://www.ebi.ac.uk/ena/; ERP number: ERP109293).
Electronic supplementary material
Acknowledgements
This study was financially supported by the Academy of Finland (M.R., 277495), the Sigrid Juselius Foundation (M.R.), the Jane and Aatos Erkko Foundation (M.R.), the Competitive State Research Financing of the Expert Responsibility Area of Tampere University Hospital (M.R.), Competitive State Research Financing of the Expert Responsibility area of Oulu University Hospital (M.R.) and the Tampere Tuberculosis Foundation (M.R., S.-K.H.), the Emil Aaltonen Foundation (S.-K.H), Foundation of the Finnish Anti-Tuberculosis Association (S.-K.H.), the City of Tampere Science Foundation (S.-K.H), the Väinö and Laina Kivi Foundation (S.-K.H.), the Finnish Cultural Foundation, the Central Foundation (S.-K.H.), the Finnish Concordia Fund (S.-K.H.), Orion Research Foundation sr (S.-K.H) and University of Tampere Doctoral Programme in Biomedicine and Biotechnology (M.O.). We thank the Tampere Zebrafish Core Facility, partly funded by Biocenter Finland, for maintaining and providing the zebrafish, Hannaleena Piippo, Jenna Ilomäki, Leena Mäkinen, Tuula Myllymäki and Sami Leino for technical assistance, Marko Pesu, Mataleena Parikka and Hannu Turpeinen for scientific advice and support, Heini Huhtala for the statistical advice and Helen Cooper for proof-reading this manuscript.
Author Contributions
S.-K.H., M.O., M.N. and M.R. designed the experiments. S.-K.H., M.O., S.T. and M.R. wrote the paper. S.-K.H. and M.O. performed the experiments. S.-K.H., M.O., S. T and M.N. analyzed the data. All authors reviewed and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28511-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization. Global tuberculosis report 2016. http://www.who.int/tb/publications/global_report/en/ (2016).
- 2.Havlir DV, Barnes PF. Tuberculosis in patients with human immunodeficiency virus infection. N. Engl. J. Med. 1999;340:367–373. doi: 10.1056/NEJM199902043400507. [DOI] [PubMed] [Google Scholar]
- 3.Harris J, Keane J. How tumour necrosis factor blockers interfere with tuberculosis immunity. Clin. Exp. Immunol. 2010;161:1–9. doi: 10.1111/j.1365-2249.2010.04146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flynn JL, et al. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper AM, Magram J, Ferrante J, Orme IM. Interleukin 12 (IL-12) is crucial to the development of protective immunity in mice intravenously infected with Mycobacterium tuberculosis. J. Exp. Med. 1997;186:39–45. doi: 10.1084/jem.186.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper AM, et al. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roca FJ, Ramakrishnan L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell. 2013;153:521–534. doi: 10.1016/j.cell.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiorentino DF, Bond MW, Mosmann TR. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J. Exp. Med. 1989;170:2081–2095. doi: 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore KW, et al. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science. 1990;248:1230–1234. doi: 10.1126/science.2161559. [DOI] [PubMed] [Google Scholar]
- 10.Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol. Rev. 2008;226:205–218. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pacheco AG, Cardoso CC, Moraes MO. IFNG+874T/A, IL10 −1082G/A and TNF −308G/A polymorphisms in association with tuberculosis susceptibility: a meta-analysis study. Hum. Genet. 2008;123:477–484. doi: 10.1007/s00439-008-0497-5. [DOI] [PubMed] [Google Scholar]
- 12.Delgado JC, Baena A, Thim S, Goldfeld AE. Ethnic-specific genetic associations with pulmonary tuberculosis. J. Infect. Dis. 2002;186:1463–1468. doi: 10.1086/344891. [DOI] [PubMed] [Google Scholar]
- 13.Redford PS, et al. Enhanced protection to Mycobacterium tuberculosis infection in IL-10-deficient mice is accompanied by early and enhanced Th1 responses in the lung. Eur. J. Immunol. 2010;40:2200–2210. doi: 10.1002/eji.201040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cyktor JC, et al. IL-10 inhibits mature fibrotic granuloma formation during Mycobacterium tuberculosis infection. J. Immunol. 2013;190:2778–2790. doi: 10.4049/jimmunol.1202722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beamer GL, et al. Interleukin-10 promotes Mycobacterium tuberculosis disease progression in CBA/J mice. J. Immunol. 2008;181:5545–5550. doi: 10.4049/jimmunol.181.8.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins DM, et al. Lack of IL-10 alters inflammatory and immune responses during pulmonary Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 2009;89:149–157. doi: 10.1016/j.tube.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Wittamer V, Bertrand JY, Gutschow PW, Traver D. Characterization of the mononuclear phagocyte system in zebrafish. Blood. 2011;117:7126–7135. doi: 10.1182/blood-2010-11-321448. [DOI] [PubMed] [Google Scholar]
- 18.Lin AF, et al. The DC-SIGN of zebrafish: insights into the existence of a CD209 homologue in a lower vertebrate and its involvement in adaptive immunity. J. Immunol. 2009;183:7398–7410. doi: 10.4049/jimmunol.0803955. [DOI] [PubMed] [Google Scholar]
- 19.Langenau DM, et al. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl. Acad. Sci. USA. 2004;101:7369–7374. doi: 10.1073/pnas.0402248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willett CE, Zapata AG, Hopkins N, Steiner LA. Expression of zebrafish rag genes during early development identifies the thymus. Dev. Biol. 1997;182:331–341. doi: 10.1006/dbio.1996.8446. [DOI] [PubMed] [Google Scholar]
- 21.Yoon S, et al. First demonstration of antigen induced cytokine expression by CD4-1+lymphocytes in a poikilotherm: Studies in zebrafish (Danio rerio) Plos One. 2015;10:e0126378. doi: 10.1371/journal.pone.0126378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danilova N, Steiner LA. B cells develop in the zebrafish pancreas. Proc. Natl. Acad. Sci. USA. 2002;99:13711–13716. doi: 10.1073/pnas.212515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page DM, et al. An evolutionarily conserved program of B-cell development and activation in zebrafish. Blood. 2013;122:e1–11. doi: 10.1182/blood-2012-12-471029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balla KM, et al. Eosinophils in the zebrafish: prospective isolation, characterization, and eosinophilia induction by helminth determinants. Blood. 2010;116:3944–3954. doi: 10.1182/blood-2010-03-267419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dee CT, et al. CD4-Transgenic zebrafish reveal tissue-resident Th2- and regulatory T cell–like populations and diverse mononuclear phagocytes. J. Immunol. 2016;197:3520–3530. doi: 10.4049/jimmunol.1600959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitra, S., Alnabulsi, A., Secombes, C. J. & Bird, S. Identification and characterization of the transcription factors involved in T-cell development, t-bet, stat6 and foxp3, within the zebrafish, Danio rerio. FEBS J. 277, 128-147 (2010). [DOI] [PubMed]
- 27.Hammarén MM, et al. Adequate Th2-type response associates with restricted bacterial growth in latent mycobacterial infection of zebrafish. Plos Pathog. 2014;10:e1004190. doi: 10.1371/journal.ppat.1004190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugimoto K, Hui SP, Sheng DZ, Nakayama M, Kikuchi K. Zebrafish FOXP3 is required for the maintenance of immune tolerance. Dev. Comp. Immunol. 2017;73:156–162. doi: 10.1016/j.dci.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Cui P. Complement system in zebrafish. Dev. Comp. Immunol. 2014;46:3–10. doi: 10.1016/j.dci.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Zimmerman AM, Moustafa FM, Romanowski KE, Steiner LA. Zebrafish immunoglobulin IgD: unusual exon usage and quantitative expression profiles with IgM and IgZ/T heavy chain isotypes. Mol. Immunol. 2011;48:2220–2223. doi: 10.1016/j.molimm.2011.06.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohi O, Parikka M, Ramet M. The zebrafish as a model for paediatric diseases. Acta Paediatr. 2013;102:104–110. doi: 10.1111/j.1651-2227.2012.02835.x. [DOI] [PubMed] [Google Scholar]
- 32.Stinear TP, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Etr SH, Yan L, Cirillo JD. Fish monocytes as a model for mycobacterial host-pathogen interactions. Infect. Immun. 2001;69:7310–7317. doi: 10.1128/IAI.69.12.7310-7317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barker LP, George KM, Falkow S, Small PL. Differential trafficking of live and dead Mycobacterium marinum organisms in macrophages. Infect. Immun. 1997;65:1497–1504. doi: 10.1128/iai.65.4.1497-1504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parikka M, et al. Mycobacterium marinum causes a latent infection that can be reactivated by gamma irradiation in adult zebrafish. Plos Pathog. 2012;8:e1002944. doi: 10.1371/journal.ppat.1002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prouty MG, Correa NE, Barker LP, Jagadeeswaran P, Klose KE. Zebrafish-Mycobacterium marinum model for mycobacterial pathogenesis. FEMS Microbiol. Lett. 2003;225:177–182. doi: 10.1016/S0378-1097(03)00446-4. [DOI] [PubMed] [Google Scholar]
- 37.Swaim LE, et al. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect. Immun. 2006;74:6108–6117. doi: 10.1128/IAI.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carvalho R, et al. A high-throughput screen for tuberculosis progression. Plos One. 2011;6:e16779. doi: 10.1371/journal.pone.0016779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dalton JP, et al. Screening of anti-mycobacterial compounds in a naturally infected zebrafish larvae model. J. Antimicrob. Chemother. 2017;72:421–427. doi: 10.1093/jac/dkw421. [DOI] [PubMed] [Google Scholar]
- 40.Davis JM, et al. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693–702. doi: 10.1016/S1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- 41.Piazzon MC, Lutfalla G, Forlenza M. IL10, A tale of an evolutionarily conserved cytokine across vertebrates. Crit. Rev. Immunol. 2016;36:99–129. doi: 10.1615/CritRevImmunol.2016017480. [DOI] [PubMed] [Google Scholar]
- 42.Zhang DC, Shao YQ, Huang YQ, Jiang SG. Cloning, characterization and expression analysis of interleukin-10 from the zebrafish (Danio rerion) J. Biochem. Mol. Biol. 2005;38:571–576. doi: 10.5483/bmbrep.2005.38.5.571. [DOI] [PubMed] [Google Scholar]
- 43.Grayfer L, Belosevic M. Identification and molecular characterization of the interleukin-10 receptor 1 of the zebrafish (Danio rerio) and the goldfish (Carassius auratus L.) Dev. Comp. Immunol. 2012;36:408–417. doi: 10.1016/j.dci.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Kettleborough RN, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nickless, A., Bailis, J. M. & You, Z. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell. Biosci. 7, 7; 10.1186/s13578-017-0153-7 eCollection2017 (2017). [DOI] [PMC free article] [PubMed]
- 46.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 47.Bristol, I. J., Mahler, M. & Leiter, E. H. Interleukin-10 gene targeted mutation. JAX notes (1997).
- 48.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-P. [DOI] [PubMed] [Google Scholar]
- 49.Keubler LM, Buettner M, Hager C, Bleich A. A Multihit Model: Colitis Lessons from the Interleukin-10-deficient Mouse. Inflamm. Bowel Dis. 2015;21:1967–1975. doi: 10.1097/MIB.0000000000000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gomes-Santos AC, et al. New insights into the immunological changes in IL-10-deficient mice during the course of spontaneous inflammation in the gut mucosa. Clin. Dev. Immunol. 2012;2012:560817. doi: 10.1155/2012/560817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ojanen MJ, et al. The proprotein convertase subtilisin/kexin furinA regulates zebrafish host response against Mycobacterium marinum. Infect. Immun. 2015;83:1431–1442. doi: 10.1128/IAI.03135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barsig J, et al. Lipopolysaccharide-induced interleukin-10 in mice: role of endogenous tumor necrosis factor-alpha. Eur. J. Immunol. 1995;25:2888–2893. doi: 10.1002/eji.1830251027. [DOI] [PubMed] [Google Scholar]
- 53.Verbon A, et al. Serum concentrations of cytokines in patients with active tuberculosis (TB) and after treatment. Clin. Exp. Immunol. 1999;115:110–113. doi: 10.1046/j.1365-2249.1999.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Del Prete G, et al. Human IL-10 is produced by both type 1 helper (Th1) and type 2 helper (Th2) T cell clones and inhibits their antigen-specific proliferation and cytokine production. J. Immunol. 1993;150:353–360. [PubMed] [Google Scholar]
- 55.van Crevel R, Ottenhoff TH, van der Meer JW. Innate immunity to Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2002;15:294–309. doi: 10.1128/CMR.15.2.294-309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Myllymäki, H., Niskanen, M., Luukinen, H., Parikka, M. & Ramet, M. Identification of protective postexposure mycobacterial vaccine antigens using an immunosuppression-based reactivation model in the zebrafish. Dis. Model. Mech. 11, 10.1242/dmm.033175 (2018). [DOI] [PMC free article] [PubMed]
- 57.Myllymäki H, Niskanen M, Oksanen KE, Rämet M. Animal models in tuberculosis research – where is the beef? Expert Opinion on Drug Discovery. 2015;10:871–883. doi: 10.1517/17460441.2015.1049529. [DOI] [PubMed] [Google Scholar]
- 58.Lin PL, et al. Quantitative comparison of active and latent tuberculosis in the cynomolgus macaque model. Infect. Immun. 2009;77:4631–4642. doi: 10.1128/IAI.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subbian S, et al. Spontaneous latency in a rabbit model of pulmonary tuberculosis. Am. J. Pathol. 2012;181:1711–1724. doi: 10.1016/j.ajpath.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subbian S, et al. Molecular immunologic correlates of spontaneous latency in a rabbit model of pulmonary tuberculosis. Cell. Commun. Signal. 2013;11:16. doi: 10.1186/1478-811X-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi C, Shi J, Xu Z. A review of murine models of latent tuberculosis infection. Scand. J. Infect. Dis. 2011;43:848–856. doi: 10.3109/00365548.2011.603745. [DOI] [PubMed] [Google Scholar]
- 62.Broussard GW, Ennis DG. Mycobacterium marinum produces long-term chronic infections in medaka: a new animal model for studying human tuberculosis. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 2007;145:45–54. doi: 10.1016/j.cbpc.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruley KM, et al. Identification of Mycobacterium marinum virulence genes using signature-tagged mutagenesis and the goldfish model of mycobacterial pathogenesis. FEMS Microbiol. Lett. 2004;232:75–81. doi: 10.1016/S0378-1097(04)00017-5. [DOI] [PubMed] [Google Scholar]
- 64.Talaat AM, Reimschuessel R, Wasserman SS, Trucksis M. Goldfish, Carassius auratus, a novel animal model for the study of Mycobacterium marinum pathogenesis. Infect. Immun. 1998;66:2938–2942. doi: 10.1128/iai.66.6.2938-2942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Myllymäki H, Bäuerlein CA, Rämet M. The zebrafish breathes new life into the study of tuberculosis. Front. Immunol. 2016;7:196. doi: 10.3389/fimmu.2016.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakaguchi S. Regulatory T cells: history and perspective. Methods Mol. Biol. 2011;707:3–17. doi: 10.1007/978-1-61737-979-6_1. [DOI] [PubMed] [Google Scholar]
- 67.Sakaguchi S. Regulatory T cells: key controllers of immunologic self-tolerance. Cell. 2000;101:455–458. doi: 10.1016/S0092-8674(00)80856-9. [DOI] [PubMed] [Google Scholar]
- 68.Pesu M, et al. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature. 2008;455:246–250. doi: 10.1038/nature07210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brugman S. The zebrafish as a model to study intestinal inflammation. Dev. Comp. Immunol. 2016;64:82–92. doi: 10.1016/j.dci.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 70.Brugman S, et al. Oxazolone-induced enterocolitis in zebrafish depends on the composition of the intestinal microbiota. Gastroenterology. 2009;137:1757.e1. doi: 10.1053/j.gastro.2009.07.069. [DOI] [PubMed] [Google Scholar]
- 71.Paul G, Khare V, Gasche C. Inflamed gut mucosa: downstream of interleukin-10. Eur. J. Clin. Invest. 2012;42:95–109. doi: 10.1111/j.1365-2362.2011.02552.x. [DOI] [PubMed] [Google Scholar]
- 72.Piazzon MC, Savelkoul HS, Pietretti D, Wiegertjes GF, Forlenza M. Carp Il10 has anti-inflammatory activities on phagocytes, promotes proliferation of memory T cells, and regulates B cell differentiation and antibody secretion. J. Immunol. 2015;194:187–199. doi: 10.4049/jimmunol.1402093. [DOI] [PubMed] [Google Scholar]
- 73.Redford PS, Murray PJ, O’Garra A. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 2011;4:261–270. doi: 10.1038/mi.2011.7. [DOI] [PubMed] [Google Scholar]
- 74. Benard, E. L. et al. Infection of zebrafish embryos with intracellular bacterial pathogens. J. Vis. Exp. 61, 10.3791/3781 (2012). [DOI] [PMC free article] [PubMed]
- 75.Turner J, et al. In vivo IL-10 production reactivates chronic pulmonary tuberculosis in C57BL/6 mice. J. Immunol. 2002;169:6343–6351. doi: 10.4049/jimmunol.169.11.6343. [DOI] [PubMed] [Google Scholar]
- 76.Cilfone NA, Perry CR, Kirschner DE, Linderman JJ. Multi-scale modeling predicts a balance of tumor necrosis factor-alpha and interleukin-10 controls the granuloma environment during Mycobacterium tuberculosis infection. Plos One. 2013;8:e68680. doi: 10.1371/journal.pone.0068680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cooper AM. Cell-mediated immune responses in tuberculosis. Annu. Rev. Immunol. 2009;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Korb VC, Chuturgoon AA, Moodley D. Mycobacterium tuberculosis: Manipulator of protective immunity. Int. J. Mol. Sci. 2016;17:131. doi: 10.3390/ijms17030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lyadova IV, Panteleev AV. Th1 and Th17 cells in tuberculosis: Protection, pathology, and biomarkers. Mediators Inflamm. 2015;2015:854507. doi: 10.1155/2015/854507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang R, Dodd A, Lai D, McNabb WC, Love DR. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim. Biophys. Sin. (Shanghai) 2007;39:384–390. doi: 10.1111/j.1745-7270.2007.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oksanen KE, et al. An adult zebrafish model for preclinical tuberculosis vaccine development. Vaccine. 2013;31:5202–5209. doi: 10.1016/j.vaccine.2013.08.093. [DOI] [PubMed] [Google Scholar]
- 82.Tuominen VJ, Isola J. The application of JPEG2000 in virtual microscopy. J. Digit. Imaging. 2009;22:250–258. doi: 10.1007/s10278-007-9090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 84.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Faust GG, Hall IM. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30:2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request. The whole genome sequencing data is available at European Nucleotide Archive (https://www.ebi.ac.uk/ena/; ERP number: ERP109293).