Abstract
p53 is a transcription factor that regulates expression of genes involved in cell cycle arrest, senescence, and apoptosis. TP53 harbors mutations that inactivate its transcriptional activity in roughly 30% of breast cancers, and these tumors are much more likely to undergo a pathological complete response to chemotherapy. Thus, the gene expression program activated by wild-type p53 contributes to a poor response. We used an in vivo genetic model system to comprehensively define the p53- and p21-dependent genes and pathways modulated in tumors following doxorubicin treatment. We identified genes differentially expressed in spontaneous mammary tumors harvested from treated MMTV-Wnt1 mice that respond poorly (Trp53+/+) or favorably (Trp53-null) and those that lack the critical senescence/arrest p53 target gene Cdkn1a. Trp53 wild-type tumors differentially expressed nearly 10-fold more genes than Trp53-null tumors after treatment. Pathway analyses showed that genes involved in cell cycle, senescence, and inflammation were enriched in treated Trp53 wild-type tumors; however, no genes/pathways were identified that adequately explain the superior cell death/tumor regression observed in Trp53-null tumors. Cdkn1a-null tumors that retained arrest capacity (responded poorly) and those that proliferated (responded well) after treatment had remarkably different gene regulation. For instance, Cdkn1a-null tumors that arrested upregulated Cdkn2a (p16), suggesting an alternative, p21-independent route to arrest. Live animal imaging of longitudinal gene expression of a senescence/inflammation gene reporter in Trp53+/+ tumors showed induction during and after chemotherapy treatment, while tumors were arrested, but expression rapidly diminished immediately upon relapse.
Introduction
The tumor suppressor TP53 is mutated in ~30% of breast cancers [1]. p53 is a transcription factor that is activated by various cellular stresses, including DNA damage induced by chemotherapy regimens [2]. Following damage to DNA, p53 is phosphorylated and dissociated from its negative regulator, Mdm2, allowing the protein to accumulate and become transcriptionally active [3].
Paradoxically, tumors that harbor a mutation in p53 have a significantly greater probability of achieving a pathological complete response to neoadjuvant chemotherapy [4], [5], [6], [7], [8]. Supporting these findings, mouse models of breast cancer have shown that Trp53 mutant mammary tumors respond better to chemotherapy than Trp53 wild-type tumors [9], [10], [11]. Both mouse mammary tumors [9] and human breast cancers [12] that are wild-type for p53 undergo cell senescence in response to chemotherapy rather than the widespread cell death seen in mutant tumors [9]. The greater the extent of residual disease present at the time of surgery following chemotherapy, the less favorable the prognosis for the patient [13]. Thus, because p53 mediated arrest and senescence is a barrier to eradicating tumors and achieving a low residual disease burden, understanding the genes regulated by p53 in this context is of the highest importance.
The fate of a cell with wild-type TP53, when exposed to DNA-damaging agents, can be cell cycle arrest, senescence, or apoptosis, which is guided by a transcriptional profile of induced and repressed gene expression [3], [14]. Which outcome occurs is decided by variety of factors such as tissue specificity and extent of damage, which are not fully understood [14].
One of the first discovered and arguably one of the most important targets of p53 is the cyclin-dependent kinase inhibitor p21, coded by the gene Cdkn1a [15], [16], [17]. p21 mediates cell cycle arrest induced by p53 after DNA damage [18], [19], [20]. Various cell types lacking p21 fail to arrest after DNA damage, similar to cells lacking p53 [20], [21]. Here, we have examined gene expression in MMTV-Wnt1 mammary tumors that are Trp53 wild-type (WT), Trp53-null, and Cdkn1a-null, following doxorubicin chemotherapy treatment.
Materials and Methods
Mouse Tumors and RNA Sequencing
All tumors used were from MMTV-Wnt1 transgenic mice that were Trp53+/+, Trp53−/− (null), or Cdkn1a−/− (null) and were previously described [9]. The doxorubicin treatment regimen and harvesting schedule for mice that formed spontaneous tumors have been described [9]. RNA was prepared using Trizol, and RNA sequencing was performed by the MD Anderson Cancer Center Sequencing & Microarray Facility. RNA sequencing libraries were prepared using the TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Human/Mouse/Rat Sets A and B (Trp53WT) and Illumina TruSeq RNA Library Preparation Kit v2 (Trp53null and Cdkn1a-null tumors). All samples were aligned to the mm10 genome using the STAR RNA-seq aligner [22]. Gene expression was quantified using RSEM [23], and differential expression was calculated using the EBSeq statistical package with a posterior probability of equal expression (PPEE) < .05 considered statistically significant. To avoid any artifacts introduced by batch effects or library preparations, only intragenotype comparisons were made.
Ingenuity Pathway Analysis (IPA)
Data analysis through IPA IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity) was performed using the lists of fold changes for significantly expressed protein coding genes after doxorubicin treatment for each genotype. IPA “Upstream Regulators” were determined by overlap between known regulators and targets in the Ingenuity Knowledge Base and the targets expressed in the experimental data (overlap P value determined by Fisher's exact test), and the activation z-score, which predicts the direction of regulation. IPA identified “Enriched diseases and biological functions” with a significant P value of < .05 as determined by Fisher's exact test.
GSEA Analysis
Gene expression data matrices were constructed for each genotype. As the Kyoto Encyclopedia of Genes and Genomes (KEGG) [24] and REACTOME pathways contained only human protein coding genes, only protein coding genes with an HGNC ID (PMID: 29126148) and a human ortholog were retained for Gene set enrichment analyses (GSEA). Analysis was performed using GSEA version 3.0 (Broad Institute, Cambridge, MA) [25]. One thousand gene set permutations were run for each pathway included in the KEGG pathway database [24] and for pathways related to cell cycle in the REACTOME pathway database [26]. Gene set enrichment was considered significant at an FDR< 0.25 [25].
CIBERSORT Analysis
The CIBERSORT algorithm [27] was used to infer immune cell populations in tumor samples. The training set was constructed using publicly available RNA-seq datasets of purified macrophages, T cells, B cells, and several breast cancer cell lines (accession numbers available upon request). The algorithm was run on a gene expression data matrix of all samples in this study. Each sample was assigned a P value following deconvolution. Samples included in our analysis had a deconvolution P value <.05.
Selection of True TP53 Targets
Riley et al. identified 129 possible direct targets through searching the literature and narrowing candidates that fulfill three out of four determined criteria, which include RNA and protein levels that are regulated by p53, presence of a p53 response element, reporter expression validation, and successful chromatin immunoprecipitation [28]. Allen et al. employed the small molecule inhibitor of Mdm2, nutlin-3, for 1 hour to specifically activate p53. They then performed global run-on sequencing and identified 198 possible direct targets of p53 [29]. A third study used both chromatin immunoprecipitation sequencing and RNA sequencing to identify 432 genes directly regulated by p53 in mouse cells [30].
Luciferase Reporter Assay In Vivo
The reporter lentiviral vector was made using the backbone CD502A-1 (pCDH-EF1α-MCS cDNA Single Promoter Cloning and Expression Lentivector) from Systems Biosciences and contains 1246 bp 5′ of the transcriptional start site of murine Cxcl5, followed by a Luciferase cassette, and another cassette with the EF1 promoter driving mCherry. Lentivirus was prepared in 293 T cells using a standard calcium phosphate transfection protocol. Viral supernatant was collected 48 hours and 72 hours following transfection, filtered (0.45 μm), and used to infect target cells in the presence of polybrene (8 μg/ml). Two MMTV-Wnt1 tumor orthotopic transplants were established and serially propagated as previously described [9]. Tumors that formed were excised, minced with a razor, filtered through a 50-μm filter, and plated in complete DMEM. One day later, concentrated virus was applied, and 24 hours later, media were changed. Two to 3 days after, cells were trypsinized and ~100,000 mCherry-positive cells were sorted by flow cytometry, washed with PBS, and resuspended in Matrigel and injected into the #4 mammary gland of C57Bl6/j mice. The tumor that formed was verified for mCherry fluorescence and transplanted again into multiple C57Bl6/j mice. When tumors formed, they were measured with calipers as previously described [9], IP injected with 10 μg/g of body weight luciferin, and imaged on an IVIS for fluorescence. Tumor volume was calculated as previously described [9]. Luciferase activity was plotted by average counts/tumor volume and normalized for each tumor transplant.
Results
p53 Dominates the Transcriptional Response to Doxorubicin Treatment in Mammary Tumors
TP53 codes for the transcription factor p53 that transactivates genes following many types of cell stress. In breast cancer, the probability of a pathological complete response to chemotherapy is much lower when TP53 is wild-type as opposed to mutant [4], [5], [6], [7], [8]. This implicates the genes and pathways modulated by p53 as contributors to the persistence of tumor cells as residual disease, promoting eventual relapse. Defining the p53 transcriptional response to chemotherapy is critical for understanding the reason tumors fail to regress and for developing therapies to target persisting cells. RNA sequencing was employed to comprehensively define the in vivo transcriptional program activated by p53 in mammary tumors after chemotherapy. Gene expression changes were determined in tumors from mice harvested 24 hours following the fifth consecutive day of doxorubicin treatment [9]. We used MMTV-Wnt1 transgenic mice that were Trp53 wild-type (WT), Trp53-null, or Cdkn1a-null. Tumors that arise in MMTV-Wnt1 transgenic mice remain wild type for Trp53 and respond to doxorubicin by undergoing arrest and senescence. If Trp53 is deleted, mutated, or otherwise null, these tumors instead undergo mitotic catastrophe and cell death, resulting in a clinically superior response [9].
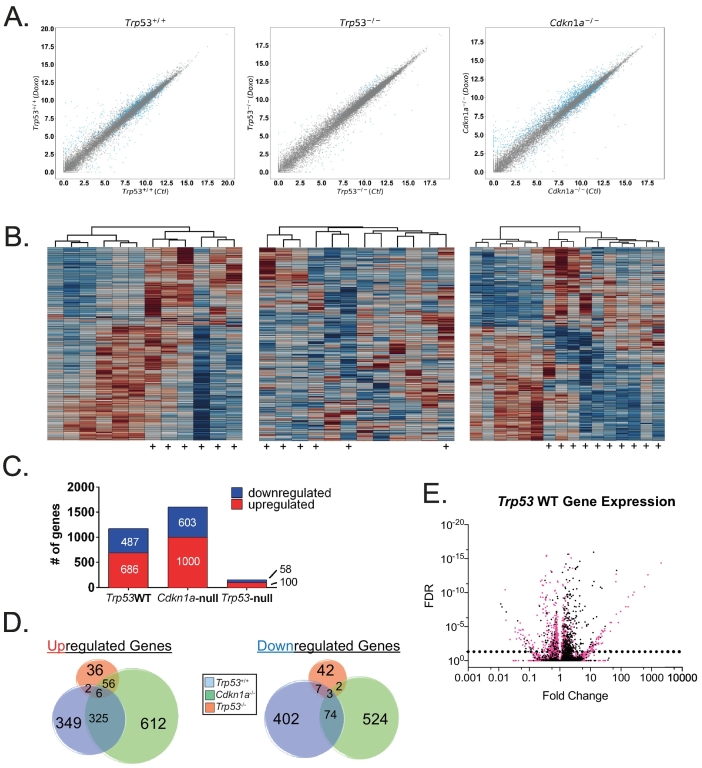
Using a false discovery rate (FDR) < 0.05, we found that Trp53WT MMTV-Wnt1 tumors had 1173 protein-coding genes differentially expressed following treatment (Figure 1, A left and C). Cdkn1a-null tumors had 1603 genes significantly changed (Figure 1, A right and C). In sharp contrast, only 158 genes were significantly changed in Trp53-null tumors (Figure 1, A center and C), despite this genotype responding better to treatment. Heat maps were generated for each genotype that included every differentially expressed gene from each of the three genotypes. Many genes were modulated after doxorubicin treatment in Trp53WT and Cdkn1a-null tumors (Figure 1B), while Trp53-null tumors had relatively few genes, and treated tumors did not even cluster entirely together (Figure 1B, center). Of note, Cdkn1a transcripts were elevated in Cdkn1a-null tumors. Aligning reads to the Cdkn1a locus showed they were all localized to the 3′ UTR region of the gene and none in the coding region (Supplementary Figure 1), consistent with the knockout strategy used to generate these mice [18].
Figure 1.
p53 mediates the vast majority of gene expression changes in mammary tumors of mice treated with doxorubicin. MMTV-Wnt1 transgenic mice with mammary tumors that were Trp53+/+, Trp53−/−, or Cdkn1a−/− were untreated and harvested (Ctl), or treated with 4 mg/kg doxorubicin IP for 5 consecutive days and then harvested 24 hours following the final treatment (Doxo). (A) After RNA sequencing, scatter plots of differential gene expression in doxorubicin-treated (doxo) versus untreated control (Ctl) were generated for each genotype. Genes significantly changed (PPEE<.05) are represented by a blue point. (B) Heat maps corresponding to genotypes in the scatter plots of (A) were generated for control and doxorubicin-treated (indicated by +) tumors using the union of every differentially expressed gene for each genotype. (C) Bar chart of the total number of significantly regulated protein-coding genes (FDR<0.05) for Trp53+/+, Cdkn1a−/−, or Trp53−/− cohorts. (D). Venn diagram depicting total numbers of significantly regulated (FDR<0.05) protein-coding genes for each cohort analyzed and the numbers of commonly regulated genes. (E) Volcano plot showing FDR(PPEE) on the Y-axis and fold change on the X-axis for genes modulated after doxorubicin treatment in Trp53+/+ tumors. Those genes not significantly regulated in Cdkn1a-null tumors are shown in pink.
We next examined overlapping gene expression changes in tumors from the three genotypes. We found that Trp53WT and Cdkn1a-null tumors shared the regulation of many genes following doxorubicin treatment, while Trp53-null had few in common with either tumor genotype (Figure 1D). Comparing Trp53-null and Trp53WT, strikingly, only 18 genes were similarly altered in both, showing that the vast majority of transcriptional changes following doxorubicin treatment are p53 dependent (Figure 1D). Interestingly, many genes (760) regulated in Trp53WT tumors were no longer regulated in Cdkn1a-null tumors (Figure 1E), which may reflect a dependence on p21-mediated effects for indirect gene expression changes. Also, while almost half of the genes upregulated in Trp53WT tumors were still expressed in Cdkn1a-null tumors, only 16% of downregulated genes remain repressed, suggesting that p21 plays a more important role in mediating gene repression by p53 as opposed to induction.
p53 Signaling and Cell Cycle Pathway Analyses in Doxorubicin-Treated Tumors
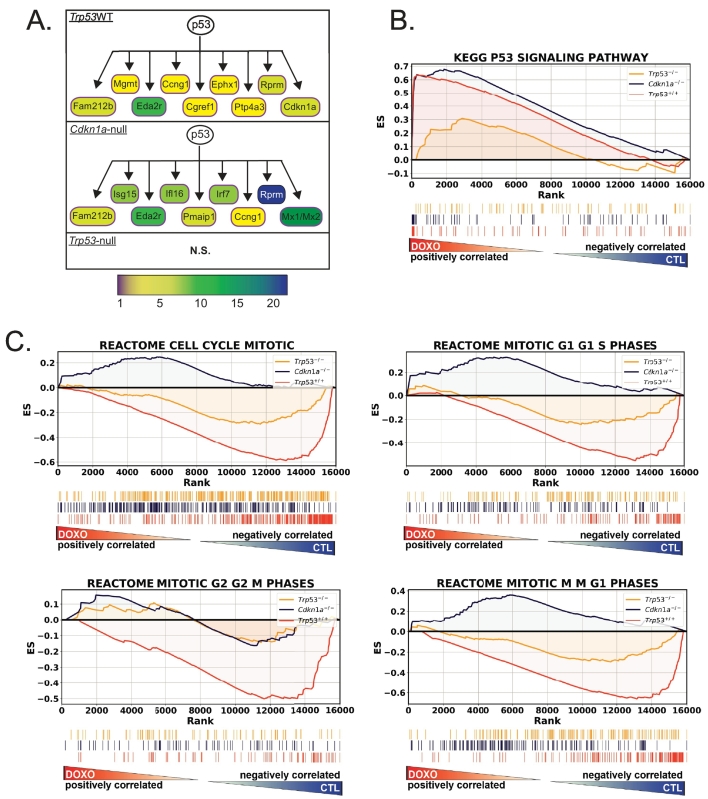
We next performed gene set enrichment analysis (GSEA) [25] on the 186 gene sets curated by the KEGG [24] as well as IPA to determine “upstream regulators” altered after treatment in each of the genotypes. As expected based on the large number of genes regulated (Figure 1A), IPA predicted significant activation or inhibition of 94, 204, and 48 molecules as “upstream regulators” of gene expression changes in treated Trp53WT, Cdkn1a-null, and Trp53-null genotypes, respectively (Supplementary Table 1). Trp53WT tumors regulated 62 pathways (50 in treated, 12 in control) in the KEGG gene set list (FDR < 0.25) after treatment, and Cdkn1a-null tumors regulated 39 (all in treated). Trp53-null tumors upregulated only 4 (Supplementary Table 2). Of the 39 pathways upregulated in Cdkn1a null tumors, 28 were also upregulated in Trp53WT tumors.
Not surprisingly, the “p53 signaling pathway” was a top hit found in both IPA as a predicted “upstream regulator” (Figure 2A) and GSEA (Figure 2B) analyses for both genotypes with wild-type Trp53. GSEA plots depict this enrichment. Many of the genes listed in the p53 pathway KEGG gene set were among the most highly expressed following treatment, resulting in a strong enrichment score in treated samples (Figure 2B) and significantly higher leftward peaks in Trp53WT and Cdkna1-null [0.61, 0.68 enrichment score (ES), respectively, both had FDR < 0.05] than in Trp53-null tumors (0.31 ES and nonsignificant FDR = 0.75).
Figure 2.
Pathways identified by analysis of doxorubicin-induced gene expression include the p53 pathway and cell cycle. (A) Schematic of top nine upregulated p53 pathway members in each genotype that were identified by IPA as being regulated downstream of p53. Fold change is indicated by color bar. (B) GSEA using KEGG gene lists was performed on RNA-seq data, and running enrichment scores for Trp53-null (yellow line), Cdkn1a-null (blue line), and Trp53WT (orange line) were plotted. Shown under the enrichment score plots are corresponding ranking of genes in the KEGG p53 pathway among ~16,000 genes analyzed. Genes stacked to the left are positively correlated and those to the right negatively correlated with doxorubicin-treated samples. (C) GSEA using the REACTOME cell cycle gene sets for doxorubicin-treated versus untreated tumors for the same genotypes as in B.
Direct targets of p53 have been identified by multiple studies [28], [29], [30]. We used these empirically determined lists to identify the direct p53 target genes regulated in Trp53WT tumors after chemotherapy treatment. Many well-known targets were regulated, including Cdkn1a, Bbc3, Bax, Ccng1, and Mdm2. Interestingly, some of these direct p53 targets were not regulated in the Cdkn1a-null genotype after treatment (Table 1).
Table 1.
Direct Targets of p53 Identified in Doxorubicin-Treated Mammary Tumors
| Riley et al. (129) | Allen et al. (198) | Kenzelmann-Broz et al. (432) | |
|---|---|---|---|
| APAF1 | *ABCA1* | 1700007K13Rik | *Man2b2* |
| BAX | AEN | Acaa1b | Mapkapk3 |
| BBC3 | APAF1 | *Acox3* | Mdm2 |
| BTG2 | ASS1 | Adrb2 | *Mdm4* |
| *CASP6* | BAX | Ak1 | *Mmrn2* |
| CCNG1 | BBC3 | *Aldh1l1* | Mybl1 |
| CDKN1A | BLOC1S2 | *Alg8* | *Notch1* |
| *CRYZ* | BTG2 | Bax | *Npr2* |
| CTSD | CCNG1 | Bbc3 | Orai2 |
| *CX3CL1* | CDC42BPG | Btg2 | *Pdcd6ip* |
| FAS | CDKN1A | Camk2d | Pdk4 |
| GDF15 | CYFIP2 | Ccng1 | *Plekhg6* |
| IRF5 | FAM212B | Cd80 | *Pltp* |
| LIF | FAS | Cdc42bpg | Pmaip1 |
| MDM2 | GDF15 | Cdkn1a | Polk |
| *MMP2* | GJB5 | Cgref1 | *Ppm1f* |
| RPS27L | ICAM1 | Cox6b2 | Pqlc3 |
| SCN3B | INPP5D | Cpt1c | *Prrg4* |
| TAP1 | ISCU | Csf1 | Psrc1 |
| TNFRSF10B | KANK3 | Dcxr | Ptp4a3 |
| TRPM2 | LAPTM5 | Ddit4l | Rap2a |
| *LYNX1* | *Ddx18* | Rap2b | |
| MDM2 | Def6 | Rhbdf2 | |
| *ORAI3* | Dennd2c | Rprm | |
| PHLDA3 | *Dut* | Scn3b | |
| *POLH* | Eda2r | Sema6a | |
| RPS27L | Ei24 | Sesn2 | |
| SESN2 | Ephx1 | Sfmbt1 | |
| SULF2 | Ercc5 | Slc19a2 | |
| TNFRSF10B | Exoc4 | *Slc4a11* | |
| TOB1 | *Fam53b* | Sncg | |
| *TSKU* | Fas | Sulf2 | |
| ZMAT3 | Fbxw9 | Svop | |
| Gas6 | Tbc1d2 | ||
| Gdf15 | *Tcirg1* | ||
| Ggta1 | Tmem19 | ||
| *Gls2* | Tnfrsf10b | ||
| Gria3 | Trafd1 | ||
| *Gtf3c1* | Trim11 | ||
| *H2afx* | Trim7 | ||
| Icam1 | Trp53inp1 | ||
| Kcnj4 | *Trp73* | ||
| Klhl26 | Tshr | ||
| Lif | Zfp365 | ||
Direct p53 targets identified by previous studies were compared to significantly regulated protein-coding genes in the Trp53WT genotype. Table lists genes that were matched, and those genes with an “*” were not found to be regulated in the Cdkn1a-null genotype.
A critical component of the cellular response to DNA damage caused by chemotherapeutics is cell cycle arrest. This arrest is mediated by p53 primarily by transactivating the Cdkn1a gene, whose product, p21, is a cyclin-dependent kinase inhibitor [15], [16], [17]. We found that gene sets representing all phases of the cell cycle and transitions for G1-S, G2-M, and M-G1 were prominently enriched in the untreated Trp53 wild-type tumors but were not enriched in either treated or untreated Trp53-null tumors (Figure 2C). This finding is consistent with the downregulation of cell cycle genes and arrest that occurs in Trp53 wild-type but not null tumors. Interestingly, Cdkn1a-null tumors did not modulate cell cycle genes consistent with either Trp53 wild-type or null tumors.
Gene Expression Differences in Cdkn1a-Null Tumors That Arrest or Proliferate in Response to Treatment
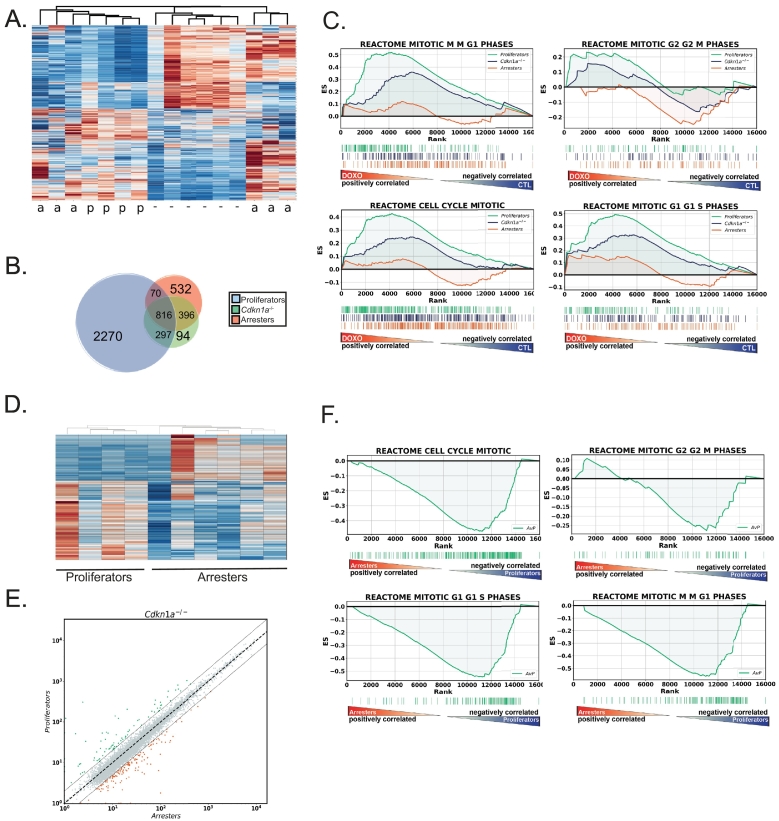
Next, we separately analyzed the Cdkn1a null tumors according to response phenotype. Unlike in cell culture models [20], [21], [31], Trp53 can still mediate a durable G2 arrest in roughly half of treated Cdkn1a-null mammary tumors in vivo [9]. To investigate this fascinating phenotype, we performed unsupervised hierarchical clustering of differentially expressed genes in all Cdkn1a-null tumors after treatment (Figure 3A). Untreated tumors and those that continued proliferation into mitotic catastrophe clustered together, while tumors that retained arrest capacity formed two separate clusters (Figure 3A). These heat maps show stark differences in gene expression despite the tumors all being the same genotype and receiving the same treatment. In fact, tumors that proliferated in response to treatment had more unique gene expression changes than they had in common with treated tumors that arrested (Figure 3B). Examining cell cycle differences in GSEA plots comparing “proliferators” to untreated tumors, we found that treated “proliferators” were actually relatively enriched for cell cycle genes compared to untreated tumors (Figure 3C). When treated “arresters” were compared to untreated tumors, relatively more enrichment of cell cycle progression genes was observed in the untreated tumors. The enrichment curve for all Cdkn1a-null tumors, i.e., the combination of arresters and proliferators versus control, showed enrichment between the two subphenotypes (Figure 3C). We next directly compared gene expression differences in only the treated tumors with either arrester or proliferator phenotypes (without untreated controls). We found that the phenotypes clustered together and many genes were differentially expressed (Figure 3, D and E). Analysis of the KEGG gene sets showed 43 pathways enriched in arresters and 33 in proliferators (FDR< 0.25; Supplementary Table 3). Analyzing the cell cycle pathways further revealed that proliferators were enriched for most of these genes compared to arresters (Figure 3F). Interestingly, Cdkn2a, coding for the cyclin-dependent kinase inhibitor p16, was elevated in arresters, suggesting a mechanism for the p53-dependent, p21-independent cell cycle arrest in these tumors (p53 dependent because Trp53-null tumors do not arrest). Other genes elevated in arresters include Wee1 and Rbl2. Genes elevated in proliferators include mediators of cell cycle progression E2f1, Cdk4, Cdk1, Ccnd2, Ccnb2, and Cdc25a. DNA replication pathway and genes were elevated in proliferators, including Cdt1, Rpa1, polymerases, and chromosome maintenance genes.
Figure 3.
Gene expression patterns differ in Cdkn1a-null tumors that respond to doxorubicin with arrest or proliferation. (A) Heat map of untreated Cdkn1a-null tumors compared to treated tumors. Those noted as “−” were untreated; those noted as “a” arrested following treatment; and those noted as “p” continued to proliferate after treatment, transiting the cell cycle, consistent with more cell death and a superior response. (B). Venn diagrams showing number of statistically significant (PPEE<.05) gene expression changes after doxorubicin treatment that were shared in the comparisons of untreated tumors to “arresters” and to “proliferators” and in the combination of "arresters" and “proliferators” (Cdkn1a−/−). (C) GSEA using REACTOME cell cycle gene sets for gene expression comparisons of untreated to “arresters” and untreated to “proliferators.” (D) Heat map of gene expression in treated Cdkn1a-null tumors that arrested and treated tumors that proliferated. (E) Scatter plot of differentially expressed genes in doxorubicin-treated Cdkn1a-null tumors that arrested (X-axis) and proliferated (Y-axis). (F) GSEA for REACTOME cell cycle gene sets using gene expression comparison of arresters to proliferators as made in D and E.
Following chemotherapy treatment, Trp53-null tumors and those Cdkn1a-null tumors that respond with continued proliferation will undergo cell death and significant tumor regression [9]. Patients with TP53 mutant tumors are also much more likely to undergo a complete pathological response to chemotherapy [4], [5], [6], [7], [8]. Interestingly, apoptosis-related gene pathways in the mammary tumors that undergo cell death and regression were not enriched compared to those that arrest and regress minimally (Supplementary Figure 2). These data suggest that an apoptosis-related transcriptional program is insufficient to induce cell death in treated tumors unless cell cycle arrest is absent. Alternatively, nontranscriptional regulation of apoptotic proteins could mediate the extensive cell death but is not detected in our study.
Senescence and SASP Genes Are Upregulated in Trp53 Wild-Type Tumors after Treatment
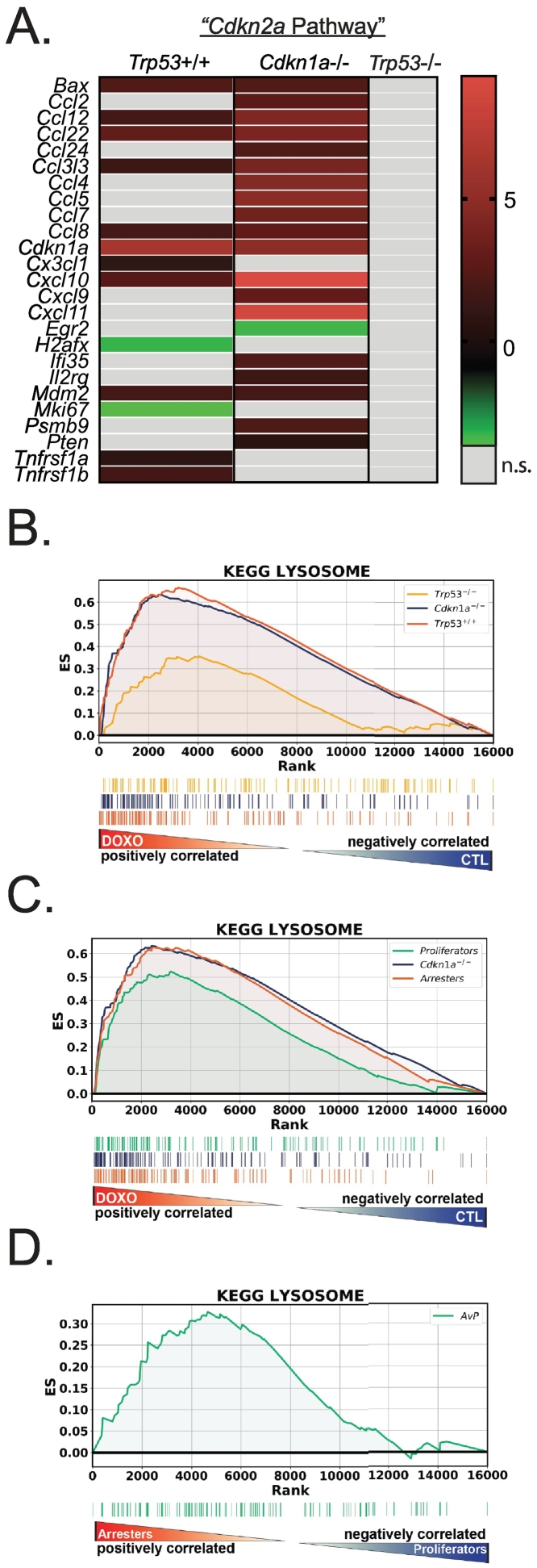
While Cdkn2a was not induced by doxorubicin, IPA identified “Cdkn2a” as an upstream regulator of the gene expression changes in treated tumors with wild-type Trp53. This was likely due to increased expression of genes that IPA associates with Cdkn2a (Figure 4A, Supplementary Table 1). Cdkn2a codes for the cyclin-dependent kinase inhibitor p16 [32], and due to the prominent role of p16 as a marker and mediator of senescence [33], [34], we examined enriched gene sets for other senescence-related pathways. Indeed, other KEGG gene sets regulated in treated tumors with Trp53 wild type (including Cdkn1a-null) included those related to lysosome, inflammation, and immune responses. Expression of genes involved in lysosome biogenesis is consistent with treated tumors having senescence-associated beta galactosidase (SAβGal) [35], [36]. The gene set for “Lysosome” was enriched in treated tumors that had wild-type Trp53 but not tumors null for Trp53 (Figure 4B). The lysosome pathway was also enriched in the Cdkn1a-null tumors that arrested after treatment (Figure 4, C and D).
Figure 4.
Senescence phenotype is induced by doxorubicin treatment in Trp53 wild-type tumors. (A) Heat map of the genes and their expression (color bar ranges from -2.2 to 8.3) from each genotype whose products were identified by IPA for upstream regulation by Cdkn2a. (B) GSEA using the KEGG Lysosome gene set was performed on doxorubicin-treated versus untreated tumors for Trp53WT, Trp53-null, and Cdkn1a-null genotypes. (C) GSEA using KEGG Lysosome gene set for gene expression comparisons of Cdkn1a-null untreated tumors to “arresters” and untreated to “proliferators.” (D) GSEA for KEGG Lysosome gene set using gene expression comparison between treated Cdkn1a null tumors that arrested compared to treated tumors that proliferated.
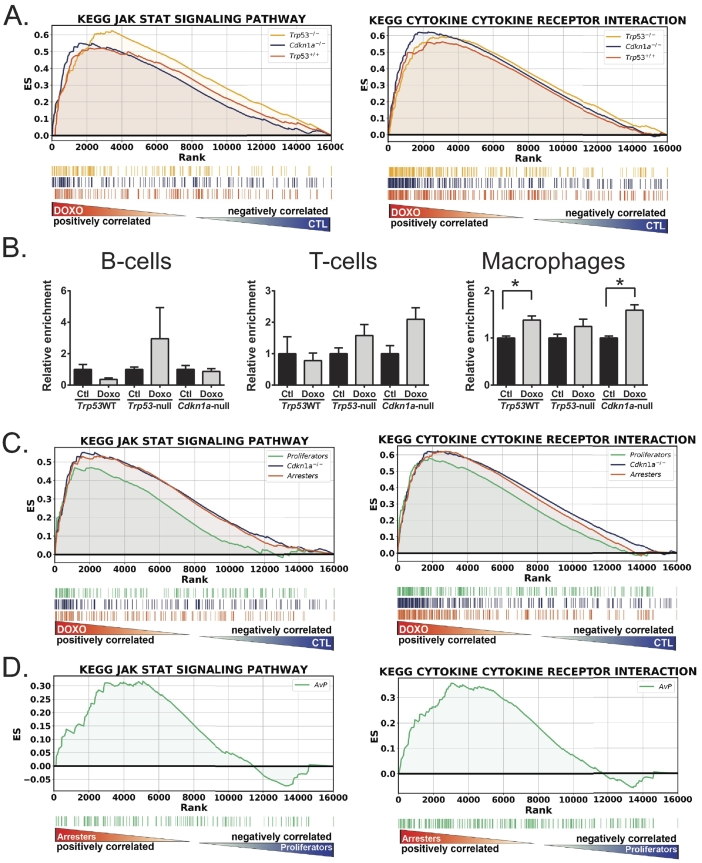
In all of the genotypes, enrichment of inflammatory genes and pathways was identified by all of the analyses we performed, which is consistent with these tumors acquiring the senescence-associated secretory phenotype (SASP) [37], [38]. Analysis using the PANTHER pathway database [39], [40] identified “Inflammation mediated by chemokine and cytokine signaling” as the top hit with greatest number of genes matched for the treated Trp53 wild-type tumors (Supplementary Table 4). IPA for diseases and biological functions identified “inflammatory response functional category” (Supplementary Table 5) as highly significant and “active” in Trp53 wild-type tumors but not Trp53 null. In our GSEA KEGG analysis, 24 out of 50 significantly regulated pathways in Trp53WT tumors were immune/inflammation related (Supplementary Table 2). Of the four pathways enriched in Trp53 null tumors, 3 were immune/inflammation related. These data are consistent with a SASP induced by DNA damage. While SASP is p53 independent [41], [42], the fewer immune-related pathways and thus diminished SASP in Trp53-null tumors likely represent a transient expression of these genes, as those cells with DNA damage adequate to induce SASP probably end up dying. Senescent Trp53WT tumors arrest, survive, persist, and generate SASP over days following treatment and thus generate stronger enrichment of these pathways. Figure 5 shows gene enrichment of the KEGG Jak/Stat signaling and cytokine-cytokine receptor pathways in all three genotypes in the treated tumors. Other immune/SASP-related pathways showed similar patterns, but enrichment of the pathway for treated Trp53null tumors did not reach significance (not shown).
Figure 5.
Inflammatory genes are expressed in doxorubicin-treated mammary tumors. (A) GSEA using the KEGG Jak Stat and KEGG cytokine-cytokine receptor interaction gene sets was performed on doxorubicin-treated versus untreated tumors for Trp53WT, Trp53-null, and Cdkn1a-null genotypes. (B) CIBERSORT analysis was performed on gene expression data from Trp53WT, Trp53-null, and Cdkn1a-null genotypes, and relative enrichment of signatures for B cells, T cells, and macrophages was determined and graphed. t tests were performed for control (ctl) and doxorubicin-treated (doxo) tumors, * represents P < .05. (C) GSEA using KEGG Jak Stat and KEGG cytokine-cytokine receptor interaction gene sets was performed on doxorubicin-treated versus control tumors for gene expression comparisons of Cdkn1a-null untreated tumors to “arresters” and untreated to “proliferators.” (D) GSEA for KEGG Jak Stat and KEGG cytokine-cytokine gene sets was performed on treated Cdkn1a-null tumors that arrested compared to treated tumors that proliferated.
Because activation of immune/inflammation pathways was so prominent, we analyzed immune infiltration by CIBERSORT [27]. Our results indicate minimal recruitment of immune cells into the tumor following treatment, as only macrophages were significantly increased (Figure 5B). This lack of immune cell presence and tumor clearance at this time point is consistent with previous findings that Trp53WT tumors regress minimally and relapse quickly in response to treatment [9]. These data suggest that the superior regression observed in Trp53-null tumors is not due to immune clearance.
We next examined SASP-related pathways in the various Cdkn1a-null tumor response phenotypes. We found that when both were compared to untreated tumors, arresters had a greater enrichment score for SASP pathways (Figure 5C). The finding that arrestors persist and produce more cytokines than proliferators that die by mitotic catastrophe is better illustrated by directly comparing arresters to proliferators (Figure 5D).
Longitudinal Expression of Senescence Genes
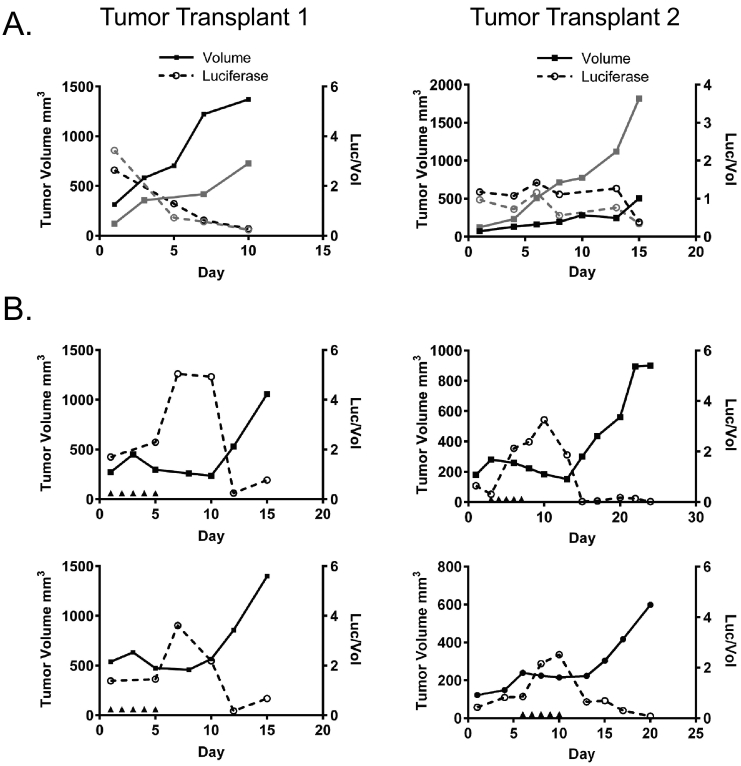
One limitation of our analysis thus far is the utilization of a single time point, 24 hours post final treatment. Senescence is induced over time, and Trp53 wild-type tumors in the MMTV-Wnt1 model system respond to chemotherapy for a period, usually 1 to 3 weeks, before they eventually relapse [9]. Thus, after observing the expression of senescence and SASP genes 24 hours after the final doxorubicin injection, we next used in vivo imaging to quantitate activity of the promoter of a senescence SASP gene starting before treatment, during treatment and response, and then after relapse.
We found that in both independent transplant lines examined, the Cxcl5 promoter was relatively inactive in untreated tumors as they grew, expressing stably in one transplant (transplant 2) and diminishing somewhat in another (transplant 1, Figure 6A). Likewise, when tumors were imaged before treatment, Cxcl5 promoter activity levels were relatively low (Figure 6B). Following treatment, however, promoter activity of the SASP gene spiked just after the final doxorubicin injection. Interestingly, as soon as tumors began to relapse, Cxcl5 promoter activity rapidly decreased to baseline levels and remained low until the end of the experiment. Thus, promoter activity of the SASP gene Cxcl5 is highly transient in the tumor, turning on during or just following treatment and then decreasing rapidly as tumor volume begins to increase again.
Figure 6.
Longitudinal expression of SASP senescence gene Cxcl5 over the course of treatment and relapse. C57Bl6/j mice were orthotopically transplanted with two different Trp53+/+, MMTV-Wnt1 tumors that express luciferase under the control of the Cxcl5 promoter region. Tumor volume was determined (solid lines, solid squares), and mice were imaged for luciferase expression (dashed line, open circles) regularly. Mice were left untreated (A) or were treated with five consecutive IP injections of 4 mg/kg doxorubicin (B) as indicated by arrowheads on the X-axis. Data are expressed graphically with tumor volume indicated by solid lines on the left Y-axis and luciferase activity normalized to tumor volume indicated by dashed lines on the right Y-axis.
Discussion
Increasingly more data from more tumor types show that TP53 wild-type tumors respond poorly to treatment compared to TP53 mutant tumors, including tumors of the ovary, lung, mesothelium, colon, bladder, and rectum [43], [44], [45], [46], [47], [48], [49], [50], [51]. p53 exerts its effects by regulating transcription of targets involved in cell cycle arrest, senescence, apoptosis, and metabolism, among other functions [2]. When p53 is mutant and unable to activate or repress these genes following chemotherapy, cells fail to arrest and therapeutic response is more robust [9]. In this study, we have used a genetic model system to comprehensively define p53-dependent genes and pathways modulated in tumors following doxorubicin treatment in vivo.
Interestingly, we were unable to identify any apoptotic pathway genes that would explain the superior tumor regression in the Trp53-null tumors. Gene set enrichment for pathways associated with “apoptosis,” “necrosis,” “survival,” “DNA fragmentation,” and “regulation of apoptosis” was not noted to be different for tumors that undergo massive cell death (Trp53-null, Cdkn1a-null proliferators) compared to tumors that arrest and resist apoptosis (Trp53WT tumors, Cdkn1a-null arresters). Many apoptotic genes were upregulated in the treated Trp53WT tumors, but these tumors undergo only minimal cell death or regression. In clinical studies, TP53 wild-type tumors also exhibit inferior tumor regression. Based on these data, the most likely scenario seems to be that the Trp53-null and Cdkn1a-proliferator tumor cells die by transcription-independent means that include apoptosis, necrosis, and mitotic catastrophe.
In tissue culture cells, when Cdkn1a is knocked out or down, cells fail to arrest and continue into mitosis and eventually die, similar to p53 null cells [20], [21], [31]. While p21 is critical for cell cycle arrest in tissue culture, the knockout mouse has a surprisingly mild phenotype [18], [52]. In the mammary tumor model, remarkably, tumors with Cdkn1a null genotypes could respond with arrest, or bypass arrest and enter mitosis, resulting in a superior response [9]. Among the 10 doxorubicin-treated Cdkn1a null tumors in this study, 6 responded with arrest, and 4 failed to arrest as evidenced by the presence of mitotic figures [9]. We analyzed these two cohorts and observed first that these two phenotypes cluster separately. Performing GSEA, we found among the top hits were “Cell Cycle” which showed that Cdkn2a (p16) was elevated in the tumors undergoing arrest. In fact, GSEA showed many differences (Supplementary Table 3). Presence of more cytokines in the tumors undergoing arrest is consistent with observations in p53 null tumors: when enough DNA damage occurs to activate the SASP, tumor cells that are p53 or p21 null (with proliferator response phenotype) fail to arrest and likely die, and thus do not persist and produce cytokines.
Our data also suggest that as a tumor relapses, expression of SASP factors such as Cxcl5 diminishes rapidly. It is not clear if this represents overtaking of the tumor by proliferating nonsenescent cells or, alternatively, if once-senescent cells lose expression of SASP factors and possibly reenter proliferation.
Gene expression analysis in our in vivo genetic model system revealed genes and pathways that are differentially activated or repressed in tumors of distinct genotypes and phenotypes, thus defining the programs that result in a poor clinical response. As more studies demonstrate more varieties of TP53 wild-type tumors that respond poorly to chemotherapy, identifying the genes regulated by p53 will be critical to overcoming this poor response.
The following are the supplementary data related to this article.
Supplementary Figure 1. Cdkn1a transcripts detected in Cdkn1a-null tumors are consistent with the targeted null allele. Coverage was visualized using the Integrative Genomics Viewer [Robinson JT et al, Nature Biotechnology 29, 24–26 (2011), Thorvaldsdóttir et al, Briefings in Bioinformatics 14, 178-192 (2013)]. Control Cdkn1a-null mice in light green, treated and proliferating Cdkn1a-null mice in darker green, treated and arresting Cdkn1a-null mice in darkest green, control Trp53-null mice in pink, and treated Trp53-null mice in magenta.
Supplementary Figure 2. Enrichment of apoptotic gene sets is similar for tumor genotypes that undergo cell death and regress (Trp53-null, Cdkn1a-proliferator) and those that undergo arrest, resist cell death, and regress minimally (Trp53WT, Cdkn1a-null arresters). (A) GSEA using the KEGG Apoptosis gene set was performed on doxorubicin-treated versus untreated tumors for Trp53WT, Trp53-null, Cdkn1a-null genotypes, and Cdkn1a-null arrester versus proliferator phenotypes. (B) GSEA using multiple apoptosis-related gene sets as indicated in the figure was performed on doxorubicin-treated Cdkn1a-null arrester versus proliferator phenotypes.
IPA of Upstream Regulation. IPA comparison analysis was run with all significantly regulated (FDR<0.05) protein coding genes from doxorubicin-treated Trp53WT, Trp53-null, and Cdkn1a-null tumors. Significance by overlap P value was determined in IPA using Fisher's exact test. Regulators with an activation z-score of ≥2 or ≤−2 were determined by IPA to be significant independent of P value.
Enriched Gene Sets in Doxorubicin-Treated Versus Control Trp53WT, Trp53-Null, and Cdkn1a-Null Genotypes. GSEA was performed using 186 KEGG gene sets, and listed are those with an FDR<0.25.
Enriched Gene Sets in Doxorubicin-Treated Cdkn1a-Null Arrester Versus Proliferator Phenotypes. GSEA was performed using 186 KEGG gene sets, and listed are those with an FDR<0.25.
PANTHER Classification Gene List Analysis. Table of top 10 pathways per genotype, ranked by number of gene matches to the PANTHER database, as determined using the PANTHER pathway analysis tool.
Subcategories and Corresponding Genes in Each Genotype for the "Inflammatory Response" Determined to be Significantly Active in Trp53WT and Cdkn1a-Null, But Not Trp53-Null, by IPA Diseases and Biological Functions Analysis.
Acknowledgements
The authors wish to acknowledge Melody C. Baddoo and the Tulane Cancer Center Cancer Crusaders Next Generation Sequence Analysis Core supported by the NIH Award Number P01CA214091, and the MD Anderson Cancer Center Sequencing & Microarray Facility, funded by the Core grant CA016672 (SMF). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
This study was supported by the Department of Defense Breast Cancer Research Program (grant number W81XWH-14-1-0216 to J.G.J.).
Declarations of interest: none.
References
- 1.TCGA-Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tonnessen-Murray C, Lozano G, Jackson JG. In: The p53 Protein: From Cell Regulation to Cancer. Lozano G, Levine AJ, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2016. pp. 173–186. [Google Scholar]
- 3.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esserman LJ, Berry DA, Cheang MC, Yau C, Perou CM, Carey L, DeMichele A, Gray JW, Conway-Dorsey K, Lenburg ME. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast Cancer Res Treat. 2012;132:1049–1062. doi: 10.1007/s10549-011-1895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Xu Y, Chen J, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Xie Y. TP53 mutations are associated with higher rates of pathologic complete response to anthracycline/cyclophosphamide-based neoadjuvant chemotherapy in operable primary breast cancer. Int J Cancer. 2015;138:489–496. doi: 10.1002/ijc.29715. [DOI] [PubMed] [Google Scholar]
- 6.Chen MB, Zhu YQ, Xu JY, Wang LQ, Liu CY, Ji ZY, Lu PH. Value of TP53 status for predicting response to neoadjuvant chemotherapy in breast cancer: a meta-analysis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertheau P, Turpin E, Rickman DS, Espie M, de Reynies A, Feugeas JP, Plassa LF, Soliman H, Varna M, de Roquancourt A. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4 doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertheau P, Plassa F, Espie M, Turpin E, de Roquancourt A, Marty M, Lerebours F, Beuzard Y, Janin A, de The H. Effect of mutated TP53 on response of advanced breast cancers to high-dose chemotherapy. Lancet. 2002;360:852–854. doi: 10.1016/S0140-6736(02)09969-5. [DOI] [PubMed] [Google Scholar]
- 9.Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, Tavana O, Yang P, Manshouri T, Li Y. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell. 2012;21:793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, Lin L, Hoog J, Goiffon RJ, Prat A. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest. 2012;122:1541–1552. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varna M, Lehmann-Che J, Turpin E, Marangoni E, El-Bouchtaoui M, Jeanne M, Grigoriu C, Ratajczak P, Leboeuf C, Plassa LF. p53 dependent cell-cycle arrest triggered by chemotherapy in xenografted breast tumors. Int J Cancer. 2009;124:991–997. doi: 10.1002/ijc.24049. [DOI] [PubMed] [Google Scholar]
- 12.te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 2002;62:1876–1883. [PubMed] [Google Scholar]
- 13.Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, Assad L, Poniecka A, Hennessy B, Green M. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25:4414–4422. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 14.Jackson JG, Post SM, Lozano G. Regulation of tissue- and stimulus-specific cell fate decisions by p53 in vivo. J Pathol. 2011;223:127–136. doi: 10.1002/path.2783. [DOI] [PubMed] [Google Scholar]
- 15.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 16.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 17.Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 18.Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 19.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 20.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 21.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 22.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 29.Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Kraus WL. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife. 2014;3 doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, Attardi LD. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 33.Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharpless NE, Sherr CJ. Forging a signature of in vivo senescence. Nat Rev Cancer. 2015;15:397–408. doi: 10.1038/nrc3960. [DOI] [PubMed] [Google Scholar]
- 35.Kurz DJ, Decary S, Hong Y, Erusalimsky JD. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci. 2000;113(Pt 20):3613–3622. doi: 10.1242/jcs.113.20.3613. [DOI] [PubMed] [Google Scholar]
- 36.Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, Kleijer WJ, DiMaio D, Hwang ES. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 37.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao SG, Jackson JG. SASP: tumor suppressor or promoter? Yes! Trends Cancer. 2016;2:676–687. doi: 10.1016/j.trecan.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, Premsrirut P, Luo W, Chicas A, Lee CS. Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes Dev. 2011;25:2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiley CD, Schaum N, Alimirah F, Lopez-Dominguez JA, Orjalo AV, Scott G, Desprez PY, Benz C, Davalos AR, Campisi J. Small-molecule MDM2 antagonists attenuate the senescence-associated secretory phenotype. Sci Rep. 2018;8:2410. doi: 10.1038/s41598-018-20000-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreno CS, Matyunina L, Dickerson EB, Schubert N, Bowen NJ, Logani S, Benigno BB, McDonald JF. Evidence that p53-mediated cell-cycle-arrest inhibits chemotherapeutic treatment of ovarian carcinomas. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberson RS, Kussick SJ, Vallieres E, Chen SY, Wu DY. Escape from therapy-induced accelerated cellular senescence in p53-null lung cancer cells and in human lung cancers. Cancer Res. 2005;65:2795–2803. doi: 10.1158/0008-5472.CAN-04-1270. [DOI] [PubMed] [Google Scholar]
- 45.Sidi R, Pasello G, Opitz I, Soltermann A, Tutic M, Rehrauer H, Weder W, Stahel RA, Felley-Bosco E. Induction of senescence markers after neo-adjuvant chemotherapy of malignant pleural mesothelioma and association with clinical outcome: an exploratory analysis. Eur J Cancer. 2011;47:326–332. doi: 10.1016/j.ejca.2010.09.044. [DOI] [PubMed] [Google Scholar]
- 46.Wu PC, Wang Q, Grobman L, Chu E, Wu DY. Accelerated cellular senescence in solid tumor therapy. Exp Oncol. 2012;34:298–305. [PubMed] [Google Scholar]
- 47.Wong KK, Izaguirre DI, Kwan SY, King ER, Deavers MT, Sood AK, Mok SC, Gershenson DM. Poor survival with wild-type TP53 ovarian cancer? Gynecol Oncol. 2013;130:565–569. doi: 10.1016/j.ygyno.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bataille F, Rummele P, Dietmaier W, Gaag D, Klebl F, Reichle A, Wild P, Hofstadter F, Hartmann A. Alterations in p53 predict response to preoperative high dose chemotherapy in patients with gastric cancer. Mol Pathol. 2003;56:286–292. doi: 10.1136/mp.56.5.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, Roth B, Cheng T, Tran M, Lee IL. Identification of distinct Basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tato-Costa J, Casimiro S, Pacheco T, Pires R, Fernandes A, Alho I, Pereira P, Costa P, Castelo HB, Ferreira J. Therapy-induced cellular senescence induces epithelial-to-mesenchymal transition and increases invasiveness in rectal cancer. Clin Colorectal Cancer. 2016;15:170–178.e173. doi: 10.1016/j.clcc.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 51.Rau B, Sturm I, Lage H, Berger S, Schneider U, Hauptmann S, Wust P, Riess H, Schlag PM, Dorken B. Dynamic expression profile of p21WAF1/CIP1 and Ki-67 predicts survival in rectal carcinoma treated with preoperative radiochemotherapy. J Clin Oncol. 2003;21:3391–3401. doi: 10.1200/JCO.2003.07.077. [DOI] [PubMed] [Google Scholar]
- 52.Martin-Caballero J, Flores JM, Garcia-Palencia P, Serrano M. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 2001;61:6234–6238. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Cdkn1a transcripts detected in Cdkn1a-null tumors are consistent with the targeted null allele. Coverage was visualized using the Integrative Genomics Viewer [Robinson JT et al, Nature Biotechnology 29, 24–26 (2011), Thorvaldsdóttir et al, Briefings in Bioinformatics 14, 178-192 (2013)]. Control Cdkn1a-null mice in light green, treated and proliferating Cdkn1a-null mice in darker green, treated and arresting Cdkn1a-null mice in darkest green, control Trp53-null mice in pink, and treated Trp53-null mice in magenta.
Supplementary Figure 2. Enrichment of apoptotic gene sets is similar for tumor genotypes that undergo cell death and regress (Trp53-null, Cdkn1a-proliferator) and those that undergo arrest, resist cell death, and regress minimally (Trp53WT, Cdkn1a-null arresters). (A) GSEA using the KEGG Apoptosis gene set was performed on doxorubicin-treated versus untreated tumors for Trp53WT, Trp53-null, Cdkn1a-null genotypes, and Cdkn1a-null arrester versus proliferator phenotypes. (B) GSEA using multiple apoptosis-related gene sets as indicated in the figure was performed on doxorubicin-treated Cdkn1a-null arrester versus proliferator phenotypes.
IPA of Upstream Regulation. IPA comparison analysis was run with all significantly regulated (FDR<0.05) protein coding genes from doxorubicin-treated Trp53WT, Trp53-null, and Cdkn1a-null tumors. Significance by overlap P value was determined in IPA using Fisher's exact test. Regulators with an activation z-score of ≥2 or ≤−2 were determined by IPA to be significant independent of P value.
Enriched Gene Sets in Doxorubicin-Treated Versus Control Trp53WT, Trp53-Null, and Cdkn1a-Null Genotypes. GSEA was performed using 186 KEGG gene sets, and listed are those with an FDR<0.25.
Enriched Gene Sets in Doxorubicin-Treated Cdkn1a-Null Arrester Versus Proliferator Phenotypes. GSEA was performed using 186 KEGG gene sets, and listed are those with an FDR<0.25.
PANTHER Classification Gene List Analysis. Table of top 10 pathways per genotype, ranked by number of gene matches to the PANTHER database, as determined using the PANTHER pathway analysis tool.
Subcategories and Corresponding Genes in Each Genotype for the "Inflammatory Response" Determined to be Significantly Active in Trp53WT and Cdkn1a-Null, But Not Trp53-Null, by IPA Diseases and Biological Functions Analysis.