ABSTRACT
The canonical role of cohesin is to mediate sister chromatid cohesion. In addition, cohesin plays important roles in processes such as DNA repair and regulation of gene expression. Mounting evidence suggests that various post-translational modifications, including phosphorylation, acetylation and sumoylation regulate cohesin functions. Our mass spectrometry analysis of cohesin purified from Schizosaccharomyces pombe cells revealed that the cohesin subunit Psm1 is methylated on two evolutionarily conserved lysine residues, K536 and K1200. We found that mutations that prevent methylation of Psm1 K536 and K1200 render sensitivity to DNA-damaging agents and show positive genetic interactions with mutations in genes encoding the Mus81–Eme1 endonuclease. Yeast two-hybrid and co-immunoprecipitation assays showed that there were interactions between subunits of the cohesin and Mus81–Eme1 complexes. We conclude that cohesin is methylated and that mutations that prevent methylation of Psm1 K536 and K1200 show synthetic phenotypes with mutants defective in the homologous recombination DNA repair pathway.
KEY WORDS: Cohesin, DNA repair, Mitosis, Meiosis, Schizosaccharomyces pombe
Summary: Cohesin methylation is a novel post-translational modification implicated in DNA repair in the fission yeast S. pombe.
INTRODUCTION
The cohesin complex is a tripartite ring in which the Smc1 and Smc3 proteins (called Psm1 and Psm3, respectively, in the fission yeast Schizosaccharomyces pombe) are connected by their hinge domains on one side, and the kleisin subunit Rad21 (also called Scc1 or Mcd1) closes the ring by connecting the Smc1 and Smc3 head domains on the other side. Rad21 interacts with the fourth cohesin subunit, Scc3 (called Psc3 in fission yeast or SA/STAG in higher eukaryotes). In meiosis, the Rad21 and Psc3 subunits are replaced to a large extent by the meiosis-specific subunits Rec8 and Rec11, respectively.
The canonical role of cohesin is to mediate sister chromatid cohesion. Cohesion between sister chromatids is established during DNA replication and this process coincides with the acetylation of two lysine residues on Smc3 by the acetyltransferase Eco1/Ctf7 (Feytout et al., 2011; Rolef Ben-Shahar et al., 2008; Uhlmann and Nasmyth, 1998; Unal et al., 2008). To allow the segregation of sister chromatids, cohesion must be removed at the onset of anaphase. This removal involves phosphorylation of cohesin and cleavage of the kleisin subunit of the cohesin complex by separase (Alexandru et al., 2001; Uhlmann et al., 1999). Cohesin is also released from DNA in a separase-independent manner, which depends on phosphorylation of cohesin and involves the cohesin-associated protein Wapl (Waizenegger et al., 2000).
In addition to its role in mediating sister chromatid cohesion, cohesin is important for other processes, including DNA repair (Schar et al., 2004; Sjogren and Nasmyth, 2001; Strom et al., 2004; Unal et al., 2004). Cohesin accumulates at sites of double-strand DNA breaks (DSBs) where it presumably reinforces sister chromatid cohesion to facilitate repair by homologous recombination (Strom et al., 2004; Unal et al., 2004).
Homologous recombination is an important pathway for DSB repair and uses homologous DNA sequences as a template to facilitate DNA repair. DSB formation is followed by end-processing reactions in which DNA ends that flank DSBs are resected to expose 3′ single-stranded DNA (ssDNA) ends that are quickly bound by ssDNA-binding proteins. Mediator proteins such as Rad52, Rad55, Rad57 and Sfr1 then stimulate the replacement of ssDNA-binding proteins with Rad51 recombinases and stabilize Rad51–ssDNA filaments. These nucleoprotein filaments then catalyze the invasion of a homologous DNA template to form joint molecule intermediates that can be resolved by various enzymes, such as the Mus81–Eme1 endonuclease (Ranjha et al., 2018).
During S-phase, replication forks may stall when they encounter DNA lesions or natural replication barriers. Several proteins involved in homologous recombination, including the structure-specific endonuclease Mus81–Eme1 and the mediator protein Rad52, play an important role in the repair or rescue of blocked replication forks during S-phase (Doe et al., 2002; Lambert and Carr, 2013; Lorenz et al., 2009; Nguyen et al., 2015). However, the mechanisms by which homologous recombination proteins contribute to DNA replication remain poorly understood. Although it is well established that cohesin is important for the maintenance of genome integrity during S phase, we are only beginning to understand the molecular function of cohesin in the repair/rescue of blocked replication forks (Gelot et al., 2016; Tittel-Elmer et al., 2012).
Mounting evidence suggests that various post-translational modifications including phosphorylation, acetylation and sumoylation regulate the function of cohesin in DNA repair. Cohesin is sumoylated during the DNA damage response, and this modification is required for sister chromatid homologous recombination (Almedawar et al., 2012; McAleenan et al., 2012; Wu et al., 2012). The phosphorylation and acetylation of cohesin are required for DNA double-strand break-induced cohesion, and cohesin phosphorylation is also required for the intra-S checkpoint (Heidinger-Pauli et al., 2008, 2009; Kim et al., 2002; Watrin and Peters, 2009; Yazdi et al., 2002).
In recent years, protein methylation has been established as a major intracellular post-translational modification, but little is known about methylation of cohesin (Carlson and Gozani, 2016; Jung et al., 2008; Larsen et al., 2016; Olsen et al., 2016). In our current work, we show that, in the fission yeast S. pombe, the cohesin subunit Psm1 is post-translationally modified by methylation on two evolutionarily conserved lysine residues (K536 and K1200), and that Psm1 residues K536 and K1200, and possibly their methylation, are required for the proper functioning of the Mus81–Eme1-dependent DNA repair pathway.
RESULTS AND DISCUSSION
Psm1 is methylated on K536 and K1200
To identify novel post-translational modifications on cohesin, we constructed functional tandem affinity purification (TAP)-tagged Rec11 (Rec11–TAP) according to our protocol described at http://mendel.imp.ac.at/Pombe_tagging (Cipak et al., 2009; Phadnis et al., 2015). We used a TAP protocol to purify Rec11–TAP together with other cohesin subunits (Psm1, Psm3 and Rec8) from meiotically induced S. pombe cells harvested around prophase I and analyzed the post-translational modifications by mass spectrometry (Fig. 1A; Fig. S1). We found that Psm1 was mono-methylated on two lysine residues (K536 and K1200) (Fig. 1B).
Fig. 1.
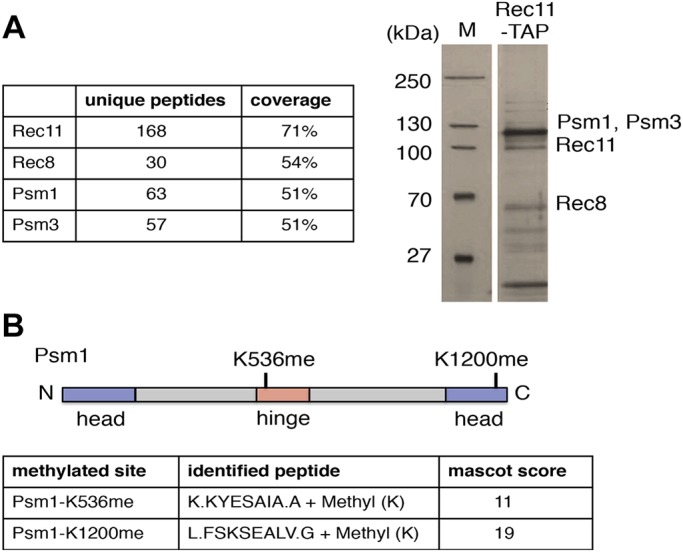
Mass-spectrometry analysis reveals that Psm1 is mono-methylated on K536 and K1200. (A) Cohesin subunits associated with Rec11–TAP were isolated by tandem affinity purification from meiotically induced pat1-114/pat1-114 mat-Pc cells harvested 2.5–3.5 h after meiosis induction. Purified proteins were separated on an SDS-PAGE gel and visualized by silver staining. Molecular mass markers (M) are shown on the left. Positions of Psm1, Psm3, Rec11 and Rec8 are indicated according to their predicted molecular mass. In parallel, samples were subjected to analysis by mass spectrometry. (B) Methylation sites identified on Psm1 and corresponding peptides are indicated.
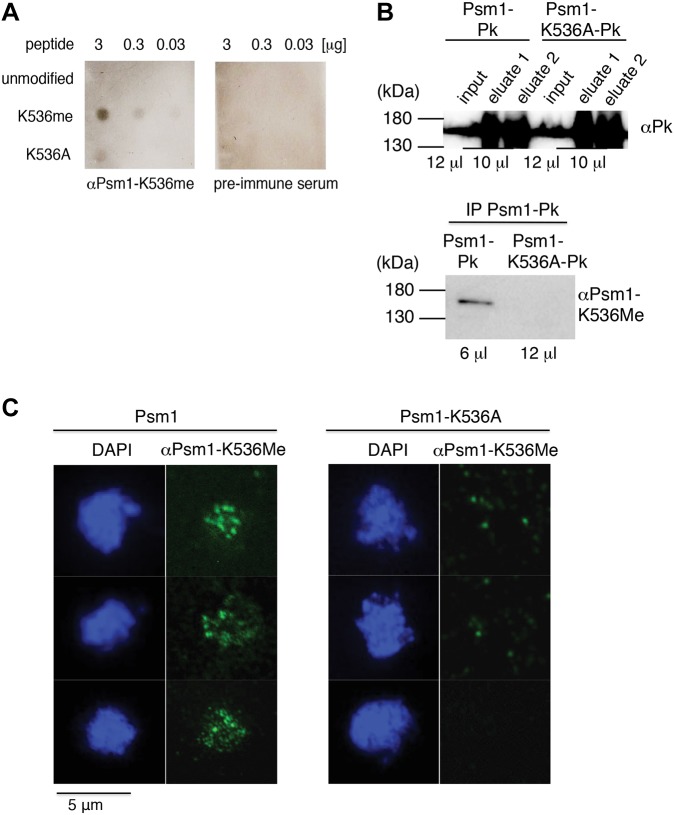
To exclude the possibility that this Psm1 methylation was an artifact introduced during the TAP purification or mass spectrometry analysis, we generated a polyclonal anti-Psm1-K536me antibody using methylated peptide [CTPTQKK(me)YESAIA] spanning the Psm1 methylation site K536me (Fig. 2A). We were not able to detect Psm1-K536me in total protein extracts (data not shown). However, upon immunoprecipitation of functional Pk-tagged Psm1 (Psm1–Pk) (Fig. S2B), our anti-Psm1-K536me antibody recognized a band of expected size in vegetative wild-type cells but not in mutant cells carrying a psm1K536A mutation that results in a replacement of the methylated lysine residue with an alanine residue, which can no longer be methylated (Psm1-K536A–Pk) (Fig. 2B). Although we cannot exclude the possibility that our anti-Psm1-K536me antibody binds to other epitopes and detects Psm1 regardless of its methylation status, given the specificity of this antibody for the modified peptide (Fig. 2A), it is likely that this band represents Psm1 methylated on K536.
Fig. 2.
Verification of Psm1-K536 methylation by anti-Psm1-K536me antibody. (A) Specificity of antibody generated against Psm1-K536me. The indicated amounts of Psm1 peptide CTPTQKK(me)YESAIA carrying methylation on K536 (K536me), an unmodfied peptide CTPTQKKYESAIA (unmodified) or modified peptide CTPTQKAYESAIA (K536A) were spotted on PVDF membrane and probed with anti-Psm1-K536me antibody or pre-immune serum. (B) Psm1–Pk protein was immunoprecipitated (IP) from cycling wild-type cells (Psm1–Pk) or mutant cells carrying the psm1K536A mutation (Psm1-K536A–Pk). Samples were eluted in two steps and both were analyzed using anti-Pk antibody. Eluate 1 from each sample was analyzed with anti-Psm1-K536me antibody. The amount of loaded samples is indicated. The same protein extracts were analyzed on two different gels. (C) Nuclear spreads were prepared from cycling wild-type cells (Psm1) or mutant cells carrying the psm1K536A mutation (Psm1-K536A). Shown are selected nuclei after chromosome spreading, staining with DAPI and after immunolabeling with anti-Psm1-K536me antibody.
Cohesin performs most of its functions on chromatin but there is also a soluble pool of cohesin present in the cell. To separate these two pools of cohesin, we prepared chromatin spreads from mitotically growing cells. Anti-Psm1-K536me antibody detected foci on spread chromatin from wild-type cells (Fig. 2C). These foci were much less prominent in spreads from mutant cells carrying the psm1K536A mutation, raising the possibility that chromatin-bound Psm1 is methylated on K536.
We conclude that Psm1 is methylated on lysine residues K536 and K1200. Our identification of methylation on Psm1 that co-purified with Rec11–TAP shows that methylated Psm1 is part of the cohesin complex. The fact that we were able to detect a specific signal using the anti-Psm1-K536me antibody on protein extracts and chromatin spreads from vegetative cells suggests that Psm1 is methylated on K536 not only in meiotic cells but also in mitotic cells, and that this methylation is likely present on chromatin-bound Psm1.
A non-methylatable psm1K536A K1200A mutant is sensitive to DNA-damaging agents
To test the potential functional significance of Psm1 methylation, we mutated the K536 and K1200 to alanine (psm1K536A, psm1K1200A and psm1K536A K1200A). Although replacing the lysine residue with alanine residues is a relatively large change, owing to different properties of these residues, alanine residue can no longer be methylated. After tetrad dissection of heterozygous diploid strains, we found that haploid psm1K536A K1200A mutant cells were viable and showed no apparent growth defect on YES plates (Fig. 3A,B).
Fig. 3.
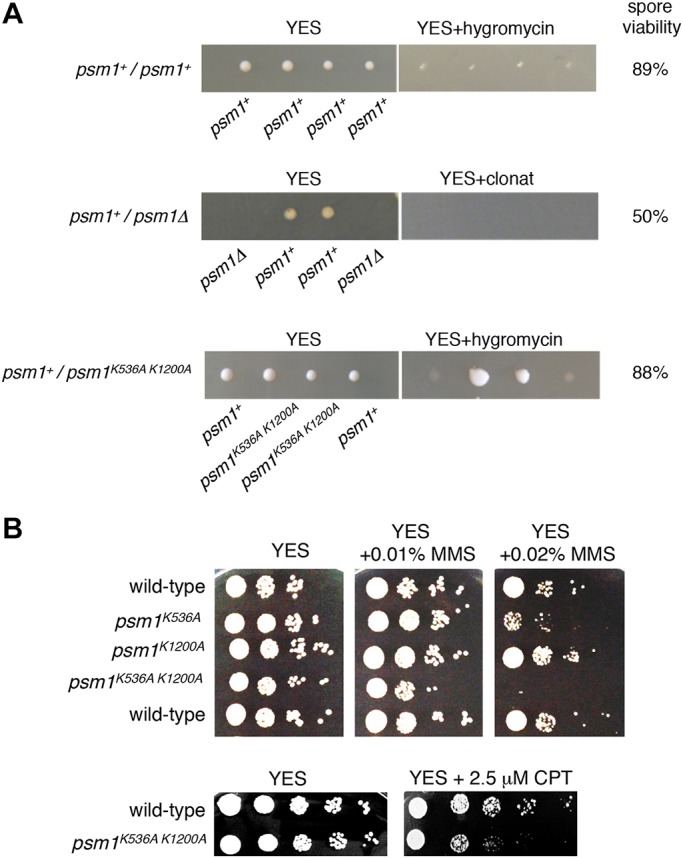
The psm1K536A K1200A mutant is viable but sensitive to DNA-damaging agents. (A) Tetrads were dissected from the indicated diploid strains on YES plates and after four days at 32°C, spore viability was determined. Colonies were replica-plated onto selective YES plates containing hygromycin B (200 mg/l) or clonat (100 mg/l). (B) Cells were grown on YES medium for 1 day, diluted in 5-fold steps, spotted onto YES plates containing the indicated amounts of MMS or CPT and incubated for 3 days at 30°C.
To test whether Psm1 residues K536 and K1200 are important for repair of damaged DNA, we induced DNA lesions in vegetative cells using methyl methanesulfonate (MMS) (Wyatt and Pittman, 2006). While the single mutants psm1K536A and psm1K1200A were only moderately sensitive or not sensitive to MMS, respectively, double mutant psm1K536A K1200A was not able to grow in the presence of 0.02% MMS (Fig. 3B). MMS induces a plethora of DNA lesions including those that cause replication forks to arrest (Boddy et al., 2000; Murray et al., 1997; Stewart et al., 1997). To test whether Psm1 residues K536 and K1200 are also required for the restart of collapsed replication forks, we tested the sensitivity of the psm1K536A K1200A to the topoisomerase I poison camptothecin (CPT). CPT stabilizes topoisomerase I-DNA intermediates leading to the accumulation of DNA breaks and collapse of replication forks at these DNA breaks (Avemann et al., 1988; Strumberg et al., 2000). The psm1K536A K1200A mutant cells were more sensitive to 2.5 µM CPT than wild-type cells (Fig. 3B). These results show that Psm1 K536 and K1200, and possibly methylation of K536 and K1200, are required to prevent the toxic effects of MMS and CPT.
We next asked whether methylation of the Psm1 K536 is regulated in response to DNA damage caused by MMS. The levels of methylation of Psm1 K536 did not change dramatically in Psm1–Pk immunoprecipitates from cells treated with MMS for up to 120 min (Fig. S2A). However, we cannot exclude the possibility that methylation of the Psm1 K536 is regulated locally, for example at the sites of DNA repair. It remains to be determined if and how Psm1-K536me is regulated.
DNA repair is compromised when cohesin protein levels are decreased (Heidinger-Pauli et al., 2010). However, we found that protein levels of Psm1K536A K1200A were similar to those of wild-type Psm1 (Fig. 2B; Fig. S2A). Therefore, it is unlikely that the sensitivity of psm1K536A K1200A mutant cells to DNA damaging agents is due to decreased levels of Psm1. The sensitivity of psm1K536A K1200A mutant cells to MMS and CPT could be caused by the inability of the mutant to efficiently repair the DNA lesions. Although psm1K536A K1200A mutant cells treated with MMS faithfully segregated chromosomes, and centromeric sister chromatid cohesion was not dramatically altered in metaphase cells (Fig. S2C), we cannot exclude the possibility that sister chromatid cohesion is defective at sites of DNA damage. Furthermore, cohesin also regulates gene expression. Therefore, another possible explanation is that expression of genes required to prevent MMS and CPT toxicity is altered in the psm1K536A K1200A mutant.
Genetic interactions between psm1K536A K1200A and mutants defective in the homologous recombination DNA repair pathway
Both MMS and CPT cause DSBs during DNA replication (Lundin et al., 2005; Tercero et al., 2003). Cohesin accumulates at sites of DSBs where it reinforces sister chromatid cohesion to facilitate repair by homologous recombination (Strom et al., 2004; Unal et al., 2004).
Genetic interactions may reveal functional relationships between genes and pathways. Negative genetic interactions (the phenotype of the double-mutant is stronger than expected) suggest compensatory, redundant pathways, while positive genetic interactions (the phenotype of the double-mutant is less severe than expected) can reflect functions within the same pathway (Ryan et al., 2013). Negative genetic interactions often involve genes with at least partially overlapping functions that can compensate for the absence of each other. While functional interpretation of negative interactions is not straightforward, positive genetic interactions are more interesting because they can provide insight into biochemical relationships between gene products and help define biological pathways. Positive genetic interactions often connect genes encoding members of the same protein complex or linear pathway (Baryshnikova et al., 2013).
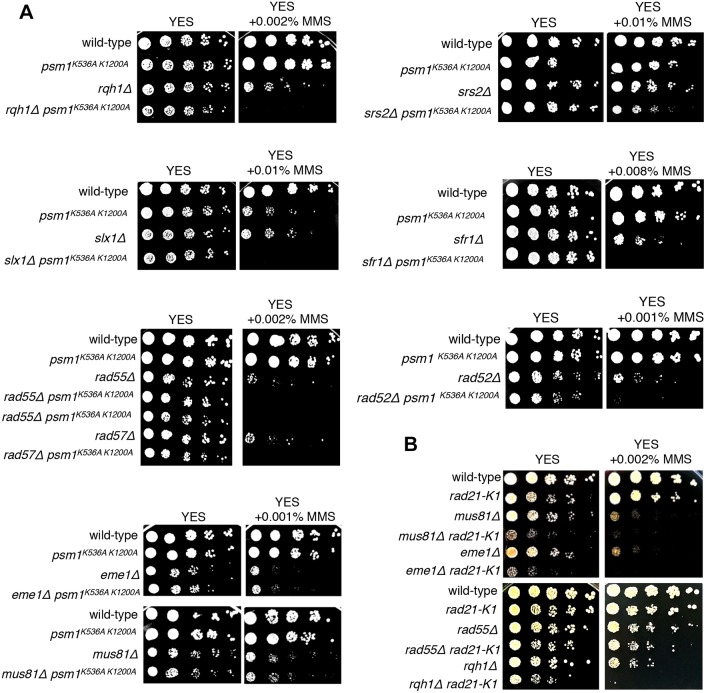
To better understand the role of Psm1 K536 and K1200, we combined psm1K536A K1200A with selected mutants defective in homologous recombination. We constructed double mutants between psm1K536A K1200A and mutations that disrupt recombinational repair of damaged replication forks including mus81Δ, eme1Δ (the heterodimeric endonuclease complex of the XPF family; Boddy et al., 2001; Doe et al., 2002), rqh1Δ (a member of the RecQ family of DNA helicases; Stewart et al., 1997), srs2Δ (a UvrD subfamily DNA helicase; Maftahi et al., 2002; Wang et al., 2001), slx1Δ (a structure-specific endonuclease, Coulon et al., 2004), and rad52Δ, rad55Δ, rad57Δ and sfr1Δ (mediators promoting the formation and/or stabilization of Rad51–ssDNA filaments; Krejci et al., 2012). In the presence of MMS, psm1K536A K1200A showed negative genetic interactions with most of the tested mutant strains (the phenotype of double-mutant was stronger than the most sensitive of the single mutants) and positive genetic interaction with mus81Δ and eme1Δ (the phenotype of double mutant was no more sensitive than the most sensitive of the single mutants) (Fig. 4A). Although these results suggest that psm1K536A K1200A mus81Δ and psm1K536A K1200A eme1Δ double mutants behave differently from other double mutants that we tested, we would like to stress that the psm1K536A K1200A mutant strain does not show any noticeable sensitivity in the presence of 0.001% MMS and this may affect the results of this assay.
Fig. 4.
Genetic interactions between psm1K536A K1200A, rad21-K1 and mutations in DNA repair-related genes. (A) Cells were grown on YES medium for 1 day, diluted in 5-fold steps, spotted onto YES plates containing the indicated amounts of MMS and incubated for 3 days at 30°C. (B) Cells were grown on YES medium for 1 day, diluted in 5-fold steps, spotted onto YES plates containing the indicated amounts of MMS and incubated for 4 days at 25°C.
Although in silico modeling suggests that psm1K536A and psm1K1200A mutations have only minor effects on the overall structure and electrostatic potential of the Psm1 (data not shown), it is possible that some of the observed phenotypes of psm1K536A K1200A mutant strains are not due to the absence of methylation but are caused by the replacement of lysine residues by alanine residues. We therefore mutated the K536 and K1200 to arginine residues, which retains a positive charge and is thus considered a conservative mutation (psm1K536R K1200R). Similar to what was found for the psm1K536A K1200A mutant, in the presence of MMS, psm1K536R K1200R showed a negative genetic interaction with srs2Δ and rad55Δ and a positive genetic interaction with mus81Δ (Fig. S3A,B). This suggests that the phenotype of the psm1K536A K1200A mutant described above is not simply due to the elimination of the positive charge caused by replacing K536 and K1200 with alanine residues. However, the psm1K536R K1200R mutant strain was less sensitive to MMS than psm1K536A K1200A (data not shown). Further studies are required to show whether and to what extent the absence of K536 and K1200 methylation contributes to the psm1K536A K1200A mutant phenotype.
To determine whether the described genetic interactions can be observed with other cohesin mutations that reduce cohesin activity, we tested a hypomorphic cohesin mutation rad21-K1 (Tatebayashi et al., 1998). Unlike for the psm1K536A K1200A and psm1K536R K1200R mutations, rad21-K1 showed negative genetic interactions with both mus81Δ and eme1Δ (Fig. 4B). We conclude that the observed pattern of genetic interactions between psm1K536A K1200A and psm1K536R K1200R mutants and recombination-deficient mutants is not observed in rad21-K1 mutant and it is unlikely to be solely due to reduced cohesin activity.
We conclude that the psm1K536A K1200A mutant shows synthetic phenotypes with mutants defective in the homologous recombination DNA repair pathway, consistent with the notion that Psm1 residues K536 and K1200, and possibly their methylation, are required for efficient repair of damaged DNA. The positive genetic interactions between psm1K536A K1200A mutant and mutations in genes encoding Mus81-Eme1 endonuclease complex are consistent with the notion that psm1K536A K1200A mutant negatively affects Mus81–Eme1-dependent DNA repair pathway. We speculate that Psm1 residues K536 and K1200, and possibly methylation of K536 and K1200, are important for an efficient repair of stalled or collapsed replication forks by Mus81–Eme1 complex. Alternatively, Mus81–Eme1 may be required for the function or recruitment of the cohesin to sites of DNA damage. A recent study showed that the Mus81-related endonuclease ERCC1–XPF interacts with cohesin and is required for the recruitment of the CTCF–cohesin complex to promoters and control regions during hepatic development (Chatzinikolaou et al., 2017).
Interestingly, we found that the Psm3 cohesin subunit interacts with Mus81 and that Psc3 weakly interacts with Eme1 in yeast two-hybrid assays (Fig. S4A). We also found one Eme1 peptide in our Rec11–TAP purification but not in other unrelated TAP purifications. Moreover, we found that Mus81–Myc co-immunoprecipitated with Psm1–Pk and the efficiency of co-immunoprecipitation was higher in the presence of MMS (Fig. S4B). This raises the possibility that cohesin physically interacts with the Mus81–Eme1 complex. This interaction may be similar to that described for the mammalian ERCC1–XPF endonuclease and cohesin (Chatzinikolaou et al., 2017). Further investigations will be required to define how the psm1K536A K1200A mutant functionally associates with Mus81–Eme1 and whether methylation of cohesin affects this interaction.
Taken together, we conclude that cohesin is methylated and that mutations that change the methylated Psm1 K536 and K1200 to alanine affect the Mus81–Eme1-dependent DNA repair pathway. Subunits of the cohesin and Mus81–Eme1 complexes interact in yeast two-hybrid and co-immunoprecipitation assays. Our results are consistent with the notion that methylation of cohesin provides a novel level of regulation that ensures efficient repair of damaged DNA. The Psm1 K536 and K1200 residues and surrounding sequences are conserved in many, but not all, species (Fig. S4C). Thus, our observations on Psm1 methylation are likely to pertain to studies of cohesin and DNA repair in other organisms. Understanding how Psm1 residues K536 and K1200 and their methylation function in DNA repair, and determining how multiple levels of post-translational modifications direct cohesin to function across various cellular processes are important future goals arising from the results described here.
MATERIALS AND METHODS
Strain construction
MMS and CPT were obtained from Sigma-Aldrich (cat. no. 129925-25g and C9911-100mg, respectively). Genotypes of strains and the figures and tables in which each was presented is given in Table S1. Mutations were introduced into psm1 gene using the QuikChangeII kit (Agilent Technologies) as described in Cipak et al., 2011; Gregan et al., 2007.
The following oligonucleotide primers were used for mutagenesis: Psm1K536A, 5′-CTTATGTACTCCTACTCAAAAGGCATATGAAAGCGCAATCGCTGCT-3′; Psm1K536A_antisense, 5′-AGCAGCGATTGCGCTTTCATATGCCTTTTGAGTAGGAGTACATAAG-3′; Psm1 K1200A_sense, 5′-GAAAAATCAGTTGTTCAGTGCATCAGAAGCTTTAGTGGG-3′; Psm1 K1200A_antisense, 5′-CCCACTAAAGCTTCTGATGCACTGAACAACTGATTTTTC-3′; Psm1K536R_sense, 5′-ATGTACTCCTACTCAAAAGAGATATGAAAGCGCAATCGCTG-3′; Psm1K536R_antisense, 5′-CAGCGATTGCGCTTTCATATCTCTTTTGAGTAGGAGTACAT-3′; Psm1K1200R_sense, 5′-TCATTTCCTTGAAAAATCAGTTGTTCAGTAGATCAGAAGCTTTAGTG-3′; Psm1K1200R_antisense, 5′-CACTAAAGCTTCTGATCTACTGAACAACTGATTTTTCAAGGAAATGA-3′. Bold nucleotides indicate the positions of introduced changes.
Transformants were confirmed by PCR-based analyses and by nucleotide sequencing. The psm1 gene, either wild-type or carrying mutations, was cloned into the vector pCloneHyg1_Ura4T (p280) using BamHI and XmaI restriction enzymes. pCloneHyg1_Ura4T was derived from pCloneHyg1 (GenBank EF101286.1) by reversing the orientation of hphMX4 and replacing the TEV terminator with that of ura4 using AgeI and NheI restriction enzymes. The resulting pCloneHyg1_Ura4T constructs containing the wild-type or mutated psm1 gene were digested using BamHI and KpnI enzymes (New England Biolabs). The fragment (∼7.8 kb) containing psm1 and hphMX4 was gel purified (Qiagen, Germany), and integrated into diploid yeast strains as described in Gregan et al. (2007). Heterozygous diploid strains were tetrad dissected to isolate haploid strains.
Induction of synchronous meiosis
Diploid pat1-114/pat1-114 and pat1-114/pat1-114 mat-Pc strains were grown in YES-Ade liquid medium to an optical density at 600 nm (OD600)=0.5 at 25°C (∼0.8×107 cells/ml). The cells were collected by centrifugation (3573 g for 20 min, 25°C), resuspended in EMM2-NH4Cl medium and incubated at 25°C for 16–18 h (pat1-114/pat1-114) or for 7 h (pat1-114/pat1-114 mat-Pc) to arrest cells in G1. The cells were resuspended in fresh EMM2-NH4Cl medium and induced into meiosis by shifting to 34°C.
To monitor the progression of meiosis, 1 ml of culture was centrifuged (6000 g for 5 min) and the cell pellet was fixed by adding 0.5 ml of 70% ethanol. Nuclear divisions were monitored by counting the number of DAPI-stained nuclei.
To monitor the pre-meiotic S-phase by flow cytometry, 0.3 ml of the fixed cells was mixed with 3 ml of 50 mM sodium citrate buffer and centrifuged (6000 g for 5 min) to obtain a pellet. Subsequently, 0.5 ml of 50 mM sodium citrate buffer supplemented with 0.1 mg/ml RNase A (Thermo Scientific) was added to the pellet, and cells were incubated at 37°C for 2 h. This was followed by sonication of the sample twice for 20 s at 40% power with a microtip sonicator (Bandelin Sonoplus HD 2070) to remove cell aggregates. To stain DNA, 0.5 ml of sodium citrate buffer containing 2 μM Sytox Green (Invitrogen) was added and cells were analyzed by FACS (FACS Calibur with Cellquest Pro software) (Cipak et al., 2014).
TAP purification
Meiotic cultures expressing Rec11–TAP were harvested 2.5–3.5 h after induction of meiosis and cells from 15 l cultures were collected by centrifugation (3573 g for 20 min, 4°C). Yeast cell powder was made from frozen pellets using a SamplePrep 6870 Freezer Mill (SPEX). Proteins were extracted using IPP150 buffer (IPP150, 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol and 0.1% NP-40) containing complete protease inhibitors (Roche) and 1 mM PMSF (Sigma). Washing steps were performed in Poly-Prep columns (Bio-Rad) by gravity flow. IgG Sepharose™ 6 Fast Flow beads (500 μl; GE Healthcare) were washed with IPP150 buffer, mixed with protein extract, and rotated for 2 h at 4°C. Beads were washed with IPP150 buffer and then with TEV cleavage buffer (TCB; 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% NP-40, 0.5 mM EDTA and1 mM DTT). Protein cleavage was performed in 2 ml of TCB buffer supplemented with 400 units of AcTEV protease (Life Technologies) for 2 h at 16°C. The eluate (2 ml) was supplemented with 6 μl of 1 M CaCl2 and mixed with 6 ml of calmodulin binding buffer 1 (CBB1; 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% NP-40, 1 mM imidazole, 1 mM Mg-Acetate, 2 mM CaCl2 and 10 mM β-mercaptoethanol). Calmodulin Sepharose 4B beads (150 μl; GE Healthcare) were washed with CBB1 buffer, added to a mixture of eluate and CBB1 buffer, and incubated for 2 h at 4°C. The beads were washed with CBB1 and calmodulin binding buffer 2 (CBB2; 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM Mg-Acetate, 2 mM CaCl2 and 1 mM β-mercaptoethanol). The proteins were eluted using one bed volume of elution buffer (EB; 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM Mg-acetate, 2 mM EGTA and 1 mM β-mercaptoethanol). A portion of the eluted proteins was separated by 8% SDS-PAGE and silver stained (Cipak et al., 2009). The remaining eluate was analyzed by mass spectrometry (see below).
Mass spectrometry analysis
NanoLC-MS analysis
The nano-high-pressure liquid chromatography (HPLC) system used was an UltiMate 3000 HPLC RSLC nano system (Thermo Fisher Scientific, Bremen, Germany) coupled to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany), equipped with a Proxeon nanospray source (Proxeon, Odense, Denmark). Peptides were loaded onto a trap column (Thermo Fisher Scientific, Bremen, Germany, PepMap C18, 5 mm×300 μm internal diameter, 5 μm particles, 100 Å pore size) at a flow rate of 25 μl min−1 using 0.1% trifluoroacetic acid (TFA) as a mobile phase. After 10 min, the trap column was switched in line with the analytical column (Thermo Fisher Scientific, Bremen, Germany, PepMap C18, 500 mm×75 μm internal diameter, 3 μm, 100 Å). Peptides were eluted using a flow rate of 230 nl min−1, and a binary 2 h gradient, respectively, for 165 min.
The gradient starts with the mobile phases: 98% A (water and formic acid, 99.9:0.1, v/v) and 2% B (water, acetonitrile and formic acid, 19.92:80:0.08, v/v/v) increasing to 35% B over the next 120 min, followed by a gradient in 5 min to 90% B, staying there for 5 min and decreasing in 5 min back to the gradient 98% A and 2% B for equilibration at 30°C.
The LTQ Orbitrap Velos was operated in data-dependent mode, using a full scan in the Orbitrap (m/z range 350–2000, nominal resolution of 60,000, target value 1E6) followed by tandem mass spectrometry (MS/MS) scans of the 12 most-abundant ions in the linear ion trap. MS/MS spectra (normalized collision energy 35%; activation value q 0.25; activation time 10 ms; isolation width 2, target value 1E4) were acquired, and subsequent activation was performed on fragment ions through multistage activation. The neutral loss mass list was therefore set to −98, −49, and −32.6 m/z. Precursor ions selected for fragmentation (charge state 2 and higher) were put on a dynamic exclusion list for 90 s. Additionally, singly charged parent ions were excluded from selection for MS/MS experiments and the monoisotopic precursor selection feature was enabled.
Data analysis
For peptide identification, the .RAW files were loaded into Proteome Discoverer (version 1.4.0.288, Thermo Scientific). All hereby created MS/MS spectra were searched using Mascot 2.2.07 (Matrix Science, London, UK) against the Schizosaccharomyces pombe protein sequence database. The following search parameters were used: β-methylthiolation on cysteine was set as a fixed modification, oxidation on methionine and mono-methylation on lysine and arginine were set as variable modifications. Monoisotopic masses were searched within unrestricted protein masses for tryptic, chymotryptic and no enzymatic specificity. The peptide mass tolerance was set to ±5 ppm and the fragment mass tolerance to ±0.5 Da. The maximal number of missed cleavages was set to 2. The localization of the methylation sites within the peptides was performed with the tool phosphoRS (Taus et al., 2011). Additionally, all of the identified methylation sites were validated by manual inspection of the corresponding MS spectra.
For better visualization, the results of the searches were loaded into Scaffold (version 3.3.1, Proteome Software Inc.), using a minimum of two unique peptides per protein and a Mascot Score of at least 10 as cutoff filters.
Preparation of anti-Psm1-K536me antibodies
To analyze the methylation status of Psm1, anti-Psm1-K536me antibodies were generated using methylated peptide [CTPTQKK(me)YESAIA] spanning the Psm1 methylation site K536me. Modified and unmodified peptides were synthesized, and rabbits were injected with modified peptide coupled with keyhole limpet hemocyanin carrier by Eurogentec (Belgium). Antibodies against modified and unmodified peptides were subsequently affinity purified in two steps from the crude sera: first by applying the serum to a column containing modified peptide, and second by applying the eluates from the first column to a column containing unmodified peptide. One volume of glycerol was added to the purified antibodies and aliquots were stored at −20°C.
Detection of Psm1 methylation
The Pk9-tagged Psm1 was immunoprecipitated from cell extracts using anti-Pk antibody-conjugated agarose (Sigma-Aldrich), and analyzed by immunoblotting with anti-Pk and anti-Psm1-K536me antibodies (see below).
Immunoprecipitation
Exponentially growing cells were harvested and the pellet was resuspended in an equal volume of lysing buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol and 2 mM MgCl2) supplemented with 0.1% NP-40 and protease and phosphatase inhibitors [50 mM NaF, 5 mM Na4P2O7, 0.1 mM Na3VO4, 1 mM β-glycerophosphate, 1 mM PMSF and Roche cOmplete Mini Protease tablets (one tablet per 10 ml of buffer)]. The suspension was frozen in liquid nitrogen and ground into a powder with a SPEX SamplePrep 6870 freezer mill. 1 ml of ice-cold lysing buffer (plus protease inhibitors) was added to 1 g of the frozen powder and gently thawed on ice. 375 units of benzonase (Novagen, 250 U/µl; Merck) was added, and the mixture was rotated at 4°C for 1 h. The lysate was then clarified by spinning at 21,130 g at 4°C. This was followed by measurement of protein concentration in the clear lysate with a Bradford protein assay (Biorad). Lysate containing 10 mg of total protein was added to 50 µl of packed anti-V5 agarose beads that were prepared according to manufacturer's instructions (Sigma-Aldrich) and incubated overnight at 4°C. Next day, the sample was spun at 5000 g for 30 s and washed five times with lysing buffer (plus protease inhibitors). Bound proteins were eluted twice with 50 µl 4× SDS loading buffer. The input and eluates were analyzed by western blotting as described below.
Co-immunoprecipitation
Cells expressing Psm1–Pk, Mus81–Myc or both Psm1–Pk and Mus81–Myc were grown in YES or YES plus 0.01% MMS for 3 h. Protein extracts were prepared as described above for TAP purification. Protein extracts were incubated with anti-V5 agarose beads for 4 h to bind Psm1-Pk. Eluted proteins were separated (8% SDS-PAGE) and transferred to a PVDF membrane (Immobilon-P, 0.45 μm, Millipore). The membrane was blocked with 2% milk in PBS with 0.05% Tween 20 (PBST) and probed with primary antibodies against the Pk and Myc tags. To detect Psm1–Pk, we used monoclonal anti-V5 antibody, diluted 1:5000 (V8012, Sigma-Aldrich) and secondary antibody goat anti-mouse-IgG conjugated to horseradish peroxidase (HRP), diluted 1:20,000 (sc-2005, Santa Cruz Biotechnology). To detect Mus81–Myc, we used rabbit c-Myc antiserum, diluted 1:10,000 (CM-100, Gramsch, Germany) and secondary antibody mouse anti-rabbit-IgG conjugated to HRP, diluted 1:20,000 (sc-2357, Santa Cruz Biotechnology). The ratio of loading between input and immunoprecipitated Pk samples was ∼1:30.
Western blot analysis
Proteins were separated by electrophoresis through 10% polyacrylamide gels containing SDS (0.1%) and transferred onto a PVDF membrane (Immobilon-P membrane with 0.45 µm pore size from Millipore). The membrane was blocked with 2% milk PBST and probed with primary antibodies. Psm1-K536me was detected using rabbit anti-Psm1-K536me, diluted 1:100 (custom made by Eurogentec, UK) and goat anti-rabbit IgG conjugated HRP secondary antibody, diluted 1:5000 (Jackson Immunoresearch). Psm1–Pk9 was detected using mouse-anti-Pk (V5) antibody, diluted 1:1000 (MCA1360 Serotec) and rabbit anti-mouse IgG conjugated to HRP secondary antibody, diluted 1:10,000 (Santa Cruz Biotechnology). Approximate positions of molecular mass markers are indicated in figures.
Dot blot assay
For dot blot analysis, peptides CTPTQKK(me)YESAIA, CTPTQKKYESAIA and CTPTQKAYESAIA were dissolved in water at room temperature to a final concentration of 100 µg/µl and spotted on a methanol activated PVDF membrane (Immobilon-P membrane, 0.45 µm pore size from Millipore). The membrane was blocked with 2% milk in PBST and probed with pre-immune serum or anti-Psm1-K536me antibody (1:100) in 2% milk in PBST. Goat anti-rabbit-IgG conjugated to HRP was used as secondary antibody, diluted 1:5000 (Jackson Immunoresearch).
Chromatin spreads
Chromatin spreads were prepared as described in Loidl and Lorenz (2009). 6 ml of exponentially growing cells were harvested and resuspended in 1 ml of spheroplasting solution (0.65 M KCl with 10 mM dithiothreitol) including 20 µl zymolyase (10 mg/ml; AMS Biotechnology), 20 µl glucuronidase (0.4%; Sigma-Aldrich), 60 µl lysing enzymes (250 mg/ml; L1412, Sigma-Aldrich) and digested at 30°C until lysis of spheroplasts was observed in 1% sodium sarcosyl. The reaction was stopped by adding 9 ml of ice-cold stop solution [0.1 M 2-(N-morpholino) ethane sulfonic acid (MES), 1 mM EDTA, 0.5 mM MgCl2, 1 M sorbitol, pH 6.4]. The cells were collected by centrifugation (720 g for 4 min) and resuspended in 1 ml of stop solution. 20 µl of the cell suspension was transferred on an ethanol-cleaned slide and prefixed with 40 µl of fixative (4% paraformaldehyde, 3.6% sucrose), lysed with 80 µl of detergent (1% lipsol) and the mixture was spread with 80 µl of fixative. The air-dried slides were washed with PBS, and blocked for 30 min with 50 µl of blocking solution (1% BSA in PBS). To detect Psm1-K536me, the slides were incubated overnight with 30 µl of anti-Psm1-K536me antibody (Eurogentec, 1:50 dilution in PBS) at 4°C in a humid chamber. Slides were washed with 10 ml of PBS (10 min, room temperature) and incubated with 30 µl of goat anti-rabbit-IgG antibody, diluted 1:500 (Life Technologies) for 2 h at room temperature. Slides were washed with 10 ml of PBS (10 min, room temperature) and mounting medium with DAPI (Vectashield) was used for microscopy. Images were taken using the following settings: exposure time, FITC, 3154.4 ms, DAPI, 2924.6 ms; acquisition mode, live speed slow; resolution, 1388×1040; binning, 1×1; pixel size, 6.45×6.45 μm.
Yeast two-hybrid analysis
Full-length coding regions for Psm1, Psm3, Psc3, Rad21, Mus81 and Eme1 were amplified from cDNA with primers that added a 5′ SfiI and 3′ SmaI or BamHI (for Fbh1) restriction sites and then cloned into plasmid pGBKT7 (Clontech) for GAL4 DNA-binding domain bait constructs, or into plasmid pGADT7 (Clontech) for GAL4 activation domain prey constructs.
Primers used were 5′-AAAAGGCCATGGAGGCCtatatcacaaaaattgttattcaaggg-3′ and 5′-AAAACCCGGGttatccttcaacgaatgccatagcctcctcc-3′ for Psm3, 5′-AAAAGGCCATGGAGGCCagtgagtctgttactactggatctg-3′ and 5′-AAAACCCGGGttaattcaaaaactcatccacaatagcag-3′ for Psc3, 5′-AAAAGGCCATGGAGGCCttctattcagaggccattctttc-3′ and 5′-AAAACCCGGGtcatagtgatgaaagtagcattccacgtttagc-3′ for Rad21, 5′-AAAAGGCCATGGAGGCCgattgtggaaatccgctgtttttacaat-3′ and 5′-AAAACCCGGGtcaagattctggaaagaaaacgcttgc for Mus81 and 5′-AAAAGGCCATGGAGGCCggacgtttactaagactggaagttg-3′ and 5′-AAAACCCGGGttactccacataaccttccaagttgattgataatgttcgtgaagaattctcc-3′ for Psm1 and 5′-AAAAGGCCATGGAGGCCactctgcagtctacagattccgc-3′ and 5′-AAAACCCGGGctatggtgcatcactggatgcttctttacataag-3′ for Eme1. Lowercase letters represent the gene-specific part of the oligos. mus81 and eme1 were cloned into pGADT7 (plasmid numbers p362 and p360, respectively) while psm1, psm3, psc3 and rad21 were cloned into pGBKT7 plasmid (plasmid numbers p351, p353, p356 and p357, respectively). All inserts were verified by sequencing. Bait and prey plasmids were introduced into S. cerevisiae strain PJ69-4A (Clontech) by standard lithium acetate-mediated transformation. Transformants were tested for bait–prey interaction by spotting onto SD minimal medium lacking appropriate amino acids according to the manufacturer's instructions. We performed at least two independent transformations and growth tests.
Supplementary Material
Acknowledgements
We thank Valeria Scholtzova, Silvia Jargasova, Richard Imre, Ines Kovacikova, Pawel Pasierberk, Peter Steinlein, Kareem Elsayad, Gerry Smith and members of the doctoral program ‘Chromosome Dynamics’ for their help, and Julia Cooper, Jan Palecek, Mitsuhiro Yanagida, Jean-Paul Javerzat and Li-Lin Du for sharing reagents.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.S., J.G.; Methodology: S.S., L.M., J.R., B.H., Z.B., S.P., I.C., K.M., L.C., J.G.; Validation: S.S., L.M., J.R., B.H., Z.B., S.P., I.C., K.M., L.C., J.G.; Formal analysis: S.S., L.M., J.R., B.H., Z.B., S.P., I.C., L.C., J.G.; Investigation: S.S., L.M., J.R., B.H., Z.B., S.P., I.C., L.C., J.G.; Resources: S.S., L.M., J.R., B.H., Z.B., S.P., A.S., K.G.-K., L.C., J.G.; Data curation: S.S., L.M., B.H., Z.B.; Writing - original draft: S.S., L.M., J.R., L.C., J.G.; Visualization: S.S., L.M., J.R., B.H., Z.B., S.P., J.G.; Supervision: S.P., A.S., K.G.-K., K.M., L.C., J.G.; Project administration: A.S., K.G.-K., J.G.; Funding acquisition: S.P., A.S., K.G.-K., K.M., L.C., J.G.
Funding
This work was supported by the Vedecká Grantová Agentúra MŠVVaŠ SR a SAV (Slovak Grant Agency; VEGA) (no. 1/0196/14 and 1/0450/18), Agentúra na Podporu Výskumu a Vývoja (Slovak Research and Development Agency; APPV) (no. APVV-0334-12) and The Austrian Science Fund (FWF) (no. P30516, P21437, W1238 and F34). K.M. was supported by an ERA-CAPS grant (I 3686). J.G. was supported by the European Community's Seventh Framework Programme (FP7/2007–2013) under the grant agreement number PCIG11-GA-2012-322300. Additional support was provided by People Programme (Marie Curie Actions) European Union's Seventh Framework Programme [REA grant agreement no. 609427, project no. 0032/01/02], Slovak Academy of Sciences, VEGA grants 2/0026/18 and 2/0070/16, and by Slovak Research and Development Agency contract No. APVV-0111-12 and APVV-16-0120 to L.C., SASPRO and the People Programme (Marie Curie Actions) of the EU FP7 under REA grant agreement No. 0070/01/02 to S.P. Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.214924.supplemental
References
- Alexandru G., Uhlmann F., Mechtler K., Poupart M.-A. and Nasmyth K. (2001). Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105, 459-472. 10.1016/S0092-8674(01)00362-2 [DOI] [PubMed] [Google Scholar]
- Almedawar S., Colomina N., Bermúdez-López M., Pociño-Merino I. and Torres-Rosell J. (2012). A SUMO-dependent step during establishment of sister chromatid cohesion. Curr. Biol. 22, 1576-1581. 10.1016/j.cub.2012.06.046 [DOI] [PubMed] [Google Scholar]
- Avemann K., Knippers R., Koller T. and Sogo J. M. (1988). Camptothecin, a specific inhibitor of type I DNA topoisomerase, induces DNA breakage at replication forks. Mol. Cell. Biol. 8, 3026-3034. 10.1128/MCB.8.8.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A., Costanzo M., Myers C. L., Andrews B. and Boone C. (2013). Genetic interaction networks: toward an understanding of heritability. Annu. Rev. Genomics Hum. Genet. 14, 111-133. 10.1146/annurev-genom-082509-141730 [DOI] [PubMed] [Google Scholar]
- Boddy M. N., Lopez-Girona A., Shanahan P., Interthal H., Heyer W.-D. and Russell P. (2000). Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20, 8758-8766. 10.1128/MCB.20.23.8758-8766.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Gaillard P. H., McDonald W. H., Shanahan P., Yates J. R. III and Russell P. (2001). Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537-548. 10.1016/S0092-8674(01)00536-0 [DOI] [PubMed] [Google Scholar]
- Carlson S. M. and Gozani O. (2016). Nonhistone lysine methylation in the regulation of cancer pathways. Cold Spring Harb Perspect Med 6, a026435 10.1101/cshperspect.a026435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzinikolaou G., Apostolou Z., Aid-Pavlidis T., Ioannidou A., Karakasilioti I., Papadopoulos G. L., Aivaliotis M., Tsekrekou M., Strouboulis J., Kosteas T. et al. (2017). ERCC1-XPF cooperates with CTCF and cohesin to facilitate the developmental silencing of imprinted genes. Nat. Cell Biol. 19, 421-432. 10.1038/ncb3499 [DOI] [PubMed] [Google Scholar]
- Cipak L., Spirek M., Novatchkova M., Chen Z., Rumpf C., Lugmayr W., Mechtler K., Ammerer G., Csaszar E. and Gregan J. (2009). An improved strategy for tandem affinity purification-tagging of Schizosaccharomyces pombe genes. Proteomics 9, 4825-4828. 10.1002/pmic.200800948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipak L., Zhang C., Kovacikova I., Rumpf C., Miadokova E., Shokat K. M. and Gregan J. (2011). Generation of a set of conditional analog-sensitive alleles of essential protein kinases in the fission yeast Schizosaccharomyces pombe. Cell Cycle 10, 3527-3532. 10.4161/cc.10.20.17792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipak L., Polakova S., Hyppa R. W., Smith G. R. and Gregan J. (2014). Synchronized fission yeast meiosis using an ATP analog-sensitive Pat1 protein kinase. Nat. Protoc. 9, 223-231. 10.1038/nprot.2014.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon S., Gaillard P.-H. L., Chahwan C., McDonald W. H., Yates J. R. III and Russell P. (2004). Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol. Biol. Cell 15, 71-80. 10.1091/mbc.e03-08-0586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C. L., Ahn J. S., Dixon J. and Whitby M. C. (2002). Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277, 32753-32759. 10.1074/jbc.M202120200 [DOI] [PubMed] [Google Scholar]
- Feytout A., Vaur S., Genier S., Vazquez S. and Javerzat J. P. (2011). Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1. Mol. Cell. Biol. 31, 1771-1786. 10.1128/MCB.01284-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelot C., Guirouilh-Barbat J., Le Guen T., Dardillac E., Chailleux C., Canitrot Y. and Lopez B. S. (2016). The cohesin complex prevents the end joining of distant DNA double-strand ends. Mol. Cell 61, 15-26. 10.1016/j.molcel.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Gregan J., Zhang C., Rumpf C., Cipak L., Li Z., Uluocak P., Nasmyth K. and Shokat K. M. (2007). Construction of conditional analog-sensitive kinase alleles in the fission yeast Schizosaccharomyces pombe. Nat. Protoc. 2, 2996-3000. 10.1038/nprot.2007.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli J. M., Ünal E., Guacci V. and Koshland D. (2008). The kleisin subunit of cohesin dictates damage-induced cohesion. Mol. Cell 31, 47-56. 10.1016/j.molcel.2008.06.005 [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli J. M., Unal E. and Koshland D. (2009). Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol. Cell 34, 311-321. 10.1016/j.molcel.2009.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli J. M., Mert O., Davenport C., Guacci V. and Koshland D. (2010). Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr. Biol. 20, 957-963. 10.1016/j.cub.2010.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S. Y., Li Y., Wang Y., Chen Y., Zhao Y. and Qin J. (2008). Complications in the assignment of 14 and 28 Da mass shift detected by mass spectrometry as in vivo methylation from endogenous proteins. Anal. Chem. 80, 1721-1729. 10.1021/ac7021025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. T., Xu B. and Kastan M. B. (2002). Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16, 560-570. 10.1101/gad.970602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L., Altmannova V., Spirek M. and Zhao X. (2012). Homologous recombination and its regulation. Nucleic Acids Res. 40, 5795-5818. 10.1093/nar/gks270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S. and Carr A. M. (2013). Replication stress and genome rearrangements: lessons from yeast models. Curr. Opin. Genet. Dev. 23, 132-139. 10.1016/j.gde.2012.11.009 [DOI] [PubMed] [Google Scholar]
- Larsen S. C., Sylvestersen K. B., Mund A., Lyon D., Mullari M., Madsen M. V., Daniel J. A., Jensen L. J. and Nielsen M. L. (2016). Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 9, rs9 10.1126/scisignal.aaf7329 [DOI] [PubMed] [Google Scholar]
- Loidl J. and Lorenz A. (2009). Analysis of Schizosaccharomyces pombe meiosis by nuclear spreading. Methods Mol. Biol. 558, 15-36. 10.1007/978-1-60761-103-5_2 [DOI] [PubMed] [Google Scholar]
- Lorenz A., Osman F., Folkyte V., Sofueva S. and Whitby M. C. (2009). Fbh1 limits Rad51-dependent recombination at blocked replication forks. Mol. Cell. Biol. 29, 4742-4756. 10.1128/MCB.00471-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C., North M., Erixon K., Walters K., Jenssen D., Goldman A. S. and Helleday T. (2005). Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 33, 3799-3811. 10.1093/nar/gki681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maftahi M., Hope J. C., Delgado-Cruzata L., Han C. S. and Freyer G. A. (2002). The severe slow growth of Deltasrs2 Deltarqh1 in Schizosaccharomyces pombe is suppressed by loss of recombination and checkpoint genes. Nucleic Acids Res. 30, 4781-4792. 10.1093/nar/gkf581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleenan A., Cordon-Preciado V., Clemente-Blanco A., Liu I. C., Sen N., Leonard J., Jarmuz A. and Aragon L. (2012). SUMOylation of the alpha-kleisin subunit of cohesin is required for DNA damage-induced cohesion. Curr. Biol. 22, 1564-1575. 10.1016/j.cub.2012.06.045 [DOI] [PubMed] [Google Scholar]
- Murray J. M., Lindsay H. D., Munday C. A. and Carr A. M. (1997). Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17, 6868-6875. 10.1128/MCB.17.12.6868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. O., Jalan M., Morrow C. A., Osman F. and Whitby M. C. (2015). Recombination occurs within minutes of replication blockage by RTS1 producing restarted forks that are prone to collapse. Elife 4, e04539 10.7554/eLife.04539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. B., Cao X.-J., Han B., Chen L. H., Horvath A., Richardson T. I., Campbell R. M., Garcia B. A. and Nguyen H. (2016). Quantitative Profiling of the Activity of Protein Lysine Methyltransferase SMYD2 Using SILAC-Based Proteomics. Mol. Cell. Proteomics 15, 892-905. 10.1074/mcp.M115.053280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N., Cipak L., Polakova S., Hyppa R. W., Cipakova I., Anrather D., Karvaiova L., Mechtler K., Smith G. R. and Gregan J. (2015). Casein kinase 1 and phosphorylation of cohesin subunit rec11 (SA3) promote meiotic recombination through linear element formation. PLoS Genet. 11, e1005225 10.1371/journal.pgen.1005225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjha L., Howard S. M. and Cejka P. (2018). Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma 127, 187-214. 10.1007/s00412-017-0658-1 [DOI] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T., Heeger S., Lehane C., East P., Flynn H., Skehel M. and Uhlmann F. (2008). Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321, 563-566. 10.1126/science.1157774 [DOI] [PubMed] [Google Scholar]
- Ryan C. J., Cimermancic P., Szpiech Z. A., Sali A., Hernandez R. D. and Krogan N. J. (2013). High-resolution network biology: connecting sequence with function. Nat. Rev. Genet. 14, 865-879. 10.1038/nrg3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schar P., Fasi M. and Jessberger R. (2004). SMC1 coordinates DNA double-strand break repair pathways. Nucleic Acids Res. 32, 3921-3929. 10.1093/nar/gkh716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren C. and Nasmyth K. (2001). Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11, 991-995. 10.1016/S0960-9822(01)00271-8 [DOI] [PubMed] [Google Scholar]
- Stewart E., Chapman C. R., Al-Khodairy F., Carr A. M. and Enoch T. (1997). rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16, 2682-2692. 10.1093/emboj/16.10.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L., Lindroos H. B., Shirahige K. and Sjogren C. (2004). Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16, 1003-1015. 10.1016/j.molcel.2004.11.026 [DOI] [PubMed] [Google Scholar]
- Strumberg D., Pilon A. A., Smith M., Hickey R., Malkas L. and Pommier Y. (2000). Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5’-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 20, 3977-3987. 10.1128/MCB.20.11.3977-3987.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K., Kato J. and Ikeda H. (1998). Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics 148, 49-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C. and Mechtler K. (2011). Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354-5362. 10.1021/pr200611n [DOI] [PubMed] [Google Scholar]
- Tercero J. A., Longhese M. P. and Diffley J. F. (2003). A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11, 1323-1336. 10.1016/S1097-2765(03)00169-2 [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer M., Lengronne A., Davidson M. B., Bacal J., Francois P., Hohl M., Petrini J. H. J., Pasero P. and Cobb J. A. (2012). Cohesin association to replication sites depends on rad50 and promotes fork restart. Mol. Cell 48, 98-108. 10.1016/j.molcel.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. and Nasmyth K. (1998). Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 8, 1095-1101. 10.1016/S0960-9822(98)70463-4 [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Lottspeich F. and Nasmyth K. (1999). Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400, 37-42. 10.1038/21831 [DOI] [PubMed] [Google Scholar]
- Unal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., Haber J. E. and Koshland D. (2004). DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16, 991-1002. 10.1016/j.molcel.2004.11.027 [DOI] [PubMed] [Google Scholar]
- Unal E., Heidinger-Pauli J. M., Kim W., Guacci V., Onn I., Gygi S. P. and Koshland D. E. (2008). A molecular determinant for the establishment of sister chromatid cohesion. Science 321, 566-569. 10.1126/science.1157880 [DOI] [PubMed] [Google Scholar]
- Waizenegger I. C., Hauf S., Meinke A. and Peters J. M. (2000). Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103, 399-410. 10.1016/S0092-8674(00)00132-X [DOI] [PubMed] [Google Scholar]
- Wang S. W., Goodwin A., Hickson I. D. and Norbury C. J. (2001). Involvement of Schizosaccharomyces pombe Srs2 in cellular responses to DNA damage. Nucleic Acids Res. 29, 2963-2972. 10.1093/nar/29.14.2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin E. and Peters J. M. (2009). The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells. EMBO J. 28, 2625-2635. 10.1038/emboj.2009.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Kong X., Ji Z., Zeng W., Potts P. R., Yokomori K. and Yu H. (2012). Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl. Genes Dev. 26, 1473-1485. 10.1101/gad.193615.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt M. D. and Pittman D. L. (2006). Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem. Res. Toxicol. 19, 1580-1594. 10.1021/tx060164e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi P. T., Wang Y., Zhao S., Patel N., Lee E. Y. and Qin J. (2002). SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16, 571-582. 10.1101/gad.970702 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.