Abstract
Children with rhinovirus‐induced severe early wheezing have an increased risk of developing asthma later in life. The exact molecular mechanisms for this association are still mostly unknown. To identify potential changes in the transcriptional and epigenetic regulation in rhinovirus‐associated atopic or nonatopic asthma, we analyzed a cohort of 5‐year‐old children (n = 45) according to the virus etiology of the first severe wheezing episode at the mean age of 13 months and to 5‐year asthma outcome. The development of atopic asthma in children with early rhinovirus‐induced wheezing was associated with DNA methylation changes at several genomic sites in chromosomal regions previously linked to asthma. The strongest changes in atopic asthma were detected in the promoter region of SMAD3 gene at chr 15q22.33 and introns of DDO/METTL24 genes at 6q21. These changes were validated to be present also at the average age of 8 years.
Keywords: asthma, epigenome, rhinovirus, transcriptome, wheezing
1. INTRODUCTION
Rhinovirus has been detected in 20%‐40% of children with moderate‐to‐severe wheezing episodes during the first 2 years of life.1, 2, 3 This etiology is of particular interest due to strong association with recurrent wheezing, prolonged need of asthma controller medication, doctor‐diagnosed asthma up to 13 years of age1, 4, 5, 6, 7, 8, 9, 10, and atopic asthma at school age.11 The suggested explanations for this association are low interferon responses (ie, impaired viral defense), early airway inflammation (ie, broken epithelial barrier; T helper2‐polarized immune responses) and genetic variations.12, 13, 14, 15, 16 Despite progress in rhinovirus research, the exact molecular mechanisms for this association are still mostly unknown.17 Furthermore, early predictive biomarkers are needed for the identification of the rhinovirus‐infected children with an increased risk for developing asthma and enabling design of effective intervention strategies to prevent asthma.11, 18 For these reasons, we examined epigenomic and transcriptomic changes in 5‐year‐old children associated with the virus etiology of their first wheezing episode and later asthma status.
2. METHODS
The detailed study protocols are described in the Data S1.
2.1. Clinical study protocol
The study protocol was approved by the Ethics Committee of the Turku University Hospital and for the first 12 months was registered at ClinicalTrials.gov (NCT00731575). At study entry, standard procedures were carried out as previously described.19 The inclusion criteria were age 3‐23 months, delivery at ≥36 gestational weeks, first wheezing episode and a written informed consent from a guardian. Selected children were recruited to regular follow‐up visits. Children were diagnosed to have current asthma at the 4‐year follow‐up visit if they met one or more of the subsequent criteria during the preceding 12 months: doctor‐diagnosed asthma, regular use of doctor‐prescribed corticosteroid asthma therapy, use of oral corticosteroids for asthma exacerbations, acute asthma attack relieved by repeated use of bronchodilator.20 Current atopic asthma was defined as asthma with laboratory‐verified sensitization (IgE antibodies >0.35 kU/L) at the 4‐year follow‐up visit. Nonatopic asthma was defined as asthma without these. The statistical analysis is described in Data S1.
2.2. Epigenome and transcriptome analysis
Total RNA (Tempus Spin RNA Isolation kit) and DNA (Qiagen's QIAamp DNA Blood Maxi kit) were isolated from whole blood. The messenger RNA‐seq samples were prepared with Illumina TruSeq RNA Sample Preparation kit v2. The reduced representation bisulfite sequencing (RRBS) libraries were prepared with a protocol adapted from Boyle et al.21, 22 For genomic localization of promoters and enhancers H3K4me1, H3K4me3 and H3K27Ac ChIP‐seq was carried out from peripheral blood mononuclear cells isolated from 2 reference individuals. Briefly, the chromatin was prepared (truChIP Low Cell Chromatin Kit, Covaris) and ChIP reactions (Auto Histone ChIP‐seq kit) were carried out with Diagenode IP‐Star SX 8G robot. The libraries were sequenced with Illumina HiSeq2000 platform. The raw data were quality controlled with FastQC23 and other tools described in the Data S1. The RNA‐seq data were analyzed with RNA Express v1.0.0 in Illumina BaseSpace cloud.24, 25, 26 The RRBS data were filtered with Trim Galore! v0.3.3,27 and reads were aligned to human genome hg19 with bismark v0.12.528 and bowtie2 v2.2.3.29 The methylation calls were extracted with Bismark. Outliers (eg, RnBeads MDS and PCA30) were excluded from the analysis. Differentially methylated regions were identified with RADMeth.31, 32 The ChIP‐seq data were analyzed in Illumina BaseSpace Cloud with ChIPSeq BaseSpace Labs tool. In addition, ENCODE ChIP‐seq data on H3K4me1, H3K4me3, H3K27me3, and H3K9me3 from PBMCs were utilized in the analysis to examine colocalization of DNA methylation changes with histone marks.33, 34, 35
2.3. Targeted pyrosequencing
The oligos were designed with Pyromark Software Assay Design 2.0. Samples were prepared, and pyrosequencing was carried out with Qiagen's Pyromark Q24 according to the manufacturer's instructions. The data were analyzed with Pyromark Advanced Software. Unpaired t test was used to calculate the statistical differences between study groups. See the Data S1 for more details.
3. RESULTS
3.1. Patient characteristics
Originally, 124 first‐time wheezing children were enrolled of whom 77 (62%) children participated in the 4‐year follow‐up visit (Table S1). Of these, 48 most representative children were selected for the transcriptome and epigenome studies according to the virus etiology of their first severe wheezing episode, and their 5‐year asthma outcome as follows: rhinovirus + asthma + (n = 16), rhinovirus + asthma − (n = 16), and rhinovirus − asthma − (n = 16). Three study subjects from the latter group were excluded after quality analysis. Further details of patient characteristics are shown in Tables S2 and S3.
3.2. DNA methylome and transcriptome changes in rhinovirus‐associated wheezing and asthma
Comparison of the children with rhinovirus + atopic asthma + (n = 11) vs rhinovirus + asthma − (n = 16) revealed methylation changes (median change ±20% and FDR ≤0.05) in 17 genomic regions associated with atopic asthma (Table 1). The strongest changes were observed in the region chr6:110720838‐110720871 located in the introns of genes D‐aspartate oxidase (DDO) and methyltransferase like 24 (METTL24), and region chr15:67356671‐67356696 located 1511 bp upstream of the transcription start site of SMAD family member 3 (SMAD3) gene, close to active promoter mark (H3K4me3). Similarly, comparison of the children with rhinovirus + nonatopic asthma + (n = 5) vs rhinovirus + asthma − (n = 10) revealed differences in 13 genomic regions (Table 1). The strongest change associated with nonatopic asthma was observed in the site chr9:139860386‐139860387 overlapping enhancer mark H3K4me1, 9.12 kb upstream of the gene prostaglandin D2 synthase (PTGDS).
Table 1.
The differentially methylated genomic regions associated with rhinovirus‐induced wheezing and asthma (median methylation difference ≥20, FDR ≤0.05)
| Genomic location (hg19) | No. of sites | Median meDiff (%) | Genomic element | Chromatin marks (±1 kb) | Closest gene (Symbol) |
|---|---|---|---|---|---|
| Differentially methylated genomic regions associated with atopic asthma | |||||
| chr15:67356671‐67356696 | 2 | 41.68 | Intergenic | H3K4me1, ‐me3 | SMAD3 |
| chr10:116273998‐116274013 | 2 | 33.70 | Intronic | H3K4me1 | ABLIM1 |
| chr2:158287048‐158287070 | 2 | 29.13 | Intronic | H3K4me1a | CYTIP |
| chr7:73456938‐73456960 | 4 | 28.34 | Exon‐intron | – | ELN |
| chr13:110384309‐110384326 | 4 | 28.07 | Intergenic | H3K4me3a | LINC00676, IRS2 |
| chr7:35302562‐35302563 | 1 | 27.40 | Intergenic | H3K27me3a | TBX20 |
| chr2:31806387‐31806402 | 2 | 25.17 | Intergenic | H3K27me3 | SRD5A2 |
| chr16:89674059‐89674095 | 3 | 23.19 | Intergenic | – | DPEP1 |
| chr17:80542056‐80542086 | 3 | 21.92 | Exon‐intron | – | FOXK2 |
| chr19:9711129‐9711155 | 2 | 21.65 | Intergenic | – | ZNF561 |
| chr7:66211672‐66211673 | 1 | 21.28 | Intronic | H3K4me1a | RABGEF1 |
| chr7:1005102‐1005133 | 5 | 21.24 | Exon, intron | H3K4me1a | COX19, ADAP1 |
| chr14:106437030‐106437045 | 2 | −20.36 | Exon‐intron | – | abParts, ADAM6 |
| chr11:123388388‐123388389 | 1 | −20.64 | Intergenic | – | GRAMD1B |
| chr1:143279717‐143279732 | 2 | −20.98 | Intronic | – | CR936796 |
| chr6:29831764‐29831935 | 4 | −24.13 | Intronic | – | HLA‐G, HLA‐H |
| chr6:110720838‐110720871 | 3 | −48.19 | Intronic | H3K4me3 | DDO, METTL24 |
| Differentially methylated genomic regions associated with nonatopic asthma | |||||
| chr9:139860386‐139860387 | 1 | 45.95 | Intergenic | H3K4me1a | PTGDS |
| chr6:144329349‐144329433 | 4 | 35.90 | Intron, exon | H3K4me1, ‐me3a | PLAGL1 |
| chr1:3292550‐3292562 | 2 | 34.38 | Intron | – | PRDM16 |
| chr19:2546695‐2546719 | 6 | 32.88 | Intron | – | GNG7 |
| chr7:138726331‐138726332 | 1 | 26.95 | Intergenic | – | ZC3HAV1 |
| chr7:48241582‐48241586 | 2 | 24.72 | Intron | – | ABCA13 |
| chr6:169977857‐169977884 | 3 | 24.09 | Intron | – | WDR27 |
| chr11:67051767‐67051780 | 2 | 23.21 | Exon | H3K4me1a, ‐me3a | ADRBK1 |
| chr18:77906226‐77906417 | 4 | 22.51 | Intron | H3K4me1a, ‐me3a | PARD6G‐AS1 |
| chr6:20320057‐20320136 | 4 | 20.95 | Intergenic | H3K4me1, ‐me3a | E2F3 |
| chr9:45439874‐45439891 | 3 | −23.22 | Intergenic | H3K4me1, ‐me3 | LOC102723709 |
| chr4:3572886‐3572887 | 1 | −33.30 | Intergenic | – | LINC00955 |
| chr4:49150172‐49150193 | 4 | −34.46 | Intergenic | H3K4me1, ‐me3, H3K27me3 | CWH43 |
Differentially methylated region overlaps with the histone mark.
Interestingly, no differences in gene expression (fold change cut off ±1.4, FDR ≤0.05) were observed to be associated with either atopic or nonatopic asthma at the time of sampling, although for example SMAD3 gene was detected to be expressed in the blood of the study subjects.
3.3. Targeted validation and stability of the DNA methylation changes
The CpG methylation changes associated with atopic asthma at the DDO/METTL24 intron (chr6:110720839‐110720905) and SMAD3 promoter (chr15:67356631‐67356721) with RRBS (Table 1) were confirmed with targeted pyrosequencing of the samples collected at the average age of 5 years (Figure 1A, Figure S1). Importantly, analysis of the samples collected from the same children at the average age of 8 years revealed that the methylation changes associated with atopic asthma in these 2 regions were still present 3 years later (P ≤ 5.08E‐04; Figure 1B).
Figure 1.
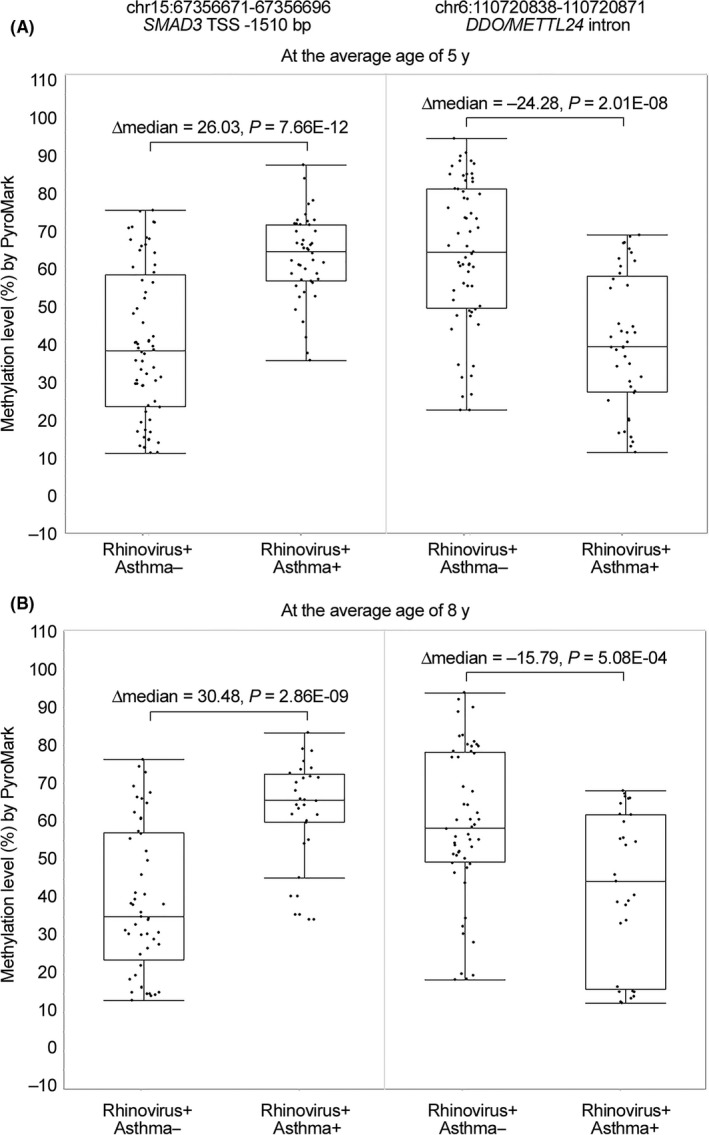
Validation and stability of the DNA methylation changes associated with atopic asthma. The strongest DNA methylation changes associated with atopic asthma (rhinovirus + atopic asthma + vs rhinovirus + atopic asthma −) were validated with targeted pyrosequencing (A) at the average age of 5 y and (B) at the average age of 8 y in samples collected from the same children. In addition, to outlier box plots, the distribution of DNA methylation levels of individual CpG sites within the indicated region for each individual is shown in the figure. In addition, median methylation differences and t test P‐values are shown in the figure
4. DISCUSSION
We detected epigenetic changes in several genomic regions in children who had suffered early rhinovirus‐induced wheezing, and which associated with later onset of asthma. Interestingly, among the strongest changes associated with atopic asthma was DNA methylation alteration in the promoter of SMAD3 gene in a well‐known asthma locus at chr 15q22.33.36 SMAD3 protein has an important function in the regulation of immune responses and together with the other components of the transforming growth factor beta (TGFβ) signaling regulate fibrosis in airways.37 While our study was in progress, DeVries et al38 reported that hypermethylation of SMAD3 promoter is associated with asthma of children of asthmatic mothers further confirming importance of this epigenetic change in the molecular pathology of asthma. In addition, we detected changes in DDO/METTL24 genes at 6q21 and in HLA locus at chr 6p21.1‐22.3 previously associated with asthma.39 HLA‐G gene, 33 kb upstream from the differentially methylated region, is an asthma susceptibility gene,40 and its expression is induced by allergens.41 Several other differentially methylated sites were also in the proximity of genes, such as ELN 42 or CYTIP, 43, 44 previously linked to lung or immune cell functions, or viral infections, therefore having potential functional significance in the atopic asthma.
Among the strongest changes associated with nonatopic asthma was DNA methylation change linked to PTGDS gene. PTGDS catalyzes synthesis of prostaglandin D2, which mediates development and symptoms of asthma by recruiting Th2 cells and inducing contraction of airways smooth muscle cells.45, 46 Prostaglandin D2 enhances proinflammatory actions of macrophages and subsequent neutrophil activation.47 Therefore, altered regulation of PTGDS may be important in the development of nonatopic asthma.
In conclusion, our results demonstrate that epigenetic changes associated with rhinovirus‐induced early wheezing and asthma can be detected in peripheral blood. The strongest changes associated with atopic asthma were localized in the genomic regions previously associated with asthma15, 16, 36 and importantly for SMAD3 promoter and in DDO/METTL24 gene were detected at the age of both 5 and 8 years. Although we did not detect significant changes in the transcriptomes at the time of measurement, it is possible that the regulation of the affected genes is altered, for example in response to antigen challenge. Alternatively, the DNA methylation changes detected in blood reflect functional changes present in other tissues, such as airways.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
Supporting information
ACKNOWLEDGMENTS
We thank the Finnish Functional Genomics Centre (University of Turku, Åbo Akademi University and Biocenter Finland) for the infrastructure support.
Lund RJ, Osmala M, Malonzo M, et al. Atopic asthma after rhinovirus‐induced wheezing is associated with DNA methylation change in the SMAD3 gene promoter. Allergy. 2018;73:1735–1740. 10.1111/all.13473
Osmala, Malonzo, Lähdesmäki, Lahesmaa and Jartti equally contributed to this study.
REFERENCES
- 1. Jartti T, Lehtinen P, Vuorinen T, et al. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis. 2004;10:1095‐1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rakes GP, Arruda E, Ingram JM, et al. Rhinovirus and respiratory syncytial virus in wheezing children requiring emergency care. IgE and eosinophil analyses. Am J Respir Crit Care Med. 1999;159:785‐790. [DOI] [PubMed] [Google Scholar]
- 3. Mansbach JM, Piedra PA, Teach SJ, et al. Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis. Arch Pediatr Adolesc Med. 2012;166:700‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jackson DJ, Gangnon RE, Evans MD, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high‐risk children. Am J Respir Crit Care Med. 2008;178:667‐672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lehtinen P, Ruohola A, Vanto T, Vuorinen T, Ruuskanen O, Jartti T. Prednisolone reduces recurrent wheezing after a first wheezing episode associated with rhinovirus infection or eczema. J Allergy Clin Immunol. 2007;119:570‐575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kusel MM, de Klerk NH, Kebadze T, et al. Early‐life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. J Allergy Clin Immunol. 2007;119:1105‐1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kotaniemi‐Syrjanen A, Reijonen TM, Korhonen K, Waris M, Vainionpaa R, Korppi M. Wheezing due to rhinovirus infection in infancy: bronchial hyperresponsiveness at school age. Pediatr Int. 2008;50:506‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Midulla F, Pierangeli A, Cangiano G, et al. Rhinovirus bronchiolitis and recurrent wheezing: 1‐year follow‐up. Eur Respir J. 2012;39:396‐402. [DOI] [PubMed] [Google Scholar]
- 9. Lukkarinen M, Lukkarinen H, Lehtinen P, Vuorinen T, Ruuskanen O, Jartti T. Prednisolone reduces recurrent wheezing after first rhinovirus wheeze: a 7‐year follow‐up. Pediatr Allergy Immunol. 2013;24:237‐243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lukkarinen M, Vuorinen T, Lehtinen P, Ruuskanen O, Jartti T. Sensitization at the first wheezing episode increases risk for long‐term asthma therapy. Pediatr Allergy Immunol. 2015;26:687‐691. [DOI] [PubMed] [Google Scholar]
- 11. Lukkarinen M, Koistinen A, Turunen R, Lehtinen P, Vuorinen T, Jartti T. Rhinovirus‐induced first wheezing episode predicts atopic but not nonatopic asthma at school age. J Allergy Clin Immunol. 2017;140:988‐995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jartti T, Kuusipalo H, Vuorinen T, et al. Allergic sensitization is associated with rhinovirus‐, but not other virus‐, induced wheezing in children. Pediatr Allergy Immunol. 2010;21:1008‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jackson DJ, Evans MD, Gangnon RE, et al. Evidence for a causal relationship between allergic sensitization and rhinovirus wheezing in early life. Am J Respir Crit Care Med. 2012;185:281‐285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baraldo S, Contoli M, Bazzan E, et al. Deficient antiviral immune responses in childhood: distinct roles of atopy and asthma. J Allergy Clin Immunol. 2012;130:1307‐1314. [DOI] [PubMed] [Google Scholar]
- 15. Caliskan M, Bochkov YA, Kreiner‐Moller E, et al. Rhinovirus wheezing illness and genetic risk of childhood‐onset asthma. N Engl J Med. 2013;368:1398‐1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bochkov YA, Watters K, Ashraf S, et al. Cadherin‐related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci USA. 2015;112:5485‐5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jartti T, Gern JE. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clin Immunol. 2017;140:895‐906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jartti T, Nieminen R, Vuorinen T, et al. Short‐ and long‐term efficacy of prednisolone for first acute rhinovirus‐induced wheezing episode. J Allergy Clin Immunol. 2015;135:691‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turunen R, Koistinen A, Vuorinen T, et al. The first wheezing episode: respiratory virus etiology, atopic characteristics, and illness severity. Pediatr Allergy Immunol. 2014;25:796‐803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. National Heart Lung and Blood Institute . Expert panel report: guidelines for the diagnosis and management of asthma. Report No.: 3. 2007. http://www-nhlbi.nih.gov/health-pro/guidelines/current/asthma-guidelines. Accessed August 28, 2007.
- 21. Boyle P, Clement K, Gu H, et al. Gel‐free multiplexed reduced representation bisulfite sequencing for large‐scale DNA methylation profiling. Genome Biol. 2012;13:R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Konki M, Pasumarthy K, Malonzo M, et al. Epigenetic silencing of the key antioxidant enzyme catalase in karyotypically abnormal human pluripotent stem cells. Sci Rep. 2016;6:22190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. FastQC: a quality control tool for high throughput sequence data [homepage on the Internet]. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 4 July, 2015.
- 24. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA‐seq aligner. Bioinformatics. 2013;29:15‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. BaseSpace [homepage on the Internet]. https://basespace.illumina.com. Accessed 3 March, 2015.
- 27. Trim galore! [homepage on the Internet]. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. Accessed 4 July, 2015.
- 28. Krueger F, Andrews S. Bismark: a flexible aligner and methylation caller for bisulfite‐seq applications. Bioinformatics. 2011;27:1571‐1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langmead B, Salzberg SL. Fast gapped‐read alignment with bowtie 2. Nat Methods. 2012;9:357‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Assenov Y, Muller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138‐1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dolzhenko E, Smith AD. Using beta‐binomial regression for high‐precision differential methylation analysis in multifactor whole‐genome bisulfite sequencing experiments. BMC Bioinformatics. 2014;15:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289‐300. [Google Scholar]
- 33. Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high‐performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O'Geen H, Echipare L, Farnham PJ. Using ChIP‐seq technology to generate high‐resolution profiles of histone modifications. Methods Mol Biol. 2011;791:265‐286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moffatt MF, Gut IG, Demenais F, et al. A large‐scale, consortium‐based genomewide association study of asthma. N Engl J Med. 2010;363:1211‐1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Royce SG, Cheng V, Samuel CS, Tang ML. The regulation of fibrosis in airway remodeling in asthma. Mol Cell Endocrinol. 2012;351:167‐175. [DOI] [PubMed] [Google Scholar]
- 38. DeVries A, Wlasiuk G, Miller SJ, et al. Epigenome‐wide analysis links SMAD3 methylation at birth to asthma in children of asthmatic mothers. J Allergy Clin Immunol. 2017;140:534‐542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kontakioti E, Domvri K, Papakosta D, Daniilidis M. HLA and asthma phenotypes/endotypes: a review. Hum Immunol. 2014;75:930‐939. [DOI] [PubMed] [Google Scholar]
- 40. Nicolae D, Cox NJ, Lester LA, et al. Fine mapping and positional candidate studies identify HLA‐G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005;76:349‐357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Contini P, Puppo F, Canonica GW, Murdaca G, Ciprandi G. Allergen‐driven HLA‐G expression and secretion in peripheral blood mononuclear cells from allergic rhinitis patients. Hum Immunol. 2016;77:1172‐1178. [DOI] [PubMed] [Google Scholar]
- 42. Ingram JL, Slade D, Church TD, et al. Role of matrix metalloproteinases‐1 and ‐2 in interleukin‐13‐suppressed elastin in airway fibroblasts in asthma. Am J Respir Cell Mol Biol. 2016;54:41‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watford WT, Li D, Agnello D, et al. Cytohesin binder and regulator (cybr) is not essential for T‐ and dendritic‐cell activation and differentiation. Mol Cell Biol. 2006;26:6623‐6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Theodoridis AA, Eich C, Figdor CG, Steinkasserer A. Infection of dendritic cells with herpes simplex virus type 1 induces rapid degradation of CYTIP, thereby modulating adhesion and migration. Blood. 2011;118:107‐115. [DOI] [PubMed] [Google Scholar]
- 45. Matsuoka T, Hirata M, Tanaka H, et al. Prostaglandin D2 as a mediator of allergic asthma. Science. 2000;287:2013‐2017. [DOI] [PubMed] [Google Scholar]
- 46. Holgate ST. Innate and adaptive immune responses in asthma. Nat Med. 2012;18:673‐683. [DOI] [PubMed] [Google Scholar]
- 47. Jandl K, Stacher E, Balint Z, et al. Activated prostaglandin D2 receptors on macrophages enhance neutrophil recruitment into the lung. J Allergy Clin Immunol. 2016;137:833‐843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


