Abstract
Using an orthotopic model of ovarian cancer, we studied the delivery of siRNA in nanoparticles of tri-block copolymers consisting of hyperbranched polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) (hyPEI-g-PCL-b-PEG) with and without a folic acid targeting ligand. A SKOV-3/LUC FRα overexpressing cell line was employed to mimic the clinical manifestations of ovarian cancer. Both targeted and non-targeted micelleplexes were able to effectively deliver siRNA to the primary tumor and its metastases, as measured by gamma scintillation counting and confocal microscopy. Stability of the micelleplexes was demonstrated with a serum albumin binding study. Regarding biodistribution, intravenous (I.V.) administration showed a slight advantage of FRα targeted over non-targeted micelleplex accumulation within the tumor. However, both formulations displayed significant liver uptake. On the other hand, intraperitoneally (I.P.) injected mice showed a modest 6% of the injected dose per gram (ID/g) uptake within the primary and most interestingly also in the metastatic lesions which subsequently resulted in a 62% knockdown of firefly luciferase expression in the tumor after a single injection. While this is, to the best of our knowledge, the first paper that correlates quantitative tumor accumulation in an orthotopic tumor model with in vivo gene silencing, these data demonstrate that PEI-g-PCL-b-PEG-Fol conjugates are a promising option for gene knockdown in ovarian cancer.
Keywords: Folate Receptor Alpha, Folate Receptor Targeting, siRNA Delivery, Polymer Micelleplexes, Ovarian Cancer, Ovarian Cancer Xenograft
INTRODUCTION
Ovarian cancer remains one of the deadliest gynecological malignancies. With current treatment options, 65% of all women who are diagnosed will eventually succumb to the disease.[1, 2] A primary reason for the low survival rates is that a majority of patients are diagnosed at an advanced stage. The disease progression of ovarian cancer produces aggressive and widespread metastatic lesions throughout the peritoneal cavity.[3] Primary treatments for these patients often include tumor de-bulking surgery along with combination chemotherapy regimens containing a platinum and taxane.[1, 4] Unfortunately, with widespread metastatic lesions, resistance and reoccurrence of the disease is the usual course.[5–7] Resistance mechanisms often include an increase in anti-apoptotic proteins, increase in drug efflux pumps, or altered drug targets.[8–15] Once patients experience chemoresistance, treatment options become considerably limited.
It is estimated that over 85% of ovarian tumors have an overexpression of folate receptor alpha (FRα).[16, 17] Folate receptor is expressed in four distinct isoforms: alpha, beta, gamma, and delta.[18–20] Both alpha and beta isoforms are cell surface receptors that are glycosylphosphatidylinositol-anchored (GPI) and linked to the membrane.[19] FRα and FRβ have a very select expression profiles. FRα is expressed on the proximal side of the tubules of the kidneys, spleen, certain lug tissues, as well as a variety of cancers.[16, 17, 21–23] Additionally, FRβ is mainly expressed on activated macrophages.[24] Both receptors express high affinities for folic acid (Kd = 1–10 nM). Therefore, folate receptors have been exploited by researchers to deliver a targeted payload to specific cells of interest by linking a targeting moiety of folic acid to either a drug itself or a delivery vehicle. This approach has been implemented in treating a variety of cancers and autoimmune diseases.[24–29] Accordingly, nanomedicine researchers have utilized this approach to selectively deliver a nanoformulated payload to target cells, while decreasing off-target toxicity caused by uptake of the payload into healthy cells.
One type of payload that researchers have been utilizing in a targeted delivery approach is small interfering RNA (siRNA). siRNA has shown promising potential in treating diseases, such as cancer, by silencing genes that give rise to a diseased phenotypes.[30, 31] Naked siRNA is negatively charged, hydrophilic, and easily degraded in vivo by nucleases.[32] Due to the properties of naked siRNA, it cannot be delivered effectively to diseased cells without a carrier to protect it and enhance its delivery. Nanoparticle delivery of siRNA has the ability to encapsulate and protect the payloads from degradation or early release, to modify the payloads bioavailability, increase circulation profiles, and modify tissue distribution profiles. However, according to a recent review by Wilhelm et al. who analyzed the tumor delivery of nanoparticles described in 232 reports between 2005 and 2015, on average, only 0.6% of the injected dose (%ID) of polymeric nanoparticles reach the tumor after systemic administration.[33] While, on the other hand, many publications describe successful gene silencing in tumor models after systemic administration of siRNA loaded nanoparticles, to the best of our knowledge, a quantitative correlation between %ID siRNA in the tumor and gene silencing effects has not yet been described. Here, we added a folic acid targeting ligand to the surface of the nanocarriers to take advantage of the FRα overexpression within the xenograft mouse model and to improve tissue distribution toward the tumor. To deliver the siRNA, triblock copolymers were utilized containing polyethyleneimine (PEI) to electrostatically condense and protect the siRNA. PEI has been documented to be an efficient carrier and transfection reagent. PEI homologues with larger molecular weight and higher degree of branching have been described to not only increase the transfection rates, but also to exhibit stronger toxic effects toward the cells.[34–36] Therefore, the polymers here were modified with a polyethylene glycol (PEG) chain to increase biocompatibility and circulation profiles, increase the stealth-like character of the nanocarriers to avoid macrophage detection, and to decrease the interaction with serum proteins.[27, 37] Lastly, a polycaprolactone (PCL) block was added in the middle as a linker between PEI and PEG to increase the hydrophobic content of the polymer, drive micelle formation, and to aid in cleaving the polymer chains and releasing the siRNA once inside the cell due to its susceptibility to hydrolytic degradation.[30, 38] Previous work performed with PEI-PCL-PEG, or short PPP, polymers has shown their ability to deliver siRNA in vitro to FRα overexpressing SKOV-3 cells, achieve a sustained protein knockdown, and display long-circulation profiles in vivo.[5, 30, 34, 39–41] Here, we used modified architectures of the polymer and their block ratios in order to determine the efficacy of FRα targeted and non-targeted formulations in vivo and ultimately, for the first time, correlated quantitative tumor uptake results with gene silencing in a SKOV-3/LUC FRα overexpressing orthotopic murine ovarian cancer model.
MATERIALS AND METHODS
Materials
Hetero-bifunctional poly(ethylene glycol) (3.5 kDa), as well as methyl terminated monofunctional poly(ethylene glycol) (5 kDa) was purchased from JenKem Technologies (Plano, TX, USA) and chemically modified based on previously published protocols.[5] Hyperbranched polyethylenimine (hy-PEI, 25 kDa) was purchased from BASF (Ludwigshafen, Germany). Dicer substrate double-stranded siRNA (DsiRNA) targeting the Enhanced Green Fluorescent Protein gene (EGFP siRNA, 25/27), Firefly Luciferase (luc), and a Negative Control (scr), as well as Alexa Fluor-488 labeled siRNA were purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA).[42] Folic acid depleted Dulbecco’s Modified Eagle’s Medium (10×) for cell culture, phosphate buffered saline (PBS), heat-inactivated fetal bovine serum (FBS), D-(+)-glucose, and sodium bicarbonate was bought from Sigma-Aldrich (St. Louis, MO, USA). The chelator pBn-SCN-Bn-DTPA was purchased from Macrocyclics (Plano, TX, USA) while arsenazo(III) was purchased from Chem-Impex INT’L INC (Wood Dale, IL, USA), and yttrium(III) chloride was obtained from Acros Organics (Geel, Belgium).
Cell Culture
The SKOV-3 human ovarian cancer cell line was obtained from ATTC (LG Promochem, Wesel, Germany). The SKOV-3/LUC cell line was engineered by stably transfecting the parental SKOV-3 cell line to stably express the reporter gene luciferase as previously reported.[43] SKOV-3 and SKOV-3/LUC ovarian cancer cells were cultured in folate free DMEM cell culture medium (Sigma-Aldrich) supplemented with 0.584 gm/L of L-glutamine, 3.7 gm/L sodium bicarbonate, 10% fetal bovine serum (Thermo Scientific Hyclone), and 1% penicillin/streptomycin at 37 °C and 5% CO2. Cells were grown in 75 and 175 cm2 cell culture flasks (Thermo Scientific) and passaged every 2–3 days when they had reached confluency.
Preparation of PEI-g-PCL-b-PEG-Fol Micelleplexes
Each polymer was dissolved in water to yield a 1 mg/mL concentration based on the 25 kDa PEI content. Concentrations were tested with a copper assay as described before.[5] Once dissolved, samples were filtered through a 0.22 µm filter for sterilization. Subsequently, micelleplexes were prepared for both in vitro and in vivo work by mixing polymer and siRNA solutions together at N/P ratio 5 based on a previously published protocol.[5]
In-111 siRNA Radiolabeling and Purification
To investigate in vitro cellular uptake and in vivo pharmacokinetics and biodistribution, indium-111 labeled siRNA was prepared and purified based upon a previously published protocol.[44] Briefly, siRNA modified with an amine functional group on the 5’ end was coupled with the chelator, p-SCN-Bn-DTPA. After purification, it was incubated with 111InCl3 for 30 minutes. Afterwards, the mixture was run though a PD-10 size exclusion column in order to separate free In-111 fractions from siRNA-DTPA-In-111 fractions. In-111 bound to siRNA was verified through gamma scintillation counting and UV absorption detection at 260 nm. If needed, peak fractions were combined for in vivo studies.
Cellular Uptake of Micelleplexes by Gamma Counting
In 24-well plates (Corning Incorporated, Corning, NY) 60,000 SKOV-3 cells were incubated overnight at 37 °C and 5% CO2. In each well, freshly made micelleplexes containing 50 pmol of siRNA-DTPA-In-111 were added. Negative controls consisted of blank/untreated cells, while positive control cells were treated with siRNA containing lipoplexes made with lipofectamine (Life Technologies, Carlsbad, CA, USA) and polyplexes made with unmodified hy-PEI. Cells were transfected for 4 hours in 37 °C and 5% CO2. Cells were washed twice with 1× PBS + 2 mM EDTA, trypsinized and spun down at 350 g for 5 min. After centrifugation, the cells were resuspended in 1× PBS + 2 mM EDTA buffer and analyzed via Packard Tricarb 2910TR liquid scintillation counter (PerkinElmer, Waltham, MA). Experiments were performed in triplicates, and analysis of the data was performed by GraphPad Prism 5.0 software calculating mean values and standard deviation.
Albumin Binding Assay
An albumin binding assay was performed to detect and mimic plasma protein affinity of siRNA and siRNA containing micelleplexes. Procedures utilized here followed a previously published protocol.[45, 46] To assess albumin binding, a concentrated stock of albumin was prepared in PBS at 450 mg/mL containing 0.005% v/v Tween 80. Further dilutions of the stock albumin were made in DMEM medium. Micelleplexes containing In-111-DTPA-siRNA were formed and incubated with 45 mg/mL, 4.5 mg/mL, 0 mg/mL albumin in DMEM media for 1 hour at 37 °C. After incubation, solutions were transferred to 30,000 MWCO spin columns and centrifuged at 735 g for 15 minutes at room temperature. Following centrifugation, the flow through was discarded and the remaining aliquot was analyzed for siRNA content using gamma scintillation counting.
Confocal Scanning Laser Microscopy (CLSM)
SKOV-3 cells were seeded in a Permanox 8 chamber slide (Nunc, Fisher Scientific, Waltham, MA, USA) at a density of 25,000 cells in 300 µL and incubated overnight in 37 °C and 5% CO2. Micelleplexes were made as described above using 40 pmol of AF-488 labeled siRNA. After incubating the cells with the micelleplexes for 4, hours, the supernatant was decanted, and cells washed with 300 µL of PBS for 2–3 minutes. Afterwards, cells were fixed with 4% paraformaldehyde solution in PBS for 20 minutes at room temperature. Cells were then washed twice with 300 µL of PBS for 2–3 minutes followed by DAPI nuclear staining at a concentration of 175 ng/mL (Life Technologies, Carlsbad, CA, USA) for 20 minutes at room temperature while shaking. Cells were then washed twice with 300 µL of PBS. The chambers were removed, the slides were blotted to remove any excess wash solution, and Fluorsave (CalBiochem, San Diego, CA, USA) was added to the slide and coverslips mounted over the samples. The samples were dried for at least 1–2 hours in the dark to let the coverslip adhere to the chamber slide. For AF488, an excitation wavelength of 490 nm was used while emission was detected with a spectral detector at 525 nm. DAPI staining was excited with a UV laser that had an excitation wavelength of 364 nm, and emission was detected at 385 nm. Images were recorded using a Zeiss LSM 780 confocal microscope and overlaid with brightfield light to gain information about cellular structures.
In Vivo Pharmacokinetics, Biodistribution, and SPECT Imaging
All animal experiments were approved by the Wayne State University Institutional Animal Care and Use Committee (IACUC). For in vivo experiments, 6-week-old female nude mice were purchased from Charles River Laboratories and injected with 6 million SKOV-3/LUC cells intraperitoneally based on previously established protocols [47–49]. Mice were monitored, and tumor growth was observed with bioluminescence imaging using a Bruker Carestream In-Vivo Extreme (Billerica, MA, USA) for 6 weeks after tumor inoculation. At four weeks post injection, the mice were placed on a folic acid-deficient diet (Envigo RMS, Indianapolis, USA) in order to reduce their serum folate to a level near that of human serum and to increase the folate receptor alpha status of the cancer cells.[46] On the day of experiments, mice were injected intraperitoneally (I.P.) or intravenously (I.V.) with targeted or non-targeted micelleplexes containing 35 µg of siRNA-DTPA-In-111. After dosing, 25 µL of blood were drawn retro-orbitally from the mice’s right eyes at 1, 3, 5, 15, 30, 60, and 120 min post injection and analyzed by gamma counting for the presence of In111-DTPA-siRNA. SPECT/CT scans were taken on a Siemens Inveon SPECT/CT (Siemens, Erlangen, Germany) at 4 hours and 24 hours post injection. After the second scan, mice were sacrificed for organ harvesting to assess biodistribution of the siRNA. Once sacrificed, the liver, kidneys, lungs, brain, spleen, bowels, and tumors were dissected, weighed, and analyzed via gamma counting for the deposition of In111-DTPA-siRNA. Results in counts per minute (CPM) of each tissue and blood sample were compared to a freshly made standard curve of In-111 and normalized to the injected dose. All biodistribution results are given as percent of the injected dose per gram of organ weight (%ID/g) while pharmacokinetic studies are represented by percent injected dose per mL of blood (%ID/mL).
Bioluminescence Imaging (BLI)
Tumor growth and luciferase knockdown of animals were monitored on a Bruker Carestream In-Vivo Xtreme (Billerica, MA, USA). Tumor growth was monitored every 2 weeks until the 6-week time point. Animals designated to BLI studies were injected I.P. with 100 µL of a freshly prepared 15 mg/mL D-Luciferin (System Bioscience, San Francisco, USA) stock solution in PBS per 10 g of their body weight. After ten minutes, mice were treated with 3% isoflurane until sufficiently sedated. Maintenance isoflurane was used during imaging to keep the mice sedated. BLI images were taken with a three-minute exposure under high sensitivity and aperture of the lens set at an f-stop of 1.1. Simultaneously, X-ray images were taken under standard 1.2 second exposure. Images were transferred to ImageJ and regions of interest (ROIs) were drawn around the tumor and metastases to determine luciferase expression. Values were normalized to day 0 luciferase expression and analysis was performed by GraphPad Prism 5.0 software.
RESULTS AND DISCUSSION
Cellular uptake
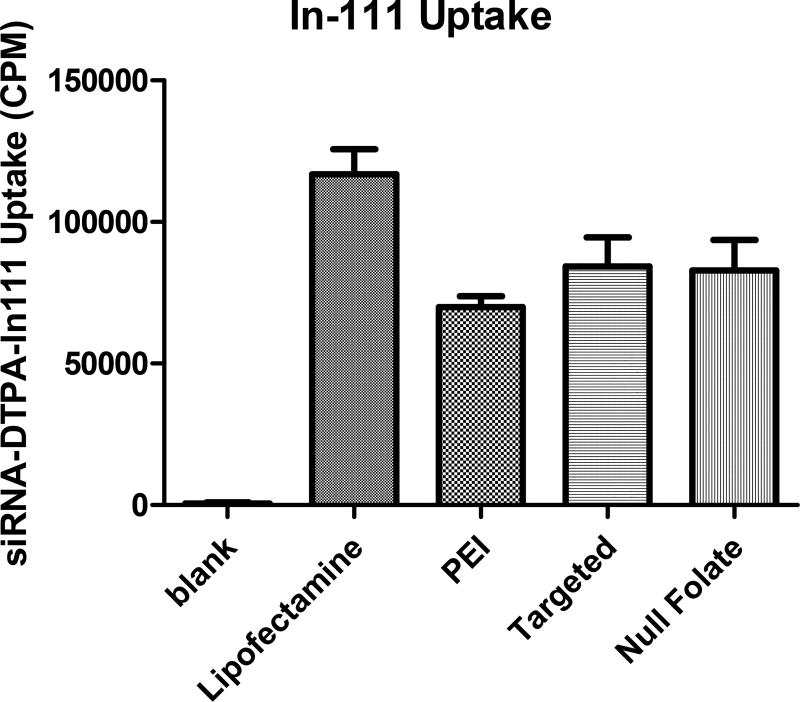
Previous in vitro studies performed with the tri-block copolymers used here consisting of polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) (PEI-g-PCL-b-PEG-FOL) with folic acid have demonstrated efficient siRNA delivery via folate receptor alpha (FRα) targeting and protein knockdown after extensive characterization of their physicochemical parameters.[5] Here, to the best of our knowledge, for the first time, we correlated the in vivo gene silencing performance of these micelleplexes by bioluminescence (BLI) with quantitative tumor uptake data using single-photon emission computed tomography (SPECT) imaging. In order to utilize SPECT imaging capabilities and to monitor siRNA tumor deposition and biodistribution, the siRNA needed to be labeled with Indium-111 (In-111).[44] To determine whether chelation of In-111 affected siRNA uptake profiles of the targeted and non-targeted micelleplexes, gamma scintillation experiments were performed and compared with CLSM results of fluorescently labeled siRNA as shown in Figures 1 and 2. After 4 hours post transfection, uptake profiles of both targeted and non-targeted micelleplexes in folate receptor overexpressing SKOV-3 cells were analyzed in comparison to lipofectamine and unmodified PEI. Of all previously screened polymers, a formulation was chosen that had similar efficacy as lipofectamine and was significantly more efficient than the delivery of free siRNA.[5] As shown in Figure 1, both micelleplexes delivered indium labeled siRNA just as effectively as the unmodified PEI but not as efficiently as lipofectamine; which was expected. As described recently, the targeting benefit is usually not observed at incubation time points as early as 4 h but results in significantly increased intracellular uptake at 24 h and later.[42] In comparison to all polymeric nanocarriers, lipofectamine demonstrated greater siRNA delivery. However, protein knockdown as a result of siRNA delivery with lipofectamine in comparison to targeted micelleplexes was previously shown to be similarly successful.[5] Additionally, the tri-block copolymers were previously shown to be better biocompatible than lipofectamine.[5] Therefore, although the uptake profile of lipofectamine seemingly demonstrates a greater payload delivery efficiency, the commercially available transfection reagent is not suitable for in vivo applications.
Figure 1. siRNA uptake studies using gamma scintillation counting.
Uptake study over 4 hours with Indium-111 labeled siRNA for 4 hrs. Samples were run in triplicates and error bars represent the standard deviation.
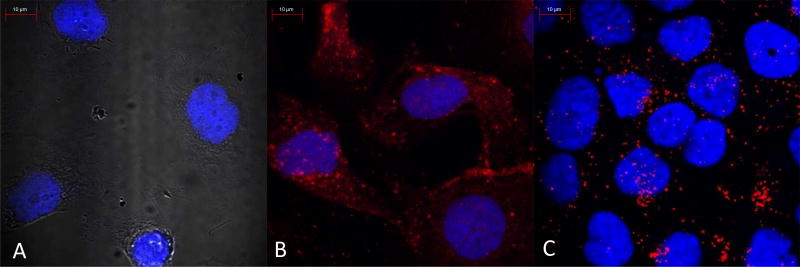
Figure 2. siRNA uptake studies using confocal laser scanning microscopy.
Confocal images were taken of untreated cells (A), targeted micelleplexes (B), and null folate targeted conjugates (C). Scale bare indicates 10 µm.
Subcellular Distribution
Additionally, uptake experiments by scintillation counting or flow cytometry do not allow for any insights in regards to subcellular distribution of a delivered isotope or fluorophore. Therefore, to visualize siRNA delivery and internalization within the cell, CLSM images were taken of fluorescent siRNA delivered by different formulations as shown in Figure 2. Figure 2B clearly demonstrates that the targeted micelleplexes are more evenly distributed within cells which overexpress FRα. Additionally, the FR targeted micelleplexes seemingly coat the outside of the cell, utilizing FR mediated endocytosis which leads to greater siRNA accumulation intracellularly over time. Conversely, null folate micelleplexes (Figure 2C) do not undergo receptor mediated endocytosis but are taken up by adsorptive endocytosis. Therefore, micelleplexes encapsulating fluorescent siRNA accumulate in dot-like structures which are assumed to be endo-lysosomes. The targeted tri-block copolymers with folic acid were designed to encapsulate siRNA into micelleplexes in order to hijack the cells’ normal receptor mediated endocytosis mechanisms, to escape the endosome, and to release siRNA into the cytoplasm.[5] Therefore, FRα targeted micelleplexes achieve a cytoplasmic targeting advantage in delivering siRNA over their non-targeted counterparts, which can be visually depicted in Figures 2 B and C. It should be noted that these confocal images are not quantitative. Therefore, the total amount of siRNA shown in Figures 2 B and C may not be significantly different, as measured by flow cytometry. However, due to the different mechanisms of uptake between targeted and non-targeted micelleplexes the subcellular distribution is different, which is in line with previously reported results.[5, 42, 50] The FRα targeted micelleplexes deliver siRNA more efficiently into the cytoplasm, whereas the non-targeted micelleplexes are entrapped within the endosomes.
Pharmacokinetics
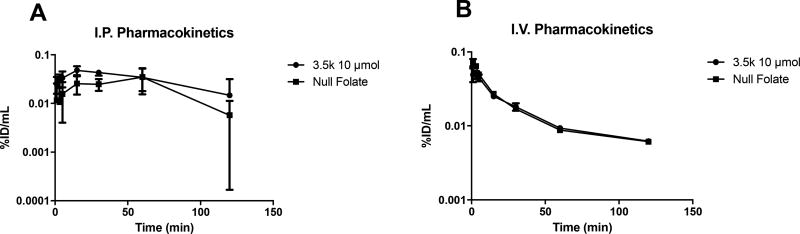
Based on the conducive stability and serum binding profiles, in vivo pharmacokinetics (PK) of both targeted and non-targeted micelleplexes formed with In-111 labeled siRNA were analyzed for I.V. and I.P. administration routes, as shown in Figure 3. Previous studies performed with free, non-formulated In-111 labeled siRNA showed an elimination half-life from the blood of approximately 15 minutes.[45] Here, PK values of I.V. injected micelleplexes were fitted according to a two-compartment model. The alpha phase half-lives were observed to be 13.2 min for the targeted and 4.95 min for the non-targeted formulations, reflecting that the distribution between compartments was very fast in case of non-targeted micelleplexes. Targeted ones, on the other hand, remained in the central compartment almost 3 times longer. Also, the beta elimination phase half-lives were clearly different with 117.8 min for the targeted and 22.31 min for the non-targeted formulation, explaining the overall lower bioavailability of the non-targeted micelleplexes. The bioavailability of the micelleplexes was analyzed through statistical determination of the area under the curve (AUC). Overall, both the targeted and non-targeted micelleplexes displayed much shorter circulation profiles than similar micelleplexes reported in previous publications.[30, 41, 45] The observation that the polymers used here with a (PCL-b-PEG) graft density of 1 form micelles that have shorter circulation half-lives than micelles formed with polymers carrying 5 (PCL-b-PEG) chains per PEI [30] is also in line with previous reports describing polymers with shorter PCL chain.[41] According to the circulation half-lives, targeted micelleplexes displayed a slightly better bioavailability with an AUC of 103.7 %ID min/mL versus 87.90 %ID min/mL for their non-targeted counterpart but a much lower AUC than micelleplexes made with PEI-g-(PCL-b-PEG)5 which had an AUC of 675 %ID min/mL.[30]
Figure 3. In vivo pharmacokinetic analysis of nude mice.
Pharmacokinetic analysis of Indium-111 injected nude mice both intraperitoneally (A) and intravenously (B). Blood samples were collected retro-orbitally at 1, 3, 5, 15, 30, 60, and 120 minutes post injection. Blood samples were analyzed via gamma scintillation counting. Error bars represent standard deviation of the mean.
For I.P. injected micelleplexes, the corresponding serum AUCs for the targeted and non-targeted micelleplexes were 125.2 %ID min/mL and 157.4 %ID min/mL, respectively. The divergence between the targeted and non-targeted formulation demonstrates the different absorption profiles as reflected in their contrasting tmax values. While tmax for the non-targeted micelleplexes was reached at 60 min, the absorption of the targeted micelleplexes from the peritoneum into the circulation was less quantitative and reached a maximum at tmax of 15 min already. Afterwards, the targeted micelleplexes were slowly distributed and excreted, while absorption of the non-targeted micelleplexes lasted until 60 min post injection. Thus, the non-targeted formulation reached a greater systemic bioavailability when injected I.P., and the AUC for I.P injected micelleplexes was about 1.25–1.5 times greater than after I.V injection.
The decreased bioavailability of both micelleplexes, when compared to previously published data using similar polymers, reflects that the nanoparticles are cleared from circulation relatively quickly. Since both clearance and biodistribution decrease the systemic drug concentration, one possible reason is simple excretion. Renal clearance causes short circulation half-lives of free unmodified nucleic acids which are likely cleaved by nucleases, adsorbed to proteins within the bloodstream, or excreted by the kidneys. On the other hand, micelleplexes have the capabilities to protect and retain siRNA but can extravasate out of the blood stream and into target or non-target tissues, which was investigated in the biodistribution experiments. The short circulation half-lives of both the targeted and non-targeted micelleplexes are therefore most likely due to fast extravasation out of circulation and into a deep compartment. Additionally, gamma scintillation-based PK analytics do not differentiate between free drug and albumin bound drug. Therefore, it is possible that the comparably low albumin binding of the micelleplexes (Supplementary Material, Figure 1) is disadvantageous for their circulation profiles.
The half-lives of both micelleplex formulations at each administration route were analyzed with a two-compartment PK model based on curve fitting. Since the micelleplexes show rapid accumulation in the liver, a two-compartment model seems appropriate with the deep compartment reflecting the accumulation in liver, spleen, and at early times points in the tumor.
Biodistribution
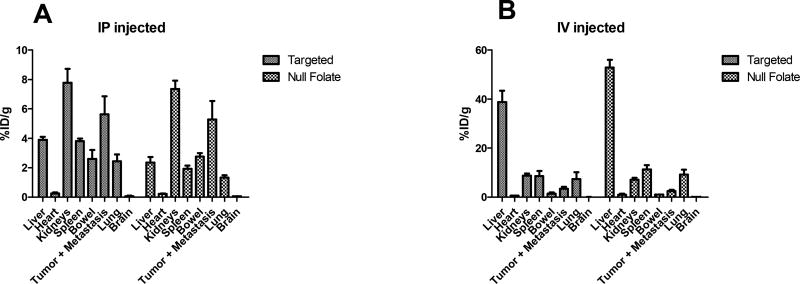
Mice were sacrificed 24 hours after I.V or I.P injection of targeted or non-targeted micelleplexes, and their organs were harvested to determine the amount of siRNA taken up into the tumor, metastases, and other main organs. The biodistribution results of both administration routes can be found in Figure 4. When micelleplexes were injected I.V., the first pass metabolism caused accumulation of the majority of the normalized injected dose per gram in the liver (38% for targeted and 53% for non-targeted), which was comparably higher than in case of micelleplexes made of PEI-g-(PCL-b-PEG)5.[30] However, it is not surprising that micelles with 5 times more PEG on the surface experience a less pronounced first-pass effect. There was a slight improvement in the tumor uptake for the targeted versus non-targeted micelleplexes (3.4% and 2.4%, respectively). However, the strong accumulation in the liver explains the rapid clearance of the micelleplexes from the circulation as described above. Unfortunately, this rapid deposition in a deep compartment interferes with circulation and deposition in the target tissue. In comparison, studies performed by Liu et al. with micelleplexes made of PEI-g-(PCL-b-PEG)5 demonstrated 17% ID/g tumor uptake with similar tri-block copolymers.[30] However, the micelleplexes used by Liu et al. showed approximately 6-fold higher systemic bioavailability and considerably slower deposition with approximately 8-fold less accumulation in the liver which allowed for slow but highly efficient tumor targeting. While the article by Wilhelm et al. pointed out that hydrodynamic diameter has a significant impact on tumor accumulation, while zeta potential does not,[33] the formulations tested here (120 nm and 4 mV [5]) were not considerably different from the ones used by Liu et al (100 nm and 7 mV [30]). However, the previously reported results were obtained in a subcutaneous ovarian cancer model. Here, we demonstrate tumor uptake in a more clinically relevant orthotopic ovarian cancer model. This model more accurately represents clinical disease and is more predicative of drug efficacy but unfortunately, in combination with the shorter circulation times of the formulations used here, did not reflect the same targeting efficacy. According to the review by Wilhelm et al. who assessed 38 reports using orthotopic xenografts, tumor accumulation in such less artificial models was found to vary between 0.0001% and 10% ID with a median at 1.1 %ID. [33] Similarly to the findings here, studies performed with FRα targeted gold nanospheres or PEG coated gadolinium achieved 5.26% siRNA and 5% nanoparticle uptake in the tumor, respectively.[54, 55] Considering that the review by Wilhelm at al. showed that inorganic materials achieve a slightly higher median in accumulation (0.8% ID) than organic nanoparticles (0.6%ID),[33] the tumor targeting results of the organic micelles shown here may in fact suffice for in vivo gene knockdown. After I.P. injection, the two strongest signals were found within the kidneys (7.78 and 7.36 %ID/g, respectively) and the tumor (including all metastatic sites) (5.63 and 5.28 %ID/g, respectively) for both the targeted and non-targeted micelleplexes. These results demonstrate that with the used formulations, the targeting effect was minimal when the siRNA loaded micelleplexes were injected I.P.
Figure 4. In vivo biodistribution analysis of nude mice.
Biodistribution analysis of Indium-111 injected nude mice both intraperitoneally (A) and intravenously (B). Animals were sacrificed, and organs harvested 24 hours post injection and read under gamma scintillation counting.
While the liver accumulated the large parts of the micelleplexes after I.V. injection, in case of the I.P. administration route, we found considerable siRNA uptake in the kidneys for both formulations. While FRα is expressed within the proximal tubules of the kidneys, it is unavailable for access via the bloodstream. Therefore, these data suggest that the uptake within the kidneys is likely due to siRNA renal accumulation rather than active FRα targeting.[22, 56, 57] Taken together, our findings demonstrate that tumor accumulation of the tri-block micelleplexes described here occurs mainly through passive targeting, potentially the EPR effect, rather than active tumor targeting. The potential role of the EPR effect was only observed after I.P. administration, however. Contrarily, these orthotopic tumors were not efficiently reached after I.V. injections due to the short circulation times of the nanoparticles. This observation reinforces the idea that the EPR effect may not play an important role in nanoparticle delivery in a clinical setting.[58–60] Compared to the results reported by Liu et al., this FRα delivery system has a greater amount of off-target uptake, especially in the liver. This could either be a result of the FRα micelleplexes described here bearing a lower amount of PEG chains on their surface, as discussed above, or it could be caused by their greater concentration of folic acid, resulting in a higher valency.[5, 30, 42] As described in the past, particles with higher valency tend to display a greater degree of off-target binding in vivo.[61] Negligible uptake was seen in the brain and heart for both micelleplex formulations independent of the administration route.
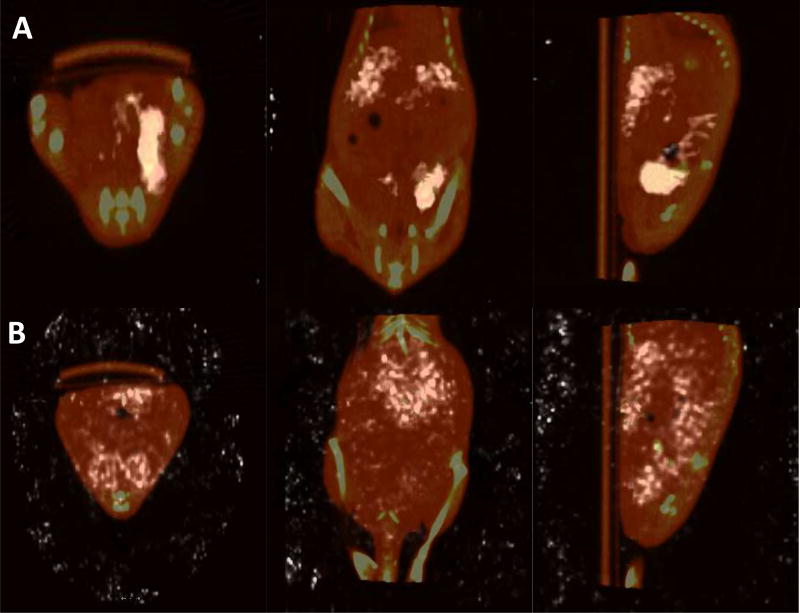
SPECT/CT images of the targeted micelleplexes were taken 4 hours and 24 hours post I.P injection. Those images are shown in Figure 5 A and B. At 4 hours, a surprising amount of localization within the primary tumor was observed which could be due to an initial effect of receptor-ligand interaction between the micelleplexes and the tumor cells. Activity within the kidneys is also visible, consistent with the biodistribution results upon necropsy. Many metastatic lesions in ovarian cancer patients occur within the peritoneum.[3] Likewise, many of the smaller metastatic tumors in the mice developed around the liver and on the caudal side of the diaphragm. As shown in the sagittal plane image in Figure 5 A, siRNA uptake was achieved within these metastatic lesions. Scans taken at the 24-hour time point did not show significant retention of the siRNA within the primary tumor at this later time point. Instead, the coronal plane image shows a strong signal in the liver which in this plane covers the kidneys. In the saggital plane image, siRNA uptake in the kidneys, and possibly in the metastatic lesions within the peritoneum located adjacent to the liver and diaphragm, can be found. This result demonstrates that early targeting effects of the micelleplexes were washed out after 24 hours, leading to excretion via the kidneys. It also emphasizes the lack of impact of the EPR effect on short circulating nanoparticles which was no longer observed at the 24 h time point.[58–60] Using an orthotopic model which is closer to a clinical representation of the disease state, we were able to better understand the fate of the micelleplexes. Overall, the SPECT/CT images display nicely that siRNA is taken up by the primary and secondary tumors initially, and that potentially, if sacrificed at an earlier time point, there may be an enhanced tumor targeting effect of siRNA localized within the tumor that was washed out at 24 h post injection. Considering that the circulation time of the micelleplexes used here was much shorter than described by Liu et al.,[30] an earlier tmax for tumor deposition is not unlikely.
Figure 5. In vivo SPECT/CT images.
Biodistribution analysis of Indium-111 injected nude mice intraperitoneally at 4 hours post injection (A) and 24 hours post injection (B). From left to right: Transversal, coronal, saggital planes.
In vivo Gene Knockdown
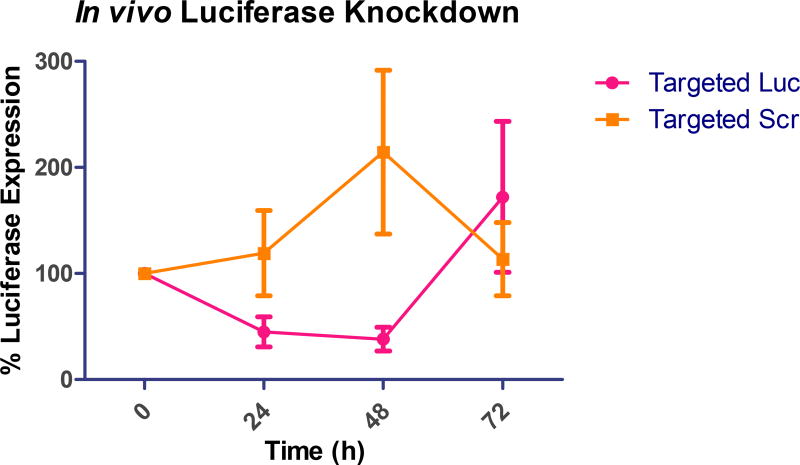
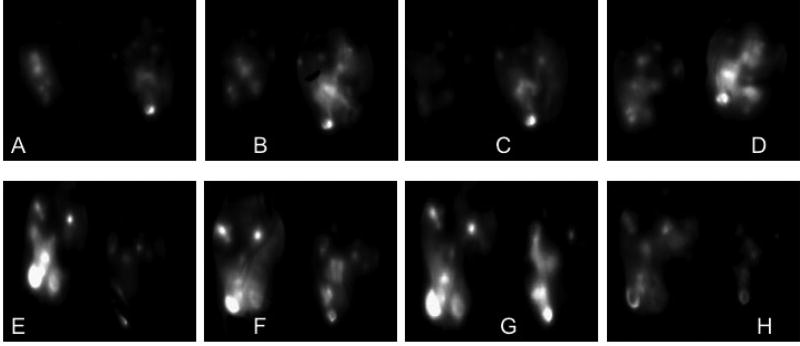
After pharmacokinetic, biodistribution, and tumor accumulation analysis, it was imperative to assess the efficacy of the siRNA that localized within the tumor for protein knockdown to correlate tumor deposition and efficacy. As the animal model was based on injection of a luciferase expressing cell line, SKOV-3/LUC, it offered the ability of measuring firefly luciferase expression in vivo. Therefore, we were able to monitor tumor growth throughout the duration of the study, as well as luciferase knockdown by firefly luciferase directed siRNA. In this animal model, as well as clinically, FRα is significantly overexpressed in ovarian tumors.[19, 21, 22] For the knockdown experiments, we utilized firefly luciferase directed and scrambled siRNA to determine RNAi effects. As shown in Figure 6, we analyzed luciferase knockdown over 72 hours after a single injection of 35 µg of siRNA. After 48 hours, the micelleplexes containing luciferase directed siRNA showed a mean 62% luciferase knockdown. Conversely, the scrambled siRNA did not show any knockdown, but rather a 107% increase in luciferase signal was measured, which was related to tumor growth. After 72 hours, we saw an increase in the initially knocked down gene expression, signaling the end of the transient knockdown. This observation was consistent with our previous data for protein knockdown in vitro displaying the most efficient protein knockdown at 48 hours post transfection.[5] Interestingly, the mice treated with scrambled siRNA displayed a decrease in the overall luciferase signal after 48 hours compared to 24 hours. This observation could be due to necrotic tissue within the already advanced tumors. Advanced ovarian cancers are known to grow rapidly with a very aggressive disease progression.[3] This in vivo trend was promising due to the greater than 60% knockdown of luciferase expression. In comparison, Bartlett et al., Gutbier et al., and Klein et al. all achieved approximately a 50% in vivo knockdown with siRNA containing nanoparticles.[36, 62, 63] The knockdown is visualized in Figures 7 A–F which are representative images of the bioluminescence recordings at time points 0, 48, and 72 hours post injection. Animals treated with luciferase directed siRNA are shown in Figures 7A–C, while animals treated with scrambled siRNA treatment can be found in Figures 7D–F. It is obvious that luciferase protein knockdown occurs after 24 and 48 hours, as shown in Figures 7A and B, but then luciferase expression increases at the 72-hour time point (7C). Similarly, the increase in signal after the treatment with scrambled siRNA between Figures 7D and E can be visually appreciated. Overall, the in vivo bioluminescence imaging data analysis shows a strong trend which indicates that folate decorated micelleplexes can efficiently knock down luciferase expression by more than 60% in an orthotopic xenograft mouse model of ovarian cancer. Considering the short tumor retention of the siRNA displayed in Figure 5, another injection 24 or 48 h after the first one would have been beneficial. Unfortunately, we were not able to follow animals beyond 48 h due to tumor growth and the condition of the animals which required sacrifice.
Figure 6. In Vivo Bioluminescence luciferase knockdown.
Luciferase knockdown in vivo after injection of luciferase (Luc) siRNA or scrambled (Scr) siRNA containing FRα targeted micelleplexes. Mean change in expression is indicated and standard deviation, n=5. Luciferase activity is shown predominantly in the primary tumor as well as in metastases.
Figure 7. In vivo Bioluminescence luciferase imaging.
Bioluminescence images of luciferase knockdown in vivo after injection of luciferase siRNA containing FRα targeted micelleplexes (A–D) and scramble siRNA containing FRα targeted micelleplexes (E–H). Time points displayed here are 0 hours (A, E), 24 hours (B, F), 48 hours (C, G), and 72 hours (D, H). Luciferase activity is shown predominantly in the primary tumor as well as in metastases.
SUMMARY AND CONCLUSION
The in vivo efficacy of FRα targeted and non-targeted tri-block copolymer-based siRNA micelleplexes was assessed in a SKOV-3 murine orthotopic xenograft ovarian cancer model. In vitro characterization results in Figures 1 and 2 demonstrated that the micelleplexes described here can efficiently deliver indium labeled siRNA to folate receptor overexpressing cells. Stability testing of both micelleplex formulations using low and high concentration levels of serum displayed acceptable siRNA retention within the micelleplexes. In vivo pharmacokinetic analysis of targeted and non-targeted micelleplexes was assessed after both I.V and I.P administration. Overall, the systemic bioavailability of I.P injected micelleplexes was about 1.25–1.5 times greater than that of I.V injected micelleplexes. Interestingly, the non-targeted formulation had a greater AUC than the targeted formulations in I.P. injected mice, but showed smaller AUCs when injected I.V. In addition to the pharmacokinetic analysis, general biodistribution, tumor uptake, and luciferase protein knockdown were assessed. When injected I.P., the two strongest signals of biodistribution occurred within the kidneys (7.78 %ID/g for targeted and 7.36 %ID/g for non-targeted micelleplexes) and tumors (5.63 %ID/g targeted and 5.28 %ID/g non-targeted micelleplexes). Conversely, when injected I.V., a noticeable difference between both formulations was observed regarding uptake in the liver. Targeted and non-targeted micelleplexes accumulated with 39 and 53 %ID/g, respectively. Tumor uptake was also affected dropping to 3.4 and 2.4 %ID/g for targeted and non-targeted micelleplexes, respectively. However, strong uptake of the targeted micelleplexes was observed 4 h after injection by SPECT/CT imaging in the primary tumor and the metastases and decreased over time due to a wash-out of the micelleplexes as seen in the images. At 24 h post injection, only a diffuse uptake and targeting was observed. Therefore, a greater overall uptake and targeting benefit would likely have been observed also by gamma scintillation counting at earlier time points.
As the most successful tumor deposition was achieved after I.P injection of targeted micelleplexes, this route was chosen to assess pharmacologic effects of siRNA delivery, measured as luciferase knockdown via BLI. After a single injection of 35 µg of siRNA formulated in targeted micelleplexes, an impressive 62% knockdown of luciferase was measured 48 hours after injection. However, after 72 hours, the transient knockdown ended and a sharp increase in luciferase activity was noted. Tumors treated with targeted micelleplexes containing a scrambled siRNA control duplex displayed a steady increase in luciferase expression after injection. While this is, to the best of our knowledge, the first report correlating quantitative siRNA uptake in an orthotopic tumor model with gene knockdown effects. Considering that over the last 10 years, polymeric nanoparticles only showed a median accumulation of 0.6%ID in tumor models, the accumulation of 2–6%ID resulting in over 60% gene knockdown needs to be seen in the relevant context.
Overall, these tri-block copolymers displayed effective siRNA delivery profiles in vitro and suitable siRNA retention in the presence of high serum concentrations. In vivo, these siRNA containing micelleplexes achieved 5–6% tumor uptake in a murine orthotopic SKOV-3 ovarian cancer xenograft model when injected I.P which yielded a 62% luciferase knockdown. Therefore, this platform of amphiphilic tri-block copolymers provides a promising option for in vivo siRNA delivery and gene knockdown in ovarian cancers.
Supplementary Material
Acknowledgments
FUNDING SOURCES
This work was supported by the Wayne State Start-Up and NanoIncubator grants to Olivia Merkel. Additionally, the Ruth L. Kirschstein National Research Award T32-CA009531 fellowship, 2015 SNMMI Bradley-Alavi Student Fellowship, and the Predoctoral Fellowship in Cancer Research from the DeRoy Testamentary Foundation to Steven Jones and the Department of Defense grant LC140278 to Anthony F Shields are gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AVAILABILITY
The raw data required to reproduce these findings are available to download from https://www.dropbox.com/s/p46utpic5x3roi0/in%20vivo%20animal%20work%20complete%20with%20AUC.pzf?dl=0. The processed data required to reproduce these findings are available to download from https://www.dropbox.com/s/6h5kk14tuncr478/animal%20data%20complete.pptx?dl=0.
References
- 1.Landen C, Birrer M, Sood A. Early events in the pathogenesis of epithelial ovarian cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26 doi: 10.1200/JCO.2006.07.9970. [DOI] [PubMed] [Google Scholar]
- 2.Scully RE. Pathology of ovarian cancer precursors. J Cell Biochem. 1995;23 doi: 10.1002/jcb.240590928. [DOI] [PubMed] [Google Scholar]
- 3.Lengyel E. Ovarian cancer development and metastasis. The American journal of pathology. 2010;177(3):1053–64. doi: 10.2353/ajpath.2010.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neesham D. Ovarian cancer screening. Aust Fam Physician. 2007;36 [PubMed] [Google Scholar]
- 5.Jones SK, Lizzio V, Merkel OM. Folate Receptor Targeted Delivery of siRNA and Paclitaxel to Ovarian Cancer Cells via Folate Conjugated Triblock Copolymer to Overcome TLR4 Driven Chemotherapy Resistance. Biomacromolecules. 2016;17(1):76–87. doi: 10.1021/acs.biomac.5b01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao LJ, Xu H, Qu JW, Zhao WZ, Zhao YB, Wang JH. Modulation of drug resistance in ovarian cancer cells by inhibition of protein kinase C-alpha (PKC-alpha) with small interference RNA (siRNA) agents. Asian Pac J Cancer Prev. 2012;13(8):3631–6. doi: 10.7314/apjcp.2012.13.8.3631. [DOI] [PubMed] [Google Scholar]
- 7.Lorusso PM, Edelman MJ, Bever SL, Forman KM, Pilat M, Quinn MF, Li J, Heath EI, Malburg LM, Klein PJ, Leamon CP, Messmann RA, Sausville EA. Phase I study of folate conjugate EC145 (Vintafolide) in patients with refractory solid tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(32):4011–6. doi: 10.1200/JCO.2011.41.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones SK, Merkel OM. Tackling breast cancer chemoresistance with nano-formulated siRNA. Gene Ther. 2016;23(12):821–828. doi: 10.1038/gt.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard Z, Paul BT, Craft B, ElShamy WM. BRCA1-IRIS inactivation overcomes paclitaxel resistance in triple negative breast cancers. Breast cancer research : BCR. 2015;17:5. doi: 10.1186/s13058-014-0512-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng H, Mai WX, Zhang H, Xue M, Xia T, Lin S, Wang X, Zhao Y, Ji Z, Zink JI, Nel AE. Codelivery of an optimal drug/siRNA combination using mesoporous silica nanoparticles to overcome drug resistance in breast cancer in vitro and in vivo. ACS Nano. 2013;7(2):994–1005. doi: 10.1021/nn3044066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creixell M, Peppas NA. Co-delivery of siRNA and therapeutic agents using nanocarriers to overcome cancer resistance. Nano Today. 2012;7(4):367–379. doi: 10.1016/j.nantod.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Sadee W. Membrane transporters and channels in chemoresistance and -sensitivity of tumor cells. Cancer Lett. 2006;239(2):168–82. doi: 10.1016/j.canlet.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, Anderle P, Bussey KJ, Barbacioru C, Shankavaram U, Dai Z, Reinhold WC, Papp A, Weinstein JN, Sadee W. Membrane transporters and channels: role of the transportome in cancer chemosensitivity and chemoresistance. Cancer Res. 2004;64(12):4294–301. doi: 10.1158/0008-5472.CAN-03-3884. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Bathula SR, Li J, Huang L. Multifunctional Nanoparticles Delivering Small Interfering RNA and Doxorubicin Overcome Drug Resistance in Cancer. J Biol Chem. 2010;285(29):22639–50. doi: 10.1074/jbc.M110.125906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F, Sethi G. Targeting transcription factor NF-kappaB to overcome chemoresistance and radioresistance in cancer therapy. Biochim Biophys Acta. 2010;1805(2):167–80. doi: 10.1016/j.bbcan.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Assaraf YG, Leamon CP, Reddy JA. The folate receptor as a rational therapeutic target for personalized cancer treatment. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2014;17(4–6):89–95. doi: 10.1016/j.drup.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Campbell IG, Jones TA, Foulkes WD, Trowsdale J. Folate-binding protein is a marker for ovarian cancer. Cancer Res. 1991;51(19):5329–38. [PubMed] [Google Scholar]
- 18.Wu M, Fan J, Gunning W, Ratnam M. Clustering of GPI-anchored folate receptor independent of both cross-linking and association with caveolin. The Journal of membrane biology. 1997;159(2):137–47. doi: 10.1007/s002329900277. [DOI] [PubMed] [Google Scholar]
- 19.Ross JF, Chaudhuri PK, Ratnam M. Differential regulation of folate receptor isoforms in normal and malignant tissues in vivo and in established cell lines. Physiologic and clinical implications. Cancer. 1994;73(9):2432–43. doi: 10.1002/1097-0142(19940501)73:9<2432::aid-cncr2820730929>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 20.Elnakat H, Ratnam M. Distribution, functionality and gene regulation of folate receptor isoforms: implications in targeted therapy. Adv Drug Deliv Rev. 2004;56(8):1067–84. doi: 10.1016/j.addr.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Markert S, Lassmann S, Gabriel B, Klar M, Werner M, Gitsch G, Kratz F, Hasenburg A. Alpha-folate receptor expression in epithelial ovarian carcinoma and non-neoplastic ovarian tissue. Anticancer Res. 2008;28(6A):3567–72. [PubMed] [Google Scholar]
- 22.Parker N, Turk MJ, Westrick E, Lewis JD, Low PS, Leamon CP. Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay. Analytical Biochemistry. 2005;338(2):284–293. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 23.Mohammadi M, Li Y, Abebe DG, Xie Y, Kandil R, Kraus T, Gomez-Lopez N, Fujiwara T, Merkel OM. Folate receptor targeted three-layered micelles and hydrogels for gene delivery to activated macrophages. Journal of Controlled Release. 2016;244(Part B):269–279. doi: 10.1016/j.jconrel.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 24.Mohammadi M, Li Y, Abebe DG, Xie Y, Kandil R, Kraus T, Gomez-Lopez N, Fujiwara T, Merkel OM. Folate receptor targeted three-layered micelles and hydrogels for gene delivery to activated macrophages. J Control Release. 2016 doi: 10.1016/j.jconrel.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Lee RJ, Low PS. Delivery of liposomes into cultured KB cells via folate receptor-mediated endocytosis. J Biol Chem. 1994;269(5):3198–204. [PubMed] [Google Scholar]
- 26.Leamon CP, Low PS. Delivery of macromolecules into living cells: a method that exploits folate receptor endocytosis. Proc Natl Acad Sci U S A. 1991;88(13):5572–6. doi: 10.1073/pnas.88.13.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esmaeili F, Ghahremani MH, Ostad SN, Atyabi F, Seyedabadi M, Malekshahi MR, Amini M, Dinarvand R. Folate-receptor-targeted delivery of docetaxel nanoparticles prepared by PLGA-PEG-folate conjugate. Journal of drug targeting. 2008;16(5):415–23. doi: 10.1080/10611860802088630. [DOI] [PubMed] [Google Scholar]
- 28.Reddy JA, Abburi C, Hofland H, Howard SJ, Vlahov I, Wils P, Leamon CP. Folate-targeted, cationic liposome-mediated gene transfer into disseminated peritoneal tumors. Gene Ther. 2002;9(22):1542–50. doi: 10.1038/sj.gt.3301833. [DOI] [PubMed] [Google Scholar]
- 29.Reddy JA, Dorton R, Bloomfield A, Nelson M, Vetzel M, Guan J, Leamon CP. Rational combination therapy of vintafolide (EC145) with commonly used chemotherapeutic drugs. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(8):2104–14. doi: 10.1158/1078-0432.CCR-13-2423. [DOI] [PubMed] [Google Scholar]
- 30.Liu L, Zheng M, Librizzi D, Renette T, Merkel OM, Kissel T. Efficient and Tumor Targeted siRNA Delivery by Polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol)-folate (PEI-PCL-PEG-Fol) Mol Pharm. 2016;13(1):134–43. doi: 10.1021/acs.molpharmaceut.5b00575. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Lu Z, Wientjes MG, Au JLS. Delivery of siRNA Therapeutics: Barriers and Carriers. The AAPS Journal. 2010;12(4):492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Li X, Huang L. Non-viral nanocarriers for siRNA delivery in breast cancer. J Control Release. 2014;190:440–50. doi: 10.1016/j.jconrel.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW. Analysis of nanoparticle delivery to tumours. 2016;1:16014. [Google Scholar]
- 34.Liu Y, Samsonova O, Sproat B, Merkel O, Kissel T. Biophysical characterization of hyper-branched polyethylenimine-graft-polycaprolactone-block-mono-methoxyl-poly(ethylene glycol) copolymers (hy-PEI-PCL-mPEG) for siRNA delivery. J Control Release. 2011;153(3):262–8. doi: 10.1016/j.jconrel.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 35.Essex S, Navarro G, Sabhachandani P, Chordia A, Trivedi M, Movassaghian S, Torchilin VP. Phospholipid-modified PEI-based nanocarriers for in vivo siRNA therapeutics against multidrug-resistant tumors. Gene Ther. 2015;22(3):257–66. doi: 10.1038/gt.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2004;12(5):461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 37.Merkel OM, Beyerle A, Librizzi D, Pfestroff A, Behr TM, Sproat B, Barth PJ, Kissel T. Nonviral siRNA delivery to the lung: investigation of PEG-PEI polyplexes and their in vivo performance. Mol Pharm. 2009;6(4):1246–60. doi: 10.1021/mp900107v. [DOI] [PubMed] [Google Scholar]
- 38.Trinh P, Atzet S, Curtain S, Ratner BD. Characterization of poly (2-hydroxyethyl methacrylate) scaffolds for tissue engineering. J Undergraduate Res Bioeng. 2008;2010(8):98–104. [Google Scholar]
- 39.Zheng M, Liu Y, Samsonova O, Endres T, Merkel O, Kissel T. Amphiphilic and biodegradable hy-PEI-g-PCL-b-PEG copolymers efficiently mediate transgene expression depending on their graft density. Int J Pharm. 2012;427(1):80–7. doi: 10.1016/j.ijpharm.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Endres T, Zheng M, Kilic A, Turowska A, Beck-Broichsitter M, Renz H, Merkel OM, Kissel T. Amphiphilic biodegradable PEG-PCL-PEI triblock copolymers for FRET-capable in vitro and in vivo delivery of siRNA and quantum dots. Mol Pharm. 2014;11(4):1273–81. doi: 10.1021/mp400744a. [DOI] [PubMed] [Google Scholar]
- 41.Zheng M, Librizzi D, Kilic A, Liu Y, Renz H, Merkel OM, Kissel T. Enhancing in vivo circulation and siRNA delivery with biodegradable polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) copolymers. Biomaterials. 2012;33(27):6551–8. doi: 10.1016/j.biomaterials.2012.05.055. [DOI] [PubMed] [Google Scholar]
- 42.Jones SK, Sarkar A, Feldmann DP, Hoffmann P, Merkel OM. Revisiting the value of competition assays in folate receptor-mediated drug delivery. Biomaterials. 2017;138:35–45. doi: 10.1016/j.biomaterials.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merkel OM, Beyerle A, Beckmann BM, Zheng M, Hartmann RK, Stoger T, Kissel TH. Polymer-related off-target effects in non-viral siRNA delivery. Biomaterials. 2011;32(9):2388–98. doi: 10.1016/j.biomaterials.2010.11.081. [DOI] [PubMed] [Google Scholar]
- 44.Jones S, Merkel O. Indium-Labeling of siRNA for Small Animal SPECT Imaging. Methods Mol Biol. 2016;1372:79–88. doi: 10.1007/978-1-4939-3148-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Buyens K, Sanders NN, De Smedt SC, Behe M, Kissel T. Stability of siRNA polyplexes from poly(ethylenimine) and poly(ethylenimine)-g-poly(ethylene glycol) under in vivo conditions: effects on pharmacokinetics and biodistribution measured by Fluorescence Fluctuation Spectroscopy and Single Photon Emission Computed Tomography (SPECT) imaging. J Control Release. 2009;138(2):148–59. doi: 10.1016/j.jconrel.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 46.Mathias CJ, Wang S, Lee RJ, Waters DJ, Low PS, Green MA. Tumor-selective radiopharmaceutical targeting via receptor-mediated endocytosis of gallium-67-deferoxamine-folate. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 1996;37(6):1003–8. [PubMed] [Google Scholar]
- 47.Vanderhyden BC, Shaw TJ, Ethier J. Animal models of ovarian cancer. Reproductive biology and endocrinology : RB&E. 2003;1:67. doi: 10.1186/1477-7827-1-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fong MY, Kakar SS. Ovarian cancer mouse models: a summary of current models and their limitations. Journal of Ovarian Research. 2009;2(1):12. doi: 10.1186/1757-2215-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw TJ, Senterman MK, Dawson K, Crane CA, Vanderhyden BC. Characterization of intraperitoneal, orthotopic, and metastatic xenograft models of human ovarian cancer. Molecular Therapy. 2004;10(6):1032–42. doi: 10.1016/j.ymthe.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 50.Jones AS Steven K, Feldmann Daniel, Hoffmann Peter, Merkel Olivia. Revisiting the Value of Competition Assays in Folate Receptor-Mediated Drug Delivery. doi: 10.1016/j.biomaterials.2017.05.034. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Influence of polyethylene glycol chain length on the physicochemical and biological properties of poly(ethylene imine)-graft-poly(ethylene glycol) block copolymer/SiRNA polyplexes. Bioconjugate chemistry. 2006;17(5):1209–18. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- 53.Zini R, Barre J, Bree F, Tillement JP, Sebille B. Evidence for a concentration-dependent polymerization of a commercial human serum albumin. J Chromatogr. 1981;216:191–8. doi: 10.1016/s0021-9673(00)82347-x. [DOI] [PubMed] [Google Scholar]
- 54.Lu W, Zhang G, Zhang R, Flores LG, Huang Q, Gelovani JG, Li C. Tumor Site–Specific Silencing of <em>NF-κB p65</em>by Targeted Hollow Gold Nanosphere–Mediated Photothermal Transfection. Cancer Research. 2010;70(8):3177–3188. doi: 10.1158/0008-5472.CAN-09-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oyewumi MO, Yokel RA, Jay M, Coakley T, Mumper RJ. Comparison of cell uptake, biodistribution and tumor retention of folate-coated and PEG-coated gadolinium nanoparticles in tumor-bearing mice. Journal of Controlled Release. 2004;95(3):613–626. doi: 10.1016/j.jconrel.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 56.van de Water FM, Boerman OC, Wouterse AC, Peters JGP, Russel FGM, Masereeuw R. INTRAVENOUSLY ADMINISTERED SHORT INTERFERING RNA ACCUMULATES IN THE KIDNEY AND SELECTIVELY SUPPRESSES GENE FUNCTION IN RENAL PROXIMAL TUBULES. Drug Metabolism and Disposition. 2006;34(8):1393–1397. doi: 10.1124/dmd.106.009555. [DOI] [PubMed] [Google Scholar]
- 57.Dhawan D, Ramos-Vara JA, Naughton JF, Cheng L, Low PS, Rothenbuhler R, Leamon CP, Parker N, Klein PJ, Vlahov IR, Reddy JA, Koch M, Murphy L, Fourez LM, Stewart JC, Knapp DW. Targeting folate receptors to treat invasive urinary bladder cancer. Cancer Res. 2013;73(2):875–84. doi: 10.1158/0008-5472.CAN-12-2101. [DOI] [PubMed] [Google Scholar]
- 58.Hansen AE, Petersen AL, Henriksen JR, Boerresen B, Rasmussen P, Elema DR, Rosenschöld PMa, Kristensen AT, Kjær A, Andresen TL. Positron Emission Tomography Based Elucidation of the Enhanced Permeability and Retention Effect in Dogs with Cancer Using Copper-64 Liposomes. ACS Nano. 2015;9(7):6985–6995. doi: 10.1021/acsnano.5b01324. [DOI] [PubMed] [Google Scholar]
- 59.Greish K. Enhanced permeability and retention (EPR) effect for anticancer nanomedicine drug targeting. Methods in molecular biology (Clifton, N.J.) 2010;624:25–37. doi: 10.1007/978-1-60761-609-2_3. [DOI] [PubMed] [Google Scholar]
- 60.Wang AZ. EPR or no EPR? The billion-dollar question. Science Translational Medicine. 2015;7(294):294ec112–294ec112. [Google Scholar]
- 61.Movahedi K, Schoonooghe S, Laoui D, Houbracken I, Waelput W, Breckpot K, Bouwens L, Lahoutte T, De Baetselier P, Raes G, Devoogdt N, Van Ginderachter JA. Nanobody-based targeting of the macrophage mannose receptor for effective in vivo imaging of tumor-associated macrophages. Cancer Res. 2012;72(16):4165–77. doi: 10.1158/0008-5472.CAN-11-2994. [DOI] [PubMed] [Google Scholar]
- 62.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. 2007;104(39):15549–54. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gutbier B, Kube SM, Reppe K, Santel A, Lange C, Kaufmann J, Suttorp N, Witzenrath M. RNAi-mediated suppression of constitutive pulmonary gene expression by small interfering RNA in mice. Pulmonary Pharmacology & Therapeutics. 2010;23(4):334–344. doi: 10.1016/j.pupt.2010.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.