Abstract
The TGFβ signaling pathway is a critical regulator of cancer progression in part through induction of the epithelial to mesenchymal transition (EMT). This process is aberrantly activated in cancer cells, facilitating invasion of the basement membrane, survival in the circulatory system, and dissemination to distant organs. The mechanisms through which epithelial cells transition to a mesenchymal state involve coordinated transcriptional and post-transcriptional control of gene expression. One such mechanism of control is through the RNA binding protein hnRNP E1, which regulates splicing and translation of a cohort of EMT and stemness-associated transcripts. A growing body of evidence indicates a major role for hnRNP E1 in the control of epithelial cell plasticity, especially in the context of carcinoma progression. Here, we review the multiple mechanisms through which hnRNP E1 functions to control EMT and metastatic progression.
Keywords: TGFβ signaling, hnRNP E1, EMT, metastasis
1. Introduction
The TGFβ signaling pathway has a dual function as both a tumor-suppressor and enhancer, contingent on the stage of tumorigenesis [1, 2]. In normal cells and during early tumor initiation, TGFβ signaling acts to inhibit growth by cell-cycle arrest; in contrast, this signaling pathway functions as a critical driver of progression in advanced cancers. In this setting, TGFβ signaling induces migration and invasion of cancer cells and modulates the tumor microenvironment promoting the process of metastasis, where cells spread from the primary tumor to secondary sites. Malignant progression is facilitated through the induction of the epithelial to mesenchymal transition (EMT), a process aberrantly activated in epithelial cancers which leads to a more motile invasive phenotype associated with metastatic spread [3, 4]. Here, we provide an overview of the regulatory mechanisms that coordinately control the process of EMT, focusing on one important downstream effector of TGFβ-induced EMT, the RNA binding protein hnRNP E1. This factor regulates EMT, cancer cell stemness and metastatic progression through the post-transcriptional regulation of EMT and metastasis-associated genes [5–8]. The multiple functions of hnRNP E1 in RNA processing and translation, as well as specific examples of hnRNP E1 targets will be discussed in this review.
2. Regulation of EMT and stemness by TGF-β signaling
TGFβ ligand acts through the type I and type II TGFβ receptors to activate the canonical Smad2/3 and non-canonical MAPK, RhoA and AKT signaling pathways regulating diverse processes such as proliferation, survival, differentiation and EMT [9, 10]. EMT normally occurs during embryonic development (Type 1 EMT); during fibrosis, wound healing and tissue regeneration (Type 2 EMT), and is thought to be aberrantly activated during cancer metastasis (Type 3 EMT) [11, 12]. The mesenchymal phenotype is characterized by a loss of cell-cell contacts and apical-basal polarity and a reorganization of actin, intermediate filament and tubulin cytoskeleton network [13]. In cancer, this transition is associated with a gain in migratory and invasive ability, enhanced chemo-resistance, and immune-suppression [3, 14]. The induction of EMT is also associated with cancer stemness, where cells exhibit self-renewal properties, which is thought to occur through the reactivation of embryonic signaling [15, 16]. These cancer stem cells (CSCs) may drive not only metastatic growth but also facilitate cancer recurrence and chemo- and radio-resistance [17]. TGFβ initiates EMT and the associated cancer stem cell phenotype through a variety of mechanisms, including changes at the level of transcription as well as post-transcriptional regulation through non-coding RNAs and RNA binding proteins.
2.1 Transcriptional regulation of EMT and stemness
At the transcriptional level, the epithelial to mesenchymal transition is induced through a number of critical transcription factors including the zinc finger transcriptional repressors ZEB1, ZEB2, Snail1 and Slug, the bHLH factors Twist and E47 and the kruppel-like factor KLF8 [4, 18]. These transcription factors directly regulate the expression of epithelial genes such as E-Cadherin, Occludin and ZO1 and mesenchymal genes such as N-Cadherin and Fibronectin. In addition, pluripotency factors, reactivated by TGFβ–induced EMT, include SOX2, Nanog, Oct4, KLF4 and BMI1 that regulate transcription of genes involved in the CSC phenotype [19]. Transcriptional regulation of EMT and stemness properties in a cancer setting have been reviewed extensively in the literature, therefore, we refer the reader to recent reviews on these topics [3, 4, 18, 20].
2.2 Post-transcriptional regulation by non-coding RNAs
Post-transcriptional mechanisms of EMT regulation involve a cohort of EMT-associated non-coding RNAs including long non-coding RNAs (lncRNAs) and microRNAs (miRNAs). Highly conserved miRNAs (~21–25 nucleotides in size), bind to specific sequences called MREs (miRNA Recognition Elements) mainly located at the 3′-untranslated region (3′UTR) of target mRNAs, leading to loss of mRNA stability or translational inhibition. miRNAs can control EMT by inhibiting TGFβ signaling by targeting TGFβ receptor II (mir-204, −302, −372), and by directly repressing epithelial genes such as E-cadherin (miR-9) and the mesenchymal genes Vimentin (miR-138) and N-cadherin (miR-194) [21–24]. In addition to these targets, miRNA-mediated repression of transcription factors is a major mechanism of EMT control. One of the best characterized miRNA:mRNA interaction in EMT involves the miR-200 family (miR-200f), clustered within two loci of miR-200b~miR-200a~miR-429 and miR-200c~miR-141 [25]. These miRNAs regulate ZEB protein expression acting in a double-negative feedback loop to control the mesenchymal transition [26–28]. In this loop, ZEB1 transcriptionally represses the expression of miR-200 family members which target both ZEB1 and ZEB2 transcripts at their 3′UTR; the consequent up-regulation of ZEBs leads to the repression of E-cadherin. The transcription factors Snail1 and Slug are also targeted by several miRNAs including mir-29b, −30a and −124, with double-negative feedback loops described between miR-34/miR-203 and Snail1 and between miR-1/miR-200b and Slug [29–35]. In addition, Twist is targeted by miR-300 [36]. Cancer cell stemness is also regulated post-transcriptionally by miRNAs; examples of such regulation include miR-200c repression of the stem cell self-renewal factor BMI-1 [37] and miR-600 regulation of Wnt signaling via stearoyl desaturase which is required to produce active, lipid-modified Wnt proteins [38].
Recently, several lncRNAs have been shown to play a role in the process of EMT. These non-protein coding RNAs are typically larger than 200 nucleotides and can regulate numerous process such as transcription, splicing, mRNA stability and translation [39]. One such lncRNA is the Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1), which regulates the transcription and splicing of a cohort of metastasis-associated genes [40, 41]. In contrast, lncRNA HOTAIR controls chromatin state by acting as a scaffold for polycomb repressive complex 2 (PRC2) and lysine-specific demethylase 1 (LSD1) leading to alterations in H3K27 methylation patterns [42]. This function of HOTAIR can regulate EMT and metastatic progression through interaction with factors such as Snail and the poycomb group member EZH2, promoting Snail mediated repression of E-cadherin and other epithelial genes [43]. One additional example of lncRNA mediated regulation of EMT is lncRNA-activated by TGFβ (lncRNA-ATB) which is up-regulated upon TGFβ and acts as a sponge for the miR-200 family thereby increasing ZEB1/2 expression and EMT induction [44]. Thus, lncRNAs are a class of post-transcriptional regulators that act to modulate the mesenchymal transition through diverse mechanisms.
2.3 Post-transcriptional regulation by RNA binding proteins
RNA binding proteins are important post-transcriptional regulators of EMT and the associated metastatic phenotype, which act as modulators of RNA splicing, stability, localization and translation. Several well characterized splicing effectors promote the epithelial phenotype, including the epithelial splicing regulatory protein paralogues, ESRP1 and 2. These proteins are down-regulated by TGFβ and control alternative splicing of EMT-associated transcripts by binding to UGG-rich motifs present downstream of included exons [45, 46]. Transcripts spliced by ESRP1/2 function in cell adhesion, motility and the maintenance of the actin cytoskeleton and adherens junctions, all pathways that are altered during EMT [46, 47]. One additional splicing regulatory factor that promotes the epithelial phenotype is K-Homology Splicing Regulatory Protein, KHSRP. This factor attenuates TGFβ-induced EMT through a number of mechanisms including regulation of miR-192–5p, a ZEB1/2 and Snail1 targeting miRNA [48]. Additionally, KHSRP controls alternative splicing of pre-mRNAs that function in cell adhesion and migration, including FGFR2; KHSRP silencing generates the mesenchymal isoform of FGFR2 known as FGFR2 IIIc [48]. Mesenchymal-promoting splicing regulators include SRSF1, rbFOX2 and Muscleblind (MBNL1). The serine/arginine-rich splicing factor, SRSF1, alters the alternative splicing of transcripts including Ron, activating expression of the mesenchymal ΔRon isoform [49, 50]. This isoform, lacking exon 11, induces EMT and promote cell motility [50]. In contrast, rbFOX2 regulates exon inclusion in mesenchymal cells through the recognition of the consensus sequence (U)GCAUG [51], which may control the invasive ability of transitioned cells [52]. Furthermore, rbFOX2 and MBNL1 have been shown to regulate pluripotent stem cell differentiation, with silencing of these factors reducing expression of Twist and N-cadherin and increasing expression of E-cadherin in human embryonic stem cells [53], such regulation of EMT associated factors may be activated in cancer cells.
In addition to the splicing factors mentioned above, the heterogeneous ribonucleoprotein family of proteins contains approximately 20 members termed hnRNP A-U and plays a key role in the regulation of EMT induction. The majority of these proteins contain canonical or variant RNA recognition motif (RRM) domains with the exception of hnRNP E and K proteins which contain K-homology (KH) domains [54]. These RNA binding proteins have been shown to regulate transcription, RNA processing and translation to promote either an epithelial or a mesenchymal phenotype. Promoters of the mesenchymal phenotype include hnRNP M and hnRNP AB. Silencing of hnRNP M prevents TGFβ induced EMT and inhibits metastasis in breast cancer cells, the activity of hnRNP M is blocked in epithelial cells by ESRP1 binding to alternatively spliced transcripts [55]. Similarly, hnRNP AB promotes EMT and metastatic progression by binding to the Snail1 promoter, leading to increased Snail1 transcription and consequently a reduction in E-cadherin expression [56]. Regulators of the epithelial phenotype include hnRNP A1, which regulates alternative splicing of Ron and Rac1 pre-mRNA, inhibiting the generation of the mesenchymal form of these proteins, known as ΔRon and Rac1b [57, 58]. The hnRNP member hnRNP E1, which regulates the epithelial phenotype, will be the focus of the remainder of this review. hnRNP E1’s role as an important regulator of EMT and metastatic progression will be discussed, as well as the multiple distinct mechanisms, including alternative splicing and translational repression, through which this RNA binding protein functions.
3. hnRNP E1 as a downstream effector of TGF-β signaling
hnRNP E1 (PCBP1 or αCP1) is a member of the hnRNP E family consisting of hnRNP E1, E2, E3 and E4 which contain three nucleic acid-binding K homology (KH) domains [59]. Of these proteins, hnRNP E1 and E2 are the most abundant and share the highest degree of sequence homology; hnRNP E1 is an intronless gene and is thought to be a retro transposition of hnRNP E2 [59, 60]. Despite their similarity, however, hnRNP E1 and E2’s function appear to be non-redundant with hnRNP E1 knockout being embryonic lethal at the peri-implantation stage in mice, whereas hnRNP E2 remain viable until mid-gestation [61]. In addition to the three KH domains of hnRNP E1 [59], this RNA binding protein contains a nuclear localization signal between KH II and KH III [62], which controls localization to either the cytoplasm or nucleus.
hnRNP E1 functions in diverse cellular processes, playing a role in embryonic development through the translational regulation of maternal RNAs in oocytes [63, 64], and controlling muscle development through its role in miRNA processing [65]. hnRNP E1 also regulates erythropoiesis via its function as an iron chaperone, facilitating the transfer of iron to ferritin in erythroid cells and its role as a translational regulator of 15-lipoxygenase (LOX) during erythroid cell differentiation [66, 67]. Additionally, hnRNP E1 modulates cellular responses to hypoxia and folate deficient conditions. In response to hypoxia, hnRNP E1 dissociates from the 3′UTR of endothelial nitric oxide synthase (eNOS) mRNA reducing its stability and leading to its degradation [68]; whereas, hnRNP E1 maintains folate homeostasis by interacting with a C-rich element in the 5′UTR of folate receptor α mRNA. Under folate deficient conditions, hnRNP E1 translationally up-regulates this transcript leading to an increase of cell surface folate receptors which promotes folate up-take [69, 70]. Numerous studies have linked hnRNP E1 to the process of tumorigenesis and metastasis, demonstrating that hnRNP E1 acts as a tumor suppressor in several cancer types [5, 7, 8, 71, 72]. Silencing of hnRNP E1 results in the transition of cells to a mesenchymal phenotype, an increase in cell motility and invasiveness, and the promotion of tumor formation and distant metastases [5, 6, 8]. In addition, loss of hnRNP E1 has been linked to the acquisition of cancer stem cell-like properties, with hnRNP E1 down-regulation observed in CD44+CD24− populations of cells, which exhibit enhanced tumor initiating properties [73].
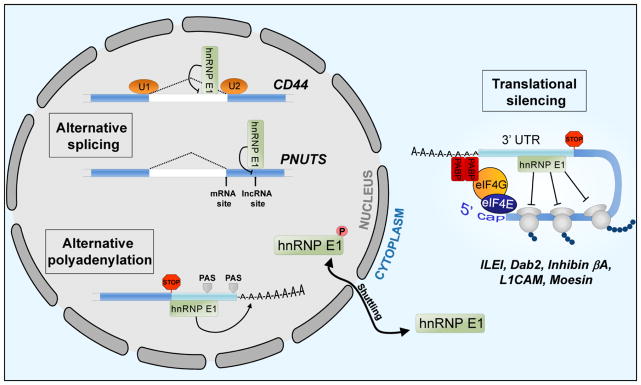
hnRNP E1 functions in a number of mRNA regulatory processes including transcription [74, 75], splicing [7, 74, 76, 77] and translation [6, 8, 69, 78], interacting with RNAs through CU rich regions such as the differentiation control element (DICE) and the TGFβ activated translation (BAT) element [5, 67]. Several studies have shown that activity of this protein is regulated through phosphorylation at a number of sites including S43, T60 and T127. Our laboratory has shown that the interaction of hnRNP E1 with elongation factor 1α1 in epithelial cells represses the translation of a cohort of mesenchymal-associated transcripts containing a 3′-UTR CU-rich element [5]. TGFβ activates Akt2 which phosphorylates hnRNP E1 on serine 43 leading to the release of this RNA binding protein from translationally silenced transcripts. In addition, p21-activated kinase 1 (PAK1) mediated phosphorylation at T60 and T127 has been shown to induce hnRNP E1 nuclear localization which is thought to be a major regulator of function leading to a switch from translational control to transcriptional and splicing regulation [7, 74]. This mechanism of hnRNP E1 regulation is activated during hypoxia as well, which induces an increase in Akt phosphorylation and nuclear localization of hnRNP E1 leading to decreased eNOS mRNA stability [68]. An overview of the post-transcriptional mechanisms through which hnRNP E1 regulates target transcripts involved in EMT and cancer progression will be discussed in the three sections below; control of alternative splicing, alternative polyadenylation, and translational regulation.
3.1 Control of alternative splicing by hnRNP E1
The process of splicing intronic sequences from mRNA is tightly regulated by four splicing elements; the 5′ splice site, branch site, polypyrimidine tract and an AG at the 3′ splice site. The role of hnRNP E1 in regulating splicing activity has been highlighted in recent work demonstrating extensive regulation of alternative splicing events via this RNA binding protein. RNA seq data has demonstrated that the majority of alternative splicing events induced by TGFβ/EGF treatment in HeLa cells are hnRNP E1 dependent [7]. Moreover, in a separate study silencing of hnRNP E1/2 in K562 cells resulted in spicing alterations of transcripts involved in cell death/survival, cell cycle and cell growth and proliferation [77]. hnRNP E1 appears to regulate alternative splicing through its interaction with splicing factors, such as U2AF which binds to the downstream 3′ splice site [7, 74], SF3B2 and U2AF65 which bind to the polypyrimidine tract [77] and the U2AF65 interacting protein RBM39 [74, 79]. Three characterized alternatively spliced transcripts regulated by hnRNP E1 are GHR [80], CD44 [7], and PNUTS [76]; both CD44 and PNUTS have a role in EMT and regulation of these transcripts will be discussed in more detail.
hnRNP E1 has been shown to regulate alternative splicing of the stemness-associated glycoprotein CD44. This transmembrane protein acts as a receptor for Hyaluronan, a component of the extracellular matrix, through interaction with an N-terminal extracellular domain, whereas, the cytoplasmic domain of CD44 interacts with factors such as Ankyrin and ERM proteins [81]. CD44 has 20 exons and alternative splicing regulates its function in cell adhesion, migration, proliferation and cancer cell stemness. Exons 1–5 and 16–20 are included in all splice variants, whereas exons 6–15 are alternatively spliced to generate 10 different variants of CD44. The standard form of CD44 (CD44s) excludes all variable exons, and a switch from variant forms of CD44 to CD44s is important for EMT induction, invasion and metastatic progression [7, 82, 83]. This pro-mesenchymal function of CD44s may be due to its role in invadopodia activity and as an activator of Akt signaling in TGFβ treated cells [82, 84]. In TGFβ and EGF stimulated cells, a shift from the epithelial variant of CD44 which includes exon 8–10 to the mesenchymal form CD44s is observed. This shift is hnRNP E1-dependent, where phosphorylated hnRNP E1 accumulates in the nucleus in a complex with T-179 phosphorylated SMAD3 which interacts with the variable exon region of CD44, leading to expression of CD44s [7].
More recently, we demonstrated that hnRNP E1 regulates the alternative splicing of protein phosphatase-1 nuclear targeting subunit (PPP1R10 or PNUTS) pre-RNA. The hnRNP E1 protein binds to a BAT-like element located near the 5′ end of exon 12, masking an alternative splice site located downstream of the canonical splice junction of the exon. The direct binding of hnRNP E1 to this alternative splice site prevents its usage and allows for the generation of PNUTS mRNA, which is in turn translated into PNUTS protein. TGFβ treatment, hnRNP E1 silencing or nucleo-cytoplasmic translocation results in loss of hnRNP E1 binding to the PNUTS pre-RNA and consequently permits alternative splicing and the generation of an lncRNA form of PNUTS. This novel lncRNA contains seven binding sites for miR-205, acting as a sponge to attenuate activity of this ZEB1/2-targeting miRNA. PNUTS lncRNA mediated repression of miR-205 activity results in increased ZEB protein expression and reduced E-cadherin levels which promote EMT induction. Additionally, expression of this lncRNA enhances cancer cell stemness and metastatic progression in breast cancer cells [76].
3.2 hnRNP E1 and alternative polyadenylation
The addition of a poly (A) tail to the 3′UTR of mRNA is a regulatory mechanism to control mRNA stability and translation [85]. 3′ processing is determined by the presence of a polyadenylation signal (PAS), typically AAUAAA, which is located 10–30 nucleotides upstream of the cleavage site. The polyadenylation specificity factor (CPSF) complex binds to this site, whereas the cleavage stimulation factor (CstF) complex recognizes and binds to a GU/U-rich region within 30 nucleotides 3′ of the cleavage site termed the GU/U DSE. Following cleavage, a poly (A) tail is added by poly(A) polymerase which recruits the poly(A) binding proteins nuclear PABPN1 and cytosolic PABPC. Alternative polyadenylation sites are present in a large percentage of mRNA and acts as a level of post-transcriptional regulation. Global 3′UTR shortening and lengthening are reported in response to physiological and development cues [86], for example, global 3′UTR shortening is observed in highly proliferative cells and during tumorigenesis [87]. 3′UTR length affects the number of RNA binding protein and miRNA binding sites on a given mRNA, and thus is a mechanism to regulate transcript stability and translation.
A role for hnRNP E1 in the regulation of alternative polyadenylation has been reported in several studies. Depletion of hnRNP E1/E2 in K562 cells affects alternative polyadenylation events; a C-rich motif upstream of the PAS was significantly enriched in mRNAs impacted by hnRNP E1/E2 silencing [85]. A second study using a reporter based RNAi screen for alternative polyadenylation demonstrated PABPN1’s role in this process and identified PCBP1 as the second highest hit in the screen, suggesting the hnRNP E1 may function as a global modulator of PAS usage [88]. The mechanism through which hnRNP E1 modulates polyadenylation is yet to be defined; however, this mechanism may be similar to that described for the hnRNP E2 target α-globulin. hnRNP E2 controls α-globulin 3′UTR cleavage and polyadenylation through interaction with a C-rich motif upstream of the PAS. Interaction between components of the 3′ processing machinery and hnRNP E2, as determined by co-immunoprecipitation studies, identified CPSF160, CPSF100 and CPSF3, which are all part of the CPSF complex that recognizes the PAS, in addition to poly(A) polymerase [89]. The regulation of polyadenylation by hnRNP E1 may act through a similar mechanism via cleavage and polyadenylation complex interactions.
3.3 Translational regulation via hnRNP E1
One mechanism through which hnRNP E1 acts to control the process of EMT and metastatic progression is via the translational repression of transcripts that contain C-rich elements in either their 5′ or 3′UTR. These transcripts have a role in the transition from epithelial to mesenchymal phenotype or in the enhanced cell migration and invasion associated with mesenchymal cells.
3.3.1 Regulation of EMT induction
Transcripts shown to be necessary for the induction of EMT include interleukin-like EMT inducer (ILEI) and disabled-2 (Dab2) which are translationally up-regulated following TGFβ treatment in an hnRNP E1 dependent manner. In epithelial cells, hnRNP E1 represses translation by binding to a BAT element in the 3′UTR of these transcripts. TGFβ treatment results in S43 phosphorylation of hnRNP E1 and loss of hnRNP E1 binding, leading to translational activation of these genes [6]. Silencing of either of these factors attenuates the mesenchymal transition in epithelial cells.
The cytosolic adaptor Dab2 plays a role in protein trafficking and clathrin-mediated endocytosis. This protein is essential for the formation of the primitive endoderm, an epithelium which is formed during early embryonic development, with Dab2 regulating E-cadherin distribution, epithelial polarity and surface positioning in endodermal cells [90–92]. The mechanisms through which Dab2 modulates EMT in cancer involves interaction with TGFβ receptor complex to control Smad activation and through the inhibition of Wnt signaling via Dvl-3 and LRP6 [93, 94]. Additionally, Dab2 has been shown to interact with β1 integrin at the cell membrane, controlling integrin activation and cell adherence during EMT [95].
The secreted protein ILEI plays a role in diverse processes such as bone morphogenesis and retinal development [96–98], and is linked to diseases such as Alzheimer’s, type 2 diabetes and cancer [99–101]. Several studies have linked ILEI to EMT induction, metastatic progression and poor prognosis in cancers such breast, pancreatic, colorectal and melanoma [101–106]. The mechanisms through which ILEI promotes EMT and metastasis during cancer progression are yet to be clearly defined; studies suggest that ILEI dimerizes through a covalent domain swap and it is the dimeric form of this protein that is active [107]. Moreover, cleavage of a 17 amino acid N-terminal pro-peptide domain and secretion of ILEI appear to be important for the pro-metastatic function of this protein [108]. Furthermore, in breast cancer, uPAR signaling is reported to promote ILEI secretion [108]. Several signaling pathways have been shown to be potential mediators of ILEI function; PDGF signaling may contribute to the metastatic progression of ILEI expressing cells in liver carcinoma. Whereas in melanoma, Jun signaling may contribute to ILEI function in metastatic progression [105]. Further research is required to elucidate the signaling pathways through which this ligand acts.
3.3.2 Regulation of the migratory and invasive ability of transitioned cells
Several translationally regulated targets of hnRNP E1 function in pathways which promote cell migration and invasion and in turn metastatic progression, this set of transcripts include inhibin βA, L1CAM, PRL-3 and Moesin. Inhibin βA is a member of the TGFβ superfamily that includes TGFβ, activins and BMP, which signals through the Smad pathway. This transcript interacts directly with hnRNP E1 and is translationally up-regulated following TGFβ treatment via hnRNP E1, leading to increased levels of the inhibin βA homodimer, Activin A. Although Activin A acts through the same Smad2/3 pathway as the EMT-inducer TGFβ, this ligand is not capable of inducing EMT itself, nor does it enhance the transition induced by TGFβ. However, upregulation of this ligand does promote the invasive phenotype of mesenchymal cells, thus increasing the metastatic progression of cells that undergo an EMT [109].
Phosphatase of regenerating liver-3 (PRL-3) has been shown to drive cancer progression in numerous cancers, promoting cell migration, invasion and metastatic progression [110]. PRL-3 functions through the modulation of several different pathways that include the downregulation of c-Src tyrosine kinase (Csk) leading to Src activation [111], increasing ERK1/2 and MMP2 activity [112], regulation of the ERM family member Ezrin [113] hyper-activation of EGFR [114] and downregulation of PTEN expression to activate Akt signaling [115], which contributes to EMT induction [111, 115]. hnRNP E1 suppresses translation of this transcript via interaction with a triple GCCCAG motif in the 5′UTR, leading to modulation of cancer progression [8]. Whether this interaction between hnRNP E1 and the PRL-3 5′UTR is sensitive to TGFβ or EGF signaling has yet to be tested, we postulate that the shift of hnRNP E1 localization to the nucleus following TGFβ and EGF signaling may lead to translational up-regulation of PRL-3, similar to other hnRNP E1 translationally-regulated transcripts, such regulation may act as a positive feedback to promote the switch to hnRNP E1 splicing events via PRL-3 dependent up-regulation of Akt signaling.
The actin-binding protein moesin, which is part of the ezrin-radixin-moesin family, is translationally silenced by hnRNP E1 in naive CD4+ T cells [116]. This repression is relieved by TGFβ treatment facilitating the differentiation of Th0 cells to in vitro-induced Tregs (iTregs). In this model, active translation of moesin was observed in iTregs, concomitant with loss of hnRNP E1 interaction with the moesin transcript. Functionally, moesin promoted TGFβ signaling in iTregs through TGFβ RII stabilization at the cell membrane, thus modulating Treg differentiation and immune suppression. The role of hnRNP E1 in modulating moesin expression specifically during EMT induction has yet to be characterized; however, it is tempting to speculate the translational control of this protein occurs during TGFβ induced EMT. Previous studies have reported an upregulation of ezrin-radixin-moesin proteins with EMT induction, with moesin functioning in actin cytoskeleton remodeling to promote cell invasion in breast cancer [117]. High expression of Moesin and reduced relapse-free survival and overall survival in cancer has also been described [118–120]; therefore a better understanding of hnRNP E1-mediated control of moesin in a cancer setting is warranted.
Finally, data demonstrating binding of hnRNP E1 to a DICE element in the 3′UTR of L1 cell adhesion molecule (L1CAM) suggests that this transcript is also translationally regulated by hnRNP E1. This transmembrane glycoprotein is a member of the immunoglobulin (Ig) superfamily, which interacts with other L1CAM molecules, receptor tyrosine kinases and integrins through six Ig-like and five fibronectin-like domains. L1CAM is overexpressed in several cancers and is linked to the induction of migration, invasion and metastatic progression through its role in integrin and ERK signaling. L1CAM has been shown to be transcriptionally up-regulated via Slug following TGFβ treatment [121]. However, this glycoprotein is also regulated at the level of translation. Similar to the mechanism of Akt2-mediated translational activation of BAT-containing transcripts, PAK1 phosphorylation of hnRNP E1 at T60 and T127 leads to loss of binding to L1CAM [74]. This mechanism of translational regulation may be important during TGFβ-induced EMT to rapidly up-regulate L1CAM and promote the mesenchymal phenotype.
4. Concluding remarks
The tumor-suppressor hnRNP E1 is a downstream effector of the TGFβ signalling pathway that post-transcriptionally regulates a cohort of transcripts involved in EMT, invasion and cancer cell stemness. Owing to the role of TGFβ signalling and hnRNP E1 in metastatic progression, this pathway is a promising candidate for therapeutic intervention, as current treatment strategies for systemic metastatic disease lack sufficient efficacy in prolonging patient survival.
Currently, several TGFβ signalling inhibitors are under investigation for the treatment of advanced cancer, including the monoclonal antibody Fresolimumab and the TGFβ receptor kinase inhibitor Galunisertib [122–124]. In combination with such approaches, blockade of hnRNP E1’s metastasis-promoting functions through the inhibition of Akt and Pak1 signalling may be an effective strategy to attenuate cancer progression. The link between this RNA binding protein and the cancer stem cell phenotype may also be targeted in this approach to control the tumor initiating ability of cells that is linked to recurrence and chemo-resistance. Additional characterization of hnRNP E1 function and regulation in metastatic cancer, as well as the function of this EMT regulator in the stromal compartment of tumors is merited.
Figure 1.
hnRNP E1 functions as a post-transcriptional regulator of EMT and cancer progression by controlling RNA splicing, polyadenylation and translation through its interaction with C-rich cis elements in target RNAs. Phosphorylation of hnRNP E1 at serine 43 by Akt2 leads to its release from transcripts under transcriptional control in the cytoplasm. Furthermore, phosphorylation at sites, including T60 and T127 by PAK1, induces the nuclear localization of hnRNP E1 which facilitates a switch from translational control to alternative splicing regulation. In the nucleus, hnRNP E1 functions as a regulator of alternative splicing of transcripts including CD44 and PNUTS and can also regulate alternative polyadenylation by binding to C-rich sequences in the 3′UTR of target mRNA. PAS=polyadenylation signal.
Acknowledgments
This work was supported by Grants CA055536 and CA154663 from the National Cancer Institute to PHH. We would like to thank members of the Howe lab for providing helpful feedback during the preparation of this manuscript, in particular Dr. Simon Grelet and Dr. Annamarie Dalton.
Footnotes
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13:616. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derynck R, Akhurst RJ, Balmain A. TGF-[beta] signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 3.Nieto MA, Huang Ruby Y-J, Jackson Rebecca A, Thiery Jean P. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Craene BD, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 5.Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MC, Merrick WC, Howe PH. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 2011;41:419–431. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-β-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tripathi V, Sixt Katherine M, Gao S, Xu X, Huang J, Weigert R, Zhou M, Zhang Ying E. Direct Regulation of Alternative Splicing by SMAD3 through PCBP1 Is Essential to the Tumor-Promoting Role of TGF-β. Mol Cell. 2016;64:549–564. doi: 10.1016/j.molcel.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Vardy LA, Tan CP, Loo JM, Guo K, Li J, Lim SG, Zhou J, Chng WJ, Ng SB, Li HX, Zeng Q. PCBP1 Suppresses the Translation of Metastasis-Associated PRL-3 Phosphatase. Cancer Cell. 2010;18:52–62. doi: 10.1016/j.ccr.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 9.Massagué J, Blain SW, Lo RS. TGFβ Signaling in Growth Control, Cancer, and Heritable Disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 10.Zhang YE. Non-Smad pathways in TGF-[beta] signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blobe GC, Schiemann WP, Lodish HF. Role of Transforming Growth Factor β in Human Disease. N Engl J Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 12.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radisky DC. Epithelial-mesenchymal transition. J Cell Sci. 2005;118:4325–4326. doi: 10.1242/jcs.02552. [DOI] [PubMed] [Google Scholar]
- 14.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Scheel C, Eaton Elinor N, Li Sophia H-J, Chaffer Christine L, Reinhardt F, Kah K-J, Bell G, Guo W, Rubin J, Richardson Andrea L, Weinberg Robert A. Paracrine and Autocrine Signals Induce and Maintain Mesenchymal and Stem Cell States in the Breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells – what challenges do they pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-β Signaling Maintains Tumorigenicity of Glioma-Initiating Cells through Sry-Related HMG-Box Factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 20.Zheng H, Kang Y. Multilayer control of the EMT master regulators. Oncogene. 2014;33:1755–1763. doi: 10.1038/onc.2013.128. [DOI] [PubMed] [Google Scholar]
- 21.Yamasaki T, Seki N, Yamada Y, Yoshino H, Hidaka H, Chiyomaru T, Nohata N, Kinoshita T, Nakagawa M, Enokida H. Tumor suppressive microRNA138 contributes to cell migration and invasion through its targeting of vimentin in renal cell carcinoma. Int J Oncol. 2012;41:805–817. doi: 10.3892/ijo.2012.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotech. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, Zhao J, Majerciak V, Gaur AB, Chen S, Miller SS. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 27.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 29.Liu YN, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L, Yi M, Stephens RM, Seng V, Sheppard-Tillman H, Martin P, Kelly K. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2013;32:296–306. doi: 10.1038/onc.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brabletz T. MiR-34 and SNAIL: another double-negative feedback loop controlling cellular plasticity/EMT governed by p53. Cell Cycle. 2012;11:215–216. doi: 10.4161/cc.11.2.18900. [DOI] [PubMed] [Google Scholar]
- 31.Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, Rowe RG, Lee S, Maher CA, Weiss SJ, Yook JI. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding X, Park SI, McCauley LK, Wang CY. Signaling between transforming growth factor beta (TGF-beta) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J Biol Chem. 2013;288:10241–10253. doi: 10.1074/jbc.M112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ru P, Steele R, Newhall P, Phillips NJ, Toth K, Ray RB. miRNA-29b suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol Cancer Ther. 2012;11:1166–1173. doi: 10.1158/1535-7163.MCT-12-0100. [DOI] [PubMed] [Google Scholar]
- 34.Kumarswamy R, Mudduluru G, Ceppi P, Muppala S, Kozlowski M, Niklinski J, Papotti M, Allgayer H. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer. 2012;130:2044–2053. doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 35.Liang YJ, Wang QY, Zhou CX, Yin QQ, He M, Yu XT, Cao DX, Chen GQ, He JR, Zhao Q. MiR-124 targets Slug to regulate epithelial–mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34:713–722. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu J, Xie F, Bao X, Chen W, Xu Q. miR-300 inhibits epithelial to mesenchymal transition and metastasis by targeting Twist in human epithelial cancer. Mol Cancer. 2014;13:121. doi: 10.1186/1476-4598-13-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RAR, Lao K, Clarke MF. Downregulation of miRNA-200c Links Breast Cancer Stem Cells with Normal Stem Cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Helou R, Pinna G, Cabaud O, Wicinski J, Bhajun R, Guyon L, Rioualen C, Finetti P, Gros A, Mari B, Barbry P, Bertucci F, Bidaut G, Harel-Bellan A, Birnbaum D, Charafe-Jauffret E, Ginestier C. miR-600 Acts as a Bimodal Switch that Regulates Breast Cancer Stem Cell Fate through WNT Signaling. Cell Reports. 2017;18:2256–2268. doi: 10.1016/j.celrep.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, Brogi E, Egeblad M, Spector DL. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutschner T, Hämmerle M, Eiβmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Groβ M, Zörnig M, MacLeod AR, Spector DL, Diederichs S. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Battistelli C, Cicchini C, Santangelo L, Tramontano A, Grassi L, Gonzalez FJ, de Nonno V, Grassi G, Amicone L, Tripodi M. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36:942–955. doi: 10.1038/onc.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan J-h, Yang F, Wang F, Ma J-z, Guo Y-j, Tao Q-f, Liu F, Pan W, Wang T-t, Zhou C-c, Wang S-b, Wang Y-z, Yang Y, Yang N, Zhou W-p, Yang G-s, Sun S-h. A Long Noncoding RNA Activated by TGF-β Promotes the Invasion-Metastasis Cascade in Hepatocellular Carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S, Fujii H, Yamaguchi A, Miyazawa K, Miyazono K, Saitoh M. TGF-[beta] drives epithelial-mesenchymal transition through [delta]EF1-mediated downregulation of ESRP. Oncogene. 2012;31:3190–3201. doi: 10.1038/onc.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP. An ESRP-regulated splicing programme is abrogated during the epithelial–mesenchymal transition. EMBO J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB. An EMT–Driven Alternative Splicing Program Occurs in Human Breast Cancer and Modulates Cellular Phenotype. PLoS Genet. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puppo M, Bucci G, Rossi M, Giovarelli M, Bordo D, Moshiri A, Gorlero F, Gherzi R, Briata P. miRNA-Mediated KHSRP Silencing Rewires Distinct Post-transcriptional Programs during TGF-β-Induced Epithelial-to-Mesenchymal Transition. Cell Rep. 2016;16:967–978. doi: 10.1016/j.celrep.2016.06.055. [DOI] [PubMed] [Google Scholar]
- 49.Anczuków O, Akerman M, Cléry A, Wu J, Shen C, Shirole Nitin H, Raimer A, Sun S, Jensen Mads A, Hua Y, Allain Frédéric HT, Krainer Adrian R. SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol Cell. 2015;60:105–117. doi: 10.1016/j.molcel.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell Motility Is Controlled by SF2/ASF through Alternative Splicing of the Ron Protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 51.Venables JP, Brosseau J-P, Gadea G, Klinck R, Prinos P, Beaulieu J-F, Lapointe E, Durand M, Thibault P, Tremblay K, Rousset F, Tazi J, Abou Elela S, Chabot B. RBFOX2 Is an Important Regulator of Mesenchymal Tissue-Specific Splicing in both Normal and Cancer Tissues. Mol Cell Biol. 2013;33:396–405. doi: 10.1128/MCB.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braeutigam C, Rago L, Rolke A, Waldmeier L, Christofori G, Winter J. The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene. 2014;33:1082–1092. doi: 10.1038/onc.2013.50. [DOI] [PubMed] [Google Scholar]
- 53.Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B, Abou Elela S, Roux P, Lemaitre J-M, Tazi J. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun. 2013;4:2480. doi: 10.1038/ncomms3480. [DOI] [PubMed] [Google Scholar]
- 54.Han Siew P, Tang Yue H, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 55.Xu Y, Gao XD, Lee JH, Huang H, Tan H, Ahn J, Reinke LM, Peter ME, Feng Y, Gius D, Siziopikou KP, Peng J, Xiao X, Cheng C. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 2014;28:1191–1203. doi: 10.1101/gad.241968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou ZJ, Dai Z, Zhou SL, Hu ZQ, Chen Q, Zhao YM, Shi YH, Gao Q, Wu WZ, Qiu SJ, Zhou J, Fan J. HNRNPAB Induces Epithelial–Mesenchymal Transition and Promotes Metastasis of Hepatocellular Carcinoma by Transcriptionally Activating SNAIL. Cancer Res. 2014;74:2750–2762. doi: 10.1158/0008-5472.CAN-13-2509. [DOI] [PubMed] [Google Scholar]
- 57.Bonomi S, di Matteo A, Buratti E, Cabianca DS, Baralle FE, Ghigna C, Biamonti G. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res. 2013;41:8665–8679. doi: 10.1093/nar/gkt579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pelisch F, Khauv D, Risso G, Stallings-Mann M, Blaustein M, Quadrana L, Radisky DC, Srebrow A. Involvement of hnRNP A1 in the matrix metalloprotease-3-dependent regulation of Rac1 pre-mRNA splicing. J cell biochem. 2012;113:2319–2329. doi: 10.1002/jcb.24103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leffers H, Dejgaard K, Celis JE. Characterisation of Two Major Cellular Poly(rC)-Binding Human Proteins, Each Containing Three K-homologous (KH) Domains. Eur J Biochem. 1995;230:447–453. [PubMed] [Google Scholar]
- 60.Makeyev AV, Liebhaber SA. Identification of Two Novel Mammalian Genes Establishes a Subfamily of KH-Domain RNA-Binding Proteins. Genomics. 2000;67:301–316. doi: 10.1006/geno.2000.6244. [DOI] [PubMed] [Google Scholar]
- 61.Ghanem LR, Kromer A, Silverman IM, Chatterji P, Traxler E, Penzo-Mendez A, Weiss MJ, Stanger BZ, Liebhaber SA. The Poly(C) Binding Protein Pcbp2 and Its Retrotransposed Derivative Pcbp1 Are Independently Essential to Mouse Development. Mol Cell Biol. 2016;36:304–319. doi: 10.1128/MCB.00936-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chkheidze AN, Liebhaber SA. A Novel Set of Nuclear Localization Signals Determine Distributions of the αCP RNA-Binding Proteins. Mol Cell Biol. 2003;23:8405–8415. doi: 10.1128/MCB.23.23.8405-8415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia M, He H, Wang Y, Liu M, Zhou T, Lin M, Zhou Z, Huo R, Zhou Q, Sha J. PCBP1 is required for maintenance of the transcriptionally silent state in fully grown mouse oocytes. Cell Cycle. 2012;11:2833–2842. doi: 10.4161/cc.21169. [DOI] [PubMed] [Google Scholar]
- 64.Shi Z, Zhao C, Yang Y, Teng H, Guo Y, Ma M, Guo X, Zhou Z, Huo R, Zhou Q. Maternal PCBP1 determines the normal timing of pronucleus formation in mouse eggs. Cell Mol Life Sci. 2015;72:3575–3586. doi: 10.1007/s00018-015-1905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Espinoza-Lewis RA, Yang Q, Liu J, Huang ZP, Hu X, Chen D, Wang DZ. Poly(C)-binding protein 1 (Pcbp1) regulates skeletal muscle differentiation by modulating microRNA processing in myoblasts. J Biol Chem. 2017;292:9540–9550. doi: 10.1074/jbc.M116.773671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. mRNA Silencing in Erythroid Differentiation: hnRNP K and hnRNP E1 Regulate 15-Lipoxygenase Translation from the 3′ End. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 67.Reimann I, Huth A, Thiele H, Thiele BJ. Suppression of 15-lipoxygenase synthesis by hnRNP E1 is dependent on repetitive nature of LOX mRNA 3′-UTR control element DICE. J Mol Biol. 2002;315:965–974. doi: 10.1006/jmbi.2001.5315. [DOI] [PubMed] [Google Scholar]
- 68.Ho JJD, Robb GB, Tai SC, Turgeon PJ, Mawji IA, Man HSJ, Marsden PA. Active Stabilization of Human Endothelial Nitric Oxide Synthase mRNA by hnRNP E1 Protects against Antisense RNA and MicroRNAs. Mol Cell Biol. 2013;33:2029–2046. doi: 10.1128/MCB.01257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Antony AC, Tang YS, Khan RA, Biju MP, Xiao X, Li QJ, Sun XL, Jayaram HN, Stabler SP. Translational upregulation of folate receptors is mediated by homocysteine via RNA-heterogeneous nuclear ribonucleoprotein E1 interactions. J Clin Invest. 2004;113:285–301. doi: 10.1172/JCI11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun XL, Antony AC. Evidence That a Specific Interaction Between an 18-Base cis-Element in the 5′-Untranslated Region of Human Folate Receptor-α mRNA and a 46-kDa Cytosolic trans-Factor is Critical for Translation. J Biol Chem. 1996;271:25539–25547. [PubMed] [Google Scholar]
- 71.Zhang ZZ, Shen ZY, Shen YY, Zhao EH, Wang M, Wang CJ, Cao H, Xu J. HOTAIR Long Noncoding RNA Promotes Gastric Cancer Metastasis through Suppression of Poly r(C)-Binding Protein (PCBP) 1. Mol Cancer Ther. 2015;14:1162–1170. doi: 10.1158/1535-7163.MCT-14-0695. [DOI] [PubMed] [Google Scholar]
- 72.Zhang M, Wang X, Tan J, Zhao M, Lian L, Zhang W. Poly r(C) binding protein (PCBP) 1 is a negative regulator of thyroid carcinoma. Am J Transl Res. 2016;8:3567–3573. [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Q, Cai Z, Chen Y, Gu M, Zheng D, Zhou J, Wang Z. Poly r(C) Binding Protein-1 is Central to Maintenance of Cancer Stem Cells in Prostate Cancer Cells. Cell Physiol Biochem. 2015;35:1052–1061. doi: 10.1159/000373931. [DOI] [PubMed] [Google Scholar]
- 74.Meng Q, Rayala SK, Gururaj AE, Talukder AH, O’Malley BW, Kumar R. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc Natl Acad Sci USA. 2007;104:5866–5871. doi: 10.1073/pnas.0701065104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim SS, Pandey KK, Choi HS, Kim SY, Law PY, Wei LN, Loh HH. Poly(C) Binding Protein Family Is a Transcription Factor in μ-Opioid Receptor Gene Expression. Mol Pharmacol. 2005;68:729–736. doi: 10.1124/mol.105.012245. [DOI] [PubMed] [Google Scholar]
- 76.Grelet S, Link LA, Howley B, Obellianne C, Palanisamy V, Gangaraju VK, Diehl JA, Howe PH. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105–1115. doi: 10.1038/ncb3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ji X, Park JW, Bahrami-Samani E, Lin L, Duncan-Lewis C, Pherribo G, Xing Y, Liebhaber SA. αCP binding to a cytosine-rich subset of polypyrimidine tracts drives a novel pathway of cassette exon splicing in the mammalian transcriptome. Nucleic Acids Res. 2016;44:2283–2297. doi: 10.1093/nar/gkw088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hussey GS, Link LA, Brown AS, Howley BV, Chaudhury A, Howe PH. Establishment of a TGFβ-Induced Post-Transcriptional EMT Gene Signature. PLOS ONE. 2012;7:e52624. doi: 10.1371/journal.pone.0052624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang SC, Zhang HS, Yu B, McMahon E, Nguyen DT, Yu FH, Ou AC, Ou JP, Benz EJ. Protein 4.1R Exon 16 3′ Splice Site Activation Requires Coordination among TIA1, Pcbp1, and RBM39 during Terminal Erythropoiesis. Mol Cell Biol. 2017;37:e00446–16. doi: 10.1128/MCB.00446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akker SA, Misra S, Aslam S, Morgan EL, Smith PJ, Khoo B, Chew SL. Pre-Spliceosomal Binding of U1 Small Nuclear Ribonucleoprotein (RNP) and Heterogenous Nuclear RNP E1 Is Associated with Suppression of a Growth Hormone Receptor Pseudoexon. Mol Endocrinol. 2007;21:2529–2540. doi: 10.1210/me.2007-0038. [DOI] [PubMed] [Google Scholar]
- 81.Ponta H, Sherman L, Herrlich PA. CD44: From adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 82.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mima K, Okabe H, Ishimoto T, Hayashi H, Nakagawa S, Kuroki H, Watanabe M, Beppu T, Tamada M, Nagano O, Saya H, Baba H. CD44s Regulates the TGF-β–Mediated Mesenchymal Phenotype and Is Associated with Poor Prognosis in Patients with Hepatocellular Carcinoma. Cancer Res. 2012;72:3414–3423. doi: 10.1158/0008-5472.CAN-12-0299. [DOI] [PubMed] [Google Scholar]
- 84.Zhao P, Xu Y, Wei Y, Qiu Q, Chew TL, Kang Y, Cheng C. The CD44s splice isoform is a central mediator for invadopodia activity. J Cell Sci. 2016;129:1355–1365. doi: 10.1242/jcs.171959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ji X, Wan J, Vishnu M, Xing Y, Liebhaber SA. αCP Poly(C) Binding Proteins Act as Global Regulators of Alternative Polyadenylation. Mol Cell Biol. 2013;33:2560–2573. doi: 10.1128/MCB.01380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27:2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kühn U, Menzies Fiona M, Vrielink Joachim AFO, Bos Arnold J, Drost J, Rooijers K, Rubinsztein David C, Agami R. The Poly(A)-Binding Protein Nuclear 1 Suppresses Alternative Cleavage and Polyadenylation Sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 89.Ji X, Kong J, Liebhaber SA. An RNA protein complex links enhanced nuclear 3′ processing with cytoplasmic mRNA stabilization. EMBO J. 2011;30:2622–2633. doi: 10.1038/emboj.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang DH, Cai KQ, Roland IH, Smith ER, Xu XX. Disabled-2 Is an Epithelial Surface Positioning Gene. J Biol Chem. 2007;282:13114–13122. doi: 10.1074/jbc.M611356200. [DOI] [PubMed] [Google Scholar]
- 91.Moore R, Cai KQ, Tao W, Smith ER, Xu XX. Differential requirement for Dab2 in the development of embryonic and extra-embryonic tissues. BMC Dev Biol. 2013;13:39. doi: 10.1186/1471-213X-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rula ME, Cai KQ, Moore R, Yang DH, Staub CM, Capo-chichi CD, Jablonski SA, Howe PH, Smith ER, Xu XX. Cell autonomous sorting and surface positioning in the formation of primitive endoderm in embryoid bodies. Genesis. 2007;45:327–338. doi: 10.1002/dvg.20298. [DOI] [PubMed] [Google Scholar]
- 93.Hocevar BA, Mou F, Rennolds JL, Morris SM, Cooper JA, Howe PH. Regulation of the Wnt signaling pathway by disabled-2 (Dab2) EMBO J. 2003;22:3084–3094. doi: 10.1093/emboj/cdg286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang Y, He X, Howe PH. Disabled-2 (Dab2) inhibits Wnt/β-catenin signalling by binding LRP6 and promoting its internalization through clathrin. EMBO J. 2012;31:2336–2349. doi: 10.1038/emboj.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prunier C, Howe PH. Disabled-2 (Dab2) Is Required for Transforming Growth Factor β-induced Epithelial to Mesenchymal Transition (EMT) J Biol Chem. 2005;280:17540–17548. doi: 10.1074/jbc.M500974200. [DOI] [PubMed] [Google Scholar]
- 96.Katahira T, Nakagiri S, Terada K, Furukawa T. Secreted factor FAM3C (ILEI) is involved in retinal laminar formation. Biochem Biophys Res Commun. 2010;392:301–306. doi: 10.1016/j.bbrc.2009.12.180. [DOI] [PubMed] [Google Scholar]
- 97.Bendre A, Büki KG, Määttä JA. Fam3c modulates osteogenic differentiation by down-regulating Runx2. Differentiation. 2017;93:50–57. doi: 10.1016/j.diff.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 98.Maatta JA, Bendre A, Laanti M, Buki KG, Rantakari P, Tervola P, Saarimaki J, Poutanen M, Harkonen P, Vaananen K. Fam3c modulates osteogenic cell differentiation and affects bone volume and cortical bone mineral density. Bonekey Rep. 2016;5:787. doi: 10.1038/bonekey.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hasegawa H, Liu L, Tooyama I, Murayama S, Nishimura M. The FAM3 superfamily member ILEI ameliorates Alzheimer’s disease-like pathology by destabilizing the penultimate amyloid-β precursor. Nat Commun. 2014;5:3917. doi: 10.1038/ncomms4917. [DOI] [PubMed] [Google Scholar]
- 100.Chen Z, Ding L, Yang W, Wang J, Chen L, Chang Y, Geng B, Cui Q, Guan Y, Yang J. Hepatic Activation of the FAM3C-HSF1-CaM Pathway Attenuates Hyperglycemia of Obese Diabetic Mice. Diabetes. 2017;66:1185–1197. doi: 10.2337/db16-0993. [DOI] [PubMed] [Google Scholar]
- 101.Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, Schreiber M, Jechlinger M, Beug H. ILEI: A cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006;10:227–239. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 102.Gao Z. ILEI: a novel marker for epithelial-mesenchymal transition and poor prognosis in colorectal cancer. Histopathology. 2014;65:527–538. doi: 10.1111/his.12435. [DOI] [PubMed] [Google Scholar]
- 103.Lahsnig C, Mikula M, Petz M, Zulehner G, Schneller D, van Zijl F, Huber H, Csiszar A, Beug H, Mikulits W. ILEI requires oncogenic Ras for the epithelial to mesenchymal transition of hepatocytes and liver carcinoma progression. Oncogene. 2008;28:638–650. doi: 10.1038/onc.2008.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grønborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Biomarker Discovery from Pancreatic Cancer Secretome Using a Differential Proteomic Approach. Mol Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 105.Noguchi K, Dalton AC, Howley BV, McCall BJ, Yoshida A, Diehl JA, Howe PH. Interleukin-like EMT inducer regulates partial phenotype switching in MITF-low melanoma cell lines. PLOS ONE. 2017;12:e0177830. doi: 10.1371/journal.pone.0177830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Halberg N, Sengelaub Caitlin A, Navrazhina K, Molina H, Uryu K, Tavazoie Sohail F. PITPNC1 Recruits RAB1B to the Golgi Network to Drive Malignant Secretion. Cancer Cell. 2016;29:339–353. doi: 10.1016/j.ccell.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jansson AM, Csiszar A, Maier J, Nyström AC, Ax E, Johansson P, Schiavone LH. The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer. J Biol Chem. 2017;292:15501–15511. doi: 10.1074/jbc.M117.782904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Csiszar A, Kutay B, Wirth S, Schmidt U, Macho-Maschler S, Schreiber M, Alacakaptan M, Vogel GF, Aumayr K, Huber LA, Beug H. Interleukin-like epithelial-to-mesenchymal transition inducer activity is controlled by proteolytic processing and plasminogen–urokinase plasminogen activator receptor system–regulated secretion during breast cancer progression. Breast Cancer Res. 2014;16:433. doi: 10.1186/s13058-014-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Howley BV, Hussey GS, Link LA, Howe PH. Translational regulation of Inhibin βA by TGFβ via the RNA-binding protein hnRNP E1 enhances the invasiveness of epithelial-to-mesenchymal transitioned cells. Oncogene. 2016;35:1725–1735. doi: 10.1038/onc.2015.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Al-Aidaroos AQO, Zeng Q. PRL-3 phosphatase and cancer metastasis. J Cell Biochem. 2010;111:1087–1098. doi: 10.1002/jcb.22913. [DOI] [PubMed] [Google Scholar]
- 111.Liang F, Liang J, Wang WQ, Sun JP, Udho E, Zhang ZY. PRL3 Promotes Cell Invasion and Proliferation by Down-regulation of Csk Leading to Src Activation. J Biol Chem. 2007;282:5413–5419. doi: 10.1074/jbc.M608940200. [DOI] [PubMed] [Google Scholar]
- 112.Peng L, Xing X, Li W, Qu L, Meng L, Lian S, Jiang B, Wu J, Shou C. PRL-3 promotes the motility, invasion, and metastasis of LoVo colon cancer cells through PRL-3-integrin β1-ERK1/2 and-MMP2 signaling. Mol Cancer. 2009;8:110–110. doi: 10.1186/1476-4598-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Forte E, Orsatti L, Talamo F, Barbato G, De Francesco R, Tomei L. Ezrin is a specific and direct target of protein tyrosine phosphatase PRL-3. Biochim Biophys Acta Mol Cell Res. 2008;1783:334–344. doi: 10.1016/j.bbamcr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 114.Al-aidaroos AQO, Yuen HF, Guo K, Zhang SD, Chung T-H, Chng WJ, Zeng Q. Metastasis-associated PRL-3 induces EGFR activation and addiction in cancer cells. J Clin Invest. 2013;123:3459–3471. doi: 10.1172/JCI66824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang H, Quah SY, Dong JM, Manser E, Tang JP, Zeng Q. PRL-3 Down-regulates PTEN Expression and Signals through PI3K to Promote Epithelial-Mesenchymal Transition. Cancer Res. 2007;67:2922–2926. doi: 10.1158/0008-5472.CAN-06-3598. [DOI] [PubMed] [Google Scholar]
- 116.Ansa-Addo EA, Zhang Y, Yang Y, Hussey GS, Howley BV, Salem M, Riesenberg B, Sun S, Rockey DC, Karvar S, Howe PH, Liu B, Li Z. Membrane-organizing protein moesin controls Treg differentiation and antitumor immunity via TGF-β signaling. J Clin Invest. 2017;127:1321–1337. doi: 10.1172/JCI89281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haynes J, Srivastava J, Madson N, Wittmann T, Barber DL. Dynamic actin remodeling during epithelial–mesenchymal transition depends on increased moesin expression. Mol Biol Cell. 2011;22:4750–4764. doi: 10.1091/mbc.E11-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Charafe-Jauffret E, Monville F, Bertucci F, Esterni B, Ginestier C, Finetti P, Cervera N, Geneix J, Hassanein M, Rabayrol L, Sobol H, Taranger-Charpin C, Xerri L, Viens P, Birnbaum D, Jacquemier J. Moesin expression is a marker of basal breast carcinomas. Int J Cancer. 2007;121:1779–1785. doi: 10.1002/ijc.22923. [DOI] [PubMed] [Google Scholar]
- 119.Jung WY, Kang Y, Lee H, Mok Y-J, Kim HK, Kim A, Kim B-h. Expression of moesin and CD44 is associated with poor prognosis in gastric adenocarcinoma. Histopathology. 2013;63:474–481. doi: 10.1111/his.12202. [DOI] [PubMed] [Google Scholar]
- 120.Li X, Roslan S, Johnstone CN, Wright JA, Bracken CP, Anderson M, Bert AG, Selth LA, Anderson RL, Goodall GJ, Gregory PA, Khew-Goodall Y. MiR-200 can repress breast cancer metastasis through ZEB1-independent but moesin-dependent pathways. Oncogene. 2014;33:4077–4088. doi: 10.1038/onc.2013.370. [DOI] [PubMed] [Google Scholar]
- 121.Geismann C, Morscheck M, Koch D, Bergmann F, Ungefroren H, Arlt A, Tsao MS, Bachem MG, Altevogt P, Sipos B, Fölsch UR, Schäfer H, Müerköster SS. Up-regulation of L1CAM in Pancreatic Duct Cells Is Transforming Growth Factor β1– and Slug-Dependent: Role in Malignant Transformation of Pancreatic Cancer. Cancer Res. 2009;69:4517–4526. doi: 10.1158/0008-5472.CAN-08-3493. [DOI] [PubMed] [Google Scholar]
- 122.Morris JC, Shapiro GI, Tan AR, Lawrence DP, Olencki TE, Dezube BJ, Hsu FJ, Reiss M, Berzofsky JA. Phase I/II study of GC1008: A human anti-transforming growth factor-beta (TGFβ) monoclonal antibody (MAb) in patients with advanced malignant melanoma (MM) or renal cell carcinoma (RCC) J Clin Oncol. 2008;26:9028–9028. doi: 10.1371/journal.pone.0090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morris JC, Tan AR, Olencki TE, Shapiro GI, Dezube BJ, Reiss M, Hsu FJ, Berzofsky JA, Lawrence DP. Phase I Study of GC1008 (Fresolimumab): A Human Anti-Transforming Growth Factor-Beta (TGFβ) Monoclonal Antibody in Patients with Advanced Malignant Melanoma or Renal Cell Carcinoma. PLOS ONE. 2014;9:e90353. doi: 10.1371/journal.pone.0090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Faivre SJ, Santoro A, Gane E, Kelley RK, Hourmand IO, Assenat E, Gueorguieva I, Cleverly A, Desaiah D, Lahn MMF, Raymond E, Benhadji KA, Giannelli G. A phase 2 study of galunisertib, a novel transforming growth factor-beta (TGF-β) receptor I kinase inhibitor, in patients with advanced hepatocellular carcinoma (HCC) and low serum alpha fetoprotein (AFP) J Clin Oncol. 2016;34:4070–4070. [Google Scholar]