Abstract
Background and Aims
Floret opening in barley is induced by the swelling of the lodicule, a trait under the control of the cleistogamy1 (cly1) gene. The product of cly1 is a member of the APETALA2 (AP2) transcription factor family, which inhibits lodicule development. A sequence polymorphism at the miR172 target site within cly1 has been associated with variation in lodicule development and hence with the cleistogamous phenotype. It was unclear whether miR172 actually functions in cly1 regulation and, if it does, which miR172 gene contributes to cleistogamy. It was also interesting to explore whether miR172-mediated cly1 regulation occurs at transcriptional level or at translational level.
Methods
Deep sequencing of small RNA identified the miR172 sequences expressed in barley immature spikes. miR172 genes were confirmed by computational and expression analysis. miR172 and cly1 expression profiles were determined by in situ hybridization and quantitative expression analysis. Immunoblot analysis provided the CLY1 protein quantifications. Definitive evidence of the role of miR172 in cleistogamy was provided by a transposon Ds-induced mutant of Hv-miR172a.
Key Results
A small RNA analysis of the immature barley spike revealed three isomers, miR172a, b and c, of which miR172a was the most abundant. In situ hybridization analysis showed that miR172 and cly1 co-localize in the lodicule primordium, suggesting that these two molecules potentially interact with one another. Immunoblot analysis showed that the sequence polymorphism at the miR172 target site within cly1 reduced the abundance of the CLY1 protein, but not that of its transcript. In a Ds-induced mutant of Hv-miR172a, which generates no mature miR172a, the lodicules fail to grow, resulting in a very small lodicule.
Conclusions
Direct evidence is presented to show that miR172a acts to reduce the abundance of the CLY1 protein, which enables open flowering in barley.
Keywords: MicroRNA, lodicule, AP2, translation, cereals
INTRODUCTION
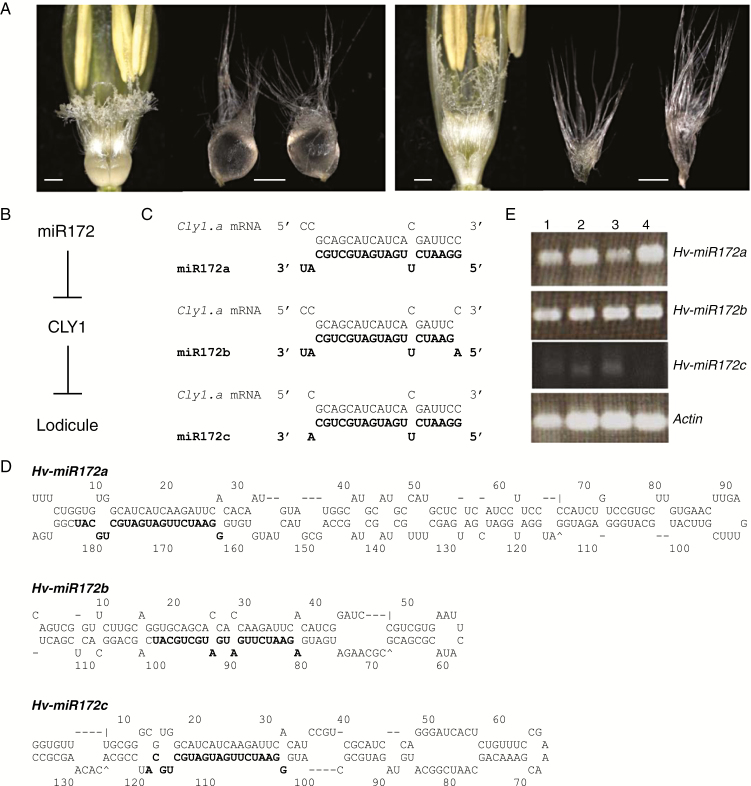
The floral morphology of the grasses differs significantly from that of other angiosperms (Ciaffi et al., 2011). The grass homologue of the dicotyledonous inner perianth whorl (which is the petal for most dicots) is the lodicule, a pair of which lies at the base of the grass pistil and stamen (Bommert et al., 2005). In a non-cleistogamous [wild-type (WT)] floret, the lodicules swell just prior to anthesis, pushing apart the palea and lemma, thereby releasing pollen and exposing the stigma to non-self pollen (Fig. 1A, left panel). In the cleistogamous floret (Fig. 1A, right panel), by contrast, this swelling does not occur, with the result that pollen is released within a closed floret, thereby forcing self-fertilization (Briggs, 1978; Lord, 1981). In the ecology and evolution of plants, it has been proposed that adaptive plasticity of cleistogamy could be driven by variation in the pollination environment, with cleistogamous flowers providing reproductive assurance when pollinators are scarce and non-cleistogamous flowers reducing inbreeding depression in offspring when pollinators are abundant (Stojanova et al., 2016). Cleistogamous barley cultivars are better able to avoid infection by Fusarium species that cause fusarium head blight, by denying entry to spores during early stages of floret and kernel development and showing coincident quantitative trait loci for blight resistance and cly1 (Yoshida et al., 2005; Hori et al., 2006; Sato et al., 2008), and they also greatly reduce the risk of gene flow through pollen dispersal (Daniell, 2002; Abdel-Ghani et al., 2004; Ma and Wang, 2004). On the other hand, non-cleistogamy is essential for the production of F1 hybrid grain.
Fig. 1.
The barley inflorescence and the Hv-miR172 genes. (A) Variation in lodicule size at anthesis. (Left) AZ (non-cleistogamous). (Right) KNG (cleistogamous). Scale bar = 0.5 mm. (B) Scheme of proposed interaction between miR172 and CLY1 for lodicule development. The Cly1.a and cly1.b alleles both encode CLY1, which suppresses the development of lodicules. Expression of Cly1.a is negatively regulated by miR172, resulting in less suppression of lodicule development by CLY1. (C) Potential interactions between cly1 mRNA and miR172 isoforms in cultivar ‘Morex’. Bold letters indicate miR172 sequences that can potentially bind with cly1 mRNA sequence. (D) In silico predicted hairpin secondary structure of the miR172 precursors. Bold text indicates the mature miRNA sequence. Precursor sequence length and free folding energy (ΔG) for Hv-miR172a, b and c were, respectively, 189 nt and −92.80 kcal mol−1, 117 nt and −58.20 kcal mol−1 and 135 nt and −63.60 kcal mol−1. (E) RT–PCR-derived abundance of primary miR172s in the immature spikes at the awn primordium stage. Lane 1, AZ; lane 2, KNG; lane 3, ‘Morex’; lane 4, ‘Golden Promise’. Actin was the chosen reference sequence.
The analysis of natural variants in barley demonstrated that cleistogamy is determined by the single recessive gene cleistogamy1 (cly1) (Turuspekov et al., 2004; Honda et al., 2005). The product of this gene (CLY1) is an APETALA2 (AP2)-type transcription factor (Nair et al., 2010). A single synonymous nucleotide polymorphism within the cly1 coding sequence differentiates the cly1.b (cleistogamous) from the Cly1.a (non-cleistogamous) sequence. This polymorphic site lies within a 21-nucleotide (nt) sequence targeted by the microRNA (miRNA) miR172 (Supplementary Data Fig. S1A), and is thought to disrupt the interaction between miR172 and cly1 transcript (Nair et al., 2010). In non-cleistogamous barleys, the assumption is that miR172 downregulates cly1, resulting in the development of normal lodicules. Besides the proposed miR172-directed post-transcriptional regulation (Fig. 1B), cly1 has been shown to be regulated at transcriptional level by an epiallele that represses cly1 transcription independently of miR172 action (Wang et al., 2015).
The miRNAs perform a variety of regulatory functions in plant development (Bartel, 2004). Their mature form is represented by short (21–24 nt) sequences able to either induce the degradation of a target mRNA or to inhibit its translation. Their specificity relies on sequence complementarity with the target. The target of miR172 is the transcript of the AP2 gene (Park et al., 2002; Chen, 2004), along with a small group of AP2-like genes (Aukerman and Sakai, 2003; Schmid et al., 2003; Schwab et al., 2005). The gene encoding miR172 is transcribed by RNA polymerase II and a full-length miRNA transcript termed ‘primary miR172’ (pri-miR172) is then processed to form a 21-bp RNA duplex consisting of the active strand and its complementary strand (Xie et al., 2005; Mateos et al., 2010). The various miR172s function in floral development, including floral transition and floral patterning (Lauter et al., 2005; Chuck et al., 2007; Zhu et al., 2009; Luo et al., 2013; Yumul et al., 2013), roles that have been retained throughout the angiosperms. The present study sought to identify the set of barley miR172 genes, and to determine which (if any) contribute to cleistogamy. A secondary aim was to explore whether the downregulation of cly1 by miR172 operates through either the cleavage of cly1 mRNA, as previously anticipated (Nair et al., 2010), or the inhibition of cly1 translation.
MATERIALS AND METHODS
Plant materials
The barley cultivar ‘Azumamugi’ (AZ, NIAS GenBank accession number JP17209) was taken as representative of the non-cleistogamous type, while cultivar ‘Kanto Nakate Gold’ (KNG, JP15436) was used as the cleistogamous type. The material was sown in the field during the autumn at Tsukuba, with an inter-plant and inter-row spacing of 20 and 80 cm, respectively. The Ds-induced mutant Ds-miR172a, which harbours a Ds element inserted into one of the barley miR172 in a cultivar ‘Golden Promise’ (USDA-ARS accession number PI 343079) background (Brown and Bregitzer, 2011) was crossed and backcrossed with the non-cleistogamous cultivar ‘Conlon’ (PI 597789), and the resulting BC1F2 progeny were raised in a greenhouse.
Measurement of lodicule volume
At least three randomly selected spikes, including the flag leaf and peduncle, were taken from each entry at the yellow anther stage (Kirby and Appleyard, 1981) and florets were sampled from the central portion of the spikes. After removing the lemma, the lodicules were photographed and the resulting images were used to measure lodicule width and depth (surrogates of lodicule volume), following Nair et al. (2010).
Sequencing and bioinformatic analysis of small RNAs (sRNAs)
An sRNA library prepared from immature spikes of the barley cultivar ‘Morex’ (Wang et al., 2015) was sequenced, and reads in the length range 17–36 nt were sorted into unique sequences, retaining a count of copy number to generate the metric ‘number of reads per million reads’ (RPM). MOODS software (Korhonen et al., 2009) was employed as an aligner to identify any set of miRNAs potentially targeting cly1, by allowing a maximum of six mismatches between the miRNA sequence and the cly1 full-length cDNA (flcDNA) including untranslated region (UTR) sequence. The sequences were then filtered based on a penalty score whereby each mismatch was assigned a score of 1, a wobble (G:U) mismatch a score of 0.5 and a bulge in either RNA strand a score of 2. The threshold score for retention of a candidate was 4.0, following the selection criterion of 3.5 as described by Jones-Rhoades and Bartel (2004). The approach based on a penalty score is used frequently (Lelandais-Briere et al., 2009; Joshi et al., 2010; Kim et al., 2012). The selected sequences were then subjected to a search against the miRBase (http://microrna.sanger.ac.uk) set of all known miRNAs (Kozomara et al., 2011) using the BLAST+ algorithm (Camacho et al., 2009), and assigned to their best-match miRNA family. To identify miRNAs that could potentially bind to an miR172 target site, we also used the BLAST+ ‘blast-short’ and psRNATarget (Dai and Zhao, 2011) as the aligner and the target prediction program, respectively.
Identification of barley miR172 genes
The selected miRNA sequences were aligned against the ‘Morex’ whole-genome sequence (WGS) (Mayer et al., 2012) through the use of Bowtie software (Langmead et al., 2009). Sequences that aligned perfectly were considered to be a putative coding locus for a given miRNA. The 400-nt context of an miR172 site was used to predict the energetically most favourable RNA secondary structure, based on the RNA folding prediction program mfold v3.5 (Zuker, 2003). These structures were evaluated based on three criteria: (1) the ability to fold into a stem–loop structure with the mature miRNA located on the stem–loop’s helical region; (2) a maximal free folding energy (mfe) lower than that of tRNA and rRNA; and (3) a predicted mature miRNA: its complementary miRNA duplex having fewer than six mismatches and a maximum of three nucleotides present in any loop or bulge (Zhang et al., 2006; Qiu et al., 2007; Xie et al., 2007). GENETYX v8.0 software (Software Development, Tokyo, Japan) was used to perform a multiple sequence alignment of barley miR172 precursor sequences with those represented in miRBase.
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from leaves and immature spikes following Komatsuda et al. (1998). The primers used to amplify and sequence the amplicons are detailed in Supplementary Data Table S1. Each 10-μL PCR contained 25 ng of template, 0.5 U of ExTaq DNA polymerase (Takara, Tokyo, Japan), 1× buffer, 2.5 mm MgCl2, 200 μm dNTP and 300 nm of each primer, and the cycling regime comprised a 94 °C/5 min denaturation, followed by 30 cycles of 94 °C/30 s, 55–60 °C (primer-dependent)/30 s, 72 °C/60 s, with a final 72 °C/10 min extension. The amplicons were purified using a QIAquick PCR Purification kit (Qiagen, Tokyo, Japan) before submission for sequencing. The Ds-miR172a mutant was PCR-validated by amplifying from a Ds-specific primer (Ds3325F) in conjunction with an miR172a-specific primer (508-3′ R) (Brown and Bregitzer, 2011). The allelic state at Cly1 was determined by a genotypic test based on the marker P101AP25′ NmuCI (Nair et al., 2010).
RNA extraction, reverse transcription PCR (RT–PCR) and quantitative real time PCR (qRT–PCR)
Total RNA was extracted using a mirVana miRNA Isolation Kit (Life Technologies) from a bulk sample of three to ten immature spikes that had been developmentally staged (Kirby and Appleyard, 1981). The RNA was quantified using a NanoDrop 1000 device (Thermo Fisher Scientific, Waltham, MA, USA), then treated with RNase-free DNase (Takara Bio, Otsu, Japan) to remove any contaminating genomic DNA. The cDNA first strand was synthesized using the SuperScript III system (Invitrogen, Carlsbad, CA, USA) primed by oligo-dT, to provide the template for RT–PCRs based on primers listed in Supplementary Data Table S1. A series of 25-μL qRT–PCRs was based on the TaqMan system (probe and primer details given in Supplementary Data Table S1). The barley Actin gene (accession number DN182500) was used as the reference. In a study conducted to identify a suitable reference gene across diverse barley cultivars and tissues, Actin was identified as the only traditional reference gene that demonstrated a highly reliable and stable expression pattern (Gines et al., 2017). All reactions were performed on a CFX96 Real-Time System device (Bio-Rad, Tokyo). The abundance of each pri-miR172 transcript was quantified in a 25-μL qRT–PCR containing 900 nm forward and reverse primers for 3′ end unique sequence of pri-miR172 (except for miR172b, for which 600 nm of each primer was used), 200 nm fluorescein amidite (FAM) double-quenched TaqMan miR172 probe, 120 nm primers targeting Actin, 120 nm Texas Red double-quenched TaqMan Actin probe and 2× SsoAdvanced Universal Probe Supermix (Bio-Rad, Tokyo, Japan). The cycling parameters were 95 °C/30 s, followed by 50 cycles of 95 °C/10 s, 57 °C/15 s. The abundance of cly1 transcript was quantified using 900 nm of each primer and 250 nm of the TaqMan cly1 probe (Supplementary Data Table S1) in TaqMan Gene Expression Master Mix (Applied Biosystems, Life Technologies Japan, Tokyo). The cycling regime consisted of a 95 °C/10 min denaturation, followed by 40 cycles of 92 °C/30 s, 60 °C/60 s and a final 60 °C/10 min extension. Actin was quantified using 300 nm of each primer and 200 nmActin TaqMan probe in 2× SsoAdvanced Universal Probe Supermix, with a cycling regime of 95 °C/30 s, followed by 45 cycles of 95 °C/10 s, 57 °C/15 s. The transcript abundance of the barley Q gene homologue was quantified using 600 nm of each primer and 200 nM of the relevant TaqMan probe (Supplementary Data Table S1) in 2× SsoAdvanced Universal Probe Supermix, with a cycling regime of 95 °C/30 s, followed by 45 cycles of 95 °C/10 s, 52 °C (Q exon 10) or 64 °C (Q 3′-UTR)/30 s. Actin was quantified in a separate reaction as described above. At least three biological replicates per sample were run, with each replicate represented by at least three technical replicates. To determine the absolute abundance of individual transcripts, the relevant fragment was cloned into pCR4-TOPO (Invitrogen, Carlsbad, CA, USA) and a serial dilution of recombinant plasmids was used to generate a standard curve. The raw CT values were converted into an absolute copy number by the use of a standard curve. Relative abundances of each transcript were calculated from (target mRNA copy number ng−1 total RNA)/(Actin mRNA copy number ng−1 total RNA).
Quantification of mature miR172
A qRT–PCR targeting mature miR172 was performed using the TaqMan MicroRNA assay (Applied Biosystems) (for details see Supplementary Data Table S1), following the manufacturer’s protocol. The small nucleolar RNA (snoRNA) Hv-snoR13 was used as an endogenous control (http://bioinf.scri.sari.ac.uk/cgi-bin/plant_snorna/home), because it is both abundant and stable, because its size is close to that of miRNAs, because its assay format is similar to that of miRNAs, and because it is unlikely to be involved in any miRNA regulatory pathway. The qRT–PCR was achieved via a customized TaqMan small RNA assay (Applied Biosystems, Tokyo, Japan), which comprised a specific stem–loop reverse transcription primer along with appropriate conventional primers and an FAM dye-labelled probe (Supplementary Data Table S1). Quantification of mature miR172s was based on the ΔΔCT method (Livak and Schmittgen, 2001), and the data are presented in the form of fold differences following normalization against the abundance of snoR13 present at the stamen primordium stage. At least three biological replicates at each developmental stage were performed, and each replicate was represented by at least three technical replicates.
RNA in situ hybridization
Immature spikes were fixed, processed and sectioned as described by Komatsuda et al. (2007). Double digoxigenin (DIG)-labelled miRCURY Locked Nucleic Acid (LNA) miRNA detection probes (Supplementary Data Table S1) complementary to miR172a, miR172b and a scrambled miRNA (control) were purchased from Exiqon (Foster City, CA, USA) for the detection of mature miR172s. Each slide was treated with 180 µL of hybridization solution containing 0.225 µL of 25 µm probe, and the subsequent hybridization and washing steps were performed at either 55 °C (miR172) or 60 °C (snoR13). To detect cly1 mRNA, the probe used was a 280-nt fragment amplified from the cly1 3′-UTR (primers are given in Supplementary Data Table S1), while for the Q orthologue the probe was a 265-nt fragment amplified from the 3′-UTR (primers are given in Supplementary Data Table S1). For cly1 and Q hybridization, each slide was treated with 180 µL of hybridization solution containing 0.1–0.25 µL of probe generated by a DIG RNA labelling mix according to the manufacture’s protocol (Roche, Mannheim). The subsequent in vitro transcription and in situ hybridization protocols followed methods described by Komatsuda et al. (2007).
Laser microdissection
Immature spikes were fixed by vacuum infiltration in cold 75 % ethanol/25 % acetate, and embedded in paraffin using a microwave processor (Energy Beam Sciences, East Granby, CT, USA) following Takahashi et al. (2010). The paraffin-embedded samples were cut into 10-μm thick sections using an RM2255 microtome (Leica Microsystems, Wetzlar, Germany), which were then subjected to laser microdissection using a Veritas Laser Microdissection System LCC1704 device (Life Technologies). Total RNA was extracted from the samples using a Pico-Pure RNA Isolation Kit (Life Technologies), following the manufacturer’s protocol. The quantity and quality of the RNA obtained were checked with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The synthesis of cDNA and subsequent qRT–PCRs were performed as described above.
Immunoblotting analysis
A synthetic NH2-LQKNGFHSLARPT-OH peptide (CLY1 positions 475–487) was inoculated into rabbits to derive a polyclonal antibody (Sigma-Aldrich, St Louis, MO, USA). The antiserum was passed through an affinity column containing the epitope peptide. A monoclonal antibody against α-tubulin was purchased from Sigma-Aldrich (St Louis, MO, USA). Immature barley spikes were frozen in liquid nitrogen and stored at −80 °C until required. A bulk of five to ten spikes was first coarsely crushed, then finely homogenized in a BioMasher II device (Nippi, Tokyo, Japan). The resulting powder was extracted in 100 µL of 50 mm Tris–HCl (pH7.5)/150 mm NaCl/0.5 % v/v Triton X100/5 mm EDTA/5 mm EGTA and 1× complete EDTA-free Mini Protease Inhibitor mixture (Roche, Basel, Switzerland) and centrifuged (3000 g, 10 min), and the supernatant was denatured by adding 100 µL of 50 mm Tris–HCl (pH 6.8)/2 % w/v SDS/6 % v/v 2-mercaptoethanol/bromophenol blue (BPB)/10 % v/v glycerol, and holding at 65 °C for 10 min. The proteins were electrophoretically separated using a 10 % polyacrylamide gel, then electrophoretically transferred to a PVDF membrane (Merck Millipore, Darmstadt, Germany). The membranes were initially challenged with a 1/5000 dilution of primary antibody in 10 mm Tris–HCl (pH7.5)/150 mm NaCl/0.1 % v/v Tween 20/1 % w/v skimmed milk, rinsed thoroughly, then incubated in a 1/10 000 dilution of horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) (for CLY1) or a 1/10 000 dilution of anti-mouse IgG (for α-tubulin) in the same buffer. Signal was detected using ImmunoStar (Wako, Osaka, Japan) and ImageQuant LAS3000 (GE Healthcare, Chalfont St Giles, UK), and its intensity was analysed using Multi Gauge v2.0 software (Fujifilm, Tokyo, Japan).
RESULTS
Mature miR172s detected in sRNA library
A total of 150 million sRNA reads (accession number DRA006260) were recovered from the ‘Morex’ immature spike library by Wang et al. (2015). After removal of adaptor sequences, rRNAs and tRNAs, and filtering by length (17–36 nt), a set of 124 196 599 (mean length 25.1 nt) was retained. These resolved into 22 755 479 unique sequences (mean length 24.4 nt) to form the population of putative sRNAs (Supplementary Data Fig. S1B). The length, distribution and abundance of sRNA reads is shown in Supplementary Data Figure S1B, where the analysis range was expanded in order to capture the existence of unusual types of miRNAs as referred by Kim et al. (2012). When the sRNAs were aligned with the ‘Morex’ cly1 flcDNA sequence, the most hits (Supplementary Data Fig. S1A) were obtained in the region between positions 1850 and 1870 conserved in plants (Supplementary Data Fig. S1C). Applying the filter within the MOODS software based on a maximum penalty score of 4.0 and RPM >1 to the miRNAs complementary to the target site (Table 1) identified three distinct sequences: miR172a (21 nt), b (21 nt) and c (20 nt) (Fig. 1C). miR172a differed from b at the 5′ end (G versus A), while c lacked the 3′ terminal U (Table 1). The sequence of miR172a is identical to the miR172 species that was interrupted by a Ds insertion (Brown and Bregitzer, 2011). The three barley miR172s were conserved in plants (Supplementary Data Fig. S1C) and the hybrids between each of the isoforms and cly1 mRNA (Fig. 1C) passed the complementarity criteria described by Jones-Rhoades and Bartel (2004). Thus, at least three mature miR172s produced in the immature barley spike appeared capable of interacting with cly1 mRNA. Use of the blast-short and psRNATarget software packages produced an identical outcome (data not shown). The RPM counts for the three isoforms were, respectively, 70.9, 21.6 and 1.3 (Table 1), suggesting a difference of almost two orders of magnitude in their abundance.
Table 1.
Mature miR172s and corresponding genes in barley
| sRNA sequence (5′–3′) | Length | No. of reads | RPMa | Target site in cly1b | Penaltyc | Mature miR172 identityd | Gened | WGS contig IDe | Chromosomee |
|---|---|---|---|---|---|---|---|---|---|
| GGAAUCUUGAUGAUGCUGCAU | 21 | 6693 | 70.9 | 1850–1870 | 3 | miR172a | Hv-miR172a | Contig_38787 | 3HL |
| AGAAUCUUGAUGAUGCUGCAU | 21 | 2036 | 21.6 | 1850–1870 | 4 | miR172b | Hv-miR172b | Contig_49738 | 6HL |
| GGAAUCUUGAUGAUGCUGCA | 20 | 118 | 1.3 | 1851–1870 | 2 | miR172c | Hv-miR172c | Contig_1561881 | 7HS |
aReads per million; 106 × number of reads/total number of reads.
bPosition of the cly1 full-length cDNA (accession number KJ363931.1) aligned with miR172.
cPenalty score of each miRNA for the cly1 is shown, calculated as 0.5 points assigned to each G:U wobble, 1 point to each non-G:U mismatch and 2 points to each bulged nucleotide in either RNA strand.
dIDs given in the present study.
eBased on miR172 isoform alignment to ‘Morex’ whole-genome sequencing contigs with no mismatches.
Identification of miR172 loci
When the miR172 isoform sequences were aligned with the 2 670 738 WGS contigs, perfect matches were obtained for miR172a on contig_38787 (chromosome arm 3HL), for miR172b on _49738 (chromosome arm 6HL) and for miR172c on _1561881 (chromosome arm 7HS) (Table 1). The location of miR172a on chromosome arm 3HL is consistent with the 3HL location as determined by wheat–barley chromosome addition lines (Brown and Bregitzer, 2011). To assess whether any of the genomic sequences was capable of forming the signature miR172 stem–loop hairpin, the selected region’s secondary structure was analysed in silico. Each of the three sequences were predicted to fold into an appropriate stem–loop hairpin, and harboured the mature miR172 sequences on their 3′ arm. Thus, the encoding loci were denominated Hv-miR172a, b and c (Fig. 1D). Similarity between the precursors was limited to the sequences of the mature miRNA and its complementary (Supplementary Data Fig. S2). [Note that, similarly, the miR172 precursor sequences flanking the mature miRNA in other plant species are all highly divergent (Supplementary Data Fig. S2).] The miR172a locus is identical to GenBank accession HM243624, but neither miR172b nor miR172c has been identified previously. A subsequent RT–PCR experiment confirmed that all three genes were transcribed in the immature spike (Fig. 1E), although, in agreement with the RPM counts, the abundance of miR172c was very low (Table 1).
The abundance of pri-miR172 and mature product
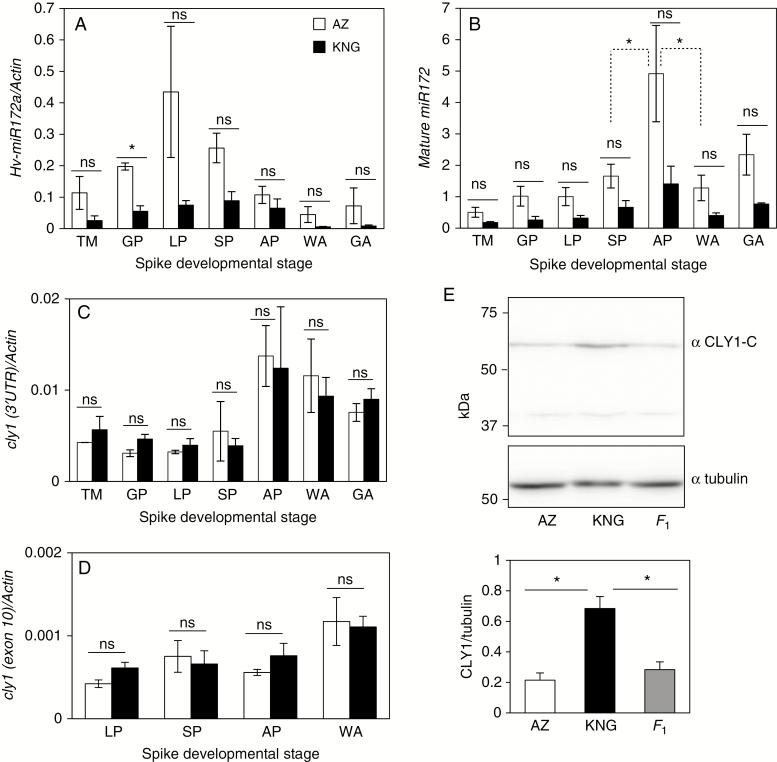
The abundance of pri-miR172 was compared over the course of spike development in cultivars ‘Azumamugi’ (AZ, non-cleistogamous) and ‘Kanto Nakate Gold’ (KNG, cleistogamous). To distinguish between the pri-miR172s generated from the three genes, their divergent 3′-arm sequences (in close proximity to the stem–loop) was targeted (Supplementary Data Fig. S3A). A standard curve (Supplementary Data Fig. S4) was used to convert the raw CT values into an absolute copy number. In AZ, the pri-miR172a RNA level was strongest during the lemma primordium stage, at which time it was considerably more abundant than in KNG (Fig. 2A). Its abundance was also higher in AZ than in KNG throughout the period between the glume primordium and stamen primordium stages. Mature miR172s determined by TaqMan miRNA assay were detected throughout the period of spike development (Fig. 2B), indicating that the miR172 sequences represented in the sRNA library were authentic. Their peak abundance occurred during the awn primordium stage, especially in AZ, in agreement with the abundance of pri-miR172s (Fig. 2A). This suggests that mature miR172s have a biological function, at least during the awn primordium stage, particularly in the non-cleistogamous cultivar AZ.
Fig. 2.
Expression of miR172 and cly1. (A–D) qRT–PCR-derived transcript abundance during spike development of (A) primary miR172a (B) mature miR172 (C) cly1 (3′-UTR) and (D) cly1 (exon 10). Actin was the chosen reference sequence. TM, triple mound stage; GP, glume primordium stage; LP, lemma primordium stage; SP, stamen primordium stage; AP awn primordium stage; WA, white anther stage; GA, green anther stage. Comparisons between SP and AP and between AP and WA for AZ are shown by the dashed lines in (B). Mean values for KNG were also significantly different between stages at the 5 % probability level. (E) CLY1 western blotting at the white anther stage (top) and abundance as determined by ratio of CLY1/tubulin abundance (bottom). α CLY1-C, anti-CLY1 C-terminal region; α tubulin, anti-tubulin. Values are mean and s.e. (n = 3 biological replications). *Means are significantly different at the 5 % probability level; ns, not significantly different.
Abundance of cly1 mRNA and CLY1 protein
A qRT–PCR analysis was used to characterize the transcription behaviour of cly1 over the period of spike development. When a pair of primers in the 3′-UTR was used (Supplementary Data Fig. S3B), the profiling showed that the gene was transcribed throughout the whole period (Fig. 2C), but that its transcript abundance was much higher (by almost 2-fold) when entering the awn primordium stage than earlier, consistent with the notion that CLY1 may regulate lodicule development. The temporal pattern of transcription was similar in AZ and KNG, and the transcript abundances were comparable (Fig. 2C). The abundance of the transcript remained comparable between the two cultivars (Fig. 2D) when a pair of primers flanking the miR172-binding site was used (Supplementary Data Fig. S3B). This result suggests that miR172-directed cleavage of cly1 mRNA, if it occurs, is minor. It was thought that miR172 functions primarily via translational repression (Chen, 2004; Chuck et al., 2007). This hypothesis was therefore tested by immunoblot analysis using an antibody raised against a C-terminal NH2-LQKNGFHSLARPT-OH peptide (positions 475–487) of CLY1. This sequence is unique to CLY1 and not found in other barley proteins by BLAST search. An immunoblot analysis showed that the level of CLY1 accumulated in KNG was ~3-fold higher (P < 0.001) than in AZ and their F1 plants (Fig. 2E). Given that lodicules swell in the F1 (Nair et al., 2010), the lower levels of CLY1 in F1 plants are consistent with this phenotype. Since the level of cly1 transcript abundance was shown to be comparable in AZ and KNG (Fig. 2C, D), the clear inference was that miR172 compromises cly1 translation efficiency. There was a difference of ~10-fold in of cly1/Actin transcript level ratio from the qPCR with the primer (Fig. 2C) and that with the exon 10 primers (Fig. 2D), but it is not unusual to see a lower signal with primers more distant from the 3′ end.
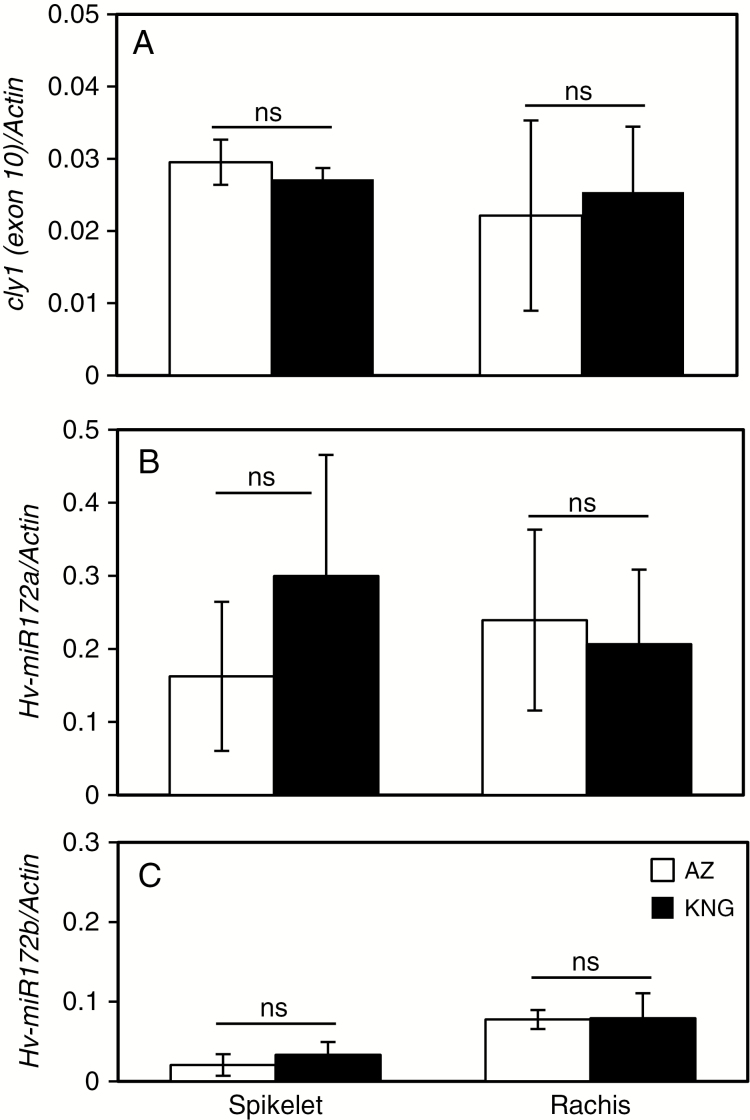
When the RNA template was extracted from spikelets and rachis separately using laser microdissection (Supplementary Data Fig. S5), once again the abundance of cly1 mRNA in AZ proved to be similar to that in KNG, even when the primers used to amplify the template flanked the miR172 binding site (Fig. 3A). The abundance of miR172a transcript was similar in the spikelet and rachis, and did not differ between AZ and KNG (Fig. 3B). The implication was that miR172a is involved in the development of both the spikelet and the rachis. The abundance of miR172b transcript in the spikelet was ~3-fold lower than in the rachis, a ratio that was consistent between AZ and KNG (Fig. 3C).
Fig. 3.
Abundance of cly1 and primary miR172 transcripts in the spikelet (flower) and rachis in immature spikes at awn primordium stage. The tissues were laser-microdissected (Supplementary Data Fig. S5). Values are mean and s.e. (n = 3 biological replications). ns, means are not significantly different at the 5 % probability level.
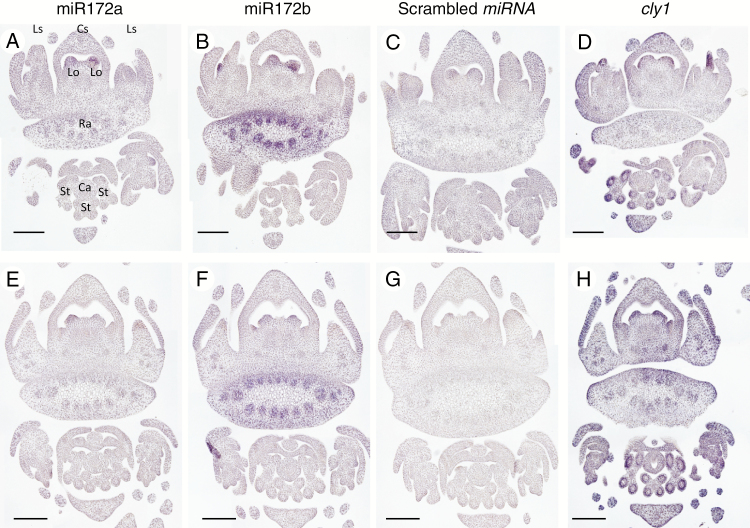
Localization of mature miR172 and cly1 transcript
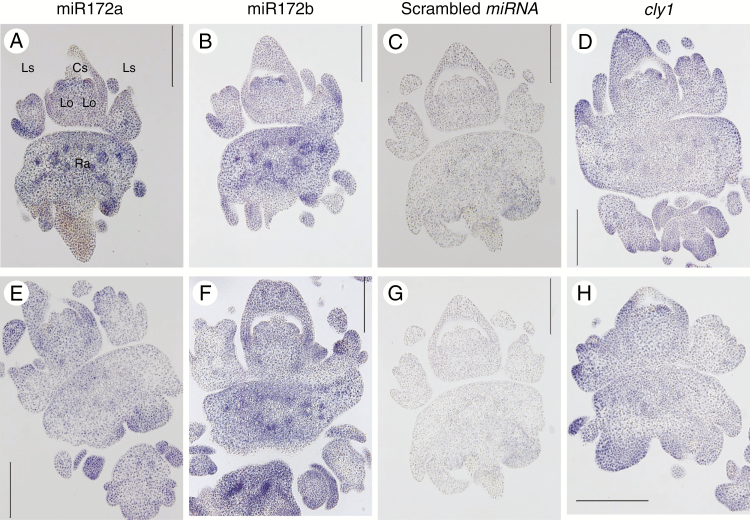
RNA in situ hybridization was conducted on immature spikes at the awn primordium stage to define the sites where mature miR172 and cly1 transcripts were deposited. Mature miR172 was detected using miRCURY Locked Nucleic Acid (LNA) probes, which deliver enhanced hybridization affinity, sensitivity and specificity. Hybridization with the miR172a probe generated signal in the lodicule primordia and rachis, but not in either the anthers or any other organs (Fig. 4A, E). The pattern of transcript deposition was the same in AZ and KNG (Fig. 4A, E). A similar hybridization profile was obtained when the miR172b probe was used (Fig. 4B, F). When the scrambled miRNA probe was used, no signal was detected (Fig. 4C, G) confirming the specificity of the two miR172 probes. The observed abundance of miR172 in the lodicules was fully consistent with its proposed regulatory role in lodicule development. The cly1 signal was concentrated in the lodicule primordia in both AZ and KNG (Fig. 4D, H), overlapping the site of miR172 deposition.
Fig. 4.
Localization of mature miR172s and cly1 by RNA in situ hybridization in the immature spike sampled at the awn primordium stage. Plant genotypes were AZ (Cly1.a) (A–D) and KNG (cly1.b) (E–H). Spike RNAs were hybridized with miR172a probe (A, E), miR172b probe (B, F), scrambled miRNA probe (negative control) (C, G) and cly1 probe (D, H). Cs, central spikelet; Ls, lateral spikelet; Lo, lodicule; Ra, rachis; Ca, carpel; St, stamen. Scale bars = 0.1 mm.
Effect of absence of miR172a on lodicule development
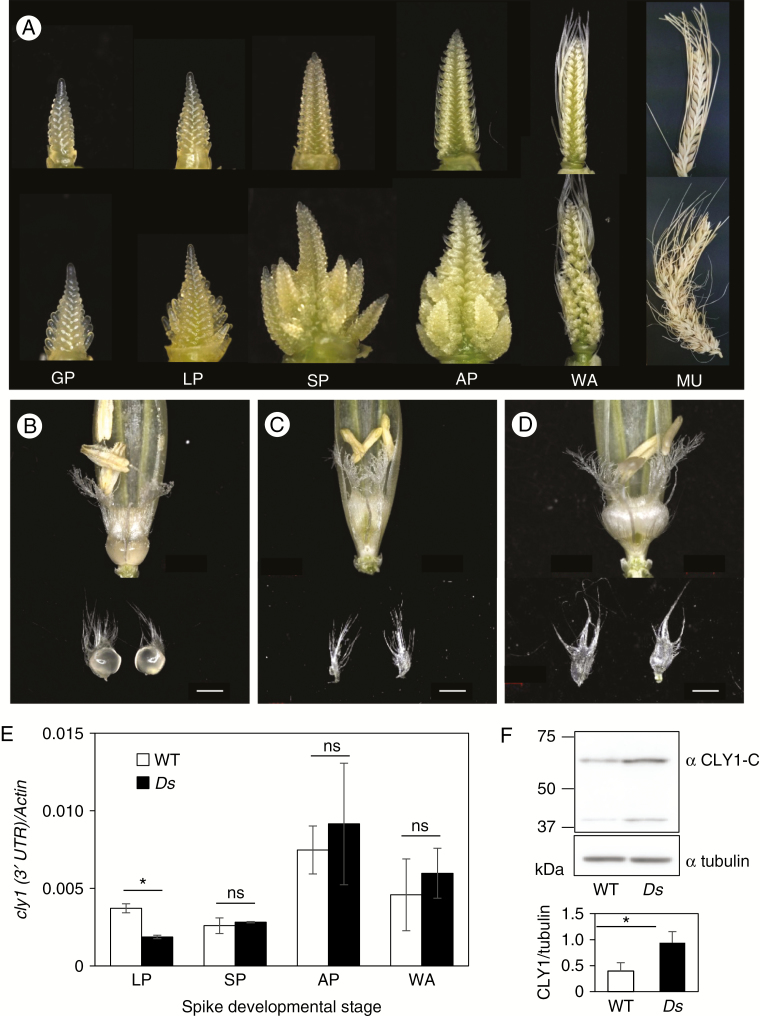
The Ds-miR172a mutant [#508–72, released as TNP 280, GenBank accession number HM243624 (Brown et al., 2014)] harboured a 3.6-kb Ds insertion within the 21-nt miR172 sequence of Hv-miR172a, lacked any mature miR172a, and produced an abnormal flower (Brown and Bregitzer, 2011). Since the genetic background of the mutant (cultivar ‘Golden Promise’) is a cly1.b carrier producing small lodicules, it is not possible to infer the effect of the absence of cly1 regulation by miR172a on lodicule size in the mutant. To examine this, the mutant was crossed and backcrossed with cultivar ‘Conlon’ (miR172a/Cly1.a carriers) to allow the selection of homozygotes in the BC1F2 generation for miR172a/cly1.b, miR172a/Cly1.a and Ds-miR172a/Cly1.a (Table 2). The abnormality of flower formation was inherited from the mutant into the derivatives (Fig. 5A). The abnormal spike phenotype of the Ds mutant was apparent as early as the glume primordium stage. Inspection of the Ds-miR172a/Cly1.a spike revealed branch-like meristems arranged in an indeterminate pattern, in contrast to the single spikelet meristem characteristic of the wild-type spike. In the Ds mutant, spikelets containing abnormal and sterile floral structures were formed alongside the normal spikelets (Fig. 5A). The lodicules of Ds-miR172a/Cly1.a were very small, even smaller than those formed by miR172a/Cly1.a (Fig. 5B, C). The lodicules formed by the Ds-miR172a/Cly1.a genotype were smaller than those formed by miR172a/cly1.b (Fig. 5D), which demonstrated the major influence of miR172a on lodicule size (Fig. 5C). Other floral structures were similar with respect to both number and size across all three homozygous genotypes (Fig. 5B–D). The temporal pattern of cly1 transcription was similar in miR172a/Cly1.a homozygotes (WT) and Ds-miR172a/Cly1.a homozygotes (Ds), and transcript abundances were comparable in the WT and Ds (Fig. 5C), a result consistent with transcript abundance in AZ and KNG and translational rather than transcriptional repression. Furthermore, immunoblot analysis showed that the CLY1 level in the Ds was higher than that in the WT (Fig. 5F), indicating that the absence of miR172a abolished the suppression of cly1 translation.
Table 2.
Spike phenotypes in plants with different combinations of Hv-miR172a and cly1 alleles
| Crosses | Line | Locus | Lodicules | Determinacya | |||
|---|---|---|---|---|---|---|---|
| Hv-miR172a | cly1 | Width (mm) mean ± s.e. | Depth (mm) mean ± s.e. | Overall size | |||
| Parents | ‘Conlon’ | miR172a | Cly1.a | 1.14 ± 0.12 | 1.12 ± 0.16 | Large | Wild-type |
| ‘Morex’ | miR172a | Cly1.a | 0.94 ± 0.08 | 1.11 ± 0.18 | Large | Wild-type | |
| GP | miR172a | cly1.b | 0.46 ± 0.05 | 0.28 ± 0.04 | Small | Wild-type | |
| #508-72b | Ds-miR172a | cly1.b | 0.19 ± 0.01 | 0.19 ± 0.01 | Rudimentary | Indeterminate | |
| Conlon*2/#508-72c | TC11-7 #5 | miR172a | Cly1.a | 0.86 ± 0.09 | 1.00 ± 0.04 | Large | Wild-type |
| TC11-7 #4 | miR172a | cly1.b | 0.35 ± 0.03 | 0.41 ± 0.01 | Small | Wild-type | |
| TC11-7 #1 | Ds-miR172a | Cly1.a | 0.22 ± 0.05 | 0.21 ± 0.02 | Rudimentary | Indeterminate | |
| Morex*2/#508-72c | TC11-9 #1 | miR172a | Cly1.a | 0.82 ± 0.07 | 0.81 ± 0.04 | Large | Wild-type |
| TC11-9 #4 | miR172a | cly1.b | 0.42 ± 0.07 | 0.40 ± 0.06 | Small | Wild-type | |
| TC11-9 #14 | Ds-miR172a | Cly1.a | 0.19 ± 0.03 | 0.16 ± 0.04 | Rudimentary | Indeterminate | |
aSpike and spikelet meristem determinacy.
b Ds-miR172a mutant in GP.
c BC 1 F 2 generation; lines were homozygous for the mentioned loci.
Fig. 5.
Effect of absence of miR172a on inflorescence phenotype and accumulation of CLY1. (A) miR172a/Cly1.a (top) and Ds-miR172a/Cly1.a (bottom) at stages from glume primordium (GP) to maturity (MU). LP, lemma primordium stage; SP, stamen primordium stage; AP, awn primordium stage; WA, white anther stage. (B–D) Variation in lodicule size at anthesis in (B) miR172a/Cly1.a homozygote (C) Ds-miR172a/Cly1.a homozygote and (D) miR172a/cly1.b homozygote. Scale bars = 1 mm. (E) qRT–PCR-derived transcript abundance of cly1. Actin was the reference transcript. (F) Western blot analysis of CLY1 in miR172a/Cly1.a (WT) and Ds-miR172a/Cly1.a (Ds) (top) and abundance as determined by ratio of CLY1/tubulin abundance (bottom). Values in (E) and (F) are mean and s.e. (n = 3 biological replications). *Means are significantly different at the 5 % probability level; ns, not significantly different.
The miR172a in situ hybridization signal was detected in the lodicules and rachis of the WT progenitor of the mutant (Fig. 6A) but not in the Ds-miR172a mutant (Fig. 6E), confirming that miR172a signal is specific to Hv-miR172a. Hybridization with an miR172b probe produced the same profile as generated by miR172a in the WT (Fig. 6B), while in the mutant some low-level signal was present in the rachis but none in the lodicules (Fig. 6F). This result implies that miR172b is not expressed sufficiently to produce signal in lodicules in the absence of miR172a. An alternative explanation could be the possibility of cross-hybridization of miR172b with miR172a in the WT. Signal for cly1 was detected in both the lodicules and the rachis (Fig. 6D) and the pattern was not disturbed by the absence of miR172a (Fig. 6H).
Fig. 6.
Effect of absence of miR172a on accumulation of miR172 and cly1 transcripts in the immature spike sampled at the awn primordium stage. Plant genotypes were miR172a/Cly1.a (A–D) and Ds-miR172a/Cly1.a (E–H). Spike RNAs were hybridized with miR172a (A, E), miR172b (B, F), scrambled miRNA (negative control) (C, G) and cly1 (D, H). Cs, central spikelet; Ls, lateral spikelet; Lo, lodicule; Ra, rachis. Scale bars = 0.2 mm.
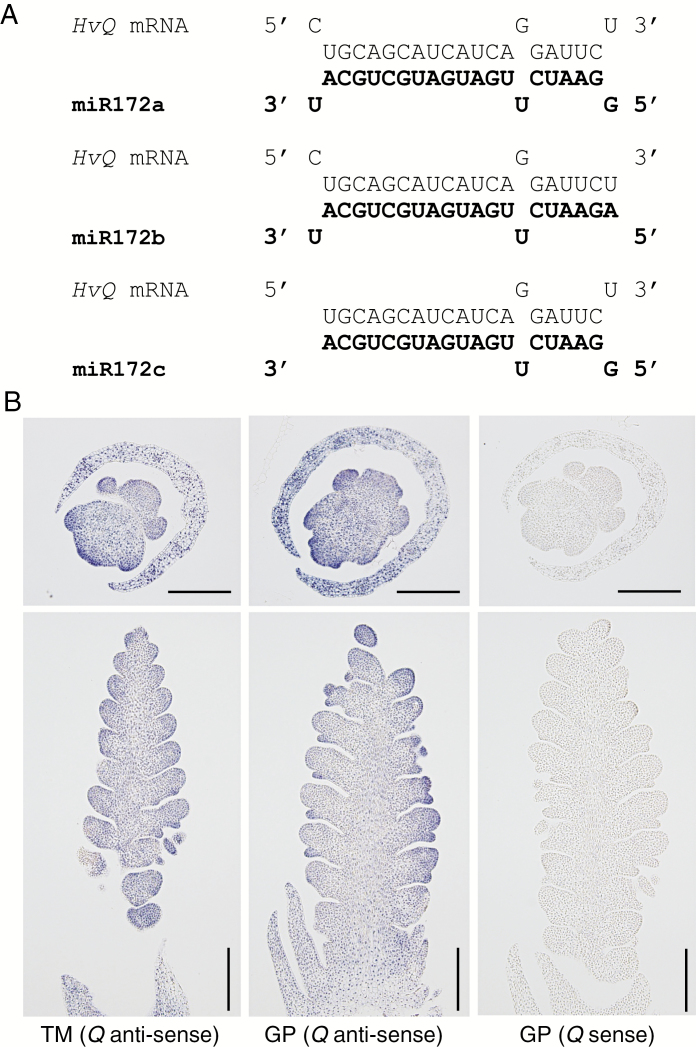
Expression of barley Q
The cly1 sequence is related to three other barley HvAP2-like genes, one of which (HvAP2L, GenBank accession number AY069953) is assumed, from phylogenetic analysis (Nair et al., 2010), to be the orthologue of the wheat free-threshing gene Q, Both Q and cly1 encode AP2 proteins of class A of the ABCDE module, which control flower pattering (Luo et al., 2016). The Q mRNA sequence has a short sequence complementary to miR172 (Fig. 7A). It was of interest therefore to investigate whether miR172a also interacted with Q. An RNA in situ hybridization experiment was conducted (Fig. 7B) using as probe a 265-nt 3′-UTR fragment of Q (Supplementary Data Fig. S3C, Table S1). The transcript of Q was detectable at the triple mound stage in the six-rowed barley cultivar AZ, coinciding with the differentiation of spikelet primordia (Fig. 7B). Transcript was initially restricted to the apex of the triple spikelet meristem, but later spread to the glume primordia and later still to the floral meristem (Fig. 7B). No signal was detected in hybridization with the sense probe for Q (negative control) (Fig. 7B).
Fig. 7.
Interaction of barley Q with miR172. (A) Potential interactions between Q mRNA and miR172 isoforms (‘Morex’). (B) Localization of Q mRNA in the immature spike sampled at the triple mound (TM) and glume primordium (GP) stages in AZ, as detected by RNA in situ hybridization with an anti-sense and sense Q (3′-UTR) transcript. Scale bars = 0.2 mm in horizontal sections and 1 mm in longitudinal sections of immature spikes.
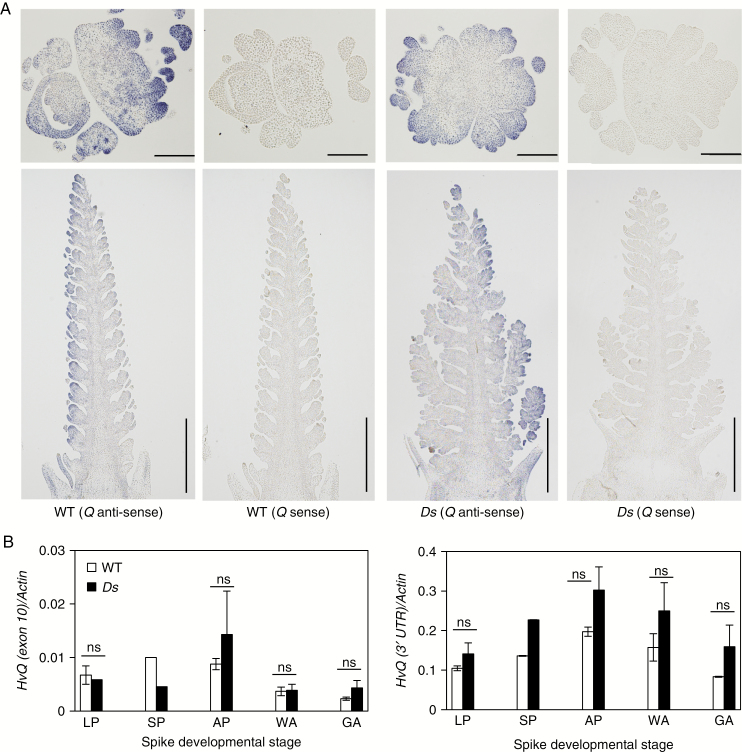
Both Ds-miR172a/Cly1.a and miR172a/Cly1.a homozygotes had a similar Q localization pattern, where Q transcript was concentrated in the spikelet and floral primordia (Fig. 8A). In a comparison of Q transcript abundance between miR172a/Cly1.a and Ds-miR172a/Cly1.a, there was no evidence for any genotypic variation at any of the spike developmental stages sampled (Fig. 8B). This result held whether the amplicon was directed at the 3′-UTR or at exon 10 of Q (Supplementary Data Fig. S3C). The spike of the Ds-miR172a/Cly1.a genotype exhibited abnormal spikelet development, including the conversion of glumes to florets in the apical regions of the spike and a branching phenotype in the basal region, with each branch consisting of supernumerary spikelets and various floral defects (Fig. 5A). However, the pattern of Q transcription did not differ between Ds-miR172a/Cly1.a and the WT (Fig. 8B).
Fig. 8.
Expression of Q in immature barley spikes. (A) Localization of Q mRNA in an immature spike sampled at the awn primordium stage of WT (miR172a/Cly1) and Ds (Ds-miR172a/Cly1), as detected by in situ hybridization with an anti-sense and sense Q (3′-UTR) transcript. Scale bars = 0.2 mm in horizontal sections and 1 mm in longitudinal sections. (B) qRT–PCR-derived transcript abundance of Q during spike development in WT (miR172a/Cly1) and Ds (Ds-miR172a/Cly1). Values are mean and s.e. (n = 3 biological replications). LP, lemma primordium stage; SP, stamen primordium stage; AP, awn primordium stage; WA, white anther stage; GA, green anther stage. ns, means are not significantly different at the 5 % probability level.
DISCUSSION
Identification of mature miR172s and their genes
Although a single nucleotide mismatch at the miR172 binding site within cly1.b is sufficient to induce cleistogamy, how cly1 regulates lodicule development has not been explained as yet. Here, it has been confirmed that mature miR172s are generated in the immature spike, as shown by Nair et al. (2010). Studies exploring miRNAs in barley through a computational approach (Colaiacovo et al., 2010) or by means of next-generation sequencing technology (Schreiber et al., 2011; Curaba et al., 2012; Lv et al., 2012; Ozhuner et al., 2013; Hackenberg et al., 2015) reported the presence of miR172s in barley tissues, but none of these studies validated the precursor structure and described an miR172 gene. We have shown experimental evidence for barley miR172s, their precursors and primary transcripts, as well as their gene function. An effort was made here to quantify the abundance of miR172 sequences, using both qRT–PCR and sRNA sequencing. The clear result was that the predominant isoform of the three identified was miR172a. Analysis of various species has shown that miR172 is encoded by multiple loci (five in both Arabidopsis thaliana and maize, four in rice), although in some cases more than one gene has been shown to encode an identical mature isoform (Zhu and Helliwell, 2011). A survey of the barley genome sequence indicated that barley harbours three miR172 genes, although the current incomplete state of the genome sequence means that the final number may be greater than this. The predicted miR172 precursor structures were as expected, insofar as the mature sequences were all located on the 3′ arm of the transcript (Bartel and Bartel, 2003; Jones-Rhoades, 2012). The length and folding free energy of the precursors also lay within the expected range, and sequence conservation only applied to the mature miR172 and its complementary sequences, as is the general case for miRNAs (Bartel, 2004; Jones-Rhoades, 2012). The indications are therefore that the three barley miR172 loci Hv-miR172a (encoding miRNA172a), Hv-miR172b (encoding miRNA172b) and Hv-miR172c (encoding miRNA172c) are all authentic.
The abundance of mature plant miRNAs generated from their various genes is known to vary across growth stages and/or tissue types (Zhu and Helliwell, 2011). All three barley miR172 genes were transcribed in the immature spike, although miR172a was the dominant isoform, thereby implicating it as the leading candidate as the cly1 regulator. It was not possible to exclude the participation of miR172b because of its presence in both the lodicule and rachis, and no miR172b knockout mutant has as yet been isolated. The abundance of pri-miR172a was particularly high at the early stages of spike development, but the abundance of the mature form peaked rather later. Examples are known where pri-miRNAs are detectable but their mature products are not, or at least are only present at a lesser abundance (Ambros et al., 2003; Michael et al., 2003; Wulczyn et al., 2007; Lee et al., 2008). The implication was that pri-miR172s remained unprocessed until required (Jung et al., 2007). The abundance of both pri-miR172a and mature miR172s was particularly high in AZ, suggesting a transcriptional difference between AZ and KNG. This is consistent with the known role of AP2 in repressing miR172 transcription (Yant et al., 2010). Thus, in addition to differences in miR172 and target association due to sequence complementarity, which influences translation between the cultivars, having more of the miR172 may also act as a feedback loop to keep CLY1 levels down.
miR172a is essential for the development of the lodicule
The abundance of miR172 isoforms varies both spatially and temporally, contributing to their functional divergence (Yumul et al., 2013). When miR172a is absent, as in the Ds-induced Hv-miR172a knockout mutant (Brown and Bregitzer, 2011), the lodicule fails to develop normally, resulting in cleistogamous flowering. Thus miR172a activity is clearly required for non-cleistogamy. In rice, the overexpression of Os-miR172b (which generates a mature product identical to miR172a) induces larger (or a greater number of) lodicules (Zhu et al., 2009), suggesting that the barley and rice orthologues have an equivalent function. Overexpressors of Os-miR172a (which generates a mature product identical to miR172b) exhibit no distinct phenotype (Zhu et al., 2009), while the A. thaliana double loss-of-function mutant miR172a/miR172b (both genes generate a mature product identical to miR172b) similarly produces a WT flower (Zhao et al., 2007). Laser microdissection was used in the present study in an attempt to quantify miR172a and miR172b in a more precisely defined site, the spikelets separated from the rachis. What emerged was that miR172a was more abundant than miR172b. As the barley miR172a and miR172b sequences differed by a just a single nucleotide, this polymorphism might be responsible for their distinct biological functions.
miR172a downregulates the translation of CLY1
The increasing abundance of both mature miR172a and cly1 transcript as spike development proceeded is consistent with their biological interaction during lodicule growth. Not only was the accumulation of the transcripts quantified here, but also their site of deposition. The localization of miR172a within the lodicules supports the hypothesis that it is a determinant of lodicule development; at the same time, cly1 message accumulated in the lodicules, as also reported by Nair et al. (2010). In the lodicules of AZ, both miR172a and cly1 transcript accumulated, demonstrating that the presence of miR172a in itself did not deplete cly1 transcript. The abundance of cly1 transcript in AZ and KNG was almost identical, and the RNA in situ analysis showed that cly1 transcript was concentrated within both the lodicules and the anthers. The inference is that the AZ and KNG lodicules harbour a similar abundance of cly1 transcript. The implication was that miR172 regulated the translation of the cly1. This hypothesis was therefore tested by immunoblotting.
miR172s have been reported to regulate not only the abundance of AP2 transcript through transcript cleavage, but also to inhibit AP2 translation (Aukerman and Sakai, 2003; Kasschau et al., 2003; Chen, 2004; Schwab et al., 2005; Jung et al., 2007; Zhu et al., 2009). It was not practically feasible to recover sufficient RNA from the lodicule primordia, but cly1 transcript abundance was successfully measured separately in the spikelets and the rachis. Lodicules are the only organs where cly1 and miR172 expression overlaps within a spikelet. The abundance of cly1 transcript in laser-microdissected spikelet sections is likely to represent the cly1 transcript abundance in lodicules. The lack of any difference in cly1 transcript abundance in these two floral parts, both in AZ and KNG, lent strong support to the proposition that cly1 transcript is not degraded in non-cleistogamous cultivars. A similar conclusion has been drawn in both A. thaliana and rice (Jung et al., 2007; Zhu et al., 2009). Comparable cly1 transcript abundances were obtained in AZ and KNG, irrespective of whether the primers flanked the miR172 target site or were directed to the cly1 3′-UTR. Nair et al. (2010) suggested that miR172 directed transcript suppression of cly1 based on the detection of cly1 cleaved products by 5′-RACE (rapid amplification of cDNA ends) PCR in non-cleistogamous barley. qRT–PCR reported by Nair et al. (2010) and the present study showed no significant difference in cly1 transcript accumulation between AZ and KNG. In situ hybridization also clearly detected signal in lodicules of both the non-cleistogamous and cleistogamous types. These results imply that cly1 transcripts cleaved by miR172 represent only a small proportion of the entire set of cly1 transcripts, and they do not act to regulate the control exerted by CLY1 over lodicule development. In A. thaliana, even though cleavage products of the cly1 orthologue AtAP2 were detectable using 5′-RACE PCR, it was undetectable using northern blot analysis, a method free from PCR-oriented bias (Aukerman and Sakai, 2003). The suggestion was therefore that miR172-directed cleavage products of AP2 transcripts are rare and their presence is not indicative of miR172 function (Aukerman and Sakai, 2003; Chen, 2004).
The alternative mechanism by which miR172a controls cly1 regulation is by the repression of cly1 translation. Many studies during the past 15 years have established translational repression as a prominent mode of action by miR172 rather than mRNA cleavage in plants (Zhu and Helliwell, 2011; Ma et al., 2013). If miR172a influences cly1 translation, then the expectation is that less CLY1 protein would be accumulated in non-cleistogamous cultivars. The immunoblot analysis indeed revealed a reduced accumulation of CLY1 in the AZ spike compared with the level in the KNG spike. The ability of miR172a to interfere with cly1 translation was further confirmed by the behaviour of the Ds-induced loss of function of the Hv-miR172a mutant, which exhibited an increased accumulation of CLY1. The increased accumulation of CLY1 always resulted in reduced (KNG) or rudimentary (Ds-miR172a) lodicule size, in accordance with the notion that CLY1 protein is the suppressor of lodicule development (Nair et al., 2010).
Perspective of miR172-directed spike determinacy
The spikelet indeterminacy and overall floral defects exhibited by the Ds-miR172a mutant were comparable with the phenotype of the SNB loss-of-function mutant in rice (Lee et al., 2006). The abnormal spike phenotype of the miR172a knockout mutant was also reminiscent of that produced by the maize miR172 mutant (ts4), in which IDS1 (the Q orthologue) is no longer regulated (Chuck et al., 2007). The glume-to-floret transition in apical spikelets of the Ds-miR172a mutant was similar to that observed in wheat with higher Q activity (Greenwood et al., 2017) or lower miR172 activity (Debernardi et al., 2017). The miR172-directed regulation of AP2-like genes is important for floral transition and floral patterning in cereals (Zhu and Helliwell, 2011; Luo et al., 2013). miR172-AP2-like (SNB/OsIDS1) modules control the conversion of the spikelet meristem into the floral meristem in rice and maize (Luo et al., 2013). The AP2-like orthologous gene Q has been well characterized in wheat (Simons et al., 2006) and has recently been shown to be regulated by miR172 in relation to spike morphology and the freethreshing character (Debernardi et al., 2017; Greenwood et al., 2017; Liu et al., 2017). However, the barley Q gene and the possible role of miR172 in its regulation in the determination of spike morphology. It was clear from the present experiments that miR172 does not compromise the abundance of barley Q transcript, agreeing with the cases of both IDS1 (Chuck et al., 2007) and SNB (Zhu et al., 2009). The present data suggest that miR172 may influence meristem fate in the barley inflorescence presumably by targeting AP2-like genes; Q may be one of the candidate genes, but interaction between miR172 and AP2-like genes for barley spike determinacy requires additional evidence.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: list of primers and probes. Figure S1: detection of barley miRNAs capable of interaction with cly1 in cultivar ‘Morex’ immature spikes. Figure S2: alignment of Hv-miR172 precursors with the top hits from miR172 precursors in miRBase. Figure S3: positions of primers and probes used for gene expression analysis. Figure S4: PCR amplification efficiency of qRT–PCR primers. Figure S5: flower and rachis tissues isolated from immature spikes at the awn primordium stage by laser microdissection.
ACKNOWLEDGEMENTS
We thank Ning Wang for help in antibody design, Shun Sakuma for help in qRT–PCR, Nils Stein for access to the barley genome sequences, Harumi Koyama for taking care of the plants and helping to sample the spikes, Hisae Kamakura for preparing material for laser microdissection, and the NIAS Genebank (Tsukuba, Japan) for the provision of seeds of the various barley cultivars. This research was supported by the Japanese Ministry of Agriculture, Forestry and Fisheries Genomics for Agricultural Innovation programme (TRS1001 to J.W. and TRS1002 and TRS1005 to T.K.).
LITERATURE CITED
- Abdel-Ghani AH, Parzies HK, Omary A, Geiger HH. 2004. Estimating the outcrossing rate of barley landraces and wild barley populations collected from ecologically different regions of Jordan. Theoretical and Applied Genetics 109: 588–595. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Current Biology 13: 807–818. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Bartel DP. 2003. MicroRNAs: at the root of plant development. Plant Physiology 132: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY. 2005. Genetics and evolution of inflorescence and flower development in grasses. Plant and Cell Physiology 46: 69–78. [DOI] [PubMed] [Google Scholar]
- Briggs D. 1978. Barley. London: Chapman and Hall. [Google Scholar]
- Brown R, Dahleen LS, Lemaux PG, Bregitzer P. 2014. Registration of the barley transposon-tagged population I: seventy lines each with a single, unique site of Ds insertion. Journal of Plant Registration 8: 226–230. [Google Scholar]
- Brown RH, Bregitzer P. 2011. A Ds insertional mutant of a barley miR172 gene results in indeterminate spikelet development. Crop Science 51: 1664–1672. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. 2004. A microRNA as a translational repressor of APET ALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S. 2007. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nature Genetics 39: 1517–1521. [DOI] [PubMed] [Google Scholar]
- Ciaffi M, Paolacci AR, Tanzarella OA, Porceddu E. 2011. Molecular aspects of flower development in grasses. Sexual Plant Reproduction 24: 247–282. [DOI] [PubMed] [Google Scholar]
- Colaiacovo M, Subacchi A, Bagnaresi P, Lamontanara A, Cattivelli L, Faccioli P. 2010. A computational-based update on microRNAs and their targets in barley (Hordeum vulgare L.). BMC Genomics 11: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Spriggs A, Taylor J, Li Z, Helliwell C. 2012. miRNA regulation in the early development of barley seed. BMC Plant Biology 12: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX. 2011. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research 39: W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H. 2002. Molecular strategies for gene containment in transgenic crops. Nature Biotechnology 20: 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Lin H, Chuck G, Faris JD, Dubcovsky J. 2017. microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144: 1966–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gines M, Baldwin T, Rashid A, et al. 2017. Selection of expression reference genes with demonstrated stability in barley among a diverse set of tissues and cultivars. Crop Science 58: 332–341. [Google Scholar]
- Greenwood J, Finnegan JE, Watanabe N, Trevaskis B, Swain SM. 2017. New alleles of the wheat domestication gene Q reveal multiple roles in growth and reproductive development. Development 144: 1959–1965. [DOI] [PubMed] [Google Scholar]
- Hackenberg M, Gustafson P, Langridge P, Shi BJ. 2015. Boron stress responsive microRNAs and their targets in differential expression of microRNAs and other small RNAs in barley between water and drought conditions. Plant Biotechnology Journal 13: 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda I, Turuspekov Y, Komatsuda T, Watanabe Y. 2005. Morphological and physiological analysis of cleistogamy in barley (Hordeum vulgare). Physiologia Plantarum 124: 524–531. [Google Scholar]
- Hori K, Sato K, Kobayashi T, Takeda K. 2006. QTL analysis of fusarium head blight severity in recombinant inbred population derived from a cross between two-rowed barley varieties. Breeding Science 56: 25–30. [Google Scholar]
- Jones-Rhoades MW. 2010. Prediction of plant miRNA genes. Methods in Molecular Biology 592: 19–30. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW. 2012. Conservation and divergence in plant microRNAs. Plant Molecular Biology 80: 3–16. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- Joshi T, Yan Z, Libault M, et al. 2010. Prediction of new miRNAs and associated target genes in Glycine max. BMC Bioinformatics 11: S14-10.1186/1471-2105-11-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, et al. 2007. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, et al. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Developmental Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Kim B, Yu HJ, Park SG, et al. 2012. Identification and profiling of novel microRNAs in the Brassica rapa genome based on small RNA deep sequencing. BMC Plant Biology 12: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby EJM, Appleyard M. 1981. Cereal development guide. Kenilworth: NAC Cereal Unit. [Google Scholar]
- Komatsuda T, Nakamura I, Takaiwa F, Oka S. 1998. Development of STS markers closely linked to the vrs1 locus in barley, Hordeum vulgare. Genome 41: 680–685. [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, et al. 2007. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proceedings of the National Academy of Sciences of the USA 104: 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen J, Martinmäki P, Pizzi C, Rastas P, Ukkonen E. 2009. MOODS: fast search for position weight matrix matches in DNA sequences. Bioinformatics 25: 3181–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research 39: D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. 2005. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proceedings of the National Academy of Sciences of the USA 102: 9412–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Lee J, Moon S, Park SY, An G. 2006. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant Journal 49: 64–78. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. 2008. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelandais-Briere C, Naya L, Sallet E, et al. 2009. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21: 2780–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Liu J, Dong H, Sun J. 2017. Functional regulation of Q by microRNA172 and transcriptional co-repressor TOPLESS in controlling bread wheat spikelet density. Plant Biotechnology Journal 16: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−2DCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lord EM. 1981. Cleistogamy – a tool for the study of floral morphogenesis, function and evolution. Botanical Review 47: 421–449. [Google Scholar]
- Luo Y, Guo Z, Li L. 2013. Evolutionary conservation of microRNA regulatory programs in plant flower development. Developmental Biology 380: 133–144. [DOI] [PubMed] [Google Scholar]
- Luo Y, Hu JY, Li L, Luo YL, Wang PF, Song BH. 2016. Genome-wide analysis of gene expression reveals gene regulatory networks that regulate chasmogamous and cleistogamous flowering in Pseudostellaria heterophylla (Caryophyllaceae). BMC Genomics 17: 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Nie X, Wang L, Du X, Biradar SS, Jia X, Weining S. 2012. Identification and characterization of microRNAs from barley (Hordeum vulgare L.) by high-throughput sequencing. International Journal of Molecular Sciences 13: 2973–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SM, Wang YF. 2004. Molecular strategies for decreasing the gene flow of transgenic plants. Yi Chuan 26: 556–559. [PubMed] [Google Scholar]
- Ma X, Cao X, Mo B, Chen X. 2013. Trip to ER: microRNA-mediated translational repression in plants. RNA Biology 10: 1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos JL, Bologna NG, Chorostecki U, Palatnik JF. 2010. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Current Biology 20: 49–54. [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Waugh R, Langridge P, et al. 2012. A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716. [DOI] [PubMed] [Google Scholar]
- Michael MZ, van Holst SMOC, Pellekaan NG, Young GP, James RJ. 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia. Molecular Cancer Research 1: 882–891. [PubMed] [Google Scholar]
- Nair SK, Wang N, Turuspekov Y, et al. 2010. Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proceedings of the National Academy of Sciences of the USA 107: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozhuner E, Eldem V, Ipek A, et al. 2013. Boron stress responsive microRNAs and their targets in barley. PLoS ONE 8: e59543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Current Biology 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu CX, Xie FL, Zhu YY, et al. 2007. Computational identification of microRNAs and their targets in Gossypium hirsutum expressed sequence tags. Gene 395: 49–61. [DOI] [PubMed] [Google Scholar]
- Sato K, Hori K, Takeda K. 2008. Detection of fusarium head blight resistance QTLs using five populations of top-cross progeny derived from two-row × two-row crosses in barley. Molecular Breeding 22: 517–526. [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, et al. 2003. Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012. [DOI] [PubMed] [Google Scholar]
- Schreiber AW, Shi BJ, Huang CY, Langridge P, Baumann U. 2011. Discovery of barley miRNAs through deep sequencing of short reads. BMC Genomics 12: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. 2005. Specific effects of microRNAs on the plant transcriptome. Developmental Cell 8: 517–527. [DOI] [PubMed] [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, et al. 2006. Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanova B, Maurice S, Cheptou PO. 2016. Is plasticity across seasons adaptive in the annual cleistogamous plant Lamium amplexicaule?Annals of Botany 117: 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kamakura H, Sato Y, et al. 2010. A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection. Journal of Plant Research 123: 807–813. [DOI] [PubMed] [Google Scholar]
- Turuspekov Y, Mano Y, Honda I, Kawada N, Watanabe Y, Komatsuda T. 2004. Identification and mapping of cleistogamy genes in barley. Theoretical and Applied Genetics 109: 480–487. [DOI] [PubMed] [Google Scholar]
- Wang N, Ning S, Wu J, Tagiri A, Komatsuda T. 2015. An epiallele at cly1 affects the expression of floret closing (cleistogamy) in barley. Genetics 199: 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, et al. 2007. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB Journal 21: 415–426. [DOI] [PubMed] [Google Scholar]
- Xie FL, Huang SQ, Guo K, et al. 2007. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Letters 581: 1464–1474. [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. 2005. Expression of Arabidopsis MIRNA genes. Plant Physiology 138: 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh T, et al. 2010. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Kawada N, Tahnooka T. 2005. Effect of row type, flowering type and several other spike characters on resistance to fusarium head blight in barley. Euphytica 141: 217–227. [Google Scholar]
- Yumul RE, Kim YJ, Liu X, et al. 2013. POWERDRESS and diversified expression of the miR172 gene family bolster the floral stem cell network. PLoS Genetics 9: e1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Cox B, Coob GP, Anderson TA. 2006. Evidence that miRNAs are different from other RNAs. Cellular and Molecular Life Sciences 63: 246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Kim Y, Dinh TT, Chen X. 2007. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant Journal 51: 840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Helliwell CA. 2011. Regulation of flowering time and floral patterning by miR172. Journal of Experimental Botany 62: 487–495. [DOI] [PubMed] [Google Scholar]
- Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA. 2009. Overexpression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biology 9: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.