All species of wood-decaying fungi occur on a characteristic range of substrates (host plants), which may be broad or narrow. Understanding the mechanisms that enable fungi to grow on particular substrates is important for both fungal ecology and applied uses of different feedstocks in industrial processes. We grew the wood-decaying polypore Fomitopsis pinicola on three different wood species, aspen, pine, and spruce, under various culture conditions. We examined both gene expression (transcription levels) and RNA editing (posttranscriptional modification of RNA, which can potentially yield different proteins from the same gene). We found that F. pinicola is able to modify both gene expression and RNA editing profiles across different substrate species and culture conditions. Many of the genes involved encode enzymes with known or predicted functions in wood decay. This work provides clues to how wood-decaying fungi may adjust their arsenal of decay enzymes to accommodate different host substrates.
KEYWORDS: basidiomycetes, decay, lignocellulose, RNA editing, transcriptome
ABSTRACT
Wood-decaying fungi tend to have characteristic substrate ranges that partly define their ecological niche. Fomitopsis pinicola is a brown rot species of Polyporales that is reported on 82 species of softwoods and 42 species of hardwoods. We analyzed the gene expression levels and RNA editing profiles of F. pinicola from submerged cultures with ground wood powder (sampled at 5 days) or solid wood wafers (sampled at 10 and 30 days), using aspen, pine, and spruce substrates (aspen was used only in submerged cultures). Fomitopsis pinicola expressed similar sets of wood-degrading enzymes typical of brown rot fungi across all culture conditions and time points. Nevertheless, differential gene expression and RNA editing were observed across all pairwise comparisons of substrates and time points. Genes exhibiting differential expression and RNA editing encode diverse enzymes with known or potential function in brown rot decay, including laccase, benzoquinone reductase, aryl alcohol oxidase, cytochrome P450s, and various glycoside hydrolases. There was no overlap between differentially expressed and differentially edited genes, suggesting that these may provide F. pinicola with independent mechanisms for responding to different conditions. Comparing transcriptomes from submerged cultures and wood wafers, we found that culture conditions had a greater impact on global expression profiles than substrate wood species. In contrast, the suites of genes subject to RNA editing were much less affected by culture conditions. These findings highlight the need for standardization of culture conditions in studies of gene expression in wood-decaying fungi.
IMPORTANCE All species of wood-decaying fungi occur on a characteristic range of substrates (host plants), which may be broad or narrow. Understanding the mechanisms that enable fungi to grow on particular substrates is important for both fungal ecology and applied uses of different feedstocks in industrial processes. We grew the wood-decaying polypore Fomitopsis pinicola on three different wood species, aspen, pine, and spruce, under various culture conditions. We examined both gene expression (transcription levels) and RNA editing (posttranscriptional modification of RNA, which can potentially yield different proteins from the same gene). We found that F. pinicola is able to modify both gene expression and RNA editing profiles across different substrate species and culture conditions. Many of the genes involved encode enzymes with known or predicted functions in wood decay. This work provides clues to how wood-decaying fungi may adjust their arsenal of decay enzymes to accommodate different host substrates.
INTRODUCTION
Wood (lignocellulose) is one of the most abundant carbon pools in terrestrial ecosystems. The decomposition of wood is a critical component of the carbon cycle and may be exploited in the production of biofuels and other technologies. The most abundant and best-studied wood-decaying organisms are Agaricomycetes (Basidiomycota) (1–5). Most wood-decaying Agaricomycetes can be placed into one of two major decay categories: (i) white rot, in which all components of plant cell walls are degraded, including the highly recalcitrant lignin fraction, and (ii) brown rot, in which lignin is modified but not appreciably removed (6–8). Phylogenomic analyses have demonstrated that brown rot fungi have evolved repeatedly from white rot ancestors (9–11).
The evolution of brown rot is associated with extensive losses of genes encoding plant cell wall-degrading enzymes, including peroxidases, laccases, and other ligninolytic oxidases, as well as carbohydrate-active enzymes (CAZymes), such as glycoside hydrolases (GHs), carbohydrate esterases (CEs), and lytic polysaccharide monooxygenases (LPMOs), coupled with reductions in cellulose-binding modules (CBM1s) associated with diverse CAZymes. Nevertheless, brown rot fungi cause rapid loss of strength and mass in wood substrates (6, 7, 12–14). The model brown rot fungus Postia placenta (also known as Rhodonia placenta) appears to attack wood cell walls in a two-step process, in which reactive oxygen species are first deployed to effect an oxidative pretreatment of lignocellulose, followed by the action of GHs that break down cellulose and hemicellulose into assimilable sugars, leaving lignin as a modified polymeric residue (15–17).
Most species of wood-decaying Agaricomycetes have characteristic substrate ranges that define their ecological activity in forest ecosystems and which can be useful for identification purposes (18–20). Brown rot species have been suggested to preferentially decay softwoods (gymnosperms, particularly Pinaceae), although many also attack hardwoods (angiosperms) (21, 22). The hemicellulose and lignin compositions of hardwoods and softwoods differ and may affect substrate preferences by fungi. Hemicellulose of hardwoods is enriched in glucuronoxylan, whereas the main hemicellulose of softwoods is galactoglucomannan, and the lignin fraction of softwood is mainly guaiacyl lignin, whereas hardwoods have a larger (but variable) fraction of syringyl lignin (23–25). Secondary metabolite profiles also differ, with softwoods containing large amounts of terpenes and other extractives and hardwoods being highly variable in extractive compounds (24).
The mechanism(s) of brown rot decay remains uncertain, but hydroxyl radicals have been implicated as the diffusible oxidants (26, 27). Fenton chemistry (H2O2 + Fe2+ + H+ → H2O + Fe3+ + ˙OH) has been proposed, and three models are often cited (4, 28). A central role for cellobiose dehydrogenase (CDH) had been proposed, but it is now clear that some brown rot fungi that efficiently cause extensive decay, such as P. placenta and Wolfiporia cocos, lack the gene for this enzyme (29). Alternatively, certain low-molecular-weight glycopeptides (GLPs) are thought to reduce extracellular Fe3+ in white and brown rot fungi (30–33). Perhaps the most widely held model involves hydroquinone cycling (34–36), which generates both Fenton reactants through the activities of a quinone reductase and oxalate-Fe3+ chelates. Several studies have suggested that laccases may also be involved in hydroquinone oxidation (37–39). Regardless of the mechanism(s), brown rot is thought to involve sequential oxidation and hydrolysis of lignocellulose.
Numerous studies have demonstrated the impact of culture conditions on transcript and protein accumulation levels in wood decay fungi (29, 40–42), including comparisons of expression on defined media (e.g., glucose, maltose, or crystalline cellulose) versus wood as a carbon source (43–47). However, there have been relatively few side-by-side comparisons of gene expression on different wood species (43, 48–55).
Several prior transcriptomic analyses concluded that wood decay fungi express similar sets of plant cell wall-degrading enzymes on both hardwood and softwood, although there may be disparities in the expression of individual genes on different substrates (43, 49, 50, 52, 55). For example, Vanden Wymelenberg et al. (55) found that 47 genes in the model white rot fungus Phanerochaete chrysosporium were upregulated >2-fold on aspen versus pine, including 13 genes encoding GHs, while in P. placenta, 164 genes showed differential expression on the two substrates (roughly equal numbers upregulated on each substrate). The P. placenta genes upregulated on pine included 15 genes encoding cytochrome P450s, which are thought to play a role in the degradation of complex wood extractives (29, 56, 57). Similarly, Gaskell et al. (49) detected three P450s upregulated on pine versus aspen in another brown rot species, Wolfiporia cocos. A comparison of the genome of P. chrysosporium with the closely related Phanerochaete carnosa demonstrated an expansion of P450s in the latter species, which is almost exclusively collected on conifer wood (53). Moreover, P. carnosa was shown to transform a higher fraction of phenolics from heartwood than P. chrysosporium (53), consistent with the view that P450s play a role in detoxifying wood extractives. Couturier et al. (43) also found that eight genes encoding P450s in another white rot fungus, Pycnoporus coccineus, were differentially expressed on pine versus aspen. Genes encoding other enzymes active in lignocellulose digestion that have been shown to be differentially expressed on hardwoods versus softwoods include those encoding LPMOs, manganese peroxidases (MnPs), lignin peroxidases (LiPs), GH10 xylanases, and GH78 α-l-rhamnosidase (43, 49, 51).
It is difficult to make generalizations from the results of previous studies comparing gene expression on different substrates, because their experimental conditions and organisms are highly variable. Most studies have used submerged milled wood substrates (43, 49, 51, 52, 55, 58), but others have used solid wood wafers (48) or sawdust (53, 54). Sampling times in studies using submerged substrates range from 3 (43) to 16 days (52), while studies using sawdust or wood wafers have sampled at 10 (48) to 42 days (53). About a dozen different tree species have been used as experimental substrates. The most commonly used hardwood is aspen (Populus tremuloides, P. grandidentata), including transgenic and hybrid lines with various lignin contents (48, 49, 58), but sugar maple (Acer saccharum) and yellow birch (Betula alleghaniensis) have also been employed (50, 51, 53). Softwood substrates include fir (Abies balsamea), three species of spruce (Picea abies, P. glauca, P. rubens), and five pines (Pinus contorta, P. halepensis, P. resinosa, P. strobus, P. sylvestris) (43, 49–53, 55). Yakovlev et al. (54) compared results on heartwood, reaction wood, and sapwood of P. abies and P. sylvestris.
The most commonly studied fungal species in analyses of substrate-specific gene expression are white rot species of the Polyporales, particularly P. chrysosporium and P. carnosa (48, 50, 51, 53, 55, 58). Other white rot species that have been studied include Pycnoporus cinnabarinus and Dichomitus squalens (43, 52), which are both in the order Polyporales, and Heterobasidion annosum, which is in the order Russulales (54). Two brown rot species of Polyporales, P. placenta and W. cocos, have been used in studies comparing gene expression on different tree species, specifically aspen and pine (48, 49, 55).
Here, we present an analysis of substrate-specific responses in Fomitopsis pinicola, which is a brown rot wood-decaying species of Polyporales that is reported to occur on 82 species of gymnosperms (softwoods), most commonly on pines and spruces, as well as 42 species of angiosperms (hardwoods), including aspen, birch, and cherry (59). This diverse range of substrates suggests that F. pinicola may have the ability to adjust its arsenal of decay enzymes according to the chemistry and anatomy of different hosts. F. pinicola, P. placenta, and W. cocos are members of the informally named “Antrodia clade,” which includes diverse brown rot species. The Antrodia clade is strongly supported as a monophyletic group (10), suggesting that the brown rot decay mechanisms of F. pinicola, P. placenta, and W. cocos are homologous.
In addition to gene expression (transcription), we examined RNA editing, which is a posttranscriptional modification that produces RNA sequences that differ from their DNA templates. RNA editing provides organisms with plasticity at the RNA level, and possibly the protein level, and it could play a role in adaptation to different environmental conditions. RNA editing has only been investigated in a few species of fungi, such as the basidiomycete Ganoderma lucidum sensu lato (white rot, Polyporales) (60) and the filamentous ascomycetes Gibberella zeae (or Fusarium graminearum) (61), Sordaria macrospora, and Pyronema confluens (62). In G. lucidum, 8,906 putative editing sites were identified without significant bias among substitution types. In contrast, in G. zeae, adenine to inosine (A-to-I) RNA editing was pronounced and shown to be stage-specific during sexual development. Furthermore, experimental evidence suggested that editing of the RNA encoding protein kinase PUK1 is important for ascospore formation and release (61). A developmental role for RNA editing seems to be conserved in filamentous ascomycetes, which is supported by observations from Sordaria macrospora and Pyronema confluens.
We obtained results at one time point on three substrates (pine, spruce, and aspen) in submerged cultures and at two time points on two substrates (pine and spruce) on wood wafers. Thus, our results enable us to assess the impact of substrate wood species versus culture conditions on transcription and RNA editing.
RESULTS
Expression profiling of F. pinicola grown in submerged culture.
Cultures of F. pinicola were grown in submerged cultures supplemented with ground aspen, pine, or spruce as the sole carbon source for 5 days. Owing to the accessibility of the substrate, rapid growth of wood decay fungi is typically observed under these conditions (29, 47, 48, 63–66). Transcript levels and patterns of expression observed here support the involvement of numerous genes in lignocellulose degradation.
The highest transcript levels corresponded to the same genes in all three substrates. Not surprisingly, putative “housekeeping” genes such as short-chain dehydrogenase/reductases (SDR), ADT/ATP carrier, and a flavin adenine dinucleotide (FAD)/NAD domain ranked among the 30 highest fragments per kilobase per million (FPKM) values (Table 1 and see Table S1 in the supplemental material). Genes encoding glycoside hydrolases and carbohydrate esterases (GH18, GH128, GH78, CE4, and CE6), broadly categorized as hemicellulases, were also highly expressed, especially those with predicted secretion signals. This sharply contrasts with white rot fungi that express high levels of cellulose-attacking hydrolases (GH6s and GH7s) and lytic polysaccharide monooxygenases (LPMOs). Oligopeptide transporter and extracellular protease may reflect the importance of nitrogen scavenging in wood, whereas extracellular lipases likely facilitate triglyceride utilization.
TABLE 1.
Thirty most abundant transcripts in submerged culture containing ground pine as sole carbon sourcea
| Pro IDb | Putative functionc | Pine |
Spruce |
Aspen |
No. of TMHsd | SPe | |||
|---|---|---|---|---|---|---|---|---|---|
| FPKM | Order | FPKM | Order | FPKM | Order | ||||
| Fompi_1030775 | AA3-3, alcohol oxidase | 12,993 | 1 | 10,068 | 1 | 11,821 | 1 | ||
| Fompi_1152048 | Aquaporin like | 8,946 | 2 | 4,319 | 4 | 7,387 | 2 | 6 | |
| Fompi_1039841 | NAD-dependent formate dehydrogenase | 6,013 | 3 | 7,407 | 2 | 5,516 | 3 | ||
| Fompi_49484 | CipC protein | 3,552 | 4 | 4,951 | 3 | 3,315 | 4 | ||
| Fompi_160683 | GH18 protein | 2,936 | 5 | 2,705 | 6 | 3,003 | 5 | Y | |
| Fompi_1084629 | Mannoprotein | 2,811 | 6 | 2,639 | 7 | 2,870 | 6 | Y | |
| Fompi_1022702 | Glutamine-dependent formaldehyde dehydrogenase | 2,588 | 7 | 2,896 | 5 | 2,452 | 8 | ||
| Fompi_1023896 | ABC transporter | 2,554 | 8 | 1,967 | 14 | 2,591 | 7 | ||
| Fompi_49935 | NAD(P) binding SDR | 2,236 | 9 | 2,257 | 9 | 1,912 | 11 | ||
| Fompi_1090124 | Endosulfine-domain protein | 2,105 | 10 | 1,338 | 20 | 2,167 | 9 | ||
| Fompi_1023614 | Acid protease | 1,816 | 11 | 2,240 | 10 | 2,129 | 10 | 1 | Y |
| Fompi_126349 | MFS general substrate transporter | 1,633 | 12 | 1,250 | 23 | 1,758 | 12 | 12 | |
| Fompi_1021547 | Lipasef | 1,631 | 13 | 848 | 36 | 941 | 30 | Y | |
| Fompi_1168917 | NAD-dependent formate dehydrogenase | 1,564 | 14 | 1,905 | 15 | 1,445 | 15 | ||
| Fompi_1022193 | GH128 protein | 1,529 | 15 | 1,514 | 17 | 1,527 | 13 | Y | |
| Fompi_1147363 | Catalase | 1,484 | 16 | 1,336 | 21 | 1,172 | 22 | ||
| Fompi_1039147 | General substrate transporter | 1,417 | 17 | 2,204 | 11 | 1,253 | 19 | 12 | |
| Fompi_159299 | CE4-CBM50 | 1,367 | 18 | 1,449 | 18 | 1,419 | 17 | Y | |
| Fompi_1023676 | Peptidoglycan binding | 1,357 | 19 | 1,448 | 19 | 1,489 | 14 | Y | |
| Fompi_1022010 | OPT oligopeptide transporter | 1,313 | 20 | 869 | 33 | 968 | 27 | 16 | |
| Fompi_129535 | FAD/NAD(P)-binding domain | 1,292 | 21 | 1,742 | 16 | 1,207 | 20 | ||
| Fompi_1131909 | Aldehyde dehydrogenase | 1,190 | 22 | 1,251 | 22 | 1,089 | 26 | ||
| Fompi_1021548 | Lipasef | 1,171 | 23 | 1,055 | 28 | 780 | 34 | 1 | Y |
| Fompi_145400 | GH78 | 1,150 | 24 | 773 | 43 | 1,320 | 18 | Y | |
| Fompi_1119394 | CE6 | 1,070 | 25 | 1,053 | 29 | 1,180 | 21 | 1 | Y |
| Fompi_98853 | GH16 | 1,064 | 26 | 916 | 31 | 1,140 | 24 | 1 | |
| Fompi_1022048 | Eukaryotic ADP/ATP carrier | 1,029 | 27 | 1,145 | 26 | 1,122 | 25 | 3 | |
| Fompi_1025554 | SDR | 1,012 | 28 | 890 | 32 | 950 | 29 | 2 | |
| Fompi_59695 | Hexose transporter | 1,012 | 29 | 2,592 | 8 | 1,429 | 16 | 11 | |
| Fompi_161955 | SDR | 962 | 30 | 1,187 | 25 | 880 | 31 | ||
Excludes genes designated “hypothetical” and those with no significant similarity to NCBI NR accessions.
Pro ID, protein identifier.
GH, glycoside hydrolase; CE, carbohydrate esterase; AA, auxiliary activities as described in reference 91; SDR, short-chain dehydrogenase/reductase.
TMH, predicted transmembrane helix.
SP, predicted secretion signal; Y, yes.
Lipase genes tandemly linked approximately 2 kb apart.
A hierarchical clustering analysis based on expression levels revealed three groups corresponding to the substrates (see Fig. S1). Transcriptome profiles of F. pinicola with pine are more similar to those with aspen than those with spruce, which is surprising, because spruce and pine are softwoods (Pinaceae), whereas aspen is a hardwood. To verify transcriptome results, we performed reverse transcription-quantitative PCR (qRT-PCR) analyses of two differentially regulated genes. Consistent with transcriptome sequencing (RNA-seq), the lowest transcript levels of thaumatin-like protein (Fompi_110401) and a peptidase (Fompi_1113467) were observed in colonized pine and spruce, respectively (see Fig. S2).
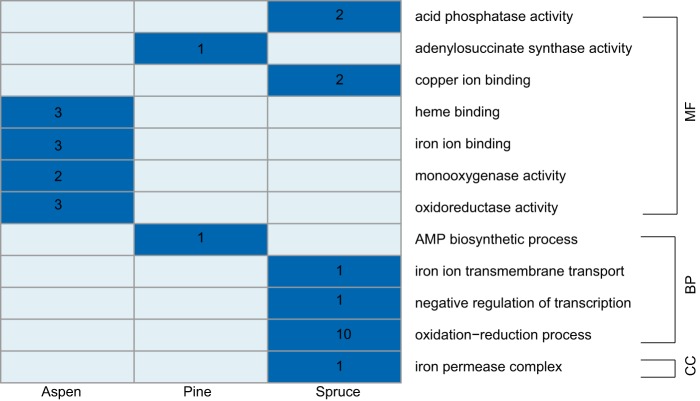
Of the 13,885 genes predicted in the F. pinicola genome, 91 genes were differentially expressed (4-fold change and P value of <0.05) among the three pairwise comparisons of substrates (aspen versus pine, aspen versus spruce, and pine versus spruce) in submerged culture (Fig. 1; Table S1). Consistent with the global expression pattern, fewer genes showed differential expression in pairwise comparisons between aspen and pine than in the other two pairwise comparisons (Fig. 1). A gene ontology (GO) enrichment analysis of upregulated genes on each substrate revealed very different terms for cellular components (CCs), biological processes (BPs), and molecular function (MF) (Fig. 2), with none shared by all three substrates.
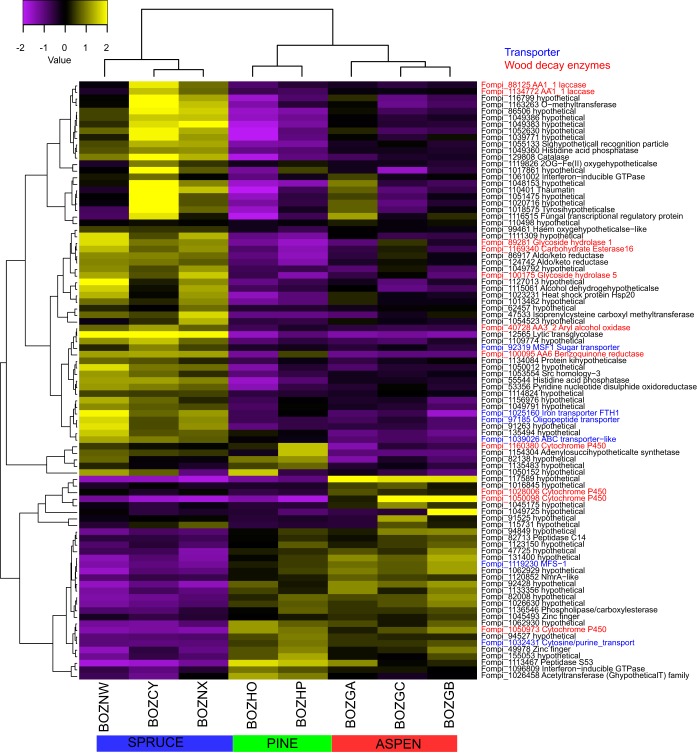
FIG 1.
Heatmap showing hierarchical clustering of 11 genes with predicted functions in wood decay with 4-fold change (FDR < 0.05) in pairwise-substrate comparisons at day 5. The scale above the map shows log2-based signals under the central row. Protein identifiers (IDs) and their enzyme names are indicated on the right side of the heatmap. Transporters are labeled in blue, and wood-decay enzymes are labeled in red. The bottom column designations refer to replicate libraries.
FIG 2.
GO enrichment of upregulated genes from each substrate after 5-day submerged incubation. Categories of GO terms are labeled on the right side of the heatmap. Substrates are labeled at the bottom. Dark blue shading indicates presence and light blue indicates absence. The number of genes in each GO term is labeled in dark blue.
Eleven differentially expressed genes (DEGs) among the three pairwise comparisons encode enzymes with predicted functions in wood decay (Fig. 1, names in red). The expression levels of these genes were roughly clustered on the basis of their putative function. These include laccase, benzoquinone reductase, and aryl-alcohol oxidase (Fig. 1), which are possibly involved in the generation of hydroxyl radicals via Fenton reactions (34–36, 39). In particular, benzoquinone reductase has been implicated in the redox cycling of Fe and H2O2, key Fenton reactants (35), and the process of hydroquinone oxidation may be augmented by laccases (39). While generally considered an oxidative process in brown rot, hydrolytic enzymes may also participate in cellulose degradation (67). Supporting this, a predicted endo-1,4-glucanase (Fig. 1) was expressed. Typical of brown rot fungi, this GH5_5 family protein lacks a cellulose binding module, and no exocellobiohydrolases occur within the F. pinicola genome. Thus, the importance of cellulose hydrolysis by conventional enzymes remains unclear. Differential expression analyses also revealed four genes encoding P450s, which are distributed among all three pairwise-substrate comparisons (Fig. 1). These may be involved in the intracellular metabolism of low-molecular-weight breakdown products of lignin and resins (68, 69).
A previous study of W. cocos (49) indicated that membrane-bound proteins are potentially important for carbohydrate metabolism during wood decay. We found that 8% to 22% of DEGs from each pairwise comparison have transmembrane (TM) domains, including six transporters (Fig. 1), which may facilitate the movement of small solutes across cell membranes. In addition, two proteins have functions related to methylation (Fompi_47533) and phosphorylation (Fompi_55544) (Table S1), suggesting that a modification of proteins might be involved during the decay process in submerged culture.
RNA editing of F. pinicola grown in submerged culture.
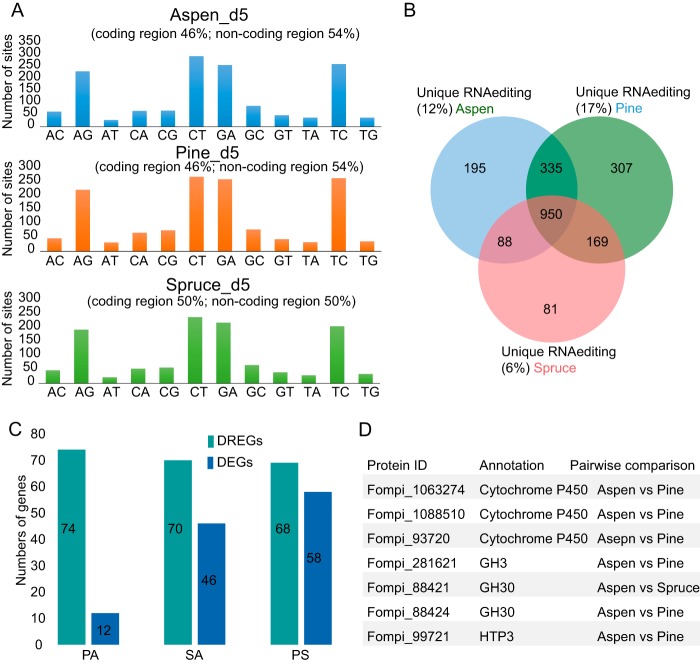
In this study, the strain used for the RNA editing analysis is identical to the one used for genome sequencing. Moreover, the frequency of single nucleotide polymorphisms (SNPs) between RNA and DNA due to different genomes should approach 100%, but we did not find any RNA editing sites with a frequency higher than 90%. Therefore, the inferred RNA editing sites are not artifacts due to differences in genome sequences. In submerged culture, we identified 1,583, 1,786, and 1,294 RNA editing sites in transcripts from aspen, pine, and spruce, respectively (Fig. 3A and see Table S2). Four primary RNA editing types were predicted, comprising conversions from A to G, C to T, G to A, and T to C, out of all 12 RNA editing types (Fig. 3A). Among these editing sites, approximately 50% were predicted within protein coding regions, with 283, 347, and 263 edits resulting in amino acid replacements in aspen, pine, and spruce, respectively. The proportions of RNA editing sites in intergenic regions and protein coding regions are similar (Fig. 3A), which suggests that RNA editing enzymes recognize targets without bias, given the similar sizes of intergenic (18.7 M) and protein coding (17.8 M) regions. Of course, we cannot exclude the possibility that the RNA editing sites in intergenic regions are in unannotated protein coding regions, unannotated 3′- or 5′-untranslated regions (UTRs) of genes, or noncoding RNA.
FIG 3.
RNA editing in F. pinicola after 5-day submerged culture. (A) Distribution of RNA editing types from aspen, pine, and spruce substrates. (B) Venn diagram showing distribution of RNA editing sites between substrates. (C) Comparison between DEGs and DREGs in substrate pairs. (D) Examples of DREGs encoding enzymes associated with wood decay.
RNA editomes from spruce, aspen, and pine substrates contain 81, 195, and 307 unique RNA editing sites, respectively (Fig. 3B). Differentially RNA-edited genes (DREGs) were defined as those showing unique nonsynonymous editing (resulting in amino acid replacement) on one substrate relative to that on another substrate. We identified 74, 70, and 68 DREGs in the pine-aspen, spruce-aspen, and pine-spruce comparisons, respectively (Fig. 3C). In each pairwise comparison, DREGs are more abundant than DEGs (Fig. 3C). There is no overlap between DREGs and DEGs, which suggests that shifts in expression levels and substitutions of amino acid via RNA editing are independent processes. There are seven DREGs encoding enzymes possibly involved in decay-related activity, including β-glucosidase (GH3), glucosylceramidase (GH30), heme-thiolate peroxidase, and cytochrome P450s (Fig. 3D). Thus, F. pinicola may use condition-specific differential RNA editing to alter its wood decay apparatus.
Expression profiling of F. pinicola grown on wood wafers.
Submerged cultures and colonized wafers share many of the more highly expressed genes (Tables 1 and 2), including some that are likely involved in lignocellulose degradation. Among these are several glycosidases, a lipase (Fompi_1021547) potentially involved in fatty acid catabolism, and an alcohol oxidase (Fompi_1030775) similar to the methanol oxidase of Gloeophyllum trabeum (70). The latter enzyme has been implicated in the generation of peroxide from methanol during brown rot demethylation of lignin. Modifications of lignin and wood extractives were confirmed by time of flight secondary ion mass spectrometry (TOF-SIMS) 10 and 30 days postinoculation (see supplemental material for TOF-SIMS methods and results and Fig. S3).
TABLE 2.
Thirty most abundant transcripts in colonized spruce and pine wafers after 30 daysa
| Pro IDb | Putative functionc | Spruce |
Pine |
No. of TMHd | SPe | ||
|---|---|---|---|---|---|---|---|
| 30-day FPKM | Order | 30-day FPKM | Order | ||||
| Fompi_1152048 | Aquaporin like | 5,523 | 1 | 6,517 | 6 | ||
| Fompi_1030775 | AA3-3, alcohol oxidase | 4,056 | 2 | 5,301 | 2 | ||
| Fompi_128521 | l-Fuculose-phosphate aldolase | 3,490 | 3 | 1,397 | 15 | ||
| Fompi_1021547 | Lipasef | 3,192 | 4 | 3,496 | 3 | Y | |
| Fompi_1131909 | Aldehyde dehydrogenase | 1,808 | 5 | 1,586 | 8 | ||
| Fompi_1090124 | Endosulfine-domain-containing protein | 1,537 | 6 | 1,561 | 9 | ||
| Fompi_160683 | GH18 | 1,493 | 7 | 1,451 | 13 | Y | |
| Fompi_1025554 | SDR | 1,487 | 8 | 1,555 | 11 | 2 | |
| Fompi_1022048 | Mitochondrial carrier | 1,454 | 9 | 1,503 | 12 | 3 | |
| Fompi_1147363 | Catalase | 1,423 | 10 | 1,478 | 13 | ||
| Fompi_148786 | CE4 | 1,383 | 11 | 751 | 50 | Y | |
| Fompi_86900 | MFS general substrate transporter | 1,367 | 12 | 961 | 34 | 12 | |
| Fompi_1044587 | Plasma membrane proteolipid 3 | 1,365 | 13 | 989 | 30 | 2 | |
| Fompi_1022193 | Glycoside hydrolase family 128 protein | 1,361 | 14 | 1,637 | 5 | Y | |
| Fompi_1022498 | Ribosomal protein S25 | 1,341 | 15 | 1,560 | 10 | ||
| Fompi_126349 | MFS general substrate transporter | 1,263 | 16 | 1,055 | 24 | 12 | |
| Fompi_1025379 | Peptidyl-prolyl cis-trans isomerase | 1,245 | 17 | 1,003 | 28 | ||
| Fompi_113951 | Ribosomal protein 60S | 1,234 | 18 | 1,183 | 19 | ||
| Fompi_1021745 | Polyubiquitin | 1,232 | 19 | 1,354 | 16 | ||
| Fompi_1039841 | NAD formate dehydrogenase | 1,208 | 20 | 1,632 | 6 | ||
| Fompi_1022753 | RNA-binding domain | 1,161 | 21 | 969 | 33 | ||
| Fompi_127599 | Actin 1 | 1,153 | 22 | 1,223 | 17 | ||
| Fompi_1112040 | Histone H3 | 1,146 | 23 | 1,184 | 18 | ||
| Fompi_1023676 | CBM50 | 1,088 | 24 | 995 | 29 | Y | |
| Fompi_1025222 | Manganese superoxide dismutase | 1,078 | 25 | 627 | 63 | ||
| Fompi_56132 | Acid protease | 1,023 | 26 | 1,138 | 21 | ||
| Fompi_1028183 | Acetohydroxy acid isomeroreductase | 992 | 27 | 692 | 58 | ||
| Fompi_1142989 | Ubiquitin/40s ribosomal protein fusion | 983 | 28 | 1,024 | 27 | ||
| Fompi_84886 | Thaumatin-like protein | 957 | 29 | 572 | 74 | Y | |
| Fompi_56163 | GH5-9 | 955 | 30 | 822 | 42 | Y | |
Excludes genes designated “hypothetical” and those with no significant similarity to NCBI NR accessions.
Pro ID, protein identifier.
GH, glycoside hydrolase; CE, carbohydrate esterase; AA, auxiliary activities as described in reference 91; SDR, short-chain dehydrogenase/reductase.
TMH, predicted transmembrane helix.
SP, predicted secretion signal; Y, yes.
Lipase genes tandemly linked approximately 2 kb apart.
Time-dependent gene expression.
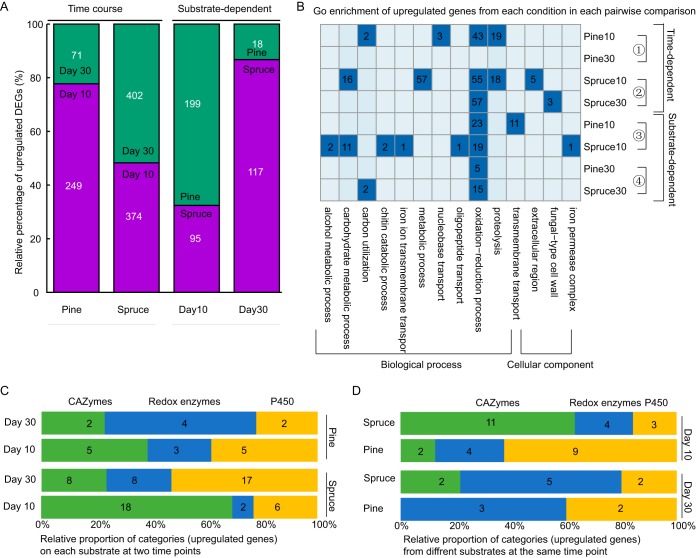
Differentially expressed transcripts corresponding to 320 and 776 genes were identified by pairwise comparisons on pine and spruce, respectively (Fig. 4A and Table S3). DEGs showed different trajectories on different substrates. On pine, there were far fewer upregulated genes than downregulated genes at day 30 than at day 10, whereas on spruce, there were slightly more upregulated genes than downregulated genes on day 30 versus day 10 (Fig. 4A).
FIG 4.
Differential expression of F. pinicola on different wood wafers (pine and spruce) after 10-day and 30-day incubations. (A) Relative percentages of upregulated DEGs on different substrates and at different time points. (B) GO enrichment of upregulated genes from each condition in each pairwise comparison. Four pairwise comparisons are labeled on the right side of the heatmap. Dark blue shading indicates presence and light blue indicates absence. The number of genes in each GO term is labeled in dark blue. Distributions in relative proportions of wood decay CAZymes (GH, CE, and PL), redox enzymes potentially involved in Fenton chemistry (AA families, HTP, POD, CRO, OXO, GLP, and QRD), and cytochrome_P450s at different time points (C) and on different substrates (D). For each category, only the upregulated genes at one time point/substrate relative to the other time point/substrate were considered.
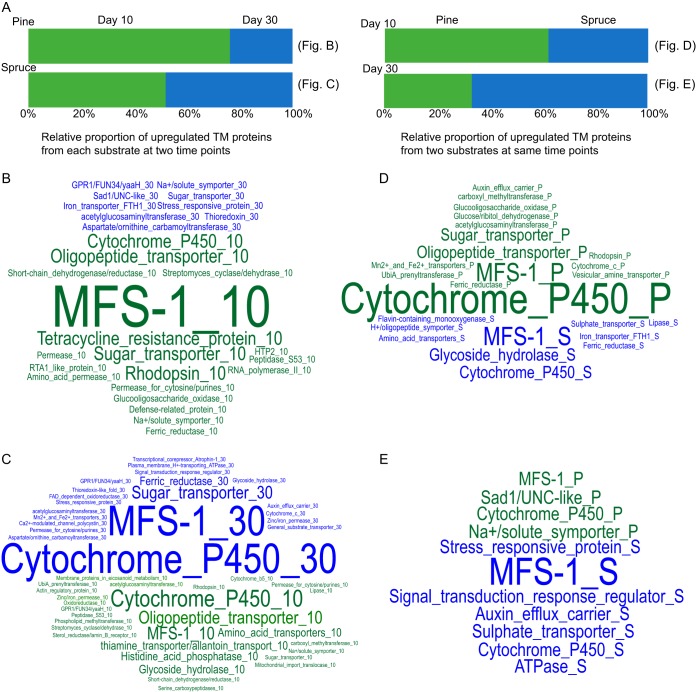
We performed enrichment analyses of upregulated DEGs from different time points on each substrate and found the enriched GO terms to be highly variable along the time course (Fig. 4B). The influence of incubation time is also pronounced among the relative proportions of TM proteins (Fig. 5). MFS-1, cytochrome P450, and oligopeptide transporter were enriched after a 10-day incubation on pine, but disappeared after a 30-day incubation (Fig. 5B). Conversely, on spruce, MFS-1 and cytochrome P450 were enriched at the later stage (Fig. 5C).
FIG 5.
Annotations of DEGs on wood wafers having TM domains. (A) Distributions in relative proportions of TM proteins at different time points (left) and on different substrates (right). For each category, only the upregulated gene at one time point/on one substrate relative to the other time point/substrate was considered. (B to E) Details for stacked bars in the panel A. The font size in the word cloud is directly related to the word (protein) frequency in each panel. The colors of words are consistent with the conditions used in panel A.
To evaluate the roles of carbohydrate-active enzymes and nonenzymatic oxidation reactions during the time course, we counted the number of CAZymes (GH, carbohydrate esterase [CE], and polysaccharide lyase [PL]), various redox enzymes potentially involved in Fenton systems (auxiliary activity [AA] family members, heme-thiolate peroxidases [HTPs], and enzymes involved in oxalate metabolism and iron homeostasis), and cytochrome P450s among the DEGs. For each category, only the upregulated gene at one time point relative to the other was considered. On each substrate, the number of CAZymes decreased with increased incubation time, while the number of lignin-degrading enzymes increased (Fig. 4C and Table. S4). In contrast, the patterns of cytochrome P450 transcript accumulation were not consistent between substrates (Fig. 4C and Table S4). On pine, the trend of cytochrome P450 is consistent with the CAZymes, whereas the cytochrome P450 on spruce is consistent with that of lignin enzymes.
On the basis of comparisons to P. chrysosporium iron-reducing glycopeptides (31, 33), 10 F. pinicola homologs were identified. Transcripts corresponding to Fompi_1027255 decreased over time in both pine and spruce. This expression pattern would be expected where the initial stages of decay relied heavily on oxidative depolymerization of cellulose, whereas later stages might shift toward hydrolytic mechanisms.
Substrate-dependent gene expression.
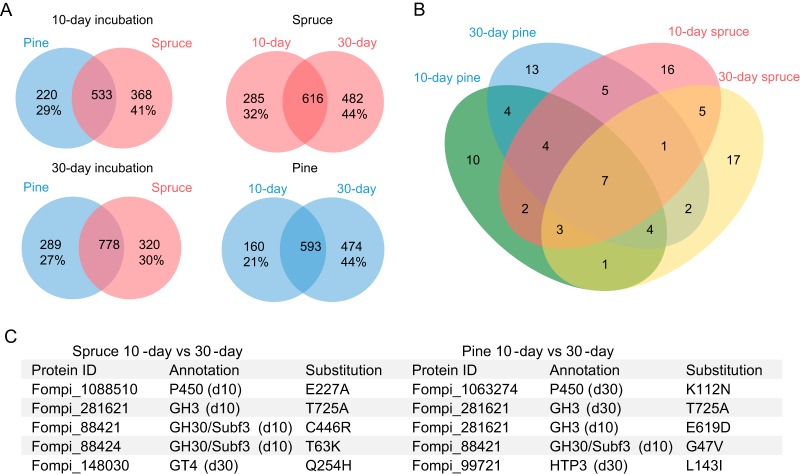
We compared the expression patterns on pine and spruce substrates at the same time points. Two hundred ninety-four DEGs were identified by substrate comparisons after a 10-day incubation, of which 199 were upregulated on pine and 95 were upregulated on spruce (Fig. 4A and Table S3). After a 30-day incubation, 135 DEGs were identified on pine versus spruce, of which only 18 were substantially upregulated on pine (Fig. 4A and Table S3).
A GO enrichment analysis of substrate-dependent upregulated genes after a 10-day incubation revealed six different biological processes and one different cellular component between pine and spruce. Most biological processes were enriched on spruce (Fig. 4B). The difference in enriched GO terms is diminished in the 30-day comparisons. After 10-day incubations, TM proteins, cytochrome P450, sugar transporter, and oligopeptide transporter were enriched on pine relative to those on spruce, while MFS-1 was enriched on spruce relative to that on pine after a 30-day incubation (Fig. 5). CAZymes, AA enzymes, and cytochrome P450s were also explored between substrates. After a 10-day incubation, spruce-dependent upregulated genes included 5-fold more genes encoding CAZymes and one-third as many genes encoding cytochrome P450s relative to those in pine. However, there was no change in AA enzymes (Fig. 4D and Table S4). After a 30-day incubation, spruce-dependent upregulated genes included slightly more genes encoding AA and other CAZyme family members than those on pine (Fig. 4D and Table S4).
RNA editing of F. pinicola grown on wood wafers.
We identified 753, 1,067, 901, and 1,098 edited sites on pine at day 10, pine at day 30, spruce at day 10, and spruce at day 30, respectively (Fig. 6A). Similar to RNA editing in submerged substrates at day 5 (Fig. 3), all 12 RNA editing types were found on wood wafers, and more transitions than transversions were observed (see Fig. S4). This observation is similar to a previous report from G. lucidum (60), which reports similar RNA editing types and frequencies with F. pinicola, suggesting a conservation of RNA editing mechanisms across the Polyporales.
FIG 6.
RNA editing of F. pinicola on wood wafers after 10-day and 30-day incubations. (A) Venn diagrams showing distribution of RNA editing sites between conditions. (B) Comparison of DREGs under different combinations of time points and substrates. (C) Examples of DREGs encoding enzymes associated with wood decay. The sample showing RNA editing is indicated after the gene annotation.
To explore substrate-dependent and time-dependent RNA editing on wood wafers, we compared the RNA editing sites in each possible pairwise comparison (Fig. 6A). Condition-specific DREGs were found, including seven genes encoding enzymes potentially involved in wood decay (Fig. 6B and C), which are the same as the DREGs identified in submerged culture. The regulation of RNA editing of individual genes showed a variation across samples. For instance, DREG Fompi3_281621 (GH3, β-glucosidase) includes one site showing the same kind of editing (T725A) on spruce and pine, but the editing occurs in 10-day samples on spruce and in 30-day samples on pine; another editing site in the same gene (E619D) is only apparent in 10-day samples on pine (Fig. 6C).
Cross-culture method comparisons.
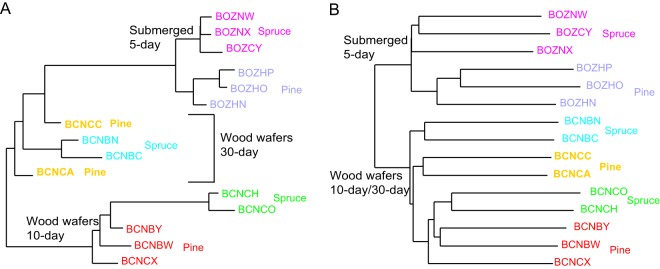
We performed hierarchical clustering of gene expression patterns using samples from different substrates (pine versus spruce), time points (day 5, day 10, and day 30), and culture methods (submerged versus wood wafers). Samples from the same time points and culture methods clustered with each other, regardless of substrate (see Fig. S5). Samples on wood wafers at day 30 were more similar to samples from submerged culture at day 5 than wood wafer samples at day 10 (Mann-Whitney U test, P < 0.00001) (Fig. S5). A neighbor-joining analysis of samples further indicated that the samples at day 5 were more distant from samples at day 10 than day 30 (Fig. 7A). We also compared RNA editing profiles between pine and spruce from different culture methods and time points. Similar to DEGs, RNA editing profiles cluster based on time points and culture methods. However, unlike DEGs, RNA editing profiles from the same culture method (submerged substrates versus wood wafers) clustered together (Fig. 7B).
FIG 7.
Cross-culture method comparisons. (A) Neighbor-joining tree with branch lengths inferred using 1 − Spearman's rho for all pairs of gene expression profiles. (B) Neighbor-joining tree with branch lengths inferred using 1 − Spearman's rho for frequency of RNA editing profiles.
DISCUSSION
Fomitopsis pinicola exhibits substrate-dependent and time-dependent differential gene expression.
Fomitopsis pinicola produces similar sets of wood-degrading enzymes on both hardwood and softwood substrates, as do other wood decay fungi (47, 49). Nevertheless, there are differences in the expression of specific genes, yielding substrate-specific expression profiles (Fig. 1 and Table S1 in the supplemental material). Genome and transcriptome analyses of other brown rot species have suggested that they employ nonenzymatic Fenton chemistry to generate hydroxyl radicals, which are generated by various redox systems (48, 49, 71, 72). In this study, differentially expressed decay-related oxidoreductases included laccase, AA3 (aryl alcohol oxidase), benzoquinone reductase, Fe(III)-reducing glycopeptides (GLP), and oxalate oxidase/decarboxylases (OXO) (Fig. 1 and 4C to D and Table S4). Glycoside hydrolases are also important for brown rot decay, and we found 15 kinds of upregulated GHs (GH1, GH3, GH5, GH16, GH17, GH18, GH28, GH30, GH31, GH43, GH45, GH78, GH79, GH92, and GH128) from different substrates and time points (Fig. 1 and 4C to D and Table S4). Along the time course on wood wafers, upregulated CAZymes were greater at day 10 than at day 30, while upregulated AA enzymes were greater at day 30 than at day 10 (Fig. 4C and D). This is not consistent with recent observations in P. placenta (17), where greater numbers of upregulated redox enzymes occurred at early stages of decay. A recent study suggests that lignocellulose oxidation by F. pinicola fluctuates for 3 months in submerged culture supplemented with spruce wood (73).
Unknown “hypothetical” proteins also contribute to substrate-specific responses. For instance, 50% (146) of the DEGs (294) between pine and spruce after 10-day incubation lack clear annotations according to InterPro prediction. Among these, 44 proteins (30%) were predicted to have secretion signals, and 47 (32%) have TM domains, suggesting that they may contribute to extracellular processes, including plant cell wall decomposition, or the transport of solutes across fungal cell membranes.
Pine and spruce “softwood” substrates elicit distinct gene expression profiles.
Transcriptomes from 5-day submerged cultures showed that global gene expression profiles on pine were more similar to those from aspen than to those from spruce (Fig. 1; Fig. S1). This was surprising, because pine and spruce are both softwoods in the Pinaceae family, whereas aspen is a hardwood. These findings suggest that it may be misleading to generalize from gene expression results on individual wood species to all “softwoods” (gymnosperms) or “hardwoods” (angiosperms). Moreover, prior studies using different genotypes of aspen (48, 58) or different wood types from pine and spruce (i.e., heartwood, sapwood, and tension wood [54]) demonstrate that gene expression may vary according to the genotype of the host species or the specific kind of wood that is used. Subtle differences in wood anatomy, physical properties, and/or chemical composition may substantially impact transcript profiles. Thus, to maximize the comparability among studies, it will be necessary not only to select a uniform set of species (perhaps focusing on plants with the greatest potential as feedstocks for biofuel production) but also to consider genetic diversity within species and the anatomical and chemical properties of wood samples.
Large differences exist in wood components for pine and spruce versus aspen. The lignin content is higher (27% to 30%) compared to that in aspen (22%), while cellulose and hemicellulose are approximately the same. However, the hemicellulose composition is largely acetylated glucuronoxylans in hardwoods and acetylated galactoglucomannans in softwoods (24, 74, 75). Other relatively minor wood components, such as extractives, may have a greater impact on specific gene expression as shown for the white rot fungus Phlebiopsis gigantea (76).
Brown rot Polyporales have elevated expression of certain P450s on pine versus aspen substrates.
Fomitopsis pinicola is the third brown rot species in the Antrodia clade of the Polyporales to be used for comparative transcriptomic analyses on different wood substrates. Vanden Wymelenberg et al. (55) and Gaskell et al. (49) analyzed W. cocos and P. placenta, respectively, and in both studies, they compared aspen and pine substrates in submerged cultures sampled at 5 days, as in the present study. Under these conditions, in all three studies, there were few genes upregulated (>4-fold, false discovery rate [FDR] < 0.05) on pine versus aspen, including 19 genes in P. placenta, 5 in W. cocos, and 12 in F. pinicola (Fig. 1; Table S1). In each species, the upregulated transcripts on pine included genes encoding cytochrome P450s (10 in P. placenta, 3 in W. cocos, and 1 in F. pinicola), suggesting that these may contribute to a common response to pine relative to aspen. The precise function of these P450s remains to be established, but the depletion of small lignin derivatives or extractive compounds observed with TOF-SIMS is consistent with a role for these enzymes in the metabolism of toxic breakdown products (see supplemental material).
Culture methods greatly affect gene expression.
Transcriptomic studies of wood decay in fungi usually employ submerged culture methods (29, 47, 49, 65, 77), wood wafers (48), or sawdust (53, 54). Of these methods, solid wafers may best mimic “natural” conditions in which the porosity of sound wood excludes many enzymes during incipient decay (78, 79). However, steam sterilization likely impacts substrate accessibility, and this pretreatment is no doubt more pronounced when the substrate is ground and dispersed, as in submerged cultures.
A global analysis of gene expression profiles on pine and spruce in submerged cultures at 5 days or wood wafers sampled at 10 and 30 days indicates that culture conditions have a greater impact on overall transcriptome profiles than substrate species, and the global gene expression pattern at day 5 in submerged substrates is more similar to that on wood wafers at day 30 than on wood wafers at day 10 (Fig. S5). The number of DEGs reflects the sensitivity of F. pinicola in recognizing substrates. Among the three sampling points, F. pinicola at day 5 in submerged culture had the fewest DEGs. Thus, F. pinicola in submerged culture exhibits the least sensitivity to substrates among the conditions that we employed. It is not clear if this effect is a consequence of the time of sampling or the culture conditions. Additional studies with overlapping time points in both submerged cultures and wood wafers might help discriminate the relative effects of time of sampling versus culture conditions.
RNA editing is involved in wood decay.
The prevalence of RNA editing has been demonstrated in only a few fungi (60–62, 80). For instance, there are 27,301 RNA editing sites in G. zeae (genome size, 36.5 Mb) (61), while there are 1,313 RNA editing sites in S. macrospora (40 Mb) (62); both studies suggested that RNA editing is involved in fruiting body development. In this study, we identified 753 to 1,786 RNA editing sites in transcriptomes (at least two duplicates for each condition) from the mycelium of F. pinicola (genome size, 41.6 Mb) under different culture conditions and time points (Table S2). Unlike G. zeae, but similar to S. macrospora and G. lucidum (60), F. pinicola has all N-to-N editing types, while in G. zeae, there are a vast majority of A to G edits. The editing types in F. pinicola are not distributed uniformly. There are four most frequent editing types (C to T, T to C, A to G, and G to A) (Fig. 3; Fig. S3), which account for over 50% of RNA editing events. In contrast to previous observations that over 50% of the genes with RNA editing sites are specifically expressed or upregulated during sexual reproduction in G. zeae (61), we did not find any overlap between DEGs and DREGs, suggesting that the two processes provide independent mechanisms for modifying gene expression and protein products.
Fomitopsis pinicola appears to be able to modify its RNA editome in response to different environments. For instance, the spruce-specific RNA editing sites in 10-day cultures can be up to 41% of the total RNA editing events relative to that on pine (Fig. 6A). Nonsynonymous substitutions could introduce amino acids with different chemical properties, which could affect protein function. Analyses of the DREGs, the genes with nonsynonymous editing sites, revealed some genes encoding proteins with roles in wood decomposition (Fig. 3 and 6), such as GH3 and P450. Accordingly, we propose that this kind of posttranscriptional modification may represent a mechanism by which wood decay fungi respond to changes in their environments.
Conclusions.
Fomitopsis pinicola, a wood-decaying Agaricomycete with a broad range of softwood and hardwood hosts, exhibits differential gene expression and RNA editing on different wood substrates (aspen, pine, and spruce) and under different culture conditions (submerged substrates versus wood wafers). Many of the DEGs and DREGs identified here encode enzymes with known or predicted functions in plant cell wall decomposition, suggesting that F. pinicola can modulate its decay apparatus according to the substrate. However, culture conditions (submerged ground wood versus wood wafers) and sampling times had a greater impact on overall gene expression profiles than did wood species, which highlights the need for a standardization of experimental parameters in studies of wood decay mechanisms. Comparisons of gene expression on pine versus aspen in 5-day-old submerged cultures of F. pinicola, P. placenta, and W. cocos, which are all brown rot species of the Antrodia clade (Polyporales), indicate that genes encoding some cytochrome P450s may be upregulated on pine. To assess the generality of these results, it will be necessary to study gene expression and RNA editing in additional species (ideally represented by multiple strains) of brown rot Polyporales on aspen, pine, and spruce substrates.
MATERIALS AND METHODS
Culture conditions.
Fomitopsis pinicola strain FP-58527 was obtained from the Forest Mycology Center, USDA Forest Products Laboratory (Madison, WI). Originally isolated from Pinus ponderosa, a draft genome sequence of this strain was analyzed by Floudas et al. (9) and the version 3 assembly is publicly available through the JGI portal (https://genome.jgi.doe.gov/Fompi3/Fompi3.home.html). Two culture methods were used to explore the transcriptome of F. pinicola on different substrates. Two-liter flasks containing 250 ml of basal salt medium were supplemented with wood ground in a Wiley mill with a no. 10 mesh screen. Each flask contained wood of quaking aspen (Populus tremuloides), loblolly pine (Pinus taeda), or white spruce (Picea glauca) as the sole carbon source. The basal medium was prepared as described in reference 49. After autoclaving at 121°C for 15 min, triplicate cultures for each substrate were inoculated with mycelium scraped from malt extract agar (2% [wt/wt] malt extract, 2% glucose [wt/wt], 0.5% peptone, 1.5% agar) and placed on a rotary shaker (150 rpm) at 22 to 24°C. Five days after inoculation, the mycelium and adhering wood were collected by filtration through Miracloth (Calbiohem) and stored at −80°C. In addition to submerged cultures, the mycelium was also cultured on wood wafers of the same three species in solid-block jar microcosms for solid-state culturing. The soil mixture for the microcosm was prepared using two parts of loam soil, two parts of vermiculite, and one part of peat moss that was wetted to capacity and placed in glass jars (473 ml). Two wood feeder strips corresponding to each type of wood were placed together on the top of the soil mixture and autoclaved twice for 1 h at 121°C with a 24-h period between sterilizations. A 5-mm-diameter plug of medium containing mycelium was placed at the corner of the feeder strips. When the mycelium covered the feeder strips completely, thin (10 mm by 15 mm by 1 mm), longitudinally cut wood wafers of each type of wood were placed on the feeder strips and incubated at 22°C. The wood wafers were removed after 10 and 30 days, snap-frozen in liquid nitrogen, and stored at −80°C. There were three replicates for each treatment.
RNA extraction and RNA-seq library construction.
Prior to extraction, colonized ground wood and wafer samples were pulverized under liquid nitrogen in a SPEX 6775 mill (Metuchen, NJ). A modified version of the method by Miyauchi et al. was used for RNA purification (81) from the SPEX-milled wafer. The samples were transferred to Oak Ridge tubes containing 12 ml of TRIzol. To disperse the material, the tubes were vortexed and the contents drawn 3 to 5 times in a polypropylene pipet. Chloroform (2.4 ml) was added with a glass pipette, and the suspension was briefly vortexed. After standing for 2 to 3 min, the tubes were centrifuged in the prechilled JA-17 rotor at 40,000 × g for 15 min at 4°C. With minimal disturbance to the interface, the aqueous phase (6 ml) was transferred to a new Oak Ridge tube, and a 0.6 volume (3.6 ml) of isopropyl alcohol was added. The contents were gently mixed by inverting several times, after which the tubes were incubated at −20°C for 1 to 2 h. The nucleic acids were pelleted by centrifugation at 40,000 × g for 10 min at 4°C. After careful decanting, the pellet was washed 2 times with 75% ethanol and air dried. The RNA was resuspended in 887.5 μl diethyl pyrocarbonate (DEPC)-treated water and transferred to a sterile Eppendorf tube. The sample was DNase treated with a Qiagen RNase-free DNase set by adding 100 μl RDD buffer and 12.5 μl DNase (3U/μl). After 20 min, another 3 U of DNase was added. RNA was precipitated by adding 330 μl 8 M LiCl and incubating at −20°C for at least 1 h. The Eppendorf tubes were then centrifuged in a prechilled JA-18.1 rotor at 12,000 rpm (∼18,000 × g) for 20 min, and the pellet resuspended in 150 μl isopropyl alcohol. The samples were then centrifuged in a JA-18.1 rotor at 12,000 × g for 5 min and washed 2 times with 1 ml 70% ethanol. Droplets of remaining ethanol were aspirated to hasten air drying. The dry pellet was resuspended in 25 μl DEPC-treated water and placed at 55°C for 5 min to dissolve. All RNA samples were quantified by a Qubit fluorometer (Thermo Fisher Scientific) and quality checked with a 2100 Bioanalyzer (Agilent).
Plate-based RNA sample preparation was performed on the PerkinElmer Sciclone NGS robotic liquid handling system using the Illumina TruSeq Stranded mRNA HT sample prep kit utilizing poly(A) selection of mRNA, according to the protocol outlined by Illumina in their user guide and with the following conditions: total RNA starting material was 1 μg per sample and 8 cycles of PCR were used for library amplification. The prepared libraries were quantified using a KAPA Biosystems next-generation sequencing library quantitative PCR (qPCR) kit and run on a Roche LightCycler 480 real-time PCR instrument. The quantified libraries were then multiplexed and prepared for sequencing on the Illumina HiSeq sequencing platform utilizing a TruSeq Rapid paired-end cluster kit, v4. Sequencing of the flow cell was performed on the Illumina HiSeq2000 sequencer using HiSeq TruSeq SBS sequencing kits, v4, following a 1 by 101 indexed run recipe.
Identification of differentially expressed genes.
Raw reads were filtered and trimmed using the JGI QC pipeline. Using BBDuk (https://sourceforge.net/projects/bbmap/), raw reads were evaluated for sequence artifacts by kmer matching (kmer, 25), allowing one mismatch, and any detected artifact was trimmed from the 3′ ends of the reads. RNA spike-in reads, PhiX reads, and reads containing any Ns were removed. Quality trimming was performed using the phred trimming method set at Q6. Finally, after trimming, the reads under a length of 25 nucleotides (nt) were removed. Filtered reads from each library were aligned to the haploid F. pinicola FP-58527 genome using HISAT (82) (see Table S5 in the supplemental material). featureCounts (83) was used to generate the raw gene counts using gff3 annotations (https://genome.jgi.doe.gov/Fompi3/Fompi3.info.html). Only primary hits assigned to the reverse strand were included in the raw gene counts (-s 2 -p primary options). FPKM normalized gene counts were calculated by Cufflinks (84). edgeR (85) was subsequently used to determine which genes were differentially expressed between pairs of conditions. The parameters used to call a DEG between conditions were a P value of <0.05 and a log2-fold change of ≥4. SignalP 4.1 (86) was used to search for secretory signal peptides in DEG proteins. TMHMM 2.0 (87) was used to predict and characterize TM domains in proteins encoded by DEGs, with default parameters. Functional categories enriched with DEGs were identified using FungiFun2 with an FDR of <0.05 (88).
Analysis of RNA editing sites.
The mapped strand-specific RNA-seq reads were divided into sense strand and antisense strand groups, and RNA editing sites were called separately for each group. Putative RNA editing sites from each sample were identified using JACUSA (89), with options to filter rare variants (ratio between reads with variants and total reads at specific position of less than 10%), variants with mapping quality less than 20, variants within 5 bp of read start/end, indels or splice sites, and filtered variants with more than 3 alleles per read pileup. In addition, we required reads to harbor at most 5 mismatches and variant sites to be covered by at least 2 reads. To further reduce false positives, a score threshold of 1.15 for variants was added. We then determined which sites had the same position and type in all biological replications, and analyzed only these reproducibly identified variants. SnpEff (90) was used for variant annotation of the sense strand and antisense strand groups. Functional categories enriched with differentially edited genes were performed using FungiFun2 (88).
Data availability.
Transcriptome data set have been deposited in the NCBI Sequence Read Archive under accession numbers SRP140951 to SRP140968 (see Table S5 for details).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation awards IOS-1456777 (to D.S.H.), IOS-1456548 (to R.A.B.), IOS-1456958 (to I.V.G.), and DEB-1457721 (to D.C.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00991-18.
REFERENCES
- 1.Gilbertson RL. 1980. Wood-rotting fungi of North-America. Mycologia 72:1–49. doi: 10.2307/3759417. [DOI] [Google Scholar]
- 2.Nilsson T, Daniel G, Kirk TK, Obst JR. 1989. Chemistry and microscopy of wood decay by some higher ascomyceyes. Holzforschung 43:11–18. doi: 10.1515/hfsg.1989.43.1.11. [DOI] [Google Scholar]
- 3.Shary S, Ralph SA, Hammel KE. 2007. New insights into the ligninolytic capability of a wood decay ascomycete. Appl Environ Microbiol 73:6691–6694. doi: 10.1128/AEM.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldrian P, Valaskova V. 2008. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev 32:501–521. doi: 10.1111/j.1574-6976.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- 5.Lundell TK, Makela MR, Hilden K. 2010. Lignin-modifying enzymes in filamentous basidiomycetes–ecological, functional and phylogenetic review. J Basic Microbiol 50:5–20. doi: 10.1002/jobm.200900338. [DOI] [PubMed] [Google Scholar]
- 6.Eriksson KE, Blanchette RA, Ander P. 1990. Microbial and enzymatic degradation of wood and wood components. Springer, Berlin, Germany. [Google Scholar]
- 7.Blanchette RA. 1995. Degradation of the lignocellulose complex in wood. Can J Bot 73:999–1010. doi: 10.1139/b95-350. [DOI] [Google Scholar]
- 8.Worrall JJ, Anagnostakis SE, Zabel RA. 1997. Comparison of wood decay among diverse lignicolous fungi. Mycologia 89:199–219. doi: 10.2307/3761073. [DOI] [Google Scholar]
- 9.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Gorecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kues U, Kumar TK, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, et al. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 10.Nagy LG, Riley R, Tritt A, Adam C, Daum C, Floudas D, Sun H, Yadav JS, Pangilinan J, Larsson KH, Matsuura K, Barry K, Labutti K, Kuo R, Ohm RA, Bhattacharya SS, Shirouzu T, Yoshinaga Y, Martin FM, Grigoriev IV, Hibbett DS. 2016. Comparative genomics of early-diverging mushroom-forming fungi provides Insights into the origins of lignocellulose decay capabilities. Mol Biol Evol 33:959–970. doi: 10.1093/molbev/msv337. [DOI] [PubMed] [Google Scholar]
- 11.Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, Lindquist EA, Sun H, LaButti KM, Schmutz J, Jabbour D, Luo H, Baker SE, Pisabarro AG, Walton JD, Blanchette RA, Henrissat B, Martin F, Cullen D, Hibbett DS, Grigoriev IV. 2014. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A 111:9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowling EB. 1961. Comparative biochemistry of the decay of sweetgum sapwood by white-rot and brown-rot fungi. U.S. Department of Agriculture, Washington, DC. [Google Scholar]
- 13.Kleman-Leyer K. 1994. Depolymerization of cellulose by brown rot fungi: comparison to that by cellulolytic systems of two non-wood degrading microorganisms. University of Wisconsin-Madison, Madison, WI. [Google Scholar]
- 14.Brischke C, Welzbacher CR, Huckfeldt T. 2008. Influence of fungal decay by different basidiomycetes on the structural integrity of Norway spruce wood. Holz Roh Werkst 66:433–438. doi: 10.1007/s00107-008-0257-1. [DOI] [Google Scholar]
- 15.Yelle DJ, Ralph J, Lu F, Hammel KE. 2008. Evidence for cleavage of lignin by a brown rot basidiomycete. Environ Microbiol 10:1844–1849. doi: 10.1111/j.1462-2920.2008.01605.x. [DOI] [PubMed] [Google Scholar]
- 16.Yelle DJ, Wei D, Ralph J, Hammel KE. 2011. Multidimensional NMR analysis reveals truncated lignin structures in wood decayed by the brown rot basidiomycete Postia placenta. Environ Microbiol 13:1091–1100. doi: 10.1111/j.1462-2920.2010.02417.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Presley GN, Hammel KE, Ryu JS, Menke JR, Figueroa M, Hu D, Orr G, Schilling JS. 2016. Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta. Proc Natl Acad Sci U S A 113:10968–10973. doi: 10.1073/pnas.1608454113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbertson RL, Ryvarden L. 1986. North American polypore vol 1 Fungiflora, Oslo, Norway. [Google Scholar]
- 19.Gilbertson RL, Ryvarden L. 1987. North American polypores vol 2 Fungiflora, Oslo, Norway. [Google Scholar]
- 20.Ginns J, Lefebvre MNL. 1993. Lignicolous corticioid fungi (Basidiomycota) of North America: systematics, distribution, and ecology vol 19 Mycologia memoir. American Phytopathological Society Press, Saint Paul, MN. [Google Scholar]
- 21.Gilbertson RL. 1981. North American wood-rotting fungi that cause brown rots. Mycotoaxon 12:372–416. [Google Scholar]
- 22.Hibbett DS, Donoghue MJ. 2001. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst Biol 50:215–242. doi: 10.1080/10635150121079. [DOI] [PubMed] [Google Scholar]
- 23.Ek M, Gellerstedt G, Henriksson G. 2009. Wood chemistry and wood biotechnology. Walter De Gruyter Inc, Berlin, Germany. [Google Scholar]
- 24.Fengel D, Wegener G. 1989. Wood: chemistry, ultrastructure, reactions. Walter De Gruyter Inc, Berlin, Germany. [Google Scholar]
- 25.Timell TE. 1967. Recent progress in the chemistry of wood hemicelluloses. Wood Sci Technol 1:45–70. doi: 10.1007/BF00592255. [DOI] [Google Scholar]
- 26.Cohen R, Jensen KA, Houtman CJ, Hammel KE. 2002. Significant levels of extracellular reactive oxygen species produced by brown rot basidiomycetes on cellulose. FEBS Lett 531:483–488. doi: 10.1016/S0014-5793(02)03589-5. [DOI] [PubMed] [Google Scholar]
- 27.Xu G, Goodell B. 2001. Mechanisms of wood degradation by brown-rot fungi: chelator-mediated cellulose degradation and binding of iron by cellulose. J Biotechnol 87:43–57. doi: 10.1016/S0168-1656(00)00430-2. [DOI] [PubMed] [Google Scholar]
- 28.Goodell B. 2003. Brown rot fungal degradation of wood: our evolving view, p 97–118. In Goodell B, Nicholas D, Schultz T (ed), Wood deterioration and preservation. American Chemical Society, Washington, DC. [Google Scholar]
- 29.Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P, Hammel KE, Vanden Wymelenberg A, Gaskell J, Lindquist E, Sabat G, Bondurant SS, Larrondo LF, Canessa P, Vicuna R, Yadav J, Doddapaneni H, Subramanian V, Pisabarro AG, Lavin JL, Oguiza JA, Master E, Henrissat B, Coutinho PM, Harris P, Magnuson JK, Baker SE, Bruno K, Kenealy W, Hoegger PJ, Kues U, Ramaiya P, Lucas S, Salamov A, Shapiro H, Tu H, Chee CL, Misra M, Xie G, Teter S, Yaver D, James T, Mokrejs M, Pospisek M, Grigoriev IV, Brettin T, et al. 2009. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci U S A 106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano T, Tanaka H, Enoki A. 1997. Relationship between production of hydroxyl radicals and degradation of wood by the brown-rot fungus, Tyromyces palustris. Holzforshung 51:389–395. doi: 10.1515/hfsg.1997.51.5.389. [DOI] [Google Scholar]
- 31.Tanaka H, Itakura S, Enoki A. 1999. Hydroxyl radical generation by an extracellular low-molecular weight substance and phenol oxidase activity during wood degradation by the white-rot basidiomycete Phanerochaete chrysosporium. J Biotechnol 75:57–70. doi: 10.1016/S0168-1656(99)00138-8. [DOI] [PubMed] [Google Scholar]
- 32.Hirano T, Enoki A, Tanaka H. 2000. Immunogold labeling of an extracellular substance producing hydroxyl radicals in wood degraded by brown-rot fungus Tyromyces palustris. J Wood Sci 46:45–51. doi: 10.1007/BF00779552. [DOI] [Google Scholar]
- 33.Tanaka H, Yoshida G, Baba Y, Matsumura K, Wasada H, Murata J, Agawa M, Itakura S, Enoki A. 2007. Characterization of a hydroxyl-radical-producing glycoprotein and its presumptive genes from the white-rot basidiomycete Phanerochaete chrysosporium. J Biotechnol 128:500–511. doi: 10.1016/j.jbiotec.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Shimokawa T, Nakamura M, Hayashi N, Ishihara M. 2004. Production of 2,5-dimethoxyhydroquinone by the brown-rot fungus Serpula lacrymans to drive extracellular Fenton reaction. Holzforschung 58:305–310. doi: 10.1515/HF.2004.047. [DOI] [Google Scholar]
- 35.Suzuki MR, Hunt CG, Houtman CJ, Dalebroux ZD, Hammel KE. 2006. Fungal hydroquinones contribute to brown rot of wood. Environ Microbiol 8:2214–2223. doi: 10.1111/j.1462-2920.2006.01160.x. [DOI] [PubMed] [Google Scholar]
- 36.Varela E, Tien M. 2003. Effect of pH and oxalate on hydroquinone-derived hydroxyl radical formation during brown rot wood degradation. Appl Environ Microbiol 69:6025–6031. doi: 10.1128/AEM.69.10.6025-6031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneko S, Yoshitake K, Itakura S, Tanaka H, Enoki A. 2005. Relationship between production of hydroxyl radicals and degradation of wood, crystalline cellulose, and lignin-related compound or accumulation of oxalic acid in cultures of brown-rot fungi. J Wood Sci 51:262–269. doi: 10.1007/s10086-004-0641-3. [DOI] [Google Scholar]
- 38.Jensen KA Jr, Houtman CJ, Ryan ZC, Hammel KE. 2001. Pathways for extracellular Fenton chemistry in the brown rot basidiomycete Gloeophyllum trabeum. Appl Environ Microbiol 67:2705–2711. doi: 10.1128/AEM.67.6.2705-2711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei D, Houtman CJ, Kapich AN, Hunt CG, Cullen D, Hammel KE. 2010. Laccase and its role in production of extracellular reactive oxygen species during wood decay by the brown rot basidiomycete Postia placenta. Appl Environ Microbiol 76:2091–2097. doi: 10.1128/AEM.02929-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kersten P, Cullen D. 2007. Extracellular oxidative systems of the lignin-degrading Basidiomycete Phanerochaete chrysosporium. Fungal Genet Biol 44:77–87. doi: 10.1016/j.fgb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Hammel KE, Cullen D. 2008. Role of fungal peroxidases in biological ligninolysis. Curr Opin Plant Biol 11:349–355. doi: 10.1016/j.pbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Benz JP, Chau BH, Zheng D, Bauer S, Glass NL, Somerville CR. 2014. A comparative systems analysis of polysaccharide-elicited responses in Neurospora crassa reveals carbon source-specific cellular adaptations. Mol Microbiol 91:275–299. doi: 10.1111/mmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Couturier M, Navarro D, Chevret D, Henrissat B, Piumi F, Ruiz-Duenas FJ, Martinez AT, Grigoriev IV, Riley R, Lipzen A, Berrin JG, Master ER, Rosso MN. 2015. Enhanced degradation of softwood versus hardwood by the white-rot fungus Pycnoporus coccineus. Biotechnol Biofuels 8:216. doi: 10.1186/s13068-015-0407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato S, Liu F, Koc H, Tien M. 2007. Expression analysis of extracellular proteins from Phanerochaete chrysosporium grown on different liquid and solid substrates. Microbiology 153:3023–3033. doi: 10.1099/mic.0.2006/000513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravalason H, Jan G, Molle D, Pasco M, Coutinho PM, Lapierre C, Pollet B, Bertaud F, Petit-Conil M, Grisel S, Sigoillot JC, Asther M, Herpoel-Gimbert I. 2008. Secretome analysis of Phanerochaete chrysosporium strain CIRM-BRFM41 grown on softwood. Appl Microbiol Biotechnol 80:719–733. doi: 10.1007/s00253-008-1596-x. [DOI] [PubMed] [Google Scholar]
- 46.Mahajan S, Master ER. 2010. Proteomic characterization of lignocellulose-degrading enzymes secreted by Phanerochaete carnosa grown on spruce and microcrystalline cellulose. Appl Microbiol Biotechnol 86:1903–1914. doi: 10.1007/s00253-010-2516-4. [DOI] [PubMed] [Google Scholar]
- 47.Vanden Wymelenberg A, Gaskell J, Mozuch M, Sabat G, Ralph J, Skyba O, Mansfield SD, Blanchette RA, Martinez D, Grigoriev I, Kersten PJ, Cullen D. 2010. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl Environ Microbiol 76:3599–3610. doi: 10.1128/AEM.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skyba O, Cullen D, Douglas CJ, Mansfield SD. 2016. Gene expression patterns of wood decay fungi Postia placenta and Phanerochaete chrysosporium are influenced by wood substrate composition during degradation. Appl Environ Microbiol 82:4387–4000. doi: 10.1128/AEM.00134-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaskell J, Blanchette RA, Stewart PE, BonDurant SS, Adams M, Sabat G, Kersten P, Cullen D. 2016. Transcriptome and secretome analyses of the wood decay fungus Wolfiporia cocos support alternative mechanisms of lignocellulose conversion. Appl Environ Microbiol 82:3979–3987. doi: 10.1128/AEM.00639-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacDonald J, Doering M, Canam T, Gong Y, Guttman DS, Campbell MM, Master ER. 2011. Transcriptomic responses of the softwood-degrading white-rot fungus Phanerochaete carnosa during growth on coniferous and deciduous wood. Appl Environ Microbiol 77:3211–3218. doi: 10.1128/AEM.02490-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macdonald J, Master ER. 2012. Time-dependent profiles of transcripts encoding lignocellulose-modifying enzymes of the white rot fungus Phanerochaete carnosa grown on multiple wood substrates. Appl Environ Microbiol 78:1596–1600. doi: 10.1128/AEM.06511-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rytioja J, Hilden K, Di Falco M, Zhou M, Aguilar-Pontes MV, Sietio OM, Tsang A, de Vries RP, Makela MR. 2017. The molecular response of the white-rot fungus Dichomitus squalens to wood and non-woody biomass as examined by transcriptome and exoproteome analyses. Environ Microbiol 19:1237–1250. doi: 10.1111/1462-2920.13652. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki H, MacDonald J, Syed K, Salamov A, Hori C, Aerts A, Henrissat B, Wiebenga A, VanKuyk PA, Barry K, Lindquist E, LaButti K, Lapidus A, Lucas S, Coutinho P, Gong Y, Samejima M, Mahadevan R, Abou-Zaid M, de Vries RP, Igarashi K, Yadav JS, Grigoriev IV, Master ER. 2012. Comparative genomics of the white-rot fungi, Phanerochaete carnosa and P. chrysosporium, to elucidate the genetic basis of the distinct wood types they colonize. BMC Genomics 13:444. doi: 10.1186/1471-2164-13-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yakovlev I, Vaaje-Kolstad G, Hietala AM, Stefanczyk E, Solheim H, Fossdal CG. 2012. Substrate-specific transcription of the enigmatic GH61 family of the pathogenic white-rot fungus Heterobasidion irregulare during growth on lignocellulose. Appl Microbiol Biotechnol 95:979–990. doi: 10.1007/s00253-012-4206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vanden Wymelenberg A, Gaskell J, Mozuch M, BonDurant SS, Sabat G, Ralph J, Skyba O, Mansfield SD, Blanchette RA, Grigoriev IV, Kersten PJ, Cullen D. 2011. Significant alteration of gene expression in wood decay fungi Postia placenta and Phanerochaete chrysosporium by plant species. Appl Environ Microbiol 77:4499–4507. doi: 10.1128/AEM.00508-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doddapaneni H, Chakraborty R, Yadav JS. 2005. Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus Phanerochaete chrysosporium: evidence for gene duplications and extensive gene clustering. BMC Genomics 6:92. doi: 10.1186/1471-2164-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doddapaneni H, Yadav JS. 2005. Microarray-based global differential expression profiling of P450 monooxygenases and regulatory proteins for signal transduction pathways in the white rot fungus Phanerochaete chrysosporium. Mol Genet Genomics 274:454–466. doi: 10.1007/s00438-005-0051-2. [DOI] [PubMed] [Google Scholar]
- 58.Gaskell J, Marty A, Mozuch M, Kersten PJ, Splinter BonDurant S, Sabat G, Azarpira A, Ralph J, Skyba O, Mansfield SD, Blanchette RA, Cullen D. 2014. Influence of Populus genotype on gene expression by the wood decay fungus Phanerochaete chrysosporium. Appl Environ Microbiol 80:5828–5835. doi: 10.1128/AEM.01604-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farr DF, Rossman AY. Fungal Databases, U.S. National Fungus Collections, ARS, USDA, Beltsville, MD: https://nt.ars-grin.gov/fungaldatabases/ Accessed 16 January 2018. [Google Scholar]
- 60.Zhu Y, Luo H, Zhang X, Song J, Sun C, Ji A, Xu J, Chen S. 2014. Abundant and selective RNA-editing events in the medicinal mushroom Ganoderma lucidum. Genetics 196:1047–1057. doi: 10.1534/genetics.114.161414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu H, Wang Q, He Y, Chen L, Hao C, Jiang C, Li Y, Dai Y, Kang Z, Xu JR. 2016. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res 26:499–509. doi: 10.1101/gr.199877.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teichert I, Dahlmann TA, Kuck U, Nowrousian M. 2017. RNA editing during sexual development occurs in distantly related filamentous ascomycetes. Genome Biol Evol 9:855–868. doi: 10.1093/gbe/evx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanden Wymelenberg A, Sabat G, Martinez D, Rajangam AS, Teeri TT, Gaskell J, Kersten PJ, Cullen D. 2005. The Phanerochaete chrysosporium secretome: database predictions and initial mass spectrometry peptide identifications in cellulose-grown medium. J Biotechnol 118:17–34. doi: 10.1016/j.jbiotec.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 64.Vanden Wymelenberg A, Minges P, Sabat G, Martinez D, Aerts A, Salamov A, Grigoriev I, Shapiro H, Putnam N, Belinky P, Dosoretz C, Gaskell J, Kersten P, Cullen D. 2006. Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveals complex mixtures of secreted proteins. Fungal Genet Biol 43:343–356. doi: 10.1016/j.fgb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Fernandez-Fueyo E, Ruiz-Duenas FJ, Ferreira P, Floudas D, Hibbett DS, Canessa P, Larrondo LF, James TY, Seelenfreund D, Lobos S, Polanco R, Tello M, Honda Y, Watanabe T, Watanabe T, Ryu JS, Kubicek CP, Schmoll M, Gaskell J, Hammel KE, St John FJ, Vanden Wymelenberg A, Sabat G, Splinter BonDurant S, Syed K, Yadav JS, Doddapaneni H, Subramanian V, Lavin JL, Oguiza JA, Perez G, Pisabarro AG, Ramirez L, Santoyo F, Master E, Coutinho PM, Henrissat B, Lombard V, Magnuson JK, Kues U, Hori C, Igarashi K, Samejima M, Held BW, Barry KW, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Riley R, et al. 2012. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc Natl Acad Sci U S A 109:5458–5463. doi: 10.1073/pnas.1119912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hori C, Gaskell J, Igarashi K, Kersten P, Mozuch M, Samejima M, Cullen D. 2014. Temporal alterations in the secretome of the selective ligninolytic fungus Ceriporiopsis subvermispora during growth on aspen wood reveal this organism's strategy for degrading lignocellulose. Appl Environ Microbiol 80:2062–2070. doi: 10.1128/AEM.03652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ryu JS, Shary S, Houtman CJ, Panisko EA, Korripally P, St John FJ, Crooks C, Siika-Aho M, Magnuson JK, Hammel KE. 2011. Proteomic and functional analysis of the cellulase system expressed by Postia placenta during brown rot of solid wood. Appl Environ Microbiol 77:7933–7941. doi: 10.1128/AEM.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ide M, Ichinose H, Wariishi H. 2012. Molecular identification and functional characterization of cytochrome P450 monooxygenases from the brown-rot basidiomycete Postia placenta. Arch Microbiol 194:243–253. doi: 10.1007/s00203-011-0753-2. [DOI] [PubMed] [Google Scholar]
- 69.Syed K, Nelson DR, Riley R, Yadav JS. 2013. Genomewide annotation and comparative genomics of cytochrome P450 monooxygenases (P450s) in the polypore species Bjerkandera adusta, Ganoderma sp. and Phlebia brevispora. Mycologia 105:1445–1455. doi: 10.3852/13-002. [DOI] [PubMed] [Google Scholar]
- 70.Daniel G, Volc J, Filonova L, Plihal O, Kubatova E, Halada P. 2007. Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Appl Environ Microbiol 73:6241–6253. doi: 10.1128/AEM.00977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algul H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arantes V, Jellison J, Goodell B. 2012. Peculiarities of brown-rot fungi and biochemical Fenton reaction with regard to their potential as a model for bioprocessing biomass. Appl Microbiol Biotechnol 94:323–338. doi: 10.1007/s00253-012-3954-y. [DOI] [PubMed] [Google Scholar]
- 73.Shah F, Mali T, Lundell TK. 2018. Polyporales brown rot species Fomitopsis pinicola: enzyme activity profiles, oxalic acid production, and Fe3+-reducing metabolite secretion. Appl Environ Microbiol 84:e02662-17. doi: 10.1128/AEM.02662-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pandey KK. 1999. A study of chemical structure of soft and hardwood and wood polymers by FTIR spectroscopy. J Appl Polym Sci 71:1969–1975. doi:. [DOI] [Google Scholar]
- 75.Sjöström E. 1993. Wood chemistry: fundamentals and applications. Gulf Professional Publishing, Houston, TX. [Google Scholar]
- 76.Hori C, Ishida T, Igarashi K, Samejima M, Suzuki H, Master E, Ferreira P, Ruiz-Duenas FJ, Held B, Canessa P, Larrondo LF, Schmoll M, Druzhinina IS, Kubicek CP, Gaskell JA, Kersten P, St John F, Glasner J, Sabat G, Splinter BonDurant S, Syed K, Yadav J, Mgbeahuruike AC, Kovalchuk A, Asiegbu FO, Lackner G, Hoffmeister D, Rencoret J, Gutierrez A, Sun H, Lindquist E, Barry K, Riley R, Grigoriev IV, Henrissat B, Kues U, Berka RM, Martinez AT, Covert SF, Blanchette RA, Cullen D. 2014. Analysis of the Phlebiopsis gigantea genome, transcriptome and secretome provides insight into its pioneer colonization strategies of wood. PLoS Genet 10:e1004759. doi: 10.1371/journal.pgen.1004759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Floudas D, Held BW, Riley R, Nagy LG, Koehler G, Ransdell AS, Younus H, Chow J, Chiniquy J, Lipzen A, Tritt A, Sun H, Haridas S, LaButti K, Ohm RA, Kues U, Blanchette RA, Grigoriev IV, Minto RE, Hibbett DS. 2015. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genet Biol 76:78–92. doi: 10.1016/j.fgb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blanchette R, Krueger E, Haight J, Akhtar M, Akin D. 1997. Cell wall alterations in loblolly pine wood decayed by the white-rot fungus, Ceriporiopsis subvermispora. J Biotechnol 53:203–213. doi: 10.1016/S0168-1656(97)01674-X. [DOI] [Google Scholar]
- 79.Flournoy D, Kirk TK, Highley T. 1991. Wood decay by brown-rot fungi: changes in pore structure and cell wall volume. Holzforschung 45:383–388. doi: 10.1515/hfsg.1991.45.5.383. [DOI] [Google Scholar]
- 80.Liu H, Li Y, Chen D, Qi Z, Wang Q, Wang J, Jiang C, Xu JR. 2017. A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc Natl Acad Sci U S A 114:E7756–E7765. doi: 10.1073/pnas.1702591114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miyauchi S, Navarro D, Grisel S, Chevret D, Berrin JG, Rosso MN. 2017. The integrative omics of white-rot fungus Pycnoporus coccineus reveals co-regulated CAZymes for orchestrated lignocellulose breakdown. PLoS One 12:e0175528. doi: 10.1371/journal.pone.0175528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 84.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 87.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 88.Priebe S, Kreisel C, Horn F, Guthke R, Linde J. 2015. FungiFun2: a comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 31:445–446. doi: 10.1093/bioinformatics/btu627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piechotta M, Wyler E, Ohler U, Landthaler M, Dieterich C. 2017. JACUSA: site-specific identification of RNA editing events from replicate sequencing data. BMC Bioinformatics 18:7. doi: 10.1186/s12859-016-1432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. 2013. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome data set have been deposited in the NCBI Sequence Read Archive under accession numbers SRP140951 to SRP140968 (see Table S5 for details).