Abstract
Sex differences are widely observed under various circumstances ranging from physiological processes to therapeutic responses, and a myriad of sex-biased genes have been identified. In recent years, transcriptomic datasets of microRNAs (miRNAs), an important class of non-coding RNAs, become increasingly accessible. However, comprehensive analysis of sex difference in miRNA expression has not been performed. Here, we identified the differentially-expressed miRNAs between males and females by examining the transcriptomic datasets available in public databases and conducted a systemic analysis of their biological characteristics. Consequently, we identified 73 female-biased miRNAs (FmiRs) and 163 male-biased miRNAs (MmiRs) across four tissues including brain, colorectal mucosa, peripheral blood, and cord blood. Our results suggest that compared to FmiRs, MmiRs tend to be clustered in the human genome and exhibit higher evolutionary rate, higher expression tissue specificity, and lower disease spectrum width. In addition, functional enrichment analysis of miRNAs show that FmiR genes are significantly associated with metabolism process and cell cycle process, whereas MmiR genes tend to be enriched for functions like histone modification and circadian rhythm. In all, the identification and analysis of sex-biased miRNAs together could provide new insights into the biological differences between females and males and facilitate the exploration of sex-biased disease susceptibility and therapy.
Keywords: Sex bias, MicroRNAs, Conservation, Disease association, Personalized medicine
Introduction
Sex difference is a prevalent phenomenon in physiology, disease susceptibility, and clinical therapy [1]. Men and women not only exhibit obvious anatomical differences, but more importantly, dozens of differences in diseases and therapeutic responses. Epidemiological studies have identified differences between men and women in disease incidence and prevalence [2]. Men are more likely to suffer from occlusive coronary artery disease (CAD) [3], autism spectrum disorders (ASD) [4], and stroke [5], [6], whereas women exhibit higher incidence of a non-obstructive CAD or microvascular dysfunction [3], rheumatic diseases [7], chronic radiation enteritis (CRE) [8], and post-traumatic stress disorder [9], [10]. As for drug response, aspirin has not been definitively proven to prevent cardiovascular events in women but men may gain greater benefit than women from angiotensin-converting enzyme inhibitors [11]. Moreover, molecular differences between male and female samples, including gene expression and somatic mutations, have also been reported from TCGA tumor datasets [12]. Accordingly, dissecting the differences between women and men is critical for precision medicine.
MicroRNAs (miRNAs) are small non-coding RNAs that play pivotal roles in a variety of cellular functions and biological processes such as cell proliferation and differentiation [13], [14], growth and development [15], as well as metabolic homeostasis [16]. There is accumulating evidence for differential miRNA expression between women and men across a variety of tissues, and the sex-biased expression of miRNAs could have functional implication [17]. For example, expression of miR-29a and miR-29c, which are involved in neuronal cell maintenance [18], is significantly up-regulated in frontal cortex of female mice in comparison with male mice [19]. Similarly, miRNAs from the miR-200 family, which target the gonadotropin releasing hormone receptor pathway, are also found to be differentially expressed between female and male rats, and this sex-biased expression pattern may partially contribute to the sexual disparity in brain development [20]. Women are more vulnerable to lupus than men, and one possible explanation is the higher expression of miR-98, miR-188, miR-421, and miR-503 in CD4+ T cells of women in comparison with men [21].
Recently, the sex-biased expression of circulating miRNAs (i.e., miRNAs in blood) has attracted much attention for their potential confounding effect that compromises the accuracy of the miRNA biomarkers [22]. For example, miR-221 and let-7g are expressed more prominently in the plasma of women compared to men, and could be sex-specific biomarkers of metabolic syndrome [23]. A more comprehensive, cohort-based survey has identified 35 sex-biased miRNAs after excluding some confounding factors like age and body weight [24]. Nevertheless, whether and how miRNAs exhibit sex-biased expression in tissues other than blood remained largely unexplored. Since sexual dimorphism of miRNAs could influence many physiological and pathological processes, a large-scale identification and analysis of the sex differentially-expressed miRNAs across multiple tissues is required for further understanding their role in human biology and diseases.
With the rapid development of high-throughput sequencing technologies, more than 500 human miRNA transcriptomic datasets and small RNA sequencing datasets across diverse tissues have become available in the Gene Expression Omnibus (GEO) database [25]. Nevertheless, no systematic investigation of the sex-biased expression of miRNAs has been reported yet. In a previous study, we developed a computational framework to identify sex-biased genes (ISBG) based on public gene expression datasets [26]. In this work, we adopted the framework above to identify sex-biased miRNAs (namely ISBM) from public GEO datasets. We finally collected 8 high quality miRNA expression datasets with gender information across multiple tissues (Figure 1) and performed a series of bioinformatics analyses on the sex-biased miRNAs identified. Our study provides some insights into biological differences between females and males and could serve as a starting point for gender-stratified personalized medicine.
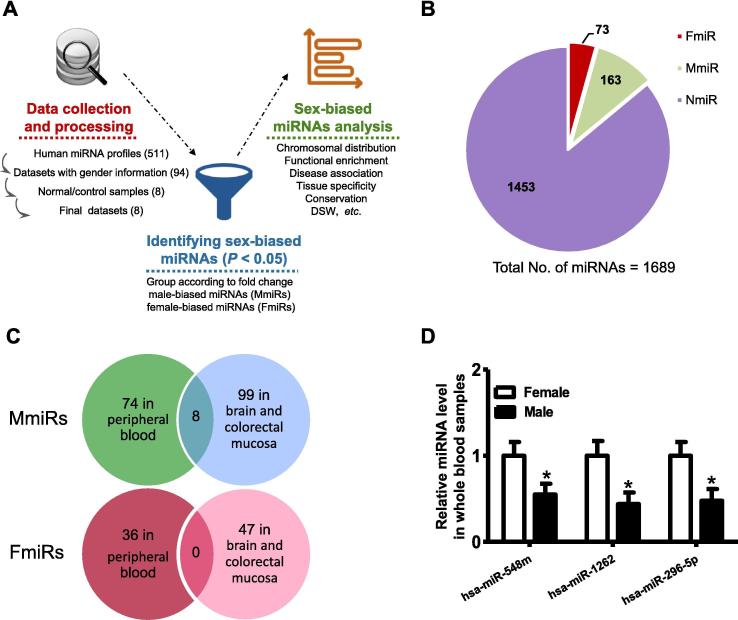
Figure 1.
The overview of the computational framework and the identified the sex-biased miRNAs
A. The overall computational framework of this work. First, human miRNA expression datasets with gender-labeled normal samples were curated from the GEO database. Second, the sex-biased miRNAs across different tissues were screened using Wilcoxon’s test comparing the expression levels between male and female samples (P < 0.05). Finally, various characteristics of these miRNAs were analyzed subsequently using different computational pipelines. B. Fraction of the sex-biased miRNAs in the dataset curated in the current study. C. The overlap of FmiRs and MmiRs between peripheral blood and the other two non-blood tissues, i.e., brain and colorectal mucosa in this study. D. RT-PCR validation of sex-biased miRNA expression in whole blood samples. The expression levels of selected miRNAs were analyzed using t-test in independently collected whole blood samples of males and females as described in the experimental procedures. The RT-PCR assay was performed on the same batch of blood sample. *indicates significant difference in miRNA expression between male and female samples (P < 0.05). N = 8–10. FmiR, female-biased miRNA; MmiR, male-biased miRNA; DSW, disease spectrum width, which is defined as the number of diseases associated with a given miRNA gene divided by the total number of diseases associated with any miRNA genes.
Results and discussion
Identification and experimental validation of the sex-biased miRNAs
Identification of the sex-biased miRNAs
To identify the sex-biased miRNAs, we first downloaded the human miRNA expression datasets across various tissues from the GEO database. Despite hundreds of miRNA expression datasets are available in the GEO database, few datasets contain normal samples with unambiguous gender labels. We discarded the datasets that had no normal samples or did not contain both male and female samples, and just retained the datasets that included normal tissues and were annotated with clear gender information. Finally, we obtained 8 miRNA expression datasets suitable for further analysis (Figure 1A).
Subsequently, we used the non-parametric test (Wilcoxon’s test) to identify the differentially expressed miRNAs between genders, with only normal samples considered). To retain enough miRNAs for further analysis, we took 0.05 as the P value threshold and did not consider fold change. Accordingly, we identified sex-biased miRNAs across four tissues, including brain, colorectal mucosa, peripheral blood, and cord blood (see Table 1 and Table S1 for details of these miRNAs). Notably, a few miRNAs exhibited conflicting gender bias among different tissues (Table S2). For example, hsa-miR-553 exhibits female bias in brain but male bias in peripheral blood. To avoid ambiguity, we excluded such miRNAs in the following analysis. Consequently, 73 female-biased miRNAs (highly expressed in females, FmiRs) and 163 male-biased miRNAs (highly expressed in males, MmiRs) were finally identified, which accounted for about 14.0% of total miRNAs in the miRNA datasets analyzed (Figure 1B). Accordingly, 1453 miRNAs that show no significant sex-biased expression in any of the tissues were grouped as the non-biased miRNAs (NmiRs).
Table 1.
Summary of the sex-biased miRNAs and GEO source datasets used in this study
Comparison and experimental validation of the sex-biased miRNAs
Previous study has identified some sex-related miRNAs in human blood [24]. Some of these miRNAs were also found in our analysis. For example, hsa-miR-45 and hsa-miR-16 exhibit higher expression level in male, which is in line with our results. Nevertheless, it seems that the sex-biased miRNAs in blood cannot fully recapitulate sex-biased miRNAs in other tissues. In our dataset, only a limited overlap of sex-biased miRNAs between peripheral blood and other non-blood tissues is observed (Figure 1C). There are only 8 shared MmiRs and no shared FmiRs between peripheral blood and other two non-blood tissues, brain and colorectal mucosa, suggesting the importance for surveying different tissues other than blood alone when analyzing sex-biased expression of miRNAs.
To experimentally validate the reliability of our computational analysis, we quantified the expression of three miRNAs in separate whole blood samples. These include hsa-miR-296-5p, hsa-miR-548m, and hsa-miR-1262, all of which show biased expression between women and men and defined as FmiRs in our dataset. RT-PCR analysis indicates that all of these three miRNAs exhibited women-biased expression in whole blood (Figure 1D), which agrees with our computational analysis. Therefore, the RT-PCR assay, at least partially, validates our computational analysis for the identification of sex-biased miRNAs (namely ISBM).
Chromosomal distribution of sex-biased miRNA genes
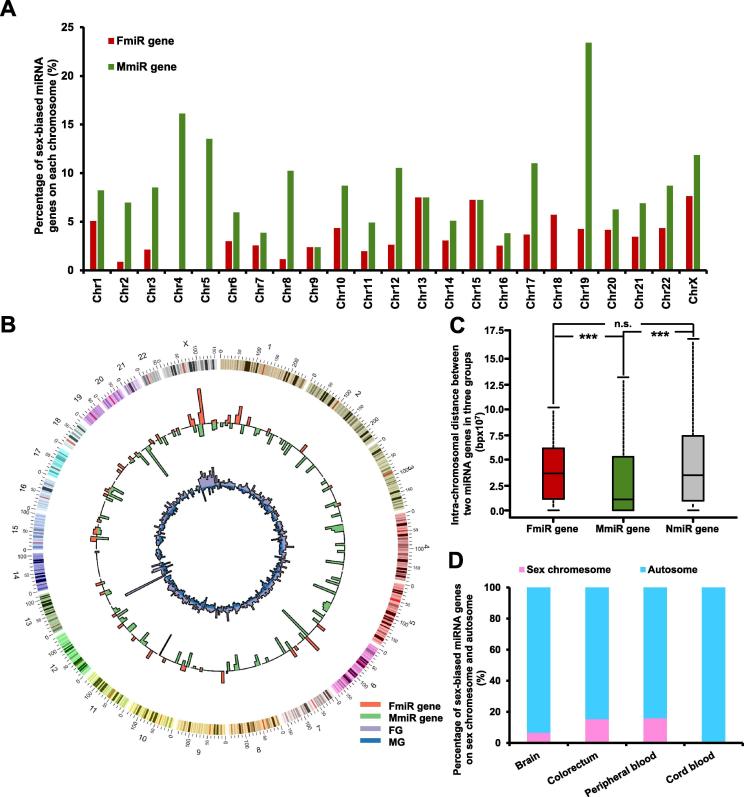
It is known that miRNA genes are scattered among chromosomes [27]. We speculate that the sex-biased miRNA genes would show non-random distribution on chromosomes. To test this hypothesis, we first compared the distribution of FmiR and MmiR genes on each chromosome. For 19 out of 23 chromosomes, the proportion of MmiR genes to the total number of miRNA genes on the same chromosome is higher than that of the FmiR genes (Figure 2A). Our previous study has demonstrated that the proportion of female-biased genes (FGs) is higher than that of male-biased genes (MGs) among most of the chromosomes [26]. It is therefore interesting to analyze if there is a correlation between the chromosomal distribution of the sex-biased miRNAs and that of the sex-biased genes. For an intuitive comparison, we first plotted a Circos graph [28] to show the distribution of sex-biased miRNAs (and genes) per million bp on each chromosome. As shown in Figure 2B, the distributions of the sex-biased miRNAs and genes are not consistent for most chromosomes except X chromosome. Indeed, no significant correlation between distributions of FmiR genes and FGs (Spearman’s correlation ρ = −0.24, P = 0.27), nor between those of MmiR genes and MGs (ρ = −0.28, P = 0.20) is found, suggesting that there seems no obvious connection between sex-biased miRNA genes and genes in terms of chromosomal distribution. We also noted that MmiR genes outnumber FmiR genes in total so that the proportion of MmiR genes would be randomly expected to be higher. Nevertheless, for some chromosomes like chromosomes 2, 4, 5, 8, and 19, the proportion of MmiRs is more than four-fold higher than that of FmiRs (Figure 2A), which cannot be simply explained by the higher total number of MmiRs. In all, our analysis indicates a non-random distribution of miRNAs across chromosomes.
Figure 2.
The chromosomal distribution of the sex-biased miRNA genes
A. Chromosomal distribution of the sex-biased miRNA genes. Y-axis shows the percentage of the sex-biased miRNA genes to the total miRNA genes on the same chromosome. B. Detailed chromosomal distribution of the sex-biased miRNA genes and sex-biased genes, the number of the sex-biased miRNA genes per million bp is plotted using the barplots embedded in the Circos graph. Red, green, purple, and blue bars represent the distributions of the FmiR genes, MmiR genes, FGs, and MGs, respectively. C. The boxplot comparing the intra-chromosomal distances between the sex-biased miRNA genes. ***indicates significant difference in intra-chromosomal distances, when comparing MmiR gene group with any of the other two groups (P < 0.001), according to Wilcoxon’s test. D. The percentage of the sex-biased miRNA genes on autosomes and sex chromosomes in each tissue. FmiR, female-biased miRNA; MmiR, male-biased miRNA; FG, female-biased coding gene; MG, male-biased coding gene; NmiR, non-biased miRNA; n.s., non-significant.
We further investigated the intra-chromosomal distribution of sex-biased miRNA genes by comparing the chromosomal distance between two miRNA genes from the same group. As shown in Figure 2C, the median intra-chromosomal distance between two MmiR genes is significantly smaller than that for FmiR genes (median distance 1.09E+7 bp vs. 3.67E+7 bp, Wilcoxon’s test P = 4.71E−7) or NmiR genes (median distance 1.09E+7 bp vs. 3.46E+7 bp, Wilcoxon’s test P = 3.43E−55), indicating that MmiR genes tend to be more clustered on the chromosomes. In contrast, FmiR genes do not show obvious clustering tendency. In order to examine the potential bias for individual chromosomes, the average intra-chromosomal distances between miRNAs for each chromosome are also examined (Figure S1A). The intra-chromosomal distances between sex-biased miRNA genes do not agree well with those between other miRNA genes, indicating that the non-random distribution of sex-biased miRNA genes. Finally, we compared the allocation of miRNA genes on sexual chromosome and autosomes for each tissue (Figure 2D). Sex-biased miRNA genes are located on the autosomes much more frequently than the sex chromosome, implying that the sexual dimorphism of miRNA expression could not totally be contributed by sex chromosomes but should be also associated with the differential regulation of autosome miRNA genes between males and females.
Evolutionary conservation of sex-biased miRNA genes
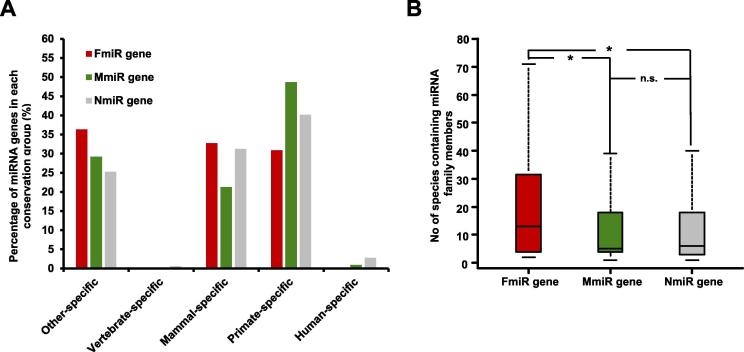
Evolutionarily conserved miRNAs are more likely to be associated with diseases [29]. Moreover, the conservation of genes is an important feature to consider when exploring gene functions [30]. To dissect the evolutionary conservation of sex-biased miRNAs, we first divided miRNA genes into 5 groups as described previously [31]. These include the human-specific group (G1), primate-specific group (G2), mammal-specific group (G3), vertebrate-specific group (G4), and the group for miRNA genes present in other more distal species (G5, which is the most conserved). miRNA genes from each group are listed in Table S3, and the distribution of FmiR, MmiR and NmiR genes among different groups is shown in Figure 3A. In the fast-evolving groups like G1 and G2 groups, there are more MmiR genes (49.6%, 56/113) than FmiR genes (30.9%, 17/55) (P = 0.031, OR = 0.46, Fisher’s exact test). We further compared the number of species in which the miRNA families of FmiR genes and MmiR genes are present, and found that FmiR families are present in more species than MmiR families (Wilcoxon’s test P = 0.050) and NmiR families (Figure 3B, Wilcoxon’s test P = 0.017). Together, these results indicate that MmiR genes have a faster evolutional rate, whereas FmiR genes tend to be more conserved, suggesting that FmiR genes are inclined to play roles in more fundamental biological processes.
Figure 3.
The evolutionary characteristic of the sex-biased miRNA genes
A. The distribution of the FmiR, MmiR, and NmiR genes in different conservation groups. Note that miRNA genes in more conserved group (e.g., primate-specific group) do not include the miRNA genes present in less conserved groups (e.g., human-specific group). B. Comparison of the number of species in which the corresponding miRNA gene family members are present. *indicates significantly higher number of species in which the family members of FmiR genes are present, in comparison with MmiR and NmiR genes (P < 0.05), according to Wilcoxon’s test. n.s., non-significant.
Expression regulation and disease spectrum width of the sex-biased miRNAs
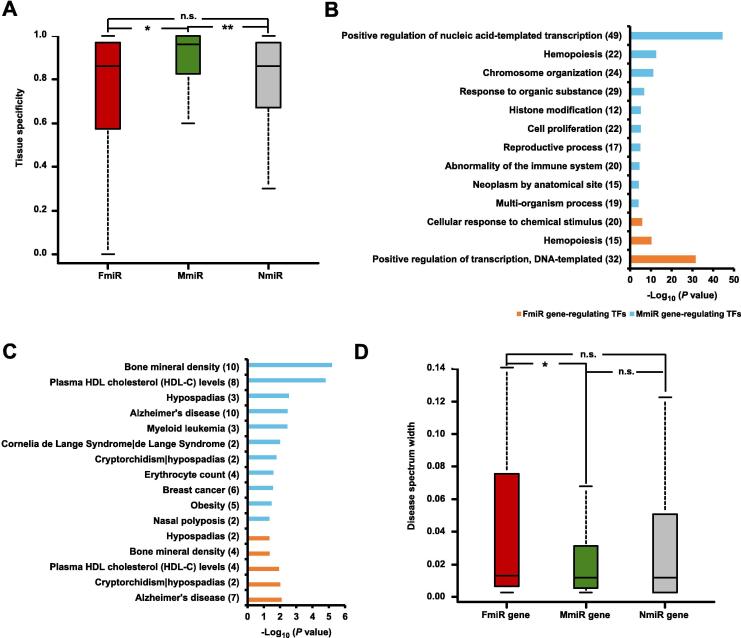
Gene expression is another characteristic implicating gene function, and miRNAs expressed in highly tissue-specific manner tend to be involved in tissue identity and differentiation [32], [33]. To understand the tissue expression specificity of sex-biased miRNAs, we computed the tissue specificity index based on the miRNA expression profiles across 40 tissues from Liang and colleagues [34]. We found that MmiRs have a significantly higher tissue expression specificity index than FmiRs (Figure 4A, Wilcoxon’s test P = 0.050), indicating that MmiRs prefer to express in a tissue-specific manner. Besides, we noted that the tissue specificity of sex-biased miRNAs is significantly higher than NmiRs (Figure 4A, Wilcoxon’s test P = 0.0036). The result suggests that the sex-biased miRNAs could play critical roles in distinguishing cell types and tissue development status.
Figure 4.
The comparison of tissue expression specificity and disease spectrum width of the sex-biased miRNAs
A. Comparison of tissue expression specificity between FmiRs, MmiRs, and NmiRs. The tissue expression specificity of one miRNA is defined as the ratio of its maximum expression level among the 40 tissues examined against the total expression from all of the 40 tissues. *P < 0.05; **P < 0.01, Wilcoxon’s test. B. The enriched functions of the FmiR-regulating TFs and MmiR-regulating TFs using the g:Profiler tool, only the top 20% significant terms (P < 0.05) are shown. Numbers in the parenthesis indicate the numbers of TFs associated with the respective functions. C. The associated diseases of the FmiR-regulating TFs and MmiR-regulating TFs using the DAVID tool (P < 0.05). Numbers in the parenthesis indicate the numbers of TFs associated with the respective diseases. D. Comparison of disease spectrum width between FmiR genes, MmiR genes, and NmiR genes. *indicates significant lower disease spectrum width of MmiR group, when compared to any of the other two groups (P < 0.05) according to Wilcoxon’s test. n.s., non-significant.
To a certain extent, gene expression is subject to the activity of transcription factors (TFs) [35]. For better understanding of the regulation of sex-biased miRNAs, we analyzed the TFs regulating the sex-biased miRNA genes. Through extensive exploration of the comprehensive ChIP-seq atlas, we identified the TFs that preferentially bind the proximal genomic regions of sex-biased miRNA genes in each tissue. On average, each sex-biased miRNA gene has 15.7 TF binding sites in vicinity, which is significantly higher than that of a non-biased miRNA gene (9.1 TF binding sites per gene on average; P = 1.47E−19, OR = 1.73, Fisher’s exact test). Moreover, some TFs are more likely to regulate sex-biased miRNA genes than non-biased miRNA genes. We identified 33 FmiR gene-regulating TFs and 56 MmiR gene-regulating TFs. FmiR gene-regulating TFs tend to be involved in functions like positive regulation of transcription, hemopoiesis, and cellular response to chemical stimulus, whereas MmiR gene-regulating TFs tend to be involved in functions like chromosome organization, response to organic substance, and histone modification (Figure 4B). These TFs are also significantly associated with diseases. For instance, both FmiR gene-regulating TFs and MmiR gene-regulating TFs are associated with Alzheimer’s disease and bone mineral density. In addition, MmiR gene-regulating TFs are also associated with Cornelia de Lange syndrome and myeloid leukemia (Figure 4C). Finally, we checked if these TFs themselves show sex-biased expression, based on the sex-biased genes identified in our previous study [26]. Our analysis indicates no significant overlap between sex-biased genes and TFs that regulate sex-biased miRNA genes (Fisher’s exact test, P = 0.53). Among 33 FmiR gene-regulating TFs and 56 MmiR gene-regulating TFs, only 4 and 9 TFs show sex-biased expression, respectively. Nevertheless, we note that some of the sex-biased TFs are likely to be associated with diseases that have known sex-biased incidence. For instance, male-biased TF CDK9 can inhibit cell proliferation and induce apoptosis in human breast cancer [36], whereas the gene encoding the female-biased TF PBX1 has a genetic and functional association with bone mineral density, one of the major determinants of risk for osteoporosis [37].
In consideration of the extensive association between miRNAs and diseases, we next examined the relationship between sex-biased miRNAs and diseases with the disease spectrum width (DSW) [38]. Intuitively, DSW describes how many diseases are associated with a particular miRNA gene, and higher DSW indicates wider disease associations of a particular miRNA gene. We re-calculated DSW (Table S4) using the updated data of the miRNA gene–disease associations in HMDD [39], which contained 578 miRNA genes, 383 diseases, and 10,381 miRNA gene–disease associations. As shown in Figure 4D, DSW of FmiR genes is significantly higher than that of MmiR genes (median: 0.013 vs. 0.012, Wilcoxon’s test P = 0.049). Combining the finding of tissue specificity and DSW, miRNA genes that are associated with more diseases tend to have lower tissue specificity, and this observation is consistent with the previous study [29] showing a negative correlation between the tissue specificity of a miRNA and the number of diseases it is associated with. We further compared the percentage of disease-associated miRNA genes among FmiR, MmiR and NmiR groups. We found that FmiR genes (66.7%, 44/66) do not have significantly more disease-associated miRNAs than MmiR genes (62.5%, 90/144) (P = 0.64, OR = 1.20, Fisher’s exact test). However, the percentage of disease-associated miRNA genes in NmiRs (38.7%, 336/868) is significantly lower than FmiR genes (P = 1.3E−5, OR = 0.32, Fisher’s exact test) and MmiR genes (P = 1.4E−7, OR = 0.38, Fisher’s exact test). These data suggest a more extensive involvement of the sex-biased miRNAs (miRNA genes) in human diseases.
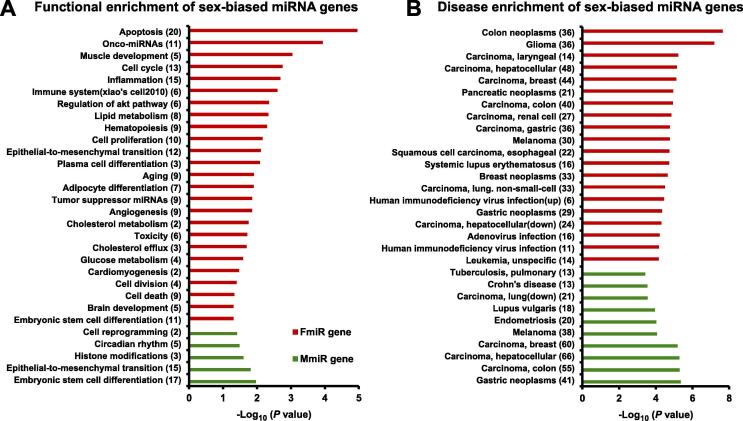
Functional enrichment analysis of the sex-biased miRNA genes
To learn more about the functions and specific diseases related to the sex-biased miRNAs, we performed the functional enrichment analysis of the sex-biased miRNA genes using the TAM tool [40], [41] to screen the enriched function terms (P < 0.05). As depicted in Figure 5A, MmiR genes tend to be enriched for terms like histone modification, circadian rhythm, and cell reprogramming. Previous studies have reported that androgen interacts with histone modifying enzymes [42] and histone modifications are sexually dimorphic in the developing mouse brain [43]. In contrast, FmiR genes are enriched for functional terms related to apoptosis, lipid metabolism, glucose metabolism, cholesterol metabolism, immune system, brain development, and a series of cell cycle-related processes (Figure 5A). These results suggest that FmiR genes tend to play roles in the fundamental processes for maintaining physiological activities, which is also supported by aforementioned biological characteristics of FmiR genes. Interestingly, among the top 25 enriched function terms of FmiR genes, 7 terms are related to the cell cycle process and 4 terms are related to metabolism, suggesting that these miRNA genes may coordinate the cell proliferation and the metabolism process. Indeed, some coding genes have been shown to link cell proliferation with metabolism [44], and it is possible that some sex-biased miRNAs could have similar regulatory roles in the cell as well. Moreover, it is well known that sex-biased gene (mRNA) expression, partially induced by sex hormone, significantly influences brain development [45]. Our result further suggests that FmiR genes could also be involved in brain development. Furthermore, we performed functional enrichment analysis on the sex-biased miRNA genes from each tissue. The top 50% significant functional terms are listed in Figure S1. As expected, when we focused on specific tissues, we found that the MmiR genes with higher tissue specificity tend to show more specified functions. For example, in colorectal mucosa, one of the significantly enriched functions of MmiR genes is carbohydrate metabolism. Indeed, the metabolic syndrome has been considered as important risk factors in colorectal neoplasm [46], which provides a plausible hypothesis that MmiRs from colorectal mucosa, through regulating carbohydrate metabolism, might participate in the disease progression.
Figure 5.
Functional enrichment analysis of the sex-biased miRNA genes
A. Functional enrichment analysis of the sex-biased miRNA genes using the TAM tool (P < 0.05). Numbers in the parenthesis indicate the numbers of miRNAs associated with the respective functions. B. Disease enrichment analysis of FmiR genes and MmiR genes. Only top 10% significant terms (P < 0.05) are shown. Numbers in the parenthesis indicate the numbers of miRNA genes associated with the respective diseases.
We further investigated the enriched diseases of the sex-biased miRNA genes. Notably, sex-biased miRNA genes are enriched in a myriad of cancers (Figure 5B). We found that FmiR genes are enriched in the miRNAs that are known to be down-regulated in hepatocellular carcinoma, and this observation is plausibly related to the enriched functional term of FmiR genes as tumor suppressor in the above analysis. FmiR genes are partially enriched in disease terms like Parkinson’s disease and Alzheimer's disease (Table S5), in coincide with their enriched function of brain development and aging. A previous clinical survey has shown that males are more vulnerable to cutaneous tuberculosis than females [47]. Interestingly, our result also indicates that a variety of MmiR genes are significantly associated with lupus vulgaris, a skin disease (Table S6). We also noted that some high-risk diseases for females like breast cancer and endometriosis are also enriched in MmiR genes (Table S6). We note that some of these MmiRs are known disease suppressors [48]. For instance, two MmiRs, hsa-miR-424 and hsa-miR-451, are shown to inhibit endometriosis [48], [49]. Thus, the lower expression of these MmiRs in females may contribute to the higher susceptibility of females to the associated diseases.
Preliminary analysis on The Cancer Genome Atlas (TCGA) datasets
For more comprehensive understanding of sex-biased miRNA expression, we collected miRNA (miRNA gene) expression profiles in tumor adjacent tissues from TCGA database, which covered 12 cancer types in 9 tissues [50]. We first compared the expression level of sex-biased miRNA genes and NmiR genes in the TCGA dataset. The expression levels across 9 tissues in TCGA dataset are summarized in Figure S2. Generally, the expression level of FmiR genes is significantly higher than that of MmiR and NmiR genes for most tissues, in line with the extensive associations between FmiRs and cancers (Figure 5B). We also tested the miRNA expression specificity across different cancer types. While MmiR genes and FmiR genes show comparable cancer type specificity (median: 0.30 vs. 0.29, Wilcoxon’s test P = 0.83), MmiR genes are more specific to particular cancer types than NmiR genes (median: 0.30 vs. 0.27, Wilcoxon’s test P = 0.014). Taken together, these results indicate that FmiR genes are associated with wider spectrum of cancer types.
We further applied our analysis framework (namely ISBM) to this TCGA dataset to identify sex-biased miRNA genes in tumor adjacent tissues. Note that the expression profiles from TCGA are available at the miRNA gene level rather than mature miRNA level. As a result, we identified 84 FmiR genes and 87 MmiR genes (Table S7). Similar to the analysis on the GEO dataset-derived sex-biased miRNAs, we compared sex-biased miRNA genes and non-biased miRNA genes in TCGA dataset for chromosomal distribution, conservation, tissue specificity, DSW, and functional enrichment. We found that the percentage of FmiR genes is higher than that of MmiR genes for 12 out of 23 chromosomes (Figure S3). FmiR genes are more clustered on chromosomes than MmiR genes (median distance 8.20E+6 bp vs. 3.03E+7 bp, Wilcoxon’s test P = 2.58E−4) and NmiR genes (median distance 8.20E+6 bp vs. 3.95E+7 bp, Wilcoxon’s test P = 3.42E−20). In terms of conservation, MmiRs’ families are found to be present in more species than FmiR families (median: 32 vs. 18, Wilcoxon’s test P = 0.016) and NmiR families (median: 32 vs. 4, Wilcoxon’s test P = 1.32E−21) (Figure S4). In addition, the tissue specificity of NmiRs is significantly higher than that of FmiRs (median: 0.94 vs. 0.79, Wilcoxon’s test P = 0.016) and MmiRs (median: 0.94 vs. 0.81, Wilcoxon’s test P = 0.016) (Figure S5A). Moreover, MmiR genes also show significantly higher DSW than FmiR genes (median: 0.057 vs. 0.029, Wilcoxon’s test P = 5.91E−4) and NmiR genes (median: 0.057 vs. 0.0078, Wilcoxon’s test P = 1.49E−16), while FmiR genes show significantly higher DSW than NmiR genes (median: 0.029 vs. 0.0078, Wilcoxon’s test P = 7.50E−6) (Figure S5B). Finally, according to the functional enrichment analysis, MmiR genes are associated with immune response, cell cycle, and cell proliferation, whereas FmiR genes are enriched for function terms like bone regeneration and inflammation (Figure S6).
Taken together, the sex-biased miRNAs found in tumor adjacent tissues exhibit noticeable distinctions in biological characteristics compared with those identified in normal tissues. The most prominent distinction is that the MmiRs identified in normal tissues seem to share some biological features with the FmiRs identified in tumor adjacent tissues. Indeed, we noticed that the MmiRs from normal tissues tend to show female-biased expression in tumor adjacent tissues (Figure S2K). That is to say, these MmiRs are more likely to show higher expression in females than males in the TCGA dataset (paired t-test P = 0.025). These results indicate that the sex-biased expression of miRNAs is context-specific and could be changed or reversed in disease conditions, even in tumor adjacent tissues without observable pathological alterations. In this study, we focused on sex-biased miRNAs in healthy samples, whereas the tumor adjacent tissues would not be good representatives of normal tissues in terms of gene expression pattern. Instead, a GTEx-like comprehensive panel of miRNA expression profiles in normal tissue samples would ultimately depict the whole picture of sex-biased miRNAs across different tissues in human body [51] in the future, although such expression panel is still prohibitively expensive and labor-intensive for now.
Conclusions
Identifying the molecular signature of sex difference has profound importance for disease studies and personalized medicine. Our previous studies on the sex-biased coding genes reveal that compared to male-biased coding genes, female-biased coding genes have higher evolutionary rate, higher single-nucleotide polymorphism density, lower homologous gene numbers, and younger phyletic age, which are highly involved in immune-related functions, whereas male-biased coding genes are more enriched in metabolic process [26]. Evidence for the functional importance of small non-coding RNAs, such as miRNAs, in diverse biological processes has been accumulating. However, whether and how the miRNAs are expressed in sex-biased fashion remain largely unexplored, which would hinder the full understanding of sexual discrepancy in physiology, disease incidence, and therapeutic response. We speculate that sex differences should also exist in terms of miRNA expression, and such sex-biased miRNA expression would carry functional implications as well. In the present study, we found 73 female-biased miRNAs and 163 male-biased miRNAs. Male-biased miRNAs exhibit a faster evolutionary rate and a higher tissue specificity, whereas female-biased miRNAs have higher disease spectrum width and are likely to be related to various cancers and neurodegenerative diseases. Functional annotation shows that female-biased miRNA genes are associated with metabolism process and cell cycle process, whereas male-biased miRNA genes tend to be enriched in histone modification and circadian rhythm.
Nevertheless, due to the intrinsic characteristics of miRNAs and the limitation of current miRNA annotations, some analyses are difficult to perform. For example, we tried to investigate the SNP density in sex-biased miRNA genes and found that miRNA gene loci (based on pre-miRNAs) currently available are too short to have sizable number of SNPs on them for further analysis. Besides, as indicated by the analysis on TCGA data, caution should be taken on the biological characteristics of samples when compiling the dataset, as the sex-biased expression of miRNAs is context-dependent.
Furthermore, miRNA expression datasets currently available clearly has their limitations. First, the sample size of our dataset is limited due to the lack of gender-labeled samples for most miRNA expression profiles in GEO. Second, the number of male and female samples could be imbalanced in a particular dataset, resulting in additional bias. Integrative analysis of sex-biased miRNAs and sex-biased genes could provide novel insights, but current tissue coverage of the miRNA datasets does not permit such analysis. In addition, many details like age and ethnic groups are missing or insufficient in current heterogeneous GEO datasets. Therefore, more rigorous pipelines, in which the sex-biased miRNAs are corrected by confounding co-factors [12], [24], cannot be performed to reduce the false positives in the current study. Therefore, we expect that more comprehensive panels of healthy human miRNA expression profiles would become available in the future, to enable more reliable analysis and provide more insightful information for understanding physiology, diseases, medicine and clinical therapy in sexual dimorphism.
Materials and methods
Identification of sex-biased miRNAs (ISBM)
We searched the human miRNA expression datasets in GEO [25]. Only datasets generated for human normal samples with gender information were retained for manual curation. The 8 GEO datasets selected include GSE15745, GSE34608, GSE41012, GSE41574, GSE48353, GSE67489, GSE70425, and GSE77668. We mapped probes to miRNA name, deleted null values, and merged redundant probes by averaging their expression values. Next, we classified the samples in each dataset into two groups according to gender information, the male group and the female group, to identify the sex-biased miRNAs. Using Wilcoxon’s test (P < 0.05), sex-biased miRNAs are identified and distinguished based on the bias direction (i.e., male-biased or female-biased) according to fold change. The final list of sex-biased miRNAs was obtained by merging the results from each dataset and excluding the miRNAs showing conflicting bias directions across different tissues.
Human blood samples
Blood samples from 9 healthy males and 9 healthy females were obtained from Ruike Donghua Translation Medical Research Center. The whole blood samples were EDTA anticoagulant, and were stored at −80 °C before use. The usage of patient-derived materials was approved by the Ethics Committees of the Staff Hospital of Jidong Oil-field of Chinese National Petroleum, Beijing Tiantan Hospital, and Capital Medical University. Written consent was obtained from all of the patients.
Bulge-loop real-time RT-PCR
Similar to our previous study [52], blood total RNA was isolated with Trizol reagent (Invitrogen). Complementary DNA was reversely transcribed in RNase-free water using 0.2–0.5 μg of total RNA mixed with 1 µl (500 nM) miRNA-specific bulge-loop RT-primers. Real-time PCR experiment was performed on Real-Time qPCR System (Agilent Technologies, Stratagene Mx3000P). For quantitative assay of miRNAs in the blood, their relative expression levels were firstly normalized to that of small nuclear RNA U6 in each gender, and then normalized to female data values using 2−ΔΔCt method. All the Bulge-Loop RT-primers for both miRNAs and U6 were purchased from RiboBio Co. Ltd (Guangzhou, China) [53], [54].
Analysis of the chromosomal distribution of sex-biased miRNA genes
The sex-biased miRNA genes were mapped onto chromosomes. The number of miRNA genes on each chromosome was counted to calculate the proportion of FmiR genes or MmiR genes to the total number of miRNA genes on each chromosome. Meanwhile, the number of sex-biased miRNA genes per million bp per chromosome was computed to depict more detailed distribution using Circos graph [28]. The correlation between the proportions of sex-biased miRNA genes with those of sex-biased genes on each chromosome was evaluated using Spearman’s correlation. We next calculated the intra-group distance of any miRNA gene pair on the same chromosome within FmiR, MmiR and NmiR groups to test if some miRNA genes tend to be clustered on the chromosome.
Analyzing evolutionary characteristics and functional enrichment of sex-biased miRNA genes
Family numbers of miRNAs were downloaded from the miRBase database version 21 [55] as one of the characteristics of conservation. Based on the species in which the corresponding miRNA family members are present, we classified all miRNA genes into 5 group according to the method described by Zhang and colleagues [31]. Numbers of species in which the family members of FmiR, MmiR, and NmiR genes are present were counted. The enrichment analysis of sex-biased miRNA genes were executed, taking into consideration of both functions and disease associations of miRNA genes, using TAM 2.0 (http://www.scse.hebut.edu.cn/tam/) with the P value threshold of 0.05 and the analysis of up- and down-miRNAs in diseases enabled [40].
Expression regulation and DSW analysis
To figure out the tissue expression specificity of miRNAs, we first accessed the miRNA expression profile described by Liang and colleagues [34], which covered 345 miRNAs and 40 normal tissues, such as brain, muscle, lymphoid, and respiratory systems. For each miRNA in our dataset, we calculated the tissue expression specificity based on its expression profile, if applicable. The tissue expression specificity of a miRNA is defined as the ratio of its maximum expression level among the 40 tissues examined against its total expression from all of the 40 tissues [26]. To investigate the regulation of sex-biased miRNA genes, we obtained transcription factor (TF) binding sites in each tissue from ChIP-Atlas (http://chip-atlas.org/) and mapped them to the proximal regions (from upstream 5000 bp to downstream 1000 bp) of the miRNA gene loci. A sex-biased miRNA identified in a specific tissue is deemed to be regulated by a particular TF if the TF binds to the proximal region of the miRNA gene in the related tissues. We further screened the TFs preferentially regulating sex-biased miRNA genes with at least 5 binding sites present among the FmiRs or MmiRs. We performed the function and diseases enrichment analysis of these TFs using g:Profiler and DAVID tools, respectively [56], [57].
DSW was presented by Qiu et al. [38] as an important measure of the relationship between miRNAs and diseases. DSW was calculated as the number of diseases associated with a given miRNA gene divided by the total number of diseases associated with any human miRNA gene. Here, we used the updated v2.0 version of HMDD [39] miRNA-disease association dataset to re-calculate DSW for each miRNA in our datasets.
TCGA data collection and analysis
We downloaded miRNA-seq data of the non-tumor control tissues from TCGA database (https://portal.gdc.cancer.gov/), discarding samples without clear gender information. We obtained data for 12 cancer types: bladder urothelial carcinoma (BLCA), cholangiocarcinoma (CHOL), esophageal carcinoma (ESCA), head-neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), stomach adenocarcinoma (STAD), and thyroid carcinoma (THCA). The expression profiles from TCGA are available at the miRNA gene level rather than mature miRNA level. Therefore, for each cancer type, we identified the sex-biased miRNA genes using the aforementioned identified sex-biased miRNAs (ISBM) pipeline. Then, the framework, which was used for analyzing the abovementioned GEO dataset-derived sex-biased miRNAs, was applied to the sex-biased miRNA genes obtained from the TCGA dataset, to analyze chromosome distribution, conservation, tissue specificity, DSW, and functional enrichment of these sex-biased miRNA genes.
Authors’ contributions
QC and YZ (Yuan Zhou) conceived the project. CC collected the datasets and JS participated in the pre-processing of the datasets. CC performed the computational analysis. WY, YZ (Yong Zhou), and JY performed the experimental assays. CC and WY drafted the manuscript. QC and YZ (Yuan Zhou) thoroughly revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
The authors thank the staff of the Recovery Medical Technology Development Co. for their important assistance in human blood sample collection and storage. The authors are also grateful to the anonymous reviewers for their constructive comments. This study was supported by the National Natural Science Foundation of China (Grant Nos. 81670462 and 81422006) to QC and the Fundamental Research Funds for Central Universities of China (Grant No. BMU2017YJ004) to YZ (Yuan Zhou).
Handled by Xiu-Jie Wang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.gpb.2018.03.004.
Contributor Information
Qinghua Cui, Email: cuiqinghua@hsc.pku.edu.cn.
Yuan Zhou, Email: zhouyuanbioinfo@bjmu.edu.cn.
Supplementary material
The following are the Supplementary data to this article:
The intra-chromosomal distance between miRNA genes and the functional enrichment of FmiR genes and MmiR genes in each tissue. A. The average intra-chromosomal distances between miRNA genes in FmiR, MmiR, and NmiR groups as well as those between all miRNAs across 23 chromosomes. B. Functional enrichment analysis of FmiR genes using the TAM tool. C. Functional enrichment analysis of MmiR genes, only top 50% significant terms (P < 0.05) are shown. Note that some tissues are lack of significant functional terms associated with FmiR genes or MmiR genes. Numbers in the parenthesis indicate the numbers of the sex-biased miRNA genes (FmiR genes or MmiR genes) associated with the respective functions.
The comparison of expression and tissue specificity in control samples (tumor adjacent tissues) across 12 cancer types from TCGA. A.−I. The comparison of expression levels of FmiR genes, MmiR genes, and NmiR genes in bile duct, bladder, esophagus, head and neck, kidney, liver, lung, stomach, and thyroid in TCGA. J. The comparison of expression specificity of FmiR genes, MmiR genes, and NmiR genes among the 9 aforementioned tissues. *P < 0.05; ***P < 0.001, according to Wilcoxon’s test. K. MmiR genes identified from the GEO dataset show female-biased expression across 12 cancer types in the TCGA dataset. n.s., non-significant.
The chromosomal distribution of the sex-biased miRNA genes from TCGA. A. Chromosomal distribution of the sex-biased miRNA genes. Y-axis shows the percentage of the sex-biased miRNA genes to the total miRNA genes on the same chromosome. B. Detailed chromosomal distribution of the sex-biased miRNA genes and sex-biased genes, the number of the sex-biased miRNA genes per million base-pairs is plotted using the barplots embedded in the Circos graph. Red, green, purple and blue bars represent the distributions of the FmiR genes, the MmiR genes, FGs and MGs, respectively. C. The boxplot comparing the intra-chromosomal distances between the sex-biased miRNA genes. **P < 0.01; ***P < 0.001, according to Wilcoxon’s test. D. The percentage of the sex-biased miRNA genes on autosomes and sex chromosomes in each tissue. FmiR, female-biased miRNA; MmiR, male-biased miRNA; FG, female-biased coding gene; MG, male-biased coding gene; NmiR, non-biased miRNA.
The evolutionary characteristic of the sex-biased miRNA genes from TCGA. A. The distribution of the FmiR, MmiR, and NmiR genes in different conservation groups. Note that miRNA genes in more conserved group (e.g., primate-specific group) don’t include the miRNA genes present in less conserved groups (e.g., human-specific group). B. Comparison of the number of species in which the corresponding miRNA gene family members are present. *P < 0.05; ***P < 0.001, according to Wilcoxon’s test.
The comparison of tissue expression specificity and DSW of the sex-biased miRNA genes from TCGA. A. Comparison of tissue expression specificity between FmiR, MmiR, and NmiR genes. The tissue expression specificity of an miRNA is defined as the ratio of its maximum expression level among the 40 tissues examined against the total expression from all of the 40 tissues. B. Comparison of DSW between FmiR genes, MmiR genes, and NmiR genes. *P < 0.05; ***P < 0.001, according to Wilcoxon’s test. DSW, disease spectrum width, which is defined as the number of diseases associated with a given miRNA divided by the total number of diseases associated with any miRNAs. n.s., non-significant.
Functional enrichment analysis of the sex-biased miRNA genes from TCGA. A. Functional enrichment analysis of the sex-biased miRNA genes using the TAM tool, only the top 20% terms are shown here (P < 0.05). Numbers in the parenthesis indicate the numbers of miRNA genes associated with the respective functions. B. Disease enrichment analysis of FmiR genes and MmiR genes. Only the top 5% significant terms (P < 0.05) are shown. Numbers in the parenthesis indicate the number of miRNA genes associated with the respective diseases. n.s., non-significant.
References
- 1.Zakiniaeiz Y., Cosgrove K.P., Potenza M.N., Mazure C.M. Balance of the sexes: addressing sex differences in preclinical research. Yale J Biol Med. 2016;89:255–259. [PMC free article] [PubMed] [Google Scholar]
- 2.Miller V.M. Why are sex and gender important to basic physiology and translational and individualized medicine? Am J Physiol Heart Circ Physiol. 2014;306:H781–H788. doi: 10.1152/ajpheart.00994.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Regitz-Zagrosek V., Kararigas G. Mechanistic pathways of sex differences in cardiovascular disease. Physiol Rev. 2017;97:1–37. doi: 10.1152/physrev.00021.2015. [DOI] [PubMed] [Google Scholar]
- 4.Zagni E., Simoni L., Colombo D. Sex and gender differences in central nervous system-related disorders. Neurosci J. 2016;2016:2827090. doi: 10.1155/2016/2827090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goto T., Baba T., Ito A., Maekawa K., Koshiji T. Gender differences in stroke risk among the elderly after coronary artery surgery. Anesth Analg. 2007;104:1016–1022. doi: 10.1213/01.ane.0000263279.07361.1f. [tables of contents] [DOI] [PubMed] [Google Scholar]
- 6.Barker-Collo S., Bennett D.A., Krishnamurthi R.V., Parmar P., Feigin V.L., Naghavi M. Sex differences in stroke incidence, prevalence, mortality and disability-adjusted life years: results from the global burden of disease study 2013. Neuroepidemiology. 2015;45:203–214. doi: 10.1159/000441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasile M., Corinaldesi C., Antinozzi C., Crescioli C. Vitamin D in autoimmune rheumatic diseases: a view inside gender differences. Pharmacol Res. 2017;117:228–241. doi: 10.1016/j.phrs.2016.12.038. [DOI] [PubMed] [Google Scholar]
- 8.Hernandez-Moreno A., Vidal-Casariego A., Calleja-Fernandez A., Kyriakos G., Villar-Taibo R., Urioste-Fondo A. Chronic enteritis in patients undergoing pelvic radiotherapy: prevalence, risk factors and associated complications. Nutr Hosp. 2015;32:2178–2183. doi: 10.3305/nh.2015.32.5.9562. [DOI] [PubMed] [Google Scholar]
- 9.Maeng L.Y., Milad M.R. Sex differences in anxiety disorders: interactions between fear, stress, and gonadal hormones. Horm Behav. 2015;76:106–117. doi: 10.1016/j.yhbeh.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akhtaruzzaman M., Hossain M.A., Khan R.H., Karim M.R., Choudhury A.M., Islam M.S. Knowledge and practices of mothers on childhood diarrhoea and its management attended at a tertiary hospital in Bangladesh. Mymensingh Med J. 2015;24:269–275. [PubMed] [Google Scholar]
- 11.Xhyheri B., Bugiardini R. Diagnosis and treatment of heart disease: are women different from men? Prog Cardiovasc Dis. 2010;53:227–236. doi: 10.1016/j.pcad.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Yuan Y., Liu L., Chen H., Wang Y., Xu Y., Mao H. Comprehensive characterization of molecular differences in cancer between male and female patients. Cancer Cell. 2016;29:711–722. doi: 10.1016/j.ccell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Y., Thomson J.M., Wong H.Y., Hammond S.M., Hogan B.L. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., Baskerville S., Shenoy A., Babiarz J.E., Baehner L., Blelloch R. Embryonic stem cell-specific microRNAs regulate the g1-s transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebert M.S., Sharp P.A. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost R.J., Olson E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai R., Ahmed S.A. Sexual dimorphism of miRNA expression: a new perspective in understanding the sex bias of autoimmune diseases. Ther Clin Risk Manag. 2014;10:151–163. doi: 10.2147/TCRM.S33517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lippi G., Steinert J.R., Marczylo E.L., D'Oro S., Fiore R., Forsythe I.D. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J Cell Biol. 2011;194:889–904. doi: 10.1083/jcb.201103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koturbash I., Zemp F., Kolb B., Kovalchuk O. Sex-specific radiation-induced microRNAome responses in the hippocampus, cerebellum and frontal cortex in a mouse model. Mutat Res. 2011;722:114–118. doi: 10.1016/j.mrgentox.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Murphy S.J., Lusardi T.A., Phillips J.I., Saugstad J.A. Sex differences in microRNA expression during development in rat cortex. Neurochem Int. 2014;77:24–32. doi: 10.1016/j.neuint.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hewagama A., Gorelik G., Patel D., Liyanarachchi P., McCune W.J., Somers E. Overexpression of X-linked genes in T cells from women with lupus. J Autoimmun. 2013;41:60–71. doi: 10.1016/j.jaut.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meder B., Backes C., Haas J., Leidinger P., Stahler C., Grossmann T. Influence of the confounding factors age and sex on microRNA profiles from peripheral blood. Clin Chem. 2014;60:1200–1208. doi: 10.1373/clinchem.2014.224238. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y.T., Tsai P.C., Liao Y.C., Hsu C.Y., Juo S.H. Circulating microRNAs have a sex-specific association with metabolic syndrome. J Biomed Sci. 2013;20:72. doi: 10.1186/1423-0127-20-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ameling S., Kacprowski T., Chilukoti R.K., Malsch C., Liebscher V., Suhre K. Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC Med Genomics. 2015;8:61. doi: 10.1186/s12920-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clough E., Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo S., Zhou Y., Zeng P., Xu G., Wang G., Cui Q. Identification and analysis of the human sex-biased genes. Brief Bioinform. 2018;19:188–198. doi: 10.1093/bib/bbw125. [DOI] [PubMed] [Google Scholar]
- 27.Sun G., Yan J., Noltner K., Feng J., Li H., Sarkis D.A. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu M., Zhang Q., Deng M., Miao J., Guo Y., Gao W. An analysis of human microRNA and disease associations. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conesa A., Gotz S., Garcia-Gomez J.M., Terol J., Talon M., Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Q., Lu M., Cui Q. SNP analysis reveals an evolutionary acceleration of the human-specific microRNAs. Nat Precedings. 2008 http://hdl.handle.net/10101/npre.2008.2127.1. [Google Scholar]
- 32.Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 33.Guo Z., Maki M., Ding R., Yang Y., Zhang B., Xiong L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci Rep. 2014;4:5150. doi: 10.1038/srep05150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang Y., Ridzon D., Wong L., Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 36.Wang L., Gao W., Hu F., Xu Z., Wang F. MicroRNA-874 inhibits cell proliferation and induces apoptosis in human breast cancer by targeting CDK9. FEBS Lett. 2014;588:4527–4535. doi: 10.1016/j.febslet.2014.09.035. [DOI] [PubMed] [Google Scholar]
- 37.Cheung C.L., Chan B.Y., Chan V., Ikegawa S., Kou I., Ngai H. Pre-B-cell leukemia homeobox 1 (PBX1) shows functional and possible genetic association with bone mineral density variation. Hum Mol Genet. 2009;18:679–687. doi: 10.1093/hmg/ddn397. [DOI] [PubMed] [Google Scholar]
- 38.Qiu C., Chen G., Cui Q. Towards the understanding of microRNA and environmental factor interactions and their relationships to human diseases. Sci Rep. 2012;2:318. doi: 10.1038/srep00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y., Qiu C., Tu J., Geng B., Yang J., Jiang T. 0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014;42:D1070–D1074. doi: 10.1093/nar/gkt1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu M., Shi B., Wang J., Cao Q., Cui Q. TAM: a method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinform. 2010;11:419. doi: 10.1186/1471-2105-11-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Han X., Wan Y., Zhang S., Zhao Y., Fan R. TAM 2.0: tool for microRNA set analysis. Nucleic Acids Res. 2018;46:W180–W185. doi: 10.1093/nar/gky509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marmorstein R., Trievel R.C. Histone modifying enzymes: structures, mechanisms, and specificities. Biochim Biophys Acta. 2009;1789:58–68. doi: 10.1016/j.bbagrm.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai H.W., Grant P.A., Rissman E.F. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;4:47–53. doi: 10.4161/epi.4.1.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 45.Miller D.I., Halpern D.F. The new science of cognitive sex differences. Trends Cogn Sci. 2014;18:37–45. doi: 10.1016/j.tics.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 46.Chang L.C., Wu M.S., Tu C.H., Lee Y.C., Shun C.T., Chiu H.M. Metabolic syndrome and smoking may justify earlier colorectal cancer screening in men. Gastrointest Endosc. 2014;79:961–969. doi: 10.1016/j.gie.2013.11.035. [DOI] [PubMed] [Google Scholar]
- 47.Kodnani A., Shah J.M. A comparative overview of histopathology of granulomatous lesions of skin. Int J Clin Biomed Res. 2016;2:6–9. [Google Scholar]
- 48.Li Q., Qiu X.M., Li Q.H., Wang X.Y., Li L., Xu M. MicroRNA-424 may function as a tumor suppressor in endometrial carcinoma cells by targeting E2F7. Oncol Rep. 2015;33:2354–2360. doi: 10.3892/or.2015.3812. [DOI] [PubMed] [Google Scholar]
- 49.Nothnick W.B., Graham A., Holbert J., Weiss M.J. miR-451 deficiency is associated with altered endometrial fibrinogen alpha chain expression and reduced endometriotic implant establishment in an experimental mouse model. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomczak K., Czerwinska P., Wiznerowicz M. The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.GTEx Consortium Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu W., Zhou Y., Xu G., Geng B., Cui Q. Transcriptome analysis reveals non-identical microRNA profiles between arterial and venous plasma. Oncotarget. 2017;8:28471–28480. doi: 10.18632/oncotarget.15310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding J., Huang S., Wu S., Zhao Y., Liang L., Yan M. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12:390–399. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 54.Yang W., Wang J., Chen Z., Chen J., Meng Y., Chen L. Nfe2 induces mir-423-5p to promote gluconeogenesis and hyperglycemia by repressing the hepatic FAM3A-ATP-Akt pathway. Diabetes. 2017;66:1819–1832. doi: 10.2337/db16-1172. [DOI] [PubMed] [Google Scholar]
- 55.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 57.Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The intra-chromosomal distance between miRNA genes and the functional enrichment of FmiR genes and MmiR genes in each tissue. A. The average intra-chromosomal distances between miRNA genes in FmiR, MmiR, and NmiR groups as well as those between all miRNAs across 23 chromosomes. B. Functional enrichment analysis of FmiR genes using the TAM tool. C. Functional enrichment analysis of MmiR genes, only top 50% significant terms (P < 0.05) are shown. Note that some tissues are lack of significant functional terms associated with FmiR genes or MmiR genes. Numbers in the parenthesis indicate the numbers of the sex-biased miRNA genes (FmiR genes or MmiR genes) associated with the respective functions.
The comparison of expression and tissue specificity in control samples (tumor adjacent tissues) across 12 cancer types from TCGA. A.−I. The comparison of expression levels of FmiR genes, MmiR genes, and NmiR genes in bile duct, bladder, esophagus, head and neck, kidney, liver, lung, stomach, and thyroid in TCGA. J. The comparison of expression specificity of FmiR genes, MmiR genes, and NmiR genes among the 9 aforementioned tissues. *P < 0.05; ***P < 0.001, according to Wilcoxon’s test. K. MmiR genes identified from the GEO dataset show female-biased expression across 12 cancer types in the TCGA dataset. n.s., non-significant.
The chromosomal distribution of the sex-biased miRNA genes from TCGA. A. Chromosomal distribution of the sex-biased miRNA genes. Y-axis shows the percentage of the sex-biased miRNA genes to the total miRNA genes on the same chromosome. B. Detailed chromosomal distribution of the sex-biased miRNA genes and sex-biased genes, the number of the sex-biased miRNA genes per million base-pairs is plotted using the barplots embedded in the Circos graph. Red, green, purple and blue bars represent the distributions of the FmiR genes, the MmiR genes, FGs and MGs, respectively. C. The boxplot comparing the intra-chromosomal distances between the sex-biased miRNA genes. **P < 0.01; ***P < 0.001, according to Wilcoxon’s test. D. The percentage of the sex-biased miRNA genes on autosomes and sex chromosomes in each tissue. FmiR, female-biased miRNA; MmiR, male-biased miRNA; FG, female-biased coding gene; MG, male-biased coding gene; NmiR, non-biased miRNA.
The evolutionary characteristic of the sex-biased miRNA genes from TCGA. A. The distribution of the FmiR, MmiR, and NmiR genes in different conservation groups. Note that miRNA genes in more conserved group (e.g., primate-specific group) don’t include the miRNA genes present in less conserved groups (e.g., human-specific group). B. Comparison of the number of species in which the corresponding miRNA gene family members are present. *P < 0.05; ***P < 0.001, according to Wilcoxon’s test.
The comparison of tissue expression specificity and DSW of the sex-biased miRNA genes from TCGA. A. Comparison of tissue expression specificity between FmiR, MmiR, and NmiR genes. The tissue expression specificity of an miRNA is defined as the ratio of its maximum expression level among the 40 tissues examined against the total expression from all of the 40 tissues. B. Comparison of DSW between FmiR genes, MmiR genes, and NmiR genes. *P < 0.05; ***P < 0.001, according to Wilcoxon’s test. DSW, disease spectrum width, which is defined as the number of diseases associated with a given miRNA divided by the total number of diseases associated with any miRNAs. n.s., non-significant.
Functional enrichment analysis of the sex-biased miRNA genes from TCGA. A. Functional enrichment analysis of the sex-biased miRNA genes using the TAM tool, only the top 20% terms are shown here (P < 0.05). Numbers in the parenthesis indicate the numbers of miRNA genes associated with the respective functions. B. Disease enrichment analysis of FmiR genes and MmiR genes. Only the top 5% significant terms (P < 0.05) are shown. Numbers in the parenthesis indicate the number of miRNA genes associated with the respective diseases. n.s., non-significant.