Abstract
Oligodendrogliomas (ODG) are brain tumors accounting for approximately 10% of all central nervous system cancers. CIC is a transcription factor that is mutated in most patients with ODG; these mutations are believed to be a key oncogenic event in such cancers. Analysis of the Drosophila melanogaster orthologue of CIC, Capicua, indicates that CIC loss phenocopies activation of the EGFR/RAS/MAPK pathway, and studies in mammalian cells have demonstrated a role for CIC in repressing the transcription of the PEA3 subfamily of ETS transcription factors. Here we address the mechanism by which CIC represses transcription and assess the functional consequences of CIC inactivation. Genome-wide binding patterns of CIC in several cell types revealed that CIC target genes were enriched for MAPK effector genes involved in cell cycle regulation and proliferation. CIC binding to target genes was abolished by high MAPK activity, which led to their transcriptional activation. CIC interacted with the SIN3 deacetylation complex and, based on our results, we suggest that CIC functions as a transcriptional repressor through the recruitment of histone deacetylases. Independent single amino acid substitutions found in ODG tumors prevented CIC from binding its target genes. Taken together, our results show that CIC is a transcriptional repressor of genes regulated by MAPK signaling, and that ablation of CIC function leads to increased histone acetylation levels and transcription at these genes, ultimately fueling mitogen-independent tumor growth.
Keywords: CNS CANCERS/Gliomas, Glioblastomas, TUMOR SUPPRESSORS/Other tumor suppressor genes, GENE REGULATION/GENE REGULATION, GENE REGULATION/Mechanisms of transcription
User-defined keywords: Oligodendroglioma, CIC, Transcription
Introduction
Oligodendrogliomas (ODGs) represent a subgroup of low-grade glioma with a distinct mutational spectrum. The vast majority of ODG cases contain chromosomal deletions of 1p and 19q and 70% harbor mutations in CIC, located on chromosome 19q13.2 (1). CIC encodes a transcription factor containing a SOX-like high mobility group (HMG) domain. At least two isoforms of CIC have been identified in humans: a short form CIC-S and a longer isoform CIC-L that shares the HMG domain but contains an extended N-terminal part (2). Mutations in CIC frequently lead to truncations or amino acid substitutions in residues that are thought to be essential for DNA binding (Supplementary Figure S1A and B). Together with 1p/19q chromosomal deletions, these recurrent CIC mutations are thought to represent an ablation of CIC function (3), being causally involved in ODG formation. Of further interest, CIC mutations are mutually exclusive with alterations of EGFR, TP53, ATRX and CDKN2A (4,5).
Most of our knowledge about CIC stems from studies in Drosophila melanogaster, where it was genetically defined as a transcriptional repressor acting downstream of receptor tyrosine kinase (RTK) signaling regulating developmental patterning (6,7). MAPK-mediated phosphorylation of Cic leads to its nuclear export and/or degradation of Cic (8) and expression of Cic target genes. In mammals RTK signaling has similar effects on CIC mediated gene repression, possibly involving MAPK and p90RSK (9). How CIC is acting as a transcriptional repressor is currently not well understood. In Drosophila, Cic-mediated repression is dependent on the co-repressor protein Groucho (6,10). Mouse studies have suggested that CIC-mediated repression is in part dependent on the binding to Ataxin1 (ATXN1) or a related factor ATXN1L (11–13).
Several mouse knockout studies have shown that germline deletion of murine Cic leads to perinatal lethality (13–15) and deletion of Cic after birth has been linked to T-cell lymphoblastic lymphoma formation (14,16). In Ewing sarcomas, a genetic fusion of CIC to the exons coding for the transcriptional activator domain of DUX4 causes de-repression of PEA3 (polyoma enhancer activator 3)-family genes (17). Furthermore, CIC deletion and mutation was shown to influence metastasis in an EGFR inhibitor-resistant model of lung adenocarcinoma, through the deregulation of the PEA3-family gene Etv4 (18). CIC may therefore act as an important tumor suppressor through its transcriptional repression of its target genes, including the PEA3 subfamily, whose deregulation was linked to cancer development (17,18). However, the mechanistic consequences of inactivation or mutation of CIC and their contribution to tumorigenesis of ODG are not well understood. Furthermore, the mechanism through which CIC achieves gene repression in mammals is not known.
In this study, we aimed at elucidating how CIC contributes to normal development, the consequences of CIC loss and how this could lead to tumor formation.
Material and Methods
Cell lines and culture
mESCs (E14TG2a.4, ATCC) were cultured on gelatine in 2i medium: 50:50 DMEM/F12:Neurobasal (Invitrogen) supplemented with N2/B27 (Invitrogen), 0.05 mM β-mercaptoethanol, 0.1 mM non-essential amino acids, 2 mM GlutaMAX, 1 mM sodium pyruvate, Pen/Strep, leukemia inhibitory factor (LIF), 1 μM MEK inhibitor (PD0325901) and 3 μM GSK inhibitor (CT-99021).
Mouse neural progenitor cells derived from mESCs were cultured on PDL/Laminin in complete NSC medium: 50:50 DMEM/F12:Neurobasal (Invitrogen) supplemented with N2/B27 (Invitrogen), 0.05 mM β-mercaptoethanol, 2 mM GlutaMAX, 0.1 mM non-essential amino acids, Pen/Strep, 1 mM sodium pyruvate, 5mM Hepes, 2 µg/ml Heparin, 50 µg/ml BSA, 10 ng/ml bFGF/EGF
Human GNS line G144 (19) and human fetal neural stem cell lines 18.5 and 21.5 (a kind gift of Dr. Steven Goldman, University of Rochester Medical Center, USA) were cultured on PDL/Laminin in complete NSC medium.
Flp-In-T-REX-293 cells were cultured in DMEM (Invitrogen) with 10% FBS (HyClone) and Pen/Strep. Cells were authenticated by measuring the expression of specific cellular markers and by observing their morphology and functionality in specific assays. All the cell lines used were routinely (every 4 weeks) tested negative for mycoplasma by PCR. Cell culture time between thawing and experiment did not exceed 5 passages.
CRISPR KO
sgRNAs were cloned into pSpCas9(BB)-2A-GFP (Addgene 48138), using the protocol described in (20) and transfected into mESCs (Lipofectamine 2000, Invitrogen), or G144 cells (Amaxa Nucleofector II, Program A033). GFP-positive cells were single cell sorted 48h post-transfection, expanded and screened by immunoblotting for Cic deletion. Gene-targeting was confirmed by PCR amplification and Sanger sequencing. Mouse Cic KO clones (KO1, KO2 and KO3) all harbored out-of-frame mutations causing complete protein depletion. Human CIC KO clones (KO8 and KO9) harbored a mixture of out-of-frame and large in frame mutations resulting in expression of an HMG domain-deleted truncated protein. Primers and sgRNAs are listed in Supplementary Table 1.
Plasmids and cDNA
The open-reading frame of human CIC-L was amplified from a cDNA library derived from human fetal brain and cloned into TOPO/pCR8, yielding a flawless 7554 bp clone encoding a 2513 amino acid protein (splice variant ENST00000572681, ProteinID ENSP00000459719).
The CIC-R215W and R1515C mutants were generated using the Quickchange II site directed mutagenesis kit (Agilent Technologies) and expressed from pPB-CAG-Dest-pA-pgk-bsd (Addgene 74918) by co-transfection with transposase pCyL43 (PBase; Sanger Institute’s plasmid repository) into mESCs and selected with 5 µg/µl Blasticidin. Primers are listed in Supplementary Table 1.
Differentiation
mESCs were differentiated into NSCs as described in (21). Briefly, 104 cells/cm2 mESCs were seeded onto gelatine into complete NSC medium without bFGF/EGF. Culture medium was changed every day. Four days after initiation of differentiation, cells were plated onto PDL/Laminin coated dishes in complete NSC medium containing bFGF/EGF (10 ng/ml). Cells were subcultured additional 10 days until homogeneous NSC populations were established.
Starvation and inhibitor treatment
80% confluent NSC cultures were washed twice with PBS and cultured in NSC medium without bFGF/EGF for 16 h. Inhibitor treatment with 1 μM MEK inhibitor (PD0325901) was performed in 80% confluent NSC or GNS cultures grown in complete NSC medium for 4 and 24 h respectively.
Monoclonal antibody generation
The C-terminal 195 amino acids of CIC (cloning primers in Supplementary Table 1) were expressed by generating a baculovirus through the co-transfection of pVL-Flag-His-CIC-C-terminus and Bsu36I-linearized Bakpak6 baculovirus DNA. Flag-His-tagged proteins were expressed in Trichoplusia ni, High Five, cells and purified by Flag–Sepharose affinity chromatography as described (22) and used to immunize mice. Spleens were isolated, homogenized and fused to the myeloma cell line SP2/0-Ag14 by addition of PEG1500. Supernatants from the resulting hybridomas were screened by ELISA, followed by further sub-cloning. Selected clones were tested for specificity and qualified for immunoblotting and immunoprecipitation using both human and mouse samples. All mouse work was approved by the Danish Animal Ethical Committee (“Dyreforsøgstilsynet”).
Immunoblotting
To prepare whole-cell-extracts, cells were lysed in high salt buffer (300 mM NaCl, 50 mM Tris-HCl pH7.5, 0.5% Triton X-100, 0.1% SDS, 1 mM EDTA, 1 mM DTT, Aprotinin, Leupeptin, 0.1 mM PMSF). Equal amounts of whole-cell-extracts were analysed by standard SDS-PAGE/immunoblotting. Primary antibodies and dilutions are listed in Supplementary Table 1.
RT-qPCR
Cells were washed once with PBS, lysed directly in RLT buffer (Qiagen) and total RNA was extracted using the RNAeasy kit (Qiagen). 200 ng of total RNA was reverse transcribed using TaqMan® Reverse Transcription Reagents (Applied Biosystems) and quantified using the LightCycler® 480 SYBR Green I system. qPCR was performed in technical triplicates. Primers are listed in Supplementary Table 1.
RNA-sequencing
500 ng of total RNA was used for library preparation using TruSeq RNA Library Prep Kit v2 (Illumina) according to the manufacturer’s recommendations. Libraries were sequenced using Illumina NextSeq 500 sequencer 75 bp Single End. RNA sequencing in G144 cells was done in technical triplicates for parental cells and technical duplicates for one WT and two CIC KO clones (KO8, KO9). Sequencing data was aligned via Tophat2 (23) and differential expression was called using DESeq2 (24). Genes were called as differential expressed for ≥2-fold change and FDR≤0.01. Data from parental cells and WT clone were treated as replicates. Similarly, data from the two KO clones were treated as replicates.
Size exclusion chromatography
293FT cell nuclear extracts were prepared as described (22) and fractionated on a Superose 6 PC 3.2/30 gel filtration column using an ÄKTA™ purification platform (GE Healthcare).
Chromatin immunoprecipitation
Cells were cross-linked (1% formaldehyde, 10 minutes), quenched with 125 mM glycine, washed twice with PBS and harvested in SDS buffer (50 mM Tris at pH 8.1, 0.5 % SDS, 100 mM NaCl, 5 mM EDTA). Cells were pelleted, resuspended in Triton-X IP buffer (100 mM Tris at pH 8.6, 0.3% SDS, 1.7% Triton X-100, and 5 mM EDTA) and chromatin was sonicated (fragment size 200-500bp). 25 µg chromatin (measured by Bradford) was pre-cleared with protein-A Sepharose beads (GE healthcare) for 1 hour and incubated with primary antibody overnight at 4°C. Protein-A Sepharose beads were blocked with 10 µg/ml BSA overnight at 4°C. Next day, beads and antibody/chromatin-mixture were incubated for 3 hours at 4°C. Beads were washed 3x with low salt buffer (1% Triton X-100, 0.1% SDS, 150 mM NaCl, 2 mM EDTA, pH 8.0, 20 mM Tris-HCl, pH 8.0) and twice with high salt buffer (1% Triton X-100, 0.1% SDS, 500mM NaCl, 2 mM EDTA, pH8.0, 20 mM Tris-HCl, pH8.0). DNA was eluted with elution-buffer (1% SDS, 0.1 M sodium bicarbonate) at 65°C overnight and purified using QIAquick PCR Purification Kit (Quiagen). DNA was analyzed using the LightCycler® 480 SYBR Green I system (Roche). qPCR was performed in technical triplicates. Primers and antibodies are listed in Supplementary Table 1.
For ChIP-seq 1-2 ng of ChIP DNA was used for library preparation, using the NEBNext Ultra II DNA library prep kit (E7370; NEB). Libraries were sequenced using Illumina NextSeq 500 sequencer 75 bp Single End.
Bioinformatics and motif finding
Sequenced reads were aligned using Bowtie2 (25) (mouse-mm10 and human-hg38). Duplicates were removed using Samtools RmDUP. Peaks were called in EaSeq (26) using standard settings and corresponding IgG control (FDR<10-5). For motif analysis 50bp surrounding the apex of each peak was used by MEME-ChIP (27) using standard settings in discriminative mode. Control peak set matching size and distance-to-TSS was generated in EaSeq (26).
Data availability
RNA-seq and ChIP-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-6681 and E-MTAB-6682 respectively.
Tandem affinity purification
Nuclear extracts (500 mg, 3x109 cells) from Flp-In-T-REX-293 cells expressing Flag–HA-tagged human CIC-L were pre-cleared and incubated with a 700 μl packed volume of anti-Flag beads (anti-Flag-M2-agarose, Sigma) overnight at 4°C with rotation. The beads were collected by centrifugation (700 g, 5 min) and washed five times with buffer A (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 0.2 mM PMSF, 1 mM DTT, 1 µg/ml aprotinin and 1 µg /ml leupeptin) and once with buffer A containing Benzonase at 4°C for 1h. The complexes were eluted with 1 bed volume of buffer A supplemented with 0.5 μg/μl Flag peptide for 4 hours. The eluate was collected by passing through a 0.45 µm, 1.5 ml centrifugal unit (Merck-Millipore). The Flag-IP elute was incubated with 120 µl of 50 % slurry HA-beads overnight. The beads were washed six times with buffer A and eluted with 100 µl buffer A supplemented with 1 μg/μl HA peptide for 2 h. The samples were boiled in SDS loading buffer and run shortly into an SDS–PAGE gel. A gel slice containing the purified proteins was isolated for mass spectrometry analysis. For comparative purposes, nuclear extracts from an equivalent number of Flp-In-T-REX-293 cells expressing Flag–HA-tagged human FUBP1 were processed in parallel.
Mass spectrometry
Proteins were in-gel digested using trypsin. Peptides were loaded on a nanoHPLC RSLC Ultimate 3000 (Thermo Scientific) coupled online with an Orbitrap Fusion Tribrid (Thermo). Peptides were separated using C18 nano-chromatography with a 60 min gradient, and acquired using Data-Dependent Acquisition with MS/MS fragmentation using Higher energy Collision Dissociation (HCD). Spectra identification was performed with Proteome Discoverer (v1.4, Thermo) using as reference the UniProt human database. Results were filtered for 1% False Discovery Rate.
Results
CIC binds directly to the promoter region of its target genes and is required for repressing Pea3 group genes in mouse ESCs
To investigate the mechanism by which CIC regulates transcription and the functional consequences of its inactivation, we initiated our studies in mouse embryonic stem cells (mESCs). mESCs can be selectively differentiated into neural stem cells (NSCs) and further to oligodendrocyte precursor cells (OPCs) (21,28). CIC deletions most likely occur in one of these immature cell types during ODG formation. mESCs therefore appeared to be appropriate to test the consequences of Cic loss. In addition the 2i growth conditions used to expand mESCs enables investigation of CIC in the absence of MEK/ERK signaling (29). Thus, we generated Cic knockout (KO) mESCs using CRISPR-Cas9 (Supplementary Figure S1C). Targeted knockout clones showed no detectable levels of CIC (Figure 1A).
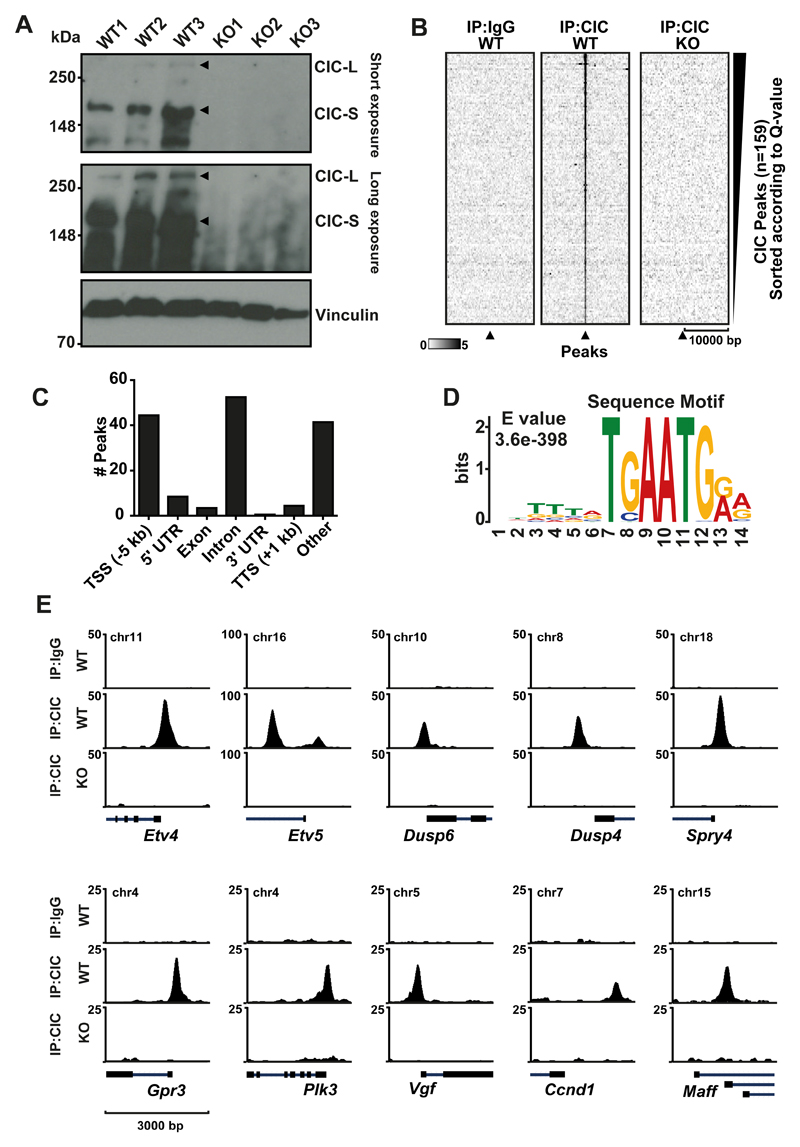
Figure 1. Genome-wide characterization of CIC targets in mESCs.
A) CIC Immunoblot of three WT mESC clones and three Cic-KO clones B) Heatmap at CIC peaks showing IgG and CIC in WT and Cic KO C) Motif analysis using MEME-ChIP. Top motif identified is depicted with corresponding E-value. D) Distribution of peaks according to gene elements. E) ChIP-seq tracks of CIC occupancy at selected target genes in WT and KO mESCs.
To obtain a comprehensive view of the genes regulated by CIC, we mapped the genome-wide location of CIC by chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) in mESCs using Cic-KO cells as a negative control (Figure 1B). We identified a strong set of 159 specific, CIC-bound regions, of which approximately two thirds were located in close proximity to a gene (Figure 1C), marking 112 unique genes.
Next, we performed motif enrichment analysis by comparing motif enrichments +/-50 bp surrounding the apex of each CIC peak to a control dataset of matching size and distance to closest transcriptional start site (TSS). This comprehensive analysis revealed the highly enriched sequence TSAATGR (Figure 1D) representing the reported CIC consensus sequence (7,17).
The expression of the PEA3 family ETS transcription factors ETV1, ETV4 and ETV5 is reported to be directly regulated by mammalian CIC (9,13). Indeed, among the most prominent targets in our ChIP-seq experiments were Etv1, Etv4 and Etv5 (Figure 1E). We also identified three members of the dual specificity protein phosphatase subfamily Dusp4, Dusp5 and Dusp6 as direct CIC targets. Other significant targets include the genes Fos, Fosb, Fosl1, Id1, Ccnd1, Rictor, Vgf, Ptpn9 and Spred1 Spred2 and Spred3. (Figure 1E, Supplementary Table 2). Interestingly CIC also binds to its own promoter, suggesting an auto-regulatory loop. Many of the CIC bound genes are implicated in the regulation of RAS/MAPK signaling, either as effectors or as members of feedback mechanisms. Indeed, we found CIC target genes significantly enriched for RAS signaling and other signaling pathways that are frequently perturbed in cancer using pathway analysis (Supplementary Figure S1D).
In agreement with the reported function of CIC to repress ETV4 and ETV5 (9), deletion of Cic led to upregulation of Etv4 and Etv5 expression. Moreover, ectopic expression of human wild type (WT) CIC, but not the HMG mutant R215W or the C-terminal mutant R1515C, reverted the increased levels of Etv4 and Etv5, suggesting that both mutants are non-functional (Supplementary Figure S1E and F). The inability of CIC-R215W to reduce target gene expression is likely due to its reduced ability to bind DNA (3). Indeed, ChIP-qPCR at the promoter region of the two target genes showed binding of exogenously expressed WT CIC, similar to enogenous levels, but no binding of CIC-R215W (Supplementary Figure S1G). Interestingly, binding of CIC R1515C mutant protein was also undetectable, which is in agreement with a recent publication showing that the C-terminal portion of CIC is critical for DNA binding (30) and mutations within this domain result in activation of Etv4 (18).
MAPK signaling prevents CIC from binding to its target genes and associated cis-regulatory elements
Previous studies have linked the MAPK pathway signaling to CIC regulation (7,9,31). To investigate the influence of MAPK signaling on CIC target gene regulation, we established NSC cultures from WT and Cic KO mESCs. In mitogen (bFGF/EGF) containing NSC medium, we were unable to detect any binding of CIC at the promoter of target genes. In contrast, removal of mitogens for 16 hours led to a strong recruitment of CIC to Etv4 and Etv5 (Supplementary Figure S2A). This was accompanied by a significant reduction of Etv4 and Etv5 expression in WT NSCs, while only insignificant changes in gene expression were detected in Cic KO NSCs (Supplementary Figure S2B).
To extend this observation to human cells and in an effort to eliminate pleiotropic effects of growth factor withdrawal, we performed ChIP analysis for CIC in primary human fetal NSCs (hNSC18.5 and hNSCs21.5) and the human glioma stem cell (GNS) line G144, after treatment with a small molecule inhibitor against MEK (PD0325901). Cells cultured in mitogen (bFGF/EGF) containing medium showed minimal CIC recruitment to the promoter region of ETV4. However, CIC binding was strongly increased in response to MEK inhibition (Figure 2A). Similarly, we observed a strong global recruitment of CIC at its target sites after MEK inhibition, using ChIP-seq (Figure 2B and 2C, Supplementary Table 2) with a significant overlap in the three studied cell lines (Figure 2D). Conversely, almost no peaks were detected in mock control samples of hNSC18.5 (n=16), hNSC21.5 (n=9) and G144 (n=8). By intersecting the set of human and mouse CIC target genes we identified 58 genes that were found in mESCs and in at least one human cell line (Supplementary Figure S2C). 21 genes were found across all samples (Supplementary Table 2). Taken together, these results demonstrate that MAPK signaling prevents CIC from binding to its target genes, and that these are largely conserved between mouse and human.
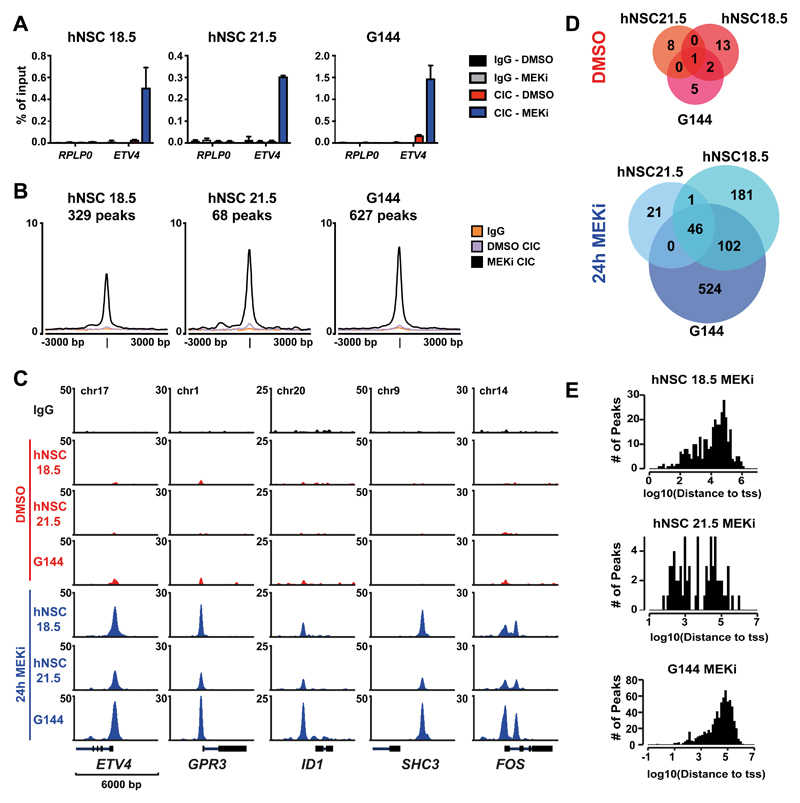
Figure 2. MAPK signaling prevents CIC from binding to its target genes.
A) ChIP-qPCR for CIC-binding at the promoter of ETV4 or control gene (RPL30) in two human neural stem cell lines (18.5 and 21.5) and one glioma neural stem cell (GNS) line (G144) after 24h mock (DMSO) or MEK inhibitor (MEKi) treatment. Data are represented as mean ±SD, n=3 B) Average intensity plot of all CIC peaks found in each cell line after 24h MEK inhibition. C) ChIP-seq tracks of CIC occupancy at selected targets genes in response to DMSO or 24h MEKi. D) Overlaps of CIC peaks detected by ChIP-seq in human neural stem cells and GNS cells with and without MEKi. E) Number of peaks found in MEKi-treated cells depicted according to their distance to the closest TSS.
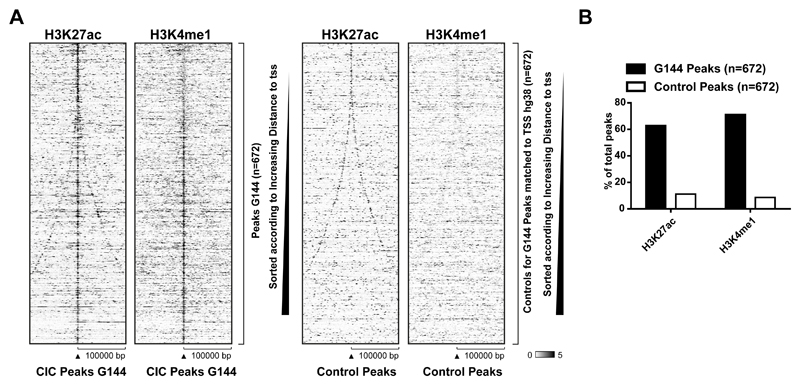
Interestingly, in human NSCs as well as in GNS cells, the majority of peaks were found >10 kb distal to a TSS (Figure 2E), indicating that CIC might act at cis-regulatory elements. To investigate if CIC binding to distal gene regions is linked to enhancer elements we performed ChIP-seq experiments in G144 cells for two histone marks, H3K27ac and H3K4me1, which are used as markers of enhancers (32). As shown in Figure 3A and 3B, CIC was found in regions strongly enriched for H3K27Ac and H3K4me1, whereas a random control dataset generated to match size and distance to closest TSS (generated in EaSeq (26)) was not enriched and consequently showed little overlap.
Figure 3. CIC targets are enriched for enhancer-associated histone marks.
A) Heatmaps showing ChIP-seq signals of H3K27ac and H3K4me1 at CIC peaks in G144 cells. A set of control regions with matched distance to TSS is depicted to the right. B) Percentage of CIC peaks or control regions overlapping with H3K27ac or H3K4me1 peaks in G144 cells.
Collectively, we have generated high confidence results for the genome-wide binding of CIC. These results show that CIC binds to a defined set of target regions in a MAPK-dependent manner and those regions are largely conserved between mouse and human. Furthermore, our results show that CIC is recruited to both TSS and to enhancers, suggesting that CIC can repress transcription via both of these regulatory regions.
CIC interacts with the SIN3 histone deacetylating complex and loss of CIC leads to increased histone acetylation of CIC target genes
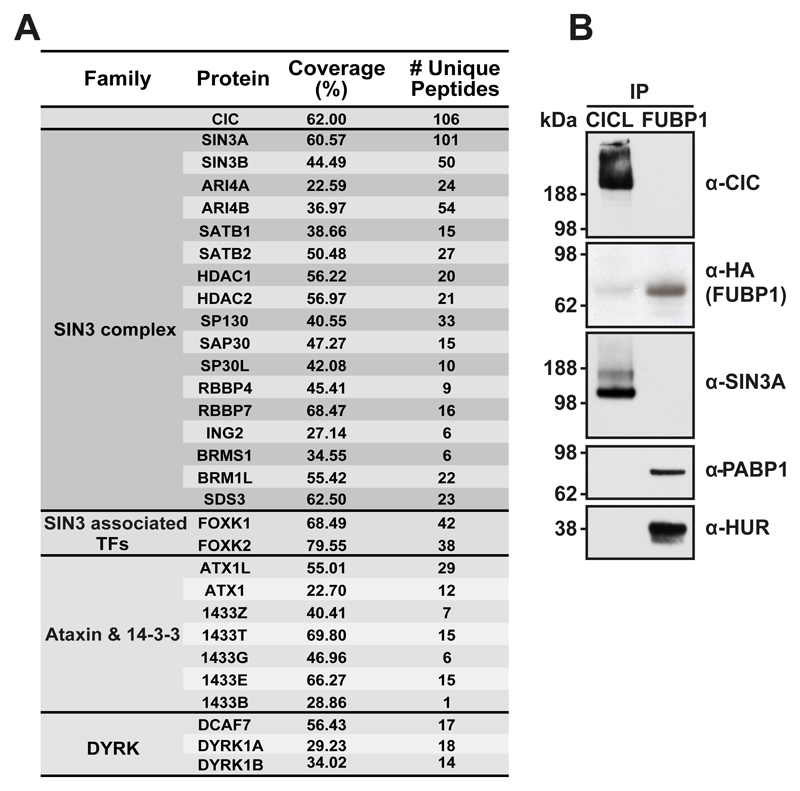
CIC is believed to act as a transcriptional repressor (17); however, the molecular mechanism through which CIC regulates gene expression is unknown. To obtain insights into CIC-mediated transcriptional regulation in mammalian cells, we purified proteins interacting with human CIC-L and identified them by mass spectrometry. Previously identified interaction partners Ataxin-1 (ATX1) and Ataxin-1-like (ATX1L) (11) as well as 14-3-3 proteins (9) were highly enriched. Among the most strongly enriched proteins were members of the SIN3 transcriptional repressive complex, including core deacetylating enzymes HDAC1 and HDAC2 and structural components like SIN3A, SIN3B and SAP30 (Figure 4A and 4B and Supplementary Table 3). This suggests that CIC could regulate transcription by recruiting the SIN3 complex. Size exclusion chromatography supported the interaction between CIC and SIN3A (Supplementary Figure S2D).
Figure 4. CIC interaction partners.
A) Table of selected CIC-interacting proteins identified by mass-spectrometry of affinity-purified exogenously expressed Flag-HA-CIC in HEK293 cells. Interacting proteins are grouped according to functionality. B) Validation of SIN3A interaction by IP-immunoblotting. An IP performed in parallel for HEK293 expressing Flag-HA-FUBP1 (immunoblotting for its interaction with PABP1 and HUR) is included as a negative control.
The affinity purification also identified the Dual specificity tyrosine-phosphorylation-regulated kinases DYRK1A and DYRK1B as well as their partner DCAF7 (Figure 4A). Size-exclusion chromatography showed that a fraction of DYRK1A also co-migrated with CIC (Supplementary Figure S2D). The Drosophila orthologues Minibrain (mnb) and Wings apart (wap) can act as ERK-independent regulators of CIC (33). Thus, our results suggest that this mechanism could be conserved during evolution.
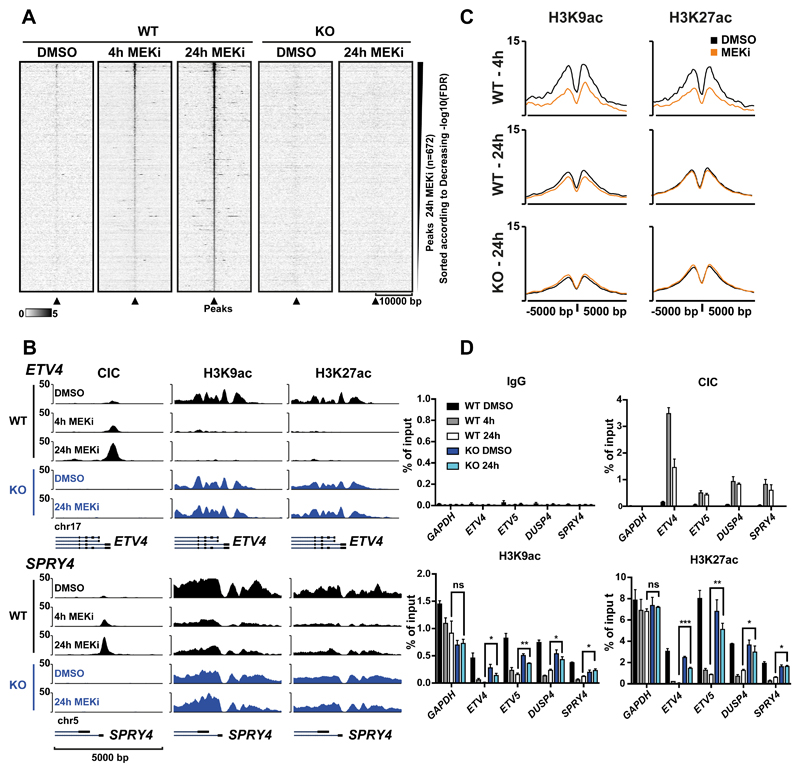
To investigate a possible link between CIC and the SIN3 histone deacetylation complex in regulating the expression of CIC target genes, we established a CIC KO G144 line and used ChIP-seq to correlate CIC recruitment and changes in histone acetylation. The human GNS cell line G144 exhibits an oligodendrocyte precursor-like phenotype (19) and represents a disease relevant models that is experimentally tractable. We detected a strong recruitment of CIC in response to MEK inhibition, whereas no recruitment was detected in CIC KO cells (Figure 5A and Supplementary Figure S3A).
Figure 5. Loss of CIC leads to increased histone acetylation at CIC target genes.
A) Heatmap of all CIC ChIP-seq peaks found in glioma neural stem cells (G144) after 24h of MEK inhibition showing CIC in WT and KO cells after 4h or 24h of MEK inhibition B) ChIP-seq tracks showing increase of CIC occupancy at selected target genes and concurrent reduction of histone acetylation following MEK inhibition. C) Average ChIP-seq signal of H3K9ac and H3K27ac at CIC peaks after 4h and 24h MEK inhibition. D) ChIP-qPCR for CIC and histone acetylation marks at selected CIC target genes with GAPDH as a negative control. Data are represented as mean ±SD, n=3 (p-value: *<0.05, **<0.01, ***<0.001).
To understand the functional consequences of CIC-recruitment to its target genes, we performed genome-wide localization studies of histone acetylation. This revealed a strong depletion of histone H3 acetylation around the promoter of CIC target genes upon CIC recruitment (Figure 5B). We observed a strong deacetylation at CIC peaks after 4h and somewhat less pronounced deacetylation after 24h of MEKi treatment. This loss in acetylation was not observed in CIC knockout cells (Figure 5C and 5D, and Supplementary Figure S3B). The effect of CIC on histone acetylation levels was further addressed in mESCs by ChIP-qPCR in wild type and Cic KO ESCs. A number of candidate genes confirmed a significant increase in H3K9ac (6/7 genes) and H3K27ac (2/7 genes) (Supplementary Figure S3C). Taken together, these results show that the recruitment of CIC to its target genes leads to their deacetylation, which is in agreement with our demonstration that CIC associates with the SIN3A complex.
MEK inhibition in human glioma cells causes repression of a subset of CIC target genes and increased proliferation in a CIC-dependent manner
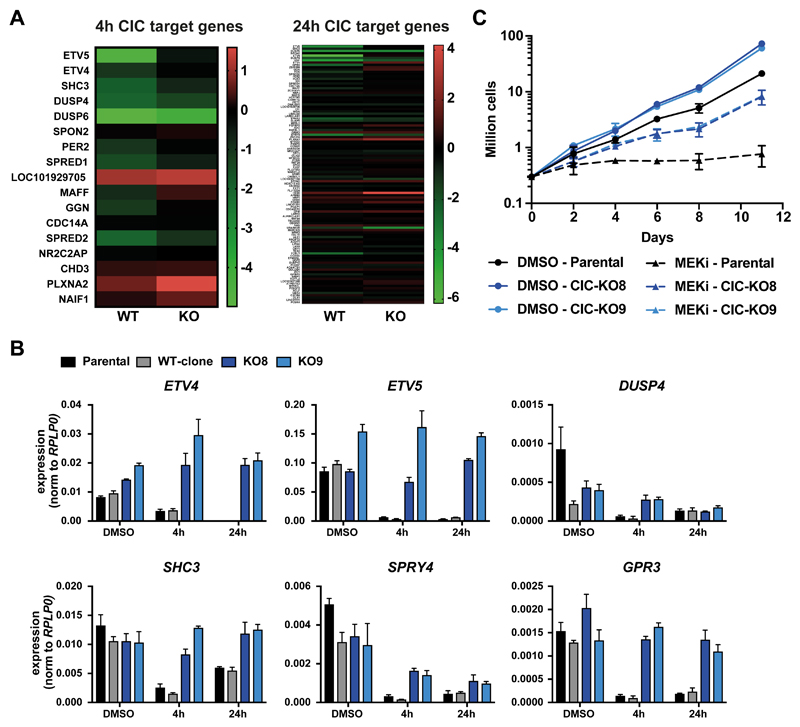
Having established that CIC binding to its target genes is ablated by MAPK activity, we investigated global gene expression changes caused by MEK inhibition and the effect of CIC deletion (Supplementary Figure S4A and Supplementary Table 4). To this end we defined “CIC target” genes as genes with a CIC peak +/- 5kb around the TSS after 4h and 24h MEK inhibition, respectively. For these sets of genes, we compared RNAseq data of parental G144 cells and one CIC WT single cell clone to two independent CIC KO clones. A large proportion of CIC target genes showed a downregulation after 4h of MEK inhibition (9/19) and to a lesser extent after 24h of MEK inhibition (14/111). More importantly, this loss of CIC target gene expression was reduced or completely abolished in CIC KO cells (Figure 6A and Supplementary Table 4), which was confirmed by qPCR (Figure 6B). Given that deacetylation of histones could be one of CIC-mediated mechanisms to downregulate its target genes, we used histone deacetylase (HDAC) inhibition (Panobiostat) to test if HDACs were involved in the repression of ETV4 and ETV5. Although, ETV4 and ETV5 expression was still decreased in response to MEKi, HDAC inhibition significantly dampened this repression (Supplementary Figure S4B).
Figure 6. Effect of CIC depletion on gene expression and proliferation.
A) Gene expression changes of CIC target genes after 4h and 24h of MEK inhibition in WT and CIC KO G144 cells (colors depict log2 fold changes between DMSO and MEKi) B) Expression changes quantified by qPCR in parental G144, one WT clone and two independent CIC KO clones. C) Growth curves measured by cell count of parental and CIC KO G144 cells during continuous MEK inhibition of mock treatment (DMSO).
Previous studies have indicated that deletion of CIC confers resistance to MAPK pathway (34) or EGFR inhibition (35). Indeed, prolonged treatment with MEK inhibitors caused a complete stop of cell proliferation in G144 cells, whereas CIC KO cells continued to grow even after 11 days of MEK inhibition (Figure 6C).
Discussion
The high frequency of CIC mutations in ODG suggests that CIC is a bona fide tumor suppressor gene. It is therefore crucial to investigate the effects of CIC (mis)regulation as this knowledge will give insight into the molecular mechanisms leading to ODG and help design future treatments for the disease. In this study, we have mapped the genome-wide binding pattern of mammalian CIC. We find that CIC binds to an overlapping set of core regulatory regions in mouse and human genomes, including promoter regions of established MAPK effector genes, as well as distal regions associated with enhancer features. Furthermore, the detection of a conserved CIC-binding motif, similar to the one previously reported for Drosophila Cic suggests that CIC function is conserved during evolution. Our results identify CIC as a key transcriptional repressor of MAPK effector genes that are controlling cellular proliferation. A number of these genes are deregulated in CIC deleted cell lines (36) and CIC-mutated ODG (37). Based on our data we propose that these genes are directly affected by the loss of CIC. A recent study suggested that CIC binds a number of genes unrelated to MAPK signaling and could function as a transcriptional activator (38). However, we have not been able to confirm this observation, because we only observed robust CIC binding after MAPK inhibition.
The PEA3 transcription factors have been shown to be MAPK and CIC regulated genes and have been linked to cancer progression (17,18). Here, we show that these genes are likely to have similar roles in ODG. In addition, we have identified new target genes of CIC whose deregulation could play an important role in ODG development. Some of these genes, as for example ID1, CCND1 and SHC3 are strongly bound by CIC and have been implicated in tumor formation. The members of the inhibitor of DNA-binding (ID) protein family are master regulators of cancer stem cells and tumor aggressiveness (39), and ID1 is directly implicated in neural stem cell identity and gliomagenesis (40). The gene CCND1 encoding Cyclin D1, which is a key regulatory subunit of CDK4 and CDK6, is instrumental during G1/S-phase transition. SHC proteins function as adaptors for translating RTK signals into the activation of downstream pathways like RAS/MAPK (41). Interestingly, SHC3 is the predominantly expressed in the brain, where it is involved in neuronal differentiation and prevents apoptosis in mature neurons (42,43). Furthermore, it was found to be misregulated in in high grade astrocytoma where it increased cell survival (44).
CIC targets also include groups of genes that are negative regulators of the MAPK pathway. The DUSP phosphatases and Sprouty Related EVH1 Domain (SPRED) containing family members are ERK specific phosphatases (45) and SOS inhibitory binding proteins (46) respectively. Conversely, Sprouty RTK Signaling Antagonist 4 (SPRY4) blocks MAPK activation by binding to RAF1 (47). This suggests that CIC fulfills a central role in a delicate feedback mechanism that keeps mitogen activated signaling in check under physiological conditions. The mutual exclusivity of CIC mutations and RTK mutants in ODG could therefore be explained not only by activation of proliferative genes in response to CIC loss, but also by the simultaneous transcriptional induction of negative RTK regulators which might protect from acquiring mutations in RTK coding genes. Importantly, CIC mutations are not found in other types of brain tumors such as high grade astrocytoma and Glioblastoma multiforme (GBM) which display an accelerated disease progression and frequently harbor aberrant RTK signaling (5). This suggests that increased RTK signaling can induce malignant cell growth through CIC dependent and independent pathways leading to mutual exclusiveness with CIC mutations and a more aggressive phenotype. Additionally, CIC mutations could have synergistic effects with the cell of origin of ODG.
We also investigated the effect of different single amino acid substitutions, which are found in human ODG patients. As expected, the DNA-binding mutant R215C was unable to target and repress genes. More surprisingly was that this was also true for the C-terminal mutant R1515C. This observation is in line with reports that a variety of CIC mutant proteins, including R1515H, were unable to suppress metastasis (18) and that the C-terminal part of CIC aids DNA-binding (30).
Based on our finding that CIC is associated with the SIN3A deacetylation complex, we propose a model in which CIC recruitment can lead to histone deacetylation. This model is supported by our demonstration that a number of genes decrease acetylation levels in response to CIC recruitment. Furthermore, we provide evidence that HDAC inhibitors could counteract CIC mediated repression on selected target genes. Interestingly, we observed that not all CIC targets changed their acetylation levels in response to CIC loss, indicating that additional factors or pathways can cause histone deacetylation and gene repression synergistically with CIC. In fact, a recent study investigating Cic in Drosophila reported that the Hippo pathway and Cic both act as brakes of the Ras-responsive transcriptional network and the perturbation of both is needed for full activation of target genes (48).
In summary, our data together with previous findings show that CIC acts as a transcriptional repressor of genes regulated by the MAPK signaling pathways. We provide novel insights into the spectrum of CIC-regulated genes revealing potential mechanisms for how loss of CIC could lead to ODG formation. Moreover, our demonstration that the SIN3A repressive complex interacts with CIC, and that CIC recruitment to target genes lead to their deacetylation suggest a mechanism by which CIC exerts its function.
Supplementary Material
Significance.
Inactivation of CIC inhibits its direct repression of MAPK pathway genes, leading to their increased expression and mitogen-independent growth
Acknowledgments
We thank the members of the Helin lab for discussion, technical advice and support. We thank Lucia Simon Carrasco and Anne Laugesen for critical comments on the manuscript.
Grant support
This work was supported by the Danish National Research Foundation (DNRF 82) to K. Helin, the Brain Tumour Charity (GN-000358) to K. Helin and S. Pollard, the European Research Council (294666_DNAMET) to K. Helin, and through a center grant from the Novo Nordisk Foundation (NNF17CC0027852) to K. Helin.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science (80- ) 2011;333:1453–5. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jimenez G, Shvartsman SY, Paroush Z. The Capicua repressor - a general sensor of RTK signaling in development and disease. J Cell Sci. 2012;125:1383–91. doi: 10.1242/jcs.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gleize V, Alentorn A, Connen De Kérillis L, Labussière M, Nadaradjane AA, Mundwiller E, et al. CIC inactivating mutations identify aggressive subset of 1p19q codeleted gliomas. Ann Neurol. 2015;78:355–74. doi: 10.1002/ana.24443. [DOI] [PubMed] [Google Scholar]
- 4.Network CGAR. Brat DJ, Verhaak RGW, Aldape KD, Yung WKA, Salama SR, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372:2481–98. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–68. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 6.Jiménez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by Torso RTK signaling: Role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14:224–31. [PMC free article] [PubMed] [Google Scholar]
- 7.Ajuria L, Nieva C, Winkler C, Kuo D, Samper N, Andreu MJ, et al. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development. 2011;138:915–24. doi: 10.1242/dev.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Astigarraga S, Grossman R, Díaz-Delfín J, Caelles C, Paroush Z, Jiménez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26:668–77. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dissanayake K, Toth R, Blakey J, Olsson O, Campbell DG, Prescott AR, et al. ERK/p90 RSK /14-3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicúa. Biochem J. 2011;433:515–25. doi: 10.1042/BJ20101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forés M, Ajuria L, Samper N, Astigarraga S, Nieva C, Grossman R, et al. Origins of Context-Dependent Gene Repression by Capicua. PLoS Genet. 2015;11:e1004902. doi: 10.1371/journal.pgen.1004902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, et al. ATAXIN-1 Interacts with the Repressor Capicua in Its Native Complex to Cause SCA1 Neuropathology. Cell. 2006;127:1335–47. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 12.Fryer JD, Yu P, Kang H, Mandel-Brehm C, Carter AN, Crespo-Barreto J, et al. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science (80- ) 2011;334:690–3. doi: 10.1126/science.1212673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y, Fryer JD, Kang H, Crespo-Barreto J, Bowman AB, Gao Y, et al. ATXN1 Protein Family and CIC Regulate Extracellular Matrix Remodeling and Lung Alveolarization. Dev Cell. 2011;21:746–57. doi: 10.1016/j.devcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simón-Carrasco L, Graña O, Salmón M, Jacob HKC, Gutierrez A, Jiménez G, et al. Inactivation of Capicua in adult mice causes T-cell lymphoblastic lymphoma. Genes Dev. 2017;31:1456–68. doi: 10.1101/gad.300244.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park S, Lee S, Lee CG, Park GY, Hong H, Lee JS, et al. Capicua deficiency induces autoimmunity and promotes follicular helper T cell differentiation via derepression of ETV5. Nat Commun. 2017;8:16037. doi: 10.1038/ncomms16037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan Q, Brunetti L, Rousseaux MWC, Lu H-C, Wan Y-W, Revelli J-P, et al. Loss of Capicua alters early T cell development and predisposes mice to T cell lymphoblastic leukemia/lymphoma. Proc Natl Acad Sci. 2018;115:E1511–E1519. doi: 10.1073/pnas.1716452115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, Mukai H, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–37. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 18.Okimoto RA, Breitenbuecher F, Olivas VR, Wu W, Gini B, Hofree M, et al. Inactivation of Capicua drives cancer metastasis. Nat Genet. 2017;49:87–96. doi: 10.1038/ng.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, et al. Glioma Stem Cell Lines Expanded in Adherent Culture Have Tumor-Specific Phenotypes and Are Suitable for Chemical and Genetic Screens. Cell Stem Cell. 2009;4:568–80. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:1594–606. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cloos PAC, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–11. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 23.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14 doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol. 2016;23:349–57. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
- 27.Machanick P, Bailey TL. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–7. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang P, Selvaraj V, Deng W. Differentiation of Embryonic Stem Cells into Oligodendrocyte Precursors. J Vis Exp. 2010;8:22–4. doi: 10.3791/1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–23. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forés M, Simón-Carrasco L, Ajuria L, Samper N, González-Crespo S, Drosten M, et al. A new mode of DNA binding distinguishes Capicua from other HMG-box factors and explains its mutation patterns in cancer. PLoS Genet. 2017;13:1–22. doi: 10.1371/journal.pgen.1006622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Y, Ha N, Forés M, Xiang J, Gläßer C, Maldera J, et al. EGFR/Ras Signaling Controls Drosophila Intestinal Stem Cell Proliferation via Capicua-Regulated Genes. PLoS Genet. 2015;11:e1005634. doi: 10.1371/journal.pgen.1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Paul S, Trieu KG, Dent LG, Froldi F, Forés M, et al. Minibrain and Wings apart control organ growth and tissue patterning through down-regulation of Capicua. Proc Natl Acad Sci. 2016;113:10583–8. doi: 10.1073/pnas.1609417113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B, Krall EB, Aguirre AJ, Kim M, Widlund HR, Doshi MB, et al. ATXN1L, CIC, and ETS Transcription Factors Modulate Sensitivity to MAPK Pathway Inhibition. Cell Rep. 2017;18:1543–57. doi: 10.1016/j.celrep.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao S, Davoli T, Leng Y, Li MZ, Xu Q, Elledge SJ. A genetic interaction analysis identifies cancer drivers that modify EGFR dependency. Genes Dev. 2017;31:184–96. doi: 10.1101/gad.291948.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LeBlanc VG, Firme M, Song J, Chan SY, Lee MH, Yip S, et al. Comparative transcriptome analysis of isogenic cell line models and primary cancers links capicua (CIC) loss to activation of the MAPK signalling cascade. J Pathol. 2017;242:206–20. doi: 10.1002/path.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padul V, Epari S, Moiyadi A, Shetty P, Shirsat NV. ETV/Pea3 family transcription factor-encoding genes are overexpressed in CIC-mutant oligodendrogliomas. Genes Chromosom Cancer. 2015;54:725–33. doi: 10.1002/gcc.22283. [DOI] [PubMed] [Google Scholar]
- 38.Yang R, Chen LH, Hansen LJ, Carpenter AB, Moure CJ, Liu H, et al. Cic loss promotes gliomagenesis via aberrant neural stem cell proliferation and differentiation. Cancer Res. 2017;77:6097–108. doi: 10.1158/0008-5472.CAN-17-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lasorella A, Benezra R, Iavarone A. The ID proteins: Master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14:77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 40.Soroceanu L, Murase R, Limbad C, Singer E, Allison J, Adrados I, et al. Id-1 is a key transcriptional regulator of glioblastoma aggressiveness and a novel therapeutic target. Cancer Res. 2013;73:1559–69. doi: 10.1158/0008-5472.CAN-12-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmed SBM, P S. Insights into the Shc Family of Adaptor Proteins. J Mol Signal. 2017;12:1–17. doi: 10.5334/1750-2187-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conti L, Sipione S, Magrassi L, Bonfanti L, Rigamonti D, Pettirossi V, et al. Shc signaling in differentiating neural progenitor cells. Nat Neurosci. 2001;4:579–86. doi: 10.1038/88395. [DOI] [PubMed] [Google Scholar]
- 43.Pelicci G, Troglio F, Bodini A, Melillo RM, Pettirossi V, Coda L, et al. The neuron-specific Rai (ShcC) adaptor protein inhibits apoptosis by coupling Ret to the phosphatidylinositol 3-kinase/Akt signaling pathway. Mol Cell Biol. 2002;22:7351–63. doi: 10.1128/MCB.22.20.7351-7363.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magrassi L, Conti L, Lanterna A, Zuccato C, Marchionni M, Cassini P, et al. Shc3 affects human high-grade astrocytomas survival. Oncogene. 2005;24:5198–206. doi: 10.1038/sj.onc.1208708. [DOI] [PubMed] [Google Scholar]
- 45.Caunt CJ, Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs): Shaping the outcome of MAP kinase signalling. FEBS J. 2013:489–504. doi: 10.1111/j.1742-4658.2012.08716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim HJ, Bar-Sagi D. Modulation of signalling by sprouty: A developing story. Nat Rev Mol Cell Biol. 2004;5:441–50. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki A, Taketomi T, Kato R, Saeki K, Nonami A, Sasaki M, et al. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat Cell Biol. 2003;5:427–32. doi: 10.1038/ncb978. [DOI] [PubMed] [Google Scholar]
- 48.Pascual J, Jacobs J, Sansores-Garcia L, Natarajan M, Zeitlinger J, Aerts S, et al. Hippo Reprograms the Transcriptional Response to Ras Signaling. Dev Cell. 2017;42:667–680.e4. doi: 10.1016/j.devcel.2017.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-6681 and E-MTAB-6682 respectively.