Abstract
Removal of a telomere from yeast chromosome VII in a strain having two copies of this chromosome often results in its loss. Here we show that there are three pathways that can stabilize this broken chromosome: homologous recombination, nonhomologous end joining, and de novo telomere addition. Both in a wild-type and a recombination deficient rad52 strain, most stabilization events were due to homologous recombination, whereas nonhomologous end joining was exceptionally rare. De novo telomere addition was relatively rare, stabilizing <0.1% of broken chromosomes. Telomere addition took place at a very limited number of sites on chromosome VII, most occurring close to a 35-base pair stretch of telomere-like DNA that is normally ∼50 kb from the left telomere of chromosome VII. In the absence of the Pif1p DNA helicase, telomere addition events were much more frequent and were not concentrated near the 35-base pair tract of telomere-like DNA. We propose that internal tracts of telomere-like sequence recruit telomerase by binding its anchor site and that Pif1p inhibits telomerase by dissociating DNA primer–telomerase RNA interactions. These data also show that telomeric DNA is essential for the stable maintenance of linear chromosomes in yeast.
INTRODUCTION
In most eukaryotes, telomeres, the physical ends of chromosomes, consist of simple, repeated DNA. For example, in the yeast Saccharomyces cerevisiae, chromosomes end in ∼350 base pairs (bps) of C1–3A/TG1–3 DNA. The properties of conventional DNA polymerases make it impossible for them to replicate the very ends of linear chromosomes. In most eukaryotes, this end replication problem is solved by the enzyme telomerase, a specialized reverse transcriptase that extends the G-strand of telomeric DNA with the use of its RNA subunit as a template.
Although existing telomeres must be maintained in all cells, there are also situations where telomeres are formed de novo. For example, during formation of the ciliate macronucleus, new telomeres are added to hundreds of chromosome breaks, the majority of which do not contain even a few base pairs of telomeric DNA (reviewed in Coyne et al., 1996). In Tetrahymena and other ciliates, this massive de novo telomere addition is telomerase mediated (Yu et al., 1991). In the ciliate Euplotes, the telomerase from cells undergoing macronuclear formation is modified by the addition of a factor that enhances its ability to add telomeric DNA to nontelomeric substrates, at least in vitro (Bednenko et al., 1997). The Euplotes activity thus acts as an antispecificity factor.
Pif1p, a 5′ to 3′ DNA helicase (Lahaye et al., 1991), modulates the telomerase pathway in Saccharomyces (Zhou et al., 2000b). Loss of Pif1p causes telomerase-mediated telomere lengthening, whereas its overexpression results in telomere shortening. Both of these effects require the helicase activity of Pif1p. In addition to its inhibitory role in the lengthening of preexisting telomeres, Pif1p has a large impact on the rate of de novo telomere addition (Schulz and Zakian, 1994). In a wild-type cell, a broken yeast artificial chromosome (YAC) is very rarely healed by de novo telomere addition. Moreover, when telomere addition does occur in this YAC assay, the new telomere is invariably added at an internal 108-base pair tract of C4A4/T4G4 DNA, a ciliate telomeric sequence that supports telomere addition in Saccharomyces (Pluta et al., 1984; Pluta and Zakian, 1989). In contrast, in cells lacking Pif1p, de novo telomere addition on a broken YAC increases ∼600-fold, and telomere addition occurs at many different sites in the YAC (Schulz and Zakian, 1994). Because Pif1p is telomere associated in vivo (Zhou et al., 2000b), its effects on telomeres are probably direct.
Telomeres have long been considered to be essential for chromosome stability, protecting ends from degradation and fusions as well as allowing their complete replication. However, in Drosophila melanogaster, chromosomes that lack telomeric DNA are readily recovered upon irradiation of mu-2 flies (Mason et al., 1984; Biessmann and Mason, 1988; Biessmann et al., 1990) or after mobilization of a telomere proximal P element (Levis, 1989; Tower et al., 1993). Chromosomes lacking telomeric DNA can also be recovered after irradiation of wild-type female flies, although such events are rare (Mason et al., 1997). A single chromosome break in Drosophila causes a cell cycle arrest, which, if unrepaired, usually leads to apoptosis (Almad and Golic, 1999). However, occasionally chromosome breaks in Drosophila acquire the stabilization functions of telomeres without acquiring telomeric DNA. This ability appears to be mediated by the sequence nonspecific binding of certain telomere binding proteins, such as HP1 (Fanti et al., 1998).
In Saccharomyces, loss of a single chromosomal telomere induces a RAD9-mediated checkpoint arrest (Sandell and Zakian, 1993). However, this arrest is transient, and cells eventually resume division, even if the broken chromosome is not repaired. In a wild-type cell, a chromosome without a telomere is lost from many cells, demonstrating directly that yeast telomeres play an active role in chromosome stability (Sandell and Zakian, 1993).
In this previous work, we focused on cells in which the chromosome without a telomere was lost. Here, we describe the fate of chromosomes that are stabilized after telomere loss. Unlike Drosophila chromosomes, there is no evidence for the stable maintenance of yeast chromosomes in the absence of telomeric DNA. In a wild-type strain, the majority of cells gained a telomere via RAD52-dependent homologous recombination. In a rad52 strain, some broken chromosomes were stabilized by de novo telomere addition, but these events were rare and occurred at a very limited number of regions on the chromosome, near tracts of telomere-like DNA. Both the frequency and distribution of de novo telomere addition were altered dramatically in a pif1 strain. These data support our proposal (Zhou et al., 2000b) that Pif1p promotes genome stability by inhibiting the addition of telomeric DNA to double-strand breaks and hence reducing the generation of terminally deleted chromosomes.
MATERIALS AND METHODS
Construction of the strains used in this study is described in detail in Sandell and Zakian (1993). Briefly, the strains were isogenic to LS20 (MATΔ his3 ade2 can1 trp1 ura3 leu2 lys5 cyh2r ade3::GALHO). The test chromosome, which contains LYS5, CYH2, and ADE3 as well as URA3 and the HO recognition site near the left telomere were introduced into LS20 by cytoduction to generate Ura+Lys+ Ade3+ Cyhs disomic versions of LS20 (Figure 1). The only accessible HO recognition site in this disomic strain is near the left telomere of the test chromosome. The rad52::LEU2 (Sandell and Zakian, 1993) and pif1-m2 (Schulz and Zakian, 1994) alleles were constructed and introduced into LS20 as described previously. We used the pif1-m2 allele because Pif1p affects maintenance of both mitochondrial (Lahaye et al., 1991) and telomeric (Schulz and Zakian, 1994) DNA. There are two forms of Pif1p in cells, a longer nuclear form and a shorter mitochondrial form (Zhou et al., 2000b). Because only the shorter form of Pif1p is made in a pif1-m2 strain, these cells have wild-type mitochondrial function but mutant telomeres and hence grow as well as wild-type cells (Schulz and Zakian, 1994).
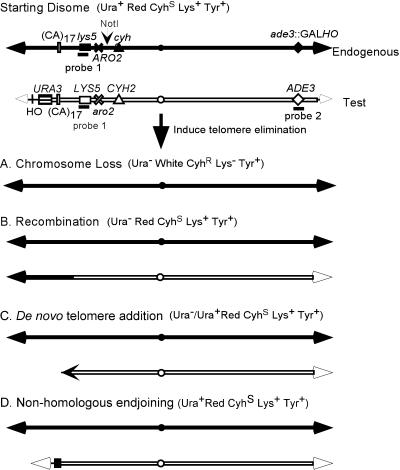
Figure 1.
Experimental assay and consequences of telomere loss. Experiments were in otherwise haploid strains that were disomic for chromosome VII. The two copies of chromosome VII, the endogenous and test copies, were constructed as described in Sandell and Zakian (1993). In addition to the markers indicated on chromosome VII, the disomic strain was ade2, ura3-52 and had no HO site at MAT. Immediately next to the left telomere of the test chromosome was the HO recognition site followed by URA3. This insertion was made within the ADH4 gene, the most distal gene on the left arm of chromosome VII, in such a way that ∼16 kb of DNA was deleted. Thus, the test chromosome is ∼15 kb shorter than the endogenous copy of chromosome VII. The HO endonuclease gene was inserted within the ADE3 gene on the endogenous copy of chromosome VII, thereby deleting a portion of ADE3. Hybridization with a probe for the deleted region (probe 2) detects only the test chromosome (Figure 2B). Hybridization with probe 1 detects both copies of chromosome VII (Figure 2, C and D). When cells are grown in galactose medium, the HO endonuclease is expressed and the left telomere on the test chromosome is excised in most cells (Sandell and Zakian, 1993). After telomere loss, the test chromosome was either lost (A), repaired by homologous recombination with the endogenous copy of chromosome VII (B), acquired a new telomere at a site that had previously been internal on the test chromosome (C), or religated in a way that eliminated the HO recognition site (D). The phenotype of cells produced by each of these events is indicated. Although most de novo telomere addition events yielded Ura− cells, in the rad52 pif1 strain, some of these events generated Ura+ cells. Theoretically, nonhomologous end joining could yield Ura− cells but no such events were recovered. To be detected in this study, recombination and de novo telomere addition had to occur in the ∼220-kb region distal to LYS5 because stabilized clones were selected on medium lacking lysine. There are four NotI sites on chromosome VII; only the one relevant for the analysis in this article is indicated. Most de novo telomere addition events that occur in a rad52 strain were near a (CA)17 tract indicated on the figure. The figure is not to scale.
YC synthetic medium is described in Zakian and Scott (1982). Telomere loss was induced as described (Sandell and Zakian, 1993). Briefly, cells were pregrown for 24 h in YC-lys-tyr + 2% glucose, which selects for both copies of chromosome VII. Cells were then transferred to YC-lys-tyr + 3% glycerol overnight to relieve glucose repression and then to YC-lys-tyr + 3% galactose for 24 h to induce expression of the HO endonuclease. Cells were plated to both YC (to assess viability) and to YC −lys −tyr + low adenine + 2% glucose (to identify cells that had stabilized the test chromosome). Cells from the selective plates were replica plated to cycloheximide and to plates lacking uracil. The disomic nature of stabilized colonies was confirmed by their ability to papillate on cycloheximide plates. A subset of the Ura+ disomic colonies was streaked on YC-lys-tyr media containing galactose to determine the fraction of Ura+ cells that still retained the HO recognition site. Only cells that had lost the HO site during the first 24 h of growth in galactose medium grew robustly on these plates. To lose the endogenous copy of chromosome VII from stabilized rad52 clones, these clones were patched onto nonselective medium and then replica plated to medium lacking lysine, to medium containing low amounts of adenine, and to medium containing cycloheximide. Lys+ clones that produced red colonies that did not papillate on medium containing cycloheximide had lost the endogenous chromosome.
Probes for Southern hybridization were usually random labeled with the use of the RTS-Radprime System (Invitrogen, Carlsbad, CA). To analyze the structure of the test chromosome by conventional gel electrophoresis, stabilized clones were grown for 48 h, and DNA prepared with the use of a glass bead protocol (Rose et al., 1990). Probes for Southern hybridization were a 1.1-kb HindIII fragment to detect URA3 or a 1.8-kb EcoRI-HindIII fragment to detect the 5′ end of ADH4. For analysis by pulse field gel electrophoresis (PFGE), yeast chromosomal DNA blocks were prepared from late log phase cells essentially as described in Rose et al. (1990). Agarose plugs were analyzed with the use of the contour-clamped homogeneous electric field-dynamically regulated CHEF-DR III apparatus (Bio-Rad, Hercules, CA). Electrophoresis was performed in a 1.5% agarose gel for 48 h at 14°C with the use of 60–120-s switching times at 6 V/cm. For NotI digested DNA, the agarose DNA plugs were incubated in TE buffer for 4 h at 4°C and then equilibrated in restriction buffer (50 mM Tris, 10 mM MgCl2, 100 mM NaCl, 1 mM dithiothreitol) for 30 min at 4°C. The DNA plugs were digested in 300 μl of restriction buffer containing 50 U of NotI for 16 h at 37°C. The NotI digested DNA was analyzed as described for undigested DNA except that PFGE was for 24 h with 25–75-s switching times. The PFGE-separated DNA was Southern blotted under denaturing conditions and probed with either a 2.1-kb HpaI fragment that contains a segment of the LYS5 gene or a 1.7-kb XhoI-KpnI segment of the ADE3 gene.
Polymerase chain reaction (PCR) amplification of chromosome ends near URA3 was performed with the use of an oligonucleotide homologous to the yeast telomeric DNA repeat and, depending on the precise site of telomere addition, either of two oligonucleotides from within the URA3 gene. PCR amplification of ends near the (CA)17 tract on the left arm of chromosome VII was performed with the use of the same telomeric olignucleotide and one of two oligonucleotides from near the (CA)17 tract. The following oligonucleotides (DNAgency) were used. Telomeric: CAX-29, 5′CGGCTCGAGC ACCCACACCA CACCCACAC3′; URA3: URA3–26, 5′CCGAATTCTT CCATGGAGGG CACAGT3′ or U3RI-28, 5′GGGAATTCGG TAATCTCCGA ACAGAAGG3′; VII-L: JLM3, 5′GCAAGCGCAT GAATTTCATC ACAATGTCAT GG3′ or JLM4, 5′GTCATCATCT GCACCCATGT GAGCG3′. PCR conditions were optimized with the use of TNK-100 buffer (Blanchard et al., 1993). PCR products were cloned directly into pCRII by using a TA-cloning kit (Invitrogen). The DNA sequence was determined with the use of a Sequenase kit (Amersham Pharmacia Biotech, Piscataway, NJ). All sequence coordinates used to describe telomere addition sites or positions of telomere-like DNA are from the Saccharomyces genome database (http://genome-www.stanford.edu/Saccharomyces).
Tracts of telomere-like DNA were identified with the use of the FindPattern program in the GCG Wisconsin Package (Accelrys Inc., Madison, WI). All permutations of TG1–3 sequence of 8 bps or longer were searched throughout the entire yeast genome. All matches were analyzed individually and the data compiled by hand.
RESULTS
Experimental Design
To assess the contribution of telomeres to chromosome stability, we constructed an otherwise haploid strain containing two copies of chromosome VII (Figure 1; Sandell and Zakian, 1993). In this disomic strain, either copy of chromosome VII can be lost without compromising viability. One copy of chromosome VII, hereafter called the test chromosome, was modified by inserting the recognition sequence for the HO endonuclease immediately internal to the ∼350-base pair tract of telomeric C1–3A/TG1–3 DNA, followed by a copy of the URA3 gene. This modification also deletes the ∼16 kb of DNA that is normally immediately internal to the left telomere of chromosome VII. Disomic cells are maintained by growing cells in medium lacking lysine, which requires the LYS5 gene on the test chromosome, and tyrosine, which selects for the ARO2 gene on the other copy of chromosome VII, hereafter called the endogenous chromosome. The strain also has a galactose inducible version of the HO endonuclease gene.
When this chromosome VII disomic strain is grown on galactose medium, HO endonuclease is expressed. As assessed by both Southern analysis and inability to grow in the absence of uracil, the endonuclease cleaves the left telomere from the test copy of chromosome VII, and the exposed URA3 gene is degraded in most cells (Sandell and Zakian, 1993). In previous work, we showed that in a wild-type strain, telomere elimination resulted in loss of the test chromosome in ∼30% of cells. The remaining ∼70% of the cells produced red Ura− colonies. Because these cells were Ura−, the telomere must have been excised during growth in galactose medium. However, the red color of the resulting colonies indicated that the test chromosome was stabilized during colony formation.
A priori there are four events that could lead to stabilization of the test chromosome after telomere loss. First, the broken chromosome might gain a new telomere via gene conversion, with the use of the endogenous copy of chromosome VII (Figure 1B). Second, the test chromosome might gain a new telomere by telomerase-mediated telomere addition (Figure 1C). Third, nonhomologous end joining, a process that joins two broken ends by a mechanism that requires little if any homology and often results in the loss of a few base pairs at the break site, might restore the telomere to the broken chromosome (Figure 1D). Finally, it might be possible for the chromosome to be maintained stably without telomeric DNA as seen in Drosophila (see INTRODUCTION). On an unmodified chromosome, homologous recombination with the subtelomeric repeats on a different chromosome is another mechanism for replacing a telomere (Dunn et al., 1984). However, the lack of subtelomeric repeats on the modified VII-L chromosome rules out this possibility in our system.
Identification and Characterization of Clones in Which Test Chromosome Was Stabilized after Telomere Loss
Cells were grown in nonselective galactose medium to induce telomere loss. Next, an aliquot of cells was plated on complete medium to assess viability and on medium containing low amounts of adenine and lacking both lysine and tyrosine. Those cells that produced red colonies on the selective plates had the appropriate phenotype for cells that had retained both copies of chromosome VII. Selecting stabilization events on medium lacking lysine underestimates the stabilization frequency as de novo telomere addition and homologous recombination events must occur within the ∼220-kb region between LYS5 and the end of the test chromosome to generate a Lys+ cell (Figure 1).
In every experiment, there was a small fraction of cells that did not undergo telomere loss during the 24-h period of growth in galactose medium and hence retained both the HO recognition site and the URA3 gene near the VII-L telomere. Although in the wild-type strain, there are virtually no chromosomes that retained URA3 after telomere cleavage (34), in the rad52 pif1 strain, the test chromosome could be cut and stabilized without loss of URA3 (described in more detail below). To identify and eliminate the small fraction of clones derived from cells in which the telomere was not cleaved, a subset of the Ura+ clones that passed the first tests for disomy were streaked to selective medium containing galactose. Cells in which the telomeric HO site was intact grew poorly on selective galactose plates owing to the cell cycle arrest induced by telomere loss during this second plating on galactose medium (34). In contrast, disomic clones that had lost the HO site during the first 24-h growth period on galactose medium grew well on these plates. Frequency of stabilization (see supplementary Table 1, online) refers to the fraction of viable cells that retained the test copy of chromosome VII after telomere loss.
To determine how chromosome VII was stabilized, we analyzed the structure of the left arm of the test chromosome from independent stabilized clones. First, we prepared restriction enzyme-digested DNA from these clones and analyzed it by conventional agarose gel electrophoresis and Southern hybridization, with the use of an URA3-specific hybridization probe (Figure 2A). For example, NcoI digestion releases a 1.3-kb fragment that contains the left telomere from the test chromosome. If a new telomere is added within this region, an URA3 hybridizing fragment smaller than 1.3 kb is generated. If a new telomere is added internal to the URA3 gene or if the chromosome is repaired via homologous recombination, the strain will not contain a fragment that hybridizes to the URA3 probe (Figure 1).
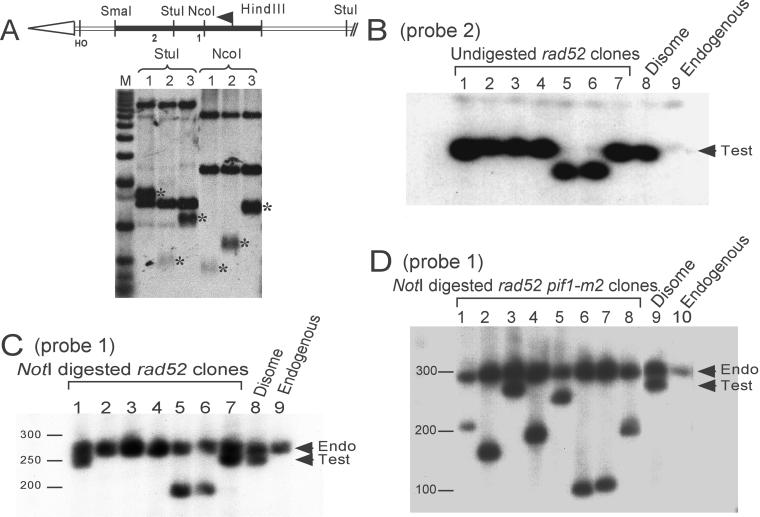
Figure 2.
Southern analysis of stabilized clones. (A) Structure of chromosomes stabilized by telomere addition within URA3. The structure of the left telomere of the test chromosome is shown. The open arrow head represents the ∼350-base pair tract of C1–3A/TG1–3 DNA. The solid black rectangle is the 1.1-kb HindIII-SmaI fragment that contains the URA3 open reading frame; the bent arrow denotes the translational start site for URA3. The region between the telomeric tract and the end of the HindIII-SmaI URA3 fragment is a 308-base pair EcoRI-HincII fragment containing the MAT locus with its HO recognition site. There are 50 bps between the HO cut site and the start of the telomeric tract. Sites for relevant restriction enzymes and for the HO endonuclease are indicated. DNA from two stabilized clones (lanes 1 and 2) or the starting disome strain (lane 3) was digested with StuI or NcoI, separated on a 1% agarose gel, and analyzed by Southern blotting with the use of a probe for the entire URA3 gene. Asterisks indicate telomeric restriction fragments. The other bands that hybridize to the probe were from either the ura3-52 locus or the portion of the telomeric URA3 that was proximal to the StuI (or NcoI) site. The sites of telomere addition as deduced from this analysis are indicated on the diagram (labeled 1 and 2, for clones in lanes 1 and 2, respectively). M, molecular weight markers (1-kb ladder). (B) PFGE mapping of stabilized clones. Positions of hybridization probes and the relevant NotI site are shown in Figure 1. Probe 1 (detects both copies of chromosome VII) was used in C and D; probe 2 (detects only the test chromosome) was used in B. Lanes 1–7 in B contain undigested DNA from independent stabilized clones isolated in the rad52 strain. NotI-digested DNA from the same clones is shown in C. Lanes 1 and 7 contain DNA from clones stabilized by de novo telomere addition within URA3; lanes 2, 3, and 4 contain clones stabilized by recombination with the endogenous copy of chromosome VII; lanes 5 and 6 contain DNA from clones stabilized near the (CA)17 tract. Lane 8 contains DNA from the starting disome strain; lane 9 has DNA from a haploid strain containing only the endogenous copy of chromosome VII. Lanes 1–8 in D contain NotI-digested DNA from independent stabilized clones isolated in the rad52 pif1-m2 strain. Each clone was stabilized by telomere addition internal to URA3. Lane 9 contains DNA from the starting disome strain, and lane 10 contains DNA from a haploid strain containing the endogenous chromosome VII.
Next, the stabilized clones were analyzed by PFGE (Figure 2, B–D). Chromosome VII is ∼1.1-mega-base pairs. Although the test chromosome was ∼15 kb shorter than the endogenous copy of chromosome VII, this difference was too small to be detected after PFGE of undigested DNA (our unpublished data). The rare-cutting restriction enzyme NotI cleaves chromosome VII into five large fragments. The NotI fragment that contains both the left telomere of chromosome VII and the LYS5 gene is ∼270 kb in a wild-type strain (Figure 1). After NotI digestion and separation by PFGE, the difference in size between the test (∼255 kb) and endogenous (∼270 kb) copies of chromosome VII was easily detectable (Figure 2C; lane 8 contains DNA from the starting disome strain; lane 9 contains DNA from a haploid strain containing only the endogenous chromosome VII). Clones in which the test chromosome was stabilized by de novo telomere addition will contain a NotI fragment smaller than the starting ∼255-kb fragment, and this fragment will hybridize to chromosome VII-L sequences (Figure 2C, lanes 5 and 6). Clones in which the test chromosome was stabilized by homologous recombination will produce a NotI fragment of the same size (∼270 kb) and sequence as endogenous chromosome VII (Figure 2C, lanes 2–4).
In Wild-Type Cells, Homologous Recombination Is the Primary Stabilization Mechanism after Telomere Loss
In wild-type strains, the frequency of stabilization after telomere loss is ∼70% (Sandell and Zakian, 1993; and supplementary Table 1 online). In a rad52 strain, the frequency of stabilization was only 0.3%, reduced >200-fold compared with wild type (see supplementary Table 1A, online). Thus, a RAD52-dependent event, presumably homologous recombination, was responsible for most stabilization events, and in its absence, >99% of cells lost the test chromosome after telomere excision. Consistent with this interpretation, with the use of a combination of conventional and pulse field gel analysis (Figure 2), 30 of 30 stabilized clones from the wild-type strain lacked a URA3-hybridizing fragment, and the left end of both copies of chromosome VII had the same structure (our unpublished data).
Stable, Terminally Deleted Test Chromosomes Can Be Recovered in rad52 Strain
Because most stabilization events were eliminated in a rad52 strain, this strain could be used to identify alternative pathways for telomere acquisition. The structure of the left end of the stabilized test chromosome was examined in 51 independent rad52 clones from the stabilization experiment summarized in the supplementary Table 1, found online. Conventional and PFGE analysis revealed that the test chromosome was shorter than the starting test chromosome in 20% of the stabilized clones (see supplementary Table 1). In 3 of 10 of these events, the test chromosome ended within URA3 as revealed by the appearance of an URA3-hybridizing fragment smaller than the terminal restriction fragment from the test chromosome (Figure 2A, asterisks; Figure 3A, circles). Two more chromosomes ending within URA3 were obtained in independent experiments with the use of the rad52 strain (Figure 3A, boxes). In seven clones, PFGE analysis of both undigested (Figure 2B, lanes 5 and 6) and NotI-digested DNA (Figure 2C, lanes 5 and 6) revealed that the test chromosome had lost ∼50 kb before stabilization. In independent experiments, three additional stabilization events that eliminated ∼50 kb were isolated (Figure 4A, squares).
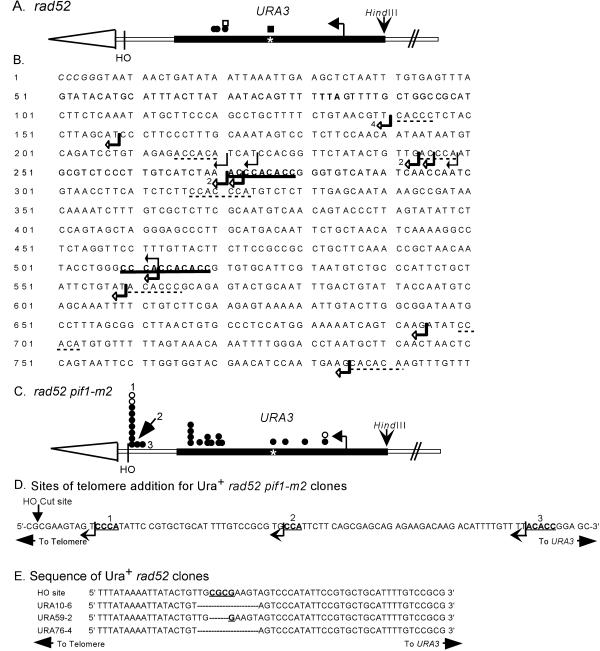
Figure 3.
Positions of de novo telomere addition sites near or within URA3. (A) Positions of independent stabilization events from the rad52 strain are denoted by circles or squares. Circles are clones isolated in the experiment used to determine the frequencies of stabilization reported in Supplementary Table 1A. Squares indicate clones isolated in independent stabilization experiments. Open symbols denote clones whose positions were determined solely by Southern analysis. Closed symbols denote the positions of clones that were mapped precisely by DNA sequencing. The asterisk marks the position of an 11-base pair telomere-like sequence in URA3. The exact sites of telomere addition for these clones are shown in B, which presents the sequence of the strand running 5′ to 3′ from the end toward the center of the chromosome of the URA3 sequence, starting with the SmaI site used to clone the gene. The telomere is distal to the SmaI site. The TAA termination site for Ura3p is in bold. Black arrows indicate the sites of telomere addition in four independent stabilization events isolated in the rad52 strain. Open arrows indicate the sites of telomere addition in 15 independent stabilized clones isolated in the rad52 pif1-m2 strain. In three cases, multiple clones had the same site of telomere addition: in these cases, the number of events at the site is indicated. The 9- and 11-base pair tracts of telomere-like DNA are in bold and underlined. Tracts of telomere-like DNA of 5–7 base pairs are underlined with dotted lines. (C) Positions of independent stabilization events from the rad52 pif1-m2 strain; symbols are the same as in A. The exact sites of telomere addition for these clones are marked by open arrows in B. The positions labeled 1, 2, and 3 are shown in expanded form in D, the sequence of the 100 bps internal to the HO cut site. Telomere addition at these sites yielded Ura+ cells. The arrow indicates the points of transition between the sequence of the test chromosome and the added telomeric DNA. The bold, underlined bases fit the consensus for telomeric DNA. (E) Compares the sequence around the HO cut site in the test chromosome with the sequence for this region in three independent Ura+ stabilized clones isolated in a rad52 strain. The dashes in the sequences indicate missing bases.
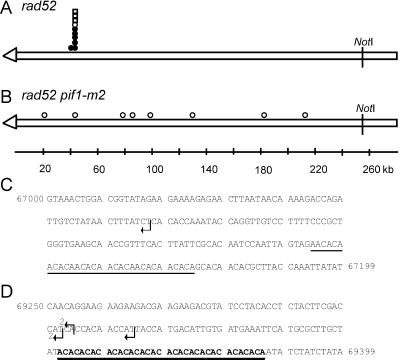
Figure 4.
Positions of de novo telomere addition sites internal to URA3. The ∼255-kb NotI fragment from the left end of the test chromosome is depicted in A and B. The open arrowhead represents the telomere. The symbols indicate the sites where new telomeres were added in independent stabilized clones isolated in the rad52 strain (A) or the rad52 pif1-m2 strain (B). Circles are clones isolated in the experiments used to determine the frequencies of stabilization reported in Supplementary Table 1A. Squares indicate clones isolated in independent stabilization experiments. Open symbols denote clones whose positions were determined solely by PFGE and Southern analysis. Closed symbols denote the positions of clones that were mapped precisely by DNA sequencing; the sequence at these sites is presented in C and D. C and D are the sequence of two sites ∼50 kb from the left telomere of chromosome VII. Arrows indicate the sites of telomere addition in five independent stabilized clones isolated in the rad52 strain. The sequence in C is 200 base pairs, starting at base pairs 67,000 from the left telomere of chromosome VII (all sequence coordinates are from the Saccharomyces genome database; http://genome-www.stanford.edu/Saccharomyces). A 31-base pair CA-rich tract that resembles but does not match the consensus sequence for telomeric DNA is underlined. The dotted line denotes a 7-base pair stretch of telomere-like DNA. (D) Shows 150 base pairs, starting at base pairs 69250. The (CA)17 tract is in bold and underlined. There were no other stretches of 6 bps or greater of telomere-like DNA in the 2.4-kb segment of chromosome VII between coordinates 67,000–69,400.
De Novo Telomere Addition Occurs at Low Frequency near Tracts of Telomere-like DNA in rad52 Cells
The terminally deleted chromosomes that were recovered in the rad52 strain could be either de novo telomere addition events or, as in Drosophila, rare chromosomes that were stabilized without addition of telomeric DNA. To distinguish between these possibilities, we used PCR to amplify the ends of the stabilized test chromosomes. For clones stabilized within URA3, we used primers from near the 5′ end of URA3 and an oligonucleotide complementary to yeast telomeric DNA. Only ends that had acquired telomeric DNA will yield a PCR product of the appropriate size. We obtained PCR products from four of four stabilized chromosomes (Figure 3A, filled symbols). These products were then cloned and sequenced to determine the precise sites of telomere addition (Figure 3B, black arrows). Each of these additions occurred at a different site within URA3, demonstrating that each event was independent. Although each site of telomere addition was novel, three were within 28 bps of each other (Figure 3B) and each occurred within or distal to an 11-base pair tract that matches the consensus sequence for yeast telomeric DNA (Figure 3B, bold and underlined). Tracts of DNA that are normally at internal sites on a yeast chromosome but that match the C1–3A consensus sequence for telomeric DNA will hereafter be called telomere-like DNA.
To clone the ends of the stabilized chromosomes that had lost ∼50 kb, we had to map the ends more precisely. From the size of the terminal NotI restriction fragment, we deduced that the ends of these chromosomes fell within lambda clone 70302 (American Type Culture Collection, Manassas, VA). EcoRI and HindIII subcloned restriction fragments from this lambda clone were used to probe a Southern blot of PFGE-separated, undigested DNA from the stabilized clones. This analysis revealed that the ends of all but one of these stabilized chromosomes were within a 2.0-kb EcoRI restriction fragment (positions 67651–69751); the end of the remaining clone was in the adjacent 1.1-kb EcoRI fragment. Based on this analysis, an oligonucleotide from near the ends of the chromosomes and a telomeric oligonucleotide were used to PCR amplify the very ends of the stabilized chromosomes. PCR products of the appropriate size were obtained from six of six stabilized ends (our unpublished data). Thus, all of the stabilized ends that were analyzed had acquired a new telomere.
Five of the ∼50-kb stabilized ends had a new telomere added at one of three sites within an 18-base pair region on chromosome VII (Figure 4D, sites marked by black arrows). Each addition event was independent, including those occurring at identical positions, because the sequence of the new telomere was different on each clone. These addition sites were localized 37–49 bps distal to a stretch of telomere-like DNA, (CA)17 (Figure 4D, bold and underlined) The sixth site was 2.27 kb distal of the same (CA)17 tract and 76 bps distal of a 31-base pair CA-rich tract that resembled but did not match the consensus sequence for yeast telomeric DNA (Figure 4C, tract underlined).
Even in a rad52 Strain, Most Stabilization Events Occur via Homologous Recombination
The test chromosome in 80% of the stabilized clones from the rad52 strain had the structure expected if the test chromosome had been healed by gene conversion with the endogenous chromosome VII: they had no URA3-hybridizing fragment (our unpublished data) and their size was indistinguishable from that of the endogenous chromosome VII (Figure 2, B and C, lanes 2–4). When DNA from these clones was digested with NotI and analyzed by PFGE, the terminal NotI fragments from both the stabilized test chromosome and the endogenous chromosome were ∼270 kb (Figure 2C, compare lanes 2–4 to lane 8). To demonstrate directly that the test chromosome in these stabilized clones had acquired sequence information from the left end of the endogenous chromosome VII, we isolated haploid subclones from three independent stabilized clones that retained only the test chromosome. Although chromosome loss is low in the wild-type disomic strain, the frequency of chromosome loss is high enough in the rad52 strain (Sandell and Zakian, 1993) that subclones that lost the endogenous copy of chromosome VII could be isolated easily (see MATERIALS AND METHODS). DNA was prepared from these subclones, digested with HindIII, and analyzed by Southern hybridization with the use of a probe that detects a portion of ADH4 that was deleted from the test chromosome during its construction (Figure 1, legend). Each of the three haploid subclones containing the test chromosome stabilized by recombination had a 5.2-kb HindIII fragment that hybridized to the ADH4 probe (our unpublished data). From these data, we infer that 80% of the stabilized clones in the rad52 strain arose via homologous recombination between the test chromosome and the endogenous copy of chromosome VII.
Frequency of De Novo Telomere Addition Is Increased Greatly in Absence of Pif1p
The DNA helicase Pif1p inhibits de novo telomere addition on spontaneous and induced double-strand breaks in a YAC (Schulz and Zakian, 1994). However, the YAC is compromised mainly of lambda DNA and has an 108-base pair stretch of C4A4/T4G4 DNA, a sequence not found in yeast. To determine the effects of Pif1p on stabilization of breaks in yeast chromosomal DNA, the pif1-m2 mutation was introduced into both the RAD52 and rad52 chromosome VII disomic strains. This mutation affects telomeric but not mitochondrial DNA (Schulz and Zakian, 1994) (see MATERIALS AND METHODS). The frequency of stabilization in the pif1-m2 strain (71%) was similar to that of the wild-type strain (74%; supplementary Table 1A). The structure of the stabilized test chromosome was determined in 39 clones isolated in the pif1-m2 strain. As in wild-type cells, most (92%) of the stabilization events involved homologous recombination. However, in contrast to the wild-type strain where de novo telomere addition was not detected, three of these stabilization events (8%) occurred by de novo telomere addition. By the criterion of Southern mapping, two of these addition events were within URA3, and the third was at a more internal site on chromosome VII but was not near the (CA)17 tract (supplementary Table 1B).
The frequency of stabilization in the rad52 pif1-m2 strain was 12% (supplementary Table 1A). Although this frequency was lower than in the RAD52 pif1-m2 strain (71%), stabilization after telomere loss was much more frequent in the rad52 pif1-m2 strain (12%) than in the rad52 PIF1 strain (0.34%). With the use of the same combination of conventional and PFGE analysis described previously, the structure of the test chromosome was determined in 36 stabilized clones from the rad52 pif1-m2 strain. In all 36 clones, stabilization occurred by de novo telomere addition (supplementary Table 1A). Thus, in the rad52 background, de novo telomere addition was ∼180-fold higher in the absence than in the presence of Pif1p. Most of the telomeres were added near URA3 (28/36; 78%) (positions summarized in Figure 3C). The remaining de novo telomere addition events (8/36; 22%) occurred throughout the 220-kb region between URA3 and LYS5 (Figure 4B). Unlike the rad52 strain, there was no strong preference for telomere addition near the (CA)17 tract.
With the use of the strategy described above, we cloned and sequenced the sites of telomere addition for 24 rad52 pif1-m2 clones that had stabilized near URA3 (Figure 3B, open arrows). Fifteen of the telomere addition events occurred at 10 different sites within URA3; three sites were used more than once. All of these addition events were independent because the added telomeric DNA was different on each clone. Telomeres were also added at three different sites within the 308-base pair region between the HO recognition site and URA3 (9/24; 38%) (Figure 3, C and D). Because no URA3 coding information was lost, these events yielded Ura+ colonies. Seven of these addition events occurred at the identical site, only 9 bps internal to the HO cut site (Figure 3, C and D). Again, these were each independent events because the sequence of the added telomeric DNA was different on each clone.
Nonhomologous End Joining Can Rejoin a Broken Chromosome to Its Excised Telomere
Loss of Pif1p affected both the frequency and distribution of de novo telomere addition events (Supplementary Table 1). Because 25% of the stabilization events (3% of the starting cells) were stabilized by telomere addition distal to URA3, Ura+-stabilized clones were relatively common in the rad52 pif1-m2 strain. However, in the rad52 strain, these events were exceptionally rare. For example, in a typical experiment carried out specifically to identify such events, 99.5% of the rad52 disomic cells were Ura− after 24 h of growth on galactose medium. With the use of the criteria described above, 120 Ura+ disomic clones isolated from this strain were tested by restreaking on galactose medium; all 120 retained the HO recognition site. Therefore, in the rad52 strain, <0.004% of the starting cells acquired a new telomere distal to URA3. From different independent experiments, we were able to identify three disomic clones from the rad52 strain that were Ura+ and lacked the HO recognition site. Cloning and sequence analysis showed that none of the three were generated by de novo telomere addition. Rather, each was missing 3–10 bps around the HO recognition site but was otherwise unaltered (Figure 3E). The amount of homology at the junctions is typical of nonhomologous end-joining events (Moore and Haber, 1996). Thus, these rare Ura+ events are most easily explained by cleavage at the HO site, followed by degradation of the ends and nonhomologous end joining of the telomere to the broken chromosome.
Distribution of Telomere-like Sequences within Yeast Genome
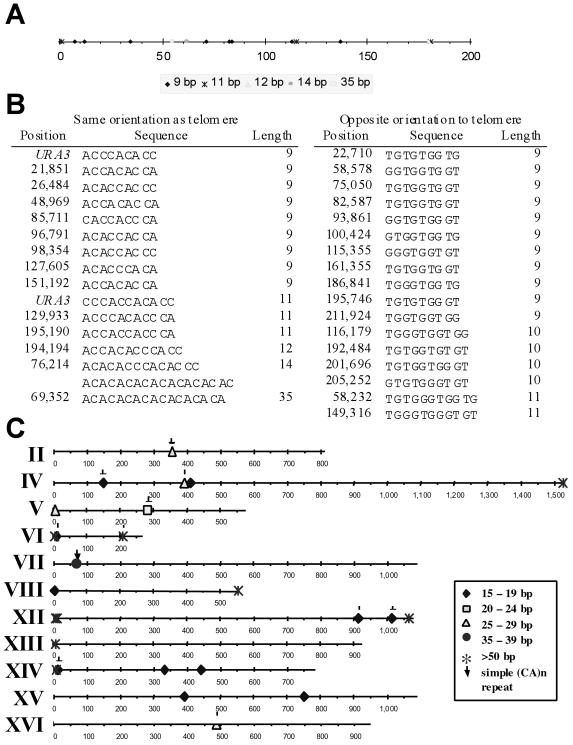
In the rad52 strain, 70% of the de novo telomere addition events occurred close to a naturally occurring (CA)17 tract (Supplementary Table 1B). Because the (CA)17 tract was proceeded by an A residue, it constituted a 35-base pair sequence that fits the C1–3A consensus for yeast telomeric DNA. However, uninterrupted CA tracts of this length have not been reported in telomeric DNA (Wang and Zakian 1990). The remaining telomere addition events occurred within URA3, in each case distal to an 11-base pair tract of telomere-like DNA that was separated from a 9-base pair tract by only 230 bps (Figure 3B). These results suggest that long tracts of telomere-like DNA or closely spaced shorter tracts might promote telomere addition after chromosome breakage.
One way to assess the importance of telomere-like tracts to telomere addition is to determine whether there were similar tracts in the ∼220-kb region of chromosome VII within which telomere addition could yield a Lys+ cell. Only tracts in which the C-rich strand runs 5′ to 3′ toward the centromere have the appropriate orientation to support telomere addition (Murray et al., 1988; Pluta and Zakian, 1989). We used 5′-[(C1–3)A]n-3′ as a query to determine the distribution of telomere-like tracts of 9 bps or longer in yeast chromosomal DNA (other than in telomeres themselves). In the ∼220-kb left end of chromosome VII, the next longest stretch of telomere-like DNA was 14 bps (Figure 5, A and B). This ∼220-kb region also contained one 12-, two 11-, one 10-, and eight 9-base pair tracts of telomere-like DNA. No two of the telomere-like tracts were as close as the two tracts within URA3: the closest tracts, a 12-base pair tract at position 194,194 and an 11-base pair tract at position 195,190 were separated by ∼1 kb (Figure 5B).
Figure 5.
Distribution of telomere-like sequences in yeast. (A) Distribution of telomere-like DNA, sequences that match the C1–3A/TG1–3 consensus sequence for yeast telomeric DNA, within the distal ∼220 kb of the test chromosome VII. Only tracts of nine bases or longer that are in the correct orientation to support telomere addition are shown. The most distal 9- and 11-base pair tracts are from URA3 and were only on the test copy of chromosome VII. When URA3 is in its normal position near the centromere of chromosome V, these 9- and 11-base pair tracts are not in the same orientation as telomeric DNA. There was one 9-base pair tract of telomere-like DNA on the endogenous copy of chromosome VII that was eliminated during construction of the test chromosome. (B) Exact sequence and position of 9-base pair or longer tracts of telomere-like DNA in the same (left) or different (right) orientation as telomeric DNA are shown. The coordinates for each tract are based on the sequence of intact chromosome VII, which is ∼15 kb longer than the test chromosome. (C) Presence of tracts of telomere-like DNA of 15 bps or greater in the yeast genome. Chromosomes I (230 kb), III (317 kb), IX (440 kb), X (745 kb), and XI (670 kb) had no telomere-like tracts >14 bps. Only tracts in the same orientation as telomeric DNA are shown. The subset of C1–3A/TG1–3 tracts that are (CA)n DNA are marked by arrows. All tracts >35 base pairs are adjacent to Y′ subtelomeric repeat sequences, with the exception of the (TG)31 tract located around position 210 kb on chromosome VI. ♦, 15–19 base pairs; □, 20–24 base pairs; ▵, 25–29 base pairs; ●, 35–39 base pairs; *, >50 base pairs; ↓, CA tracts.
In the entire yeast genome, there were only nine additional tracts of telomere-like DNA in the correct orientation to support telomere addition that were as long or longer than the (CA)17 tract on chromosome VII-L (Figure 5C). All but one of these tracts was within 13 kb of a telomere, and each of these was adjacent to a subtelomeric Y′ element. These telomere proximal tracts ranged in size from 52 to 159 bps, and each had the heterogeneous sequence characteristic of yeast telomeric DNA. C1–3A/TG1–3 tracts next to Y′ elements were described previously (Walmsley et al., 1984). The longest internal stretches of DNA with the complex sequence of telomeric DNA were five tracts, each of 15 or 16 bps, distributed among chromosomes IV, XIV, and XV (Figure 5C).
Chromosome VI was the only chromosome other than chromosome VII that had an internal, very long stretch of telomere-like DNA. In this case, the tract was (GT)31 at position 210,332 (this tract was on the right arm of chromosome VI where the correct orientation for telomere formation is 5′-TG1–3-3′.) Additionally, there were three internal (CA)n tracts of 26 or 27 bps (on chromosomes II, IV, and XVI), one internal tract of (CA)11 on chromosome V, and eight internal (CA)n tracts in the 15–18-base pair range (Figure 5C). Seven of the 16 yeast chromosomes, chromosomes I, III, VIII, IX, X, XI, and XIII had no internal tract of telomere-like DNA >14 bps in the same orientation as telomeric DNA. Although long telomere-like tracts were rare, given the 38% G + C content of the yeast genome, they were not under-represented in the yeast genome. In fact, (CA)n tracts where n is >4 were much more abundant than expected from sequence composition alone (our unpublished data).
In the entire yeast genome, there were 779 tracts of telomere-like DNA of 9 bps or longer in the same orientation as telomeric DNA. Of these, only 4.5% were ≤230 bps from another telomere-like tract of 9 base pairs or longer. We conclude, that tracts as long as the (CA)17 tract on chromosome VII were exceptionally rare, and two moderately long tracts as close as those within URA3 were relatively rare.
DISCUSSION
We determined the mechanisms that lead to chromosome stabilization after telomere loss in Saccharomyces. Three pathways were able to stabilize broken chromosomes: homologous recombination, de novo telomere addition, and nonhomologous end joining (Figure 1). Regardless of the mechanism, each stabilization event resulted in acquisition of a telomere. Although previous work from our lab demonstrated that loss of a telomere often results in loss of the affected chromosome (Sandell and Zakian, 1993), it was not clear from this or other previous studies (Kramer and Haber, 1993) whether stabilization ever occurred without telomere acquisition. Thus, even although double-strand breaks in yeast bind several telomeric proteins, such as the Ku heterodimer and Sir proteins (Martin et al., 1999; Mills et al., 1999), unlike the case for HP1 binding in Drosophila, this binding is not sufficient to confer telomere function in the absence of telomeric DNA. Together with previous studies (Kramer and Haber, 1993; Sandell and Zakian, 1993), these data argue that telomeric DNA is required for the stable maintenance of linear chromosomes in Saccharomyces. This dependence is likely to apply to other eukaryotes, which, like yeast, maintain their telomeres by a telomerase mechanism. Because telomeric DNA in Drosphila is not telomerase generated (Biessmann and Mason, 1997), Drosophila's ability to maintain telomere function without a special telomeric DNA may be related to its unusual mechanism of end replication.
As expected, in a wild-type cell, the majority of the stabilized test chromosomes acquired a new telomere by homologous recombination with the endogenous copy of chromosome VII (Supplementary Table 1). However, even in the rad52 strain, most (80%) stabilization events involved homologous recombination. Surprisingly, these events were not eliminated in a strain with a complete deletion of RAD52 (rather than the insertion allele used for most experiments) nor in a rad52 strain that also lacked Rad59p, a Rad52p-related protein whose function partially overlaps that of Rad52p (Bai and Symington, 1996; Bai et al., 1999) (Teng and Zakian, unpublished data). Extremely rarely, occurring at <0.003% of the broken chromosomes and accounting for <1% of the stabilization events, stabilization was due to nonhomologous end joining (Figure 3E). In contrast, in mammalian cells, at least half of induced double-strand breaks are repaired by nonhomologous recombination (Liang et al., 1998).
Although 20% of the stabilization events in the rad52 strain were due to telomere addition (Supplementary Table 1A), these events were relatively rare, stabilizing <0.1% of the broken chromosomes. Indeed, healing a broken chromosome in yeast by de novo telomere addition is so rare that a previous study that analyzed fewer cells did not observe any such events, leading the authors to speculate that de novo telomere addition does not occur on natural yeast chromosomes (Kramer and Haber, 1993). Telomere addition anywhere in the ∼220-kb region between the telomere and LYS5 would have been detected in our assay. Nonetheless, telomere formation occurred at a very limited number of sites. Thirty percent of these events were within a 300-base pair portion of URA3, <1 kb from the site of chromosome breakage (Supplementary Table 1B and Figure 3, A and B). URA3 had one 9- and one 11-base pair tract of telomere-like DNA, separated by only 230 base pairs (Figure 3B, bold and underlined). Telomere-like tracts of this length spaced this close together were relatively rare in the yeast genome. Most (70%) telomere addition events occurred ∼50 kb from the end of the test chromosome (Supplementary Table 1B). Six of six sequenced ∼50-kb terminal deletions were within a few kilobases of a (CA)17 tract (Figure 4A, closed symbols). Four other telomere addition events were mapped to this region by Southern hybridization (Figure 4A, open symbols). After the (CA)17 tract, the next longest tract of telomere-like DNA in the ∼220-kb region between the telomere and LYS5 on chromosome VII was a 14-base pair tract at position 76, 214 base pairs (Figure 5B). Based on the absence of telomere additions events near this 14-base pair tract (Figure 4A), we estimate that telomere addition was ≥10 times more frequent near the (CA)17 tract than near the 14-base pair tract. However, other features of a telomere-like tract, in addition to its length, such as its chromosomal context, might affect its ability to promote telomere addition.
Tracts of telomere-like DNA as long as the (CA)17 tract were extremely rare in nontelomeric regions of the yeast genome. For example, seven yeast chromosomes had no internal tract of telomere-like DNA longer than 14 base pairs in the proper orientation to promote telomere addition (Figure 5C). We propose that long tracts of telomere-like DNA, such as the (CA)17 tract on chromosome VII, increase the probability of telomerase-mediated telomere addition. This proposal was also made in a previous study in which a 78-base pair tract of C4A2/T2G4 DNA, a ciliate telomeric sequence not found normally in yeast, promotes telomere addition at an HO-induced double-strand break on chromosome III (Kramer and Haber, 1993). In this system, no telomere addition events are detected in the absence of the ciliate tract, consistent with our finding that chromosome III had no long tract of telomere-like DNA (Figure 5C). This model suggests that de novo telomere addition is rare in wild-type cells in part because there are few internal tracts of telomere-like DNA long enough to promote telomere addition.
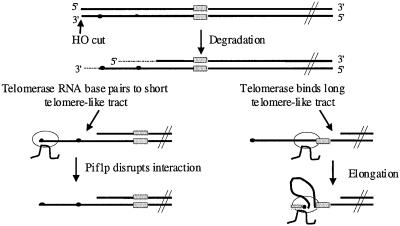
How might long tracts of telomere-like DNA increase the probability of telomerase elongation? After HO induces a double strand break, the 5′ end is degraded much faster than the 3′ end (White and Haber, 1990). This degradation will expose the G-rich strand of telomere-like tracts (Figure 6). Because there was a minimum of 37 bps between the (CA)17 tract and the start of the newly added telomere (Figure 4, C and D), the (CA)17 tract was not itself the site of telomere addition. Previous studies also found that several base pairs can intervene between the site of telomere addition and a telomere-promoting sequence (Murray et al., 1988; Wang and Zakian, 1990; Kramer and Haber, 1993), but our analysis is the first to document the ability of a naturally occurring tract of chromosomal, telomere-like DNA to promote telomere addition.
Figure 6.
Model for de novo telomere addition after chromosome breakage. The striped and spotted rectangles represent, respectively, the C and G-strands of a long stretch of telomere-like DNA, such as the (CA)17 tract on chromosome VII. Small closed circles represent the G-strand of very short, 3–9-base pair stretches of telomere-like DNA. After the HO endonuclease excises the telomere, the break is degraded, with degradation of the 5′ end proceeding faster than the 3′ end (denoted by dotted line). These short TG1–3-tracts can recruit telomerase, either by simple base pairing with telomerase RNA (left) or by first binding Cdc13p (Lin and Zakian, 1996; Nugent et al., 1996), which then recruits telomerase (Evans and Lundblad, 1999; Qi and Zakian, 2000; Zhou et al., 2000a). The TG1–3/telomerase RNA association is disrupted by the Pif1p helicase, thus inhibiting de novo telomere addition. In the absence of telomerase lengthening, this 3′ single-strand tail is channeled into recombination. The G-rich strand of a long stretch of telomere-like DNA (dotted rectangle) can recruit telomerase by binding the anchor site of telomerase (right). The 3′ end of the DNA break is then brought to the active site of telomerase by looping out of the intervening DNA allowing elongation of the 3′ end by addition of TG1–3 DNA (spotted rectangle).
These data argue that telomere-like tracts do not act simply by increasing the probability of primer pairing to the template region of telomerase RNA. Rather, we suggest that telomere-like tracts recruit telomerase by interacting with the anchor site (or another site) on telomerase (Figure 6). To explain the processivity of ciliate and human telomerase in vitro, DNA primers are proposed to make contact with telomerase at two sites: the catalytic site where the primer is base paired to telomerase RNA, and the anchor site (Greider, 1991; Morin, 1991; Hammond et al., 1997). Binding at the anchor site can retain telomerase at the telomere during the translocation step, when the primer is released from the active site. Consistent with a model in which the single strand (TG)17 tract binds telomerase, TG1–3 and (TG)n but not C1–3A or duplex oligonucleotides bind Saccharomyces telomerase in vitro (Lue and Xia, 1998). Yeast telomeres are clustered in vivo (Gotta et al., 1996). This clustering might facilitate the ability of a shorter tract of telomere-like DNA that is close to a telomere, such as the 11-base pair tract within URA3 (Figure 3B), to recruit telomerase from another telomere, whereas the same length tract far from a telomere would be less effective.
Another reason telomere addition at double-strand breaks is rare is that it is inhibited by the Pif1p DNA helicase. When cells lacked the Pif1p helicase, the frequency of telomere addition increased almost 200-fold (Supplementary Table 1A). This increase probably underestimates the inhibitory effects of Pif1p because a pif1-m2 strain does not have as severe a telomere phenotype as a pif1Δ strain (Schulz and Zakian, 1994). In addition, the distribution of telomere addition sites was altered in the absence of nuclear Pif1p (Supplementary Table 1B). First, a larger fraction (78%) of the telomere addition events were within 1 kb of the telomere in the rad52 pif1-m2 strain than in the rad52 strain (30%) (Supplementary Table 1B). The most common site for telomere addition in the rad52 pif1-m2 strain, used in 30% of the stabilization events, was only 9 bps from the HO cut site (Figure 3C, site 1). At this site, new telomeres were added directly to a CCCA tract, the very first telomere-like DNA internal to the HO cut site (Figure 3D, site 1). Telomere addition at this site was never detected in the presence of Pif1p, even although we searched specifically for these events (Figure 3E).
The distribution of de novo telomere addition events internal to URA3 was also affected by Pif1p (compare Figure 4B, rad52 pif1 to 4A, rad52 strain). Whereas all nine of the large terminal deletions in the rad52 strain were within a few kilobases of the (CA)17 tract, only one of eight such events was near this tract in the rad52 pif1 strain. Nonetheless, telomere addition was not random in pif1 cells because in 23 of the 24 sequenced telomere addition events, the new telomere was added to a 3–9-base pair stretch of telomere-like DNA (Figure 3, B and D). A similar spectrum of addition site sequences was seen in the rad52 strain where 7 of 10 new telomeres were added at 3–11-base pair tracts of telomere-like DNA (three were added to nontelomeric DNA; Figures 3B and 4, C and D).
How does Pif1p inhibit the frequency of telomere addition and alter the distribution of these events? Because its catalytic activity is necessary for Pif1p to inhibit lengthening of existing telomeres (Zhou et al., 2000b), the simplest possibility is that Pif1p also acts as a helicase when it affects telomere formation at a double-strand break. We suggest that telomerase interacts with DNA breaks by base pairing of telomerase RNA to very short stretches of telomere-like DNA, stretches that are very common in yeast DNA (Figure 3B). However, in PIF1 cells, these interactions are rarely productive because Pif1p dissociates them (Figure 6, left). Thus, in wild-type cells, telomerase action at a double-strand break requires an additional mechanism, such as a long stretch of telomere-like DNA that can retain telomerase after Pif1p-mediated displacement of its active site from the DNA primer (Figure 6, right). This model explains both the increased frequency and the altered distribution of addition events in pif1 cells because in the absence of Pif1p, telomerase would have a higher probability of productive association with very short tracts of telomere-like DNA.
Some chromosomal rearrangements isolated from both normal human fibroblasts and human tumor cells, once thought to be terminal deletions, have been found to be subtelomeric translocations (Meltzer et al., 1993). These data suggest that de novo telomere addition is much rarer in human cells than originally thought. If the human Pif1p-like protein (Zhou et al., 2000b), like its Saccharomyces counterpart, inhibits de novo telomere addition, it could contribute to genome stability by limiting the number of terminal deletions, thereby reducing events that result in loss of heterozygosity.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Taggart, Y. Tsukamoto, L. Vega, and J.-Q. Zhou for comments on the manuscript. This work was supported by grant R37 GM-26938 from the National Institutes of Health and a pre-doctoral fellowship to M.K.A. from the Howard Hughes Medical Institute.
REFERENCES
- Ahmad K, Golic KG. Telomere loss in somatic cells of Drosophila causes cell cycle arrest and apoptosis. Genetics. 1999;151:1041–1051. doi: 10.1093/genetics/151.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Davis AP, Symington LS. A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics. 1999;153:1117–1130. doi: 10.1093/genetics/153.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Symington LS. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- Bednenko J, Melek M, Greene EC, Shippen DE. Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-assisted de novo telomere formation. EMBO J. 1997;16:2507–2518. doi: 10.1093/emboj/16.9.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H, Carter SB, Mason JM. Chromosome ends in Drosophila without telomeric DNA sequences. Proc Natl Acad Sci USA. 1990;87:1758–1761. doi: 10.1073/pnas.87.5.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H, Mason JM. Progressive loss of DNA sequences from terminal chromosome deficiencies in Drosophila melanogaster. EMBO J. 1988;7:1081–1086. doi: 10.1002/j.1460-2075.1988.tb02916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H, Mason JM. Telomere maintenance without telomerase. Chromosoma. 1997;106:63–69. doi: 10.1007/s004120050225. [DOI] [PubMed] [Google Scholar]
- Blanchard MM, Taillon-Miller P, Nowotny P, Nowotny V. PCR buffer optimization with uniform temperature regimen to facilitate automation. PCR Methods Appl. 1993;2:234–240. doi: 10.1101/gr.2.3.234. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Chalker DL, Yao MC. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu Rev Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- Dunn B, Szauter P, Pardue ML, Szostak JW. Transfer of yeast telomeres to linear plasmids by recombination. Cell. 1984;39:191–201. doi: 10.1016/0092-8674(84)90205-8. [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW. Telomerase is processive. Mol Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond PW, Lively TN, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer KM, Haber JE. New telomeres in yeast are initiated with a highly selected subset of TG1–3 repeats. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- Lahaye A, Stahl H, Thines-Sempoux D, Foury F. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis RW. Viable deletions of a telomere from a Drosophila chromosome. Cell. 1989;58:791–801. doi: 10.1016/0092-8674(89)90112-8. [DOI] [PubMed] [Google Scholar]
- Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF, Xia J. Species-specific and sequence-specific recognition of the dG-rich strand of telomeres by yeast telomerase. Nucleic Acids Res. 1998;26:1495–1502. doi: 10.1093/nar/26.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Mason JM, Champion LE, Hook G. Germ-line effects of a mutator, mu2, in Drosophila melanogaster. Genetics. 1997;146:1381–1397. doi: 10.1093/genetics/146.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Strobel E, Green MM. mu-2: mutator gene in Drosophila that potentiates the induction of terminal deficiencies. Proc Natl Acad Sci USA. 1984;81:6090–6094. doi: 10.1073/pnas.81.19.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer PS, Guan X-Y, Trent JM. Telomere capture stabilizes chromosome breakage. Nat Genet. 1993;4:252–255. doi: 10.1038/ng0793-252. [DOI] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin GB. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- Murray AW, Claus TE, Szostak JW. Characterization of two telomeric DNA processing reactions in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4642–4650. doi: 10.1128/mcb.8.11.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Pluta AF, Dani GM, Spear BB, Zakian VA. Elaboration of telomeres in yeast: recognition and modification of termini from Oxytricha macronuclear DNA. Proc Natl Acad Sci USA. 1984;81:1475–1479. doi: 10.1073/pnas.81.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Zakian VA. Recombination occurs during telomere formation in yeast. Nature. 1989;337:429–433. doi: 10.1038/337429a0. [DOI] [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley RM, Chan CSM, Tye B-K, Petes TD. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984;310:157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]
- Wang S-S, Zakian VA. Sequencing of Sacchromyces telomeres cloned using T4 DNA polymerase reveals two domains. Mol Cell Biol. 1990;10:4415–4419. doi: 10.1128/mcb.10.8.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:633–670. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G-L, Blackburn EH. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;76:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]
- Zakian VA, Scott JF. Construction, replication and chromatin structure of TRP1 RI circle, a multiple-copy synthetic plasmid derived from Saccharomyces cerevisiae chromosomal DNA. Mol Cell Biol. 1982;2:221–232. doi: 10.1128/mcb.2.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Hidaka K, Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol Cell Biol. 2000a;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J-Q, Monson EM, Teng S-C, Schulz VP, Zakian VA. The Pif1p helicase, a catalytic inhibitor of telomerase lengthening of yeast telomeres. Science. 2000b;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.