Abstract
Premature termination codon (PTC)-bearing transcripts are often degraded by nonsense-mediated decay (NMD) resulting in loss-of-function (LoF) alleles. However, not all PTCs result in LoF mutations, i.e., some such transcripts escape NMD and are translated to truncated peptide products that result in disease due to gain-of-function (GoF) effects. Since the location of the PTC is a major factor determining transcript fate, we hypothesized that depletion of protein-truncating variants (PTVs) within the gene region predicted to escape NMD in control databases could provide a rank for genic susceptibility for disease through GoF versus LoF. We developed an NMD escape intolerance score to rank genes based on the depletion of PTVs that would render them able to escape NMD using the Atherosclerosis Risk in Communities Study (ARIC) and the Exome Aggregation Consortium (ExAC) control databases, which was further used to screen the Baylor-Center for Mendelian Genomics disease database. This analysis revealed 1,996 genes significantly depleted for PTVs that are predicted to escape from NMD, i.e., PTVesc; further studies provided evidence that revealed a subset as candidate genes underlying Mendelian phenotypes. Importantly, these genes have characteristically low pLI scores, which can cause them to be overlooked as candidates for dominant diseases. Collectively, we demonstrate that this NMD escape intolerance score is an effective and efficient tool for gene discovery in Mendelian diseases due to production of truncated or altered proteins. More importantly, we provide a complementary analytical tool to aid identification of genes associated with dominant traits through a mechanism distinct from LoF.
Keywords: dominant negative, antimorphic, nonsense-mediated decay, frameshift alleles, stopgain variants, NMDEscPredictor, NMD escape intolerance scores, WES analysis, bioinformatic tool, genotype-phenotype correlations
Introduction
Translation-dependent nonsense-mediated decay (NMD) is an evolutionarily conserved mRNA surveillance mechanism that ensures dynamic regulation and high fidelity of gene expression in eukaryotic cells. It is a well-established “rule” that multi-exon transcripts that harbor termination codons out of their normal reading frame context, generally termed premature termination codons (PTCs), are likely to be subject to mRNA degradation by the NMD mRNA surveillance machinery and thus result in a predicted loss-of-function (LoF) variant or null allele. PTCs can be introduced into transcripts by various mechanisms including protein-truncating variants (PTVs; stopgain and indels), mRNA isoforms, and alternative translation.1, 2, 3
In mammalian cells, NMD requires an exon junction complex (EJC) that is comprised of a dynamic group of proteins that are positioned 20–24 nucleotides (nt) upstream of exon-exon boundaries by the splicing machinery in the nucleus.4, 5, 6 After an mRNA is exported from the nucleus, EJCs are removed during the pioneer round of translation by a translating ribosome. According to the EJC-dependent model for governing NMD, if a PTC is located more than 50–55 bp upstream of the last exon-exon junction, the transient interaction between the downstream EJC and the terminating ribosome is predicted to elicit NMD and degrade the mRNA harboring a PTC, i.e., NMD+ transcripts.3, 4, 7, 8, 9, 10, 11, 12, 13 On the other hand, a truncating variant that results in a PTC located within the last 50–55 bp of the penultimate exon, or the entire last exon, is predicted to escape from NMD (NMD− transcripts). The EJC-dependent model is well supported by a preponderance of experimental data that examine NMD efficiency, and the 50-bp rule alone accurately predicts NMD sensitivity in ∼85% of cancer-related mutations14, 15, 16, 17 although a number of exceptions have been reported.
Approximately one-third of mRNAs containing pathogenic variants in genetic disorders and cancer are subject to frameshift or nonsense mutations that result in the generation of PTCs.18, 19 Transcripts with PTCs located in the penultimate and last exon of genes can be NMD−, giving rise to stable mRNA translated into mutant proteins that can have a potent dominant-negative activity, thereby leading to human disease traits responsible for a broad spectrum of clinical phenotypes.2 Importantly, such PTVs that escape NMD (PTVesc) may be erroneously interpreted as LoF alleles when in fact they behave as gain-of-function (GoF) alleles; examples include DVL1 (MIM: 601365), causing Robinow syndrome autosomal-dominant 2 (DRS2 [MIM: 616331]); DVL3 (MIM: 601368), causing Robinow syndrome autosomal-dominant 3 (DRS3 [MIM: 616894]); and CRX (MIM: 602225), causing Leber congenital amaurosis 7 (LCA7 [MIM: 613829]). Moreover, some of those genes cause disease only when carrying NMD− variants, so current tools to predict variant pathogenicity that rely on LoF intolerance or haploinsufficiency scores20, 21, 22 will fail to inform probability of pathogenicity due to transcripts escaping from NMD and being translated into mutant proteins.
The systematic application of the 50-bp rule for NMD prediction of transcripts with a truncating variant requires the identification of the precise location of the predicted PTC as well as the relative position of the last EJC. Notably, there may be an unexpectedly large distance between the location of a given frameshifting insertion-deletion variant (indels) and the next predicted PTC in some transcripts, sometimes surpassing the last EJC, that can result in NMD− transcripts. As a result, those transcripts may translate into proteins with GoF properties.
Here, to investigate the potential role of escape from NMD for variant alleles implicated in human disease, we designed an efficient tool, NMDEscPredictor, to predict whether a given frameshifting indel will lead to NMD+ or to NMD− transcripts based on the relative location of the variant within the gene. Using this algorithm, we computationally classified PTVs, including frameshift insertion-deletions (indels) and stopgains, as NMD+ and NMD− in two control databases, the Atherosclerosis Risk in Communities Study (ARIC)23 and the Exome Aggregation Consortium (ExAC).20 We then developed an NMD escape intolerance score to rank each multi-exon canonical mRNA transcript in the genome based on the disequilibrium between expected and observed number of NMD− variants relative to the NMD+ variants in a given control database. Our analysis revealed a total of 1,996 genes significantly depleted (i.e., ranked in the top 5%) for NMD− variants in either control database, a relevant (98%) portion of those are likely to be tolerant to LoF. The resulting list includes genes for which C-terminal truncation does not lead to haploinsufficiency, for instance, DVL1 and REST (MIM: 600571), leading to hereditary gingival fibromatosis (HGF [MIM: 617626]), that provide poignant examples.24, 25
These findings support the hypotheses that mapping the location of PTV mutations within a gene is relevant for assessing the variant pathogenicity as well as providing information concerning disease mechanism as haploinsufficiency or potentially GoF. Moreover, we show that ranking genes based on the derived NMD escape intolerance score is an effective and efficient tool for gene discovery and may facilitate elucidating the underlying biology of Mendelian disease traits due to production of truncated or C-terminally altered proteins.
Material and Methods
Whole-Exome Sequencing and Annotation
Whole-exome sequencing was performed at the Human Genome Sequencing Center (HGSC) at Baylor College of Medicine through the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) initiative. DNA was obtained from the subjects and unaffected family members after written informed consent and the study was approved by the institutional review board at Baylor College of Medicine (protocol no. H-29697). Using 1 μg of DNA, an Illumina paired-end pre-capture library was constructed according to the manufacturer’s protocol (Illumina Multiplexing_SamplePrep_Guide_1005361_D) with modifications as described in the BCM-HGSC Illumina Barcoded Paired-End Capture Library Preparation protocol. Pre-capture libraries were pooled into 4-plex library pools and then hybridized in solution to the HGSC-designed Core capture reagent (52 Mb, NimbleGen) or 6-plex library pools using the custom VCRome 2.1 capture reagent (42 Mb, NimbleGen) according to the manufacturer’s protocol (NimbleGen SeqCap EZ Exome Library SR User’s Guide) with minor revisions. The sequencing run was performed in paired-end mode using the Illumina HiSeq 2000 platform, with sequencing-by-synthesis reactions extended for 101 cycles from each end and an additional 7 cycles for the index read. With a sequencing yield of 11 Gb, the sample achieved 92% of the targeted exome bases covered to a depth of 20× or greater. Illumina sequence analysis was performed using the HGSC Mercury analysis pipeline,26, 27 which moves data through various analysis tools from the initial sequence generation on the instrument to annotated variant calls (SNPs and intra-read insertions/deletions, i.e., indels). Variants were called using the ATLAS2 variant calling method and the Sequence Alignment/Map (SAMtools) suites and annotated with an in-house-developed Cassandra annotation pipeline that uses Annotation of Genetic Variants (ANNOVAR)28 and additional tools and databases.
Variant Prioritization
Two types of variants were selected for analysis: indels and stopgains. First, indels and stopgains were retrieved from unfiltered vcf files of the Baylor-CMG (N = 6,109 individuals) and Atherosclerosis Risk in Communities Study (ARIC) (N = 10,940 individuals) databases for further analysis; individual personal genome raw data, not “massaged” by joint calling or forced calling methods, was utilized. For frameshifting variants, further examination of the insertion to deletion ratio revealed that this metric is similar among the databases: Baylor-CMG (0.42), ExAC (0.43), and ARIC (0.39). The retrieved indels and stopgain variants were reannotated using ANNOVAR28 against the Gencode v.19 transcript reference set including 95,379 transcripts. Second, off-target indels and stopgain variants were removed. Then, we performed variant prioritization as follows. If an on-target indel or stopgain variant has a variant read number (vR) greater or equal to 5, it was retained (Figure S1). Third, the internal database frequency was computed for each indel or stopgain variant in the Baylor-CMG and ARIC databases. Rare indels and stopgain variants were extracted for further analysis (Baylor-CMG database frequency ≤ 0.01 and ARIC database frequency ≤ 0.01) (Figures S2–S4). At this step, in-frame indels were removed from the analysis. Version 0.3.1 of the Exome Aggregration Consortium (ExAC) dataset was used for our analysis as another control dataset. Variants in the ExAC database were included in further analysis according to the following rules: (1) it has to meet the high-quality variant criteria set by ExAC, (2) its minor allele frequency (MAF) should be less than or equal to 0.01, and (3) it has to be present in the Ensembl v.19 canonical transcript of the gene.
Prediction of NMD Incompetent Frameshifting Indels and Stopgain Variants: NMDEscPredictor
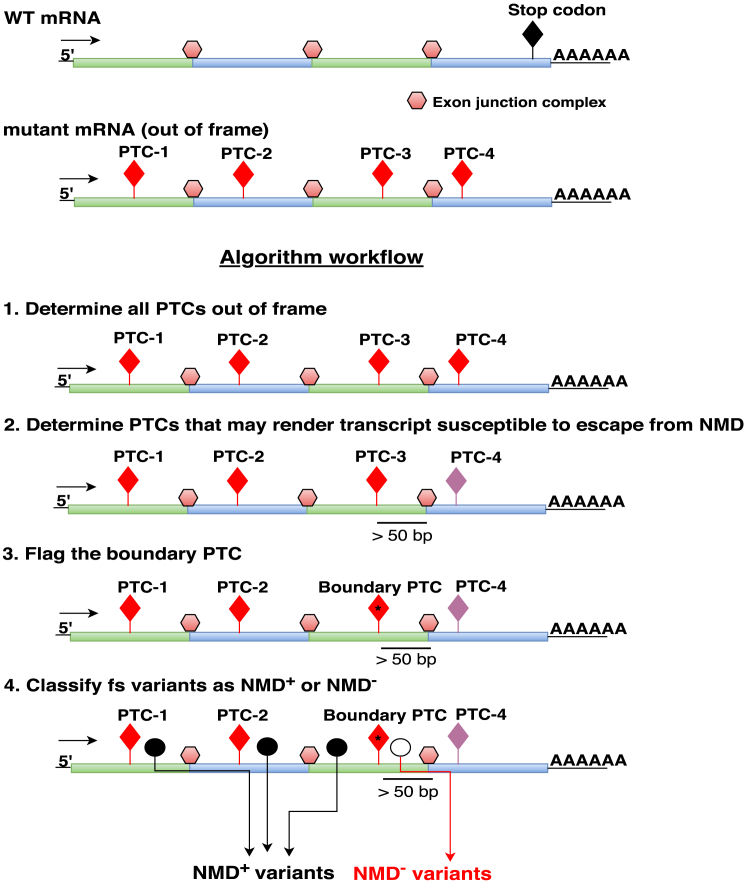
First, to predict frameshifting indels that potentially lead the transcript to be subject to degradation (NMD+) or to escape degradation (NMD−), we designed an algorithm for computational prediction of frameshifting indels as NMD+ versus NMD−, NMDEscPredictor. For each Ensembl transcript in the v.19 transcript reference set consisting of 95,379 transcripts in total, coding sequences were retrieved using ExtractTranscriptSeqs function in Genomic Features Bioconductor Package. We first removed 7,471 single-exon transcripts from further analysis. For the remaining 87,908 transcripts, we predicted each possible PTC in the −1 frames and +1 frames. For instance, −1 frame indels lead the reading frame to shift one base ahead, due to the removal of one base pair or the addition of two base pairs. Likewise, +1 frame indels lead the reading frame to shift one base behind, due to the addition of one base pair or the removal of two base pairs.29 Using the 50-bp rule,14, 15, 16 we classified PTCs into two categories, those that may lead the transcript to escape from NMD (NMD−) or not (NMD+) (Figure 1). Then we flagged the PTC right before the first PTC that may lead the mutant transcript to escape from NMD: that is the boundary PTC that separates the transcript into two regions regarding the likely NMD fate of a variant allele. For this prediction we include only frameshifting indels and stopgain variants annotated according to any of the canonical transcripts retrieved from the table placed in the ExAC database repository (fordist_cleaned_exac_r03_march16_z_pli_rec_null_data.txt). In addition, we removed from further analysis frameshifting indels in which their annotated transcripts did not present with any PTC or boundary PTC in either the −1 or +1 frame.
Figure 1.
NMDEscPredictor Algorithm Workflow
Horizontal lines denote transcripts with alternating different exons shaded green or blue. The exon junction complex is demarcated by a light red hexagon and the position of stop codon is shown (lollipop structure with filled black diamond on top). For each multi-exon transcript in the Ensembl reference set (version 19), we first determined all potential PTCs (shown as lollipop structures with filled red diamonds marking the map position in transcript) in the −1 and +1 frames. Second, we identified PTCs that may result in the transcript escaping from NMD based on the 50-bp rule (lollipop structures with filled purple diamond).16, 17, 18 At the third step of the algorithm, we flagged the PTC upstream of the first PTC that can escape from NMD and labeled it as the boundary PTC (lollipop structure with filled starred red diamond). Transcripts with frameshift (fs) variants located upstream of the boundary PTC are predicted to undergo degradation by NMD (NMD+ transcripts and NMD+ variants) (filled black circle), whereas variants located downstream of the boundary PTC are predicted to escape NMD (NMD− transcripts and NMD− variants) (open white circle).
Using this algorithm, we constructed a publicly available webtool, NMDEscPredictor to predict whether a frameshifting indel will lead the transcript to escape or be degraded by NMD (Figure S5).
Development of a NMD Escape Intolerance Score
After classification of frameshifting indels and stopgain variants as potential NMD+ and NMD− transcripts, we designed an NMD escape intolerance score to rank each gene based on the probability of depletion of NMD− versus NMD+ variants in the control databases, ARIC and ExAC. NMD− versus NMD+ variant numbers were normalized by rare synonymous variant count in each region. Normalization based on the synonymous variant count was preferred to the coding sequence length because it has a higher performance compared to the coding sequence length normalization. For instance, the overlap between the top 5% depleted genes for NMD− variants in either control database and our control OMIM list of genes (Table S1) that cause disease via potential gain-of-function (n = 39) drops significantly using coding-sequence length normalization (28.2%) compared to synonymous variant count normalization (33.33%) (Figure S4). The probability of depletion (ratio of the observed/expected # of NMD− variants) per canonical transcript was computed separately for each mutational type (−1 frameshifting indels, +1 frameshifting indels and stopgain variants) given a Poisson distribution. The expected number of NMD− variants were defined as the total number of variants in NMD− and NMD+ regions multiplied by the ratio of the number of rare synonymous variants (MAF ≤ 0.01) in the NMD− region to the total number of rare synonymous variants. The expected number of NMD− variants per canonical transcript for each mutational type was calculated as follows (Figure S6):
The NMD escape intolerance score as the probability of enrichment per canonical transcript was precomputed for each mutational type, separately given the expected number of NMD− variants calculated for each category. To generate a combined NMD escape intolerance score for each canonical transcript, we used Fisher’s combined p value test. This test integrates all p values for a particular canonical transcript into a statistic, χc, according to Fisher’s method,30 where
pi is the p value obtained via Poisson distribution for the ith subgroup mutational type, and k the number of subgroup mutational types for a canonical transcript. The final p value for the entire canonical transcript is calculated as the probability of observing a value no less than χc, based on a chi-square distribution with 2k degrees of freedom.
Protein-Protein Interaction Analysis
To measure how connected a gene is to its neighbors in a physical protein-protein interaction network, we calculated a degree centrality measure, i.e., the number of edges that a node has in a network, for each gene using the physical interactions network data provided by GeneMania in a R/Bioconductor package named SpidermiR.31 Then, genes were annotated with their PFAM protein domains extracted and their structurally resolved interaction interfaces.
Statistical Analysis
In summary, we performed permutation tests (the number of permutations = 10,000) for allele-specific expression analyses. We performed Fisher’s exact test for the pLI comparisons and the Mann-Whitney U test for tissue-specific expression analysis and degree centrality measure analysis in protein-protein interaction networks using the physical interactions network data provided by GeneMania in a R/Bioconductor package named SpidermiR. We also performed binomial test to compare the ratios in proportions of genes that have their NMD− regions overlapping with Pfam protein domains and structurally resolved interaction interfaces.
Results
NMDEscPredictor Algorithm
For any given frameshifting indel, there could be a large distance, in base pairs, between the variant location and the genic map position of the next “out-of-frame” PTC. Therefore, an accurate annotation of the resulting PTC based on the 50-bp rule14, 15, 16 is crucial for frameshifting indels. To accomplish this, we designed an algorithm to classify frameshifting indels into two categories based on annotation of regions in the transcript: NMD competent (NMD+) and NMD incompetent (NMD−) regions (Figure 1). The algorithm workflow is described in Figure 1 and Material and Methods. In this algorithm, we retrieved all of the putative PTCs in the −1 and +1 frame of each Ensembl transcript and determined which PTCs can escape from NMD based on the 50-bp rule.14, 15, 16 For each transcript we flagged the last NMD-competent PTC. This “boundary PTC” establishes a transcript position, or boundary, between potential NMD+ and NMD− gene regions given the assumption that any frameshifting indel placed after the boundary PTC is predicted to utilize the next PTC, which is predicted to escape from NMD. In aggregate, assessing if any frameshifting indel will fall into the defined NMD+ or NMD− region in a transcript allows a rapid prediction of whether an aberrant transcript will likely be subject to nonsense mediated mRNA decay (NMD+) or escape from NMD (NMD−) and produce an aberrant prematurely terminated protein (Figure 1). We developed a web-based tool, NMDEscPredictor, to enable rapid query of NMD competency for any given frameshift allele (Figure S5).
Evaluation of NMDEscPredictor using Experimental Data
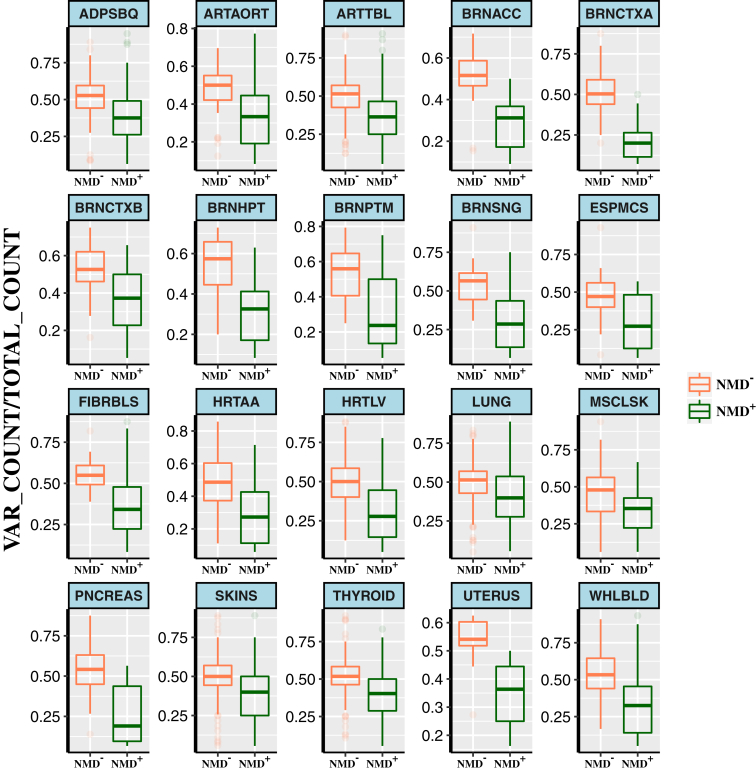
To evaluate the prediction accuracy of the algorithm using experimental data, we applied NMDEscPredictor for frameshifting indels to multi-tissue RNA-seq data and WES data from 1,634 samples in 173 individuals generated by GTEx.14, 32, 33 We used the allele-specific expression (ASE) values assessed across multiple tissues from the GTEx RNA-seq dataset that detected expression levels of two haplotypes of an individual. For variants that elicit NMD, NMD+, we anticipate that the average ratio of variant read (vR) count to total read (tR) count across all samples harboring those variants should be close to 0. Conversely, for variants that escape from NMD, NMD−, we expect the average ratio of vR count to tR count across all samples carrying those variants should be close to 0.5. Using NMDEscPredictor, we predicted NMD+ and NMD− mutant transcripts of 344 distinct frameshifting indels across 43 tissues available in the GTEx data. Our analysis revealed that NMD+ frameshifting indels present with a significantly lower ratio of vR count to tR count than NMD− frameshifting indels across 20 tissues out of 43 tissues examined in total (permutation test, Bonferroni-adjusted p value ≤ 0.05, Figure 2, Table S2), supporting our hypothesis. Our analysis also shows that the p values change significantly according to the number of data points available for a given tissue (Table S2 and Figure S7), so tissues with very few variants could not be used to evaluate our tool. The observed p values are also impacted by the fact that the variance in ASE values of NMD+ variants is higher than the variance in ASE values of NMD− variants.
Figure 2.
Evaluation of the NMDEscPredictor Algorithm Performance using GTEx Data
To test the algorithm performance in an independent dataset, we used GTEx multi-tissue RNA-seq and WES dataset to predict NMD incompetency of 344 distinct frameshift variants available in GTEx. The frameshift variants predicted to be NMD+ by NMDEscPredictor have a significantly lower ratio of variant read count (VAR_COUNT) to the total read count (TOTAL_COUNT) compared to those frameshift variants predicted to be NMD−. This is consistent with the hypothesis that NMD+ variants will lead to mRNA degradation. VAR_COUNT to TOTAL_COUNT values were extracted from allele-specific expression available in the GTEx dataset. Tissue abbreviations are denoted as follows: ADPSBQ, adipose, subcutaneous; ARTAORT, artery, aorta; ARTTBL, artery, tibial; BRNACC, brain, anterior cingulate cortex; BRNCTXA, brain, cortex; BRNCTXB, brain, frontal cortex; BRNHPT, brain, hypothalamus; BRNPTM, brain, putamen (basal ganglia); BRNSNG, brain, substantia nigra; ESPMCS, esophagus, mucosa; FIBRBLS, cells, transformed fibroblasts; HRTAA, heart, atrial appendage; HRTLV, heart, left ventricle; LUNG, lung; MSCLSK, muscle, skeletal; PNCREAS, pancreas; SKINS, skin, sun exposed (lower leg); WHLBLD, whole blood.
Of note, further examination of the NMD− transcripts uncovered the frameshifting indel variant chr6:26,465,566_CAA>C that is located in exon 4 of BTN2A1 transcript ENST0000042938 (7 coding exons in total). Although this variant maps nearly in the middle of the gene, the transcript was predicted by NMDEscPredictor to escape from NMD, on the basis of the location of the boundary PTC. Indeed, the average ratio of vR count to tR count for this variant was quantified as 0.478 across different tissues from GTEx RNA-seq data, which supports the computational prediction (Figure S8).
Up to 34% of Frameshifting Indels and Stopgain Variants Lead to NMD− Transcripts
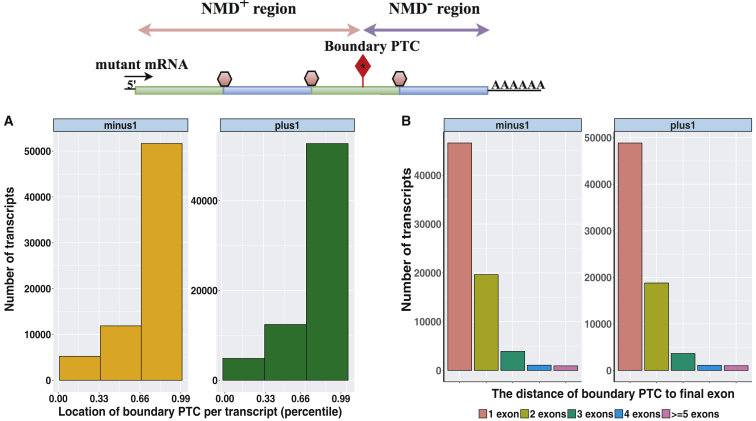
Prevailing thought assumes that NMD− gene regions form only a small portion of any transcript based on the 50-bp rule;14, 15, 16 therefore, most PTVs are considered likely to fall into larger-sized NMD+ regions (Figure 1) and thereby result in LoF alleles. However, the analysis of Ensembl GENCODE v.19 transcripts using NMDEscPredictor indicates that 36,880 out of 72,132 (51.1%) and 37,655 out of 76,506 transcripts (49.2%) with at least 2 exons and with a boundary PTC present have NMD− regions located downstream of 85% of normalized transcript length in the −1 and +1 frame, respectively. Remarkably, there are still 17,094 (23.6%) and 17,281 (22.5%) transcripts that have NMD− regions that encompass at least one third of the coding length in the −1 and +1 frame, respectively (Figure 3A). Similarly, further investigation of exonic locations of boundary PTCs revealed that 25,532 (35.3%) and 24,540 (32.07%) of transcripts have their boundary PTCs located at least one exon upstream of their penultimate exons, in either −1 or +1 frame, respectively (Figure 3B).
Figure 3.
Distribution of the Boundary PTCs in All Ensemble Multi-exon Transcripts
Above demarcates an individual transcript (horizontal rectangular structure) with exon junction and boundary PTC (lollipop with red filled diamond) shown as in Figure 1. Horizontal lines with double arrowheads demarcate NMD+ (pink) and NMD− (purple) regions. Each transcript is partitioned into two separate regions, i.e., NMD+ and NMD−, based on the location of the boundary PTCs.
(A) The bar plots show the relative distribution of boundary PTCs per transcript (percentile). About 51.1% and 49.2% of all Ensemble multi-exon transcripts have their boundary PTCs located within 85% of the normalized transcript length in the −1 and +1 frame, respectively. However, there are still a quarter of transcripts that have their NMD− regions encompassing more than a third of their coding sequence length.
(B) The bar plots demonstrate the distribution of boundary PTC locations with regards to the distance to the final exon of a given transcript. About 35.3% and 33.4% of all Ensemble multi-exon transcripts have their boundary PTCs located upstream of their penultimate exon in the −1 and +1 frame, respectively.
In summary, despite the observation that NMD− regions span 15% of the coding sequence length on average, there are still 22%–23% of transcripts that have NMD− regions encompassing more than a third of their coding sequence length.
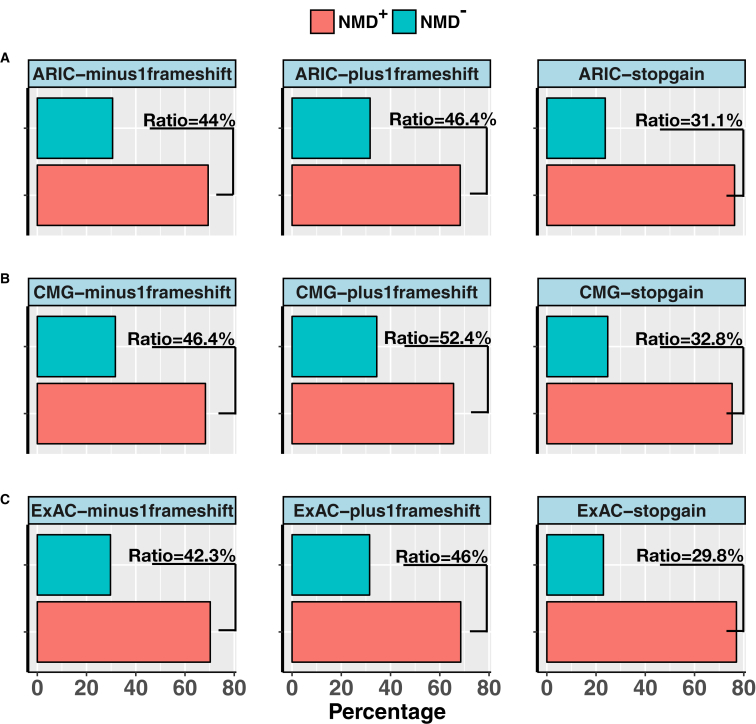
The algorithm for predicting whether any frameshifting indel would trigger NMD (NMD+) or escape from NMD (NMD−) was implemented on WES data from 6,109 samples in the Baylor-CMG database, 10,940 samples in the ARIC database, and 60,706 samples in ExAC database (Figure S9). As a control, the algorithm was applied to 32,273 and 84,507 frameshifting indels retrieved from the ARIC and ExAC control databases, respectively. We then removed variants that could not be annotated to any canonical transcript in Ensembl v.19 as well as variants mapped to transcripts without a predicted PTC, without a boundary PTC, or mapped to single-exon canonical transcripts. The algorithm output led to the computational prediction of frameshifting indels obtained from the ARIC database that were classified as: 69.41% and 68.30% as potential NMD+, 30.59 and 31.70% as NMD− in −1 and +1 frames, respectively (Figure 4A). Concordantly, the frameshifting indels retrieved from ExAC database were as follows: 70.26% and 68.51% as potential NMD+ and 29.74% and 31.49% as potential NMD−, in −1 and +1 frames, respectively (Figure 4B). As a database including personal genomes from subjects presenting with diverse Mendelian phenotypes (the “disease cohort”), the algorithm was applied to 25,351 frameshifting indels available in Baylor-CMG data. We identified 68.28% and 65.59% as potential NMD+ and 31.72% and 34.41% as potential NMD− (Figure 4C) in −1 and +1 frames, respectively. In summary, these results suggest that the fraction of frameshifting indels that are predicted to be NMD− versus NMD+ in the CMG database, highly enriched for genome variant data of individuals with diverse “disease phenotypes” (i.e., a disease population), is significantly higher than that observed in the ARIC and ExAC control databases in −1 frame (p values 0.043 and 0.0001) and +1 frame (p values 4.47e−5 and 3.48e−5). Of note, the fractions of frameshifting indels that are predicted to be NMD− versus NMD+ in the ARIC and ExAC databases are not significantly different from each other (p values 0.07 and 0.69) in −1 and + 1 frames, respectively (Fisher’s exact test).
Figure 4.
Classification of Protein-Truncating Variants in ARIC, Baylor-CMG, and ExAC Databases
Bar charts display the percentage of −1 frameshift, +1 frameshift, and stopgain variants as predicted to be NMD− and NMD+ according to NMDEscPredictor in the (A) ARIC database, (B) Baylor-CMG database, and (C) ExAC database.
Likewise, when we grouped stopgain variants into potential NMD+ and NMD− categories on the basis of the 50-bp rule14, 15, 16 in each of these three different datasets, the algorithm outputs were highly comparable to each other; i.e., 76.26% and 75.26% of stopgain variants were classified as potential NMD+, whereas 23.74% and 24.74% were predicted to escape from NMD (NMD−) in the ARIC and Baylor-CMG databases, respectively. Importantly, both datasets ARIC (population control) and Baylor-CMG were generated using the same WES experimental platform (Nimblegen capture/Illumina sequencing)34 and computational algorithm (Mercury pipeline) that generated binary alignment/map (BAM) and variant call files (VCF).26, 27 Similarly, in the ExAC database, we predicted 77.01% of stopgain variants as potential NMD triggering (NMD+) and 22.99% of stopgain variants as escaping from NMD (NMD−) (Figure 4). The fraction of stopgain variants that are predicted to be NMD− versus NMD+ in the Baylor-CMG database is significantly higher than the ARIC and ExAC databases (p values 0.015 and 3.88e−6; Fisher’s exact test), respectively. On the other hand, the fraction of stopgain variants that are predicted to be NMD− versus NMD+ in the ARIC database is significantly higher than the ExAC database with p value 0.01 (Fisher’s exact test).
Genes Depleted for NMD− Protein Truncating Variant (PTVesc) in Control Databases
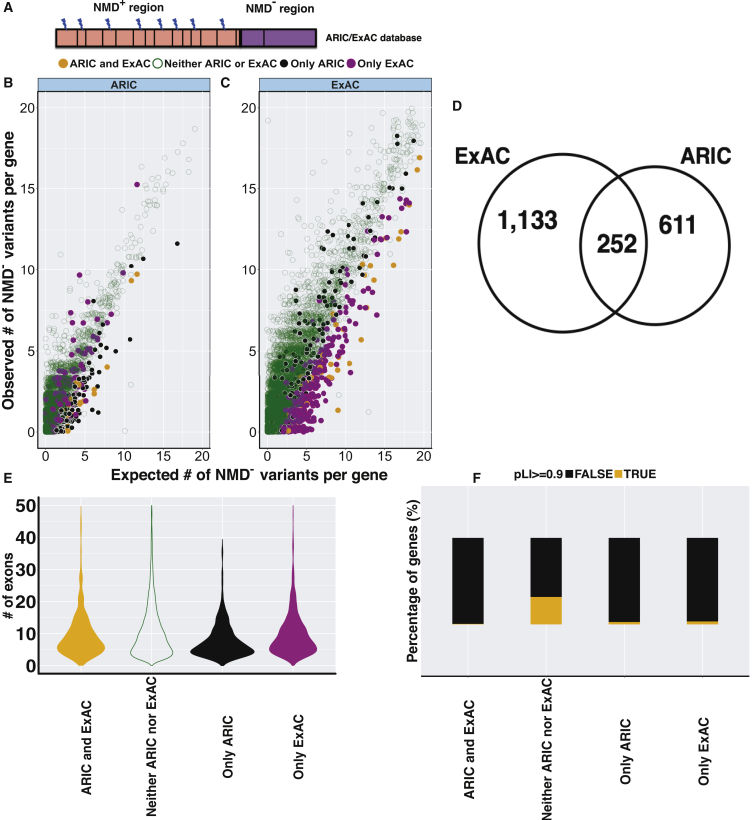
For a given gene under negative selection, the observed number of PTVs is expected to be less than the expected number of PTVs under a neutral model. A number of studies have developed LoF intolerance scores for testing a gene’s tolerance to haploinsufficiency under a dominant disease model.20, 22, 35, 36 pLI score based on the comparison of observed and expected number of LoF variants, i.e., nonsense and canonical splice site SNVs, per gene is defined as the probability of a gene’s falling into the haploinsufficiency gene category. Therefore, genes with high pLI scores, i.e., (pLI ≥ 0.9), are classified as LoF intolerant.20 It is important to note that the LoF-intolerant gene classification can be biased toward identifying genes that can cause human disease due to haploinsufficiency, mainly amorphic (i.e., null alleles) or hypomorphic mutations.37 These statistical inference methods are perhaps not applicable to the discovery of genes such as DVL1 that may cause disease by the presence of antimorphic or neomorphic mutations; i.e., GoF versus LoF alleles.37 To find variants potentially leading to antimorphic or neomorphic alleles, we compared the expected number of PTVesc to the observed number of predicted PTVesc in both ARIC and ExAC databases (Figure 5A). These data were used to develop a NMD escape intolerance score; a publicly available online tool. This analysis showed that 863 and 1,385 genes are significantly depleted for PTVesc in the ARIC or ExAC databases, respectively (i.e., ranked in the top 5%) (Figures S10, S11, 5B, and 5C). Of those, 252 genes display depletion for truncating variants in NMD− region in both ARIC and ExAC databases (Figure 5D, Tables S3 and S4). In order to quantify how often those levels of overlap might occur by chance, we performed a permutation test which shows that the observed number of overlapping genes (252) stands at 14.6 standard deviations away from the average number of overlapping genes in 10,000 permutations (115) (Figure S12). This shows that the overlap between ARIC and ExAC datasets is higher than expected by chance. This percentage of overlap (252/863 = 29.2%) is also aligned with the 29% overlap between ARIC and ExAC frameshift variants and the 46% overlap between ARIC and ExAC stopgain variants (Figures S13 and S14). Among these genes, the top five genes that show the most depletion in ARIC or ExAC databases are shown in Table 1.
Figure 5.
Features of the Top 5% Depleted Genes for NMD− Variants in Control Databases
(A) General structure of a transcript displaying NMD+ (pink) and NMD− (purple) regions; lightning symbols represent variant location. Vertical black lines represent potential PTCs.
(B and C) To identify genes that were depleted for truncating variants in NMD− region compared to NMD+ region in control databases, we compared the expected to the observed number of NMD− variants per gene (please see Material and Methods section) variants in the (B) ARIC database and (C) ExAC database. Genes depleted for NMD− variants in both (ARIC and ExAC) were shown as orange filled dots; genes depleted for NMD− variants only in ARIC database were shown as black filled dots; genes depleted for NMD− variants only in ExAC database were shown as purple filled dots and genes not depleted for NMD− variants in either control database were shown as dark green empty circles.
(D) The Venn diagram shows 1,385 and 863 genes as the top 5% depleted genes for variants in NMD− region in the ExAC and ARIC databases, respectively; 252 genes were common to both.
(E) The violin plots show that genes depleted for NMD− variants in both databases (ARIC and ExAC; filled yellow violin) do not significantly differ in number of exons from genes not depleted for NMD− variants in either control database (neither ARIC or ExAC; open green violin).
(F) Stacked bar plots indicate that genes depleted for NMD− variants in both (n = 252) or either database (n = 611; n = 1,133) have significantly higher proportion of genes with pLI < 0.9 compared to genes not depleted for NMD− variants in either control database (n = 14,425) with p values = 1.49e−25, 1.09e−46, and 2e−78, respectively. Orange bar shows the percentage of genes with pLI ≥ 0.9 and black bar shows the percentage of genes with pLI < 0.9.
Table 1.
Top Five NMD− Intolerant Genes, −1, +1 Frameshifting Indels and Stopgain Categories, in ExAC/ARIC Databases (DB)
| DB | Cat. | Rank/Total Transcripts | Gene | pLI | Chr | # of Exonsa | # of NMD+PTVs | # of NMD−PTVs | MIM # (Gene/Phenotype) |
|---|---|---|---|---|---|---|---|---|---|
| ExAC | −1 | 1/9,319 | TRIM22 | 3.15e−12 | 11 | 7 | 6 | 0 | NA |
| ExAC | −1 | 2/9,319 | TGIF1 | 0.09 | 18 | 3 | 5 | 0 | 602630/142946 |
| ExAC | −1 | 3/9,319 | MRPL15 | 1.33e−5 | 8 | 5 | 6 | 0 | NA |
| ExAC | −1 | 4/9,319 | NPHS2 | 9e−3 | 1 | 8 | 5 | 0 | 604766/600995 |
| ExAC | −1 | 5/9,319 | NXPE4 | 2.74e−8 | 11 | 5 | 8 | 0 | NA |
| ExAC | +1 | 1/8,814 | CLCC1 | 0.026 | 1 | 10 | 10 | 0 | NA |
| ExAC | +1 | 2/8,814 | CLN6 | 0.381 | 15 | 7 | 4 | 0 | 606725/601780 |
| ExAC | +1 | 3/8,814 | CPA4 | 3.54e−7 | 7 | 11 | 6 | 0 | NA |
| ExAC | +1 | 4/8,814 | SLFN12 | 5.03e−10 | 17 | 3 | 10 | 1 | NA |
| ExAC | +1 | 5/8,814 | SLX4IP | 2.33e−5 | 20 | 7 | 9 | 4 | NA |
| ExAC | sg | 1/12,086 | CNPPD1 | 1.63e−5 | 2 | 8 | 8 | 0 | NA |
| ExAC | sg | 2/12,086 | POP1 | 2.42e−8 | 8 | 15 | 16 | 0 | 602486/617396 |
| ExAC | sg | 3/12,086 | SLC3A1 | 7.54e−16 | 2 | 10 | 16 | 0 | 104614/220100 |
| ExAC | sg | 4/12,086 | LY6G6F | 2.55e−6 | 6 | 6 | 5 | 0 | NA |
| ExAC | sg | 5/12,086 | PUS3 | 1.08e−5 | 11 | 3 | 9 | 0 | 616283/617051 |
| ARIC | −1 | 1/5,769 | IFNAR2 | 0.005 | 21 | 8 | 4 | 0 | 602376/616669 |
| ARIC | −1 | 2/5,769 | NBPF20 | 0.0004 | 1 | 110 | 8 | 0 | NA |
| ARIC | −1 | 3/5,769 | PPIP5K1 | 0.102 | 15 | 29 | 3 | 1 | NA |
| ARIC | −1 | 4/5,769 | TM7SF3 | 0.089 | 12 | 12 | 5 | 0 | NA |
| ARIC | −1 | 5/5,769 | ZNF669 | 0.06 | 1 | 4 | 3 | 0 | NA |
| ARIC | +1 | 1/5,264 | TET2 | 7.05e−26 | 4 | 9 | 13 | 0 | 612839/614286 |
| ARIC | +1 | 2/5,264 | PPIP5K1 | 0.102 | 15 | 29 | 3 | 0 | NA |
| ARIC | +1 | 3/5,264 | CSF3R | 1.41e−26 | 1 | 15 | 5 | 0 | 138971/617014 |
| ARIC | +1 | 4/5,264 | UBXN11 | 1.2e−10 | 1 | 14 | 5 | 0 | NA |
| ARIC | +1 | 5/5,264 | ATP5J | 0.1 | 21 | 4 | 2 | 0 | NA |
| ARIC | sg | 1/7,931 | PRAMEF1 | 0.098 | 1 | 3 | 6 | 0 | NA |
| ARIC | sg | 2/7,931 | SSC5D | 0.35 | 19 | 14 | 7 | 0 | NA |
| ARIC | sg | 3/7,931 | CCDC173 | 1.084e−15 | 2 | 9 | 6 | 0 | NA |
| ARIC | sg | 4/7,931 | NUPL2 | 3.67e−5 | 7 | 7 | 3 | 0 | NA |
| ARIC | sg | 5/7,931 | USP6NL | 0.102 | 10 | 14 | 3 | 0 | NA |
Abbreviations: DB, database; Cat., category; PTV, premature truncating variant (−1, +1 frameshifting indels, sg:stopgain)
Number of exons based on Ensembl v.19
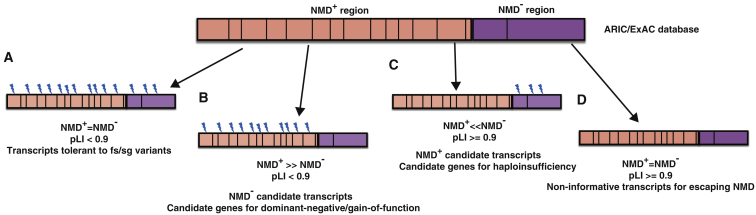
We found that this set of genes depleted in both databases (n = 252) does not differ in number of exons relative to genes not depleted in either control database (n = 14,425) indicating that depletion of variants in NMD− regions, are not due to a size bias (Mann-Whitney U test p value = 0.35) (Figure 5E). Further examination of the 252 genes revealed that they have a significantly higher proportion of genes with their pLI < 0.9 (proportion = 0.996) compared to genes that are not depleted for truncating variants in NMD− region in either ARIC and ExAC databases (n = 14,425 genes, proportion = 0.781, Fisher’s exact test p value = 1.49e−25). Likewise, the top 5% of genes depleted for truncating variants in NMD− region only in the ARIC database (n = 611 genes) or only the ExAC database (n = 1,133 genes) have significantly higher portions of genes with their pLI < 0.9 (proportions = 0.98 and 0.976) compared to genes that are not depleted for truncating variants in NMD− region (n = 14,425, proportion = 0.781) in both ARIC and ExAC databases with p values 1.09e−46 and 2e−78 (Fisher’s exact test), respectively (Figure 5F). Therefore, this set of 1,996 (252 + 611 + 1,133) genes depleted for PTVesc in either control database, are candidates for causing disease through escaping from NMD and termed “NMD− candidate genes” (Figure 6).
Figure 6.
Classification of Transcripts Based on Truncating Variant Density in NMD− versus NMD+ Region in Control Databases Allows Development of NMD Escape Intolerance Score
Transcripts were classified into four groups based on truncating variant density in the NMD− versus NMD+ region in ARIC/ExAC control databases. Vertical black lines represent potential PTCs. Lightning symbols represent variant location.
(A) Transcripts tolerant to frameshift(fs)/stopgain(sg) have truncating variant densities in NMD− versus NMD+ regions that do not differ significantly from each other. Those transcripts mostly presented with low pLI scores.
(B) NMD− candidate transcripts: transcripts in this category present with a lower NMD− region variant density compared to NMD+ region and often display low pLI scores < 0.9. The genes corresponding to those transcripts are candidates for causing disease through dominant-negative or GoF effects.
(C) NMD+ candidate transcripts in this category present with a lower NMD+ region truncating variant density compared to NMD− region and may present with high pLI scores. The genes corresponding to those transcripts are candidates for causing disease through haploinsufficiency.
(D) Non-informative transcripts: this category of transcripts includes transcripts currently with no truncating variants in the control databases, therefore was considered non-informative.
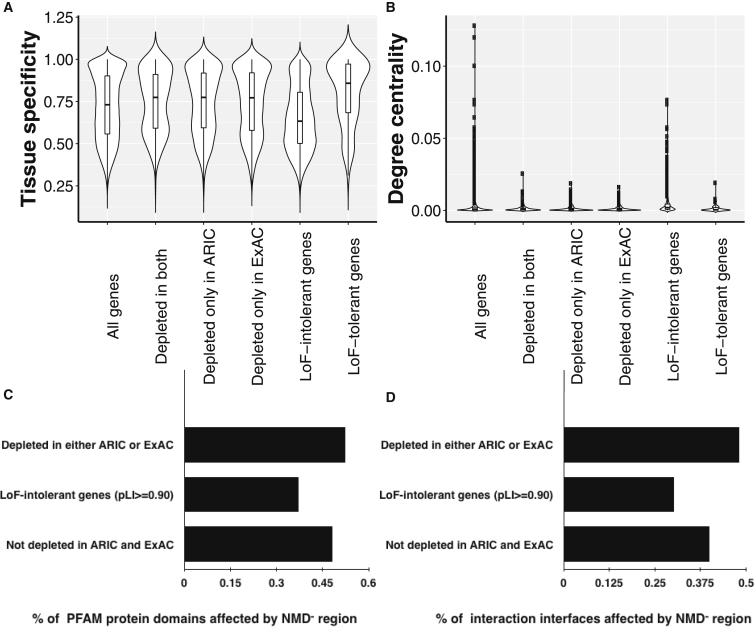
We examined further characteristics of this discrepant pLI gene set. The total set of 1,996 NMD− candidate genes that are predicted by NMDEscPredictor are more likely to show tissue-specific expression (Mann-Whitney U test p value = 5.89e−8) and their gene products have fewer protein-protein interactions than the average number of protein-protein interactions per gene in the genome (Mann-Whitney U test p value = 1.67e−12). Furthermore, disruptions in NMD− regions of those genes are more likely to be affecting their annotated protein domains (binomial test p value = 0.0003) of their encoded proteins than the genome average (Figure 7).
Figure 7.
Tissue Specificity and Protein Characterization of 1,996 Genes (Top 5%) Depleted for Truncating Variants in NMD− Region in the Control Databases
For all the genes analyzed in the genome (n = 16,411), genes depleted for truncating variants in NMD− region in both databases (n = 252), in only ARIC database (n = 611), in only ExAC database (n = 1,133), LoF-intolerant genes (pLI ≥ 0.9) (n = 2,959) and LoF-tolerant genes (n = 98), we calculated the following.
(A) Tissue specificity values using tau measure.38 The tau measure takes values between 0 and 1; when a gene’s tau measure is closer to 1, it is annotated as more tissue specific. The average tau measure of depleted genes for truncating variants in NMD− region in either control database (N = 1,996) is significantly higher (0.744) compared to the genome average (0.719, Mann-Whitney U test p value = 5.89e−8) and compared to LoF-intolerant genes (0.651, Mann-Whitney U test p value = 7.45e−62).
(B) To measure how connected a gene product is to its neighbors in a physical protein-protein interaction network, we calculated a degree centrality measure, i.e., the number of edges that a node has in a network, for each gene using the physical interactions network data provided by GeneMania in a R/Bioconductor package named SpidermiR.31 This analysis revealed that the genes predicted to be intolerant to truncating variants in NMD− region in either control database by NMDEscPredictor (N = 1,996) are significantly less connected to their neighbors in the physical protein-protein interaction data compared to the genome average (Mann-Whitney U test p value = 1.67e−12).
(C and D) Those genes were annotated with their PFAM protein domains and their structurally resolved interaction interfaces. Transcripts depleted for NMD− variants show a higher fraction of their annotated PFAM protein domains (0.525) overlapping with their corresponding NMD− regions compared to the average of all transcripts (0.483) with binomial test p value = 0.0003. In a similar way, these transcripts present a higher fraction of their structurally resolved interaction interfaces overlapping to the NMD− regions (0.46) compared to the average of all transcripts (0.407) with binomial test p value = 0.045.
We further examined this set of 1,996 genes that include potential candidates for causing disease through escape from NMD in terms of literature evidence for association with disease. As anticipated, and acting as a positive control dataset, this group significantly shares 13 known genes out of 39 OMIM genes (33.3%, Fisher’s exact test p value 0.024) associated with a human disease phenotype whereby it has been specifically shown that NMD-escaping PTC variants, NMD− variants, act via a dominant-negative or GoF mechanism including ALX4 (MIM: 605420) causing parietal foramina 2 (MIM: 609597), DVL1, F10 (MIM: 613872) causing factor X deficiency (MIM: 227600), FAM83H (MIM: 611927) causing amelogenesis imperfecta, type IIIA (MIM: 130900), FGA (MIM: 134820) causing afibrinogenemia, congenital (MIM: 202400), GHR (MIM: 600946) causing growth hormone insensitivity, partial (MIM: 604271), HBB (MIM: 141900) causing thalassemias, beta (MIM: 613985), IFNGR1 (MIM: 107470) causing immunodeficiency 27B, mycobacteriosis (MIM: 615978), KAT6B (MIM: 605880) causing SBBYSS syndrome (MIM: 603736), KIDINS220 (MIM: 615759) causing spastic paraplegia, intellectual disability, nystagmus, and obesity (MIM: 617296), NOTCH2 (MIM: 600275) causing Hajdu-Cheney syndrome (MIM: 102500), and RHO (MIM: 180380) causing retinitis pigmentosa 4 (MIM: 613731) (Tables 2 and S1).
Table 2.
Top 5% NMD− Intolerant Genes in Control Databases Reported to Cause Disease via Dominant-Negative or GoF
| DB | Cat. | Rank/TotalTranscripts | Gene | pLI | Chr | # of Exonsa | # of NMD+PTVs | # of NMD−PTVs | MIM # (Gene/Phenotype) |
|---|---|---|---|---|---|---|---|---|---|
| ARIC | +1, combined | 197/5,264, 185/6,064 | ALX4 | 0.23 | 11 | 4 | 1,3 | 0,0 | 605420/609597 |
| ARIC | +1 | 185/5,264 | DVL1 | 0.157 | 1 | 15 | 3 | 0 | 601365/616331 |
| ExAC | +1, combined | 322/8,814, 256/10,542 | F10 | 0.16 | 13 | 8 | 2,4 | 0,0 | 613872/227600 |
| ARIC | combined | 252/6,064 | FAM83H | 0.884 | 8 | 4 | 2 | 0 | 611927/130900 |
| ARIC | −1, sg, combined | 53/5,769, 396/7,931, 175/6,064 | FGA | 6.34e−10 | 4 | 6 | 3,2,6 | 0,0,0 | 134820/202400 |
| ExAC | sg | 253/12,086 | GHR | 2e−4 | 5 | 9 | 4 | 1 | 600946/604271 |
| ARIC | sg | 303/7,931 | GHR | 2e−4 | 5 | 9 | 3 | 1 | 600946/604271 |
| ExAC | −1, +1, combined | 32/9,319, 244/8,814, 112/10,542 | HBB | 1.02e−8 | 11 | 3 | 4,3,10 | 0,0,1 | 141900/613985 |
| ExAC | −1 | 394/9,319 | IFNGR1 | 0.32 | 6 | 7 | 2 | 0 | 107470/615978 |
| ExAC | combined | 524/10,542 | KAT6B | 0.99 | 10 | 16 | 3 | 0 | 605880/603736 |
| ExAC | +1 | 372/8,814 | KIDINS220 | 0.029 | 2 | 29 | 3 | 0 | 615759/617296 |
| ARIC | +1 | 228/5,264 | NOTCH2 | 1 | 1 | 34 | 3 | 0 | 600275/102500 |
| ExAC | +1 | 192/8,814 | RHO | 0.0005 | 3 | 5 | 2 | 0 | 180380/613731 |
Abbreviations: DB, database; Cat., category; PTV, premature truncating variant (−1, +1 frameshifting indels, sg:stopgain); combined, combined rank for −1, +1 frameshifting indels and sg: stopgain categories; subj., subject
Number of exons based on Ensembl v.19
Candidate Genes Parsed by Enrichment of PTVesc
With the aim of accelerating gene discovery by the identification of candidate genes conveying Mendelian disease traits via a potential GoF mechanism, we scanned a WES database from a disease cohort, Baylor-CMG, for PTVesc in 1,996 NMD− candidate genes. A subset of 387 genes have more than one truncating variant predicted to escape from NMD in unrelated individuals with Mendelian disease. We further investigated for evidence of two distinct potential PTVesc in the same gene in probands with a similar clinical phenotype captured in PhenoDB.39, 40 To accomplish this, we assessed the phenotypic similarity between any two affected individuals with PTVesc in the same candidate gene by generating a pairwise phenotypic similarity score among HPO term sets recorded for those probands in the PhenoDB database.39, 40, 41
This analysis revealed the top-ranking gene as RE1-silencing transcription factor, REST, with a perfect phenotypic similarity score of 1. REST was found to have two distinct heterozygous final-exon-truncating variants in the Baylor-CMG database in two families with HGF (HPO term HP:0000169), the most common genetic form of gingival fibromatosis. This observation was arrived at by two independent approaches: family-based personal genome analysis and the predicted NMD incompetency-parsing algorithm described herein. Ultimately, identification of this candidate gene significantly depleted for PTVesc, REST, and GeneMatcher42 identification of additional cases, delineated phenotype-genotype correlations in 11 individuals from 3 unrelated families who presented with autosomal-dominant HGF.24
In addition to REST, our analysis uncovered two distinct heterozygous potential NMD− frameshift variants in the Ferm and Pdz Domains-Containing 1 (FRMPD1) (MIM: 616919) in two individuals (BAB7358 and BAB7598) who presented with immunodeficiency. FRMPD1 encodes a protein that is involved in the regulation of subcellular localization of the activator of G-protein signaling 3 (AGS3) that displays an immune function. This was demonstrated by studies that showed that AGS3-null B and T lymphocytes and bone marrow-derived dendritic cells presented with defects in chemokine signaling.43, 44 Besides variants in FRMPD1, we identified a homozygous potential NMD− final-exon frameshift variant in a female individual (BAB8983-PED3272) who presented with hypogammaglobulinemia and natural killer (NK) cell deficiency in the ZBTB24 gene (MIM: 614064) that was previously associated with immunodeficiency-centromeric instability-facial anomalies syndrome-2 (MIM: 614069).
Since the NMD escape intolerance score was designed based on the probability of depletion of NMD− versus NMD+ variants in control databases, it is not applicable for genes that are devoid of truncating variants in control databases. In order to overcome this limitation, we scanned the Baylor-CMG database for genes with potential PTVesc. This analysis revealed 3,035 genes that have more than one potential PTVesc in unrelated individuals with Mendelian disease; from those, there are 129 and 183 genes from ExAC and ARIC databases, respectively, that do not have any PTVs. One of those genes is the proteasome maturation protein (POMP [MIM: 613386]), for which there are two distinct frameshift variants in its penultimate exon in unrelated individuals, both of whom present with immunodeficiency. Segregation analysis and further characterization of the POMP function uncovered this gene as potentially causative of immunodeficiency when truncated.45
Another candidate gene, dual-specificity phosphatase 4 (DUSP4 [MIM: 602747]), was observed to have a homozygous potential NMD− frameshift variant in an individual (BH8977-1) with microcephaly, agenesis of the corpus callosum, lissencephaly, and epilepsy in the Baylor-CMG disease cohort; no truncating variants were observed in this gene in either control database. The DUSP4 protein has been shown to play a crucial role in neuronal differentiation in mouse embryonic stem cells.46
In addition to POMP and DUSP4, the PTVesc screening in the Baylor-CMG revealed Complexin 1 (CPLX1 [MIM: 605032]) with a homozygous potential NMD− stopgain variant in two affected siblings, who presented with epilepsy in the Baylor-CMG disease cohort. In a recent study, this gene was shown to cause myoclonic epilepsy and ID in two families in an autosomal-recessive inheritance pattern47 (Table 3).
Table 3.
NMD− Intolerant Genes Identified in the Baylor-CMG Database
| DB | Cat. | Rank/Total Transcripts | # of Subj. | Gene | pLI | Chr | # of Exonsa | # of NMD+Variants | # of NMD−Variants | MIM # (Phenotype) |
|---|---|---|---|---|---|---|---|---|---|---|
| ExAC/ARIC | NA | NA | 2 | CPLX1 | 0.795 | 4 | 3 | NA | NA | NA |
| ExAC/ARIC | NA | NA | 3 | DUSP4 | 0.54 | 8 | 4 | NA | NA | NA |
| ExAC | −1, sg, combined | 184/9,319, 326/12,086, 320/10,542 | 2 | FRMPD1 | 3.53e-10 | 9 | 15 | 8,13,22 | 5,8,17 | NA |
| ARIC | −1, sg, combined | 50/5,769, 290/7,931, 126/6,064 | 2 | FRMPD1 | 3.53e-10 | 9 | 15 | 4,3,8 | 1,1,2 | NA |
| ExAC/ARIC | NA | NA | 2 | POMP | 0.861 | 13 | 6 | NA | NA | 601952 |
| ARIC | +1 | 18/5,264 | 2 | REST | 0.972 | 4 | 3 | 2 | 0 | 617626 |
| ExAC | +1 | 316/8,814 | 3 | ZBTB24 | 0.0002 | 6 | 6 | 3 | 0 | 614069 |
PTVs on those genes are predicted to be pathogenic due to GoF. Abbreviations: DB, database; Cat., category; PTV, premature truncating variant (−1, +1 frameshifting indels, s: sg:stopgain); combined, combined rank for −1, +1 frameshifting indels and sg:stopgain categories; subj., subject
Number of exons based on Ensembl v.19
Discussion
Classic Mendelian medical genetics implies that a specific gene is associated with a disease trait that displays a consistent pattern of inheritance, and indeed this holds true for most recognizable gene-disease associations reported to date. However, it is becoming increasingly recognized that different variants in the same gene may cause an identical or similar disease as either a monoallelic autosomal-dominant (AD) trait or biallelic autosomal-recessive (AR) trait.48, 49 Some variants may lead to LoF (null or hypomorphic) alleles while others in the same gene may result in GoF (antimorphic or neomorphic) alleles. PTCs that trigger NMD often give rise to disease by reducing the expression of the transcript below necessary levels, whereas PTCs that escape from NMD can produce defective protein products that have detrimental effects in various ways; e.g., antimorphic or neomorphic mutant alleles.
Importantly, NMD is intimately connected to human health by giving rise to the presentation of distinct traits, making it an important consideration for variant interpretation (Figure 6). For example, in a study that investigated the position-dependent effects of PTVs in SOX10 (MIM: 602229) and MPZ (MIM: 159440), the NMD− variants led to a more severe neurological phenotype than NMD+ variants through the dominant-negative activity of truncated proteins.50, 51, 52 Moreover, NMD+ and NMD− variants can lead to distinct modes of inheritance for disease traits, for instance, ROR2 (MIM: 602337) bi-allelic NMD+ variants cause autosomal-recessive Robinow syndrome (RRS [MIM: 268310]) while heterozygous NMD− variants give rise to autosomal-dominant brachydactyly, type B1 (BDB1 [MIM: 113000]).53 Considering potential effects of NMD is critical for variant interpretation, delineation of genotype-phenotype correlations, and potential explanations for phenotypic differences caused by PTVs in the same gene.51, 53 In summary, not all PTCs result in LoF alleles and thus this imposes important limitations to the statistical haploinsufficiency inference scores and highlights the need of complementary scores, such as the NMD escape intolerance score presented herein, which also consider potential dominant-negative and GoF alleles.20, 54 Furthermore, the catalogs of common and rare human genetic variation enable an estimated number of PTVs per an individual genome. The 1000GP phase 3 analysis outlines that 149–182 sites are presented with PTVs per a typical genome.55 On the other hand, an individual genome is reported to carry an average of 85 heterozygous and 35 homozygous PTVs in the ExAC database, of which 18 and 0.19 are rare (<1% allele frequency), respectively.20 In general, a strong negative selection is anticipated to act against PTVs, thereby reducing the number of the sites harboring PTVs relative to other sites carrying neutral mutations in an individual genome (Figure 6).
Ranking the genes according to the NMD escape intolerance score led to the identification of 252 genes that are significantly depleted for truncating variants in the NMD− region in both control databases; a gene set that includes genes in which experimental data have shown that disease-associated mutant alleles are NMD− variants that act through mechanisms other than LoF. Importantly, examination of their pLI scores indicate that these genes are highly tolerant to PTVs, but the score we propose here indicates that such PTV tolerance is localized to the NMD+ gene region. Further analysis revealed that a higher fraction of the protein domains and interaction interfaces in those gene products are translated by those NMD− regions compared to all other gene products in the genome. Taken together, these observations support that this set of NMD− intolerant genes may convey disease phenotypes by mutational mechanisms other than haploinsufficiency and any disruption in their encoded C terminus translated by the NMD− regions are more likely to result in an interruption in their interactions with other proteins.
Excitingly, identification of human gene defects is increasingly capable of providing biological insight beyond disease relevance. Yet PTVs, the most abundant pathogenic variants, are impulsively classified as LoF mutations from the outset of their discovery without characterization of the biological effects, including NMD. Despite the fact that there are a number of recent reports highlighting the scale and implication of inter-individual and tissue-specific variation,14, 32, 33 NMD is still highly under-recognized in genomic analyses and certainly has a large impact on the presence and presentation of a wide variety of human disease. In this context, ranking of genes in terms of their depletion of NMD− relative to NMD+ variants in control databases provided a list of genes that could underlie Mendelian phenotypes due to escape from NMD. Our analysis is thus very useful for prioritization of candidate genes for NMD-specific disease. Indeed, integration of our analysis method with systematic assessment of phenotypic similarity between probands presenting with PTVesc in the same candidate gene in the Baylor-CMG database proved to be a powerful tool for identification of candidate genes that could give rise to Mendelian phenotypes due to escape from NMD.24
Manifesting a phenotype only when a variant escapes from NMD has ramifications beyond genotype-phenotype correlations. It is still widely believed that meaningful biological insight comes from the study of mouse models. This is particularly evident with the International Knockout Mouse Consortium’s ambitious goal to generate homozygous null alleles in virtually every protein-coding gene of the mouse genome,56 which does not recapitulate the phenotypes of escape-only, i.e., NMD−, genes like DVL1, PPM1D (MIM: 605100) causing intellectual developmental disorder with gastrointestinal difficulties and high pain threshold (MIM: 617450),57 and ZIC1 (MIM: 600470) conveying the phenotype of craniosynostosis 6 (MIM: 616602).58 As genomic sequencing as a diagnostic tool continues to expand with the enabling of precision medicine, the human population is truly analogous to a “Petri dish” becoming the most powerful genetic screen for gene discovery and for establishment of an allelic series with genotype/phenotype correlations and containing variant alleles with GoF as well as LoF mutations in the series. To fully utilize this screen, amid the scientific euphoria of gene-discovery researchers and clinicians must not antiquatedly deem all pathogenic mutation as “LoF,” rather a specific perturbed function, which includes altered expression. This screen has the power to identify a nearly unlimited number of discoveries, not only gene discovery but detailed biology of gene function.
Acknowledgments
We thank all individuals and their families and the referring physicians who submitted samples for testing. No additional compensation was received for these contributions. This work was funded in part by the US National Human Genome Research Institute (NHGRI)/ National Heart Lung and Blood Institute (NHLBI) grant number UM1HG006542 to the Baylor-Hopkins Center for Mendelian Genomics (BHCMG), the National Institute of Neurological Disorders and Stroke (NINDS) R01 NS058529 and R35 NS105078 (J.R.L.), and the National Human Genome Research Institute U54 HG003273 (R.A.G.).
Declaration of Interests
Baylor College of Medicine (BCM) and Miraca Holdings have formed a joint venture with shared ownership and governance of the Baylor Genetics (BG), which performs clinical microarray analysis and clinical exome sequencing. C.A.S. is an employee of BCM and derives support through a professional services agreement with the BG. J.R.L. serves on the Scientific Advisory Board of the BG. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc., and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered in the Baylor Genetics Laboratories. The other authors declare no competing financial interests.
Published: July 19, 2018
Footnotes
Supplemental Data include 14 figures and 4 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.06.009.
Contributor Information
James R. Lupski, Email: jlupski@bcm.edu.
Claudia M.B. Carvalho, Email: cfonseca@bcm.edu.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
Atherosclerosis Risk in Communities Study (ARIC) Database, http://www2.cscc.unc.edu/aric/
Baylor Genetics Laboratory, http://bmgl.com/
ExAC Browser, http://exac.broadinstitute.org/
Gencode v.17, https://www.gencodegenes.org
Genemania, https://genemania.org
GTEx Portal, https://www.gtexportal.org/home/
NMDescPredictor, https://nmdprediction.shinyapps.io/nmdescpredictor/
NMD Escape Intolerance Score, https://nmdprediction.shinyapps.io/nmdescintolerancescore/
OMIM, http://www.omim.org/
Pfam, http://pfam.xfam.org
Three-dimensional protein networks, http://www.yulab.org/DiseaseInt
Supplemental Data
References
- 1.Kervestin S., Jacobson A. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurosaki T., Maquat L.E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 2016;129:461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 4.Le Hir H., Izaurralde E., Maquat L.E., Moore M.J. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh G., Kucukural A., Cenik C., Leszyk J.D., Shaffer S.A., Weng Z., Moore M.J. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saulière J., Murigneux V., Wang Z., Marquenet E., Barbosa I., Le Tonquèze O., Audic Y., Paillard L., Roest Crollius H., Le Hir H. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat. Struct. Mol. Biol. 2012;19:1124–1131. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- 7.Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 8.Ishigaki Y., Li X., Serin G., Maquat L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 9.Le Hir H., Gatfield D., Izaurralde E., Moore M.J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim V.N., Kataoka N., Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 11.Gehring N.H., Neu-Yilik G., Schell T., Hentze M.W., Kulozik A.E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 12.Schweingruber C., Rufener S.C., Zünd D., Yamashita A., Mühlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta. 2013;1829:612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Lykke-Andersen J., Shu M.D., Steitz J.A. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 14.Rivas M.A., Pirinen M., Conrad D.F., Lek M., Tsang E.K., Karczewski K.J., Maller J.B., Kukurba K.R., DeLuca D.S., Fromer M., GTEx Consortium. Geuvadis Consortium Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–669. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindeboom R.G., Supek F., Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 2016;48:1112–1118. doi: 10.1038/ng.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Z., Yau C., Ahmed A.A. A pan-cancer genome-wide analysis reveals tumour dependencies by induction of nonsense-mediated decay. Nat. Commun. 2017;8:15943. doi: 10.1038/ncomms15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mort M., Ivanov D., Cooper D.N., Chuzhanova N.A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 19.Frischmeyer P.A., Dietz H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 20.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrovski S., Gussow A.B., Wang Q., Halvorsen M., Han Y., Weir W.H., Allen A.S., Goldstein D.B. The intolerance of regulatory sequence to genetic variation predicts gene dosage sensitivity. PLoS Genet. 2015;11:e1005492. doi: 10.1371/journal.pgen.1005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gambin T., Jhangiani S.N., Below J.E., Campbell I.M., Wiszniewski W., Muzny D.M., Staples J., Morrison A.C., Bainbridge M.N., Penney S. Secondary findings and carrier test frequencies in a large multiethnic sample. Genome Med. 2015;7:54. doi: 10.1186/s13073-015-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayram Y., White J.J., Elcioglu N., Cho M.T., Zadeh N., Gedikbasi A., Palanduz S., Ozturk S., Cefle K., Kasapcopur O., Baylor-Hopkins Center for Mendelian Genomics REST final-exon-truncating mutations cause hereditary gingival fibromatosis. Am. J. Hum. Genet. 2017;101:149–156. doi: 10.1016/j.ajhg.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White J., Mazzeu J.F., Hoischen A., Jhangiani S.N., Gambin T., Alcino M.C., Penney S., Saraiva J.M., Hove H., Skovby F., Baylor-Hopkins Center for Mendelian Genomics DVL1 frameshift mutations clustering in the penultimate exon cause autosomal-dominant Robinow syndrome. Am. J. Hum. Genet. 2015;96:612–622. doi: 10.1016/j.ajhg.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Challis D., Yu J., Evani U.S., Jackson A.R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R.A., Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth J.R. Frameshift mutations. Annu. Rev. Genet. 1974;8:319–346. doi: 10.1146/annurev.ge.08.120174.001535. [DOI] [PubMed] [Google Scholar]
- 30.Fisher R.A. Genesis Publishing Pvt Ltd.; 1925. Statistical Methods for Research Workers. [Google Scholar]
- 31.Cava C., Colaprico A., Bertoli G., Graudenzi A., Silva T.C., Olsen C., Noushmehr H., Bontempi G., Mauri G., Castiglioni I. SpidermiR: An R/Bioconductor Package for Integrative Analysis with miRNA Data. Int. J. Mol. Sci. 2017;18:18. doi: 10.3390/ijms18020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consortium G.T., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium G.T., GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bainbridge M.N., Wang M., Wu Y., Newsham I., Muzny D.M., Jefferies J.L., Albert T.J., Burgess D.L., Gibbs R.A. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12:R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartha I., Rausell A., McLaren P.J., Mohammadi P., Tardaguila M., Chaturvedi N., Fellay J., Telenti A. The characteristics of heterozygous protein truncating variants in the human genome. PLoS Comput. Biol. 2015;11:e1004647. doi: 10.1371/journal.pcbi.1004647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itan Y., Shang L., Boisson B., Patin E., Bolze A., Moncada-Vélez M., Scott E., Ciancanelli M.J., Lafaille F.G., Markle J.G. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. USA. 2015;112:13615–13620. doi: 10.1073/pnas.1518646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller H.J. Further studies on the nature and causes of gene mutations. Proc. 6th International Congress of Genetics. 1932:213–255. [Google Scholar]
- 38.Yanai I., Benjamin H., Shmoish M., Chalifa-Caspi V., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- 39.Hamosh A., Sobreira N., Hoover-Fong J., Sutton V.R., Boehm C., Schiettecatte F., Valle D. PhenoDB: a new web-based tool for the collection, storage, and analysis of phenotypic features. Hum. Mutat. 2013;34:566–571. doi: 10.1002/humu.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobreira N., Schiettecatte F., Boehm C., Valle D., Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posey J.E., Harel T., Liu P., Rosenfeld J.A., James R.A., Coban Akdemir Z.H., Walkiewicz M., Bi W., Xiao R., Ding Y. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Branham-O’Connor M., Robichaux W.G., 3rd, Zhang X.K., Cho H., Kehrl J.H., Lanier S.M., Blumer J.B. Defective chemokine signal integration in leukocytes lacking activator of G protein signaling 3 (AGS3) J. Biol. Chem. 2014;289:10738–10747. doi: 10.1074/jbc.M113.515031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.An N., Blumer J.B., Bernard M.L., Lanier S.M. The PDZ and band 4.1 containing protein Frmpd1 regulates the subcellular location of activator of G-protein signaling 3 and its interaction with G-proteins. J. Biol. Chem. 2008;283:24718–24728. doi: 10.1074/jbc.M803497200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poli M.C., Ebstein F., Nicholas S.K., de Guzman M.M., Forbes L.R., Chinn I.K., Mace E.M., Vogel T.P., Carisey A.F., Benavides F., Undiagnosed Diseases Network members Heterozygous truncating variants in POMP escape nonsense-mediated decay and cause a unique immune dysregulatory syndrome. Am. J. Hum. Genet. 2018;102:1126–1142. doi: 10.1016/j.ajhg.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S.Y., Han Y.M., Oh M., Kim W.K., Oh K.J., Lee S.C., Bae K.H., Han B.S. DUSP4 regulates neuronal differentiation and calcium homeostasis by modulating ERK1/2 phosphorylation. Stem Cells Dev. 2015;24:686–700. doi: 10.1089/scd.2014.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Redler S., Strom T.M., Wieland T., Cremer K., Engels H., Distelmaier F., Schaper J., Küchler A., Lemke J.R., Jeschke S. Variants in CPLX1 in two families with autosomal-recessive severe infantile myoclonic epilepsy and ID. Eur. J. Hum. Genet. 2017;25:889–893. doi: 10.1038/ejhg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harel T., Yesil G., Bayram Y., Coban-Akdemir Z., Charng W.L., Karaca E., Al Asmari A., Eldomery M.K., Hunter J.V., Jhangiani S.N., Baylor-Hopkins Center for Mendelian Genomics Monoallelic and biallelic variants in EMC1 identified in individuals with global developmental delay, hypotonia, scoliosis, and cerebellar atrophy. Am. J. Hum. Genet. 2016;98:562–570. doi: 10.1016/j.ajhg.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harel T., Yoon W.H., Garone C., Gu S., Coban-Akdemir Z., Eldomery M.K., Posey J.E., Jhangiani S.N., Rosenfeld J.A., Cho M.T., Baylor-Hopkins Center for Mendelian Genomics. University of Washington Center for Mendelian Genomics Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am. J. Hum. Genet. 2016;99:831–845. doi: 10.1016/j.ajhg.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inoue K., Khajavi M., Ohyama T., Hirabayashi S., Wilson J., Reggin J.D., Mancias P., Butler I.J., Wilkinson M.F., Wegner M., Lupski J.R. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat. Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- 51.Khajavi M., Inoue K., Lupski J.R. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 2006;14:1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- 52.Inoue K., Ohyama T., Sakuragi Y., Yamamoto R., Inoue N.A., Yu L.H., Goto Y., Wegner M., Lupski J.R. Translation of SOX10 3′ untranslated region causes a complex severe neurocristopathy by generation of a deleterious functional domain. Hum. Mol. Genet. 2007;16:3037–3046. doi: 10.1093/hmg/ddm262. [DOI] [PubMed] [Google Scholar]
- 53.Ben-Shachar S., Khajavi M., Withers M.A., Shaw C.A., van Bokhoven H., Brunner H.G., Lupski J.R. Dominant versus recessive traits conveyed by allelic mutations - to what extent is nonsense-mediated decay involved? Clin. Genet. 2009;75:394–400. doi: 10.1111/j.1399-0004.2008.01114.x. [DOI] [PubMed] [Google Scholar]
- 54.Kosmicki J.A., Samocha K.E., Howrigan D.P., Sanders S.J., Slowikowski K., Lek M., Karczewski K.J., Cutler D.J., Devlin B., Roeder K. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meehan T.F., Conte N., West D.B., Jacobsen J.O., Mason J., Warren J., Chen C.K., Tudose I., Relac M., Matthews P., International Mouse Phenotyping Consortium Disease model discovery from 3,328 gene knockouts by The International Mouse Phenotyping Consortium. Nat. Genet. 2017;49:1231–1238. doi: 10.1038/ng.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jansen S., Geuer S., Pfundt R., Brough R., Ghongane P., Herkert J.C., Marco E.J., Willemsen M.H., Kleefstra T., Hannibal M., Deciphering Developmental Disorders Study De novo truncating mutations in the last and penultimate exons of PPM1D cause an intellectual disability syndrome. Am. J. Hum. Genet. 2017;100:650–658. doi: 10.1016/j.ajhg.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Twigg S.R., Forecki J., Goos J.A., Richardson I.C., Hoogeboom A.J., van den Ouweland A.M., Swagemakers S.M., Lequin M.H., Van Antwerp D., McGowan S.J., WGS500 Consortium Gain-of-function mutations in ZIC1 are associated with coronal craniosynostosis and learning disability. Am. J. Hum. Genet. 2015;97:378–388. doi: 10.1016/j.ajhg.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.