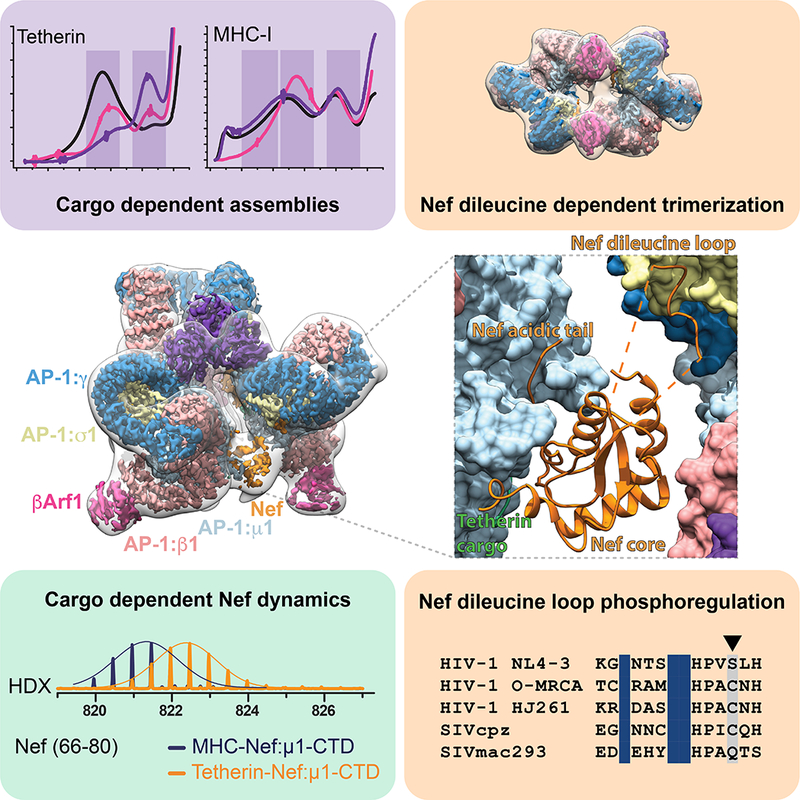
Summary
The HIV accessory protein Nef counteracts immune defenses by subverting coated vesicle pathways. The 3.7 Å cryo-EM structure of a closed trimer of the clathrin adaptor AP-1, the small GTPase Arf1, HIV-1 Nef, and the cytosolic tail of the restriction factor tetherin suggested a mechanism for inactivating tetherin by Golgi retention. The 4.3 Å structure of a mutant Nef-induced dimer of AP-1 showed how the closed trimer is regulated by the dileucine loop of Nef. HDX-MS and mutational analysis were used to show how cargo dynamics leads to alternative Arf1 trimerization, directing Nef targets to be either retained at the trans-Golgi or sorted to lysosomes. Phosphorylation of the NL4–3 M-Nef was shown to regulate AP-1 trimerization, explaining how O-Nefs lacking this phosphosite counteract tetherin but most M-Nefs do not. These observations show how the higher-order organization of a vesicular coat can be allosterically modulated to direct cargoes to distinct fates.
Keywords: HIV, clathrin, adaptor protein, trafficking, cryo-EM
In Brief
Structural and dyamics measurements of HIV Nef interacting with vesicle adaptors show how differences in cargo dynamics can allow the same adaptor system to direct cargoes to different fates, expanding the regulatory capacity of a limited number of adaptors.
Graphical abstract
Introduction
Human and simian immunodeficiency viruses (HIV and SIV) encode a variety of so-called accessory proteins that are frequently dispensable for viral spread in cell culture. However, they are critical for efficient viral replication and transmission in vivo because they promote viral evasion of innate and adaptive immune responses and counteract antiviral host restriction factors (Kirchhoff, 2010; Tokarev and Guatelli, 2011). One well- characterized example is the multi-functional factor Nef, which is encoded by all primate lentiviruses. HIV and SIV use Nef to downmodulate their primary CD4 receptor from the cell surface (Garcia and Miller, 1991) and to counteract the restriction factor SERINC5 (Rosa et al., 2015; Usami et al., 2015), thereby preventing superinfection and promoting efficient release of fully infectious viral particles. Another major function of Nef is the downmodulation of MHC-I molecules from the cell surface to prevent recognition and destruction of virally infected cells by cytotoxic T-cells (CTL) (Schwartz et al., 1996).
Nef-mediated downmodulation of MHC-I is conserved between all primate lentiviruses and generally involves targeting of MHC-I at the trans-Golgi network (TGN) by hijacking the clathrin adaptor complex AP-1 and the Golgi-specific small G-protein Arf1 (Coleman et al., 2006; Roeth et al., 2004). The ultimate fate of MHC-I, however, is cell-type dependent. In T-cells, Nef typically directs MHC-I into post-Golgi clathrin-coated vesicles (CCVs) that sort MHC-I to the lysosome for degradation (Roeth et al., 2004; Schwartz et al., 1996). In other cell types, Nef and MHC-I accumulate at the TGN or at endosomes (Lubben et al., 2007). These different fates of MHC-I were reported to be a function of phosphoregulation (Kasper et al., 2005). Thus, AP-1 engagement by Nef can have different consequences when parts of the system are differentially phosphorylated.
In addition to down-modulating CD4 and MHC I, most SIV strains use their Nef protein to also antagonize the restriction factor tetherin that blocks the release of progeny virions from infected cells (Sauter et al., 2009; Zhang et al., 2009). In contrast, pandemic HIV-1 group M strains evolved the accessory protein Vpu as potent antagonist of human tetherin (Neil et al., 2008; Van Damme et al., 2008). Vpu is only encoded by HIV-1 and some closely related SIV strains and counteracts tetherin by hijacking AP-1 (Jia et al., 2014; Kueck et al., 2015) to drive TGN accumulation (Schmidt et al., 2011) and/or lysosomal degradation of the restriction factor (Kueck and Neil, 2012). In comparison, SIV Nef proteins remove tetherin from the cell surface via interactions with the plasma membrane clathrin adaptor complex AP-2 (Serra-Moreno et al., 2013; Zhang et al., 2011). Remarkably, epidemic HIV-1 group O strains that resulted from independent zoonotic transmission of SIVs infecting greater apes apparently evolved a third distinct mechanism to overcome tetherin restriction. Their Nef proteins induce accumulation of tetherin at the TGN, presumably by hijacking TGN-specific clathrin adaptor AP-1 (Kluge et al., 2014). In contrast to pandemic group M and epidemic group O HIV-1 strains, the very rare N and P groups of HIV-1 have not evolved an effective tetherin antagonist (Sauter et al., 2009; 2011; 2012; Yang et al., 2011). On the other hand, FIV and HIV-2 use Env itself to target tetherin (Celestino et al., 2012; Chen et al., 2016; Hauser et al., 2010; Le Tortorec and Neil, 2009; Morrison et al., 2014), and do so using a sequestration rather than a degradative mechanism (Hauser et al., 2010; Le Tortorec and Neil, 2009). Thus, the acquisition of potent anti-tetherin mechanisms was most likely a prerequisite for the efficient spread of HIV/AIDS in the human population (Sauter et al., 2010). It is currently unknown, however, which structural and biochemical mechanisms are responsible for the differential ability of HIV-1 group M and O Nef proteins to antagonize human tetherin.
AP-1 is the heterotetrameric clathrin adaptor complex devoted to the biogenesis of CCVs at the TGN (Stamnes and Rothman, 1993; Traub et al., 1993). AP-1, like other clathrin adaptors, binds to membranes and transmembrane protein cargo on one face, and to clathrin on the other, thereby generating cargo-loaded CCVs (Owen et al., 2004; Traub and Bonifacino, 2013). AP-1 consists of two large subunits, β1 and γ, a medium subunit, μ1, and a small subunit σ1 (Owen et al., 2004; Traub and Bonifacino, 2013). The large subunits consist of ~600-residue helical solenoids followed by a linker and an appendage domain. β1 contains the clathrin-binding domain, and both β1 and γ bind to the small G-protein Arf1 (Ren et al., 2013). AP-1 binds to cargo bearing either YxxΦ (where Φ = hydrophobic) or (D/E)XXXLL dileucine (LL for short) motifs. YxxΦ motifs bind to the C-terminal domain (CTD) of μ1 (Owen and Evans, 1998), while LL motifs bind to a hemicomplex formed by σ1 and γ (Doray et al., 2007; Jackson et al., 2010; Traub and Bonifacino, 2013). In its inactive cytosolic state, the helical solenoids block the YxxΦ and LL cargo sites on μ1 and σ1- γ, a conformation referred to as the locked state (Collins et al., 2002; Heldwein et al., 2004; Jackson et al., 2010). Arf1 triggers the recruitment of AP-1 to the TGN membrane, where it has further profound effects on AP- 1 structure, driving both its allosteric unlocking and its dimerization (Ren et al., 2013).
HIV-1 Nef targets Arf1-unlocked AP-1 via both of its two cargo sites. The cytosolic tail of MHC-I contains an incomplete YxxΦ signal, which is not competent to bind AP-1 on its own (Noviello et al., 2008). Nef forms a ternary complex that stabilizes the binding of the MHC-I tail to the YxxΦ binding site of the μ1 CTD (Jia et al., 2012). Multiple regions of Nef interact with the MHC-I tail and μ1-CTD, with the proline-rich region (aa 72–78) being most prominent among them. Nef can also hijack the LL binding sites of the AP-1 and AP-2 complexes (Chaudhuri et al., 2007; Doray et al., 2007; Roeth et al., 2004). The structure of Nef bound to the α-σ2 hemicomplex of AP-2 was determined (Ren et al., 2014). The interactions with the most central acidic and LL residues of Nef with the γ-σ1 complex of AP-1 are conserved with AP-2, while peripheral residues are more variable. Vpu targets the LL site on AP-1 in a similar manner to Nef, promoting AP-1 unlocking and making the μ1 CTD available to bind the YxYxxΦ motif in the tetherin tail (Jia et al., 2014).
Three years ago, we discovered how in the presence of Arf1 and cargo, Nef induces the oligomerization of unlocked AP-1 into trimers of two types, open and closed (Shen et al., 2015). This ability of clathrin adaptors to trimerize was completely unexpected. In the same year it was reported that Arf1 could stabilize trimeric contacts between the COPI F-subcomplex (Dodonova et al., 2015), a process that does not depend on Nef. Thus, Arf1-mediated trimerization of coat adaptors is a widespread principle that positively regulates cargo packaging into clathrin coated vesicles. In the presence of Nef and MHC-I tails, AP-1 and Arf1 can open trimers, which in turn can form higher order lattices whose symmetry matches that of clathrin. Lattice formation by AP-1 and Arf1 appears to be a fundamental and agent mechanism of coat formation predating the evolution of HIV, yet can be hijacked and promoted by Nef in the case of selected cargoes such as MHC-I. The ability of cargoes and Nef mutants to promote open trimer formation correlates with their ability to promote clathrin basket polymerization in vitro.
We also showed tetherin, but not MHC-I, promoted the formation of a different type of trimer, which was closed and incapable of forming lattices (Shen et al., 2015). It seemed clear that this trimer was not a positive regulator of clathrin assembly, but otherwise, it was not clear whether it had a function or what its function was. We previously determined the structure of the closed trimer at 7 Å resolution, which was inadequate to visualize amino acid side chains. Our previous inability to visualize interactions at the side-chain level prevented us from determining which interactions drove closed trimer formation. Thus it was not possible to mutate these side-chains, eliminating the principal tool for assessing its biological function. We wondered if the closed trimer might be related to the inhibition of tetherin by the Nefs from those HIV strains that depend on Nef to block tetherin, but we lacked tools to test this idea.
In this study, we sought to determine whether differential open and closed AP-1 trimer formation by MHC-I and tetherin could account for the different fates of these cargoes. We determined the structure of the tetherin-bound closed trimer at 3.7 Å resolution by cryo-electron microscopy. The structure provided the critical missing insights into the determinants of trimer formation. It showed how Nef uniquely stabilizes the closed trimer by bridging the μ1 CTD (YxxΦ binding site) of one AP-1 to the σ1 (LL binding site) of another. We verified this structural concept by showing that the Nef LL motif is critical to the ability of Nef to induce closed trimer formation. We used hydrogen deuterium exchange coupled to mass spectrometry (HDX-MS) (Engen, 2009; Englander, 2006), a powerful technique to probe protein dynamics, to determine whether different cargoes lead to different dynamics in Nef. We showed that the ability of the Nef LL motif to bridge between AP-1 copies correlates with the dynamics of the YxxΦ-μ1 interaction, explaining why tetherin was shown to promote the closed trimer formation whereas MHC-I does not. Analysis of these structures and a related structure of an Arf1 -mediated trimer of COPI (Dodonova et al., 2017; Dodonova et al., 2015) led to a model for cargo-regulated alternative trimer formation via the differential bridging and differential use of the Arf1 binding sites of β1 and γ. Collectively, these data explain how the closed trimer forms and that, unlike the open trimer, it is a Nef- and infection-specific state.
The most curious aspect of the finding that Nef and tetherin drove closed trimer formation was its occurrence in vitro for the M-Nef of HIV-1 NL4–3 despite that M-Nef is essentially inactive against tetherin in vivo. O-Nefs inhibit tetherin biologically, while M-Nefs do so minimally. Here we explain this by showing that the lack of activity by M-Nefs is due to phosphorylation of the dileucine loop. The M-Nef samples used in our in vitro studies were generated in E. coli and are dephosphorylated, explaining their O-Nef-like behavior in the in vitro and structural studies. Here, we show M-Nef phosphorylation prevents it from forming closed trimers. Our model thus provides a structural explanation for the differential activity of HIV-1 group O- and M-Nefs against human tetherin. More broadly, these results show how differences in cargo dynamics can allow the same adaptor system to direct cargoes to different fates. This insight, which was derived from the high resolution cryo-EM structures and the HDX-MS analyses, may have profound implications across normal biology, not just in HIV infection. This idea suggests a mechanism whereby the limited number of cargo adaptors present in cells is versatile enough to target the vast assortment of cellular cargoes to a multitude of destinations.
Results
Atomic model of the closed AP-1 trimer
The complex of NL4–3 Nef fused to the cytosolic tail of human tetherin was reconstituted with Arf1-GTP and the heterotetrameric core of AP-1. The complex was purified by size exclusion chromatography (SEC), the peak corresponding to a trimer of AP-1 complexes was isolated, and the sample subjected to cryo-EM (Figure S1-3). Both AP-1 trimers and monomers were identified, and their structures reconstructed (Figure 1A-B and Figure S1, 2D, 3). The AP-1 monomer resembles the previously determined hyper-unlocked conformation determined in the presence of a tetherin-Vpu fusion (Jia et al., 2014), with the addition of Arf1 molecules bound to both the β1 and γ sites (Figure S2D). The AP-1 trimer consists of hyper-unlocked monomers mutually arranged in the “closed” conformation previously described at 7 Å resolution (Shen et al., 2015). The structure is quasi-C3 symmetric (Figure 1C) and was resolved to 3.7 Å (Figure S1A, 2A-B, 3A-F) using localized reconstruction of the monomeric subunit (Ilca et al., 2015), revealing many new aspects of its organization and Nef, Arf1, and cargo interactions.
Figure 1. Atomic model of the AP1:Arf1:tetherin-Nef closed trimer.
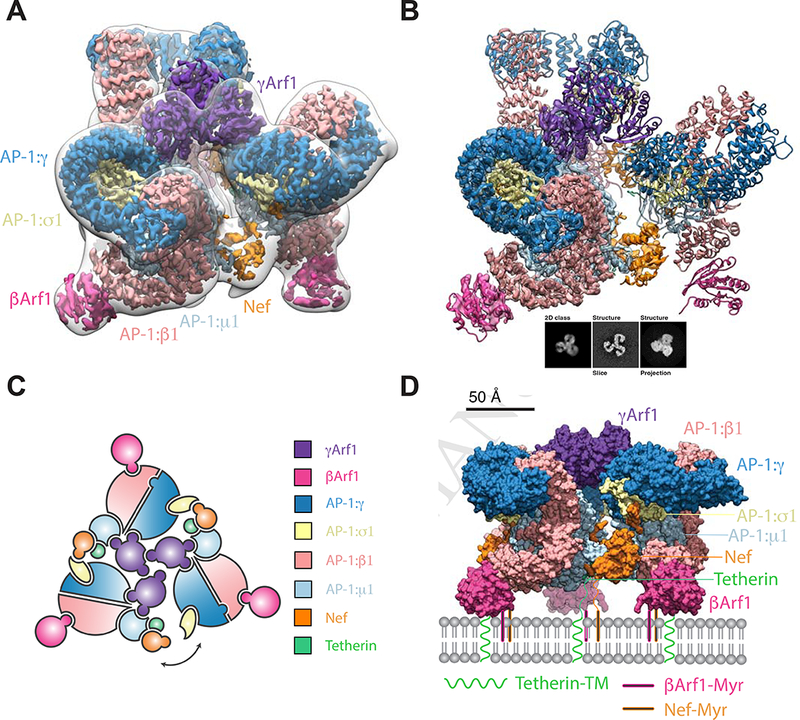
(A) The closed trimeric state consists of three copies of γΑrf1 assembling three AP-1 cores with one copy of HIV-Nef at each AP-1 dimer interface. The subunit reconstruction is shown in the context of the transparent whole trimer reconstruction. The AP-1 core and γArf1 subunits are sharpened at −50 Å2, Nef and βArf1 subunits are unsharpened and the transparent whole closed trimer low pass filtered to 18 Å is shown for clarity. (B) The trimer subunit contains the AP-1 core, two copies of Arf1 (γArf1 and βArf1) and one copy of Tetherin-HIV-Nef. A 2D class, corresponding volume slice and projection are shown for reference. (C) A subunit schematic for the closed trimer showing interaction interfaces and the break from perfect three-fold symmetry of the complex allowing movement of the cargo Nef trans AP-1 bridge. (D) A model for the closed trimer docked at the trans golgi network. See also Figure S1-3.
The overall trimer is approximately a prism with triangular edges of ~190 Å and a height of ~140 Å. AP-1 molecules do not make direct contact with one another. Instead, the trimer is bridged at the top (in the view shown in Figure 1A) by a trimer of Arf1 molecules bound to γ1 (hereafter, “γArf1”), and at the bottom, by Nef. Another three Arf1 molecules are bound to β (hereafter, “βArf1”). These molecules are at the periphery and are not involved in inter-AP-1 bridges.
Two of the three inter-AP-1 gaps at the bottom of the assembly are ~30 Å across and spanned by ordered Nef molecules (Figure 2A-E), while the third opening, at ~40 Å, is too wide to be spanned. The inability of Nef to simultaneously span all three gaps accounts for the quasi-C3 symmetric nature of the assembly. Nef makes extensive contacts with the μ1-CTD and the tetherin tail, which resemble contacts (Figure 2A-B) described for the μ1-CTD:Nef:MHC-I tail complex (Jia et al., 2012). The Nef LL motif is well-defined in density and engages the γ-σ1 (Figure 2C) in trans with respect to the μ1 interaction. The Nef sequences 149–157 and 168–179 are disordered over gaps of 22 and 25 Å, respectively between the LL motif and the Nef core. The Nef core loop consisting of residues 92–94 could make electrostatic contact with the N-terminus of β1, in cis with respect to the LL binding site. Collectively, these observations show how Nef promotes the closed trimer assembly of AP-1 by bridging in trans between the μ1-CTD and LL sites of two pairs of AP-1 monomers.
Figure 2. Nef bridging via the dileucine loop stabilizes the closed trimer.
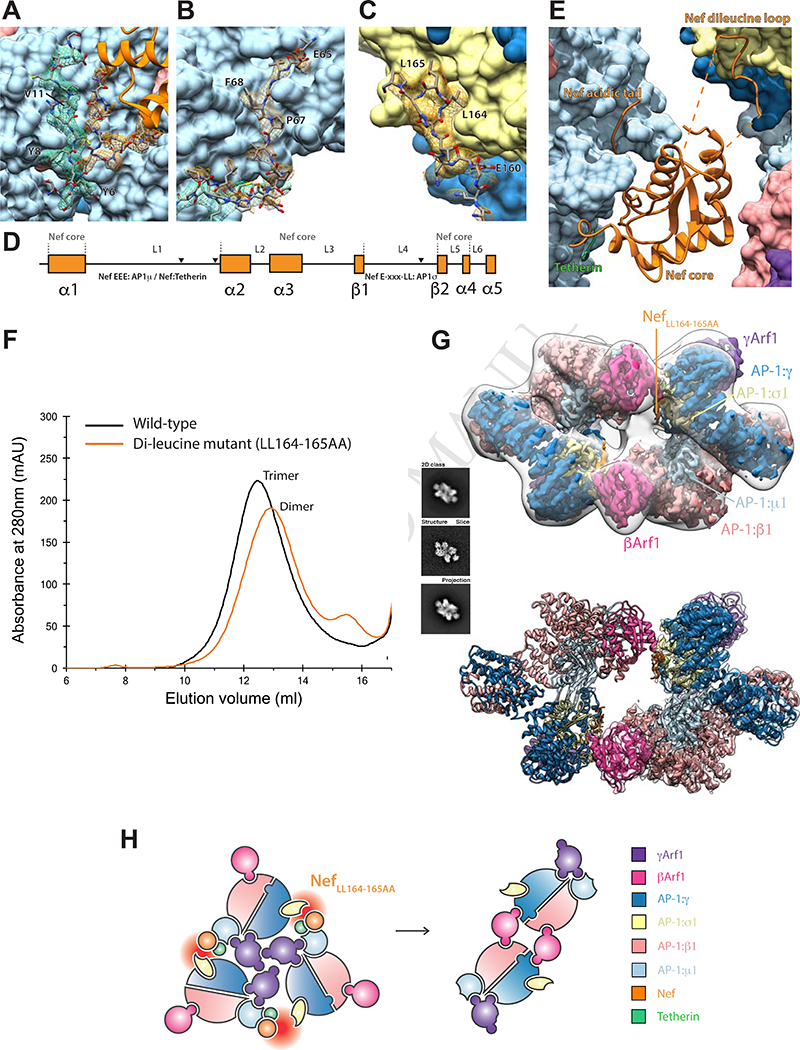
(A) An AP-1:µ1:Tetherin:Nef sandwich stabilises the Nef core to AP-1:µ1. (B) The canonical Nef acidic patch binds directly to AP-1:µ1. (C) The ExxxLL motif of Nef binds to AP-1σ1 (D) These binding regions of Nef are found on the unstructured loops L1 and L4 but are stabilized in the trimeric assembly. (E) Consequently, the trimer architecture is closed by HIV-Nef at the AP1 trimeric dimer interface. (F) Mutating the dileucine loop, tetherin-NefLL164–165AA, responsible for binding AP-1:σ1 results in closed trimer destabilization and dimer formation as indicated by SEC. (G) The dileucine mutant adopts a dimeric oligomeric state as confirmed by cryo-EM. A 2D class, corresponding volume slice and projection are shown for reference. (H) The unstable trimer created by the knockout of the AP-1:σ1:LL interaction leads to a rearrangement into dimers driven by new βArf1:AP-1:γ interactions. See also Figure S1-3 and 4.
The γArf1, as well as the AP-1 core, are well-defined by the density (Figure S2B). The three γArf1 molecules contact one another through their β2-β3 turn, α4-β6 connection, and C-terminal helix (Figure 3A). The structure revealed multiple contacts between γArf1 α2 and α3-β5 loop with the μ1-CTD (Figure S4A), confirming a previous report of a non-switch Arf1 region interacting with the related μ4 domain (Boehm et al., 2001). The three βArf1 molecules bound at the periphery of the complex are in an appropriate geometry to insert into the membrane via their N-terminal myristates, and the μ1-CTD binding site for MHC-I, tetherin, and other YxxΦ based signals is an unobstructed ~20 Å from the membrane (Figure 1D). The γArf1 molecules are membrane-distal in this geometry, but Nef is poised to bind the membrane via its own myristoyl modification. Thus, at least nine points of contact between the closed trimer assembly and membranes are evident, showing how this assembly could bind cargo at the TGN.
Figure 3. γArfl and βArf1 control AP-1 oligomerization in a cargo dependent manner.
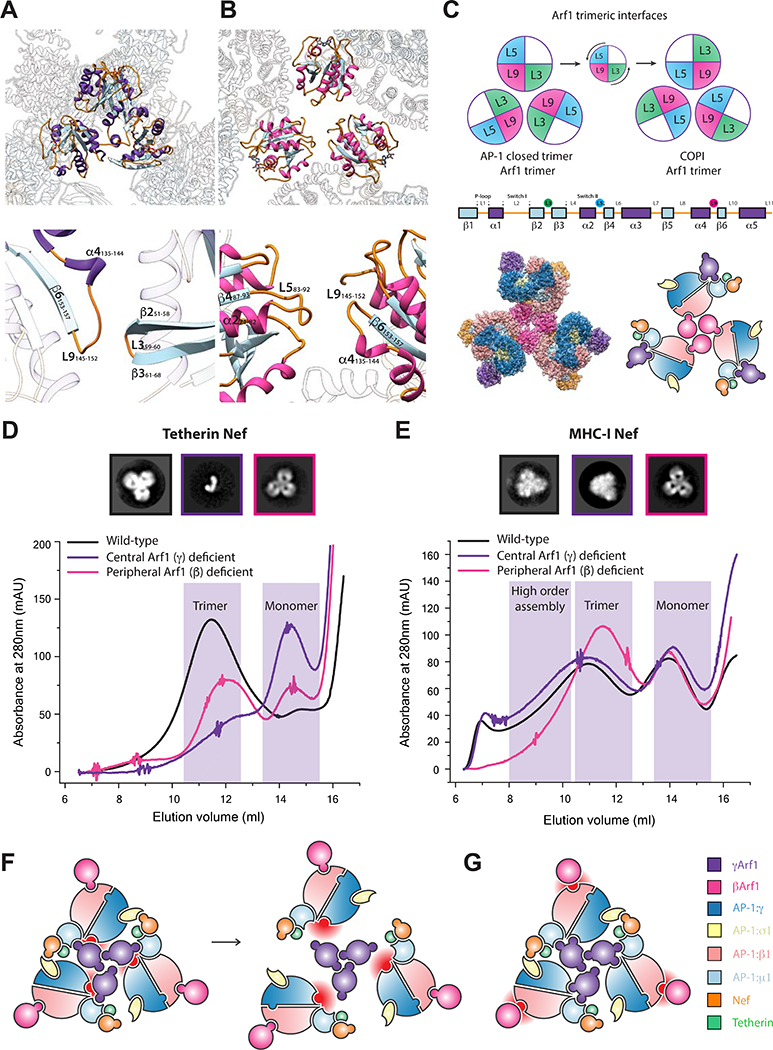
(A) The binding loops of the closed AP-1 trimer are mediated through γArf1 loop 9 - loop 3 contacts. (B) Conversely the trimerisation architecture observed in COPI assemblies exhibits a rearrangement to utilize loop 9 - loop 5 contacts. (C) The counterclockwise turn of the Arf1 assemblies allows these new contacts to be made, yet both still utilize loop 9. In the COPI-like Arf1 L9–5 trimeric architecture, assembly via βArf1 is sterically plausible. (D) In the presence of tetherin-Nef cargo, Arf1 dependent trimerization of AP-1 is dependent on γArf1 but not βArf1. (E) In the presence of MHC- I-Nef cargo, AP-1 may trimerize via both γArf1 or βArf1. Negative stain EM 2D class averages and SEC support that an assembly in the presence of MHC-I-Nef cargo thus may be assembled via βArf1 trimerization. (F) The knockout of the γArf1:AP-1:γ interface completely destabilizes tetherin trimers to monomer, but does not disrupt MHC- I assembly in the same manner. (G) The knockout of the βArf1:AP-1:β1 does not affect the tetherin trimer assembly. In the case of MHC-I trimers, assembly via γArf1 leads to a closed trimer like state. Critically, MHC-I trimers can be seen assemble via both γArf1 and βArf1 oligomerization. See also Figure S4.
Nef bridging uniquely stabilizes the closed trimer
The structure described above suggested that the strength of the μ1-CTD-LL site bridge might control the stability of the closed trimer. The properties of the tetherin-NefLL164– 165AA assembly were therefore investigated. AP-1:Arf1:Tetherin-NefLLAA migrated on SEC as an apparent dimer (Figure 2F), confirming that the Nef dileucine sequence is essential for closed trimer formation. The structure of the AP-1:Arf1:Tetherin-NefLLAA dimer was determined at 4.3 Å resolution by cryo-EM (Figure 2G, Figure S1B, S2C, S3G-I). AP-1 is in the hyper-unlocked conformation, as seen in the closed trimer, thus the change in oligomerization is not driven by a change in AP-1 monomer conformation. As seen previously in the crystal structure of the unlocked AP-1:Arf1 dimer determined in the absence of Nef or cargo (Ren et al., 2013), two βArf1 molecules cross-link the dimer, while the AP-1 complexes themselves do not contact one another. However, the hyper- unlocked conformation of AP-1:Arf1:Tetherin-NefLLAA cross-links through a unique βArf1-AP-1:γ contact that is not seen in the crystal structure of the unlocked conformation (Figure S4D). This new contact allows for dimerization of the hyper- unlocked conformation as the γArf1-AP-1:γ interaction present in the unlocked conformation is no longer available for crosslinking. Residues ~160–166 of NefLLAA were observed bound to σ1-γ despite the absence of the Leu side chains (Figure S4F). Thus, NefLLAA is still capable of binding to AP-1 at the high concentrations used for reconstitution, but the interaction is too weak to tether two AP-1 subunits in the closed trimer assembly. These data confirm that the closed trimer assembly is induced by Nef dileucine-mediated bridges.
Alternative Arf1 trimers lead to alternative assemblies
We previously reported that AP-1:Arf1:Nef complexes could form open trimeric assemblies that strongly promoted clathrin basket formation (Shen et al., 2015). The open trimers are only connected by Arf1 trimerization at the center, and the ensuing dynamics and heterogeneity of these structures has limited attainable resolution to a 17 Å reconstruction that is presumably a superposition of multiple conformations. Recently, the cryo-electron tomography (cryo-ET) and sub-volume averaged 9 Å structure of an Arf1 trimer bound to the related coat complex COPI was reported (Dodonova et al., 2017). The AP-1-γArf1 closed trimer described above is distinct from the COPI-Arf1 trimer, although they share the use of the “L9” (α4-β6) loop (Figure 3A-B). We modeled combinations of βArf1 and γArf1 bound to hyper-unlocked AP-1 trimerized via either the closed trimer or the COPI Arf1 trimer geometry. AP-1 bound to βArf1 in the COPI geometry was sterically possible and competent to bind membranes in the context of known AP conformations with hyper-unlocked AP-1 subunits (Figure 3C). This suggested to us that the closed trimer was driven by γArf1, and the open trimer by βArf1.
We disrupted the two Arf1 interfaces on AP-1 using the previously described mutants AP-1ΔβArf1 and AP-1ΔγArf1 (Ren et al., 2013). In the context of tetherin-Nef, which forms predominantly closed trimer assemblies under the conditions tested, AP-1ΔβArf1 migrated almost normally on SEC, while AP-1ΔγArf1 shifted sharply towards a monomeric distribution (Figure 3D), as predicted by the closed trimer structure. In contrast, in the MHC-I-Nef sample, which is known to consist of open trimer and higher order structures (Shen et al., 2015), AP-1ΔγArf1 behaved identically to wild-type (Figure 3E). In contrast, AP-1ΔβΑrf1 bound to MHC-I-Nef (Figure 3E) migrated in the position expected for the closed trimer (Figure 3D). Thus, MHC-I-Nef AP-1:Arf1 trimers are not dependent on yArf1 for their assembly. Furthermore, the AP-1ΔβArf1 mutation switches what would have been open MHC-I trimers into closed trimers. These data strongly support the concept that alternative Arf1 trimerization and the use of alternative Arf1 contact sites on AP-1 controls whether the assembly formed is open or closed.
Nef polyproline strand dynamics mediates cargo selectivity
The Nef core is inherently mobile in the closed trimer EM density (Figure S5), yet its presence is critical for bridging and stabilizing the trimer. The observation suggested to us that Nef mobility might positively regulate formation of the closed trimer. We used HDX-MS (Figure S6) to compare the dynamics of Nef in the context of complexes of the tetherin-Nef and MHC-I-Nef constructs with µ1-CTD. Consistent with the crystal structure of the MHC-I-Nef: µ1-CTD complex (Jia et al., 2012), which showed extensive interactions between the polyproline region and MHC-I, peptides overlapping with this segment (residues 66–80; Figure 4A, B) were well-protected (< 20 % exchange at 10 s; Figure S6B). This region was less protected in the tetherin complex (~50 % exchange at 10 s; Figure S6A). Elsewhere in Nef, there were relatively few differences in protection (Figure 4A). These data show that Nef is held rigidly to the µ1-CTD in the MHC-I complex, while it is bound more flexibly in the tetherin complex. This flexibility could facilitate formation of the asymmetrically bridged closed trimer (Figure 1C, 4C) by lowering the energetic cost for Nef to bridge from μ1-CTD in one AP-1 complex to the σ subunit of the adjoining AP-1 complex.
Figure 4. Cargo modulates Nef dynamics.
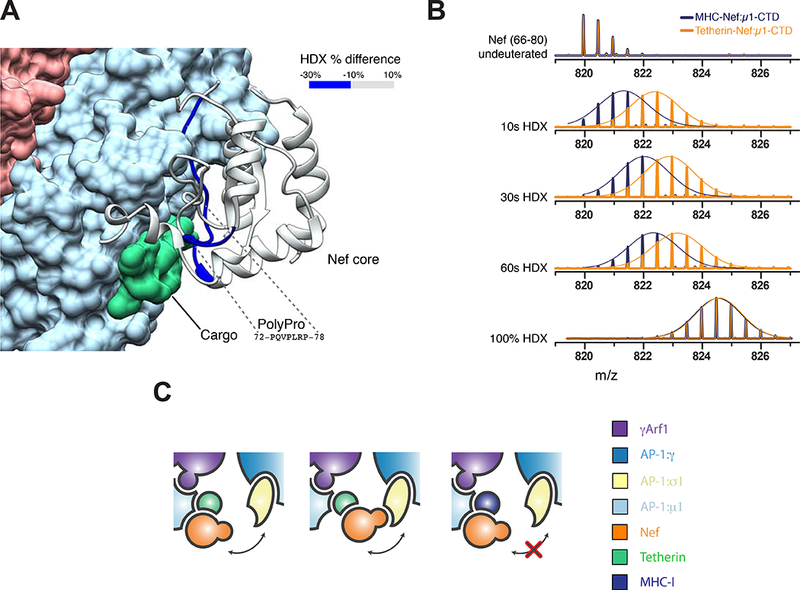
(A) Mapping of the changes in deuteration levels of Nef between tetherin-Nef:AP1:µ1-CTD and MHC-Nef:AP-1:µ1-CTD. The HDX-MS Difference at 10s is overlaid onto the cryoEM structure of the AP- 1:Arf1:tetherin-Nef trimer. (B) Mass spectra of the peptide from Nef (66–80) in tetherin- Nef (orange) or MHC-Nef (blue) upon AP-1:µ 1-CTD binding. A Gaussian fit is used to represent the distribution of peak heights of the ion peaks across the m/z values, and overlay with mass spectra (straight line). (C) The stronger binding of Nef to AP-1:µ 1 in the presence of MHC-I cargo may prevent Nef from reaching its AP-1:σ1 binding site. See also Figure S5.
Phosphoregulation of the closed trimer
We sought to determine whether the closed trimer is functionally important in tetherin downregulation. HIV-1 NL4–3 Nef is considered functionally inert in tetherin counteraction, while HIV-1 group O Nefs are quite effective (Kluge et al., 2014). This functional difference has been mapped to a handful of residues surrounding the dileucine signal (Figure 5A). We used GST pull-down assays to determine the ability of NL4–3 and O-MRCA Nef proteins to interact with a μCTD-deleted form of the AP-1 core (AP-1 coreΔµCTD), which is capable of interacting with cargo via the dileucine motif but not the YxxΦ motif (Jia et al., 2014). NL4–3 Nef bound to the truncated construct (Figure 5B), consistent with the observation of LL binding to the σ1- γ in the cryo-EM structure of the closed trimer. O-MRCA Nef bound robustly to AP-1 coreΔµCTD as well (Figure 5B). This is potentially consistent with a functional role for an O-Nef:AP-1 interaction in tetherin downregulation, but begs the question why a greater difference between NL4–3 and O- Nefs was not observed. Given the presence of a Ser adjacent to the LL motif in NL4–3 and most other M-Nefs, but not O-Nefs (Figure 5A), we considered whether this residue might be subject to phosphoregulation. Ser169 is predicted to contain a consensus phosphorylation site for casein kinase I (Hornbeck et al., 2015). We found that CKI phosphorylated Ser169 uniquely and stoichiometrically in vitro (Figure 5C). CKI phosphorylated NL4–3 Nef does not bind to AP-1 coreΔµCTD (Figure 5A-B). Consistent with the dependence of closed trimer formation on LL engagement, phospho-Nef does not manifest a closed trimer peak on SEC, but rather redistributes to aggregate, open trimer, and monomer peaks (Figure 5D). These data show that closed trimer formation by NL4–3 Nef is subject to phosphoinhibition via a site which is absent in O-Nefs.
Figure 5. Phosphorylation of NL4–3 Nef by CK1 disrupts the interaction between AP-1 σ1 and Nef dileucine motif.
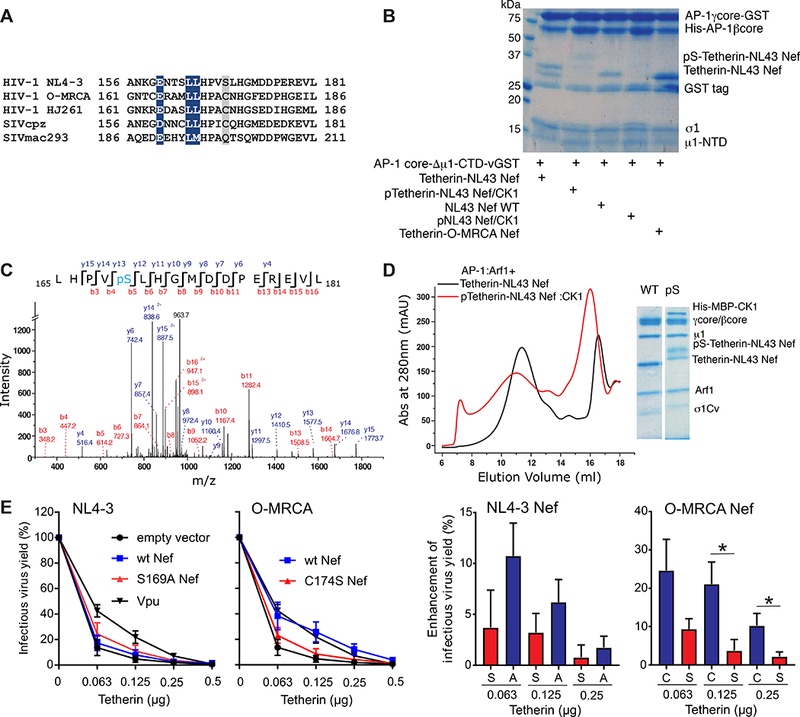
(A) Amino acid alignment of Nef proteins in the D/ExxxLL motif region. The dileucine motifs that are conserved in all Nef strains are colored in blue. The residues aligned with NL4–3 Nef Ser169 predicted CK1 phosphorylation site (Hornbeck et al., 2015) are highlighted in gray. (B) Collision- induced dissociation (CID) fragmentation spectrum of the phosphorylated peptide in tetherin-Nef treated by CK1. Ser169 in NL4–3 Nef is phosphorylated. (C) GST pull down result suggest that Nef phosphorylation by CK1 largely reduces Nef dileucine motif binding to AP-1. AP-1 coreµCTD-GST is selected to represent the interaction with the Nef dileucine motif. The phosphorylation of Nef largely reduces the binding to AP-1coreµCTD- GST on the beads. (D) Gel filtration and SDS gel analysis showing the contents of trimer peak. The phosphorylation of NL4–3 Nef disrupts the closed trimer assembly. (E) The effect of Nef differing in the Ser phosphorylation site on infectious virus release in the presence of tetherin. Infectious virus yield from 293T cells co-transfected with an HIV-1 NL4–3 ΔVpuΔNef construct and vectors expressing the indicated nef alleles or human tetherin was determined by infection of TZM-bl cells (right). Data show mean percentages (± SEM) relative to those detected in the absence of tetherin (100%) obtained in five independent experiments. Results obtained for NL4–3 Vpu are shown for comparison. The bar diagrams show the increase in virus production in the presence of Nef compared to the vector control. Stars refer to the difference from the EGFP control panel. *p < 0.05, **p < 0.01.
To directly determine the relevance of this Ser phosphorylation site for the anti-tetherin activity of Nef, we introduced mutations of S169A and vice versa C174S in the NL4–3 and O-MRCA Nef proteins, respectively. The parental NL4–3 Nef only marginally increased infectious virus yield from 293T cells after cotransfection of a ΔVpuΔNef HTV-1 NL4–3 proviral vector with fixed quantities of Nef expression construct and different doses of tetherin expression plasmid. Mutation of S169A increased its capability to counteract tetherin, although it remained substantially less effective than Vpu or the O- MRCA Nef (Figure 5E). This is in agreement with previous data showing that S169C only conferred full anti-tetherin activity to the HIV-1 JC16 Nef in combination with a S163I change in the third variable residue of the ExxxLL motif (Gotz et al., 2012). Mutation of C174S, however, almost fully disrupted the anti-tetherin activity of O-MRCA Nef (Figure 5E) supporting a role of phosphoregulation in the ability of Nef to counteract tetherin.
Discussion
Nef is an important determinant of HIV infectivity and pathogenesis, and exerts many, if not most, of its effects by hijacking the coated vesicle trafficking pathways of infected cells. We previously showed that NL4–3 Nef hijacks AP-1 in a startlingly complex manner (Shen et al., 2015). Here, we build on this work to show how different AP-1 hijacking mechanisms are used by cargoes undergoing different fates, and by Nefs of the M and O groups of HIV-1 groups that are known to counteract the host restriction factor tetherin by divergent means. Our previous low and intermediate-resolution structural studies revealed that tetherin and NL4–3 Nef stabilize a closed AP-1 trimer, while MHC-I and NL4–3 Nef promote an open conformation. Both structures are capable of docking onto membranes but only the open trimer has a geometry matching that of the clathrin lattice (Shen et al., 2015). The role of the closed trimer was mysterious, with progress stymied by the limited resolution available. Here, we obtained a near-atomic reconstruction of the closed trimer for the first time, making it possible to interpret molecular contacts, design mutations, and elucidate its function.
The atomic model of the closed trimer, and comparison to related structures, clarified two points critical to functional analysis. First, a large section of the Nef LL loop was visualized in the density. This loop could be uniquely assigned to a Nef molecule bound in trans to the μ1-CTD of a different AP-1 complex within the trimer. This established that Nef binds in trans between the LL binding site and the μ1-CTD, and suggested that the ability of Nef to bridge these two sites in trans could be the driving force for closed trimerization. Second, the improved resolution, together with the publication of a higher resolution structure of an Arf1-mediated trimeric interface in the related COPI coat (Dodonova et al., 2017), suggested that the open and closed trimers use Arf1 contacts differently. These higher resolution insights made it possible to functionally dissect the determinants for closed trimerization.
The LLµ1-CTD bridge model predicted that the closed trimer must be held together by strong contacts on both the LL and μ1-CTD sides, with sufficient affinity to drive formation of an oligomer that would otherwise be energetically unfavorable. We thus expected that mutation of the LL sequence to dialanine (AA) would block formation of the closed trimer. Confirming this idea, we found that tetherin and NefLLAA forms a mixture of monomers and dimers, but not closed trimers. We went on to determine a near-atomic cryo-EM structure of the tetherin-NefLLAA dimer. The LLAA loop density could be visualized in this structure, showing that the residual affinity of the LLAA mutant loop was adequate to bind at the high concentrations used to generate the sample, but inadequate to drive closed trimerization. Subsequently, we found that CK1 phosphorylation of Ser169, which immediately follows the LL sequence in NL4–3 Nef, also weakens binding to the LL site. In this case, a mixture of aggregates, open trimers, and monomers replaces closed trimers. While it is still unclear what controls the population of dimers and open trimers when closed trimerization is blocked, these data firmly establish LL- μ1-CTD bridging as the mechanism for stabilizing closed trimers.
These and previous data (Shen et al., 2015) and the recent 9 Å cryo-ET structure of the COPI coat (Dodonova et al., 2017) highlight the conserved and ancient role of Arf1 as a building block and integral component of vesicular coats. The conformations of the open and closed AP-1 trimers are so different that it seems impossible to rationalize them in terms of a single mode of Arf1 trimerization. Differential Arf1 driven trimerization was confirmed using a set of mutations in the Arf1 binding site of the β1 and γ subunits of AP-1 (Ren et al., 2013). Blocking the γ site converts the tetherin complex from a closed trimer to a monomer, while the while β1 site mutant had little effect. Disruption of the γ site has little effect on the MHC-I complex, but strikingly, blocking the β1 site converts it from open trimer to closed. Previously, we showed how the presence of multiple Arf1 binding sites on AP-1 could result in the cross-linking of AP-1 into larger polygons (Shen et al., 2015). We now find that the multiplicity of binding sites leads to the ability to trimerize in ways that lead to sharply different functions.
SIV/HIV is believed to have made multiple independent crossings of the simian/human species barrier. SIV downregulates tetherin via Nef hijacking of AP-2, a mechanism that depends on the presence of the sequence (G/D)DIWK in simian tetherins (Zhang et al., 2009). The inability of SIV Nefs to downregulate human tetherin represents one of the main barriers to interspecies transmission (Neil et al., 2008; Van Damme et al., 2008). The pandemic M-group HIV-1 uses Vpu to downregulate tetherin (Neil et al., 2008; Van Damme et al., 2008), and this effect appears to occur through retention of Vpu and tetherin in the TGN (Dube et al., 2009; Jia et al., 2014). However, O-group viruses and a few others appear to have evolved a Nef-dependent but (G/D)DIWK-independent mechanism that is entirely distinct from that of the AP-2 dependent SIV Nefs, a key facet of this mechanism is the retention of tetherin in the TGN, the main locus of AP-1 activity (Kluge et al., 2014).
The closed trimer structure is competent to bind membranes but does not match the higher order geometry of the clathrin cage, nor does it appear capable of promoting clathrin assembly. This makes the structure ideally suited to retaining and sequestering cargo at the membrane where it first encounters AP-1. AP-1 predominantly acts at the TGN, thus the behaviour and structure of the closed trimer, and subcellular localization of AP-1 suggest a structural mechanism ideally suited to retaining tetherin in the TGN (Figure 6). Thus, the closed trimer binds to tetherin, Nef, and Arf1 at the TGN membrane, but does not progress efficiently to clathrin coat formation because the geometry of the closed trimer does not match that of the clathrin lattice. In contrast, MHC-I promotes formation of open trimers, which in turn promotes CCV formation that directs MHC-I to lysosomes for degradation.
Figure 6. Model for allosteric regulation of Nef target fate.
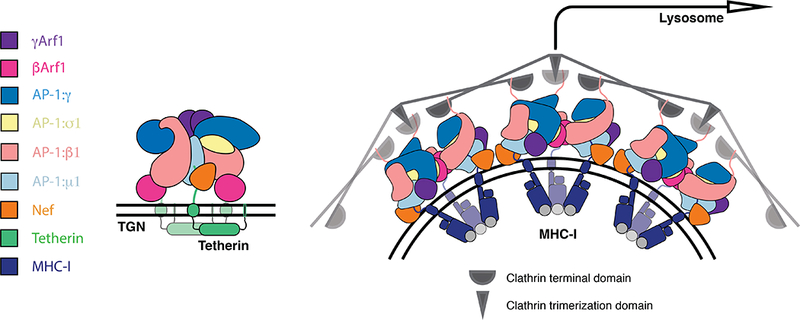
Tetherin cargo loaded AP- 1:Arf1 trimers are retained at the trans golgi network in a trimeric architecture not competent to assemble into higher species and thus recruit clathrin. MHC-I cargo loaded AP-1:Arf1 trimers may assemble through allosterically induced architectural and assembly changes that permits higher order assembly, clathrin recruitment and trafficking to the lysosome. See also Figure S6.
One apparent contradictory aspect of this model is that we observed closed trimer formation with both NL4–3 and O-MRCA Nefs, despite that NL4–3 Nef is known to be ineffective in driving TGN retention of tetherin. We noticed NL4–3 and most other M- group Nefs, but not O-Nefs, are predicted to be phosphorylated on the LL loop by CK1, and confirmed the phosphorylation of NL4–3 Nef experimentally. Notably, the Ser phosphorylation site is found in most HIV-1 M Nefs but generally absent in Nef proteins of SIVcpz, the direct precursor of pandemic HIV-1 strains. In agreement with the present results, it has been shown that substitution of S169C is critical for effective tetherin antagonism by HIV-1 Nefs but does not significantly affect other Nef functions (Gotz et al., 2012; Kluge et al., 2014). Our results thus explain how M-Nefs were functionally inactivated with respect to TGN retention of tetherin because HIV-1 M strains evolved Vpu as potent tetherin antagonist during adaption to the new human host.
Considerable effort has gone into characterizing how the allosteric unlocking of single AP monomers regulates their ability to bind to cargo (Jackson et al., 2010; Kelly et al., 2008), membranes (Collins et al., 2002; Ren et al., 2013), and clathrin (Kelly et al., 2014). We have now shown that in the context of HIV-1 infection, allosteric regulation also occurs at the level of alternative trimerization of AP-1 complexes. It remains to be clarified why TGN retention, as opposed to lysosomal degradation, seems to be advantageous for HIV-1. Possibly, retention in a subdomain of the TGN might help prevent the tetherin sorting pathway from intersecting that of its substrate, HIV-1 Env. Beyond the implications for HIV-1 Nef, the larger implication of these results is that higher order assemblies of vesicle coat adaptors are remarkably plastic, and that this plasticity can be harnessed for regulatory purposes.
STAR METHODS
CONTACT FOR REAGENT AND SOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contacts, Xuefeng Ren (snowren@berkeley.edu) or James H. Hurley (jimhurley@berkeley.edu).
METHOD DETAILS
Plasmid construction
The His6- and GST-tagged AP-1 constructs were previously described (Shen et al., 2015). MHC-I (338–365)-NL4–3 Nef (no linker) (Jia et al., 2012), Tetherin (1–21)-10aa linker- NL4–3 Nef and Tetherin (1–21) −10aa linker-O-MRCA (Kluge et al., 2014) Nef were expressed as TEV-cleavable N-terminal His6 fusions. The rat CK1δ (1–317) gene was codon-optimized, synthesized using gblocks (Integrated DNA Technologies, Coralville, Iowa), and then subcloned into LIC 2C-T vector, which was expressed as TEV-cleavable N-terminal His6-MBP fusion. Bicistronic CMV promoter-based pCG expression vectors coexpressing parental or mutant Nef proteins were generated as described (Kluge et al., 2014). All constructs used in this study are listed in Table S1.
Protein purification
The AP-1 complexes were expressed in BL21 (DE3) pLysS (Promega, Madison, WI) strains and induced with 0.3 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 20°C overnight. The cells were lysed by sonication in 50 mM Tris at pH 8.0, 300 mM NaCl, 10% glycerol, 3 mM β-mercaptoethanol (β-ME), and 0.5 mM phenylmethanesulfonyl fluoride (PMSF). The clarified lysate was first purified on a Ni-nitrilotriacetic acid (NTA) column (Qiagen, Valencia, CA). The eluate was further purified on glutathione-Sepharose 4B resin (GE Healthcare, Piscataway, NJ). After TEV cleavage at 4°C overnight, the sample was concentrated and then loaded onto a HiLoad 16/60 Superdex 200 column (GE Healthcare) in 20 mM Tris at pH 8.0, 200 mM NaCl, and 0.3 mM tris(2-carboxyethyl)phosphine (TCEP). The sample fractions were pooled together, adjusted to 30 mM imidazole, and passed through 1 ml of glutathione-Sepharose 4B and then onto a Ni-NTA column (Qiagen) to capture the residual GST- and His-tag fragments. The sample was adjusted to 20 mM Tris at pH 8.0, 200 mM NaCl, and 0.3 mM TCEP by buffer exchange in the concentrator.
His6-tagged tetherin-Nef and His6-tagged Arf1 constructs were expressed in BL21 (DE3) Star cells and induced with 0.3 mM IPTG at 20°C overnight. The cell pellet was lysed by sonication and purified on a Ni-NTA column in 50 mM Tris at pH 8.0, 300 mM NaCl, 20 mM imidazole, 5 mM MgCl2,3 mM β-ME, and 0.5 mM phenylmethanesulfonyl fluoride (PMSF). The proteins were eluted with 300 mM imidazole and then loaded onto a HiLoad 16/60 Superdex 75 column (GE Healthcare) in 20 mM Tris at pH 8.0, 200 mM NaCl, 5 mM MgCl and 0.3 mM TCEP. The sample fractions were pooled and proteins were quantified by molar absorption measurements.
AP-1:Arf1:tetherin-Nef complex assembly
The recombinant AP-1 core was mixed with Arf1 and tetherin-Nef proteins at a molar ratio of 1:4:6 and then incubated with 1 mM GTP at 4°C overnight. The mixture was then subjected to a Superose 6 10/100GL column in 20 mM Tris at pH 8.0, 200 mM NaCl, 5 mM MgCl2, and 0.5 mM TCEP. For the purposes of EM analysis, fractions corresponding to AP-1:Arf1:tetherin-Nef complexes were pooled and concentrated for cryoEM analysis.
Cryo-electron microscopy
Samples for data collection were prepared on 1.2/1.3 C-flat holey carbon copper grids that were prepared by plasma cleaning for 10 s using a Solarus plasma cleaner (Gatan Inc., Pleasanton, CA). Reconstituted complex at 0.7 mgmL−1 was applied in a 3 µL drop to one side of the grid and plunge frozen into liquid ethane using a Vitrobot mark VI. The humidity was controlled at 100%, 22 °C and the grids blotted with Whatman #1 paper using a blotand a magnified pixel size force of 8 for between 3.5 and 4.0 seconds. 2,200 micrographs were collected on a Titan Krios (FEI; BACEM UCB) at a nominal magnification of 22,500X and a magnified pixel size of 1.067 Å pix−1. The dose rate was 6.57 e−/Å2/sec at the sample and achieved with an illuminated area sufficient to cover one hole and the surrounding carbon support. Data collection was carried out using SerialEM with one exposure per hole and focusing for each exposure, the defocus range was set to collect between 0.75 and 2.00 µm defocus. Movies were acquired on a K2 direct electron detector (Gatan) in superresolution counting mode with a dose fractionated frame rate of 250 ms and total collection time of 9500 ms.
AP1:Arf1:tetherin-Nef (L2A) cryo grids were prepared identically to the WT AP1:Arf1:tetherin-Nef sample. 1,989 micrographs were collected on a Titan Krios at a nominal magnification of 22,500X and a magnified pixel size of 1.067 Åpix−1. The dose rate was 7.09 e−/Å2/sec at the sample. Data collection was carried out using SerialEM as reported for the WT AP1:Arf1:tetherin-Nef sample except with a defocus range between 0.75 and 2.50 µm and a dose fractionated frame rate of 200 ms and total collection time of 5600 ms. Full details of data sets are listed in Table S2.
Image processing
Movie data were processed using MotionCor2–1.0.0 (Li et al., 2013; Zheng et al., 2017) with internal 2-fold Fourier binning to return the super resolution pixel to the nominal magnification pixel size of 1.067 Å pix−1, discarding the initial 2 frames, 5 by 5 patch based alignment and dose weighting up to the total exposure of 62.4 e−/Å2. The contrast transfer function of full dose non-weighted micrographs was estimated using Gctf-v1.06 (Zhang, 2016). 252,212 particles were picked from non-dose-weighted micrographs using Gautomatch-v0.50, particles were extracted in a 384 pixel box and subjected to three rounds of 2D classification at 3-fold binning in Relion-2.0 (Kimanius et al., 2016; Scheres, 2012) resulting in 64 classes with 209,816 selected particles. Ab-initio 3D classification was performed in cryoSPARC (Punjani et al., 2017) extracting a monomer structure with 75,442 particles that could be refined to 4.23 Å and over three 3D classifications and subset selections leading to a closed trimer structure with 61,948 particles reaching 6.25 Å. The closed trimer particles were refined in Relion-2.0 to 4.27 Å in C1, low occupancy density was observed for one subunit attributable to complex mobility. The closed trimer particle dataset could be further 3D classified in cryoSPARC to obtain a structure from a stable subset of 11,108 particles which refined to 6.83 Å in C1 using Relion-2.0 (Table S2), exhibiting high occupancy density for all three subunits (Figure S1). To obtain a high resolution structure of the monomeric subunit of the closed trimer from the 61,948 particle set we used localized reconstruction (LocalRec) methods (Ilca et al., 2015). This accounts for the quasi-C3 symmetry present in the complex due to local monomeric subunit movement by extracting monomeric subunits of the closed trimer as new subparticles for further classification and refinement (Figure S2). The trimer subunits (containing the AP1 core, two copies of Arf1 and two copies of Nef) were localized, recentered and extracted in new 224 pixel boxes using LocalRec, Scipion (de la Rosa-Trevin et al., 2016) and in house written scripts. For each subunit extraction, a vector describing the center of the whole closed trimer to the center of mass of the AP1 subunit was defined in UCSF Chimera (Pettersen et al., 2004). Model coordinates of each subunit were rigid-body fitted to the subparticle described by the defined vector and used to segment the map using scolor by selection within 25 Å of Cα atoms. The remaining density was softened by multiplication with at 3 σ mask extended and softened by 4 and 9 pixels, this was used for signal subtraction to leave only the subunit of interest. Using the Relion refinement parameters and vectors, LocalRec was used to localize the subparticles in every whole closed trimer particle image with and without signal subtraction. These were extracted as new subparticles, with recentered origins. The subparticle extraction was repeated for each monomer of the closed trimer structure resulting in 185,787 extracted subparticles. Two rounds of 2D classification cleaned the subparticle image stack to 139,172 particles. The subparticles were refined in Relion-2.1 to 3.93 Å with a measured map B-factor of −164 Å2. A masked 3D classification into 3 classes with Tau 10 and no angular searches produced a particle stack of 53,841 subparticles that were refined to 3.73 Å with a measured map B-factor of −137 Å2. For focused 3D classification of the Nef density a mask was created around the segmented density of Nef and 3D classification performed on the density inside this mask without sampling of the angles obtained from the 3D refinement, into 3 classes with Tau 16. This successfully characterized the high, partial or low occupancy of the Nef density.
For the AP1:Arf1:tetherin-Nef (L2A) sample, movie data were processed using MotionCor2–1.0.0 and the contrast transfer function of full dose non-weighted micrographs was estimated using Gctf-v1.06. 162,309 particles were picked from nondose-weighted micrographs using Gautomatch-v0.50. Particles were extracted in a 384 pixel box and subjected to a round of 2D classification at 4-fold binning in Relion-2.1. Seventeen selected classes were then used for template based Auto-picking in Relion-2.1, which yielded 399,032 particles. These particles were subjected to 4 iterative rounds of 2D classification in Relion-2.0, resulting in 51 classes with 162,509 selected particles. Ab-initio 3D classification was performed in cryoSPARC, extracting two dimer structures with 17,994 particles and 42,902 particles that could be refined to 8.64 Å and 7.80 Å, respectively. The two dimer particle angles and reference volumes were further -refined in Relion-2.0 to 7.31 Å and 6.72 Å, respectively, in C1. Similar to the wild-type AP1:Arf1:tetherin-Nef, the L2A dimer structure presented symmetry mismatching due to local subunit movement. Localized reconstruction subparticle extraction was performed in Relion on the of the 6.72 Å dimer subunit as described above. Subparticles containing the AP1 core and two copies of Arf1 were localized, centered and extracted in new 192 pixel boxes as shown in Figure S2. 85,804 subparticles were subjected to a round of 2D classification leading to 85,529 particles. The subparticles were refined in Relion-2.1 to 4.27 Å with a measured map b-factor of −151 Å2.
Local resolution estimation was performed using Relion. The particle orientation distribution was assessed for all maps by calculating cryoEF scores (Naydenova and Russo, 2017) and reconstruction resolution isotropy using the 3DFSC algorithm (Tan et al., 2017).
Modeling
Modeling for the closed trimer monomeric subunit was performed in UCSF Chimera (Pettersen et al., 2004) using starting models from PDB entries 4P6Z, 4HMY and 4EN2, followed by manual adjustment in Coot (Emsley et al., 2010). Structure refinement using Phenix real space refinement (Adams et al., 2010) was used to improve the model geometry and fit to the 3.73 Å LocalRec density B-factor sharpened at −136 Å2. GTP nucleotide modeled into γArf1 was constrained during refinement using ligand-protein distances from 1O3Y. Map-to-model comparison in phenix mtriage validated that no overfitting was present in the structures and emRinger scores were calculated (Barad et al., 2015) to identify regions requiring remodeling in Coot (Emsley et al., 2010) before a final 5 cycle real space refinement in Phenix. Atomic displacement factor refinement was used to calculate the residue B-factors. The final model map cross correlation was 0.76, with a map-vs-model FSC0.5 of 3.85 Å and an average model atomic displacement factor (ADP) of 29.3 Å2 for the AP1:Arf1:tetherin-Nef trimer. The entire closed trimer was modeled by fitting of the monomeric subunit atomic coordinates back into the whole trimer reconstruction at 6.83 Å (b-factor sharpening −100 Å2) and optimising using Phenix real space refinement. For the mutant AP1:Arf1:tetherin-Nef (L2A) dimer modeling was performed in the B-factor sharpened map of −151.2 Å2 and the final model map cross correlation was 0.77, with a FSC0.5 of 4.35 and an average ADP of 83.2 Å2. The entire dimer was modeled by fitting of the monomeric subunit atomic coordinates into the whole dimer reconstruction at 6.72 Å and optimising using Phenix real space refinement. Model geometry was validated for all models using MolProbity (Davis et al., 2007). All map and model statistics are detailed in Table. S2.
Negative stain electron microscopy
AP1:Arf1 was reconstituted with either MHC-I-Nef or tetherin-Nef as described and 4µL applied at 0.07 mgmL−1 to glow discharged continuous carbon 300 mesh Cu grids for negative stain. The sample was incubated for 1 minute, blotted and stained by twice by 1 minute incubation with 4 µL 2% w/v uranyl acetate. All samples were visualized on a FEI Technai 12 microscope operated at 120 kV, using a Tietz 4K camera, at a nominal magnification of 68,000 corresponding to a pixel size of 1.6 Å using an approximate dose of 35 e−/Å2. Data were collected manually and image processing carried out in Relion-2.0.
HDX-MS
After the AP-1 trimer was separated on a Superpose 6 column, the trimer fraction was pooled and concentrated to 20 μM. Amide HDX-MS was initiated by a 20-fold dilution of AP-1 trimer complex into a D2O buffer containing 20 mM HEPES (pD 7.2), 200 mM NaCl, 2 mM MgCl2 and 0.2 mM TCEP at room temperature. After intervals of 10 s-60 s, exchange was quenched at 0 °C with the addition of ice-cold quench buffer (400 mM KH2PO4/H3PO4, pH 2.2). The samples were then injected onto an HPLC system (Agilent 1100) with in-line peptic digestion and desalting. Desalted peptides were eluted and directly analyzed by an Orbitrap Discovery mass spectrometer (Thermo Scientific). Peptide identification was carried out by running tandem MS/MS experiments. A Proteome Discoverer 2.1 (Thermo Scientific) search was used for peptide identification. Mass analysis of the peptide centroids was performed using HDExaminer (Sierra Analytics, Modesto, CA), followed by manual verification of each peptide. The deuteron content was adjusted for deuteron gain/loss during digestion and HPLC. Both nondeuterated and fully deuterated complexes were analyzed. Fully deuterated samples were prepared by three cycles of drying and re-solubilization in D2O and 6 M guanidinium hydrochloride.
Nef phosphorylation by CK1
40 μM Tetherin-NL4–3 Nef or NL4–3 Nef was incubated with His-MBP-CK1 (6 μM) in the sample buffer consisting of 4 mM MgCl2, 1 mM ATP at 30 °C for 2 hr. 5 μl of 20 μM sample was diluted into 195 μl of 0.05% TFA, then injected into Agilent 1100 HPLC with in-line peptic digestion. Digested peptides were identified and characterized by tandem mass spectrometry (MS/MS) in an Orbitrap Discovery mass spectrometer. Proteome Discoverer 2.1 software was used to search the phosphorylation site in NL4–3 Nef.
GST pull-down assay
20 μg of recombinant AP-1 coreΔµCTD-GST was immobilized on 30 μl glutathione Sepharose and incubated with the wild type or phosphorylated Nef samples at 4 °C overnight in 20 mM Tris pH 8, 200 mM NaCl, 0.1 mM TCEP. The beads were washed 4 times, mixed with 40 µl of SDS buffer and boiled for 3 min. 15 μl of each sample was subjected to reducing SDS/PAGE.
Tetherin antagonism
To determine the capability of Nef to antagonize tetherin, HEK293T cells were seeded in six-well plates and transfected with 4 μg of NL4–3 ΔvpuΔNef IRES eGFP, 1 μg Vpu or Nef expression plasmid and different amounts of tetherin expression vectors (6 well). At two days posttransfection supernatants were harvested and the yield of infectious HIV-1 was determined by a 96-well infection assay on TZM-bl indicator cells.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Stars refer to the difference from the EGFP control panel. *p < 0.05, **p < 0.01. Statistical calculations and group comparisons were performed using a two-tailed Student’s t test implemented in the Prism package version 4.0 (GraphPad Software).
DATA AND SOFTWARE AVAILABILITY
Image processing and modeling software
All software is available as detailed in the key resources table.
Cryo-EM map, raw data and model accession numbers
The entire wild type AP1:Arf1:tetherin-Nef dataset post 2D classification was deposited as EMPIAR-10176. For the wild type AP1:Arf1:tetherin-Nef sample, cryo-EM maps for the closed trimer were deposited as EMD-7458 (closed trimer), EMD-7563 (closed trimer stable subunit subset), EMD-7457 (trimer monomeric subunit) and EMD-7456 (trimer monomeric subunit after nef focused classification) and EMD-7455 (monomer). Particle stacks associated with EMD-7458 (closed trimer) and EMD-7457 (closed trimer monomeric subunit) were deposited as EMPIAR 10178 and 10177 respectively. PDB model coordinates were deposited as 6CRI (closed trimer), 6CM9 (trimer monomeric subunit) and 6DFF (monomer). The wild type AP1:Arf1:tetherin-Nef monomer map was deposited as EMD-7455. For the AP1:Arf1:tetherin-Nef (L2A) dileucine mutant sample, maps were deposited as EMD-7454 (dimer) and EMD-7453 (dimer monomeric subunit). PDB model coordinates were deposited as 6D84 (dimer) and 6D83 (dimer monomeric subunit). See Table S2.
Supplementary Material
Figure S1, related to Figure 1. Cryo-EM classification workflow. (A) The wild type closed trimer, monomeric subunit and monomer 3D classification workflow. (B) The Nef dileucine mutant dimer classification workflow. In both cases after cleaning by 2D classification in Relion, initial model generation and 3D classification was performed in cryoSPARC (Punjani et al., 2017). Closed trimer and dimer particles had their monomeric subunits extracted (as described in Figure S3) as subparticles for further refinement.
Figure S2, related to Figure 1. Subparticle extraction and refinement by localized reconstruction. The extraction of monomeric subunits from whole quasi-C3 symmetric complexes as subparticles (Ilca et al., 2015) permits further classification and refinement of the minimal monomeric subunit of the closed trimer. This permits the resolving of high quality structures despite the apparent local variability in trimeric and dimeric oligomeric states. (A) Using a fitted PDB to segment the initial reconstruction masks are created for signal subtraction to leave only the desired subparticle. Vectors describing the sub-particle locations are then used in conjunction with Relion refined parameters to extract and recenter the desired subunit. The subparticle extraction of the closed trimer monomeric subunit increases the particle count from 61,929 closed trimer particle images to 185,787 monomeric subunit subparticles, which could be further classified and refined (Figure S1). (B) The quality of the monomeric subunit density for the closed trimer after localized reconstruction, classification and refinement shown at the measured map B factor of -137 Å2. (C) The quality of the monomeric subunit density for the dileucine mutant dimer shown at the measured map B-factor of -151 Å2. (D) Monomers were identified in the AP-1:Arf1:Tetherin-Nef sample having both γArf1 and βArf1 bound but no resolvable Nef core. The monomeric subunit of the closed trimer was determined and has a resolvable Nef core. The AP-1 core and γArf1 subunits are sharpened at -50 Å2, Nef and βArf1 subunits are unsharpened for clarity. Both structures have the same AP-1 heterotetrameric core conformation. Thus both the AP-1 core of the monomer and monomeric subunit of the closed trimer are found to be hyper-unlocked when compared to the hyper-unlocked structure of a tetherin-Vpu fusion AP-1 structure (4P6Z) (Jia et al., 2014).
Figure S3, related to Figure 1. Map and model validation. (A) Local resolution estimation of the whole closed trimer complex (transparent low pass map filtered map shown for clarity), (B) the closed trimer monomeric subunit refinement and (C) the monomer refinement with angular distributions of the final refinements, cryoEF scores and 3DFSC sphericity. (D) Whole reconstruction gold-standard FSC resolution estimation of the closed trimer compared to the trimer monomeric subunit and true monomer. (E) emRinger and (F) map-vs-model validation of the closed trimer monomeric subunit atomic model structure. (G) Local resolution and angular distribution for the dileucine mutant dimer and (H) dimer monomeric subunit. (I) Whole reconstruction gold-standard FSC of the dileucine dimer compared to the monomeric subunit. Map B-factors are as reported by Relion whilst maps are shown without sharpening for clarity.
Figure S4, related to Figure 1, 2. Arf1 architecture and binding interfaces. (A) γArf1 contacts to AP-1:μ1 as well as (B) AP-1:γ in the closed trimer. (C) βArf1 contacts to AP-1:β1 in the closed trimer. (D) In the dileucine mutant dimer a novel βArf1:AP-1:γ interface is present (left) when compared to the crystal structure βArf1:AP-1:γ dimer contact in 4HMY (right). Both AP-1:σ1 and μ1 cargo binding sites are occupied by the LL motif of Nef and YxxΦ of tetherin respectively in the closed trimer (Figure 2A-C) as well as in (E) the wild-type monomer and (F) dileucine mutant dimer.
Figure S5, related to Figure 1, 2. Resolving Nef in the closed trimer by focused classification. (A) A mask (transparent orange) was used to enclose the segmented density of Nef and focused 3D classification without angular sampling determined 3 classes that could be refined to between 4.06 – 4.19 Å. (B) The three structures reveal a high, partial and low occupancy state for Nef in the closed trimer monomeric structure. (C) The high occupancy state shows a very good agreement between the Nef model and the refined density after focused classification. All volumes are B-factor sharpened by −50 Å2.
Figure S6, related to Figure 4. Supporting data for HDX data. Deuterium exchange data for Nef when bound to MBP-tagged AP-1: μ1-CTD in the case of (A) tetherin cargo or (B) MHC-I cargo fusion. HDX-MS data are shown in heat map format where peptide coverage is represented as rectangular strips above the protein sequence. (C) The difference map between bound MHC-I-Nef and tetherin-Nef reveals greater areas of protection on Nef when bound with MHC-I cargo.
Movie S1, related to figure 1. The closed trimer high resolution map and atomic model. The closed trimer monomeric subunit reconstruction shown as an assembled AP-1:Arf1:tetherin-Nef complex. The Nef trans AP-1 bridge, the γArf1 trimer, γArf1:AP-1:γ and βArf1:AP-1:β1 interfaces are highlighted. The AP-1 core and γArf1 subunits are sharpened at -50 Å2, Nef and βArf1 subunits are unsharpened for clarity.
Highlights.
Cryo-EM structure of the AP1 :Arf1 :tetherin-Nef closed trimer at 3.7 Å
AP-1 closed trimer is controlled by the Nef dileucine loop
Arf1 trimerizes in two ways depending on cargo and Nef dynamics
Nef dileucine loop APσ1 interaction is phosphoinhibited in M-Nefs
Acknowledgments
We thank D. Toso and P. Grob for cryo-EM support, G. Stjepanovic for training and assistance with HDX-MS, D. Krnavek for technical assistance, J. Huiskonen and P. Afonine for helpful advice and discussion, and D. Sauter for comments on the manuscript. Access to the FEI Titan Krios was provided through the BACEM UCB facility. This research was supported by NIH grants R01 AI 120691 (X. R.) and P50 GM082250 (J. H. H.) and an Advanced ERC grant ‘Anti-Virome’ (F. K.) and DFG- funded SFB 1279 and SPP 1923 (F. K.).
Footnotes
Author Contributions
J.H.H. and X.R. conceived the overall project. K.L.M., C.Z.B., X.R., C.M.S., E.H. performed research. K.L.M. and C.Z.B. performed wild-type and dileucine mutant EM analysis respectively, X.R. performed HDX-MS, MS/MS and mutational analysis, C.M.S. and E.H. performed in cell experiments. J.H.H., X.R. and F.K. supervised research. J.H.H., K.L.M., C.Z.B. and X.R. wrote the initial manuscript draft with editing by F.K. All assisted in manuscript preparation.
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D-Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barad BA, Echols N, Wang RYR, Cheng Y, DiMaio F, Adams PD, and Fraser JS (2015). EMRinger: side chain directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Aguilar RC, and Bonifacino JS (2001). Functional and physical interactions of the adaptor protein complex AP-4 with ADP-ribosylation factors (ARFs). EMBO J. 20, 6265–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celestino M, Calistri A, Del Vecchio C, Salata C, Chiuppesi F, Pistello M, Borsetti A, Palu G, and Parolin C (2012). Feline tetherin is characterized by a short N-terminal region and is counteracted by the feline immunodeficiency virus envelope glycoprotein. J Virol 86, 6688–6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R, Lindwasser O, Smith W, Hurley J, and Bonifacino J (2007). Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol 81, 3877–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shingai M, Welbourn S, Martin MA, Borrego P, Taveira N, and Strebel K (2016). Antagonism of BST-2/Tetherin Is a Conserved Function of the Env Glycoprotein of Primary HIV-2 Isolates. J Virol 90, 11062–11074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SH, Hitchin D, Noviello CM, and Guatelli JC (2006). HIV-1 Nef stabilizes AP-1 on membranes without inducing ARF1-independent de novo attachment. Virology 345, 148–155. [DOI] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, and Owen DJ (2002). Molecular architecture and functional model of the endocytic AP2 complex. Cell 109, 523–535. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, et al. (2007). MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rosa-Trevin JM, Quintana A, del Cano L, Zaldivar A, Foche I, Gutierrez J, Gomez-Blanco J, Burguet-Castell J, Cuenca-Alba J, Abrishami V, Vargas J, Oton J, Sharvo G, Vilas JL, Navas J, Conesa P, Kazemi M, Marabini R, Sorzano COS, and Carazo JM (2016). Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J Struct Biol 195, 93–99. [DOI] [PubMed] [Google Scholar]
- Dodonova SO, Aderhold P, Kopp J, Ganeva I, Rohling S, Hagen WJH, Sinning I, Wieland F, and Briggs JAG (2017). 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodonova SO, Diestelkoetter-Bachert P, von Appen A, Hagen WJ, Beck R, Wieland F, and Briggs JA (2015). A structure of the COPI coat and the role of coat proteins in membrane vesicles assembly. Science 349, 195–198. [DOI] [PubMed] [Google Scholar]
- Doray B, Lee I, Knisely J, Bu G, and Kornfeld S (2007). The gamma/sigma1 and alpha/sigma2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol Biol Cell 18, 1887–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M, Roy BB, Guiot-Guillain P, Mercier J, Binette J, Leung G, and Cohen EA (2009). Suppression of Tetherin-Restricting Activity upon Human Immunodeficiency Virus Type 1 Particle Release Correlates with Localization of Vpu in the trans-Golgi Network. J. Virol 83, 4574–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. Sect. D-Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen JR (2009). Analysis of Protein Conformation and Dynamics by Hydrogen/Deuterium Exchange MS. Anal. Chem 81, 7870–7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander SW (2006). Hydrogen exchange and mass spectrometry: A historical perspective. J. Am. Soc. Mass Spectrom 17, 1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J, and Miller A (1991). Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350, 508–511. [DOI] [PubMed] [Google Scholar]
- Gotz N, Sauter D, Usmani SM, Fritz JV, Goffinet C, Heigele A, Geyer M, Bibollet-Ruche F, Learn GH, Fackler OT, et al. (2012). Reacquisition of Nef- Mediated Tetherin Antagonism in a Single In Vivo Passage of HIV-1 through Its Original Chimpanzee Host. Cell Host Microbe 12, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser H, Lopez LA, Yang SJ, Oldenburg JE, Exline CM, Guatelli JC, and Cannon PM (2010). HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology 7, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EE, Macia E, Jing W, Yin HL, Kirchhausen T, and Harrison SC (2004). Crystal structure of the clathrin adaptor protein 1 core. Proc. Natl. Acad. Sci. U. S. A 101, 14108–14113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, and Skrzypek E (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilca SL, Kotecha A, Sun XY, Poranen MM, Stuart DI, and Huiskonen JT (2015). Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nature Communications 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L, Kelly B, McCoy A, Gaffry T, James L, Collins B, Honing S, Evans P, and Owen D (2010). A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 141, 1220–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Singh R, Homann R, Yang H, Guatelli J, and Xiong X (2012). Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat. Struct. Mol. Biol 19, 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Weber E, Tokarev A, Lewinski M, Rizk M, Suarez M, Guatelli J, and Xiong Y (2014). Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper MR, Roeth JF, Williams M, Filzen TM, Fleis RI, and Collins KL (2005). HIV-1 Nef disrupts antigen presentation early in the secretory pathway. J. Biol. Chem 280, 12840–12848. [DOI] [PubMed] [Google Scholar]
- Kelly BT, Graham SC, Liska N, Dannhauser PN, Hoening S, Ungewickell EJ, and Owen DJ (2014). AP2 controls clathrin polymerization with a membrane-activated switch. Science 345, 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BT, McCoy AJ, Spate K, Miller SE, Evans PR, Honing S, and Owen DJ (2008). A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature 456, 976–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SHW, and Lindahl E (2016). Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff F (2010). Immune Evasion and Counteraction of Restriction Factors by HIV-1 and Other Primate Lentiviruses. Cell Host Microbe 8, 55–67. [DOI] [PubMed] [Google Scholar]
- Kluge SF, Mack K, Iyer SS, Pujol FM, Heigele A, Learn GH, Usmani SM, Sauter D, Joas S, Hotter D, et al. (2014). Nef Proteins of Epidemic HIV-1 Group O Strains Antagonize Human Tetherin. Cell Host Microbe 16, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueck T, Foster TL, Weinelt J, Sumner JC, Pickering S, and Neil SJD (2015). Serine Phosphorylation of HIV-1 Vpu and Its Binding to Tetherin Regulates Interaction with Clathrin Adaptors. PLoS Path. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueck T, and Neil SJD (2012). A Cytoplasmic Tail Determinant in HIV-1 Vpu Mediates Targeting of Tetherin for Endosomal Degradation and Counteracts Interferon-Induced Restriction. PLoS Path. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tortorec A, and Neil SJ (2009). Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 83, 11966–11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XM, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, and Cheng YF (2013). Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubben NB, Sahlender DA, Motley AM, Lehner PJ, Benaroch P, and Robinson MS (2007). HIV-1 Nef-induced down-regulation of MHC class I requires AP-1 and clathrin but not PACS-1 and is impeded by AP-2. Molecular Biology of the Cell 18, 3351–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JH, Guevara RB, Marcano AC, Saenz DT, Fadel HJ, Rogstad DK, and Poeschla EM (2014). Feline immunodeficiency virus envelope glycoproteins antagonize tetherin through a distinctive mechanism that requires virion incorporation. J Virol 88, 3255–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenova K, and Russo CJ (2017). Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nature Communications 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJD, Zang T, and Bieniasz PD (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–U421. [DOI] [PubMed] [Google Scholar]
- Noviello CM, Benichou S, and Guatelli JC (2008). Cooperative binding of the class I major histocompatibility complex cytoplasmic domain and human immunodeficiency virus type 1 Nef to the endosomal AP-1 complex via its mu subunit. J. Virol. 82, 1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D, Collins B, and Evans P (2004). Adaptors for clathrin coats: structure and function. Annu Rev Cell Dev Biol 20, 153–191. [DOI] [PubMed] [Google Scholar]
- Owen DJ, and Evans PR (1998). A structural explanation for the recognition of tyrosine-based endocytotic signals. Science 282, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF chimera - A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Ren X, Farias GG, Canagarajah B, Bonifacino JS, and Hurley JH (2013). Structural Basis for Recruitment and Activation of the AP-1 Clathrin Adaptor Complex by Arf1. Cell 152, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Park SY, Bonifacino JS, and Hurley JH (2014). How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. eLife 3, e01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeth JF, Williams M, Kasper MR, Filzen TM, and Collins KL (2004). HIV-1 Nef disrupts MHC-1 trafficking by recruiting AP-1 to the MHC-1 cytoplasmic tail. J. Cell Biol 167, 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, et al. (2015). HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526, 212-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Muench J, Kim K-A, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, et al. (2009). Tetherin-Driven Adaptation of Vpu and Nef Function and the Evolution of Pandemic and Nonpandemic HIV-1 Strains. Cell Host Microbe 6, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Specht A, and Kirchhoff F (2010). Tetherin: Holding On and Letting Go. Cell 141, 392–398. [DOI] [PubMed] [Google Scholar]
- Scheres SHW (2012). RELION: Implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Fritz JV, Bitzegeio J, Fackler OT, and Keppler OT (2011). HIV-1 Vpu Blocks Recycling and Biosynthetic Transport of the Intrinsic Immunity Factor CD317/Tetherin To Overcome the Virion Release Restriction. Mbio 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Marechal V, LeGall S, Lemonnier F, and Heard JM (1996). Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med 2, 338–342. [DOI] [PubMed] [Google Scholar]
- Serra-Moreno R, Zimmermann K, Stern LJ, and Evans DT (2013). Tetherin/BST- 2 Antagonism by Nef Depends on a Direct Physical Interaction between Nef and Tetherin, and on Clathrin-mediated Endocytosis. PLoS Path. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QT, Ren XF, Zhang R, Lee IH, and Hurley JH (2015). HIV-1 Nef hijacks clathrin coats by stabilizing AP-1:Arf1 polygons. Science 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnes MA, and Rothman JE (1993). The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell 73, 999–1005. [DOI] [PubMed] [Google Scholar]
- Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, Carragher B, and Lyumkis D (2017). Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat Methods 14, 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarev A, and Guatelli J (2011). Misdirection of membrane trafficking by HIV-1 Vpu and Nef: Keys to viral virulence and persistence. Cell Logist. 1, 90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM, and Bonifacino JS (2013). Cargo Recognition in Clathrin-Mediated Endocytosis. Cold Spring Harbor Perspectives in Biology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM, Ostrom JA, and Kornfeld S (1993). Biochemical dissection of AP-1 recruitment onto Golgi membranes. J. Cell Biol 123, 561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Wu Y, and Goettlinger HG (2015). SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526, 218-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, and Guatelli J (2008). The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, and Hatziioannou T (2009). Nef Proteins from Simian Immunodeficiency Viruses Are Tetherin Antagonists. Cell Host Microbe 6, 54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FW, Landford WN, Ng M, McNatt MW, Bieniasz PD, and Hatziioannou T (2011). SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion Release. PLoS Path. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. Journal of Structural Biology 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1. Cryo-EM classification workflow. (A) The wild type closed trimer, monomeric subunit and monomer 3D classification workflow. (B) The Nef dileucine mutant dimer classification workflow. In both cases after cleaning by 2D classification in Relion, initial model generation and 3D classification was performed in cryoSPARC (Punjani et al., 2017). Closed trimer and dimer particles had their monomeric subunits extracted (as described in Figure S3) as subparticles for further refinement.
Figure S2, related to Figure 1. Subparticle extraction and refinement by localized reconstruction. The extraction of monomeric subunits from whole quasi-C3 symmetric complexes as subparticles (Ilca et al., 2015) permits further classification and refinement of the minimal monomeric subunit of the closed trimer. This permits the resolving of high quality structures despite the apparent local variability in trimeric and dimeric oligomeric states. (A) Using a fitted PDB to segment the initial reconstruction masks are created for signal subtraction to leave only the desired subparticle. Vectors describing the sub-particle locations are then used in conjunction with Relion refined parameters to extract and recenter the desired subunit. The subparticle extraction of the closed trimer monomeric subunit increases the particle count from 61,929 closed trimer particle images to 185,787 monomeric subunit subparticles, which could be further classified and refined (Figure S1). (B) The quality of the monomeric subunit density for the closed trimer after localized reconstruction, classification and refinement shown at the measured map B factor of -137 Å2. (C) The quality of the monomeric subunit density for the dileucine mutant dimer shown at the measured map B-factor of -151 Å2. (D) Monomers were identified in the AP-1:Arf1:Tetherin-Nef sample having both γArf1 and βArf1 bound but no resolvable Nef core. The monomeric subunit of the closed trimer was determined and has a resolvable Nef core. The AP-1 core and γArf1 subunits are sharpened at -50 Å2, Nef and βArf1 subunits are unsharpened for clarity. Both structures have the same AP-1 heterotetrameric core conformation. Thus both the AP-1 core of the monomer and monomeric subunit of the closed trimer are found to be hyper-unlocked when compared to the hyper-unlocked structure of a tetherin-Vpu fusion AP-1 structure (4P6Z) (Jia et al., 2014).
Figure S3, related to Figure 1. Map and model validation. (A) Local resolution estimation of the whole closed trimer complex (transparent low pass map filtered map shown for clarity), (B) the closed trimer monomeric subunit refinement and (C) the monomer refinement with angular distributions of the final refinements, cryoEF scores and 3DFSC sphericity. (D) Whole reconstruction gold-standard FSC resolution estimation of the closed trimer compared to the trimer monomeric subunit and true monomer. (E) emRinger and (F) map-vs-model validation of the closed trimer monomeric subunit atomic model structure. (G) Local resolution and angular distribution for the dileucine mutant dimer and (H) dimer monomeric subunit. (I) Whole reconstruction gold-standard FSC of the dileucine dimer compared to the monomeric subunit. Map B-factors are as reported by Relion whilst maps are shown without sharpening for clarity.
Figure S4, related to Figure 1, 2. Arf1 architecture and binding interfaces. (A) γArf1 contacts to AP-1:μ1 as well as (B) AP-1:γ in the closed trimer. (C) βArf1 contacts to AP-1:β1 in the closed trimer. (D) In the dileucine mutant dimer a novel βArf1:AP-1:γ interface is present (left) when compared to the crystal structure βArf1:AP-1:γ dimer contact in 4HMY (right). Both AP-1:σ1 and μ1 cargo binding sites are occupied by the LL motif of Nef and YxxΦ of tetherin respectively in the closed trimer (Figure 2A-C) as well as in (E) the wild-type monomer and (F) dileucine mutant dimer.
Figure S5, related to Figure 1, 2. Resolving Nef in the closed trimer by focused classification. (A) A mask (transparent orange) was used to enclose the segmented density of Nef and focused 3D classification without angular sampling determined 3 classes that could be refined to between 4.06 – 4.19 Å. (B) The three structures reveal a high, partial and low occupancy state for Nef in the closed trimer monomeric structure. (C) The high occupancy state shows a very good agreement between the Nef model and the refined density after focused classification. All volumes are B-factor sharpened by −50 Å2.
Figure S6, related to Figure 4. Supporting data for HDX data. Deuterium exchange data for Nef when bound to MBP-tagged AP-1: μ1-CTD in the case of (A) tetherin cargo or (B) MHC-I cargo fusion. HDX-MS data are shown in heat map format where peptide coverage is represented as rectangular strips above the protein sequence. (C) The difference map between bound MHC-I-Nef and tetherin-Nef reveals greater areas of protection on Nef when bound with MHC-I cargo.
Movie S1, related to figure 1. The closed trimer high resolution map and atomic model. The closed trimer monomeric subunit reconstruction shown as an assembled AP-1:Arf1:tetherin-Nef complex. The Nef trans AP-1 bridge, the γArf1 trimer, γArf1:AP-1:γ and βArf1:AP-1:β1 interfaces are highlighted. The AP-1 core and γArf1 subunits are sharpened at -50 Å2, Nef and βArf1 subunits are unsharpened for clarity.


