Abstract
Antibiotic resistance is a serious and growing threat to human health. The environmental microbiome is a rich reservoir of resistomes, offering opportunities to discover new antibiotic resistance genes. Here we demonstrate an integrative approach of utilizing gene sequence and protein structural information to characterize unidentified genes that are responsible for the resistance to the action of rifamycin antibiotic rifampin, a first-line antimicrobial agent to treat tuberculosis. Biochemical characterization of four environmental metagenomic proteins indicates that they are adenosine diphosphate (ADP)-ribosyltransferases and effective in the development of resistance to FDA-approved rifamycins. Our analysis suggests that even a single residue with low sequence conservation plays an important role in regulating the degrees of antibiotic resistance. In addition to advancing our understanding of antibiotic resistomes, this work demonstrates the importance of an integrative approach to discover new metagenomic genes and decipher their biochemical functions.
Introduction
Antibiotics are indispensable therapeutic agents for human health against infection from pathogenic bacteria. The emergence of resistance against antibiotics, however, has outpaced the discovery of new antibiotics. The Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO) declared that we are in the post-antibiotic era and are facing an antibiotic resistance crisis. This critical situation urges for immediate actions in politics, research, and health care systems1.
Tuberculosis is a widespread infectious disease, which is caused mostly by Mycobacterium tuberculosis2,3. Currently, nearly one-third of the global population is infected by the pathogenic bacteria. Rifampin, also known as rifampicin, belongs to the rifamycin family, and is a first-line antimicrobial agent against tuberculosis. Rifampin binds to the β subunit of DNA-dependent RNA polymerase, and inhibits the RNA synthesis of bacteria through steric occlusion4,5.
Since rifampin is an effective anti-tuberculosis agent, molecular mechanisms underlying the resistance to rifampin have been studied extensively6–9. A few mutations in bacterial RNA polymerase attenuate the binding affinity of rifampin10,11. Alternatively, rifampin can be modified by glycosylation12, adenosine diphosphate (ADP)-ribosylation13, monooxygenation14, or phosphorylation7, which also results in decreased binding affinity to RNA polymerase. The glycosyltransferase Rgt1438 confers rifampin resistance by converting rifampin to 23-O-glu-rifampin12. Rifampin ADP-ribosyltransferases (Arr) attach an ADP-ribosyl group to the hydroxyl group at the C21 position of rifampin to form ADP-ribosylrifampin (Additional file 1: Scheme 1a)13,15–17. Rifampin phosphotransferase (RPH), recently identified from Listeria monocytogenes7, is a kinase that modifies rifampin to phospho-rifampin at the C21 position7,18. The diversity in the chemical origins implies that antibiotic resistance can readily be acquired by multiple organisms. Therefore, antibiotic resistance genes and related proteins should be explored as immediate targets of research to understand comprehensive antibiotic resistomes19,20.
With the advances in sequencing technology, metagenomic approaches have been applied to identify antibiotic resistance genes from environmental samples21–24. Since many antibiotics originate from secondary metabolites of bacteria25,26, resistance determinants are ubiquitously found in microorganisms, even in ancient environments27–31. As such, reservoirs of resistance determinants have been investigated to profile abundant resistance genes and their effects on microbiome composition32. Nesme et al. analyzed antibiotic resistance gene determinants from 71 environmental shotgun metagenomic samples from a global environment survey33. Li et al. applied network analysis to identify 18 types and 260 subtypes of antibiotic resistance genes34.
In order to deduce the functions of particular genes and proteins, sequence similarity has been widely applied as an important measure. For example, resistome profiling35 or functional metagenomics36,37 have been applied to reveal the genes related to the antibiotic resistances. Studies of conserved residues alone, however, might not be sufficient for predicting the function of the genes38. An inherent limitation of sequence similarity-based approaches becomes more pronounced in determining the functions of newly discovered genes39. Recently, Balskus et al. have developed a profiling method by combining protein sequence similarity network analysis with quantitative metagenomics to discover an unidentified enzyme, suggesting that additional information can greatly improve the quality of metagenomic data40.
Within this context, we have developed an integrative approach of combining sequence analysis and molecular interaction analysis of the protein structure with highest sequence similarity. Potential arr genes (arr-wd), obtained from the sediment microbiome, were analyzed to locate conserved functional sites. As an experimental validation of our computational prediction, in vitro and in vivo resistance activity assays were carried out with rifamycins to assess the ADP-ribosyltransferase activity of expressed proteins. This work represents an example of using microbiome sequence data to discover proteins related to the antibiotic resistance, to construct detailed three-dimensional models to extract functionally important structural features, and to validate computational predictions by biochemical experimental studies.
Results and Discussion
Novel rifamycin ADP-ribosyltransferase genes in the sediment microbiome
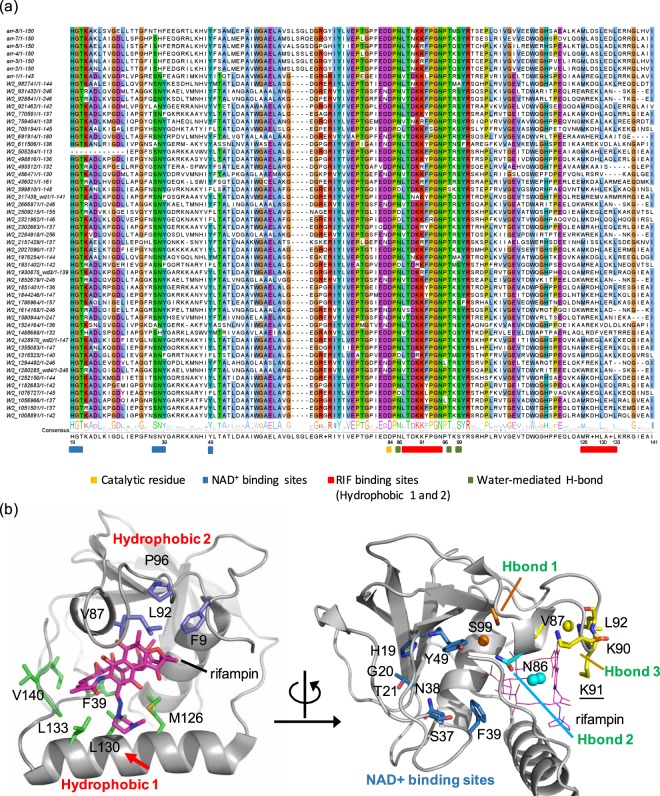
A total of 45 rifamycin ADP-ribosyl transferase (arr) homologous genes (arr-wd) were obtained from the sediment microbiome with the query of seven arr genes annotated in the comprehensive antibiotic resistance database (CARD) (Additional file 1: Table S1)41,42. The similarity between the seven known arr genes and 45 arr-wd genes ranges from 48.23 to 71.01%. These 45 putative arr genes were aligned with the seven known arr genes to find the conserved functional regions in the amino acid sequences (Fig. 1a). ADP-ribosyltransferases exhibit multiple conserved motifs that are presumably related to rifampin binding and nicotinamide adenine dinucleotide (NAD+) binding through hydrogen bonding, hydrophobic interactions, and cation-pi interactions (Fig. 1b). Our results include the rifampin and NAD+ binding sites reported in the previous studies of arr-ms from Mycobacterium smegmatis, arr-sc from S. coelicolor, and arr-2 and homologous genes (arr-3, arr-4, arr-5, arr-7, and arr-8) from several gram-negative pathogenic bacteria13,43–46. In particular, H19 and Y49, which were identified for NAD+ binding13, were highly conserved in our alignment.
Figure 1.
Integrated analysis of rifamycin ADP-ribosyltransferase (Arr). (a) The sequence conservation of rifamycin ADP-ribosyltransferase (Arr). The residues that are conserved more than 78% of the sequences are highlighted based on the clustalw color scheme. The genes selected for experimental validation are shown as wd1-wd4. (b) Proposed key residues and their molecular interactions in the rifampin-binding sites of Arr-ms structure (PDB 2HW2). In Arr-ms, the variable X residue in M[R|K][D|E]XL motif is Gly129, and its position is highlighted with a red arrow. Water molecules are depicted with spheres.
Structure-guided conservation analysis
It has been proposed that NAD+ binding sites in ADP-ribosyltransferases are formed by a long flexible loop between β1 and β3 strands (Fig. 1b; Additional file 1: Figure S1), and several residues constituting the loop are essential for the function of Arr13. A HGT motif (H19, G20, and T21 in Arr-ms) at the end of β1 strand and a tyrosine (Y49 in Arr-ms) in β3 strand are spatially adjacent to an aspartate (D84). Site-directed mutagenesis studies have demonstrated that these amino acids, H19, Y49, and D84 in Arr-ms, are critical for the catalysis through hydrogen-bonding interactions with NAD+13. Indeed, these residues are highly conserved in the putative rifampin ADP-ribosyltransferase genes of the current study: 98.0% for H19, 87.0% for Y49, and 100% for D84 (Fig. 1a).
In addition to the HGT motif, we found that SN[Y|F] motif in the flexible loop region between β2 and β3 strands (Figs 1a and 2b; Additional file 1: Figure S1) is highly conserved (92.0% for S, 87.0% for N, 79% for Y, 21% for F). Although the X-ray crystal structure of Arr complexed with NAD+ is not available, a docking simulation indicates that the SN[Y|F] motif is likely to be located at the entrance to the active site, providing the shape and/or charge complementarity to the sugar-phosphate group of NAD+ (Additional file 1: Figure S2). Presumably, such non-covalent interaction with HGT and SN[Y|F] motifs polarizes the nicotinamide portion of NAD+ to help generate the oxocarbenium intermediate.
Figure 2.
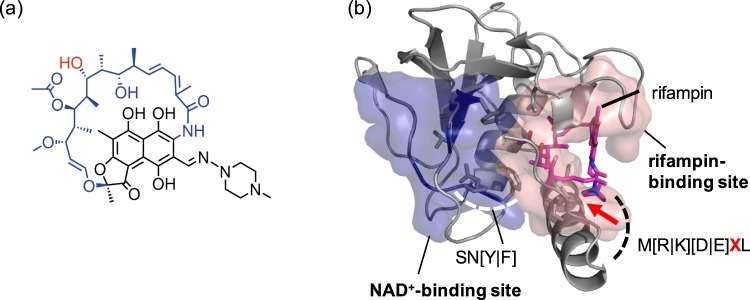
Structural analysis of rifamycin ADP-ribosyltransferase (Arr). (a) A structure of rifampin. The aliphatic ansa chain and the site of modification by ADP-ribosyltransferase is colored in blue and red, respectively. (b) Two substrate-binding pockets depicted in the structure of rifamycin ADP-ribosyltransferase from arr-ms (PDB 2HW2). The variable X residue in M[R|K][D|E]XL motif is highlighted with a red arrow.
The X-ray crystal structure of Arr-ms (PDB ID 2HW2)13 displays a large binding pocket to accommodate rifampin (solvent accessible surface area = 621.95 Å2), comprised of amino acid residues from α1 and α2 helices and a loop between β5 and β6 strands. We categorized the residues into four types, depending on their molecular interactions, hydrophobic 1, hydrophobic 2, cation-pi, and water-mediated hydrogen bond (Fig. 1b). High conservation was indeed observed in the arr genes that we predicted from the metagenomic data. All key amino acid residues comprising the rifampin-binding site were well-conserved (Fig. 1a).
The hydrophobic 1 region is a main hydrophobic pocket for rifampin-binding. It is comprised of hydrophobic bulky residues, F39, M126, L130, L133, A139, and I141 in Arr-ms (Fig. 1b). The three residues M126, L130, and L133, surrounding the piperazine ring portion of rifampin, are located in helix α2. The piperazine ring of rifampin is likely to be inserted between L133 and M126 by a hydrophobic interaction. L130 also possesses hydrophobic characteristics and is highly conserved in our metagenome sequences [L(88.0%)|V(10.0%)|I(2.0%)], thus contributing to the formation of hydrophobic pocket that is positioned close to the piperazine ring. In helix α2, M[R|K][D|E]XL residues seem to be essential for rifampin-binding; they are highly conserved for both the reference and metagenome sample sequences. Since the methionine, the fourth amino acid (labeled as X), and two leucine residues are positioned toward the pocket in Arr-ms, a variation of these key residues would regulate the binding affinity and/or activities. In the aliphatic ansa chain of rifampin (Fig. 2a), two methyl groups (C31 and C32) and diene units are surrounded by F39, A139, and I141, which allow hydrophobic interactions.
The hydrophobic 2 region is a shallow and rather surface-exposed pocket. This region consists of F9, V87, L92, and P96 (Fig. 1b). While the residues F9, L92, and P96 are positioned in the loops, and directly exposed to the aqueous environment, they are spatially close to engage in hydrophobic interaction with a methyl group (C14) of rifampin. The residues L92 and P96 are located in the loop structure between β5 and β6 strands, and are part of the rifampin binding loop13. The residue V87 is positioned between β5 and β6 strands, generating a small α-helix structure.
K91 is located in the flexible loop structure between β5 and β6 strands (Fig. 1b). The residue is positioned above the naphthoquinone ring portion of rifampin, and the positively charged ε-ammonium (NH3+) of K91 makes a cation–pi contact. The residue is highly conserved in most of the Arr protein, constituting 85% of lysine and 15% of arginine. This spatial arrangement effectively shields the aromatic rings from exposure to the aqueous environment. The adjacent amino acids are also highly conserved as [L|V]TK[K|R]P: 73.0% for L; 98.0% for T; 94.0% for K; 85% for K; 96.0% for P.
In addition to the rifampin–Arr-ms interactions discussed above, we also observe three water-mediated hydrogen bonds, which might be necessary for rifampin binding (Fig. 1b, Additional file 1: Figure S1). In the rifampin binding loop, a water molecule (numbered as 1226) forms hydrogen bonding networks between a hydroxyl group (O2, 3.07 Å) of the rifampin naphthoquinone ring and backbone amide groups from V87, T88, K90, and L92 residues (2.89–3.04 Å). As discussed above, these residues are also involved in the hydrophobic interaction with rifampin, which might further contribute to the binding affinity. Another water molecule (numbered as 1335) mediates hydrogen bonding between the carbonyl oxygen atom of the O-acetyl (O8, 3.26 Å) group of rifampin and the hydroxyl group of S99 (OG, 2.77 Å). In addition, two water molecules (numbered as 1230 and 1381) form hydrogen bonding networks between N86, V140, and the N–H group of rifampin. These conserved residues are represented as DPN[V|L]. Here, D84 denotes the catalytic residue for the ADP-ribosylation function in Arr-ms13.
Phylogenetic relationship of arr genes
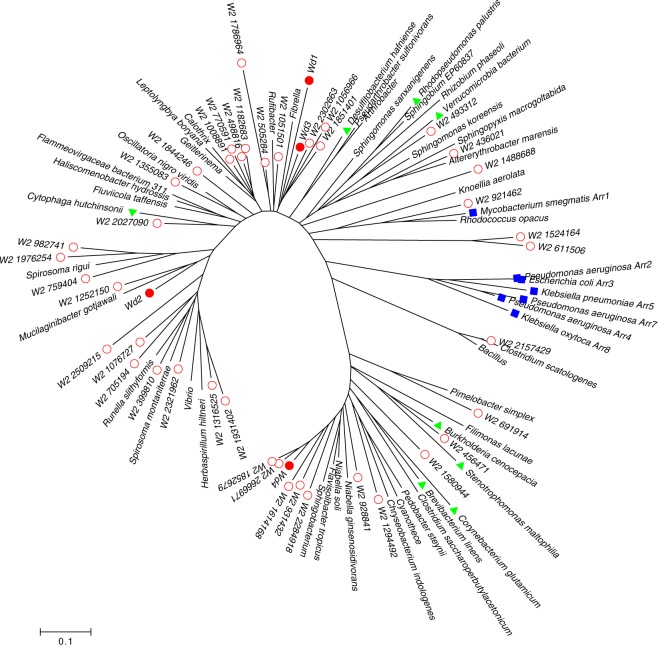
In order to find the phylogenetic relationship of the newly found arr genes, arr-wd, with the known genes, a neighbor-joining tree was built. In the phylogenetic tree, four arr-wd genes (arr-wd1-4) experimentally validated in this study were analyzed with the known arr genes (Fig. 3). In addition to arr-ms and known arr genes from gram-negative bacteria (arr-2, arr-3, arr-4, arr-5, arr-7, and arr-8; denoted as blue filled rectangles in Fig. 3), arr-wd1, arr-wd2, arr-wd3, and arr-wd4 (denoted as red filled circles in Fig. 3) are placed with the homologous arr genes identified from the complete genomes. In particular, arr-wd1, arr-wd2, and arr-wd3 are clustered with the arr genes from Rhodopseudomonas palustris, Desulfitobacterium hafniense, Verrucomicrobium spinosum, and Cytophaga hutchinsonii, which were used in the phylogenetic tree of a previous study13. On the other hand, arr-wd4 is clustered with the genes from Brevibacterium linens, Corynebacterium glutamicum, Burkholderia cenocepacia, and Stenotrophomonas maltophilia. While the previous study showed a wide distribution of arr genes in the phylogenetic tree13, our study expanded the phylogenetic tree by incorporating diverse arr genes from an environmental sample and validating them experimentally.
Figure 3.
A phylogenetic tree of the metagenomic arr genes and well-known arr genes. Red circles denote arr genes collected from the metagenomic sample in this work. Red filled circles denote arr genes validated experimentally in this work. Blue rectangles denote previously known and experimentally validated arr genes. Green triangles denote arr genes referenced previously but not validated experimentally.
To find the origins of the microbial genomes, the arr-wd genes were investigated by using homology search. We found that the arr gene W2_2157429 shows the highest homology (98.54% protein similarity) to Clostridium ljungdahlii. A total of 13 out of 45 arr-wd genes show high homology (>80% protein similarity) to the known species (Additional file 1: Table S2), implying that arr-wd like genes might be prevalent in various organisms, although they are yet to be annotated.
Biochemical analysis of representative arr genes
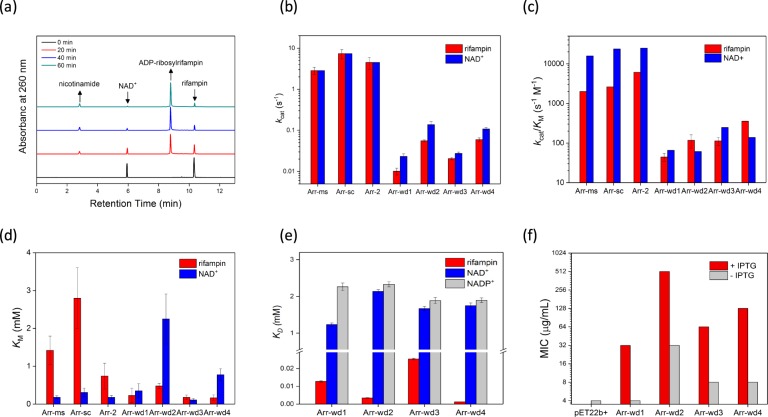
To evaluate whether the newly discovered genes display ADP-ribosyltransferase activity, we selected four metagenomic genes (arr-wd 1-4) and expressed the corresponding proteins (Additional file 1: Table S3 and Figure S3). All four putative arr genes possess the conservation of the essential functional residues (Fig. 1a), comprised of NAD+ and rifampin-binding pockets (Fig. 2), and exhibit rifampin ADP-ribosyltransferase activities (Fig. 4; Additional file 1: Tables S5–S6 and Figure S4). The activities are relatively lower than the previously characterized arr genes13 by about 1–2 orders of magnitude in both turnover number (kcat) and catalytic efficiency (kcat/KM). The data, however, definitively indicate that all Arr-wd proteins discovered herein are ADP-ribosyltransferases that modify rifampin upon the reaction with NAD+, and the metagenomic sequences can be denoted as arr genes that induce the resistance of rifampin. Interestingly, Michaelis constant (KM) and the dissociation constant (KD) of the Arr-wd proteins for rifampin are significantly lower than those of the previously reported Arr proteins (Fig. 4d,e; Additional file 1: Table S7). These results indicate that Arr-wd proteins exhibit tighter binding to the rifampin than other Arr proteins, and the arr-wd genes could start to develop resistance even at low concentration levels of antibiotics when sufficient kcat is provided.
Figure 4.
Biochemical analysis of Arr-wd 1–4 proteins. (a) A representative HPLC trace of rifampin ADP-ribosylation by Arr-wd proteins. (b–d) Steady-state ADP-ribosyltransferase activities of Arr-wd proteins together with the reported values of Arr proteins. (b) Turnover numbers, kcat (s−1) (c) Catalytic efficiencies, kcat/KM (s−1 M−1) (d) Michaelis constants, KM (mM) (e) Dissociation constants, KD (mM) (f) MIC values (μg/mL). Values reported in (b–d) and (e) are derived from the fit to the Michaelis-Menten equation and Stern-Volmer equation, respectively.
The binding constants for NAD+ and rifampin were also measured with Arr-wd3 protein without the His-tag (Arr-wd3 + Δ6His) (Additional file 1: Table S7). The protein yields only slightly perturbed values, indicating that Arr-wd proteins expressed with the His-tag are valid for evaluating the biochemical activities.
The in vivo activities of the proteins were determined by the minimum inhibitory concentration (MIC) values (Fig. 4f; Additional file 1: Table S9). Their MIC values are slightly lower than other arr proteins, presumably due to their lower in vitro activities of Arr-wd proteins. Their MIC values, however, are considerably higher than the background level, again suggesting that the arr-wd genes can inactivate the antibiotics in considerable rates. The ADP-ribosyltransferase activities measured both in vitro and in vivo conditions, therefore, convincingly demonstrate that we have discovered a novel set of arr genes from environmental metagenomes, and have predicted their biochemical function.
Biochemical analysis of NAD+ binding sites
Our structure-guided conservation analysis and in vitro activity assay described above suggest that putative Arr-wd proteins are likely to have structurally well-defined NAD+ and rifampin-binding sites, which are essential for the dedicated chemical reaction. When NAD+ was replaced with nicotinamide adenine dinucleotide phosphate (NADP+), NADH, and NADPH, no ADP-ribosyltransferase activity was observed, indicating that Arr-wd proteins exhibit selective interaction with NAD+ over other analogous cofactors.
As described above, the SN[Y|F] motif is highly conserved, composing the putative NAD+-binding site: SNF in Arr-wd1 and SNY in Arr-wd2-4. The variation of the third residue between Phe and Tyr seems to be insignificant because there is no clear correlation between the sequence and binding affinity (KD) or ADP-ribosyltransferase activity (Additional file 1: Tables S6–S7 and Figures S4–S5). Therefore, only the hydrophobic interactions with the benzene ring of the side chain seems to be sufficient for retaining the interaction with NAD+. Since the Tyr or Phe residue is located at the intersection of NAD+ and rifampin-binding sites, it is possible that this hydrophobic residue also protects the oxocarbenium ion from other side reactions (Additional file 1: Scheme S1b and Figure S2).
Our sequence analysis also demonstrate that a large portion of the previously reported genes have SH[F], instead of the SN[Y|F] motif, with histidine (H) rather than asparagine (N) in the second position. To evaluate whether the sequence variation is related to the action of rifampin ADP-ribosyltransferase, we carried out site-directed mutagenesis of one of the metagenomes, arr-wd3. The N34H mutant of Arr-wd3 yields 6-fold lower kcat/KM and slightly elevated KD values than Arr-wd3 (Additional file 1: Tables S7–S8). The results imply that the residue is involved in NAD+-binding and/or positioning, and the N34H mutation perturbs the interactions with NAD+. Interestingly, the KD value for rifampin has been also altered by the N34H mutation, suggesting that the mutation might have allosterically influenced the thermodynamic parameters associated with the rifampin-binding event.
Biochemical analysis of rifampin-binding sites
Steady-state activity data demonstrate that the Arr-wd proteins are kinetically competent in rifampin ADP-ribosylation, although their activities are lower than those of the previously reported Arr proteins. Notably, however, the Arr-wd proteins exhibit considerably lower KM and KD values for rifampin, implicating that Arr-wd proteins can interact with rifampin in a thermodynamically favorable manner.
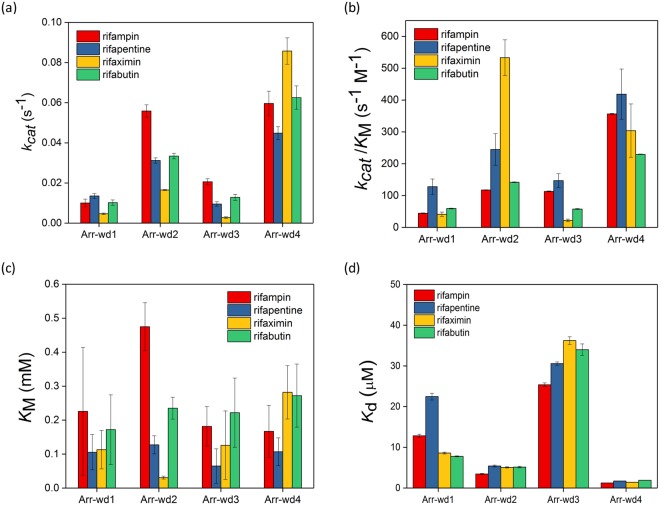
We evaluated whether the proteins can react with other semisynthetic rifampin analogues that are clinically approved and prescribed as drugs, such as rifapentine, rifaximin, and rifabutin (Additional file 1: Table S6 and Figures S6 and S7)47. All Arr-wd proteins react with these analogues to afford the corresponding ADP-ribosylated products (Table S5). However, their kinetic parameters are markedly altered depending on the substrates (Fig. 5), implying that modified chemical structure at the C8 position of the naphthoquinone core significantly changes the degrees of interaction with Arr-wd proteins. In general, Arr-wd2 and Arr-wd4 exhibit higher steady-state activity than Arr-wd1 and Arr-wd3. But Arr-wd2 and Arr-wd3 exhibit noticeably high and low kcat/KM for rifaximin, respectively, implying that specific interaction with the protein environment is operative in the ADP-ribosylation of the rifamycins. In contrast, rifapentine seems to be easily modified by all Arr-wd 1–4 proteins, indicating that the substrate is more promiscuous than other rifampin analogues.
Figure 5.
Kinetic and thermodynamic parameters of Arr-wd proteins from the steady-state reactions with rifampin derivatives, rifampin, rifapentine, rifaximin, and rifabutin. (a) Turnover numbers, kcat (s−1) (b) Catalytic efficiencies, kcat/KM (s−1 M−1) (c) Michaelis constants, KM (mM) (d) Dissociation constants, KD (μM). Values reported in (a–c) and (d) are derived from the fit to the Michaelis-Menten equation and Stern-Volmer equation, respectively.
To evaluate whether the altered catalytic activities are originated from the perturbation in the initial step of the reaction, i.e. association of the substrate to the protein, we measured the rifampin binding constants by monitoring intrinsic tryptophan fluorescence. The KD values are in the order of Arr-wd4 ≈ Arr-wd2 < Arr-wd1 < Arr-wd3. This trend is similar to the inverse of the kcat/KM, implying that substrate binding is likely to be the part of the rate-determining steps in catalysis. However, no significant difference was observed between the KD values of the rifampin analogues. Presumably, fluorescence changes of the tryptophan residue might not be sufficiently sensitive to differentiate the structural changes of the proteins that entail ADP-ribosyl transfer during steady-state reactions.
The chemical origin of the different activities across rifampin analogues is yet to be delineated. We tentatively conclude that amino acid residues comprising the rifampin-binding sites could be responsible for this differentiation. As described above, the α2-helix composed of M[R|K][D|E]XL residues at the C-terminus makes direct contact with rifampin (Figs 1b and 2b), and is conserved throughout the metagenomic sequences (Fig. 1a). Although the fourth residue X is one of the least conserved residues in our metagenome sequences as G(15.56%), K(17.78%), H(40%), A(8.89%), R(4.44%), K(6.67%), M(2.22%), S(2.22%), and F(2.22%), our structural analysis suggests that it is located in proximity to the complexed rifampin, directly pointing towards the substituent at the C8 position. Therefore, we suppose that it may play an important role in determining the ADP-ribosyltransferase activity.
Site-directed mutagenesis of rifampin-binding sites
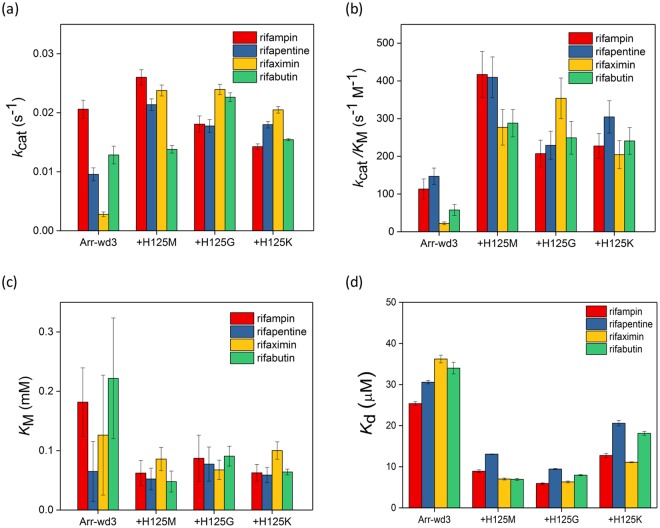
In Arr-wd 1–4 proteins, the specific residue X is Met, Gly, His, and Lys, respectively. To evaluate whether the least conserved residue in the M[R|K][D|E]XL motif plays a role during the enzyme catalysis, we prepared three Arr-wd3 variants, H125M, H125G, and H125K (Additional file 1: Table S4); Met, Gly, and Lys are the amino acids that other Arr-wd 1, 2, and 4 proteins natively have at the analogous position, respectively. Although turnover rates (kcat) (Fig. 6a; Additional file 1: Table S8) and MIC values (Additional file 1: Table S9 and Figure S8) are marginally perturbed by the H125 mutations, the mutants H125M, H125G, and H125K display enhanced kcat/KM for all rifampin analogues. The data demonstrate that this residue is involved in the enzyme activities, and the relatively low activities of Arr-wd3 are at least partially attributed to the steric hindrance with little flexibility of the histidine residue at this position.
Figure 6.
Kinetic and thermodynamic parameters of Arr-wd3 H125 variants from the steady-state reactions with rifampin derivatives. (a) Turnover numbers, kcat (s−1) (b) Catalytic efficiencies, kcat/KM (s−1 M−1) (c) Michaelis constants, KM (mM) (d) Dissociation constants, KD (μM). Values reported in (a–c) and (d) are derived from the fit to the Michaelis-Menten equation and Stern-Volmer equation, respectively.
It is also supported by the KD values of the H125 variants (Fig. 6d; Additional file 1: Table S7). H125G mutation yields the largest decrease in KD value, followed by H125M and H125K, for all rifampin analogues, implicating that the size and the flexibility of the side chain is correlated with the degrees of favorable interactions with the rifamycins.
The most dramatic enhancement of the kcat/KM is observed from the reaction with rifaximin. When Arr-wd3 protein is compared with the H125G variant, the kcat/KM is increased by 16-fold, indicating that the removal of the imidazole ring presumably alleviates the steric clash with the bulky and strained 5 and 6-membraned rings fused to the naphthoquinone core of rifaximin. In contrast, all Arr-wd3 mutants exhibit the least degree of enhancement upon the reaction with rifapentine or rifampin. The reactivity pattern with the rifampin analogues are consistent with our observation described above that rifaximin and rifapentine are the least and the most promiscuous substrates, respectively. These data suggest that the combined effects of the chemical structures of the antibiotics and their dynamic interactions with the enzyme active site may determine the enzyme catalysis, and the X residue in M[R|K][D|E]XL motif plays an important role in the reaction.
Even with the low sequence conservation in the X residue of the M[R|K][D|E]XL motif (Fig. 1a), the role in the reactivity and biochemical function was revealed only after the examination of protein structure. Therefore, the current study suggests that integrative analysis of 1-dimensional sequence and 3-dimensional structural information can provide us comprehensive information to elucidate the function of unidentified genes.
Conclusions
Using metagenomic analysis on the sediment microbiome, we have identified new ADP-ribosyltransferase genes that are involved in the development of antibiotic resistance. These proteins share low sequence similarity with known enzymes, but have highly conserved amino acid residues around the enzyme active site, which catalyzes the transfer of NAD+-derived ADP-ribosyl group to the antibiotic rifampin.
To gain a detailed understanding of this process, we carried out extensive analysis of gene sequence and protein structure to locate functionally relevant regions. For experimental validation of our finding, we expressed four representative genes into the proteins: Arr-wd1, Arr-wd2, Arr-wd3, and Arr-wd4. These proteins are kinetically competent in the ADP-ribosylation of rifampin using NAD+. Steady-state kinetic analysis and binding assays revealed that the Arr-wd proteins exhibit much tighter binding interaction with rifampin and its analogues than previously reported Arr proteins. The steady-state activities of the Arr-wd proteins are altered depending on the substructures of the rifamycin substrates, and site-directed mutagenesis studies provided important chemical insights into how the steric interaction between the protein and substrate might affect the enzyme activity.
This work nicely demonstrates the advantage of an integrated bioinformatics approach to discover new enzyme functions from environmental microbiome data. Taking a step further from simple sequence analysis and classification, we have constructed three-dimensional models to identify structural features important for enzyme activity, and validated computational predictions with biochemical experiments. Our studies indicate that chemically guided integrative approach can improve the quality of the metagenomic data and can provide greater knowledge to predict structure-function relationships, thereby facilitating the annotation of protein function with high accuracy. Sequence alignment and site-directed mutagenesis also suggest that even a single amino acid residue with low sequence conservation can alter the in vitro and in vivo enzymatic activities, which further highlights the importance of identifying key enzyme residues to understand and combat antibiotic resistance.
Methods
Data collection
A microbiome sample was collected from the soil sediment (latitude 33°49′65″N longitude 126°96′84″E), and sequenced using Hiseq™ 2000 platform (Illumina, San Diego, USA). The sample was prepared according to the Illumina protocols. For a 350 bp insert size, one microgram of genomic DNA was fragmented using Covaris. The raw sequencing data described in this study is available at European Nucleotide Archive (ENA) with the accession number PRJEB25358. The 151 bp paired-end sequencing reads were assembled by using MegaHit48 with the default option. The genes predicted by FragGeneScan49 were analyzed to find the rifampin resistance functions. Seven arr genes (i.e. arr-1, arr-2, arr-3, arr-4, arr-5, arr-7, and arr-8) were obtained from the comprehensive antibiotic resistance database (CARD)41,42. These genes were used as query to search homologous genes in the microbiome sample by blastp with the threshold of 50% sequence similarity and e-value < 1.0e-10. As a result, 45 homologous genes were obtained (Fig. 1a; Additional file 1: Table S1).
Phylogenetic analysis
In order to delineate the phylogenetic relationship between the putative arr genes and the annotated genes from the sequenced genomes, a phylogenetic tree was constructed (Fig. 3). The known arr genes were obtained from the CARD database and NCBI repository. In order to find homologous arr genes in the microbial genomes, 45 arr genes were searched against a set of 29,574,723 protein sequences of 8,369 bacterial genomes (downloaded from NCBI ftp server on 12/05/2017). In the phylogenetic tree, we included 45 arr-wd genes, 7 known arr genes from CARD database, 8 arr genes from the previous study13, and 37 putative arr genes that were searched from the microbial genomes. For each arr gene, five most homologous sequences (evalue < 1.0e-10) were obtained, and a set of non-redundant sequences were retained to build the phylogenetic tree. Neighbor-joining method was applied to build the tree with the bootstrapping value of 1000. MUSCLE50 and MEGA51 were used for multiple sequence alignment and phylogenetic analysis, respectively.
Integrated analysis of protein structure and sequence
Our integrated analysis of ADP-ribosyltransferase exploits rifampin and NAD+ binding sites. For this purpose, both structure-guided and sequence-based approaches were taken. The structure-guided conservation analysis reveals the amino acids that are sterically and/or spatially conserved and physico-chemically similar, but relatively low in sequence conservations. The analysis consists of three tasks: (1) multiple sequence alignment (MSA) of the reference arr genes; (2) ligand-protein complex binding analysis between rifampin and Arr-ms; (3) mapping conserved residues from MSA with featured amino acid residues interacting with rifampin. A previously reported X-ray structure of Arr-ms complexed with rifampin (PDB ID: 2HW2) was used to find amino acid residues involved in the ADP-ribosylation activity. Both intermolecular (i.e. between rifampin and Arr-ms), and intramolecular interactions of Arr-ms are considered in the rifampin binding sites. The molecular interactions are determined from the distances between the atoms that participate in hydrogen bonding or hydrophobic interactions. The results were incorporated with sequence conservation data obtained from metagenome analysis.
No crystal structure is currently available for Arr proteins complexed with NAD+. We thus performed Autodock52 simulation to visualize possible orientations of NAD+ when bound to the protein. The size of grid box was set to 22 × 24 × 28 Å3. Structural models of Arr-wd were generated from the Swiss modeling53 to simulate their possible three-dimensional structures (Additional file 1: Figure S2). PyMol54 was used to visualize the X-ray crystal structure of Arr-ms (Figs 1–2; Additional file 1: Figures S1–S2 and S5). Discovery Studio Visualizer55 was used to generate the 2D diagram of molecular interactions between rifampin and Arr-ms. The sequence alignment figure was generated by JalView56.
Protein expression and purification
Among 45 homologous genes from the metagenomic analysis, four genes denominated as arr-wd1, arr-wd2, arr-wd3, and arr-wd4 were selected as the targets for biochemical characterizations (Fig. 1a; Additional file 1: Table S3). The codons of the DNA fragments were optimized for E. coli. expression prior to the gene synthesis (General Biosystems), and were inserted into pET22b(+)/ampR vector using NdeI and XhoI cut-sites. Each plasmid was transformed to either DH5α or BL21(DE3) competent cells for sequencing or protein expression, respectively. All proteins were expressed with a His-tag at the C-terminus, and Arr-wd3 without the His-tag (Arr-wd3 + Δ6His) was additionally prepared to ensure that the presence of His-tag yields no significant perturbation in their activities. Single variants such as N34H, H125M, H125G, and H125K were prepared by site-directed mutagenesis. The PCR was carried out with KOD DNA polymerase (Toyobo) and primers (Additional file 1: Table S4). After PCR, the reaction products were digested with DpnI and transformed to DH5α competent cells. The plasmids were extracted by using miniprep kits, and sequenced at Macrogen, Inc.
For protein overexpression, a single colony of BL21(DE3) was picked and inoculated in 10 mL autoclaved LB media containing 100 mg/L ampicillin. The culture was grown in 200 rpm orbital shaker at 37 °C overnight, and inoculated in 1 L autoclaved LB media containing 100 mg/L ampicillin. When the optical density at 600 nm (OD600) reached ca 0.7, 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at the final concentration was added. After 3 h of induction at 37 °C, the cells were harvested by centrifugation at 5000 rpm (4715 × g) for 15 min at 4 °C. The cell pastes were frozen in liquid nitrogen and stored at −80 °C for further usage.
The protein purification was initiated by thawing the cell pastes in lysate buffer (20 mM sodium phosphate buffer, pH 7.4 with 500 mM NaCl). The cells were lysed by sonication for 15 min in an ice bath (on/off = 3 s each). The lysates were centrifuged at 13000 rpm (18800 × g) for 30 min at 4 °C, and the supernatants were loaded to Ni affinity column (HisTrap HP column, GE Healthcare Life Sciences). The column was pre-equilibrated with the lysate buffer at 4 °C using ÄKTA Protein Purification Systems. After loading the cell lysates, elution buffer (20 mM sodium phosphate buffer, pH 7.4 with 500 mM NaCl and 500 mM imidazole) was applied in a linear gradient (5–50%) (Additional file 1: Figure S3). All proteins were eluted when ca 100 mM imidazole was applied. The protein expressed without a His-tag (Arr-wd3 + Δ6His) was purified by Q anion exchange chromatography by varying the concentration of NaCl (0–1 M) in 20 mM sodium phosphate buffer, pH 8.0 at 4 °C. Relatively pure fractions (ca 80–90%) were concentrated using Amicon stirred cells (EMD Millipore) or centrifugal concentrating devices with 10 kDa cutoff membrane filters; the protein was further purified by size exclusion chromatography (S75 column), eluted with 20 mM Tris/HCl buffer, pH 7.0 with 150 mM NaCl. The purity of the protein was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The mass of the protein was confirmed by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometer (MALDI-TOF; Bruker Ultraflextreme TOF/TOF) with sinapinic acid as matrix solution. The protein was concentrated up to ca 0.5–5 mM and stored at −80 °C until further usage. The protein concentration was determined by UV–vis spectrophotometer (Agilent Cary 8454). The extinction coefficients at 280 nm were estimated from the sequence (Additional file 1: Table S3)57.
In vitro activity assay
The assays were carried out in dark due to the light-sensitivity of rifampin and its analogues. Rifampin was dissolved in methanol, and diluted with the buffer used for the assay (Fig. 4). Initially, we followed the previously reported procedure (Additional file 1: Figure S4a)13. In short, 10 μM protein in 100 μL 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer, pH 7.5 was mixed with various concentrations of the substrates, NAD+ and rifampin. The solution was left for 5 min at 22 °C, and the reaction was quenched by the addition of 100 μL methanol. The reaction mixture was analyzed by HPLC (Agilent 1260 Infinity II, C18 column with 2.7 μm, 120 Å, 4.6 × 100 mm). We later modified the quenching method by increasing the volume of methanol to 400 μL because 100 μL methanol was not sufficient to completely quench the reaction (Additional file 1: Figure S4b). In addition, we modified the HPLC method to accurately detect the formation of nicotinamide from the reaction mixture as follows: 100% H2O and 0.05% trifluoroacetic acid (TFA) for 3.5 min, followed by a linear gradient to 90% CH3CN with 0.1% TFA over 9.5 min (Fig. 4a). The ADP-ribosylated products were confirmed by electrospray ionization mass spectrometry (ESI-MS; Thermo Finnigan LTQ) (Additional file 1: Table S5).
Steady-state kinetic parameters of Arr-wd enzymes were obtained by varying the concentrations of one of the substrates, either NAD+ or rifampin, when the other was fixed to be 1 mM. The product yields were measured by detecting the concentrations of nicotinamide by HPLC. The standard curve of nicotinamide was measured independently (Additional file 1: Figure S4c). The kinetic parameters, kcat, KM, and kcat/KM, were determined from non-linear iteration curve fits to the Michaelis-Menten equation (Figs 4 and 5; Additional file 1: Table S6 and Figure S4). Activities of Arr-wd3 variants were also measured by the same procedure as described above (Fig. 6; Additional file 1: Table S8).
In vitro activity assay was also carried out with NADP+, NADH, or NADPH, instead of NAD+, and with rifampin analogues, such as rifapentine, rifaximin, and rifabutin (Fig. 5; Additional file 1: Figure S6). Representative HPLC traces, Michaelis-Menten fits, and kinetic parameters are included in the Additional file 1: Table S6 and Figure S7. Rifampin analogues and the corresponding ADP-ribosylated products were eluted at 10.6 min and 9.1 min for rifapentine, 10.3 min and 8.6 min for rifaximin, and 9.9 min and 8.4 min for rifabutin, respectively. The reaction products were further analyzed by ESI-MS (Additional file 1: Table S5).
Determination of binding constants by intrinsic fluorescence changes
Intrinsic tryptophan fluorescence changes were monitored with microplate reader (Biotek Synergy H1m) at 22–25 °C. Arr-wd1-3 and Arr-wd4 proteins possess two and four tryptophan residues, respectively. Sequence alignment together with structure homology modeling suggests that two tryptophan residues are located at similar positions in Arr-wd1–4 (Additional file 1: Figure S5). Conformational changes induced by the addition of substrates were monitored by using 295 nm excitation to avoid any possible interference from tyrosine and phenylalanine residues. The fluorescence emission was observed at 325–450 nm, and the fluorescence emission at 325 nm was used to obtain the dissociation constants (KD). Because rifamycins exhibits strong absorption at both excitation and emission wavelengths (Additional File 1: Figure S5b), the concentrations of rifamycins were kept at 1–4 μM so that the absorption at both excitation and emission wavelengths remains less than 0.05, thereby minimizing the inner filter effect58. The plot of F0/F versus the concentrations of rifamycins yielded a linear correlation (Additional File 1: Figure S5c), which is fitted to the Stern-Volmer equation (Additional File 1: Figure S5d) to determine the dissociation constants (KD) for rifampin and its derivatives (Figs 4e, 5d, 6d, and Additional file 1: Table S7). The protein samples (5–20 μM) were prepared in 100 μL of 50 mM HEPES buffer, pH 7.5.
Determination of MIC values
Minimal inhibitory concentration(MIC) values for BL21(DE3) cells containing arr-wd genes were measured by the procedures reported previously13. In short, the plasmids containing arr-wd genes were transformed into BL21(DE3) cells, and were grown on the LB/agar plate containing 100 mg/L ampicillin overnight at 37 °C. The cells were mixed with 0.85% saline to adjust the OD625 to be 0.1. The aliquots of 100 μL cell culture in saline solution were mixed with 18.9 mL Mueller Hinton broth media and 0–512 μg/mL rifamycin dissolved in methanol. Either 0 or 100 μM IPTG at final concentration was added to the solution. After the solutions were incubated at 37 °C overnight, optical cell density (OD625) was measured with the microplate reader to determine the minimum inhibitory concentrations of the antibiotics (Fig. 3f; Additional file 1: Table S9 and Figure S8).
Electronic supplementary material
Acknowledgements
We thank Youna Cho for sequence data processing, and Dr. Dongwhan Lee for helpful discussion. This work was supported by a grant from the Collaborative Genome Program (20140428 to W.J.S. and M.R.) funded by the Ministry of Oceans and Fisheries, Korea, a NRF grant from Korea government (NRF-2016R1C1B2007898 to W.J.S.), and a grant (2017NER54070 to M.R.) by Research of Korea Centers for Disease Control and Prevention.
Author Contributions
J.H.S. and M.R. conducted the metagenomic sequence analysis. H.E. and W.J.S. conducted biochemical experiments. J.H.S., H.E., W.J.S. and M.R. designed the experiment, analysed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
The raw sequencing data described in this study is available at European Nucleotide Archive (ENA) with the accession number PRJEB25358.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jae Hong Shin and Hyunuk Eom contributed equally to this work.
Contributor Information
Woon Ju Song, Email: woonjusong@snu.ac.kr.
Mina Rho, Email: minarho@hanyang.ac.kr.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-30547-x.
References
- 1.Ventola CL. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 2.Chakraborty S, Rhee KY. Tuberculosis drug development: History and evolution of the mechanism-based paradigm. Cold Spring Harbor perspectives in medicine. 2015;5:a021147. doi: 10.1101/cshperspect.a021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sensi P. History of the development of rifampin. Rev. Infect. Dis. 1983;5(Suppl 3):S402–406. doi: 10.1093/clinids/5.Supplement_3.S402. [DOI] [PubMed] [Google Scholar]
- 4.Marriner, G. A. et al. In Third World Diseases Vol. 7 (ed. Elliott, R. L.) 47–124 (Springer-Verlag Berlin Heidelberg, 2011).
- 5.Campbell EA, et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–912. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 6.Alifano P, Palumbo C, Pasanisi D, Talà A. Rifampicin-resistance, rpoB polymorphism and RNA polymerase genetic engineering. J. Biotechnol. 2015;202:60–77. doi: 10.1016/j.jbiotec.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Stogios, P. J. et al. Rifampin phosphotransferase is an unusual antibiotic resistance kinase. Nature Comm. 7, 11343, 10.1038/ncomms11343, https://www.nature.com/articles/ncomms11343supplementary-information (2016). [DOI] [PMC free article] [PubMed]
- 8.Goldstein BP. Resistance to rifampicin: a review. J. Antibiot. 2014;67:625–630. doi: 10.1038/ja.2014.107. [DOI] [PubMed] [Google Scholar]
- 9.Floss HG, Yu T-W. Rifamycin mode of action, resistance, and biosynthesis. Chem. Rev. 2005;105:621–632. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad S, Mokaddas E, Fares E. Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Kuwait and Dubai. Diagn. Microbiol. Infect. Dis. 2002;44:245–252. doi: 10.1016/S0732-8893(02)00457-1. [DOI] [PubMed] [Google Scholar]
- 11.Jamieson FB, et al. Profiling of rpoB mutations and MICs for rifampin and rifabutin in Mycobacterium tuberculosis. J. Clin. Microbiol. 2014;52:2157–2162. doi: 10.1128/JCM.00691-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spanogiannopoulos P, Thaker M, Koteva K, Waglechner N, Wright GD. Characterization of a rifampin-inactivating glycosyltransferase from a screen of environmental actinomycetes. Antimicrob. Agents. Chemother. 2012;56:5061–5069. doi: 10.1128/AAC.01166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baysarowich J, et al. Rifamycin antibiotic resistance by ADP-ribosylation: Structure and diversity of Arr. Proc. Natl. Acad. Sci. USA. 2008;105:4886–4891. doi: 10.1073/pnas.0711939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu LK, et al. The structure of the antibiotic deactivating, N-hydroxylating rifampicin monooxygenase. J. Biol. Chem. 2016;291:21553–21562. doi: 10.1074/jbc.M116.745315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dabbs ER, et al. Ribosylation by mycobacterial strains as a new mechanism of rifampin inactivation. Antimicrob. Agents. Chemother. 1995;39:1007–1009. doi: 10.1128/AAC.39.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agrawal P, Miryala S, Varshney U. Use of Mycobacterium smegmatis deficient in ADP-ribosyltransferase as surrogate for Mycobacterium tuberculosis in drug testing and mutation analysis. PLOS ONE. 2015;10:e0122076. doi: 10.1371/journal.pone.0122076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morisaki N, et al. Structure determination of ribosylated rifampicin and its derivative: new inactivated metabolites of rifampicin by mycobacterial strains. J. Antibiot. 1995;48:1299–1303. doi: 10.7164/antibiotics.48.1299. [DOI] [PubMed] [Google Scholar]
- 18.Qi X, et al. Structural basis of rifampin inactivation by rifampin phosphotransferase. Proc. Natl. Acad. Sci. USA. 2016;113:3803–3808. doi: 10.1073/pnas.1523614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 20.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 21.Subirats J, Sànchez-Melsió A, Borrego CM, Balcázar JL, Simonet P. Metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes. Int. J. Antimicrob. Agents. 2016;48:163–167. doi: 10.1016/j.ijantimicag.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 22.Flórez AB, Vázquez L, Mayo B. A functional metagenomic analysis of tetracycline resistance in cheese bacteria. Front. Microbiol. 2017;8:907. doi: 10.3389/fmicb.2017.00907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo J, Li J, Chen H, Bond PL, Yuan Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Research. 2017;123:468–478. doi: 10.1016/j.watres.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Chen B, et al. Metagenomic analysis revealing antibiotic resistance genes (ARGs) and their genetic compartments in the tibetan environment. Environ. Sci. Technol. 2016;50:6670–6679. doi: 10.1021/acs.est.6b00619. [DOI] [PubMed] [Google Scholar]
- 25.Crofts, T. S., Gasparrini, A. J. & Dantas, G. Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 15, 422, 10.1038/nrmicro.2017.28, https://www.nature.com/articles/nrmicro.2017.28supplementary-information (2017). [DOI] [PMC free article] [PubMed]
- 26.Wright PM, Seiple IB, Myers AG. The evolving role of chemical synthesis in antibacterial drug discovery. Angew. Chem. Int. Ed. 2014;53:8840–8869. doi: 10.1002/anie.201310843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pawlowski, A. C. et al. A diverse intrinsic antibiotic resistome from a cave bacterium. Nature Comm. 7, 13803, 10.1038/ncomms13803, https://www.nature.com/articles/ncomms13803supplementary-information (2016). [DOI] [PMC free article] [PubMed]
- 28.D’Costa, V. M. et al. Antibiotic resistance is ancient. Nature477, 457, 10.1038/nature10388, https://www.nature.com/articles/nature10388supplementary-information (2011). [DOI] [PubMed]
- 29.Bhullar K, et al. Antibiotic resistance is prevalent in an isolated cave microbiome. PLOS ONE. 2012;7:e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segawa T, et al. Distribution of antibiotic resistance genes in glacier environments. Environ. Microbiol. Rep. 2013;5:127–134. doi: 10.1111/1758-2229.12011. [DOI] [PubMed] [Google Scholar]
- 31.Forsberg, K. J. et al. Bacterial phylogeny structures soil resistomes across habitats. Nature509, 612, 10.1038/nature13377, https://www.nature.com/articles/nature13377supplementary-information (2014). [DOI] [PMC free article] [PubMed]
- 32.Allen HK, et al. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 33.Nesme J, et al. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr. Biol. 2014;24:1096–1100. doi: 10.1016/j.cub.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 34.Li B, et al. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. Isme J. 2015;9:2490–2502. doi: 10.1038/ismej.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu, Y. et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nature Comm. 4, 2151, 10.1038/ncomms3151, https://www.nature.com/articles/ncomms3151supplementary-information (2013). [DOI] [PubMed]
- 36.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wass MN, Sternberg MJE. Prediction of ligand binding sites using homologous structures and conservation at CASP8. Proteins. 2009;77:147–151. doi: 10.1002/prot.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rost B. Enzyme function less conserved than anticipated. Journal of molecular biology. 2002;318:595–608. doi: 10.1016/S0022-2836(02)00016-5. [DOI] [PubMed] [Google Scholar]
- 40.Levin, B. J. et al. A prominent glycyl radical enzyme in human gut microbiomes metabolizes trans-4-hydroxy-L-proline. Science355, 10.1126/science.aai8386 (2017). [DOI] [PMC free article] [PubMed]
- 41.Jia B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McArthur AG, et al. The comprehensive antibiotic resistance database. Antimicrob. Agents. Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houang ETS, Chu Y-W, Lo W-S, Chu K-Y, Cheng AFB. Epidemiology of rifampin ADP-ribosyltransferase (arr-2) and metallo-β-Lactamase (bla(IMP-4)) gene cassettes in class 1 integrons in Acinetobacter strains isolated from blood cultures in 1997 to 2000. Antimicrob. Agents. Chemother. 2003;47:1382–1390. doi: 10.1128/AAC.47.4.1382-1390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tribuddharat C, Fennewald M. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents. Chemother. 1999;43:960–962. doi: 10.1128/aac.43.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chowdhury G, Pazhani GP, Nair GB, Ghosh A, Ramamurthy T. Transferable plasmid-mediated quinolone resistance in association with extended-spectrum beta-lactamases and fluoroquinolone-acetylating aminoglycoside-6′-N-acetyltransferase in clinical isolates of Vibrio fluvialis. Int. J. Antimicrob. Agents. 2011;38:169–173. doi: 10.1016/j.ijantimicag.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 46.Fonseca ÉLd, Freitas FdS, Amorim JC. d. & Vicente, A. C. P. Detection of new arr-4 and arr-5 gene cassettes in clinical Pseudomonas aeruginosa and Klebsiella pneumoniae strains from Brazil. Antimicrob. Agents. Chemother. 2008;52:1865–1867. doi: 10.1128/AAC.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aristoff PA, Garcia GA, Kirchhoff PD, Hollis Showalter HD. Rifamycins – Obstacles and opportunities. Tuberculosis. 2010;90:94–118. doi: 10.1016/j.tube.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 49.Rho M, Tang H, Ye Y. FragGeneScan: Predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38:e191–e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.PyMOL. The PyMOL molecular graphics system, version 1.8 Schrödinger, LLC.
- 55.BIOVIA, D. S. Discovery Studio Visualizer, 2017 R2, San Diego: Dassault Systèmes (2017).
- 56.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 58.Lakowicz, J. R. Principles of Fluorescence Spectroscopy. 3 edn, (Springer, 2006).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data described in this study is available at European Nucleotide Archive (ENA) with the accession number PRJEB25358.