Abstract
Cell surfaces are critical for diverse functions across all domains of life, from cell-cell communication and nutrient uptake to cell stability and surface attachment. While certain aspects of the mechanisms supporting the biosynthesis of the archaeal cell surface are unique, likely due to important differences in cell surface compositions between domains, others are shared with bacteria or eukaryotes or both. Based on recent studies completed on a phylogenetically diverse array of archaea, from a wide variety of habitats, here we discuss advances in the characterization of mechanisms underpinning archaeal cell surface biogenesis. These include those facilitating co- and post-translational protein targeting to the cell surface, transport into and across the archaeal lipid membrane, and protein anchoring strategies. We also discuss, in some detail, the assembly of specific cell surface structures, such as the archaeal S-layer and the type IV pili. We will highlight the importance of post-translational protein modifications, such as lipid attachment and glycosylation, in the biosynthesis as well as the regulation of the functions of these cell surface structures and present the differences and similarities in the biogenesis of type IV pili across prokaryotic domains.
Keywords: archaea, protein secretion, cell surface, S-layer, pili, protein targeting
This review discusses molecular pathways required for the biogenesis of archaeal cell surface proteins.
INTRODUCTION
Archaea, which were initially mainly isolated from harsh environments and thus were long considered extremophilic species, are now known to be ubiquitous, with some playing key roles in vital ecological processes such as the carbon and nitrogen cycles (Chaban, Ng and Jarrell 2006; Falkowski, Fenchel and Delong 2008; Madsen 2011; Martens-Habbena and Stahl 2011; Martínez-Espinosa et al.2011; Koskinen et al.2017). Indeed, recent studies have shown that archaea are much more common in the human microbiome than had previously been known (Bang and Schmitz 2015; Koskinen et al. 2017). Moreover, recent identification of the Asgard, an archaeal superphylum, has revealed that archaea have an even closer evolutionary relationship to eukaryotes than was previously thought (Zaremba-Niedzwiedzka et al.2017). Yet, compared to bacteria and eukaryotes, most basic processes in archaea have not been adequately characterized. For example, while cell surfaces play key roles in cell biology and archaeal cell surfaces are, at least in part, distinct from those of bacteria and eukaryotes, much remains to be revealed about the composition, biosynthesis and functions of the cell surfaces in archaea.
While the cytoplasm of both bacteria and archaea is enclosed by a cytoplasmic membrane composed primarily of glycerol phosphate phospholipids, the lipid composition of these membranes is distinct and specific for each of these prokaryotic domains (Jain, Caforio and Driessen 2014; Caforio and Driessen 2017). Rather than fatty acids linked to the (sn)-1,2 positions of glycerol via ester bonds, the typical archaeal lipid core consists of C20 isoprenoid units linked to glycerol via ether bonds in the (sn)-2,3 positions (archaeol, C20 2,3-diphytanyl-sn-glycerol). However, many of the proteins integrated into the cytoplasmic membranes of archaea and bacteria are conserved, such as the core of the Sec protein transport machinery, and the energy generation machineries (Pohlschroder, Gimenez and Jarrell 2005). Subsets of some archaeal phyla also produce additional membrane proteins that share homology with bacterial counterparts, including many families of transporters, euryarchaeal membrane-bound chemotaxis proteins or the well-characterized bacteriorhodopsin of photosynthetic haloarchaea (Oesterhelt 1998; Lee, Dodson and Hultgren 2007; Schlesner et al.2012; Quax et al.2018), which has subsequently been identified in oceanic bacteria (proteorhodopsin) (Beja et al.2000; Pinhassi et al.2016).
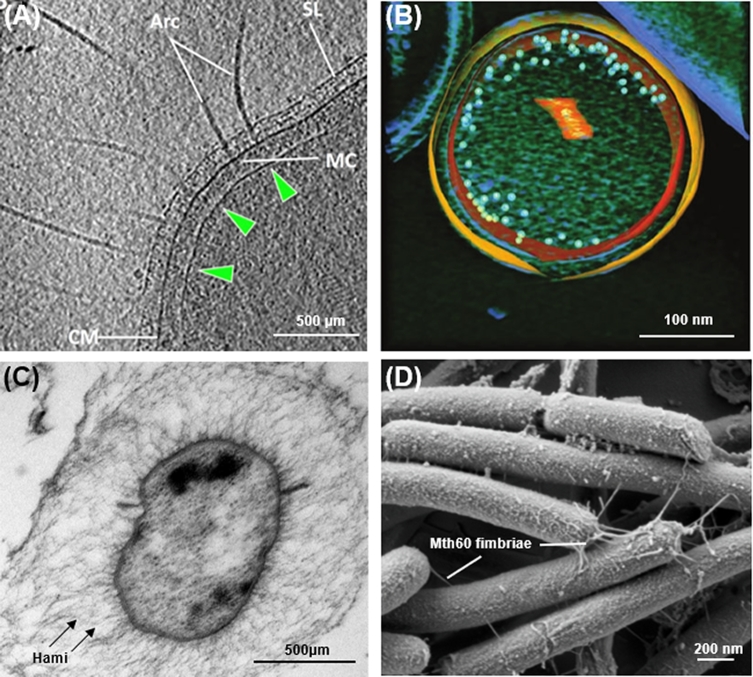
Most well-characterized archaea have a pseudocrystalline proteinaceous surface layer (S-layer) that envelops the cytoplasmic membrane (reviewed in Sleytr et al.2014). This S-layer appears to take on roles similar to those of the bacterial peptidoglycan cell wall for such functions as maintaining cell stability and morphology (Fig. 1A). While a peptidoglycan layer has not been identified in any archaeal species, a few euryarchaeal species produce pseudopeptidoglycan, which is outside of an S-layer and contains N-acetylglucosamine and N-acetyltalosaminuronic acid, rather than N-acetylmuramic acid as in bacterial peptidoglycan cell walls (Albers and Meyer 2011; Klingl 2014). Other surface structures observed in some archaea include the proteinaceous sheaths identified in Methanospirillum hungatei and Methanosaeta concilii, which, distinct from the S-layer surrounding each of these cells, enclose linear cell chain communities. Moreover, polymers like methanochondroitin, a polysaccharide observed in some Methanosarcinae, glutaminylglycans identified in the haloalkaliphile Natronococcus occultus, or halomucins in Haloquadratum are archaeal surface structures (Albers and Meyer 2011; Klingl 2014). Also, double membranes have been identified first in the crenarchaeote Ignicoccus hospitalis, and later in Euryarchaeota, such as Methanomassiliicoccus luminyensis and the Candidatus Altiarchaeum hamiconexum euryarchaeote, as well as in representatives of the Archaeal Richmond Mine Acidophilic Nanoorganism (ARMAN) group (Fig. 1B and C). These archaeal outer cell membranes, as well as non-S-layer cell walls, were recently discussed in detail by Klingl (2014). However, little is known about the biosynthesis of any of these structures, which will therefore not be discussed in this review. Finally, as in bacteria, archaea have a diverse set of cell surface filaments, some of which have only been observed in a few archaeal species. These include such structures as the grappling hooks (hami) of the as yet uncultured Candidatus A. hamiconexum (Fig. 1C) (Perras et al.2015a, b), Pyrodictium abyssi cannulae or the Methanothermobacterthermoautotrophicus Mth60 filaments (Fig. 1D) (Chaudhury, Quax and Albers 2018). Presumably, these structures play an important role in cell adhesion to biotic or abiotic surfaces or both. Unfortunately, characterization is restricted to electron microscopy for many of these structures. This lack of biochemical characterization is typically due to an absence of genetic tools for these organisms. Conversely, type IV pili, which are required for early steps in biofilm formation including surface adhesion and cell aggregation, are conserved across a broad range of bacterial and archaeal species, allowing for detailed characterization in model archaea (Szabo et al.2007; Imam et al.2011; Makarova, Koonin and Albers 2016). Many species from diverse archaeal phyla also produce type IV pilus-like filaments that are required for swimming motility, archaeal flagella or archaella (Fig. 1A) (Albers and Jarrell 2018).
Figure 1.
Archaeal cell-surface components. (A) Electron cryotomography of Pyrococcus furiosus; displayed is a tomographic slice through a frozen-hydrated P. furiosus cell, showing archaella on the cell pole. Arc, archaella; SL, S-layer; CM, cell membrane; green arrowheads, polar cap. MC, (archaellar) motor complex. Image and modified legend adapted from Daum et al. (2017). (B) A 50 voxel-thick slice of a central section of an ultrasmall archaeal ARMAN cell with the segmented inner and outer membranes in orange and yellow, respectively. Ribosomes are represented with light blue spheres drawn to scale. Low mass densities in the volume-rendered cytoplasm are in green. Segmented orange label in the cytoplasm corresponds to the cross section of a tubular organelle structure. Image and modified legend adapted from Comolli et al. (2008). (C) Electron micrograph (ultrathin section) of Candidatus Altiarchaeum hamiconexum; the cell is surrounded by an EPS matrix and cell surface appendages (hami), which extend beyond the matrix. The cell has two membranes with a faint periplasm. FtsZ aggregates are located at the inner membrane. Image and modified legend adapted from Probst et al. (2014). (D) Scanning electron micrograph of M. thermautotrophicus containing Mth60 fimbriae grown on gold electron microscope grids. Image courtesy of Gerhard Wanner, Ludwig Maximilian University, Germany.
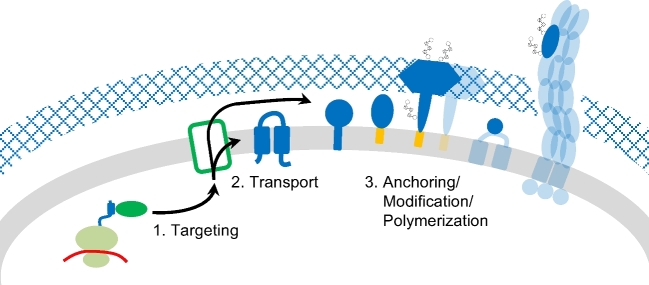
Cell surface biogenesis requires that specific proteins are translocated from the cytoplasm, where they are synthesized, to the surface. In eukaryotes, upon translation initiation, proteins destined for the cell surface are first targeted to the endoplasmic reticulum (ER). Following co- or post-translational protein transport into or across the membrane, via the universally conserved Sec machinery, cell surface proteins are delivered to their final destinations via an ER–Golgi secretory route requiring sequential budding and fusion of vesicular carriers (Colombo, Raposo and Thery 2014). In contrast, prokaryotic proteins that are to become part of the cell surface are targeted directly to the cytoplasmic membrane, where they are either inserted into the membrane or are transported across it (Fig. 2). In addition to the co- and post-translational Sec-dependent transport of unfolded proteins, many prokaryotes also use the Twin arginine transport (Tat) pathway to translocate proteins post-translationally across the cytoplasmic membrane in a folded conformation (Dilks, Gimenez and Pohlschroder 2005; Pohlschroder, Gimenez and Jarrell 2005; Goosens and van Dijl 2017). Furthermore, surface proteins may require additional post-translational modifications (PTMs) to function properly, and/or for secreted proteins to be surface anchored (Eichler and Maupin-Furlow 2013). Finally, those secreted proteins that are subunits of surface structures must be correctly incorporated into the corresponding structure (Fig. 2).
Figure 2.
Schematic overview of common steps in the biosynthesis of archaeal cell surface proteins. During translation, hydrophobic domains of the nascent polypeptide chains are recognized, targeting the ribonucleic protein complex to the membrane (1). The cell surface proteins can be integrated into or transported across the membrane (2), followed by anchoring via transmembrane (TM) domains or covalent linkage to lipid anchors (3). Further post-translational modifications (PTMs) of cell surface proteins include the removal of the signal peptide as well as N- and O-glycosylation. Protein–protein interactions can lead to the binding of soluble proteins at the cell surface or to the polymerization into larger structures such as type IV pili or the S-layer. Proteins involved in distinct pathways associated with these processes (green), the cell surface proteins themselves (blue) as well as their various membrane anchors (orange) are not specified here but discussed in detail in this review.
In recent years, the development of model systems representing species from various archaeal phyla has facilitated the study of cell surface biogenesis in this domain of life (Leigh et al.2011). In this review, we will discuss various aspects of archaeal cell surface biogenesis including how proteins are targeted to the membrane, and then transported into or across it, in either an unfolded conformation, via the Sec pathway, or in a folded conformation via the Tat pathway. We will also address some of the diverse array of mechanisms known to anchor these secreted proteins to the membrane. Finally, we will discuss the biosynthesis of cell surface structures. As noted above, to date the information about the biosynthesis of archaeal filaments that are part of the cell surface is limited to the S-layer, type IV pili and archaella. Since archaella biosynthesis was recently reviewed by Albers and Jarrell (2018), we will focus on the S-layer and type IV pilus biosynthesis.
Membrane lipid structure and biosynthesis
The cytoplasmic membrane plays a critical role in serving as a cellular barrier between the cytoplasm and the extracellular environment so that cellular homeostasis can be maintained. Even prior to being identified as a separate domain of life (Woese, Kandler and Wheelis 1990), it had long been known that archaeal membranes have a distinct lipid composition differing from that of bacteria and eukaryotes (Kates, Wassef and Pugh 1970). The archaeal membrane composition might be an ancient trait which has enabled archaea to inhabit a variety of extreme environments (Hartzell, Millstein and LaPaglia 1999; Cavicchioli 2006; Chaban, Ng and Jarrell 2006; Pikuta, Hoover and Tang 2007). The archaeal lipid glycerol backbone consists of glycerol-1-phosphate (G-1-P) (Caforio and Driessen 2017), which is synthesized by G-1-P dehydrogenase from dihydroxyacetonephosphate, an intermediate of glycolysis and gluconeogenesis, prior to being coupled to the lipid chain (Nishihara and Koga 1997; Nishihara et al.1999).
Initial analyses of cytoplasmic membranes from Thermoplasma acidophilum, Sulfolobus acidocaldarius and various halophilic archaea revealed that the archaeal membrane lipid chain is composed of isoprenoids rather than the fatty acids found in bacterial and eukaryotic membranes (Langworthy, Mayberry and Smith 1974; Mayberry-Carson et al.1974; Kates 1977; Caforio and Driessen 2017). The basic building blocks of the isoprenoid chain, the 5-carbon unit isopentenyl pyrophosphate and its isomer, dimethylallyl pyrophosphate, are synthesized via the mevalonate pathway and variants thereof (VanNice et al.2014; Vinokur et al.2016).
While the typical archaeal cytoplasmic membrane is composed of lipids based on archaeol (Villanueva, Damste and Schouten 2014), some archaeal cytoplasmic membranes are based on C40 glycerol dibiphytanyl glycerol tetraethers (caldarchaeol), forming an atypical membrane monoloayer structure (Langworthy 1977; Komatsu and Chong 1998). This monolayer membrane structure is predominantly found among thermophilic crenarchaeota, such as T. acidophilum and S. acidocaldarius. Additionally, for a subset of thermophilic archaea, the isoprenoid chains of the monolayer can also be cross-linked, forming an H-shaped tetraether lipid (Jacquemet et al.2009) or contain cyclopentane rings. These might play important roles in maintaining functional membranes and cellular homeostasis under extreme pH or thermal stress by increasing membrane dense packing, reducing the rotational flexibility of the isoprenoid chain and decreasing overall membrane fluidity, potentially providing an adaptation to the high temperatures these organisms inhabit (Uda et al.2001; Macalady et al.2004; Chong 2010; Koga 2012; Schouten, Hopmans and Damste 2013).
MEMBRANE TARGETING
In all cells, a significant fraction of proteins synthesized in the cytoplasm are destined to become integral membrane proteins or proteins that are otherwise anchored to the extracytoplasmic side of the cytoplasmic membrane (Table 1). To ensure that these proteins are properly localized, cells have evolved mechanisms that can target proteins to the bacterial and archaeal cytoplasmic membrane or to the ER membrane, in eukaryotes.
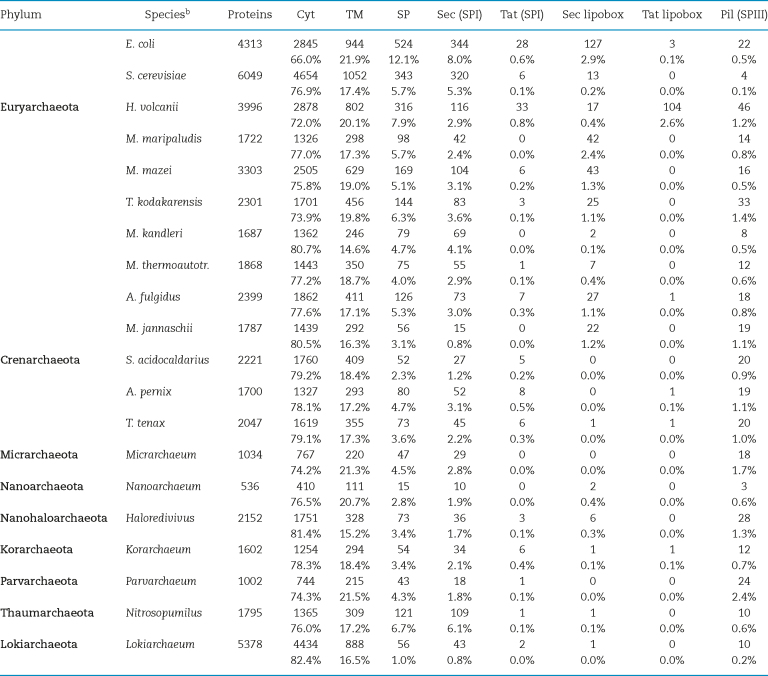
Table 1.
Prediction of archaeal signal peptide and TM domain-containing proteins.a
|
Co-translational targeting of proteins to the cytoplasmic membrane
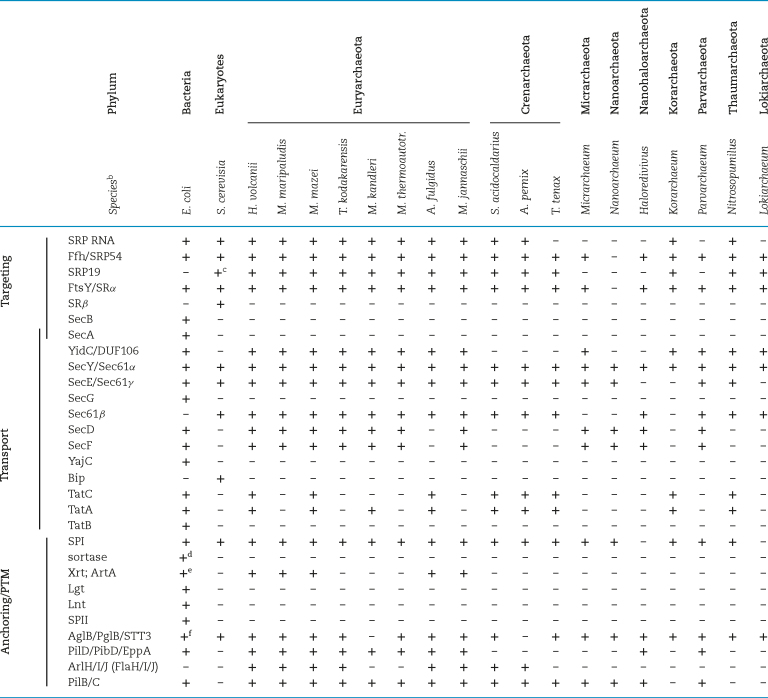
Co-translational protein targeting to the cytoplasmic membrane is performed by an essential, evolutionarily conserved pathway found in all domains of life (Table 2). As a nascent peptide chain emerges from a ribosome, the signal recognition particle (SRP), a ribonucleic protein complex, recognizes highly hydrophobic domains that are transmembrane (TM) domains of integral membrane proteins or part of the N-terminal signal peptides (Yosef et al.2010). Such signal peptides contain a conserved tripartite structure that consists of a set of positively charged amino acids followed by a hydrophobic (H) domain and a processing site. SRP arrests translation of the ribosome-nascent chain (RNC) upon binding of the H domain or TM domain, then targets the newly formed RNC-SRP complex to the membrane-bound SRP receptor (SR) (Halic et al.2004; Grudnik, Bange and Sinning 2009; Akopian et al.2013). This coupling of translation and translocation prevents newly synthesized proteins that are targeted for secretion from misfolding in the cytoplasm, prior to secretion. Also, it alleviates the need for chaperones to maintain proteins in an unfolded conformation prior to transport.
Table 2.
Cell surface biogenesis components.a
|
While the number of SRP components varies between archaea, bacteria and eukaryotes, all SRPs contain an RNA backbone as well as SRP54, whose primary role is signal peptide binding to the SRP via its M domain (Romisch et al.1990). SRP54 also guides the nascent peptide chain to the membrane via interaction of its N-terminal NG domain with the SR (Zwieb and Eichler 2002; Lichi, Ring and Eichler 2004; Kuhn, Koch and Dalbey 2017). While the bacterial RNA backbone varies in size from species to species, most archaeal and eukaryotic SRP RNAs share a similar size and secondary structure (7S RNA). However, circularized structures of SRP RNA found in some Thermoproteus species may provide increased stability at high temperatures (Plagens et al.2015). The eukaryotic SRP consists of six protein subunits, of which only the SRP19 subunit is conserved in archaea. As found for other systems, the eukaryotic SRP is thus more complex than the archaeal counterpart. Both the eukaryotic and archaeal SRP19 interact with SRP54, thereby inducing conformational changes that increase SRP54 binding affinity to the 7S RNA (Diener and Wilson 2000; Rose and Pohlschroder 2002; Tozik et al.2002; Zwieb and Bhuiyan 2010).
While deletion of the gene encoding SRP54 has profoundly deleterious effects in all organisms, archaea do not appear to require SRP19 for protein transport, in contrast to eukaryotes (Rose and Pohlschroder 2002). Archaeal SRP54 may have the capability to bind to 7S RNA independent of SRP19 (Diener and Wilson 2000; Rose and Pohlschroder 2002; Tozik et al.2002; Yurist, Dahan and Eichler 2007). Nonetheless, studies of the archaeal SRP, particularly those that have included structural analyses, have significantly advanced our general understanding of the molecular mechanisms underpinning the function of the SRP in general. For example, the crystal structure of unbound and signal peptide bound forms of the Methanocaldococcus jannaschii SRP revealed that signal peptide binding to the M domain of SRP54 induces a coordinated folding mechanism that leads to the repositioning of the NG domain, thus facilitating binding of the RNC-SRP complex to the conserved SR (Hainzl, Huang and Sauer-Eriksson2002).
The archaeal and bacterial SR is each comprised of a single protein, FtsY, which is homologous to the eukaryotic SRα proteins (Miller, Bernstein and Walter 1994). Membrane-bound FtsY/SRα has been proposed to play an essential role in guiding RNC-SRP to the cytoplasmic membrane in all organisms, and, ultimately, to the Sec translocon or YidC insertase (Fig. 3A) (Lichi, Ring and Eichler 2004; Haddad, Rose and Pohlschröder 2005; Egea et al.2008; Hainzl and Sauer-Eriksson 2015). Alternatively, data by Bibi et al. suggest that at least the Escherichiacoli SRP receptor FtsY mediates ribosome targeting to the membrane during its own translation in an SRP-independent manner. mRNAs encoding SRP-dependent substrates are then targeted to the membrane-bound ribosomes and hydrophobic nascent polypeptides are recognized by SRP as they emerge from the ribosome, to facilitate proper assembly of the RNC on the SecYEG translocon (Bibi 2012; Bercovich-Kinori and Bibi 2015).
Figure 3.
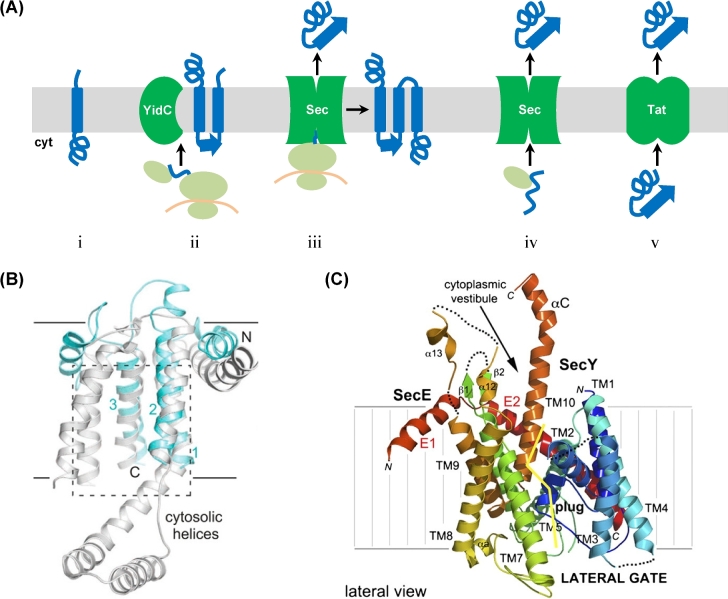
Translocation of proteins into and across the membrane. (A) With the exception of spontaneous insertion of membrane proteins (i), four main routes for the translocation of cell surface protein into or across the membrane exist in archaea: co-translational YidC-dependent insertion (ii), co-translational insertion or translocation via the Sec complex (iii), Sec-dependent post-translational translocation (iv), and post-translational translocation in a folded confirmation by Tat (v). See the main text for more details. (B) Archaeal Mj0480 and bacterial YidC share key structural features. Structure-based alignment of M. jannaschii Mj0480 (light blue; 5C8J) and Bacillus halodurans BhYidC (gray; 3WO6) showing views from the plane of the membrane. The proteins superimpose with a root-mean-square deviation of 3.9 A° over 105 equivalent residues (out of 141 visible) (Borowska et al.2015). (C) Structure of P. furiosus Pfu-SecYE and the crystal packing. SecY is colored using a rainbow pattern. A ‘clam shell’ structure is formed by the 10 transmembrane (TM) helices with a lateral gate opening between transmembrane helices TM2 and TM3 (in the N-terminal half of the ‘clam shell’) and TM7 and TM8 (in the C-terminal half of the ‘clam shell’). The yellow line delineates the lateral gate on the SecY subunit. Structure and modified legend adapted from Egea and Stroud (2010).
The interaction between SRP and SR is regulated by GTP hydrolysis, whereby the GTP-bound SR-SRP complex is stable, but dissociates upon GTP hydrolysis, allowing the delivery of the nascent peptide chain to the protein transport machinery for transport into or translocation across the cytoplasmic membrane (see below) (Egea et al.2004; Focia et al.2004; Wild et al.2016). While the structures of several of these archaeal components have been resolved, remarkably, in vivo studies of co-translational protein transport in archaea are limited to the Halobacterium salinarum bacteriorhodopsin. In vivo analysis of the membrane insertion kinetics of this well-known archaeal membrane protein, which uses light energy to generate a proton gradient across the cytoplasmic membrane, suggested its co-translational targeting to the cytoplasmic membrane and insertion into the membrane via the Sec translocon (Gropp, Gropp and Betlach 1992; Dale and Krebs 1999; Dale, Angevine and Krebs 2000). Interestingly, when bacteriorhodopsin is fused to dihydrofolate reductase and a cellulose-binding domain of Clostridium thermocellum, this polytopic membrane protein construct appears to be inserted into the Haloferaxvolcanii cytoplasmic membrane in a post-translational manner (see below) (Ortenberg and Mevarech 2000).
In bacteria, co-translational membrane targeting by the SRP can also deliver proteins to YidC, a membrane protein insertase that can insert small proteins into the membrane (see below). A corresponding mechanism has not yet been described in archaea.
Post-translational targeting of proteins to the cytoplasmic membrane
Rather than being stalled at the ribosome, many proteins containing an N-terminal signal peptide are translated to completion prior to transport even though they are destined for transport across the membrane. Most of these proteins are not recognized by SRP due to the relatively low hydrophobicity of their H domains. However, even some signal peptide H domains that meet the necessary ‘threshold’ of hydrophobicity are not recognized by the SRP pathway (Lee and Bernstein 2001; Huber et al.2005; Akopian et al.2013) because SRP recognition and binding can be affected by other factors such as the reduced elongation speed or translational arrest during initial protein synthesis (Fluman et al.2014; Pechmann, Chartron and Frydman 2014), or by the presence of helix-destabilizing residue(s) within the N-terminal hydrophobic domain (Adams et al.2002).
In this case, cytosolic chaperones, such as the bacterial SecB, might help maintain a protein in an unfolded conformation. SecB also subsequently targets proteins for delivery to SecA, the Sec translocase. This ATPase can facilitate protein translocation across the cytoplasmic membrane via the Sec translocon (Fig. 3A) (Woodbury et al.2000; Banerjee, Lindenthal and Oliver 2017; Tsirigotaki et al.2017). While an H.volcanii chimeric protein in which dihydrofolate reductase has been fused to the S-layer glycoprotein (SLG) signal peptide appears to be translocated post-translationally, chaperones that can help proteins in an unfolded conformation in the cytoplasm have not yet been identified in this species (Irihimovitch and Eichler 2003).
Additionally, proteins can be post-translationally targeted to the bacterial and archaeal cytoplasmic membrane in a folded conformation and translocated across or inserted into the cytoplasmic membrane via the Tat transport pathway (Fig. 3A) (see below). Proteins that are targeted to the Tat pathway include, but are not limited to, those that must fold properly to bind a co-factor prior to secretion across the cytoplasmic membrane or those that are pre-assembled into heterooligomeric complexes. Tat substrates are targeted to the Tat translocon-associated membrane protein, TatC, via an N-terminal signal sequence that has a tripartite structure resembling that of the Sec signal peptide. However, the Tat substrate signal peptide contains a distinct, nearly invariant, pair of arginine residues, from which its nomenclature originated, within a consensus motif that precedes the H domain (Goosens and van Dijl 2017). Substitution mutants in which either arginine of this pair is replaced with another amino acid residue are not targeted to the Tat translocon (Sargent et al.1998; Buchanan et al.2001; Rose et al.2002). Moreover, intrinsic properties of the Tat substrate signal peptide prevent mistargeting of these proteins to the Sec translocon. Amino acids adjacent to the twin arginines and a relatively low degree of hydrophobicity of the Tat signal peptide play a critical role in modulating the targeting of substrates to the Tat translocation pathway (Cristóbal et al.1999; Rose et al.2002; Dilks et al.2003; Huang and Palmer 2017).
PROTEIN TRANSPORT INTO OR ACROSS THE MEMBRANE
While a few low-complexity membrane proteins appear to be inserted into the cytoplasmic membrane spontaneously, either insertases or translocases catalyze the insertion of the majority of membrane proteins into the prokaryotic cytoplasmic membrane or the eukaryotic ER membrane (Fig. 3A). Translocases are also required for protein transport across these membranes, although a subset of proteins might cross the membrane via membrane vesicles, a topic that is discussed in the review by Forterre et al. (submitted manuscript).
YidC-dependent membrane protein insertion (YidC insertase)
The first member of the YidC/Oxa1/Alb3 insertase family identified was Oxa1, which is involved in the membrane insertion of the N-terminal tail of cytochrome c oxidase subunit II in mitochondria (Cox II) (He and Fox 1997; Hell et al.1997). Later, using Oxa1 in a comparative sequence analysis led to the identification of Alb3 in chloroplasts and YidC in bacteria. Oxa1, Alb3 and the YidC homologs found in Gram-positive bacteria share a conserved five TM domain topology that folds to form a core hydrophilic groove that is critical for insertase activity (Jiang et al.2003; Mathieu et al. 2010; Kumazaki et al.2014a). Based on X-ray crystal structures obtained for YidC, it is believed that this groove interacts with a substrate at an amphiphilic protein–lipid interface, thereby allowing the TM segments of the substrate to slide into the lipid bilayer (Kuhn and Kiefer 2017; Kuhn, Koch and Dalbey 2017). The YidC homologs of Gram-negative bacteria contain an additional TM domain at their N-termini that may serve as a membrane targeting signal (Sääf et al.1998; Kumazaki et al.2014b). However, this additional TM segment does not interact with the conserved TM domains, and it is dispensable for insertase activity (Jiang et al.2003; Kumazaki et al.2014b). Moreover, despite differences between the TM domains, YidC/Oxa1/Alb3 homologs can all complement the functions of each other in deletion mutants (Jiang et al.2002; van Bloois et al.2005; Dong et al.2008; Funes et al.2009).
An archaeal DUF106 domain protein was found to be YidC-like even though it has only three TM domains and might therefore be the simplest known member of this family of insertases. These archaeal proteins share only weak sequence similarity with YidC insertases, and, to date, none has been able to replace the function of YidC/Oxa1/Alb3 (Luirink, Samuelsson and de Gier 2001; Zhang, Tian and Wen 2009; Borowska et al.2015). However, a recently determined crystal structure revealed that the DUF106 domain protein MJ0480 from the archaeon M. jannaschii shares an intriguing structural similarity with the core region of the E. coli YidC insertase (Borowska et al.2015). Analysis of the crystal structure of MJ0480 showed that the locations of the three TM domains correspond to the locations of TM1, TM2 and TM5 in the YidC of Gram-positive bacteria (Fig. 3B) as well as to TM2, TM3 and TM6 of the E. coli YidC. Furthermore, a more detailed analysis of the M. jannaschii YidC sequence revealed additional hydrophobic segments showing distant similarity to the E. coli YidC TM4 and TM5 domains (Kuhn and Kiefer 2017). Additionally, the MJ0480 peptide sequence is predicted to form a coiled-coil motif near the cytosolic interface of its hydrophilic groove, which also contains a motif known to be essential to YidC function (Chen et al.2014; Borowska et al.2015). Hence, despite showing only distant homology to other YidC/Oxa1/Alb3 insertases, archaeal YidC homologs have a similar conserved core hydrophilic groove structure, suggesting that these archaeal proteins are indeed bona fide YidC insertases. Moreover, while direct proof that MJ0480 can insert a substrate into the membrane is lacking, using a photocrosslinking assay Borowska et al. (2015) showed that MJ0480 can bind E. coli RNC complexes during translation, similar to E. coli YidC, supporting the view that DUF106 containing proteins belong to the same insertase family. A comparative homology analysis of archaeal proteins identified additional DUF106 domain containing homologs in some members of the Crenarchoeta and Korarchaeota phyla, indicating that a wide variety of archaeal species have YidC homologs (Kuhn and Kiefer 2017) (Table 2).
Sec-mediated membrane protein insertion and protein translocation across the cytoplasmic membrane
The Sec translocon, including SecY (Sec61α in eukaryotes), is highly conserved across all three domains of life but archaeal SecY is more similar to the eukaryotic Sec61α than to the bacterial SecY (Tsirigotaki et al.2017). The high degree of conservation of this pore component extends to functionality as expression of archaeal SecY from Methanococcus vanielli can rescue the phenotype caused by the deletion of secY in E. coli (Auer, Spicker and Bock 1991). The second pore component, Sec61γ in eukaryotes and SecE in bacteria, lacks significant sequence conservation between eukaryotes and bacteria. Archaeal SecE shows sequence conservation to eukaryotic Sec61γ, while the genomic localization is conserved between archaea and bacteria. This clearly demonstrates an evolutionary link between these pore components in all three domains (Hartmann et al.1994). Finally, the third component of the archaeal and eukaryotic Sec protein-conducting channel is Sec61β. However, these subunits share no sequence conservation with their bacterial counterpart, SecG (Pohlschroder et al.1997; Pohlschroder et al.2005).
Insight into how these protein components are assembled into a functional Sec translocon, and hence the mechanism by which proteins are inserted into, or transported across, the membrane, was revealed through structure determination for an archaeal complex (Van den Berg et al.2004). The high-resolution structure of the Sec translocon from M. jannaschii revealed that the three subunits SecY, SecE and Sec61β are assembled into an hourglass-shaped protein-conducting channel. This channel funnels toward a pore opening that is occupied by a plug domain. The hydrophilic interior of the channel and its hourglass structure are proposed to reduce interactions with the translocated peptide, thus facilitating its movement through the hydrophobic barrier imposed by the cytoplasmic membrane, while the plug domain serves to maintain the seal of the channel pore (Van den Berg et al.2004). This study, as well as the subsequent determination of the crystal structure of the Pyrococcusfuriosus SecEY channel (Egea and Stroud 2010), also confirmed that, in addition to having a controlled opening across the membrane for the transport of proteins through the membrane, the Sec channel adopts a ‘clam shell’ structure (Fig. 3C). This structure allows the channel to form a lateral gate opening over its entire length at the interface to the lipid bilayer. This facilitates the insertion of the H domain of the signal peptide or TM segments of a translocated protein into the cytoplasmic membrane (Van den Berg et al.2004; Egea and Stroud 2010). The main part of the Sec channel is composed of SecY whereby its 10 TM domains form two halves that are connected by a hinge domain. These establish the ‘clam shell’ structure as well as the overall structure of the hourglass-shaped channel. SecY also has an extended cytoplasmic loop that interacts with the protein targeting machinery. Meanwhile, SecE serves as a clamp that holds the SecY channel together and modulates its lateral gate opening (Van den Berg et al.2004). The P. furiosus Sec channel crystal structure also indicates the fidelity of the Sec channel, as the lateral gate opening does not disrupt the plug domain position in sealing the channel pore (Egea and Stroud 2010). The structural features described here are also consistent with the subsequently determined crystal structures determined for the bacterial Sec translocon (Tsukazaki et al.2008; Tanaka et al.2015; Li et al.2016).
The mechanism of protein translocation through the Sec translocon can be described as follows: as the protein carrying the signal peptide is targeted to the Sec translocon, its positively charged N-terminus remains on the cytosolic side of the membrane via the charge interaction with the negatively charged phospholipid head groups. The signal peptide binding destabilizes the close state of the channel to allow protein translocation into the channel, which occurs as a loop. This displaces the plug domain, and opens the pore for the translocation across the cytoplasmic membrane. While the H domain of the signal peptide is detected within the channel, it is laterally inserted into the cytoplasmic membrane and positioned in the groove just outside the channel lateral gate. As the protein translocates across the channel via the pore and the TM spanning domains are inserted into the cytoplasmic membrane, the signal peptide remains inside the binding pocket of the lateral gate. This pocket is located at the interface between the channel and lipid. It was postulated that upon complete translocation of the proteins, the C-terminal part of the signal peptide undergoes conformational changes that would expose the cleavage site to be processed by a signal peptidase (see below) (Li et al.2016; Rapoport, Li and Park 2017).
For co-translational insertion, upon being targeted to the membrane by SRP and its receptor, the RNC-SRP binds the cytoplasmic domain of the SecY/Sec61α channel to initiate translocation of the nascent peptide chain in a GTP-dependent manner (Mitra et al.2005; Jomaa et al. 2016). Since H. volcanii SecY and SecE homologs also bind to the haloarchaeal RNC-SRP, co-translational translocation in archaea likely occurs in a similar manner (Ring and Eichler 2004).
In bacteria, post-translational translocation of proteins across the Sec translocon also requires the SecA ATPase, which not only helps targeting fully translated proteins to the Sec translocon, but also provides the energy needed to translocate the proteins across the channel (Oliver and Beckwith 1981; Jungnickel and Rapoport 1995; Osborne, Clemons and Rapoport 2004; Park and Rapoport 2012). Conversely, in eukaryotes, Bip, an ATP-dependent chaperone in the ER lumen, drives post-translational translocation of the eukaryotic Sec substrate by binding to the emerging protein, thus providing directionality of the transport (Rapoport 2007; Rapoport, Li and Park 2017). While previous studies have indicated that haloarchaea also perform post-translational translocation of Sec substrates, proteins which substitute for SecA have yet to be identified in archaea, and an ATPase-driven ‘pulling’ of substrates through the pore, as observed in eukaryotes, would require an extracellular source of ATP (Ortenberg and Mevarech 2000; Irihimovitch and Eichler 2003). This suggests that as yet unidentified proteins are required to provide the energy necessary for post-translational translocation through the archaeal Sec translocon (Nguyen, Law and Williams 1991; Pohlschroder et al.1997; Rusch and Kendall 2007; Calo and Eichler 2011). For example, a crystal structure along with in vitro analysis of the two membrane-associated proteins of the Thermus thermophilus SecDF complex revealed that this complex can facilitate translocation of unfolded proteins across the Sec translocon. The energy of this ATP-independent translocation is provided by the proton motive force that regulates the conformational changes of the complex that drive protein translocation and prevent backsliding into the cytoplasm (Tsukazaki et al.2011). SecDF complex from the halophilic bacterium Vibrio alginolyticus had been shown to utilize the sodium gradient across the membrane for facilitating its protein translocation across the Sec translocon (Tokuda, Kim and Mizushima 1990; Tsukazaki et al.2011). In bacteria, SecD and SecF form a complex with YajC (Duong and Wickner 1997). Some of the archaea within the euryarchaeota phylum and from several other phyla encode homologs of the SecD and SecF proteins, while a YajC homolog is not present in any sequenced archaeal genome (Pohlschroder et al.2005). The archaeal SecD and SecF proteins were shown to form a complex and removal of these proteins in H. volcanii resulted in a cold-sensitive growth phenotype and a secretion defect, phenotypes that are similarly observed upon SecDFYajC mutation in bacteria (Hand et al.2006). Hence, it is possible that the SecDF complex utilizes an ion gradient to drive the post-translational protein transport in archaea, instead of relying on an ATPase like SecA. Clearly an effort to understand the Sec post-translational translocation mechanism in archaea is needed. These findings indicate that the archaeal Sec protein-conducting channel comprises properties of bacterial as well as eukaryotic proteins (Pohlschroder et al.1997; Kinch, Saier and Grishin 2002; Rapoport, Li and Park 2017).
Tat-mediated protein transport across the membrane
In stark contrast to the Sec transport pathway, the Tat pathway is dedicated to the translocation of folded proteins across the cytoplasmic membrane (Fig. 3A) (Goosens and van Dijl 2017). This translocase only allows translocation of properly folded proteins due to the presence of a critical structural proofreading domain that recognizes misfolded proteins and prevents their translocation (Matos, Robinson and Di Cola 2008; Rocco, Waraho-Zhmayev and DeLisa 2012). Much of what we know about the mechanism underlying transport via the Tat pathway stems from studies in bacteria and chloroplasts. The components of the Tat transport system are comprised of up to three functionally distinct proteins, TatA, TatA-like (TatB) and TatC (Table 2) (Goosens and Dijl 2017). The TatB and TatC proteins are preassembled at the cytoplasmic membrane (Berks 2014; Palmer and Berks 2012). TatBC substrate complexes trigger the oligomerization of TatA proteins, which form the translocase of the Tat system and facilitate substrate translocation across the cytoplasmic membrane (Fröbel, Rose and Müller 2011; Goosens and van Dijl 2017). Once Tat substrate translocation is completed, and its N-terminal signal peptide is processed by a signal peptidase, the TatA subunits disassemble from the TatABC complex (Mori and Cline 2002). Despite this apparent elucidation of the molecular mechanisms supporting Tat substrate translocation, the mechanistic details of the translocation process require clarification. While some have suggested that oligomerized TatA forms a protein conducting channel (Gohlke et al.2005; Walther et al.2013), others propose that the role of TatA in translocation is to destabilize the local membrane bilayer (Bruser and Sanders 2003; Beck et al.2013; Rodriguez et al.2013). While energy required for protein translocation of Tat substrates is typically drawn from a proton gradient across the membrane, in some haloarchaea such as Haloarcula hispanica a sodium gradient provides the energy required for translocation of Tat substrates (Kwan, Thomas and Bolhuis 2008; Goosens and van Dijl 2017).
Analyses of archaeal genomes have indicated that some encode multiple homologs of TatA and TatC, while none encode an identifiable TatB homolog (Pohlschroder et al.2004; Yuan et al.2010). For example, the H. volcanii and the S.solfataricus genomes each encode two paralogs of TatA and TatC. However, there is no correlation between the number of Tat component paralogs and the number of the Tat substrates encoded by the organisms: S. solfataricus encodes only five predicted Tat substrates while H. salinarum NRC-1 encodes more than 50 putative Tat substrates, despite encoding only a single TatA and two TatC homologs (Dilks et al.2003).
In fact, in bacteria, as in archaea, the Tat pathway is used to varying extents. In silico analyses have revealed that the haloarchaea use the Tat pathway to transport nearly half of their secreted proteins (Table 1) (Dilks et al.2003; Storf et al.2010). In all non-haloarchaeal genomes, the Sec pathway is predicted to transport the vast majority of their secreted proteins, even though several of the genomes encode Tat pathway components as well as Tat substrates (Dilks et al.2003). This implies that halophilic archaea have evolved a preference for the Tat pathway as the primary pathway used in protein translocation, probably as an adaptation to their high salt environments. The proteins produced by haloarchaea have highly negatively charged surfaces that provide hydration shells, which prevent precipitation and improve stability upon exposure to a high salt environment (Danson and Hough 1997; Madern, Ebel and Zaccai 2000). Hence, folding of proteins in the cytoplasm, where ATP-driven chaperones can efficiently facilitate protein folding, may prevent the accumulation of misfolded and aggregated proteins in the extracytoplasmic environment, which is devoid of ATP. Therefore, since the Tat pathway transports proteins in a folded conformation, it appears to provide the best means by which to transport secreted proteins across the cytoplasmic membranes of the haloarchaea. This does not apply to the subunits of surface structures, such as the SLG and the pilins, where cytoplasmic folding might lead to premature polymerization. Consistent with the extensive use of the Tat transport pathway, three of its four components are essential to the viability of H. volcanii (Dilks, Gimenez and Pohlschroder 2005).
PRE-PROTEIN PROCESSING AND PROTEIN ANCHORING
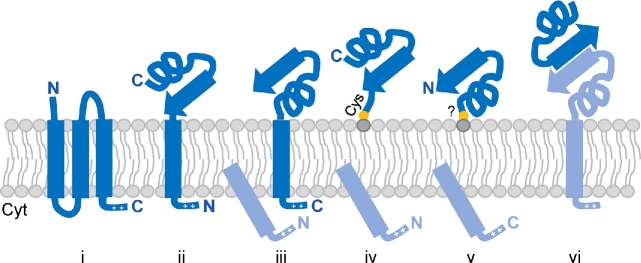
For integral membrane proteins, the signal-anchor sequence, recognized by the SRP as it exits the ribosome, is its first translated TM domain. Conversely, for many proteins that are translocated across membranes, the H domain of the N-terminal signal peptide is recognized. This is typically removed from the protein by a signal peptidase subsequent to translocation. Hence, proteins that are not released into the extracellular environment must be anchored to the cell surface using mechanisms other than intercalation of an N-terminal TM domain into the lipid membrane. In archaea, these anchoring mechanisms include intercalation of a C-terminal hydrophobic domain into the membrane; covalent attachment of a lipid moiety to either the N-terminus or C-terminus, which is then embedded in the cytoplasmic membrane; and strong interactions with other surface anchored proteins (Fig. 4).
Figure 4.
Anchoring strategies of archaeal surface proteins. Proteins can be anchored via multiple TM domains (i), single N- or C-terminal TM domains (ii and iii, respectively); N- or C-terminal covalent lipid interactions (iv and v, respectively); or interactions with other surface-anchored proteins (vi). Cleaved signal peptides and interacting surface proteins are colored in light blue.
TM anchoring of proteins processed by signal peptidase I
Many proteins targeted to the Sec or Tat pathways have an N-terminal signal peptide that targets substrates to the proper transport pathway and contains a conserved recognition site that is cleaved by signal peptidase I (SPI) (Eichler 2002; Tuteja 2005; Dalbey, Pei and Ekici 2017). The processing site recognized by SPI is located just downstream of the signal peptide H domain (Dalbey, Pei and Ekici 2017). Archaeal SPI activity was first demonstrated in the euryarchaeon M.voltae, where it was shown to mediate the removal of the SLG signal peptide, a well-characterized Sec substrate (Ng and Jarrell 2003). The archaeal SPI is more closely related to the eukaryotic one than to the bacterial SPI, having protein domains and the catalytic residues required for its processing activity in common with the eukaryotic homolog (Eichler 2002; Ng and Jarrell 2003; Bardy et al.2005). Furthermore, some archaea have multiple paralogs of this enzyme. For instance, H. volcanii has two SPI paralogs Sec11a and Sec11b, of which only Sec11b is essential (Fine et al.2006). Perhaps the additional paralog is required for processing either a specific subset of substrates or is required to cope with an increased amount of secreted proteins under certain growth conditions (Fine et al.2006; Ng et al.2007).
SPI processed proteins other than membrane-embedded proteins must be anchored using a different mechanism in order to remain membrane bound. Proteins that are transported via the Sec pathway may carry an H domain at the C-terminus that is inserted into the cytoplasmic membrane via the Sec translocon lateral opening. Examples of archaeal Sec substrates that undergo signal peptide processing but are TM anchored to the cytoplasmic membrane via intercalation of a C-terminal TM domain are the SlaB subunits of S. acidocaldarius and S. solfataricus S-layers (Grogan 1996a,b; Veith et al.2009; Albers and Meyer 2011; Sleytr et al.2014).
A subset of bacterial Tat substrates is anchored to the cytoplasmic membrane via intercalation of an N- or C-terminal TM domain into the membrane (Hatzixanthis, Palmer and Sargent 2003; Bachmann et al.2006). While Tat substrates anchored to the membrane by its N-terminal TM domain have not yet been identified in archaea, the H. volcanii Tat substrates halocyanin 2 and halocyanin 3 are known to use C-terminal TM domain intercalation for membrane anchoring (Gimenez, Dilks and Pohlschroder 2007). In contrast to Sec substrates, it is not yet known whether the TM domains of Tat substrates exit the Tat translocon laterally into the bilayer, or rather emerge on the extracellular side with a subsequent re-insertion into the cytoplasmic membrane. However, the membrane insertion of Tat substrates is independent of the membrane insertase YidC (Hatzixanthis, Palmer and Sargent 2003). It has been suggested that the N-terminal signal peptide of Tat substrates is intercalated into the cytoplasmic membrane by an unknown insertase prior to binding to the Tat(B)C complex (Bruser and Sanders 2003; Hou, Frielingsdorf and Klösgen 2006; Shanmugham et al.2006; Bageshwar et al.2009). Meanwhile, other studies have shown that TatC does not only serve as the binding site of the Tat substrate signal sequence (Holzapfel et al.2007; Zoufaly et al.2012) but also functions as insertase in mediating the hairpin-like membrane insertion of the Tat substrate signal sequence into the cytoplasmic membrane (Fröbel et al.2012).
Archaeosortase-dependent C-terminal covalent lipid anchoring
In both bacteria and archaea, a subset of Sec substrates has a C-terminal TM domain that is only temporarily anchored to the membrane (Fig. 4). The best-studied system is the C-terminal anchoring of proteins to the cell wall of Gram-positive bacteria, which is catalyzed by sortases. These transpeptidases recognize a conserved C-terminal tripartite structure containing an H domain, preceded by a conserved amino acid motif, and followed by a highly charged region. In Staphylococcus aureus, an LPXTG motif-containing tripartite structure is recognized by sortase A (SrtA), which subsequently processes the substrate, and transfers it to a peptidoglycan precursor (Schneewind and Missiakas 2014). More recently, a subset of Sec substrates in Gram-negative bacteria was identified that contain a tripartite structure reminiscent of those of sortase substrates, but having PEP as their motif (Haft et al.2006; Haft, Payne and Selengut 2012). However, the genomes encoding these proteins do not encode sortase homologs (Haft et al.2006). By using the partial phylogenetic profiling method to analyze the genomes that encode proteins with these conserved C-termini, a protein was identified, and termed exosortase, which was proposed to be responsible for the C-terminal processing and anchoring of its substrates (Haft et al.2006). This finding also led to the discovery of a distant homolog of exosortases, termed archaeosortase, in a subset of Euryarchaeota (Table 2). Archaeosortases can be categorized into different subfamilies: Archaeosortase A (ArtA), ArtB, ArtC, ArtD and ArtE (Haft, Payne and Selengut 2012). The genomes that encode archaeosortase also encode putative substrates, with a C-terminal tripartite structure albeit with a distinct motif for each subfamily. ArtA-encoding genomes have from just one to as many as 52 predicted substrates containing a C-terminal PGF motif rather than the PEP motif (Haft, Payne and Selengut 2012). Despite lacking homology to the sortase, the exo/archaeosortase is proposed to carry out a similar proteolytic cleavage reaction. Amino acids of the putative catalytic triad of sortases are conserved residues in archaeosortases (Ton-That et al.2002; Haft, Payne and Selengut 2012; Abdul Halim et al.2018).
While exosortases have not yet been experimentally analyzed, the ArtA-dependent anchoring mechanism has been studied in H. volcanii. Unlike many Euryarchaeota that encode two or three archaeosortase paralogs from different subfamilies, the model archaeon H. volcanii encodes only a single archaeosortase, ArtA (Haft, Payne and Selengut 2012; Abdul Halim et al.2013, 2018). The associated tripartite structure with its PGF motif is present in nine proteins, including the H. volcanii SLG, which was long thought to be anchored to the cell surface via intercalation of a C-terminal TM domain (Lechner and Sumper 1987; Sumper et al.1990), similar to the Sulfolobus SlaB S-layer subunit (Veith et al.2009).
While an H. volcanii ΔartA strain is viable, it exhibits a severe growth defect, particularly under low salt conditions, as well as impaired swimming motility, abnormal cellular morphology and lower mating efficiency, all consistent with an impaired S-layer function (Abdul Halim et al.2013; Banerjee et al.2015). Mass spectrometry indicated that the C-terminal hydrophobic peptide is absent from SLG in wild-type cells, precluding it to function as a C-terminal membrane anchor. However, C-terminal peptides could be identified in a ΔartA strain, supporting the view that the C-terminus is proteolytically processed in the wild type (Abdul Halim et al.2013). The importance of the conserved PGF motif could be validated by its permutative conversion to PFG, which precludes proteolytic processing (Abdul Halim et al.2015). These results are consistent with detailed mass spectrometric analyses of proteins from M. hungatei and Methanosarcina barkeri containing the conserved tripartite structure. All such analyses showed an absence of C-terminal peptides in protein extracts of wild-type cells (Haft, Payne and Selengut 2012).
ArtA substrates may be translocated via the Sec as well as the Tat pathway. Direct evidence of ArtA-dependent processing was obtained using a H. volcanii Tat substrate that has a conserved PGF-tripartite structure located towards the center of the protein, rather than near the C-terminus. This unusual placement could be attributed to a strain-specific gene fusion, which couples an ArtA substrate to a downstream gene with a NifU domain. Unlike processed short C-terminal fragments from typical ArtA substrates that are difficult to detect, the C-terminal fragment of the fusion gene is long and thus amenable to experimental detection. For this substrate, ArtA-processed N- and C-terminal fragments could be readily identified, validating that archaeosortase has proteolytic activity (Abdul Halim et al.2017). Using this ArtA substrate in combination with site-directed ArtA mutants confirmed the importance of predicted active site residues, which are conserved in ArtA and resemble those of sortases (Abdul Halim et al.2018).
However, unlike sortase substrates, which are anchored to a peptidoglycan precursor prior to insertion into the membrane, ArtA substrates are anchored to the cytoplasmic membrane via a covalent lipid attachment to their C-termini (Haft, Payne and Selengut 2012). This explains previous results showing that the C-termini of H. salinarum and H. volcanii SLG are modified by covalently linked lipids (Kikuchi, Sagami and Ogura 1999; Konrad and Eichler 2002; Abdul Halim et al.2015). While lipidation is ArtA dependent (Abdul Halim et al.2015), it remains to be elucidated if ArtA is directly responsible for the lipidation reaction. ArtA-dependend lipidation was the first description of a C-terminal lipid-anchoring mechanism for secreted proteins identified in prokaryotes (Fig. 4).
Possibly, the C-terminal processing and lipid anchoring of ArtA substrates minimizes overcrowding of the cytoplasmic membrane, since a lipid anchor takes up less space in the membrane than a TM domain. This is especially true for SLG, which is one of the most abundant proteins in haloarchaea (Sleytr et al.2014). It should, however, be noted that two fractions of the H. volcanii S-layer protein could be separated, which were proposed to represent a TM-anchored and a lipid-anchored fraction (Kandiba, Guan and Eichler 2013). Currently, it is not clear if the TM-anchored fraction is an assembly intermediate or if a minor subset of the SLGs retain their TM domain. If retained at all, the amount must be minor as peptides from this region were below detection limit in wild-type strains (Abdul Halim et al.2013). In Sulfolobales, which lack an archaeosortase, the S-layer is composed of iterations of two subunits, only one of which is C-terminally TM anchored (see below) (Veith et al.2009). Finally, it may be that lipid-mediated protein anchoring allows for more rapid protein shedding when local conditions change drastically.
N-terminal covalent lipid anchoring
N-terminal lipid anchoring of secreted proteins to the cytoplasmic membrane has been studied in detail in bacteria and partially in archaea (Fig. 4) (Gimenez, Dilks and Pohlschroder 2007; Narita and Tokuda 2017). Such lipoproteins contain a highly conserved motif known as lipobox that is located at the C-terminal end of the signal peptide H domain (Buddelmeijer 2015). The lipobox contains a canonical LAGC motif with the cysteine being completely and the glycine highly conserved (Narita and Tokuda 2017). Upon translocation across the cytoplasmic membrane, by either the Sec or Tat transport pathway, the cysteine sulfhydryl group side chain is modified in bacteria with a diacylglycerol moiety by prolipoprotein diacylglyceryl transferase (Lgt), resulting in the anchoring of these lipoproteins to the cytoplasmic membrane via intercalation of two acyl chains (Mao et al.2016). Subsequently, signal peptidase II (SPII) processes these lipoproteins, removing the signal peptide immediately upstream of the diacylated cysteine residue of the lipobox motif to ensure membrane-anchoring of the processed lipoprotein. In some bacteria, these lipoproteins are further modified by the addition of a third acyl group via amide linkage to the amino group of the cysteine residue, catalyzed by the apolipoprotein N-acyltransferase (Lnt) (Lu et al.2017; Noland et al.2017; Wiktor et al.2017).
While genes encoding homologs of SPII, Lgt or Lnt have not been identified in any archaeal genome, in silico analyses of euryarchaeal genomes have readily detected putative signal peptides containing the canonical lipobox, especially in the haloarchaea (Storf et al.2010) (Table 1). While replacement of the conserved lipobox cysteine in the H. volcanii Tat substrates, disulfide bond formation protein A and maltose-binding protein, results in unprocessed mutant proteins that are released into the supernatant, some cysteine replacement Tat lipoprotein mutants remain bound to the cell surface, likely due to membrane-anchoring mediated by the hydrophobic stretch of the unprocessed signal peptide (Gimenez, Dilks and Pohlschroder 2007).
Reminiscent of sortase and archaeosortase substrates, while the signal peptide tripartite structure and, in this case, the amino acid sequence of the lipobox motif are conserved, the anchoring machinery of lipobox containing lipoproteins appears to be distinct to each domain. While both bacterial and archaeal substrates are membrane anchored, the distinct archaeal machinery might be required because of its unique membrane composition. Unfortunately, neither in silico nor in vivo studies have led to the identification of the archaeal lipobox containing lipoprotein biosynthesis components, thus far (Szabo and Pohlschroder 2012).
SURFACE STRUCTURE BIOSYNTHESIS
Electron microscopy, and to some extent, biochemical analyses have revealed that archaeal surfaces are decorated by a highly diverse array of proteinaceous structures, including some that appear to be specific to certain species or phyla, as well as structures that are conserved across all phyla (Fig. 1). However, to date, a detailed understanding of the biosynthesis of archaeal surface structures remains largely limited to the S-layer and type IV pilus-like structures.
S-layer
Most well-characterized archaea have a pseudocrystalline proteinaceous S-layer that envelops the cytoplasmic membrane (Sleytr et al.2014). S-layers typically consist of a single SLG, but co-expression of S-layer protein isoforms (Lu et al.2015) as well as S-layers composed of heterodimers have also been found (Veith et al.2009; Albers and Meyer 2011). Such paracrystalline protein layers are found in nearly all cultured archaea and their geometry has been characterized for species from a wide range of the archaeal phylogeny (Baumeister and Lembcke 1992; Trachtenberg, Pinnick and Kessel 2000; Arbing et al.2012). The S-layer covers the whole cell and is in most cases the only constituent of the cell envelope outside the cytoplasmic membrane (Albers and Meyer 2011; Sleytr et al.2014; Engelhardt 2016; Rodrigues-Oliveira et al.2017). Thus, it forms the outermost cell layer, being directly exposed to the environment, which for many archaea is harsh in their natural habitats. As S-layers are not only found in archaea but are also common in bacteria, they may represent cell wall structures, which emerged very early in cellular evolution (Albers and Meyer 2011; Sleytr et al.2014). The archaeal S-layer appears to take on roles similar to those of the bacterial peptidoglycan cell wall for functions such as maintaining cell stability and morphology. In addition, it creates a pseudoperiplasmic space around the cell by acting as a permeability filter (Baumeister and Lembcke 1992). The importance of the S-layer to cope with cell turgor has been reported (Engelhardt 2016). Two notable functions have been proposed for the S-layer: its ‘antifouling’ property (nano-lotus-effect), which minimizes unspecific binding and could be important for functions such as nutrient uptake. This is reminiscent of the self-cleaning properties of the lotus plant. Due to their structural properties, S-layers may also reduce flow resistance (a type of nano-sharkskin effect), which may facilitate archaella-driven motility (Sleytr et al.2014). This is reminiscent of sharkskin properties supporting fast movements. The S-layer, like other cell surface structures, may also be relevant for virus/phage susceptibility (Kandiba et al.2012). Remarkably, the SLGs of the Haloquadratum walsbyi strains HBSQ001 and C23 are highly divergent, despite a very high overall genome similarity. Detailed analysis showed that the divergence is not due to an enhanced mutation rate. Instead, one strain must have incorporated a novel SLG from foreign DNA at the expense of removal of the original SLG and its encoding genome region (Legault et al.2006; Dyall-Smith et al.2011). The Haloquadratum pangenome contains a set of genomic islands that code for diverse SLGs (Martin-Cuadrado, Pasic and Rodriguez-Valera 2015), perhaps allowing SLG switching to disguise the organism from being targeted by viruses. However, while the peculiar square shape is strictly conserved for all strains, it is unlikely that the SLG defines the overall cell shape, since a diverse set of SLGs can be used as an S-layer (Legault et al.2006).
The SLG, which forms the S-layer, is one of the most abundant archaeal proteins, making up around 10% of the cellular protein content. Accordingly, the S-layer is a metabolically expensive product that places a strong demand on the protein biosynthesis and protein secretion machineries, a cost that indicates that the S-layer must confer a strong selective advantage (Sleytr et al.2014; Engelhardt 2016). S-layer proteins make two types of contact: one between subunits to allow lateral assembly and the other to underlying cell structures, which in archaea is commonly the cytoplasmic membrane (Sleytr et al.2014; Engelhardt 2016). The 2D crystals formed by S-layers have various symmetries, which can be summarized as oblique (p1, p2), square (p4) or hexagonal (p3, p6), with hexagonal assemblies found to be predominant in archaea. S-layer proteins have been studied in many Euryarchaeota (halophiles, methanogens) and Crenarchaeota (for listings see Albers and Meyer 2011; Sleytr et al.2014; Rodrigues-Oliveira et al.2017). It should, however, be kept in mind that archaeal diversity is much wider than had previously been anticipated with the recent discovery of a considerable number of new archaeal phyla (Asgard group, DPANN group, novel sister taxa to Crenarchaeota in the TACK group) (Hug et al.2016; Zaremba-Niedzwiedzka et al.2017). These newly recognized phyla are frequently only defined by culture-independent methods so that they have not yet been analyzed with respect to their S-layer (Rodrigues-Oliveira et al.2017). Thus, it is uncertain whether what currently seems typical for the S-layer of a limited number of characterized species will hold true when the full archaeal diversity has been inspected.
Archaeal SLGs are predicted to have an N-terminal SPI processed Sec signal peptide and are thus transported via the Sec pathway in an unfolded configuration. N-terminal processing by signal sequence cleavage is evident from N-terminal sequencing, which has been done for many archaeal SLGs (Lechner and Sumper 1987; Sumper et al.1990). For M.voltae, signal peptide cleavage has been attributed to SPI (Ng and Jarrell 2003). The SLG is then subjected to various types of PTMs (see below). Transport by the Sec pathway implies that the SLG is folded outside the cytoplasmic membrane where it assembles spontaneously into the highly ordered structures that are typical for S-layers. Thus, folding and multimerization are intricately related. This would not be compatible with usage of the Tat secretion pathway because large complexes or aggregations might form in the cytosol prior to transport. The multimerization pattern is defined by the protein sequence and the lateral contacts made by the folded protein. It is independent of the underlying cell structures (Sleytr et al.2014) as was shown for bacteria but is likely to also hold true for archaea. By analyzing the S-layer proteins from the bacteria Thermoanaerobacter thermosaccharolyticum (with square lattice symmetry) and T.thermohydrosulfuricus (with hexagonal lattice symmetry), it was shown that the species-specific symmetry is regenerated upon in vitro refolding after denaturation (Sleytr 1976). Studies on self-assembly of archaeal SLGs are rare (Mark et al.2006). However, 3D structure analysis of the M.acetivorans SLG (MA0829) reveals a plausible model for the self-assembly into an S-layer (Arbing et al.2012).
The S-layer of two of the best-studied model archaea, the halophilic archaeon H. volcanii and of the thermoacidophile S. acidocaldarius, differs in several ways. As is typical, the H. volcanii S-layer is made up of a single protein (Sumper et al.1990). In contrast, the S-layer of S. acidocaldarius is composed of heterodimers, with a small, membrane-anchored subunit SlaB and a large subunit SlaA, which is not anchored to the cell membrane but rather to SlaB (Grogan 1996a; Veith et al.2009; Albers and Meyer 2011). Protein modeling revealed that the SlaB protein structure consists of multiple beta sandwich domains, which are followed by a coiled-coil domain and then a C-terminal TM domain (Veith et al.2009). Based on this structure, it was proposed that the SlaB protein serves as a membrane anchor for the other S-layer subunit, SlaA. The SlaB coiled-coil domain forms a rigid structure that protrudes outward and interacts with the SlaA subunit, thereby forming an S-layer dimer that is anchored to the cell surface (Veith et al.2009). The small subunit SlaB is anchored to the cell membrane via a C-terminal TM domain (Veith et al.2009; Albers and Meyer 2011). In contrast, much, if not all of the haloarchaeal SLG is only temporarily embedded in the membrane via its C-terminal TM domain as this is subsequently removed and replaced by a lipid anchor in an ArtA-dependent process (see above).
Besides signal peptide cleavage, and, in some species, lipidation, glycosylation of the SLG is a PTM that has been demonstrated for all SLGs, across various archaeal phyla, that have been characterized biochemically, indicating the importance of this PTM in S-layer biogenesis. Halobacteriumsalinarum was the first prokaryote in which both O- and N-glycosylation of proteins were detected (Mescher and Strominger 1976). In the former, glycans are covalently linked to serine or threonine residues, while in the latter glycans are attached to the side chains of asparagines. In contrast to bacteria, where O-glycosylation seems to be the predominant form of protein glycosylation on surface structures, archaeal cell surface proteins appear to be mainly modified with N-linked glycans (Messner, Schäffer and Kosma 2013; Nothaft and Szymanski 2013; Kaminski et al.2013b; Schaffer and Messner 2017). Similar to eukaryotic N-glycans that are sequentially assembled as lipid-linked oligosaccharides (LLOs) on the cytoplasmic side of the ER membrane, archaeal and bacterial N-glycans are assembled on the cytoplasmic side of the cell membrane. Mature LLOs are enzymatically flipped across the membrane in reactions catalyzed by flippases and then transferred en bloc onto an asparagine residue within the consensus sequence N-X-S/T. This transfer is typically catalyzed by an oligosaccharyltransferase (OST), which is the widely conserved AglB in archaea. AglB is distantly related to the bacterial OST PglB as well as STT3, the catalytic subunit of the eukaryotic OST complex (Table 2) (Matsumoto et al.2013; Kaminski et al.2013b; Wild et al.2018). However, in contrast to eukaryotic N-glycoproteins, which are modified by N-glycans having a common core, the structures and compositions of N-glycans in archaea are highly diverse, resulting from the distinct glycosylation pathways found in each species (reviewed in Jarrell et al.2014; Albers, Eichler and Aebi 2015). Some N-glycosylation pathways add monosaccharides to the glycan after it has been transferred onto the protein, such as the attachment of the terminal mannose in H. volcanii N-glycans (Abu-Qarn et al.2007). In combination with the transfer of incomplete LLO precursors, this can result in microheterogeneity, where different glycoforms of one glycan type can be found at the same glycosite (Kaminski et al.2013a; Esquivel et al.2016). Furthermore, multiple N-glycosylation pathways can be present in a single species and can simultaneously modify the same glycoprotein, as was shown for the SLGs of H. salinarum (Wieland 1988) and H. volcanii (Kaminski et al.2013a; Parente et al.2014) (Fig. 5A and B). This extended complexity is intriguing, especially since N-glycosylation of the SLG by an AglB-independent pathway in H. volcanii increases under low-salt stress conditions (Kaminski et al.2013a). However, little is known about the regulation of these alternate N-glycosylation pathways, and the reason why certain N-glycosites are typically modified by a distinct pathway has yet to be determined. Thus far, characterization of archaeal O-glycosylation has been performed only for haloarchaeal SLGs (Mescher and Strominger 1976; Sumper et al.1990; Lu et al.2015). However, characteristic serine and threonine-rich clusters are present in most SLGs, making it likely that they are also O-glycosylated. Unfortunately, the components of the biosynthetic pathway supporting this PTM have not yet been identified.
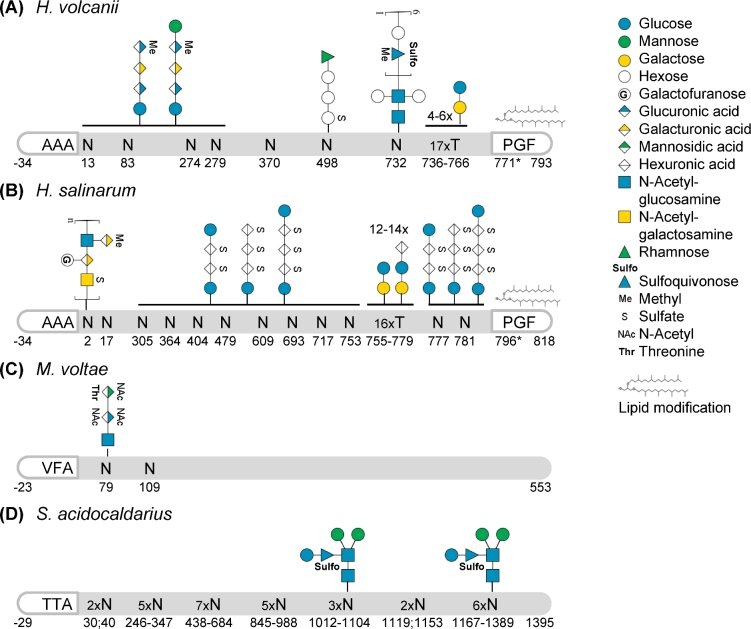
Figure 5.
Post-translational modifications of S-layer glycoproteins. Schematic representations of S-layer glycoproteins from (A)H. volcanii P25062, based on Sumper et al. (1990), Kaminski et al. (2013), Parente et al. (2014), Kandiba et al. (2016), Abdul Halim et al. (2017); (B)H. salinarum B0R8E4, based on Mescher and Strominger (1976), Wieland (1988), Kikuchi, Sagami and Ogura (1999), Jarrell et al. (2014); (C)M. voltae Q50833, based on Voisin et al. (2005); and (D)S. acidocaldarius Q4J6E5, based on Peyfoon et al. (2010). Their signal peptides (last three amino acids before the cleavage site, defined as the peptide bond between positions –1 and +1, are indicated), N- and O-glycosylation as well as lipid modification are highlighted. The position of the PGF motif, conserved for ArtA substrates, is indicated but the processing site has not been resolved yet (*). Glycan compositions are given for confirmed glycosites; solid horizontal lines indicate that all subjacent glycosites were identified with the glycans given above the line. It should be noted that for H. volcanii and S. acidocaldarius shorter glycans of the corresponding N-glycosylation pathways were identified for several N-glycosites and that the extent and type of N-glycosylation can depend on the growth condition. Monosaccharides are depicted according to the Symbol Nomenclature for Glycans (Varki et al.2015). While this is a selection of well-characterized SLG N-glycosylation, a more comprehensive summary can be found in Jarrell et al. (2014).
Since for most archaea, the S-layer represents the last barrier between the cell and its environment, it is tempting to speculate that the extent of SLG glycosylation corresponds in some way to the local environmental conditions. Indeed, it has been reported that the potential density of N-glycosylation, i.e. the number of putative glycosites (N-X-S/T sequons) in relation to the protein length, is greater in (hyper-)thermophilic archaea as compared to mesophilic archaea (Claus et al.2002; Jarrell, Ding et al.2014). For example, the SLG of the thermoacidophilic S. acidocaldarius contains 31 N-glycosylation consensus sites, while the SLG of the mesophilic M. voltae contains only two sequons (Fig. 5C and D). Within the halophiles, H. salinarum shows more extensive SLG N-glycosylation than H. volcanii, which thrives in less extreme salt concentrations (Fig. 5A and B). Glycosylation might, therefore, increase the thermostability and negative charge of the cell surface, with the latter being favorable for an extensively hydrated shell. The hypothesis that N-glycosylation is involved in adaptation to environmental conditions has been further supported by the impaired growth of N-glycosylation pathway mutants under stress conditions for various archaeal species (Abu-Qarn et al.2007; Meyer et al.2011; Meyer and Albers 2013). Additionally, changes in N-glycosylation patterns have been observed for different growth conditions (Guan et al.2012; Ding et al.2016). However, it should be noted that the occupation of putative N-glycosites, as well as corresponding N-glycan structures, is known for only a few species.
Besides the importance of N-glycosylation in affecting the role of SLG in maintaining cell morphology in H. salinarum (Mescher and Strominger 1975), the effect of altered N-glycosylation on SLG assembly and function has been studied mainly in H. volcanii. The deletion of aglB in H. volcanii leads to increased shedding of the SLG, while an altered protease susceptibility is observed in different N-glycosylation pathway mutants (Abu-Qarn et al.2007; Tamir and Eichler 2017). Furthermore, decreased secretion of reporter proteins into the culture medium was observed in cells having impaired N-glycosylation (Tamir and Eichler 2017). All of this points towards important roles for N-glycosylation in promoting the correct folding of SLG, and in the assembly of SLG into an organized S-layer.
Recently, SLG N-glycosylation was also found to be associated with mating in H. volcanii (Shalev et al.2017). However, while the deletion of genes encoding N-glycosylation enzymes results in reduced SLG N-glycosylation and reduced mating success, a direct role for SLG in mating has not been shown. Therefore, the specific cell surface N-glycoproteins involved in mating remain to be elucidated, especially since mutations in glycosylation pathways have pleiotropic effects.
Type IV pili
Type IV pili are generally composed of major and minor type IV pilins, with the major pilin comprising the bulk of the structure (Hansen and Forest 2006; Giltner, Nguyen and Burrows 2012). The type IV pilus biosynthesis pathway shares conserved core components that have been identified in representative species of most bacterial and archaeal phyla, suggesting that their common ancestor had these surface filaments (Szabo et al.2007; Imam et al.2011; Makarova, Koonin and Albers 2016). While type IV pili are required for effective adhesion to biotic and abiotic surfaces in both domains, type IV pilus-like structures have also evolved to carry out a variety of distinct functions, which is why these surface filaments have been referred to as ‘prokaryotic Swiss army knives’ (Berry and Pelicic 2015; Pohlschroder and Albers 2015; Chaudhury, Quax and Albers 2018). These type IV pilus-like structures include a diverse variety of surface structures such as the DNA-uptake structures of certain Gram-positive bacteria as well as the archaella, which are required to propel archaeal swimming motility and are discussed in a recent review (Albers and Jarrell 2018).
In bacteria, type IV pilins, the structural subunits of the type IV pili, are targeted to the Sec pore via the SRP and are co-translationally transported across the Sec translocon (Francetic et al.2007). While it has not been established whether archaeal pilins are co- or post-translationally transported, to date, all in vivo characterized archaeal pilins have predicted Sec signal peptides (Pohlschroder and Esquivel 2015). Prepilin peptidases, which are homologous in bacteria (PilD) and archaea (PibD/EppA), process pilins and pilin-like proteins. However, unlike other Sec substrates, these proteins are processed N-terminally to the H domain of the signal peptide, thus leaving an N-terminal hydrophobic segment that is likely transiently inserted into the cytoplasmic membrane (Albers, Szabo and Driessen 2003; Bardy and Jarrell 2003). While the bacterial prepilin peptidase also methylates the N-terminus of its substrates, this was not observed for archaeal prepilin peptidases. However, the significance of this PTM of bacterial pilins is enigmatic (Giltner, Nguyen and Burrows 2012).
Two distinct subfamilies of prepilin peptidases have been identified in archaea. One, PibD, has a broad substrate range, including pilins, while the other, EppA, processes a specific subset of pilins (see below) (Szabo et al.2007; Ng et al.2009; Nair et al.2014). The identification of putative pilins, which often lack significant sequence homology, has been aided by in silico analyses that can detect conserved prepilin signal peptides (Table 1 and Table S1, Supporting Information) (Szabo et al.2007; Imam et al.2011). The N-terminal H domain of processed type IV pilins serves as a structural scaffold at the core of the type IV pilus.
Other components required to assemble a functional type IV pilus, in addition to the prepilin peptidase, include PilB, an ATPase that provides the energy required for assembly, and PilC, a TM protein that anchors the pilus to the surface of the cell membrane. Each of these components form large families of homologs (Table 2) (Pohlschroder et al.2011; Melville and Craig 2013; Losensky et al.2014; Berry and Pelicic 2015; Pohlschroder and Esquivel 2015). These components, and their homologs, are typically encoded by pilB/pilC gene pairs in gene clusters that often also contain type IV pilin genes (Szabo et al.2007; Nair et al.2014). Interestingly, M. maripaludis contains two pilC paralogs in its pil operon, both of which are essential for piliation. Additionally, the characterization of this operon also revealed the importance of three minor pilins for piliation (Nair et al.2014). However, the major pilins, such as the M. maripaludis major pilin MMP1685, are often encoded by a gene having a distinct genomic location, possibly because prominent structural genes must be more extensively transcribed compared to the genes for the pilus biosynthesis components (Szabo et al.2007; Ng et al.2011; Makarova, Koonin and Albers 2016). Nair et al. (2013) also identified an additional minor pilin outside the pil operon that is required for piliation. Both bacterial and archaeal genomes frequently contain several pil operons, and prediction programs have identified genes encoding as few as three and as many as 50 prepilin-like proteins in various prokaryotic genomes (Szabo et al.2007; Imam et al.2011; Makarova, Koonin and Albers 2016).
As noted above, the archaeal swimming motility structure is evolutionarily related to type IV pili rather than bacterial flagella, leading to the renaming of these ‘rotating type-IV pili’ as archaella and its subunits as archaellins (Albers and Jarrell 2018). Unfortunately, the highly mnemonic gene names (fla for flagella, flagellins) were retained. To resolve this inconsistency, we propose to also rename the genes encoding the archaellins to arlA (previously flaA or flgA). Similarly, gene names starting with arl (previously fla) should be used for the operons of archellin-related (previously flagellin-related) genes. The archaellins are processed by PibD (Albers, Szabo and Driessen 2003; Bardy and Jarrell 2003; Tripepi, Imam and Pohlschroder 2010) and PilB and PilC homologs, ArlI (FlaI) and ArlJ (FlaJ) for archaella biosynthesis. ArlI is also the energy-providing component of the ATP-driven archaellar motor. Moreover, ArlH (FlaH), encoded by arlH (flaH) that is seemingly always co-localized with arlI and arlJ, is also required for archaella motility (Chaudhury et al.2016).
The M. maripaludis S2 genome encodes only two type IV pilus-like systems, the pil operon encoding PilB/PilC orthologs, EppA as well as three pilins processed by EppA, MMP0233, MMP0236 and MMP0237 (Ng et al.2011; Nair et al.2014) and the arl cluster. In addition to the arl cluster, S. acidocaldarius has two pil operons: a UV-inducible operon (ups) that encodes components of a type IV pilus involved in cellular aggregation and DNA exchange, and another operon that encodes components of the archaeal adhesive pili (aap), the traditional pili adhesion system. The ups operon includes two homologous genes that encode major pilins, UpsA and UpsB (Fröls et al.2008). In the presence of the PilB/PilC homologs, UpsE and UpsF, respectively, each of these pilins can independently lead to the assembly of functional pili (van Wolferen et al.2013). However, despite the absence of Aap pili in ΔaapF cells, which lack the PilB paralog AapF, these cells can still establish biofilms, although at a reduced capacity and with less complex architecture, suggesting that the UpsE homolog can, at least to some extent, catalyze the assembly of this pilus composed of non-cognate pilins (Henche et al.2012).
Studies performed in H. volcanii, which has five pil operons (pilB1C1–pilB5C5), as well as an arl operon, have revealed still greater complexity in the diversity of type IV pili in archaea. The assembly of type IV pili containing any of the adhesion pilins, PilA1–6, which are required for H. volcanii adhesion under laboratory conditions, involves PilB3/PilC3, which are encoded by an operon composed exclusively of these two genes (Esquivel, Xu and Pohlschroder 2013). Each of these adhesion pilins has a completely conserved pilin H domain that not only provides scaffolding for assembly of the type IV pilus core but also plays a critical role in regulating archaella-dependent motility (Esquivel and Pohlschroder 2014). Although the pilins PilA1-A6 can each be assembled into a type IV pilus that can mediate adherence to an abiotic surface, each of the pili thus formed appears to play a distinct role in the initial stages of biofilm formation. Recent studies of a second H. volcanii Pil system encoded by pilB1/pilC1, which are also associated with predicted pilin genes, revealed cross-complementation between pilB/pilC paralogs. For instance, the adhesion defect of a ΔpilB1/pilC1 deletion strain can be rescued by expressing pilB3/pilC3 in trans. However, pilB1/pilC1 cannot rescue the ΔpilB3/pilC3 adhesion phenotype (Legerme and Pohlschroder, unpublished data).
To help understand functional and evolutionary relationships between somewhat disparate biosynthesis systems, Makarova et al. used in silico analyses to phylogenetically group the archaeal PilB ATPases into four clades and to associate those with type IV pilin genes in the same gene cluster. Clade 1 includes PilB homologs involved in the assembly of type IV pili containing pilins processed by EppA. Clade 2 is comprised of euryarchaeal PilB homologs, including the H. volcanii paralogs PilB3, PilB4 and PilB5. Clade 3 includes ArlI homologs, and clade 4 encompasses the PilB homologs of the TACK superphylum (Makarova, Koonin and Albers 2016). Interestingly, the haloarchaeal PilB1 and PilB2 are assigned to a different subclade than the associated genes in the cluster: PilB1 and PilB2 belong to a subclade of Clade 4 (Clade 4C) while the associated pilin genes belong to Clade 2. This led to the hypothesis that the PilB and PilC components encoded by some euryarchaeal pil operons were acquired from the Crenarchaeota via horizontal transfer, while the pilin genes associated with these pilB/pilC genes are the result of duplication and diversification of ancestral euryarchaeal pilin genes. This proposed hybrid origin led the authors to further hypothesize that PilB ‘can work with different pilin sets resulting in modular evolution and extensive combinatorial diversity’, which is consistent with the in vivo complementation results of H. volcanii pilB1/pilC1 and pilB3/pilC3 (see above). The euryarchaeal major pilin may have required adaptation to be recognized by the crenarchaeal PilB1 upon horizontal transfer, while the modification of these pilins was minor enough to not prevent their recognition by PilB3 (Makarova, Koonin and Albers 2016).
While PibD, PilB and PilC are the only components of the type IV pilus biosynthesis pathway which, to date, are known to be conserved across all archaea and bacteria, it is likely that additional components are required for the biosynthesis of functional archaeal type IV pili (Table 2). Such additional components might be unrelated between bacteria and archaea and may depend on the functional role played by the specific type IV pilus and the particular environment inhabited by the cell. For example, many bacteria have a second type IV pilus-related ATPase, PilT, which is required for the retraction and disassembly of type IV pili (Merz, So and Sheetz 2000; Aroeti et al.2012; Burrows 2012), providing cells with the ability to perform twitching motility, and thus allowing movement along moist surfaces. While twitching motility has not been observed in archaea and a gene encoding an archaeal PilT homolog has not been identified in the large and diverse array of archaeal genomes sequenced, it cannot be excluded that archaeal type IV pili might be able to retract, while using components distinct from PilT. The in silico studies of archaeal pil systems by Szabo et al. and Makarova et al. have led to the identification of other potential components that might be needed for type IV pilus biosynthesis in some clades (Szabo et al.2007; Makarova, Koonin and Albers 2016). These components might form supporting structures or might play regulatory roles in type IV pilus biogenesis. For example, FtsZ-family GTPases (FtsZ3) are associated with Clade 1 loci in most Thermococci, as previously described (Szabo et al.2007) as well as with the halobacterial 4F subclade in Halobacteria (Makarova, Koonin and Albers 2016). Moreover, a FleN/MinD family ATPase is associated with type IV pili of Clade 1, Clade 2 and the halobacterial subclade 4F. In bacteria, the FleN/MinD family ATPases are involved in regulating flagella assembly and managing the localization of type IV pili as well as flagella (Dasgupta, Arora and Ramphal 2000; Schuhmacher et al.2015). Interestingly, archaeal chemotaxis genes are often co-localized with fleN/minD genes. In fact, screens for adhesion mutants in a transposon insertion library recently led to the identification of mutants having transposon insertions in chemotaxis genes, indicating that chemotaxis is important for the proper function of type IV pili (Legerme et al.2016).
Finally, many bacterial and archaeal type IV pilins are glycosylated. In H. volcanii, pilin glycosylation appears to play several distinct roles in regulating the assembly and functions of type IV pili (Esquivel et al.2016). Of the six adhesion pilins, all but PilA5 are glycosylated in an AglB-dependent manner. Expression of PilA2 pilins in a ΔpilA1–6 ΔaglB strain results in the formation of pili bundles, while PilA4 expression in the same background results in non-adhesive pili having a curled appearance. The aggregation of pili into bundles on the surfaces of cells lacking AglB-dependent glycosylation might promote interactions between cells, and, hence, promote cell aggregation and possibly biofilm formation. Interestingly, since H. volcanii archaella are glycosylated with the same oligosaccharides as are the pilins and the SLG, and since archaellin glycosylation is required for archaella biosynthesis and/or stability (Tripepi et al.2012), decreased glycosylation might promote biofilm formation via two distinct pathways. Similarly, archaellin N-glycosylation is required for the assembly of functional archaella, and therefore for motility, in M. maripaludis and S. acidocaldarius (VanDyke et al.2009; Meyer, Birich and Albers 2015). Furthermore, while N-glycosylation of S. acidocaldarius type IV pilins remains to be elucidated, cell surface attachment of type IV pili is impaired in M. maripaludis N-glycosylation pathway mutants (VanDyke et al.2008). However, the N-glycans attached to M. maripaludis type IV pilins have an additional hexose in comparison to the N-glycans of its archaellins (VanDyke et al.2009). Furthermore, SP processing is a prerequisite for the N-glycosylation of type IV pilins, but not archaellins, in this species (Nair and Jarrell 2015). The diverse array of type IV pilins and their differential N-glycosylation potentially allows for various regulatory mechanisms to control the distinct functions of type IV pili.
CONCLUSIONS
There is still significantly less attention given to understanding the cell-surface biogenesis of archaea, compared to bacteria and eukaryotes. However, protein crystallography, an increased number and diversity of model archaea systems that are amenable to biochemical and molecular biological techniques, as well as the availability of more efficient sequencing tools and an expanded repertoire of in silico approaches, has significantly advanced our ability to study archaeal cell surface biogenesis.
Using novel high sensitivity mass spectrometry strategies in proteomics approaches will allow us to accurately determine differential protein expression as well as PTMs under a variety of conditions. Improved mass spectrometry will also allow us to confirm N-terminal processing sites and will lead to the development of better subcellular localization prediction programs for archaeal proteins. Furthermore, the application of cryoelectron microscopy will allow to obtain more detailed information about the structures of the cell surface proteins and protein complexes (Poweleit et al.2016), especially as technical limitations due to the high salt concentrations have been partially overcome (Bollschweiler et al.2017). Moreover, the expansion of available tools for in vitro studies will aid to address basic questions related to the energetics of protein transport. Combining these approaches with the use of genetic screens will allow us to identify additional as yet unknown components, such as an ATPase required for Sec transport or a SPII analog involved in lipoprotein processing. Finally, developing culturing techniques for species of the novel and only recently identified archaeal phyla will be invaluable to getting insights into the evolutionary history of protein targeting and transport. Here, the Asgard family is a particularly intriguing archaeal superphylum that not only appears to have a distinct holotranslocon composition but also contains the closest known prokaryotic relatives of the eukaryotes.
A more complete understanding of archaeal cell surface biogenesis might also lead to insights that clarify the roles played by various cell surface structures in adaptations to the diverse ranges of environments inhabited by the archaea. It could also aid in the development of novel biotechnical applications. As described in the review by Kerry et al. in this thematic issue, many secreted archaeal proteins are used to degrade various polymers and greater understanding of the mechanisms supporting their secretion, processing and PTMs would be critical for efficient heterologous expression of these proteins but also for high levels of expression of active enzymes from the native host. In fact, advances in our understanding of the mechanisms supporting the biogenesis and functions of the archaeal cell surface clearly have important implications for the development of a vast array of potential biomedical applications. For example, archaeosomes, which are vesicles composed of archaeal membranes, are more efficient drug delivery vehicles than are conventional liposomes (Benvegnu, Lemiegre and Cammas-Marion 2009). These can be further stabilized by adding an S-layer to their surface (Pum and Sleytr 2014). Hence, a deeper understanding of the mode of archaeosortase action on its substrates, including SLG, could lead to the development of better drug delivery vesicles.
Supplementary Material
Acknowledgements
We thank the Pohlschroder lab for helpful discussions and Christine Moisl-Eichinger and Gerhard Wanner for providing us with image 1C and 1D, respectively. MFAH was supported by a University of Pennsylvania Research Foundation grant. MP was supported by National Science Foundation grant MCB-1413158. SS was supported by Deutsche Forschungsgemeinschaft (DFG) grant 398625447. FP was supported by the Max Planck Society (MPS).
REFERENCES
- Abdul Halim MF, Karch KR, Zhou Y et al. Permuting the PGF signature motif blocks both archaeosortase-dependent C-terminal cleavage and prenyl lipid attachment for the Haloferax volcanii S-layer glycoprotein. J Bacteriol 2016;198:808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul Halim MF, Pfeiffer F, Zou J et al. Haloferax volcanii archaeosortase is required for motility, mating, and C-terminal processing of the S-layer glycoprotein. Mol Microbiol 2013;88:1164–75. [DOI] [PubMed] [Google Scholar]
- Abdul Halim MF, Rodriguez R, Stoltzfus JD et al. Conserved residues are critical for Haloferax volcanii archaeosortase catalytic activity: Implications for convergent evolution of the catalytic mechanisms of non-homologous sortases from archaea and bacteria. Mol Microbiol 2018;108:276–87. [DOI] [PubMed] [Google Scholar]
- Abdul Halim MF, Stoltzfus JD, Schulze S et al. ArtA-dependent processing of a tat substrate containing a conserved tripartite structure that is not localized at the C terminus. J Bacteriol 2017;199:00802–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Qarn M, Yurist-Doutsch S, Giordano A et al. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J Mol Biol 2007;374:1224–36. [DOI] [PubMed] [Google Scholar]
- Adams H, Scotti PA, De Cock H et al. The presence of a helix breaker in the hydrophobic core of signal sequences of secretory proteins prevents recognition by the signal-recognition particle in Escherichia coli. Eur J Biochem 2002;269:5564–71. [DOI] [PubMed] [Google Scholar]
- Akopian D, Shen K, Zhang X et al. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem 2013;82:693–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami M, Lüke I, Deitermann S et al. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell 2003;12:937–46. [DOI] [PubMed] [Google Scholar]
- Albers S, Eichler J, Aebi M. Archaea. Essentials of Glycobiology. Varki A, Cummings RD, Esko JD et al.Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press: 2015; 283–92. [PubMed] [Google Scholar]
- Albers SV, Jarrell KF. The archaellum: an update on the unique archaeal motility structure. Trends Microbiol 2018;26:351–62. [DOI] [PubMed] [Google Scholar]
- Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol 2011;9:414–26. [DOI] [PubMed] [Google Scholar]
- Albers SV, Szabo Z, Driessen AJ. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J Bacteriol 2013;185:3918–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Wootton JC, Gertz EM et al. Protein database searches using compositionally adjusted substitution matrices. FEBS J 2005;272:5101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbing MA, Chan S, Shin A et al. Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans. Proc Natl Acad Sci USA 2012;109:11812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroeti B, Friedman G, Zlotkin-Rivkin E et al. Retraction of enteropathogenic E. coli type IV pili promotes efficient host cell colonization, effector translocation and tight junction disruption. Gut Microbes 2012;3:267–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer J, Spicker G, Bock A. Presence of a gene in the archaebacterium Methanococcus vannielii homologous to secY of eubacteria. Biochimie 1991;73:683–8. [DOI] [PubMed] [Google Scholar]
- Bachmann J, Bauer B, Zwicker K et al. The Rieske protein from Paracoccus denitrificans is inserted into the cytoplasmic membrane by the twin-arginine translocase. FEBS J 2006;273:4817–30. [DOI] [PubMed] [Google Scholar]
- Bageshwar UK, Whitaker N, Liang FC et al. Interconvertibility of lipid- and translocon-bound forms of the bacterial Tat precursor pre-SufI. Mol Microbiol 2009;74:209–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Tsai CL, Chaudhury P et al. FlaF is a beta-sandwich protein that anchors the archaellum in the archaeal cell envelope by binding the S-layer protein. Structure 2015;23:863–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee T, Lindenthal C, Oliver D. SecA functions in vivo as a discrete anti‐parallel dimer to promote protein transport. Mol Microbiol 2017;103:439–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang C, Schmitz RA. Archaea associated with human surfaces: not to be underestimated. FEMS Microbiol Rev 2015;39:631–48. [DOI] [PubMed] [Google Scholar]
- Bardy SL, Jarrell KF. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol Microbiol 2003;50:1339–47. [DOI] [PubMed] [Google Scholar]
- Bardy SL, Ng SY, Carnegie DS et al. Site-directed mutagenesis analysis of amino acids critical for activity of the type I signal peptidase of the archaeon Methanococcus voltae. J Bacteriol 2005;187:1188–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Lembcke G. Structural features of archaebacterial cell envelopes. J Bioenerg Biomembr 1992;24:567–75. [DOI] [PubMed] [Google Scholar]
- Beck D, Vasisht N, Baglieri J et al. Ultrastructural characterisation of Bacillus subtilis TatA complexes suggests they are too small to form homooligomeric translocation pores. BBA-Mol Cell Res 2013;1833:1811–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beja O, Aravind L, Koonin EV et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 2000;289:1902–6. [DOI] [PubMed] [Google Scholar]
- Benvegnu T, Lemiegre L, Cammas-Marion S. New generation of liposomes called archaeosomes based on natural or synthetic archaeal lipids as innovative formulations for drug delivery. Recent Pat Drug Deliv Formul 2009;3:206–20. [DOI] [PubMed] [Google Scholar]
- Bercovich-Kinori A, Bibi E. Co-translational membrane association of the Escherichia coli SRP receptor. J Cell Sci 2015;128:1444–52. [DOI] [PubMed] [Google Scholar]
- Berks BC. The twin-argnine protein translocation pathway. Ann Rev Biochem 2014;84:843–64. [DOI] [PubMed] [Google Scholar]
- Berry JL, Pelicic V. Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol Rev 2015;39:134–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibi E. Is there a twist in the Escherichia coli signal recognition particle pathway? Trends Biochem Sci 2012;37:1–6. [DOI] [PubMed] [Google Scholar]
- Bollschweiler D, Schaffer M, Lawrence CM et al. Cryo-electron microscopy of an extremely halophilic microbe: technical aspects. Extremophiles 2017;21:393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowska MT, Dominik PK, Anghel SA et al. A YidC-like protein in the archaeal plasma membrane. Structure 2015;23:1715–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruser T, Sanders C. An alternative model of the twin arginine translocation system. Microbiol Res 2003;158:7–17. [DOI] [PubMed] [Google Scholar]
- Buchanan G, Sargent F, Berks BC et al. A genetic screen for suppressors of Escherichia coli Tat signal peptide mutations establishes a critical role for the second arginine within the twin-arginine motif. Arch Microbiol 2001;177:107–12. [DOI] [PubMed] [Google Scholar]
- Buddelmeijer N. The molecular mechanism of bacterial lipoprotein modification—How, when and why? FEMS Microbiol Rev 2015;39:246–61. [DOI] [PubMed] [Google Scholar]
- Burrows LL. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 2012;66:493–520. [DOI] [PubMed] [Google Scholar]
- Caforio A, Driessen AJM. Archaeal phospholipids: structural properties and biosynthesis. Biochim Biophys Acta 2017;1862:1325–39. [DOI] [PubMed] [Google Scholar]
- Calo D, Eichler J. Crossing the membrane in Archaea, the third domain of life. BBA- Biomembranes 2011;1808:885–91. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R. Cold-adapted archaea. Nat Rev Micro 2006;4:331–43. [DOI] [PubMed] [Google Scholar]
- Chaban B, Ng SY, Jarrell KF. Archaeal habitats — from the extreme to the ordinary. Can J Microbiol 2006;52:73–116. [DOI] [PubMed] [Google Scholar]
- Chaudhury P, Neiner T, D’imprima E et al. The nucleotide-dependent interaction of FlaH and FlaI is essential for assembly and function of the archaellum motor. Mol Microbiol 2016;99:674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury P, Quax TEF, Albers SV. Versatile cell surface structures of archaea. Mol Microbiol 2018;107:298–311. [DOI] [PubMed] [Google Scholar]
- Chen YY, Soman R, Shanmugam K et al. The role of the strictly conserved positively charged residue differs among the gram-positive, gram-negative, and chloroplast YidC homologs. J Biol Chem 2014;289:35656–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong PL. Archaebacterial bipolar tetraether lipids: physico-chemical and membrane properties. Chem Phys Lipids 2010;163:253–65. [DOI] [PubMed] [Google Scholar]
- Claus H, Akca E, Debaerdemaeker T et al. Primary structure of selected archaeal mesophilic and extremely thermophilic outer surface layer proteins. Syst Appl Microbiol 2002;25:3–12. [DOI] [PubMed] [Google Scholar]
- Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Bi 2014;30:255–89. [DOI] [PubMed] [Google Scholar]
- Comolli LR, Baker BJ, Downing KH et al. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. Isme J 2008;3:159–67. [DOI] [PubMed] [Google Scholar]
- Cristóbal S, de Gier JW, Nielsen H et al. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J 1999;18:2982–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey R, Pei D, Ekici Ö. Signal peptidase enzymology and substrate specificity profiling. Methods Enzymol 2017;584:35–57. [DOI] [PubMed] [Google Scholar]
- Dale H, Angevine CM, Krebs MP. Ordered membrane insertion of an archaeal opsin in vivo. Proc Natl Acad Sci USA 2000;97:7847–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale H, Krebs MP. Membrane insertion kinetics of a protein domain in vivo. The bacterioopsin n terminus inserts co-translationally. J Biol Chem 1999;274:22693–8. [DOI] [PubMed] [Google Scholar]
- Danson MJ, Hough DW. The structural basis of protein halophilicity. Comp Biochem Phys A 1997;117:307–12. [Google Scholar]
- Dasgupta N, Arora SK, Ramphal R. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 2000;182:357–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum B, Vonck J, Bellack A et al. Structure and in situ organisation of the Pyrococcus furiosus archaellum machinery. eLife 2017;6:e27470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener JL, Wilson C. Role of SRP19 in assembly of the Archaeoglobus fulgidus signal recognition particle. Biochemistry 2000;39:12862–74. [DOI] [PubMed] [Google Scholar]
- Dilks K, Gimenez MI, Pohlschroder M. Genetic and biochemical analysis of the twin-arginine translocation pathway in halophilic archaea. J Bacteriol 2005;187:8104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilks K, Rose RW, Hartmann E et al. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol 2003;185:1478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Lau Z, Logan SM et al. Effects of growth conditions on archaellation and N-glycosylation in Methanococcus maripaludis. Microbiology 2016;162:339–50. [DOI] [PubMed] [Google Scholar]
- Dong Y, Palmer SR, Hasona A et al. Functional overlap but lack of complete cross-complementation of Streptococcus mutans and Escherichia coli YidC orthologs. J Bacteriol 2008;190:2458–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J 1997;16:4871–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith ML, Pfeiffer F, Klee K et al. Haloquadratum walsbyi: limited diversity in a global pond. PLoS One 2011;6:e20968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Shan SO, Napetschnig J et al. Substrate twinning activates the signal recognition particle and its receptor. Nature 2004;427:215–21. [DOI] [PubMed] [Google Scholar]
- Egea PF, Stroud RM. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc Natl Acad Sci USA 2010;107:17182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Tsuruta H, de Leon GP et al. Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: implications for the targeting step at the membrane. PLoS One 2008;3: e3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J. Detection of genes with atypical nucleotide sequence in microbial genomes. J Mol Evol 2002;54:411–5. [DOI] [PubMed] [Google Scholar]
- Eichler J, Maupin-Furlow J. Post-translation modification in Archaea: lessons from Haloferax volcanii and other haloarchaea. FEMS Microbiol Rev 2013;37:583–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt H. S-Layers. eLS. Chichester: John Wiley & Sons, Ltd, 2016;1–12. [Google Scholar]
- Esquivel RN, Pohlschroder M.. A conserved type IV pilin signal peptide H-domain is critical for the post-translational regulation of flagella-dependent motility. Mol Microbiol 2014;93:494–504. [DOI] [PubMed] [Google Scholar]
- Esquivel RN, Schulze S, Xu R et al. Identification of Haloferax volcanii pilin N-glycans with diverse roles in pilus biosynthesis, adhesion, and microcolony formation. J Biol Chem 2016;291:10602–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquivel RN, Xu R, Pohlschroder M. Novel archaeal adhesion pilins with a conserved N terminus. J Bacteriol 2013;195:3808–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive earth's biogeochemical cycles. Science 2008;320:1034–9. [DOI] [PubMed] [Google Scholar]
- Fine A, Irihimovitch V, Dahan I et al. Cloning, expression, and purification of functional Sec11a and Sec11b, type I signal peptidases of the archaeon Haloferax volcanii. J Bacteriol 2006;188:1911–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluman N, Navon S, Bibi E et al. mRNA-programmed translation pauses in the targeting of E. coli membrane proteins. Elife 2014;3:e03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focia PJ, Shepotinovskaya IV, Seidler JA et al. Heterodimeric GTPase core of the SRP targeting complex. Science 2004;303:373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francetic O, Buddelmeijer N, Lewenza S et al. Signal recognition particle-dependent inner membrane targeting of the PulG Pseudopilin component of a type II secretion system. J Bacteriol 2007;189:1783–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröbel J, Rose P, Lausberg F et al. Transmembrane insertion of twin-arginine signal peptides is driven by TatC and regulated by TatB. Nat Commun 2012;3:1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröbel J, Rose P, Müller M. Early contacts between substrate proteins and TatA translocase component in twin-arginine translocation. J Biol Chem 2011;286:43679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröls S, Ajon M, Wagner M et al. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol Microbiol 2008;70:938–52. [DOI] [PubMed] [Google Scholar]
- Funes S, Hasona A, Bauerschmitt H et al. Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc Natl Acad Sci USA 2009;106:6656–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol R 2012;76:740–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez MI, Dilks K, Pohlschroder M. Haloferax volcanii twin-arginine translocation substates include secreted soluble, C-terminally anchored and lipoproteins. Mol Microbiol 2007;66:1597–606. [DOI] [PubMed] [Google Scholar]
- Gohlke U, Pullan L, McDevitt CA et al. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc Natl Acad Sci USA 2005;102:10482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosens VJ, van Dijl JM. Twin-arginine protein translocation. Curr Top Microbiol Immunol 2017;404:69–94. [DOI] [PubMed] [Google Scholar]
- Grogan DW. Isolation and fractionation of cell envelope from the extreme thermo-acidophile Sulfolobus acidocaldarius. J Microbiol Methods 1996a;26:35–43. [Google Scholar]
- Grogan DW. Organization and interactions of cell envelope proteins of the extreme thermoacidophile Sulfolobus acidocaldarius. Can J Microbiol 1996b;42:1163–71. [Google Scholar]
- Gropp R, Gropp F, Betlach MC. Association of the halobacterial 7S RNA to the polysome correlates with expression of the membrane protein bacterioopsin. Proc Natl Acad Sci USA 1992;89:1204–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudnik P, Bange G, Sinning I. Protein targeting by the signal recognition particle. Biol Chem 2009;390:775–82. [DOI] [PubMed] [Google Scholar]
- Guan Z, Naparstek S, Calo D et al. Protein glycosylation as an adaptive response in Archaea: growth at different salt concentrations leads to alterations in Haloferax volcanii S‐layer glycoprotein N‐glycosylation. Environ Microbiol 2012;14:743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad A, Rose RW, Pohlschröder M. The Haloferax volcanii FtsY homolog is critical for haloarchaeal growth but does not require the A domain. J Bacteriol 2005;187:4015–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Paulsen IT, Ward N et al. Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol 2006;4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Payne SH, Selengut JD. Archaeosortases and exosortases are widely distributed systems linking membrane transit with posttranslational modification. J Bacteriol 2012;194:36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl T, Huang S, Sauer-Eriksson AE. Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature 2002;417:767–71. [DOI] [PubMed] [Google Scholar]
- Hainzl T, Sauer-Eriksson AE. Signal-sequence induced conformational changes in the signal recognition particle. Nat Commun 2015;6:7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 2004;427:808–14. [DOI] [PubMed] [Google Scholar]
- Hand NJ, Klein R, Laskewitz A et al. Archaeal and bacterial SecD and SecF homologs exhibit striking structural and functional conservation. J Bacteriol 2006;188:1251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JK, Forest KT. Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. J Mol Microb Biotech 2006;11:192–207. [DOI] [PubMed] [Google Scholar]
- Hartmann E, Sommer T, Prehn S et al. Evolutionary conservation of components of the protein translocation complex. Nature 1994;367:654. [DOI] [PubMed] [Google Scholar]
- Hartzell PL, Millstein J, LaPaglia C. Biofilm formation in hyperthermophilic Archaea. Methods Enzymol 1999;310:335–49. [DOI] [PubMed] [Google Scholar]
- Hatzixanthis K, Palmer T, Sargent F. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol 2003;49:1377–90. [DOI] [PubMed] [Google Scholar]
- He S, Fox TD. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol Biol Cell 1997;8:1449–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K, Herrmann J, Pratje E et al. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett 1997;418:367–70. [DOI] [PubMed] [Google Scholar]
- Henche AL, Ghosh A, Yu X et al. Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius. Environ Microbiol 2012;14:3188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel E, Eisner G, Alami M et al. The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry 2007;46:2892–8. [DOI] [PubMed] [Google Scholar]
- Hou B, Frielingsdorf S, Klösgen RB. Unassisted membrane insertion as the initial step in ΔpH/Tat-dependent protein transport. J Mol Biol 2006;355:957–67. [DOI] [PubMed] [Google Scholar]
- Huang Q, Palmer T. Signal peptide hydrophobicity modulates interaction with the twin-arginine translocase. MBio 2017;8:135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D, Boyd D, Xia Y et al. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J Bacteriol 2005;187:2983–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K et al. A new view of the tree of life. Nat Microbiol 2016;1:16048. [DOI] [PubMed] [Google Scholar]
- Imam S, Chen Z, Roos DS et al. Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria. PLoS One 2011;6:e28919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irihimovitch V, Eichler J. Post-translational secretion of fusion proteins in the halophilic archaea Haloferax volcanii. J Biol Chem 2003;278:12881–7. [DOI] [PubMed] [Google Scholar]
- Jacquemet A, Barbeau J, Lemiegre L et al. Archaeal tetraether bipolar lipids: structures, functions and applications. Biochimie 2009;91:711–7. [DOI] [PubMed] [Google Scholar]
- Jain S, Caforio A, Driessen AJ. Biosynthesis of archaeal membrane ether lipids. Front Microbiol 2014;5:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell KF, Ding Y, Meyer BH et al. N-linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol Mol Biol R 2014;78:304–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang FL, Chen MY, Yi L et al. Defining the regions of Escherichia coli YidC that contribute to activity. J Biol Chem 2003;278:48965–72. [DOI] [PubMed] [Google Scholar]
- Jiang FL, Yi L, Moore M et al. Chloroplast YidC homolog Albino3 can functionally complement the bacterial YidC depletion strain and promote membrane insertion of both bacterial and chloroplast thylakoid proteins. J Biol Chem 2002;277:19281–8. [DOI] [PubMed] [Google Scholar]
- Jomaa A, Boehringer D, Leibundgut M et al. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat Commun 2016;7:10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, von Heijne G et al. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 2009;12:1652–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell 1995;82:261–70. [DOI] [PubMed] [Google Scholar]
- Kaminski L, Guan Z, Yurist-Doutsch S et al. Two distinct N-glycosylation pathways process the Haloferax volcanii S-layer glycoprotein upon changes in environmental salinity. MBio 2013a;4:e00716–00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski L, Lurie-Weinberger MN, Allers T et al. Phylogenetic-and genome-derived insight into the evolution of N-glycosylation in Archaea. Mol Phylogenet Evol 2013b;68:327–39. [DOI] [PubMed] [Google Scholar]
- Kandiba L, Aitio O, Helin J et al. Diversity in prokaryotic glycosylation: an archaeal-derived N-linked glycan contains legionaminic acid. Mol Microbiol 2012;84:578–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L, Guan Z, Eichler J. Lipid modification gives rise to two distinct Haloferax volcanii S-layer glycoprotein populations. Biochim Biophys Acta 2013;1828:938–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L, Lin C, Aebi M et al. Structural characterization of the N-linked pentasaccharide decorating glycoproteins of the halophilic archaeon Haloferax volcanii. Glycobiology 2016;26:745–56. [DOI] [PubMed] [Google Scholar]
- Kates M. The phytanyl ether-linked polar lipids and isoprenoid neutral lipids of extremely halophilic bacteria. Prog Chem Fats Other Lipids 1977;15:301–42. [DOI] [PubMed] [Google Scholar]
- Kates M, Wassef M, Pugh E. Origin of the glycerol moieties in the glycerol diether lipids of Halobacterium cutirubrum. BBA-Lipid Lipid Met 1970;202:206–8. [DOI] [PubMed] [Google Scholar]
- Kikuchi A, Sagami H, Ogura K. Evidence for covalent attachment of diphytanylglyceryl phosphate to the cell-surface glycoprotein of Halobacterium halobium. J Biol Chem 1999;274:18011–6. [DOI] [PubMed] [Google Scholar]
- Kinch LN, Saier MH Jr, Grishin NV. Sec61β–a component of the archaeal protein secretory system. Trends Biochem Sci 2002;27:170–1. [DOI] [PubMed] [Google Scholar]
- Klingl A. S-layer and cytoplasmic membrane - exceptions from the typical archaeal cell wall with a focus on double membranes. Front Microbiol 2014;5:624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga Y. Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea 2012;2012:789652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H, Chong PL. Low permeability of liposomal membranes composed of bipolar tetraether lipids from thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochemistry 1998;37:107–15. [DOI] [PubMed] [Google Scholar]
- Konrad Z, Eichler J. Lipid modification of proteins in Archaea: attachment of a mevalonic acid-based lipid moiety to the surface-layer glycoprotein of Haloferax volcanii follows protein translocation. Biochem J 2002;366:959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen K, Pausan MR, Perras AK et al. First insights into the diverse human archaeome: specific detection of archaea in the gastrointestinal tract, lung, and nose and on skin. mBio 2017;8:e00824–00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G von et al. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol Biol 2001;305:567–80. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Kiefer D. Membrane protein insertase YidC in bacteria and archaea. Mol Microbiol 2017;103:590–4. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Koch HG, Dalbey RE. Targeting and insertion of membrane proteins. EcoSal Plus 2017;7:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazaki K, Chiba S, Takemoto M et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature 2014a;509:516–20. [DOI] [PubMed] [Google Scholar]
- Kumazaki K, Kishimoto T, Furukawa A et al. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci Rep 2014b;4:7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan DC, Thomas JR, Bolhuis A. Bioenergetic requirements of a Tat-dependent substrate in the halophilic archaeon Haloarcula hispanica. FEBS J 2008;275:6159–67. [DOI] [PubMed] [Google Scholar]
- Langworthy TA. Long-chain diglycerol tetraethers from Thermoplasma acidophilum. Biochim Biophys Acta 1977;487:37–50. [DOI] [PubMed] [Google Scholar]
- Langworthy TA, Mayberry WR, Smith PF. Long-chain glycerol diether and polyol dialkyl glycerol triether lipids of Sulfolobus acidocaldarius. J Bacteriol 1974;119:106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Sumper M. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J Biol Chem 1987;262:9724–9. [PubMed] [Google Scholar]
- Lee HC, Bernstein HD. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc Natl Acad Sci USA 2001;98:3471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YM, Dodson KW, Hultgren SJ. Adaptor function of PapF depends on donor strand exchange in P-pilus biogenesis of Escherichia coli. J Bacteriol 2007;189:5276–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault BA, Lopez-Lopez A, Alba-Casado JC et al. Environmental genomics of Haloquadratum walsbyi in a saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species. BMC Genomics 2006;7:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerme G, Yang E, Esquivel RN et al. Screening of a Haloferax volcanii transposon library reveals novel motility and adhesion mutants. Life (Basel) 2016;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA, Albers SV, Atomi H et al. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol Rev 2011;35:577–608. [DOI] [PubMed] [Google Scholar]
- Li L, Park E, Ling J et al. Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 2016;531:395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichi T, Ring G, Eichler J. Membrane binding of SRP pathway components in the halophilic archaea Haloferax volcanii. Eur J Biochem 2004;271:1382–90. [DOI] [PubMed] [Google Scholar]
- Losensky G, Vidakovic L, Klingl A et al. Novel pili-like surface structures of Halobacterium salinarum strain R1 are crucial for surface adhesion. Front Microbiol 2014;5:755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Xu Y, Zhang K et al. Crystal structure of E. coli apolipoprotein N-acyl transferase. Nat Commun 2017;8:15948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Lü Y, Ren J et al. Identification of the S-layer glycoproteins and their covalently linked glycans in the halophilic archaeon Haloarcula hispanica. Glycobiology 2015;25:1150–62. [DOI] [PubMed] [Google Scholar]
- Luirink J, Samuelsson T, de Gier JW. YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett 2001;501:1–5. [DOI] [PubMed] [Google Scholar]
- Macalady JL, Vestling MM, Baumler D et al. Tetraether-linked membrane monolayers in Ferroplasma spp: a key to survival in acid. Extremophiles 2004;8:411–9. [DOI] [PubMed] [Google Scholar]
- Madern D, Ebel C, Zaccai G. Halophilic adaptation of enzymes. Extremophiles 2000;4:91–98. [DOI] [PubMed] [Google Scholar]
- Madsen EL. Microorganisms and their roles in fundamental biogeochemical cycles. Curr Opin Biotechnol 2011;22:456–64. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV, Albers SV. Diversity and evolution of type IV pili systems in archaea. Front Microbiol 2016;7:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Zhao Y, Kang X et al. Crystal structure of E. coli lipoprotein diacylglyceryl transferase. Nat Commun 2016;7:10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark SS, Bergkvist M, Yang X et al. Self-assembly of dendrimer-encapsulated nanoparticle arrays using 2-D microbial S-layer protein biotemplates. Biomacromolecules 2006;7:1884–97. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Stahl DA. Nitrogen metabolism and kinetics of ammonia-oxidizing archaea. Method Enzymol 2011;496:465–87. [DOI] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Pasic L, Rodriguez-Valera F. Diversity of the cell-wall associated genomic island of the archaeon Haloquadratum walsbyi. BMC Genomics 2015;16:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Espinosa RM, Cole JA, Richardson DJ et al. Enzymology and Ecology of the Nitrogen Cycle. Portland Press Limited, 2011. [DOI] [PubMed] [Google Scholar]
- Mathieu L, Bourens M, Marsy S et al. A mutational analysis reveals new functional interactions between domains of the Oxa1 protein in Saccharomyces cerevisiae. Mol Microbiol 2010;75:474–88. [DOI] [PubMed] [Google Scholar]
- Matos CF, Robinson C, Di Cola A. The Tat system proofreads FeS protein substrates and directly initiates the disposal of rejected molecules. EMBO J 2008;27:2055–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Shimada A, Nyirenda J et al. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc Natl Acad Sci USA 2013;110:17868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberry-Carson KJ, Langworthy TA, MAyberry WR et al. A new class of lipopolysaccharide from Thermoplasma acidophilum. Biochim Biophys Acta 1974;360:217–29. [DOI] [PubMed] [Google Scholar]
- Melville S, Craig L. Type IV pili in Gram-positive bacteria. Microbiol Mol Biol R 2013;77:323–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature 2000;407:98–102. [DOI] [PubMed] [Google Scholar]
- Mescher MF, Strominger JL. Bacitracin induces sphere formation in Halobacterium species which lack a wall peptidoglycan. J Gen Microbiol 1975;89:375–8. [DOI] [PubMed] [Google Scholar]
- Mescher MF, Strominger JL. Purification and characterization of a prokaryotic glycoprotein from the cell envelope of Halobacterium salinarium. J Biol Chem 1976;251:2005–14. [PubMed] [Google Scholar]
- Messner P, Schäffer C, Kosma P. Bacterial cell-envelope glycoconjugates. Adv Carbohydr Chem Biochem 2013;69:209–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BH, Albers S-V. Hot and Sweet: Protein Glycosylation in Crenarchaeota Biochemical Society Transactions, 2013;41:384–92. [DOI] [PubMed] [Google Scholar]
- Meyer BH, Birich A, Albers S-V. N-Glycosylation of the archaellum filament is not important for archaella assembly and motility, although N-glycosylation is essential for motility in Sulfolobus acidocaldarius. Biochimie 2015;118:294–301. [DOI] [PubMed] [Google Scholar]
- Meyer BH, Zolghadr B, Peyfoon E et al. Sulfoquinovose synthase–an important enzyme in the N‐glycosylation pathway of Sulfolobus acidocaldarius. Mol Microbiol 2011;82:1150–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, Bernstein HD, Walter P. Interaction of E. coli Ffh/4.5 S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature 1994;367:657–9. [DOI] [PubMed] [Google Scholar]
- Mitra K, Schaffitzel C, Shaikh T et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 2005;438:318–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Cline K. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J Cell Biol 2002;157:205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair DB, Chung DK, Schneider J et al. Identification of an additional minor pilin essential for piliation in the archaeon Methanococcus maripaludis. PLoS One 2013;8:e83961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair DB, Jarrell KF. Pilin processing follows a different temporal route than that of archaellins in Methanococcus maripaludis. Life 2015;5:85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair DB, Uchida K, Aizawa S et al. Genetic analysis of a type IV pili-like locus in the archaeon Methanococcus maripaludis. Arch Microbiol 2014;196:179–91. [DOI] [PubMed] [Google Scholar]
- Narita SI, Tokuda H. Bacterial lipoproteins; biogenesis, sorting and quality control. Biochim Biophys Acta 2017;1862:1414–23. [DOI] [PubMed] [Google Scholar]
- Ng SY, Chaban B, VanDyke DJ et al. Archaeal signal peptidases. Microbiology 2007;153:305–14. [DOI] [PubMed] [Google Scholar]
- Ng SY, Jarrell KF. Cloning and characterization of archaeal type I signal peptidase from Methanococcus voltae. J Bacteriol 2003;185:5936–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, VanDyke DJ, Chaban B et al. Different minimal signal peptide lengths recognized by the archaeal prepilin-like peptidases FlaK and PibD. J Bacteriol 2009;191:6732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Wu J, Nair DB et al. Genetic and mass spectrometry analyses of the unusual type IV-like pili of the archaeon Methanococcus maripaludis. J Bacteriol 2011;193:804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Law D, Williams DB. Binding protein BiP is required for translocation of secretory proteins into the endoplasmic reticulum in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 1991;88:1565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara M, Koga Y. Purification and properties of sn-glycerol-1-phosphate dehydrogenase from Methanobacterium thermoautotrophicum: characterization of the biosynthetic enzyme for the enantiomeric glycerophosphate backbone of ether polar lipids of Archaea. J Biochem 1997;122:572–6. [DOI] [PubMed] [Google Scholar]
- Nishihara M, Yamazaki T, Oshima T et al. sn-glycerol-1-phosphate-forming activities in Archaea: separation of archaeal phospholipid biosynthesis and glycerol catabolism by glycerophosphate enantiomers. J Bacteriol 1999;181:1330–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland CL, Kattke MD, Diao J et al. Structural insights into lipoprotein N-acylation by Escherichia coli apolipoprotein N-acyltransferase. Proc Natl Acad Sci USA 2017;114:E6044–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. Bacterial protein N-glycosylation: new perspectives and applications. J Biol Chem 2013;288:6912–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D. The structure and mechanism of the family of retinal proteins from halophilic archaea. Curr Opin Struct Biol 1998;8:489–500. [DOI] [PubMed] [Google Scholar]
- Oliver DB, Beckwith J. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 1981;25:765–72. [DOI] [PubMed] [Google Scholar]
- Ortenberg R, Mevarech M. Evidence for post-translational membrane insertion of the integral membrane protein bacterioopsin expressed in the heterologous halophilic archaeon Haloferax volcanii. J Biol Chem 2000;275:22839–46. [DOI] [PubMed] [Google Scholar]
- Osborne AR, Clemons WM, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci USA 2004;101:10937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer T, Berks BC. The twin arginine transport (Tat) protein translocation pathway. Nat Rev Microbiol 2012;10:483–96. [DOI] [PubMed] [Google Scholar]
- Parente J, Casabuono A, Ferrari MC et al. A rhomboid protease gene deletion affects a novel oligosaccharide N-linked to the S-layer glycoprotein of Haloferax volcanii. J Biol Chem 2014;289:11304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys 2012;41:21–40. [DOI] [PubMed] [Google Scholar]
- Pechmann S, Chartron JW, Frydman J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol 2014;21:1100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perras AK, Daum B, Ziegler C et al. S-layers at second glance? Altiarchaeal grappling hooks (hami) resemble archaeal S-layer proteins in structure and sequence. Front Microbiol 2015a;6:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perras AK, Wanner G, Klingl A et al. Grappling archaea: ultrastructural analyses of an uncultivated, cold-loving archaeon, and its biofilm. Intra-and inter-species interactions in microbial communities: Frontiers in Microbiol 2015b;5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G von et al. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 2011;8:785–6. [DOI] [PubMed] [Google Scholar]
- Peyfoon E, Meyer B, Hitchen PG et al. The S-layer glycoprotein of the crenarchaeote Sulfolobus acidocaldarius is glycosylated at multiple sites with chitobiose-linked N-glycans. Archaea 2010;Article ID 754101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikuta EV, Hoover RB, Tang J. Microbial extremophiles at the limits of life. Crit Rev Microbiol 2007;33:183–209. [DOI] [PubMed] [Google Scholar]
- Pinhassi J, DeLong EF, Beja O et al. Marine bacterial and archaeal ion-pumping rhodopsins: genetic diversity, physiology, and ecology. Microbiol Mol Biol R 2016;80:929–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, Daume M, Wiegel J et al. Circularization restores signal recognition particle RNA functionality in Thermoproteus. Elife 2015;4:e11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschroder M, Albers SV. Editorial: archaeal cell envelope and surface structures. Front Microbiol 2015;6:1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschroder M, Dilks K, Hand NJ et al. Translocation of proteins across archaeal cytoplasmic membranes. FEMS Microbiol Rev 2004;28:3–24. [DOI] [PubMed] [Google Scholar]
- Pohlschroder M, Esquivel RN. Archaeal type IV pili and their involvement in biofilm formation. Front Microbiol 2015;6:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschroder M, Ghosh A, Tripepi M et al. Archaeal type IV pilus-like structures–evolutionarily conserved prokaryotic surface organelles. Curr Opin Microbiol 2011;14:357–63. [DOI] [PubMed] [Google Scholar]
- Pohlschroder M, Gimenez MI, Jarrell KF. Protein transport in Archaea: Sec and twin arginine translocation pathways. Curr Opin Microbiol 2005;8:713–9. [DOI] [PubMed] [Google Scholar]
- Pohlschroder M, Hartmann E, Hand NJ et al. Diversity and evolution of protein translocation. Annu Rev Microbiol 2005;59:91–111. [DOI] [PubMed] [Google Scholar]
- Pohlschroder M, Prinz WA, Hartmann E et al. Protein translocation in the three domains of life: variations on a theme. Cell 1997;91:563–6. [DOI] [PubMed] [Google Scholar]
- Poweleit N, Ge P, Nguyen HH et al. CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pilus. Nat Microbiol 2016;2:16222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AJ, Weinmaier T, Raymann K et al. Biology of a widespread uncultivated archaeon that contributes to carbon fixation in the subsurface. Nat Commun 2014;5:5497. [DOI] [PubMed] [Google Scholar]
- Pum D, Sleytr UB. Reassembly of S-layer proteins. Nanotechnology 2014;25:312001. [DOI] [PubMed] [Google Scholar]
- Quax TEF, Altegoer F, Rossi F et al. Structure and function of the archaeal response regulator CheY. Proc Natl Acad Sci USA 2018;115:E1259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 2007;450:663–9. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Li L, Park E. Structural and mechanistic insights into protein translocation. Annu Rev Cell Dev Biol 2017;33:369–90. [DOI] [PubMed] [Google Scholar]
- Ring G, Eichler J. Extreme secretion: protein translocation across the archaeal plasma membrane. J Bioenerg Biomembr 2004;36:35–45. [DOI] [PubMed] [Google Scholar]
- Rocco MA, Waraho-Zhmayev D, DeLisa MP. Twin-arginine translocase mutations that suppress folding quality control and permit export of misfolded substrate proteins. Proc Natl Acad Sci USA 2012;109:13392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Oliveira T, Belmok A, Vasconcellos D et al. Archaeal S-layers: overview and current state of the art. Front Microbiol 2017;8:2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F, Rouse SL, Tait CE et al. Structural model for the protein-translocating element of the twin-arginine transport system. Proc Natl Acad Sci USA 2013;110:E1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romisch K, Webb J, Lingelbach K et al. The 54-kD protein of signal recognition particle contains a methionine-rich RNA binding domain. J Cell Biol 1990;111:1793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RW, Bruser T, Kissinger JC et al. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol 2002;45:943–50. [DOI] [PubMed] [Google Scholar]
- Rose RW, Pohlschroder M. In vivo analysis of an essential archaeal signal recognition particle in its native host. J Bacteriol 2002;184:3260–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch SL, Kendall DA. Interactions that drive Sec-dependent bacterial protein transport. Biochemistry 2007;46:9665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sääf A, Monné M, de Gier J-W et al. Membrane Topology of the 60-kDa Oxa1p Homologue from Escherichia coli. J Biol Chem 1998;273:30415–8. [DOI] [PubMed] [Google Scholar]
- Sargent F, Bogsch EG, Stanley NR et al. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J 1998;17:3640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer C, Messner P. Emerging facets of prokaryotic glycosylation. FEMS Microbiol Rev 2017;41:49–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesner M, Miller A, Besir H et al. The protein interaction network of a taxis signal transduction system in a halophilic archaeon. BMC Microbiol 2012;12:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Missiakas D. Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochim Biophys Acta 2014;1843:1687–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten S, Hopmans EC, Damste JSS. The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: A review. Org Geochem 2013;54:19–61. [Google Scholar]
- Schuhmacher JS, Rossmann F, Dempwolff F et al. MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc Natl Acad Sci USA 2015;112:3092–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev Y, Turgeman-Grott I, Tamir A et al. Cell surface glycosylation is required for efficient mating of Haloferax volcanii. Front Microbiol 2017;8:1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugham A, Wong Fong Sang HW, Bollen YJ et al. Membrane binding of twin arginine preproteins as an early step in translocation. Biochemistry 2006;45:2243–9. [DOI] [PubMed] [Google Scholar]
- Sleytr U. Self-assembly of the hexagonally and tetragonally arranged subunits of bacterial surface layers and their reattachment to cell walls. J Ultrastruct Res 1976;55:360–77. [DOI] [PubMed] [Google Scholar]
- Sleytr UB, Schuster B, Egelseer EM et al. S-layers: principles and applications. FEMS Microbiol Rev 2014;38:823–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storf S, Pfeiffer F, Dilks K et al. Mutational and bioinformatic analysis of haloarchaeal lipobox-containing proteins. Archaea 2010;2010:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper M, Berg E, Mengele R et al. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J Bacteriol 1990;172:7111–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Z, Pohlschroder M. Diversity and subcellular distribution of archaeal secreted proteins. Front Microbiol 2012;3:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Z, Stahl AO, Albers SV et al. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J Bacteriol 2007;189:772–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamir A, Eichler J. N-Glycosylation is important for proper Haloferax volcanii S-layer stability and function. Appl Environ Microbiol 2017;83:e03152–03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sugano Y, Takemoto M et al. Crystal structures of SecYEG in lipidic cubic phase elucidate a precise resting and a peptide-bound state. Cell Rep 2015;13:1561–8. [DOI] [PubMed] [Google Scholar]
- Tokuda H, Kim YJ, Mizushima S. In vitro protein translocation into inverted membrane vesicles prepared from Vibrio alginolyticus is stimulated by the electrochemical potential of Na+ in the presence of Escherichia coli SecA. FEBS Lett 1990;264:10–12. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Mazmanian SK, Alksne L et al. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. J Biol Chem 2002;277:7447–52. [DOI] [PubMed] [Google Scholar]
- Tozik I, Huang Q, Zwieb C et al. Reconstitution of the signal recognition particle of the halophilic archaeon Haloferax volcanii. Nucleic Acids Res 2002;30:4166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg S, Pinnick B, Kessel M. The cell surface glycoprotein layer of the extreme halophile Halobacterium salinarum and its relation to Haloferax volcanii: cryo-electron tomography of freeze-substituted cells and projection studies of negatively stained envelopes. J Struct Biol 2000;130:10–26. [DOI] [PubMed] [Google Scholar]
- Tripepi M, Imam S, Pohlschroder M. Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. J Bacteriol 2010;192:3093–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripepi M, You J, Temel S et al. N-glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. J Bacteriol 2012;194:4876–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirigotaki A, De Geyter J, Sostaric N et al. Protein export through the bacterial Sec pathway. Nat Rev Microbiol 2017;15:21–36. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Mori H, Echizen Y et al. Structure and function of a membrane component SecDF that enhances protein export. Nature 2011;474:235–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Mori H, Fukai S et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 2008;455:988–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja R. Type I signal peptidase: an overview. Arch Biochem Biophys 2005;441:107–11. [DOI] [PubMed] [Google Scholar]
- Uda I, Sugai A, Itoh YH et al. Variation in molecular species of polar lipids from Thermoplasma acidophilum depends on growth temperature. Lipids 2001;36:103–5. [DOI] [PubMed] [Google Scholar]
- van Bloois E, Nagamori S, Koningstein G et al. The Sec-independent function of Escherichia coli YidC is evolutionary-conserved and essential. J Biol Chem 2005;280:12996–3003. [DOI] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM Jr, Collinson I et al. X-ray structure of a protein-conducting channel. Nature 2004;427:36–44. [DOI] [PubMed] [Google Scholar]
- van Wolferen M, Ajon M, Driessen AJ et al. Molecular analysis of the UV-inducible pili operon from Sulfolobus acidocaldarius. Microbiologyopen 2013;2:928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDyke DJ, Wu J, Logan SM et al. Identification of genes involved in the assembly and attachment of a novel flagellin N-linked tetrasaccharide important for motility in the archaeon Methanococcus maripaludis. Mol Microbiol 2009;72:633–44. [DOI] [PubMed] [Google Scholar]
- VanDyke DJ, Wu J, Ng SY et al. Identification of a putative acetyltransferase gene, MMP0350, which affects proper assembly of both flagella and pili in the archaeon Methanococcus maripaludis. J Bacteriol 2008;190:5300–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanNice JC, Skaff DA, Keightley A et al. Identification in Haloferax volcanii of phosphomevalonate decarboxylase and isopentenyl phosphate kinase as catalysts of the terminal enzyme reactions in an archaeal alternate mevalonate pathway. J Bacteriol 2014;196:1055–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M et al. Symbol nomenclature for graphical representations of glycans. Glycobiology 2015;25:1323–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith A, Klingl A, Zolghadr B et al. Acidianus, Sulfolobus and Metallosphaera surface layers: structure, composition and gene expression. Mol Microbiol 2009;73:58–72. [DOI] [PubMed] [Google Scholar]
- Villanueva L, Damste JS, Schouten S. A re-evaluation of the archaeal membrane lipid biosynthetic pathway. Nat Rev Microbiol 2014;12:438–48. [DOI] [PubMed] [Google Scholar]
- Vinokur JM, Cummins MC, Korman TP et al. An adaptation to life in acid through a novel mevalonate pathway. Sci Rep 2016;6:39737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin S, Houliston RS, Kelly J et al. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J Biol Chem 2005;280:16586–93. [DOI] [PubMed] [Google Scholar]
- Walther TH, Gottselig C, Grage SL et al. Folding and self-assembly of the TatA translocation pore based on a charge zipper mechanism. Cell 2013;152:316–26. [DOI] [PubMed] [Google Scholar]
- Wieland F. Structure and biosynthesis of prokaryotic glycoproteins. Biochimie 1988;70:1493–504. [DOI] [PubMed] [Google Scholar]
- Wiktor M, Weichert D, Howe N et al. Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis. Nat Commun 2017;8:15952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild K, Bange G, Motiejunas D et al. Structural basis for conserved regulation and adaptation of the signal recognition particle targeting complex. J Mol Biol 2016;428:2880–97. [DOI] [PubMed] [Google Scholar]
- Wild R, Kowal J, Eyring J et al. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science 2018;359:545–50. [DOI] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990;87:4576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury RL, Topping TB, Diamond DL et al. Complexes between protein export chaperone SecB and SecA Evidence for separate sites on SecA providing binding energy and regulatory interactions. J Biol Chem 2000;275:24191–8. [DOI] [PubMed] [Google Scholar]
- Yosef I, Bochkareva ES, Adler J et al. Membrane protein biogenesis in Ffh- or FtsY-depleted Escherichia coli. PLoS One 2010;5:e9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zweers JC, Van Dijl JM et al. Protein transport across and into cell membranes in bacteria and archaea. Cell Mol Life Sci 2010;67:179–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurist S, Dahan I, Eichler J. SRP19 is a dispensable component of the signal recognition particle in Archaea. J Bacteriol 2007;189:276–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 2017;541:353–8. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Tian HF, Wen JF. The evolution of YidC/Oxa/Alb3 family in the three domains of life: a phylogenomic analysis. BMC Evol Biol 2009;9:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoufaly S, Frobel J, Rose P et al. Mapping precursor-binding site on TatC subunit of twin arginine-specific protein translocase by site-specific photo cross-linking. J Biol Chem 2012;287:13430–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C, Bhuiyan S. Archaea signal recognition particle shows the way. Archaea 2010;2010:485051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C, Eichler J. Getting on target: the archaeal signal recognition particle. Archaea 2002;1:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.