Abstract
Mammalian nutritional status affects the homeostatic balance of multiple physiological processes and their associated gene expression. Although DNA array analysis can monitor large numbers of genes, there are no reports of expression profiling of a micronutrient deficiency in an intact animal system. In this report, we have tested the feasibility of using cDNA arrays to compare the global changes in expression of genes of known function that occur in the early stages of rodent zinc deficiency. The gene-modulating effects of this deficiency were demonstrated by real-time quantitative PCR measurements of altered mRNA levels for metallothionein 1, zinc transporter 2, and uroguanylin, all of which have been previously documented as zinc-regulated genes. As a result of the low level of inherent noise within this model system and application of a recently reported statistical tool for statistical analysis of microarrays [Tusher, V.G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121], we demonstrate the ability to reproducibly identify the modest changes in mRNA abundance produced by this single micronutrient deficiency. Among the genes identified by this array profile are intestinal genes that influence signaling pathways, growth, transcription, redox, and energy utilization. Additionally, the influence of dietary zinc supply on the expression of some of these genes was confirmed by real-time quantitative PCR. Overall, these data support the effectiveness of cDNA array expression profiling to investigate the pleiotropic effects of specific nutrients and may provide an approach to establishing markers for assessment of nutritional status.
Keywords: nutrition‖microarray‖micronutrient
Nutrients play a significant role in health promotion through participation in many diverse physiological mechanisms. With growing awareness of entire genomes and the resources available to investigate this information, there are increased opportunities to examine the impact of nutrition on gene expression. Regulation of gene expression by minerals, vitamins, and specific macronutrients is well established but, in most cases, this influence has been clearly documented for only a very small number of genes (1). Although nutritional science is beginning to embrace the tools of genomics, including cDNA arrays, few attempts at large-scale or global evaluation of nutritional gene regulation have been reported. Consequently, it remains to be established whether the currently identified nutritionally regulated genes are unique, or whether others are similarly modulated. Alternatively, there may be populations or families of genes similarly affected by changes in nutritional status. An additional challenge confronting global gene expression profiling in integrated physiological systems is the detection of relatively subtle, albeit metabolically significant, levels of modulation expected compared with the more drastic alterations found in the pathological changes associated with cancer or infectious disease or resulting from acute stress.
The molecular function of zinc affects a wide variety of physiological systems, including those necessary for proper immune function and growth (2, 3). However, the specific pathways that become dysfunctional when dietary zinc supply is inadequate or excessive, because of dietary supplements, have not been identified. Furthermore, specific indices or markers for zinc status assessment of individual human patients or populations are lacking (3, 4). Metallothionein (MT) genes are very sensitive to zinc status and have been well characterized, providing a good model for highly responsive zinc-regulated gene expression (reviewed in ref. 5). Zinc exerts its effects directly on transcription of MT through metal-responsive gene promoter elements and their associated metal-sensing transcription factors (MTFs), such as MTF-1 (reviewed in ref. 6). Interestingly, null mutation of the MTF-1 gene is lethal at day 14 of gestation (7), whereas MT gene deletion is not lethal (8, 9), suggesting that direct zinc regulation of gene transcription via MTF-1 extends beyond MT regulation to other essential physiological systems. In addition to direct effects on gene transcription modulated by zinc, there are likely to be indirect, or secondary, effects on gene expression produced by the alteration of zinc-dependent functions that influence other gene regulation pathways (10).
Our laboratory has successfully identified dietary zinc-regulated genes from the small intestine by using subtractive library hybridization as well as mRNA differential display (11, 12). Unlike cDNA arrays, these methods do not rely on existing genome database information and therefore can detect previously unknown genes that are differentially expressed and, in the case of differential display, with high sensitivity for low-abundance mRNAs (10, 12). Differential display has allowed us to identify intestinal uroguanylin precursor as an up-regulated gene in zinc deficiency (13). Subsequently, we confirmed these mRNA screening data and extended them with measurements of uroguanylin peptides by both Western analysis and immunohistochemistry (14, 15). However, differential display requires extensive experimental manipulation; consequently, limited numbers of genes can be screened in a short time span. In this report, we have tested the feasibility of using cDNA arrays to compare global changes in expression of genes of known function that occur in the early stages of a single micronutrient deficiency, specifically zinc deficiency, to complement our ongoing analysis of zinc-regulated mammalian genes.
Materials and Methods
Animal Protocol.
Sprague–Dawley strain rats (male, 150–200 g; Harlan Breeders, Indianapolis) were individually housed and fed a modified AIN 76A semipurified diet, as described previously (12). The rats were divided into three groups (n = 4/group): zinc normal (ZnN) (30 mg of Zn/kg diet), zinc-deficient (Zn−) (<1 mg of Zn/kg diet), and a third group pair fed (PF) to the Zn− rats. After 2 wk, the animals were anesthetized and exsanguinated by cardiac puncture to determine serum zinc concentrations (12). Small intestine (scraped to recover the mucosa) and a kidney were excised and processed as described previously (12). These experiments were approved by the University of Florida Institutional Animal Care and Use Committee.
RNA Isolation and Probe Labeling.
Tissues were immediately homogenized in TriPure reagent (Roche Molecular Biochemicals), and total RNA isolated. Kidney MT mRNA levels were used as an index of dietary zinc status, as previously described (12). The quality, integrity, and quantity of the RNA from intestines of individual rats were determined (i.e., intact ribosomal RNA bands on electrophoresis and UV absorption spectrophotometry) and pooled by using an equal contribution from each animal. The RNA was DNase-I (Roche Molecular Biochemicals) treated, phenol-chloroform extracted, ethanol precipitated, and poly(A)+ RNA was isolated by using OligoTex mRNA Maxi-Prep (Qiagen, Chatsworth, CA). Complex probes were generated by reverse transcription using the poly(A)+ RNA, freshest possible 33P-labeled dATP (NEN) and an Atlas gene-specific mix of oligonucleotide primers (CLONTECH). Unincorporated radiolabeled nucleotides were removed with Nucleospin Extraction spin columns (CLONTECH), and probe yields were quantified by liquid scintillation counting.
Hybridization and Imaging of Atlas Arrays.
CLONTECH Atlas Rat 1.2 arrays and reagents were used for analysis of gene expression according to the manufacturer's protocols. These membrane format arrays were chosen not only for initial investment considerations, but also because they are limited to known genes. Furthermore, array cDNA spots are short regions of cDNA selected for low homology to other genes, and gene-specific primers are used in probe syntheses. The array data in this report were generated from two identical experiments, the first using six new membranes and the second using four new and two reused membranes. Three membranes for each dietary condition were separately prehybridized in Express Hyb buffer (CLONTECH), and the 33P-labeled probes from above were denatured, diluted with carrier DNA, and an equal amount added to each of the membranes. Hybridization at 68°C was allowed to proceed for ≈40 h, after which the membranes were washed and exposed to the same phosphor screen for 1–10 days. Phosphor images were acquired using a Storm 840 Fluorescent Imaging System (Molecular Dynamics). For membranes that were reused, the previous probe was removed by boiling, as described by CLONTECH.
Analysis of Array Data.
Phosphor images of each array were first imported into atlasimage software (Ver. 1.5; CLONTECH) for densitometric measurement of gene- and array-specific parameters such as intensity and global background levels. A preliminary analysis of zinc-regulated genes was also performed by using arbitrary threshold settings for fold change (ratio) and absolute intensity differences. The values for all genes with background subtracted intensities >1 were then exported to an excel (Microsoft) spreadsheet to generate a sorted data matrix for all arrays within each experiment. At this point, data were filtered to eliminate genes detected fewer than two times in each condition and replicate array values normalized by the sum of the intensities method. Total intensities from the two conditions were then scaled and mean intensities calculated for scatter plot visualization of the fold changes.
Primary statistical analyses of the array data were performed by using the Significance Analysis of Microarrays (sam; excel Add-In version) procedure developed by Tusher et al. (16). These analyses were performed on actual and log-transformed mean signal intensities from both experiments. The lists of genes identified as significantly modulated from all four sam runs were compared for shared members and merged before ranking analysis. Specifically, all genes appearing in a list from only one sam analysis were eliminated, as were genes detected in only one of the two hybridization experiments. Also culled were any that appeared on the up-regulated list in one experiment and the down-regulated in the other, as well as any with no absolute difference (0 or 1) in one of the two hybridization experiments. Finally, genes with a 20% or smaller fold change and an absolute change smaller than 1.5 × the median intensity of the experiment were removed from the list. The remaining genes are the most consistently detected significantly changed genes and therefore the best candidates for genes modulated by this nutrient deficiency.
Confirmation of Zinc Modulation by Real-Time Quantitative PCR (Q-PCR).
Real-time PCR primer and probe sets were selected for each cDNA by using primer express software (Ver. 1.0; Applied Biosystems; Table 1). Sequences for cDNAs were obtained from GenBank for use in primer design, although in some cases information for the Atlas-specific regions of cDNAs was purchased from CLONTECH and the primers designed from within that region. Q-PCR reactions were performed with either SYBR-green or TaqMan chemistries by using one-step reverse-transcriptase PCR reactions on a GeneAmp 5700 Sequence Detection System (all from Applied Biosystems). The Universal 18S rRNA primer/probe-set used to normalize all assays was also from Applied Biosystems. Before quantitative measurements of experimental samples, assays were validated by ability to produce a three-log or greater dynamic range and, in the case of SYBR assays, produce a single prominent peak in dissociation analysis. Standard curves for candidate cDNAs were prepared from a five-point 1/3 serial dilution run in duplicate (slopes for all assays reported were −3.3 ± 0.3), whereas samples were run in quadruplicate. cDNA quantities were normalized to 18S rRNA quantities obtained from the same plate, with the ZnN sample used as the calibrator.
Table 1.
Q-PCR primer and FRET probe list
| Rat mRNA | Forward primer | Reverse primer | FRET probe |
|---|---|---|---|
| Metallothionein 1 | TGTGCCCAGGGCTGTGT | GCAGCACTGTTCGTCACTTCA | CACGTGCACTTGTCCGAGGCACCT |
| Zinc transporter 2 | CAAGACCTGAGGGCAGAATAGTC | CCTAGGAGAGTCCCATGGTGTAGT | AGCCGGGACCCCAGGATCAAGA |
| Uroguanylin precursor peptide | TCCCCGATGTGTGCTACAAC | TTCCTGGGATGCACAAACAG | CCGCCTTGCCCCTGGACCTC |
| β actin | TTCAACACCCCAGCCATGT | GTGGTACGACCAGAGGCATACA | |
| Intestinal fatty acid-binding protein | CACACAGGAAGGAAATAAATTCACA | GACGCCGAGTTCAAACACAA | |
| Mitochondrial ATP synthase, α subunit | TTTGGTGGTGGCTCTTTGACT | TTGGAATGTAGGCGGACACA | |
| Glutathione S-transferase, subunit 8 | GATGGATGCCTGCTTTTTGG | GATGGCTCTGGTCTGTGTCAGTAG | |
| Ribosomal protein L36a | CGCTACGACCGGAAGCA | TGGTTTTGGCCTTCTTTCGA | |
| Aldehyde reductase | CAATGACGGCCTCCAGTGT | TCCATGTCCCCAGACCAATC | |
| Proteasomal ATPase (TBP1) | GAGTTTCCAATGCCCAATGAG | ACTGACGTTCATCTTCCGTGAGT |
FRET, fluorescence resonance energy transfer (probes are 5′ FAM and 3′ TAMRA).
Results
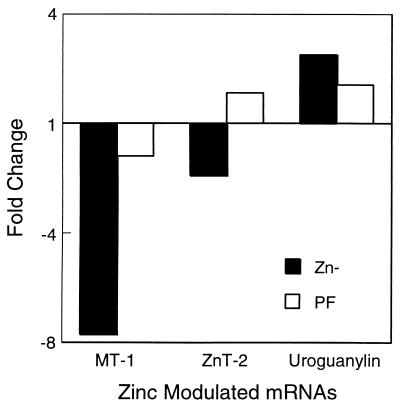
Rats in the 2-week Zn− diet study displayed the expected physiological responses. Specifically, the Zn− group compared with ZnN and PF groups, respectively, had decreased weight gain (29 vs. 79 and 45 g), depressed serum zinc concentrations (0.3 vs. 1.3 and 1.2 μg Zn/ml), and decreased kidney MT mRNA levels (0.50 vs. 1.00 and 0.74 arbitrary units). Relative expression of three genes known to be strongly regulated by dietary zinc in mucosal cells of the small intestine is shown in Fig. 1. Both MT and zinc transporter 2 mRNA levels are markedly reduced, 7.7- and 1.9-fold, respectively, by zinc deficiency. In contrast, preprouroguanylin mRNA levels are increased by 2.5-fold in the Zn− rats. Taken together, these results clearly demonstrate the effectiveness of the dietary protocol.
Figure 1.
Zinc-modulated expression of MT-1, zinc transporter 2, and uroguanylin precursor mRNAs. Expression of MT-1, zinc transporter 2 (ZnT-2), and uroguanylin precursor mRNAs in rat small intestine as measured by Q-PCR. Pooled total RNA from Zn−, ZnN, and PF rats was analyzed by the relative quantitative method. Data are expressed as the fold change with the ZnN values set at 1.0. The same RNA was used for subsequent array profiling.
33P incorporation into the reverse-transcribed probes derived from Zn− and ZnN poly(A)+ RNA samples was comparable and exceeded the array manufacturer's criteria, including low background signals. Array hybridization times were lengthened to 1½ days, which appeared to improve signal strength and sensitivity without increasing the background signal. Phosphor image exposures were performed at varying times; however, beyond 3–4 days, the signal-to-background noise ratio became too low to effectively distinguish low intensity signals, so 3-day exposures were used for all analyses (Fig. 2). The two reused array membranes showed no qualitative or quantitative differences from new membranes.
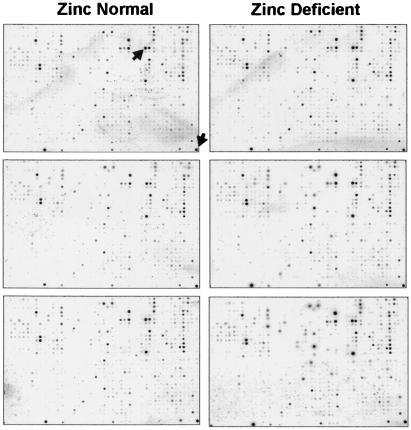
Figure 2.
Phosphor images of Atlas Rat 1.2 arrays. Images from one experiment obtained from a 3-day phosphor screen exposure are shown on an exponential contrast scale to aid in visualization of low-intensity spots and background relative to the very high intensities of a few genes. The images on the left are from three membranes hybridized to the rat small intestine ZnN probe, whereas those on the right are from the small intestine Zn− probe. Overall, a very similar pattern of expression is seen in all arrays. On this array, the most highly expressed genes in the rat small intestine, aldolase B and 40S ribosomal protein S29, are indicated by arrows on the first array. For gene grid assignment, refer to CLONTECH's Atlas Rat 1.2 Array web site (http://www.clontech.com/atlas/genelists/7854-1_Ra12.pdf).
As determined by atlasimage software, the global background pixel intensity for all arrays was between 3 and 6, whereas some mRNAs produced signal intensities of 10,000–15,000. Image quality permitted use of global background subtraction for cDNA signals; however, a few cDNA spots required the use of local or regional background subtraction. Data from two experiments detected above-background 483 and 365 cDNAs, respectively, of the 1185 cDNAs on the arrays. A list of all genes detected and their relative intensities, as well as the original digital images, is available at fshn.ifas.ufl.edu/Cousins/Lab_Data/Rat_Arrays.htm.
Initial evaluation of differentially expressed genes by using atlasimage software was based on two arbitrary criteria: an absolute difference, initially set at approximately eight times the intensity of minimum detection threshold, and a fold ratio change, set at 2. Within these criteria, there were no outstanding perturbations of expression with the genes on these arrays, and only one gene, intestinal fatty acid-binding protein, exceeded both absolute and fold change parameters. Reducing the fold ratio difference criterion to 1.5, however, generated a preliminary list of 13 candidates for zinc-regulated genes that were consistent in signal intensities and magnitude of change between experiments. However, it also became apparent that there were some signals with little variation that just missed the arbitrary threshold criteria.
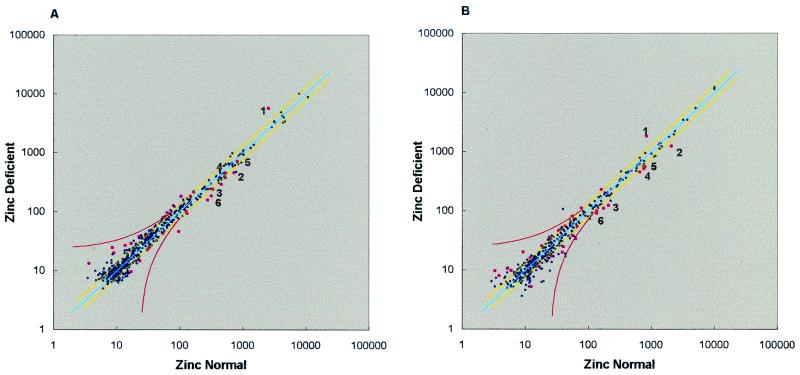
Different methods to calibrate the ZnN and Zn− conditions were explored, as the Zn− intensities were consistently higher than the ZnN intensities. After calibration, the cDNA intensities ranged from 1 to 12,711; however, the average intensities on each array ranged from 199 to 231 and 212 to 253 (experiments 1 and 2, respectively), whereas the average median intensities were 23 and 24. After calculation of the standard deviation and coefficients of variance for each gene's averaged intensity, data were prepared as a scatter plot to visualize all relative changes (Fig. 3). This plot showed that, compared with scatter plots from many other array data sets, this data set had a very low variance, also evidenced by high correlation coefficients (r = 0.96 and 0.99) on regression analysis. In addition, the slopes of the best fit lines (0.98 ± 0.01, 1.12 ± 0.01) were very close to the ideal slope of one.
Figure 3.
Scatter plots of array data. Average ZnN and Zn− intensities for each gene were plotted for the two experiments. The line of identity is blue, whereas yellow lines indicate a 1.3-fold change up or down, and the red curves indicate an absolute intensity change equal to the average median intensity value for each experiment (23 and 24, respectively). Pink dots indicate genes identified in Table 2, whereas numbered dots correspond to the genes numbered in Table 3. Note the very tight distribution of all data points around the line of identity, which is further evidenced by correlation coefficients of 0.96 and 0.99, respectively, from regression analysis.
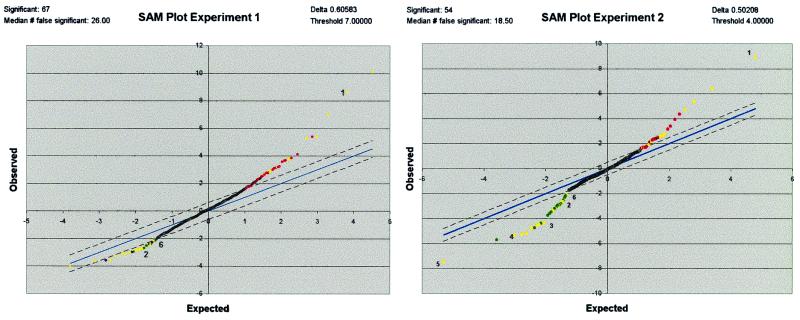
Because the degree of changes is small in this system, yet there is an observable precision, on the basis of the replicate membranes and experimental design, the sam method (16) was used to identify potential zinc-regulated cDNAs. This method does not examine “fold changes,” which do not measure statistical probability, but rather uses the variance of repeated measures compared with the difference of the mean values to assign significance to array data. Hence, sam should be able to identify small but reproducible differences in this data set. Each experiment was analyzed separately by sam to produce a list of potentially regulated genes, on the basis of the degree of significance (Fig. 4). As array data often exhibit a large range of gene intensities, sam analysis was also performed on log-transformed values from both experiments. These four individual sam analyses were executed with δ values selected to produce initial sam lists with at least 25 up-regulated and 25 down-regulated genes. Compiling the top 25 of the four sam analyses resulted in a list of 62 up-regulated and 63 down-regulated gene candidates. However, after prioritization (ranking) according to number of top 25 appearances in the four sam analyses and exclusion of those that failed to meet the rules described in Materials and Methods, the list was reduced to 16 up-regulated and 16 down-regulated candidates (Table 2). Thus, the ranking analysis eliminated 75% of the genes collectively identified by sam. This reduction empirically suggests that genes remaining on the list, after application of the ranking analysis rules, have a much lower false discovery rate (FDR) than the predicted FDR from any single sam analysis (30–40%) and are, therefore, more likely to be true zinc regulated genes.
Figure 4.
Significance analysis of array data. sam plots for the actual intensity data from experiments 1 and 2 respectively, are shown, where the observed sam score is plotted against the “expected” sam score. The line representing “observed” equals “expected” is blue, whereas the dashed lines indicate the significance threshold specified by the sam δ value. The plot shows significantly different up-regulated genes as red or yellow (Upper Right) and significantly different down-regulated genes as green or yellow (Lower Left). Yellow dots indicate genes that are listed in Table 2, whereas numbered dots correspond to genes in Table 3.
Table 2.
Results of sam output ranking analysis
| Gene | GenBank accession no. | Atlas grid | Experiment
|
Ave. rank | Ave. fold Δ | |||
|---|---|---|---|---|---|---|---|---|
| 1 | 1Log | 2 | 2Log | |||||
| Induced by zinc deficiency | ||||||||
| Fatty acid-binding protein, intestinal | M35992 | B14f | 2 | 4 | 1 | 1 | 2 | 2.2 |
| Growth factor, schwannoma-derived | X55183 | E02d | 1 | 5 | 4 | 2 | 3 | 2.1 |
| Nucleoside diphosphate kinase B | M91597 | A11f | 3 | 6 | 10 | 12 | 8 | 1.6 |
| Pancreatic secretory trypsin inhibitor I precursor | M27882 | F11c | 9 | 1 | 4 | 5 | 2.8 | |
| Adipocyte fatty acid-binding protein (FABP4) | U75581 | B14h | 4 | 3 | 16 | 8 | 2.1 | |
| Ecto-ATPase precursor; cell-CAM 105 | M92848 | A02f | 6 | 11 | 11 | 9 | 1.3 | |
| Cytochrome P450 4B1; P450-isozyme 5 | M29853 | B01f | 22 | 2 | 7 | 10 | 2.9 | |
| Thioredoxin peroxidase 1 | U06099 | A14b | 20 | 9 | 8 | 12 | 1.6 | |
| CC chemokine MIP3 α exodus | U90447 | E02l | 23 | 13 | 6 | 14 | 1.7 | |
| CD4 homologue, W3/25 antigen | M15768 | A03q | 11 | 16 | 18 | 15 | 1.5 | |
| Signal transducer CD24 precursor | U49062 | A04f | 16 | 17 | 17 | 1.3 | ||
| LIM domain protein CLP36, homologous to rat RIL | U23769 | A06a | 21 | 17 | 19 | 1.4 | ||
| Max; c-myc dimerization partner and coactivator | D14447 | A05g | 2 | 10 | 6 | 1.3 | ||
| Fructose (glucose) transporter | D13871 | B02f | 3 | 9 | 6 | 1.3 | ||
| Somatostatin | M25890 | E03f | 20 | 13 | 17 | 1.5 | ||
| CD3, γ chain | S79711 | A03h | 24 | 25 | 25 | 1.4 | ||
| Suppressed by zinc deficiency | ||||||||
| Glutathione S-transferase Yb subunit (GSTM2) | J02592 | A14e | 1 | 4 | 5 | 11 | 5 | −1.7 |
| Mast cell protease precursor 8 (RMCP-8) | U67911 | F07g | 15 | 6 | 4 | 2 | 7 | −2.0 |
| Sodium/hydrogen exchange protein 3 | M85300 | B08g | 20 | 2 | 20 | 1 | 11 | −2.1 |
| Arginase 2 | U90887 | C09f | 5 | 21 | 9 | 20 | 14 | −1.4 |
| Kidney aminopeptidase M | M26710 | F08j | 2 | 22 | 22 | 17 | 16 | −1.5 |
| Mitochondrial ATP synthase, α subunit | X56133 | C04d | 14 | 23 | 24 | 10 | 18 | −1.6 |
| Cytosolic acyl-CoA thioester hydrolase | D88890 | C04e | 7 | 17 | 3 | 9 | −1.7 | |
| Microsomal glutathione S-transferase (GST12; MGST1) | J03752 | A14d | 11 | 10 | 6 | 9 | −1.5 | |
| Cytochrome P-450 4F1, hepatic tumour | M94548 | C07i | 4 | 15 | 10 | −1.3 | ||
| Prothymosin-α | M20035 | A08e | 12 | 15 | 14 | −1.4 | ||
| Mitochondrial ATP synthase, D subunit; ATP5H | D10021 | C04a | 22 | 8 | 15 | −1.4 | ||
| Glutathione S-transferase, subunit 8 | X62660 | A14m | 11 | 7 | 9 | −1.5 | ||
| ATPase, sodium/potassium, γ subunit | X70062 | B10j | 13 | 9 | 11 | −1.5 | ||
| 60S ribosomal protein L36A | M19635 | C11d | 3 | 19 | 11 | −1.3 | ||
| Aldehyde reductase; 3-dG-reducing enzyme | D10854 | C10m | 1 | 24 | 13 | −1.3 | ||
| ATPase, proteasomal, liver, TBP1 | U77918 | F10k | 25 | 14 | 20 | −1.6 | ||
This table lists the top ranking genes from the 62 up-regulated and 63 down-regulated genes identified collectively by the four sam analyses. Solid lines separate groups of genes, on the basis of the number of their occurrences (four vs. three vs. two) in the four analyses. Those below the broken lines distinguish genes that occur only in the analyses from only one hybridization experiment. ave., average.
It is of interest to note that, whereas there are generally larger fold changes higher on the list, there are multiple exceptions to this trend. Within the up-regulated gene candidates, there are three genes that appear no lower than position 12 in all four sam analyses and would be predicted with the highest level of confidence. In the down-regulated group, there are six genes that appear in the top 25 of all four sam analyses, but their position in the ranking had a larger spread. The lowest confidence candidates are those that appear in both normal and log-transformed analyses of only one experiment; however, as will be seen below, this may still identify a real change in expression status for these genes.
Independent confirmation for select array-identified candidates was performed by Q-PCR, which allowed other RNA samples to be included in the comparison. Therefore, RNA from the PF group of rats was also analyzed for the expression levels of the candidate cDNAs. Of the six genes for which real-time confirmations were performed, all but one confirmed the expression change observed on the arrays (Table 3), although with smaller magnitudes. Additionally, three showed no effect of food restriction, and two showed food restriction producing an effect opposite of zinc deficiency, whereas one indicated there might be an additive effect of food restriction with zinc deficiency (Table 3).
Table 3.
Summary of Q-PCR confirmation for selected genes
| Differentially expressed gene | Confirmation of zinc modulation | Array expression | Influence of food restriction |
|---|---|---|---|
| Intestinal fatty acid-binding protein (1) | +++ | ⇑ | ⇑ |
| Mitochondrial ATP synthase, α subunit (2) | − | ⇓ | None |
| Glutathione S-transferase, subunit 8 (3) | ++ | ⇓ | ⇑ |
| Ribosomal protein L36a (4) | + | ⇓ | None |
| Aldehyde reductase (5) | ++ | ⇓ | ⇑ |
| Proteasomal ATPase (6) | ++ | ⇓ | None |
Confirmation of array (+); no confirmation of array (−); multiple (+) indicates the degree of confidence between array and Q-PCR changes.
Discussion
The rat model of zinc deficiency has a well established history with good biomarkers of zinc status and generally utilizes a second normal zinc control group that is food restricted to compensate for the zinc deficiency-induced anorexia observed in this model (12). Although some expression profiling techniques, such as differential display, can accommodate three or more groups, microarray analysis becomes significantly more complex with more than two groups in the comparison. As concerns about discerning the difference between zinc-regulated gene expression and regulation effects produced by food restriction were easily addressed during the confirmation stage (Q-PCR can easily accommodate three or more treatment conditions), only two dietary conditions were used for the array phase of this project.
Our experimental design using three replicate arrays for both treatment groups allowed us to observe experimental variation and labeling and/or hybridization artifacts. In this way, consistent changes across replicates and experiments aided in identifying the relatively small magnitude of expression changes anticipated from an in vivo micronutrient deficiency study. The data from the two experiments respectively identified 41 and 31% of the 1,185 genes as detected consistently above background levels. This percentage appears to correspond with other reports for arrays using RNA from a metabolically active tissue such as the small intestine (17–19). The magnitude of the changes reported here is less than that of those that occur for some genes that have been documented as zinc regulated. Responses of MT, zinc transporter 2, and preprouroguanylin (uroguanylin precursor) genes to the zinc depletion used in these experiments were presented (Fig. 1) to show a maximal response, representing both up-regulation and down-regulation, that could realistically be expected on the basis of past findings (12–14, 20, 21). Clearly, of the genes that constitute this array and were expressed in the small intestine, few exhibited a change on zinc depletion comparable to these three genes.
Because our goal was to identify genes that were altered across all individuals in the model population, RNA samples were pooled to reduce signal noise in expression introduced by individual animal variation. This approach has previously been used successfully in this nutrient deficiency model for mRNA differential display (12, 13). With only a single sample from each treatment, this pooling also reduced the variation introduced by poly(A)+ selection efficiency. In addition, a batch-labeling approach for triplicate membranes minimized variance between and subsequent degree of normalization required among replicate membrane arrays.
To compensate for any small differences in labeling efficiency between samples of RNA from the two dietary conditions, total signal intensity was calibrated between the two conditions. Although this calibration appears to work well in this project, it does have the potential to mask a suppression or induction that occurs across a large population of genes evaluated. Although a better method would be use of a battery of genes known to be unaltered by the treatments, such a list is unavailable at this time. Previously, data from Northern blot analysis have shown no obvious change in intestinal mRNA levels during zinc deficiency for β-actin (12), a commonly used housekeeping gene that traditionally has been used as a normalizer for such experiments. However, our array experiments indicated that actin levels are slightly elevated in zinc deficiency, and subsequent analysis by Q-PCR confirmed a 15% increase in actin mRNA levels. This observation makes actin unsuitable for normalizing these data, especially given the small magnitude of the fold changes observed. Previous conclusions based on actin-normalized data should retain their validity, because they report changes of a much larger magnitude.
On the basis of array data statistics, it appeared that changes as small as 1.3-fold could be distinguished in our rat nutritional deficiency model. To confirm this analysis, a small random selection of genes was analyzed by Q-PCR (Table 3). Results for 60S ribosomal protein L36A, aldehyde reductase, and liver proteosomal ATPase confirm not only the small level of gene suppression evidenced in zinc deficiency but also three of the genes in what empirically appears to be the lowest confidence group according to the ranking analysis. An additional note on confirmation is the identification by sam of two mRNAs previously defined as zinc regulated. Intestinal fatty acid-binding protein and CD3 γ-chain mRNAs were previously identified as regulated by dietary zinc by using subtracted library hybridization and mRNA differential display analysis, respectively (10, 11). Overall, these data support the use of array analysis, particularly when coupled with the sam procedure, for the investigation of relatively small expression changes resulting from altered dietary conditions.
Collectively, array data thus far have tended to identify differentially expressed genes that initially do not provide a clear mechanistic explanation for phenotypic effects (16, 19, 22). Rather, array studies have shown how a given condition, treatment, or response evokes expression differences in a plethora of seemingly unrelated genes. The results presented in this report are no exception. Although cluster analysis can begin to address this issue, it is most effective when performed on many more mRNAs than were analyzed on this array system. Now that the detection of small changes in our model has been demonstrated, a larger-scale array project, including cluster analysis, would be in order.
A deficiency of zinc could influence gene expression through dysfunction of the catalytic, structural, or regulatory roles this micronutrient plays in biology (2, 3). For example, whereas a number of the identified differentially regulated genes code for enzymes, only one, kidney aminopeptidase M, is a known zinc metalloenzyme (Table 2). Thus catalytic dysfunction of zinc enzymes, if this occurs in mild zinc deficiency, does not influence regulation of their synthesis in a way that produces noticeable change in the specific mRNAs. Similarly, only a few of the genes identified, e.g., LIM domain protein CLP36, produce proteins that require zinc occupancy for a structural function.
Although the regulatory function of zinc is most associated with an MTF/metal-responsive gene promoter element-linked mechanism of transcriptional control, a regulatory role for zinc clearly extends to other venues. For example, Lck (p56 lck) requires zinc for cytoplasmic binding of the CD4/CD8α receptor (23, 24). Although not present on these arrays with rat genes, murine zinc deficiency has been shown to up-regulate the Lck gene (21). A surprising number of genes identified in our experiments are associated with maintaining the redox state of the cell. A key hypothesis to explain aspects of zinc function relates to the oxidant-induced loss of zinc from MT to other sulfur-rich domains such as zinc fingers and ring fingers of regulatory proteins (25). A number of the identified genes that participate in redox-responsive pathways may dysregulate as a consequence of a shift in cellular redox during zinc deficiency. Furthermore, as more information regarding upstream elements for specific genes becomes known, some of these and other genes that are involved in zinc-requiring systems may be shown to use MTF/metal-responsive gene promoter element-controlled transcription. An example of the use of such information has been the characterization of zinc-regulated genes in yeast (26). By using the extensive yeast genome database, it was possible to characterize genes regulated by the zinc-responsive transcription factor Zap1p. The response of yeast to zinc deficiency demonstrates that differentially expressed Zap1p-regulated genes include many proteins that are unrelated to zinc metabolism and include enzymes, regulatory proteins, and many proteins of unknown function. It is of interest that a majority of the yeast genes that increase in zinc deficiency do so by a factor of approximately 2- to 3-fold. This observation is particularly curious for a single-celled organism, known to be extremely responsive to the environment. Also relevant to what has been found in mammalian zinc deficiency (10, 12–15), many up-regulated yeast genes in zinc deficiency appear to do so by indirect mechanisms.
One known limitation of array analysis is the lack of sensitivity to detect low-abundance mRNAs. This drawback is evidenced in these experiments by the lack of detection of zinc transporter 1 (ZnT-1), which is present on these arrays. We have characterized ZnT-1 zinc regulation and localization in the rat small intestine by using Northern blot analysis and immunohistochemistry (20, 27). For this reason, more sensitive large-scale screening methods, such as mRNA differential display, are still necessary to identify those genes that are involved in the response to zinc deficiency yet are expressed at levels below a detection threshold.
The very low level of noise in the fold ratio change suggests several points for future investigations. The most obvious is that systems with low inherent variation between the two conditions will allow accurate identification of regulatory events with changes well below the 2-fold threshold emphasized repeatedly in the literature. In particular, this sensitivity supports the use of array analysis in studies of an integrative nature, including those related to nutrition, where the goal is identifying the key regulatory genes involved in a particular physiological function. Furthermore, because markers for analysis or assessment of the dietary status of many nutrients or for health-related dietary patterns are lacking, array profiling may provide a tool for such evaluations for both individual subjects or patients as well as for selected populations.
Acknowledgments
We thank Drs. James G. Booth and George Casella of the University of Florida Department of Statistics for helpful discussions of statistical analyses. The research reported here was supported by National Institutes of Health Grant DK 31127 and by the Boston Family Endowment Funds of the University of Florida.
Abbreviations
- MT
metallothionein
- MTF
metal-sensing transcription factor
- PF
pair fed
- sam
Significance Analysis of Microarrays
- Zn−
zinc deficient
- ZnN
zinc normal
- Q-PCR
real-time quantitative PCR
- ZnT-2
zinc transporter 2
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 2, 2000.
References
- 1.Cousins R J. In: Modern Nutrition in Health and Disease. 9th Ed. Shils M E, Olson J A, Shike M, Ross A C, editors. Baltimore: Williams & Wilkins; 1999. pp. 573–584. [Google Scholar]
- 2.Mills C F. Zinc in Human Biology. New York: Springer; 1989. [Google Scholar]
- 3.King J C, Keen C L. In: Modern Nutrition in Health and Disease. 9th Ed. Shils M E, Olson J A, Shike M, Ross A C, editors. Baltimore: Williams & Wilkins; 1999. pp. 223–239. [Google Scholar]
- 4.Panel on Micronutrients, Subcommittees on Upper Reference Levels of Nutrients and of Interpretation and Use of Dietary Reference Intakes; the Standing Committee on the Scientific Evaluation of Dietary Reference Intakes, Food and Nutrition Board. Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc. Washington, DC: Natl. Acad. Press; 2001. , in press. [Google Scholar]
- 5.Davis S R, Cousins R J. J Nutr. 2000;130:1085–1088. doi: 10.1093/jn/130.5.1085. [DOI] [PubMed] [Google Scholar]
- 6.Andrews G K. Biochem Pharmacol. 2000;59:95–104. doi: 10.1016/s0006-2952(99)00301-9. [DOI] [PubMed] [Google Scholar]
- 7.Günes C, Heuchel R, Georgiev O, Müller K-H, Lichtlen P, Blüthmann H, Marino S, Aguzzi A, Schaffner W. EMBO J. 1998;17:2846–2854. doi: 10.1093/emboj/17.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michalska A E, Choo A K H. Proc Natl Acad Sci USA. 1993;90:8088–8092. doi: 10.1073/pnas.90.17.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masters B A, Kelly E J, Quaife C J, Brinster R L, Palmiter R D. Proc Natl Acad Sci USA. 1994;91:584–588. doi: 10.1073/pnas.91.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchard R K, Cousins R J. J Nutr. 2000;130:1393S–1398S. doi: 10.1093/jn/130.5.1393S. [DOI] [PubMed] [Google Scholar]
- 11.Shay N F, Cousins R J. J Nutr. 1993;123:35–41. doi: 10.1093/jn/123.1.35. [DOI] [PubMed] [Google Scholar]
- 12.Blanchard R K, Cousins R J. Proc Natl Acad Sci USA. 1996;93:6863–6868. doi: 10.1073/pnas.93.14.6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanchard R K, Cousins R J. Am J Physiol. 1997;272:G972–G978. doi: 10.1152/ajpgi.1997.272.5.G972. [DOI] [PubMed] [Google Scholar]
- 14.Cui L, Blanchard R K, Coy L M, Cousins R J. J Nutr. 2000;130:2726–2732. doi: 10.1093/jn/130.11.2726. [DOI] [PubMed] [Google Scholar]
- 15.Cui L, Blanchard R K, Cousins R J. Kidney Int. 2001;59:1424–1431. doi: 10.1046/j.1523-1755.2001.0590041424.x. [DOI] [PubMed] [Google Scholar]
- 16.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. . (First Published April 17, 2001; 10.1073/pnas.091062498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iacomino G, Tecce M F, Grimaldi C, Tosto M, Russo G L. Biochem Biophys Res Commun. 2001;285:1280–1289. doi: 10.1006/bbrc.2001.5323. [DOI] [PubMed] [Google Scholar]
- 18.Toulouse A, Loubeau M, Morin J, Pappas J J, Wu J, Bradley W E C. FASEB J. 2000;14:1224–1232. doi: 10.1096/fasebj.14.9.1224. [DOI] [PubMed] [Google Scholar]
- 19.Hooper L V, Wong M H, Thelin A, Hansson L, Falk P G, Gordon J I. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 20.Liuzzi J P, Blanchard R K, Cousins R J. J Nutr. 2001;131:46–52. doi: 10.1093/jn/131.1.46. [DOI] [PubMed] [Google Scholar]
- 21.Moore J B, Blanchard R K, McCormack W T, Cousins R J. J Nutr. 2001;13:3189–3196. doi: 10.1093/jn/131.12.3189. [DOI] [PubMed] [Google Scholar]
- 22.Cao S X, Dhahbi J M, Mote P L, Spindler S R. Proc Natl Acad Sci USA. 2001;98:10630–10635. doi: 10.1073/pnas.191313598. . (First Published September 4, 2001; 10.1073/pnas.191313598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin R S, Rodriguez C, Veillette A, Lodish H F. J Biol Chem. 1998;273:32878–32882. doi: 10.1074/jbc.273.49.32878. [DOI] [PubMed] [Google Scholar]
- 24.Huse M, Eck M J, Harrison S C. J Biol Chem. 1998;273:18729–18733. doi: 10.1074/jbc.273.30.18729. [DOI] [PubMed] [Google Scholar]
- 25.Maret W. J Nutr. 2000;130:1455S–1458S. doi: 10.1093/jn/130.5.1455S. [DOI] [PubMed] [Google Scholar]
- 26.Lyons T J, Gasch A P, Gaither L A, Botstein D, Brown P O, Eide D J. Proc Natl Acad Sci USA. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMahon R J, Cousins R J. Proc Natl Acad Sci USA. 1998;95:4841–4846. doi: 10.1073/pnas.95.9.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]