Abstract
Background:
Epigenetic machinery plays an important role in genomic imprinting, a developmental process that establishes parent-of-origin–specific monoallelic gene expression. Although a number of studies have investigated the role of 5-methylcytosine in imprinting control, the contribution of 5-hydroxymethylcytosine (5-hmC) to this epigenetic phenomenon remains unclear.
Objectives:
Using matched mouse blood samples (from mice at 2, 4, and 10 months of age), our objective was to examine the effects of perinatal bisphenol A (BPA) exposure ( diet) on longitudinal 5-hmC patterns at imprinted regions. We also aimed to test the hypothesis that 5-hmC would show defined patterns at imprinted genes that persist across the life course.
Methods:
Genome-wide 5-hmC levels were measured using hydroxymethylated DNA immunoprecipitation sequencing (HMeDIP-seq). Modeling of differential hydroxymethylation by BPA exposure was performed using a pipeline of bioinformatics tools, including the csaw R package.
Results:
Based on BPA exposure, we identified 5,950 differentially hydroxymethylated regions (DHMRs), including 12 DHMRs that were annotated to murine imprinted genes—Gnas, Grb10, Plagl1, Klf14, Pde10a, Snrpn, Airn, Cmah, Ppp1r9a, Kcnq1, Phactr2, and Pde4d. When visualized, these imprinted gene DHMRs showed clear, consistent patterns of differential 5-hmC by developmental BPA exposure that persisted throughout adulthood.
Conclusions:
These data show long-term establishment of 5-hmC marks at imprinted loci during development. Further, the effect of perinatal BPA exposure on 5-hmC at specific imprinted loci indicates that developmental exposure to environmental toxicants may alter long-term imprinted gene regulation via an epigenetic mechanism. https://doi.org/10.1289/EHP3441
Introduction
DNA methylation is an epigenetic mark that typically occurs at cytosines (5-methylcytosine; 5-mC) in cytosine-phosphate-guanine (CpG) dinucleotides. Research has shown that 5-mC plays a role in X chromosome inactivation (Cotton et al. 2015), regulation of gene transcription (Medvedeva et al. 2014), and genomic imprinting (Bartolomei and Ferguson-Smith 2011). In addition to 5-mC, recent studies have shown that the oxidized form of 5-mC—5-hydroxymethylcytosine (5-hmC)—is a stable epigenetic mark present in a variety of mammalian tissues (Globisch et al. 2010; Hahn et al. 2014; Wu et al. 2011). Active processing of 5-mC to 5-hmC occurs via a Ten-Eleven Translocation (TET) methylcytosine dioxygenase–mediated oxidative pathway (Shen et al. 2014), and previous studies have shown that exposure-induced oxidative stress can alter both TET enzyme expression (Coulter et al. 2013) and global 5-hmC levels (Delatte et al. 2015). In addition to these characteristics, 5-hmC has a complex role as both a positive and negative regulator of transcription (Hahn et al. 2014; Wu et al. 2011), suggesting that it may represent an important secondary genomic regulator in the methylome.
During early embryonic development, DNA methylation undergoes a distinct wave of demethylation and de novo methylation (Reik et al. 2001; Smallwood and Kelsey 2012), processes that assist in the regulation of stem cell proliferation and differentiation (Messerschmidt et al. 2014). After differentiation, there is a second phase of DNA methylation reprograming that occurs to establish a baseline methylome in primordial germ cells prior to birth (Smallwood and Kelsey 2012). Given these multiple waves of reprograming during development, DNA methylation has been proposed as a mechanism driving the developmental origins of health and disease (DOHaD) hypothesis. The DOHaD posits that exposure to environmental factors (e.g., diet, chemicals) during critical periods of development influences developmental plasticity, thereby altering disease susceptibility later in life (Bateson et al. 2004; Heindel et al. 2015; Jirtle and Skinner 2007). Of particular interest in the DOHaD field is genomic imprinting, an epigenetic phenomenon by which parent-of-origin–specific monoallelic gene expression is established during development (Bartolomei and Ferguson-Smith 2011; Das et al. 2009). Genomic imprinting is critical for proper placental maturation, embryonic growth, and development, and it also plays important roles in postnatal development—especially in parent–offspring interactions (e.g., milk suckling, maternal care) (Plasschaert and Bartolomei 2014). A vast amount of research on genomic imprinting has shown that differential DNA methylation at imprinting control regions (ICRs) plays an important role in establishing genomic imprinting during development, and that these regions are not demethylated during postfertilization reprograming (Arnaud 2010; Kelsey and Feil 2013; Kim et al. 2017; Pidsley et al. 2012; Smallwood and Kelsey 2012). Given that imprinted gene expression is controlled via developmentally programed epigenetic marks, imprinted genes have been investigated as potential targets of developmental environmental exposures in both human and rodent studies (Haycock and Ramsay 2009; Heijmans et al. 2008; Nye et al. 2015; Susiarjo et al. 2013).
However, virtually all of the existing studies on genomic imprinting have relied upon sodium bisulfite treatment for the generation of DNA methylation levels from biological samples. Although bisulfite treatment is a useful quantitative tool for measuring the methylome, it also has an inherent weakness: It does not distinguish between 5-mC and 5-hmC. This is because both 5-mC and 5-hmC are resistant to sodium bisulfite-induced cytosine deamination (Huang et al. 2010). This means that bisulfite treatment–based measurements of the “methylome” actually represent combined levels. In recent years, methods to specifically measure 5-hmC, including hydroxymethylated DNA immunoprecipitation sequencing (HMeDIP-seq) (Tan et al. 2013), have been developed. Despite these recent advances, however, little work has been done to identify the role of 5-hmC in genomic imprinting. One recent study showed enrichment of 5-hmC in imprinted regions in the human brain and placenta (Hernandez Mora et al. 2018), but no study has yet investigated the contribution of 5-hmC to imprinting control in animal models. Furthermore, existing animal studies have not investigated effects of the environment on 5-hmC levels at imprinted genes.
Developmental exposure to a number of environmental factors, including endocrine disrupting chemicals (EDCs), has been linked to changes in DNA methylation. Bisphenol A (BPA) is a commercial EDC that is found in a variety of consumer products (e.g., receipt paper, metal can linings) and has near ubiquitous human exposure across the world (Calafat et al. 2008). BPA can activate growth-related transcription factors, bind nuclear receptors involved in cell growth and maturation, and also alter DNA methylation levels across the epigenome (Krüger et al. 2008; Manikkam et al. 2013; Singh and Li 2012; Sui et al. 2012; Watson et al. 2007; Wolstenholme et al. 2011; Zhang et al. 2012). Developmental BPA exposure in mouse models has been shown to alter the methylome (Anderson et al. 2012; Kim et al. 2014; Singh and Li 2012), including specific effects on DNA methylation at imprinted loci (Susiarjo et al. 2013). Despite these results, however, it remains unclear whether developmental BPA exposure can specifically affect DNA hydroxymethylation.
Here, we used the HMeDIP-seq method to measure epigenome-wide 5-hmC from longitudinal mouse blood samples in an effort to examine DNA hydroxymethylation at imprinted loci throughout murine adulthood. HMeDIP-seq is an antibody-based high-throughput sequencing method that measures the genome-wide distribution of 5-hmC. Although this method has some inherent antibody inefficiency and fails to provide base-pair resolution sequencing data, it does provide cost-effective amplification of high-frequency, low-signal 5-hmC marks within gene bodies (Skvortsova et al. 2017; Tan et al. 2013). Given the discovery nature of this novel study and the lack of a priori knowledge regarding 5-hmC levels in mouse blood or in response to toxicant exposures, HMeDIP-seq was chosen to measure genome-wide 5-hmC from longitudinal blood samples. From this data, we tested the hypothesis that perinatal BPA exposure alters longitudinal 5-hmC levels at imprinted genes. We also tested the hypothesis that imprinted genes would show defined 5-hmC levels that persist across the life course.
Materials and Methods
Study Animals and Blood Collection
Mice included in the longitudinal analysis were offspring sourced from a genetically invariant viable yellow agouti mouse colony maintained by sibling mating and forced heterozygosity for more than 220 generations (Waterland and Jirtle 2003). Within this colony, the allele is passed through the heterozygous male line, which has a genetically constant background 93% identical to the C57BL/6J strain (Waterland and Jirtle 2003; Weinhouse et al. 2014). Two weeks prior to mate-pairing with males, 8- to 10-wk-old wild-type dams (Control: ; BPA: ) were placed on one of two experimental diet groups: a) Control (modified, phytoestrogen-free 7% corn oil AIN-93G), or b) Control diet (see Figure S1) (Anderson et al. 2012; Weinhouse et al. 2014). For each mouse, the experimental diet was assigned using an online random number generator. After randomization, any dam litter mates assigned to the same exposure group were switched to different diets. All dams in a diet group were co-housed for the first 2 wk of exposure, prior to mate-pairing. During mate-pairing, dams were individually housed and a sire was immediately added to those cages; both the dam and sire had access to the experimental diet until pregnancy was confirmed, and then the sire was removed. In an effort to limit cage bias, all exposure groups were in the same room with the same environmental conditions, and cages were rotated on the shelves so that the same mice were not always closest to the lights or the floor. However, during preconception, gestation, and weaning, different colored chow pellets were used for the Control and BPA chow to ensure that the correct diet was placed in the correct cages. BPA dose was chosen based on a previous dosing study from our lab that showed intake of diet produced, on average, liver (Anderson et al. 2012). This exposure level is within the range of human exposure levels measured in human fetal liver samples (range: nondetect to liver) (Anderson et al. 2012). Dietary exposure was continued through pregnancy and lactation. At postnatal day (PND) 21, BPA-exposed offspring (total ; used for these analyses) were weaned to the modified AIN-93G Control diet and followed along with Control offspring (total ; used for these analyses) until 10 months of age. All diets were provided by Envigo. At 2 and 4 months of age, tail-vein blood samples were collected from all mice (see Figure S1). The mice were sacrificed at 10 months of age, and blood samples were again collected, this time using cardiac puncture. Six mouse blood samples from each exposure group were selected for inclusion in next-generation sequencing analyses. In accordance with methods established by the National Institute of Environmental Health Sciences (NIEHS) Toxicant Exposures and Responses by Genomic and Epigenomic Regulators of Transcription (TaRGET) II consortium (Wang et al. 2018), we selected male () and female () blood samples collected from six separate litters of Control and BPA-exposed mice. All animals were housed in polycarbonate-free cages with ad libitum access to food and drinking water, and were maintained in accordance with Institute for Laboratory Animal Research (ILAR) guidelines (National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals 2011). The study protocol was approved by the University of Michigan Committee on Use and Care of Animals (UCUCA PRO00004797).
DNA and RNA Isolation
To allow for matched analyses of DNA hydroxymethylation and gene expression from the same set of mice ( per group), genomic DNA and RNA were co-isolated from frozen mouse blood that was collected at 2, 4, and 10 months of age using the Qiagen Allprep DNA/RNA/miRNA Universal Kit (Catalog #80,224; Qiagen). Yield and purity of all DNA and RNA was measured using a NanoDrop spectrophotometer. All samples were stored at prior to DNA and RNA isolations.
Real-Time Quantitative PCR
The Bio-Rad iScript cDNA Synthesis Kit (Catalog #1,708,890) was used to reverse transcribe complementary DNA (cDNA) from of RNA for each sample. In preparation for real-time quantitative polymerase chain reaction (RT-qPCR), cDNA samples were diluted 1:2.5 in RNase-free water, then mixed with forward/reverse primers, nuclease-free water, and Bio-Rad iQ SYBR Green Supermix (Catalog #1,708,880). RT-qPCR was then performed using a Bio-Rad CFX96 Real-Time System C1000 Thermal Cycler (Bio-Rad). The preprogramed 2-step curve protocol was used for all qPCR reactions: 95°C for 3 min, [95°C for 10 s, 55°C for 30 s, plate read] X 40, 95°C for 10 s. The melt curve for each plate was 65–95°C with a 0.5°C increment for 5 s and a plate read at each temperature. RT-qPCR analyses were performed for the Gnas gene in triplicate for each cDNA sample. Three housekeeping genes—Actb, 18S, and Gapdh—were included as internal controls in all RT-qPCR runs. In addition to the housekeeping genes, an inter-plate calibrator control of brain cDNA was included for calculation of relative gene expression across multiple plates. Expression levels were calculated following the method (Livak and Schmittgen 2001). RT-qPCR primers for the Gnas and Gapdh genes (see Table S1) were designed using the online Genscript Real-time PCR Primer Design software (https://www.genscript.com/tools/real-time-pcr-tagman-primer-design-tool). The and 18S gene primer pairs were sourced from the literature (Dolinoy et al. 2010; La Salle et al. 2004). Primer pair specificity for all designed primers was checked using the National Center for Biotechnology Information (NCBI) Primer-BLAST online tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
Next-Generation Sequencing of 5-hmC
Epigenome-wide hydroxymethylation levels were quantified in blood from mice using HMeDIP-seq (Tan et al. 2013). Unlike some other methods, HMeDIP-seq is an antibody-based approach that does not provide base-pair resolution data. This method is also subject to the inherent bias of a multistep, immunoprecipitation-based sequencing method, including potential for antibody binding inefficiency, as well as library preparation bias, read mapping bias, or PCR amplification bias (Meyer and Liu 2014). Despite these weaknesses, HMeDIP-seq is a cost-effective method to measure genome-wide 5-hmC, including at regions of high-frequency weak 5-hmC signal (Tan et al. 2013). Sequencing data were generated by C. Lalancette at the University of Michigan Epigenomics Core Facility for a subset of Control () and BPA-exposed () mice (Figure 1). Blood samples from the six mice (3 male, 3 female) in each exposure group were sequenced at three time points across the life course (2, 4, and 10 months of age), for a total sequencing data sample size of 36 samples. The six mice for each exposure group were selected from different litters to minimize litter effects. Sample quality and quantitation were assessed using the Agilent TapeStation genomic DNA kit (Catalog #G2991AA; Agilent) and Qubit broad range dsDNA (Catalog #Q32850; Invitrogen), respectively. Indexed adapters and PCR primers were synthesized by Integrated DNA Technologies (IDT). Enzymes used for library preparation were sourced from New England BioLabs (NEB).
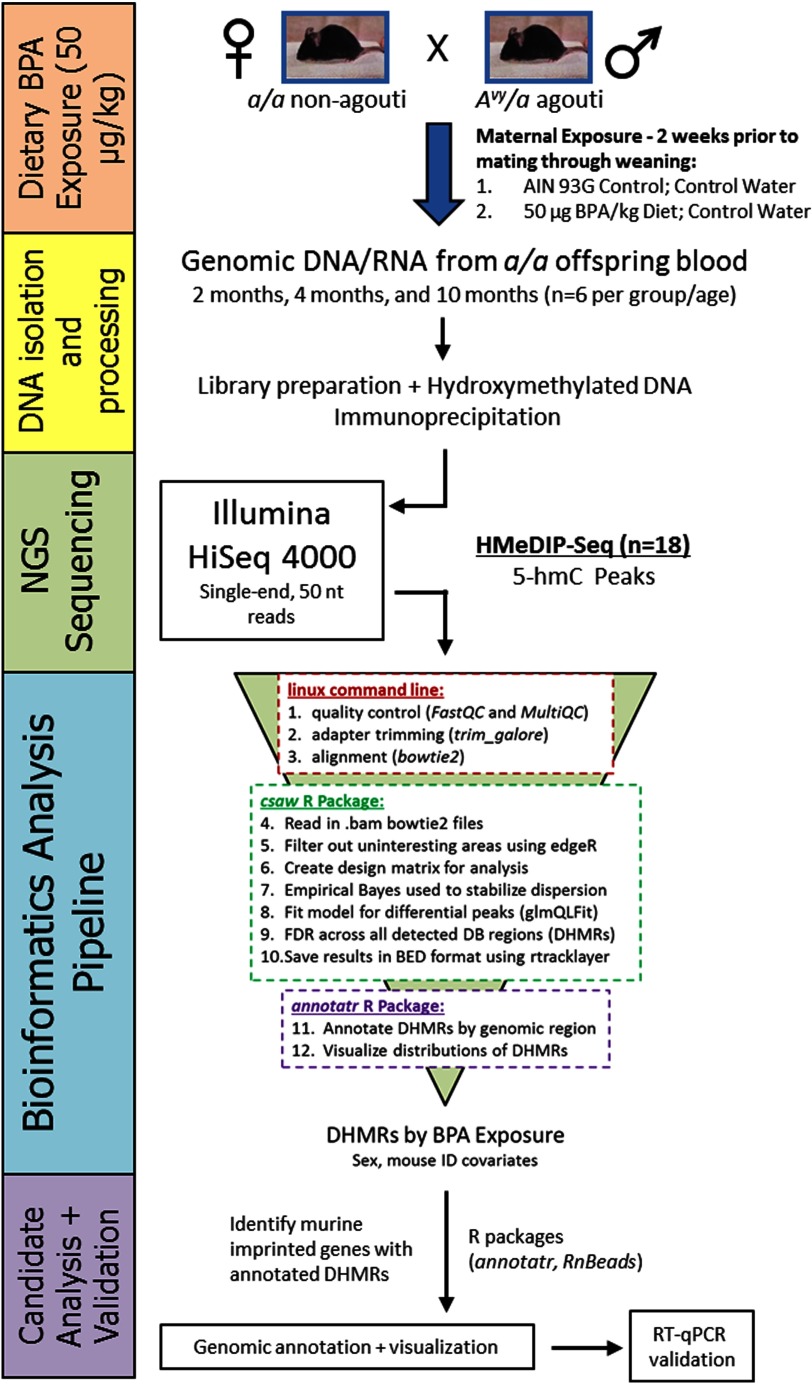
Figure 1.
Sequencing data collection and analysis workflow. Two weeks prior to mate-pairing with males, 8- to 10-wk-old wild-type dams were placed on one of two experimental diet groups: (1) Control (modified, phytoestrogen-free 7% corn oil AIN-93G), or (2) Control diet. Dietary exposure was continued through gestation and lactation until weaning at postnatal day 21. Genomic DNA was isolated from matched wild-type offspring blood samples at 2, 4, and 10 months of age. DNA was isolated from a subset of Control ( per age group) and BPA-exposed ( per age group) mice, then processed in preparation for HsMeDIP-seq. All processed samples were amplified and sequenced on an Illumina HiSeq 4000 sequencer using single-end, reads. BPA-related differentially hydroxymethylated regions (DHMRs) were identified and annotated using a bioinformatics pipeline. Annotated DHMRs were then visualized in the genome browser. The target gene region—Gnas—was then validated using RT-qPCR on available RNA from the blood samples.
A total of of genomic DNA (gDNA) was sheared by adaptive focused acoustics, using the Covaris S220 (Catalog #4,465,653; Covaris). This sheared DNA was next blunt-ended and phosphorylated. A single adenine nucleotide was then added to the 3′ end of the fragments in preparation for the ligation of the adapter duplex with a thymine overhang. The ligated fragments were cleaned using Qiagen’s MinElute PCR purification columns (Catalog #28,004; Qiagen). DNA standards for HMeDIP (Diagenode 5-hmC, 5-mC, and cytosine DNA standard pack for HMeDIP; Catalog #AF-107-0040) were added to each sample before denaturation. Resuspension was then performed in ice-cold immunoprecipitation buffer ( sodium phosphate pH 7.0, sodium chloride (NaCl), 0.05% Triton X-100). A 10% volume (input) was retrieved before of a 5-hmC–specific antibody (Active Motif, Catalog # 39,791) was added for immunoprecipitation overnight at with rotation. Dynabeads Protein-G (Catalog #10003D; Invitrogen) was used to pull down 5-hmC–enriched fragments. The 5-hmC-enriched DNA fragments (from immunoprecipitation) were then released from the antibody by digestion with Proteinase K (Catalog #AM2548; Ambion). After cleanup with AMPure XP beads (Product #A63880; Beckman Coulter), the percentage input enrichment (%input) in the IP was evaluated by qPCR, using hydroxymethylated, methylated, and unmethylated primers for spike-ins. Samples with high %input for the 5-hmC spike-in—typical inclusion threshold was —were then PCR amplified for the final library production, cleaned using AMPure XP beads, and quantified using the Qubit assay (Catalog #Q32850; Invitrogen) and TapeStation High Sensitivity D1000 kit (Catalog #G2991AA; Agilent). Single-end, reads were obtained for each library by sequencing on the HiSeq 4000 system (Illumina). Each HMeDIP-seq sample was run on a single sequencing lane.
Next-Generation Sequencing of 5-mC
Enhanced reduced representation bisulfite (ERRBS) was performed at the University of Michigan Epigenomics Core as described previously (Akalin et al. 2012; Garrett-Bakelman et al. 2015). Briefly, of genomic DNA was digested using MspI, a restriction enzyme that preferentially cuts CG-rich sites. The digested DNA was then purified using phenol:chloroform extraction and ethanol precipitation in the presence of glycogen, before blunt-ending and phosphorylation. A single adenine nucleotide was next added to the 3′ end of the fragments in preparation for the ligation of the adapter duplex with a thymine overhang. The ligated fragments were cleaned, then processed for size selection on agarose gel. Selected fragments were treated with sodium bisulfite to convert unmethylated cytosines to uracils, which are then replaced with thymines during PCR amplification. After cleanup with AMPure XP beads (Product #A63880; Beckman Coulter), libraries were quantified using the Agilent TapeStation genomic DNA kit (Catalog #G2991AA; Agilent) and Qubit broad range dsDNA (Catalog #Q32850; Invitrogen). Single-end, reads were obtained for each library by sequencing on the HiSeq 4000 system (Illumina). ERRBS samples were multiplexed, with three samples per sequencing lane.
Sequencing Data Quality Control, Trimming, and Alignment
HMeDIP-seq data for all Control and BPA-exposed mice were compared using a suite of bioinformatics tools (Figure 1). First, the Sartor lab mint pipeline was used for data quality control (FastQC and MultiQC), adapter trimming (trim_galore), and alignment (bowtie2) (Cavalcante et al. 2017). For quality and adapter trimming, we required a minimum overlap of and minimum quality score of 20, along with special ERRBS trimming of from the 3′ end. For bismark methylation extractor (ERRBS data), the minimum threshold to consider a CpG site in analysis was five reads. Although the default minimum overlap in trim_galore is , we selected a less stringent minimum overlap of in an effort to include all legitimate genomic sequences and improve read depth. Despite these benefits, the less stringent minimum overlap length increases the possibility for adapter contamination in our data (Krueger 2017). The quality score cutoff was selected based on previous research showing an optimal tradeoff between correct read mapping and read survival at quality scores between 20 and 30 (Del Fabbro et al. 2013). The lower end of this range was selected to maximize read survival after trimming. Default parameters were used for bowtie2.
Bioinformatics Pipeline for Differential 5-hmC
After data quality check, trimming, and alignment, the csaw package was used to test for differential 5-hmC by BPA exposure (Lun and Smyth 2016). Aligned HMeDIP-seq data were read into R (version 3.4.0; R Development Core Team) using the window Counts function in csaw. When reading in the data, extension was set to 52, window width was set to 100, and sex chromosomes were removed. The data were then filtered twice—by count (average count ), and by local enrichment using the filter Windows function ()—to remove regions of negligible binding. After filtering, normalization factors were calculated for each sample using the normOffsets function on binned data (). Normalization factors were linked to filtered data using the asDGEList function, and then the estimateDisp function was used to generate dispersion factors based on a multifactorial design matrix. The glmQLFit function was used to fit a model [] for differential 5-hmC binding; an empirical Bayesian method used to stabilize the QL dispersion estimates. Contrast statements were used to extract modeling results for the variable of interest: BPA exposure. To correct for multiple testing, filtered data was clustered using a window length of ; the combineTests function was then used to compute a combined p-value for each cluster. Multiple testing correction was performed using the Benjamini-Hochberg method (Benjamini and Hochberg 1995, 1997). Differential hydroxymethylated region (DHMR) length cutoff was set at , and significance was set at a false discovery rate (FDR) of .
Bioinformatics Pipeline for Differential 5-mC
The DSS R package was used to test for differential 5-mC in ERRBS data by BPA exposure (Feng et al. 2014; Park and Wu 2016; Wu et al. 2013, 2015). Within the DSS package, the DMLfit.multiFactor function was used to test for differential methylation by BPA exposure using a multifactorial modeling approach according to the following formula: . The DMLtest.multiFactor function was used to test the null hypothesis that the BPA exposure coefficient was equal to 0. Differentially methylated CpG sites (DMCs) by BPA exposure were then sorted and filtered according to a FDR cutoff of .
After testing for differential hydroxymethylation using csaw, the annotatr R package was used to annotate all DHMRs to the mm10 genome (Cavalcante and Sartor 2017). The annotate_regions function was used to generate genomic annotations, which include classifications by region class (e.g., intron, exon, promoter) and annotated gene identifiers (IDs). A list of mouse imprinted loci was then sourced from the MouseBook online database (Williamson et al. 2013). All available imprinted genes were manually cross-checked with the generated list of BPA-related DHMR-annotated gene IDs.
Sequencing Data Visualization
5-hmC levels were visualized using the csaw and GViz R packages (Hahne and Ivanek 2016; Lun and Smyth 2016). Using csaw, data was first read into R as a GRanges object using the extractReads function. Next, the GeneRegionTrack in the Gviz R package was used to define regions of interest for visualization. Separate blue and red genome tracks were used to represent the forward and reverse strand reads, respectively. The plotTracks function was then used to plot 5-hmC levels for these regions. Reads-per-million was used for the y-axis in all graphs, and scale was adjusted to individual region coverage. 5-mC levels were visualized using the RnBeads R package (Assenov et al. 2014). Within this package, the rnb.execute.import function was used to import the aligned ERRBS data. Next, the rnb.sample.groups function was used to group samples by age, and the rnb.plot.locus.profile function was used to plot relative 5-mC level in a heat map output for defined regions of the genome. A custom color palette was chosen from RColorBrewer for the heat map gradient.
BPA-related DHMRs at imprinted loci were visualized using the GViz R package to determine directionality and magnitude of differential 5-hmC. Plots generated in R (version 3.4.0; R Development Core Team) were formatted for publication in Adobe Illustrator CS6 (version 16.0.5).
Pathway Analysis
Pathway analysis for DHMRs was performed using ChIP-enrich (Welch et al. 2014). Within the ChIP-enrich online interface (http://chip-enrich.med.umich.edu/), the gene set filter was set to 2,000, peak threshold was set to 1, and adjustment for mappability of gene locus regions was set to false. The genome used for pathway analyses was mm10, and the ChIP-enrich method was used for enrichment testing. DHMRs were split into hypo- and hyper-hydroxymethylated regions prior to separate analyses. Only regions from TSS were included in the ChIP-enrich analysis. All pathway analyses included Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. After correcting for multiple testing, only pathways with an were considered significant.
Results
Epigenome-Wide Differential 5-hmC and 5-mC
Using the csaw R package, multivariate models were constructed to test for differential 5-hmC by perinatal BPA exposure ( diet). Based on the csaw models, we identified 5,950 DHMRs by BPA exposure (Table 1). Comparing the directionality of DHMRs by BPA exposure, we found more hypo-hydroxymethylated regions (; 71.4%) than hyper-hydroxymethylated regions (; 26.2%) (Table 1). We also identified a small fraction of BPA-related DHMRs (; 2.4%) that showed both hypo- and hyper-hydroxymethylation. Taken together, these epigenome-wide results show that the directionality of differential hydroxymethylation varies by region, with a skew toward decreased 5-hmC by BPA exposure. Using the annotatr R package, BPA-related DHMRs were annotated to the mm10 genome. Compared with a random distribution, BPA-related DHMRs were depleted at CpG shores, CpG shelves, promoters, 3′-UTRs, 5′-UTRs, exons, and introns, but slightly enriched at regions greater than from a CpG island (interCGI) (see Figure S2).
Table 1.
Differential 5-hmC in mouse blood by BPA exposure.
| BPA | |
|---|---|
| DHMRsa | |
| Hyper-hydroxymethylated | 1,559 |
| Hypo-hydroxymethylated | 4,247 |
| Both | 144 |
| Total | 5,950 |
Note: The csaw R package was used to examine the effect of BPA exposure ( diet) on 5-hmC levels across the epigenome. Models included a paired mouse ID variable to account for within-individual effects, as well as a sex variable. Directionality of DHMRs (relative to Control) is indicated in separate rows. BPA, bisphenol A; DHMR, differentially hydroxymethylated region; ID, identification; , direction of DNA hydroxymethylation change (BPA exposed relative to Control).
DHMR-calling significance threshold was set at a false discovery rate (FDR) of .
To investigate differential DNA methylation by BPA exposure, ERRBS sequencing data were generated from the same blood DNA samples used for HMeDIP-seq data. Unfortunately, due to biased and limited genomic coverage inherent in the ERRBS method, overlapping DNA methylation data were available at only one of the identified imprinted gene DHMRs: Kruppel-like factor 14 (Klf14). At the Klf14 DHMR, there were no significant changes in ERRBS data by BPA exposure (see Figure S3). However, in our differential methylation analyses of the ERRBS data by BPA exposure, we identified a number of DMCs annotated to each of the top six imprinted loci that had DHMRs (see Table S2). Of note, we identified three CpG sites within the guanine nucleotide binding protein, alpha stimulating, complex locus and the neuroendocrine secretory protein antisense (Gnasxl/Nespas) ICR that demonstrated significant hypomethylation with BPA exposure (see Table S2). None of the identified DMCs at the other top imprinted genes—growth factor receptor bound protein 10 (Grb10), pleiomorphic adenoma gene-like 1(Plagl1), phosphodiesterase 10A (Pde10a), Klf14, and small nuclear ribonucleoprotein polypeptide N (Snrpn)—fell in known murine ICRs.
BPA-Related DHMRs at Imprinted Genes
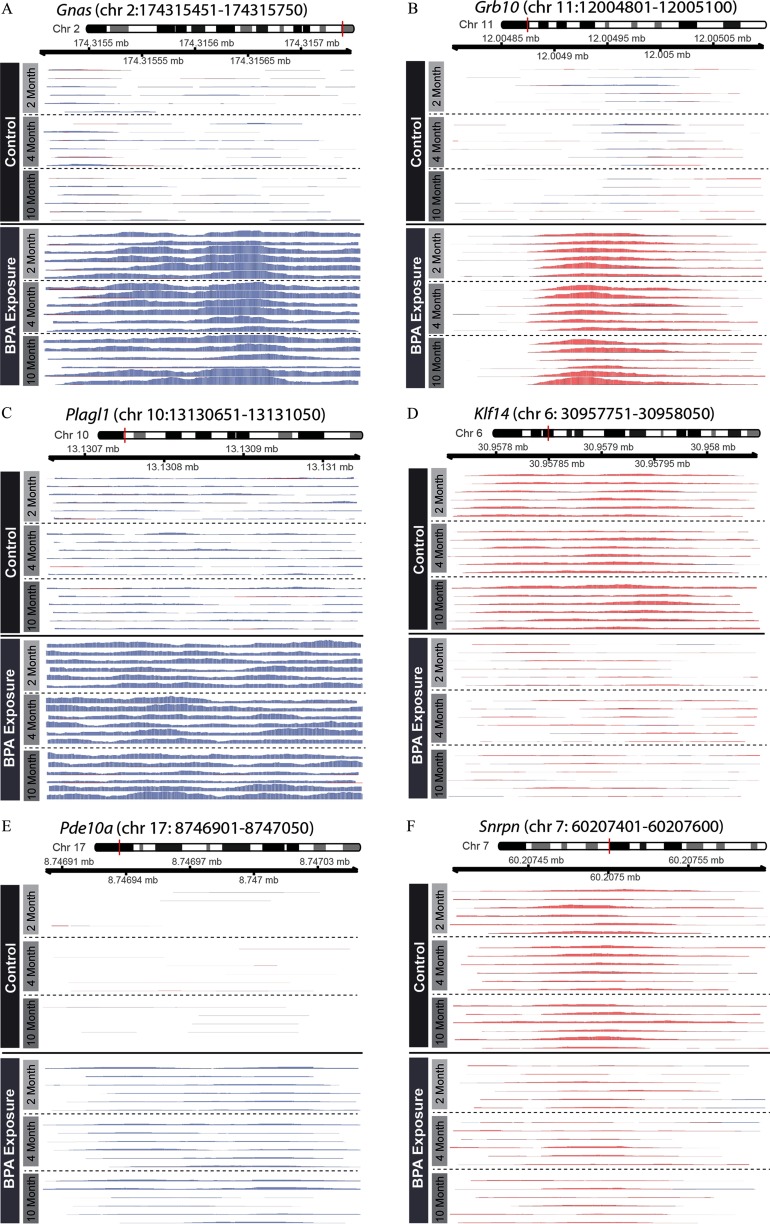
Previous research has shown effects of developmental exposures on DNA methylation at imprinted loci (Gallou-Kabani et al. 2010; Susiarjo et al. 2013), but it remains unclear whether exposures can also alter DNA hydroxymethylation at imprinted genes. Further complicating this question, despite recent research in humans showing enrichment of 5-hmC at imprinted loci (Hernandez Mora et al. 2018), the distribution and potential function of 5-hmC at murine imprinted genes is unknown. In an effort to broadly identify BPA-sensitive regions of 5-hmC at imprinted loci, we cross-checked the BPA-related DHMR annotations with a database of known murine imprinted genes (Williamson et al. 2013). In total, 12 of the 151 known imprinted genes had annotated BPA-related DHMRs (Table 2). Five of these 12 genes had increased 5-hmC peaks by BPA exposure—Grb10, Pde10a, phosphodiesterase 4D (Pde4d), Plagl1, and Gnas. The remaining 7 imprinted loci—protein phosphatase 1, regulatory subunit 9A (Ppp1r9a), phosphatase and actin regulator 2 (Phactr2), Klf14, potassium voltage-gated channel, subfamily Q, member 1 (Kcnq1), cytidine monophospho-N-acetylneuraminic acid hydroxylase (Cmah), antisense Igf2r RNA (Airn), and Snrpn—had decreased 5-hmC peaks by BPA exposure. The number of CpG sites within these DHMRs varied by region, with 5 of the DHMRs showing zero CpG sites (see Table S3). In an effort to confirm the DHMRs, we visualized the 5 most significant imprinted gene DHMRs and 1 DHMR at the well-characterized Snrpn gene, showing marked changes in 5-hmC peaks by BPA exposure that persisted across all of the three matched sample ages (Figure 2). We also found striking changes in 5-hmC peaks at the remaining 6 imprinted gene DHMRs, despite their decreased significance (see Figure S4). In an effort to determine whether this was a shared trait at imprinted genes, we visualized 5-hmC peaks at insulin like growth factor 2 (Igf2) and H19, imprinted maternally expressed transcript (H19), 2 well-characterized imprinted genes that did not have any annotated DHMRs with . Despite their lack of significant DHMRs, Igf2 and H19 still had specific 5-hmC peaks that showed nonsignificant changes by BPA exposure (see Figure S5).
Table 2.
Imprinted genetic loci with BPA-related DHMR.
| Chr | Start | End | Gene ID | Gene Name | Location | Direction with BPA exposure | csaw score | FDR |
|---|---|---|---|---|---|---|---|---|
| chr2 | 174315451 | 174315750 | 14683 | Gnas | Intronic | 113.4 | ||
| chr11 | 12004801 | 12005100 | 14783 | Grb10 | Intronic | 70.5 | ||
| chr10 | 13130651 | 13131050 | 22634 | Plagl1 | Exonic | 60.7 | ||
| chr6 | 30957751 | 30958050 | 619665 | Klf14 | Exonic | 55.9 | ||
| chr17 | 8746901 | 8747050 | 23984 | Pde10a | intronic | 50.1 | ||
| chr17 | 12828001 | 12828550 | 104103 | Airn | Intronic | 16.2 | 0.02 | |
| chr13 | 24335001 | 24335150 | 12763 | Cmah | Intronic | 13.1 | 0.05 | |
| chr7 | 60207401 | 60207600 | 20646 | Snrpn | Intronic | 13.0 | 0.05 | |
| chr6 | 5079501 | 5079850 | 243725 | Ppp1r9a | Intronic | 12.8 | 0.05 | |
| chr7 | 143348051 | 143348350 | 16535 | Kcnq1 | Intronic | 11.5 | 0.07 | |
| chr10 | 13402051 | 13402250 | 215789 | Phactr2 | Intronic | 11.3 | 0.07 | |
| chr13 | 108963951 | 108964200 | 238871 | Pde4d | Intronic | 10.1 | 0.10 |
Note: List of imprinted loci with annotated, significant () DHMRs generated from csaw modeling results. For csaw results, a transformed FDR is used for the score, such that ; higher score indicates a larger degree of significance (lower FDR value). Additionally, direction of effect by BPA exposure is shown, with a () indicating increased 5-hmC with BPA exposure and a () indicating decreased 5-hmC with BPA exposure. BPA, bisphenol-A; Chr, chromosome; DHMR, differentially hydroxymethylated region; FDR, false discovery rate; ID, identification.
Figure 2.
Differential imprinted gene 5-hmC peaks by BPA exposure. 5-hmC coverage was visualized at six imprinted loci with significant BPA-related DHMRs: (A) Gnas; (B) Grb10; (C) Plagl1; (D) Klf14; (E) Pde10a; and (F) Snrpn. 5-hmC levels are shown for matched 2-, 4-, and 10-month blood samples, as indicated by y-axis labels. Blue and red peaks represent forward and reverse strand 5-hmC enrichment, respectively. The Gnas, Plagl1, and Pde10a DHMRs occurred on the forward strand, whereas the Grb10, Klf14, and Snrpn DHMRs occurred on the reverse strand.
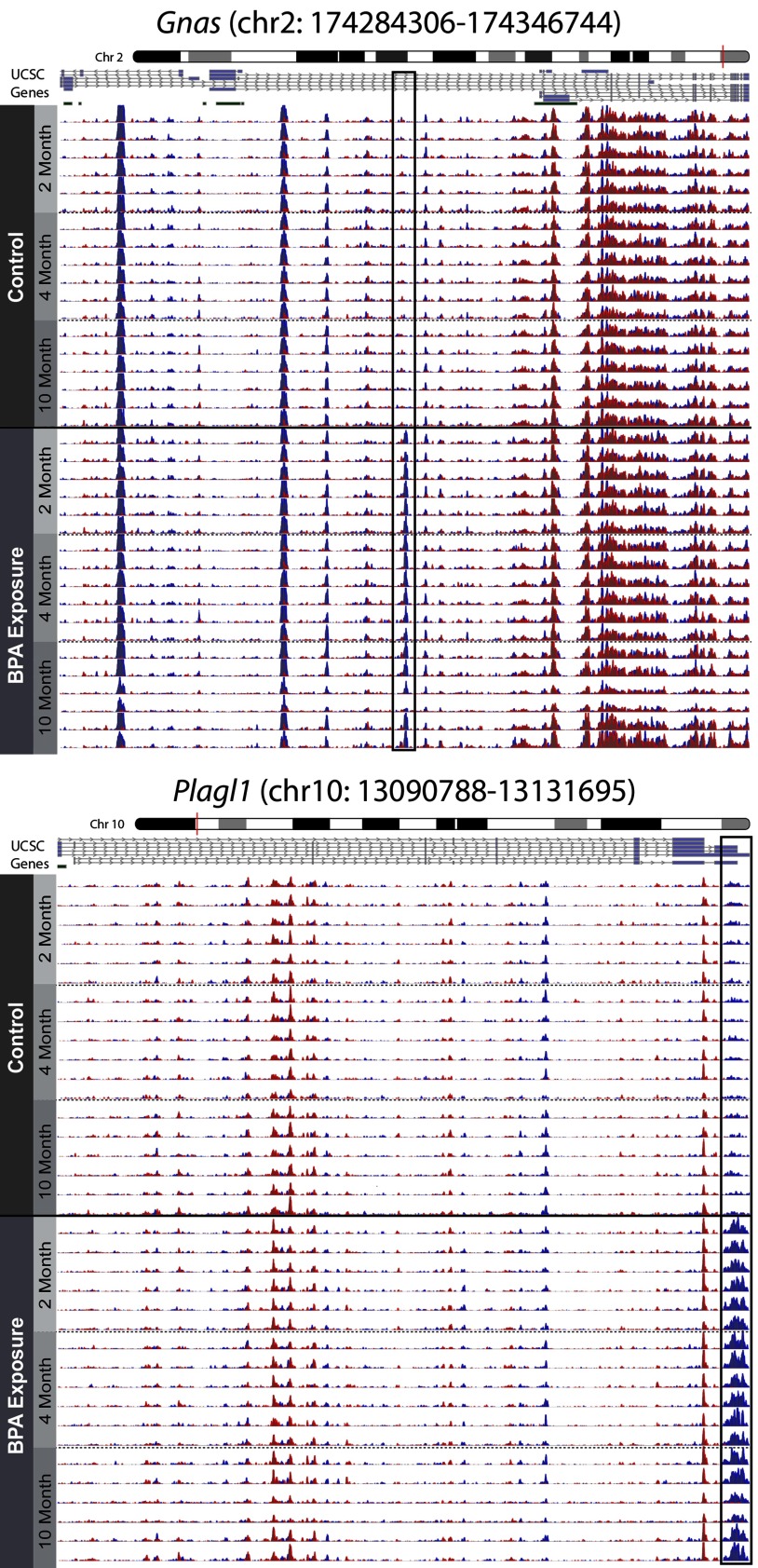
In examining the imprinted loci visually, two of the DHMRs—Plagl1 and Gnas—had 5-hmC peaks that were cut off by the DHMR boundaries, suggesting larger scale 5-hmC patterns at these genes. To provide a complete picture for these two loci, 5-hmC levels across the entire gene were visualized. Both Plagl1 and Gnas had widespread 5-hmC peaks along the entire gene-coding region, with only the annotated DHMRs showing apparent modifications by BPA exposure (Figure 3). Of note, 5-hmC peaks across these two genes were consistent between mice and across time, a pattern that was also observed at the Igf2 and H19 imprinted genes (see Figure S5). Together, these data show that imprinted genes have predictable 5-hmC peaks along the length of their gene bodies.
Figure 3.
Genomic context of Gnas and Plagl1 DHMRs. The DHMRs annotated to the Gnas and Plagl1 genes were visualized in the context of their respective imprinted genes using the csaw and Gviz R packages. 5-hmC levels across the complete Gnas and Plagl1 imprinted loci are presented, showing distinct 5-hmC peaks along the length of both genes. 5-hmC levels are shown for matched 2-, 4-, and 10-month blood samples, as indicated by y-axis labels. Boxes indicate significant DHMRs that were identified using csaw differential hydroxymethylation models. Blue and red peaks represent forward and reverse strand 5-hmC enrichment, respectively. Bars below University of California, Santa Cruz genome browser gene tracks represent CpG islands.
RT-qPCR Gene Expression Data
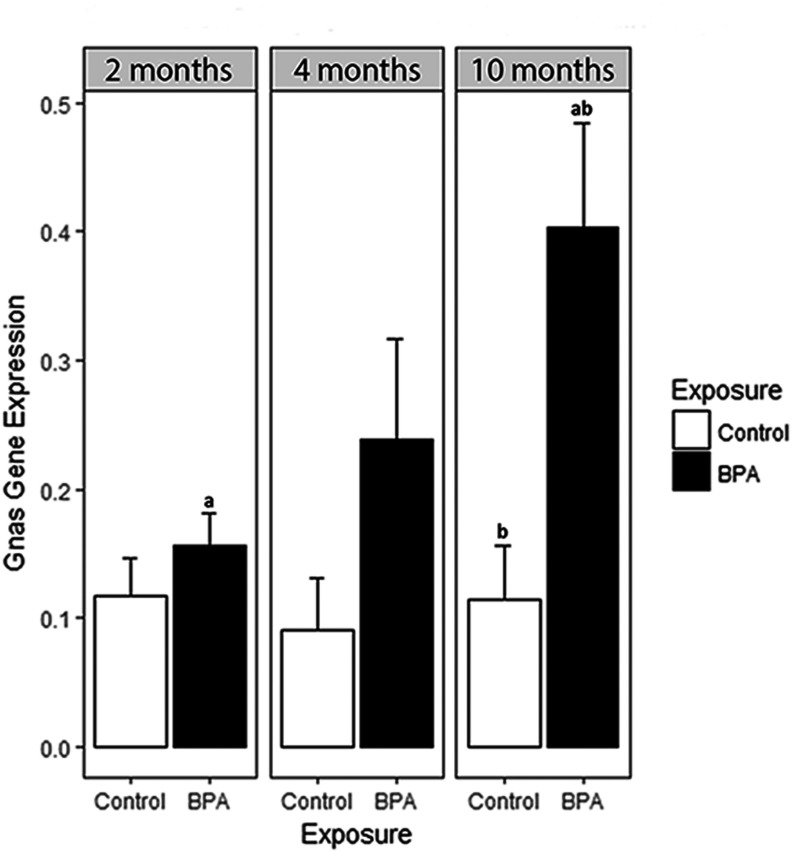
To follow up on the DHMR annotated to the Gnas gene, we performed RT-qPCR on RNA from matched mouse blood samples to examine longitudinal mRNA expression. At the Gnas locus, there was a significant increase in mean mRNA expression from 2 to 10 months of age in mice exposed to BPA () (Figure 4). This increase was not significant in Control samples. Additionally, mean Gnas expression was significantly lower in Control mice than BPA-exposed blood at 10 months of age ().
Figure 4.
Gnas expression by BPA exposure and age. Based on BPA-related DHMR annotated to the Gnas gene, real-time quantitative polymerase chain reaction (RT-qPCR) was used to investigate longitudinal blood Gnas mRNA expression levels. RNA was isolated from the same longitudinal Control ( per age group) and BPA-exposed ( per age group) mouse blood samples used for DNA hydroxymethylation analyses. RT-qPCR was performed on the Gnas gene in triplicate. Three housekeeping genes—, 18S, and Gapdh—were included as internal controls in all RT-qPCR runs. In addition to housekeeping genes, an inter-plate calibrator control of brain cDNA was included for calculation of relative gene expression across multiple plates; all expression values are shown relative to this inter-plate calibrator. Expression levels were calculated following the method. aMean Gnas expression in BPA-exposed mouse blood showed a significant increase between 2 and 10 months of age (); this pattern was not found in Control samples. bMean Gnas expression was significantly lower in Control blood than BPA-exposed blood at 10 months of age ().
Pathway Analysis for DHMRs
The ChIP-enrich pathway analysis was performed on separate lists of hyper- and hypo-methylated DHMRs. In an effort to maximize biological relevance and limit number of comparisons, DHMR datasets were restricted to regions from the transcription start site. After correction for multiple testing, a small number of gene ontology (GO) terms were enriched in hypo- and hyper-hydroxymethylated DHMRs (see Table S4). Of note, enriched pathways showed no overlap between hyper- and hypo-hydroxymethylated DHMRs. In the BPA-related hypo-hydroxymethylated DHMRs, the only two significantly enriched pathways were hippocampus development and spinal cord association neuron differentiation. In the BPA-related hyper-hydroxymethylated DHMRs, the two most significant enriched pathways were l-ascorbic acid binding and oxidoreductase activity. These pathway analysis results indicate that only a few GO terms were enriched in the significant DHMRs, and that enrichment was specific to directionality of BPA-related DHMRs.
Discussion
Epigenome-Wide Differential 5-hmC
We found statistically significant effects of perinatal BPA exposure on DNA hydroxymethylation across the epigenome. Although the exact mechanism driving this relationship is unclear, it is possible that BPA-related DHMRs are a result of BPA-induced oxidative stress (OS) (Gassman 2017). Based on the available literature, free BPA is thought to induce OS via enzymatic formation of BPA phenoxyl radicals (Sakuma et al. 2010), which can then be further processed to other reactive oxygen species (ROS), including superoxides and peroxides (Babu et al. 2013). In addition to potential ROS-induced cytotoxicity (Gassman 2017), the pro-oxidant activity of BPA also has the potential to modify 5-hmC levels across the epigenome. Research has shown that Tet enzyme activity is activated under oxidative conditions (Chia et al. 2011; Coulter et al. 2013; Zhao et al. 2014), indicating that active processing of 5-mC to 5-hmC may be affected by ROS production. Further supporting this idea, recent research has shown that OS-inducing exogenous factors—buthionine sulfoximine and fine particulate matter with an aerodynamic diameter of —can alter DNA hydroxymethylation levels (Delatte et al. 2015; Wei et al. 2017). These prior results support the hypothesis that BPA exposure could be triggering changes in 5-hmC via the induction of OS. This idea should be further explored in future studies examining the effects of endocrine disrupting chemicals on 5-hmC levels across the epigenome.
BPA-Related DHMRs at Imprinted Genes
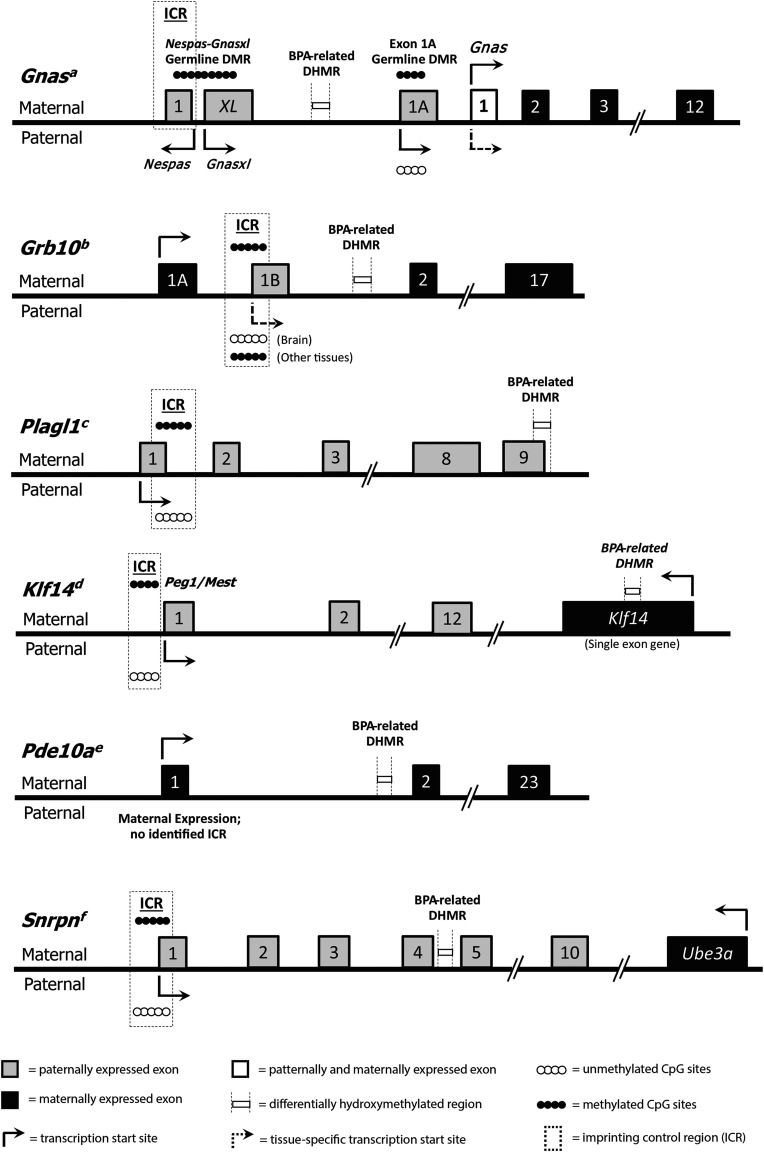
In addition to examining the effects of BPA on broad-scale DNA hydroxymethylation, we also specifically investigated whether BPA exposure altered 5-hmC levels at murine imprinted genes. Of the 151 interrogated imprinted loci, 12 had annotated DHMRs (7.95%) (Table 2). As a broad-scale comparison, of the estimated 24,360 known protein-coding genes in mice (Blake et al. 2017), 1,616 unique gene IDs had annotated DHMRs (6.63%). These results indicate that imprinted genes show slight enrichment for DHMRs compared with all known genes combined. Although none of the 12 imprinted gene BPA-related DHMRs overlapped with known ICRs (Figure 5), expanded visualization of Gnas and Plagl1 showed widespread 5-hmC peaks across imprinted gene regions. The directionality and magnitude of the imprinted gene DHMRs was also gene specific. Of note, 5 of the imprinted gene DHMRs had zero CpG sites (see Table S3), a result that could be driven by several possible scenarios: a) DNA hydroxymethylation at noncanonical methylation contexts (i.e., CHG sites, where , C, or T); b) inexact DHMR definitions due to inherent lack of specificity in pulldown method (HMeDIP-seq); or c) false positives in our data. Supporting the first option, recent work in mice has shown intragenic enrichment of non-CpG methylation at particular domains in clusters of genes related to embryonic development (He et al. 2017). Further fitting with this idea, 8 of the 12 imprinted gene DHMRs had at least 10 CHG sites within their chromosomal ranges (see Table S3). Nevertheless, it remains difficult to distinguish between the three outlined scenarios using the data available in this project. The most significant imprinted gene DHMR was annotated to the Gnas locus, which encodes the G-protein alpha subunit protein, a key component of G-protein coupled signal transduction (Plagge and Kelsey 2006). Gnas is an imprinted gene that has a complex expression pattern, four alternative promoters, a number of isoforms, and may be involved in energy homeostasis (Peters and Williamson 2007; Plagge and Kelsey 2006). Given the complexity of transcriptional control at this locus, the identified intronic Gnas DHMR may represent a long-range regulatory region. This hypothesis is supported by research demonstrating that 5-hmC is enriched at enhancers (Ehrlich and Ehrlich 2014; Stroud et al. 2011; Sun et al. 2013; Wen et al. 2014), regulatory regions that can be quite distant from the gene promoters they activate (Pennacchio et al. 2013; Shlyueva et al. 2014). Based on this idea of distant regulatory regions, we hypothesized that the documented increase in intronic Gnas 5-hmC would show a corresponding BPA-related change in Gnas expression. Using RT-qPCR, we found increased Gnas expression by BPA exposure across all three time points, with the magnitude of this increase reaching significance at 10 months of age (Figure 4). Given the complex regulation of this locus, the age-specific effect of BPA exposure on Gnas expression may be a result of changes in alternative splicing, which is dynamic during the aging process (Li et al. 2017). During aging, Gnas splicing could shift toward specific isoforms that are controlled by 5-hmC at the BPA-related DHMR, leading to a late adulthood effect of developmental BPA exposure. Future functional work should explore this idea, as well as test for potential effects of BPA exposure at the human GNAS locus.
Figure 5.
Organization of the Gnas, Grb10, Plagl1, Klf14, Pde10a, and Snrpn imprinted loci. The thick black midline represents the gene sequence. Maternally expressed exons are indicated by black boxes, and paternally expressed exons are indicated by gray boxes. Exon locations for each gene are relative, but not to scale. Exon 1 of the Gnas gene is both maternally and paternally expressed and is therefore indicated by a white box. Arrows show directionality and start sites for transcription. Parental origin-specific expression patterns are represented by the location of the arrow above or below the midline, and context-specific expression is indicated by dotted arrows. Filled and empty circles represent methylated or unmethylated alleles, respectively. Imprinting control regions (ICRs) are indicated by dotted line boxes, and BPA-related DHMRs are indicated by boxes bordered by dotted vertical lines. None of the identified BPA-related DHMRs are located in ICRs. Figure for Gnas adapted from Tibbit et al. (2015); Grb10 from Hikichi et al. (2003); Plagl1 from Iglesias-Platas et al. (2012) and Smith et al. (2002); Klf14 from Parker-Katiraee et al. (2007); Pde10a from Wang et al. (2011); and Snrpn from Sanli and Feil (2015) and Shemer et al. (1997).
In addition to Gnas, three other imprinted genes—Grb10, Plagl1, and Pde10a—showed significant increases in 5-hmC by BPA exposure. The first of these additional hyper-hydroxymethylated genes, Grb10, encodes the growth factor receptor-bound protein 10 (Grb10). Grb10 is involved in a number of biological processes, including cellular proliferation, apoptosis, and metabolism (Holt and Siddle 2005; Kabir and Kazi 2014; Plasschaert and Bartolomei 2015; Riedel 2004). Grb10 is maternally expressed in most tissues, but it is paternally expressed in the brain; this complex imprinting pattern has been established through tissue-specific alternate promoters (Sanz et al. 2008). The tissue-specific maternal and paternal expression patterns of Grb10 have been linked to fetal growth and adult social behavior in mice, respectively (Garfield et al. 2011). This complex expression patterning is thought to be controlled via epigenetic modifications (Sanz et al. 2008). Like Gnas, the Grb10 DHMR is intronic, meaning any functional effects of differential 5-hmC at this region would have to be through long-distance contacts. The second additional gene, Plagl1, encodes the pleomorphic adenoma of the salivary gland gene like 1 (Plagl1) protein. Plagl1 is a zinc finger transcription factor that regulates other imprinted loci involved in embryonic growth, including H19 and Igf2 (Varrault et al. 2006, 2017). As such, BPA-related alterations in 5-hmC at Plagl1 have the potential to affect an entire network of imprinted genes. Like Gnas and Grb10, Plagl1 has several alternative transcripts in the mouse that are controlled by alternate promoters (Iglesias-Platas et al. 2013). The Plagl1 DHMR overlaps Exon 8 in some Plagl1 isoforms, but not others, suggesting that this region may play a role in alternative splicing for this gene. The third additional hyper-hydroxymethylated imprinted gene, Pde10a, encodes phosphodiesterase 10A (Pde10a). Pde10a is a member of the cyclic nucleotide phosphodiesterases (PDEs), a family of enzymes that regulate intracellular levels of cyclic AMP (cAMP) and cyclic GMP (cGMP), endogenous molecules involved in signal transduction (Bender and Beavo 2006; Soderling et al. 1999). Pde10a shows its highest expression levels in the brain, with minimal expression in peripheral tissues (Bender and Beavo 2006; Soderling et al. 1999). Recent research shows that pharmaceutical inhibition of Pde10a leads to increased energy expenditure, decreased food intake, reduced adiposity, and improved insulin sensitivity in mice with high-fat diet–induced obesity (Nawrocki et al. 2014). As such, altered regulation of the Pde10a gene has the potential to modify murine energy homeostasis. Although we found a Pde10a DHMR in blood tissue, where expression is minimal, our developmental BPA exposure occurred throughout tissue differentiation, so it is possible that the BPA-related DHMR annotated to Pde10a is present in multiple tissues, including the brain.
We also visualized two hypo-hydroxymethylated DHMRs annotated to imprinted genes—Klf14 and Snrpn. The first of these genes, Klf14, encodes the maternally expressed Krüppel-like factor 14 (Klf14), a member of the Cys2/His2 zinc finger transcription factors (Small et al. 2011; Wang et al. 2017). Research shows that KLF14 acts as master regulator of adipose gene expression in humans (Small et al. 2011), and may be involved in metabolic disease risk. In addition, recent results in mice suggest that Klf14 may interact with peroxisome proliferator-activated coactivator (), a transcription coactivator that regulates a number of metabolic pathways, including hepatic gluconeogenesis (Wang et al. 2017). Based on these prior studies, changes in 5-hmC at Klf14 have the potential to not only alter murine energy homeostasis, but also to modify the risk of metabolic disorders. The second imprinted gene to show BPA-related decrease in 5-hmC was Snrpn, a paternally expressed gene that encodes the SmN protein, a key component of the spliceosome in the brain (Shemer et al. 1997). Snrpn has multiple alternate promoters and splice variants, is directly related to neurological function, and is located in the human Prader-Willi and Angelman syndrome (PWS/AS) imprinted domain (Wu et al. 2012). The PWS/AS imprinted domain is highly complex, containing a large number of C/D box small nucleolar RNAs (snoRNAs) and two ICRs: Prader-Willi Syndrome Imprinting Control (PWS-IC) and Angelman Syndrome Imprinting Control (AS-IC) (Wu et al. 2012). Research suggests the snoRNAs play a role in the etiology of the murine PWS phenotype (Relkovic and Isles 2013; Skryabin et al. 2007), which is often characterized by weight gain, decreased activity, and impaired attention (Relkovic and Isles 2013). Here, we identified a BPA-related hypo-hydroxymethylated region in an intron of Snrpn. Although the functional relevance of this region remains unknown, previous work has shown that developmental BPA exposure alters DNA methylation at SNRPN/Snrpn (Faulk et al. 2015; Susiarjo et al. 2013), providing support for the idea that BPA can affect the epigenome at this locus. Additionally, the human SNRPN domain is enriched for 5-hmC across the expressed allele in brain tissue, indicating that 5-hmC may have a functional role in control of SNRPN gene expression (Hernandez Mora et al. 2018). Therefore, it is possible that BPA-related changes in Snrpn 5-hmC could alter gene transcription, thereby modifying risk for PWS, a neurobehavioral disorder.
Remarkably, BPA-related DHMRs at the described imprinted loci persisted throughout adulthood despite BPA exposure ending at PND21. These data indicate that perinatal exposure to BPA can have long-lasting effects on 5-hmC. Given the complex role of 5-hmC in regulating transcription activation (Hahn et al. 2014; Wu et al. 2011), the identified BPA-related DHMRs may reflect programed changes in gene regulation. Supporting this idea, a number of recent studies have shown that 5-hmC is a stable epigenetic mark that has an important role in gene regulation (reviewed by López et al. (2017). Additionally, previous work has shown that there is a unique binding protein (Mbd3) that recognizes 5-hmC (Yildirim et al. 2011), suggesting that 5-hmC has its own epigenetic function. Expanding on this idea, a review of the 5-hmC literature suggested that 5-hmC may be maintained across DNA replication via a complex of the DNMT1, Tet, and UHRF1 proteins (Shen and Zhang 2013), although the validity of this theory has not yet been determined. Building on these ideas, our results show differential 5-hmC at imprinted genes with complex regulatory patterns, indicating that 5-hmC marks at these genes may play a role in the establishment of alternative splicing in response to BPA exposure. Though limited, our RNA expression data support this idea, showing that BPA exposure has long-term functional consequences at the imprinted Gnas locus. Based on these results, a new hypothesis has emerged—that differential 5-hmC at imprinted genes could be an important mechanism driving the developmental origins of adult disease.
In addition to the BPA-specific DHMRs, our data separately show that widespread 5-hmC peaks are established across entire imprinted genes during development, and that these patterns persist throughout life. This was apparent at genes with annotated DHMRs—Gnas and Plagl1—and well-characterized genes without annotated DHMRs—Igf2 and H19. These results match recent research in humans, which showed 5-hmC enrichment at the H19-IGF2 locus in brain and at GNAS A/B in placenta (Hernandez Mora et al. 2018). Combined, the available data indicate that 5-hmC may be involved in imprinting control. As technologies for measuring genome-wide 5-hmC continue to advance, efforts should be made to further examine the regulatory role of 5-hmC in genomic imprinting.
Due to the biased and limited genomic coverage of the ERRBS method, we were not able to compare 5-hmC and 5-mC data at most of our identified DHMRs. At the Klf14 imprinted locus, we showed that 5-mC levels at the identified DHMR did not significantly differ by BPA exposure (see Figure S3). This lack of significance may be a result of inconsistent coverage of ERRBS data across samples at this region; alternatively, 5-hmC at this region may be uniquely sensitive to BPA exposure. Future studies should utilize alternative methods—such as oxidative bisulfite treatment—to coinvestigate 5-hmC and 5-mC at identified imprinted gene DHMRs.
Given that the identified DHMRs had almost no coverage in our ERRBS data, we also looked for differential methylation at known imprinted gene ICRs. Of particular note, we identified three CpG sites within the Gnasxl/Nespas ICR that showed significant BPA-related hypomethylation (see Table S2). In previous studies, hypomethylation at this ICR has been linked to increased expression of the Gnas locus (Coombes et al. 2003; Williamson et al. 2006), a pattern that matches our expression data. These results, combined with the previously identified DHMR, suggest that developmental BPA exposure has a dynamic effect on the epigenetic landscape at the Gnas imprinted gene. Future studies should utilize chromosomal conformation capture techniques to examine whether our identified DHMR interfaces with the ICR to assist in gene regulation.
Pathway Analysis
GO terms showed specific enrichment based on directionality of differential hydroxymethylation. This suggests that BPA exposure is associated with both up- and down-regulation of several biological processes. In the BPA-related hypo-hydroxymethylated DHMRs, the only two significantly enriched pathways were hippocampus development and spinal cord association neuron differentiation. Although it is difficult to interpret these pathways from blood samples, 5-hmC has its highest levels in brain tissue, where it is suspected to play a role in neuron development (Kinde et al. 2015). As such, it is possible that these enriched pathways reflect a predifferentiation effect of developmental BPA exposure on 5-hmC levels in stem cells. Previous work has shown that epigenetic marks at imprinted genes are maintained during postfertilization reprograming (Plasschaert and Bartolomei 2014), suggesting that the epigenetic effects of developmental exposure could be maintained at imprinted loci during cellular differentiation. Building on this idea, our results indicate that 5-hmC establishment at imprinted genes could be involved in regulating neural plasticity. Additional work in predifferentiated cells and brain tissue could help determine whether BPA actually alters neuronal differentiation via 5-hmC.
Limitations
Little is known about the stability of 5-hmC at imprinted loci across blood cell type proportions, but previous studies have shown that epigenetic marks can vary across blood cell types (Houseman et al. 2012; Skinner 2016). As such, it is possible that the documented effects of BPA on genome-wide 5-hmC in blood are simply a reflection of shifts in blood cell type proportions. Counter to this idea, however, previous studies have shown that DNA methylation is often conserved across different cell types at imprinted loci (Skinner 2016; Talens et al. 2010), suggesting that investigating differential methylation from whole blood should still be valid at imprinted genes. Although this evidence supports the validity our BPA-related DHMRs at imprinted loci, the dynamics of 5-hmC at these imprinted genes across blood cell types remains to be elucidated. To further explore the idea that altered 5-hmC in blood impacts imprinted gene regulation, future studies should also investigate allele-specific expression in individual cell types using new single-cell RNA-seq methods (Santoni et al. 2017). Despite lingering blood cell type questions, the use of matched blood samples allowed for direct measurement of 5-hmC from the same mice over time, reducing the potential confounding of interindividual variability. Additionally, matched blood samples provide greater translatability to human epigenetics studies, which typically rely on peripheral blood samples.
Due to low RNA yields from the longitudinal blood samples, it was not possible to examine gene expression at all identified imprinted loci with annotated DHMRs. RNA yields were diminished due to the small amount of blood collected during in vivo tail vein collection at 2 and 4 months of age. Despite the low amounts of blood RNA available, the use of longitudinal samples allowed for direct measurement of expression at the Gnas imprinted locus from the same longitudinal samples used for HMeDIP-seq data generation. As such, BPA-related changes in Gnas expression directly coincide with BPA-related alterations in DNA hydroxymethylation at this locus.
DNA hydroxymethylation has only recently become recognized as an important consideration in the field of genomic imprinting, meaning its role in imprinting control is poorly defined. As a result, the exact functional effects of the BPA-related changes shown in this paper remain undetermined. So far, the available research suggests that 5-hmC can have very different regulatory effects depending upon its genomic context, so the relationship between BPA-related DHMRs and gene expression could vary in a gene-specific manner. Similarly, the phenotypic effects of BPA-related DHMRs remain unclear without additional measures of murine biology throughout the life course. Despite these limitations in interpretation, we identified a large number of BPA-related DHMRs, indicating that DNA hydroxymethylation is sensitive to environmental factors during development.
Conclusions
We measured 5-hmC in matched blood samples collected from isogenic mice at 2, 4, and 10 months of age, and then examined the effects of BPA on the longitudinal DNA hydroxymethylation. Across the epigenome, we identified a number of exposure-related DHMRs, suggesting that perinatal BPA exposure can alter this DNA modification throughout life. At 12 imprinted loci, including the Gnas locus, developmental BPA exposure significantly altered 5-hmC levels in blood across all three measured ages. Echoing this result, we showed that BPA exposure modified Gnas expression throughout murine adulthood. These results suggest that BPA-related increases in Gnas hydroxymethylation may have long-lasting effects on gene expression at this complex imprinted locus, possibly through shifts in alternative splicing. In addition to BPA-related DHMRs, we also found stable patterns of 5-hmC along a number of imprinted loci, including Igf2 and H19. Combined, our data indicate that 5-hmC may play an important regulatory role in imprinting control, and that establishment of DNA hydroxymethylation at imprinted loci may be sensitive to developmental BPA exposure. Future studies should examine the contribution of DNA hydroxymethylation to the methylome at imprinted loci, as well as the impact of additional environmental exposures on this recently rediscovered epigenetic mark.
Supplemental Material
Acknowledgments
We thank C. Lalancette at the University of Michigan Epigenomics Core for HMeDIP-seq DNA library prep. We also thank the University of Michigan Bioinformatics Core for sequencing data generation and quality control.
This work was supported by the University of Michigan (UM) National Institute of Environmental Health Sciences/U.S. Environmental Protection Agency (NIEHS/EPA) Children’s Environmental Health and Disease Prevention Center (grant P01 ES022844/RD83543601), and the Michigan Lifestage Environmental Exposures and Disease (M-LEEaD) NIEHS Core Center (grant P30 ES017885) as well as by the UM NIEHS Institutional Training Grants T32 ES007062 (J.J.K., E.H.M.) and F31 ES025101 (E.H.M.), and the University of Michigan School of Public Health Regents’ Fellowship (J.J.K.).
References
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, et al. 2012. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13(10):R87, PMID: 23034086, 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson OS, Nahar MS, Faulk C, Jones TR, Liao C, Kannan K, et al. 2012. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ Mol Mutagen 53(5):334–342, PMID: 22467340, 10.1002/em.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud P. 2010. Genomic imprinting in germ cells: imprints are under control. Reproduction 140(3):411–423, PMID: 20501788, 10.1530/REP-10-0173. [DOI] [PubMed] [Google Scholar]
- Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. 2014. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods 11(11):1138–1140, PMID: 25262207, 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu S, Uppu S, Claville MO, Uppu RM. 2013. Prooxidant actions of bisphenol A (BPA) phenoxyl radicals: implications to BPA-related oxidative stress and toxicity. Toxicol Mech Methods 23(4):273–280, PMID: 23193990, 10.3109/15376516.2012.753969. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Ferguson-Smith AC. 2011. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3(7):a002592, PMID: 21576252, 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson P, Barker D, Clutton-Brock T, Deb D, D’Udine B, Foley RA, et al. 2004. Developmental plasticity and human health. Nature 430(6998):419–421, PMID: 15269759, 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- Bender AT, Beavo JA. 2006. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev 58(3):488–520, PMID: 16968949, 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57(1):289–300. [Google Scholar]
- Benjamini Y, Hochberg Y. 1997. Multiple hypotheses testing with weights. Scand J Stat 24(3):407–418, 10.1111/1467-9469.00072. [DOI] [Google Scholar]
- Blake JA, Eppig JT, Kadin JA, Richardson JE, Smith CL, Bult CJ. 2017. Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res 45(D1):D723–D729, PMID: 27899570, 10.1093/nar/gkw1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. 2008. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003–2004. Environ Health Perspect 116(1):39–44, PMID: 18197297, 10.1289/ehp.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcante RG, Patil S, Park Y, Rozek LS, Sartor MA. 2017. Integrating DNA methylation and hydroxymethylation data with the mint pipeline. Cancer Res 77(21):e27–e30, PMID: 29092933, 10.1158/0008-5472.CAN-17-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcante RG, Sartor MA. 2017. annotatr: genomic regions in context. Bioinformatics 33(15):2381–2383, PMID: 28369316, 10.1093/bioinformatics/btx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia N, Wang L, Lu X, Senut M-C, Brenner C, Ruden DM. 2011. Hypothesis: environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics 6(7):853–856, PMID: 21617369. [DOI] [PubMed] [Google Scholar]
- Coombes C, Arnaud P, Gordon E, Dean W, Coar EA, Williamson CM, et al. 2003. Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus. Mol Cell Biol 23(16):5475–5488, PMID: 12897124, 10.1128/MCB.23.16.5475-5488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton AM, Price EM, Jones MJ, Balaton BP, Kobor MS, Brown CJ. 2015. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum Mol Genet 24(6):1528–1539, PMID: 25381334, 10.1093/hmg/ddu564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter JB, O’Driscoll CM, Bressler JP. 2013. Hydroquinone increases 5-hydroxymethylcytosine formation through Ten Eleven Translocation 1 (TET1) 5-methylcytosine dioxygenase. J Biol Chem 288(40):28792–28800, PMID: 23940045, 10.1074/jbc.M113.491365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Hampton DD, Jirtle RL. 2009. Imprinting evolution and human health. Mamm Genome 20(9–10):563–572, PMID: 19830403, 10.1007/s00335-009-9229-y. [DOI] [PubMed] [Google Scholar]
- Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. 2013. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One 8(12):e85024, PMID: 24376861, 10.1371/journal.pone.0085024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B, Jeschke J, Defrance M, Bachman M, Creppe C, Calonne E, et al. 2015. Genome-wide hydroxymethylcytosine pattern changes in response to oxidative stress. Sci Rep 5:12714, PMID: 26239807, 10.1038/srep12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Weinhouse C, Jones TR, Rozek LS, Jirtle RL. 2010. Variable histone modifications at the Avy metastable epiallele. Epigenetics 5(7):637–644, PMID: 20671424, 10.4161/epi.5.7.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Ehrlich KC. 2014. DNA cytosine methylation and hydroxymethylation at the borders. Epigenomics 6(6):563–566, PMID: 25531248, 10.2217/epi.14.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Kim JH, Jones TR, McEachin RC, Nahar MS, Dolinoy DC, et al. 2015. Bisphenol A-associated alterations in genome-wide DNA methylation and gene expression patterns reveal sequence-dependent and non-monotonic effects in human fetal liver. Environ Epigenet 1(1):dvv006, PMID: 27358748, 10.1093/eep/dvv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Conneely KN, Wu H. 2014. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res 42(8):e69, PMID: 24561809, 10.1093/nar/gku154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, Lesage J, et al. 2010. Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One 5(12):e14398, PMID: 21200436, 10.1371/journal.pone.0014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfield AS, Cowley M, Smith FM, Moorwood K, Stewart-Cox JE, Gilroy K, et al. 2011. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature 469(7331):534–538, PMID: 21270893, 10.1038/nature09651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Bakelman FE, Sheridan CK, Kacmarczyk TJ, Ishii J, Betel D, Alonso A, et al. 2015. Enhanced reduced representation bisulfite sequencing for assessment of DNA methylation at base pair resolution. J Vis Exp 96:e52246, PMID: 25742437, 10.3791/52246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassman NR. 2017. Induction of oxidative stress by bisphenol A and its pleiotropic effects. Environ Mol Mutagen 58(2):60–71, PMID: 28181297, 10.1002/em.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, et al. 2010. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5(12):e15367, PMID: 21203455, 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Szabó PE, Pfeifer GP. 2014. 5-Hydroxymethylcytosine: a stable or transient DNA modification? Genomics 104(5):314–323, PMID: 25181633, 10.1016/j.ygeno.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne F, Ivanek R. 2016. Visualizing genomic data using Gviz and Bioconductor. Methods Mol Biol 1418:335–351, PMID: 27008022, 10.1007/978-1-4939-3578-9_16. [DOI] [PubMed] [Google Scholar]
- Haycock PC, Ramsay M. 2009. Exposure of mouse embryos to ethanol during preimplantation development: effect on DNA methylation in the H19 imprinting control region. Biol Reprod 81(4):618–627, 10.1095/biolreprod.108.074682. [DOI] [PubMed] [Google Scholar]
- He Y, Hariharan M, Gorkin DU, Dickel DE, Luo C, Castanon RG, et al. 2017. Spatiotemporal DNA methylome dynamics of the developing mammalian fetus. bioRxiv 166744, 10.1101/166744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. 2008. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 105(44):17046–17049, PMID: 18955703, 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindel JJ, Balbus J, Birnbaum L, Brune-Drisse MN, Grandjean P, Gray K, et al. 2015. Developmental origins of health and disease: integrating environmental influences. Endocrinology 156(10):3416–3421, PMID: 26241070, 10.1210/EN.2015-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez Mora JR, Sanchez-Delgado M, Petazzi P, Moran S, Esteller M, Iglesias-Platas I, et al. 2018. Profiling of oxBS-450K 5-hydroxymethylcytosine in human placenta and brain reveals enrichment at imprinted loci. Epigenetics 13(2):182–191, PMID: 28678681, 10.1080/15592294.2017.1344803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikichi T, Kohda T, Kaneko-Ishino T, Ishino F. 2003. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res 31(5):1398–1406, PMID: 12595547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Siddle K. 2005. Grb10 and Grb14: enigmatic regulators of insulin action—and more? Biochem J 388(pt 2):393–406, PMID: 15901248, 10.1042/BJ20050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86, PMID: 22568884, 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. 2010. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5(1):e8888, PMID: 20126651, 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Platas I, Court F, Camprubi C, Sparago A, Guillaumet-Adkins A, Martin-Trujillo A, et al. 2013. Imprinting at the PLAGL1 domain is contained within a 70-kb CTCF/cohesin-mediated non-allelic chromatin loop. Nucleic Acids Res 41(4):2171–2179, PMID: 23295672, 10.1093/nar/gks1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Platas I, Martin-Trujillo A, Cirillo D, Court F, Guillaumet-Adkins A, Camprubi C, et al. 2012. Characterization of novel paternal ncRNAs at the Plagl1 locus, including Hymai, predicted to interact with regulators of active chromatin. PLoS One 7(6):e38907, PMID: 22723905, 10.1371/journal.pone.0038907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. 2007. Environmental epigenomics and disease susceptibility. Nat Rev Genet 8(4):253–262, PMID: 17363974, 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir NN, Kazi JU. 2014. Grb10 is a dual regulator of receptor tyrosine kinase signaling. Mol Biol Rep 41(4):1985–1992, PMID: 24420853, 10.1007/s11033-014-3046-4. [DOI] [PubMed] [Google Scholar]
- Kelsey G, Feil R. 2013. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci 368(1609):20110336, PMID: 23166397, 10.1098/rstb.2011.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, He H, Kim H. 2017. Inversion of the imprinting control region of the Peg3 domain. PLoS One 12(7):e0181591, PMID: 28719641, 10.1371/journal.pone.0181591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Sartor MA, Rozek LS, Faulk C, Anderson OS, Jones TR, et al. 2014. Perinatal bisphenol A exposure promotes dose-dependent alterations of the mouse methylome. BMC Genomics 15:30, PMID: 24433282, 10.1186/1471-2164-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde B, Gabel HW, Gilbert CS, Griffith EC, Greenberg ME. 2015. Reading the unique DNA methylation landscape of the brain: non-CpG methylation, hydroxymethylation, and MeCP2. Proc Natl Acad Sci U S A 112(22):6800–6806, PMID: 25739960, 10.1073/pnas.1411269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F. 2017. Taking appropriate QC measures for RRBS-type or other -Seq applications with Trim Galore! Babraham Bioinformatics. https://github.com/FelixKrueger/TrimGalore/blob/master/Docs/Trim_Galore_User_Guide.md [accessed 25 June 2018].
- Krüger T, Long M, Bonefeld-Jørgensen EC. 2008. Plastic components affect the activation of the aryl hydrocarbon and the androgen receptor. Toxicology 246(2–3):112–123, PMID: 18294747, 10.1016/j.tox.2007.12.028. [DOI] [PubMed] [Google Scholar]
- La Salle S, Mertineit C, Taketo T, Moens PB, Bestor TH, Trasler JM. 2004. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev Biol 268(2):403–415, PMID: 15063176, 10.1016/j.ydbio.2003.12.031. [DOI] [PubMed] [Google Scholar]
- Li H, Wang Z, Ma T, Wei G, Ni T. 2017. Alternative splicing in aging and age-related diseases. Transl Med Aging 1:32–40, 10.1016/j.tma.2017.09.005. [DOI] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408, PMID: 11846609, 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- López V, Fernández AF, Fraga MF. 2017. The role of 5-hydroxymethylcytosine in development, aging and age-related diseases. Ageing Res Rev 37:28–38, PMID: 28499883, 10.1016/j.arr.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Lun ATL, Smyth GK. 2016. csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res 44(5):e45, PMID: 26578583, 10.1093/nar/gkv1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. 2013. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One 8(1):e55387, PMID: 23359474, 10.1371/journal.pone.0055387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedeva YA, Khamis AM, Kulakovskiy IV, Ba-Alawi W, Bhuyan MS, Kawaji H, et al. 2014. Effects of cytosine methylation on transcription factor binding sites. BMC Genomics 15:119, PMID: 24669864, 10.1186/1471-2164-15-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Knowles BB, Solter D. 2014. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 28(8):812–828, PMID: 24736841, 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CA, Liu XS. 2014. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat Rev Genet 15(11):709–721, PMID: 25223782, 10.1038/nrg3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. 2011. Guide for the Care and Use of Laboratory Animals. 8th Edition. Washington, DC:National Academies Press. [Google Scholar]
- Nawrocki AR, Rodriguez CG, Toolan DM, Price O, Henry M, Forrest G, et al. 2014. Genetic deletion and pharmacological inhibition of phosphodiesterase 10A protects mice from diet-induced obesity and insulin resistance. Diabetes 63(1):300–311, PMID: 24101672, 10.2337/db13-0247. [DOI] [PubMed] [Google Scholar]
- Nye MD, Hoyo C, Murphy SK. 2015. In vitro lead exposure changes DNA methylation and expression of IGF2 and PEG1/MEST. Toxicol In Vitro 29(3):544–550, PMID: 25596546, 10.1016/j.tiv.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Wu H. 2016. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 32(10):1446–1453, PMID: 26819470, 10.1093/bioinformatics/btw026. [DOI] [PubMed] [Google Scholar]
- Parker-Katiraee L, Carson AR, Yamada T, Arnaud P, Feil R, Abu-Amero SN, et al. 2007. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet 3(5):e65, PMID: 17480121, 10.1371/journal.pgen.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. 2013. Enhancers: five essential questions. Nat Rev Genet 14(4):288–295, PMID: 23503198, 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Williamson CM. 2007. Control of imprinting at the Gnas cluster. Epigenetics 2(4):207–213, PMID: 18094621, 10.4161/epi.2.4.5380. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Fernandes C, Viana J, Paya-Cano JL, Liu L, Smith RG, et al. 2012. DNA methylation at the Igf2/H19 imprinting control region is associated with cerebellum mass in outbred mice. Mol Brain 5:42, PMID: 23216893, 10.1186/1756-6606-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagge A, Kelsey G. 2006. Imprinting the Gnas locus. Cytogenet Genome Res 113(1–4):178–187, PMID: 16575178, 10.1159/000090830. [DOI] [PubMed] [Google Scholar]
- Plasschaert RN, Bartolomei MS. 2014. Genomic imprinting in development, growth, behavior and stem cells. Development 141(9):1805–1813, PMID: 24757003, 10.1242/dev.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert RN, Bartolomei MS. 2015. Tissue-specific regulation and function of Grb10 during growth and neuronal commitment. Proc Natl Acad Sci U S A 112(22):6841–6847, PMID: 25368187, 10.1073/pnas.1411254111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293(5532):1089–1093, PMID: 11498579, 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Relkovic D, Isles AR. 2013. Behavioural and cognitive profiles of mouse models for Prader–Willi syndrome. Brain Res Bull 92:41–48, PMID: 21971015, 10.1016/j.brainresbull.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Riedel H. 2004. Grb10 exceeding the boundaries of a common signaling adapter. Front Biosci 9:603–618, PMID: 14766395. [DOI] [PubMed] [Google Scholar]
- Sakuma S, Nakanishi M, Morinaga K, Fujitake M, Wada S, Fujimoto Y. 2010. Bisphenol A 3,4-quinone induces the conversion of xanthine dehydrogenase into oxidase in vitro. Food Chem Toxicol 48(8–9):2217–2222, PMID: 20594952, 10.1016/j.fct.2010.05.051. [DOI] [PubMed] [Google Scholar]
- Sanli I, Feil R. 2015. Chromatin mechanisms in the developmental control of imprinted gene expression. Int J Biochem Cell Biol 67:139–147, PMID: 25908531, 10.1016/j.biocel.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Santoni FA, Stamoulis G, Garieri M, Falconnet E, Ribaux P, Borel C, et al. 2017. Detection of imprinted genes by single-cell allele-specific gene expression. Am J Hum Genet 100(3):444–453, PMID: 28190458, 10.1016/j.ajhg.2017.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz LA, Chamberlain S, Sabourin J-C, Henckel A, Magnuson T, Hugnot J-P, et al. 2008. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J 27(19):2523–2532, PMID: 18650936, 10.1038/emboj.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer R, Birger Y, Riggs AD, Razin A. 1997. Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci U S A 94(19):10267–10272, PMID: 9294199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Song CX, He C, Zhang Y. 2014. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem 83:585–614, PMID: 24905787, 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Zhang Y. 2013. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Curr Opin Cell Biol 25(3):289–296, PMID: 23498661, 10.1016/j.ceb.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15(4):272–286, PMID: 24614317, 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Singh S, Li SS. 2012. Epigenetic effects of environmental chemicals bisphenol A and phthalates. Int J Mol Sci 13(8):10143–10153, PMID: 22949852, 10.3390/ijms130810143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. 2016. Differential DNA methylation analysis optimally requires purified cell populations. Fertil Steril 106(3):551, PMID: 27349925, 10.1016/j.fertnstert.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin BV, Gubar LV, Seeger B, Pfeiffer J, Handel S, Robeck T, et al. 2007. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet 3(12):e235, PMID: 18166085, 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvortsova K, Zotenko E, Luu P-L, Gould CM, Nair SS, Clark SJ, et al. 2017. Comprehensive evaluation of genome-wide 5-hydroxymethylcytosine profiling approaches in human DNA. Epigenetics Chromatin 10:16, PMID: 28428825, 10.1186/s13072-017-0123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, Kong A, et al. 2011. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet 43(6):561–564, PMID: 21572415, 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Kelsey G. 2012. De novo DNA methylation: a germ cell perspective. Trends Genet 28(1):33–42, PMID: 22019337, 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Smith RJ, Arnaud P, Konfortova G, Dean WL, Beechey CV, Kelsey G. 2002. The mouse Zac1 locus: basis for imprinting and comparison with human ZAC. Gene 292(1–2):101–112, PMID: 12119104, 10.1016/S0378-1119(02)00666-2. [DOI] [PubMed] [Google Scholar]
- Soderling SH, Bayuga SJ, Beavo JA. 1999. Isolation and characterization of a dual-substrate phosphodiesterase gene family: PDE10A. Proc Natl Acad Sci U S A 96(12):7071–7076, PMID: 10359840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Feng S, Kinney SM, Pradhan S, Jacobsen SE. 2011. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol 12(6):R54, PMID: 21689397, 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui Y, Ai N, Park SH, Rios-Pilier J, Perkins JT, Welsh WJ, et al. 2012. Bisphenol A and its analogues activate human pregnane X receptor. Environ Health Perspect 120(3):399–405, PMID: 22214767, 10.1289/ehp.1104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Terragni J, Borgaro JG, Liu Y, Yu L, Guan S, et al. 2013. High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell Rep 3(2):567–576, PMID: 23352666, 10.1016/j.celrep.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. 2013. Bisphenol A exposure disrupts genomic imprinting in the mouse. PLoS Genet 9(4):e1003401, PMID: 23593014, 10.1371/journal.pgen.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talens RP, Boomsma DI, Tobi EW, Kremer D, Jukema JW, Willemsen G, et al. 2010. Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. FASEB J 24(9):3135–3144, PMID: 20385621, 10.1096/fj.09-150490. [DOI] [PubMed] [Google Scholar]
- Tan L, Xiong L, Xu W, Wu F, Huang N, Xu Y, et al. 2013. Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method. Nucleic Acids Res 41(7):e84, PMID: 23408859, 10.1093/nar/gkt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbit CJ, Williamson CM, Mehta S, Ball ST, Chotalia M, Nottingham WT, et al. 2015. Antisense activity across the Nesp promoter is required for Nespas-mediated silencing in the imprinted Gnas cluster. Noncoding RNA 1(3):246–265, PMID: 29861426, 10.3390/ncrna1030246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Dantec C, Le Digarcher A, Chotard L, Bilanges B, Parrinello H, et al. 2017. Identification of Plagl1/Zac1 binding sites and target genes establishes its role in the regulation of extracellular matrix genes and the imprinted gene network. Nucleic Acids Res 45(18):10466–10480, PMID: 28985358, 10.1093/nar/gkx672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, et al. 2006. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell 11(5):711–722, PMID: 17084362, 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Wang L, Tong X, Gu F, Zhang L, Chen W, Cheng X, et al. 2017. The KLF14 transcription factor regulates hepatic gluconeogenesis in mice. J Biol Chem 292(52):21631–21642, PMID: 29123026, 10.1074/jbc.RA117.000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Pehrsson EC, Purushotham D, Li D, Zhuo X, Zhang B, et al. 2018. The NIEHS TaRGET II Consortium and environmental epigenomics. Nat Biotechnol 36(3):225–227, PMID: 29509741, 10.1038/nbt.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Soloway PD, Clark AG. 2011. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics 189(1):109–122, PMID: 21705755, 10.1534/genetics.111.130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. 2003. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23(15):5293–5300, PMID: 12861015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CS, Bulayeva NN, Wozniak AL, Alyea RA. 2007. Xenoestrogens are potent activators of nongenomic estrogenic responses. Steroids 72(2):124–134, PMID: 17174995, 10.1016/j.steroids.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Feng Y, Liang F, Cheng W, Wu X, Zhou R, et al. 2017. Role of oxidative stress and DNA hydroxymethylation in the neurotoxicity of fine particulate matter. Toxicology 380:94–103, PMID: 28153600, 10.1016/j.tox.2017.01.017. [DOI] [PubMed] [Google Scholar]
- Weinhouse C, Anderson OS, Bergin IL, Vandenbergh DJ, Gyekis JP, Dingman MA, et al. 2014. Dose-dependent incidence of hepatic tumors in adult mice following perinatal exposure to bisphenol A. Environ Health Perspect 122:485–491, PMID: 24487385, 10.1289/ehp.1307449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RP, Lee C, Imbriano PM, Patil S, Weymouth TE, Smith RA, et al. 2014. ChIP-Enrich: gene set enrichment testing for ChIP-seq data. Nucleic Acids Res 42(13):e105, PMID: 24878920, 10.1093/nar/gku463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, et al. 2014. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol 15(3):R49, PMID: 24594098, 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson CM, Turner MD, Ball ST, Nottingham WT, Glenister P, Fray M, et al. 2006. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet 38(3):350–355, PMID: 16462745, 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- Williamson CM, Blake A, Thomas S, Beechey CV, Hancock J, Cattanach BM, et al. 2013. World Wide Web Site - Mouse Imprinting Data and References. https://www.mousebook.org/imprinting-gene-list [accessed 25 June 2018].
- Wolstenholme JT, Rissman EF, Connelly JJ. 2011. The role of bisphenol A in shaping the brain, epigenome and behavior. Horm Behav 59(3):296–305, PMID: 21029734, 10.1016/j.yhbeh.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. 2011. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25(7):679–684, PMID: 21460036, 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M-Y, Jiang M, Zhai X, Beaudet AL, Wu R-C. 2012. An unexpected function of the Prader-Willi syndrome imprinting center in maternal imprinting in mice. PLoS One 7(4):e34348, PMID: 22496793, 10.1371/journal.pone.0034348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wang C, Wu Z. 2013. A new shrinkage estimator for dispersion improves differential expression detection in RNA-seq data. Biostatistics 14(2):232–243, PMID: 23001152, 10.1093/biostatistics/kxs033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Xu T, Feng H, Chen L, Li B, Yao B, et al. 2015. Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res 43(21):e141, PMID: 26184873, 10.1093/nar/gkv715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung J-H, Chen PB, Dong X, Ee L-S, et al. 2011. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147(7):1498–1510, PMID: 22196727, 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Zhang LJ, Feng YN, Chen B, Feng YM, Liang GJ, et al. 2012. Bisphenol A exposure modifies DNA methylation of imprint genes in mouse fetal germ cells. Mol Biol Rep 39(9):8621–8628, PMID: 22699882, 10.1007/s11033-012-1716-7. [DOI] [PubMed] [Google Scholar]
- Zhao B, Yang Y, Wang X, Chong Z, Yin R, Song S-H, et al. 2014. Redox-active quinones induces genome-wide DNA methylation changes by an iron-mediated and Tet-dependent mechanism. Nucleic Acids Res 42(3):1593–1605, PMID: 24214992, 10.1093/nar/gkt1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.