Abstract
Many cancer type-specific anticancer agents have been developed and significant advances have been made toward precision medicine in cancer treatment. However, traditional or non-specific anticancer drugs are still important for the treatment of many cancer patients whose cancers either do not respond to or have developed resistance to cancer-specific anticancer agents. DNA topoisomerases, especially type IIA topoisomerases, are proven therapeutic targets of anticancer and antibacterial drugs. Clinically successful topoisomerase-targeting anticancer drugs act through topoisomerase poisoning, which leads to replication fork arrest and double-strand break formation. Unfortunately, this unique mode of action is associated with the development of secondary cancers and cardiotoxicity. Structures of topoisomerase-drug-DNA ternary complexes have revealed the exact binding sites and mechanisms of topoisomerase poisons. Recent advances in the field have suggested a possibility of designing isoform-specific human topoisomerase II poisons, which may be developed as safer anticancer drugs. It may also be possible to design catalytic inhibitors of topoisomerases by targeting a certain inactive conformations of these enzymes. Furthermore, identification of various new bacterial topoisomerase inhibitors and regulatory proteins may inspire the discovery of novel human topoisomerase inhibitors. Thus, topoisomerases remain as important therapeutic targets of anticancer agents.
1. Introduction
DNA topoisomerases are responsible for DNA unlinking and these ubiquitous enzymes play critical roles in many biological processes involving DNA [1–7]. There are two types of DNA topoisomerases, type I and type II. Type I topoisomerases break one DNA strand of duplex DNA to allow either the passage of the other DNA strand through the break or the rotation of downstream DNA duplex about the break, and then reseal the broken strand. As a result, they alter the linking number in steps of one [1–7]. There are three subtypes of type I topoisomerases: type IA, type IB, and type IC. Type IA topoisomerases require a nick or a single-stranded region to bind to DNA [8,9]. They cleave one strand of duplex DNA, covalently attach the active-site tyrosine to a 5′-phosphoryl group, and utilize the ‘strand passage’ mechanism to alter DNA topology [10,11]. In contrast, types IB and IC topoisomerases cleave one strand of duplex DNA, covalently attach its active-site tyrosine to a 3′-phosphoryl group, and utilize the ‘swivel’ mechanism to relax DNA supercoils [12]. Bacterial topoisomerase I (Top 1) and topoisomerase III (Top 3) are type IA topoisomerases. Eukaryotic Top 1 belongs to the type IB subtype, and topoisomerase V found in the archaeal genus Methanopyrus is, thus far, the only member of the type IC subtype [13]. It is noteworthy that type IB topoisomerases are found in some bacteria [14]. Type II topoisomerases alter the linking number in steps of two by breaking both DNA strands of duplex DNA [referred to as the ‘Gate (G)-segment’], passing another duplex DNA [referred to as the ‘Transfer (T)-segment’] through the break or the ‘DNA gate’, and then resealing the broken DNA strands [15–18]. Type II topoisomerases go through a series of large conformational changes when catalyzing the reaction by the ‘strand passage’ mechanism. When type II topoisomerases cleave two DNA strands, they form phosphotyrosyl bonds between the two active-site tyrosines and a pair of 5′-phosphates to ensure the integrity of DNA can be restored. There are two subtypes of type II topoisomerases, type IIA and type IIB. Topoisomerase VI belongs to the type IIB subtype [19], while all other type II topoisomerases belong to the type IIA subtype. Each subtype of topoisomerase is structurally and functionally conserved and forms a protein family [1–7].
Upon its discovery [20], DNA gyrase was identified as the cellular target of the coumarin and the quinolone antibacterial drugs [21–23]. Since then, DNA topoisomerases, especially type IIA topoisomerases, are recognized as therapeutic targets of many anticancer and antibacterial drugs [6, 24–31]. Several antibacterial agents, including fluoroquinolones, target the bacterial type IIA topoisomerases, DNA gyrase and topoisomerase IV (Top 4). Both human Top 1 (hTop 1) and the two isoforms of human topoisomerase II (hTop 2) are the cellular targets of clinically important anticancer drugs. A large number of small molecules have been identified as inhibitors of hTop 1 or hTop 2. Many of them have been tested in clinical trials, but only a few have enjoyed clinical success. Manipulation of enzyme activities to achieve therapeutic effects has been accomplished by the following mechanisms: (1) suppressing enzyme activity by catalytic inhibitors; (2) activating enzyme activity toward the bona fide or a surrogate substrate; (3) turning the enzyme into a poisonous agent that is toxic to cells. Interestingly, many clinically successful inhibitors of hTop 1 and hTop 2, as well as bacterial type IIA topoisomerase inhibitors, utilize mechanism 3, which is often referred to as ‘topoisomerase poisoning’ [6, 24–31]. These topoisomerase poisons convert a topoisomerase into a cellular poison by trapping a covalent topoisomerase-DNA catalytic intermediate as a topoisomerase-drug-DNA ternary complex. Topoisomerase poisoning is often caused by the inhibition of the religation reaction [32], which is also supported by the structural studies of topoisomerase-drug-DNA ternary complexes [33,34]. However, some topoisomerase poisons, including spontaneous DNA damage and fluoroquinolones, have reported to stimulate the strand breakage reaction [35–38]. Thus, mechanism 2 appears to be used by some topoisomerase inhibitors to poison their target. Ternary complex formation triggers cytotoxic events, which include the inhibition of DNA replication, the generation of double-strand breaks (DSBs), and subsequent cell death [6, 24–31]. The catalytic inhibitors of either hTop 1 or hTop 2 that utilize mechanism 1 have not seen much success in clinical applications. One possible explanation is that elevated levels of topoisomerases in cancer cells would reduce the efficacy of catalytic inhibitors. In contrast, elevated levels of topoisomerases would increase the potency of topoisomerase poisons [39]. We recently discovered novel quinolones that inhibit both hTop 1 and hTop 2 without poisoning them [40]. These compounds exhibited promising in vitro and in vivo anticancer activities. Thus, catalytic inhibitors that are capable of inhibiting multiple human topoisomerases may be developed as potent anticancer drugs [27,30]. The development of enzyme activity modulators can benefit from a detailed understanding of the catalytic mechanisms of the target enzymes. For topoisomerases, structural biology has proven particularly helpful in elucidating not only the enzymes’ catalytic mechanisms [1,3,6,41], but also validating the actions of clinically important drugs that specifically target these essential enzymes [33,34,42–44].
New bacterial topoisomerase inhibitors (NBTIs) [45,46], as well as toxins and regulatory proteins that target DNA gyrase [26], are also described in this review because their novel binding sites and modes of action to modulate the activities of bacterial type IIA topoisomerases may inspire the discovery of new anticancer drugs that act on hTop 2. In addition to the α and β form of hTop 2 (hTop 2α and hTop 2β), type IIA topoisomerases, and two type IB topoisomerases found in the nucleus (hTop 1) and mitochondria (mTop 1), human cells also have two isoforms of Top 3 (hTop 3α and hTop 3β), type IA topoisomerases [1–7]. However, no inhibitor of human Top 3 has been reported. Thus, we excluded type IA topoisomerases from this review. Our review focuses on recent structural studies of topoisomerase-drug-DNA ternary complexes that have advanced our understanding of the molecular basis of topoisomerase poisoning. Thus, we would like to refer readers to other outstanding reviews on topoisomerases [such as 1–7,41] and topoisomerase inhibitors [such as 24–30,44,45] for comprehensive overviews of the many essential studies that have broadened our understanding of the cellular functions and catalytic mechanisms of topoisomerases, as well as topoisomerase-targeting anticancer drugs in discovery and development stages. We apologize to the many colleagues whose work is only briefly discussed or excluded due to space limitations.
2. Cellular functions of topoisomerases
(1). Type IB topoisomerases
The swivel function of eukaryotic Top 1 is essential for removing DNA superhelical tension that arises during both DNA replication and transcription [47–52]. Interestingly, Top 1, but not its catalytic activity, is required for activation of transcription [49,50,53]; however, the topoisomerase activity of Top 2β is required for transcription [54,55]. In human cells, activities of hTop 1 and RNA polymerase II are coupled [56]. Studies on the effect of camptothecin, a Top 1 poison, have demonstrated that efficient expression of long genes requires both Top 1 and Top 2β [57–59].
mTop I is the only topoisomerase exclusively targeted to mitochondria [60]. However, a long Top 3α isoform produced by an alternative, upstream translation initiation site [61], as well as Top 2α and Top 2β, have also been found in mitochondria [62,63]. mTop 1 plays critical roles in transcription and DNA replication of circular mitochondrial DNA [64,65]. Although cells manage to survive in the absence of mTop 1, loss of mTop 1 severely affects mitochondrial integrity and energy metabolism, [66,67]; however, the presence of Top 3α in mitochondria likely contributes to this survival [61,63,68].
(2). Type IIA topoisomerases
DNA gyrase is the only topoisomerase that is capable of introducing negative supercoils [1–7], while Top 4 is primarily responsible for decatenating daughter chromosomes [69–71]. Bacterial chromosome is kept at a certain negative superhelicity by the balance between the relaxation activity of Top 1 and the supercoiling activity of DNA gyrase [72]. Lower eukaryotes have one type IIA topoisomerase, Top 2, whose structure (note that Top 4 and Top 2 differ in their C-terminal domains) and function are similar to that of Top 4 [1–7, 41]. One distinct difference between bacterial and eukaryotic type IIA topoisomerases is that bacterial enzymes are A2B2 heterotetramers, whereas eukaryotic enzymes are homodimers. Bacterial type IIA topoisomerases have been shown to maintain activity when the two subunits are fused into a single peptide [73,74]. Thus, a heterotetrameric type II topoisomerase can be converted into a homodimeric type II topoisomerase by gene fusion.
Unlike lower eukaryotes, mammals have two isoforms of Top 2, Top 2α and Top 2β [75,76]. These isoforms share roughly 70% amino acid sequence similarity and exhibit similar catalytic activities, but they have distinct cellular functions [1–7]. Top 2α is required for DNA replication and chromosome segregation [77–80], and its expression is significantly elevated in proliferating and cancer cells [81–84]. Thus, hTop 2α serves as a biomarker for some cancers. In contrast, Top 2β plays critical roles in transcription in normal mitotic and postmitotic cells, as well as in cancer cells [54,55,81]. In addition, it is also required for development and the survival of some neural cells [85–87].
3. Structural studies of topoisomerases
(1). Type IB topoisomerases
Compared to relaxed DNA that is in the lowest energy conformation, both positively and negatively supercoiled DNA molecules are in higher energy states due to the existence of torsional strains in these structures. In a simple three-step process that does not require additional input of energy, type IB topoisomerases effectively harvest the free energy stored as DNA supercoils to return these energetically strained DNA molecules to their relaxed forms [1–3,6]. This relaxation reaction starts with the catalytic tyrosine-mediated nucleophilic attack on a scissile phosphodiester bond in a DNA duplex, which results in the formation of a transient single-strand break composed of a 3′-linked phosphotyrosyl bond and freed 5′-OH [88–94]. The tension resulting from DNA supercoiling can be dissipated via rotation of the DNA molecule about the break (Fig. 1). Finally, the cleaved phosphodiester bond is resealed and the catalytic tyrosine is regenerated, resetting the enzyme for the next relaxation event.
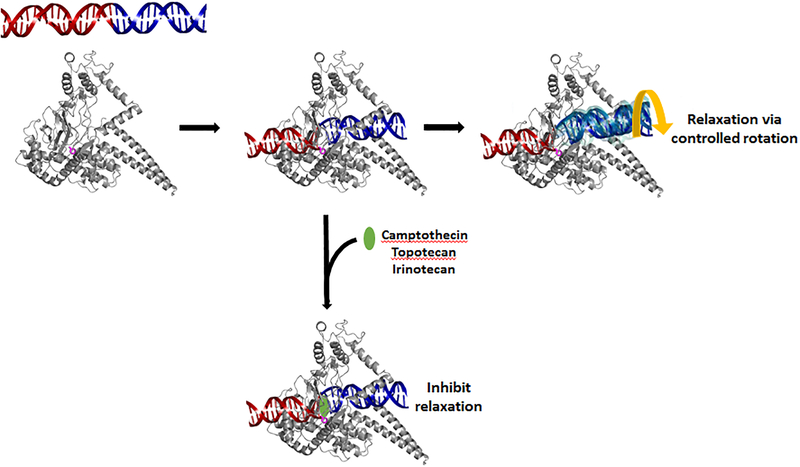
Figure 1. Relaxation reaction catalyzed by hTop 1 and the effect of camptothecin class of anticancer agents.
Type IB topoisomerases cleave one strand of supercoiled DNA via the formation of a protein-linked phosphotyrosyl bond between the catalytic tyrosine (purple sticks) and the 3′-phosphate group. While the DNA duplex upstream of the break (red) is bound tightly by hTop 1, very few interactions are present between the downstream DNA duplex (blue) and the enzyme, which allows both positive and negative superhelical tension in DNA to be released by the rotation of downstream DNA duplex about the break. The insertion of camptothecin class of anticancer agents (green ellipsoid) stabilizes the hTop 1-mediated DNA strand break and inhibits the enzyme’s relaxation activity. The cartoon representation of hTop 1 (PDBid: 1RRJ [137]) was generated using PyMol (http://pymol.org).
Structural analyses of hTop 1 in covalent and noncovalent complexes with DNA suggested that the postulated DNA rotation during the catalytic cycle is likely achieved via a “controlled rotation mechanism” [89,95,96]. Briefly, the catalytic core of type IB topoisomerases is composed of a Cap domain and a catalytic (CAT) domain, which was first observed in tyrosine recombinases [88,97], and these two domains clamp around the DNA duplex upstream to the cleavage site. The DNA upstream of the single-strand break is tightly bound through several protein–DNA interactions between the core of Top 1 and DNA, primarily to the phosphate backbone of the DNA, and is thus unable to rotate. In contrast, the DNA downstream of the break is only loosely held by the nose cone of the Cap domain and linker regions of CAT domain. Conceivably, this weak protein-DNA interface can be easily ruptured by the stored superhelical tension, and the downstream DNA can then rotate about the cleavage site by one or more turns until all torsional strains are released or the break is resealed. Given the limited number of DNA-interacting residues, it appears to be difficult for the enzyme to capture the downstream DNA in the presence of high superhelical tension. Nevertheless, this interface may be re-established at lower superhelical density, which halts the rotating DNA and places the 5′-OH in a favorable position for engaging in the religation reaction [90,98,99]. The rotation and orientation of downstream DNA is ‘controlled’ by the enzyme, as opposed to random rotation, which would complicate the religation reaction [89].
Types IA, IB, and IIA topoisomerases have distinct requirements of Mg2+ for their catalytic activities [100–104]. Structural analysis of the enzyme-DNA covalent complex revealed why type IB topoisomerases do not require a divalent cation for their activity [88]. The transesterification reaction catalyzed by topoisomerases is known to proceed through a high-energy bipyramidal transition state in which two oxyanions are attached to the pentavalent phosphorus. Unlike types IA and IIA topoisomerases, which utilize Mg2+ as an essential cofactor for electrostatic stabilization of the transition state [100–102], type IB topoisomerases and members of the tyrosine recombinase family employ positively charged Arg and Lys residues for this purpose [88,97,105].
(2). Type IIA topoisomerases
Even before the first crystallographic visualization of type IIA topoisomerases was available [18,106,107], a series of elegant biochemical studies had established that type II topoisomerases change the linking number of DNA through a ‘two-gate’ mechanism [108–112]. According to this mechanism (Fig. 2) [1–6,41], the catalytic cycle of type IIA topoisomerases starts from the binding of the G-segment DNA to the enzyme, a process corresponding to the assembly of the so-called ‘DNA gate’ [44,113]. The formation of such a topoisomerase-DNA binary complex allows both strands of the G-segment to be cleaved via Mg2+-dependent transesterification reactions occurring between the catalytic tyrosine residues and the DNA backbone’s phosphodiester bonds [113–115], which introduces a reversible DSB into the G-segment. When a DSB is formed, the DNA backbones are unlocked and the two halves of the G-segment can move apart from each other. This potentiates the DNA gate to enter an open state in which the gap between cleaved DNA ends is spacious enough for the T-segment to be passed through. The incoming T-segment approaches the DNA gate by going through the entry gate located on one end of a type IIA topoisomerase (the N-terminal end of eukaryotic Top 2) and moving along the DNA-conducting path that is used by the G-segment to reach its binding surface. After passing through the DNA gate, the T-segment is directed toward, and eventually released from, the exit gate at the other end of the topoisomerase (the C-terminal end for eukaryotic Top 2) [1–6,41]. This unidirectional DNA transport mode is referred to as being ‘two-gated’ because the entry and exit of T-segment is mediated via two distinct protein gates of type IIA topoisomerases.
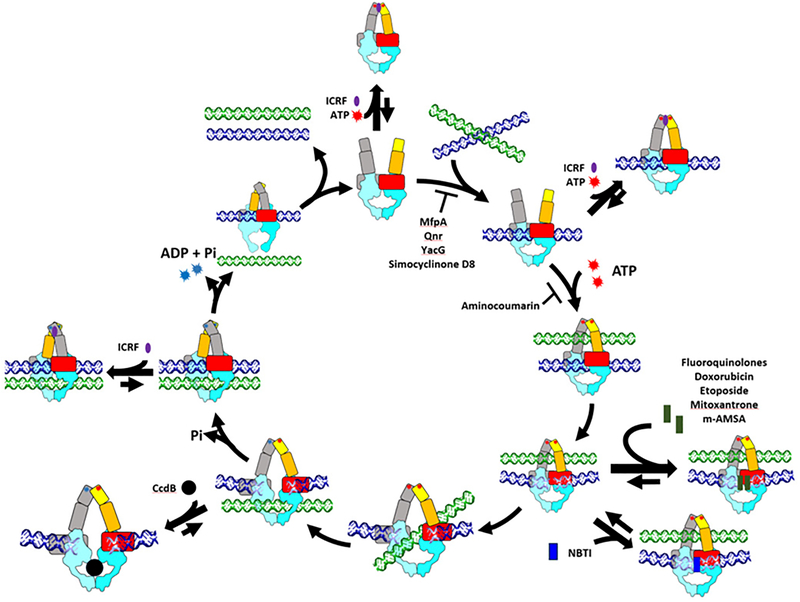
Figure 2. Catalytic cycle and inhibitors of type IIA topoisomerases.
The catalytic cycle of type IIA topoisomerases starts from the entry of G-segment DNA (blue) through an opened N-gate. Binding of two ATP molecules induces closure of the N-gate to facilitate the capture of T-segment DNA (green). The Cleavage of the G-segment followed by pulling the broken DNA ends apart creates a path that allows the T-segment to go through. This duplex passage event is likely driven by the hydrolysis of one ATP and a clockwise rotation of the N-gate, which not only pushes the T-segment through the DNA-gate but also causes the upper cavity enclosed by the N-gate to collapse to prevent the T-segment from moving backward. The religation of the cleaved G-segment is expected to shrink the bottom cavity and thus induces opening of the C-terminal exit gate and the release of the T-segment. The second ATP hydrolysis event then takes place to reset the enzyme for next catalytic cycle. The turnover of type IIA topoisomerases may be blocked by arresting the enzyme at different intermediate states upon the binding of small molecule inhibitors or regulatory proteins. The ATPase, transducer, Toprim, and the remaining C-terminal fragment (from WHD to C-gate) are in yellow, orange, red, and cyan, respectively. The domains of the second protomer are colored grey and light blue. The design of this figure was inspired by Larsen et al. [345].
A more detailed depiction of the catalytic mechanisms of type IIA topoisomerases would require the following aspects to be addressed in structural terms: (1) the enzyme’s overall architecture and spatial domain organization; (2) the assembly of a protein-linked DNA gate; (3) strand breakage of the bound G-segment to unlock the DNA backbones; (4) capture of the T-segment; (5) conformational changes associated with the DNA gate; (6) release of the T-segment and resealing of the cleaved G-segment; and (7) resetting the enzyme conformation for the next catalytic cycle. Thanks to a large number of crystal structures obtained for various fragments of type IIA topoisomerases and two catalytically competent type IIA topoisomerases, which represent structural snapshots of different regions of type IIA topoisomerases at distinct stages of the catalytic cycle, we have obtained valuable insights into each of these steps [1,3,6,41].
All type IIA topoisomerases exhibit a two-fold symmetric architecture with the constituting domains appearing in pairs [1,3,6,41]. The entry gate is formed by two GHKL ATPase domains [116], which are known to dimerize upon ATP-binding, but dissociate in the nucleotide-free state [106,108,117,118]. With gate closure and opening being controlled by ATP, the gate appears to act as a nucleotide-operated protein clamp suitable for T-segment capture. Structural analyses revealed that each nucleotide binding pocket is formed by elements from both ATPase domains; ATP would thus promote dimerization by ‘gluing’ the two domains together [106,108,117,118]. Although each type IIA topoisomerase possesses two structurally equivalent ATP binding sites, it has been shown that one ATP is sufficient to lock the entry gate in the closed state [119–123]. This finding suggests that, once closed, the entry gate would not re-open until the end of the catalytic cycle when both bound ATP molecules are hydrolyzed (Fig. 2).
The DNA gate is compositely formed by a type IIA topoisomerase and G-segment DNA. The protein part of the DNA gate is composed of two copies of each of the following domains: the topoisomerase-primase (Toprim) domain, winged-helix domain (WHD), and a so-called ‘tower’ domain [124]. The Toprim domain adopts a Rossmann-like fold and contains a Mg2+-coordinating DXD motif that is essential for the transesterification reaction [125]. The WHD domain is central to type II topoisomerase functions by harboring a helix-turn-helix DNA-binding motif and the catalytic tyrosine residue [18,126]. The tower domain provides additional DNA-binding residues and is an integral part of the DNA gate [124,127]. Crystal structures of the DNA gate in its closed state revealed that all these domains contribute to the formation of a G-segment binding groove, with the WHD domains at the bottom and the Toprim and tower domains lining the sides [44,122–124,127–129]. The bound G-segment is primarily anchored by interacting with the helix-turn-helix motifs of the WHD domains and is bent into a U-shape by a pair of conserved isoleucine residues that intercalate into DNA at sites 12 base pairs apart [126]. Basic residues from the tower domains interact with the DNA backbone, presumably to stabilize the bent conformation of the G-segment. The DNA duplex enclosed within the two intercalating residues is distorted toward the A-form, which likely places the scissile phosphodiester bonds, the active site tyrosine residues, and the catalytic Mg2+ ions at an optimal orientation for engaging in the transesterification reactions [44,123,124,129]. A structural feature unique to the DNA gate is that, to perform strand breakage and religation reactions, the catalytic tyrosine of one subunit must team up with the Mg2+ presented by the Toprim domain of the opposing subunit [44,123,124,127–129]. Therefore, closure of the DNA gate is a prerequisite for the phosphotyrosyl bond to be reactive, and the decoupling between these two catalytic modules would protect the covalent tether from being attacked by nucleophiles present in the vicinity.
The exit gate corresponds to the C-terminal dimerization interface that is usually observed in the closed state [33,42–44,113,123,128,129]. It is expected that the opening of exit gate would be a transient event during the catalytic cycle and the conformational equilibrium associated with the gate should be biased toward the closed form, because premature gate opening would significantly alter the overall architecture of a type IIA topoisomerase and may prevent the cleaved G-segment from being religated. Nevertheless, opening of the exit gate has been visualized by crystallography in the absence of a T-segment [34,124,127].
The structures of a fully active type IIA topoisomerase showed that the DNA gate is sandwiched between the entry and exit gates, an arrangement that agrees fully with the proposed two-gate mechanism [113,129]. In addition, a cavity is observed on each side of the DNA gate, suggesting that the T-segment may be temporarily held within the enzyme during its transportation. These structures also revealed that the DNA gate is connected to the entry and exit gates via the transducer domains and coiled-coil regions, respectively [113,129]. These bridging elements are thought to mediate allosteric communications between the gates. For example, the conformational changes induced by ATP hydrolysis may alter the structure of a DNA gate via the rearrangement of the transducer domain. Also it has been shown that conformations of the DNA and exit gate may be coupled by adjusting the kink of the coiled-coil region. However, more conformational states of type IIA topoisomerases, such as the opening of DNA gate, would be required to understand how the opening and closing of these gates are coordinated.
4. Mechanisms of type IB topoisomerase poisoning
hTop 1 is the cellular target of the quinoline alkaloid camptothecin and its clinically active derivatives, topotecan and irinotecan (Fig. 1) [130–133]. Similar to Top 2 poisons, these drugs trap the Top 1-DNA covalent complex and convert the enzyme into a cytotoxic covalently-linked protein adduct on DNA [6,28,30], which causes the inhibition of DNA replication and the generation of a DSB [134,135]. Crystal structures of the camptothecin and topotecan-bound enzyme-DNA covalent complexes revealed the intercalation of a drug molecule between the cleaved DNA ends [136,137]. Specifically, the A~D rings of the pentacyclic drug stack against base pairs flanking the strand breakage site, and the quinoline nitrogen of B-ring and polar/charged groups of E-ring form hydrogen bonds or salt bridges with nearby amino acid residues. It has been demonstrated that the E-ring exists in equilibrium between lactone (closed-ring) and carboxylate (open-ring) forms under physiological condition [138]. Given that E-ring is not involved in base-stacking interactions, and that both forms can be modeled equally well into the electron density of topotecan to provide favorable protein-drug interactions, it appears that opening and closure of the E-ring does not alter the drug’s efficacy [139,140]. The strand breakage site-specific drug binding physically blocks the religation reaction [136,137]. The Top 1-catalyzed relaxation reaction is also impaired [141] because the stacking interactions between the bound drug and 5′-base pair about the scissile phosphate would increase the threshold energy for the downstream DNA to rotate.
The anticancer agents indolocarbazoles and indenoisoquinolines also act as hTop 1 poisons [142–147]. Structural analysis showed that the planar polycyclic cores of these compounds intercalate at the strand breakage site, resembling the binding mode of camptothecin [148]. The branching moieties, on the other hand, form distinctive and chemotype-specific protein-drug interactions. Collectively, the observed changes in protein structure and drug-interacting amino acid residues in response to different drugs revealed the landscape and flexibility of the drug-binding pockets. This information should facilitate the development of new type IB topoisomerase poisons. Interestingly, no catalytic inhibitor of type IB topoisomerases has been reported thus far. The structure of hTop 1 in complex with a DNA duplex containing an 8-oxoG lesion suggests the enzyme may adopt an inactive conformation in which the catalytic tyrosine bends away from the active site [149]. Any compounds that bind and stabilize this conformational state of hTop 1 may act as effective catalytic inhibitors. However, the clinical relevance of developing such Top 1 inhibitors has remained to be elucidated.
5. hTop 1-targetting anticancer drugs
Camptothecin (Fig. 3) is a plant alkaloid that poisons hTop 1 [130–133]. Clinical development of camptothecin was discontinued because of its intolerable adverse effects and low therapeutic index [150–152]. However, derivatives of camptothecin, topotecan and irinotecan (Fig. 3), are currently used in the clinic [153–155]. Unfortunately, their clinical use is limited due to their dose-limiting toxicity, especially neutropenia, myelosuppression, and diarrhea [156–161], as well as chemical instability due to the rapid opening of the E-ring [162–164]. mTop 1 is inhibited by camptothecins [60] and mTop 1 can be targeted by topotecan, but not by camptothecin, in vivo [165,166]. It is not clear, however, if the effect of camptothecins on mTop 1 contribute to the anticancer activities of these drugs.
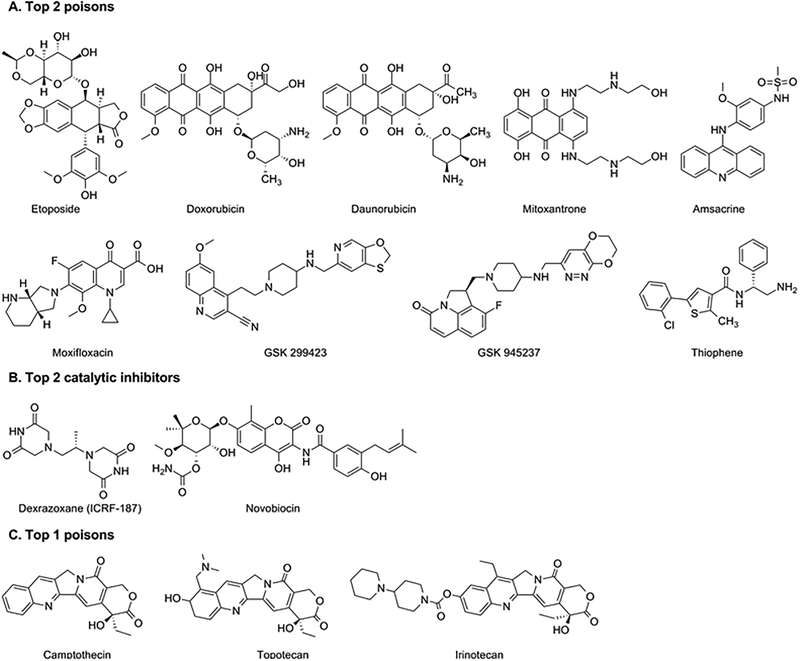
Figure 3. Structures of topoisomerase inhibitors.
A. Type IIA topoisomerase (Top 2) poisons. B. Type IIA topoisomerase (Top 2) catalytic inhibitors. C. Type IB topoisomerase (Top 1) poisons.
(1). Camptothecins
a. Topotecan
Topotecan (Hycamtin) (Fig. 3) is a semi-synthetic water-soluble derivative of camptothecin [167]. It was the first hTop 1 inhibitor approved for oral administration [168,169]. It is often used to treat ovarian and small cell lung cancer [153–155,169].
b. Irinotecan
Irinotecan (CPT-11, Campostar) (Fig. 3) is another water-soluble derivative of camptothecin [170,171]. It is a prodrug and is converted to a biologically active metabolite, ethyl-10-hydroxy-camptothecin (SN-38), by a carboxylesterase [172,173]. Irinotecan, together with fluorouracil, is often used for the treatment of advanced colorectal cancer [153–155,174].
(2). Non-camptothecins
Camptothecins are the only hTop1 inhibitors approved for clinical use. Despite their effectiveness, these drugs have limitations due to their instability and severe side effects, as well as drug resistance caused by P-glycoprotein [175,176]. To overcome these limitations, non-camptothecin hTop 1 inhibitors have been developed and investigated [177–180]. Among them, indolocarbazoles (NB-506) [142–144], indenoisoquinolines [145–147], and dibenzonaphthyridinones (ARC-111) [181–183] are under clinical development.
a. Indolocarbazoles
Indolocarbozoles are synthetic analogs of antibiotics isolated from several actinomycetes [184]. NB-506 is a DNA intercaltor that can poison hTop 1 [142,143]. NB-506 and camptothecins share the binding site on hTop 1 but hTop 1 is not the only target of NB-506 [185]. Edotecarin (J-107088) is a derivative of NB-506 that does not intercalate into DNA [144]. Similar to NB-506, it poisons hTop 1 but its effect on an unidentified target(s) also contributes to its promising anticancer activity [144].
b. Indenoisoquinolines
Indenoisoquinolines are synthetic non-camptothecin analogs that poison hTop 1. Since the first indenoisoquinline, NSC 314622, was reported in 1978, many derivatives have been synthesized and tested for anticancer activity [186]. A Phase 1 clinical study of indotecan (LMP400) and indimitecan (LMP776) in adults with relapsed solid tumors and lymphomas was recently completed although study results have not been published [187].
c. Dibenzonaphthyridinones
ARC-111 was selected among dibenzonaphthyridinones for preclinical studies of its anticancer activity [181–183]. More recently, Genz-644282 has been the subject of both preclinical and clinical studies [188,189].
6. Mechanisms of type IIA topoisomerase poisoning and inhibition
The ability of type IIA topoisomerases to adopt a spectrum of conformational states is consistent with the notion that these enzymes must undergo extensive structural rearrangement during the course of strand passage (Fig. 2). In theory, modulating the activity of topoisomerases can be achieved by trapping them in any of these conformational states. However, two modes of action are utilized most successfully by type IIA topoisomerase inhibitors: (1) trapping of a cleavage complex by topoisomerase poisons or toxins and (2) inhibition of ATPase activity that leads to the blockage of enzyme turnover. Structural analyses of type IIA enzymes bound to topoisomerase poisons, toxins, or catalytic inhibitors have revealed their mechanisms of action. The mechanisms utilized by bacterial type IIA topoisomerase inhibitors may provide useful insights to the development of novel anticancer agents that target hTop 2.
(1). Type IIA topoisomerase poisons
Several type IIA topoisomerase poisons are used successfully in the clinic as anticancer and antibacterial drugs (Fig. 3) [6, 24–31]. These drugs stabilize cleavage complexes that contain a DSB. Structural analysis of the catalytic core (the 55-kDa DNA breakage-reunion domain of the ParC subunit plus the 30-kDa Toprim domain of the ParE subunit) of Streptococcus pneumoniae Top 4 complexed with DNA and fluoroquinolones (including moxifloxacin, clinafloxacin, and levofloxacin) provided the first visualization of drug-stabilized type IIA topoisomerase cleavage complexes [33]. These structures, although determined at sub-optimal resolution (~4 Å), revealed that two fluoroquinolone molecules intercalated four-base pairs apart at the sites of strand breakage and are stacked against the flanking +1/+4 and −1/+5 base pairs [33]. The intercalated fluoroquinolone molecules physically prevent the 3′-OH from approaching the enzyme-linked 5′-phosphate and block religation of the broken DNA strands, which effectively trap cleavage complexes that contain a DSB. Thus, the strand breakage site-specific drug insertion provides a basis for topoisomerase poisoning by quinolones. In addition, the quinolone-stabilized cleavage complex is in a closed conformational state where the two halves of the cleaved DNA strands are tethered by base paring formed between the two cohesive ends [33]. Transition of a cleavage complex from a closed conformational state to an open conformational state is expected to be suppressed by the bound drug molecules; opening of the DNA-gate would disrupt the stacking between quinolone molecules and DNA bases, and expose the aromatic quinolone core to solvent. The interactions formed between amino acid residues and fluoroquinolone molecules in ternary complexes are not defined clearly because of the limited resolution. Nevertheless, the general vicinity of the quinolone-binding pocket matches with the quinolone resistance-determining region [190,191].
Although this initial set of structures of the quinolone-stabilized cleavage complex were determined at lower resolution [33], the key structural observations described above, including the intercalation of drug molecules at strand breakage sites and the cleavage complex being observed in the closed conformation, agree with subsequent high resolution structures [34,44,191–195]. A clearer picture of the exact mechanism of quinolone-topoisomerase interaction came from the higher resolution structures of the Acinetobacter baumannii Top 4 cleavage complexes stabilized by levofloxacin and moxifloxacin [34]. Importantly, the A. baumannii Top 4 structure revealed the chelation of a Mg2+ molecule by the C-3, C-4 diketo moiety of moxifloxacin (Fig. 3), an observation consistent with the involvement of Mg2+ in quinolone function [196,197]. Moreover, two conserved amino acid residues, Ser-84 and Glu88, located on helix α4 of the WHD domain form hydrogen bonds with the Mg2+-coordinating water molecules. This Mg2+-water bridge between the C-3, C-4 diketo moiety of floroquinolones and the conserved serine and glutamate/aspartate residues in the WHD domain of the GyrA/ParC subunit appears to be the only direct interaction between a fluoroquinolone and a topoisomerase [34,193]. Two conserved amino acid residues that correspond to Ser-83 and Asp-87 of E. coli GyrA are hot spots for quinolone-resistant mutations [26,198]. Thus, the presence of the Mg2+-water bridge also provides the molecular basis for the quinolone resistance conferred by amino acid substitutions at these location [29,34].
The crystal structures of drug-stabilized cleavage complexes of hTop 2α and hTop 2β showed that the Top 2-targeting anticancer drugs and the antibacterial quinolones most likely act in a similar manner [42,43]. The tetracyclic aglycone core of etoposide (Fig. 3), the tricyclic dihydroxy-anthraquinone moiety of mitoxantrone (Fig. 3), and the tricyclic acridine moiety of m-AMSA (Fig. 3) were observed to intercalate between the base pairs that flank the strand breakage sites, which prevents the religation of cleaved DNA strands. The interactions formed between the topoisomerase and each drug may explain the drug’s specificity toward Top 2-mediated DNA breaks. Notably, a loop region and several residues involved in drug-binding were found to display extraordinary conformational flexibility, which reveals why the cleavage complexes formed with either hTop 2α or hTop 2β can be targeted by drugs with structurally distinct protein-interacting moieties (namely, the E-ring and glycosidic group of etoposide, the hydroxylalkylamino arms of mitoxantrone, and the methanesulfon-m-anisidide group of m-AMSA). The majority of the drug-interacting residues are conserved between hTop 2α and hTop 2β, consistent with the finding that both isoforms are the cellular targets of these drugs [24,25,27,28,30]. It is worth noting that the co-crystallization approach had been applied successfully to the structural analyses of etoposide- and mitoxantrone-bound cleavage complexes of hTop 2α [199], thus the obtained crystal structures of m-AMSA- and mitoxantrone-bound cleavage complexes of hTop 2β are expected to be functionally relevant. In contrast, the crystal structure of doxorubicin-bound cleavage complex was only determined using a post-crystallization drug-replacement procedure, and therefore the functional significance of this structure has remained to be elucidated [43].
To develop antibacterial drugs, it is preferable that one compound can target both DNA gyrase and Top 4. Dual targeting of DNA gyrase and Top 4 may increase the potency and reduce the appearance of drug resistance [200,201]. However, the accumulation of hTop 2β-induced DNA breaks are associated with deleterious chromosome translocation and rearrangement events, resulting in frequent occurrence of therapy-related leukemia [202,203]. Therefore, it would be clinically desirable to have an anticancer drug that preferentially targets hTop 2α. Structural studies revealed the presence of an hTop 2α-specific methionine residue that may be exploitable for this purpose [124,199]. Given the reactivity of methionine toward platinum(II) and the recent finding that the formation of a single coordination bond between methionine and platinum(II) can irreversibly trap the cleavage complexes formed by hTop 2α and 2β [199], the development of isoform-selective drug may be achieved by targeting this hTop 2α-specific methionine.
(2). NBTIs and allosteric modulators
In addition to type IIA topoisomerase poisons described in the previous section, new classes of small molecules with potent antibacterial activities have been identified as NBTIs in recent years [191,204–206]. GlaxoSmithKline has also reported a series of cyclohexyl-amides, amino-piperidines, and tricyclic compounds as NBTIs [206–210]. Among them, Gepotidacin (originally GSK2140944 [211,212]), an analog of GSK299423 (Fig. 3), and AZD0914 [213–215], an analog of quinoline pyrimidine trione-1 (QPT-1), are in clinical trials.
Structures of GSK299423- and QPT-1-stabilized DNA gyrase cleavage complexes have been reported [191,192]. Two QPT-1 molecules bind to the same sites in the cleaved DNA as moxifloxacin in a ternary complex although the interaction of QPT-1 with DNA gyrase is completely different from that of moxifloxacin with DNA gyrase. As described in the previous section, fluoroquinolones interact with the conserved amino acid residues in the WHD domain of the GyrA/ParC subunit. In contrast, QPT-1 interacts with the conserved amino acid residues in the Toprim domain of the GyrB/ParE subunit [192], which may explain the activity of QPT-1 against fluoroquinolone-resistant strains of bacteria [216].
The GSK299423-stabilized, DNA gyrase cleavage complex has revealed a novel binding site utilized by this class of NBTI [191]. GSK299423 inserts its planar quinolone-carbonitrile moiety between the central (+2/+3 and +3/+2) base pairs enclosed by the strand breakage sites and the oxathiolo-pyridine group into a hydrophobic pocket formed by surrounding amino acid residues. It induces the formation of ternary complexes containing a single-strand DNA break, not a DSB [191]. Notably, this NBTI binding site falls on the dyad axis of the enzyme, suggesting that one type IIA topoisomerase-DNA complex can accommodate just one drug molecule (Fig. 4), in contrast to the binding of two drug molecules per cleavage complex for fluoroquinolones and QPT-1, as well as etoposide [192]. Upon the binding of an NBTI, the two cohesive DNA ends can no longer slide against one another. Thus, a single molecule can effectively lock the enzyme-DNA complex in its closed conformation [191]. As was the case with topoisomerase-fluoroquinolone-DNA ternary complexes [217], topoisomerase-NBTI-DNA ternary complexes could arrest replication fork progression in vitro [31].
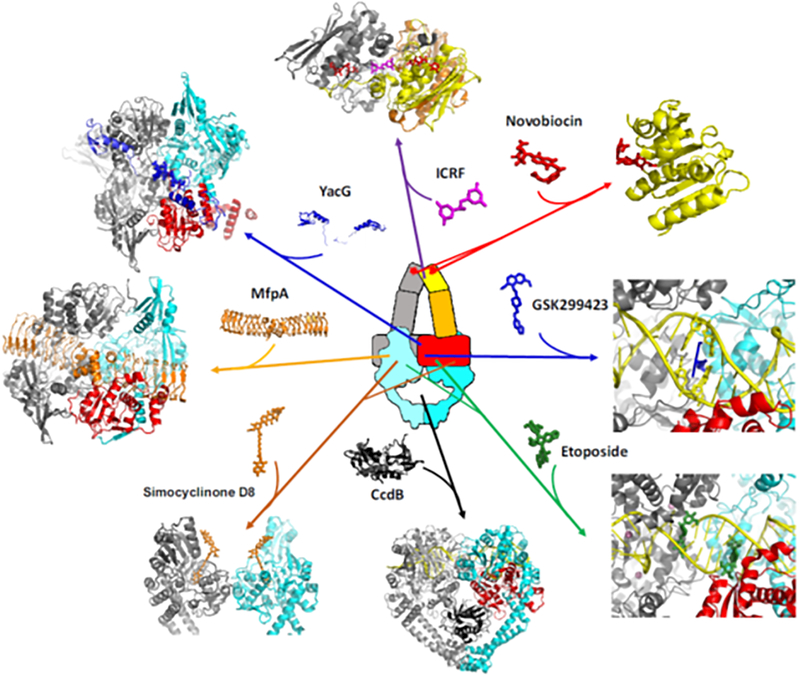
Figure 4. Targeting sites and binding modes of selected type IIA topoisomerase inhibitors and regulatory proteins.
Functional modulation of type IIA topoisomerases can be achieved by the binding of small molecules and regulatory proteins to various sites in the enzyme. The crystal structures of type IIA topoisomerases in complexes with small molecules {ICRF-187 (PDBid: 1QZR [117]), novobiocin (PDBid: 4URO [346]), GSK299423 (PDBid: 2XCS [191]), etoposide (PDBid: 3QX3 [42]), simocyclinone D8 (PDBid: 2Y3P [233])} and regulatory proteins {YacG (PDBid: 4TMA [228]), CcdB toxin (PDBid: 1X75 [240])} were generated using PyMol (http://pymol.org). The structural model of MfpA-bound GyrA was constructed by manual docking of the MfpA (PDBid: 2BM4 [222]) to the N-terminal fragment of GyrA (PDBid: 1AB4 [107]) followed by stereochemical idealization.
A more recent work by GlaxoSmithKline has led to the discovery of thiophene antibacterials (Fig. 3) that are capable of inhibiting DNA gyrase via stabilization of the cleavage complex [218]. Co-crystallization of these novel inhibitors with DNA gyrase and DNA has revealed that this new class of compounds binds to a previously unrecognized pocket formed between the Toprim and WHD domains, away from the bound G-segment DNA which harbors the bindings sites of other Top 2 poisons described above. A major incentive for developing new compounds against bacterial type IIA topoisomerases is to overcome the growing resistance to quinolone antibacterial agents. In this regard, the development of NBTIs, including thiophene antibacterials, whose binding pockets are distinct and non-overlapping with the quinolones [218], may provide a new approach to conquering quinolone resistance and also inspire the discovery of novel hTop 2 poisons. Interestingly, the thiophene compound-bound structures of DNA gyrase-DNA complex were obtained in the presence of GSK945237 (Fig. 3), indicating that these two distinct classes of NBTIs target the same conformational state of the enzyme-DNA complex and may act synergistically [218].
(3). Toxins and regulatory proteins
The catalytic activities of type IIA topoisomerases are modulated through their interactions with various proteins. Several toxins and regulatory proteins that target DNA gyrase have been identified and characterized in bacteria [26,219,220]. An effective approach for blocking type IIA topoisomerase function is to prevent type IIA topoisomerases from binding to the G-segment DNA. Two types of proteins have been identified that act in this manner.
The pentapeptide repeat protein family has over 500 members [219,221]. Although the majority of them are found in bacteria, they are also found in Arabidopsis, zebrafish, mouse, and humans. MfpA and Qnr adopt a quadrilateral β-helix fold, with overall dimensions and distribution of negatively charged groups on their protein surface markedly resembling features of B-form DNA [222–225]. Biochemical and structural studies suggest that these DNA-mimicking proteins may interact with the primary DNA-binding groove of DNA gyrase (and perhaps Top 4), thereby blocking the binding of the G-segment (Fig. 4). Since the antibacterial activity of fluoroquinolones relies on the formation of gyrase-fluoroquinolone-DNA ternary complexes, the expression of either MfpA or Qnr antagonizes the actions of fluoroquinolones and, thus, may contributes to antibiotic resistance [220]. Despite the supposedly tight association between MfpA/Qnr and gyrase (Kd in the low μM to nM range), no structures of MfpA/Qnr-gyrase complexes are available. Therefore, it remains unclear how the binding specificity is defined to ensure the assembly of these inhibitory complexes in vivo, especially given the high local concentration of DNA and the presence of other DNA-mimicking proteins in cells. Identification of MfpB, a GTPase that modulates MfpA action and quinolone resistance [226], indicates that additional factors may regulate pentapeptide repeat protein function.
The chromosome-encoded and zinc-finger containing protein YacG is another bacterial type IIA topoisomerase regulator that inhibits DNA gyrase by interfering with the binding of the G-segment [227,228]. Rather than mimicking B-DNA and competing with the G-segment for the primary DNA binding groove of gyrase, a crystal structure of the YacG-DNA gyrase complex revealed that the zinc-finger of YacG interacts with the Toprim domain (Fig. 4) and protrudes into the G-segment-binding groove, thus occluding the binding of the G-segment [228]. Moreover, the C-terminal tail of YacG fits into a hydrophobic pocket on the surface of GyrB that is also targeted by the oxathiolo-pyridine group of NBTIs [191], which further stabilizes the inhibitory complex.
Simocyclinones, such as simocyclinone D8, are antibiotics that contain an aminocoumarin moiety [229,230]. The aminocoumarin antibiotics are known to be ATPase inhibitors of type IIA topoisomerases [26,27]. Although simocyclinone D8 can bind to the GyrB subunit of gyrase [231], it binds primarily to the GyrA subunit of gyrase (Fig. 4) [232–234]. As a result, this small molecule is capable of inhibiting the supercoiling activity of gyrase by preventing gyrase from binding to DNA, rather than by inhibiting the ATPase activity of gyrase [232]. Simocyclinone D8 also exhibits modest activities against Top 4 and hTop 2 [235–237].
Blocking the transportation of the T-segment by the F plasmid-encoded protein CcdB represents another mechanism of gyrase inhibition [238,239]. Structural studies indicated that CcdB binds to the large cavity enclosed by the two GyrA subunits by interacting with cavity-facing residues from the C-gate region of gyrase (Fig. 4) [240]. Since this cavity serves as a temporary storage for the T-segment DNA before its exit through the C-gate, the presence of CcdB would render gyrase inactive. Structural modeling further suggested that CcdB cannot be accommodated in the cavity if the cleavage complex adopts the closed conformation, a state essential for resealing the cleaved DNA [240]. Hence, CcdB-binding would promote the formation of a DSB [241].
(4). ATPase inhibitors
The transfer of the T-segment by a type II topoisomerase is driven by the conformational rearrangement of the GHKL ATPase domain in response to ATP binding and hydrolysis [121–123,127,128]. Therefore, inactivation of a type II topoisomerase can be achieved by restricting the nucleotide-dependent structural changes or inhibiting ATPase activity. Small molecules useful for both purposes have been identified [242,243]. The bisdioxopiperazines, such as ICRF-187 (dexrazoxane) (Fig. 3), inhibit eukaryotic Top 2 by arresting the two ATPase domains in the ATP-bound, dimerized form, effectively locking the entry gate in its closed state (Fig. 4) [244–247]. Since neither the G- nor T-segment is able to go through the entry gate in the presence of ICRF-187, this compound not only renders Top 2 inactive but also compromises the action of Top 2 poisons by suppressing the production of topoisomerase-induced DNA breaks [248]. Given that ICRF-187 on its own does not exhibit appreciable affinity toward the ATPase domain, it appears that the ATP-induced closure of the entry gate is required prior to the binding of ICRF-187. Indeed, structural analysis performed on the ATPase domain of yeast Top 2 revealed that a single ICRF-187 bound at the dimer interface without overlapping the ATP binding pockets [116]. Importantly, a glutamine residue that interacts with the γ-phosphate of ATP, and has been implicated in ATP hydrolysis, also forms hydrogen bonds with ICRF-187. This observation suggests that ICRF-187 may restrict the movement of residues involved in ATP hydrolysis and explains the compound’s ability to inhibit ATPase activity. Interestingly, while the closed entry gate is structurally two-fold symmetric, ICRF-187 (Fig. 3) is an asymmetric molecule with only one methyl group present in the 2-carbon linker that connects the two piperazinedione moieties [242,243]. As a result, only one of the methyl group interacting surfaces in the binding pocket is exploited. In contrast, ICRF-193, the most potent bisdioxopiperazine, contains two methyl groups and, thus, binds tighter to eukaryotic Top 2s [249].
Unlike bisdioxopiperazine compounds that act as uncompetitive inhibitors, the aminocoumarin antibiotic novobiocin (Fig. 3) is a competitive inhibitor of the ATPase activity of bacterial type IIA topoisomerases [250,251]. Structural studies established that the sugar and coumarin moieties of novobiocin compete with ATP for the nucleoside binding site [252–254]. However, rather than occupying the triphosphate binding site, the benzoyl moiety of novobiocin extends toward the opposite direction without contacting the neighboring subunit and, thus, would not induce closure of the entry gate like ATP does [252,255]. It appears that eukaryotic type IIA topoisomerases are not targeted by novobiocin due to divergent evolution of drug-binding residues. It has been shown that the efficacy of novobiocin can be significantly reduced by the occurrence of a single amino acid substitution in the drug-binding pocket [256,257]. Unexpectedly, anticancer activity of novobiocin has been reported, likely achieved via targeting Hsp90 [258–261]. Since Hsp90 also harbors a GHKL ATPase domain, whose structure is very similar to that of GyrB [106,262], this finding suggests that novobiocin may have a broader target specificity than previously realized.
7. hTop 2-targetting anticancer drugs
As listed below, hTop 2 poisons are successful anticancer drugs used in the treatments of various cancers. However, two serious side effects, therapy-related cancer [203,263–266] and cardiotoxicity [267–270], associated with these drugs limit their use. Etoposide (Fig. 3) and other hTop 2 poisons cause the development of secondary malignancies, especially therapy-related acute myeloid leukemia (t-AML) [271,272] and therapy-related acute promyelocytic leukemia (t-APL) [273–276]. t-AML is caused by hTop 2-mediated, and more specifically hTop 2β-mediated [201], DSBs and chromosome rearrangements in the mixed lineage leukemia (MLL) gene [277–281], and t-APL is caused by the translocation between the promyelocytic leukemia (PML) gene and the retinoic receptor α gene [273–275].
Doxorubicin (Fig. 3) is a potent anticancer drug that can be used to treat many cancers. However, its full potential has not been realized due to the cardiotoxicity associated with its use [267–270]. The primary mechanism of doxorubicin-associated cardiotoxicity appears to be oxidative stress, due to increased levels of reactive oxygen species (ROS) and lipid peroxidation [282–285]. The main pathway for ROS formation by anthracyclines is the formation of semiquinone radicals resulting in redox cycling [283,285]. Another source of ROS generated by anthracyclines results from formation of doxorubicin-iron complexes that catalyze the Fenton reaction [286–288]. Cardiomyocytes are particularly sensitive to oxidative stress. hTop 2β is the only topoisomerase expressed in myocytes [289,290] and its poisoning by doxorubicin, in addition to increased ROS, may contribute to the cardiotoxicity of doxorubicin [291,292]. Further studies are required to determine the exact mechanism(s) of the cardiotoxicity of doxorubicin but the current strategy to prevent cardiotoxicity is to limit the cumulative dose of doxorubicin.
In addition to these side effects, P-glycoprotein (multidrug resistance protein 1) confers resistance to epipodophyllotoxins and anthracyclines [293,294]. Despite the clinical issues described above, hTop 2 poisons are among the most widely prescribed anticancer drugs. Some Top 2 poisons currently used in the clinic are described below.
(1). hTop 2 poisons
a. Epipodophyllotoxins – Etoposide and Teniposide
Podophyllotoxin was originally isolated from the podophyllum plants, American Podophyllum peltatum and Podophyllum emodi [295]. Easily accessible podophyllotoxin is used in the synthesis of etoposide (Etopophos, VePesid, VP-16) and teniposide (Vumon, VM-26) [296]. Etoposide (Fig. 3) is widely used in the treatments of solid tumors, such as testicular cancer and small cell lung cancer [297]. It is also used to treat lymphomas and nonlymphocytic leukemia. To improve its water solubility, a prodrug etoposide phosphate (Etophos) was developed for intravenous use [298,299]. Teniposide is an analog of etoposide and is approved for the treatment of acute lymphoblastic leukemia (ALL) in children [300]. It is also used to treat other cancers, including Hodgkin’s lymphoma and some brain cancers. Teniposide is less water soluble and has higher plasma protein binding than etoposide [301,302].
b. Anthracyclines – Doxorubicin, Daunorubicin, and Epirubicin
Doxorubicin (Adriamycin, Doxil) (Fig. 3) is an anthracycline antibiotic originally isolated from Streptomyces peucetius [303,304]. It is used to treat a wide variety of cancers, including ALL, acute myeloblastic leukemia (AML), breast carcinoma, ovarian carcinoma, Kaposi’s sarcoma, Wilms’ tumor, thyroid carcinoma, gastric carcinoma, and Hodgkin’s disease [305,306]. Doxil is a form of doxorubicin contained inside a liposome (liposomal doxorubicin) for a slow release to treat AIDS-related Kaposi’s sarcoma, multiple myeloma, and ovarian cancer [306–311]. Doxorubicin is one of the most potent anticancer drugs but its use is limited by the cumulative dose-dependent cardiotoxicity [267–270].
Daunorubicin (Cerubidine, DaunoXome) (Fig. 3) has similar therapeutic effects to doxorubicin [312,313]. It is used to treat ALL, AML, chronic myelogenous leukemia (CML), and Kaposi’s sarcoma [314–317]. DaunoXome is liposomal daunorubicin that is used for the treatment of AIDS-related Kaposi’s sarcoma [315,318].
Epirubicin (Ellence) is an active isomer of doxorubicin [319]. Although epirubicin has similar therapeutic effect to doxorubicin, epirubicin is preferred over doxorubicin in certain chemotherapy regimens because it has fewer side effects [320,321]. Epirubicin is eliminated faster and has reduced toxicity compared to doxorubicin. The spatial orientation of the hydroxyl group at the 4’ carbon of the sugar moiety differs in doxorubicin and epirubicin. Epirubicin’s 4’ hydroxyl is in an equatorial position allowing for rapid conjugation of epirubicin with glucuronic acid, which could account for its faster elimination and lower toxicity [322,323]. Liposomal epirubicin has shown to be an effective agent to treat brain glioma [324,325].
Anthracyclines act as Top 2 poisons. In addition, they can intercalate into DNA [326] and may influence the functions of various proteins besides hTop 2 [327–329]. As previously described, anthracyclines also generate ROS [283,285–288].
c. Mitoxantrone
Mitoxantrone (Novantrone) is a synthetic anthracenedione (Fig. 3) developed as an alternative to anthracyclines to reduce cardiotoxicity while retaining antineoplastic activity [330,331]. It is used for the treatment of multiple sclerosis and AML [332–334]. While mitoxantrone has higher tolerability than the anthracyclines, cardiomyopathy is still a concern in long-term treatment [331,335,336].
(2). hTop 2 catalytic inhibitor
a. Dexrazoxane
Dexrazoxane (Zinecard, Cardioxane, ICRF-187) is a bisdioxopiperazine (Fig. 3) and is a derivative of ethylenediaminetetraacetic acid [242]. Dexrazoxane is the only drug approved to treat cardiotoxicity caused by anthracyclines [337–339]. Its cardioprotective mechanism is attributed to the chelation of iron by dexrazoxane, thus decreasing the production of ROS during anthracycline treatment [340,341]. Dexrazoxane is orally active as a prodrug that is hydrolyzed, by sequential ring openings, into the active metabolite ADR-925 [340,342,343]. Recent studies have indicated that dexrazoxane’s cardioprotective effect may be associated with catalytic inhibition of hTop 2 [290,344].
8. Prospective
Both hTop 1 and hTop 2s are targets of current anticancer drugs. New inhibitors of these enzymes are also in the pipeline. Thus, topoisomerases continue to be important therapeutic targets of anticancer drugs.
Clinically used Top 2-targetting drugs act on both hTop 2α and hTop 2β. The expression of hTop 2α is often elevated in cancer cells and the anticancer activity of these drugs is mainly mediated by the poisoning of hTop 2α. Furthermore, a significant number of studies suggest that both the development of therapy-related leukemia and cardiotoxicity are caused by the poisoning of hTop 2β. Thus, it seems reasonable to develop hTop 2α-specific poisons as anticancer drugs to prevent the therapy-related leukemia and cardiotoxicity. The development of either isoform-specific hTop 2 poisons or potent hTop 2 catalytic inhibitors may provide safer therapeutic options for cancer patients.
In contrast to topoisomerase poisons, topoisomerase catalytic inhibitors have not been successfully used in clinic. Despite the lack of structural similarities between hTop 1, a type IB topoisomerase, and hTop 2s, type IIA topoisomerases, some small molecules, including our novel quinolones, have been shown to act as dual catalytic inhibitors of hTop 1 and hTop 2s. Thus, catalytic inhibitors of multiple topoisomerases may become successful anticancer drugs.
Human cells also contain two type IA topoisomerases, hTop 3α and hTop 3β. Although some small molecules that inhibit bacterial type IA topoisomerases have been identified only in recent years, hTop 3 inhibitors may be discovered and developed as new anticancer drugs in the future.
Acknowledgements
We thank Dr. Robert Kerns for his support and Dr. Lisa Oppegard for her critical comments on the manuscript. Studies from the authors’ laboratories were supported in part by National Institutes of Health grants GM59465, AI087671, and T326M008365, and Ministry of Science and Technology grants 106–2113-M-002–021-MY3 and 104–2911-I-002–302.
Abbreviations:
- ALL
acute lymphoblastic leukemia
- AML
acute myeloblastic leukemia
- CML
chronic myelogenous leukemia
- DSB
double-strand break
- G
Gate
- h
human
- m
mitochondria
- MLL
mixed lineage leukemia
- NBTI
new bacterial topoisomerase inhibitor
- PML
promyelocytic leukemia
- QPT-1
quinoline pyrimidine trione-1
- ROS
reactive oxygen species
- Top 1
topoisomerase I
- Top 3
topoisomerase III
- Top 4
topoisomerase IV
- t-AML
therapy-related acute myeloid leukemia
- t-APL
therapy-related acute promyelocytic leukemia
- T
Transfer
Footnotes
Declarations of Interest
The Authors declare no competing interests associated with the manuscript.
References
- 1.Champoux JJ (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem 70, 369–413 doi: 10.1146/annurev.biochem.70.1.369 [DOI] [PubMed] [Google Scholar]
- 2.Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev. Mol. Cell Biol 3, 430–440 doi: 10.1038/nrm831 [DOI] [PubMed] [Google Scholar]
- 3.Corbett KD, Berger JM (2004) Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct 33, 95–118. doi: 10.1146/annurev.biophys.33.110502.140357 [DOI] [PubMed] [Google Scholar]
- 4.Nitiss JL (2009) DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9, 327–337 doi: 10.1038/nrc2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vos SM, Tretter EM, Schmidt BH, Berger JM (2011) All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol 12, 827–841 doi: 10.1038/nrm3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen SH, Chan NL, Hsieh TS (2013) New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem 82, 139–170 doi: 10.1146/annurev-biochem-061809-100002 [DOI] [PubMed] [Google Scholar]
- 7.Pommier Y, Sun Y, Huang SN, Nitiss JL (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol 17, 703–721 doi: 10.1038/nrm.2016.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirkegaard K, Wang JC (1985) Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol 185, 625–637 doi: 10.1016/0022-2836(85)90075-0 [DOI] [PubMed] [Google Scholar]
- 9.DiGate RJ, Marians KJ (1988) Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem 263, 13366–13373 [PubMed] [Google Scholar]
- 10.Brown PO, Cozzarelli NR (1981) Catenation and knotting of duplex DNA by type 1 topoisomerases: a mechanistic parallel with type 2 topoisomerases. Proc. Natl Acad. Sci. USA 78, 843–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lima CD, Wang JC, Mondragon A (1994) Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature 367, 138–146 doi: 10.1038/367138a0 [DOI] [PubMed] [Google Scholar]
- 12.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH (2005) Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434, 671–674 doi: 10.1038/nature03395 [DOI] [PubMed] [Google Scholar]
- 13.Taneja B, Patel A, Slesarev A, Mondragon A (2006) Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J 25, 398–408 doi: 10.1038/sj.emboj.7600922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogh BO, Shuman S (2001) A poxvirus-like type IB topoisomerase family in bacteria. Proc. Natl Acad. Sci. USA 99, 1853–1858 doi: 10.1073/pnas.032613199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown PO, Cozzarelli NR (1979) A sign inversion mechanism for enzymatic supercoiling of DNA. Science 206, 1081–1083 doi: 10.1126/science.227059 [DOI] [PubMed] [Google Scholar]
- 16.Liu LF, Liu CC, Alberts BM (1980) Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell 19, 697–707 doi: 10.1016/S0092-8674(80)80046-8 [DOI] [PubMed] [Google Scholar]
- 17.Mizuuchi K, Fisher LM, O’Dea MH, Gellert M (1980) DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc. Natl Acad. Sci. USA 77, 1847–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger JM, Gamblin SJ, Harrison SC, Wang JC (1996) Structure and mechanism of DNA topoisomerase II. Nature 379, 225–232 doi: 10.1038/379225a0 [DOI] [PubMed] [Google Scholar]
- 19.Bergerat A, De Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P (2007) An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386, 414–417 doi: 10.1038/386414a0 [DOI] [PubMed] [Google Scholar]
- 20.Gellert M, Mizuuchi K, O’Dea MH, Nash HA (1976) DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. USA 73, 3872–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gellert M, O’Dea MH, Itoh T, Tomizawa J (1976) Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. USA 73, 4474–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugino A, Peebles CL, Kreuzer KN, Cozzarelli NR (1977) Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74, 4767–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gellert M, Mizuuchi K, O’Dea MH, Itoh T, Tomizawa J (1977) Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74, 4772–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deweese JE, Osheroff N (2009) The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res 37, 738–748 doi: 10.1093/nar/gkn937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitiss JL (2009) Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 doi: 10.1038/nrc2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collin F, Karkare S, Maxwell A (2011) Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Appl. Microbiol. Biotechnol 92, 479–497 doi: 10.1007/s00253-011-3557-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailly C (2012) Contemporary challenges in the design of topoisomerase II inhibitors for cancer chemotherapy. Chem. Rev 112, 3611–3640 doi: 10.1021/cr200325f [DOI] [PubMed] [Google Scholar]
- 28.Pommier Y (2013) Drugging topoisomerases: lessons and challenges. ACS Chem. Biol 8, 82–95 doi: 10.1021/cb300648v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aldred KJ, Kerns RJ, Osheroff N (2014) Mechanism of quinolone action and resistance. Biochemistry 53, 1565–1574 doi: 10.1021/bi5000564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pommier Y, Kiselev E, Marchand C (2015) Interfacial inhibitors. Bioorg. Med. Chem. Lett 25, 3961–3965 doi: 10.1016/j.bmcl.2015.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiasa H (2018) DNA Topoisomerases as Targets for Antibacterial Agents. Methods Mol. Biol 1703, 47–62 doi: 10.1007/978-1-4939-7459-7_3 [DOI] [PubMed] [Google Scholar]
- 32.Osheroff N (1989) Effect of antineoplastic agents on the DNA cleavage/religation reaction of eukaryotic topoisomerase II: inhibition of DNA religation by etoposide. Biochemistry 28, 6175–6160 doi: 10.1021/bi00441a005 [DOI] [PubMed] [Google Scholar]
- 33.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, et al. (2009) Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol 16, 667–669 doi: 10.1038/nsmb.1604 [DOI] [PubMed] [Google Scholar]
- 34.Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang J, Kranz M, et al. (2010) Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol 17, 1152–1153 doi: 10.1038/nsmb.1892 [DOI] [PubMed] [Google Scholar]
- 35.Robinson MJ, Martin BA, Gootz TD, McGuirk PR, Moynihan M, Sutcliffe JA, et al. (1991) Effects of quinolone derivatives on eukaryotic topoisomerase II. A novel mechanism for enhancement of enzyme-mediated DNA cleavage. J. Biol. Chem 266, 14585–14592 [PubMed] [Google Scholar]
- 36.Kingma PS, Corbett AH, Burcham PC, Marnett LJ, Osheroff N (1995) Abasic sites stimulate double-stranded DNA cleavage mediated by topoisomerase II. DNA lesions as endogenous topoisomerase II poisons. J. Biol. Chem 270, 21441–21444 doi: 10.1074/jbc.270.37.21441 [DOI] [PubMed] [Google Scholar]
- 37.Kingma PS, Osheroff N (1997) Spontaneous DNA Damage Stimulates Topoisomerase II-mediated DNA Cleavage. J. Biol. Chem 272, 7488–7493 doi: 10.1074/jbc.272.11.7488 [DOI] [PubMed] [Google Scholar]
- 38.Anderson VE, Zaniewski RP, Kaczmarek FS, Gootz TD, Osheroff N (2000) Action of Quinolones against Staphylococcus aureus Topoisomerase IV: Basis for DNA Cleavage Enhancement. Biochemistry 39, 2726–2732 doi: 10.1021/bi992302n [DOI] [PubMed] [Google Scholar]
- 39.Kreuzer KN Cozzarelli NR (1979) Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J. Bacteriol 140, 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerns RJ, Towle TR, Hiasa H (2017) Quinolone-based Compounds with Anticancer Activity. U.S. Patent Application Serial No.: 62/432,430
- 41.Schoeffler AJ, Berger JM (2005) Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans 33, 1465–1470 doi: 10.1042/BST0331465 [DOI] [PubMed] [Google Scholar]
- 42.Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, et al. (2011) Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science 333, 459–462 doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 43.Wu CC, Li YC, Wang YR, Li TK, Chan NL (2013) On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs. Nucleic Acids Res 41, 10630–10640 doi: 10.1093/nar/gkt828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR (2010) Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One 5, e11338 doi: 10.1371/journal.pone.0011338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayer C, Janin YL (2014) Non-quinolone inhibitors of bacterial type IIA topoisomerases: a feat of bioisosterism. Chem. Rev 114, 2313–2342 doi: 10.1021/cr4003984 [DOI] [PubMed] [Google Scholar]
- 46.Ehmann DE, Lahiri SD (2014) Novel compounds targeting bacterial DNA topoisomerase/DNA gyrase. Curr. Opin. Pharmacol 18, 76–83 doi: 10.1016/j.coph.2014.09.007 [DOI] [PubMed] [Google Scholar]
- 47.Champoux JJ, Dulbecco R (1972) An activity from mammalian cells that untwists superhelical DNA – a possible swivel for DNA replication. Proc. Natl. Acad. Sci. USA 69, 143–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R (1987) Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature 326, 414–416 doi: 10.1038/326414a0 [DOI] [PubMed] [Google Scholar]
- 49.Zhang H, Wang JC, Liu LF (1988) Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc. Natl. Acad. Sci. USA 85, 1060–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D (1993) DNA topoisomerase I is involved in both repression and activation of transcription. Nature 365, 227–232 doi: 10.1038/365227a0 [DOI] [PubMed] [Google Scholar]
- 51.Shykind BM, Kim J, Stewart L, Champoux JJ, Sharp PA (1997) Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev 11, 397–407 doi: 10.1101/gad.11.3.397 [DOI] [PubMed] [Google Scholar]
- 52.Zhang CX, Chen AD, Gettel NJ, Hsieh TS (2000) Essential functions of DNA topoisomerase I in Drosophila melanogaster. Dev. Biol 222, 27–40 doi: 10.1006/dbio.2000.9704 [DOI] [PubMed] [Google Scholar]
- 53.Kretzschmar M, Meisterernst M, Roeder RG (1993) Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc. Natl Acad. Sci. USA 90, 11508–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ju BG, Rosenfeld MG (2006) A breaking strategy for topoisomerase IIβ/PARP-1-dependent regulated transcription. Cell Cycle 5, 2557–2560 doi: 10.4161/cc.5.22.3497 [DOI] [PubMed] [Google Scholar]
- 55.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. (2006) A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 doi: 10.1126/science.1127196 [DOI] [PubMed] [Google Scholar]
- 56.Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan-Salis KY, et al. (2016) RNA polymerase II regulates topoisomerase I activity to favor efficient transcription. Cell 165, 357–371 doi: 10.1016/j.cell.2016.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ljungman M, Hanawalt PC (1996) The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogenesis 17, 31–35 [DOI] [PubMed] [Google Scholar]
- 58.Solier S, Ryan MC, Martin SE, Varma S, Kohn KW, Liu H, et al. (2013) Transcription poisoning by topoisomerase I is controlled by gene length, splice sites, and miR-142-3p. Cancer Res 73, 4830–4839 doi: 10.1158/0008-5472.CAN-12-3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.King IF, Yandava CN, Mabb AM, Hsiao JS, Huang HS, Pearson BL, et al. (2013) Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62 doi: 10.1038/nature12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Barceló JM, Lee B, Kohlhagen G, Zimonjic DB, Popescu NC, et al. (2001) Human mitochondrial topoisomerase I. Proc. Natl Acad. Sci. USA 98, 10608–10613 doi: 10.1073/pnas.191321998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Lyu YL, Wang JC (2002) Dual localization of human DNA topoisomerase IIIα to mitochondria and nucleus. Proc. Natl Acad. Sci. USA 98, 10608–10613 doi: 10.1073/pnas.192449499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Low RL, Orton S, Friedman DB (2003) A truncated form of DNA topoisomerase IIbeta associates with the mtDNA genome in mammalian mitochondria. Eur. J. Biochem 270, 4173–4186 doi: 10.1046/j.1432-1033.2003.03814.x [DOI] [PubMed] [Google Scholar]
- 63.Zhang H, Zhang YW, Yasukawa T, Dalla Rosa I, Khiati S, Pommier Y (2014) Increased negative supercoiling of mtDNA in TOP1mt knockout mice and presence of topoisomerases IIα and IIβ in vertebrate mitochondria. Nucleic Acids Res. 42, 7259–7267 doi: 10.1093/nar/gku384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang H, Pommier Y (2008) Mitochondrial topoisomerase I sites in the regulatory D-loop region of mitochondrial DNA. Biochemistry 47, 11196–11203 doi: 10.1021/bi800774b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sobek S, Dalla Rosa I, Pommier Y, Bornholz B, Kalfalah F, Zhang H, et al. (2013) Negative regulation of mitochondrial transcription by mitochondrial topoisomerase I. Nucleic Acids Res 41, 9848–9857 doi: https://doi.10.1093/nar/gkt768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Douarre C, Sourbier C, Dalla Rosa I, Brata Das B, Redon CE, Zhang H, et al. (2012) Mitochondrial topoisomerase I is critical for mitochondrial integrity and cellular energy metabolism. PLoS One 7, e41094 doi: 10.1371/journal.pone.0041094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khiati S, Baechler SA, Factor VM, Zhang H, Huang SY, Dalla Rosa I, et al. (2015) Lack of mitochondrial topoisomerase I (TOP1mt) impairs liver regeneration. Proc. Natl. Acad. Sci. USA 112, 11282–11287 doi: 10.1073/pnas.1511016112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu J, Feng L, Hsieh TS (2010) Drosophila topo IIIalpha is required for the maintenance of mitochondrial genome and male germ-line stem cells. Proc. Natl. Acad. Sci. USA 107, 6228–6233 doi: 10.1073/pnas.1001855107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H (1990) New topoisomerase essential for chromosome segregation in E. coli. Cell 63, 393–404 doi: 10.1016/0092-8674(90)90172-B [DOI] [PubMed] [Google Scholar]
- 70.Hiasa H, Marians KJ (1996) Two Distinct Modes of Strand Unlinking during Theta-type DNA Replication. J. Biol. Chem 271, 21529–21535 doi: 10.1074/jbc.271.35.21529 [DOI] [PubMed] [Google Scholar]
- 71.Zechiedrich EL, Khodursky AB, Cozzarelli NR (1997) Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev 11, 2580–2592 doi: 10.1101/gad.11.19.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drlica K (1990) Bacterial topoisomerases and the control of DNA supercoiling. Trends Genet 6, 433–437 doi: 10.1016/0168-9525(90)90306-Q [DOI] [PubMed] [Google Scholar]
- 73.Lavasani LS, Hiasa H (2001) A ParE-ParC Fusion Protein Is a Functional Topoisomerase. Biochemistry, 40, 8438–8443 doi: 10.1021/bi0155201 [DOI] [PubMed] [Google Scholar]
- 74.Trigueros S, Roca J (2002) A GyrB-GyrA fusion protein expressed in yeast cells is able to remove DNA supercoils but cannot substitute eukaryotic topoisomerase II. Genes Cells 7, 249–257 doi: 10.1046/j.1365-2443.2002.00516.x [DOI] [PubMed] [Google Scholar]
- 75.Tsai-Pflugfelder M, Liu LF, Liu AA, Tewey KM, Whang-Peng J, Knutsen T, et al. (1988) Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21–22. Proc. Natl. Acad. Sci. USA 85, 7177–71781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Austin CA, Sng JH, Patel S, Fisher LM (1993) Novel HeLa topoisomerase II is the II beta isoform: complete coding sequence and homology with other type II topoisomerases. Biochim. Biophys. Acta 1172, 283–291 [DOI] [PubMed] [Google Scholar]
- 77.Carpenter AJ, Porter AC (2004) Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIα mutant human cell line. Mol. Biol. Cell 15, 5700–5711 doi: 10.1091/mbc.E04-08-0732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McClendon AK, Rodriguez AC, Osheroff N (2005) Human topoisomerase II α rapidly relaxes positively supercoiled DNA - Implications for enzyme action ahead of replication forks. J. Biol. Chem 280, 39337–39345 doi: 10.1074/jbc.M503320200 [DOI] [PubMed] [Google Scholar]
- 79.Li H, Wang Y, Liu X (2008) Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIα in cell cycle progression. J. Biol. Chem 283, 6209–6221 doi: 10.1074/jbc.M709007200 [DOI] [PubMed] [Google Scholar]
- 80.Ramamoorthy M, Tadokoro T, Rybanska I, Ghosh AK, Wersto R, May A, et al. (2012) RECQL5 cooperates with topoisomerase IIα in DNA decatenation and cell cycle progression. Nucleic Acids Res 40, 1621–1635 doi: 10.1093/nar/gkr844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sandri MI, Hochhauser D, Aytonm P, Camplejohn RC, Whitehouse R, Turley H, et al. (1996) Differential expression of the topoisomerase IIα and β genes in human breast cancer. Br. J. Cancer 73, 1518–1524 doi: 10.1038/bjc.1996.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Isaacs RJ, Harris AL, Hickson ID (1996) Regulation of the human topoisomerase IIalpha gene promoter in confluence-arrested cells. J. Biol. Chem 271, 16741–16747 [DOI] [PubMed] [Google Scholar]
- 83.Goswami PC, Roti Roti JL, Hunt CR (1996) The cell cycle-coupled expression of topoisomerase IIalpha during S phase is regulated by mRNA stability and is disrupted by heat shock or ionizing radiation. Mol. Cell Biol 16, 1500–1508 doi: 10.1128/MCB.16.4.1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Turley H, Comley M, Houlbrook S, Nozaki N, Kikuchi A, Hickson ID, et al. (1997) The distribution and expression of the two isoforms of DNA topoisomerase II in normal and neoplastic human tissues. Br. J. Cancer 75, 1340–1346 doi: 10.1038/bjc.1997.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang X, Li W, Prescott ED, Burden SJ, Wang JC (2000) DNA topoisomerase IIβ and neural development. Science 287, 131–134 doi: 10.1126/science.287.5450.131 [DOI] [PubMed] [Google Scholar]
- 86.Lyu YL, Wang JC (2003) Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIβ. Proc. Natl Acad. Sci. USA 100, 7123–7128 doi: 10.1073/pnas.1232376100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lyu YL, Lin CP, Azarova AM, Cai L, Wang JC, Liu LF (2006) Role of topoisomerase IIβ in the expression of developmentally regulated genes. Mol. Cell. Biol 26, 7929–7941 doi: 10.1128/MCB.00617-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG (1998) Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 279, 1504–1513 doi: 10.1126/science.279.5356.1504 [DOI] [PubMed] [Google Scholar]
- 89.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ (1998) A model for the mechanism of human topoisomerase I. Science 279, 1534–1541 doi: 10.1126/science.279.5356.1534 [DOI] [PubMed] [Google Scholar]
- 90.Redinbo MR, Champoux JJ, Hol WG (2000) Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA. Biochemistry 39, 6832–6840 doi: 10.1021/bi992690t [DOI] [PubMed] [Google Scholar]
- 91.Krogh BO, Schuman S (2000) Catalytic mechanism of DNA topoisomerase IB. Mol. Cell 5, 1035–1041 doi: 10.1016/S1097-2765(00)80268-3 [DOI] [PubMed] [Google Scholar]
- 92.Yang Z, Champoux JJ (2001) The role of histidine 632 in catalysis by human topoisomerase I. J. Biol. Chem 276, 677–685 doi: 10.1074/jbc.M007593200 [DOI] [PubMed] [Google Scholar]
- 93.Krogh BO, Schuman S (2002) Proton relay mechanism of general acid catalysis by DNA topoisomerase IB. J. Biol. Chem 277, 5711–5714 doi: 10.1074/jbc.C100681200 [DOI] [PubMed] [Google Scholar]
- 94.Tian L, Claeboe CD, Hecht SM, Shuman S (2003) Guarding the genome: electrostatic repulsion of water by DNA suppresses a potent nuclease activity of topoisomerase IB. Mol. Cell 12, 199–208 doi: 10.1016/S1097-2765(03)00263-6 [DOI] [PubMed] [Google Scholar]
- 95.Lesher DT, Pommier Y, Stewart L, Redinbo MR (2002) 8-Oxoguanine rearranges the active site of human topoisomerase I. Proc. Natl. Acad. Sci. USA 99, 12102–12107 doi: 10.1073/pnas.192282699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB Jr., Stewart L (2002) The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. USA 99, 15387–15392 doi: 10.1073/pnas.242259599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng C, Kussie P, Pavletich N, Shuman S (1998) Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell 92, 841–850 doi: 10.1016/S0092-8674(00)81411-7 [DOI] [PubMed] [Google Scholar]
- 98.Lisby M, Olesen JR, Skouboe C, Krogh BO, Straub T, et al. (2001) Residues within the N-terminal domain of human topoisomerase I play a direct role in relaxation. J. Biol. Chem 276, 20220–20227 doi: 10.1074/jbc.M010991200 [DOI] [PubMed] [Google Scholar]
- 99.Interthal H, Quigley PM, Hol WG, Champoux JJ (2004) The role of lysine 532 in the catalytic mechanism of human topoisomerase I. J. Biol. Chem 279, 2984–2992 doi: 10.1074/jbc.M309959200 [DOI] [PubMed] [Google Scholar]
- 100.Goto T,Wang JC (1982) Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. J. Biol. Chem 257, 5866–5872 [PubMed] [Google Scholar]
- 101.Wang JC (1971) Interaction between DNA and an Escherichia coli protein omega. J. Mol. Biol 55, 523–533 doi: 10.1016/0022-2836(71)90334-2 [DOI] [PubMed] [Google Scholar]
- 102.Domanico PL, Tse-Dinh YC (1991) Mechanistic studies on E. coli DNA topoisomerase I: divalent ion effects. J. Inorg. Biochem 42, 87–96 doi: 10.1016/0162-0134(91)80035-G [DOI] [PubMed] [Google Scholar]
- 103.Been MD, Champoux JJ (1980) Breakage of single-stranded DNA by rat liver nicking-closing enzyme with the formation of a DNA-enzyme complex. Nucleic Acids Res 8, 6129–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Champoux JJ (1981) DNA is linked to the rat liver DNA nicking-closing enzyme by a phosphodiester bond to tyrosine. J. Biol. Chem 256, 4805–4809 [PubMed] [Google Scholar]
- 105.Sherratt DJ, Wigley DB (1998) Conserved themes but novel activities in recombinases and topoisomerases. Cell 93,149–152 doi: 10.1016/S0092-8674(00)81566-4 [DOI] [PubMed] [Google Scholar]
- 106.Wigley DB, Davies GJ, Dodson EJ, Maxwell A, Dodson G (1991) Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351, 624–629 doi: 10.1038/351624a0 [DOI] [PubMed] [Google Scholar]
- 107.Morais Cabral JH, Jackson AP, Smith CV, Shikotra N, Maxwell A, Liddington RC (1997) Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388, 903–906 [DOI] [PubMed] [Google Scholar]
- 108.Roca J, Wang JC (1992) The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell 71, 833–840 doi: 10.1016/0092-8674(92)90558-T [DOI] [PubMed] [Google Scholar]
- 109.Lindsley JE, Wang JC (1993) Study of allosteric communication between protomers by immunotagging. Nature 361, 749–750 doi: 10.1038/361749a0 [DOI] [PubMed] [Google Scholar]
- 110.Roca J, Wang JC (1994) DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell, 77, 609–616 doi: 10.1016/0092-8674(94)90222-4 [DOI] [PubMed] [Google Scholar]
- 111.Roca J, Berger JM, Harrison SC, Wang JC (1996) DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl. Acad. Sci. U.S.A 93, 4057–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kampranis SC, Bates AD, Maxwell A (1999) A model for the mechanism of strand passage by DNA gyrase. Proc. Natl. Acad. Sci. U.S.A 96, 8414–8419 doi: 10.1073/pnas.96.15.8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM (2010) A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature 465, 641–644 doi: 10.1038/nature08974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Noble CG, Maxwell A (2002) The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J. Mol. Biol 318, 361–371 doi: 10.1016/S0022-2836(02)00049-9 [DOI] [PubMed] [Google Scholar]
- 115.Pitts SL, Liou GF, Mitchenall LA, Burgin AB, Maxwell A, Neuman KC, et al. (2011) Use of divalent metal ions in the DNA cleavage reaction of topoisomerase IV. Nucleic Acids Res 39, 4808–4817 doi: 10.1093/nar/gkr018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dutta R, Inouye M (2000) GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci 25, 24–28 doi: 10.1016/S0968-0004(99)01503-0 [DOI] [PubMed] [Google Scholar]
- 117.Classen S, Olland S, Berger JM (2003) Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc. Natl. Acad. Sci. USA 100, 10629–10634 doi: 10.1073/pnas.1832879100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bellon S, Parsons JD, Wei YY, Hayakawa K, Swenson LL, Charifson PS, et al. (2004) Crystal Structures of Escherichia coli Topoisomerase IV ParE Subunit (24 and 43 Kilodaltons): a Single Residue Dictates Differences in Novobiocin Potency against Topoisomerase IV and DNA Gyrase. Antimicrob. Agents Chemother 48, 1856–1864 doi: 10.1128/AAC.48.1856-1864.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harkins TT, Lewis TJ, Lindsley JE (1998) Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry 37, 7299–7312 doi: 10.1021/bi9729108 [DOI] [PubMed] [Google Scholar]
- 120.Baird CL, Harkins TT, Morris SK, Lindsley JE (1999) Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl. Acad. Sci. U.S.A 96, 13685–13690 doi: 10.1073/pnas.96.24.13685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baird CL, Gordon MS, Andrenyak DM, Marecek JF, Lindsley JE (2001) The ATPase Reaction Cycle of Yeast DNA Topoisomerase II. SLOW RATES OF ATP RESYNTHESIS AND PiRELEASE. J. Biol. Chem 276, 27893–27898 doi: 10.1074/jbc.M102544200 [DOI] [PubMed] [Google Scholar]
- 122.Williams NL, Howells AJ, Maxwell A (2001) Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol 306, 969–984 doi: 10.1006/jmbi.2001.4468 [DOI] [PubMed] [Google Scholar]
- 123.Schmidt BH, Osheroff N, Berger JM (2012) Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity. Nat. Struct. Mol. Biol 19, 1147–1154 doi: 10.1038/nsmb.2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wendorff TJ, Schmidt BH, Heslop P, Austin CA, Berger JM (2012) The structure of DNA-bound human topoisomerase II alpha: conformational mechanisms for coordinating inter-subunit interactions with DNA cleavage. J. Mol. Biol 424, 109–124. doi: 10.1016/j.jmb.2012.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aravind L, Leipe DD, Koonin EV (1998) Toprim – a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res 26, 4205–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Harrison SC, Aggarwal AK (1990) DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem 59, 933–969 doi: 10.1146/annurev.bi.59.070190.004441 [DOI] [PubMed] [Google Scholar]
- 127.Dong KC, Berger JM (2007) Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature 450, 1201–1205 doi: 10.1038/nature06396 [DOI] [PubMed] [Google Scholar]
- 128.Fass D, Bogden CE, Berger JM (1999) Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat. Struct. Biol 6, 322–326 doi: 10.1038/7556 [DOI] [PubMed] [Google Scholar]
- 129.Laponogov I, Veselkov DA, Crevel IM, Pan XS, Fisher LM, Sanderson MR (2013) Structure of an ‘open’ clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport. Nucleic Acids Res 41, 9911–9923 doi: 10.1093/nar/gkt749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wall ME, Wani MC, Cook CE, Palmer KH, McPhail AT, Sim GA (1966) Plant antitumor agents. 1. The isolation and structure of camptothecin, a novel alkaloidal leukemia and tumor inhibitor from Camptotheca acuminata. J. Am. Chem. Soc 88, 3888–3890 doi: 10.1021/ja00968a057 [DOI] [Google Scholar]
- 131.Wall ME, Wani MC (1995) Camptothecin and taxol: discovery to clinic-thirteenth Bruce F. Cain Memorial Award Lecture. Cancer Res 55, 753–760 [PubMed] [Google Scholar]
- 132.Hsiang YH, Hertzberg R, Hecht S, Liu LF (1985) Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem 260, 14873–14878 [PubMed] [Google Scholar]
- 133.Hsiang YH, Liu LF (1988) Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin. Cancer Res 48, 1722–1726 [PubMed] [Google Scholar]
- 134.Hsiang YH, Lihou MG, Liu LF (1989) Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res 49, 5077–5082 [PubMed] [Google Scholar]
- 135.Tsao YP, Russo A, Nyamuswa G, Silber R, Liu LF (1993) Interaction between Replication Forks and Topoisomerase I-DNA Cleavable Complexes: Studies in a Cell-free SV40 DNA Replication System. Cancer Res 53, 5908–5914 [PubMed] [Google Scholar]
- 136.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB Jr., Stewart L (2002) The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. USA 99, 15387–15392 doi: 10.1073/pnas.242259599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chrencik JE, Staker BL, Burgin AB, Pourquier P, Pommier Y, Stewart L, et al. (2004) Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol 339, 773–384 doi: 10.1016/jmb.2004.03.077 [DOI] [PubMed] [Google Scholar]
- 138.Thomas CJ, Rahier NJ, Hecht SM (2004) Camptothecin: current perspectives. Bioorg. Med. Chem. Lett 12, 1585–1604 doi: 10.1016/j.bmc.2003.11.036 [DOI] [PubMed] [Google Scholar]
- 139.Lavergne O, Lesueur-Ginot L, Pla Rodas F, Kasprzyk PG, Pommier J, Demarquay D, et al. (1988) Homocamptothecins: synthesis and antitumor activity of novel E-ring-modified camptothecinanalogues. J. Med. Chem 41, 5410–5419 doi: 10.1021/jm980400I [DOI] [PubMed] [Google Scholar]
- 140.Lavergne O, Demarquay D, Kasprzyk PG, Bigg DC (2000) Homocamptothecins: E-ring modified CPT analogues. Ann. N. Y. Acad. Sci 922, 100–111 doi: 10.1111/j.1749-6632.2000.tb07029.x [DOI] [PubMed] [Google Scholar]
- 141.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH (2007) Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 448, 213–217 doi: 10.1038/nature05938 [DOI] [PubMed] [Google Scholar]
- 142.Yoshinari T, Matsumoto M, Arakawa H, Okada H, Noguchi K, Suda H, et al. (1995) Novel antitumor indolocarbazole compound 6-N-formylamino-12,13-dihydro-1,11- dihydroxy-13-(beta-D-glucopyranosyl)-5H-indolo[2,3-a]pyrrolo[3,4- c]carbazole-5,7(6H)-dione (NB-506): induction of topoisomerase I-mediated DNA cleavage and mechanisms of cell line-selective cytotoxicity. Cancer Res 55, 1310–1315 [PubMed] [Google Scholar]
- 143.Arakawa H, Iguchi T, Morita M, Yoshinari T, Kojiri K, Suda H, et al. (1995) Novel indolocarbazole compound 6-N-formylamino-12,13-dihydro-1,11-dihydroxy- 13-(beta-D-glucopyranosyl)-5H-indolo[2,3-a]pyrrolo-[3,4-c]carbazole- 5,7(6H)-dione (NB-506): its potent antitumor activities in mice. Cancer Res 55, 1316–1320 [PubMed] [Google Scholar]
- 144.Saif MW, Diasio RB (2005) Edotecarin: a novel topoisomerase I inhibitor. Clin. Colorectal. Cancer 5, 27–36 doi: 10.3816/CCC.2005.n.014 [DOI] [PubMed] [Google Scholar]
- 145.Strumberg D, Pommier Y, Paull K, Jayaraman M, Nagafuji P, Cushman M (1999) Synthesis of cytotoxic indenoisoquinoline topoisomerase I poisons. J. Med. Chem 42, 446–457 doi: 10.1021/jm9803323 [DOI] [PubMed] [Google Scholar]
- 146.Cushman M, Jayaraman M, Vroman JA, Fukunaga AK, Fox BM, Kohlhagen G, et al. (2000) Synthesis of new indeno[1,2-c]isoquinolines: cytotoxic non-camptothecin topoisomerase I inhibitors. J. Med. Chem 43, 3688–3698 doi: 10.1021/jm000029d [DOI] [PubMed] [Google Scholar]
- 147.Kummar S, Chen A, Gutierrez M, Pfister TD, Wang L, Redon C, et al. (2016) Clinical and pharmacologic evaluation of two dosing schedules of indotecan (LMP400), a novel indenoisoquinoline, in patients with advanced solid tumors. Cancer Chemother. Pharmacol 78, 73–81. doi: 10.1007/s00280-016-2998-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D, Stewart L, et al. (2005) Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem 48, 2336–2345 doi: 10.1021/jm049146p [DOI] [PubMed] [Google Scholar]
- 149.Lesher DT, Pommier Y, Stewart L, Redinbo MR (2002) 8-Oxoguanine rearranges the active site of human topoisomerase I. Proc. Natl. Acad. Sci. USA 99, 12102–12107 doi: 10.1073/pnas.192282699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gottlieb JA, Guarino AM, Call JB, Oliverio VT, Block JB (1970) Preliminary pharmacologic and clinical evaluation of camptothecin sodium (NSC-100880). Cancer Chemother. Rep 54, 461–470 [PubMed] [Google Scholar]
- 151.Moertel CG, Schutt AJ, Reitemeier RJ, Hahn RG (1972) Phase II study of camptothecin (NSC-100880) in the treatment of advanced gastrointestinal cancer. Cancer Chemother. Rep 56, 95–101 [PubMed] [Google Scholar]
- 152.Muggia FM, Creaven PJ, Hansen HH, Cohen MH, Selawry OS (1972) Phase I clinical trial of weekly and daily treatment with camptothecin (NSC-100880): correlation with preclinical studies. Cancer Chemother. Rep 56, 515–521 [PubMed] [Google Scholar]
- 153.Dancey J, Eisenhauer EA (1996) Current perspectives on camptothecins in cancer treatment. Br. J. Cancer 74, 327–338 doi: 10.1038/bjc.1996.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Garcia-Carbonero R, Supko JG (2002) Current perspectives on the clinical experience, pharmacology, and continued development of the camptothecins. Clin. Cancer Res 8, 641–661 [PubMed] [Google Scholar]
- 155.Martino E, Della Volpe S, Terribile E, Benetti E, Sakaj M, Centamore A, et al. (2017) The long story of camptothecin: From traditional medicine to drugs. Bioorg. Med. Chem. Lett 27, 701–707 doi: 10.1016/j.bmcl.2016.12.085 [DOI] [PubMed] [Google Scholar]
- 156.Negoro S, Fukuoka M, Masuda N, Takada M, Kusunoki Y, Matsui K, et al. (1991) Phase I study of weekly intravenous infusions of CPT-11, a new derivative of camptothecin, in the treatment of advanced non-small-cell lung cancer. J. Natl. Cancer Inst 83, 1164–1168 doi: 10.1093/jnci/83.16.1164 [DOI] [PubMed] [Google Scholar]
- 157.Grochow LB, Rowinsky EK, Johnson R, Ludeman S, Kaufmann SH, McCabe FL, et al. (1992) Pharmacokinetics and pharmacodynamics of topotecan in patients with advanced cancer. Drug Metab. Dispos 20, 706–713 [PubMed] [Google Scholar]
- 158.Rothenberg ML, Kuhn JG, Burris HA III, Nelson J, Eckardt JR, Tristan-Morales M, et al. (1995) Phase I and pharmacokinetic trial of weekly CPT-11. J. Clin. Oncol 11, 2194–2204 [DOI] [PubMed] [Google Scholar]
- 159.Rivory LP, Robert J (1995) Identification and kinetics of a β-glucuronide metabolite of SN-38 in human plasma after administration of the camptothecin derivative irinotecan. Cancer Chemother. Pharmacol 36, 176–179 [DOI] [PubMed] [Google Scholar]
- 160.Schellens JHM, Creemers GJ, Beijnen JH, Rosing H, de Boer-Dennert M, McDonald M, et al. (1996) Topotecan, an active drug in the second-line treatment of epithelial ovarian cancer: results of a large European phase II study. J. Clin. Oncol 14, 3056–3061 [DOI] [PubMed] [Google Scholar]
- 161.Schellens JHM, Creemers GJ, Beijnen JH, Rosing H, de Boer-Dennert M, McDonald M, et al. (1996) Bioavailability and pharmacokinetics of oral topotecan: a new topoisomerase I inhibitor. Br. J. Cancer 73, 1268–1271 doi: 10.1038/bjc.1996.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fassberg J, Stella VJ (1992) A kinetic and Mechanistic Study of the Hydrolysis of Camptothecin and Some Analogues. J. Pharm. Sci 81, 676–684 doi: 10.1002/jps.2600810718 [DOI] [PubMed] [Google Scholar]
- 163.Mi Z, Burke TG (1994) Differential Interactions of Camptothecin Lactone and Carboxylate Forms with Human Blood Components. Biochemistry 33, 10325–10336 doi: 10.1021/bi00200a013 [DOI] [PubMed] [Google Scholar]
- 164.Burke TG, Mi Z (1994) The structural basis of camptothecin interactions with human serum albumin: impact on drug stability. J. Med. Chem 37, 40–46 doi: 10.1021/jm00027a005 [DOI] [PubMed] [Google Scholar]
- 165.Croce AC, Bottiroli G, Supino R, Favini E, Zuco V, Zunino F (2004) Subcellular localization of the camptothecin analogues, topotecan and gimatecan. Biochem. Pharmacol 67, 1035–1045 doi: 10.1016/j.bcp.2003.10.034 [DOI] [PubMed] [Google Scholar]
- 166.de la Loza MC, Wellinger RE (2009) A novel approach for organelle-specific DNA damage targeting reveals different susceptibility of mitochondrial DNA to the anticancer drugs camptothecin and topotecan. Nucleic Acids Res 37, e26. doi: 10.1093/nar/gkn1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kingsbury WD, Boehm JC, Jakas DR, Holden KG, Hecht SM, Gallagher G et al. (1991) Synthesis of water soluble (aminoalkyl) camptothecin analogues: inhibition of topoisomerase I and antitumor activity. J. Med. Chem 34, 98–107 doi: 10.1021/jm00105a017 [DOI] [PubMed] [Google Scholar]
- 168.McCabe FL, Johnson RK (1994) Comparative activity of oral and parenteral topotecan in murine tumor models: efficacy of oral topotecan. Cancer Invest 12, 308–313 doi: 10.3109/07357909409023029 [DOI] [PubMed] [Google Scholar]
- 169.O’Reilly S (1999) Topotecan: what dose, what schedule, what route? Clin. Cancer Res 5, 3–5 [PubMed] [Google Scholar]
- 170.Sawada S, Matsuoka S, Nagata H, Furuta T, Yokokura T, Miyasaka T (1991) Synthesis and antitumor activity of 20(S)-camptothecin derivatives: A-ring modified and 7,10-disubstituted camptothecins. Chem. Pharm. Bull 39, 3183–3188 doi: 10.1248/cpb39.3183 [DOI] [PubMed] [Google Scholar]
- 171.Sawada S, Yokokura T, Miyasaka T (1996) Synthesis of CPT-11 (irinotecan hydrochloride trihydrate). Ann. N. Y. Acad. Sci, 803, 13–28 doi: 10.1111/j.1749-6632.1996.tb26372.x [DOI] [PubMed] [Google Scholar]
- 172.Satoh T, Hosokawa M, Atsumi R, Suzuki W, Hakusui H, Nagai E (1994) Metabolic activation of CPT-11, 7-ethyl-10-[4-(1-piperidino)-1- piperidino]carbonyloxycamptothecin, a novel antitumor agent, by carboxylesterase. Biol. Pharm. Bull 17, 662–664 [DOI] [PubMed] [Google Scholar]
- 173.Rivory LP, Bowles MR, Robert J, Pond SM (1996) Conversion of irinotecan (CPT-11) to its active metabolite, 7-ethyl-10-hydroxycamptothecin (SN-38), by human liver carboxylesterase. Biochem. Pharmacol 52, 1103–1111 doi: 10.1016/0006-2952(96)00457-1 [DOI] [PubMed] [Google Scholar]
- 174.Rothenberg ML (2000) Irinotecan (CPT-11): Recent Developments and Future Directions - Colorectal Cancer and Beyond. Oncologist 6, 66–80 doi: 10.1634/theoncologist.6-1-66 [DOI] [PubMed] [Google Scholar]
- 175.Brangi M, Litman T, Ciotti M, Nishiyama K, Kohlhagen G, Takimoto C, et al. (1999) Camptothecin resistance: role of the ATP-binding cassette (ABC), mitoxantrone-resistance half-transporter (MXR), and potential for glucuronidation in MXR-expressing cells. Cancer Res 59, 5938–5946 [PubMed] [Google Scholar]
- 176.Bates SE, Medina-Perez WY, Kohlhagen G, Antony S, Nadjem T, Robey RW, et al. (2004) ABCG2 mediates differential resistance to SN-38 (7-ethyl-10-hydroxycamptothecin) and homocamptothecins. Pharmacol. Exp. Ther 310, 836–842 doi: 10.1124/jpet.103.063149 [DOI] [PubMed] [Google Scholar]
- 177.Meng LH, Liao ZY, Pommier Y (2003) Non-camptothecin DNA topoisomerase I inhibitors in cancer therapy. Curr. Top. Med. Chem 3, 305–320 doi: 10.2174/1568026033452546 [DOI] [PubMed] [Google Scholar]
- 178.Pommier Y 2006. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 66, 789–802 doi: 10.1038/nrc1977 [DOI] [PubMed] [Google Scholar]
- 179.Pommier Y (2009) DNA Topoisomerase I Inhibitors: Chemistry, Biology, and Interfacial Inhibition. Chem. Rev 109, 2894–2902 doi: 10.1021/cr900097c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Teicher BA (2008) Next generation topoisomerase I inhibitors: Rationale and biomarker strategies. Biochem. Pharmacol 75, 1262–1271 doi: 10.1016/j.bcp.2007.10.016 [DOI] [PubMed] [Google Scholar]
- 181.Li TK, Houghton PJ, Desai SD, Daroui P, Liu AA, Hars ES, et al. (2003) Characterization of ARC-111 as a novel topoisomerase I-targeting anticancer drug. Cancer Res 63, 8400–8407 [PubMed] [Google Scholar]
- 182.Kurtzberg LS, Battle T, Rouleau C, Bagley RG, Agata N, Yao M, et al. (2008) Bone marrow and tumor cell colony-forming units and human tumor xenograft efficacy of noncamptothecin and camptothecin topoisomerase I inhibitors. Mol. Cancer Ther 7, 3212–3222 doi: 10.1158/1535-7163.MCT-08-0568 [DOI] [PubMed] [Google Scholar]
- 183.Zhou W, Dai Z, Chen Y, Wang H, Yuan Z (2012) High-Dimensional descriptor selection and computational QSAR modeling for antitumor activity of ARC-111 analogues based on Support Vector Regression (SVR). Int. J. Mol. Sci 13, 1161–1172 doi: 10.3390/ijms13011161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sánchez C, Méndez C, Salas JA (2006) Indolocarbazole natural products: occurrence, biosynthesis, and biological activity. Nat. Prod. Rep 23, 1007–1045 doi: 10.1039/b601930g [DOI] [PubMed] [Google Scholar]
- 185.Urasaki Y, Laco G, Takebayashi Y, Bailly C, Kohlhagen G, Pommier Y (2001) Use of camptothecin-resistant mammalian cell lines to evaluate the role of topoisomerase I in the antiproliferative activity of the indolocarbazole, NB-506, and its topoisomerase I binding site. Cancer Res 61, 504–508 [PubMed] [Google Scholar]
- 186.Pommier Y, Cushman M (2009) The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol. Cancer Ther 8, 1008–1014 doi: 10.1158/1535-7163.MCT-08-0706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. https://clinicaltrials.gov/ct2/show/study/NCT01051635?term=LMP400%2C+LMP776&cntry1=NA%3AUS&rank=1.
- 188.Kurtzberg LS, Roth S, Krumbholz R, Crawford J, Bormann C, Dunham S, et al. (2011) Genz-644282, a novel non-camptothecin topoisomerase I inhibitor for cancer treatment. Clin. Cancer Res 17, 2777–2787 doi: 10.1158/1078-0432.CCR-10-0542 [DOI] [PubMed] [Google Scholar]
- 189.Houghton PJ, Lock R, Carol H, Morton CL, Gorlick R, Anders Kolb E, et al. (2012) Testing of the topoisomerase 1 inhibitor Genz-644282 by the pediatric preclinical testing program. Pediatr. Blood Cancer 58, 200–209 doi: 10.1002/pbc.23016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yoshida H, Bogaki M, Nakamura M, Yamanaka LM, Nakamura S (1991) Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother 35, 1647–1650 doi: 10.1128/AAC.35.8.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bax BD, Chan PF, Eggleston DS, Fosberry A, Gentry DR, Gorrec F, et al. (2010) Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 466, 935–940 doi: 10.1038/nature09197 [DOI] [PubMed] [Google Scholar]
- 192.Chan PF, Srikannathasan V, Huang J, Cui H, Fosberry AP, Gu M, et al. (2015) Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin. Nat. Commun 6, 10048 doi: 10.1038/ncomms10048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Blower TR, Williamson BH, Kerns RJ, Berger JM (2016) Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A 113, 1706–1713 doi: 10.1073/pnas.1525047113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Laponogov I, Pan XS, Veselkov DA, Cirz RT, Wagman A, Moser HE, et al. (2016) Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones. Open Biol 6, 160157 doi: 10.1098/rsob.160157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Veselkov DA, Laponogov I, Pan XS, Selvarajah J, Skamrova GB, Branstrom A, et al. (2016) Structure of a quinolone-stabilized cleavage complex of topoisomerase IV from Klebsiella pneumoniae and comparison with a related Streptococcus pneumoniae complex. Acta Crystallogr. D Struct. Biol 72, 488–496 doi: 10.1107/S2059798316001212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Palu G, Valisena S, Ciarrochi G, Gatto B, Palumbo M (1992) Quinolone binding to DNA is mediated by magnesium ions. Proc. Natl. Acad. Sci. USA 89, 9671–9675. doi: 10.1073/pnas.89.20.9671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Noble CG, Barnard FM, Maxwell A (2003) Quinolone-DNA interaction: sequence-dependent binding to single-stranded DNA reflects the interaction within the gyrase-DNA complex. Antimicrob. Agents Chemother 47, 854–862 doi: 10.1128/AAC.47.3.854-862.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yoshida H, Bogaki M, Nakamura M, Nakamura S (1990) Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother 34, 1271–1272 doi: 10.1128/AAC.34.6.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang YR, Chen SF, Wu CC, Liao YW, Lin TS, Liu KT, et al. (2017) Producing irreversible topoisomerase II-mediated DNA breaks by site-specific Pt(II)-methionine coordination chemistry. Nucleic Acids Res in press doi: 10.1093/nar/gkx806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ince D, Zhang X, Silver LC, Hooper DC (2002) Dual Targeting of DNA Gyrase and Topoisomerase IV: Target Interactions of Garenoxacin (BMS-284756, T-3811ME), a New Desfluoroquinolone. Antimicrob. Agents Chemother 46, 3370–3380 doi: 10.1128/AAC.46.11.3370-3380.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Strahilevitz J, Hooper DC (2005): Dual targeting of topoisomerase IV and gyrase to reduce mutant selection: direct testing of the paradigm by using WCK-1734, a new fluoroquinolone, and ciprofloxacin. Antimicrob. Agents Chemother 49, 1949–1956 doi: 10.1128/AAC.49.5.1949-1956.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Azarova AM, Lyu YL, Lin CP, Tsai YC, Lau JY, Wang JC, et al. (2007) Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc. Natl. Acad. Sci. USA 104, 11014–11019 doi: 10.1073/pnas.0704002104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pendleton MJ, Lindsey RH Jr., Felix CA, Grimwade D, Osheroff N (2014) Topoisomerase II and leukemia. Ann. N.Y. Acad. Sci 1310, 98–110 doi: 10.1111/nyas.12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gomez L, Hack MD, Wu J, Wiener JJ, Venkatesan H, Santillan A Jr., et al. (2007) Novel pyrazole derivatives as potent inhibitors of type II topoisomerases. Part 1: synthesis and preliminary SAR analysis. Bioorg. Med. Chem. Lett 17, 2723–2727 doi: 10.1016/j.bmcl.2007.03.003 [DOI] [PubMed] [Google Scholar]
- 205.Black MT, Stachyra T, Platel D, Girard AM, Claudoon M, Bruneau JM, et al. (2008) Mechanism of action of the antibiotic NXL101, a novel nonfluoroquinolone inhibitor of bacterial type II topoisomerases. Antimicrob. Agents Chemother 52, 3339–3349 doi: 10.1128/AAC.00496-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Miller AA, Bundy GL, Mott JE, Skepner JE, Boyle TP, Harris DW, et al. (2008) Discovery and characterization of QPT-1, the progenitor of a new class of bacterial topoisomerase inhibitors. Antimicrob. Agents Chemother 52, 2806–2812 doi: 10.1128/AAC.00247-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Miles TJ, Barfoot C, Brooks G, Brown P, Chen D, Dabbs S, et al. (2011) Novel cyclohexyl-amides as potent antibacterials targeting bacterial type IIA topoisomerases. Bioorg. Med. Chem. Lett 21, 7483–7488 doi: 10.1016/j.bmcl.2011.09.114 [DOI] [PubMed] [Google Scholar]
- 208.Miles TJ, Axten JM, Barfoot C, Brooks G, Brown P, Chen D, et al. (2011) Novel amino-piperidines as potent antibacterials targeting bacterial type IIA topoisomerases. Bioorg. Med. Chem. Lett 21, 7489–7495 doi: 10.1016/j.bmcl.2011.09.117 [DOI] [PubMed] [Google Scholar]
- 209.Miles TJ, Hennessy AJ, Bax B, Brooks G, Brown BS, Brown P, et al. (2013) Novel hydroxyl tricyclics (e.g., GSK966587) as potent inhibitors of bacterial type IIA topoisomerases. Bioorg. Med. Chem. Lett 23, 5437–5441 doi: 10.1016/j.bmcl.2013.07.013 [DOI] [PubMed] [Google Scholar]
- 210.Miles TJ, Hennessy AJ, Bax B, Brooks G, Brown BS, Brown P, et al. (2016) Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases. Bioorg. Med. Chem. Lett 26, 2464–2469 doi: 10.1016/j.bmcl.2016.03.106 [DOI] [PubMed] [Google Scholar]
- 211.Ross JE, Scangarella-Oman NE, Flamm RK, Jones RN (2014) Determination of disk diffusion and MIC quality control guidelines for GSK2140944, a novel bacterial type II topoisomerase inhibitor antimicrobial agent. J. Clin. Microbiol 52, 2629–2632 doi: 10.1128/JCM.00656-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jones RN, Fedler KA, Scangarella-Oman NE, Ross JE,Flamm RK (2016) Multicenter Investigation of Gepotidacin (GSK2140944) Agar Dilution Quality Control Determinations for Neisseria gonorrhoeae ATCC 49226. Antimicrob. Agents Chemother 60, 4404–6. doi: 10.1128/AAC.00527-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jacobsson S, Golparian D, Alm RA, Huband M, Mueller J, Jensen JS, et al. (2014) High in vitro activity of the novel spiropyrimidinetrione AZD0914, a DNA gyrase inhibitor, against multidrug-resistant Neisseria gonorrhoeae isolates suggests a new effective option for oral treatment of gonorrhea. Antimicrob. Agents Chemother 58, 5585–5588 doi: 10.1128/AAC.03090-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Huband MD, Bradford PA, Otterson LG, Basarab GS, Kutschke AC, Giacobbe RA, et al. (2015) In vitro antibacterial activity of AZD0914, a new spiropyrimidinetrione DNA gyrase/topoisomerase inhibitor with potent activity against Gram-positive, fastidious Gram-Negative, and atypical bacteria. Antimicrob. Agents Chemother 59, 467–474 doi: 10.1128/AAC.04124-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Alm RA, Lahiri SD, Kutschke A, Otterson LG, McLaughlin RE, Whiteaker JD, et al. (2015) Characterization of the novel DNA gyrase inhibitor AZD0914: low resistance potential and lack of cross-resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother 59, 1478–1486 doi: 10.1128/AAC.04456-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Chan PF, Huang J, Bax BD, Gwynn MN (2013) Recent Developments in Inhibitors of Bacterial Type IIA Topoisomerases In Antibiotics: Targets, Mechanisms and Resistance, Gualerzi CO, Brandi L, Fabbretti A, Pon CL (Eds.) John Wiley & Sons, Hoboken, NJ, pp. 263–297 [Google Scholar]
- 217.Hiasa H, Yousef DO, Marians KJ (1996) DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J. Biol. Chem 271, 26424–26429 doi: 10.1074/jbc.271.42.26424 [DOI] [PubMed] [Google Scholar]
- 218.Chan PF, Germe T, Bax BD, Huang J, Thalji RK, Bacqué E, et al. (2017) Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase. Proc. Natl. Acad. Sci. USA 114, E4492–E4500 doi: 10.1073/pnas.1700721114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vetting MW, Hegde SS, Fajardo JE, Fiser A, Roderick SL, Takiff HE, et al. (2006) Pentapeptide repeat proteins. Biochemistry 45, 1–10 doi: 10.1021/bi052130w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hooper David C., Jacoby George A. (2016) Mechanisms of drug resistance: quinolone resistance. Ann. N Y Acad. Sci 1354, 12–31 doi: 10.1111/nyas.12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bateman A, Murzin AG, Teichmann SA (1998) Structure and distribution of pentapeptide repeats in Bacteria. Protein Sci 7, 1477–1480 doi: 10.1002/pro.5560070625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, et al. (2005) A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science 308, 1480–1483 doi: 10.1126/science.1110699 [DOI] [PubMed] [Google Scholar]
- 223.Hegde SS, Vetting MW, Mitchenall LA, Maxwell A, Blanchard JS (2011) Structural and biochemical analysis of the pentapeptide repeat protein EfsQnr, a potent DNA gyrase inhibitor. Antimicrob. Agents Chemother 55, 110–117 doi: 10.1128/AAC.01158-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Xiong X, Bromley EH, Oelschlaeger P, Woolfson DN, Spencer J (2011) Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: conserved surface loops direct the activity of a Qnr protein from a gram-negative bacterium. Nucleic Acids Res 39, 3917–3927 doi: 10.1093/nar/gkq1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Vetting MW, Hegde SS, Wang M, Jacoby GA, Hooper DC, Blanchard JS (2011) Structure of QnrB1, a plasmid-mediated fluoroquinolone resistance factor. J. Biol. Chem 286, 25265–25273 doi: 10.1074/jbc.M111.226936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tao J, Han J, Wu H, Hu X, Deng J, Fleming J, et al. (2013) Mycobacterium fluoroquinolone resistance protein B, a novel small GTPase, is involved in the regulation of DNA gyrase and drug resistance. Nucleic Acids Res 41, 2370–2381 doi: 10.1093/nar/gks1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Sengupta S, Nagaraja V (2008) YacG from Escherichia coli is a specific endogenous inhibitor of DNA gyrase. Nucleic Acids Res 36, 4310–4316 doi: 10.1093/nar/gkn355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Vos SM, Lyubimov AY, Hershey DM, Schoeffler AJ, Sengupta S, Nagaraja V, et al. (2014) Direct control of type IIA topoisomerase activity by a chromosomally encoded regulatory protein. Genes Dev 28, 1485–1497 doi: 10.1101/gad.241984.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Schimana J, Fiedler HP, Groth I, Süssmuth R, Beil W, Walker M, et al. (2000) Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tü 6040. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot 53, 779–787 doi: 10.7164/antibiotucs.53.779 [DOI] [PubMed] [Google Scholar]
- 230.Holzenkämpfer M, Walker M, Zeeck A, Schimana J, Fiedler HP (2002) Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tü 6040. II. Structure elucidation and biosynthesis. J. Antibiot 55, 301–307 doi: 10.7164/antibiotics.55.301 [DOI] [PubMed] [Google Scholar]
- 231.Sissi C, Vazquez E, Chemello A, Mitchenall LA, Maxwell A, Palumbo M (2010) Mapping simocyclinone D8 interaction with DNA gyrase: evidence for a new binding site on GyrB. Antimicrob. Agents Chemother 54, 213–220 doi: 10.1128/AAC.00972-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Flatman RH, Howells AJ, Heide L, Fiedler HP, Maxwell A (2005) Simocyclinone D8, an inhibitor of DNA gyrase with a novel mode of action. Antimicrob. Agents Chemother 49, 1093–1100 doi: 10.1128/AAC.49.3.1093-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Edwards MJ, Flatman RH, Mitchenall LA, Stevenson CE, Le TB, Clarke TA, et al. (2009) A crystal structure of the bifunctional antibiotic simocyclinone D8, bound to DNA gyrase. Science 326, 1415–1418 doi: 10.1126/science.1179123 [DOI] [PubMed] [Google Scholar]
- 234.Hearnshaw SJ, Edwards MJ, Stevenson CE, Lawson DM, Maxwell A (2014) A new crystal structure of the bifunctional antibiotic simocyclinone D8 bound to DNA gyrase gives fresh insight into the mechanism of inhibition. J. Mol. Biol 426, 2023–2033 doi: 10.1016/j.jmb.2014.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sadiq AA, Patel MR, Jacobson BA, Escobedo M, Ellis K, Oppegard LM, et al. (2010) Anti-proliferative effects of simocyclinone D8 (SD8), a novel catalytic inhibitor of topoisomerase II. Invest. New Drugs 28, 20–25 doi: 10.1007/s10637-008-9209-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Oppegard LM, Hamann BL, Streck KR, Ellis KC, Fiedler HP, Khodursky AB, et al. (2009) In vivo and in vitro patterns of the activity of simocyclinone D8, an angucyclinone antibiotic from Streptomyces antibioticus. Antimicrob. Agents Chemother 53, 2110–2119 doi: 10.1128/AAC.01440-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Oppegard LM, Nguyen T, Ellis KC, Hiasa H (2012) Inhibition of human topoisomerases I and II by simocyclinone D8. J. Nat. Prod 75, 1485–1489 doi: 10.1021/np300299y [DOI] [PubMed] [Google Scholar]
- 238.Miki T, Chang ZT, Horiuchi T (1984) Control of cell division by sex factor F in Escherichia coli. II. Identification of genes for inhibitor protein and trigger protein on the 42.84–43.6 F segment. J. Mol. Biol 174, 627–646 doi: 10.1016/0022-2836(84)90087-1 [DOI] [PubMed] [Google Scholar]
- 239.Miki T, Park JA, Nagao K, Murayama N, Horiuchi T (1992) Control of segregation of chromosomal DNA by sex factor F in Escherichia coli. J. Mol. Biol 225, 39–52 doi: 10.1016/0022-2836(92)91024-J [DOI] [PubMed] [Google Scholar]
- 240.Dao-Thi MH, Van Melderen L, De Genst E, Afif H, Buts L, Wyns L, et al. (2005) Molecular basis of gyrase poisoning by the addiction toxin CcdB. J. Mol. Biol 348, 1091–1102 doi: 10.1016/jmb.2005.03.049 [DOI] [PubMed] [Google Scholar]
- 241.Smith AB, Maxwell A (2006) A strand-passage conformation of DNA gyrase is required to allow the bacterial toxin, CcdB, to access its binding site. Nucleic Acids Res 34, 4667–4676 doi: 10.1093/nar/gk636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Herman EH, Witial DT, Hellmann K, Waravdekar VS (1982) Biological properties of ICRF-159 and related bis(dioxopiperazine) compounds. Adv. Pharmacol. Chemother 19, 249–290 doi: 10.1016/s1054-3589(08)60025-3 [DOI] [PubMed] [Google Scholar]
- 243.Ishida R, Miki T, Narita T, Yui R, Sato M, Utsumi KR, et al. (1991) Inhibition of intracellular topoisomerase II by antitumor bis(2,6-dioxopiperazine) derivatives: mode of cell growth inhibition distinct from that of cleavable complex-forming type inhibitors. Cancer Res 51, 4909–4916 [PubMed] [Google Scholar]
- 244.Roca J, Ishida R, Berger JM, Andoh T, Wang JC (1994) Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc. Natl. Acad. Sci. USA 91, 1781–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Chang S, Hu T, Hsieh TS (1998) Analysis of a core domain in Drosophila DNA topoisomerase II. Targeting of an antitumor agent ICRF-159. J. Biol. Chem 273, 19822–19828 doi: 10.1074/jbc.273.31.19822 [DOI] [PubMed] [Google Scholar]
- 246.Olland S, Wang JC (1999) Catalysis of ATP hydrolysis by two NH(2)-terminal fragments of yeast DNA topoisomerase II. J. Biol. Chem 274, 21688–21694 doi: 10.1074/jbc.274.31.21688 [DOI] [PubMed] [Google Scholar]
- 247.Morris SK, Baird CL, Lindsley JE (2000) Steady-state and Rapid Kinetic Analysis of Topoisomerase II Trapped as the Closed-clamp Intermediate by ICRF-193. J. Biol. Chem 275, 2613–2618 doi: 10.1074/jbc.275.4.2613 [DOI] [PubMed] [Google Scholar]
- 248.Sehested M, Jensen PB, Sørensen BS, Holm B, Friche E, Demant EJ (1993) Antagonistic effect of the cardioprotector (+)-1,2-bis(3,5-dioxopiperazinyl-1-yl)propane (ICRF-187) on DNA breaks and cytotoxicity induced by the topoisomerase II directed drugs daunorubicin and etoposide (VP-16). Biochem. Pharmacol 46, 389–393 [DOI] [PubMed] [Google Scholar]
- 249.Hasinoff BB, Kushchak TI, Yalowich JC, Creighton AM (1995) A QSAR study comparing the cytotoxicity and DNA topoisomerase inhibitory effects of bisdioxopiperazine analogs of ICRF-187 (dexrazoxane). Biochem. Pharmacol 50, 953–958 doi: 10.1016/0006-2952(95)00218-O [DOI] [PubMed] [Google Scholar]
- 250.Mizuuchi K, O’Dea MH, Gellert M (1978) DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc. Natl. Acad. Sci. USA 75, 5960–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Sugino A, Higgins NP, Brown PO, Peebles CL, Cozzarelli NR (1978) Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc. Natl. Acad. Sci. USA 75, 4838–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Lewis RJ, Singh OM, Smith CV, Skarzynski T, Maxwell A, Wonacott AJ, et al. (1996) The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J 15, 1412–1420 [PMC free article] [PubMed] [Google Scholar]
- 253.Lamour V, Hoermann L, Jeltsch JM, Oudet P, Moras D (2002) An open conformation of the Thermus thermophilus gyrase B ATP-binding domain. J. Biol. Chem 277, 18947–18953 doi: 10.1074/jbc.M111740200 [DOI] [PubMed] [Google Scholar]
- 254.Lafitte D, Lamour V, Tsvetkov PO, Makarov AA, Klich M, Deprez P, et al. (2002) DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5’-methyl group of the noviose. Biochemistry 41, 7217–7223 doi: 10.1021/bi0159837 [DOI] [PubMed] [Google Scholar]
- 255.Gillbert EJ, Maxwell A (1994) The 24 kDa N-terminal sub-domain of the DNA gyrase B protein binds coumarin drugs. Mol. Microbiol 12, 365–373 doi: 10.1111/j.1365-2958.1994.tb01026.x [DOI] [PubMed] [Google Scholar]
- 256.Holmes ML, Dyall-Smith ML (1991) Mutations in DNA gyrase result in novobiocin resistance in halophilic archaebacteria. J. Bacteriol 173, 642–648 doi: 10.1128/jb.173.2.642-648.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Contreras A, Maxwell A (1992) gyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol. Microbiol 6, 1617–1624 doi: 10.1111/j.1365-2958.1992.tb00886.x [DOI] [PubMed] [Google Scholar]
- 258.Allan RK, Mok D, Ward BK, Ratajczak T (2006) Modulation of chaperone function and cochaperone interaction by novobiocin in the C-terminal domain of Hsp90: evidence that coumarin antibiotics disrupt Hsp90 dimerization. J. Biol. Chem 281, 7161–7171 doi: 10.1074/jbc.M512406200 [DOI] [PubMed] [Google Scholar]
- 259.Burlison JA, Neckers L, Smith AB, Maxwell A, Blagg BS (2006) Novobiocin: redesigning a DNA gyrase inhibitor for selective inhibition of hsp90. J. Am. Chem. Soc 128, 15529–15536 doi: 10.1021/ja065793p [DOI] [PubMed] [Google Scholar]
- 260.Burlison JA, Avila C, Vielhauer G, Lubbers DJ, Holzbeierlein J, Blagg BS (2008) Development of novobiocin analogues that manifest anti-proliferative activity against several cancer cell lines. J. Org. Chem 73, 2130–2137 doi: 10.1021/jo702191a [DOI] [PubMed] [Google Scholar]
- 261.Radanyi C, Le Bras G, Marsaud V, Peyrat JF, Messaoudi S, Catelli MG, et al. (2009) Antiproliferative and apoptotic activities of tosylcyclonovobiocic acids as potent heat shockprotein 90 inhibitors in human cancer cells. Cancer Lett 274, 88–94. doi: 10.1016/j.canlet.2008.09.001 [DOI] [PubMed] [Google Scholar]
- 262.Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP (1997) Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell 89, 239–250 doi: 10.1016/S0092-8674(00)80203-2 [DOI] [PubMed] [Google Scholar]
- 263.Ratain MJ, Rowley JD (1992) Therapy-related acute myeloid leukemia secondary to inhibitors of topoisomerase II - from the bedside to the target genes. Ann. Oncol 3, 107–111 [DOI] [PubMed] [Google Scholar]
- 264.Felix CA (1998) Secondary leukemias induced by topoisomerase-targeted drugs. Biochim. Biophys. Acta 1400, 233–255 doi: 10.1016/S0167-4781(98)00139-0 [DOI] [PubMed] [Google Scholar]
- 265.Kudo K, Yoshida H, Kiyoi H, Numata S, Horibe K, Naoe T (1998) Etoposide-related acute promyelocytic leukemia. Leukemia 12, 1171–1175 [DOI] [PubMed] [Google Scholar]
- 266.McClendon AK, Osheroff N (2007) DNA topoisomerase II, genotoxicity, and cancer. Mutat. Res 623, 83–97 doi: 10.1016/j.mrfmmm.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L (2004) Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev 56, 185–229 doi: 10.1124/pr.56.2.6 [DOI] [PubMed] [Google Scholar]
- 268.Ferreira ALA, Matsubara LS, Matsubara BB (2008) Anthracycline-induced cardiotoxicity. Cardiovasc. Hematol. Agents Medic. Chem 6, 278–281 doi: 10.2174/187152508785909474 [DOI] [PubMed] [Google Scholar]
- 269.Menna P, Salvatorelli E, Minotti G (2008) Cardiotoxicity of antitumor drugs. Chem. Res. Toxicol 21, 978–989 doi: 10.1021/tx800002r [DOI] [PubMed] [Google Scholar]
- 270.McGowan JV, Chung R, Maulik A, Piotrowska I, Walker JM, Yellon DM (2017) Anthracycline Chemotherapy and Cardiotoxicity. Cardiovasc. Drugs Ther 31, 63–75 doi: 10.1007/s10557-016-6711-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ratain MJ, Kaminer LS, Bitran JD, Larson RA, Le Beau MM, Skosey C, et al. (1987) Acute nonlymphocytic leukemia following etoposide and cisplatin combination chemotherapy for advanced non-small-cell carcinoma of the lung. Blood 70, 1412–1417 [PubMed] [Google Scholar]
- 272.Pedersen-Bjergaard J, Philip P, Larsen SO, Andersson M, Daugaard G, Ersbøll J, et al. (1993) Therapy-related myelodysplasia and acute myeloid leukemia. Cytogenetic characteristics of 115 consecutive cases and risk in seven cohorts of patients treated intensively for malignant diseases in the Copenhagen series. Leukemia 7, 1975–1986 [PubMed] [Google Scholar]
- 273.de Thé H, Chomienne C, Lanotte M, Degos L, Dejean A (1990) The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature 347, 558–561 doi: 10.1038/347558a0 [DOI] [PubMed] [Google Scholar]
- 274.Kakizuka A, Miller WH Jr., Umesono K, Warrell RP Jr., Frankel SR, Murty VV, et al. (1991) Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell 66, 663–674 doi: 10.1016/0092-8674(91090112-C [DOI] [PubMed] [Google Scholar]
- 275.Reiter A, Saussele S, Grimwade D, Wiemels JL, Segal MR, Lafage-Pochitaloff M, et al. (2003) Genomic anatomy of the specific reciprocal translocation t(15;17) in acute promyelocytic leukemia. Genes Chromosomes Cancer 36, 175–188 doi: 10.10022/gcc.10154 [DOI] [PubMed] [Google Scholar]
- 276.Hasan SK, Mays AN, Ottone T, Ledda A, La Nasa G, Cattaneo C, et al. (2008) Molecular analysis of t(15;17) genomic breakpoints in secondary acute promyelocytic leukemia arising after treatment of multiple sclerosis. Blood 112, 3383–3390 doi: 10.1182/blood-2007-10-115600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Super HJ, McCabe NR, Thirman MJ, Larson RA, Le Beau MM, Pedersen-Bjergaard J, et al. (1993) Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA-topoisomerase II. Blood 82, 3705–3711 [PubMed] [Google Scholar]
- 278.Domer PH, Head DR,Renganathan N, Raimondi SC, Yang E, Atlas M (1995) Molecular analysis of 13 cases of MLL/11q23 secondary acute leukemia and identification of topoisomerase II consensus-binding sequences near the chromosomal breakpoint of a secondary leukemia with the t(4; 11). Leukemia 9, 1305–1312 [PubMed] [Google Scholar]
- 279.Aplan PD, Chervinsky DS, Stanulla M, Burhans WC (1996) Site-specific DNA cleavage within the MLL breakpoint cluster region induced by topoisomerase II inhibitors. Blood 87, 2649–2658 [PubMed] [Google Scholar]
- 280.Strissel PL, Strick R, Rowley JD, Zeleznik-Le NJ (1998) An in vivo topoisomerase II cleavage site and a DNase I hypersensitive site colocalize near exon 9 in the MLL breakpoint cluster region. Blood 92, 3793–3803 [PubMed] [Google Scholar]
- 281.Felix CA (2001) Leukemias related to treatment with DNA topoisomerase II inhibitors. Med. Pediatr. Oncol 36, 525–535 doi: 10.1002/mpo.1125 [DOI] [PubMed] [Google Scholar]
- 282.Myers CE, McGuire WP, Liss RH, Ifrim I, Grotzinger K, Young RC (1977) Adriamycin: the role of lipid peroxidation in cardiac toxicity and tumor response. Science 197, 165–167 doi: 10.1126/science.877547 [DOI] [PubMed] [Google Scholar]
- 283.Davies KJ, Doroshow JH (1986) Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. J. Biol. Chem 261, 3060–3067 [PubMed] [Google Scholar]
- 284.Julicher RH, Sterrenberg L, Bast A, Riksen RO, Koomen JM, Noordhoek J (1986) The role of lipid peroxidation in acute doxorubicin-induced cardiotoxicity as studied in rat isolated heart. J. Pharm. Pharmacol 38, 277–282 doi: 10.1111/j.2042-7158.1986.tb04566.x [DOI] [PubMed] [Google Scholar]
- 285.Berthiaume JM, Wallace KB (2007) Adriamycin-induced oxidative mitochondrial cardiotoxicity. Cell Biol. Toxicol 23, 15–25 doi: 10.1007/s10565-006-0140-y [DOI] [PubMed] [Google Scholar]
- 286.Myers CE, Gianni L, Simone CB, Klecker R, Greene R (1982) Oxidative destruction of erythrocyte ghost membranes catalyzed by the doxorubicin-iron complex. Biochemistry 21, 1707–1712. [DOI] [PubMed] [Google Scholar]
- 287.Keizer HG, Pinedo HM, Schuurhuis GJ, Joenje H (1990) Doxorubicin (adriamycin): a critical review of free radical-dependent mechanisms of cytotoxicity. Pharmacol Ther 47, 219–231 doi: 10.1016/0163-7258(90)90088-J [DOI] [PubMed] [Google Scholar]
- 288.Xu X, Persson HL, Richardson DR (2005) Molecular pharmacology of the interaction of anthracyclines with iron. Mol. Pharmacol 68, 261–271 doi: 10.1124/mol.105.013383 [DOI] [PubMed] [Google Scholar]
- 289.Capranico G, Tinelli S, Austin CA, Fisher ML, Zunino F (1992) Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim. Biophys Acta 1132, 43–48 doi: 10.1016/0167-4781(92)90050-A [DOI] [PubMed] [Google Scholar]
- 290.Deng S, Yan T, Jendrny C, Nemecek A, Vincetic M, Godtel-Armbrust U et al. (2013) Dexrazoxane may prevent doxorubicin-induced DNA damage via depleting both topoisomerase II isoforms. BMC Cancer 14, 842 doi: 10.1186/1471-2407-14-842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lyu YL, Kerrigan JE, Lin CP, Azarova AM, Tsai YC, Ban Y, et al. (2007) Topoisomerase IIbeta-mediated DNA double-strand breaks: implications in doxorubicin cardiotoxicity and prevention by dexrazoxane. Cancer Res 67, 8839–8846 doi: 10.1158/AN-07-1649 [DOI] [PubMed] [Google Scholar]
- 292.Zhang S, Liu X, Bawa-Khalfe T, Lu LS, Lyu YL, Liu LF, et al. (2012) Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med 18, 1639–1642 doi: 10.1038/nm.2919 [DOI] [PubMed] [Google Scholar]
- 293.Hait WN, Choudhury S, Srimatkandada S, Murren JR (1993) Sensitivity of K562 human chronic myelogenous leukemia blast cells transfected with a human multidrug resistance cDNA to cytotoxic drugs and differentiating agents. J. Clin. Invest 91, 2207–2215 doi: 10.1172/JCI116447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmoller J, Johne A, et al. (2000) Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glyco-protein expression and activity in vivo. Proc. Natl. Acad. Sci. USA 97, 3473–3478 doi: 10.1073/pnas.97.7.3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sinkule JA (1984) Etoposide: a semisynthetic epipodophyllotoxin. Chemistry, pharmacology, pharmacokinetics, adverse effects and use as an antineoplastic agent. Pharmacotherapy 4, 61–73 [DOI] [PubMed] [Google Scholar]
- 296.Stahelin HF, von Wartburg A (1991) The chemical and biological route from podophyllotoxin glucoside to etoposide. Cancer Res 51, 5–15 [PubMed] [Google Scholar]
- 297.Hande KR (1998) Etoposide: four decades of development of a topoisomerase II inhibitor. Eur. J. Cancer 34, 1514–1521 doi: 10.1016/S0959-8049(98)00228-7 [DOI] [PubMed] [Google Scholar]
- 298.Witterland AH, Koks CH, Beijnen JH (1996) Etoposide phosphate, the water soluble prodrug of etoposide. Pharm. World Sci 18, 163–170 [DOI] [PubMed] [Google Scholar]
- 299.Greco FA, Hainsworth JD (1996) Clinical studies with etoposide phosphate. Semin. Oncol 23, 45–50 [PubMed] [Google Scholar]
- 300.Grem JL, Hoth DF, Leyland-Jones B, King SA, Ungerleider RS, Wittes RE (1988) Teniposide in the treatment of leukemia: a case study of conflicting priorities in the development of drugs for fatal diseases. J. Clin. Oncol 6, 351–379 doi: 10.1200/JCO.1988.6.2.351 [DOI] [PubMed] [Google Scholar]
- 301.Clark PI, Slevin ML (1987) The clinical pharmacology of etoposide and teniposide. Clin. Pharmacokinet 12, 223–252 doi: 10.2165/00003088-198712040-00001 [DOI] [PubMed] [Google Scholar]
- 302.Petros WP, Rodman JH, Relling MV, Christensen M, Pui CH, Rivera GK, et al. (1992) Variability in teniposide plasma protein binding is correlated with serum albumin concentrations. Pharmacotherapy 12, 273–277 doi: 10.1002/j.1875-91114.1992.tb04460.x [DOI] [PubMed] [Google Scholar]
- 303.Brockmann H (1963) Anthracyclinones and anthracyclines. (Rhodomycinone, pyrromycinone and their glycosides). Fortschr. Chem. Org. Naturst 21, 121–182 [PubMed] [Google Scholar]
- 304.Arcamone F, Cassinelli G, Fantini G, Grein A, Orezzi P, Pol C, et al. (1969) Adriamycin, 14-hydroxydaunomycin, a new antitumor antibiotic from S. peucetius var. caesius. Biotechnol. Bioeng 11, 1101–1110 [DOI] [PubMed] [Google Scholar]
- 305.Arcamone F (1982) Doxorubicin: Anticancer Antibiotics. Academic Press, San Diego. [Google Scholar]
- 306.Rivankar S (2014) An overview of doxorubicin formulations in cancer therapy. J. Cancer Res. Ther 10, 853–858 doi: 10.4103/0973-1482 [DOI] [PubMed] [Google Scholar]
- 307.Airoldi M, Amadori D, Barni S, Cinieri S, De Placido S, Di Leo A, et al. (2011) Clinical activity and cardiac tolerability of non-pegylated liposomal doxorubicin in breast cancer: a synthetic review. Tumori 97, 690–692. doi: 10.1700/1018.11082 [DOI] [PubMed] [Google Scholar]
- 308.Munshi NC, Anderson KC (2013) New strategies in the treatment of multiple myeloma. Clin. Cancer Res 19, 3337–3344 doi: 10.1158/1078-0432.CCR-12-1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Staropoli N, Ciliberto D, Botta C, Fiorillo L, Grimaldi A, Lama S, et al. (2014) Pegylated liposomal doxorubicin in the management of ovarian cancer: a systematic review and metaanalysis of randomized trials. Cancer Biol. Ther 15, 707–20. doi: 10.4161/cbt.28557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Gbabe OF, Okwundu CI, Dedicoat M, Freeman EE (2014) Treatment of severe or progressive Kaposi’s sarcoma in HIV-infected adults. Cochrane Database Syst. Rev 13, CD003256 doi: 10.1002/14651858.CD003256.pub2 [DOI] [PubMed] [Google Scholar]
- 311.Bulbake U, Doppalapudi S, Kommineni N, Khan W (2017) Liposomal Formulations in Clinical Use: An Updated Review. Pharmaceutics 9, pii: E12 doi: 10.3390/pharmaceutics9020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Dimarco A, Gaetani M, Dorigotti L, Soldati M, Bellini O (1964) Daunomycin: a new antibiotic with antitumor activity. Cancer Chemother. Rep 38, 31–38 [PubMed] [Google Scholar]
- 313.Behal V (2000) Bioactive products from Streptomyces. Adv. Appl. Microbiol 47, 113–156 doi: 10.1016/S0065-2164(00)47003-6 [DOI] [PubMed] [Google Scholar]
- 314.Bassan R, Lerede T, Rambaldi A, Buelli M, Viero P, Barbui T (1995) The use of anthracyclines in adult acute lymphoblastic leukemia. Haematologica 80, 280–291 [PubMed] [Google Scholar]
- 315.Petre CE, Dittmer DP, (2007) Liposomal daunorubicin as treatment for Kaposi’s sarcoma. Int. J. Nanomedicine 2, 277–288 [PMC free article] [PubMed] [Google Scholar]
- 316.Estey EH (2013) Acute myeloid leukemia: 2013 update on risk-stratification and management. Am. J. Hematol 88, 318–327 doi: 10.1002/ajh.23404 [DOI] [PubMed] [Google Scholar]
- 317.Raut LS (2015) Novel formulation of cytarabine and daunorubicin: A new hope in AML treatment. South Asian J. Cancer 4, 38–40. doi: 10.4103/2278-330X.149950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Gbabe OF, Okwundu CI, Dedicoat M, Freeman EE (2014) Treatment of severe or progressive Kaposi’s sarcoma in HIV-infected adults. Cochrane Database Syst. Rev 13, CD003256 doi: 10.1002/14651858.CD003256.pub2 [DOI] [PubMed] [Google Scholar]
- 319.Arcamone F (1987) Clinically useful doxorubicin analogues. Cancer Treat. Rev 14, 159–161 doi: 10.1016/0305-7372(87)90002-8 [DOI] [PubMed] [Google Scholar]
- 320.Cersosimo RJ, Hong WK (1986) Epirubicin: a review of the pharmacology, clinical activity and adverse effects of an Adriamycin analogue. J. Clin. Oncol 4, 425–439 doi: 10.1200/JCO.1986.4.3.425 [DOI] [PubMed] [Google Scholar]
- 321.Smith LA, Cornelius VR, Plummer CJ, Levitt G, Verrill M, Canney P, et al. (2010) Cardiotoxicity of anthracycline agents for the treatment of cancer: systematic review and meta-analysis of randomised controlled trials. BMC Cancer 10, 337 doi: 10.1186/1471-2407-10-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Tjuljandin SA, Doig RG, Sobol MM, Watson DM, Sheridan WP, Morstyn G, et al. (1990) Pharmacokinetics and toxicity of two schedules of high dose epirubicin. Cancer Res 50, 5095–101 [PubMed] [Google Scholar]
- 323.Robert J (1993) Epirubicin. Clinical pharmacology and dose-effect relationship. Drugs 45, 20–30 [DOI] [PubMed] [Google Scholar]
- 324.Todorov DK, Ninjo SS, Zeller WJ (1995) Antiproliferative activity of liposomal epirubicin on experimental human gliomas in vitro and in vivo after intratumoral/interstitial application. J. Cancer Res. Clin. Oncol 121, 164–168 [DOI] [PubMed] [Google Scholar]
- 325.Ju RJ, Zeng F, Liu L, Mu LM, Xie HJ, Zhao Y, et al. (2016) Destruction of vasculogenic mimicry channels by targeting epirubicin plus celecoxib liposomes in treatment of brain glioma. Int. J. Nanomedicine 11, 1131–1146. doi: 10.2147/IJN.S94467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Frederick CA, Williams LD, Ughetto G, Van der Marel GA, Van Boom JA, Rich A (1990) Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin. Biochemistry 29, 2538–2549 doi: 10.1021/bi00462a016 [DOI] [PubMed] [Google Scholar]
- 327.Pang B, Qiao X, Janssen L, Velds A, Groothuis T, Kerkhoven R, et al. (2013) Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat. Commun 4, 1908 doi: 10.1038/ncomms2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Pang B, de Jong J, Qiao X, Wessels LF, Neefjes J (2015) Chemical profiling of the genome with anti-cancer drugs defines target specificities. Nat. Chem. Biol 11, 472–480 doi: 10.1038/nchembio.1811 [DOI] [PubMed] [Google Scholar]
- 329.Agudelo D, Bourassa P, Bérubé G, Tajmir-Riahi HA (2016) Review on the binding of anticancer drug doxorubicin with DNA and tRNA: Structural models and antitumor activity. J. Photochem. Photobiol. B 158, 274–279 doi: 10.1016/j.jphotobiol.2016.02.032 [DOI] [PubMed] [Google Scholar]
- 330.Fox EJ (2004) Mechanism of action of mitoxantrone. Neurology 63, S15–S18 doi: 10.1212/WNL.63.12_suppl_6.S15 [DOI] [PubMed] [Google Scholar]
- 331.Evison BJ, Sleebs BE, Watson KG, Phillips DR, Cutts SM (2016) Mitoxantrone, More than Just Another Topoisomerase II Poison. Med. Res. Rev 36, 248–299 doi: 10.1002/med.21364 [DOI] [PubMed] [Google Scholar]
- 332.Thomas X, Archimbaud E (1997) Mitoxantrone in the treatment of acute myelogenous leukemia: a review. Hematol. Cell Ther 39, 63–74 [DOI] [PubMed] [Google Scholar]
- 333.Voltz R, Starck M, Zingler V, Strupp M, Kolb HJ (2004) Mitoxantrone therapy in multiple sclerosis and acute leukaemia: a case report out of 644 treated patients. Mult. Scler 10, 472–474 doi: 10.1191/1352458504ms1027cr [DOI] [PubMed] [Google Scholar]
- 334.Cocco E, Marrosu MG (2014) The current role of mitoxantrone in the treatment of multiple sclerosis. Expert Rev. Neurother 14, 607–616 doi: 10.1586/14737175.2014.915742 [DOI] [PubMed] [Google Scholar]
- 335.Yeh ET, Bickford CL (2009) Cardiovascular complications of cancer therapy: incidence, pathogenesis, diagnosis, and management. J. Am. Coll. Cardiol 53, 2231–2247 doi: 10.1016/j.jacc.2009.02.050 [DOI] [PubMed] [Google Scholar]
- 336.Damiani RM, Moura DJ, Viau CM, Caceres RA, Henriques JA, Saffi J (2016) Pathways of cardiac toxicity: comparison between chemotherapeutic drugs doxorubicin and mitoxantrone. Arch. Toxicol 90, 2063–2076 doi: 10.1007/s00204-016-1759-y [DOI] [PubMed] [Google Scholar]
- 337.Seifert CF, Nesser ME, Thompson DF (1994) Dexrazoxane in the prevention of doxorubicin-induced cardiotoxicity. Ann Pharmacother 28, 1063–1072 [DOI] [PubMed] [Google Scholar]
- 338.Jones RL (2008) Utility of dexrazoxane for the reduction of anthracycline-induced cardiotoxicity. Expert Rev. Cardiovasc. Ther 6, 1311–1317 doi: 10.1586/14779072.6.10.1311 [DOI] [PubMed] [Google Scholar]
- 339.Langer SW (2014) Dexrazoxane for the treatment of chemotherapy-related side effects. Cancer Manag. Res 6, 357–363 doi: 10.2147/CMAR.S47238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Hasinoff BB, Kala SV (1993) The removal of metal ions from transferrin, ferritin and ceruloplasmin by the cardioprotective agent ICRF-187 [(+)-1,2-bis(3,5-dioxopiperazinyl-1-yl)propane] and its hydrolysis product ADR-925. Agents Actions 39, 72–81 [DOI] [PubMed] [Google Scholar]
- 341.Malisza KL, Hasinoff BB (1996) Hydroxyl radical production by the iron complex of the hydrolysis product of the antioxidant cardioprotective agent ICRF-187 (dexrazoxane). Redox Rep 2, 69–73 doi: 10.1080/13510002.1996.11747029 [DOI] [PubMed] [Google Scholar]
- 342.Buss JL, Hasinoff BB (1993) The one-ring open hydrolysis product intermediates of the cardioprotective agent ICRF-187 (dexrazoxane) displace iron from iron-anthracycline complexes. Agents Actions 40, 86–95 [DOI] [PubMed] [Google Scholar]
- 343.Hasinoff BB, Herman EH (2007) Dexrazoxane: how it works in cardiac and tumor cells. Is it a prodrug or is it a drug? Cardiovasc. Toxicol 7, 140–144 doi: 10.1007/s12012-007-0023-3 [DOI] [PubMed] [Google Scholar]
- 344.Vavrova A, Jansova H, Mackova E, Machacek M, Haskova P, Tichotova L, et al. (2013) Catalytic inhibitors of topoisomerase II differently modulate the toxicity of anthracyclines in cardiac and cancer cells. PLoS One 8, e76676 doi: 10.1371/journal.pone.0076676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Larsen AK, Escargueil AE, Skladanowski A (2003) Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol. Ther 99, 167–181 doi: 10.1016/S0163-7258(03)00058-5 [DOI] [PubMed] [Google Scholar]
- 346.Lu J, Patel S, Sharma N, Soisson SM, Kishii R, Takei M, et al. (2014) Structures of Kibdelomycin Bound to Staphylococcus aureus GyrB and ParE Showed a Novel U-Shaped Binding Mode. ACS Chem. Biol 9, 2023–2031 doi: 10.1021/cb5001197 [DOI] [PubMed] [Google Scholar]