The second messenger c-di-GMP is a key regulator of bacterial physiology. The V. vulnificus genome encodes nearly 100 proteins predicted to make, break, and bind c-di-GMP. However, relatively little is known regarding the environmental signals that regulate c-di-GMP levels and biofilm formation in V. vulnificus. Here, we identify calcium as a primary environmental signal that specifically increases intracellular c-di-GMP concentrations, which in turn triggers brp-mediated biofilm formation. We show that PAPS, a metabolic intermediate of the sulfate assimilation pathway, acts as a second messenger linking environmental calcium and sulfur source availability to the production of another intracellular second messenger (c-di-GMP) to regulate biofilm and rugose colony formation, developmental pathways that are associated with environmental persistence and efficient bivalve colonization by this potent human pathogen.
KEYWORDS: PAPS, Vibrio, biofilms, c-di-GMP, extracellular signaling, metabolic regulation, second messenger
ABSTRACT
Poor clinical outcomes (disfigurement, amputation, and death) and significant economic losses in the aquaculture industry can be attributed to the potent opportunistic human pathogen Vibrio vulnificus. V. vulnificus, as well as the bivalves (oysters) it naturally colonizes, is indigenous to estuaries and human-inhabited coastal regions and must endure constantly changing environmental conditions as freshwater and seawater enter, mix, and exit the water column. Elevated cellular c-di-GMP levels trigger biofilm formation, but relatively little is known regarding the environmental signals that initiate this response. Here, we show that calcium is a primary environmental signal that specifically increases intracellular c-di-GMP concentrations, which in turn triggers expression of the brp extracellular polysaccharide that enhances biofilm formation. A transposon screen for the loss of calcium-induced PbrpA expression revealed CysD, an enzyme in the sulfate assimilation pathway. Targeted disruption of the pathway indicated that the production of a specific metabolic intermediate, 3′-phosphoadenosine 5′-phosphosulfate (PAPS), was required for calcium-induced PbrpA expression and that PAPS was separately required for development of the physiologically distinct rugose phenotype. Thus, PAPS behaves as a second messenger in V. vulnificus. Moreover, c-di-GMP and BrpT (the activator of brp expression) acted in concert to bias expression of the sulfate assimilation pathway toward PAPS and c-di-GMP accumulation, establishing a feed-forward regulatory loop to boost brp expression. Thus, this signaling network links extracellular calcium and sulfur availability to the intracellular second messengers PAPS and c-di-GMP in the regulation of V. vulnificus biofilm formation and rugosity, survival phenotypes underpinning its evolution as a resilient environmental organism.
INTRODUCTION
Bacteria can exist as free-living, planktonic cells or in surface-attached, multicellular biofilm communities that are composed of bacteria encased in a matrix of polysaccharides, DNA, and proteins (1). Biofilms are more resistant than their planktonic counterparts to environmental stressors such as antibiotic exposure, oxidative agents, ionic stress, and desiccation. The process of biofilm formation is tightly controlled at multiple stages, from approaching and probing a surface, to irreversible attachment and maturation, to biofilm dispersal (2). The second messenger c-di-GMP is a key biofilm regulatory molecule (3), and bacteria can sense diverse environmental signals and modulate their intracellular c-di-GMP levels accordingly. It is synthesized by diguanylate cyclases (DGCs) that bear a GGDEF domain and is degraded by phosphodiesterases (PDEs) that harbor EAL or HD-GYP domains (4). The GGDEF and EAL/HD-GYP domains of DGC and PDE are often associated with sensor domains that regulate their function in response to external cues such as oxygen level, light, nitric oxide, temperature, nutrient limitation, and surface contact (5). Additionally, DGC and PDE activity can be regulated at the level of gene expression. The downstream effects of c-di-GMP are mediated by effectors (proteins or nucleic acids such as riboswitches) that bind the molecule and elicit a change in gene expression or protein function (6).
In Vibrio vulnificus, elevated levels of intracellular c-di-GMP trigger biofilm formation by inducing expression of the brp-encoded exopolysaccharide (EPS) (7, 8). Excessive production of EPS and matrix proteins promotes development of the rugose phenotype, which is characterized by a distinct wrinkly colony morphology, and increased biofilm formation and stress resistance (9–11). brp expression is also dependent on the regulators BrpR and BrpT (7, 12). BrpR shares homology with VpsR of Vibrio cholerae, which has been shown to bind c-di-GMP (13, 14). BrpT binds directly to promoter regions upstream of brpA and brpH, but unlike its V. cholerae homologue VpsT, it does not depend on first binding c-di-GMP to do so (12, 15). However, the expression of brpT is regulated by BrpR (12). It was also recently shown that c-di-GMP, via BrpT, regulates the expression of the cabABC operon (16), which encodes a system for the secretion of a calcium binding matrix protein CabA that is required for biofilm and rugose colony formation (17).
The V. vulnificus genome encodes nearly 100 proteins predicted to synthesize, degrade, and bind c-di-GMP (18, 19), but relatively little is known regarding the environmental signals that regulate c-di-GMP levels and biofilm formation in response to changing environmental conditions. Vibrio vulnificus and the bivalves (oysters) it colonizes are autochthonous to estuary ecosystems (20, 21). These partially enclosed bodies of water are in constant flux due to the varying flows of freshwater (rainfall and snowmelt) and seawater (changing tides) that enter, mix with, and exit the water column (22). The salinity can range from 5 to 30 ppt, vary between estuaries, and change daily depending on the tides, weather, and anthropogenic factors. It was previously reported that calcium increased phase variation and biofilm formation in V. vulnificus (23, 24), but the mechanism remained nebulous. Here, we identify calcium as a primary environmental signal that specifically increases intracellular c-di-GMP concentrations, which in turn triggers subsequent brp-mediated biofilm formation. A screen for transposon (Tn) mutants that failed to respond to increased calcium revealed that sulfate assimilation is genetically linked to brp expression. We show that a specific intermediate of the sulfate assimilation pathway, 3′-phosphoadenosine 5′-phosphosulfate (PAPS), is required for calcium-induced brp expression. Moreover, PAPS production was also separately required for development of the physiologically distinct rugose phenotype. These results support the notion that PAPS is an intracellular second messenger linking environmental calcium and sulfur source availability to the production of another intracellular second messenger (c-di-GMP) to regulate the physiological responses of biofilm formation and rugosity, phenotypes that underpin the evolution of V. vulnificus as a successful environmental organism.
RESULTS
Environmental calcium regulates brp expression by moderating the intracellular c-di-GMP level.
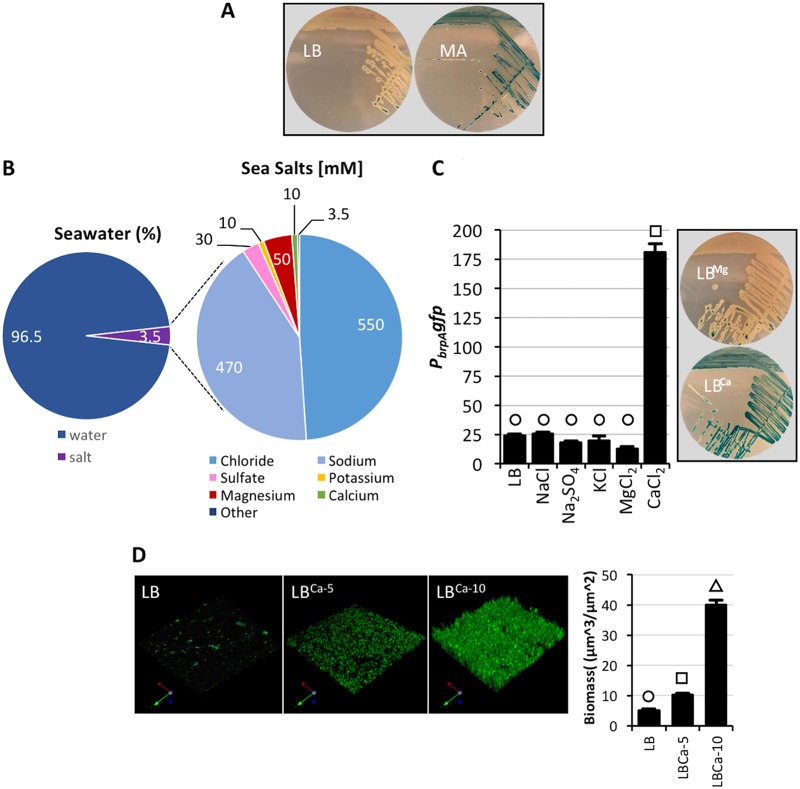
Our previous investigations indicated that brpA expression (monitored using a PbrpA lacZ reporter construct) was low in wild-type cells grown in LB. Conversely, the same reporter strain exhibited high-level induction when grown on marine agar (MA) (Fig. 1A), a medium that better reflects the composition of seawater. To identify the component(s) of MA that triggered brpA expression, the reporter strain was grown on LB that was systematically supplemented with the major mineral constituents of MA. While the major mono- and divalent cations present in seawater (Fig. 1B) had a negligible effect on brp expression when added to LB at typical seawater concentrations, strikingly, the addition of CaCl2 to 10 mM strongly induced PbrpA expression (Fig. 1C). These results suggested that calcium specifically regulated brp expression. Disruption of either brpF or brpJ (which encode a glycosyltransferase and a transport protein, respectively) did not affect calcium-induced brp expression (see Fig. S1 in the supplemental material), suggesting little or no feedback inhibition of PbrpA expression due to incomplete polysaccharide biosynthesis or failed export of a completed polysaccharide. Notably, while increasing Ca2+ concentrations enhanced V. vulnificus biofilm formation (Fig. 1D), it repressed V. cholerae biofilm formation under the same conditions (see Fig. S2 in the supplemental material). These results indicated that environmental calcium can have different effects on biofilm formation by closely related species.
FIG 1 .
Environmental calcium specifically regulates brp expression. (A) Wild-type V. vulnificus cells bearing a PbrpA lacZ reporter were grown on LB or marine agar (MA) plates containing X-Gal. (B) Pie charts showing the typical composition of salt in seawater (left) and a breakdown of the concentration (mM) of the major sea salts (right). Data are from the National Oceanographic Data Center (https://www.nodc.noaa.gov/). (C) PbrpA gfp expression in V. vulnificus grown in LB or LB supplemented with the major sea salts to concentrations found in seawater. Shown on the right are wild-type V. vulnificus cells bearing a PbrpA lacZ reporter growing on LB agar–X-Gal plates containing 50 mM MgCl2 (LBMg) or 10 mM CaCl2 (LBCa). (D) Fluorescent imaging of V. vulnificus biofilms growing in LB or LB containing 5 or 10 mM CaCl2 (LBCa-5 and LBCa-10). An axis indicator is shown in the bottom left corner of each panel. Statistically significant differences among the samples (P < 0.001 as determined by one-way ANOVA followed by pairwise comparisons with a Bonferroni adjustment) are indicated by different symbols above each bar.
Calcium-induced brp expression is not dependent on Brp production. The indicated wild-type (WT) and brpF and brpJ mutant strains were grown on LB and LBCa plates containing X-Gal. Download FIG S1, PDF file, 0.8 MB (789.2KB, pdf) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inverse effect of calcium on biofilm formation by V. vulnificus and V. cholerae. Wild-type V. vulnificus (Vvu) and V. cholerae (Vch) were grown in LB or LB containing 10 mM CaCl2 (LBCa). (Left) Images of the biofilm ring in culture tubes. (Right) Quantification of biofilm formation by crystal violet (CV) staining in 96-well plates. Statistically significant differences among the samples (P < 0.001 as determined by one-way ANOVA followed by pairwise comparisons with a Bonferroni adjustment) are indicated by different symbols above each bar. Download FIG S2, PDF file, 0.3 MB (366.6KB, pdf) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of the brp locus is regulated by the intracellular concentration of c-di-GMP (7, 12). To assess if the increase in brp expression in response to extracellular calcium correlated with an elevation in intracellular c-di-GMP levels, wild-type cells were grown in LB or LB supplemented with 50 mM MgCl2 or 15 mM CaCl2 (LBMg and LBCa, respectively), and biofilm formation and the intracellular c-di-GMP levels were quantified. Biofilm formation was low for cells grown in LB or LBMg but increased 7-fold in LBCa (Fig. 2A). The c-di-GMP level in cells that were grown in LBMg was half that of cells grown in LB (Fig. 2B). Conversely, the c-di-GMP level in cells that were grown in LBCa was 60% higher than that of cells grown in LB. Collectively, these data suggested that the intracellular c-di-GMP level in V. vulnificus increased in response to environmental calcium, which in turn regulated brp expression.
FIG 2 .
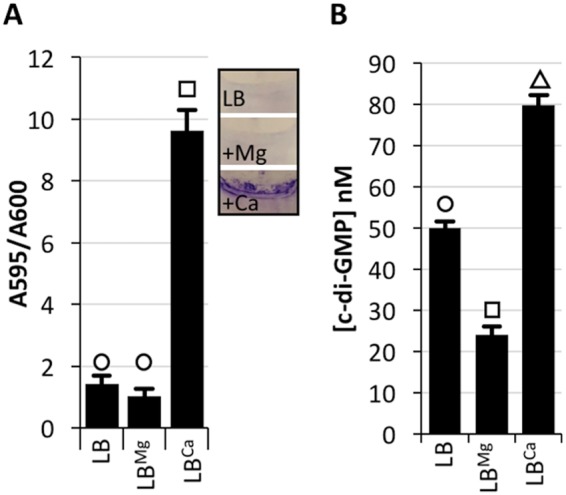
Calcium regulates brp expression and biofilm formation by increasing intracellular c-di-GMP levels. (A) V. vulnificus biofilm formation on coverslips during growth in LB or LB supplemented with 50 mM MgCl2 (LBMg) or 10 mM CaCl2 (LBCa). (B) LC-MS/MS quantification of c-di-GMP from whole-cell extract of V. vulnificus grown in the same medium. Plots show the mean, and error bars represent the standard deviation. Statistically significant differences among the samples (P < 0.001 as determined by one-way ANOVA followed by pairwise comparisons with a Bonferroni adjustment) are indicated by different symbols above each bar.
Cysteine biosynthesis and calcium-induced brp expression are genetically linked.
A V. vulnificus transposon mutant library was screened for the loss of PbrpA lacZ expression under calcium-inducing conditions to determine how an increase in exogenous calcium concentration resulted in increased brp expression (see Table S3 in the supplemental material). Multiple insertions mapped to brpT. This was anticipated since brpT is absolutely required for brp expression (7). Of particular interest was an insertion that mapped to cysD (aot11_12580), the gene coding for sulfate adenylyltransferase, which participates in cysteine biosynthesis (25). Our previous transcriptome sequencing (RNA-seq) analyses (16) indicated that cysD transcript levels increased when intracellular c-di-GMP levels were elevated but failed to do so in a brpT mutant. These data suggested that cysD expression was positively regulated by c-di-GMP and dependent on BrpT.
A cysD null mutant bearing the PbrpA lacZ reporter was constructed and grown on LBCa agar to verify that a functional CysD was indeed required for calcium-induced brpA expression. PbrpA expression was elevated in wild-type cells but not in the cysD mutant, and this defect could be complemented by expressing an ectopically integrated copy of cysD (Fig. 3A). Accordingly, PbrpA lacZ expression was low in LB but increased 4-fold in wild-type cells grown in LBCa (Fig. 3B). In contrast, a mutation in cysD prevented calcium induction of brpA expression, and this could be complemented by ectopic expression of cysD. Thus, a functional CysD was necessary for PbrpA expression in response to calcium.
FIG 3 .
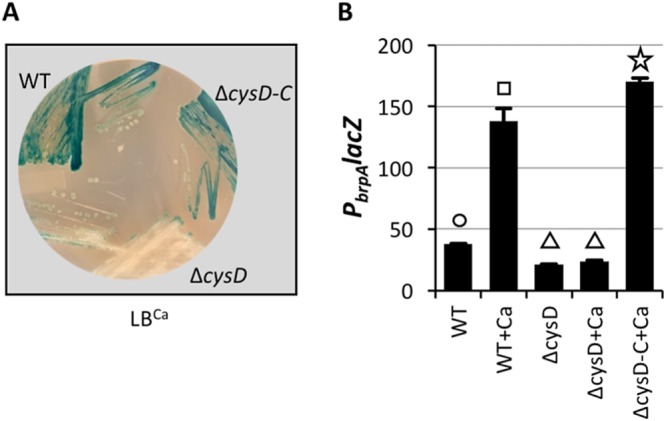
Deletion of cysD abrogates calcium-induced PbrpA expression. (A) The wild-type (WT), cysD mutant (ΔcysD), and complemented strains (ΔcysD-C) were grown on LBCa containing X-Gal. (B) The same strains were grown in LB ± 10 mM CaCl2 (+Ca) for quantification of PbrpA lacZ expression. Statistically significant differences among the samples (P < 0.001 as determined by one-way ANOVA followed by pairwise comparisons with a Bonferroni adjustment) are indicated by different symbols above each bar.
The sulfate assimilation metabolic intermediate PAPS mediates calcium-induced brp expression.
Disruption of cysD is anticipated to render cells auxotrophic for cysteine. However, the Tn libray screen that unearthed cysD was conducted on LB agar, which contains up to 0.3 mM cysteine (26). Previous reports of mutant screens on LB agar have resulted in the recovery of cysteine-auxotrophic mutants (27). To determine if the V. vulnificus cysD mutant was indeed auxotrophic, the ΔcysD PbrpA lacZ reporter strain was grown on minimal medium containing (MMcys) or lacking cysteine (MM). The wild-type strain grew well on both types of medium (see Fig. S3 in the supplemental material), while the mutant grew well on MMcys but failed to grow on MM. This confirmed that the cysD mutant was indeed auxotrophic for cysteine but that cysteine was not limiting for growth under the conditions used for Tn mutagenesis. Notably, these results suggested that the failure of a cysD mutant to induce brp expression in response to elevated calcium was unlikely to be rooted in its inability to synthesize cysteine itself. Rather, they implicated an intermediate in cysteine biosynthesis as requisite for calcium-induced PbrpA expression.
Auxotrophic phenotype of the cys mutants. The wild-type (WT) and cysD, cysC, and cysH mutant strains were inoculated onto minimal medium with (MMcys) or without (MM) 0.5 mM cysteine. Download FIG S3, PDF file, 0.7 MB (756.7KB, pdf) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The cysDNC operon participates in assimilatory sulfate activation in bacteria (see Fig. S4 in the supplemental material) (25). Environmental sulfate is transported into the cell, where CysD, together with CysN, then uses ATP and GTP to convert the acquired sulfate to adenosine 5′-phosphosulfate (APS). CysC, an adenylyl-sulfate kinase, uses another molecule of ATP to convert APS to 3′-phosphoadenosine 5′-phosphosulfate (PAPS), better known as activated sulfur. CysH is a sulfotransferase that converts PAPS to sulfite and adenosine 3′,5′-bisphosphate (PAP). Sulfite is converted to sulfide by the action of cysI and cysJ, which can then participate in sulfurylation reactions or be utilized in cysteine biosynthesis. To identify the sulfate assimilation intermediate regulating PbrpA expression, null mutations that systematically targeted specific steps of de novo cysteine biosynthesis were constructed. Deletions of cysD, cysN, cysC, and cysH were generated in the PbrpA lacZ reporter strain, and the mutants were assayed for growth in medium that included or lacked cysteine. All of the strains grew equally well on MMcys, but the mutants failed to grow on MM (Fig. S3), confirming their auxotrophic phenotype.
Part of the sulfate assimilation pathway leading to cysteine biosynthesis. Environmental sulfate is taken up and converted to adenosine 5′-phosphosulfate (APS) by the action of CysD and CysN. CysC converts APS to 3′-phosphoadenosine 5′-phosphosulfate (PAPS), which is then processed to adenosine 3′,5′-bisphosphate (PAP) and sulfite by CysH. PAP is converted to AMP by CysQ, and sulfite is reduced to sulfide by CysI and CysJ. CysK catalyzes the formation of l-cysteine from sulfide and O-acetylserine. Alkane sulfonates that are taken up can directly converted by mono- or dioxygenases to sulfite. Download FIG S4, PDF file, 0.2 MB (230.8KB, pdf) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Calcium-induced PbrpA lacZ expression was monitored in each mutant background. PbrpA expression in the ΔcysD, ΔcysC, and ΔcsyH ΔcysC mutants was reduced relative to wild type on LBCa and LB that lacked calcium (Fig. 4A and B). Conversely, PbrpA expression increased in the ΔcysH strain on LBCa and LB, and cellular c-di-GMP levels of the ΔcysH strain were nearly twice that of the wild type and the ΔcysC, ΔcysD, and ΔcysC ΔcysH mutants on LB and LBCa. These results suggested that mutations that interfered with the conversion of sulfate to PAPS (ΔcysD and ΔcysC) inhibited calcium-induced PbrpA expression, while a mutation that favored the accumulation of PAPS by restricting its conversion to sulfite (ΔcysH) had increased intracellular c-di-GMP levels and increased PbrpA expression.
FIG 4 .
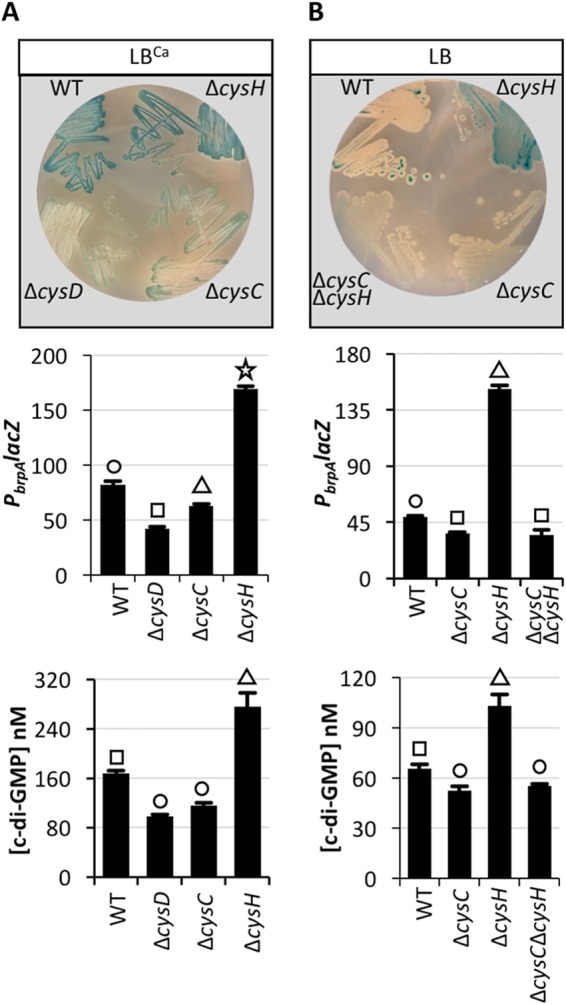
PbrpA expression is reduced in a ΔcysD or ΔcysC mutant and induced in a cysH mutant. The indicated strains were grown on LBCa (A) or LB (B) agar plates containing X-Gal (top), and the same strains were grown in LB ± 10 mM CaCl2 for quantification of PbrpA lacZ expression (middle) and the intracellular c-di-GMP concentration (bottom) in the WT and cys mutants. Statistically significant differences among the samples (P < 0.001 as determined by one-way ANOVA followed by pairwise comparisons with a Bonferroni adjustment) are indicated by different symbols above each bar.
Deletion of cysD does not prevent c-di-GMP-mediated PbrpA expression.
To determine if brp expression was absolutely dependent on PAPS production, the diguanylate cyclase, DcpA, was expressed in wild-type and ΔcysD cells to increase the intracellular c-di-GMP concentration and PbrpA lacZ expression was monitored. An equally robust increase in PbrpA expression was observed in both the wild-type and ΔcysD strains (Fig. 5). This suggested that elevated intracellular c-di-GMP levels could bypass the input of calcium, cysD, and PAPS in regulating brp expression.
FIG 5 .
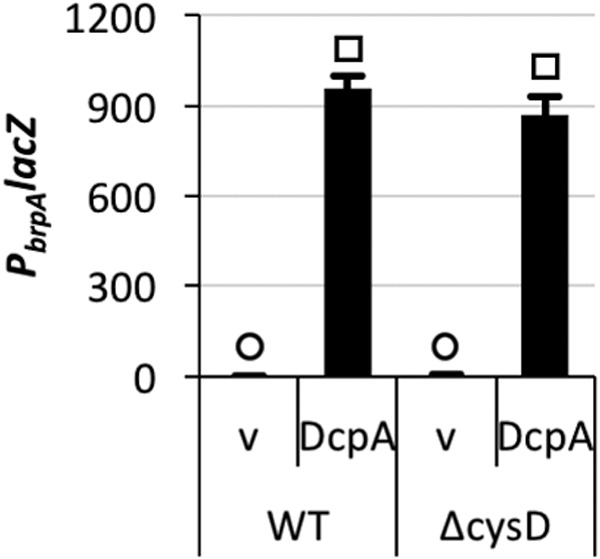
Elevated intracellular c-di-GMP levels alleviate the need for a functional cysD for calcium-induced PbrpA expression. PbrpA lacZ expression in the wild type and cysD mutant under unaltered (v [empty vector]) or elevated (DcpA) intracellular c-di-GMP levels. Statistically significant differences among the samples (P < 0.001 as determined by one-way ANOVA followed by pairwise comparisons with a Bonferroni adjustment) are indicated by different symbols above each bar.
PAPS production is required for rugosity.
The extent of rugose colony development in response to increased intracellular c-di-GMP concentrations in the wild-type and ΔcysD strains did not correlate with their comparably equal levels of PbrpA expression. Rugose colony formation and an associated increase in surface area were evident in wild-type cells expressing DcpA (Fig. 6A and B). A lesion in brpJ abrogated this phenotype, indicating that rugose colony development was dependent on the production of the brp polysaccharide. While there was little effect on rugose colony development when cysH was deleted, the ΔcysD and ΔcysC strains failed to become rugose when intracellular c-di-GMP levels were increased. Moreover, PbrpA expression increased in a ΔcysD background when intracellular c-di-GMP levels were elevated (Fig. 5), yet colonies failed to become rugose, and deletion of cysD abrogated pellicle formation at the air-liquid interface relative to wild-type cells (Fig. 6C and D). These results suggested that the ability to produce PAPS (which is reduced in ΔcysD and ΔcysC relative to wild-type and ΔcysH cells) was required for rugose colony development and likely pellicle formation in a manner that was distinct from its role in regulating PbrpA expression.
FIG 6 .
Mutations in cysC and cysD inhibit rugose colony formation in response to elevated intracellular c-di-GMP levels and calcium-induced pellicle formation. (A) Rugose colony development by the indicated strains under unaltered (v [empty vector]) or elevated (DcpA) intracellular c-di-GMP levels on LB agar plates supplemented with 0.5 mM cysteine (LBcys). (B) Quantification of the surface area of the rugose colonies in panel A. (C) The wild-type (WT) and cysD mutant strains were grown in LBcys or LBcys+Ca, and pellicle formation at the air-liquid interface was imaged. The left image in each panel is the top-down view of the pellicle, and to the right is the 3D rendering, where the dark-to-light transition indicates low-lying to elevated surface area regions. The scale is to the right in each panel. (D) Quantification of the pellicle surface area in panel C.
c-di-GMP and BrpT regulate the expression of genes in the sulfate assimilation pathway.
Our previous comparative transcriptomic analyses under unaltered and elevated cellular c-di-GMP conditions (12) suggested two groupings for the genes of sulfate assimilation: the expression of those involved in the conversion of sulfate to PAPS (cysDNC) increased when intracellular c-di-GMP concentrations were elevated, while those required for the production of cysteine from PAPS (cysHIJ) were downregulated (see Table S4 in the supplemental material). Moreover, the expression of cysDNC appeared to be positively regulated by BrpT. To confirm this, quantitative reverse transcription-PCR (qRT-PCR) was performed on RNA isolated from wild-type and ΔbrpT cells expressing DcpA or carrying the empty vector. In agreement with our RNA-seq data, the expression of cysD was strongly induced in wild-type cells under elevated intracellular c-di-GMP conditions (Fig. 7), while the expression of cysJ decreased 2-fold (see Fig. S5 in the supplemental material). Moreover, the c-di-GMP-mediated increase in cysD expression was negated in a brpT mutant, indicating that cysD is positively regulated by c-di-GMP and BrpT. Thus, not only does the production of PAPS trigger brp expression, but the activity of BrpT, the transcriptional activator of brp expression, biases the sulfur assimilation pathway to favor the accumulation of PAPS (i.e., the expression of genes directing PAPS production is increased while the expression of genes that would deplete the PAPS pool is repressed).
FIG 7 .
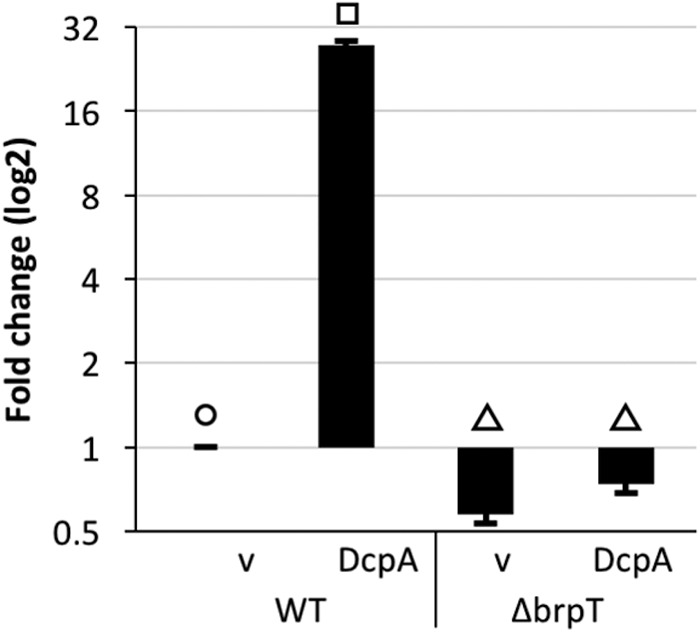
The expression of cysD is c-di-GMP and BrpT dependent. qRT-PCR was used to determine cysD transcript levels in wild-type (WT) and ΔbrpT cells under unaltered (v [empty vector]) and elevated (DcpA) intracellular c-di-GMP levels. Expression values are relative to those of WT under unaltered conditions. Statistically significant differences among the samples (P < 0.001 as determined by one-way ANOVA followed by pairwise comparisons with a Bonferroni adjustment) are indicated by different symbols above each bar.
The expression of cysJ is c-di-GMP-dependent. qRT-PCR was used to confirm that cysJ transcript levels decreased in wild-type cells when intracellular c-di-GMP levels were elevated (DcpA) relative to unaltered (v [empty vector]) conditions. Expression values are relative to those under unaltered conditions. Statistical significance was determined by the Student t test (two-tailed distribution with two-sample, equal variance). Download FIG S5, PDF file, 0.1 MB (22.2KB, pdf) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
c-di-GMP regulates the expression of the brp-encoded extracellular polysaccharide that promotes V. vulnificus biofilm formation and rugose colony development (7). Temperature is one of the few primary environmental signals known to affect brp expression in V. vulnificus (12). Here, we identify calcium as a primary signal regulating biofilm formation in V. vulnificus. Calcium, second to magnesium in divalent abundance in seawater (28), specifically triggered brp expression. Genetic screening revealed that an intermediate of the sulfate assimilation pathway (PAPS) was part of the regulatory circuitry altering brp expression in response to changes in environmental calcium concentration. Our genetic data suggested that genes required for the production of PAPS in response to extracellular calcium affected brp expression in V. vulnificus. Mutation of cysD, cysC, or cysH is known to alter the intracellular level of the PAPS intermediate in other bacteria (29–33). We showed that cysD and cysC null mutants, which cannot produce PAPS, could not induce PbrpA in response to increased environmental calcium, whereas a ΔcysH mutant, which accumulates PAPS, exhibited increased PbrpA expression even in the absence of added calcium. Although we did not directly quantify intracellular PAPS levels (the modified nucleotide is highly unstable) (34), intracellular c-di-GMP (and presumably PAPS) levels are elevated in a V. vulnificus cysH mutant relative to the wild type. Thus, mutations in genes that decreased (cysD and cysC) or increased (cysH) PAPS production influenced calcium-induced brp expression. Moreover, elevation of intracellular c-di-GMP levels increased the expression of genes required for PAPS production (cysDNC), and this increase was BrpT dependent. c-di-GMP also repressed the expression of cysHIJ, the products of which drain the PAPS pool. Thus, this feed-forward signaling network biased the sulfur metabolism pathway toward PAPS accumulation, which in turn increased cellular c-di-GMP levels, which increased brpT expression, which ultimately increased production of both Brp polysaccharide and PAPS. Disruption of brpF or brpJ (which encode a glycosyltransferase and a transport protein, respectively) did not affect calcium-induced brp expression, suggesting little or no feedback inhibition of PbrpA expression due to incomplete polysaccharide biosynthesis or failed export of a completed polysaccharide. It was recently reported that lesions in cysH, cysJ, cysK, and cysN negatively impacted rugose colony development in Vibrio fischeri (35). However, the defect was most severe in the cysK mutant, while disruption of cysN had little impact, suggesting that the mechanism must differ from that in V. vulnificus.
A second messenger is an intracellular signaling molecule whose concentration increases or decreases in response to the binding of an extracellular ligand to a cell surface receptor. Thus, PAPS behaves as a second messenger in V. vulnificus, akin to c-di-GMP. We propose that elevated environmental calcium increases PAPS production, which ultimately mediates brp expression by altering the cellular c-di-GMP pool. PAPS production may regulate eps production in Vibrio parahaemolyticus as well; lesions in cysD and cysC prevented pellicle formation at the air-liquid interface, a phenotype that is dependent on expression of the cps polysaccharide locus (36). A V. vulnificus cysD mutant exhibited the same defect. This is noteworthy since V. vulnificus and V. parahaemolyticus occupy similar environmental niches (37). Our data suggested that PAPS levels also affected the production of a biofilm matrix component, in an addition to the Brp polysaccharide, that is essential for rugosity. A candidate is the calcium-binding matrix protein CabA, which is required for biofilm and rugose colony formation (17). It was recently demonstrated that cabABC expression in V. vulnificus is c-di-GMP and BrpT dependent (16). Although elevation of the cellular c-di-GMP level (conditions under which CabA should be produced) overcame the calcium-induced PbrpA expression defect of the V. vulnificus cysD and cysC mutants, it was not sufficient to support rugose colony development. PAPS may influence CabA activity, or PAPS may regulate the production of another unidentified matrix component.
APS and PAPS are modified nucleotide intermediates of sulfate assimilation that have both been shown to have regulatory effects on gene expression. In Escherichia coli, APS acts as a signaling molecule that is directly bound by the transcriptional regulator Cbl. This results in the inhibition of genes required for organosulfonate utilization and thus helps to establish the hierarchy of preferred sources of sulfur (38). Expression of the csgBAC locus that governs curli production in E. coli is controlled by the regulator CsgD (39), and csgD expression is regulated by the PDE YciR (40). Hence, csgBAC expression is c-di-GMP dependent. It was recently demonstrated that csgBAC expression increased in a ΔcysH strain, while a ΔcysD ΔcysH double mutant or overexpression of cysH decreased csgBAC transcription (33). Since, csgBAC expression is affected by the intracellular concentration of both c-di-GMP and PAPS, and csgBAC expression increased on a ΔcysH strain, this suggests that PAPS accumulation must lead to altered cellular c-di-GMP concentrations and suggests that the signaling link we identified between the two second messengers in V. vulnificus may be evolutionarily conserved in E. coli and potentially other Gram-negative bacteria. Ultimately, PAPS appears to mediate the expression of genes for cell surface modification in response to environmental stimuli (curli in E. coli and EPS in V. vulnificus and V. parahaemolyticus). Our study clearly demonstrates interplay between PAPS and c-di-GMP signaling in V. vulnificus and PAPS was recently shown to impact cAMP receptor protein (CRP)-cAMP regulatory networks in E. coli (41), suggesting overlap in the pathways regulated by these three modified nucleotides (cAMP, c-di-GMP, and PAPS). Collectively, these studies highlight the widening regulatory influence of PAPS on bacterial physiology and its intersection with other modified nucleotide-regulated pathways positions PAPS as a potential global signaling molecule.
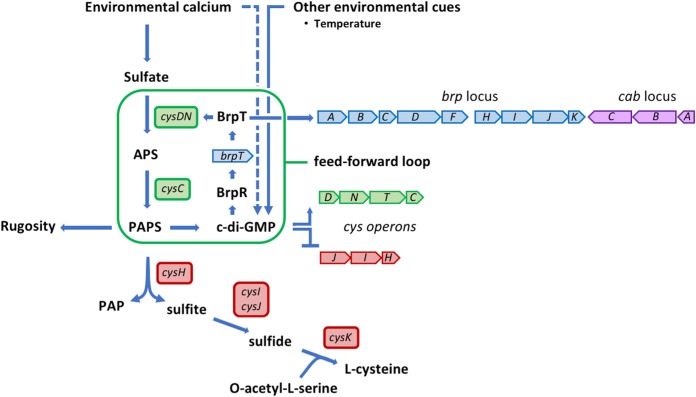
We propose the following model for the integration of environmental calcium signaling and PAPS production in the regulation of brp expression (Fig. 8). Environmental calcium upregulates sulfate assimilation. Sulfate that is taken up is converted to APS by the action of CysD and CysN and converted to PAPS by CysC. The accumulation of PAPS triggers an increase in the intracellular c-di-GMP level, which in turn increases brpT expression via the regulator BrpR, while simultaneously repressing expression of the cysJIH operon that normally drains the PAPS pool by processing it into sulfide and eventually cysteine. BrpT activates expression of the brp exopolysaccharide and cysDNTC operons, which generates more PAPS, c-di-GMP, and BrpT. Thus, a feed-forward regulatory network is established in which calcium, c-di-GMP, and BrpT bias the sulfate assimilation pathway toward the accumulation of PAPS and increased brp expression.
FIG 8 .
Model for the integration of environmental calcium signaling and PAPS production in the regulation of brp expression. BrpT and c-di-GMP bias the sulfate assimilation pathway toward the accumulation of PAPS in a feed-forward regulatory network (framed in green). Points of activation and repression are indicated by solid blue arrowheads and flat lines, respectively. Environmental calcium stimulates sulfate uptake. The sulfate is converted to adenosine 5′-phosphosulfate (APS) by the action of CysD and CysN and converted to 3′-phosphoadenosine 5′-phosphosulfate (PAPS) by CysC. The accumulation of PAPS triggers an increase in the intracellular c-di-GMP level, which increases brpT expression via the regulator BrpR and also represses expression of the cysJIH operon that drains the PAPS pool by processing it to PAP and sulfite and, eventually, to cysteine. BrpT activates expression of the brp exopolysaccharide, cab matrix genes, and cysDNTC operons (blue, purple, and green block arrows, respectively), which generates more PAPS, c-di-GMP, and BrpT, thus establishing a feed-forward signaling network that biases the sulfur metabolism pathway toward accumulating PAPS to increase brp expression and biofilm formation. PAPS also has a separate and distinct impact on rugose colony development. The operon structure of cysDNTC and cysJIH (red block arrow) allows for their simultaneous regulation by c-di-GMP and, for the regulation of cysDNTC, by BrpT. Environmental calcium may also influence cellular c-di-GMP levels independent of sulfate assimilation (dashed blue line). Other environmental cues (e.g., temperature) can directly affect intracellular c-di-GMP concentrations that may then feed into the PAPS–c-di-GMP loop.
Oysters and other bivalves have the capacity to locally concentrate calcium for shell growth and repair (42). Conceptually, the ability of V. vulnificus to sense a change in environmental calcium concentration and initiate biofilm formation at the transcriptional level makes it a perfect cue to promote niche (oyster) colonization by this pathogen. Calcium can repress Vibrio polysaccharide (VPS)-dependent biofilm formation by V. cholerae (43), although V. cholerae can also form distinct Ca2+-dependent/VPS-independent biofilms in which calcium is proposed to interact with negatively charged groups in the O antigen or capsule and physically mediate surface adhesion (44, 45). The different physiological biofilm responses of V. vulnificus and V. cholerae to calcium under certain conditions may help explain how these closely related species use similar biofilm networks yet evolved to occupy distinct niches in a shared environment. The adoption of calcium as a primary signaling molecule for V. vulnificus biofilm formation is ideal, and it is intriguing to think that V. vulnificus may have in fact evolved to exploit calcium as a molecular beacon to indicate preferred environments for colonization.
MATERIALS AND METHODS
Media and strains.
LB, LB agar, Bacto agar, and 2216 marine broth were purchased from BD Difco. Marine agar plates contained 1.5% Bacto agar. Antibiotics and additives were purchased from Sigma and used at the following concentrations: ampicillin, 100 µg/ml; chloramphenicol and gentamicin, 35 µg/ml; isopropyl-β-d-thiogalactopyranoside (IPTG), 100 µM; X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), 100 µg/ml; and l-arabinose, 0.05%. Minimal medium plates contained 40 mM HEPES (pH 7.2), 0.4% galactose, 1× M9 salts, 0.02% Difco yeast extract, and 1.5% Bacto agar. Strains are listed in Table S1 in the supplemental material.
Strains and plasmids used in this study. Download TABLE S1, DOCX file, 0.1 MB (116.5KB, docx) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fluorescent reporter fusion analysis.
The wild-type strain bearing the PbrpA gfp reporter was grown overnight, diluted 100-fold in fresh medium containing the indicated salts, and inoculated in triplicate into a 96-well plate. After 16 h at 30°C, the optical density at 600 nm (OD600) and florescence measurements were read on a BioTek Synergy H1 microplate reader (excitation, 485 nm; emission, 528 nm). Data of three technical replicates from three biological replicates were collected for all experiments. Plots show the mean, and error bars represent the standard deviation.
β-Galactosidase assays.
Quantification of β-galactosidase activity was carried out following the original Miller protocol (46) with the following amendments. Briefly, 1 OD600 unit of cells was pelleted by centrifugation for 10 min at 12,000 × g. Cell pellets were resuspended in 1 ml Z buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4, 0.05 M β-mercaptoethanol, pH 7.0). Next, 50 µl on 1% SDS was added and the mixture was vortexed. Chloroform (100 µl) was added, and the sample was vortexed for 10 s and then centrifuged for 2 min at 12,000 × g. A 150-µl aliquot of each sample was transferred to the well of a 96-well plate containing 50 µl (10 mg/ml) of o-nitrophenyl-β-d-galactopyranoside (ONPG) and placed in a BioTek Synergy HT microplate reader, where A420 and A595 measurements were taken in 5-min intervals. Modified Miller units were calculated using the formula described in the original assay. All assays were performed in triplicate.
Tn mutagenesis.
The mini-Tn10 delivery vector pNKTXI-SceI (47) was used for transposon mutagenesis. Cultures of V. vulnificus bearing a PbrpA lacZ reporter and E. coli bearing our Tn on a replicating plasmid were grown at 30 and 37°C, respectively. After overnight growth, cultures were diluted by 1:100 and grown to an OD600 of 0.5. One milliliter of each culture was washed of antibiotics by pelleting with centrifugation and resuspended in 100 µl of LB. The E. coli-V. vulnificus mixture was then placed on a nitrocellulose filter disc on an LB plate and placed at 37°C for conjugation. After overnight growth, cells from the filter disc were resuspended in 5 ml LB and plated onto LB with rifampin, 120 µg/ml X-Gal, 2 µg/ml chloramphenicol, 160 µg/ml kanamycin, and 15 mM CaCl2. After 48 h of growth, white colonies (representing loss of brp expression in the presence of calcium) were rescreened on plates containing a 10 mM mixture of rifampin, X-Gal, chloramphenicol, kanamycin, and CaCl2. A total of 30,000 mutants were screened on LBCa plates containing X-Gal for isolates exhibiting decreased PbrpA lacZ expression. Identified mutants were pooled, and collective genomic DNA was prepared and subjected to whole-genome sequencing at the Indiana University—Bloomington Center for Genomics and Bioinformatics (IUB-CGB). Geneious (Biomatters) was used to map the reads to the mini-Tn10 reference sequence and recover flanking V. vulnificus genomic sequences that were mapped back against the whole genome of strain ATCC 27562 (19). In-frame markerless deletions of target genes were generated by splicing by overhang extension (SOE) PCR (see below).
Mutant construction.
Allelic replacement of the target gene was achieved by SOE PCR as follows. PCR fragments corresponding to 3 kb upstream and downstream of the target gene (for primers, see Table S2 in the supplemental material) were amplified and stitched to a central trimethoprim or kanamycin antibiotic resistance cassette. Recipient strains carrying the pMMB-TfoX expression plasmid (48) were induced overnight in LB containing ampicillin and IPTG at 30°C. A 10-µl aliquot of cells was added to 500 µl of Instant Ocean at 20 ppt (IO-20) containing IPTG and 25 µl of the SOE PCR product. The transformation mixture was incubated statically overnight at 30°C. The next day, 1 ml of LB was added to the tube, and the cells were allowed to outgrow for 6 h before being plated on LB with selective antibiotics. Allelic replacement of the target gene was confirmed by PCR.
Primers used in this study. Download TABLE S2, DOCX file, 0.2 MB (158.9KB, docx) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rugose colony morphology assays and crystal violet staining of biofilms.
For visualization of PbrpA lacZ expression, strains were grown overnight on LB plates that included the appropriate antibiotics, supplements, and X-Gal at 30°C. Images were captured after 48 h using a Leica MS5 dissecting scope equipped with a Leica DC300F camera. To assess rugose colony morphology, 2-µl aliquots of overnight cultures were spotted onto the indicated LB plates and incubated at 30°C for overnight, followed by an additional day at room temperature. Colony morphology images were captured as described above. A fixed radius was used to determine the surface area for colonies with the SP package (49) in R (50). Corresponding 3D elevation maps were created with ImageJ (51). A representative image of samples done in triplicate is shown.
For crystal violet staining, plastic coverslips were placed into 12-well tissue culture plates containing 3 ml of LB medium plus amendments and inoculated with an exponentially growing culture of the indicated strain to an OD600 of 0.025. After 16 h of static growth at 30°C, coverslips were removed and placed into 4 ml of 0.1% crystal violet for 10 min to stain. The coverslips were rinsed 3 times in 4 ml VPBS (130 mM NaCl, 5 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4 [52]), and the violet stain was solubilized in 4 ml of 4:1 isopropanol-acetone. The A595 reading was normalized to the OD600 for each sample. All assays were done in triplicate.
qRT-PCR.
The primers used to monitor expression of the indicated genes are listed in Table S2. Strains carrying an arabinose-inducible plasmid expressing the DGC dcpA (7) were grown with antibiotics overnight at 30°C. The next morning, an aliquot was diluted 1:100 into fresh medium and grown to an OD600 of 0.5. The culture was then diluted into 5 ml of LB containing 0.1% arabinose (to induce DcpA expression) to an OD600 of 0.025 and grown for 5 h at 30°C. RNA was extracted from 1 OD unit of pelleted cells using the Bio Basic Total RNA miniprep kit. qRT-PCR was carried out using the SensiFAST SYBR Hi-ROX one-step kit. Reactions were performed on an Applied Biosystems StepOnePlus real-time PCR system. Data were analyzed using double threshold cycle (ΔCT) analysis. All assays were done in triplicate.
Monitoring biofilm development in microfluidic chambers.
Polydimethylsiloxane (PDMS)-glass flow cell devices containing eight 40- by 5- by 1-mm chambers were fabricated and sterilized (53). Mid-log-phase wild-type cells (OD600 of 0.1) were seeded into separate flow cell chambers. Control chambers for background fluorescence contained medium only. Initial attachment (no flow) proceeded for 20 min, followed by a flow rate of 0.25 ml min−1 in LB or LB supplemented with 5 or 10 mM CaCl2 for 16 h. The chambers were then flooded with the same medium containing 2.5 µM Syto21 (Molecular Probes) for 10 min. Biofilm images and z-stacks (20 × 1-µm slices) were captured with an Olympus IX83 microscope using a UPLSAPO 40× silicon oil immersion objective (NA, 1.25; WD, 0.3 mm). Background fluorescence was subtracted from each sample, and quantitative analysis to determine biomass was performed using cellSense (Olympus) and Comstat (54). Data from three biological replicates were analyzed for each strain. The images presented are from a single representative experiment.
Quantification of the intracellular c-di-GMP concentration.
c-di-GMP was extracted from whole cells as previously described (12). Briefly, the indicated strains were grown overnight in LB or LBCa with antibiotics when appropriate. The next day, cells were diluted 1:100 in fresh medium and grown to an OD600 of 0.5. Cells were again diluted to an OD600 of 0.025 into fresh LB supplemented with the indicated cations and l-Ara where indicated. Cells were grown for 12 h, when peak brp expression was observed in the presence of CaCl2. Six OD units of each sample was centrifuged at 4°C for 10 min at 12,000 × g. The pellet was resuspended in 200 µl of 40% (vol/vol) acetonitrile–40% (vol/vol) methanol–20% 0.1 N formic acid. This suspension was incubated at −20°C for 30 min and then centrifuged at 4°C for 5 min. The supernatant was recovered, and 10 µl of each sample was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a Quattro Premier XE mass spectrometer (Waters) coupled with an Acquity Ultra Performance LC system (Waters). c-di-GMP was detected by electrospray ionization using multiple reaction monitoring in negative-ion mode at m/z 689.16→344.31. The MS parameters were as follows: capillary voltage, 3.5 kV; cone voltage, 50 V; collision energy, 34 V; source temperature, 110°C; desolvation temperature, 350°C; cone gas flow (nitrogen), 50 liters/h; desolvation gas flow (nitrogen), 800 liters/h; collision gas flow (nitrogen), 0.15 ml/min; and multiplier voltage, 650 V. Chromatography separation was reverse phase using a Waters BEH C18 2.1- by 50-mm column at a flow rate of 0.3 ml/min with a gradient of 10 mM tributylamine plus 15 mM acetic acid in 97:3 water-methanol (solution A) to methanol (solution B) with the following parameters: t = 0 min; 99% A:1% B, t = 2.5 min; 80% A:20% B, t = 7.0 min; 35% A:65% B, t = 7.5 min; 5% A:95% B, t = 9.01 min; and 99% A:1% B, t = 10 min. A c-di-GMP standard curve for calculating the c-di-GMP concentration in each extract was generated by dissolving synthesized c-di-GMP (Axxora) in extraction buffer to 250 nM, followed by 2-fold serial dilutions to 0.975 nM. Data from three biological replicates were analyzed for each strain.
Statistical analyses.
Statistical significance was determined by the Student t test (two-tailed distribution with two-sample, equal variance) when directly comparing two conditions or a one-way analysis of variance (ANOVA) followed by pairwise comparisons with a Bonferroni adjustment when comparing data with multiple samples.
Tn insertions that disrupt calcium-induced PbrpA expression. Download TABLE S3, DOCX file, 0.1 MB (91KB, docx) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sulfate assimilation pathway genes regulated by BrpT and c-di-GMP. Download TABLE S4, DOCX file, 0.1 MB (53.8KB, docx) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENT
We thank Ankur Dalia for the pMMB-TfoX expression plasmid.
Footnotes
Citation Chodur DM, Coulter P, Isaacs J, Pu M, Fernandez N, Waters CM, Rowe-Magnus DA. 2018. Environmental calcium initiates a feed-forward signaling circuit that regulates biofilm formation and rugosity in Vibrio vulnificus. mBio 9:e01377-18. https://doi.org/10.1128/mBio.01377-18.
REFERENCES
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.O’Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 3.D’Argenio DA, Miller SI. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- 4.Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou SH, Galperin MY. 2016. Diversity of cyclic di-GMP-binding proteins and mechanisms. J Bacteriol 198:32–46. doi: 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd CD, O’Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Y, Rowe-Magnus DA. 2010. Identification of a c-di-GMP-regulated polysaccharide locus governing stress resistance and biofilm and rugose colony formation in Vibrio vulnificus. Infect Immun 78:1390–1402. doi: 10.1128/IAI.01188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrison-Schilling KL, Kaluskar ZM, Lambert B, Pettis GS. 2014. Genetic analysis and prevalence studies of the brp exopolysaccharide locus of Vibrio vulnificus. PLoS One 9:e100890. doi: 10.1371/journal.pone.0100890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Natl Acad Sci U S A 102:16819–16824. doi: 10.1073/pnas.0505350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A 96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y, Rowe-Magnus DA. 2011. Overlapping and unique contributions of two conserved polysaccharide loci in governing distinct survival phenotypes in Vibrio vulnificus. Environ Microbiol 13:2888–2990. doi: 10.1111/j.1462-2920.2011.02564.x. [DOI] [PubMed] [Google Scholar]
- 12.Chodur DM, Guo L, Pu M, Bruger E, Fernandez N, Waters C, Rowe-Magnus DA. 2017. The proline variant of the W[F/L/M][T/S]R cyclic di-GMP binding motif suppresses dependence on signal association for regulator function. J Bacteriol 199:e00344-17. doi: 10.1128/JB.00344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yildiz FHF, Dolganov NAN, Schoolnik GKG. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol 183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava D, Hsieh ML, Khataokar A, Neiditch MB, Waters CM. 2013. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol 90:1262–1276. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chodur DM, Rowe-Magnus DA. 2018. Complex control of a genomic island governing biofilm and rugose colony development in Vibrio vulnificus. J Bacteriol 200:e00190-18. doi: 10.1128/JB.00190-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JH, Jo Y, Jang SY, Kwon H, Irie Y, Parsek MR, Kim MH, Choi SH. 2015. The cabABC operon essential for biofilm and rugose colony development in Vibrio vulnificus. PLoS Pathog 11:e1005192. doi: 10.1371/journal.ppat.1005192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Römling U, Gomelsky M, Galperin MY. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 19.Rusch DB, Rowe-Magnus DA. 2017. Complete genome sequence of the pathogenic Vibrio vulnificus type strain ATCC 27562. Genome Announc 5:e00907-17. doi: 10.1128/genomeA.00907-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamplin M. 1995. The ecology of Vibrio vulnificus, p 75–86. In Proceedings of the 1994 Vibrio vulnificus Workshop. Food and Drug Administration, Washington, DC. [Google Scholar]
- 21.Oliver JD. 2015. The biology of Vibrio vulnificus. Microbiol Spectr 3. doi: 10.1128/microbiolspec.VE-0001-2014. [DOI] [PubMed] [Google Scholar]
- 22.Cloern JE, Abreu PC, Carstensen J, Chauvaud L, Elmgren R, Grall J, Greening H, Johansson JOR, Kahru M, Sherwood ET, Xu J, Yin K. 2016. Human activities and climate variability drive fast-paced change across the world’s estuarine-coastal ecosystems. Glob Chang Biol 22:513–529. doi: 10.1111/gcb.13059. [DOI] [PubMed] [Google Scholar]
- 23.Garrison-Schilling KL, Grau BL, McCarter KS, Olivier BJ, Comeaux NE, Pettis GS. 2011. Calcium promotes exopolysaccharide phase variation and biofilm formation of the resulting phase variants in the human pathogen Vibrio vulnificus. Environ Microbiol 13:643–654. doi: 10.1111/j.1462-2920.2010.02369.x. [DOI] [PubMed] [Google Scholar]
- 24.Williams TC, Ayrapetyan M, Oliver JD. 2015. Molecular and physical factors that influence attachment of Vibrio vulnificus to chitin. Appl Environ Microbiol 81:6158–6165. doi: 10.1128/AEM.00753-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kredich NM. 2008. Biosynthesis of cysteine. EcoSal Plus 3. doi: 10.1128/ecosalplus.3.6.1.11. [DOI] [PubMed] [Google Scholar]
- 26.BD Biosciences 2006. BD bionutrients technical manual: advanced bioprocessing. BD Biosciences, Franklin Lakes, NJ. [Google Scholar]
- 27.Oppezzo OJ, Antón DN. 1995. Involvement of cysB and cysE genes in the sensitivity of Salmonella typhimurium to mecillinam. J Bacteriol 177:4524–4527. doi: 10.1128/jb.177.15.4524-4527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atkinson MJ, Bingman C. 1997. Elemental composition of commercial seasalts. J Aquaric Aquat Sci VIII:39–43. [Google Scholar]
- 29.Gillespie D, Demerec M, Itikawa H. 1968. Appearance of double mutants in aged cultures of Salmonella typhimurium cysteine-requiring strains. Genetics 59:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krone FA, Westphal G, Schwenn JD. 1991. Characterisation of the gene cysH and of its product phospho-adenylylsulphate reductase from Escherichia coli. Mol Gen Genet 225:314–319. doi: 10.1007/BF00269864. [DOI] [PubMed] [Google Scholar]
- 31.Haverkamp T, Schwenn JD. 1999. Structure and function of a cysBJIH gene cluster in the purple sulphur bacterium Thiocapsa roseopersicina. Microbiology 145:115–125. doi: 10.1099/13500872-145-1-115. [DOI] [PubMed] [Google Scholar]
- 32.Townsend GE, Forsberg LS, Keating DH. 2006. Mesorhizobium loti produces nodPQ-dependent sulfated cell surface polysaccharides. J Bacteriol 188:8560–8572. doi: 10.1128/JB.01035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi E, Motta S, Mauri P, Landini P. 2014. Sulfate assimilation pathway intermediate phosphoadenosine 5′-phosphosulfate acts as a signal molecule affecting production of curli fibres in Escherichia coli. Microbiology 160:1832–1844. doi: 10.1099/mic.0.079699-0. [DOI] [PubMed] [Google Scholar]
- 34.Sekura RD. 1981. Adenosine 3′-phosphate 5′-phosphosulfate. Methods Enzymol 77:413–415. doi: 10.1016/S0076-6879(81)77055-1. [DOI] [Google Scholar]
- 35.Singh P, Brooks JF II, Ray VA, Mandel MJ, Visick KL. 2015. CysK plays a role in biofilm formation and colonization by Vibrio fischeri. Appl Environ Microbiol 81:5223–5234. doi: 10.1128/AEM.00157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enos-Berlage JL, Guvener ZT, Keenan CE, McCarter LL. 2005. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol Microbiol 55:1160–1182. doi: 10.1111/j.1365-2958.2004.04453.x. [DOI] [PubMed] [Google Scholar]
- 37.Blackwell KD, Oliver JD. 2008. The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina estuaries. J Microbiol 46:146–153. doi: 10.1007/s12275-007-0216-2. [DOI] [PubMed] [Google Scholar]
- 38.Bykowski T, van der Ploeg JR, Iwanicka-Nowicka R, Hryniewicz MM. 2002. The switch from inorganic to organic sulphur assimilation in Escherichia coli: adenosine 5′-phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol Microbiol 43:1347–1358. doi: 10.1046/j.1365-2958.2002.02846.x. [DOI] [PubMed] [Google Scholar]
- 39.Mika F, Busse S, Possling A, Berkholz J, Tschowri N, Sommerfeldt N, Pruteanu M, Hengge R. 2012. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol Microbiol 84:51–65. doi: 10.1111/j.1365-2958.2012.08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mika F, Hengge R. 2014. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol 11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longo F, Motta S, Mauri P, Landini P, Rossi E. 2016. Interplay of the modified nucleotide phosphoadenosine 5′-phosphosulfate (PAPS) with global regulatory proteins in Escherichia coli: modulation of cyclic AMP (cAMP)-dependent gene expression and interaction with the HupA regulatory protein. Chem Biol Interact 259:39–47. doi: 10.1016/j.cbi.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 42.Sillanpää JK, Ramesh K, Melzner F, Sundh H, Sundell K. 2016. Calcium mobilisation following shell damage in the Pacific oyster, Crassostrea gigas. Mar Genomics 27:75–83. doi: 10.1016/j.margen.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Bilecen K, Yildiz FH. 2009. Identification of a calcium-controlled negative regulatory system affecting Vibrio cholerae biofilm formation. Environ Microbiol 11:2015–2029. doi: 10.1111/j.1462-2920.2009.01923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kierek K, Watnick PI. 2003. The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca2+-dependent biofilm development in sea water. Proc Natl Acad Sci U S A 100:14357–14362. doi: 10.1073/pnas.2334614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kierek K, Watnick PI. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl Environ Microbiol 69:5079–5088. doi: 10.1128/AEM.69.9.5079-5088.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller JH. 1972, Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 47.Nakhamchik A, Wilde C, Rowe-Magnus DA. 2007. Identification of a Wzy polymerase required for group IV capsular polysaccharide and lipopolysaccharide biosynthesis in Vibrio vulnificus. Infect Immun 75:5550–5558. doi: 10.1128/IAI.00932-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalia AB, Lazinski DW, Camilli A. 2014. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. mBio 5:e01028-13. doi: 10.1128/mBio.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bivand RS, Pebesma E, Gómez-Rubio V. 2013. Applied spatial data analysis with R. Springer, New York, NY. [Google Scholar]
- 50.R Core Team 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.r-project.org/. [Google Scholar]
- 51.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.CFSAN 1998. Bacteriological analytical manual, 8th ed. Center for Food Safety and Applied Nutrition, United States Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- 53.Pu M, Rowe-Magnus DA. 2018. A Tad pilus promotes the establishment and resistance of Vibrio vulnificus biofilms to mechanical clearance. NPJ Biofilms Microbiomes 4:10. doi: 10.1038/s41522-018-0052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Calcium-induced brp expression is not dependent on Brp production. The indicated wild-type (WT) and brpF and brpJ mutant strains were grown on LB and LBCa plates containing X-Gal. Download FIG S1, PDF file, 0.8 MB (789.2KB, pdf) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inverse effect of calcium on biofilm formation by V. vulnificus and V. cholerae. Wild-type V. vulnificus (Vvu) and V. cholerae (Vch) were grown in LB or LB containing 10 mM CaCl2 (LBCa). (Left) Images of the biofilm ring in culture tubes. (Right) Quantification of biofilm formation by crystal violet (CV) staining in 96-well plates. Statistically significant differences among the samples (P < 0.001 as determined by one-way ANOVA followed by pairwise comparisons with a Bonferroni adjustment) are indicated by different symbols above each bar. Download FIG S2, PDF file, 0.3 MB (366.6KB, pdf) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Auxotrophic phenotype of the cys mutants. The wild-type (WT) and cysD, cysC, and cysH mutant strains were inoculated onto minimal medium with (MMcys) or without (MM) 0.5 mM cysteine. Download FIG S3, PDF file, 0.7 MB (756.7KB, pdf) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Part of the sulfate assimilation pathway leading to cysteine biosynthesis. Environmental sulfate is taken up and converted to adenosine 5′-phosphosulfate (APS) by the action of CysD and CysN. CysC converts APS to 3′-phosphoadenosine 5′-phosphosulfate (PAPS), which is then processed to adenosine 3′,5′-bisphosphate (PAP) and sulfite by CysH. PAP is converted to AMP by CysQ, and sulfite is reduced to sulfide by CysI and CysJ. CysK catalyzes the formation of l-cysteine from sulfide and O-acetylserine. Alkane sulfonates that are taken up can directly converted by mono- or dioxygenases to sulfite. Download FIG S4, PDF file, 0.2 MB (230.8KB, pdf) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The expression of cysJ is c-di-GMP-dependent. qRT-PCR was used to confirm that cysJ transcript levels decreased in wild-type cells when intracellular c-di-GMP levels were elevated (DcpA) relative to unaltered (v [empty vector]) conditions. Expression values are relative to those under unaltered conditions. Statistical significance was determined by the Student t test (two-tailed distribution with two-sample, equal variance). Download FIG S5, PDF file, 0.1 MB (22.2KB, pdf) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in this study. Download TABLE S1, DOCX file, 0.1 MB (116.5KB, docx) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S2, DOCX file, 0.2 MB (158.9KB, docx) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tn insertions that disrupt calcium-induced PbrpA expression. Download TABLE S3, DOCX file, 0.1 MB (91KB, docx) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sulfate assimilation pathway genes regulated by BrpT and c-di-GMP. Download TABLE S4, DOCX file, 0.1 MB (53.8KB, docx) .
Copyright © 2018 Chodur et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.