SPF1 and SPF2 are nuclear-located SUMO proteases that contribute to balancing the levels of SUMO conjugates downstream of SIZ1 and are involved in the regulation of plant development.
Keywords: Plant development, PTM, SIZ1, sumoylation, SUMO proteases, transcriptome, ULP2
Abstract
Post-translational modifiers such as the small ubiquitin-like modifier (SUMO) peptide act as fast and reversible protein regulators. Functional characterization of the sumoylation machinery has determined the key regulatory role that SUMO plays in plant development. Unlike components of the SUMO conjugation pathway, SUMO proteases (ULPs) are encoded by a relatively large gene family and are potential sources of specificity within the pathway. This study reports a thorough comparative genomics and phylogenetic characterization of plant ULPs, revealing the presence of one ULP1-like and three ULP2-like SUMO protease subgroups within plant genomes. As representatives of an under-studied subgroup, Arabidopsis SPF1 and SPF2 were subjected to functional characterization. Loss-of-function mutants implicated both proteins with vegetative growth, flowering time, and seed size and yield. Mutants constitutively accumulated SUMO conjugates, and yeast complementation assays associated these proteins with the function of ScUlp2 but not ScUlp1. Fluorescence imaging placed both proteins in the plant cell nucleoplasm. Transcriptomics analysis indicated strong regulatory involvement in secondary metabolism, cell wall remodelling, and nitrate assimilation. Furthermore, developmental defects of the spf1-1 spf2-2 (spf1/2) double-mutant opposed those of the major E3 ligase siz1 mutant and, most significantly, developmental and transcriptomic characterization of the siz1 spf1/2 triple-mutant placed SIZ1 as epistatic to SPF1 and SPF2.
Introduction
Post-translational modifications (PTMs) are able to rapidly and reversibly reprogram protein activity, and are involved in development and responses to environmental challenges. Among the many types of PTMs, one of the most well documented mechanisms is the attachment of small peptides structurally similar to ubiquitin (ubiquitin-like peptides, UBLs) (Miura and Hasegawa, 2010; Vierstra, 2012). Small ubiquitin-like modifier (SUMO) is a UBL family member that is mainly involved in nuclear-associated functions such as the regulation of transcription, chromatin-remodelling, mRNA biogenesis, nuclear–cytoplasm trafficking, and DNA repair (Gareau and Lima, 2010; Mazur and van den Burg, 2012; Cubeñas-Potts and Matunis, 2013). Briefly, sumoylation is achieved by an enzymatic cascade that involves maturation of the pre-SUMO peptide by specific SUMO endopeptidases, followed by three enzymatic steps (SUMO E1 activation, E2 conjugation, and E3 ligation) that drive the transfer of the maturated SUMO to a specific lysine residue, normally within the consensus ψKXE (ψ, large hydrophobic residue; K, lysine; X, any amino acid; E, glutamic acid) (Gareau and Lima, 2010; Cappadocia and Lima, 2018). The attachment can be reversed by specific SUMO isopeptidases, which counteract sumoylation and also contribute to the recycling of the SUMO peptide (Hickey et al., 2012).
SUMO conjugation can exert different effects on a target protein: (1) changing conformation, (2) aiding in protein–protein interactions (PPIs) via SUMO-interacting motifs (SIMs), and (3) blocking of PPIs, for example by competing with other PTMs (Wilkinson and Henley, 2010). Target proteins can be the subject of mono-sumoylation, poly-sumoylation (SUMO chain formation), or multi-sumoylation (multiple sumoylated sites) (Hickey et al., 2012; Hendriks and Vertegaal, 2016). Specificity of sumoylation may be determined by the large number of SUMO proteases, rather than being determined by the conjugation machinery, which is usually encoded by a limited number of genes. SUMO-specific proteases that belong to the C48 family of Cys proteases have been annotated as Ubiquitin-Like protein-specific Proteases or Sentrin/SUMO-specific Proteases (ULPs/SENPs) (van der Hoorn, 2008). These have been described as modulators of sumoylation through their action on SUMO moieties, namely by (1) processing the pre-SUMO (maturation), (2) removing SUMO from the modified target proteins (SUMO deconjugation), or (3) editing SUMO chains. ULP/SENP cysteine proteases are a heterogeneous family, which contribute to the specificity and complexity of the SUMO machinery (Hickey et al., 2012).
In plants, sumoylation seems to be essential for embryonic development, organ growth, flowering transition, and hormone regulation (Elrouby, 2015). In addition, SUMO plays a role in stress-associated responses to stimuli such as extreme temperatures, drought, salinity, and nutrient assimilation (Castro et al., 2012, 2015). During such stresses, the profile of SUMO-modified proteins changes dramatically, with greatly increased SUMO-conjugate levels and a decreased pool of free SUMO (Miller et al., 2013). After the imposition of stress, SUMO conjugates slowly diminish by the action of ULPs. ULPs are fundamental players in the fine-tuning of the SUMO conjugation/deconjugation levels and, consequently, are essential to balance plant growth and stress responses (Conti et al., 2014; Yates et al., 2016). On the other hand, knowledge regarding the importance and functions of SUMO proteases in plant physiology is very limited and many ULPs are yet to be extensively characterized. ULPs fall into two large groups (ULP1s and ULP2s), by homology to yeast ScULP1 and ScULP2. The Arabidopsis genome includes eight predicted ULPs, six of which have been shown to function as SUMO proteases in vitro (Chosed et al., 2006; Colby et al., 2006; Conti et al., 2008; Novatchkova et al., 2012; Kong et al., 2017; Liu et al., 2017a). Each of these ULPs is likely to contribute individually to specific functions within the plant, judging from the functional characterizations available to date. For instance, ESD4 loss-of-function results in a pleiotropic phenotype (severe dwarfism), while the closely related ELS1 does not have such a severe phenotype (Murtas et al., 2003; Hermkes et al., 2011). OTS1 and OTS2 act redundantly in flowering transition, plant growth, and photomorphogenesis, as well as in pathogen defence, and salt and osmotic stress responses (Conti et al., 2008, 2014; Sadanandom et al., 2015; Bailey et al., 2016; Castro et al., 2016). The function of SPF1 (also designated ASP1) and SPF2 has been recently associated with the control of flowering time, and gamete and embryo development (Kong et al., 2017; Liu et al., 2017a).
In the present study, we performed a structural and phylogenetic characterization of plant ULPs, which pointed to SPF1 and SPF2 forming a key subgroup within ULP2-like SUMO proteases. Complementation assays indicated that Arabidopsis SPF2 is functionally homologous to the yeast ScULP2 gene and that SPF1 exerted a dominant negative effect, while SPF mutant plants constitutively accumulated more SUMO conjugates. Accordingly, we demonstrate that the SPF1 and SPF2 catalytic domains reacted with SUMO activity-based probes. Arabidopsis T-DNA insertion mutants showed diverse developmental defects, and microarray analysis provided evidence for a specific transcriptional signature that suggests the involvement of SPF1/2 in secondary metabolism, cell wall remodelling, and nitrate assimilation. The spf1-1 spf2-2 (spf1/2) double-mutant also displayed an antagonistic morphological phenotype with respect to the well-characterized SUMO E3 ligase mutant siz1. Most significantly, the spf1/2 siz1 triple-mutant was phenotypically siz1-like, which places SPF1/2 as epistatic and downstream of SIZ1.
Materials and methods
Plant material and growth conditions
T-DNA insertion mutants were used to evaluate loss-of-function in Arabidopsis thaliana SUMO proteases SPF1/ASP1/ULP2b (At1g09730) and SPF2/ULP2a (At4g33620). Mutants were obtained through the NASC European Arabidopsis Stock Centre (http://arabidopsis.info) or the Arabidopsis Biological Resource Center (https://abrc.osu.edu). All mutants were SALK lines in the background ecotype Columbia-0 (Col): SALK_040576 (spf1-1), SALK_022079 (asp1-2; also designated as ulp2like2-2 by Liu et al., 2017b), SALK_090744 (spf2-2), SALK_140824 (spf2-3), SALK_023493C (spf2-1), and the previously characterized line SALK_065397 (siz1-2). Single T-DNA mutant lines were inter-crossed to obtain the corresponding combination of double-mutants. The spf1/2 siz1 triple-mutant was obtained by crossing the double-mutant spf1-1 spf2-2 (i.e. spf1/2) with siz1-2. The genotypes were confirmed by diagnostic PCR, following the instructions for SIGnAL T-DNA Primer Design (http://signal.salk.edu/tdnaprimers.2.html) and using the primers listed in Supplementary Table S1 at JXB online. Synchronized seeds were stratified for 3 d at 4 °C in the dark. Surface-sterilization was performed in a horizontal laminar-flow chamber by sequential immersion in 70% (v/v) ethanol for 5 min and 20% (v/v) commercial bleach for 10 min before washing five times with sterile ultra-pure water. Seeds were resuspended in sterile 0.25% (w/v) agarose, sown onto 1.2% (w/v) agar-solidified MS medium (Murashige and Skoog, 1962) containing 1.5% (w/v) sucrose, 0.5 g l−1 MES, pH 5.7, and grown vertically in culture rooms with a 16/8 h light/dark cycle under cool white light (80 µE m−2 s−1) at 23 °C. For standard growth, 7-d-old plate-grown seedlings were transferred to a soil/vermiculite (4:1) mixture and maintained under identical growth conditions, with regular watering. Mutant lines were morphologically characterized according to the developmental map for Arabidopsis described by Boyes et al. (2001). Morphological parameters were measured using the ImageJ software (https://imagej.nih.gov/ij/).
Pigment extraction and quantification
For estimation of the chlorophyll and carotenoid contents, plant leaves were incubated in 80% (v/v) acetone for 1 h in the dark. The plant material was spun down and absorbances at 470, 645, and 663 nm were measured in a microplate spectrophotometer (SpectraMax 340PC; Molecular Devices). Total chlorophyll was calculated as 20.2A645+8.02A663 and total carotenoids were calculated as [1000A470−1.82(12.7A663−2.69A645)−85.02(22.90A645−4.68A663)]/198 (Arnon, 1949; Lichtenthaler and Buschmann, 2001).
Anthocyanin extraction and quantification was adapted from Ticconi et al. (2001). Plant leaves were weighed (fresh weight, FW) and incubated at 100 °C for 5 min in extraction buffer composed of 1-propanol (37%, v/v), HCl, and H2O, in a 18:1:81 ratio. Samples were subsequently incubated overnight at room temperature in the dark. The plant material was spun down and absorbance of the supernatant was measured at 535 nm and 650 nm in a similar microplate spectrophotometer. Total anthocyanins were calculated as A535−A650 g−1 FW.
RNA extraction, cDNA synthesis, and RT-qPCR
For reverse-transcription quantitative real-time PCR (RT-qPCR) analysis, RNA from plant tissue was extracted using an RNeasy Plant Mini Kit (Qiagen). RNA quantity and quality were assessed using both a Nanodrop ND-1000 spectrophotometer and standard agarose-gel electrophoretic analysis, and RNA samples were treated with Recombinant DNase I (Takara Biotechnology). Synthesis of cDNA was performed using SuperScript II Reverse Transcriptase Kit (Invitrogen). SsoFast EvaGreen Supermix (Bio-Rad) was used in the RT-qPCR reaction mixture according to the manufacturer’s indications. The reaction was performed in a MyiQ Single-Color Real-Time PCR Detection system (Bio-Rad). Primers for semi-quantitative RT-PCR and RT-qPCR (Supplementary Table S2) were designed using NCBI Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/) (Ye et al., 2012) to ensure specific amplification within the Arabidopsis genome, and obeyed the following guidelines: 100–250 bp PCR amplification product size; 50–60% GC content; ~60 °C Tm. Primers were designed to span an exon junction when possible. ACT2 (At3g18780) was used as a reference gene (Lozano-Durán et al., 2011).
Microarray analysis
Genome-wide transcription studies were performed using an ATH1 microarray chip (Affymetrix) with three independent replicates per genotype, with each replicate representing RNA from a pool of four different MS plates containing 10-d-old seedlings. Plants were grown in a plant growth chamber with 16/8 h light/dark cycle under cool white light (80 µE m−2 s−1) at 21 °C. RNA was extracted as described above, followed by a column cleaning step using an RNeasy Plant Mini Kit (Qiagen). Microarray execution and differential expression analysis were conducted at the Unité de Recherche en Génomique Végétale (Université d’Evry Val d’Essonne, France). The method to determine differentially expressed genes (DEGs) was based on variance modelization by common variance of all genes (Gagnot et al., 2008).
Plant protein extraction and western blotting
Plant tissue was ground in a microtube in liquid nitrogen with the help of polypropylene pestles. Protein extracts were obtained by adding extraction buffer [50 mM Tris; 150 mM NaCl; 0.2% (v/v) Triton X-100] supplemented with Complete Protease Inhibitor Cocktail (Roche) according to the manufacturer’s instructions. Following incubation with agitation for 1 h at 4 °C, the microtubes were centrifuged twice for 30 min at 16000 g. The supernatants were recovered and stored at −80 °C. Protein was quantified spectrophotometrically using Bradford reagent (Sigma; Bradford, 1976). Equal amounts of protein were resolved by standard SDS-PAGE in a 10% (w/v) acrylamide resolving gel, using a Mini-PROTEAN Cell apparatus (Bio-Rad). For western blotting, proteins were transferred to a PVDF membrane using a Mini Trans-Blot Cell (Bio-Rad). The membrane was blocked for 1 h at 23 °C in blocking solution [5% (w/v) dry milk powder in PBST]. The primary antibody anti-AtSUMO1 (Abcam) was added in a 1:1000 dilution and incubated for 3 h. The membrane was washed three times with 10 ml of PBST for 10 min, and then incubated with the secondary anti-rabbit antibody (Santa Cruz) at 1:2000 in blocking solution for 1 h. The membrane was washed as described above and developed using a chemiluminescence reaction with an Immune-Star WesternC Kit (Bio-Rad) and a ChemiDoc XRS system (Bio-Rad) for image acquisition. PVDF membranes were incubated for 15 min with Ponceau S solution [0.1% (w/v) Ponceau S; 5% (v/v) acetic acid] to stain for total proteins.
Plasmid construction
Arabidopsis SPF1 and SPF2 coding-sequence (CDS) PCR products were purified and cloned using the pGEM-T Easy system (Promega). Final constructs for pGEM-SPF1 and pGEM-SPF2 were confirmed by sequencing. The SPF1 sequence was shorter than the one annotated in TAIR (www.arabidopsis.org), implying the existence of two additional introns. This shorter SPF1 isoform sequence displayed a complete match with the protein sequence NP_001184951.1 in the NCBI database (http://www.ncbi.nlm.nih.gov/). The SPF1 and SPF2 full fragments were excised by restriction using NotI and AscI and were then subcloned into the Gateway Entry vector pENTR. The LR reaction for recombination between the attL (entry clone) and attR (destination vector) recombination sites was carried out in the pMDC43 vector (Curtis and Grossniklaus, 2003). Recombinations between the pENTR constructs and the pMDC43 destination vector were performed using LR Clonase II (Invitrogen), following the manufacturer’s instructions.
To generate pCM190-SPF2, primers with the restriction sites PmeI-NotI (Supplementary Table S3) were used to amplify the SPF2 CDS from pGEM-SPF2 and, after digestion with PmeI-NotI, the product was subcloned into pCM190. pGEM-SPF1 was digested with NotI and the resulting fragment was cloned into pCM190 to yield pCM190-SPF1.
The cDNAs encoding the catalytic domains of SPF1 and SPF2 (hereafter referred to as cSPF1 and cSPF2) were amplified from pGEM-SPF1 and pGEM-SPF2 using primers listed in Supplementary Table S3. Amplification products were cloned into pNZY28-A using the NZY-A PCR cloning kit (NZYtech). cSPF1 and cSPF2 were then respectively excised using the restriction enzyme combinations EcoRI + NotI and BamHI + NotI (NEB) to clone into the expression vector pGEX-5X-1 (GE Healthcare).
Yeast complementation assay
The yeast (Saccharomyces cerevisiae) mutant strain ulp1-ts (temperature-sensitive) has been described previously (Li and Hochstrasser, 1999), and the ulp2∆ mutant and an isogenic ULP2+ strain were obtained by sporulation of the EUROSCARF diploid strain (Y21424, Mat α/a; his3D1/his3D1; leu2D0/leu2D0; lys2D0/LYS2; MET15/met15D0; ura3D0/ura3D0; YIL031w::kanMX4/YIL031w). Both mutant strains were used for the complementation assays with SPF1 and SPF2 from Arabidopsis. Yeast strains ulp1-ts and ulp2∆ were transformed (Gietz et al., 1995) with the constructs pCM190-SPF1, pCM190-SPF2, or the empty vector (pCM190), and plated at 25 °C on minimal medium (yeast nitrogen base, YNB) supplemented with the appropriate amino acids according to each strain genotype, and doxycycline (10 g ml−1; Sigma). The constructs expressed SPF1 or SPF2 from a tetracycline-regulatable promoter, so the tetracycline analogue doxycycline was added to the plates to inhibit SPF1 or SPF2 expression (Garí et al., 1997). A 10-fold serial dilution of three independent colonies for each transformation was made, and 5 μl of each dilution was spotted onto minimal medium (YNB) supplemented with the appropriate amino acids with or without doxycycline (10 g ml−1). The plates were incubated at 25 °C or 37 °C for 5 d.
Covalent labelling with HA-tagged HsSUMO-VME probes
Vinyl methyl esters (VMEs) are probes that react irreversibly with the ULP catalytic cysteine and establish a covalent bound that can be detected by SDS-PAGE followed by western blotting using an anti-HA antibody (Borodovsky et al., 2002). Recombinant glutathione S-transferase (GST)-SPF expression constructs were transformed into the E. coli strain BL21(DE3) pLysS, and expression was induced at an A600 of 0.6 with 0.1 mM IPTG at 16 °C overnight. Cells were harvested by centrifugation for 20 min at 4000 g. Bacterial pellets were resuspended in buffer A (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 1 mM BME), disrupted by sonication, and cleared by centrifugation at 34000 g for 40 min. Recombinant GST-SPF protein was purified by batch affinity chromatography using Glutathione Agarose beads (ThermoFisher Scientific). The beads were washed with buffer A, and proteins were eluted on gravity columns with buffer B (50 mM Tris-HCl, pH 8.0; 10 mM reduced L-glutathione). Eluted proteins were stored at –80 °C.
The SPF activity assay was performed in a reaction buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% (w/v) glycerol, 2 mM EDTA-NaOH, 0.15 µg µl−1 BSA, and 2 mM DTT (Pinto et al., 2012). Human SENP1 catalytic domain (cSENP1) and UCHL3 were obtained as described by Pinto et al. (2012) and Grou et al. (2015), respectively. Human SUMO (HsSUMO) probes (Borodovsky et al., 2002) were obtained as described by Pinto et al. (2012). The reaction was carried out at 25 °C for 2 h, using 100 ng of human influenza hemagglutinin (HA)-HsSUMO-VME and 200 ng of either protease in a final volume of 20 µl. The reaction sample was mixed with Laemmli sample buffer and incubated at 65 °C for 10 min followed by 5 min at 95 °C. Proteins were separated electrophoretically in a 16% SDS-PAGE gel, transferred to a nitrocellulose membrane, and probed with monoclonal anti-HA antibody (16B12, Covance) and phosphatase alkaline-conjugated secondary antibody anti-mouse IgGs (A2429, Sigma-Aldrich).
Transient expression in tobacco
Agrobacterium tumefaciens EHA105 containing constructs-of-interest was co-infiltrated with a suppressor of gene silencing, the p19 protein of tomato bushy stunt virus (TBSV), to prevent the onset of post-translational gene silencing (Silhavy et al., 2002). The pellet was resuspended in 1 ml agroinfiltration buffer [10 mM MgCl2; 10 mM MES, pH 5.6; 19.6 mg ml−1 acetosyringone] and grown in non-supplemented medium until a final A600 of 1 was obtained for the empty or transformed strain, and A600 of 2 for the p19 silencing vector. The resuspended pellets of both the transformed strain and p19 were incubated for 2–5 h and subsequently infiltrated in a 1:1 ratio with a 5-ml syringe in the abaxial side of 3-week-old Nicotiana benthamiana leaves. Expression of each transgene was monitored 4 d after transformation with an Olympus FluoView FV1000 confocal laser microscope, using excitation wavelengths of 488 nm (green fluorescent protein, GFP) and 635 nm (chloroplast autofluorescence). Bright-field images were detected using transmitted light. Detection specifications were maintained between different biological samples.
Phylogenetic and bioinformatics analysis
The automated gene family annotation resource Plaza (Van Bel et al., 2012) was used to retrieve amino acid sequences of ULP gene family members across 30 phylogenetically representative species, based on queries using the search terms At4g15880, At1g09730, At1g60220, and At3g48480. Phylogenetic analysis was performed using maximum likelihood (RaxML) with 1000 bootstrap iterations, as previously described (Castro et al., 2017). The final output of the tree was produced using the SeaView v4.4.0 software (Gouy et al., 2010). Protein sequence alignment of the catalytic domain of Arabidopsis SPFs with homologous proteins from eukaryotic organisms was performed using PRALINE (Simossis and Heringa, 2005). Gene ontology (GO) term functional categorization was performed in VirtualPlant 1.2 (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/) using the BioMaps function with a 0.05 P-value cut-off (Katari et al., 2010). Redundancy exclusion and scatterplot analysis were performed using REVIGO (http://revigo.irb.hr/), with a 0.7 C-value. The scatterplot presents the cluster representatives in a two-dimensional space (x- and y-axis) derived by applying multidimensional scaling to a matrix of the semantic similarities of the GO terms (Supek et al., 2011). MapMan was used to plot spf1/2 deregulated genes in the Metabolism overview pathway map (http://mapman.gabipd.org/web/guest/home) (Thimm et al., 2004).
Results
Plant ULP2 proteases are phylogenetically and topologically diverse
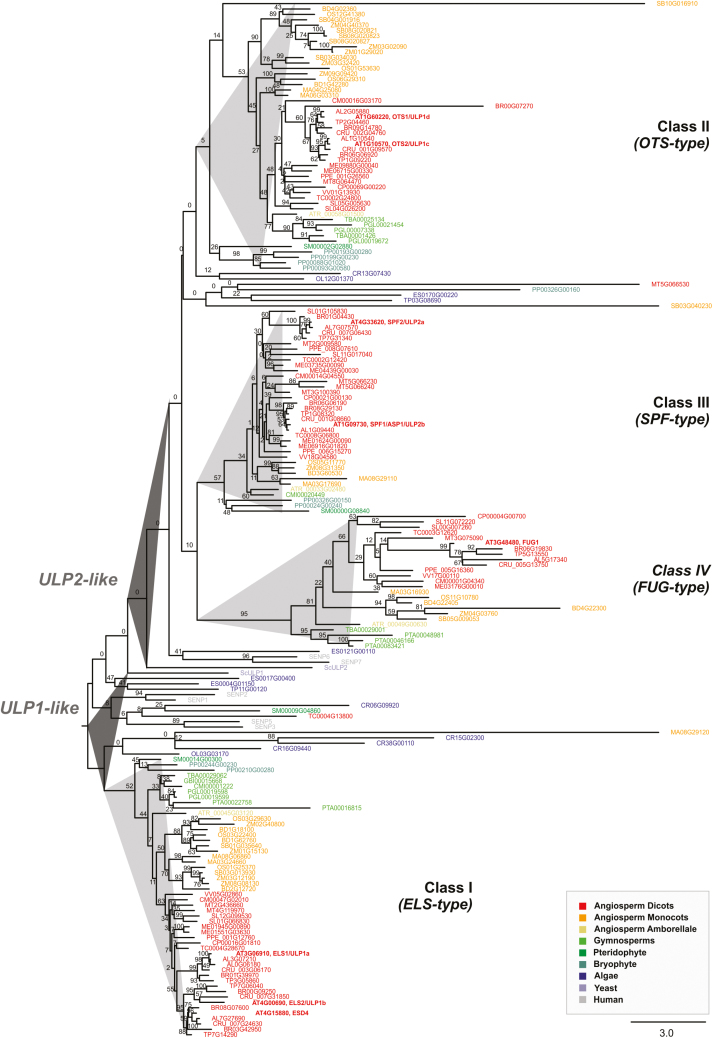
Previous predictions for Arabidopsis ULP SUMO protease family members have been scarce in scope and, above all, inconsistent as to the relationships between the main existing phylogenetic subgroups. For instance, they have missed inclusion of the At3g48480 protein, or placed OTS1 and OTS2 (also termed ULP1d and ULP1c) in either ULP1- or ULP2-related clades (Miura et al., 2007a; Lois, 2010; Novatchkova et al., 2012). To resolve this issue, we performed a significantly more comprehensive ULP phylogeny. A plant ULP ortholog search in 30 representative genomes was carried out using Plaza (Proost et al., 2015), and was based on homology searches with the seven consistently annotated Arabidopsis ULPs and the putative family member At3g48480. The phylogenetic reconstruction displayed two major branches that resolved ULP1s (yeast ScULP1 and human SENP1-3, -5) and ULP2s (yeast ScULP2 and human SENP6-7) (Fig. 1). Both branches contained algae and plant ULPs from all major taxa, demonstrating the polyphyletic origin of plant ULPs. Our analysis uncovered a series of interesting findings. ULP1s encompassed Arabidopsis ESD4, ELS1 (also termed ULP1a) and ULP1b, whereas Arabidopsis OTS1 and OTS2 are most likely ULP2s and not ULP1s. Plant ULPs could be further categorized into four phylogenetic subgroups or classes (Fig. 1), which we have named based on the classification proposed by Novatchkova et al. (2012). Class II (OTS-type; OTS1/2) and Class III (SPF-type; SPF1/2) contained paralogs from all major taxa all the way to briophytes, suggesting a very ancestral duplication and subsequent subfunctionalization that remained conserved across plant evolution. Arabidopsis ULP At3g48480, which was often absent from ULP annotation (possibly due to its smaller protein size), showed up as an independent subclade/class across at least the flowering plant taxa, and was named Fourth ULP Gene Class 1 (FUG1).
Fig. 1.
Phylogenetic analysis of the plant ubiquitin-like protease (ULP) family. The phylogenetic reconstruction includes ULPs present in representative plant genomes, as well as human SENPs and yeast (Saccharomyces cerevisiae) ULPs. Phylogenetic analysis was performed using maximum-likelihood with bootstrap analysis (1000 trees; numbers on each branch represent the percentages of bootstrap).
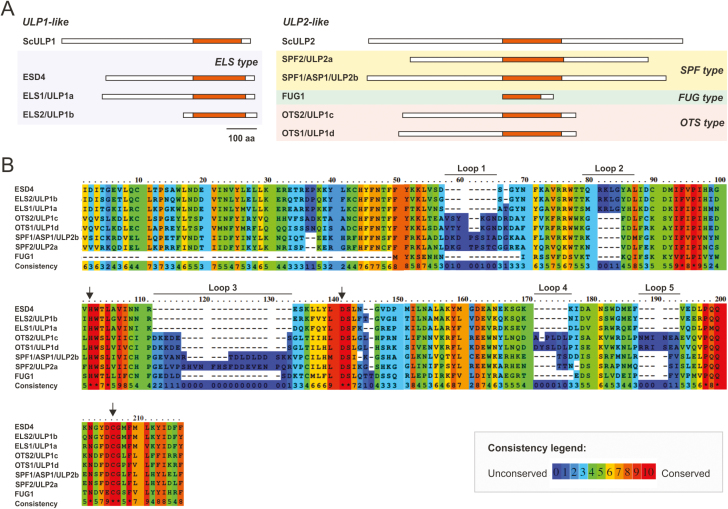
The present study specifically addressed Arabidopsis SPF1 (also termed ULP2b or ASP1; At1g09730) and SPF2 (also termed ULP2a; At4g33620). Their proteins displayed 30.5% identity, as well as a highly conserved region that matched the catalytic domain and possessed 46% identity (Fig. 2A, B, Supplementary Fig. S1). For both proteins, topological analysis revealed the catalytic domain to be located within the centre of the protein, while ULP1-like proteins were located in the C-terminal end (Fig. 2A). Analysis also demonstrated that At3g48480 was restricted to the catalytic domain and lacked both the N- and C-terminal ends of ULP2s (Fig. 2A). Remarkably, the catalytic triad (His-Asp-Cys), essential for protease activity, was conserved among all Arabidopsis ULP members (Fig. 2B). Within the catalytic domain, it was possible to discriminate five main extensions (loops 1 to 5; Fig. 2B). Loops 1, 3, 4, and 5 were common to SPF1/2 and OTS1/2, and absent in ESD4, ELS1, and ULP1b, while loop 2 was specific to ULP1b. Loop 1 and in particular loop 2 were larger in SPF1/2, whereas loops 3 and 4 were larger in OTS1/2 (Fig. 2B).
Fig. 2.
Topological analysis of the plant ULP proteins. (A) Schematic representation of Arabidopsis and yeast ULP protein topology with the catalytic domain highlighted in orange. The scale bar indicates 100 amino acids. (B) Protein sequence alignment of the catalytic domain in Arabidopsis ULPs. The arrows indicate the three conserved catalytic residues. Consistency between sequences indicates the levels of conservation of each residue. Five main extensions can be discriminated within the catalytic domain (loops 1–5).
SPF1 and SPF2 differentially complement yeast ulp1 and ulp2 mutants and react with activity-based SUMO probes
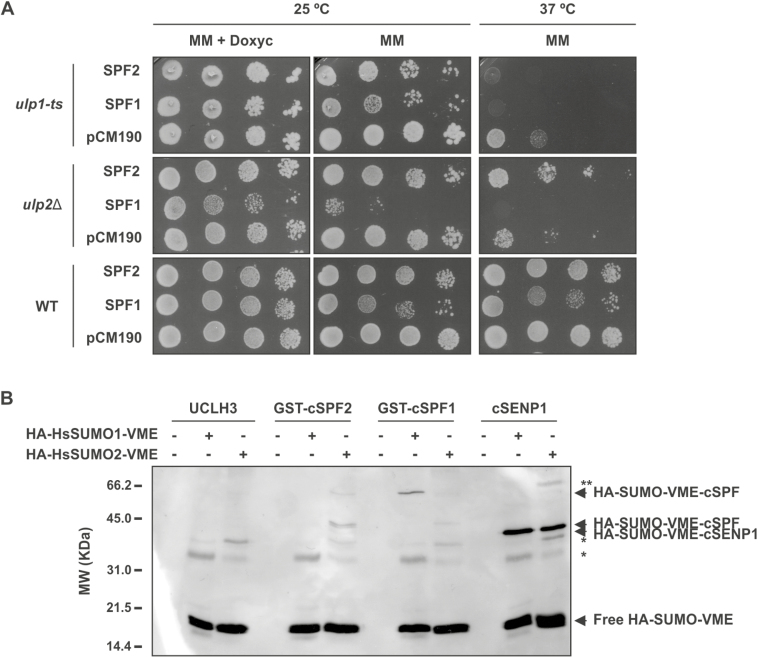
SUMO proteases may display different activities, breaking endopeptidic bonds that are important for SUMO maturation or having isopeptidic activity for SUMO removal or SUMO chain editing (Hickey et al., 2012). Phylogenetic analysis placed SPF1 and SPF2 closer to ULP2s from non-plant models (yeast Ulp2 and human SENP6/7; Fig. 1). To validate this hypothesis, yeast complementation of the ulp1 and ulp2 mutants was performed, expressing the Arabidopsis genes from a multicopy plasmid (pCM190; Garí et al., 1997). Yeast ULP1 is an essential gene, so the complementation assay required the use of a previously described temperature-sensitive mutant (ulp1-ts) (Li and Hochstrasser, 1999). The deletion of the yeast ULP2 gene is not lethal, but ulp2Δ mutants show sensitivity to a variety of stresses, including elevated temperature (Li and Hochstrasser, 2000; Schwienhorst et al., 2000). Hence, the temperature-sensitive phenotype of the ULP2 deletion allele ulp2Δ (Y21424, EUROSCARF) was used for the complementation assay with Arabidopsis SPF1 and SPF2. Both yeast mutants were transformed with the vector (pCM190) or the plasmid expressing either SPF1 or SPF2, from a tetracycline-regulatable promoter, so that expression was inhibited in the presence of doxycycline (a tetracycline analogue) (Garí et al., 1997). The temperature-sensitive ulp1-ts mutant was not able to grow at 37 °C when any of the two Arabidopsis genes were expressed (Fig. 3A). However, SPF2 could complement ulp2Δ temperature sensitivity, whereas SPF1 could not. Remarkably, the ulp2∆ mutant was sensitive to SPF1 expression and yeast growth was clearly diminished at both temperatures (Fig. 3A). The toxic effect of SPF1 was doxycycline-dependent and more severe in the ulp2∆ background than in an isogenic wild-type strain or in the ulp1-ts mutant (Fig. 3A), suggesting a dominant-negative mutant effect of the presence of Arabidopsis SPF1 in the absence of the yeast ULP2 ortholog. Collectively, these results suggested that (1) Arabidopsis SPF1 and SPF2 were not ULP1 proteases, (2) SPF2 was functionally homologous to the yeast ULP2 gene, and (3) SPF1 function was related to ULP2 SUMO proteases.
Fig. 3.
SUMO protease activity analysis of SPF1 and SPF2 by yeast complementation assays and reactivity of SPF1/2 catalytic domains towards human SUMO (HsSUMO) vinyl methyl ester (VME) probes. (A) Transformants harbouring the vector pCM190 or the constructs to express SPF1 (pCM190-SPF1) and SPF2 (pCM190-SPF2) were plated on selective minimal medium (MM) with doxycycline (10 g l−1) and incubated at 25 °C for 4 d. Ten-fold serial dilutions were made for three independent colonies (a representative colony is shown for each transformation) and 5 μl of each dilution was spotted onto MM or selective MM with doxycycline. Plates were incubated at 25 °C or 37 °C as indicated, and photographs were taken after a 5-d incubation. (B) In vitro SPF1 and SPF2 catalytic domain (cSPF1 and cSPF2) activity was tested against HA-HsSUMO1-VME and HA-HsSUMO2-VME. Human deubiquitinase UCHL3 and SUMO protease SENP1 catalytic domain (cSENP1) were used as negative and positive control enzymes, respectively. The arrows indicate free HA-HsSUMO-VME probes and their conjugated forms with ULPs, as labelled. * Indicates unspecific bands; ** indicates a possible adduct between an SDS resistant dimer of cSENP1 and SUMO2-VME. Molecular weight markers (MW) are displayed.
To examine SPF1 and SPF2 SUMO protease activity in vitro, we used activity-based irreversible inhibitors in the form of vinyl methyl ester (VME)-derivatized HA-tagged HsSUMO1 and HsSUMO2 (HA-HsSUMO1-VME and HA-HsSUMO2-VME, respectively). As previously stated, VME probes bind irreversibly to the ULP catalytic domain, which can be detected by western blotting (Borodovsky et al., 2002). For the activity assay, we expressed and purified the SPF1 and SPF2 catalytic domains (cSPF1 and cSPF2) coupled with a GST-tag at the N-terminus. cSPF2 reacted positively towards HsSUMO2-VME, while cSPF1 reacted mainly with HsSUMO1-VME (Fig. 3B). In addition, SPF2 revealed other bands with lower molecular weight that were probably the result of sub-products of SPF2 expression reacting with HsSUMO2-VME (Supplementary Fig. S2). The negative control UCHL3, a specific protease for ubiquitin, did not react with any of the probes while cSENP1, a human ULP (Kolli et al., 2010), reacted towards human SUMO, revealing a shift for the expected size of SENP1-HsSUMO-VME (Fig. 3B). Collectively, results supported the roles of SPF1 and SPF2 as SUMO proteases.
SUMO conjugate levels are modulated by SPF1 and SPF2 in planta
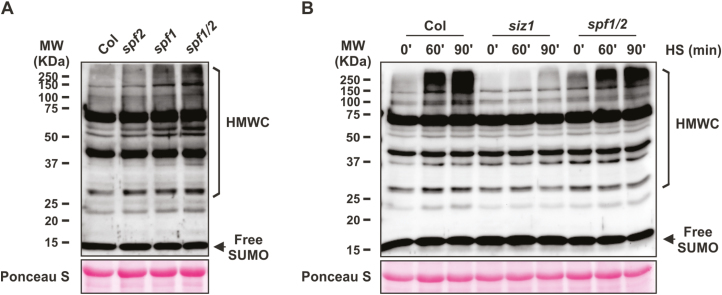
To verify whether SPF1 and SPF2 had an impact on SUMO-conjugate levels, we examined the sumoylation profiles in Arabidopsis SPF1 and SPF2 T-DNA insertion lines (Supplementary Fig. S3A). Given that SPF1 and SPF2 are phylogenetically and topologically close (Figs 1, 2) and that functional redundancy has been displayed by other gene family members (Castro et al., 2016), we also generated a double-mutant spf1-1 spf2-2 (hereafter referred to as spf1/2). We confirmed abolished gene expression in the mutant backgrounds using semi-quantitative RT-PCR (Supplementary Fig. S3B). Sumoylation patterns were analysed by western blotting of whole-plant protein extracts using specific anti-AtSUMO1 antibodies, thus covering the predominant SUMO1/2 peptides (Saracco et al., 2007; van den Burg et al., 2010). When compared to the Col-0 wild-type, high molecular weight SUMO conjugates constitutively accumulated in the spf1/2 double-mutant and also to some extent in the single mutants (Fig. 4A). To further characterize the lack of SPF1 and SPF2 in Arabidopsis, we examined the level of SUMO conjugates of the Arabidopsis spf1/2 double-mutant subjected to heat-shock (HS) stress (Fig. 4B). SUMO-conjugation increased in response to stress, and this increment could be regulated by an altered balance between conjugation and deconjugation activities, in which ULPs play an important role (Pinto et al., 2012). Here, although HS stress induced SUMO-conjugate accumulation, no major changes were observed in spf1/2 compared to the wild-type, as the conjugate levels in Col-0 in response to HS were close to those in the conjugate-overproducer spf1/2 background. As expected, SUMO conjugates failed to accumulate in the siz1 mutant that was used as a negative control.
Fig. 4.
Immunoblot analysis of high molecular weight SUM1 conjugates (HMWC) in Arabidopsis wild-type Col-0 and SPF mutants. (A) Analysis of leaf protein extracts from 1-month-old plants grown in soil. (B) Analysis of plate-grown 10-d-old plants subjected to heat-shock (HS, 37 °C) for 0, 60, or 90 min; the siz1 mutant was used as a negative control of SUMO conjugate induction after heat shock. Protein extracts (50 µg per lane) were analysed by immunoblotting using anti-AtSUMO1 polyclonal antibodies. The larger subunit of Rubisco stained with Ponceau S was used as a loading control. Molecular weight markers (MW, Kaleidoscope, Bio-Rad) are displayed.
SPF1 and SPF2 are localized in the nucleus
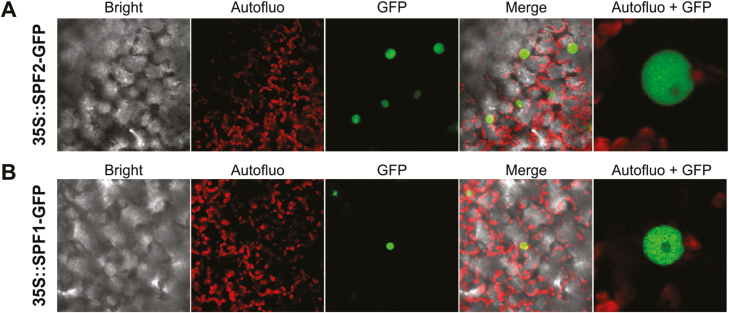
Differential recognition of SUMO substrates by SUMO proteases has been partially attributed to differences in subcellular localization (Hickey et al., 2012). Since localization of ULP proteins is crucial for their biological function, we investigated the location of SPF1 and SPF2 within the plant cell using transient expression of GFP-fusion proteins in N. benthamiana. Expression was visualized by confocal microscopy 3 d after agroinfiltration. Both SPF1 and SPF2 were localized specifically within the nucleus (Fig. 5A, B), showing no signal at the nucleolus, which was suggestive of specific subnuclear localization for both proteins.
Fig. 5.
Subcellular localization of Arabidopsis SPF1 and SPF2. SPF2 (A) and SPF1 (B) were N-terminally fused to GFP and transiently expressed in N. benthamiana leaves. The confocal microscopy channels depict a 60× magnification of bright field, chloroplast autofluorescence (red), GFP fluorescence (green), or an overlay of these channels (Merge). Autofluo+GFP represents a digital magnification of the cell nucleus.
SPF1 and SPF2 mutants are developmentally compromised
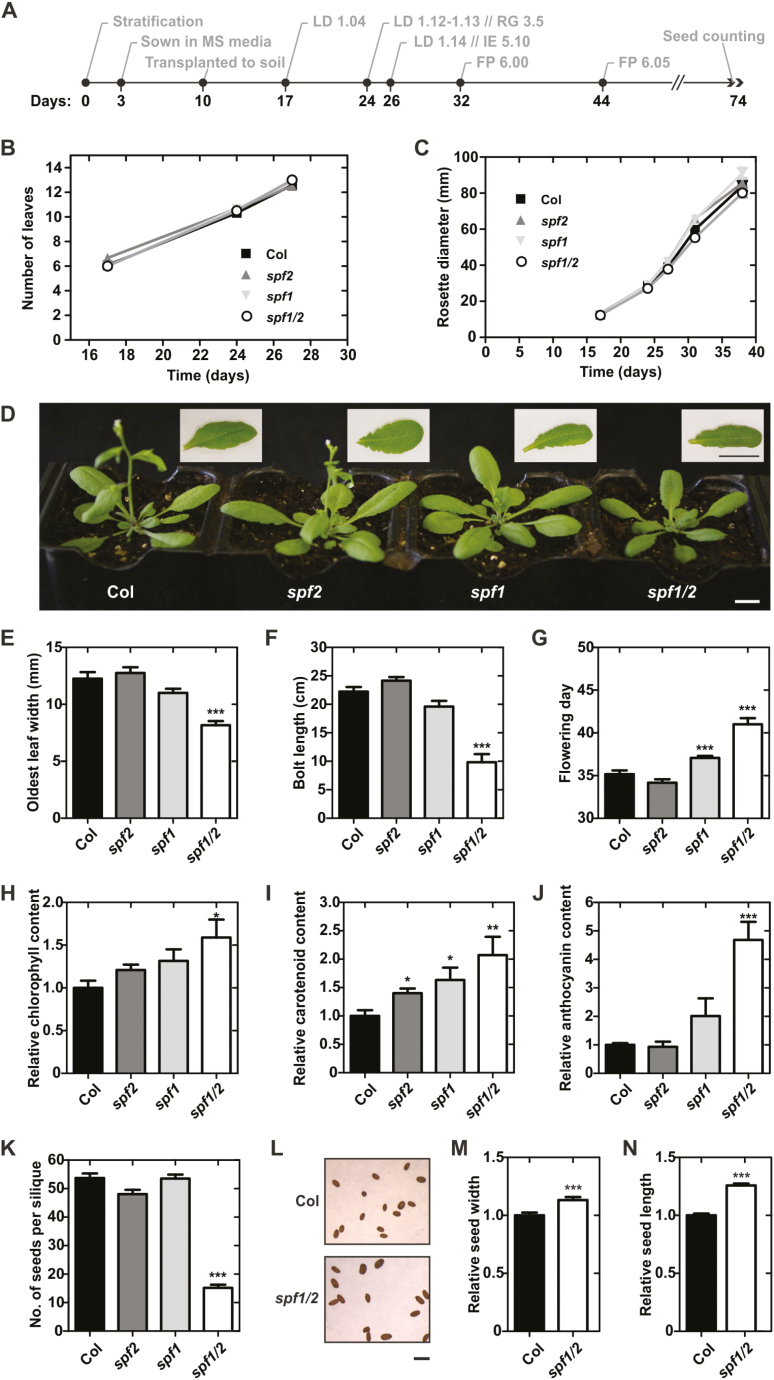
Sumoylation has been shown to modulate many aspects of plant development, as well as key mechanisms in various stress responses. Many of the previous findings regarding the role of SUMO in plants have been based on reverse genetics approaches (Lois, 2010). To investigate SPF1 and SPF2 function, a systematic characterization of morphological/developmental features of null-mutants was carried out (Fig. 6A). In the earlier stages of development there were no severe phenotypic differences between genotypes growing in soil (Fig. 6B, C). However, at later developmental stages, morphological analysis suggested that, in comparison to Col-0, both the spf1 and spf1/2 mutants displayed altered leaf morphology and late flowering times (Fig. 6D). Although spf1/2 rosettes displayed a slightly smaller diameter (not significantly different), the most interesting aspect was that spf1/2 leaves were significantly smaller in width (Fig. 6E). Overall, spf1/2 plants showed a clear delay in development that included late flowering and a shorter bolt length at that developmental point (Fig. 6F, G), but taller plants at the end of the life cycle (Supplementary Fig. S4A). Another striking feature of the double-mutant plants was the darker colour of the leaves, and hence we measured pigment contents in leaves of 1-month-old plants (Fig. 6H–J). The results indicated that spf1/2 accumulated relatively more chlorophyll, carotenoids, and anthocyanins than Col-0. Finally, we observed that spf1/2 seed production and morphology were also severely affected, resulting in a low number of seeds per silique (Fig. 6K), but seeds were bigger compared to Col-0 (Fig. 6L–N). No differences were observed for silique size between spf1/2 and Col-0 (Supplementary Fig. S4B). To genetically confirm the present results, second allele mutants were examined and displayed similar phenotypes (Supplementary Fig. S5). Collectively, the results indicated that the spf1/2 double-mutant aggravated various single mutant phenotypes, indicating at least partial functional redundancy between SPF1 and SPF2.
Fig. 6.
Developmental characterization of Arabidopsis wild-type Col-0, and the spf1, spf2, and spf1/2 mutants. (A) Chronological scheme of Col-0 development, with selected stages based on phenotypic analysis of soil-grown plants (Boyes et al., 2001); LD, leaf development; RG, rosette growth; IE, inflorescence emergence; FP, flower production. Number of leaves (B) and rosette size (C) of plants at key developmental stage points. (D) Morphology of 1-month-old plants grown under long days. Insets show a representative leaf of each genotype. Scale bars indicate 1 cm. (E, F) Morphological measurements of 1-month-old plants. (G) Age of plants at flowering. Chlorophyll (H), carotenoid (I) and anthocyanin (J) contents in 1-month-old plants relative to Col-0. (K) Number of seeds per silique. (L) Seed morphology in Col-0 and the spf1/2 mutant; the scale bar indicates 1 mm. Seed width (M) and length (N) relative to Col-0. Error bars represent standard error of the means (SEM): n=12 (B, C, E–G); n=6 (H, I); n=5 (J); n=6 (K); and n>36 (M, N). Significant differences with respect to the wild-type were determined using unpaired t-tests *P<0.05; **P<0.01; ***P<0.001.
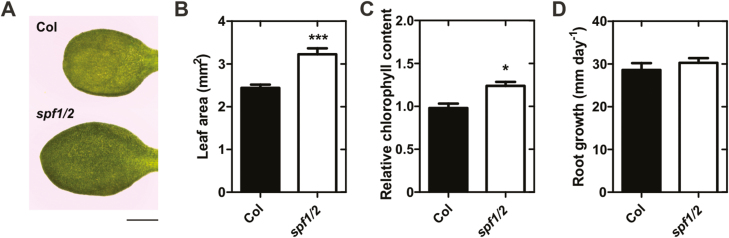
In addition to the phenotypes displayed in plants growing in soil, we noticed that the leaves of plate-grown, 10-d-old spf1/2 mutants were bigger and darker than those of the wild-type (Fig. 7A). We therefore characterized spf1/2 seedlings growing in MS media for 10 d. Compared to the Col-0 wild-type, spf1/2 seedlings displayed a greater leaf area and higher chlorophyll content (Fig. 7B, C), but no differences were observed for root growth (Fig. 7D). In summary, we observed a series of developmental phenotypes in spf1/2, at both earlier and later stages, which revealed that these proteins were important for multiple steps in plant development.
Fig. 7.
Morphological differences between 10-d-old seedlings of wild-type Col-0 and the spf1/2 mutant grown on plates. (A) Morphology of one representative leaf of each genotype. Scale bar represents 1 mm. Leaf area (B), relative chlorophyll content (C), and root growth (D). Error bars represent SEM: n=9 (B); n≥5 (C); and n≥12 (D). Significant differences with respect to the wild-type were determined using unpaired t-tests: *P<0.05; ***P<0.001.
Microarray analysis implicates SPF1/2 in the control of development and secondary metabolism
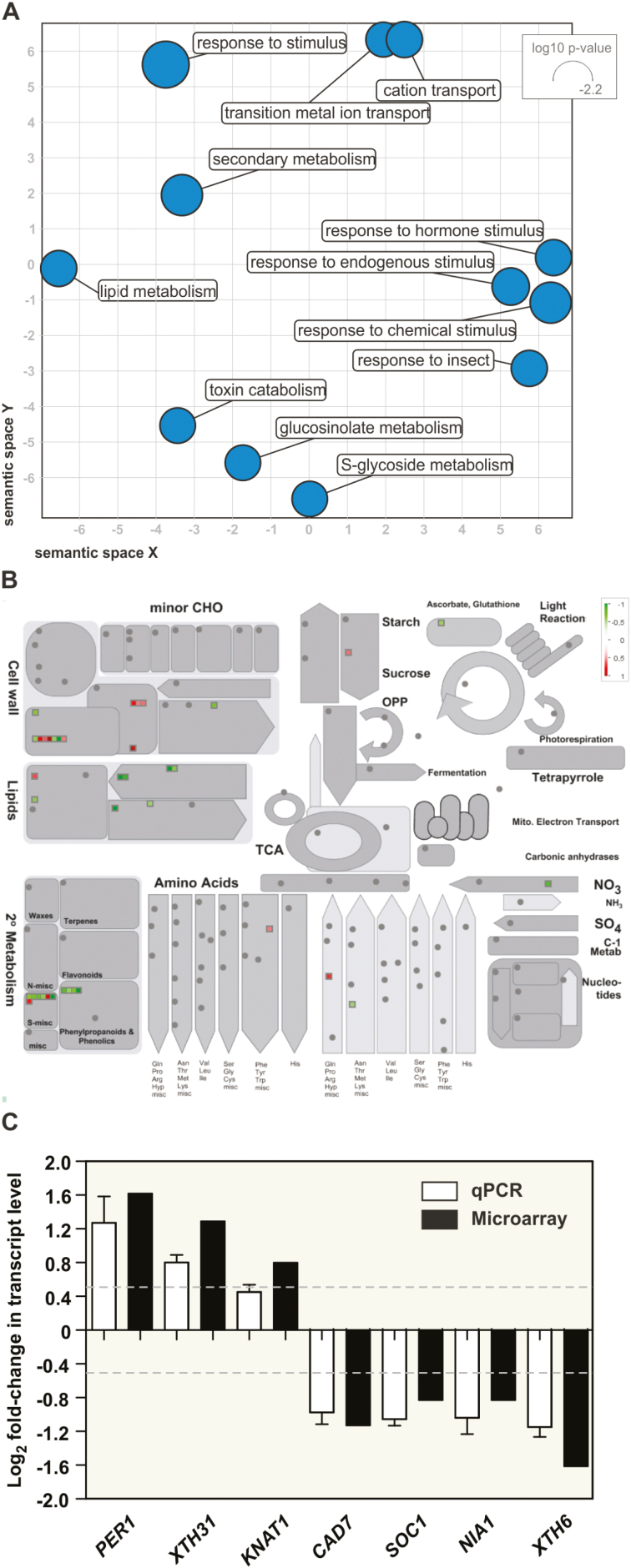
Sumoylation is strongly involved in nuclear mechanisms, particularly in the control of gene transcription through the regulation of chromatin remodelling complexes, co-repressors, and modulators of transcription factor (TF) activity (Mazur and van den Burg, 2012). In light of this, SPF1 and SPF2 might modulate gene expression by promoting desumoylation and counteracting SUMO-dependent control of transcriptional regulators. To determine whether the transcriptional profile correlated with SPF1/2 function, we performed a microarray analysis of 10-d-old wild-type and spf1/2 plants. We had already demonstrated the presence of altered plant morphology (Fig. 7) and SUMO conjugate levels (Fig. 4B) at this developmental stage. Microarray analyses indicated that 115 genes were down-regulated and 100 were up-regulated. Gene ontology (GO) and MapMan analyses were used to compare differential expression against biological processes and the overall metabolic pathways of Arabidopsis (Fig. 8A, B). The results revealed that many differentially expressed genes (DEGs) were involved in cell wall and secondary metabolism, including genes pertaining to the biosynthesis of phenylpropanoids (particularly lignin biosynthesis), glucosinolates and lipids (Fig. 8A, B; Table 1). The majority of these genes were found to be down-regulated. In contrast, one GO category particularly enriched in spf1/2 was the response to hormone stimulus. Although no specific hormone signature could be highlighted, we could observe the up-regulation of genes that are functionally associated with auxin, brassinosteroid, cytokinin, gibberellin, jasmonate, and salicylic acid hormones (Table 1).
Fig. 8.
Transcriptomic analysis of 10-d-old plate-grown spf1/2 seedlings. (A) Scatterplot analysis of enriched gene ontology (GO) terms for spf1/2 differentially expressed genes. The size of the circles indicates the frequency of the GO term. (B) MapMan analysis of spf1/2 deregulated genes using the ‘Metabolism overview pathway’ map. The colour gradient indicates down-regulated genes (green) to up-regulated genes (red). (C) RT-qPCR analysis of differentially expressed genes in the spf1/2 mutant compared to the Col-0 wild-type: PER1 (At1g48130), XTH31 (At3g44990), KNAT1 (At4g08150), CAD7 (At4g37980), SOC1 (At2g45660), NIA1 (At1g77760), and XTH6 (At5g65730). Error bars represent SEM of three independent biological replicates. The dashed lines represent the threshold for log2 fold-change that was used to set differential expression in the microarray experiment.
Table 1.
Genes constitutively deregulated in spf1/2 compared to the wild-type Col-0
| AGI ID | Gene name | Log2 ratio | P-value | Description |
|---|---|---|---|---|
| Hormone metabolism | ||||
| Auxin | ||||
| At1g77690 | LAX3 | –0.65 | 2.41 × 10–4 | Auxin influx carrier |
| At5g35735 | 0.58 | 9.44 × 10–3 | Auxin-responsive | |
| At1g56150 | 0.59 | 6.19 × 10–3 | SAUR-like auxin-responsive | |
| At4g14560 | AXR5, IAA1 | 0.88 | 2.49 × 10–10 | Aux/IAA protein |
| At5g18060 | SAUR23 | 0.96 | <0.01 × 10–11 | SAUR-like auxin-responsive |
| Brassinosteroid | ||||
| At3g30180 | BR6OX2, CYP85A2 | 1.30 | <0.01 × 10–11 | Brassinosteroid-6-oxidase |
| Cytokinin | ||||
| At1g22400 | UGT85A1 | 0.64 | 5.00 × 10–4 | UDP-glycosyltransferase |
| Gibberellin | ||||
| At2g14900 | 0.65 | 2.58 × 10–4 | Gibberellin-regulated | |
| At5g25900 | KO1, CYP701A3, GA3 | 0.71 | 1.45 × 10–5 | Kaurene oxidase |
| Jasmonate | ||||
| At1g52070 | 0.61 | 2.07 × 10–3 | Mannose-binding lectin | |
| At5g42650 | AOS, CYP74A, DDE2 | 0.81 | 2.26 × 10–8 | Allene oxide synthase |
| At1g52100 | 1.09 | <0.01 × 10–11 | Mannose-binding lectin | |
| Salicylic acid | ||||
| At5g38020 | 0.70 | 2.23 × 10–5 | SAM-Mtases | |
| At5g37990 | 0.82 | 1.61 × 10–8 | SAM-Mtases | |
| Secondary metabolism | ||||
| Phenylpropanoids (lignin biosynthesis) | ||||
| At4g37980 | CAD7, ELI3 | –1.13 | <0.01 × 10–11 | Cinnamyl alcohol dehydrogenase |
| At5g66690 | UGT72E2 | –0.81 | 3.20 × 10–8 | UDP-glycosyltransferase |
| At4g39330 | CAD9 | –0.66 | 1.29 × 10–4 | Cinnamyl alcohol dehydrogenase |
| At4g36220 | CYP84A1, FAH1, F5H | –0.56 | 2.57 × 10–2 | Ferulic acid 5-hydroxylase |
| Lipids | ||||
| At1g06080 | ADS1 | –1.51 | <0.01 × 10–11 | Acyl-lipid/acyl-CoA desaturase |
| At5g14180 | MPL1 | –1.50 | <0.01 × 10–11 | Myzus persicae-induced lipase |
| At5g04530 | KCS19 | –1.02 | <0.01 × 10–11 | 3-ketoacyl-CoA synthase |
| At1g06350 | –0.91 | 4.48 × 10–11 | Fatty acid desaturase | |
| At3g08770 | LTP6 | –0.91 | 4.48 × 10–11 | Lipid transfer protein |
| At4g34250 | KCS16 | –0.62 | 1.47 × 10–3 | 3-ketoacyl-CoA synthase |
| At3g11670 | DGD1 | –0.60 | 2.80 × 10–3 | UDP-glycosyltransferase |
| At4g38690 | –0.56 | 1.92 × 10–2 | PLC-like phosphodiesterase | |
| Glucosinolates | ||||
| At3g14210 | ESM1 | –1.72 | <0.01 × 10–11 | Epithiospecifier modifier |
| At4g13770 | CYP83A1, REF2 | –0.74 | 2.35 × 10–6 | Cytochrome P450 |
| At2g43100 | LEUD1, IPMI2 | –0.68 | 5.43 × 10–5 | Isopropylmalate isomerase |
| At5g23010 | IMS3, MAM1 | –0.64 | 5.52 × 10–4 | Methylthioalkylmalate synthase |
| At1g07640 | OBP2 | –0.60 | 3.01 × 10–3 | DOF transcription factor |
| At3g44320 | NIT3 | 0.75 | 1.26 × 10–6 | Nitrilase |
| At1g54010 | GLL22 | 0.90 | 6.97 × 10–11 | GDSL-like lipase/acylhydrolase |
| Cell wall | ||||
| At5g65730 | XTH6 | –1.61 | <0.01 × 10–11 | XTH |
| At1g67750 | –0.66 | 1.31 × 10–4 | Pectate lyase | |
| At5g47500 | PME5 | –0.63 | 8.57 × 10–4 | Pectin methylesterase |
| At4g28250 | EXPB3 | –0.59 | 6.49 × 10–3 | Beta-expansin |
| At3g23730 | XTH16 | –0.59 | 6.24 × 10–3 | XTH |
| At1g20190 | EXPA11 | 0.57 | 1.08 × 10–2 | Alpha-expansin |
| At1g55850 | CSLE1 | 0.57 | 1.49 × 10–2 | Cellulose synthase/transferase |
| At3g29810 | COBL2 | 0.59 | 4.76 × 10–3 | COBRA-like protein precursor |
| At2g06850 | XTH4, EXGT-A1, EXT | 0.63 | 6.61 × 10–4 | XTH |
| At3g28180 | CSLC4 | 0.78 | 1.94 × 10–7 | Cellulose synthase/transferase |
| At4g30290 | XTH19 | 0.88 | 2.09 × 10–10 | XTH |
| At5g33290 | XGD1 | 0.95 | <0.01 × 10–11 | Xylogalacturonan xylosyltransferase |
| At3g44990 | XTH31, XTR8 | 1.29 | <0.01 × 10–11 | XTH |
| Other | ||||
| At2g45660 | SOC1, AGL20 | –0.83 | 6.01 × 10–9 | AGAMOUS-like transcription factor |
| At1g77760 | NIA1, GNR1, NR1 | –0.83 | 7.34 × 10–9 | Nitrate reductase |
| At4g21680 | NRT1.8 | 0.61 | 1.81 × 10–3 | Nitrate transporter |
| At5g50200 | NRT3.1, WR3 | 0.62 | 1.11 × 10–3 | Nitrate transporter |
XTH, Xyloglucan endotransglucosylase/hydrolase; SAM-Mtases, S-adenosyl-L-methionine-dependent methyltransferase.
The categories were chosen taking in consideration the enrichment of gene ontology (GO) terms, and the list was gathered using Classification SuperViewer (Toufighi et al., 2005) and The Arabidopsis Information Resource (TAIR) (Lamesch et al., 2010).
Interestingly, some genes previously described as being deregulated in siz1 mutants were inversely expressed in spf1/2 DEGs. Examples included nitrate reductase NIA1 (At1g77760), the AGAMOUS-like transcription factor SOC1 (At2g45660), and the xyloglucan endotransglucosylase/hydrolase XTH31 (At3g44990) (Jin et al., 2008; Miura et al., 2010; Park et al., 2011), which are involved in N-assimilation, flowering time, and cell growth, respectively. In spf1/2, the observed deregulation in transcript levels for these and other genes was confirmed by RT-qPCR (Fig. 8C), thus validating our microarray data.
Co-expressed genes tend to be controlled by identical transcriptional regulators, and share common cis-elements in their promoters. Given that sumoylation often targets regulators of transcription, we identified statistically over-represented cis-elements in the promoters of spf1/2 DEGs that may act as binding sites for SUMO target candidates. In our DEGs, we were able to observe an enrichment in MYC2-like binding sites (Supplementary Table S4) in both up- and down-regulated genes.
SIZ1 is epistatic to SPF1/2
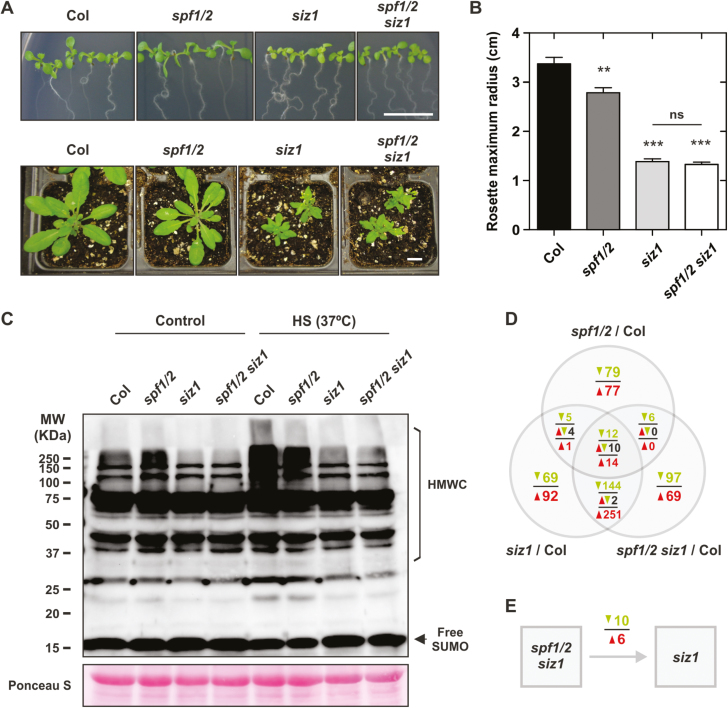
When we compared spf1/2 to mutants of the Arabidopsis SUMO conjugation pathway, it become clear that spf1/2 displayed antagonistic phenotypes to those of siz1. SIZ1 is the major SUMO E3 ligase and has been the subject of most functional studies in the pathway. In contrast to SPF1/2, loss of SIZ1 function induces diminished accumulation of SUMO conjugates, early flowering, and decreased pigment content (Catala et al., 2007; Jin et al., 2008; P.H. Castro et al., unpublished results), suggesting an epistatic relationship between SIZ1 and SPFs. To further examine this, we generated a spf1/2 siz1 triple-mutant and determined its phenotype characterization. Morphologically, the triple-mutant resembled siz1 and was similarly affected in the accumulation of high molecular weight SUMO conjugates, even after HS treatment (Fig. 9A–C), suggesting that SIZ1 was acting upstream of SPF1/2.
Fig. 9.
Characterization of the spf1/2 siz1 triple-mutant. (A) Morphology of 10-d-old plate-grown and 1-month-old soil-grown plants. (B) Rosette maximum radius. Error bars represent SEM, n=7. Significant differences for mutants compared to the wild-type Col-0, and for siz1 compared to spf1/2 siz1 were determined using unpaired t-tests: ns, non-significant; **P<0.01; ***P<0.001). (C) Western blot analysis of high molecular weight SUM1 conjugates (HMWC) in 10-d-old Col-0, spf1/2, siz1, and spf1/2 siz1 subjected to heat shock (HS) for 1 h. (D) Venn diagram representing differentially expressed genes in each mutant genotype compared to the wild-type Col-0. (E) Differentially expressed genes in the spf1/2 siz1 triple-mutant in relation to the single-mutant siz1. In (D, E) the colour scheme represents down-regulated genes (green), up-regulated genes (red), and anti-expressed genes (black).
Transcript profiling was carried out on the spf1/2 siz1 triple-mutant, and this was compared to the siz1 and spf1/2 transcriptomes (Fig. 9D). We identified DEGs in all three mutant genotypes in comparison to the wild-type, and we then cross-referenced the three data subsets. A total of 26 genes were similarly differentially expressed in all three mutant backgrounds. These included the bHLH transcription factor FBI1/HFR1/REP1/RSF1 and the putative phytochrome kinase substrate At1g18810, both of which are involved in phytochrome signalling (Fairchild et al., 2000; Schepens et al., 2008). The results showed an extensive overlap in the differential transcriptome of the siz1 and spf1/2 siz1 mutants (Fig. 9D). When we compared expression values of spf1/2 siz1 directly to siz1, only 10 genes were down-regulated and six were up-regulated, indicating that their transcriptome virtually matched (Fig. 9E). This transcriptomic data reinforced the notion that SIZ1 was upstream of, and epistatic to, SPF1/2.
Discussion
Sumoylation is essential for eukaryotic organisms, mainly because it regulates the activity of vital proteins. It is therefore crucial that SUMO homeostasis is tightly controlled, and in recent years some studies have shed light on SUMO protease activities and their essential roles in many aspects of cellular homeostasis (reviewed by Hickey et al., 2012). In plant genomes, as in other organisms, SUMO proteases seem to be more abundant in number than the E1/E2/E3 components of the conjugation machinery, making them prime candidates for the regulation of SUMO conjugation/deconjugation homeostasis. In the present study, we performed a functional characterization of SPF1 and SPF2, two ULP2s that form a separate phylogenetic subgroup within Arabidopsis ULPs. Our results support a conserved evolutionary role for both proteins in plant growth and development.
Previous phylogenetic studies singled out SPF1 and SPF2 as homologs of yeast Ulp2 and mammalian SENP6/7, making them natural candidates for poly-SUMO chain editing proteases in Arabidopsis (Hickey et al., 2012). Here, we report a more thorough phylogenetic and comparative genomics approach that suggests the presence of three ULP2 classes within plant genomes. These display a topological pattern of specific loops within the catalytic domain that separate them from plant ULP1 SUMO proteases (Fig. 2B). In humans, it has been shown that the catalytic domains of the ULP2s SENP6 and SENP7 create loops for SUMO recognition (Lima and Reverter, 2008; Alegre and Reverter, 2011). For example, SENP6/7 loop 1 is essential for activity and SUMO isoform discrimination, but it is not conserved either in yeast or plant ULP2s, highlighting the caveats that must be placed on functional inference based on ULP homology. An interesting characteristic that is intrinsic to the SPF-type of plant ULP2s is that the catalytic domain is located in the middle of the protein (Fig. 2A), a feature shared with yeast and algae Ulp2 paralogs, suggesting that this may be the most ancestral group, as opposed to the OTS-type of ULP2 proteases. With regards to the function of the N- and C-terminal ends, the model proposed for yeast ULP2 is that the N-terminal domain acts mainly in nuclear targeting (Kroetz et al., 2009), whereas the C-terminal end contains motifs for PTMs such as phosphorylation (Baldwin et al., 2009). In agreement with this, the Arabidopsis SPF1 C-terminal end was previously identified as being a phosphorylation target (PhosPhAt database; Durek et al., 2010). It is important to note that other ULP2-like proteases have previously been proposed by Kurepa et al. (2003) and Lois (2010). However, these putative ULP-like genes are part of transposon elements (Hoen et al., 2006) and were designated Kaonashi ULP-like (KIU) sequences. Although they potentially have catalytically functional domains, their SUMO protease activities have never been studied. Nevertheless, KIUs also belong to a phylogenetically distant branch from the remaining ULP family members and are strongly silenced (Hoen et al., 2006), suggesting a minor contribution to SUMO regulation if it is the case that they do function as SUMO proteases.
SUMO proteases can have a dual function as both maturases of the pre-SUMO peptide or as isopetidases that remove SUMO conjugates from targets, and it is important to establish the individual contribution of the different ULPs to each biochemical role. Loss of SPF1/2 function resulted in the constitutive accumulation of high molecular weight SUMO conjugates (Fig. 4), implicating them as SUMO isopeptidases. This is consistent with their phylogenetic proximity with yeast and human ULP2 proteins, both of which display major isopeptidase activity (Lima and Reverter, 2008; Eckhoff and Dohmen, 2015). Here, we further demonstrated that SPF2 was capable of complementing ulp2∆ but not ulp1-ts, placing this plant protease as a functional homolog of the yeast Ulp2. The observed dominant negative effect of SPF1 on ulp2∆ also suggested a functional correlation with its yeast ortholog, in which the existing topological differences (Fig. 2) may accommodate the observed phenotype. SPF1 and SPF2 both displayed reactivity of their catalytic domain with human HA-SUMO-VME probes, albeit with separate affinities for different SUMO isoforms (Fig. 3B). Our data, combined with recent studies demonstrating endopeptidase activity of SPF1 and SPF2 (Kong et al., 2017; Liu et al., 2017a), make a definitive case for SPF1/2 functioning as SUMO proteases.
In planta, SPF1 and SPF2 loss-of-function mutants coincided in a series of developmental defects. Several of our results supported the existence of unequal redundancy, tending towards SPF1 as being more important: (1) SPF1 seemed to be much more expressed than SPF2, as shown by semi-quantitative RT-PCR (Supplementary Fig. S3) and by publicly available transcriptomic data (Supplementary Fig. S6); (2) compared to spf2, spf1 mutant alleles displayed more prominent phenotypes in leaf morphology, flowering time, pigment accumulation, and increased SUMO conjugates (Figs 4, 6, 7; Supplementary Fig. S5); (3) several plant genomes display a single-plant SPF1/2 subgroup member (e.g. Selaginella moellendorffii, Oryza sativa, and Amborella trichopoda), and the Arabidopsis SPF1/SPF2 duplication seems to map to a dicot-specific event. Previous functional reports also support this claim (Kong et al., 2017; Liu et al., 2017a).
SPF1/2 control a series of development features, making them interesting candidate genes for crop improvement. The spf1/2 mutant phenotypes included (1) late flowering, indicative of a delay in development; (2) altered leaf morphology; and (3) severely impaired seed production (Fig. 6). However, seeds were also bigger, which may provide an interesting potential for increasing seed size in crop species (Fig. 6L–N). We have shown that SPF1/2 controls several genes involved in secondary metabolism (Fig. 8A, B; Table 1), which may explain the observed developmental defects. For instance, genes involved in glucosinolates and lignin deposition, such as Ferulic acid 5-hydroxylase (F5H), were down-regulated in spf1/2, suggesting that SPF1 and SPF2 act as positive regulators of lignin deposition. Down-regulation of lignin biosynthesis may cause net flux changes through the phenylpropanoid metabolism that could explain why spf1/2 displayed increased anthocyanin content. In support of this, the metabolic interaction between lignin and anthocyanin biosynthesis has been previously reported (Ring et al., 2013). The observed differences in leaf morphology displayed by both plate-grown and adult spf1/2 mutants may have reflected changes in either life cycle or cell expansion. Both factors have been associated with SUMO pathway mutants (Murtas et al., 2003; Miura et al., 2010), and both factors contribute to the multiple and complex regulatory modules regulating leaf morphology (Gonzalez et al., 2010). Indeed, several components of the cell wall remodelling apparatus were affected in spf1/2, including members of the xyloglucan endotransglucosylase/hydrolase (XTH) family such as XTH31, which has previously been observed to be down-regulated in siz1 (Miura et al., 2010) and was over-expressed in spf1/2 (Fig. 8C). Most significantly, we have found substantial evidence that many phenotypes displayed by spf1/2 oppose those of siz1, including SUMO-conjugate accumulation, late flowering, higher pigment contents, and reduced accumulation of reactive oxygen species (P.H. Castro et al., unpublished results). Here, the spf1/2 siz1 triple-mutant morphologically resembled the siz1 single-mutant, suggesting that SPF1/2 are epistatic to SIZ1.
Both mammalian SENP and yeast ULP vary in their subnuclear localization (reviewed by Wilkinson and Henley, 2010) and contribute differently to SUMO dynamics within the nucleus. In Arabidopsis, ULPs display a variety of sub-cellular localizations: ESD4 in the nuclear envelope, OTS2 in speckle-like bodies of the nucleoplasm, OTS1 in the nucleoplasm, and ELS1 in the cytoplasm and endomembranes (Murtas et al., 2003; Conti et al., 2008; Hermkes et al., 2011). Recently, SPF1 and SPF2 were both localized in the nucleus (Kong et al., 2017; Liu et al., 2017a). In addition, we observed that SPF1 and SPF2 were both located in the nucleoplasm and in nuclear bodies (Fig. 5). In accordance with this, plant SUMO conjugates are mainly nuclear-targeted proteins and ULPs contribute to the regulation of nuclear SUMO dynamics (Saracco et al., 2007; Elrouby and Coupland, 2010; Miller et al., 2010). Among SUMO targets are transcription factors, co-repressor complexes, histones, mRNA biogenesis proteins, and many other components associated with nuclear processes (Mazur and van den Burg, 2012). In addition to previous reports that SIZ1 and OTS1/2 significantly influence the plant transcriptome (Castro et al., 2016; Catala et al., 2007), SPF1/2 were also involved in transcription regulation, and seemed to mainly influence secondary metabolism, N-assimilation, and flowering time. Some of the DEGs that we found such as NIA1, SOC1, and XTH31 (Fig. 8; Table 1) have previously been associated with SIZ1 regulation but with the opposite behaviour. As previously noted, the spf1/2 siz1 triple-mutant phenotypically resembled siz1 and, accordingly, the transcriptional profile of spf1/2 siz1 was superimposed on that of siz1 but not spf1/2. Taken together, SPF1/2 function seemed to take place downstream of SIZ1. The simplest model is that targets of SIZ1-dependent sumoylation are subjected to SPF1/2 desumoylation. Most bona fide candidates include transcription factors such as PHR1, ICE1, ABI5, HSFA2, and MYB30 (Miura et al., 2005, 2007b, 2009; Cohen-Peer et al., 2010; Zheng et al., 2012). Cis-element enrichment analysis also highlighted MYC2 as a potential target for SPF1/2 regulation (Supplementary Table S4), and in support of this MYC2 has previously been shown to be sumoylated in vitro (Elrouby and Coupland, 2010).
Sumoylation of target proteins is largely under the control of SIZ1 E3 ligase activity (Miura et al., 2005; Catala et al., 2007). Although many SUMO machinery components are sumoylated under normal conditions, SIZ1 is the only heavily sumoylated protein under stress conditions (e.g. HS, ethanol, and H2O2) (Miller et al., 2013). One possibility is that SIZ1 may be one of the major targets of SPF1/2. In accordance with this hypothesis, yeast Siz1 and Siz2 are high-copy suppressors of ulp2Δ phenotypes, suggesting that the requirement for yeast Ulp2 is bypassed by SIZ1 overexpression (Strunnikov et al., 2001; Hannich et al., 2005). However, plants might display higher complexity, since spf1/2 and siz1 revealed opposing phenotypes in our current study and their transcriptomes were not significantly co- or inversely expressed (Fig. 9E).
An often-neglected aspect to consider when addressing Arabidopsis ULPs is a possible functional redundancy between different ULP subgroup members. For example, esd4 and ots1/2 mutants have been shown to accumulate high molecular weight SUMO conjugates under non-stress conditions (Murtas et al., 2003; Xu et al., 2007; Conti et al., 2008; Castro et al., 2016) and ESD4, ELS1, OTS1, and OTS2 have shown SUMO1/2 isopeptidase activity in vitro (Chosed et al., 2006; Colby et al., 2006; Conti et al., 2008; Hermkes et al., 2011). On the other hand, we have previously reported that the triple-mutant ots1/2 siz1 showed accumulative defects, which partially place OTS1/2 and SIZ1 in different pathways (Castro et al., 2016). The esd4 siz1 mutant, like spf1/2 siz1, resembles siz1 (P.H. Castro et al., unpublished results), but SIZ1 and ESD4 are also likely to function in different pathways since the siz1 pleiotropic phenotype is largely reverted in the NahG background (expressing a bacterial SA hydroxylase that hydrolyses SA), while esd4 is not (Hermkes et al., 2011). However, more recently Villajuana-Bonequi et al. (2014) reported that a mutation in the ICS1/SID2 gene, a key enzyme in SA biosynthesis, is able to partially suppress esd4 developmental defects, suggesting that ESD4 and SIZ1 may overlap in some functions. Discriminating desumoylation targets for each ULP will be an important step towards dissecting the circuitry of regulation via SUMO removal, and ultimately identifying the origin of specificity within the sumoylation pathway. This goal can be achieved by combining mutant backgrounds of ULPs with previously demonstrated high-throughput strategies for identifying sumoylomes (Miller et al., 2010).
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Protein sequence alignment of the catalytic domain of SPF1/2 subgroup members.
Fig. S2. Purification elution of recombinant proteins with the SPF2 and SPP1 catalytic domains with an N-terminus GST-tag.
Fig. S3. Schematic representation of Arabidopsis T-DNA insertion mutants for SPF2 and SPF1 and semi-quantitative RT-PCR.
Fig. S4. Plant and silique size of the wild-type Col-0 and spf1/2 mutant.
Fig. S5. Morphology of 1-month-old plants of the SPF2 and SPF1 second-allele T-DNA mutant.
Fig. S6. In silico analysis of SPF2 and SPF1 expression patterns.
Table S1. List of primers used for genotyping Arabidopsis T-DNA insertion lines.
Table S2. List of primers used in semi-quantitative and quantitative RT-PCR.
Table S3. List of primers used for plasmid constructs.
Table S4. Cis-elements over-represented in the promoter region of differentially expressed genes in spf1/2.
Acknowledgements
We thank Mark Hochstrasser (Department of Molecular Biophysics & Biochemistry, Yale University, New Haven, CT, USA) for kindly providing the ulp1-ts yeast mutant strain. This research was funded by FEDER (through COMPETE), and by Fundação para a Ciência e Tecnologia (FCT), within the scope of project SUMOdulator (FCOMP-01-0124-FEDER-028459 and PTDC/BIA-PLA/3850/2012). PHC was supported by FCT (SFRH/BD/44484/2008). HA and FF were supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (FEDER) (NORTE-01-0145-FEDER-000007 and Norte-01-0145-FEDER-000008, respectively). The work was supported by FEDER through the COMPETE 2020—Operacional Programme for Competitiveness and Internationalisation (POCI), Portugal 2020, and by Portuguese funds through FCT, within the framework of projects ‘Rede de Investigação em Biodiversidade e Biologia Evolutiva’ (POCI-01-0145-FEDER-006821) and ‘Institute for Research and Innovation in Health Sciences’ (POCI-01-0145-FEDER-007274). This research was also supported by a grant from the Spanish Ministerio de Ciencia y Tecnología (AGL2016-75819-C2-1-R) and FEDER (PCQ, AGC, ERB).
References
- Alegre KO, Reverter D. 2011. Swapping small ubiquitin-like modifier (SUMO) isoform specificity of SUMO proteases SENP6 and SENP7. The Journal of Biological Chemistry 286, 36142–36151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. 1949. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology 24, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M, Srivastava A, Conti L, et al. . 2016. Stability of small ubiquitin-like modifier (SUMO) proteases OVERLY TOLERANT TO SALT1 and -2 modulates salicylic acid signalling and SUMO1/2 conjugation in Arabidopsis thaliana. Journal of Experimental Botany 67, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin ML, Julius JA, Tang X, Wang Y, Bachant J. 2009. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle 8, 3406–3419. [DOI] [PubMed] [Google Scholar]
- Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. 2002. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chemistry & Biology 9, 1149–1159. [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. 2001. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. The Plant Cell 13, 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Cappadocia L, Lima CD. 2018. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chemical Reviews 118, 889–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro PH, Couto D, Freitas S, et al. . 2016. SUMO proteases ULP1c and ULP1d are required for development and osmotic stress responses in Arabidopsis thaliana. Plant Molecular Biology 92, 143–159. [DOI] [PubMed] [Google Scholar]
- Castro PH, Lilay GH, Muñoz-Mérida A, Schjoerring JK, Azevedo H, Assunção AGL. 2017. Phylogenetic analysis of F-bZIP transcription factors indicates conservation of the zinc deficiency response across land plants. Scientific Reports 7, 3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro PH, Tavares RM, Bejarano ER, Azevedo H. 2012. SUMO, a heavyweight player in plant abiotic stress responses. Cellular and Molecular Life Sciences 69, 3269–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro PH, Verde N, Lourenço T, Magalhães AP, Tavares RM, Bejarano ER, Azevedo H. 2015. SIZ1-dependent post-translational modification by SUMO modulates sugar signaling and metabolism in Arabidopsis thaliana. Plant & Cell Physiology 56, 2297–2311. [DOI] [PubMed] [Google Scholar]
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH. 2007. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. The Plant Cell 19, 2952–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosed R, Mukherjee S, Lois LM, Orth K. 2006. Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. The Biochemical Journal 398, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Peer R, Schuster S, Meiri D, Breiman A, Avni A. 2010. Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotholerance. Plant Molecular Biology 74, 33–45. [DOI] [PubMed] [Google Scholar]
- Colby T, Matthäi A, Boeckelmann A, Stuible HP. 2006. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiology 142, 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Nelis S, Zhang C, et al. . 2014. Small ubiquitin-like modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Developmental Cell 28, 102–110. [DOI] [PubMed] [Google Scholar]
- Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A. 2008. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. The Plant Cell 20, 2894–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas-Potts C, Matunis MJ. 2013. SUMO: a multifaceted modifier of chromatin structure and function. Developmental Cell 24, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX. 2010. PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Research 38, D828–D834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhoff J, Dohmen RJ. 2015. In vitro studies reveal a sequential mode of chain processing by the yeast SUMO (small ubiquitin-related modifier)-specific protease Ulp2. The Journal of Biological Chemistry 290, 12268–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N. 2015. Analysis of small ubiquitin-like modifier (SUMO) targets reflects the essential nature of protein SUMOylation and provides insight to elucidate the role of SUMO in plant development. Plant Physiology 169, 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N, Coupland G. 2010. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proceedings of the National Academy of Sciences, USA 107, 17415–17420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. 2000. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes & Development 14, 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, Balzergue S, Aubourg S, Renou JP, Lecharny A, Brunaud V. 2008. CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Research 36, D986–D990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nature Reviews Molecular Cell Biology 11, 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garí E, Piedrafita L, Aldea M, Herrero E. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13, 837–848. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, et al. . 2010. Increased leaf size: different means to an end. Plant Physiology 153, 1261–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27, 221–224. [DOI] [PubMed] [Google Scholar]
- Grou CP, Pinto MP, Mendes AV, Domingues P, Azevedo JE. 2015. The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Scientific Reports 5, 12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M. 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. The Journal of Biological Chemistry 280, 4102–4110. [DOI] [PubMed] [Google Scholar]
- Hendriks IA, Vertegaal AC. 2016. A comprehensive compilation of SUMO proteomics. Nature Reviews Molecular Cell Biology 17, 581–595. [DOI] [PubMed] [Google Scholar]
- Hermkes R, Fu YF, Nürrenberg K, Budhiraja R, Schmelzer E, Elrouby N, Dohmen RJ, Bachmair A, Coupland G. 2011. Distinct roles for Arabidopsis SUMO protease ESD4 and its closest homolog ELS1. Planta 233, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CM, Wilson NR, Hochstrasser M. 2012. Function and regulation of SUMO proteases. Nature Reviews Molecular Cell Biology 13, 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen DR, Park KC, Elrouby N, Yu Z, Mohabir N, Cowan RK, Bureau TE. 2006. Transposon-mediated expansion and diversification of a family of ULP-like genes. Molecular Biology and Evolution 23, 1254–1268. [DOI] [PubMed] [Google Scholar]
- Jin JB, Jin YH, Lee J, et al. . 2008. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. The Plant Journal 53, 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, et al. . 2010. VirtualPlant: a software platform to support systems biology research. Plant Physiology 152, 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolli N, Mikolajczyk J, Drag M, Mukhopadhyay D, Moffatt N, Dasso M, Salvesen G, Wilkinson KD. 2010. Distribution and paralogue specificity of mammalian deSUMOylating enzymes. The Biochemical Journal 430, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Luo X, Qu GP, Liu P, Jin JB. 2017. Arabidopsis SUMO protease ASP1 positively regulates flowering time partially through regulating FLC stability. Journal of Integrative Plant Biology 59, 15–29. [DOI] [PubMed] [Google Scholar]
- Kroetz MB, Su D, Hochstrasser M. 2009. Essential role of nuclear localization for yeast Ulp2 SUMO protease function. Molecular Biology of the Cell 20, 2196–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD. 2003. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. The Journal of Biological Chemistry 278, 6862–6872. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Dreher K, Swarbreck D, Sasidharan R, Reiser L, Huala E. 2010. Using the Arabidopsis information resource (TAIR) to find information about Arabidopsis genes. Current Protocols in Bioinformatics 30, 1.11.1–1.11.51. [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. 1999. A new protease required for cell-cycle progression in yeast. Nature 398, 246–251. [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Molecular and Cellular Biology 20, 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Buschmann C. 2001. Chlorophylls and carotenoids: measurement and characterization by UV-VIS spectroscopy. In: Wrolstad RE, Acree TE, An H, et al. , eds. Current Protocols in Food Analytical Chemistry. New York: John Wiley & Sons, Inc, F4.3.1–F4.3.8. [Google Scholar]
- Lima CD, Reverter D. 2008. Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. The Journal of Biological Chemistry 283, 32045–32055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Jiang Y, Zhang X, et al. . 2017a. Two SUMO proteases SUMO PROTEASE RELATED TO FERTILITY1 and 2 are required for fertility in Arabidopsis. Plant Physiology 175, 1703–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yan X, Kong X, Zhao Y, Gong Z, Jin JB, Guo Y. 2017b. Transcriptional gene silencing maintained by OTS1 SUMO protease requires a DNA-dependent polymerase V-dependent pathway. Plant Physiology 173, 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois LM. 2010. Diversity of the SUMOylation machinery in plants. Biochemical Society Transactions 38, 60–64. [DOI] [PubMed] [Google Scholar]
- Lozano-Durán R, Rosas-Díaz T, Gusmaroli G, Luna AP, Taconnat L, Deng XW, Bejarano ER. 2011. Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. The Plant Cell 23, 1014–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur MJ, van den Burg HA. 2012. Global SUMO proteome responses guide gene regulation, mRNA biogenesis, and plant stress responses. Frontiers in Plant Science 3, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. 2010. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proceedings of the National Academy of Sciences, USA 107, 16512–16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Scalf M, Rytz TC, Hubler SL, Smith LM, Vierstra RD. 2013. Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Molecular & Cellular Proteomics 12, 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Hasegawa PM. 2010. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends in Cell Biology 20, 223–232. [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Hasegawa PM. 2007a. Sumoylation, a post-translational regulatory process in plants. Current Opinion in Plant Biology 10, 495–502. [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, et al. . 2007b. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. The Plant Cell 19, 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. 2009. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proceedings of the National Academy of Sciences, USA 106, 5418–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Miura T, Hasegawa PM. 2010. SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant & Cell Physiology 51, 103–113. [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, et al. . 2005. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proceedings of the National Academy of Sciences, USA 102, 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–475. [Google Scholar]
- Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G. 2003. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. The Plant Cell 15, 2308–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M, Tomanov K, Hofmann K, Stuible HP, Bachmair A. 2012. Update on sumoylation: defining core components of the plant SUMO conjugation system by phylogenetic comparison. New Phytologist 195, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Song JT, Seo HS. 2011. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nature Communications 2, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto MP, Carvalho AF, Grou CP, Rodríguez-Borges JE, Sá-Miranda C, Azevedo JE. 2012. Heat shock induces a massive but differential inactivation of SUMO-specific proteases. Biochimica et Biophysica Acta 1823, 1958–1966. [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K. 2015. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Research 43, D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring L, Yeh SY, Hücherig S, et al. . 2013. Metabolic interaction between anthocyanin and lignin biosynthesis is associated with peroxidase FaPRX27 in strawberry fruit. Plant Physiology 163, 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Adam E, Orosa B, Viczian A, Klose C, Zhang C, Josse EM, Kozma-Bognar L, Nagy F. 2015. SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 112, 11108–11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD. 2007. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiology 145, 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepens I, Boccalandro HE, Kami C, Casal JJ, Fankhauser C. 2008. PHYTOCHROME KINASE SUBSTRATE4 modulates phytochrome-mediated control of hypocotyl growth orientation. Plant Physiology 147, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienhorst I, Johnson ES, Dohmen RJ. 2000. SUMO conjugation and deconjugation. Molecular & General Genetics 263, 771–786. [DOI] [PubMed] [Google Scholar]
- Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyán J. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. The EMBO Journal 21, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simossis VA, Heringa J. 2005. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Research 33, W289–W294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Aravind L, Koonin EV. 2001. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics 158, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Abel S. 2001. Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiology 127, 963–972. [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. 2005. The botany array resource: e-Northerns, expression angling, and promoter analyses. The Plant Journal 43, 153–163. [DOI] [PubMed] [Google Scholar]
- Van Bel M, Proost S, Wischnitzki E, Movahedi S, Scheerlinck C, Van de Peer Y, Vandepoele K. 2012. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiology 158, 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg HA, Kini RK, Schuurink RC, Takken FL. 2010. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. The Plant Cell 22, 1998–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RA. 2008. Plant proteases: from phenotypes to molecular mechanisms. Annual Review of Plant Biology 59, 191–223. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. 2012. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiology 160, 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villajuana-Bonequi M, Elrouby N, Nordstrom K, Griebel T, Bachmair A, Coupland G. 2014. Elevated salicylic acid levels conferred by increased expression of ISOCHORISMATE SYNTHASE 1 contribute to hyperaccumulation of SUMO1 conjugates in the Arabidopsis mutant early in short days 4. The Plant Journal 79, 206–219. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. 2010. Mechanisms, regulation and consequences of protein SUMOylation. The Biochemical Journal 428, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I. 2007. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. The Plant Cell 19, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates G, Srivastava AK, Sadanandom A. 2016. SUMO proteases: uncovering the roles of deSUMOylation in plants. Journal of Experimental Botany 67, 2541–2548. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Schumaker KS, Guo Y. 2012. Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 109, 12822–12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.