SUMMARY
Triple-negative breast cancer (TNBC) is an aggressive subtype that frequently develops resistance to chemotherapy. An unresolved question is whether resistance is caused by the selection of rare pre-existing clones or alternatively through the acquisition of new genomic aberrations. To investigate this question, we applied single cell DNA and RNA sequencing in addition to bulk exome sequencing to profile longitudinal samples from 20 TNBC patients during neoadjuvant chemotherapy (NAC). Deep-exome sequencing identified 10 patients where NAC led to clonal extinction and 10 patients where clones persisted after treatment. In 8 patients, we performed a more detailed study using single cell DNA sequencing to analyze 900 cells and single cell RNA sequencing to analyze 6,862 cells. Our data showed that resistant genotypes were pre-existing and adaptively selected by NAC, while transcriptional profiles were acquired by reprogramming in response to chemotherapy in TNBC patients.
INTRODUCTION
Triple-negative breast cancer (TNBC) is an aggressive subtype that constitutes 12–18% of breast cancer patients (Foulkes et al., 2010). TNBC patients lack the estrogen receptor (ER), progesterone receptor (PR) and HER2 receptor and therefore are not eligible for hormone or anti-Her2 therapy. Deep-sequencing studies (Balko et al., 2012; Balko et al., 2014; Shah et al., 2012), multi-region sequencing analysis (Yates et al., 2015), and single cell sequencing studies (Gao et al., 2016; Navin et al., 2011; Wang et al., 2014) have shown that TNBC patients harbor high levels of somatic mutations, frequent mutations in TP53 (83%) and complex aneuploid rearrangements (80%) that result in extensive intratumor heterogeneity (ITH).
The standard of care for many TNBC patients is neoadjuvant chemotherapy (NAC), which includes a combination of taxanes (mitotic inhibitors) and anthracyclines (DNA intercalators). While NAC is effective in some TNBC patients, about 50% evolve resistance, leading to poor overall survival (Foulkes et al., 2010; Liedtke et al., 2008). The genomic and molecular basis of chemoresistance in TNBC patients remains poorly understood, in part due to a lack of methods that can resolve ITH and detect genomic information in rare subpopulations. A major gap in knowledge is whether chemoresistance arises due to the selection and expansion of rare pre-existing subclones (adaptive resistance), or, alternatively, through the induction of new mutations that confer a chemoresistant phenotype (acquired resistance) (Navin, 2014). This question has been studied for decades in bacterial systems (Luria and Delbrück, 1943) but remains poorly understood in most human cancers.
Previous genomic studies of therapy resistance have reported acquired resistance (Ding et al., 2012; Kim et al., 2015; Kolodziejczyk et al., 2015; Patch et al., 2015) or adaptive resistance (Ding et al., 2012; Kurtova et al., 2015) to systemic chemotherapies in different human cancers. In acute myeloid leukemia, whole-genome sequencing identified different modes of clonal evolution, with some patients acquiring relapse-specific mutations and others selecting minor clones (Ding et al., 2012). In high-grade serous ovarian cancer, platinum-based chemotherapy induced new somatic mutations, consistent with acquired resistance (Patch et al., 2015), while resistance to cytotoxic chemotherapy in bladder cancer was associated with the selection of pre-existing subpopulations (Kurtova et al., 2015). In glioblastoma, treatment with temozolomide induced many new mutations in post-treatment tumor samples, consistent withan acquired model of therapy resistance (Kim et al., 2015; Kolodziejczyk et al., 2015).
Previous work on chemoresistance in TNBC patients has focused mainly on in situ hybridization methods (Almendro et al., 2014) and bulk genomic profiling techniques (Balko et al., 2012; Balko et al., 2014). With targeted cytogenetic markers, one study showed that genetic diversity did not change in response to NAC but instead selected for mesenchymal phenotypes (Almendro et al., 2014). A study in TNBC used next-generation sequencing (NGS) to profile residual disease in post-treatment chemotherapy samples and identified a number of clinically actionable mutations (Balko et al., 2014). In another report, authors identified JAK2 amplifications as a potential therapeutic target to overcome resistant disease in TNBC patients (Balko et al., 2016). However, these studies were based on targeted markers or bulk genomic tissue profiling and had limited ability to reconstruct clonal evolution during chemotherapy.
Single cell DNA (Navin et al., 2011; Wang et al., 2014) and RNA (Gao et al., 2017; Islam et al., 2014; Tirosh et al., 2016; Yuan and Sims, 2016) sequencing methods have emerged as powerful tools for resolving ITH, reconstructing evolutionary lineages, and detecting rare subpopulations (Grun et al., 2015; Habib et al., 2016). The application of single cell DNA and RNA sequencing methods to solid tumors has enabled phylogenetic reconstruction of tumor lineages (Navin et al., 2011; Shah et al., 2012; Wang et al., 2014), resolved rare subpopulations (Lohr et al., 2014; Martelotto et al., 2017) and provided insight into the phenotypes of stromal and tumor cells in different cancers (Johnson et al., 2014; Meyer et al., 2015; Patel et al., 2014). We reasoned that these technologies could overcome many of the technical hurdles that have previously challenged bulk genomic studies of chemoresistance in TNBC patients.
Due to the extensive ITH reported in TNBC patients, we hypothesized that genomic aberrations associated with chemoresistance are pre-existing in the tumor mass and adaptively selected in response to chemotherapy. In this study, we analyzed longitudinal frozen samples collected from TNBC patients during NAC treatment. We identified two classes of clonal dynamics in response to NAC, in which the mutations, CNAs and expression profiles were eliminated from the tumor mass, or alternatively persisted after NAC. In the clonal persistence patients, we applied both single cell DNA and RNA sequencing methods, which further showed that genomic mutations and copy number aberrations were adaptively selected in response to chemotherapy, followed by transcriptional reprogramming to evolve the resistant phenotypes. Importantly, this integrated evolutionary model would not have been elucidated by profiling only the genomes or transcriptomes of the resistant tumor cells independently.
RESULTS
TNBC Patients Treated with Neoadjuvant Chemotherapy
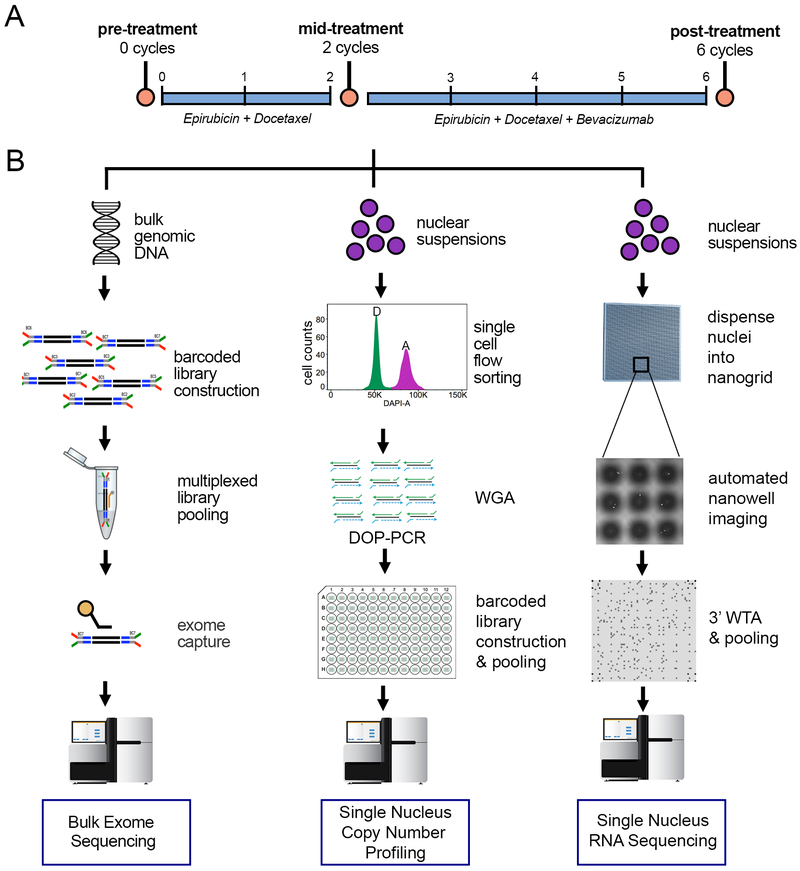
We focused our study on 20 treatment-naive TNBC patients with local disease who were treated with NAC (Table S1, Methods). All patients were classified as TNBC based on the absence of ER staining, PR staining and HER2 copy number by FISH. NAC treatment included an anthracycline (epirubicin) and a taxane (docetaxel) for 2 cycles, after which patients received 4 cycles of the same chemotherapy in combination with an angiogenesis inhibitor (bevacizumab) (Figure 1A). Frozen core biopsy samples were collected pre-treatment, after 2 cycles of therapy (mid-treatment) and during the surgical excision that occurred after 6 cycles of NAC (post-treatment). To mitigate spatial variation, two core biopsy samples were collected from each time-point, and large surgical tissue sections were used from the post-treatment samples for genomic analyses. Exome sequencing was performed on matched longitudinal samples from 20 TNBC patients, while a more detailed analysis using single cell DNA and RNA sequencing was performed on a subset of 8 patients (Figure 1B).
Figure 1 – Overview of Treatment Schedule and Experimental Design.
(A) Timeline of chemotherapy treatment schedule and sample acquisition. Core biopsies were obtained prior to NAC at 0 cycles and mid-treatment, after two cycles of NAC (docetaxel and epirubicin). The surgical sample was obtained after four additional cycles of NAC in combination with bevacizumab. (B) For each longitudinal time-point sample, three experimental procedures were performed, including bulk exome sequencing, single-cell copy number profiling and 3’ single-nucleus RNA sequencing using a nanogrid platform (methods).
Clonal Extinction and Persistence of Mutations in Response to NAC
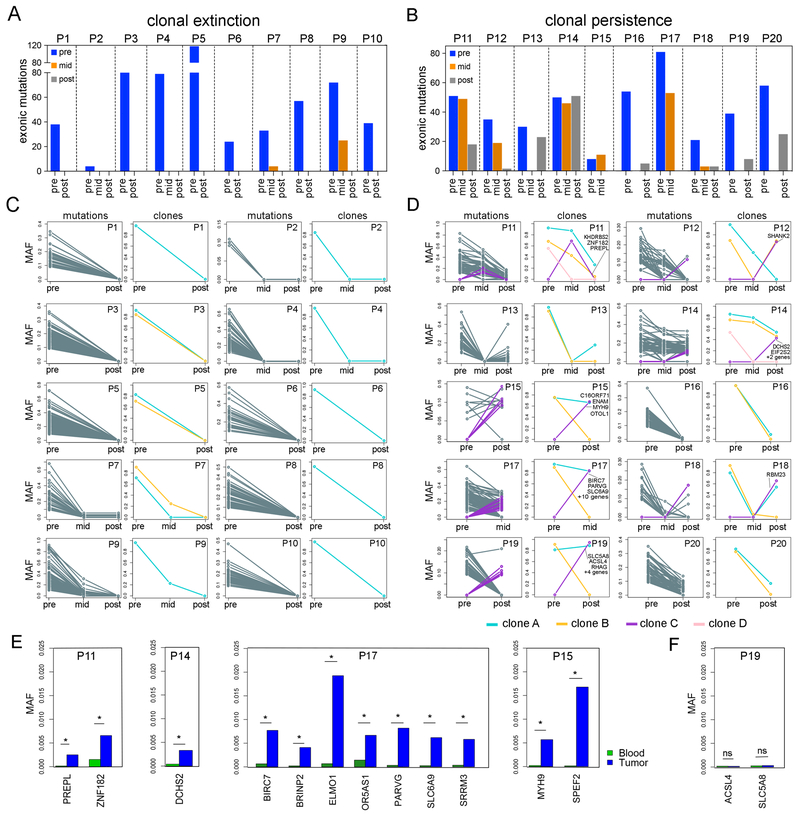
To investigate mutational evolution in the bulk tissue samples, we performed exome sequencing (mean depth 180×) on matched pre-treatment, mid-treatment and post-treatment samples from 20 TNBC patients (Table S2, Methods). Matched blood samples were sequenced in parallel (mean depth 125×) to distinguish somatic mutations. Our data identified TP53 mutations in 60% of the TNBC patients, consistent with frequencies reported in TCGA (Cancer Genome Atlas, 2012). On average, 49 nonsynonymous mutations (range 4–118) and 3 indels (range 1–6) were detected in each patient. Among the 20 patients, 10 showed no detectable mutations after treatment (Figure 2A), while 10 showed residual mutations after treatment (Figure 2B). Notably, we did not observe an increase in mutation burden in response to NAC in any of the TNBC patients.
Figure 2 – Mutational Evolution and Clonal Dynamics in Response to NAC.
Bulk exome sequencing of matched longitudinal samples from 20 TNBC patients. (A) Total number of exonic mutations detected in the clonal extinction patients with no residual mutations after NAC. (B) Total number of exonic mutations detected in clonal persistence patients with residual mutations after NAC. (C) Line plots of raw mutation allele frequencies (MAFs) in left panels and inferred clonal subpopulations in the right panels for clonal extinction patients. (D) Line plots of MAFs in left panels and inferred clonal subpopulations in right panels for clonal persistence patients, with mutations that expanded in resistant clones after NAC labeled with purple lines. (E) Targeted deep amplicon sequencing of pre-existing resistance-associated mutations in four clonal persistence patients, with stars indicating mutations that were statistically significant (p <0.05) in the pre-treatment tumor samples by DeepSNV. (F) A single patient (P19) in which the resistance-associated mutations in the post-treatment tumor sample were not statistically significant (not mutated) in the pre-treatment tumor (ns, p >0.05).
We compared mutation allele frequencies (MAFs) before and after therapy (Figure 2CD) and applied PyClone2 (Roth et al., 2014) and CITUP (Malikic et al., 2015) to estimate clonal subpopulations after copy number (Sathirapongsasuti et al., 2011) and purity normalization (Oesper et al., 2014) (Figure 2C-D). This analysis detected 2–4 major subclones in each TNBC patient and two distinct responses to NAC: 1) clonal extinction, wherein clones were completely eliminated (Figure 2C), or 2) clonal persistence, wherein clonal frequencies shifted but remained present in the post-treatment samples (Figure 2D). The classification of clonal extinction or persistence patients was also supported by indel frequency data (Table S3), tumor purity estimates from histopathological slides, and computational purity estimations with ThetA using the exome sequencing data (Table S4). However, in two clonal persistence cases (P12 and P16), the histopathological estimates were 0% for tumor cellularity in the post-treatment tumors, while the exome data identified residual mutations at low frequencies.
In the clonal persistence patients, most mutations were detected both pre-treatment and post-treatment, but had decreased frequencies in response to NAC (Figure 2D). However, we also identified a number of new mutations (N=33) that emerged in response to NAC (Table S5). In P17, for example, 13 new nonsynonymous mutations were detected that emerged after NAC at the mid-treatment time point, including nonsynonymous mutations in the apoptosis inhibitor, BIRC7, the actin binding protein, PARVG, and the solute carrier, SLC6A9, which had significant functional impact scores based on SIFT (<0.05) (Ng and Henikoff, 2003) and POLYPHEN (P>0.85) scores (Adzhubei et al., 2010). Similarly, in patient P19, we identified 7 new mutations in the post-treatment sample, including a significant nonsynonymous mutation in the solute carrier SLC5A8. While none of the resistance-associated mutations recurred across multiple patients, they did share common biological functions, including cell proliferation, apoptosis, solute transport, and cytoskeleton regulation (Table S5).
We next asked whether the new mutations detected post-treatment were spontaneously induced due to acquired resistance, or alternatively, existed at very low frequencies pre-treatment, but were not detected due to the limited coverage depth of exome sequencing (mean 180×). To address this question, we selected a subset of apparently new post-treatment mutations (N=14) and performed targeted deep-amplicon sequencing (mean depth 1,671,000×) of the pre-treatment bulk tumor DNA (Table S6, Methods). We applied DeepSNV (Gerstung et al., 2012) to detect rare mutation frequencies that were statistically significant compared to matched normal blood (sensitivity 1e4). The amplicon data showed that in 4/5 patients, the suspected de novo mutations did in fact occur at low frequencies in the pre-treatment tumor (range 0.02–2%), consistent with adaptive resistance (Figure 2E). However, in one patient (P19), the de novo mutations were not statistically significant compared to the matched normal sample (Figure 2F). The mutations in this patient may have arisen de novo after the tumor cells were challenged with chemotherapy, or alternatively may have not been sampled due to insufficient sequencing depth, or sampling from different spatial regions.
Copy Number Evolution and Clonal Extinction in Response to NAC
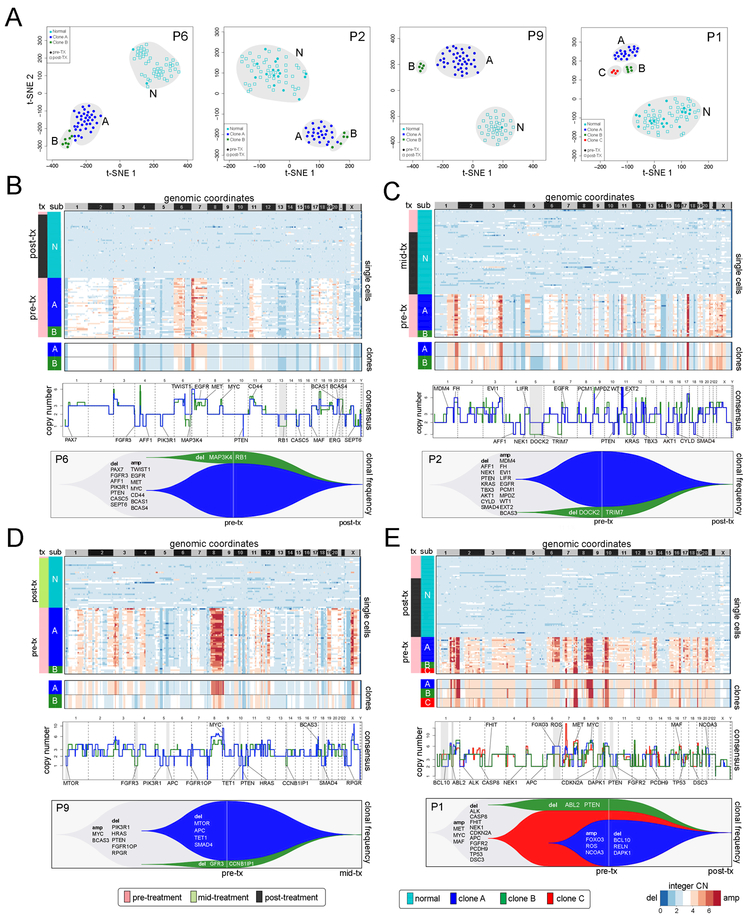
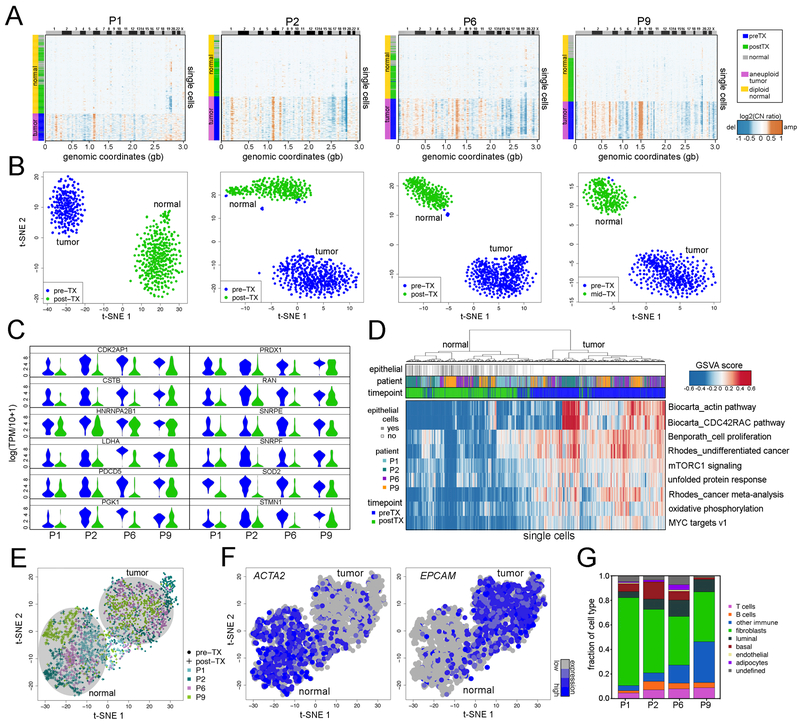
To investigate copy number evolution in response to NAC, we performed single-nucleus sequencing (SNS) (Gao et al., 2016; Navin et al., 2011) on 900 single cells from matched longitudinal samples of 8 TNBC patients (Methods). We selected 4 clonal extinction patients (P1, P2, P6, P9) and 4 clonal persistence patients (P11, P12, P14, P15) for this analysis, based on their classifications from the exome data. Single nuclei were isolated from aneuploid-gated distributions with FACS and used for sparse (~0.1×) whole-genome sequencing to quantify genomic copy number at 220kb resolution (Methods). FACS analysis of DAPI-stained nuclei showed that the 4 clonal persistence patients had aneuploid distributions in both the pre-treatment and post-treatment sample, while the 4 clonal extinction patients had low or undetectable aneuploid distributions post-treatment (Figure S1).
To delineate the clonal substructure of the 4 clonal extinction patients, we identified common chromosome breakpoints across all the cells in the population (Nilsen et al., 2013) and performed optimal clustering (Hennig, 2015) and t-SNE projections (van der Maaten and Hinton, 2008) (Methods). This analysis identified 2–3 clusters of aneuploid tumor cells and one cluster of normal diploid cells in each patient (Figure 3A). The aneuploid clusters were found exclusively in the pre-treatment tissues, while the diploid clusters were mainly associated with the post-treatment tumor. Next, we computed consensus integer copy number profiles for each subclone and inferred clonal lineages using maximal parsimony (Methods) (Figure S2A). The clonal frequencies were then plotted with Timescape (Smith et al., 2016) to visualize clonal dynamics in response to NAC (Methods). These data identified two major clones in three patients (P2, P6, P9) and three major clones in the fourth patient (P1) in the pre-treatment tumors (Figure 3, Figure S2A). Consensus profiles indicated that the multiclonal tumors shared common evolutionary ancestors, as evidenced by shared CNAs, including early events in MET, MYC, and PTEN in P6 (Figure 3B), MDM4, EGFR, and PTEN in P2 (Figure 3C), MYC and PTEN in P9 (Figure 3D) and MYC, MET, TP53, CDKN2A and ALK in P1 (Figure 3E). These tumors also had CNAs that emerged in the later stages of tumor evolution after diverging from the common ancestors. However, irrespective of the number of clonal subpopulations, NAC resulted in the extinction of tumor cells in these patients, as evidenced by the presence of exclusively diploid copy number profiles after treatment.
Figure 3 – Copy Number Evolution in Clonal Extinction Patients.
(A) t-SNE plots of single cell copy number profiles from the pre-treatment and mid-treatment or post-treatment samples of four clonal extinction patients with normal cells (N) and tumor subpopulations (A, B, or C) labeled. (B-E) Clustered heatmaps of single cell integer copy number profiles and consensus integer copy number profiles of the clonal subpopulations. Consensus line profiles show annotated cancer genes and subpopulation-specific differences indicated with grey bars. Lower panels show analyses of clonal dynamics calculated from optimal clustering results and maximum parsimony trees, and plotted in TimeScape with cancer gene and clonal frequencies annotated.
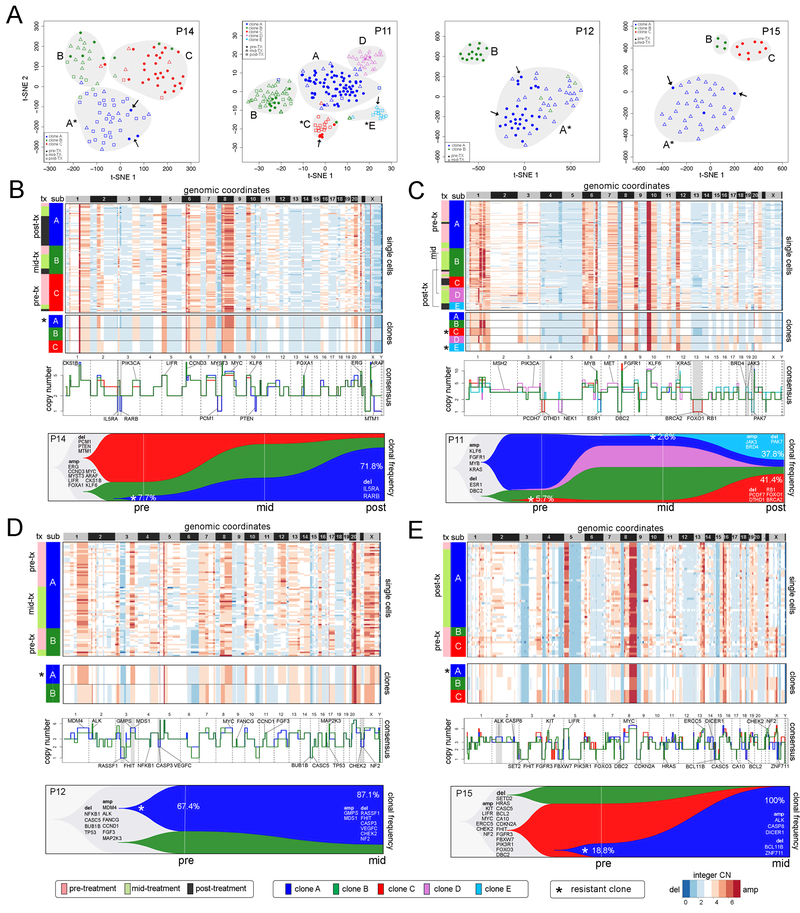
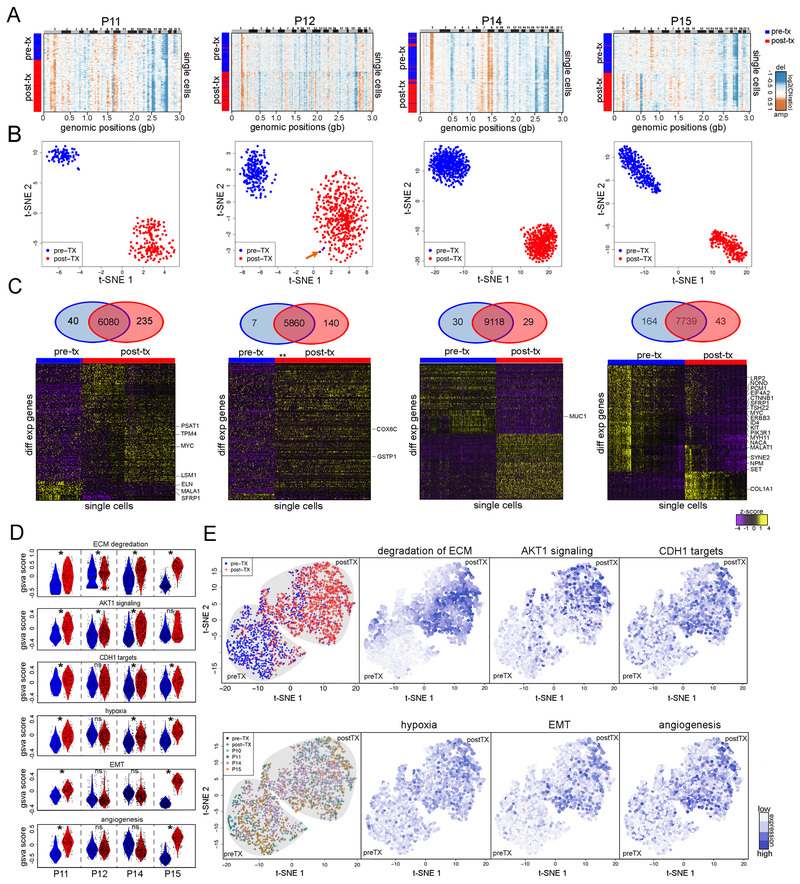
Adaptive Copy Number Evolution in Response to NAC
To delineate copy number evolution in the 4 clonal persistence patients (P11, P12, P14, P15) we detected 2–5 clusters of aneuploid tumor cell copy number profiles (Figure 4A) and constructed maximum parsimony trees from event matrices (Figure S2B). Strikingly, we found that in all 4 patients, a minority of pre-treatment tumor cells (indicated with arrows) clustered with the post-treatment tumor cells, suggesting that they shared a resistant genotype. To identify specific CNAs in the resistance-associated clones, we computed consensus copy number profiles from the single cells (Figure 4B-E). While most CNAs were shared between subclones, we also identified specific CNAs that occurred exclusively in the chemoresistant clones. In P14, resistance-associated clone A displayed two focal deletions on chr 3p, including a 5.3mb hemizygous deletion of 3p26 (IL5RA) and a 14.3mb hemizygous deletion of 3p24–22 (RARB). This clone expanded after NAC from 7.7% to 71.8% post-treatment (Figure 4B). In P11, two resistance-associated clones emerged after NAC, including clone C that expanded from 5.7% to 41.4%, and clone E that emerged mid-treatment at 2.6% and expanded to 37.8% (Figure 4C). The resistance-associated clone C had a 22.9mb hemizygous deletion on chr 4p15 (PCDH7, DTHD1), a hemizygous deletion of a 5.8mb region on chr11q21–22 (MAML2) and chr13q (RB1, BRCA2, FOXO1). In contrast, resistant clone E had a 23mb gain of chr 19p (JAK3, BRD4) and a 20.0mb loss on chromosome 20 (PAK7). Expansion of the two minor clones with different genotypes suggests convergent evolution towards a resistant phenotype. CNAs specific to the resistant subclones were also identified in P12 and P15 by comparing consensus profiles (Figure 4D-E). However, our data did not reveal any recurrent CNAs in the resistant clones among the 4 TNBC patients.
Figure 4 – Adaptive Copy Number Evolution in Clonal Persistence Patients.
(A) t-SNE plots of single cell copy number profiles from the pre-treatment and mid-treatment or post-treatment samples of four clonal persistence patients with tumor subpopulations (A,B,C,D,E) labeled. Arrows indicate pre-existing single cells from the pre-treatment samples that share the post-treatment chemoresistant genotypes. (B-E) Clustered heatmaps of single cell integer copy number profiles and consensus profiles of clonal subpopulations. Consensus line profiles show annotated common cancer genes and subpopulation-specific differences are indicated with grey bars. Lower panels show analyses of clonal dynamics calculated from optimal clustering results and maximum parsimony trees, and plotted in TimeScape with cancer gene and clonal frequencies annotated. Stars indicate the chemoresistant clones that were selected and expanded in response to NAC.
Transcriptional Programs of Tumor Cells in Clonal Extinction Patients
We investigated phenotypic evolution in response to NAC using a high-throughput nanogrid single nucleus RNA sequencing (SNRS) method (Gao et al., 2017). SNRS performs automated imaging and selection of up to 1,800 single nuclei in parallel for 3’ mRNA profiling. We profiled the transcriptomes of 3,370 single nuclei isolated from two matched longitudinal samples per patient from the 4 clonal extinction patients (P1, P2, P6, P9). An average of ~500 nuclei were selected from each time point for SNRS, which resulted in an average of 1.2 million reads and 4,107 genes detected per cell. To distinguish normal stromal cells from aneuploid tumor cells, we calculated copy number profiles from RNA read counts (Patel et al., 2014), using a set of 240 diploid normal breast cells from a different patient as a baseline reference (Gao et al., 2017) (Methods). Clustered heatmaps identified a large number (90–99%) of aneuploid tumor cells in the pre-treatment tumor, but only diploid normal cells post-treatment, consistent with the single cell copy number analyses (Figure 5A). We detected differentially expressed genes using MAST (Finak et al., 2015) between the pre-treatment tumor cells and the epithelial cells in the post-treatment samples, by excluding other cell types based on their RNA profiles. With MAST, we regressed out the batch effects caused by the single cell gene detection rates (Methods) and performed high-dimensional analyses to determine if tumor cells were present post-treatment (Figure 5B). To determine if the batch effect regression affected the biological signal, we compared the corrected and uncorrected datasets, which revealed a high concordance (72 – 99%) (Figure S3A). We predicted the intrinsic breast cancer molecular subtypes (Gendoo et al., 2016) and found that all 4 TNBC patients were dominated by the basal-like subtype of tumor cells, however there were also a small number of tumor cells present in each tumor with other subtype signatures (Figure S3B). This analysis identified distinct clusters of pre-treatment and post-treatment cells, and further showed that no tumor cells were identified post-treatment, suggesting that they were eliminated by NAC.
Figure 5 – Transcriptional Profiles of Clonal Extinction Patients.
(A) Clustered heatmaps of single cell copy number profiles calculated from single cell SNRS data from pre-treatment and mid-treatment or post-treatment samples from 4 clonal extinction patients, clustered with 240 normal breast cells from a different patient. (B) t-SNE projections of single cell RNA profiles from pre-treatment and mid-treatment or post-treatment samples from each clonal extinction patient, with immune cells excluded. (C) Violin plots of single cell RNA expression data for 12 cancer genes that were significantly upregulated (FDRadj p-value < 0.05, log2(foldchange) ≥1) in the pre-treatment tumor cells across all four clonal extinction patients, in relative to post-treatment normal epithelial cells. (D) Cancer gene signature analyses and clustering of GSVA scores for single tumor and normal cells from all 4 clonal extinction patients. (E) t-SNE projection of combined single cell data from the four clonal extinction patients, with immune cells excluded. (F) Expression of fibroblast marker ACTA2 and epithelial marker EPCAM in the tumor and normal cells. (G) Normal cell type classification and frequencies of cell types in the post-treatment tissue samples.
Clustering of differentially expressed genes showed that a number of genes were upregulated in the tumor cells relative to the normal epithelial cells (mean = 530 genes) in the post-treatment samples, including several known cancer genes (NRAS, MYC, FGFR2, TP53) (Figure S4A). While most cancer genes were unique to individual patients, this analysis also identified 12 known cancer genes that were upregulated in all of the clonal extinction patients (Figure 5C). To determine if cancer phenotypes were shared across the TNBC patients, we performed GSVA analysis (Hanzelmann et al., 2013) and clustering on a set of cancer-specific signatures, which showed that the actin pathway, CDC42RAC pathway, cell proliferation, undifferentiated in cancer, mTORC1 signaling, unfolded protein response, cancer meta-analysis, oxidative phosphorylation, and MYC targets were upregulated in the pre-treatment tumor cells (Figure 5D). The gene signature analysis also showed that the pre-treatment tumor cells had increased proliferation and apoptosis (Figure S4B-C). These genes and pathways may potentially play a role in conferring sensitivity to chemotherapy in the clonal extinction patients.
We combined all the tumor and normal cell RNA data from the 4 patients, and performed a high-dimensional analysis of the gene signatures with t-SNE (Figure 5E). The data indicated that normal cells and tumor cells formed two distinct clusters. Importantly, these results did not show any clustering of single cells by patient or batch, suggesting that batch effects were minimal. Within the normal cell cluster from the post-treatment samples, we found high levels of the ACTA2 fibroblast marker, and within the tumor cell cluster from the pre-treatment samples, high levels of the epithelial marker, EPCAM (Figure 5F). We further classified all normal cells in the post-treatment samples by 8 major breast cell types using cell-type specific markers and showed that fibroblasts were the most abundant cell type (mean 51.1% ± 14.9% SD) present after NAC, followed by other cell types such as T-cells (mean 6.9%) and other CD45+ immune cells (Figure 5G, Methods).
Acquired Transcriptional Reprogramming of Chemoresistant Tumor Cells
To investigate phenotypic evolution in the clonal persistence patients (P11, P12, P14, P15), we performed SNRS oñ400 nuclei from each matched time point sample, resulting in an average of 1.2 million reads and 5,166 genes detected per cell. We calculated copy number profiles from the single cell data (Methods) and performed 1-dimensional clustering (Figure 6A). In contrast to the clonal extinction patients, a large fraction of aneuploid cells (85–99%) were detected in both the pre-treatment and post-treatment samples, consistent with the single cell copy number data. The few normal diploid cells that were detected and removed from expression analysis.
Figure 6 – Transcriptional Reprogramming in the Chemoresistant Tumor Cells.
(A) Heatmaps of single cell copy number profiles calculated from single cell RNA data from pre-treatment and mid-treatment or post-treatment samples from 4 clonal persistence patients. (B) t-SNE projections of single cell RNA profiles from pre-treatment and mid or post-treatment samples from each clonal persistence patient, with an arrow indicating two cells from the pre-treatment samples that cluster with the post-treatment expression profiles in patient P12. (C) Venn diagrams and clustered heatmaps of significant differentially expressed genes between the pre-treatment tumor cells and post-treatment tumor cells with cancer gene annotations. (D) Violin plots of single-cell GSVA scores for the pre-treatment and post-treatment tumor cells from all 4 clonal persistence patients. Significance * indicates FDR adjusted p < 0.05 and |mean GSVA score difference| ≥ 0.1. (E) t-SNE projection of all combined single cell data from the four clonal persistence patients, labeled by pre/post treatment, sample origin, or GSVA signatures.
To determine whether any of the tumor cells with chemoresistant expression profiles existed at low frequencies in the pre-treatment tumor, we performed high-dimensional analysis (Figure 6B, Methods). In three patients (P15, P14, P11), we did not detect any pre-existing transcriptional profiles that clustered with the post-treatment tumor cells, despite profiling hundreds of cells. To identify genes upregulated in the chemoresistant post-treatment tumor cells, we performed differential expression analyses using MAST (FDR adj p-value < 0.05; |log2 fold change|≥1) and identified a number of differentially expressed genes (N=59–275) in each patient (Figure 6C). A few significant genes included known cancer genes (MYC, ERBB3, KIT and PIK3R1), but were not recurrent across the TNBC patients (Figure 6C).
Molecular subtyping showed that the tumors had many basal-like cells, with the except of one patient (P12) where a number of tumor cells were classified as HER2-enriched, luminal A- and luminal B (Figure S3C). Interestingly, P12 was HER2+ by IHC protein levels but did not have copy number amplifications by FISH (Table S1). These data show that the subtype composition of single cells did not change drastically in response to NAC, suggesting that subtype-switching did not occur (Figure S3C).
To identify common phenotypes of the chemoresistant tumor cells, we performed single cell gene signature analysis using GSVA for a set of cancer-related signatures and clustered the normalized scores from all 4 patients together (Methods, Figure 6D-E). This analysis determined that gene signatures including, degradation of ECM, AKT1 signaling via mTOR (Creighton, 2007), CDH targets (Onder et al., 2008), hypoxia (Harris, 2002), EMT and Angiogenesis, were upregulated in the chemoresistant tumor cells after NAC (Figure 6D). We used t-SNE to cluster all the single cell data from the 4 patients in high-dimensional space, which separated the pre-treatment and post-treatment tumor cells into two distinct clusters. Notably, the single cells from different patients were intermixed within each major cluster, suggesting that batch effects were minimal (Figure 6E). We labeled the chemoresistance gene signature scores of single cells in the high-dimensional plots and showed that they were highly enriched in the post-treatment samples (Figure 6E). Next, we used an extended cohort of breast cancer patients with chemotherapy (N=412) from the METABRIC cohort (Curtis et al., 2012; Pereira et al., 2016) with gene expression data and long-term clinical follow-up data to determine if any of the chemoresistance-associated signatures correlated with patient survival. This analysis showed that two signatures (AKT1 signaling and Hypoxia) were associated with statistically significant (p<0.05) worse survival (Figure S5).
Although the full transcriptional programs of chemoresistant cells were not found to be pre-existing in the clonal persistence patients, we further investigated whether subsets of chemoresistant genes were expressed in single cells in the pre-treatment samples by integrating the single cell DNA and RNA datasets (methods). First, we determined if subclonal CNAs in resistant clones of the single cell DNA data were present in the RNA copy number profiles, to classify each cell as having either a resistant or sensitive genotype. To decrease technical noise from transcript dropout and sparse coverage, we rescued missing values in the single cell RNA data using SAVER (Huang et al., 2017) before calculating copy number profiles. We then compared the tumor cells with resistant genotypes in the post-treatment tumor to the sensitive genotypes in the pre-treatment tumors to obtain the top variable genes using MAST (FDR adj p-value < 0.05; |log2 fold change|≥1.58) (Finak et al., 2015). By performing random forest regression and classification, we defined ‘primed cells’ in the pre-treatment tumor that expressed a portion (17~ 60%) of the chemoresistant genes (Methods). In three patients (P11, P12 and P14) we detected pre-treatment cells (4–33 cells) that expressed a subset of the chemoresistant genes (Figure S6A-B). The frequency of the primed tumor cells was lower than the pre-existing chemoresistant copy number profiles of single cells, suggesting that CNAs alone were insufficient to confer the full resistant phenotypes of the tumor cells.
We further performed an integration of the exome mutations and the 3’ single cell RNA datasets to determine if expressed mutations could be identified in the last exons of the expressed mRNA transcripts. This analysis showed that the expressed mutations were detected exclusively in the pre-treatment single cell RNA data of the clonal extinction patients, while the clonal persistence patients showed evidence of expressed mutations in both the pre-treatment and post-treatment single cell RNA datasets (Figure S6C). These data further confirmed our classification of the clonal persistence and extinction patients.
DISCUSSION
In this study we investigated the genomic and phenotypic evolution of tumor cells in TNBC patients in response to NAC, which revealed two distinct classes of clonal dynamics: extinction and persistence. In the clonal extinction patients, NAC eliminated the tumor cells, leaving only normal diploid cell types after treatment, including many fibroblasts and immune cells. In contrast, the clonal persistence patients harbored a large number of residual tumor cells with genotypes and phenotypes that were altered in response to NAC. Using single cell DNA and RNA sequencing methods we performed a detailed analysis of 8 patients, which showed that the CNAs that emerged in response to NAC were pre-existing and adaptively selected, while the expression profiles were acquired through transcriptional reprogramming. Our data further suggests that a small fraction of genotypes selected by NAC were primed for transcriptional reprogramming and had subsets of chemoresistant genes expressed prior to treatment. Collectively, our data support a model of chemoresistance in which two modes of evolution (adaptive and acquired) were operating to establish the resistant tumor mass (Figure S6D).
Our data contrast with previous genomic studies that investigated chemotherapy response in AML, glioblastoma and ovarian cancer, which reported large increases in mutation frequencies in the post-treatment samples (Ding et al., 2012; Johnson et al., 2014; Patch et al., 2015). In our exome data, we observed decreases or no changes in the mutation frequencies of post-treatment samples in response to NAC. The differences in mutagenicity may be due to the chemotherapeutic agents that were used to treat the glioblastoma and ovarian cancer patients, since telozolomide and cis-platinum have been shown to be highly mutagenic, while taxanes and anthracyclines are not known to be highly mutagenic. However, our results are consistent with a previous study in TNBC, which reported no significant increase in somatic mutations after chemotherapy (Balko et al., 2014).
The genomic data generated in this study is consistent with a punctuated model of copy number evolution in TNBC patients (Gao et al., 2016), wherein hundreds of chromosomal aberrations are acquired in short evolutionary bursts at the earliest stages of tumor progression. We speculate that early bursts of genome instability generate rare chemoresistant clones with genotypes that do not expand significantly until the population is challenged by chemotherapy. Interestingly, we often identified more clonal subpopulations in the mutational data compared to the copy number data, suggesting ongoing mutational diversification in the tumor lineages, which is consistent with reports that copy number and mutation rates represent two distinct molecular clocks during breast cancer evolution (Wang et al., 2014). Collectively our copy number and mutational data showed that chemoresistance-associated mutations encompassed diverse biological functions and were pre-existing in the tumor mass.
In contrast to the diverse mutations, the transcriptional programs converged on a few common pathways identified across multiple patients. The gene signatures associated with chemoresistance included EMT, CDH1 targets, AKT1 signaling, hypoxia, angiogenesis and ECM degradation. The CDH1 targets signature includes genes and transcription factors that are upregulated after the knockdown of E-cadherin (eg. Twist, Snail), and together with the EMT signature, relate to the transition of tumor cells to mesenchymal phenotypes in response to NAC (Liberzon et al., 2015). This observation has previously been reported in post-NAC samples from breast cancer patients (Almendro et al., 2014) as well as in studies that demonstrate mesenchymal cells in post-treatment tumors desensitize tumors to cytotoxic agents (Zheng et al., 2015). AKT1 signaling is a survival signal and has previously been implicated in paclitaxel resistance by inhibiting apoptotic pathways (Kim et al., 2007). Similarly, hypoxia has also been shown to enhance chemoresistance in tumor cells via HIF-1 (Cosse and Michiels, 2008; Doktorova et al., 2015; Petit et al., 2016). Studies have also shown that ECM degradation is important for invasion, migration and metastasis (Lu et al., 2012; Oskarsson, 2013). In most patients, we found that the chemoresistant transcriptional programs were not pre-existing and were acquired via transcriptional reprogramming after treatment, however a small fraction of primed tumor cells with subsets of resistant genes were identified in some patients.
Our data has several important clinical implications. First, the pre-existence of chemoresistant genotypes in the tumor mass indicates that there may be diagnostic opportunities for detecting chemoresistant clones in TNBC patients prior to the administration of NAC, to predict which patients may benefit from chemotherapy. Second, the stratification of TNBC patients into clonal extinction and clonal persistence groups may have prognostic applications in patient outcome or survival, beyond histopathological techniques. Third, our data on chemoresistant phenotypes raise the possibility of therapeutic strategies to overcome chemoresistance, such as targeting EMT signaling (Marcucci et al., 2016), using PI3K/AKT1 pathway inhibitors (Liu et al., 2009; Owonikoko and Khuri, 2013), or inhibiting hypoxia with HIF-1 inhibitors (Hu et al., 2013; Semenza, 2003) to re-sensitize the tumor cells to chemotherapy.
Limitations of our study include the small number of patients (N=8) that were analyzed at single cell resolution. Future work will need to be performed in a larger cohort of TNBC patients to understand the generalizability of our evolutionary model and the chemoresistant gene signatures. Functional studies will also be needed to validate the chemoresistant signatures and understand their mechanistic roles in conferring resistant phenotypes. Although batch effects have been identified as a major confounding effect in single cell RNA data (Tung et al., 2017), we mitigated these errors by processing all samples in parallel, using identical reagents, and normalizing our data. Another potential source of error is spatial bias in the core biopsy samples, which we mitigated by using two independent core biopsy samples from each time point and large surgical specimens from the post-treatment time points.
In closing, we expect that the combined single cell DNA and RNA sequencing methods used in this study will provide new insights into genomic and phenotypic evolution in response to chemotherapy in many human cancer types. In most human cancers, chemotherapy remains to be the first line of therapy and standard of care, and tumors often respond well initially but frequently develop resistance within the first few years. The use of both single cell DNA and RNA sequencing methods was critical in our study, since the application of only one technique would have led to the false inference of either adaptive or acquired evolution. Other important future directions include an analysis of matched metastatic tumors, to understand whether the chemoresistant clones in the primary tumor seed distant metastases and also confer resistance at these organ sites. These studies will become more realistic as single cell DNA sequencing technologies increase in throughput and decrease in cost (Wang and Navin, 2015), and the ability to perform multi-modal single cell DNA and RNA profiling in the same cell becomes technically feasible (Zahn et al., 2017) for analyzing larger cohorts of TNBC patients.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Nicholas Navin (nnavin@mdanderson.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fresh frozen triple-negative breast cancer (TNBC) tissue samples were obtained from the Predict Response of Preoperative Treatment of Breast Cancer Early (PROMIX: NCT00957125) trial in collaboration with Dr. Theodoros Foukakis at the Karolinska University Hospital and Internal Review Board (IRB) approval at the University of Texas MD Anderson Cancer Center and Karolinska University Hospital. All patients were enrolled under informed consent. The triple-negative status of all tumor samples was determined by immunohistochemistry and FISH copy number. The PROMIX trial recruited patients with localized, primary breast cancer, including inflammatory breast cancer, suitable for primary medical treatment and/or patients with regional lymph node metastases. Frozen tumor tissue samples were collected <2 weeks prior to treatment initiation (14G core biopsy), after 2 three-weekly cycles of epirubicin 75 mg/m2 and docetaxel 75 mg/m2 (14G core biopsy), and after 4 additional three-weekly cycles of epirubicin 75 mg/m2 and docetaxel 75 mg/m2 in combination with bevacizumab 15 mg/kg (surgical excision). Core biopsy samples were collected prior to NAC and after 2 cycles, while surgical samples were collected after 6 cycles of NAC.
METHOD DETAILS
Bulk Exome Sequencing
Bulk genomic DNA was extracted from frozen tumor samples following the manufacturer’s instructions outlined in the DNA Mini Kit (Qiagen, #51306). Germline genomic DNA was extracted from matched patient blood samples with the DNA Blood Mini Kit (Qiagen, #51106). Extracted DNA was sonicated to 250 bp using the S220 Sonicater (Covaris). KAPA libraries (KAPA Biosystems, #8502) were constructed from the sonicated DNA according to manufacturer’s instructions using 10–250 ng of input DNA and unique NEXTflex-96 barcodes (Bio Scientific) for each individual sample. Barcoded libraries (8–12) were pooled together for exome captures using the SeqCap EZ v2 kit (Roche, #06953212001), which targeted 44 Mb of the genome. Final v2 exome capture products were quantified with the dsDNA HS Fluorometric Assay (Invitrogen, #Q32854), Agilent’s High Sensitivity DNA Chip, and KAPA Library Quantification kit (#KK4835) and sequenced at 100 paired-end cycles on the HiSeq2000 or HiSeq4000 system (Illumina).
Exome Data Processing and Analysis
FASTQ sequences were mapped to the human assembly NCBI build 37 (hg19) using the Bowtie2 alignment software (Langmead and Salzberg, 2012). Samtools (v0.1.19) was used to convert SAM files to compressed BAM files and to sort the BAM files by chromosomal coordinates (Li et al., 2009). PCR duplicates were marked with Picard (v1.56), and the sorted, marked BAM files were realigned with the Genome Analysis Toolkit (GATK1.4–37) (McKenna et al., 2010) at intervals with indel mismatches. Mutect2 was used to detect single nucleotide variants (SNVs) and to perform local realignment for indel calls (do Valle et al., 2016). Germline variants were removed by filtering out variants in the matched normal blood samples. Gene annotations and function prediction scores were performed with Annovar (Yang and Wang, 2015), dbSNP build135 (Sherry et al., 2001), 1000Genomes (Genomes Project et al., 2015), Polyphen, Avsift (Ng and Henikoff, 2003), and COSMIC (Forbes et al., 2017). Mutation sites were selected if there were at least 30 reads across all time points; and MAF < 0.01 in the germline blood; and variant read count ≥5 for a depth of 30, MAF ≥ 0.09 for depth >30 in at least one time point. Mutations were then identified in the other time points if at least 5 variant read counts were detected.
Estimation of Clonal Dynamics from Mutation Data
Non-synonymous somatic mutations were identified in the bulk exome sequencing data for 20 TNBC patients with 2 or 3 longitudinal samples. Bulk tumor genomic copy number profiles were estimated from the pair-end exome sequencing depth using the R package ‘exomeCNV’ (Sathirapongsasuti et al., 2011). Tumor purity was estimated with THetA2 (Oesper et al., 2013). The variant allele frequencies from each point mutation were normalized with both exome-derived copy number profiles and estimated tumor purities, and initial clusters were identified using PyClone2 (v0.12.9) (Roth et al., 2014). Mutation clusters with only one mutation were excluded from further analysis. The PyClone cluster frequencies were calculated as the mean variant allele frequencies of mutations within each cluster. The clonal frequencies were then adjusted using CITUP (Malikic et al., 2015) by joint calculation of the cluster identifications and optimal trees across the tumor time points from same patient.
Detection of Rare Variants by Targeted Amplicon Sequencing
Forward and reverse primers were designed to amplify ~150 bp regions containing the targeted mutation site identified by bulk exome sequencing for the tumor sample and its matched blood sample. Amplicons were PCR-amplified, purified, eluted in ~20μL H2O, and run on an agarose gel to confirm fragment sizes. Fragments were pooled together in equimolar concentrations, and the larger amplicons were sonicated. A New England BioLabs kit was used for end repair of amplification products (NEB, E6050L), and amplicon products were subsequently purified with the DNA Clean and Concentrator-5 Kit (Zymo Research, #11–303). NEBNext DNA Library Prep enzymes were used for 3’ adenylation (#E6053L), adapter ligation (#M0202L), and PCR amplification (#M0541L) to barcode the library with a P7 unique NEXTflex-96 barcode. The final barcoded library was quantified by quantitative PCR using the KAPA Library Quantification kit. The sample was sequenced on the MiSeq system (Illumina #MS-102–3001, 150 single-read). DeepSNV (Gerstung et al., 2012) was applied to determine whether mutations were statistically significant and present at higher levels in the tumor samples compared to the matched normal blood samples at a p-value < 0.05, with 5 bp flanking regions around the mutation site of interest to establish error rates.
Multiplexed Single Cell Copy Number Sequencing
The highly-multiplexed single-nucleus-sequencing (HM-SNS) protocol published by Gao et al. 2016 was used to multiplex single nuclei for copy number sequencing. Briefly, nuclei from frozen tumors were isolated by mincing the tumor in DAPI-NST buffer (800 mL of NST [146 mM NaCl, 10 mM Tris base at pH 7.8, 1 mM CaCl2, 21 mM MgCl2, 0.05% BSA, 0.2% Nonidet P-40]), 200 mL of 106 mM MgCl2, 10 mg of DAPI, and 5mM EDTA) and filtering through a 37-μM nylon-mesh filter (Lake et al., 2016). Single nuclei from the aneuploid distribution were flow-sorted into 96-well plates through the FACSAria II flow cytometer. Each individual well contained 10 μL of lysis solution from the Sigma-Aldrich GenomePlex WGA4 kit. The diploid population of DAPI fluorescence intensity was initially established with a control diploid cell line.
Whole-genome amplification (WGA) was performed as described in the Sigma-Aldrich GenomePlex WGA4 kit (WGA4–50RXN). The DNA concentration was measured (Thermo Fisher Scientific, Qubit 2.0 fluorometer), run on gel electrophoresis for size distributions, and sonicated to 250 bp using the S220 Sonicator. The products underwent NEB end repair and were subsequently purified with the DNA Clean and Concentrator-5 Kit. The NEBNext DNA Library Prep enzymes were used for 3’ adenylation, adapter ligation, and PCR amplification to barcode each single-cell library with a P7 unique 8-bp identifier (NEXTflex-96 barcodes) and common P5 adapter for sample multiplexing. After adapter ligation, libraries were purified with AMPure XP beads 0.5x, and PCR-amplified (8-cycles). Final library concentrations were measured by Qubit, and libraries were pooled in equimolar concentrations. The final multiplexed pooled library was quantified by quantitative PCR using the KAPA Library Quantification kit and the QuantStudio 6 Flex Real-Time PCR System. Size distributions of the pooled submission were evaluated on 2100 Bioanalyzer (Agilent Technologies). Pooled 96-libraries were sequenced using 76 single-end or 76 paired-end cycles on the HiSeq2000 or HiSeq4000 (Illumina).
Single-Cell Copy Number Data Processing
Sequencing data was processed into a master FASTQ file using the CASAVA 1.8.1 pipeline (Illumina Inc.) and then demultiplexed using an in-house perl script (Gao et al., 2016) into individual FASTQ files representing the sequencing reads from each single cell. Sequence reads were aligned to the NCBI build 37 (HG19/NCBI37) using Bowtie2 (Langmead and Salzberg, 2012) and converted to sorted BAM files. Poorly mapped reads (MQ<40) were filtered out using Samtools (0.1.19) (Li et al., 2009). PCR duplicates were identified as sequencing reads with the exact same starting position and were also removed from read counts via an in-house Python script. Single cell copy numbers were calculated using the ‘variable binning’ method followed by the Circular Binary Segmentation (CBS) (Olshen et al., 2004) as previously described (Baslan et al., 2012; Navin et al., 2011). Sequencing reads were counted into 11,927 genomic bins with variable start and stop coordinates to simulate mapping bias across the genomic positions, where the median genomic length spanned by each bin is 220 kb. Loess normalization was used to correct for GC bias (Baslan et al., 2012). Copy number segmentation was performed using the CBS method (Olshen et al., 2004) followed by MergeLevels (Willenbrock and Fridlyand, 2005) to join adjacent segments with non-significant differences in segment ratios. The parameters used for CBS segmentation was alpha=0.0001 and undo.prune=0.05 respectively. Default parameters were used for performing MergeLevels, which successfully joined false positive detections of erroneous chromosome breakpoints.
Copy Number Clustering and Phylogenetics
Clonal subpopulations in single cell copy number data were defined using an optimal clustering method as previously described (Gao et al., 2016). To minimize the effects of parallel association in the clustering results, we first obtained a copy number event matrix by applying the population segmentation method using R package ‘copynumber’ (Nilsen et al., 2013) to find common breakpoints across all cells in the population. The optimal number of clusters was identified using the short event matrix and determined by the average silhouette distance width using R ‘pamk’ function in the ‘fpc’ package (Hennig, 2015). Integer copy numbers were calculated by scaling the absolute ratios with the population average ploidy and rounding to the nearest integer values. Following identification of clonal subpopulations, the consensus copy number profile of each subpopulation was calculated as the median copy number of all single cells within the subpopulation. Clonal lineage analysis was performed on aneuploid single cells that had genome-wide copy number aberrations. The relative clonal frequency of each subpopulation was calculated as the number cells that belonged to each specific sub-cluster divided by the total number of clonal cells. The tree structures of the clonal subpopulations were inferred using the maximum parsimony tree method in R package ‘phangorn’ (Schliep, 2011) from the annotated event matrix where copy numbers larger than ground states were labeled as ‘gain’ and those smaller than ground states were labeled as ‘loss’. The ground states were labeled as ‘neutral’. Finally, clonal lineages were analyzed with TimeScape (Smith, 2017) using the MP tree structures and the clonal frequencies across the treatment time points for individual TNBC patient samples.
Nanogrid Single-Nucleus RNA Sequencing
Single-nucleus RNAseq was performed as previously reported by Gao et al. 2017. Briefly, frozen tumor sections were minced in DAPI-NST lysis buffer and filtered through a nylon-mesh filter to further remove cytoplasmic components. The final suspension was diluted to 1000 μL of 1 cell/50 nL with 1x PBS and D-RNase free water (0.35x PBS in the final dilution). Single nuclei were dispensed into the WaferGen SmartChip™ (5000 wells), and nanowells were selected (~500 cells/chip) and visualized with the Wafergen CellSelect™ software according to the manufacturer’s instructions to confirm that selected wells had 1 cell/well. Chips were placed in freezing chambers and stored at −80°C until reverse-transcription (RT). Frozen chips were thawed, and 50nL of RT solution (56μL 5M betaine, 24μL 25 mM dNTP mix, 3.2μL 1 M MgCl2, 8.8μL 100 mM DTT, 61.9μL 5X SMARTScribe™ First-Strand Buffer, 33.3μL 2X SeqAmp™ PCR Buffer, 4.0μL 100μm RT E5OLIGO, 8.8μL 10μM Amp Primer, 1.6μL 100% Triton X-100, 28.8μL SMARTScribe™ Reverse Transcriptase, and 9.6μL SeqAmp™ DNA Polymerase) was deposited into each selected well. After RT, complementary DNA (cDNA) products from selected wells were pooled together, purified, and underwent PCR amplification. The amplified product was purified with 0.6x AMPure XP beads and eluted in 11μL D-RNase-free water. The eluted product was quantified with Qubit and Bioanalyzer. The cDNA was then diluted to 0.2 ng/μL and used to construct Nextera XT DNA libraries with i7 index primers. The final libraries, containing ~500 barcoded single-nuclei transcriptomes, were sequenced at 100 paired-end or 76 paired-end cycles on the HiSeq4000 system (Illumina).
Single-Nucleus RNA Sequencing Data Processing
After sequencing, the BCL2 intensity files were processed into a master FASTQ file using the CASAVA 1.8.1 pipeline (Illumina Inc.) and then demultiplexed into individual fastq files with each file representing one single cell. Sequencing reads were mapped to the human transcriptome (HG19) using bowtie2 (Langdon, 2015), and gene expression levels were summarized into TPM values using RSEM (Li and Dewey, 2011) using uniquely mapped reads.
Copy Number Calculation from Single-Nucleus RNA Data
Single cell copy number based on the RNAseq data were calculated from a [log(TPM/10+1)] matrix using a sliding windows of 50 genes as previously described (Gao et al., 2017; Patel et al., 2014). To perform the calculation, genes were sorted by their genomic coordinates spanning from chr1 to chr22, chrX and chrY. We excluded genes that were detected in less than 30% of the cells, which resulted in 3,000–7,000 genes per cell that were used for the copy number calculations. We used a set of 240 normal breast tissue single cells as diploid baselines as previously described (Gao et al., 2017). Relative gene expression was obtained for each gene by subtracting the baseline levels. To mitigate the bias caused by extreme gene expression levels, we replaced the relative gene expression values ≥ 3 with 3 and relative expressions ≤ −3 with −3. We then obtained a ‘moving average’ of adjacent 50-gene relative expression values to represent the [log2(copy number ratio)] of that genomic location. We further normalized the [log2(copy number ratio)] to their mean values for each cell to center around zeros. Single cells without genome-wide CNAs were defined as normal cells and were normalized to diploid cell baselines to remove minor CNAs caused by cell type variations among normal cells.
Differential Gene Expression Analysis
Single cell RNAseq data has a bimodal gene expression distribution, and as a result, the number of genes detected was variable. We applied MAST (Finak et al., 2015), a two-part generalized linear model, to adjust gene detection ratios across cells and to compare the gene expression in post-treatment cells to their matched pre-treatment samples, which resulted in fewer false positive discoveries (Jaakkola et al., 2016). Single cells that did not express either GAPDH or ACTB were excluded from the analysis. We also excluded a list of 217 curated house keeping genes, which included 98 genes from a previously published list (Tirosh et al., 2016) and genes that were detected in <30% cells in both the pre-treatment and post-treatment groups, which resulted in 7,318 genes per patient on average for differential gene expression analyses. For clonal extinction patients, we compared all pre-treatment tumor cells to post-treatment normal epithelial cells in each patient. For clonal persistent patients, we compared all pre-treatment tumor cells with all post-treatment tumor cells. The significant differentially expressed (DE) genes were defined as having FDR adjusted p-value < 0.05 and |log2(fold change)| ≥ 1. Clustered heatmaps of significant DE gene expression were generated with R function ‘heatmap.3’ based on z-scores of log(TPM/10+1). Single cells from both pre-treatment and post-treatment tumors were projected to 2D space using the tSNE with R package ‘tsne’ (Donaldson, 2016).
Breast Cancer Subtype Prediction
Single cells were classified into 5 established intrinsic breast cancer molecular subtypes (normal-like, basal-like, luminal A, luminal B and Her2 enrichment) using the R ‘genefu’ package (Gendoo et al., 2016) with [log(TPM/10+1)] data matrix. Single cells with low prediction confidence (< 0.7) were undefined. In the pentagon plots, the total number of single cells within each tumor was scaled to 500 single cells per tumor to balance the visualization across different tumors.
Gene Set Variation Analysis
Gene signatures of single cells were quantified by applying the single-sample gene set variation analysis (ssGSVA) (Hanzelmann et al., 2013), which calculated the signature enrichment scores of individual single cells independently without normalization across cells. The gene expression [log(TPM/10+1)] matrix was used and single cells that did not express either GAPDH or ACTB were excluded from the analysis. We performed unbiased analysis on a set of 50 hallmark signatures (Liberzon et al., 2015) and a set of 4725 curated pathway signatures (MSigDB,C2 sets). The resulting GSVA score matrix was organized as having single cells in the columns and the signatures in the rows. Comparisons of single-cell enrichment scores of pre-treatment and post-treatment single cells were performed using the R package ‘limma’ (Ritchie et al., 2015). Differentially enriched signatures were defined as having FDR adjusted p-values < 0.05 and |mean score difference|>= 0.1 as described previously (Gao et al., 2017). Projection of all single cells from the extinction group or the persistent group tumors to 2D space was performed using t-SNE (Donaldson, 2016) based on the top differentially enriched signatures.
Normal Cell Type Prediction from Single Nucleus RNA Data
Prediction of known normal cell types in the clonal extinction post-treatment tumors was performed using a semi-supervised approach. We first classified immune cell types using established markers: T cells (CD4|CD8A|CD8B), B cells (CD19|CD20) or other immune cells (CD45). For the remaining cell types we performed a decision tree using Gini coefficients to evaluate the variability of known cell type markers across all the remaining cells. Larger Gini coefficients indicated stronger unevenness of the marker gene expression across the remaining cell populations, and therefore the cell type with the largest Gini coefficient was determined first. The remaining cell types with smaller coefficients were identified subsequently in an iterative process. The markers for the remaining cell type classification in sequential included: fibroblasts (ACTA|CAV1|FAP|FN1), luminal epithelial cells (CK8|CK18), adipocytes (ADIPOR1/2), basal epithelial cells (CK5|CK14) and endothelial cells (PECAM1|CD34). We used the gene expression [log(TPM/10+1)] cutoff=1 to determine whether a cell marker was expressed in a single cell. In our datasets, we observed a subset of cells that were co-expressed both fibroblasts and epithelial cells markers. Since ‘fibroblast’ cells were defined before epithelial cells, we reclassified certain ‘fibroblast’ cells as epithelial cells if they expressed higher levels of epithelial markers on average.
Survival Analysis in Extended Patient Cohorts
To determine whether the chemoresistance-associated gene signatures were associated with patient survival, we obtained the METABRIC dataset (Curtis et al., 2012; Pereira et al., 2016) with mRNA gene microarray expression data and long term clinical follow-up data. We analyzed a total of N=412 breast cancer patients that received chemotherapy. To test the association of selected gene sets with patient survival, we first performed gene set variation analysis (Hanzelmann et al., 2013) and stratified patients into low enrichment (ssGSVA score ≤ −0.1) and high enrichment (ssGSVA core ≥ 0.1) groups. We then performed a Cox proportional hazard regression of survival month and survival status over gene set enrichment status using the R ‘survival’ package (Therneau 2015) and the log-rank test p-values were used to determine significance.
Integration of Single Cell Copy Number Data with SNV Mutations
To integrate single cell copy number with the bulk mutation data, we used Samtools (0.1.19) (Li et al., 2009) to perform ‘mpileup’ of sparse single cell copy number reads and quantified the number of variant reads at specific mutation sites that were detected by exome sequencing. Mutation sites with ≥ 1 variant read counts in at least one cell were called as mutated sites. Mutations in single cells were then mapped to clonal subpopulations in maximum parsimony trees constructed from the single cell copy number data. With this approach, a subset of clonal mutations were successfully mapped to the truncal lineages of the copy number trees, however the limited coverage at subclonal mutation sites did not permit accurate mapping to the subclonal lineages.
Integration of DNA Mutations and RNA Single Cell Data
Since the single nuclei RNA expression data had coverage only at the 3’ ends, we mapped all exome mutation sites to the last exons of genes to identify a subset of mutations with these criteria. We used Samtools (0.1.19) (Li et al., 2009) ‘mpileup’ to obtain sparse 3’ single cell RNA sequencing read counts for mutations detected in the last exons of genes. Mutation sites with ≥ 1 read count coverage in the RNA data that matched the mutation variant base were considered as positive for having the expressed mutation, while sites with ≥ 1 read count coverage and only reference bases were considered as negative for the expressed mutation. Sites with no read count data in samples were indicated as ‘low coverage’ for detecting the expressed mutation.
Identification of Primed Cells by Integrating Single Cell CNA and RNA Data
We identified ‘primed’ cells in the pre-treatment tumor samples that expressed a subset of the chemoresistant genes by integration of single cell CNA data with single cell RNA data. Due to the sparseness of the single cell RNA data, we performed imputation using Single Cell Analysis Via Expression Recovery (SAVER) (Huang et al., 2017) to improve copy number estimations. Next, we identified the CNAs associated with resistance in the single cell DNA copy number profiles that expanded in the resistant clones from the persistence patients. To find the preexisting single cells that had resistant CNA genotypes, we clustered all pre-treatment single cells with post-treatment single cells using hierarchical clustering with the calculated RNA copy number profiles. Single cells with resistance associated CNAs that clustered together were defined as having a resistant genotype, whereas pre-treatment single cells that did not have resistance associated CNAs were defined as having a sensitive genotype. We next obtained a restricted list of significant top variable genes (N<50 genes) by comparing the transcriptional profiles of the post-treatment resistant cells (those that had both resistant genotypes and phenotypes) to pre-treatment nonresistant cells (those that did not have resistant genotypes or phenotypes) using MAST (FDR adjusted p < 0.05; |log2(fold change)| ≥ 1.58) (Finak et al., 2015). Next, we performed random forest regression with the R ‘randomForest’ package (Breiman, 2002) on 70% of the data using the top variable genes followed by prediction of resistance/sensitive classification for all single cells within each tumor. We defined the predicted probability of classifying a resistant single cell to be the resistance expression score of each cell. To determine the cutoff of the resistance expression score and define the primed cells, we calculated the cutoff score that separated pre-treatment cells within each tumor into two groups using the least square method and took the median values across the 4 patients as the universal cutoff (cutoff=0.172). We manually set the resistance cutoff as 0.6 to account for RNA dropout noise in the single cell RNA data. As such, single cells that had a resistance expression score >=0.6 were defined as resistant cells, and single cells that had resistance expression scores between 0.172 and 0.6 were defined as primed cells.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Frozen breast tumor tissue samples | Karolinska University Hospital | P1-P20, this manuscript |
| Critical Commercial Assays | ||
| GenomePlex WGA4 kit | Sigma-Aldrich | Cat#WGA4–50RXN |
| DNA Clean & Concentrator-5 | Zymo Research | Cat#11–303 or 11–306 |
| Qubit dsDNA HS Assay Kit | Invitrogen | Cat#Q32854 |
| NEBNext end repair module | NEB | Cat#E6050L |
| NEBNext dA-Tailing module | NEB | Cat#E6053L |
| T4 DNA ligase | NEB | Cat#M0202L |
| NEBNext High-Fidelity 2X PCR Master Mix | NEB | Cat#M0541 L |
| KAPA Library Quantification Kit | Kapa | Cat#KK4835 |
| Nimblegen’s SeqCap EZ Exome V2 kit | Roche | Cat#05860482001 |
| Ampure XP beads | Beckman Coulter | Cat#A63881 |
| DNA Mini Kit | QIAGEN | Cat#51306 |
| DNA Blood Mini Kit | QIAGEN | Cat#51106 |
| KAPA Library Preparation Kit | Kapa | Cat#KK8502 |
| DNA Clean and Concentrator-5 Kit | Zymo Research | Cat#11–303 |
| WaferGenlCELL8v2 | Takara/Wafergen | Cat#1565 or Cat#640003 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131–1096 |
| Oligonucleotides | ||
| Primers for amplicon validation, see Table S5 | This manuscript | N/A |
| Software and Algorithms | ||
| Burrows-Wheeler Aligner vO.7.12 | Li H. and Durbin R. 2009 | http://bio-bwa.sourceforge.net/ RRID:SCR_010910 |
| SAMtools v1.2 | Lietal.,2009 | http://samtools.sourceforge.net/ RRID: SCR_002105 |
| GATK | McKenna et al.,2010 | https://software.broadinstitute.org/gatk/ RRID:SCR_001876 |
| Picard Tools | Broad Institute | http://broadinstitute.github.io/picard RRID:SCR_006525 |
| MuTect2 | Cibulskis et al.,2013 | https://software.broadinstitute.org/gatk/ RRID:SCR_000559 |
| ANNOVAR | Wang et al., 2010 | http://annovar.openbioinformatics.org/en/1atest/ RRID:SCR 012821 |
| exomeCNVv1.4 | JF Sathira pongs asuti JF etal., 2011 | https://cran.r-project.org/src/contrib/Archive/ExomeCNV/ |
| THetA2 | Oesper L et al.,2014 | http://compbio.cs.brown,edu/projects/theta/ |
| PyClone2 vO.12.9 | Roth A etal., 2014 | |
| CITUP | Malikic S et al.,2015 | https://github.com/sfu-compbio/citup |
| R package TimeScape’ | McPherson A etal, 2016 | https://github.com/shahcompbio/timescape |
| deepSNV version 1.16.0 | Gerstung et al.,2012 | https://bioconductor.org/packages/release/bioc/html/deepSNV.html RRID:SCR_006214 |
| R package ‘DNAcopy’ | Shah etal., 2006 | https://bioconductor.org/packages/release/bioc/html/DNAcopy.html RRID:SCR_012560 |
| R function MergeLevels | Willenbrock and Fridlyand, 2005 | https://www.rdocumentation.org/packages/aCGH/versions/1.50.0/topics/mergeLevels |
| R package ‘copynumber’ | Nilsen G et al.,2012 | https://bioconductor.rikenJp/packages/3.1/bioc/html/copynumber.html |
| R package ‘phangorn’ | Schliep KP 2011 | https://github.com/KlausVigo/phangorn |
| R package ‘tSNE’ | Donaldson J, 2016 | https://CRAN.R-project.org/package=tsne |
| R package ‘fpc’ | Hennig C, 201 | https://CRAN.R-project.org/package=fpc |
| RSEM | Li B and Dewey C, 2011 | https://github.com/deweylab/RSEM |
| R package ‘MAST’ | Finak G etal., 2015 | https://github.com/RGLab/MAST |
| R package ‘GSVA’ | Hanzelmann S etal., 2013 | https://bioconductor.org/packages/release/bioc/html/GSVA.html |
| R function ‘heatmap.3’ | Murtagh and Legendre 2014 | https://github.com/obigriffith/biostar-tutorials/tree/master/Heatmaps |
| R package ‘randomForest’ | LiawAand Wiener M, 2015 | https://www.stat.berkeley.edu/~breiman/RandomForests/ |
| R package ‘SAVER’ | Huang M et al.,2017 | https://github.com/mohuangx/SAVER |
| R package ‘survival’ | Therneau T, 2015 | https://github.com/therneau/survival |
Acknowledgements
This work was supported by grants to N.N from the Lefkofsky Family Foundation, NCI (1RO1CA169244–01) and the American Cancer Society (129098-RSG-16–092-01-TBG). N.N. is a T.C. Hsu Endowed Scholar, AAAS Wachtel Scholar, Andrew Sabin Family Fellow and Randall Innovator. This study was supported by the MD Anderson Sequencing Core Facility Grant (no. CA016672) and the Flow Cytometry Facility grant from NIH (CA016672). The study was also supported by grants from the Breast Cancer Research Foundation, the Swedish Cancer Society (CAN 2015/713), and the Cancer Society in Stockholm (154132) to T.F. The study was also supported by a KI-MDACC grant from the Sister Network Institution Fund and GAP program at MD Anderson to NN, and the StratCan at the Karolinska Institute to N.C., T.F. and J.H. Additional funding support includes the ALA Fellowship to C.K., Susan Komen Postdoctoral Fellowship to R.G., and AACR Basic Cancer Research Fellowship to R.G. The PROMIX clinical trial was supported by grants from Roche, the Cancer Society in Stockholm and the Swedish Breast Cancer Association to T.H. We thank Helen Piwnica-Worms, Gloria Echeverria, Funda Meric-Bernstam, Debu Tripathy, Ken Chen, and Russell Broaddus for useful discussions and Sohrab Shah for assistance with software.
Footnotes
DATA AND SOFTWARE AVAILABILITY
Single cell copy number and data are deposited in NCBI Sequence Read Archive under accession SRP114962. Software used in this study is listed in the Key Resources table.
Declaration of Interests
Thomas Hatschek received an institutional grant to Karolinska University Hospital from Roche to support the PROMIX trial.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, and Sunyaev SR (2010). A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendro V, Cheng Y-K, Randles A, Itzkovitz S, Marusyk A, Ametller E, Gonzalez-Farre X, Muñoz M, Russnes HG, Helland A, et al. (2014). Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep 6, 514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko JM, Cook RS, Vaught DB, Kuba MG, Miller TW, Bhola NE, Sanders ME, Granja-Ingram NM, Smith JJ, Meszoely IM, et al. (2012). Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med 18, 1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, Owens P, Sanders ME, Kuba MG, Sánchez V, et al. (2014). Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov 4, 232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko JM, Schwarz LJ, Luo N, Estrada MV, Giltnane JM, Davila-Gonzalez D, Wang K, Sanchez V, Dean PT, Combs SE, et al. (2016). Triple-negative breast cancers with amplification of JAK2 at the 9p24 locus demonstrate JAK2-specific dependence. Sci Transl Med 8, 334ra353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslan T, Kendall J, Rodgers L, Cox H, Riggs M, Stepansky A, Troge J, Ravi K, Esposito D, Lakshmi B, et al. (2012). Genome-wide copy number analysis of single cells. Nat Protoc 7, 1024–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, and Weinberg RA (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L (2002). Manual On Setting Up, Using, And Understanding Random Forests V3.1.
- Cancer Genome Atlas N (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosse JP, and Michiels C (2008). Tumour hypoxia affects the responsiveness of cancer cells to chemotherapy and promotes cancer progression. Anticancer Agents Med Chem 8, 790–797. [DOI] [PubMed] [Google Scholar]
- Creighton CJ (2007). A gene transcription signature of the Akt/mTOR pathway in clinical breast tumors. Oncogene 26, 4648–4655. [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. (2012). The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. (2012). Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Valle IF, Giampieri E, Simonetti G, Padella A, Manfrini M, Ferrari A, Papayannidis C, Zironi I, Garonzi M, Bernardi S, et al. (2016). Optimized pipeline of MuTect and GATK tools to improve the detection of somatic single nucleotide polymorphisms in whole-exome sequencing data. BMC Bioinformatics 17, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doktorova H, Hrabeta J, Khalil MA, and Eckschlager T (2015). Hypoxia-induced chemoresistance in cancer cells: The role of not only HIF-1. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 159, 166–177. [DOI] [PubMed] [Google Scholar]
- Donaldson J (2016). tsne: T-Distributed Stochastic Neighbor Embedding for R (t-SNE). R package version 0.1–3.
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. (2017). COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45, D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Smith IE, and Reis-Filho JS (2010). Triple-Negative Breast Cancer. N Engl J Med 363, 1938–1948. [DOI] [PubMed] [Google Scholar]
- Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, Tsai PC, Casasent A, Waters J, Zhang H, et al. (2016). Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet 48, 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Kim C, Sei E, Foukakis T, Crosetto N, Chan L-K, Srinivasan M, Zhang H, Meric-Bernstam F, and Navin N (2017). Nanogrid Single-Nucleus RNA Sequencing Reveals Phenotypic Diversity in Breast Cancer Nature Communications (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendoo DM, Ratanasirigulchai N, Schroder MS, Pare L, Parker JS, Prat A, and Haibe-Kains B (2016). Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 32, 1097–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstung M, Beisel C, Rechsteiner M, Wild P, Schraml P, Moch H, and Beerenwinkel N (2012). Reliable detection of subclonal single-nucleotide variants in tumour cell populations. Nat Commun 3, 811. [DOI] [PubMed] [Google Scholar]
- Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, and van Oudenaarden A (2015). Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255. [DOI] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, and Regev A (2016). Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann S, Castelo R, and Guinney J (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AL (2002). Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer 2, 38–47. [DOI] [PubMed] [Google Scholar]
- Hennig C (2015). fpc: Flexible Procedures for Clustering. R package version 21–10.
- Hu Y, Liu J, and Huang H (2013). Recent agents targeting HIF-1alpha for cancer therapy. J Cell Biochem 114, 498–509. [DOI] [PubMed] [Google Scholar]
- Huang M, Wang J, Torre E, Dueck H, Shaffer D, Bonasio R, Murray J, Raj A, Li M, and Zhang N (2017). Gene Expression Recovery For Single Cell RNA Sequencing. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lonnerberg P, and Linnarsson S (2014). Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 11, 163–166. [DOI] [PubMed] [Google Scholar]
- Jaakkola MK, Seyednasrollah F, Mehmood A, and Elo LL (2016). Comparison of methods to detect differentially expressed genes between single-cell populations. Brief Bioinform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. (2014). Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343, 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, Grimsby J, Sougnez C, Muller F, Hu J, et al. (2015). Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res 25, 316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Juhnn YS, and Song YS (2007). Akt involvement in paclitaxel chemoresistance of human ovarian cancer cells. Ann N Y Acad Sci 1095, 82–89. [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, and Teichmann SA (2015). The technology and biology of single-cell RNA sequencing. Mol Cell 58, 610–620. [DOI] [PubMed] [Google Scholar]
- Kurtova AV, Xiao J, Mo Q, Pazhanisamy S, Krasnow R, Lerner SP, Chen F, Roh TT, Lay E, Ho PL, et al. (2015). Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature 517, 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. (2016). Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon WB (2015). Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, et al. (2008). Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 26, 1275–1281. [DOI] [PubMed] [Google Scholar]
- Liu P, Cheng H, Roberts TM, and Zhao JJ (2009). Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov 8, 627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, et al. (2014). Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 32, 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Weaver VM, and Werb Z (2012). The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol 196, 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE, and Delbrück M (1943). Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics 28, 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malikic S, McPherson AW, Donmez N, and Sahinalp CS (2015). Clonality inference in multiple tumor samples using phylogeny. Bioinformatics 31, 1349–1356. [DOI] [PubMed] [Google Scholar]
- Marcucci F, Stassi G, and De Maria R (2016). Epithelial-mesenchymal transition: a new target in anticancer drug discovery. Nat Rev Drug Discov 15, 311–325. [DOI] [PubMed] [Google Scholar]
- Martelotto LG, Baslan T, Kendall J, Geyer FC, Burke KA, Spraggon L, Piscuoglio S, Chadalavada K, Nanjangud G, Ng CK, et al. (2017). Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nat Med 23, 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Reimand J, Lan X, Head R, Zhu X, Kushida M, Bayani J, Pressey JC, Lionel AC, Clarke ID, et al. (2015). Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A 112, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. (2011). Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin NE (2014). Tumor evolution in response to chemotherapy: phenotype versus genotype. Cell Rep 6, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, and Henikoff S (2003). SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31, 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen G, Liestol K, and Lingjaerde OC (2013). copynumber: Segmentation of single- and multi-track copy number data by penalized least squares regression. R package version 1120.
- Oesper L, Mahmoody A, and Raphael BJ (2013). THetA: inferring intra-tumor heterogeneity from high-throughput DNA sequencing data. Genome Biol 14, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesper L, Satas G, and Raphael BJ (2014). Quantifying tumor heterogeneity in whole-genome and whole-exome sequencing data. Bioinformatics 30, 3532–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, and Wigler M (2004). Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5, 557–572. [DOI] [PubMed] [Google Scholar]
- Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, and Weinberg RA (2008). Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res 68, 3645–3654. [DOI] [PubMed] [Google Scholar]
- Oskarsson T (2013). Extracellular matrix components in breast cancer progression and metastasis. Breast 22 Suppl 2, S66–72. [DOI] [PubMed] [Google Scholar]
- Owonikoko TK, and Khuri FR (2013). Targeting the PI3K/AKT/mTOR pathway: biomarkers of success and tribulation Am Soc Clin Oncol Educ Book. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, Nones K, Cowin P, Alsop K, Bailey PJ, et al. (2015). Whole-genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494. [DOI] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, et al. (2016). The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 7, 11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Gouel F, Dubus I, Heuclin C, Roget K, and Vannier JP (2016). Hypoxia promotes chemoresistance in acute lymphoblastic leukemia cell lines by modulating death signaling pathways. BMC Cancer 16, 746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, Ha G, Aparicio S, Bouchard-Cote A, and Shah SP (2014). PyClone: statistical inference of clonal population structure in cancer. Nat Methods 11, 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathirapongsasuti JF, Lee H, Horst BA, Brunner G, Cochran AJ, Binder S, Quackenbush J, and Nelson SF (2011). Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 27, 2648–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP (2011). phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL (2003). Targeting HIF-1 for cancer therapy. Nat Rev Cancer 3, 721–732. [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. (2012). The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, and Sirotkin K (2001). dbSNP: the NCBI database of genetic variation In Nucleic Acids Res, pp. 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M (2017). timescape: Patient Clonal Timescapes. R package version 1.1.0.
- Smith MA, Nielsen C, Chan FC, McPherson AW, Roth A, Farahani H, Machev D, Steif A, and Shah SP (2016). E-scape: Interactive visualization of single cell phylogenetics and spatio-temporal evolution in cancer. bioRxiv. [DOI] [PubMed] [Google Scholar]
- Therneau T (2015). A Package for Survival Analysis in S. R package version 2.38.
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung PY, Blischak JD, Hsiao CJ, Knowles DA, Burnett JE, Pritchard JK, and Gilad Y (2017). Batch effects and the effective design of single-cell gene expression studies. Sci Rep 7, 39921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton G (2008). Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605. [Google Scholar]
- Wang Y, and Navin NE (2015). Advances and applications of single-cell sequencing technologies. Mol Cell 58, 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, et al. (2014). Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbrock H, and Fridlyand J (2005). A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics 21, 4084–4091. [DOI] [PubMed] [Google Scholar]
- Yang H, and Wang K (2015). Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR In Nat Protoc, pp. 1556–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. (2015). Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, and Sims PA (2016). An Automated Microwell Platform for Large-Scale Single Cell RNA-Seq. Sci Rep 6, 33883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn H, Steif A, Laks E, Eirew P, VanInsberghe M, Shah SP, Aparicio S, and Hansen CL (2017). Scalable whole-genome single-cell library preparation without preamplification. Nat Methods 14, 167–173. [DOI] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, and Kalluri R (2015). Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.