Summary
TERT promoter mutations reactivate telomerase, allowing for indefinite telomere maintenance and enabling cellular immortalization. These mutations specifically recruit the multimeric ETS factor GABP, which can form two functionally independent transcription factor species – a dimer or a tetramer. We show that genetic disruption of GABPβ1L (β1L), a tetramer-forming isoform of GABP that is dispensable for normal development, results in TERT silencing in a TERT promoter mutation-dependent manner. Reducing TERT expression by disrupting β1L culminates in telomere loss and cell death exclusively in TERT promoter mutant cells. Orthotopic xenografting of β1L-reduced, TERT promoter mutant glioblastoma cells rendered lower tumor burden and longer overall survival in mice. These results highlight the critical role of GABPβ1L in enabling immortality in TERT promoter mutant glioblastoma.
Introduction:
Telomeres maintain DNA integrity by protecting the ends of chromosomes but progressively shorten with each cell division (Blackburn et al., 2006; Counter et al., 1992). Telomere length is maintained by telomerase, a multi-subunit complex that binds and elongates the telomere ends. Telomerase Reverse Transcriptase (TERT) is the catalytic subunit of telomerase, and its expression is the rate-limiting step in telomerase activity across a wide range of tissues (Bryan and Cech, 1999; Counter et al., 1998). While normally silenced in somatic cells, over 90% of human tumors reactivate TERT expression, allowing cancer cells to gain replicative immortality by avoiding cell death and senescence associated with telomere shortening (Chin et al., 1999; Kim et al., 1994; Saretzki et al., 1999; Shay and Wright, 2000). Two activating mutation hotspots in the TERT promoter, termed C228T and C250T, are found in over 50 tumor types, and are the most frequent mutations in several tumor types, including 83% of primary IDH wild-type glioblastomas (GBM) and 78% of oligodendrogliomas (Arita et al., 2013; Killela et al., 2013; Zehir et al., 2017). These mutually exclusive mutations exist predominantly in the heterozygous state, acting as the drivers of telomerase reactivation (Horn et al., 2013; Huang et al., 2013; Killela et al., 2013). In high-grade gliomas, TERT promoter mutations correlate with increased TERT mRNA levels and enhanced telomerase activity (Spiegl-Kreinecker et al., 2015; Vinagre et al., 2013). Furthermore, in tumor cells bearing TERT promoter mutations, these mutations are necessary – albeit not sufficient – for achieving replicative immortality (Chiba et al., 2015; Chiba et al., 2017). Both TERT promoter mutations generate identical 11 base pair sequences that form a de novo binding site for the ETS transcription factor GA-binding protein (GABP) (Bell et al., 2015). The presence of either promoter mutation allows GABP to selectively bind and activate the mutant TERT promoter while the wild-type allele remains silenced (Akincilar et al., 2016; Bell et al., 2015; Stern et al., 2015). GABP has no known role in TERT regulation outside of TERT promoter mutant tumors.
The GABP transcription factor is an obligate multimer consisting of the DNA-binding GABPα subunit and trans-activating GABPβ subunit. GABP can act as a heterodimer (GABPαβ) composed of one GABPα and one GABPβ subunit or a heterotetramer (GABPα2β2) composed of two GABPα and two GABPβ subunits (Rosmarin et al., 2004; Sawada et al., 1994). Two distinct genes encode the GABPβ subunit, GABPB1 encodes GABPβ1 (β1) and GABPB2 encodes GABPβ2 (β2). β1 has two isoforms transcribed from the GABPB1 locus, the shorter GABPβ1S (β1S) and the longer GABPβ1L (β1L), while β2 has a single isoform (de la Brousse et al., 1994; Rosmarin et al., 2004). Whereas β1S is able to dimerize only with GABPα, both β1L and β2 possess a C-terminal leucine-zipper domain (LZD) that mediates the tetramerization of two GABPαβ heterodimers (de la Brousse et al., 1994; Rosmarin et al., 2004). Although β1L or β2 can form the GABP tetramer, GABP tetramers containing only the β1L isoform are functionally distinct from β2-containing tetramers and may control separate transcriptional programs (Jing et al., 2008; Yu et al., 2012). Furthermore, while abolishing the full tetramer-specific (β1L and β2) transcriptional program impairs the self-renewal of hematopoietic stem cells in mice (Yu et al., 2012), inhibition of the β1L-only tetramer-specific transcriptional program has minimal phenotypic consequences in a murine system (Jing et al., 2008; Xue et al., 2008). Thus, if the GABP tetramer-forming isoforms are necessary to activate the mutant TERT promoter, disrupting the function of these isoforms may be a viable approach to selectively inhibit TERT and reverse replicative immortality in TERT promoter mutant cancer.
However, it is currently unclear whether the GABP tetramer-forming isoforms are necessary to activate the mutant TERT promoter or whether the GABP dimer is sufficient. Two proximal GABPα binding sites are required to recruit a GABPα2β2 tetramer, and, interestingly, the TERT promoter has native ETS binding sites upstream of the hotspot mutations that are required for robust activation of the mutant promoter (Bell et al., 2015). These native ETS binding sites are located approximately three and five helical turns of DNA away from the C228T and C250T mutation sites, respectively, which is consistent with the optimal spacing for the recruitment of the GABP tetramer (Bell et al., 2015; Chinenov et al., 2000; Yu et al., 1997). Here we tested the hypothesis that the C228T and C250T hotspot promoter mutations recruit the tetramer-specific GABP isoforms to the mutant TERT promoter to enable telomere maintenance and replicative immortality.
Results:
The GABP tetramer-forming isoform β1L positively regulates TERT expression in TERT promoter mutant - but not wild-type - tumor cells
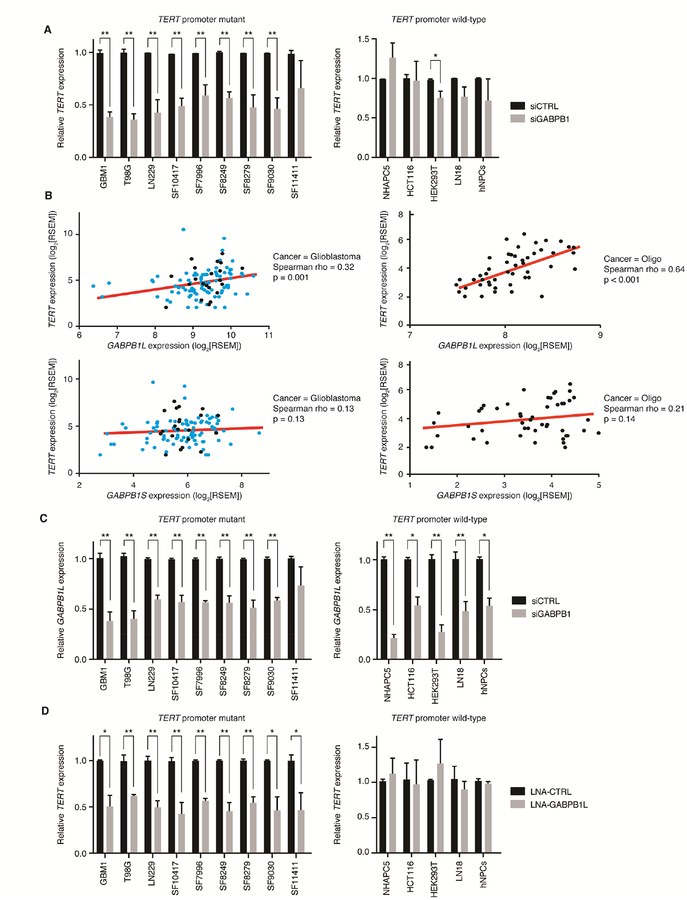
To determine if the GABP dimer-forming isoform (β1S) or the tetramer-forming isoforms (β1L and β2) regulate the mutant TERT promoter, we performed gene knockdown experiments in vitro and expression correlation analysis in primary tumors. We used siRNA-mediated knockdown of β1 - affecting β1S and β1L - and β2 in three TERT promoter mutant glioma cell lines, six early passage primary cultures and five TERT promoter wild-type and TERT expressing cell lines. Knockdown of β1 significantly reduced TERT expression in eight of nine TERT promoter mutant cell cultures, but had limited effect in the TERT promoter wild-type cultures (Figure 1A). In contrast, siRNA-mediated knockdown of β2 had a less robust and more variable effect on TERT expression in TERT promoter mutant cells (Figure S1A).
Figure 1.
The GABP tetramer-forming isoform β1L positively regulates TERT expression solely in TERT promoter mutant tumor cells. (A) TERT expression following siRNA-mediated knockdown of β1 (siGABPB1) in TERT promoter mutant (left) or TERT promoter-wild-type (right) cell lines and primary cultures. *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to a non-targeting siRNA control (siCTRL) in each respective line. (B) Correlation of GABPB1L (top graphs) or GABPB1S (bottom graphs) expression (log2[RSEM normalized counts]) versus TERT expression (log2[RSEM normalized counts]) from 109 TERT-expressing GBMs (left graphs) or 49 TERT promoter-mutant oligodendrogliomas (right graphs). Red line indicates trend line. Black points indicate Sanger-validated TERT promoter mutant GBM and oligodendroglioma samples, teal points are GBM samples that were not tested for TERT promoter mutation status. Spearman’s Rank-Order Correlation was used to generate Spearman rho and p values for each correlation. (C) GABPB1L expression following siRNA-mediated knockdown of β1 (siGABPB1) in TERT promoter mutant (left) and wild-type (right) lines. *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to a non-targeting siRNA control (siCTRL) in each respective line. (D) TERT expression following LNA-ASO knockdown of β1L (LNA-GABPB1L) in TERT promoter mutant (left) or wild-type (right) cell lines and primary cultures compared to a control LNA-ASO (LNA-CTRL). *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to LNA-CTRL in each respective line. Values are mean ± S.D. of at least three independent experiments (A, C, and D; two independent experiments for SF10417). See also Figure S1.
We also tested whether the expression of TERT correlates with expression of specific GABP isoforms in clinical samples, including TERT promoter mutant GBMs and oligodendrogliomas. This analysis revealed a significant positive monotonic association between TERT and GABPB1L mRNA in both cancer types (Figure 1B), but no significant correlation between TERT and GABPB1S (Figure 1B) or GABPB2 (Figure S1B) mRNA levels. Analysis of GABP isoform and TERT expression data in the predominantly TERT promoter wild-type colorectal cancer revealed no positive correlation between TERT expression and GABPB1L or GABPB2 expression, although a positive correlation between TERT expression and GABPB1S expression was found (Figure S1C). Due to the significant positive correlation between GABPB1L expression and TERT expression in glioma, we specifically looked for depletion of the tetramer-forming GABPB1L isoform mRNA in our β1 knockdown study and confirmed that this isoform mRNA was significantly depleted after siRNA-mediated knockdown in 13 of 14 cell lines (Figure 1C).
We further explored this potential dependence on the β1L isoform for activation of the mutant TERT promoter by directly knocking down β1L with a degradation-inducing Locked Nucleic Acid Anti-Sense Oligonucleotide (LNA-ASO) targeted to the GABPB1L-exclusive 3’ UTR of the GABPB1 transcript. This LNA-ASO specifically depleted GABPB1L transcript levels with no reduction in GABPB1S transcript levels (Figure S1D). LNA-ASO-mediated knockdown of β1L reduced TERT expression across all TERT promoter mutant cultures and had no effect on TERT expression in all TERT promoter wild-type cultures (Figure 1D). Taken together, these data support that the GABP tetramer-forming isoform β1L positively regulates TERT expression in TERT promoter mutant glioma.
CRISPR-Cas9-mediated disruption of GABPB1L reduces GABP-mediated activation of the mutant TERT promoter
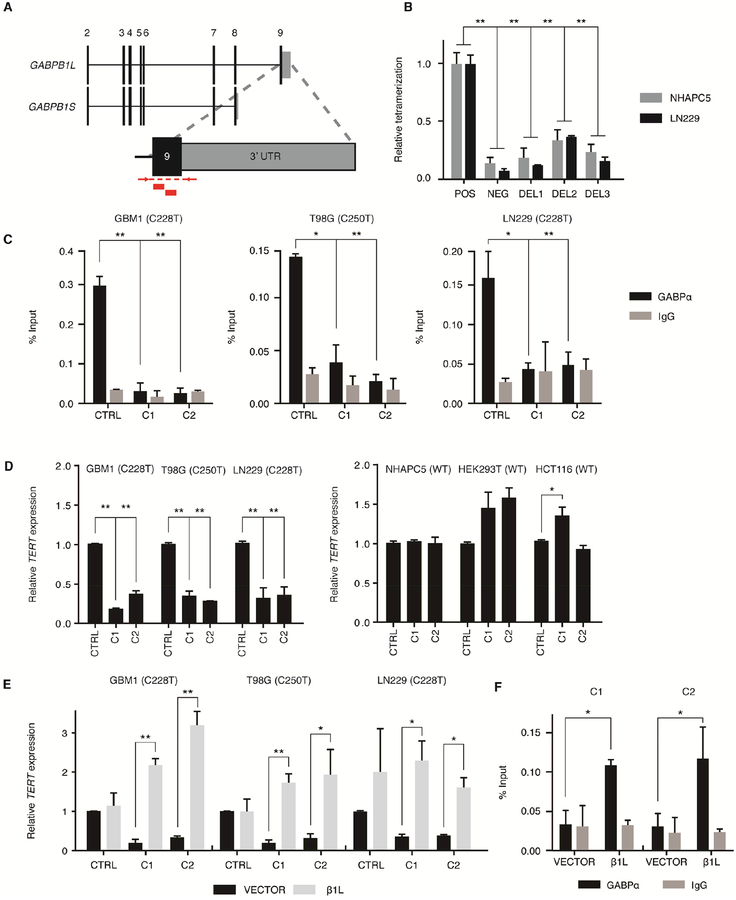
We then directly tested the necessity of β1L for mutant TERT promoter activation by generating clones with reduced β1L function from three of the aforementioned TERT promoter mutant GBM cell lines (GBM1, T98G, and LN229) and three TERT promoter wild-type control cell lines (NHAPC5, HCT116 and HEK293T) using nuclease-assisted vector integration (NAVI) CRISPR-Cas9 editing (Brown et al., 2016; Gapinske et al., 2018) (Figure 2A). We isolated two independent GABPB1L-edited clones (C1 and C2) and one isogenic CRISPR control clone (CTRL) for each parental line using one of two non-overlapping sgRNAs targeting GABPB1 exon 9 or a sgRNA targeting an intergenic region of chromosome 5, respectively (Figure S2A and Table S1). GABPB1 exon 9 contains the coding sequence for the LZD, and disruption of this exon is sufficient for ablation of the β1L-containing tetramer while leaving β1S intact (Chinenov et al., 2000; Sawada et al., 1994). Each GABPB1L-edited clone had the disruption of at least one allele via integration of a puromycin or hygromycin resistance cassette with most remaining GABPB1L alleles containing indels in the LZD (Figure S2B and Table S2). Analysis of cassette integration and locus integrity at predicted off-target cutting sites in coding regions (Hsu et al., 2013) via PCR and Surveyor assay, respectively, showed no aberrations outside the target regions (Figures S3A–F). GABPB1L-edited clones had reduced β1L protein levels with no measurable reduction in β1S levels, further confirming the specificity of our editing approach (Figure S3G).
Figure 2.
CRISPR-Cas9-mediated disruption of GABPB1L reduces GABP-mediated activation of the mutant TERT promoter. (A) Exon structure for the GABPB1 locus, depicting the GABPB1S and GABPB1L isoforms. Inset shows targeting strategy for CRISPR-Cas9 editing of GABPB1L. Red blocks indicate sgRNA target sites. Red arrows and dashed lines indicate primer locations and target amplicon for PCR validation of editing. (B) Quantification of β1L tetramerization in the wild-type (POS) or mutated (DEL1-3) state. The negative (NEG) state consists of one β1L vector and one β1S vector, the products of which are unable to form a tetramer. *p value<0.05, **p value<0.01, two-sided Student’s t-test of DEL1-3 or NEG respective to the positive control (POS). (C) GABPα or IgG control ChIP-qPCR for the TERT promoter in CRISPR control (CTRL) or β1L-reduced clones (C1 and C2). *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to respective CTRL. (D) TERT expression relative to CTRL for β1L-reduced TERT promoter mutant (left) or wild-type (right) clones. *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to CTRL. (E,F) TERT expression (E) or GABPα occupancy (F) in β1L-reduced clones relative to CTRL 48 hr following transfection with empty (VECTOR) or β1L expression vector. *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to respective VECTOR control. Values are mean ± S.D. of at least two independent experiments (C and F) or three independent experiments (B, D, and E). See also Figures S2–S3 and Tables S1–S3.
We next examined whether the indels in the remaining GABPB1L alleles (Figure S2B) were sufficient to generate β1L protein with reduced tetramerization activity. Using PCR-mediated site-directed mutagenesis, we replicated three mutations (Table S3) in GABPB1L and assayed the ability of the mutant β1L to form the GABP tetramer (Figure 2B). DEL1 and DEL2 are in-frame deletions in the GABPB1L LZD-coding region and DEL3 is a putative loss-of-function frame-shift mutation in the same domain (Figure S2B). Each of the tested mutations reduced the ability of β1L to form the tetramer compared to the wild-type control, thereby indicating that the CRISPR-Cas9-induced mutations in the GABPB1L LZD-coding region are sufficient to produce variants of the GABP tetramer-forming isoform β1L with reduced function. Thus, all GABPB1L-edited clones will be referred to as “β1L-reduced” to encompass reductions in both protein levels and protein function.
Chromatin immunoprecipitation of GABP followed by quantitative PCR (qPCR) at the mutant TERT promoter revealed the loss of GABP binding in the β1L-reduced TERT promoter mutant clones compared to the control lines (Figure 2C). Furthermore, analysis of TERT expression via RT-qPCR confirmed a significant reduction in - but not complete loss of - TERT mRNA across all TERT promoter mutant clones, whereas no decreases in expression were detected in clones from TERT promoter wild-type cells (Figure 2D). Additionally, overexpression of exogenous β1L in each β1L-reduced clone was sufficient to rescue both TERT expression (Figures 2E and S3H) and GABP binding at the mutant TERT promoter (Figure 2F). Taken together, these data confirm that the GABP tetramer-forming isoform β1L is necessary for the complete activation of the mutant TERT promoter.
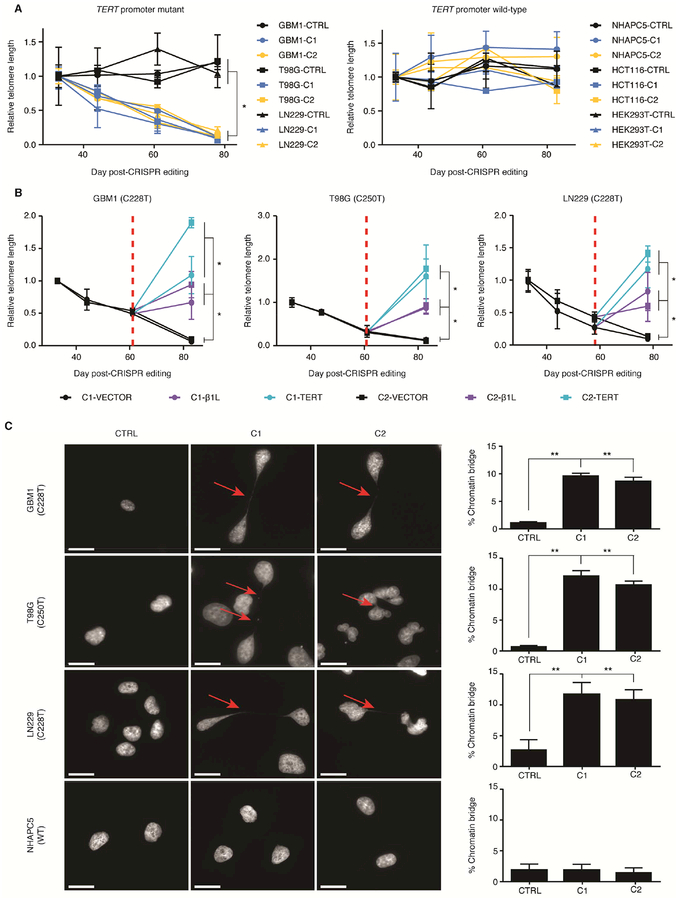
β1L-mediated activation of the mutant TERT promoter is required for telomere maintenance in GBM
As TERT expression is closely linked to telomere maintenance, we next investigated the effects of reducing β1L function on telomere length in the TERT promoter mutant cell lines. Measurements of mean relative telomere length at four time points following CRISPR-Cas9 editing uncovered significant telomere loss only in clones from TERT promoter mutant cells with reduced β1L function and TERT expression (Figure 3A). Expression of exogenous β1L or TERT was sufficient to halt this telomere loss in all clones (Figure 3B). Uncontrolled telomere shortening and uncapping can result in end-to-end fusions of telomere-deficient chromosomes and the formation of chromatin bridges (Capper et al., 2007; der-Sarkissian et al., 2004; Hackett et al., 2001). We identified chromatin bridges in a significant proportion of the TERT promoter mutant, but not TERT promoter wild-type, β1L-reduced clones 70–75 days after editing, indicating widespread telomere dysfunction following telomere loss (Figures 3C and S4A). Likewise, telomere dysfunction was readily rescued by expression of exogenous β1L or TERT (Figures S4B and S4C). These data support that disrupting β1L function is sufficient to induce telomere loss and dysfunction in a TERT promoter mutation-dependent manner.
Figure 3.
β1L-mediated activation of the mutant TERT promoter is required for telomere maintenance in GBM. (A) Telomere length at days 44, 61, and 78 in TERT promoter mutant lines or days 44, 61 and 83 in TERT promoter wild-type lines post-editing relative to day 33 post-editing for CTRL or β1L-reduced clones. *p value<0.05, two-sided Student’s t-test comparing values between CTRL and β1L-reduced clones at day 78/83 for each respective line. Values are mean ± S.D. of at least three independent assays. (B) Relative telomere length after transfection of an empty (VECTOR), β1L, or TERT expression vector in TERT promoter-mutant lines 78 or 83 days post-editing. Red dotted line indicates time of transfection (at day 58 [LN229] or 61 [GBM1 and T98G] post-editing). *p value<0.05, two-sided Student’s t-test of values of β1L or TERT versus VECTOR at day 78/83. Values are mean ± S.D. of at least three independent experiments. (C) Representative DAPI images (left images) and quantification (right graphs) of chromatin bridges (arrow) in CTRL or β1L-reduced clones at days 70–75 post-editing. Scale bar = 20 μm. *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to CTRL. Quantification values are weighted mean ± S.D. of at least ten independent fields of view. See also Figure S4.
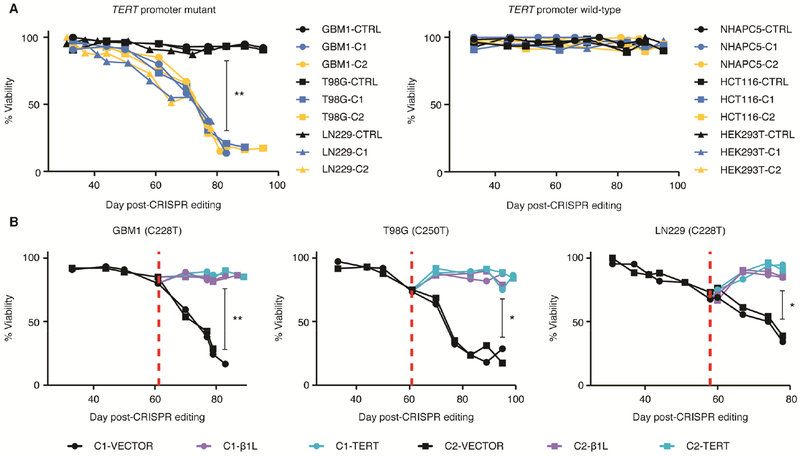
Disrupting β1L function is sufficient to induce short-term and long-term growth defects in TERT promoter mutant lines in vitro
Previous studies have reported that TERT depletion and telomere dysfunction result in both immediate and long-term growth defects (Cao et al., 2002; Fitzgerald et al., 1999; Iwado et al., 2007; Shay and Wright, 2006). Thus we sought to determine whether reduction of β1L results in a growth phenotype as a result of reduced expression from the mutant TERT promoter. Monitoring cell growth prior to significant telomere loss (days 45–48 post-editing) revealed a growth defect in all TERT promoter-mutant β1L-reduced clones (Figure S5A). We further inhibited β1L in the β1L-reduced lines with an LNA-ASO to deplete any residual β1L function and observed no further changes in cell growth (Figure S5B) or TERT expression (Figure S5C) regardless of TERT promoter status. Interestingly, LNA-ASO-mediated knockdown of β1L in TERT promoter mutant control lines significantly reduced cell growth compared to the LNAASO controls, suggesting a short-term growth effect following reduction of β1L and TERT levels.
Long-term changes in growth and cell viability may occur due to telomere dysfunction in the TERT promoter mutant, β1L-reduced clones. We monitored each β1L-reduced line throughout the process of telomere loss and identified a progressive loss of cell viability in β1L-reduced clones from TERT promoter mutant cells, a phenotype that was absent in the clones from TERT promoter wild-type cells (Figure 4A). We observed complete growth arrest in both GBM1 β1L-reduced clones, and substantial but incomplete arrest of the cultures of T98G and LN229 clones. β1L-reduced clones derived from T98G underwent complete growth arrest in all cases except one instance when a surviving population emerged following long-term culture. Unlike GBM1 and T98G cells, both LN229 clones consistently had a population of viable cells emerge following the period of massive cell death. The underlying cause of this heterogeneity in cellular response among the three lines is unknown, but could reflect residual function of β1L in β1L-reduced clones, potential β1L-independent mechanisms of activation of the mutant TERT promoter, or other factors. Importantly, overexpression of either exogenous β1L or TERT was sufficient to counteract the loss of viability (Figure 4B). This gradual loss of viability signified the loss of replicative immortality in TERT promoter mutant β1L-reduced clones.
Figure 4.
β1L reduction induces loss of replicative immortality in TERT promoter-mutant GBM lines. (A) Cell viability of CTRL or β1L-reduced clones measured approximately every 7 days from day 33 to day 99 post-editing for TERT promoter mutant and wild-type lines. **p value<0.01, Welch’s t-test of CTRL clones versus β1L-reduced clones at day 83 post-editing. (B) Cell viability measurements following transfection with an empty (VECTOR), β1L, or TERT expression vector. Red dotted line indicates time of transfection. *p value<0.05, **p value<0.01, Welch’s t-test of vector transfected cells versus β1L and TERT transfected cells at the final recorded time-point for each line. Values are median of three independent experiments. See also Figure S5.
β1L regulates a subset of GABP transcription factor targets in GBM cells
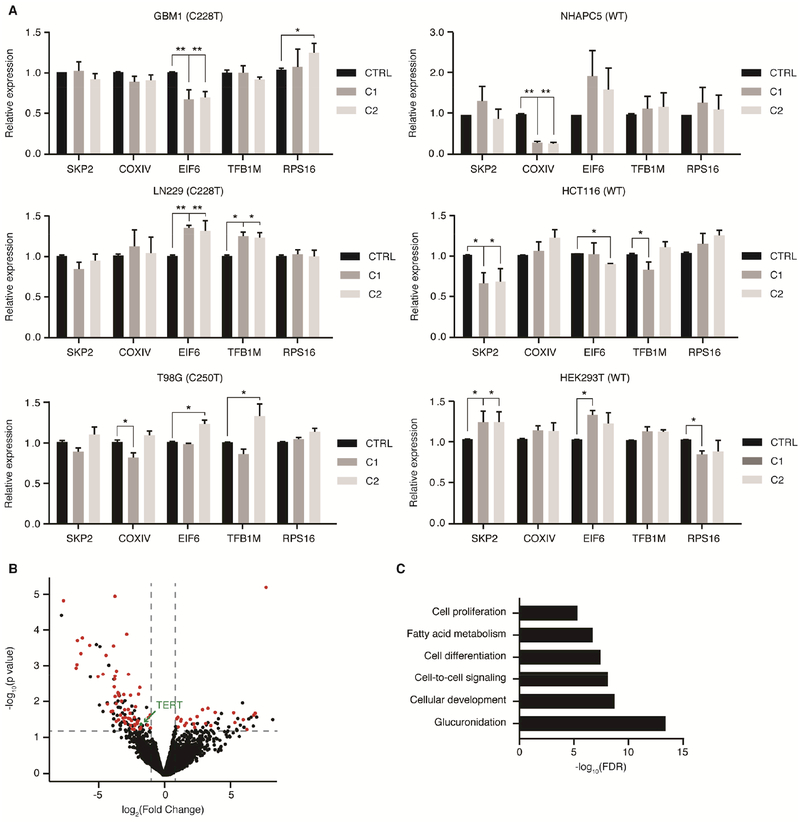
We next explored whether the observed changes in growth rate and cell viability are sole consequences of TERT depletion, are mediated by changes in levels of GABP target genes, or are a combination of both factors. The four targets selected for preliminary expression analysis (COXIV, EIF6, RPS16, and TFB1M) are essential for cell growth and have been previously identified to recruit the β1L-containing GABP tetramer via two ETS binding sites in their promoter (Carter and Avadhani, 1994; Donadini et al., 2006; Genuario and Perry, 1996; Yang et al., 2014). SKP2 contains only one ETS binding site in its promoter and should be unaltered by changes in β1L (Yang et al., 2007). We identified minimal differences in the expression of each of the five targets between the CRISPR control and β1L-reduced clones (Figure 5A).
Figure 5.
β1L regulates a subset of GABP transcription factor targets in GBM cells. (A) Expression of one GABP dimer target and four GABP tetramer targets relative to CTRL for β1L-reduced clones derived from TERT promoter mutant and wild-type lines at day 45 post-editing. *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to CTRL. Values are mean ± S.D of at least three independent assays. (B) Volcano plot of expression differences between CTRL and β1L-reduced TERT promoter mutant lines (GBM1, T98G, and LN229) as determined via RNA-seq at day 45 post-editing. Maroon-colored points represent putative GABP-regulated genes that are differentially expressed (log2 Fold Change>1 & FDR<0.05). (C) GO-terms analysis of 161 genes that are commonly differentially expressed genes between CTRL and multiple β1L-reduced TERT promoter mutant lines. See also Tables S4 and S5.
To further interrogate the effects of β1L reduction on global gene expression, we performed RNA sequencing (RNA-seq) for our TERT promoter mutant CRISPR control and β1L-reduced lines 45 days post-editing (Figure 5B and Table S4). We identified 161 transcripts, including TERT, differentially expressed (DE; FDR<0.05) after β1L reduction that were common to all three TERT promoter mutant lines. A majority of these DE transcripts (55%) were transcribed from genes with GABP-bound promoters, as determined from ENCODE ChIP-seq data from TERT promoter wild-type and mutant cancer cell lines (see STAR Methods). Interestingly, however, the vast majority (99%) of GABP-bound genes were not differentially expressed between the control and β1L-reduced lines. Gene ontology analysis of these DE transcripts identified enrichment in genes involved in development, cell-to-cell signaling, and proliferation (Figure 5C and Table S5). This global transcriptional analysis further validates that we have significantly inhibited the function of β1L in the β1L-reduced cell lines. These data, in combination with our qPCR analysis of canonical GABP tetramer targets, supports previous studies delineating specific transcriptional programs that different GABP species may control (Jing et al., 2008; Xue et al., 2008; Yu et al., 2012). The basis for the differential sensitivity between the effects of disrupting β1L function on the mutant TERT promoter and selected down-regulated GABP loci relative to other GABP targets is unknown, but may be due to compensation by β1S, β2, or other ETS factors at certain GABP binding sites and not at other sites, or due to cell type specific differences in the GABP transcriptional program. These data suggest that the GABP binding site created by mutations in the TERT promoter and a subset of GABP binding sites are more sensitive to inhibition of the β1L-containing GABP tetramer, while other GABP-bound sites are less sensitive.
β1L-reduced GBM lines accrue DNA damage and undergo mitotic cell death in a TERT promoter mutation-dependent manner
The direct correlation between telomere shortening and viability loss (Figure S6A) suggested that the loss of viability is a consequence of cell death or senescence induced by telomere dysfunction. The formation of chromatin bridges after telomere dysfunction induces breakage-fusion-bridge cycles that lead to the accrual of significant DNA damage in telomere-deficient cells (der-Sarkissian et al., 2004; Hackett et al., 2001). While canonical apoptosis and cellular senescence have been widely observed as results of significant DNA damage after telomere dysfunction, both mechanisms are dependent on functional p53 and RB pathways (Saretzki et al., 1999; Whitaker et al., 1995). However, these two pathways are commonly mutated in TERT promoter mutant GBM, including the GBM1, T98G, and LN229 lines (Table S6), making apoptosis and senescence unlikely to occur at high levels. In p53- and RB-deficient cells, mitotic cell death has been implicated as a primary phenotype following telomere dysfunction (Fragkos and Beard, 2011; Hayashi et al., 2015). Mitotic cell death can result from chromosome fusions, high-level chromosomal rearrangements and DNA damage, oft-described consequences of breakage-fusion-bridge cycles during telomere dysfunction (Hayashi et al., 2015; Vakifahmetoglu et al., 2008; Vitale et al., 2011).
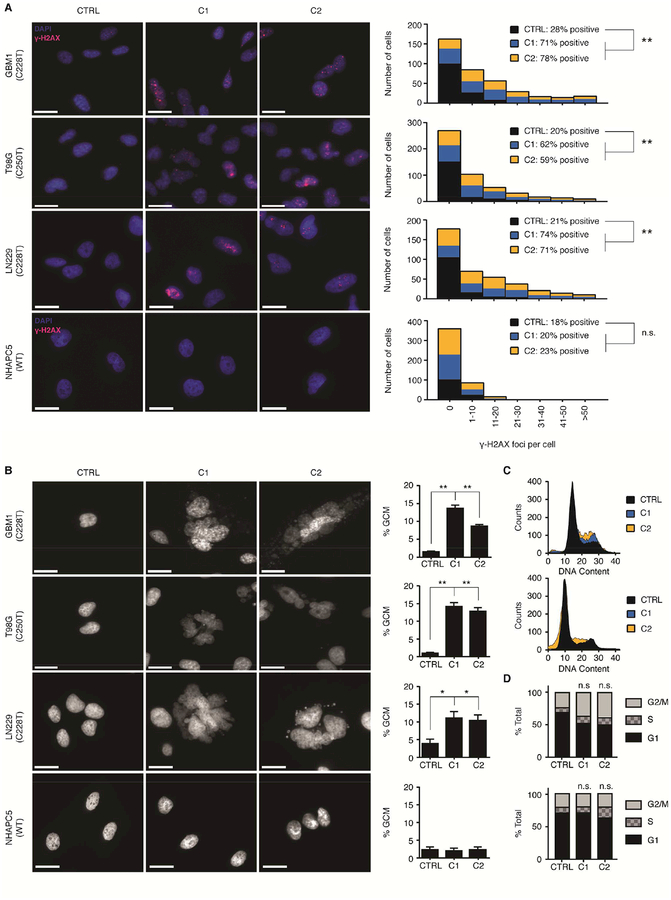
Indeed, we observed a significant increase in the amount of the DNA damage marker γ-H2AX exclusive to the β1L-reduced clones from TERT promoter mutant cells by day 73 post-editing (Figures 6A and S6B). Likewise, we identified giant cell micronucleation, a prominent feature of mitotic cell death (Ianzini and Mackey, 1997; Vakifahmetoglu et al., 2008), in β1L-reduced, TERT promoter mutant – but not wild-type – cells at this same time point (Figures 6B and S6C). Overexpression of exogenous β1L or TERT was sufficient to fully rescue both the DNA damage (Figure S7A) and mitotic cell death phenotypes (Figure S7B). Additionally, chromatin bridge formation, γ-H2AX staining, and giant cell micronucleation accumulated over three time points (days 45, 61, and 73 post-editing) in the LN229 β1L-reduced clones, thus supporting that these phenotypes may be dependent on telomere shortening (Figure S7C).
Figure 6.
β1L-reduced GBM lines accrue DNA damage and undergo mitotic cell death in a TERT promoter mutation-dependent manner. (A) Representative images (left images) and quantification (right graphs) of γ-H2AX staining in CTRL or β1L-reduced clones at day 70–75 post-editing. Scale bar = 200μm. **p value<0.01, two-sided Student’s t-test compared to CTRL. Quantification values are sums of at least ten independent fields of view. (B) Representative DAPI images (left images) and quantification (right graphs) of giant cell micronucleation (GCM) in CTRL or β1L-reduced clones at day 70–75 post-editing. Scale bar = 20 μm. *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to CTRL. Quantification values are weighted mean ± S.D. of at least ten independent fields of view. (C,D) Histograms (C) and quantification (D) for cell cycle analysis of CTRL or β1L-reduced LN229 (top graphs) and NHAPC5 (bottom graphs) lines at day 75 post-editing. See also Figures S6–S7 and Table S6.
Moreover, cell cycle analysis of the β1L-reduced TERT promoter mutant cells between day 70 and day 80 post-CRISPR-Cas9 editing revealed a modest G2/M enrichment, another hallmark of cells undergoing mitotic cell death (Deeraksa et al., 2013) (Figures 6C and 6D). Cytometric analysis of senescence and apoptosis/necrosis markers identified a modest increase in apoptosis in TERT promoter mutant β1L-reduced clones, thereby implicating non-apoptotic mitotic cell death, with modest contributions from canonical apoptosis, as the primary driver of cell death in these lines (Figure S7D). Therefore, TERT promoter mutation-dependent telomere dysfunction induced by reducing the function of the GABP tetramer-forming isoform β1L and reducing TERT expression culminates in a loss of replicative immortality characterized by a profound loss of cell viability primarily driven by a mitotic cell death mechanism.
Reducing β1L function impairs tumor growth and extends mouse survival in vivo
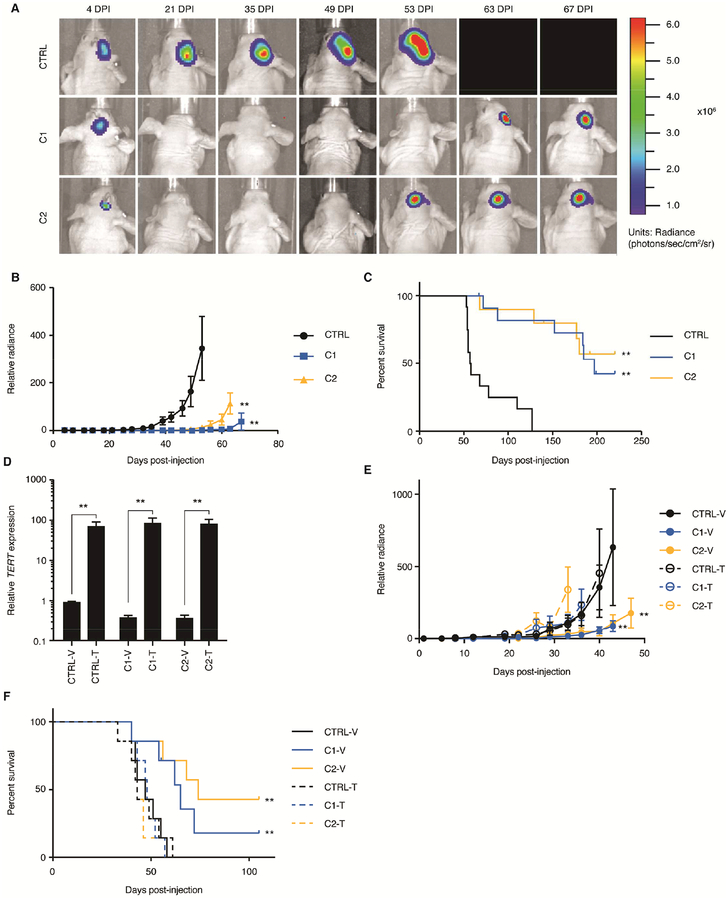
In order to determine the effects of β1L disruption in a TERT promoter mutant setting in vivo, we orthotopically injected CRISPR control or β1L-reduced LN229 cells expressing luciferase into nude mice and monitored tumor engraftment and growth via bioluminescence imaging (BLI). A proportion of the mice injected with β1L-reduced tumor cells did not show evidence of tumor formation over the time course, and those that did form tumors showed significantly decreased tumor growth when compared to mice injected with control cells (Figures 7A and 7B). Importantly, mice injected with the control lines had a significantly shorter median survival compared to mice bearing the β1L-reduced lines (Figure 7C). All mice were validated for tumor burden post-mortem via visual inspection. Despite LN229 C1 and C2 having an attenuated growth arrest phenotype compared to the other lines (Figure 4A), β1L disruption and reduced TERT expression in these lines were sufficient to significantly inhibit tumor formation and growth and extend survival in mice injected with them. Furthermore, lentiviral transduction of LN229 C1 and C2 with a TERT expression vector was sufficient to rescue both the tumor growth and survival phenotypes in an independent cohort (Figures 7D–F). In conclusion, inhibition of the mutant TERT promoter through disrupting β1L function is sufficient to prolong survival in mice bearing LN229 GBM xenografts.
Figure 7.
Reduction of β1L impairs tumor growth and extends mouse survival in vivo. (A) Representative IVIS bioluminescent images of CTRL or β1L-reduced LN229-derived tumors at 7 time points post-intracranial injection (injected on cellular day 51 post-editing). DPI = days post-injection. (B) Relative tumor bioluminescence quantified twice per week for each group (CTRL: n=12, C1: n=12, C2: n=10) until first recorded mortality. **p value<0.01, two-sided Student’s t-test compared to CTRL peak luminescence. Values are mean ± S.D of all mice in each group. (C) Kaplan-Meier survival curve displaying disease-specific survival of mice (Simonsen Labs, see STAR Methods) injected with LN229 CTRL or C1 and C2 β1L-reduced cells over time. **p value<0.01, log-rank test compared to CTRL. (D) TERT expression 4 days post-transduction of CTRL or β1L-reduced LN229 clones (41 days post-editing) with either a control (V) or TERT (T) lentiviral expression vector. **p value<0.01, two-sided Student’s t-test relative to respective vector (V) control. Values are mean ± S.D of three independent experiments. (E) Relative tumor bioluminescence quantified twice per week for each group (n=7 mice per group) following stable transduction with a control (V) or TERT (T) lentiviral expression vector. **p value<0.01, two-sided Student’s t-test compared to vector control peak luminescence for each respective line. Values are mean ± S.D of all mice in each group. (F) Kaplan-Meier survival curve displaying disease-specific survival of mice (Envigo, see STAR Methods) injected with LN229 CTRL or C1 and C2 β1L-reduced cells following stable transduction with a control (V) or TERT (T) lentiviral expression vector. **p value<0.01, log-rank test compared to CTRL.
Discussion:
Telomerase reactivation occurs in more than 90% of human cancers and is fundamental for tumor cell immortalization. While the occurrence of TERT promoter mutations early in GBM evolution suggests they are important for tumorigenesis, their role in maintaining telomere length, replicative immortality, and cell viability at later time points has been relatively unexplored. We have identified the tetramer-forming β1L isoform of GABP to be a necessary component for full activation of the mutant TERT promoter and replicative immortality in TERT promoter mutant, but not wild-type, GBM cells. These results add to recent studies showing that TERT promoter mutations are necessary but not sufficient for cellular immortalization in TERT promoter mutant tumor cells (Chiba et al., 2017; Li et al., 2015). Our results also suggest binding of the β1L-containing GABP tetramer to the mutant TERT promoter is necessary to maintain maximal expression of TERT.
Telomere shortening and loss of cellular proliferation has been previously observed in brain tumor cultures after sustained inhibition of telomerase (Barszczyk et al., 2014; Castelo-Branco et al., 2011; Marian et al., 2010). One difference with these studies and ours is that in addition to potently reducing the expression of TERT, our β1L-reduced clones had concomitant deregulation of a subset of GABP-regulated genes that may influence the observed TERT-dependent phenotypes. Although overexpression of exogenous TERT rescued cell growth of the cells with reduced β1L function, expression of TERT at more physiologic levels through activation of the endogenous wild-type TERT allele may allow for more precise analysis of phenotypes. Thus, we cannot fully rule out that other β1L target genes may contribute to the in vitro and in vivo phenotypes we observed.
The growth decrease occurring as early as 48 hr after LNA-ASO-mediated knockdown of β1L raises the possibility that, in addition to the gradual and protracted loss of viability, β1L and TERT reduction also could have immediate effects. As telomere length is heterogeneous within tumor cell cultures (der-Sarkissian et al., 2004; Wang et al., 2013), cells with shorter telomeres may be more vulnerable upon reduction in TERT expression. Conversely, we expect that the subset of GBM cells with longer telomeres – and not those with critically short telomeres – would preferentially survive through the cell expansion required to establish the clonal cultures of β1L-reduced cells, and then succumb to gradual decreases in telomere length at later time points. Overall this ongoing process could contribute to the gradual loss of viability detected in the bulk population assays. The more immediate effect in our LNA-ASO cell experiments is consistent with an acute telomere-mediated cell death phenotype in NRAS-mutant melanoma due to dependence on TERT expression from the mutant promoter (Reyes-Uribe et al., 2018). However, due to the limitations of our CRISPR-Cas9 experimental design and focus on later time points, further studies to investigate the mechanism of immediate cellular effects following reduction - or elimination - of β1L function in TERT promoter mutant GBM will require inducible systems and single-cell analysis.
β1L tetramerization activity and TERT expression were reduced but not eliminated in our experiments. Attempts to further suppress TERT mRNA expression in the β1L-reduced clones through LNA-ASO-mediated knockdown of β1L had no effect. Therefore, a low level of expression of TERT from the mutant promoter may be maintained independent of β1L function. Although our data strongly support β1L as the main driver of TERT expression from the mutant promoter and the primary factor enabling cell immortality in TERT promoter mutant GBM, they also support the existence of a secondary mechanism contributing to the overall TERT expression level in TERT promoter mutant tumor cells. Secondary mechanisms could involve an activating structural change in the mutant TERT promoter G-quadruplex or activation through recruitment of other ETS factors (Chaires et al., 2014; Li et al., 2015; Lim et al., 2010; Makowski et al., 2016). Additionally, the GABP tetramer-forming isoform β2 may be able to partially activate TERT expression at the mutant TERT promoter. β2 knockdown significantly reduced TERT expression levels in a subset of TERT promoter mutant GBM lines. However, the absence of a positive correlation between GABPB2 and TERT expression levels in glioma tissue samples and the near total loss of the occupancy of GABP at the mutant TERT promoter after disruption of β1L suggest that β2 plays a more minor role, at least when β1L is present. We cannot however exclude the possibility that β2 plays a role in regulating the mutant TERT promoter in a small subset of cells. Therefore, to fully eliminate TERT expression in TERT promoter GBM, it may be necessary to jointly inhibit β1L alongside one or more secondary mechanisms of TERT expression.
Overall, the present study gives credence to β1L as a potential therapeutic target for tumor cells with the mutant TERT promoter. GABP is recruited to the mutant TERT promoter in multiple cancer types (Akincilar et al., 2016; Bell et al., 2015; Stern et al., 2015). The prevalence of identical TERT promoter mutations across a large number of cancer types (Bell et al., 2016; Zehir et al., 2017) highlights the potentially widespread role of the β1L-containing GABP tetramer as a dominant factor responsible for enabling replicative immortality in cancer. This is particularly relevant as direct telomerase inhibitors block tumor cell immortality, but can also affect TERT in normal stem and germ cells (Jager and Walter, 2016; Shay and Wright, 2006). Although GABP is a transcription factor, it is an intriguing target due to its dual function as a dimer and tetramer. β1L is not required for normal development in mice, and in GBM cells the majority of GABP target genes do not seem to be as sensitive to reduction of β1L compared to the mutant TERT promoter. Thus, inhibiting the dispensable tetramer-forming β1L isoform while leaving the dimer and other cell-essential GABP isoforms unperturbed could be a viable strategy to block cellular immortality in TERT promoter mutant tumors, including glioma.
Star Methods:
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joseph F. Costello (joseph.costello@ucsf.edu).
Experimental model and subject details
Cell lines and primary cell cultures
GBM1 (male), T98G (male), LN229 (female), and LN18 (male) cells were cultured in DMEM/Ham’s F-12 1:1 media, 10% FBS, 1% Penicillin/Streptomycin. The GBM1 primary culture was previously described in Bell et al. 2015 (Bell et al., 2015). HEK293T (female) and NHAPC5 (male) cells were cultured in DMEM H-21 media, supplemented with 10% FBS, 1% Non-Essential Amino Acids, 1% Glutamine and 1% Penicillin/Streptomycin. The NHAPC5 culture was previously described in Ohba et al. 2016 (Ohba et al., 2016). HCT116 cells (male) were cultured in McCoy’s 5A media supplemented with 10% FBS and 1% Penicillin/Streptomycin.
SF7996 (male; passage 6), SF8249 (male; passage 4), SF8279 (male; passage 4), SF9030 (male; passage 3), and SF11411 (female; passage 4) are TERT promoter-mutant, IDH1-wild-type patient-derived early passage glioma neurosphere (GNS) GBM cultures and were previous described in Fouse et al. 2014 (Fouse et al., 2014). SF7996 (GNS) and GBM1 (serum) are derived from the same piece of tumor tissue from one patient and differ only in derivation conditions. SF10417 (male; passage 9) is a TERT promoter-mutant, IDH1-mutant patient-derived early passage recurrent high-grade GNS oligodendroglioma culture. hNPCs (male) are human Neural Precursor Cells derived from human induced pluripotent stem cells as previous described (Xu et al., 2016). All GNS cells and hNPCs were cultured in Neurocult NS-A (Stem Cell Technologies) supplemented with 2 mM L-Glutamine, 1% Penicillin/Streptomycin, B-27 without vitamin A (Invitrogen), N2 supplement, 20 ng/mL EGF, and 20 ng/mL bFGF, and 1% sodium pyruvate. SF10417 was additionally supplemented with 20 ng/mL PDGF-AA. hNPCs were additionally supplemented with 5 ng/mL heparin. Cells were grown on 1.6 ug/cm2 laminin-coated flasks and dissociated with StemPro Accutase (Gibco). All cells were maintained at 37° Celsius, 5% CO2. LN229, T98G, HEK293T, LN18 and HCT116 were acquired from ATCC through the UCSF Cell Culture Facility and validated for cell identity via STR testing. The GBM1, SF7996, SF8249, SF8279, SF9030, SF11411, and SF10417 cells are patient-derived cultures validated to be tumor by exome-seq and/or RNA-seq. hNPCs (Xu et al., 2016) were a generous gift from Haoqian Xu and Michael Oldham at University of California, San Francisco. All cells tested negative for mycoplasma contamination.
Animals
Mice and Animal Housing
Athymic (nu/nu) female mice at 5 weeks of age were purchased from Simonson Laboratories (Figures 7A–C) and Harlan Laboratories (Figures 7D and E). Five mice were grouped per cage. Humane endpoints for sacrifice were established as >15% body weight loss from last weighing and/or the presence of gross neurological symptoms such as hunching, asocial behavior, or spastic behavior. All protocols regarding animal studies were approved by the UCSF Institutional Animal Care and Use Committee (IACUC; protocol AN111064-03B) for Dr. Theodore Nicolaides at the University of California, San Francisco.
Orthotopic xenografting and in vivo imaging
144 hr prior to orthotopic xenografting, LN229 control and β1L-reduced lines were stably transduced with Firefly Luciferase Lentifect™ Purified Lentiviral Particles catalog # LPPFLUC-Lv105 (Genecopoiea) with MOI=5. Separately, 240 hr prior to orthotopic xenografting, LN229 control and β1L-reduced lines were stably transduced with either EF1a-TERT-RFP-Bsd catalog # LV1131-RB (GenTarget) or EF1a-empty-RFP-Bsd catalog # LVP-427 lentiviral particles with MOI=0.5. Transduced cells were selected in 5 μg/mL blasticidin (Sigma-Aldrich) for 72 hr, validated for TERT and RFP expression via RT-qPCR and fluorescent imaging, respectively, and stably transduced with Firefly Luciferase Lentifect™ Purified Lentiviral Particles catalog # LPP-FLUC-Lv105 (Genecopoiea) with MOI=5. All cells were verified for stable luciferase expression prior to injection. 30,000 LN229 CRISPR control or β1L-reduced cells 51 days post-editing per mouse (CTRL=12 mice; C1=12 mice; C2=10 mice) or 50,000 LN229 stably transduced TERT (T) or empty vector (V) CRISPR control or β1L-reduced cells (7 mice per group) were injected into the frontal cortex. Animal’s body weight was measured 3 times per week, tumor size via bioluminescent imaging (BLI) on a Xenogen IVIS Spectrum Imaging System was evaluated 2 times per week, and general behavior and symptomatology was evaluated daily. All BLI images were taken with small binning and a normalized exposure of 30 s recorded 12 min after intraperitoneal injection of 5 μL/g of 30 mg/mL D-Luciferin catalog # LUCK-100 (GoldBio).
Method details
TCGA expression data set
The collection of the data from The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Research, 2008) was compliant with all applicable laws, regulations, and policies for the protection of human subjects, and necessary ethical approvals were obtained. Analysis of all data analysis was done in R project version 3.3.2 (http://www.r-project.org/). RSEM normalized RNA-seq expression data for GABP isoforms (GABPA: uc002yly; GABPB1S: uc001zyc, uc001zyd, uc001zye, uc001zyf; GABPB1L: uc001zya, uc001zyb; GABPB2: uc001ewr, uc001ews, uc001ewt) and TERT were downloaded along with clinical information from TCGA (level 3 normalized data, December 2015, http://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm) for 143 GBM (109 TERT-expressing and 34 TERT-non-expressing) samples, 49 oligodendroglioma (49 TERT promoter-mutant samples), and 249 colorectal cancer (249 TERT-expressing) samples. TERT mutation status was obtained, when available, from Ceccarelli et al for the glioma samples (Ceccarelli et al., 2016). GABP isoforms were analyzed for monotonic associations with TERT using Spearman’s correlation. H0: Spearman’s Rho=0; H1: Spearman Rho≠0; α=0.05. A linear trend-line was generated using the PCA orthogonal regression line.
Transcriptome sequencing and analysis
Total cellular RNA was isolated from GBM1, T98G, and LN229 CRISPR control and β1L-reduced clones 45 days post-editing via standard TRIzol protocol (ThermoFisher). Prior to library synthesis, RNA was treated with DNase (Roche), scored on an Agilent 2100 Bioanalyzer for quality control, and quantified on a Qubit® Fluoremeter using the Qubit RNA HS Assay kit (ThermoFisher). Only the samples with RIN >7 were used for RNA-seq. RNA-seq libraries were prepared with the KAPA Stranded mRNA-Seq kit for Illumina platforms (KAPA Biosystems) according to manufacturer’s instructions. Briefly, 1 μg RNA was used for mRNA capture. After fragmentation, first strand synthesis, second strand synthesis, and A-tailing, Illumina adaptors with dual indexes were ligated. The libraries were amplified 11 cycles before pooling with 8–10 samples/lane for sequencing. All libraries were sequenced at the UCSF Center for Advanced Technology on an Illumina HiSeq4000 sequencer with paired-end reads and an average read length of 50 base pairs.
Adapter and polyA sequences were removed from reads using cutadapt v1.8.1, with the minimum overlap between adapter and the 3′ of the read set to 1 nt. Reads shorter than 20 nts after adapter trimming were discarded. Reads were aligned with TopHat (v2.0.14) using a GENCODE V19 transcriptome-guided alignment with parameters −r 200 −library-type fr-firststrand, --prefilter-multihits genome. To estimate transcript abundance, aligned data was processed with FeatureCounts (v1.4.6) with parameters -s 2 -B -p -O -T 24 using a GENCODE V19 GTF reference.
EdgeR was used to determine differential expression between the six β1L-reduced clones and three CRISPR control clones from TERT promoter mutant lines. All three CRISPR control clones were used as a reference (“REF”) in comparison to the six β1L-reduced clones (“TEST”). Genes with <1cpm/3 samples were discarded from the analysis prior to library size calculation. The Beyer-Hardwick Method was used to determine genes significantly altered between the “REF” and “TEST” with FDR<0.05. Non-directional GO-TermFinder was used to determine GO-enriched processes for differentially expressed genes. GABPA-bound genes were determined from ENCODE GABPA ChIP-seq data for all available cancer cell lines (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegTfbsClustered/wg EncodeRegTfbsClusteredV3.bed.gz). BEDOPS closest-features was used to determine transcription start sites within 3 kb of called GABPA peaks presented in ≥2 samples. These transcription start sites are referred to “GABP-bound genes” throughout the text.
siRNA and LNA-ASO knockdown
Non-targeting, GABPB1, and GABPB2-directed siRNA pools were obtained from Dharmacon. Scrambled control and GABPB1L 3’ UTR-directed Locked Nucleic Acid Antisense Oligonucleotides (LNA-ASOs) were obtained from Exiqon. 100 μL of cells were seeded at a density of 30,000 cells/mL in a 96-well plate and transfected 24 hr after with a final concentration of 50 nM siRNA or 25 nM LNA-ASO and 0.1 uL of Dharmafect 1 reagent (Dharmacon). At 48 and 72 hr post-transfection, cells were lysed and cDNA was generated using the POWER SYBR Green Cells-to-Ct kit (Ambion). Quantitative PCR was performed to measure the expression levels of GUSB, TERT, GABPB1L, and GABPB2 as described below. All siRNAs and LNA-ASOs were independently validated at 48 and 72 hr post-transfection for >50% knockdown of target transcript in all cell lines.
RT-qPCR
Quantitative PCR was performed with POWER SYBR Green Complete Master Mix (LifeTechnologies) to measure the expression levels of GUSB (forward primer: CTCATTTGGAATTTTGCCGATT; reverse primer: CCGAGTGAAGATCCCCTTTTTA), TERT (forward primer: TCACGGAGACCACGTTTCAAA; reverse primer: TTCAAGTGCTGTCTGATTCCAAT), GABPB1 (forward primer: TCCACTTCATCTAGCAGCACA; reverse primer: GTAATGGTGTTCGGTCCACTT), GABPB1L (forward primer: ATTGAAAACCGGGTGGAATC; reverse primer: CTGTAGGCCTCTGCTTCCTG), GABPB2 (forward primer: CGCCACCATCGAGATGTCG; reverse primer: TCCAGAGCTATGTCAAAGGCT), SKP2 (forward primer: ATGCCCCAATCTTGTCCATCT; reverse primer: CACCGACTGAGTGATAGGTGT), COXIV (forward primer: CAGGGTATTTAGCCTAGTTGGC; reverse primer: GCCGATCCATATAAGCTGGGA), EIF6 (forward primer: CCGACCAGGTGCTAGTAGGAA; reverse primer: CAGAAGGCACACCAGTCATTC), TFB1M (forward primer: GTTGCCCACGATTCGAGAAAT; reverse primer: GCCCACTTCGTAAACATAAGCAT), and RPS16 (forward primer: TCGGACGCAAGAAGACAGC; reverse primer: AGCAGCTTGTACTGTAGCGTG). Each sample was measured in triplicate on the Applied Biosystems 7900HT Fast Real-Time System. Melting curves were manually inspected to confirm PCR specificity. Relative expression levels were calculated by the deltaCT method against GUSB.
CRISPR-Cas9 editing
Plasmids encoding spCas9 and sgRNAs were obtained from Addgene (Plasmids #41815 and #47108). Oligonucleotides for construction of sgRNAs were cloned into the sgRNA plasmid as previously described (Brown et al., 2016). Target sequences for sgRNAs are provided in Table S1. Targeting vectors PuroR TV and HygroR TV were acquired and incorporated at target loci as previously described (Gapinske et al., 2018). In brief, LN229, NHAPC5, HEK293T, HCT116, and T98G cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions in 24 well plates. GBM1 cells were transfected by electroporation using a Gene Pulser XCell (BioRad) in PBS at 140 Volts, 950 μF. Each cell line was transfected with equal amounts of Cas9, target sgRNA, targeting vector PuroR TV (GBM1, LN229, HCT116, HEK293T, and T98G) or HygroR TV (NHAPC5) and universal sgRNA. Cleaving of the targeting vector by the universal sgRNA-directed Cas9 allowed for integration of the PuroR or HygroR cassette at the control or GABPB1L target loci. Integration only occurs post-cutting of both the targeting vector and target genomic locus. Clonal populations were selected with Puromycin (0.5 μg/ml HCT116 and T98G, 1 μg/ml GBM1 and LN229, and 2 μg/ml HEK293T) or Hygromycin (0.5 μg/ml for NHAPC5).
Analysis of on-target and off-target editing
Analysis of on-target and off-target mutations was conducted as previously described (Gapinske et al., 2018). In brief, genomic DNA from each clone was isolated using the Animal Genomic DNA Purification Mini Kit (Earthox Life Sciences). PCRs to detect integration of the targeting vector at on-target or off-target sites were performed using KAPA2G Robust PCR kits (Kapa Biosystems) according to the manufacturer’s instructions. The DNA sequences of the primers for each target are provided in Table S1. PCR products were visualized in 2% agarose gels and images were captured using a ChemiDoc-It2 (UVP). Indels at off-target sites were analyzed with the Surveyor Mutation Detection kit (IDT) by first amplifying the target locus using PCR with KAPA Robust2G DNA polymerase. The resulting PCR products were melted and reannealed according to manufacturer’s instructions, and 18 μL of the reannealed duplex was mixed with 1μL of Surveyor Nuclease and 1 μL of Enhancer Solution and incubated at 42° Celsius for 1 hr. Final product was loaded onto a 10% TBE polyacrylamide gel and run at 200 V for 30 min. The gels were stained with ethidium bromide and visualized using a ChemiDoc-It2 (UVP). On-target editing of GABPB1L (Figure S2A) or control locus (Figure S3B) was evaluated by PCR to detect the integration of the targeting vector. DNA sequencing of the alleles without integration was used to detect indels (Figure S2B). Analysis of off-target mutations was performed by testing integration of the targeting vector at predicted off-target sites (Hsu et al., 2013) in coding regions for each sgRNA used in each cell line (Figures S3A and S3D-F). For predicted off-target sites within coding sequences we performed Surveyor assays to detect indels (Figure S3C).
Immunoblotting
Immunoblotting for Cyclophilin B (loading control) and β1 (β1S and β1L) was performed using a rabbit anti-Cyclophilin B antibody PA1-027A (Pierce antibodies; 1:1,000 dilution) and rabbit anti-GABPβ1 antibody 12597-1-AP (Proteintech; 1:500 dilution) using the NuPAGE system (Thermofisher), according to the provider’s instructions. Detection of primary bands was done using the Li-Cor goat anti-rabbit 680RD secondary antibody (1:15,000 dilution) on the Li-Cor Odyssey Fc imaging system.
NanoBiT protein-protein interaction assay
Full-length GABPB1L or GABPB1S was cloned into either the pBiT1.1-C [TK/LgBiT] or pBiT2.1-C [TK/LgBiT] vectors (Promega; N196A and N197A, respectively) using In-Fusion HD Cloning (Takara). In accordance with the manufacturer’s instructions, the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent) was used to introduce three separate deletions (DEL1-3) into the pBiT1.1-C-GABPB1L vector (see Table S3 for mutagenesis primers). Mutagenized plasmids were validated using Sanger sequencing and purified for use in the NanoBiT assay. Prior to use, 1 volume NanoBiT vector was diluted into 3 volumes of pCMV6-Neo control vector (OriGene) to a final volume of 10 ng/μL. 100 μL of LN229 or NHAPC5 cells were seeded at a density of 30,000 cells/mL in 96-well plates 24 hr prior to transfection. Cells were transfected with a total of 100 ng of plasmid DNA and 0.3 μL X-tremeGENE HP DNA Transfection Reagent (Roche) according to manufacturer’s instructions. The following combinations were used to assay β1L tetramer formation in LN229 and NHAPC5 cells:
POS: pBiT1.1-C-GABPB1L-WT + pBiT-2.1-C-GABPB1L
NEG: pBiT1.1-C-GABPB1L-WT + pBiT-2.1-C-GABPB1S
DEL1: pBiT1.1-C-GABPB1L-DEL1 + pBiT-2.1-C-GABPB1L
DEL2: pBiT1.1-C-GABPB1L-DEL2 + pBiT-2.1-C-GABPB1L
DEL3: pBiT1.1-C-GABPB1L-DEL3 + pBiT-2.1-C-GABPB1L
24 hr following transfection, Nano-Glo® Live Cell Substrate diluted in Nano-Glo® LCS Dilution Buffer (Promega; N205A and N206A, respectively) was added directly to the cells and luminescence was assayed 1 hr later on a GloMax® 96 MicroPlate Luminometer (Promega) according to manufacturer’s instructions. All data were normalized to the positive control (POS) for each cell line.
Cell proliferation and viability assays
100 μL of cells were seeded at a density of 5,000 cells/mL in 96-well plates. At t=0, 48 and 96 hr post-seeding, MTS (Cell titer 96 aqueous MTS, Promega) was incubated for 2 hr at 37° Celsius in a ratio of 1:5 in media, according to manufacturer’s instructions. Plate was read on the Bioplate Synergy 2 microplate reader at 490 nm. Cell proliferation of individual samples was calculated by normalizing absorbance to their corresponding absorbance at t=24 hr. Each time point was analyzed in triplicates. For cell viability, cells were trypsinized, collected and counted on a hemocytometer with trypan-blue exclusion approximately every 7 days from day 33 to day 102 post-editing, or until the minimal sensitivity limits of the assay were reached. Between viability time points, cells were split prior to confluency and replated at 1/8th density to ensure consistent growth conditions. The ratio between viable and dead cells was used to determine cell viability. It is important to note that trypsinization of cells undergoing telomere dysfunction may have influenced to the viability phenotype in the GBM1 and T98G clones after day 85 post-editing.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) for GABPα was performed using the ActiveMotif High Sensitivity kit. In brief, GBM1, T98G, HCT116, and HEK293T CRISPR controls and β1L-reduced clones were grown to 80% confluency in 15 cm plates and fixed with 4% formaldehyde. Chromatin was sonicated to a size range of 200–1200 bp by the Diagenode Biorupter. 12–18 μg of chromatin was used per GABPα (Santa Cruz Biotechnology: sc-22810) and IgG control (Cell Signaling: 2729) immunoprecipitation for each cell type. Enrichment at the TERT promoter was determined by qPCR with the ssoAdvanced Universal SYBR Green Supermix (Biorad) supplemented with Resolution Solution from GC-RICH PCR System (Roche). The following primer set was used for qPCR: TERT+47 (forward: 5’-GCCGGGGCCAGGGCTTCCCA-3’; reverse: 5’ CCGCGCTTCCCACGTGGCGG-3’; Tm=74° Celsius). PCR was carried out on the Applied Biosystems 7900HT Fast Real-Time System. Three replicate PCR reactions were carried out for each sample.
Telomere length measurement
All telomere length measurements were conducts using the telomere qPCR protocol initially described in Cawthon 2002 (Cawthon, 2002) and later modified in Lin et al. 2009 (Lin et al., 2010). DNA was collected from CRISPR control and β1L-reduced cell lines at days 33, 44, 61, 78, and 83 post-CRISPR-Cas9 editing using Phenol:Chloroform:Isoamyl Alcohol (Invitrogen) according to manufacturer’s instructions. DNA was diluted to a final concentration of 2 ng/μL prior to analysis. Telomere length was measured by qPCR with POWER SYBR Green master mix on the Applied Biosystems 7900HT Fast Real-Time System using the following telomere (TEL) and single gene control (SGC) primer sets: TEL-qPCR, primer forward: CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT, primer reverse: GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT; SGC-qPCR, primer forward: CAGCAAGTGGGAAGGTGTAATCC primer reverse: CCCATTCTATCATCAACGGGGTACAA (Cawthon, 2002; Lin et al., 2010; Xie et al., 2015). The following PCR conditions were used: 95° Celsius for 10 min followed by 40 cycles of data collection at 95° Celsius for 15 s, 60° Celsius anneal for 30 s and 72° Celsius extend for 30 s along with 80 cycles of melting curve from 60° Celsius to 95° Celsius. Relative telomere length was determined as the linear relationship between TEL and SGC (T/S). Three independent RT-qPCR reactions were carried out for each sample, with each independent experiment performed on distinct days with distinct populations of cells.
Exogenous β1L and TERT overexpression
GABPB1L human cDNA (OriGene) was cloned into pCMV6-Neo Vector (OriGene) using the Cold Fusion Cloning Kit (System Biosciences) according to manufacturer’s instructions. The pCMV6-Neo-GABPB1L plasmids obtained were validated by Sanger sequencing using the manufacturer’s primers. 2 μg pCMV6-Neo (empty vector, for control purposes), pCMV6-Neo-B1L or pCI-Neo-hEST2 (Addgene) were transfected into each GBM1, T98G, and LN229 CRISPR control clone (CTRL) or β1L-reduced clone (C1 and C2) using 6 μL X-tremeGENE HP DNA Transfection Reagent (Roche) according to producer’s instructions at day 61 (GBM1 and T98G) or day 58 (LN229) post-editing. C1/C2 and β1L/TERT refers to the clone number and cDNA transfected, respectively. Overexpression of exogenous β1L and TERT mRNA was confirmed by RT-qPCR as described above. Clones were maintained in 100 μg/mL G418 (Invivogen) and validated for continued GABPB1L and TERT expression three weeks post-transfection. Lentiviral TERT rescue is described above under the “Orthotopic xenografting and in vivo bioluminescent imaging” subheading. pCI neo-hEST2 was a gift from Robert Weinberg (Meyerson et al., 1997) (Addgene plasmid # 1781).
Fluorescent imaging and quantification
CTRL and β1L-reduced clones were seeded at a density of 25,000 cells/mL on day 70 post-editing. Cells were fixed in 4% formaldehyde and permeabilized in 100% methanol before co-staining with DAPI and anti-γH2AX AF647 conjugated antibody (EMD Millipore 05-636-AF647) at 4° Celsius overnight. All images were taken at 63× magnification on an AxioImager M1 upright fluorescent microscope (Zeiss) with 2.8 ms exposure. Post-processing and signal normalization of images was done using the on-board ZEN2 software. Quantification of extent of chromatin bridge formation and giant cell micronucleation was performed as follows: each slide was assigned a randomized number to blind the quantifier prior to counting. Ten computationally randomized unique 40× fields of view with a cell number of n>20 were used per slide. For each field of view, total cell number, number of chromatin bridges, and number of giant micronucleated cells were counted. Only nuclei completely in the field of view were counted. A chromatin bridge was defined as a solid strand of nuclear material linking two distinctly independent nuclei. Two nuclei linked by a chromatin bridge were counted as one cell. A giant micronucleated cell was defined as a single cell containing n≥5 uncondensed nuclei. The weighted proportion of chromatin bridges and giant micronucleated cells was determined per field of view and summed into an aggregate proportion. All methods and quantifications were verified using the same parameters as described above by an independent party. Quantification of γH2AX was performed similarly to chromatin bridge and giant cell micronucleation counting with the following differences: n>10 cells per field of view was used as a threshold and individual visible γH2AX foci were counted per cell per field of view. This procedure was likewise followed to quantify LN229 clones at day 45 and day 61 post-editing (n=4 fields of view).
Flow cytometry
On day 75 post-editing, 300,000 cells/line were stained with a combination of Hoechst® 33342 (Thermofisher; 10 ng/mL), AnnexinV-PE (BD Biosciences #51-65875X; 1:1,000 dilution), and C-12-FDG (Setareh Biotech; 33 μM final concentration) for 45 min at 37° Celsius in the dark. Samples were run for 10,000 counts on a Sony SH800 cytometer and analyzed on FlowJo®. The same gating strategy was used for all experiments. All data were collected ONLY after a stable flow of cells had been established. Then, FSC-A vs. FSC-H gating was used to select for singlets along the positive diagonal. Next, FSC-A vs. SSC-A gating was used to remove all cellular debris (FSC-A/SSC-A low particles). Finally, non-specific antibody/fluorophore uptake was used to gate against dead cells with compromised membranes.
Quantification and statistical analysis
All statistical analysis was done using GraphPad Prism 7. Non-parametric Spearman correlation was used for GABP isoforms versus TERT and telomere length versus viability analysis (α=0.05). Adjusted p values after multiple comparison correction are reported for each correlation. A non-parametric Spearman correlation was chosen due to the failure of a subset of data sets to meet the homoscedasticity assumption of the Pearson test. Mouse survival data for the orthotopic xenograft experiments were analyzed with the Kaplan-Meier Log-Rank Test (α=0.05). The non-parametric Welch’s t-test was used as listed for samples with unequal sample sizes (α=0.05). A two-sided heteroscedastic Student’s t-test was used as listed for all other assays (α=0.05) after confirming differences in variances between tested groups. All error bars shown are mean ± S.D. A sample size of 3 independent experiments (biological replicates) was used for all experiments, unless otherwise noted, in order to ensure appropriate statistical power to detect a statistically significant change of at least two-fold. 3 technical replicates per biological replicate were used for each experiment as noted.
Data and software availability
All data used for GABP isoform and TERT expression correlations are available for public access from the TCGA (level 3 normalized data, December 2015, http://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm). All raw data used for RNA-seq analysis has been deposited in the European Genome Archive (EGA) under ID code EGAS0000100258.2. Scripts used for RNA-seq analysis are available at https://github.com/UCSF-Costello-Lab/Tert-gabp.
Supplementary Material
Significance.
TERT promoter mutations are the third most common mutation in human cancer, and the single most common mutation in glioblastoma. Understanding how the promoter mutation leads to tumor cell immortality could uncover potential targets to undermine immortality and reduce tumor growth. TERT promoter mutations selectively recruit the transcription factor GABP to activate TERT expression across multiple types of cancer. Our results suggest that the normally dispensable β1L isoform of GABP is a key to tumor cell immortality in TERT promoter mutant brain tumors. Inhibiting GABPβ1L may be an approach to reverse tumor cell immortality while sparing TERT promoter wild-type cells.
Highlights.
The β1L tetramer-forming isoform of GABP activates the mutant TERT promoter
β1L disruption induces telomere loss and death only in TERT promoter mutant cells
Disruption of β1L reduces tumor growth and prolongs survival in xenografted mice
GABPβ1L is a potential therapeutic target for TERT promoter mutant glioblastoma
TERT promoter mutations generate a binding site for GABP and reactivate TERT expression. Mancini et al. show that GABPβ1L, among GABP subunits, is specifically required for the function of TERT promoter mutants, reducing GABPβ1L causes telomere loss and cell death exclusively in TERT promoter mutant cells.
Acknowledgements:
This work was supported by a generous gift from the Dabbiere family (J.F.C.), the Hana Jabsheh Research Initiative (J.F.C.), NIH grants NCI P50CA097257 (J.F.C. and J.A.D.), NCI P01CA118816-06 (J.F.C.), T32 GM008568 and T32 CA151022 (A.M.), and NCI R01CA163336 (J.S.S.), and the Sontag Foundation Distinguished Scientist Award (J.S.S.). C.F. is supported by a US National Institutes of Health K99/R00 Pathway to Independence Award (K99GM118909) from the National Institute of General Medical Sciences (NIGMS). Additional support was provided by Fundação para a Ciência e Tecnologia SFRH/BD/88220/2012 (A.X-M.) and IF/00601/2012 (B.M.C.). J.A.D. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Financial Interests: R.J.A.B and J.F.C are co-founders of Telo Therapeutics Inc. and have ownership interest.
References:
- Akincilar S, Khattar E, Boon PL, Unal B, Fullwood MJ, and Tergaonkar V (2016). Long-range chromatin interactions drive mutant Tert promoter activation. Cancer Discov. [DOI] [PubMed] [Google Scholar]
- Arita H, Narita Y, Fukushima S, Tateishi K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP, Kawahara N, et al. (2013). Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta neuropathologica 126, 267–276. [DOI] [PubMed] [Google Scholar]
- Barszczyk M, Buczkowicz P, Castelo-Branco P, Mack SC, Ramaswamy V, Mangerel J, Agnihotri S, Remke M, Golbourn B, Pajovic S, et al. (2014). Telomerase inhibition abolishes the tumorigenicity of pediatric ependymoma tumor-initiating cells. Acta neuropathologica 128, 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RJ, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, Choi S, Hong C, He D, Pekmezci M, et al. (2015). Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348, 1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RJ, Rube HT, Xavier-Magalhaes A, Costa BM, Mancini A, Song JS, and Costello JF (2016). Understanding TERT Promoter Mutations: A Common Path to Immortality. Molecular cancer research : MCR 14, 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW, and Szostak JW (2006). Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med 12, 1133–1138. [DOI] [PubMed] [Google Scholar]
- Brown A, Woods WS, and Perez-Pinera P (2016). Multiplexed Targeted Genome Engineering Using a Universal Nuclease-Assisted Vector Integration System. ACS Synth Biol 5, 582–588. [DOI] [PubMed] [Google Scholar]
- Bryan TM, and Cech TR (1999). Telomerase and the maintenance of chromosome ends. Current opinion in cell biology 11, 318–324. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Li H, Deb S, and Liu JP (2002). TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene 21, 3130–3138. [DOI] [PubMed] [Google Scholar]
- Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, and Baird DM (2007). The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes & development 21, 2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RS, and Avadhani NG (1994). Cooperative binding of GA-binding protein transcription factors to duplicated transcription initiation region repeats of the cytochrome c oxidase subunit IV gene. J Biol Chem 269, 4381–4387. [PubMed] [Google Scholar]
- Castelo-Branco P, Zhang C, Lipman T, Fujitani M, Hansford L, Clarke I, Harley CB, Tressler R, Malkin D, Walker E, et al. (2011). Neural tumor-initiating cells have distinct telomere maintenance and can be safely targeted for telomerase inhibition. Clinical cancer research : an official journal of the American Association for Cancer Research 17, 111–121. [DOI] [PubMed] [Google Scholar]
- Cawthon RM (2002). Telomere measurement by quantitative PCR. Nucleic acids research 30, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al. (2016). Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 164, 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaires JB, Trent JO, Gray RD, Dean WL, Buscaglia R, Thomas SD, and Miller DM (2014). An improved model for the hTERT promoter quadruplex. PLoS One 9, e115580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Johnson JZ, Vogan JM, Wagner T, Boyle JM, and Hockemeyer D (2015). Cancer-associated TERT promoter mutations abrogate telomerase silencing. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Lorbeer FK, Shain AH, McSwiggen DT, Schruf E, Oh A, Ryu J, Darzacq X, Bastian BC, and Hockemeyer D (2017). Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science 357, 1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, and DePinho RA (1999). p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97, 527–538. [DOI] [PubMed] [Google Scholar]
- Chinenov Y, Henzl M, and Martin ME (2000). The alpha and beta subunits of the GA-binding protein form a stable heterodimer in solution. Revised model of heterotetrameric complex assembly. The Journal of biological chemistry 275, 7749–7756. [DOI] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, and Bacchetti S (1992). Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. The EMBO journal 11, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Ellisen LW, Caddle SD, Haber DA, and Weinberg RA (1998). Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene 16, 1217–1222. [DOI] [PubMed] [Google Scholar]
- de la Brousse FC, Birkenmeier EH, King DS, Rowe LB, and McKnight SL (1994). Molecular and genetic characterization of GABP beta. Genes & development 8, 1853–1865. [DOI] [PubMed] [Google Scholar]
- Deeraksa A, Pan J, Sha Y, Liu XD, Eissa NT, Lin SH, and Yu-Lee LY (2013). Plk1 is upregulated in androgen-insensitive prostate cancer cells and its inhibition leads to necroptosis. Oncogene 32, 2973–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- der-Sarkissian H, Bacchetti S, Cazes L, and Londono-Vallejo JA (2004). The shortest telomeres drive karyotype evolution in transformed cells. Oncogene 23, 1221–1228. [DOI] [PubMed] [Google Scholar]
- Donadini A, Giacopelli F, Ravazzolo R, Gandin V, Marchisio PC, and Biffo S (2006). GABP complex regulates transcription of eIF6 (p27BBP), an essential trans-acting factor in ribosome biogenesis. FEBS Lett 580, 1983–1987. [DOI] [PubMed] [Google Scholar]
- Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, and Shippen DE (1999). Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proceedings of the National Academy of Sciences of the United States of America 96, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse SD, Nakamura JL, James CD, Chang S, and Costello JF (2014). Response of primary glioblastoma cells to therapy is patient specific and independent of cancer stem cell phenotype. Neuro-oncology 16, 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkos M, and Beard P (2011). Mitotic catastrophe occurs in the absence of apoptosis in p53-null cells with a defective G1 checkpoint. PLoS One 6, e22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapinske M, Tague N, Winter J, Underhill GH, and Perez-Pinera P (2018). Targeted Gene Knock Out Using Nuclease-Assisted Vector Integration: Hemi- and Homozygous Deletion of JAG1. Methods Mol Biol 1772, 233–248. [DOI] [PubMed] [Google Scholar]
- Genuario RR, and Perry RP (1996). The GA-binding protein can serve as both an activator and repressor of ribosomal protein gene transcription. J Biol Chem 271, 4388–4395. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Feldser DM, and Greider CW (2001). Telomere dysfunction increases mutation rate and genomic instability. Cell 106, 275–286. [DOI] [PubMed] [Google Scholar]
- Hayashi MT, Cesare AJ, Rivera T, and Karlseder J (2015). Cell death during crisis is mediated by mitotic telomere deprotection. Nature 522, 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. (2013). TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, and Garraway LA (2013). Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianzini F, and Mackey MA (1997). Spontaneous premature chromosome condensation and mitotic catastrophe following irradiation of HeLa S3 cells. Int J Radiat Biol 72, 409–421. [DOI] [PubMed] [Google Scholar]
- Iwado E, Daido S, Kondo Y, and Kondo S (2007). Combined effect of 2–5A-linked antisense against telomerase RNA and conventional therapies on human malignant glioma cells in vitro and in vivo. International journal of oncology 31, 1087–1095. [PubMed] [Google Scholar]
- Jager K, and Walter M (2016). Therapeutic Targeting of Telomerase. Genes 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X, Zhao DM, Waldschmidt TJ, and Xue HH (2008). GABPbeta2 is dispensible for normal lymphocyte development but moderately affects B cell responses. J Biol Chem 283, 24326–24333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA Jr., Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. (2013). TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proceedings of the National Academy of Sciences of the United States of America 110, 6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, and Shay JW (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhou QL, Sun W, Chandrasekharan P, Cheng HS, Ying Z, Lakshmanan M, Raju A, Tenen DG, Cheng SY, et al. (2015). Non-canonical NF-kappaB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat Cell Biol 17, 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KW, Lacroix L, Yue DJ, Lim JK, Lim JM, and Phan AT (2010). Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J Am Chem Soc 132, 12331–12342. [DOI] [PubMed] [Google Scholar]
- Lin J, Epel E, Cheon J, Kroenke C, Sinclair E, Bigos M, Wolkowitz O, Mellon S, and Blackburn E (2010). Analyses and comparisons of telomerase activity and telomere length in human T and B cells: insights for epidemiology of telomere maintenance. J Immunol Methods 352, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski MM, Willems E, Fang J, Choi J, Zhang T, Jansen PW, Brown KM, and Vermeulen M (2016). An interaction proteomics survey of transcription factor binding at recurrent TERT promoter mutations. Proteomics 16, 417–426. [DOI] [PubMed] [Google Scholar]
- Marian CO, Cho SK, McEllin BM, Maher EA, Hatanpaa KJ, Madden CJ, Mickey BE, Wright WE, Shay JW, and Bachoo RM (2010). The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clinical cancer research : an official journal of the American Association for Cancer Research 16, 154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, et al. (1997). hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90, 785–795. [DOI] [PubMed] [Google Scholar]
- Ohba S, Mukherjee J, Johannessen TC, Mancini A, Chow TT, Wood M, Jones L, Mazor T, Marshall RE, Viswanath P, et al. (2016). Mutant IDH1 Expression Drives TERT Promoter Reactivation as Part of the Cellular Transformation Process. Cancer Res 76, 6680–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Uribe P, Adrianzen-Ruesta MP, Deng Z, Echevarria-Vargas I, Mender I, Saheb S, Liu Q, Altieri DC, Murphy ME, Shay JW, et al. (2018). Exploiting TERT dependency as a therapeutic strategy for NRAS-mutant melanoma. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosmarin AG, Resendes KK, Yang Z, McMillan JN, and Fleming SL (2004). GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein-protein interactions. Blood Cells Mol Dis 32, 143–154. [DOI] [PubMed] [Google Scholar]
- Saretzki G, Sitte N, Merkel U, Wurm RE, and von Zglinicki T (1999). Telomere shortening triggers a p53-dependent cell cycle arrest via accumulation of G-rich single stranded DNA fragments. Oncogene 18, 5148–5158. [DOI] [PubMed] [Google Scholar]
- Sawada J, Goto M, Sawa C, Watanabe H, and Handa H (1994). Transcriptional activation through the tetrameric complex formation of E4TF1 subunits. EMBO J 13, 1396–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, and Wright WE (2000). Hayflick, his limit, and cellular ageing. Nat Rev Mol Cell Biol 1, 72–76. [DOI] [PubMed] [Google Scholar]
- Shay JW, and Wright WE (2006). Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov 5, 577–584. [DOI] [PubMed] [Google Scholar]
- Spiegl-Kreinecker S, Lotsch D, Ghanim B, Pirker C, Mohr T, Laaber M, Weis S, Olschowski A, Webersinke G, Pichler J, and Berger W (2015). Prognostic quality of activating TERT promoter mutations in glioblastoma: interaction with the rs2853669 polymorphism and patient age at diagnosis. Neuro-oncology 17, 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JL, Theodorescu D, Vogelstein B, Papadopoulos N, and Cech TR (2015). Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes & development 29, 2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakifahmetoglu H, Olsson M, and Zhivotovsky B (2008). Death through a tragedy: mitotic catastrophe. Cell Death Differ 15, 1153–1162. [DOI] [PubMed] [Google Scholar]
- Vinagre J, Almeida A, Populo H, Batista R, Lyra J, Pinto V, Coelho R, Celestino R, Prazeres H, Lima L, et al. (2013). Frequency of TERT promoter mutations in human cancers. Nat Commun 4, 2185. [DOI] [PubMed] [Google Scholar]
- Vitale I, Galluzzi L, Castedo M, and Kroemer G (2011). Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol 12, 385–392. [DOI] [PubMed] [Google Scholar]
- Wang F, Pan X, Kalmbach K, Seth-Smith ML, Ye X, Antumes DM, Yin Y, Liu L, Keefe DL, and Weissman SM (2013). Robust measurement of telomere length in single cells. Proceedings of the National Academy of Sciences of the United States of America 110, E1906–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker NJ, Bryan TM, Bonnefin P, Chang AC, Musgrove EA, Braithwaite AW, and Reddel RR (1995). Involvement of RB-1, p53, p16INK4 and telomerase in immortalisation of human cells. Oncogene 11, 971–976. [PubMed] [Google Scholar]
- Xie Z, Jay KA, Smith DL, Zhang Y, Liu Z, Zheng J, Tian R, Li H, and Blackburn EH (2015). Early telomerase inactivation accelerates aging independently of telomere length. Cell 160, 928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, et al. (2016). Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 22, 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue HH, Jing X, Bollenbacher-Reilley J, Zhao DM, Haring JS, Yang B, Liu C, Bishop GA, Harty JT, and Leonard WJ (2008). Targeting the GA binding protein beta1L isoform does not perturb lymphocyte development and function. Mol Cell Biol 28, 4300–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZF, Drumea K, Mott S, Wang J, and Rosmarin AG (2014). GABP transcription factor (nuclear respiratory factor 2) is required for mitochondrial biogenesis. Mol Cell Biol 34, 3194–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZF, Mott S, and Rosmarin AG (2007). The Ets transcription factor GABP is required for cell-cycle progression. Nature cell biology 9, 339–346. [DOI] [PubMed] [Google Scholar]
- Yu M, Yang XY, Schmidt T, Chinenov Y, Wang R, and Martin ME (1997). GA-binding protein-dependent transcription initiator elements. Effect of helical spacing between polyomavirus enhancer a factor 3(PEA3)/Ets-binding sites on initiator activity. J Biol Chem 272, 29060–29067. [DOI] [PubMed] [Google Scholar]
- Yu S, Jing X, Colgan JD, Zhao DM, and Xue HH (2012). Targeting tetramer-forming GABPbeta isoforms impairs self-renewal of hematopoietic and leukemic stem cells. Cell Stem Cell 11, 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.