By crowdsourcing, via an international competition, algorithms for seizure prediction based on long-term human brain activity recordings, Kuhlmann et al. show that clinically relevant seizure prediction is possible in a wider range of patients than previously thought. The data and algorithms are available on epilepsyecosystem.org to support further development.
Keywords: epilepsy, seizure prediction, intracranial EEG, refractory epilepsy, Open Data Ecosystem for the Neurosciences
Abstract
Accurate seizure prediction will transform epilepsy management by offering warnings to patients or triggering interventions. However, state-of-the-art algorithm design relies on accessing adequate long-term data. Crowd-sourcing ecosystems leverage quality data to enable cost-effective, rapid development of predictive algorithms. A crowd-sourcing ecosystem for seizure prediction is presented involving an international competition, a follow-up held-out data evaluation, and an online platform, Epilepsyecosystem.org, for yielding further improvements in prediction performance. Crowd-sourced algorithms were obtained via the ‘Melbourne-University AES-MathWorks-NIH Seizure Prediction Challenge’ conducted at kaggle.com. Long-term continuous intracranial electroencephalography (iEEG) data (442 days of recordings and 211 lead seizures per patient) from prediction-resistant patients who had the lowest seizure prediction performances from the NeuroVista Seizure Advisory System clinical trial were analysed. Contestants (646 individuals in 478 teams) from around the world developed algorithms to distinguish between 10-min inter-seizure versus pre-seizure data clips. Over 10 000 algorithms were submitted. The top algorithms as determined by using the contest data were evaluated on a much larger held-out dataset. The data and top algorithms are available online for further investigation and development. The top performing contest entry scored 0.81 area under the classification curve. The performance reduced by only 6.7% on held-out data. Many other teams also showed high prediction reproducibility. Pseudo-prospective evaluation demonstrated that many algorithms, when used alone or weighted by circadian information, performed better than the benchmarks, including an average increase in sensitivity of 1.9 times the original clinical trial sensitivity for matched time in warning. These results indicate that clinically-relevant seizure prediction is possible in a wider range of patients than previously thought possible. Moreover, different algorithms performed best for different patients, supporting the use of patient-specific algorithms and long-term monitoring. The crowd-sourcing ecosystem for seizure prediction will enable further worldwide community study of the data to yield greater improvements in prediction performance by way of competition, collaboration and synergism.
Introduction
International team-based medical science and online data and algorithm ecosystems (Wiener et al., 2016) offer a new large-scale alternative to standard collaborative or multicentre studies. Such ecosystems facilitate reproducibility and comparison of findings through standardized procedures (Wiener et al., 2016). For example, analysis of standardized high quality experimental data can yield improvements in predictive algorithms for diagnostics and treatment of patients. The most promising algorithms, or ensembles of multiple algorithms, can be selected from a large pool using standardized evaluation procedures. This paper presents a crowd-sourcing ecosystem for epileptic seizure prediction. The ecosystem arose from an international seizure prediction competition, a follow-up evaluation, and creation of an online platform, epilepsyecosystem.org, for yielding further improvements in seizure prediction performance.
The randomness and uncertainty of seizures significantly affects the safety, anxiety, employability, and overall quality of life of people with epilepsy (Schulze-Bonhage and Kühn, 2008). Seizure prediction offers the possibility to provide warnings to patients so they can move to safety or activate an intervention, such as acute, fast-acting medication or an implantable control device that uses electrical stimulation to avert seizures (Mormann et al., 2007; Stacey and Litt, 2008; Freestone et al., 2015). The field of seizure prediction is largely based on the search for predictive EEG features or preictal biomarkers that can be input into machine learning algorithms. These algorithms can be trained to output the probability of an impending seizure and provide warnings to patients.
Over the past 20 years, there has been a concerted effort to develop reliable seizure prediction algorithms (Mormann et al., 2007; Kuhlmann et al., 2010, 2015; Freestone et al., 2015; Gadhoumi et al., 2016). However, progress was limited by the best data available at the time, which were typically short human intracranial EEG (iEEG) recordings obtained during epilepsy surgery evaluations (Mormann et al., 2007). These recordings are rarely longer than 2 weeks in duration. Therefore, they do not provide sufficient data and numbers of seizures for rigorous statistical testing of seizure prediction algorithms (Snyder et al., 2008). Data collected during inpatient monitoring are also associated with antiepileptic drug dose tapering, which shifts a patient’s brain state away from its typical regime (Duncan et al., 1989).
In 2013, the world’s first-in-human clinical trial of an implantable seizure prediction device, the NeuroVista Seizure Advisory System, demonstrated the feasibility of prospective seizure prediction in long-term iEEG recordings (Cook et al., 2013). Fifteen patients with drug-resistant epilepsy were implanted with the continuous recording device for a period of between 6 months and 3 years, thus overcoming the shortcomings of previous iEEG recordings. Some patients in the trial achieved very high seizure prediction performance, with the best case obtaining seizure sensitivity of 100% for 3% time in warning. However, for some patients, especially those with greater seizure frequency, sensitivity was as low as 17% and time in warning as high as 41%. To encourage new prospective clinical trials for seizure prediction devices, it is necessary to demonstrate that improvements in performance relative to the original trial can be achieved and satisfactory predictive performance is possible for a wider range of patients.
The first attempt to crowd-source prediction algorithms was the 2014 American Epilepsy Society Seizure Prediction Challenge (Brinkmann et al., 2016). The contest involved data from long-term (up to 1 year) dog iEEG recordings acquired with the NeuroVista device and short-term (<14 days) human iEEG recordings. After the contest was completed, a follow-up evaluation of the top algorithms was performed using held-out data (Brinkmann et al., 2016). The held-out evaluation with unseen iEEG data from the same dogs revealed that the top algorithms performed better than random prediction. However, a pseudo-prospective evaluation was not performed. (Here a pseudo-prospective evaluation refers to a special case of retrospective evaluation that mimics real-life application of the algorithms and simulates all aspects of a prospective evaluation with continuous data collected on an ambulatory device but the evaluation focuses on data collected in the past.) Nor were long-term human data available for evaluation. Therefore, the potential clinical insights were limited.
To overcome these shortcomings and seek improvements in retrospective seizure prediction performance for patients in the NeuroVista trial, the Melbourne-University AES-MathWorks-NIH Seizure Prediction Challenge was completed in 2016. Herein, we describe the results of this crowd-sourcing competition, which includes data from three patients with the lowest seizure prediction performance from the NeuroVista trial. A follow-up pseudo-prospective held-out data evaluation highlights the clinical utility of seizure prediction for the most difficult patient cases. We present this study as one application of crowd-sourcing on a unique dataset. More importantly, the data are made available to the global community to advance epilepsy research.
Materials and methods
Subjects and data
IEEG data were recorded chronically from humans with refractory focal epilepsy using the NeuroVista Seizure Advisory System implanted device described previously (Cook et al., 2013) (Human Research Ethics Committee, St. Vincent’s Hospital, Melbourne – approval LRR145/13). Sixteen subdural electrodes (4 × 4-contact strips) were implanted in each patient, targeted to the presumed seizure focus (Fig. 1). The electrode leads were tunnelled to a subclavicularly-placed implanted telemetry unit. A rechargeable battery powered the implanted device. Data were sampled at 400 Hz with signed 16-bit resolution and wirelessly transmitted to an external, hand-held personal advisory device. Recorded iEEG from the 16 electrode contacts were referenced to the group average across all electrode channels and continuously saved to removable flash media. The electrode contacts were made of platinum-iridium and embedded in silicon.
Figure 1.
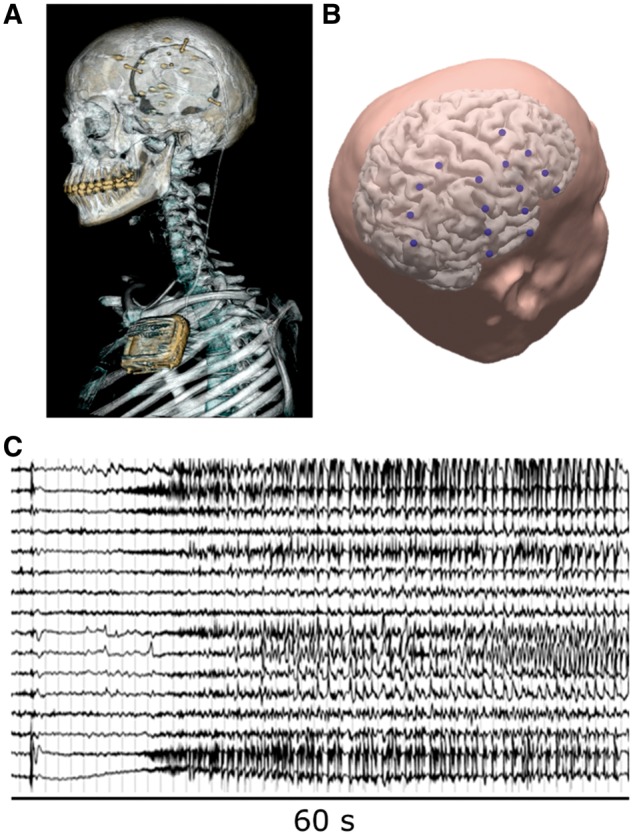
The NeuroVista seizure advisory system. (A) Example CT scan of the NeuroVista seizure advisory system implanted in a patient. (B) Imaging reconstruction showing example locations of electrodes (blue) used by the implant to record iEEG. (C) An example seizure recorded with the 16-channel device.
The three patients with lowest seizure prediction performance in the Cook et al. (2013) trial were selected for the contest. Patient selection was motivated by the opportunity to show the maximal benefit of crowd-sourcing solutions. Furthermore, it was anticipated that a demonstration of accurate forecasting in the difficult patients would show that seizure prediction is viable for a much wider range of patients than initially thought. Many seizures (∼35) per patient were made available, providing the opportunity for machine learning algorithms to be patient-specific. Given the nature of competitions run on kaggle.com, there are limitations on the amount of data participants can download as this can affect participation rates and clog the platform bandwidth. The decision to study only three patients met this constraint while providing an adequate amount of data per patient such that accurate and reliable algorithms could be developed by contestants. Adding more patients to the study would have limited our ability to achieve this.
Patient 1 was a 22-year-old female diagnosed with parieto-temporal focal epilepsy at age 16. She was treated with the antiepileptic drugs carbamazepine, lamotrigine, and phenytoin. She had resective surgery prior to the trial. Patient 2 was a 51-year-old female with occipitoparietal focal epilepsy. Her epilepsy was diagnosed at age 10. At the time of the trial, she received carbamazepine medication. Patient 3 was a 50-year-old female. She was first diagnosed with seizures of frontotemporal origin at age 15. She was receiving levetiracetam, oxcarbazepine, and zonisamide for treatment and had undergone resective surgery prior to the trial.
Only lead seizures, defined as seizures occurring without a preceding seizure for a minimum of 4 h, were used for each patient (see Supplementary material for more details about seizures). The data were divided into labelled training and unlabelled testing sets for the contest. Held-out data were used for follow-up evaluation of the top algorithms after the contest had finished. To mimic practical application and avoid in-sample testing, testing data followed training data and held-out data followed testing data in the recordings. To avoid signal non-stationarities in the immediate period following implantation of electrodes, the contest data came from the period between 1 and ∼7 months after implantation, with the remaining data used as held-out data.
For the contest dataset, preictal data clips were extracted from the 66 min prior to lead seizures in six 10-min data clips (allowing a 5-min gap to ensure preictal data were used). The preictal data clips were separated by 10-s gaps. Interictal clips were selected similarly in groups of six 10-min clips with 10-s spacing beginning from randomly selected times, ensuring a minimum gap of 3 h before and 4 h after any seizure. The mean of the signals was subtracted from the signals. Data clips were stored as ordered structures including sample data, data clip length, iEEG sampling frequency, and channel indices in MATLAB format data files. Training data files also included a sequence number indicating the clip’s sequential position in the series of six 10-min data clips. The temporal relationship between interictal and preictal data was not made available to the contestants. The training and testing data were selected to provide an adequate number of lead seizures for both training and testing.
The held-out dataset consisted of continuous 10-min data clips, in the range of 4 h after the previous seizure to 5 min before a lead seizure, without any overlap. This kept the number of held-out clips to a minimum to make it easier to pseudo-prospectively evaluate the top algorithms within a tractable amount of computation time. Clips between 60 min and 5 min before a lead seizure were considered preictal and all other clips interictal. The timing of the clips relative to seizures was also noted. A summary of the data for each patient and the number of clips in the training, testing, and held-out datasets is shown in Table 1. Note there are much more held-out data than contest data to assess out-of-sample algorithm performance with confidence, and the held-out data were more continuous to facilitate pseudo-prospective evaluation and mimic real-life application of seizure prediction.
Table 1.
Data characteristics for the seizure prediction contest and held-out data experiment
| Patient | Recording duration, days | Seizures | Lead seizures | Training clips (% interictal) | Testing clips (% interictal) | Held-out clips (% interictal) |
|---|---|---|---|---|---|---|
| 1 | 559 | 390 | 231 | 797 (69.0) | 205 (74.1) | 12003 (91.2) |
| 2 | 393 | 204 | 186 | 2027 (89.2) | 994 (94.0) | 22630 (96.8) |
| 3 | 374 | 545 | 216 | 2158 (88.3) | 689 (91.3) | 25079 (95.6) |
Table structure mimics a similar table from the 2014 American Epilepsy Society Seizure Prediction Challenge publication (Brinkmann et al., 2016) to facilitate direct comparison.
The contest and algorithm evaluation
The contest data clips were bundled and made available for download on the contest page at kaggle.com (https://www.kaggle.com/c/melbourne-university-seizure-prediction). The contest ran from 3 September to 2 December 2016. Contestants were permitted to develop algorithms in any computer language and using any features, machine learning, and data processing methods they wished, but classifications were required to come directly from an algorithm; classification by visual review was prohibited. Contestants uploaded preictal probability scores (a floating-point number between 0 and 1 indicating the probability of each clip being preictal) for the testing data clips in a comma separated values file, and immediate feedback on classification accuracy was provided on a real-time public leader board on kaggle.com. Contestants were permitted up to five submissions per day. Public leader board scores were computed on a randomly sampled 30% subset of the test data clips. Official winners were determined by the private leader board score, which is based on the remaining 70% of the testing data. Classification scores were computed by Kaggle as the area under the receiver operating characteristic (ROC) classification curve, created by applying varying threshold values to the probability scores. This score is referred to as area under the curve (AUC) (Fig. 2). Prizes were awarded for first (US$10 000), second (US$6000), and third (US$4000) place finishers as determined by the private leader board scores. Winning teams were required to make their algorithms publicly available under an open source licence. Performance for the held-out data was also scored using AUC.
Figure 2.
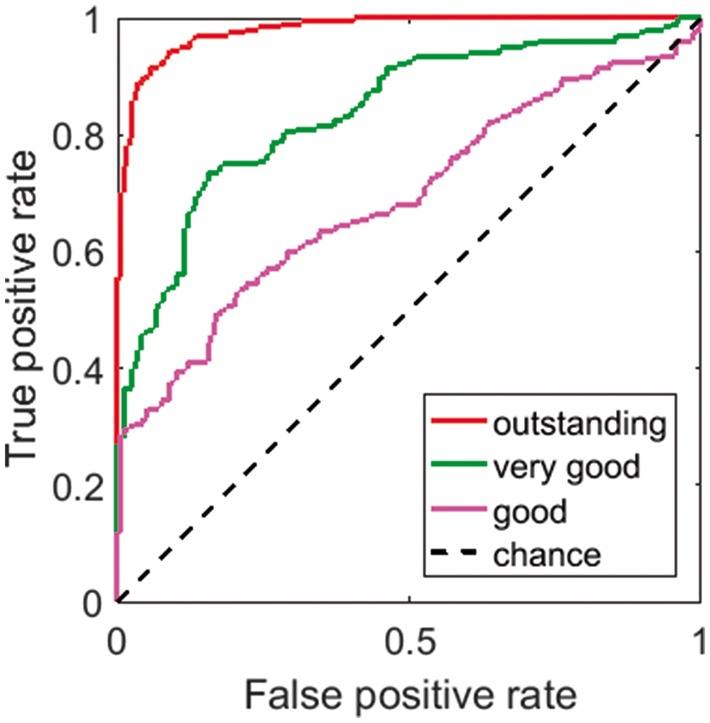
AUC-based evaluation of seizure prediction algorithms. The performance of algorithms can be assessed through ROC curves that plot true positive rates versus false positive rates and computing the AUC to quantify performance. A perfect predictor has an AUC of 1. True positive rates refer to the proportion of preictal data clips correctly classified and false positive rates refer to the proportion of interictal data clips incorrectly classified as preictal. In this paper, each algorithm provides a preictal probability for each data clip and a threshold applied to a given preictal probability is used to determine if the algorithm predicts that the corresponding segment is preictal or interictal. This is determined for all data clips. Different threshold values give rise to different points on the ROC curve. The legend and coloured lines indicate example performance levels of different algorithms.
The AUC analysis of algorithms presented here largely mirrors that used in the 2014 American Epilepsy Society Seizure Prediction Challenge (Brinkmann et al., 2016) in order to facilitate comparison.
In addition to the machine learning oriented AUC-based analysis, a clinically relevant pseudo-prospective evaluation of the algorithms was also considered for the held-out data. In the original trial of the NeuroVista Seizure Advisory System, the device displayed three coloured indicator lights to indicate the likelihood of impending seizures. A red, white and blue light indicated a high, moderate or low seizure likelihood/risk, respectively. A similar approach was applied here. Each algorithm considered here provides a preictal probability for each data clip and two (one upper and one lower) thresholds are applied to this probability. When the probability is above the upper threshold, high-seizure-risk predictions are generated and high-risk (i.e. ‘red light’) warnings are given for a specified warning duration (Snyder et al., 2008). When the probability is below the lower threshold, low-seizure-risk predictions are generated and low-risk (i.e. ‘blue light’) indicators are given for a specified duration. The times of prediction were taken to be the end time of each ‘threshold-crossing’ data clip.
Pseudo-prospective seizure prediction performance was evaluated with the metrics of sensitivity (proportion of seizures correctly predicted, i.e. number of seizures occurring during high-seizure-risk or ‘red-light’ warnings divided by the total number of seizures) and proportion of time in warning (i.e. proportion of time in red-light or, more specifically, the amount of time spent in warning assuming a fixed warning duration generated after each prediction divided by total recording time) (Snyder et al., 2008). The goal in seizure prediction is to maximize sensitivity and to minimize proportion of time in warning. For high-seizure-risk warnings a warning duration (Snyder et al., 2008) of effectively 1 h was used, consistent with the definition of ‘preictal’ in the contest training data (see Supplementary material for more details).
While the aim of assessing high-seizure-risk periods is clearly important to seizure prediction, patients also want to know when they are not going to have seizures. Therefore, as described above, low-seizure-risk prediction (i.e. ‘blue light’ activation) was also assessed. In particular, if patients are told they currently have a low-risk of having a seizure (i.e. the blue light is on), then they will be perturbed if a seizure does occur during this period. Thus, low-seizure-risk prediction accuracy was assessed by the proportion of seizures occurring during low-risk and the proportion of time in low-risk. This effectively characterizes the false-negative rates of algorithms. Time in low-risk was determined assuming a 3 h low-risk period duration, consistent with ‘interictal’ data in the contest training data being at least 3 h before any seizure.
Seven of the top 10 teams, including the three winners, along with the special case of the top performing team for Patient 3 agreed to participate in the held-out data evaluation. The top performing team for Patient 3 was included because the other teams’ algorithms included in held-out evaluation could not outperform the original trial performance for Patient 3 when evaluated on the held-out data. Each of these teams provided detailed descriptions of their algorithms.
Statistical analysis
The Hanley-McNeil method (Hanley and McNeil, 1982) was used to statistically assess if algorithm AUC scores were above chance for the contest and held-out data. Moreover, a different statistical test designed to compare AUC scores derived from the same data (Hanley and McNeil, 1983; Mayaud et al., 2013) was used to assess if the AUC score for the top contest algorithm for a given held-out dataset (i.e. all the data combined or the data from each of the individual patients) was different from the AUC scores for the other algorithms involved in the held-out data evaluation, as well as a circadian predictor (Karoly et al., 2017). The circadian predictor was based on pseudo-prospectively evolving seizure probability as a function of the time of day, where a single seizure probability was given for each hour of the day and this distribution of seizures as a function of time of day was updated each time a new seizure occurred. For a specific data clip, time of day was determined and the corresponding seizure probability was then treated as the preictal probability to make predictions.
Pseudo-prospective seizure prediction performance was compared against the performance of three naïve predictors: random analytic Poisson (Snyder et al., 2008), periodic (Winterhalder et al., 2003), and circadian (as noted above). The random analytic Poisson predictor is an analytic method that determines the sensitivity of a Poisson process-based random predictor for a given proportion of time in warning. The periodic predictor was based on making predictions using two parameters: a fixed prediction interval and a first-prediction time relative to the start of the recording period of interest. Performance of the periodic predictor was evaluated over 12 prediction intervals spanning 40 min to 60 h and all possible starting times that were multiples of 10 min. This ensured that a large range of proportion of time in warning values were considered.
Unless stated otherwise, statistical tests with the Hanley-McNeil, AUC comparison and analytic Poisson methods were assessed with a significance level of 0.05 with subsequent Bonferroni correction for multiple comparisons for the 10 082 algorithm entries.
Algorithms
Algorithms are summarized in Table 2 and listed in order of AUC-based performance on the contest private leader board. Algorithm categories in Table 2 are sorted based on the processing stages of the algorithms, which begin by analysing windows of data within each iEEG data clip to derive features that are fed into machine learning algorithms to make estimates of the probability that a given data clip is preictal. Detailed information regarding these algorithms is provided in the Supplementary material.
Table 2.
AUC scores for the held-out data experiment compared to scores on the public and private leader boards
| Team | Window (overlap, s) | Features | Machine learning algorithm | Ensemble method | Public leader board | Private leader board | Held-out data | Per cent change | Sensitivity at 75% specificity |
|---|---|---|---|---|---|---|---|---|---|
| Team A notsorandomanymore (1st place) | 20, 30, 50 (0) | Spectral power, distribution statistics, AR error, fractal dimensions, Hurst exponent, Riemannian autocorrelationa,b, cross-frequency coherence, correlation, other featuresc,d | Extreme gradient boostinge, k-nearest neighbours, generalized linear model, linear SVMf | Ranked average | 0.85276 | 0.80701 | 0.75275 | −6.7234 | 0.58322 |
| Team B Arete Associates (2nd) | 60 (30), 600 (0) | Correlation, distribution statistics, zero crossings, complexity, mobility, maximum frequency, total summed energy, entropy, normalized summed spectral energy | Extremely randomized treesg | n/a | 0.78328 | 0.79898 | 0.73364 | −8.1773 | 0.56306 |
| Team C GarethJones (3rd) | 80, 160, 240 (0) | Spectral power, distribution statistics, RMS of signal, first and second derivatives, correlation, spectral edge | Polynomial SVM, random under-sampling boosted tree ensemble | Weighted average | 0.81524 | 0.79652 | 0.65523 | −17.7388 | 0.41632 |
| Team D QingnanTang (4th) | 75 (0) | Spectral power, correlation, spectral entropy, spectral edge power, square of features | Gradient boostingh, extreme gradient boosting, radial basis function SVM | Weighted average | 0.7965 | 0.79458 | 0.71805 | −9.6319 | 0.52086 |
| Team E Nullset (5th) | 30 (0) | Spectral power, correlation (and eigenvalues), spectral entropy, Shannon entropy, spectral edge frequency, Hjorth parameters, fractal dimensions | Adaptive boosting, gradient boosting, random forest, extreme gradient boosting, gridsearch | Voting classifier | 0.81423 | 0.79363 | 0.62929 | −20.7074 | 0.46132 |
| Team F tralala boum boum pouet pouet (6th) | 60 (0) | Spectral power, spectral entropy, time/spectral correlation (and eigenvalues), Petrosian fractal dimensioni, Hjorth parametersj, variance, skewness, kurtosis, convolutional neural network outputsk, long–short-term memory network outputsl | SVM, random forest, extreme gradient boosting | Weighted average | 0.80493 | 0.79197 | 0.71822 | −9.3118 | 0.49742 |
| Team G Michaln (8th) | 600 (0) | Mean, SD, spectral edge at 50% power, skewness, kurtosis, Hjorth parameters, spectral entropy, maximum cross-correlation, spectral correlation, Brownian, Petrosian and Katz fractal dimensions, wavelet singular valuesm | Decision trees | Simple average | 0.80396 | 0.79074 | 0.73441 | −7.1238 | 0.56327 |
| Team H Chipicito+Solver World (Special case: best team for Patient 3) | 15, 60 (0) | Spectral energy, spectral energy ratios, and absolute spectral amplitude | Extreme gradient boosting, random forest, convolutional neural network | Weighted rank average | 0.80334 | 0.73968 | 0.76632 | 3.602 | 0.62635 |
Additional information on algorithms is available in the Supplementary material. Table structure mimics similar table from the 2014 American Epilepsy Society Seizure Prediction Challenge publication (Brinkmann et al., 2016) to facilitate direct comparison. AR = autoregressive; RMS = root mean square; SVM = support vector machine.
aBarachant et al., 2013; bCongedo et al., 2017; cTemko et al., 2011; dTemko and Lightbody, 2016; eChen and Guestrin, 2016; fCortes and Vapnik, 1995; gGeurts et al., 2006; hFriedman, 2001; iPetrosian, 1995; jHjorth, 1970; kLeCun et al., 1998; lHochreiter and Schmidhuber, 1997; mBrinkmann et al., 2016.
Circadian-weighted algorithms
To assess whether or not circadian information can be used to improve the performance of the contest algorithms, circadian-weighted versions of the algorithms considered in the held-out evaluation were also evaluated on the held-out data. To achieve this, for a given contest algorithm and for a given data clip, the preictal probability of the circadian-weighted algorithm was generated by multiplying the preictal probability of the algorithm with the preictal probability of the aforementioned circadian predictor for the corresponding time of day. The circadian-weighted algorithms were then evaluated in the same way as the original algorithms under the pseudo-prospective evaluation framework.
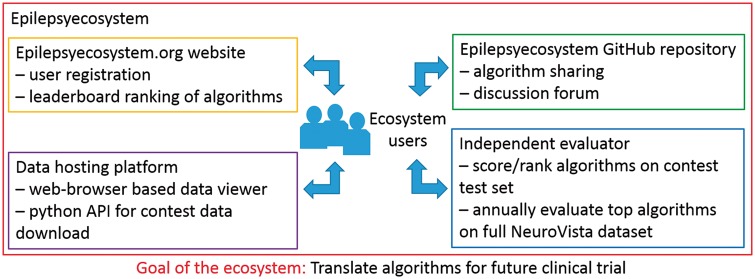
Epilepsyecosystem.org: ongoing crowd-sourcing ecosystem for seizure prediction
Epilepsyecosystem.org (https://www.epilepsyecosystem.org) is an evolving data and algorithm ecosystem focused on bringing people together to solve the problem of seizure prediction. Leveraging off of the results of the contest described here, Epilepsyecosystem.org provides an online environment for people to use the contest data and the source code for the top performing algorithms to further develop improvements in seizure prediction algorithms. The core aspects of the ecosystem are captured in Fig. 3. The main initial features of the ecosystem are to provide (i) a Python programming language-based API to enable users to download the contest data and independently train algorithms; (ii) an iEEG data viewer to enable users to visualize the data in an efficient manner; (iii) a GitHub organization at https://github.com/epilepsyecosystem where people can access the code from the top algorithms from the contest as well as share and discuss code and algorithms; and (iv) a benchmarking procedure, involving an independent evaluator, for comparing the performance of new algorithms to the performance of the top algorithms from the contest. As incentive, the top algorithms from the ecosystem will be invited to participate in an evaluation of algorithms on the full NeuroVista clinical trial dataset involving long-term recordings from 15 patients. Such an evaluation will help find the best seizure prediction algorithms for the widest range of patients and provide further justification for a large-scale prospective clinical trial of seizure prediction. To find out more about how the ecosystem works, visit https://www.epilepsyecosystem.org.
Figure 3.
A schematic illustration of the main features of Epilepsyecosystem.org.
Data availability
The contest data used in this study are available for download at https://www.epilepsyecosystem.org. Top performing algorithms as determined through participation in Epilepsyecosystem.org will be invited on an annual basis to be evaluated on the full NeuroVista Seizure Advisory System clinical trial dataset.
Results
AUC-based evaluation
Altogether, 646 individuals in 478 teams entered the competition and submitted a total of 10 082 algorithm entries. AUC scores for the top scoring teams are listed in Table 2. For the private leader board, 291 teams and, for the held-out evaluation, all considered teams, showed statistically significant performance that was greater than that of a random classifier (P < 0.05/10 082 for significant cases; 95% confidence limit AUC of a random classifier following correction for multiple comparisons was 0.6033 and 0.5292 for the contest and held-out data, respectively). Moreover, for all the data in the held-out evaluation combined, statistical testing found that the AUC score for first-placed Team A was greater than the performance for Teams C and E, but not necessarily for the other teams or the circadian predictor (P < 0.05/10 081 for significant cases; the AUC score for the circadian predictor was 0.7433 and the AUC scores for all teams are presented in Table 2). A less conservative Bonferroni correction based only on comparing Team A to the other of teams in the held-out evaluation plus the circadian predictor found that Team A performed better than Teams C–F (P < 0.05/8 for significant cases). The ROC classification curves used to derive the AUC values were qualitatively similar for the contest and held-out evaluations (Supplementary Fig. 1).
When considering individuals, AUC scores for the held-out data differed across patients (Supplementary Fig. 2). For Patient 1, statistical comparison of AUC scores found that the best contest algorithm for Patient 1 (Team F) performed better than Teams A and C and circadian prediction, but not necessarily the other teams (P < 0.05/10 081 for significant cases). For Patient 2, similar testing showed that the best contest algorithm for Patient 2 (Team A) performed better than Teams C, E and F, but not necessarily the other teams or circadian prediction (P < 0.05/10 081 for significant cases). Likewise, for Patient 3, it was observed that the best contest algorithm for Patient 3 (Team H) performed better than Teams D, E and G, but not necessarily the other teams or circadian prediction (P < 0.05/10 081 for significant cases). A less conservative Bonferroni correction based only on comparing the top team for each patient to the other of teams in the held-out evaluation plus the circadian predictor found that the top team for each patient performed better than Teams A–D and circadian prediction for Patient 1, Teams C–F for Patient 2 and Teams C–G and circadian prediction for Patient 3 (P < 0.05/8 for significant cases).
AUC performance for individual patients did not appear to show time-dependent trends when AUC was computed on quarterly (3-month) blocks of held-out data (Supplementary Fig. 2). Sub-models of first placed Team A gave varying AUC scores (Supplementary Table 1), demonstrating the benefit of using the combination of complementary models to boost performance.
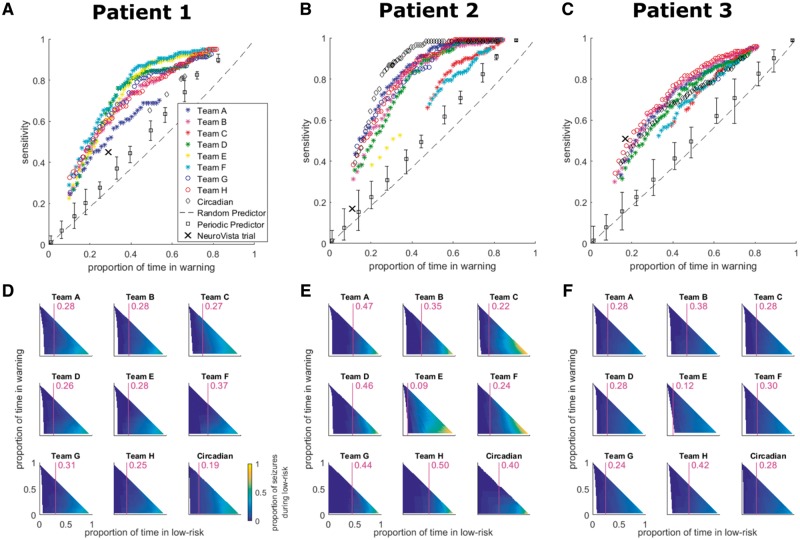
Pseudo-prospective evaluation
Pseudo-prospective evaluation of seizure prediction performance on the held-out data (Fig. 4A–C) shows different contest algorithms performed best for different patients. Many algorithms performed significantly better than both random prediction (when corrected for multiple comparisons based on the 10 082 algorithm entries) and periodic prediction. Algorithms also performed better than circadian prediction for Patient 1. The algorithms performed significantly better than the algorithm from the original clinical trial of the NeuroVista device for Patients 1 and 2, and performance was comparable for Patient 3.
Figure 4.
Pseudo-prospective seizure prediction results for the held-out data. (A–C) Seizure prediction performances for Patients 1–3, respectively, for all competition teams considered in the held-out evaluation. The results are compared to circadian, periodic, and random prediction, and to the original NeuroVista trial performance. The y- and x-axes correspond to sensitivity and proportion of time in warning (i.e. time in ‘red light’ or high-seizure-risk), respectively. For the different teams, data points correspond to different preictal probability thresholds and only data points surviving correction for multiple comparisons are plotted. The legend in A applies to A–C with teams listed in descending rank on the private leader board. Error bars for the periodic predictor indicate the ranges of performance over all phases. (D–F) Low-seizure-risk advisory performance for Patients 1–3, respectively, for all teams considered in the held-out evaluation as well as circadian prediction. The y- and x-axes correspond to proportion of time in warning and proportion of time in low-risk (i.e. time in ‘blue light’), respectively. The colour bar in D indicates the proportion of seizures occurring during low risk and applies to D–F. The purple vertical lines and numerical values overlaid on the subplots in D–F provide, for a proportion of time in warning of 0.25, the maximum proportion of time in low-risk for which the proportion of seizures occurring in low-risk is at most 0.05.
Many algorithms demonstrated impressively high proportions of time in the low-seizure-risk state, with zero or close to zero seizures occurring during low-risk (i.e. ‘blue light’) periods (Fig. 4D–F, deep blue regions). This effectively indicates low false negative rates for the algorithms.
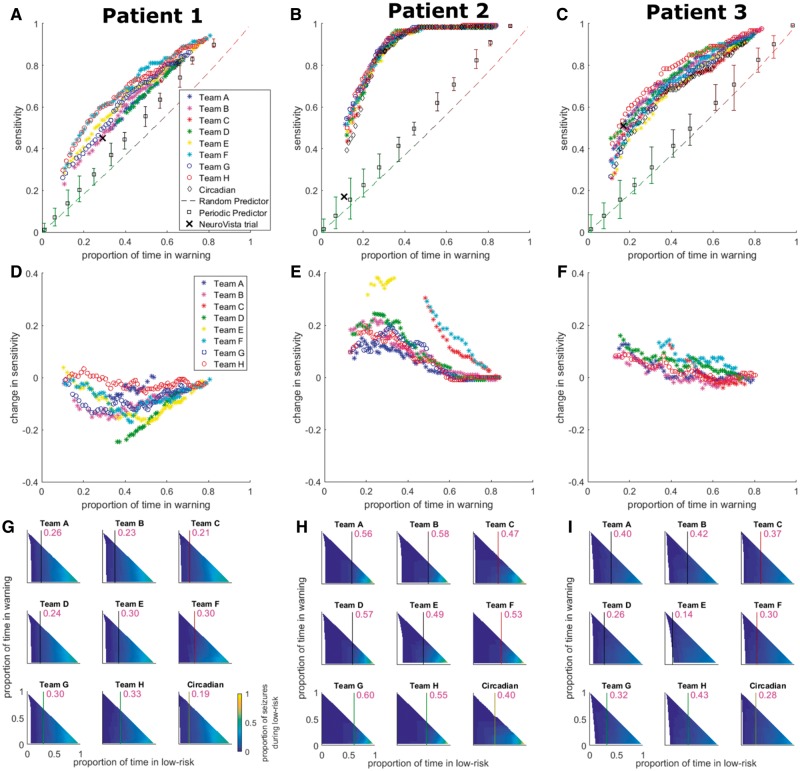
Pseudo-prospective evaluation of seizure prediction performance on the held-out data using circadian-weighted algorithms (Fig. 5A–F) revealed that improvements in performance relative to that of the original algorithms were achieved for Patients 2 and 3, whereas the performance for Patient 1 decreased. Comparing the analysis of low seizure-risk prediction for the original (Fig. 4D–F) and circadian-weighted (Fig. 5G–I) algorithms demonstrated that when circadian weighting improved seizure prediction performance (i.e. for Patients 2 and 3), it also generally led to an increase in the proportions of time in the low-seizure-risk state with zero or close to zero seizures occurring (Fig. 4D–F, deep blue regions in both figures). This can also be recognized by noting, for a proportion of time in warning of 0.25, the maximum proportion of time in low-risk for which the proportion of seizures occurring in low-risk is at most 0.05. The values for this are given for each algorithm in Figs 4D–F and 5G–I, and generally increase for Patients 2 and 3 for the circadian-weighted algorithms relative to the original algorithms. Thus, circadian-weighting can help to reduce false negative rates in the appropriate patient.
Figure 5.
Pseudo-prospective circadian-weighted seizure prediction results for the held-out data. (A–C) Circadian-weighted seizure prediction performances for Patients 1–3, respectively, for all competition teams considered in the held-out evaluation. The results are compared to circadian, periodic, and random prediction, and to the original NeuroVista trial performance. The y- and x-axes correspond to sensitivity and proportion of time in warning (i.e. time in ‘red-light’ or high-seizure-risk), respectively. For the different teams, data points correspond to different preictal probability thresholds and only data points surviving correction for multiple comparisons are plotted. The legend in A applies to A–C with teams listed in descending rank on the private leaderboard. Error bars for the periodic predictor indicate the ranges of performance over all phases. (D–F) Change in held-out data performance when subtracting sensitivity of the original algorithms from the sensitivity of the circadian-weighted algorithms for Patients 1–3, respectively. The y- and x-axes correspond to change in sensitivity (positive/negative change indicates an increase/decrease in performance when adding circadian-weighting) and proportion of time in warning, respectively. The legend in D applies to D–F. (G–I) Low-seizure-risk advisory performance for Patients 1–3, respectively, for all circadian-weighted algorithms (i.e. teams) considered in the held-out evaluation as well as circadian prediction. The y- and x-axes correspond to proportion of time in warning and proportion of time in low-risk (i.e. time in ‘blue-light’), respectively. The colour bar in G indicates the proportion of seizures occurring during low risk and applies to G–I. The purple vertical lines and numerical values overlaid on the subplots in G–I provide, for a proportion of time in warning of 0.25, the maximum proportion of time in low-risk for which the proportion of seizures occurring in low-risk is at most 0.05.
To further assess properties of the original and circadian-weighted algorithms, the distribution of true predictions relative to seizure onset (65 to 5 min before seizure) was also evaluated for each algorithm for the held-out data. It was found that many algorithms tended to produce more true predictions between 35 and 5 min before seizure for Patient 1, while for Patients 2 and 3 there was no clear tendency for true predictions to be closer to seizures; however, these results are very dependent on the time in warning and the algorithm (Supplementary Fig. 3).
Discussion
We present a crowd-sourcing ecosystem (Wiener et al., 2016) for seizure prediction involving an on-line competition, follow-up evaluation and an on-line platform, Epilepsyecosystem.org, to obtain further improvements in seizure prediction performance. To date, this has been a major success based on the large number of participants and algorithms submitted for the competition and the improvements in human seizure prediction observed in the follow-up evaluation.
The key clinical finding of this study is that seizure prediction algorithms were found that could provide good results for patients that previously had poor seizure prediction performance. This suggests it may be possible to provide seizure prediction to a wider range of patients than previously thought. Patients on whom prediction algorithms had unsatisfactory performance in the earlier trial (Cook et al., 2013) could achieve pseudo-prospective performance better than random and periodic predictors and as good as, or better than, circadian predictors when considering either the original algorithms or their circadian-weighted versions. Moreover, using the same benchmark dataset, different algorithms demonstrated best performance for different patients, again suggesting that seizure prediction algorithms should be patient-specific.
Pseudo-prospective sensitivity of the algorithms was much better than the results from the original NeuroVista trial (Cook et al., 2013) for the same time in warning for Patients 1 and 2. Performance for Patient 3 was comparable to the original trial. This is possibly due to Patient 3 having a much higher seizure frequency (43.7 seizures/month) than the other patients (<20.9 seizures/month). A greater seizure frequency may indicate a greater number of different preictal states, making it more difficult to distinguish preictal and interictal states. Seizure frequency appears to be an important factor that affects seizure prediction performance given that patients with the lowest seizure frequencies gave the best prediction performance in the original trial (Cook et al., 2013). In addition, electrode location and sampling of the epileptic network may affect prediction performance. The difficulty to predict seizures for the patients considered here may be due to complex epileptic networks, as indicated by poor post-resective surgery outcomes for Patients 1 and 3, and widespread seizure morphology for Patient 2 (Cook et al., 2013).
To see if performance for Patient 3 could be improved, the special case of the best team for Patient 3 on the private leader board data (Team H) was also considered. Figures 4C and 5C show that the best algorithm for Patient 3 on the private leader board was also the best for Patient 3 on the held-out data; however, performance only slightly exceeded that of the original trial for the circadian-weighted version of the algorithm. It should be noted that the original trial only evaluated performance on a 4-month period and was able to optimize evaluation parameters on the training data. Here, the algorithms have been evaluated on much more data (>9 months) and evaluation parameters, such as the warning duration (Snyder et al., 2008), were restricted based on the format of Kaggle.com competitions. This format essentially required that a fixed preictal period be defined for all patients, whereas for each patient, the optimal preictal period may differ. These differences mean that the original trial had more flexibility to find an optimal result. Overall, the new results showed significant improvement over the original trial results.
Comparison against naïve predictors was generally favourable for the contest algorithms. One exception was circadian prediction, which achieved better sensitivities than the original algorithms for certain proportions of time in warning for Patient 2 (Fig. 4B). However, in this case the low-seizure-risk advisory performance was worse for the circadian predictor (Fig. 4E). For example, for a proportion of time in warning of 0.25, the maximum proportion of time in low-risk resulting in zero seizures during low-risk was 0.32 and 0.04 for Team A and circadian prediction, respectively. Statistical comparison of AUC scores with a very conservative correction for multiple comparisons based on all the submitted contest algorithms showed that the best original contest algorithm for Patient 1 performed better than circadian prediction, whereas the performance of the best original algorithms for Patients 2 and 3 could not be statistically distinguished from circadian prediction. On the other hand, when using a less conservative correction for multiple comparisons based on all the teams considered in the held-out evaluation and circadian prediction, the best algorithms for a given patient performed better than circadian prediction for Patients 1 and 3. Moreover, when comparing the performance of the circadian-weighted and original versions of the contest algorithms, circadian-weighting boosted pseudo-prospective performance for Patients 2 and 3 but not Patient 1. Taken as a whole, these results indicate that circadian prediction and circadian-weighting are important factors to consider in the design of seizure prediction algorithms; however, this is dependent on the patient and the statistical evaluation parameters (Karoly et al., 2017). It is also likely to be dependent on the circadian-weighting method used. In addition, circadian prediction is only possible in patients with high enough seizure frequencies to ensure seizure probability as a function of time of day can be characterized. On the other hand, the machine-learning-based prediction algorithm applied in the original NeuroVista trial was shown to produce high prospective seizure prediction performance for patients with low seizure frequency (Cook et al., 2013). Thus, the machine learning-based contest algorithms shown here are potential candidates that may also work well for patients with low seizure frequency.
Considering technical implementation approaches, the 2014 American Epilepsy Society Seizure Prediction Challenge demonstrated that useful predictive features included spectral power in discreet frequency bands and also in time domain and/or frequency domain inter-channel correlations, while support vector machines were most commonly used to estimate the preictal probability of data clips based on the predictive features (Brinkmann et al., 2016). For the current contest, similar features were also predictive of seizures (Table 2). In addition, less common features were used, such as fractal dimension, Hurst exponent, and Hjorth parameters. Contrasting the methods used to estimate the preictal probability of data clips with methods of the previous contest, a greater number of decision tree-based machine learning algorithms, such as extreme gradient boosting, were employed by the top teams, although support vector machines and deep learning approaches, such as convolutional neural networks (Kiral-Kornek et al., 2018; Truong et al., 2018), also played a role.
As was observed for the previous contest, AUC performance remained reliable for six of the eight teams considered for the much larger held-out set (less than a 10% change in performance relative to the contest test set). In a clinical setting, stable performance is a promising indication that algorithms can remain reliable for long-term use in seizure advisory systems. Differences in training (such as training separate algorithms for each patient versus training an algorithm on the data from all patients combined) or overfitting the contest data may be the reasons for the reduction in performance for the other two teams. To make reliable algorithms, this suggests care should be taken to make algorithms patient-specific and not to overfit data during training by using appropriate techniques (Witten et al., 2016).
The strength of the presented results for the patients whose seizures have previously been the most difficult to predict, strongly suggest it is time for a larger scale trial. These results underscore the importance of long-term monitoring for reliably evaluating a patient’s epilepsy and predicting their seizures. The outcomes suggest a clinical solution where an army of algorithms is trained for each patient and the best algorithm is chosen for prospective, real-time seizure prediction. This could be achieved by recruiting crowd-sourced solutions into the pool of best algorithms deployed on a seizure prediction platform. This work presents pseudo-prospective seizure prediction results, which should be taken to be hypothesis generating, i.e. providing an indication of which algorithms to implement in a larger prospective clinical trial of a seizure prediction device. Future work is needed to assess prospective performance of the considered algorithms and the role these algorithms may play in multimodal systems that seek to improve seizure prediction performance by going beyond considering EEG (Dumanis et al., 2017).
To aid in the push for a larger scale trial, this paper sets forth a crowd-sourcing ecosystem (Wiener et al., 2016) for seizure prediction that will yield further improvements in seizure prediction. This will be achieved by making the contest data and the sharing and benchmarking of algorithms available through the aforementioned https://www.epilepsyecosystem.org, described in the ‘Materials and methods’ section and Fig. 3. The algorithms from the contest provide an initial benchmark to find even better seizure prediction methods. Moreover, annual evaluation of the best algorithms in the ecosystem on the full NeuroVista trial dataset will ensure the best algorithms can be found for the widest range of patients. Evaluation of performance on the full trial dataset will also include weighting of algorithms by (time-varying) factors, such as circadian and other multidien information (Baud et al., 2018), interictal spike rates and/or temperature of each patient’s local region of residence. This will help to find new ways to tailor patient-specific algorithms. In addition, performance will be evaluated on different seizure types within a patient (Payne et al., 2018) to give insight into the role seizure types play in seizure prediction. This ecosystem provides an excellent approach for raising the standards of reproducibility and comparison of findings in the field of seizure prediction by way of encouraging worldwide competition, collaboration and synergism on a unified platform using high quality long-term human iEEG data.
Supplementary Material
Acknowledgements
We thank kaggle.com for helping to run the competition and all of the 646 contest participants. Research was supported by Melbourne Bioinformatics on its supercomputing facility hosted at the University of Melbourne (VR0003). We thank Robert Kerr and Shannon Clarke for contributing to the creation of the Epilepsyecosystem.org platform.
Glossary
Abbreviations
- AUC
area under the curve
- iEEG
intracranial electroencephalography
- ROC
receiver operating characteristic
Funding
This work was supported by American Epilepsy Society, The MathWorks Corporation, National Institute of Neurological Disorders and Stroke (NIH 1 U24 NS063930-01), The University of Melbourne, and National Health and Medical Research Council (APP1130468). A.T. received support from Science Foundation Ireland Research Centre Award (12/RC/2272). T.P. was supported by the National Institute of Neurological Disorders and Stroke (NINDS) (R01NS079533 - Truccolo Lab) and U.S. Department of Veterans Affairs, Merit Review Award (I01RX000668 - Truccolo Lab). G.W. and B.B. received support from National Institutes of Health (NIH) (R01-NS92882 and UH2NS095495). B.L. received support from NIH (UH2-NS095495-01, R01NS092882, 1K01ES025436-01, 5-U24-NS-063930-05, R01NS099348), Mirowski Foundation and Neil and Barbara Smit. L.K. and D.T.J.L. were supported by James S McDonnell Foundation (220020419).
Competing interests
G.W. reports equity in Cadence Neuroscience Inc. and NeuroOne Inc. B.L. declares licensed technology to NeuroPace Inc. through the University of Pennsylvania. All other authors declare no conflicts of interest.
References
- Barachant A, Bonnet S, Congedo M, Jutten C. Classification of covariance matrices using a Riemannian-based kernel for BCI applications. Neurocomputing 2013; 112: 172–8. [Google Scholar]
- Baud MO, Kleen JK, Mirro EA, Andrechak JC, King-Stephens D, Chang EF et al. . Multi-day rhythms modulate seizure risk in epilepsy. Nat Commun 2018; 9: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann BH, Wagenaar J, Abbot D, Adkins P, Bosshard SC, Chen M et al. . Crowdsourcing reproducible seizure forecasting in human and canine epilepsy. Brain 2016; 139 (Pt 6): 1713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Guestrin C. Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2016. ACM; 2016; p. 785–94. [Google Scholar]
- Congedo M, Barachant A, Bhatia R. Riemannian geometry for EEG-based brain-computer interfaces; a primer and a review. Brain-Computer Interfaces 2017; 4: 155–174. [Google Scholar]
- Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995; 20: 273–97. [Google Scholar]
- Cook MJ, O’Brien TJ, Berkovic SF, Murphy M, Morokoff A, Fabinyi G et al. . Prediction of seizure likelihood with a long-term, implanted seizure advisory system in patients with drug-resistant epilepsy: a first-in-man study. Lancet Neurol 2013; 12: 563–71. [DOI] [PubMed] [Google Scholar]
- Dumanis SB, French JA, Bernard C, Worrell GA, Fureman BE. Seizure forecasting from idea to reality. Outcomes of the my seizure gauge epilepsy innovation institute workshop. eNeuro 2017; 4: ENEURO. 0349–17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Smith SJ, Forster A, Shorvon SD, Trimble MR. Effects of the removal of phenytoin, carbamazepine, and valproate on the electroencephalogram. Epilepsia 1989; 30: 590–6. [DOI] [PubMed] [Google Scholar]
- Freestone DR, Karoly PJ, Peterson AD, Kuhlmann L, Lai A, Goodarzy F et al. . Seizure prediction: science fiction or soon to become reality? Curr Neurol Neurosci Rep 2015; 15: 73. [DOI] [PubMed] [Google Scholar]
- Friedman JH.Greedy function approximation: a gradient boosting machine. Ann Stat 2001; 29: 1189–232. [Google Scholar]
- Gadhoumi K, Lina JM, Mormann F, Gotman J. Seizure prediction for therapeutic devices: a review. J Neurosci Methods 2016; 260: 270–82. [DOI] [PubMed] [Google Scholar]
- Geurts P, Ernst D, Wehenkel L. Extremely randomized trees. Mach Learn 2006; 63: 3–42. [Google Scholar]
- Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982; 143: 29–36. [DOI] [PubMed] [Google Scholar]
- Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 1983; 148: 839–43. [DOI] [PubMed] [Google Scholar]
- Hjorth B. EEG analysis based on time domain properties. Electroencephalogr Clin Neurophysiol 1970; 29: 306–10. [DOI] [PubMed] [Google Scholar]
- Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997; 9: 1735–80. [DOI] [PubMed] [Google Scholar]
- Karoly P, Ung H, Grayden DB, Kuhlmann L, Leyde K, Cook MJ et al. . The Circadian profile of epilepsy improves seizure forecasting. Brain 2017; 140: 2169–82. [DOI] [PubMed] [Google Scholar]
- Kiral-Kornek I, Roy S, Nurse E, Mashford B, Karoly P, Carroll T et al. . Epileptic seizure prediction using big data and deep learning: toward a mobile system. EBioMedicine 2018; 27: 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann L, Freestone D, Lai A, Burkitt AN, Fuller K, Grayden DB et al. . Patient-specific bivariate-synchrony-based seizure prediction for short prediction horizons. Epilepsy Res 2010; 91: 214–31. [DOI] [PubMed] [Google Scholar]
- Kuhlmann L, Grayden DB, Wendling F, Schiff SJ. Role of multiple-scale modeling of epilepsy in seizure forecasting. J Clin Neurophysiol 2015; 32: 220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE 1998; 86: 2278–324. [Google Scholar]
- Mayaud L, Lai PS, Clifford GD, Tarassenko L, Celi LAG, Annane D. Dynamic data during hypotensive episode improves mortality predictions among patients with sepsis and hypotension. Crit Care Med 2013; 41: 954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mormann F, Andrzejak RG, Elger CE, Lehnertz K. Seizure prediction: the long and winding road. Brain 2007; 130 (Pt 2): 314–33. [DOI] [PubMed] [Google Scholar]
- Payne DE, Karoly PJ, Freestone DR, Boston R, D’Souza W, Nurse E et al. . Postictal suppression and seizure durations: a patient-specific, long-term iEEG analysis. Epilepsia 2018; 59: 1027–36. [DOI] [PubMed] [Google Scholar]
- Petrosian A. Kolmogorov complexity of finite sequences and recognition of different preictal EEG patterns. In: Computer-Based Medical Systems, 1995, Proceedings of the Eighth IEEE Symposium on; 1995. IEEE 1995; p. 212–7. [Google Scholar]
- Schulze-Bonhage A, Kühn A. Unpredictability of seizures and the burden of epilepsy. In: Seizure prediction in epilepsy: from basic mechanisms to clinical applications. Weinheim: Wiley; 2008. [Google Scholar]
- Snyder DE, Echauz J, Grimes DB, Litt B. The statistics of a practical seizure warning system. J Neural Eng 2008; 5: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey WC, Litt B. Technology insight: neuroengineering and epilepsy—designing devices for seizure control. Nat Clin Pract Neurol 2008; 4: 190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temko A, Lightbody G. Detecting neonatal seizures with computer algorithms. J Clin Neurophysiol 2016; 33: 3994–402. [DOI] [PubMed] [Google Scholar]
- Temko A, Thomas E, Marnane W, Lightbody G, Boylan G. EEG-based neonatal seizure detection with support vector machines. Clin Neurophysiol 2011; 122: 464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong ND, Nguyen AD, Kuhlmann L, Bonyadi MR, Yang J, Ippolito S et al. . Convolutional neural network for seizure prediction using intracranial and scalp electroencephalogram. Neural Netw 2018; 105: 104–111. [DOI] [PubMed] [Google Scholar]
- Wiener M, Sommer FT, Ives ZG, Poldrack RA, Litt B. Enabling an open data ecosystem for the Neurosciences. Neuron 2016; 92: 617–21. [DOI] [PubMed] [Google Scholar]
- Winterhalder M, Maiwald T, Voss HU, Aschenbrenner-Scheibe R, Timmer J, Schulze-Bonhage A. The seizure prediction characteristic: a general framework to assess and compare seizure prediction methods. Epilepsy Behav 2003; 4: 318–25. [DOI] [PubMed] [Google Scholar]
- Witten IH, Frank E, Hall MA, Pal CJ. Data mining: practical machine learning tools and techniques. Cambridge, MA: Morgan Kaufmann; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The contest data used in this study are available for download at https://www.epilepsyecosystem.org. Top performing algorithms as determined through participation in Epilepsyecosystem.org will be invited on an annual basis to be evaluated on the full NeuroVista Seizure Advisory System clinical trial dataset.