Abstract
Our aim was to develop and apply a comprehensive noninvasive prenatal test (NIPT) by using high-coverage targeted next-generation sequencing to estimate fetal fraction, determine fetal sex, and detect trisomy and monogenic disease without parental genotype information. We analyzed 45 pregnancies, 40 mock samples, and eight mother-child pairs to generate 35 simulated datasets. Fetal fraction (FF) was estimated based on analysis of the single nucleotide polymorphism (SNP) allele fraction distribution. A Z-score was calculated for trisomy of chromosome 21 (T21), and fetal sex detection. Monogenic disease detection was performed through variant analysis. Model validation was performed using the simulated datasets. The novel model to estimate FF was robust and accurate (r2= 0.994, p-value < 2.2e-16). For samples with FF > 0.04, T21 detection had 100% sensitivity (95% CI: 63.06 to 100%) and 98.53% specificity (95% CI: 92.08 to 99.96%). Fetal sex was determined with 100% accuracy. We later performed a proof of concept for monogenic disease diagnosis of 5/7 skeletal dysplasia cases. In conclusion, it is feasible to perform a comprehensive NIPT by using only data from high coverage targeted sequencing, which, in addition to detecting trisomies, also make it possible to identify pathogenic variants of the candidate genes for monogenic diseases.
Keywords: Cell-free DNA, next-generation sequencing, trisomy, noninvasive prenatal test, fetal fraction
Introduction
The discovery of cell-free fetal DNA (cffDNA) in the maternal bloodstream (Lo et al., 1997) has revolutionized prenatal diagnosis. Initially, cell-free DNA (cfDNA) was used for detecting qualitative traits, such as fetal sex (Lo et al., 1998; Rijnders et al., 2001; Wright et al., 2012) and Rhesus D status (Faas et al., 1998; Finning et al., 2002). More recently, next-generation sequencing (NGS) technologies have provided the means for noninvasive detection of fetal aneuploidy with high sensitivity and specificity (Fan et al., 2008; Ehrich et al., 2011; Palomaki et al., 2011; Sparks et al., 2012a; Gil et al., 2015; Norton et al., 2015). Noninvasive prenatal testing (NIPT) using NGS of cfDNA is now being widely used as a screening test for the most common aneuploidies in the prenatal setting. Chromosome Y read count has been used for accurately determining fetal sex (Chiu et al., 2011; Koumbaris et al., 2016) and estimating fetal fraction (FF) (restricted for male fetuses only) (Fan et al., 2008; Hudecova et al., 2014; Xu et al., 2016).
High coverage targeted sequencing allows the accurate detection of fetal alleles without requiring parental genotyping (Liao et al., 2011). In addition to aneuploidy detection, this strategy enabled the identification of variants associated with monogenic diseases, especially de novo variants (Lam et al., 2012; New et al., 2014; Chitty et al., 2015). This method also enabled the development of methods for FF estimation by using single nucleotide polymorphisms (SNPs) from sequencing analysis of maternal plasma cfDNA, thus avoiding the need of parental genotyping, and reducing laboratory steps and turnaround time (Jiang et al., 2012; Sparks et al., 2012b; Koumbaris et al., 2016). FF estimation is crucial for test accuracy, because insufficient fetal cfDNA may lead to false negative results. Thus, measuring the presence of fetal DNA (independently of fetal sex) in maternal plasma in any test (e.g. trisomy detection) should improve its reliability. The aforementioned analyses are already being performed in clinical settings, although not within one single test. The development of parameters to perform all these analyses simultaneously by using only maternal plasma sequencing data may further reduce cost and turnaround time.
NIPT in Brazil is currently offered by private laboratories, and is performed by outsourcing the technology or the test itself. In the present report, we propose the implementation of an in-house NIPT by using high-coverage targeted NGS in order to estimate FF, determine fetal sex, and detect trisomy and monogenic disease without the need for parental genotypes. We used skeletal dysplasia (SD) as a monogenic disease model.
Subjects and Methods
Subjects and samples
Peripheral blood samples were collected from pregnant women (N=45) and non-pregnant individuals (8 mother and children pairs, N=16), the latter being obtained to establish a proof of concept of the test. Pregnant women were at least 18 years old, with singleton pregnancies, and at 10 to 36 gestational weeks. This study was approved by the Research Ethics Committee of Instituto de Biociências (Universidade de São Paulo - Brazil), and informed consent was obtained from all patients or legal tutors.
Blood samples were collected in EDTA tubes, and plasma processing took place within six hours. Blood samples were centrifuged at 1600 x g for 10 min, and recentrifuged at 16000 x g for another 10 min. Plasma cfDNA extraction of 2-4 mL was performed by using the QIAamp Circulating Nucleic Acid kit (Qiagen) following the manufacturer's protocol. cfDNA was first eluted in a total of 150 μL and then concentrated to 60 μL by vacuum centrifugation.
Proof of concept – mock samples
To establish a proof of concept of the test and validate the bioinformatics pipeline, we generated 40 in silico pregnancy mock samples by mixing the fastq reads from both mothers and children (five different FFs for each one of the mother-child pairs). We mixed the fastq reads with different fractions in order to simulate different ‘fetal fractions’ for each pair, mimicking the progressive increase in FF during pregnancy from the first to the third trimester. Among these samples are pairs with children affected and unaffected by Down syndrome.
High coverage next-generation targeted sequencing of plasma samples
For cfDNA library preparation, we used the NEBNext Ultra kit (New England Biolabs) according to the manufacturer's protocol. Libraries were indexed, multiplexed, captured for a gene panel using Nextera Rapid Capture (Illumina), and quantified by real-time quantitative PCR by using the KAPA Library Quantification kit (KAPA Biosystems). Libraries were then sequenced in a MiSeq system (Illumina) using the MiSeq Reagent kit v3 (2x75 cycles), as well as in a HiSeq system (Illumina) with a HiSeq Rapid SBS Kit v2 (2x100 cycles).
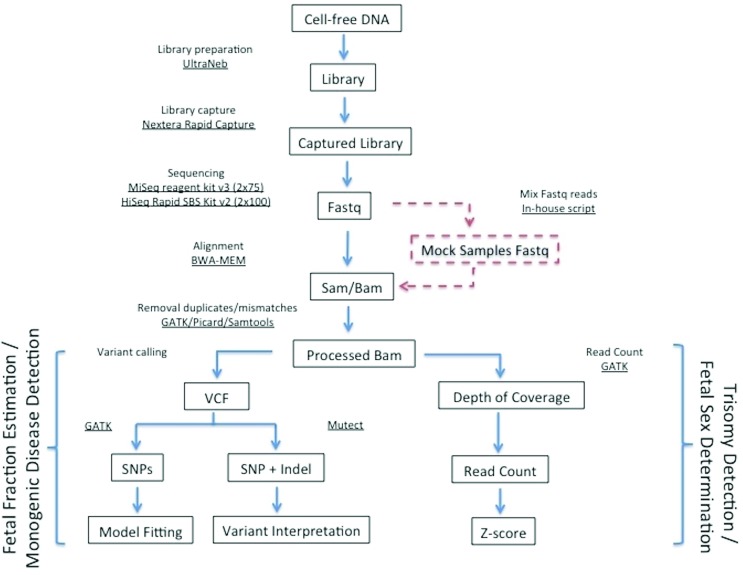
The fastq files were aligned by BWA-MEM (Li and Durbin, 2010), duplicated reads were removed by Picard (http://broadinstitute.github.io/picard), realigned based on known local indels with GATK (McKenna et al., 2010; DePristo et al., 2011; Van der Auwera et al., 2013), and reads with more than two mismatches were removed using Samtools (Li et al., 2009). The mean coverage of BAM files was determined using Samtools Depth. For FF estimation, we performed variant call with all patients with GATK. For aneuploidy detection, we generated a Depth of Coverage file for each sample with GATK. For monogenic disease detection, the somatic variants were called by using Mutect (Cibulskis et al., 2013). The workflow is outlined in Figure 1.
Figure 1. Test workflow. Red: performed only in the mother and child fastq files to generate mock samples.
Gene panel
For targeted sequencing, we used a panel of genes of clinical interest used in the routine diagnosis performed at HUG-CELL (Human Genome and Stem Cell Research Center). The panel consists of 497 genes of clinical interest (Table S1 (157.1KB, pdf) ) belonging to the following groups of disorders: Hereditary Cancer, Skeletal Dysplasias/Craniofacial diseases, Neuromuscular/Neurodegenerative, Intellectual Deficiency/Autism, and Recessive Diseases Screening (http://genoma.ib.usp.br/pt-br).
We used SNPs to estimate FF, and read count to detect aneuploidy. To estimate FF, we used 6739 probes distributed in 388 genes across the autosomes (minus chromosomes 18 and 21, which were excluded because they are the most common trisomies and can affect minor allele fraction – MAF – estimate), comprising approximately 1.5 Mb. For the detection of trisomy on chromosome 21 (T21), variable regions were excluded to minimize variance in the read count analysis. These regions were frequently observed as false positive CNVs calls in Hidden Markov Model (HMM) analysis of NGS data in more than 100 patients, who were sequenced by the same panel for genetic diseases. After this correction, we used 240 probes distributed in 19 genes (approximately 33.5 Kb) across chromosome 21 for T21 detection.
Fetal fraction estimation / Model fitting and evaluation
Maternal plasma had a mixture of cfDNA from the mother and fetus. For any biallelic SNP, there are four possible combinations of maternal/fetal genotypes. As we do not know the genotype combination at each interrogated locus a priori (Table 1), we generated a model so as to fit our data and to estimate FF (developed with R, v. 3.2.3) based on each locus MAF (Supplementary Material Text 1 (162.1KB, pdf) ).
Table 1. MAF given FF for different maternal/fetal genotype combinations in maternal plasma.
| Possibilities | Origin | Genotype | Minor Allele | Mean of B allele fraction (MAF) |
|---|---|---|---|---|
| 1 | Maternal | AA | B | 0 |
| Fetal | AA | |||
| 2 | Maternal | AA | B | FF/2 |
| Fetal | AB | |||
| 3 | Maternal | AB | B | 0.5-FF/2 |
| Fetal | AA | |||
| 4 | Maternal | AB | B | 0.5 |
| Fetal | AB |
To evaluate the model and fitting accuracy, we used 2255 simulated samples (with different mean coverage and SNP number values), as well as 40 mock samples (from which we had the expected FF from fetal specific alleles), and 16 non-pregnant samples (either mother or child in which FF is expected to be 0).
Detection of trisomy 21
To detect the frequency of T21, we used the read count generated by GATK Depth of Coverage. For each chromosome, we calculated the chromosome proportion defined as the sum of reads on that chromosome divided by the total reads of autosomes minus the chromosome of interest.
The reference dataset of each chromosome consisted of pregnant samples (including mock samples) with a fetus unaffected by trisomy. To normalize the reference dataset, we calculated the median and standard deviation for the reference dataset and removed the samples falling outside of three median absolute deviations. Importantly, the reference dataset has to contain samples sequenced with the same platform (MiSeq or HiSeq).
We then used a Z-score approach to calculate the genomic representation of the chromosome of interest compared to the reference dataset for each test sample:
Z-scoretest sample = (Ptest sample – Pmean reference samples / SDmean reference samples
(P = proportion of the chromosome of interest; SD = standard deviation)
Fetal sex determination
For fetal sex determination, we used the chromosome Y read count. The reads covering the SRY gene were counted with GATK Depth of Coverage, and the proportion of chromosome Y was determined as the sum of reads of chromosome Y divided by total sum of autosome reads. The reference dataset consisted of female fetus pregnancies (including mock). It is important to note that the reference dataset has to contain samples sequenced with the same platform (MiSeq or HiSeq). Normalization was applied and a Z-score was calculated for T21 detection.
Detection of monogenic disease and variant interpretation
Skeletal dysplasia (SD) is a group of bone and cartilage disorders that affect fetal development in utero or postnatally. Prenatal onset SDs are clinically detectable through gestational ultrasound presenting limb defects or reduction. Many of the prenatal onset SDs are autosomal dominant and lethal, but some of them are non-lethal. The molecular confirmation of the lethality of the fetus prior to birth would certainly help to manage the pregnancy.
The availability of probes for several genes of clinical interest in our panel (including several forms of SDs) allowed us to perform a specific analysis for this disease, with the aim of performing a proof of concept analysis in our data for the prenatal detection of monogenic diseases.
For the analysis of possible pathogenic variants, we performed variant call individually by Mutect (Cibulskis et al., 2013), and annotated it by Annovar (Wang et al., 2010) and several public databases (ExAC, Exome Variant Server, 1000 Genomes), including our in-house database of 609 Brazilian control exomes. We screened for rare variants (minor allele frequency < 0.5%) that are present only in genes related to dysplasia/craniofacial disorders (Table S2 (110.3KB, pdf) ). For de novo variants in the fetus only, we expected to detect the MAF variant at approximately half of FF.
Blind dataset for validation
For blind validation of our methodology, we used eight pregnant samples comprising controls and fetuses affected by T21 that were not previously known to test results. Library preparation and sequencing (by using HiSeq) were performed for the rest of the samples used in this work, as described above.
Results
Sample characterization and sequencing
A total of 61 peripheral blood samples were collected, 45 being from pregnant women and 16 from non-pregnant individuals (8 mothers and children pairs). The pregnant women were aged between 20 and 46 (mean: 32.5, SD ± 5.86) and at 10-36 gestational weeks (mean: 20.4, SD ± 9) (Table S3 (212.9KB, pdf) ). Among the eight non-pregnant pairs used to generate the mock samples, we collected two children affected by Down syndrome (T21) and six unaffected by T21 (Table S4 (125.3KB, pdf) ).
Sequencing of 47/61 samples was performed using MiSeq (33 pregnant women and 14 non-pregnant individuals), and yielded an average of 15.2 million raw reads per sample (ranging from 8,695,612 to 33,517,518). Mean coverage in BAM files was 191.65 X (39.99 X - 294.3 X, median: 208.6 X).
Sequencing of 14/61 samples by using HiSeq (12 pregnant women and two non-pregnant individuals) yielded an average of 106.9 million raw reads per sample (ranging from 35,316,152 to 246,692,132). Mean coverage in BAM files was 519.92X (201.6X – 928.38X, median: 522.75 X). Total average coverage (MiSeq and HiSeq altogether) comprised 267 X (Median: 222 X).
Mock samples
Mock samples generated by a fastq file mixture instead of cfDNA mixture of mother and child have the advantage of allowing files with multiple FFs with low cfDNA input and cost, since it only requires sequencing mother and child once. However, there are intrinsic differences of PCR duplicates in the fastq files of both mother and child, which can be a confounding factor when estimating FF directly from the mock sample. In order to have an accurately expected FF for the mock samples, we employed two methods: fetal-specific alleles by using maternal genotype information (Chiu et al., 2010; Liao et al., 2012) (using only SNP positions with a coverage of at least 100X), and chromosome Y read count for male fetus pregnancies (Chiu et al., 2011; Hudecova et al., 2014). The estimated FFs in both approaches (fetal-specific alleles and chromosome Y read count) are strongly correlated, especially for high coverage samples (Pearson correlation r2 –all samples: 0.846, p=2.527e-06; coverage ³ 100x: 0.951, p=1.506e-08; coverage ³ 150X: 0.997, p=8.449e-07; coverage ³ 200X: 0.996, p=0.05119). Since the Y read count is only applicable to male fetus pregnancies, we used fetal-specific alleles to predict FF.
Fetal fraction estimation
The fitting of the samples was performed, as explained in the Methods section, by comparing the MAF values distribution between the test sample and simulated samples for the specific MAF range (0.02–0.25) (Figure S1 (109.6KB, pdf) ). A higher mean coverage and SNP number of simulated samples positively affect the model fitting, as expected (Figure S2 (123.2KB, pdf) ).
We tested the model fitting accuracy for mean coverage and SNP number values obtained for our MiSeq sequenced samples (150X and 2000, respectively), which were lower than the samples sequenced by HiSeq. We found a high correlation between the expected and fitted FF values (Pearson correlation r2 =0.999, p < 2.2e-16), with median degree of deviation of 0.000 (-0.033–0.050), calculated as: (Expected-Fitted)/Expected (Figure S3 (111.4KB, pdf) ).
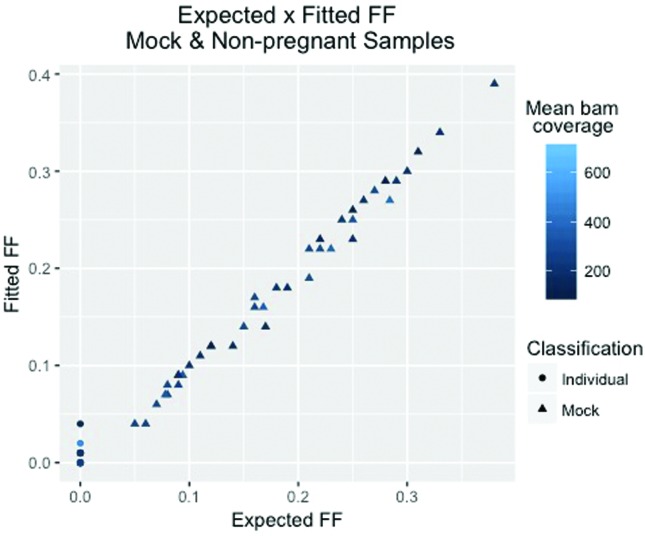
The use of our mock and non-pregnant samples indicated that the model fitting is also accurate when using non-simulated samples (Pearson correlation r2=0.994, p < 2.2e-16) (Figure 2, Tables S4 (125.3KB, pdf) and S5 (215.7KB, pdf) ).
Figure 2. Evaluation of the modeled fetal fraction (FF) and the mean coverage effect. Individual: Non-pregnant sample. Shape and color incorporate both individual classification and mean coverage value, respectively.
The developed model was then used to estimate FF for all samples (mock, pregnant, and non-pregnant). After VCF filtering, our samples had average SNP numbers of 4162 (11-5529, median: 4423) and 3990 (3514-4327, median: 4018) for MiSeq samples and HiSeq samples respectively (Tables S3 (212.9KB, pdf) , S4 (125.3KB, pdf) and S5 (215.7KB, pdf) ).
Mean fitted FF for pregnant samples was 0.12, varying between 0.02-0.30. Correlation analysis showed a strong positive correlation between FF and gestational age (Pearson correlation r 2 =0.5, p=4.4e-04). We did not find a significant association between FF and maternal weight (Pearson correlation r2 =-0.137; p=0.38) (Figure S4 (112.6KB, pdf) ).
Fetal sex determination
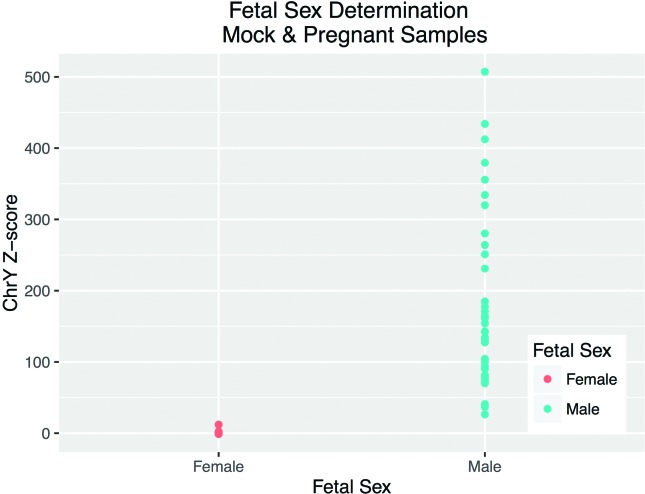
For fetal sex determination, the normalized reference dataset consisted of 33 and 10 samples for MiSeq and HiSeq, respectively. Chromosome Y proportion and Z-score were calculated for each mock and pregnant sample (Tables S3 (212.9KB, pdf) and S5 (215.7KB, pdf) ). Male fetus pregnancies have an average proportion of 8.9e-05 (1.27e-05 – 2.38e-04) and Z-score of 185.35 (26.27 – 507.2), while female fetus pregnancies have an average proportion of 5.05e-07 (0 – 6.05e-06) and Z-score of 0.2 (-1.05 – 12.09). The groups do not overlap and, as such, they can be easily distinguished from one another. For the 81 samples for which we had confirmation of fetal sex (40/40 mock samples and 41/45 pregnant samples), we observed 100% accuracy (Figure 3), with 100% sensitivity (95% CI: 90.51%-100%), and 100% specificity (95% CI: 91.96%-100%).
Figure 3. Fetal sex determination in 44 females and 37 males. Chromosome Y Z-score according to fetal sex.
Trisomy 21 detection
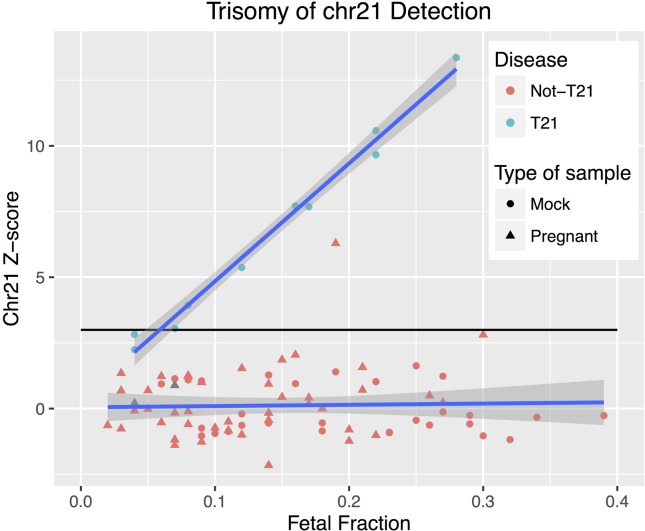
After normalization, the euploid reference dataset for T21 detection consisted of 54 and 16 samples for MiSeq and HiSeq, respectively. We then calculated chromosome 21 Z-score for all 83 samples (10 mock T21, 30 mock not-T21, and 43 non-affected pregnant samples) that had confirmation of the fetal diagnosis (two were excluded since they did not have confirmation), independently of FF. For the mock T21 samples, 8/10 had a positive Z-score (threshold 3.0) and 2/10 had a negative Z-score (false negatives) (Tables S3 (212.9KB, pdf) and S5 (215.7KB, pdf) ). The 43 non-affected pregnant samples had only one false positive (Z-score =6.29, FF=0.19) (Figure 4).
Figure 4. T21 detection. Chromosome 21 Z-score as function of FF. Color and shape incorporate both disease status (blue = T21, red = Not-T21 samples) and type of sample (circle = mock, triangle= samples with pregnant women), which represents 10 T21 samples (all mock), and 73 non-T21 samples (30 mock and 43 pregnant).
Using a threshold of FF0.04 as inclusion criteria, we had 76 out of the 83 samples (8 mock T21, 30 mock not-T21, and 38 pregnant samples) estimated with 100% sensitivity (95% CI: 63.06% to 100.00%) and 98.53% specificity (95% CI: 92.08% to 99.96%). By considering T21 frequency as 1:800 births, and the specificity and sensitivity values shown above, a false positive rate of 1.5% and a false negative rate of zero for our test were estimated.
We observed a positive correlation between FF and Z-score values for the T21 affected samples (Pearson correlation r2=0.994, p=6.013e-09), while this was not observed for unaffected samples (Pearson correlation r2=-0.033, p=0.7821).
Dataset blinded to diagnosis for validation
We also performed sequencing (by using HiSeq) and analysis of eight additional samples comprising affected (T21) and unaffected samples with pregnant women. The sequencing of these samples yielded an average of 16.5 million raw reads per sample (ranging from 10,338,602 – 23,619,856). Mean coverage in BAM files was 56.8X (32.7X – 83.5X, median: 57.6X).
The analysis of these samples revealed that five were from male fetuses and three from female fetuses. Regarding T21, five out of the eight samples were from T21 fetuses while three were from unaffected T21 fetuses. After disclosure of the original data, we verified 100% accuracy of our results regarding fetal sex determination and T21 detection (Table 2).
Table 2. Test results for blind dataset.
| Sample | Trisomy | Test Result T21 (Z-score) | Fetus Gender | Test Result Fetal Sex (Z-score) |
|---|---|---|---|---|
| FD1500110 | T21 | T21 (12.9) | Male | Male (55.7) |
| FD1500068 | T21 | T21 (8.5) | Male | Male (27.41) |
| FD1500092 | T21 | T21 (7.8) | Male | Male (101.9) |
| FD1500073 | T21 | T21 (6.8) | Male | Male (51.44) |
| FD1500098 | T21 | T21 (4.4) | Female | Female (-1.05) |
| FD.15.00142 | Not-T21 | Not-T21 (0.2) | Female | Female (-1.05) |
| FD.15.00141 | Not-T21 | Not-T21 (0.17) | Female | Female (-1.05) |
| FD1500107 | Not-T21 | Not-T21 (-0.55) | Male | Male (98.8) |
Monogenic disease detection – skeletal dysplasia
Seven samples from pregnant women had prenatal ultrasound findings suggesting SD. After VCF analysis, we detected known pathogenic variants in 5/7 samples (Table 3).
Table 3. Samples with women who were pregnant with fetuses diagnosed with SD by ultrasound.
| Patient | FF 1 | GW 2 | Gene | Pathogenic Mutation | AF 3 | Confirmed on child |
|---|---|---|---|---|---|---|
| F10177-2 | 0.22 | 32 | FLNB | NM_001457:c.605T > C (p.M202T) | 0.11 | ✓ |
| F10775-1 | 0.12 | 29 | FGFR3 | NM_000142:c.742C > T (p.R248C) | 0.07 | ✓ |
| F10609-1 | 0.2 | 30 | FGFR3 | NM_000142:c.742C > T (p.R248C) | 0.08 | ✓ |
| F11247-1 | 0.13 | 26 | FGFR3 | NM_000142:c.742C > T (p.R248C) | 0.034 | ✓ |
| F11077-1 | 0.07 | 23 | FGFR3 | NM_000142: c.1108G > T(p.G370C) | 0.031 | NA |
| F10951-1 | 0.07 | 33 | × | × | × | NA |
| F10774-1 | 0.11 | 33 | × | × | × | NA |
FF: fetal fraction.
GW: gestational weeks.
AF: allele fraction.
Not found. NA: Not available.
We detected a pathogenic variant in sample F10177-2 located at the FLNB gene (NM_001457:c.605T > C:p.M202T), and associated with a lethal form of SD (Ateleosteogenesis type 1/Boomerang dysplasia) (Daniel et al., 2012).
Pathogenic mutations in the FGFR3 gene were identified in four patients. FGFR3 mutations are associated with thanatophoric dysplasia (TD), an autosomal dominant disorder, which was the initial diagnostic hypothesis (DH) for the three samples in which we detected the mutation NM_000142:c.742C > T:p.R248C, the most common mutation associated with TD type I (Tavormina et al., 1995; Wilcox et al., 1998). The other sample harboring an FGFR3 mutation (F11077-1) had an initial DH of campomelic dysplasia, with unspecific ultrasound findings (short bent bones, brachycephaly, and narrow thorax). This patient (F11077-1) had a rare mutation in FGFR3 associated with TD type I as well (Rousseau et al., 1996).
We were unable to detect pathogenic mutations in two samples (F10951-1 and F10774-1) that had an initial DH of osteogenesis imperfecta (OI). We did not have the child's genomic DNA to verify whether this was due to methodological reasons (unability to detect it noninvasively), or whether the mutation was not present in our panel. Although we did not find a pathogenic mutation for these cases, we did detect a VUS (variant of unknown significance) for patient F10774-1 (NM_001235.3:c.580C > A:p.R194S) located at SERPINH1, a gene already associated with a recessive form of OI (Bonafe et al., 2015).
The MAF of these variants is about half of FF, as expected for de novo variants associated with autosomal dominant disorders. No other variant within the expected MAF was classified as pathogenic or probably pathogenic by using ACMG criteria (Richards et al., 2015). Results were confirmed by sequencing the child's genomic DNA after birth, when available.
Discussion
We have developed an NIPT for genetic diseases by using NGS that incorporates the following analysis: FF estimation (using only maternal plasma sequencing data), fetal sex determination, trisomy detection, and monogenic disease detection. A key strength of this study was the incorporation of all analyses in one single test, which was performed with the same gene panel used for the regular clinical genomic diagnosis in our center. This strategy requires only minimal modifications if the panel is up to date. To our knowledge, this is the first work to create a comprehensive test, and it has the advantage of allowing samples with different diagnostic purposes within the same laboratory workflow. While high coverage exome sequencing generally is not yet financially feasible for prenatal testing, this approach opens up the possibility to test hundreds of monogenic diseases with NGS, by targeting all coding sequences instead of solely relying on the investigation of mutational hot spots.
FF determination aids in avoiding false negative results and improves the detection of point mutations. Therefore, we developed a model to estimate FF that uses only plasma sequencing data regardless of fetal sex. Other groups use SNPs from targeted sequencing data to predict FF through a statistical binomial mixture model, relying on several mother-child genotype combinations to correctly predict FF (Jiang et al., 2012; Sparks et al., 2012b; Koumbaris et al., 2016). We showed that it is possible to perform FF estimation with simpler statistics (MAF values vector comparison in R) by using only the most informative genotype combination (mother homozygous, child heterozygous).
Since FF is an important factor in NIPT accuracy and has been correlated with different maternal traits in other populations, we investigated its correlation with factors such as gestational age and maternal weight in Brazilian pregnant women. We found a positive correlation between FF and gestational age, in accordance with other reports (Lo et al., 1998; Zimmermann et al., 2012; Hudecova et al., 2014; Rava et al., 2014; Zhou et al., 2015; Xu et al., 2016). However, we did not find a significant correlation between FF and maternal weight as reported by others (Ashoor et al., 2012; Wang et al., 2013; Hudecova et al., 2014). This lack of correlation may be attributable to our small sample size.
In this work, we established a threshold of 0.04 FF for T21 detection, as reported in the literature, for better accuracy. Literature data indicate high sensitivity and specificity for T21 detection by NGS varying between 94.4% - 100% and 97.95% - 100%, respectively (Chiu et al., 2011; Gil et al., 2015), and our test sensitivity and specificity values were within these ranges, showing that we have high sensitivity and specificity for T21 detection. We also performed the test in a blind dataset for validation, resulting in 100% accuracy for fetal sex and T21 determination.
The two false negative samples for T21 detection have fitted FFs of 0.04, which is the detection limit found in the literature (Ehrich et al., 2011; Norton et al., 2012; Palomaki et al., 2012; Sparks et al., 2012b), so they were expected to present low Z-scores (Ehrich et al., 2011; Norton et al., 2012; Palomaki et al., 2012; Sparks et al., 2012b).
The one false positive result for T21 observed in our sample can be due to several factors: confined placental mosaicism, fetal mosaicism, vanishing twin, or even maternal malignancies (Osborne et al., 2013; Grati et al., 2014; Bianchi et al., 2015). We had a high positive correlation for the T21-affected samples, and the false positive sample did not fall onto the correlation line, as reported by others (Hudecova et al., 2014). Corroborating the estimated high sensitivity and specificity of our test, we verified that the NIPT test in one of the pregnant women (F10117-1), who was referred to us with a positive diagnosis for T21 from a different clinical service, was negative for T21. The follow up of this case revealed that the child was born normal, in accordance with our NIPT screening test.
As shown previously, it is possible to perform fetal sex determination with targeted NGS (Chiu et al., 2011; Koumbaris et al., 2016). In this work, we demonstrated that it is possible to determine fetal sex with 100% accuracy by only one probe on chromosome Y instead of multiple probes (Koumbaris et al., 2016).
Monogenic disease testing for SD was performed on seven cases, with a detection rate of 71% (5/7), thus demonstrating our test's capacity to incorporate detection of monogenic diseases, especially in de novo or paternally inherited variants. By using children's genomic DNA (unpublished data from our center), our noninvasive SD detection rate was similar to the postnatal detection rate (75/125 = 60%) by using child's genomic DNA (unpublished data from our center).
Thanatophoric, achondroplasia, and osteogenesis imperfecta are among the most common types of SD (Unger et al., 1993; Milks et al., 2017). For patient F11077-1, who had an initial DH of campomelic dysplasia, we confirmed the diagnosis as TD type I. Despite the existence of clinical overlap, this differential diagnosis is important because TD is lethal, while campomelic dysplasia is not always lethal. This differential diagnosis is also important for the medical team in the postpartum management as well as for the family's psychological preparation.
In the two patients for whom pathogenic mutations were not detected (F10951-1 and F10774-1), the DH was OI. For these patients, we cannot discount the hypothesis of the pathogenic mutation being in a gene absent from our panel, since the number of genes associated with SD has grown at a fast pace in recent years, especially due to the advent of NGS (Bonafe et al., 2015). Another possible explanation is that the mutation could reside in an intronic region, or it could be a deletion, which would be overlooked by the currently available tools. For patient F10774-1, we detected a VUS in a gene that is already associated with a recessive form of OI. It is possible that this patient had a recessive form of the disease, and the lack of identification of the second mutation may be a limitation in identifying mutations that are present in the mother.
Chitty et al. (2015) have recently demonstrated the effectiveness of NIPT to detect FGFR3-related SDs. However, they targeted hotspots in the gene, which has the pitfall of possibly overlooking pathogenic variants. Dan et al. (2016) showed the feasibility of using targeted sequencing for 16 genes. However, they used maternal and paternal genomic DNA sequencing for variant detection. Comparatively, our test might be more advantageous because we are covering the entire coding sequence of hundreds of genes associated with monogenic disorders (therefore covering all exonic variants and many differential diagnoses), and also because we are able to perform the detection by using only plasma sequencing, thus lowering costs and turnaround time. These results suggest the possibility to expand our approach for detecting other monogenic dominant diseases, particularly those caused by de novo or paternally inherited variants. NIPT as a screening test for dominant disorders should be considered in the near future, particularly with the increase of the reproductive age in most populations together with the burden of de novo paternal mutations with aging, and the effect of selfish mutations in paternal gonads (Maher et al., 2016).
It is important to note that for the five SD cases in which we detected the pathogenic mutation, the AF is about half of the FF, which is expected for autosomal dominant disorders. This also demonstrates that our FF estimation model is accurate and helpful for detecting pathogenic mutations, since we can target the mutation within the expected AF according to the disease inheritance model.
We showed the relevance of using targeted sequencing to develop an integrated NIPT (using only maternal plasma) by combining all analyses (fetal fraction estimation, fetal sex determination, trisomy, and monogenic disease detection). Further reduction of sequencing costs will enable an even higher coverage, thus improving the ability to detect autosomal recessive or X-linked mutations more accurately, when the mother is heterozygous for the variant.
To our knowledge, we are the first group in Brazil to develop an in-house, non-invasive prenatal test performed with NGS. NIPT is presently available for patients in Brazil, but the test is either performed abroad or through outsourcing technology. In this work, we demonstrated that it is indeed possible to perform NIPT for several fetal diseases by using only plasma sequencing data, a practicable amount of targeted sequencing, and relatively simple statistics.
Acknowledgments
The authors are grateful to Dr. Clarice Savastano and Dr. Gerson Kobayashi for helping with the manuscript. The present work was supported by grants from CNPq (573633/2008-8) and FAPESP (2013/08028-1; 2013/14996-0; 2015/11998-8).
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Maria Luiza Petzl-Erler
References
- Ashoor G, Poon L, Syngelaki A, Mosimann B, Nicolaides KH. Fetal fraction in maternal plasma cell-free DNA at 11-13 weeks' gestation: Effect of maternal and fetal factors. Fetal Diagn Ther. 2012;31:237–243. doi: 10.1159/000337373. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, Garber JE, Wilkins-Haug L, Vora NL, Warsof S, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314:162. doi: 10.1001/jama.2015.7120. [DOI] [PubMed] [Google Scholar]
- Bonafe L, Cormier-Daire V, Hall C, Lachman R, Mortier G, Mundlos S, Nishimura G, Sangiorgi L, Savarirayan R, Sillence D, et al. Nosology and classification of genetic skeletal disorders: 2015 revision. Am J Med Genet Part A. 2015;167:2869–2892. doi: 10.1002/ajmg.a.37365. [DOI] [PubMed] [Google Scholar]
- Chitty LS, Mason S, Barrett AN, McKay F, Lench N, Daley R, Jenkins LA. Non-invasive prenatal diagnosis of achondroplasia and thanatophoric dysplasia: Next-generation sequencing allows for a safer, more accurate, and comprehensive approach. Prenat Diagn. 2015;35:656–662. doi: 10.1002/pd.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RWK, Sun H, Akolekar R, Clouser C, Lee C, McKernan K, Zhou D, Nicolaides KH, Lo YMD. Maternal plasma DNA analysis with massively parallel sequencing by ligation for noninvasive prenatal diagnosis of trisomy 21. Clin Chem. 2010;56:459–463. doi: 10.1373/clinchem.2009.136507. [DOI] [PubMed] [Google Scholar]
- Chiu RWK, Akolekar R, Zheng YWL, Leung TY, Sun H, Chan KCA, Lun FMF, Go ATJI, Lau ET, To WWK, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: Large scale validity study. BMJ. 2011;342:c7401–c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan S, Yuan Y, Wang Y, Chen C, Gao C, Yu S, Liu Y, Song W, Asan, Zhu H, et al. Non-invasive prenatal diagnosis of lethal skeletal dysplasia by targeted capture sequencing of maternal plasma. PLoS One. 2016;11:e0159355. doi: 10.1371/journal.pone.0159355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel PB, Morgan T, Alanay Y, Bijlsma E, Cho T-J, Cole T, Collins F, David A, Devriendt K, Faivre L, et al. Disease-associated mutations in the actin-binding domain of filamin B cause cytoplasmic focal accumulations correlating with disease severity. Hum Mutat. 2012;33:665–673. doi: 10.1002/humu.22012. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, Tim R, Lu V, McCullough R, McCarthy E, Nygren AOH, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011;204.205:e1–205.e11. doi: 10.1016/j.ajog.2010.12.060. [DOI] [PubMed] [Google Scholar]
- Faas BH, Beuling EA, Christiaens GC, von dem Borne AE, van der Achoot CE. Detection of fetal RhD - specific sequences in maternal plasma. Lancet. 1998;352:1196. doi: 10.1016/s0140-6736(05)60534-x. [DOI] [PubMed] [Google Scholar]
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finning KM, Martin PG, Soothill PW, Avent ND. Prediction of fetal D status from maternal plasma: Introduction of a new noninvasive fetal RHD genotyping service. Transfusion. 2002;42:1079–1085. doi: 10.1046/j.1537-2995.2002.00165.x. [DOI] [PubMed] [Google Scholar]
- Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. 2015;45:249–266. doi: 10.1002/uog.14791. [DOI] [PubMed] [Google Scholar]
- Grati FR, Malvestiti F, Ferreira JCPB, Bajaj K, Gaetani E, Agrati C, Grimi B, Dulcetti F, Ruggeri AM, De Toffol S, et al. Fetoplacental mosaicism: Potential implications for false-positive and false-negative noninvasive prenatal screening results. Genet Med. 2014;16:620–624. doi: 10.1038/gim.2014.3. [DOI] [PubMed] [Google Scholar]
- Hudecova I, Sahota D, Heung MMS, Jin Y, Lee WS, Leung TY, Lo YMD, Chiu RWK. Maternal plasma fetal DNA fractions in pregnancies with low and high risks for fetal chromosomal aneuploidies. PLoS One. 2014;9:e88484. doi: 10.1371/journal.pone.0088484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Chan KCA, Liao GJW, Zheng YWL, Leung TY, Chiu RWK, Lo YMD, Sun H. FetalQuant: Deducing fractional fetal DNA concentration from massively parallel sequencing of DNA in maternal plasma. Bioinformatics. 2012;28:2883–2890. doi: 10.1093/bioinformatics/bts549. [DOI] [PubMed] [Google Scholar]
- Koumbaris G, Kypri E, Tsangaras K, Achilleos A, Mina P, Neofytou M, Velissariou V, Christopoulou G, Kallikas I, Gonzalez-Linan A, et al. Cell-free DNA analysis of targeted genomic regions in maternal plasma for non-invasive prenatal testing of trisomy 21, trisomy 18, trisomy 13, and fetal sex. Clin Chem. 2016;62:848–855. doi: 10.1373/clinchem.2015.252502. [DOI] [PubMed] [Google Scholar]
- Lam KWG, Jiang P, Liao GJW, Chan KCA, Leung TY, Chiu RWK, Lo YMD. Noninvasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: Application to β-thalassemia. Clin Chem. 2012;58:1467–1475. doi: 10.1373/clinchem.2012.189589. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao GJ, Chan KC, Jiang P, Sun H, Leung TY, Chiu RW, Lo YM. Noninvasive prenatal diagnosis of fetal trisomy 21 by allelic ratio analysis using targeted massively parallel sequencing of maternal plasma DNA. PLoS One. 2012;7:e38154. doi: 10.1371/journal.pone.0038154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao GJW, Lun FMF, Zheng YWL, Chan KCA, Leung TY, Lau TK, Chiu RWK, Lo YMD. Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clin Chem. 2011;57:92–101. doi: 10.1373/clinchem.2010.154336. [DOI] [PubMed] [Google Scholar]
- Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. Quantitative analysis of fetal DNA in maternal plasma and serum: Implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YMD, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CWG. Early report presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- Maher GJ, McGowan SJ, Giannoulatou E, Verrill C, Goriely A, Wilkie AO. Visualizing the origins of selfish de novo mutations in individual seminiferous tubules of human testes. Proc Natl Acad Sci U S A. 2016;113:2454–2459. doi: 10.1073/pnas.1521325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks KS, Hill LM, Hosseinzadeh K. Evaluating skeletal dysplasias on prenatal ultrasound: an emphasis on predicting lethality. Pediatr Radiol. 2017;47:134–145. doi: 10.1007/s00247-016-3725-5. [DOI] [PubMed] [Google Scholar]
- New MI, Tong YK, Yuen T, Jiang P, Pina C, Chan KCA, Khattab A, Liao GJW, Yau M, Kim SM, et al. Noninvasive prenatal diagnosis of congenital adrenal hyperplasia using cell-free fetal DNA in maternal plasma. J Clin Endocrinol Metab. 2014;99:E1022–E1030. doi: 10.1210/jc.2014-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton ME, Brar H, Weiss J, Karimi A, Laurent LC, Caughey AB, Rodriguez MH, Williams J, Mitchell ME, Adair CD, et al. Non-Invasive Chromosomal Evaluation (NICE) study: Results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;13:e1–137.e8. doi: 10.1016/j.ajog.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, Tomlinson MW, Pereira L, Spitz JL, Hollemon D, et al. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015;372:1589–1597. doi: 10.1056/NEJMoa1407349. [DOI] [PubMed] [Google Scholar]
- Osborne CM, Hardisty E, Devers P, Kaiser-Rogers K, Hayden MA, Goodnight W, Vora NL. Discordant noninvasive prenatal testing results in a patient subsequently diagnosed with metastatic disease. Prenat Diagn. 2013;33:609–611. doi: 10.1002/pd.4100. [DOI] [PubMed] [Google Scholar]
- Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, van den Boom D, Bombard AT, Deciu C, Grody WW, et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med. 2011;13:913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- Palomaki GE, Deciu C, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, van den Boom D, Bombard AT, Grody WW, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med. 2012;14:296–305. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rava RP, Srinivasan A, Sehnert AJ, Bianchi DW. Circulating fetal cell-free DNA fractions differ in autosomal aneuploidies and monosomy X. Clin Chem. 2014;60:243–250. doi: 10.1373/clinchem.2013.207951. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnders RJ, van der Schoot CE, Bossers B, de Vroede MA, Christiaens GC. Fetal sex determination from maternal plasma in pregnancies at risk for congenital adrenal hyperplasia. Obstet Gynecol. 2001;98:374–378. doi: 10.1016/s0029-7844(01)01480-6. [DOI] [PubMed] [Google Scholar]
- Rousseau F, El Ghouzzi V, Delezoide AL, Legeai-Mallet L, Le Merrer M, Munnich A, Bonaventure J. Missense FGFR3 mutations create cysteine residues in thanatophoric dwarfism type I (TD1) Hum Mol Genet. 1996;5:509–512. doi: 10.1093/hmg/5.4.509. [DOI] [PubMed] [Google Scholar]
- Sparks AB, Wang ET, Struble CA, Barrett W, Stokowski R, McBride C, Zahn J, Lee K, Shen N, Doshi J, et al. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat Diagn. 2012a;32:3–9. doi: 10.1002/pd.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. Noninvasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: Evaluation for trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012b;319:e1–319.e9. doi: 10.1016/j.ajog.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Tavormina PL, Shiang R, Thompson LM, Zhu YZ, Wilkin DJ, Lachman RS, Wilcox WR, Rimoin DL, Cohn DH, Wasmuth JJ. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nat Genet. 1995;9:321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- Unger S, Scherer G, Superti-Furga A. Campomelic Dysplasia. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH, et al., editors. GeneReviews. University of Washington; Seattle: 1993. [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high-confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat Diagn. 2013;33:1–5. doi: 10.1002/pd.4119. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox WR, Tavormina PL, Krakow D, Kitoh H, Lachman RS, Wasmuth JJ, Thompson LM, Rimoin DL. Molecular, radiologic, and histopathologic correlations in thanatophoric dysplasia. Am J Med Genet. 1998;78:274–281. doi: 10.1002/(sici)1096-8628(19980707)78:3<274::aid-ajmg14>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Wright CF, Wei Y, Higgins JP, Sagoo GS. Non-invasive prenatal diagnostic test accuracy for fetal sex using cell-free DNA a review and meta-analysis. BMC Res Notes. 2012;5:476. doi: 10.1186/1756-0500-5-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XP, Gan HY, Li FX, Tian Q, Zhang J, Liang RL, Li M, Yang XX, Wu YS. A method to quantify cell-free fetal DNA fraction in maternal plasma using next generation sequencing: Its application in non-invasive prenatal chromosomal aneuploidy detection. PLoS One. 2016;11:e0146997. doi: 10.1371/journal.pone.0146997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu Z, Gao Y, Yuan Y, Guo Y, Zhou L, Liao K, Wang J, Du B, Hou Y, et al. Effects of maternal and fetal characteristics on cell-free fetal DNA fraction in maternal plasma. Reprod Sci. 2015;22:1–7. doi: 10.1177/1933719115584445. [DOI] [PubMed] [Google Scholar]
- Zimmermann B, Hill M, Gemelos G, Demko Z, Banjevic M, Baner J, Ryan A, Sigurjonsson S, Chopra N, Dodd M, et al. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenat Diagn. 2012;32:1233–1241. doi: 10.1002/pd.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.