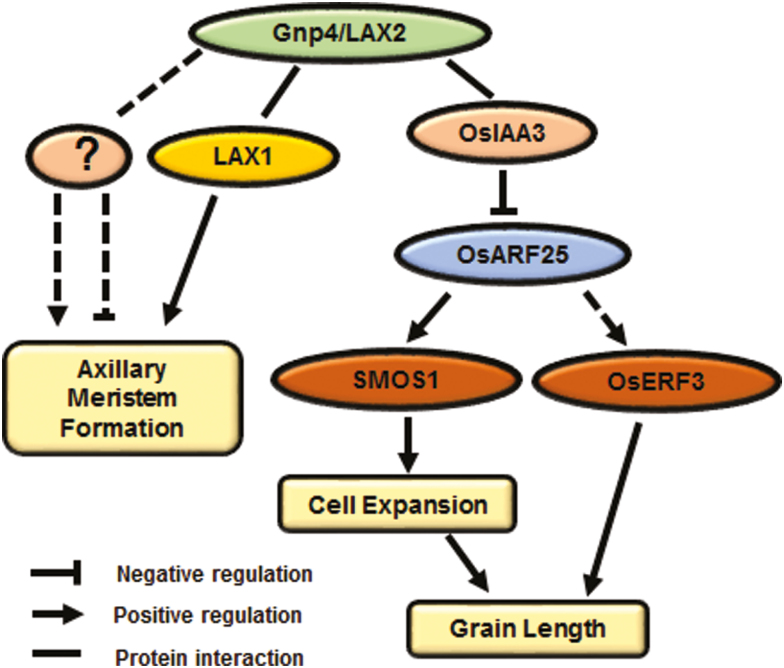
Gnp4/LAX2 interferes with the OsIAA3–OsARF25 interaction to activate the expression of OsERF142/SMOS1, and hence regulates grain length in rice.
Keywords: Auxin, Gnp4/LAX2, grain length, rice, transcription regulation
Abstract
Grain length is one of the determinants of yield in rice and auxin plays an important role in regulating it by mediating cell growth. Although several genes in the auxin pathway are involved in regulating grain length, the underlying molecular mechanisms remain unclear. In this study we identify a RING-finger and wd40-associated ubiquitin-like (RAWUL) domain-containing protein, Gnp4/LAX2, with a hitherto unknown role in regulation of grain length by its influence on cell expansion. Gnp4/LAX2 is broadly expressed in the plant and subcellular localization analysis shows that it encodes a nuclear protein. Overexpression of Gnp4/LAX2 can significantly increase grain length and thousand-kernel weight. Moreover, Gnp4/LAX2 physically interacts with OsIAA3 and consequently interferes with the OsIAA3–OsARF25 interaction in vitro and in vivo. OsIAA3 RNAi plants consistently exhibit longer grains, while the mutant osarf25 has small grains. In addition, OsARF25 binds to the promoter of OsERF142/SMOS1, a regulator of organ size, and positively regulates its expression. Taken together, the results reveal that Gnp4/LAX2 functions as a regulator of grain length through participation in the OsIAA3–OsARF25–OsERF142 pathway and that it has potential value for molecular breeding in rice.
Introduction
Grain weight, grain number per panicle, and panicle number are major factors influencing yield in rice (Oryza sativa). Among them, grain weight is least affected by environmental factors (Sakamoto and Matsuoka, 2008). Nevertheless, grain weight remains a complex quantitative trait that is affected by multiple genes. Grain length, grain width, grain thickness, and grain filling rate are contributory factors that determine grain weight (Xing and Zhang, 2010). Molecular studies of each of these characters are essential for a complete understanding of their potential roles in yield improvement.
To date, many genes contributing to grain weight in rice have been isolated, such as GS3, GW5, GS5, GL3.1, GW7/GL7, GW8, GLW7, OsGRF4, Big Grain1, XIAO, SLG, OsLG3, and OsLG3b (Weng et al., 2008; Mao et al., 2010; Li et al., 2011b; Jiang et al., 2012; Wang et al., 2012, 2015; Zhang et al., 2012; Duan et al., 2015; Liu et al., 2015; Feng et al., 2016; Si et al., 2016; Yu et al., 2017, 2018). While they all ultimately affect cell expansion or proliferation, they can be classified into several groups according to the pathways involved, including the G-protein signaling pathway, the proteasomal degradation pathway, the transcriptional regulation-related pathway, and the plant hormone biosynthesis or signaling transduction pathways (Zuo and Li, 2014; Li and Li, 2016). Although these genes have been cloned and functionally characterized, knowledge of the underlying molecular mechanisms and genetic interaction networks remain elusive and fragmentary. Consequently, it is important to isolate novel grain size-associated regulators in order to understand the molecular mechanism behind grain weight in rice.
The RING-finger and wd40-associated ubiquitin-like (RAWUL) domain is a new member of the ubiquitin superfamily and has been found in the same polypeptide chain as a the RING finger domain in the polycomb repressive complex 1 (PRC1) RING family, and in the same polypeptide domain as the WDR48-p80 protein family (Sanchez-Pulido et al., 2008). In Arabidopsis, two families of RING-finger proteins have been characterized as RAWUL domain-containing proteins, namely AtRING1A/B and AtBMI1A/B/C (Xiao and Wagner, 2015), and play roles in developmental phase transitions, cell proliferation during organ growth, and water-stress responses. However, little is known about RAWUL domain-containing proteins in rice and other crop plants.
Indole-3-acetic acid (auxin) plays an important role in growth and development of plants by regulating many biological processes (Gallavotti, 2013; Ljung, 2013). The molecular mechanisms of auxin perception are relatively well understood in different plant species (Salehin et al., 2015). The auxin signaling transduction pathway consists of four components, namely the auxin receptors, the AUX/IAA repressors, the auxin response factors (ARFs), and the downstream target genes (Salehin et al., 2015). Signaling transduction is initiated by the perception of auxin by the TIR1/AFB receptors that encode F-box proteins and are components of an E3 SCF ubiquitin ligase complex (Dharmasiri et al., 2005). AUX/IAA co-receptors function as repressors of the pathway by directly binding to the ARF transcription factors, and together with the co-repressor protein TOPLESS they repress their activities (Szemenyei et al., 2008). Auxin triggers the formation of the AUX/IAA-SCFTIR1/AFB co-receptor complex and the degradation of the AUX/IAA protein in a 26S proteasome-dependent manner, which in turns results in the de-repression of the ARFs and thus the transcriptional activation of their target genes (Salehin et al., 2015; Dezfulian et al., 2016).
There are 31 AUX/IAAs and 25 ARF protein genes in the rice genome (Jain et al., 2006; Wang et al., 2007). To date, several rice AUX/IAAs have been associated with specific phenotypic effects, including root development, plant architecture, and biotic and abiotic stress responses. OsIAA3 (referred to as OsIAA31 by Jain et al., 2006) was the first functionally characterized AUX/IAAs protein in rice and its gain-of-function causes growth defects in leaf blades and crown roots (Nakamura et al., 2006). Functional analysis of ARFs have been mainly based on studies of loss-of-function mutants. For example, loss of function of OsARF16 and OsARF12 lead to iron-deficiency responses (Qi et al., 2012; Shen et al., 2013), while OsARF23-OsARF24 has been shown to promote cell growth and morphogenesis by regulating RICE MORPHOLOGY DETERMINANT (RMD) expression (Li et al., 2014). These studies highlight many aspects of plant growth that are controlled by auxin-related pathways, but the underlying regulatory processes have only been identified in a few cases. Thus, it is still not clear how these AUX/IAA and ARF networks achieve target-specificity, and whether other factors or signaling proteins participate in this regulatory process.
In our previous studies, a natural mutant with defective development of lateral spikelets on the secondary panicle branches and increased grain length was characterized and the candidate gene was designated as Gnp4 (Grain number per-panicle 4), which shares the LAX2 (LAX PANICLE2) locus (Tabuchi et al., 2011; Zhang et al., 2011b). Here, we report that Gnp4/LAX2 encodes a RAWUL domain-containing protein and has a hitherto unknown role in regulating grain length. Gnp4 functions as a regulator of grain length by participating in an OsAUX/IAA–OsARF25–OsERF142 pathway.
Materials and methods
Plant material
Seeds of Oryza sativa subsp. japonica cv. Nipponbare and transgenic lines used in this study were generated within our laboratory. All the transgenic plants used for phenotypic evaluation were more advanced than the T2 generation. Rice accessions used for haplotype analysis were selected from the rice mini core collection (Zhang et al., 2011a). The T-DNA insertion mutant osarf25 and its wild-type (Hwayoung) were provided by Dr De’an Jiang (Zhejiang University) and the OsERF3-overexpression plants were provided by Dr Rongfeng Huang (Chinese Academy of Agricultural Sciences, Beijing).
Plasmid construction and rice transformation
To construct the overexpression plasmid Pro35S::Gnp4, the full coding sequence of Gnp4 was amplified from the cDNA of Nipponbare, digested with AscI and SpeI, and cloned into the binary vector pMDC32 (Curtis and Grossniklaus, 2003). For construction of the GUS (β-glucuronidase) plasmid, the 2-kb promoter region of Gnp4 was amplified from the DNA of Nipponbare, digested with PacI and AscI, and cloned into the binary vector pMDC162 (Curtis and Grossniklaus, 2003). For construction of GFP (green fluorescent protein) plasmids, the coding region of Gnp4 was inserted into ProSuper1330::GFP vector, and OsIAA3 and OsIAA17 were amplified and digested with SpeI and AscI, and cloned into the binary vector pMDC83 (Curtis and Grossniklaus, 2003). To construct an OsIAA3-RNAi vector, a 235-bp fragment containing part of the coding sequences and the 3′-UTR region was amplified from the cDNA of Nipponbare, digested with SacI and SpeI, and cloned into the pTCK303 vector (Wang et al., 2004) to generate the forward insertion. Next, a dsRNAi fragment obtained by digestion with BamHI and KpnI was cloned into the same vector to generate the reverse insertion.
All plasmids were introduced into A. tumefaciens EHA105. Rice transformation was conducted by the Agrobacterium-mediated method as previously described (Hiei et al., 1994). A full list of primers used in this study can be found in Supplementary Dataset S5 at JXB online.
Phylogenetic analysis
The amino acid sequence of Gnp4 was used to BLAST search its closest homologous proteins from other plant species against databases in Uniprot (http://www.uniprot.org/). Multiple-sequence alignment was optimized with the Megalign program in the DNASTAR software package (http://dnastar.com). A neighbor-joining tree for homologous proteins was constructed using MEGA5.0 (Tamura et al., 2011).
GUS staining
Tissues of transgenic plants containing the ProGnp4::GUS vector sampled at different growth stages were fixed in GUS-staining solution [50 mM Na2HPO4, 10 mM Na2EDTA, 0.5 mM K3Fe (CN)6, 0.5 mM K4Fe (CN)6, 0.1% TritionX-100, 1 mg ml–1 5-bromo-4-chloro-3-indolyl β-D-glucuronic acid]. After 12 h at 37 °C, the stained tissues were dehydrated in an ethanol series of (100%, 95%, 85%, 75%) to remove the chlorophyll, and photographed using a digital camera (Nikon D900).
Total RNA extraction and qRT-PCR analysis
Total RNA was extracted from different plant tissues using RNAiso Plus (Takara). First-strand cDNA was synthesized in 25 μl of reaction mixture containing 2 μg Dnase I-treated RNA, 200 U M-MLV reverse transcriptase (Takara), 40 U Recombinant RNase Inhibitor (Takara), and 0.1 μΜ oligodT. Quantitative RT-PCR was carried out in total volumes of 20 μl containing 10 μl SYBR EX Taq preimix (Takara), 0.4 μl Rox Reference Dye II (Takara), 0.2 mΜ gene-specific primers, and 2 μl of first-strand cDNA on an ABI 7500 real time PCR system. OsActin1 was used as a reference.
Subcellular localization
The ProSuper::Gnp4-GFP, Pro35S::OsIAA3-GFP, or Pro35S::OsIAA17-GFP plasmids were transformed into A. tumefaciens EHA105 and together with the p19 strain and mCherry marker were suspended and mixed in a solution containing 10 mM 2-(N-morpholino) ethanesulfonic acid, 10 mM MgCl2, and 150 μM acetosyringone. After storing at 28 °C for 2 h in darkness, the mixed solution was co-infiltrated into epidermal leaf cells of Nicotiana benthamiana. After 3 d of incubation at 25 °C, the leaves were sampled for confocal microscopy (OlympusFV1000). The GFP and mCherry markers were excited with a 488-nm and 543-nm laser, respectively. Emission spectra were collected at 500–550 nm for GFP, and 565–615 nm for the mCherry marker.
Yeast two-hybrid assays
The full-length and truncated fragment series of Gnp4 were amplified and recombined into a linearized pBGKT7 vector digested with NdeI and EcoRI according to the manufacturer’s manual for the Seamless Assembly Cloning Kit (CloneSmarter, C5891). pGADT7-OsIAA3 and pGADT7-OsIAA17 plasmids were extracted from the positive clone selected from the IRAT109 cDNA library constructed by the Takara company. The bait and prey were introduced into yeast strain AH109 by the polyethylene glycol-mediated method. Experimental procedures for screening interacting candidates and plasmid isolation were conducted according to the manufacturer’s user guide (Clonetech, PT3024-1).
Bimolecular fluorescence complementation (BiFC) assays
The full-length coding sequence of Gnp4 without a stop codon was amplified and recombined into linearized pSPYCE(M) vectors (Waadt et al., 2008) to construct Gnp4-YFPC. Similarly, the full-length coding sequences of OsIAA3 and OsIAA17 were cloned into pSPYNE173 to construct OsIAA3-YFPN and OsIAA17-YFPN, respectively. These plasmids were transformed into A. tumefaciens EHA105. For transient expression the strains, together with the p19 strain and mCherry ER-rk CD3-959 (Nelson et al., 2007), were co-infiltrated in 5–6-week-old N. benthamiana leaves. Tobacco epidermal leaf cells were observed with a confocal microscope (Olympus FV1000) 3 d after infiltration.
Co-immunoprecipitation assays
The coding sequences of OsIAA3 and OsIAA17 were amplified and cloned into the ProSuper::Myc vector to construct ProSuper::OsIAA3-Myc and ProSuper::OsIAA17-Myc, respectively. The full-length coding sequence of Gnp4 without the stop codon was amplified and recombined into a linearized ProSuper::Myc vector to construct ProSuper::Gnp4-Myc. Similarly, Pro35S::HF-Gnp4 was constructed by recombining the full-length Gnp4 with the Pro35S:HF vector. Co-immunoprecipitation was conducted as described previously (Zhang et al., 2017).
Yeast three-hybrid assays
To construct the yeast three-hybrid plasmids, full-length OsIAA3 and OsIAA17 were amplified and recombined into the MCSI location of the pBridge vector in the EcoRI-BamHI site, resulting in BaitI. For construction of BaitII and BaitIII, the truncated fragment and full-length Gnp4 were amplified and recombined into the MCSII location of BaitI in the NotI-BglII site, respectively. OsARF25-pGADT7 was used as prey. Bait and prey were co-introduced into yeast strain AH109 and incubated at 30 °C for 3–5 d, when equal optical-density yeast cells were plated out on selective medium. Qualitative evaluation was made of the interaction activity between bait and prey.
Determination of β-galactosidase activity
β-galactosidase activity assays with minor modification were conducted as previously described (Kippert, 1995). Yeast cells were collected and re-suspended in 800 μl Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0), and placed on ice. β-galactosidase assays were conducted after equilibration at 30 °C for 15 min; 160 µl of 4 mg ml–1 o-nitrophenyl-β-d-galactoside (ONPG) was added and the mixture was thoroughly vortexed before incubatation at 30 °C. The reaction was stopped by addition of 400 μl 1M Na2CO3. The OD550 and OD420 values were determined. Three replicates were performed, each with five technical replicates.
Scanning electron microscopy
Samples were fixed in 2.5 % glutaraldehyde and vacuumized for 30 min, then stored overnight at 4 °C. The samples were subjected to dehydration in an ethanol gradient series: 50% ×2–3 times, 70%, 80%, 90%, and 95%, each for 15 min, followed by two 20 min treatments with 100% ethanol. The dehydrated samples were treated with a mixture of equal volumes of ethanol and isoamyl acetate for 30 min and with isoamyl acetate for 1–2 h. After critical-point drying they were coated with gold using ion-beam sputtering for deposition and observed using a S-3000N scanning electron microscope (Hitachi, Tokyo, Japan).
Transient transcriptional activity assays
The effector, reporter, and internal control plasmids were transformed into rice protoplasts using the PEG-mediated method. Total proteins were extracted using lysis buffer (Promega, E4550) after incubation at 25 °C for 12–16 h. GUS and firefly luciferase (LUC) activities were assessed as previously described (Zhang et al., 2017).
Transcriptome analysis
High-quality total RNA was extracted from 10 young panicles (1 cm in length) of Gnp4-overexpression and wild-type plants. Illumina sequencing was performed using a HiSeq2000 system at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. Gene Ontology (GO) analysis was conducted by searching the differentially expressed genes (DEGs) against the AgriGO database of Oryza sativa subsp. japonica (http://bioinfo.cau.edu.cn/agriGO/index.php). Analysis of the significantly enriched pathways was conducted using the KEGG database (http://genome.jp/kegg/). Protein domain analyses of DEGs were conducted at DAVID (https://david.ncifcrf.gov/tools.jsp). The file containing all the 2-kb promoter sequences of the rice genome was downloaded from RAP-DB (http://rapdb.dna.affrc.go.jp/) and filtered to obtain sub-files for AuxRE cis-element analysis using Perl script.
Yeast one-hybrid assays
To construct the ProOsERF142::LacZ and ProOsERF3::LacZ reporters, different promoter regions were recombined into the EcoRI and XhoI sites of the pLacZi2μ vector, respectively. For construction of pB42AD-OsARF25, full-length OsARF25 was recombined into the EcoRI and XhoI sites of the pB42AD (pJG4-5) vector. The pB42AD-OsARF25 plasmid and the reporter constructs were co-transformed into the yeast EGY48 strain. Transformants were grown on SD/–Trp–Ura drop-out plates containing X-β-gal for blue colour development to detect the interaction. The ProFHY1::LacZ reporter and pB42AD-FHY3 were used as positive controls (Li et al., 2010).
ChIP-qPCR assays
The young panicles of Pro35S::OsARF25-FLAG plants were harvested and cross-linked, and then assays were conducted using a ChIP Assay Kit (P2078, Beyotime, China) according to the manufacturer’s instructions. The enriched DNA fragments were analysed by qRT-PCR using ABI7500 system and Software v2.0.5.
Haplotype and nucleotide diversity analysis
The data for single nucleotide polymorphisms (SNPs) used for haplotype analysis were downloaded from the rice 3K project (RFGB, http://www.rmbreeding.cn/Index/) (Zheng et al., 2015). Different haplotypes were obtained using DnaSP5.10 (Librado and Rozas, 2009) and a neighbor-joining tree for haplotypes was conducted using MEGA5.0 (Tamura et al., 2011). Nucleotide diversity analysis was conducted as previously described (Sun et al., 2018).
Accession numbers
Sequence data from this work can be found in the GenBank/EMBL databases under following accession numbers: Gnp4/LAX2 (Os04g0396500, KY673700), OsIAA3 (Os01g0231000), OsIAA17 (Os05g0230700), OsARF25 (Os12g0613700), OsERF142/SMOS1 (Os05g0389000), and OsERF3 (Os01g0797600).
Results
The gnp4 mutant shows longer grain length
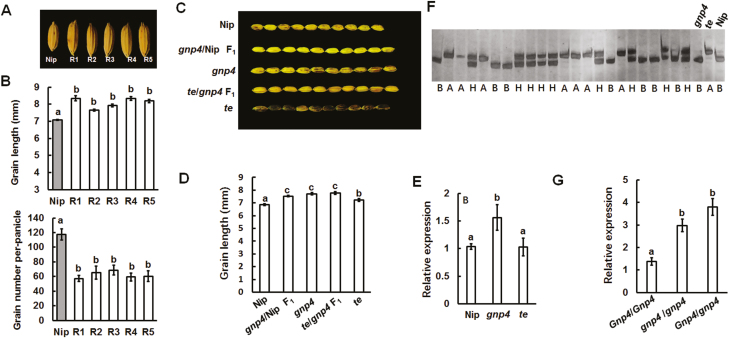
In our previous work, Gnp4 was narrowed to a 10.7-kb region on chromosome 4, in which there was one predicted ORF (Os04g0396500). However, there was no DNA sequence difference between wild-type (WT) Nipponbare and the gnp4 mutant except for different DNA methylation levels of several nucleotides in the promoter region (Zhang et al, 2011b). We found that the increased grain length and reduced grain number were closely linked. (Fig. 1A, B). The F1 seeds from the cross between gnp4 and the WT or gnp4 and te, a rice tillering mutant (Lin et al., 2012), showed similar grain length to gnp4 (Fig. 1C, D). The expression level of Gnp4 was consistently higher in the gnp4 mutant than the WT and te, and in the gnp4/gnp4 and Gnp4/gnp4 individuals compared with Gnp4/Gnp4 individuals (Fig. 1E–G).
Fig. 1.
High expression of Gnp4 correlates with longer grain length. (A) Grain morphologies of wild-type Nipponbare (Nip) and five recessive F2 segregates (R1–R5) of the cross between Nip and gnp4. (B) Grain length and grain number for Nip and the five recessive plants. Data are means (±s.e.m.) (n=50 grains from three panicles). (C, D) Grain morphology (C) and grain length (D) for Nip, gnp4, te, and F1 plants (n=50 grains). (E) The relative expression of Gnp4 in Nip, gnp4, and te (n=4 plants). (F) Genotype analysis of gnp4/gnp4, Gnp4/gnp4, and Gnp4/Gnp4 individuals using SDS-PAGE. (G) The relative expression of Gnp4 in gnp4/gnp4, Gnp4/gnp4, and Gnp4/Gnp4 individuals. Data are means (±s.e.m.) (n=5 plants). Different letters indicate significant differences between means according to LSD tests (P>0.05).
Next, we examined correlations between the expression levels of Gnp4 and grain length and grain number per panicle in 17 japonica accessions randomly selected from the rice mini core collection (Zhang et al., 2011a). We found that grain length but not grain number per panicle was correlated with Gnp4 expression levels over 3 years (Supplementary Fig. S1A, B). These results indicated that the gnp4 mutation was an epigenetic change that might be correlated with longer grain length with higher expression levels of Gnp4.
Overexpression of Gnp4/LAX2 increases grain length
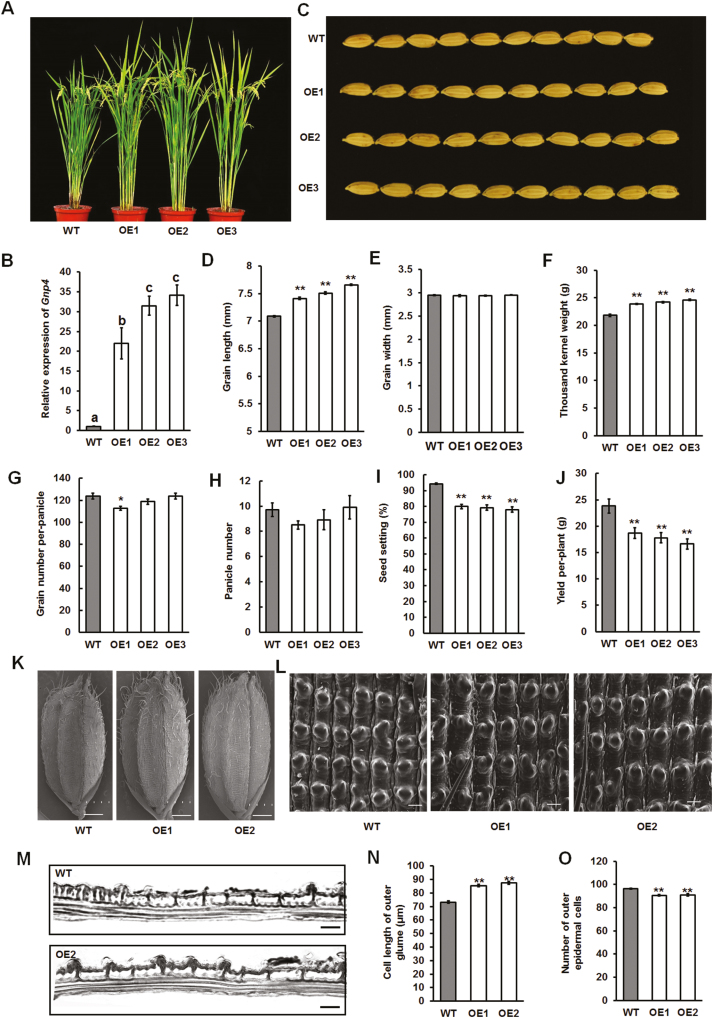
To confirm whether the expression level of Gnp4 correlated with the grain length, an overexpression construct (Pro35S::Gnp4) was introduced into the Nipponbare wild-type (WT). Three overexpression lines (OE1, OE2, and OE3) showed significantly increased grain length compared to WT plants (Fig. 2A–C). The mean grain length in the lines OE1, OE2, and OE3 were about 7.41 mm, 7.51 mm, and 7.66 mm, respectively, compared to 7.09 mm in WT plants (Fig. 2D). In addition, thousand-kernel weight was increased by 9.5%, 11%, and 12.8% in the OE1, OE2, and OE3 lines, respectively, compared to WT plants, but there was little change in grain width (Fig. 2E, F). However, there was reduced yield per plant as seed setting was reduced in the Gnp4 overexpression plants compared with the WT and there were no significant differences in grain number per panicle (except for OE1) and panicle number (Fig. 2G–J).
Fig. 2.
Gnp4/LAX2 is a regulator of grain length in rice. (A) Morphology of Gnp4-overexpression and wild-type (WT) Nipponbare plants at the mature growth stage. (B) Relative expression levels of Gnp4 in the WT and three transgenic plants. Data are means (±s.e.m.) (n=3 plants, each with three technical repeats). Different letters indicate significant differences between means according to LSD tests (P>0.05). (C) Phenotype of grains from Gnp4-overexpression and WT plants. (D–J) Grain length (D), grain width (E), thousand-kernel weight (F), grain number per panicle (G), panicle number (H), seed setting (I), and yield per plant (J) of Gnp4-overexpression and WT plants. Data are means (±s.e.m.) (n=20 plants). (K) SEM of glumes of WT and Gnp4-overexpression plants. Scale bars are 1 mm. (L) Enlarged images of the outer surfaces of glumes. Scale bars are 50 μm. (M) Longitudinal sections of the lemma before flowering. Scale bars are 50 μm. (N) Cell length (N) and number (O) of outer glumes of Gnp4-overexpression and WT plants. Data are means (±s.e.m.) (n=12 grains). Significant differences compared with the WT were determined using Student’s t-test: *P<0.05, **P<0.01.
The spikelet glumes in rice set a limit to the final grain size, and this is determined by co-ordinated cell expansion and cell proliferation (Li and Li, 2016). We found that the mean cell length in Gnp4 overexpression plants was significantly increased compared to the WT, but the number of epidermal cells in outer glume region decreased (Fig. 2K–O). Thus, the longer grains in Gnp4 overexpression plants mainly resulted from enhanced cell expansion in the spikelet hulls. In addition, the expression levels of several genes conferring larger grain size by cell expansion were much higher in Gnp4 overexpression plants than that the WT, including GL7, GLW7, POSITIVE REGULATOR OF GRAIN LENGTH 1 (PGL1), and POSITIVE REGULATOR OF GRAIN LENGTH 2 (PGL2) (Supplementary Fig. S2; Heang and Sassa, 2012; Wang et al., 2015). Taken together, these results showed that overexpression of Gnp4 could increase rice grain length by promotion of cell expansion.
Subcellular localization and expression patterns of Gnp4/LAX2
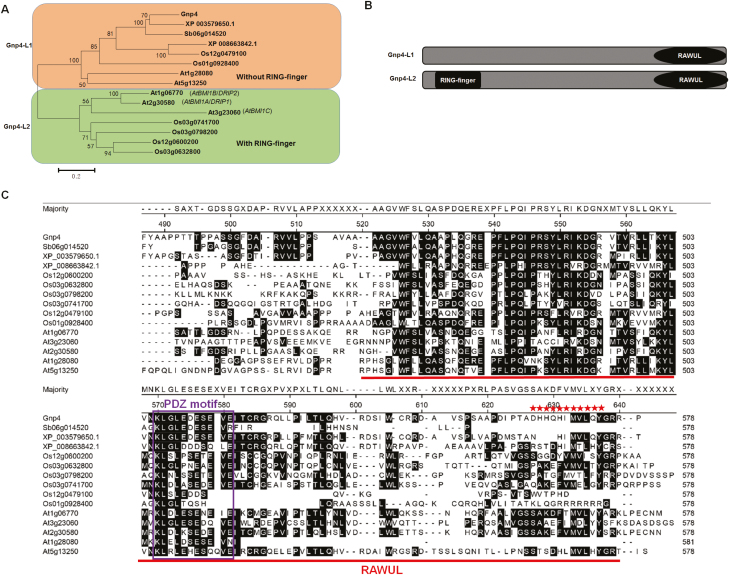
Gnp4 is predicted to encode a 394-amino acid protein. We identified six Gnp4 paralogs in rice, and five orthologues in Arabidopsis, one in maize, one in sorghum, and one in brachypodium distachyon (Fig. 3A). In all cases, the proteins contained a conserved RAWUL domain at the C-terminus, and they could be classified into two groups, namely Gnp4-LIKE1 (Gnp4-L1) and Gnp4-LIKE2 (Gnp4-L2) according to the amino acid sequences at the N-termini (Fig. 3B, C). A C3H4-type zinc ring finger was present in the Gnp4-L2 group, whereas the Gnp4-L1 group was characterized by numerous stretches of the same amino acid residue, such as Arg, His, and Ser. Gnp4 belonged to the Gnp4-L1 group (Supplementary Dataset S1).
Fig. 3.
Phylogenetic tree and analysis of amino acids of Gnp4/LAX2. (A) Phylogenetic relationship among Gnp4 and homologous plant proteins. The tree was generated using the MEGA6.0 program by the neighbor-joining method. (B) Schematics of the Gnp4-L1 and Gnp4-L2 protein groups. The RING finger and RAWUL domains are indicated. (C) The amino acids of RAWUL in Gnp4 and homologous proteins. Alignment was conducted using the Megalign program in the DNASTAR software package. The RAWUL domain is indicated in red, the purple box indicates the PDZ binding motif, and the red stars indicate the amino acids required for its interaction with OsIAA3 and OsIAA17.
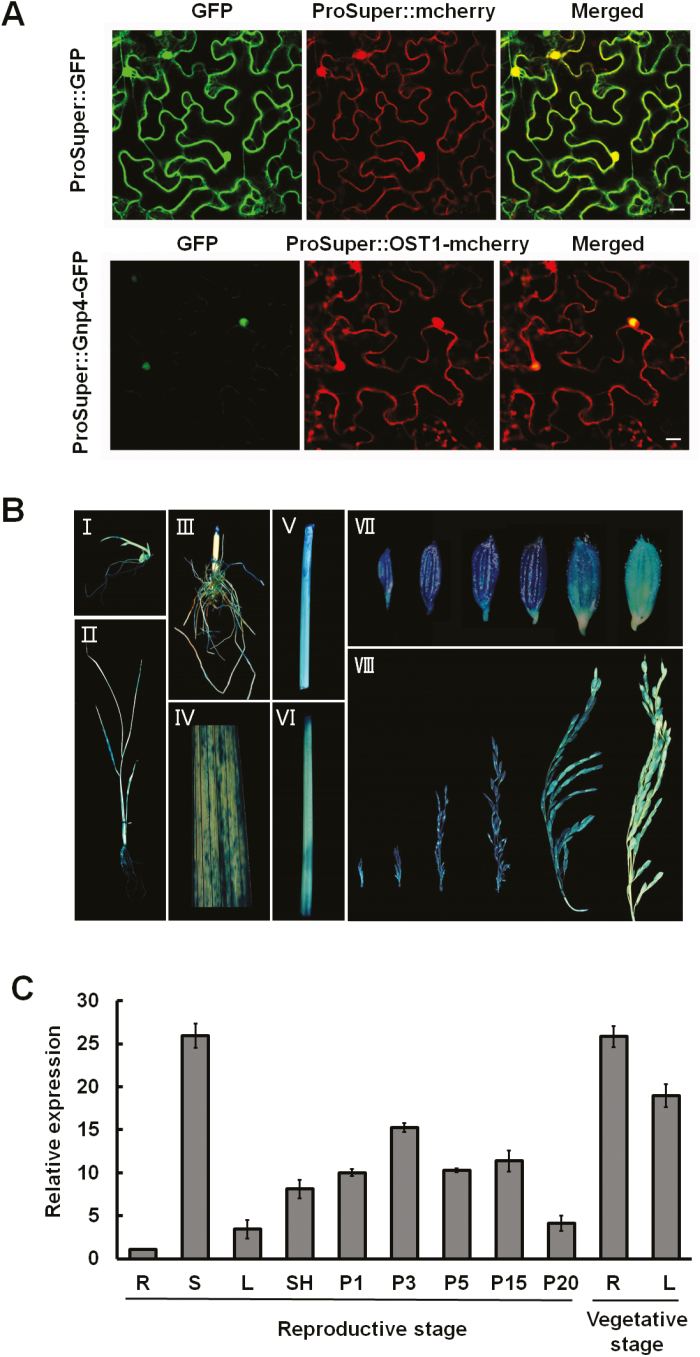
Nicotiana benthamiana leaves infiltrated with Agrobacterium harboring the ProSuper::Gnp4-GFP construct showed clear GFP signals in the nuclei, indicating that Gnp4 is a nuclear protein (Fig. 4A), which was consistent with the subcellular location of LAX2 in rice root cells (Tabuchi et al., 2011). Histochemical analysis of different tissues from ProGnp4::GUS transgenic plants showed that Gnp4 was widely expressed in both the vegetative and reproductive tissues, but was especially higher in stems and young panicles (Fig. 4B), consistent with previous results (Supplementary Fig. S3). RNA from roots, leaves, stems, sheaths, and panicles of different lengths were isolated and used for quantitative RT-PCR analysis of Gnp4 expression, and the results were in agreement with the histochemical analysis (Fig. 4C). The expression pattern of Gnp4/LAX2 was thus consistent with a role in regulating grain length.
Fig. 4.
Subcellular localization and expression pattern analyses of Gnp4/LAX2. (A) Subcellular location of ProSuper::GFP and ProSuper::Gnp4-GFP in tobacco leaf cells. ProSuper::OST1-mcherry is a mCherry marker localized in the nucleus and cytosol (Ding et al., 2015). Scale bars are 20 μm. (B) GUS staining of various tissues of transgenic plants containing the ProGnp4::GUS construct. I, bud; II, seedling; III, root; IV, leaf; V, stem; VI, sheath; VII, glumes; and VIII, panicles at different stages. (C) qRT-PCR analysis of relative expression levels of Gnp4 in different tissues from wild-type Nipponbare. R, root; S, stem; L, leaf; SH, leaf sheath; and P, panicles at sequential lengths, where the number indicates the panicle length (cm). Data are means (±s.e.m.) (n=3 plants each with three technical repeats).
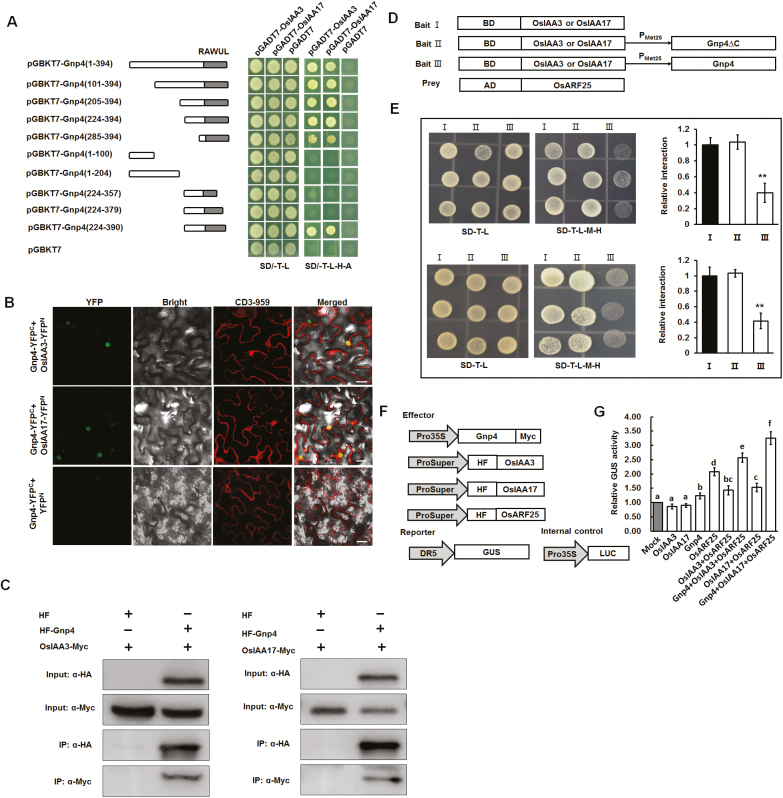
Gnp4/LAX2 interacts with OsIAA3 and OsIAA17 in yeast and plant cells
To elucidate the potential mechanism by which Gnp4 influences grain length, a yeast two-hybrid assay was performed to identify interacting proteins. Auto-transcriptional activation activity was not detected with the full-length and truncated fragment constructs of Gnp4 (Supplementary Fig. S4). Consequently, the full-length Gnp4 was used as a bait to screen two yeast prey cDNA libraries. A total of 23 candidate interacting proteins were isolated, among which OsIAA3 (Os01g0231000, referred to as OsIAA3 by Jain et al., 2006) and OsIAA17 (Os05g0230700), the nearest homologous protein of OsIAA3 in rice, were identified (Fig. 5A; Supplementary Fig. S5; Supplementary Table S1). Moreover, subcellular localization showed that OsIAA3 and OsIAA17 also localized in the nucleus and showed similar expression patterns to Gnp4 (Supplementary Fig. S6).
Fig. 5.
Gnp4/LAX2 interferes with the OsIAA3–OsARF25 and OsIAA17–OsARF25 interactions. (A) Gnp4 interacts with OsIAA3 and OsIAA17 in yeast cells. The RAWUL domain of Gnp4 is indicated by the gray boxes. SD/–T–L, selective medium lacking Trp and Leu; SD/–T–L–H–A, selective medium lacking Trp, Leu, His, and Ade. (B) BiFC assays showing the interactions between Gnp4 and OsIAA3 and OsIAA17 in tobacco leaf epidermal cells. CD3-959 is a mCherry marker (ER-rk CD3-959). Scale bars are 25 μm. (C) Co-immunoprecipitation assays showing that HF-Gnp4 interacts with OsIAA3-Myc and OsIAA17-Myc in plant cells. (D) Schematics of baits and prey used for yeast three-hybrid assays. (E) Yeast three-hybrid assays showing that the OsIAA3–OsARF25 (top panel) and OsIAA17–OsARF25 (bottom panel) interactions were suppressed in the presence of full-length Gnp4. Relative interaction activities were evaluated using β-galactosidase assays. Data are means (±s.e.m). Three replicates were performed, each with five technical replicates. Significant differences were determined using Student’s t-test: **P<0.01. (F, G) Transient assays showing the effect of Gnp4 on the OsIAA3–OsARF25 and OsIAA17–OsARF25 interactions. Relative GUS and LUC activities were measured and normalized to LUC activity. Data are means (±s.e.m.), four replicates were performed, each with five technical replicates. Different letters indicate significant differences between means according to LSD tests (P>0.05).
Next, we found that constructs containing the RAWUL domain of Gnp4 (amino acids 101 to 394, 205 to 394, 224 to 394, 285 to 394, and 224 to 390) interacted with OsIAA3 and OsIAA17, but no interaction was observed with the constructs lacking the region corresponding to amino acids 380 to 390 (Fig. 5A). These results suggested that these 11 amino acids at the C-terminus were required for interaction between Gnp4 and OsIAA3 and OsIAA17. Furthermore, Tabuchi et al. (2011) identified the last 15 amino acids of LAX2 as being important for its interaction with LAX1, indicating the potentially critical function of the RAWUL domain for protein–protein interaction. However, LAX1 was not pulled down by Gnp4 in our yeast two-hybrid assays.
To further characterize the interactions between Gnp4 and OsIAA3 and OsIAA17 in plant cells, a BiFC assay was conducted by transient expression in tobacco leaf cells. As anticipated, fluorescence signals were observed in the nuclei of leaf cells where Gnp4-YFPC was co-expressed with OsIAA3-YFPN or OsIAA17-YFPN, but not with YFPN alone (Fig. 5B). We also found that HF-Gnp4 proteins interacted with OsIAA3-Myc and OsIAA17-Myc proteins in tobacco leaves, but not HF proteins alone (Fig. 5C). Collectively, these results indicated that Gnp4 interacted with OsIAA3 and OsIAA17 in plant cells as well as in yeast cells.
Gnp4/LAX2 forms a dimer and interferes with the interaction between OsAUX/IAA and OsARF
Although Gnp4 is a nuclear protein, no transcriptional activity or recognizable DNA binding domain were found, indicating that it might not regulate transcription directly. We found that Gnp4 could form dimers in yeast and plant cells (Supplementary Fig. S7). Aux/IAA proteins are well established as transcriptional repressors of the ARFs that play a key regulatory role in plant growth and development (Salehin et al., 2015). Thus, we speculated that Gnp4 might function by modulating the interaction between OsAUX/IAA and OsARF. It is well documented that OsIAA3 and OsIAA17 interact with eight OsARF activators (Shen et al., 2010), among which OsARF25 showed a similar expression pattern to Gnp4 and high expression levels during inflorescence development (Supplementary Fig. S8). Next, a yeast three-hybrid assay was performed to explore the effect of Gnp4 on the OsARF25–OsIAA3 and OsARF25–OsIAA17 interactions. We constructed three kinds of bait (Bait I, II, and III) to test for interactions with OsARF25 as prey. Bait I contained only the full-length OsIAA3 or OsIAA17, Bait II contained the C terminus-truncated Gnp4 (Gnp4∆C), and Bait III contained an entire Gnp4 (Fig. 5D). Transcription of Gnp4∆C and Gnp4 can be conditionally regulated from the Met25 promoter (ProMet25), which is actively repressed in the presence of methionine but not in its absence. We found that yeast harboring Bait III grew much more slowly than that containing Baits I or II on selective medium lacking methionine (Fig. 5E). Moreover, β-galactosidase assays showed that the OsARF25–OsIAA3 and OsARF25–OsIAA17 interactions were suppressed when Gnp4 was expressed, but not when Gnp4∆C was expressed (Fig. 5E), suggesting that Gnp4 functioned as a ‘blocker’ of the interactions, at least in yeast cells.
A transient transcriptional activity assay was then conducted to confirm that Gnp4 suppressed the OsARF25–OsIAA3 and OsARF25-OsIAA17 interactions in planta. We found that the relative GUS activity increased significantly when the reporter was co-expressed with Pro35S::Gnp4-Myc (Fig. 5F, G). Together, these results indicated that Gnp4 interfered with the OsIAA3–OsARF25 and OsIAA17–OsARF25 interactions, which may have de-repressed OsARF25 and enhanced the transcription of its downstream target genes in plant cells.
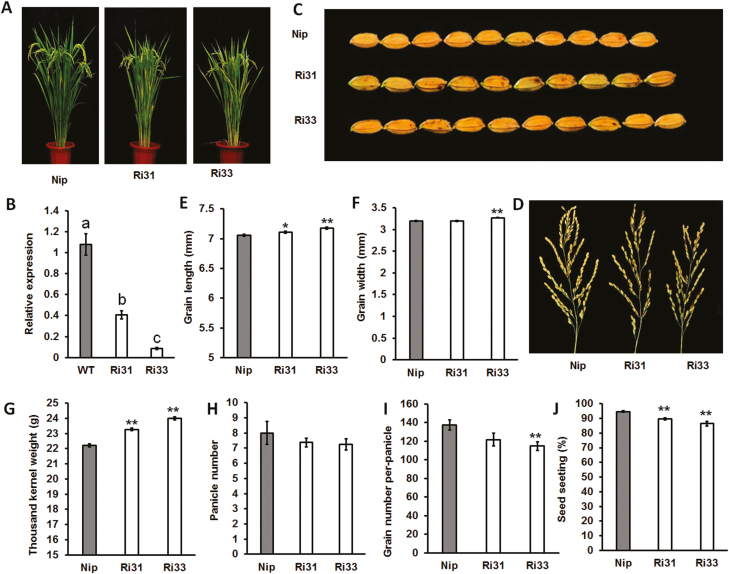
Characterization of grain length in OsIAA3-RNAi and osarf25 plants
We next reasoned that the reduced expression of the OsAUX/IAA genes might mimic Gnp4/LAX2 overexpression. We constructed a RNAi vector of OsIAA3 and transformed it into the Nipponbare wild-type (WT). Two independent OsIAA3-RNAi plants, namely Ri31 and Ri33, were selected for detailed phenotypic analysis (Fig. 6A, B). We also checked the expression levels of several near-homologous genes of OsIAA3 and found no significant differences between the WT and Ri31 or Ri33, except that OsIAA19 was down-regulated in Ri31 (Supplementary Figs S5, S9). Compared with WT plants, grain length significantly increased in Ri31 and Ri33, and wider grains were observed in Ri33 (Fig. 6C–F). The thousand-kernel weights were increased by 4.7% and 8.0 % in Ri31 and Ri33 plants, respectively, compared to the WT (Fig. 6G). Similar to Gnp4-overexpression plants, Ri31 and Ri33 showed significantly decreased seed setting, but similar grain numbers per panicle (except for Ri33), and panicle numbers (Fig. 6H–J). These data suggested that the knockdown of OsIAA3 expression had a positive effect on grain length.
Fig. 6.
Characterization of OsIAA3-RNAi Plants. (A) Morphology of OsIAA3-RNAi plants (Ri) compared with wild-type Nipponbare (Nip) at a mature growth stage. (B) Relative expression levels of OsIAA3 in Nip and two RNAi plants. Data are means (±s.e.m.) (n=3 plants, each with three technical repeats). Different letters indicate significant differences between means according to LSD tests (P>0.05). Phenotypes of (C) grains and (D) panicles from OsIAA3-RNAi and Nip plants. Grain length (E), grain width (F), thousand-kernel weight (G), panicle number (H), grain number per panicle (I), and seed setting (J) of Nip and OsIAA3-RNAi plants. Data are means (±s.e.m.) (n=15 plants). Significant differences compared with wild-type Nip were determined using Student’s t-test: *P<0.05, **P<0.01.
To clarify the function of OsARF25, we obtained a T-DNA insertion mutant, osarf25, in the variety Hwayoung (HY) background. Homozygous mutant plants were identified by genomic DNA and mRNA levels (Supplementary Fig. S10). Phenotypic analysis showed that osarf25 had smaller grains and panicles. The thousand-kernel weight and grain numbers per panicle were significantly reduced in osarf25 relative to HY (Fig. 7). In addition, we found that the outer glume cell length, but not cell number, was significantly lower in osarf25 than in HY (Supplementary Fig. S11). Hence, we concluded that OsARF25 modulated grain length by cell expansion, as does Gnp4.
Fig. 7.
Characterization of osarf25 plants. (A) Morphology of wild-type Hwayoung (HY) and osarf25 plants at a mature growth stage. Phenotypes of (B) grains and (C) panicles from HY and osarf25 plants. Grain length (D), grain width (E), thousand-kernel weight (F), and grain number per panicle (G) of osarf25 and HY plants. Data are means (±s.e.m.) (n=15 plants). Significant differences compared with wild-type HY were determined using Student’s t-test: *P<0.05, **P<0.01.
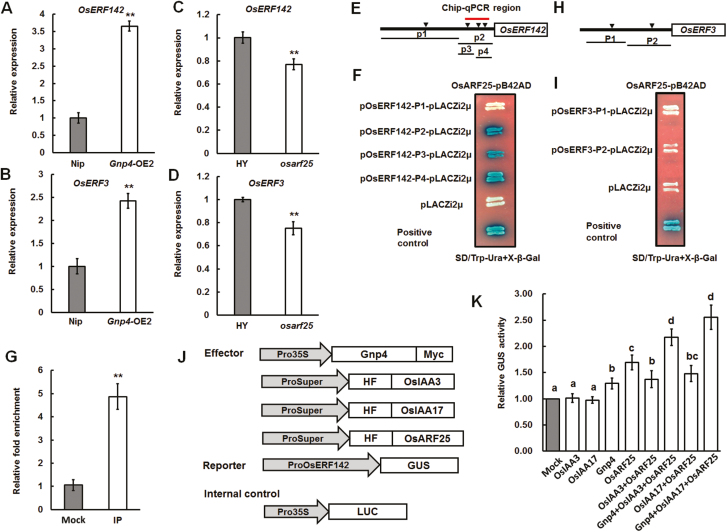
The grain-size regulators OsERF142/SMOS1 and OsERF3 function downstream of Gnp4/LAX2 and OsARF25
To further investigate the downstream targets regulated by Gnp4, transcriptome analysis was performed for Gnp4-overexpression and wild-type plants. A total 846 and 449 genes were up- and down-regulated, respectively, in Gnp4-overexpression plants compared to the WT (Supplementary Dataset S2). The expression levels of several genes were quantified by qRT-PCR to confirm the RNA-seq results (Supplementary Fig. S12). Gene ontology (GO) analysis showed that genes affected by the overexpression of Gnp4 were significantly enriched in 40 GO terms (Supplementary Table S2). Among these terms, several specific GO keywords were consistent with the molecular function of Gnp4, such as GO: 0045449 (regulation of transcription), GO: 0003700 (transcription factor activity), GO: 0030528 (transcription regulator activity), and GO: 0005634 (nucleus) (Supplementary Fig. S13). Two KEGG pathways were enriched, namely ko04075 (plant hormone signal transduction) and ko04626 (plant–pathogen interaction) (Supplementary Fig. S14). These results were consistent with our proposed role for Gnp4 in the regulation of grain size.
We also analysed the gene functional categories of the up- and down-regulated genes using the DAVID database. Five terms were significantly enriched for the up-regulated genes and one term was enriched for down-regulated genes (Supplementary Table S3). Among the up-regulated genes, the most significantly enriched was the AP2 (APETALA2) domain, which plays roles in various biological processes of plant development and abiotic and biotic stress responses (Licausi et al., 2013). These results provided hints on the downstream targets of Gnp4 that are potentially involved in regulating grain length. A total of 28 genes encoding AP2 domain-containing proteins were found to be up-regulated in Gnp4-overexpression plants and were selected for further investigation. Interestingly, the AuxRE (TGTCTC) cis-element was found in the 2-kb promoter region of 18 of these genes (Supplementary Table S4). Among them, OsERF142 (also known as SMOS1 and SHB) is well known to influence organ size through the auxin signaling pathway, and to modulate root meristem size by influencing GA biosynthesis (Aya et al., 2014; Li et al., 2015), and OsERF3 is known to be involved in root development, drought tolerance, and defense responses in rice (Lu et al., 2011; Wan et al., 2011; Zhao et al., 2015). A recently isolated novel AP2 domain-containing protein, OsLG3/OsERF62, has been shown to be a positive regulator of grain length (Yu et al., 2017). Next, we generated OsERF3-overexpression plants in the Nipponbare background and found that several independent transgenic lines showed significant increases in grain length and thousand-kernel weight compared to the WT plants, indicating that OsERF3 was a positive regulator of grain length (Supplementary Fig. S15). We thus speculated that OsERF142 and OsERF3 might be common targets of OsARF25 and act downstream of Gnp4.
To test the hypothesis, we first checked the expression levels of OsERF142 and OsERF3, and found that the mRNA abundance was much higher in Gnp4-overexpression plants than in the WT (Fig. 8A, B). In contrast, reduced expression of OsARF25 led to significantly decreased expression levels of OsERF142 and OsERF3 in the osarf25 mutant in the HY background (Fig. 8C, D). Next, yeast one-hybrid and CHIP-qPCR assays confirmed that OsARF25 binds to the P2 region of ProOsERF142. (Fig. 8E–G). However, we could not detect any interactions between OsARF25-pB42AD and fragments of ProOsERF3 in yeast cells (Fig. 8H, I). In addition, we found elevated GUS activity driven by the OsERF142 promoter when Gnp4 was co-expressed compared with controls (Fig. 8J, K). These results collectively indicated that OsERF142 and OsERF3 function downstream of Gnp4 and OsARF25, and that OsARF25 binds to the promoter of OsERF142/SMOS1 and positively regulates its expression.
Fig. 8.
Gnp4/LAX2 regulates OsERF142 and OsERF3 expression. (A–D) Relative expression levels of OsERF142 and OsERF3 in Gnp4-overexpression and wild-type Nipponbare (Nip) plants or in osarf25 and wild-type Hwayoung (HY). Data are means (±s.e.m.) (n=3 plants each with three technical repeats). Student’s t-test. **P<0.01. (E, F) ProOsERF142 used in yeast one-hybrid assays showing the binding activity of OsARF25 to the P2 region of ProOsERF142. AuxRE cis-elements are indicated with black triangles. (G) ChIP–qPCR indicating the binding of OsARF25 to the P2 region of ProOsERF142. Data are means (±s.e.m.) (n=3). Student’s t-test: **P<0.01. (H, I) ProOsERF3 used in yeast one-hybrid assays showing no interaction between OsARF25 and fragments of ProOsERF3. (J, K) Effectors, reporter, and internal control used in the transient assays to determine the effect of Gnp4 on transcription of OsERF142. Data are means (±s.e.m.). There were four replicates, each with five technical replicates. Different letters indicate significant differences between means according to LSD tests (P>0.05).
Nucleotide diversity and haplotype analysis of Gnp4/LAX2
In previous studies, several quantitative trait loci located near Gnp4 conferring grain length have been isolated in different rice cultivars, such as GWT4a, qLWR4, and qGL4b (Lin et al., 1996; Ying et al., 2012; Segami et al., 2016). To investigate the natural variation of Gnp4 in different germplasm types, we further analysed the sequences of Gnp4 in 259 cultivated and nine wild rice types and found 17 haplotypes (Hap) based on 46 SNPs. Hap1-Hap5 and Hap8 were mainly present in the indica subpopulation (sub-I), Hap6, Hap7, Hap9, and Hap10 were mainly present in the japonica subpopulation (sub-J), and Hap11-Hap17 were present in wild rice (Supplementary Fig. S16; Supplementary Dataset S3). This analysis showed an obvious diversification of the Gnp4 locus in the indica and japonica subspecies. Furthermore, we found that the nucleotide diversity of Gnp4 in japonica (π=0.00016) was much lower than that in indica (π=0.00093) and in wild rice (π=0.0021). Significant Tajima’s D and Fu Li’s D values were also observed in japonica (Supplementary Table S5). These results indicated that Gnp4 alleles in the japonica subpopulation might have been selected during domestication.
Discussion
Gnp4/LAX2 functions as a subset of the auxin response pathway
Although the biosynthesis, transportation, and signal transduction processes of auxin have been well studied, the regulators of components of the auxin pathway are largely unknown. In this study, we found a component of auxin signaling involved in the regulation of grain length in rice. Gnp4/LAX2 physically interacted with OsIAA3 and OsIAA17 in yeast and plant cells, and affected the interaction between them with OsARF25 (Fig. 5). OsIAA3-RNAi plants displayed a phenotype with longer grain length similar to that of Gnp4-overexpression plants, while the loss-of-function osarf25 mutant had small grains (Fig. 7), leading us to propose that Gnp4 interfered with the OsIAA3–OsARF25 interaction and increased the expression of OsARF25 target genes, including OsERF142 and OsERF3. The rice smos1 (oserf142) mutant has small organ size due to decreased cell size (Aya et al., 2014). SMOS1 (OsERF142) interacts with SMOS2 (also known as Dwarf and Low Tillering, DLT) to from a complex that regulates the expression of its direct target, OsPHI-1, which is involved in cell expansion (Hirano et al., 2017). In addition, expression levels of OsERF142 and OsPHI-1 were decreased in plants overexpressing OsIAA3 (P58L, a constitutively active form of OsIAA31 named by Jain et al., 2006) (Hirano et al., 2017). We found that OsERF142 was up-regulated in Gnp4-overexpression plants compared to the Nipponbare wild-type (WT), but was down-regulated in osarf25 compared to the HY wild-type (Fig. 8A, C). In addition, up-regulation of OsPHI-1 and three OsPHI-1-like genes in Gnp4-overexpression plants were detected in our RNA-seq data (Supplementary Dataset S24). Moreover, microscopy showed that the cell length increased significantly in Gnp4-overexpression plants but decreased in osarf25, resulting from changes in cell expansion (Fig. 2K–O; Supplementary Fig. S11). Based on our results and previous reports, we propose that Gnp4 might function in an OsIAA3–OsARF25–OsERF142 pathway to regulate grain length as shown in Fig. 9. It has been reported that LAX2 interacts with LAX1, which is involved with the auxin and brassinosteroid signal transduction pathways, to regulate the process of axillary meristem formation; moreover, LAX1 interacts with LAX2, and LAX1-overexpression plants also show decreased seed setting, similar to Gnp4/LAX2-overexpression plants (Komatsu et al., 2003; Tabuchi et al., 2011). So Gnp4/LAX2 may function as a subset of the auxin response pathway to regulate axillary meristems and pollen formation or grain development through different interacting protein-dependent pathways.
Fig. 9.
A proposed model for the functions of Gnp4/LAX2. Gnp4/LAX2 encodes a RAWUL domain-containing protein. It is involved in axillary meristem formation and acts in the LAX1-dependent and LAX1-independent pathways (Tabuchi et al., 2011). In addition, Gnp4/LAX2 interacts with OsAUX/IAAs, such as OsIAA3 and OsIAA17. AUX/IAAs function in plant cells as transcription repressors of ARFs. With elevated mRNA levels, the abundance of Gnp4 possibly functions as a ‘blocker’ of the OsIAA3–OsARF25 interaction and thus enhances transcription of downstream target genes. OsARF25 acts upstream of OsERF142 and OsERF3 and directly binds to the promoter of OsERF142, which positively regulates cell expansion, consequently affecting phenotypic variation in grain length.
Gnp4/LAX2 functions as a regulator of protein interactions
Several studies on Gnp4-L2 proteins have been carried out in Arabidopsis. Overexpression of AtBMI1C accelerates flowering by repression of the FLOWERING LOCUS C (FLC) gene (Li et al., 2011a) and AtRING1A/B regulates cell-fate differentiation by suppressing expression of Class I KNOTTED-like homeobox (KNOX) genes (Xu and Shen, 2008). These proteins function as transcriptional repressors. Post-translational modification of potential targets of some of these Gnp4-L2 homologs have also been described, suggesting a possible E3 ligase activity of these ring finger domain-containing proteins. AtBMI1A/B, also known as DREB2A-INTERACTING PROTEIN1 (DRIP1), and DRIP2 negatively regulate the response to water stress through interaction with DREB2A and mediating its degradation (Qin et al., 2008). Here, we found that Gnp4 functions as a regulator of grain size in rice, indicating that the Gnp4-L1 and Gnp4-L2 proteins may use different mechanisms to regulate plant growth. Compared to Gnp4-L2, Gnp4-L1 proteins do not have a RING finger domain at the N terminus, which is required for the commonest class of E3 ubiquitin ligases, suggesting that they lack this activity. Moreover, a PDZ (post-synaptic density protein PSD95, Drosophila disc large tumor suppressor Dlg1 and tight junction protein ZO-1) binding motif (KLGLEDSEV) has been found in the RAWUL domain of Gnp4 (Wardell, 2013). This motif usually functions as a regulator of protein–protein interactions or dimer formation (Fanning and Anderson, 1996; Short et al., 1998) (Fig.3). We also found that 368 proteins in rice contained the core sequence ‘LGLE’ of the PDZ motif, including OsIAA3 (Supplementary Dataset S4). Together, our results showed that Gnp4 forms a dimer and functions as a regulator of protein–protein interactions (Fig. 5; Supplementary Fig. S7).
Manipulation of the expression level of Gnp4/LAX2 has the potential to improve grain yield
Our results demonstrated that Gnp4 functions as a regulator of grain length. Overexpression of Gnp4 significantly increased the grain length and thousand-kernel weight of rice, indicating its potential value for breeding. However, we found that seed setting was decreased in Gnp4-overexpression plants (Fig. 2). When we evaluated correlations of Gnp4 expression levels with seed setting and yield per plant in 17 japonica accessions, we found that accessions with 7–10-fold increases in Gnp4 expression levels relative to Nipponbare exhibited high yields, implying that there is an appropriate level of Gnp4 expression that might be required for yield improvement (Supplementary Fig. S1). Constitutive overexpression of CBP1 (CLUSTERED PRIMARY BRANCH 1) under the control of the maize ubiquitin promoter has been shown to increase grain length but not yield per plant, a result of other unfavorable agronomic traits; however, optimized expression of CBP1 using a panicle-specific promoter did result in improved yield in rice (Wu et al., 2016). In a similar way, it would be interesting to test whether controlled expression of Gnp4 in specific tissues could be used to improve yields in rice.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Analysis of the correlation between Gnp4/LAX2 mRNA levels and several agronomic traits in 17 japonica accessions.
Fig. S2. Relative expression levels of several genes related to grain length in rice.
Fig. S3. In silico expression analysis of Gnp4/LAX2.
Fig. S4. Auto-transcriptional activation activity analysis of Gnp4/LAX2 in yeast cells.
Fig. S5. Phylogenetic tree of Aux/IAA proteins in rice.
Fig. S6. Subcellular localization and expression pattern of OsIAA3 and OsIAA17.
Fig. S7. Gnp4 forms a dimmer in yeast and plant cells.
Fig. S8. Expression pattern analysis of OsARF25.
Fig. S9. Relative expression levels of nearest homologous genes of OsIAA3 in wild-type Nipponbare and OsIAA3-RNAi plants.
Fig. S10. Identification of osarf25.
Fig. S11. Scanning electron microscopy of glumes of wild-type Hwayoung and osarf25.
Fig. S12. Validation of transcriptome data by qRT-PCR.
Fig. S13. Gene ontology analysis of DEGs.
Fig. S14. KEGG pathway analysis of DEGs.
Fig. S15. Phenotypic analysis of OsERF3-overexpression and wild-type plants.
Fig. S16. Haplotype analysis of Gnp4/LAX2.
Table S1. Gnp4/LAX2-interacting proteins isolated by yeast two-hybrid assays.
Table S2. Enriched GO terms in significant DEGs.
Table S3. Significantly enriched protein domains of up- and down-regulated genes in Gnp4/LAX2-overexpression compared to wild-type plants.
Table S4. DGEs containing AP2 domains in Gnp4/LAX2-overexpression compared to wild-type plants.
Table S5. The nucleotide diversity of Gnp4/LAX2.
Dataset S1. Amino acid alignments of Gnp4/LAX2 and its homologous proteins.
Dataset S2. DEGs in Gnp4/LAX2-overexpression compared to wild-type plants.
Dataset S3. Details of Oryza sativa varieties and wild rice genotypes.
Dataset S4. Proteins in rice containing the core sequence ‘LGLE’ of the PDZ motif.
Dataset S5. Primers used in this study.
Author contributions
ZZ designed and performed the research and wrote the article; Jinjie Li contributed to supervising the research and revising the manuscript; ZT contributed to the transcriptome experiment; XS contributed to helping with the transgenic experiment; HZ supervised the research; JY contributed to helping with the haplotype analysis; GY, GL, HG, Jilong Li, WH, HH, YX, and ZY contributed to the preparation of samples or reagents; YQ contributed to the identification of the osarf25 mutant; RH contributed to the Pro35S::OsERF3 transgenic formation; WY contributed to transcriptome experiment; and ZL conceived the research and assisted in writing the manuscript.
Supplementary Material
Acknowledgments
We thank Shuhua Yang (China Agricultural University), Jose M. Alonso (North Carolina State University), and Robert A. McIntosh (University of Sydney) for critical reading and suggested revisions for the manuscript, De’an Jiang (Zhejiang University) for providing the osarf25 seeds, Hong’en Xu (Technical University of Munich, Germany) for help with cis-element and PDZ binding motif analysis, Dr Mingliang Xu (China Agricultural University) for providing the pTCK303 vector, Dr Zhijun Cheng (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences) for providing the BiFC vectors and the te mutant, Dr Lizhong Xiong for providing the DR5:GUS vector, and Dr Qingchang Liu and Dr Jigang Li (China Agricultural University) for providing the pLacZi2μ and pB42AD vectors. The research was supported by grants from the Ministry of Science and Technology in China (2016YFD0100300 and 2015BAD02B01), the National Natural Science Foundation of China (31171521), and the China Postdoctoral Science Foundation (2015M581206 and 2017T100117).
References
- Aya K, Hobo T, Sato-Izawa K, Ueguchi-Tanaka M, Kitano H, Matsuoka M. 2014. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant Cell Physiology 55, 897–912. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezfulian MH, Jalili E, Roberto DK, Moss BL, Khoo K, Nemhauser JL, Crosby WL. 2016. Oligomerization of SCFTIR1 is essential for Aux/IAA degradation and auxin signaling in Arabidopsis. PloS Genetics 12, e1006301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. [DOI] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S. 2015. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Developmental Cell 32, 278–289. [DOI] [PubMed] [Google Scholar]
- Duan P, Ni S, Wang J, Zhang B, Xu R, Wang Y, Chen H, Zhu X, Li Y. 2015. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nature Plants 2, 15203. [DOI] [PubMed] [Google Scholar]
- Fanning AS, Anderson JM. 1996. Protein–protein interactions: PDZ domain networks. Current Biology 6, 1385–1388. [DOI] [PubMed] [Google Scholar]
- Feng Z, Wu C, Wang C, et al. 2016. SLG controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. Journal of Experimental Botany 67, 4241–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A. 2013. The role of auxin in shaping shoot architecture. Journal of Experimental Botany 64, 2593–2608. [DOI] [PubMed] [Google Scholar]
- Heang D, Sassa H. 2012. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. Plos ONE 7, e31325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hirano K, Yoshida H, Aya K, et al. 2017. SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING form a complex to integrate auxin and brassinosteroid signaling in rice. Molecular Plant 10, 590–604. [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP. 2006. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Functional & Integrative Genomics 6, 47–59. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Bao L, Jeong SY, Kim SK, Xu C, Li X, Zhang Q. 2012. XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. The Plant Journal 70, 398–408. [DOI] [PubMed] [Google Scholar]
- Kippert F. 1995. A rapid permeabilization procedure for accurate quantitative determination of β-galactosidase activity in yeast cells. FEMS Microbiology Letters 128, 201–206. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J. 2003. LAX and SPA: major regulators of shoot branching in rice. Proceedings of the National Academy of Sciences, USA 100, 11765–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Liang W, Zhang X, Ren H, Hu J, Bennett M, Zhang D. 2014. Rice actin-binding protein RMD is a key link in the auxin–actin regulatory loop that controls cell growth. Proceedings of the National Academy of Sciences, USA 111, 10377–10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, et al. 2010. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. The Plant Cell 22, 3634–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhao Y, Chu H, et al. 2015. SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation. PLoS Genetics 11, e1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Li Y. 2016. Signaling pathways of seed size control in plants. Current Opinion in Plant Biology 33, 23–32. [DOI] [PubMed] [Google Scholar]
- Li W, Wang Z, Li J, Yang H, Cui S, Wang X, Ma L. 2011a. Overexpression of AtBMI1C, a polycomb group protein gene, accelerates flowering in Arabidopsis. PloS ONE 6, e21364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, et al. 2011b. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nature Genetics 43, 1266–1269. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P. 2013. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytologist 199, 639–649. [DOI] [PubMed] [Google Scholar]
- Lin H, Qian H, Zhuang J, Lu J, Min S, Xiong Z, Huang N, Zheng K. 1996. RFLP mapping of QTLs for yield and related characters in rice (Oryza sativa L.). Theoretical and Applied Genetics 92, 920–927. [DOI] [PubMed] [Google Scholar]
- Lin Q, Wang D, Dong H, et al. 2012. Rice APC/C(TE) controls tillering by mediating the degradation of MONOCULM 1. Nature Communications 3, 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Tong H, Xiao Y, et al. 2015. Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proceedings of the National Academy of Sciences, USA 112, 11102–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K. 2013. Auxin metabolism and homeostasis during plant development. Development 140, 943–950. [DOI] [PubMed] [Google Scholar]
- Lu J, Ju H, Zhou G, Zhu C, Erb M, Wang X, Wang P, Lou Y. 2011. An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. The Plant Journal 68, 583–596. [DOI] [PubMed] [Google Scholar]
- Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q. 2010. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proceedings of the National Academy of Sciences, USA 107, 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Umemura I, Gomi K, Hasegawa Y, Kitano H, Sazuka T, Matsuoka M. 2006. Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. The Plant Journal 46, 297–306. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Qi Y, Wang S, Shen C, Zhang S, Chen Y, Xu Y, Liu Y, Wu Y, Jiang D. 2012. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytologist 193, 109–120. [DOI] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LS, et al. 2008. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. The Plant Cell 20, 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Matsuoka M. 2008. Identifying and exploiting grain yield genes in rice. Current Opinion in Plant Biology 11, 209–214. [DOI] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M. 2015. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. The Plant Cell 27, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Sung ZR, Calonje M. 2008. RAWUL: a new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics 9, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segami S, Yamamoto T, Oki K, et al. 2016. Detection of novel QTLs regulating grain size in extra-large grain rice (Oryza sativa L.) lines. Rice 9, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Wang S, Bai Y, Wu Y, Zhang S, Chen M, Guilfoyle TJ, Wu P, Qi Y. 2010. Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.). Journal of Experimental Botany 61, 3971–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Wang S, Zhang S, Xu Y, Qian Q, Qi Y, Jiang D. 2013. OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant, Cell & Environment 36, 607–620. [DOI] [PubMed] [Google Scholar]
- Short DB, Trotter KW, Reczek D, Kreda SM, Bretscher A, Boucher RC, Stutts MJ, Milgram SL. 1998. An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. The Journal of Biological Chemistry 273, 19797–19801. [DOI] [PubMed] [Google Scholar]
- Si L, Chen J, Huang X, et al. 2016. OsSPL13 controls grain size in cultivated rice. Nature Genetics 48, 447–456. [DOI] [PubMed] [Google Scholar]
- Sun X, Zhang Z, Chen C, et al. 2018. The C-S-A gene system regulates hull pigmentation and reveals evolution of anthocyanin biosynthesis pathway in rice. Journal of Experimental Botany 69, 1485–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386. [DOI] [PubMed] [Google Scholar]
- Tabuchi H, Zhang Y, Hattori S, et al. 2011. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. The Plant Cell 23, 3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. 2008. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. The Plant Journal 56, 505–516. [DOI] [PubMed] [Google Scholar]
- Wan L, Zhang J, Zhang H, Zhang Z, Quan R, Zhou S, Huang R. 2011. Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PloS ONE 6, e25216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y. 2007. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. [DOI] [PubMed] [Google Scholar]
- Wang S, Li S, Liu Q, et al. 2015. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nature Genetics 47, 949–954. [DOI] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, et al. 2012. Control of grain size, shape and quality by OsSPL16 in rice. Nature Genetics 44, 950–954. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K. 2004. A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Molecular Biology Reporter 22, 409–417. [Google Scholar]
- Wardell B. 2013. Evolution of barren STALK2/LAX PANICLE2 (BA2/LAX2) in angiosperms. Masters Thesis, California State University, Long Beach, CA. [Google Scholar]
- Weng J, Gu S, Wan X, et al. 2008. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Research 18, 1199–1209. [DOI] [PubMed] [Google Scholar]
- Wu Y, Fu Y, Zhao S, Gu P, Zhu Z, Sun C, Tan L. 2016. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnology Journal 14, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Wagner D. 2015. Polycomb repression in the regulation of growth and development in Arabidopsis. Current Opinion in Plant Biology 23, 15–24. [DOI] [PubMed] [Google Scholar]
- Xing Y, Zhang Q. 2010. Genetic and molecular bases of rice yield. Annual Review of Plant Biology 61, 421–442. [DOI] [PubMed] [Google Scholar]
- Xu L, Shen WH. 2008. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Current Biology 18, 1966–1971. [DOI] [PubMed] [Google Scholar]
- Ying J, Gao J, Shan J, Zhu M, Shi M, Lin H. 2012. Dissecting the genetic basis of extremely large grain shape in rice cultivar ‘jz1560’. Journal of Genetics and Genomics 39, 325–333. [DOI] [PubMed] [Google Scholar]
- Yu J, Xiong H, Zhu X, et al. 2017. OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biology 15, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Miao J, Zhang Z, et al. 2018. Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnology Journal. In press. doi: 10.1111/pbi.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang D, Wang M, et al. 2011a. A core collection and mini core collection of Oryza sativa L. in China. Theoretical and Applied Genetics 122, 49–61. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang J, Huang J, et al. 2012. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proceedings of the National Academy of Sciences, USA 109, 21534–21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Pan Y, et al. 2017. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nature Communications 8, 14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Yao G, Zhang H, Dou H, Shi H, Sun X, Li Z. 2011b. Fine mapping and cloning of the grain number per-panicle gene (Gnp4) on chromosome 4 in rice (Oryza sativa L.). Agricultural Science in China 10, 1825–1833. [Google Scholar]
- Zhao Y, Cheng S, Song Y, Huang Y, Zhou S, Liu X, Zhou D. 2015. The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. The Plant Cell 27, 2469–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Yu H, Zhang H, et al. 2015. Rice functional genomics and breeding database (RFGB)-3K-rice SNP and InDel sub-database. Chinese Science Bulletin 60, 367–371. [Google Scholar]
- Zuo J, Li J. 2014. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annual Review of Genetics 48, 99–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.