Abstract
Environmental health science aims to link environmental pollution sources to adverse health outcomes to develop effective exposure intervention strategies that reduce long-term disease risks. Over the past few decades, the public health community recognized that health risk is driven by interaction between the human genome and external environment. Now that the human genetic code has been sequenced, establishing this “G × E” (gene–environment) interaction requires a similar effort to decode the human exposome, which is the accumulation of an individual’s environmental exposures and metabolic responses throughout the person’s lifetime. The exposome is composed of endogenous and exogenous chemicals, many of which are measurable as biomarkers in blood, breath, and urine. Exposure to pollutants is assessed by analyzing biofluids for the pollutant itself or its metabolic products. New methods are being developed to use a subset of biomarkers, termed bioindicators, to demonstrate biological changes indicative of future adverse health effects. Typically, environmental biomarkers are assessed using noninvasive (excreted) media, such as breath and urine. Blood is often avoided for biomonitoring due to practical reasons such as medical personnel, infectious waste, or clinical setting, despite the fact that blood represents the central compartment that interacts with every living cell and is the most relevant biofluid for certain applications and analyses. The aims of this study were to (1) review the current use of blood samples in environmental health research, (2) briefly contrast blood with other biological media, and (3) propose additional applications for blood analysis in human exposure research.
Overview
Environmental health sciences employ various tools for linking environmental chemicals and other stressors to preclinical and ultimately adverse outcomes in humans. These include the traditional studies of chemicals from external exposures found in media, including air, water, food, dust, consumer products, and other sources from commerce, transportation, agriculture, and manufacturing (Araoud et al. 2016; Arayasiri et al. 2010; Duarte, Pena, and Lino 2011; Freire, Koifman, and Koifman 2015; Funk et al. 2014; Ghio, Sangani, and Roggli 2014; Käfferlein et al. 2005; Little et al. 2012; Liu and Pleil 2002; Marion et al. 2014; Mo et al. 2009; Pleil, Funk, and Rappaport 2006). In the past several decades, the focus has broadened to include analysis of human biological media, primarily blood, breath, and urine, to represent the cumulative dose integration of inhalation, der mal, and ingestion exposure pathways (Amann et al. 2014; _Pleil, Stiegel, and Sobus 2011a; Rappaport 2012a; 2012b; Roos and Jakubowski 2010). These chemicals measured in biological media are broadly termed biomarkers (or bioindicators) in the literature and comprise the human exposome (Burger and Gochfeld 2001; LaKind et al. 2014; Pleil 2012). Although a somewhat fluid distinction, the term bioindicators is used more specifically here as representing that subset of all biomarkers that is more closely related to indicating toward an adverse outcome. In addition to in vivo biomonitoring, contemporary exposure science has embraced suites of in vitro tests for metabolomic activity and toxicity testing based on static designs using specific adverse outcome pathways (AOP) and dynamic/kinetic designs using cell bioreactors (Angrish, Madden, and Pleil 2015; Baranska et al. 2015; Latterman and Buchs 2015; Pleil 2016; 2015; Pleil, Williams, and Sobus 2012; Tarrant 2010; Valcke and Haddad 2015).
In the diagnostic medical and clinical sciences, blood analysis has always been the “gold standard” with urine and breath measurements as important but secondary media. In public health, however, blood sampling is more difficult to implement because study participants, in contrast with medical patients, are generally representative of a healthy population and are therefore less likely to submit to an invasive procedure involving needles (Holland et al. 2003). Further, population-based studies require large numbers of subjects; thus, sample collection generally does not occur in a clinical setting and is more amenable to noninvasive techniques (sampling breath and urine). Nevertheless, similar to medicine, blood is an invaluable biofluid for public health assessment because it directly interacts with every living cell and is representative of all the chemicals circulating in the body (Barr et al. 2002; Levitt 2004).
The aims of this investigation were to (1) contrast the relative value of blood as a diagnostic biofluid with respect to breath and urine, (2) examine existing environmental science applications of blood analysis, and (3) propose future research and development for incorporating blood into routine public health and environmental exposure assessments. The purpose of blood (and other biomarker media) analyses is to develop a series of measurable chemicals that inform the progression from environmental exposure to internal dose to chemical metabolism and then to eventual health outcome (Pleil 2009; Valcke and Haddad 2015). The ultimate goal is then to use empirical measurements to explore the current state of the human exposome and deduce how perturbations from “normal” might interact with the genome to affect health state. Although blood biomarkers may also be useful for diagnosing short-term health effects for individuals, the general focus of environmental health science is to understand and mitigate long-term population-based disease.
Blood versus other biofluids:
As noted earlier, blood sampling is invasive for human subjects. In contrast with breath and urine collection, blood sampling tends to be more expensive, requires certified health professionals, creates more infectious wastes (e.g., needles, swabs, bandages), increases risk of infection, and needs to be performed in a controlled clinical environment (Holland et al. 2003; Vaught 2006). Further, blood samples need to be processed rapidly (before freezing) if analytical protocols require separation of plasma/serum from red blood cells or isolation of genetic material (Holland et al. 2005; Holland et al. 2003; Li, Todor, and Luo 2016). Finally, the amount and number of samples allowed for in environmental studies are limited by institutional review board (IRB) rules. These logistical factors tend to restrict the number of subjects and frequency of sample collection.
Existing blood applications:
Some blood-borne biomarker measurements for exposure/dose assessment are used to validate the interpretation of routine biomarker data from less invasive matrices, such as breath and urine. This is especially true for semivolatile and nonvolatile organic compounds from the environment, such as pesticides, polyaromatic hydrocarbons (PAH), and a variety of manufactured chemicals used in consumer products and industry (Hoet et al. 2009; Pleil 2009). The more prevalent applications of blood measurements for environmental health assess downstream biomarkers, including adducts (e.g., protein or DNA) and conjugates that are unique to blood, which reflect exposures over longer time periods (Johnson et al. 2010; Rappaport et al. 2012).
Future research for incorporating blood into exposure science:
There are four related research paths for expanding blood biomarker applications in the environmental health arena. The first is the continuous development of novel biomarkers that might bridge the gap between exposure and health outcome, including epigenetic markers (e.g., miRNA), ultra-trace-level adducts on specific protein peptides, and endogenous response markers (e.g., plasmaand cell-derived mediators, such as cytokines and chemokines). The second is developing postprocessing methods for efficiently extracting specific blood components from previously frozen samples, especially human serum albumin, hemoglobin, and DNA. The third path is developing less intrusive sampling methods that are more amenable to population-based studies outside of the clinical environment, including micropuncture capillaries, real-time measurements, and sample stabilization via dried blood spots. The fourth is incorporating highthroughput techniques, such as “omics” technologies that are capable of assaying thousands of proteins or metabolites simultaneously to identify differences in expression levels and patterns.
Method Issues for Using Blood
While there are many advantages to using blood for biomarker analysis, several methodological issues serve as drawbacks. One issue is the need for a medical team for sample collection, which increases the expense of the analysis and makes it difficult, if not impossible, to collect blood samples in remote field locations (Vaught 2006). Blood analysis also creates biohazardous waste that needs to be properly disposed of. All blood samples need to be considered infectious and handled with caution (Holland et al. 2005). Another major disadvantage to blood analysis is that subjects are more reticent to provide blood samples than breath or urine (Benyshek 2010; Holland et al. 2005). In a study of childhood leukemia, only 50% of the subjects provided blood samples while 95% gave buccal-cell samples (Holland et al. 2003). Blood sampling is more invasive and may cause brief pain. Some individuals, such as children, the elderly, the chronically ill, and pregnant women, are not included in blood analyses for ethical reasons, while others think it is simply unnecessary and choose not to participate. Blood might also have lower analyte concentrations compared with other matrices. Urine typically has concentrations in the part per billion (ppb) range, while blood concentrations tend to be part per trillion (ppt) (Barr et al. 2002). For some compounds, such as phthalate esters (PE), urine is the preferred matrix because metabolites levels are 10-fold higher in urine than serum (Alves et al. 2014). However, as analytical methods continue to improve and become more sensitive, the concentration issue is becoming less challenging.
Exposure Science Framework
Exposure science has traditionally focused on characterizing how an individual’s interaction with his or her environment leads from stressors (chemical or otherwise) to adverse health impacts (Cohen Hubal 2009; Lioy 2010; Lioy and Smith 2013). The manner in which an individual interacts with his environment then needs to be analyzed to determine the risk of exposure (duration and frequency of contact, activity level, and pathway of exposure) (Klepeis et al. 2001). Chemical-specific biomarkers and their mixtures are then used to quantify the exposure, demonstrating how much of the stressor entered the body and tracking its movement to different biological compartments (Sarigiannis and Hansen 2012). This information is then passed on to toxicologists in order to assess potential health effects based on levels of chemicals partitioned throughout the body (Calafat et al. 2005). Exposure science relies strongly on experimental data; however, studies are increasingly using predictive models in an attempt to characterize potential exposures in a more expeditious manner (http://www.epa.gov/tsca-screening-tools; Kavlock et al. 2008). Biomarkers are known to link exposure science and toxicology research to explain the entire dose-tooutcome continuum, including both exposure information (absorption, distribution, metabolism, and elimination [ADME]) and indications of biological effects (Pleil et al. 2007; Teeguarden et al. 2016) (Figure 1).
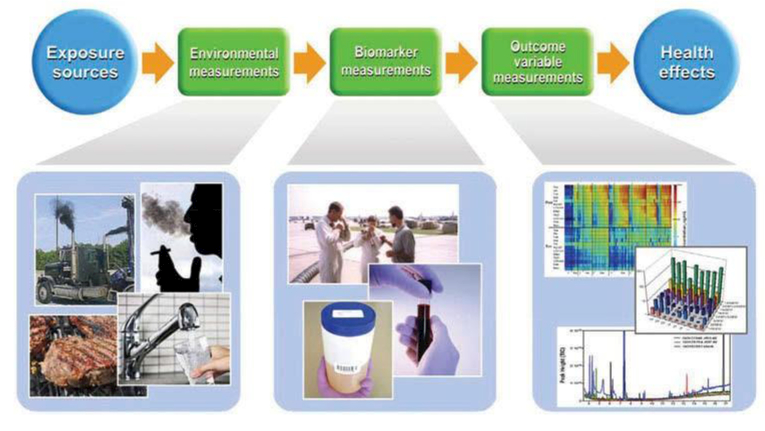
Figure 1.
The role of environmental and biomarker measurements in explaining the exposure source to health outcome continuum. Environmental measurements from water, air, smoke, dust and food are linked to biomarker measurements from blood, breath, and urine and are interpreted using different types of observational and mathematical tools, such as heat maps, bar graphs, and charts.
Biomarker Categorization
Linking exposure to potential adverse health effects requires analyzing multiple biomarkers that demonstrate an exposure has occurred and that show a change in the biological system that is indicative of an adverse effect (Pleil and Sheldon 2011). Biomarkers have typically been categorized broadly by function as biomarkers of exposure, effect, and susceptibility (Boffetta 2010; Chen et al. 2005). For the purposes of this review, the exposure/effect/susceptibility paradigm is used as follows:
Biomarkers of exposure are chemical-specific (the stressor itself or a chemical metabolite). These biomarkers are considered exogenous in origin and used to assess the quantity of the chemical within the biological system.
Biomarkers of effect are not necessarily structurally similar to the stressor; they are considered to be of endogenous origin, resulting from interactions with the environment and generally linked to an AOP in human systems biology.
Biomarkers of susceptibility demonstrate an individual propensity for a sensitized reaction to a stressor of interest. These biomarkers are typically related to genetic, epigenetic, or enzymatically induced traits and describe the likelihood that an environmental interaction might trigger a cascade leading to adverse effects.
Certain biomarkers from each of these categories may be more amenable to analysis in one biofluid versus another, while other biomarkers may be equally well determined in multiple biofluids. In this review, the focus is on the different categories of biomarkers that can be analyzed in blood.
Classes of Blood Biomarkers
There are four main types of biomarkers in addition to exogenous chemicals that are commonly found in blood: endogenous chemicals, inflammatory, molecular modification, and damage bioindicators.
Endogenous biomarkers reflect the earliest effects of external perturbations to the human organism. Typically, these compounds represent housekeeping chemicals from hydrocarbon and oxygen metabolism, as well as volatile oxygenated species, including alcohols, aldehydes, ketones, organic acids and proteins. The probative aspect is not their presence but a shift away from what is considered a normal (or unremarkable) pattern (Pleil 2012; Pleil, Stiegel, and Risby 2013). For example, increased levels of reactive oxygen species (ROS) are indicative of oxidative stress (Amann et al. 2014), which might be assessed by monitoring levels of glutathione (GSH) and glutathionine disulfide (GSSG) in blood (Dalle-Donne et al. 2006). Stress genes are often induced after exposure to an environmental stressor and may indicate toxicity. Heat-shock proteins, particularly Hsp70, are often expressed as a result of exposure to heavy metals or pesticides, and these exposures were found to produce recognizable heat shock protein expression patterns (Gupta et al. 2010). Clinical markers, including cholesterol, triglycerides, and liver enzymes in blood samples, may also be considered endogenous biomarkers and indicate adverse health risks when abnormal levels are observed (Pagana and Pagana 2009; Watkins and German 2002). Circulating mitochondrial DNA and microRNA (miRNA) were also identified as biomarkers of environmental exposure to pesticides and indicators of early cancer development (Budnik et al. 2013; Liu et al. 2012).
Inflammatory bioindicators represent the first level of biologically relevant parameters that react to an external stressor, which is often the first step in an AOP. The inflammatory response serves as a recruitment mediator for repair function stimulated by external stressors, such as air pollution, and generates macrophages to release chemokines and proinflammatory cytokines, which might be employed to track the biological response to the stressor. In some scenarios, anti-inflammatory messengers are also released, which reflects a depression of repair function and may be considered adverse. Cytokines were identified as biomarkers of exposure to traffic emissions and smoke, which have been linked to cardiovascular diseases (Brugge et al. 2013; Channell et al. 2012; Ghio et al. 2012; Kendzia et al. 2012; Stiegel et al. 2015; Zuurbier et al. 2011). The cytokine interleukin-6 (IL-6) stimulates chemokine expression as well as C-reactive protein synthesis and secretion from liver in response to inflammation. C-reactive protein is a serum biomarker of acute-phase response and an indicator of cardiovascular diseases (Devaraj, Xu, and Jialal 2003; Meier-Ewert et al. 2001; Yudkin et al. 1999). While these may be useful biomarkers that some exposures have occurred recently, cytokines (and other protein markers) are nonspecific, tend to have a short serum half-life, and are difficult to detect. In fact, the acute response may be delayed by a few (or more) hours and may recover within the same time frame (Angrish et al. 2016; Nelin et al. 2012). However, these endogenous markers are useful to assess health state and indicate an overall level of stress on the biological system. Inflammatory biomarkers also lack tissue specificity and recognizable toxin-specific expression patterns and are therefore difficult to interpret when more than one toxicity mechanism is possible (Tarrant 2010).
Molecular modification bioindicators are typically defined as large molecule adducts and conjugates. These include protein adducts, especially those on hemoglobin (Hb) and human serum albumin (HSA) proteins, which together comprise approximately 92% of all blood-borne proteins (Funk et al. 2008). Protein adducts are considered long-term biomarkers: Hb has a half-life of approximately 60 d, while HSA has a half-life of 30 d (Rappaport et al. 2012). HSA functions as a carrier for proteins and fatty acids (Fanali et al. 2012), while the primary function of Hb is to transport oxygen from lungs to tissues (Schechter 2008). However, exogenous chemicals and their metabolites might attach to the active sites of these proteins, forming adducts that interfere with their normal functions (Boysen and Hecht 2003). Similarly, exogenous compounds, such as PAHs, form adducts with cellular DNA, inducing DNA mutations that affect reproductive functions (Kwack et al. 2014). DNA adducts reveal how much of a carcinogen has been adsorbed, metabolized, and bound to DNA but not repaired (Gallo et al. 2008; Porru et al. 2014). Other stressors may affect DNA through epigenetic attachments, often in the form of methylation (Ouyang et al. 2013).
Damage bioindicators comprise a wide range of organic molecules and cellular fragments from adverse biological effects. The most common example of clinical damage markers is liver enzymes, which are essentially leakage proteins from injured cells. Two liver enzymes, aspartate aminotransferase (AST) and alanine aminotransferase (ALT), are released into blood circulation upon hepatocellular damage or cell death. Blood tests are performed to assess the potential for liver disease and other viral infections due to high AST or ALT levels (Kew 2000; Kim et al. 2008). Circulating blood cells provide a convenient source of DNA to assess DNA-specific damage indicators. Cytogenetic damage indicators include chromosome aberrations (CA), sister chromatid exchanges (SCE), and micronuclei (MN). CA are characterized by chromosome breaks and rearrangements greater than normal 1% occurrence. In SCE, entire DNA strands are separated and exchanged between two sister chromatids, while MN are entire chromosomes with centromeres left outside of the main nuclease due to structural damage or abnormal mitosis (Holland, Zhang, and Smith 2000; Watson and Mutti 2004). DNA singlestrand breaks (SSB) are another type of damage bioindicator that may be detected using a comet assay. Other indicators of DNA damage include physiological cell death, termed apoptosis, and accidental cell death, known as necrosis. Apoptosis suppression is a common biomarker of cancer (Ward et al. 2008), while necrosis is the result of severe injury and is characterized by cytoplasmic and mitochondrial swelling prior to membrane breakage (Kroemer, Dallaporta, and Resche-Rigon 1998). Cell count assays might be utilized to assess apoptosis and necrosis and determine the consequences of toxicity and damage (Ortiz et al. 2000). DNA damage bioindicators were shown to vary based on environmental exposures, as these often include fragments or whole molecular structures of the exogenous chemical (Gallo et al. 2008; Poirier 2004).
Environmental Exposure Continuum
The human exposome describes the counterpart to the human genome that represents chemicals, both exogenous and endogenous, that interact with human systems biology (Pleil and Sheldon 2011; Wild 2005). This concept was further elucidated to begin to explain the gene–environment interaction that appears to drive health outcomes from autoimmune diseases, cancer, and chronic conditions, including cardiopulmonary disease, neurological disorders, and asthma (Pleil 2012; Rappaport 2011; 2012b; Rappaport and Smith 2010; Schetter and Harris 2012; Smith et al. 2011). Circulating blood contains essentially all of the chemicals that comprise human systems biology and thus also represents the human exposome (Barr et al. 2002; Levitt 2004; Rappaport 2011). Other biological media may also reflect portions of the exposome, especially for those compounds eliminated via breath and urine or those sequestered in adipose tissue (Amann et al. 2014; Barr and Angerer 2006; Lehr, Hartwig, and Sell 2012; Pleil, Stiegel, and Sobus 2011a; Roos and Jakubowski 2010; Trayhurn 2005). However, blood is truly the central compartment for human metabolism. Figure 2 demonstrates how exposure to the health effect continuum is interpreted via biological measurements and the relationships among the major biomarkers and bioindicators and biological fluids within which these are found. The less invasive media, breath and urine, might certainly overlap in some suites of biomarker compounds and generally display higher concentrations, but with more sensitive contemporary instrumentation, the lower levels in circulating blood are now becoming accessible (Pasikanti, Ho, and Chan 2008; _Zhang et al. 2013b).
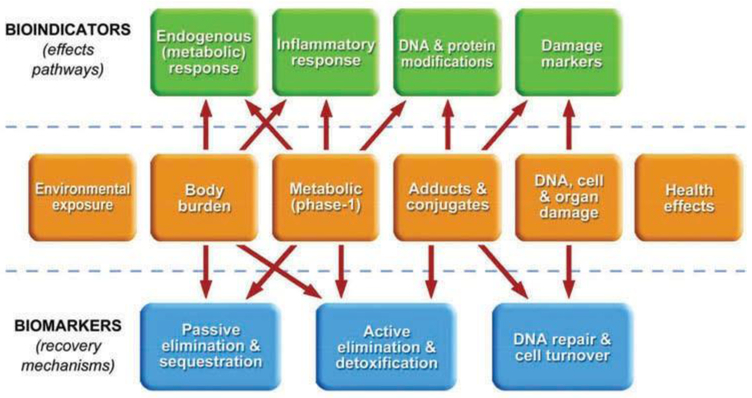
Figure 2.
The center sequence from environmental exposure through health effects is represented by the biological progression of biomarkers and responses of increasing complexity that are assessed in human biological samples. The upper entries represent bioindicators of effect (or at least preclinical effects) generally found in circulating blood. The lower entries represent biomarkers of recovery and repair functions; those labeled passive and active elimination are typically found in breath and urine or sequestered in adipose tissues. DNA repair and cell turnover are not generally directly observable as biomarkers but are inferred from a reduction in damage biomarkers.
As shown in the conceptual model proposed in Figure 2, markers representing effects pathways (top progression) are expressed in a series of groups depending upon their source along the internal exposure to effects progression (middle progression). All bioindicators are present in blood; some specific classes may also be measured in other biological media with emerging methodologies. For example, endogenous markers of carbohydrate metabolism, such as ketones and aldehydes, are detected in exhaled breath (Ligor et al. 2008), markers of systemic inflammation, such as cytokines are found in urine (Stiegel et al. 2015), and markers of DNA damage, such as DNA SSBs were identified in oral buccal swabs (Basu et al. 2005; Holland et al. 2008).
The primary advantage for using blood as the medium, however, is that all of the different bioindicator classes are available in one medium and are measured by applying the appropriate processing and analytical methodologies. Further, there is another class of biomarkers specific to circulating blood that are not on the effects pathway (upper) nor the recovery pathway (lower). These are the native compounds of exposure that are found in the “body burden” box and are measured prior to biological activation or removal (Pleil 2009). In the following sections, the typical blood collection and processing procedures are described, an assessment of the currently utilized blood biomarkers of exposure, effect, and susceptibility is provided, and also some of method issues for using blood as a biological medium are discussed.
Blood Biomarkers: Environmental Applications
Blood Composition, Collection, Transportation, and Storage
Blood is composed of many different cell types, including lymphocytes, granulocytes, and red blood cells (RBC) as well as serum and plasma (Holland et al. 2005). Blood lymphocytes are typically utilized to analyze CA and SCE (Holland et al. 2003). Neutrophils, a type of granulocyte, may be isolated for DNA analysis, while RBC are a good source for studying Hb adducts (Vaught 2006). Whole blood may be advantageous for evaluating certain biomarkers. For example, heavy metals, such as manganese (Mn), lead (Pb), and cadmium (Cd) are typically measured in whole blood due to their low levels in serum and plasma as well as a lack of epidemiological data from exposure assessments of these biomarkers in serum or plasma (Kim et al. 2015; Sommar et al. 2014). Different blood cell types and liquids require distinct sample preparation and storage conditions, which may need to be optimized prior to collecting samples for the study (Holland et al. 2003). Recently, a multicolor flow cytometry assay was developed that is capable of distinguishing major leukocyte subsets in human blood, bronchoalveolar lavage, and lung tissue, and this technique may be used to visualize the proportions of different blood cell types without requiring prior separation. This flow cytometry technique may be utilized in exposure science to compare the proportions of cell types present in samples from exposed and unexposed individuals, as demonstrated for smokers versus nonsmokers (Yu et al. 2016).
Blood sample collection for environmental applications is identical to methods used for clinical evaluation. Typically, a phlebotomist or other trained personnel is needed to draw blood samples, especially when samples are being collected from children (Holland et al. 2003; Vaught 2006). Blood is drawn from the antecubital vein (Levitt 2004) in volumes of 20 ml or less (Polkowska et al. 2004). It is vital that blood is collected in a sterile environment to avoid contamination and fungal or bacterial infections. In addition, the procedures utilized for blood sampling need to be standardized for all participants to ensure that differences in sample collection do not influence the results of the study (Holland et al. 2003). Samples for case and control subjects need to be drawn at the same time and in the same manner for consistency (Holland et al. 2003; Holland et al. 2005). Sterile evacuated plastic tubes are typically used for sample collection. Collection tubes need to be free of trace metals, which interfere with analyses of metal exposure (Vaught 2006). Anticoagulants and preservatives for sample tubes need to be selected based upon the target of interest, such as RNA or DNA, as different targets require certain stabilizers (Holland et al. 2003). However, great care needs to be taken when choosing anticoagulants and other processing additives, because some of these are known to affect the levels of particular biomarkers in serum and plasma samples. Biomarker levels reported in different investigations may not be directly comparable if proper sample collection and preparation procedures were not adhered to (Gillio-Meina et al. 2013).
After blood collection, samples need to be immediately centrifuged to create a Ficoll gradient, which separates blood into plasma, white blood cells (WBC), and RBC (Holland et al. 2005; Li, Todor, and Luo 2016). Blood samples need to be processed shortly after collection, and all samples need to be cryopreserved within 24 h of sample collection (Shabihkhani et al. 2014). Different cell types and components require unique storage conditions. For example, cells need to be separated from serum immediately after sample collection for proper analysis of cytokines. If the desired target is unknown, blood samples may be kept at 4°C for several days to protect cells against enzymatic degradation while maintaining cell viability. Depending upon the target of interest, protease inhibitors may be added to the sample to slow degradation. However, these inhibitors are toxic to live cells and should not be used if cell viability is necessary. If novel targets are being analyzed, it may be necessary to conduct a preliminary experiment to determine optimal storage conditions and times before gathering samples from subjects (Holland et al. 2003).
If possible, blood samples need to be collected at the location where samples will be analyzed to avoid sample handling issues that may affect sample integrity. However, this is typically not feasible, because samples are frequently collected in the field, and sample transportation is thus unavoidable. Shipped samples need to conform to proper guidelines and restrictions for biological media (Holland et al. 2003). Depending upon the blood component and what the sample will be used for, blood samples may be shipped at room temperature, on ice, or frozen (Belloni, Meschini, and Palitti 2008). For example, serum and plasma samples need to be stored at −80°C for analysis of soluble molecules, but plasma may be stored at room temperature for immunoglobulin analyses, as these are more stable under these conditions (Gillio-Meina et al. 2013; Holland et al. 2003). Lymphocytes need to be stored in liquid nitrogen vapor at −150°C if viability is required (Vaught 2006). When considering storage conditions, it is important to remember that apoptosis generally increases with time and temperature (Belloni, Meschini, and Palitti 2008). Therefore, transportation conditions need to be specifically selected based upon cell type or liquid and the component of interest to ensure that the sample will be properly preserved for analysis. During transportation, measures need to be taken to ensure that samples do not undergo any freeze–thaw cycles, which might damage cells and decrease sample integrity (Gillio-Meina et al. 2013; Holland et al. 2005; Holland et al. 2003; Shabihkhani et al. 2014).
Storage conditions for blood samples depend upon cell type and preservation method. Samples need to be maintained at either 4, −20, or −80°C, depending upon the medium and compound of interest (Holland et al. 2005; Vaught 2006). Blood samples may be stored in either plastic tubes or glass tubes with polytetrafluoroethylene (PTFE) or aluminum cap liners (Polkowska et al. 2004). However, the exact tube utilized depends upon the medium of interest, such as whole blood, plasma, serum, or RBC, all of which have specially designed tubes for optimal stabilization and preservation (Shabihkhani et al. 2014). Cells may be cryopreserved in small volumes for long-term storage. For cryopreservation, dimethyl sulfoxide (DMSO) is added to cells as a cryoprotectant, and samples are stored in liquid nitrogen vapor to prevent cell death (Holland et al. 2005; Vaught 2006). Ellis and Venturini (2013) also showed that blood samples may be frozen in larger volumes and that frozen aliquots removed from the stock solution without thawing the samples and without a significant loss in sample integrity, thereby decreasing the number of sample tubes needed to store multiple aliquots. While high levels of reproducibility were observed for cholesterol, triglycerides, glucose, and immunoglobulin G, the effectiveness of this technique for other analytes in blood may vary (Ellis and Venturini 2013). Blood samples are commonly stored in biorepositories, such as the European Cancer Study biorepository, which contains more than 380,000 blood samples. Nanobarcoding technology was employed to uniquely label samples in small surface areas without using removable labels that may fade or fall off with time (2005; Holland et al. 2003). Efficient sample storage and electronic databases are necessary to maintain sample integrity and ensure that samples are easily retrieved for future analyses.
Traditional Environmental Exposure Assessment
Environmental applications of blood analysis have historically focused on measuring exogenous compounds in blood circulation to detect environmental exposures and estimate relative body burden. Levels of compounds, such as heavy metals, that an individual has been exposed to may be assessed in healthy populations to detect biomarkers associated with low-level exposure, in addition to providing a baseline to which affected individuals may be compared (Pollack et al. 2015). Placental and maternal blood was examined to evaluate fetal risk to Cd exposure from dietary and tobacco sources (Piasek et al. 2014). Blood Pb levels were also measured to determine the extent of Pb exposure, and these levels may be tracked in populations to identify potential contributing factors, such as smoking status, alcohol intake, red meat consumption, geographical location, and tap-water contents (Bellinger et al. 1987; Counter, Buchanan, and Ortega 2015; Lopes et al. 2015; Ngueta et al. 2016). Trace levels of PAH from environmental exposure to diesel exhaust and markers of exposure to toluene, ethylbenzene, and m-xylene were also determined in blood samples from volunteers in controlled chamber studies (Marchand et al. 2015; Pleil et al. 2011b; Pleil et al. 2010). In addition, volatile organic compounds (VOC) released from blood were analyzed to detect changes in cellular respiration, and this technique may be utilized to detect alcohol content in blood to assess the amount of alcohol consumption (Amann et al. 2014; de Lacy Costello et al. 2014; Pleil 2016; Pleil et al. 2014).
In many cases, biomarkers of exposure are present in multiple biofluids. In such cases, investigators have predominantly turned to less invasive biofluids such as urine or breath. However, there are a number of situations where blood remains the best source of biomarkers of exposure. For example, biomarkers with long elimination halflives exhibit higher concentrations in blood than urine. Ochratoxin A, a nephrotoxic mycotoxin found in contaminated vegetables, has a serum half-life of 35 d in humans. In addition, ochratoxin A has poor elimination kinetics, and therefore the concentration of ochratoxin A in urine is much lower than found in blood (Duarte, Pena, and Lino 2011). Mercury (Hg) and Pb also have long halflives of 30 and 70 d in blood, respectively (Brodkin et al. 2007), and Cd levels in blood reveal exposure over preceding months (Kim et al. 2015). Perfluorooctanesulfonate (PFOS), perfluorohexanesulfonate (PFHS), and perfluorooctanoate (PFOA) have serum elimination half-lives of approximately 5, 8, and 3.5 yrs, respectively (Olsen et al. 2007). Therefore, urine levels of biomarkers with long elimination half-lives will be lower than the actual concentration in the body, which might lead to an erroneous conclusion that an exposure has not occurred. For biomarkers with long elimination half-lives, blood is the ideal biofluid in which to measure the concentration.
Blood biomarkers may more accurately reflect chemical levels that an individual was exposed to than urinary metabolites (Arayasiri et al. 2010; Käfferlein et al. 2005). Hemoglobin adducts of aromatic compounds have average lifetimes of approximately 120 d, whereas the respective native compound concentrations in blood display halflives ranging from minutes to a few hours, depending upon the exposure history (Käfferlein et al. 2005). Thus, Hb adducts reveal the mean exposure to a chemical that occurred over the past several months. N-Methylcarbamoylated haemoglobin (NMHb) levels, a blood biomarker of N, N-dimethylformamide (DMF) exposure, were evaluated in workers from a polyacrylic fiber plant. NMHb was capable of distinguishing between three different levels of DMF exposure with statistical significance. While urinary metabolites N-methylformamide (NMF) and N-acetyl-S(N-methylcarbamoyl)cysteine (AMCC) may identify DMF exposure, these biomarkers were not able to distinguish between high, medium, and low exposure, but this was possible with the blood biomarker (Käfferlein et al. 2005). In addition, some compounds, such as 2-butoxyacetic acid, display inconsistent levels of conjugation in urine excretions, which skews the results when measuring the level of the free compound to determine the extent of exposure to 2-butoxyethanol from inks, varnishes, and cleaning fluids (Jones and Cocker 2003).
Blood may also be the best biofluid to use for analysis when urinary biomarker metabolites are nonspecific. For biomarker analysis, measuring metabolites of the parent compound is not always sufficient for exposure assessment. A parent compound may generate several nonspecific metabolites that are excreted in urine, but analyzing these metabolites may not reveal which compound the individual was actually exposed to. Some pesticides and other contaminants have common urinary metabolites that do not reveal the identity of the parent compound (Barr et al. 2002; Needham et al. 2005; Smolders et al. 2009). The identification of 3,5,6-trichloropyridinol in urine samples might be due to exposure to either chlorpyrifos or chlorpyrifos-methyl pesticides (Barr et al. 2002; Needham et al. 2005). Chlorpyrifos metabolites are also found as natural degradation products in the environment and may not always be indicative of direct exposure to the parent pesticide (Smolders et al. 2009). Further, some biomarkers do not have any apparent known metabolites, and therefore the parent compound needs to be quantified instead; in such cases blood is frequently the best sample for measuring the most relevant biomarker (Barr et al. 2002). Other biomarkers and metabolites, such as organic Hg, are predominantly excreted in feces instead of urine, making blood the ideal biofluid for analysis (Brodkin et al. 2007; Needham et al. 2005). Thus, knowing the likely exposure route and identity of the biomarker might help the investigator or clinician select the most appropriate biofluid to use for exposure assessment.
Environmental Biomarkers of Effect
Traditional exposure assessment has been broadened philosophically to include forward mapping toward health outcome. In this case, indicators of the presence and levels of specific chemicals that an individual was exposed to are statistically linked to subclinical and preclinical effects. The measured compounds, whether exogenous or endogenous, are not necessarily toxic themselves but might trigger perturbations that lead to increased adverse health risk. These compounds are known as biomarkers of effect or bioindicators. As stated previously, biomarkers of effect are of endogenous origin and are altered by interactions with the environmental stressor, which then becomes involved in an AOP.
Biomarkers of effect include protein indicators, DNA indicators, and adducts of both proteins and DNA. Protein indicators of effect include alterations in protein expression levels and patterns. Normal protein expression may be altered by exposures to stressors that interfere with endogenous protein levels (Merrick 2008). Exposures to air pollutants and other harmful substances might lead to an inflammatory response affecting cytokine levels. Diesel exhaust, nitrogen dioxide (NO2), and ozone (O3) exposure were shown to acutely elevate levels of WBC and interleukin-8 (IL-8) in plasma samples from human volunteers (Channell et al. 2012; Steenhof et al. 2014). In a study of the effects of O3, fibrinolysis markers, such as plasminogen activator inhibitor-1 were measured in human blood samples and found to decrease after O3 exposure at moderate temperature, while the markers rose after exposure at elevated temperature, indicating that the effect of O3 on the fibrinolytic pathway is temperature dependent (Kahle et al. 2015). Ozone was also found to elevate levels of stress hormones and lipid metabolites in serum samples from subjects exposed during intermittent exercise (Miller et al. 2016). Exposure may induce protein overexpression. For example, the glycoprotein mesothelin and soluble mesothelin-related peptides become overexpressed in malignant mesothelioma as a result of asbestos inhalation (Marini et al. 2011). Some proteins may maintain normal expression levels but experience a change in activity due to exposure. The activity level of the protein β-glucuronidase was increased as a result of low-level organophosphorus insecticide exposure (Ueyama et al. 2010). Protein modifications are another common effect of exposure to stressors. Common modifications include protein oxidation and lipid peroxidation, which are signs of oxidative damage (Rossner et al. 2009).
DNA indicators of effect include changes in gene expression, mitochondrial DNA, DNA modifications, DNA damage, and miRNA (Boffetta 2010). Gene expression patterns were found to alter based upon exposure to environmental contaminants, such as ultrafine carbon particles (UFP), arsenic (As), and benzene (Huang et al. 2010; McHale et al. 2009; Mo et al. 2009). Increased or decreased gene expression in the presence of an environmental stressor may indicate that an AOP was triggered or that DNA damage repair responses and defense mechanisms against exposures were activated (Mo et al. 2009). Mitochondrial DNA was labeled as a useful biomarker of damage and exposure. Unlike genomic DNA, mitochondrial DNA does not have histone protection or repair mechanisms and may therefore reflect more severe damage (Hou et al. 2010). Exposure to airborne particulate matter (PM) and halo-alkanebased pesticides was linked to damaged mitochondrial DNA (Budnik et al. 2013; Hou et al. 2010). Environmental contaminants, such as As, nickel (Ni), and ultrafine PM, were reported to produce increased DNA SSB in exposed individuals (Avogbe et al. 2005; Basu et al. 2005; Glei et al. 2005; M’Bemba-Meka, Lemieux, and Chakrabarti 2005; Popp et al. 1997). Levels of DNA SSB may be used to distinguish affected versus unaffected populations (Basu et al. 2005). Telomere lengths have also been reported as biomarkers of DNA damage. While exposures to traffic pollution, PAHs, fine particulate matter (PM2.5), pesticides, and Pb have been associated with telomere shortening in chronically exposed individuals (Hoxha et al. 2009; Wong et al. 2014; Zhang et al. 2013a), individuals exposed to low doses of persistent organic pollutants (POP) or As exhibited telomere lengthening (Shin et al. 2010; Zhang et al. 2013a). Although telomeres tend to shorten with age and exposure, telomere lengthening in healthy individuals may be a biomarker of earlystage carcinogenesis (Shin et al. 2010). Increased MN and binucleated micronucleated cells (BNMN) in blood lymphocytes are biomarkers of exposure to genotoxic agents. The frequency of MN increases as a result of chromosome breakage or loss, indicating that DNA damage occurred. Elevations in MN and BNMN frequencies were attributed to air pollution, pesticides, and As (Martínez et al. 2004; Pedersen et al. 2006; Ramos-Chavez et al. 2015).
DNA modification through methylation is another biomarker of environmental exposure. Some common aberrant methylation patterns include global hypomethylation and gene-specific hypermethylation/hypomethylation (Bollati et al. 2007). DNA methylation mediates gene expression through protein–protein and protein–DNA interactions. Deviation from normal methylation patterns may be indicative of oxidative damage (Franco et al. 2008). While one study observed increased DNA methylation as a result of low-dose benzene exposure (Kim et al. 2010), reduced methylation was noted in a study of traffic particle exposure (Baccarelli et al. 2009). DNA methylation levels change based upon the type of environmental exposure and exposure time (Baccarelli et al. 2009). Whether an exposure results in elevated or decreased methylation may be used in the future to determine the nature of the exposure that occurred. miRNA, small noncoding RNA that regulate gene expression, were identified as biomarkers of exposure and cancer due to altered expression levels in plasma and blood samples in affected individuals (Bollati et al. 2010; Liu et al. 2012; Zhang et al. 2010). Upregulation of miR-222 has been associated with metal-contaminated air particle exposure (Bollati et al. 2010), and miR-122 was identified as a specific blood-based biomarker of liver disease (Zhang et al. 2010).
Blood levels of environmental compounds often reflect the dose to which an individual was exposed. For example, high levels of compounds are often found in blood samples of individuals exposed to higher concentrations of toxins than for those exposed to lower doses (Käfferlein et al. 2005; Lope et al. 2010; Lunder et al. 2010; Turyk et al. 2010). While some compounds with long half-lives in blood may be directly measured, other compounds have short halflives in blood, and thus the exogenous compound cannot be directly analyzed to verify exposure. For example, cyanide (CN) is rapidly metabolized and has a half-life of less than 1 h in whole blood. Thus, the level of CN cannot be accurately measured by evaluating the native compound (Fasco et al. 2007). For cases such as CN, evaluating DNA or protein adducts isa useful way to assess the effects of environmental compounds. DNA and protein adducts have longer half-lives, typically 3–4 mo, depending upon the protein lifetime. Adducts are also stable and reflect longer term exposures than urinary or breath biomarkers (weeks or months vs. hours or days) (Johnson et al. 2010). Protein and DNA adducts also rise with relevant exposure dose and may be utilized to assess total amount of exposure (Beach and Gupta 1992; Bechtold et al. 1992; Binková et al. 2000; Käfferlein et al. 2005; Magagnotti et al. 2000; Topinka et al. 2007). Benzene exposure was assessed by monitoring levels of Hb and albumin adducts with benzene oxide, as well as albumin adducts with S-phenylcysteine (SPC), a benzene metabolite (Bechtold et al. 1992; Lindstrom et al. 1998). Protein adducts are typically favored over DNA adducts for exposure assessment because they are not repaired; the concentration of DNA adducts at any given time depends on the repair rate and may not linearly rise as the dose is increased (Watson and Mutti 2004). In a previous study of policemen exposure to traffic emissions, the levels of c-PAH increased by threeto fourfold, but the amount of benzo[a] pyrene DNA adducts only rose by approximately 20%, indicating that DNA repair mechanisms were eliminating some of the adducts (Topinka et al. 2007). These biomarkers of effect indicate that an environmental stressor induced alterations in the chemical makeup of the individual. While the presence of DNA modifications and DNA and protein adducts can be verified, the long-term effects of these biomarkers may or may not lead to an AOP. Whether an AOP is triggered may depend upon the susceptibility of the individual. Some individuals may possess more efficient DNA repair mechanisms than others, allowing them to avoid some of the deleterious outcomes of DNA adducts. Other exposed individuals may display reduced DNA repair after chronic exposure (Arayasiri et al. 2010). The concentration of the compound, length of exposure, and consistency of exposure (once versus daily) all affect the severity of consequences from exposure and whether an AOP might be triggered.
Assessment of Susceptibility
The eventual path to an adverse outcome is thought to involve both environmental component (exposome) and genetic component (genome). Thus, there is some form of intrinsic individual susceptibility to different environmental stressors (Boffetta 2010). For instance, not all smokers contract lung cancer; not all sunbathers suffer from melanoma; not everyone on high-fat diets has the result of heart attack development. In general, susceptibility is difficult to assess directly but might sometimes be determined by examining trends in exposure of larger populations. In the case of asbestos, inhalation was correlated with malignant pleural mesothelioma (MPM), but some individuals with high asbestos exposure did not develop the disease, while others with lower exposures were more susceptible. Recently, three Fe metabolism genes were identified with protective properties against MPM development, explaining some of the differences in individual susceptibility (Corvella et al. 2016). In some cases, a specific genetic sequence or polymorphism was used to assign relative risk in conjunction with a specific exposure, or a host factor (e.g., age, weight, or health state) was associated with increased risk in the presence of environmental chemicals (Boffetta 2010). In general, however, it is difficult to predict individual susceptibility. Blood measurements of various endogenous biomarkers might be employed to estimate relative susceptibility via comparisons to onboard dose. Signaling chemicals, such as cytokines and various expressed proteins, may be directly interpreted to establish relative risk, and other chemicals present in blood might serve different levels of protective functions.
Biomarkers of susceptibility include antibody levels and genetic polymorphisms. Levels of Cdinduced metallothionein (MT) mRNA were used as a biomarker of renal dysfunction susceptibility in peripheral blood lymphocytes. Several studies showed that high plasma MT antibody (MT-Ab) levels correlated with a higher risk of renal tubular damage, while low MT-Ab levels indicate decreased susceptibility. MT is a small protein that binds Cd, leading to detoxification. Renal damage occurs as a result of excess Cd that is not bound to MT within the cell (Chen et al. 2005; 2006). Genetic polymorphisms, which are defined as sequence variants that affect approximately 1% or more of the population, also influence individual susceptibility to compounds found in the environment (Kelada et al. 2003). In an analysis of blood gene expression levels and clinical biomarkers using a decision-tree approach to discover correlations among asthma endotypes, polymorphisms in the gene HNF-4a that are known to affect diabetes were also implicated in enhanced asthma susceptibility (George et al. 2015). Further, single-nucleotide polymorphisms in catechol O-methyltransferase (COMT) were found to initiate increased susceptibility to neurobehavioral issues due to Hg exposure among children (Woods et al. 2014). Polymorphisms that affect DNA repair genes may also enhance susceptibility by lowering the individual’s ability to correct mutations to the genome. Polymorphisms were detected in the x-ray repair cross-complementing (XRCC1) gene, which functions in base excision repair, and these polymorphisms were found to induce deficiencies in DNA repair mechanisms (Feng et al. 2014; Topinka et al. 2007; Tuimala et al. 2002). XRCC1 mutants exhibit accumulation of SSB in DNA (Thacker and Zdzienicka 2003), and XRCC1 variant 399Gln leads to increased DNA adducts and SCE levels (Da Silva et al. 2013). Polymorphisms also produce proteins with decreased activity compared with wild-type protein. Methionine synthase reductase variant 66GG protein exhibits fourfold reduced activity and cannot perform its normal function to catalyze conversion of methionine synthase into its active form. This variant was also shown to lower genomic stability, thus increasing susceptibility of the individual to environmental exposures (Zijno et al. 2003).
Finally, vitamin and mineral deficiencies are also bioindicators of susceptibility to environmental stressors. Many vitamins and minerals are critical for cell survival, and a lack of these micronutrients leads to significant damage. Iron deficiency was linked to increased susceptibility to Mnand Cd-mediated toxicity after exposure (Kim et al. 2015), while folic acid and vitamin B12 deficiencies produced SSB and double-strand breaks in DNA, as well as oxidative lesions. Folic acid deficiency resulted in uracil incorporation into DNA instead of thymine, thereby enhancing the risk of chromosome breaks (Ames 2001; Fenech 2001). Unfortunately, approximately 10% of the U.S. population have a folate deficiency and approximately 4% suffer from a lack of vitamin B12 (Ames 2001). Vitamin and mineral deficiencies may be due to genetic defects or a lack of dietary consumption, both of which enhance susceptibility. Knowledge of these types of biomarkers might prove useful to prevent AOP in individuals who are prone to genetic damage or dietary deficiencies.
Blood Biomarkers: New Developments
Advances in Sampling Techniques: Dried Blood Spots (DBS)
Dried blood spot (DBS) sampling has become more accessible and applicable for blood sample analysis in both clinical and environmental settings. DBS sampling was initially developed in 1963 by Dr. Robert Guthrie to detect phenylketonuria in newborns by pricking their heels and collecting a blood spot on paper. Newborn DBS sampling is still performed, and several other metabolic disorders are also currently tested for (Guthrie and Susi 1963; McDade, Williams, and Snodgrass 2007). DBS sampling is less expensive than collecting large volumes of liquid blood samples, and it is less painful and less invasive for the subject. Therefore, more individuals are likely to provide DBS samples (Benyshek 2010; Ostler, Porter, and Buxton 2014). Individuals who have chosen not to provide DBS samples for investigations stated different reasons, including medical conditions, use of blood thinners, fear, or thinking it unnecessary for the study (Williams and McDade 2009). DBS samples can also be analyzed after several months or years for quantification of certain analytes. DNA has been extracted and amplified from DBS samples stored for 19 mo, and even after this length of storage at room temperature, comparable DNA levels were achieved (Kline et al. 2002).
DBS Sampling and Storage
Dried blood spot sample collection does not require a trained phlebotomist, as anyone can be easily trained to acquire the samples. In one study, more than 2000 DBS samples were collected by trained field interviewers without a medical professional (Williams and McDade 2009). Spring-loaded finger-stick lancets were developed that cut to uniform depths of 2 mm to enhance blood flow and reduce pain and bruising (Benyshek 2010; Edelbroek, Van Der Heijden, and Stolk 2009). The first blood drop is wiped away and is not applied to the filter paper. Subsequent drops are placed on the filter paper by touching the drop to the paper without the finger contacting the paper, to avoid smearing (McDade, Williams, and Snodgrass 2007; Ostler, Porter, and Buxton 2014). Filter paper with preprinted circles of approximately half an inch in diameter is utilized to control the sizes of drops collected. The circles can hold approximately 50–80 µl blood, and blood circles of 3.2–6 mm in diameter are required for most DBS assays (Benyshek 2010; Edelbroek, Van Der Heijden, and Stolk 2009; McDade, Williams, and Snodgrass 2007; Williams and McDade 2009). The quality and type of paper needs to be considered prior to sampling (Edelbroek, Van Der Heijden, and Stolk 2009; Mei et al. 2001). RBC transport easily within hardwood fiber sheets, providing higher clarity assays compared with softwood fiber papers, which have more complex pore structures (Li et al. 2014). For assessments of metal exposure, blood collection cards need to be treated prior to sample collection to remove background levels of heavy metals in the filter paper, which were found to significantly skew results (Funk et al. 2015).
The collected samples are allowed to dry for at least 15 min and for as long as 4 hr (McDade, Williams, and Snodgrass 2007; Ostler, Porter, and Buxton 2014). Once the samples are collected, these do not need to be centrifuged immediately and stored at cold temperature like traditional blood samples. DBS samples are typically stable for at least two weeks at room temperature and easy to store because they take up less space than liquid blood samples (Benyshek 2010; McDade, Williams, and Snodgrass 2007). In fact, some light-sensitive compounds demonstrated better stability stored as DBS samples than as liquid plasma or whole blood samples (Bowen et al. 2010). For best results, DBS samples need to be assayed or frozen within 2 wks after collection (Ostler, Porter, and Buxton 2014). Storing DBS samples in airtight leakproof plastic bags may generate heat and moisture accumulation due to a lack of air exchange, which leads to sample degradation (Edelbroek, Van Der Heijden, and Stolk 2009). Degradation of DBS samples might be reduced by adding vapor-phase desiccants to the plastic bags in which the DBS samples are stored or by adding humidity indicators. These storage bags can then be placed in waterproof bags for shipping and storage (Benyshek 2010; Edelbroek, Van Der Heijden, and Stolk 2009). While liquid blood samples need to be shipped on dry ice, DBS cards can remain at room temperature, lowering shipping costs (Amsterdam and Waldrop 2010). A lab freezer can store approximately 8,000–10,000 DBS samples, which is greater than the number of liquid samples that can be stored (Benyshek 2010; McDade, Williams, and Snodgrass 2007). Neonatal DBS samples can be stored at −20°C for several weeks or yrs, but sample loss was noted after 5 yrs (Edelbroek, Van Der Heijden, and Stolk 2009). Biobanking of liquid blood samples is also expensive, but DBS samples do not require cryopreservation and do not need to be stored with the same stringency, thus severely decreasing storage cost (Barbareschi, Cotrupi, and Guarrera 2008; Riegman et al. 2008).
DBS Challenges
While DBS samples have many advantages, there are several method issues that need to be addressed. The protocol for DBS sampling is not as well controlled as liquid blood sampling, which may cause some samples not to meet quality control standards (Edelbroek, Van Der Heijden, and Stolk 2009). Some sampling issues include blood spots that are too dilute, contaminated, smeared, spotted multiple times, or have too little volume (Ostler, Porter, and Buxton 2014). Samples that suffer from these issues cannot be used for analysis, as heterogeneity in sample spots may lead to erroneous results and conclusions. Differences in the type of filter paper utilized and in the paper batch may also affect the amount of blood that is absorbed (Chaudhuri et al. 2009). Thus, DBS sample analysis data may not be directly comparable to liquid blood samples using either serum or plasma due to differences in sample collection and blood composition (Benyshek 2010; McDade 2014). Since DBS samples only contain approximately 50 µl blood, assays that require more volume may not be compatible (McDade, Williams, and Snodgrass 2007). Therefore, individual methods for compound analysis may need to be developed for DBS analysis. The newly developed assays may not be immediately compatible with automated clinical assays using serum or plasma, which would delay the applicability of DBS for clinical analysis (Benyshek 2010; McDade, Williams, and Snodgrass 2007). However, some investigators have already been working to solve this problem. An immunoassay for C-reactive protein that is compatible with serum, plasma, and DBS was developed, and C-reactive protein measurements were highly correlated in the three media (Brindle et al. 2010). Multiplex analyses for DBS were also prepared to increase the number of proteins that can be assessed using a single DBS. A 25-plex multianalyte method with xMAP technology was developed to analyze inflammatory markers and growth factors (Skogstrand et al. 2005). Further, how the findings of DBS and liquid blood results relate also needs to be thoroughly assessed. Correction factors may be applied to the data to overcome some of these issues, but these factors may depend upon the method being used and population being studied (Benyshek 2010).
Current and Future DBS Applications
Several commercial assays were developed for DBS analysis, including inflammatory markers, growth factors, C-reactive protein, blood lipids, metabolism, immune response, and biomarkers (Benyshek 2010; McDade, Burhop, and Dohnal 2004; Skogstrand et al. 2005). DBS assays were also developed for a proinflammatory cytokine, IL-6, immunoglobulin E, leptin, total cholesterol, high-density lipoprotein cholesterol, and anti-Müllerian hormone with accuracies and precision similar to those of established serum-based assays (McDade et al. 2012; Miller and McDade 2012; Miller, Sharrock, and McDade 2006; Samuelsson et al. 2015; Tanner and McDade 2007). For analysis, a portion of the DBS is punched out and placed in an elution buffer to reconstitute the sample into hemolyzed liquid whole blood. For new assays, elution times, mixing times, temperature, and buffer constituents needed to obtain optimal concentrations and results need to be determined (Skogstrand et al. 2005). Recently, mass spectrometry (MS) analysis of DBS samples with off-line and on-line extractions has become more popular with improved instrumentation for surface analysis MS (Wagner et al. 2014). The first multiplexed assay using DBS was also recently reported. In this assay, 60 endogenous proteins were targeted using tryptic peptides and isotopelabeled analogs with multiple reaction monitoring (MRM). This method was found to be highly reproducible, and measured concentrations for 80% of the proteins were consistent after 10 d of storage at either −20, 4, or 37°C (Chambers et al. 2013). While these advances in DBS analysis hold promise for extension of current liquid assays to DBS applications, development of additional validated DBS enzyme immunoassays for use in field research will take time (McDade 2014; McDade, Williams, and Snodgrass 2007).
DBS samples may be employed to monitor exposure to environmental compounds over time. A study was conducted to measure the presence of perfluorinated compounds (PFC) in newborn DBS samples collected during 1997–2005. The presence of PFC in commercial products, such as surfactants, insecticides, and carpet, decreased in 2000 due to suspected liver toxicity. The presence of PFC in newborn DBS sampled during 2005–2007 was significantly decreased compared with earlier exposures, indicating that the population exposure to PFC had been reduced as a result of decreased commercial production. This analysis showed for the first time how DBS analysis might be used to measure temporal environmental exposure on the population level, as DBS samples are collected from more than 98% of newborns in the United States (Spliethoff et al. 2008). Exposure of infants to dichlorodiphenyldichloroethylene (p,p’-DDE), a breakdown product of the organochlorine pesticide dichlorodiphenyl-trichloroethane (DDT), was also assessed using DBS analysis (Burse et al. 1997). Highthroughput methods for Pb, PFC, and perchlorate analyses were also developed using MS (Hsieh et al. 2009; Kato et al. 2009; Otero-Santos et al. 2009; Schreinemachers et al. 2015). DBS samples were also utilized to measure exposure of pregnant women, newborns, and children to heavy metals, including Pb, Cd, As, and Hg (Chaudhuri et al. 2009; Funk et al. 2013; 2015; Michniewicz et al. 2015). Organochlorine pesticide and polychlorinated biphenyl (PCB) exposures have also been tracked in DBS samples from newborns to determine environmental exposures transferred from pregnant women to fetuses (Ma et al. 2014). Similarly, PCBs, chlorinated pesticides, and brominated flame retardants were investigated in a population study using DBS measurements to estimate exposure to environmental toxicants (Batterman and Chernyak 2014). Hemoglobin adducts of benzene oxide, a carcinogen, were also found in DBS samples at quantities similar to those seen in whole blood analyses (Funk et al. 2008; Rappaport, Funk, and Chaing 2008).
DBS analyses show potential for use in clinical trials. Clinical trials require multiple blood draws from patients over the course of several months to years. These repeated samplings may be tiresome for individuals and extremely costly. As DBS analysis becomes more common, expansion of the technology into clinical trials looks promising (Amsterdam and Waldrop. 2010). Another significant advantage of DBS sampling is the ability to obtain samples from populations in remote areas with lower levels of technology. Since a trained professional is not needed for sample collection, samples may be taken in areas where access to a hospital, clinic, or other sterile environment is limited (Benyshek 2010). Patients trained in proper sample collection procedures and quality control standards might also take their own samples at home and ship them to a lab or clinic for analysis to monitor their biomarker levels without needing to travel and be admitted to a hospital, saving them both time and money and increasing sampling efficiency (Edelbroek, Van Der Heijden, and Stolk 2009). These sampling advantages hold promise for further expanding DBS technology in the arena of environmental exposure assessment to reach remote populations, allowing for inclusion of isolated and chronically ill individuals who may otherwise have been overlooked. DBS sampling provides an avenue by which to signficantly increase population sizes of studies without further raising the cost of the study, since individuals can easily collect their own samples.
New Developments: Incorporation of “Omics”Technology
The range of blood analysis applications has significantly increased with the growth of “omics” technology in the past several years. The phenotype of an individual is the sum of the genome, transcriptome, proteome, and metabolome, and each “ome” branch is interconnected with the others, explaining the different traits of individuals (Titz et al. 2014; Trifonova, Lokhov, and Archakov 2013). An advantage of “omics” technologies is the ability to perform large-scale, high-throughput discovery screens without searching for a particular target compound. The ultimate goal of “omics” research is to identify every gene, protein, and chemical present within a sample (Vineis et al. 2009; Yin and Xu 2014). While the genome has been thoroughly studied, much remains to be discovered regarding the other three branches (Smith and Rappaport 2009). A current research objective is to parse out biomarkers of exposure from biomarkers of disease in “omics” experiments (Rappaport 2012a). The influences of an individual’s environment strongly affect the chemicals that are present in that person’s body. The effects of outside stressors, such as diet, pollution, pathogens, gut microflora, and drugs, can be measured in the blood metabolome using these techniques (Rappaport 2012a; Schnackenberg and Beger 2008). Data from “omics” experiments may also be applied to analyze toxicity pathways. Pathways of toxicity are cellular processes that link toxicant interactions with adverse outcomes (Bouhifd et al. 2013). These high-throughput “omics” techniques can be used to detect patterns (signatures) that are indicative of particular toxicities (Hartung 2009).
Of the four main “omics” branches, metabolomics and proteomics are perhaps the most developed and relevant for blood analysis. Although the human genome has been well studied, only 15–50% of disease was linked to genetic factors in genome-wide association studies (Vineis et al. 2009). Transcriptomics, which evaluates changes in mRNA expression levels, typically yields short term information instead of long-term. mRNA is heterogeneous and unstable, making it difficult to analyze for biomarkers of exposure (Vineis et al. 2009). Proteomics provides information regarding function, but the size of the plasma proteome is large (Anderson and Anderson 2002; Watkins and German 2002). The plasma proteome was estimated to contain approximately 50,000 proteins in different glycosylated forms, 500,000 tissue leakage proteins in a variety of splice variants with varying posttranslational modifications, and around 10,000,000 unique sequences of immunoglobulins (Anderson and Anderson 2002). While advances in characterizing the plasma proteome were accomplished, plasma consists of 55% albumin, which dominates protein detection, making it difficult to identify less abundant proteins in blood samples without first removing highly abundant proteins (Anderson and Anderson 2002). However, automated techniques that deplete the sample of highly abundant proteins prior to LCMS/MS analysis elevated throughput and coverage, helping ameliorate this issue (Dayon et al. 2014; Such-Sanmartín, Ventura-Espejo, and Jensen 2014). The metabolome is smaller than the genome or the proteome, with approximately 3,600 distinct confirmed serum/plasma metabolites (Trifonova, Lokhov, and Archakov 2013), but still contains vital information regarding exposure, creating an advantageous biomarker for discovery (Watkins and German 2002).
Blood is the predominant carrier of small molecules, such as hormones and nutrients within the body, making it an ideal biofluid for proteomic and metabolomic analyses (Trifonova, Lokhov, and Archakov 2013). Metabolic profiling is often conducted using either plasma or serum. Serum is utilized to transfer small molecules between cells and tissues, and thus contains a wealth of information about chemicals circulating within the body (Rappaport 2012a). Researchers have debated whether plasma or serum is the optimal biofluid for proteomic and metabolomic analyses. However, evidence indicates that the two biofluids are comparable in the number of detected compounds and display similar subject variability (Wedge et al. 2011; Yu et al. 2011). The reproducibility of results using the two biofluids may be analyte dependent, as some studies demonstrated serum to be more replicable than plasma for metabolite analysis (Breier et al. 2014), while another investigation obtained greater reproducibility with plasma (Yu et al. 2011). However, both studies demonstrated that metabolite concentrations are higher in serum than plasma (Breier et al. 2014; Yu et al. 2011). While this may make serum more advantageous for some applications, serum preparation requires more postprocessing steps than plasma and is more difficult to standardize. In addition, some important metabolites were only identified in plasma and not serum (Wedge et al. 2011). Therefore, selecting serum or plasma for analysis depends upon the compounds of interest, goals of the study, and preference of the researcher.
Current Challenges of “Omics” Strategies
While advances have been made in improving the applicability of “omics” sciences, several key challenges remain to be addressed. Due to a lack of standardized procedures, “omics” results using MS may be difficult to reproduce between labs (Vineis et al. 2009). However, recently, a large-scale analysis of 1000 human plasma samples was conducted using an isobaric mass tagging quantification procedure, which provided consistent results for proteins with stable expression levels across samples (Cominetti et al. 2016). Different sample handling procedures, including temperatures, timing, and humidity levels, were shown to produce variation in proteomics and metabolomics experiments, and steps need to be taken to reduce inconsistencies in sample preparation (West-Nørager et al. 2007; Yin et al. 2013). Samples are sensitive to storage conditions, and incorrect storage temperatures might lead to increased enzymatic cleavage of plasma samples (Pinto et al. 2014; Yin and Xu 2014). Improved nuclear magnetic resonance (NMR) methods with optimized sample preparation procedures for metabolomics analyses were recently developed, improving quantitation and coverage of different protein classes, such as amino acids, organic acids, and carbohydrates (Gowda, Gowda, and Raftery 2015). Further, there is a wide range of “normal” levels of peptides, proteins, and metabolites among healthy individuals, making it difficult to establish a baseline for nonaffected, healthy individuals (Bischoff and Luider 2004; Enroth et al. 2015; Vineis et al. 2009; Watkins and German 2002). However, as more metabolomics data is being published, this limitation is decreasing due to the abundance of information regarding normal and abnormal ranges of metabolites (Li, Todor, and Luo 2016).
Despite the large number of experiments performed, few new proteins have been introduced as biomarkers (Liotta, Ferrari, and Petricoin 2003), and only approximately 10–20 blood metabolites are currently assayed in clinical diagnostics (Trifonova, Lokhov, and Archakov 2013). However, some exposures are unlikely to be described by only a single protein or metabolite change; instead, these exposures may induce changes in several proteins or metabolites, resulting in characteristic patterns (Liotta, Ferrari, and Petricoin 2003). Currently, “omics” techniques still suffer from coverage issues, making it challenging to identify relevant expression patterns (Anderson and Anderson 2002). A method for analyzing the low-molecular-weight plasma proteome using top-down MS was developed, making it possible to quantify proteins that may not be detected in a typical mass spectrum (Cheon et al. 2016). Assaying all of the proteins or metabolites in a sample and correctly identifying them is extremely challenging, especially considering the (1) number to be characterized, (2) influence of mass defect on identification, (3) incompleteness of spectral libraries and databases, and (4) instrumental limitations (Pleil and Isaacs 2015; Trifonova, Lokhov, and Archakov 2013; Veenstra 2007; Yin and Xu 2014). Improved separation techniques and faster instruments might alleviate the coverage issue, but not without significantly increasing the expense of the assay (Pasikanti, Ho, and Chan 2008). Despite the current expense, “omics” research might help reduce health care costs in the long run (Anderson and Anderson 2002; Smith and Rappaport 2009; Vineis et al. 2009). The slow transfer of biomarker data from the research lab into clinical utility might seem daunting, but implementation should improve as assays become more developed and faster instruments are utilized. The main challenge facing “omics” techniques today is the lack of validation strategies for novel biomarkers. It is difficult to know if altered expression levels are a direct indicator of exposure or a downstream result due to activation of an AOP. Affinity-based tests might be used for biomarker validation when available, and single-reaction monitoring may be employed to assess a single protein in a protein mixture (Veenstra 2007). As high-throughput discovery techniques continue to evolve, their use for clinical and environmental applications is certain to increase.
Range of “Omics” Applicability and Future Directions
“Omics” strategies have been used in clinical settings for several years. Metabolite differences between controls and patients with known diseases were utilized to identify signatures of disease. Investigations of metabolomic biomarkers of diabetes, macular degeneration, asthma, Parkinson’s disease, and tuberculosis have all been conducted in clinical research (Li, Todor, and Luo 2016). NMR analyses using blood sera were found to predict a type of coronary artery disease with greater than 90% specificity (Mayr 2008). Metabolomics was also utilized to identify differences in patients with small-cell lung cancer, and metabolites that are associated with worse survival times were identified in plasma but not serum samples (Wedge et al. 2011). Further, plasma metabolites released after exercise-induced myocardial injury were also detected by metabolomics, and an elevation in compounds in the citric acid cycle were noted in myocardial ischemia, a condition in which blood flow to the heart is reduced, causing the heart to receive insufficient oxygen levels (Mayr 2008). An analysis of plasma metabolites in control subjects and patients with autosomal dominant polycystic kidney disease was conducted to determine whether mealtimes and sampling time of day exerted an effect on metabolite levels. Interestingly, higher metabolome variability was observed after morning rather than evening mealtimes (Kim et al. 2014). This study demonstrates the importance of standardizing sampling procedures during experiments to ensure that consistent and relevant data are obtained across different studies for comparison.
Recently, “omics” techniques have been utilized for environmental exposure applications. Proteomic profiles of subjects naturally exposed to different levels of As in drinking water were collected, and surface-enhanced laser desorption/ionization-timeof-flight-mass spectrometry (SELDI-TOF-MS) was utilized to detect differences in 20 proteins in serum proteome profiles between low-, medium-, and high-exposure groups (Zhao et al. 2010). Similarly, SELDI-TOF-MS was employed in a different study to determine the impact of benzene exposure in shoe-factory workers. The human serum proteome was examined, and platelet factor 4 and connective tissue activating peptide III were consistently downregulated in benzene-exposed workers (Vermeulen et al. 2005). Metabolomics was also used to investigate serum biomarkers of workers exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Hosnijeh et al. 2013). Blood is also a reliable medium for metabolomics studies in nutritional research, as blood metabolites include fat, lactate, and glucose (Savorani et al. 2013), and blood samples were employed to characterize lipid metabolic profiles after high-fat meals and to identify biomarkers of sucrose intake (Beckmann et al. 2016; Bijlsma et al. 2006). These types of studies demonstrate how “omics” experiments might be utilized to detect proteins and metabolites that are sensitive to low levels of exposure, revealing early biological effects of AOP. A strategy to incorporate metabolomics data measured in three different biofluids, plasma, urine, and saliva, was also implemented to obtain a more comprehensive picture of the complex interactions that occur within the human body as a whole (Do et al. 2015). This valuable information might be utilized to gain a better understanding of environmental exposure science.
Although not discussed in detail here, new bioinformatics tools are becoming increasingly important to assess complex data streams from metabolomics studies. Currently, large databases, computational tools, and cloud computing are addressing data interpretation challenges from an ever-increasing volume of biomarker and bioindicator information (Bonvallot et al. 2014; Chen et al. 2013; Judson 2010)
Summary
Blood is clearly an invaluable biofluid for analysis of environmental exposure, despite requiring a more invasive collection procedure than for excreted biofluids, such as breath or urine. Under the assumption of sufficient analytical sensitivity, it should be possible to characterize the majority of human exposures (regardless of source or exposure pathway) in blood as it comes in contact with all living cells. Exogenous compounds, endogenous compounds, and inflammatory, molecular modification, and damage biomarkers and bioindicators can all be analyzed in blood samples to assess exposure, effect, and susceptibility. In cases where compounds that have long elimination halflives, nonspecific urinary metabolites, or protein adducts need to be investigated, blood is the best biofluid for analysis.
One of the more promising new research areas for blood analysis is implementation of DBS as a surrogate for whole blood samples. Here, there is essentially no volumetric limit for sampling (as encountered for 10 ml whole blood samples) as each sample contains only ~50 μl. DBS sampling can be self-administered, and once dried, the sample is stable and may be shipped and stored at room temperature for a couple weeks. Although the DBS approach loses volatile constituents, including water and dissolved gases, there are advantages in that the remaining materials such as proteins and inorganics become highly concentrated. DBS assays have been performed to detect pesticides, develop multiplex protein assays, and identify protein adducts.
Metabolomics and proteomics analyses are also increasing in frequency and application to environmental assessment of blood samples. Improvements in MS have led to faster, more sensitive instruments capable of detecting small molecules and proteins in blood, serum, and plasma. High-throughput experiments are being performed to identify patterns in expression levels that are indicative of different exposures and disease states. While “omics” techniques still suffer from the overabundance of HSA, coverage issues, and need for standardization and validation, their usage in clinical and environmental fields is on the rise as technology continues to improve.
In conclusion, blood sampling in the environmental health field remains a viable and important methodology. Newer analytical technologies that require smaller volumes are closing the gap in logistical concerns in regards to acceptance, invasiveness, and need for trained personnel. Due to these advancements, blood sampling is expected to remain a standard measurement of environmental exposure assessment in the future.
Acknowledgments
The authors are grateful for advice and encouragement from Myriam Medina-Vera, Jon Sobus, Michael Madden, Timothy Buckley, and Michelle Angrish from the U.S. Environmental Protection Agency (EPA). The U.S. Environmental Protection Agency through its Office of Research and Development has subjected this article to agency administrative review and approved it for publication. Mention of trade names or commercial products does not constitute endorsement for use.
References
- Alves A, Kucharska A, Erratico C, Xu F, Hond ED, Koppen G, Vanermen G, Covaci A, and Voorspoels S. 2014. Human biomonitoring of emerging pollutants through noninvasive matrices: State of the art and future potential. Analytical and Bioanalytical Chemistry 406:4063–88. doi: 10.1007/s00216-014-7748-1. [DOI] [PubMed] [Google Scholar]
- Amann A, de Lacy Costello B, Miekisch W, Schubert J, Buszewski B, Pleil J, Ratcliffe N, and Risby T. 2014. The human volatilome: Volatile organic compounds (VOCs) in exhaled breath, skin emanations, urine, feces and saliva. Journal of Breath Research 8:034001. doi: 10.1088/1752-7155/8/3/034001. [DOI] [PubMed] [Google Scholar]
- Ames BN 2001. DNA damage from micronutrient deficiencies is likely to be a major cause of cancer. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 475:7–20. doi: 10.1016/S0027-5107(01)00070-7. [DOI] [PubMed] [Google Scholar]
- Amsterdam PV, and Waldrop C. 2010. The application of dried blood spot sampling in global clinical trials. Bioanalysis 2:1783–86. doi: 10.4155/bio.10.158. [DOI] [PubMed] [Google Scholar]
- Anderson NL, and Anderson NG. 2002. The human plasma proteome. Molecular Cellular Proteomics 1:845–67. doi: 10.1074/mcp.R200007-MCP200. [DOI] [PubMed] [Google Scholar]
- Angrish MM, Madden MC, and Pleil JD. 2015. Probe molecule (PrM) approach in adverse outcome pathway (AOP) based high-throughput screening (HTS): In vivo discovery for developing in vitro target methods. Chemical Research in Toxicology 28:551–59. doi: 10.1021/acs.chemrestox.5b00024. [DOI] [PubMed] [Google Scholar]
- Angrish MM, Pleil JD, Stiegel MA, Madden MC, Moser VC, and Herr DW. 2016. Taxonomic applicability of inflammatory cytokines in adverse outcome pathway (AOP) development. Journal of Toxicology and Environmental Health, Part A 79:184–96. doi: 10.1080/15287394.2016.1138923. [DOI] [PubMed] [Google Scholar]
- Araoud M, Gazzah N, Douki W, Najjar MF, and Kenani A. 2016. Rapid multi-residue method for the determination of pesticide residues in human serum. African Journal of Biotechnology 11:12579–85. [Google Scholar]
- Arayasiri M, Mahidol C, Navasumrit P, Autrup H, and Ruchirawat M. 2010. Biomonitoring of benzene and 1,3-butadiene exposure and early biological effects in traffic policemen. Science of the Total Environment 408:4855–62. doi: 10.1016/j.scitotenv.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Avogbe PH, Ayi-Fanou L, Autrup H, Loft S, Fayomi B, Sanni A, Vinzents P, and Møller P. 2005. Ultrafine particulate matter and high-level benzene urban air pollution in relation to oxidative DNA damage. Carcinogenesis 26:613–20. doi: 10.1093/carcin/bgh353. [DOI] [PubMed] [Google Scholar]
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, and Schwartz J. 2009. Rapid DNA methylation changes after exposure to traffic particles. American Journal of Respiratory and Critical Care Medicine 179:572–78. doi: 10.1164/rccm.2008071097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranska A, Smolinska A, Boots AW, Wallinga JW, and Van Schooten FJ. 2015. Dynamic collection and analysis of volatile organic compounds from the headspace of cell cultures. Journal of Breath Research 9:047102. doi: 10.1088/1752-7155/9/4/047102. [DOI] [PubMed] [Google Scholar]
- Barbareschi MS, Cotrupi S, and Guarrera GM. 2008. Biobanks: Instrumentation, personnel and cost analysis. Pathologica 100:139–48. [PubMed] [Google Scholar]
- Barr DB, and Angerer J. 2006. Potential uses of biomonitoring data: A case study using the organophosphorus pesticides chlorpyrifos and malathion. Environmental Health perspectives 114:1763–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr DB, Barr JR, Maggio VL, Whitehead RD Jr., Sadowski MA, Whyatt RM, and Needham LL. 2002. A multi-analyte method for the quantification of contemporary pesticides in human serum and plasma using highresolution mass spectrometry. Journal of Chromatography B 778:99–111. doi: 10.1016/S0378-4347(01)00444-3. [DOI] [PubMed] [Google Scholar]
- Basu A, Som A, Ghoshal S, Mondal L, Chaubey RC, Bhilwade HN, Rahman MM, and Giri AK. 2005. Assessment of DNA damage in peripheral blood lymphocytes of individuals susceptible to arsenic induced toxicity in West Bengal, India. Toxicology Letters 159:100–12. doi: 10.1016/j.toxlet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Batterman S, and Chernyak S. 2014. Performance and storage integrity of dried blood spots for PCB, BFR and pesticide measurements. Science of the Total Environment 494-495:252–60. doi: 10.1016/j.scitotenv.2014.06.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach AC, and Gupta RC. 1992. Human biomonitoring and the 32P-postlabeling assay. Carcinogenesis 13:1053–74. doi: 10.1093/carcin/13.7.1053. [DOI] [PubMed] [Google Scholar]
- Bechtold WE, Willis JK, Sun JD, Griffith WC, and Reddy TV. 1992. Biological markers of exposure to benzene: 5-Phenylcysteine in albumin. Carcinogenesis 13:1217–20. doi: 10.1093/carcin/13.7.1217. [DOI] [PubMed] [Google Scholar]
- Beckmann M, Joosen AM, Clarke MM, Mugridge O, Frost G, Engel B, Taillart K, Lloyd AJ, Draper J, and Lodge JK. 2016. Changes in the human plasma and urinary metabolome associated with acute dietary exposure to sucrose and the identification of potential biomarkers of sucrose intake. Molecular Nutrition & Food Research 60:444–57. doi: 10.1002/mnfr.v60.2. [DOI] [PubMed] [Google Scholar]
- Bellinger D, Leviton A, Waternaux C, Needleman H, and Rabinowitz M. 1987. Longitudinal analyses of prenatal and postnatal lead exposure and early cognitive development. The New England Journal of Medicine 316:1037–43. doi: 10.1056/NEJM198704233161701. [DOI] [PubMed] [Google Scholar]
- Belloni P, Meschini R, and Palitti F. 2008. Effects of storage conditions of human whole blood on the viability of lymphocytes. International Journal of Radiation Biology 84:613–19. doi: 10.1080/09553000802203630. [DOI] [PubMed] [Google Scholar]
- Benyshek DC 2010. Use of dried blood spots: An ideal tool for medical anthropology “in the field.” Journal of Diabetes Science and Technology 4:255–57. doi: 10.1177/193229681000400205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, Van Ommen B, and Smilde AK. 2006. Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Analytical Chemistry 78:567–74. doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- Binková B, Giguère Y, Rössner P Jr., Dostál M, and Šrám RJ. 2000. The effect of dibenzo[a,l]pyrene and benzo[a] pyrene on human diploid lung fibroblasts: The induction of DNA adducts, expression of p53 and p21WAF1 proteins and cell cycle distribution. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 471:57–70. doi: 10.1016/S1383-5718(00)00111-X. [DOI] [PubMed] [Google Scholar]
- Bischoff R, and Luider TM. 2004. Methodological advances in the discovery of protein and peptide disease markers. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences 803:27–40. doi: 10.1016/j.jchromb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Boffetta P 2010. Biomarkers in cancer epidemiology: An integrative approach. Carcinogenesis 31:121–26. doi: 10.1093/carcin/bgp269. [DOI] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun H-M, Jiang J, Marinelli B, Pesatori AC, Bertazzi PA, and Yang AS. 2007. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Research 67:876–80. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, Pegoraro V, Motta V, Tarantini L, Cantone L, Schwartz J, Bertazzi PA, and Baccarelli A. 2010. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environmental Health Perspectives 118:763–68. doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvallot N, Tremblay-Franco M, Chevrier C, Canlet C, Debrauwer L, Cravedi J-P, and Cordier S. 2014. Potential input from metabolomics for exploring and understanding the links between environment and health. Journal of Toxicology and Environmental Health, Part B 17:21–44. doi: 10.1080/10937404.2013.860318. [DOI] [PubMed] [Google Scholar]
- Bouhifd M, Hartung T, Hogberg HT, Kleensang A, and Zhao L. 2013. Review: Toxicometabolomics. Journal of Applied Toxicology 33:1365–83. doi: 10.1002/jat.v33.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen CL, Hemberger MD, Kehler JR, and Evans CA. 2010. Utility of dried blood spot sampling and storage for increased stability of photosensitive compounds. Bioanalysis 2:1823–28. doi: 10.4155/bio.10.142. [DOI] [PubMed] [Google Scholar]
- Boysen G, and Hecht SS. 2003. Analysis of DNA and protein adducts of benzo[a]pyrene in human tissues using structure-specific methods. Mutation Research/ Reviews in Mutation Research 543:17–30. doi: 10.1016/S1383-5742(02)00068-6. [DOI] [PubMed] [Google Scholar]
- Breier M, Wahl S, Prehn C, Fugmann M, Ferrari U, Weise M, Banning F, Seissler J, Grallert H, Adamski J, and Lechner A. 2014. Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS ONE 9:e89728. doi: 10.1371/journal.pone.0089728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle E, Fujita M, Shofer J, and O’Connor KA. 2010. Serum, plasma, and dried blood spot high-sensitivity C-reactive protein enzyme immunoassay for population research. Journal of Immunological Methods 362:112–20. doi: 10.1016/j.jim.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodkin E, Copes R, Mattman A, Kennedy J, Kling R, and Yassi A. 2007. Lead and mercury exposures: Interpretation and action. Canadian Medical Association Journal 176:59–63. doi: 10.1503/cmaj.060790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugge D, Lane KJ, Stewart A, Tai AK, and Woodin M. 2013. Highway proximity associations with blood markers of inflammation: Evidence for a role of IL-1β. Journal of Toxicology and Environmental Health, Part A 76:201–05. doi: 10.1080/15287394.2013.752325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik LT, Kloth S, Baur X, Preisser AM, and Schwarzenbach H. 2013. Circulating mitochondrial DNA as biomarker linking environmental chemical exposure to early preclinical lesions elevation of mtDNA in human serum after exposure to carcinogenic halo-alkane-based pesticides. PLoS ONE 8:e64413. doi: 10.1371/journal.pone.0064413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger J, and Gochfeld M. 2001. On developing bioindicators for human and ecological health. Environmental Monitoring and Assessment 66:23–46. doi: 10.1023/A:1026476030728. [DOI] [PubMed] [Google Scholar]
- Burse VW, DeGuzman MR, Korver MP, Najam AR, Williams CC, Hannon WH, and Therrell BL. 1997. Preliminary investigation of the use of dried-blood spots for the assessment of in utero exposure to environmental pollutants. Biochemical and Molecular Medicine 61:236–39. doi: 10.1006/bmme.1997.2603. [DOI] [PubMed] [Google Scholar]
- Calafat AM, Ye X, Silva MJ, Kuklenyik Z, and Needham LL. 2005. Human exposure assessment to environmental chemicals using biomonitoring. International Journal of Andrology 29:166–71. doi: 10.1111/j.13652605.2005.00570.x. [DOI] [PubMed] [Google Scholar]
- Chambers AG, Percy AJ, Yang J, Camenzind AG, and Borchers CH. 2013. Multiplexed quantitation of endogenous proteins in dried blood spots by multiple reaction monitoring-mass spectrometry. Molecular & Cellular Proteomics 12:781–91. doi: 10.1074/mcp.M112.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channell MM, Paffett ML, Devlin RB, Madden MC, and Campen MJ. 2012. Circulating factors induce coronary endothelial cell activation following exposure to inhaled diesel exhaust and nitrogen dioxide in humans: Evidence from a novel translational in vitro model. Toxicological Sciences 127:179–86. doi: 10.1093/toxsci/kfs084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri SN, Butala SJM, Ball RW, and Braniff CT; Rocky Mountain Biomonitoring Consortium. 2009. Pilot study for utilization of dried blood spots for screening of lead, mercury and cadmium in newborns. Journal of Exposure Science and Environmental Epidemiology 19:298–316. doi: 10.1038/jes.2008.19. [DOI] [PubMed] [Google Scholar]
- Chen CJ, I Hsu L, H Wang C, L Shih W, H Hsu Y, P Tseng M, C Lin Y, L Chou W, Y Chen C, Y Lee C, H Wang L, C Cheng Y, L Chen C, Y Chen S, H Wang Y, M Hsueh Y, Y Chiou H, and M Wu M. 2005. Biomarkers of exposure, effect, and susceptibility of arsenic-induced health hazards in Taiwan. Toxicology and Applied Pharmacology 206:198–206. doi: 10.1016/j.taap.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Chen J, Qian F, Yan W, and Shen B. 2013. Translational biomedical informatics in the cloud: Present and future. Biomedical Research International 2013:1–8. doi: 10.1155/2013/658925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Jin T, Huang B, Chang X, Lei L, Nordberg GF, and Nordberg M. 2006. Plasma metallothionein antibody and cadmium-induced renal dysfunction in an occupational population in China. Toxicological Sciences 91:104–12. doi: 10.1093/toxsci/kfj053. [DOI] [PubMed] [Google Scholar]
- Cheon DH, Nam EJ, Park KH, Woo SJ, Lee HJ, Kim HC, Yang EG, Lee C, and Lee JE. 2016. Comprehensive analysis of low-molecular-weight human plasma proteome using top-down mass spectrometry. Journal of Proteome Research 15:229–44. doi: 10.1021/acs.jproteome.5b00773. [DOI] [PubMed] [Google Scholar]
- Cohen Hubal EA 2009. Biologically relevant exposure science for 21st Century toxicity testing. Toxicological Sciences 111:226–32. doi: 10.1093/toxsci/kfp159. [DOI] [PubMed] [Google Scholar]
- Cominetti O, Galindo AN, Corthésy J, Moreno SO, Irincheeva I, Valsesia A, Astrup A, Saris WHM, Hager J, Kussmann M, and Dayon L. 2016. Proteomic biomarker discovery in 1000 human plasma samples with mass spectrometry. Journal of Proteome Research 15:389–99. doi: 10.1021/acs.jproteome.5b00901. [DOI] [PubMed] [Google Scholar]
- Counter SA, Buchanan LH, and Ortega F. 2015. Blood lead levels in Andean infants and young children in Ecuador: An international comparison. Journal of Toxicology and Environmental Health, Part A 78:778–87. doi: 10.1080/15287394.2015.1031050. [DOI] [PubMed] [Google Scholar]
- Crovella S, Bianco AM, Vuch J, Zupin L, Moura RR, Trevisan E, Schneider M, Brollo A, Nicastro EM, Cosenzi A, Zabucchi G, and Borelli V. 2016. Iron signature in asbestos-induced malignant pleural mesothelioma: A population-based autopsy study. Journal of Toxicology and Environmental Health, Part A 79:129–42. doi: 10.1080/15287394.2015.1123452. [DOI] [PubMed] [Google Scholar]
- Da Silva ALG, Da Rosa HT, Karnopp TE, Charlier CF, Ellwanger JH, Moura DJ, Possuelo LG, De Moura Valim AR, Guecheva TN, and Henriques JAP. 2013. Evaluation of DNA damage in COPD patients and its correlation with polymorphisms in repair genes. BMC Medical Genetics 14:93. doi: 10.1186/1471-2350-14-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Colombo R, Giustarini D, and Milzani A. 2006. Biomarkers of oxidative damage in human disease. Clinical Chemistry 52:601–23. doi: 10.1373/clinchem.2005.061408. [DOI] [PubMed] [Google Scholar]
- Dayon L, Galindo AN, Corthésy J, Cominetti O, and Kussmann M. 2014. Comprehensive and scalable highly automated MS-based proteomic workflow for clinical biomarker discovery in human plasma. Journal of Proteome Research 13:3837–45. doi: 10.1021/pr500635f. [DOI] [PubMed] [Google Scholar]
- de Lacy Costello B, Amann A, Al-Kateb H, Flynn C, Filipiak W, Khalid T, Osborne D, and Ratcliffe NM. 2014. A review of the volatiles from the healthy human body. Journal of Breath Research 8:014001. doi: 10.1088/17527155/8/1/014001. [DOI] [PubMed] [Google Scholar]
- Devaraj S, Xu DY, and Jialal I. 2003. C-Reactive protein increases plasminogen activator inhibitor-1 expression and activity in human aortic endothelial cells: Implications for the metabolic syndrome and atherothrombosis. Circulation 107:398–404. doi: 10.1161/01.CIR.0000052617.91920.FD. [DOI] [PubMed] [Google Scholar]
- Do KT, Kastenmüller G, Mook-Kanamori DO, Yousri NA, Theis FJ, Suhre K, and Krumsiek J. 2015. Network-based approach for analyzing intraand interfluid metabolite associations in human blood, urine, and saliva. Journal of Proteome Research 14:1183–94. doi: 10.1021/pr501130a. [DOI] [PubMed] [Google Scholar]
- Duarte SC, Pena A, and Lino CM. 2011. Human ochratoxin a biomarkers-from exposure to effect. Critical Reviews in Toxicology 41:187–212. doi: 10.3109/10408444.2010.529103. [DOI] [PubMed] [Google Scholar]
- Edelbroek PM, Van Der Heijden J, and Stolk LML. 2009. Dried blood spot methods in therapeutic drug monitoring: Methods, assays, and pitfalls. Therapeutic Drug Monitoring 31:327–36. doi: 10.1097/FTD.0b013e31819e91ce. [DOI] [PubMed] [Google Scholar]
- Ellis HJ, and Venturini DS. 2013. Demonstration of a frozen sample aliquotter to prepare plasma and serum aliquots without thawing frozen parent samples. Biopreservation and Biobanking 11:153–60. doi: 10.1089/bio.2012.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enroth S, Enroth SB, Johansson A, and Gyllensten U. 2015. Effect of genetic and environmental factors on protein biomarkers for common non-communicable disease and use of personally normalized plasma protein profiles (PNPPP). Biomarkers 20:355–64. doi: 10.3109/1354750X.2015.1093546. [DOI] [PubMed] [Google Scholar]
- Fanali G, di Masi A, Trezza V, Marino M, Fasano M, and Ascenzi P. 2012. Human serum albumin: From bench to bedside. Molecular Aspects of Medicine 33:209–90. doi: 10.1016/j.mam.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Fasco MJ, Iii CR, Stack RF, O’hehir C, Barr JR, and Eadon GA. 2007. Cyanide adducts with human plasma proteins: Albumin as a potential exposure surrogate. Chemical Research in Toxicology 20:677–84. doi: 10.1021/tx6003425. [DOI] [PubMed] [Google Scholar]
- Fenech M 2001. The role of folic acid and Vitamin B12 in genomic stability of human cells. Mutation Research/ Fundamental and Molecular Mechanisms of Mutagenesis 475:57–67. doi: 10.1016/S0027-5107(01)00079-3. [DOI] [PubMed] [Google Scholar]
- Feng N, Li Y, Long C, Xia ZL, and Brandt-Rauf PW. 2014. Effects of DNA repair gene polymorphisms on DNA damage in human lymphocytes induced by a vinyl chloride metabolite in vitro. Biomarkers 19:281–86. doi: 10.3109/1354750X.2014.907345. [DOI] [PubMed] [Google Scholar]
- Franco R, Schoneveld O, Georgakilas AG, and Panayiotidis MI. 2008. Oxidative stress, DNA methylation and carcinogenesis. Cancer Letters 266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Freire C, Koifman RJ, and Koifman S. 2015. Hematological and hepatic alterations in Brazilian population heavily exposed to organochlorine pesticides. Journal of Toxicology and Environmental Health, Part A 78:534–48. doi: 10.1080/15287394.2014.999396. [DOI] [PubMed] [Google Scholar]
- Funk WE, McGee JK, Olshan AF, and Ghio AJ. 2013. Quantification of arsenic, lead, mercury and cadmium in newborn dried blood spots. Biomarkers 18:174–77. doi: 10.3109/1354750X.2012.750379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WE, Pleil JD, Pedit JA, Boundy MG, Yeatts KB, Nash DG, Trent CB, El Sadig M, Davidson CA, and Leath D. 2014. Indoor air quality in the United Arab Emirates. Journal of Environmental Protection 5:709–22. doi: 10.4236/jep.2014.58072. [DOI] [Google Scholar]
- Funk WE, Pleil JD, Sauter DJ, McDade TW, and Holl JL. 2015. Use of dried blood spots for estimating children’s exposures to heavy metals in epidemiological research. Journal of Environmental & Analytical Toxicology S7:002. doi: 10.4172/2161-0525.S7-002. [DOI] [Google Scholar]
- Funk WE, Waidyanatha S, Chaing SH, and Rappaport SM. 2008. Hemoglobin adducts of benzene oxide in neonatal and adult dried blood spots. Cancer Epidemiology, Biomarkers & Prevention 17:1896–901. doi: 10.1158/10559965.EPI-08-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo V, Khan A, Gonzales C, Phillips DH, Schoket B, Györffy E, Anna L, Kovács K, Møller P, Loft S, Kyrtopoulos S, Matullo G, and Vineis P. 2008. Validation of biomarkers for the study of environmental carcinogens:A review. Biomarkers 13:505–34. doi: 10.1080/13547500802054611. [DOI] [PubMed] [Google Scholar]
- George BJ, Reif DM, Gallagher JE, WilliamsDeVane CR, Heindenfelder BL, Hudgens EE, Jones W, Neas L, Cohen Hubal EA, and Edwards SW. 2015. Datadriven asthma endotypes defined from blood biomarker and gene expression data. PLoS ONE 10:e0117445. doi: 10.1371/journal.pone.0117445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghio A, Sangani R, and Roggli V. 2014. Expanding the spectrum of particleand fiber-associated interstitial lung diseases. Turk Toraks Dergisi 15:1–8. doi: 10.5152/ttd.2014.3950. [DOI] [Google Scholar]
- Ghio A, J. J, Sobus R, Pleil JD and Madden MC 2012. Controlled human exposures to diesel exhaust. Swiss Medical Weekly 142:w13597. [DOI] [PubMed] [Google Scholar]
- Gillio-Meina C, Cepinskas G, Cecchini EL, and Fraser DD. 2013. Translational research in pediatrics II: Blood collection, processing, shipping, and storage. Pediatrics 131:754–66. doi: 10.1542/peds.2012-1181. [DOI] [PubMed] [Google Scholar]
- Glei M, Habermann N, Osswald K, Seidel C, Persin C, Jahreis G, and Pool-Zobel BL. 2005. Assessment of DNA damage and its modulation by dietary and genetic factors in smokers using the comet assay: A biomarker model. Biomarkers 10:203–17. doi: 10.1080/13547500500138963. [DOI] [PubMed] [Google Scholar]
- Gowda GAN, Gowda YN, and Raftery D. 2015. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Analytical Chemistry 87:706–15. doi: 10.1021/ac503651e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SC, Sharma A, Mishra M, Mishra RK, and Chowdhuri DK. 2010. Heat shock proteins in toxicology: How close and how far? Life Sciences 86:377–84. doi: 10.1016/j.lfs.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Guthrie R, and Susi A. 1963. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics 32:338–43. [PubMed] [Google Scholar]
- Hartung T 2009. Toxicology for the twenty-first century. Nature 460:208–12. doi: 10.1038/460208a. [DOI] [PubMed] [Google Scholar]
- Hoet P, De Smedt E, Ferrari M, Imbriani M, Maestri L, Negri S, De Wilde P, Lison D, and Haufroid V. 2009. Evaluation of urinary biomarkers of exposure to benzene: Correlation with blood benzene and influence of confounding factors. International Archives of Occupational and Environmental Health 82:985–95. doi: 10.1007/s00420-008-0381-6. [DOI] [PubMed] [Google Scholar]
- Holland N, Bolognesi C, Kirsch-Volders M, Bonassi S, Zeiger E, Knasmueller S, and Fenech M. 2008. The micronucleus assay in human buccal cells as a tool for biomonitoring DNA damage: The HUMN project perspective on current status and knowledge gaps. Mutation Research/ Reviews in Mutation Research 659:93–108. doi: 10.1016/j.mrrev.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Holland NT, Pfleger L, Berger E, Ho A, and Bastaki M. 2005. Molecular epidemiology biomarkers—Sample collection and processing considerations. Toxicology and Applied Pharmacology 206:261–68. doi: 10.1016/j.taap.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Holland NT, Smith MT, Eskenazi B, and Bastaki M. 2003. Biological sample collection and processing for molecular epidemiological studies. Mutation Research/ Reviews in Mutation Research 543:217–34. doi: 10.1016/S1383-5742(02)00090-X. [DOI] [PubMed] [Google Scholar]
- Holland NT, Zhang L, and Smith MT. 2000. Cytogenetic markers and air pollution. In Relationships between acute and chronic effects of air pollution, ed. Heinrich U, and Mohr U, 65–78. Washington, DC: International Life Sciences Institute (ILSI). [Google Scholar]
- Hosnijeh FS, Pechlivanis A, Keun HC, Portengen L, Bueno-de-Mesquita HB, Heederik D, and Vermeulen R. 2013. Serum metabolomics perturbations among workers exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Environmental and Molecular Mutagenesis 54:558–65. doi: 10.1002/em.21802. [DOI] [PubMed] [Google Scholar]
- Hou L, Zhu Z-Z, Zhang X, Nordio F, Bonzini M, Schwartz J, Hoxha M, Dioni L, Marinelli B, Pegoraro V, Apostoli P, Bertazzi PA, and Baccarelli A 2010. Airborne particulate matter and mitochondrial damage: A cross-sectional study. Environmental Health 9:48. doi: 10.1186/1476-069X-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoxha M, Dioni L, Bonzini M, Pesatori AC, Fustinoni S, Cavallo D, Carugno M, Albetti B, Marinelli B, Schwartz J, Bertazzi PA, and Baccarelli A. 2009. Association between leukocyte telomere shortening and exposure to traffic pollution: A cross-sectional study on traffic officers and indoor office workers. Environmental Health 8:41. doi: 10.1186/1476-069X-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh H-F, Chang W-S, Hsieh Y-K, and Wang C-F. 2009. Lead determination in whole blood by laser ablation coupled with inductively coupled plasma mass spectrometry. Talanta 79:183–88. doi: 10.1016/j.talanta.2009.03.027. [DOI] [PubMed] [Google Scholar]
- Huang Y-CT, Schmitt M, Yang Z, Que LG, Stewart JC, Frampton MW, and Devlin RB. 2010. Gene expression profile in circulating mononuclear cells after exposure to ultrafine carbon particles. Inhalation Toxicology 22:835–46. doi: 10.3109/08958378.2010.486419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NM, Qian G, Xu L, Tietze D, Marroquin-Cardona A, Robinson A, Rodriguez M, Kaufman L, Cunningham K, Wittmer J, Guerra F, Donnelly KC, Williams JH, Wang JS, and Phillips TD. 2010. Aflatoxin and PAH exposure biomarkers in a U.S. population with a high incidence of hepatocellular carcinoma. Science of the Total Environment 408:6027–31. doi: 10.1016/j.scitotenv.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K, and Cocker J. 2003. A human exposure study to investigate biological monitoring methods for 2-butoxyethanol. Biomarkers 8:360–70. doi: 10.1080/13547500310001600941. [DOI] [PubMed] [Google Scholar]
- Judson R 2010. Public databases supporting computational toxicology. Journal of Toxicology and Environmental Health, Part B 13:218–31. doi: 10.1080/10937404.2010.483937. [DOI] [PubMed] [Google Scholar]
- Käfferlein HU, Ferstl C, Burkhart-Reichl A, Hennebrüder K, Drexler H, Brüning T, and Angerer J. 2005. The use of biomarkers of exposure of N,N-dimethylformamide in health risk assessment and occupational hygiene in the polyacrylic fibre industry. Occupational and Environmental Medicine 62:330–36. doi: 10.1136/oem.2004.017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle JJ, Neas LM, Devlin RB, Case MW, Schmitt MT, Madden MC, and Diaz-Sanchez D. 2015. Interaction effects of temperature and ozone on lung function and markers of systemic inflammation, coagulation, and fibrinolysis: A crossover study of healthy young volunteers. Environmental Health Perspectives 123:310–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Wanigatunga AA, Needham LL, and Calafat AM. 2009. Analysis of blood spots for polyfluoroalkyl chemicals. Analytica Chimica Acta 656:51–55. doi: 10.1016/j.aca.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Kavlock RJ, Ankley G, Blancato J, Breen M, Conolly R, Dix D, Houck K, Hubal E, Judson R, Rabinowitz J, Richard A, Woodrow Setzer R, Shah I, Villeneuve D, and Weber E. 2008. Computational Toxicology—A State of the Science Mini Review. Tox. Sci 103:14–27. [DOI] [PubMed] [Google Scholar]
- Kelada SN, Eaton DL, Wang SS, Rothman NR, and Khoury MJ. 2003. The role of genetic polymorphisms in environmental health. Environmental Health Perspectives 111:1055–64. doi: 10.1289/ehp.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendzia B, Pesch B, Marczynski B, Lotz A, Welge P, Rihs H-P, Brüning T, and Raulf-Heimsoth M. 2012. Preand post-shift levels of inflammatory biomarkers and DNA damage in non-bitumen-exposed construction workers— Subpopulation of the German Human Bitumen Study. Journal of Toxicology and Environmental Health, Part A 75:533–43. doi: 10.1080/15287394.2012.675304. [DOI] [PubMed] [Google Scholar]
- Kew MC 2000. Serum aminotransferase concentration as evidence of hepatocellular damage. Lancet 355:591–92. doi: 10.1016/S0140-6736(99)00219-6. [DOI] [PubMed] [Google Scholar]
- Kim K, Mall C, Taylor SL, Hitchcock S, Zhang C, Wettersten HI, Daniel Jones A, Chapman A, and Weiss RH. 2014. Mealtime, temporal, and daily variability of the human urinary and plasma metabolomes in a tightly controlled environment. PLoS ONE 9:e86223. doi: 10.1371/journal.pone.0086223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Kim DS, Lee SK, Lee IK, Kang JH, Chang YS, Jacobs DR, Steffes M, and Lee DH. 2010. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy Koreans. Environmental Health Perspectives 118:370–74. doi: 10.1289/ehp.0901131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WR, Flamm SL, Di Bisceglie AM, and Bodenheimer HC Jr. 2008. Serum activity of alanine aminotransferase (ALT) as an indicator of health and disease. Hepatology 47:1363–70. doi: 10.1002/hep.22109. [DOI] [PubMed] [Google Scholar]
- Kim Y, Lobdell DT, Wright CW, Gocheva VV, Hudgens E, and Bowler RM. 2015. Blood metal concentrations of manganese, lead, and cadmium in relation to serum ferritin levels in Ohio residents. Biological Trace Element Research 165:1–9. doi: 10.1007/s12011-014-0223-1. [DOI] [PubMed] [Google Scholar]
- Klepeis NE, Nelson WC, Ott WR, Robinson JP, Tsang AM, Switzer P, Behar JV, Hern SC, and Engelmann WH 2001. The National Human Activity Pattern Survey (NHAPS): A resource for assessing exposure to environmental pollutants. Journal of Exposure Analysis and Environmental Epidemiology 11:231–52. doi: 10.1038/sj.jea.7500165. [DOI] [PubMed] [Google Scholar]
- Kline MC, Duewer DL, Redman JW, and Butler JM. 2002. Polymerase chain reaction amplification of DNA from aged blood stains: Quantitative evaluation of the “suitability for purpose” of four filter papers as archival media. Analytical Chemistry 74:1863–69. doi: 10.1021/ac015715e. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Dallaporta B, and Resche-Rigon M. 1998. The mitochondrial death/life regulator in apoptosis and necrosis. Annual Review of Physiology 60:619–42. doi: 10.1146/annurev.physiol.60.1.619. [DOI] [PubMed] [Google Scholar]
- Kwack SJ, Kim DY, Kim YJ, Roh TH, Choi SM, Lim DS, Shin HS, Kim HS, and Lee BM. 2014. Potential application of benzo(a)pyrene-associated adducts (globin or lipid) as blood biomarkers for target organ exposure and human risk assessment. Journal of Toxicology and Environmental Health, Part A 77:1491–501. doi: 10.1080/15287394.2014.955904. [DOI] [PubMed] [Google Scholar]
- LaKind JS, Sobus JR, Goodman M, Barr DB, Fürst P, Albertinin RJ, Arbuckle TE, Schoeters G, Tan Y-M, Teeguarden J, Tornero-Velez R, and Weisel CP. 2014. A proposal for assessing study quality: Biomonitoring, Environmental Epidemiology, and Short-lived Chemicals (BEES-C) instrument. Environment International 73:195–207. doi: 10.1016/j.envint.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latterman C, and Buchs J. 2015. Microscale and mini-scale fermentation and screening. Current Opinion in Biotechnology 35:1–6. doi: 10.1016/j.copbio.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Lehr S, Hartwig S, and Sell H. 2012. Adipokines: A treasure trove for the discovery of biomarkers for metabolic disorders. Proteomics—Clinical Applications 6:91–101. doi: 10.1002/prca.201100052. [DOI] [PubMed] [Google Scholar]
- Levitt DG 2004. Physiologically based pharmacokinetic modeling of arterial–antecubital vein concentration difference. BMC Clinical Pharmacology 4:2. doi: 10.1186/14726904-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Grigoryan H, Funk WE, Lu SS, Rose S, Williams ER, and Rappaport SM. 2011. Profiling Cys34 adducts of human serum albumin by fixed-step selected reaction monitoring. Molecular & Cellular Proteomics 10: M110.004606. doi: 10.1074/mcp.M110.004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Huang X, Liu W, and Shen W. 2014. Control performance of paper-based blood analysis devices through paper structure design. ACS Applied Materials & Interfaces 6:21624–31. doi: 10.1021/am506618c. [DOI] [PubMed] [Google Scholar]
- Li S, Todor A, and Luo R. 2016. Blood transcriptomics and metabolomics for personalized medicine. Computational and Structural Biotechnology Journal 14:1–7. doi: 10.1016/j.csbj.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligor T, Ligor M, Amann A, Ager C, Bachler M, Dzien A, and Buszewski B. 2008. The analysis of healthy volunteers’ exhaled breath by the use of solid-phase microextraction and GC-MS. Journal of Breath Research 2:046006. doi: 10.1088/1752-7155/2/4/046006. [DOI] [PubMed] [Google Scholar]
- Lindstrom AB, Yeowell-O’Connell K, Waidyanatha S, McDonald TA, Golding BT, and Rappaport SM. 1998. Formation of hemoglobin and albumin adducts of benzene oxide in mouse, rat, and human blood. Chemical Research in Toxicology 11:302–10. doi: 10.1021/tx9701788. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Ferrari M, and Petricoin E. 2003. Clinical proteomics: Written in blood. Nature 425:905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- Lioy PJ 2010. Exposure science: A view of the past and milestones for the future. Environmental Health Perspectives 118:1081–90. doi: 10.1289/ehp.0901634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy PJ, and Smith KR. 2013. A discussion of exposure science in the 21st century: A vision and a strategy. Environmental Health Perspectives 121:405–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JC, Weschler CJ, Nazaroff WW, Liu Z, and Cohen Hubal EA. 2012. Rapid methods to estimate potential exposure to semivolatile organic compounds in the indoor environment. Environmental Science & Technology 46:11171–78. doi: 10.1021/es301088a. [DOI] [PubMed] [Google Scholar]
- Liu R, Liao J, Yang M, Shi Y, Peng Y, Wang Y, Pan E, Guo W, Pu Y, and Yin L. 2012. Circulating miR-155 expression in plasma: A potential biomarker for early diagnosis of esophageal cancer in humans. Journal of Toxicology and Environmental Health, Part A 75:1154–62. doi: 10.1080/15287394.2012.699856. [DOI] [PubMed] [Google Scholar]
- Liu S, and Pleil JD. 2002. Human blood and environmental media screening method for pesticides and polychlorinated biphenyl compounds using liquid extraction and gas chromatography-mass spectrometry analysis. Journal of Chromatography B 769:155–67. doi: 10.1016/S1570-0232(01)00640-7. [DOI] [PubMed] [Google Scholar]
- Lope V, Pollán M, Fernández M, De León A, González MJ, Sanz JC, Iriso A, Pérez-Gómez B, Gil E, Pérez-Meixeira AM, De Paz C, Cisneros M, De Santos A, Asensio A, Astray J, Martínez M, García JF, LópezAbente G, García-Sagredo JM, and Aragonés N. 2010. Cytogenetic status in newborns and their parents in Madrid: The BioMadrid study. Environmental and Molecular Mutagenesis 51:267–77. [DOI] [PubMed] [Google Scholar]
- Lopes ACB, Navas-Acien A, Zamoiski R, Silbergeld EK, Carvalho MFH, Buzzo ML, Urbano MR, Martins AC Jr., and Paoliello MMB. 2015. Risk factors for lead exposure in adult population in southern Brazil. Journal of Toxicology and Environmental Health, Part A 78:92–108. doi: 10.1080/15287394.2014.942125. [DOI] [PubMed] [Google Scholar]
- Lunder S, Hovander L, Athanassiadis I, and Bergman A. 2010. Significantly higher polybrominated diphenyl ether levels in young U.S. children than in their mothers. Environmental Science & Technology 44:5256–62. doi: 10.1021/es1009357. [DOI] [PubMed] [Google Scholar]
- M’Bemba-Meka P, Lemieux N, and Chakrabarti SK. 2005. Nickel compound-induced DNA single strand breaks in chromosomal and nuclear chromatin in human blood lymphocytes in vitro: Role of oxidative stress and intracellular calcium. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 586:124–37. doi: 10.1016/j.mrgentox.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Ma WL, Gao C, Bell EM, Druschel CM, Caggana M, Aldous KM, Louis GM, and Kannan K. 2014. Analysis of polychlorinated biphenyls and organochlorine pesticides in archived dried blood spots and its application to track temporal trends of environmental chemicals in newborns. Environmental Research 133:204–10. doi: 10.1016/j.envres.2014.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magagnotti C, Orsi R, Bagnati R, Celli N, Rotilio D, Fanelli R, and Airoldi L. 2000. Effect of diet on serum albumin and hemoglobin adducts of 2-amino-1-methyl6-phenylimidazo[4,5-b]pyridine (PhIP) in humans. International Journal of Cancer 88:1–6. doi:. [DOI] [PubMed] [Google Scholar]
- Marchand A, Aranda-Rodriguez R, Tardif R, Nong A, and Haddad S. 2015. Human inhalation exposures to toluene, ethylbenzene, and M-xylene and physiologically based pharmacokinetic modeling of exposure biomarkers in exhaled air, blood, and urine. Toxicological Sciences 144:414–24. doi: 10.1093/toxsci/kfv009. [DOI] [PubMed] [Google Scholar]
- Marini V, Michelazzi L, Cioé A, Fucile C, Spigno F, and Robbiano L. 2011. Exposure to asbestos: Correlation between blood levels of mesothelin and frequency of micronuclei in peripheral blood lymphocytes. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 721:114–17. doi: 10.1016/j.mrgentox.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Marion JW, Lee C, Lee CS, Wang Q, Lemeshow S, Buckley TJ, Saif LJ, and Lee J. 2014. Integrating bacterial and viral water quality assessment to predict swimmingassociated illness at a freshwater beach: A cohort study. PLoS ONE 9:e112029. doi: 10.1371/journal.pone.0112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez V, Creus A, Venegas W, Arroyo A, Beck JP, Gebel TW, Surrallés J, and Marcos R. 2004. Evaluation of micronucleus induction in a Chilean population environmentally exposed to arsenic. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 564:65–74. doi: 10.1016/j.mrgentox.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Mayr M 2008. Metabolomics: Ready for the prime time? Circulation: Cardiovascular Genetics 1:58–56. doi: 10.1161/CIRCGENETICS.108.808329. [DOI] [PubMed] [Google Scholar]
- McDade TW 2014. Development and validation of assay protocols for use with dried blood spot samples. American Journal of Human Biology 26:1–9. doi: 10.1002/ajhb.v26.1. [DOI] [PubMed] [Google Scholar]
- McDade TW, Burhop J, and Dohnal J. 2004. High-sensitivity enzyme immunoassay for C-reactive protein in dried blood spots. Clinical Chemistry 50:652–54. doi: 10.1373/clinchem.2003.029488. [DOI] [PubMed] [Google Scholar]
- McDade TW, Williams S, and Snodgrass JJ. 2007. What a drop can do: Dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography 44:899–925. doi: 10.1353/dem.2007.0038. [DOI] [PubMed] [Google Scholar]
- McDade TW, Woodruff TK, Huang Y-Y, Funk WE, Prewitt M, Kondapalli L, and Gracia CR 2012. Quantification of anti-Müllerian hormone (AMH) in dried blood spots: Validation of a minimally invasive method for assessing ovarian reserve. Human Reproduction 27:2503–08. doi: 10.1093/humrep/des194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale CM, Zhang L, Lan Q, Li G, Hubbard AE, Forrest MS, Vermeulen R, Chen J, Shen M, Rappaport SM, Yin S, Smith MT, and Rothman N. 2009. Changes in the peripheral blood transcriptome associated with occupational benzene exposure identified by cross-comparison on two microarray platforms. Genomics 93:343–49. doi: 10.1016/j.ygeno.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei JV, Alexander JR, Adam BW, and Hannon WH. 2001. Use of filter paper for the collection and analysis of human whole blood specimens. Journal of Nutrition 131:1631S–1636S. [DOI] [PubMed] [Google Scholar]
- Meier-Ewert HK, Ridker PM, Rifai N, Price N, Dinges DF, and Mullington JM. 2001. Absence of diurnal variation of C-reactive protein concentrations in healthy human subjects. Clinical Chemistry 47:426–30. [PubMed] [Google Scholar]
- Merrick BA 2008. The plasma proteome, adductome and idiosyncratic toxicity in toxicoproteomics research. Briefings in Functional Genomics and Proteomics 7:35–49. doi: 10.1093/bfgp/eln004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz A, Samà S, Martucci M, and Parlato G. 2015. Analyzing dried blood spots to monitor lead exposure in Calabria, the southernmost region of peninsular Italy. International Journal Environment Sciences 5:891–903. [Google Scholar]
- Miller AA, Sharrock KC, and McDade TW. 2006. Measurement of leptin in dried blood spot samples. American Journal of Human Biology 18:857–60. doi: 10.1002/(ISSN)1520-6300. [DOI] [PubMed] [Google Scholar]
- Miller DB, Ghio AJ, Karoly ED, Bell LN, Snow SJ, Madden MC, Soukup J, Cascio WE, Glimour MI, and Kodavanti UP. 2016. Ozone exposure increases circulating stress hormones and lipid metabolites in humans. American Journal of Respiratory and Critical Care Medicine. doi: 10.1164/rccm.201508-1599OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EM, and McDade TW. 2012. A highly sensitive immunoassay for interleukin-6 in dried blood spots. American Journal of Human Biology 24:863–65. doi: 10.1002/ajhb.v24.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J, Xia Y, Ning Z, Wade TJ, and Mumford JL. 2009. Elevated ERCC1 gene expression in blood cells associated with exposure to arsenic from drinking water in Inner Mongolia. Anticancer Research 29:3253–60. [PubMed] [Google Scholar]
- Needham LL, Patterson DG Jr., Barr DB, Gringer J, and Calafat AM. 2005. Uses of speciation techniques in biomonitoring for assessing human exposure to organic environmental chemicals. Analytical and Bioanalytical Chemistry 381:397–404. doi: 10.1007/s00216-004-2975-5. [DOI] [PubMed] [Google Scholar]
- Nelin TD, Joseph AM, Gorr MW, and Wold LE. 2012. Direct and indirect effects of particulate matter on the cardiovascular system. Toxicology Letters 208:293–99. doi: 10.1016/j.toxlet.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngueta G, Abdous B, Tardif R, St-Laurent J, and Levallois P. 2016. Use of a cumulative exposure index to estimate the impact of tap-water lead concentration on blood lead levels in 1to 5-year-old children (Montreal, Canada). Environmental Health Perspectives. 124:388–395. doi: 10.1289/ehp.1409144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GW, Burris JM, Ehresman DJ, Froehlich JW, Seacat AM, Butenhoff JL, and Zobel LR. 2007. Half-life of serum elimination of perfluorooctanesulfonate, perfluorohexanesulfonate, and perfluorooctanoate in retired fluorochemical production workers. Environmental Health Perspectives 115:1298–305. doi: 10.1289/ehp.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz A, Lorz C, Catalan MP, Justo P, and Eguido J. 2000. Role and regulation of apoptotic cell death in the kidney. Y2K update. Frontiers in Bioscience 5:D735–D749. [DOI] [PubMed] [Google Scholar]
- Ostler M W. J Porter H, and Buxton OM 2014. Dried blood spot collection of health biomarkers to maximize participation in population studies. Journal of Visualized Experiments 83:e50973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero-Santos SM, Delinsky AD, Valentin-Blasini L, Schiffer J, and Blount BC. 2009. Analysis of perchlorate in dried blood spots using ion chromatography and tandem mass spectrometry. Analytical Chemistry 81:1931–36. doi: 10.1021/ac802419n. [DOI] [PubMed] [Google Scholar]
- Ouyang B, Bernstein DI, Lummus ZL, Ying J, Boulet L-P, Cartier A, Gautrin D, and Ho S-M. 2013. Interferon-promoter is hypermethylated in blood DNA from workers with confirmed diisocyanate asthma. . Toxicological Sciences 133:218–24. doi: 10.1093/toxsci/kft079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagana KD, and Pagana TJ. 2009. Mosby’s manual of diagnostic and laboratory tests, 4th ed. Philadelphia, PA: Elsevier. [Google Scholar]
- Pasikanti KK, Ho PC, and Chan EC. 2008. Gas chromatography/mass spectrometry in metabolic profiling of biological fluids. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences 15:202–11. doi: 10.1016/j.jchromb.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Pedersen M, Vinzents P, Petersen JH, Kleinjans JCS, Plas G, Kirsch-Volders M, Dostál M, Rössner P, Beskid O, Sram RJ, Merlo DF, and Knudsen LE. 2006. Cytogenetic effects in children and mothers exposed to air pollution assessed by the frequency of micronuclei and fluorescence in situ hybridization (FISH): A family pilot study in the Czech Republic. Mutation Research/ Genetic Toxicology and Environmental Mutagenesis 608:112–20. doi: 10.1016/j.mrgentox.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Piasek M, Mikolić A, Sekovanić A, Grgec AS, and Jurasović J. 2014. Cadmium in placenta—A valuable biomarker of exposure during pregnancy in biomedical research. Journal of Toxicology and Environmental Health, Part A 77:1071–74. doi: 10.1080/15287394.2014.915779. [DOI] [PubMed] [Google Scholar]
- Pinto J, Domingues MRM, Galhano E, Pita C, do Céu Almeida M, Carreira IM, and Gil AM. 2014. Human plasma stability during handling and storage: Impact on NMR metabolomics. Analyst 139:1168–77. doi: 10.1039/c3an02188b. [DOI] [PubMed] [Google Scholar]
- Pleil JD 2009. Influence of systems biology response and environmental exposure level on between-subject variability in breath and blood biomarkers. Biomarkers 14:560–71. doi: 10.3109/13547500903186460. [DOI] [PubMed] [Google Scholar]
- Pleil JD 2012. Categorizing biomarkers of the human exposome and developing metrics for assessing environmental sustainability. Journal of Toxicology and Environmental Health, Part B 15:264–80. doi: 10.1080/10937404.2012.672148. [DOI] [PubMed] [Google Scholar]
- Pleil JD 2016. Cellular respiration: Replicating in vivo systems biology for in vitro exploration of human exposome and microbiome biomarkers. Journal of Breath Research. 10:010201. doi: 10.1088/1752-7155/10/1/010201. [DOI] [PubMed] [Google Scholar]
- Pleil JD, Beauchamp JD, Miekisch W, and Funk WE. 2015. Adapting biomarker technologies to adverse outcome pathways (AOPs) research: Current thoughts on using in vivo discovery for developing in vitro target methods. Journal of Breath Research 9:039001. doi: 10.1088/17527155/9/3/039001. [DOI] [PubMed] [Google Scholar]
- Pleil JD, Funk WE, and Rappaport SM. 2006. Residual indoor contamination from World Trade Center rubble fires as indicated by polycyclic aromatic hydrocarbon profiles. Environmental Science & Technology 40:1172–77. doi: 10.1021/es0517015. [DOI] [PubMed] [Google Scholar]
- Pleil JD, and Isaacs KK. 2015. High-resolution mass spectrometry: Basic principles for using exact mass and mass defect for discovery analysis of organic molecules in blood, breath, urine, and environmental media. Journal of Breath Research 10:012001. doi: 10.1088/1752-7155/10/1/012001. [DOI] [PubMed] [Google Scholar]
- Pleil JD, Kim D, Prah JD, and Rappaport SM. 2007. Exposure reconstruction for reducing uncertainty in risk assessment: Example using MTBE biomarkers and a simple pharmacokinetic model. Biomarkers 12:331–48. doi: 10.1080/13547500701246334. [DOI] [PubMed] [Google Scholar]
- Pleil JD, Miekisch W, Stiegel MA, and Beauchamp J. 2014. Extending breath analysis to the cellular level: Current thoughts on the human microbiome and the expression of organic compounds in the human exposome. Journal of Breath Research 8:029001. doi: 10.1088/1752-7155/8/2/029001. [DOI] [PubMed] [Google Scholar]
- Pleil JD, and Sheldon LS. 2011. Adapting concepts from systems biology to develop systems exposure event networks for exposure science research. Biomarkers 16:99–105. doi: 10.3109/1354750X.2010.541565. [DOI] [PubMed] [Google Scholar]
- Pleil JD, Stiegel MA, Madden MC, and Sobus JR. 2011b. Heat map visualization of complex environmental and biomarker measurements. Chemosphere 84:716–23. doi: 10.1016/j.chemosphere.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Pleil JD, Stiegel MA, and Risby T. 2013. Clinical breath analysis: Discriminating between human endogenous compounds and exogenous (environmental) chemical confounders. Journal of Breath Research 7:017107. doi: 10.1088/1752-7155/7/1/017107. [DOI] [PubMed] [Google Scholar]
- Pleil JD, Stiegel MA, and Sobus JR. 2011a. Breath biomarkers in environmental health science: Exploring patterns in the human exposome. Journal of Breath Research 5:046005. doi: 10.1088/1752-7155/5/4/046005. [DOI] [PubMed] [Google Scholar]
- Pleil JD, Stiegel MA, Sobus JR, Tabucchi S, Ghio AJ, and Madden MC. 2010. Cumulative exposure assessment for trace-level polycyclic aromatic hydrocarbons (PAHs) using human blood and plasma analysis. Journal of Chromatography B 878:1753–60. doi: 10.1016/j.jchromb.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Pleil JD, Williams MA, and Sobus JR. 2012. Chemical Safety for Sustainability (CSS): Human in vivo biomonitoring data for complementing results from in vitro toxicology—A commentary. Toxicology Letters 215:201–07. doi: 10.1016/j.toxlet.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Poirier MC 2004. Chemical-induced DNA damage and human cancer risk. Nature Reviews. Cancer 4:630–37. doi: 10.1038/nrc1410. [DOI] [PubMed] [Google Scholar]
- Polkowska Ż, Kozłowska K, Namieśnik J, and Przyjazny A. 2004. Biological fluids as a source of information on the exposure of man to environmental chemical agents. Critical Reviews in Analytical Chemistry 34:105–19. doi: 10.1080/10408340490475911. [DOI] [Google Scholar]
- Pollack AZ, Mumford SL, Mendola P, Perkins NJ, Rotman Y, Wactawski-Wende J, and Schisterman EF. 2015. Kidney biomarkers associated with blood lead, mercury, and cadmium in premenopausal women: A prospective cohort study. Journal of Toxicology and Environmental Health, Part A 78:119–31. doi: 10.1080/15287394.2014.944680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp W, Vahrenholz C, Schrell C, Grimmer G, Dettbarn G, Kraus R, Brauksiepe A, Schmeling B, Gutzeit T, Von Bülow J, and Norpoth K. 1997. DNA single strand breakage, DNA adducts, and sister chromatid exchange in lymphocytes and phenanthrene and pyrene metabolites in urine of coke oven workers. Occupational and Environmental Medicine 54:176–83. doi: 10.1136/oem.54.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porru S, Pavanello S, Carta A, Arici C, Simeone C, Izzotti A, and Mastrangelo G. 2014. Complex relationships between occupation, environment, DNA adducts, genetic polymorphisms and bladder cancer in a case-control study using a structural equation modeling. PLoS ONE 9:e94566. doi: 10.1371/journal.pone.0094566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Chavez LA, Sordo M, Calderon-Aranda E, Castañeda-Saucedo E, Ostrosky-Wegmean P, and Moreno-Godinez ME. 2015. A permethrin/allethrin mixture induces genotoxicity and cytotoxicity in human peripheral blood lymphocytes. Journal of Toxicology and Environmental Health, Part A 78:7–14. doi: 10.1080/15287394.2015.956025. [DOI] [PubMed] [Google Scholar]
- Rappaport SM 2011. Implications of the exposome for exposure science. Journal of Exposure Science and Environmental Epidemiology 21:5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- Rappaport SM 2012a. Biomarkers intersect with the exposome. Biomarkers 17:483–89. doi: 10.3109/1354750X.2012.691553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM 2012b. Discovering environmental causes of disease. Journal of Epidemiology & Community Health 66:99–102. doi: 10.1136/jech-2011-200726. [DOI] [PubMed] [Google Scholar]
- Rappaport SM, Funk WE, and Chaing S. 2008. Hemoglobin adducts in dried blood spots as measures of carcinogen exposures. Epidemiology 19:S62–S63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, Liu H, Grigoryan H, Funk WE, and Williams ER. 2012. Adductomics: Characterizing exposures to reactive electrophiles. Toxicology Letters 213:83–90. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, and Smith MT. 2010. Environment and disease risks. Science 330:460–61. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegman PHJ, Morente MM, Betsou F, De Blasio P, and Geary P; the Marble Arch International Working Group on Biobaking for Biomedical Research. 2008. Biobanking for better healthcare. Molecular Oncology 2:213–22. doi: 10.1016/j.molonc.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos PH, and Jakubowski N. 2010. Methods for the discovery of low-abundance biomarkers for urinary bladder cancer in biological fluids. Bioanalysis 2:295–309. doi: 10.4155/bio.09.174. [DOI] [PubMed] [Google Scholar]
- Rossner P Jr., Milcova A, Libalova H, Novakova Z, Topinka J, Balascak I, and Sram RJ. 2009. Biomarkers of exposure to tobacco smoke and environmental pollutants in mothers and their transplacental transfer to the foetus. Part II. Oxidative damage. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 669:20–26. doi: 10.1016/j.mrfmmm.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Samuelsson LB, Hall MH, McLean S, Porter JH, Berkman L, Marino M, Sembajwe G, McDade TW, and Buxton OM. 2015. Validation of biomarkers of CVD risk from dried blood spots in community-based research: Methodologies and study-specific serum equivalencies. Biodemography and Social Biology 61:285–97. doi: 10.1080/19485565.2015.1068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarigiannis DA, and Hansen U. 2012. Considering the cumulative risk of mixtures of chemicals—A challenge for policy makers. Environmental Health 11 (Suppl. 1): S18. doi: 10.1186/1476-069X-11-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savorani F, Rasmussen MA, Mikkelsen MS, and Engelsen SB. 2013. A primer to nutritional metabolomics by NMR spectroscopy and chemometrics. Food Research International 54:1131–45. doi: 10.1016/j.foodres.2012.12.025. [DOI] [Google Scholar]
- Schechter AN 2008. Hemoglobin research and the origins of molecular medicine. Blood 112:3927–38. doi: 10.1182/blood-2008-04-078188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetter AJ, and Harris CC. 2012. Tumor suppressor p53 (TP53) at the crossroads of the exposome and the cancer genome. Proceedings of the National Academy of Sciences 109:7955–56. doi: 10.1073/pnas.1205457109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg LK, and Beger RD. 2008. The role of metabolic biomarkers in drug toxicity studies. Toxicology Mechanisms and Methods 18:301–11. doi: 10.1080/15376510701623193. [DOI] [PubMed] [Google Scholar]
- Schreinemachers DM, Ghio AJ, Sobus JR, and Williams MA. 2015. Perchlorate exposure is associated with oxidative stress and indicators of serum iron homeostasis among NHANES 2005–2008 subjects. Biomarker. Insights 10:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabihkhani M, Lucey GM, Wei B, Mareninov S, Lou JJ, Vinters HV, Singer EJ, Cloughesy TF, and Yong WH. 2014. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clinical Biochemistry 47:258–66. doi: 10.1016/j.clinbiochem.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JY, Choi YY, Jeon HS, Hwang JH, Kim SA, Kang JH, Chang YS, Jacobs DR Jr., Park JY, and Lee DH. 2010. Low-dose persistent organic pollutants increased telomere length in peripheral leukocytes of healthy Koreans. Mutagenesis 24:511–16. doi: 10.1093/mutage/geq035. [DOI] [PubMed] [Google Scholar]
- Skogstrand K, Thorsen P, Nørgaard-Pedersen B, Schendel DE, Sørensen LC, and Hougaard DM. 2005. Simultaneous measurement of 25 inflammatory markers and neurotrophins in neonatal dried blood spots by immunoassay with xMAP technology. Clinical Chemistry 51:1854–66. doi: 10.1373/clinchem.2005.052241. [DOI] [PubMed] [Google Scholar]
- Smith MT, and Rappaport SM. 2009. Building exposure biology centers to put the E into “G × E” interaction studies. Environmental Health Perspectives 117:A334–A335. doi: 10.1289/ehp.12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MT, Zhang L, McHale CM, Skibola CF, and Rappaport SM. 2011. Benzene, the exposome and future investigations of leukemia etiology. Chemico-Biological Interactions 192:155–59. doi: 10.1016/j.cbi.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolders R, Schramm K-W, Nickmilder M, and Schoeters G. 2009. Applicability of non-invasively collected matrices for human biomonitoring. Environmental Health 8:8. doi: 10.1186/1476-069X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommar JN, Hedmer M, Lundh T, Nilsson L, Skerfving S, and Bergdahl IA. 2014. Investigation of lead concentrations in whole blood, plasma, and urine as biomarkers for biological monitoring of lead exposure. Journal of Exposure Science and Environmental Epidemiology 24:51–57. doi: 10.1038/jes.2013.4. [DOI] [PubMed] [Google Scholar]
- Spliethoff HM, Tao L, Shaver SM, Aldous KM, Pass KA, Kannan K, and Eadon GA. 2008. Use of newborn screening program blood spots for exposure assessment: Declining levels of perfluorinated compounds in New York State infants. Environmental Science & Technology 42:5361–67. doi: 10.1021/es8006244. [DOI] [PubMed] [Google Scholar]
- Steenhof M, Janssen NAH, Strak M, Hoek G, Gosens I, Mudway IS, Kelly FJ, Harrison RM, Pieters RHH, Cassee FR, and Brunekreef B. 2014. Air pollution exposure affects circulating white blood cell counts in healthy subjects: The role of particle composition, oxidative potential and gaseous pollutants—The RAPTES project. Inhalation Toxicology 26:141–65. doi: 10.3109/08958378.2013.861884. [DOI] [PubMed] [Google Scholar]
- Stiegel MA, Pleil JD, Sobus JR, Morgan MK, and Madden MC. 2015. Analysis of inflammatory cytokines in human blood, breath condensate, and urine using a multiplex immunoassay platform. Biomarkers 20:35–46. doi: 10.3109/1354750X.2014.988646. [DOI] [PubMed] [Google Scholar]
- Such-Sanmartín G, Ventura-Espejo E, and Jensen ON. 2014. Depletion of abundant plasma proteins by poly(Nisopropylacrylamide-acrylic acid) hydrogel particles. Analytical Chemistry 86:1543–50. doi: 10.1021/ac403749j. [DOI] [PubMed] [Google Scholar]
- Tanner S, and McDade TW. 2007. Enzyme immunoassay for total immunoglobulin E in dried blood spots. American Journal of Human Biology 19:440–42. doi: 10.1002/(ISSN)1520-6300. [DOI] [PubMed] [Google Scholar]
- Tarrant JM 2010. Blood cytokines as biomarkers of in vivo toxicity in preclinical safety assessment: Considerations for their use. Toxicological Sciences 117:4–16. doi: 10.1093/toxsci/kfq134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeguarden JG, Tan C, Edwards S, Leonard JA, Anderson KA, Corley RA, Harding A, Kile M, Simonich SLM, Stone D, Waters KM, Harper S, Williams DE, and Tanguay RL. 2016. Completing the link between exposure science and toxicology for improved environmental health decision making: The aggregate exposure pathway framework. Environmental Science & Technology. doi: 10.1021/acs.est.5b05311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker J, and Zdzienicka MZ. 2003. The mammalian XRCC genes: Their roles in DNA repair and genetic stability. DNA Repair 2:655–72. doi: 10.1016/S1568-7864(03)00062-4. [DOI] [PubMed] [Google Scholar]
- Titz B, Elamin A, Martin F, Schneider T, Dijon S, Ivanov NV, Hoeng J, and Peitsch MC. 2014. Proteomics for systems toxicology. Computational and Structural Biotechnology Journal 11:73–90. doi: 10.1016/j.csbj.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topinka J, Sevastyanova O, Binkova B, Chvatalova I, Milcova A, Lnenickova Z, Novakova Z, Solansky I, and Sram RJ. 2007. Biomarkers of air pollution exposure—A study of policemen in Prague. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 624:9–17. doi: 10.1016/j.mrfmmm.2007.02.032. [DOI] [PubMed] [Google Scholar]
- Trayhurn P 2005. Endocrine and signalling role of adipose tissue: New perspectives on fat. Acta Physiologica Scandinavica 184:285–93. doi: 10.1111/aps.2005.184.issue-4. [DOI] [PubMed] [Google Scholar]
- Trifonova O, Lokhov P, and Archakov A. 2013. Postgenomics diagnostics: Metabolomics approaches to human blood profiling. Omics 17:550–59. doi: 10.1089/omi.2012.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuimala J, Szekely G, Gundy S, Hirvonen A, and Norppa H. 2002. Genetic polymorphisms of DNA repair and xenobiotic-metabolizing enzymes: Role in mutagen sensitivity. Carcinogenesis 23:1003–08. doi: 10.1093/carcin/23.6.1003. [DOI] [PubMed] [Google Scholar]
- Turyk ME, Anderson HA, Steenport D, Buelow C, Imm P, and Knobeloch L. 2010. Longitudinal biomonitoring for polybrominated diphenyl ethers (PBDEs) in residents of the Great Lakes basin. Chemosphere 81:517–22. doi: 10.1016/j.chemosphere.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueyama J, Satoh T, Kondo T, Takagi K, Shibata E, Goto M, Kimata A, Saito I, Hasegawa T, Wakusawa S, and Kamijima M. 2010. β-Glucuronidase activity is a sensitive biomarker to assess low-level organophosphorus insecticide exposure. Toxicology Letters 193:115–19. doi: 10.1016/j.toxlet.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Valcke M, and Haddad S. 2015. Assessing human variability in kinetics for exposures to multiple environmental chemicals: A physiologically based pharmacokinetic modeling case study with dichloromethane, benzene, toluene, ethylbenzene, and m-xylene. Journal of Toxicology and Environmental Health A 78:409–31. doi: 10.1080/15287394.2014.971477. [DOI] [PubMed] [Google Scholar]
- Vaught JB 2006. Blood collection, shipment, processing, and storage. Cancer Epidemiology Biomarkers & Prevention 15:1582–84. doi: 10.1158/1055-9965.EPI-06-0630. [DOI] [PubMed] [Google Scholar]
- Veenstra TD 2007. Global and targeted quantitative proteomics for biomarker discovery. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences 847:3–11. doi: 10.1016/j.jchromb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Vermeulen R, Lan Q, Zhang L, Gunn L, McCarthy D, Woodbury RL, McGuire M, Podust VN, Li G, Chatterjee N, Mu R, Yin S, Rothman N, and Smith MT. 2005. Decreased levels of CXC-chemokines in serum of benzene-exposed workers identified by array-based proteomics. Proceedings of the National Academy of Sciences 102:17041–46. doi: 10.1073/pnas.0508573102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineis P, Khan AE, Vlaanderen J, and Vermeulen R. 2009. The impact of new research technologies on our understanding of environmental causes of disease: The concept of clinical vulnerability. Environmental Health 8:54. doi: 10.1186/1476-069X-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Tonoli D, Varesio E, and Hopfgartner G. 2014. The use of mass spectrometry to analyze dried blood spots. Massachusetts Spectrometry Reviews 9999:1–78. [DOI] [PubMed] [Google Scholar]
- Ward TH, Cummings J, Dean E, Greystoke A, Hou JM, Backen A, Ranson M, and Dive C. 2008. Biomarkers of apoptosis. British Journal of Cancer 99:841–46. doi: 10.1038/sj.bjc.6604519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins SM, and German JB. 2002. Toward the implementation of metabolomic assessments of human health and nutrition. Current Opinion in Biotechnology 13:512–16. doi: 10.1016/S0958-1669(02)00363-4. [DOI] [PubMed] [Google Scholar]
- Watson WP, and Mutti A. 2004. Role of biomarkers in monitoring exposures to chemicals: Present position, future prospects. Biomarkers 9:211–42. doi: 10.1080/13547500400015642. [DOI] [PubMed] [Google Scholar]
- Wedge DC, Allwood JW, Dunn W, Vaughan AA, Simpson K, Brown M, Priest L, Blackhall FH, Whetton AD, Dive C, and Goodacre R. 2011. Is serum or plasma more appropriate for intersubject comparisons in metabolic studies? An assessment in patients with small-cell lung cancer. Analytical Chemistry 83:6689–97. doi: 10.1021/ac2012224. [DOI] [PubMed] [Google Scholar]
- West-Nørager M, Kelstrup CD, Schou C, Høgdall EV, Høgdall CK, and Heegaard NH. 2007. Unravelling in vitro variables of major importance for the outcome of mass spectrometry-based serum proteomics. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences 847:30–37. doi: 10.1016/j.jchromb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- Wild CP 2005. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiology Biomarkers & Prevention 14:1847–50. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Williams SR, and McDade TW. 2009. The use of dried blood spot sampling in the National Social Life, Health, and Aging Project. Journal of Gerontology Series B: Psychological Sciences and Social Sciences 64B:i131–i136. doi: 10.1093/geronb/gbn022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JYY, De Vivo I, Lin X, and Christiani DC. 2014. Cumulative PM2.5 exposure and telomere length in workers exposed to welding fumes. Journal of Toxicology and Environmental Health, Part A 77:441–55. doi: 10.1080/15287394.2013.875497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods JS, Heyer NJ, Russo JE, Martin MD, Pillai PB, Bammler TK, and Farin FM. 2014. Genetic polymorphisms of catechol-O-methyltransferase modify the neurobehavioral effects of mercury in children. Journal of Toxicology and Environmental Health, Part A 77:293–312. doi: 10.1080/15287394.2014.867210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Peter A, Franken H, Zhao X, Neukamm SS, Rosenbaum L, Lucio M, Zell A, Häring H-U, Xu G, and Lehmann R. 2013. Preanalytical aspects and sample quality assessment in metabolomics studies of human blood. Clinical Chemistry 59:833–45. doi: 10.1373/clinchem.2012.199257. [DOI] [PubMed] [Google Scholar]
- Yin P, and Xu G. 2014. Current state-of-the-art of nontargeted metabolomics based on liquid chromatography-mass spectrometry with special emphasis in clinical applications. Journal of Chromatography A 1374:1–13. doi: 10.1016/j.chroma.2014.11.050. [DOI] [PubMed] [Google Scholar]
- Yu Y-RA, Hotten DF, Malakhau Y, Volker E, Ghio AJ, Noble PW, Kraft M, Hollingsworth JW, Gunn MD, and Tighe RM. 2016. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. American Journal of Respiratory Cell and Molecular Biology 54:13–24. doi: 10.1165/rcmb.2015-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Kastenmüller G, He Y, Belcredi P, Möller G, Prehn C, Mendes J, Wahl S, Roemisch-Margl W, Ceglarek U, Poloikov A, Dahmen N, Prokisch H, Xie L, Li Y, Wichmann H-E, Peters A, Kronenberg F, Suhre K, Adamski J, Illig T, and Wang-Sattler R. 2011. Differences between human plasma and serum metabolite profiles. PLoS ONE 6:e21230. doi: 10.1371/journal.pone.0021230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkin JS, Stehouwer CDA, Emeis JJ, and Coppack SW. 1999. C-reactive protein in healthy subjects: Associations with obesity, insulin resistance, and endothelial dysfunction: A potential role for cytokines originating from adipose tissue? Arteriosclerosis, Thrombosis, and Vascular Biology 19:972–78. doi: 10.1161/01.ATV.19.4.972. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lin S, Funk WE, and Hou L. 2013a. Environmental and occupational exposure to chemicals and telomere length in human studies. Occupational and Environmental Medicine 70:743–49. doi: 10.1136/oemed2012-101350. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fonslow BR, Shan B, Baek M-C, and Yates III JR. 2013b. Protein analysis by shotgun/bottom-up proteomics. Chemical Reviews 113:2343–94. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, Fei M, and Sun S. 2010. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clinical Chemistry 56:1830–38. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- Zhao L, Gao Y, Yang Y, Wei Y, Li Y, Feng H, Wei W, Ding Y, and Sun D. 2010. Serum proteomic profiling analysis of chronic arsenic exposure by using SELDI-TOF-MS technology. Toxicology Letters 195:155–60. doi: 10.1016/j.toxlet.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Zijno A, Andreoli C, Leopardi P, Marcon F, Rossi S, Caiola S, Verdina A, Galati R, Cafolla A, and Crebelli R. 2003. Folate status, metabolic genotype, and biomarkers of genotoxicity in healthy subjects. Carcinogenesis 24:1097–103. doi: 10.1093/carcin/bgg064. [DOI] [PubMed] [Google Scholar]
- Zuurbier M, Hoek G, Oldenwening M, Meliefste K, Krop E, Van Den Hazel P, and Brunekreef B. 2011. In-traffic air pollution exposure and CC16, blood coagulation, and inflammation markers in healthy adults. Environmental Health Perspectives 119:1384–89. doi: 10.1289/ehp.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]